class: center, middle, inverse, title-slide .title[ # Spatial Prediction Models in <a href="">R</a> ] .subtitle[ ## Workshop ] .author[ ### Gabriel Carrasco-Escobar, MSc, PhD(c) ] .institute[ ### UC San Diego ] .date[ ### 2022-01-10 (updated: 2022-11-29) ] --- <style type="text/css"> # .remark-slide-content { # font-size: 28px; # padding: 20px 80px 20px 80px; # } # .remark-code, .remark-inline-code { # background: #f0f0f0; # } # .remark-code { # font-size: 24px; # } .huge .remark-code { /*Change made here*/ font-size: 200% !important; } .small1 .remark-code { /*Change made here*/ font-size: 80% !important; } .small2 .remark-code { /*Change made here*/ font-size: 65% !important; } .tiny .remark-code { /*Change made here*/ font-size: 50% !important; } .tiny2 .remark-code { /*Change made here*/ font-size: 55% !important; } .tinyq .remark-code { /*Change made here*/ font-size: 5% !important; } .big1_t { font-size: 120% } .big2_t { font-size: 150% } .big3_t { font-size: 200% } .small0_t { font-size: 90% } .small1_t { font-size: 80% } .small2_t { font-size: 60% } .tiny1_t { font-size: 40% } .tiny2_t { font-size: 20% } .text_right { vertical-align: bottom !important; text-align: right !important; } .text_left { vertical-align: bottom !important; text-align: left !important; } .pull-left2 { float: left; width: 35%; } .pull-right2 { float: right; width: 60%; } </style> # Packages and Data - [Data processing]() ```r library(tidyverse) ``` - [Spatial data processing]() ```r library(sf) library(terra) library(raster) library(mapview) ``` - [Model construction]() ```r library(tidymodels) library(spatialsample) library(randomForest) ``` --- # Packages and Data These data were downloaded from the [Malaria Atlas Project](https://malariaatlas.org/) data repository and were originally collected as part of [a study](https://link.springer.com/article/10.1186/1475-2875-10-25) conducted in 2009. Data can be downloaded in this [link](https://raw.githubusercontent.com/HughSt/HughSt.github.io/master/course_materials/week1/Lab_files/Data/mal_data_eth_2009_no_dups.csv) (Sturrock, UCSF). ```r malaria_eth <- read.csv("/mal_data_eth_2009_no_dups.csv", header=T) ``` <div id="htmlwidget-f2849f82ac0dde560480" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-f2849f82ac0dde560480">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","45","46","47","48","49","50","51","52","53","54","55","56","57","58","59","60","61","62","63","64","65","66","67","68","69","70","71","72","73","74","75","76","77","78","79","80","81","82","83","84","85","86","87","88","89","90","91","92","93","94","95","96","97","98","99","100","101","102","103","104","105","106","107","108","109","110","111","112","113","114","115","116","117","118","119","120","121","122","123","124","125","126","127","128","129","130","131","132","133","134","135","136","137","138","139","140","141","142","143","144","145","146","147","148","149","150","151","152","153","154","155","156","157","158","159","160","161","162","163","164","165","166","167","168","169","170","171","172","173","174","175","176","177","178","179","180","181","182","183","184","185","186","187","188","189","190","191","192","193","194","195","196","197","198","199","200","201","202","203"],["Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia","Ethiopia"],["ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH","ETH"],["Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa","Africa"],[6694,8017,12873,6533,4150,1369,5808,8922,19345,634,3416,10882,19611,1636,18330,18624,7095,19825,19645,7075,13347,6256,7066,1283,11973,17425,2831,2706,21932,3655,16669,7522,9026,9160,5565,6378,19625,7721,58693,58695,58723,58691,58697,58696,58719,58699,58700,58701,58734,58732,58698,58714,58721,58720,58703,58718,58704,58717,58733,58706,58705,58738,58737,58716,58713,58715,58727,58731,58711,58742,58728,58735,58729,58730,58707,58736,58709,58740,58741,58708,58739,58712,58710,58763,58762,58802,58725,58726,58797,58798,58756,58799,58801,58744,58804,58757,58758,58694,58722,58849,58848,58851,58845,58692,58702,58724,58800,58759,58761,58760,58754,58794,58796,58743,58745,58795,58746,58793,58755,58747,58753,58803,58748,58752,58751,58750,58766,58765,58749,58685,58805,58768,58769,58767,58681,58680,58683,58679,58783,58782,58786,58785,58784,58787,58781,58778,58777,58779,58780,58789,58775,58774,58776,58772,58791,58773,58788,58792,58771,58790,58770,58818,58816,58819,58815,58817,58812,58814,58806,58813,58807,58810,58811,58808,58809,58823,58824,58820,58822,58825,58827,58829,58828,58821,58826,58850,58847,58846,58839,58835,58840,58838,58836,58841,58837,58844,58842,58843,58834,58833,58830,58831,58832],[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,9.5635,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.1959,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.712,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0271,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],["UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","UNKNOWN","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL","RURAL"],[2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009,2009],[4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,6,7,7,7,8,7,7,8,7,7,7,7,7,7,7,7,7,7,7,8,6,6,7,7,8,8,8,8,6,7,6,6,7,7,7,6,6,6,7,5,7,7,7,8,7,7,7,7,8,7,8,8,9,7,6,7,7,7,8,7,7,8,8,6,7,7,7,7,8,7,7,7,7,7,7,7,7,7,7,7,7,8,7,8,8,8,7,7,8,8,6,5,8,8,7,7,7,7,5,7,8,6,5,7,9,7,7,7,8,8,8,7,7,7,7,8,6,8,8,8,8,7,7,6,8,7,7,7,6,7,6,6,7,6,6,6,6,7,7,8,6,8,8,9,7,6,7,7,6,7,8,6,7,9,7,7,8,7,8,7,6,6,6,7,6],[15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,15,16,16,16,15,15,16,16,15,14,16,16,15,15,16,15,16,16,15,15,18,16,13,15,15,16,16,16,15,15,16,16,15,15,16,15,14,15,15,15,14,12,16,14,15,16,16,15,14,14,17,16,16,15,16,16,16,15,16,14,16,15,16,16,16,17,15,15,15,16,15,15,16,16,16,15,16,14,16,16,17,15,16,16,13,15,16,15,16,16,16,15,16,15,16,16,16,16,16,16,17,16,15,15,16,16,16,16,16,16,16,16,15,15,16,16,16,15,16,16,16,16,16,16,15,15,15,15,15,16,15,14,16,15,15,15,18,15,16,16,16,16,16,15,16,16,16,15,16,14,16,16,16,16,15,16,16,18,16,16,16,13,15,16,16,15],[220,216,127,56,219,215,220,129,214,220,215,215,219,207,209,216,215,212,107,198,198,105,216,106,109,209,220,220,220,218,164,37,221,212,211,71,206,218,109,101,109,107,110,110,110,110,110,108,108,104,109,106,108,110,109,110,110,110,107,110,102,106,110,110,101,110,104,105,101,110,110,102,106,104,109,108,109,80,106,110,110,109,110,104,110,110,110,110,110,108,106,110,110,110,110,110,110,110,107,108,101,98,103,107,101,109,109,110,110,110,110,111,110,110,109,109,108,108,109,108,110,110,110,110,110,110,105,110,110,112,110,110,110,101,109,109,108,105,104,109,110,104,110,108,109,110,110,110,110,108,110,110,111,110,107,110,110,110,110,110,110,43,110,107,100,109,105,109,105,80,103,104,108,110,97,110,100,105,110,99,102,94,108,109,104,109,106,107,103,104,104,43,108,110,110,107,104,106,105,94,106,110,101],[0,0,0,0,0,1,0,0,1,0,0,0,1,1,0,0,0,0,0,0,0,1,0,4,0,0,0,8,0,0,0,0,1,0,1,1,0,1,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,4,0,0,5,0,2,5,1,0,3,5,14,4,0,1,0,8,0,2,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],[0,0,0,0,0,0.004651163,0,0,0.004672897,0,0,0,0.00456621,0.004830918,0,0,0,0,0,0,0,0.00952381,0,0.037735849,0,0,0,0.036363636,0,0,0,0,0.004524887,0,0.004739336,0.014084507,0,0.004587156,0,0,0.009174312,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0.009090909,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0.036363636,0,0,0.046296296,0,0.018348624,0.046296296,0.009090909,0,0.027272727,0.045454545,0.127272727,0.036363636,0,0.009090909,0,0.071428571,0,0.018181818,0,0.00990099,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0.00952381,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0],["Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy","Microscopy"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>country<\/th>\n <th>country_id<\/th>\n <th>continent_id<\/th>\n <th>site_id<\/th>\n <th>latitude<\/th>\n <th>longitude<\/th>\n <th>rural_urban<\/th>\n <th>year_start<\/th>\n <th>lower_age<\/th>\n <th>upper_age<\/th>\n <th>examined<\/th>\n <th>pf_pos<\/th>\n <th>pf_pr<\/th>\n <th>method<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"className":"dt-right","targets":[4,5,6,8,9,10,11,12,13]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- # Packages and Data ```r cases <- malaria_eth %>% st_as_sf(coords = c("longitude", "latitude"), crs = 4326, remove = F) %>% mutate(pf_pos = ifelse(pf_pos > 0, 1, 0), pf_pos = factor(pf_pos)) %>% dplyr::select(pf_pos, longitude, latitude) ``` <div id="htmlwidget-dc1ff843a1b7dd7cb492" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-dc1ff843a1b7dd7cb492">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","45","46","47","48","49","50","51","52","53","54","55","56","57","58","59","60","61","62","63","64","65","66","67","68","69","70","71","72","73","74","75","76","77","78","79","80","81","82","83","84","85","86","87","88","89","90","91","92","93","94","95","96","97","98","99","100","101","102","103","104","105","106","107","108","109","110","111","112","113","114","115","116","117","118","119","120","121","122","123","124","125","126","127","128","129","130","131","132","133","134","135","136","137","138","139","140","141","142","143","144","145","146","147","148","149","150","151","152","153","154","155","156","157","158","159","160","161","162","163","164","165","166","167","168","169","170","171","172","173","174","175","176","177","178","179","180","181","182","183","184","185","186","187","188","189","190","191","192","193","194","195","196","197","198","199","200","201","202","203"],["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.712,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0271,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,9.5635,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.1959,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],[{"type":"Point","coordinates":[38.9412,5.9014]},{"type":"Point","coordinates":[39.8362,6.3175]},{"type":"Point","coordinates":[40.7521,7.5674]},{"type":"Point","coordinates":[38.765,4.7192]},{"type":"Point","coordinates":[37.3632,4.893]},{"type":"Point","coordinates":[39.7891,6.2461]},{"type":"Point","coordinates":[40.649,7.1657]},{"type":"Point","coordinates":[41.1559,6.7542]},{"type":"Point","coordinates":[39.2547,5.9471]},{"type":"Point","coordinates":[41.0551,6.8386]},{"type":"Point","coordinates":[38.2213,3.8966]},{"type":"Point","coordinates":[39.1915,5.3172]},{"type":"Point","coordinates":[37.8933,4.1376]},{"type":"Point","coordinates":[41.2297,6.8855]},{"type":"Point","coordinates":[39.5655,5.3182]},{"type":"Point","coordinates":[40.3708,6.6748]},{"type":"Point","coordinates":[38.3667,5.7924]},{"type":"Point","coordinates":[40.7487,7.0833]},{"type":"Point","coordinates":[38.0833,4.8833]},{"type":"Point","coordinates":[39.2865,5.6295]},{"type":"Point","coordinates":[41.4464,6.6522]},{"type":"Point","coordinates":[38.263,5.7115]},{"type":"Point","coordinates":[38.2671,5.3957]},{"type":"Point","coordinates":[38.231,5.124]},{"type":"Point","coordinates":[37.993,5.505]},{"type":"Point","coordinates":[38.6116,6.0332]},{"type":"Point","coordinates":[40.6187,7.1968]},{"type":"Point","coordinates":[37.3569,5.1092]},{"type":"Point","coordinates":[38.3261,4.8138]},{"type":"Point","coordinates":[40.0778,6.8246]},{"type":"Point","coordinates":[38.8319,4.7372]},{"type":"Point","coordinates":[37.396,5.147]},{"type":"Point","coordinates":[40.2445,6.8403]},{"type":"Point","coordinates":[39.3276,5.836]},{"type":"Point","coordinates":[39.1084,5.9359]},{"type":"Point","coordinates":[38.136,5.625]},{"type":"Point","coordinates":[40.704,7.3082]},{"type":"Point","coordinates":[39.0175,5.9245]},{"type":"Point","coordinates":[34.6281,9.0283]},{"type":"Point","coordinates":[34.6469,8.9695]},{"type":"Point","coordinates":[34.712,9.5635]},{"type":"Point","coordinates":[34.7542,8.567]},{"type":"Point","coordinates":[34.7948,8.9661]},{"type":"Point","coordinates":[34.8295,9.0556]},{"type":"Point","coordinates":[34.9654,9.8265]},{"type":"Point","coordinates":[34.9704,8.7288]},{"type":"Point","coordinates":[34.9801,9.0055]},{"type":"Point","coordinates":[34.9999,8.9174]},{"type":"Point","coordinates":[35.0271,8.1959]},{"type":"Point","coordinates":[35.0395,8.2834]},{"type":"Point","coordinates":[35.0599,9.0309]},{"type":"Point","coordinates":[35.078,8.9772]},{"type":"Point","coordinates":[35.084,9.6416]},{"type":"Point","coordinates":[35.1159,9.7045]},{"type":"Point","coordinates":[35.1556,8.8416]},{"type":"Point","coordinates":[35.159,9.8058]},{"type":"Point","coordinates":[35.2034,8.8657]},{"type":"Point","coordinates":[35.2913,9.5869]},{"type":"Point","coordinates":[35.327,8.1959]},{"type":"Point","coordinates":[35.3308,8.9826]},{"type":"Point","coordinates":[35.3445,8.9457]},{"type":"Point","coordinates":[35.3963,8.6219]},{"type":"Point","coordinates":[35.3968,8.622]},{"type":"Point","coordinates":[35.4004,9.674]},{"type":"Point","coordinates":[35.4173,9.1255]},{"type":"Point","coordinates":[35.5107,9.6234]},{"type":"Point","coordinates":[35.5277,8.353]},{"type":"Point","coordinates":[35.5489,8.385]},{"type":"Point","coordinates":[35.5591,8.837]},{"type":"Point","coordinates":[35.5865,8.6584]},{"type":"Point","coordinates":[35.5915,8.2419]},{"type":"Point","coordinates":[35.6191,8.4416]},{"type":"Point","coordinates":[35.6542,8.2087]},{"type":"Point","coordinates":[35.6542,8.2088]},{"type":"Point","coordinates":[35.6907,9.2143]},{"type":"Point","coordinates":[35.7183,8.343]},{"type":"Point","coordinates":[35.7437,9.0024]},{"type":"Point","coordinates":[35.7507,8.5789]},{"type":"Point","coordinates":[35.7586,8.5789]},{"type":"Point","coordinates":[35.8027,9.1884]},{"type":"Point","coordinates":[35.8033,8.7131]},{"type":"Point","coordinates":[35.8197,8.8835]},{"type":"Point","coordinates":[36.021,9.0327]},{"type":"Point","coordinates":[36.1707,8.642]},{"type":"Point","coordinates":[36.4795,7.4853]},{"type":"Point","coordinates":[36.5047,9.9551]},{"type":"Point","coordinates":[36.5301,8.431]},{"type":"Point","coordinates":[36.5301,8.481]},{"type":"Point","coordinates":[36.5692,9.2721]},{"type":"Point","coordinates":[36.5827,8.9215]},{"type":"Point","coordinates":[36.602,7.8568]},{"type":"Point","coordinates":[36.6266,9.4378]},{"type":"Point","coordinates":[36.6292,9.6796]},{"type":"Point","coordinates":[36.6729,7.7164]},{"type":"Point","coordinates":[36.6994,9.5265]},{"type":"Point","coordinates":[36.7281,7.7457]},{"type":"Point","coordinates":[36.7529,7.6636]},{"type":"Point","coordinates":[34.5418,9.0147]},{"type":"Point","coordinates":[34.5584,9.4807]},{"type":"Point","coordinates":[41.2514,9.1806]},{"type":"Point","coordinates":[41.2528,9.4228]},{"type":"Point","coordinates":[41.3133,9.0366]},{"type":"Point","coordinates":[41.4976,9.1427]},{"type":"Point","coordinates":[34.5888,8.479]},{"type":"Point","coordinates":[35.0732,8.8923]},{"type":"Point","coordinates":[34.6097,9.3239]},{"type":"Point","coordinates":[36.7579,8.7646]},{"type":"Point","coordinates":[36.7957,7.7957]},{"type":"Point","coordinates":[36.7973,7.8636]},{"type":"Point","coordinates":[36.7999,7.7734]},{"type":"Point","coordinates":[36.854,8.4486]},{"type":"Point","coordinates":[36.8562,8.7554]},{"type":"Point","coordinates":[36.8836,9.0549]},{"type":"Point","coordinates":[36.9107,7.5704]},{"type":"Point","coordinates":[36.9335,7.7056]},{"type":"Point","coordinates":[36.9438,9.1567]},{"type":"Point","coordinates":[36.9891,7.6245]},{"type":"Point","coordinates":[36.9941,9.0856]},{"type":"Point","coordinates":[36.9957,8.1466]},{"type":"Point","coordinates":[37.0276,7.7082]},{"type":"Point","coordinates":[37.0569,8.442]},{"type":"Point","coordinates":[37.0951,9.9101]},{"type":"Point","coordinates":[37.1103,7.7261]},{"type":"Point","coordinates":[37.1853,7.7618]},{"type":"Point","coordinates":[37.2193,7.7477]},{"type":"Point","coordinates":[37.2206,7.6222]},{"type":"Point","coordinates":[37.3451,8.8988]},{"type":"Point","coordinates":[37.3466,8.6991]},{"type":"Point","coordinates":[37.3792,7.8775]},{"type":"Point","coordinates":[37.41,5.12]},{"type":"Point","coordinates":[37.4124,9.7905]},{"type":"Point","coordinates":[37.6791,8.4993]},{"type":"Point","coordinates":[37.8315,8.597]},{"type":"Point","coordinates":[37.9556,8.4754]},{"type":"Point","coordinates":[38.02,5.56]},{"type":"Point","coordinates":[38.14,5.6]},{"type":"Point","coordinates":[38.25,5.07]},{"type":"Point","coordinates":[38.263,5.7117]},{"type":"Point","coordinates":[38.3136,7.3651]},{"type":"Point","coordinates":[38.3987,7.3123]},{"type":"Point","coordinates":[38.4259,7.3283]},{"type":"Point","coordinates":[38.4823,7.3304]},{"type":"Point","coordinates":[38.4903,7.2976]},{"type":"Point","coordinates":[38.5255,7.1648]},{"type":"Point","coordinates":[38.6353,7.3525]},{"type":"Point","coordinates":[38.6811,7.9926]},{"type":"Point","coordinates":[38.6886,7.7633]},{"type":"Point","coordinates":[38.697,8.1747]},{"type":"Point","coordinates":[38.8453,8.3568]},{"type":"Point","coordinates":[38.8844,7.9231]},{"type":"Point","coordinates":[38.8953,8.694]},{"type":"Point","coordinates":[38.9027,8.5766]},{"type":"Point","coordinates":[38.988,8.5048]},{"type":"Point","coordinates":[39.179,8.4592]},{"type":"Point","coordinates":[39.2998,8.2064]},{"type":"Point","coordinates":[39.3785,8.5253]},{"type":"Point","coordinates":[39.501,7.2264]},{"type":"Point","coordinates":[39.5139,8.3486]},{"type":"Point","coordinates":[39.7774,8.8836]},{"type":"Point","coordinates":[39.7803,7.8223]},{"type":"Point","coordinates":[39.8374,8.994]},{"type":"Point","coordinates":[40.3698,8.3784]},{"type":"Point","coordinates":[40.3708,8.8757]},{"type":"Point","coordinates":[40.4205,8.5641]},{"type":"Point","coordinates":[40.4641,8.8181]},{"type":"Point","coordinates":[40.53,8.82]},{"type":"Point","coordinates":[40.6098,8.6852]},{"type":"Point","coordinates":[40.6467,8.7469]},{"type":"Point","coordinates":[40.6572,9.1998]},{"type":"Point","coordinates":[40.7848,8.8762]},{"type":"Point","coordinates":[40.7868,9.1864]},{"type":"Point","coordinates":[40.8317,8.9247]},{"type":"Point","coordinates":[40.8815,8.7661]},{"type":"Point","coordinates":[40.8815,8.7662]},{"type":"Point","coordinates":[40.882,9.1064]},{"type":"Point","coordinates":[40.9838,9.1895]},{"type":"Point","coordinates":[41.0049,9.1873]},{"type":"Point","coordinates":[41.0365,9.3063]},{"type":"Point","coordinates":[41.07,9.28]},{"type":"Point","coordinates":[41.1138,9.1352]},{"type":"Point","coordinates":[41.1456,9.0751]},{"type":"Point","coordinates":[41.15,9.03]},{"type":"Point","coordinates":[41.1509,9.034]},{"type":"Point","coordinates":[41.191,9.405]},{"type":"Point","coordinates":[41.205,9.1511]},{"type":"Point","coordinates":[41.3146,8.9945]},{"type":"Point","coordinates":[41.3965,8.992]},{"type":"Point","coordinates":[41.4014,9.4602]},{"type":"Point","coordinates":[41.5294,8.8262]},{"type":"Point","coordinates":[41.5532,8.7281]},{"type":"Point","coordinates":[41.5791,8.8865]},{"type":"Point","coordinates":[41.622,8.9874]},{"type":"Point","coordinates":[41.6346,9.1368]},{"type":"Point","coordinates":[41.7083,8.782]},{"type":"Point","coordinates":[41.7384,9.1384]},{"type":"Point","coordinates":[41.7534,9.4243]},{"type":"Point","coordinates":[41.826,8.903]},{"type":"Point","coordinates":[41.8785,8.9745]},{"type":"Point","coordinates":[42.2162,9.1429]},{"type":"Point","coordinates":[42.3192,9.2356]},{"type":"Point","coordinates":[42.3604,9.3663]},{"type":"Point","coordinates":[42.38,9.3166]},{"type":"Point","coordinates":[42.4915,9.3658]}]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>pf_pos<\/th>\n <th>longitude<\/th>\n <th>latitude<\/th>\n <th>geometry<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"className":"dt-right","targets":[2,3]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- # Packages and Data ```r Oromia <- getData("GADM", country="ETH", level=1) %>% st_as_sf() %>% filter(NAME_1=="Oromia") ``` <div id="htmlwidget-2ebaf619c3e6b014a28e" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-2ebaf619c3e6b014a28e">{"x":{"filter":"none","vertical":false,"data":[["1"],["ETH"],["Ethiopia"],["ETH.8_1"],["Oromia"],["Oromiya"],[null],["Kilil"],["State"],["04"],["ET.OR"],[{"type":"Polygon","coordinates":[[[37.20056915,7.28216076],[37.1827507,7.26485777],[37.15417862,7.24066639],[37.12858963,7.22844601],[37.11060333,7.22527552],[37.1048317,7.22449589],[37.06813049,7.23524618],[37.05447006,7.23752642],[37.03951263,7.24001646],[37.00329208,7.23443651],[36.96464539,7.22473288],[36.96579742,7.24192429],[36.95546341,7.26049089],[36.93113327,7.2827158],[36.91413116,7.29824209],[36.87915421,7.32969427],[36.84093475,7.32598972],[36.80350494,7.26934481],[36.79111099,7.25228357],[36.78422546,7.2552948],[36.76252365,7.26590776],[36.73939133,7.28471851],[36.71987915,7.29850149],[36.71418381,7.30213976],[36.69407272,7.3131299],[36.67256165,7.3280406],[36.65174484,7.34501171],[36.63805389,7.37120247],[36.63295364,7.37786293],[36.6227951,7.37922287],[36.61294556,7.37287283],[36.60596466,7.36836243],[36.59833527,7.36903238],[36.5749855,7.37933302],[36.54123306,7.39304352],[36.50655365,7.4058342],[36.48392105,7.4160099],[36.47015381,7.41539764],[36.45316315,7.41742468],[36.42707443,7.41320467],[36.40629578,7.40854454],[36.37463379,7.41557503],[36.36164474,7.43141556],[36.34866714,7.44541597],[36.32370377,7.44695616],[36.301754,7.44987631],[36.28693771,7.45972681],[36.27952957,7.46328688],[36.26541519,7.46932745],[36.24089432,7.47729778],[36.22469711,7.48438787],[36.21631241,7.49078178],[36.20677567,7.49870872],[36.20059586,7.50363874],[36.18088531,7.52359962],[36.15655518,7.54262066],[36.12761688,7.55564117],[36.08694839,7.55389118],[36.06219864,7.55748129],[36.04506683,7.56548166],[36.03532791,7.57258224],[36.03248596,7.59145308],[36.03979874,7.6140337],[36.04712677,7.63432503],[36.05190659,7.65390587],[36.04189682,7.67436647],[36.0205574,7.69201756],[36.01823807,7.69684744],[36.01216888,7.70948839],[35.99723816,7.74511957],[35.98635864,7.75465679],[35.9784584,7.7581706],[35.96108627,7.76640081],[35.95201111,7.76966143],[35.9416275,7.77392101],[35.92255783,7.78670168],[35.897789,7.80691242],[35.87505722,7.82179308],[35.85998917,7.82933378],[35.84861755,7.83896399],[35.85112762,7.84795427],[35.85908508,7.86013794],[35.8742218,7.87578392],[35.88687515,7.89253807],[35.89531708,7.90936708],[35.90242767,7.92620802],[35.90512848,7.9460187],[35.89878845,7.97108984],[35.89176941,7.99202108],[35.88803864,8.00052166],[35.87801743,8.04023266],[35.87271881,8.05877399],[35.86878967,8.06529522],[35.86087036,8.07723427],[35.83733749,8.07949448],[35.81634903,8.08242512],[35.80958176,8.08137608],[35.79808044,8.07913494],[35.78284836,8.08017445],[35.76576996,8.08123493],[35.75334167,8.06784248],[35.74399567,8.05094147],[35.72800446,8.03956985],[35.72518158,8.03063107],[35.72285461,8.02246666],[35.72987366,8.00587559],[35.73851395,7.98882389],[35.76741409,7.9691124],[35.77137375,7.96289158],[35.78251266,7.95395088],[35.79527283,7.94802046],[35.79992294,7.94066954],[35.79972076,7.93260908],[35.7922554,7.9206996],[35.78355789,7.91022968],[35.77606583,7.8991704],[35.76214981,7.88260603],[35.7395401,7.86662531],[35.71643829,7.85547495],[35.70346832,7.85910511],[35.6937294,7.86435509],[35.68098831,7.85633469],[35.6764183,7.8534646],[35.6669693,7.84489441],[35.64159012,7.82889366],[35.62812042,7.82060146],[35.6182785,7.81474304],[35.60622025,7.80957794],[35.58961868,7.80863285],[35.57017136,7.81292295],[35.55463028,7.82021332],[35.54439926,7.83099365],[35.53620911,7.84938478],[35.5331192,7.86733532],[35.53052139,7.87998581],[35.52072525,7.89560652],[35.51218033,7.89234638],[35.50479889,7.88470602],[35.49719238,7.88029623],[35.49023056,7.88256598],[35.4849205,7.879776],[35.47431183,7.87350559],[35.45973206,7.87251568],[35.44466019,7.87797594],[35.43353271,7.88275623],[35.42873383,7.88082504],[35.41842651,7.86413527],[35.40948868,7.8433609],[35.40057755,7.82957363],[35.38914108,7.82702351],[35.3719902,7.83247375],[35.3580513,7.84299421],[35.34985733,7.83487415],[35.34202957,7.82136869],[35.34141922,7.80339718],[35.34087753,7.77661085],[35.32489777,7.76533079],[35.31080246,7.76098061],[35.2930336,7.7519002],[35.28988647,7.74723434],[35.2755661,7.74600792],[35.24794388,7.72651196],[35.22273254,7.70310259],[35.22016525,7.68024254],[35.22032166,7.66259718],[35.21232224,7.65149689],[35.17293549,7.65422916],[35.11603546,7.64509201],[35.07709503,7.73100519],[35.03850555,7.73951292],[34.99547195,7.7479744],[34.93197632,7.72816849],[34.9371109,7.7647934],[34.93227768,7.82795429],[34.9442749,7.8793931],[34.95068741,7.89028645],[35.00061417,7.86232185],[35.01309967,7.88245773],[35.00848007,7.92350197],[35.04833984,7.9544363],[35.10855865,7.95711327],[35.14102554,7.9890275],[35.14071655,8.02168941],[35.11793518,8.0825882],[35.08908844,8.11392212],[35.10387802,8.11511803],[35.1432724,8.14285278],[35.15765762,8.13671017],[35.13813019,8.18182755],[35.11891174,8.20060921],[35.073246,8.22018528],[35.01825714,8.21964359],[34.96479034,8.24734688],[34.9259491,8.24904251],[34.88258362,8.25093079],[34.85509491,8.25064754],[34.82642746,8.26193523],[34.78580475,8.30363941],[34.79507065,8.32690907],[34.78525162,8.35524273],[34.76763916,8.36337948],[34.72875214,8.39002228],[34.66742706,8.39503956],[34.63033676,8.40200043],[34.57427216,8.40347958],[34.55652618,8.45513439],[34.522686,8.47554493],[34.47119522,8.44757652],[34.43635178,8.42177391],[34.39491653,8.40078449],[34.34727859,8.376647],[34.3291893,8.38696003],[34.31329727,8.38992882],[34.31520844,8.40679264],[34.33491135,8.43860626],[34.34405899,8.4692421],[34.35743332,8.49992847],[34.35822296,8.52204323],[34.35358429,8.55567265],[34.34547043,8.60009956],[34.31242752,8.68209934],[34.27938843,8.70883656],[34.25573349,8.70263863],[34.23517609,8.68614769],[34.22351837,8.66976738],[34.1984787,8.65617085],[34.18964005,8.65163231],[34.15405655,8.65118313],[34.14157486,8.65195179],[34.14363098,8.66451168],[34.14448929,8.66976929],[34.1437912,8.67536545],[34.14220047,8.68807125],[34.14214706,8.69543171],[34.14105606,8.70179749],[34.14143372,8.71088028],[34.14009857,8.72303009],[34.14041138,8.73007488],[34.14156723,8.73600006],[34.14112854,8.74314022],[34.14080811,8.7486105],[34.13996124,8.75638008],[34.1395874,8.76471996],[34.13949966,8.77110958],[34.14044952,8.77692032],[34.14048004,8.78394032],[34.14012909,8.79158974],[34.14053726,8.80109024],[34.14152145,8.80982971],[34.14135742,8.81783009],[34.14114761,8.82404995],[34.14092636,8.83399773],[34.14092636,8.84142017],[34.14055252,8.8499403],[34.14061737,8.85908794],[34.14080811,8.87215042],[34.14076233,8.88268471],[34.14110947,8.89016056],[34.14107895,8.89585972],[34.14103699,8.90114975],[34.14060974,8.91096497],[34.14382553,8.95114231],[34.14471436,8.96223831],[34.22148132,8.96336269],[34.26724243,8.96426392],[34.3053894,8.9650116],[34.37595367,8.96638298],[34.43065262,8.96743679],[34.50370789,9.00413418],[34.48366547,9.04714584],[34.45832825,9.07318306],[34.42683411,9.07179546],[34.39326859,9.11633015],[34.34087753,9.16877937],[34.40174103,9.18663502],[34.43955994,9.21557522],[34.45238876,9.25748062],[34.45172882,9.30870247],[34.46627045,9.36581039],[34.45180511,9.44911098],[34.51297379,9.44800091],[34.49873352,9.51423073],[34.52323151,9.5411129],[34.53214264,9.59056473],[34.54846573,9.62966251],[34.58539581,9.63035965],[34.63201523,9.58802128],[34.66147614,9.57498646],[34.6859169,9.56368637],[34.73513794,9.54530144],[34.78298569,9.55333138],[34.79205704,9.6033144],[34.81085587,9.63011837],[34.81045532,9.66238403],[34.80995941,9.70224094],[34.79614258,9.73623943],[34.80731201,9.76105404],[34.83166504,9.80311584],[34.85446548,9.81668377],[34.86972809,9.82066727],[34.8770752,9.8454361],[34.89215469,9.864604],[34.89768219,9.88175678],[34.86679459,9.90226078],[34.88778305,9.90821362],[34.90441895,9.95587826],[34.92765427,9.93527985],[34.94299698,9.93356705],[34.96611786,9.9659071],[34.98818207,9.99676704],[35.01501846,9.99519062],[35.01730347,9.96483898],[35.01965332,9.928792],[35.04851532,9.91774082],[35.08102036,9.92192078],[35.08081818,9.93900776],[35.08834076,9.95048904],[35.15958405,9.9484024],[35.22079849,9.92542553],[35.22288895,9.91025639],[35.23074341,9.89325237],[35.2784729,9.90707874],[35.29745483,9.92248344],[35.31098938,9.91123676],[35.32469177,9.88479614],[35.34980011,9.86607552],[35.3637886,9.81304359],[35.38689041,9.80189323],[35.40803909,9.79452038],[35.41962814,9.78514481],[35.37882996,9.76540661],[35.38302994,9.75899601],[35.37536621,9.74995041],[35.36508179,9.72746658],[35.36839676,9.71928978],[35.38938522,9.7200489],[35.40027618,9.71613884],[35.4107666,9.71367741],[35.42450333,9.70074844],[35.44120789,9.68501186],[35.45537567,9.67223072],[35.46842194,9.67037582],[35.48488235,9.67800331],[35.50510788,9.68870258],[35.5332756,9.69041824],[35.53397369,9.69641876],[35.53939056,9.69763851],[35.5537262,9.69611835],[35.56108856,9.69201851],[35.56922913,9.67807865],[35.57851791,9.66551876],[35.59442902,9.65603828],[35.62475586,9.65116882],[35.63472748,9.6514883],[35.65343857,9.6349287],[35.65054703,9.61302853],[35.64358139,9.59455013],[35.64524841,9.5776577],[35.64749146,9.55331039],[35.66503525,9.52709675],[35.67139053,9.5172348],[35.68648529,9.51041508],[35.71076202,9.48677158],[35.73185349,9.48149872],[35.75289154,9.48169422],[35.76499176,9.47335052],[35.77013397,9.4594717],[35.77732468,9.44013977],[35.77841568,9.43069935],[35.76592255,9.42759991],[35.75400162,9.41654778],[35.75012207,9.40308285],[35.75517654,9.36478806],[35.74919891,9.34861279],[35.75055695,9.33112812],[35.7567482,9.32363796],[35.7739563,9.32432365],[35.79719162,9.30861759],[35.80845261,9.30887604],[35.85987091,9.3249712],[35.91108704,9.31595802],[35.91787338,9.28563881],[35.9307251,9.26723194],[35.94932175,9.24915409],[35.97541428,9.24290752],[36.03157806,9.23342609],[36.04396439,9.19506264],[36.05361176,9.11943722],[36.03402328,9.10741711],[36.02807617,9.05597687],[36.04244995,9.03217793],[36.04215622,9.01504803],[36.06394196,9.01990795],[36.08654404,9.01261711],[36.09697342,9.03241444],[36.07553864,9.06263733],[36.08417511,9.05842113],[36.09464264,9.0649786],[36.10855484,9.08003139],[36.128685,9.05111694],[36.13142776,9.02078819],[36.15963364,9.0206604],[36.16708755,9.02799225],[36.17679214,9.04452515],[36.22594833,9.03096771],[36.24647903,9.04096222],[36.23580933,9.05094624],[36.20764923,9.06715298],[36.18458557,9.07195187],[36.16147232,9.10535145],[36.17191315,9.10829735],[36.20394897,9.15734673],[36.2289238,9.22903061],[36.22633743,9.27172852],[36.19489288,9.28897381],[36.19459534,9.32628918],[36.2009964,9.34027195],[36.20589066,9.35474014],[36.23426819,9.3470068],[36.21458817,9.33241653],[36.21476364,9.31487083],[36.21837234,9.29711819],[36.25317764,9.30182362],[36.27022934,9.32837009],[36.27511597,9.34383297],[36.28963852,9.32153034],[36.29548264,9.30169201],[36.31899643,9.30585003],[36.3261795,9.33077335],[36.33376694,9.33979893],[36.34926224,9.34489155],[36.37471771,9.35801888],[36.40823746,9.35778427],[36.43635178,9.35797215],[36.46141052,9.35715675],[36.48410797,9.36786747],[36.47518921,9.39463711],[36.48364258,9.40515995],[36.49913025,9.41473675],[36.507267,9.4193449],[36.51491165,9.44413376],[36.49866486,9.47288418],[36.49443054,9.50936604],[36.46116638,9.52805805],[36.41901016,9.51585388],[36.40063095,9.51098824],[36.38030624,9.48048019],[36.35513687,9.49472237],[36.33595657,9.51199436],[36.31417465,9.54068851],[36.30102539,9.55551529],[36.26626587,9.56869507],[36.23177719,9.57032299],[36.22913742,9.5798254],[36.20077896,9.6149292],[36.19261169,9.6584959],[36.19186401,9.68787766],[36.17749786,9.70131397],[36.16188812,9.73046684],[36.15905762,9.7469368],[36.14341736,9.75775146],[36.13249207,9.76259613],[36.10528564,9.77332401],[36.13647079,9.80744743],[36.13241959,9.82983589],[36.12820816,9.85221386],[36.15234375,9.84893131],[36.15219498,9.86636353],[36.13402939,9.91588974],[36.11980057,9.96133327],[36.12064743,9.97157288],[36.13990021,10.00970554],[36.16732788,10.03384781],[36.1845665,10.04966164],[36.21892548,10.05321217],[36.25891876,10.07845211],[36.2678833,10.08848858],[36.26946259,10.10361385],[36.27189255,10.11308765],[36.28990173,10.1211977],[36.33231735,10.13839436],[36.36942291,10.16165257],[36.39527512,10.18709755],[36.41619492,10.21676445],[36.44535065,10.24584389],[36.49091339,10.23426914],[36.50977707,10.23936939],[36.51548767,10.23993111],[36.52244186,10.25043583],[36.53791428,10.26316261],[36.54507446,10.2692585],[36.54934311,10.27882671],[36.55448151,10.28507233],[36.56200409,10.28369904],[36.5692749,10.28157711],[36.57584763,10.27923584],[36.5754776,10.27103138],[36.57468414,10.26417351],[36.58802414,10.26186752],[36.610569,10.25922203],[36.64340591,10.27113342],[36.65969086,10.27704048],[36.67146683,10.28401566],[36.68494797,10.29199886],[36.69219208,10.29628944],[36.68871689,10.31141567],[36.6853981,10.31759548],[36.6859169,10.32312965],[36.68227005,10.32948208],[36.68090439,10.33461475],[36.68349838,10.33930492],[36.6881485,10.34499836],[36.68917465,10.35184669],[36.69165421,10.35731983],[36.6999588,10.35750961],[36.7098999,10.35773659],[36.71801376,10.35792255],[36.72351837,10.36079979],[36.73139572,10.36135197],[36.73879623,10.36147594],[36.74571609,10.35997009],[36.75241852,10.36598396],[36.76176071,10.36730957],[36.76661301,10.36234093],[36.76938248,10.35779572],[36.77300262,10.35255241],[36.77659988,10.34596825],[36.78330231,10.34413242],[36.79195786,10.3433857],[36.79873657,10.33784103],[36.80659866,10.33537102],[36.81181717,10.33727551],[36.81700897,10.34174156],[36.82473755,10.34246445],[36.83358002,10.34247017],[36.83961105,10.34080124],[36.84531021,10.34077168],[36.85059738,10.34102726],[36.85939026,10.34113026],[36.86923981,10.34069252],[36.87582016,10.33376884],[36.87878036,10.32761574],[36.88311005,10.32065487],[36.88873672,10.31551647],[36.89796066,10.31284809],[36.91203308,10.31366444],[36.91917419,10.314188],[36.92463684,10.31379414],[36.93077087,10.31093979],[36.94185638,10.31172752],[36.95367432,10.31250286],[36.96474838,10.31188202],[36.97165298,10.31509304],[36.97991943,10.31402683],[36.98960876,10.30878067],[36.99710464,10.30296898],[37.00160599,10.29533482],[37.00720596,10.28884983],[37.0132637,10.28493881],[37.02169418,10.28482342],[37.03079987,10.28610325],[37.04009247,10.28535843],[37.04587173,10.2883997],[37.05076981,10.28360367],[37.05588913,10.28274345],[37.06184006,10.28173447],[37.06970596,10.27608395],[37.07434464,10.27073002],[37.07810211,10.26337433],[37.08710098,10.25949383],[37.09417343,10.25822544],[37.10084915,10.25587368],[37.10779953,10.25323296],[37.11460495,10.25069714],[37.12050247,10.2492609],[37.1277771,10.24767399],[37.13483429,10.24598122],[37.14101791,10.24507618],[37.14917374,10.24464321],[37.15755081,10.24425697],[37.16622543,10.244133],[37.17453384,10.24330711],[37.18181229,10.24126434],[37.19036865,10.2362709],[37.19599915,10.23091125],[37.19989777,10.22329617],[37.19748688,10.21387577],[37.19342804,10.20601082],[37.18940735,10.19976616],[37.18582916,10.19364071],[37.18252182,10.18800545],[37.179142,10.18388081],[37.17518234,10.17841721],[37.17095566,10.17396069],[37.16830826,10.1685009],[37.16770172,10.16327381],[37.17182922,10.15531254],[37.17598343,10.1513176],[37.17977524,10.14437866],[37.1821022,10.13734436],[37.18413925,10.1303606],[37.18628693,10.12399673],[37.18839645,10.11668873],[37.19073868,10.11155605],[37.19550323,10.10452557],[37.20103073,10.0999651],[37.20650864,10.09503746],[37.21331406,10.08790779],[37.22050095,10.08391285],[37.22509384,10.08164787],[37.23018265,10.0796833],[37.2358551,10.07710171],[37.24074554,10.07561016],[37.24716187,10.07611752],[37.25369644,10.07669353],[37.26188278,10.0766716],[37.27164078,10.07363892],[37.27737808,10.06839275],[37.28250885,10.06489086],[37.2877121,10.06287956],[37.29259491,10.05793476],[37.29653931,10.05284977],[37.29874802,10.04479313],[37.29972458,10.03877544],[37.3021698,10.03403664],[37.30561066,10.02665329],[37.31287003,10.02170086],[37.3178215,10.01805019],[37.32040405,10.01238346],[37.32325363,10.00810146],[37.32954025,10.00584888],[37.33930206,10.00451851],[37.34474945,10.00581837],[37.34930038,10.01194859],[37.35192871,10.01955128],[37.35359192,10.02715492],[37.35520935,10.03395748],[37.35698318,10.0402317],[37.36219406,10.04317856],[37.36754227,10.04356098],[37.3735733,10.04219913],[37.38040161,10.0430994],[37.3873024,10.04654408],[37.39827728,10.04615021],[37.40514755,10.04258823],[37.41220856,10.03984547],[37.41836929,10.03720379],[37.42435455,10.03378677],[37.42928696,10.02947807],[37.43424606,10.02503681],[37.43984604,10.01778793],[37.44446945,10.01455879],[37.45321655,10.01233006],[37.46133041,10.01211357],[37.4672699,10.01317787],[37.47574997,10.01493931],[37.47807312,10.00918293],[37.48313522,10.00350857],[37.48854446,10.00304508],[37.49332428,10.00091171],[37.50160217,9.99794769],[37.51083374,9.99943638],[37.51972961,9.99991417],[37.52923584,9.99617672],[37.53677368,9.98895359],[37.54104233,9.98340511],[37.54616165,9.97743988],[37.55136108,9.97342777],[37.55168152,9.96684456],[37.55292892,9.95855427],[37.5572319,9.95386124],[37.56334305,9.94989967],[37.56997299,9.94516468],[37.57432175,9.94194794],[37.57988358,9.94085979],[37.58693314,9.93939781],[37.59063339,9.93298244],[37.58966827,9.92684937],[37.59239578,9.91876411],[37.59931564,9.91779041],[37.60579681,9.91311836],[37.6128273,9.90941048],[37.61745453,9.90169048],[37.6182251,9.89388943],[37.62356567,9.88903999],[37.63073349,9.88700962],[37.63662338,9.88554955],[37.64138031,9.88248253],[37.64490128,9.87786102],[37.64756393,9.87349987],[37.65158463,9.86910915],[37.65641403,9.8648243],[37.66185379,9.86048412],[37.66692734,9.85946465],[37.6744957,9.86191559],[37.68039322,9.86231804],[37.68511581,9.85751438],[37.68971634,9.8506918],[37.69456482,9.84270287],[37.70065308,9.83671474],[37.70530319,9.83931065],[37.71065903,9.84261417],[37.71700668,9.842556],[37.72434616,9.84454441],[37.73069763,9.84485912],[37.73600388,9.84079456],[37.74289322,9.840662],[37.74822998,9.84666443],[37.74853134,9.85593891],[37.75395584,9.85707664],[37.76096725,9.85192966],[37.76908875,9.85144138],[37.77459335,9.85650253],[37.78189087,9.85725212],[37.78808594,9.85962772],[37.79405594,9.86037445],[37.80058289,9.86182404],[37.80699158,9.86529064],[37.81602097,9.86715031],[37.82325363,9.87389088],[37.82724762,9.88076115],[37.83082199,9.88463783],[37.83641434,9.88789177],[37.84268188,9.89670277],[37.84841537,9.89757824],[37.85593796,9.89594078],[37.85922241,9.90022087],[37.85876083,9.90582848],[37.86038971,9.91278839],[37.86446381,9.91286182],[37.86993408,9.90753365],[37.87446213,9.91116428],[37.87592697,9.91630554],[37.88015366,9.92150784],[37.89302826,9.92725849],[37.90139008,9.93037415],[37.90932465,9.93044853],[37.91440201,9.92357922],[37.91878128,9.91872692],[37.92340469,9.91474819],[37.93007278,9.91458702],[37.93507767,9.91898632],[37.93532181,9.9259634],[37.93078613,9.9305172],[37.93228149,9.93577862],[37.93283844,9.94127941],[37.93874359,9.9440794],[37.94424438,9.9411335],[37.95193481,9.93839645],[37.95706558,9.93863773],[37.96414185,9.94296646],[37.97038269,9.94538021],[37.97842026,9.94908142],[37.98603439,9.94973946],[37.99341965,9.94908619],[37.99923325,9.95358181],[38.00255966,9.95963573],[38.00845337,9.96413517],[38.0160141,9.96677876],[38.02558136,9.96831226],[38.03387451,9.96752262],[38.04358673,9.96977997],[38.04563904,9.97477722],[38.04811478,9.9815321],[38.05321503,9.98188972],[38.05993271,9.98180389],[38.06232071,9.98815632],[38.0643158,9.99789524],[38.06979752,10.00152397],[38.07492447,10.00221157],[38.08120728,10.00217533],[38.08271027,10.007514],[38.0802803,10.0152235],[38.08756256,10.02025127],[38.09408188,10.02021694],[38.10071945,10.02225685],[38.10626984,10.01940632],[38.11312866,10.01599121],[38.1182785,10.01775074],[38.11708832,10.02361774],[38.11832809,10.0289917],[38.12097931,10.03477001],[38.12530136,10.04315662],[38.13093948,10.04745007],[38.13695145,10.04741383],[38.14215851,10.04325199],[38.14513016,10.0366869],[38.1505661,10.03512192],[38.15544128,10.03985691],[38.16035461,10.04410267],[38.16733932,10.04608345],[38.16990662,10.05065823],[38.17061234,10.05775261],[38.17662811,10.06310558],[38.18429947,10.06683731],[38.18809509,10.07047653],[38.19424057,10.07386112],[38.20011902,10.07807636],[38.20374298,10.08178329],[38.20946121,10.08445072],[38.2163887,10.08636379],[38.22197342,10.09225273],[38.22737885,10.09506035],[38.23355865,10.09889317],[38.24042511,10.10263348],[38.24778748,10.10194969],[38.25151825,10.09647942],[38.25237656,10.08842754],[38.25811005,10.08765316],[38.26418686,10.08927536],[38.27175903,10.09180069],[38.28077316,10.09092712],[38.28612518,10.08853817],[38.29527664,10.0892477],[38.300354,10.09540558],[38.30226517,10.10344601],[38.30860138,10.10795307],[38.31447983,10.10715008],[38.32180786,10.10718632],[38.32651901,10.11454773],[38.3237114,10.12343407],[38.32596207,10.13311958],[38.33094025,10.14309216],[38.33332062,10.15136242],[38.34004974,10.15622234],[38.34827805,10.15884113],[38.35407639,10.15507412],[38.3570137,10.15084267],[38.35806274,10.14431095],[38.36630249,10.14048576],[38.37562943,10.14035511],[38.38989639,10.1432066],[38.39749908,10.14427757],[38.40093613,10.14864159],[38.40205383,10.15470505],[38.40631104,10.16037273],[38.41187668,10.16452694],[38.41822815,10.16717529],[38.42572403,10.17175579],[38.42589569,10.1778307],[38.42105103,10.18075275],[38.4262619,10.18666649],[38.43004227,10.19313335],[38.42664719,10.200037],[38.42358398,10.2070055],[38.41939163,10.21322918],[38.419384,10.21984673],[38.42417526,10.22270775],[38.42721176,10.22682858],[38.43071747,10.2316227],[38.43681335,10.23546314],[38.44160461,10.23783493],[38.44807434,10.24025154],[38.45419312,10.2466898],[38.45574951,10.25389194],[38.45533752,10.2611351],[38.46267319,10.26767254],[38.46088028,10.27596378],[38.45787048,10.28200912],[38.46006775,10.2899704],[38.46590042,10.29088497],[38.47253036,10.29180431],[38.469841,10.29852581],[38.46609116,10.30522442],[38.46792221,10.31109047],[38.47372055,10.31460857],[38.47530746,10.32239532],[38.47506714,10.33066654],[38.47797775,10.33880234],[38.48120117,10.34436703],[38.47954178,10.35112572],[38.47608948,10.35622597],[38.47667694,10.36326027],[38.48348999,10.36544895],[38.49037933,10.364007],[38.49619293,10.36357307],[38.50284195,10.36326408],[38.51529694,10.36693096],[38.52378464,10.36781025],[38.53439331,10.36650467],[38.54115295,10.37029362],[38.55088043,10.37328339],[38.56021118,10.37592316],[38.56595993,10.37431335],[38.57173538,10.37512493],[38.5779686,10.37717247],[38.58486938,10.37511444],[38.59211349,10.37438583],[38.59911346,10.37410831],[38.60466385,10.37460041],[38.61343384,10.37706089],[38.62384796,10.37835026],[38.63027573,10.38217449],[38.63594818,10.3803463],[38.6444397,10.37697506],[38.65015411,10.37649345],[38.65648651,10.3736105],[38.66415787,10.372756],[38.67134476,10.37409496],[38.67601395,10.37742043],[38.68230438,10.37904072],[38.69015884,10.37871361],[38.69739914,10.37939167],[38.7025528,10.38122368],[38.70747375,10.38433933],[38.71263504,10.38686943],[38.72071457,10.386199],[38.7295723,10.3859272],[38.7382431,10.38562679],[38.74612045,10.38374615],[38.75292969,10.37955284],[38.76123428,10.37558651],[38.7675972,10.37054443],[38.77214432,10.36616039],[38.77743912,10.36562061],[38.78462982,10.36672211],[38.79069138,10.3665247],[38.79844666,10.36807251],[38.80646896,10.36883545],[38.81564713,10.36432076],[38.82290268,10.36376286],[38.82845306,10.36303139],[38.83432007,10.36182499],[38.83974457,10.36102867],[38.84572983,10.3610239],[38.84506607,10.35476208],[38.84843063,10.34501362],[38.85186005,10.33907509],[38.85323334,10.33377743],[38.85456848,10.32540131],[38.85534668,10.31426525],[38.85541916,10.30805874],[38.85560989,10.30015182],[38.85567856,10.29473782],[38.85947037,10.28602791],[38.86084366,10.2779808],[38.85565567,10.26976109],[38.85224152,10.26541042],[38.8480835,10.26129913],[38.8445816,10.25606728],[38.84444809,10.25065231],[38.84762955,10.24432659],[38.84463882,10.23816204],[38.84072113,10.23438454],[38.83527374,10.22573376],[38.83268356,10.21211338],[38.83367157,10.2039547],[38.83810043,10.19806385],[38.83648682,10.19089317],[38.83440018,10.179245],[38.83491516,10.17166233],[38.83374405,10.16185284],[38.8358345,10.1556406],[38.8392067,10.14895248],[38.8443222,10.14430809],[38.84721756,10.13873482],[38.84741592,10.13105106],[38.84859085,10.12389088],[38.8526001,10.11805344],[38.84739304,10.1130867],[38.84260941,10.10992527],[38.83908844,10.114398],[38.8339653,10.11294365],[38.82899857,10.10966396],[38.82361603,10.10968113],[38.81883621,10.10495663],[38.81664276,10.10011482],[38.8109436,10.09440899],[38.80529785,10.09349632],[38.79953003,10.09027386],[38.79197693,10.08936024],[38.78448486,10.08591461],[38.77726364,10.08280182],[38.77268219,10.07723808],[38.76494598,10.0708046],[38.75792313,10.06825161],[38.75124359,10.06333065],[38.74098206,10.06108761],[38.73385239,10.05632877],[38.7277298,10.05434608],[38.72135925,10.05144405],[38.71473694,10.0527916],[38.70565796,10.0530014],[38.70146179,10.04936218],[38.69572449,10.05090046],[38.68868637,10.05023193],[38.681633,10.05277824],[38.67446899,10.0532856],[38.66880035,10.05542183],[38.66266632,10.0573616],[38.65723419,10.05145168],[38.66048431,10.04546833],[38.66144943,10.03912449],[38.66081619,10.03381157],[38.66526413,10.02591801],[38.66949844,10.01917171],[38.67381668,10.01514912],[38.68111801,10.01342392],[38.68648911,10.00800514],[38.6941185,10.00782013],[38.69966888,10.004282],[38.70985031,9.99839401],[38.71862411,9.9960165],[38.72544479,9.99134159],[38.73357391,9.99287987],[38.740242,9.9875288],[38.74585724,9.98565674],[38.75212479,9.98471832],[38.75805664,9.98067188],[38.76487732,9.98064518],[38.77121735,9.97810078],[38.77932358,9.98238468],[38.78475571,9.97834206],[38.78960037,9.97190189],[38.79686737,9.9677248],[38.80057144,9.96219635],[38.80266571,9.95355511],[38.80142593,9.94651127],[38.79732132,9.94077492],[38.79639435,9.93371487],[38.79782486,9.92778301],[38.80133057,9.92217541],[38.80247498,9.91491032],[38.80246353,9.90946388],[38.80071259,9.90334797],[38.79912949,9.89647388],[38.79597092,9.88906384],[38.79085159,9.88262653],[38.7935257,9.87624836],[38.79863739,9.87012482],[38.80302811,9.86211395],[38.80548477,9.85443211],[38.80571747,9.84866142],[38.80654526,9.84289932],[38.80835724,9.83531666],[38.8082428,9.82748222],[38.8077774,9.8195467],[38.8114624,9.81182194],[38.813694,9.80620193],[38.81822586,9.80272961],[38.81999969,9.79454041],[38.8215065,9.78449726],[38.82663727,9.7754221],[38.82889938,9.76910591],[38.83582687,9.76298809],[38.84026337,9.75700951],[38.84599304,9.75289249],[38.85298157,9.74953175],[38.85848618,9.74493504],[38.86260986,9.73991585],[38.86654282,9.73363304],[38.87199402,9.72957802],[38.87963486,9.72564125],[38.88594437,9.72112274],[38.89184189,9.71824074],[38.89674759,9.71429348],[38.90441895,9.71033859],[38.90934372,9.70873165],[38.91775513,9.71152496],[38.92314148,9.71799946],[38.92816162,9.72509193],[38.93473434,9.72794342],[38.94026947,9.72516346],[38.94474792,9.72115803],[38.94984818,9.71709824],[38.95381546,9.7138052],[38.96025085,9.71075726],[38.95909882,9.71851826],[38.95516968,9.72623158],[38.95271683,9.73268318],[38.9520874,9.73866272],[38.95767975,9.74374199],[38.96413803,9.74905396],[38.96794891,9.7561655],[38.97441864,9.76110554],[38.97766113,9.75627613],[38.98075867,9.75198174],[38.98587036,9.74904346],[38.99010468,9.74597073],[38.99714279,9.74801064],[39.00447845,9.750597],[39.0121994,9.75220299],[39.01659012,9.75772572],[39.02361679,9.75785637],[39.0305748,9.75548363],[39.03496933,9.75280476],[39.0359993,9.74701309],[39.04298782,9.75084686],[39.05045319,9.75343609],[39.0587883,9.75346947],[39.06887436,9.75295639],[39.07653809,9.75458431],[39.08209229,9.75697803],[39.08710861,9.76151085],[39.09221268,9.7630806],[39.09738541,9.75824356],[39.10177231,9.75505447],[39.10453033,9.74859619],[39.09740448,9.74382019],[39.09768677,9.73544216],[39.10292816,9.7336998],[39.10960388,9.73247623],[39.11700058,9.73327637],[39.12269211,9.73494053],[39.12747574,9.73853111],[39.13347244,9.74138546],[39.1393013,9.74065781],[39.14303589,9.73531628],[39.14489746,9.72864342],[39.14432907,9.7207098],[39.14774323,9.71535778],[39.14595413,9.70844078],[39.14631271,9.70022297],[39.1453743,9.69334602],[39.14909363,9.69077015],[39.15568924,9.68869495],[39.16178131,9.68877792],[39.1662674,9.69268417],[39.17422867,9.69484997],[39.17863464,9.68928719],[39.18402481,9.68575764],[39.18427277,9.69390202],[39.18410492,9.7001524],[39.18455887,9.70668221],[39.18351746,9.71483707],[39.18417358,9.72006226],[39.19057465,9.71844482],[39.19735336,9.71635246],[39.20012665,9.70873547],[39.20489502,9.70655632],[39.20863724,9.71090984],[39.21139145,9.70474625],[39.2167511,9.70839787],[39.22349548,9.70520973],[39.22500992,9.69891071],[39.22806168,9.69371223],[39.23280716,9.69916153],[39.23902512,9.70239067],[39.24477386,9.70182133],[39.24990463,9.70490646],[39.25231934,9.71238613],[39.25519943,9.71979809],[39.25769043,9.72456932],[39.26004791,9.72931004],[39.26358795,9.73522377],[39.268116,9.73974228],[39.27355194,9.73744392],[39.28039169,9.73318672],[39.2875824,9.73396492],[39.28939056,9.7387867],[39.29174042,9.7451334],[39.29785538,9.74803257],[39.30311203,9.74600887],[39.30809784,9.74401665],[39.3111496,9.73865509],[39.31335068,9.73143673],[39.32044983,9.72412491],[39.3287735,9.7243681],[39.33514023,9.72584248],[39.34178543,9.72745991],[39.34671783,9.72514153],[39.34076309,9.71709538],[39.33802795,9.71021366],[39.33934784,9.70391846],[39.34448624,9.69900417],[39.35031509,9.6999321],[39.35770416,9.70064449],[39.36413574,9.70145226],[39.37011337,9.69937706],[39.36972427,9.68956757],[39.36479187,9.68457603],[39.36098099,9.68027973],[39.36696625,9.67528629],[39.37129974,9.66996765],[39.36818314,9.66303349],[39.36212921,9.66152859],[39.35774994,9.65686417],[39.35724258,9.64785862],[39.35555649,9.63984585],[39.3533287,9.63410854],[39.35269547,9.62823677],[39.35204697,9.6220541],[39.35439301,9.61768341],[39.36168671,9.61891556],[39.36600113,9.62200642],[39.37102509,9.62569523],[39.37545395,9.62922955],[39.3805542,9.63050938],[39.38586044,9.63082123],[39.38832092,9.62242126],[39.38882065,9.61574078],[39.38530731,9.61072445],[39.38418961,9.60290241],[39.37966156,9.59626675],[39.37335968,9.59202766],[39.37325287,9.58289242],[39.37792969,9.58016396],[39.37852859,9.57309246],[39.37647247,9.56733418],[39.37262344,9.56403828],[39.36931992,9.55775738],[39.36581421,9.55290699],[39.36060333,9.54606342],[39.35284424,9.54920197],[39.34770584,9.54761028],[39.34140015,9.54539204],[39.33834076,9.54125595],[39.3342247,9.5334034],[39.32969284,9.52655697],[39.32294464,9.52573395],[39.31814575,9.52940178],[39.31293869,9.53555965],[39.3086853,9.53078365],[39.30213165,9.5226965],[39.30254364,9.51199627],[39.30519104,9.50524426],[39.29953766,9.50276375],[39.29362106,9.50059319],[39.29409409,9.49174118],[39.30106354,9.48778725],[39.30404663,9.48113537],[39.30553055,9.47374344],[39.30969238,9.46768093],[39.31484985,9.46700382],[39.32359314,9.46998119],[39.33155823,9.47299576],[39.33856583,9.47478104],[39.3437767,9.47230053],[39.35038757,9.47196102],[39.35561752,9.47423363],[39.36308289,9.47386074],[39.36941528,9.46993732],[39.37506866,9.46705723],[39.37692261,9.46328831],[39.37334061,9.45799351],[39.37290955,9.4521637],[39.3727417,9.44683075],[39.3783989,9.44608021],[39.38790131,9.44978428],[39.39392853,9.45504856],[39.40528107,9.45026207],[39.41125107,9.4474802],[39.41834641,9.44195747],[39.42456818,9.43712425],[39.43032455,9.43387604],[39.43772507,9.43413544],[39.44770432,9.4324913],[39.44932175,9.4252739],[39.44763184,9.41880512],[39.44308853,9.41438007],[39.44722748,9.40947056],[39.44956589,9.40471077],[39.45387268,9.4012022],[39.4522438,9.39391518],[39.44859314,9.38972282],[39.44564438,9.38390636],[39.44048309,9.38138962],[39.42593765,9.37941551],[39.42041016,9.37145138],[39.41188431,9.37445068],[39.40386581,9.38043785],[39.39485168,9.39190483],[39.37628555,9.383955],[39.37301636,9.3757391],[39.35670471,9.36903191],[39.3466835,9.37850571],[39.33715439,9.38050747],[39.32511902,9.38051891],[39.32009506,9.37404823],[39.31958771,9.36558056],[39.30654526,9.36608982],[39.30302429,9.35662842],[39.29298782,9.34916496],[39.29146957,9.3317318],[39.28543854,9.31579685],[39.27828217,9.31225014],[39.26441574,9.32419109],[39.26620483,9.28845215],[39.284729,9.27753735],[39.2661705,9.24448681],[39.26042557,9.25032425],[39.25758362,9.25611401],[39.23366165,9.29361248],[39.20718765,9.29194069],[39.177742,9.2710743],[39.19408798,9.2524128],[39.19974136,9.24996948],[39.21368408,9.22812176],[39.18858719,9.20925999],[39.20161438,9.19032383],[39.21463776,9.17337894],[39.23367691,9.15792465],[39.23968506,9.14845657],[39.24952316,9.13558578],[39.26953888,9.14172745],[39.2941246,9.1492691],[39.31445694,9.1489315],[39.32316971,9.14878654],[39.32016373,9.14022255],[39.32884216,9.13720608],[39.33504486,9.12102318],[39.3372879,9.11409473],[39.34012604,9.10830498],[39.34292984,9.09964275],[39.34576797,9.093853],[39.34860229,9.08806324],[39.34846878,9.07658005],[39.3542099,9.07074165],[39.3570137,9.06208038],[39.34702682,9.06417465],[39.31529999,9.06022739],[39.29675293,9.05326939],[39.29335022,9.03171444],[39.29210663,9.02382183],[39.28818893,8.99897861],[39.26963425,8.98056126],[39.25859833,8.96263599],[39.26538086,8.92238522],[39.25908279,8.91001701],[39.24101639,8.88892269],[39.23749161,8.86202526],[39.26052475,8.85055161],[39.26552582,8.84307575],[39.24398422,8.83661461],[39.24385452,8.82768154],[39.25448608,8.81319523],[39.25898743,8.80372715],[39.26649094,8.79375839],[39.27199173,8.78304482],[39.27298355,8.76685429],[39.26195145,8.74693584],[39.27044678,8.72451305],[39.28581619,8.71779156],[39.29898071,8.71851349],[39.30530548,8.72172546],[39.30844116,8.72582722],[39.31585312,8.73552132],[39.324543,8.74688721],[39.33154678,8.7409029],[39.34204865,8.72694588],[39.36106873,8.72095013],[39.3771019,8.73089695],[39.37461853,8.75281811],[39.37363052,8.76676655],[39.37464523,8.77971745],[39.37816238,8.79117107],[39.38618469,8.79863548],[39.40548706,8.80996132],[39.41775131,8.81454182],[39.42424774,8.8030777],[39.4312439,8.79011726],[39.4397316,8.76769161],[39.44922638,8.75223827],[39.47024536,8.74175167],[39.49225235,8.72528458],[39.5007515,8.71481228],[39.51475525,8.73311234],[39.51925278,8.73898888],[39.53377533,8.75796032],[39.55178452,8.76875019],[39.55693817,8.79144669],[39.56198502,8.81783962],[39.59149933,8.79687309],[39.59900665,8.79287529],[39.61702347,8.78686905],[39.63406754,8.7987957],[39.65010071,8.80275345],[39.66213608,8.81319237],[39.6791687,8.81565285],[39.71672821,8.81608009],[39.73327637,8.82624054],[39.73133087,8.84419155],[39.73688507,8.85720921],[39.73849869,8.87206078],[39.74801254,8.91027832],[39.74829483,8.94674301],[39.74711227,8.97199631],[39.7466774,8.9889183],[39.74651337,9.0097599],[39.74616241,9.0152607],[39.74115753,9.02302456],[39.740242,9.0395422],[39.74367905,9.04800129],[39.74616623,9.05515766],[39.74749756,9.06313515],[39.74245071,9.09125805],[39.76391983,9.09512997],[39.78024673,9.10435104],[39.78635406,9.10844612],[39.79380417,9.11274338],[39.79713058,9.12146664],[39.80195999,9.12605095],[39.80773544,9.13544559],[39.81742096,9.13631439],[39.82406998,9.13680935],[39.83330917,9.13747978],[39.839077,9.13838005],[39.84900665,9.13812923],[39.86449051,9.1410799],[39.87765884,9.14472961],[39.89841843,9.13893986],[39.90901947,9.1317997],[39.91003799,9.12524986],[39.91560745,9.09733963],[39.91874695,9.06840992],[39.92839813,9.05414963],[39.94245911,9.04768944],[39.95186996,9.02998924],[39.94904709,9.0134697],[39.94762802,9.00359917],[39.96817017,9.00355911],[39.9995575,9.00715923],[40.02193069,9.00227928],[40.03482056,8.99444962],[40.04745865,8.97788906],[40.05109787,8.96135902],[40.04388046,8.94231892],[40.04058838,8.92304897],[40.03661728,8.90652943],[40.01926804,8.88912964],[40.01091766,8.87077904],[40.01041031,8.85608959],[40.02421951,8.8450489],[40.04034805,8.84110928],[40.05118942,8.84062958],[40.06695557,8.84396935],[40.08096695,8.85134888],[40.08997726,8.85866928],[40.09449768,8.86216545],[40.10503006,8.8769989],[40.12054062,8.89577866],[40.1409111,8.91890907],[40.15224838,8.93195915],[40.16169357,8.9485836],[40.16989136,8.96449852],[40.17440033,8.96701908],[40.18745804,8.9743185],[40.20204544,8.97950363],[40.22232819,8.9877491],[40.23780823,8.99733829],[40.24225998,9.0131588],[40.2460289,9.03540897],[40.24427032,9.05559826],[40.24549866,9.0748682],[40.25431061,9.08607864],[40.2714386,9.10139847],[40.30060959,9.12813854],[40.32702255,9.15465832],[40.34954071,9.18714809],[40.36952972,9.21826839],[40.3923111,9.25510788],[40.40812302,9.28118801],[40.42576981,9.30427742],[40.4496727,9.32962799],[40.47102356,9.35475731],[40.49882126,9.37504768],[40.52841187,9.38545799],[40.56626129,9.38779736],[40.60596085,9.39104748],[40.63597107,9.39594746],[40.65955353,9.40430737],[40.67554474,9.41751671],[40.68382263,9.43074703],[40.68889236,9.43835735],[40.69356155,9.44438267],[40.69856262,9.44764709],[40.70709229,9.44622707],[40.71953201,9.44387722],[40.74860382,9.44486713],[40.7832222,9.44788647],[40.81576538,9.45273685],[40.84236526,9.46358681],[40.86218262,9.46301651],[40.88999557,9.44864655],[40.91752243,9.4218359],[40.92751312,9.40513611],[40.91385269,9.39719677],[40.89121246,9.38953686],[40.85822296,9.38536644],[40.84313202,9.38923645],[40.82806396,9.39309692],[40.79764175,9.39555645],[40.78882217,9.38689709],[40.79262161,9.36739635],[40.80032349,9.34512711],[40.80851364,9.32675648],[40.81256485,9.31092644],[40.82475281,9.30535698],[40.84410477,9.30479717],[40.87410355,9.30944633],[40.90296555,9.31821632],[40.92697525,9.32312679],[40.95355225,9.33557606],[40.97071457,9.35150623],[40.97634125,9.35196495],[40.99608612,9.35410595],[41.01407623,9.35720634],[41.0300827,9.3724556],[41.05648422,9.3910656],[41.08354568,9.40441608],[41.11471558,9.41405582],[41.14544678,9.4246254],[41.17045593,9.4414053],[41.19480515,9.46229553],[41.21157074,9.48099327],[41.21496582,9.48609543],[41.21988678,9.49071503],[41.23058701,9.49567986],[41.23931503,9.49879551],[41.25225067,9.50038052],[41.26050568,9.50018501],[41.26755524,9.49936485],[41.27967453,9.50153542],[41.30869675,9.50307465],[41.33649445,9.49463463],[41.35903549,9.4901247],[41.39308548,9.48575497],[41.43261719,9.47585487],[41.45887756,9.46687508],[41.47903824,9.46002483],[41.4909668,9.45742416],[41.50665665,9.47050476],[41.52206802,9.48532486],[41.54060745,9.4995842],[41.56415558,9.50694466],[41.59772873,9.51141453],[41.63273621,9.51332378],[41.6425972,9.51514435],[41.65415573,9.4812336],[41.67019653,9.47063351],[41.70227814,9.46665382],[41.72194672,9.46753407],[41.73461914,9.47148418],[41.75452805,9.48457336],[41.77299881,9.49105358],[41.79254913,9.4895134],[41.81519699,9.4767437],[41.83867645,9.47791386],[41.85970688,9.48801327],[41.88241959,9.48118305],[41.90829849,9.47389317],[41.91337967,9.47246361],[41.93874741,9.47954273],[41.95448685,9.48911285],[41.98252487,9.50849247],[41.99000931,9.50662327],[41.99535751,9.50646305],[42.00233078,9.50771236],[42.01910019,9.50817299],[42.03171921,9.50440311],[42.04390717,9.49831295],[42.05213928,9.4919529],[42.05781937,9.48508263],[42.06761551,9.47480774],[42.07477188,9.47371674],[42.0810585,9.48816299],[42.09626007,9.50131226],[42.10968018,9.50796223],[42.12186813,9.51922226],[42.13890076,9.53469276],[42.15259933,9.55637264],[42.14612961,9.57063198],[42.13341141,9.58249187],[42.13005066,9.59210205],[42.12784958,9.59837246],[42.12485886,9.61452198],[42.14225006,9.62029266],[42.15959167,9.63068199],[42.18038177,9.64434242],[42.20053101,9.65314198],[42.22052002,9.65616226],[42.24077225,9.65664196],[42.25863647,9.65158176],[42.26264954,9.65690136],[42.27185059,9.66695213],[42.28594208,9.67614174],[42.30276108,9.69113159],[42.3112793,9.69909191],[42.31814194,9.70566177],[42.32834244,9.71338177],[42.33412933,9.71599102],[42.33977127,9.71961784],[42.34619141,9.72136116],[42.35576248,9.71786118],[42.37657166,9.71383953],[42.39530945,9.71545219],[42.42425537,9.71918678],[42.44853592,9.72198868],[42.47468185,9.72105503],[42.49895859,9.72105408],[42.52977371,9.72759056],[42.56805801,9.73692799],[42.59513474,9.74813271],[42.61848068,9.75840473],[42.64742661,9.77521229],[42.66379547,9.77654934],[42.68104172,9.77427769],[42.69411469,9.7686758],[42.70345306,9.75840378],[42.71372604,9.74066257],[42.7305336,9.72105408],[42.75014114,9.71078205],[42.77908707,9.69864368],[42.79029465,9.69304085],[42.82297516,9.67436504],[42.85379028,9.66222668],[42.89767456,9.65008736],[42.90794754,9.65008736],[42.91728592,9.65755749],[42.9256897,9.67249775],[42.93502808,9.67996788],[42.95090103,9.67249775],[42.963974,9.65568924],[42.97798157,9.62861061],[42.97984695,9.60713387],[42.96864319,9.5921936],[42.94343185,9.58752537],[42.93844223,9.58348656],[42.92102051,9.56511497],[42.90888214,9.56324768],[42.89207458,9.5744524],[42.8799324,9.58472443],[42.86405945,9.58659172],[42.84071732,9.58285713],[42.81363678,9.57258511],[42.80149841,9.54924107],[42.78749084,9.52309608],[42.77628708,9.51189137],[42.74734116,9.50162029],[42.73613358,9.49508381],[42.7286644,9.48294449],[42.71465683,9.47454071],[42.70064926,9.47173977],[42.68197632,9.46987247],[42.66610336,9.4586668],[42.65583038,9.45586586],[42.64088821,9.44746208],[42.61754608,9.42691994],[42.60353851,9.41664791],[42.58486557,9.40917873],[42.56712341,9.39610577],[42.55218124,9.38209915],[42.5374527,9.37298203],[42.55158234,9.34524155],[42.55628967,9.3210516],[42.54695129,9.30523109],[42.55136108,9.2865715],[42.56108093,9.27030182],[42.57099152,9.25818157],[42.57960892,9.23750114],[42.58359146,9.21538162],[42.59548187,9.19359112],[42.61429977,9.17396164],[42.62477112,9.16603088],[42.63470078,9.15852165],[42.63510895,9.14397144],[42.6259613,9.13208103],[42.61903,9.12643147],[42.60942841,9.11846161],[42.59934616,9.10617447],[42.61930084,9.08928108],[42.64434814,9.07298183],[42.66228867,9.04896164],[42.67406082,9.02847862],[42.68013,9.01502132],[42.70307159,8.98091125],[42.73064041,8.94802094],[42.76118088,8.91771126],[42.7840004,8.89284134],[42.80516052,8.87003136],[42.82807159,8.85579109],[42.84954834,8.84429073],[42.85977936,8.8318615],[42.86532974,8.82511139],[42.86536026,8.80435085],[42.85754395,8.78512287],[42.84391785,8.75792122],[42.83712769,8.72783089],[42.83198929,8.69523144],[42.83842087,8.66439152],[42.85987854,8.63630104],[42.88402939,8.61584663],[42.89878845,8.60267162],[42.90531921,8.58222103],[42.90760803,8.54903126],[42.91236877,8.5221014],[42.91082764,8.4987812],[42.89702988,8.45775127],[42.89857864,8.42917156],[42.89884949,8.40910149],[42.88440704,8.39851189],[42.85809708,8.39169216],[42.83091736,8.38115215],[42.79818726,8.37007236],[42.77937698,8.37326241],[42.7681694,8.36317253],[42.75394821,8.33712196],[42.74753189,8.3227272],[42.73487854,8.29437256],[42.71914673,8.24566269],[42.71115875,8.20676231],[42.70159912,8.17961216],[42.67851639,8.14732265],[42.66135788,8.1302824],[42.65140533,8.129673],[42.63062668,8.14043236],[42.61597824,8.15960312],[42.59477615,8.1978426],[42.5780983,8.22853279],[42.56702805,8.25561237],[42.55999756,8.29199314],[42.55007553,8.35050297],[42.53872681,8.39722252],[42.53451157,8.42900372],[42.52976608,8.45483303],[42.52013779,8.47871304],[42.50255585,8.4920826],[42.32303619,8.53677368],[42.30714798,8.55964279],[42.29468536,8.58604336],[42.27499771,8.61510372],[42.26307678,8.63595295],[42.23966599,8.63988304],[42.23267746,8.63503361],[42.21561813,8.61120319],[42.0977478,8.51898861],[42.06972504,8.49397373],[42.04130554,8.44746399],[42.02600479,8.42583466],[42.02705383,8.32302475],[42.00878906,8.29474926],[41.96260452,8.2236042],[41.94840622,8.20089436],[41.9374733,8.14357471],[41.92981339,8.10139465],[41.92634583,8.0805645],[41.92544556,8.03974533],[41.92216492,7.97810507],[41.91888428,7.93592501],[41.91713333,7.91287518],[41.91516495,7.88334513],[41.9130249,7.85381508],[41.91240311,7.84439516],[41.95714188,7.77478504],[41.94249344,7.72746515],[41.92946243,7.69701529],[41.90480423,7.63592529],[41.88835144,7.59112549],[41.87807083,7.56261539],[41.86779404,7.53817558],[41.86710358,7.5327754],[41.87275314,7.50846577],[41.88273239,7.46548557],[41.89360046,7.41868544],[41.89612198,7.41120577],[41.96523285,7.38411522],[42.07074356,7.34275532],[42.11126328,7.32635498],[42.13358307,7.31733513],[42.14599228,7.31323481],[42.15550232,7.30872488],[42.17493439,7.30011511],[42.19911194,7.29144001],[42.20985413,7.26400471],[42.20984268,7.22690487],[42.20838165,7.21760511],[42.20345688,7.19463301],[42.204422,7.15922499],[42.20470428,7.14103508],[42.18696213,7.09639502],[42.15761185,7.05132532],[42.16264343,7.00711536],[42.14758301,6.98062515],[42.13994217,6.96726513],[42.1341629,6.95416546],[42.1060524,6.93021536],[42.09873199,6.91573524],[42.07623291,6.88673544],[42.07552338,6.86398554],[42.06692123,6.84108543],[42.06288147,6.82592297],[42.05963135,6.79182959],[42.05553055,6.74875546],[42.02548981,6.70641565],[42.01464081,6.68044567],[42.01346207,6.65860558],[41.99917984,6.64161587],[42.00038147,6.6286459],[42.00648117,6.61438608],[42.00403976,6.59762573],[41.99954224,6.58984566],[41.98791885,6.58239603],[41.96469879,6.57027578],[41.936409,6.55317593],[41.91368103,6.54401588],[41.90845108,6.54225588],[41.88798141,6.53534603],[41.86072922,6.53534126],[41.8536644,6.56890917],[41.81966019,6.57047653],[41.7987709,6.55746651],[41.75788116,6.5405364],[41.73892975,6.51990652],[41.70449829,6.45240641],[41.69033051,6.42941666],[41.68416977,6.42413664],[41.65612793,6.40917683],[41.63335037,6.39138699],[41.61968994,6.36171675],[41.57188797,6.41553688],[41.52800751,6.46493721],[41.47626877,6.51025724],[41.43778992,6.54396725],[41.38003922,6.56552744],[41.35838699,6.56552744],[41.31853867,6.58315754],[41.28304672,6.5988574],[41.22846985,6.62300777],[41.17432785,6.65892792],[41.15628815,6.67688751],[41.13102722,6.69125795],[41.0720787,6.69432783],[41.07120895,6.68789768],[41.01553726,6.64455795],[40.98996735,6.60423803],[40.97223663,6.58707809],[40.93614578,6.55115843],[40.90005493,6.51163816],[40.8856163,6.47211838],[40.86035538,6.43619823],[40.85674667,6.3859086],[40.86100769,6.3611083],[40.8644371,6.35227823],[40.86194611,6.34571838],[40.86450577,6.32025862],[40.87299728,6.29085827],[40.87969589,6.26650858],[40.88162231,6.24434328],[40.88002777,6.21722841],[40.87797546,6.18766832],[40.87350082,6.16987848],[40.86787415,6.15128851],[40.86642838,6.13365364],[40.866436,6.11376858],[40.86951447,6.09449816],[40.86667633,6.07663822],[40.8711853,6.05576849],[40.87840652,5.98751831],[40.87753677,5.94434834],[40.87645721,5.89048815],[40.87573624,5.85464811],[40.87479401,5.80789852],[40.90127563,5.74011803],[40.92229462,5.68337822],[40.86500549,5.66901827],[40.83220291,5.65589857],[40.81148529,5.6349082],[40.78344345,5.61464834],[40.75288391,5.5943985],[40.69437408,5.56716871],[40.6795845,5.54063845],[40.63305283,5.53006887],[40.61596298,5.50689888],[40.60538483,5.49028873],[40.58104324,5.47594881],[40.56350327,5.45390892],[40.54898453,5.42767859],[40.53289413,5.41064882],[40.5011673,5.40572596],[40.45589447,5.39869881],[40.36950302,5.38640928],[40.3103714,5.37630892],[40.27667236,5.36816931],[40.2406311,5.36607933],[40.20835114,5.35554934],[40.17443085,5.35337925],[40.13459015,5.35786963],[40.09794998,5.36487961],[40.06340027,5.37854958],[40.03367996,5.39678955],[39.99266052,5.34660578],[39.98897171,5.34209585],[39.94229126,5.28498983],[39.90729904,5.21960974],[39.8783989,5.14031982],[39.85871506,5.0930748],[39.83359146,5.03276968],[39.81285858,5.00206947],[39.80445099,4.95147943],[39.78398895,4.89058971],[39.77141571,4.86151695],[39.76506805,4.84684944],[39.75851059,4.80106974],[39.74356079,4.72347927],[39.7458992,4.66657925],[39.73970795,4.65472937],[39.72512817,4.6330595],[39.7170105,4.61567926],[39.70982742,4.59736919],[39.70326996,4.56816959],[39.6901474,4.54328918],[39.67464828,4.50518942],[39.65559006,4.46888924],[39.64205933,4.43593931],[39.63145828,4.40270948],[39.61882019,4.36846924],[39.60320663,4.33376932],[39.58577728,4.29653931],[39.56696701,4.26458931],[39.53968811,4.23516941],[39.52878952,4.16673899],[39.50909805,4.11441898],[39.47827911,4.04032898],[39.49967575,3.96572876],[39.48284912,3.94199896],[39.4318161,3.85303879],[39.39961624,3.79565883],[39.28469467,3.62307858],[39.22049713,3.6024785],[39.09286499,3.5339787],[39.08083725,3.53862858],[39.07385635,3.53452849],[39.06570053,3.52791357],[39.06116104,3.52638125],[39.05148697,3.52311587],[39.04681396,3.5215385],[39.04119492,3.52262449],[39.03775406,3.52328968],[39.03139496,3.52451873],[39.02487183,3.52216864],[39.02421951,3.52193356],[39.01528549,3.51278853],[39.00709915,3.5172987],[39.00167465,3.52092862],[38.98322678,3.52031875],[38.97015381,3.52561879],[38.96412277,3.52774334],[38.96271515,3.52823877],[38.95585632,3.52128363],[38.94886398,3.51473284],[38.94687653,3.51286864],[38.93405151,3.51130867],[38.92602921,3.51089382],[38.9203186,3.51051021],[38.90764999,3.50965881],[38.89591599,3.51151872],[38.88492584,3.51559877],[38.87062454,3.52066875],[38.856884,3.52585888],[38.83628464,3.53176379],[38.82889557,3.53220892],[38.80182266,3.54067898],[38.79409409,3.54315877],[38.78110504,3.547189],[38.77539063,3.54992414],[38.75982285,3.55447173],[38.74259567,3.55950403],[38.73444366,3.56251907],[38.72914505,3.56395888],[38.72377396,3.56535888],[38.71829605,3.56680894],[38.71240234,3.56909895],[38.70850754,3.57494402],[38.70578003,3.58249402],[38.69987488,3.590204],[38.69541931,3.59691906],[38.69062042,3.60358405],[38.68676376,3.61020422],[38.68354416,3.61730409],[38.68020248,3.61980343],[38.67786789,3.62154913],[38.67037582,3.61977911],[38.66923904,3.61465907],[38.67050171,3.60946608],[38.67055893,3.60922408],[38.66944122,3.60606718],[38.66864777,3.60382915],[38.66702271,3.60099721],[38.66592026,3.59907413],[38.66241837,3.59487915],[38.65518951,3.59400415],[38.64928436,3.59557915],[38.6428833,3.59692907],[38.63572311,3.59908915],[38.63049316,3.60030913],[38.62504196,3.60141921],[38.61845779,3.60162425],[38.61037827,3.60382915],[38.60974121,3.61112928],[38.60641861,3.60928583],[38.60329819,3.6075542],[38.59614944,3.60447907],[38.58864212,3.60418415],[38.58018494,3.60382676],[38.57243729,3.60336924],[38.56824875,3.6067543],[38.56108475,3.60962415],[38.55320358,3.61255932],[38.55132294,3.6174593],[38.55201721,3.62347436],[38.54790497,3.62955427],[38.54668808,3.63242507],[38.54388809,3.63901687],[38.53724289,3.64815927],[38.53275299,3.65270925],[38.52700424,3.6516993],[38.52151871,3.65033937],[38.51359558,3.64697766],[38.51689529,3.6397593],[38.51675797,3.63247418],[38.51735306,3.62632418],[38.51100159,3.62291431],[38.50231171,3.6197927],[38.49638367,3.61800599],[38.48750305,3.61499929],[38.48129272,3.61307931],[38.47618866,3.61090422],[38.44875336,3.62645578],[38.43856049,3.60424113],[38.43719482,3.60125875],[38.42982483,3.6008594],[38.42220688,3.6014843],[38.41319275,3.60257936],[38.40400314,3.60304928],[38.39599228,3.60410428],[38.38788223,3.60421443],[38.38264465,3.60425925],[38.37674332,3.60461926],[38.36878204,3.60512948],[38.36144257,3.60551929],[38.35236359,3.60555434],[38.34168243,3.60617948],[38.3348732,3.60718942],[38.32943344,3.60752439],[38.32235718,3.60916615],[38.3119812,3.60973954],[38.3029213,3.61000443],[38.29410172,3.61066437],[38.28391266,3.61142945],[38.27761078,3.6119194],[38.27127838,3.61200452],[38.26275635,3.61148953],[38.25342178,3.61200953],[38.24529266,3.61326957],[38.23710251,3.61372447],[38.22558212,3.61392951],[38.21917343,3.61487961],[38.21307755,3.6156745],[38.20331192,3.61657953],[38.19696045,3.61741948],[38.18845749,3.6179595],[38.18096161,3.61745453],[38.17076111,3.61546469],[38.16212082,3.6131146],[38.15385056,3.61172962],[38.14383698,3.60933447],[38.13870239,3.60922956],[38.13300323,3.60924459],[38.12950134,3.60801315],[38.12617111,3.60684204],[38.12028122,3.60849953],[38.11582184,3.61323452],[38.110672,3.61803961],[38.10668564,3.62222457],[38.10244751,3.62655973],[38.09680557,3.63139963],[38.09187317,3.63521957],[38.08744049,3.63814974],[38.08272552,3.6418047],[38.07539749,3.64767218],[38.06900024,3.65291977],[38.06430054,3.65682983],[38.06033325,3.66021299],[38.0550499,3.66435981],[38.05083466,3.66747642],[38.04486084,3.6695447],[38.03910446,3.67567492],[38.03453827,3.68150973],[38.02858734,3.68585491],[38.02229691,3.69246984],[38.01880264,3.6969049],[38.01385498,3.70218229],[38.01298523,3.70863318],[38.00947189,3.71231484],[38.00526047,3.71525979],[38.00253677,3.72222996],[37.99589539,3.72632003],[37.99048996,3.72768998],[37.9851532,3.729105],[37.9790802,3.73202991],[37.97224045,3.73516488],[37.96615219,3.73846006],[37.96080017,3.74168992],[37.95460129,3.74541497],[37.94828033,3.74908996],[37.94316101,3.75186014],[37.93672943,3.75538993],[37.92938232,3.75909996],[37.92329788,3.76306009],[37.91657257,3.76851511],[37.91078949,3.77152014],[37.90578079,3.77423],[37.90019989,3.77769017],[37.89332962,3.78243017],[37.88587952,3.78736019],[37.8782692,3.79274011],[37.8722496,3.79718018],[37.86685181,3.80087018],[37.86037064,3.8052702],[37.85354233,3.8098402],[37.84706116,3.81389022],[37.84066772,3.81827521],[37.8336525,3.82331038],[37.828022,3.82683039],[37.81797409,3.83316541],[37.8123703,3.83657026],[37.80731964,3.84034038],[37.80160522,3.84443521],[37.79338074,3.84934044],[37.78268051,3.85557032],[37.77774048,3.85903049],[37.77273941,3.86305046],[37.76739883,3.86674047],[37.76099396,3.87156796],[37.75466919,3.87627053],[37.74956131,3.88002038],[37.74433136,3.88394046],[37.73995972,3.88692045],[37.73498154,3.89006042],[37.72827911,3.89345551],[37.72151184,3.8975606],[37.71678925,3.90073061],[37.7098999,3.90487051],[37.7044487,3.90899062],[37.69813919,3.91381049],[37.6906395,3.91911054],[37.6841507,3.92304063],[37.67642593,3.92798805],[37.66973877,3.93297052],[37.66007996,3.93918824],[37.65309143,3.9435606],[37.64865875,3.94678068],[37.64331055,3.95003557],[37.63712692,3.95310569],[37.63412094,3.95771074],[37.6309967,3.96246576],[37.62123871,3.96880078],[37.61195374,3.97443581],[37.60579681,3.97919583],[37.59717941,3.98503089],[37.59170151,3.98743749],[37.58536911,3.99061084],[37.58010483,3.9944675],[37.57701492,3.99862576],[37.56894684,4.00263596],[37.56121063,4.00776577],[37.5546875,4.01167965],[37.55371857,4.01226091],[37.54325867,4.02044106],[37.53467941,4.02745104],[37.53047943,4.03133106],[37.52542114,4.0343008],[37.52022934,4.03664112],[37.51596832,4.0394311],[37.5097084,4.04663801],[37.40679932,4.11423111],[37.40110779,4.11818123],[37.39447784,4.12206888],[37.38459778,4.12802124],[37.37960052,4.13111115],[37.37251282,4.13567114],[37.36531067,4.13995123],[37.35932922,4.14339113],[37.3536377,4.1468215],[37.34949875,4.15030146],[37.34420395,4.15376616],[37.33903503,4.15774155],[37.32947922,4.16425133],[37.32043457,4.17026615],[37.3146286,4.17362118],[37.30886841,4.1767664],[37.30357742,4.18054152],[37.29750061,4.18492651],[37.28952789,4.19013643],[37.28173065,4.19462156],[37.27388,4.19951153],[37.26738739,4.20369625],[37.25969696,4.20938921],[37.25288773,4.21415138],[37.24808502,4.21752167],[37.24121857,4.22166157],[37.23444366,4.22559166],[37.22874832,4.22902632],[37.22169495,4.23333168],[37.21389771,4.23882151],[37.20700836,4.24330139],[37.20108795,4.24731159],[37.19653702,4.25025177],[37.19042206,4.25416756],[37.16144943,4.27018309],[37.12368774,4.28277016],[37.1042366,4.29878998],[37.08478165,4.32548904],[37.05846405,4.34303474],[37.03939438,4.36172438],[37.01354599,4.37183857],[37.00481415,4.375772],[36.99704742,4.37890959],[36.99104691,4.38203716],[36.98538208,4.38528728],[36.98143768,4.38922215],[36.97540665,4.3911171],[36.96973801,4.39449692],[36.96372604,4.39552212],[36.95614624,4.39668703],[36.94973755,4.40421534],[36.94155121,4.40738201],[36.93497849,4.40893221],[36.92815781,4.41191721],[36.92148209,4.4148221],[36.91426849,4.41824722],[36.90460587,4.42174721],[36.89552689,4.42489719],[36.88751602,4.42640734],[36.87894058,4.42822886],[36.873909,4.42957211],[36.86647797,4.43179226],[36.85734558,4.43552208],[36.85098267,4.43862247],[36.84370804,4.44161224],[36.838974,4.44386816],[36.83879852,4.44395208],[36.83215332,4.44372654],[36.8320694,4.44372416],[36.82291031,4.44346237],[36.81545639,4.44319248],[36.80639648,4.44315481],[36.79842758,4.44300222],[36.79223633,4.44276237],[36.78242111,4.44197989],[36.77559662,4.44159222],[36.77002716,4.44136238],[36.76206207,4.44097233],[36.71820068,4.43944407],[36.71258163,4.43924809],[36.69149399,4.49550819],[36.65753174,4.58037376],[36.68083954,4.6043005],[36.64640808,4.71075726],[36.82847977,4.70917463],[36.85474014,4.73605967],[36.8634758,4.79303932],[36.86085129,4.93506527],[36.8647995,4.94419861],[36.86071014,4.9787674],[36.86753464,5.00701618],[36.87144089,5.02906036],[36.88218689,5.0749836],[36.89297104,5.10978794],[36.9212265,5.14513063],[36.94896698,5.19468641],[36.98310089,5.19241238],[36.99257278,5.1958909],[37.00331116,5.19908285],[37.02255249,5.19647312],[37.05222321,5.19177294],[37.08211899,5.18638229],[37.11222839,5.18627214],[37.14530945,5.18894243],[37.18182755,5.193923],[37.19715881,5.21031475],[37.2169075,5.21268463],[37.23810959,5.19943333],[37.26316833,5.19700336],[37.2923584,5.19436312],[37.31724548,5.17353106],[37.34073639,5.15959978],[37.36717606,5.15509939],[37.41721344,5.16655064],[37.44151306,5.17998171],[37.4655838,5.19317293],[37.49127197,5.20291376],[37.51786423,5.22047567],[37.55135345,5.23392677],[37.57785416,5.24238873],[37.57983017,5.24767733],[37.6002121,5.25523853],[37.62973404,5.26600981],[37.63728714,5.28316689],[37.64670563,5.29928732],[37.65501022,5.30181313],[37.67422867,5.30764341],[37.70088196,5.30588341],[37.72893906,5.29218197],[37.74712753,5.27661085],[37.77282715,5.28335142],[37.79261017,5.27422047],[37.81007767,5.26830006],[37.82712936,5.24973822],[37.85265732,5.2346468],[37.88508606,5.23431015],[37.90081787,5.24485397],[37.90206146,5.25429726],[37.9024353,5.25938559],[37.90438461,5.27937651],[37.90477371,5.30395794],[37.90103531,5.32691908],[37.89637375,5.34987974],[37.90121078,5.37324762],[37.91927338,5.39597702],[37.93522263,5.42303324],[37.94021606,5.44992447],[37.94681549,5.4742856],[37.94925308,5.50759697],[37.94781494,5.53768873],[37.94153595,5.56844997],[37.94112778,5.58510542],[37.93710327,5.59739113],[37.93017578,5.61667204],[37.94351959,5.63946438],[37.96373367,5.64569426],[37.97746658,5.64940453],[37.99964523,5.64994907],[38.00801468,5.65015459],[38.02407455,5.66006565],[38.04081345,5.67364693],[38.05550385,5.68033743],[38.07516479,5.68894053],[38.08046341,5.70103598],[38.08708191,5.72125673],[38.08888245,5.74560785],[38.0720787,5.76255894],[38.05415344,5.76414871],[38.03921509,5.77238941],[38.01987457,5.79004002],[38.0122757,5.80427074],[38.00901413,5.8265419],[38.00713348,5.84927273],[37.99630356,5.86694336],[37.98247528,5.88782454],[37.97899628,5.90757561],[37.97758484,5.92777634],[37.96857452,5.95601749],[37.95269394,5.96334791],[37.94465637,5.9629879],[37.92282486,5.96007776],[37.90397644,5.96050787],[37.8949852,5.97449875],[37.88048553,5.98503923],[37.86553574,5.98662901],[37.85654449,5.99900961],[37.84296417,6.01094007],[37.82709503,6.00999022],[37.81332397,5.99778986],[37.80373764,5.99317884],[37.79148483,5.99590969],[37.78317642,6.01060009],[37.78636551,6.0287509],[37.78977585,6.04944181],[37.78513718,6.07263327],[37.77772522,6.0981245],[37.77573776,6.11357546],[37.77469635,6.12385559],[37.77809525,6.14959669],[37.79053497,6.17246246],[37.80979919,6.19609404],[37.82556534,6.22073174],[37.84026718,6.24825096],[37.84804535,6.27882242],[37.85141754,6.29946804],[37.85215378,6.30478382],[37.85028458,6.32684469],[37.84820557,6.33166504],[37.84541702,6.35187578],[37.84008408,6.37874699],[37.81959534,6.39342785],[37.80783463,6.41017866],[37.79745483,6.42923927],[37.79628754,6.44946051],[37.79992676,6.47818136],[37.80777359,6.50065517],[37.81459427,6.51057196],[37.82339859,6.52053308],[37.84247589,6.5416851],[37.85442734,6.55777645],[37.87148285,6.58222866],[37.89277649,6.59607077],[37.91261292,6.59066916],[37.9147377,6.58365345],[37.91846085,6.57437563],[37.93270111,6.55720663],[37.95266342,6.54962015],[37.96876526,6.54895449],[37.98340607,6.55694008],[37.99685287,6.5756135],[38.00577164,6.57536554],[38.01592636,6.5756135],[38.03351212,6.56149292],[38.05159378,6.53473902],[38.07339096,6.52904129],[38.09073257,6.52606821],[38.09519196,6.52160931],[38.10113525,6.5126915],[38.11450958,6.49956179],[38.13928223,6.5060029],[38.14968491,6.49658918],[38.1464653,6.47602797],[38.14348984,6.45819187],[38.15522385,6.43925619],[38.17061996,6.44982672],[38.19293213,6.45695734],[38.21549988,6.46346807],[38.22145081,6.46109819],[38.24559021,6.46822882],[38.25365448,6.45983839],[38.26007843,6.45215702],[38.2828598,6.43747568],[38.29658127,6.41570044],[38.29275894,6.39592218],[38.29322815,6.37203979],[38.2882309,6.3598423],[38.27251434,6.34788513],[38.24860382,6.33627987],[38.23136139,6.33110476],[38.21389389,6.33017445],[38.20526123,6.32426786],[38.19757843,6.31900311],[38.18842316,6.30425358],[38.18177032,6.28427219],[38.17833328,6.27577114],[38.17374039,6.26681042],[38.17191315,6.25532961],[38.17905045,6.23833799],[38.19330978,6.22617674],[38.19792175,6.2064352],[38.187397,6.18221045],[38.17103195,6.16688728],[38.15839386,6.15952682],[38.14138412,6.15445662],[38.12506485,6.15512657],[38.11700439,6.16705704],[38.10230255,6.18044853],[38.08672714,6.1610446],[38.08305359,6.13719893],[38.0798645,6.11698675],[38.09207153,6.09632492],[38.09859467,6.08707905],[38.10014343,6.07727337],[38.10498428,6.06603241],[38.11649323,6.05020094],[38.11904144,6.03320932],[38.12159348,6.01667786],[38.13883209,6.01026726],[38.15034103,6.00040627],[38.15932083,5.98502493],[38.16255188,5.97561407],[38.17015076,5.95702219],[38.18097305,5.94165087],[38.20373154,5.93502045],[38.20669937,5.92826557],[38.21179199,5.91666889],[38.21480942,5.89324665],[38.20888519,5.87905979],[38.1996994,5.86480045],[38.19839096,5.8494525],[38.20799255,5.84019184],[38.22338104,5.8386116],[38.23923111,5.84437227],[38.25450134,5.85850859],[38.27043152,5.86852455],[38.28858566,5.86406422],[38.30374908,5.861444],[38.31039047,5.86371279],[38.32532883,5.86881447],[38.33979034,5.88283587],[38.34941864,5.89225674],[38.35813904,5.90213728],[38.38087845,5.90399742],[38.39017487,5.91317034],[38.39608383,5.92005253],[38.38842773,5.93300104],[38.37480927,5.94304323],[38.36248016,5.94712973],[38.35578918,5.94897175],[38.35037613,5.95120668],[38.33825302,5.95558357],[38.32752991,5.95537233],[38.3247757,5.96322346],[38.31895065,5.97506285],[38.30454254,5.97998047],[38.29312897,5.97333956],[38.28226089,5.9707036],[38.27901459,5.97473478],[38.27170944,5.98657942],[38.27597046,5.99772549],[38.27923965,6.0062871],[38.28553391,6.01041651],[38.29878235,6.01909113],[38.31388092,6.02783346],[38.31813049,6.03920031],[38.31987,6.0474205],[38.32256699,6.05182695],[38.32806015,6.06079674],[38.33320236,6.06917191],[38.3370285,6.07481289],[38.35150528,6.09623098],[38.35497284,6.10375452],[38.36251068,6.12019491],[38.36528397,6.14279985],[38.3769722,6.16451693],[38.39421844,6.18177271],[38.41328812,6.18912363],[38.43468094,6.19876575],[38.43897247,6.21687937],[38.43598175,6.24051046],[38.4329834,6.26553154],[38.42539215,6.29009247],[38.42746353,6.30019283],[38.42493057,6.30661345],[38.42354202,6.32566404],[38.42177582,6.34822655],[38.42249298,6.3546586],[38.42375183,6.36555433],[38.42764664,6.38594484],[38.44103622,6.40536308],[38.45329285,6.41676903],[38.46253586,6.42236471],[38.4705162,6.42737865],[38.47528839,6.4303751],[38.50379562,6.43634558],[38.51574707,6.43651772],[38.5375824,6.43679571],[38.56019974,6.43957758],[38.56654358,6.44045591],[38.58331299,6.43701601],[38.60631561,6.4228363],[38.62674332,6.41703415],[38.64501572,6.42317915],[38.65472412,6.42136669],[38.66145325,6.41908407],[38.67201233,6.40898323],[38.69062424,6.40622282],[38.69591141,6.40117264],[38.70257568,6.39600849],[38.70716095,6.39245176],[38.71750259,6.39703226],[38.73865128,6.40895319],[38.75001907,6.40952349],[38.75795746,6.40906811],[38.76648331,6.40320253],[38.771492,6.39976263],[38.78343201,6.38805151],[38.79552078,6.37252045],[38.81212997,6.35591841],[38.83202362,6.34321642],[38.85253906,6.32033539],[38.86321259,6.31068659],[38.87411118,6.3008337],[38.88557816,6.28477192],[38.89773941,6.25860977],[38.90364075,6.25095606],[38.92091751,6.22855711],[38.92547989,6.2203064],[38.92899323,6.21394014],[38.93650818,6.20033455],[38.94221878,6.1919136],[38.95005035,6.18037271],[38.96059799,6.15927076],[38.98812866,6.14709949],[39.01131821,6.14226913],[39.03448868,6.13743877],[39.0450325,6.14166689],[39.05561829,6.14590931],[39.07971573,6.15300035],[39.10289764,6.15734053],[39.13185883,6.1587863],[39.13041687,6.16599655],[39.11320877,6.17655706],[39.10359573,6.2022481],[39.09970856,6.21852875],[39.09398651,6.23388958],[39.08596802,6.25201035],[39.09085083,6.2708025],[39.10944748,6.28733253],[39.11954498,6.29899406],[39.12574768,6.32189369],[39.12691116,6.33607006],[39.127388,6.34435463],[39.12464905,6.36063528],[39.12122726,6.38058615],[39.11665726,6.39778709],[39.11185837,6.4179678],[39.10752869,6.43768883],[39.09996796,6.45557976],[39.09539795,6.47209024],[39.09588623,6.4945612],[39.09661865,6.52161264],[39.08621597,6.5451684],[39.06978607,6.55329418],[39.05256653,6.55698442],[39.04844666,6.5698247],[39.0529213,6.58404779],[39.0645256,6.59881687],[39.0593071,6.60878658],[39.03219986,6.61180687],[39.01129913,6.60909653],[38.98325729,6.60385656],[38.9596405,6.60087919],[38.94167709,6.61033678],[38.94445801,6.63027763],[38.94942474,6.64380312],[38.93442535,6.65561152],[38.91785049,6.65920877],[38.90176773,6.66565943],[38.87694931,6.66820955],[38.8551178,6.66869926],[38.83774948,6.67491579],[38.83353806,6.6916604],[38.81792831,6.7036109],[38.8098793,6.70132113],[38.77930069,6.70021105],[38.75424194,6.69703102],[38.7422905,6.70094109],[38.72735977,6.71197128],[38.72093964,6.73010254],[38.70555115,6.74204302],[38.68854141,6.74436283],[38.67842102,6.74896336],[38.67200089,6.76227379],[38.65821075,6.76962423],[38.64348984,6.76872396],[38.63566971,6.77171421],[38.63580322,6.78557301],[38.63639069,6.79970551],[38.63985443,6.81268692],[38.64769363,6.81810999],[38.6553154,6.82338047],[38.66221237,6.82773113],[38.66759872,6.83227539],[38.68315125,6.84033203],[38.69834137,6.84857273],[38.71408463,6.86057758],[38.71099091,6.87120867],[38.70479965,6.8854394],[38.69976044,6.90334034],[38.70185089,6.91503048],[38.70463181,6.93063116],[38.70120239,6.9450922],[38.69477081,6.95749283],[38.70053101,6.9686451],[38.71116638,6.97375727],[38.71438217,6.99141407],[38.71049881,7.01298523],[38.69765854,7.01383018],[38.68938065,7.00086737],[38.67741394,7.00535488],[38.67438126,7.0164752],[38.67807388,7.02879715],[38.68445206,7.03395128],[38.68087006,7.04835653],[38.66799927,7.05985737],[38.65351105,7.0617075],[38.63695145,7.06609774],[38.64042282,7.07801819],[38.64617157,7.09554291],[38.63034058,7.11084938],[38.61149216,7.12418032],[38.58364105,7.12629032],[38.56636047,7.11231947],[38.54611588,7.09087706],[38.52813339,7.09102869],[38.50510025,7.08783865],[38.48002243,7.09039879],[38.46391296,7.0998292],[38.46002197,7.11818981],[38.44254684,7.12681246],[38.42084122,7.1225214],[38.40063095,7.12147045],[38.37807465,7.12976074],[38.35691452,7.14585161],[38.3421936,7.16445208],[38.3218956,7.16174221],[38.31343079,7.14710951],[38.3052597,7.12161255],[38.28918457,7.10710955],[38.27189255,7.08347845],[38.25442886,7.0598979],[38.24634171,7.03903008],[38.24155426,7.02824831],[38.23791885,7.02005482],[38.216362,7.01440573],[38.19379425,7.01234531],[38.17007446,7.000875],[38.14658356,6.97746372],[38.12929916,6.97382975],[38.11711502,6.98091412],[38.10894012,6.99962997],[38.10191345,7.01077557],[38.08787537,7.02294588],[38.07336426,7.02960634],[38.06415558,7.04338694],[38.06300354,7.06543779],[38.06142807,7.07221317],[38.06646347,7.08770895],[38.06947327,7.12239075],[38.07639313,7.15408182],[38.07801437,7.18026304],[38.08696365,7.19494343],[38.1002121,7.20709515],[38.10433197,7.21087551],[38.1248436,7.22188616],[38.14397049,7.2322073],[38.16379166,7.24504852],[38.18107986,7.25789976],[38.19859314,7.26154995],[38.20577621,7.26547909],[38.22003174,7.27554131],[38.23363113,7.2879324],[38.24654007,7.29664326],[38.2652092,7.30672407],[38.27567291,7.31894159],[38.28152847,7.33868361],[38.2771759,7.36118364],[38.27944183,7.3805995],[38.2924881,7.39141178],[38.30886841,7.4014926],[38.31891251,7.4141674],[38.32519913,7.4338932],[38.32821655,7.45638323],[38.3272438,7.46168184],[38.34122849,7.47857952],[38.36175919,7.48773003],[38.37835693,7.49482107],[38.39172745,7.50192165],[38.40688705,7.50346041],[38.41385651,7.50417185],[38.42675018,7.51472139],[38.42954254,7.53389883],[38.42566299,7.55386972],[38.41891479,7.56945801],[38.40104294,7.57870054],[38.4008255,7.59530306],[38.40856171,7.60514307],[38.42026138,7.62434292],[38.41822433,7.64248371],[38.41088486,7.66223431],[38.41243362,7.68155336],[38.4258194,7.69676065],[38.43865204,7.71886683],[38.44262314,7.73882771],[38.44484711,7.75953388],[38.46833801,7.77766609],[38.49244308,7.79539347],[38.49996185,7.81122112],[38.50346375,7.83095217],[38.51290894,7.84891033],[38.52815628,7.85843325],[38.53239441,7.86128378],[38.54948425,7.8708744],[38.56308746,7.88519621],[38.5666008,7.90536499],[38.57575607,7.92428017],[38.58650589,7.93826008],[38.58867264,7.96106768],[38.59193039,7.97459841],[38.59736633,7.99058914],[38.61219788,8.00684071],[38.60996628,8.01158047],[38.62454605,8.02615547],[38.62770081,8.03359509],[38.61268234,8.04330158],[38.61611176,8.05666161],[38.62185287,8.06841278],[38.60706329,8.07919312],[38.59341049,8.09088326],[38.58394623,8.10810471],[38.5884819,8.12723541],[38.58799362,8.14242554],[38.5872612,8.15623665],[38.58146286,8.17187691],[38.56802368,8.19392776],[38.55159378,8.21298885],[38.54255295,8.22584915],[38.54205322,8.24610996],[38.54925919,8.26268291],[38.56074524,8.26608944],[38.57922363,8.26360893],[38.59354401,8.26019955],[38.60072327,8.24962807],[38.60354614,8.22293568],[38.61284256,8.19465351],[38.62375259,8.16291046],[38.63002396,8.14405918],[38.63513565,8.13233757],[38.64368439,8.1247673],[38.6540947,8.11374569],[38.66659164,8.0956049],[38.67284393,8.08986378],[38.67926025,8.09581184],[38.68451309,8.12245464],[38.6814537,8.14845562],[38.67864227,8.1712265],[38.68218613,8.19042778],[38.69315338,8.2033844],[38.71601868,8.21944809],[38.71592331,8.23115921],[38.70205307,8.2444706],[38.68240356,8.25800037],[38.6620636,8.2708416],[38.65095139,8.28508186],[38.65761185,8.30443287],[38.66602325,8.32696056],[38.66723251,8.35048485],[38.66164398,8.37601566],[38.65445328,8.39463711],[38.63964081,8.41209793],[38.61675262,8.43113899],[38.60427094,8.43156815],[38.59088135,8.42601871],[38.5841713,8.43267822],[38.57578659,8.44489193],[38.56567383,8.44827938],[38.54810333,8.45306969],[38.52684402,8.45761967],[38.51322174,8.44951916],[38.5014534,8.44396877],[38.47788239,8.44735909],[38.46828842,8.43500519],[38.46526718,8.41614246],[38.46160507,8.39979172],[38.45170593,8.38502979],[38.4478035,8.36536598],[38.44072723,8.33526611],[38.43246841,8.3122139],[38.43021011,8.29875088],[38.42163467,8.28902245],[38.40086365,8.27951241],[38.38240433,8.27347183],[38.37549591,8.26593685],[38.3669548,8.2628212],[38.34734344,8.25424099],[38.33317566,8.25855064],[38.32188416,8.27234173],[38.30703354,8.29969311],[38.30787277,8.3298645],[38.31220245,8.3524456],[38.31146622,8.36994648],[38.30082321,8.37635612],[38.28925323,8.3806963],[38.28204346,8.39955711],[38.26631546,8.40663815],[38.23833466,8.40930748],[38.21754456,8.40877819],[38.1983757,8.40133762],[38.18003082,8.39286709],[38.16418076,8.40805054],[38.15675354,8.40924835],[38.13895416,8.40941811],[38.12254715,8.4065876],[38.11846161,8.40195274],[38.11000061,8.39036846],[38.10295486,8.38900757],[38.08653641,8.39009762],[38.08132553,8.38482094],[38.07709503,8.38061714],[38.06670761,8.37596703],[38.05252457,8.38243103],[38.03892517,8.38921738],[38.02804184,8.39028263],[38.00823593,8.37250614],[37.98816681,8.35952568],[37.97335815,8.36291599],[37.9483757,8.36741638],[37.92798615,8.37816715],[37.9099884,8.37048626],[37.88762665,8.34987545],[37.86294556,8.3357048],[37.83914566,8.33214474],[37.81625748,8.33180523],[37.7965889,8.33471489],[37.77899933,8.33969498],[37.76545334,8.34711647],[37.74796677,8.35176563],[37.74258804,8.34905529],[37.73088837,8.34315586],[37.71704865,8.334095],[37.70454788,8.33703518],[37.69221878,8.35356617],[37.68364716,8.36931229],[37.67870712,8.37770748],[37.6660614,8.38353729],[37.6306572,8.36455631],[37.61196899,8.35408592],[37.59803772,8.36531639],[37.5872879,8.38969803],[37.57608032,8.41406918],[37.55797958,8.42550945],[37.534935,8.42359161],[37.51221085,8.4169693],[37.49856949,8.40361881],[37.48582458,8.3798399],[37.47138214,8.35070705],[37.46615219,8.33131504],[37.46338654,8.31169319],[37.46257401,8.305933],[37.46408463,8.28242111],[37.47202301,8.26608944],[37.48817444,8.25236607],[37.50103378,8.24526787],[37.51726151,8.23636723],[37.53370285,8.23253632],[37.55106354,8.22917652],[37.57351303,8.2249155],[37.59597015,8.21812534],[37.61033249,8.21335506],[37.62168121,8.20903397],[37.62388611,8.20004368],[37.61697006,8.17720127],[37.60576248,8.15039921],[37.59917068,8.1270771],[37.59543228,8.11172581],[37.59213638,8.09313393],[37.59180069,8.07401276],[37.59442139,8.0569706],[37.59729004,8.0359993],[37.59992218,8.01987743],[37.60158157,8.01066685],[37.60139084,8.00167561],[37.60189056,7.99499512],[37.60107803,7.9741087],[37.60282898,7.96485758],[37.58870697,7.95957851],[37.57276917,7.95223808],[37.55751801,7.94833803],[37.54943848,7.94925785],[37.53927994,7.95385838],[37.53281021,7.95960855],[37.52841949,7.9660387],[37.52495956,7.9720192],[37.52172852,7.97706938],[37.52033997,7.98350954],[37.5184288,7.99185991],[37.51399994,7.99967051],[37.50679779,8.00747108],[37.50168991,8.01182079],[37.49704742,8.0145607],[37.49079895,8.01706123],[37.47763824,8.0142107],[37.45964813,8.00765038],[37.44601822,8.00526047],[37.432621,8.00334072],[37.42453766,8.00282097],[37.41669083,8.00116062],[37.40944672,7.998312],[37.40412521,7.99359035],[37.39178848,7.99290037],[37.38981628,7.98472452],[37.38773346,7.97438335],[37.38981628,7.96197224],[37.39897537,7.95402098],[37.41661453,7.94358063],[37.4288559,7.93070936],[37.43671417,7.91852856],[37.43440247,7.89806652],[37.42424393,7.87760496],[37.41177368,7.85392284],[37.40484619,7.82726049],[37.4037056,7.800138],[37.40209579,7.77047539],[37.40002441,7.74105263],[37.40164566,7.70955992],[37.40119553,7.68266726],[37.40697479,7.65760517],[37.41984177,7.63380337],[37.42800522,7.6104908],[37.43540573,7.58405876],[37.44510269,7.56431675],[37.43865585,7.54613543],[37.4252739,7.52382326],[37.40721893,7.49218035],[37.40062332,7.46059752],[37.3967247,7.433465],[37.39028549,7.40242243],[37.37973404,7.38000059],[37.35659027,7.36093187],[37.33815002,7.35264158],[37.33115768,7.34970856],[37.33122253,7.34344482],[37.28763962,7.33442068],[37.26226807,7.32543039],[37.23783112,7.31205988],[37.21155167,7.29271889],[37.20056915,7.28216076]],[[38.76155853,8.85361481],[38.77299118,8.85746765],[38.77746582,8.85242462],[38.77838135,8.84180832],[38.78059387,8.83591938],[38.78665543,8.83528042],[38.79202652,8.83577633],[38.79758453,8.8334856],[38.80549622,8.83724403],[38.81254578,8.84001541],[38.81748962,8.83929348],[38.82439423,8.84133244],[38.82383347,8.8464489],[38.82390594,8.85275841],[38.82627869,8.85761833],[38.82704544,8.86325836],[38.82941055,8.86878204],[38.82693481,8.87366581],[38.82600021,8.88018703],[38.83298492,8.88291168],[38.8406868,8.87985134],[38.84707642,8.88002014],[38.85298157,8.88178349],[38.85838699,8.88363838],[38.86380768,8.88285065],[38.86871719,8.87303543],[38.86983109,8.8807745],[38.87341309,8.88829231],[38.877491,8.89578724],[38.87331009,8.9050951],[38.87718201,8.91252708],[38.87944794,8.91831398],[38.87349319,8.92375469],[38.8680687,8.92761898],[38.86083221,8.93217754],[38.85555649,8.9382925],[38.86141968,8.9426775],[38.86949158,8.93962002],[38.88208008,8.93619728],[38.89142227,8.93433189],[38.89675903,8.93702698],[38.90110016,8.94614124],[38.89623642,8.9548378],[38.90278625,8.96241188],[38.9040947,8.9695425],[38.90624237,8.9774189],[38.9048996,8.9841156],[38.90433884,8.99215698],[38.90605545,8.99753189],[38.90500259,9.00949955],[38.90116119,9.01958466],[38.89665985,9.03049755],[38.89334488,9.03722668],[38.88632202,9.04681683],[38.8829422,9.05210495],[38.88297653,9.06047726],[38.88195801,9.06855869],[38.87911224,9.07553005],[38.87631989,9.08158398],[38.86822128,9.08490849],[38.85675049,9.0835228],[38.84952927,9.07816887],[38.84257126,9.07861328],[38.82473755,9.07588863],[38.81337738,9.08002186],[38.80512619,9.0823288],[38.7999649,9.07856369],[38.79131317,9.07768154],[38.7781868,9.09365177],[38.77124786,9.09819508],[38.76521301,9.09815025],[38.75928879,9.09589481],[38.75085449,9.09052849],[38.73802185,9.08686256],[38.72710037,9.08571339],[38.71664429,9.08775043],[38.70674896,9.08183193],[38.70067978,9.08051205],[38.69455338,9.07773209],[38.68540955,9.07302666],[38.68369293,9.0663166],[38.68370056,9.0595417],[38.68354034,9.05003738],[38.67676544,9.04316711],[38.67259598,9.03867245],[38.67208099,9.03314495],[38.66722488,9.02623272],[38.66653824,9.01673222],[38.66703796,9.01002789],[38.66007614,9.00933552],[38.65194321,9.01028442],[38.65105057,9.0024929],[38.65779114,8.99481869],[38.65938568,8.98540211],[38.65464401,8.97693634],[38.64850998,8.97599602],[38.6394043,8.97403622],[38.64281845,8.96585369],[38.65336227,8.96290016],[38.66540909,8.9610815],[38.67622757,8.95760441],[38.68096161,8.94966507],[38.68064117,8.94108963],[38.68790054,8.93974209],[38.69384384,8.94189739],[38.69682693,8.93270779],[38.70196533,8.92562866],[38.70842743,8.92091465],[38.71914291,8.91971588],[38.72333908,8.9145813],[38.72722244,8.91084385],[38.73107147,8.90509129],[38.73651505,8.90129089],[38.74358749,8.90258598],[38.74447632,8.89211178],[38.74316025,8.8804121],[38.74729538,8.87715912],[38.74520111,8.86949062],[38.74551773,8.86342335],[38.75411606,8.85859299],[38.75646591,8.85324097],[38.76155853,8.85361481]],[[42.25291061,9.18970203],[42.25944901,9.19227219],[42.2647171,9.19788742],[42.27203751,9.21566772],[42.26871872,9.22960281],[42.26226044,9.24362278],[42.25632858,9.25255203],[42.25365067,9.26153278],[42.24934769,9.27003193],[42.25030899,9.285532],[42.25991058,9.29883194],[42.26604843,9.30977249],[42.26496887,9.32132244],[42.25851822,9.33441257],[42.26461029,9.34905243],[42.27031326,9.36166477],[42.26613998,9.36598206],[42.26126862,9.37351227],[42.25299835,9.38380241],[42.23807907,9.38494205],[42.22187805,9.37947273],[42.20478058,9.37023258],[42.18743896,9.36168289],[42.1685791,9.36283207],[42.1574707,9.37632275],[42.14661789,9.38751221],[42.12915039,9.38867283],[42.11422729,9.37251282],[42.10870743,9.36412239],[42.09962845,9.35035229],[42.08525848,9.32911301],[42.0704689,9.30394268],[42.06467819,9.28489304],[42.06050873,9.26587296],[42.05887985,9.24943256],[42.05973816,9.23765278],[42.0613327,9.2264595],[42.07368851,9.22096348],[42.09084702,9.22512341],[42.11240768,9.23074245],[42.12061691,9.22844315],[42.12734985,9.22655296],[42.13726807,9.21582317],[42.14855957,9.20626259],[42.16571808,9.21042252],[42.18214798,9.21689224],[42.19746017,9.21987247],[42.20848846,9.21193218],[42.22078705,9.19637299],[42.23371887,9.18591213],[42.2460289,9.18700218],[42.25291061,9.18970203]]]}]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>GID_0<\/th>\n <th>NAME_0<\/th>\n <th>GID_1<\/th>\n <th>NAME_1<\/th>\n <th>VARNAME_1<\/th>\n <th>NL_NAME_1<\/th>\n <th>TYPE_1<\/th>\n <th>ENGTYPE_1<\/th>\n <th>CC_1<\/th>\n <th>HASC_1<\/th>\n <th>geometry<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- class: inverse, center, middle # [_**Data Exploration and Processing**_]() --- # Spatial Distribution ```r ggplot() + geom_sf(data = Oromia, fill = NA) + geom_sf(data = cases, aes(col = pf_pos)) + scale_color_manual(values = c("gray", "red")) + theme_bw() ``` <img src="index_files/figure-html/unnamed-chunk-11-1.png" width="50%" style="display: block; margin: auto;" /> --- # EO Covariates .pull-left2[ [Bioclimatic variables](https://www.worldclim.org/data/bioclim.html) .tiny2[ ```r bioclim <- getData('worldclim', var='bio', res=0.5, lon=38.7578, lat=8.9806) %>% crop(Oromia) %>% mask(Oromia) ``` ] ] .pull-right2[ <div id="htmlwidget-7094c2f306f3022a8ba7" style="width:504px;height:504px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-7094c2f306f3022a8ba7">{"x":{"options":{"minZoom":1,"maxZoom":52,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["CartoDB.Positron","CartoDB.Positron","CartoDB.Positron",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["CartoDB.DarkMatter","CartoDB.DarkMatter","CartoDB.DarkMatter",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenStreetMap","OpenStreetMap","OpenStreetMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["Esri.WorldImagery","Esri.WorldImagery","Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenTopoMap","OpenTopoMap","OpenTopoMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addRasterImage","args":["data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAyEAAAJvCAYAAABsy95bAAAgAElEQVR4nOy925Ml2XXe91s7b6equrrnAoCkKEu0IiyHww9+dfjN/8J0V/giC1AAARGUKMqAKSA4GPvMmAPQgghSMEUJEIIISgjQMmcGfwKe/OZHR/jBEbJpmSRIEBhMT1dXnXPysvyw99q5MytPXXoGYHfX/ror6tQ5eTLzZO6Tub691vct+d73vvc6GRkZGRkZN8TrJ19bp3+XuPi4Y4h/S/itDPFxiUMQAD7/1mfe+KnscEZGRkbGUwN39SIZGRkZGRlTzAnIHOWe20tFEeiH4AIJ+crJ1y9dV0ZGRkbG8wfJmZCMjGcLn7j/hfW/eucf55njjJ8K1if/NBIEQVA0Pk6RZj2UAYCCgp4eweEQBpSKYu86DC55/lff+sU81jMyMjKeQ2QSkpHxlOLjr3x+XUpJpx2VVPH5SkrOhnP+9Xe/koOzjJ8KXj/52jolDi78dIFsVJSAL7cqKOL7jHgA8fc8Q3IdIgKZjGRkZGQ8b8gkJCPjKcR/88o/WisDjTQx6Guk4Vw3APQMFDg2uuE73/3qG3/rlf9uXVBQSYWiFDi2ustEJeMDY33yT9eWASlCRkMSItHSRRKSkpMSF0mKvTagT0xCIBORjIyMjOcJWROSkfGUIp1RbqRhJRWVeDlvgaOjA+CT919dl1LS0wNwKAc00lBJxd+5/2u51j7jA0PRiYbj4uuebAxoJB4pAYGRoCytOyMjIyPj9qH8y96BjIyMi7BgraenoKCj41Q7Wu3iMiUlIo5aKnp1iPjMyIGsONcNiuIQPv7K59fputLsyMdf+fy6korfe+dLeYY54wJSPYiitPTUVDGroQxUlJGEGBzCSit6GWgDObbn59iXCRnC+M3IyMjIeD6Ry7EyMp5CWDnWSlZ0dDErIgiVlLwgx5zrlpYODf9a7Wi15WPFSwA8HE7pGei048gdArDTllZbqpBVabXjQFZsdcs333kzE5GMCV47+a11kSTMa6pos2sEYUgyGfZcRUGh/n29DPQzkpISj30i9X0EJJdkZWRkZDwfyOVYGRlPKSwDouqDtDoQhxfkGIdwVw5x0ZHIkxPwehHBjXoSKYJLkQ8EnXhC01DHbQ25JCZjAW++9bkY8BeJ65VDotXu0g/ATjp20lGomxCMy8q6DDkDkpGRkfH8I5djZWT8lPDJ+6+ua6n4+ttvXDqT+6n7X1zXUiEIrbaICCtpcLgoDAZ4qI8BT1YKHFt2NFJzOpxxxx3G5QocAwOqPoA8lBUVJbVU1FRR7P7LD3593dPHrIohl2rdTsyzIFYe5bMhypYuZkVGTYhf3vQgJc4TERzYeAwZEmRgCFqTFGmGJZORjIyMjOcXOROSkfFTwCfuf2FdS0VJyS89eH1RLP6ZB+v1p+5/cQ0+63FHDqM1r8NRSYETiTX2lr1YSYWtu5EGgFZ7BpRG6jA77RhQaqlwOFZSU+DoGfystpTUUlJTTfbJIdg+ZdwevHbyWxfOuRGClv6C6NygDBN9SMdAhy/HGtCRgMDkcYohrCUjIyMj4/lGJiEZGT9hfOL+F9ZlSDpWUlx4/bMnX15/5sF63WpHJSV35NCLf8UHfWaHutWWQcfgzNY1X3cp498rqYP+o6CWioLCu2xR+KZxOsR1pCVbKSop+cT9L2Qicosg4V8Ks+bVSBP8WJmTjvlzhiUXrEqLSG6Wsh5zMpL1IBkZGRnPD3I5VkbGTxC//ODX18dyh3PdxDKoAeX1k6+tO3pKCja6o6HmnrvDRnd8VO7SmfeQKKd6xla3FFJQSRGteGsqWnq27Oh1CPaoYxlVqz21lByG7MiLcsz7esaKGkHY0FJKGUqzSnbaMoQZa3M/KqTgjhwwoHzq/hfXL7l7vDe8z4DmMq3nHJ40yOy5qdsVTAXlRiSMOKfEotgz51Woi6VZGRkZGRm3B5mEZGR8AHzl5OvrSgsOqWi04O+887ffgFFfIQiNVHThq1aEUiiHUAbHq5qKA2lYUVGIb/B2QM05O16Qo7itLTt22lHOsim+9Mr/S1EFQXpDGUuvaimR0GekoeScIjznKETp1RduiZSJa5HXoZRS0+G7tw8MfObBeu2374mPEZdMTp59LGUt5uTD4GYkJG1kOCEhe8qv7LVBltdvyFmQjIyMjOcLmYRkZDwhvnTyu+uKgjJx+/ntB99c/3/yQ1ZS0SZBl8xmhAeUJvRbUDniMRtccKuy7tPWIM5nPwZKLbHsCUyFuz0ay7LmmJfVrKjZ0cbnJRCSM3pWUrEJ8adlVXp6SkpWUgUNiYuuXJ6UlMm6spD4WcarJ19dw/6sxRypUcIc1lQTwOnl46K9hIBk8pGRkZHxfCKTkIyMJ8CrJ19dV/jSKCfCoEojJWVwDwIoQsZio7voKNSHLAdYQ7cSxAfvDRUNFZUWdDIgCD0DK2rO2AKwkjrOUvf0sYSlCBmNmoqOjlb7SAj8doOInZpHnEcr1btyyIYdQ/DY6uhCaVYXdCIORVkFPQlM6/SL8Fl6Bjq6RU1JxrMFO+dGIOycpsQkkotZ+ZU9N6BUQWMkCH0Yz5FYa0Er/YWMSyaxGRkZGbcHmYRkZDwBfkbvxsC+DiJvgBUVKyrO2bGjBcaMhsECry0thbgpEVG/njYSDQsAfUGVEQVDOgvt/bEcSkEvIxlIMyYO766lCAUlDi44YlmndcvCFBSxt4MglKFUy0iQovQhluz18pKajKcfDoljGy4Sg6Xsx7wkywhIxxBGmR9X9vdlmY+MjIyMjNuB7I6VkXFDvH7ytbUvNQlN2eh5JFt2IROwS0gCQCGOQhw1ofcHPW0I4c/Y0oZZ4nKhfCslD2kGwtyHCvHvMb8iCQFg+n4jD/ZcQ+k7WuMoIxWZOhrZPpdh2TIEnmkzOgkz5imyHuTZhpHUudh8SffhyakLI2k6NoyAwGjTm9r6avg3x9Jzv3nyjezMlpGRkfEcIpOQjIwb4NWTr657Bs6kpZWBQTSSkZaBI61jT4QUqc7DenMAcdlSXVwP2GyyL4jqGXzZFxKaw43i30E1ZjoaythEbkVNxajVMEveOkl+ulAeY4+ndMWXlZmAXsLvlNTASJZ8CdiUfGU8e+jC2HShFNB+ljAkY2e+jn19RJ4EWROSkZGR8Xwil2NlZFwTXzr53fVH9S6KckwTsxkVBU3gHD+UM2AMzGoqdrS0dNT4/hxt1HIMFDg27HAiHOkKmBIVK20ZUEopJoFfH5YtsKxFQRfIyoByzIpNKAkrKKK9ak05cTGyINOIihfNTy8Nd1hxT1f8UE7Z0gUCI3QobSjBylmQZx9WVgieZMwF6ikBhikBSXuE2DKWXbP+IoKbdFPPyMjIyLi9yCQkI+MaeP3ka+smiMvrEKiZ7a3lEJwKgyg1JTu6kDcYxeFWApXC2+UWtHTsJOnxETqFGOlIS6Wq2dfWmhruAjmwQHIb1mFZki0dFZ4YleoYRNlCUABozHrYZ0q3d6gVHxuO6NzAe3LGGbt4DAaUf/b2G5mAPOMYzRaGqAkZS++WBeOp3iglGS68f0CDTmmYkJeMjIyMjIx8V8jIuAKvnnx1vQsBfRFKmDa07JIgvwrlVC8NB/yc3mNFxQsc+SxJ4iAVMxc4VtTBEaukCXoR8IJ1041sAh3x277YbX0f6qDlsACxpaeh5FAbXtQDjmm4FzIvKTHShdnvO1pTU/ICNU1otmgQHE6EX3rweq7bfwbw6slX12bDO3/ezruR35SAjFm5i/NWHQMtHV0oG0zXYa8DzDutZ2RkZGTcbsj3vve91/+ydyIj42nEZx6s1/fkThR7p1mGI13R2HNqmRH/+iPZ8mM5Z0sby6G27CLJOOaAFfXePgqD+HKsc3YXhO4W5Fn2JHXesnUe0HBXGwaUh3Iel28ouacrVuq1I2fS8iM5o6eP27DsjQsZmkOt+U+Gj9LgOHKO/50fccqOH8kjtuE9Zgnc0fH1nBF5avHqyVfXY2ZtSjathG+u77BlzEEtLa/yJYH9BSH7HHNhe4qU1KROWvP1OSRrQzIyMjKeM+RMSEbGDJ89+fL6sydfXh/KauIyBeOsrlngCkIrPa309ChberZ0kYCYsLfAcUhDQRE1FTDqMbbSBRn4GHw1VKyoJ88V4Z9hX/B3rE3oil5FPQiMXauF0W0rDRBtf+y5Qyr+vbrmIS1/PGyo1LcpXIW9TTEX42c8fTBXqvm4sXM3/51iFzJ06XvGLMf+c7+PgKSYO2lll6yMjIyM5x9ZE5KREfD6ydfWHT0NNVt2cZbWbEvNqragwKlEZ6uGEqfCubRoyD6YBqTEhcCtCCVYFY/ZXHAP2tLSMO087vAz0RUFOzqseWHqTmWogtgc/Gx1K8NEg9ImwaLNelu9vuVZrBO7ZUF+ZjimoeD/2J1yKjs6lANKXtJDChynso2zGH3SyT3j6YQRi6saAqbL9QzUlLTBfnpeqjfvKbJEcNIMxxyXvTbfp+uQmYyMjIyMZwe5HCvj1uPVk6+uHTIhBqmNrpVjmR7CE48qlmIdUlFrSStew/FINoAPsCpKNkHEbbPI5qiV2uUq6jUiWtKFLIsRn56Bx2xiUGgBWR9q8cGTEAsQKwo+qsc4hFPxTRPt9SJkMvx+KJ0MoXmh3+aRrrhDzfFQ04tyKjseyiZu9642nIsPSB9yhjUtNIesLbtckvWUwWylrWO5mQnY+DaDBfsNRkRd+CnYhayejbHUcMH+XiIghn1E4zKxelquCPD5tz6Tx1VGRkbGc4ScCcm41Xjt5LfWFnylJSDprKui0Y7XMh9GRg6poiakVG+VO9qT9nFdbXCgsvcLQkc/CbIGlEF8zxBBULFAcdRp9Iw19i2pQHxaUmVWwOm6OzqcjLoW27c7WsfP+5Ie8rI2vE/LGR2P2bFhF2fHH4o1nhs/T8+AE6FXH8B++v5r62++82YOGH+K+EcnX4mlSv/krc8vHvs0a7FUgpf2A/FGCESDhDnBWCqXuizD8iS2vL/21t/LYygjIyPjOUYmIRm3GibETgN2mNbEW1O/VI1R4jjUKpQ8+RnbNmRP5gTA1mEEYkCpcJOCLCMGaaYi1aNYRmRuC2zrTIO7mpLH7CJB2kkbBe0ucbcaXb28yLjRkpe14a8UK34wnHMmO7bSTY6F9TiZawZ6HSbPZyLy08OvnvxPkYAIji+c/JN1eo5tHM31HkvlTQVuQi5TpDqmeVnWPCNij5fef90SrIyMjIyM5xuZhGTcWnzh5J+sYQy8YKw9t2Z9vh5+zGhYkHWkvuWfGY6u1IHAhmlw95gNBS6UZhV0SQBX4OI6xuyJRvrikuCxiE0EvbDdsjJpV/QVVSwVs/U7po3ijLwY+jDj3WjJz+ghH3UNDvj39Zh/h2NHz1bauPw8OO3pIwGJ+yy+w/ovPXh9bbqaQhwb3fEv3n49E5MPCZ87+Y0LQm3HQM+0zMlKCzs6JIwJP85HDdCcVKZj3WDfjSVR+5zQpOVac3OHKRmx5cf9/eJbfz+PkYyMjIxbgExCMm4l1if/dG0EYz7jCz5AaiiDtHvstWFB1VgzP1rMWTDnS1hGW10TqcNYjmJ/+6AwKYORMeir1NFK2jm9jO9NBeq2LxYMDij3dDVZlwTC86Ie8DBoVgp1lOH9NQV3qRhUGUR4N6gAurD91No17uvEaUkZop7A/5vPlleUsZ9IJiMfDEsERBk7cNiYMkJr1ggjgdCoOfLLj+d4JCTTXiH+3Mtk2ZR8yGzZpfKsNAuSvjcTj4yMjIzbh0xCMm4VfvvBN9c76ThiFRr4DWzpIsEwpAFSTRV0FuWFTuZOx0ZufSAbfbKuYhZwWZBohMEHdNNgvdGpy1QRsiwW9JvjVrqOUZR+8Svtl++QRAti25CgCTlS6wMBhcA2UDNzyhoSUjWfOdcYkE73oZBpE0TrDt8zLVXLuBmWCEivXpdj56JfIAF1KLvzr48ZuTSTMSfk6RjrmTYxTEnJfFvzLApwwRHO1vPaW/8gE5CMjIyMW4hMQjKee7x58jtrm4E9wjfrcwh3qDmTlgGNzfp8VqCMOgkgKX0q6HA0mO2t7w/SJM5UaRDnXbXGQM/Iw1yMXlHQaEErA4362eoylHA1WoQSKKi0oBfrnj46GRkpuqsH4b1jQLiiYksficEjtjTqmxDe04at9BTqScWRG/fsZzngfS1ptWcju3hsxiXGzteeXAhOBLMi9s+7CeHwx9FTqX2aEcuUVIl25Z+9/d/f6iDVSMdvvfVrb9jjdGwKQiEuNhQEYumVjePUic3GjS/98+MvJZVpeaIhfX3J6ndfnxBb5vW3/uEb4G2w7bVMPjIyMjJuNzIJyXhuYZqEn5EXUfo4E1tRcCo7X7K0ILJu6VlRs6MNYnSHQ6nUMYgvz9pIsMZVRx1ITaESZ/+NJLiEcNzTA86knZVqDbhAYhr1X8c0uLN+Ib5JoFBqwVZGjYqRGIdwSIUgHGqFonQa9B6M7+lk4N6woqZgRUGhwss0bBmoRNgOAwfiOKfnNNqy+qzJippTRrveaOcqaW8TicvPS7iMkPig+WJPkU/ff21dShn1L+l5hNtZwmWkQ1E+e/LldTo25tmoMoykEsdZor9YUWG6IICxd81IsFOk9tQp9gnNl7CkCYGRjGRkZGRkZGSLkoznEr/84NfXR7KilhINovAyUIJGvRbkoWzYsMN6J1hjQOu/YcFZGwiM6TMq9fqLO1rzoh5wRyvuqG8XaGVO8xKpO7riWBtqLS+UTDU66k5s5tocr2w9RjgkbN+WtX0eUCr1GRXToFQ4GgqOdTUJGI+15kBLSjwBEeBYSlrVSChelNAHhYI7WvMzepdjbYCRVKQZnVEPcPGSMg9ezX3pVx68GWfFP33/tb3dsI3UGBm5Ldin+9BAAFPxuB3jKqh8VlSsqKjD3wYrHhy/DzL5met4lmDLXoZ9ndkzMjIyMjIMuVlhxnOHX3nw5vpFucMhDdvQqM8IRUPJzw93OZUd35eHcTY/LafahkxFiaMOncgrypDx8Ouo1Mt8D3XsVX4mHQ7h37n3aOmoKDnUikOtaaXnhWHFu+6cnoFT2cXMwLGuYnfzef+GNFPTMlCFUpkOZSddFPrWWnJPV1RhHQc6fuYB5c/dKefSUWnBXx/u8VEaVuJisVQjwlaVO85RiHDa9zgRHg89LQN/wYZ33Tk/kEdJZsMfq7TZXQrfX6WjC40M0wwQTPUrbegIX1EuGgVYGZcRkp7+uc2MfPbky+teh4mmxjC3wE0fH9JwQHPhPanLmqEP/T/SYz0nNUslVmaAsLT+pX2FnA3JyMjIyFhGLsd6Anzq/hfXv/fOl/KN9ClFIzWHNAjCihogGJfCYRBgr7SM7lG2nIm957Bu4weUMegeRKmDXqNjoA+NBc9oYzBdq7fPHURZaclBICUDBC2KL5EZYHEGOnUgSm2CB+Ag6EhsGV+K47MnoqMLkVdpTGv3H0vLS1pzWDh2g39/FUjIVhVU2alyKMJLRckP+pYymT2HsbTsIGznlE3cxtj87mKwbMdz/rwFqUsExH9uF923bHnLjJjYvaB45vUjnzv5jfWgy13ILbtgmJORLmTw5pm2JRIydrwZ1+VF5+N7+oXvwtL3Yx/26UTePPmdNWRNSEZGRsZtRyYhT4COjk/c/8JaVfnX3/1KvpE+RfhHJ19ZrxKBbqmOTqadoDsG+sQqVBKR9+hYNVrx+hIqX/rUBFtfIzMmJu/CTP25tLGDOvhArFDf2LDBsdKSnfRx3aYambsWmfjcnhuAMpAR3/NhtO4tcJTqSYIobKRjpWmWYYjlWmbJe0rHR6mondArgQjB2dDTiN9+q4oADY6PsWKrPS9wSInjY8MRHQNn0npCJRoD4LPoreVRiIvZEICO3tM6EXodnbWsJ32qo0kxf96C7yGIs591pPqPqzIScDHT0IdxXc2W29ccMNXu+PKuEUtkY97v44Mik5GMjIyM241cjnUDfPyVzy/WpGci8nTgMw/W65KSlVS8wFHMgvRJkPqR4YgqkI4/ce9jTfwOteaxbGKmA3yw1VBFN62akkaLC25CTRCLb6TjlB2+GZwXXK2ouKsNKy1oKPiRbDiTNvbqUJSX9BCnoRdJICj2GvjgzzIh8wDVYPoNs1C9ozUrLdhIzxktZ7JjQHlRD1hpyS/IEUeF41E/RCKwcs7rQoA2PFeLoOFv39hQKER4uSr4k92OFqXB8aecxeD03xbv8ojz4DrW0ql1WR+CXsVniJwINus/7z9yFYyspL1JDM9iNuRzJ7+x7jUpjZJlYXh6nJowvv3jMhLbeZZjqRkheFMEIGYA9wnSbdtLuIqQLL1v3rTw4ntc7huSkZGRcQuQMyHXxD4CkvF04DMP1vH87LSjlgqnEsqmvAi7xmc0dsFa14IwDcHxlg5zGQJiSRVMA2MTpquAKCHY7mOvkSKYnMIYAG6kp1LfMLAMfT/Al4UVQWjeyzAJNG2bpvPwkEiqbN1mtTrvRWL7WohwT1eciS8V60U5dMFmWIdg0+o4G8aSr0KEVgd2OpZVHbuCrSr3ioIXGmHbV/y47yhFuKs+IC5CZqampKNnOwt+42y6TH8Pk8WuJiFGOpayJr/84NfXzwIRSYXnvQ6RWC4RELiorTDNElwsrzKkmb3puobZ3xe7ni9t/yJunhVJ92Vpm8rAmye/s84ZkoyMjIznG9kdawGZcDx7+Prbb7zx9bffeMMhHMqKUp0PiENX8BWV77PBwFY6zkPpUEVJTcX7ch6DrDYQCrPHTdHKwEY6etFYEtXKQCdDzFbAVOOwCc0Qe5RjrTik4kBLDtRrTfpQLuZ0rO3vGMvD0tImX042kpSGMpCrgkoLVuqzNYVKzFzc0yaWjz2UjV9HyEDccQUrKahlbEhnpEOBbchfrMTRqbILs/U/2oxh7Kn28TjtGEJp2PL8RklBISaBHgPQQtxITJ6D0qqrsOR8tS/bMD9WNmZ9P5tikYCkjlfzJoFzLGVJLpKW8e+5TfC+zNVVJXI30ZdkZGRkZDx/yJmQPfj4K59fW5lVSkpEBNWrb572nkoqKpkKcftQntJqm0u5PmS0dHR0nMmWF/SQJnHz6cXL04cw899oQ8/AuZgblg/muqT0qU30JI16XYW5TDngVIaQVTGb3zGIbhmotaQJfTxaBhpKDrWkljK6X7X0sSO6kZ+xl4OLJVZgZVejVsQ0IiaAL9SXZTUUyfLCj2XHOR29DDxiyx8PG/rBkxSHcEzJGT13KKnEk5GVODY6IMAmkI9KhLNh1A+sxJdw/dXygHe7joe0bKWLM+2jviVYDcs0oL74eMwgpZiXqKVYcmsy+98B/Ustz/rsyZcnZEMQOu0ppZiU25kTVlqSZbAMSBkMEqwfyNi5fIh0OR0nlxGE+frnuKwsy8wR5kjHabrem+p10p4mGRkZGRnPL3ImZIaUcHz8lc+v51mR6xAQACcFpUxLeZZu9jnr8uHiX7z9+hs77Thjl5RVBa1E0F2UQQNyR2vu6Yo7WnMQzHjtPQW+Y/mWjm3IZLwvW86kZUPLY9nGBoDgA62DYNdr/TuMcFTqWKnv21HhWIWshWEIpWDWTT0VEZvYOM2ypLPclrWxuv5W/PLndFGQ3qKchq7nQ3j8R+4h33en/En4+Qu2/JgtP2bHmfa0qpxpzzk9Kxm7vgNshiGSkvfUb6d08EJZcExJGz6LLV+Ii5mOqwJSWzaFlSgtZVDmeBqyKHPiMajGHyMZseROR8crRSfHSJJ//nVfsmUleCnSMQFjhuIyEpLm+a6TDZFk/fP+Ipdhn7B+//KZgGRkZGTcBmQS8gEhInzy/quToOPjr3x+rYkYWnWYBAUioft1IClLZCfjyWFB8EPZeEKgLpZngTUE9OSk1oJD9Z3GrdGbhFlkIyVGCizYqihYhZKqhoJ7QxNJRa0lZehubs8NIQzzDls+IzIPzEyLYoSpC+NHFgK9eWPDDvUuVWHNgvDI7fiRO+fHsuFUWs7YxfcPYfkukJtTdvype8RDt+X77jF/xnkow/L7fKcoOBAXy7tO6TjTnkdDzyNazuj4UdtTiXCvKCflQQXFxJL3OrDMQKofsceXkZm/bALy2ZMvr42ApETEiSdRqcXukExmpEQExszHvMzKi8c94dzEUTSW7YF3wtpHPlLSsNRUMr1G7Xv/ZfjLPv4ZGRkZGc8WsjtWgichAk6KSY+Hjg5VpZRyDKLmpSXiULVeCv53p74kyMqzPnX/i3FfaqlYSc1vv/VqLt26Jt48+Z31C3rIy3o4mZW3wBpgRx+1HGfS0tJT4tiGAK+i5ECrWLJkGQ7wjlgFjioQjlYGtrGXB+G3+H4kIUPxsq4A2NBzKjtOg2NVarVr5VXmVuT3eSxRMuKxxTdGPJcuCOIH6kCMKgrOaH0zxeCg9GM5B6YlMmUQxHche3KsDRUFHxsOOaTkkIKVOEoROvValFPt2QZ9C8B5aEDYhA7tCvxYNvzf7kfsAhkEYh+PPsnoXAf7mvbZa3Myd5ml7f/89ms/ke/PPPMxR1pKlhIOI1q9DlEPY+SjD+Mg1QNNu6ILd2gWycQ+GJk2kqGBhO7rzTL/DOl1LCWB+/Qi1ynHWrIONmRhekZGRsbzjZwJ+RCwFPTMxZvpb03qvo2slFLipOBT97+4NgJSSUkjdZxJvirYyRhhfcwtozA+74nDRjrOpeWRbHksuxjIp/03LJtVUUw6mpfqgoDbn8flHgyOOpRg1RTcUU99ejTujffQ0khurDdIGvB1pK5X6Uw1nMqODTt2tNGZ61w6TmVHK37vBpRNeL2ljzPlJtDfykgUrDTtLpVvhhg6qm90oHGOI+cpWBlJkidXDuFUWr7vHvMDOeOhbOJ3wo6lNXy8KfYRkKXX5hmUOUwn8mHisu/kvHQs3b+UmNjnSMuuYFoOlRKQioJDakw7cZPypXmW47rZKRuDS9m5edmYLb+vyeJ8nXPchFhlZGRkZDy7yFf7BE8iEp8HAAieL6AAACAASURBVAUFTi661cyJyGR2W4owA+oWSm/2B1UZ+3HGlkey5fvu0aT/geKtddPzYMTDgjoJhMBnMPqo2dAQvHdBjA7EdaXN3tLO4r6/iKOhYMCL1bf0bIOYHaDSgkJ975JKvV7EyqrKEMx5QkHM2OykY8PuQpd3RdmE0itPeoboptXv+Wf7/DE94K7WsWSsVaXVgVocB4VQCBxLyc+6ho+xosLxoja8SEOljntDw7HWPJTzSNCMYFkzvZ8kLJjfZ3H7YeMqArIPpm/ZhyGG7RczCQfU0RXLv+auFbRbl5YpAdnvYrYEIx9V2P6SHqQM6xz3b/y3j4wsbyvfmjIyMjKed2R3rA8B0f1HClQH5JIb7ZJnv0NAislrdbiZl1JMHHM+d/Ib699669dymcIVeMyGDa0ndk441MrPIGsVO6a3Iegv8V3VHUqHBVJNzC5sxPdDRy2T4UMkEWFLT0Ph8yiikUz0hKDYv41TWjbBhWsrfSQ1q1hcI6iWITMiNFrSSj+xVz2THedsqaWMxGc0PRDOA/mwXiWtDFGc3geyko4xh3CkKz6ih1TquBMcsopAes7o+airKMW0TXBcFJQO/nS35R4VB0Gj8YKWlCK0qjwadvzQPeaM7URQf9NSrMtgwa+5TC05bqWaEtNgfPbky+tehw9UmrWPfFzm4LW0nAX1Rs6WSssMBWPZZ2pUsEoaFsJ4fbFyLgvmjVjPcR0dh+1HGWjHWNYlUbVS4HVXdSDgNm6tJ48vcZzuYzoOMzIyMjJuH/J00zUhIkgyw5r+ndqRijiKhUzIEtLymrnrDHir2F4HKilIy3RyWdbVOFOvXNiw4xHnPJTNpEt5gXeqigQgBEQWYDVacFcbTKRuug+z4x2AHZ0PCEOfEHPf6pLgyjIfA8pD2bCVsamhLASgrfRRAJ52brcMx4Byxo4dXSytsqDO9sfes2EXtjydAbfHEj5/owUvUFOLeB2JK/zzOA4Lx1aVboBOlc0w8INdx/thXwAaET7alNwtHXfLgkN8T5IqsZGFyxvq3QTjN8FRYFnE6cy8uWxZ+ZOJw/3nfvLv0NL7rnLsuuxz2PvnsFK9ObogSrdzvWEXMmI+M7aljeV5V1ndjiL1qwXnqVmD10IVrLTiMPk5puGuNhxpzct6yD1dcYzvUVMEclIm2d6lCZmMjIyMjNuDnAm5BLJQ1mF9QlQ1vj51A3KhbOd6MH3IIG4SjFnpwo4WVaWRmhrnQ89r2gTfZijKRltgPI+xxCpkQqwXhzULTI0EBoiCdKcJUbGgNqxPQkbClx1Zc7ig89CxUZzKSE48ASpi2U0sDRNPTAbRxabhVtp0zo42KTFLZ5Sb8JVuA0mBjoKCjj72QRmPkc+UPHRb7gwVO7X1QC3eQWw3KL0qZ8MQe4T8mJ0vI0QoRTgfBrqdcq8qOO96Gi040pr3LIPE2JHbvhdpACqhcOwqjEG7u+L5Pp7LJTG8kREjFPsMHz578uW1vXYT0mJj5arg3kiCPy7jCV/SzmjIro0WvVNjC9vWjm5ioHAZ0omPfmnAcTEbA76pppk7FDhWQe+UljyKev1Vj6MVTz5qrfG6pZb0fM/7i2RkZGRk3A5kd6wEn7j/hRvPjtpMLPibcCm+pKYNbldzzMthJuuSYvGGLIjvkCwVysCZbtnqjt9750u5LGsBv/LgzbUgVFJQ48uw7ukhxzTANHCzDuUW3A0wCSAV5SiUvKSEIQ3yrHGhuVelegjDI9kyoLFz+ViSQnzvVvrottXh9SjmluUQ3/tDen7Eo+iklW7D4TvDg2ld+tjUzv6uKWPvkSNW/PxwlwFlpSW/IHfiZ29E2KrigDP1AeORFIFgCQ+14xfqhlKETe+Py8oJj3vlf9M/Z0PH+3LOLrhnzQmXD3z7cB6u15zuOjqBJnzi9NhsYr/6D0bepyR1JFeXkY356/O/LbBPxej7rhH2+U23ZK/1Ieth48/GwJIVr607zRqdB1IJTMrn0u0XOA5poitcmqVrZj1vxm1LMEDo2dGxoqKl55Fs2dJOyrKmQne/7i++9ffz9S0jIyPjOUbOhNwANqua/p67YF0V6FxWfnDZa22w/m3Ey1KReu+ytx0dPY1UoUdFEbMLzVDE/hheYOsgCZaGEIBZ53ELGMdgbwywzN7WwjwjHxLekzaV8yVZHXe0ieYDANUsk7FS3yldFDrpJpmOIpROnYUCK0nI6jjmhA0tFcWkzGvAl5nt6NjRYSYI5uD1WFoeui3oHQRhFUqXCvVrPpSCR9rxWHsacXy0Lul3yt1a6NVv99124EddR4HwbvGYjiF2nifunc+e2H6lAfKTCJHTgHrUUpWsqKMZQRs+85OqUVIdxhJuQkDS5+eZijSLk05EFBSTz+h/u0ji7H02rryTW7lX/5GuB4glVraeucmBfcaUjFQUMTtYzsawLWMmDgClOkpqVpScilJriYhf5xnbyT76sZ4bFmZkZGTcBmQSEnCdLIjVnqe/5+i1v7YmZLJuGWcWl6BxZrunlpJOPzyR7/OGjh40zOqKD4q8SPoA64Q+zyDsQtBT4VBVevF6i0qL2EvBpLWC4P+bXS8U2Oz+ELQQLjQDHDgX/86JVbC6EPzBRnoKIyDAFtOdjIHdjo4zaTkPQVvq+JWSHnvNnvdlZ9NAtkpmsh9Ly5ns6FCqUKb0Ql3wfut7VxRAKcJpLxxJQSlC4eDYFZzulPPeMkOwZeCP3PtR+L9kwLBvxv1JYbP/XiYtNFSYWsQfgwrYTI7TdWDH57pZmjnSLuhpBsWIrA+2x+vEnBxYZsNydEPYj56pyNxycF5vMTW3mCMtv7J9GGZkMCUkLdNsrm+SKJPja8dpSM5no94UwZ6TQPhtjDdSUKrP9iHTcZGRkZGRcXtwq0nIk5RfzZ135vAuSiMR6a9JFq5zE1aUTvswM+n4lQdvrh/reS7LmkF1YBBhYGBHS0ON2e1WWlAGwlepC8GShjKWgkYL3pctZ7R4Z6w21tlXmjalDHkOKWJn9JgV0dGV6Vxar0GRIQiKfSf1Q1YxYEwJSI/SiycK2yQIbBnwxU3DlYGmBbNGTOZ5gNQK+sfuPD5fIRwUQuWgdoIbvC3w+eCD8OOy4LjyJOTFxtEOcNb2/FB3rCjY0POunPlzcAkBSUt25pmM68LWdcwKwdHSLfYiucoWOF1+vuw0UzP9fs4/yxzznhnj+sa+NVNyM1ruTsu9bHyO+zTus4vif5/Zmk5+7CvrckhsyAkj8fDLpGYCJbvgMFcHcpd+LjM12MrYYNO0ToUKvdj59d81FaLhgn9euaMrOjEnrTaMW//3l05+d51LsjIyMjKeX9xad6ybEpB9zj5LGFBUBz6s2b3UX3/sSeHX3UjNp+5/cf3p+69lxywgPQ4ON+mq3QbBsmUwBojEwiFU6mL2wLQiW9qQ0eiTcpfpjLMg7OjZhWC/F08k0v4hvteIt+Y9l5aHsvUaD4Yo8m0ZYuf1eU+HM9lGMjHMxoJlHeYz3PZ6F36WHLK2gU45FXpVb7M7+AtDp77Dd+McP1fWHJXCnQbqApoS+kGpRfiI1BxLwc9Iw3/QvxzJgAXHcwKS7sd8dv6mqChpKL1eIZlTsWNiuhPbhzmZGH2apoQj3cdUf2Gf4br9OdJtABeOR4r5c6lewpfjjce1xHFITROaFy717UiPbTpevTlDH8bG9Pjs2y+HUKgvCay0iGQizcb5ZYTAwWPJXxWunanddBmeX1FyoCWVev1WCmXgzZPfyde1jIyMjOcUt5KEPEkGBK72/08hlzQj2xdwzQM0Wfg3MO2mXOG7qheh2/q1d/A5REpAfPDmLVqtpV8bCIFlLlrpQy8NTw68gLanCTX1YyvAgZ6eLjk/1kEdoA8WvRCIRHiXzazPnYd6Bt6Vcx5LSy/KWehyfiYtWzrfiDD0FLH9tm7n+zpPpyJfWyYtibL9NdG2feYS39/hiJpKhHZQnMBZP3CuviTrhdpxt/YCdIBNB04IrxUcOt8rpBbhbxZHYWbd/1tRY/bS6b6mGYhU5HxTTNar0+xEqjvxYbNL9uxiNmK+n9PtuBvt376gXiavXCRAtv/z96Ud6NNtpIH9/PqxzwDDxoiNTyOo86yOMpYtpoTbX4f8+NoxminYsoVKJPUlbswMJsdWAqG3ddr38iaTPRkZGRkZzzZuJQm5KZ5ENKs6xFKsVCNyWSCjOkTL3un27eY93Y8NO19uJNWNy1meN8wzQapDnAXvdWAXMhADXvcxyBiIOYSNdJzRsg22sm0QcPd4UblZ+abaCwu2rWyqQOhkoMdsgDUK48F/2XzjwzGYeygb7xYkvov6RrqYebF1PJbNBQ3L0jjSPQRkLNdxE/euHW3ct0Z9R/fNoLy/8+8UfHbkrPM/73cD/QDnbfj8DtpBfQmXCJUTftx3k22+rIe8pEe8FETv6f7b/ixlKC5D+tn39UAxLDVITLdvZxTm5VcXv/PXJSIu5gUulkjZ63OdydLn9/tYxH1Jz6nPrHWxXwx4DUeq45gfCyMa82V6hsXjJDhW1LGrepTOi5EXpZUh9v4wsuGzHI571DRa0IRjEDNjupyxAWi0fKLrbUZGRkbGs4dbrQm5CqmPf4rr9AC4UHKSEJGlUq0405hkUOaz3l6eOtaElxS02oLU9Nrfam3IN9958w0YyYiEfh69DjGr8ZgNOzmkUq8ROaPFensMIdjfUbINzd5gFFC74AiUlkABrKhGF6BwXrbB2crKXuw8nokvgtlKm+RRrNeCLys6p6PC61cE6+x+MUCcB95pWYxtz5fw+ONQhoDYngM/A76jRaXklB3/b3/OC1R8pDSbXxBg0w8cFo5KhD8+HXg89LzfOY4Kx25QXlo5dgOcDQMPafmP+4/yAznnsWxZBSckRfmYHnOeZHiMEJ2xjV22bxp+muZggEgs5xa4dkz8ORmzLv69Ixnw5GDMkCyRDv/+Pnm+WCAUl3+Ky3Qk83fOg3VNtjaE/ejCGGvpY3bBoAx0WE+YPhLruQtWz0DDNJNULnwOW3YQtSc4D8TdO6r57EyD72RfJPs/iunHrN00W+OPekURjSIyMjIyMn5y+Nb9b08mcFsZ441ffPvv/MRjyltHQm5SimXhm+FJ+gzMa973BSCqAyKXz7TGkgxroqh+ffuard1GGBn5zIP12po9NqHPh3WS3gSScCobzMbXB8R96DjdzYIjq2/37kadDAx4Ua4RhJ4hNGHzJVklEp2xLDA8mzhbdagMVJTUWjLgy1gaKdiGYLGimDhy7RNZp5kEC+ats7U9b8LiQn3plA9Ex87aZ9LyJ+4Rj4cV9/QulQiHoTnh3dKXY8kOznvlVHve145yEFYUHHWOzTB4TQnCISWlCn8GUbS8peclPeQ0FLm1DBxShexTSxcDe3OEut53bZ4VmcNm6VN75amdbDEpNbLMRR/2xwjcSEa9eDvd/mXUaZ/wfk43zLbX+tSMy81tmKfWubaNgT7qL9Jj0kYasuxIlpanmYbG9tlRTrbbykCh9onH8isbl9bgcwiZQEOPtwG2Xi1Dsu0lOHxfpB3++3RTgfpvP/jmGuCzb3/6jes8N38+IyMj43nH7wfyYZN4hifVZz4pbh0JMaSNv67CeFKCmxHuQlCx7z2XaUP2b2cZFnT0oWSr1fanPmCeJfT48rZWOg5pEBzvy5aHnHNPD2ioOGeHMvhmkKGZ2hCCHzvXdoyNgHgS4WfYd/RxtrcP5VODKJWWvC/nwbL3gDO2CSEtRh0KHcdywIYWB5zKjiY0FDyV3ST4XSKyc/Gxjc2UgFSUXogvGktpDGnWYEvHu+6cR/0R94qCLjQrfHFl4w4+duCoNg2nfc8f6zkPaWELL5QFZ91Ah/JnnIeAVrGuJmlGpw8kzeCJ0+iw5H/fjPRbBiQtSQOCrcBFpy6DuTn1gbj59/SRjHgReDEJ4NP9vC4um4RIX0sJ0/S1y9c99n4pMK2IZT/2lbvZ5xiNEbq4/IAw0FJF37aw7zJQqs8RgS/NKtUsoEek5GsrPaKjvsn0U9NeNslnVgnW2iNB/NLJ767heg0M7fynJMOw9NwHbWJp+M2Tb6x/9a1fzGQmIyPjqcY3H/yrtVn1m3mOwelPtxz2VnVMT7Mg8+7D9niphjtt1nUT7MtsTEohAqFIyUr6nqXuyjaj+PW338g3vEuQ6kQa8d3Sa/FB1V28VuE9OYu2tytqFOWUDeaOZX0/vIVvHRsBFnhBdyfDpHFhK/7crLTk+/KQniF2ik67eW9oWYXsxIqaY20ocfyZvM8hDYrykLMLZVZpd3cIrl4Tge/YG8Rq+a0kKw22bNa7Y+CAGkE41Iq/MbzAPSqOi4JHfc+Rc/z1uwWq8HADLxzAroPzDt5vBx71PbUI72nHI1q20nOsFY+kpVLHQ9mylY4tPXeoA/nquasNW/FBbysD7/F4sm/p731BYoHjHod7z/85u9CocCretqBcUe5ySMu0MWQXMlFpU8FNIKZzXEeLlZIA25d915P5eZpbDs+3nWbq0t82Rmz8AheIlG0rvcakZM73XSnjOLL1F+o1H3Z87LsgCHe1juuPGhQZ2IRsTCdTMppmiVLiOYjGsrH5Mb6MiPzmyTdGdzy9XGuUbgugCN/jm2RFfvvBN9dzYp+JSEZGxtOCbz74V2vw17f0WmVuhx2+gmF+rbR7VYHjb7/zt35i17RblQlJyYaRj2nd9ygivlj+MK1t/qAZiHF2fYSVZC3v+5SY3ETImwFbHTszV1LwiA13Qo8Jh05KsAp8d3EwwjcSVd+TO2gqTKArFwPLNs729pzSUVJGW1RvtttDsCS10p/3ZRuExn0kK1UyG1xOAs1hsr9+BjtoI7A+IaNjV9p52wI/SdZTBDLzc67hbBhnRlpVuh6aCg6Cg6oTOChh1wt9sDreak+N4z223KOmUF+q9T7TUqSW3tu84kBLWum5ozXvyUhCDGkWYB8ROWfHQSi3g6kj1G4hCzJdt//M6fk1jYj1zihxPOJ8cR22nsuyGzCSQ9N0pM/Nida+z7m0nXQboxPbxZzJZe9bauA4LQe76NxFyAZ6S2tfClaG3MiYFfQZECuts+9I6spl60szP0YIhkCC/PpGC+oBXSzP+srJ19eff+szb/zqW7/4xm89+JfrQXxG0sTyS4i6luRzfxjX1d88+ca6UJdLvDIyMp4Y37r/7fUn3/nbl15DvnX/2+smZOrHMld/z4xX7li9f/He0l16fxxNbL5z/w/Wf+ud//oncj27VZkQgE/ef3Xdhzr4OZGYzwimzy8JyS8jIvs6oC8FLJdlQ4xwpM5YcfYzLH/ECoA33/pcvuktIBWrmzailIIXOAL8OTHSYX+PQV1PrwOF+EzIioqGajFYmQdXj9mwYRffn2aygKhHsdn8UzaTdVjGxWaO0yxMWqZlKdV01jzNjthy6TbTPhpNIENHWvM3hhf4q8WKQuC9rudeUfDSylGIL8eqkpj0va3SDbAdBt4dOnb0nNPzSHaB+HjXsW3INKS6lhUVd7Xhx3LOoVb8W/fDcWZ64Tsy14lY8HxEE/cfiGVIguNx6JR+sQmhxAzSEauoPUi7owsulsU94vzSDu9z/ZCVwxnSkjmb2Z9fF9J9nJsLLL33cnJ1OdL3p8sv6aAOaSbvTcd9o770qw69PgocTbS/HrMYG+kmWY02nE2vHxndtpzKBWLg1GcX55/bIXzhrV96AzwBSQlEpcVknfuIxTyDkS6bEgjLrswzHFbaNV+PrQvgc2//3XxNzsjIuBHmYvE5HKNGz7IZjjEeSO9X8xYBQJigcXFdMM2Op9bs6T3iJ0FEbh0JsZKsfWVXhvmM5U1gZVijK4+dUG/bK+ImxGNJlD7PfMBo0ZuSEEGCiaajDI40b7z13145UL5z/w/WB1qyo+e//O5/9VzfKI2EFFJQRrtR4SU5DsGod8SqKGlDJsKyFaa/MRJyzMEFQa7ByptM/2DWp7HLvVw8pytqDqnD9padsJYsZA3zchxD6pKUitbNIcsuNG3IuvQMHGrFi3rAf1q+xGZQelUvSm9GTcigsAu72A7Kea886nv+hDNaGWi04IfujJWW8YK4oaOTgbOgewG4ow2HVGzpqCj4v9wPLmQFlixm53oMI3BpCZdpIdKyHzsO6QV2RRVLjWxi4ioSMg9orewpLblKt2HnqKSIpgEAOzrqsO15yViqOds3iZEG3enjm16rbCwskZpU6G7HKT32lfrM8SEVB1peyKoIEhpw9vH7ACS6qVGIbzdFexz3Qd3ErWUOGw/pMXez7McSCZlvx7YFywQkxa++9YtvpNqSJRKSrq+XIZdoZWRkXIlvPPj9tdnlG4xkwJQwTA1WXGy8DMRmyen9aYh3t/GeYdrM+TUy1aAa5veID4uQZBIScFka/sY3dhlnoRWNnYB3tLF3CCwTkMv2w0SnRZIxsaC6CEGTreP1t/7hhQGSsmvFC1JtcF+V9nuW8en7r61FXAgEx/N+T47ijLi5VO2k45QNmxA2ddqHLIbPolj2BIgkI0UbCcwQtTv9Qu+XUopJZ3H7gm/Y7f0cjqmJ65LA2lBTRlcnI9JFCITBB5AVLlrmKsqR1tzRmv+sfJnNoNytHE7g3ip8th66AX60GTjrvQj9THu2DPxAzuKs9s8ORwiwY+A86EF2YTvndDjgWFc0jJ3nUxJin22eWbLn0+XucRhn2Mfz0k80HGmQOgmi8V26G0o27CgoIhkB4vfpPR7vzTykBGd+nbC/j1hdICktXSS9Z+ziPl416bGYSZ2RnjnmGQTL1KT6j30lZWnWyJaxzJPpQ5pwZbImoGkWogu9dbZJW8WlLEUvQ/w7JQgpMbks+zzPBF6lB1kiDsXkJr7fCqCYCTcvIyHpZ8lEJCMjYx9+//631zaRlxIGK7dKJ+DSSacBjY1hJ/dB0XjVteVS10IgmpIskQ5ZuOX1ocS1j9dn+cCT2LeuK9RSh+J9N6ulE3MT2KxgtzCLKiGwvQ4BSTUgxUwzYpoF611hlqJzF5ivnHx93csQg8GtdLwvWzbSspFl0e3zgrE0zk2O3zlbHrPhIecxvLesQJoFSZHW3psbkXUzNzguCnpT2D5Y0GQdqy9DRRloxbQT91I38jHILheDU8FrMubZkoqCX9BjROBu5bhT+4aE563PgBTO/z4sHAqcqncGO6Tg5/UOP6uHHGrFz0rDy+J7sWwC6ah1PBYVxSRws1Id0wssBcXp50q/k+chgE9xmQZiKdvoxduWuSmira/NqKeajvk1wb6bl4nnGy1jgG43kDqU9FWUMdtwHQKiSSCeZmuNiO0rZZsj1aBcdqztdcssDck2DSYwh9Fjvg8ZEI37Nd3uPFuRlmLF8azT66JLnp+/fz7O51mOvwxkApKRkXEdmF1uFQw+0uuZEQ/LTFfhHlXiKFQiAUkJh0MQHXWk4ySUJx5R88kweR08+bDL5/y+V+jFe9R37v/BpaVjV+FWCdPhalHmh4XUYca7GvXxZlrKmLFI3Wz27VM5m+VOYb77VfK5Kkp6GSIROZMdDuHUZtmFMM8/ZkN++8E317dBSGnNCwcGOu0pKail9D1ExBM6K72x89DrAOIzWY8gul3NezKAD4asg/qAMuj+8ZXObuybybbA+JgVFSUdA4/ZxAtTqhNZmikusO4T49hv1F/IjrXhjHYiGt4y8Ge7lv/obo0qqEI7QFP6x048MVk5R6EVlQgP+46XCj87fqcveS8oMzoGjkNjyFYGHtLRUHCoVaxT9QROeUnvTIJW03NcBpvNn8NfkKcZkyWC75BgS8tkFsnIWydDPMbgz2eJC3bK07KwOV01nVa8CYhGm+Q0MyF4d7T+is+bZkqWsiZXkdh0Penv62RdfHtGS9/bOAu1xkK06B3ESq5cJETp+pdKoOx5QXDJS2kmIz3H6fO2/JMQjiX9yYclTDc8DUQoIyPj6cXvz7Qf6URbGhHYtX5ODszZ04Vr8/x+IAqljASlovBEQnzvLsuEmPNmicPmd0T36zPn+M79P1ibCci+zMg3Hvz+WMLKwN97+5NvwC0kIQa72fwkXKbm606JhXXyNmJhc8BLteY3ccGy4ATMWca72Hg7TR+o7pJu1RZoDxSxbOBrD765/gt59NwJ3O2L6UunfEBp/STsK/woCey7hETE9+qo9TCysgkzE0WYWTDyscN3RO/oLgTJgjCoUoiEs79sYGAwDcehNlQhuNvIDhcDZtu+xNIrg5X8mA+cbf+QmuPBu3wV4ntCEPb/j9z7/IfDCziBTe91IAJ0oeqsdL4sqxKhEkEEDrXgz/sdDjiSkocaunOIn9k5lZYzaWkC8bEysQ1dvAgeUtHr2LPjsWzi/tr5SlEneaGbjIEUlj2EMVtpz5sge2AU81vZk5V6zcuc0ucOtJqI/waGeP7mSAnkvCRtKUuxRCCWPt/S++fYlzWaP28TJrKH7E61TKkNb7LOJCgfknR+uo2lx+n7FrVyevOA3793lkG7wk3LML9eLxGa65C8jIyM24Nv3/9OzHY4hK30FyahDYUKQ1K1Uc1KQI141BQJwXCkTXnjtUeBsK6UxJSM2Y1WLlYgFMm1q99zfU2ft0unfU6HTHQj8/vRP3/wrfXfe/uTb9xaErKEfTf4m2DJYnfJ979jWWyZZkQMi042qjgZSzhsxhVGy1cfQFl9vQlFe9oQLAtTV6UBOKDhH5/8izXAI86fC0IyndHtZ68NXoquvpStEBfLsPxxnH5FFGWnVr7WozLtMwPQaQ/CYjlXkbimyeScTWvabZl4bkRjzJTqSAoKTzjDWmC0l7X9mY/llZbcpeb9kBlLgyiHsJIC09CbK9ag/ndd+LKs0vnMyADcLR3/Z3sGwJFWsbxMlEhANrQc6yrO3gCcypYX9QCHZeaGeBFdUcWMgz9P0+9ROTvm6TnYXmLNmx6PfR3oe4Y4w1ThU90xixEa6V0Gx3g8NfnummOU14KrrgAAIABJREFU3SCuKiO7sM6Fx9PXr7++dJl917o5yfHXkf5CmaF3ZlGqUJqoyf5ZpvayjPNVEy1WejXImAG6+P79GZbrPn9d+HGwvN/XzUhlZGTcDvybV/6XdbugGzPthwqLmeOl8uOla3Ua+xVcbHY74CdXLJPfxXumxRAyeb5QiYQiJTL7iIhh6f7z7fvfWfty6+l90fDPH3xr/dSSkE/ff209oPzeO1/6UIPg69x8fhqzV9YhGq7ep1REmqJM6snNfccvX9DSIbiQduvinL+GAMu/39cE1vh+DYJER6BKCw6kme/KM4l/+fb/+Man77+2HmSshUxn2HsdwvkYg/thNkOeOpOlDef840BadKyv7PQiyZzO4i70X5iFlsI0S7IN5S4W2MGUVFm5jBkYmORL8I0IKwp2dHxMD2JQ7AXHPZUKf224R0MRg+y68E5YtYOqgL7z+pDTNlivDkrB2OumUsdWes7p6IPA7izojSoKdtKhOo7jNjhmASE7Eo6feDOHVFw+D7zTrF9KSMyd7IPCGhhOSGD4vQquXsr+AH7DjhX1SO4WZstHLcy0ad8+7LtBDeHaUCVjasllLd2Gkd7rEJD5/k6XKf14URcbT6ZaDgvYl6x3r5O9sBFsBYPXzVbA5WThw8K8xGzpPGY9SEbG7cN3X/nDmBFIMxnptbXE0am3sy+Y6S10uvxyFn1aFryi4HGYZE61HPb+GHOGMq20bMuJxGur6Hgti+u4BhFJMS0Z1lhxMcdTKUz/1P0vxpP3yfuvrj95/9UPJHy5CT4oAdHZ7Pdy87BhstzSMiZaNTF1KUW05BV8Td8oUvIlRkX4Z/X4O9pIQCQw3VRAbX/vpKNjYMOODTt8nXpJQ8mXTn73p3bsf9JQHYMoSz2aXseLonu2anPyFh76f33418VHng5staXV3v8Ee999KIIJwb4xNs/SpMtt2PGI82gX24WfNvkBT1zS91k36xf1gJeHAz6iR3zE1RThwrhSH7o6hJ+TA/5aseLQOfrBEw8ncFhDXXpSsumV7TDQDsqfDhv+Ymj5/rCsZziVHbWWrNST5XM63yU9sVw9p+NU/JhLXZPSjOA+4f3SLL4nYS4SCPu5KWySYBRWj6TH94upJ8unrmswluYZLIuS7m9Pz442umMZ0s941fXIjoGZUtg1Yf6+lFDBzQJzK5Gz78uOjh3dJKvayzD2BhG9UHqVCudN1zEXl1+Fy5bf99oS0bmOFvCqfbPjt7T+9NgbAfnNk2+sl+x+MzIynj/84Sv/Jn7XSxx1mCSuGQ1K5j+2jP3YcvZ3PbvHpBUyDS4Izq3B61z/PDph2bpEp+Ww9njfpe+y2GVOkHR2z0zXMcdTmQlZ2tFP3n91/a13vvwTmVH6IMRjqcHg3Lc/veEN6NgjJAZY49/juoKTk/WpmA1AC4JNJFoFe1GYzoIqQ6xpt6Zx8wFzzo5DGh6zoQp8fEPHZsF56FmH6hBrLcfHwxhU7JlFjyL1MGstyAXSUEQyONZ5Ts/ptARrjn3HekA5W7DunZfsFQvj2KnE2ZJDShr1sx8VwhEVpTrOZIeiPNIOhpJD5zhr4dh5QXoRFMCFE1aF0BQF21452pUciIvi+1YGWrzKpVSfjyln4y0SJktBSx+DaOsJUYZZo5pynKlJvlvp8RzL0uyYuLC+KUHpk/fsw5xI+FI3iSTNUKmjovYld/TRUctKuOy5Sh1LaXg7X0uYE6zLNCFzmMuXhiyHhHUs9Ti56mYyf32ia0se2/JR+yKjgxZ7Pntq2nFdWEZk3+UoFbZflWHZJ5Cfr2/vxri8vCxFJh4ZGbcH333lD9eXxUyTzELApMx+z3V/39VoXN7D3wMcQ5hEs/LnsZzWLH6XMb93XmcSbP546og4jagWG9JeuoWnAKN5pcSmc0+K9P3zWdUlLOk75q/bDKPtZYqlG1WBo5Qy9hIp8b0KKkoaGTtHQCivCf0k5rPb9lz6vM9mjCUs5uozkqNxOMz3zUpLNrTsaHlXTic1+c86vvnOm5HAqo6ZKMuO7Asq0hlcI5BLgeHSeCrETX6uElHvW89liPsVLwADbaKJsC98oz6kPqfnffWz10eUvETDi3qA1Ye2OlA7YVDYdlCXSl16HYjhrFO2vXLPlRy5ghLhQEs+qivu6pghqHBeDyJd0Ap4l6xW/DjVZL/tdSB2yF5qKJp+bvttZYbxuC9cZtOsSGoYkWZaGqr4UyUEyI5xakULcEdrDmm4ozUrLbmjdSSaTWJJbJay87KdkeTc3OVqaXxY/5GCImZr/LXFE8I+uRndBJctn2ZI5t+h9Htj+2e/Td9x+XYv4jLiErPECxa+F9Z9wyzMh4FclpWR8fzi7Vf+1/V1NHZLWWpgcq20MmO7WykXM//pYytltutjhbtQ4pXeW/ZdeR2y2B9kvn/74CtLxrtxqg3sZdiTOX7KsEQ0nuTGedl6ryIeV5GPfevp8R3R7aZsjQld8s9KpmywTINUiSU7RbKsBU52HMylJy2vsBIdqw83WGmWBQv7BtGWljJoSc7YccaWDTtee+sfPDc3zm++8+Yb88AImJyzFEY60p/LOtsbLCCL5zj8uwoXE7T7x2ka+KXbmpfcDSHwB3jIju+7x/xFsIN9yZV8pKi4p94pq8HRiOOwlNig0Ak09UDh/OPzTjnvB7bqO6r/Rb/jnJ6PseKjruEwkOOt9GxCmV+Hd0tKL0DbWRfsHo1ZEDs37cxdbF+QO8eoiRkxL3GyiYP584tkMgTw6b5GkbhKXN8mNrrcxeXmje1SmN97HaYe5l3Wr4P5cum1oUkS3WlJ1dI69pWALYvAx3XaOGyln1g9z/fJLCAVpUvKtm5qkX6dK3NKRuZIXQf3vX4dXOZc+LxljzMyMq7G/H6U3oftdctyRPOW5LX5ckYk5ven+bYulvISS7PGCbTp8kZyYk+sZL377j1LepCbxOX77tdPXTlW2kV8CZ++/9o6ndW+Ka46aGkqbK7vmO/nOAs7Lm/7byGWJw9TgXFKFFIHpXSGNnVRSgPk0dmqimuy/TWxaTq7fN2GeLGsgoKt+hr9Suq9yz+r+L13vvSGaY6A6bmWKVGYn38jIOl5mpfwfFi4LvHeV98+T/H+uTuNwfRKSo6l4Kj0Y6PvfeDaiOOocBw3cGelFLtAbDph13mHrELgqHD0Cu92HW1I/x67ggE4xPcBGdAJ0SgYhecwDfZ6Bvrko877f+wrS7LPZr8tI3KdIHDpuJmlcbr+edmQBc+lOnp8pqnVgU4GHnEel9tKF7MhS4J0+17a9gY0frev+uy2//vms9Jzv8GX2l2nRGAfQRlmv/1nGMnv0o1r6dja+tO+Kwg4NYFlut39sNeush/YZ8Prrh4eGRkZGTfCUintvvJWe91sdZdKtIbkKppeyS5ea21ibbSySc1z0mt2+v7LCk6vc89dWtbHu3ZPHiEIpbo4UWU9QuApJCEOASn2BldIwWcerC9kS77+9hs3IiaS6ALSx+nvdDlDOhs+PxmF7bdMyz2WPqNbEBmlW7IuFgMO6yOdDj5zSEprDNN1FowkxoTp6fZT28x0H1dUqAzs9HKb02cZRkTmn28+o5BiTkCWZtvTMZuSyJ8GbLbZMmgwHVNb6XHBhrhnwIUx+rjzzhwvDweUIry8cqh6HchRo+y6MAsTPsZBJXQD/Gjbs6GnDaP0x4PvjL4JDklL+5ciPVY2Y6QMUcRsuYbUwcka5s1dqazm1ZotXeWOlZLIeL5wSe8cxXdlGWegbP483lwCsdjS0ccmfSM27OIy1jMkPs9YamYCRNN0FbiJucG+MWpELX19XoZmFgtLY3AfiRuSYzx/PV0uXaalo6GarN8IlT/OLulh08eeK+m+9ixrmi6DV3NdscxCZ3YYieG8TG6im7mmg1dGRkbGg+/+F29895U/XHfJNdeumw3WF06jQHwsoRp7clj2o90TfQnjtdvuULaeJr7XIj67xrpwz5Gg2ZRY/GzrEy7ea+aIInaZagfT2DTtCwLwrfvfXo+TRv7++em3PzFZRr73ve+9fumWf4owcrEkQr0u9pGRv/vgf4jEZU48TJ+R3oiNiBRS7D3gc8wJR1pXbu83FDNyM8+IWA+Q9DUnY8OwdF2pQ5Ztp6CIs8K27NjUppyUushkXSXvc8a5buNrX3v7i89NSdY+fOr+F9eTMpSQ0bLH4+yBXQBG/p6erz6Om9EuOYWV7lh5zD7YcvteS7edWuGZXgim9rWpUcKxNvznxcfYDcrZMMTg/qG2/HzRIAJ/8yNCUw9sd46yUIYBHm8d755BPyh/tNtySsdZKLkyIryliy5JqUFCz3CBnIyNAhMSknxf/AXbf5Z5aVaVHFcjH9bF3pyp9jWC2ldGk5oPHHMwOb7ptuwS31DyiPOY2ZifTzvPL+oRg/jzfRpK4YwwmhFEiWMbbj2XkRDw59D6qKTXppqSKth2A5yxZUcXj8Nl420I+2Prmm83vVaMpHCIf4/NHb3L3JJFsKGmQhliZ3prlnpTEuK392QwXUqhLrqyARfK5/o94npbbv76vtlDyJqQjIzbgj985d+s03ixwpc7w3jNamfxmZ/8KryWMtxp7DXrBmalVeDteBt8hUGHcjfcTfy91+5lGrfl+3FpmAoa12MZkXn52GWw0ixR/3hOPm6CpyoTslR/fFMi8pkH63VKRIzYTMpTEgKw5G7lmHarTLFEQGw/09m9pUBnH4EZkt4J88/baR/1In79vgmdbz5ojLqPJMTPErsLAV26/zbXvLSfLR27RNz+YfRceBZg/WhMO5Rmw+YEZH7MUsJoGYZ9kBBAPmb7Ye36ZN1W1jNe2MZ6/8msucBWlUqEF4qC97qeMzr+n37gr7gVXS/UA5SF+qaEOmZOdoNSh7FXqNCFmRHrCXIVzKJ1rgnw49l0VPvLe1Jydlk266YwIpGe5zbYMVumxbIPS6WVKSFMRdht6Ixr58W+UwPKNpAEO3Jzi+f0vNlj+/wSjlhKjFNy5knoaGxhWZh911RN1p2WxM0nUpauY7bMlqvHgB0HL6KvL2RFPmwsZUysVGtJv3KTfblutiQTkIyM24O5vq7FVxw0FNQJsdiFK5Pv1+biRIyJykvG2MIbj4zEwrIbA46B4f9n711jbEuu87BvVe19zunue2eGQ+oBKwpg5EeQGAES6EeQ5E8QID8CW3ncuZewI8sRJAiWBSOKiEiCaQqtQSQxMRHRMQjHAkFGCiEH0DyoBynFECAEMAQYAhwFhpWfCSI5CmlTQ87ce7vPOXvvWvmx6qtau84+3X3vvHrIuxqNPn32q3bt2lXrW+tba+ESUwE7PVCAiO3DWOI6f3u61zEAckxnFbVUviqH3o8nlVsFQijXWSqvkyW6lpebUGRaft4xaRXTanEOBwrRVeepyuucchWFGX1qBWxxigIVzGN9s8QzJJ1iSZEYC23CZMKEv3b/Z87/x1d/5ltiEf3saz/78g+/9IlzD04PQMeCh2LJUnwMBA6YjirLLdXlOlna7yolkfLH+z16BJyFgEHNOvKdssGlJuw1YT9GdDEgBgMhQcx5G0UwasKdEPGNZJPqLjt3r1LI2J61ViV5wry6K/fzSncd80wxuOxNIRjgvUf3/vnn1/ZX+35M4DOyLFME5NPCMdw2M164v5RL7NGjgyAgOZpSpaNNi+8j76ttb6UEGlDoc+KDFoCYBa4rwGKDVfFStP1YqViVPkDQtWSwaNu1FM+yJJ7O6GWShHBFEP8xuQkli/thYd9ZxrKnyJh1VWwO8Ax8PJNn8q0kr9/71XOCDTIEhuZvyGDjBBEDyESwOZeUqj6vjTXLVKXfc8ZRGNDgOaYyW5uRsNJl6xra53b4+ZsxJccMeku6Cqu8vxNya7Jj/bX7P1OoWF7Ja2kv/offvZ8yCybPsGBJSfX7L1nSl7bXO6XlsxZiY2YdT8USzCk8VymiXnEKpe1m7Y2IJWMXz/vX7/83VwK7byb57Gs/+3J1gy4/Sx90Nmkqv1VhO6xGTcV4ewNr8bFxfQzIts+z/b4NBP+/5CG+ii2CCIakeKGP+M5Vj+/oerylI97cAg8vBY+2oXhBgpgHBQDeTOPBZCbNdVvaWNSADXps0B/sW/cLZRzyt417AGrWN/8dr0mF3v+2fbX0y3bSA8IsVz6173VZrFoLGAHlZfZ8XWVpTwvb2vMB9vxGJKzRl1S8a+1qvEPuLw9ACMxW+ZjjXlK/OC0DtiWjx4SbVapnPA/nLnrFrsoi9l7J0tz8pOKf00++8iPPAMgzeSbfIvLr9145NzARcBcdTtGhR8ApugI6rIaHeSd6kTKLniDijkTcDRGbYEliVlmTbOP6eMyAVAAIUIEKv6M3ZevgidW8OpTrjJa0z9D7we1v1wsC3FJPCDPPXyd+wbjK2vlei4WTH6FzCeMGrvf2VGqHAYPqRuN1fJE2Kfx/uO3AfIAt7cP4ERZdo4JAS2+XQcm3kvy9V19+2ccRefHP9++8+onZS/hf3v/ZcxFBhCnUPimA8f7Hg/MtyU28dZRjCr21VQ+oNUad2mOQCX9WT7EJAS+sBUMCHo+W4eqtvdn0VxG4e5ItKhPwaEy41IRHGBczrl3lDekzCFtrxCAV5EUcVhg/Zplp789b1s2TMS0q7q1X5CowR2+VBzPtMVNzrmM0p/niUWMofP70dv8l71BL9bKiVM7zkSuye/EApP2+zUDm+3apj1saWr0nhjdeLzxPbNod9boKOijtWgZPTx8b8m7IM/DxTJ7J25Pfvfdr5wHAv//6f/qBeZdexAoXmLLPW3IAesIpIrawmAzSrdZSwUiEYIOI52KEKnCREnZaiyKbB8TWWg842rWid7PoACtIHNz/yEcyutLPpEt0LD/fvpv5OW4FCGkD0luhd8Qrf75atbdg+YX6prSWp93PBoivyWDtu4rywerSV8lSIKl3jbVA4qYKqz9u6TyKhDNskJzltmbx6fBTDz51foIVfuaVH/vATAxvR4IbR14Uis+8+tOLffB3Xv3Eyx978MlF8HIsWLf1GFzn7fB0HFpJWo/NkieG4sfV19KA57XDd4QOqw54cyf47s0Kbw0JUYBVNCoWMWgUmzSZd7wNNvf0lJgnYQu468Bg/F1uVq+1jTs5BCFX9ZMHB3NPSZx9R4f2VRS5pWvEK/rPeyzm7ThuAGnBjz/m2L5tu0j1OgApGfgxs9j8mGOAMM7G4xIIWKHP8Rvzdi55bK4S9uWIGm9Tk2QYHLopkHlSae+rpWV50PxOxKY8o189kw+y/M69Lx6sXVSc/4N3EBD8/ku/eQ7YezhkhbswC7LF/mkSVbxT8sV7v3rOmlf0+E9Q/CevPzjog39w7/VzwDwgHawg8GU2hlmMZsAGIWetqnOQQrHJ69VaAnoRQIBBBZMqtsUQBozFux1mgGHnjG4x07tsrp0wZIAD1Pl2xCFLAbiajlWSYul8/XwnvCDALcmO9SP3z889kOjAdGZUIA4VgsMAzjlFYo9hBkyWxFtRCWquAiMtCGkBR0slY1uWxFOd2v2JX1sPCPdfN9jRK0w+doTiPSLczhoi/G7KcQonWGGE1TwYc/lD4xJ2pfLyW7jAp1/5+LfEYsuxSblpbMxPPPhbV9LXfDYlb9E+Jn4ckRrUIcyerZeWxkgFq1M/woA7usZH9AT/+uoM3/0C8MZjwXMb4KsPFWcrwfMnis1K0XUJX3/Y4SsPFW+OE/5Et7gQ5oqac0+ZeSioYBSbFDfaYY8Jg9TMSdUroHhTtrP2L43hq8A235MWWPvMWzc5zzHxtnq+K+1kvmRNajN67VzSgKcVel3OsEFEQJepTN4D5YMPlxYYBtzb/aTynvusXT26XGhynCnox9L+HpMNVogIuGiSMSgUp1jb9d4mHeuYJ+TYWQkYBXJldqyZt8j1L/fjWAeeAZBn8sGVf3jv188DjLpTADrsPelRPbc9BKfB1qvvefUvPNV4/0cv/ea5NwZMqiUFfNIaeE0F+t97/T++8XV+996vnU9Q/Iev/2dP/S5+8d6vngcITovhTGeGn+99/X459z+49/p5WVszYNmhUqQEwCk6PC89Js0peiVgLYJBtcxDKwn48Mo8IQ/HhK9O+wJ+TvNaTsPfFlNDwVr2YjNw3RuuasxkXXv9NmC+jhGAJLy9DFhXya0AIYDFhJgikWbpZkeX+ek6UGEPOGLS5Ir6HXo5OGj6nHsAsIWVPzcRxoBMLpC7BUJX0UeCqyXS7u8VHqsf4BZJ1IrqPH7OaZ8HsE8uCHWVc/kThPD4zqFrnxJ1iwEjRpzk9JtbDAXgXeru4Hk8aa2WD5I8aXD+dSBknRU8n5HoKqFr1n9ulWAqjcDc6+VBSJ8Dw0M+z6mucIoeH9EN/t2PrPFoKzhZARd7YErAC6eKF+5MSAq89Tjif//agC0SHmLApfNe0MtBS49/i7xXcicjWLnbFjirNN7SsVoQckz8u8K0r168B/HYOVvaHM91jCTUpr6mHMswcoz2dFMP7JLUwPOIDVboNZRUy8fSynrAOpSZwVIptx5kpnomLZPpj63mx3jjdgssfa8pOPvSR7YwJms7IqJeFUl3k/5YlqvO6T1XBM1XeUKWwMrSdz/+6g9/086Dz+SbS/7hvV8v61Qngp3SWGPvzgSd0XwYy0DZBHtvd6o3AiX/KHtAkM9f/eYmg9bcnRGCIMC/89r1IOR/u/dr557iKpADz81v3Xvt/CqPxu/d+43zRxiLUt6jFtjdYZqtqd/7+v2Xf+vea+dkBXhQtcPkgtAFp4h4Xkzb7KT230Wy9fJCJ5xIwLf3HS4nS5v/dR2wRsAjjLjjPDI7pFlq3wlaUtTvXRupT5ICTfF9fYbO1mTM40oYUJ+g31og5Efvv3zeVhbvJGKv1XLITFE+L79fNEY1QOCrRXqPClAH6KmsC/VowIRL3eWAz2XOfrs4LVHE/DVbENKCD0oFE3Pw4GkhSwHlvQNmS+41CjMxWcaEruzHDDldtqv7YFbrp4RH2BVFZ0LCDiN2ul/sU/79ZgYiTyJXgRCf1ewm9KAl8OG3AzZm1uiOBgf78csMVbR4BwjuphX+7fULeOEEiHk4P9oBHzpV3DmdsN1FvPFI8H88uiwBcVuZZ/ka8tW94jdl4OHbSkV0nwH8JAaK6/s6p1JdJ3x3jPY1D3qvGajm56rlnFJ5326a7eNYu3zb5/sf98S+nfg1D0TOdAPNi46vVA8sx+kQEPhYD84TbNcGfal1tMNQCjgOOR7oWJyIl4iAdR5lnGsBFFCyQo81urcNQqw/5nKT8/kCmNdRsQg4PFjxXr/6nppwjUp4BkyeyaH833/pt89Peou1++5f+Y/e8/HhAQhgIGTUqtxSwQYqIPDfCQQbsRpZgyqGnPIdALapKsr0ZPzB/S+dAwY07HjTh2i0mtQAQsq0ny6nvt+qzRk9AqIYVQmg0TlgUK9EV6HnoFYKr9u9cu4Vdm+q8kHfA1KZHQIEL2KFIZ+7rffB9ZGe+QjBXfS4IxErkdLPCmCrFv8xQfFi6JEA7DWVjJWPMZbA9i0m7JCwc3Po5OZgv8ZV/dGC39l2RTVGbVALLvv78IY74O3XAblObh0I8QvaSjoMOs0U9xV6TJhwgnWujxHKgrjHcFAsbsgDuC4IqQCQM2zQacAb8ggX2GHUqdC8WuVgHvx7vIZIlDArPMhjW/BBZfKYtdUrnJ76wYHRZukBDoPOAeAiZ/gxq2OPHjX1ZrVuzq3pdv8JD7HFXWyww4g9RlM+dMKAcVaIkUCM9I5vlXS+18kxINJmc1qS6jY9fKZtvAKL3V0lHsgwFqNHRK/2/DsIvl1P8G+cnmLdAWdrxTAJnj+dsB8DvvFY8M8vE/7ZuMMWEzaI+DozPkn1hIzNe7PPlC1/X1T4BqSZJZnKsk9hfJ33Yt5XcbG/qCy3YN6/Ty0IacFEmx77KiCyfPycttl+d1Pxi53vy9Os6lOZp5U/iaLXUGJ3GLx+IbvSzxxPbYYrgtQNVgUkCswTelMQAqCcg8Ln/BCXWOUqJp6SVcHo1UDC98O8j7j9evFP8boAd47VpYKGBCF+iwchwDMg8kyqfOWv/Pb5fjLlcMwD5M/+L+8dEPm9e79xzvemk6qwEgQAjP8zGTMoWYlgr1Xh3zi9ZtuAEEovltodWIjJyteZMohpQQiA4p0xL0ylwI9q+9B7wvP6eXXEIR01uPeUaW8JPjw48Yo5jyaN6Xn02EjAI53wOM+GPcIMtOwxYYVYAs97BJxKhEKx04RTidipXe3CgY2YgdlDNePdmXTY6oSL4rn2BuAKMFpaFenaBCgJilXW/7jfKreLIAcA7r3+0fd0nnpfAtOZ7pXUKQ6CVvY6wseKFA8JOqwzVaDy0c2qysJ+lF5iUeYnNWWZygozzBQlRkLxV3kF2659uKQdWwQPYkcc9Yr0m2NWV28NbgOz7LgaiORFUd1ylBGHaTNH90r58yxZAld5eBDwXGqlcFH49BLSwfHPZFnoybvOgnyMyvekcQ0RYdEa7pXhnST8C2zxf14EvCAd/hXp8PxpQt8njJMgBsFZF9CNxic9RcSbsHzogybsckE+Zt+YX6MNlq7xI75d/Dv3Js4pUnbOtmbOcoFD3wZ7l6rFaolD2x6/BISOxVtddf3aDyZ8Rzln3RSMtCCJfRkgpUhgEsWJ2mI25CUZqP3sxxBQx5kHae0+QQW9zOOWbgpAgEMvlAHQMfeBVUmxOdlKYJriwPucg4PbmKOvzUrmx5b3SX36/mfPAQMtz+JHvnWEQdibEPChVUCfX6UowOPBlO+7fcAffvTL53/uV//8uz4ufu/eb5wDBj6SwrI5iSCpooNgFC2mSaaRVTWwEESwgs0qW00I2dibtGbqBDDzmlgWqJyUx4GWpFquC2RAoHVua+dGBd//PEPlU0XIbM3hnGGUezOwjVl519KeANLBvFLPeTO4/9s53utlHWoJD+beAAAgAElEQVRcRYBPG2/77DHhDH0mvE7o8lyxQ8KkRoWKeR5/hBE9AnoVrCUUwOMpU0uUX0+n94CL+qD3dB8abDR7ecJ7Dj4o7zkIIQAJkFmmKC58xxZlU95rc9nBzJnPGI8JEzY5uFqbTu+FiTylWPwvZJ+vO7eDVUXo+LLnrajlu3xPbUpbr4SQyOXFAxq/iHnLKrn0HJiTu8clqzm9OrR21lew9iG53xPmFI6IiLW7rvfysBCjtxE8STaybxX51Cs/+fIxb8h1CusSADl2DCHg0jEMRO8RoKoH4wlS27LHhP9XHuErCFg/egFdDFitElarhDtJMEy1iuuAuUXeX9ODjk4DdCFgmgtDhEB0HsewFCcF1IKBrXJHL4j1hToLtM6UbIKkmyjOs3cah8keWmk9FNcJ22sZTcYDY8Gxd6nS8+Zzj9ElB3Sw/j5Vi/+i1b5HyFlo5tcgVWqf/SLVQ0uKUih0owIWMR9HbX+190DQFYunY379+TNWjJLQa53Nljwi7dh4WnmS9L6HYMOOXaLqXncNAhLgmYfkgy6//9Jvnp+GgK9PBqx9kbkeplACNg4UFmsHmMIdBRgUGCaFqlGW/q2nDPi+ifzB/S+d91LpVhDzXnQilYiuwDoEpEwZSjAwIsBMZ4tZh+M36xAKVYrApkf2VDjAQaNsyLSkznlJgtjczuMnRem/ck4CmWSzjwiw1uqFIMUSsP5n+6q3QzIoqO8qj2XQN803gjrX+Nl4hOJCa4A4gLKP31fFsn8BARMMaLBS+j6fnyCmZJyE4AwRnUSMUDxy15mvS0bZYgwHj/frjwcnS5RjzqF/wQXbv9fynhuWPvPqT78cEQ6yQ7GwVuvJaAv50WuyzcGS/L9mKOhwgjXuYI2THPQ4YcKIESv0OMMGp1iVh0JaiiId0Kj8Plf9euHw8wXO2u/tXo7TsCi8Nw82SKMakTJFaiq/FBZ58xlwGEPCY32fXqVgUVltPVIEQv77hPSMiuXkuuD0JWFQsLgfyrExtzSWmDWpzzB6jQ4v6AlO8ztRJ9b5xEUvxf8zXeKP30p481GHNAke7wQXucbiAMWbsNigS4wHaXrb+yHly98LLVH868VAypyyRqDB+I2Yf3j/dDnT+2eK/TQD6XzXb+qx4zFd/rREtWqfyU3mBrbnVNfYqBUPnBspDmv+8Nw0oCxlVOPcsMOAnUwHwK7SO+1ca/TYaI+1xpLBim2mkaZDqFQjtV/OR+19efFxFiv0dixMkQBgxRUhONUeJ2rtAJDpY3NzytL48Eartp/9dW4i7bWOLYrtGF7a/6pCllynWvGA5Jl8MOUizZU9CpV4AFgFwZSA7ajYT8CkddtbYyqxEvRUvJPyj+9/6ZwxGQKUWXAlAUEEXRD0wZT8kxCwCfUePJ2Ko5fAhCIwQLUKBhL6TOWKUovyJQc4/LkJcGK+Tod6jk0IeC5GnIVY2rUK8xVvyjSwTfYeWDtpHK3zNemStj6k4hmh0CvQggnTdUzG7JF4iAFbTIXuJW77IEbFolzKiB0mPM7HbPMMussr1B4Jlxixx4Q9EraY0GVgOKjiUT7OFwdeMsC0dbtULJ7D798e99HX/+LL75cHhPK+eLcJLGJ+AQg+CEyCzBcZwChYtPZTedrlLC02eOzzEtXCHP19XsAj7upJbUv+Lj6BcnKVkBJmn33qXCnfUVo++hKP/BhP3fjbk8Pt1UI5osa22P3XQGhPCaOicxPLLS21fiGWfIdUI58FpVe5DoBcRcdr5SovCI85UMzUvB8EHGuNeE4ti9JaTYn1blweR7nEiLemCY93wDjZ4imCHJpM1/ihsk0vh59YehynnpVzaP0FKohvz+3fbW6vPjm7l7GM1ZzBCIfGhaU+vqnwPVuiYl0FUNprxNzCXmNR2IFKrfRGjLb96/Jk53MdvQuUSVK2sqWcC7CCjFPtZ5lviscCoaQB99eIkFKLhGPSA+Yl6RDKHn6MRkiOBqn0hg5yZYB6O879Pbfj5Lp35rCdh+BiuQ3L2+fJGFIBhK0cK+T5TD548vsv/eY5qVa0mJOGRPFU85i/ntQUclWjORGM8BewbE/vZDvZRgFywDdmmZpIt+ozIEmgHlaleAYKvdw+x6wwl/dT7Nw+6BzuuOB+O7H9uS/ydyVORYBNqO0Mec5cBcE6hAJcJmgBSsjX9N6Dzs0HpG6xbgc1Pz9feA8I6Vr7rG15+pZfj6aif6VZVqkAwSD2nQcng1Tw4fcHDNQS2DJOhW0Z8l4ESR5o+TYd84pQ/uLrf+lW6GvvS0xIXZAsmDnInM4TEQAxR7cfGOTS65HFxSgXwEXO6sRgSm/5TVCsEaHZ6uDrFgSXeeFJ7sXLYQC6VYRmOt829We7eLbABDC6zVTONa9+Xa+jRdmqHqJ6LU6G5AiahbKbXY/7HlP8VugBAUaMGHQq1lKLrbkVdS9vhbQAxCtLfHatpbsdR+U9aM5R96/Paq6sS7Fcr9BhrRE9Iu7oquyfAJzpyiwrMhQQYNvrM/+KbvEvTT2SCmKwhWrIyuxQri3oFNi5TFl1JOnBWOK7m5r7a5c7H8NgE3xy5619ZPdD5Znph41a0CGg14hBpoOYGD/XtLEQS9RCuw/zmPr6LK0sWaj8/3bfEavsCfCxGWwZ39WIDmkhW9+ECWs1GDLmzGKMsWCQd1vHQnTOZe41zg0RCpxijUsxd1dUMwoNsOjZdfZmbfN1IgJOsc7K9pgLG07Y59gUjrM+Z77yfcExsNEeIzSDVD7r4xSrq2hPfmzMjSS136+SpX3bINrrpMtjhu9kWwwyQRD0yQDvM7l94utcADWwGrBgaaCOnR7BLPQiGBTo89gy4MHPRldaLmX7ZPJH3/fbZe3ZjorHk8vq1HguKL0DBxKsbYVWlUECYHNEDWKv27zy36pPFuehJdiawv9F6jG9yxolYtfgPMJjqLusg1G1eM6LlCDuveUxAhY/PNSz4mxdqVm/yB7wqWt5HNe8ddaY2nllZoxqgEiC5kxiLsawmQq47xvYQwCsEbFGxIWrL+XBRRsPsiRLFP+Pvv4XbwUAAd4nEBIdKODD94uw7UPMa8JO9FZNAg3SqQA+oErD8IrOHkNROiwrQMQ3cIkL7LHT/RPdA5WUli7m28pig+K6mUCppUD5Y2yAxdn/3qq6P5JGmGl3jULFrGCmiPAYy1TTO4WtApWlQWxWBNt3lVPAjhhzfE0GVk8I3L6Z5acefOr8mKX9qgxWcxd+Vb6PiafjMO6DWXpiVoI6NYX0VCNOEfEN7PN7ZcHLnSSoev5sjh/ISuMJImIALnaCiz2wnSxAbnTvbQG1Whe5aim393iHqaYwlfoOM/WzV/rq4mEKW5l4pVKJWkqL7wsP8kgDW6m9fyEfRjDis5QdxrUcF9ZjOVZQkYBecWgx8xLLHSs2WJW+SBjL8+/RFYDB+xuRsJMRL+pprok04VFzeoLJMR81SUKXn0EHmc0g9FYESInP6WCUN9Ix+xxnd6o9dhm4nmhncT1ima06BOxlKADlIS4P+gd50Z8ydGy9REvA2y+4Sx5jSjv/chyyP24CRNjLbV/WwHg5eq72+q0QiDCY9TYH23+rC4v3eQU7ALM0tPQE9BmE9CLYMSaUc6rzLoxJse5CLloHjFOlwCqq4v8kxfkoBB+COnpFaiwH6VDF4+AzjuYBSKW+xt5hDjKCfWZ8eYCgC/X9GBPKvTF2JGSvRnQB6BSedszt4jmn7CWya1eQEgUlc9eoWt4hziN7TXntqwbnPVi7iol+QgErbVYoCtetHpjBEBY46LOh52KW5ueQ2eIlqq19CTpLoV4Akcps3rrMMzSzZvlztt6Npet6UDVJBnaKWwU+KO8LCPnUKz/58ice/ML5DiMYXVD53YIxf+/Fo7n6MOxhMsIjgAHbtaKxgtkL5t4Af942KPtJxFD+4TJSB8qEM2ywLU60+fbWjdahw4gRrcdkqT+8cEmbMH8Z2j4T1CDew+NjVlwY9E9KWT3fPl9nlYk5b6fvvhmltYRXOmGYKbdLKVuf1kI6SipVs6m8rnO6P46BRxihYpNhhLmHo+bFATbxEnys8vj4jrDGpgPe2hoAmZQxSlUR3MF5QMrqV+8rQQ+UXmApTbGWvxPq8UBOgTpT8OY1Lfg9jQ6H9Kt5AHEgr2C2z9V1QupzjVfuRwByoj0uZUDAPIc8/+5lLN6FABSF3uhT82xfS20bMOICQ6G6rdEVb5ZRppD7tCoVCSw8JrDZM4Ge4llmmwJWaurcXmPpszu6wWUGG0BCp3E2v6zRI2pAL/T4zMVn5zs25o+BNmYHvAmF1O9XVbGnEyo6x4HK3Nu11P4fywHo/0OOAQnN98/k/ZHfzdSnOxJn6V6DHFrVVyKFMbEO9CIejgVvYN2pYpwU/+ZCwPkffvTL50GB73nte9/WGODo7mMNfO+DYJ8MHA1JD1ga1taa+IG0qAjFpBUkBJBKlgG4zClnlU6mxYMRc1A5UEGMvz7HfhSBioEK7pdQrx0F5TyMmekF6KPgMq9JFZAomGmKMrq5BvBxtooe9KajXLdcH7XQ3x4+IRATD0xZd5zrcVTyf+Wlv38+u6YcGlWAvGZK9oh475hkw14GSTHrZQHWX6I5Nb7OM18FCCZRl3GsXuv7Xn/3an28HXlfjTBcAFvaAz0lS7UwSDPiL70h9BhMXinKg4ixEwCww4ivySNcYMDX5DEeY/tUba88z2pF9G30n7fZR1GPbZWqKgyC5X4EV1wAW09QCzSOLYDsT4I0io9b8W1fsjiaB6nDCj1OscLOZfW5iVLwrSCfeuUnZy96Ozb4Xfv/Aa1vBmTqmDBKXjzYjxmMgKxsao0HYbCc56r6onE+ZmOFiDN0eBErfHgd8aE7CUHMysWYEP4u3UsrNcDc1VlAKAUT7Z5qfAGAWRYihc7iRbjNK7LH3iXGXLR0GsbMLJ33mPh3zVMd/XMhAOkQsEGHU10Vb0Ir/v1iTMUaEWuNqDn85gsn9+UMeCH72XxAKyByXw9Sa1Tw/nxsl21XRA3YaJfrx1Swxu/YGlKMTnO9IbumG2eScIp1iTU5001R3pfk2Jhv3xn+PyEdfL/0nI79f91YvW4/eYJz/eirPzibBzzQ4Ocfe/WHX34GQG6XBFr4JXuUhR5d8ypQkbZg7apACRhTEfIx5gFJmdo0HGEL/Llf/fMvv51sWH/0fb99/iff/9vnqzydMnaPXoQWAAQAXZC8zT5r9mL3ORhd1VGPXMxGFAakoxyv2noJ6zvs+4dC5ZgelnUwLxEzYU2qGfTk2TUHZ3vaVgz2JqYMQCbUBABLc21tk8kIFjFMxTjl1wjGffhihXWuRjEnK6onoqU4tcX9yBYQtW3f99p//jIBBY+fRMv/gAGRBC3B8D7IvD33NcvXu1ps8O3K+0bkN6tczI3oDhYkT0dhDqY2FoLbazrO+bYlK/2ECW/gER7KtgCWNivWTaVasI+n8GQ7WpeZBxa0AtNyHBHK4A4gpcssk9tcfNDz39knTPtr2yb3vcwoK+wr30Z6Qbg/21uDfjWDwojT/HSsnd868rEHnzwnBe0mMTAtparNJsZ9Wqljw6ZNKvKMc5gpQ9nNy3MRHMRyTEIntaDniGSpEovlqAKQE3S4gw4vhB4v9rEEUr5wqjhd2YJw8Y0O39CxWIjYXqC+e72GMmG2wXCMzahWJ74Lldval7Z7K3aVBEvhaveZAwJdvAfrotT+XAYiS/3u78PPOa0yeyxtr7f7M/ZsaiqY2zsVsM8WLrZ5oz0GmdDliBuf3nhO9ZkbIxKBWh5bpHVUz48UEOGTXTBWw2h81VCB8hzi/L5zvRCBUbGWPBL07pjHue2z2q6w8L3v56UYjTpeUdrHuD9aCa8SnrNtc3vcTWJJrtrnr7/6QwVkHGvLM/Dx/gqrhVu8QB0DMSvPgAGNhKwsMzYiexRWoUy6QJor+TtVBJlTod5pE52nX0Ux8ES6Ev9XqR4MQItHQbXGgRBYSD6GHhC+EX3+UEsPmBciKQEI9S67R/NcVC+IPxZuH8D6ahWBUS34fJ8UA4B1BjwQ275LUgLnB1UMSYGczWs/zTPODWW+rnN+zPPR0DwFmk/Ne14zYnFNIghpWSvch0a91hNG4ZwU1Qw+13kiqgdO5h4SVI+H3VueI7M3ROhBkfk4+6AYht83EGJB5l0JNp9b9KWhTM0XM1KsEhTiLPuthR8LIITCYEoGUraBqK1w+0wBZGYJpMUH7hVJ0jLMMsog0q7cz1KlZ78g+/6grdRXPI75PDXg1pSBNr0uFQvPx/dUBa+IUeG2vk0FyNDaewdrAMAFtje2Mr6f8uMPfv4cAD79ysefSAH42INPnrdK6DHx4+SYpdaPo/a5e0/g3BsyB5LcV/IkRy8Dn+++GfulOCAUyABykjqx9RrwIlb4jr7H82vB17YJD0fg9HHAR54f0UfFn77V4SQEPJzMRSyuHYf3WF3YhbdbUr1Wr0gL0tgndqzO3v2ShlvN01AV2Gk2+5KGNlduvcXOpIUQtY6FzrJAtfMCDQtLxgdfvyjC2ukDx+u9pEzXkpytrHpCtjl2jV7e9tr8NGUoEsCsZfW9Zj8OqMWw2mQH3gNGix/jQ3h9845wvq3zyKmuMIrFdVhqZEWndR73ufr9M2iph0ue2/YdaRMceLDNd+I6ihyF/TWjRCy8py3tauld9vvwMwHIM7m98jv3vni+zk+U42glRrUhcABQUqQKagD1JgaoAutgQdSsdh7AOAVWEZcCVmjh/8f3v3T+PW+zBsgffvTL53d79w5HxnsAXYCl/k1AiEAMRslCUqjSg2pUI5+Byq9AXQCS8bABmNdB1cV4uH1jYK0QWMKFkEHDCExT9XgE1HoiCTX2JDkwksT2TUmRctwJj48CPJ7SLIOWHWcV3Ac1gxVrT3nDkSVSsffTDBYo85P9oqyqxQPhZnzqmR5sqKCkyw2Qo+Di+1/7vhs96zbRScKcOmfrUKU7R3q1pbbH7wsYM+IqffY2yftCx/rpB58+56Cgld3TD3zq2GqRdxY8+EDUZQ/ETWIVZpSqBb6kuB/WLwnZJevrmVxnWfNt5MDtHP5rF0FbgFnZvS6yBAZegfUAjp/5Q043AOxzdoWIOLPim6IRD5RhUsbae5scJ5tghP1zm4UApP18nfzXD/7bbHGqCpsff20aVfb/knXV/1Srcz3OwGFXPi8Jt5mFxR2vFvBKYDlKwiipTEbeKt3l59ZrsLiQHCeykYBNJ+gj8NVxwJvTiK9d2PPfjwHjlItLobHIuHYxW0ebidQs8YdAnv1BzyjHFhXp0JwjALmiRe3HNTqs8pvBbct9d7MJr9C1UL0LrQcruut4ShbBl88WttIOG6zKe9cm3OB90Gq2yaNgzYB6Nx74/vJcg0zw8S70DFSrWaaXYF5V2EvMiQ3ogbU4kGgZshAOkiBYwoMKJjiLR9dHS0aVJWUfOKR+HoIXN6cxPgU4yLp11XvTnqu1Kl61v1+bju0DPAMgHxQhCA2otShYC6MXwSYGnMZQArs3cT6Wo1NI6XUcnJK+V0UfqkcF4Bz59PJPH3z5/J8++PJ5zBQvnjOpgY7W28J9GNdC+hPbtE85liL/aj5PDAY8+lI3JJ8P9W9SYEiK3aQYJvP6rCKw6YA+2Dl4vZBnu87FHrKvDNxkL0r2rBigyhGqUr9H/l7cd1FQapuQPleC5OGzhSYw9mNeWd3F/+TvFIfzEcV796m3tvGHTyLFkK6HFNTiNc7bNHtFWrDi11mej23z/99meV80xx4R6+wH6PNy7uM/vGLiO/CYx+FpxVsnkyqk+SHg8HVNWmWkFXo/DpVQXzOEMR/HXf0+/35rBfTXOmZtbxdbHme5d1JRVpeOLxzFbBPgi+YLwe1ylu0OEVECVujxsQefvFa5/6kHn7oxAHi3pPWEHKvp8RMP/tZ5Cz7aQoIt8AM8B7X2/ZJnYwYo3NnbZzbbVw18sJ4Cf6kAUoH31MU2r3nrXi4ASAQPh4TtaOf8Bgb88/2IRxem9J2sFKcx4EQCNohYZzDT3u9Vf3vUFNO+P9iHtOQvKZU9rH6FLzjoAffa1RJqr7v8nhwXApFj4p9b+xyn7KNgv5+ixx1dYaPzCioeKJzpqtDROheHQcMDgAJMTrUvxf4AzIoSck71wIFza1sLg8+uFvbKiplGKxiGClTZToJJ3wcJBgw5p7NdBLtLhSevU/79mOfnXmOJR4o6vx++i21tmWPPjvuzXk77TNq+u+qcV60Hz+T2yO/c++L579z74jnHcMxeD3ImIixGYhMFmy4bVYLFLRCkBJjiS8UZMIV7FaTU34iNQdPHjTyJ/JMHXz7/g/tfOv8nD758jty2dRBMqRYpoCdmSjUgHagKvgDFg8BigiHfO2uUUNjsPlRKl/+e+6v7TahgaJjm5yvgATV1scWUSAFEivk8HEWwV7WYk+xFsrZIAS+kj0UhcwC2JoWAtdi62Ll1IMEzAeZr0uT0m1bB93+p7PsA8/aYpxUPLtrz+fohxSPj1k/SsZZAjJ+fb7O8L3Ssu3qCvZgiy87yWW3YmZ5aMGXLO7A8WN6uvF1L/qGVvHKjaTed3CDxQZbtAsf/j1G8biptfwqY+QpIWTkwWk5VWu048yLRU1VByDQ771KbCER+4ZW/8TJgXodJEzqxBAQr9Dh/8LfPV+jxFi7w373yE++69dB7Po4BEP5lYLkHJnymS9Zdnb3sh6CuBYJ+P56XXqclsFksy4Uq5MfaYdKG9vpM37uTqQQZ986CTDrCGgF/knaISRBlk61JCQ8x4Ctv9fiXO0EXFd92FnCy64AtcKmmaFs+9ep5LKlUtcZq+MmwR8Cg5qUhsBollX280temcvV1Pdj/vp9ID/L/V3rXkteSfXcodo6ahWvJaLD8PubUtpIArXELG/SuIm+9D1OwQyloZeAjYZKxbFcINljhFH32VsjMo1ljtmqNEEgt5krF24+RuUegxlycaodJatrISqurmWSAmk0tyDxNMs/nlfOl7HDtPMdr8F56zVFuktBrwKka6WwnE9aITR2O9t25el3wYJXtYzv85/k7dpx22waiP5PbLQMU0IQBTLkK9Dnb1aYzhfpOX2MbTjopyTlUadlXZ/G3c7aFWoF6zE2UwT+4/6Vz77Hws0uJz0ClYGmjxYvbxqEqAqxijqsImW4KzKqXMyaEoqigxjuCSDsr+6llTqy7CLZj9qwQNLj9kypivpCI1TIp2ccEWEPwUBPGZGBoSsDlpNilVN72XuwZvbWzY866gH1SBLX3dXTxEw+RimfEz2WCOu/s8nwaIDODnReCAR97JhD8wGt/+W2/96lpm58Xl+YblawPq3lnGEPi2Tw0qQP2vD97/5fPf/jV/+JWzlHvKQg5f/C3zyMCnscJVtohSshVz2tsAhdKChcl4GYUq/dClhROZsHhQm4xFLaILimNcs0iyessXfdp2uut9GNuW7UUWN+3mbdSntKOXZegpM3WlKD48Qc/f35IUzJgdop1ueb7KddVNaf4Z+0nCG+ltedu21rF2Pe9B6s+4HxJlsBogBUhtAlzwlq7ouhPsIDt0cVbrBDRa8BD2ZvXUeXg/PxrCrLgK8MeD3Os1ABz21/uA9a9LS6rCJyEgDEpxlyZnTEi2px7STgS2jHF/5kxr41X6kAPUHaL54xfIS/8S55DuOuQzrV0zevkSfi1ZX7Ifb3HVNI93tFVAfcXMpQ28zi+mwMmbLTLVYstbmSFOW2y14gkWsCAeb2m4vUYmnP7+/VjkR6oBEWnWrKyrBGP9p3nT68XlpH23b7qf57XB8XPtuVYkz4/eQZ6TrmPCRBvWo18aWzy/jjmlkxSTBHcyjMAcrvld+59cTbPLz1b0oD2CRgmwdnKFPm9e9yrCOxyvERSBoHn6FNn9T84t9br/sH9Lx1dcwQVdFiGKgAuDmJQ4Cxa/Mp2zO9xpmfRcyFS6VkEMiw0O+mc/jRCXcHBCkJI8RrVtqfmtfLejpDPNfHeXSwI53iriwNL0pH3T2JxKgymn5yXo5xb7P9HKWFSxSZk6nlu06aTQiEbU43l2GR6/aRWpJU5AmlqXVTsMaczXaVr8fubxnw8iQRITb/b6JgJmuM/aSBxoKOZ0ubrYMAPvPr9t3aOek9ByENcIiJiLyOewyme1w026HAhQ148AavHPc8mNeF4oC/w5N6QqzwN1x0DILPP9ci2UKx8FUAdgpbrAEW7/e0AkM5x7f15vJeD0lq0GcyWkIoC1CpkVAJIKNpn9SdwWhWjoG0y6/9MV9jKiB6xgBV6Tt4N+fQrH3+Z3hAPjtp6KQGCn3rwqfO23+v+lUq0RPmh+JSF7bk4djwF5Fj1dBYe9BlcekScqD3RCwE26Irm758r6Sp02c5pNbyf+h7sUPmzj3IuLSAHHSdgO1jLLgdgSMBzq4C0t5vdacJp7stdsfTn+3WTppeIUBaepb6aK4I5eFS7UpDRC59RmxCg9ZjQQDCXWp9kSYGw7GOhJHg45hVpv+N1JuTYnLzN0tfae+IXd7r5uQjRs7DJz3qdK49PYkAzSh6L6hXnuTfAj02CuQqYw8GcxPedBWB7WNaYav3zlkSb2wZJB96pY9Iab+YA59A7w36MuW1syyT0lsjs2e9Q004zeYm/7rwv5tJpsMryjQHhqnt4J6nBz+TdkyEDe2AOQAJqMcJeBOsQcJbdDScrxTAJVl1DdcrLowcWwHyuIU0I5RCOqauljUu1lOiCVTDdvg/mjVl3KN4EO27uxYihpupl1qxJgSm3vcvpr+jRKF4XqXEZvEemx6X45B7cnxm2ggJ7ek9s2rRMWagnZYarFcFPEKSUs1GpeTwIqoZJcZlBTS36WNu2jgByYP7FaH3FawAGKrs8FwAGuryhrN5TNagsfQ+gZKuivBMAhPNse60ScJ7XJHqGuZYX3ZKgcWEeukkl9dsi7ykIGXXCiBkA/10AACAASURBVAlRAi6ww3fr86UD3xQmQ5t7QVg0D6hK4yGl4sliRap6c7VnpQUevDYXxjlQqko6F6mrrKfHBsZcSajt89fy/1/Xfk9toYJhnxP2SC4talcssW0xR/4cEw54Wk49fWnK51ujwx1sSrvWWZG6qQLzduXTr3z85Y89+OS5v4+2ICRlSZmr/egVsWXFN86+hzvXYU0ZYO4ho4JtcQ5S0pD62BNSVe5ghY1GcH5UmNV9yv1NSVCcag/AJjhPx5ryxOxdwXRJ+3n361vF2ST4ynbCWgR/5k7AKgZ8fSt4OE04RcQjrZCWVWhXiEU5ZFvqfVeKVsjWMS9moZ+n22U7hxz43SGUiuhbGeCDo4EapGfH1kwo7XWWqFrcFnNaYf7v/9pVDt8PhRZPjcVV2fYLGRA14DldYy3V0zBgwh1dleM5Gz6vm9k7vM2xa0GNQtpSpEIeN0Cm8EnOmOYyqHG/ci1JGBQW4K8RWxmxw4TnsSrWTALYlGlaEYK9TIX6leR4EcF2zmrBj/+u/Z5j9SQX39zDMu0MUgGurxpM8V4VPl+BlKKewGGRT7sv64+l+/irr/5AUTx+8f4vnR+jZz2T2yO/de+18za5BVABSESd5zZR8OKpUbG6CAgtzgHYDdnqTrd1AEQtRSzPB1QwEIuCmD0ceggyjkmUmkK3D2LB4mrnfjwoxNG1OPqKQUMz5YrAwp2TfzPrDCv3rtH7wc8xmLFpyKiE6YoZEwPQ65IzOalm33nW20SwjoLdlKnHGQgNqRqoJgVOzd2DKWkGLJz7gMdjwptpxIuxL/EsfbDCi5gE6w7Y9Nbu/ZSfT24zr8Mq6RaQPk/DS1HU+dYbbq6ay94toWGY45Rrsc/g9aRG9LcTPP9eyHsamO6pBxMSLnPGpud0nYNNa6A6AwZb66g1+jCotx0cVw0WOt2OSbFI5k+08Pvgeb9fCxB88HJrMb9pG4+9AC2v+ybnaQchY0OGkuTT+mTIz4NCBYqW1uuACNvCZ+jbdoJ1yWoD1IwVfHbMQvVuyi+88jde9spj66UA5mO0jgA56G8DCWF2n/zl9/zL768acxRmeSJdjlQZ3x5KXxSoej8qpkzS8yFq9BUGCItWi0+Cwo8c74r2T/oiJeyTYj8Be00YVPFob5P/nV7wQhetkjDqhNKjvics0uT7GWDsB2YKolcSgZq5ic+k1wj/bnnvXFvXwl+PgKaVwkfOyrsP9vfC9LPHxI8ZfvbgZOZ9lIRBppKgw4DmXKHtNczGUZ2TqhfLjwue29dlWbLSsk/a7yZnoKheHJ2loiwgVdKid2vpOsf67EovMAtIQkoa6TWCM3IINrlK+5BJGIMcvlt8f7oSCxVn7xezfLWAkm334gGIvz8A+Mz9z51/5v7n3vX565k8ufhkCvXdpl/Qnnaf0/PGAOxHYNUpxpzqtgafm0egC5jV0RBB+Z6/USwGo9bDqMltrlK4WACw1iep5+vNnoCTTko9D3o5GODtPRgt3iHlqo/MgIXiVZic92NSAxZTYipdRyHm9QK9LYpBgV3Skna3gCM1MNMHKW3xlc93eU2p3wHbVPWNSRWP0lT253Pog52zC+atisGeCzN2JdjfvSp2akQszgwEIEtzUgtAjulrATKjNL8bwtTq7Rzp6WLXARC/fYTi8y994dbOT++pJ4QT/B4D9hjwR+ENfJe+gA+lDT6cTvGmbJFEMeYq5h1ijjsIbuGsNKenjRFZWiy9tBls2tiGJWm51620C78Plm0VfHpRrqMRtAodwPS+8yJrVF4noFzTW4gVrMpJfn2cbfftj0fub6ldXqmdMGHIwf9bjLiQXVF6lrKJvRcSFp5ZbftcgfXej2PHWs+ZrMF4D+uFpeKGBeRoe15y4C1NqilZ1lcEFyErUp4akqBYZ+Xs0NtibvHObWnH1xzw132+lvZ4UXtsk+JSEzYS8HCwqf3DG+PfbpMU+s4OFk1kC30N9PVvHa/lE1L4PvDexOieCZABhwO0tFzzzjoNJdB9wIQRWlz1rejsnq/2iIRsMm37bEmJbaW1YF1gwAt6gnWhsU1GbVLJ3wVAE0apXOAOAQMz9cHobCxE6WVAwgo1o9YEy7Hfu/eaNNFSPFIUgyZEMdBTA+jn77P/HPMTGpBm37NvgvOe+P7ysvTe8zlEBKw1YgMDD55WMyC5lJkLBhtHPRQI1hnc9tk7BKC8P8eO5X5LAZ1/9dUfePnv3v/8Ob3Az9Lz3i75X++9dg7UOYVxCREyC8he5fSuBBJVwUahYu1HYJyyESQC02gAQJED1DUHB4t9PyWLH3nMzFXKNTeDg2bIlZgoZoxSAgbBKlbAkSbgrCdQmJ8jLkxvs0DzDEwYzM5gc2bQUtTMV0apyp5fIb3M2sN9GVsyJi0V1VO+10lrEH0UYK/1c1IDFQlA79pJSlcHwQ5WF2RCpWlZR9b+jlFw9zTh8WXAfjRARM/MoJoNKLWjh7wmEYByjps/h8OMrF5uGHL2ROINPLO5E/P4mKU23dQrclV69tsg76knhHKpO+x1xFu4wFflLexkwpn2VoBMBXewwSnW6NFhhQ6nWGHV1LZoZQk1Xietx4O/cy/LcU9Ge572u6X2+H15fv5d+vXeFQ9O+N2xWiErdCUGo29ydhxaQRPGbFEUCLY5dJZpfP09+aDrY7zp1qoAGN3kAjskKB7JtlzD7/NTDz51zt+jnf02Zf5s55ZzrwR7j5wHI/Qo+OPWeW+fGpX9s3Y1HaR5bmZVCcUyS+9Hubaa9XeDztKmZkXK0qEaZ99T5/rcKp6LvyfoiteDPPq2L/z//MxzTlD8C93jq2lXrnUxJbw1TdiOluHkLAruxojnpMMGluL1pNDy7P7oEaFCTWEgNS2VMXtEGBNTx/eyMCUxLehsP8EjgINn5vvMyxLNiu3yyvSSQu09oH4M8Po8JyUAONEOz+sKJ+gwImEntYo66WbVQyE41d4C1iHYoMNKu4P7SiVmIpQYlKCCO7rCWmOpC0Qvkz9ul3szwBZuBsqzsGFUKV6aXkMuZmjPjIUNOw0zD9a8ev31aXpJGyPtq4PgIQZsc9LwCTobxwBmz973PcfeRjtsMm2vjcFq6Trt3GbUq0P50Vd/8GWBPAMgt1B8XJ5XwjoRrCRgldPtT2BwuWWP6qJakb9s4xgnwThJoSsxUN17P4IYZWoV7ZeB0lqUbynKvLWpejxqulop2zad4LQTrCNKvMeUapuSVg8MpQVRBCBGgap0LQao09MBVFAxsXYIdAY+CEBWETjp675UlIekVnckKbb5+5LWt3kWNIQWD7FWYLQJFpcTILjQCev8nF7sIl7sI7og2OZGxwAMg11k05vn6bQTbILgNAachYgPhQ7Phw53pK6XfVn759rLMX1tSd5JRwj1LM8UEK1rot+P230K/mPna/WaANxab8h7GxPiPBfGXDd1K0GxldEqB0tEUqMQXciAHYwq5DuWVjh+BoDQoNqbCNWawwd2GFC8JDdBoX5fT9M45hZspc0Ys5Qmdkmsj6w6e0QNOOcgjlm5vEoEy4qfpwVdFczFfWutkXBQOfqYfOLBL5zzXl9+5b96xxb5Y6psS3XyQK99qb0QYFh/c9ELB2O19SxR1ll1ndNoDkGBV7o8tYfAgv9TwRJ3vDTn5Wcee2wcmCehTn4JilNE7FTxWEesEPCN/YS9Kj6UV8g7MWKcFKoEq8tnJ0ea90KFEflOEub1KzyBsgUKPq6IHhBuA8wSRBveocfn8LkueTgALFr2rbVatrexI/zMdnI/0qmY8elUO1xiLPSj8rwVWEuHQRJWGmsfaX52JTB87hUekLBBh4S5UWANu05CQgDjS6zdRoFygZoyL4ZlWbtSvVdUbxYzlvlU0v55LcX8HPS76z/KViaLZwG96OYxopfLvx8kllYgWr0pXmJONtBWo7ejD6mnAYJfvP9L50uUrKXvnsntkARggxrLUOdVAwaTKvzIWHfAfrR5YtUhW9htG+lFrShMIU46r9nh09JGwUxzZWpyKupU2gkK1rF+3mavC70YAsyyWC2FmbSpdVXt8qU4YD53lwGOvy9SsNaheniSGj1r7+8nV1LfBOBi0lJZflKreG7tsGxYY6ZJTWr7FdADxZS37RNpVVoAwt1gwCPle1nDiuVuOsGmA954FLDugU2vOFtJvm/BMFgbxwTs8vlOEXGRjRgAipflmIe+/Ywj+7wdqSu+HgCbGmNM1o+tit57exUrxVPwjxmLb4u8L3QsoAZ6RwS8KdsMQio+HWAxC63S2uZpLm7DGyrnwBL96lD5XFJIOWiv8nrMH7gpHoc847nyZXseKu/8/qbAw4tm1c28FzcPoLwqiInnoPX4mALn9w1lEbCFfZ+fp1dgfNva9LaApXZm2372lY+9K4v+0jOsnqgaYE/x9Cp6Now2Y3SPqowpT4qQM/B4+hWvx3PT2xEhWGnECTqbOGXe/x5YkCYXmifiPx9m4Fq+93afdtQEiMWFwILn3kj2PO+miEEVj9OEDoK70uGRWmrafR5TpGVx/GwdMAb85B5mE247Plrrvf/sz8W5YkmpLMcsvBfS3DmVGCq6QefXJv2LnwEDQ61CyxTKAsEdXeNUzdY1IGUKVoeLTOAjmI0QXOoICLCTCZsMRCIEW5nKvv6+Ux4v0Hk9Ip90gilto8rsfqvHdT4muODFTIta6rtDw80cON4UiAC5RpTUZ0+Qzuww/r5YAHNCckU8SWes7xbHA8/r7zvAFBMGq7ftvInB6JncHgnlrxSgoaCnKyu7uRDxlD+PiZmwBEF05gHROo3P0tmWauXq0vRmShJT4SJ7FQg0gmbqVm4XY0j2k9XQUJgCPaJSmKJkcJEPoofEV0a3a9fv/BDm6GVbCVBisKDnaXTzXPbqJJV8XusHXqOPQEjmYdl0tj4mAA/HXEdMFWOScm/rmPtWFb0I9hmArFDrsNCjQvC2kYBVqAHtlE20tmxH80ptR0saIGLtWUXgTKV4olJKhZ5l3l1eb3keIr3yOqrTr7z092dehQR94oxZJVGIhpJ8JkFzcUnOcUCC5KyQZoThmsJ6L0vGzTn1vyaAuY31Qt5TOtaoVjyrR4cTWeMOTjAhYY8JO4x4JHs8lB22MmIvY7YQW0YtnzGgBlPfTLFeogB4t7sP/Fw61ltFgzuuPY+nVvEvMPfcUNp0t+L25/etwkW5CbLNRAi0aWiXjl/itNPl5++PfUSX31X8dy9tH3oARrpTCZx2/beUvu4mVdlvIkugg/dHWo3fty3GRiGthRQioAIQ254VJQ2lZoenSiXUbBgr7YqC2SNm2hXPaW07UaM71bYyoFqKq3m5nXZH/PUiR37bbQDwCCMuMGKE4iEGPMaIPSZ0wQIL39QBXaYX9DBKluTrbzKNbI2IU3RYq93HWiPWaqmH+V31XszjcwAbo5Mk+21AM5VYRc6c5TxVS7JkdOD/Xdnmv2+MFlmJ7xz9KMKqe29ydfOVdoUCReX2FL1LG0oF3wKumbVulUHICtY3QI4dyXdzqh3WMJrRWruDwHyOW3pevCwZNgqNS6sCD9QUp+tMs7MClZOlyhXGlDlPDwiGDqlOV1WhZ99ESDZATTNKqNGq7B1aZTA3n0v4Xhktcq02zgAUxYIAxHtBGLxKIMmUyjexij6T2ye/fu+Vc64jAqCXgF5CnoeAfa5X0YtgFey3C6bQjply9Xgn2A45HiRTl0oweLDsWatsh+KITqoFgADVw+HT2hKMRP+bM2CZNV+xHSuQILVKHQgolci1ghEGjFMU1SPShepNIQCjB0Vg2cCGZMHfm05w0lt/3Fnb7wuniudP7PNJX2lfbMemq1XlE8z78HCasE0WGN7l1Lp3uoCzGMCMe3vV8u4FABdpwkO1+e1uZ+stY212k2I75UQCEXi0V1yOikeD4utbS5TyeFA8HqydDOifwPpX82yVfL+PvdVLRujr5Asv/cqibnLsey9+fhlhXm/OY8wQOYoV+CV4GbPuwHupDIm5kFIMzHWT2yLvmSfkh176m+dcpldicR59rmDBxSc56/2IhBPtIVKL6PmiM4Lri8oc91AcWkM96vXn9Bb7MaPQlorUUq2852MJgHA/H5RbQVIsWal4TqZp9cdeJ5XKcujZmHuSqreiDdL18SfWF/N9rkv9tvTyzq2PUgq3AdbHfPF4busjs4PSGvqxB588f6fqirQ0LAoDdyn0fAwYc4xNVTxL0CEY19COpar0UVH0Hj2CrpNyDaOZrNy4CVmBi7BMQbs8RqqnaW6FTs0YbD97ucriY31R6V1VuZTCzbcgaeRzAFvNXgERXKgW5dAMEGEGMEiV4b3Sys7nwszuZiyw8zOdIverVLccZI1aE6TXCHXzy7E+aLd5Dwa9mbUfbXHzHq2ZdwYZZOR728to76/W+9thxPOwlLwEIzEr+G0hxw29DwpcymiAXa3N6+wZ2WfaEq1ofrFh2860xx6TccyBGW0JuQYJYN4GVuM1kDzhrvboYQVM6dFKmT8+ii1yNX6mnpfxJa1HpPTVYpatKrwLZsYqAfPi57hqLOkQsked3lebNwZJJVV0DVKu15oWsmt5eUa7uv3ym/dePQfmoJ5iFdEjepFci0KB7AXhuxCgWOXAjfHIcGAlck/PKv7bTF1i/Qxri4mnXkHoccjre6jeFQZ/87ykXPmMVzEcBqInPbQoT2r37cEL28rzjckqnrOi+5TyXB9ruxmU30XFfhRcDJl+pordmONbmD530lx+Wkohx1EtZoRCD047DzPeCzCq1z4fR6qXJRSQEhS/S3aGVZ7QmEXrcjBQJwKsRaCwmky7YsjmvDH3EF0nV60dlC+89CvnzFrpMwj+0ktfOE8AfvC1w6KB5uWYJ2jxthqvCyZo0Ts097O1bT5vcm3mMaT1P3nQwrsv7wkI+ZH75wZASjCYUa1OsZ4VbNFsARQkTJiwk6qI0GIMVGrP0qBoFez2+2OyRLOi8uEVjRbYeABChdDupSp+Xvw5SPWxa1VLuwUDT7PvBPO4hLZ4GnBIufEAhEqjlxbtE2TQinQVyFjynszPNQdo9JxYn9Zja8VrmVHXKGv05dmH0t9Ph+Z9wPsSyPL345+Tvx7BCBVMvvze/V/pXNWlym1tWtVCKSleFEGf7/mOy2XFTB8bmNJ5Jwebb1ELyfFa3kLsr7H0v+/Jm8QJLZ13QMI3xqko2BfZmmXWcwNYa1QqlimTNgpOEDFAi9dhQvW42f6x9MsgFikSAUih7QRsZZ4fKkDQZc8Bs4rx+xYEt/c1f8dY7Op4Fe1aiOywj3lcrxEi9j89M8xUBVgmtWpwCDlDjI8Ds3QZAVICx/0d1zig7LHMGbbWGnEplc46izty3hGjAVQDzw4jolRPrUDK8+wRsMrjhBmqGHuxRB31vl07f+tJmvfnkpdXcz/vpBosvIeEQAcw8MQzEHTY+KieZQYp38SYs7QuPJPbKf4pERD3eRyvQx2JJyFg1AqYqdyvolQvAyqQAFAyX/GrpHPFtQAEMZDdZiMqa0BWzI0OZlZ91uWonoUcM0FviACjOuVdKwhpR2aQGn7CgG9PpZoSMgXN3pXtZJ6bKPNsVVMCdmOl5Nr1ZOZt6YPgcrA1h23t87kVmmNALAPZkOND/Oww5X0YlE9ZI2CbEi6SveO9BGyyRySptSsAeCONCAC+LfR4PJk63kfB46GmDB5zHEpy77xf4/xTaj2fx4xz7fdL+7WFBJc0KX+8b1OAo5+Wtce1TdS1nEh4rjslqQOUdcvYhs+99D+f/9Brf+XWGFXeNRDyQy/9zaLw9dIhirnP6f04y4XrvNSwSBt8F7BsPN5SDrQPvQ6WY1bt64TKCZVSKhz0fvC6tFhzQfPZq3iepbZ6LnaryiyTfOaghOfhd9Y/1pe+4N51tU+uS2jcBmG3lDFr/3zbMaDi+8WDOQ9uWj47Exd4qzSze60hGBxf/mmEVJk2JWzb3vaevfgYlpq9QmdKWEINEPfnXWs3G1tGazPF3KxHirt5KdjC4ik+3Hf4xhjwUO3evy30lm41u7j/eL93rav0v6U+Kovukf6r9KOrvIvI15gvgG+otWOC4jIHAI5QfFhWCHl1fT50uNMF7CbF16YBz2NVvDoXOT/bBMVGY1E6fTHGTgP2mLBCxD6PlUFS8aaMMFf1PDhamkVnPl94Y0F739672f7PfkwOZHoQv8WAXmLOTtUVr8FZLhopqF6RAAucDBA8hz5b++fWrRUieih2GVjshFZ984pwDFXvnLXWL4IcFwQdobm/kIEekPnbqDE5b8oee01gIoMEq83hPb1XU0erF6ndw0vrBQmQ+qyd17QVeuggE7ZNHOEErckE3OE+hafFktj/44JX5DZyqZ/JspRinc3Y6nMijCgCUft/00mhSAGVwmRAoXoj+mCKdgtGqhekXsfHjXlhFi2Lc8jW+rztcrBzsHYHaV/7qX7vQYmna3kRtX0VNTOW96Cspcav7CfFblKsoxUW7ELNmrWbtBQljALEjueooCHlPtymhMuxpsYIAB4pK5/HUodkr6mZUw/niwkJK+nw9TSih+A0RGxYoBAW7L51XpUdElSBN9KAF0Nfiivukxm59qp4lOeDmI07V80hFJXsTdDDhDDFwXFEDVGpIIJrCufyz7/0hXNvGON5LT7P0891dizPZfds90NWhR3PItTWuF5jTZKih+vabZrP3nEQ4sGHF4FghR5nYoljs8MOQOVvW4NWuMR+FpNAaQcDlYGnCdxe8npQCV6qXXDT8/jvDb9e7wBjMFT7ed6+ONu2ZKX3n5eu6z0nxwLhj90Plee2P66zJnrA5j1Hx7wQ9JpQBbvADsygxj54WqnXOwQI/vpXPXNLqWsLXR131RvRAuIIU57W2rlq0/X61TvCSQSZcmUFADdRcFcDhtGsz2ddwDqnSxymeQ5w0nkO7zu33YGUJWljL7xSTgoLzx7deVP5mzOkgAXmzPCwypWJV8HSKKoCmxRxGgJ2KWFSKbUmVhnYD0gIKsVTwlbxbwly11DoQ4MMs2rwFE7Ix95p3qf3knhp9/e0LH81quLtfqQH7TAZmEJACS4vwNuoAwKZWbAuYIH+vsCknwcYtyGwvjLv2GjKSAbGVOZVzALba8AgCSPm9Kn2ntv+uJQRUeXAylfoWU3fXUfzWxIPvDi38357xLIAe/He5xFXU6s8/ctn41pSGp/JB0e+5KhYheopns5KRZ6KdS0m6Ot7ABV4lL9alf7i+ZbqkfDZpRT0RkgJsmahQnHX6yRTvtwEImKB1bPA8XyML1BY9kfdhoU2T6nGg3A7kLN5TRkUiQWOA8gxIdzPepExKTHYelO8IcnOv7FUWdilhDEDj71731dhbnZlUpMEq9jOLGXI/RoRsFfzsnawZCchGdWqU8Ggteo61wyuS34WiyJ4nKasN5i+wbmWaedT1l68MbQYENUonDepi0Zp57tjutyx464SA7ZSsj9SrzJ6chMXuaCjHax573LBxSeRdxSEtADEK2SksQRUS/ScblQVYy6exyL/Ka0n4knkKkXcK8gB85oGHFTkHy8BDR7LgjjVszAPHPUeksPc9XVALy3y7Lslq7V9N/eOCEJ52P4eWlDSgr6EOR3Ny9IxfpunsGywwoAxt/v6NL18/gyB95STp33mzEAxp8OhnHeJTlKfb7Wu+4XORoCj0mnEINOMt7mGBZz7x9RrqOlYVZCkKt8CZkUyK89HNgGnwwrbyZTG5zfANqf56LNVbyqtnFu3gXkQcssdrd9L2c7/xX3Xxilw8rfPOjsHhYaEQRUnEoznm41Ad4JlVOnEgMedHHNAuQAQoXheelzolN83236ZY2ZKgHT2MrBAH+89QSvtSFGek7d0y8J9t/1y+O5VL+D8fdbZ6PHbfK0NSgcL1idF4EInrBDQiwAsrteMxx4BvQIXWXkexGJFWLcDsCxaKiiAl/06IqGT+fl8W3a5X4LWZAfq7j9AZjznMr9rVmicJwWoBoi2L68TegqpGIxIV+bm55hemtOvEsansFjjkniL7Wfv//I5QdCz2iC3Uzg/t3Ug2lS2DAgXoGSw8hQoylL1cV3Yz9flAEz532dFv8SDSA1GDxmEjKl6LnhuL70jRHiwE2T+1x8bxDwgrFeyivWeGWTvaVje+wLYcScyv+/9VK9/OdRto2oOAg/QyebAxzkm0ICTdeAqCFIKGLSaGFk0UtWC2beYiqeVc/1WJwwq6CVYbIiaue5hGquOkEs6PE4JL4SIKAJVxUYCvq4jnpc+p2P2dE4WSj6MB2znqJsaUTwDoZ3rWpp+e1yv0a2nh9elV7/LNZp2mWK6ZDSjZ8UXovVGFjJC/t79/+k8QfGjr/7g+zqXPb1ZeUE+99rPlZsJEESJiFJdTEEEW+yxw4hdznnlaSuSFxxBwMolIm0Vg6WCcU8ipAW1yjOLxHGfLrdjgxVWeWsHowf5v23b1uhnKVx7dIWG1u67JLQA0grYcvyp2vvjfV/M4zFC2Z+fO3fvvE9/7zy2ffm0+Wmv7alVta2CDVZFqWir3EvTbk/VYhaufR4vBBFPI4wHIcBcUjZ9W9p0vN5D0juPDJ/TSq1wnMCyXPmCgkalYXFBuzY5+5tc4fxEu5Ky1dpjaQrXHfCdL0z4175rwL/67YqPnAm6aL0/JuAsRKwlOH5/LfDHX7hz8l751/dFAArnn2217Ex2/uCOie5YX6TPDA6heE7+VPclmxNgi1cfBCfRrHCWN16xRsDd0OFMIu6GDqe5/z7UdfiOuMKLocN39j0+3HX49rDCn+lW+IisMqXL9mV2Lb7f5lmx1hEESJ7I27nFvw8ltsI989YYwWfv9+ezZZ9YZqyqRfCdYjxFgvGfSXsyWlq21omUZ8lFO8BiaNr3fpA0S917muNhHrmeFzXgOyIdZIYCKoe9TR/N8bMti14VFp/kZ9+finnmFn8uLzedwz0QukoMJ4FlcQAAIABJREFUWNx8Xbhuf1LAOBcBxw1jv3j/l86PFTZ8Ju+etF6QCMGJBCt8J1YAzwOHKKYAk1bli/eZB0HLX3oz+MQZk+F/S+YpMaVW8jljkFLAcBUzpTRYVq0u1n1ZIZ3nt8rf1TPjQQx/AZTga6tLYp/XnV1r01XwwX15/oeDYkzAOqfA3eV0t1My8AJYGxmnMqaaEpdGpP2keDwmPBoTHo/J5nWHWhRW+PDhlLBP9t6ehoAT4bqZqdGqeJRS8Vqs8xv+nHS4GyI2ErERMx8xTmebUvEMKxQP04Qxz5+q9HoJzmLAXbFkBHdjxF3pZgV0WwMwjb3MpAfMg8uvk3Z98PMlV/bW2LEMStr/qc/VdYoxhf7Xz+s0vvnaVt7YklDjWj9z/3Pv65z1jtOxPvfaz738gy99/BwQTDohSjQFXiwOZMpudQCFZuPdS4ChVFrg5wr1k1m6vLT8b1q9mfEK8IjYFE8qEx4he4We4gvWHSrZpghW70r1BLXto3gXoFeCaoYDZ+lFjZ0IqLURKle7KvW+Tfye3hy7LpCaOh5LbfSZgo55Qbz4gPNiUXXPgh6vNguYT4vrKXhPKwR1S9IjzjxTXhK0KLcB4viWaabA+GJunNAEUihDa+1xIVUxXBfLvbsOauD6C100i9kk+LbnttisO5ydRHztzd4tWtUDElAnGFqUDilTc2+Oj5fwVnd6PnzV1hJ3UN7X+bm5rw/K7CA4FaOWnbiULn0017rAilK9OU2z4lgBgufFRvAqCFYQDKoYklo2FFjA4ykitjqVt+QS46zv2ZcAPRdmUQpqfbDE/6e0FEUC9iXqkd8fADbawyiosQA2wJ65JeaoYgCJ84d5RBjDYbQEyaYZGmomRDWvRMzWLp8NhV4iLrQjEiCYxc7wWbPeS5/nJd9n/n/ActQzBW4rXo/37/f8u0OPcTun+W3IfdCjhmVGjdhLjR2r+9dx6D0cN5GWdja/dpwFwbfzz2fuf+48uGej0KOFDT8I8nfvf/4csPFLWcroc1uEAARA9jSiGE8AU4wDXMxHrlUBOIqVEIjU+RM51W6fi/IlVKBQfWMVgLTS56mUXg4GnwM14HvpeD+6GIgd3P+5aYveGRFg1VkGK8lDelQ1CloCdhOwHS3jVBRgOwGrYGuIXVjQh+pRYXriIRkFKgDYw+bnPlgA+pADvxkv04mtd5ZiWwFNiBJKe7s8v0cERBFsk2LQhF2e5wCj0Z0E296rYqeKTTDq1j7ljHxurXuMMRuJTJdg1fYuCE6yx53PrhfBoILBPUNKgmXdI82Xa/kSJWvJbnGVx0Ty+uy1Q+pRS+efjnwfYJ6gJb0ZwCzRE73DrbfEM1hu6p1+N+VdCUz//Gs//zJT8nboEIScvAGnJS2lWd69dZsK/xYT2kDmLtMWrnJreVnmfR9ub+k9fc46RG/BfFssA8cvOlXJqelb2UYLHM1cfMdL57V9MHMr/tohL4asjtzSRDzA8hWMjw2w1hvAnr0Jj9EP7NainNxz6/LznZCw1g4rdLiQ/axdHvixjwlEPABtPRVPIh9/8N+f34TGtQRALAlAwCAT7irrm0sJWKuWCVpTHJjMKVONY2qT2UYrLVHAat4hK5sBp9ljQs7yh+8o7pwaMJRgvwBycSab1MippRTeMrwirQffcV9Kap6pLRh5P8mu93w0vSGmbM7fB2BuzRnUAFs3CWICTjo7RxSrsB4FeFE641fDggo1AachlGJVTL8IVIumD4REXoQGBANmrqp5e18BlRDox+0xmYMRAWOA5u72uRLrqVdUjoBacwNIM0WJ/TxAcYERlxgLpeoO+pyYodbrAFAKG/L6/l0ssSJOY6JVz4NN/5mAmhnBSHgoRhoxhQGgB7COo+LtIOceFSywD9s4MD/vA2k2fso+Khnc2xMLIsVg0C6uS0CE7V4SP3cvVWsnmOLCTjASIPjM/c+dcz5+Wo/8bZJfvP9L5z77pJfPv/SF89sGRDz4ALhG2Bzb58Qd7WzuYw8CGOuhc04TUAKp/Rym6uYbtzurpaPZxqDzGMzDwFiPIZEKBoRU41EkX5fFEQFPy8n7o4KZ4oVB9cZYO6W0V1A9GSMMgEwKnERLp1vamv+yfgnPTQAyJQMup9E8SoNagLtPEzwmC1hfBTNyXqQJpFwFSPZEKbY5JmQPAyv7DEA8nXanCWcZuHQiSCl7hgV4rHWGqLGQARuYlyXAqF0AME6KdTCTs5ceAReFLFzFrxMEInuZECSinUKOrRec/6Zm/fG669I5Wlp8Ei0ZINtxXLz5IthhPFh7AMsK2WfveJuQqNRj0urxfz/lXcuO9bnXfu7lH37pE+cjRkzKgKCEIIIzrIv3gEr6IFPJAQ/UBYqd5JVsYJnX7mWJFnSogBlI8mKLq1lNN9rNFraI/HBRA9fLQprvATA31ySpWPhrxcqlmiGHYKl8dkGn3DZKcnSqeSarFlVfHU8zt2b6DFtLyngLTJbS6bKNAsFzemKB2BlkPpIt7uoJgDmXvlUQvVTQxes8XfX4Y+e/iXhQSeug5gmTVmp6O7xwQoXWJ6yo3g8WGYwIOEUoE/FHYo+zLuCkt6JLd08H7MeAf/b/3cGUeb5vPBY8GnLqwQaA+FhHP9ZpiTmkIFrPtE8ySHaZiwX5Daro8qFJa2paX6l9cM+SY2OE4i0MgAJ/Ohmd6LtkjdPOgu77lS26xjmuVsq7XZeteLVNQ7aK9fkzlYoeAZdZUfTFIZNrowUZhrIgsI9U67tVg5QPFVIAJetWBThzkOe9kI9lX6630QwsYYHo3ksTINjlhfghBuzEDAyXWel+LmcQI8mMWdMYZMm4IuvrVMAh6w15OgE9fUue1DpGDhdYH+idxGgWQJotzrExDapgRkXzMsg8loZe27XGMs8w2YDFB9bFnNSotLCoLy3WuTWL7WCiiHLveghYjJqBGQDxz87/z0rrCeYNAT4Y9UU+/9IXzoNUZaXeX137Pnv/l88B4DZk1FkCIExzTYt4m/aV0yRrV1QPci3U11dNvtz34BCGxXIYYqAHYxUrHQqwcctsW6oW0xFDjs+YKmBQrRmvktp5CmhuhiubRe+GBypmhGJCkzmFK+Q2f2OfMKniJAacdEYR64PNuatIoGHnDpEB6DkYPQH7ZIahsy7ToaaEADMQncWAiynhYZpwFiKSKi6zocJ7QyfV/5+6t/uVJcvuhH5rR0Rmno97b310uXG7DQyDJdC88AIv/AVIvFBdZdkYMwbUzIwEGAY8IMtwusAgzYzHg/EA4ykG3LI8Hrm6etSALBCSX+aJF0ACWWJGlmYs+sN2dVXde889JzMjYi8e1v7tvfbOyDzn3vrsdXWVeTIjI3bs2B/r47d+C7cxprWjjCvm9Q0IuEzQfQ/rGsSS/LODSh1UKuk7D8V24RnAdYyp9ovlhIyzYVN4TtL1ljWPBmftRPFr5JQdR+UZL0mAZMfRcc2rSNmfLE+RCAv/OftqeW3TtNdKzkf0qJ5RZogW/dbD3Qd0n5sI5ydaJ+Ttd3/xra9+5ReuBimh3RETtiBe20jGKsaSpJitMWCHEQN6jMkn5+VFlMolT7pPLBcUmtVBTf0k9q5DwB4TQBq1tPkPCV9uMBUuhKkYGEhnyYI0pxWd7MnM3kSf/Gu+Qt5HryFrt+b5DIDUsIa8OaYhfCzach8GB38e4NAo4fkvsQFxuT7KFaH4UJ4dPIOl58hoCDf5FsbxtXd+9l6T5z988y9ftUbLEq68VtYDSHuco0MpyXwrY4GiuK6kAZIND5TFzUNxBAWyRQOE+OUVChnD43FG1ID9LDhbdQgBuN4KrvfA+ZCwvqMtqPs2kxFl0Wqf0PL4r1+ZLNiGgjOMCfbsJyjOpctJ4bb5m8ctQFF8/3WBJIHg2RzxZFL8yFmPPnv1Du9jCIL9XML9lL1G/JEaffcjDNghYp/N/AIl85tFrsMCwY2rnVH3iVuH3Pxsx0xRNoty7KFa5bOy6JuDJVSQPSr82wRd8Eo2gCpXhHVheK6qEJu70Tp6KhmuRVnymp2SpXA+gFQh3eBzS4GG1iESUv+Qbtd7DDnPffL+pDF/9lj2TvE4vVYtjftjRiVQR0OWDBE+K8IQGcH3uWsAcrS7PefnWX7zK3/rKkNDE6EG4aYsLrrMs/jZSWuAAHSk2No0iOQ1rBXmbTC3gLG0OXnpGXX1tSty9LiJcOQIhhSFHziEeMVkiOwn1ugobeFv/NrWuQHsr0HxLF4sqEhjou+dcZIcO6O7n3WCls3RjJBVqlGyTxXT2W9TtH5hwjqvz2MUwLM54qKzaMU6BMzRcjXE9QFg0fNtjFgFy825net5s0vvSGAyiHMdKHAbzekRIdiE2vinjKrYpEZuNeZaJTc6Yy1WE4blTLkmrRHSGibY4zhtL2BrQC8hr3NLxxIeCzBWXh8jaX/UhbXIR0GWoiOnpDhpbZ62bh9/ThosBvO9q2DDpyefSrHC3nUNlUp6JDs1NbHaPIUwLFPhqcz7cNWL5od4RUHAZGnk91QeNunvczXe/gsd8DgAHSK2mDCkdhe6VS3GAYohERKEq1NL1iWcwcMF2C+Et3jFldZuTJYuFSB6fXxl5E5D5os+BX9Y6pPCmLVMK7yUgM8SYK3MiBhFcIMdhnQ3PO4UI9kp4TPb34NZ65T4/uC1fb/QAOnTqARKaNPDFWZnSHjvsl8yaHRQuAB6A4QUuwYjMAaQbbT4+RAF3/4wYN3ZRnEzKaIK5qiJ2rYopdZH7b0efsbr9Thc3HpSxDrhZtwlmt1ZSb+bEtXFcjuCAFBGRNzvUysmWL5LD8HjaM/w6dghOq/RPgIrNymYTEr7hNEPK5Bri+tTTHnR32LGTmas1UBzuwTrBIBB/BqE5f5ySXx5/mryfGpJzBcEqGrOrdg7Vpe2OCJhIjQ+OnRYoUswHzM4rjHlmhuEQRkLlo2tGcZ172t0eMPIb1xU8vkd7ZMeATtMUJQ8N0Ya235p5ZgSz3a0XkIe6t9zDSM00feRaDHg5+acbf7NsTyQqr3N56egWT4awmPRXIPRDmMgIyuhvXJtAJCi38+/J32a8vYbX7+ymihIcGjLk+IaUu8bNrM+L/e0ZIAAZdwyCgIg55dRprRuqSpmqdcUqx8C3M56kHdBA4QKugA5OZxnP1spbvfc74th0qclh4aBh24tUu46o4YosSpfROtkdCaOj7EYE7xeEEse30dNNaUsAkLIVT4nDG5l66xCRLCLRot7uTLIFufTblaErsytfbQaI5a7Yrls59KlyHmZgZMGrJyRGBIl+6gx6XUJgSK2vu0VGGCV7ffJgDpPz4iV1v2Y3CHiIjnEdohYocdGBI91QkhOsh6SoyEBSFCnUriwhddmSBY6bJOmk8lfnMHBv9vP/XvvqBpwCHenbtfmJ04SnXPjcL/mutwnp0gEKgdnhFbnZBvo/Ppv3vjvrz5rZizgUzBC3n73F9/6c2987conbwsUT3CDhzjHOQZu6/n7KXX4CkPalGNKaO8xu3yIgHCQyNwmF/v6G4Ap2es0EDxLFFBT/t7IiH9yfiVvSROsKNpTmbBBj0td4YnscKkrXIv5YdcOfuCTRNm2QTvcyui8eaVKOKMtrSXrWY/s1RYLAAjOAOEZOWgZ3juVeOuhbbWRUU+UU74wb0TwufAZAZYotUlgkiXLviT6NhjtvCjUk+h55C++83NvAZYTwnO2cLy2Pd4YLREMg8GJmiJuoc5SJK/AMlCdm4xSAKr7s3GG1D8Rk1o0ZBfN4B4R8cRWW3x/Al7tezydTUVdzQHPomc3sna2FXpLO0oPkqp1wOGCxsrCfhR4SIO/j1XyhnkDhRERnp8h7hkF5rNDDbn8/hjwR6PiYeiwDgHfnfY4lw4PumAQAQWu5xmDGF3xjc5WCFHpwZozBInJyxGKW5nyM+Em4w3mOrpYpOQHpO+JmVX7bkyFELloUuFZocuRlm36bIZB9i51wCo5IsgaNojgiU54X3apvkms6rvwvDT0eU8kOGCdD1EbZ6TF9YZFj2D5FHRaKDCGmOvcbGXKx66PwKaK/3A5GmJ9WI+liKL8+ff+/kqCvG2yr3/zx6uN8Juv//bVHuZV9ZCO5UguvzseAQTuhma195Dbit4q3i/MGQBYJbjDXqiolJXgo5BofBJCSBXFP5OdmzMUy2nrMGE6GU36NORbr79ztZSo26Os4YJiLMww5cZ78fPapmXcdElB38e6cncVfUBR3oeuwK0AQqsk537QSAhSksSBkpSejXIp+R2Tgz95Sl5GPTzEqoMdQ+NlnEttj8c7B+URizi/tq57bXBGjObrCDQqtinhfDsn2LyWUT+rYgRwIaEUKFRFH+q1gzAokot0EDzou+ww415guW9zhjpFKJ7qlB02q2gQ4K1GPEgOpNuGF7mNjnKtGDWiFzILKtZiczImJ9ZNcm8rUDl1jomvPk5p4ad3fQ4UZEyv4cA4oMFD4dpbGyI8D38n+VxWQ8kGkhU+nrOzdEC9fn3eDJFPJRLy337ja2/9uTe+dtWhw63ujK5WaL1pCsHbQNknpoNOjTFmd+BZ7NNiXyvH9OgteewBw/ZTsfbYXlt0Qz4/vUQzNHsVX8UG35Vn2MoEgWCtPTbaYyfmeSXGvLASaM57WJF/ymHPtbq2bQbrdF/tZPAKscdu2z17JZGTKSkwCULEyAml9SqWjYeLdzHa7oqO+P6dqmfR5Wr3KwxpgoxHN3J6SP05Dj2SHw+Lw30gKD58adA8emWL4TJoyAqc95L4dprxWtikvNfZPrdzdrBkPFZPj6kNzCd6Ms+Iah6fnUaMUAwo1K3dCSMEqAsaAmWjnlSr6Md9DJA+K5jLUiCI7Thm35Rn8AH2CDCPF6IlWj/VETfTgB/qmH9j3soOVlMDAJ5Uta4N0hN1RqtE0fNDKfBEVMewf4I7Mhv1aW5M0Fw/w/flOin7hBudJaX0iezxkq4T65RihxnrBO0cXVh/TkUDvXA83cqUN6MZEXuZcaEDdqkqeCeS1w0a8yNhnzAP407mFJlEho56KFFMeOxWWGW8cxGhGjbLVznASB+LXFBao+PU93/n9d/OivNPv/tTbwHAr3/lNxY94na92hBpo14H97GgNAxqRuNOpoMNfEkKFvvzZXQcE1/7xPdVh1Dq6iShlzZ+Du+P6zPnyAhfNwIZvsOk8jJmD4WJz1z2PJ1vFyz6PKSkOA+T8hXLWdiP7+dYGycAUpHAcow/X74v997XLlk11+J5achsOsnOm+0cLY8v1IYH2zxFM2C6YPvAqMkbnxK/maCucFA0kWzM0fHEhHRCrXZqnnhf3NYjbc9DwIek+U06kl+zBwSsJGAGlelU+TxF/426vENQycQcAxIZSboO96h9coACyXmb+nbUiC2MUdHrHH7fbz8zh82ys+a+4qMhx3QZ1hnjvCtOoGWp89PKXKYOYcfEysAp+/DnwxD5VIwQwAwRvv+33vjPrqARO5lwIyNe1j4re5oSH3NISQEVxYxUy0ID9jLleiLMNpncINH8XVclQXvP4trdujcSzjGk30R8GLYYtMOFDjjXARGKGxmxQZ+pVulN7OC5m+dUoM4GG3/LtpBuk/kYZaPTqp1AGUhkxTomtG5pmi0N3JCUJZ/85D3+QJkEvh0sutiKN/j4fot9dW0qSQdtaRafVmqIVIGfLEHKGOmYEXP0w8t/8c6//9bVm//lUcXFt4NjJ7dDaR7TFNMM9xk0YJOgNeV+A0ZoGo98xjRKiieen5tRqtil9x6DThlV8+fcTBlZiZAMqTiEYxUPIaMf1bNObCJ3Yb5zxCP93bJxlfNJ/p7GGKNBMRtnHM+lZkZMninKU4zYzjNeE2PSCy7SMrl+4pjbJThUhOaCeXw+syzVl/CV4AteN9fKyP2X7j89JfafuOfpcxvWToG71AEvYcAeER9gn44HZtQb3JLxaH0Rqz6Jothoj1EsKsw5PGKf1646/0QxCvLxvA9/bfaBLzA4aMgGCO+xFY67gNMexCX5qXf/5efa7P6lBYPlZ9796be8IXIqAkLx3sO2cNeS0Nl0X2lz9FoD+PMgOQIvCZKsthfl2kkp8sd+ZL6ON+A+bfmfXA0QL0XpMoOKaxMhm1SYFYRioVKeGTURMWanVTDGKF8XhPkjm4BS9Txdh1ArES3kFuINFAfXCZKP89XLGQ1hRXIK4Vj591LySzrUORc8BwCcDcBaBfsZ2O2T44fXQDEhVUvxwaCWezelnA7Mdo+bRMG7n8ueNaT+6oNgnM3Qu02GwZAKPzLPhmQhgJ2Xv52j4gI9JoyO7Y85kVb3is9xl/LCehHcOmgWnXPUT5iUTqHTauL+ookoQwR7tTxC1mqiDgYg0/N6hzElQit46V1y11RZa4ed1NByQvz9Ne25FaeBF7/mmXHTweNscltAhyZz2bzj2Y7+LA2RT80I8fLXvvEfvwUA/9Gbv3S1xR7flT2+qA8BSQVUpIS51+jxOG2Kxnhjx3QJPsJiUj2KYksl/1JXuEnwp8EllFOYTzELufJDStATbNSgYI90jT/RneG9ecSkZjD8sJ7jQ+xxmyxxRkxy0qkAD3UNUYM92DlNGeBg8REKwqbW2ucw3RIXPymCgcNcCnpLkRTSzKOdMYoFbiMqgINw+M3FvPylsrSPJAFwRsmEpQgJ2+1hcXuMlWHVVppfqho/IWZD0RKhZ6zQH0yxX3jzl/MmNcOS0ZcMkbfe+Xerz7725q84JaYoqWTD8lGxERErdLjQdYbKXOoKBPisXbvt+bKny7m9As3tgJ/7O5qa78wbVOOySaHKKEpdIwL5937ZIt1uw0ZZFIy2w458BiSjpGE58bJyVbmNzaREdrhwzlR8UDYubxiPiPhj3RtlsYZi4KA2oIESAs843ipPygwDbjD8XTGSSk2R4Po4wCuT1vYSSQl53dm7HCxjeinRxccYESA5GrGD1ffgZnYsyTqK5rniHSSArSdtUmGXlEp+PohtRt7LR9ywv39jeStOAg/t8m3J79PvCBFgRSHvjfssxBuN3GiPjV2CcmnEn5LnMSI6CDrt8DOfE8aZJSkOLUJKZ6y1zzmGfk/ZYkrGOaGOignAb3zlN68GDfiJb/7kp3KfS0/AQ7CAoqwJBDc6p7kbsHWPdy2CTQhYBeDZbLWHBrF6RVakr2YaXHVWMyOq5VyQBQtISd9TiUIABpEyeJVWhgFQGyqUwZ2PuSKOnCtHOgjPimqVyv1v+67kjqwSJOtmb9XQtylZ++neot1dANYuR2VMxgWp4FcBGFVwHWNO8t7PipuZriNL3PfGDFkTVY1SHaCDAtkA2GLG+1HxUjCa8Qhg69YlOuMGFAMEsH1jhOIcAU+jOZm2mDGoGTdnacdTWAT9PDkDZ0RMKM48yXus4NV1h/d25vBaJfbEKiqhh3kZXNO47yxBApdkKRrsa6VFFAIeXwS1EBq5c9GR1kQpl8T2s1gxGkJtJyvsjYVy3JNsfFaGyLG1+lO6uORK2NeJAcWKe5nyd84K1OgMAoUBl6mWetCS4A6YbRdh3kLak36zJjOCQWhs4+WD4iav0IyVDgBe1g2+iA1e2QS8FHq8jDV+VC8QAKwQ8EhXeEnXOMeATVoaachcqnF2bNK1dkklGLQriYzJOKLi7xPSqch4PCA98i3EhPkW5T5DDonS+KA1nM+lptwQtuXPy/etUsHkTKAYJSX1tn7vDQpGqFjp/hi8y0cg/O/Z7j2mo4rbKYYvb3C0QhgNq9rzeoWhzBLINW3aAzpcqnnoN+gwpP4ms0+m7EWtGJe+8HkixyNEjBK0jBn2u2Ks1NGVco52qRJISqY8HEPL/XJc/GZNPn7bzCQnsLNtxdfCiAITvAuMxV7r6BgNkWtMeB97PNbJCmHBIlMDDgtPnsT1oo6GtH/fpUDXBtJc/U0D60ZG3MiIa9njViZsMWcqXR91sLXneKbVMTjTHvMiq4lFhsrnc9pwfP8IJBnzMa9HnON3RSV5TkVh7pokZtiAb8dnLc+ThxFFn+v4u+TzbIB48WvorjFqFZbM6pnhAB9pN/nW6+8cXVM/TvkXv/lG7tOQ9rT7GIctzW0E6xUhFb5LEdZo61nJ8RB0oUCzTOFX3I6aK5AzIdyONyNDRDF0dVTEC4+h4cJICSl829ojXuZocKvdZP/H2SqYT3OhB851hiIj0aW2kq9ZwurnGSoF+8wn8e8TRTFhYKOqQaQUuSghqY73GrFN/wcxxqlZNTvmqOh3sIhUVDIAJtgXmKxdjMDKiQrNtL6MrvgoBoC8Tw5iuuINpsy6Zeeztn6wLxTBdIItyWHkfJlx8T7iDZCY/xfxc5EVzT0BThW9TnuGEaDExf0jNPOU4g0g74DjXsb/LFb6aYr87u/+7tc+7Yt6+bk3/9LVgA4X2OALeoFX41n2xq21w4eyw4CAP5ZneE0vAFjS+B4G5eJD4oO70JVt1qm43AjL2yDt4B6lQjq9m6ReZA6FQPCSrvEAA77Ur/BDZwH/4LoAJz7QER9gBxXgZV1hTMrWNUacoceIiAcY8AxTTrwiLSgVFg/9IK51wvKmGB2kpN30WyubQqV8BhkojofzjlHBKcqk8IZO1bZGSW5Vqw5dYpjY5888cMxPmfa3/G7EhKlRvn7xnT+fN6eff/OvXMWm7/7iOz/31s+/+Veu+FxbTzb7Z0B/oMyuHAxjndofYQll5xhyYq0ZHiXRvO1DjjVWLmckpPVmeAPSP1N6X45tvFwovWGzZIC0rFfFCCpjgIYEWWV8j7Sc+x6O5b/jJk9Mtk9Wt/skVIjX99+Vp9/mlHDsP0hh9G2CX41pbLfQRfZDa5SwwvgopV7GkuFSYJP0HId8TqB2bhzLQTgmPK+H6dXf13lwFL+JnBIbL4cMLMc+A+qo6jEFz2+Wvj+YGxUgCWJQ2Ll8hCVCnxuKdZecyg05Jr4H2qjyKTkF9+Iz/7wbIb/6xn93xbW3fUViAAAgAElEQVSQaxqF0DNf66pL+2KGdabDzVFoz3QJLvdJyO+8/u5iYjpwGBlhFHPlyDboHNl0aa0WYN1JjggAFhWJavCnlQMi3CbUTCcGefJGyHrQCmKlatCrNq9jSWiwiCjmKAeGC2AGxzS7Wh4oxgRZrtZ9YfO63gMf7GdsNWIjAa+uu2xQrbpihNxOFtnY9ILtZGvvPhoz44Ouy/kns1oF8m2M2ISQDbhtqq3y/lyYo85Dh8exlFMgo98jDLgMHaZEKX+NCc8S/dBDDLgQFvAt+8k2RtxgTvDmiA2MdetD3VdrLa/xKtYYxGouXWPK+ZWX6KsaIeziZ5iyQTMhZuOCaztrHI0SsU2627kS4yB3wrI8e9axukYA8FS2+T0LedMw8HPPR8N9mQfO58nBbgHTkSmWX03ncRnYrM8HADvHOvqz3/jqp7qOfSZwLC9/+Z2/8NYvvPnLVw91jR+ND8yC1pJYs9FSgOwlXcNCcOaV3miPR7pBBPBYtrlDzzHgRoGX9QwjZjzGNid/E0rhoyXnukpJmAFn6LHWDl+WM3ygI7437RFu1wgieBZnvIdd9saOiPi+7HJNi5d1gwfoM5abBcqGdE87zDhL8IdW0TB63QnBKQYjYq4vQhgEgMyIsNYOt2mCtIpEiagwQFeKM3lL2OBbdaKUxxl2jqKU1/fKim1Gc2VQ1NkpJmQ6K9cwP0gxomqDpE1498Zmh4CvvfkrV94zS+wyN1jmiVBaRSNHmhq10TNXAVagMFPqqqRIDvIYMISlAjAYloVZJZ/N13TgeT18yrfN7qdAhDLtK3QxJY7PYajOVd9ja0BUxoX7jMe1HPvt7+MRA8SfOyS4VhCDRlJGjVjl56hZlWfgnDARH/bmXIrJ+Aiw/qUitMWcl8916m+v/OZ2o+RvMFfkuOeeeUCHXn7vJSMMxN971VcLfdNCyZZ+w+inQPKaVsbv3dGepahgxj3n/g2Vo4LS5ly1NYbaPuN479SuS7gZ1w3A3v/kpwTfoXiYgRdvTNC7fzyGevy3rdz3HJ+lmNGr6f2UHWAhjTM/JjdpH1yneUY68esEQZ5PjMOPU/6X19+9AmoSFqCmPvdCuCyP9vNTYYo2/54VGZIFGFxpTkaITzyftdDeqgKbgc4WMyTy3BBkS81T8VJamFbfJWdTNBgXUBsmdrxg6ICzdNx2BDYD8HgLPB3NENjMRqv7YBA8myJuUg7GKpiBsemT0tyVGiHPoLieIkQ6jKp4Ohvl+6tDj01nBtHNpBjEoGuP54hzGBweams/64J4McOgMJiuEXAZOpgH38hVAFtPHiTXL5PGvbBPb0GWJ9sTLtDjBjP2iNimtZ4G8aT1PPRjdFTFB9gn9IKt7oSV9wi5OCz1JEl5hVwve61rQBHiS2mNkk4F+2ZOtXIsCktHkl9ruf63hWNPsXG11+I5iMTh+yjFKXFXHaZPQj5TOJaXKQ1YhubWCHgoPTYpD2JOytxZ8kz3Sb3+Yb3ApatkbXkcm4TRNcPgXFemrGnAA13jXFdgzsg57LugZixstMMqTbabZC2/P03YRmOxWSHkUOCgAbtkTRskxxbpHSL+GFvcYsoVQW9kqqg019rhPHmf/IOfUEKNZUG1TWDQLkdqBu2MYUmPP8Lgfm9/F0iXv66HZbRKCAe9x323Cj3hTIRaUUgOwGfjpfVCexhXm/BuUCmyXBzebzjRNv+3P7eHX/lj2+TLG9njieywxYS9zBXMxW/OvlW8qyH3c71pt3fA7wmTqvuF6WMJztOMFU1GylKftEYChWfoxZL+jh23JLzSXb/heY+NTkaEOhwe48d+CxUi7G2bNqKlBdsbl8eiHZ2WRPZ2IV8yTvx5/Ptu4drlff16X/FwuVPze0nuipRMRzxyFIucuo0Oh1EpCtdM9vcKVgPF8+bfd5N8EWnzV7zcB2blo4B3CedvC2v9QRZTDA+jiECKcqE40RhtWCNkR4w3DP7X17959b+9/neuficZDR+X3Ac6WtqM3K7B5aV5KnEe14kcJH8HMQW9Zb66WNl/Hx0hrKrvNNP0Vm1pmsy/W6OESemqUp2Dn3fBYFyk890kJp6owIPBohKPx9nYsCJwMyfFXAQvrayqOSM7vrL7JpiL5XZWnHfFMbROkQ7ADLZRi+G2U6s7ImJRbgXzFw6hrTT6OoQUAYnYq8Hcn2LM6zip1VkvxO7N8lh8rmWp8cHCpnWktVy75BeOMOrzW405SZ7QLZ7jLLGmDhoyeyBAh2DMhgfHYHaaSoKKpWNUcPB/ybHLcxNl0soxI+DYusMoyF3iofQ8D1/p5Absef3VN97+VCFZn2kkxFe0HhDwGCO+gDUAPkAb4LeY8XIyLOgBWWmPSfb4tjzDpQ62UOqACYqblBD6XrjBn4wv4ct6ib+Xanmca49HWOE93OJl3eBWJzAh82VdYYMuW+tn6HCNCR9gj3P0uEnW8jMZc/Jrm2xExWgWzZMraqlcbNAwsnSVge3XsCiKUROsQUl3R6+r9+6mweVqg7TGAg0PWvQ0OBT1ouGHMft4RKyUchpAUYqnA6g9+b5w3ymM+BKefwmzX/JUjAltdHA6n7tCr3Xnnol5hWf0yWj1YrVMyvBnWHOUGaIlPOzvg+FYv/n2KPCqFvbG51+uqQkcJm7c1PfuPRb8bfvqxcrPGbsWsbfAoZFARpFyv4fSKmPHDI37Gi0ByFQvEWaYEJMr8BE5AeNm9Zgpo9RDzwjFGlO/tZElT3RAo9G/ZgNCC/zNs1zdfV+H99/CQqs+ONU/qBVhRvMo99lgltrG6Gw91up782Nwqb1LkDZ/Dfa5KIkkCB8oZ5kQ8ePf/IlPJAry1W/86bdaSFaBx3k2mLb9RXJxuyMbfD6vSw61Ypb+yE/O0PqkhesPUDtzaKQDNbNeQIcBmuIolhvSQky/+fpvX91Fw3wf+buvf+tqjZD3Y6A8Ly/+M+98oVFCo4MwrADBKti9d8kqyJS8oTBc9Z1imgVnK6v3wUjEfhKsei25G0JIVQ3VApahWDymTV7P9yAlWV1VciL8ymlrD9f8veAmhZS3s8GdZijWqQ8uVrabsDiiAthOim20tXVUxS5qrr0EpH0rWr/dzjGvy6QxDqjvsbBQ2Ro+IuIcPbrk0Jhh1csNFVCi/ApXmV6T4p6KwBoTmB2zQZcZzgCOx8N6XmySgAaQJEddIUcx+JXiDB1GIO2dBXWx0g6TTNnBN+T21tI6pZakZiOsIxhAvVfwXhiN8EW1/W8O2nDkc3/+QtaRarSk9IQpRXsI6/JCQ+Tf+xSgWZ8NO9Ybf/PqRkrdiA4BW5kMoqTAOTo8xohROzzG3vCG6PGFMCAC+HI8xx/IM+xh0JlrMfalcx3wfbnFE9mlMLJFU/4+nmCFHpc6YI0OPxzWQAS+IGtchjN8e97hse7xx7LNhcBudMKX5QzQUpeAbFgBgm3ikPcPcp0GzjXGXDHzXHtcJ6NlULvPDgNUkBlIOEHIluA9IoKCHacxQimKix1PfnsAuV0c5oReechP671ovZ6FFaU2NmiMhMYK9xCtVrwy1OY+2D2URDO2wxRMwkhKRMYbFLUii3RsWTToKW+ljYJo6hMm8jPxmaHLDoI1LEpWFJf6XqkgUyZ3H75f2pwHno99HVAS4Zak/ZzREXqVOne+pVyegLrex5Ki/FENEH+twjkpEEfj24a3vTLERdN72UZE3AC4RKL0xiEE6VidIMIggTo3witehBPle+Xzkrb/aogSXNu94lvDHg+lVCxfNjSi1NCvY+IJOsiu4g34ZRrgsi0y2unr4SApD23bB3TYZCPdlNWLpBacocvwidH1wW995beufvLdTw6OtWREHevzVlqCh8poa5973qzrzf/f+Ma/+olv1J+kkOKbvbdJpCoBgnN0uX8DgLVYdGSrMTvlyArHyAnw0QyRv/v6t67WYvuTotCec10ElvcYAFX0tZNSJXsVjJp3EwRBBH2CWW16ixT0XV1scDOYx3/VG+Rp1ZtivOqBZzsBYMYJfxOStUNlHziMhuR6G8kIGZvUL+77XWfXMhYrywnZTvafbFlnCfu76oCXVh3mqPjjdMIHocdZCmcwejOmJPY5WvSD+RlzilDcYMaldIklzI6JsOjHjc54OfSOgj1FiuAdkZbQ/jQ5gDcIuIbiCUaca49bzHm8rNElREupyUahAQIURwL3NOYCwro6r+OF9anoSVuwACCZA+0Y/n4QQdSiW/k9IbOcArhNREU+l2JpLb4rAr0kXKOoR3imLA+pfx5ZglRxPvhcRhY4nBGxSzmSwfXtpwnK+lThWL/yxttXf+2Nv3k1I2KLfYZU8WFc6oBBBGsJOAcpejuco8caARddwA+fdfjRzQpDUoS5IW60xxf0LCt0MyJGzPh/8TgnBO9kxjVGbDXiH+3O8KVNj4ergGeYcIYeg5YE8huZ8KFOxsyFDtcp0SdAsHOPmQYIwA0sRR6a8bOXuSQ/SWGp4YbnB0gHORgENCHsexdqRp3DQE97rcjFrPDwP5UUg3qFatBzA1qCN/E++dqnEOZhOLaOavj2+uMoPkm2xd0DqDzYXmhk+P9cMAZ0VT2Y9ndeWKjL2oVsgHg4wJieWOnjZdH0nwxlS9du8Z4+qmPn0MXfHhMq7ITzaVrM+Zo377SRMCLxcS0APlfEX+ewnelVGK3zIX2bt7sU7aInk/+BVGEehfcf8PkO5Th/9dY49Asu+eZZ1Kodu0CJmPD9MYhX9ZtmbSv3fwh9bI/JY0HL2tJrOGiXv5Y/v7hzH6UBXvhslFhBnPwxfl7xOp1abo7ADMNz6fAIA1ZJGWU/vcgGfV+xmjBsUy10LNwlHhrB90sexvvCgj6vcheMjGNlK2POWeS87NPdlyJ0mhN7kf7eJYgk5Zuu0OSLyEoEGyFsp8zhpXmwNG8HFxFZhzTvRLJiHsSSumctLFVWtbywZfllrU0295EM5ozMzcTyyyDhV6rl+KVkdACYZsnFDvvOjI4pAs/2iie7pAek9q46IIjgQWfKs1UGt/saZ6sJMkV7HaOiD0YBuxKbozs1qPvDzgxOViYPKCQkAqAPJRletZ5virKnck1fp/ViBysOWPQXc1KQOCBqcaR5AwSggWt7v+XJFmcBowV8DAYv1Oz8K5GFct3cv4zOp3auE5TUi//7k4BgEj3zPMcvvffCOeDX/XnhOjNidiYDtX71IsbPR5EDDe1f/8rPXwEGVwGAU1Ui/aI8SJ8/owe2S+rtWiwH44t4hBkznsgtJsw4wzrBYgQvR2N+PksD8wE6bBOGcIMOKxG8cmbX++7tnJVEw/YZLe5rssIfwConnOsKO5nwVHYYUksA5KSkL18GrAfF731/xgMMeEl6PNEO38ENnsoOl7rK9JpW8XjCefo1UMOwzHJmuLEkPPG3THyiaUHjg5jcDpIrHXvxHqhWaPn7yVFBKVQA8WG4rlJ+/XMqHunl63jPAFBX4DZlKQ+IanIcKHOoQ4wZ2oCSVF7D23y0IGSPh5elKAewXGfFy4wZHYbqM4GYVzjB5URKzRQasYMGrICkBDchU9d2u18aFvwcrq/1oF9Lv9V5PH6BPyU+IpSZQKqoGnLInN6O+0RBWtjQ0jjRhd+dbKuHIqDc75DNbcLSDscTi1xZcvWcr7/0SgWGfW+KlJ2JCtOAUiGEz6eN1HVaVjtGOf1z9+NSF34PlEhDdePu7lqDIY8BxUFftOLnNdvkox2nZKkIHTdoCouqAoVAIUgHqB03QgEtBTrZBuD5CxQ+j3jHAYVsg22E9D6yZHwsXfM+x33exNqsOdp79DiwEKZh6c/TKmvOAlNMx7RSZ2UxFwg9NUqPC3NJLMpp9SeGVIeC0VCv5AJlD8wOgiYKktFSjECIrX9dsOKku9mUaRolq15zpOGYjLPkY3ajYD1oVT+kFR8Z8d9bpMPyUvpO83u/hM6xGEFBgEcbu/6TvWI/FQrhIGak9CHgO1OCgavdr6LkgpwNQDczEhAwKhBmwftxxAOxSIetkybP4owbzBhgxQiBwoA4JeIRgPpRzKQgawQzsFLbP8AOgDmUFUg1PgT7xOBVorjF8FiqKs69kJFW/z0he+18X8rHDBA8c2xQ3hHGpZC1uPLzkjKwn4dV71TyOdtD3ccbAa2ecxf0qz62juhOEqtkdF6Dn8/iI+kxGXjLEfRPQvqvfuUXriIUEyZ0jaL6UaTGxlvA6T15itf0Ye6cS13lAfIjiX53nxaf19Y9nk0RY7TCO4/6Dk93wKsXwIdxxD/dX+L/mC3Ze60dHmLAZdehi6X4mK94ywTK80TZdrZSbFaKHznvcb4L2EfFrc641AE3MmblkK/nOhisCiFT/fpq7BzYZP/xg6ZTS5QrEAVCUZDjH0FrpaVW/I55lX3korasaRxEl9AFxIwFBIpR4NvrJ4A3BAjNKs+2tDXDsLQu+Na2k+/99Vm0sTamlidgcIat/+75/AklCf6+YjApAJgxosMzTHhwAsnYtr8ohsUI5SI3IBzkNLQ5IXcpUl5ZtN8c3iE35VFZ2OvQAFkyIrznq/1srj57vjWDlYtb8ZEn3z80IAAmFxbjw9PtOns4RVqN2vdpKpgJWMVcbpoA4KFDxwwLfu+/O7YhHECYjih9NGb8+Q1mUJ9XmvmzdN2lDTEsGC5L0KVDjpvlc7J3aYwXZdXUUTPKkck67NyKv/36b109DqaI/Jlv/MzHZpC8/cbXr049o1Pi+8FDrE55A2kY+/Xvq9/405+YgfVZCCPme8yZFXHEPjujOk25cygMj34POVV/4ZRwLO4QMaBUnApia9ZGOuy1PDXSjrczS4BMMe7rWQDAWS/Yz5oVcirogBkgrgZrfvW0uDwuiOWFtHCqJTFIlWBITFg0Prqg1ZrqrxsVqaaI4HpXoiVcdwOMNrgPdn6ORgWyAbdKsLIxRUH6kJL2k1EUYcbkOkVD1m795h5xrQZ/uhDbc/dRU+V2xajlNwFG4sP1kzmldPys0GGT/u8xY4WAjQS03TepVuuGpnMbs2mB5PF7uPcl4lFrIG2RQTr+xmRk09ghVa8fy9S92M42t847C49Ja2z4zzwVPKP7dPQVqO5p8cYLzx1Q0+8CBrlVlewkEkjOPdnogBuxEgoTtNJs/uobb18xh+STyg+pNCkqdMfrzS5L7QVUhPSPKU2FJ7nDY7nJdGBbmbDSHmt0+PvhAwzo8B3t8c/JK6mKp+CL5wH72bCb/+OT9/DP66v4YrfCH0xbvIx19pz84+cDbiZFmAWPsEGA4NbREP4jeo4/klvsEfFhHPHe9QavXNhi9MHOsI9rdLjGmKIgE74vt3hVz7DDnOFYe8wZ+0p6XKRrXMs+80uTBhQoLC4FPlKgNsSQs5AiUAYgiybynKxz4N97JUagGFDyKCKACTYg/flpFZMlouSPFKo2Gi31pl7jLr0Uxblcm8JcFn7mrXIaSwFdqe6erkcq4ogZFsupYVIZypYmm8d18mqlqGKtyHUaKg9ASJurMUWUCAvZJwa1fnkmIy5SgjoXDnphPGa59db5ay8tXoQ9+L8pXgFvz+epKn1uiYd2cSP2z6RV0VojonN/Zm8ccABT8MbyfcW3hW1kTIrJ5nZMMcZK5K/Q+wpqXLI3yknj20Pw6tBjN0ZMmMCCV8QI+6gVUOeLLNUSaaMjwGHOCOcljY+AQypoyq7aijnXDrUb71xY8jUvKd9Lx95nZeeYWqc8GkJFPV22PZOIXfIuj3nrLP01Ju76u6KSLyoxGw6S+7qsoSYekrW0NvGY1mhZinBCyjP/QY2GAHUO0LF5OyYmwK1M+ThNCiUAXOiQawvYfOuSMlXw5ffJCfnm6799FaE5z8vnCEx5jTGvuV+hogIr8dQVJoRgrUPIdTCimv5gs0Hw6ExT8rdgM2g2JqYTRoXP/ZhjMVoIqzpWIwRg/ofk95naV0uOB6MVmwGpOKEZOTQagJQXMiumqHh/nDGIRSgeDMGMEQEupcP7uscPocd+Bp5Nia63k5zLIQpcJypfKuKr1OjtHPE4GqvnJXo86qyBkpaSqOYUBsyIGcTyKCPIEmpRiscacQ3TmR5gwCthwKiKR9JjG2MqPGtOZ0YhCLXq095VEtkLuxVn8IBglOxi8LMbTBn+NUCwc2u71foyxZtwXlLyso7bhJjXcW94eKPEO4fo9L3LQRhQUCOedIZCpITPz5th/by0braGyRIdu98r1K1j0rzGpNv0GnI+covO4FrxSTIC9t7iYlc8r2e5kzbUE9KQ1KzYdWJdvTe1GFatcUIUxR4THukG5zrgUle4jhGstfK9Z4rfn29wiwnXYY/f39/iT67O8GVZYxVsEl6PEd++nfBd3WIjPX4MD/BYJ3xPbrDWUovioa7wCAMuQodHZ+aR2E3AwyHY4jUB7yEVi0lesV3yBpxrj1Ei9pgPanx4C3cvhmen0lEMEBvY/liDPBQc90G/YpndheJRFOTlZxjdTw7ySbcKI3NPvPLG1+g2b1rXxQCplTNbMErtFSqRlAFdVQzHjilsUrmt3rOO4wqc9a+Dj2mHUebquKk5vrp2ohb1fdsew810lNlNWjOQJhhOdY+YKZvJp68oixNvh/3pKZftdksFVIHHcx5OeG+07holleenZ3ApnLsk9LqcqiXixR9miYnuN67J9109Wt786lpucfbjtr0rKov+NITBnUnAg9Blg6qNGLGgVQv7AwpG9hASUI9VRjpb8Z/Vlcvr+XOuVldoj7naBHnuct2SdK44HoU5FaG5r3BPIJGG/xxIha0EEF0hpo2L/RUgqRp8MV783P/rb/wPVwDwZ7/xr72QV+3tN75+BRRIq8/lYgVgi5h6h47/vsTLuEkvMZDdFVGh8+TtN75+9YMYDSEtZ1GyyjzzxAocc4DtI3udc+S6wDML/n79nGiKDbqM4++w7FluPymQ6Fq6RDkukpLLe8kUtQLgbAVMW1Pw14NiM1hUZCmXY0l4rJcpGRd9Z1S6qlT2DWJFaSMrVUQyrYNkxGJxQUZfpsmKFV6PEaNqvs+tKm7niJdWAVGAOVqEYoUO+1RM0JLqrU/2c4FobYKdfKvGXAWYsTOD8MpyDPcHVUZc0r6dDIlz7RJpSCnOHKHZEbOhUyPBvS6CRSt4Dy1cir1GuLrpM8kRm+b9CqUmVGsIjM26LUhQ67Tme8OCY6lHADQeZZtacqPcb48tEY7nqeth9zEfGCJeXz+lG07icQFsbzkH28/17657eV6WxueR3haPubHH7i9L0JMZhy6FWS1uSAaAM6wQYfRrg4ZUeHCNL8oGgwjOu4CnU8T/ru/hve4ZNmrF7v5heIqb/Yx/4bXLRKWneP9ZwPduzNg51x5dsMrPD3WFS/S4xoQH0uMVMR/fy6sus1NEBZ4ll8teI3ZSbMsIxffDLQbt8KpuwJrmNxirxXLM8AQAKDkiTADfYaoGE61Nr9h7RqOlMFw7dtsoSEvxWykwWud7hOb9jIKtZtFGtnbOA7RERqjsthCuwr/dZSWexoyPANQUut7jWIQW+BJOPrhXLkIjamkNl2xgOTq6TgM8wxdzXWxzLZW4B3SVF2OUiKc6Ju+IJcPPaaH0IVUfdmXb23vxvjzfF0BZJFks8ZQQLtemL+dqvkCVPC4oC/7SmVuk1HOirQ5kCX7VJiEC5sWqWdQsxM7F3B+vVfvNmKVhEQBchC7XA3g59PggAmcS8EQLdIBCQ5AtaSuMe/z5kmLaGvK+ngyAHAZnVIvwAsoaHXZK8oraXZChAcn73s6/j1s4/m9kdNGDci1+9kx22GDAFlM2AMi44r1/UTTDLgGLWvzaG79+9VGhWTRAmOfHz/yzY34IxW+6QevN9RgEomzatUJwisHu8ywhwTIserHkSzXheN/JVPolTZmdzMkDbkIs+fP2hyeNoLLJJ8D5DRzqGUGQPe8VJa+QCcsp82IRhv1kyIftaFEQq8FxvG1t1HeOZZ9lBIR0voyCkGqXOR4xGQKEejHqQrrdLmiOkvBaxrRlBg2CJc+vAagGPB5nPOjt1bJfBc9GxVkvULE6ZWuEFD3R5Ki1c+9nxaozSJpIiSSPiLiZI9YhYFbNkf0h1XqiE4dRFDPGjNaXELlL9Ck3ZM7rMPfmPuX2cB5ehA5RFTtVjFpXEw8w6PqsZEOzlY4RG0awvUbTp11iBytmu0NdQoDR85jaR+bLnVhEfJ2MKMpSLh0d6n4P93tSnX6wtD8AcJHx+84TOnI8QuWU8XHsu6W8P4G4fUpyNMTn/PqI7y+/8TeuYjIsP86q6v0pw+MUdt6HdJAs6U66gwfgQ92imleVW+yxgYV0O6zwo3qJV8OAf+xhh289fg/vTTcw+jBT9kaZ8E/NP4SXscZrYai4vPtgg7yDYIMBD/uAP3F2hsdb4HIFfLhTrDtjxTgbgPWguDibsd11eHwj+N64x2OMGXI1IOBhXOGJ7DFLxF4mPMYuw2+YVzEkD3xRwEv0wu65+JeojHN4E/IwN5PJs2QBnsbtcNB6q3p2hodPPOxQVBofGvRRFlPG03PVGrpTJTFK8UD6aI+kAcz3PrIzuGN8hIVt9dW+Bzhqz1ShnUbLoF3Bc6Ykfipjnn98iWqTEhKbj12rw5kO2Mlkyh8sndi3B3CecZfcxerQKxhWdpOiIP63mTsdhyFYoEQ72Dt8X9g/fEK61Tsp4eZDCBYArBoThDAsvvqRVY/OWjw2+kUkn7PdxR09L9s366FiT6WkbEzFa+NXK/b3Ch22Lg+BY+gP5lvLAYPgC/2AV9BjHQKupxmbNLOeoC716J9UW/CwRLSKkttG6vh7eu4CCg4XMPrxKY99pPMb494lBowaM0Nfmfu2uY4uUkKHxyclEzQn/QOoKvVSOg24xh43cggTYDv5+QZ93gi5xr0IF/1Xv/Gn3/K5IISZ0injGbtm0bSHFGaeTNSRoJ5cy7gW8nVE6etjEvGDmRPC9Wh3wgBp5RRGnTyt71YAACAASURBVAaxjdF4NAeqlb/9+m9dvYx1ZcDbHK8dTK20RVD5npCifUw0+pPibBCsetMTcp2NvRkg0dz65TzNpfi3z33za6NFLw7n4BwF+8kiGfsUBKRBxXPtkkE0R8nUuYx8bEfJBg60REpEgAd9MKhOMOawfVR8Z9zjC3HARW8nOJcOmyDYRoWqQqYEA4NFSyKs/gdSm3YwCNA2RryvlhswIODpPGMTAkZVvNSb43afbj4qcKMR5yFgB8VGAhCBJwAeY49RjO58huJ7us171hk6xBmZdKAVu+USnbPXOmpN2J+PVPPRjVCMErFRY7zyqAGu37cw1lP+nvs0nbptTl5GekjIJCGto9E7WI9HqYsx1J4fsByrYwbK8xr3PieEToelHNPWQToknWaXHO4TSr/dJ2LyInKyTggTyk/JID1mPQ6m7OE45aVs6oyWzIgIIvgj2UKi4NFth/fCDbbYOx7oGS/hAsR6P+hDwmdayPHx1ipsPtAel6HDF84t8fx8bdjN87VgmpELDF2czbi8GPHwcsRmPeD/eYIc2ttoj7OUZ3KLkBdf5n+E5MmbgGyAeCWF9weUjfmUF9vDjloIFIBSxdMvlkcGailwswyJ4XeHtKGH73lcidgkg0QMFNMq13kQJ7YFvnZpMrRQr2P3QUNDcFiE0YydgFHNM7JKwJsVOuzVrjE5DGOVB+IiIOtExxyba/p+9Pfkqex4H6tkeNj5muhDevWzokfBuLJ9vAZT6cgMxbwFn0y3SxFEq49R2lsWj3px6ETMAHWuPir9LfzKGyPVJus+98wtLwL58eOx8xfJhkr53htorD/Aew1pjhDba+eej8wHO/dGgjHJBGv3eYKA+Cgkg9fsmfZsbWRkKcejHgOHizU3Kj6rETGPCw8pa6Oc+ffOK+Vxy5+UHFtHKNkw0WNHWuxg7RjmKBz/e5kWfnd/GdNawzWSDDwWnSzjgspBdoJkhaPNe1uWA0/8R2r1Zyvtej+mdfr0b5aVD/8r2xOsD0/lg/zO6+9ejYh4CavqPFN6Nj2W3DY1bPQwh80M/T4A606wS3UuGFm43ZtOAABnK8V2tIKDx2QpQsL8jfHIpCD8irkl+yk5expDJyZfbF5PpeSbAMVg8H07R+CsB/ZzcdA8mWKqtwT84bzHl2Sdj2fke4Zt2QGSHLe29vrvAwRbndM+Y7kgF2Ir3KyKTYomt7fN9jJBfRDBWrucpN6jlFrg2jciZjr0TgSj0vjXFAWv+4vJ0yNiNkTIakioOx12ZlAZKoV7JOf3hFL7a48WTu/pfOu7bJmwWKjUx2K8EX7XvtjCmlqH0l1QxmPft06CJfjUlEic/G+OXatAhg9xTaNE/PIbf+MKAP78N/7Nj+yEeaFiha1CJhKgGjHrvJAf4rzDOmeliJsCqR+Zs/B/7Z4iduaVnhLsSZJltpMZH+gef2q9xm60kGYXFJte8E+8Jvj+0xXmCLxyOaPvFdMk6HvFHDsMnWI9RAxDxHoVcXFxixg7iCguZIVRV4iqOEOPcxiTw1OMOdy/E9vSOBHW2i1w6pPNx8TnSVDWjVHn8Ym+SmYu0ie2WGgKhVKoBLF/d5irDdVHP/xz02oQh4PBuyQVlCtHIDRNyJTwlQwNhh17CJAWAyigDQuQv+c6odzXx0jKrtsgBYIN+hQ2LAYbMaIs2OgnoTcigMJWdK49FIqdTJmtiiFKoND8MgrCitB8JsRAdzDP1aRk9KiLtbHdnjXEt4dRHBpUANK1Sv8zisXRw0XW80H5Zx1wPz957umFoEXr5KMhYt81hozoyehJq6aWKMjdOSSePSwbhunVF4RsjVsqNCFd69kcM/XnAMG11p6nEoGpo1FdvmY9TwRo+nx5QSdUbkBAXx1vhrCNXVsLvSECoF5jDoomlnPdxyh8XuPx3l7yRGud3wMZ0snood/8IHd7Db0wYkLHCO+b89Mi5TExN3lMeVK2YPkpnjzDJ67vkjHox88SdLJdK3/m3Z/+gYuCAMcjoKfEE13cda4f/+ZPnOwXixoX5ZSf0SifkkK6ZBzSg15F60WwCgYb2qQE7CGIVRCfSzRimq3uRoRmA4QJ4gAQunostnkiTB73uR5AMSymucCvjEXqyP2LGTQlOlHyQXguMwBSdCStuVMi6AEMxPsQFgmRGfhQJ/zhZFGMxzohTnaOG51xrgZLfdCFCpKrAB7rmNcxrqNDch6cd2WuWWV1ZzCIedd3WiBJt1oojQYNuESfyUEiSiQjiM1Re5al9gfnaola1snghMxyXaa+06PkSTIKojjMraTjifs39SzqD88OQN0FMu3lWG7HXWvZuGAYLMkp2K//3gujiPa+GDczIiaJ6LTNLSn5uMcS4D1BCiMrHjb3ceWJfKSK6RGKqJNFQ1CgWTREMoYehBd02Gt6nxLVZwAf4hlCEPy+vI8dxgoGwE2nR8DLusY/++AcT3aKf3A947V1jy89Al57oIgRePVBmiRRIBHJABGcb2Z0QXF+PmIYJlxc3uA7334Vz247bEfBFzeCL6LHdlrj+9OED9SMn04FGzGF1xLcQy7o1WnIXn77ziYRoQY+wsFXP6haBaJHyLUIRsfa5BO9uoVzZViCGn3tKPVGGdJ9sK5BhKKuc1Af2w58bzTZvXFzEOMKT9+daZ+fVVHqeN7acG1DmJwEc1LPvUSUwnW+zzZVjY+yeDIpX/R4BO860dFFIOUALUOmfL+wfeRoEQAXsMJsgySOcSAFMXUhpFuIAHx1+PKd9aSxm9V1WwLqUHTnFpwVDtWJYwWwAFT4aaBUTm8NiHlh/csRnoVoij7HeuQ3wogaQ5uLVqW/SdHLPqGyUsaTb583bu39LWbcxrnSqPtUKbeNfrVY5oiSJ8LNj23S/N45WdJ1S6ieCpeHRwpWYoZRVBsntxoTe0vBPPM+Tok0Y6jtB6DuW86jVu5lwLhNdyl5k8ZHPpdE9Cq4kT1YZd0X/ypz6v7xnLZfWJB2lNmqfIuxF46IOY+BeSqkFSZcaG7mPL8HilOA1+IGv3KU7z+IBkhbUyXC5d45QxIgjPe4ge0jJxzv3kA8Jv/z69+48vOh5DdZkrqf2wCjpgWCFWCed79z+dfLNXC9M+V9jOYY2k6Ch5vUVgXUGRGEQDF/g2vnfkp9Je533kMfy+/HaAZCzvuI5ZgulPwRAFgPZoBsx+RUVMsTWfXlmCAlidwvs7cTDSHNFc1HVWw1YoOSY3aNETs1bP9rYY11MtJY66Nz8z3ncYrgRg2mOkiJyookYyZa1GROa9akijMJ2KnpJ0GALWY8lj0iFK/qOsOUexGspEv7vUG6LOJR9IARig0C1hLSfcW85u9SRJO6EFB0prXaNagznaU5uoMRCJ2p7QTUDbiH8njCsIIWNsPo8oLNofhiUX8iQVooaxttaf8m++cxR8HSHGOUqZwjZBhqBzg6bWRWUDpsPRsr176l3BIPcSdhz196869f/YV3/uxHWgtPGiFL3bCkrI1ah9R9RIQGCFB3HhPV6V28lq19njYWygYDhhSgpbX7e/sb/DNnF3jtQeqQGdiOARfriBDKxJ0mQWQ1c1EMw4Rx7LHbbrBazbjddQX3GYHLleAfjvbg/gi3WCXYBh/QDM2GB9JmNqgdQ0989tI5il4AlfIPFE8nN3QqPOX4Ej70x/PYVpYgTjRACFVo2+OfCyckozFL0hpWNBa5+bT3xwJWfG2v51mivKftPh6AnM/iNr6iOByKr3MC2LjrIdjogFHmHFmxyXyoGAWY55VdSMgeAFx05pEyKJXgVk21rvtKwbyRllayVaipDNMr6Jmi6t8cPnP+HjiELAxHanNUbZGafrdLnseYilO1BbXyNe9xbkp7FD2Gdb2UenxyI2gLl1XncePjlFFJQ5EQKBo3APuuROe8Z50bIIkCYjWeBNy6B5RcJW2OCQKchZD6mX3LyHCsDBzg0Cg7Jsfud5kMoZal2j6teILUNkJ2jKrWcuisn/cyY5+UiceyxQzFjezuvG5pY912W48tw26C1bTw+XgzIraJffGm8XByQ2aUZm4KxTKC7emFuQGfGlefdylOkEOhskIp7Fd33693opwyaFmQkMeVdbsY81mBc8Yf1zHfdq5NJb8MGDrJ0CYeQ+TFOJvy7w0CSpeSwFsDJKpBartwaIBkUo25NmQY0djP5VgWEyTjFX/LCM2sdZv2c4Fs+fV21QExAM9GWNXz5AxioehdjHiq9syK48bpQ+kmJjcABgRcY8KtRuRIhJZ9gHToHXAQJxgTrGuH5HBIsVxGPewZWKQp8jxNLgipeQtzFTIds1fEIzTl8dnN0MDIdOLa4TY5FAMk51d4yQaMaqYPB5JzVUxn8eN3dseckntBVl/gHMfWGt5jSPtQq68QWNxrqBAobE+EEYVwTbV7ZpTLo3vSGNIS9fCGCI2WX3rz164A4D9458+8kDHykXNCnkfGVBCx9tTFpAB2WCfv9h5TVkrPdZ1vvEfA//10hyeyx8XqAqtecbsX3O4Fj84ViVEOfR+hKthPSYFPYdabm3VirRB0neJsPSPGDnMUvD/O2MwFGx4geCYjGIUxOEWXH3hISVcCe0CbZIFTKVapsd0ZxpMGED1yXUoaGsUUEB5/hn4x5OeVlEN1rd4EmKRJ2bvcHRoN+disCEsbjKh+0ype+TfuGEkLIaFWLd54ltrb6+FIdg6TAi+rjQ8fPmRUhufy1LselmP9Vdq70r4wQECwTomzTLi/FasJbEmvoTr/OhmngYaFiNFBzubdvtV4UEfFR8SoDBwTBeEJtRe7R60M+T4MMCYTKg+D+GdSxBsmxBPTExlwGBFZBWCebTOeVHPkBACmaKQQQEmivI+wDX4zosfT309tKB8u1CSDKMdINt7vUp4KQcOh4SsoOSCtAWLfC9Yo0Ml8Dyh0kx5+V743prMpcV3OADZBMKkASSG+dRuKvwPCHO+7qZXfnTbsX0TazbGiZ3ZOFSTv+piiE0yU3CWjn/KLb/5qVk5/4Z1/+2AjMyPB+rI1BNgfWy3wUG6YfkPtEA6o1X30poWkLnkrZ4kWcQHwG1/5zSsA+Ol3f+ojeQE/TSFcljA14Pi4aJ/xXVh1f41WvvX6O1eA5c6xn9uaRt7oqBiI1AyMNuqRk9MTg1OfoiNRSwX0/SxVEcBxTnAn1Ep/EACJUnfv/KlUxH39EEZEaEgUlivg2d6uvZ/LUigAzlfIJDqs/0HIlk9I99eYIxC6Yjx1oZx7neiqrOghsJ1S8cAQcDvHHC3ms1BNRAtpX97FmPJBSoT9aXKZXMIiigHI6xQAtG45D3UyQyJWNN2e9QxIsFmtoZI0lrxsdc57kfVf8fDTUNnB1o9LDIniObrrlHofgxLrUWCaHGd27dIWM0Q6tP6UDjVcPN/zPfKoWlmC59v1a+lxCP1sZQkyWuqxFf1yglRUvx56TB3mmLHjdZUqgAC3xrun9aJRkaNGiNxjIz8lpzxGrUebhskzbNOGNeEcazzAGX4svoIJxhizQYf/T66x0g5Rge89DvhgF/HFc8Gqj9hPIXFjm6dxP5VFYrfvzDAZA7a7Drt9wGY9IwTge/sRPQR/b7pJbTKDJ6ik6unsdBu2xDfvUiVJCpUOUvIGKYp+gBkkPu8DQDES1GBYOxgL0qahjQMKFd2ppxIguZpxi2Ena4zCQsGK5WI3x2ocTLDoFaM6HboD6mD/u7VawUiVwkDTIeSFzdMRdyhWumdqaMVju62tHkIQsJdD+lX2NxX3LuWsZG92ao9nLFsvTI1OCxPSCgFnKdwcYcwsL3XGdPJsF1Po91B59lC0Vny7S05J+T3vu2X+KgZO8h7JcijXn4sbOICcJ8FN17f72azmZYR5sMa0Ma0CsgEC1B47vg/pXJ3/IsnUGiDuNSamO24ahDW1ClMbjQCANQoTyJK6TiWHMK+2D9seaz9lXgeZ8kg3CdhGrWmzRW573Y69Kp65jfZZtPu8DJ3x/CNin44tikQpXPqiyejtfPg4ZTm6ktb3VHzrCXaVAXUfOvhffPNXr4yxrsMaPXaYKjjVzrGI5erAcixSXDtgrA1pnVFbhbieh+Y4nq9HyLAlbt6/8ZXfvPpBMUTIRklHze09SAHqiIXm9fOU/OZX/tYVYDl3ftwtEZl4WnTS/BJ6a7kDheEPKMYH155ZteK2uB0tb2I7meJ/XnLf7R40rZPBjItxBrpQlHXO3QPejLSmsfo44VhBDFo1p21k5yboqjN63fb6PO8c7XxdQFV9PQhwOytmFWz69JtYoiurrhhnYwRWncHPd7MaLXkUPMaY96DRGRMW6dBsFLDIH5PFh+QkEZHMhOX7fUZRQv36+RRjnpsPk6bhHUm8b9YF4Xq+c+sA3xdKekMDMJk8JgNkn/NPDR1zjh43aX7vpZRGoNOQxaUZqWmJaNgvI2YMUqKjdo/FoXkfOeYk8ux7fk6NC+tScZAdlyrH1cHZDa+T6Iid04WGCBldC1StntutWI23+Wg0xPa3giL5z9/8r69GTPjaOz977zXxqBHycUdBKDNmdDB6NEnc0aNM2KdrWXipdPIHssUEzXRrEZZY9H8+2YJVPX/sbMhRj9x+LWHUeRbECGx3AXMUbPeCJ7eCi7HHdgRudJ893Cy4Q2H0g94yb3XTADFIVsRKuzwJgPRgxSIDLNzTpYkziy4qVgHICZZ+svQnpgGtfbtxnucw4Xbpdy38w0troPQI6NVw1zSeJi31EKjQe1anW1jFd5+r4vHySwp6K/5e2igIoQDkEYf22Cc8eJt7AhSjxWBO5fsAyZXQP2xgIhxZTGijknuLGWfocDtHqxzbiW06Oz6PZYODhQzbp8L7LPAgzeeJ1XGEx5SFI9SPv5JBJCv9Jbzt2iPJ23FkIOwjvfspSuOodvneR0I8XGspgkGw2lJbauKEIr6ifN0XNfxwBY/3P7yhu+A0tdFXpEfxPHGD7dL4O++6YqDBIli8j1G5wKOZj/aeSbiENXjxG8OpAoXPK8fgbMeOZVue5/hjElPfDOgrqC7FR0X8OW8xHY2ktBXmWwOnNcn9eLHvBVtXC4O/btfnNir6M+/+Kz8Qxgel04C19JgRcSOkJS9OsvvKkhe4jYafEn7P5HMa6h5+00FK4T6YJqIwZ8/tXCLN3OfHqNhF4KW15EgCYFS4ibk2RxyiAjuHLfKUu5S2MKGPgOzdcNtPJaJBWaUIxrrHYn6ezxOJqthNNLjKitUFMTpdNcfWBOSq6LyWwiqoAylXI8FmVyI41w7vY04EKrXQAJmhuMGMl7A6YFf0VMleuEaNMEfbOWw8HSN7NvupXjs8/I7C63CdXaaTLYrvkL7xziRCwYCIIUG1irPbK/415D+jUhCqwqp0iLbU8Lk9joTjLmG9E0JF1e0n/nq9OxeNiTYisuScpRRDppZSPNtH2rm3nIaZMi/4JkXL+FuiBTSNBT9a/tM3/6ur/+Sdf+de6+NJOFa7WJ+SdmM7dWM0RAbpMOqMWS2Dn0XfgIIxvZY96N0nTdstJjzFiE0aiAuOVogA11uDaQFAVMHNzsKXH9xYgcIPdgYp4YSyAmIhD/zrBMca0OEWE4hvphfU3yPvfZUME+ZBxGT1W2jM/J+kj+TvmCTlIwP0AgM1FKTtc4ZevbRWbTuh26eyrK4t05ACqCBe3gApBcLINV3Yh1pl0U+1fC5HyUtGK/ZxTF5KP4lY9bdm1HJJ3vlZzPn8/plxEp3rkD0mzMVooS/Ey+7EFnZOzFvM6GSFTapKezsa+wd0ycBMm6/U+SkAMm7WC5VdbsL1d3XScftsfcSDSn/dluNY1i5FRLKxmLDXDL52sE2/F9s575sL4q+NdE7em48isB88VW+sRu1yXggxrO29n2xHI8y/oePDPGR1siwArCRgnaAgq2A0wLtoBby6CMzuSZAv38MfaIjyuT6LNkYL7XC9sWYD5gUgWcv3vuwBO4SYhbTm3Y9Kt6IRdu/Z5jpR/n4FciMUUYqHr4bpWXHcUzWt7pIalrUstZJt4+rzGAV5+42vX1EpapUkK4hpkLmhGUdLxczuK+0TZMTYHA2+aFwptLlGyOvKwe/TbwYhBM/+E2LKV0ZIWBdEpBgJimJUzLMZBoQ+MarBZYtJ4Dy+C/V6SegUxRscPFcLqfKUu5RpLjl1QVKbkpGxn81YojESxHIwpsi1ERhdscEuFKjZrOqqmRfYEmCMVXRSkkCF9+mj7a1MKTpO8cYM9SXAvO+EkJ5pn1kdW6IWoN6TyHTFv02pFfdaDBgeN0q0YokoqAHep6fdrdZLZ3gAxem6tH+QsrZdX4xw5Hi04L7GCBmnCBkupWCL8y1H/lActs8j/H2rKy0ZHLxfOsLuEupbvhg26ZPt3BEDeuhztvoeOSEAMjjiSOMWHuhdHkczRDbopMMeI3bYI2CdlfM9JkyYcY4Ba10hQPBeuMVaO6y0w5flDCsR/KkfCrjd27X2E3C5SRugaAp12t9PbwXfuY740mXAdlZs54g/jPuMZ7xEnwvM0dNN2NVWphyVCCjYuyqPAqhodMnEQiWd4S9f5CyHDBOTAQdDj1B/D8KEOFBRXbcNLwYU2FCNZTehF16bz4DjHs94oDYffk/RdF0aSNcY8VCt+GPdEkCqCZ+gMuphNnbsOhEAeKrPndhuU4wKuy4NVvtMECrIXHm/TrS7APAIK2gaCyvtIBJdCPtwMZihWLtPiUOeO0E31vMhpnacSTm+zzVzCs/6ThXbGFONjxJtOOa5XqH008hxl1iXCMlqoU4UkWTIaH1/fnNuoyVAUZrZmolREucI8Oc4Jt444nWWKC1bBSW4a/sNYcoKvrtHSNo8luXw3Dyf5PsMEKzRYYBgh5jHtR0FXHSCVWfOjZUKxmgezH2kcRaw1ZhXUP8czyRgHQJ2MeIP1aJv3FRbz2JbXNQaUJTnJSGU65SYAVjO46OG5uQIuTgrpRx/KD4C1UEqbS5KHU02J8BpWaPPhpc5AsqGZ20IR6Mqp8Q7WNgeOi2Weiwbxo5F6vSKuCxvv/H1Kyozz1Po8D7Jn7/2xq9fLeVVUAqkL5GsuPWjPd5j0r1xdt/cEIqgwAqpYG5o3GtRgttoPZXmgDq/DTAoFh0lm07QB1PMmeA9zkWRz21ICeRzygvh/cxq30dnaGgscCfKEAh9AvYo69wQ6rVviVXQM2sRDhbVIGOdAF1n574dFUMnOXITADwbnUItgnlKrFguAmJ0wLVDaoMOt5jxHnboIFbRXMuTm6F4jDE5gbyBnZL7s6JaDEefgzdDsceMGzGoz0q7XBPE8jhoEi1HQjwDKscFi+2SpnjXaB1e52L+Ce/XCx0nHHc0VrwutdE+l12gc9HrFqyJ5uFbA5bJGu6bG1KiGnMmWaIBYwystiodgzBSWke9/7zLe6Dm8x5bE0gZfMoB4Z8dnc+qhf1zlBkrDNgn2gKuwxF672jInb3X3WGAvKjYQJ2wx2hMWQC22IPFDfcYMcGq3d7IHk9lh0dxjZd1g8uUwP7SqsPNTvC9p4opLTLk/w6d4nYEPnxmeSJnK8UH82QeBxF8P46OeUYzRzkXTQFyFfIBRmVGb3vxUhY6XS7oHtdvVqbDPDaMDRFGD+f5o8+0z3/7JCr+rmsmFb0exvZU/pd+Lv+BBKdxShSvw2P8tSjBHc//VMaKv7e0lcoWixaxlsZSJV0WY6TC077yv793fme0wJJwo6XPAwTn2mdsqT+nv2YrAkvkN27zgDNno+c2JC/fJt29wBL9PthyU7DNgoZEWTQV+zR5/ZLse4SedS74QNlI7xWN5GaIsilFXfZLcM9qqw9zcyU1o2+n/1uQDJkFqix6JY9JRB2d8efObHULCl6f+vNcujz++BTWi3d5tzPkPsIzrxHKexGsQ7lmJ8C3byd8bzfhyRSxcspJJ1InwsISafeqeDxPiU3to7RPFv+/yHmWDJCAQxikvd4tgsMk1VPCir0DOssFUXvWaxYkRA23MijARyt4CKDK7buvfFR43NtvfP3qeX/zS2/+2hUNkmNtmiQuRsu8AcGI3H3kbnjVaSHpClASYme3h7bX6tM6GLVmxvLXC7D5RcjTnJwpTD7vQiKYcFAs1cPoBI0PbzxENeOCwvVwCBaxYK7Hqquhp3w/znZsVXQvGTZTLFAvhRkfYzRYVfaK06HUCc76FO3XQiRihkf97PiXdzq9jFWGp95gzmAaGiADAh5VBSPdvpXz81L7odhpyXPMfZX0kFXaIUPz+akx5nfCyY8JLcyCLdU9nYaMtFFa1AZ1K6BOhm9pbL0ib0ZJ2ndhxoVfSyfEbLTcdW+t1NGV+v6ZQ+xzPo/NuVNz0XRq1kspRp5Fiqmb1bXiWE39rjZ7uauuHH/7scCx7iMvstnZ7wJmrS1dG3SsttljjR4vxzO8ohsMCPiCrDN044c3PboA/OG1Yp+wky+fR8zRktStCJFi1YsVNFwB56HDd29mfOm8w7TTPCENczynNtDKNgOil8PF2m/CHo3MgS9qUREfIeH3hF+RvzqoWeE0cmioELpRrllDNLxRYgu7tURQQmTFJ1mEBkLvBiSP8ZGR2ho/rBjN8/A3K3TWX2lx4GTfYU7YzBSV0fQ7sagR80y8ISYKyxwEHMOYQZjqCWB3kPtC68lLaBzkkMaXXg9uxgPMe9ND8AADVmqVWZcrnCaDA4yaAe9PM+bbDpcDn4tBcnhuCj1sPgLiw9SDCLapKJSHCLTPJJ/PPWG/mDDCEGChdV85nZGKbCxooXyc9TBCEt25+Pc6hOwplIae9y6mLLbJC5my+DmVFcoMxaVYNGwjAR0sB8OewbLR8qLSLrE1fS8yTIRdup/LzAmwiNbQuXynaAnpvUhWQASSfXk7HMIAyk/98w05EvBJC73fnpmKQs8oIycHiqQj4/A1bfj57IYxlQcWrQ0Q9K4exwpWp4l5IEv5HqcgXfeBadGZUO69vPO05Fx3fU7EEwyFYQAAIABJREFUr3/lN67uWzOEURA780cbrzREOi2GMfvP2rlsVHFd9utof8ST6+sGPI+J1iqED3TANUZ0WOWxznyq+wI3urSfqtrWMKTFJ4jVz+AxFQzKGSBd8p5VdT+Aqt4Hoyj+XEDJBaEh40k4eMwcy3EsVJhSxLJRQWrcOS2mXD/H2YyKdQhF6U/HmqPHzjmmMMoYFUOo11xf/DUkGlwV4GVd4Y+MrBqPEXGRsktvZMIX9aw4fxrHAp1YFk0I1T4ksEgL52SH4pS7j5RCsyWy6dma6Fyd3D5LYTtvE8GCEYWkGiJqlqdfU9p9gefmuQa1XGAmcU9NvRxrW9H0Yt5ZT+dmtNek+IKC0elt/MzuNx7M30UYmFufCK0a0xXpVFHRytlu60N5X697NQlFe43nkWNQ/iXpHp39yNdOHRAQTg6uEnRrFFcmrB75bamWm3BwIvnzc6xxgQ0e6Rle1XO8gjUUimtMeJISxx9PEe+PM57FiC9uenzhgSV4DZ2i6y0Z/fcfR0gMuFgJNuuIIfb4ve0tvtCt8Now4GYqxXCuZazuJUIN756GzQDLdKAiXOBSRXlfaYco5hWYJGKfJqgkg2ZGxBkGM7YSU9ZeYo5qrLXHRYryrJPKQ32uSx4N8/zbA/YKP9vABHYOIrL5+Pofg3um3lPJUGBEiXj4JOoOtVeTZ/Hf+9/wWmuwamq5lwEBK3R4gCHVzkD+jOF6bxANrs/5v0fIxh7xih7KtkKX75U1V9hHq9SGM3S4RI+1BPRihQfZMzmSJWUcRzmsL8FE5FmBjSSPgxQKQPaFjXOGvO17bxwo0mbiPg8i+fOQGKz4v8ye4v2iGH+8nUvceSR914USrShz7/+n7u1arVmSM7EnMqtqfez9fp0+rVYjmfEYjH1jPBf2pcF/wDfWafvSxqIZydYgC2wNRoK3D565sBEMYmTPCCGDrwZ8zuAfYBD4BxgM9p1gMPJIstStc96Pvddeqz4yfBEZmZFZWWvvffq0mImXl71WraqsrKzMyPh4IiJa6GKHO+1LvG5mxuBd9PSkW0mleC49I2TmAEM2NZmX0ZIXr1MFR953Hlf9izQX5dhADnvvcOOzZVXn6xZ0i5G9aPZzPe/tnLdzTO+t/zty6KJ3w8X5MjFwMQH8LzodtfyeAOABARcEjBDPq65dVfDrjXddg8MYDTbqc5RnP07qrgfyfLJ8MJBsRrYtKq7JfCrDCaInJfJNWY8dQkOx1z7oZvcSUlnOw2HHHSZacKa52LApvZfrSllr/2ltkIE4wcUA45kz79AKHZrxjwD8rf/43/7fr3YCApVqvYv/43/5P//9f+c/+ltXr7/m+bC7swhYmmEq7qucceaAQIvVeMTp+vUY6RG1mqbU88iWWoVRWRlBsia6og29npDnR24rr7eOnGSUhHhDDr7MBrlX6CMRei9ejsAixO86wrEvPbBpD3KAi0pGiMaVJbRjOzS2QuM2tL2ZcyYroIR6ARWMlfP9VWnR76r8KK8NDDgncS2axYpAmAJj5tzWxCK0z5zbDfE/mb0i8Xndx4hwRIcbdDhhxkeacEcTXvMuFbpV2UD7FSAit3rt6yx/KsifYzG8lzykfXeLbGB4jr8ExmgsVf6vo3oxCoTCobRAnpY60HmjMNwFsoYnLPDReMzpPREeaE791KLTjoQHB9KAcZH7lB/I82dYVy201wU6lSfb8cxjEGIgd75e5AQx3lojYr0mr8nRdf0zC5XKXmJKsquVGZhy8WOVye299N3nO2XPV47XZXjzb0Foplrfoqvqiq9c8NcmWamAPE0LcnBpUCypRf1MM+5ownuMOKIDA/ib/ojvOlFKeji8cB6vD8BlEsbU94wlmtoWAB+mgIeR8PXHDnej9POPH85YAvCp7/GJ6/Ed7HDLfSp2Y6FFNvA6Y/qEpmjlV2u6TviJyuwLPTvkolfZa6CLWCcRE1IGMKCM59Bqn5byBMkLW0dShfZ6UqnSkoPSyomfBXaBONlNo54DVP1mN3cHikFqcv5L6iK23uEIn4R/ICslCnNSpamusLzVDx2DVgYx7UdNDOAY+7OjvEEuYOzIYR/7MsAXMDL7WRm0kt7l0BH2jnB05XhYrC2AhHkeXPl3FYRuIFNWEbQwH81wVT57eb1W4L0GlQIMrKCytO3r6EvzbD4pX+17WIasLK1uw/4FaguYKEU75zA4wjlINhjZNNuidks8rT0sPQSLrP8tzEsVlYEcagX8wozTEsQLG9vaOen9d3qPh5nFcgl5r7Y6sH0+VYJ2yHWMr1nJZdP76am1jmqLl/K8Ok1ltuZd2w/Ke9SZpyzZQElA+Gr6HNNJ5tgFFX9/NrQV9K+8NOHQeft5rlE95gzG//jZ/7SpZFxti6+PRDDzS5V97QOw9gA9Rq14kC2Ya92qWq4fu58zfzU5hsSAUBSyo1fE/AWQgrtr8gR0PkK24jkaH6JklQKFcaXf4lJUyBdQXquKQCpeyLFuR6XkaF+0r1Psh650O67KNxZjpNFrQ8yAxUD0BOdj+T4K6S3pO9jhDQv8SlPyatxmLhbIZt2v+VCx3/F6329RndI3KQzV8d4I0NZjuIMreIh6I5U/a6yeejYmCrijMQnMNn7EVkoHsvGn9gZeq2quEHHlCTVZnlmTnf/S9zaUVIuxPqVYYn1vQPimZiGU9pbIVdcwsonWz5p5vrZb8rrMj6UoooWzDtGQ/lR6FhzruRjStoXFCmFr4ZBAOGLAnjt4OPwNvsUeHkfn4INYdw+ecB49XnmPm85h3zO8Y3Q+KyDMwPeGDj8ZZ/xfXy145T08SYDWe0y4n0Oyyj5AanOAJSOWQnXSBI1Khj6/A6WXlIRSEmzfDh00jkRqnixwlCebDZZSpj4hx5So5XMyBe+ysNJm4LrZ2PHXtlXp0M/qZcgadJnPWz+XKW65ul/O766KSByCShHJdHAOHDq8RoeHGKirSQEk1Z9PbQcoQwqptonNwqBFDAM4BbFvKUeaaUWKGuVxVAt6AOPMAXtSL4mMU0eihJ1iVjNtfyFOdVTUgqNjzwDGANx64OWOMJ15pUDYNxg4ZznxRCnIO29Mayw0Vt8JIHHn743SYzPJJAyqsci1FJEtBWCK6R+PPgd+W8ufwstqIkIKmiQgeVV0/izUzqyVBL1q/g/RuHE/h5TDXhWlrTErn0/mlaaZdMVvWUlsZe1RRStZg1gsj6clYArZkzsyY+Kc1hiQSsZZaDDjipzcwHr+LJ/VTbX1LGwCCreqlus7teu7fY56lnKl8DINrqlgXSgUpaL4GNkAyWR8KQSPgBMJqGBhxplmnFPllDU9NcPWU0l4t1A9Vt8UnrRFjJxR8Klkg0gJWkhs3a5mF8zvKsOwnivYWANBHYTbUkzqGbaAk0FJslBmXpzaNUHo6tnsnPBEFxud2cRlsvAdrfPBnAV1vb8jKfTHLEHgltVMDYUlQbEUYmXOrxWKFun5h47wMDOYKcGplByV6VM17a6nmG7XyRgskYcMpi+qfACy9hgoslY9JvppwLqtA7YFY7UKg33jARneM9KCWx6erMbqXqlV0uusnqoIWXlDihMG7OExc05xC8hefOEleTeEty8pK5+uU02tLzLEgomo4IuLieu1AnhG6+geZLw5VQxZycNa4xmKv9omkJEcW3TN2NM6N8Ezq4QiQAyAj2J/a+0+RjofSiUlFgmOGbme2+ZKCfEVeyUQPHks3Fi1kbZual38+Vy3ukaYpYem7tXsHT0IHzBFZuXwb33Sg1mCz78z9jjPkgf87kx4fcMgBziFRHjgF14zfpE8HkbCn30UyMieHB7Y4Y+X+5TnekSpHSs+UJhudv/p8/TscYjsULMcaHrhAC4qpQMiNCfsHknBHd3orYBrr2NI+ldrgbPKkFUc1GOicR7qvlRvkbxXsRhYCAqgzEwWoAoIGZ+p76j0OmiMQ4+cdUE3FR2jAVpVXO6984TOeRHOggikHmKF1iwpPTmceEntUGRHPj6TbtjWy6NjZRUP7acG5embGGCE9DgTA3JQ3I3zKTsVcY736SFxKzWl2iRmLKcYfzEuwOvBYTozRs6jaDO9jDHwwhlh3BNhiC71KWJ86yrnel76jBKGYFewQ4ZJWSyzff/arr2PytCqxGhxsN7cx3KEWvjXtJFAqSBMZtOsFRDLwFubY2DGWQPuzfVp/SQFek0ZaljG6KgXbOecvK8lxnMw8G5cMMb+SiCpKGM61yeW8blPMTxiVfx6nouYF5si2ypBgCbBKK1j+nbUgmfXfWmRMzx0YxNTy/5jG4NdT9biaIuJ6m9PFWLVCHPEgDOmaLQo+X6d/Ub+xrg40+VrNn/7m1VIWte0rLaFYcwIFzZIX6kex/+sEQ/ye5/94dtf+/KXV8eV9D3a+f57n/3hWwCw1/3OD37/rWNK7FehVev2tG8ZXiVzTQTOM81pfT713bXmQ0CusNyj3MdVsK1hOQ9R5L5Fn5K/1HxToaNA5o/K+wYnAo4nqRTOEBiWBn4vUfmY4+chej68A05j9o4sIfMhVSg0aH2lNJkB1c8L5wyIsxlCjdfwlP8DwmfvZzFQHDozv3yGLi4hVyUfg6TEHQPjbObFOab9rt8amSB1HUNWXhXP0Wt8GlM58nM44Ctc8AYDqFolS7Ee9TtBUQLi+RBZ4kXMWtqq6WHJzvOp4n1pHOO8CFD5qOQL+v2jG4vjNgWvQtUHeJxjYVPHAimeSeHVnHzO1qvpoPCrLG9ZIV1kIonVqL34qsTI+Fklo4ynmFDK0Nr3Gq1Sk614/lTKmf6ybA1kSGmWZ9cKUDCfWtxii4Ood+q5tOLIdsK0mHCLimsegWIFBPTkVxqjuLh80tJOdMGJJnxNF9xhxnd2Hq9vF7w4BNwcAo47xndfMF7sclpeIlFEdkMAEadgsVfHBW92hD+ZzzhzwPf9Dgf4qDjkCRkgKWB37HHDPV7xgAM67NnjlnvsucPAOTMPIMI2U/aaKCwrb6iZFBNYaOFRGenZpet6dniBPgkfHUqoVT3mAGIGKo2HkEw+e/gCQqIMw76h3vy+TnVHq/9rq5dmW8jfPRx2ENjVHh6vXIcbI70enMOeJA2qhyiGWjujN+Oq0C1lTDb7l7qBLQRLnf2qkOS0fDImNld4LTTPzPi4LGnzG5lx4iWdTxAlxj69wtZ0rBTydD9Fyz9rAHeOnZmiNUst+WrNstQ7CSxsZZ6q4z6UnGnHsjo9xSogqJ89lBaROUiV9CHGjQwxJeZjMC5Lej+HvMlfE3+2FBCddzI/cjt2822RVUi1KJrGKvVxvu1iDNCt9zh4wi56sDonAsfR4C5mzgqIJQtZU2FBz9P/U/zPKAsSqhKu87RDzi4HlDV4HqOfFUDpm0ACatLg1RbVm68VACwE66n0XKjWc+BIQClk9HD4J7/0T962zvu9z/7wrSoWStajYN+pXXv1NWkP3lBA6vb1HIUESwxItDKvkmzU10f4U1Rg6j47SNXkum5VDcuxXrcdPG6iAqLzP8T9QrPd2XsDwjd6E/+291TwEc009TBnxQDxd6Ls/dBzFfakcSDB8F4l+3nU9Lchf9f7tNKIqyKSxpGiJz3GepwXKRegmbG0r95FiGlMM6zGop4IOyoNhUqaGevMAQtnbwIrvzEQrnScS37qADzQXMRdtFZ4DaOqiRvnXqPSC7y+o10T1gNhs4Su2iTGA82geI2tEg4glT5IBmS4aq8JKRbVm2xZeq+cbSrLbQOXtnuV41p80h6z/bfvaPcIoG0L9lVTidaxfct1lKxHJsdubSmP34yupRFvEf0rb/7d1dOpN4RQWpavkQOByCVG2PKCaNsKw1IPCADcYAcge09usMen4QZ/g28BAP/6YY9ffBPgnWTCclHh8I4xTg5dJ78pRvTh4nEeCeMsOfzPE/B/f7jgI2b8otvjLiw4YUmwonvMGLHg53AAoG5AWTbWZa5Wd0snmpNly8KZpJ3t7AL6m3pc1Aty5A636GFdn1mIUiajC1PHVRdPFtxqmsw7bBWnyULe2vJSC+22AFE9N/qohARIfIUGEU+B8ar3IMqVXj/OOdx04twSxf46AOe4dFRQm6tn14wr9RPpufWmuYtKpATFZUFVx6AnUQKswHjBYhi+KCSaAliv3ZPM5qN3OHqHziFtQFPcPKyF3NFaZFKhWj0yzyHrzfCQDS6NhclNWbrXRenQ3PuX+F7mmK1FLXsWUpBcsrqhmk3YbnjMSHEbep216tlz7RxqrTGFbMjvXAgDGuBp29kS9HQYOkjw6z4+Y+/sWMnfu0nqtlxYahrYDbur1pidK6LwbvNLXcc6L1UwA8p4LYYU4VpMsCPwdGs2sMb7AiiKCFpq8XgLSbA8LXshr6eqVG/KBVk4kMQcofheQ7QsFPa5kCWbHWfrN6Waa+wqmEUtlKhw7tklj+yJphV0TaklOF0brwWh+c4eq0XgTF/1vj18snJeqMSe114129d6/1bDoGOKqVirPd0oIfYXNaIl5R8CPVYj0R4eOzgMUfBmQILR4/AMTnhk50SA3xnZbw6yToVnxftFD8muFy8IADyYyujp2aO3uo7/UI8GkPdCLRAIlLEgaczU8wHjkSfgfgKWINDMOXAy5ux8Nkil+wbZG5WHKv/RfUP6xgX/ZHD6zcKcxLNLiUdqlilN/qIC+Z/SPV7wgNvIhQLWMoHlYRYae45z/QONuOUMArPG2fRsaMeXtGQGS6OZmwFSOHZCwF+5h3R8H5UBjQUBcszHiGXF447cF4ZfMQrnorS69nRdaAoHDVw/suSfs4WUAcmo9VSevCAUitIlZoM9mnGsIXD1ut6i1ppeIq8doxnAgTCgL2BZQBnr+lRua4s06ufnKiDAE+BYT9HAlGqIVU0EKuJA7L0YjAFSDqZHhwN3OKDDv3nc4zu3DEecPBwPZ4/dPoCZUjFCjorJw0ULqki7f37HeFhCysRDAP4iXBLOUCFKr9BjTkE8HAXf0t2nG5gVlHUBeIgSccZcBFRLe2UqNSX1gJhBKCxKakmyGEXNx60T1WIsgVIB6UmgTglza5hW/c7ssxJKSE+GZmUhzJ5bpKCN56t79MKCvdL+zsy4n0IKcpa0hLIpnM3sDwAQ3cevacDIwXgychBbDwGDWRe/nkegYoJn68Z6rJRmSD50pSKvOHLqYxVA5N3GJH4xUFBNaH10xY8QiN45ZCVj76iwnumGMkQcdK2AFEypgmcpnvqCrCxq/EVLdLFMrifAR0ujIwI7gQYwkFIFCySC0vktCyrDWN+SQvC07EzXsKn2t6UxfwHEAmBri3ruX4Z75CB9hZlJfzVG7BxijSBkZeeEBT0ozW0flds6hz2glrs8b0JUNmz/xHtGTWugrrWAHCOiBgrr88vnX98y7AahgqzDBnyLQjHeakyhKwJrvWFaJRDQuAWuzrEKMUXIZDmWT03x2Fd7SB8zFD5FKKj3KAfJmmPT3lrKtYJyIg0gxmlQ2Q7QmKdP3Eu3lEQRxNaw6LQ+4jNr3MoFc5F9r14TdUsEwoF7XGhGH2Mb7XXHKupAlQ/11ufZKe/0HlNKRpKRAxSTqOR+zWCAxUPupCMpzixZdFnS0k4hZruKQz4HYBengI/Kyt1F1rQNVE+JISjHb1kFA5B7qhfEO8LdFHCIhUi3qK4FotXWPQFOXSTRI/JhDtgHKjysgCggAeoByscHJ2M8BkbvXPLSz8zoYppyADhH490CxgeeowIqCp8qDALHXpJi+Ib3CGD8hM54xVorxKVzdQ9X5dbG8Mieu677tTW/WwqIPULmnPo3te5roWPLz1SZAEpjS8+SZe3CC84xvszGuImXMKRzLWpFkj34lClvinKgGjQWBICe7p2oyXo2H2iKMmCOh629X/XUq/fK2qtTnpvv16PDQ4yt03HQGGZ7v2tk5c7C+LyRzOOpVCghtQJS0x/+07//OQD88i/91tuyc1TAsKxlS0m8KvkcLUq4i5EMewzYxeJzmtZxzx7jArw/ado5D+8YD6ND5xn73SLeEM8YusiAPWOeCZeJ8DASvn8LvD97/OlZJuMBHq9dj/uw4KXzGGNA7zkE9ER4H6SW5wCHE+bVosjPYzHmAhUZeYkpVgOuYadtRpEyhiGn5a3Pl7+qfWcLumbOz/hCUSCS5Y6cORfpWC3QJewjUSE5qnKShQqdhKXbr+xvtppoAPiOCHdhwV+OjO/0HZglYLd3hMGJ9WsxG6Z4I2LAExFekMPHJWDkkAKUB4j3pLai2JiYljKcvUfr33pQwRDzNXLuDl6C35KguPZmjMzoAsN7UTQOcWcZY0FDT4TbIY5RAO5nUab0nXROdmAL9UljWyknKkzrOMlzra+1niyHMrBcFZBU4NDcR2M7bBCdxT3b2zjIHLOekS1y2gDwqKZiPYyPWYLW9zHzlnIWMsRnHINkudpFHqDVzR0BFw6Y4iZoDaqPYaBr0sQQatFTr0lv1j6wzhgG5PnWeqba4yp9qgWD0kPT+qxnaJXg+j6ihpSCrn0flsslSI0Zm8U8/1PeY01P9YaoZdTCEup2tqhnH2uibCvDPSgpKPpMgBozMr+vYSXXnrXlKV/byqPle0MByWOdPWU5yHY9J2wfa2HqgaYk3Nl5plZkGwNS72FFLAjNMStk3n/s7y6uMQJh4pDSzI7Ra9BHhUTlda0LMvgyTsM7YN9LnOjDSCm4PHlf67gzlpivJQg/UI+Kkiol97MR7szUaykk+jvrZ3NO7wkuIBUb1D7U/FF/q6vDA5lfSTminDhDPfZJwaMOJxaVXtEDAwTSrDBshWgnfs6inKixU2M5VQEhM8fTGEA8m2ea0bEr9nul+l1bhbZemapA2dT+eu41QT8bDIzcwNlQ3MNhjPdWSOg104b8VvLkWsi38VH2+FPIwrY8shcntVPVKNnq63OVH/V69OgwYYamlyGsk/o8RrZPHTvMJh7mOe1YenJ2LFVA9PMv/9Jvva0nmtKWAtKTxxBhRrtYhCqAsUeHPffYwae/n/I+ZUz6k3ux6DxMHqeF8aIHAI/AhKEP6HxIRcC8Y5wXh/MkjGbogGMP/E3f492F8fU84/sHjzn4lGHjwyTQl7N6DSCLzrr1OzNFE4NrTIY6w4IeA7LmrkF89QZAKDfKWshRC6rdclQpqK2yllEpaeYRFToByT8OZLfyCmtvmi3zWLfJKih6uUMpFPcETKZpFV4HA4lRJr13YsWWYlUeAQ7nkFMSqnCo0Kt0j2oJW+GnVpJyvyQu5bE1rkqOUoInIUMNLswYopfCFp4CgJcD5cxSLo8HGCnLy8vB4a8uS2F50HfZEiBb3gnbt1oBcQBC9RKzNU42Nx1jDZ4EZFNdOMKXmFdtXLMQJcWjoVw9RnZTqN+b/T0XkCzPmcHoGNHjJsHr6hGbwXhY5tLDwev76vMpafIM9ZC0+mPJ4pFbDNtCuiypZbl1vIZKte7peNvS9VQblvI1FXql/2GlHKSYBJTGARWs9a+19j/Fe9GCWKV6ScXGTQku9hwoF8c+XYu7EKWvJFUM1LPQeu9bG30Ln237YmkrE9ZWf68FyGZlSXl6znymz2LPu+FcVbu2fud+ZLpgwYGzJyXHlgp1UJ4ue9SepGbEJfKFHaROxr6jaDyI9yCBVzmK1coh8R9Dx5gWSQaysPHEbvAZ5cc5NqQcRYnf4Fgr5HHBSj0wluo7dw7Yw+EUjPJqmtYChBNnHt0iuxcE5PWrf/soI6jhUJvxyAkL0j0j/7qjBR27NAesB2T1rIYXKSog980qppkCyv33sYxcck2pgKjHYkceF0YqGKqKtg243sGjZ0EGeJIRU2+shxhgZxPjscV3cn/12Z/mnd1qS4lAyUOTfxeyMMjWev+mVKcCVtOpxhJ9k2fTfUAUy3ZK56fQphJil6VVQOpjP/yl335bB6PXFmiHCF3hgBe0j4rHgAkL7nHGiBkTScaoCQsOfAsG49Z5vJ8W3PMiwVgXmUxHP+DHd8CHswOzw+1Ogs++92bG1x87fDyLK/bVHnj/ABx64NVBXK9vlh69Bz5cBK/vIBkojs6hZ6lWPcWArxfocIkLTQPraiFLrQjqyZgiw9eFLBOpXFC1O83WCCBkYUW9LH1a2CVnUmERkCJuVthXRaPIohQFwGsBzxqHEarjue8lHKvOqrQwF73sVKA1G/HZ1E8AgPslw36YkYKy9XeBZgnTvukczhdbtiwz3h4aH1IKV/pds4dZKJv2UWIXBIpls4O1rNO19VdJ8KUujcO7eSme7ybinb++CDzwpnPY+bwheZ+ffxxlDHxsq6WAtL7X763FWlKsiIEo2HMHl4M5vZljgPStN14T2z5RhPFxzG5GgBVPrGJjyc5ReYb1WmkJd1bRqBmgzTwiEMeccezCATtyCXZnlZEWRMq2v1Y2rlm5ufis245myQIoxTO1nh3I2fd8JZQudN1KaMmvVoTe65HrDHxrooA9d6JwKbwLLkEWynbLOKzsyc3eAgIhImnzZ7OPbAkGVrGwVto+8uAdupXn6DkZsfR5r5H2TDyiGYe+NVd1z3iqxfS6IvQ0waQ2TljFw5Iqi/XesiBg4G6VBcuSHVeGZMJSOLFmIuzNX00Q4SgL0XvnEm+TWMHcT/V63Eq4KO7HaLRxwIsDY14Ejn13plQ1fVzq9VkaYSzcyRHhEiQlsxr05iBFRwdH6B0leFUal5C9IdYDguocNVbqOeKBFaH4tIiMoXyYAJyZcT8HOAIOsRiqpkY/xNTrYjwidPYZAhCSQZHh4NIeZhEPAFJdLgCxLIDAbGcEvKMLDvF92/VUG/MsH1QEiJKYJdaVuGvDZIuscRdPOF8NAuoNkfvkAsWp9kj0xp1IOHsPSgrIOt00VskbrNHoRFPkgdtpaK23d4vUSKLRkWeaILD+Mol/PfbPoXqdM0JSPHTsbEzIYsbxWpv2DJt58ZsqIMATPCEtBaSm+oW00wjKy3mPh4TAFa5MAAAgAElEQVRZ7dHhiAFzVD6kxoacq208REjECTNmMF5Sh0tgnJeA3eJiNiLgkwNhmkTI8w64TIyHiRIutPOE+1Em+vtLzGITGG92DgEOD0tUNkwGnFP01FjBRH+z3pGLmZSl6xHx3Lgg4BK+m3itjNQCld0AtoRK693whhnZCWOVBD2/9d2hhPQAdfBwPi9RAx7koXUWhImqhUTbvl9CqkwucCxKdRUuUVClqt/J2xBEmE8VZnWTM0xbxLtM+t2ZdgorOXOKn0nPkX4vs2nk91sK4cK4qRjPhTkpIACKwnYA8GEOcDPw6S4LQfezwJ9kg1YFIEOuarLvZMsrUh/TGJwCLx6PWeWilZSbuZAV43Nm74rCsdL4UYYebLVZPk9bWdfvuk62PB7A2sMVkNMk6++XGGgezLEtiEq+v7ZXtlX3XQWaredTXqieDxuMvr7nui9XUso3qRZ+s1ejtO4tEDiAXSdqbe2NQoKIy1Z89nPw0bYndbpeK4Zcg7Mq350gGPeJAnbR8n6SkNTVuZaemxFLaUFIa8Zu2NYbpQJKPZceU0Bqb8XWet+6dotqIbxFWrtDx1zhwgJPK+NuslJfC3C8ioXUzIEao6D72C4ax/Rsu4Wo4QMQwwxFg0jvRSnpo2LSe04JKZS/TEved2tLfG0ss2RjOew1U5A6TvU5s1EyCFl5sW3onVpGl7136ALjLsbc2TT38ggBnikZfGbl1w3eqwlHFO61QNKOK/xK56uOgfUY6+fXvMOEgHua0POQFJHa67ygNCZeaMFtY9lbWQbIMCdGO1nENbIeUIcyjq6lMBRQs2gs1PMYjDsaoal2y2vzefUexGY9n2mOisjao92CcAHl2q4Lr6qHolbcnquArGP28nOJYSZ7QlQJqT20z1VE1Jv902ZPTEpIHQ/yFOXjKVQz/BkzgA4jJPhtSCj8ULwshqShe+CQYjN6OBzI4a+mWYTORUSao3f4eFExUxhTT8CHkSVD0QiMC+G8MPZeFvcYGDfe4X5izEHTb6pAoDhwnbzZQ2GzttT0EDOQ2EIubcGxLNTE5pgGwOeaB5QEbBtcWzBV89ky2hbLVW/I1vdamLVMTK3yltZu13iccjCzMll9DmXcQN5ArCLCEGG1qzh6gDD1m46AGTgQ4UPlBz+QS4qIXFMG1Zd9jzE9pIqMAzisIBe5bkmOyVkSI1HlI49EwupW76TlnQGyO9+TKMZ6ba20XGMP2dInc6NzuS6I/gXkcw0x8A5AEKtfB8FJHzyABUU2r1Y/kpBanZNuET1Y12JE1nPqcSGxThddKyQ2bkMTN1jlUxWQOr6j3lxqN7o+1DUsrc62a8K5elDre9Sklv86zsCbYGjPlAIz6z4JD3ErIbcNtXAIhnfZvzUJPMK2VWKDn4tbBrCCIC20ztRkSXnwAqmQ3CIya7f9HNtekDozFmAEDVqKTFrfFA9tSd+RttrO0r8WslsKW71GW9R61mKOVAKJ9cbV8z9bwPPcl8QgGb5nC7c6SN0P5XsOAvVklqyCDrKP73v5SyTQ6iFuCkTZ8DEuoiQs0XjDjAImeg39qb+pkuEjY35YgnhDIl/0rvSC6LbTNTwlgW1GLcIS+biNwRsc4RX5FGwOAJ48TiHgPiwxYx4QWEbvEBmqwsSYRQFJ+xyJV2lalsIQqLKC00HjNVpggHiuenbJoGrXTCvpCyDrtZZjWqsgQzG3reYtL4iVsZQPikIrg14XxvOcFSc912bv0wxx16BHWxD7A3dJvnvs/C1aEFJBP/tMHdwq7e9z6JoSYA1/U1REenRX647UiUxq0rupBztQ5tF//wf/w1sHwn/zxX/+ZP2B/uiP/uhHTz15i37ls7dvi0YL4aDMN35LBwzo0sD18NhjwBljrIxB+F64xSuWWh4nLJghRQU/Jdm2tcL1fazjsCepnv5yIHwYJRXqzhPeTQt+ft9hYeBhljoQBIGjnELA686jjxhxhwxlmphxFwO8RDEQbU89GjZ9nF10EwIW4ujZyQt9F+NA7OY8oLbfynn1a9dAMutJqC06tZVHBVEbD6KC7WOCbP37ZBiZDVa219R9UbJB03q9xndoHniOlizNSuQM/Ef7cjDRgA7A7UC4RH5wCZy8IvZe1hshkKI434yL+loGKn32tUWT03zQef6K1naLydzDeqRqJS9gHYxYW6L1+gpFVbQDXE/Du3Pi5ZhYPivZzdXoDQAkXe9dzBqlnraDL+eQi5Y6vacKAtp3WzncrjFUn7fYaA3TaikHrUDxa+7hzXth7VUB0PSElu1t34vN7y2YDqBKbm6rjqmyND7qS8qekrqtp1Irnq11TuERolC9k9KqaNvcUsa3aKLtZ27BJZ/i6WjBMVpPahURnX8eLikvLetpXYjwWoISG0ReK4taRHbaaOGpttJrHlK9Vy2UWOVM9y9pI4+trQtihVE1GO7gsYdLsKOBRMFaALzscpDwoRdlZFqEP73eEV7sBW6ldBgYfccIAXAOOI+EaSG8e5DrloonqrAOlHDPnB0v8yrlhzbo/McXgdrdeId9R0m5UG+xBspbnnmepb5S78sZmOqOVFO1td9Kn8QbfmHG+yCFJm/Ip33CNtMTFXx/DLkuSD1nynS/XBhidL7+JT2ggyQFOqJLCkgPSdTjQDhhxh2N8HC45R42zjL1yzyZtq+ekMc40oglyUdWseH42zkqA6eYXcqzS3NUYarquR1pwYMpoaDzXOd3HUuqvGYy6BZd93c0pjUJ5EQY10j5ZNoPacakBnjuMNKcFBAxkj2+qp+bol2zYF0wJ0UEEGUEKGFZAIqK89KvNj9V/rlUKYonzPjtL/7Ok5WQ5/l8Nugff/l5uuHaxW42WxKGOmOBBm8vkBzG8lkmgFhWHV7TkATwBSJUnzmgI8K7mIrOg/DKe7zaEW53SEXxPswSsPR+DLifAwYXJ0EUUIUxIfYrwoCQMe87OLwgydal2U+ANfxCF2gPV7jaykHOY7KDT2kLc6Xz/FmP36CTAjpGAfFVjIeSCtN9FPId1oKtFYb1J7Um6WdnftNjNpOQbUv/6/30nnqsT8yeUnC5niPxQfl+gpWNHqHCi2HHMPdJXd+BOaWMtff2VZ9agsqWa7692YuHymYiU+ypixanyTB+nV/1u1LlyD6fM/9R/dV3os/N3FZAdBOq27LWwikqAz2VEIGl8cAUjzvkDcCOaTFe8X21lCu5rgzA1PejbcJcU1N9rMUM60QDViHRd1H0t3EfvbbeSPO7KD0uZXt5dtn7KxGyMsDI0CutDqzCACGPtb1frd4O8I+mr/2myofCJjXW4znBivX99PqsaJWGhNZc2qKuYZFzoJSvX4kMnwZE0Og5Z3nSdlrttfpu22pBF1RguaZwXBMYijpR8V654Gw+Jt/X9BRR5CnnyP4a0n3tczqgqYBI25zmt72uQxkrcOGQ6iSNLFCQZJhxlALN+2jgmBbgYaRUaBBASoUbwyMwzoSH6PxSZcIanlpkFRULEW2PCWJ9IC6ycU1BFJEW31Sarrl9q3soaf/tfryLXy4IuOcFY+yP7jWtHWyoDFGFwSj+9XEP1jgdb977jj0eMKfskpopS5WRACkIeKY5ve8WbLZFj5kG6t/tKGqckey7PqfmxZLhVoRiHksR6Ww4qPu3UDYs61q2lcYBxOrtwqt3MWbjOUpAiG0r3CuA8dtf/J3P/+4Xv/q5gygyT+GFVl5+Lu02lA2Vt/X59L94OPLbuLaftHPHAn/vB//wbfOHBn0rSkiLbEpeb6AqM2aZOGZAVUPt0WHPHV7zDrfosCexqswqAMZ4jQcO6EG4cR5vfIefvyW8OjB2vTCMzonwx2CcQsDCwP3CeOFdEqY4tne/hFgoiJMwOTFjoLzAHMq8/5SeRRbnCXPKx22vUSFzRgBT3DiRq3wf0eEAjx6Uip95xCJOlAP5gHIzqYVbB6QKq1aR8NHbAKBQYrJrvPxs/3rKGZxUgWiRVQD0XCX9LEWgpDAcRaFUT1PruW1Dj7eUqY4I0yIWJ41rOPjsJbL98SSCaLbUl8JvLfhvKSD181pSCJ1er/PIWt4ChMG2YFnav5ZVDFgrhfX1qoDoX6rG1pljAVjFgiQIQvU9MOMc64UUSlK83rZTB7bbe9fxQLViad/VFtnf7RxOm4lRPGos8rV5a8+xlZvbf/P96qB3tdgptZSRmlVbRaLlIbHZ+Aa4Qhmphb6aPFP6X7d77f81qs/zVZ+ssF/0xQjSW8rNY5swRYVDM/i4uEnqcVVEOnYxz3/13JzP0+DPrVS8157fUguO9xwFpB4HO752nOx5z92sWwq4FRy3lMTcpzYkLbeXPXx2TuqcPWNJAdADuaiwRLhtnPJ3U8A5Gmc7h5Te+zRp7AfQecbQMbyX/yFIliyFYSnfrWmJXvYxxDi7yJc15k73+tMScFpCqqi+hLzH3l/TNuKzMEpjjqPs2WyJaAvne+v/et9QuiUp6DhB4On3vKQ3wZC9Y2GBlJ+XUHieU3/i/xYSojaqHiHB6Wea8YBsVFWexhDB/IYHHLlLsgyne23Pk9bxLPiW59aCr/4+QKBRlrepRf4S44b1t5YHWj2gS5FaFgkmpcqIrjsNcj/TXBQZtHFeW4K4LTR9pglnKT2bfv/1L3/4uVWeW2vwmyoelhxnftcbRaT0uJe1mmzxWE5zYN2Pdj1AeY6/94N/+PZHP/jdR5WRbw5Ea1CdEUv+qvCQB1jdUftY9mbEnD4rE1vA+HGYUo59AvAVT1jAOMLjE9/jRefwC68Z08IYZ8K8REsFgGMUeN/NC07LBM0WAQCnEHBBSNlx6qG9xHehmY4kUEsyTqj7ToP5lAIYN9znwCvSZ13SyyMAn2BXFGpa0j2yRULp1vnELHX0FGbliAoBXT8yy5jrcDumIpjNCtw9ZSuSZg2ZzHrqKMcVdBtCInOJ+V9YCitpXAIgrnHbx3q81Vo+gLDwOhZirhjrzhFOcx6TQ0/S8RDhbqYvAHB0voB4WYG//quUvWVrK0BLGJgQAHYFfraAsDWu0+/c+A3IXg+N76gVT4sJVm9DjuHJ0IGuUjoUfrWYqWq/A8AY1ljqLkK6UnrnmFbSelas10MrEOt4E1F+l0lB5dU7WcU86TNTOyuZjvfK89TwbnjkjdcqHyjOtRtz6f3QGBNbVBBYW/HsJsrmmFJ9vs1SpL/ZxBT2WqvAPBbo2bPDQrxSSACkSuyr32h9zMKRivVAebOt64zoeanKOLLVrVW8VedBi2w8jk2lq4pF0deI4dBAy3VRwthxQ1uB8C2hSj0YVhCxlIWfpykO9t46Lrc8iBWWpJXte9lnWh+3lLNgZchXrWTYjGhWSblG6p1TI0AHwg11BQTXQWIWPAGnKCwrvPNhZkxBYkTe7CkFny8B2PWM3jO8YxAx5tklb4jGZTgIf53NvtEaD4YkRXnZldbucwi4sMB6tY83nnAOUmx3Cpz4GJC9yjqHrBHnHDh6m7GKvdN+EMm+rLCrmixM9egc9pD99E/DOcVDvCDhAoxYWyV+rgPhk+Er8lkLbNR1q94OzSp3yz3+wklF9doT+4HGAp6nT0horxUgp7RXnmmT+9ReZuVn9jc19Gk7AZx5V+zfHY3YsceROzBlD4YqGIB4JCYsKTV4qZATwNlbYTP+DfAYecE+isvnCO+SsZMK6LbiuaWJluhRWtCjw9/94lc/t7//xpc//Px3P/uDt21J9KejpeKVCyT5kwPhgqkYe+XtDMZIc+KrY4S+tRSROjHAFv3oB7/79kdf/PrnW79/a56QdlG49bEBfVJABvSY48s+8g49e9zwDq8w4A4zLlET101XLCuCsft0L8emRVyzgASuBQgjmKMlVwd3AmPigImzBqywL3U71v9VkPBEOMLjCI9aW7VwLEIWEHSR2s3WQWBeO5L/Cs9QBcSSNwKXWjGA7BmwWZgEcmO9GNn6MbgS/qACvxVeS7iM/Ndjg0OCPNWWbr13MO0MjtJfQJUUKrJ4WGpNYytcW8u+vV4Fa8Xm3gyEmufbDVAhYltxMRZiVkOP1HW9RRZa99Ok1QPWz6twKj2u3y0O2HqFUp9ipV/9a9NKEpVBlvVfsQSWmyFRO/9+u/AWYnpo+V6PCDXmE4Di/VzzjKDxexOmuLEpZoglrc61iotHG4Jmr32KkNayyG3FrLRay5WK13NLPavXqKWA6HGNVSss8Q0FBBAoqcAiSq9HwV+qmh3rZ2n31Voet5R1AI/W8pC2KFn/tqyUW314Km0Vh7S09UsLGlIfaxUmvFZs7TFxYCtlcsvTaz0guf1ScANQeOsoXS8KiK7hHZHEVTiHQyc84UXncPCEId7i0wPhphcPtyMJPj/u5H/vGc4BzjOmKRoRL7ko4RRjAuv+19/tHqCkXvkb73B0Dh/Dgo/LkrwfnjRlDhJ8WBWdAKuMlJSguQ0Nw+5rDuWepV4Rax3XtOaeCAd4SB2PNWR5y/OSa6GsyVaydwBu0SfDao5TyLGsZyw4Y8aBu1UmtGtUe1sIa09iQJmFNCNAsjxmYawTFlMBXZJEaIyGBnsLFCqkLG0LSgVDlJmAusaajE2GfAFIz2sVkDPNONN8lcfo2LUUEKVf//KHn39rgngkQV/Ue1yuAC/nbKcVt8rIN6FayrrmEfnWnv0fffmjTU1n4ZJF6oZzj7PUBcEOHoSXvMMt9wnapJNSteyHGNa0gPEnpxk/GWf86XvAyGPwBLzsHX7uIIFcn3QdXjqBPE1xAZ+NBeCCkIK/lVT5mGCZm9aOyGQ3fw3Es6/MV9CAE00FPOcuYi8v0Upg712MV+RUO7J9LIVkVQLUnW1Jma2vxik9R8Pi3BEVOdX1elVSakGylhn19/q+ZNqYWKxfvc/wHk+lopLjdrKCBaCwyM8BqfCkjncfN8Aa5lW2tY5rAfKG5avrlWnX/59KtZBcw7KAMu5DYUe1RdVC5R6R1WH3Se/MXDANJsyz6dD9EtJz6y3sewzMSQEtn7H92fbdbr4taNyWQLV1vBVjIm01oDmNYzW1CwOuBZzH3r3CF1pruv6+pbzqBtrF7WDrjo8pIg6UlIcWDEuVGVUyNHZNY596ZBip8jopClbDikQRySl9r493C561FoAz0Wp7q59T26D077FxsX18ilLZ6mPd5tY11vPQR6PWLkLZdmyhEmtL4zeFZdRQPtsPK4ioAlnAtKp4EJ23OhcUOij1sdSbX0J4cxV0YN8Jv9e/aiQBxGCiRsWhY/Q+7iPRcj3OhPNImBdKXhD1WKuxa3BUGGuAkkdaJcS+wb0TyNiJlwTh1hi+85KTMMwR+qR7W4gxI4HLDFgAipTnQLmHar8sL9S09UC+N5A9HK+d1LG/xJlgk7HUWQwno3ykwr5FX7JHU4yg2Ru3g18lg0jCPy3o4RJE1EKOWlTz0noeWUjWFsw0y2OS+GehPIfP0aK/Q5cg7wsCHkwA9gNJHbpWEgbLh3u4pMhs8QPHa8/rwN0qxbr0NSTFZ0sB+VlRzZ0mWnCJ0dgWZ9KCytn3ATye6KN+hwDAeDqv+lbhWJZWeFkOkMB0SdH7DvfJIzJhxp0KlyT4IAdKGi0jT+YFjPcYceEFbzBIZqQlM7KbnjB4YQY3ngRWAsaOHFyM9fjI+irExd1HxrmYvquy00MyK7nEYAkfMOGOBCp2y31yc6s70zJ9ZqSg9QUBf4wPKbWjLrpPMETsbNycKMeUWMVDPQpzLKZ0Doy95jE3QulgZmDvyyC6OtWg/qYeBfub5jm0UJ1UYwIGbmEgQArBcZHJaQ2JXSUgTtFtTbF9/VUdywtTwdTr6rK6AXiQpHQkUWgW1irW6q6173Q7aFHHt05XDJJ89ppxrFWcrrZKtei52r7G81ihXOelVfAc0eqdWtKCX/X3pFg4Owfk7wKzWVPMVhbEFJiUBiNkWOiXVRJtBhodA02d6Tgr9QqzShAsswFbD0c5FmhqYK0Mbnbj28Inl9Y6Ja6+l+dmHrL2dNSb0rX7tpQZF3mSKAFiQCFkSAOwhmJpykdbRMzWM9JzlPSudnyUz/VwKQugFp6TzDB+rUDFgmATaS7/JR1XYbq+R8sSWrSJNTzKfrNrrmW5B4Cn2GpLj46oeVvvCo37XINjta7L6ds1aF5ozx16lgxEisnPePzyHlv8Zqry/K89SmWwO1DCr2w/fZV+VVNAH6KQJlwa0crrMMRMSlLrgrGL0NRPqE884a8uUh9qcMBxT9hHBMvdRRSUmwG43QdMC6HrGLshglQYmCaHH3/MRiclzfqn+8XU4O8Oku1Hf2rx4gCBPk3R+PLPpxEvKMJ4mNFFRWpkxi5CttK1sWGFmk6BV3tJH3l1+g7JBEaUeWGRFTLyvRMH3EY4MQNJBgkRln0xigg3FJHaMJM9LHkQtSTBO4wJ5nTLA040i9ANhxPN+EgXvA77glvtqlXWKh7bqoGUIFoGEvoUA8AFc1KYevgk5GsdjH1MP6sZ5dTzoWtoH5PJW0WiN2vTKuCqZocKmqX9vZCamVptAPcYn5Ul6tsm7VOApAbuo3LJYAwgnHDBhBkOPRgBAdnrxYlPPU0BqYngVorIFizr2/YCAQDqmiMA4Cljc22asAkLxvg9IMOWLDYQyN6QGQETiVvuIyacQoCPQmjnGYde0vmRCmiUrSU9EW69BHop8xRtOls6Ss1Y0/NyYrAPERKmdESXFiJDsIgXLCnAK1kOo7vQQSpvTiQLcAePM2L2CRO8oZurtXAoX1PI095RCr4bXIZOARmeU8fVSeB+dqF7J991jOvfVLDt4nkq9Hq3hgDJey4tQJ7WnhZVIuoA6XERq9JgODxRDli0zSjkZ/Dx3Tvg2AN7H5USY1HZ2nxaAdHWQwJkF34r81d9nbWWW2vgY8HX9rg+p3i0bNAijMWsVMp0/PSQvkd9r/q+fOO7o6x8JEudR4RL5OeUmJbstdJ2rKXPzgeNLVLlolb+6qHYYkStujSu8fkaXRMqgZa1zl6bz6lhAU+5R13jw1KGFpQwhC4qIB4uxaXZc3R91qS8Bhu/63Gty1t7ALq4AWsbWj8gwbQgnkCbctPCMkTp2B5LvY/9Wx9XWqoxseNijysPf4ye4gH7JufWdG2u1RAuW3lerLmCaQdEOaj7sBXUnz5H+FlrXTShcVX8x1agfDI2cZ5buptrxqQOAhdST8gDFrygDqcg8R8PS4gw3TLebOikltfQyf7d94zjPmDoAi6jw4d7j3F0uEzi/UgeEMMXJs77Zf3u7L5R7yFADhKfgwj1n3Y9bp0khfnIErE5Q+DdzKKo1Lw3jWc0HDFEIZljwptaAQHK7+pRbu07DoS7sOAjL5K5MxlIERWT6971rYD3ot/mXd9yj9sY32DrbkxYkpeAsTGfTDuPUas3tTV9i1QB0fucI3cUg0eGW9WexC3oof7Wqp2h8qglLYeqx+sUtZpG/TkKyK9/+cO/NmWl5ZWnuI6tAtJv+ChqL9bW+77us870M1FCHiMplqJxIR1usMeRB7zkHV6HPTTwSDdF64rbwae6IXt43HgHR9kSfBgYS5DgNu/EFeyRq0+/W6yFKVuadGO1AVqqgAQIdOoOM06Yk1fDAfhIEwB5IXszie2LYSBv8OzQG2viAsFaWhiRXJ+tTblNpFTDakUZXE5LDGTmVriBXVY2avy/VUTqTEl2amlrqqTUpNdZ5pogVqZNhfEM0Q0/B3l3zigvc8jMtI9KjP5P96s6wSxt7TvxiGSX/Lbg78xnC8nS4yqE10qFvd6hhAO1lJg0HqadLaiYvjbdNCzsSjdZ+T0GfpuxZWTFQwM7070JCbaony2MUT8v6vSgMgj86Bz2cdCtgmMVj9oTM0VIg4WY2VM6J549uwnb8bRUj+tT6CnwrKfUE7HxYRZ+EJBjx/RYDv6tPCMUkjW5phqKtY8wqDLTF8F6YJYrzN9S7floKQEqUKrSocc65Kx9SvZ57d07c129ma+K3j1BcbBxCx4OL3hXCB7621OEnrW343Ha2lxb1edbtJUpy6bkFXhLhMBwhr9ZRdB6L9bPVf5V+Jm9czvrVd5jVAHpbeaxeo2Q1F1QCJbuZZ0Z/yN53JAY3l5Qh+/7HfZOnvEuLBE6RDj0hFc7ikaVqNyoEUT3JBJ0w93Z4e5MeH9yKe5zCowpSLxnHZMRsFY61s9cG530bwlp1uyUCxgXDhiZ8X7JFvE21DTHQF44Ky6PEXP5Ln3cB9U7s9OEOlgSRFz3hwsH3PE6fui5wD0Pwi16LGDcRZlmIcYFko3rQgt6eBzQoRUPVyfVeYzqM9Uj8ng/Y6xHFTOlCod6QCbzvV57DnQ1qNryq1Z2uAG+UDxUfszX0DdSKr5tRSTX4nPV8QUjpvSdoYijbYMZ0IbQ2eO2vX8h4Fhb5CMOVrXZG94DAF7yDm9YXH260V8qXKIIASLIc/RkvJsXvL44TAF4cYjF76JCIsWPZDjGJWZViOOlAekBSBuCHrd5sQVeoBM64EQzbrnHe7ogAHjAnBZgckUS48Rz2si07SM6aKphzdCggVo9IuzECK9as2RixsG7BG/po9DWRWYlQvxaEVHr+Na0asF3WsdVmK0zKNXn1YHRrTbDErNuIXtldBOq8bPavI3/SMJ4VG7GRZ5xGIDzmN87kCuF63shAhBU4I2MpcLJ5tS6Jb5X3eu2mKK+q5blvLTYronMi3G5qVVMiOXW6g3Rv8+xIGhAek11vI6dLFq9/egcBhc3+I32tXKwd/KOtW8zZ4iEWrIdyk3SbsI19Wa8Fa7VCkJv0Wa9AJTB5xlOJeSqc2ujxWOWutX9zAZrhdvObG2MPLYBwg86IjykDH5aOLUky1+2qPXOCmsxykxcSoMZiQkBY2OTUqFUDUdbpGkxi2MoIVf1syQPAXc4co/v8AE/pgfc06XhcVkrflv0lHMTXAnravNKP03qzLoo4YWWAgIFyJhpVejH7pV4JbJiYZ+lJUzVgekWEtaBYCsAACAASURBVGPn6cHEqmRFJEIowRjg8MJp9W+fYjMAYBdc2sde7QiHHhhn8XwoeqGLBsPeMzofwEw4XxweRuHvgMb9aZC4Ue5DKZTnGLM8lzLMM/NA3Usl4JzSdyArEkeSZDRfhQkXVkHTYwwx82M1fdSQogxcfxavfXmuZvFUxUniaWI7nFOjK/WglOC1A+E9JnyCATtyipSNxkmpo2Zjp9p7UrnWlCf2cBhj0b8JC26xx0eacMtDgpHXxWH9lfXUOma9unaebWbrM/3tIx+pZUO5PgCGzyjcXknlPL1fMgox4DbAm7L2yj4A4nUECW+wAeAeDr/25S9/q8rEt0kCxeohxbcz2qhPv2cvyDXI7BbvZNNmrQpvBfB/q0rIP/ryR5//6mc/elsfVyiWej+O2OGEC4aYjeGTcEiQpgvKSpU1XWImhHcY8YAFt+jxz06MN77DV3cONzvhAeMsgurr3uOraYkWaZeySinM5YSywKBVPKzyAcjiO3CHEwkusYdU0fwIxhE97jAnL8cdTcm6ZN1W6mXZsceFFrynC3Z8xAUCCdtFLfwUQrKovGhoC6qApO8NKU6FQiAzwcCCv92qp2SZsZ5v23BejlllhCHVtR2QqsUmBhxy3+aQvR0ItYckV86VA3EOMJcKTcifJZtKVkbmIPft40MkNqXPDtkkOsoKSg0VK/D5lUXNI1ei143VocQh29dQB11rWzonrPJhf5dryHyW9xLM99q9T8jKXOsdvRsD9k4skZ6ADxdhFy+GHOw/LjI3ApcwQE/Zc2VJi0YSSphETtdcnr8YJc56KTTOKT0/Zw/SxJI2U8cbeJ6VT4UPuS5vnrrp1gqI/VzHfBTPUikkDIVXZd5lhXIb4NvyJBBUWKRYFyCs8O2t7DH6PFu05elZ11ShVZY+Ec4kJm6KT1Zbyi3crGeXNvYyK2Bcew1FpKbWpndEj+9gBwLhwgMA4ETj1evrDdQK5dlC6JvCfR2/8ZgCYj1hTyHbtxAVEGCRxCXsskAZFZCLgS8/RrWgafvvQAUES9/XjrejZ6xyaiFY8lkMgWcsuLDEJ3omvBkkJundKHvY3jnsHWHfCb/47suAh5EQosHl5Y3xMATCh3uPj2fCw1RmoLJvaqpi1ADlN0YZMUpF8iKypBXX9lIMGzLv7YhSQggA+BgIJyw4QhStoeZXyP0gKvs8seDvLyaYva7ubtuRMc8xocorj+SBgCSbfI0Rf8YP2EMUpcC5BMEDh6iIZjidFaZrkqukAwMcPAZMHPB1FPQ1rkzX+i1yOtprGQhbsSDyfDI/VAgO4KtzUM8BkILSdxxjoKKRIMlwRgGx3hL1zrZhZFk5aXlJrLFEx9ODQCzK/RizZBEIv/nFr/xUCoh6Q37nB7//VnnBU6jmWUmMMu9A0jtLlfYFS3qnnPYj8YZo3J+ovNn7zMjRMRZqVXs9Wmnjr2UQ+9Y9Ia14EBtd/wnfAAAmmlOwjAMlS+AQ4VbKIC2D10nlUgVInyb6KQT8+YnxbxwcxoVwP0aBR4PFohCjC3visNqgBeKQBRW7kFIufKNIyDUBZ5pSoBJYLZnykurNUIO8tHjMp3zAAMF+93AFNAgAzhwwBjKVy3N/a28DkL0VtddCyRkmqAyxhXFdxRyY6xwBDwsXQe+9IyyBV0tcLeS2T8mrEftrvepkrptDKWzXz6PPP5gpp4qJq7wa4vGS2AbmfKwm62GorfWqtCh+uCYrOFu4l1UstD+I9VusnGnnClGO79FnrZ/fxoEACqOSz3oucyzqxfLOAKDfEbyjlM9e0116l612HREu4AgZawwU6nvncddsONazEmJfJnASCmyaacDUQ7FGAeMZtAHqrQ3VeljUW3LNY9LygJTKSKm02GMtD8Q1PLMqJNpOXwQQZ4E0WxXX/WRz/tPE3TVlwTS3oHEnKRUwZUFVoA/Zo9u6r25m9X3keUpvh60lor/LM65Xo/b1ghlfY8QbDPhAI6YEh20LLn7zl/y7tu/g075yLUtXht+1vSL6PPbax7wXi5l5pfLVbvOpZPtXeztqL0lfQeXsuV31/AT1Zsre+II6nHiROAWWWh977+Ad8GAKTg1O1vl5BoZ4WODSkn7Xu/IZ+2hEuRgvR085IcvMUekPDO+pgHpqKt060YTjNTy3VRurJmbgSB4XDjgjYM8OCAapoFZ8cOI5ngiXELAjGTP1TgHbxj9L6nG2w6JwNkBkiCO6VCRZDRfgGFfKIcK2NLnOdVHWG172ERN6uATJkmfLsJ5DlL2ekxVSKVR8TNv2EFnPzsWn1kJS4XbCAltFXdtuxXnUNEVDSe2Pb6XJ1j16Qhlz8m1nv/qvvvjbnwPA7372Byujfot0HD3cpvIH5D6rp8OmK9YsdwuWpGS0DCz6m/V6XMvO+Fia9L92ONZXdI8b7PGC9zhigGfJBKKFcTQji8ZPqNvNM2EiYaB3NGLPHe4wJQ/KiUVrA7ILF7AaYRmodd54UbmQoFg/lOxG+5IHfO3O8DE9pVqzJgg2qGfJp6+CxwkTjtwXyksHl76PZpE7iED3kTP+c14Yn1CHibNw9dJIqHWmqzruw1HGulpSYbwmPVd/t58Da1YU8X70MUherPCEcRFFYPAU2WDZl2vkxNQKR1kwllZLr47NBEXI79u7aIFZsvKhpB4PZk7Sc92l0DiWFJJ4TQGVqshWRG9aJOMl4n6Pip09n3MiBevtqN9nTRnOZvoSlbgPoyQBsLCmKQBfjwtuuyzOquCgHg+FPuzdOvmAvYfOOfVwqVIrWbFyhxZmBCK4qKQvofRwWUuibr7XRDirwHi0s+I8FbalZD0gQKlo1Ja9a8J4uibCGraolRlLPbBZASjv+VwFxCq21pKt3/XZ9LsqfYpL1y3cQi9qw4wlfeZaIA+Jr5cKZN7c2kK7h+4BI06YUsGx58SBPCbIq/faWo3ra/M82GZkLTjZcyBbAQw84il6jGoFRGmdSrl8Rk0woPNXvzOQEqw4UDruAIwckgL7jmcc2eNF52Klc8LAshcwS+ziEoCZgPcnh1fHgL6PcyJ2RbNgfX0vBhdbpJaceGeth4Ih53SOYKsAhPhj7XXwyHBWa4DTrdRm8kv3JYm70Hd75oB9fGbrLR4qXqPQakb2ANeka1wVl1byDsnOGUsDmHNk3DuMCDjFjKNH+KINyz9qRUTh5urp1XU5I+Auehn33OEDjbjQgluWeBDCdQUkQZ3MSJbemDKWwGZjew6psD2gi4UessKhqXstbSkj6hkU+fBpa8/2f8KC3/riv/hWFRBLNk7kqQqJTZ3cIh3zCSMIhBkLtID4jIADhiTrSpzIdtHSWkF8rHhui34mSshWhzUr1oKAA+3wIshy/kAXECjlSwdKDWzCkpSNDjkHvWKRVZgPDPyzrx04BoUBwCc7sQWdA+MUQrEULdZbPR8X5IrXvlo8HVxaRDuWjeuECUf06NnjGLF2dzTihLEIpPxLuscOHRYEfBIO8CDcsASC7aNH5y9wBliyKwxxY/RRbPizecQeDj8/9FgY+H8eRqmB0rskGN/PjJtOcLeW9PdxkcBtVSrq6SKMX4TLLjLpKV4DyPf7UVILHjvJQtW7KFQbgVSzW01L6WmxmbhUkaixslYB2XW2zSxk2zaAtUA4eCrSOALZUq8VvuuNSIVoK0xb93maN7RWQLT2i24qPahQ3NKlFGEQ0auhaWx9HNuUWlk3zvjiwpK9PoT2uNl6LETyHqdF8/OL9uiJcB935RvvsIvjpOO8cPZaPSzqRaAC6tc7UWKUdMvTvvZOvI+iTEhqTHD2ZCxxvDTGSZ9dlbzHUijXVeOhY67Cs2kjmGtqQdSmkqw9X9bzoVRXSM99yB4Cro7VpMdsim7lMVYRuItpWvW+tq3n2h+vxX7szWf1wMp3Gbf3wcIZHFzVF21rQoZrSJxVVFaMQF2n3BUDTrbMqVci8+QMtQCAk/GeqEBR47239p2WUlGfaxU1hY1tVYDX++XPuS/rPuTzJIB17f2Q4ypY5TG3YxFWra37YZ8lCWQR0lHHhNQeEPtsgxEz1euRC2YK7MdBgqYB4CdLiMYUxhiEV59nKSK4I8KhIzhC2pdudozj3jwnIyXK+PN3HvdjLtgXmHGMvOrDDDwsAgu08ZIdqIB0Aki8Sz+n2kqUlZSA0nBjSTMSLpUH4h4zPjKwZ8myyYgGnhCzxbkyff2kG63L+4blN5q4owWb1Fi5HYnlXfmcFDCUAso9HN5hxHuMAAZRRCJPu5j1ccIildchtdFc9NHYWmh7iKeDIRXIfbz+F8KtJOTBkuJatxSRVrCyPZPQ9hhbWGDLql63uyCvT+UTWQFaq/7WU2IprfPqM9A2IlhIfc8ev/nlTwe/eg5tBa7/g6ic1E93rc6QKG0dgBkzJEPtiBl79BCA1jXvkyTztQrHY8qHgDfbi+2v3RMCZIZ4ojHmdBbrHxNjz10xQY/c4RI3tD13ONOMPTqxtnHeUEQrDWDO8RSWUqo6xPR3THEoy8Xa0uSVdAEBukjlOXTx3HKPE814xXssyNlwPDu8wSF+JtzRlK65ia/AVtzew+NsYlU0TuaCBdMoDGYC4928SL2NCGP5GCRsrYZpTYtsDA8Tg1mEytMk7u4P84LvDl1yfZ+XgKOX6raaYUyFzodJhPS9k3S4rYJ3mmVLv+8pX28tXi3S4ypkazyKenLqeJXFnGvb0DgUFai9Q5Jy5TjBGSVASa1VAivKgrVSnX64IKPU6dQbKl5aWP5dFoQn9Sg5+bwFf4q3Sc9bw+lU8VHlp/eEaZYUmFOQTHEKWdA+7roMnfAk90+/c4zJqBQRfTc6nnViBCAqZCRpl4ESqqa/a9895fFzoLxr8XNybKxJlcJvQi1FRNrcVkSa7RhvSOJTBnqgFkmlJLxAa26EbwR9sH2zV+v9rNKzdy4pbvvIS6xnSVOVqxekft6t55cg0tLqmYSHJ9A1iBawhks9hbbOs4LIY3ErzeuRA9if4v1owckynptXx5TEyHHdG+PhNjP81MqHkiqQtgih9X5ZJU0t+6cQcOIFFwTcosPRuZSiVnnNbe/wai+pePe9GCfmhTDODkNXekIuo/RNveiXhZM9O9WpqsbisZm0ZdRovSEb3M4s/P8U5MwjPF54j58swClmyQQ6gF2RCVF1jgVyrSpqzMA5BOzNpnnN4LL1k8ayIt7jPcY0o0+YcYRk1VOkh/Va7qvRmoxgr4ZWKTEw44IZP8fHNAeOlajYUkSuZRq01MHhgebC+/HU9VvczxgwJvMcz23Lrvdra98aCCZafur4j2+LfsMoJ7/32R++fQr/UaXAw+MGHvc44waSJCrvQW3FrRV0/pR7bdG3qoT82mf/7ZPcRWdMIEi9DEQ28x5naFG/V2EXO2esN5wDkQAZKGWculkvYDyEkDJiAcBXF5k4Fzb5+FkrX68tnFZL1wBRFUj0+jua4qYqQZOWGaZc+SSacgDjNe/wgBkHdKmS5x1NOMS6IT0kK8Vd/E3dpIB4RZTNzCjrE9xhgguEXXB4gR7/6lFMTQTgsgB/dp5xG4N6L8wYOSCMjE99j7sQUsGiSxCr1ftlxtFJ9o9bJ4GD9xMnKNMYgFc7UWh6Mzdt8HoNCQNyILyFdqnHw/lSubACrr02BQAin9v7tkKjShBhHbBvT1ePREuZkr6oUqLPyUWmrvz8pUdDlQKpAFx20FrM9bMG89t7arv1MaD0Ttnn0+dmFvz1aeaUujhZ+Um8NLvYtyUQPswBB++SVc6eu7DWhzEeLaNMLqEc02DWnt3ULeygbkfJ6IppfJzpTw1JeAo5IAbDmz4+cbOU668oGJWi8pStLxk5Kgx0neLxuVR7kOu2xIPgckyKsWgDMLFohAvHJBnkcOEMo1JjjW1T4/m2SAUNzQZooVlbz2FhGkqtWIznChu18rMYI1F9jv2trh5t+1n2QT3o6sXKClvttQFKqFxrjpWZwx4nVUBsEcSAtdejZWkesVRpgdcCokKV984lyLDun+rd7En4ToAUC361B14eApwDdsOCJRCIsgICAM4zLqPDn7/ziZ8pX3cA3s1LMwkIIGtuMwuehSZFeFTKlLVxLrMUJJyY8THM2JPHrXPpTRzJwzFwxhJT68sgH6Mif44emiN5nHjBxNn4ol5Zy89t15/K1ixE9A0GXBDwNS64IL8PqSdCOGHGBClpYL0earRV6M4u+j2O6HCOcojN+tmqjP5UpaOOp0syFolydMs9rJK79bx1inOFQ9XX1t5E6xl8Cllvi9LFoHEu+BdHAalJM3P9gwq6ZeGmlhYsyYtxjzMGoxIoxE0LGWrgeisG8Cn0+Rf/ZXPMvjUl5Fc/+9Fbb+BHW6TudHX/iPXF40dfSiXF//4H//jtLQ0pXWEAxwXCuOUeI0JMVRdwiLVClEYEfMWi1Gjg98c45uq6tBAsQAR+RS9NcXvUBfceuQaI/u0gm9IbPkQvzZLyqOui7iBM+oIFdzTBg/B9HLAjh3/OAd/lQ5oUF2j+GOAVZRzV992QihJ+mEWxuovFkzwI95jxr/UH7DvCjy8LAjP++HTBnjw0ANumNL0hYGGP94vkbP/e0IkVm4HzwngzOPycH4pMVoGlBslfPMii/t7BY/Cl0C4W/TW8qvMqQAPjTGlT0VSxOx+9EpwFd+tFYRYrPSCTdBWUbSz5F5M8Ru/fd0g1L1qknhKFIjEy1ElJlQR9ti3hsENOOQzkjF3Sn3VWqfo+NbRKaY4ZxOz5S8gKCFB6nWDOGy/y4OqNYZYChDcxG9Z5lvH5EDd4ZrFeyjUSXC7zj9DrvY2CZeFfdqwQCEsIyZpuIQgLSigCuGzDekQsORjlFapUcPF7anODtmBZ10iVDLUSbpENuFQBr44NAdCMD9FgwNq6Xae/rGmLz25Zyev2FjB2kHG58a7I9qPvSYSlmJefl4JXWp4JyPvfRSjI+EjMhn1frYxZrc2ylWIWKBWIlhWzVjzsN2d+a80dBxRV4FtK0HpTz8pSqQpkxaT8W/XX3GMraN9m97LUw6fYD+1HX1mGFdcPlN5+jfWwHnm97qjQrqionoMYw25jRfELB7zyHseOcOyB92eJ93q1J3z6akbv8xh1Q0iV0AGJAZkWwjQ5jAvw7sJJ2Rgc4cY5fDUtiZfkdYyULSTFkBkBX/mO5RMc8VhaWbyGcCk5RLgq9egiLwSyonIkGWetHdbDIQSPCYxXrkv3DHFsjs4XCT5sn5SvnU2yE/Xg1spVTsyh2TtFuSUQXmLAAsZf4gwHwgv08o7Q469iOb9b9LiLgeeKrtAAd6W/pAecaCw8Ck+tSbRF9RpRAfYSFd9LzAKn2URrGuBFSTbZ99RrrtXR9T4Wuij86nGZdKuflnbIVdj/ZaDf+PKHn//OD37/LSDPxWhneZSafRIqccaUCocDQGf4i1VWuijjamata1GKet71WLqfMXla36JOb2hLuf/mF7/y+Qe64L27FJYiXRSSCUIG545GvKcxeUEI2UJxikN0MbAmtfTppm8DeC5xUWtbWs3Ypgx2oCJQXhl1bWkqM8/kDF7qSv8kom4HY5mU64AzL5g44OXgsO+kQvX3DsLEeoiFZQLjBh32MS7jdedwHwsWnXnBPS8484LXncdtT1JJ3RHe7CnXHYlSxKc3wHf2kj7x0JceDkA8Ed87xGDE+Fg5v7vpuwOIGLte/usx/dvHyub6WY8nBcSX59fGKu9sUHg+vgRRJDq3ttR7kv7rffW/9tp6EWzVeCVVTGwVeUtUfZZny4oDmd+0HVsQUnHQ05L/W7KFIWsPU+eNd6haYkvIGV+mqMzd9pKeVxXPOcic0IKJe69xGuL9uPGxRkN86UTtyvW26KGOQUsBsUH7NnNYAa8z1s80rpQ3aku20KN1Dj8WU3LNXd9iho95JSwEoJ4ja4GZkkLimZLCspVZ5Nqm2PJ4PMWDon3y0eOhpIUjH5ZQvC8V+npQ+g/kjFr1xkYoq6grbQkYwNoT4Vmt+VT876s0nqsiiI98v7bZ1b8pT9c26k3UeszrOVXGfqyv/WlJs+DUMSpH7lPaYS1AaOMnO7hi/6zjhdI4x73Rg5ICogVWJT4h4Ege3x069ER44zvc9IRDB9zupU+v9oTDsD1/AWCeCf/vTzr8f191eBgJ372V88/RM39aAj7OOY5T4jPyZ+UjGZ5UvseaF9SC/Rav6AzPmwMnD/jMgn4gqLwg/0+Y8TEKb6cYR3XHi3gYyCUvjhrctliUjWVTmqJXpvYAeUhM1w26lFlTDbIXWvATOqc4jpcYMFFIyoZCKqVEgASenzDjPY040Yg3fMCn4fgsg81zqOZ3DoQDd1czLG15SK7VGbLnbT2L/vaUZ9V1/Bt/jdXNfxrSDFtPpT16dNHgvzdpmGeElefjKRnL+phLT4uabtG3Csdq3WhLS1oQ8N998V83B+lvf/mffv4Hn/3PbzW9XF1czAYnAgJtkoBCOUc3wTM0pW+5uFX5UEYbjPJhU/OqIiLPIXSBwMWUkR9jhoqcTaDMPKHtAsJkXpj0wnbiTxD3ti7D05zzihNEcfi06/FxkXOmqHQ5Al7uCJ8ufSpqCGQMqnfAy47QeeDuIsIlM/AwAa8PojgMHaHzEi8CiPdgDjn17RyA286lwoKd5wSbIuJVLZI46CkdsHccIVu5IFNHjHmhHBQdMrwq1btQy9FWgvP4nPNicry7NZMvyoxwFpaTh6Py7FiIVu2BUYu//WvhXzW16rfUMR19hKRNoX1v6+1Qr8S8iCJi4WtzHKeFJXHAEgRqp2328VltMLoqIFKpPntCDh2le1ulIxofC1I43hz7bxWCnkS49SSCt6aatnU/7FikMQIK6JWeun63VMSOqPC8JWAkSMTGhrVFFnplr23zvLZHJLVVZYKZG54Qpa1+1jCeax6T+jdVJI4uZkSKSQuSUmsUEBt3p8f+vX/6H3wOAP/bf/i/vq37lhR8ZJe99QgF4w1Si69D2ythvSRPjSMpgsqLcXictD9KU2xHg+hrWFbtzVh7QBr9e6Qn7bm1fc0aEpY9Iuq5BzJv0j3J3s8qpvk4Usp4hxhHGeNAeohg/eNxxtE5vBwk7sM7YNczPr2Rex4GLrwggXOcPzngx+963I8Sg/YwE14fgJsOGEe5ZmSGVTtVEJf6WSFloAIAGAXFvkf72SbGEOWl3Btq3qYs5Mwca+WINVmVvNsIcZI4iiXClwT+fMaCF+jgo9HPI2eVBIC9uVHNC1tUK1B2/QeU/Ek9BhMFvMeY+M0DiRJyprlqm3GKMtQr3qNnSQH803g/Mgxxzbskk2iGdx24ezTNaweXPKwy5wXuDgICFtxiwCVCzwCRr1prrYZbblHL57Eg/EujgChtKWX23Yo8re8AGAGcMGKIx2rKsu66bT0mHv756jtV+pkHptfuGgfaVD4s/fDL/+Tz3/nB7799wTvcssCzvoN9EvBf8VBMZPVatEiVij3JkEh+7zw4ipVUN6UqFQGMM834GFMCn2jCCRN6HKUf7HCLHnt4fEIHfOQ5KS/aL7VEfYURR3R4RR0unINNrUIEyIb2ynWYY4YhQLwU399L1oq97/DjUZ7hz8+yCdx0Dr/wwhWQqGkBxsXj0EtQ4PsHCTR/7Qkv9gyAMc4EZsKu11gAOTZ02a2tQe3HnvD+zHgZa0yQY5MZS2IL6nzvD2N+78wEIkbvke6rnpR5kc/zQkV2LFtwTzcI5cMqgOt5QD6381kgV1JlgDnHmbhGXIumCU5zZ2MNafC7BGXna2rFSa+3cTGW9PvC2WtjA/RtP1QB0f1qCfIM1su0BODDWX6/HeR9///svWuIZVmWHvatvfd53Bs3MvLVr3lrZJmBkWWLAaOfxmAGzUOWsh8eJATGYIwweGxGWEg2zk6wBxsjyeMH88NgjIxeXd05M8wDhKAx+I+QEQKhEbJgJI2mq7uruiozIyPi3nsee2//WHvtvc6550ZEVmXVdFfVgiAi7uO87j3nrG+t7/uWBofWMH0tAzACrsaAE2eg1R1torNphzIdWqMjx0+Ox4kl+FhoDBVxZXTcFsqXPD4/FtcBEVmv6FbmyQYwvVlrNy392swJj7fjNc8drOb6jZuWMb+Zz28M+lqkQYOmgs2BhGg8lsDHEqCpZq9riDnsa2My7dNHYGUN1gmAvugDKiL8xNM/sXi9/vee/qnJ47/26I3HS68DMBHna2qSbOl8uKGEiWZyG7zNwEO55hYS1O1Df08q1QWZG5UsDSuUd0qSOgeQtxGuHwOcS2YIQAEcbTxMGuvZt0MKX6WQZ/JvCU6sA1qyaBSlSX4sEdbG4OHKYJfMTWoL3Dv1IIoYBoPTVcC69TB2us3/7Bt1Lmz90KcG3FkHPNsavBg9+kB40QPfd2LwckC2yx2SMFw6n7orIM9Xqj2bQS2YTphpW0B+jby/UYm/pXSeqSKGXGvWxmAbgLOk+5BPbx940vQpOVzEMQvWhQ4l55iADKGytWbeVeNt1OvWupXrvu0mlcOEvXEPTc5NWKDOTBChp79Le5yFJhcJRCS+ig47GrFNpjnPqcO92LxnIKKLIyGlprqQyyDXTl4roRNfXcwxoKxXCZgWBQSAGGBSxZ93TyWyNe/S5PWFc9uAjrpTfTeH5N9aHO7hYVAMoObHv4bLcgn5fynkeibr0KBkPmPluoLfawEhS1PSj8VtAIhEhzF/cxtYDDFkoDAg4Jw63Ius6JeKjgjW5QbdQ+hVEedhzDQCYFrpq2CwTfxOuShvMaKOFn302FIRjW8if8mZZ+lQwTCNKn15NcC4j3oyk6Q1BjYSLsKYHWfWsBkInRK3uIn4wts6wj4VLnwALsaAE2twmjLS73Qe3eBxd2UnmgxxtpKuwkk9ragDDE4yaAkMFFY1dzmqRLfygTB6Bg5A4owHoKkYfMiFWgDIUsXaUELfVLoeAHdG+Devm92h1HvidFljUEl7uttIByV9uQyLZAAAIABJREFUCdS+TTsouVuhkmc/28bJ9h55Dph2a+bgQ2JOa9Mhx0wfq8kQyrTM4Zq7j16+D8idLAEgteX9cwD6kY+HgD5niE0BqHCvVxWw9ex3LyGanKUBmPp4WhTAaA2gnF0n+1gbrobvZwNE30voqudcI6LBx3WzQvj8vxlE8HpK5VELs/OyXqGzwjq48mXVN2i9jKUkMVNLj6TXU0Azr24XMbVN1BoioHVyvSrgWt7/Khzof//pF/O1/emjrzzW+yiGHLobsqR5uI271G27InNb4NuGPoZzatWxmCdrXkEgPXn9NhPab7N8eY+AD08l0ZSYAxCJQg8u360GBmticbIU6yTpFwrvHWdREdt37wbghx8E/O673CF3NsAHgnMpacuawFRoGg0enET8i+f8/+Zlle8/PkZsY0RrDL55FfL5qsXoWhMCINmoT7tqACbC7/wJzK4B0qENQB5imCldEw1JWcbamHyt6dS1JQMLGAwos78qlG2QfdHOgPOuC3czZCjhtJCi3RiXOl/6M7UJ5Mt53yadFsCDCA1ITTzn70SdzqcrRHQYcS4HMzYLXbmbI7s+zr7PukisuxQd+dwNmcf8Ma0BqaLFBfENT141X8JSV/S9xAdFTfswYqljIRQ0sdvV+zem43sMfNx2HQCDkZsmW722TsjSpPR5/JU3/uIrIUkHrvA00eKSeqxjlU8gvkEzsn8QWwYaFNJQwwDEcuIUmzoGFmdgNwa+GBmYzCm12Q1rP0NyMiSrAYvRN6i4I2O45boPqUqTELsWzJ+Sg49xYgV8ahwQyikm1ZR99EAA7jubKTEAJ+DblAFaotz+XlcW7+xiroQTSbJPk09kVUfuarhlig0ntTF3NiSsiblrYYi7KbU7/MJJYh0jrxtYTuxlO8v7Cg1MugqDnwo2BSyIpkIvTyfjk6FXaQGVTctTV6eolifHVlfhr7PIPbZf8vd1y9EDI4/RizR9q7Y3O6ZoXU6mbNkCIAzxZ76qY5pNwlS8Mch+E+40hKu+dC0qy8npZD8XgJ7e/nm3RG64woPej8ii+Mue8ne5MiWpkGM0nyo8p1gtaUQsEf7IGz89ub78/c//+mOtG3kdMafLaFCyVLG+TTv62NdtSbAOHOt0mNn/ZZuAcgyLw1FJ6JwpVLpNzeC3G5cHQN42Hj390pOnj77yGEh0MyUqvc4R6jahu15Lz83F3a8KRObbd9O7jzv63H69ukuiZ57ox+R1moq8tG7RfmjAqrVL8hg7/phcABPAob9Jn2qKY9XzLuBuY2AGXsbFjgHIScP3HAYeU/ARIiF4wotLh9875+V0MeKtizKs1KoE3Ud2cpS5QgGHhQSj9nveVdD7IfQr/T4AQFpfwLQbokNWp4sd6a1owEBEHpNtuAjj5PPgdcRMe+zSRPMg2wha/IYE9f759ulCgt4vyTf0sZksK/21idVB99aAcJlMeDaxxpaGpC0q3YyyvOtF6ksdWx2eYi4OmNTFA5LzZ8rfbgI8AkQCgNPYYks9wkIXBDgsWMyvHTfRcpdc+T4K4Y+whvTZ32M8CkQiREtTbHyvO1Za9z2P9w1CjnVBLBl4JXx8VQACAH/hjT/35H/44i8/ljbbt8wFHsYTVJHFLndjg2+aC6yjQw2LTRr+twO3FQfwl5o1ImVbzjHgDBUqKv74lxghPtlaiO4RsUGFc3T5/Q0sGlg8NBVOLHNA18ag8yZTvwySi4bi0oaUbJ/kLNxN2rsPTYXzMMLHiCsfsHJMp7rouO1tibBOF+7zPSepg08Vy8g6CwC46tIFukKmQAECQHTSmm5ICVRYG+H99AJwtWea1xUPUcXLPXcw7q2ZulVZTIDL6CknpJqi1Y8MZKSjAjCgEMqWgAVg6pglFrEabGgQxJaPunrF4CvrLsy0q8Fdg/I3UChHjkqFv7HLNKSlvEx0KFmXog7hEp1rqbOyNLxR09F0tNX09T5wZ+Oim76ussgmATHy8vqRO067MaJ1hDtpY0XI7iOwHwLOWrO8/WFZoyOfn8wZqYgQqEw9tgb49J2AN58bdCFmty5xtMnVyxin809iXm2uXr6KRe9NIUUI4S9L9X/eBZhTsW6aI7LULbkpNDdf9HC3ec+xdVRJFAukgWeq82SJMu3qbBWxqiMudgbPtjgYdvo6IiDmhGOurZnfuF4FOMyP0CIP/JbL09a5vJ1l1tOx+RrzOMbBluXx/9PtngIP/q3pXFO3K3swgFD7+Vf5/TT5/ojdqtjBb+HxkNgKfw2+J4lWYogRdyqTr4V3mGyAyhg8OA1wLuJia3DvdER96XCySuDSRhiKMOmaHwLB2QBY4OHdgLcvGmxHPgAvx4Btovu1yUb+mWcnyYoM6xmpDNCUjgMA7FOhcCnZBpY7ePr/+fVjSec2/39OB5YBghJDjDhDhQ4sAP8OOhDSfI208nMMuBfrvP5TW25q+xAwpOMvxwNA7rro8EprUSiZx68zrHO1BwBhD89Oo8SaEKH1tdHhmdnhOe3RptxKBio/pz3uxTZ/t64zw9BAuAdb3HbR49zssY4Vd0URM1Xz2DmmiYYhxgRkbO6srGONC9pnJyxNxwRenZLJ7/3e7XxcF8cE5Xoil4PNVKyb3K+AQr16r9S9D1wT8lff+EuvDD50/IU3/tyTueexjnWs4SlijNxWsulLKHxBnyY2A8gOWgDrQBogJz9NLHM6hMMIADUM7qHGm2AE3qrZHifWYFMTLnteHoOa4rcuN32p5pjIiUANQkV8IetQePWWCC1Z+BgzUJGq9qklbIdSKa8tV6D6kXD+MuLNi4gTZ3B/zcno3RUnhVzVnibtc+0GUC6u8wty7ZiWtaoJu54T2P0APN8yhcckYbo8pzsDmZ4T+feYAM4u7cdu4IRn7gzlbPliij3v4Kf0MkASZKm4yWNMIdNC+0oBCqno69kmZVnLsURHmj+XaV7qtcdCOkI69H7lbVeUs6VtKr8TrSEQdgMvv63iBNwMno//mDQ+bNPLQKS23AFpjEHjkAcn6i7V0rr1dsnxrMAIgh24YnaaqQzw7NKwfiXRC4st5eHslXlcRw36w7MOSN5Otczrquf8vBaeH8a8cqiBxsF6jwCUm0IDHfn7p55+/mDf/s6jp/l6ONdlzOMffOE3Hsv+iClAa9h1b5VOslUdsesJ/cjFjq4r14M/+tWfec/X70dPv/TkK4/+1uTaHRAnAxyB23dG5vSq11mfvK7aWc10KbeNJZvh6+hYgGiC2B1Mc94FlGgAorslABfHhE7cwmCPYoqgrXctEU5Tjy6kHwEgAcBpAiC1Bdq20D3P1kBVle0fvcFm7Zm6K4MH1b3FGAYlIRKMifjB+wH/8h2eJQKPLDBHKNTBOe1RtklTqCx4BkcDyvSlfJyOXEf0a4RSJVj7tozQpe6rpk0JEyK/HsCVcqSSzyCkXEFv24AARJOvg0LfEn2IV4WSvP5ZsVQ/tmzHyiY/ASycJwB3UGMffXbre9tc5e/TlgZ05LMml4Ft0XHwdcoe1azJNsiWaDqWLKcG091vClmGp6i6eum7QWzL2yaa/G3Ax3X3k3l8lLohrAVi0CGaoOtGAgvAkI7IdUMHB/hcENHH67ouCG/T+4xf/uqXnyx1Q94v+NBxnSPB//H5/2sRoNyL7eSCwC1p7nIIqr+MzIW1VKx/s5MV2O+ewHZ8knjsacQmVpkOljUYMxEsV3QoXzgrIlhjJhNTHYslcmVMLqgDuBMSYNCOPKchRuC04cS9tlwND5FBwlltcN4HDCHi7SvgMxtCZTkxveooi7iJgKYOGEea0IEEeHjPj0tHxKqbS1UBg7cwBJw0QO0J2x4AKLmg8Lpqx8BIwIh0PgZfXJlWVUmMNaVKJ7dLAwRFU0LEXR39Hn0eCRCZxwR0zJYNHArCBVAszeHQ711ys9KhqWhzSppQ0PR7WB8Tkaw/YFSXaE5lC5FBWzcQ+vSaMR2/RlG1+tFktzJEBqd+ZIrW8yFpe0KENYSTumw36PBYynfeo9zANfisLLH3+MCOW7UhDKFQfoSCxSAEeQp9ZQkIdDDJPq8XJWHyiDdS1X7iqz/z5B984TeOFjDeT9zUxn9VICIGFbqatARAAOAnnz669bXVALma3JoytPGs5cSysqU4oGlwUol+v/Glpz+XgYgkD69aZZxW9Mvf84T+JleqV6VlWZjJ3BoBBrfv0hxzNjvcXg0odCceKMdNRPlNrCCc7iZaDBTQRAYgeh4WT7oHWpic9Mo51KhkdxuKGUAGrKnI5ey0MGVNxOgNVnUsE89F/7FQ3AIKNevenQ4vrlY8IPUyJOqXwWX0kHkmHQIaVREXahYwTS4bGHQIWMNOP43UWdXJ+zyC+h2AnOHKWpe6IMeWIZ0K6ejYWOEiFvepPnVGKrCbVge2N25m21a6BmXlLREuYpgUUWXdRv1fkQHi3LL5+uuPposLgOjI4yy2mfq+JSY9XVKfWSaihxXGiZ69Nl+e/BY9GMAgQsACMLX0XpovdGwwniTHngK26KedRaF9XtO9DOp1h89NO5fzv7+XQq4T+jhq0KFnfxyL22pDlq6xxwYU6ngtnZBf/uqXn+hp6f/zV//r1wZAbor/6Gt/9ui6nj76yuMd8byQMTLqC4gHAnQDdotBBCyqDF568LDB54mKdSc2GBFwDzVaY3DlA75zyW5an7E1HlTsaiUTygGuPu4DJ1Un1uDKp8qN4RkMd2pi/nWIGDxfODdkU3ucE4PTlhPBGIEfeOAxjpQF4yEC908iTmqmTV32wIsd8AP30vqTBkRE5ONIOemoXMx/y/VQLsBN7XNi6VzEOBLWDb9IgMnpyHa/b79MlTgHPLui3L7XcdKUC9XzLeWhh/pxHc4W0TuljRoVVUyASt7eqojogSntjP8vXaAieKejwEQAiLFTwKCpXfq9c1eteadjrmGR7lTtMNuesgK2T5YuAymQE6fbbSLWTcTLvcHZiv8fPAMP6WS1VcRVV4ZGrhywcoR39xGtpUx5Ezc1HbKt+niJIUAGR4SJaUBESXp9BHZDRB+4oilDwmpDuesiI9SsQQYiOiwxZUwDkTHGo10QHXNR6Px2UmZhlErlbYYTzuMmO92l9/6pp1/K2/8rj77yWIu7X0f80a/+zJN//MXffCxdkBNH6H1EPwIAYZsofLuRgcnLDrhKX7TXSXtzC8kKkCr/0m0j/bg9eP28InkbQfdtQi9zT2PWr7TRped4+Tzl3k6Sl/mNV+Z0AMuga97B0BOftdZDr8OA0EbHepDo8uBEMTgQAHKaiCvsJGVy90A68ifWoM+ui6VrcNfY0v0EO+PdWZXXaboua8wC2iYy1Uq2cTaIM0RCCITf+06L/cDn9WUvJibAg4b3dTtGWE84D2PSXJpU9AN8rpDbSSckgAuDHhH7xDzQs4eWtEx6YnlQjxmojgaVeR7XhbxfH1+Zrs6fI8Gm1GpAn1yrapxjYBOayDBC6z08IrbwMLECVAGgIja9KfTOElkAnrQmFocDWXWNTvKaeedinyjn8jbRjQyxwbfMJfYY8G1zlYHwnga0scJAHqvgMkVLgI0u/uq5NNztAfZmwDrw8RH6u3Zq0tdL0YoAKHOWiAtg57THWWyxphqX6HFBbAF0Ftsb+xYM4g7jtkYU3ysh2o+5SQAgNrpFOH7Mele7X81dr+bxKsZTEvT1r3/9y6/6pu+l+Ouf/xuPB3gWuKcOiAj32sQhPEuOVAHAO6FHD7bZHRHycEKg3Oza6LIV3oCABhancPiBps6C2z4wEDmzafhLno9Qtq0x7I1uCHjrMuabf52HyHGV+P5aEu6IOyceY0r+JSkXpyNrgHcuOcH/9F3e7pdXNieR6yZiSJ2JzZqrE6M3GJVOw7mYdCRx4nASImFUHQ4fuNsyeMK+s7Am4nxr8rAqXnZJ1MVpC2C3pl1PaCtg3YSipXBhQv8ROtC+K/bDu56yq4qIsgVcEPExOSaWFmCh6VAhYnGdx+K6GSJzmtWSm1Q+zmrWivyOkTIAmL9PdyMEhOjqJMB0J+lKyX6u6nJcth1TbnapMLVOeiPRAKxrZF//+fZqoKSPhVC+xvT3GPRzMYGQkozpG7xYw1aWDljNsu4uFPAATPUaP/a3f+pWF7x/+IXfeCy78hML9KK/9/lffwxMk26eEaS2R22dvlGWqlm5if7M0y8crEPb1wbECfj4MOJ3fu63HgPAqqL8Ga1c+S4MnjtR2zFipz7490PHknj66CuP5fMV6/NjgE3TtG7bMdHuU7d5nY55Z2VeIRUqlgGDEAao0/Vd5+51XQfmmNh88ppYCmU61qmafAZuW1YwuatvwG5OMjfqbsPnV1vxfaaaV0jARattx53wfgTub0LuimuwUSs6lnQ+lgCIxD/856w76VMHW2y5d74UI7qUxHdgytIefiK0zvtMpeuxj3xUPSLWsJPBqPJ5ia3vEpjWwnR5jV7GsVj6JPchYBt9NrMZwbQsWWsPn6v991GjA7tqtslxCwDOw4ht2u81lW+lULwsCFv4pPnBBERo84k5CJHr0oDjA/nmCaps+45GnCetxRYsChWqjSSjUlw4jQ3uxeZgvboj4xGxoxHv0BXuxzUvL8/tiHmgqVFJsWhGRFM2UGGleISk/eXvwgb1RFdyXZFiUOe4Pqd5/5cT7e9Fi16J//aL/8tj4FATMqaCxlz3sUuf9ypdXwAGIz59Jr/4xi+8tmPxgWtCfr/jz3ztTz/5PxNlS042HiNv0aeTW6okQ4wZgPQQGziT7Xk9hfwlv8KY24cbVLjvHEJksCEWpAxsYq4IV+kiXFvKlBmuLEVsagL6csNrHWGY2QQtJce7nrIGoG0CTnqLO2umXDnH08ulYxJjAR/FzSTkToPM8pCkNniCc4G5vYgI3k7eayzzau1qTKDEoKnLl9wlkGTFGhYifOeUhNfH4KN0ANQFNAkbK0egBC64sj89BtbEbA3J+yHvXa5qzeeLEB1PdjQo4WNTNDayHXLclj6f6VR5mgjKrSrLy6yU8lpZP68nxOkwSZv2s1L7LRQ1IuBkNaXdMXCZbl+M3L1qHG9PjHqfptsh2yk3Mr2v1gC9L3Qr1gEVACIi6IuhJB5AoWUdh3x8vgwRuSuok4zbhiTSN1Gz+BqgK9Bl1+ezO141XneH41Vj5yPu1AZjAF72ASvL3dROt/Z8RGMIO/96wIcEqd9CO5N4L/SsY/Gq3O3DKejJXWimAfEUgHTdr6JlS+iFREUPK9TAYv7KJSH+wbIU+Mi0oRQVDDry+HRcpY4+oSGTxeVyztyvDVrH53Q/Au9cRTxcM20zxOK+t2n5mn//TqqaDgZtk8Sm+T7B9wMdc/Ahrxt7g7rmuSGf2hC+ecFMgBejz+CoixFVLLoHiT246i+/NVtB/y5/U0rS7Xu6NgCHHT9d6Jivc+l9FbHN/jZ1LPQYgDldaZu+DQFx4t4l15dz9FhjlV/PE9l9LpjOt0Vfiwod/Pg5taS/0loRnSNdUg8Lg7PQIKT78z7J8X3qRHkEnMUWTbTZTEPPCNEhOdU6gQUNKKQbKkmygDa9rQIcpttuUzexxJDyNLke3JaKeVOV/6MUWnC+JDyXYzafATIHbK8jPvIgBAD+w6/92Sd/8/N/87EIo5o4HZLTQazhgBM4vEAPA8KWBhYJwmR+JAiwKAMJN7FKnFsWezoDmJFQUcwUkjsN4WXHYt1NTdg0kigj6zYA4OG6dDYqC+yTHgTg167qmLsgACegXOXmm8p2b3D/zohxNCDipPlkNSbbW8pVbj1Eqm08QqBspQgUupWE3GzaZmSRobJfhAX63gKGAc/FlsXHbRNgbcygRlfQ+sGg8QZdb2CTCFtX3QXIOEQET1ilG2I/8sHQybWEaFia2rNjljpOgJ7NUd44Byi5+zC3vb2hOiu2yPK+Veszd7rrTdp2BgK10+8rCYJ0QETgLcMb9XYK+GG7Xe7+yDq9J5yuAva9dIUinFqXuJ7p2SO7kbshsu6isykga/SFwqWdyPh3GS4pAKQLEVUsgMEDuNcQNm1Es2fB63bgivveMxVsKQr4IZgY8/CyGSZ6pVjqggDAH/vazz75e5//9cc3TVKfd0PmcUzD8d0QXYx40SdaUYzoxpirxQIMe5RK9QcRx6CGPuZatH5bgKKTjNsCkaXXHCa4CYAuDEicu1fpfeHfNt+wb6KN5Up2NAfJlKRFWhy/ijxorEfAv96sJlRKl7vXgBhW7HrCSROxqvmxe5vkYmUPJ5o7F9HUIRegdLg6TIDHfBjhO89avLgyeNkBD0/4sWdb4Efv8zVi+4xy16NN1X4DJAF92s98rIQSGbL+chs9WrKTQkE+VpGTYqFEAQm8zS7yUhRhHeh0CruubYUjf8+jy4k/8JDq9FjAGfHFdBs9tqK5SBpTADgFDy1u6PB7KPQqk45HCy56nqSZZBKilZnPSAKQiqshdyLmgGCuN9KFgQHMAKkiO44GRNwLbZpCTnjTXAIAPhfWeG72eEY7XFKPHwx3slZEz6XZpQGOAwW8oB3uRgZZVdKxzB3oRJ+wdP7rLmQAcIkengJOUxdmUIXi+XskrjunP0pULB0CXx0Y6BEoO2D1GDONVJyx5Dho570PCqR9LEAIgNSyi/mAskc63yj28NjAoibCBhYbrHAZPZ5FpmxdYsAaFfY0TpbZRDs52ZyqcA+BOx4VOJk7qQvA8IETPyKaJHNEot+Qm0nqFEgnwUoCWLbBJQF6N3CnoO8Nqoo7DlEBhnnoFruEsZz0GxMRAmV3k8lr9E1IgZN95+BsxLYzsCZi3xlULsLOrG5NAiSGxD445vVKSBfHUIRRlf4hgyQcWAnzceHE39kAW3s4F7HvbN7f0RuQn84y0baLS45Wc0rVdYJ2oVYJqKLJsTpcvgjo5xQuns1Snj/cT/1dSDdRGUq5ioudHQ2saqtF5GX7gLJcrRFifvbh9ggVK8bi2iYABEidjoA0/JK7JTIQz6R91XNR5pQsicoQhlBccF610nmb+GNf+9kbAcTfffQrRzsp380ABNBV0uTKFyPuOJMpWPIazZl/3XHs870uXmen5KaQyehCvTp4XiU3c20IcJjY6ddcB46WKFdBLUf2fwWHDiP+zfo0zzaSeUAyQ0ls2OU8jZGLFgBbh5+krjWASdeaQUc5zgeFKFOAi7xexzffXuFyz+58z3sPHwysIezGiN99zufwWWVxPnisyeIyemwjU6xa4i6CuE4Os4q6dpYSAKJpPvxI+Q7f5LQ3d7SSv28bunPSJBq3/uwIxeVqDYsGLV4msXoHj1VKuzzKNHWAAcclBt6vyF0C0b7sMGZDnaJdmxpPzIs0FZkDwKbBiFffL/mtC7XitqaHW1oQ7oU2a2q7WGFPA3p4fNtcYR0rnNMePxLOyjGCxdtmix5jplVJLFlgjwioU+dHnmNAHg6AwgY1zrHHHkPusJR9La+dn8363L0JeCyBuO+1kGOpYYQIznuwHmsuQBdLcNGt+QRe3ovu47r42IAQSq3deRtyQMhVida4bNt3n9jfWyoyA/rJwK1VdHiIBp9yFfqU0ckU79rycLYI5l2bdLMYPYCUlI9+KopuHFOqfCCs24BuIDQVO03JTQTApPKtE8uqChhHw5qP4dDhY6kLAiQgYcB0q0Bw6ca0CEAUWLBJv0EUMQyWKVVJCB1T1Vr0J0LdaeoABN5WYyKMNwx6ZmBjHM3huk3EqmE9zOA56fYzuhfASbkP3D0xJuJkJYk+AxR+jdjgTsFIPsamGAHw/ziIJUG7BH825f95x0XWpdeh1z3/7IimNsPavWz6ukKlk/3SYHM/2DzfBSizX/h1pQOS9yMU+tdSGOKLmk/gq7YAqwa5c9El4EAEnG+Zgtj70tXIMysU0JsnqvKcABHgvVEuXkfcZIf7vRRiAV5bvjZejUAIr1eMflO8V1/5m+KmbsjSc6IHya+ZVUo1AFmqoi5OsSafE6H5OudD5+YdPq+oJ+tYpYG9iQoTgZOKSRS1RZ7VxFq5lFRPDC7Sdls9VLDEUtfjmNNVft5GWBsQ0jXi3UumW4oxy7uDZ8v5RO8zSDoQIj6/oyThXPVfJyCylA7qjohcHQ4tYYVCxVSvRs0AA5ZBhtCw9f/VwutuOifm3yanQIElUu5bZdlCy6pgJp2MBjYPNd6k9Ey0JgwQbM5RrrsGloINHQB5AXpTIBdxiQEDheyAJfmRTDSXkIHRATxfRAYEbjFgIB7orJ3+yrDogJOkMRCdB4CsBTnagZZCW9qrS+pxFlvshZ0Cpnh1GGFRHxyX91tUISwPlfxeiiUQokMDkqWzMNOwP4CC0McGhOxpzHxGEHAaG+wxwsLgNFaowMOQxFbXGcIm0bZK9cVgiwGnscbnsML3txXadASlCr5K1rkPbbFMBTiZGtM8DGeQppdzF8M0LPAWy12i4qQ0emTLW4AT+KlGIZbnrMeqmboZCQ3LJIGh3DTkN0wRnjsb0I82AwB9s7Ji1VuH3CVJs6NQVR5d57BuRwAuWzcOnrIlMADULvGMTbll2PrwSx0iTfzoASAZiqBtRlTBpOF7LIrXyTfAyfOYqFt17XN1f92O2HUFHcRIIP0ZJfrY6XrAxbbCOJqJIxd/jvPEv/w9fy1QaFXi3qWXI/9rQKOrkd4XtywiZF2MsyFrNwwVmpoATRG5Oxuys5kh4DP3uBq37yy6gdC6su6io2GKl3xmIoIv+zZNbkJyzJp2RAgmINvujoFB+TDTgrgjd9FjQERPU/9evyl82CHXNeCwQNE64Eo1eV83wJvz0I+BjyVK1lLchnJ1XXVTUzqOVUoNACyADQAHzlXzmD92bFuXhu1p8PEgrHBJXCn4NFr8EE5QEeFeQ7nr0Vbl+qCvFcBcazc1zZhshz081roIJIyhKRUz0VAN30t6D+w83z9rIrTWYOcD3h35i/UiXWfnm7BHYMvwtJIqdUOABFJUlZ4LgvPo//tVAAAgAElEQVRjiJy0NzCTYzqnOxl9cMDFo4swYk02A5AlIKK/Add964594wK4I8KdDY+X1KOLYksbcAKHd7DPJgNbjNiggtgsi0HOvdjAKvCquzgiSK8W6F283YRmcvTL5yva2Duo0UURv/O2DmD7ZwNaTD9/MJzm8/YZ7dGRx3PaIRi2jz6LDVOu4BEoogoW56bDALYDFoq7HjIIlO+Jg4GLyAZBz2gL0a7I7yoaVNHAKhE1cFhYeJWYf+YflfvNXJgeEztIwIdoaCQGFEvy190BkfjYgJAt+nz4DYALYm/K+3GNK4y5G1KlC1IIwGXkk9AjZiqWBU9rr4hdqGRatg6erwDUQAYirAegTLmSSddEMVehB1+ukbUr1eE6Wek6G3NyPVlXCk0Hci6wviBtm7PM57VVxDAwZYkMJ/fGRlBkcNAYTn6lc7BalbmwIW2vMeUGZExACPy6cbRoo8/V95PKIwSDk7XHfl/l7glvn4cxpVNCatIuULYLSDc9A1AwIIogw8vv+ikAkQTe+FK973uLuvapChhxZ5M6RgNX1QZ1w2nqgKb2iAFYNR69iUBv0zYefqfmyZwWlmcgRgSbdA++V61wNTRSu3vJ/lrD34t9Z3MnR9Y5epOpbGVbphtYu1CEot6gdgGVjdglACKT45kayO/RGhP5n98vXZUpkMrzJUJxx6pMccgyxOYKtSX4WTulUVcers7y38MsWYko5wFTHOLRzswncTwqInxqZdCnYZXbdFqvKtYkVQTsYnytgvSlIFzfAbmtBuRVY6l7dlOS4rM1qMn/23h83UuuaTrm3Q+9Ht1dEQBSJTpEEy0+W9V4dxhx19lsu14lG3N97ZBCCoBJgQpAnmgOHHY6tM4vzCigc/AhdF15LKZjIt21H9wYvHMVsUfpWGyT0Hye0A1g/cQ2+onmaum4ahA7pARdBhxqi1f+narwM7rVUkI5BytzICLvERvfY+FnnRVNgZRtFIfOLQ04S5b/V0lDcYUxfzMKFS1mKkzuHCSaloC1Jd3LbUNcQmWdml6u59Uc9hemz4ckYL5MrkoDPDYJFDSwWMca++S2BQAPRBeysL6ldcj2ncUWHXlcUYch0bb4uUTxxdT44rrI83dwqAXJnzkF/Bdv/Cff8x3w+bwQEZyLNuS6os0HrZX5/WA1/L7E3F5NPhS2eCs3oyFGPA8jvuW75JLFlYo2OmxinVxPDD7XliyKiMFIiJxEne9F18HVW52c6snYUj22hp2tKotciY6RdR7rJqJtPFatR1UFOBcmrfPBU/4BEhCxMdskNnVgqpY3CJEQA9A0I6wLIIpo2gHWBjjn4UcD5zhhl86H9wYhEIbeMoXKRBjDdCpxhDImJCG0R+08DEWsVj2M4ceIItp2SMcq5h+XOiMCQGQ5dT2gSu4qVnvRp+flByiJcl0FrNoBq3bAyUmPk/WI002f992YAOsCjAmo6xEnJz2cC1i3HqvGo6kDAzMqx053JVjoP/2Zhwx3bGqfO1sCFHRXQ2+3/C12mDp84I7QyWqEOXKmDp4mgERXNQ2VjoZ8P2LkiffORqzqiNNVQFPFPJdF7+983zU9i4HT4fbsRmRXt5AE5c4AD08URcEA3SjbWN47n2I/D3nO4GN04XpN8Yff+Okn1gAPNxH3TiI+dxazNbOPbKG6DR/MjUZPdgamblkSOqG5iab1KjdEASyvWs0cZgAkr/sIEFkCFzrmAwhtNJPl6O7KAJ87IPdigx+mNdaOcGYtTitCWzG4cOl+4VzM1x5nuftR2QJAxMnwmD5QP25tZLpseo9doGnpgpFco/+1z45YOcJZQ3h4NiaKJgvAxWKWLe0Pt4GLfcvbpkGG1ibwbyRHpjLoMNvBxgJK9Ot1+BixJrsIfkQfpTVSN+lGOqW/mNh9x6gSXsp0pwEBlzRghxEdPK5SLiJ5xzZ1IvSwP5sAmN5mfexuC0B0x8lCpqGbfK5K50nPUzsW8tnsacxdlDuxyQZAz2iPbfo+b1CjgsFL6rAlbWTM3Y75+U9gq2CJERE2GqxjDUrgbB3rtB3L8zDKPk/3RVv9SswLHH/+IwBAgHLOEEymVPUYszgdOF7c+aDF+h+bTgjAk9f/6hf+98fyv00akRfUoY4GFzTg07HFOYaJR/Vz7DGmbkgFi1NUGLmLjDoJbt/ZB7zlC8n+B4YGn1qzK1CIxWVIuhrA1G7Wp05HCKx3ELoW60dMpgnxXA+Dyno4F3JnZN5SF+qVJOtNHeA9u2BhNNz9yJWt1LnY8BeyrtlhKwSDqhpZk1B57HcVTFtAEOsyDKwN8J47C3YVmJbjAvRlfxxNTqS1GFzAifcmuVfFtF9QugqTt1EH61OYPiaAJwQGTU0zpO1IlohheoJZGw6qgUIz04J+u4l5OGSddDf8eZrUXVrWTEgVUjpSkjYL/Wvu3KX9+HXyIMHgE6mDw92jXs136UfCumX3Mh8IddLEiCtZ11tEqQqaafLv3HQWgA800dxM94tpDD4UUb1QJMQhq7aJLmK546IHUlYEdAmkuEB5GrwG53ONyHS6vMyq+aQd8qpxf8P0PC56eLzYNvgX5x6fXR8OBnzdIfxzXbW+bo15gCEA0LSzcJtOSMi/NXWqVDd5HUvV3WWworsVSxStYwDk2OPHpjVX0aBBg3Ws8MNmhbu1RWUYyJ/VJg95ZZpk6VzqjrBcdybbMaFlTalWch8IvtwvZJnyW4MOXt60OLRqPH78hxNbwEb8Wz8S8Nv/qsa+D2jJcuITkZgFLHCV6FOaw5VaWvx8JbnNFJz0Gg+eCJ+BgtIhaN2IdDcmuh8ibIPPz2uQUQjD0xBQ4mfdkjB7XijcBoTn6NHAYp9AxZhoL1sasI4VDGiSaG9pYOOcaLFPw5bXscoAZZtq2TUsTuAy1eu9hKa+BYg7Wcjg76bJ67KPDSw+HdYYEPBtc4mX1OX86h1zhTo6PAxr3g+q4BGwo3HSHayimcxUWVqrp4AujUeoYbFS09fnuqrj+2wmnaX5te+jQr3S8V+98Z8++e+++L89BoAaFXoMk+f17JdjM4s+qPhYgRCAgcgvJSAiCG+DOlVSmDFHYPHV27QFcOiUcoUR3+4NToxFiAbf6UbsYphcIOt0RkiCum5inuath+w1SvvgZjMfmjqg6w2ayoMMsO9ctlGUEO2EVKlF3zFvqQvFKkTKyW1VeR56ONoMBspyR3T7coJLRyJGysBDh/5/HE0GFXIznDh6uTEty0zeTxTy486FvJ55F1znn1rnMo420bxwsE4RUQqVIASabHNMQEG6LFGqRCbA1nyz9p47E3ysj1+qnEsuYzZ1eKjMG1kSk2c6GhXxv/4NIIsRYzx8Pz9ebJZt6ljJ/jZ1wL6zGEda1K1ICDDJ3vVqW2W1U8F+TPNhCgULQJ6BAwD3N5zwXm7TzIT0Xj0wrXVF3A6UjohMXl/e1vd20/04h/lff/oJ/vPfeEwU8e4Lrh5GAJ/9a3/8yWff4zL/70e/+lgqkD/59NFi1fBnnn7hyW88+urjpeduEzIlWWJpYOA8lhKS2yQXGqTM49jjy+u//vspgGY6WC7gLDa4hwYNDM4qiztp/ps1PGxQ6IpV6n7MHfjygNmZw6EeIKhDv1+6HnI9knDOHwx05d9IrwdOTjoMg8N2W+F3365w7yTiagzoYshEaDl6NEtsR/W3fuV0kGShYslci4hij1xl8KHBqhLkxjIdXUdFBhV4+OF8zvZ1n3aHMptEb6PQynZgbUUEWymL6HuybliswUOU22jLMsAdkZ5YRwEAiMAlBsjogE2sebCholK9X6MHLWYXJyygALubwIgF4TJ1My6ow55G2GhQR4dPxxOso8vLIBBW0WGAz0dRzoU+OZeOmJ4fAlQsauzNFohlxoVQsbJV95GO5UfVgvdVQ4MOiaXO0IcRH0tWw89/9T9+oulZA3y2pusyGjQ4S97TWjDVRJ6Oftc6PGg40X4WB2wxTlqBOx/xzi7ifEfZNhHgFvq64XkR6yZkQCHJurMhd0V8Enb3I1fdmY6lTkrDXQPWlXArXrta6Ym2Yq+oq2AhJBtfE+G9AVHIPzEanGy6SUWtaYfU8SihE3kBEc6FRPFKNxM3wqWfpdfLOvXjROEA6PCyR1TViGFweX9kn6vKH6VK8XKZOlBVHlXlYW3IP84xTYufG1HVPtMRhMZliGlyhmLSjwScnPR88yepQKrjO0v4dWVRU8qc5WVJAuFcmHxezoZMp6pnFU5tUqCjT/Q5gIHUyWpMHY+Y9SDWMPVCHMUkKjvdvun61DpGSoMJ02djpl0LDUyaiufmtI6npK8qoL2hBHKMmtWFmEXun8SrRfifiubjs3/tjz95PxqQ/+fRr02AxW89+tpRoPEzT7/w5HUNbBQeNzBNKuaiyleJg5kBM7rUbeK698hzxxIjj4BNrHOi+ilX4ayVrge/ztmIO6uYZzDpqBTlakn3IY9bed2CGF3ipuGtS/Httzf4V99a41++XcFH4M0XhG1g56saJk07LzQioUmFRA6J0MOEU7chHZcCOA81CMAUeGwTvSTrJW7R4TNq2cfCx5inrAvl6SqOuIgjnscBV5Envl9ixC4l0VfgKegEFljfi012mVpFh3uxwT3UmU4GcP4hs8k0deiCelzRgEsa0MDhHuqs5TgWS/sux9zPfiT0ZPXJIMRbHUemmj0Ma9TRwSOgpxEb1LCRhe36OLNjqc1dCU9xYkhx02enuyC8jepacOQ8m/IzPp4RZ+BOQl87PyxbdOBj2AnRoYHIrzz6yq0qdQTCBUbsR48zX2EbfRaURUgXxeG5ZxenFyNgyeFszVXkEIsNr1BgmiqCDDJ/X0IqXeNo8OJljc3JeHCD0ZSZDDDU+VdVDKr8yFV+EQCXRBiw1oMowGj9BXkMQwWigLoZU5LP3YYYGbA45zGOItzmlVo3pgQ60aC8hbEegezksRIFjUsnQ6p2hvhv64AwAz9VNbLofdPh8mKdtCwF1ACAsQHBm7xffixfd5u6McEbxGhg3YjgDYaBXyPdFakoxtRBMioLD4HQdQ5Lgs9AlPeDOdaUaF2T3ZhMpwfYzSt/BojZIKCycQJEuFNEE2es3H0BsGoHhGAyBW+9HtAGj2EwGH2dt6uaJSSyHZWNag4ITTog0qkIEdh3yPM+el8cr8aQZoOMgDUG33rBdD0CD+yUNS7NKtQdEac6ItIdadILfufnfusxwANBf+xv/9RHgrv7YUT/l//Ek/rml71SMF3m5huXAJGn6Xor183r4phT1hxw3KaCZwAYZbUuFdO5+FxTtoZU3c3rvca+99jj+nlZ5jza6PBj9hQPGoOzFdKsIf59tg6Ztqn1cNmMY6HwlPdZzfZY6nzo9wF8XRMaKyBFHd0ZCZNuOFHA5z5zia6rcLV1+EffcLgYp5/GLhbLVqHlzcOmpFqeG9XrXqLDndQPls99QMhCagEiDSx6eJhZgr7kJgUgTy2X5+UTm3dMrOIQGyKYG8DueRxwmrZXuhVbeDynDqvosIbDGYRixbPKCCWRvyR24XwYLc6pxyZWeJ7cp3ZRFzxtBlHMxJieK0Z1NOZAQihnk/18n90Um6hZPxruYksj3qUtLtFjTRX6yODMRoIhNSmdmE5W0pcp2NSxpzE7PL2kDmexPbov827IkDqPVVymn2qaH8Csmfd1ML7LQtRNZSp9seWVkLkgQOmufVCuWBIfaxCi4089/dIrHeivPPpbjykSrlBOCpd4q2fG4e3Qo4l8AXjZMzzh38CdmtCPFpXlyeaGgLt3BjR1wDCYyaRxAIjKMtXZcDC1/MBSVxyGDCZAQd9YdMRIsI4TdnmNsR6UQME4WkSjxeQMUAAP5/jvqprSuQR8GOtBJsIosDEHKBJ1+ttgDlQwAUgA0NgefVfnbbI2wLoRfnSK5jUFVtLhAdRN23mEWGhgRKVDZJNQXd6jnck03S14grix1LXPXYz82RAPpuT9lc8NEz2J/D/ZZ9UpCZEHX+ZuSbLp7UeTRemsP6HcHdKXZmMivLJhbRt/sJ45ZcOn4zD6MqGeiJ3LhkS5WtWcJHUjJgBLd0gA4O464u0LynqU/QjYV7j6EArNa+qWBSBG/PaXfjMXEX78Kz/9kbp5fLdGGeL16gMOHz390pNfefSVx7ept91k2fuqMdEFKPCBhceXAMjS624b0lWvop0AkT8Y7mIFC0t8rsg5ZwiIVOyzdXD3tYCP7KB3ROdhXaFpynvyskyhYg2Dy9c+foxfMxnsOiv4ODdiv6swDCZ1iyl1D8pxlQq8Tornw/Mm+5c0CQMC7qHBHh47jFjDLYIYA7abPdBxiCj8FhTOktofhgYvnPQf1xCdUZWvfx0Cen3/U8m1aDBqpXm5xJA7JmtYIAmvCZQB7Db6TFtiO9/jsdTFuB018dXPOQ00u5TcylBo+awdDEaZ5i7fU/X8ca1GRJsoXXKs+qSlaaM7ACJyrKo4pazp825OSRJw91EDIMDUpEFHDZeBiC4dfFBznObxsaRjvY740tOfewIANbiqNhALzjwi3godqtSCbokvyi/7iJ2PGANw2Udse+DljtANUt32sCZRrqRyH2hqiToW3YeEoUIZ0hSaqvYT0MHP89/6t030LwATAKJBQN30sCbAuTEDC3mdSV0UH0zSm/j8Y90IMjH/nocAFB35PRWvS/QjvH2F1mUo5ufqekTd9Pn91o2lG0KFouCcR1UNC4DJICQ6mqaA6Rsx/7+wD4nSJI4yozcYvWEKlwKHmV6VAUOa3eLK/0Jf0/Q5ef9kOQo0WFOWK98dodeJFXIMSXRvI/qhdE4qccIR17NkCrBE1eC5LOlzsEhT0GOmoclz83cKsBIR/W6I2KeP1N9wj5P3WlM6IVC/dVgqScY//Q9+6/E//uJvPv5HXyzA5JN4/fHvPP2TT4CS0FSga6fKLwWhVOY+jJue1nwsaTzmXY5j27RErTLq8WPgZE4H8Qj4g+EuNnBYkcHnTspsItZclcGqujg172ACKC5YykVw7m5VnAjF5XB6jRtHixcva+x2NWKkiS6Q36//5iIPa0YMqtrjZD2iNsD9iiv0Pmki5sdStB2TY5pAh/xojSULsU0elMfHslCL9PG9TKnkARhJlKr5EEO5blzFcq/Rr9NOWZPjvbCOfGzUvjZJOL9FAbRDom4BDEQ2cPl4NLCwKN3idUri78QaKzhU0eI7ZottEna/i30GOefo8TZ2i9s0/d7JEOaYn5t/L99LSJKrP5NL6uHUZy2id7Za9vlH07GEFi//63kVL6nDXsTp0aFOgK1oWtI+pPNYg455Ev5hJdrfDfHfvPGfvRKw+rAoWfT1r3/9yx/Kmj7C8dc//zceA1zlcjBoYdHCYkMW+xiwSh7kLl3wfIyoDZ+Ed2o+CR5sIj77YM/UmyrgalvKxKM32dHGB0LbjAieUNX+QIAOMPWqSUJz5wL26UayWnUwaV5IiAQ/ukyrAgBbjYiBQCbCD44HGC4AiChD9hIIGLomAwp5fwyEGA1iIFQNz2SR5RAFBG8Rgs3LkMflpjj0dX6etzNmStYSoPGDg63G/Pd1UeheJZkXWhZQOh668ifdJACZ5jQP0WfYbAc83c7drjroNtTOox8tgqcyz8QE+ORgBpSqo2yDWCTLY11vky3ykET3Bk0zYL+r0PUWVcUdtsutw+AJJyufAYhzgUGK2q4YCdu9VX8bBQCKu9tVx3ahIQLnO5qAERnc6QOLaSOKqBbg/1eudEyy8P3IdS9E7oTM6Ts+8BDEeRUsABhDhDOEMZRK/R9545MuyeuOrz/61cdy7AfEGyfL/9qjNx4Dhze5iLmlanl+3glZukHqxOlYCjWnWwGHdCp2IipLm9M6NrHCNk0vn2tT5D3zBHUOSeT7+jCu8Blq8H0rh9qWmT3OxjwJXSy8ZfhgXSVqlhHNHc9qWnIQXOp2zEMDize/tcGzS4Mf+5ErvP3OCpd7gx/9gatM6wWKiYjcN3ww2F612G4rfOdFjd8752R/H1iYLpayLcyBvex8Bog8t4fP91GPuPg66aSIvawccxn8J4/x51QoPgCys9Uk8Y4RlxhxRlPgdVPIoMAtPDZw+W+xQu0QsKURn4srvIMuU5aEpiVJ+WXuHJRvyyXGPDldjs0WHu/SLs8uq6PDCg5ttHjbbFFFi7uxwV0sky7nnZE5BUtcuK4bKipLOJbCX2JAJH7tN8w5vi/cQRUNPDEFSz5L2YdL6rGJNethYhk6KVoR3bHwiOhoxAV1ed/r6BboVwxuKrA7mwBDPexUu2R9lLsgOr78xV/KhaKlDoi+tv6Pb/yXH/ix+ISO9Rriz3ztT1/7Qf39z//64wBgFzzuWofalBOs9wRneCaIdTxAkEXQuvIc0hBCTpz73mZXqMwBNqXTwbM5ptsgNxHtlNKuS8VEgIUk+PIaMhHG+MlzcQZ8pDMigMKPjjUWwSLSdLmyrcYEeC9J9gjv3aQqRybCmjEvK4ZEGQvLYjxbjTDGI4Rl7YmrhwxiDIBxZJAVI8GaADOhmS0MIlS0NmMCYhBAVG7ufhRdTJmjokNT5wAoS14eIsnrTW5ZRlMeuLOhO1uyTiKTP1N2ALM8E8UyENpf1DxV3qQJ9mk9IkgnijAugtL++DSVPaaZMiYJ2QFO+Fd1+V76UFzZKsMUrbKvDDp6z4BkSEPybNKOdCP/neYPZsCnp6Jbmi5zCYDI6+QJnQQ6U3Qs8tgn8cHFbWqnN7lkLfHA30uIWw5wODvguiGEOomRpNbGkuBKtXquRxEu9dJ3rCTC06Ffm1hjDYdPNS4DdgEdlS0OWHkdioopbn3yvFwPjlGs9LUoRsJuV2O9Zkeruha3Ql5/UwHfenuNyz1le3axYXduRIyss/PpmhEDYX2yh7UB6/WAq+4E7+zTdoEndesZHnONwlL1vE3dgHIMpzoBee4letxDk97Lj62PpDXaYWtJQwDw53seh1sDkSFpXbbJDesCBZy+Cy6+fQotTiNTpiwob5/Y33pENGTQRIOGDMZYgJreF9HI1GDTHHnNQB49RmxhMwARgLO0/9cJz0W8bUD4Nm2xidUB/S0eWQap51dwGGLIXc5ntMUZWrxjtvhcOIVRRa996ujI+kHIM0YMDi2CLQhNdLlbck573Md64rQl75VBj0sD97IgPnfqPh7x5Td+/smXv/hLj8VdTh8fPh4pn/uQrHo/Lsf99z320aNNHRFpAVuiPNAtRKDrXE5Ceehdck1SCbG4ZAHJljXNAhEtA7CcQANJqJ2q/s5x0j4HGEDpJMhryMTJ8/K//AiFKgZKYKHQsFw98N92hLWl62GMh7UjnBvy3/k5O6KqOlRVN9k+WabebvnbGI9xqHlZiZLl6iFTw+bCdqF0STVRU8iqasxULz2c0DmfLIDTzTS5ZumQqfCaGue94YnxRlMhplQJm9y3gqfM49Y/Yo0sP4Pq9mhwI9sof59uBnzq4RZN5dlpx0bUibpn1DYSRQYdeZuLTfDomTYYcwepTEtfcssRcBBT90I6IUKp8oEF5yK6zd+r2fdWbHtNep0108qbdg4C2PbXYFqF/jBdPj4JThC+/uhXb0XJksTjgMI3q6++qh5kiYozjzkAORToJtoiikuRSdu2hsv6v4le5ACY6Er8FLw8iC3TcMiidQI+pNCU3qNE6E3lE30zZM2HdE11zJ33ymPl/Hpx3uLkpAMRcHFZo+/les8uei92wPMrwr1NwMOzqaOhtlWX672xpaAxDBaXfZzQnAQEVvmY0mTauU+0q+ksGJokoNX8O6GE6MD0PBdnKn58+nod+Tk12FDWcx6Hg9dP1h9LEh4QsYFDA4NTVKhhcYkBDSweoIVMipfvTwODBibpJkJypALWZOFAqGj6PdL7cI4eMpfjYVzhXmzRJKF1RyMexBYncBM62HRZ8eBYSKcpQFzJNJVqSPS2OHmdXt7c5Uyel8/wNDbY04hvmYuc6AbEbA6hhzHy9ky/0wHx4NyidEwB1oNIx+PYHB+9LFmHhZmcv++NgPa9GaNQ4NRez4+HgJEPOj4BIR9C/Ntf+9ncKVk64Ja4Mix8fZuARl2PedaGiI6r2qOuh1wpd47/B0oVjN1NWCfBbXP+otVNj7rp0ay6DBTa9SVc1cO6Efd/+P/jJL4a0a53qJpuIhx3boBzqdKTQENVdSAT4aoexnrUzR5V1cHaEU17harq8ntkGcZ4VO0Vgwc7wlY9XNWhWb+Eq/dw9Q5VewUyAVXdZ0CSgUkCDK7quVOTqGCa2gUwvUsAiYAS/VOv9hPx/BLNaymWuNZA6YDomSrjaJlalQCPVboOMQSQjggZ1vJUySFLAAcnEOrCr6hgdT1ic9KlIYvsGgYwpYwI2Gw6tKsuD6lzNqBtk0WxK3oQInZoC5EwjqxjGoZCCRs8V2j7kXDV0YRatVf360oBAx8Pp2MLmIjAgVMYgDTkkM8Jcc7Sc0PmywJYBFsZmgwwlPOsIoLLncdP4oOIf/fpn3wi+hCAk5glbch1Fr5A+Vzn08XncRMVSxI+n2gl+TULHRD5XcGyyDdW2MQKTbRoEthwYOvfz8Y17iXb9nV0WEeHJlqs0m8HgwYODVwe+NXA5Zu7g8EqOl4OavyAWeHTjXLsM8Cq5gG2GpA3lUdTe9TOJw1XyIYcUhSRYa25u2kK8JjOaAr4nW9X2Zr8Uw+v8DvfWOPlRYthsBi9wbri8/NiZxIVLB03WyzcAS5qaeOPpu2xXvfY1EyB1BoKoRg16VgMs6RWwJ7+sSph1yGCdV6ezba82m5W9Jn8fZme+wGH+g4ePmhQkcEdcljB4jwOOI8DhhgOfvQyir1uyECmg8cDNGqOx9T+tktbNN8/D6ChMr18QMAVRuxzdZrwLbrCO7TDOXV4QR068ljHChYGb5tt1uAM6Rh4dawvMOBNc5mPgwYVHhEv0eM7tMOb5hJX1OGKOh7YTB3eNBd4Tl3e1qdupHcAACAASURBVG6WpM6plAIsN7HG58IpfijcxWls8IJ2GIitl980L9HTiCZKd2hqs80DLM3itWATG9yJDdaocUl91pYARQciy+oxQts98/7qgYUcP/8Rp2JJaDDG10mfKKgfftHuExDyIYXcpL/h97gYPfrAdoCVpTwtet9ZfPs7K5y/bIrAXAnVrSSLBAYiKZGV6pRU76vK55tDu95nwXXf1ei7GuvNOdanz9G0V2g/+w1Yy52I7vmnM+jwo8ugQwCHsSODBjvm7oXQo4wJDDDsCDIBxvjUth9hXV86FnZE1V4BAOrVBarmCtZ1sFWX1jGb5Ol62KrP6za5exLyOmX7iUIGEsZ6BjDp5xjAEHACcGWviMlDBm1100+AhQ4z624IdWocLcbR5uRefozhSuakC6Kqm5xAhDzRvc3aHn/QdRB6WOlqIP3PCUHd9LljdLVzaOqAupLjJglMyN8jWU6ILCQnEncdttsVy17dmYuR5+DUjgEIULoXAh4ElIj2ozLF7erg8zDL4ORYyGsFgAhwWbL+/UQP8uHFktXnTz39/LXHf+kMfT/C0et0IfOoooGNpLoePBitiganscKD2OaERkTVkgyLiJoiO3nJ7yZaUEQW5VIEztJ8hzvG4sTyBPRVhTTQNHVcLc8hcjagdly40NeP6bmLyayjJQMoeV7i++8FPHu2ToAiJWGe8NZ3TnD3bD/RaT18wAN7BXwI8Jh3lqWw0rYd7qxiphwDqZt0jTOVHEv5rG3qlFSYCtXn3yl5bEvT4pOAGkmE2U3peEjXxseIPgYMkYcNAgwS+kniOk3qddKduygYsIKbbK1R2yXL7RAycNFxGTwq4uOxStqRlxhwTn2eI3Ivtmgjf7O+L2xwL7ZYRYcOI94mBiL9whlQgU0T3kqDmOfRwOJOrPEgrNCiQkDEM9piSwN2GPGMttx1gT8YvnhseQRCGx2qaLCJNc5ph7fMJb5lLmBAOItt+ixNBv3zY5t1G6pwEKjQ8gZ4PKMrPKOrSYW/fKfMYkdFU7E+TsnwL77xC98198JPNCEfYmyjx12qsbYGtSFcjQGbOvmce2DX8wmz7yzu3d3luRm7XZVtWvvOoV0Nk2R4GMpgOgBlOGA9IAZC3fQYx0Kx8mON9Y//UwCA/8aDRPVxsN6xjWxKXEOwaNYv4Qeu/k0GCpoAZ3lZVdUhBBaakwkwdkSctZRtxe5VoqUAAGN7jP2agQsFGNsj+BrG9vBjC1fv8nsiDChpMUwCRVofwsl+Am51P1n3OB7n92qdi2hI5nbAsu/OFdG6cx4hlAnrwCFAyfs5o0zI/9NhjEVHIomFhPCx+TOOeTtjMg4YBgc53MYG1LZP2qE0FTsa1C5kIXpV+QPbzTylPRLqKqT5IEAATcTolU2Jfyzgoh8L2PCBOxn9rJMr1rzymrmFrzwe4nQS83wZSwBl0gFJIhPRlcjLdVL0SXwwYcDVXuG6HwudTEpy8UHW33QH5JgzlgifY96+Uomeb6sOoeNUMFnMewCcIid/99HAgdCSQWuZqiXgA2DNhzVMs3I25O6lVVq5w/kdh3HscbmmtI3Hd17UWK8bhGBwf+Px/MLBB8LQW5w0ERcdYd1EdJ3DyeaQmjS/Rm63baaMhghsPSfzFVEGIDKnY67R0Z0NB8oT1LU4+WD9Sjt0LzbYKutemdullwEcfu/0/0MsXQMLwgYuA40BIb9WBPMdQqZU1bP0tUfIM0KmNDEz2XfdARliQEMmn0MxBoyIeIE+L2cTK/5OKtvje7Et648NXlKHkQJ2cczr1tsn+1Elwf88xDJ4QMAeAzpigb2mTL1jdjgNNS6oQxVNpsTpYyvLigDeMpf4TNgwqIeFhcU6VuhgcEV7bDFgpICz2OZZISJg12Fh8oVCrK4FlNyLK1wSHyuhWs3td6vkMibnuizz4zpFfQrKpmD4r7zxFz80kPIJCPkQ476tUBEPRWotwZHFWzuPB41FbYFtR3CWE72LyxYxsHVr04xZGK3BRlV5DIOFTAJ3zmf3K+u4Y9GPLYK3uUvi6gHW9ej+2R+Aay8REsCwdoQf53aMEa7mrsWYXhejYcAitCzXw1Yd/NAgeHbbEqAh3Q0A8EMDW3UgV8AGb88WwdewFQMOa3bp8Sv4YTXjIJe/yYTJpT9Gk7ohmgtd5oLo5+RxAV8Sc2tiHcaGNPCxJMda+CkAYS4C5dfN/z90szFpZ5YGgaVXJOE5TYeM2YDaDApMFNHoOBpcXjVoao/BE+qauyxC8RNAZUyExoxCC+xHQu14nzRgMIQDtujoS/JTaFL8W947pGGGQ+DXtK4Ai8MhjrxMiawzAXdR5q/XgvSAIla/yQb4k3i9IQDkWFtfJ48Ry8LTDyqWwIjEnkZUsHDxcOSi/H9sO7PjkqKNaAATEPEw1YRbMthYi9YSeh9hU9ew1lbdCYAIhVOot8WMgtcr1wh9vbAzcKCvgfL3wwdbvPnOGf75N07w8GzExc7imy8jNjWw3Ts8vyJ0PsLZiPXJvuwnxQNzDYnffXODZ1eEIQAvh4B9AiCiC6kNYas2TYvEpfMxBx1yr9TfGQ1wNcBYw2GLER3K7Ax5vV7nTcP45HnRa/JjBpcYsYJNeo/iYrWFx27hmzHXsOh9XlqnHqbIQCRgB48aBitYIO0PpeO1h8cGFRqYDNqqRAfcY8Bb5hJ1Eoffi43S4zBo2GFk7QwNcDA4izW2GHOSfm467GhEE212sLoXV9hjwDntEUxxrOqSKF+HttitosEL2uEuVqiiwfeHOwCAUxA8BYwUcJJmoQiokO6h6MHkO6ANBebQ4U5sEBBxST2qaCfgiDUrMWlJKAMRvv58POO/f+PPP/lLX/zLmSIrx+XDBCDAJyDkQ4nf+bnfenxSEx62BrshwoFwNfLJ9ZmVzcPbTipOvM63hP1Q7PUe3LG4c9pl16sYC92m9hbbqxZVxSJsl2hX2tUKQBaO+9Gh71a48+Db8Feb3OXw3sHaEXXDN51xrBKgsNwFqDrUq3P0uzM07VUGHtKtaE6eIfgaw34DV+8mQMPYHtbt8/9kPGx7kTsbzuyAuQ3waGGT37kfVrn7EaNB8BUIAXPzhrHn6amSuJMJQGDdClA6J8YGRF/AyThU2c3LmhFm5hTmB5eoVck3RAER0WtYG5IOI+YuCTB1LZOOBsBaDq8oDfJcGZIoAMdMOiwHlKzk5iXrFxB6tW1xcdFgu7fY7SzOTns07ZAoG+lCn87+cTS546bDBwYimoIVZvfRwTNoZs1IAQ6WphbHALBZ8W/pkogwXVx5dJdEnp/ft/WtXZIEM1uRSY/5yEleHwpV7ZP44EI473Jz/zuPnj7+yaePjt7Q5CORxHLh4wYwBS436UGWQoMOrv5NH8uDCeFhQRhRZlMAMsfElARV1dl1Uizv0cmmCNnPjENrDE4qQmp+407DRaem4oS/rkKagp6E3JWAjngAQMSlSv6W0EWbfB3UHWxi17x/4w+9xFtvn+DllcU/eTbi043DDz4ccX7pcNlH3G0I9+/us3FH3h+Ki+6Df+gPPE/DXgPe/OYZnAM+9fAK/+8/OcPVwJa9NRGuok/HinL3SOhaJ9agDxHbwJ9Tlyr+HlPB9NxdS8DIBhU2KR1eEmbLZ5XfP2+1oojndbeBl11SpRVs3p7T1H1Z/l5O9QY7eKxSB0ILqx3RxBGL9SMBK1hUae8MCBcYUKfHWHpegG6PgBepM4Hk7NVhxJaAHh5nsclOW1U0eMtc4pz28BRgIuFbBARJ+NP95of8GWrYPBSxg0dLDn302FKPS+rR2XM00cLCqOnlEW+ZS4jzFGuARlxSj0+HDdYJUI1Jj2Bh0ESLc9pnm96AeOD/a9IxCyiGEgE8AR2ELE5vosUF7XEF4Cyu8neBPyOBIoddto+LHkTHL77xCxMg8mFY8s7jExDyAcdvf+k3H9+pDB5suLL1nQuDdQ2c74A+lWkHzzmUVG2dmSZvTRIqx0ho2i4N1mOQ0bScqIuzk9jn7rcrYA24qs+PVU0Haz3GsUL/4gEn6WAAIol6vbqAH5uc9FPbweyBetWD6h6Nexf91T0GJvUOY79CvToHnIdxO1C/hrH8WpNYUVT3iH0Nu7qcHpxmAAUCjnQf+GB4YMAM1IiGo1C0gBn4SMGzRpJwH5hVBQWUTDsgInAPwebHtUWvrkBKcAekbPYxT34dc7DB6yQYpRkJnv/uuxrDsGC76Clv1/pkn+e66KTFBzY0YMAU8pT7Qi0LGAaCswFj2ifp6OiOA1OyUiKgOhhGbdZ1yX438KDD1jHoFvDB6zvsoHgwmJkDH/k3gM+ZhlhbxXSuNBMklhu6M7SYcHwSrzeWqsw3ARGJ6zAiRQaV70c0eR0VS4enCMTScQOmtrB6H/XQtbmd7ICQJ0VvwLbsdTJdAPj3qo6wBhmAkBpQ6tSg1HnhYd7t0NSo4KePSWd0HnXT4+5Zhcu3WjyoLF4OAW++62AJ+HY/4P7/z96bxNqWZOd534rYsfc55977unzZVFZVVsciKaoMiqQalgc2DUMwKMMDFQTLgAfUQBzYAwkWBBvwwA/pgWAIhgxL8MBqANmAAEuQKBg2VCA4oiCARVKiRIkUiyyySpVVle17+d7tzjm7iQgPVkTsOOee+/IlqyozYd4FXNzT7LP7Zv2x1v//KzfL/lagoxYqyd9ppV0lzgE+8YknZF8nEb1GfZyrp/WoeX59p7EcOeE7mxlQZfBRti0B3OsqCvn765ShnlYJqeVrnRhVrdxbzu6xn4/zDKKl2rI5at+NMSXd9XBPiLtVEp/aZPKWZJDV7jTP7O6HQKSLyqpwUcHdhQycS89aIqN4XgxHRFTxapKglY04spEpuYpHbsdFAQbL5HmyJJsBCmsZuRsWBIm0seGhXBJEqwyeQJ8I4VsGjuKCW3SsouOhrNnIyJn0HIW09QIjE/fD7SLesM/ZqGNHPS1dz3U1LItBBBR89OJLNWef8J7np21pfzDbsXJ82PyQGxDyfYrX/ssvP9hOWpJuLawWHmsil73hpecGji4aXn9iGPz8YGrM3CMc4pyMXa4b2tYnjshEPy2AgLE++TkIzWJIRHDPYrVhGhxj37E6PmNKVZWmGYnRsFidY+zENCwLp0NMIKTWJNv0jP1R2RZZpLaqIGBCAQHGDrRL5W8wLWhWZ9hmbsES46GdIIjOIwiYiLRzj7GsEldk68AbyICiSbfYIKVVC0/hjvhprnpk2mFdLclhjC9tCMZOBN8QfLNTXcj7TfenoWlGxsynSADOB5NavmYJ3no0Mm3N/GrvWbffLlGPToZgr0hqZn+UOoGo1c9y5BY9Y5IBpAkYPG03suj0Zq+tWKkdLxtTNhP58s8VHNd6hrXFB+Fk6Zm8sBnmyk9TeRYYgSgzAKmlcq/jbvioS7RmJo4f4n1kAnxdWamTwgJWSECl8Ek0aWmqVgon8Ln/60/9gRvh+rCjThieFYjkX0wHkgKpT4D3GfsmZjlyIuMlQPECiToiHDXpO7TIuuVK39dtQrvJ4V1a2sSLMCKlQtgkz5wdACJaaWgOgIb5fjWrVWWi+JX1S9VQeLoi3O3bF5xdOB5dNqxHz6Mt3O2UMG2NmtteXi45uXWR5uu5OD/iyemC5+9f4txVrogPalB7uV6yXjsuhsSriJFjY1hEbTsaqgv/yGgFZNvH0pK0M09mnlEGd4di9prY5SWpcEC+X1wFIrma8LRWrX3wmSNUn+1zoTxxp1Uqr6NWCOb57PKOdpex6zI/TzOixPOJUEjiA54X46ooaxFbRgn0TGyYeGg2mCg8NhtuxY6T0NKbiZfCMb14TBCOoqPBlGtw/2gcp7apVXSJQ6WAacvIWsbS0HU7LrkTlzsAHnbdywEEw1oGTgmlkqLbGHe2vY5cDSn7uapygnqUTARcNJzLNgENT5vOm9qs8CY+/LgBId+neOXv/fSrv/Nn/8kDgN5rNePec+fcv2+SEd8xw9Ty8CI90IwqDOXky5qID2q2drq2DNOSu7cG1perlLg1DNsly6MLVifnNK22UeXqgBx5xv4IMYHj2w9Zn9/FLS5pVmfIsieer/BTx+L4XYzrmbbHjNujHfABwGSRow3STsShgSjIJmhVw0QYGty9h7rsiyX26Fxfb7sZvGTgkUf2VgNMBpowA5PFqEAkhw2pnWru0c7tWblFzLZzvzLsVkDq8GNXyUpWI21TDc4yl8MXIrsxnljJLM0mjUbbt7KDe2lv2q1qyDUjrvujlLnlKrdk5QrF/uWZRybbNrdu7YKdxWpTlK6ePD6mHwy3b/XaLpbWN49c+iRUIKKE9c2mKwBptfBMk9CPlmHS9pUxEdPbBmKMKenX83OfYJ7/75gJMq/rMCVQkd5nJa3c1lU4N9X3uQaTlzXV/dM+YkQYy3pEmtSO9Qd7jOuDjzp5qHuvcxwipee4rh0rx/s1MqwrHvvVj0OgJDCPcDdV0jiTeesRfP2sEyVi1+RrTZQb7tgm8SJg2QjHnZ7L2Q09mxHavVbUKYlQ5DtfJnzXJrS59Wln/0hMpqtpe6LsDmJURrU5Pv7yEz72ouEXfu0uY4y8tp5UuWmER4+Oef1Ryxc+P9EttoiJHB1vWK22vPnmHf75Ny0/9LzwQ59/i+Atw9DStgNB1OvofN1wfyWEKLx4d+LNdy0v3Pb4YPidd/TeMcbIEz/fk4W5AlK7dueKgyb68zZlqdu1TEjUStb9Kpm9DliYAwCgjkZk52Q8BF48sZDL9wFIX8239pnpU/vZ/tlXr8V8vs0tQ7XBoieyRg3+lliW2NK6VnM/GoQu6PP8QkbekHMQeCEcJUlf4aVwDCi4KOAlRU3Iz7wPE5MQQ7p+7qR9fSwtp2wRhOPYXgEQK1q62PBI1ldUtUKqwNTtefnz/chEcgOY6hoOAGW+czXsdlxyKhsWtPgYyjyAJEgb/0C2YX2U4gaEfB/jB/++jsA+/HNffuBcGp2ygWl0xKj99qBJ2sLNRlXWaCJpZL459aNwfqktNMvlxDQFnBsZ+gWu7TF2xDZbxv6E4JtCQM/k8NXJY5r2EmknBRSA684R4zEvP8KdbjBPRsbtcUnmg28Y17doT9aagJsIXrBH54xnd2mffwu6IX1ukOMNcdsii9SHlQFG4xVU5AdkEGgC0o3ETVumwwT9qyMYXd9pd/TL2AExnhgsxiYFEd8WADA7ms/zyzyW/ThEJodd9a38QM8Vihx5NLK4sdtQWiLm92bn/aHXGTzsrxdA40a8nzlCdRWjdjGOQYhIquYExtEQgrBcDtqm1SoYmQaVEPVTo9WdJnuLWLa9ZdF5dWIe59HaGVyoUpY1CiZEoLX62X7bVA1EMrCoyeZTSO1WzBWQPF1+L5KoPwmgGIExxKTcVY2KVSOoPkIjOu/NDTP9AwtP3AEeNQA55BtSt9ZcBzDqz95PNeRp4PMQADm0bsLhJNYmPoOCY8EJszFfVKWsVoTWCJ2RdI3oOZ1dz1UB73DVA7TaOSV/ITUp9DuqeUUud48s7oPBubFqI520QrovLZ77/yXSdAP/wR++5Dd+75iLQXhn9FyO8Oa7es/5rd+7w4/9e2/o9NYTRXj5449Yf+M+//pt4duPX+K5ozS4EOEnvvCQyRt+8+3AbWexBr71sKGf4PKhmjNqRQeIWYI17/d5pB9q9bTIiCpRCVJI6HfQdexiq0cswkPZsoxNUajK862P7T742D/OgtCIDnbsk9vz9D2h+IDstkZREur8XQYU+ZzP4PZ6sYPdys6YqkD1NuT1zOujalSRNV7J2dhS0TiXHnVrb0uir9sVqnldT54HPSZZPjdHl1W2IqxwOFSWulR8JBZuSFbYWsvIKuqxiYTSyibI9yQhDTvHIrKg1apnqZCFg9PexIcTNyDkA4ijTt2orZsIgyuEcW1vkZ1WFkjcD6cXR9cGztc6wboXjBh8cCw6HVHqmpFx6GhcR9NeKtG73egI9+JCSdwSaG6dIkcb4ukxcrJBFgMCyP0zpAnQTimxD4h4mnbN2J8AMLz1Eu2dR8jtNfGyQ9qRpl0XgBEnq4AiCGxbDsbY6Oh3OynPY7LEyczgA5CWAkhKVSRVQ+r2rCIL33gEnRfMylpAUdaKwagq1wG+SA6VxT2cDBTfkfSwrwHIVfAxH8im8eXBX1c+6qhJnn5qrjjeh5CqI1NTEd91PnMrhMPYgLUTrusZ+47TJ8fak713f62FCvzUME2GbjEkTxjP2dkCdT02hYfU2EjTxGoEdiac5/+1k3pupcreN3mJ+2T+GlJOYRew+KgJp5WrKlmC8j9MUDCS1ynvjRjnyosR7cO/iQ8m/uTP/elXa7DxrB4fT2u9+F6GjYYg/tqqSOZwdPF6jlo20LMy9/+36eIYEvka4HZjWTYq+dtZHWRSud3k49OGKwAkS2TXACHHfP0pqKlbrurpmmbmtxmJO6/LZxUgyfcfawNf+NwFv/F7xzjb8J3tyNY3DCHy6du1QqEgRud711neHia+1XveHS3KKoCLX7vP+RSAyPkUkkCEFJnsizHSGOGy6tmsR8DHNEI9VQlibv7JiXiP5whHVjs6VGGr24AUGMhBAHIoDODEsCWUqivMFZGBcPCcfa85O9SosZ5Xvc552fugKB+B7P2x2GtJy/su77OJyKlclv06ErgXl2qkGeeKR/a8ydfg/r6pDQkDkenAduf36luyuz0uGrxopcPKQIyRNWMhp7O3rdfxeXIc4m8EtM3LIPSpQhSqfbxlYBTPCcsrsr838eHHDQj5AOL5uwP37p8COjI1+FZVrCblizi7m6RZE3nxfpKstYGPv9zjp4a+12R0vW7pB8OTx8f4qSmE5X57VNzJu7tvI92AOV8xDanFamyQVtue5PYaTEDaACEZ9T13hn1Tk3c/LTB2xN16rL9dptYqNyGtxy4H4vlSOR8mKJgIwj//M3/l2tLmH/vyX9IExQb9G5sCMoA0H2bQEUSnSZErKeV2nZZJrrBU1ZJMZLftrMoFFM6KVovmFrZCDE8qYVNwO34p+djtjyq6VsUAQpRZwz9J5NbcjqxilZfTLXalL2kmnItst1ky+erNNrd7rU4uZ9C0XpVEwpjA+dkx02S0pUOSfLANxVnemECwFttMeN+maohoy0cCQdMkOCfJLA2GyWBtZNubwlVSA8PI6JUQns/f7AdS9k9lcpirG/vAAnbfN/l0kLn9yooCjXzsQ1K+qgsdU8ijzEIfIqvmanXmJr6/8Sd/7k+/+vNf+rkrVQ+4KpeaP/tuRyNzi8azVD+aaAgSWUXHWvRe6SUkMq9nNjWrEvUEPHLi50RTpdyCFVEJWodwlE5eZ/Q8zm22C1dVQLKpaZDSihUihEmr3zm/zIAk+CpJk7jL+UjXLoA1oWoZvSq4ke9b3/rWXbo2cOvWlsWyp2kmjk4uOT895gc+3vPGw47WOr56ke75p47TX/kY944in3vlnNXRGpHA/ZXwqbsN/+adwJMwlnapda+j206EMcSyJ5t8f4mRJJLFiPphZPI2KBgc986JzIEQNOE+wrHCXklafQKRG5loo9lphZordXNkraTMLMw+HE4MfaqC2CQVnM+L3C7mDlQyMljorzkbDwkbgArwZlCyEG3E6+Ps+t6n7d/3JJmXq7K9l4y8YS6IRBY0PB+OiKKV7zGd54tEe9fqyf567XuZ2OJ8nkUYcpUwsts+uV/NLBWWqNK7NhomE9gylParicBaRha4QqjX/fFso0eHDBPrdWixuLhkInIpW4I4XLSJy3IDSD4KcQNCPoAIUUePGjcUkniI6sGQEz9rIuteUtInjKMtD6lbd86RRU/bOrbbjsYGvE/9+EE4PlmzWF0UIz+3SCpUYyaab7EvPS7rE0eLOA+dR44i8YnF3L8knu1K3Bo7aIKfWwC6Een0wR3OF1qxuFxAN/Ar//Fff8++yl/96b/26h//Z//VAwBpdVyFsVFgYyI0ESYhjuiZOVgFJjbskNZz5CpMiUSCB/1tbtmyZkMMluDbQr7X7RtnI8SK7BnjrAITvFWAZr0m8niogIj3prgF5+Ma/AxqbDNhJNIt+jJtni+k6tikrVGew+CjDttMjH2XlM6mRKjXKowxHueuSu2W9ShtdroeMVJ8Zoa+YUz+INpuFZkQ1lurbYJp18fUEpO5IEaS/HPUh3tdITnEFWEPhIho61RWh9t3Oq+rI/VnptbxT0/E1miyUHNbbzghH3xk+dB9SdUcdZtJfr8rh/v+4lkBCFDSp3qEdz/qBEgldud7TA1AGsnttFpxG6O2XWUOk6tasDL4gKuiFVcqlnsVDjG7SnL7caiVFOZ7TAYjuQryqVfe5Zuv3eP8vOPf/O4JX/yxN8t8Li4bvR944VNdRx8ib44D/dbydi/4cItPfczRNJ5hgt8+9YwxIKk9KR97h8rO1v4e/Z7sbSaaAzvJ5/6xGdNccstRTehu9yoCFuEuHW+zwTG34JS25qpFao4DfI9UvRGUHyLANsZqftcD53orD1UNPJFODFPyUsnzLmprImxDKNPmdb0OgOj2KH/jidHBLUE4iR1HOIh6Pq7jNAPpvbaqeT5XP8vHpAYg+zGk76/uC51fk1rFngsr3kyDeyOBFR0jnibugp9c8XiameDT9nMdWi3Ttqwt2qY2S/TexIcdN80KH1A0bsCYgG0mhsERgrBwuwlTW0HCJ2dJ63swnD05wVjP8Z3HNM2Ea30Z/XLtiEhIPcKTenQs11rxyF4Vqw3hdIUcT+BSUm+iHv0J4vmC+O4KOe5n00C3QdoBOdlgbq8xJ1v9jQvQRKQJyMlGOSHT9e0L+yHNXH0RExXYmJiqGmiLl/MzN8RN8+vEGZHFoLwTozwTaWoSRkxEeAUgVN8ZO2DsVP5ss0VM2DFVNHaicSplbIxWEJp2VAJ5akUw1hfgkR/s5c8bXDuph0unXAzr1BHetSPW1e1cvoxc5mMI4NxY2q2cDMkHxQAAIABJREFUG7FN+r0baTuVXLZ2Umnlrsc2Y6p2KDg1Rsmuy6UaNC6PLnGuL5UdMTGpfeXKhy3r71NVZ5qErk0P7CCEfapOaa2Khce0QyQXktrP7u+czbynqy1ajZkTLVedUj7OFZRadcvI1QeJgplIm6Y1QBaIuIkPJuo05bsZbXyvCoknPFVec5/7cZ1ZYT1dk+jguU2lJh7bPQAi6TxvjZ6LJ04rGY3R87exakKoSljztthKZc6HvSTVXiWP7xuwztNW97ZqEGTn873XTaMDF5/+9CNef9TSNvCbX30RgKPjNZ/+9COevzvy3JG2jsWoSeh59JyGid98PPLPfrvj137nmDfWnnX0pWpR8yDGxGMIZI7H7PWR/8YEHifiTitW/hvSXw1Q6hH5GujWUYOU686h/TaoeZ3mVi4fVcWqNRXgLJWL3fc58jYeWpZFaEU4Ek2COzGlolCknkVYB79TGarb0jrMwYR7InLGUEwIcwVDquVnVbH9ikVuf6v3R369/75ecn6dBw7y9PXf7m+zt8nEWgYahGVsGJnYyMgo/op61tOu7/eTvNZqWNtUAb0BIB+NsD/zMz/zUx/2Svz/PcY/+cO/+MK/+JWfyn4cq6MNQ79kGPVmVPp90eqICnMIRCEE4fyyZdEKIvqZ9/o7lWwV2m7EtQMgmpA2o94gvCFODln2mJfOYJMUkQBWHmkgjnnY2hAvldwndiJODrPsYdNhXrxU8OEi2KhViaFBjgd+5T/83159/Qf/k1981n3xnU/8Z7/4ie/8vz+FSfPq0nxjGj7MD+sg6uKdFaicV+5K2i8lTADRqohYNQaUxmvVJD+8TdSX0WCaQTkgxhOj3fEKAaP7N6lmicytWCFYXW7mPzQBmx3qi0u9p3FaMVGwEhKnZD7GxnhNNLzFNAFjolawTCQEQ9uNNG6iaTMQ0WV2yy2uUyAbg0ngU6V5XdvTOAWjw3bJODRKem08t25f0C22Kk8cGkCSMILBNmGWhA6GYbTJ/V3Bb4iCa3T9fZiTrlwByYpW6vFRt4zsjtrWj+RsUFjzSvbDGq285GmnXA0J83JD1Mdmec9ue0BE6Bohp8SP/uHXfuq5P/P5Zz5Pb+L3H5/5sz/8i9/4+1/9qaujwJRR6Zx41eZ++/3o9XnjJerll98/Zdr95cXyfrfFKkrExlmpyGGJaL+9qgtpO08nBieGpVHCuTVCZ1V2d9UYFsmA0IhWQlyj56+zMwckg3E97yXJ9Cpwb5MpoamuG9FTN/E/Yrqep3KvKZXaMFdXS7tl8vPIfLZcEYnRFD6HCLx0f836Ysm/e1f4/CcvdODFeo6Pt7xw/5JPfWzND7y8oX9ywo9/PPKpY8vDC1WhezR6zuPEBl8AQwYVpGMayEBRX2/JDtszUIloEjsT0Cm8g1z56LDlWLQpCW+q1iA9H3ReGzwCyRXcl8TbV0lxfq33i5mPlO8f+ZzM52eMMJDduoXMfDDM4CSfY4Lei3LC7dL9x2FYGqvTy241IQOxyFwdGiuwVp+zaqa5e7ZfMnEqPaemxyA8H464FTtEMtlbgXWTzulaGaw2SPTVOuxvD9U2ZeCUTtFUV5iJ6Pkxmf+86PzGVCm7EOWEGAy9THjR1jGLoYnmfQ1chL0qYN7GvJ3zMddKkRFtAWuYQckX//OfuHkufIhx0471AUbTjIgJGJkIQRgGQ+e0DQuyF4OOai26wMnxoGXvvmG7dcBRSjbBtZ5xsAx9g9xWYrUYlXvFRFj2yNhgbl8Se6etVkGQox6OJsQmABJSa1SPckbuXhDfua3rMzTacuWFHUdzidBd1Yh/5sg3jqaap6uSiihIk/w4Mlk9x0EZ3jSf3DudOCU7U5iIkDxLEpld8EVZaxqOSuUIcltWVtpKwGNH2WqefwxClKuSmbW6VpmP0XmHKDiTEwVX1Ldcp1UZayf6QS3G2+WWaXAq7WynklRkQn2Mhn6z0l3a+GJ02HUDruur1rCB7fqY4C2uHfFjoxyXcFUdTKshc9uW9rHDmPrWQ9TKXWMjTYTNoMlVVnYDirrb/ohV/v0hF/b8Wc0vqc0LI4lbkoXW4KCh4Zj6xqyBfrop+X7YUe//Q2Tcp4U/0G50qE0jL+PQOXcoMgDJ0wciTdXukwGIRYrUbv7fpr5BZyneHyq/m8HErCiXIwMMa2O53oyASddWrgoWQYs0TYyy44p+qAKysx9S+1VNcq/jG998DlDwc7o2OAPj4OgWmVvmd3bgT/74G+X1vbu3+NprR4hYvn5mIOh+Wiflo6mqJuT7siatsTjPByIju6139VrWLT/ZFyRXSiQBkJec49wHLoNnqtqpFklcIBDZyMRJdFdG5GFXbasWRrBp/hkomXTstR00ptYsKXyQrKJl4kzq3le9chiOjMGj96X8zVi1qwkUQBNSa+v+9eHSel2mfd2LRyIMyZSvxxfCd4vBRqEvgra7VZtMWI/EHf5KXu+cqOc1yOtiKlC/f8zq6/Rgq1fy44mEYiyYzQ2XSS3re8XTKMR4LGMCvwG95pc4NjLS3aS/H4m4OQofUNhmnP0k2g1N43nh/oZhaADH5IXJq8zhwqlj7mq1pWk8xyee0ycnKts4Ovre4b1gbWS5HNSxtk0eGtGkFqUAZkwtVBNxsJrYTwbebXXUZ7DQp1H/e+eYo74oVmVzQTnaQG+QY1/IhJCAy/B9SO1GrWDEye5UMjIQ0W1I0+5xRLBBfxPl6neHogIqMs4O8BnQhcnOACQBhtySlduhcuTEIE+fAUitSBWwhGAxxhdn8zL/tK1NMzJNrhBQbTMqlyNx60W0PUxfawve0C8LAX4cXTEvXKy2RaigbGdqL7ONtnlZNyGbLlXVZvnixsbU2jQD5F1FrASam5DAirZNZQBSPEJMqlJUVY+6BStXN8r6VcDEpt9n7Bf22rHyvBK2u1Jez2R4J7uckpv4/sYvfOkfP/heJBOHwEcdu4Zj1QDBM87fXPkvrGJTPBYgj/6mEV/J1+pVAJLbB+uWRJiJ6JCvl5QcJSBibeKTpW2dKt4YKB8k88TqQZBpakpF5JCTua6Dzuf8fMVrb6xSi1hk0XkuNyrPvZ20qthvW779nbt87nNvXJEht81U7lN3nzvlx0/WWDfx8ref43deX3AxRt6ZSGAgFkByKBHNwOM6Kdg8sg4zNydLvGY+wy1jOZ082xhUHCDqsutqh0sj4ZdMHB1Ic54wFHnfukLWpOOd2/BcIqUbSBUxnXaKkUUirwuSBkKy/K4OmLhUubAiuDTy4pnbmJxIYb/UKl6k33ai/h99qgz1qdq0kYm1jJpcC4wJji9peC4sS/WnrypBWfmNMv8Z1B2K6/gdmfsRiDt8nGfhcuW2R0kDCPX1e9161AMK82DB4fv9daHcmvk3qm52A0I+KnFzFD6gsE2f5G915Pq5Fx7StFsev/MiPpyw2Vg2g6Vt1BiuKJ64iZO7b+O6nvXFMePgkgqTxRhNAJdWjQmbdoNttoSLFbJtMc+rIpe0nrhuiZuOeKoGhriJuOk0s2s8nK5gsZewpnasaCJxGJHbWjWQo0C8NMhy4k/8+p9/8Ms/+rdffT/74pf/2N98FeBP/MuffYCJiDqEKSEdSiUEtIIBlLYsoLQZzAKw6WeDne9Yk91p21IeigHr9fPJzpWPDBx8g3Vz5UBdxId5HnV1JEoxOQwyE9Gz67o1045SDSgvKPt5GBuYBpdc0eftmCanFTMJtN22LMtVvJXgDbbR9Rr6JZdnJ0xTQ9NMKmjgDSe3LlisLsp65+gWl2zXx4x9R7fcsF0vq+2LWKv/sxww6IjpeptbOjSxWnWB2ycDPhguN0KT2eGQ/D1mMJKBSKlo7LVz5SqGT306+2aFh2LfBf33/ot/8oAIU5XbWDM/8PcJ7zfx/Yks0XsdKT2TevfJvfu+EHC1d/26z3S+1xNYnyVhuRsXpd2lS60/C6NJpwGayvPDGVg6diodWS1u/7MMQDL4EIk4G1l0yu2TqtoxTRZnQhGIyNPPVZO5vSq/npLpKHuqfXWL1tHxhj/0+Vm+PIOa3/qd53npdmDdG371q7d41Htef/cTNAa++BPfLvfZzeWKxXKjrVyTlGrtw1PHZ1+cWHSebW95dNbw248nfAylZajD7LQP7buZA6USsV+RyP9zwpwrD+fBp7atXYf1/fPiudixwfOWrLEYnos6yCIId9DW4zrZPhbLFJU03kdN/NsEQloRXlrVSmN6vxrS6MbZFIpZavYFsQj3G8cYow7QAOdR0/XnjWOIkWVSwnocpgJ8M2k9b1uHYSTyhlwySmDLyCq2nNAx4LnNAhcNyzKHua3MIhzTMFb7eD8O+ZXUvI7a8yOrbAEM0eujFFXdytPtX9c1V2iQGaB6yUpZ+gzKlRJd/3SuSyyfeQnpeRvL/0ORQet14YmcoMtc01873U18MHEDQj7AKEpMJtAu1Vk8eEvwwmYwWBPpR+WFDNOsruTuvIsfO8a+Y7HccH56C1BlJoDF8hKAKbXvNO0lmEBct+pODshqILyzRFZJGtbb1GKVLtYsf3u5QLpBv/cpUQ+CvTNoAuog9oJ0yvqNnt8XEAEQl7PN+sNUychKMqTqQn3DqWV9r4vMC6m9OUzQz0dtM2O6mpnGYHYUtHKyH3yTHNyVvD7zRiLWempAFJMKTAYnhdButMgexGDtUEwSYzQ7HiIqmWtLFWWaOjYXxzvrOfTLypDMM44N45hd0JWTUnujjP2SJgGZzFnJBonD4Mpy6/+grSO3TnqmSZXTCjfEQLcYCcHQD4YeQ2Nn1avclpUrEIeqHSRAY5iNCPfbqm7/nZ9+X+dVY4QpxHJK3RRAPrj4hS/94wd1u4sg79leEZn5IfujqdeZEz4NiMBhMut1QCSPqmbVpQ7DSmwZ8c7g1YkCD9IqZa4HzKIM+wCkSQT03H6VjQqdC7hW+RtZdnuaDE2T1Paq1tfrlK9yNM3ENDWl9SpLg5eWrMq0EOoBHOhc5OGZ4bMv98ijlhAtb28CR1b49msvcHw80HUD3/zWHUZ/j3u3Rz7xylvlvvL83ZF/9e8c91cN374I9GFiswdA2gQ361YlVa3yV+Rq8//azG//e60yXAU1sEuMz687LCZ2PJYtF0xMBO7SXRl5z/LCJ9ZykVQ4sgDBc51yhazAW+tQ7jHrELjVWLY+KNCo+BVtAjh9CFwG5V48osdhuGCkD57bOM6jyu4qDyRFtIWYnkfuc/RMLHDcToBqayaI0FRwKgIbJpY0xUzwOi+esbqSDgER2JXBrauT2YhQj4uqFV7laEj534tnxDNm00IGXOKC7Mckytqx7IpI5Pk/DYDk/1oBCeVY19wWVcibcDcp8IceN63SH2CI0fYZSKPqwbBedzRNKG7p+h20TeDycsk0OHAT3dG7uE4JyKujtSo3ScR7QwgWP6WeymYL7aRVjMwd6EbiOjmTB1MUp+Roo+aBAH1LvFxoG5S3yNFW/xqPtB46BSBYkC7xRACxIIvf3/74yhf+9qslOwg6L9qQFLzmVixpA1n1ShcaueKuHg5lK1rxKf08NlxV8soKYsmoUffhUP7EaGuWW6g3h3W7lRGRME9bZH5jIY3rvP3O57Z6r8ubK1DO9em3nhC0fStLBNcqXOuLY7abJZfnJ8VbpAYPm8tVWfcYDI3rZ85QiqFf7FRr8vplE7Vl51PbX+BoNXG0mjS5atRobb3umCZL6wJtU/V3p/W4LoHK57qR3apH7UEioq2Jzxq5MpIrNd1N6eMDjxqAhPR/P7HJ75X8upvEHIrvpYmh2Xudz9i7cbEz4g4KNFqjfIDWCM5mLkhul42p8pHI5UlQxDUR10SaRCavlbCMjTinwMPagHPTjhKe2eO7SVqd3Jp5KHJVpPBHSlV3vs9loDJNzc7njy8MY4DFYuSHf+BdfuSTA5+9YzAi/O4bLf/qa8f869++x1ffgTdOha+/0fLaN14ieKv3n97yyTvwW6cjp37iPPpCBr+FS9WpuT1qWXE7auCwX/WwFQCpief18buuncthdo5jnuf9uGQiMMjVtjVdztx6NaZBpD4qCGitGk6+tQ5sQuDJ5LkI+t2lD2xiKPLD9fm9xXMWPKeMpUVtJDBK4FxGLplY41njk8fHxEDgMQMbPBdMvClrLpl4Wzblmumi+nysRSGEi2ZHvlfbyWzZh1OVmOfIICdXTGouSBYK8BKvtEQe4teAApVD7ZMZAAiSTBJNai3zxTl9GR2jeDyhKGT5vA7P3HQ1x5Bo/p7ZgwZIey+W7zqawk25iQ8vbmDgBxA/+nf/6gMxQQFCCj8tePzOx3hy2u4oCx0tAq0L3Lu3Znm05s7zryGrHmk8J+0Ek2U1NQT/WTabNBqyPmKx0iR57E9oF706lAcDW6emginitoVtiywG4uVSeR8n65kbcrTR36ZKjBxtFcScGeT5dEMxAkzERKiXDn7yaz/7gBD5yg+9z4qITX9QbK/FpWJs8g0higIRuFK9iCFotcNQVLHiYAu4kHZS7kp+iBsde42DmytBwRCjpWkv8eNSW7KCxU8dtplli5v2six32NzR2VltnfKTNh4ApZKiybRWJGqSeoyCT6aIOr0tSYaYQPSmyAgH3+BarZY4o2Tydrll2CxUWnfHZT0ldCayOr5kuz4py2hcTwxKns/tXsN2Sd+3tO3ENGklzgdh8obVYqJpNGnKFZZ+SEphXgipZbCZhHv31mz6Y5psIpIfxEHKaHDe7nq/NJbKJ0cPRa6M6ISHTpjr4wf//p969at/NrVlBVikXe6sXOGS3MT3PmaZT6r/WmvYBxMhARWYz5gmkYTr5Ob9VkPgvSsiOVaxUTMzTGnBakWBh0sA5LiVonq16nR5t44m+kGTfzFaCXEuEAM7pp8+SAHnbeuLYl3XDSxWW1zbE5JRqg+Gcdj199HBiN1WrNq/qGxT9ZlrpysCGZk/AvDWW3d47a2WL/7Et7m9Cjx8KLz1zoquXbDeWn70C2+wXS955+ERv/em4+5xYPCGpYPXzyPf+obAN16iEWEbA2P0VSsdHNNgRfAxskqj8BOxjIjbA8ds35jOHThfXPU+jVkBu1KwdSWkfp9BzZKG4+h4KFuei1oNmdWT4DyoslY2BbzfOG6l4y8C95eGweuanY2Bcx8YkkeKSb9b44tMrkV4RF8S+gsZ0lqnFi7p6aLFYjiXHosaZtZk6uPYciEDaxk5iR1ttCxjw4DnOLbcjl0BExl8BCIrZtGFWrJ4f9/sX1r7juf153Vr1n7kz7poC9m8XuYgE23U58jtuORctpzEBcexYxRfzXOet9vjsbxXZNBynf9JkLkidKj6chMfTtyAkO9h/Ojf/asPNHnMLTwtdnnByJ0CQPy0oGkvi8xqiMJUOeKGoA+f5dFa/SGML74ecrQhnq8wqw3Pvfx13nrt84D6PDSuL+T0sFlhlmudYe804QZtRcqVABNUovdQZSAYBSXdgHl+DS3EHsQ6MEIc00h+FyummA5t/+TXfvbBVz7/t54diNR9OiaW0b9ZjUvmFi1QYFIz04JUzOWU5LaeuE29pUOz47peTzcvJxRjRmvWEAQflrju/NrVFvGlqpXfYyiGkTFqwp/buHZveaaAAwDXDnhvsdaXykoO6wYFD0OryXubqiduUpd2dJSz9psRiWzXC3Vzd0LbbUtFJEZTeCfTZNluXeIWzeeBTcehbSdCMHgv9IMlRApZvWmU4GpNKCO5wBWTwkOx45mQXk9+ljHOhPVl+/6bqYrCUHkswyt/7/21dN3E7y/2FX1md+qATY/9wNVR7H0+AAfnc/VcuC5hetr61ddhVl1S/wmNRqQQz23if2QVuGUbaJpI00S6dsSHWX1KBFznWa8d1sSkHpeBg1ZE2nak7Ua6hXr75PbI7foYawLhQLWjribuk9DDnmpW9iAq3kNJ+CKLYAC8+OITXnhe+OVf+wTWwEsn8PaZ4IPh9hK+9rUXefmlC17+2BPu3F5w++4pPxCSoem/eJm21/vEpY+EAK2oCpP2/gtHRjkOkvbjqZ9YJo4D7FZAMl+gJpTXx7WOuu1qPp67x31f4jkvJ8+/S8c5EHnCwDKlPwtsqhaEMp8WwxM/YcemKKE1Rv8uBhhSy+eRtZz7wDZB3kyefyI9x9GVhHzA74Di49gSiGwTR2IRHRFVZ2uxjFi2MnI7dqnVStu7bsU2gfZ5b2zSftSqj1Cj9ny+H3KgPxR536sy1pzQ7w8G7JsJ5qS+T1WM/SQ/EAsfBGYiewYg9fVfv37WSuj+ubDPEcvtW8WhPpqDbus38cHHDQj5HkSudERMcemOQR2746AVBnXhHmiOlSzOFoa+TaADjpaBcZRCZuwWG5p2i3nuTJPmbkQaA6xVXrYd6JY9y6Nz+s2KzeUt3NjNYCS1XkUscWoU/CwuoPFq9OcmrYRAqYIAKtN750KrJyFVGFYNsqIABnGW6IJmm40oG9jHNBRjf39VEWcS1wOdb85r2wpg5CpIzpdNqpCkz3daizLHJYiS0b2d/5vkEzJZdV0fGq0MBQPGw9DocWyTEWNuWRuaNE9LBptS1K+WKhubgEgejYxRUyjZaR1j9idJbVdNM+6AGqAAB4C22zJNjhhjGQV1SSWLzUxQLe1NNuC9wfsFMRqO2k3i8JiimHV5scQmADKOtozgdq1PCY5nmmC7VbUuBTFp3VLrVojCdtslh/XdyJyT9+prz1FzQnK71sXPfvnB8d96dhBRP4w+eQM+PrSogYOOxuYe7Vh9fpWkfAhw5AToaWDkvYBIBiD7yUlOSpZStQWJtuY4KzuqaovO41xg8oaTkzSolFQKQ1DfpnG0CtS9lAEVa2YA0nYD3UIHiLQ9d8TYJU07Mo46WHRIVhcoAKTmeWRBip39VXNKzO68jPVg4Y/8yEN+47fvc//2yLa3/O47wvkIR43haLng9r3HfPO1e3ztmyf82BfepF1u+fE//Iihd5yeLnly0fDa44a1D6waVasakrnfrcbgjHD/CH73scUkcv+Uvgc4DXN70tOPWyQbE1qk+GfUkcFMjlzJ2Ac3Og84io5LGSFqxWKLL+RvXWYyW4yRR9PEPZkrVIMHn25SToR3J22hCqgPyiJVQBpm5/dLGVhFx91wzKWMDHgshlVqqwI4iW2qGgWOcDySDcexLdtlUjuTlG2ZI4OpQ2dNOLCP68rJdWEQ7FPI3xlkzECySviBUB2TzOk4lS2r6EoLVK78TETWMqi3SQVEDoGZq9t3FXzMIMYQnnJ+ufj0ed/EBxM3IOS7jC/87f/1AY3yA2w7t1tJO5TR/LKT20mT3klbfRbLDcerE7ZpdCm3r+Q+4nb5RBPl5aCgwBt1KUeld+9+8mtMF7cJvmG7XrG5OMa6BSd3e0KvACN4h7GjAhATMLcvoQma1GeJ2kz0NgEkEi8XiYMRiGeN8iqigpHCvszDhL2f3+dr2qmW6rNURaRLiEKS/rhN7V6xerBOqYTdsNuOVfxG0luCbhsQXe5FRdu1pkTEn0w1UthAMArKAAlJGrjxCF75Mst+lvzNQGKy2JB4NQsFKhYwVrdl7E/KKjbttgCJorplApIqJblKUldPMmDJ1RUAa4Y9PkrlFWDuMvZdAhNNSf51usj56THLozOd1k5855uf5PbtC6bJMHltX/NBWC18Icq6NKraNB4xrvTYhMoLZNvb9Bq6NuCDMO5V1nbdnmMi3eu6tU0sbVJZ+jcDkWWro8nDBA//3JcfxAjP/x/vDSr+8D/4T2+Ax4cUgbjTSmN2vqNIjYJez4fUdPR3VyscBrkyGnu47eJw0tEgJcGxGFw0HKc0aSWaOLaJiNwabceyomT0xkYWLvL8C2e0nV6DbbdlHFqO77zDNCy5PLtDv12wWGiFscm3NaPX0u17pzTNyPLkHcxqw3R+G+s2TMORViLj4ZTQe4Nzyb8jGRCaSgTDFkPUGaTsyIIHS+OG5Ac0X5vdYsuP/sjb/MZXn+d8q2PrPkYuRviXrzl+7ZuvMMbIwkRe/85zfPbz32J1fMHQ32W1GvnEK+/whWo9v/3a83zzbU3Wf+JH3mV5tMZYT/tvXuGbj9MxiErofuyn4tJdt2rVKlpuL8mFmWsUUl+/IOnVHEMavR9SZSbPtwYpCyyraBmJPBZ9Zq+iY1WlQ0PexxG+fj4/iwIwRjVnnFA54myaeCYDIToswoUMep5h+Fg4xqZpLlGjvpPoMAiLmEjv6VrIFY7n4txC3WG5H2fi5exKP5vuZX+Tej0hc2l2QXrdtvasteY8gJBflzY2CcQkU7wfeRn5mjyKLZcyFDPKU9kSCdyJK25X22eQHa5Gfbzrysh1rVfFZJKZR2QqwFHfIW5qIR9+3ICQ7yL+yP/5Pz0Idh4lyRWQMnKOjshHwKw2JbGXBpYfe434nU8jRsv1Mc79884Flifv6AzGRkf7F6N6fTivZoPOExuP6waGb5/QtCNtd4afHNOwxHVhTmrFzwnzctCkPNhdYnd+XY+iNQG5NWm1YxOJIerl3XtwBukabc/aTAo8xtStm6RjvvL5v/neCWHuwVktkPWW6ANxCrMDndFKi1iUrJ72UfT1TS/34Jjk6C5I5pMAmGrszIjyYJpARDk2s2JWmKV88/7YcWdPICW1cElbkd4bj0y5SqKVhgI+8miWnUpLVOaJNHZ20ssSwFOvFS2trCVPkFQVqSsqygE6Ikaj0sCjK8n80CehgrS/prHDdZvkzh548viYySsPZEz/mybQtppEiUScmxjHhmnKvI04t09NQtto1WQcG20n8U1xWB8zvk27T1u5ZmBUJKjNrIyVR51zG8vkpVRGROCdn/nyg2cBIjfxwcfPf+nnHuQ++Hy15cRkX45V/8+Rk4bMDbm2GrIXz9qOlcmw9Trk3nmLtgo1MrtfN8mQ0JrK1bxVp/LF6gI/OZp2q39332V4/VMApeonEsuJ3zSebjFwcvd1pJn2u16lAAAgAElEQVTYnj3P8tYFtlvrPWMMNK6n3yxVdtTvJnNNswtA6tj3B1HTVCEwV1kzAMnv9+OVj635t9844o9+rufJWcvFVujHfOsVHm8jX3/L4dzH+eSnv8O95x8RginiGpm39vLHH/HKZ0aCt3SLdbonTfzmOwErqpx0GZQM3VcABHbBR9mW6vvstO2rY6mSszpN5nEAhY8RiIx784P5nJyI9HjuxgUjgQsZGAkcR8eFjBxHV87e06ij8nUL2SljcW/fpBqwi4YzGXBRR+NXZT20HbE+/7Z4VjuaVqT12/2vLYNCZm/UVY18nczO8nNbYS2RfQiI1Ms7JNFbxyE+ly4jFO+P62KQqUw/EnDRskhpZy+egVD26XXX8zywEDF7+/Hp6ywErp7z9RrH97h33MT3P25AyHcZmRuQOQUMdsetu3AuUgIry0mfqI3HLS5pG18qIaB68nn0WBbDXJ0YLOZkmwAIxKVHlmt4suDoE19n88YrFSE6FF6KNFOR5ZWTTTIZrNa/G4kpYWW/J7kb4djO1Q4fiFttu5JVMraz1fduvry/8tlnACAwZ6leM1ExVpfjsyX23k0iJ+wmquP7zvqm1qy8Gpk/AgragmgrWzZthB3Dwv2QbtTjV3FK9p3YMUqMlzY9Mgc7q2TZkDxDKi1yC37swMyruVP9IOxI6+bYByAwV1ZiVO3+frtI85vbsnziWoxDR7u4YH15Bz+ZStIzgYEkI7o62jJNWZErsDnTVr3JGxobivqU9sZra0pDoO0mFoMlRqEfmytu6DXI0HWMVw5txqPOwnaUa7+/iY9W/JMv/aMHtZdDHYeSm2d98D9Lm9X+dIed1OXK+/wHCjqWJlUpE/iwRsdBJi8cL0IB33kgoWnXeo+dLP12xXazxFcAQlXwAs552m5AFj3m9iULE1QcxGg1euyP6Dcr+u0iSWvrNWsSmBGZfT3K+lv1SzB7CZZ16k0UgxSzQWMC09juAJDMFbHNxL37j/kTyXzwydmLLFzkznHgM595m83lil//7dt8+9Jz992WGD/O/ftnNO3I5dkJxgbefPM2rQs8fNLyg597xPGts2L0+vidFxijStka0UpYVnianpIYw1VgMhUQMauqBeaR/E3VDJpN+up5T+W4z6P5GTQHhOPYciZ9aT+6kDG5rQOE1KoVeMLAEY4Bz4DnDi09Kj97HB2eyCiBW0lCV9uNtCExxzI2hTvyLGERFmIYYkiCtqouldsHj1Mal/lWtkrmn+X6eVpLXA108jwhVyHeO+rWy1GUTJ/DRcup7LbS7atV7fNF3q9aVs1Z2f+tUkpvQMiHHTcg5LuIaVhim77wB9SpPJlCWZ+I4EvM3bOUqIZUCYlgIu0nv8PJm5/gfO2S+Z1eEOcXLd/5+h/i+Y+9zuLlb0EbMPcvU1KtFQI5BnxE3IY4blmOrxcDwmmtPiKy6JF2BBuQxahgZtMot8FE5KjfI2lXo+xJGle6BrqGeDHAJl2wufiT9NTlJAGSGIn9xFc+/b8/EwD54nf+6wel2gGw6GDbE0OcAU12sKuz2vQ8lW73BhJHIJOZewudV1ASBGxERkPsRj0WGUwkHxRMrl7kVq04t62lFowdgnsCktKmNGuyyEL3hxnmqpifMjCY923TbvBTB1ZNLNVLZAYZIe3g7CuTw6SxvabVnvLgW1x3mUDnyObyCEmN6NqWpWBksehZHp1xcXqfJ+/e0l51D41VEGGdAgrnJrw3dIueGIXN5YoQhHE0+CDqoRKFxsx8EJP+O+c5Oem5uOx2AMeuP8hcAYGZxJ5HkK0oALlMqmtGIGHS9yS738SHF/seDzn2k5t99Sz9bDdk77P9RHWfvHooybI718wMNmyU1Oqho8bZF6R4GaQWLIGiiNS5WMDvu++qHPbRiarkDf2CaWzptx3TZHZ8ekABQONGVsenxG3HtFlhXI+crJNRrNCtHjP0S2wzsd10xEBR3XLOJyAzlipIBhPGKjcsxl3QEcXsXGPT6HYcz+tqSq6itEsdpPr8598qy4jRcHa24MW7nnvHwutPhF9/aDn5+nN84sRgDbxxHrmcAs91jje2E21zD7jH6drwzTM9E/qoo93E+XyYSiKrVYw8Eh6ZCcu5zShvSV7rfT6DQOJ02MLrqBPfrXgWiXvhE3CpDRFt4g5YIrdjx1qmqp0nJ8G6nIB6Y5wxqJwsgS0To8xysovYFC0VXae8BF3m3djxWHqG6FM74NNHVnqUuN1EdYzPgCADj32Qb5l5V/tg/1DFqf5duPILvWYsUraxjrqisK3Ur0bxuGgLET1L5daR52UQHsoF9+MxAfXuOORk/l7KXE/bj4d4JU+r/NzEBxs3IOS7iN/483/x1R/463/ngchdlsdndC+8rl/sjZZLciKP50t1Ak9tWSRjKWsikxdi0BFma1SaMUaj/AzOFICQEm/LzMnw2hMsR1s1xWsn4oVBFtXoezLCi5cLZDHqAynMruS4q4YMctQjCw8VKQ9ldmoL1rpHMj+kcyXbFPtsIzxffPsvPMCI/lYMxAoAJRvtmO22p+urFddG4YtEGETbqtJrWU4wyVwN2Sdy5uMDSJN62CdzpVKkxzJAcGQX9hyx6r/OlakMSACMHUtrli439egGM3/HDEyadk3wbnZ5RyshttniFp7gXSGqj4NDJpKDesM4Ki/Ij02S/NRHsTFqmga593yi7QZWJ0+4PLtbEp7sxJw5S60LpYLS2KCHryKgNzbuyE7ncBbGhE6ykWFuX4lRTToPVT+sgeDh/t+9acX6KMa+sdyhyP38sJs01MlIUeJ5ynzKwKVcVeyBWeY3zxM0karXU5WEVMLUVWpYOs1cBQEKZ2kcDa0LjKOl33T4sWEYWqbJJK8mU25hEcGkNkgjURXyFr02iCz7uQpyoTLfGUzsbGfxEKrUrxKYyEAi/6b2ENH22/Q6T1ddi/vKWTMPTcVQpuQ39ea3n6MfLI/OLMME944ip4PhzHv+9eOcXCpJ/Gyjo/K/8Rb0IdCIVj+2abRoqqofO9uY/rsKcGQ1q30wknkE+yPzcef3+xBWJWMjmsxnBan6/IjVa0/kOOr2P5QNRnTaW7FlJDDg2aaKS4tlhWNMUrlrxpI8r0V5HwOek9jS7Y3uG4SjNND0LJUKvXZ2Wxuz0ILKDB8OQd6T+J/jUFJu9vaVQQ1Fc0Uhr0Ogar0U/T5KZEwEfJv2dBftlXmPTHTsSlOrieDVPOJpYOO6766rut5UQD46cQNCniHsf/P/PJgmg/yNXdLrvf/hHz24aI9x7UiMhu7u2wWASBNUdamtOlO7YeaFOE94V8lnPkjpu88D5ufnC07udNQ+gGKBTmY1qjJcp6CByRDr0frUfhQHp98Dv/If/Y1XAf7Yl//SgzihRoQ1ByKHibAU4mUFZjpJUjcCF564VGkvcRYSN+aXXvhf3jNR/OLbf+EBAK6hSMiECGHcqXiINSoH3KRMdMygSXYqIjv7x+SKyN72JJnfMhKYwUYMM7gYm1QV0c/0QW3ATYhpiENVIckAxoZCfI/T7E9SFlu3xsnM04lRAUZ+DWo0WQMVUPDhx678HqMyz7bZJqlnm0jqjsXyMs0/4qdGvUXcRAiWy7Pn1F9kaNlunXoaiMwjq+n4D31Lt2gZB1dGdutYdp5+MBgby4it91IUtICkIGSveHPkoleIu4R1mJM9H3arJ85GRq/8k5v4aMfTRhf3U6H95Ou9+rvr7w+Zoh3qJ68nq1WG9gHToa72Mag3CFAk1IfRIKJAvu9VGt0nv5x8PocgNDYot8OlFqmo1dY4tHrNn2yIJuJWZ/Tnz6lKor9+8Ma6qYCRDB5AQYle59pyuc8ZqSP/LgSrlY7qtZjIN37vYzQ2sFqNbLaOlz/+EJGA+/YL/Oo3Gl5fa9XiR+44fvfUE2JkZRWU9FGrAuvUQttFU9y+64R1/1jlOHQu7B/PfQPCujKSlbAOzUuq36xlwkXDglmVaqrS1AxQDMJRdIwSkpKVMIj6oSxocNEURaqLBIZfZJW4JSNNNHTR0mDYit9Jvrfo+/fTWZr3YI/snKv719T+9Ze37FC1cXe6w7HT5hjT67pxYmfZCjxqdavcArVIYO26yC1amXxeR/7dvgJajvp4H6qWPAvIu4kPN24e7c8QL3/slNffuE333/3fDxobeOWzr3H2+B4bq1KGbbehO3mkAMRNOkLe+CLpCsovwPqq+hCQO1rWv39vTYzCo8dLghd8uhCHrcrkimMGACLQpZEfl25uPsDxRDxTIPDrf+6/fRXgj/7D//5BTO1GGXyU9VmMxK1TPw0Tdio05vmeeJGqEes8PQoEfJiVsDZRqwsrneaXnnsGAPL4Lz3ANZB6sHeqHMZoi1eViUrX6HuZHzyazbJ7FwyxtN2KgR292E65DNqWBYy5+hDBVFK+VUUomyOK64mJs6OALbV3LQd9sPdOgUjILV46TQwWMR7DLONrzQY/KvDMvjHBtwWoxDjzSSIGt7gonCMRj3HDToUlBss0rGjaNdnNfRoW+Kkp8zE2ME0NF2fHhCBMk8W5iUksxij5PCco3bLHGM84djx5fKz8jsEWfkmM2gYiRsHBajVgTOTiQttR8shsjMrnUK+E2cU8xhlzFs5KuhyKp4LM0zZ2/uz9yPTexPc/fuFL//hB/YB/P2lVnRYcShJi9d2uEs9uhSO/3h8FrSsfsOvK3aXmDEHopJI8Tbe0XBXJQDhGYfRa2ROxjKNhmoR+VPEGPV9jMfpsmlj4U7AnG77otWX2fMlweh8RzzR2BUgApcXRuQnXjrh23KmG6ECEKfPOIKKOemADZnAiJmJNVVkxs7HhZz73Bn5qaNzAV3/rk/zKv3wZgM0IHzvStrTjReCzn3uDH0+/vzw/4vR0wTtPOoYJfucsVUhixGJT4mjLscik9NwOlyMDlkziPpRs1kWvWoI3q07NZohzxWAfcJrUjrdNwCK3gNXciXxGHOG4YFQfj0iCLVIASK7anKTKiUG4kLGsf96GLtqd83O/KvKsEZjb2fZNGQ9dA5mYPZVsYibv59ifD+y2w03Vd8rpuFpVyZ/MnItqsCBxMbaM1MpWtVDEKnacSU+DYRGbcuQzeD1UQdtZJ8KOKlgNxGZAdBMf5bgBIc8Q1igxcZqE5UIN5ZZHlwx9i58cAyDmLou7r+lDp/EgEVn1Kq2bCIMl0QdoImIm7r/879icP4cxnvPzVxgwBC+0rY5+lUqKaAItXUMc/QxAdAXhSJBxQtrZYO+f/5m/cm3iFrepBJoBCGhy7QLxwiK3kmRRALk1/+4rP6zeHz/5b//8g/r9+wpX+ZIkXgmmSeR0C42FsUYRWhERa7RFy+SKDFfJB7k+XN+4pvQAs4nM7vKwe2a5K1+kRBMT50OrJRIFzEgcU6nFTbODe5iQLPtrUR+SxiPpYS8DBZAAxVtEwYcvVRIAEY9Y/c66Xt9XQCbPJwORGO3cxhUNY79kuz5KBmWeaVJXZ9eODL2blXsAk3gdWX0npKqIa5PRZhA2/W6biAhsNrYougH0vdN2raCO6ra0iughqY0Jh0lKm0uO3NFX8z5ye2I+pDcA5KMVP/+ln3uwP5qd+9Dfq8cdnj46ud9hdYgPUlc4VIhKrlXPsnuJaNY8yt81ySHdytVzM8YMkrUNcZw0sRsmk85rScaeov49EeIouHR/cQfaXDERGk8MhnFcsrlQsJ/bumAG6rvy1kGrJRUAqdutCtDYByB77+sQCaUCCgHnlJ/WukDbGC56lSj+oU9fcPf+Y0TCjp9Rt9hw73n4dBT+6S99hs8dz+nEw41yaYag0r8CvJsucE/ASfaKEIgkTsVsD3VlXavX+xyk3eM7u6TvR/6sF88qZjL3vI9HQgEJl4ysGZXjgZ4vy9jssDji3vk+g4SqFZfdUfz3UwHZj1wpyOT+HL58HtJ2XiVh63rtgo6Zo7MLNurPirKVXAU99RLy8vzevGz5PwMlnxq5siDxwEjEMkkoYATUiX665vhkAFKv436UVKDafzCDk5v4aMQNCHmPeOF//AcP/GK+ifvJYF1PCywWK/ptx+p4pF0+0YTVxPKUlOVEXAfonY6yNyE1HefEG+ydJxwt1wyn92cJUwPjaAnezG7nPpZG5R0AAmAEWTiiGfnln3g2UniJkC7VigcRBwPnEVmm9U1PxVrx6is/8v7BxxdP//KDUu0AfaDW76V6WLqmYiN7rYhMYeahGJUM3iGs59cS5ypJSN/pUBviItFTCP4l6tHEAHQxTQ+x81BGTf28DEA6T2TQbuQgCkJI88+JQTY9hB0wUkcNMPa/y7+RPYlNEU8MHdOwQkxgHDpEAj5m40JDiAFr5j7x7Dlgbdh1W07tZ027LR4kwUsaAU7rGLU1xaXtUjPEJCtqta1QCezVdsW56GVyNSRVO+ppChiJFH7ITXz04ue/9HMP4Pp2qv0R5f3v92O/7UaYhw/2Wyuua+Pe/7wmHl/3udlLRXLCsp+uj34+N0evvKZ8HeTqB4HyJA3EHbPBuv1TssDFZHGLC4JvaNqR9Xqh19JkiioWqBmpMb6Ahbpl65AhYZ6ugJkD72tAYvZacENyRv/M597gM1A4I2233TFSndtHB/ykA0r//h/9diG6+8lx9uSE7dbhnOe1N1a8fgbvDrrvl1gahM4YfIyAoY/zKD+8t4khzDyFOvLv9qsD9Xm0jE2R4d3/XY5LGXEYjpLvR4jxyjkzVfPv8YziWcYGG7UqAuws47uNnLr7PQBA+jxXk+AqEfzQNs7zvTrtDjirAIhP1a2A30nyHbZwQHRZvoAxVwEHmInimc+STQuztldLk6b9/T0Dau5ZjkicXdNvHi0fqbgBIe8RTRNYHV+weHJECOqC2ywucLce4xYXbC/u0S7PkeMNskycDxtTUg+y6tXbI0caJcvKTnK8xT+5w8WTe7Stx0VhvdUR7HF0hLEjPGwxr6REPBO/60qBtRAmfvnHnm4MWMev/vRfexXgj//CX3wAaNKMUdK2DYUIn+OZJXefEr90+39+9Yunf/kBjVUVJ5MEE/Ozta+lbC0l4c8tDU3FH0EQJzBVPBFQDonREUvyvIOfswwnyBiJY0SOovJHYFbRMjH5qKD8kl77NKQL2taVZYFN+o2ffycmKiAxQTkiwcyeMe3EjhLZXkjaVsmHtRhJxnkfTJYYLDHaYkKZw4+deoVEwdqgrRpJVaddbPDBMA6OpplK9UO9ZbQtLEuPbtdqtDiMSQmryYBDwYiCkkDb6LJCAh7GRvpBj09jY0ncTJXVSaqMFFlgEwvIiXsPhraJDNP1++smPvioKyC1+k42jatTtEPqPO8VdRWkTiLfC8Dsf+YwZfS1/mynClL5guRwRnb4SDCfl5kXoiA6Aa7/j713j5Vly++7PutRVd299z7vc993Zu4dex6O48R2nMS2jIIiIYGSEI0w/MFfkUDiFcCAFKIkurlKLGIIQQpPgcR/ICGDMYa/QIRA5Jn4Rcx4PGOP7537fp17XvvV3VW1HvzxW6tqVe/e++wznnvvjNO/o9bp3V1dVd1dVb2+6/d9RDV0+7Kmr6rELc55TddVVLWTwNJ1M2T9EBTd6gpdK9RJ73WiMAoVSyYIwqQz2Xc17XrGg/sHPP+Zd4b9894UVt3STclgQwBIQc3UYZjvkddaTAIXPtm7G+PkZvvB8j0DEKUC3tUTQGJsR/CW2aId9uG1V5/h9TsVrYPD3tNHnwbssMDQKM2t2tJY8Va5u4rsoVmHiA8yVG4wtPgJcOwR7Un+PuGsDmJTO7JNp6SQ1PSIdEUyfa9X4naVww33Yz100vK68tpOU89GI85Ry6QFqTC0aRJnbwPknE0FeXSdBfNs/J3PkbHcZPA9br+sIYxxo7yKg4tcIE70V7nL0W55bX6uiRa3QdsqJyYyFUs6GT5R2uZExMJ3SXtm3eV3mtdVXlsugquDI1/xPnai9O+u2oGQR5Rzmm4959r1E9arRmaVkvbDXD1Er67IRT7oCY1HLaL0lvc74mklg/s8Y1Vay6qINh1V08qALs2wgcxEHd59luvzJWp+D/WkB6X4ylP/2XeOnpJtaHUcB8npPf7DH/w2qFaXKGXOOeyq3PVJP3IZcJkNetYmrTZE6Zz4BNRyhyP9H/OMX35dJcMmkE4HIV2sOpVskOP4mZRTo6UOpRegErtpjshQ1o8J7CChla7okmyrRNUYvovyOyldsYJoRzIty7tmHDAk0WnEYCqX8gq8hA/WPUYHbN0nMWvA+0ztUoSg6VYzjo8X9L0e0tNrG2gxw7EZo6JzhplxhCggZHlcFeJylbIW4rAOSUo/+5ZLUJJ1JEqJGL1sbu3qk68znYk0QNomKC3dsB5VmzPWaTrkzDLlfpy3f5n2UYafZQCSOyBlVUpAsFFJj5Qe36Rm5SrpWTE1WAmJlpVPVa/QKe/JewH2tl4O53/spHtQisxDygYBEm0yTDocfduI3e5G98IYj3PjteH8bohQXHL3I4OTDD7yssP7dNXkfgZeMSoJa0zULXkfBQXHW5o68OSVwP0TzYu34ZU7hlMfUUExU4o9q/nhF1fcfuoe2nh++Vc/TW0V7xxFDoMSVivxjH4iMrpnDe//gmH9aAk9HZSXx1qPx6lx0F1hqIBZHGfvSyF8XtccywonOpAEOvapcQT2YsVD1dIme2CK1z6eJP1ytQnSM/2rP/PM9to0evAqQpT/Bz1WAn9ntqU8ObeliXa4DpSUsIvO5ZxqHlD0uLGTkmx+N2vzO9127Zlsawc4vutrO1l0V0PpNJP8xAvfQOuIdxpV96j9NWre0SweYKqVWC82ftARoJXMmvdarG4BtRcFgAQI9yvCO3uEu1fBerr1PM0Ij7NxXVfRJj95ATjf+QvYRLCu06+qivzKH/loAMhXrv7tcb1hY5iS6VmlZiRXZeW2+Xe+aSVgxZjx7/xBVslzs7w1KlkdM0LxOggosXHsBKVdjK0eKF0DACktftNN2SCdJBPEiKDuh0wStdndmLz3kbaxFaRky+CCknUe11ubIF2RoCRpWYUBgACENEAJwRC83Ly3MtjQkdPTGqXFCUu2mUzEclp9gNOlpe9FqDvuz0itKtPVYcpv33yufH0uH9KA54Lu0a4+/soUmPK2bRBw0cCgHIhtF9QyAJH89zbqzeY+5fXmmdNyFrvcplHiMqSYXlJLdqbfODy35dSEWBos5E6JdAWtGSmQ3lmx1+5qYlfj3QzvqoFelbsgPmiMDSJ4N6K/MFY6q8vljOPjGT6M4CKX2PKO2rDN7kcJNCafW3LnyzdjfKJ/RTbd62QSYfywXHLsy9v3rsb1DX1X8/xn3uEH//AbPHPT8cILH/K5pz0zrXiyttxsDH/mn3iT5154i/n+EV/5tU/zYB156yjQx4hFvpvy+2rxtIXA2mw5Xjbvg3QEHuXCVkcziMebaFhEyyLayfFdvu5E9SxTu/pYd/TKY6MeaFgwAtlmYxC9LYfjonpU3+QiU4fuAgByXhckrzOL0M86jakz1rkSWhgH4X5+7Lx1l8AkXzvyOhexSZ2lKQApuziOmL7XKcVqW1320/4Pf/q/eulSC+7qI6ldJ+QRde/+guu37qFqx80n7vDh+09K8vjcQXCYJ+9LsnhOHdcR1QBB/o/ay49RzXB1Cg8r4vGc9bvP07VzXF+zWs7EEcgGXEq0bmadJNDurWFlJnkc38n61T/9d4Wa9X/9xZcAfuXHfv/Uq4vqy83fGNb/E+1fO3MBiN5BM/7I0Zc2x7V0TNKoQM0MsXzeO+maGDO6ZfWOwRvWB9GSoIAgdscKsfe18rCax8FdK54UblQnSWuRHbJANCAp8wVIQCSls+fxuTOSIzI4aDEBGmLtq4bXbwMiefZ0s4I/yznWxmNsT1W3aOMI3o4zrlGBl3mqPNMagsG3luPjBW1r6b1i1nisCZjEp7JWOhaZD1/XgcPjilnjyQJdUDg/itFzfog1YaItGd7TAFpEiF5a+AJnwg139cnV//Gl/3k4T78TtpebYuKQuO4wdlzyMirNfgcitpg328yKyFkS82KglF2Q8mXTMtKw8v8mZYXEgg+WOyGmwPmjccL0MZWO26wTEWqWQXeS92RMoF/v4bQ449l6Rd+JK1bfV0MXxKfQT2vFGcvW67QRePu9fda9Yl5Hnke6D7/x65/lR374NYzJg0qZcHCumoCPqmqH/A8YOy2ZjpXPM7mZyXlXdkByiGH+u++aobOitcdYoXFJFyfwuS+8jtKBz39hyQsvjKGt1vZ07ZzFwX2u7QXePpF8kS6OTkhltyNTs/L983RHF9V5eghdDJwbtl9j8548UOtBBB5UZBYt82hxhWDaFhNDWdzdKj9Qs1za+21C9cd5P5vdnbyti87MEnyU3Y+yq3DeuS3zb+JEZRkF4wY9vOfcEcq1GRrqEbH52KEUHUkGJgrFIjYsVQuKc4FILqfCsK5tHZFHdUFKyujP/fR/+dJf+vl/9SMd9+xqe+1AyCPq3pHFvvosX9h7SD0/Zja/RjzcQ115AICiFQ3IngTgSceC4WowDG4rJULnhYW7crK4vmF5sg+IEB0k/K2uPV1nOD5c0HcVezfeJVovg/HFjI+qNm18P6lSxgoQyWXsSNHKpZXQupRGZceYtpVOSlPLZ1VV8n/ukuABydEYghBDYcuUgUcWrm/UAD68gfzDnzJFJvkgmyDCpiyWlFWSFpJ1OpNAS9a7lFOzW/Yh5tnTKQVjuK9lgFE3q/ScRyk9zLp6Z8E6oVUFhfPjD+/paY0PisqM4tq6CoQooOR0ZQddRxatO6/pe1XoPGRdVRXoOk1ViWWp0MIi3m/8UESFUUJlyQBEKzFIA/C7qPTvqXpcHUjZpSgHnuUM9OZsd3F5HdahYKBgqTSbnpetC/RbFffz3c1+YjnPE+KUmpXNFcoqwYsPKlmAB2ggBEXX1tSzKmkyNH3XELzGOc1Y26IAACAASURBVDNMOMWoMDoQgkKbgLFuCC1t2z3mM0/vDSdrxbtvPk3nDG2veP1bz/H0M/doZqu0L3oAJfm64Fw16EWMcQlQjJStqQPe9u9PrICDdIBTFhGArVq8qwdKVt6mUh5br5J2zTFbnAwTIkoF5vWKGDRf/IG3eOfBp/FRc6/z+CBAxKTvsCdOAEiFOtdB67LguBy056qSpXCpI8o6kJ7ASkkyeu5u5G4JTAF1vmcT+D2b3THmkWy+9jK1+R4vA0Ac29LOpx2ey5TesryNU7n+ZhdETz6b7d3MUsOS17WNipWrBByBSEidk02N2uPWDoh8MrUDIZcoHxQnD29z5eY73H72VaIzxOMKddATnUbN82VRSRcEEu0H6etXKv3ipalfDXjN3hNvofUzHN6/MfwQGBuoKgEhLv9IOEvsLOHdFfr7PuY3/xHXpCvSvTTMtm4FIkMqexFyWNVgLbRrsK4INEwzf0qPTlv5O9AKpY2I2IcRRkG/GgYSWyoHHGanGuOTIH1aJd9bXpe9dzI6FUCibF4+aUY2dSCpy5Kds8qcEABTtfi+mVAytA6DCFYbh9KBZrGkbxvoK7yzg3g9RIXrK0La5rWra9Zr4Z4rDfip8LZJ1sRdLzalfS/rKMXl2aJUktajBCMipg4lEMmdkPw6U9itZBctc87XsKuPp8oOCDx60HJZHUhZmtFqNQ9SSpcjGAdveeuqeC0b9zVTMWulxPbBwCQdHThjzXvR8ZYzbMq/c/V+zMTxQVFB0mpY5vOOvm0SsPD0bUPX1sNEQiiuE1qLdkupwOnhTVanC05P51y70nL/aI/OwdffnDOz0Hm4c79m1lzlqWdXeG+KrghY2xOCwfVyLSwdtUrKFozgYwQRAjbG7B+DilPQYm0vAKS49sh2wtChVTqQZdNZt2aqdph0mS2O+Kd+6jX+93/wAoYxXR2gZbRhbTA0aAGRMdBv0XuADFC3OUNtGh1s6hu2HUf5WYMagNB+rFGMOSR5+U0RfA5BNKgJLes80PG41tZxy2PbahNs5VKbJxIZJIzrLF9Znhbl45vdh83uh4kar6bn8rie3NHKDlmBTrkzwYaZ8rYpeC+3A2PnQ35Cz/9cLjLN+Nmf/s9f+is//6/vgMjHWDsQ8oiqEg/+g/duUdUd+9/3u8RVnehYoOZuTDLPNrqVFoE0EFuH0mk0laZ5zecsrE/wrxnq0wPmezO6Ttry83lL3XSsV1VyWLHcf/8FbupX4OYJX7n+d/7AniBfrl9+eQAixo4C9tLSN4RROwKwf1X+NxUqBFCaGFN3wzuZUg/yswHF63qXlolkC9/opVs15IfAePUtr3/ZYtmIqD+6ESwAg7PVmZ+V/LybjnRiCrUsBZ6xq0CncEQdIFn4Gi0znjn0MCevaxyWsUPSt/NhnTFo6maF62uqqqfrapanM4yJ9L3BOUMMcPXqCqXg4GA9iF37vhaBbZDZT+fGHASlBJQs12YQ5krnRN5DbQN9r9AJfMSQKVpCXclhhGlcIbPOWgZ0KkJj41Yx+64+nvp7X/rFl7Y5DJWDifP432VdlKSe1zGEu6En/v/5tYrtA7UcepcHQw2amTKFB4Wi0gIKNqvaACC2OC2zJsSWgzS1nQ1rtAAcEapLZ895TQhCrV2tamJUzOYrbCUTVjHq4dzL61YarHVo7ambFYf3bxGj5v7Dht4pFk3k9jXHax9UnHYyEL2/hNUbC159+0X+5B97Y9gnAQo66Uumg7pM04pRHPMA6mY9oWNpk37fFIN172bHJBR5RRML36IrrJQfAImx7eR5pQO/+pUf4p2Hirudo4vjTDZAM6HuKG4Yy8mmjpDzgcfkOyqOnZw3caJ6qnS9zLkh5fHe4nGEQQNiop50SmS/poPu7OC1SDSt3J1ziDvWZQwWNmukS0ltAouL3vlFHY8sPL/MOfztTC5kAHIRUBJNyegZUwrTS+DxKAAyccs6B4A8yrHvPMC2q4+2diDkEbXuFYcfVjx51WDefprn6zW+r1m4t4khDQCffYDaV8SjgLquYO1hVoHWY2ckV1PBIkWMK4dpluxdvcfRwytUtWe+t2Rx8AClI/fuXBPai9fEYPiVP/mYGSDfg/Xl+uWxMxL+xksoDSH565fgQ03/V/MD4loS6FUMRBxgoU+vzSNjn36Qc+hhnwWdMLhhZTDglNCnMukbipDCACZKmnrIy4SRdrWtKicApw4jkPFF+FWZrhxGulZZQxK76QiJSqWUWBCrGIgJQGW+dwySvO46OeZM5QhrOSg3A9K6bhyqNU0/zNA2jWOdUuNDVIMYPUZYt9IRqaow2JX6BGCyDsSY5BQUNfocetVIwxJui9FxADTnyyh39VHV3/vSL74EZ+lV5cApFLSITaBRDjzO55nHNHADh8eiOCEk3v20tg3YzEC9KuhWaPGaSC2LRsvAr1KjjWluuGUAstkdmWx3S+djarIwFa1rUncviK11KLQV870T6Uw4m7oHqRMSFVaHITw022afHC+4eu1EdILesOoUx6uKeSVdkLWDPkTWDmqjUuciJFcuW4jNR31ItvCFRN2yPa6vB4BR2vYOFKsNA4xNypYAFJ1yT9YTEFKuJ5d3Df/wVz7Hc0+s+L17kWPv8VHS58mX4HRkla88b4gYGDsdyCrOHC3bjsCrsR6O26xLyoLpFk+r/NABmWOZRTPki2xzWsui7kCc6FjyvpWgBNi6jrPvLW4FH1saGee+/iIAcv7rHr82uxW98o98fwZNwCcnrzg89ijReRXN0GGBR+s/dvXdWzsQ8oj64FQSX+eVBhqeSjPM0RnuvPZFtIrc1l9D2wec/qMfZP70m6hZj65WqIN5mj7TxS+YhtWa+P4SqAl9w8nD21gbiDFSN2vqW3eYdXPgGg9/9ksvPwTe/ITe/ydZX9Z/7eWfiD/70uCWNUylB6FgmWwGoIm+F8qW0tIByVSuMgwRyD6wcWMwrCqGzBBVFTNPBSNsACCNh9aMGSHDAnEEMOW6bfnjkaw3B/et8adAdTYFWqbHeouyCYjk7kran5yk7t1s4GorNQ4wrB2Z00p52tXeFOQoGTzMZrJcBiTe6aFboVN3wzkzcamS/JAwdEuMjkPHUNYtmpH83OSz0OD6sz9MmVfvvGJvnjntEaN2IOSTqsex2c0zyNs469uqDDfMgvNMYSltUXM+xOZgZpPWkjNAqiQyr5Si1mqybDk0zvczALHTcbO8boN+5QND924bOIHUOYhisuC8ZmY983knLnU64PtGzBqsT92Q9HmU56YOrE6vsG4t8cEBlYmchkz1EgCiFOxVcC9p19c+8uv/6Hmu7Xu+8MWxI1LmiMj+haQbcSmfRGOs463Xn+Hq1RVV1TPfP5F9SsBlBC3TCRGtPdq4IbBQrITn2Kod3oc87of7mSJ6uIIPXp2hgavG0IbAOrljKaBReaJDXpc1IiY918ftV4UIQ9ehPELycbYJALKgXMNEFL9OAMQnHUgVx3yLpXIcxOqMQD5b4o7rHAFN/jsf35elX+Xlt1GxNu9v1jYtSLluea/ndRimQKQc8JcUK1MeW+n96qgeCxRkd6yl6hOAq+hxbO9fjlXux7ba/H6+HZ3Irj762oGQR9SR8xilJnaM3/hX/rVhtv65v/XfvRSdAad48OETHN6/yXxvyfUbv4ZapF+LrEnQAkjC6w+Jh3Lhdt2C4KU1X1VysVZ7LUoHHv7sl/7Adz4eVV9Wf+XlnzA/JxStEOSmtNyMRTVzcI7oOwEhoRupXCZfpN35rmJWiUMWSNfKJFqdQzoj+cciX1SjSsGG6WEbRsG6jkCcABNVFz+WJqJKtycVwYBa9MTWoA7WQu0KClV7Ym9Fc0LhoGX9JH09DyqUDqgotCwYBxAD79wblI50bY21LolLw/B/XXucs1SVJwRF31vq2jGb9azXFXtzx+lK1llXIYGLxPXNM8pGwEdprStC9w0RqBU3rKz7AAknLF8jeFOdK5Td1UdfFwGQywpaL7vcJhViU4A+DTw7S60BmKlxQLmtuZFP7WEbxUKR7UCkrEHIrs4+fl4GjlKRvrcs9gQtVFVLp2do43GrsU1uTcCYMACA1emCttO8d7emzx4YSgDIaR+ptAILLkSsVrgQOVwpZlV2s+snjljD+0zXg02dyJNP3+fNN24TInzu8ycb2SCjNkQXFuFKhwGA5OWVimOgYQBlhI6lk5zcuwZjW+YVfPYpx6dffJdvvfIsv/GWpg9yrVtow0Jrag0PnYCPSimWYXRRmnzOBYUqP5O7DjAeQxFYKsciWiLS/Xja1DzwjhvWcs85VngMiv1Y0SqHBw5iPQxkLZpFtKzTcplilZ+fJYE7jHTCUjtS0rIuAiIZgDwO9epRdSYP5IIzfNszeZ9LitWmfDyDkmxX7NX2fJHzSjMNXlzSsmCTTiJ1Eai4TCd2W+30IB9/7UDII+qP/09/9uXf+Of+t5eu/7f/9MsA39h4/u1//198+e38x4/BU3/zf3ipbizhzh6s1mAj+vOzNNXiISTLXhuIbUV9cI+mndP30x+Mr/1L/9buZMiVOx7lFa/Qi8Tox2wRbSBkylXuiFgGF6yUH6K0Er1OZaCC2BZgIeMOA9FGAR2QuhfpgpZn/nXqTjR+dL7Ky+sITRiv6BqUCaI9KQXoOg5dFtUi+hQXUY0W8BHD2BHZACLaCN0s+BpjW7xrRAMyP8Z1c2LUtMvrgNC9jA6yH1pEsr63MnM70LikI2KtF/F6ZOiWCGfcD4OuqhoBQ84QCSQAkZZpaqFUSfih6EKASUghFNStJHZXijPgZVffnVUOBr4d151xPWPlgVw5TNsEICADyRmamdIDBeuK1ROAMRGTn7Nbk1nzKNFCm3XePEYGIGectPI54RX1bCUdBQ3aeJanC8kHKdziZvNWQEvXoI3nZGkwGh6sxu30qYO79pGll06VC5E+jt/C137rMzz3zDHXbt4DmICRGFSiTmls3af9Mxjr+NSnPxyAx707t1gs1gMoAulMblKryi6JsXIt8q6egJj8Gu/GweSP/9gbaB1oFkc8OP40EHimrrjfexZa8/xVxYcn4GN6jzGyjuGMVes2PUgO6usKMCA0PTVoP2YYbuiK569oPqNrOgecWt53gSWeD9UKDcw2XLAW0U7WV1ZpsOCJ1JjBvjcP4BV6AqxNse4MpPL/2+hX+fHSqnoTqDxKg/GoZR5n8qG05d10tMpdis3XnAd+TNTD+qo0NJ1v2CYPXacNqlfWe5yXT3QRYJGui74QFO7qo6sdCLlE/ej/+GcuDQje/6v/wssA1/77v/4SQTI+4oNj1MGCePcYWsZBaOVQ85a9a+/RtTP6rtqBj22lto0INh7LlKts6RRFwK6MFX2I1gIEBypQHMwDhtFDEqmruSVWwn9QRAEcFmJbXKQyOFEREn2LKo+w8z4V4CUoVBWJffrfI+DDIKGWyN/SccmvC+NscJFDUgIRZR3RWbTphiBD75o0e6mHjoh8NBGdqAHBGwKGvq8k2LBoq1vrByGqtWGQ0czn3SCk7XuTwg/lcy27GGHT8rNISx858tOvLwMQGL/artNo/Wje866+s5X1IJetbUOKx5n9vEyVs8mbNIuOwL4ah4RVQgL53BnP+PEPX+y0hGsWf6vRY2TY/iUOwkyXGpy/vQIjjoc5T6PvGmaLJacnC/re4oNGq8hir0sULSVdSxVpqsiqk3X2qRPfxzikvMcwpc3s13DtSs8r79Z07gpfqDvm+ydDVyTTMbXxaOMH226lAsvjPb7+yg3+xB97jb5r+M3fO+D2wT5NfYPPf+HNIVsk19hpTR2CRMHKblnGuiHQUKmpZTAwZBgBvHEY+ENPwMF+z2+/WfOj37/k5LTiw5OKSin6GAer3KlGZDtdME8V5cH5ZvJ6Sc976kbHzZsnrFYN3/itmmXSJ/TKczPMWSm3tWuxTc/RoAmMIMKgBjbv1Kr24jqPflWCjW0i6kc7112sA5Flzq/vxDldgqDSqQwEKFTR0Cs/gIkeR71ByyqpnI9KTL8MBUslQPeXf35kuOzq46udAeZHVL/5az/Km7/+J3j4W3+UeAfie8fEE0VcGuLD2Zi27Q3d6hoAVX2eA/o/3vXl9t9+GWuZ3JRG2YbYt/KL7zPQSJ9r7pRsAhhr5NakC5sPYtULMv3ZGNGL9GGgaeVfetVEVCU3dJSE9aa4VcXyGhGg64iq5LXoUW+iDAIm8lU/IKDGq/HYCGqq47AbPGjrwXrUrEXNWrAiWLf1Et9PW9iblsGmcsSgqKp+GGBY62iajjodh96rQfch1KlIVbn0v09uPkHyDbzM6jqnJra7IF2QDDqMmf4olJqR/BprQ2pcKZzfQZCPs/7+l37xpUf9KEwHSB8NXW5bKvY2e1ON4pquqLW4YOVuSF6yfIUqbnYDdJQVtzx22doE27m6dsx3atfzAUT4oKhrP4T9eW/xzvLwwT4xwqpTdHl+Rcl+9TEOVMY+xqFb0Hu497DiygweLhXf+L1bPLh7E+8N1vaSIVScb9r4ARjM95a0PXz1q5/h7gfX+cwTPfdOFHtzN1wfxHrXTic2JpQtnRLjXQIjQs1y3ZzgK5QOmARWtJEcFIA/9YeW/NEf+Sbf94VXCDFS147nnruHLSYgkoGh3EccneIGGM3goycMwZUN5sIsjg8e1Lzx1jXeeHePB3SDIP2JsKBCsx+rCYDI4Zf9ht7CoJilbZUZIJsAqKwSFPx+zqPLgAu4KMl8vE2WV2G4PW5t64KU25+4WaX7FfpMWnqFZUnLmm5Ytvy3CUDix3Bt2tV3tnadkI+o/H/yZ1/+1C/+pZfUwQoC+NevEU/mHN17nuANN774VWh6+oc3aFd7afZrRz85r758+m+8/JNX/5uXAKFfgYjR+24AHso2xG4FVQOzPXmuW4lr1BBI6McEdh+gMklzkehaMY7gA2CuhLIl/CCkb0/ykU0DntxR8QGWbryi54HO5u9QIXaPvRLHrHL5zZBCHQbBu6qDULEyINl0qqkd0RmMr4b8kBg1VdVibY82bkg67pWIZOkt1bwleIP3QtVoZt24j1ENVp45VFPCCvXQAclC3DIzJFv5wlmBeowwa/wAcmaN52DRp2BDWLdm8vyuPt7KxrcX2es+Srg+plE/3mBgk4JVVjlz2qCp0cy0GkIIa6uozdgFATjHkK1wu5o+vglALtMF2dyG0XGgY2kdme8dDwP1vqto2wofNM4pnMlAP83+9hXHJxWdU8wqeHJPcXUR8EFx2mruLiMrn2eDx/pwFXjQKhoNSx/wQbO4s8eH9xasW8Ni5vn+z48WJ85VE1Dyoz94F60ib71znVfet/RBAMHr33qOT7/wzsQla/pblXVndtCMZGpWtghfnV7hG7/zNIfL1L3y4AI8cRD543/iG4N970/90BE3n3wPU7Ws3JPE9B5rpWijGjQWbRq65u8507Qy4JinoU3+6kogEoE1njsh8sH9yAKLQ0TxBsWKgEZoV0vl2I/jTHyevd90uSpLnpOgxUyzyvqUFj/pKPSJWpWnlza7fOedO9uyTi6qTT3IuJ7L1bbt9PhBVD7d1hSgbYKFETSFs1S6SUq6rH9OzSHLQrAun2Xedl6/Qg0ZIyVd7aJyhJ0O5BOu3S/8R13WEzvDydsvcvft7+fhvWucHElKOjpiF0csDu4Tg+JbP/MXdifDBfXLh//yy1iLUka6H916fDI7ZKVuiGrm0MwEkBiLqoqWblVJN2TRZJso4V/4BEAaA41BzS1qVslyixnMZ3K/MqjKoKxGWQ2zRm5Gw8xIOGWTbvmaWoxkYkyUPCh0Iem5brwIx/MG4BmAmDB0QyREUQnFr+6x9SmmEspDnsUUqoc9k6icK9v1VrWjqnu0kuR078a5Cms9fW8GAOKcWPZmbUiukl5VVeFMQyoP/PLM8d7cUTeOvT3hxRs90sF29fHU399CwyoTzTfrcZyzNmsb/3rzEcVm12WT1qJolGZhNLWRzA+t5HX5fwXD49sE5dsE7P4R47ptoOY8YboxkfliTT0/QqmA6yrRVqUuiFZMjvEYxDnridunWEPK6IG7x5oHp4pVmj8xCtoQ6IsNtzFy6gNLL1qukID+Bw8s904UR6eGd998Ou1vpkWO254tVtTzNVf2exYVVFrxzgcLjk8sb7z2LN/47U8Pr5HriR6MLcrHRRMiYYxjKOEJVw96rswjz9/uuDKD2sBr9+E3f+MLw7puPf0Opmq5/8GzgxbEx7O6jzwAz7kcMB6rZcdr8h2lWx4Er/Ec0XOHNXdZs9wY8msU83h2nja7Xm1W/iSzdfRlXLCyNXC28D1PUH0Z+pVXcQAbJejo1EYHffJe8mvD8P8256tttW2gv61rUuZzjF0iPQk2LCsvWwKcPWY4Av3gwZfojSnLJb+ux11a27HLBfnuqB0I+Qjr1/78z7389i//FOH9Gxw9uM4HH1zlvQ/2uH7rHurqGtV41N4ae+Met17clLzvalv98r2/8HKMXgBIObJVWjofyTkr9jKzRlWPnZIMPqwBY86OJiqN2q9Rs0rARyaK927MF6kqqGvpisxquc1nss5ZI69tDGpmpYNit4x+cgVGMXuA2OmJcladp6ItKlMslPXTEMXaDUDE1iuCtxNed6ZR2Ep443WzxiSHnk1wEoJQrOR/NfytlWR5ZP3INulOrhKgZEqW0XEYqGkV0Ql49CmN3Tm164T8Y1ZlB2Pz6C/PInEmSnqE5IBu0nk2uLUV4CMPSi/T1SjLX2KcUl5GyvtV5VksWjk3lKdd7xGjxjtL5wzOqWH53CFROmKsw5jIEzfHSZa9RsBR52HlIz5KBopBKFmnwdNHEaiv00pnFj58WPPENU/v4cFScXhc8+Zrz8o2k0i9rBgUN28/YF5DY+HOseKDI8233qs5WedMoTChZMl7qgjBpOuMXEN8IYhXKvDCZ9/kj/yR13nx+99kngwrliGw6hT33n+Ow7tPD+u984GE0EakG7CKZWLGWJtUKZgGCE6+p4K2lIeyvQosVU+frnmt8hOqldp43WZtOrUNn0nRBSmX20xm32a2UG7PJWrZ5vMl4CjXl99Tfn2n/EBfO/t5yK0EILlKIGLidtG2QtEVbf3+HLBjGEMa9cbnkM/h8rs9L5gwElKyukel/zerzGS5qHrcBMzs6pOrHR3rI67fe/0K6/UP8/Co4d6RYdWD/fqneObBdbTxPPNP/j/4V5+E2j16ZbuSateQtSDWgm3E6na2J6DDVLA+JboWnIN6NupGcvXpfufAaNRBI9OZdZ1S1hM9yxqhcDVJY6GV6E2qSkbWfS/P54pRwItPFLB8jcu2vyTXrQBDyGHqiigdpfuRBKPoMbQtbnOK8slpRQeUDYNmJDqDajqoeyrX4Y6vYmw3zE7m+0pHoWC5KnG5PbYSGkXfjpqSqu6JQeGcpe8N1gS0iXSpa2N0HD6CnB/SFNbEuSNSJi5bG6irkCx8I1XtRXjbW9atQWlxxwpe7Zi9H1P9qV/48y9v64bAOIh6HGrVyHef/r1NZL5Z5XY2Z5QzAFHAFWuojQzOKw2LeupWBSTAnNZb3N/W0ciAZes+BbkcTN5jlMcFaESq9Pys8cxmPXXTcfXWeyyPnmB1umB5OiMkGpYPitpOgX8M4lRX1x3rdcXnX3yAtZ4QFa986yYhwg9cdVy/tuT0pOE3Xp1x7ITL2Q1IXxNi5N5aYVoI0bBykvljbeT2E4cSZph0Yq6vMdbRtTWr5Yzrtx7wQ1/4kBAV77x7ja++o3lirnnhuRNe+ebzfN/n3href6Z7+iJTRCdrbe8tIWiqetSBhGBYr/b4gT/0Fl+Mmv/zy8/z1mGk/cZNbl9zrNvb3DlSLF3k2HnaGFJ+jHQ98uy5J06CBofvD3XmuCvdpDKUkSBCR4WhjgaN4ki1rJWTsLzIEJqZaV+lHXR5HOb75T6s8FjyQH880KpiP/L7YGOZcr8fVRk8aRQmqgmQiEUg4eZ5NvwsPYIC/qjE80AcgIhCUWG2Apbxe5tm/mQjboWiV55IpI5jj6UtROriYBXQmAl4EMexfCzYR3agSmB3WdrWrj662n0DH3Ht/df/zMtdZ9KPjvxgtb1mva5o1zXYSAyGX//Sf7CjYl22Tg9hvZRbPRfw0XcCOpJAfQAgrh2csgABDKu1jDa8F8Cwv4D9fTg4kK4GMARMGiuPlXyOkMBH36dRiBsfc/IlDxoTSK5bjIL18846HUcAkv8ebmGiDcHrsfNRpqvn7kke8FuPaZaSI5LFgmnAkMWqxvbYqsNYhy5oFmWFkCyBdcQUQYUhKoyJMphKoYV1tf2HbbDsTHSrLnU8rA1UlYCWtjPolLBudKSpH/1DvKuPry4S+Z5XZwdAFwOZbVsoxb6yjGJfG641KnVBZOY+3ze66IKUt2Kd51runrdf26hbQQI2yzyb2oYUCeWpmhZtekLQxKDwTnNyWuES9dGkY13uB0KQPB9TOfYPliwOTqnna6x1fN+L9/jcZ++xWhleef0qx8uKGwvFntXMlGJPizDax4gHTn3gYe/xQe5LfkrkG9+8LV2QoAhB7HlFB7Zmvljzj/6/5/jtb97iN79+i3fvWfaspvOkcEM1TCbIfZ0AWChyh1KX1fZnriPG9rz5xm2++TvPMd97yF4NXYjcW0Wc07x/qHjQyn6vYqAvAEi+EqyTg5VCBvWK0a1qk86UB5sl1elItYPeoNqY5d+LNVfDOAGTAUi5zlJM7pja7GY6mE2ULGCiH1kUQYZZQL8JNrY9trnt/Hd5v9/sYJ+rJ5G6DAC5qEqKVSkYP2+75wnF8xFSRTPRhGSAVnY88ueypNAsFp+VBEKeDTfNr80dkM317eqTq10n5GOok7/151/WwK3isXvp/zv88/DDn8BOfS9XVRPXK9AKFYIADpDQQtVKUGG27HWp/ZCDDnM5L5SsSsn9zCWa6C2T1yYyCV0EzgAAIABJREFUolHGEtsEdPrCyawv0tl1mJLKs8pVxWn0t978ecuPh9GOt6iSmhUJw3KlY5ayntidPaVV3aOWPtGyqiFgDPJAQQ0fUTmjGckWnhGtPc5VaB0GQFJVgb7XgxNQ7ngoFdFq+0yzUpG9uaOqPMtlhTERa0IauCjms562mzp7zf69X3pp/bf/3A6kf8xV2r9OH388q84SuJSzlOYx1rNJ44hErlVmOL1mdmqzC8kqNxZ+D6kLMnRImHZFLuqCwHbdx/icABFrRJBe156qTnos5anqlqqeU9UetbaUW8qDdu812oznc91Mu5LWOpSOPPvMIa+8fp3379vUAVLcaAytj8yCYh2EnkWaIX7nWAbwxy7w7r2KysDD+9e4cfvesC2bOpfNbM3nX3zA/Qf73HtY4deKlYucEvnt1/aoDbyQKFcxapnIUBFj/JBHIpRNjdaBw/tXuXHbYWxP8Jb1ao/jpeHDY8XqH/7g0FmqtKKpHYedaFtyJkjugmRNyDppAMqcjrK7UAq+80DfojlSqRuTXpO7H+Vj+7Ee/t7M4thWGWTkbBCQNHejFDoolumCX3ZsWs66RpV12bOqPAcvCh3crEcBkMuej3EL5SmDPklW2aaZOTsZsUlhcyoMQKS0OM5VYelxzKhoccw2ckR6XDo+puU2qFffaQvxXX37tQMhu/req+WJgIL5PvjsdNUnz8ue6B10OeErEJenqKoinp6MI5CqKsjjGrXYk/uZtmWsgIq+G+1+tQbdQ9QMMcaVmYIbEBpWOc06hCN6MfwHVJXARBamRwUBVOWJIUinA9JIMGlFUiq7yra5hImGJAY1UrK8hqCJnUVZTzU7xvdzYhxd2M7MUho/SF+0CSgd6bOQNuqBagWjmLaqAjFAh9C0nNcDvWyybj3qQA4O1nivmc8VxowZCkrJbHBthablg5IU9t1vxcdWJY8dLgYicLF7z7Z1b66/Qm9dx6b7z2YHxqAwWpy0awO1PQt6BxqWGsFI4Kz7lQ/ymNFToLHZ+bAb9tJ5eyGK/0RlJJxzPnM0jaOerTDG062ucnp0jdVyhnNmEtI5GjTI3sWgBpqUtl7+N/J/31f8zis3AFg0kcUs8ODYcLSW8MITFzBKyb4UO38SApUSWqML8MLTLafLmmtF17W0754t1jzVdDz1lDh1eWe5e29B30e+9r7iH/zK89w6CHzhC+8WExgiUC/BiPeaw8M5H977FA9PDD/2I29gq44re477pxWv3QcfA6fBo3v43XcrDn0/dD0y/Up0Ewwp5QYzoWAdKEsXA0vcMKiNwJHqhgH6lShArkvAYFObcVH1w8CaCYDOlW17a6VotOZarXnYKQ4wtCFwHEcwcl43YDOEsawBbKjLWfFuq4H2dY5L1kWV3aekIzXSo7a9l7xMXQwtN5fzxfea6WzSyUkAsrDpraJY3fnUyRodsQIVhh6HT/fzemAMINwsRxiOCYPmr//8Lpftk64dHWtX33MVl0u5061TKno/Kp/XSwEgzkG7Jq6X4B2x1G5kgXquqko6j1octUzKItF6BCAxgZNNDscmSRxQCXQoLfa+KnNCjJLOC4iGpUJAhE0Xac0IKkyQ7knZFcnPWQEfQ3ckdyASdSsGNQEgJVCx9RJTdUOK8WClmcTlxni0DhJopj1axTEnQMXBQSu/ZqCS6Di4aek0GyyPQV0Fmlo0INZm4W1gb3+F1oGqkvkuASRZqD6+7b7XmJ/5X7dqFXb1na+LHLHy89vufydrc5b6vH2o0+mX6Xv5mCtpWDD+vY2CtRlWKOu7eP9Gx2+V1p00FyYwn3c0TTeEAa6X+3iv6TpLDIVrnInD67ZuIxhCMGjtB/3Wc0+uOV4r3nuo+eBBWr+PuAD7VnNgNY2WmWgfZYa+j4F1mih52Ea+9W7Dyalldbrg6OEV7n14Y7I9pQK27jHW0XcVb797wIcPK+4dy/beW3oRq3/rKd547Vke3L1JjHqYTCjr6WcecO1Kiw/wwTtPEKPm+U/d5coMrs+EShYiHHrPw97TFgCkQjNLs+oycJXBfoWmQXNVWWZo+hhoh66JAJAOzzxarsSGRaxIWJSGaQfkcQf1WdCeB9YNmpkyVEqxMJo9I3qWPkZCjGilJjSlTY3I49S21122CxJUvBCAbBPfZ0CRXan6IZ0l/d5snJuhWM8mhWoqto+0aV0KNRCkzvtcqmhook2gIzk7olnSIQYA+sz+i06k7LKM2pkKQ4U5FxDu6uOtXSdkV99T9eN3/s2XMAZWa2LToCo3diuauaSWZ5esZobSmtgfTrsVaSSiZvMkZM/J6WkZm/42xekRglDASipWlRy2es+gSNUKdLo/dERyjsn0wjihbVUB+uw+UwQL5h/2ITE9jJ2R3B1Jy8agJENkeG0UnQhA7TBId0ibKv0vg//s7T8OhvRkZlSrqYeI1vJTH9IySoPVAWs9fq3R+eJuEECTgEnfG65ePUWrSDQeowN106dQs3EUaM3YcdEmUiUB+64+2voHX/pftgK97xQt67za7I6c91hZnjh0NCozpVllQD1QrRi7IlmadV537VHg46KyJmKtgAptBMjHqPDODjbXXdEJ0RudlRjF/GHToS4EQ99VaOM5uHLK1fmM01ax7OGoleDCuRWL4lUaAC+05jR41lEGgJ7IMoBRintr6LzG2qvceWhwAX7q9v3J9nJyxd7BKS/OWr756i2OVoqDSrN0kTsrz+o9g1aWg6bmJ37skDvvPcGNWw/Se5HOiK06rt88RL+z4LdenzN/53meu9Xzhc/e53e/dR2rFbNeswye4zAOXG0CHI3S9FGG2rn7sa8MtyqLC9C6wCkOR6QjJHcrP3TSDmJ9rluWzMSPz2V6YOR88JvXK7kiY3ejSnReH0V/Y0A0LRkI5k4EWU/y6ApEojoTBSXbuSz44NGhfeedw4rRijh3IvLymrMgRIDWtP/QJ/1ODnis0Nj0+eW/t11fNrNF8r5kINLj2Ee6W6VFb/maTNvbpGEB/NWf/4u7Dsh3Se1AyK6+tyrzhWazUTiOE+Dg+9EBKwahbRmL2jsgdmvU1atE72G5FFChNEoLWKFdj/QrEEBSJb6pd0LxAummOC++oDESj1di5wvp9Q4qC60AlbjswSjJE1EKtZB1xuNWOByFtGQEIqMry+TnKujtv0iplM7xcRpiQNkkVk9AJQaDmrWYsB7SioNPPO4witGj10OXxVQO34+Xib39FdY61qs5fW+SQZlnsVizXjeDJalzhqpyNLMOax3aBNp1M/Dc1UY4Z0yUE2sDSldUVaCqpAtibUR7MP/uL73U/cc7bchHUb/8pV96aRNUlI5Y5cAiFM+Vj09pXGcBSknhukynJQORckAYiFxVFVeNobFCw6qtdD9yT9IHQI00PtmXfJ/su3opml8GN5sdlBjFaCRE2X5toak9VeUHcK51GFyo+t6wbq1oqLzYWTuniEETkkC8gpSaLu4VkieSActohf3ZTx8B4Jzmw7sL3r5vMFocwmojyfHzCh6sFYfe08eAT4O+Qx9pgubYwfJ9w4nzXLF60HkBifrV4JOY/rU3r7O/cNy67lmtLW98aPnMnuab9yJt8Nzt4MrXnkMpeOeDBctW8dnnVtx64h7eyfv/ge97iDae19+4zut3Kn7n3RtUWjpZTzSGzmtWPmKU4th7TqJ0Rdoognqh7QVqDC5GjlzgODjWeJYp5bzHs4jV4GDV4Vkrz6zQfwilJ9NZ5RgrDwOdjrn8eKZgjddjJsLzPWXpk4NXCNAR6WIYO2XEgiI2XtG3uWGV5VWhubgkMC4Bh0pg4HHTzs8DLNv0HJvQLqgo82KJPtclcGiTNicSBzrco7a7adNro550WJqk+ug3VCN5PTlh3aYuTt7/Zjfk/a6r3Teyq++tygRs74QK5YV2hXNyv8gFiW0LdkPaphXq4EDASNvKpck7lMoD8HSR9E4E7jEJ2p2HroXeEVsnwCPGFHQYiGsZ1KtZJUBpGL2kDkiIqJmlUGJC60Ub0kcmKemJlqUGPJWmwyqXdCEb3RGK5YIe9STD+jJdy0unBFCDp3s1BIplsenkpdrjsWgTCF7EsVXT0q5n0uXQUFUOW/ewbpjPW5wzKBWZz1ts3aONR6lA3SiheBlPtBqXZneDNxgdQINzBms91nradrw8aRN5zN/TXV2yfvlLv/RtU93K4dlF3YtteQ5lmWLwt+25XIMlqoJ5PVKvJsuLQ/Z2W17G0/Yy09GZwrWtQ1KCmBgZHK9sst0dnguKPgF5sfRV2MK2Oh/zpfNUpjqGaHBOY6N0VryzGOtEBF/ByXqf527IuXzn0LB2Ii972EbaIDPyWmmhB6WOyHGUC0vsBRDuo/na15/i+194QN104s6lHTpdk5596oT9g1OUjvje8t7925ysFU/OFa3XvL92/NoHjmvWYJSiC4FrDxquXm0IUVHVPUdHc+nYzDw3o+LVe3CU9HG1li7OfqW50zqWMes2xu9fcKOiS7C0DzKgzYL1svuRgYNF06aZ+NJad7PysZePv/z6TUvpTZqiRtHFwExpKqVYxySij9KVKSuL6zMAeVQQYV7/41KwRspVnFiin1nunIPfcdZZ6rztTd25xu3ryAR4ueK5zddWUQsV6xFalQxKygBDoWIZGQ6kvcwUMiAZE/hh2W1i+V198rXThOzqe6q+8ux/Mc6EOy+6j74fbXi1HpyuhpT0toW+J3btoAtRdYPa30cdXEUpTex7WU/hohVdJx2TvP4Q5XbkkzdngHUg9h5Oo9zyMkDs3EBMV+LZOXke2M5zyY9nyhVM/w+6oGelbTm9FYAM7lmZlmWnM1Ex6EEfMnldMeLPjj3WemzdY23PfG+ZlpPHlQrMF+v0vKdueqqmpW7W1M2Kqm7FAtj45KgjvPNsRzpuKxCCYr0ebUyHfdpdrT6S+slf+HMv/+QvSIdpu8f/+FimT5xXl01Hv+xry9fn/9fRp1RwdUbLMayreLy0zy21IZcJLvRhXL4c1OUQQx+gc4reS2dDqZhAwnj+dG1Nm2zagy/XUdz30hHJ1rklIHHOsFw29F3Fel2lbA8xjHjx+WOefuqEZ54+5tlb/ZBEDuI4tW8MC63Z05qZEnrTQhkCcBydDJhD5Gil6LrcFc00S9GdZQACsFo17M8in3mq5Ue++JCDBp5bWIxSPHSeD/ueZQi8elfxwQdXqWqZnLl584iu19w9rHh4qrg1V1ypNEbBkfPc6z3vtT3rpO2zKGYYDpRlpgwLZYaOVk9ILlnSZThRHYHIldgMmpEMGJpoJqF+OSCwBCublQME87ryY+X9nIiugFoprFLsa10Ircdb7k/L49PB9jZBegYplwUgeTubXY9tA/vz1kvaz20AZPt+bHY5x4T1oAq73mIfhueL17YpG+TbpXWW32fugmTRe95/XwCi9ozf1q4+6dp1Qnb1PVdfeeLvvvzjD/6dl4hBgMVmrZdw7Tbq+lNC0VqfjgGG9+8S12vpVswaWJ7KfaXHQEKQ0cVqLZc1rWDdQu8FcGiIhx1UitgqaIHsWHXHoeZRHtcRfSsBIZNctLJ+ZOSKiHVvBhmNT/QphiwQpQE3/jjEyslzm1a+GUzoAO1oXThY9yYAoqwjJo660gFFwIepLa5Y8UK26c089brJuhLP1RuHKBUI3lA3a6q6kyDEph4GYYvnX5NQxXWNObk25JVU6QepX4sr2SrsE7xBq0jTOLSO9J2hy776Kg5WwLv6aOonf+HPvbxJy7KpBTAMrorDdqQ4jVQX+PbE6ufN+uaBX9YIZG64B94+ilypFVfnY0hgWRmIBD8CkawVCen5/LJ+gyWSG5mVSY5Z6S1lGtbmsrK9yGJvPWyra2dDF6S2HuesnMtB5jSUka5JXj6EMXdjeSzARWmheX3ttTl9mPPFZzuMqalrRwyKw8M5dw8rahtZdYqDeeD6QeQZ4N37Bqvhg2XSR6QPxIXIE8byTtezJnDfOQ58ze++fsAT1+Y89dQhVdXjnMVax1e//hQ/8Lm78n1Ujhc/dURdd/ig+aEv3GN5OuOPNY4P7+5x97DiVx+s6bvAnVcV5lu30UAbAy8uKl54suf6tRVf/b0r+CB6liupO3y9UTxoI2sf0EpACsCtPfjgOHLcje5XR0roNlXULGLFSfo7dxXycWNQ6GiGLsnmMTccK1uOWZtAz2ZQ56YmwirF3Ci0UgQHXdL65cFv7tZs0rAu44hV1iYAKf+6DO2qzDeR/ZraEEunbHMb+T0zeW1QEeL4GZxHpyrBwXn79KguyHldDFV0psRQfnTU67aAjUBkthvyftfV7hvZ1fdkfeX63zlfGzCHn3A/91LstNCzZsl+NwbU9RvEB/eTbkN+uAYq1bqVEUWVTovsphUjcZXEGy5dMAPQptFJBgjJZjeugNag9t24/gBUGpVHRlqBFU5I/v2IHlQDsRUaFzYBkgxSkpBV9Rp0svLt7eCUFTNVK2jO5I1suGQp7UUjkihYpmrx/VkgIm9fE5N3ac4aqaqWrp3RLFYEb9HGiUOO6annhwRf47o5atYPAK1yx1CndF0dcMdX0cbRtfNhmz5oVisBUBl06PQjpR/xY7Wr33+NLEL5MQ9RxMxjlbPKU30ITAdPJT3rPKrWRV2SkgZTLjdLLbGVDyyCoXOKylzu2MgDfo2a+EKAAI7SY6LENT6MNK/ysRhFkF5boQx6Z7FpAA8Qim5eXQXWbbISjcm/Jyr63gxhnWUIqPMa1ypWK0Mf4KgPvP5BzVOt4dbNFcYErlxZs1j0GBP4+rf2uXesUUreQ21g1cs+Xqs1Nu1Klx290n71RI77wGmvOGkt792/yf4s4rxi0QQeLOEbv3eT/Vng9q0ls0SzVOk9Xrl6ilKBZ5/pcO4G1x5arFIcBkeI8r0v8by10izfsfzppzr2GtmPAPzkD5zw9nv7QOSog720o1ca+T4eLGEdIsuU9gDjgDyqyDxa9mNNm/Qf8p7GL0ox2uzK9/p4IHnb8nLcp89P4tXpUxdHJ6pf6fgUmdKuzjtat4UOwsUUrMfRfYwuXSMQKScAzutI5KM4MKayBxUxUZ3peJwJVFTj/c3zvQQo24BGplPl7+48SlWOrJT7gZzensFIjWVNT4+/dGd2Vx9P7UDIrv7gVgiwPAZtZDrUO6hnqOs3JNhw+aEABCezOrEPotvACdUKBIC0XkjomVxu0y+MA2XSZdSpUTTutSwzV8R7HUlZOepIrEFpl0ADsqxRqDTNrBqGqafYIt2R1oxWvgSIybndJ7BhA8rpEYj4DSCSAUh+XgPddPo460Gki5HTkXUCKrKs7xuq2QnGttLV0IG6PpZVp6BDvVihWWG6pehYsOACatHKtk2Atjqz3RAVwRt8oqyEoAbgkdPVd/XRlmL8zHPXw0cRDGtAK0XGyFAOTpjMSuZq0MNM8OPUZo5DXvc4Oy1agmaw6N2+nhhHULHZ7VBqzAXJy2zrqOTKACREeV2MnAEySkWquid4Q5e6kV1X0TlD3+the9bGyfEco8J7jTEB5wxdJ7k7XW+T+5WcJ0ddpLtnWLZ7XNnz3LyxZN1aEbsrWPajkL71yTnLKOZWnLT6KJ2Q8jPuCTxMEy4Bw1EPy15z2keuNprDLtB5RXVqeHh6gA8HfOrJlttPHA4ZJpC6owcdB3bGZ67D80/3fPkbexy5gAmKw9jTdYH/+/+9xaduBPZmsGwVH95dsO4VH55E3uv6IePkTgcLLZqBw+DwRFbKMY9ynbkeZ5Pumy7StrMbU36fm/F5ZXejDDssdUkykB0/o0Cc0K3y65chTNzZ+oICBNsByO8nqfv3I43bBjJK291HlcgVR2Dtz+lklN1Rn0BBTJ9vvn9RbYYhjh2u7Sdoeb3wabkhmBI7gI+/8fM/c/7k5a4+kdqBkF39gawv+7/0cjbi+PHlX35JKQ3GCn2rbYWC1VSoppKOR4zgAvG0h1rL6KJKU59Zx5FpGY6k2Ugbi3E8k3SELlGzHirUTckKAaBzY4xzbVHTsFehe5Vk8wDZuyXiBehk0EIEp1CNEz2I1yK6t0E6D/NOKFj5ByJrQnQc76fK1Kls2RuVxrtaxK+2x7kKo0vdRjdMA5pqhe/n2INDsB5/eBX9/H1UE4ke4mGNanowHtqK2FbEVUNYLYb1VVWLdxatojR98my1iRNdyOo/+md3PyAfQ400E/k7RJnlzcOGkgRY2o8OzMJiQKBQWPLpMvK04exgSgaLTJ7L/H7NFIwA3JppKlN0LopBvQ9qGPBf3e85WUpKeQYiSmXqhtRFaehllQGFWdxeGahtGLoxITm/5S6jc5q203SdxgcBCzFEYpoyD0G0IsZKDk/Wjdw9rGh72ecuGfL1Tjoipw8Vs2PLqx9coTEyZ/L0tYBZau4v5TzyEbogeRVHDtYx4Io3qlE06VvJAYFvOznPKydp6287+b6UFzqcXSlmSnO4bvjRep+r104GYT3AlWunPHdljlaRtrX88Gc6FouOr75ywDdPOnoCv92e8tZ7FY5Ig04z5JL1cUyPjpJY3uKHAyFrMhbR0uK5HmfDYyEBh5zhkUFF/l9CD6e6ogZDg2aJG453zUg7lGUkA2Rfa1yMnIbR7jgnnxsUa/zQ2gjI5zUK5jcF6tsByHkdEDi/C/LtULC2gZBvR5MxaF0uACDj/ayfGTNT4OznUFLDvh2QNm7XDFBllv4vuyK7+u6qHQjZ1R/8CpHYLlGLBRwfC90qBNGJZEK3S45WDqhJXY8AqwJgdIwjsPLMKTUdQQ0dC2WidGG0Iq576YRkRaw2ohPJyezege1R2doX0j6mbfSAnQab5cu/ChFsMd9cywVcWT92QhK1KXbVQPNS2pOBmnQ8clz69OObWOlGPVj9yj55DCvU3hoAs3eC2ouohZWp59CjGqGchfsIAHKG4Kt0s0MnRBtP7GqMDYSgh/xJgJ0178dfw+As2d0qzgpSy78DUBUD+9HaV01ea4aB43Q90xDEKegYBpFK0sANMvCeVWNQ4Zn9V7Lvs5ln3ZpkmpdmtvOhnja52dHYrHNzRRBhelONVK++syn0L9J1Fmuld1NqmrSJZ9YZEzUrb+/avsfoSO8VhyeWzkkn6l4b6UJgnd6DVQqjFOpQDw3cYyfDShcjq3QiqbSsQtGk18i24mCcehjEw6knTNLHQRyfJPxQEzp49e0Ff3ixJkZNn7ohq1XD7Wsd79ytOXxnxqKJ6Ac1T1wJrPqaPkTe73se0A0D+gwkfDEYrtDUGGr04DS1VG6Ytd+P1XDMZDqRzLKPQYfjvp8tT6BnSu9RG4Pn3HGzSUQ00xofJYdF9lWqpF3lv7dt+6IOiD+Havr7ASCyzSn42NRvbd6/TD1u+vo0RFCd6XKU9SjwsU0fstlVmdKtsh5I1vs3f/o/fWmXEfLdVTsQsqs/8PWV/b/18o+3P/NSXC4FgDgPnRNgUBmx2A0R+nQB7KQLwSp3ERAGVKa3Ngr6OPzaiM3uuD2V+emW0StUJa2JybyPStSpthCpKz2Q8pVWRKMGQKCqjQt+6pLEPPiqg+xbzhoJihxmGHsjFCivUXUvQCSVNh3Bi5B8GDIqLwJzJ62aMgm5nh+hrENZLx0RQB0sJcXdJA3LzIAR/YuqE2WsD6JdsR7qHn/cTAGI9vgg9qPWK5yDgMYSdoL0T7hy5kA541xW2b3IM6QllaWsixywysHKZtdj8/UR2G8uSkFPEwEKFosWaz33H84ATZ80XKVrVnbOdl5dKj8E5HpgNFw/cEOGR98blNL0vYQOei+dkNIVCyB4NQkrDFERnBKrasB5y2LmODqRbsgDMaNj5SKVkjT0iHzefRAh93o1rt8lYBHTwLxSOtn1CgBpjOJao1gXzFOlwHQSshdimp0nUqVuV5cyOWR/Ha8fKdqv3+BgFvncC4d8eHePB8cW5xUnXfL2GAw1FM9ei7z1QEDkEzQcxn7oKFgk5FQ6a4F5gqgtYTgK5tEOYupsuWtQKaxwvEZVyR0pu1f9/+y92a8s23kf9vvWUNXdezrn3HMnkiIly4oVxEGQB0kU4H/ABkwnBmk7iG0EiTUZDGBasgQbVrYOhEiQFUkPMazJL0HyJgFBKFt800MQQKRpIAgCR05oWcMVqTudYQ/d1VVr+PLwrbVqVe3ew7n38OpS7O/gYO/dXV1VXd279/db32+YawA0ckp3TA5Yoz10X2kkclaIjwI8gLEBrwMQWyj0FQUrBx6OIXq3112AQN2ea1Z3AiI5bf6uJTGVN29/V/Cxq26iYd1l+iEBhFOb3psqX1eCQpPa3T0Q+XDVHoTs65uifvulX3j0vY8/d4reicNVFEJ3oT8lXUjhjgDSXS2UdBu9/EFDBAop3kC0IpFBKuk3FIMDpW6MgV6oVHQI8OUWdLwUIJJT13WaysQKrEC4HmSqkYRFOm8IAGqTRqUHqI0yvWnlfjZJQxLlfEA8OmcFNXHLIg+oFOwE5EmH2PZqI7dnsJDpWiJyj6BFD1r2ovWAHEu9sgYdHQrA6gY5501EfNKA1wsgEvzFSaKppOR2FYsdqOQjoNweosL5T/3n+z8YL7j+z0//i9P8ff2nf06HKrdT1czx1YlI/dg6XW3qXnP19lq4Xh87T1Rqeoys4iM5E0ktG4bRfK17mtaM8/MljIn4lo8+xRtfvQ9AJZG5OF3VVrlE1TW4pr/JgvQMgM7XGqs2om1EzyEhgwCzBkegHxSUZugoYm9mOWYOLDTpMyemx4VIaG3Av/tai/RpAB9TJBEDfYzomRHSijzS9corxJYIlggnWmEbGQ8Xou9oNaHRcu0uB8a7HZeV/fyqBYyNa4cAA8IZ/ETYvUVAj4BN9Hj30gCXwFeeHGHLjJwhvuGAFWkcKY0uWZ6/0xMcM3qOhcLXVu8aC13C7QDgEh49BaySBmQBXVKyN4la01cULAawSkkfeZrj0/0ZWOTEbnnPKRySxoKfzBc5AAAgAElEQVQUGkVwcaRcIV33niMWZR0dBZwRgJZUyVyJGHNAgKsN9U0Ndl8ym66v96MDAa6Ci6uOdqoCX9NReE2fvKt2JNc0lHE3Deu2muew50WO5wU0e0H6h7P2IGRf3zT12y/9wqPvfeOHThEZtLSjMtVHsHcpHpekaTdUnLDIahTCQgYgETIhyR+EmsZphedCo2JAJgALCywbASAhAE0rAnVSYxo7INSswtumUWWKqrVrAfYRUIA6suDej8vBfZDGJcpEhLcaCDMxn4oCRLwWQJEE6tp24KgR/ELATJ6MqKRPoSjTj0iA1zLVUBEYTAE7vDYCsFYroEvuY47kHGa5HzGdV/BW9q1EUhjSdjGqyWrxvl5M/Zu/9i9PLVFpPuvK77Hagldur8EAoHikwNxUmbZiZk1Pril/fJojYKqj5smISgi1VQqtFRqW1rmRSqvVoQYVjH5Q0AnotomqOAyqgAytuACRbN9brsHs6c3teXP44KZXODp0k4kHx6vZNvWxAAk4NCaIG5anFF5I+N3HGtvEEcuTik2MCMxoKpOADObO2GEJjQ4BD6mBY8aZDxiYoXsBJpFFnA6IVqSvXv+BJZ08QoCBQ8SAiAHynljBgMFlNZkhjlcXkMWUy+iQ08bz9KHjgMsgdBgDwhFMAkgGjiU0sUco7yEHRlPeA0BHHgc8Tm0PYfAfHDb46ibgMspnZguNHgFtap0PyKBN75F3o4NPn9sriJZk/GQdaX6WCC8tlFD0enG7ytv1fH0zS5hSsUyasOSj3PbJdZMOBLjZEet5ag4q6lIF1t9cN2lAnreus+OuK1TJ6lOdzw3OEal2ARB+QddyXy++9iBkX99U9dvf8ouPAOB73/zsKZSSZtkNwDYBAZvDBRgASxgh4nSJWM2+AhjByPR2OiRgYPDgQYdLoG2RRfJyvEZAQwhyHm0jTbyrgEm2DI5upGsZVTxFqTXldgaE+uQEFFATwF4aA04DDzIBbD3Ia7H4jR7UOMTNEqQCtNkKECki3gjSEdr2o9WvCcn5CuLM1TqwVxJqZrSAqWUjOSyZ535+WJ6S0l6sTJ1Ft16l20IBJsyEftB7R6wXXP/2r//mqaZEqdpxaVWlE5gDkVzFQYuFj39T3gEgvw61tmO+/TyFmiaPHcGBbJsmIjQODufNvmgwxmpshDYR3UZkqjGQYH0/2u5mOlV9rMC7rXmBqRsXEeNgGWFtQPDTho6jAI8+Af1dLl4+UZayCcPlVuH+CnjjrFqpJsIqaRIodb/5tXoWPI7JwICgmfBuHKarvmmNgjxhQQLkCr2IpT3bcCjUqA18aRTzpCLTo/JkpkfEUVpdd4njX2syAKFY5YZcgWBZ4Yg0Xl1qvNUFbJmgYXCGIVHLuEzAnlKPA7YF0Cyg8WcPGqwHxsCxGBasSAOMFGpIWGmFMy9HzeeRKVdIgCVnzWS64FITDlvgfAustKTLu8gYmNHOUGR27uo5ouNRwyIif40eIqjP05KbdCA3AZAPS9XThhdFw9qlS5lXlpY7eNgdbeqVXJI9yPiGrT0I2dc3Zf32a/9UwMjmH54mxaoAkHrpUyVuRkj6jxpgtCR2vrljUSQTlcGDHp5IBok14LN1ctRiARymAWwD7i4FbJw/AxYpJ8M2oKYFUyeaEedkcqISaNHJ1UtXq0ENxpDFbSduX90gIY0rBmIE9QHsPCgSwATeahATEAN4TSATQAc9KCpgMCAVUqJ5N7Ht1bYDLQZQ60AHPTgSqIkFkFHrZEpCSgBV74SOttXgrQWpAD8cIAYLjqpMPtrFFpv1KtmUilXpMGg8ObNoLe+H6C+ofvdv/OZpdpoGpKHyzBN8nUsRIfIoHufZ/RmIRJ7y48ccB06rzEnzk+4PqbHNtJbJJCR/mwIFc2kQmmoCcmAJKwtYE8sUBEiOVwQYPZ4pKcBa+VkpxtHhFsACzikYgwI8nMstI+DSNEMroLUiPN+VzJ4zQhaW0dqAvjfl2AeHPTabFv0g9KzGRnhPCJGglVjnZvDETNj2ku+x6RW2TqYfJt2XgYtQpYBnPsAh4oA0thxxrHR5vY6MBhzK9cqv5SYGDAhwrLBS4+cHQZLTNQgPqZEUdYj4u4WChsIJyRzrI0uDBweMN88J7wweH10aPO0juhix5jBZ4RbK1ggW1onS9TQy/IZLCGbWf/QQGtRj6nHIFks2OIGFhcKSFO5Zjc4xHruAjiP+bLtAHxkXPuBjpsXLK8IblwGPvUfPcQp+0rsvA6qjNC15ZaGxdgLC3rkUHU1m0Soi5BlMH2N5/6v0Hs7nDx4htJsdMwvt5yv/EYybZG71BOSm1vouupC7TB3q86przBR5MTSsXM8DGCwMBjg06dVwmFLXDFRKj5F9pvSdK9/v68NbexCyr2/q+u3VT4ve4ER+/uS///7T+n5aGPDWCwCJiWZlFWjVjKJyl9SdmSh+dilC97gdO6vZZzgtVmBsgPO1NOurhYAJ2wBDj9K22fSn0Ae5P3i5LbI0+gDEmrcRy89OjkkHFjBKqFpBXLBoGcE9palI3r8XBy2dPtwbL/QqiGidjEfok52uCSJwb7w4dWlJjyctTZU4g0Xwk0vQg0OgtWAexDZ44QDXwy4G8GAR+hWCb9EA2MaDIuLdbFp4Tzi7tLjcErqBcP/9vsj7KqUgb8X8pzw3r4yrFCxVNbIZkNRgpAYi4/6n3VWt3xDJFY02zLPJQxbA61mDRtW5LDQhDwG15jF8kEaheQ1MjJZEcmsDtPHouhVUctMyNFKjqB5RpN+NpokYBoX7Rw6901h3qjA4QyQ0lmGTE96211Cace/YI0aF9WWLUIWFGh0RosaqCWLX6wWIqcjwkcp51JMXWyGxtY+4CLEE4kUAaw44UgYrLefVBQlwlEmXgMx8Bi0pLKDwhB2GGHFPWbwTRculocSOVim4RJ/aIhb7Xk2EhSK0BvjYaxs83Rxg4RW+46MDHj+z+INnGg9IYx24vHaPPeEo0bN6iKvWOYSytWaPe2wn76NLctBMWLGBhcIhFF5SFi8vNIiAp33EOkRsWc7r3hJoDPBvHgOvHhDOxJwvgVsBrho0ST0/VKKTu2c0XloIuNt68SHJDlzFRpqvtq+BR/CgMbpnldcY+VNbTaZ9eVpYADo9vyXudXVXgfpNdR1QoTLLmX5/Uz3P87orQBJ7XbeTiuWSXiQDm6lpZUhUwNvpZvv6k6s9CNnXvqr64p/5lUfz2z75le87pVbLXz2lRl1HCvjirRt5FoHBzo+ZIobK9/zWY+D1h/JnLniQNgIcFAn48EG+xihAZLEU6pW1YOdA2sqxrbhWcdaKNAtAyZ9PVn1Kg3cCQHL1ChwjyLLQs5oIUBDAsZBmhNoB3MlURdtOaFcqQrcbIBJiv4Q5WYv2Y6sl56SNeXmwlAQ+Atj0oEMCwwtNKxLi00OwN8mVS5LWm3aL7WYJrSOYpaHbOhHw+ts1m/u6YzWpQxyChNblykAE2C1MF2fbKU0rb3Ndc9KSQs9xAiAUhG+fbXs1jROTGqjURZg2040WkNKYccuasqcVTyYhAGBMgDHiYHVw0GEYLJgJXWcLYMk6EmO4AJpFG2C1xmIRYAyDSGNIAMUFwBoWPwklonJiFGvpftCFIpapXq0N0Fosd3NkQeQxzyT7ZeQQRJ0+OlxgPPEehgj3E43zMkYMHPHaQmPVSDJ6F8bGWQa4sigSK+3HChpbRJxFX8TKCyg0RGgUoYmEI2PwtpPQwIYUjrSCVYTDBaPbWnziZY/vXHi0rUfbBLz+ENj2YkX89LzB2YbQbC0OLOFpH/HYewQonMMVK9rIjA5+bOqZ0FRN5gIa3/VxRmSPP3xHAhtXSsEFxkIpNIbx9gXhONmabULE6yuNP1wzAhMOlIaLolfRIDw0Fg+XBBc1dKLx5Y/HwPX7J733eQTl+d46kHBOq8vGCzqtv9dtr0sULeD5Ach1RhHPU3dp9q/bRgTrfKfA0ZumH7dtf9t2+RNiBBsj8FDVfnZNWerH7OvDV3sQsq993VJf/I5fffTJf//9pwQvgnBgGiwQeaRtAcUxCw7SUUQAluQv3LtPwZCwMtlPVp0+BjUGzFGmGTnIQCe5bbcVsbltQK0SoJKOzcMWcA6UaV29k+lNx6XB4d6AYkBcK9DSgxZBBjdHW/CQtCGK5X/jRYyeAshoOYC3FmrNiBcryTKxHsAAeAUOCvAa3DVQr57LBObZJeAZvCHZxifbYIhrkB9W4roVVRKgBwQvdJZuGJOlX/6f/uIVULiv91bbwLBKNAGWhD6i0tuz1oIAKFMPTQIWtjllu2pqc8uo6Kptb4Dw6U3VrWVAUVftirWrBMfnaYXIndr0V0urcRICiPajbT1eeuUJNhcHGAYLIsbh8RpKBayOngIALp6+jM16BWtjebxN+iYi4Pg4aQmsNLHD0MBYB3p6CEUGg1OwmXqjgLYJ8EHAx/llcnwjwCZHusUiwHsJKux6DT/TjVDV3ALjr37+OBki8NBYZLO8IUqTa0mmQocLxtlWXqcjo8Wel8VaNlOMMpVoQTL1uOSAQ9LQMDjQCprEsnehDTrPWJLCidGwivDtrwYcHwzQJoKZsGgFWMVI4mangdXKQesIayMenCh8G8Zt3nzc4neeKFhWcEn8Lg2+RsvZilfE5VmH0hDBaDmeUcB//DrjX39NaGb3jMJLxx7HB/K+OF9r/PnXGH/wWK7Pt69kkSZ2jDUH3FMGL6+oCPwvB4ZK124b5fo4ZrREOG4JQyA87gM8iy5Ek6S9G1DRzViWydI2TXlsus+kZr5HLHklGYDEip6Vf95VuwDBdfos4MVMQ24qSsDqRa0HPQ9FrK6cEkOgCdgI1ZRp17FyOQRY6CuUrn39ydYehOxrX3eoL/6ZX3n0yd//gZGqZRXQzz7Mshh9lcTVLQN9ADvIJEAz2EUBKTmLBJIxQhpg70B2m24LoJaBRSO0KxeAFgI++sQ98EF0I4qAEASkAEImDwmAeEq5IWqSZQKVRKZtFHesNPWgNlG8mACbJhjpP622JWwQUQn1yivAGfC2gbu8h0ZHwFyALyzUvQHotGSXBIV4dgBEheBaBNegWx/Ce/kIikEjFpAijVlrsa8XWMyV8RqkiR1mAvSagqWQsiOybfSObYBM1aJJkzRfCW2UrPg75gJE8lQk731OkzczBy/h6ueEci5N6phiTohRIQaN4/vP4L2F1h4HD74Gahz8xQlcf1D217YOzknad7uQ971tHNpFl5zgPGIwsP1CLKqpon9VOKLIwvQobjdGggaNibDWg6OBc6Y8j/w107t0ZYrHGG2AtQKWhsr3IQIDGCsltrGdH6OIjq3C8WLUsJxtCZsQk12wTBC2MZYQySUpKCLcbxV6D7x6EvFsrcAdoY+EpRFtzr3jHtZ6eK/LVId3iBqIGEpF2CrTiIjx2kvAwxOFr/zxAmvPWKeFl7Po8THb4B3ncQbgITU4Y497yuJjKw1mh35QeP1BgNERC6WxUApDlGudPx6OD+Rar6wGtnJNP3o/YPO2xjAwHrQaWknqvI9jkjyla1xDwsjAxjM2URLcD0nDMWOZ7BMaKDgw1vDFbYswNrsZeGzz5AMBvmqS66b4usb5rtSnF1nXJakDKBqh28BODS5ustB9HhDCNwCMXDe7iE3fp3sA8uGrPQjZ177uWF/81l9+VGtGvvjn/vm1q/Tf+8YPndJhC9g0NdjGtIybdCR5WuIBRAJ7yOREpcyNkDouNwDbdFtkoN+AjU6K1ih6ErH3ke11ootRAjb157NP4KhjgJO+w4wOWhjEspdMBHcNaCGohWwAvJLJSOuAxoO3qQXoG/C2wbC+j+Aa2LMD8FEHfnaIGDrwxRJQEYgK22evwrZrxGDhvYX3BjEoxKjhnIb3Gv2gk9SGdzY6+3pvlUXpubJA3SiCj3xlGhGRkqJTLRRhSN3+/E++SQL3I61wEbIolHZOPuxsMlJPSmoqESBv6Sywzg07pduleR81JRkceK8qIODQrs6AbEcNYOiX6LcLGOMFWCQqV7vYwlgHbRwWh++CFgOgGMOTh2iWF1ifP4Q2UX7FNE9sgDMIysdtbQApcelarXooHcR6N4jmAhgpZHNgqEjWEPI05NkQEZlxYBSIZQrVpNdliIwQZbHh5UPGomG8dH8L5zT+4M1WPgqitIPHRmOhCes+FDOBVqkC8j52n7FsA/7oiYJnxkorHDT5WjN8/uzY8TuZbYkdm8lkKgYCKYJSjEXr8ec+JnbJNlHgBm+w2QJvvs24TxYHWmGTnMIakwbMmtGYCB+UZJ8sCA8OY3EUY6ZyLXsvlK2XjxhnGw0fxc640XKfj+NHr2MGx+l7eRMjfCfTD0VCXQNGOmAZXu9ojOt0+WwvW6/W1xk75bE3fLzdNg2Z07TuAhCet/3OFKxszXubUWGstp+nmk/3O+pvbgMYc3pVzrC5S6gicD1A+cef+flTQOhmj37t7+2n7X+CRb/1W7/1E3/SJ7Gvff1pre9987OnvHXAJgglq2fJzfAEpMyCHCzIgwY1AdAMWkZgpUBGgfsgOos8GlBU3LP4vBu7mJj0GFaDz7ZAkCkLei1CdJetfgl01MuEQjEwKHCfKFnWCwhZt2K5ayLoXicZH54QvvZApiIqIm6E/uX6IwTXgFmhWV7AD8ui94jBIAQt4YQpqT3b8EYmhDQJOTtbout1cQpyATj4lb+0/+PwPuv3/osvnNY/R+YJ9Sqk6URdksh9lfdeAvqq29YhYqnFOlZhnFzkiUc9TQl8fcNR36/TgSdc/bSvk5bw4JBxsPQT8JH1F40JODrZgCNB6YjFqpscx/UtXDJeaJd9uf34wZvQBxeggw76E5fgtTxRfrYAn62wffwaHr/1Oi4vWwyDxuBVASKqOocCQHTEgwcXaFcbrI6e4N0//jguzlYYvMZ6YwqbkmhMaQ9RphghSsP8bhdxaFXRIpRfc2ZoRVh7SfdeGsKBBb7zW9dgJlyuLd58YrEeZD+NluOsHeNZQjcM4NhoHFkRaP/5b9vg97+2wlknj2kNcH/FOFwFPLi3Ha/fcFUcHJnKNcilNCMGKhSuGKiESXL5KlqcbS/6mbeeanx1HfCJY43XHzgBDFWuyvna4GgVJkDHB1VAyLNLjWXD5ef/9y2F45YKfc8HmYZsPWObwFumhvkU2lgbNeR5RIDQ2gBgi1isfefVzxrePAnoEHbaWN/Fone+cn9T4F72iLoOjEiuz/XHnGs68gTkJmvemLa5a93FGaueftxlCpJdvHZVBjo14KmvYaZ41fUzv/YP9n93PsDaT0L2ta+vY2Ur4Fzf83/9nVNaMvhcgSzAPRUwQG1amjOJI+AieIgCXgARwxOJMF0bwAcBKZ3PBH5wiNf/mUoTCSgGDwrk07EDiV5DRRGs2wiKovdA0ELnWgbwYEX7ETVCaIp9r+g6NGwrNJahT9oUb8HZ9ccbDH0DpSOGvgEz0C4GeK/gvfDknRu1IHsA8vWpufYDENCRwUOeVOSpSd0yaBpvz/s5JJVE1AIiyuN43DYDCEUjZWvX5EXPUM/8Z0BWyCUhXQDIPAwwA1vvFZrGoVuvYExACEqMDxI4iSFbREMmIAcXUEcb0IMNaGUAePm9c/J7oJ45hCA0pJC0EHkpOzIVgbtoGCIUMY5fegfNwVOol89Ab34LBq/hnCqPkeuQJjh1yGF6ae63qgCIzonLd0waDxcE5Ml0SkCjMQFEwCoQvvX1gHeftni6Jjw8inj3QhXrWY0ReGbr4XefLKAIWBg53oPDiJfu92IjXE1C6nOX94HsJzuBZYAQ06SC4vh8RUNSgykqFsqKgJOVAqDx8MSXjFZSEkLpgyoAJL8GgEx6Q3odjlai8cmA58GSIBmxES4QLrcjdc8SFU2UPI/0HpUnUUwUyvNMx7Dla3pe6fVzldbDJT1Ivv+2HJ271BiQeD0IUel4msdfihqQ5GDCu0wQYqLsuVuyQV40AKmPf5fSUCWQ8qb91GAjgmCgrgVQP/aZnz3dA5EPrvYgZF/7+gDrS/+JULg++bvfd4pIwLgYC1ox+LEBlExDeENABNTLWix8AQEiuYyWLkLJhEUsXSKuhBrYCPIk62TZNjRSErwbmb4oBpxMTEhHoA2gNoA7lokIAtDbkrDOrMBRgVQsYESmHw7GOLihET0JAO8ahKDgnIHvZNqx2Vi8shhEbOxkZZlZVoJ3Bbrt6/nrjf/yC6e7gva0QjEJAGQaYol2tgjXZYjkMkTgHXamhqiAiDng2CVEnx/nOrF6Y0RnYUycNMO5fFDYJqqgqvgjwRt4BygVJu9LpQOUSlqn1skCwKoVy97Wgi83oKMtlB6d5hZtwLozV5y1tGJYG6FVRNN6NEePoV66kPwcSMPuPZUpCIDKInhKPyQILatJoCDrPoZACBV97rgRxyijKGk1gMXSIQSFg6XB1hm4IL/rVhEWaRuG2B0PQY7z4KTHAwB/9PYSqzbiwUkv5wSCc6bQOqlywssARJE4Uu2qmg4aOSU3MCWdSq3nAVob8er9kapWJ8trxZNt874jUwqdFEDq/biQcbzk8lmiFbBsGMdL4N1LQudGh7g8DbGV6cIEfIOAmRV1XXNTBpssenPbmwHErqDO2+ou4CMXgSDwuZog7gAku6hM1zX9X2/R+/ut20BQfp4EKha/HnHnBKSuH/vMz54C+6nIB1F7ELKvff0J1Be//VevfLh98ivfd8pdA1I9Slunkkg9J5tZI5OQpBXhrRttdDyAhsQZK4vQ8x9yNSb4AhipWYrBAyTL42AYhew2AgpQDxz4mQFf2DEtPe+iPwBRhPcWSkXEYOC2B3BDcqcJGpvLA8QoAYRZ2Hq5tnBO4c03T4qrECWb08jAwS/vpyDvt776NwWAME/TxOd/dhVRovtwuXOe0wFk8S2glWw/BzZ50lE/IK96WzU2d3k/9fEBTO7P+wuz81kYeQd7rzAoRmPHk6gPnTUKfT86G2QqGSDUR6UCOBLMYhCAkbRQpCG8HaMA58FnDfhC6IXWehA1iJHgPcEYLk02p07WewJZeb+fv/WtWG2eYOiOcHF+UGx7oxcgkJvaTMfiRMk67xkXPuKVZdIkaIZqCZc9gCDXM09IrBLtjiLg2dkCByuPphGnqvv3Oty/B/zb3zsqx/rIocJmEFqSItGSAPIeUSriY690IIUyUQAw0b/MF50zECwTkEgTehYTFZesGAjQco149rg8IarLewIMSgilvI5U7suv+eAJjcEEOeRrKZa7ohs5XDCMYZx3Bp2bHqs2ZuBsV11NCAmEmJywanergDHMT1dAoa3e5Xnf/ezi2QlAYOj03IqN8Q5txXXC9kw3ykCkXMO0TaCR4jRvvneFFNZ1HRC5y7TirvSrep932W+mYOVzvUlfQuk1q+lYd5kGASMYASS9/ed/7R/u/za94NqDkH3t60NS8Y+t2OH2BugBOhykG1sz0AyjDc8K40SEKPE0MP1tVihZBGWpDwxYDz5bgVbDSM8CZAW4DQB0+grAE7gnccDqE7Uq0TKUdlDao+8kyJBZwdoeMeqi8/DeQKmAi4sDGBPRDxqKGP2gsekJx1pWhgdPWPzSX3pkAewNsV5MffR/EWvjr/2tLxRBeojXa2EzGKkrTugo023ysC3EKWipQYXQr3hyey06nwqyS5dZbtOEwtFvNGFpZTV7F/igHbSRDBCc09BaWsXswKYUkmBcySTPa3l/bzT4j3sB257A5ydwTx5iuznGMFjESAImaFyJl+dL8F5Amg8KWjO69Qp+sOi3ktAeeXqe+akSCY1tSBQmRYSFVljmTNIg1701go3OtoyTpWzbDbIfF0QzsVqMExulGFpHfMfH1/jDP17h2Ybw0ZcHPDmzeHwpFK6DZcS2V+gHjUXLRcNxlwpR7bzuavYcuVwjAOm5DAXY0DjRYMCocSLkg9C1xNlKgEQWpGctjSJOeUIEU4VUZttjjWqK5BUiM/r0OiHenKeddSB9Nc5RqH5PmIrTWCbBZvG4ntC1OG1H1zbLutDzqlyX2ctQKGE87ttRxJLT5y0iHMUZcEm0O1aINFoFX1e1vuK2Nv15U9R372M8yl3pWvPj0g2ggirQmF+TXVOgm7Q2df39z/z0KYA9GHmBtQch+9rXh6ToZAAtAJwxuDPg8wWodeA2ABcOWAj1ShZ0NWAtKDKYHfiSQW36IE2xwaWfi1T+opBisAnptmzNKwCELEpyOvckgnlALHiHlCNiAuANYrAI3oIUw/UtmkUHZoUQTLHd9c6i7005j67T6J3CchHw6kFAl/a/+KX95OODqAlwqL6/fvsxSK/QVBgTsFJTu0p+SLpt8rU6VmEL7pqo7NKBaMLCCDVp0YYZCLlbI1RvZ8zYqGfNEiKBnxyBrRdXuEjgrcXw7ivo1vewPj9C1zUIgdJKPtA5hdZKYGFkeZ650Q6BsL5cwpgGQ28QgkwCau2HXLMp7QgADhsAQ6Z45WmWiNFDHEMnV20EESFuZRKgSEIDm9aX58tMWCwcGgM8OGAcHW7x7Nzi4y9LCODB0sM5W5LlAUDrmMDaKCaf12inTRPa222V3z+NiULBrIBcyXutgCKQph5pyjTXoxAJRc8FqqZdo53z1mX3LAEPvSPcWzGerCkZMEyBNoA02RjLkkoJ6kmLVBrWCmzd4AiVAzkjeJIbUpcqIIZKqF5u0Hc1ySdosIFoAVtoaAggWrODowjLqkxUnqeeJ0DwtroNVLwXAAKM9r8q2YIrULr6aZK0y5UMV8HHrm3uWp/7zE+dGhj87K/96P5v1/usvTvWvvb1Iavv+fL3n9IyIj5uwZsW6mQDOvRAA8ASaJHmBasFcLEBbzy4I9AhUEISI8C12HVthHKlAO4MEBV43YKWg1BRVm506xqUbJNzDbpGVooHC3X/AvFiBQwGQ3cPwTXYdgfQxoMoJgrWIbZbW1Y0zy4aMAst479f5KsAACAASURBVHKjcbgKCL/wl/cf3l/nevNvf+H0Oh57TuUGpkAga0jmwCE7QM//XNe7rwHK1bXY8fZdpzQHKPU5rawE8i0bxsnRyKNhHnUfRRgeR/dqYKQZyTYRSnH6H9AueygVoHTAwck7ULYHNU4MGFRE6A5x+exlbC4PMQwGZ+ctnJPQQR+EDqQVVxMMYNFGGB2LcN7aCOcUeqfhHF1JSM9VO2T1XpysXjtKq/9plT//Dl1sCYetgBCtGL1TCDGBkvTcX3l5PeohdIRzGkoxtp2FbeTAv/dHh/iW1zp0ncFy6WFSOGkIYnOcr+EuIboPI5UovwaxgNHqc6d6HfIUY3zOKoneFWKU95gxM2pQlAlG28RkDDAFGvV7IcTRrjcywc96/Vpbc9YR1gNjHXaDkJB0ThmkZN1HDTQcx0LHyvkg5b6q4c2PrW+bC9Ztsiio4U1Hvmwr5zdOQu6jxQZe0tgp4JibybZ535c0lKM4CjudpOqfw+Qcx5rTsW6jM329AMj4+KvPoabJZVCYz7OeQF137vU+QgUUdQrRrG9jMALHcvsv/vpP7P+evcfag5B97etDWJ/8f/7OKR0a8KUADD5fADqAjpyEGIqSUlLJLwlkGThUkkXiJAYEkfCl/1S0J9/z5e8/RSBJRweAoMUS2ETkoEIAoNaDL0QEz72VLJE8CfGSI4KoQIsBvGkxdPegtMPls5dx/uwIJ/fPsL44wsXFAtteY3Cq5CoYE/fg4wOqd/+rL5z62d/a6wCAmzVr161xEiQosK4Qr05HgKlGo24WcyOYK9vP7jq+Jtn+4SFjuQhoTMRiEabZNxA72HH1HpMwvQxAVEpXtzakIMII27jikLU6egpmDW16DN2RZNcMLS7ODjEMFl1nii1vNlBgllX4raMEOgREGS3AoTERSkuT7QOV560q6hUg1z9rRDI4fNYzXj0YpyF6x7TBB8KyYbRWksqb9BWQIMYYCe1igFYRKk04N+sVjPHw3mAYDIwJcM5cmSjV18+5ZKkdaScAUIonAKSmY3mv0vMSAFWDl1whTYgUMazlAjzypEMAMFcWv5mCRlfeP/X7cN2nyZFBySbJ1Q2Esy1w4XOTOq0uTlv12jAhg5PRAWucdpTrh9HiltKUImtI5DHj4+3s6A4RgRiWFXq6qiNRkMTwQJwCExkNdKF7tayLOL4jXyYvjnKA4vWi9LuAkLvY8t4ELOYkuPcCQmQ/15/D/DneJkTfVQ4eFgbZBJig4OGvTLwCRwxwsDAwJL8re7rW3WsPQva1rw9xffIr33eKwIhfXYG7BurBJbAMSQPCZWpBi4Avffcv3/jB9z1f/IFT7hqhmgwG3LWgg07csfz4h5B9ygzRIkTn9QKxX0LpAbQYEDdLkPGgxiGsD6Fsj+35y+jWR9huliLKvRAe/GarYf/Znm71QdbT//oLp/Vqe93UF2chjKBkDkLmlbedAxBgBCC7gAiqY8yBRj1tqRvJulGXfBHgYw892kbsZwVEyDTDez2Zcsy1DKP+QwCIgBBfAEgupQKadosYxTjBDQ28k5XlZ0+P4L3CttfoBzlhHygBh0wj4sk552oti3NUpJL/EeKoKcjgI2eNApLRAQAXA+P+gor5Xa13qF+HEOU4q0XAcunLc8yam6Z1aNohPT+Fy7MTaOOLjTEgz6O24M3PKXhV8j6yxe68CjCoJh55GgUgPVaed0waj7o45imQ7CdPPHwCe0ZzeS5EKM5iedKRJ3S5uoEwjGw7rFqhULk0FbZaQONmkLyQIeJKfk0E0KcD7WpbsxA9a0XyNETPm9MKcOyahvQIUKArj8uPVaAyDQFGAPIqFngHPdbkEJIORKXr10DDQOGILRwiLslVQYoMR+F9T0JuAyFzUFGDjuSPduP2d62vNwjJNb0+Vz8stzzAwkARTQDKHojcra5zQtzXvvb1IagvfsevPvrid/7zR3TUiz5k3QKdFr1GnzI8nEF8urx1X1/65C8/gvXgiyX47EBujAq8FdE5ey0Tj97K16BHSlbUCG4JHiyYdeLMNyAKYG9g2zWWBxc4OL7A+cUCXScN3R6AfLB19t984TQ3w0TSsBol/wFp2LRKLlVV76PVDf9pbHxNemx+aKZoaRpvr//nYyiafm91Fp6Pt+XzsHpsLpfNOAXIQCJ/b0wo9Kr6/no7AJPmmStHohg03GDhXYOhX8A7Czc0cH2LbrNAt1mgH7Q00Gl3o5NVvc8MMgRo5JX6TBHyYbw/gw4XxikSIG5VGYANQaYh9TWzWlb0GyPXQyuZgByuAhbJSIIjkgB/1L0YIwBLqYAYNKx1k0mFc+YKAJkXKaG75SnLLgvtevpUwgln+hdSgHNTqpY44o2icuemFsbdMFLfYrU9MDqU19VaccGKLEPhiy2lfcj1zdOR+ePqcswwRDBEJTdn1za5dgEQQDJFHip7LdCwUDtvB4Q+NJ+C5Me0ND4uIjlKkfwPiNAsVsCc9pOfqqMwsap9r/Vem3na0W4+LwDh6h8gYGMOOKZAit8XAAGmmhGd/tVlk7Q6zsDsZz/9k6ef+8xPnb7nA3+T1F6Yvq99fSMUsUwlvAK/fSK0KOtFaL5e4Mv/2c/cqdn/V3/hFx991xf+/imigju/j6Z5GzxYyfsYRm8qvz2EiZcAgGF9H9r0iMGgf/o6lBb9B6kIpR202eLy2ctlJTmvnA4/96k9APmAqvuB3zwFpDEDkALdABBg0t/Q3HyJjkCaaUNTmlY90cgNMtE4CYk8fq2raEFopOqUx1fbahp1J3MqTXbZEvqMhMwdrtIUJIOoBDiU4mKlOq9M/SnnVkAKQGlF1w0WMShQmqh4v0TwpmgUhsGW1f1+UCWDI/I4CZk/B2agtbLqngFGbqAzOKmBR5idfmCgc7JOvNBU3J3qPrgxEcZwsa21NhaaWYwpkJGB1dEam4sDKB2wOn4bAOD9a9DWixjfye+6MUHeK4kmVV+3fM0nov4EckJUE0E9pYkPKuCxS49k9Ei30qrK8UgUtgwwfBhBcHa/qvUguypragDREPlAWA/A5SDgOd/X+eRGVk1BappZnRdSV4CkqsdqLZ/BJcCwboUVkMTo43a7qtYxoGx79f4Fa2wpoGOPr3KHDXlpwClPY0btw5Y8NmkvfdKBNGxgWT4cMnULADxFxBvC/uZ1myC9BhXzicfNXmS3F08mErdrVG5LiAdGkftNddtz1qSKRiTvi0DFjetzn/mp01/4tX+0/1t4Te1ByL729Q1QX/quXykfYt/9v//dUwCIbz4AmwAyd/8jAgBf/os//+i7Pv+jp6Qi+HIJmAC/PgGpAI46fVXg5HJ18fQhtPGSMp3Spp2zWB5s0C7W4KigdEC/XWDbtdj+D596dEOvsK8XXMMP/eap0an5T00uERdAkkvPpiHZvrReoJyI1Kmicqk0nUBq2Hg6vchVgxANWfGv789Ne6YZ1QAlN5mKgPtHHi892EwsXsdjjI3jLhyS7zc3/F5oHRG8AQeUBjxPTIbeIEQFX6YXItLODkz5OtXi/sg5QoPKz5mCJXSg9BwhDbGf3Q7IvhYJMW49lzC/fPm0YigtzX6bphLWBmgd0bYDLi+WYAYOj9dYHT2BT4sK1Axwl/fgXQNtPLr1Kl0DBiB6rRBGu91R7O+LSH33Na5AiBJp2S5a3iSEMTuCawHCc61GeX2q96rkgPDkvTQ/xuCvnmPnZKpUZ7KEKEAisIBDkxyycrCmZ0ZMk5Cs/ajDCPUMNFzniDXPE9nVCl9n15v32LIuTf2WgnxPSTOCWM5rYqtLDMcRMSfZp8cP5NGwSanwVO6/zhlq1/ne1TlLzun9AY7pvsZMEP2CyTs3AZAsVL8pgyQXgaBJFWtyYGpX/tlP/+TpP/31H98DkR21p2Pta1/fYJUTzumgA7UDvvypf/LcH25f/tQ/eWQOz9Cdv4LYSxhbv76HoTsCRw0/LOH6I3QXLyMEhRgUhr7BdrOAcxb9tsH6/AiP33odflgWbv7b/91f23/QfoAVP/svT43mQtHJzf38f03nMVoE1VlIvYuWlR83p17l+6wet83Ur3we9Wq11dPzoNn3dWX6WGtlP0ox2sUApRhN46CUpKXrWR7EfB91XdWJ6DQpiRgGA+c0YiQ4pxGCEnpSEADig4JzYiObqVTACEAAofwMQUBFYAEVWy/b+DBmeDBGwJKb4hrwMQAX5doJFYsKJUteFy5ftRIgYm3EcjmgbQc07QBtYko3J5jVObTxaNotOIWHEjH8YKFVxGLVoWmGktReO4eJ5iZOJiC3VaZeaTWlwtWp6OW2CGx6SmJ9KrfNxfd1Bszgx0T4+aStvj6Tx6drbdRIj1NEODAKS0MylSNgoQjtDkGTgoASnehUWWCeAwAVbm6gFkph4HijBex19ymQ2PlSQE8BHWTyMSAUABIpT2WmEwJP8QpNKYIxkEeAiNnz4wJuPr/68cDNTXsGTLcBkJxYflcqVqjoVLvoV9eVStSzu6TNz2vMSuHquUdM52DTylOVyCNdzHEo5/vZT//knpq1o/aTkH3t6xusvvTJX3703f/HD51ixx/e56l//Vd/+tF/9Cv/42l38RKUCth2BwjOYHN5DDfIR4NzBu1igG179NsFNpu28PHXly0A4OLiW/CxT3wNf/gP/tYegHwApT/3G6eZcsSV0pdIVrF1JXTODZtSo70qAPROl2Y4TzjqiUV2atIYKVjZkra2Qs1gokw/VHYyGvMeavFwLTJmBrQZH0sEvHLP4eiox8FBh2a5BVFE0w4AgKFv0lSCoNSo7+BKNF3vW8ARl9uZxYmJSOOia8sEJLs/AXLO2YY3i6azqHnraAI+6muVAUOWEXNN08JIBxry60Fjc5yF6SXlmyQGKO9bkbx+xoglr7UBzilYG3BwfIGm7dAszxHDJzAMDUgx6GCLdrlBszyHOr6E3XicNBtcPP0IurWF71oBLImKlbUk8jqIuN05k6YiqSGLIyQtbliBJudJahSjZwAy0c8kQLewjG4gXG4JR0ue5KfMp1tWAy4huKcbwsmSEdMEKeAq0JTXUQD3xslrs7Ly1ceRfmirHJzxuVcgZ75PnjpgefAEhGQnLDEuVLiIHn3xVhrb97zCHsBX2mMFwiEMOgRcJFE5J8Dh8h5YwdMoNq8rIocnjk10nWGShekqgSqdUjaAMVH9urqLruImAPI8YYTPM3W5qfJzzWYC+TpclxrPiNDQO5/HTeekoREQoKvP412Uvn1drT0I2de+vgHrX/2FX3whDT+piOBaeGdF0KtYKBsbsen1XqOFWHUuVh3Wly0YhM3QYLkUh6Fnp3/10e+8iJPZ162lP/cbp0BNOZq6QoVAJeRNJ/49kMTFiid2qUZXwXAJYOSGu0m0nwxEMgBRJJa45XzUuP/c+GeqF5DSr2d0LLoGPC8s4+BgwOHRBs2igzEO2g7ouwMwq2ItO2/diHjMCUmAhChRrlKD7Sv3t+3WYhjMBLDU2RYjdWcUm8vP6X+639f6jsgY/bakwXXpTqsIvc/7HlGIiNG5HDO3PZmulivEJMoOBNYEawOsDRJC2HZYHL8Ddf8Cy4uXQJsDrA7PABWxfPg18LYBdASttuDzQ7i+hVYRg2+gtQQt7tLW5MDRqcA/WfUmcXgtPC86opDAcVRXJiDOyesiDlajxuZsQzgRhhh6J+5Whwue5Knk9+zaMU6SB8d14CNXl16QpRHQoQhotEyu8jnn1yhTZ5iBJoGTkKg1jjkBC/leQ4TfZkbTEYG61IIIgEbPEf2OBj8kwTQwteiNYFzCF0tfAGXikasGIHltvp5mTPUlXICJ7DOt1qftDRQs6ys5IM9b/oYJwV2LZ+d9Xd1FxzGvDPp2PS7DxPx9nr08T+167ned2Hyz1x6E7Gtf38S1uTiWBs8OIjZP/9/8x3+9gJzHAP7DX/pnp0oFHB5JMvrTZysQ8Z5+9QHW4kc+fwot2oSaflI3XvOwN50+4fP2VjMu1waNEbpQBhRZUxKZC83Kh3Gikbn7c/1Hzm/QiktTX1OXgDwt4cnPu2q5CGhbD6UDlIpgViAKsE2PoV+WyUfWJWWRee14NQckACYAxHsNV6ZABA5TO9msAcmp6Hk/Loz6gpAoPuLAJM+7CJAzsKlU57nRnQjR48gdj+m+mhKkSUDO4PMkQMBliIRhMGgaD2ZCtz7C8uHXgMbj4JU3MLzxHVDaQd3vgKMteN0iPj1C9+xV9NsVjB3ArKCtR3BGrG6dnVyj/DzmlQHtPOtjrgPJgEHTCHKB0WY4Z63m7TY94WxDsHp0DqtLUaITeTF47QYq+7iu8jmt7KhrmrtixUTPqlesTXoiw44k9fI48LWuVgxGALCOcZxizBpaxlzAPf25Tjq/DhzcNi3IE5B6IpKfSwYl+fae/I374mua969X3da8v8hzmV+f+fe3VaFfVfa8uyYgcQ9Irq09CNnXvr6JKwZZu/vKf/t9N4KJ3/nBv/sIAD7+s//zaQzA5mf+yqPNB3B+3+zV/PDnT4FpkzcX89aTibz6nLc3JhZq1rbXiCxZDINTaBJgGbwqDSIRoW2E9uM9oXcSGrcLfOSJi9WjfsCnffmQm8opeJlrNhQxmkb+QK8WHiEQ3GDRLjoQRfhhtJ4miqPo04QqvI7L+7iuLJ6uKTZ5AhIqwIGQJhHJncl5Ks5Wg5f7eg9s/Hidt16AAxHg8zQDKMLU/FVn7g+m/H+hZY36mVp/QBAA0iQdj2hkgPONwvEKaBuxJjbWwdhBFOFeQx1tcP+j/w7UpKTtiwWoCYiuhV2ssXrtDbhnD+CHJfpuhd638N6kyRGnycgUfAEi3I8xifd3TB/q92ZxycK4j2IkoNPPiSqVL4zVjLONXOs8pXj3cjwPShOMhQVePiRcbHEFhAx+fI/JazdaRAceJ1jOj+/JwLXVrojTywQkvZ4yQdhdOR+krvzzWZqL7Wpo57SkAQH1Ze3gJ/fv0mNcnXZM319TAIIJ6Jg+Pk4mC/Pnk6lgN9VtU5Db09Mr0PUCmvWrU6e4U9CeqXMCKvXODJDrzrMGGgJEZBpyHYD5IEHcN1rtQci+9vVNXL//I3/7uSYZe93HB1fND3/+dGcAYBIT7xION0bcjgChY9W6CO8JlHbYJHelrPHIK/hGc7FhVRrQYWzkgRF8KGKQSpOHRNOKDLCW44/akalGY8z8qPZHYv1agylmShOPnFNDIMWInmCMn4AOIi6J4PXtY24FUs4ElQlIBiAZtDk3DSKUfI+pqJyrppVZQIMhQs/T5iQLlgNzWW1XABy42BDnUsmlKQITiVcW++epVGMieqexHQjHh0KPPHnwLkhFQAfEswPojzyBMhHUOsTHB+CuBa8BZXso2wNRBPluaBG8pKVnilUWpOfbQpieqGg5RlveeQZIOe/cvd9QcyAaWUIFrRZqVtbbBB4nQiEKvUor4LAFvnbO+Mgxla/Z9rnedwYg5XrmiU2iW0UwwONrqTUV5zeMX6YW1uk7VzWVc3BQU6nm5WYXJ9D12waaNrU3fT+fzGgoZOtdOb+arjUFJbWGRFXbANn96+YG+v3QsN5vc15fh10uVhnU5K/j8xyBl1C1rgcgV/QjfHUydB142k9Abq89CNnXvva1rw9ZZQCyC2hka9XrKlOypPlOf2gTaMi1aAO0iugHDWtHzURjY6JWEaBkSuG9gk/AQrQWowNSdmuSA+YVdYxASI10LZV0I9lqtjwfNT23nGYeg0kajyRAjzQR12cqGFA9Tx0KEKlDCkNQ8F6a6wxOsj6Gs91u2j7EUa+QxcwusKycJ9SSwUhN28mV77MJfGRtAZAE7Gk7oqsNpAsMq5MOpRruGMN4+d6A87UttsXBN1gcPsHmq98Ku1hDDWeAV4jbFcKT++CoodsN6GgDWvWA1whPX0HTdlAqors8hFIB2rBkh+TrqgIADV/Z3mbgVt+2CyD7mVXuXRy2MtCS48jXkyWX12ndTx3Ihk6u8VlHWJoxkwWYOpfVVVMWFQEgIIaUCUKAZkIXIhZKoWeexNGZpAUBxkY3Y61MucrOWTdReVy1Uq5ABYDUt+Xvr6Nh1fvPupS8yh/SVAOYNs4aVFLVIzEsq2v3Wa4RbsWS6Rz87RvdsZ53CnIbAJlvc9PtdOPMKz0mL0DMQcm1xxjtCH7x139iv3h3Te1ByL72ta99fYiq+eHPny7aMM1XIMYuIW5NWdEqIkQFY2J5TK7NRowHGhMxeJWabgVrJfxus5XGtm1El+GcxjDoZPs60rV0ZQecKTw13SnEEYhkkKDAaGwsPP3irFV0JnnfKdOgb8RalgkhmNIgk2IohPI1Bj0BJXOqUBamA2kKEsfpiGR0jCnmdeJ5nVzeewEeQ2T0zJOk7EzDUCQr6bnmlqcamRokt5cmt5qSAECkcb3aahQxO0D4yKtbtO2A1arFO4+X6AeNzeajOD5+CQ9eeQvd5T3Y7k0gKlDjoR8+Rnj3JQzr+2iSfTYtHJqDp3CbY6hgoHPYaSRAoXyfr2PThBJAmhPRI9EVDciuUEJgt3Ccb+gztWIcLtI1JADJZe1wkbQwyfr4cmBYImw8ow+Ms0He/wtFOG4r4MkZaKZjp9v7NB4JDGyDUPz6GNEqlRywgHeiwwIKC9rdsOenYUA72/D5xAMQAfoluSu3hx3r5ToBhVilpmsoqAq0mORwVYvk57Sr+rwjMQxnwMKFhvVeLGzvWnMq1ougJRWgNtn3brNkLZ8UO+/LExxKV3X3NrsnXgDgE2hRUDsnHnsK1t1qD0L2ta997etDUosf+fwpKZbmer6KXK3855rnIxgTS2YLIDQbawO8V4i9ZGRoxeh6jbaJAMSStqbXDMmeOVQNKTBOQPL/DEAy+Mn9mlaiCVGaoZDpVwyb6FvG8AQ8WRtgTEj7lf1NnnZqjpUKYBJBeojSRLsUyFeoV/EqENlubZmm5MrPzSc72RxE6JL4fAhpCsJctAM5XTvwdPXa89WGT4HK9tlhCdVEpH5E4PS8QRKSl47faFn93zrg2dkCi9bCmIh7xwMWC4ehN+h7g2fvPsTqaA04A/XgEnToEd44QQwWSju4y3uw8Ry0ENcsuzqHaTZwri3ZIZnORhpA0KWlUirAewutI2Ig9KwK9e46g4H8euwCJ7vev9nymWf71NVyvNJC17rc1noTcbKKALoQ4SLjuN2lDZpOR0x6DQxVpgZKFYCZX981PLascELXt0kevLOR37UyvwuYXLftdVXTo+pj+vL43fsxydY3p7hHYpESUYRh9dyOU3dxw7qrJS9w9ynIHIDkn+tXPd+3C4DU5zz+Du/Wctx2PUYdTdwDjvdR+7DCfe1rX/v6kJTSPAnju3I/CUDRaTtSmP6v6UqKoZNo3Nop1SA7QHWdxpByMUIkdL1GPyj0gyrUmkzByo/LeRBGxwIY8nFtNd2oHbkaG2GMTF3EHjjKfxPLOWrNMEbWlUMwCMGAKBZ6FSkujlgZmGgdoUi+nwOQyfNNmobs9iWaj1EzkjM6mCUDJLCsmA8pYTsiu1iNAMSB0UOC48KsCZkGyEmLsutVzUAlN8mNrjJVMAbuPbvUeHZu8eysgTER7WLAg4dneOmVJ1istji89xbUKxfAgRca3uEW9tU3YZo1YrAyZrEeZESoPnQnMMbBNj1M46D0tFkzJoBZgVkVvUiIqmiJarCwC4yUjJQ8QQmjpW/c0W/6MAO813QmywZ4sJoe8H5LODQCd5/1DBflNbxuQjOeo4C/t/yAs+DxLDo8iw5bjrhPFg+oQY8gNrvXjHBURYfaVQwBHxmA5LC93Bw7xApAjBUo3mqbm9O8DejaM3AU4SgWW98ALlkhniSo0NOYRVL/9xSv2AMD792O97pG/b2K0echjfXt9SLB/DHzgMf3m0lyFwDyg58+PX1fB/lTXPtJyL72ta99fYgqBILRgDYRMaoJhSVPG/I0oi6VaU4FGIwUp7b1MCbgrR8X6+XVj/1vp97LtCK4SpPgFHwCHcW+N2WR5PNYtB4073qCAA+OIibOTbVpIjgKYMmPsVbsZSWVO4ElI+cHAIvVWqYePNrwKiXXAkoheAOlxPMGMYK0/CHLTax3o7A6RnF2ClEl213C4FSiYKVTj6ML1pCE6H1g+AhsQk4OGKlY86Yxlq8Mu4PakgMh5ftEmalpWBivWeQUVpiS6VdtFu6LhbFzCheXDZzTODlZ48En/j+oo42AsHUDDB58thKHrKiAxiOeG/TrB1hcrMFegyhAaQfvWsRYXePkXqWVTJjy7SYlsUtIooYxcaKtAaZAZOKOFfPznupLakOEkcp31QI6C87zdQSEqvbyAeGiB7aecT4w7i8IfqNw6SP6QHh5qYorVq4+MFotk6v67XuiUjArK2w4YAMPxwoLaLxEDR7zgBVMeTJuB+DMORTzqQZhpGFlLcb4ftndfM8F6fOar/BTSnQHxiT1+tzq8jNgU2eRAFcBwRxc3xWATC2IXyz42FVyVmECOsJswvE8wOk2YOFv0I9cF4r4g58+Pf2lX3+014bMag9C9rWvfe3rQ1K5aWtaDyJGv1U771fmKgApVqg0/1n8cN768b9R/gBufuavPGp++POnWjFghWqjNBeBOkcghwKSkp8jE7RmnK8bHB8M0Amc5MdI2GWiuqhYXKlIM7SZUsTyXCBPPgRURbTLDsb2QpOJgNIeBkgr8rXGQ0vjXQOVtFofZxORnNLNPGpAJvfnJpjTynXgYtUaADiOZUUbGEFI5uTXFXB9hsS8igUsp+aVGD4FLQaWkL3eCSA9OXRYtAH373Xotxbb3uDyYgl4De5a0Mlavl+3gBG3LHgNKAazQvCNuGUNDYJfIPj2ipifKIJ5SmdiVkXUn18nZkouWKNLG09ARvUcZ/SrbEwATJ3S5lSsXHMnrVxWAycLIHaEbWA83TIOLGHTi+Zm44DWpLybCLy1lVctOmkMX7Fj66OJcBZ8M9xMSgAAIABJREFUabg9GJRWy6XhZ5icOH4NbUd8BGjStGdAsk26DnfHQMC7rMzPgUhudnXWhjCV96FLoKYGSHVzTOn26xpvjwgD9UIBSD7ue6Ex5ee9i8ZWT0CyBW++/b1MPGrL7XE/19Ovrrt9Dz6urz0da1/72te+PiQllCQuzfZdqgYcqjR20tQrHaB0LJz/uoaf+1T5wyiJ55UQPgXjZYrXvJk8XzeVyLuiiCUa1mrVp+lLLFQwpcQCVusIY3z6L3oQRQylI4ztQUoCM7UdQCpC6Tw5CVcoV9I4q5Line/PNKD5dcwgrgYiYdZX5R+z1iOAk7PRdAri0+3zmq8ez+umNs4zl4lItqptjDT5zik0rcPqYMBq5RCZ8OzNb8P2ndfBlwtw1wB9A14v4S7vwW2P4DbHZd/901cwdCdw24NifayUaIioTNGuvk+cG4FJjGNaen5vzGmAz1P1JGReWScybpvdy8apyWEj9wUG1m7ceO1j2RYAHliNl6zGoVLoOeJtN7bKgRmWVEo2F3DJM13PZSU/HxAwpJXwLCrf5Zg0gpqaKvT+iqt/8+Pl0qwKAMmOXTfpTm4KI8znfhd9h0cs290WcBjuoKOYAwdf0dhyzelW9WPrba/bblcVrUey4o3ME+BzFwDyQQc8fiPXfhKyr33ta18fkvJeJVciBeYI2wS4ocrEUCi0pVqAnjUbMekkiKJkPqTG7Y0f/Zs7V+KGn/vUI/2539jJV870KijaqVO5uJQO8GApjj81RYxImtdXXnuCoW/gnYVS4mxlrYM2Ht41ErYHWXFfHpzDNF16ngExWHBUiGzSY6VZ1sqLi1OWo8aYbIFn4vP0c4gSuOh51B1kXcjgRx3IECTIbogMF8cpiEvNkNBt0utwS1OTG9Dc/ASWMLxyf/o5T0PkGsiqfU6vP1lKSOGDE4flwuHoaAPTONz/+P+Nzdsfw7Y7SCDTAUGDlj1gIrBuYRcX8MNBei1iucYZfIwUrCjXOKoUJqiglaSph5xQrwQUxyBTGgWGMgJMCz1QAZxCH/OEI8wnUjsmUPNt6ilKNg3YVZGBkAIKFYAhcrmWEUAXI766GX8/Xl3I844AXlMN3nEOXx0GKBAOlIYrVKtRoxHA6BCgQdgiQAFoMIr2h6rNlceMJ+sQp8nntzTxWf9xW3L5bjtdAqrJSP1boEHQrMWid0JVGgFSnnJcf9y7AZC7Nt13oWHVAvQIgoG6EXjddX+7alcOiCKqrne+VgFqtm6fqVe7zi1f7/erOfnTXnsQsq997WtfH4Ja/MjnCxgIgWDt1RXm7CJVVx1ImAFKLkV8ayBl+IW/PE5E0tfijvT3/sUpR1zVgGBsIJ/993/16v7/0f96CgCbywMY66CUTGS0jrCt0K2aRYcYNGwzpHBCJ02w2aad2DIVCdGCEvWKWRU6FiW6UX6uuXHOFKGYXK+yhiE3wtm6VWhXSQvC8rw50bKEjjVvLqQ0RoBxV5vTDDzmlzKL3hVSdggz7i8JRysR7MdAaFuPw3tPYds10DqYpsPRYo3u8h6CbxEvV1A6TAxjlXZQeoA2W9BCwgrd5T24/qBcVwAgHUVTE/UEyOVrLXockclnJ7Qr4GEHYLgu0LBcjx1GAjVQidXPY4bLCFQiy2TrsCGsnQC4WLmY1XbKF2mstFCE8xCxUhpn0WMLDxcjFtBoIdOQTL3bwCf3q7zKr9BANB4AsE0ARexuRwjiEwDJK/mjk9P1dZcm+TYx9U12tJbVTjqYwv/P3rvESpOt6VnPt9aKiMx9/S9VdarPOXa77Ua2jIVA6usIMYZBNxgxRCCDkYWNaQZc1KIohNs2AouLQALsAQxBbTxh4AGy5AFtt4eA5TZuu9t9+pw6Vf9tX/ISEWutj8G3IjIyd+79779uXW7HW9onc2dGRkas3P/J743ve99X7iUgj9VQvAsBeSwOR82OHeO7uIqZZuc4BtIxbivHyYOWUazh/vTxY+/Xa6SSgD7kSz1jHseaMWPGjG8Chk7CZhvYbAMxerbbavf8ZOxl2nEYxOfTq9tVHa0odV+8OHAPuHXdh+tf+vmPwYr/2FeEkKhq64A4l6mqFu8ToeptbMwl03/Uq739pL4pug8dnbIGu15gdMUaiudh7EwPhNBTy9jBCWt8j2xZINu0G/85VjYcaj12TjvTx/a3vZPgPCmM05E5JBOlW2q7ZthsPZvWE6Njszo3LUdI1M9eoNkR+xofWsQlUIGSmE5IOG9dJnd5g/vgDe7pLdXiBh86RDLiMqHejCNvzqXJ31AaCe2wloOuZ1yjNF3ju+t1OMI3xX0jWA9tczg2N8AJPGmEy0Z43ng+XASeBE89KSzfxMSbmLiJmV6VbS6OUBjhuCWyJu0FCq5L2ZqPFJxmiyDjqB7sXLCUu6NExw59cMB6mwvWu+gZHnLqum/fA3TyF/1YAtKP3m8P49AV7LHHtDu24+9xOGb10Dl47to3D2NTvabxBwYDiSMkuRzHYy46DERkxsOYSciMGTNmfAMw6EFOlyZCHgTfPmQL/XN3v8CPE5B+/P0f/MK/+oUEkfm/+hcefP1Dz1//0s9/3Pdh1BT4EPGhLzoCuxJfL24J9YZQb/BVGcUaxepWKHftcndV3qeRXDm/0zJ4n/eK3pTcqGXJoxVvIQYTbYG9D1ROCAffhlNCMRQkFY4KN7FGlUcL0cEK5umnmIpTU1IlOKFywjJAHaCLjsuzntOTSLPoCFWHZod2Ad3WVGdvePqdv0e1uEEWHXKxLm+i9vu5paW757dm0VtQjWu+xlcbfGjHz6BZrixR3Wd8FQsZySMhyVpGsgphzknupKSrCjG50UbaHmO8PTaG9RDyhDROyY6buGcNnayrLvOyTdwkG6saytGBINzkxFbTqPGBYr08KfR9KTEF6Mjjc9tCUqav9eXz17K9R2iLBe5QdN9Xdh+zi713Dd6y7X1akfvgcSX8cKID+xzl4GNJz2OK9vvOrzpCHqavGTBIxqc/qahP7lu34bg8jloGl7R0x1nri+B/+uX/bBalP4B5HGvGjBkzviGwxHGZjFfl0RYVrLiejlwN7kSH+Pv/7r/2pX3xTce13gXP/uNf/mg4tpPzG1LyhGD6Eects8L5iK82pH4JIYFTyEKKC3zY2thWsyFFS3wfiFhKrtgGT4hHtK+zIWxxRzrseTuWfXesEp5NlxQvjCMbDhu5qoo7kgOmWddVCX07lv7xECm5rxgTIGalqoSq1FyLJtHUCR8yzaKlqlvq5ZWRjLq322ULyw59dY4sI2hCtx4JCbqAPFtD76APaBcgJCQnxCdk0SJNh98kc9JiCUSc7/FdQ06euunoAOd2lseadSQZMQ5Bg7I3JjXFdL1FGPNZDkMEYWfRa9vtHg9ex9dN9zV9TVZovHDhHZ9sI1mhEtMSjEGT5fr41E3qbUYCgwjdI/RlNKg5KIyHgaSE0qinlfuvgL+t8/E2HOvMfJ6COZGPjjQ9lCA+Rf+ATe30PfwRPce74CG9ynD8eg9pMIczIyj3kawgu8/yoZE2jyey/7ne5/DVa+Qv/fKfmcnHIzB3QmbMmDHjGwInSs52JdmJ7tnfHmLQgpgLVvkR5df/nX/9G/Hl9+o//Zc+Bji/uMWHrgTjbfBVh/P9eCUesNvS1dCuJtQrpIwHDQSk70wI33eNbTchIDH60RELbFRIswn16yqPj8d7RogqJ/S636UYnh5e0eCoJp0Ph5ERx+6L9KEv1IcSxoc59FWvXLdw28LVbeDlm4bN1jI7+q7h5vW3Wf/Wj6HbGlm2yJMNErLdbzI0CfesQ560uG+twIFuajQOyeCKhGg/TTd2TQjJuiLVBs0OXzpOsEuw935nPBBK52lqz3vs/O52SY7fn2IgF3XQMS/FEu7vbpsyex2sk2AdrQ8XgQ/qwPPKU8sun2Pa/YCd2PuhIt6Nn7fQc1zgDUZSjmsXyraFfHh9fNn1tkL/bcc+PedDvEs3ZHC+mv48FodF/X1kYcD0eB6yDn7bPgfsTHX3dSZJM0kzURNRbaxsqg05JBwmkd/Zch9eUBiOodf4Tm5c/7hjJiEzZsyY8Q3DkHkxWN+aW5aMFrywG8UayIcTxVffrBnklx//0Y9ltA3ORjxCa2LpEK37UUcrhKFckYecavrteRFGFzvUaCnqzidiV+3pXXLyZh+b9ztDwWe63o2OTUNwoBPLmhB2xbOXnbtSJcLCOWoRGnEsxOMRKhwNbhzDmf4Mjx8iY4GDsldcGWR6v4xqXS7g6Ym5TK1ax2bjuX5zQbs+oapaTr79m7jn10bahq7Ce2s4c8ggIeoc2jrrgriM1LtiVk43yOmO9OGzrX9dxuBcxocO5zKh6vBVHA0RhrUdRtpGm14Zxq7udiuGui4meXAMK+u+YUBfxO6H5GMYzeqi3b9p7XcncN0pr7aZVa8svBBEeF57vlPXfLeq+XZVj59JIlPheE8alpPOxrTLESdX2DNKV8To941HVTi6g9C8vWN/hAZkwNu6HPcV53nyM4xoPdTteRcdybuMfR0jH9O1PLaGaUJwPu841H0uWqPjluZx5GoYu4okWu1HTchAQIbbXP6bnvvhOkw1ILMj1uMxj2PNmDFjxjcQozNQ2lnkjmFxnzOX4XcC087MT/yvv/iRL5oPgtnuDsWwlBwQccmGZLIvOpBIToFmubHU9IIhrNCuyE/NbvfT5mMyEpKTUHmlTzKO8CiTpO4sIDailVFQIyO9lpGOSV052m8efAz3OfYcXjUdOycie797gU1vo0jL2o73yWXHB9/+Pr5qCSfXIEq+ORnXTpYdbDw8VfCgrdjBZqwDIgqlmyHL1kiJU7PzHdZyWyEhoV3Ahy2pb8rf2v0FqpN9m94US8fuAUH6MciEBMbE2HrSCQFxk1G5ISckadHYqLIqHDZpEZvnTNtah+uictwmZZMzlUjJAREywgKPopxKAC3WuoWc9OW2Le88fKo9Zmt7eJZDoR/KCF8+6GJ4dY+y4rX3uJ/IDHibo5Yge+npD6HCf6k6iHdNQz8WvPh5zn9KEKb/5qZWusfdrByDEuTu+zzO/WsmHp8PcydkxowZM74BuP1zP3dnjGpIAE/3zMPL5KrqkHz9TcVP/OX/8CPTMmSk7k23EJIVyIPoPjs0e3KyK9Yimdg35uDkkqWmx4pQ9+PzUPItfLbkbbcjalmFpkp4Z6L/vlj29qU+rP1OUzBAxMhHJXYlvXGOUIrXISXdl1sn+2NITrBtxUTrw08q1rGDEH2ABeUVcawICy9jgd1UNo50dVNz9eo9bt+8z/UPfx/dZ9/i5pMfNQJy1iIXCflWgjaZcKV3aOd2I1hlXWXRjz+EvFv3yvQ5wK4jVdZ26pY1wIdchOm7oMpjdrxTZ9L7ks+nmIrPp+TDXj/VQewIwfCapNDl/dGjTpVVynyyjdzkRKfKRo2InIinQmjJbDSz1d35xXuKyUEbstMhDB0V276V3fNTW97qoNfwNvIwjGC9q9PVFFnebYxpmkHykB7kMQLzh97ni+73IYH+4flOCcn01h/4jQ9djsP70+ePdVcOuzqp/A1VEqhkvr7/WMwrNWPGjBnfENz+uZ/7+Ow/+CsfDb+PdqiPcBP6MsXoXzZ+8q/8+x8RFN3WpfjVsQBm6G5ET+4bnO9I3YJ+e0qoN1SN6UZiWpBSGAXp06v0g3uWyP5aDeNsKVkhfLpM5uiUhG4ocHvBiwUWqppF7lAED4Qgs0vPTmpX3wGGkG4VSl7ExIVKdlkVU6KSKaL3UpkvvU3iLyvhR57k8bhDUG7WnovTjqfv/4Dq7I2RBJ/hh0AW5CJD46FN6BYbw+oGpfZk1n+5Ixd2cFOPYof2pivRLuBONlTq6bdnuOxxLhFTjUimacqYind0bShrZE5kftBupH0XsgHe6fFwwmENp12PcniL2myTYxLWLXc+l3HfZbep6HoGd6uBWgyfQzshge1BcWkjTEPBukON3yWki3XI1hKp1I3GBRmlUsetdNTqR3Iy/D00BFoiibvdsulV+qnW4lAPcoxUHNKFPbJzTxckHxTlsBN/P0RAPk8Y4TS076HtfVnHx+DwGOUB4fve5zsRIQ1OWFHtE0mPENnDZO3GcEulkjASEGC25X1HzCRkxowZM75BGDoiUzIyhYjyyS/+K99YwnEMf+vn/vzHAD/5f/x7H9EH8GmnSQBIzsTRp7fk9ZKwuC3uWJ529YRqscIHR4wVPkS6zQJfWbK395mcdkGGA3aiftMueJS+3z0/JJMvKqUToVzIJpZxn21UKi+4TAnBs26FF9tmwHAWotwpZQaicadYFMGJsHDW/VgEaCpYNonz89b0GCFztm6I0UbSZNki5yZEr9330dUSFKQJdgxXh29SxtxChiabPqRJ45pAKfxDgk2GANJs0Jsl/vQWzR7ne9rN+6bnCYmu82NXTpx1O7zLqHd0ZW2P5YNo0eJ4pyW3xe57oIs2Hue94hz0Ufa6U1WV2XR+j3QcOnFldqTElRNsRGjzbvxpKkrvD6TKdx2i9u9PU8UHwtFLhjK6l8RcsSo8XflDyqJUKmPq9325E3Zs+QuNX+22MUxHv6Y4NiZ1n8h8+vgXcbf6sseUBvH8Tjj+lpwVvUu6ACpCoYUcfd7hSkr65KJGuT/9dz4I0d8lPHHGDjMJmTFjxoxvOILfWdO+/PiP/iNFQKb4W//8fzke+0/+1T/90dgFySagJph1rG4btFyNV3XEbolmN7pl+SpaSGF2BJdG8iFCEe/vXJtSspErFQihzO0Hs5c1tywheLtKHzwMtc2yKsWyBxBSv7OVDaW1IQpdqYRDSUOfXnFVivj9YB08dvU+KZw1sKyVOih1nchZqCrlyfMXPPsgcnv1jNs373PhO1xIyNMN7nyL1gk5rfaFKUP2yYTgaXTIMkGVkYZBeW/btYomkIvOSEoUqCOaPL5Zo9uz4ryWSFFJm6ass9klp+zwIRc3N+uEhJBHAgiMnYxQSEaKdsgecM4+G+/suaZOgCeWsblhKZe1crOdzvjv9p3HW+WidmSFTVRqB7Wz8cRVv19sumK3a0t2t/uxwBMQNiQqHAErQDdE1mJFp1dhI3EkKA2eSh2t7Jf1ZhMrVHii6h452A80fLcC9qHS+xhpeFedxhfB0AEyo4fdsRy3BJa3HtuxtXnITnd8nereqNmAikAiEcSz1e6eV5eOhzw84joQEy+ePHdB3hkzCZkxY8aMbyCmV6uzzfvgw++OK20/+Vf/9EfiFG13X0G6rUcyMojT6+UVKS5MKO0zXh192yBOiTEgosTo97ogg1uTc0LXBULIJW1dcIsyRtU7vBNCHsL2XMm7sCvzTTmsYTSoT/D+qbApYSFdGp4XbvrJCE1W/CA2l+kVVNjkzHt1GN25lqX7cbbINHUeR5WqKtEszPIp1Bsu3/sBzY//BgS1TkbrkYuIuxRY1rBuy/ySAwdS745HFglu7WTEK3hBTnZFlfqMqEJr+9YeiA7aCjnZUtU9T0qKfbd5ApgTWd8FMwkoBgBgmTa5d6MwXdXInojSmMTH9B7ZUU07HUEtkLFObLeWiaPqUC1dkK1n0+3WMmVKgKGO75OxLlWblPNa6BJcR5vuH0fihs+I44XxdMZ/PXl+O0lOP9FAS6KXzOA+3JdAvGEU60Qr1mLDVMOIUSLRFUIz0KF36S7sG8wef346Cmj379//YffjcO/vYsF7CBl0UGUfh5a7h3Alc+d+t68pqd8d10MEZByXnNjshuJ6FkmsdDtqPw7F6rv3u99a+Nj9rHEkPF9Ez/OPG2YSMmPGjBnfQNz82Z/7+OI/+t8/Gq7ov/kz/+I/sh2QQ4hTC89zGUTR1RKNAXe2RotNLyGRN0v67ak5QWHaj6ppUXWkvsx1J1f0IBnvTaQeoydnR13H0dbY+5Inkq04VlUC4J05bOVsBOTyRImpkJFK6Uq1ebaw8ayssOlsJCsr1G5XdBsPKF0GsQDCxgtJ4UQcy9K4WIRd92Mo1J9cdGw2gbrpqBcbFmcvcRe3SNPDMhmJCJBvbKxKThawXKCfrtDVbtSK1tZPFslIyfNolXcjSOPtwIJD1x1SkhE1p8KUQFcL3LNb8s3CROtND8lROyV2C7p2Qd/ZGJxzECPUtdL3nveer3nx8gSdBBkGn2nqREw2suWcERYnjCNcMQrPn6/ZbC7Gtasq66h00VLah7I6TTokh1CFbbTyceEctymRVFmIp9d8J9dj0HMc7m7aHdkvgs3Ct1E/kpEKT0sck733ClTZERuzcD3eBXkb3iZmz6J7043vgscQkPtC+Y7B3dPZeFsWy2M0Icf0LGCkYyD9vR4fsZq+Nj04hiUkTeQDAfugA7nvGHb7y/ylX/6zv2v+v/qrxkxCZsyYMeMbiutf+vnfdV9mP/XX/uRHBJBylVKjRxYdeht2wvU6Qhac7wj1BlXPdn1O3WzwPtF3fhzJCkDXeYZr3TYyJVRVJCWHc7uwwhASMXqCy3vuTRdVZtt6nHPU1ZAtYvuqg1J5aOpM23vOFtZ1sK4JgIx6hEqFOKmQK2/ajyDQZiuka28ExDsbRzpZRJbLnuXJlqfPe7q2pqpb3NkauVgj570REAcsHLKMyALoOuh7Gx8bcjsWwMJE6toLclEOpAGqcfYJgkeqBHUAVUQEXfXoqqz/MuGCGQLoqjaRexVZ9K/ouw+p6mhEMDlCSNRNT9105ORRPR1DCps6UVWZpunZbipbV680daYOiaZ23KwqE63HQMoWOBlCpvJK19mo1sUy0772nDTKy5XsjbxNzZkrZ2NcjRNWMZv2BuXMOTYZtprpCyEYxqQcJlK3fT08OhQx297h+eExsI6IFzcmkQ+6kb2cCtnt+3FS6N053vvcQHTewbb7If3CfZ2LodPyefQhX4ZW4m2i+UO77KnLlcffcb16aw6L3v2bgIfXbnivGY/HTEJmzJgxY8bXgp/6P//URyAmmg4Z7coXdhbc5Q16c0LcXhAWtzaWFSJ04HxPs1iN+xGne5WZD3F0zErJOiDAGHQ4huxlIyIDchZ8MNen5dJxu2pwolRVpu8dMQpddASvnC4jMQpVlanj0O2w22E8KzjoklB5G+GqvXVMgoM6CwosKhPF1yFTVZnlsmexbKnqnvOnn5JTxXZ9zrKKyGmPVGqEwik4wX2nNvaTFd12RjC2tob0ilwEIKLrslbNpChyAk2NnJyi7Us4OYG+R69v0JV9Lu7pBmnKeNbKIU2PthVEj4glqVeutUwX8cQYaBYtOTnWq8XY3WhqI3NnZ3ZuuWh4fFSePlkjkrm+PinjV4IPsSSxO6qSzr5cJM6c0va+TJ4Jtd9ZLA9YBOtQVYU4bnqlLZbItThqJ9yWv5emkIi+lKVmo2u5IOMyFRvmY5huF8q+dhkbO9IxTVoftuGefT6ELyIKn2KqoRhzbu4ZjxrwZZCHL0us/bb97EhF3vt9eOxwHcfcEH2I4nHv89Pu0NBZ+Z9/+c//rrto9FVjJiEzZsyYMePrQUim5lZhyK6gC2gfCjFJBFa2HdjIFtBtzknJUzcbVMWuuGcrzL2L0AdwphNYLC0vZV8nMtx3hGDCjpwtZb2qe07ObqnqlvrFezRLK7Bfvbik7fw4LrVY2OsuL1dsNg0pOlabivXW741vNZVlZ8QkZJXRbhagKqGTwSvLZWLRRBbLlsVyQ7NcUz17gTQ99c0J7r1b04FkkEsT0EshEaSMbnuICn0ZvRq6HSLISQVtX2LhbfwKQJ49s3XdbpD3nqM319BHSCBLRaoeloKc1Oi2EKA3GVpGt61mOWhEFjifCUROL15y8/p9NhsTf5wsEhcX2/G8RZRm0VHXPTGGcTsnRu5OTjqWp2vew0IoQ0j4KvLqxRNevm5Yt47b1sje9Ip3X7oetYdlZev6Zm1F9Ll3bLOOBgALETwOBVrNeGTsYqQD4jC13p1ibzRLdk5ZjZgF75jKzd2U8oEEDJqJ4fe3FdfHcksONSCPxTEiwpExsgHVSNi+WsH1Q/qOww7Ifba7YHqPY6TtPqG+Q/Di9yx2H4PpyNlMQL4YZhIyY8aMGTO+FvzqP/vffQzwU3/9T3xEFojBgvMmYXhyuYI+oF1AQsSlSL28AWBz+wTnMj70OO/QLORsRVTKNhqUksP5hIiSot3XLDifxsedKCkrdUkPb5YrRBIXT9/QdzWuStRNJOsurf701MaTTi9uWK2WLJY9i2VP98MzgofTZaYOmS46Fk0y+1qvtJ0nRksWN31EHvUSZ+drmsWWZrmmXl5bbkrI+A9fo+sKCR3q1MhHVhulygpVgDYyio9tpghZlsEkJ8jTCu2ivaYK1jlJETl/glQ1enNlBARg4UpCIkZaFs34nnpbgiFDwp1sqEOkX19w8fwzNrcXo3vZ7c0pXed59qQdbXxj9KgKfe/HDpSqsNnY6JUTIyg5C5vVCXXTEaqObrtku16OmpAhwFAwV7GenSVvVLP97aLp9L0zZ7NVryycUHuhz0pwNkL3Mk7FytbxsCJ1Z6d8jIAMSAdjT0kUr0Ilvrhu2Xn2Bxa5mV0eyJAh4ot/1MN6ibvjWMc0II/J5Li774PclkeNaD2+O/NFuyD3BRPCzvlqwEBAHkPQEhnVPOo+/MQB610JCcwE5ItgJiEzZsyYMeNrw0/99T/xEWDdjuzMjUlNdizL1saKSqChdgFfbcYU9RB6cvZ4n0gJUq5HggHW8XAi1m0BXNUT6p7YVYVQZCMuJVF90JXUly/ImxNUrYPiq46z84auvWR52tFuLaX9/fMbTi4/odsuCVWHOGW7qVgse9brmqpKuG3F+dmWGD3eZ2JccHrej/kZVZWoKhlHxgYC4rztjyrCacQ10eri/sgipoQEh67zrkp1oLcROa8Yotwl1EZAvLcuye0KjakEfAS4vIDNFsmKthF5egbrLSwXAOirW/S6Rk6LjalT9Mrhqxb/3kvq1+ekfslh7Je9AAAgAElEQVTN62/hnPL8+QrnE9tNQ98H+t6RktDU0E6c0Lat3yN4fm2dke2mwfvMatWw2gTazkhI29speQ+3nY66j70lUbhamzNWXTJgVinjxREzY2ZI1F2XYhi5kr0iO0/C+/ZR40FNBzKQjIilgLjSWXnIcnZqF/tY/6Rje/KlwzfY/d7XsRn0C1MScZ/I/KERrUHnoI/o3HyZONTOTI/7GAGB/c9yus3e45pH4pE0vdWG9xDTLshMQL4YZhIyY8aMGTO+Fvz0//VvfUStkAWNzgpu5yALUicjJICWPArAskMieLehBlJs6NslzmWcS0gwUXPwPXnogjgboVI1oXPVmOVtzh4JSuwqqqalbrbk7HHna2TZ4vsb68okT7s55enzG1a3S+omcvbkM3zY4r77gvcuV0aU+sDJ+St++x/8AT78zmds10ve+9YLgNFK+Ox8harpJcC6Iz7EPW2K852loTtbGzoHJ2p6jqe1dSy8M3/atY05aZ+gmRTi/aQ47NPO49mrTdzkDIsFUlW25uLQrrXqftEgTy+hbY2Y9D00DfI04/JqTGXHb3BNj9usoemR+g28jsib9zk5XZOTZ7NpuLpu2Lae261jUSneBa43jsqb0PzFrY1QnTZwcZrYtJ7V+oQuOpwon904Vp1y0QjbaFkqfb/T3oARjWenyuu1hT3edvCmyyRVelUqEbaaaftBGyJ0hYAMJORw1Kk/UvIf04dUOJzKKDzvJdOoJ8tunKudlM974vSCt9ngWoL64x2jDu/f974P4b5OSh4LfFcG2qY6ly+OwdJ2SE9/276HMSwvjqQ7vYfHH00/NyK2n0lybN0Oz396PPuPv3u3ZMZxzCRkxowZM2Z8PRhmSEKZr8liguflUIBXdtt0iPdISEZYAKInp4DzPVUDqa+pm11QoWlErMMxjGCJFOLh1FK/q55cQg/rZkuobcSKOiJ1NL/ZkNBthaojVB0XTyLb9cIIyPM36JtT3O95Y+/56Snh7IrlyZZmsSKEHucjms2VK2dn+hSn1NHTd9VIQJzfaVdSv8Q7RVxxw6p0p+doewsUSRNb0T4VRXUpkEpWiZwFe12YXNntownQAakLubt8DlcvEe9R1yBlLEWbBqkXaLc1QrJukWUwFy3vQHtk2aPXmfzyDFLRE2RPu12QkkNVSFnGDoaNXQm3LWPq/CYqz07tOe+U9dYbv2qFT9ewSmn0oFolJaljGXbWvJWzzzZl4XKpbDob2VpnIyEZpdfBxUjpMMF7QkcnrHEtjxT7gxai426S+ZSoeBWy2Psk0Uk3YeeONX2fx+IxBORt+zvsDLyLxe4UHjeOmA2P6NgJOI6h2/JQd+UYjhX8x4jFKELXQ9J1TwdqHL3aCdH33K+GEayDjshUQzM9NvNcy3MX5EvATEJmzJgxY8bXgr/5M//DxwA//at/3MIKwQgIRiIICXy2XJBO0bbkjWdnHRGXrcD3PVpC8lKsEcng3CQKjpF4iNM9ciCieJ9YnL0EIKcaRJGQobJkcUQ5OX/F5vYJZ09+m5P2nM3N+yzU43wH7Y295nKLA5qFdSeW558RuxNyqnA+Ii6zun5K8B3NomV5usZ7G8PK2eEXiWpxi0iyzgoYCTs5KMbaMpM1JLUHty/YrcpMlgi6apHzpY1hOWczTNsNcnoG1ZAamO2+c8hwv9sitZEVNlvoOjQrMth5udKJaW2N46vn+GpD7E6pmy3tpiFnT98HcjISkhRykWBc95ltUZWfesemE04bLSRFCZXtvstKpzbWdBMzG83kTqlK+rntQ+mzcNXC73tq+09Z8Vhh3LMf75fHx/bHeaQE5dnvO0Qy4aAcP9YlYe/5hMeNBXdDwGumJ5MkProQP5bwfQyDJe/bLGM/r4gdjEx4dSTJVOqJksmksQh/aLRrKsx/FzJSE/Y6EB0RDojIYQdkwLFxrTHX4xEZLUnTURKW2H/tTEC+PMwkZMaMGTNmfL0QhUqR3llXRBRRMbenKqPFjUmWHbpajAV6WNyS+wbNfgwwDPUW6a24Note04zY74JIwocOX7WWvO6yWc1e3iAnHb7oT6jKrTO3qer9Twn1Cs2e5R/5Nbb/74/jL6+QJyvkaUZfO/LrJUTPkx/7NeTU9Cxue4OulrQ3z+nbpdkHq8OHnvP3vmeZKOuGnOq9MSw531gwIJieowmMiuxBmD5YQ4kgwZG/r+iqQTc12x9+h+b0FXK+xr1/i3zQIpenRjAqTAMyoCsdIB9sVKuqoVnaY9uVaUKaBjkHUmERfY8sKiOOF4luc46uL4mxIoSeqo5cX5/w8o11W+qgfP+mvF1WXiQ7twrhWeWJGRa1crKI5MaIyM2mIZVi8TNt8WrF7LkGbtb2ma6IVDgy8EQC37/y1L7IYkouyBLHbbm6PRSQPTvBeUZLt8PWMyBj56Ebt89FknOXfAxi9CTmjpVEaYlE0ijRGboZASFoRU+mlXinqJ3isSNY92Vc3B+g9/l0HJV6ekkjiRl+fzi/3XCfNmOAx+GLlsYh1FQANOppMLextfQ4hO1BL2baAYnlmXvP/YjF7n02xINpwHB802MdiFH8ip3C/nHDnC0/Y8aMGTO+VvzNn/wfP0Zl7DoUv1P7PShyGgsh0JIXkkYLX+e7EmK4xocW5/ux6wAQQo8P1l1xLhVdiH3VhXqz2/6kQ5oeOemRJhkZCmpWtYuEe7rC/97PcE+vwSn1M9N6yGlEzorF7OUG9/4t7ve8sX0tOyMT5+vx/UVyKdJbUmudBvfdF/jTW+TpjXV4Llf2+rMSRJjyjnAMAuxl6WIMKed9Qm8WrH79n+CTv/1PUy+vrJPkCqk7LyNYVWVjWGFCQnI2cfrwkyKkHvoWxJmg3Ym9rqrKa9TySVJGe0qIpKBZ6NpFMQWw0airtRCT0GXlJibabB2BCuHMeUIx+Xpx7fjkpelHYnS82UCryloTGyJbEpHMmsSaxKtCEW6J9GRWmriOVkaeVmJuWOIQhAZHg6PGU+HKj1DhCDg68vgTUfpyH6wTErFjHnM+RMexq8NbrzImph/qTWTyX6OBpd699vu24LzDbaddkK8K0zEuRQv5uKtpmZ7f9LUDyfA4Qlkbd/B4Q8UJDZe6xE2cw3yxSDa/sd3awr4r1n2WvOO29zx37PHD/STyHllMJPqhC6Sfj9TNuIu5EzJjxowZM75W/PTf+jc/GmsWnQioVaA1ciJNj64a8KXYkYi2tRXtWSGCluJEXC7TSDbKFfumEJAyfhVafLCRqZxqwvmVieIH8pEFOVFQkNMKrZMNlCi45Q3iFXl2Y8fTWxaH+1BMmxHtdfiIJpDWo5th7tzhfRpJUU6WCi/JIe9dI07JVye491uIIM+LmxXs7HOd2GPiIDjLB0mZ9HcuoA9cvXrG8nSNe/4GqRNy2iFNtkT1ywuoF9B39vr7kPOOmACEgLgS6R67cTPtkwUiiq15SgFxSuocfV+x2lRcbUz7sSlNnYgS1Yr8Riq8CN6Zw/BtVl5s4McrR/CZNpmgXFT2AgQTyjkVLYnWDFbpy/ONOq474dnCkRR6VXrN45+XdSYED7jSQRn+d9h/x/1X9ofOyTTxfPq4/QFOxdtCkrxzsCpX0K3bsivQ8+TavnVMHsZIVN4hGf2LQFE62b/qP+2CHBsDe2jsK3DXgarCc64NDhmF/L0kXrMZz7ehIpGpCHRlqA72CcgxIpIn+o9DHCNv92ln8kDD1P7u3WSEb8YXx0xCZsyYMWPGV46f+dt/7CMy6MZZkQzo+iDNe/h2zwKn0boOw/MbT/70EqoiIg8Jd7NE1wvi9oxMhSOOupGBmJz8nn8AbYV2lY0qlWwQqc3eVk4EKod4Z8fw9BLpe7hMyNZctbSNuNNogvHzUxtjCi0SJ2LxdYckRTtFatN6+FThfMnZkISqN6H9zRK5WEOd8T92bcVtpnRAbHu96smvanTV4J6u0DaYYP7mEu1q2vVTfvPXf5QYhe+cbHHnW+QsQgD5ve+VdSwF1flTIxjtZrfe4nakA6xT0ndGRgYC0ncwOUeikj+tyS8u6TbnxM66JKvbE77/yRmr1tElZZvNRrfNmayM5eKVRtrkaNeOrgjIF+L4jVee87ridR+NRJBpSWhxW06i3Gg/OlFV6vBjV0NovHDdWTDhpfess/3VbPNAYqyrUU+0BeZ4Zd2RTelfdOW5MNF2DB2PQ2S00CGzkR0cmOzDc2PnYKFhb6SrJxPK82BFr+PtXZAo+a36jy+C+9y6Am5iWfx2XcVj0VDRqP37v6Xb6zooSlu8qYduCUA38at+iIBAIRrvmP8xEhw9PnKV1Y7yf/nL//msB/mSMJOQGTNmzJjxleJnfu2PfTTcl2aSbVFlSG4kH9p5s+pVQTxoL8gFZj+7SLjqtW0fxboS0ZN7K1BM3G23w3iWhITUEU127VMW/U53AYhX8A45acx9qgpWdIuzb0dfivHbDnl+ClmRk1O0763Ab2ojJMnIkn66LcfnxrExADlp0eiRkgCv2yKGr28tfa9yyFLInyi6qtB1jdSR+Ol7xO6EevOCeHuJ8z03rz7k+s0FfR+4vq3wXnn14gnN//NPEuoNy5/4u3ZyKaKvb+zYumLru90gp+c7guGLeP38KXTbHQkZF6gQlb63x3vGztX65pxUzAG6ksNi2YiCJitXh+I8oUQy2/L7Vs3aNiCgsIpGCFrNbNkJjiMl9wUL/4tlRMaJ0Gsudrs11zHTq/IseLpsRX2rysI5Os30at2HjkRk59okWBeiKuNZDrPdRe+6Yt2HfEQh0UsiqKOV46X9kJr+UBjfFI8Z13pX3ccgvh/uD2chkyn9IfvE9p/3Xvt50FCNWguHsKC6c04D+ZiSkmGM6/PiMWNZw/rN9rtfL2YSMmPGjBkzvjL8zG/88Y8IFkaIg1HX6YafDFGMeAQjKLotSuMmoy8LSfGm2SCBRktLxynOd+RU7UayXEY0m2bk9Ba9WRoBWHTgLKxQgmlRNJmtL029O+D12joSgF51lsWhwLqFyqM31+jt1mxxHbjvfmip4s5BDXptxyaL3RgTTQddcZ4KCVl0yKIHX7JAAESIf+9DXn3yY1RNi/eR7fqUxcmKOnrWN88Ioefm6oIYPZtNoI+CKry5bvA/+JAf+b3/kPSbl3heQLZ1lGZD/q03Jk4PYs5XgDx7vt/1mI5rpbjfKcmTIi56iB5fRfr1cnzYO+VsAet+V7zbR7wbX+lLR8KX7I005k2YUH1LZlv6CwOmWReHLkUVRjKuNFHhOFdn8SgKSQeXLWhwtOQyjnU3cE+h9EWMHCTR8ucqiJrg/fD9p6NHDiNJg5PU1JHqWOI5cGck6yFE+XxF/33o9/4R7hOMKRkZgg7T3ujY249lIHAVoYzORRqqcfxqKPjbchzTsa9hnXvSXhfGjmmfGL6NeL2LM9buPexvKH+O5PQZ746ZhMyYMWPGjK8EP/vJv/2RpXd7tKRjs027qsypEZCg0EvJCrGnNDrY1BYe2FYQso0ydQGSQ7c1uV1aB6TAnK8yPpQxqm3RkAzBgF2AOqJtBVXEPenMcvZ2DWcnuwM/WaCvbki/dWajW5db8g897mkLtGgvNjIGpNsf2HvdVEhdMknOt2jIpjtJ3vQtix69XZiuBdAuWElVrvSnv1Pz8vu/n76vuHpzxs1tTYzCcvmEb3cN7bah3dZsNjUiZm277YVaYdMJZyeBl598m9P1OU9O/m9k2ZFfnJM/cejWfGxl0SGnW9x31hAs64S6ge26uG9lpKqs0wPQtbtxrJTR1tZfYyg5LJm2rXEu8/xpy8vXDSAEZ6L0zK7oW+JZF6F5ZGrb6skZ3mRoScVFakdEBk2FCYV1DA5MojTquaYnYVkgL6PwYVWBg9c5j+NV58V5SdgFD04NXlf0u+6Imu7CT0afhtv9zIz9YnaqAckTPcVIwGS/qA3qUNmd4xfBYTE+FO5h0j041tHYkZFj+xxG2e7HtFvUUI0kIahjQWAt/UhkjMxk1vSEAyH74Lo17AdgTUtPYkPLEDJ4eJ73WQTDXU3I28nHjjTaA34mIl8DZhIyY8aMGTO+dPzs61/4COfMzSlmu7Aes40fTa+sF2H4AC2p6bTFlSlWRjyiGpkoj2tXo9njqg6RXEToRTTrh+wRu95OSEYIBjTmiKVJ4BZwPeK3Zk3bJ+TZOfrpNWRHXjVm5tpWZJ+RkHfHCLAux3OzhGVXUuAV3dRIFmh607fEhDTRujg3S+uEADSVdVaAvq9YrxtidFyvPH0SbreO5eKS5bIlJUeMQtuHsUkRk5RbR9971jenPL3YGmHb1rRX75NTQFxmET6xNazd7jPoWtBqt/59X9yy0p4exATxdt7iEn1fkbMnZ2G1qsfd1d5CBJNaUjmYLW8+uI49jCL1ZgGwlyvhVcar/0Y8ir1uscQdCtt1KaKH0SJPzTplFt6V9zUJels6MGB2vNNjSCg1nk3J8qgKmRi0J1M8RBZSOd4pcRlGiKbOUsMqxEn2xhT3dU7uwzHHqsPfw5GOx+fFVJfiymDZlOw4hFPqvddMR6l6SZMO2O54phqQ4TWKsmXXUTwMJ3yoE3KfKP3e7Q8F6YWASLlIMDtifTWYSciMGTNmzPhS8bOvf8E0IE7ABYYZLAEIoJjL0oi+FCnOQgbH4MLo0ehNh9B7tGR6aNFW+MqE1pbDsR13pyVUULPfdUGgdFqcOW71zjQOTpFGik6iJ7/qkfSJHW9I5G1D2ja4kw1sanRMfbfjA+tq6Hphj28rZNETXz/DVxvkbGMp6EtFQ4KrMn4V3c4ZrMtoF+h7s6q9XQUr5LONAqkKOXu6LhCTY7114/KmbNIV75Rm0XF+eUX+5AKA3C7pWhuZqpsN0nS45y3S1LtRrKxGNoI37QeMXRFSNvIB0KuNd4WEvH/F8uWa9c0pfd/w4qrifJloezvmqEqafLwJE5sPDw3Wtrbb/avVQ5HbsxtrSuTRFWoY5wLHRiIXWtNKwqniyuRczEqDwwlsNLOaXPGvCFA6KoMjU0diqYFWEkl0HAPqJO0V0A9dTR9Gse7LARnOw09I0GPGrB4stB+pA/m8Go5jeMhZ6nC7KYnrxTpEU1IxxTQJxCEmXKdijXU1j4UT3odj43zvgun42Uw+vlrMJGTGjBkzZnyp+JWnf+Hjn339Cx9RVVbg+p1LjbYRaTzaRxOfJ0ygHgVSyQ7JE6esIjDXLkC7u8IqCwsHJPoxVFBcsltJOyvfaRek5I6QBeqMnCRYCrpWWEeIkH79ffy3XwHexraA7e1zfNtRx5e4p0Vnkh399dMxBLHbnsGrkoTuEydPf0C3eorvl1TnGzjpkScKmwTaQnbo1qOfrOFUkLMtdd0joiwWPeFqQYzCzcbz2z9ccnlWcXoSefOyRgt5SdmmuZpK2bae9WrBYrlBLtfE3/w21y+/TewqQt2jdYuclA7RVYecVzA0QHLeERCwPJABq10RJnVCkzl8LU5u2K6XeK8s68zLG891C7d95jZnkirrUswNxb4vV87bSZE3FKcDbJxp11kAhVLM5tINQR12PV3GDkdGudbItVq348NQs0qZgPCe1CzEcZsTn9FS4ViU/JDBNUuwDgyyL9pe01OVXItDmNFvOW4Zxp2UCk9fciUObW5NIB+pS/mlewXzPhR9tB7kkGhYornb+/1dMF2DQ0w1Hx5HpY6GQF0yUF659eh81csubyMWrccxDI/3pDtC9EMCMtjmHsMwhvUugv+j5yjeenUzCflKMYcVzpgxY8aMLx2/8vQv3LWxHEYbYrak7rDnomnfSEmQIagwpP3OyLhdGeEq25juI6K55IaEgXRk63xkt/e71MkyQhzoraIbgQ70OqDbBu2Kle5t6SIsb/ChNbF507N9+SHd1Xv07SlvPv0um9UT2s2Sze0Zsa/puxo5sc5M7JZGWlpfHLUyUqUxBV63Hr0GvV1w/uSKuulZnuy6Ot6Z/uPqNvDiVTNmF06fV4W2c2PHJH/vPV794EfZrhd0nZGWnALpB8/JPzhF3wTyq5789yO6ao3NdHH80T7tfhQbW3OYOQCgm9qyV3zGuUwXrQOSstKWMLmMdSPsYxWU3VXp+xKr+0JAprCuyX5RGWXXGRke78XCBldECzdMmXPveR4CT3ygEuHEeTzCip6WRFOIyHA0Fc4sgcmImj7EF6KTC7mYwo5r/1h3iex+tPqd/nTFiwpMO/GQ69PgovUuGN7p8LHHYghqPLw/fX5AX84loVRqZ7ISS3JZSz8SkJaelp6eiB75ry8mzolMJNLRs6FlQ3uUgNz9K5mc6yMJCOw6OEPnI2okakRVZz3I14S5EzJjxowZM74S/Mrlf/ExwM9++qdsPGuaBA4Tpyy1b6MIOEGjERElQ8hI0WDoSCJ6G9UayEW5lRD39CV7iKZNoelhmawLs3JFFO/QjZA/vWR9/QGnL1rce1fE7hSAfntKqDfE188IVST2DX3XoCpcvTnn/HxNjJ62rTk7X/Pk/X+IrhfktLPklbRBt9hYVq8Wcgjo1cJGzpJnef4SEaVrF3in3HTecjZy0VlkQUQRUVQFEVAV+qRUAbzPvH55yXb9h1mvzbq4rnu26yV9V3Hy9AfmFBaU/MkZulriq5fItzI6fC4pl8wStc9jWM+MrdO6Rrc165tnrApJS9kCCC8aocvOpt1QbjQVbcROR9DfU0AePv62kRoLD8xkNR1HK7sgw47EK3Vcii/vDbUIt3GnkVgTR32IQ9iMTk123X4lPQvCqEUZDmHocgxIksdOjUPoUarRAlhoSrdgXcaQDsnU58FDOhDBcczt6rE4lkUyfY9DUjQkuPckricanV0ei7+XFOzE8iV9/qBLMpCUAQMBuY+EPNbyeO8YSiZIkpl0/E5gJiEzZsyYMeMrxa988N8YGfntPzHRiuxsewXsKru3q/oSxbofUEapgqWaN/3ucdFdOdSXYj/6YgNcbt2kIClaEgE0Ods2i5GcZxt0ZVa+fddw88mPcrr5DM2OFBtirNiszlie3uKv1lTNhnazZHV7QtcFXr06x4myWPasVwva3/hDPP/WD8a3bq/ep3GKrFsLWiyuWQCETP7sKd3mknZ7ivcRH/rS2TCSEYYOhEohH0ZE/DTBO8OnL05QhUVTjR2TtvNcnG959sELAK7+/h9i8ckNOQWuXj3n2fX3qX7rNf67r5CztLNRntZ5G0/69JLf/tv/DIvlBh8iLz59zg9fLOii0CdrconAMghER4VyExNhMjJVFRFzSxqJw/jxcHccacAwzjPtRjik6DB03P9mMvq0VM82WwL7sE4Os+sdOjQ9ainsxY2rUU+tHiQRgS2RGs+WyEo6TrTay/jwJXTwMAt80LaAEZGWWFLT3Xh/gGcXjHhYXOuRonogQPeNSn0RAjIc+/Dex7Qe00I/4IrDVmBNbzqbkqo+jExNP7spgYlli50L2oTYHRGVx3FQ7348JpBwwOEo1rGxq2OPzUGFXy5mEjJjxowZM74W/Mp3/vvxC/xn/r9/wwhJJXbVvStjP1Cse91u7KoQD3m2RoqIfUgQH5H8bvthH2AWv2DakuGx6EeHKxn23fTglBQDObkxe8SHFucjm9UpXbtgEQOhXlM1pzQxcHO7QLNQVZmuCObPL9+Mh+V8NMvgLOjVKSnVhA9emOVwFaEPuLM10p6zvjmlWbbUzZamznTRATo2j0R2RZErJmODPqRPjH6q3jmqKtP3rsR/LFD9NiF8iEimbxuu3pxxdVPjXWa5PeOs7nH+CmlKEZqE/P0L3IfXaBbS6oxPPj0jeLMyTlm43ghtqfuTQuNtJCupjWUBxYx1p04YitFW0t5V98PycnhuWsTCjpC4CRkB051URRTui9YDsAR2hYUTLoLnXJfcpEQtwie5pSJwooFt8WzqySw0kERpC/nwRahux3C3MO3LuUyPZ9f5uZux0ZPvjHZNi+IxnFDuvtewLsfE5l/U/equrmSiBzpCSHbbR7wYmarUs5Z2j4AMSONnrfcSkMPuh+397QSkvyfl/BjiA9bED2EmIF8+ZhIyY8aMGTO+fvS6+wYysyMbwepl18U4GK3S1YR05MmVYDWiMgrY6+LG1XSFnETbvu53hGbR455a5ofeVORPnrJ++R00C4qNeonLOB/JCSMn2bO9fc7y2Q9YnFzbe7+4KFoM61CcnG7xwUTeIpmcArFbEKKNR1XNjY1EAdo1pk/JQn36mpy/Q+oD23TKxcWWtjshJldcsGQyhqV7BAR2Vr3eKVmFzdaK3ICy2Xja7oSmzjx9sma9WnB1U9P3jpcvL3iSHcurSxDFfXBjy/vpOS//7h/h2erXcU9vuHn1Ieutwzs3ZDmyrO14Pt1YwnhW2SsVh3GnoWc1FKECLNWC7IYuRECIB8WnK38YiTwW3/sWuCVVHaEpHQswuVEG2pxpnONV6mmyI+5d2Xb0ZX9uUmLvuhzCgrAnrJ7mlICNYtkIVqIjs5gkgg9kaNCFJMyk+L5Sejj3YbxpOMdDvEvI4efFMcH21GL4sEsyCM49DndAXI7ta8hSOSQfx/BlE5DPi5mAfDWYSciMGTNmzPja8Tf+8F/8GOBnfu2PWaBhKEWOUzQqUmdLTh/E59kZociyn/lRhOeaHXR+FLBLSIWATMZTttXu9y6Qs9uRkmXLyfvf4+rVP0VV9WzXF6g6Fic31Ms31E3H7fUpN1dnuE8/4Nn7L1ievaGpP2CzCWQV1uuKrrMgv+q9dtSEDAGK4uzYVv/wDyCSibEi9jWahb6viNHR90u8z9R1z9mpjbq0vRszEowAHBmTUYtkyQptv/98yoJP5qC1bc/RDJvOUXnldhU4Pa343q//QZ5/8ILz+tfQtuL2e7+fH3z/GZ/+8CknJx1vrhbEJLQ9o05l6MZUItykzE1r40+92jV56yxocbGaFpyGfsh1QcrWeSzGB8tbC7obdAU70XFPsrA/DaPLVYCsAxIAACAASURBVBYduwSfsuEFwnfTCQvxbDWRUK7pUSBqpiol8630VOruZIMksZErxAjBWjrOtRnJyLhtWe4h6RugJux1caZOXgN60mS8bJeF8i6YJpw/FoPz1fT2sfa3w3aH27cl8LGSwBmLUZDeHXQdIvvC8bv5HI8ToY9dlEeMYE3f413sdw81KjO+fMwkZMaMGTNm/M7jPhOgLPtdD6c7MnK4ncumBxmctHxxzlr0to/oIGR0XbP63u9nef6ZuWrFQE414fINpxc3XL++JEZPs2zxobVQRJ/YbMxpyrvM7fUFz5Y3hJCAYKNPokiGrqu4ffMUcUqoOkLVk1OgKy5aLz57QlUlqirR957FoqfrArmcUy4uX6pGIAaC0b+lJuoieCemqxHLzeiTFD8AKSRifwxIBD59ccqiSdRvLjl9fY5c3o7C+89eV+RXNYtK2ZSIh+7gOPpSzG01k3FjSGE/KVgHojAt+w6v9MuBrHogIg8Jm2t2ova2FLxVIS0ZZUXkR/2SE3Wsc6bXzC3R9CgKrSQa9bSSypjUjoxMNQyDIH1bdCde98un4T2HPJDB8WnoGAxjRoIUncxuEUPp9kwJyLFzfjgz5PGjWPcRiS+KjNLSj90gV0bxjo2aHX297kaxHuOC9RgcIyCz7e43BzMJmTFjxowZv2P4G3/wLz445vBTf+1PfjRa7Q63gJy3SBTyzYLp84M1LlW0USeXkRMbj9J1Ze5ai56uXRDjt9EsnJy/NitddeS0K9J9iKTYkGJD31XE5NAMUgltcYhqmo7NpqJvPbVXvFP6PtB1NaHq0dywvj2jWSx59eIS55XVJuC2gUWTWCx6qqpHVbi9LSNbVaJtK05OOlbrgBPZMxU7xNCZqPzdx4caLOV9ElN5G+FStVvvtHShAhI9zWLNyUnP915U4zZgBGR6LAqskpWLESVqoiouUfa8EtmJxwcdQUfaG7E6ZkWbDtyjjhXmh0nlsJ/E/VJaTmPgqQ88CZ6+DzaGpcpWIhGzAB66EOtypFuJRijENCID+cgoi0nCfDp4X4eQmDo+7Y6l2hv82iGW0a2sd895ivvGmz4PBhL0Ll2Qh3A4rjXY+z46YPCAgEz3eWfbg+3edkzw+QnIodvYjC8XMwmZMWPGjBnfbIgiF1uziG3L11a2vAowC9wh1NCec6OFr4Rko1reRrp01RBfP0NViF2FquP1Zz9i+3nzlOXpGu8zbVtzenFDqNfkVOFEaepE3zticnRdxZuXTwghIwKVN31EykB09L2nbQNNE3lztSSnU9LQ6UgCXmk7R10Lb16fkVXYtt40HVk4Oek4O19xfb2g7XdOWVMMZMDJLnwd7o5sHSMwAyFJWVjWSozC7c0JfO/HqT/rOL14jYiyrJV1K7xaW6GYdIx7QRW87DohAFsSN6Wj0ODGZIjDUaSAGzsR9nHqnel/P4475T2iMi1SB63FTrBehPrl9wbPC1pWKXKaApc+sC2iFqf7OpN1oUjpIGCvlbhXIA9OXqE4ZQ3hi9MOTFceW0zGsqwL4sb9DhkZ07V5W2F9SGKmRfIv/m9/8kFC/5/8y//1R4ePfR4CciwRfuh+6NgN8TR7KSy78bPh3KcNtTtjWUcIzPi5vyUL5PB1X6QDchj6OOPLxUxCZsyYMWPGNxa/+s/9t2Nh9dO/+sc/Yjoz79TGr0p3RGPRgri8b89b7eeH+GrDydkVXbukbxtyCqg6RDKb1Ql973GuBPFtLsy29+YELWNNYNa3zmU2G3OY6nE4P+g2hL4ridHOE6NlfIz2uqWmSVnoujA+X1e74q6ue6qmBXZF/yAIz0oRqw/rYI/JhJQ81DkZXqeFUBh3c1zfVrTdOctFT7tpeH1lFrzeQXBw20NSJRT/X1UjIG0pGBd4ejItZn1rbr/7BzL85hC6kusxxUNWvbttZBx9OnSsmpIVxzAiZP0KQXgmgR/xNa9S5Kq8piVxKzZrdqJhjxRsJZaugYwi6bX0JkE/CBscBNeDPgWMuA3EYRpeOHSIwuScP29H4m3kY4ov2vU4RkAOHx8IZYXHH7iAdcSRiAlCLKvqxe1Z8x4Vxz9yBGv/uB6v67hv2/+fvXd9liVLz/p+a628VNXe+5zu0z0zPZKRrTDGYAjCRGBZEvwD4vLB1owEMWET2Ca4gyRsIQnEpkHI3IUDHGFCvgQQhIHWYAg0Mx8clj/hQEEQRGBky4ExSDPT09N9bvtWlZnr8vrDu1ZWVu3a+9z6Mt2dT0d17arKzMrMqrP3++T7Ps8zd0LeO8wkZMaMGTNmfCjws9/2V3eKrW/733//aXHGAlTc7nLBUCx5m60wVm14E+aox335CBcaoktIvuJeN4GryyUxWqoqcvH4Lu2yJ0WnDlhixgRzZ0VF6AJD0EIrRUNV5fEguT52Uyx2jdGxLdCkc1DiULlEu/A4JyxWHSk6qirhrBCihhNqcCFgt2SjkIopKXkWhGionQrXh8Hm+4p1Z9noJBtD1OLZGAipdAXya7l483msqMXRE/GogLuQgEMowuhDuG00qeCmUS4o4YEJawxBNE3dmQVBZJTCD5lU9Cbm1O/dlPKFVCxyqdSb9fieTiwJxhJb3bo0G97ulbNFa3IogLHcF83MkzDVkvyRN37vMzk2/fE3/uDrp5/9S9e6IYdQ3L32n3saaJdDxnMC+jlbDE0+l+qI5ghYOgZ9bJyOsb1gWvlUh1M6RYe6IJE4kqSbCMjf/Lt/YXbFeg9hfuZnfuaPf9A7MWPGjBkzZrwIfu3f+tHT6j94C1M7Tf9eR+TSMe2cyOMl4e1X6S51HMsPLTFU9F072t/2fUUMlqYNNI1W4E07cHF+TAiOGM1WG2LBHnAzMoaRjABsNrvX+3ycdGUyGbEG7tzpePnVhyxW5zx465t5+OgY7y1db+m8CsvLGJWzE9LxjBDJGSNkd2QDVa6mrYGmklEQ/+AKQh7DGpKMZenOCJYk1hLpSfREKiybfMX7mhuS2flIckL5LqnZOVd7Sdb7hXBPpM2FZBmLanE4LLVsr8JXGGocr8qCjsiGwNqofDzk8asipF5lzYfF0BHwmaD0JmRCpeRqRaMjXUbGeyCPWEW8SVjRbdZ7Bf0+4SgkZP94x/PGbjbIsxKQKW4jItPRMTsZc3se3JRgXufPZzs6p+e0whFJXNERJOLzMF/ZxlToPnXFcsaNNr3HZpW3Geikv3UUq8Ql3oSZgLz3mDshM2bMmDHjQw/3K74GPdAYjDNwr4bFAENCepCzhu4r34Lvl8RYUdc9ddMz9A0pGWJ0xGhJ0eAygVisOoxJeVRLRgICSkCc1YT3fZRl2zbQ99f/zNZ5bEudq7QLUUjL5dldDRM8O6Lr3TgCZs12LAuuE5AyWvUkiIDfW9caCLmmqxx02eI3Jh3DKoGEtckZGsZwGdL2OUEta0shnYnHlIDEbJ/rSbRmSw7SDWXgNAAQbr8Kv1/o9kRWuegvXZmAEAjcN92YWj61CLbZ/QqEKzOMZEQ7OTKSAwt4owyuuHcBoz1wKZrL3ipBmRzXgeOwe/fb57camHeLgAC8/sb3vQ7Xycj0HJduxoviUOp7JNFSsUDJXo1jyITDYjhhyZXpRmJRzsPStCxpsRjOWRMyeWmoc3I71MbRULOhp5P+4D6VDkgxEZjxwWEmITNmzJgx40OP9OYdTBOw7QaCYO46zLJGfK9hhPfv4vslfmhJ0RKD/vkr9wWjXiMvszrWUEJjjnFOENmOY5XuybiuAZdzSIzVbQBYJypG30PtNHQwSRnFGmgXPWeP7rDutgRER7DMTjYHPD3xKJgSpimJsUYF5tPnp8uuKn1vn0pIompDXM4EmRarKXcJpmNWca9bVEaoSrF+SAcyTVPfRyn2x+OajP3AbjG/v40NxeUq4cRiYZuwbvSqfNGZrI3nWIoJ8N4xZCJSoHkiKS+plsStONbGH8wI2T2ebcDi9JiKucBN42YvipvIyBSHHLmm5+KmfdsaDeiyh1LKOwYcjoYqj2ZFGiqGLF2vqRjw434cswT0M1qalkjEYBnEU5sKL4Er6UhG6GTI6x12t5ozQL4xMEv+Z8yYMWPGhx7/+Lv+4utmOSAPLHJeXJVygZS2uhERQ932bK5WnD8+Yb1utaiOlhAsy+XA6qhjdaRdkG5zRLc5wvsqkw7dZEzaFalconLqkLVYeBZLj6sSVRWpqkjbBo6Peuq6aEW22R9JtJtyfLRdx5iEc0lF7JbsilXe87rgvLxmzHbZ6a0so8RBb7BTP28JiOwu65Penw/FBldf61PSgEIJRBHWBDrixKJ213a3EIpSiA/ErBnR8aNSIpZuw23ZILrvpdsgo/i9jGKVoibmba+NP7iNCsMdaXhFlvwSORpHuvQ4tYAeCDwyGyJpFFrXk+UGE+gybQElIg67M2rkxCK5k+JNpOSFFNTYa6Na5fVSxD/JMetF8fob3/d6IST7uI2AlMeHbgVFY7OvLxnE4yXSycC5rDnjigvZ8IhLHsg5nQwMmdIV0rmh54Gcs5aOc7niSjouZc1aNmPXpJeBs3RJLwOxfBMnjLqQj6dxvPrcf/yHnko/M+P5MWtCZsyYMWPGRwb/4T/6nafY6yL2gn/vr/63p8YkHrz9SQ0IjCYH+VmaKvLyKxe4KuAqj4glDDXdZjlqRYbgsEbyiJSM5AJguRzyaJclZTcsgKPjDRdnK4agTllTWANNo2Tl6GiDqwP9puXBg2N6rxqUdW9G+99p1seTHEejbAlGeQxZkL2XGbK/XCE7IcGFz3oJox0Qn7Q0vUhxtLXtiAS2KeSHtCCluA+5KK/FjfdwXf+xcyyTq9n+hqvYpasC7JCFog+xsFMQvywtnzANVxL5RXPJxoSx8D80/tVSEUiTwELtpBxLuyOG7s32qn/ZVp/Hy9wkkb2Mr01F6YWAleK/pJonhB9+4/e8rxqFP/rZv3g67YQ8LxlSG+MwdiQiCX9AfF4MmsNOqnnuLJrrpGGqCznkphWJ17Qgz9IBmTUh7z3mcawZM2bMeBfwzz/7hfGq2a964zfOf7w+IPzstx8mH1Ok5KiqQMx5H7AViPddi6sci6VqQUKoiJNRKmuEwVsdYaplotnQ9UXA2kRKbhzBcjZRN3F00ZrCOsEaIUaL9zUhOKxLLJdBxfPGUAfHZmAchSpdj4L4FGRkiieVkvvdlpWzvDMETpzTTgkqTK+NYSGONZEVjj53PEoQ4c4287uWVHSLGUnHkIMB4bAr0z7qUb+xi+la0+3EPGoFsJKapVQ0WFY4rDF0EsfOTYQdrcB0XGyzN1K0T0CAndTzAiVB192+9ke0pvqJUvwXMvVBja2UztZtuE2vo6oaQ5vLzTX9uN1dt7A0kpBDVrzxBR2zDmEOIvzgMZOQGTNmzHhBTAlIeXwTEflneVnLTFY+CKwvThiGhhAsITgql5QciDpOhWCBiuAbJSl9M45igWo3CkIwBBzOCVhUuJ6JRhmnAnj86JiY7EhKrFFRuqarW/rBUjvh8qqmbRLL5YBziapK9MP1P9P7HZDSxSiCc1um0GT3HrYdEGu0Lpf8ehJo9jhSSOCjkKex+FoYqDFPdT08IQwm6nhSDgUsHYFWHHXuCPQm7hCQcv8kMnITEZm+P6RrLk9r41HrVsOleN5OhmiEhVRA4GVZcGkG1uWc7pXY5Yq7EzsW2HC4izPNAykEw+1tp6w/7YaU/VMtSXxhIfrz4sfe+IHXQTsi0+ef5Jg17TYUojENM5x2QQrxSBwewdt/7mlyTm5yxNofxZoJyAePmYTMmDFjxgvgn+0RkOnzv3qPZEyXTdxOVma8N7AuEqPB+wpJEKLFGlFdh1VrXWsjksepnEvqdhW2BXHJAInJUFWaneGcjATEWsEP2/DCfk/8PiUF5edCSrre0TRWx6/ilsiYwhieAoe2H5OuHpNgjRmTz0XUAaugkJRQ8kjQEazaGB3FIlHlwlnD9szOWFGxzI25XOwJRGOuJX0XTMmGw44FbiERT+qKxCyEP3geKJbCu69vTGAlNQ2Oc5OvzBuTrWMNx9KwkIqHdrOznqB2vlPrX7jeydnu/7bz44t9bx4/S0Z2RrPK9nfe6xukSC5kBOBHPvsXdn7f7es/9kfZDILDUWHpCaQJMdgOnmXdxxPCCJ+FgNz2/GzL+42DmYTMmDFjxnPin37mp0/dLfZEh4jIFDMR+eDgvd0p8kO03DnqaJoBY2V0xzJGsFbYTEapproOZ9Vet3RKYrAlwH3H7ncfReMx3Y41sFx52jZwednuOG+JPDkX5LakdCUg0CWhsVAZc62jcoiAJKCyhoc+TJyOZLxavybSjGNPU9crwQi0phofOyyt6DaLSFvXy8LuLOqeFvLXB9gYXwMdfzJys8z4UEfETajJHWk5Nz0bE8ZcEIATabJrVhzHsYLZ7VREEoMJhCzIv37OtQtigVosfe6W3FRMHxrl+qC6IDfhx9/4Q9f257/67J89VRH4dUczPQchi8R3CcqzEJBnxV/7/J/Z2c+nEZnPBOT9x0xCZsyYMeMFECeV3D4hiSL808/89M4fv/1lEk8mKzPePfx/3//bXwc4/qG/d+qc6juqSjshMRqqxhOGmhgqQrAjwWiqOArLi0Wv3hLWJuo64v31kjkGOwrZbyIItVO73+Uy0DSBqvZ0/YoQ7Uh4jFGSUToYcF0Xcmj7MW1dr8pymyicVIYkek3YRDOOZ02XTaJuYBZYGstG4kRILTS56JwmZKe9EtsIO40Il0v6VhzepJwwbq6NVhUtxH54Huxa9JZuwlSUvo9iG7zVfei414UZqHHckZZ3zFoJRz65l3gqsTv7XtYt2R8BcOO+bM9FQ5XfIXfC8ghawX43pRzLlKiApox/GPDn3vjB1wF+4LP/9Y2Fviah73ZBymf7bhGQ//HzP/7cv0NnAvLBYHbHmjFjxoznwD652IczWuTdNEBwqIPiDPzKvzOTkfcLxz/0904L0QB1qVqtOqxLxFCx2TTjsiIG79XG10dDUyXaJlI3MZOQQN83hOAwRrK2JAvZg8suXLqtmEr4oORltvtUVYnl0nN23hKiHd2xhqCdkyFutRvDDZKIMKnp1BJY32dIKihv7G4XxJhddyxdVnNAhrxuEuEi6fXsi5zjMO16TL/nZYwoIWNI4RQ7TldmsuyeFqQ4aY3nZpKYvR9yOM0aOVS6FwJTyExJ367EciINa+PxJuGz1fB4/vbeZ2ofXIkdt1lhxn1tcxBeQq2DD+lF9ke6tudDOwZ/+I3f/aH9PfD9n/3x00I4DulHyhhWkOvZIdeXzYYPt4xiPS35ONQNmcnHB4uZhMyYMWPGM+JJBORpsU9ESiE4E5H3F/UP/INTY4S2SZycdFS1J/ga7x1dV2OMjlx1faVjXMmwaCOV0w4IgMuC9RDcSEAKSsp6ISIlX6QI1KcJ6tboaNYQLJJDCjeDdl+GcDPxGN9rr+bzcVtEXwUt6ZbOEkVG1yyLjlyFJFSZEQ17bRUvwjpFBjSQb5MdsHZseGFHxzAlGAXXXKvyclMtQekolJEnh2Ug4rK9bhzlzFuUM34bEdHnzU5XpcZpV4aSWSI7XZl9UXo6QKoqsbnTkh9P9nNjDhfat5GQH3zjd30k/v3/vs/8yWu/J9P439ONYD2JhLxI92PGB48PR69vxowZM75B8G4RENgd5bJsg+L23bZmvLeoXOJoFajrSLvocS7hqoC1CWcTKRVXKxkDBq3J1rr7hGPyOOS8kP1skIKyrRJgKHlky0fD0TJg7Hb0yllhUWsXZFHpbZ9wlMdFz+EzyxjDC/NyQ5KdTkjKz5X7IQlehD4lvMh4K8uayVgUHNbL30RAprhJdF6IwnSZUuRPi9HDHY/bC5tRSzLRo/g8YrXv5HRt3QNictDzMd23wG5a+LPgJnLyYcRf+akfvUYQyjl62hT425abCciHH7MmZMaMGTOeE/tXXQ/Npe+PjOwjilzriBijRGT6tDOGX/63f8P8R/c9gBb7QtNodoiIIUXHZtNQ15F+cHjv6Acdq3J1Gt20rE3YnDEScpBh6Xxo/od2N0JQklFGsMoyxshIRLw31BpbQtc7zSBJsGqFEA3rHvp8YT1J1nDkL1gpd+OEiCRU41E6HF6EJrtcYcxILHR7wjD5Luq4jLpfhbzcOqecF6hTVrE73e2C7OsbDo3l6PjVtig91OTZJx0bE0Z9RdwhOGXU6snYhu8pglFheY0bheyH9qWkn+9jf9yqFpeJyfWr+AZzK9H4/p/6HR+pf+N/5ad+9PX9jkghmJWptuYEN+SATPU/Mz56mDshM2bMmPGU+Cef+enThBYv05Jq+nj/5+f9JXvIdOvnv/eLc4fkXYb/PV88dVaF4c4lhr4hRTc6U3mfc0BQwlDseYtew1pdr4xhJdFk8a14fGvnm4QdLUhBEbr7qF2TMoaVxNB5Q+91FCtlUbqzu92HfQISROjTdtyqjFrVmXgoOSnFn+gNLfi8JLykkXiEcbb/cNcjkMYbFA2FjJoRXXeXgBTtBxx2hNoe13ZbBa248d+VwWgHI9+e9t/aVsciuRMSx+fKfpVwvbLN0cnJyFNdxfekg/sjaFp8uY3Lm3gtzPCjgkMdkYKSh+LMdWJ26LkZHy3MJGTGjBkz3gOUcmLnavMNy0bZvTK7b51anjv0/IznR/y9XxgJSFVFjEmIKPEoKembThPTTe5wSNZ0DF5HrYoWxDkd20rJ7HxWVZVGQXrttONR3LBKF8RZyeNYORHdalckJWgq2AyGy06JyJ2F5nq47JRViMZoqTvRehSEib6jyuzIZ/JR4EW7EkPRaSB4JHc/ZEKsdZmbvooWDf/b13kwrr+9TTsZU/JSCMJN76Lajd31ptt+WkzXnepR4oRUjcuarR7kacasyj4upRqv/JdtTq/sFyLytONJH3a4G02Xr+N5xtlmfLgwC9NnzJgx4ynxTz7z06fxBiZwU17IIQvf267+1JPtTDd5k1j9X/4W7Y4cNQZn4BN/7bs+UuMc7weG3/3F01df7mnbgDFC1+lMlHPC2UVDP1iq3A2pqsSyjbnbISwWfjLCZVQHIiYLzyXb7F7/xKe675h03ctOl6sddH6arg7rQZetchfER+iijKQiTL5nozZkjwDHrO2IyDZNfWekSYmHRXUN/oaSfp2zNPaD6spzsQTz7Q01xdxJuHYuJkTgSQRkuq2w1zmoxI4F/5OusO7rPuxETO4novubnLEKbhoV2u6HGcXuQhoHxups7lvGs6JJ/Jdv/M6P9L/d6VhWOdcOO54HT9ixHCgjWvsjWbMW5KODWRMyY8aMGe8Tiv7jaca0EuxcM/y57/nC6ZSIlNEsZ7bbepJz0ozDqJxwcVkjYlguB5wTus7hvXYkSiAhaBdDR7CEtvW585ELcDHjuJbNxXaSw+S02PL6qNa/ItBn4tH7iSC9EsDgbLbbndS8ItCL4Mx+ZshuYVwIiIrKGTMzNF5u25EYNRK3zOELjMno0QhOtuQDGAnIFDEXmTcRkH1XrKe5Al4crqbLlvyOssQ+psTjUOdhKiYvBGR6Hg7tV7EMnnZOqklhPS2gp4qV8jp8tByxbkMZy/p9n/mTp9YYopBzWQwVFTUOL5EBjyA444hSCOHTJabP+HBhJiEzZsyY8S7gkMC8wHDYQegm3DRS8nPf84XTk9qyDrq1Rfb0XVZwZykYA5e/40unxz85d0OeBUMw3LsbcDbRdTXD4Nj0jhANlVPCsVpsGV5dR+o64lxCsltSCHYMF2yb7SdojdDltPUqj27Zva9JVelo1itOeHDh6ALUFqrMQsuo1RA186OehHoUV7WRQKRp0byF3yMmkd0xJplc9S9J3/rzLkqxbTFYMXiTRiJyExxmdMs6/LrNqoynIyD7mOaVjNxC9H33lwPtQnjiDikZg/P23v82zcpNKOeo2P922aZ32gkJWdRvDpChjzrqPa1HRUVLRU8g4mmox++CMWYnT2Tugny0MJOQGTNmzHhK3DSKNX39EBGRvWWAMZl5CoMWi6NDUV5xvCpuNLCutYbGMV4dr/PfdGdhtYh0v/uLp8tFJP7Eb37hP9i//lt/+lS6S4gBjEU255i6BVfr4/P7auEE/B/9930oC4SmElI01KtICI6qSrhgaerEchHG1HTnEjFa2jbQNAPe1wxDTSxjWNHQ1GkUsKs9rwrYqypdIx9QdCKqCbFOePWukp23Hzsue2iCGT9fn4Q2E5B+kv8hot+bMsp3E/mYCtJLN6TPxf+UdExJ87RjMv0e74xx7RXqU6tbz36U4fXlpqnohThsuyNblO5BuXdi6UxHg47PTbsbyci4w9NtFsJTX9MmbMeDzOTo484Sux9g2f+biIRHxeZupwNSjnVLQD4OXZApfuKNH3n9+z/746eFjLSTUrQ853AMkjPpjYBEfvLzf+pjdZ4+DpiF6TNmzJjxHuJpr3MaVDRcZwvVMj4DWzKycIYq/9ZOoqNYLrslOatjReVqe4oG+33Pn2ny6z79t05/3af/1qn4HobN9oWUEN8j3SV0V9vnQ893yp86/c7hdHzP73jw/aff8egHvuEdvcoYVErqTDV4i/caEJiSitJXRx2rVcdL985pmgHrVMhurWo/mipqEGGw+CxaL6NYxV0r5ryRMtpl3TYjJEXD5dpxtbGsO0tTwbKGPioJrSysKsM6COsgo/XukLYC8+J8VYjHPmneGiDoVWaHocXmuXyt2QtlmIrEYZeAlCv9h9LQ9x9brncTDgnCp7gpCySZrUNV+W864nRtX8yuBmWab5I4nPmxj1ostVhacdRiWUnNSuqnWlfyeNe+NkaPy4zH8HHET7zxI68DHLHAYalwrGioMiExWI7NkpD1Rz/5+R+bCchHEDMJmTFjxoynxLd9XjsL6ZZbIRAFhzoeZRsFhYCULIf9NcomGqddj8blK9Zm2wUpwmmAutIr8ccr/1zH+eu+5X85fN5ztwAAIABJREFUNc1SOxy+145HdwmhB2vz8wMSvf5srXZGAOqG73R/5vQ71j98ijXQD8+1D+8nkkA/WLx3DMERoqXzKi5v24C1wur4kpc++SbLowte/Tf+BauTc6oqC2fFjOsNwY4akQJjhNqpC1ZBVaXRFQuUiHRe09HXvTpllc96CJlk5r/YIck4djV21vaOqUtbiW8hJ4ecncoybtKF8GzzOw6VyDeNTO0/f4hgHAovPERk9tccsz3M9j4Z2eloPI3D1G4HJ49NiR2F6VO4TNAcliaPDDksrVTU2FvfrxzB9SPZ7mvg+QINPyooRMTlc2kwHNFiMLR5WG3GRxvzONaMGTNmPAOe1gL0No1IeR2j13EXzvLq0vDWVSlctssUEuPGe2hyve9yzVQ54fgo4L3l7p2et++vaOrnyxz49d/606cSAeugbpWESMK+/Gkk9PrYZ2IRO4hePWXrVke2ANlcIm89gHVCvOHbfu4PnMpQQbT849/0518H+FX//X9zmmJFCDXDZkHd9vyL3//BBLUtaqH3lhMYiUKIqu2wVlgse5p2g6s6quYKSY7FnXcAGIZGjzkHEYoYhuyGlUTtfKe5INYJdZ3w3hKToXbCECxVJSxq4WxjRg0IwPkgvLo0hKhdkZCTzS2MXY8gwtLasQOyTpHW2Jz/od2W/WLXj52IXWJSY4n5yn3cW2faAYEnW6iOBEfsWPBPxeRlNGp/W248uolo/jm0GVMUglEK23LvxJJgDEAs+3Lt2EdnLNWd2DwO5lAyU3Qmh3CIiPh8hf/j7iXxY2/8wOs/9tm/fFpG6iKJE5asGfhzb/zg3P34iGPuhMyYMWPGewyDWu+a/PMhavLWVRrdi6brGaMEpM4vGKMJ2stGhehNJZysNMG7bSJXay2KnRWG4dnCvn79L/tfx9EpWZ+DxG3no6oxzWp3hZRLqBh0ZCsva1yFubPEfHKFaQUpFrUu8Wt/6kdOf83f+PHTur3CmERVeZbHl9TNwL/zl3/yAxndGoLBR/De4mziaBmonHYpSp5HDHpeh81LupJNpFjRNMNo7VsyR0IwIwGBbT5IQQk1LMsVknLZqc3ysoZVA20Fx7W6CBkD6yA01rDI4pIu7ZLWNo/ytWYyorRHQEr+x36RPYXLV+mnIYTlvgjMn2yh+/Qk+NAV75Ijsj9S9aydAyUKh0sd7YAwWhKn0dFKz0Fx9AomUeXtlPGqhNDiqA9s+6bux4zD+KNv/P7XQYmg+5iK9T+umEnIjBkzZjwDvv3zv/n1b//8kwXfhl0Resr35WYoqc8wpG3ZYidkpd5TMhf9R1MJlYNlIxyv4s6V9hBUBF3XCeue8epx9FC3mGYJkiCEUXQu3SVy9VhF6dbqaFYM2hUp980Cqko7I4sl+AArh1l4zMKDTZhlT3Vyhju6YHnnbVZ336JdnbO5Oubi7A7HP/T33ncisvyrv+F1yMnlWYQeohm7In2nBKS7fIUwLDDNdsQsJkvfV1izHa2q6zQSENhmgiRRrU7RgJT3BCUixwsZ9T0xQR/g0gv3N4lHnXARIkPWgUzL3ARsUuIiRTpJO8/v54CAjlsdutpfnotoKnghB2VsaGpfWzDNB9nd1nXsa0FuIypF+7GPfVveMqa1T2SKluMQSZhiSlDK3lR5NMvCqP3wJh4IXswdHbE72g4zl1bPjB9843e9/gd/Sjuhf/yNP/j63AX5eGAex5oxY8aM58C3f/43v/6Pvvsf3FgwT8snh45dTEuThbP4JCycZUi7xVY1GeMqV71rZ3j1JFE54eTYc7WpaJvE8VFPCA4Rw2LhOTLCo8dLTQF/VhICSkRCHrECsFa1H4/f1sd+0A5IyLaZk6vuxACLI+iu1E2prTF1hfsVQO+Riw1y7qBNmGOQS0hv3qG6Euq25/juQ+72S67+9N88/coPfe59LUKaCu6f1VyuK3xUHUY/WJytcFWiW6+wLtG0Hf7yJR6982lEDFUVuXP3ihgt1q1GYfoQLLUT1p2jcmrLm0Rv0++B9zqKBdDW28+rrSGutTNyPwQkKjG1YtRNi+3IX8yPa2NwxtDLtkx3k6J9WkLX2bnKk6gmBXz5aUouErJjsVtcpCyGnrhT6N9ELJxYuCFFvbxf2inkbze23nemSpk4bfejdDXSjURk6r81jrflx4PxNFJNlt0dW/OT9fRcGmrcTtbIbShEZe6a7OLj5hT2ccdM12fMmDHjXUAZ6zj4WtZ+lBKpsVosHlVP/hXsJ/VM5w3GqotTUyWOVgOrVcdi4WkaTe42hp2cimdC8Ei/Ue0HaLcjJe1+GKtuWN0VDJ0Sjhh21+/WW8csSbBaQV1h7r0Cr7yE+ZZXsb90gXnZalvHW6giVJG67nG1ai+qKvLaj/3t97UjctmpFmMIhiFoOOCmc1xtamIeJyuC8/OHn6LbtHSblqFv8lhZwOWUdJdF6En0sygNLWf1+RC2xXJdJ7pe80VC3D5/sTGEBF2SawMqEfC54zE6Y5HoJdFJGnUehWTsw2LG1HMVSN+ez2ExVBP9RrHe7bMF7RTFoGEf3sR3TYRdtBc3aTBg66QF14lRjcPlUawpxtBPE0gIQ873qMXRSrXTHRrfp2xTnN6yUH6/GxL29mFq1TtjxscV87d/xowZM54TltvJB2gXZGHN6IBVZvqPKsOqun32uTbbEqWIlbtenylkxNWBGLep3c6lcTzr/Mf/o2e6qigycboaNjqKFXqkn1j0NguoG12ukJCh05ux2iEJmiliqgazWCJnj+H8AtO04EoCn2AWEXtvjTna0CwvqJorJSLLjuWq49/683/9fR/N8rmuNQbqWmibOGkKqS3v1eWSGC0xWrqupu8WeF9jbdppDIGOW607R5IsXrf6XBnDSlH1KCGacWxrCIZNgAsveqXdGBbGEhH8xH1taqPbkxgy6SgjVUVX4Q8U/6UTEjNZOUQPSuGcEIwcJhdO7I1kZ+f95Lo+6bqd7yFtyC5B2loG797vL+vkZm3BYdeu7fGVbQQSazOwNgPJyKgJAa51V2p09Gsh1wdM9nU1BXMXZMbHHfM41owZM2a8ByhuVlXWg9RWCUhlDHcXcHeViMkwPDY7KdcFxsAikxSD2rWKqG3smLxtr6/31R/+rS82ztBdjYJzyc5YoxuWq7di9DyOJeePwKsVsLnzEsSAeL8lKS7/mRkC6c23MKUtkARaMEeCO7nELAfoa+p+Q+tbjEkE3/DK6U+dPnj9M+/LiIY1Ooalgn/w3sBC9RopOlwVSEmLaRFD1+uxOZeoRImg93bMB7EGYjTUtdrxlrErIIveofeW3qv71RD0fY2B1kHjDF0wmAAPQ8BhOJOt7XKVuxgVJhMBaHF6FZ+S2m1yDoiWvH1WgjS5iC5lepgQiQqrj/e6HGXb0xGuMqqVENYMrKTZWSfujXEVTPUlN+lKnFiSiTuvle5H4Hrg4D7RKTSh6D4iCZuPQc+NG8fAynbVKWtbGvW5K9LhCcjEt0sh+XyM+4BlJQ1rM2CwE6veMj62HcMqMuwZMz6umEnIjBkzZryHSKitamUNrTOsajhZCos20Q+WxhnW8Xo6OmwLRJenl9paOFoG6kYLpndOP/v6O+/mzvYbiF7JR0ra3Siko271ubpSm97VMcSAcZUGGG6ukG6Daduto5arYL3W17MgQpJAFK3Q+63lqVlE7RQ0gerRBmOVhADc+2OfP334J777PSUiIjCUQ80ajrpWO926DhpOaIV+syQER9dX2U0rXzUPVtPWXWIIjpgMPhp8MKwWKY9oJZIUW96tOD2J8jhn1WxgM6gjVm01J+TBsHW8WkskIqN7E2jaeY1lyHoEi6HB0hHxQMgFdotD0NGtorgYXa8mhfRt+RU2l9Fh8nrReyyl3lk2PqWT1r6e46auyoCnocYTDxbv20L/cGFfyEi590TuSktnIi0VGwI+k55a3M4I2SFdTRHPW9H31G6IG0fXevbGFVFr3jLYldCckz/72f/udNZCzPg4Yh7HmjFjxoznxG05IPsomhBnYbVQR6vjo8Arx6L2vSaPAOXUdIA7Cy1ClYDodmIyOvbzgrkJNyIHERKDjl3V7fa1qlLnrIJmCasT/dlV4D3S9zp2ZS30vTpkASaHmpgqM6ooUJOr/fynyAgYoTo5w5hIu9ggYgjB8tIf+bvv6WhWGXdr8qU5Z4Wm0kR0EYOrApIMKerVdu8t/WBZLnPeQyzZIEounBXWvcVH6HrH4C3rriKV1HSXCHkUy1lY1HDU6mfa+e3+lPv979p0BKrcly7FdGToKg9jCdoJCHtko8DudTbiLd+vmyxvDQZvIo/NhsdmQ4cfC/Fns+yd/jwVzWfL49xDgK0Wo7z/lIDsv+MO0clk48IMbAhsDhCGaWfl0DFPgxNh6/i1IYzHXc5x2S9BCGzjGHsCH9fU9BkzZhIyY8aMGS8Alx2JDsGiIvTKqsvRq8fCveOoHZA6ceekwxpYVmYkH2UEqzaGygmvvRRZNnB3FVm0kdUiF/XvAQmRzblu++iukpGcmG6OXlKS4Spk2CgxcbUSjbqBdoE5OoHKkf7VQ9L/+1XS//UIeedCK+y2oaizZRPAJ2VkPVmgnpDBghXwFZIs1ulxfuqbvs5r3/wOn3jtwXtKRGI+nctGePkkcu+O586djpdfOePlVx8SQ0XwDd7XnF00hKDEsaq2FskhWKwRqmpS6EfD2dqx6TRRfd1VufOhYYVnGx3FOut0+bO1YYgwRBk7M9M/1EV7MHW3EmCdS1tQUmIxrHB8kiUJ4dJ4NkTWJjBMzHnXRnsapUC/jXyUsaVIuiZIL1a2FsMJLSe0NFTUOAYCfc7WOJTavj02HU+aFvxO7A4Z8MRrRbsgBzUn+2SmpJ9PBeaFHHgTR2JSjs+beG3ka/987OpVdt9ver+/vzr+ppYA/gABmjHj44CZhMyYMWPGc+ImUfqUlFigtYbaGRZNGe/RJO6qCrR1onbbTBCDakGWlbo0hWg0mNDCso246umvKD8rzPKOkg3rNO8DoF2CtZh2uSUmAG0WqIegHZGqwtx5GZJBzh1y1UCA3ApAiuLbZsFD63QEzSewBtPquBOtx56sqV/7Oqu7b1HVPe3iisXqEoDVH/777zoRees//dKpy52ozWA4WgYWuVvlKk/TbvC+JgRHCDqCtclE8vKyoe9qRAwpqfDcWhlzQlSQrh0S73PuRtzqRo5zo8lHHcMC6KJeJz8fhCHKjkHBaFSwY2fLSCQKAVGr2C3CpFy2OYxwf+xpfyxqH9PXCunYx1RfUdBQMZjAlRkOrlPWe5puyVRjYTHs54kc+jdZbHMjiVrUGWu6/9We61UhF6WzcdvoV1m+/NcTd8a47KQDMmPGjF3MJGTGjBkznhPbTIVtR6QQkPLLtbaGxuUL/96w6RwpGobBMQw1n37tcidNuyxbOwgRNsP217QSmEhVJZ5hEuzpUTocvlenK1dhqu04llkcYY7vKvkAJSSbi/xivhL+776K/WYLdb66a7W6N22FaStwBlM71YfcXyFrg1zmN6gEYwVJBgkOc7yhPXlAe/SQZnnG8cmG46P+XQ80rGwehXPw6t1AVSWaxrM66rA2MfRLYqgY+hrvK7peCdTVxvHooqbPyfQ2J6a7ST7Lolar3s1gsHt/cetKWNSCT9qJqZzuRxTYRC2EN5mQJFBnrIlGoYxZlXdLk1I3AQMRC6yyCmEo1rZGS+QiKIddJ6xnRemKRJNGm1pPZJhc4S9diIE47sc+pu5T01NVCnzQkazSKUkcDjTcX3+6/SnUPSxd05BE0s6+7wcrHgpSDCaNz01Jy74j1qFjmjHj44pZmD5jxowZzwlrNDgOGIc24uTnymjeRCEZzuoE02awatUarRa8WQhdRcPxQui9rleIhronWdraUdeBt/7oCzpg3YacEWJcFqHkyll8j3E1cnUG6wvVghiLPLwP9Rn0QyYcFqxB7p8gzWNkE7CvOSQmiAmzapDzHrk0SLCYBESLXLQ6jgXgJ3+aqghJHaJe/aYvE33L4wefYPUn/s7p23/se174PHz5c186vbuEk6UWiy/d7Tg+WWNdRJKh36gGpl30u6cpWIbB8bWHNc1G97f3jrbWArtyiUVjOF9bshxGtfpOHbNS0tyXplLimYDLHkLSAvosBlbWjQTXAo0x9GLGHseUMJRit5oU8oW8OAwvS8sD043LX5mty9bGPN040LTDUcr5UmIXImIBxILZFv1rPMfSqIOW8aykpiMQRtKyda8qBGYKDVxUy901/agJSci1jkziMAHZH6saiRM59HHixOWysLwI+ve3l8x2DGsqhldStB0XO2TBO81agduzTmbM+Khj7oTMmDFjxgvAGrN7I4/GGNWCFKjFblkHmipxcrJWC1e3XWbZCJWDVSs6hmWEo0WirfUKexFAvyfoLpVENEskekQmBVJKSkD6DbLOoYVXZ8qwNh30Xn+OuXB77THxK/eQx0vERxWmF0bmgSrrPx6uVA+SycaNaLRQ7tbHxFDRNP7mZZ8Sv/i5L53WTrsVbRO591Kfc1YSflASZqzgqjDmghiTWK067txZs2gDTSWcrS1ff9TQVImud/Re9R8pkW149T2sVQICZJcsWPdGnbkyU10HUc2+saxT5DJNCn9jdsawCtxe4nlxwNIRrSfjNp3GFIf0EZbtaNY+ISgdgTZ7eQGssoNWMnJQ31GISNm27t8WK9pxpGp/TOq2zJ6BQER1HvuuXcGoBuSmUbSbAhin+34ot2T/tWk3pXRXynOnn/1L76nxwowZ34iYSciMGTNmPCdu+gW6X8o4qyM/daVdDtDRqiSGug5UTnBGNdqgBauzQuWUiJTxqxDe41/ZktQVK2jXAz8gvlfL3u5K9SKSMKsj6LttfoiPiI/IZaduWG2NeQ3ca48Ib7+KvGVJb0XoYs4HMRgH9lPnpEcnyMUSgoMqIoNDukZv64VqTIYG6Vok5Svm0fBP/+9778ohOwOVE46PPC/fO8N7x8X5MQDdZqmOWDkbpKqUCKXkSMnhqjQSSADrVPPjveo/fDQ8uBpP0Qg/SUwfAry9SdypLSFBbfTK/PST9rK9pt5mUXqLw2FwmJ2y12d3Jth2R0qCejXpOMSJJqT8XMTjt56vG771tdgxO8Tmx/vrTO8LGSli8K2O5bDQex9PO8o0Hd0qBf+h8MN9InbTccpkuf0xrrJPU/erp92/GTM+jpi//TNmzJjxHPjnn/3CKeymppd069IRsajgGLTL0Xu96m0MrFY9TTuwOrnieJFYtUJbQ1UJd489dZ1YLYLqQJxeqT86Gnjp3jn//l//0+/dVdOS+zFsNDU9Bjh/oK9dPIIQkPUVcv8hcv+h5oZYozefkD4g968wzmI+FWl+zS9w+c9/Jet/9iuIP/8KcuWRS5C1Qx4vSb4l3H8VAHl0DJusQbECVST1S0QcMSyomjVN23H33hkvHQnv/LYvvdB5qCxsAjy4sHz160u+8pVXuP9wyb/+6hG/8Iv3aBc9xsrYFQmhyjdL31d4rwwkRO12fO1+w6bTjJDLzvLg0rDMuTBNte2EQc4lyWRkSIJPgo+C36utPcI6RdYp0qWEw1BPnJeEw4F/xTmrI6pYelLg6ygT43LTcjmSuDQDfR4T0nXjODZkMddDArNOw0wcqOCweH1adLd5IrzsW8jH4W+RqJfuSTDXs0zSgVshWg5Lk9+vBBTuZpPsHmc5Lth1zgrmyeRCUCvkQ3qQNBJCxxELFjQ02RdsxoyPG+Zv/YwZM2Y8I37+e794epsw3LLVc9jJgiqLUCLifUW7WHN0575OQFWaTRGjoapkJB6atJ2oKsHm0aAwLPnV/9NfeNeJyD988Ntf/4f3f9vr0l0pAek7uDpT0vH4wTY/JG5F5/K1c6TzyKOkeR9RIKBuWPnY68UVfXeEP38ZikFSArlakGKN74+0G1JFJFnC+o52RoIjxZoUlQTEsKBeXFHXPXeOPG3NCxGRb/ob3/X6t/zN73rdJ7jsDF9/VCNZRN57w9A3SDLEUBFDRbdp6btmHImzRrjqVb/TVKrpKK5XRWAOjN0vEcYRLZPDJ+9vhHVKPM6tkjKW5WVKJvTn5sCXburClBCikZ2k85JCUYrxYk9b3/DnfyQnJo5EpKAWSy3bkL1SOO+PMcXxva6nmBeUbkgju9LUmzox0+OcEoFnEXdvU9EzGZvY/06JVSFw03MG265Ned/y3ht2Xb+m5EMQTO7yTFHcvXo8nnCrK9mMGR9VzCRkxowZM54BP/+9XzyFZwsqBB35CdEQxyJU2FwdI2JHMXNTQVUlnFUb33bhaRu1803R0LYDMVRcnb/E+YNPvPsHV9BdqfVusXPKAYbSbZCLi3HkSjUeIOd5vSGBF7AgZ4KcaZFWNTqTFIaldgOKerjS4zYm4c9fRrpGx6+SJXTH25+HJaYIiX2LqwcWi8jxItHW8Og/e7GOSEjQheJGpiRxIuehXXSs19qh6buaGCwxqM2uNbru1aB6jt4brnrDZlDS+bjXkTpQIXrntxsWUbLhjGHIpCOKkhLQLsiULBRCO9WF7LyOwUkpmBPeJNaEnQR0J7rMtCiusSypaFGR+DiuZa6LpksHoYxb3Zajoft0OCtjvyjfD14s2Ndd9Pgd29syElVjEeTgPt+EaTr69Fj2CVra24dDxOcmF6yyThHSl+Ne0OScENkxE5gx4+OE2R1rxowZM54B0/KksoaYi8fpqM2Un7R5WilEw8brCFDvDX1f0XcLrs5eISYNJmzqxNHSq9bARk7uXuKc0PcVIoZhqFmvF9R1YLncdWt6N/Cd/Y+eSgw6kmUsMvSaen61UYY0BCRorgd9VIE5IFcV5iggm9wFqPM5OW8IX/0EYVjhey3ij3ot9KRzmJMNNfdJ73yafn2Hfn2H5YmOfoVhubNvYVji6h5X63F/8tNvc3xyzNnjI37+ywtefoHjvvLCUW3YBNjkEalVhIuLBRcXC1IyXK0rFq2jbSJD2Kamv3Zv4OF5zTuXSl6+diF88ki3lUQ4qlQqLqKmA+veIrK17X25MSyzWP2rg5alVS6tpwW5B3wSVsbRS9F1CAZ4lZb79KTc5SjhdxaDN4laVEPiJU2cqLbjSE60i+LQUcKV1KyNH0XihTOOAXyZ6LS5ePdPGFGaOk+Nz2WyM5gw7u+JLHZGqEDHn6pMeErHoRTyZmecKo1uWwXjOFUeznLYvXErx9oMW/IhbhR0bfUjt6Psu7CbuzIVo6fJz+P70o/L1Dj+2Bt/4L1zvJsx4xsUcydkxowZM54S//K3fPF0KiCHbUekjNgUAuKMobaG2urySbZWvTHB4C2L1Ya60cLKOXXCqpvIYuGxNmGdFkyLhccYIeTit10MeF+/q8f2nf2Pnor3ECJyeYmcP1YC0vVKPGJCygH0ul/FPMs0CbnK17QSSG9U02GEGBZcnb9CjJZ2cYUMascrj4+RroY6YCYFand5D2MT1unYWQwtKVbIRF8QfUtVeZp2oG0D1sCb/8nzd0OMUSIC+tmAEoSLy5qvP2h5eNbQe0vXO84umnG9wVu+er/h4VXuUIh2VYYAm6DbO2pU+1FuXSZupSvmrP4hFtH6NyL0JIbJKFCcdES8KPkIE4H0NpvG7FyR3w/Mm17hb8Vh9COiZus0pY8dC6mvicSnSeO34Vn0DSeiEnuH48J0rM3AsGcZXMavBKHl5u/9/uiX3zkX17ssu8uq7mNfF7Lf9dgXozscVSY3+92QQkB2Qh6zdmZKTjpe3OltxowPI+ZOyIwZM2Y8JXZyQbIX6qGyxhlDXa6oii570WsXRASaSnSMZ73EDzUxGWLUzAhjhHbRQbfgzitfJoZv5dGDu6RkSNFgnRB8jbXvTr7Ad6x/+JS+h6MTJR2VGxPOzSqLxIeElPdbJyUaUYmG9AY5W2i4YHW13XAetWq++U3C2980OkvRaxEpwcGwwlSR+vgxQ3fM0C+wNlK3lhT1z9P0PnrdHxGLdQFrI31fUVkt/l8EUYTzQTiqLH0UKmt4cOHwUU0FnIXlQmiqxGUmXOvesvH6GfdRRgL6uFe3s2WtRKPzZMJhxswQ0OaSNaoTCgkWxnJstJC+TFqmOgw9gidxx1RsZGvrGtHwwscyUGM5xnLOfvG96/ok6OhQjSVlbYjF0OJIaOK3EzN2BKYjS4E0Omw96/hQ0axMUbohC1F61BvLFR2CBh7ui8AjiR7PkuYaGfAmjiRk52cSlu0I2zSno5CN/fvp+T0UhljeW0aC4bIjloxEpIj0IV3LAgnEiW7m9nG2GTM+yphJyIwZM2Y8AV/+3JdOhSyByPVE7Qw16n5lMaOQeB9d0DC6mPQetOjsB8v5+ZLlciAEDS0EJSEF/dU9NlcrJR9WVIOA8OaP/JZ3ZXTjO9Y/fEoS6AfSwzcxjf5JkCySlkdrzKoGA/IgYWpBOgcuYVpRh6vBIlcLhsev0PAW2IS9u4HeYaqEZHV2t1myOjlHNo2uv+yRsyMEkEG7C9ZGYqg4f/gpXBWwLtIurohBXxexxFBzeX5Hz2OoSLmATwL/6rd+6fRb/+fveqFzcxEStTFsvLDJF6iXtbpchWC5f1YxZD61CToiFQXaPJpXhOUWuPJbEto6Qwpwd6F2vU2lpGXdb4vpl2pHZQzrmDjGYZPhpcpxHi2PxHMmukNhr5g/z6kgYpRgTAv+UWRtEkuptqnppBvF6WsTtJDPwYE1jmgSl3iO5XAnYt+dqxCM25YpWFHjiaxEtSm9iZyZNQAN9TgudcSCbk8EDluiNLX7He1y8zjXSGjE4nJ6fHMLAYjZgWuKaVdkSoKmnY6BMLpwHTpWHaOb7Dtxdsaa8bHFTEJmzJgx4wlwVkmEoOQDcsq1ABQbJENCr6iXLkgSJShlxCflgrTzcLQwauV61ebcCEuKwtd/9HvHIvre//ATp2kvwO/hn/jud3d2PCUVoCf0snwaGdkUAAAgAElEQVQSWCfMyw1iI7L2EEE2FfK4xpz0+vNGx7CMFbi7pg4O/84ncVWHWQ5gRUnIkG1sg2XolyyGWhPQF4N2S4LDTLo6rgr4viUMNa42xNBgXSCGmhhqri5OePDgBO8tlUs8PNeieIh6vn/ue75w+iv/zm98qnO0+Z1fPO284dWlwVkVmX/5XMdkNlE7Wo2FjdfP164tTSVcZuJQCIgIXIVEZQ0nleUiqO1tSPp9EFMiBGGTU9KBkczcWaiw3SUzWjo3Vr8TD0NgyIShI47lbosbOyHjR7knci4koDdxq+coyd65u1Ln5aelcimJa3HjiFKNI5nIJVwjIrXYURey3/F42gZVITwmi94XRhPWh+wetaLdKdZ3ROl5P8tzCZ1tGzNAJsQ+mETI+xkljdssdsF91ng4DE7ceFxbUheoqcYzVh5X2NxhcuN+mB1y4mluGCW72ZB4xoyPNmYSMmPGjBlPAZdHfkpZUVmt2a+GTEyiaJ1pzGjNWrQj01GhKLCqs11rFiQPwZAkj/1M3vP//M+//z0Tq37ncHoqIULXKRGptRiTmGBllXzUlnLZ1rQR6WvthPj8p6PqoU6YasCGKy7f/Le5++qXlXxsGiQF5OyYYVBb2xiqMXxQT4wjxQZjItZGmjYiYknJ6Zha3+CqAKEmhkp/znh86aidYwgGH3eNAX7ue75wWhvDL/vbv+Hg+fvy5750eu9IGIKaBWy8cNQY2gqWTj+/QbRQH5ISkaL1aCpYVKojaZxhiHAV9LPfxERIhnVKvFQ5omi3IyGEZHBZe7KsDbXLzmHZNe2ogbMud9ickpHSVUkwBgnaSWm7wNERSeUZySRgb4QokrB5ZKjY+BYxus8jVsPk+rzDUl/LK4k3OmHtX+0/NHq1j/308P1uQCEl+vO2VFmw1eQIQszC+yk0UPD6/k2Jy/7+9YSRiEz3JQlEk2jF0ZuIG0evpqYB+r0sRGSYWO7uEwzNQwlUk8ySGTM+rphJyIwZM2Y8JQrxKOSicrCsYeOLfaqMOpHaMBaT1hiCCFUyWKcFc0yGIVgNKc/5IcD7NyFeVXoAGcZZSIJxFklxVNPLucOsJkLdTYOpEunRMeZiCXVANkWrYbh8/Bp3+wfI1UJF54uBlHScTPIxmmbQHBBAkiNGtd3VxxZJK2xWTV+dn+CqsNWUAJuN4/HaUFk4apUgCjoq54whJL0S/ouf+9KpAY6ztOWyZyw9H2Qxeek8lETzo0aJiTPwOCR0DZvvDX5teHklrFpNvP/yQ6udL2uoMLTWcDkkjirDVdDvQZdk6z4F9EE/76I1KXr/8v3KnwhHYke7XtCr84U09ETuUtNi6UlcZHHztcwOk2BSpBeheyxEZY8rxAmBGcecsq5i7BaYODpjvQh2Oya7JIgsmMfAFR0VS0wWxp+x5i6rsesxLfP3heRT4vEkOLG0psKJ1fMGtEAv4VpmCDj6fM6LTmabUK8y/v3PYsjLN9R4Ai7rcGYiMuPjipmEzJgxY8YTUKaUYNcZa9kIi1oJyNUAoPoEn5R4bMew1KJV0GI3idb/1hjON5Y+KJm5QVbynkDuv4NcdpjaadejGPSsrD534ZGNQTYtskEJxUnP8PPfquunPMbyS76CbFr6s09weX7E8R0YvvzNVItL0sUJm4tXaNqBriShA6lf0l3eQ8TgKn3joV8iySBiWR5d4n1LDBXWRUKo6DZLFqgOpG0SbSZQX7+QHcOA2mgeYhTN7XBG3bpeWsKrx4KPhrONdjZC/lwAuijUzvCpu4mLjQUM9spxFRKN1QiUTxxvz1/vLXUlvHIkXHTaEQHtlLXG8FYXuFs5lrXh5Uq/H5de6JJwtRGSCK21vHZs6DO/Oh+EVaXkinwMK2tpxFCbmkGEr8pmLHY9wrFxrLL9rs9lvRODQ8Xl5Up+L7vuWGNwn7E04sar+KU74k0cLXpLqnpCWGBpxT2x2xFJ3DRkNHWGKo9vQo3jDisG/OgidcKSNWrVvKIdyZLJWpAyIjUVkE+x1abo/xNsh73EjveFiNRZKH9BQj/hNIr0SzK6vu9WcL6v/QDy0JbJSwuROBKRGTM+jpjVUDNmzJjxFLBGp5OmNrwleLByWwelKYpmoIxnxUmtFaJhM1h1TTJKTmJ6f66Ifvv/81+c0nvMska6gKwN4vUGKkwXD1SCab3qOvqa9OCI+lNvUR2f4fujMcncHHUYmwjB0a2XXJ29QuiOSVE1HFeXmvlhrNCffYLoW5Yn72CMkGLF0OvrMWgeis8uWK4K1G1P3XhCsKTo8N7RDxaR7ZhbNTnvm9LZ2MtvuezhzlHkpePIp+7svgbqbrVq4N5LPd/yqQ0vHUfureBOY3l5aTAGvnouPN7Ao7Xhfs4FWTaa+VE6KkeNCsw7STwOkSsvHC+Ee0eaRWLzvgnQpcRbl2X9rC3xQnZExote5Rd0Ks6ZbdK5J+2MUE1tdpf5+mIhE1McEkrDbtheSTpfXNN+bHNBnmTXe5vYus5GtU9aNiI4KcerAm6HvSZOtzeQjVGcnonCzv5lklGe3dexxIkWpBzrSmpq0X2wuS9T5ZvBqLUxNYuJmL6MaunxxHEca8aMGTMJmTFjxoynQrX329JNRmem6dqVhTo/UdvrRZqgBWaSnBcyum2Bs+/TFdEe0tsBQkIuNWwQKxoyuE7gE/QONg6s0H3l3wTANAGSxSx7muUFAOGtT0C0tJ98k8XC03Wq3zAmEX1Lu+hYLntCUJ1H8C1Dd0yKDa4aEJkUo1VAkiEMNWGokWRo2g2uCiyWPUkMMVrN7Ahk+9wsuDYatFcCJMvHddxoZ2FRwcXacf9MdSRVzm8pnS1j4M0L4eyi4WpT4zMh+/Rd4ZWTSGsNXUq82QXud4krr8GDm2H7/n0UHmyEtoKXXUUvifPMlFat8Om7wqtLm41b9bZOia9vIldBR7bOYtTk9bQlU8XwIIrQ5uLWYtgQeSSeXhItlhUVJzeIn72Jo8jaye73MpC4sINa9YrLRfb1AuGONCylGkexbiMiNo+e3YTbSEoR3AezJVELmvEWs+3tNlBQdmxwp4RjGtwIRS+iWhJXuh47+71FSTifbr/GUYsdj93kbggwitHL/jk0xb2MYckBAvK02SszZnwUMY9jzZgxY8YTUFLP0x5H8FGvrreNYZNdoPI4+0ha/KCFbhQdy0oxF6xhqwGI+Uq4s4bVH/77p13vSH/pN70novRv+9/+wGkc7qhAfLPGnIimnb99hFhR56qTblxeNg0ilot/8cs5uvcmfn2HFGvao4ekWHP5+FM0mzu4qufuvYfw8B6PH93h6M5jrs5f4u6rX2OxWtJe3qXftHzlF76Jl16+xA8tdbOb+m6s4KpADBUh31JyWBtJUQXrR8cbPpUsnV/y4ErPqc8Wucsa8OoqVUTd50N2m3KGB5eGLocI3l0YXs3jVW+ewS95yVA5+NkvC3XW97zUwKqF5TLyS18Tzi4rHq3hX28Gojg2g2OI220e1Wrt+/VN5CLpVe91ivzCI8snjyxtrR0RHx0P+khEuyGgZGRlLVGEBz6wsvoFssBaIjU2d0IMd2k4Y8CTOGPgCssRFe2khF5ScSw1XzNXYwG9Nn4s/gsR0nwOdcBaG8+rssSKYWMCa+N3ivtDxfKTCujyPofWq3FjkT+19HVjcOKWqnhjxu7FEQs8gSt6zihWvhWCHZ2qNgyjVkPyaFTKXYubjqNoXTrj835olkhPHHNUyohaLY4L04+OZIWI/P/svduvLNt13vcbc9alu9dlX84hzyElUbfQQeDASB6C5ClQHiXZkGDAIgwBUWAnDhJACOAEcRAE3maQBzkIkhh+CWxADwKCQLJhRw5E5YmA/gQHfhAUBrBFHvrwnLP32uvS3XWZc448jDmrqnttiqRuPOau76CxenVXV82qXuQeX43xfV8hOWXlDfVEQuA0Y6Rsu5KQFW8r1k7IihUrVnwHFAJyPm5lo0P2ouS76m7OeQNmAvImqC60Jnnf9w8VIfzJFSXDB58n3VzTffjD9F/7cSMjr7bmfBUcGjy6b+350ZyI+uOO/rhl/+rzjP0FYWzp989prl7ifWDot4z9xXSMEBwfffPzDH1Dt3+Kr3s22z3ttme367m/3XHz8sm0fQwV3s93iFM0Qlc3I5okj2k5qnqk3XQ8efLAtrGOQ+1NiN6nOdfjorLrd4yJY56BexjMVnlUG9m67ZR/8Rr2vfD5J/DsKrBpI5UI+5QYVKfU88onxtFZan2CWowQlAyYxgtj7mw92QiNEwKWcr4RRyXm4NWPwr43Jywnp9+xEZa5XD+kNHVBahwjiU5noTjYWFQZI6qx/Ivy6Ik8ME7jTPa5kqPB9HOUnCOS79wfssKhXYT9gWV5HOXxGFEgnYjZvxcsbYPL8/MOybkNcdm+puKa3fTakYFAOhl/Wo5hWcLOQuSfCU3ZdyEYBSORLp99FLs+w3dpprs8n7KeqDNJWY6NBcLJmleseJuwkpAVK1as+C7gxEiDJVxDKEJkgaZKeDdrRQqaMwOhQkaSKmO0wriMZoVo+SHdKIT4J0NC/t1/+h+/aH7kA/rDM8LYcrh/hr7eTO8P//LzSBPQoYKxQgePDp7N7gFfBfrjFucDYbSxqv72M1O4Ygg1bkEk6jqwvThMwnPnA03bcXm9Zwie7da6ICX9PcaKato24kSJY0WKHnFKjM6S5AFfBy62ga3lKFI5IyJd1KkD9axxuaMBISkxKV12qwIjIiEZGelG4dB5PrmtaJywcY5RlY03c4Ehd7nMStnO12EjdHe9MuQv1jt7LSo8dxWfqSvebSouG7jeKk1l2+0HywFxmM7D9iczuSgjQKqT4BysozDmovwS0ydE0RNyEDP5WYrQl9a65xoRr45ORmo8bc7FOMjIjZu7YTtqdlp/TwLqKaPje9i+EJATi1yZU8uLTsPjJl1J0WBsqDkyPEonrxb78vhp32UcSxZECKwrVNa9XPso6cT6uDsjO/BmC+CaioY/+Nqda1lWrHhb4H/pl37pp77fi1ixYsWKTzM2P/fF3+n/ydd+CowwqM7utk1ldrDd4Ey0Tg41dJYp0eUMkALFiIzL4XgCk3OWd/Pol/zfv/dT8jN/5nf+uM7h3/vn/+kLd1Uj24H64iX17pbN7o7XX/s36D5+j9oFNFW8/hc/iR6uqNJopCp6wuGKGGsOD5eW3J48Q7dB1ZOipzvszLVqe6Td9Giq2T9s2GxGms0RX42ZoAh3N8/p+hrvlapKiGh2wUo5Ad2DmkuW85GY7Bi+KkF9QhwrwHJHBOFhsOunmMtVH3PHwsnUTeizY9llY3P7DzHlcSjlple+fq/cDcpDTIRMNO5j4luHxOv7itY5QoLDiG0DfHMY2UcL/GudcAjZflfh4zTyKgX6CJ+78IRo4ZSH0b7f1gu1OPYxmZNaLkWL5eyYfxZhepXDMI9EBhIDaSqYyZ875Pv6EeVI4CAhi6/n0R+wgnqHhQEGSRwYrf8hsNOaW9fTYba0z3TDU91YqKHoRG6Kk1ZJal+uo7hF2bdlAYFeHSqadRjWKagwlwdBJtG5fcY6SQ4bzRIEXRCApebC4aaflncSuOPAhuaEgJQeRJGUA6jMo1Dz6JZO5wUz8Zn2IkovkSjppPMyEBjztximK66ZFs3XyotjzB0tJzJ1SH76F/79P7b/ra9Y8a8K1k7IihUrVnwPKA5X808Lnqv8XCTl3L9Htr7nd1fPYYF4Rlqq6o/57ughomNEdib4ZqzQ4EnJ03cbXn/0w7bG6Amhprv7DPH+ivH1c8LY4n1gsz2yv7/CuYivTbsB4F2i7xobyxoaxtHT9RWvPnnC4eEJKVaEwRywQnA4Ueo60h22jP3CujfNd+zrtrc09uX1CVUmIND3FfdHx11n41dDsnA/6x4oXUonv4M5YFWuuJbZNuX98juYLe4mazIsQD5x28FNpzzfCj+yrbl0jp2YTkAEXo6RjReOUXlIiQsxz6QDkdvOsmTGxU36mEwTtFm0z0qBX/5hLqNI9XS3HhocmzekydhIVaInsqMiik46Bpi7C6UYPsg4aT6e65YkyoGBXiIdRl6iJC61mce8zgTtb7q7/6aBpToLwJdC8GXxcR7sNxKn98/3lnK3wSE2CpYdp8p+mtx5CMRpHEsm2uMyJVgQGnm83tJ5Wr63/MwyLf38vG1sa5weUROjzon1hYB4cY/2t2LF24aVhKxYsWLFd4FzUXrRcKjC3b6iqR7b9MZvwzjiWWfkXGsSovzx60ISRkQOs0hWh4btxT3P3/sAgNtP3qfZdnSHLf1xx+0nP8T9zWep6p6h3xBCRd8bCdAkk3bD+Ti/ljxNMzIExxA8D3ezVkQX+oT9Q8v9/YbjsSVFb7qPJOaOFSqO+91EOFLMIzSZ9ITgGbIW53pjzlcX3nHhS6FtrlKHlPAi0+8JIxL3+YvZOMcXdhVPK8/n25pNJh8JE4yXzIwuJT7sgmWPjJZ4/yPXjitv234crGxtKxuzqkW48o53Xc0Oz9ePI/vBujNlnK90bqpMQi6dx4vw1FUTGSmEoZUypjRb9J6TglESoxgJOeTiuFZHrX56LHFgWIwMzda7H8v+ZOzoToZH3YKytjfhD3K9ehMB8eoeibNr/AlhPycKRdReUHoXS1es6uz95eeWXaGy/1MBPhNpmvdx2iFZdkCODGyyK5lD2NLSUNNQs5WWC9ngxRHzf7V4oqapC7JixduK1R1rxYoVK74Dbv/qb78ozwthiAmch+MgXG0TMQne2V1xJ4J3cByYNAQwOwUl5lwJw2wZW3ll19p7l//N//ni4Vd+/o/kkvXv/PZffyGbEd6z0D4eHFIl5PkDctFz0X6I9jXvfOYj0s01rz/+Aik6Dg9mHdV1NeNQUzcjVRVQFfb3Vzx95yXj0HD3+pq6DtR14PBwyWZ3nI4dgjCOFcf900l4fjiY2H0IlvdxtXDi6o45LyQKdR0IwRNChfeJoa/YbHrGsSZGx92+ovLKEISbzvJYxoVeo5SUUZWNCEPudHS57vvXriu8y9a5XvnkTgDPq0W74spZIfuk9nyzH3k5Bl6OwBGeVxWfu3C8vrXE8oNG+lDxjXHgUjxdgkOKHPJKPhzgR3zNrp5tmceoXDeObRRqL9wNiV6VJ97Tx1l0fqMzcRwWoz0VDtSE4cvwwajKE20YSNPf3CgxBw/GKXyvY8Th+ZY8TPsvBKRWl/MuSnemuDqdIsqpw5Mlsn/3iDlzpFYb9yodhXZBIpI6OglGCk7E+B7wJ+5TilLjuWXPBZsTclDcrqpvQ5SW16Z0Y5bncqBni/39BhIDgSaXUeW4cErQlmvzeAKBo/Y0eV2rO9aKtxlrJ2TFihUrvgNKJkjphsxJ6EyC9OU4VskAGcvI1qJym0ZtzkIMy/bHQejHuSjRX/6tF/wh8W/92q+8OH7wo6SXV6RXNeTugQ4e7XOeRB55ks2IuzxwcX1Du+3Z7I60m46rqwN3ry/oDluGvmEYPCE4xqFhs3ugrgN3dztUHSG4qZMRo9APntv7hk++9c58/k4ZRkeKQtd77u9NGK9JqOqRV6929F1NyDqRJcaxRiSRkvD8ycgPfabjc89HNpVMmR9FjO5FppEqt3itPGKC613iydXI1cVIU+Wke4y4eBF23vGk9pkgSh53ShyIfCsMfOMh8UDAAZfi+UY/EFF6TVx4x875SXB+IHI3JFRnw4Ly/Zesk+vGBPEP6fS8lw5RhYCUcaSCSXBN4kY6bqRnL7O7U0GxxS3PfXbVOkcv8cTlqeBN5fIyS+MPm/5dVli0IcuOikNOuhJ1TmyfHbXsvyoL1j1u6jKUlHWX09Srxb4t1fxN41iPaZTH0Wbx+4EeRScCstym7LegkA2AWjzV4jNp8Z2uWPE2YiUhK1asWPEdYBkef8D7QXj3+ZHKm21viDCELEA/t8zi9P94SyFaUrLBrFz7Ubjf14xBuP2rv/1i2Y35bvCTf+dXX2hyvProvfnFMmYTvVnzdjW6N02G3lsXon3nWzRth68CVTPi64CvEre3W4ahNk3HQqtRVZHr6wPHY8Pt7ZYQKpp2oG0iIRONrq/44Pc/x93NM+o637FXaJtE13tevbqyTJA8dhWio6qsc+J9wlcB7xMxOlTNKrepIu1m5P33X7PNdV0tlmyemMlIISJ2rXUatemCXeemiiQVKq9sq9Pvah8TSZXLNutExLMTz080ds0+ifNd7hsduc9i7o5E1KLFyK5LRB5S4mWneDEiUjvLGBmjTlkjzys/ffbcnrY8N1eouVPg9TFZWBbY50VuKZaLw1QJ4CuJ4EVWPRK5keOJ5qKgPC9C+nKs787ElvxZG3ny6k5cu97UGSidkUJGdlrj1dGqn7oX5b8az1Za7tQyRAoRmexy8xmO2R43oVOQ4zkBKetbrquEEi4hCFdseUeveMoFT7lgS8NImLZdjl+ZJXAkEIjfU+9oxYofHKzuWCtWrFjxHbD9uS/+zvB/fe2nksKzX/3pL+9+/ou/s/v5L/5O95tf+6lCTlJy7DvPGK2joRh5SfpYT7JECYwTMUF6cdjqR0FVJqG6At1vfu2ntj/3xe/oomMERNhcPHD76jn3L9/jyY/9c8SDdh49thAdemwZPn6PdH9F6rb4zRENnqY+EIYLwthk7UdFCB4RuHsw4flxv+PwcM07732ECLTtyPG44dXrHUTPs2d7LneBYax42FeMwXN336LJoUA/eFISqkrZbgOaHCmZc9HL25bLXSBFj3PK65sLxtHGsrxXUhLqOiEC41hx0ZoG4L1rJQRHn2s6L/P4UCVCJcLGOy4rxzsXsG2VY1fRNIn9sZoS7BsnXHrHdeN4uhVeHZTP7EwvogkuasePXVU8qyq+eF3hg0eT8MRViAp7AvtkfwQjOqkvOiL7lHi/rawTliAoDEkREcZk330twp1GGhyN/XVMRCDkZ2P+KYuSOEiaUroLFBO8FxcpBCr1bHKoX4OnwXNBQxBz3YqSaLQiFvcrhCRKgzcNE7Obl0e4dz3+rJ9yvo7l33vIdreaH2UkK0xHmwP9igsWlJEq858zgXreZ3bdErGRqwrPSKCVmj0dDRUDgQpPyv8pEDIdqTIZW55BJ8FCCCURJE7nYiNX1ona0XKhDZe0tJhTW5SEIFPHZViQjEgiZFIy6oiIEDWiAn/hF/6D1R1rxVuHVROyYsWKFd8FLv/+Tz/SZjz71fm1CMhf+8oLkJOAQu/msawlEnZnXrNnbz0VbMIQrYsCpwSm+S4ds/6//+KvfPmLf/fvv/jw93+cj/7mL3x59zd+80X8/ffwX/gWjJYBkh52SDMgLmcvVB3j6+dUzT4TkBpfBYbjhqYdcuEf8X5DP3j6wXOhwuH+mhAqqirQtnanuesrQjRB+tVuZBwdh15MuO8dMQpVlaagx2NX01SOzXYk5G7IPndohtFx+1DR1sp1Ei4ueiRntkDuPAXH5S7y9LonxB0Pg1nzepE8WmVWvA4Ykl33998ZaZrIJ682vHxd041GQKYOSmU6nYvWrH2bSqm9sKsdH+4T1xvHk61Seet2vb+puB8Tl3hiUm4ZpwDB+W/EFt0FIwdJlVpgyO+XlPduMY61E59F9Y57Am5BSEpS94nI+qwTUfpANZZI7tRNWzzTFofwUjp6yvhVXkvOHrEAw4FaNyf6hRKgeJCRb4dlCvpyPcv7/mWf5x2I88+V9z1uIu7LLd7kctUTiJroJdBS0TGyoaZjPBGujwQatbHATgIbtdJozNbC5Sucx9iqaRSu6EC6xfH3dNlmOE6akNIFKeNYtVT2va1ykBVvMVYSsmLFihV/TLj4ez/z5eGv2NhU0Td/e4esrMXIRcioFqQX8vbL2uTb7eMPwv/7y//JRJAOf/vnvnz3dz968ey9V+ixsTBCzB1LJNHtn9LuIAxbwrAlhtrschNsLx8Y+g1OFF8HPvcevHx5wf2hous9+4cddR0YYsN226PZsWkYXbYvdrlrkUetgoUx7ruKplKeXPeT49army0xCapwc9fg89hXTEI/wt1DTVLh6rIjBE9VRcaxYrcbp9BE75Tr1rHPlX2dbXPBCEbCCMwnrxueP7EslKRGNoYcEvlsY2Ndzy8TdZ0gF5pXW/v9w73jXz4oP/Hcxu6e78yC97OV42WnbFxDHxL3jJN2YXZVyp2MaGL6BJm0mnC+V0vxNicsYZfHyQ4p0amb7GPL30iVaUGLnwL0lsX/8vlOa3qJ7NTzLpuJFF1rQ43jhn4KukkYEYkLorHVKndfjFD1EuklstFZ97DR6ixB/TERKSgp5Uv6YZkfp+L2fmHZG3OnxmW73/EN5KOgpcKJcKd7nFxMgnPydRuJmSxk/YjkLkUWvzfqGCRMgvaCjmEaYRuJHGRko/XkslXGv8A0IQMjXhxpadVLWK15V7z1kK9+9at/6/u9iBUrVqz4QcfvfekrRk70ceHhF7oRS9GGxgvXGytyCypv3ZCLv/czf2jHrM/+97/x4smzW3ZXN9x8/DnrYGyPVHXPtz74IUSUvqt5+uwBXwfuby9pmpEqaydSdBwOG17ftYQgkytYXSeOnWe7iWzbSNsG9vuGITiaKrFpA3f7hkPn8M6E/CGaFuPuYK8V4hYibJt5zcfByJp38PQisW0jY9abhAhf+Pye47Hm0FW8vPNTx6j28Gpv7lmjKhfeXKgOQfnXP6uM2Qb5OMh0nUXgyU652IZMiIQYjTidf3Wv9sLzi0wuolkrlzyQ3z12JJQfrzcMSfkojrxmwCN8TjbUIlxki9+Q80x6VbqSI5FHnp77mts4/xHcMp4QkUJwChmJKP1ZV+GSmofcmXlGQwJeMzAwF8UHGaeCvojMI2b7G0lTNscPpWseZKBf6Cue6GbqHiy1IoWMvEnovTzWMmPDXjvVTZVta3XT84iNnpXXymcjkRrrMuyx76Bj4KAdO9mwye5WRUdiz7gFE5AAACAASURBVCsOmGNVheOYe1OKTttH4kRahnzuDdW07UigpmJDjSDT68MZISnYa0dY5Iz8b//wy38kF7wVK/5VxCpMX7FixYo/BfyZX//2xCG+gZi8KT8kJtg2f7S7px/9zV/4cumS7B+2HPY7AMQlnjy7ZbvtadrA65tL7l5fTZ2Ncazpji3jWNE0gXeeHbm+HBmCFfEPh6yHGR33+5qU5s7HGAVfJZoq0VQW7FhVibZJtE1k2ygiSu2NODSVEY+H3h4wC/fHIAzBnLgqr3Sj8PLGRPWVT/QBDn3utvQ2Gte4+R+7XQ3vbBb5GtEI0dMLe3z26Vwwl3N/EwGJCbZzTUmdp3tKMOVzV/Oj9YYE3MZIR5yIxY2O3KbAPqaT774WYSN+ssUFG82qcxepo4z0uOwE5dhhuSIX+ecGzzU1XxAbh7ukxiPsqHgXG3ErnRZg6ma8qaPgcRO50EwWPnB39EWAL9ZD2Gh10vEZJZ10Q4pj1TLbo+A888Nem3NPlusqz4tQvJCkIvLucuFv3Z+I5bwLB+2mMahxsV3pgBQidKSfzrWQiI6BjiGHHkrWeKS8vXVEBsZJ6N7l59fsHp3XUXsGRo7aP7oOK1a8jVjHsVasWLHiTwlv6oIUFFtYMI2B8OYxrPujo/prX3lxtUuk//XP/6Hvnv7T/+i//jLA9X/7j1/ox+8gAu++/yEAut8yhIX7kjdRuIgyjvM/G20bqH0zWRKDERKA9lDhBDZtpOs9n7zacrELPH9iBVjdRI7HmsonLi9GXt+1jNma+NWDY9sYObg/ClGtuB+iJcqDY9/LFPx3c++5vhg49j5fy9kmuangQk140XihqWBT60Qqam9EJOYgwa53xAQPh4rtJqJqBGTf29pKl8XlzswQrBMUk+T3bJTL9dA6oU/KzjluF99lh5GSQ4p8QTb0qtQiJyXrThyHrA0ZF65KLZ4Njo7EJo8uPZGKJmehgJGZXq078lwaoi5D9mZb2BY/Ja3PDlCGz6qR0xvpTUuCcBAz4b2VbsraGMVyN8r+l+SjdCeKLfCyH7LsapRtyrhUwpwYfNadRJmdvgKJtugpsEG5pTPXhiYTh3EiFw01CWXQQFPGH6fVVNP4VJM7KMvxsY6RLQ13HLhmt+janI5TCUK9KKmO9FO3aJmYHnXW8Ei2DV6x4m3FSkJWrFix4lOA5UgWFOHy/HtUK7jszr29tvsbv/ni8Ld/7o80xtH1nqqquLi0kMGUzLWq6y1McNtGDodmEp2rCpqYBOSffd7hq0TfV3zrVYOqIGJFeVDh0Dna2kaf7h8qmsbRNvmOvk+2n2jPd5vEMDq2g2PXJjZt5PoCXt2ZcPyiARHl7mji/ZiMCIQEv/v1LW3+F62MdSU1/YcXuG6FXascehux8k6m7ot3sNskbu5tlMuIBUjvqXwJoCy2yzZCVkiOdzNBCcnE7E1OcB+T8u5W+Gd3gToX6sV2twjKP4ojz9wcnxczIalFeOI9H8SeZ1IzqlJnB6daHDvJlrX5b2XInythmLUIPyq7kz+okYRHuGU4Gdl6RzfcSMezLM4GeJCRg1ghX+O50pYrbafy/Fa6rBmJfN3d8a4ujoXpK+pMbEbe3GkBHhX91jGR6XUHOHVsqKbOR8fAgYFI4oosKM+i8zI2BbCjoWNkFE9QE4kHDTTUdDJQU7Gnz12Thnu6iTCVMMchPxpqbtlPxKGhZq9dHluLbKQhoQQCe+3YSstRe2rx1Pm7OmpPRTU5ZK0EZMXbjnUca8WKFSs+RfALLlK6C1Oo4cJxCyDFP7q1jhPbz+X1HSlWOGcFXFMlhsFGq0J0hOAtqfyhztqOiqRCiI4YHG0buNrlu9VRGEdHCNatGIOFA15s47TuIXicV5om0jT2ehGzl+BH60zMGo9to49G1NziEvSByc446Szy74I9DotuRky2ThFbl4hytU0MwY5fiEzpmFReaapZo1NGtQrRUbVOTUjzCFmXlONo+o7zQMCquEsRuEmBY7K09D53PXo1ncgllaWxk6ZSfdTEIRUNBIvXlbh4lNdgJrkO2OX7j89ouVQb1ypjV2AEpBTyDuGJbmjVs9Oabf5si+ep2hjcJQ1j7pCA6UsODDiEBxmm/S5dvEro4puyTN7kmFUISOku7Gio8bnwn4MJO8apm1Keb2ioxLOTmWQFjYwL+9xCJhzCXjsO9OaaRU3UxMDIqJFB7fh77Rb7CowaOWpv2+SRq4C9HjVN7lhLHciKFW871k7IihUrVnyKcB5uuExb9zLftQeoqsT7/8Ovv/jwv/vS99QNGf/zr7wohX59mXjvhz4iRvvnoO82xOAYo3B1GXh9VxOCAH5yq+oHYwIiStukqSsSwlyY96NpQWo/O1w5r9TApg2E6PDe0s9VhavLgXH0DIPn6eVcqFWVCdivt8qmTcQoHAfh2CmNF8ZoI1VvymKpzuyRo9prJbvFiYngi1VwwaE3cmKkxUhJ7W1bMPJSUEiJd5k0eiMir0KkFUGBzzc1H4+R52LuUh+kuYAdSAwMdOq5ykSgS4mNswGejXN8kvIIG46YOyoOS2WsxTFqos26kWL97Jc+xsx3HEtuybOsD9lRMZBoctKHYuGHiOdZ2nAnA1u1MaUb1+VjKFfacpCRK93wkTxQi5EURdmquVK9lKMRlNwFOTBmEtRM+pGl8xW4k6GtZQJ5EZ/bVbHOQpOJRscwCchnXYjP41yBEaYxrYqKgZGttPN3kMe0Bg0glmxeyEYg4sUxaqTXnlZaIxZEggaqxWiYbR9y5kkgarRclrNSq3RDVqx427GSkBUrVqz4PqLUiUGVyglJddKEgBGPUkj7PBZUxpiOvefhV743AjL8Z195seQ5h85z++oZF1d7AOpm5PLqyKvblnF0U0egH0wMDqb7KOQCIGSh+JOrERFLed/3JTleCAkuW9gfKi52AXFQYQnoxVp3HEt4YXZCqhPHY0VTzSwiRqEbZCIDYMV/UuuCLFFGpQQjBYV8HEfyyJh1EcYok7ajqZRto2wb+/2Yb+IPQSa3rjGBRPveKm/dmUMv7AfoonIfSgciMSrQw20KtOL4MA14HC2eHR4n8IHaGNyN9KDwo9VmyjSJGCExtyr7+ZD1C23OBu/UROJRPZs89mOakNPxp4jm3BJld5KRkajzCFRH0SvUFGlHq7ZtQnknbdlnXQhArZ4oiQpnY0ty6tw1SrS/50w4Lmn4SB5OLH0d8CADl9oQc7fnPAMETIx+kH4iYJHEgYFeByrxPOgRL24KOiydiIaaSjx3ejgp/O/Snu2iM1JIR/kZiXg8xyxqD2pRin0WlZvGxbocgnDUbtLOlDU4LIywdFv8NJbVUctafq1YsY5jrVixYsWfEv7sb/zsCWF4k069dEJCmseJliNH3pHHo773USz95d96IVnbUO7mD4Pj4aFlHGr29xfs7y+oqoiIbbNtdLLhrarEoTc729J5cLlaTQr7oxVWQ16bLHJPYjJRfYrCy5uWu31DisI4+iz+Fo69535fc7+v2R+qab/AFHAoMmerlOsE0Fb2qBb/qsV8Db2DTTVvW0jdEO1RcByMPO17Gx+rvG1bhO7eQe0WxDF/tsquXiJw4UsOhDKidJq4ZeSlDmzw9ERuGbhh4K4UtlJGghJDUtqsCbFifP4jObfePRB4YKQnciDSacwuVkolMrlqgY2EHYiUFPRCFsqY2IhOQvfNgqS0eJrswjURYxytei61nrQiNRWCYy/d9HAIB0Y+kT2v5MDD5Dg1cmAmM0qaxrYOYkRtlMhYrotEPpa7rNEIfKJ3RBK9mlfVoAEvji22jh0tjVRspSUQOSzscCsqBh1P9BjlveVPXRCOQkB0MUIWiajOv486kjQ+GjErD8AIica5S1L+TvX0e12x4m3BSsVXrFix4vuANxEQVaZOSOWsqF7e9S/FcMnncP/LX3jUBfmx/+nXXgDsru55uLtm6Bvu7jccj34KAgyLMSnnrJMx9A0xOsbB8/HLHapWVB8Hoc43rs2NSiatyjg66my3G4KjHxyq0NZGXMgF+1JL8dHr2sa6BmG3sQ5KIRr7o5/3Haqpm1G6MTvcJESPC81H4x93Q5Jawnk8Ix7lsi/JRzcayfJOaBf5JWHqkphl8LbJ12QREt7Wtk1bwSZrYGJItDh6EncaJjH6A2EiAV3RIiwE2z7fQx9VCfnvYCMuhwfOrlZQOhjVSefhQMCrTERie6ZEKa5a5RN9Fn6XbsiYrXvjwjmroLhoteonN6xCnjyORiueUvOJ7FESgqNjYJQ4uV51E9HI4Y3aWLcEE90fxETgMY9W3XPkOZcmbCdOJKGRivvc+bhmR8fAJVsCEYenmoaxBMTcqcrnbPSqZtCRgx5ppaGimroTBz0+SnpfisjLT48nSiTlTofHW8dDpw+RNFJLPX1vy9BKMPKxTKFfseJtw0pCVqxYseJPEX/2N372y//sL/3Wi/PXvcgkSi93853kzIxF0VuK6hBZRJ99e7is4/ihz99yd7fj4VAzRkc32h3/J1lM/vHLHVe7kbqJcKgtKXw0q9y7vZ+sbIcAhxGimjbDO+VuX3G5C1kHYunj17vEGOx36+TYyFKVBeFJ4YOPW0RspOmdpz2bNjIMnpd3FftepvOOi47QGOcuy3zt5k5FTNAnpXVCF9R0NNhI2Bhna1SAIaeW106ovJEJ7+BiG6YgxCEY2dkPRlLA6sxDHruqByOMqtZtAXi39XzUQ62C4LnUagoZLN2MXuJJZ8Or8FlpGbIgHbIbmibepaUj0RHZY6GCmhdS4zhImIrfJUnZE3iX9o1EZLltzDkbhYAAeUTLTY5a5TUwHUvpXBQS9a7u+EQOvKsXcwAiAw8y0FJxpY4HGdhpw2vZE0ncyWFa1zWePTbS9E15zcBIRcUreVhY6Npf/JYGFZ1yO55yQUOdyUogEHnKBRZc6DlIP5GERiqMdlTTmFQgUIt1SCqpiBpPR6pIyBlRKEQGgTqvq2g/TKieZ/mKFbTY+pJGRGYyso5lrXibsf71r1ixYsWnAGI6YzZeJqF10RY3Hh4GaBdBEklh81/9kxfvvLMnBEddl1EST7vt8dVAu+moqsBzFzkcNiYGd4oTOAYjOP04C30PXQVdNaWEX2wD+2PFq73w7qVyHGQaDStdh5f3nqayETFzlTJyQy85j0NyvscsBJ9yP8KchA5MmpB3rgOHzhMf3HSccv845BGqKrOQytm5AFOZqGr6DNPQzMVj7Y2YHIOyqebXo9o/hjHZmirvps5N7WFTw34w8rWrjcwU8fcY55GxbW3rSw5aEW5Tyg5YcSrop7XknIzlnfCDxqnQL1kZMIcUbvA8LDQZBwJ97iTAMsxvPrcbBp7kwMKCzVnXw66vTl0ac6aayUdc3MUvqotrbRmIbBTu3UCtjp1YOOBGqynI0NZjyfA1ngqhoabOpMP2n7jnSJtteEu44ZGefRajezxbGgYCFY6GmgM9NT4HCRqheKI7Rokc6HOsYCFaMXeO5m7KUMbqiuOWbE1YnolBQlFNVAuiUOFx+TvaiieRcFkTEzVOuo+iIyldklHnOwk+/+9tmS6/YsXbiJWErFixYsWnDJMOIo8jdbkg7qMVxPdHN9nYLqHqcD5bt4YGcUrVjLy+uebjlxuaJnG393kca97/TC5OE573x8rSwoGbg0zjT2BC+nEgZ27A/uimPA3II0wqU6ZJQRfsmKdWxGbpW1V6onVpqtMwwduj5H0otZMpF6RoP8D23WgeXcPGskgyEx2g/jbTL2VU7dDnXJA4j4LVecE3nZ5kejiRSfheRtjARqrGPPqUUO4ZCQsSklC8Ckly4S/CrY48oc4uWIUUzNuXfBE4TQsHTp77TBgKaehJPJH6JPSwngiI5M+UY6YTgnI+RpSywH3MjloAV8nGp3ZaT9vGPOK104bLnNdRqXUhrtWOd0nDjRzNrYrZreoiZ3/UeHoCm0UHZFiIy3e0E5Gx4zeTjmRJQDY0DIx4HD4/T+g0nrV0ryodDy3nLW5y6SpDXoXYJlVq8URNJ6/7PK5WuirLcS0/Dd0VornqQVa8vVhJyIoVK1Z8n1EtlOc2MlRC73RKIBfsxvhdZ6F9QxCeJiEEx+3tlt1upK4DF5sOXwW6w46qGamqkXffe8nh8D6vH+z/8kuHpRT2TuwufrGt9Y5JExETvH9tAYH1outQOyEm5X5U9uP8WlvBu5fWEekCuEWngHwOTuZuiKoRlm++bLhoLb/j2VXg8iLw5Eq5ezC3LYBPHoQ+d0/GpBDsOhXxebl+c66KsqscfVRStK5ILTAqbLPWJGGjVpXLJCuCjDLdmx7j3I2y8xBe9nPheNMnGic838pEFmOChynHw/bU4GhwDCQC6SRZHCwJ/EjgSOB9tiedE587E0VHUtAvCvLyTimYHWIkgIoaR6fxUVZJISt+IiLWcSg2uedC+NKlKZ2RgpId4nTWuVxkt6uCJBW12upa/JT5sdOGZ2wZiSeZIrV6fphnHBjoJJyMR9VUk2uXSstIMN2JRGr1k87kgg0jgYHABRsimSzk8z7ST8drpCJoZLL/zd2PYdHNaKQ6SUVP5TuUQFLriizJj+Trad2XmImhidI9HhHJY2FvDnJcseIHHas71ooVK1Z8iuDECv8yhrRtyrjMjCGPNO2Pnv2+tdyNYIGCvgpoEqpmRJMQQs1w3HDoLH9j1yrvPhlpawv+K8X1foD7zrQaY5x1G0WIftEqlxvls1fKj76TuKjn7kDpOoyZbXSjuWrV2SL3oimidvv9PNPDOhBQV8q+lyx0t+5IISBjtLUGNeenUZWkyhD1hICco3I24nbdmv1xQUzzqFYJFSzXtrd4B7PzzedXgggrNwf/Jey5yPw+GCFSYCOeGqHFZ1veanKaehOiKCo2QgVW9Jd/pP2Za9WbUC/eL4Vwl3M4EuaAFc8eyyIgLlyzgGndBSNpIlXn4YJF3F46PLW6aT2jpImAlOsGTF2SGj89CkwwnwXlWp1YBQN5DG3uXABUaq5aA+NEkiocOxoEmToqpbvTUJtGZDF+1Ug1JZzX2HtlfKoEHJ5DEGoxQfyGhq1spuPXMneHzr8bVZ1ctVaseBuxkpAVK1as+FPGv/kPTq16w6IqT7m4rpwRgeNgd+iLOFvJImyxoj2pcHvf8PK1jbOMfcvYt3gfEKfEUOHrwE/+xCd84cc/5As/csOT6553nw48v4yWi1HPY1RGOiQX6UwBhUUQbuJv4clOuWptHUuReB/gvrNRKsvRsGJ+jHPwoonEbTRrTPZoKrh5cFy01v0oieSbNk3Hrb0RioIuKaMa+VgSEJ/1NZods4qbVuNlWsMQizDYtjsGJeWOB5j+o0/K/Zg4mv6YQ7BOTS0yFdIi1gGyazdbK18421EhIjC7UFkKefNIC7BM/z4fx6oWxbaNFbk86mUoBbxDiKJEsSDAtNgHeftCSAq+XSFQXl+SkUJCyqjWclyrdEjOtQ4brWjxbLRioxUO08Q4wGkRvXsUZUdrYYfI1LXYLSwYmgWBc9Nr9n4hJA01G2pGApK3KhbCZZsN9bT/ct0KGWkWxzNb3kQt1glZdoCW5wrgxUhYGfcCGHWcnLrOr82cGr+SkBVvJ+SrX/3q3/p+L2LFihUr3kb8P3/pt1445vEo8vNahE0l7Gor4Cdb2kU12VZwvbHU8kMvtDV8/t2eyid8lajryHbXMQ4Vm53NzTet/QyhJoaKcaj54JtPubk3O1wL8jPNxbPtrOe4tTw9nMBnr/XEXvhmLzwMRgYKmWqc8KSVHPpXsk0e29sqsy5lme/xbKcMQdi1yqaNk1B+3znuOjiOylDyR3LI4xIhlfGq+b1lAF7UQkDMkWwflY2za56yy9VxhH1I9Gq5HUGVMqxzVTnuQ5p+dyI8bdw0CuYFXo52pz6q0ucOQukypGz02hO5k4Eax1arE81IKfoLaSl6ENN4RHqJky4ErKhfjho5ZLLrfUa7yANJ+XqU4l8mDUjBcpsSkjieUJnHo1rL4xaUQnz5XkLpJEz2vkXjEVBGibTqp45FxzjZ+97LrP240g2dhEl7sTzGwIjgps/t1PQqg4R8buV4iaGMcRHZ0XBgmPQpIzHrSPwUiHjFFo9jT8+GGgtM7KcOyTKssGSLFEF6JbNQfXltlmv/P/7R//zIcnvFih9krJ2QFStWrPg+4c/9g5/9cgKGZLkQYAWJF7u7X7IsYiYgQU/vnO/axBBkIjEikNQCAA+HhsN+w/HYcntzzcPdJao2nqVJiGNlGSJ7j2YtyEVj2pDaCzdH5faYx6S8kaAuqLlbLerRJzvlopltc8dsMXsY4TBYhwPsZxn/qku4H7OWo3QswITyxxEO/ezGdZ6rIgKNK4GI5lZVyEV5rXIyvbYsoVWzFiUpx2gjSUNS+qg0Hl73Sp+UPn/2mBJd7lBFTFNiY1qzne4SQ7LrEFU573b4xf34o4Qpp6OXyDXNpK8onZAudzOKUHxZ/J93OOIZYYgy321fhvPFxfO0eK1kgZQ79nFBRs5Hs5bPbT+Px7S2zJ2D8yyMa2251Gbq6owSpw7IEmMmXA01DTU7tY6fLEacKnVTR6UQkFo9O22osjbmSlsaLd0QRzXpWxJXbBEcF2xoMrkIBOq8/tI9qfFEEhtqdrSzYF0cilLhaagpoYZePLXUVFJN+/D5vzdduxUr3jasJGTFihUrvo/4c3k0S7HCVbJweszCcdMj5BGt4r7jjDQcBwsfPI627aHz1LUVqZvNyH7fst83vHq94eOXOz74/c8xHDeMvRVyKQmXm0TllcrnY0fYVlZIP4zKy6PpJUqI4hjtcRyEbjSSUMbDVK27oAqHmNgPmkezbN91Hs9qfNZW5A5IeT5GzWRnkUnSeUQs/FDEdCZFYF7g8zE1d2M0a1SGxZhbnEieGQGUQEAw0tJ6oRKZtDitE3bOme4kbzdkYtFlFibMgvAxWjeooJbZ3hY46SQ4rMNxofVUyPZEbqQnomypptGsQOKBkbucuXGQkU7srntcBB1GSXnMKk37S1jo4IHAHSMxO1tN123hgLXE7LLl3vh6QfWGEmLusJy+V34v7/eESYgeMc1Ig7ln1eonPUgvlgJfNCH+jNTU6qlxbKhQ1GyA1XGtLdfaTt2hHktgbzOx2NCwo5m6KQMWlHiRB7WKQ5eRCVv7ckyrkLtCKBpqqqKB0XBy/UwzcqoDOh/lAvjLf/Gvv3h0QVes+AHG6o61YsWKFd9n/Nv/8M9PYxhlRGubtQ+NhyjC3ZioBTaVTEX8SXp5hI9fV9ReaZrIy5stYOL1IYu7P3xV8Y2PNmxq5f3PHPE+4bzyuXd72jbw6vUGny18P7ObC6SPD1ZwbfIxp6yQPCr27qVyMQjferDCf1RLwO6SQjAnryHMKeWzMaw9r7KAvfbCGC2ErvZGwGKCrvcnnRDvoBtngbGI5YYsx6/A1jIkpeRdn1vzll0+a61Y3I/WDQHoYmJQ01NsnLOuB5YFHjEiUQhIUmXMd+JHVQ4xWSdkUbQvA//K6xs8G/XciLk0lTErKETFc6k1G/wkCi/FdMFyBCpOOSFzwX8QS1Fv8l38cu/RxqxOR6eKZqW8X15fosIR8qtvSgEvxy2vtPipqzNfizmRvCeyo84Z6dCqOWdZV4eJfJT3IkqQRKWOHbV9Xht6CVzpbPPbT+NXs/dUrZ6D2MhVn122GhyRObm8EJFi/buhZmBpO+ypsdG5jpFIJC6sj0sHBOZ09fJ6SU/Pb+ZrQV5neERKVqz4QcfaCVmxYsWKTxFSvkNfRn4sr2IeKTIRto0uDcEKdcUKfOsgmLuUEyvep3GnMW+r8NAJ3/xoy4cf7Xh1V3Fz2xCjbaiLoLvSXWhyBsao8PpoBKDL+zsOMo2ELTGqdSS6pGwb5dmFnUMZuVJmTUhIdo5lFzE7dJW0dc2/F7viytmaCgEpBKUQkJD0RJ+iizWdj081zsIUYzIiIwJ97n5YpsjZeJCm6R9Ol3/v8j5Lt2WXL3ot8sgWF+Bcf7HTx/cDrasRuc3dkYHTQve7GeHpiYxEWjwN7qSzUUhHPNvPkjAsE9XLWoXTkL3J6WlxmsvX3zSmtbSk3VFPRGPIqpiybZsF3Ut4hFb9ZC+8peIo48m5le5GQo3Q5MeSvEWSEQhiJhNpWtc9JoLaULPJzlo1noTSUM+Ce+oTy97SMSmi+mmMSx6PmdVSTw9g2s/aDVnxNmHthKxYsWLFpwyK3YnfVZ79YLqFYgsbdS7kyyjTEOeCvus9Xb/l2fVgInWvPFHh9r7m1YNpLYZo2hGw37vR8dDteH4daGsjD+NxPp4TofVwjDaKtA/CZe2ydmW2sl3CRN9WKL4+CJetjWMNObOjwEa8TgmKJZ3nDokwOXU5W7I5amkR8cNC687GC0fNd6QXa1mSj+XzISkXtXDTJw7JOhjF/cphxKIWwYnQL05yVKXPd8BbEY6LES0nJuhGoRU3Ha90QUoaedF3HOSx7SvMrlIHAi2eHjkZwVoW+Msid9kRaamsC1T0Cwgjsxj6TWShkI56UcynBREpx2qwtPIyOlaJm/4Qlp950/39tFgD+fmVtoySGImmEyFml6t4QlzK+jxm3+vzZ8p+IolOArXO6fPl/Tj9TFNuSPnd5W6RrcleHxh5T5/YucssKldM2D6wTEK3lt6g40SJyhhWGdE6hyW0z2Nc53qRFSt+kLF2QlasWLHiU4TnrZ8IxxiNgBQRdrHDLXkX/WLEaYw2/HEchENvpKOuE8PouLzoeXY9cNFaQThG5bZT7nNWWx9nm90hhwGWDorPz6eUcEwg/3qIdNFyOo5B6YKedBpE5hTx4wjfutfs9DWTm6icZHcsn+8aS4dfNi5iMv2JqpGNNyWfd/l6FSxF/8vNe53Xux9NhB4xwlHyM2qRRwV0Wc5S8zHZ3qp9B0lnq9pCVErmR+mCLPUZXmcHr3M4hKMEHmTkn86QmQAAIABJREFUTvpHHZBSTC8L7XmtypHAnfR8JHZ3v2MOLSxkonRElLnA/3adEuVc3yJTF6S4ey0Jy/LnrBex0t+rjWX1EqnVTanvxa7X1jtOny0Pn8Xm1QmBkOxWNdLlgbGyv3Jdxvz6vL7IMatnyrpDvp5DDjl8ojv00X9p0pGk6W/LckdGjdN5Lv96zjshIbt0VVQkjSSNNDlTZMWKtwVrJ2TFihUrPiX4+i/+9guAd1rHIShdsgLXqQnFG2dFvOZuSFKddBSQk7p7+zkExxBsRn5/vGTTRkSU6y30UegzgchZiJNupPazK1VT5VTx3A3ZOEjRHKKSKq/HyNPaM+R1xrNRp6Kv6JPm0aX5/fOCe0hz8XqMyhAlu27JZOPbVHC5SdzsHffHfLdeTnNWoNxlz2RKZOpmlC4HzK+Pqnw8BnbOTdke5HPZejed0+sQcxdDkQSXbs5+uE2BK/EkmBy1rp03gqNx6h5E5g7BeXFfim8TUVs5f+t6bunZasWd9LTqzUVLzUL2mG1qYXbG6rPtLNid+43WJKxrUDow5fqXbkRZyTQOtxjDCgsCUT671IKcj4ZZMGFek+hkiVuIVpW7QcUa1+XxqkJryn5tpGomVU1OSU/YmNYUxiiBpoyzCVMHZUR5yInop85gM9Ep2pQuh0MmPJE4jUbVeL4ltyffUUs1kZpLtnQy0uuAF9tXLR7USIaiUwfknFyo6qMWkb7BaW3Fih9krJ2QFStWrPiUwInVJW1ld/odwoW3saYqE5CS1XeeEu6d5CDB7Drl4e5oDlZXu8Dd3vPQ2e9ejCCIZP2Fwn0P4+gYgjAubqg3lXVDghopUuA2BTo14XZxm1r+Y3KaKGFOU0uUWmsqPHNeRynqKydTxwSsk3IcjWDdHhx9OCUflZNJD+JFJgJiv8/jWMuk87JtnR/2PtP1bRbnpEq2r806A5RDKsJq+zmcFZARTvoSS/KxDPWzwR+hwj0KF6yzViLk7T2OZ2nDTiuutDk53lITsRxfKqNZFY6X0k3djbK9CeCFOjtl2b6s0+Dzusr6l+eyPCc4K/QX1sClYG8xFyvBOibLNPRe4rTON2ldyrmMYnqcQl4clsxe+hNFC7I8P2Cy9126W0nunJR9DWcZHucdFMsBiRzoGfMY1q3up29r0MCd7hk18qbwwe+k4VkSul/8i//lqgtZ8VZg7YSsWLFixacAv/elr7yondBW1uXwWXxd+xL0BnUZkco/3WLuqBTPpdTRPO5Ue7O5jcmS14fe9l8+WjoYY4Lff+V4vrPPwDyaVfC0cdwNiZ14DhppkaxR0UfE4xDT1HXYhzR1JYqDVdl+VKixkL8SWDgknVLIi6uWJIj5s4egk/3uqOVcjWSInN5gLtsJs41w6QKk3AnZZCteyHesxYr+Iq4fVbOG43S8qMVNz3fiZxtgse5RgU4jW/Mxyho9wrDoOJyHANr28H66nPQkJVfk3JmqdBfglCi43CEBGEg0ed2O05Gh6fyZr+Hy3dLRUQHUiM2QiVNAaTORqnFcUHOXr1jMa44yn//5WXp1RJn1JiOnOpdp1Eyqyf2rI5DkdAzNyIhwZMzEzS/2EaYuzvKaRRtSJGjMHY34SJsRdD6GFzc5Yg0aTr6DmAfbokZEFsRQR2qpCYTps6OOiBTnrXn/a2jhircFKwlZsWLFiu8zfu9LX5nufDa59jmMcNFYV2JXWXE9NQeyNmLZ+VjeZ40J9oO9ftfB7dFNdreVs/GoqDCmxM47gjK5QR0HG8Uasxhc8uhXH5WHMJu61ggHIl2ye85Ly1q7Wz3/AxNUQcF7O48uppPuB1nr4rBz/uRoOg2A69bSyi8qa1N80qUTYkGyTkhZV8gWwU0eWytjV8rjwMNl9yJiY2/vbk2EftcrQxI6TYSs8bAiey6UCykpGpIDJb1cpuPaKJQj5c+ZMHwecRpIVDga9Qz5rvxBxkUSuudd3U7nKxTxeDohNQVlXGipeyhdBgsHtGNu8LkbcCpML65TcbHvpW1vvxBnO4r1r51/nwvwQrKKnS7qOYiddY3jICOVuhNtynknxE1na0ShjJkllCQxj1wtReZ2rGVHZsds2ftARyDS5DyRkilivSGlFk+nA0k9jVREIoOmSW9SiEJNNXU76pznsiQtJkqXPPI1k61aakYtParSPZITHRSsBGTF24WVhKxYsWLF9xG/+6WvvChdjGXyeXG+KkGAITHdlvYO4iK5vBCRgpBKjkjuagikmPM1otnXOky/sOyIgInUY+6U1M7ICDCNM114oYrKTZyL4FF1UQbnNeb9zy5BQhcTldgI0vKcoxYBvJx0Q3wmA4eYeLbxk2VwLcIhGSEq+SAA3SKvoRAVn126yjpCDl08F8S3YmSnj9B6aLzd7j8kIyhljKkjssFT53GgESuce01Ui27JXP6WUMCSaq4nJKSQivL7QRZuS+ooieo1biIPAGhFQBklTp2ByRULlzUgp/2GXiIv6XiW8zRsHEwnbchybUu8SSzdSzzJ8ChdmF4SdXIcJNBLoNWKCiGgRLFsDYcQJCHqKeniBWX0qug1ylhVy9wBKS5VLdW0vUMRNTLjNFvq5s7LQ878kHy+5eeSyI2LTsegltlRiWfQEc66KQWFgCyJz7I7de6IVUsNCinvo2y3Eo8VbytWErJixYoVf0r43S995YWNVdnYVbnbX/IpGm+ko3b2fIj20wlT4V46Isdcqy7JhzDb3MJs22vCcvu58cI+WPlTyi6fi3JV63gMSRmT8tmtn0hLLULrhcYLr8NcNiYtReyMcRptmkXihajU+WSGNHcrInBRWUF+GJk0MFFhHy207hsPkfd3ntYLD/kkldmVCmZhfJ4WWojQmVLUS9DgsBCpRwA1wf0Y58DDLpnNb8JcpfpctHZEKoRxuqMN9wTapS7j7A536YYsi/xl8V0cokoxX6vLnQOfOwyZ7BBPOhJFN6IoToUkSqVuKsDLcRyCU6HCAgy3UwK5rWs5plTW1RNPNCFFhxFyd6CsYSIjAgdGPnEH2iwW7yVAfl5IUVlzlHRCBM5R0s6Lc1aN6T8ORBOWq1ni2ufFAv9UFiNmRaifOxuYDa+eHW85ChUI+fP+ZATrZBsNVIsE9FEDInJGamz8KpJIemrtC5A08uv/+O+s5GPFW42VhKxYsWLFnzC+/ou//SK+oc5qF85WtTOiEBV8vuNvQYX204uJzZ1AWysPgzzqhBQtyRiVkGTKEEn57n/Bxjtze1LlZoxcV56NCLchUiehEmFQ5RuHQHFWArhPShhMmF2K6X0qQmfhifcT2SinWwhI6UYUy9xKhCHplAy/q41YveojG+8yWbK9HFK+33yYNSwPGm0EKp7e7fe5UCYf98K5nFdiqDPhKh0SsOtTyEntLbxwTMUhqtjSnrpaHYnsqOizogCYSEqbE85L56NY8y7zOaCklisq5h7Vy2keRhFhP2HLQMq6ETc5Si0bFIUsjGoFOmqjTw8yTNtcacOWij0jguWULLscMh1XpvOIC8F0lG+jXckfLKNLgcRdToGvcY/Sy0uoX0kgX7pgFZT3bB92PopyJ/1EIkaJDIRJkN4sCEa/6Jb0hGlttlwhEPPrceGTJYvjz7QjEE5crkSMDA2MxAVRKVqXovUIBFR1InBlhOt//0f/40o+VqxgJSErVqxY8SeKD/9Ds901l6ZZEK0qeeRKOI7KmMS6IP6xdmGJeDYtsxyUKQQkAagRkbQYPypEpBbYOUeXEr0mboJSYWNSClOuRim4y1BJ0PN7yHAod7cRRCzJutPEJof0LZdb8jRKB6OEAXoxQjVk+98uJhxuGtlyCDvn6dTCBAPz+oquYXknfxm2F2HyRHL5HCKPk9A9JvTvo+llhqT0qQiIdRJzB9I0/x9zoV5crebztDvzZV1jJmnL70vz+spnlyTxUWcpX8UGz17GqfNQyIJDptT1iOdBBlurpCy8zsRBIjutuKRmmYx+fs2WFrw+kwqwjkeSQkgW4YEq03dSCAFYp6PSWdsxSpqyQey8HpOPN6GEDx7zX2KUNJGgZefDxO+Px8ngVIBfnLRsvWbLG4i43BXqdZiubyUVUU/JWnm+FNjb2NZizTprSSxXxI73aysBWbFiwkpCVqxYseJPCL/3pa+8uG5l0mh4J7nYtY7HLjthbWsbSxrLvJDPepBcT02Bgbkz8tDlgnahqygouSGlkE3YXf1ikzsmzU5V89hSIQu1CH3WR8BMOsp2AlOxWTDf1bd9HzNR2OQ1bUQYmDUn+xTZSnajEjvvLiVC8pbOntczJOVJ43gIs+3uVmyYZtTZhvW8mD4v4E/Xemqb2zjTqZS1OOAuJPbJuhu1OEZN7Bd32wuqSZxebFzTJOAumFy4cjFfOinLK6iytO9N03WaLXRnYfhAYqsVoyREoVYL7OslTqNTZq/rqfHccORKWxSll8iF1tO1Kk5XywT08rrPJXt538agTrUrxc2qhBomsXEwp/aHVeNQ1Wl0zKuz986+j2lEK4+hlRGsEj4oeezrSJiOV7oagrChmtZd9CWFZBSBfiEZMJOR5bdgYnpHLZ6oktNFcqbIGbkASGdaj6IzKQbLS+JRfoe4aj9WrDjDSkJWrFix4k8QRbsRdbbYbSsjE97BRauM0Yrx/ZBF4czdECdz7oeq5XYMuQYq5MQzW/LGZHf0HUYKyijUmG10TfBtBfjGeV6NZeRoDhw8LOxqe9IkLK6YnZ3ehNcp5HEc5SYFLuX/Z+/tfmTLtuyu31x774jIPOfUqbr94XarLWEhIcMDfwH8ARgkRLVlsCUEkoV4ByHAYB1ZMggjwC9+QyAh0RLmum/TYHfL8gvCQjzwgpD8YmHZ+Ku77+1bp85Hxsfee63Jw1xzrRU7I09V365zgao1jvJmZGTEjrVXRN2cY885xhhK1wNsnTsJzJlEoHAEDhJ4525YY+BHS2RRS2K/NG2hnzsMnFflx0tkQjLZMaLgxb4Xym6f+70wckyJZ0Mo2pakSlJlTteuYgB7EaIEflfn8ssLkT21CLcMjUpK5qsRqjoO5t2PhSpsb4lM28nwonsLL/L3DOwIvJGZOx1Zc3fgIpGgwmccEOC9LCxEjrLwXHfs1fI5Xmi9gn9iZZ+DEX3PLkQWSex1KL+TfK7F6Yo2iDCBBpDEycX04p8jLeRpRbmwsJfxSvPiJMu7IzYa5sJ0e5yTlrdyKWJ0xQIQJ8ayvw7J+5iACytaRthozsjQJrKMjKbpUCMMO6l5It7hMK1IuHqO6z2CDKyNK1ZLPLr2o6PjaXQS0tHR0fGR4KNXbkc7Nla6d5ONXt3vU043N0emUzIy0eb7Ja2uWUMw0uIidg84VDHHLPJr+usHzD3KczrAjuXuU5MII7BqXptkx6SmOvfux4oyEZ4kIjZ2VMeHIqHM8nvqesC+7xDeaSwE5xzNLngaBBa3ojVSdB9yJyDa3gjwoLF0CAbqOJBfuY/UbI6B2pWAOnK2L+NpPl6THcPy496z8DwPc71nqTkXWsdx2v24NUXnwX9tx6Z0oJp9fEqcPebOgGNS+2nPwF4HTroyS+TLrJXYYXa4FyLIzHPdZTIZS9K4/+wdDgHuGFmo+pGWqDiBAop1sGsutsTJ77+FJTtXRa5zMcoIVXYDiySOzFl0ft3Zql2jp19nK7AHHhGQlcTYlECRSBBB1UasnHwMMhA1lrEtVS0idCVZzofG0vnYrqsTkI6Op9ET0zs6Ojo+Ev7IX/qjf9aTzIdgXYwxZJvZTECmKRFycatasz9aIbsTElWISQrxAFiiHdd/Nvesqv0wu9+slcgdgFXN/crzM1xncVG7jrtoFZ6bVkSviuhblq01ZboWpa4t8eP52JddWc+PIa8nf71bnMDUbtA5dzpWVd5FKy29OPZj3jEwNgW/XyX/0brwNtVS2QMK9zmg0DUisfn9W12vivX2HC3tu2Za3GV9yIVYXvNM5FzGtK61Iq3bVJu54WNJ/kc55q7DXofaqSA9IiW7PHplI1cr7+TCe5mJYmngQi3G2/fwXZiZSXwpFwLCW5mL9qLVuJxYy0jVJedznBub2mFTRvhIVjt65TkfQD7OykkWlizEL89pjrNIyqNr/tkzJ62FtXRb/LbmLt3Y7It3I8bN+vw4kcSFmZmF93oiaiJqylL1xF527GQq5ycIqv7fqbLoXEIH6/7m91OtA9LHrzo6PozeCeno6Oj4iPCRqSnA3U7ZjcppFu73yn4XGULN6hgHIw2ndJ247WL0Idgolom4632X1ciIv545PQkXd3jK9rtwrYkgO1V5gR8QTrmwmvIo1j7Pyp80lSK2vcJccyYep29PErioFe2ChRsOKtxJHY0JWFdAMUvcQ7HHTURVCwdU2499FM4pFeepKRORPzju+GJdSU1H5JhJwQLcE3i9rjwLQ7ny73sRsT+Enra+ZiL1CRN3DLxmZiWxY7AxLtGSNg7C69w9UIFFq3NUYijEx8vgO4Zy/Fb8vU0QD1CcoWyddc9dzE5+j+8ZeculjETNuXR/qXc8113pDE0E3mTHque648hMEOG9XPhM9+x14E0wF6mdDpxlJaJl/GpS056YZsNIkRMLd6CyoMRqpfu2qCsMC5G9jjxnV4mcaPksmE3xULQf7qplz12vRtZ8pOvH8pD3YY/rQeydCaXAGZvRuRUPnhxzWOHMnexzkrkyMjAyFA3JrAtrPs8xhxhK7vBZZ0QK4emko6Pj94beCeno6Oj4iHBB+TiYte40JQ6TEsQcsmIShqAWKtg8p8VutOfGVLsk7fDOErPjVKMjgfp/8HU8hTKCBLUI93yNeEUu8rExBy2oadotvID0ADjHRCivvzbkJWL2ulG1EJ1SkDYdmkQVoPs63kTP6vD12+++jHa1fslXxK+v/Nuj32ssmSaaX8vXV8a01Do/OwbesnChis3dStevwr/Ptq8e8OfdCicInifie+MkzY8XNnu5HXB7rhN3uYwuXSaxZ7iDVUI55uL8QWYeZLaEcrzgH3L3onXHmljzHr6RMxHltVxKN8aJRURzyrm97iJVwL7m126JkhMRJyDtWJYH/E2Ye5qRmepqFaB0bpZNGvrCWjoeS7O3ipauhe31jAvEt2S4duns+RdW1twNcaczH2H011t0YdW1fKYtclGKLsSP2QMHOzp+cvROSEdHR8dHhNvMTgP8zGdnVIXTaWSJTkDgNFthepiUh4vpR1wXItSAwtSIzz1JHSph8K7LmqpjVgCSZJF044LlYXoD9gJJlZ0EVo25wDM8LUOvSKUorexpzALhUSSnRF+Pc4UsZPGU8U9FiKrMV7kKXuh5snfVJvjoU0Q5qlENt8Ntx5WGrNnwjgv5/FsqFfJ9MY9prUlZNPAuey6BFfBtXgZac0GA0jEAisC73Q9p1ltGslSrzqSs5THRGwnMuYA+yconuiM2hf5A4KAjF4l8onsTkGer3qMs7PUud5uU1+FcRpi80/DAhedqo0dnVszgSriwlEyPdtQpyrWjFlzb7Z6aDoanss8s+bmhcf7KafSNrW57HB9val2vEpRsEO+MOCEZGApBgxqw6D/b4zzLZC3322ifEY5V16vnClK0I5FUfu8CdSXxKz/4zzsB6ej4CdBJSEdHR8dHxGGC3ag8v4/sdmsdi3qYuCzCOQbm1Qq6l/eReR0sIT3WgMK7nbLm2iyYdyrzogxD1Yd492QKVUtxNQ4k+X8yEfGOyOsUmSQwehGei3bPtnCScKtt3o76xE3hfCJy0nj1WKgdkvcaeS4DJ43c5VGnRdXsizNhuc+/Bx+jqdg3xeaFNov68RohcJ9pS0lJz6835SBDyfsY1K2J9UpT4BkVraOUQpNcHjjLava46kJ0u+I/NALv8lZg5MKvqntcXpt3Ah5ymLUgal2JL+WS75NS0L/UAyi8yyNX9zpxpyNv5MxvhXfsGXmh+5wh4h2HNY89LUzBehOLRN5w5sJazs3HugBWSVeF/T6XEeYWZp2JNrHduiU7zsyc8+jagR0XqXvpZGNoOiuaOxX1ffTPwZpF4jXiMJbHWBdjZmXHWL7H8vlwbYkR7YHAWRcjcbLjpLZ3o4wlFb0dx1q1Ehd3xeoEpKPjJ0cfx+ro6Oj4iPjeJyuHXS6uY0AEdruFwz5aNkg00nCYlP0uIZIT0HMdepiyDewqpQMSkzlpDaFJuc7dkvMKu+swbTwk0LsjPnqyqnIfhnLbHltF3VPWWPgo0XbMZWvZ60Sk/b4VtJu4O5MHtZLOh14SWtbh63ZxMsA2FNC7JX7F2v+gLaQrQbljycL8pGb961qWKbuXzck7RUasPHdjt7m63q6iWta61e1axo08vd1HtlrYeSe2XQ9/DSc9fs6PuyNSxqK8y7RIYsdIQnkjZ2ZMTH6UhTdy5ofygGJjVD+XnvNC7/KxAqf8Lgz59jGHIpaRsywo9/W1VsTt+wG16zNlNy4f23JB+DkLwv17Kzpv4QQroewZ8/sxsEVLPs5ki+JMeGzkqo7ADfkYIyOLGu1bWDnpJQvS94WA+PNVtYjSXXQeZOgjWB0dv0/0TkhHR0fHR4Sq8PLFzDgmUhKGAXb7hWUZmSYfyzJycZkDu9G0IlBJgwvTfRRrjsoQhEngEutjvBtyXq3IbW1pHWMQ1qhlHMuLcAic1YpnL3fNghbauDYvfqFqHdrgu+21rWvdQy3k9wwsmB1uQMw+WCuBScBRYzl+qwNpuwoD0gi1Da14m+Z5K57jQSll15yjMidby0OKvNW1HD/k/RgJZZRHkGzxat0Pz1Bp81B8bW2XyLsnXqzvGDg1Y0EtWqH7IuaUtd1LxXQbPmZUk8VnPtN7/t7whmMjDj/JzMzO1p7HrBTlSNaS6D2DBt7JyYiGCAcdWXJY4J7rpPay1hskyR/nazU9jGWEeIdi+9iJkZmFHRNCIOZPnne5vBvio10jY+lSgH0evatyYSYyXoUfmhZkraREhAM7VrUwwx0TkVgIyKwLbsOrqr3r0dHxDaOTkI6Ojo6PiMsceHYnfPrZW5ZlYhwjw7hyPu0Zh0SQQEywRuGy2GiW2+46sYiZqHjYYUj5yn3MKeYpB62rWQDHLfPIcJesKQhNw6GMJSmBVZUTEQ8qDNwmHvCYYGx1HF783XpcmzQO8JCsZK0BdPX1XFtxvOoOhCsy0trmlvOlJpUvJF7kELo1WxKf1Ny/7rAi+JzT3sdMjTxXA+CgA0cxb62jLFedDdNXWOhewLojR1kRhecl7q8W5Qspl8iBSUPRRLS06cKKMJbnnKWSFdecXKQKsx9ktr3SgfeS+JG8e7QfYHoKe626XwHhzMJZ3hAQfk4/4UHOvOEIcg8KD3Im6KHseUswql7Dhf+Wnr6W83L65d0rW4d3SNou246JNmQQyFqVx8nlJ71kAmt2y+/lZGvK2Twh76DvZ8C0T0jioguLLowyYvkqUzmPs84UzYfauFgnIB0d3zz6OFZHR0fHR8R+l1iWgGpgt5+5e/ae/eFIShaMJkJxxrLbeVQqVm2Hd0puidHnJlCkHeMaQw43bCZmvOOgSsnlaDsiPgrlegu/gr9uCnu47moA2TuIMoL0IbTuVSHrTtrCtl2vNutooZvXv/WaZVQtj1QJlYC4k9ggwhiM1HnXxdMnDvlPZNysYdBAlFS+VnEtR16bwF4H7jKJaHM+WlKwSCLKYyE9zXH89y0Bgeugw4Bwr0Z2jjm00PdmujG+1L6Wi7JnljIi9TvypuhY3nDkImZLmyDnpJiLljtp+ZgTmGi9db5aiETi1To848M7G0MeffPCf8lDeO3j233ZMZmon6xrURO++6jd3BAWd9qactjgQXYkt+LNmg97nHltLbpa+KBqSVDv6Oj4OOidkI6Ojo6PiHUVPv1k4XQ88OLlWwCWZU9Koeg72kwQv+3f15iF1KkSD9eN3E0WXugajyA5GDE/cApCTK63sOep2u0hC9HXfNUYTAvhI0DgV7ndmYqr+70wnq6uZVXnqrZIdgE7VBLjz3dNiJAT3JFiCbyTwKmkk9fXruQoljEo73i03RcfSQvAfRhKKGMbxqiZPMypEjK3mXU9yjHrPLx7MxFQ1XKlf9SQiYiPgq0INm4VqBqbLZlqnbVSJhtKJSWtJW3I3ajiHiWw19GIEDlMMBf2LnKvY18jKxEp7+V1UT8x0iZ9zyz8iDfsmJhZWDgwYmNariOZZeWg4yN9iHdDrCNk4qQFrkID7TyN/AwEVmJZ3y13MO/etOuLpHI+Tjpa8mHOY/FK3G5dEIoTViEgupZkdO8s2WfDntu7IB0dHwedhHR0dHR8RPziL7xnnkcr/tPAuzcvuZx3LEsoXZLLIiS17kdMzVXxZrRqGmz8aokmPH9YlPNqgYT3oz3fuiLCLj92N+TgQ5oskNxNiZmMKO34ymN41oY0heb2+7Ybsc1Q8JTv9r74iMAYIRgksMdCDqttLaXQdavdrQjeyYnrMFJz26/KnzRxydoJvy8pHGNiHwL3IfBFmrkQOTBwz8iCC85jcaOKpEJAJrVyXnTIP9sYV5QsgM/HclzyGJbv14plcGzF562zWQIWeSy0fy+X8rjLplDfMfJS73gvM/e6Y5aVF7rnjZx5x6k8biDwjANvOZY9cftaL+rfccqdhMQiMXeWprz3ibDptjzIman0xijPdW2IINnhauHUaFacGA25D9MSCIevaZIBGDjrYwG6P25W05eEbEG9sDLrksXm9dgWVFgtd3v4YEfHTwedhHR0dHR8RNzdn4EDw2BFznyZOB53JBXevbf/C77fK+/Pwm5UliglbBAqCdkmqK+pIRYJlvyANY9kTQGWrxHy4VetPbNhi5jHk6ZMItq8iPYY7fjMVgsi+AiOF3o5/wSyQD3bvma9yjFlcbEm7nIfISAcGLLmpBbtTjxaobqvwZ24Jich2Zx1IRVicCHxjMCiyuu0lpEpBd6x2BhVHrtC7ap9e677LLZ2snAWTw6lM8zrAAAgAElEQVSPjb6Fsg9wPcp2a9wsbW5Huf1G+r4nqVbCmkv4z/SOKSefP2dHSEZA3IHKrWt9r1w4vmfkPbXY906D/7xjYmLgXnel5C8hgFL3ZslWukPuKlV7XBP0W7dIiFr1Ir5BkcSO6dH5RiJj7tooWjqA7urlnY2d1OfOLOzUfnbh+0AACVl4ft318PelE5COjo+PTkI6Ojo6PiKGcWUYEmGIxGUkpcDpMrCugaSmzyjuV0myJkRKcKHjMMFpqaGFd6PwsOYk7E2NekVY8u2WFLgT1DYxvKwZsW5E6RhoKfSgWrCSf2PCdUhNUXtrrGYiPLL0vRCL69JCImbL272Eqw5OwnJD3mgNENza33pmiZ9Ddcuy0TPlegwLTP8yZdZXimVRLmpy6jZILzZCa9dbtGNPbmW8bOicr9jXfZKVkZp6/nvFkumD+qiXQshp5CLCc92V8MKDjhyZOcjEkgMXJwY+03sScJSZe504ysBL7gE4ZwoFjUNUFoCvWT8SRLhnbyNXUI7d2uyavHwoo15+rpesv1CsA+cjgKLKJMMjG97UjE4NYlqNIHLV0dJMcgZCCRQsLlcsV7a7ax6/atdkZMjsFDoB6ej46aCTkI6Ojo6PiGFcuH/+nt3+zLs3nwLw4n7hfBkLWfid1xOHSRGxPJDdWMXpAPvJ8kLOi5j7lcJ5qbqMtWEdKWsoVOG8ainkBxE0eXBh1YN4eN9Zr8kBkIMLr8em2s6CO2GNeTa/fW7bA9hmZLRQKN2HiJSOjusy/LkRJaryUiZ+V+sIj9vx2uOriDyhfMJUitQvdGbHUFLOL5k8XEi8i8JehCOxZHc4AYntz9k1LJIKARnyFfuikdHrETMnXm6zu0jK5+WF+WNsyWFLalo74in/CffYvue657lej2491z1L1kYIwmd6XwIGT6zc6cRLPVjHRHe8lQsDgU95RiTyNhOvsy5W/COMMvCMA2dmTjKzeicE6wJJ83kgj3ZttRYhD2XZe5u7hNhrDMSiUwKKQDwSObAr3Y1WWC95VM7DDAVh1qXpiK2lcwIw6/lqzwcJzBq53Q/s6Oj4GOgkpKOjo+MjYrc/EdcdEhLzZcdut7LMA2FQTkfTikwDJS/EicgQzKbX7Hq9eLOfh2BF6CB2xX/OxdUYpHRVXIzuiGokxzsjg5hT1jElBszd6kHbYtfsayeES+6EXEhX41jmJmW/cwetIra+qTBp1oOHDFYiseZRqwEpmSWOKXdm0Hg1vtRa/Vb1R31915CsuRBl83vHMdl1exUr+oNcdzRCHlc6s+RU9KGkggeEQeWaeWXcImB1HCvvRbb3VUwz0Y5ZBa1dpdaJzIMK2y7MkjsfqI2tTTqwSCzdrElt3Gqn9qf/IisXSex15KB2/8/qiAa14+S1efCfY9XIl/KQR+QmTsxEzEHLiciZmTv2uNA+5TUAbNVHOxk5505HSYRpyJyTDDD90qprduF6/BmTq08BhYBA1Y20WhO34L0VgtjR0fFx0S16Ozo6Oj4Sfuk/+ZVXqoFxd2a+3DHtFvaHMy9eHnn+7MIahePFXLLmFYZQNR7zWse0XCMi+XZMt6+gr0mZo5ZQwzF3PFQpX4W8ZEvag+TcBq3p5suNEaEqWlbcUcpJgq/Fn9UK0p9Cq+m4FcK3sLHFzR2INyxXj2/JhKsL/OtCKp2IEbl6LXf28tc4bpyo6toMbdbFQuS9XIr9qx9zULka33IUW93cWfE9U2oavJMO/+cEZ71xvHYEqZK+oRTlu9wL8G7SqNYx2DNwYS17tmc0okLkhR5KR2unY04WD+XY7R4DnJmb9zAyq41YRSJHLiwbpym38d1aKdszaghkItnxWEoSOsCSScolE6KAsOrKqvaJ9BEsyxixf1thu//O1xD18d72UayOjp8eeieko6Oj4yPgD/2n/+2rcRcRScRlR1xHpmkhDImJhS9fP+fF/Yo+jLw9WedjHEBEGYIlrZ+zBuThYhqRqNYpcSH5QtVMtFiSEY82I6TNMveRpyDCKMIeOKMcdMhjSlZ8XzZFnBe17TG9oL3kUSaHj0htdSBQGwZOelxc7qSmuiRdj30NBMuFyATCw+hudRtsrEr4IhfL+yLctuOOCO/y0Z4z8D5Tr9r9SCRRJpXSVdgSAnvsZMXu1V4/tuK9lSruRKO9fXXfZozJ4e9BOxrXjoslTEi/kNipUcFJp3ycqVgmLwj3+WeAk1iaeQCOsuRRp8h50wkxjcb66JzfpYfy8yQT7/XEXnZMDCUJfWZF8idMCEWYbuQv+M4TtZJCzUQnadPNys5XPg54S1vjoYdbPcjarGFBSRr5lV/rNrwdHT9tdBLS0dHR8RHw5stnTFPk/vlbUhoIISKjEteRMCQOhwWRkekyADXB3IXpMdn3pHA3mWuWQ8QIBDcICJhDluTpIH/INs0jkX8pdl39WRh4F2vuxiWPRTlBuA4pTLmMq8nnt+A2vK1Qfdv1cPjj3EXLNR4egDhJ4JjF4u3V9O0ElP9eAU/3HrL243qMy1LLAY55xMtJlOs3yjGlujs5WuF9OxaU0Cs9w6BubSxFu3ErfLFde/saH1IotF0gz3I5ycKdTlfOV8k7O5IIKkWz4aJyxTJPzJVqLOnkC5GX3FvXoen42FpTISKzrnWMKmPRBQR2JM65U5JEWTWylx2LRgYxS2brSPi557E0yeemlnvi+++jXEoqjl1B5WrUDmzMyvbadtA7JfUcYu96dHT8v4xOQjo6Ojq+QZz+zd94BXB4MXB/P7Mse0QSkketUhxY5pHD3YWH444g1rVYkgnR7/dKCHCejZhYfoiRkZZzlIBChAXN41VGIKJqLmVv2+m241MBGEIVhAu1a7LN4vDb1k2ox2hHbrZWuZHbpMMf78Skpq3X9Qk2UpbUzulILSTdUcu1Je2YlR+77bj479pRK082txyORgCfOxs+JuQF8ELkIpG9Dnl0SDNhuRZYe2Cgn8OgQrzR9fA1aSZAY/bX+iq0CfYmMq8FeEA4yVJutyJ1wZhpUNPy1ByPAEXQXsnLOXedEsqd7DnphSGTgwddgMisS3GkEglX+RuLLhyBvUwkUsn0GPKY1aJVXF8/C/75CXlnataI77Eg1103NU3KKOMNstFHrDo6/r+KTkI6Ojo6vkG8OQkvDkpSSEkYhpUYR+I6cnq453d/bDaon7488+XbifNSi87zInz2IvHuODKvPvefNSI5fDApXKJ1QkJOgHaxuQvPvUnixffSuGC1HREnJJeYcJcq14PY1edUSEGbn+HdhRZLLqK9aE3UcMM2zdwJAFAKySjKUVf2OSBwxdLJFxL3bgHrr53Jws/owIoy565J25Hx15o3V+cvEssarIuzcmDIeSBZeyCRU759x2ji7XxsH4M6MJWxraDXIuh6OzIxoAJyzb3y+cSrPfH9WDdr3sJH4jw8ctLAgQNHsR0asjjcHjtchRiOWQOT8mfBydNJLGk9Zvp5ycRlYGBhNiE+g3UtSCXpXPMI1aILIoEdE3/xV/9MKfj/jV/+D18tunDR+arD44+392DGwwsvemGQgUXX/P7lrBJVRKqOQ0llvOq/+dU/3wlGR8f/T9FJSEdHR8c3DFVhjYFpikiuQOMycj5PLIvlg/z49R1LNAE6mOPV1iVrCJBizf2wVHT7viQtHYuEXd1uecGiphdYboxsTY1F70Vzca9VkO7jUJ57AdfHuJVv8dR9/t0JClyTEvLRo5gF77kpzt+zcJ8DCqvZLuUYu1wIH5vCvT1uux7XN9zKL7mIBRJ6h8PWpFyyiLkI0rPTlHc8JgIvdPdIjF7HyoyIBKnnu+SOypaAOLH4ENzdycepvIMTVBg11LG5JvTPMeVQRb99+/imvTgzX3UoziwIYkL2bH974kIgsJeJlWjJ5Jv1/5e/+ucKQfhTv/wfvHJtRz7h60BCMYveUa9dzHysyjscAL/yg67f6Oj4NqCTkI6Ojo5vEHOEhwu8uLOfl9lE6efznuNxIqmwrMJ5HtiNcJqNbIC5Y6UomZCYtkDExrR8HKstnzUTj6TK0MSs+2jVo+TtnBfSQskjTXlMx+EFL1RhuCM04zDaEh8SonZfyQvR+vitW1aU6+OeZGVWc3Hy1zxn694xF/A+vgQwSeBeR9Y8tKPATCyFeUsOJq3OTNYNUAYNHHOnw7UoXrivOSE9Zr2EOXat7HIXxI+1JSCto1bN+6hkTLgeHauOW+lJW+Nb5MQTzv04Q9HdVPh7dMulbGwsgR3e3ZoYSSgPnPP9wUbRsjeapY7ba+/ZGWmRgb/w/T/9JDn4r371P/qzAP/aL/+7r/w+73QA5YPtLlgi0rscHR3fcnQS0tHR0fEN4rwqL/aCBFjXwG//o59jmiLzPHC6DIxj4jwPJQ09JtjtLIxwGpV5DYyDoirsp8RpFlSFmHUjMcE5ZrcosU5HxAlEHb8CCuFYctfDxex+exLhrFqyP9qCdMlX8oWaddG6WAUGVlITNJi1BCKlaB0IUHQXykWi5WlklHGspnSOInyme96xcJaVH5G409E6I1nr8FwnnsnAi3FgToFnOjBr4qiRH+Ur7T5SVDoQYgGDlhth2oMkMbs/Lezz6NU5Z1IoylEupdguV/CpxGPQUNbvY2buoFWSxL0bgrBQR7pcb1JGn7SOQW2xDUAESlZJcTyTxy5aDh/J8kdrs05fj4vgL+JmxKl0T/y4F2bL2dDEneyZdWFl5RN5xp///r/ztQhDJxYdHR2OTkI6Ojo6vkEk4G6nvDsGRO7ZjYnzZUdMYgLzVAvKICYp3o1GQFSt6+GdkXFU1lNgiTZhc1qVKdiVdFU4p3TlxASmCSn+P6o2ZpU1IZYRIpw1kZKRmFnbGLna9dheRXcBuo8QtZoQdzWqdrLXBW5bIO+ZmJtOAlQRsgfwPRRBtHkleZ6HH28kcD8EBoG7QUirspPAuhk98+7GPpOPM2spvqsg3ZyxjiwcGFkbobntxzUx2I4ctcnnWwvfm1kujQC+7aLEfDuoXAnXff+gkpGxIRORx7khbRerhXc+1kwwwIigEyKwDsmyIUMxvxtjLhn89YIIOyb+4+//251YdHR0/J7Rwwo7Ojo6vkEE4A/9wQc+eRa520dUBRFlXYXdmFiiMK/mfBVTDg4MEJrCU3IgYYwuxjUxesK6IBbUZgVoUi1fASmuWQCr6lUhvJTHetZDKuJuDyHcWu7aiJKLza2bsd645t4W5+24kWsjtmiP0GZoKMp7MRclH8H6sZyZiWaVK0aAlqTMSVkT7EMgZW2LSvu6Jqz3UEGoHYo9IyPCPud/rJI4s5Y1bNdqxGu4Op8o5uVU3cTqOFd7jFgK/Xq+W2G/40O6EA849GMmsY7Gkjs6ayaBW6Lkz22Pfb0+zR2tOpJlva5qh+u5Gq5xcW3I1UhVR0dHx+8BnYR0dHR8p/FXPv/Lr776UV8fCfjx6zt2Y2JeAksUTueBeRViagp1rV+q4LEISxQu2TErJRgHZTfAfjDCktAygpVyl0OxxPNFlXPT/YjAotf5GJE6piWQ8zH0UeHtmHNxe5FYCnwVG9tx29kt0WhtgB2ai+OVhMq1lsS7Hj4mdJSFdzKzkDjJQpTEWYz67HWwoD2t3aBjSpw1cWoE32B/4GwEyxO7rRs0qblGrXimiHUPLrI+SUCsw2BJ3rEhNWdZmGW97mqQio5EM3FzMucdn1vJ6lA7Ik/9bs177Xu+TW2HD5GNuqY13x4JRYuySMzvg51xOybnWR1+G0w78p99/9/rXZCOjo6fCH0cq6Oj4zuFW6Tjr33+g1cR5Y/+4Jd/3wXVL30iHM+B1+8GhgB3u8T7c+DFXSIm4c3RCsS5aQ6cFynhbO041mkO7CcFTDj+fjYC0aaku7A8KcQUrdvhnQi10L41ExdPl3Z71ytReB6RCtRgQv+uUrMuEvVK/C20uohCBtTdqiJR0lWRPOIp7MoijVVtU4zvdSii8aMs3Otz636AkQ9N2XUqFc1JlFQKZ9e8uCYk5iGrRSLvcvfDx65C1oY4vGyfMoWp41k56FDINrnVtreMOuVzOLNwEPu9ooVobMemXD8CXFnr+vsyEBgbHQlUXYrnprT6nIFQMkViFtrXHJXaeVqwbspORyNUDckK2MjVmWqz665WX1cH0tHR0XELvRPS0dHxncFTXQ8vtn/j81/9fXdFfuud8uP3FrA3r/DmGHiYLXDwNAtrskTzFkO2pboswhIhJiEI+bZpRzwDxAnIPgvLwe11UyEa3onwTA+zXrVSf1Ubw4Jrp62WfNQOiT3Oux/2nHol3r+80E08DuJLaE4cr3oT/weejVGzQ6AlJoaTWNp2KwA/p2RuX0j5QzaT2OVsDi+2Jw0sJN7LzGs58aWcuIhpQ5wAXFhwHUwlKde5IwMDsTlvqMSlHZMCSuaGfy2Syn1b8fmF9Wo/25Gu9vVXEhfWq+c7uRu5TqOvLmAfzhzx4L8p2/4e8xick9QtWkvdrgPp6Oj4/aJ3Qjo6Or4zaKM02qJXqbPwTlTcEaq1Tf0XfvDHvrLw+uG8Mi3Ci3ng+eRdD+WLB0/+ttGqSyYYQzDSEZOyRFvLZYHnB3vOYbKRrQDcjUJc6trnRvPhFq3lyncz+gPW6QC4Y+DSaAcirmswIiP5Grlne7REpH61e+qvR3mc378dC0pNJ2XSwD67LynKjoFZTYFv2oQ8IpS7A17Av9CRNyzsNJAasuKkydcc81kEEQ6M7HTgt8I76yiossjMpANbPcrSdCDcqnbF7GpbW1whlG5SaAib40Isn5xIHScDmn2v42zemfhQWKE/z3Utvr9rfn5NaTerAT9S6bxIKp0Th4vSfVyuHbkaqESmJSWdgHR0dHwT6J2Qjo6O7wz8KnortParzH4/bKP5Kr6OfiShvNOV31pn/s5p5u+dFn64rPzDY+S0KIMYAdkNRkDGAMcZTnPtfiwJjpdAUktgP83C+3MehQmmDTkmc7Vyd6vnYeDlMOT1e0FbNQ8LiZnEe1a7Op+/LtSuiIfdLbkw9Q5I1R6kRyWyi7r9q92H9vYi17oFW591QbzL4UQw5GK/JTwuVl+IPMjCOf/monYOr+XCURbeyoWzrGa3Kytv5Mw7uRjpUSMzs6ycmTnLcjV65cdfiMysXFiYs2Wv76t3LdpuiSCcsy6kfrlmxI5/wV4z5o6Gd13avfg6ny3J/3abwEHrNNUxry0BYfNze+9J6nouLOwfRUNuggU7Ojo6vgH0TkhHR8d3Ar/++fdfgc/4V3ip1VrS+u1bIylfhXMu5vZNVyIp3AlMg+V9HFdlNwj3k41mxWSjW4p1SoLaiNaaTNMRkx1jGkAR5lxBzuquTVLSzwNw2YzleMFpxKN2fSrxqgTA4XtQOyFfD7677dX9+rM7SVlux6Ahu0pZsT40Nr9+W1EemEvnxs9rQDir2et6yvoi16NMNd9i5YtwAq4F1t71cMLVhgfaucfynFbU7WnlE6O93uZj0nZnWgepdmTq6+1l7ULVvBJzqLplJPB1P6/eEXFXL6ife+u23M4rgd4F6ejo+ObQSUhHR8e3Ek46WmwJCNQRLb/fRpLq41sMCP/L57/+6i0LC4l/6Qd//FFBlrBci4lAEJvXj1ihvURlUViTvaoq3E3w9mxC9TWPZ51X5fXRiEWKsNsJywpxhZiqbuMWEvaaVaehHBg4spZORzuKVgvjcFXo6+YFQnN8JxPb3XTXpfaK/PX+Bcac+WGBhmZbG1RAQtOVqsRnInAUt4G1kbM7HTmylvyQ7f7XMak6bnbJovgqNLcAxBNzOY+nHMJ87eumOI/EbGXrP6er3yFVz9He73tc1Szt8FTF2nxinYQktIjMyzGbMasrN6wPOG05FnHiYeN3I4GFNa/R99Hw577/b3UC0tHR8Y2hk5COjo5vDX7j8199detK87bodpSCe/P4lpjEUgTaY983V89/7fP//oroTNm9yPHJMDCIsCbrUjxEc6naSxagC4wDfHIwIrKqcl5sROuLc2IQYRA4r6YrOSdlFywlfR8Cp2hrGUUYs2uWEQTJ32tx/RzTUrRdEKhBeL4JMZOodq9qp6DdIxuj0s3efkgMvbV8vdeRuSSG530TO7YHC45qf6ZabcqDLAy6I6C8YOR38qCTw5PcPaRwkUjbofHvAbnqfHnHo33cIY8hbcfLfMP2Wa8CJnBvsTKXvfLj2ykGfBgwbdalm/VZV2286nJcWJlkuLqvJSa35qxv5Yfc2pOt3bJ3pn6SrmBHR0fHh9BJSEdHx7cC//Pn/8Mrv0Lf4vZVf8PtULenx1yuU77l5rFWlGcI9yEUEjENQoo0JKGmn4+rdUGGPJcUVUnJHrMTmIIwx/o6c7LAwXPrhKU2gjWJEDVw8Q4Dkm/LlYIgNvtQRpNyl8DJwJCTu1vXrNA+X66fD6an2Fr/buFC6IQWvUSbCO4FsDs0uV7B93cmMhPZE9gxsJcAWsXYj9+nwJhdtFwcXu14r9fa6iD8GFP+M3lhKWuS3GHx498KY/QCX5quzNC8C+3+t49tn992mw46lU5OyroVF+z7mmjGrFrcJOale1Lf41RetTpneTdp23Hq6Ojo+P2ik5COjo5vFH/7X/mNV892wsOs/OP/3R/96OMb/+vn/+OrayerdOUQBY+v9LYagC1uCYR3DHmEyErCW89tNRXPQ57fV+UwhJxyDkPIRV0OEyTBabXnneacAK4112JOtfsQqInnkwiL1nXaWFMoa9jnUScfnVqyjqAtzlfSVWBesZ0VBW3Hh9qr9bUDYq97bWNbiuHN3rSvM6k9alTvttTnXyQWS1u/z/UUAxMeKPhaTvxSfM5E7grld6YN7dvn812IDAh7xrwnyoJrRezY2zEsH4OaGDgzM2QXrjbTw+9zc91b8I6J5t5a+7iWfNTP0uPxNt9nH63y7k3bdfKRqWhs7JED1lOIzT47OWs/1+173zNBOjo6vml0EtLR0QHA3/qXf6OMFv0Tf+knIw+/+6//5quQsyuCCH/nT/zmqyVrGAbhGyUlf+PzX3/lo0NeXC+5UFw3xVQRHXviN/7dijBRSvr3TWi1sHUU9yiqPiPkMagv0sJE4E6tG5L5BgdxhyvhnHKy+VpHXS6ZZIwihXCMKgwiHIJky1Q4aWIvpodISskIGVAGsS7MMZlw24TYdiV7l8mJ70n7vS2y21BCdxRLogSV4oC1HbtyDcG2G9I+KqgJ0lF4Lxee6z6/b7EU1RepmomQr8V7Unlri/sP5D2f6YGXTJzyiJyRlshE4KXecSGySCznFVQsVFCcZjwWo/te+LocHiwomQQ4FtarPWxJbnusp8bUbr1++9oWkii858wdO9wdayKUnJCTLMU62N6/p13KqllBPX//uV2jv8aZpROQjo6Oj4JOQjo6vqP4O3/iN18BJH1cADkh8ZLkj9wgJX/zj//VVy+mQExwScopJn7p2cD9lLMwRnN18hJdBN78qd98lRQ++6//ud9XUfPXP/+1VwcCBxmIquXae0sS2rMyMe9jXUgp0G6Mu98iMa2F7HaUZ8RsU10UPolwPwTOMRGxjoRIM26jpgEJ0iSUN7kfkwjvkuVciCpRTeCe8BEf674MApdMki6aeDmM5VjLVZFJFh/X17t1ztsr/lADCH0869YYWttlko0mxWFjTLHs92s5ld+5U9ZCZJ9dpyR3D9ri2cefkihvufA93RUjgPckjnLhmR6Km9WKZX/sGFgkMunAnokjl0f6j22RXs/1cXjgtpvW5m2Qv7fjVe1YVqu1uUV4WixE7ooqpBkXU8n78+ExqccaE8p62/dn283pGpCOjo6PjU5COjq+g/i7f/I3X331oyr+5h//q9cC7JzWndTyLqy4DiwJnu3MblYEDgcQUWIS1gi7UZlX4R/9q7/5ak3w5Zz4p7//z38tQvJXPv/Lr0K+OrvPV8Q9Pdw7G3Ctd4DHBKTFrXGsxyMydZxoVCv8pzwO0z77BRMvhmbmP69tziQpiHDK2o67Qbj42tV1HZpHvpzoNMfJnZEBbGOBc0rMzdX2HaGI0wV7jwaVXBybO5SLtSOeH1L3wQvaNe+m/9yOS9m6KhHz+5x02OMoxa4FKCqa9SVP7XskESWVIjtRgwfb5wQs6wRMnB2Y+F0uPGfkhzkXxHsclyaXxDNKVizLpA0k3HYA2lLcCUhqRqDs0xYePffWXnmh7+cR8+7Uz9jXs+09M3Ngh+en+J67RqTuTyNU3xCsmD8JLcmydddjtMJ8f0zvgnR0dHwsdBLS0fEdw9/9k7/5Koh1K8CK41Ufj84Apci/hZd74RLh03vlk2eRZQl8+SC8v5jt7DjAi7vIZ59emOeBN+8mnt+vvH43kdQeN8lXX239a5//4FVb7rvG4YIVuSNSAve80HQk9EqP4PdBHZ1pLWe38BL4wsrEwEzkU3bFXhbsov5I4Od3I/vBOhM/PEcWVb5cVw4yGGHDCIWRk8Csls0RRJg1kafWLDQu+ZqU2JCtQxDuxbJGolrnA1yrYZoRJxDPQ2BQ4U3yfJBYAgp9H/Y6cMnuVCkTEHdZUq6zQ5x0+LhVu59t38S7RAHhoGM+/sqJlSSx6EHWnOnhRboX+HtGLhZhWNbtXQTvjoAJ1O/ZkVD+fnjPAzNHLvZYUd7phaEQq8SF5UoYvu1S1NGk6y6P3xcarUvbUWhHmqCOd7VdCv9M3iK4W3ex9rUdMyszKztGnnN4RDjGTB7cLLglIG4jvGbK40L7NgtkOxK2PX5HR0fHx0AnIR0d3zEEoRS87SjWLQLiv95yBRELzjtM9v0P/+Ef8vbLF/DDZ8gxsOb6RoJ1QpIKu13isgwEMSeoIcAnO9ONHGMqpGjROpr0Ru2K9dToPJxwmBaEq6LayvtWj/D4KnObQfHUubf31yv8yh0jB4YSjudYSVyicorKZ7tgWo/c4VjU+gxJbe8HhEuyYnon1o0o70cu/mb1oEBtROKBT3pn2CYAACAASURBVEMgNqNXE9J0BlzfYo5HpwR3IeTH1BKzdgdMm2HC7RrGB434vFnXU8Xy9ue2Y7JnJCnMshbi4d8B9tl+98DIpThWQZKBmgxy/e54gX+UC/dMREaOzFzEPiF37EkoZ5kRQhlkMjF6vDrO0NDPp0bUfG/cjcqx1Xhsye01mbl+3HaE6qs6IY45J60DXJpwx5Yg2uvVUTHrKNVzbgMQ6/qEYjpQujtfT9ze0dHR8ZOik5COju8YkpoeIeUi2UXTbdMjoeX+1Lg1DSJGQIIwBPjsRWQ/RTQLZMdReX7QnAAuaIJlGRiCMgTldA4s0V5rEJiTkZEQ6+hPyjqJQYSglSBsZ/CXPCbjV2x9lOlKaCu3i71bpKOd4b9lmTpi7lMHGThqLWYnLBf8i2iDQC/SngQciQRgxvQcCUtCP8jArImdBIIYIfG1jVL1FEu283WSEVWJOvAQE5MIewmlE1LOl2oBrGqaHNdoeMdmJZXwvypgNietVvvRfm/3roUX4tt9s5E4KZqPtdnbdk8PjDxX62YkMVF5ajoISuI5BwYNnGUpblZOmI4sICbeTjmTY2HlzJw/F7Fc8d9a3l6akaxbsDGlwDZJvu61Ff/bLseHsO0afdVj4WltRh0Vs8e1eSdb3Y6PqAlSOkuxeawTMNfggJHTr3NOHR0dHT8pOgnp6PgOoXWrglqMq16LolvUMR/TMuwGIQjsJ+WTZzPTLvLD3/4e9/cXfvZ7R378+o4UhZ//3sL748Q8D+x2tWj358ZkY0VrsnA+0Tp2FDAHqXu1gn/dEIzQFL1F1I1elWtPdUG251hGZEQJShnnASMNEeteHHTgOSMHsdtHVgSzgnW9xTMZ+e154aSJC7GQt4R1JBJw1GhrVTgmGKgdEMGcrc4pZacvI1lHVi7A319sxGhAeDmMpOQC9FhE6AmzrAU4a+R9VnkobmF8jYRpJrb71OZfbLUGrXA9oUUjs17t9/XIke9plHrl3lLMz+zynyK30j3ns0kofyA9Z68D/zC8axLIL0SUL+Q9Ryae6QElmeYjk4Nl03EQ3Ka3kpLrfWhVHdd70T6mprDX0auv6mTcErz7bVuLx0tWtKT7ev8j7hrm41aKEJo/560O5CpBnVY8X4mZj9htnbM6Ojo6PiY6Ceno+I6gteB1qF53PVrccs16vrMOiOPhNDFcRsYxcTrtWGNgGpT7FzOHw4Kq2dMuS0BViCkH+sV2Ht86KyGZgNktfn08a0BYsPGl4jxFyIV/LdR9Yv+pK8dPEZCrq/2ZiLQ/g2VbjNmB6SHVsaWQ1+Zk45wJxkIq4u+Uy2ny6Iyfx4IS1Cx1/XETJvhfREoGhu2BjaMt+er8hBEVRTnIwEVTvoZdu0Brc9V9xaxvV00blzCDW722e9G6NbU6G+9ktNqQpRng8W7DkEmXi6fbInfQAFJHziY8vXwsx71gwvQ7Hdkz8InuUZSTLFeF+YWVRR7K+ykIzzgws2RtyXZM6mm0x92SAqjC8raQ/71gOypo6/L9uU1Enl5DJUJ54O/qPPx7q01xXYkfeynvlpbuVXtuXZTe0dHxMdFJSEfHdwAtAdl2P8r9jdPULQiwG+Fut+kuJEGiEKOwxsCL5zPTtLKugZiEyxwIYo9LSvlaoo1kqeSr9KMQIsRcEK9qBZRb0kItxqGOkbSFmmBX+2eu7UcjNcDwStvQpH67RiA1GRk1Ndu6HcJUQgDrsfPz8nGcgPiVfu8SWKDg4y5OzIWpjWIZDkEgBRaNRfPiV/cTifcoU7a0dRLWahqWfEa7TUDhRW7bwFZXKu8HPM678A6ANPetNz4tiprmo4QR1j0e1fbXx8A8tdv1FguRi0SWTB7WvKIB4YXusrNV5MDEMY9feaigF9y2T6aXGLlOM9+GK9Y113d0x1R+f4u4OAl4itC0x2pJRUvEHhMQKT/7c54Sh7ciff/Zzm0tLlx+jFuJ8DOus7oWsdvr185iJyAdHR0fG52EdHR8h5CANV0XT+6M5eWZW7wGqSWUAKMIP3pQ5Ai/9BJ+7rOZ02UgReHhNHHYWzGz263M88g8D5wvA5c5cNhH1lWIychITCZoX2IWvSsseSzrbrAxrSVa1kZQTwC3K/vW7TC0xfcOsz69y4X3JVOB7ZhQksdFpRdqsiks20J8JfEPOfIL3HGfC7jXXEoXZs/AQuJM5CSr3d8cbs/AgBEHKwZtxwe1Vx1yF8SJ4V6ERQKicJDAF7rk7g/F9WkgcM4z/uS98f3w4EQnKYMKiI/wVNF9yN0lyw/x6+mPC2wnJrc0EnUvtYjO1ybhu5AWqdqEdo2tqLp9PxTlh+GBo+54qXsmHXjGjkECx0Zu/4wDRy7WPSIyZzezkaFoP57qXrQWtX6e9n756FbV7LgQ/KngQYAl64UGCY+6G7cE/atG9rJDScy6shP7s/wUGdHyPoSr+4Ci9wgN8XuqY2NJ8MNNsvN1hfIdHR0dvx90EtLR8R2Aj/VsJ6xW9fTwCiccbisL1rGIamNTSZW3JyHpjt1YBdC7MbHbRc6nifNl5LBfuczmlOVdEaA6Z4npQ8AIUBDThwxBSMnGlM4pcpDBCi9NzERzl8pdBV+fh9V51+CegQsDSWtRfAttwbstxAJCyPkWCbWgOwbe6cqz7OYUxa4c77W6JnmRnlAQc8dyUbtfRR+AXc7riJkkLarsch7Ikse0ppy/sg8hi/cpxy4CduoV/qpZUBKxkKVWX3CrgHYi0O7FU4LnD6ElF08VsiupXIW3fU1MGkqux0piYCjrfBAjG3vGUlBXcmgF+EGnEqS4YGJ475j4Wtyq9qvgugnLIZGm2E95bytuaUiKOF8TKpZyv33d7fNaIuTfB26TFv/MBuTqvfR1tsqWrSXw1uFM8r7QuH51dHR0/LTQSUhHx7ccHjToBKT4IWm+Iu5X3kNgEOtI7EPg2SRcsjBjTRZKmFSZsjD9R++Fz+6y1e6zyBKFhzc7ROy1lijMK6gKZ5ucyboQIzS7wca7lgjk0Swv8Na8yJ3kq8Faiysv51p71btcRLk0PVLF2W79uhXo2l48fcW3LfTmxlHovSxc8tXuEoyHezH5iFXNs2gft+TCzzsUEbPW9XW8kIEvo125/zSMvIkrewlMIhwIHPMcf9UtSNbMuFvYtS7kgo2GnVhZpI5atR2IdKOUNt3K44JdvoKEAIzqsunbBNA7Ei2R8D1zUbnteyj3HWXhh7znue6vtCX+fCOhw5Xd8JSJzDP2PORPwW3rBUPboUm4nqamnLdjaI7251Ufj7pt98vHrUpnKD9n1urUtWruZojtYnV/c4vqmsWyTVlviUi7xqe6Ie4cFrk9ptfR0dHxMdFJSEfHtxwt+XDi4UhNJ2QXhBeT8G5RPttLzhMxovH6BM8mmKN9jQO8HOD9BZ7v4XdeD4yDJaJfFmEc4O2pFrEPF2Eaah7GEinlWUwWbjg3C/POi9OMkNPZrXj3eXf7X9db7EqpSPn9leOPuzNtxm8cntDd2s160ra7R83ASx3KuJXtq5bMjYTZAlc/Khgl2JX+hiRsXaq80P0yrpyI7Am8i5EHVmYNpAgvh5HfiieiWOflnL2P7nNuSYuRwElWHtToz0XiB0eI2uTtLbb3b6/Ab9Fa8d7qiAwEZhaEUIrfs7uRIQxM7HVgkbUQhxU4y8o+/8myPRyuxPH3OnEUGDlwoYrXn+uBRSJn3ElKr8jBUwV6+zkwotRqJ9w2OT35fMVGrXy8KjZrvUVYts9dNDKIrdbHpqImkEq2fS9aQuL/fbiL2VcRbs3HFYRBejZIR0fHTw+dhHR0fAfgY1ctnJDYqJOL1KUQgN0Iw2qdjl/4JLvrrMJxhuNsxGEI8PZs341gCEushKIQDYV1qbeTWrfD13S3U+aTZ5HYqJbkYD9piNOQSUfJLSljRhTNSDk/rAvShvC1nYItPLW7dZSCp0eLllzSVnvaxCwxC6Kvi0QnKWDfBjzTo54HwCkXySZ2r8GMD7qiaSjp7wllRyidl3adUcx1K2G5GzYy5pkWlYg5Hl+td0OAp66ep7LnH8ItYtN+CrdHqP2Tahvr57onMBCyE1YqnY7UnE+r+dgzccxU5LU8oHmvb5kTbNfQnmcVoF+v23UfH3LI8n2N+Dje8ETf6TZ8zfb8sdzntKSuM+L2vGvuwFnX7ToP5EPrbB20FOUvfP9Pd1F6R0fHR0cnIR0d32L8H3/sr9goVv55bRywEtfuWJrJwX4Q5ghTTjWPCV4+j0yD8vrdyPO9cJqry9VhhNOaXa5WmjDERvORdSO+hoD9PATruIQ8wuVJ7GsSO14OLgzAgfCIaGgpUKtDVCDnmKiNLwFXjldbtDqIW+FzcXPl3IXOpRjWLFYWc4taWHmmEyqU8aIVu4I9qDsZXV+d3jGyJ3DC8z7c1Nf0LyuWkO7LsDUadalxg/X8LhJzJ6cSMDcxDsBF6khX0OuRHe8IXbtH1WNsHbFabc728dv7licCAvdMpXMyZFetbadj0vrOP9cdR1nK+JWfb0I5Z9esVj+x3Z8tih21JHa5wG8LeCcOPkr1de15286CqXTSTetrgJXIuNFmCIGoa+5+1OdtheqOkWvh/HZcq5zvjfX7fZ2AdHR0/LTQSUhHx7cYt8hHzJ0PRztQEnIH4rKaNuRuMjKxGxPzGviZTxe+eDPx4g6Ol1zgm/6a86qFVAzZdndJlVysqiX7w0nQEq0o/gdv7N4pWCdlN4CIcE7KIHAQIerAm2TWq0ot5F+GkZ/dD8xR+eGyWrBgDvw7ZjVG2hRw66Oy2bMuniou6/1nWa8KVE8fTygv9QBAFGVQYSKwZ+Aomq9SBxvXyofz7BEfAvPsBtd4DM3PLUmKkniv5rhlnRV7dkRBE0k8HTs1ov1YRtRGDUWvkcSK6nZPtkSsEg99RDq2LlvgNrIu5PaC+OmEckG4U/tztGPky5y0bj9PV489qKWsC8Iq1hGZdOCNHIv9rL8ffg5P2d066n8B1Qr6wMQ7zlfHClf/tXwYPuY0SMCF5zb+eKurtpbvU/Nn+aJGqI56LvdFTcyyMObhwaiJKJEhk7bb53WNNkXefrb36L/4/r/fCUhHR8dPDX0AtKPjW4xEHcV6ioB4psWiyrtFWZORhpiMjKwR3j5MqFrmx4v7aEQjGNloEZMdf0m1E+IuWKO4mDjfl28P+XdRYY5qXYzROixTvj/lcyl2swj3MjAgrKp8coCffWYhgL6ks94eQ9nqAbYOQl8HW+FvOxZUil/xx8KdjuVKfkJRMaJSi1s4ZhLko1k+ZuUp4ACiFMF4FFvFqeQ+BHa0V95zxyk/d8jibXfWavGhLlEbeujHGR89n3IejjF3iFyjMNA6iHm3oQqpfSxor77GwI6pjFqZ2YCV3gHhTifudcekQ3k/tonfflzNY0pfJaxPqswsT47gRZLpMr4mfA2LxqvPWostOVtYSxZOWdeGSEdNrKzluV81chUbgt0eRxAWjR8kaB0dHR0fC70T0tHxLcX//sv/0ysvN9zK9BYB8d+vqkXEviTTYaxJuJvg3UmY18BhUu4OeVZfc0cjkw1POp9jfZEpCCJVkB7ISehaiQhkQqNVL3JaMnEJkJKNjU0i7CVw0cTzMLALgurA67jyt75ceTEO3A9WoB4G4bdi5MJazrGFF5ltUboN5rs9snKdHt6KnMc8FnTQsR5HLCtE8+8DwqwRlVowe/djbYrENqQvICySmD1/QoVV3KlrtqwQNevi1tHKnbNaYrRsroC7DXHIx/PxsWoTG/DMiW2hq+Uxggc1+uO9S+UaHj8vu29ogvJiISSu83gvM/c6Feves1g3y7U3/rXLKeqrJH436z7cMa19b9fcDWvD/T4kJk9q3aGVeNUxaAX32+d/qNPi3Y904yU/JBav++bapHA1stWSoaTK9uXbY6dyCeL6c+26k4TyF//yn+ldkI6Ojp8qOgnp6PiWor2W6tqPdphkK5GNWFdiicolqoUIoogElgjvZ+FnnsE4ZF8qv9KfCcVuyAJ0rc5WlzxOpVoJiWCBiVJes3ZIIvB2sUJ4EDiMwn4wq+BTVA4hMKjwM3srxHYDvHuwMaBTTDwfA/tB+IVPlP/zd7KN7hM2sVCdhDyx2/Uerp+4dX3Zk8/b43ghN+ljR6WhKfz2DOwZisWvHy/lK/0rqYjPwYrLKji/LqLXfF9QG8/a6XC1JseQxd72HmvRxwSESU3wveQRrpqLUTUrgnCWerVetZIMx0i1sg3UzI9bxX61CrBj7HUkifJc9/m1Fl7qPV9wZGJgryPvZWaRxEGNvlwkEkilazIxMLOU801NcV3Hsq7F+KUD1vy34eL293riXg5XLlZum7vFlmRt978VfLfPSVf/NT7G1jbXH992SUImtoOY05hb+hrpeqwH2ZIeP/dOQDo6Ov7fQCchHR3fYjwOK7smH+0V3lkT72ItoPYIgxgZyLyD3QiSnbDipkL35HNPNY96Pa6VVNkPwnl9XHglrdqCiJGmReFZtuadM5kJeIAf7DPpGRHuQuBuyFfkFd6fJRf3NhaUREvwILSZGrHoFiauRcEfGnDxtXox6yLyXc6msOwKG/9ZskvUSLCODuFK+C3lPXm8L+dsDezjSGtTvHvOSBLTGswSCSrsc0jjRepV/LUhBEEFNzNuU7f9dezYRlxujfms1JExH5MLXBffQaWQJoeSmBiLSsfO3VYSiWXvXurhqnNzYCrvnWeQLBIZNPBaTiySihgdKpkwMhIfkRHfk1u3WyJxq/MTnxCVt/CO0IcIxlcRkFvYkhkX8l+v3YIHv07ux4fshTs6Ojp+GuiakI6ObymiKmtO33ZNiI9g+T8f0/JC9ZgiUdVSnvM41BCs4wBwf4gsi7BGKeMl7ZjJmP8fRSRb02YxelQbTdo3db6qFXvnpJxWLQGFpRAWeFiUh1UL+RCBl3vhYbFzOozwYhzYD8InexvDelgTv3tUTrIWktU6K7WjM5FURpRaUe/2Sv8tlBG3psBfcicjUEddlnzt3wvhPYHnjbbhPguM27WZS5SJ0C+sRTQfxToW1s24XsuKcs75Jd7leDQ2lJ/bZmB4X2JbdF+J+VXK15gLfBu9ctcr01ws2AjcLf2F6z/awnliYM5dljdyvrIt3jPm17CxsqBG8lzI79/9ffT9/VAuxlO6DF/Xtabk62k/UvNve371Mdev+U0V/yu141RJSjUVcHH89rz9vt+LDqqjo6Pjm0bvhHR0fAvxNz7/9VetFqLtfFyPYKXNNdOBnxsHXkzCECzv47N7m2dfz5JteQURxYfQp4ZYDAIn6pXxRZUlJYIIz4Lw5lIdslata1k02bgWUvJBBrF1z5nl7IKRjEHgMNhalgi/+EL48gTnFV7Pif8rPnCRWEaIBs0dkjyG5J0RP38nE6ZPkPIzfHVCeDuKMwBnWfIVeLLGooq6W53FRE36/sd2e/7+PPOAEYiUMz5GguV8oGW9V68rqRSgZZkaOOXj7Bg4s14VxUMmJk7OfKTLbV0HxvI76yBUHcT2ipVrMFJTBi83Rt8crlepYXrXwYGWaZI4sfBSD9zrxEnWMiLn3RfFuloPMhfi4eNzLQHxbkDrkvXo/dMb+5px1qXcflLvcePYtcN2nXZuZ/50/srvFbXLmbhoyq5aiopTQKkXG1yTJdfPBR6J4Ds6Ojp+WuidkI6ObyH+2R/8i1cz3h5qd331XMs4jX+Nudj83jPl03vlboTDTolJ2A2g2ozcPFGf+9y8lILHir0lGgER8S5CdciCrCfJ1dKQbX4DlPGrReHFHp7t4WeeWRdkjjANyvfu4Rc/TfydeOS9zFwaa14fC6qFthWClywx11zkJ6ldIv/51r/tvm7LbssMsY6FBfY1vxPlHQtnIhOB74WJf+oXI39gnHLiO/w8B54zlQT4YVMAe8nfdli8+I6SOMtqxbvYvUt+xpozQ/zc2vV6Id92dvz45k51nUeyfV0PN2w7Jk/BX2PKXmajhmLP6+TtnNfv63McZSl7shC5SCxZIt6dqWuMV85TjxzBvsZoVbvnW6H3beOC2oVIpKsC30nRN+1E1Y5ptZ0PqOdYyYheaWDa53d0dHT8tNE7IR0d31K03Y/r++so1vV99v0hJmIaGIJyt4O7feQ8S9GFOFxkDkZI/GrrboA5CsFDCTHC8S7PW+1DPdCiTjq8A+AiZ+t2DL701QjKZZUccggv7rTklXzvk5V5DsVu2MkEuJBcyrjVWdYrl6i2IPfbpcMhj8XGodnOlPNA2sdsi90oCdRcoyQ3LU5E7nPXQ0T5Q58EeLvni3XgD0wTv50vwh915bzRs7Tru/7ZR6JScZJqzyGS0LajIvW5bUfnlobCzs8fXwMK206Gn3v7eWvHvp5a+4GRmNfu1r+X3MHZ6nQmHZiJtu8YeZkYOMuSj2sjeHa8kHtS16/bdgjatX+dDsVTI17t7ZaIbJ9zq+ugm71v9+yrOnG31ubZIS2pDDffh69vNdzR0dHxMdBJSEfHtxB//fNfewW3Z9HXq6LJ4EXnGxZI8FvvAi92VgL9kz//lml6zo9e7xhyBT4Nlh/iI1vTACnXV7uBkqZer55Xhy5NqZRWTkKkWcs52vjWEOHTveWE7Abhi7Py43NCFX7uzkriF/crz+7g4TTyt38M72W+coDyK/aL1OLPx3t8RMvxVBGqVCtj4ZoMBEyArZqvxKtneGx0CZIIaoL0u6awVuB/+78D/8wfnjlME68fdpxW+BkdiQqv41BE9Ci5W3Nbu1FfC0StBG0fkzYFacp6glYLsXXHcnhqfMjnplekKJMkP+5mf3xRKXdXpiYxRJv3yIX+VRQubMP3RoRLHh876ISivJVLWfOYBeHW2bFeWy3y1ycJ0zepjXjcLYtP/g4gatstgVHG3NGIDFLT27/qNZ1UGEl0ewjyfZVcfmitHR0dHT9N9HGsjo5vKbZuQE5AFMpXomZSOBaUOSmnBS4RlnnicFjYjco4JIIo46DsJy2BhfP/0967xUiSZvd9//NFRGZV9W1mdnZml8ubRFIg6QfDL7Yf/WbAsCFruU0LFggTBAjaoiVepCVNcunelknKS0oUJZEiKNsCJRmwoJ4ZG9D7PumJDzYoULbMy5rc1XB3Znr6WpfMjIjv+OGLE3Hiiy8iI7Oqu6urz6+QqMrMuGdOz/nHOf9z6i7koaabVpFJZqBPydyGZfJeja4kTNh4bqe23zoMLYMzIuSO8HDN+OoD4CsfFHjrk0/x0VPCn5SrEGirbMYwYyAlP+OlNVPlV1NBW91kIJx6roPcGqFj1bpZbgmHBRFuZA73Hxc4XoVp82/fYHz7a4QbuWtLscSI3T+X8eMuqW49Fvo45Li6Y+queDCz9zMP7b6i0rKBR4WG1yvu3CT70uVDQDdxXh+bLqsSMrg2GJeBiae0wSltsMIGa5S9GS6x0dxFAkS329XLjqEzB9qIftGEgYhNKRp3QwbjzzEm9X0FMBCZcuzyTf3Nd75o7XkNw3ghWCbEMK4g8d3ySoUncW27wOr5xjMcGKiB4+Ml8syjKDxqH4zjeR6WW5chcC6b3RGAoummJZ4RRzRobeqaZXOiNhvSvkeEAxdeP94w3r7JuH5U4aPjBdBs/7RiHBaETQ1sNgUerxmbpuxKzn9YhqZKX4jheCjU5iKlMrqEpufNUMJO7vBX5LH0obtT6CaV4RPLDIsM+MYTxsp7HDjCpg6lb5nrl3hNlTTp19tSM8oG2Y+uI9IwgJYsj3TiErRgi9vu9vZN0wGymO3Tx422FEvfsWd1B79AmIzuOHhCMjicUYVu7kiXrYpLysJzKQ7cjdjnIX/r9+Lz1GVVu8LRRHZmj5qa15vMyFipFrVFaN2Aw7ESsjCd5mL9KYZhGLtgIsQwriAywRroiwtEr/fXCfMf1qix4gzehwD6gwcL/Dt/7j483wAAVLXDIvdYb8I/H3p+m2zzcME424Tynaoxw8YeAkYwnVfKLOuoKflh4LWFw9ECOFg2d4U9o2i6Yr1+QPjkzRq//6HDo8cHuFYQfNWVHMXZHW1AF2SI4XlLUkLQKfvuXtNlUnkTli6R4dNuiac+BM6rmpE5wknNWHuPA5fhq2cl1uyxhsdpUzqmWwaPCRApl5L3dSlTra77VIcmKVXTXcOmxEV8PMO5FR0iQFKBb0k1HGeDuSXdc99+d2S+yTFtsGlt97qsquuWJfsTMRNLkNT086mypW2ZMQ8Gs4cnNyK3+vudWwYmwkTK1qbQM0gk46FFiT52y4IYhvEiMRFiGFeQMQEir8XLyd/y/JRrVMwoyOHpirA8OsWbWY3Hj27gYBkCmaoOvg9JZLjGqH64YFw7bOzLJ90+61ZoMNYMZAgixDXZEAmPFs1k9U+/FjIvj44zHK9y/MlmjSMKd8O/e1mg9oQPNxWqr+X45BHw1mqJD+l04COIg+l9sx8aCSIloPbEYJaSpf7skNAK1zdeCMKbywzHZzW+bbnE937zBn/0jUV77u9vSnyMNdaoUZJ0pqJeh69tbV61ENIBa6buzvczHcPsDTCe9ZjuqtQJjG3XNr6GJflm2GWXYRIKZCg4az0fBTswcdMDLfZgjGdr4msXZ+l02ZKcq4PbOvyv7SrG3awQ7fWIySgbvK+zNvuiv3OxYR7o+1NMgBiG8aIxEWIYV5C49GpbVkQHQCU8ziSUY+CsBP7wDz6D7/6eP25ESIXNJmunpnsOAkSmo9eecLpyeHCi7eZdeZZnoKAQAGZErYrxAIiDH+V64fD0jPBvn3o8qmrcyjI8RYkTDnfqPzrJ8f5T4CE2yGrC06fAp5cFvrIusKauLeuYhyGc2v4ZkNQd8ArcTgrXDQAKVQq1Ro3fP1shA+E7P1Xh5s0V3jwucLwhVExwlGHtCxwhR8ke9+ms3c7QWD4MMqvBcXWdrBzSJTwp38zYtdHiChh2vwotkN2oEJCWvJKFkuedQNT+ERlUb29B5wAAIABJREFU2GVQzlBhiQxM/ePW2Q5W12dOCVvqGOPzT70m1PCDEqopAZJ6P3VsWVNOF297Djr7AUwfv2EYxovCRIhhXEHiid+DDkrqdT3SwTO3QgQId88flzUeH+f48Oufwnod/snYlA61b/wXzSYJQWisSqBeE1Z1KE8qm1IsMVlX8FhzM1MCIUPStXwFTr3H03WNj9aEBxxmapR1AQA4Q4UcDie1x4o9ajD+lM9whAzHqxpv4gBPad34GsbbE6cYC1LjO9Nj60u2Rdrk6sBPtrFGjYdgfBqH+OBhjg8e3sThgpERcCN3KDLC47MKSzjcpBwPsALAOOQFTlGCm+2HWSeqzXC7n/icUu6PfotdtEH7ePmVtNAF+uVUTq0rxzKVidDryTEXcPAcjPt1kw0J2/TtdPYStRrI2J3nEkUjWmoA1DvO2K8RZzPiTMz08Q6zIZKV2kckaFL/TUpXLAdqfSB62bFj7n82/TKsbdkcwzCM542JEMN4BUgF2B6MOg44qYkBuSkDAuOMPb7yiPDw9Dr+7NsVNqVDVROqul+KxejKs1ZVECc1hzA3WIKD54QR7tiXqPHYM5ZwOGoCLQ/CI96ESd6QAYuM+1hjiawpUWJ81Z/hBgqcNjM/jhECsE/yARacty154xKsKQEyZfaOy3Q0bRaCPB5j1c70cCwG+G7exYaCzbtmxr96ssb7dIJv4WsAgG8/WKKsGTkIT1Biwx5EhCPO8RYO8HUCTrnzsggVuNc5Swf44g8QI7eIMC04iJVpfcL/MVUmNFUipsWY7CNr5qbIuUiLXY/Q2as10jPBI3TPqpuStpp8ey4HHM78MZ2pfalJ8lsIXbLC75rne4S4yVA8i7A+7ujVFxZz1neqpM0NxJhhGMZlwUSIYVxRtmU/poJuyVqsUWPJDie+htsAReEbERIEh5jSPYeOWIsc2FR6W4GMCGsfzNYlxDjtW5HhOWRKlnADAaIfQMiknKFCCd92cpLwUQJybUwW9hEgsuVwn90N2rzGdfdhRknTnakJ6DO43rDBU5T4Cj1phuxV+BpO4EC4scnx6cMMN6oM3jMeYgMHwhHyIOCYQDQcOpdH4iD2VIjQkPkbrRCh/rEP2u6qfWjDeDw4UW9jVyTrpgcV6v3ooYs6OPeqJ1atznnXLJegp4xPobMnROGanCcTMvq9Y99tX2U/prI2uvGAEK5LfxuGYRiXBZsTYhhXkM++9/13pUtUTTx4SPcoedTw7aN7LYQua3hUzKiYwT4Yy9tSLBV/FVkwpTMDx6XHqvbNTBARHZ2wCALCt3+vULcP3V5VH39JnXH6lCo8pnUnWMijpLrtJqUZC05jcaPRYW5nNA/7T00B76/bL0kqUWNNFVYo8ZhWeEJrPKQVHjQm+qe0ximVeODLxltD+MxigU/RAZac4RAZDpDhJopmDskwkJRZEA40MGs7DoHzunlHD2lMCRK9zZiMg8DLOAxmdEztgEZBB8siXjK43pwP6GNoZpBU0XWT7E0GF4zpyHrfVVkmlMC5NmsgLXr1vhjTQmF21kSdg1zvjDJklLWiIWZK5E4tX3ONkivUXIdHdA4U/bjmJ6NOuKUaEOhWw4ZhGC8Sy4QYxhWlptSwvvQd7F7HoOZ3BY+suXt+QOGfiixjkGt8Hx6omlgmcyEbAgBnFXDmPTbczIYgQkHUZDdC+FNGR1I2RTd1I0rkWONzEOOzBM6uKU+SAKsk364/1QI1FRiy2m98rfolMrvd8V/1jPJdWVKJbp7FIed4n45x6yTHW8scb14DPo0C1ceMI9eE8r7AEy6DeCNV3gTGQROgh33UqBoZJVQUpkI4HgoLXd4VC4/h8z4e6TvrUsoHoGc81/NK4ozKprlO2qcCAEvkyNhhQ2HaTVz6lbELHdbQGdNL+N4xNO6j5LHOJVfXOGxzficwoF/WJ8/n0C7HwRfTy0pFn0jW+Gv6+2lWZw9PhJIr/C/v/qJ1xjIM44VjmRDDuKLsKkB8+xh2Blo6h1tFBiI02ZAwJb304TcQvCBnG8KDdY0Nc1duxeIJ8Y0XRJVhqYxHpbIjkgGRO/US5AWTcgjCxT8QZx2Wjf9CB+rxdZgSINvKs3YJHnWmSc5F/pY7+lLe4wG8z2coPeNsA9y8VuPtIrhJHvoKD7BBDoebvBjsS8RF3mQlHJRwoi5wr6i7dqNG/GZbKcEyh1RnKZ0ticWHXIv4GFofDncCUDIj/ZKjeZ/befHtpz8s86u5PldZFqP/PU+RUX+avQiyTiCnxXdbikUOJQ8zhYZhGC8Ky4QYxhVnqhypTr5Og2Ue1CUO3AInZzmKwuNo6fDhaVimIGBTE2oPfPS0xlNf93wfJRirusYa4q/oB4ziUSEGNqixaAzonYk6hJCF8mCkAtdu23Xvjrs+F/07vN8fAjgamEfXJNUmd1sXLcYwUJZtrZq7/CXV+L0N8NbmAGdVgZI9HvkKj7EB0HUYE8T7oo34EpxKu17HTWlSZGjXGYVBxqNZR8z1gh56OIWPlstBALvpietRpkAEBxBmlkjnK/2d0PMv9PWOvxsZMvhEqd4uHpIpATKHXbIgIkz1PBFdbja2Tomq7YSl/zuu4VFzPficDcMwXiQmQgzjiqKD+P7r052QMrmr3hQeZU1Zy9PaY5F75LnHtcMa1cchQFzmDidlt59Nc0+2RmdCF++HDsIH5vgmPtKHFc+9qFQ2Qbarg+k1Vcnz3laXv0/dvv4NjAeZ/W5V6X3U8DgjxhkqOEeAB766OsHbfIgjyoL3AoT33XEycK7gkcO1gxr1dWv9GdwF9ECXkZDyp4xdd9d8JFjVXb/0NnTQnzJIB+M5AHatYAoCQyaZ6zbDFErEGFhyEKRrVDijqmdAF4cLNeZ1yTAVzVjGOTNCxuZnTE1N18wVIJo5AkTIGkERZ0HCMWrvR5cFkVa8uiPZhsvm8zARYhjG5cFEiGFcUeJSq97rCQHSBYHhd5jy7XCIDNdchtdyh6enDuuymXDdBGCrmnFc18ipmxkh3o/O1B22B/TFhwOBGL1WwSJWdKmSLAsM2+5qn4UOZrcFoXEJ1i7sImg8uN0XJSpgpbzIg5Gzw7/LbyBzhBNf440sx2FGWG0KvE+nPZ+DnKOU5Mj8EBEE3ZBCOQ701tO/5e/YdJ/8nuhSKupEi+7KlamshwMaj0bIiGzUvkX41CSlbuFaFJy151U29/UdCMdYgRuhsUbVtvatULcCbIF8cJ3C8yH7dPR6FqTEkBbb2gfSZQP94DX9XMrXSo4dQoZhGJcDEyGGcQX57e/7p3fGJl+PtWKVZSvyWHDWZi8IwJII1xaEP/q4M2ZXzZCQlQ9ej41n5ekIW9MCJM58eHCbdQE603kwl9dNu9tw552a4NpBMi3jQmNbZf7cEqw562c9Q3hXGqTRZUZ6Xd3yV9rRAsC3XsvxcMW4mTvcXBAyB2RuiYerDdao2+yFDjTDtrvzKJT0SJWOEaSrFLXBridqPSVT80JSODSmd5LnnQ3cI2RfSvTLrSryKDgb7Eva9cr3T1hyhuNm9TWqVqSW0fWuMOwktW1Q39zMSYwul9pGqrvVNiquBsJQCAI0zAHRrYMZ3Hk/SAlOZtRU4x+/+yUzpRuGcSkwEWIYV5AyUXu/3Zgud9WBBRw2CGbxB9jgtK5RnS5CFyJm1Kr1bsWMkrltlyvG8/Z9+F6XKx2Ur5oSG2qCzqJ5XQQIgGa4HSGeAj8suQpM+Q7iazEVbMbzNsa2o2eJ6NfjbY+11pXg8QAFVijxu8dnKODwdl4AIBxvgJtL4NPrQ9yns14pWphEH2WWQMjhmu5mrs1KCRk7ZBheJ/GkjAXH8fFLWZS+S5+6PplaVn8n0ByDLn3i9oo32TY1uJBAWKLAGmUrcmIBgua1YQnieDkWEBot1NwtkxJuY+vvIkQ0WlRMCRJZLmUql30XlKPmGjnl7RX04Ka1b92ey//67i+bADEM49JgIsQwXkE4CgaF1iiNGiV5EIeBhQfI8MhXeCMrWgFSqqCthG/9Gjpsq+B7gqjrEkVtEA3o4YgVluqfpZ4JnFI+j36Lv7mtUufe6dbdh1KMdV0aC4L1thi+yTQ5ZOSx4fBpPEWJx7TGo+oAB5vr+MamwldXTZZFGbTjPTtQKwj6GQaZRp46/nkG6TEBJW2XM7hBRiOUZFUoKZjGxbdSqU++RsjV6Oso30nJ8sgxbpqGA7INbVTX1AkRMnV+AOB5+/Jj/hERIkDaIyKCYGx9WSbVJELQMz5kn3p/JVftMWRwADlsuOxlQS6iQ5hhGMZFYiLEMF4B4lKQqSAtg8Na3YG+hQVuUg7PwNp3AwgrMA7JhUwIOg+IFjZ1r7NSV0aljdMSckr52BpVz+xcQ8zLXXDK0fqxh0Gfr77Lvu3c42uUT9zlTy2/z/sOwXgNCsd9353iFCXOqESxcShA+IhW2DS+h7CdsK0Crg3K5bpncFhAyunC51g0WZZ2ZkQjHqYCY6hlxwzNbTAM3z53kUASAdIZpv1oiZo8X1EV5n80AXwo06t611AM6Hp4YXdNh8by4Tl14uO83pA5pVViDE91ddtG1WRBOjE73Ldk1foeqRrMu5ccGoZhPA9MhBjGFUUH63PLU4AQUC45Q9F033nTLUKpCjHOuMt4dAEt2i5YEibqu9myTbn/nW4X3GU7pMRGH2NJ3fRyeU8H1GMehuE6uxvQ57Y1nbPtKSGywgYEh1OUWFOFNSr8gXuAN/gQp1S2c1aApoxJ+WnEwN2W7jSfYVeeRc01HAbr8R34sXbOsny8fni/m3VRR5+HLlkLLWTHWwPL8itsAAJWyHDAYVL8AedtZ6288dNo47rQ+XTGS6QIhJp98lzHmLMskUvOC4k7hQ2zN9PCRF/DsHw4z3iZmutWiDA8uBVZlgUxDOPyYSLEMK4Yv/W5376Tas0rbAt4StQ4Qo7XqWinoOtQUUp0PDwec9lM5+7X+3twa0SXY5luDdwXKzFaQMSFJXGL2Hi9sM78IYPC2GyQ1HGdZ/6CPi4H3wouIIivD+kEOTtkjZgAAOauja32ZbSZKKrgOEfRyJDWAN/us/PhaMYM2g7Td/vju/zSzUq2kSEDNcJA+0P0LIuuVCu8niO0py0p+FqkxTDBYYOy3XPqeo4F3dq8vU1U7JM9yODA1C+Vat/b0zsCpEWEfO/iDmeGYRgvCyZCDOOS8c8++7/d0UbuGn7UaD5WhjJmQpf1Uoi/IoNDAYebWQYplV8DyDmUUS2awO9UbakG97Ifcvz6+GIjtD4OHbSOMXb8U6KqnRS+x53gseOJjzv8Hgbzu9DL+qACKVEBMJgYvskGAGF2S4XQYcqDUXDfHxMM6aGV7QpVG3jLOYnhO752cRAbZyykhCs13DAWl21LZoTWu7Kdflld3MVK59cC0lw3JzfocpUSrdsE51zfUMyc0jVZDgiiw4PbzAiBQOTg2Ce/L1Pblu9DP7tRg4nAzCAigLtrSnDtclaKZRjGZcVEiGFcEv73z/7zO0CYowEMuz9pYl+Efn1MgEyJjzZDwa7tUFUQwROw8V03LG7X6bdP3ajgMBYgXq2TQosLGWDXbkvtoyurms82b8Au6LIu4Nncde6Cfrl2nRjxYIDUBHSo2SgUBEbBefPbtaZxnVlpy7n2LM8Rc3kQPNOZkdDFSrIhVZuFGLt+VUJgBNEi/qRwLXRHLPG9PA+kHS4w7Gw1algHgakbsKgFiV5nmzF97HvcCpKm/a78zaqk0UqxDMO4rJgIMYxLwnUU8GCctv2D0h6KsZIlYCrLMVaa1S+PErFyjBIrv8BrC4dHqxp1Yz4Psx6AtQps4i2XqFXw2exnpG1uLI7iUqCq2V/3WnT8M9rxnpeUsJvbVWrf/QkiRvRUcJCDODB0uZTjbu7Iiio5EBSc4ZQ2zfFws71h2ZX4R6bObzQ7FHlAUn/r7c4RcBI+D2eF97etJ8U/a1wrCn3vNZ2hia8tNXmMuBRLLxdKtc73XeZEhy8TIIZhXGZMhBjGJaFo6uoPOccTWrd3RqU17VQ51VhmRN7ToYhM0k75M2TGRA1GRsBhDhw5h7X38ExYw+OgCQtFHDzCBkyA3lTPv0FdJyZ5LV6uf6z9u/VTYVScKYl9HHPvBKfmQsSZD2FuBiQ1B2LubIh4nyJGKvW+BL4E13pCumsWtl2RR9F8XqwEXcEZNtSfO1EhLCvdrWr1zfFNIN3NTum+Q1kjkMaOPUZml8y9jvK5itdIEBEWC6Mm5E9uSwf+21rmTp2LiJGue1fWCpMcGape3vBiGJv6PgfC/C5vhmEYzwv7l8kwLgn/0Xv/+d01PBZNyOmJUTVzIUJnJN8GYvKQ9+rodckilAh+klo/5C56wiCeK8MyM0AUyrJcY7Ytmq5En3AFblHRBr/d8LkQtoYgllsB1Zaj9I7PD85BP7aRyoL0RUz4GfOD6OB1ynR+EaSM0HO9Cfpahdki1cAb4UDtta7Uvkp4rBoTdwUZRjkMvsNrYQ/SdUvvo0aNElW777LZixyj/my7HF43Z0VmmGRwbWerOei2sxuUvc9D2vM+ryxI6tjkd5hfkiFvBXr/f63iByFyvXWFspmMrn/GIKLgAdnxWM/TPMEwDONZYCLEMC4R//F7n727gUcOGoiLFGPv1WDU5JNCQ8RNLFq6dUMA+W/rFZ6ugad1KMfKiXBIrr3H6zkM1lOdYptSrC4DEmdw4jvZHB1D/476NFOm/NT24mVTE8CfN2NdmnYJGrXZu1bXNLwXEGGyphprqttWtzEV0t+ZsWXlHFIZLSFrfuJz2qVcLR7WF/8dM+fanSdTsU9AL+cvQgRAKzhSn3fq+uiMxhwhIls2DMO4jFg5lmFcMgjAgjMc02Z0mfHSq+m5GWGZqS483d1dAHhc1jhw4b6uTEh/yOswtBAemybTwGCU5Af7n2qdu2/Qr7ef6oxETY2+LgnTAd1U6dWcfe+K3vec9dtgtSkrisvCUrM6PBgEbnspExMyhIF+rZCNRKE+vnCNwuTydTPLI4U+BoZvy8OkA1acHZD3xNOitzGHqVI4PZxRf8/C9RgvyRLmdLvaN3uwrXSq7/vpJq57rsZWmbXd1PJAECz/+N0v3d1hVcMwjGeO3SIxjEvGX3jv++9KdkEH66x+quYhpU/yOI8AAdCWzTgQFshQA7iWEQ6zEEpnRHidCpTwOFZZkNClyI8KhDg7MVVuJaVcY0iJWmq74Tp1d82nfTK7i5DnyViJWEzZysB+eZRsI2uKgzJ2yHn4T74Y0rVoiMu99P77n2MozQL65W/BX5KjaO5zyfZ0dm+uOV3/lKjbx1x/TUybfdixpOlZIQIE6JevAd2xxrRLzTwHEyCGYVxGTIQYxiVk3c4eH5YtVSPB274CRAfr0hb1CBn+zHKJP3szg2s8ISUzTnzdHN9IkBp5QGSbeh/bgs+xMrJtjBnQ4yDuPBmYZ0Hq+CTATwXr8fFTIxpF3Ik/pGzEAIBumKESiPKeeEB2uesfCz/tXYmPM2t9Ep05P3Utp4RhbErX7NNu+Fl9lnP2F5de9UrXaKwXWId83jrLMYajDP/kvV82AWIYxqXEyrEM4xISD5LrRrhNiIyRwH3bXecuWOzmSCyQ4foC+M5vOcGjJ0v4mkD3c3x9XeIxl8jhwACuc4bryPENnGFFFXLOGjN83QbF/eBRgtbtd0Dmd6HqrlMbWF+wCfc8QevUuqm73IxumrgWbrJs3BEqb3IZslzGricAa/SvR+hmFUTklAdEl15NlUWFZfvvBdN43ZZoyf4z9alvm3Ivx6evyRj6+sz5rLoStHkDCM9LfGwZZcDI9PTWvM5dtk+T+m5TM7RQ/01kZnTDMC43JkIM4xIipVbCtnkYU1mO+P2+OVyarwYOkWPJ4W7swxVjuazw7/0Hv4vV8Zug3/sOuI8KYBX8IQfk8KmDHKuK8bgscdp0YZJ9pHwffuTv5DlRvO688h1gupzrshC3to1JdbAabiOL3tPeF32Nm/a8jUApKGs6S8UjAjt0W2X99xhaOHdm9RrclhaFcX8O2aAkL3Ud5pZs6f2j2WPwrFzcoMp9SAmcnlgcyXq0niVy7XDDWfuj7joTXbwQNwzDuGisHMswLhn/4HP/6M4Jrdvn27IYqUBrrPNULEBijrjAAec4ogyFIywWG7hvvo/F4RN827d+jKMC+K4bBd4qCnzvGzk+eZ3xoKpxghIl+ab7UpWcAzEnnIo7LU11uZoTYO47MDA+pouGQCiaHMauy7teUB/+XnKmyqr6aDHCCJ3RZJsiYmJ0KWDKk5QiPpe4s1suJWEjn2e8/12v+y7NDlLiQH9XLuJ7E7fpvQgyxK1/x59nKkNmGIZxGbFMiGFcIv7R9/3TO6CuLGqbz2Ou8bq/jh88l+DllEI24ylXuFVn+OCDW6Df+Q/x6OENfPuf+zc4KN6CZ6BmhmfCVx4wPuAVnrquk9c+cxvajM1I16t42diP8Cx41mU6IiiGHo/uDvoBFiibblUhyM9ak3fKVyBCpOtypqbWq++SCIQM/Wnf8faA+dPNU5+7lFE5ZG1pVfCOjH9H9hEg8XFuEyRyjR0RPKM1qftmFXk93q5edxvhs00vOyYOdImYzoKMZc26TmDdcrKdGn4gWgzDMC4T9i+UYVwifujdH7jbK2lJzflAN+AvxT7tbyWgPaYNjmmD+7QCEfCvvlbgX/6fb+F3/uAAf/rH34EnK+BfPyzxtWqF//thhX9TP8VjWrf7nGodHNMLGolbH8kcUtmS1PWYunP/IgkdzvptZeMZGgTCkvvZkjDwLwNFQemaanhiZBhmQ9qOatQvf6ob10aKZ1HKw0h3xbpIEalN/XNx1Ak3pwZzOiJk5JCpuR7yPKN5WYZdsyHxNolc28J37DORIZDxYEgC4X969xfMlG4YxqXFMiGGccmQwXIpAbKtE9A2ATLWQUrWKbg/Af1J5fGoDGHj//X7N/FHJxs8RokNPB7xMVZUtf6LsWMrm+A3i9rDemI47p+XNhinjz82SO/eGUlv50WWq0hQDowH/WdUokCOEhUqeBzxAjyRLZLMivzOmlxISb6XdZF5E2PB+q7zKKaOR97TwyTH9inL7erp2VZmOMW2MiwRhPq7mcGBKPx3Wu/g29jpuBoBUqN/fjoTozMd8VGYJ8QwjMuOZUIM45LxE+/88F3d6layHrrGPn7IbIa66as09hiD4XGdF7jBB7jBSxxyjqdVjZIZH/EGj7nC/3OywkdY4xglzqjCKVUoybdzO4ZmY3k050G+9/CN0KooLg8behBij0t8PtuyHfF25q73LInbzmbIWt+HTBiPP7NbfIBrvGjvfMeMCQGnxtrHvpvUNqqJ78oYU5+X7pI21p5Wnp83MzJ3/bEZHJo2sxBlRKjJSD0rqMlmAF23rLZrVnvkw6GVGsuCGIZx2bFMiGFcQnTZyr7ZDU0F3wZU2rwq6xIcrvEC38RHKOFRwOGNIkOREcozjw08PsYapRYc1AWW8W/tP9DnFDOWPUmJg30C48vKWPArd76l1p8ROlhJwLtEhiUfgsGtfweQ6xUyCJnKeMjcEP1adwxu9LsTZ6S2ibVt3dnizEbKV7GvFwRItxE+D9r8H557IPGZzcmCnLcNsBYXtXotowz1SJtfEyCGYbwMmAgxjEvItvalwi6lJ7KdbGK911yBmhklMw4Lwts3PXK3wLoCjjYZ/hBP2xxEfFydaJpnMH/RSOlPirHWqqn3dmVMgMi10n4PfbfdgfCQzvAZfwMPEpvw4BAnM1pviBS5AcG07olbGRrO3bUTz/V+oDIt+zQa2MYuJVdzSuYuIqPV9+RMd6ECQinbHIERGhN35Ya7sG37pLIzYmTXrxmGYVxmTIQYxiVk3kyMeQIkNnG7xHoMjw1qPPYVbrkcK/Y4LIBbN0qsygWqmnB9UeAPj7t19JRuvZ/nWeIUn5sc15z1JHsQ+0NSAiSHa70Z5wnKUwF1N0ncA8iwRI6yaXKsPRUAsKLQKWuBHGeNe2hwbsQg1iVPw7pbPf8jhWTM5hq8w3cBzXls//zHsntT12eKGlpQZu1/G3NFcJwhnIN016p5fB/dMbnmOHf/7ujvJUWzQ7QnhJXB3jAM42XAbpkYxkvGNn+HZpca+zVVeIA17vsN1vC4fsC4fn2NRXOr4sYh4ztxA7d4iWVzh17q/lNZEHl//DzmZ0tSxuP43HYJfuNSN3ltzC9RIMcB5ygu8L6N7mYkTBnFhbLx70x5QmJfTao8TrYrRzG236JxqMw9H+nWRNGPJv6s4u5OqWszhZ6JIR3E5Gdsu/qxqwDR6PPbJgDm+FCmcCAQufCIRPN5t20YhvG8sUyIYVxCfu7ej94FgF+8/Rt39Ou7dv4ZC/BTfoA1apTkccIV3sACB8sam02Og2WNPHPwHvhEnuNhmeEUVU8EiCxKTXZPdaISr4JsYyrY1PtJCZCUt2AsQB921+qe6/KgkD3ogjwPRkl+ULo0l13u8K+waYPi1Ll87M5wyOGfbpkfEXsiPDGypvxq7nGlAvHutXo0UxRvR5gK64PxWk93p142I/7ezvF86GxMf07Kxd9r0//t6BkjU/NFwvHsnhGJtzMYCsk1MgpT6P/hO3/DvCCGYbw0WCbEMF4SdOAzN8ORw7WTqoWpu75LzhoTM7DIPT5+eIAHT3J4DzgH1I3f4Ihz3OQliuZOs0dTnqLKgAoe309sZJ9aLtUNa6yrkgxpE3RHsV3KxHRnJwBYo8QxVnuXYqUCyanPYaqc6AQbnFEnhlwbcKugnrsysznB+/SxTwvffe6+p7IdU9uZuw99LSQXs0tGZS7JLmRqvoj8/SzR33WP/nBDwzCMlwETIYZxifm5ez96t27Gykkb3vEGvMOHBLriaRBRkidKcHIQzqjCQ1rhkByOT3Pcf+Lw/z6q8P4jhyenDt/+OvCWW+Bb3RG+h27ghg/tYlNzTVLmdaALyCt1pPHMdJK7AAAgAElEQVRxxy2H4/f7+xn6UuL3xtobyzJjxylipG5kTGofuyJBsZ5UP0Xse/HEWKHEkrOe6JNtt8tuyYLsy1iZVWrg4i5sEwpzhYSLjomUGHmeXERp1PjwT+4JD2ZvHbEMw3jpsHIsw7jkxOZhKRuiLSUqEkCLj0Havo5xRhUe4BQL5MiZUNaEPAPW7PG1sxJPNhm+5y3g1HssiJARganJNkg3LNUVq6aqV06kg/j47nroNNQvEZub7Ymvz7ZgU46pRsqyPUT7XeS5LteaQ1ceNAyG45bJ24SJlIZd58XoNdIC5CLa185ZPzX/Y6qEa862PFjNRCH4PTJRsq1dByCeFynN2rdRw6SfyrIehmFcASwTYhiXnLv3fnznO5zamCzZBPktWYaYDUqsqcYRF6jAcAQscsbrWY5jVPj/6jP88ceE93GKr/EZ/jU/wRNaY90EhjqzIPtPBepj9fCxMJnXISwRgG9ZT5vPB1mGLaJOX9d9SF2PKYN0LFo4Ot4MbpANmbNfvZ1tYoqQHow4tv200CLoAXzj+xpmdp53BuMicWRmccMwjDEsE2IYLwH7CJE7t3/tDjV3/B2yVni4iQC0RI2ntMb7Pse1+9dx4Agf1Bs8xBorqvC7G48H7gwAcNaYtPXQwni+Sbz9uQMH59653/UOf6p7VJzVSBm9hdRyqbv9c4zo3SC8/uv6uo2JFiA0Eijg2kGEej05djmPOBslbJuervepGwnExxIfZ+qc5qDN5anj7bWk3bE8bsyFs2+WaO4QQkcE5rThfh8uYhuGYRiXARMhhnFFSQmXO7d/7U4NbtquZoOsRI0aj2mF2nmU3uPt+hAf0goAsESOP3VP4UAoUffEBxCCwgq+NcLrYN6BWo8HMH33fW4Zls5MpLaRep5afkyIjG23H9B3WYwSdRskp7Y1JSji56lMQiyMavIo2M0+7im2lWxJqZxebqy0TDJaqU5Xwac0Hbz3z2f8PbTbnz/LBNHxh8/z+QX2XWnY/lgplmEYVwUTIYbxCiHC5Bdv/8adG7xECY9N02lJB2cVGAU7HFGGt/gAR5ThmGsccY77dIaS6qQAkXBe8E05GBrBEwd7Y610U3ATwApjg/Zk2fj4ptAulbmeEikvkjKqLJH5IThc5wKntB5sZ6zUaJfyoxIeRXR/v1+ylX79PMRCSYRHKs+QEpS7HMeca7Gr5yUuHdvHayL7nJsN2ZW54towDONlxkSIYbyCyByS3/rcb98pUeMT/ghHKHBMGxTsUJLHZ+gQDsAxKoCBpyhRwYNAWCLHGnUbLHUCpCu54ia0C+9UgzviUwJkLAjbFpyNzZmYgzagzxEiGQgZMhzxAhuq2vPXQXkOh2tYQA5nTVUvGySejpKmg2ARSTJ4EAilSTV5gENLXtDwvKeul1N/z0FnQ1LvjTEnU3MesiaXcZ7t7/q9CT4Zv/f+tJieml0zB+uKZRjGy4qJEMN4hfmRd36wDWD+xWffuUMMXEcBMPDmIsfXNht8w50gZ4c1VShRo1DT0gVtKt+lw1VKPMydfK7p1kmLh13uWEuZVXxcqeXqxpfBnKGirgzJQ9ogh/Xf5utYo8YDOm07gQFBPMw1LksmKD6Pmvwsc7ped8r3Mr2N7b1MUiVZGhFTF4kMbcSe282UJNtdiGw/n/hzy5Gp5ggi2ud5c85TymUYhnGZsO5YhmEACEZzB8IGHq9jgdcPCG9mRXiPyuADAWONGis1OXzMf6H/HutipR8V/KxAbK5QOQ9zA9EaHk9p3QoQ6UKmt7FBhYIdbvICjqltmUygIGB2OJecHQrO2gyKPObMBImN3HO9N3PpzwhJ/68lawRsPGNkDtvWuYguWtu6d8XIeW47l6kuWa75kfetm5ZhGK8KlgkxDAMA8P3v/cU2K/I73/cv7hwuGIcrwmc21/EhnWFFJWpUqMn3hgNqr0eqg9KcIHvK36GJBUi8r5QJfB/RMuURiZ+vsOkFr9ozwWCsqcZ9d4pP++sgEBacA1Qh+Gi4FTBzaOeNcByszhcUqfbEF8HwOuXKE9QtkzKez/l8pOHB1NT6XTxG27axS0evAE1+1+K5IcNOZpJXkRlAZkA3DONqYyLEMIwB//67/9ldAPguAF/77P9x5zN8DSV7/JF7OLiTPnZHfVvL2n0Y624lx9MV1XRB3nmGxcnU92132UVEFcjau+O65OmYNijh8SYf4bTJKjEYJXW+mjklYyXVKDgbXE/Jhkx7QDpx9Cy8GXHr3s6r0XVhq1Hv5dvpsgShAcBYsJ+pa9+x27nuWppFPQE6XZ61bYChQ78F8bPO+BmGYbxIrBzLMIxJHmCNE1R4i5a4wQcD/0EqEzH2PMXcQGuOt0T+rhGyNecNtlOBYLxNXSakA1LpngQABQcPwHfjJm7yEjn3y28KztqOW3q7qeNJmdi3iTwtFvVwxIskJUiltfA2L8nU8Wetv8Y1zzPkcJArlkKf477nu2tpFjCvPMvR/OPYp2zNMAzjZcFEiGEYk3zuvf/i7kfuFBkRrnO41w90pVip4G7OZPFd7vRuK8PSr8frTGVr5jJW9gWEMqElikHQmsO1Ho5P8CEWyPDJZY4lZ+01BILPY46nQ7bPYNQ09M/k7HrLaXSZnBYi+6Cv6Bhzrve2/WeN0MhUdknQz7d5KbrtuLal8pzHnGPc57zmLh8L0tT5WWcswzBeZqwcyzCMrfzIOz94959/9p/dAYAbfIA1nbRekDiIKpCjRDd7JEO/s9PcO7s1fNv1aGyy9xxhcRHlR6n2vSHTwSA4FKrbkUAIJVK3eIlbvMR15FhkUJPru1kTjtEKEbk+IiqA8F5ow+u6DA/1vSHt3BK1XOoazCkvOy/yuff37yaFi6ZICI8p5pq6RfzVM5oguKb4q0LngZozCV66ZtUT259TeufgBsNECQQih3/4zt8w8WEYxkuPZUIMw5jFmmqsqMJrvGzLi4BhIFajRo5uknfZhnxddmRuF6wSNYId3iczGlPbkXKdi/Q/pIYglqhQo5sUL9TwOOAcb/kjLOFwgAzHG0YprXyZWqERhEgU4BK3d8DlPcc0WSY0p1Uv0GUqzitGYlGht6e9Lg7UllSNiQvq/YRl4msa70u+ZxeVfUghpV+yTtyEYGxuyi4lWbscz7MWkIZhGM8LEyGGYcziB979S3crcHs3WVrNiqiQu8UlaqybwDzMNegC1ToSE1MPWZ4TP3qZMZExx8+RYtsy8f7DOVc4YLke/fM9QNYGrn9SrnFKJTz6JTZFE7T2sh9q+73wvBEv6bvwu4mQfdg2mFAH5vIZtF292oxFJzLiMigtPOKyt/g4KFo/Po7xc+j/bEOEiBYjY+etH1PbHzQXSIoZExyGYVxdTIQYhjGbH3r3B+7++fdu3wWGQwm1EIkDde1JmPOzrWXvnGzKHN+Ifj5H2MT7l/M5wAIZHEpUvenxK6rwgNY4bvphPUXojJVHweURL5JCJMW2wNkT90zxQiowP48QGRMjqf2khFtKRMQmdlk+b98Zn9auRcwcM/qUqNh2XnNM61mTixvd3sTskG6fw/2YMDEM46pgnhDDMHbmJ9754btfvP1378jzTjRwM1E9BEqb5vUaHlWTGdHCRNjX7yFI0D/XR3BRJVpSMnaKNVa0abM/eh+PaYVTZLjOBU6obI6T2n98KzAOkTfDID28EiI1eWQjoiTuprWNbdd7H/R09LDddOA/lXnZ9pnpyfNj7X3lOLSI4ej7Nn4O4WeXdrjhODq/0hhhOGM96g9xRKi539o45S2SffzmO180L4hhGFcG+vKXv/zFF30QhmG83Pzs7b/dChK5S+xAWKELuuW1bUgQNn96uk8KGwlMdzE4b2OsZEbfIS+QYYkwab6CxxEv4AC8zocokOExrdpjl9/f5V/HQ6xx352i1OVcEyJkzOTP4Nbkrq9lKtC+CDHS7cO3mQ0gLXr22V/Rs7iHc6mw3eAu4mjOsgB6Wbxd0KWD49uue8vqfdQ8XC/lszIBYhjGVcNEiGEYF8YXbv9qK0aoESFyx36uANklCJQ2wSnkrnIWBbHnZUqILJCjQI5DLlBS3ew/BOZLzvAZfxMVPEryeErr9hy+17+BD7HCfXeKtTqflNCQEYBTtN28VOvflAC4yGF4cbYjZSrfx4siIkQ+TxEh2zI7IkJ2EUBzSgHH8BPraqEWCxHP6e+8Pt5ff+fnTYAYhnHlME+IYRgXxi/c+8k2WIoDsm0B6K4CJEUqMJ3TFnZMIM3tvCSeh6rpDJZhWD7km+C5gEPBDgvOseBQbPQQGxw3pVqaVKZjTogcd9UaO7ddDdpTpOZsxOziwZizrylfR+wVmbPf81yHSf8HOkN+93fzGc3whhiGYVxFLBNiGMYz4/O3f/kOEJdJDct1tt19TpVbhfXqnrCJa+pr9ihot5kTsg29rwVybBrL+bZg9gBFW/p0DQdYcoYajAIO13iJZTNTpGCHFVVYUYVv9jfwIZ1hQxXKJhNSTQiyHDT5vqYmNS+kESXxcMRdSuDOA4F2nmRfJAze8eegtzneLc1vFcJ6e2Gdiy/NiksIu+PjQWmWB1sWxDCMK4sZ0w3DeGb8yr2fugsAP337V9oyLT34DxgKhzlm4rD+MKBMBXa7khIaS+TI4HCGzeBYY8QHIxzhBpYIk9NzUGhhTDUKZCgpzEC5hQIf4gwycNCDkWMoRHJ0Aw4x89wcU090xOueYo0DLGZta1+6a7ZbOZZ0usrY9cVUdN2ziSK11Oe17RjE17SrGOmOy00KkSCKE4MI0bU27m/PMAzj6mHlWIZhvBCk/j5VtjXGrp6CjNLzNPbhCMv2b2k4G881iY/zFBs8oJNgUEeBRZMVKVE3XbNKMBg5dbkaXWoUM9dbE68zxb4eiF3Z9bPTXbEYPGrQF3SGLT2zI/yVIdupja/uQja3bGrs85P39O/e/sgho65QzjAM4ypjmRDDMJ45X7r3+UFGZN+OSQw/YkUfsm8L25hVM99D6MrD9GDCNE+xgifGKZX4tL8BAuGQCzx1x3Ag5Fzg67zCmzjEhziDQ5gvAqCdJ5IqlyoQT6WfR3xXP8xzl3PvStH0FndpYZtadpeuaMBwLognhpvY/dR3SAf93XfORe0Kpr5RhAxdhiz2Lo3tO1OfXeraBX9IujRNJqr7Z1sdZxiG8UIxEWIYxnMjJUZ2IVVLP3c97QvZ1rq3Rt3rqlWi7okQYc6xOHicYI0VShSU4Vv8Tayp7gXFD2mF7+CbeIwMNXziznsn2CSoPuTgPZEuXIAfFSJSkiWDDLtj68q/4vOKsy5jsy5SQxE9OrEj29+Wdtf7SmURYh/LcJ+hE9mUHBubYSKf9VQTg7SY4VZsCH3BEcTOBlXyeFNlWb3lyLIhhmFcXawcyzCM546IkV3YVYDEd6u7winfPo+R7V/kbBHZVw2PR3SCb7hjnFKJgjPUqHFGFRbIcIoa17lApYRGqnRHgv4bvMB1LnDYdNgKM0rSx607Ze3jM0gN0Gu3Gz26a9iVFOU87MWVRf25pNPVnGnkKfSslm0dv8Y6ec393FPrdmVf3X6nSuvi4zYMw3jVsEyIYRgvBDGtSwetKeYKkKmSodTd/izxfjd7pOvKVCBDBocVysm75VP7dvBYocQ36DFyZLjFR1igwAobXOMCb9ACG/Z4iBXADqCu7Iyi8NiBgpBpjusGL3FCm9GMQ+qIpxoAxM0DUu9LoB0b+XN2bcbFNeKHQCg4a9dtmRF/p7wgYlKXbWvTupybHNs24rbO8RT4uesOO3j1s0GyfW06l/2NlWUZhmFcZSwTYhjGC0XESAqdudjGPp2wxoLNOIh9g6+DmqBx32BRxM0aFU6wRklBYtTwOKYNTrjCp4oCS+StF0QfS3xnf02hRKyGx2u8xBEXo3f4W9M79wPubbD6GXtPtuXBrShwTD0B4ppjcAheibiEaQqXeGTs4DhsZ+p/Yi66ZnMyI2E5t3NWJLWtlFmekPps+7NOzJRuGMargGVCDMN44Wgh8tdv/48D8/ocPDMcETw3d5qJ2jv2OojWHZdqMKi9W52pZfoB6ALZoK5fs0tbYdf4UTYokTUzQz6mMxTI8Im6wAFnAAFrdL6R1PblnAiEAoQ3+AAZHB7TSp0Ht4VoGahtODv3eHelV6bF/Y5VPpJ8Ux4PjUcnojK45rwZkkaZO7xxDvF12ZYVmXpfREWtPic5D50lkWUJOapmFg0A/J17P2vzQQzDuNJYJsQwjEvF37r3391t76xHd5mnEAES37mP7+Kn3ucmS1GPmM83I8b0fZAg8xQbnGCFEjVWVOIUJb7iT/Gt7ggZE5acYcF5ciq4XJ81hUzII2yQg3CLF+2yS87xlr+GN/gIR80cEMkihL/TLW2B3SaHj7W61RmQbtmONpvBbvQR70e2q/cr2yqax9jxyd8FZ+35aT/K2PmkG/46FCP38KQNsD5m+Vt7aGJvDLdGdcMwjFcDy4QYhnHp+NV7P9P6RcLd6fEshCACZF9k3fjONsHhAR0n32v3vcd+66bV8AI5lpyjIo8TlDhwhLf8AR7yBjUYT7CRAxk0J67hURIBqABe4Ag58iYQP0KB6yjgOUxnX7sKNTwcAJayrOZXPNCwO/f5rXnHSGVdulB72wyTPtqL0pre1bECoa1tWyZG3J6bLg/L2aESX4kImV6eQv5Kn7sWFEDWilcpqwotjtPrSWYk5SF5llkqwzCMy4bddjEM49Iy5ReJOW+wLMgARck2MDxW2Ay8Kefdn2wrg8MbfIiCs5CTYeBTxQJvNsMROy9EHLRqIz3jCUpsUOMIBTKETMqqCXlvokCOrvOU3IV3TD0fRxz85uxaQ7leb1v3qSn0UMatyzJB+phpP44uZ4qPQ/tOeh4V7g8d7GVIlFdGn5teJodrH3IdJeuUKXeKB6PaUiSWOnfpohX7QwzDMK4qJkIMw7jUfOne5+9KZmSMixIg8Tb1VHcP7v2c5xgkoyLbPOAcb/EhCIQ1M77tNWBJDgfIcMB5W3YV+whkvxV5rKjCKWoQA0vkbTC8Ro0SjIK7bl/yD/8wgI+njJ9PcMg5jgXVXRCfmAvSHJ9kaDw68eEwL/ukRUd8/LHwSLUwllK4PLoa8T7GStKmjmkMh7gXmmEYxtXE/qUzDOOlQIRIytMhZnThWXQXmuoQ1ZVyzSsJ03NL1qjwdXeMx7TBa7zEB36Nj44Jr+UZchAkVtZ31wtlohchc0YVntAaDoQKHmdUtdmDM1S4xgVu8LIN+LcOD1RBes79u/Nzg26dD4iXj4cmprYVX1ctPrTPo3fcUVYkfgg6Q6Kfp7a57fsk3o6Ul0R/Vqltp9glA2gYhvGyYp4QwzBeGkSI/MTtX7rT1v2f0wvyopBgukKFx3SKkmoc1W8gQ4aTkpE7tJkNiVu1KZvVXfm6yRGslK9j2fzzXhNDXr7GRZMx2TTb6YSCdLSCeo74vT21nQerYLyfBQqzMmRCe//atFmQxtuh0XNK4tfj7YyRymrgHN8l2XfcYSs1FV17P+R3juH0esMwjKuKZUIMw3jp0O1Lz1sa9SzYZf8hU7HBCht83R3jCBlW3uNhWeMJrVETY8kZ8uafa/mdKgPqOmaFUiwmoESNTTOTxIPxuj8YneA9VXqlMwVzz1u31g3Pu25QqUyG3rcDUDQdtLSfI0WuzsdF57BLh7XzoD0qu2ZSWAkywzCMVwUTIYZhvJRsm6PgmXuP583c0iwAqFHjFBt8nR7j99zH+F1+iPu8QUkeD+gUp1S2MzJ0QC/lVrGJmkChc1bjJMngcEIlKngcINtbpM0RIiniDlQiFApkg3KlGGmoG0+MF4pm1opsJy7zao97phDRJVlz19Fdu1JiIh5SmDo+4Zfu/TUrxTIM45XAyrEMw3hpESHyVz/3C3fi9+JA2zdPHV2+zkPBRB5a6H5AT3BKh/AeWHKGY9qgRNUr35EZGp66Mh9dmuUotIhdcoYaYUCitABeqYF4+0AgZEyoadgBaizQFnO6GPx3QQ8rTFE35V3yOz4eaUsspVa6BC2FnB+A5DmOrQN037mxNrtjLY+ldW/qHAzDMK4qlgkxDOOl5++984Wtd493yUy8CMSsvkGFU6zxkTtuzM7AmiqsG2v6CqUyaveH8Ml2JAMCBD/FmsKwxTMqcUyb53ZOcQYgDsyrRpTk0f+KYnN5jN6OZHpi9Da9+uznhvn7flcq+Emz/LNommAYhvEyYiLEMIwrwRwhAgzLtJ51udYu4ifM/KhxgjUe4gTvuyeowShR4SmtcEIrnFGFY1q3pm3dqteB2uC7RI011VgjCBhpNRzfbU91jprD2GRzvd1YgKSyA6nXdw3UUxkWaq7FEnlbsiXHNQftKZmLdDAbGt773cXEF7Pr9g3DMK4SVo5lGMaVQYRIqjxLGBcEzzYY1L6BKUKwXIPh8QSnOKQlyqa7EjdZDkeHOOUNjrDo1qN+dkCW3aXEZ1up0i7b2YWUONllcrgsJ9dYey1SJVJj5xkfd2rq+RgiAlNZEClHk/2Lx6VWXcH8SKmWYRjGVcUyIYZhXDnmZkU0L8K8PoUHY4UNzrBGibotsZJshmQ5UusBzaC/PYLaff6noDMiev3zBNW7DgHUYkPTD/yjSenqeFPn3Z3T9v1X8IPOX3r/KWRZGYgIAD99+1dGBbRhGMZVwjIhhmEYkCD2+bRy3aXcqOpNSg/rnWKNazjABnWvZa/Qv+vfte+Ng2MxfXsAuVouB6HqdZnaj+DX2P+a9s9pGNDrcxtrOyzvD7fXZSfG1nQAkGgCkCKPSq40HOVd5DtQNOuMlaoZhmFcZUyEGIZxJYmzIVMlWoJnHnTPqtkjo4kAN7HOGHoCODBnErdrujvFd9aDMCngWh+C3OEf81fEHg3JAmixoVvhXrY0eTwIUIvGqWnrY4hpPiVuxpbHxLJ6eOKcErhQvuVaATIlogzDMK4i9q+eYRivBHM7aGk8T9+hPu+09l3XnTyW6G563HEqNR08mLf7gfhFdW96FkMC42Obcz22iYZt59sOXEyUZqX+3mXooAjIuHmBlWQZhvEqYCLEMIxXhjlCZKpbVs3pspqLJg7gJZiWALeCxxk2OKYNntI6eZd+biBMjScBmJ+hmct5t6M9HPKQAYfZRPmTMPcaZHDJR3cc4aGFiG6PPLcFb8q0nhpk+KV7n7eBhYZhXHnoy1/+8hdf9EEYhmE8b7aVZ0kJVsk1CspQc2gDW1A34XtbqdZcYj8Hjdwf0kHsQlXT3sIRCo4D592N6VL6JUP+zsO+5UXnzZ5o78t5fRapa+AxPcRQyrH0vrUnRHt8CmQ44iXWVGGFML/l7r0fNwFiGMYrgWVCDMMwGnT5js6GiACRv+X9OQJk165bYwJEjk9g9bPCBiuqsFFNZfcN5vVgv4smj0RS6nFenuXcDY95JXSSpUkdS+q1AhkWKFAgG7xnGIZxVbFMiGEYrzSSEUndNc8aY3iMZBl0VmQbUwZ2nTWYEiHxwDs5Dvk7g0PRZEiu86INarW4mpMh2TcLMpb90KVLz7sL1Nj+Ul3DYpE3tm65JRMi6wNAmWijLF6QvJEqIlh++t5/Y1kQwzBeGSwTYhjGK83fe+cLd8eCzbFgfJ9AeuoOus4F7ILMEmGwmogefp7QutexCUDrqYhN63OOMWafcqsX0YZ2W5ZFC6O8Jwb7gm8OaSE7LlQzJRJNgBiG8aphIsQwjFeeX3/n5+/++js//0yDwFTplnSo2mt7TXvXBYp2WwDUdHWPvPGJFM0YPQm4x/a5SymWXi42cgPbg/85DIcLnt/ontq+fi91zPE1EYP6PvuM35MeXoZhGK8aJkIMwzAadhUiU36PVKBP0c95ELEhwkIHyjJh/b47wX06wQplr6PUmDiQid+pO/+yviBlYMu2qOj83o54G/rHqd/neeRw7WDBrk2xGzQHkPI2oOsgJs8dEBoBTIgR2XYFP5rhkrKsL9z7K5YFMQzjlcOGFRqGYSi0EPlvP/c/THbQij0egzkWjQ/kolre6rKquD1wEAUBCZZLVMiQoSQfunw1SzgmFJTBg1sxExMLkeCPCf6GZ2Gg3tbZS851365bU6Vg/ZkhelI8Ib5X19+/H7l6HTlcK0TijEeN+pka6Q3DMC4zJkIMwzDOgQSwqTKmue17eeJu+fR6XVjrENrqijm9bsLjNUo4EE6pxLLp7HULB1g3AbDOXMTT1wfnAzeQH7v6PHYVERcVpKeEzZhRfpf5Ip6nW/ZqYiHiwfjv7/1Vy4IYhvFKYt2xDMMwZpDKiizo4u7jTBmYU8TBuc6KSHmRU3f4j7AMJUbscISit75jgidujfhzMjdzO2iNDRWUfcavbSNeZ1926RA21VmMlaujJp/cbjw3RDpmffHej5kAMQzjlcU8IYZhGDN41ub1Xc3JqWBX5obEHhH5e8lB6MQComru5IsnBNg9AzEnw+GY2geBes/ltW0/c4TKHLad37Z2w4L4RdzI+/JaT/SdwzdjGIZxVbByLMMwjD3Y15swxj7lWHrGRVwOpj0iAJArE3XZlGJpo7e+2y/rTzE2Q0W/r7fnmJBFgfhwgsZ2Yt/GHAp2ydke2861+4yHWZHkshyW9DR0isRzScyMbhjGq46JEMMwjB2QbMhP3P6lSdP6FHHpVY16b1+IFiIxJToz+hM6Q4EcBTJc50UYttgICREjoWwoZEzWiUBazPDyO2Ve1ziVuSAQPEL6XQTQmJiIr4KPXnc7tMiV45AskKDPb9sQRTmvMdHVF3HD6yfXrPucbDK6YRiGlWMZhmHswd+597N73ckWAaI7TGXI9hIgwlQAXaJG3RRobVC2GRHfDDjUAbYEyytK5yjk/TnGbREgcTtij3mek/625v3Paqwtr/wUyLq/2bVlYHrdXZl7PeJjNAzDeNUxY7phGMY5+cnbf3M0K5IhaztVZY3rAgBu4AArlFjvVZQ0RJvQU0GuiIAk57UAAAn9SURBVJ4DLLDkvBUUN7AcBNE1PApkyeBaSrm2laPl7EbFhmRgxoq5dpFj+lx3ETdaPNTqPD1Ni4rYZF7Dt8eQwbUtj2tw67WR/Qk2Hd0wDMNEiGEYxoWghYgWHlKqkyHDEjkOmy5Vp1hjhbK3jX1LsmQ/Yd/T6x9hgQoeRdNDS8RJwQ5HWLQZEgdCwVkvkAa6IYm5GuSXbOerSqakBCs1G1wbuseC/6nMwZTw2Ob5EGGg3/fN6ykxooVLJ2C6M9LnINsBOuP/T937r018GIZhNJgnxDAM4wL41Xs/cxcYZkX6AW4oiRJTdxx4n1eACLrMKmbV7F+OQ5bz6Eqy5Pgq8sjZDYRI5+no71uER2puBm3xXaTe09mdKbaVN40JnO4cumNwADwIGRPQnEcsPPr77MrZxgYsbvOcGIZhvIpYJsQwDOMZEGdGBMk8pILWfRkLwseEiBwDgXCIBRacI2+EhWQ4Cs5wRuVgOrpkQpaJe1hahOhMSA5CNdOErs9Hiw8dyOfofC16+W0dvrZdbz3zQ84jtX7KoC4ZpO55R00ef/3ej1gWxDAMQ2HGdMMwjGeAZEa0AJEOVHMH/U0RTzs/LxLwV404qshjiRwMbs3sJWqU6GcHgOkMiN728Bx056vufCRD4xpRlEd/EwPE3TrbnsePbddAjPAZO2TsULDrHWOBDEXj8JFHrv6WbcTXxzAMw+iwTIhhGMYz5vO3f/mOBNdi+paMArD9Dj2w+/BAYHsmRJa5xYfwCIZxvb/YgL5GhRpBnGgvSCxCPBhF0xLXNfsTQ7vONOQJU3l8ntmOwwnj9VMtjGtiVCNCcEogylT04etdhiTMP+mEpgfwE+/8sGVBDMMwIuz2jGEYxjPmV+791N0v3fv8XQBYIMcbfL0X3I9N2Z56fw5jAXXIaAQR5MF4TGdYUdlmOeR1AmGNCqfYYI0KZdPRSgfwU1kQOUMRXhlc74j0JHS937Du/CzPVJYjbZqnNruyC5IdGduHnjiv1zEMwzCGWCbEMAzjOXLn9q/deY2vYYkM9+kYJ1g/k/3Mnei+UN6ON/n64P0lchxjg1PaoECGW3zQy4DE/g8AKKlGwVnrBVkia83ypZJGxcgxOjGGT3BRZWg1iQ+kEz9z/Tqxd0RnRErU+DHLgBiGYYxiN2kMwzCeI3fv/fjd682sDt0i9yKJBchcD8pjWuEprXsZkVNsACDRXHeY/SipxoaqpLHcg7FujsS1j/2ExEUO+8sSwwozuNb3oR9Df4lsYzg5PmXcNwzDMDrsX0nDMIznzAolzlyFW3yIinzPH/IiKVEhh8OKAHCBojFaP6XTdpmUABAxEjwfQbgc0QIlQkZEzq8N2lV3q22zPC5KcMxtkzu1XNxyNwO1IxczdvBUD5Y1DMMw0lg5lmEYxgvib97+B3c8GE9wun3hHZhbigWgNZ8TCEVzX0qyHm/wNaxR43EjQt7iG6OZG8kGrKjEY1rhTb7WBvNLzlQXLLTT2McEiHTAusiMRw6XnPcBhJIsgRgYqwSTsi0tVHTWqGxEyI+884NWhmUYhrEFK8cyDMN4QfzMvb/83ILVuCRLlx/V8GAwSlRgeFTN4z4d4ymdAQCu4yBZ1tW1rwVOadO0IK7xmFZtBuS+O8UH7mkwdrctbKkX1Gsy3r318Fg7Xn2eYwb2jKl9iB8lfogoEkO7bCNXZVkOZALEMAxjJlaOZRiG8QL5uXs/ehcAfvr2r9zZtuxFksFhgQJnjTG+hkeOrNf5Siaod+VHIdxeNu1311SjpP7ysswKG3hilFiB4VvTe2hj2w1sDL/74mQO8XK7iJY4mzEX0ouT/CKgmbR+OYrqDMMwXg4sE2IYhnHFSGUsZEaJXmbVmM63cYsPUXAwZ4fOT0GcaEP2mmqsm3KkCh4LFNigBMPjm/xrOODEhPXGc7Kmuj22OWJi2wBCGnnE68cUUdvebQMO49cLG0poGIYxG/OEGIZhXBIkG6InbwNoSqV2v8+uhUf8N5AYDBjNueBmVsghFrjBy3booIgRAmGDCgUyEAj36QQOhDXKNtNQIMdn/E0AaDtjCQUyrFHjEZ3hDT5sp6SPBfO6rEqOkhEExphQyECoVcYjlf2I1y0TgkgmyQN9D4l8Lj/47g9YGZZhGMYOWDmWYRjGJUEGGv7s7b99R7eLlbkTuxJnPmJiP4aUZAFBgMjvM2ywphJv8Q1kcPjhd/6ruwDw9z/3P985pk0rOMKMjRoFcpSoADTT0smjYNd6Quqmp5Sc0xmVyPja5LlIRoLU89Q5aLL2GvaFiF4/RaFM7PHy+5ZyGYZhGH0sd2wYhnHJ+KV7f629qy7dqxZ73DOKMypzqFCjQj0QLWIi18brM6rgwW2WoECGN/haK0Dk+At2WFGFY9q0x3WIHE9p3Zs1MjXPJEsIkPhvWU4e8evAsORqjJTnpBVCpj8MwzDOjZVjGYZhXFLu3P61O9ISt0SNFcqd1hcBEgf3WphMBeSynIT/X7z3Y4OSo1+8/Rt3pKXvARY4xRoZMpUJCeVYp1Q2PbgYN3iJDA5fdY/arMItPsAJbXCLD/CGP+zto4Bry67GjjuIoGhyO/zoVHadGUllSvR29fbktZoYf+nd/9JKsAzDMPbERIhhGMYrzBdu/+qgK9cv3PvJvYPrL97+u3d0OVYOhy/c+yt3AeBv3f6tOzU83uAjAMB9OhmUNd3kJd72oZNWXPZUjIgnWSbfoUdWCQ+nyrUAbBUiJTy+/72/aMLDMAzjAjARYhiGYTwXZDjjIRdYIMMJbXotgYEgJL7J30QNjyMumnK0kKnIQGCEMrDrXAy2n8qGaJwSGyV8r0RLKBMlYX/+vdsmPAzDMC4YEyGGYRjGc+UXb/9Gm33hRNAvk9szOHxX/QZqMM4oZFayZpz5Y7fGIec44HwgPHK4VtyIB0R7RGowVqjbrlp6/bYDVvPbBIhhGMazwUSIYRiG8dzRXpIYgkPeCIPvrt9EDcZH7rTNjHgwVlRhhRKf9NewaHwzYd0AI7TVPWoEjYiNEr7NhEhGRZdleTD+0/c+Z8LDMAzjGWPdsQzDMIznzs/d+9G74hVJIZmMJ7QBAVihxCM6wymF3wecowLjI3fSCg/JlpTw2KDudbTS7XozEJ7QGse0wRkqVPBthy8TIIZhGM8Hy4QYhmEYl4JfuP337wAhE8JtOVWOIy5wSl1nMD148RovcEIbvMaho9ZR4xU5pRJHXOAtPuzNMfFg1GB8SKftpPZPN0b4v/De95sAMQzDeE6YCDEMwzAuHSJIipnzURYcCrje4CM8pTXWqPE6H+AzfL1dRgQIAPwn733f3d/63G/fqeHxl9/5IRMfhmEYzxkTIYZhGMal50u3f/OO7qSVR9XEGRwydnizESElarztr+MQeesLiUXI8zt6wzAMI+b/B+u/gk6Oi7t5AAAAAElFTkSuQmCC",[[10.38333333333,34.14166666667],[3.50833333333,42.9833333333367]],0.8,null,"bioclim$bio17_27","bioclim$bio17_27"]},{"method":"removeLayersControl","args":[]},{"method":"addControl","args":["","topright","imageValues-bioclim$bio17_27","info legend "]},{"method":"addImageQuery","args":["bioclim$bio17_27",[34.14166666667,3.50833333333,42.9833333333367,10.38333333333],"mousemove",7,"Layer"]},{"method":"addLegend","args":[{"colors":["#040404 , #27123E 11.1111111111112%, #5D1161 24.1830065359479%, #951474 37.2549019607846%, #C63370 50.3267973856214%, #ED5F47 63.3986928104581%, #F69B23 76.4705882352948%, #F9D15E 89.5424836601315%, #FFFE9E "],"labels":["20","40","60","80","100","120","140"],"na_color":"#BEBEBE","na_label":"NA","opacity":1,"position":"topright","type":"numeric","title":"bioclim$bio17_27","extra":{"p_1":0.111111111111112,"p_n":0.895424836601315},"layerId":null,"className":"info legend","group":"bioclim$bio17_27"}]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[34.14166666667,3.50833333333,42.9833333333367,10.38333333333,true,"bioclim$bio17_27","Zoom to bioclim$bio17_27","<strong> bioclim$bio17_27 <\/strong>","bottomright"]},{"method":"createMapPane","args":["point",440]},{"method":"addCircleMarkers","args":[[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,9.5635,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.1959,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.712,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0271,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],6,null,"cases - pf_pos",{"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"pane":"point","stroke":true,"color":"#333333","weight":1,"opacity":[0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9,0.9],"fill":true,"fillColor":["#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#FDE333","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#FDE333","#FDE333","#4B0055","#FDE333","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#FDE333","#4B0055","#FDE333","#FDE333","#FDE333","#4B0055","#FDE333","#FDE333","#FDE333","#FDE333","#4B0055","#FDE333","#4B0055","#FDE333","#4B0055","#FDE333","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#FDE333","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055","#4B0055"],"fillOpacity":[0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6,0.6]},null,null,["<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>1 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.9412 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.9014 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>2 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.8362 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.3175 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>3 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.7521 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.5674 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>4 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.7650 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.7192 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>5 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3632 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.8930 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>6 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.7891 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.2461 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>7 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.6490 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.1657 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>8 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1559 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.7542 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>9 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.2547 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.9471 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>10 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.0551 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.8386 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>11 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2213 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>3.8966 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>12 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.1915 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.3172 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>13 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.8933 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.1376 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>14 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.2297 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.8855 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>15 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.5655 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.3182 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>16 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.3708 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.6748 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>17 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.3667 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.7924 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>18 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.7487 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.0833 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>19 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.0833 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.8833 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>20 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.2865 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.6295 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>21 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.4464 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.6522 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>22 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2630 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.7115 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>23 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2671 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.3957 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>24 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2310 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.1240 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>25 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.9930 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.5050 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>26 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.6116 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.0332 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>27 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.6187 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.1968 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>28 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3569 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.1092 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>29 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.3261 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.8138 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>30 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.0778 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.8246 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>31 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.8319 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>4.7372 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>32 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3960 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.1470 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>33 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.2445 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>6.8403 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>34 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.3276 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.8360 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>35 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.1084 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.9359 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>36 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.1360 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.6250 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>37 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.7040 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3082 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>38 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.0175 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.9245 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>39 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.6281 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0283 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>40 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.6469 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9695 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>41 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.7120 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.5635 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>42 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.7542 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5670 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>43 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.7948 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9661 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>44 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.8295 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0556 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>45 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.9654 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.8265 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>46 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.9704 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7288 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>47 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.9801 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0055 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>48 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.9999 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9174 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>49 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0271 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.1959 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>50 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0395 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.2834 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>51 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0599 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0309 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>52 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0780 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9772 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>53 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0840 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.6416 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>54 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.1159 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.7045 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>55 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.1556 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8416 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>56 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.1590 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.8058 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>57 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.2034 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8657 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>58 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.2913 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.5869 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>59 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.3270 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.1959 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>60 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.3308 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9826 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>61 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.3445 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9457 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>62 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.3963 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6219 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>63 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.3968 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6220 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>64 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.4004 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.6740 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>65 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.4173 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1255 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>66 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5107 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.6234 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>67 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5277 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3530 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>68 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5489 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3850 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>69 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5591 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8370 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>70 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5865 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6584 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>71 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.5915 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.2419 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>72 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.6191 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4416 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>73 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.6542 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.2087 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>74 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.6542 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.2088 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>75 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.6907 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.2143 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>76 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.7183 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3430 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>77 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.7437 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0024 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>78 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.7507 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5789 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>79 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.7586 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5789 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>80 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.8027 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1884 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>81 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.8033 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7131 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>82 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.8197 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8835 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>83 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.0210 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0327 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>84 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.1707 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6420 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>85 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.4795 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.4853 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>86 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.5047 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.9551 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>87 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.5301 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4310 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>88 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.5301 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4810 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>89 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.5692 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.2721 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>90 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.5827 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9215 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>91 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.6020 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.8568 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>92 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.6266 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4378 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>93 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.6292 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.6796 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>94 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.6729 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7164 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>95 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.6994 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.5265 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>96 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7281 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7457 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>97 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7529 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.6636 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>98 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.5418 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0147 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>99 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.5584 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4807 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>100 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.2514 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1806 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>101 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.2528 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4228 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>102 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.3133 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0366 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>103 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.4976 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1427 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>104 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.5888 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4790 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>105 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>35.0732 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8923 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>106 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>34.6097 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.3239 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>107 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7579 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7646 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>108 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7957 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7957 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>109 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7973 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.8636 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>110 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.7999 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7734 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>111 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.8540 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4486 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>112 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.8562 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7554 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>113 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.8836 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0549 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>114 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9107 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.5704 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>115 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9335 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7056 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>116 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9438 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1567 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>117 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9891 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.6245 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>118 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9941 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0856 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>119 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>36.9957 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.1466 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>120 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.0276 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7082 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>121 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.0569 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4420 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>122 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.0951 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.9101 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>123 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.1103 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7261 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>124 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.1853 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7618 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>125 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.2193 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7477 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>126 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.2206 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.6222 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>127 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3451 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8988 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>128 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3466 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6991 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>129 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.3792 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.8775 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>130 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.4100 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.1200 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>131 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.4124 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.7905 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>132 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.6791 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4993 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>133 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.8315 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5970 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>134 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>37.9556 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4754 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>135 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.0200 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.5600 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>136 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.1400 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.6000 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>137 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2500 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.0700 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>138 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.2630 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>5.7117 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>139 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.3136 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3651 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>140 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.3987 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3123 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>141 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.4259 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3283 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>142 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.4823 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3304 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>143 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.4903 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.2976 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>144 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.5255 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.1648 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>145 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.6353 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.3525 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>146 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.6811 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.9926 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>147 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.6886 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.7633 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>148 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.6970 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.1747 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>149 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.8453 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3568 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>150 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.8844 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.9231 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>151 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.8953 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6940 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>152 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.9027 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5766 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>153 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>38.9880 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5048 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>154 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.1790 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.4592 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>155 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.2998 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.2064 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>156 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.3785 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5253 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>157 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.5010 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.2264 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>158 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.5139 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3486 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>159 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.7774 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8836 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>160 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.7803 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>7.8223 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>161 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>39.8374 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9940 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>162 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.3698 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.3784 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>163 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.3708 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8757 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>164 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.4205 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.5641 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>165 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.4641 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8181 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>166 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.5300 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8200 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>167 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.6098 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.6852 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>168 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.6467 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7469 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>169 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.6572 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1998 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>170 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.7848 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8762 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>171 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.7868 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1864 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>172 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.8317 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9247 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>173 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.8815 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7661 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>174 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.8815 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7662 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>175 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.8820 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1064 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>176 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>40.9838 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1895 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>177 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.0049 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1873 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>178 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>1 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.0365 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.3063 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>179 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.0700 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.2800 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>180 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1138 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1352 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>181 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1456 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0751 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>182 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1500 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0300 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>183 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1509 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.0340 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>184 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.1910 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4050 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>185 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.2050 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1511 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>186 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.3146 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9945 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>187 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.3965 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9920 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>188 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.4014 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4602 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>189 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.5294 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8262 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>190 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.5532 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7281 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>191 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.5791 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.8865 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>192 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.6220 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9874 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>193 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.6346 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1368 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>194 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.7083 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.7820 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>195 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.7384 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1384 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>196 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.7534 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.4243 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>197 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.8260 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9030 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>198 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>41.8785 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>8.9745 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>199 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>42.2162 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.1429 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>200 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>42.3192 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.2356 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>201 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>42.3604 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.3663 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>202 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>42.3800 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.3166 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>","<div class='scrollableContainer'><table class=mapview-popup id='popup'><tr class='coord'><td><\/td><th><b>Feature ID <\/b><\/th><td>203 <\/td><\/tr><tr><td>1<\/td><th>pf_pos <\/th><td>0 <\/td><\/tr><tr><td>2<\/td><th>longitude <\/th><td>42.4915 <\/td><\/tr><tr><td>3<\/td><th>latitude <\/th><td>9.3658 <\/td><\/tr><tr><td>4<\/td><th>geometry <\/th><td>sfc_POINT <\/td><\/tr><\/table><\/div>"],{"maxWidth":800,"minWidth":50,"autoPan":true,"keepInView":false,"closeButton":true,"closeOnClick":true,"className":""},["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],{"interactive":false,"permanent":false,"direction":"auto","opacity":1,"offset":[0,0],"textsize":"10px","textOnly":false,"className":"","sticky":true},null]},{"method":"addHomeButton","args":[34.5418,3.8966,42.4915,9.9551,true,"cases - pf_pos","Zoom to cases - pf_pos","<strong> cases - pf_pos <\/strong>","bottomright"]},{"method":"addLayersControl","args":[["CartoDB.Positron","CartoDB.DarkMatter","OpenStreetMap","Esri.WorldImagery","OpenTopoMap"],["bioclim$bio17_27","cases - pf_pos"],{"collapsed":true,"autoZIndex":true,"position":"topleft"}]},{"method":"addLegend","args":[{"colors":["#4B0055","#FDE333"],"labels":["0","1"],"na_color":null,"na_label":"NA","opacity":1,"position":"topright","type":"factor","title":"cases - pf_pos","extra":null,"layerId":null,"className":"info legend","group":"cases - pf_pos"}]},{"method":"addHomeButton","args":[34.14166666667,3.50833333333,42.9833333333367,10.38333333333,true,null,"Zoom to full extent","<strong>Zoom full<\/strong>","bottomleft"]}],"limits":{"lat":[3.50833333333,10.38333333333],"lng":[34.14166666667,42.9833333333367]},"fitBounds":[3.50833333333,34.14166666667,10.38333333333,42.9833333333367,[]]},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n if( strip !==null) strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null},{"code":"function(el, x, data) {\n return (function(el,x,data){\n var map = this;\n\n map.on('keypress', function(e) {\n console.log(e.originalEvent.code);\n var key = e.originalEvent.code;\n if (key === 'KeyE') {\n var bb = this.getBounds();\n var txt = JSON.stringify(bb);\n console.log(txt);\n\n setClipboardText('\\'' + txt + '\\'');\n }\n })\n }).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> ] --- # Data Extraction ```r data <- bind_cols(cases, extract(rast(bioclim), vect(cases))) %>% st_drop_geometry() %>% drop_na() ``` <div id="htmlwidget-3d14110bb495c5c07f31" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-3d14110bb495c5c07f31">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","45","46","47","48","49","50","51","52","53","54","55","56","57","58","59","60","61","62","63","64","65","66","67","68","69","70","71","72","73","74","75","76","77","78","79","80","81","82","83","84","85","86","87","88","89","90","91","92","93","94","95","96","97","98","99","100","101","102","103","104","105","106","107","108","109","110","111","112","113","114","115","116","117","118","119","120","121","122","123","124","125","126","127","128","129","130","131","132","133","134","135","136","137","138","139","140","141","142","143","144","145","146","147","148","149","150","151","152","153","154","155","156","157","158","159","160","161","162","163","164","165","166","167","168","169","170","171","172","173","174","175","176","177","178","179","180","181","182","183","184","185","186","187","188","189","190","191","192","193","194","195","196","197","198","199","200","201"],["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>pf_pos<\/th>\n <th>longitude<\/th>\n <th>latitude<\/th>\n <th>ID<\/th>\n <th>bio1_27<\/th>\n <th>bio2_27<\/th>\n <th>bio3_27<\/th>\n <th>bio4_27<\/th>\n <th>bio5_27<\/th>\n <th>bio6_27<\/th>\n <th>bio7_27<\/th>\n <th>bio8_27<\/th>\n <th>bio9_27<\/th>\n <th>bio10_27<\/th>\n <th>bio11_27<\/th>\n <th>bio12_27<\/th>\n <th>bio13_27<\/th>\n <th>bio14_27<\/th>\n <th>bio15_27<\/th>\n <th>bio16_27<\/th>\n <th>bio17_27<\/th>\n <th>bio18_27<\/th>\n <th>bio19_27<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"className":"dt-right","targets":[2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- class: inverse, center, middle # [_**Predictive Model**_]() --- # Split Data - [Traditional CV]() ```r bad_folds <- data %>% vfold_cv(v = 5) ``` <div id="htmlwidget-20a0b033ea790ace39e0" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-20a0b033ea790ace39e0">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4","5"],[{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[3,5,6,7,8,9,10,11,12,13,14,15,16,17,19,20,21,22,24,25,26,28,29,30,31,32,34,35,36,37,38,40,41,42,43,44,46,47,48,50,51,52,53,54,55,56,57,58,59,60,62,63,65,66,68,69,72,73,74,75,76,77,79,80,81,82,83,84,85,86,87,89,91,92,93,94,96,97,99,100,102,103,105,106,107,108,109,111,112,114,116,118,119,121,122,124,125,126,127,128,129,130,131,132,133,134,135,136,137,139,140,141,142,143,144,145,146,147,148,149,150,151,152,154,155,156,158,160,161,162,163,165,166,167,168,169,170,172,173,174,175,176,177,178,179,180,182,183,184,185,186,187,188,189,192,193,194,197,199,201],"out_id":null,"id":{"id":["Fold1"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,4,5,6,7,8,10,11,12,13,15,16,18,19,20,21,22,23,24,27,28,29,31,32,33,34,35,36,37,38,39,40,41,42,43,45,49,50,52,54,57,58,59,60,61,62,64,65,66,67,68,69,70,71,72,73,76,78,79,80,81,82,83,85,86,88,89,90,91,92,94,95,97,98,99,100,101,102,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,128,129,130,132,133,134,135,136,137,138,139,142,143,144,145,146,147,148,149,150,151,152,153,154,156,157,158,159,160,161,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,179,181,182,187,188,189,190,191,192,194,195,196,197,198,200],"out_id":null,"id":{"id":["Fold2"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,8,9,12,14,17,18,19,21,23,24,25,26,27,29,30,31,32,33,35,37,38,39,40,41,42,44,45,46,47,48,49,50,51,52,53,54,55,56,57,61,62,63,64,65,66,67,69,70,71,72,73,74,75,76,77,78,79,81,83,84,85,87,88,89,90,91,92,93,94,95,96,98,101,102,103,104,105,106,108,110,111,113,114,115,116,117,118,119,120,121,122,123,124,126,127,128,131,132,133,135,136,137,138,139,140,141,142,143,144,145,147,149,150,151,153,154,155,156,157,159,161,162,163,164,165,166,167,168,169,171,172,173,174,176,177,178,179,180,181,182,183,184,185,186,188,190,191,192,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold3"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,5,6,7,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,25,26,27,28,29,30,31,33,34,36,39,42,43,44,45,46,47,48,49,51,53,55,56,57,58,59,60,61,63,64,65,67,68,69,70,71,72,74,75,76,77,78,80,82,83,84,85,86,87,88,89,90,91,93,95,96,97,98,99,100,101,103,104,105,106,107,109,110,111,112,113,114,115,116,117,119,120,123,125,126,127,129,130,131,133,134,135,136,138,139,140,141,142,146,148,149,151,152,153,155,156,157,158,159,160,162,164,165,166,168,170,171,172,173,175,176,178,179,180,181,182,183,184,185,186,187,188,189,190,191,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold4"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,5,6,7,8,9,10,11,13,14,15,16,17,18,20,22,23,24,25,26,27,28,30,32,33,34,35,36,37,38,39,40,41,43,44,45,46,47,48,49,50,51,52,53,54,55,56,58,59,60,61,62,63,64,66,67,68,70,71,73,74,75,77,78,79,80,81,82,84,86,87,88,90,92,93,94,95,96,97,98,99,100,101,102,103,104,107,108,109,110,112,113,115,117,118,120,121,122,123,124,125,126,127,128,129,130,131,132,134,137,138,140,141,143,144,145,146,147,148,150,152,153,154,155,157,158,159,160,161,162,163,164,167,169,170,171,174,175,177,178,180,181,183,184,185,186,187,189,190,191,192,193,195,196,198,199,200,201],"out_id":null,"id":{"id":["Fold5"]}}],["Fold1","Fold2","Fold3","Fold4","Fold5"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>splits<\/th>\n <th>id<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- # Split Data - [Traditional CV]() ```r bad_folds <- data %>% vfold_cv(v = 5) ``` 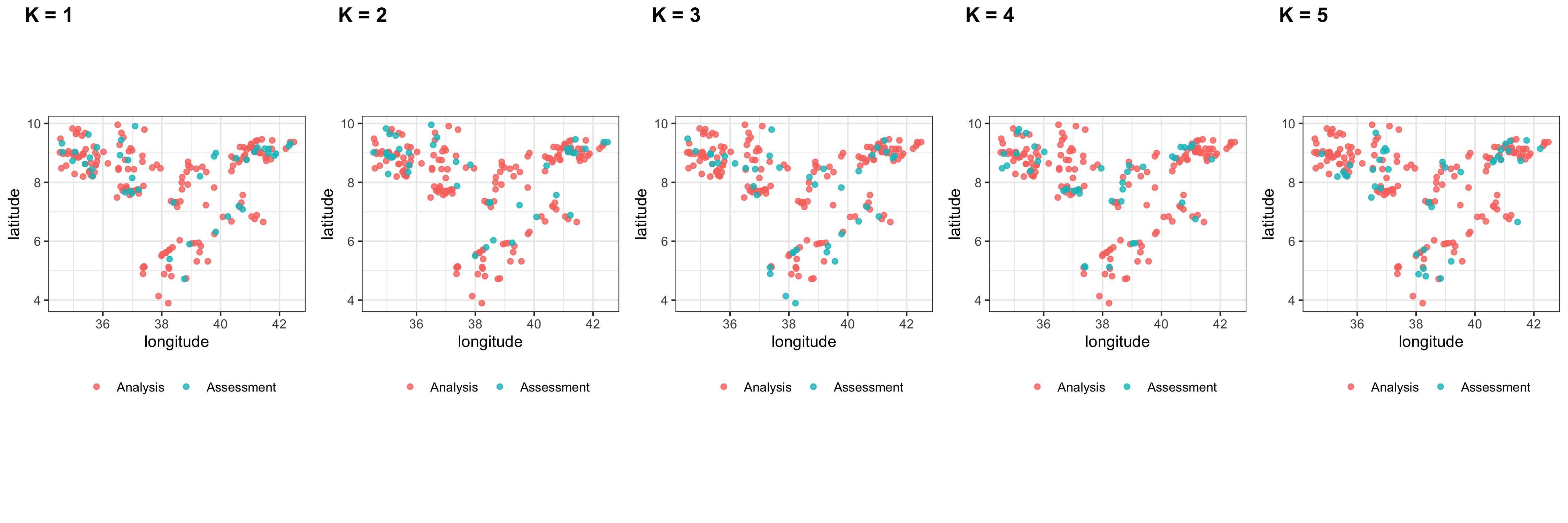<!-- --> --- # Split Data - [Spatial CV]() ```r good_folds <- data %>% spatial_clustering_cv(coords = c("longitude", "latitude"), v = 5) ``` <div id="htmlwidget-a41436f62bed00a86bd4" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-a41436f62bed00a86bd4">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4","5"],[{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,4,5,6,9,11,12,13,15,16,17,19,20,22,23,24,25,26,28,29,30,31,32,33,34,35,36,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159],"out_id":null,"id":{"id":["Fold1"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,84,96,97,98,99,100,101,102,103,104,128,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold2"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,82,83,85,86,87,88,89,90,91,92,93,94,95,98,99,100,101,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold3"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,17,18,19,20,21,22,23,24,25,26,27,28,29,31,32,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold4"]}},{"data":{"pf_pos":["0","0","0","0","0","1","0","0","1","0","0","0","1","1","0","0","0","0","0","0","0","1","0","1","0","0","0","1","0","0","0","0","1","0","1","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","1","0","1","1","1","0","1","1","1","1","0","1","0","1","0","1","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","1","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0","0"],"longitude":[38.9412,39.8362,40.7521,38.765,37.3632,39.7891,40.649,41.1559,39.2547,41.0551,38.2213,39.1915,37.8933,41.2297,39.5655,40.3708,38.3667,40.7487,38.0833,39.2865,41.4464,38.263,38.2671,38.231,37.993,38.6116,40.6187,37.3569,38.3261,40.0778,38.8319,37.396,40.2445,39.3276,39.1084,38.136,40.704,39.0175,34.6281,34.6469,34.7542,34.7948,34.8295,34.9654,34.9704,34.9801,34.9999,35.0395,35.0599,35.078,35.084,35.1159,35.1556,35.159,35.2034,35.2913,35.327,35.3308,35.3445,35.3963,35.3968,35.4004,35.4173,35.5107,35.5277,35.5489,35.5591,35.5865,35.5915,35.6191,35.6542,35.6542,35.6907,35.7183,35.7437,35.7507,35.7586,35.8027,35.8033,35.8197,36.021,36.1707,36.4795,36.5047,36.5301,36.5301,36.5692,36.5827,36.602,36.6266,36.6292,36.6729,36.6994,36.7281,36.7529,34.5418,34.5584,41.2514,41.2528,41.3133,41.4976,34.5888,35.0732,34.6097,36.7579,36.7957,36.7973,36.7999,36.854,36.8562,36.8836,36.9107,36.9335,36.9438,36.9891,36.9941,36.9957,37.0276,37.0569,37.0951,37.1103,37.1853,37.2193,37.2206,37.3451,37.3466,37.3792,37.41,37.4124,37.6791,37.8315,37.9556,38.02,38.14,38.25,38.263,38.3136,38.3987,38.4259,38.4823,38.4903,38.5255,38.6353,38.6811,38.6886,38.697,38.8453,38.8844,38.8953,38.9027,38.988,39.179,39.2998,39.3785,39.501,39.5139,39.7774,39.7803,39.8374,40.3698,40.3708,40.4205,40.4641,40.53,40.6098,40.6467,40.6572,40.7848,40.7868,40.8317,40.8815,40.8815,40.882,40.9838,41.0049,41.0365,41.07,41.1138,41.1456,41.15,41.1509,41.191,41.205,41.3146,41.3965,41.4014,41.5294,41.5532,41.5791,41.622,41.6346,41.7083,41.7384,41.7534,41.826,41.8785,42.2162,42.3192,42.3604,42.38,42.4915],"latitude":[5.9014,6.3175,7.5674,4.7192,4.893,6.2461,7.1657,6.7542,5.9471,6.8386,3.8966,5.3172,4.1376,6.8855,5.3182,6.6748,5.7924,7.0833,4.8833,5.6295,6.6522,5.7115,5.3957,5.124,5.505,6.0332,7.1968,5.1092,4.8138,6.8246,4.7372,5.147,6.8403,5.836,5.9359,5.625,7.3082,5.9245,9.0283,8.9695,8.567,8.9661,9.0556,9.8265,8.7288,9.0055,8.9174,8.2834,9.0309,8.9772,9.6416,9.7045,8.8416,9.8058,8.8657,9.5869,8.1959,8.9826,8.9457,8.6219,8.622,9.674,9.1255,9.6234,8.353,8.385,8.837,8.6584,8.2419,8.4416,8.2087,8.2088,9.2143,8.343,9.0024,8.5789,8.5789,9.1884,8.7131,8.8835,9.0327,8.642,7.4853,9.9551,8.431,8.481,9.2721,8.9215,7.8568,9.4378,9.6796,7.7164,9.5265,7.7457,7.6636,9.0147,9.4807,9.1806,9.4228,9.0366,9.1427,8.479,8.8923,9.3239,8.7646,7.7957,7.8636,7.7734,8.4486,8.7554,9.0549,7.5704,7.7056,9.1567,7.6245,9.0856,8.1466,7.7082,8.442,9.9101,7.7261,7.7618,7.7477,7.6222,8.8988,8.6991,7.8775,5.12,9.7905,8.4993,8.597,8.4754,5.56,5.6,5.07,5.7117,7.3651,7.3123,7.3283,7.3304,7.2976,7.1648,7.3525,7.9926,7.7633,8.1747,8.3568,7.9231,8.694,8.5766,8.5048,8.4592,8.2064,8.5253,7.2264,8.3486,8.8836,7.8223,8.994,8.3784,8.8757,8.5641,8.8181,8.82,8.6852,8.7469,9.1998,8.8762,9.1864,8.9247,8.7661,8.7662,9.1064,9.1895,9.1873,9.3063,9.28,9.1352,9.0751,9.03,9.034,9.405,9.1511,8.9945,8.992,9.4602,8.8262,8.7281,8.8865,8.9874,9.1368,8.782,9.1384,9.4243,8.903,8.9745,9.1429,9.2356,9.3663,9.3166,9.3658],"ID":[1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,42,43,44,45,46,47,48,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203],"bio1_27":[198,222,198,200,215,229,176,238,191,232,219,228,227,234,192,231,165,207,184,200,242,186,203,197,178,169,172,222,198,203,180,230,219,191,194,193,182,197,205,205,200,205,211,209,211,196,210,200,196,199,203,200,208,204,207,195,200,199,199,203,203,194,203,189,199,204,198,199,197,208,195,195,198,196,202,194,194,195,193,190,220,190,193,178,207,212,209,188,197,214,206,165,214,180,186,202,207,149,208,217,162,218,207,205,207,187,193,185,200,209,188,189,189,182,190,193,202,192,206,161,194,197,195,192,197,181,194,232,208,193,179,181,195,192,199,186,189,198,195,196,195,188,186,192,195,190,197,191,187,197,204,208,187,212,148,201,247,158,251,214,207,207,206,202,200,195,221,208,225,197,210,210,194,201,194,201,197,190,168,165,159,219,190,223,225,200,205,222,207,190,151,230,210,167,215,176,216,200,187,204,200],"bio2_27":[145,143,142,136,109,142,141,141,146,141,143,136,132,141,130,143,134,142,124,138,141,142,135,129,136,138,141,107,128,145,135,109,144,143,146,140,141,146,121,122,121,124,124,124,129,125,128,126,125,127,123,123,132,125,133,124,130,132,133,136,136,125,133,126,135,138,136,138,133,141,133,133,133,137,137,138,139,133,137,134,142,141,154,139,151,151,142,134,156,144,143,140,145,150,153,122,116,129,132,134,127,127,129,116,147,154,156,153,149,148,141,154,155,142,155,145,155,156,153,144,157,157,157,155,153,149,155,110,151,149,144,141,140,141,128,142,144,144,143,143,143,145,141,138,137,141,145,138,150,148,147,146,143,144,151,144,155,141,154,145,143,144,142,141,141,139,143,140,141,138,139,139,136,135,134,134,133,133,131,131,131,134,132,135,134,129,132,135,131,128,126,135,127,124,131,124,131,129,128,132,136],"bio3_27":[75,75,78,78,72,75,78,73,76,74,76,77,77,74,75,74,77,75,72,76,73,77,74,72,74,76,78,72,76,76,75,74,75,76,76,75,78,76,74,75,80,75,72,69,78,77,75,80,77,76,69,70,77,70,77,72,81,77,78,78,78,71,76,73,75,76,77,76,76,74,75,75,76,74,74,74,75,77,76,77,61,74,78,73,73,71,67,77,76,67,70,79,68,77,76,76,67,81,73,77,84,77,76,67,72,76,75,76,79,73,76,75,74,76,74,74,75,73,77,75,73,73,72,72,75,79,73,75,72,77,78,79,74,76,73,77,75,75,74,75,74,75,74,76,74,76,75,74,75,74,74,73,71,72,78,71,69,81,67,75,73,76,73,74,76,76,70,75,69,77,77,77,75,75,75,74,75,78,81,81,82,71,79,76,77,75,81,79,81,83,84,78,80,80,80,82,75,74,75,73,71],"bio4_27":[618,714,820,1124,1329,730,751,813,654,832,1414,854,1262,808,862,813,636,815,1170,661,776,729,875,1100,982,524,746,1319,1137,832,1092,1290,844,634,622,732,776,617,1222,1164,1074,1181,1284,1421,1120,1165,1179,1009,1151,1227,1355,1383,1144,1408,1159,1401,943,1220,1183,1038,1038,1422,1272,1424,961,963,1129,1073,945,1043,981,981,1349,1048,1228,1095,1106,1323,1109,1139,1786,1148,751,1125,1146,1219,1476,1261,1010,1439,1367,768,1429,801,835,1190,1438,755,1326,1108,646,1178,1197,1384,1178,869,961,841,846,1162,1103,866,887,1059,866,1083,924,948,975,917,1004,1005,1032,993,1173,943,966,1231,1371,1076,1092,1096,934,784,1057,729,732,731,760,708,751,717,800,720,816,831,1073,840,1103,1212,1279,1467,1348,1547,864,1523,1871,720,1969,1294,1412,1267,1297,1287,1166,1128,1580,1261,1581,1108,1116,1116,1144,1205,1109,1274,1178,1037,785,769,735,1483,960,1135,1098,1202,759,849,750,686,626,841,852,809,736,715,952,908,971,1111,1289],"bio5_27":[296,316,284,299,303,324,263,330,288,323,321,323,321,324,286,323,257,296,284,295,334,284,304,299,283,261,258,308,294,294,281,316,310,287,292,293,268,294,298,297,287,300,310,311,306,287,307,288,287,294,304,299,305,305,305,293,289,296,296,299,299,294,303,286,294,300,296,297,290,309,289,289,297,292,303,292,292,292,289,284,331,290,289,279,313,318,317,285,297,322,313,254,323,274,283,293,307,229,297,300,238,313,304,305,312,285,293,283,299,314,287,287,288,281,290,294,304,293,308,260,296,299,299,296,300,277,297,318,315,291,273,273,302,294,298,284,279,290,286,286,286,280,274,277,282,277,291,278,283,294,301,308,285,310,240,301,355,241,360,305,303,298,300,295,290,283,321,299,323,285,296,296,284,290,282,292,285,275,248,245,239,311,273,306,305,286,283,300,284,267,226,308,285,246,293,256,297,282,266,285,283],"bio6_27":[105,127,103,125,153,135,83,139,97,133,135,147,150,134,114,132,83,109,112,114,143,100,123,122,101,81,79,161,126,104,103,169,119,100,100,108,88,104,135,136,136,136,140,132,141,125,138,132,125,129,128,125,135,127,134,122,129,125,126,126,126,120,129,115,116,120,121,117,115,119,113,113,123,107,120,106,107,120,109,112,99,101,94,91,107,107,107,111,94,110,109,78,111,81,83,134,134,70,117,126,87,150,136,134,110,83,86,82,112,112,103,83,81,95,82,100,99,82,110,68,82,85,83,82,96,89,87,172,108,98,90,96,113,109,124,100,87,98,94,96,95,88,85,96,98,93,98,93,84,96,103,108,86,111,47,100,131,67,132,113,108,109,108,106,106,102,118,113,121,106,116,116,105,110,105,111,108,105,87,85,80,124,107,130,131,115,122,131,124,113,77,137,128,92,130,105,124,109,96,105,94],"bio7_27":[191,189,181,174,150,189,180,191,191,190,186,176,171,190,172,191,174,187,172,181,191,184,181,177,182,180,179,147,168,190,178,147,191,187,192,185,180,190,163,161,151,164,170,179,165,162,169,156,162,165,176,174,170,178,171,171,160,171,170,173,173,174,174,171,178,180,175,180,175,190,176,176,174,185,183,186,185,172,180,172,232,189,195,188,206,211,210,174,203,212,204,176,212,193,200,159,173,159,180,174,151,163,168,171,202,202,207,201,187,202,184,204,207,186,208,194,205,211,198,192,214,214,216,214,204,188,210,146,207,193,183,177,189,185,174,184,192,192,192,190,191,192,189,181,184,184,193,185,199,198,198,200,199,199,193,201,224,174,228,192,195,189,192,189,184,181,203,186,202,179,180,180,179,180,177,181,177,170,161,160,159,187,166,176,174,171,161,169,160,154,149,171,157,154,163,151,173,173,170,180,189],"bio8_27":[205,229,206,205,219,237,183,246,198,241,226,233,233,241,196,239,168,215,187,205,250,189,206,199,181,175,179,226,202,211,184,234,227,198,201,197,189,204,195,195,191,196,197,198,197,183,197,186,182,186,191,186,196,188,195,180,188,186,188,192,192,179,191,172,188,193,187,189,187,199,185,185,184,186,190,185,185,180,183,176,213,181,188,166,200,206,199,173,194,204,196,156,205,174,181,192,197,148,213,220,158,208,195,195,198,183,190,181,189,200,176,184,186,170,187,183,197,189,198,152,191,194,192,188,187,170,189,236,199,184,168,170,198,195,202,189,183,192,189,191,190,182,182,189,193,186,195,190,182,193,202,208,187,212,147,203,256,160,261,219,210,211,210,206,203,197,227,211,231,200,212,212,196,204,197,205,200,192,167,164,158,226,191,226,228,204,203,223,205,186,146,231,209,166,213,169,217,200,187,207,207],"bio9_27":[197,218,190,185,198,226,170,232,188,226,200,218,209,227,183,224,169,200,169,202,237,191,211,183,188,169,166,205,184,196,165,213,211,189,191,198,175,195,212,212,206,213,220,216,219,203,217,208,203,208,209,205,216,208,216,202,205,207,208,208,208,200,211,195,200,206,204,202,197,209,196,196,204,194,206,193,194,201,194,193,216,189,190,179,205,209,207,193,194,211,204,166,212,178,183,210,216,138,189,201,154,226,216,213,207,179,189,178,200,208,191,185,179,186,181,194,195,182,201,159,183,186,185,184,191,181,187,217,206,189,177,179,203,199,186,191,183,192,188,189,188,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,227,148,230,195,187,188,187,184,183,181,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,171,201,187,173,186,181],"bio10_27":[205,230,206,215,233,238,183,246,198,241,237,240,242,242,205,239,170,215,201,209,251,191,215,213,190,175,179,240,214,211,195,247,227,198,201,201,189,204,222,221,215,222,230,230,226,212,227,212,212,217,223,220,224,224,224,215,212,216,216,218,218,215,221,209,213,218,215,215,211,224,210,210,218,211,221,210,211,214,209,206,240,206,204,195,224,229,230,205,212,234,226,176,235,191,198,218,228,158,223,229,170,235,224,226,224,200,207,198,212,226,203,201,201,196,203,208,215,205,221,175,208,210,210,206,213,193,208,249,228,207,193,195,207,200,214,191,199,207,205,205,205,198,197,200,205,200,211,200,203,213,220,227,204,231,158,220,267,165,273,228,224,221,221,217,214,208,239,223,242,210,223,223,208,215,207,216,211,203,178,175,168,236,202,235,236,214,213,231,215,199,159,238,220,178,224,185,226,211,199,215,214],"bio11_27":[191,212,185,185,198,219,164,226,183,220,200,218,209,222,183,219,156,194,169,194,232,178,192,183,164,165,160,205,184,190,165,213,206,184,187,185,169,190,191,191,186,192,197,195,197,183,196,186,182,185,189,184,195,188,194,178,187,184,185,191,191,177,188,171,188,193,185,187,187,198,185,185,182,186,188,183,183,179,181,176,206,178,186,166,197,202,195,171,186,200,192,156,201,172,178,188,193,138,189,201,154,203,194,190,196,178,182,177,189,197,175,180,179,169,181,181,193,182,196,152,183,186,185,183,185,169,185,217,194,181,165,167,182,182,186,178,182,190,187,188,187,182,177,183,184,179,183,179,174,181,187,188,168,191,137,180,219,147,222,195,187,188,187,184,183,178,199,190,202,181,193,193,178,183,178,184,180,176,157,155,149,198,177,206,208,183,194,208,196,182,144,216,198,157,204,169,201,187,173,186,181],"bio12_27":[958,738,999,546,622,673,1138,593,988,644,540,507,443,623,675,677,1329,901,657,754,531,971,653,608,1092,1533,1144,557,525,888,647,478,785,927,993,873,1134,962,1407,1411,1421,1409,1349,1402,1372,1583,1407,1471,1607,1553,1529,1609,1478,1599,1506,1710,1624,1647,1652,1634,1634,1753,1635,1837,1708,1660,1762,1760,1734,1665,1761,1761,1810,1778,1766,1839,1836,1893,1874,1934,1522,1875,1654,1781,1761,1674,1423,1862,1753,1341,1431,1830,1319,1836,1748,1455,1278,1173,899,806,1040,1238,1456,1330,1570,1859,1879,1841,1894,1512,1536,1594,1709,1538,1569,1377,2062,1618,1691,1613,1542,1482,1426,1354,1240,1421,1370,460,1106,1243,1242,1240,916,877,572,971,923,861,861,839,858,1035,865,923,728,951,884,772,915,879,847,811,870,761,925,832,605,1175,573,910,1080,1001,1075,1099,1064,1110,942,1006,884,1076,951,951,1081,1011,1046,994,1013,1031,1125,1134,1152,846,1003,765,739,901,805,699,788,857,1023,630,754,936,694,825,656,708,736,708,728],"bio13_27":[198,159,196,163,129,151,250,151,207,159,174,148,130,156,191,157,233,206,162,188,141,192,154,153,189,246,240,105,150,175,176,91,167,206,207,178,238,200,275,274,234,264,246,261,228,302,248,228,303,285,285,300,259,305,269,322,260,297,299,286,286,339,310,354,283,284,325,313,282,292,285,285,358,295,342,319,320,376,336,357,283,336,237,352,325,311,302,369,289,292,304,281,291,306,286,295,240,252,176,133,216,195,255,252,317,330,331,323,399,309,320,260,307,330,268,293,412,293,344,361,283,269,259,238,271,296,247,90,283,249,271,263,178,173,150,192,132,122,124,122,122,159,140,157,134,180,201,142,230,223,216,207,186,191,161,187,141,181,140,140,193,153,182,182,162,174,163,155,156,170,144,144,184,179,188,185,189,187,216,216,225,163,184,123,118,182,119,101,118,143,215,90,124,204,95,136,96,112,129,117,125],"bio14_27":[18,8,7,4,15,8,7,4,18,4,5,6,4,4,7,4,20,4,8,11,4,17,14,8,24,22,8,12,4,6,6,12,5,16,19,19,7,18,5,6,11,6,6,0,11,7,8,19,7,8,2,2,11,1,10,4,25,8,9,16,16,4,6,5,26,22,11,16,29,18,30,30,4,26,7,18,18,4,13,9,5,14,31,9,18,17,8,11,29,6,6,31,6,24,25,5,2,10,9,7,9,10,9,4,14,21,24,22,8,14,13,24,19,12,21,14,21,18,13,12,17,17,16,17,10,9,17,12,3,10,10,9,22,19,6,17,13,14,13,13,14,18,10,6,5,6,7,5,5,6,8,8,6,5,12,5,4,16,5,5,9,5,7,6,5,6,11,8,10,8,7,7,10,10,10,10,10,9,10,10,10,9,9,7,6,9,7,5,7,8,9,5,7,8,6,7,7,8,4,8,8],"bio15_27":[72,83,74,102,71,85,84,105,70,102,102,106,95,103,111,91,61,91,79,93,109,74,81,82,61,58,79,63,93,73,96,63,82,77,70,73,83,71,77,77,67,78,81,93,77,79,81,67,81,81,91,91,82,93,84,91,68,84,84,78,78,92,87,91,71,73,84,79,69,74,69,69,92,72,88,78,78,92,82,85,88,81,54,90,78,78,91,84,63,94,95,59,95,64,62,77,83,73,66,59,70,70,82,81,82,68,69,67,80,80,83,61,67,85,64,83,79,68,79,92,69,70,70,66,82,80,70,63,100,80,83,86,68,72,85,74,55,54,54,54,53,53,55,61,64,68,84,65,99,98,98,99,80,95,56,83,85,50,84,60,64,61,63,62,61,62,62,61,61,62,59,59,64,63,64,65,66,65,68,67,68,65,64,58,58,67,58,56,58,60,70,56,60,70,57,62,58,59,64,60,63],"bio16_27":[428,367,444,304,303,345,565,346,430,368,293,288,234,358,377,364,544,479,330,386,319,454,325,309,473,613,544,250,282,415,346,211,395,426,434,413,550,428,691,692,654,686,659,764,662,778,700,658,805,779,827,875,752,893,774,940,756,860,860,820,820,979,875,1024,820,806,928,902,822,827,831,831,1016,858,971,930,931,1072,980,1041,818,985,689,974,923,883,817,1022,822,783,835,804,776,840,792,717,621,594,422,344,498,571,731,648,853,889,917,871,986,821,846,717,813,857,729,760,1104,780,901,947,757,734,707,653,690,757,682,206,692,684,697,719,422,415,295,454,377,354,357,349,353,427,375,416,341,466,501,370,575,547,529,514,485,468,404,475,334,475,313,389,495,439,482,488,461,487,425,437,395,477,400,400,498,465,487,465,477,477,539,540,556,391,463,320,308,429,324,264,320,364,481,236,321,451,273,341,269,301,332,303,313],"bio17_27":[65,39,46,23,53,35,49,23,72,26,19,23,16,25,27,30,94,36,50,43,20,64,56,46,99,95,52,63,26,46,35,54,38,64,71,66,48,66,38,42,61,37,27,7,41,44,33,67,41,39,12,12,40,9,38,18,83,35,38,56,56,17,26,22,80,74,41,54,90,69,93,93,20,82,26,61,60,20,49,38,21,50,121,30,65,60,25,45,105,20,18,117,19,107,110,48,14,66,56,50,58,41,37,19,52,101,102,103,61,55,49,106,94,45,98,51,84,88,66,45,82,79,75,79,50,50,69,50,17,48,45,38,78,67,39,64,62,63,62,61,64,80,60,48,42,43,36,37,24,29,33,34,35,29,66,35,29,97,31,54,68,58,66,66,62,65,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,46,35,38,38,37,37],"bio18_27":[301,238,444,85,142,223,565,210,430,225,95,58,93,216,64,364,351,479,152,263,193,291,104,119,179,583,544,113,105,415,107,93,395,305,434,285,550,428,123,126,186,118,97,74,115,130,97,191,124,114,86,92,93,85,93,104,204,113,116,154,154,101,103,112,199,195,140,167,210,198,215,215,102,203,111,180,179,107,162,145,76,168,448,317,359,336,250,162,412,229,237,479,222,461,444,131,94,335,289,264,323,143,98,103,160,450,438,451,239,164,160,416,426,155,402,156,258,403,218,273,380,233,352,230,164,200,223,91,177,186,183,189,146,130,112,291,190,179,179,247,254,241,242,273,191,256,192,201,174,163,184,168,221,172,275,208,211,356,196,306,350,334,356,369,363,375,288,336,287,356,325,325,342,314,323,300,307,323,344,350,351,267,314,253,246,263,277,256,269,284,321,231,252,284,237,178,218,225,248,246,255],"bio19_27":[245,169,64,23,53,158,80,45,111,47,19,23,16,46,27,49,298,61,50,62,43,164,62,46,186,406,81,63,26,61,35,54,55,255,110,117,77,103,658,592,619,671,659,764,662,721,670,658,755,738,822,875,728,893,749,927,746,827,828,804,804,970,846,1008,820,806,898,881,822,817,831,831,983,858,936,915,915,1033,952,1041,790,944,234,974,881,80,800,963,223,777,835,804,770,835,204,595,618,66,56,50,58,553,702,630,810,195,206,196,986,779,806,109,173,824,98,721,184,158,866,947,82,136,129,275,652,754,274,50,676,644,672,663,129,119,39,164,99,96,95,92,99,427,60,48,42,43,36,37,24,29,33,34,35,29,101,35,30,146,33,54,68,58,66,66,62,66,65,63,60,67,59,59,67,64,65,63,62,62,66,67,67,55,60,48,45,54,47,41,46,50,58,36,43,50,38,341,35,38,38,37,37]},"in_id":[3,7,8,10,14,16,18,21,27,30,33,37,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,129,130,131,132,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201],"out_id":null,"id":{"id":["Fold5"]}}],["Fold1","Fold2","Fold3","Fold4","Fold5"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>splits<\/th>\n <th>id<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"pageLength":5,"scrollX":true,"dom":"tp","columnDefs":[{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false,"lengthMenu":[5,10,25,50,100]}},"evals":[],"jsHooks":[]}</script> --- # Split Data - [Spatial CV]() ```r good_folds <- data %>% spatial_clustering_cv(coords = c("longitude", "latitude"), v = 5) ``` 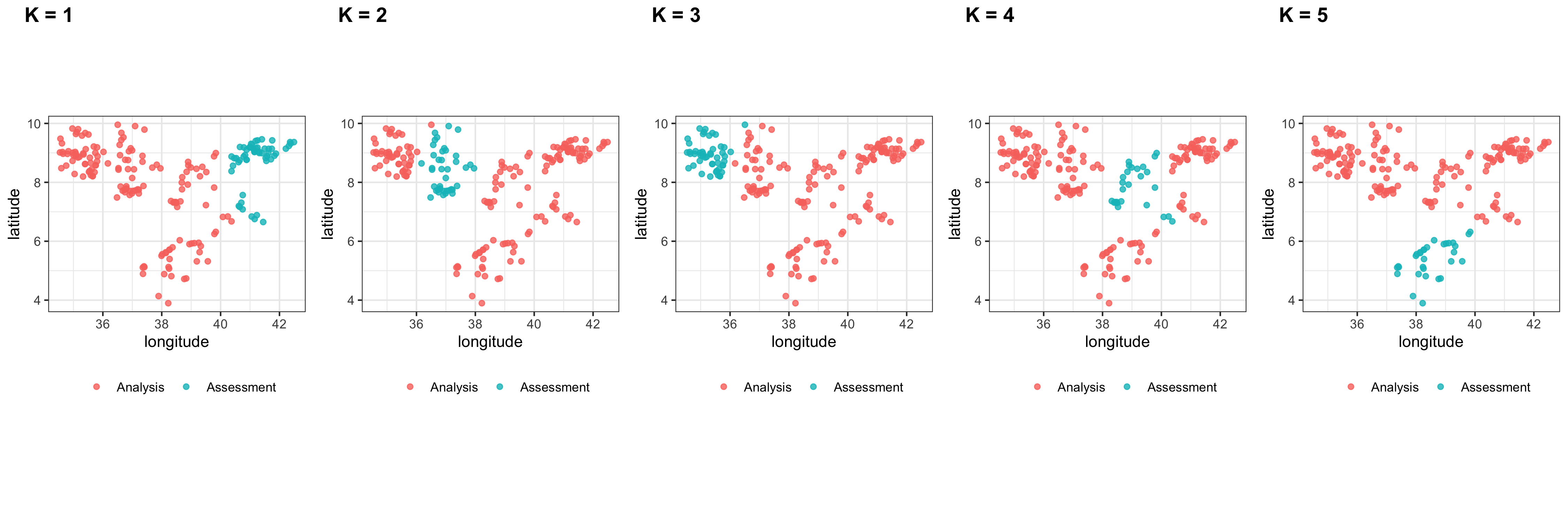<!-- --> --- # Parametrization (Recipe) - [Logistic Model]() ```r lr_recipe <- workflow() %>% add_variables(outcomes = pf_pos, predictors = starts_with("bio")) %>% add_model(logistic_reg(mode = "classification", engine = "glm")) ``` <br> - [Random Forest Model]() ```r rf_recipe <- workflow() %>% add_variables(outcomes = pf_pos, predictors = starts_with("bio")) %>% add_model(boost_tree(mode = "classification", engine = "xgboost")) ``` --- # Training <br> - [Define performance metrics]() ```r metrics <- metric_set(roc_auc, accuracy, recall, precision) ``` <br> - [Train models]() ```r regular_lr <- fit_resamples(lr_recipe, bad_folds, metrics = metrics) spatial_lr <- fit_resamples(lr_recipe, good_folds, metrics = metrics) spatial_rf <- fit_resamples(rf_recipe, good_folds, metrics = metrics) ``` --- # Performance - [ Logistic Model (Traditional CV) ]() ```r collect_metrics(regular_lr) ``` <div id="htmlwidget-e4bd493d85a3509ee56b" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-e4bd493d85a3509ee56b">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4"],["accuracy","precision","recall","roc_auc"],["binary","binary","binary","binary"],[0.845365853658537,0.895449346405229,0.932248803827751,0.797526457421194],[5,5,5,5],[0.0332004216123972,0.0291610238312152,0.0365963690354252,0.060855905095842],["Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>.metric<\/th>\n <th>.estimator<\/th>\n <th>mean<\/th>\n <th>n<\/th>\n <th>std_err<\/th>\n <th>.config<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-right","targets":[3,4,5]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> --- # Performance - [ Logistic Model (Spatial CV) ]() ```r collect_metrics(spatial_lr) ``` <div id="htmlwidget-a6c8db2b959f1f23b56a" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-a6c8db2b959f1f23b56a">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4"],["accuracy","precision","recall","roc_auc"],["binary","binary","binary","binary"],[0.759634812192416,0.865202722838191,0.824679245283019,0.469596136567835],[5,5,5,5],[0.0778170303806387,0.0606771431655374,0.0630349793446286,0.0797016947032687],["Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>.metric<\/th>\n <th>.estimator<\/th>\n <th>mean<\/th>\n <th>n<\/th>\n <th>std_err<\/th>\n <th>.config<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-right","targets":[3,4,5]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> --- # Performance - [ Random Forest Model (Spatial CV) ]() ```r collect_metrics(spatial_rf) ``` <div id="htmlwidget-88b5ce566e9d6c02fddf" style="width:100%;height:auto;" class="datatables html-widget"></div> <script type="application/json" data-for="htmlwidget-88b5ce566e9d6c02fddf">{"x":{"filter":"none","vertical":false,"data":[["1","2","3","4"],["accuracy","precision","recall","roc_auc"],["binary","binary","binary","binary"],[0.794427784197369,0.853429508756605,0.935666666666667,0.313148023360288],[5,5,5,5],[0.0642276815306426,0.0675236324536032,0.0545170717400624,0.118785955044038],["Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1","Preprocessor1_Model1"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>.metric<\/th>\n <th>.estimator<\/th>\n <th>mean<\/th>\n <th>n<\/th>\n <th>std_err<\/th>\n <th>.config<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-right","targets":[3,4,5]},{"orderable":false,"targets":0}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> --- # Final model - [Logistic Model]() ```r fit_lr <- data %>% dplyr::select(pf_pos, starts_with("bio")) %>% glm(pf_pos ~ ., family = binomial(), data = .) ``` - [Random Forest Model]() ```r fit_rf <- data %>% dplyr::select(-c(longitude, latitude, ID)) %>% randomForest(pf_pos ~ ., data = .) ``` --- # Spatial Prediction .pull-left2[ - [Logistic Model]() .tiny2[ ```r pred_lr <- predict(model = fit_lr, object = bioclim, type="response") mapview(pred_lr) ``` ] ] .pull-right2[ <div id="htmlwidget-007e0324f838fdd720c2" style="width:504px;height:504px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-007e0324f838fdd720c2">{"x":{"options":{"minZoom":1,"maxZoom":52,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["CartoDB.Positron","CartoDB.Positron","CartoDB.Positron",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["CartoDB.DarkMatter","CartoDB.DarkMatter","CartoDB.DarkMatter",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenStreetMap","OpenStreetMap","OpenStreetMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["Esri.WorldImagery","Esri.WorldImagery","Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenTopoMap","OpenTopoMap","OpenTopoMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addRasterImage","args":["data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAyEAAAJvCAYAAABsy95bAAAgAElEQVR4nOy9eZxkV3Xn+T33LbFn5FabaldJSAgBEsMmIUC2p932NB4bLRgbe7y1l8bTNu1u/LHHS1g9Tbt77KZt7Hb3GLvby3gBIQEGSbYxMsJaAAHal1JVqdas3CMz9hfvvXvP/PEiQoUQtAGhkkrvq08qK2PJiHjvRuT53fM758jtt9/+a+Tk5OTk5HyNmHd9vHHZa+5DxLJ84kKaG1XWNwN2be9z4E2f5qb//p0c2J6QWkNvYIhT4VTH8e2Xt3n/vSEGeOsBn8FvfPcNZ/u15OTk5OQ8t5iz/QRycnJycl54XLH4042vdv3Goy9/xstv0iPc//g093lLDHH81tENBj9561f9XTk5OTk55x6SZ0Jycl5YXNH9hcY91f+Q7xznPCc88rZbGj/2f38Y+9gcj9xzJfsPnMB4KeVXPIYOfNa+8Bqc9fDDhHu+cB6nepaSZziU9DnorfNyu4Vl6VPXArOEHJUOA0lJsMy7Mj6GMj4Pe6v8QHEvnsBj3SFrDKkR8E9vviZf6zk5OTnnILkIycl5nnJl4bcaBCFSKIFzqE2gtY62NzB7LuKu1o/nwVnOc4L7P29p/PLy43gYvs3uZ9r4rLqEAGGNIUumy7wrc8RrEpMQElDVEAckYkmw/GTlfB5uD/m0d4LX2p18wVviUruVtsRYHAV8+iT8+O5Zbj0xZIAlwdEyQy7TWV5z03fl6z0nJyfnHCIXITk5z0PeUH9/g2IFXTkOzoFN0dYmMjsPvo9Mb0U3V5Dprdy1/iM3vGHbnzcwBnvnJ5H5OjI1A+q42/xKHrjlfEN8+JoPNn761Y73fV64VR6gLCVezm7KGtCXhEvdLJ/yTmYZDQ0AaMmADhEhPgEel9rtPOCd5tvsfv7aOwxAkYB9bpZZLdKUiBSlJREBhvPdNIvSA6CmIXukggFemwuRnJycnHOGvCYkJ+d5iNoEXTsJUR+KFbS1ycn3vxqqdUhitLWW3bBY4crwvQ0dtCEeYPbuwh1ukfztKiQxV8aN3Guf8w2xlSI/et9p/kYewqFUpcSytHnELNKUPk9Kh+2uxgY9TkiTviR4eIT4JGRZEABBOCk9yoRYHAArpkeC45hp0pMhJXzOczW+6J3mMpllq5ZJxHFK+2xocjYPQ05OTk7Os4x/tp9ATk7OM6AOmd4KM9vBeDDosPudD6DLBtm+BzZX0WgAYQk9fTzLkGzfj2zbA24B/40lCIvg+7xh/s8bxAMwBooV7lp422Q3+cr0hobsvJC7lr8/32HO+TJOvOO2xpPS4SfCl7IZO/6bfp6uDmjTw+GYlSpHzBoxKTVKKI4FXSfFUpESQ42pSok7vSPs0jkeMqepUqCnESUJWaND4ln6xMxTZaerMUvIk4Tcyyp1KVLXkG/fVube5fRsH46cnJycnGcR74d+6IeuPttPIicn50vZM/XI1TiHbiyBs5AmmJ3ng0vRU8d46D1XMNs/Da1jyHwRUBCDhEVW/mQXHDaEl6VgU6RQwh18IMueuIQ90wev3u0+dfWuY39ytRRD7J2PsnfXw1ef5I13nO3XnfP8YuOmw1fb1LCn4vNH8SFUlJeykxO6SkJKhwE9Inzx6RHR1A4WS4CPJ0JEzJCEshRpSY8SIS36JKTsYpahpPSJqVKgI0PKFLjXW0BRZrXMmvQ5bjZY6/hsSszxDxy8+iXfe0m+TnNycnLOAXI7Vk7O85E0ndSCSFjKLFmnDqOtDR75rW9hbTPkntuvQmYVgOGHy7jHHgUvYGb3IR667yLSv1vB3ncC3Vwh/UwRCoXs9yyfwD34KPY4UJ9j8PBFEJbO7uvNeV6y78+/84YF6fFLnc9S1zKJWg7LCikpAR4hAYrS0wEDHeLh4ePT1h6ObG0mmlAmZIoyAAE+DkdTeiiKj2FV27zC7qAlEfNaJSRg2XQxwDadwoqS4mhJfBaPRk5OTk7Os0lux8rJeY64YuVnGhjhnvnf/qrWpysHv9RQV8x+qM1AEKKP3kf7lpfSas5y4KWPsX7PZVz+qkNI6GMfHHLy8MXsm93Anbqb8G37qX8hRipDvEu3Q78LQYq9t4f/ep/07k2gRHRsH+XpR6m+YycUdhD9yK2NN//YRzDbDOaVr8Edewyc456Z9+ZWrRch91/38cYj2uYJswIKT7DANplhwa1m3a9MmYt0B/fqYXaYOdraZ1ZqtOmzTWZxKB4eIsJxt0wg2Z8bq5aCFFjRTXbKHKe1SYDPQW8Nh9JhwD6dY136dCSioD7HzAYzWiJQ7ywflZycnJycZ4s8E5KT8xxwZeV3G7rWJ/l0mdfd+xPPWCx+xcbPNa6s/l5D1aHdDsu/sxOiHtrZgHKR0tQKlakOt//95bz04kXKFz6BfdIQH9/N3oseBGvQZg2iPhe+5rM8+YlvI/6oZrUjiQ+phzu4iNkT8aE/+N84dXRf9gkQhCR/fpjLLztG54uvRLZvwx0/CP0BUqvz+kM/nhe3v8h48PpbGr4IM1pgVqvEmlCRIl0GlKSILz5DTViRDp54lChQlRJr2sYgbNM6FssOmaUsBTzxiDQm1gSH0tU+Ax2yqm0AKlKkw4ABQyyOfW6KRW1S1xItibBY6lpEkLN8ZHJycnJyni1yEZKT803mSn1Pwz3yELJjCm/vGlJzX3L9rv/wZ40r2j/foDvAPXg/rG3gDm0we+BR3JEnkWIFbXYwu9aov+whLtjdo9uucvzTb8ItzvLBW15BEtVoHrmE9un9aDTA7G2SxB4Yh31kwPLBl+NdnLL0t2/is7/93SxECZ99ooRbLqGnTuFd1MIYy+rSVtLPrcBmGxKLO7HA4K4Lef3Bf54LkRcRazbhCdujjMdVuh2ArUwTaUKKRRAKEuDhMStTtOljcfwTdzE7dY5tWiXWlB4RsaaEBMyaKXzxSUkxCB6GjvYoSkiAx4AhQ1IqFNgkJhCPwyzTpMsmPUThR2/6wTwrl5OTk3OOkNuxcnK+ibzmln/diIchwZvLxLdNcfDBN+FUaK3d1vjCxpAYy6/85L24Qz0+9ftv5Vv+5c08/oHvoNf32bVrnfnC/ZhtxzDnbYXeMiuffAPlSoRNfTZbJbw9q3zvW+6n8PqjlItHwQh6LMYtTXHhVXeSrG6l/8A+iuUBT/z+tzKIAk6se1SMsJ5Yjt95FXu9O/jsjd/D666/hQNXPoDsmcXeG2F29kHgA7ddyvd5Ka/r/3jj7j/4Hq78iQ8jVeEzB96fB4TnKCekw1AsD8hp5rWK1azrVUWKzFGjqiEonO/qPOytcUJXKUjIkulxWjZoSpehGxJIjYSUVFMsFqs2qxsRH1XHUBM6rkdfIgqEILChXe4xXaw6DEKNIiva4h7zJG8/2wcmJycnJ+dZIxchOTnfAH927Z837GjOgYfHL9340zcAvO7un2qkx7bjv7FP+oCgnT4nD1+BCMzPtfn4oRpV8XjNeQHD07v58P94KQOrHLnhrYjATMHwyaOzlO/6J/yL8DaIfbxX1ZjdeYTmwgGiQYFde5YZHt5HuHsB2TmPO7QKTjDbC0gzIj25gySqUCj2+Ls7Lmbv1oSXvOwQdx+5iESVWB27L3wYtzQLgA5Cjn3kzew8/zHa6xey5aoTxB+z7K5DsPcU9uB2HlpWLr3nVVS2nuTCW9/fmHnlfdzzgX+GU+FEE87fosjv/LNcnLzAKarPUCxbtMaFbobP8Bh9BlzrLiUQwSks0KeMzxIbVKXEeTrNIVkCoK19RAwruomiWBxOEwyCiMeUlGlpl4KE+OITa0IoASkpnhimqWBxrGtn8px++0O/lK+rnJycnHOIXITk5HydfO7ajzUMwqLpYRDWafPRa25sXLevyNre17LlW+4mfXgKs7OFFAsYo6xuhEzPCOcXC3gCH1jYxLmLuXR3wt8egYIYFnRA1yZcJFNsL3osfe71zG0/iRw8gTkvwlvey0NHqrxhuoOzARhHeseQ1pOvBaBU2eTU0fPZbBfYv3+F3//UHJdPQz/yeOCLF7PpEgpiqBmP3uYOjh7exQOLSvrRb6dctOz7nlO4T+6BsIj/quPEtwobD1zOnffu4vLzLMunt+FObicspEz3ijyyAj/2o3/Hzntfj6qwepbPS87Xz/Qv3dz474+lRJKyLG0iEjrekH/qLmeXVlliQJeEC5kC4FPecWaosahNErHs1jlOyQZd7WBxhBLgjVy/ZlTPYRDa2seNxIlVR1lKlKRATCZUYlISLCUp8Ms3/stcfOTk5OScg+RzQnJyvg7a//y2xgVblFYrADWsmR4RCS0zpNqaZVfd8IEbX87OYIZKaYO1f3gl/V6JY8shrVaZYQpfGHQ4bTqknRJPbCh3e6fpYFmWHiJCXQu8bN5w1xMl9k4HVHaeYv0Lr2FlaY6Fps/5u7rUX3M/3k5w6x4bx/cxjEq0mrO02kVEwBNhi1fi/rWUrSWPR1aU7YWA2MGKG3LoyVl2T8Nax6PkCzO1hOHx3fhBSjE+yKMf+A7KBeWD95Xx1eOBzZjDKx4VLXLrUWWv7qAiIe3Tuzm+WCb0IP32i/M5Di9QPvV7h68+KT1qGpKKskun6UhWLP6AWeS4WUfEcLccZdl0WNUWPSIGmn1tSp+O9hAEHwMoCSkFCSlIiCceiSaAYsTD4thqZhkSU5MSu3WOSFJCfHw8PAzf+rYr8vWUk5OTcw6SF6bn5HwdfGRjnU+cSPikd4zHvGXWtM00FZZ0g0gdIsr5dQ/POEh86vMLlEpDLt0/wDoo+XChV6FIwENmnZPSA6DDkFQc3+pvpSIeX1xyDJ3jMw9u4djt38LK0iz7X3IIT2B5cRa3VmHwqX0ky9s5vVRncalKc7NImgqXXf0PHDkxRbmY8rodHk+sK02XshlbYnVU8DmpA4axR985Dm6kpKlh+96DtDdrbD50GeubIY8tBKwxpJNapsSnj+XW9RazJuD2h6rcsZDQ6QVUCo7zdq2f5TOT843wPZdnbXHnKZCirMuAFn2OyxqzVJljimU2sVg2XAerlr5GOBRBJt2vPPFwKP6oLe9Qs/keFovFIWLwMExJhRIh01KlR0SAx5BsgGGKZVarZ/Nw5OTk5OR8E8lFSE7O18ifXPv/NWa0yMNmjX1uFkHwxNAhYq9spa+W4wtZ8DSz7RQYZWN5D71eCWuF3VuHeAZO2YjXMc92V2VTBuy3MzhRDMIn0iV6aonU8ZbL+uzbmrBz/2HO273EZz9zKZ6BUimhdfgS0rhEMN1k395VrBM+djxm3751MI7ZqYTPH/f59OmEw7ZHhKXtLDXjMe8HVPC5e8Hy+j3KvqrPetvniYcuw1rDpz+7G88ondTRl4Qj2uWo9tjpFdgnFWYDj/mi4TvONxxaEXpDw5Gf/dHcOvMC5iP3lZnRUiY6ZUBTujS1TVcHXGhnadMnVctQ48xKxVOd3lJSQglQHCUKWX0HHgUJmTVTeGIoSWE0P8SQalb/0dQOXR2QquWUbABQo8Sc1gD4t9e/L+/MlpOTk3MOkouQnJyvgZuv+WDjtf4sh0yTrsScMBu06FGnQqwJx3WFt14MW2djlruKFwxYePhViDiiyOPxk0UGQ4+5mqVNQj3w8BBqWmDN9BmSToK0WS+gZjyCIOWJxQDjxRRKPVp94bILO0RRwNTcScovOUi0uoOTJ+e55fSA794XsrFe4/Bnr8Q64eXnKRXx2JCILgkbxCzamI61BBjqnsfpdZ/dWxJCP6sdaXVCFvqWOxYTjto+dS1wWVCnSgDAmsYMnfJkL+GWJy2bqeUl+9pn+ezkfKO8ct5jTfosmS5LusFpt4ZVyyvYyxe8U/RGtiuLRXHoSIQoDsEQa4KPT0xCXWrEJAT4WNxkovqMqSEIIganSlVKhOJTkgIJlgt0KwkpTenSl5hfvfFncmGbk5OTcw6SF6bn5Pwj+X+v+6PGstdmwZY5zCJlsl3dCkWWdQOLo0jI548U2DcLV72sy80fu5LLL+hRnx3Q6gbc1l9lx/EK113oU8BjugSzw5C6BhyhTSQpBfVYlQ5HbBkfw/LKFJcd6HPy8CXsOnCQ472UmcUq3UgolV7O/n2fpN+tc2zF56ppn05f2X9gkQ/+/Xl0nKWPBeBipomwhBi2BAGRdXQ0oWVjHu8oB3oVui6hZjxWXcJJ6bJpBkxrieNmnYOuSOTFtO0W1kyfHbbAZfM+j64bElWWfvl782DxBc49qzGh8ViWNi3bZs7M0NU+HRmypi2s2kxMiEEwXyZE7Oj3lKSAh0eITzK6T0hArAlDYhxKkRAdCe5EsyL0nTrNFldm1eswpxVaMjh7ByMnJycn55tKngnJyflH8OlrPtpoSp8NuhyVVQoS4FD6GtHRATNSIySgLhVOJTHdSLj9oQonBwl3PlamMn+Km1ZarJkefVKOL4UkOO7a7BGIYYsfIAjn2xm2uSrTWqYjMV1J2Oj47Nh1mtsfqqDO4/JtHtVyylzNIqK4pWlElB3TjjiFu1dibvmHXeypeViUHikbDLnPrCIIiwy4w65wixynjM+y6TGQlBUX80Wzwh0sMi0+M1pgl5tiWrMd6gRLhwgfQ0kDTrmIT672abmU19/0XbkAeYHzyNtuadxvVtiUPuvaxhefgUYAHNWs9a4jswuOBQiAYLCaZUJCCZgxNaw67EiSeGIoEFKWIkUJKUmBumR2xbpU6BFRlEyQLMgmRTwutltpyYAuw7NwJHJycnJyngtyEZKT8z/h09d8tDFlvMnP01RwqmyhzrzU6WqfnkaT1qN3eidY7il/qwv0sSynMesnLuI76tO82e3iSW+D27rrLJkuD3kr/L05yd1ujSfMMj1J6EjMnJbZpmVqGhCn0GvXecVOZeXUflQhSQx79zTZdeAxWgsH6LSqrHcMp/uWS6oFluKER1oxl04ViLGoQF8SFqXPCa/FSWmyThuHUtMCJc2SouM2qks6ZEOGdCTmPm+BnkajLkhD2hLTkogiHjtMkcvqBe677uO5b/8FQPLOWxsn3nHbl52r1o/d1vgbt8CG9DjsTtNybcpSwhMPQYhcRDSqAxGRif0KINUUX3wMQlVK2ZBBEXbrPBZHqhZFaWsPD8MMVQKeej8pSkzCpvboMqBPSoChy5DiyP6Xk5OTk3PuIbfffvuvne0nkZPzfMT+9C2NhU3D5wZZsP6ot8x2naJPwjFW8MWjTIF1bVOUEKuOFEtRQl7mdnKxTrM19HnV/piZ2Q6e51An/M7dZR7z1ibdgAACfFZpUSRkVquUCdjpagyxXFGe4qF+n6vmyogoj6xbLpjy2bNtyNZtG0SDEotLVVbbHtbBHd0WiTia0ud1uo0NTVCUz3inCPBZp42HR40ir7O7aMqQeS1igHWGOJRl02W3q/Owt4SHx4KuUZSQWapZAb7O8RrdQsEYXjInHG1CvSCc96ffmWdEnqcc+/7bGv81eZiElJbrEmnET/Em/kDu5jp9DTfJF5g3dbo6wKolkIAUm3W8Uosv/kRwxKPBg0ay9eth8MRjj2zBw6OqWabvGCsAFCSkpwN2yxbWaWfZE7LOWgDzUicixh85hGeoMCShSEibPtNU+Dc3/mS+tnJycnLOIfJMSE7O00jeeWvj0NtvbSRW+ES0Sopj2XSZ0wqh+rRlwLxMYdVNrCRd1yciJhkF/BUNuKAaELtsQOHKcp3/emeVhdMzXBSUs9tiKRICEKg38skHFPGpaMhBb52D3hq1Iuz2i9y63mIQC5fM+tRKShhaSpUu/V5If2jYiJRTfYtBeMws0ZeEDU0AKOIxpxUiYs5jlh06jYfHPd5JyupTE58UJcCwT6pc4uZZMl026DKnZcpSoEKRJd2kpxGL0qKnlvmiodkT7rVNKuHZPGs5/zMuPzBg0a6yYpu8kYvxxedD5mGsOm4xD5FozIptAjDQIQaDVYtTO6r8UCqmTKpZF6xAApxatps53sxLsWpZ0HWO6zIDSXmSZRxZ4TnAq/V8hiR0dcBAhww0E7yJpgCEBCjKQIfUtMAeN0OfIVfZvcy7Mv/tuv+RZ9tycnJyziHywvScnBGN63+rcYXdTYqyxyvx4Kpjm5aZk5BIU2a0yMPeCl0GWBwVKbKhHSpSRMRQkSIRMRfqdmKxHO2lbAt9HluzzHcChJSHFjzeeEmP+x/YSiQpT5p1LI6XuC30vax4tysxAR4dBiRYNgcQOeVN1TpzNcdKy1ArOT75aIHvrk3TbBVIrLB7WjnSFHa4EvdiadHnqGmxy9U4ZDaY1hJr0qVJl4EMqVJim05x0rSZddnOtQJH6dKXhFOygVXHUVnDJ7PbbJU6DmVN2zxgVjnRL7FLq7ypOMeHN9f44bN9EnO+Inc8XqBoivTdgPvkBOqUNW3iUDqum00w1xjrLFbtZHq5xeKNMnZTUmEgmXhQ7MSGdZ85QUVLxCTEmnCUJYLRn5d4lNF40qxSJGB61DXLqsGI0B4NN9ymNY7JKgBr0qUoIZfYbVw5W2apU2I9Tc/ascvJycnJefbJJ6bnvOj5m2tubix+4Imrt2iNFGVThhyny04p09QYH8Oi6RGJpScxu3SGVTqcxwwbdIlJcWpRoCghFYqk4hii3KmLhC7kVDpkmoBEoZAWONZP6EnCy9wWqpRIxLEhfTwMG/SoUaROiTIhD9sWu7XKF5M2W122q/x3a30WibisXmR2ZsATC0VOdRwXzhqWB46D0uQCt5VNE7Fq+pQJaZoBJQ1JxVGmQJFwVGIstEmZo8gj3ipL0mZD+gwYcp7MMa9VtlCjREhXhkQkBOKzQ+usmh5tiSknRfpY5KYTV2+9/iX5hOvnEWs/fFvj4T99/OqyZ/isLjDUIW/gIv5XLuTzHOcKcwl7ZRuJUTqul3WsEg+nDicOh8MTD1B8fBISBCHAJyWlr9FkXkhfB0xJFYcjFJ+ElL1sZbdOc4QVEGFTOxQkoEdETcqUpUhXB+zXeVQMe5jjKCt0GHCxbuVlMz6pNbz0L9+S27FycnJyziHyTEjOi5rfve4PG2tel9fbXRggwlFQj0ukTt9ZyngkOPa4KQIMbw62cIddZatMs0aHMoXMQ49DsPjqMZR0ZDOxDEk4adrUtcguSrxii8enVgZEYtmvU/RISbAMxVLVIgPJ7FwnpMkuneGUbDBNhYunfY5sgFU41U/Z75XpWMvtjxp+8DtPMLh/Cgv4HrRH9hYPoaQ+Q7FsdxWOe+tUpMhuN0NLIgIMa9IFYBtTfN4ssk4260MwTFMhJmG7VhGERCxzWpncZ930eYXdwoPeKp/wjrJVa0SuhvzMxxv6vjxgfC759et/r3HYLeCJx+9/6N9+ybEvhUogwmNJl0iyyeVlDdgWBLwlfTVDTXmVmQWFRVlF1WXWQAnwxJt0vnJqMUZAoSwlBhpRlxoJKYoy1BhVpUt/Min9lezlsKxkMz9cZmM0GNrapyLFUX5NaNHFIKzSwohQosBv3Pjzk9ex/bk8mDk5OTk5zwm5CMl5UXNc1gEY4pghpE3Cdko8oBvEJuX1bKHpsm49p6QLCTijrNFiqAllKWSFuyippkQCJ3WVC9iBQ6lrmQI+r/NnOJXEfGFFOW46JFg8FfZJhdOSZVO8UVYiIRMRPoYSIWUNuLcZMUuBv+6v8VLqLNuYCEvLCb9/6wE2dECAYRAXSXDs0zkeMYvMUxvVmnjMa422DDhiVjlPp1mVDoIwpxW6EtMjyrplUWJIyn43x7LJrFltGTKlBZqSWcSusvuIyIYdljVgu1apasjF84aFDSH4odsaW/44L1J/LvhX1//7Rm+cjVB4/3V/3HhUFmi7Hu+pXMVH+suckCabpkviMhFypzlMNXkpV5WmuWuwyUOuxayWeK15KY/qcTquixVHqilTpkrX9fDFx6qjIGEmUDB0tIfF8i1cyoPmFFOU6TEk1oS26/GQnMSo0KHHJWYvJ3WNoWbPoSQFZrXKsrTYJfO0iLKZO2wS5l2xcnJycs55chGS86Llrmv+qlH0Al5mt7PXlDjpIqoEDHEkYimqzx2yyDaqk0nhj5g1AIaa0NcIM9rJLUuBSLOZBgbDirRxolxoZ1kyPeoFoZl6RGo5z1VJcCyZLodknaqG2FFRuEGoUCQiYUBKiXBUO9Km7gpEktB2Ka+dLtEdwrFBzMtnA/52PeJxs45tznHMtChrSEF8UhwzWuJRs84WrZCIJcAnxlKjxBodWjKgriVO0WePbKHDAB8zETFdiVGUQD0iSQhG2aFZQj7vLfMqu43dYciTcYRnYKoIq13wfuS2xt+0N1kwbTw17HN13nrz23Jh8izxU9c1GlbtJKh3KCqGL3KUoYvxMLxv8CindY3UpdmUc81sU+tug4+bh7kzrjJvajzOKfaabSML3jxbZD+Pc5qOdkk0Yd6boUyBzqio3MNjRqqcz1Y8DAvSZooye9wsB80Sb3QXsGDadCRiXqt8Tg9xhEXqUqUkBbbqFA+7Y1jjqFEEIMER4PHeG38xXyM5OTk5LwJyEZLzouTXr/+9Rs0UKOCzanrMuSIDUl5fqvPgoEdBPYrqc9h0GHpZx6m6FqkQYpCJ4OjrAF98klEXKquW0JQ4oFsI1LAmAzoy5K5uh4GkE3Fz2FvLbFikFMSnwyCbKE1CTBYwLsg6+3QLLYk4IU1mTIUaBVZkwMlOgZ6zbJJweMOjPCocPi191qRLS3oUCLFY6lrkkFllXsus0mKGLCMC2VyQDhFdGeJUsTgiksmguSEpAR51LbJkOrzEbWVN+hz01nm13U6C5YveMrPpTl5SKPHgasr5NZ+lOOHOuMNx06RJl2mpcNQof3HtXzT+j71Vkvfmww2/Ed553Q0NAIsbFYmPvqujoz0gO7fH7elRuwFQdYgYVB1WLU27QUvaRN48JS2w3dV43PTYq3P0JeGAbscIHOQ0AT5zWpvY9QzC9byEx12HJ7xV1unQ04iBiWm6Fge9NTbpsUdn2eOmaHt72Gen+XvzOK9iP4/JaUICpsk6tstdBAEAACAASURBVN1w47vy9ZCTk5PzIiMXITkvKn702v+rcb7ZwYq2WATKFHGiHDEeQ1IODQocMRsUCVgxPVIcbbL2u00JMAg+Hk6zwM5iGcV4EzzGAmaVWa3wSruVY6bNsrTZojU2JCIkoEiIo0+TLhExRSwDMjGgKIKwKh12aJ0lSVmTDih0JcYwzUAdS6ZHxfkcNpvZ7U130pVoSEyVAg5lh9bJRs0pA4akOIoj0VOiwIAhe2TLqJNRwHBSsN5nu06zIh1iUpZM9lyrWuRRs84OV2NeS3Sw2AS6anm4bfm8t8waHQY6ZKtMY7EMJWWXK/PhY0Pe8hyf93OJX7j+NxsGQ4ol1RQlExUA4/+LGCxMMh8AIlmmDfGwakfF5rDuNtlhtrAkHd6hl3DY9ehKzJQWEGCrVBloykGzBApbzAwGYdOmlPBpMcDHR9WxrE1STTnmFjOLokkZeClTWmDZdHkZu9mQATVKVKRIUQN+8UPvzAVITk5OzouQXITknPP87HXvaUxJGYsjlIAFXZ9080k0xUiNWVdihgJLMqCAz7K0RxkBNxkouKEdSlJAxzvLOOpSo6UdVBUjHh5CrAnHTROLY1naWKOsSYeImBrFzAOvMzQl27GOiIk1xeJISDEYqlLCx7CpPfoSUySgrX1C8alrmUO2x/9SquEPpgGIsSzKJmVChqRUKbGsG8xIleOmyQE3z4p0KRKSYKlQICIhUYuVATNUR49v6RExQw2ATXqUCbA4ioR0yI7PtJZ41CywkzleLjN8Uk6RYNkuNTbMgCd1CR8PTwwFfPpYvou9hIFwIAwp/pu/akS/+b9/WfC59sO3NR5uD3nArLLD1Sjj85abr3tRB6k/c92/a7S0wx/f9B9v+LXrf7vR1QHeaMSTO0MBi2SXeXiko8ycL8Ekq+XhYcmyeuP7qjqKUuSAbqFPzHE34FFvmYiEMiFFQpqjovFdOkNbyizrBlulzue8JRIsLc2uH2qMEY9QAhIsdanSdj1ezT7ecdP33wDwp9f+WaMpPYqE/OKNufjIycnJeTGTi5Ccc5aPXfOhxmGzwYBs/kYBH0FINZ1Mg4ZsjsGC6bBGH4ey201xxFulRhEPQ0t7eGKINaEqZVrapSxFhhrT0wE6yoo4tag4jAgRI3sWjqZ0CfFZcuvEkjJNhVOyMZrB4EadhbLZCwZBxJAS0NXMFtV3A84z84Ti09QOZSmwaLpYVyMUwxO02KIVjrOKkSzAbGmPkIAl3WC7zABQJqRMyCY9Anx6DPHEEOAT4GOxVCiy383xmFmiREhKSksiltigSNZ+OCFl1WQdsprS5bO6QlkDTkoHDKzTxmCy7I6GbEqPCkXmSwbroBk5tg3Gkyee4oL3/WFjVc+jbnw6ErGDGjGOD13zgUaA4btvvv5FF7T++vW/1xAED48fuObdjZqpYMkKxsdZkKxgPBMXY2vWOMuhPJUJUdzkNgVTIHIRgvBGLibB0ZWYFdNnp5vm8/IkSxoTSkCsWUteZ5RZqvjicUrXCAnwR48TEuCbrGWvIPR1MBEnR8365Dn84E3veNGdw5ycnJycZyafmJ5zTnLzNR9sHDctOpLtGg81ZkhKrAkJFh+PEgWqUqZKCStZ1kJRamS2qxkts65tSlKgSglF6WqfeDQVXVESTTDiEUiAjARAldKoS1AEZDvOy7qRWaE0oqkdfAwWR0xCT6OndrHFQ9VNWpiWpABAn3iUxUhp0qWgPrcliwA8adapaFY4XxrVgXgy3il3bNIjwbIhAyISKhRHBfAFzmOWItluuYw6XY2HJQJsZZo2fVK1kyyJxTHvKjg0y6aMdtenKE923Q2CVYsvHhExLfoUfNg6pWwpGbqR4a+vuWkyAfuC9/1hA+BoN2XBZXa13VJmyWRD9C4NK9x/3cdfVBOz/+jaP21cZOcnwtIXn4EOiVyUCYPR5emoJbOckR0ZZ0jG3z28L7ncqpu00T1i1liQTTbo0JKIvsRMSYVYE7ouW+8pKcuuyQldZdN16LsBETGRxqSa0htlZ8bvjZIUGY7qpla1/RwdsZycnJycFxJy++23/9rZfhI5Oc8mH73mxsZnvRO0tU9KSkiAIChKTwcMNaYkBWqmgqLMU6dHRFezOQY73TQVDVg2XQ5ymoqUJuLDEy/7jiEZBYLj4E/J5imUJBM3BmFDOwCTnessA5NSlxplKWJxDHQ4miBtR79H8cXHw1CUkIEOEYSihAw1mdi1AOap0WGAICxpczL4rSolrLqJGPHwJpmgAj4JlsvseTzkLQGQjMRDlSIpjpIGeAiRpJPaDodjn2yjTxZcdhng4WXB58jaNW4vrCgBPqH4XKDb2OlqfMf2Mv2h8OBGTEEM98gSkWQ75+/evpt7TzvulEWmNROFRoUDOkWTmPNNmS1Fj7v6LYZi2a0Vrr75e87JXfV3XndDo+t6BBIwa+psuvaXdMAaC71x7Y/FTmo8xtm0M21aY55+ueGptTteJyUpMitT9DRiw7WwWIqjdVqVMm3XmaxNX3xSzebceBg88YhcRCABBQlRdCJKxvdJNeX9N/27c/K85eTk5OR8beR2rK+Dt7/1XY2//PBv5X9In6fc4R2m43qTug8RGQXz8eSyzNJiqUqJAUO6OmCgET0psFMrLEgPi1KUkK72gSywVh3vJ2cBnVFAHKqKiOA0Ky6PNaEkRazakeUqe9xx4FeWIjEJ4SjrMr7Ol0woOFxmkRoNjgOdFKwDWZtUMZzUNSpSpKdZ1iXVrA1rn4iaVDK7Dn4mEhBqFEeBrMOitOkzRXn0vIRAPZwoiVj6pOxwdVZNi2mpZPUpDDEIs1qhK4OReMpeW1mK1CixopuUKGBGAm3aFRGgGwmq8Ihp4lD6klDVkFe6Lay1DSe0yy6dYs30M1EkcIIudS3whOuy2i+wYnrMaIk+luSdtzYeW1X2ThlE4NbmJm+/+fte0O/Lf3HdrzX6OpiIhRW3jo+PEY9U05GFL2Ms+Mbrx2pWkP50oXFmVuTpP3uj26Sajla1oU3/S+pGDEI6ytqNRXRZShMLoYfJpqxjKJsSsSb0tD+qTUkpmiIzUqOlmYXvPdf/l8YGHX7zxl94QZ+rnJycnJxvjNyO9XXy9re+q/F91/zci8oe8kLgh6/9xUZfo1FAJZOd2HGQHkpmW0o1JRQfS2Z9EoRpM0VCyoPeKl0Z0pchqVr6o9kIFktMgupTu9FZce9TlhcRQXEkmtAdZWKc2sltEiw6yn5EmtmeBhqRjgrTxyLDx6c3CkatWsJRcbiMgkNFseoIxMt2oiWTEQkpVVOeBJpbmaZIwBRlAJp0J3a0E6ZFZSRK9ro5LI5F2cTDsMwmgqElEdNUEARLNmNkRstUdVQfopb9bKMsRYpkBclTUiaUgIEO2UadE16LjiSkFi7a1+V1bJnYvQI8TkqPI92UAh5dSdjhqtmxEse6DDhp2nQkZlF6VDWkoB7bTIHVjlDyhAv3dljvQdMMvunr65vJX1z7F40pqTzV8QqXBfLjNYSbnFdVN/l6pqzHmLGIOPPnMWZ0Tsfiw4w6ogHMStaYoCCFbM2qTtaoYBholL0XeEoYy0jgjO1hBQkJJMjaAbsWA81aPRTI3ne/fP17Gz973Xvyz9CcnJycFym5Hetr4O1vfVcDmPTaH/v4/+Lm9+Y7es8D/tX1/75RpcSatvDwaLsOscZMeVMTv/o4E+LUUjYlzEiHJ5pQNWXqVGjRw6rLCsFdm1RTfPGJRx2HzkRHgeKY8e1CCbL8x8hiJSITH77iKEiBqpSINJ7sGjuUUAJ8PAJ8ElICfDa1TXlkvwrwJwX1RQkBiDRmi9TpMEBRCoR0dUAoPiEBilKhQG9ko/IwhPhUKVLWgI4Ms+nXo+a9KSlbqE9qCKoasiH9kQApTe6zLG02tccVeoBl6dKWAV0GRCO72oAhO2SWN9t9VMVje9HHOjg6HPK4abIhAy60cxzy1pnREgX1ScVRUI++ZLUmkWQBrUMJ8Jh1ZQIM52mZEMPeUsiWqnJw3fFX8iRvsntekDatn7juVxslCnS1T18zMeWNuovZUfZNMF8ycHD8+QM842VfjTMzIpmhLhOwoQRUpExBApquhWAyG9Uo6zK+/VhoGPEmGbyQgI52Jxa9sT1s/H4YZwXrUqWjPYpSoKd9Uk2pmSp9l73uKVPjdz/0Ky+4c5iTk5OT87WRZ0L+kYwFSM7zk3df//80wlEb27E1JAuqQmJNJgIk0Rin2e5v1/Xx8ehp1hUr0hj/DI98VweTYOvpO8hjxp56EZlkX868/ZnFwAaZiBarloEOGWo8ESBFCbO6kdFOsY+X7VNrFsgF+HhiMBgKo4xOFu6ZSRG8T9ahaDzCrq092tojJp1kc3oakWLpjpoQZ8fKUtcyVQpMU8HDUFA/q5WRePKaWxKxZvoMxSIIu2WeQA0DSRgQs5VphjrMjv2oFqdufPpqOR2l3D/ssCC9rHUxljXTp6QBTemxYrrsdjWK6rPT1djppugwIBkNb1ylTdP0SXCclC7LRCRWeXjNMh16vNxuxwG3X/ORF8R79Z5r/6rxmWs/1viV6/9zY6hD1t0mfR1MBEVW65F1tPLwspV0xtyPM9fhmDOv/0rXjS2BZ2Y+REaPoZo1cThDcKekWfZPXfY1mUZiJ++PWJNJIXpBQrzRLJIzH3O8/hNSChLSdT32mO3UzRQFQsqmhIgw0IifuO5XXxDnMCcnJyfn6ycXIc/AMwmOf+wOY87Z4Tdu/Pkb/uON776hrwP6bsCmtqlKeRIojesyviTjID4DjfDxqUiJGanRpMsUZSoURx57mdR6ABNLjDce/MZTAVx2fdYidVyI7omZ7GYDkxoPT7xsrsIZgWSsSfacyIrA3ejRiqY4qdlwml3mVCmTdc4a37arA7rap6sDpiSzUPl4VKQ4EUhTlClKOLJXOVoyoM+Qgma1KN7ov4iYoaTMUsViKRBkmQ1Xo65FCupRp0ygHse8TSJipkY/y0gojetalt0Qi1I1hnkKLJoOAR59Yoajie41LY7EkGPV9CjikUgmxLoMqWjxqeMkKR3JAt7YZQP6VoZZMNwipixPb/77/OPnrv/1xifMMe4wp3i53YFgGLfQHQ8VHAftT+90NeaZZoTAl4uTM38e//vprXvHj+/IOr5NsnMjK+G49W82GDG773jNj69LRh3WYk1IJnNvss5pfTeYWBkjHaIoVVOhS5b9MCIUCJk1dYYaE7noGz7GOTk5OTnPb/LC9K/AmcXnTxclX2238cz7j21bk/udsVMOWTvWP7v5P+W2g2eRmqkQu4RZU8/qQDSzSCUa40tW4OtGRbhZbUaaFdOSsKlZrYRIVog77hhl1VEzFXquj5EQp3YSiI2v90ZD2rJC3ae6FI0DuKcHhmNrl4xCO48vtdY4kYnXPiTASVYjMM5wGBH6DDOxJJZULUVCBhpNun6lailJgZ5GFCVkSsqTx8gGHwaTIPGpzIpht6tzwmwSqs9WrVDQaY55m8xqlTWTFenvsXUWTAcfoU3WntXHMKNFZs0UfY0YaMxAhgyxLEqPBWfYMAPW6TBNhQI+VUJSyYLvQD0KGFoy4AnTpKaZyCrgY8WxhSm6REQSU9Mi62bA37kO+3SaaQISHPukwoIOeM/1/6UR4LHD1c7qbIpPX/PRxl+a+/HxSEYzNFquzVZvjuNuOQvQvSGeM7jRumHU7WosLFQTzvwseeryL/8cOrMwfWwZHQ8lFPnyAYcTIaF2kh1xKKhO7uuJwSi4kVDxzhQ8OrrvqBYqE9lPiXNVRUdC3sNjyJACISEBMclEKA90yLTUaGr7K3b3ysnJyck5t8i395/GmYLj7W99V+Or2bDGhaFf7fqvxPgP/I9f+8u57eBZ5L03/uINdanRdr1J955MNIx2dDUr4h5nKxQ3sWtFOpzUW4x3ogtSYNpMoShFk+3WjwMueGpHOcu22EnGxBf/qWDwjF3ncQA3trGMrWOT4XJnBo6jIt9wNM9h3IUIsiLgcWF7OrKXlaRAQQrEmlmvAEJ8HI4hKT0i1rVDZ9Tty4zaFof4pDg6DNikR1uGbNCjJX2OmnUMQl2LrEg7myNCxKLpEEuaFdYTM6cVZrVMUf1JAf2UqdDRAUfNJifNBofMKsvSJiahRZ+IZDK7pCsxTemSoJS1QIBHUwZk/8oGHzbp0mNIlyGrMp5pkZDg6JDSkZgntcuGDNnhsnO2UyvfpJX2lfnkNR+evKeXGNDXiIisM1s0Oi/j8zPUmKP29DP+njM/X57+WfJ0Ufv0TO2ZomN8/TPZtyxPFbxnWRiZrH03GoA4tmCNMyZnMrYbWnVPDe0cXRYSjJ6LTupIfPERMfR0kGVLNFsrIVnXrYoUqZgyZVP6Soc3JycnJ+ccIRchT+PrsV09Xai845p//YzC4swsyDioGOiQH7jm3Y0fuObduRh5lrhCDxBKQEmKk13VcZEsMPG3jxmLBEFINKFNf2SJ0smgNmBkM0kmBeZZO91M0IwLz8fdhsZWGiPelwduIyE0zn5M/PPoqCNSdt2Zg+Wy56mT75HGKEpnVAsyLVmwXZICFfn/2XvzGEmy/L7v83sRkZF3nV3dMz3dszOzMzt7cbXkUlwetiwJNHRYgnd2CZKwLVs2YEj/mDQtyxeg5gCSfMmCCMMHYJiATRMSseSuBcowBNgybZkiTXK11N6zu3P29Fl35RUZx/v5j/deZFRNVXf17MzubHd+G42qysyKjIyIzPr93u97tOlKm5bEtGkxlF7tRjWQDiWVOz60yCnIcK95V4/Y1xE7MmGTQe3OdNdMGNgWj+kKcyo6tNiWkaPueE1Jy2ePHJisdhHLNGdOzutsU1Kxpj0u6LAuOCt/tCq0tpu9bo6YScGckqnMa0euVe16J7E5Ka7By8hZ0TZtP0E4lIzXowOmkpMScdWu8ljceoevrrPxD1/43LVf+fSvXptS8Q9e+PVrAGMp+KC5WmsmSi0RDFM7c9eS/zfXnGYGyFkIE5H7TQpOo2eF5uBkQxMmIIvf9Za/VHVTEqAsmo3FdWnr7TftpoMJBLj3jhFH9Zurs3luiTNNmJM75yx111LlaWlLLLHEEks83Fi6YzUQLHfPojmE1cUmLeJejz0vRExNi/hfPvdfvhj25eQ2ltkk58e/8en/6FqYcgDencdz2cWFpkVEdRhbnbWAazB60q5tdK1WJJKQeZvfWOJ6W4HqFZqRgBDkFktcZ4iE5wshgrFnQ4aJh/GULoC2bxBg0bwmxGQ+OX1FeuzoIX3potg6byT2k4nE6zratAjks4lm9KVDsDBelR5HOiWVFl1aTHEalXX6XrrsJiCRGiYyZ1N73DBHdQNgkHrCMqBDlxTB0cRu2G1EDFuyypFO6UqbkpIhXQrffBzplE0ZkhDXxy4iIiFihxErOKvhjiYcyYwSl9je84GKbRI+Vj1GguFNM+KGOMrYthyxqQOetxuMKVjz2pmf/Nyn3pX3z2+98L9euyUzIhVGktMloasxYykYaosWhn8a3eYle72evp28XgJCYGVAmGCc1jyE++Gtk4/zoG7Qaz2Tc20LmSHhMaHBbr5vYKEFaf7cpFKl0nLGC2J8rogLXAyNRnDUmmlWB3gCHNgjRIT/6Tf+8+Xn3RJLLLHEQ4zlJKSB+9GrwmPeDYQi+Gdf+IVr/8oL/96101YClw5d50dmZ8yto7youqIv0LCC+LqiIpa4Lo4CKq1c8+GLrCDWXdwf1pQXK7bNgrI5+QiZInWh11hFbq5QB/F7y3lg1RQst23XpOQUvqmoyCn5KFfp0qJPB4syIWNGjiC0SWqB+VQzWj69PDzfmrgsjk0Zsk7fk558s6MRU+Zs2R4btkMhTjze14Sr1Qp9bRF5YTnAJdYQhB1GbHNUT3RULRMyIjG+oSvYZ8zEu3Il3t515ilWYRIQqUtgn/t2BWDC3DVddHCkOcuMnB/qDMixTP1E57rsYFH2ZEyFcsOMuCFjssZ04p3Ef/pT/+21V2TkNERiOZSMXZlyy4zpakyCMJSYD1SbfELez6oZnqoREo6HDMKiAQmTigdtMtRfY06b9FZnrbDdcC3HEteTvCbC9V7/3KAsiqcnwvH3gAs49JRDDXYNWtOxQv7NVGe0xF2rY50y0VmtJVliiSWWWOLhxrIJaeBek4a3UBjuMe14kEYlpHAHG1J4qwuOsvyD/KDoGifCTsWtgodpRaUVuRZ1EeS46sVbkqYTfMaGL4ZEwmxgsYIdchsCNSs0N4Dnunu6lW94QusSCsNmOGEQsDsdi+XQjusJSJiYGAwdSRlIlx4pT9sVWiQkxLRJSHHUoy0d1jSpXEuG0qXy05IYQ9tPWwoqpuS0NWFExoA2PT81uGrXGUvOt6JdphRsaoe7MmXPzLhsB8SeghUcsgzCCp06iK4tKZFEFFoxb+SzgMs1CZSzZoJ9SObekzGVWkoqVrTDoUxpk9CXTt0sVVS0iPmDmaOFrWsHxTLWmU+ijzmSnA4xt8yIXZnx+9Gb7+g19iuf/tVre4x5w+wx9CGKhTgdxV0Z8Y1ohxLFKsykpKst/xpi2qZdXx+ppMebWFlohMI11rS7DY85TazeRNhGoCO+RTfCgtYlmJqmeNqEBtw1Gvb5NDpYs/kO05PwHJU6vVSuBZXXmoQmpdSypqvNNacrnaUmZIkllljiEcDSHesdxEn3qwf63RONSyBlnGxAmq5dS5yNCEMsSS0UdzQXt3J/msDW0UwMIBRUjHXqV20dxSmsRDtBuyXxmR6BptJ0wWo2KbAQqRukFqZHRLUGBY7nilReKzCQXl2wixiG0uNj9nLtXHVbpnyk2uK6OSKTss44GZCyw4iLOiSXghLLkU5IJWEOzLVg6oMLV8XRlzq0aNPigAldEgqpyCgZaEpXnetXTxMyhK7/2PiA3QJcM7OlPT4kQyzwm/Iyl2SNm7p77HwoSuyzThIipprRFkvpG5Cpp5EFrUxGzr5MmZDRp+O0FOS1TuRQJ/yW+SZX5QJzynplf6oZA+nwqtlFcBkqlak40Ak/95m/ce2ybPBXP/uX3vZ76Jc+8z9cW9E2l7TLSCeMmfL7kc+e0RmRGDqkHDHlZZPwmHU0o0va5UCHPGU2uSNHvGl2avtaWFCqwpTktGwQWDQcBsGeMR0JtKiztBWBhmVOTDjCRCZp0KrC+8VRCoVKF5PbJu0w5a2200MzqC2ynetWhYqt3y9Bq7UQx1ufnbNcH1tiiSWWeNix/KQ/BeehPdzTFesBJhf3alqaTU1zm0ta1v1R2+s2hL6ppG4FubYajY6FEc4bFKy5zuusgpo25W1MgZqK1bThPbmyDG5FuPkcwSkouF8ZiTB+xTqVVk1bATcxML54jzD8YPUEAM9XmyQYHtMelS80n6iGZF5Dsi8zLuqQkWRsMGROTkxEpjmZ5pRUlFQYXEhhRk6XlEgNbRJWtM2OTJhJwWU7YEVTKtRNUTTmNXMEwJSCQ8noa4uPmRU20oheZEiIOWJKWAe32NqeVrH0aFNQ0ZW2yyMhJ6eop0MzchJfnB4xpYfT7Ez8lGPqnaUCfWebI+7ovrvfT7laJNzQXW7pHnt6xB5j9xwi5BRv25Xu9z79m9dcsOKE0p+nCstdPeCO3cOibFf77OghMTH7MuXlaI8VbTGmYCYFOSUr2uFJuXhMA9JsQBJp1ddME2+x/W6EATbR/L2zGpLTKGF1mGbj/RMS1cNU0CAkktA1bjKVSqs+d0HMHrQv4XOr1LLWl4Ttngz97HsL6VyL2j1siSWWWGKJhxfLJuQUnJdOdVqz8k5Sp0IDclojssS9EYqj2OsgUuMEt+odrCpdcN8jcSvONbfd007CanEIHoRGsJsXmwcdw2kUlmaDAtRJ7c3Hh0Ymlrg+v4Fr3zzfghM935IREcLjdsBQYiKEy3bATTNiypw9xox8ANyIjIKSQitWpFdTzMJzlpR1YvseYwAyCgaa0NGElJhbZsxYCsbimoSplNySQ9oa0yamrTFHMufL9oj1LtwucjqakHnaTaZzutKudSK5P46uMTIUXgcTUttHOmPinZIMQpeUgde8GAylVgylx0SdC1dfulzUFSzK1N+m6pqCueYc2RFZLYiOSGmRUdCRNr/86V+59suf/pUzm5G9v/i/1/f9Py/8/Wu/+cKvX/sKB9zSPd7QbW6YMR/nqVpzkUhcU4ym1lnzzsiZMudNM+IPoxtc17t8leu8Jtt8vLpEIglDM6ipfOGaCzqN03Ay7yO4ZZ2Vpn4adSo0Js1px1nPV7u0+cZEREh94GXsX3OwuQ4I76+ZZvWCgJGIFRnUjbiI+PeC236gNTqB/HfP1WyJJZZYYonvDZZ0rAbumQlyCtXq3RKp34vWFe77V1/4968FJ60ljuNf//R/UJ/H0Dw46tRi1TnQQAIFJdeCliTH6CTNyUZdgKmpf440qgu3xK8Gz3VeryiLd8YK2wqIiZ09q1R1k5OeWPk2LLQkcy0RMXzT3EEQnmGNEstmEnOYF3zL7HNT9vy0ICORmEOZkWlep6+PmdGSmEwLF37o9SdrZg2AETMuaZ8N7bAjM563G3wt2mFGyZGZ82y1zp64ydCWOnrRH01W+Vo+8RqIKeM5rJjYWevqorgMae/uGEYcMaVPhzEzhtIl91SqHKcdCc5khVbEYmsh+mI7lqnO6mnTLTlgSJexzz8BNyUJ5x6cRieRqLb33ZIVEjXkVPzup3/z2khLvmy2+UG7xWsy4ovyBj9un+H/+Mxfu6Zq+aQ8x20z4q4ekNmMucy5ZQ55xm7ynFzmm3KDXF34XkdSZjrnUEdMdMbjZpNdmXLT7lD4pm8mGX8Q3WCLVXJKVqMe1+2dRkPhrreICJXjk4zm9yIJpRaoD8xs0vqaX09+37ymQ+MTphNh+hGmIs4+Oap1T8HMIeh5gptW03whTHUcXdFN+XIKP41avG9UFfz9M//eaUubJZZYrGmv6wAAIABJREFUYoklHn4sJyHvIN6uHuS8225u/355Ao8yWjg73rmdL5oIX9QFIwBwxXEotgziRLNUdWaFYOrpRSiwmpqP0MAIhpYkiN8e+OKx4fAT4/QlzqGrkVdS017sIrAQ9Q2Ry5UoqGqKSpcWY3Erzt/Ip1w3Y+7IkdsHEj7MFXK88B5LTsGGDJlrUWsmhtIlZK9XuJBCRdmXGTNx2pgUwzPVGpkUPF2tUWDZNVNaRPxzZouCiot94SOtHo+bNj8o6/zOeEShyi6j+lg6AXqE9c2FEx87l6/CNyoznXvBsq0dvFJp1UXuCBdoaFESiSgJ+h3XbNypdki8ta+q0jdd1syAnunSM12sVszJa1vlwlsC3DFjCql4mRG7ZMyl5FvmkDfMPgPp8HvRa/W1cigZd3SfQx3X7k139YDXzR4r2uYDPEFfugxNr56+qTqK37YeMGJWNw7hnF+3d9lQl+HS9sF+x8TdGLqmQywxCSGwMqonZe5aOVuYHiY0zanIWRqR8B44+fxhX4/ng/hcEy82X+hLpP6d5vMETUipJZnmdKVNT7q14QJASUmheb0fF836qfu5xBJLLLHEw4PlJMTjO9VZfCei9JO413Ycr17hXZrCPAyIxNFuQq7Bae1aaEqa1BZVS+ydoyIxPsNgwU2P8PoNXWQkBATKloj4lHJdFGhivGWt1hSURJLaL6tt2rX+xHr9B1A7XLWlRUxMKi0u21UySiox3JARhVQM6BAxp0uKQeiQMmHOhrjAQUdNS0iIeD9P8BK3nEhdc2eXG6hPmrCiKX0SuhJRqvI+u8Yn2kP+8XyPH6gu8HTSoRUJK/M2N0fKUVnxmk54n/S4YUa8xiETn6cSEdE3XVrE9KVDoVV9vHc5wmKJiRop9eF1lyQ+6X2mcyosHUnJKbz/VnCGckQfEeGuHjhXLtOmQ0pGXhfuTdvZ4HE2JSejoM86XzW3eExXSNUFLu4z9q5e83rq0CZmKF0EYd8eAi7t/EAmWKO0adUGBiH3xPqJ2NTOMMa85X1tUd6UfTLN2ZABl80WE83YtQdO6C0plVbHgjWb9KnKH8+T9KyTj4WF6P3khGRB/bNvoYOFhiQ8JtC32v7aCQ5YgU4YTCCa2w7baR7LVNz7KvINpXuPuulLpZZCKmYsNSFLLLHEEg87Hukm5Gc+9fPXzkN9Ou/tD/qY7xQ/86mfvxZLwpKWdRyLsLT02LRBxNTNm4ibcgR73oBQjLkVXed8FYo0l3a9oMbYRpEWaFhGwYprQBbTKmetO2VGC1fAhUsjUMFgMVlxNBaXam4Qp5kQw/P2Emva5pVonzEZG9qjq21iNewacSJvcfa0OQWb2qfAWd4+q5dY0TY3zFGdtt6RNoUvHCc6o68tLkuHuVqsQoFlXVPuzkv2ZMqf6q9TKcQGZC5Uquxozp6ZMdWChIjrskdLEzqmz6E6q91MCgqfCdGRNoIw0zkr0kdR2p6K0xanJbFYT78yXktS1YnvJSVW1QvtlY1oyKEdk2tBW1IU5UDHxEREYjiyE4JQPUxZDMJYZ6TS4lXZYdvuMzNzntGLXLVDXonugsCRzUkkoaDiDdljTo4RJ8qea06hBYd2zEQy+tLlAitsc1g7oDUnAgkxY53U106lFqTkVrWNkYgNGdAiZoc5fdN19EASjjQ7cY0eb6mb4YD3ej+cDDUMv1cbKGCONTiuGTn+PCIu5TzXon4N4TUG4flJXUnYTmiYci3qJrVD6s6tiSm0PJbXs2cP+Qsv/NVr//Pn/ovlZ9sSSyyxxEOKR5aO9dOf+rlrNS3G/wvfN782cZYw/KSA+N0Wkp+kZf3sC79wbemY5fDvfOavXws5BFuyCjhR+mk4rUm0aIOO5RqA5qpwWPkNE45QsDVpLGECkkjrGC0rlXRB1fGTlITIuW55UXzLT0gqLG1atEjqa2ouJa9E++wwYk279DSlozFTyRmqm360fVZIi4QSpU3MgA4ZJalG3OWAEIEXmqSSkkgiBiREIhxScCGJybFMKJlqxY/qJT7w5JROAjsz5RPRGoPE8MmVLh+263xM1njSDvigvVRT20L+zUzn5BQ+/8LU4XQhcT28TzJf3ALMmHsDWa3F9JVWddMUzpXBFcbrZqV+XYV3yJprQeQDES2WQiuvLnEr+j1S9tVRxw7tmExKusR0SF2mihddl1py1+5xZCfescztY4GjygnCgDapxqzSqzNiwuO6pkNLkmNTCCegd1/DBCGnZGIndaM780L70DQEaligWIXJRvP2Jk6mqJ9GxWoK0sP3zWakeawD5S9MNYJ9cHh8eN+cRJM6WlGR2YxK3bTjitnCYEgkrps3t+9um3/hhb+6/FxbYokllnhI8Ug2IT/9qZ879Q9bc3pxWoFaU6FYNC7fjYnHvdBMQv7ZF37hkf6D/dd/6r++ForcVFJG6lyiVmVYW5uK12RYPb6iHIS9ocALPPaiUUAF5x5YcORDIRwEvaHANUjdXPT8yjb4UD5ZZGG4VWo5th8AA+kRifEhlq7ke0P2qLBs6ZC+phzIrLaK3ZEx6/QpvNtQj3YdnNfTFq+zzZejW1xghTXpM5ReLfautOIxWef9rTY3bcbVqMNuWbIvGZukPNNJeXoQc/Nuh8rXpS0DK20oKmiLYRgb/uSlDj8crfuU9lndcOUUBJvhQqvaanfGnKlm9cq4xdb2wQBGFs186Y97EK9b/3NMTE/atU4kwdOYcLqFltdalFQYkTokMVDVtmS1NgW4wyFvikt037OHrlHThf4nuED1fRBmnZVB5GhcMiH1U6uu6dCVTj1hKClpeapfwMnmtW7A1DVgwTI6PDY85mSw5mkI1/NJNKcgzf04+RxnaUdyinoyGLZ38jWd9n3Yp/DchRZkNiMlIfNNTeyIdo5q5q/6cA0tscQSSyzx8OGRbEKaOCn4hvNNPN4LOE+eyaOC//Cn/ta1Q2Z10RhhGOkE8bkQx8S5DQF4EKqHwi44DKXSYq5z2tKiUnssIyQUj6HZEHH/gxYgPGbhMuT+lVqyIn3A25ESuyRrzDFbYPGr+4VWlFrSEbcyP9WMi7bPRe1zwxzQJWHddp0uAWVFXZ7GRR3WNr0AQ01rAXqJ5aIOSYn5MFfqJgfg6/mUV8w+c2uZqSXB0BbD7aykncDv3y3YmSqrqZBEQqelXLlQMIgjjAjjTHisZ+ppTKklGzLkJ/Q5npBN5pq7/BWd11qHmWbO3tZrQEqq2tUqnMuMnJnOaUmCqq2F97G4dPaIyGWJBLqQb3pcnsXCUKBDSptWbQecehXHplnFoox1ylfNLb9f89puN+xLpbYOl2zCuT6V5JSMyRz1i8XkI7Ou0Qr0uiZC5sZf7j7LyDt+FX7aEtyowhSkSakKX5u336shOHm7m9DZ+ho+63EBYeqXa+FCPDENN6wFLeu0/Ts5kQnbFhG2OaQlic8Vcbk5lQ8DBY45nC2xxBJLLPFw4ZFcZjo50WjiJLWqeVvz95o/n9acnHyOd2ta0lzh/7uf+9uPLH/6P/vsX3kR4C9/5hevqadUBcrLnLy2IF1YmXq6lbTIPRdd1YKfhrRMwlRn9KVDP+qw48XIUYMp74S5C+pIwKKoc4+d2hlGIucKRMq6ucIN3aUjkSvCG7SXEFbYtJh1blWxn4xYDsk4YEKfNmPjVvu3tM+ckoSIribMmJNLyhTjNCq4IvouB8ykw4SMVXr0vdj6QCd8I9plzXaogARhSzsMk4ivF2M+Yvo802vx+rRgt4SWGC4boSgMH37cTS62D2K+epjzAh/hD6I7bHPEU3aTriYMtM1Vc5GJp1kd6phNGfK6OmeulsRM7IyedLCSMNMMK85GeMX0XeGsETkFLT8lCMem7+lTLVxTMsbUk6x5o2nIfCK7xelGKulyqGNWZeDE6jpjrjmptOibLiM7fst1ZjHMtVg0m1rVU50Q2pcQ1ecvCNTnOm9kgSwyZ/BNxpdGGa3INSkFekwf4dLKFza6Im7iU4jbTkJMQbmY9J1hWuEoYramUoXHWnG3u9cX9E7HQzfd+0MRcfsdjAIijaiExnbkTN++0Ogb3ziqKpnmtUlEMGWwjUnM0gVwiSWWWOLdw1nMIIBf+/wvves15SPXhNzrgMNbG43mbfeahJxF32rinW5GQtHx9z7/dx7Z5uMk/rtf/8UXAf7tz/y1a4IB8eGEHNcShPPQDHwLP7e8UBpcEZYSH6Nv1bQVPf4zUIewBapWKq266PsAj9O3Tp+ybQ7rEL2AULBarWoRu4jLz4gxDOhwV8YUlKzTZ0dGJEQIQkbBXCoOmFAZpzUZM3eZIUwptaLQgkQSdvSQFglvyLZfsbfMfaje3JR81K6xEsU8HiXczAt+qDOgsvBHnh1x8U6Pf3BrilVldKuFBf7l55VXbrbppe44rJqYP6mX+aJ2uWNGJNYwlpwPVRfZNxkVltdFuWrX2DfOiSrXksRTb4KVryvI3QQq0xnBPSs0gladYH1O6UIW6XCHAx6Tde5yyEyzmtplUUpPjyu8DmXHHjDVGYWUtaWwwWVehGYlXBthJT+COuAxUIZKLSmkpFXb6BqfBj9nrvO6+S0piSXG6HE61FxzPm++ws9FP8T1KueX+e1j+opAj2oGXIYpW3htzcbjrAlps9lN/KQvlqR+b5wMMAxTGLd99cfFoIijLtr82Dabz3+aPiXcF0vs80NMncvjJo+5f8wipwd44EykQEttLsoEzVzzs7L5t+C78cd2iSWWWOK9gvvVwt8tPHJNyIOgOQE5rTkJaDYW34mj1tvax6VV76lQtfSly8e4ym/rNwgOQU0NDUB5gl6jaklNqw6+y7WglLJ2rorEUHknoOCOFYTrBqmD2CzKhlml7Yurvgw4lBljyckpeFq3eF126UmHXXtAJW5qE4mgIsTErJqhowVpxVTmtS5iS4dECCPJGNDhgAlHEjNjTuybkkIrEJgyZ4LTXCSSOG0ErhAv1WWGbGtBhGFfXEPwiq7zpO2RWaFrIv7YD90F4Pe/tMXTj2d8/KDPUW75WjViSMLrt7tcXCv5wnXh6W6L/3O2S1tjclOSkTOWnIqKQixdTTiQGav0mEtFhxRkXjd9GTko3pXK6SgmfloSKD9BD1JR0afNDiPW6bPPxIfrVSQSMVdnLDDBCfULT5laocuMOTMcxSvTrKYouWuipGDRIIZCPTQCzUBLWDShgmFXR7QkpivpsWnGIq+mISL31L+QXv+NfEriqXjzE1SpkL+RSlpPWJrUvzMbj0aIYaUVsSR0fQZLc6p3L+pW0264pKytg0WESN1k56RNcLMRaX6fEPvpiqUrHXde1Yn7gxHByWNqcOGsBuE8blmhafzZF37h2snPx3fzD+9Pf+rnri2bmSWWWOK9jvdKAwIg/+gf/aNf/F7vxHcLzQN/r6aiifC4+zUh97P6DY97J7GcgNwb/+5P/c1rT+sWMyn5mn3dFbR2AlAnep92TgamXzchT0aPs233yTRrpEXP68yQpnOWYok9JUixXDKbrNLjlu7xhGzybb2JQRhIj+ftJb5ubiIYZjpnrFOsVvRNrybNXJEL3NZ9t7+4xO8NBkREpBozFlc8d3DTlilz+rSdHoYZ+zr2TZMLAWyRcMfu1YVtJBGbssItu+upRIaPcoUP2jXmWO7KjD+3tsYPfexNVi6+zmc//+P84Ptn3N1tY0T56l2IBK4MDa8cVPyh7jPUlB9sD3glm6MoRxS8aY54Q/b4kepJSixvRId8sNrkS9EdHrND9syUr9s3COJ1i7MPnvn095CSftyZyVG1PhQ9XWtEnAtYzm3dpy0tdu2Bbz4q1syAQiu2ZIUpOUc6YaLTBk3PaRRCQGVFdXzF3rqpQTNPo/n9ulkBnLVs33QptKTQAhE59rvNHI6QRh4RkUqLQgv6psdUMwovSj+WZK4VbdPBalUnjJ+cYjSbjnBbM2n9glmnpGLfHr5FoB6eI4jXwTUBzWlHuO4jTK0lCva8ALnmb7EDBrzmZyE0T4gYmB6Z5mSNLJ6F69ZCU9U81okk/I+/8TfO/Nw7rfFo4jyf+Q/SSJz2x3zZiCyxxBLvFbwTDce7+Zn2yE5CzqJcnfXzee9b4r2DCMPrssMqPVqS8Jxc5g/5trtPFoGFwLHCqaBiYPp1w5JIzAXzGEc6IdN5LRgO9rHhdyutSCTGqPOdKih5plpnzXSYaUlP2gBcZIVCqjoxOvEBiG3TrilH4Fb629KqKVeKMtA2N+WAljjrXYNgVDgU1zQVlPS0SyUplVgOdEJFxTpO99CXLgUlXUk50ilHOnU6DH8NP21XmFJywaS0rWF3AmXR4p/+k0/y8adn3LzbprLC4Uz4kfflfPNmiyRS7ticRCJeifb5ETPgqTRlvQtvjlq8YQ+dVoWYHZnxo9VjPN5KmOTrqMD7qlW+Jq8TEtZTadVmArHPPIlxqetOyO9yQnItuKsHPMkF7sghsRoqsQyk64pkH3znLIFd0bypfb6or1DghPFBZI24ZtLRuxZUJwiajDAFc7ct3KEEvCboUEc1RWuqLiE90qj+XVWh0hz1dCMRd+4q/1sWZaJTYmJKXBhj0ChVLLJtRBwdKvbC+tBAlVpi5XjAYMgVCc1IKi0irx+xcrp9b1PfEV5/aAKMRE6rVFtNW28rXdXaqGbzUW9DxKfvuH3ItWDqXdHguABdZNEcLSYxLm+k0OJUeta/+en/+Nov/8bffPHvfu5vv/i9tisPf/SXzcgSSyzxdnGeyep3a6Lxbk55H6lJCLiDeV5qVcBZDctZwvWzpifvCh2rsf3lH73T8Zc+c+1aSotV6fFqdROAnumyXx3Uj0mNCzYMK9OXog0OvTD5cbMJwC3dq9OdwRVOibRoSwtBnCWsD3MThAtmjT9SXWYiBTMp2LRd9szMF4jCgcyYU/Cm7tCXDrnfdrCV3ZJVMnK21FG5wqp+oBW1iFnTLjsyxonxKy4wpK8uhO+irvAKd6io2JAhc0pSYiZk9GhzVw+IiehJh0MdYxB+gCfZsj2eMj1GtuTjaynPPjniznaPp5/a4bXXNhjNIr6+V/LJq3BnP2ZjYPnybbCqvGqnHIoTn3+AIbc04xvRDivapqctciouapcVEtpi2FY3x/j78sX6XKyaAQd2hPiCNWhZCp/7UWlFJBFjO+VqdIm2P14ZBW1czsqUnLt2151bSengggyf5RJf5TpjO6UlLoOlpETVOTMlRMfSwCMx3plsVk+8Ap3NIHVhvm6GLngQ52qVN2h+LUmOWf02U8qBetoRrIX/ov4ov8w/qa+z5nQilpiOtF2Yoc8eca2IYaTjepITruNm7kdXOnR8I7xr99/yPglTlNCcB+e2ZoMSjos7rq3aQazU8pg2JVCwYu8IFhzpEj9NDPQ7F1SZLUI8GwgNz0ndSTh2v/q5/+pFcJqRcGzD6zgL510wan6WntVUnKcAWH4mL7HEEg+K9xJdqol34/PskZuE3I+GdRZF535uV+dpXr4TnEbpOrn9B1mBe5REmRbXXNzSvZpCdRJVo3CpqOrsirnmrGuPHRnXiekdaTO1M1+s5TQ1IeCKvU0Z8gG7xRvRocujsH0u0eGK7fGSHGKAHi32ZIyqZShdDmUCCh1JSXwB6Qpey5g5W5oylbm3g3U6jh0ZM2NeP3+FZUfGhHDDPh3nEuVzNFyz5GhCoXidqdOSxETsyJifbF0CYCe3RAaOjtq0Ekt/ZY8rVwzffHmDq72YvSPot5UsF75qD9nUNnMq3jQHDLRNV1bZtRl/zD7BREu+Ynboa8rvRm/wp6v3M4gjqrLFjs59We7slF16utNUuAyOgsxnZwB1YjlAjzY5ruBvk5D7AMEDe1TTnJwup+CCrDCnorJV3Qg2bXjDtWLEBUiGZiFkxYQGIpH2MUenxB/DUPSHoMa4kQvStLBtPlco/MU/m6rysdWUHz/6EN+Qm8y14MiOa2rShlklwrmEPc0Wd+SICkuPlCuyyQ3dpW863La79fEKBf+mWWXPHtbHCxY0LoNgxUngw5TQNS8V6psti4Jq/f4JupLjU4xGGntjmtLMxakT1/1xDRPBWpDuRfBBJn8aLMrPvvAL12oXLX8+7mVb/p00IM3vf+3zv/TieYuE5VRkiSWWOC/eq81HwLtRNz5yTcj9/hDdT99x1vbeKXrWyecOPz/IFOWs0dlZF/ijIKgMLkmBbz5jfvz+xsq1iGFip3RNh+fMFeZUFLVGoKLQ0tNw8Knojurjsj1KnwFiSNTQ1cQ5W2mLMSXPxF0ulB2umyMA9nVER9qMmRF73vycnD4DMgpKLHMpiTGMJUPROrAvwnDElISYHikjMmbk5N6qd0JGQkQaijxf6ralRa4lxoudFaUjzrVrSk43Fjb6yuqsQze1XL6ywz/7ymNs33qcG7f6JJHywffNiOOKL7w0IK+UrsZk/hh/Wp5hrpbH+4buYUwkcEjBM3aNfXGajpUoolJlpCWFp1mFYnauOW1JSSSuU8mNb/4ycuJGw5d5AfmEjHX6jHTmJiE6oysdhtLDovSkXU+SAvVpUSA7R7OSEkWOFb7hegmIiFzToJ6i5MMvJ3Za3x+LBGWDv7bc6wpNR2hpAlw+jaHQgrZp85WDnD/duch2PmJXRn4/3cSlS0qLmFQGpNpw4yL1mouIdfpsygArymt6t57MhADHs2xvg/A+0Liablm20awFfcex/ccJ1AONzbDQSiXiplMGOTbBAWf8EOEc4JrHWbGUYapyQpcSktqbifFhgnPWFORBPpPvVwi8nULhUfiMXWKJJd4+3usNyEm8Uwssy7S7Bs4q9O839XjQhuXtPPZBG5uTF/T32wX+TqK5WhpWWUNOguPrR295vEV5Ui7SpkVGTp+UzGYUWjDVWV0w2aADQWrL3QpnlfsH0XW2bI9n7Cp9EsYUfLOckCCMJedApr6AMr4pkLpYDtSigDklE+ZkFAzp0qNN6RuIUFCH1facgj5tWsRkfmLiVuxjCr+iXnkHqeBOFOOE61d1HRH48Adv8yN/5DaXtiasP/cVuqnlay8POZoZVgYFcVzx+o0+7RiurMFn3tfmT2x2+YAZ8C98dMSPPVNx9eKcDjEjrfhI0mdKSUdjnrcX6SeGSWUpcP8DnQeotRGB4hY0MZGn+DSnFzt6yNiHVO4xPhZMGUTQc83Z0AE5BVPmfqHBNkL3DIkX7jdT0hdOT24SkBD7ML2KvukSrGvDvrsMD6mL72AN3dQeue2ZY//BFdp906NNizdlSmzgSbvuWg6JaUlC4gv856oLzCn4ptxmX0dMNCMj51W9zSo9AAakRGpYkz6XZYOh6THVOV3TqfevGVboRN+ter+bqezBIvi0UMPQ2DbdxWBh7xv794b1DljhOg2W16WWxxqQky5dp4UpnuYGdpa7V3M/362spvNg2YAsscQSZ+H7uT77Tvf9kZuE3A/nDRhsTigetLlo/s5Z9r4nvzb37bT7T9uHB7k4HuaVOts4p7AoTlNJMQizhkAWFgFtFuW67tCWFs/Zi+xGI7arfZfwbRKf2O0mIwBznbNqhl4j4gtRUeZaUWDpk3AoOammbNgub5g91s0Kj+sqL3vtRkJE4lfsu5qCwC5HTDSj7YvECwyZNCY5bRJ2GTWuCVdY97XNROb1a0+IGFExU+dcFehZkUR+ddyJsr86yfjUP/87bP/Wj1FVhtmNJ3nfk3uMR13iuEJEGY3b9LsVK33l0qVDLlx+ma//s49zYQ1u3u4TR0rasnwg6bHZEUZzuGVGXLUrPKF9nt6qeOWNik1pMdCYP86HOZAZMylJNeIL9ltvoUpNNKtT0nMKNsyqE+LTZsSM3Kdur0mfibjE+JnO6UuHQ6Y8Z7f4/+SVensV1uknBLp0GDOtHZ9C8RuK40DRst762GhVTwROUoaKhrYh0Jzqa+sUulDIEVFJnXOVzPj6pM2QFld1nZnM6fkp0BO6xoHJ2NURB3qEYFwwpaexVWLZ5oib4lzQ2rQo/dRsXQYc6ZS+6TpNjHHakoHpOSvqE9kkdfPUcNsK+xtoWEYSn4uzcLIKxyvC1JM7oN5+pW62Eqm77uLGn6GayuWnHyezS85Kd7+fVfnSPGSJJZZ4r+H7uflo4jw0rZOvNTxu2YQ8IN6NJPTzNDxn3fZO/nF9WPnLoUC5Yrb4dnWdhORYM5gQU0lVFzpPRBeZk7OpPWbiVs7fMHtc0U0kknrleKRTp1XwVKJUUlakzxo9xmQ8oWusa8qr5pAf1E3GWrHPnDYR69oGu84Nc0Dln7fCOvoVJT1S5uJW2Mc+qM+qYkRcICEVGwy5pXtcZpWXZFZTcrq0OGQKAkO6HDFlnT5HTCm9NuSyXOQuB3UhXaklI+drekRqInSny/bdNZKkIp93UBVW147oDQ/49jeeZu8o4aMfvsVLL10iaRX87v/7R9kbCz/68duMvr3FLDe8sh3x0cslf/im4QvVPh/TC6TBhczClJIn4hYvFXM+Kiu8ah11bUbpQwVdU1bgHK5ULYm0mOmEp8zjdPzrbJNgRV2BLR3vT+UmFJG4VishJlFDLgWKHhOPK7amC2ljZT/oh8K0o242FGY+wyQ/kTMD1Na7J6/BpgNbE2HqMFVnPvCm2WcvmnBBB3zArrETjck91UkQXpFt0OM0r7nXfbxptyn8PqXSIjIrHOiEkpK2tFiXAfs6Row7Jgh+ijM/9lqauTqBntWcVEl9zXp7Y0/ta1rqqqdfRbV73ELjIX4615w2uWOzaDZCDs9p9LGmnfASSyyxxHsdD0vDcT+cVzcSFr6XTcg9cB6R+nluv99jmlONs0Tvb/f53g5+5lM/fy3sx8OQRVLVhY/wVHSZm3aHp81jvG7vMNcCIxGPmQtMNOPAHrkSSpUP2TVej3aJEG7aHZ7nw0QIm9rjjoy5yAq3zIFb1ZWIDi0e0xXWbJtDM6erCU+32kzykkudmC/N5rxsdlitUg5lzmPa4zWURrL5AAAgAElEQVR2eYU7tCUh04KeGGIMuU/1tigpbgLS9ta1odguKFmRHnMtaztaRx3roFgmZPRpU1ExZV5TsSKJWNMOu3JESxKOdILFUlLRkTYfsRtMXn0/q2tjxqMuK+97iYMv/Ajbd4c8v3GH/VHCB5/dQVUwBtQKv3l7zDot+OIlKoXEQGzgi9cN36wmlKJcjFvMraUXG17fFTIqRpVlXzIuaMpdM2WoKfsy41l5gm/ZN4GopiFZv6puEDa0x1hyVumxx9hpcLxt7V09oNSS2LjMlAkZ69rnlhnRkZRMc1ZkwJFOEBFSSQkBiE6fsKDahedtFs8huBKobXtjiUn8sW1OxyAknC9wMlG82aBUWA50TCYtuqQcUbCuPe7IET9RPcXLZp+Wb5oDra65jWbwYhDvu+T4kjta8ax5giOZIip0xYn6d607Xs39cMciohL3GtHFBCTQzELwoAjgbambCE1NsEBG9di0pH4OT0kM2SDuOCyauebrDLgf/QrevYDYJZZYYonz4lFpPk7DefR179km5N1alT+NvhSK+fMU9A+q1XhQO+CwzbMakQcVzj8o3q3G5nuJX/6Nv/niv/Xp/+TaDbvNFbMFwIp26PpVc+uPZ4ThQrTGvo64KhfIsFzUIZmUXDBrFLaiTcKKtoh0QIzhCYYkGBIMCqyQ0BFDZjvskbNbVLSJeG2Wc0emrGmXbZnxutnFWOGiDhl7+10rWtvoJr74HpHRkbR2OGqRUIkryI6Y8riuc0sOWfOTDld0z0mJKbRiT8ZOY0FGTExKzJN2g5vmoHY36kjKWGc8IZs8ay/w/l6LKMnZuPQm+fwpdJayeXGb/qDLfDoE4OJP/DZf/Oyf5crlA2wV0cLUwvRXJwVTKtoYvmEOuEyfrsa8Wc5ZNwlHpWWUl2ySsmcLBrSoUNa0TSYlUym4oAO+BeQUuKQUQ0lZF/d3vWB7XXvMJWcOvvlyU4F6Zd0X5PsyoU2LDikzmdOR1An8vZjZitOfBCvZYMOb1KL+4wW2m56EotzRnZqp4cdsbTGIHM/QqKcM/muEa6D60uX9XGSXCRHCnmQYhC4pj0mb60T1+zMEG1qtyL1Av05m92X8jj1grnk97XlN7wDOqCHxLlsul6SsAxUhCOxjLHEd3hjcv8LrqKQC9VQ1FrokwTUU4Ri4iZTXvnjqVvOYKpZK3ddAw2pOW5qoqZXN5Pn3OB62yfISSyxxfzzKzceD4D05yz5pi/jdOJmnaTBONif3KvzP0xQ0G4jziuDhdOet8zZND4KTz60o3+vgr3cKj5tNLpkNbuougnBDDkgkoisdWpJwoCOe5AI/Wj1Ni4Qfri5zSyZ8yG7yL+oVerTpa4sV61ykLtBmLhU/ubrCY9LhCdPmgneYmqjznHoq6vJNDplT8XXjchku2yHbxuWPfM3cZioFqU88b+HExxalR7sugGPPrY8wdGgRE5OR0yLhrhxRUvKk3SAmpofLgejRJpHIJ1Ln3jEqY1P7PK2D+riIp5a1SPhT9ik+dWGFpy8WpFs3EbH0hxOO7ryPg90NNp7/Mr2t6zz//h32fv8T/M7rhjt3h3zlpQvkWFpE9FKYY9lnzqsy4kdkE4vy451VrsuYia3YsTkvyxFtidiRjA1SplT0SShRNw2QGUEOXVDV75nE06i2dECEYU8mCEKHlJSYNgkD6aOqdHDno+0nSRMyfzzjurkBpxuZ2pkrkDme53EyUbyp/6jUMvep5XPNmdgJYzt1SekNXVH4nZN6hqaFbUdSVkyfVemxom1iDIVYpLEPvyO3eVP2GanTu3QldXbOkpw6XamoyDSri/oKl5R+ZMdEGG9aELFi+rSlTdd0HKXQDEm8vXBoLNRTqZpZIGGbzYlRsIo+Oamoc1JOaRpCc9M8Zs1zEJ4/vK4HwbvxOfkgCA3Id+vv2BJLLPG9x/K9fn68ZychJ/FuCqfPojqd53Hnua/5mAcRu98rE+Q8eLs0rodxGrJIf3ZWr4oypMuP2Uv83+Zb9X19EobSZUTBiqYMJOaGzfgBLvLD/R6jPKzEwqyomOYw0pI7WvKBpMcXyhFTKbAoH6nWKcRpDSbkXKDHXCraGjsNA1Ne1duu+fAr8C0S1ulTYWlrTE9SCio6ktZuWV1adVOSM2eLVfbMtHbB6pGyph0qqTiQCSE9vCtturZFhgtE3GSFlJhX9DZdSZlby63DmCSKGF9/hu7wLnHiVtjjpEDiimK8QpSUzLM2iQhlCV/fK1khIcXw5pFSYPmBZEhqhElpeX/S9ecA9smZUWJFuWMzXo526FaXeMJ0eN1OmUpOVxPmUrFpVjjSKZVWhCT5Og/CN08j31gYhBLr7IqlzQFHbMkqN3QHi7KCo69Nccnucy381CR38nxZvNeCHqF2ffJUoWZh3ZyMhOakmasBpzs9ndQwhODF0JBUWL5sXKDmiIyxybikQwzCm7JfBzAO6DtXL2JEhEzDHGohhg/J5yf1KJWE42eYasZQurSM0+PsckRbWnQl5W615/VCx92ymg1PeM3NVPX6tTVoa6ERc6YQptbYnDZhMmccv+9HLIuRJZZ4dLB8vz843pOTkLPwnZ7gn/7Uz107S48RcL/i/H5oTjlOm66c9Txn5Y18p5kj97r9Xq/9YeNSv/jZn3/xju7TkZRP6PsAnw8BXJFNPsjjFFIxIKZFTJeYDyY9FGVKyUfSHhdXLE+uwXNblk4s5Fj+4WyHFEOFUlqlozGbtkNHY34nuuWmLmZEzzcOGaXTY3BEh5QWiRc4O5enGXMSNUw9pSoloU2rttuFEJDn6FptWnQ0ZocR4FLDFeVQnDbAGf7GDKVLSsxdM+Zbfioz1JQPVBu0pUVExJfNLl+YH/HarlDkKdKeUxYtvvKVq0zGHSavvp/XXnqOg70h23eHvG/F8PQzdxnEEc912jzVS3hyFRIM7UjYGihfqY5Qhd+eHZASM6MkJeKy7ZNiiIh4zRywZ10mypp2SIjoalJPMBKJaXlHs1CYbsuoTi131KmKkoojprV+IGSoZOSUWHrarqlTBkOuRS1AF4ynfbk8jpZ3faqbVzlOr4IFfegkguBa1R6zwQ1oruaHQMPMZox1xh27x649YKxTZjpnR4/YlSkDdU2oqjMRGOmYCjcpMb75PDkNOdnwuGbL3Vay0FhMcW5pBRW5Fkw1Y4MhPdPF2ewmx/bb5aos0uObx6KqbQHeemzCZKt5HpuJ7uH2IFpvPu6s1/T9hCUta4klHl4sG5C3h/fcJ/q7dSKb2z1PsGBoAM6iTj1okd7czjsx5XhQ3Etjci88DML0gP/+1198cUCHH47WaZPwkWqLLjE/YR/nk1zgh6qLbDPnkg55Ku5weQUGJuKTnRW2BkpZCeNM+PZdw868Yiw5HWIEWKfFrSqnS8QlaXNV3TQjk4KBpsRqKFE3AZE5bVrMcNarKTEdSRnQYUiXQ5kxJWdXphQ+Hd0gZOR1kwFOxJyRc0NcgV9SskqPgoo7HLiAxZAJQsSckj3GvCrbbNkeq9ph1RfeM537YMOIZy9ZRCzZ3iUmox5fvWv56qs9bt94jHkesfXYXV662aLTUq6/scnFnrDZVzb7br9WTMzLWcbv7szpkfCH1WHdKIxMTp+EXclo+WZjU3uMKTg0LsjQTTTc/uZaIEhNKRuaHutmBYAuKYKwxoAWMXNKjnRSGxHs2UMScVSzYBPbDHoMx7ElybECvWkTC86xSdVNRELuRUhTP8u56aTIO+AkrSg8V0HJyI6Z6oxSS2Y6p6RirnNu6R5tDfvo3q+5Fsw0q1/rolkyb8kliSTyCe4ub2TFDI4dk7vVHtt2n227T6YZmeYcMuUxWefxaIt1s+Jpi65ZDccrNFjN1w2LhiRMiJoNSaH5MUpWPV1hEUh4Vphi87idBw/bQsoSSyyxxMOG91wTcj+8293myabjXlSt84YW6ol/98N3oyl5mKce98KLn/35F1tGuGrX+fOXu2yaFlc6MR+6YJhSceQTvb9cHlFWwoWOobJwYa2grGB3pnwrn1GqsqUdL/ouGVEypaRNxFQrDnHF81QKepqwYybsmAlzSg4lY0CnLvwPdEKPlDEZPdocMEGxzJjXq+qCs1SNMKxrr6ZeRUTMfXPSIWVTe6zTx3i6TUlFx6ep90ldYjUJl7TLJ5M12mKYa05Gzobt8NF2jw9/9Ft86SuXSdIJ02mLjz8OlzdKVtePaLcrxkdDLg2VQbfk5m7CxrBCBNqp5erjE57oRnxirU2CIfJFduI/auaU7JKxI1NeFZcaP5I5K7SYkvO62SMTl5KeelF0qSWZzim1ZIMBW7JCl5RUg/1uVKeqD6VHpY7Ss2L6WJTYTzkArCqFL9IjzMI+lorMZnUBfEzn0LCPDU5Nb0lc90V3EHTXIYQnxOonQwqbVr6hIC8o69ccSURbEroac8S0fh6ATDNy3prv0Wx+gsVuatzULZWUgfRqKlfsbYhnmtU2vwC37W4dmNmVNn3p0jUdVo0zJhAvxG9OeELjdnLaEY5v87ambsQpqE7XzYRp0snb7oWHjUq6xBJLLPEw4dc+/0svhv/fN5qQJk5rRN6JUffbKcbv9zvn1WScdL867/Pey3mrub17TXzOCkx8WPHs3/szL770wq9fG/YL/i+5w58vrrAlMKfiX7u0zt+6+wZzUxFHQ24eWtrG8O2bLYxAbi2PR04PcbtU3jSHRNUqxtOunrVrZFTsm4wN26UUl/1x1a5yW8ZMJGfEjApbh7RZLEdM6dGuz9UWa4xwAuQtHfCqbNf3gzu/Gc6i1uCyQwZ02JEJW9pnLB1nW0vMmna4I0eO1iUJXVoMSLiyCgfb8CEuY1EGJPzxT9zlzo3L7IyFO9efxohy+yDiwtAyGfVYWz9kdNRnnAmRiXhlVLI9jVhJhbVBxeFRyjfHOR9bb1FgmVNRUNGnwwg3zXjeDFi1Lb4UbdNXJ8r/ptmn8pQgoWQqczKcW5WIAbVcijYA6iDHW3JIQsQ2rpmpqOiSUklMKi0qLJVap5kg54qucN2YxRREXQOR2Yye6TInp6LyIXxR3Yg0EYrpOkskTCd8VkZHUi/YXgi5A5qNS6BrnZX8bfxiyLq4on/VGxIEuN91OSUnG4EmRMKEwZBTEGnEIWNUrRO1E9eZKWGfEnHBm29Wd0gkoSNtZpoREtBb4oI6J3byltcFC/2Hu04XoYNBaB4QGrpmQ3KStnaSYvYgWDYjSyyxxHcLv/b5X3rxUadknazDz1Orv6eakO/kBD6Ipe/JxPKzwgdP3ncenIfqda/9an49TbtxXkvg0yyIz2pwzrpPEH7mUz9/7WGiZAX8S5/7zIsj4K/4n7/0U//btZSIcWb40epxSpRZLiQitCNhL7NMrXVibFW+rTNeifaJMBwax6m/I4d0TUJHYyI1PKl9XmPEXCquaI+vmdskRJSUTgMibQoqFBdIuCYD16CopUvCbdmnS8pUCnItGYphzIypFHWRaxAGPlkdYJ8RiDunFuWi9uhri5HMMSq10P3xNOEbOxUjrfgT8UW2i5LXZMyXv77FV/ZKnunDb395hfdtWv7hZJc/YzY4fGXI1YspIsoXDzO6RxEvcUhmSz462YDbLf5wMuUNc0Sxt8a3zAHP2lXuypS2usnNWHI+uiWsH3T5Ql5xR4543l7k981rGJzL1Q3dISZips6eOGgfruiG14JUdMXNI1JvOzz3x9SIEBPTlQ4TXzg7LU7OLTOqn2PsG0FwYukWST1RmDN3MxxvJyvElOTEEtfBfKHxABZULWI60maiM1qSUGqJPRFaKH76UnmBeEAI34sk8poL5wTWw9H1XjJ7jnLnLYrrCYu6VxHE42c1NosgwgK1loHp1+/3JjULFiGIubo5S4WtpyRjnbJh1liVHm+wmBzBovk42VA0G5Hm45poJqOfFNKH1/UgzciyAVliiSW+22i64X2v9+W7ibPq7vPU4993dKz74eTJP8sa8axJwv2E6c3f+06F4+dpLh50KnG/1yONfye3fy/r4IcdP/DZP/vin/vcZ178x/sTjihIMfzu0YTLA8PWQNmvSm7ojH1bckfn/F70Btd1hzEZt+SQKQWXdZVUncVuJS7+7rYZMZGcr5k9XwwXFOpCBTPNmWhGiwSDYYdDRjqjIynXZa/et5u6RyIRMx9EGCg8fTocMuW2HNar72sMOGDiJyWusFzRFi116w1dUiZkHBSufExxYXsVyl2Z8N8cvMYbOuHyZkEnga/ehq4mRAaGHcutnZTxNOG5TptnBwlDTXnOrjGi4OuTjC4RLY3dNMf2asvdV2VEIW7iszd2FLeJp58FV6tcXZr72E6YqXO9qrDkWpBKi1EttnfUqk0GdEkpsXRpkYgT6h/oiMQnpQM+gb7NPhM6pBzqhExzl+1BcIma1VoToKZWhQJaMPVUJZb42O0iTh+yblbI/LQn4v9n791jLMuu877f2uece8591aur3z1PznA41NCiRIlyDFmWo9iIbEnIDIcWJRJSYMGGYwQRIYGSIycpT5D8YQkKZMCw4QQGHMWGJUCioiiSkTimRcViLJGUSA85HD7m0d3Tz3reqvs8j73yxz771Knbt7qr+jFTM1NrMOjuW/eee27dc/Ze31rr+z6DqZ1DdUxx40qeV+H/r3MjQgmJS7nnERMWbZPXZZUuTk66zp0oKKrrYT8ein/vehRYBuU4VyRRJQcdyK4XiOeSZKWCmAcBE50wIZ/ZvZj28fCdjukRLU9er4OPekzLDR8mjuIadkxKP47jOI53Ytzr2nakOiH3K+6EQg8ycnS7EaVZif5+5PWDdC5mHXPWOUy/551iVldnliLWrHOoP++d2g2ZFT/06edf+IfP/9OVRXuSsyZhdaCMreN99GRCrE6C9HuKh3kl2CRSp4x1wc7xhtmmJ2P6kjKvCWMKFrVJQ0NSyUmIuK5bVeLpK/QdaRIRV7pCAcZV8cWBwpSME8yRY0sZ1R06OKPFiKACGye0SyaOSzGnTXJxfhvvZ5ET2uSy6TGvCQj8Eat8SJfZIWOcFzzdTtgcLBBj6JPz6OPXybJzQMATwzaXdyzfd3aCCZQgsDzz7Zf4/T94Px9odDjVFm4OlEvphFdlBy/F2yKkR+pI5lKwIUMe0RO80s85GQXM06LAkmGZo0UgbnxsznTJyZmXDuDASkNC8jK5DzCMSnM+gJt2g1Nmib4d0jaJI3prUV3D3tRwQwY8Yk/wFTMm05xQdjsLAA3CEoZ7vs3uKNOc6ZBpVnZhkkpdyuqui3hM6MaZSgNDg3Njn+ikSsJ9hDW1Lzf2VWDFdUkSiYklqj7DNdNjohkjSfG+Ib5zYu+gFjUryRdxymC55iSlq3zLNEk1I9XsFmL49LhVSsaW3aZlmntGsvz7Ae6z1AR7d2WP7S2A416j3smtF1eOQjfkGHwcx3G8e+Jjz37yFgXW47h9HJlOyJvdvrpXzw/glg1vPyCxH5F9etRrP5neu42DAqD9/l2Pd4pp4UHiv/yNn3rhiU7EU4sBr2YjXiuG5GVSmmGJCXjMtHimWOZ99gQntU2E4XVuMpacK7pGSsFLwSrvt0uMJeO0bTMsOQdeVlUQwtIB248GtcoRrbY408EWMadkgRxLQsSYjJFOyCgYMCYiYIE2FmVeE9bZISFiKBMMQqIh1xghCAPGVXL8LbnB/20uAXDCREQGvj3p8OH5FptmTNLaodks+IHv/RZJYBhay82NhJNnVhmNIr718hP8+94QVViezxnkliEF3zA3uGkGXDM79CRlkZgJOYmGDJnwPTjH+kGhvL84ySo9UsmJCEoZYtehaIlLjs/pAnPSokXMnDaZp0VaVv43tc+mOkf4a3aVjII17bnqPTkBgZPo1ZSx5GyqM4lMaNCWhKB0DPdJek8HjtxuOlX1P5aYJTPPeVnmpFlkufx7gCGRBm3TIi5VozIKzpgTnDZLtKRJKGEJKOJKKcorQ6laYmnQlBgjAbHENMQplbXEqX5FErCo7rt9SE5W9/MsE0WYLV87K+H3/AxPxndjY0pHWtWxpnkaHkQZpBrt6khzz3E9CT4o/wsl3AO2po9TPz8/bnU3/I/DyKy/mXEMQI7jON49McsC4p0e92ONe0d2Qu429uOA7NeROKynyH4g43ZVu8MQ1vfjutzunOrncLvnvNvi9K/+4AuXP/6vVr5ltviwnuKGTtiSEae0SZ+MyAoNAv7jT/9n1U34LPC/PP+/rtyQBpdZ4wRdLLCoTV43PdokDjiUYzpWlZAAQRjqmJCQM9JmlR4RDcY45adTdokvyyVCMY6sXRKrC7XsyJjHdJkTtEqfEzfeVGBpEFGgvGG2adNgiQ7zmnBVArZ1QCghT8XOSHCpY1k28H9cGfIk83zuc6c4d3KMGEtulW/Q4y92uqTjJl+7HPPSaESHkM/l68yvLfNZ3qAwyhIdBowx0qRPymN0WJUdNmTAB4qztELDxXzERgEBwmN60ik1aQDixoT6OqRDiwFj1sQRqgXDpgyICckomKPFSFKaNOhpv0x7DeMSPIQoeW1kqMeQpsR8Ta4x0ZScojQ2bDHRlDnTZpEOa8CSdBnKmKYknJUlJmQ0NULF0qVZkdebktAqjS9TaZCScUYXKMSW7uvKlg6q7oUPR1g3FFiaxAwZkYj7HoY6JiBggYSb2gPgvcVJembCRnlrBgRY8l3StuwaBNZj1nhW3cDQiiES17XJbUrDRHu6Hh4QeOBQNywssIw1reB0dT5YjDT2VLfq/BkHgG7trtQ5IHcTtyvkvNlxDD6O4zjuLXzR890ygfFujiMBQo4Kied2ClIwW753VvJ+u9GqgwKKWaT0WcecBYr2A0D3qoL1blsYvjDa4RvmGk/rEtuS0tWYMQWXTI9nf+NvzPwd/I3f+MkXfuGjv7xyw24wb9rckBGbMmIsGSMmdKVJjkWwFFKUPAhnnwcuWR6WnY6sVInqSIO5Mkmdo8WYjLGmNEvewJr0edKe5Lr06WpCX1JGDIk0oCdDWsRsMWJeEx7VLl/jOsvi3MjfmKRcY8QPffsqq9dP8x0bXf7ih6/xh188SxznfO2rjxOIkKOMJwEmKGgEMKYgw7IhIz7b77EZDBmR8l57mpZEpBQYFV7XPud0gcftPBFCViajwxIgnLRt5mhwxfTZoE+AYdF0ybQgEEOvTOLB/TvAMNIJKruO4XPSpqc7e+5Fz8tolP4pRTnytckO89Lmhm4SERJLhC3BwIAJTYkZk9GSBEFY1CavyYBFWpzULgNSXmeDljSrDtYSHS7YeT4n36QnQxSn8nVBF1mjdwsfQ1UxNfPDeekyL21GTHbHvFDmpIWo0CbiZbnJnDYrNa5A3QiYBwazuiD7PTYNTkYl8KlL9NaPOa1qBY68PkQJJay4Napagi9bkd09l8aDE8VidC9PxpPq7zWOwgjWMQA5jrdz+D1+lird/dz3f+y5n6lyvumu637r2ZsZ+41Vzbq/j0r++mbH/Vrr5DOf+czfux8Hupd4u32J04Biv5juStyuq3Gnbkj9eAc5v9upY836c/rz3OmzvVuAyN/4yH+z8rQ8zJrs8L3Fo/yVT3/kQJ/7v3j+760AvJfztIi4Ks48sEHIqBzLGuqkcjPv6YCWOEdvr9JUqKUhIR3c2EuK45Xs4EjbUTlOFBLybfYsV802kQYMZcI6O5ygS5+Jk+TVJi0iOtrgmtmhz7jiVQQY/s78+3n49IhGnLNwYovV68t05wb8uy8tszEp+Jru8F3RPB+4kLO5E/KvVvt8ObjKkrbJxG1WfcZ0SHimWOZrZoOn7RItQk6GEUuJ8Mf9Ac/EbW6kOZd0wJOmwxt2zAWT8Adc45r0qs/V0wEdadLXUQU+TjBHohGvcJ1IgurzxYTc1C0GdogR5yQPLpFulKTrOWnRpckNtjjBHGv0iEsS+xNyjlf0Gt/GQ6QULGiTl8wV5nBjYEaFR+0C7w3aXCrG/FFwuRqLW5Y5utrkCbvIvw2+SYAjdg91wgmZ46pdI5BgD3eiIZHzsRfHQVkQ52eSlkaUZ1jkiq5zWhaZ05gbsk2bhK7G9GTM6/YaBbYi78Ot4031jb0OOuoqXPXuTERIJBFDHc28nqeBix+tiqVBIAGFFsTSYKyTqsvhQU39fWKJsVpUXisBAblmdyXFu9/6+lbFMQA5jrdrfOzZT67U78Hb3Y+BOMGOf/7pX7qr6/0Tz31qZZYh6fQ65ZXyDpNreHDzLz/9P931veiB2H5rSf0+f7vlrvcSD2p9OxIgBI72lzk95uRjFql7VkfkfmySh3ntnZ67Hwipx910St7JwOQTz31q5TCL7qc++osrp5mnpRE7MuEb9g2WzHzpbG7Z1gGKliNTloKCEzJHSMCACRNNq+TbyfO2GJPSJCahgcFxPAaMmafNe+wyr5n1KgHcpE+HZjW+1MA5k78hm8zT4jqbPKVn+bpcY5EuD9l5/s6HQm6stllaGJM0JyyfvcQrX3ua33wZ/jS4wccaj7CVWjJVfkdeocCS0KClDQYypqUx2zLi48GTfD0dsiwxHWPYKHKWgpBclYG1XGbAlhnzPrtIhtIj5ZvBuuv8kKIo2zrgtCyyzZCAwBGhpUlKRlsSBjrmfZxnU4YVQf+ivVFxa8A5oXun8PNyAsGww4gWDcZkNAjpMeB9eo6XuMLfNB9kp7A82g751dFFPhI8zMU0Zd6EFKqcb4aMc+U3i0tcZ5MLeoIuMfPWySj/iVykKXHJ2XFA0ncY/HfsgKVT91oyc2zanUqFKpKIEzLHkna4Lls0aRATcVFvsCxOeevP2Av8sbxGgGHNblRdhNuBkHpMg5C64SLs+qAcpDvhFcRMOQ42J21yCoY6RrEV16R+Ll6Fq274WJcXPmwcBfAxHcdg5DjeLlHvSNRj+l6c7qD6ezogKIsPY/7Fp3/5jtf9x5/72RW4latWBz/+3/49D5JXzOrgTL+uzmuddUkvI/8AACAASURBVMwfffanVw5a8H03eoIcg5AjEvsBEpiduN+pA/Kgz3H6sYN2Ou423slA5DDx8x/9pZWn7Rl2ZMIV2WLdbnHaLGFRtnVITo6USVxReoWYkuzsZXStapVwn5Q5bmqPliQs4hyvN+hX7xdgWNQ2r3OTC7LMFV2nKTGndI6+jOlowkTyKvnz40KvyipdEvpM+Ptn34dV4bFH1knTBo1GShDl/I+fbTJHgw+2W3x+0CfC8KeB8zy5ziZLuEp+V5sUYvkL9hxbmnNF+ixqwutmi++ypzkZhXymuEGA4SHb4YoMsKJEaviauc5DusSOTCpwtUiH9VJut2fdZ/VjUA0JK/+QBs6A8ZpuMNRRxQ9pluT+lIxzcoKERuk6viuL2y9J+wkNvqt4iB863eHKlvDaZMKfP9Pg9Q24kqZMsDyVNOk04Ld3VglwpP+BZFzQDgXKnwRXsSjXdZOJpiyZefc96TaqlkCCirNSUHDGLHPdrqGqxNIgkogzslh1umJCEiJuaq8i0D8mZ7iumzQkYsP2mKjrqk0nCDCblD4dvnPjiPU5EWF1jfioA5f6cf1rvX9KYhLmpMWG3a4Alwc1/vN7DklGfgtQutdOyFGOY2ByHNPxn3/kv17xXdF/+Bv/7Zt+fdST8uku536+QxEhGXn1eESIiFCoJdesWiMEU/3bg5NPPPepFdgtdEyHP+bt1oWDdmvqn+lO68pB1pG3wxrzIONBrl/HIOQQMavrsZ9872E5IPfz/Orncrc8kHsBKcdAxMWvfuSfr7xorjLQEYk0mMPxOnoMKpWkpsT0tF+Jl7YkcdwF7TMnLTIKJqSOI1ES0i/IMgahx7CS502IOK1z9GTMo3aRb5ib7DDivXqGIRkRhm1x4zGL2mRHJmwyYIkO6zg+xZP2FN8fLfMdj0/Y7DUQga/dhJ3C8lefmZBlAf/s5YxXg01+QM/z+1xjXXY4qd1KlnZJW/RkzGuyxjldZF4TIgxntEWC4cvGKYd9uz3JvwleJSXjJPOs0uO8nuBxu8CXg2uOn0GDHgNiGvR1RErGaVmkKGVet3SHZqkk5RPiNdurOg0NovKZyrLM45wplDYJY1KnpFX+zAOzf/mh83z2S4tkCu1QWJ8UfEk3GUrGU3aRRxsx30xHnDQxL+s2fUlZlR3eX5ymRcBFs8O35CYbtsdJs0iXJtd0oxqxyzSvgENHWgx1hGDomBYtSejSZFV75BSEBHwHj/KSXCHTgolOiMrP5rtofR0y1vFMwHE73xAffsP3ycWuqO5ed3gPQnxyIaUEb0FR/b670sFiGeukIu97PxX/+noXxgMecJ2Xu4np9fUoJwvHQOQ4fPzUR/7uStVNkICxHfOrn/7FN+36+LHnfmZluvtQ533Viw6CqXhhkTTISkEKf+/npdx5qlm1DtdfA7tCFH7N8DGrI1Jfs+odkWnQcZDu6WE7rPuNsc/6+Ts13ux16i0hpr8dAMft4nbdEB/+4r3fI1j7tQtnAaT9JHgP0hE5Vsm69wjKhbglux4gfjTKijPd6zPCYAgJoEwAU3Ia4kDFN7nOonTYYAeAhoTsMCIub92JpoQSABGvyyqP6yk2ZEibhCEpozL5HUlebQgTCh62C/SN6zh4t+zXzBrzaczll2P+0qOWf39JeFm3+WAwz06/wfnzm5xkgVWNec9CwO/1Un6weIINUl41m/RkRFdjztsuG8GAECEsr+F1xpynxdN2iZfMOl8MblSmg2vsEBCwLjs8zgLbDAF3bXpFsabE5OoI5u+xJ/lTuUiDCEGICNnRIYk0CEqCf645QemH4Qz4lBynbpWRM2BcPifght2ouib/9kuLvJ5O+HPLMa9vWS7bMWvBkMu6Ri8Ys54ucdMMELtITMCLZg2APw2u8D3FQ4RqiCUkKqVpXaLhwOWOdSN4fhOeaFqONIApSfbz2uKy3kQQmuJGvSRwf+/rkIlNSzPDBoEYTssSfR2xYXt3rDD684G9poG3EOfLJGLPtVyNb+1WQH03A3YB31gnLiGx2dSxij3vCU41KySkUEtsQiZ2wkHjfqyvb3bU971jQPLuiZ947ueqEaRHg7Ps6IigVEgsNGNsx7RNi5947udW3gwg8vHnftYBoKkOR3398OuFLyKA8zayWiA4P6NCCydGUZPd3vUDEigBh+tL22qt87+L/cJ3TYHqHGcVU/YDF3XgcVjQ4j7z/VlTDsvlfavjrVqT3nQQctQByGEunIMQ0+8lmZ91DgcBH4c95v2Kt1rR4qjFx3/zx1+49tF/srJBn76OQKgSfj8yNCk9F+qLsvMjKdgpvT5axPRkgK0t+ODSusC3nMvXb8iAHiMCDMt0uSqbPKIn6DEk8r4ZkiGWiluypQMiCcixfD1Y4//VLf5T8wEG1nKCmBfzbXZe7/DM9ik6xm1UX9pIGQUTzicRX0hvYMvzKVB6MuGcXaAnY3oyZqFUdVoOI75QbPKwneea6TNHTKQBr5k1hq7fw+eDywQ4CWErrtqfqUtgYxqs2k2WTIdUM0ICMs1B4ITMMWJCQyImmpJIjMFwwSyUnQhn9DivCRdlHUV5yJziYbtIT8a8wg1CCfifi69wzizyiYfm+N01pS8ZRgUjwhYDXgoyJyxgLPOa0CFmVbdJxfAnwVXWdJsWCY0SYA2ZEBLQImaHQeWx4aug3j9kqGPa0qQnQwotXHKO5fPBJRZou++JbUbk1TEUZUk6RBIwkBE5uQNqZVehGm2YmuOuh69cVv8WQ16OUXnwsnvNFXsARKBB5QAPDlTl6gC0P46wy1epJwZ+rfC8kbC2FR2owrlPovB22fCPAck7Jz7x3KdWqoKHH6+tXeMRYVmCsoxJGeqIUEImmlb7tX/OgwYif/0jv7DiuRymdt+HEoLu7kNRuVb4bkZAUAKSogQgtuqaVj5FmF2eV62LWi9E1Llg+wER/3ubJaxxkJjVNfGP3ymm1U7vRzH27VAoeSvXoDc9a3w7LLi3k9id/vl+f5/12EEv6P2eV+9u7NfluN25TB/jMOd00LgXVYp3Wvz9j/7jlSETxqSkZOzglIf66kDCNkNCAkKC0hE8d0l3uYBv0CckZJshhdpK1lVRdhgzxm1iPuHt0Kxkbg3CadshIeKGbAOUnZGUlkasmWGVlJ+UOZrEpGTc0E0CMaxcfIMPLUf8yGMRT5ouD7dCLpwasWYztmTEF81NmsT8evY6GzJgXhNaxJzWFhMpeFS7nNAmfUnZkQkdIs7P+Q3XsGxbJBryqHZZoA1AVibPJ+jieRsjndCQkEgCEmnQlIQb9BxQ0wwpQVqLmA5N5qRNInFFgO7SpIkjiw+Z8Iw9QU5Om4TTOsdTzHPWdvnL9n1VQr4uO9xcbbNIzNPMc852mafNjg65qutcK1b5sr7Gl81lHrFL/Fl9nGfsBV611xiTMtIJIx3T077zApGYpPxefbUQdsedvGt6QVFdI0MdsWm32LA9MgrO2YWSD+MAwkQn9O2QbxSXuWrXiCTkhFlwycJtQId/vAIGZSLik41CbQ1omOo59dEKg1SdJlWXXE00xY+7jTUtvUzccTxJfzpUtaymWsZ2VHv87rxC3q5x1Atzx3HnUL21o7jbGdjlUPV1hIghV3dP+Htmy25Xr/PcifsZP/bcz6z8xHM/t+I7Ex4YeGNRi+N0GYRATDX26Q1bQwnRqptR8shqwKAau6wBjXrnU7F7fj91AY36n3UAUjcvPcyaILfpntxNHCaHmzWBcpTj13/rH7zwVufkx6XrGXHQC2kW3+Igkr2zXn+7ucOjcFEfBMkfA5Dd+MmP/PxKpAFXdd21utWRzwMCYhpkJRm9KXGZpDaq7znElPwPJ9c7wSV8VrVyR/d+GYVa5mmRY4kIy8qTEhFww/Qrd3GfAPcYkIlTkxoyYUJBWxNS8koSuEuTVXp8cS1jYX7Mjs3pxDAch7yvmdDSqBy1ckKtEQHXzDZ9xvTJaGnEvAk5X7p9RwSsy5iXNnOaGtIpLRoNwipj3l8s0yLmrCwhCNsMnZEgCQvSplCnwjXWlIa4d20QlR4ZzuPjum6yoTukZNWoUCJOeSyRBiekS19H/PBT8APFe2kQMiBlW3N2JOXH3xfwoeIh/lLxXjIK/uCNgoeimMfbEam4bTXTzPEcpCS12yE3TJ+z2uSkNkmkQYOIHXVyvIIwYoIgrOo2Uakk5SuGsDsOVVCwYweMNa0SGrd5Kyk5Y3GqYFCTr8Q5w080JdWsVFrbO4ftnz+9idfBybSRYp0X4mM6mQJI1ZHli9KhPtXMPU93q523i/3Aye1Cy/+O4zje6vj4cz+7Uld78l2/WeONAC3TLO+Xony+lKIk7ufeA0ixlYrU/Yi6AlZRqgX6MOLWJA8SjASEhE5ZsFwLGhKVa8juPe27n35N8B3N+mNexnt6PZkFRmZxwmYZre4X08T6OoDZ73gHjTsVfGedy6zXPOgC8GHjrQYfPo6EWeFRijttcA9qA5xFrLzTBXu7i/h+j4m91TfM2yn+q+f/h5UAZxQ4smNOygIihiYxa9qjLYkj9RFVZGDHCAnoaZ+2JHsSVd8hCWpSrC0abBPQFTfqlJNzTTdoiGOXtHEJq5d5va6bLEqHmAaXdY05aaEoW7LrX2HKGf0WMQGGL5gb/FjUJcJwYi6n1w8xAm2NSSXHknLBzmEQ3jA9OiRcNTu0aXDJDvm2qMMwO01CwJqM+X/M6yxoi8gawtrnizG0SXisWOSVYINthrQlYULOIm3G4pLdERMMSWVk2KHpXOMZVb+bNeu8RpoSc06XaBFxkZs8wSmuscH/962IpxoNXioaTCTn+acMw/EiqgU//nCXJC4YfvMRFoOQuYYwKSDRkIQGHdOqqvwL0uaaXecK61wyq2TkdKXNut0qgUpETsHEppw2rsvUlIRMB9WGPw0ERqWvhogg6sYXUs3Y1gFXS06Grzh6x3NwybxV5Wpxc5dAPrXh7rcBT5+DB0T7KdXAXnWbaSPD3aQirUZT/EhYPSm4127HMRA5jrcqPvbsJ1fCslOwy49wtX+REFFDUN4S9XFEoCJx18cQvdAGAKoHAvD7xd9+/oWVlsSMNWOoo6owAOy+b8nrAJewe1J5qhkRARkFI50QS0xB4QojeIkPi+9SA+XjZs+aUAcqvuPiOyjT68V+5oR3GsG6Uw4zSxlrvzGtBxGHUfF6K+KoABA4BiG3xJ0keOvPm378fmyMd8szOSiwuB8yvQ9K3vedEP/k+X+20ldHrP5GcamqjDnuRZ8QJ9PakRY97RPTAHE8kABDJAEdmoxISUgYk9EmJiUno6DHsFrEO9IkLDsaISFtCcpOQECohr6MOaFtbsg2TYlRlA4xi9KmoSFhuQE4M7yYsZ/tJ6NFzPfqWYJwi+88GXLqZI+13gK/M77OUJzaVkcbLGpMh5CXcR4dVpT32tMUKFt5wRNhi88XG8xrTIOIpka8FKzyTHGK97HAF8xNXpENYkJumD4ZOSeZI6PgJlus1qrlTWImmmGxxOq6G0NJ6avjQ9SraW1JWLYtvigXaUuTF/UyAP8bL9EuYjqa8Je5QJZZzp/bYm1tjiwTiiLkrz1q6LQn/J9fDZkPgvLzuinqRBrM0+a8LnCVNfp2iIihI82KkyEizucFryDlulRBfWMqZ6ShNvuMZVQSu930tXtsy25zIpijKMFL5p+vllCiagQCbgUf/vh18y9kd5Z7uno5PYrlkii7h2fila1s+d344xay6x3izrHY8/O7NSTcT1DjOI7jQcXHn/vZlTrAgF3w7UnTUSmEkZcCDQVUSb8fcfLABKBvhzQlLn9umOgIS1Ddh35U6m4mCv7W8ysrHXGmtv5+96p1fvTLn7+IENS6pf65dZUrVaVpWgzsEMRWHRSrTvXOmYz6kbLdUSuLVmDHx/Qo56xRrP3U/Q4Tfn2pH2u/NedefYmq43Cr/cKdwMdbmTsdJfDh4y0BIW93o5f6hXcYecj9fn47kuVBJXYPQkg/bFvxOA4ff2hfwuLHrBImOmFM5pI4gXnpsK0D+jqsuhup5vQZ0ZakMi+sV6S64iRlvWlhRMA6OyzSIaMoVbgsp5lnq9S7WtYWN2UbI0K3dFzvM2ZMxlI5JtXRBmvGEdZPaZeLsl7xSYZM+Nj3rvHZL5zh8dM5GxstNkcwImciOSOU83aOb5gNRpKRkhPgOhwXzTrvs6d5Q0d8u+kQFIZLpkeohpFkJBrxwWabm5OCOY3ZEcfXWKRFiCHRkEIsMQ28mlRWKoZlmmNKsDZkF4AYDGNSWpKUACDgktnkPyoe51vBGmMJ2dIBaWlSKAg/9p9c4pVvXmA8ilndjEhz4T0XhlxfbZLEBSeikOtZyivBWnn/GM6ywIAxL3KZUEKXhGjOCGdIKCIlwLCoKla0EiMYl9KWdb5EPaoRBy2o80UMwo6666Nj2gx0iFNay6mTyqerjDB7g58GHNMAYY9EJnbmue4HLKYlOA9ienjQOIxa4FEMnwBM731HMTF4N4UffarLz/p7UfVWIQfPhYpK0D+xzlh2v0vQJ+SzzPw82Llb93EfjouVYTDk5bhXR5pso7tgoeyytCRhbMfE4saC3Zq62xERDKEYJpoCVCAMdgUp9iT6tY4HFNXvbJd34mK6W+JlfOsx3Rm5G6AwDXTq73BYkvusuN24vLcn+Nizn1y5H+91P+OorjNH5zd0BEOn/qvHrM3vXjoL9ePu14m5l2Pfb7WH+rGPYzd+9dO/+IKb7TXE0gBgpBNEDGNN2bA9hupIuJ7fAG5xLNQy0DHrus2EnAkpkQSVZG19wW4SM2BCh4ScggXaDJnQJHau6hqySBvBkJEzIq3c2q9Jj1XZYVPGrLLNgDGT8tiNsi7x3uIkSWvIN0cTOu2MIFC20oIn7CJnbZdT2qEvKUn5fIvSKI8/r46jklKQWhiT09WYFhEtbWAAI/CibtLQgJZGdDThrO0SELAhgzLBdjLGWcmn6euQrrToSotUc4Y6piFOVaYtTQIMc9KmKQkjnXDWzvNMMMeCOrf5jjRpk1BgiQm4dvkM3U7KpasdLrspLqIop9PKyTLDk8vKggl5WJcIMDymyyxqsxqfCAkqnoaPQi1Wiyp5zzSt1L3Adbxgb/WwTlSvGwX6DlooYXXNtKVJUxISk1S/9+mYxfeodzjqG/u0ctX05l91T6bCe4VMV/38zLu7nos9x7rXmLW+HlWOyPSGX/+3//tRIIUehwsvDuFjmlSuZTez+rm4Duf0te27D8Ae4vZ+8S8+/csv3AsA+fmP/tLK337+hRWASTnOacvVJ9O65O6uGl+DyHU1cNxCL7+t6takqIQNAQaR3fstK/lq1ecrmX1eIa8St8CNe1l0z+/AcxZ96IzfH+xyOg46klZP9KeT/unObv097iZuJw5Uv5d/7bd+5YX9gNSv/davvPBrv/UrL7yZ69ZRXmeOx7EeYByW33FQ75HDnsODindbJ+Xvf/Qfr3y1eI0PmSfpyYjvKs5zTQb8IS/TkIi+HdIxLRKTkGvOuDRrsygDOyApk+NYXIXfcxucIolzrC6wnJUlhqQ0iauNw5EGhYSIHcYkRIxJaWuDHRnh5B8zTmuTU3S4Yfp0NSbRiGtmm5SURZ0jk4IlbdOXlFRyx3EQw5YMaNLghLZZ1jY/uLjAF/9kngsRXLwmvLpVEIrw194T8e9eS3hJeySEDCQlLLsnLWL6jFjWFic0oU/Gl/MeUTnCIwhNQtCEXx9f5gN6kkuyg/dTeVhafJmrLGuHVdmhQVjJB4O73qoNUposa5c+Y1JyIgJiaZCS8R36CJfNJu+18ywlhh8Pz/GZXocrZpuejPi+4mHGFHzxlYQ/+/SAk4spvWEMwPZ2wmAU8siFbUbXuhTAd3OC2Aasy4gCS4iXV57Qx2na+7Er2DteJZgKQIQl6IylwUgne7oY+3Us6vPc87T4lr1CKCGF3krqnj7GNEG9eqyWdFgxM8cG7lSJ3G+GOy/lg/eTyDzoGMR+XeCjCDimow4y7vSc43hrYpr47Tt4FVeiWnN2R5ECgkoe269DkUQ1LpWlQYRXvGrSqHSi7nf8redXVgBakqBlQcRIVBU5LJYRE1LNiKVRjoZliDj+YEOikt+2ex+nZHjp+AJLzi5h3vPR/NiYd0o3SmVKOk1aDwgqMrvzFNqbbtY7sbA76lkVZabI6nuKHmWhz4d/jRsFzfc8ZqdAiu/CwP5k+Fnr4R7PkQPmPgFO+l7EHGjM7u2yxt3POAYhB4g7jUtNb5C36xTM4pLMqu4dloh+HHeOjz37yRU4vKP7Tzz3cysW5bHgHHOmy6rs4CUI182Q6T1G1ZJIzBDFas6W3SaSRpWAukq8cfwNDasEHVz1aVLOGAcYusQViT3AEJdSrwGGOVqsSZ8T6rgRSzhJ3kmZlCcackKbtIqQS0GPHRnTIGJDBnRpMmTCOVliyIQAw7J2+B5OcbYV0BvDMIf3n7X8ozdW2ZQRHyhO8d1EjK1lbHLO2DY9GdPQkIaEjEk5xQLrMmJeY5ZJ+EzwKk1iztguBZYrZpv3FIsoyjUZ4EmOS9pkQ1OWtMNYcprERASoKF2arJVmjUMmJDR4TJeJ1PA+e4KvmyYZBQPGnGSeD8kS36lL9MgZZcoHHh/yJAG/+5UzpNbyse8acWO1zcob3+LPmdNs9yOiAG4MlOV5w0ZfOD2OGIwNc6FyOU1pEfENc5OQgCt2FaCs+mcOFJTfiVeYyTUvrxBbmxN3m+ycaZPZoqy8FrXxLFdJ9F2UamRBLU0T4/xmhpUkc90w0CcJfmOdNZZVaIEVr1az/wZfv46nf14fMZh+fn304V60+uH2MuVHeZM+BhdHP37suZ9ZqSetvmKf15SjkjJxn6jzHvIJc8e08LLofR0y0ZRIQsbl6FJGQVR6FXkfJ1V3f378uZ9dmTWSdZj4m8//dysdaRKXACfXXYETACNunfFrTkdaRBK4LjtO5ATKzrVE7nkqRCWPY6IT2qaNte7ziEip6GerLohbtQpEd40MK6lvLRDxPiSl2lf1ultHr2bbq1K+9+764bs100aE06aKRgJMrfM8fbxZ/57VGbYzOifF1Ot87Acu7gQ6bqeI+m6Kt2Qc6yjzQQ5jgDXdlttPlq1+jDuR22cd5zBx0It5eozsnX4TeAAy/fc7Rb1idtWucUK6vFK8wZb2ecVs8nV7mVxzZ5qHW4SHOqJX6r77xbkjLSxKLA3mpI3FlsZuIV53pCMtCpxx3UDH9HRAjyEZBdsMmdcWO4wYk5Vtc7dhdmjg5FoLtmTEUDK6GvOKWeOLwRXmcGNhS9pmUV3CPialq02W1J1XV5s8bueZC92S8K/Hq6xnu/K+p2yHEOHyaoNXZYdYA141mwwlI5OChAandY7Hi0VaRLxutsiwNAhpaYObpk9LI4wKl0yPeY2ZiBuzamnEhox41fQIy2T9hLaINOAhXaKlEV0SWiWpE9wm+jVznctmh+/jLH+mOMkzXOCZ4hQDa4lE+O3gJX45/zyqQpqGfPR7evzI0wVXrnVZmJvwPcUFTp69ThAojRBWs4xWUmAErtxo8uX1jD9NtxmSkxDQLkffWtKkY1oEEhCKU87yc9e+ghrUNkc/ZuXVrzzB3T93l1juRym0em1d8nOg4xJMuITAvx52TcH85l+vXHoA4jfP6WRglpzl7f5df359Q5618d+vOOrgA44ByNslvGRsgJOn9RwPx18wNCSiKU5Io0FEq0b4jmmQSKPyLFKUkY6rY0cEVafBv4eI896YJX190PjrH/mFlZ/6yN9dAUg1J5EGbUlYlA6BGNcJQRjoGAdRHMDoSBOrjj8WSUhbWoDrXqiWtHgxlVmolADGx7S6F+zyOfxIl//d1DsXlWdIKd/tP7s/Xt10t+5rVD1vqoBRaFGNadWjAiTlPpuVYHD6/fzx65/Br5X18/B8oOnnhhLe0nm525GuWXFQW4h3YhxzQsq424tgGlxMb5TT/iH78Tzu1wV40OM8CJL6QcDlYQDAg4rpTsh+5/SxZz+54hfWAgcu3rCrxBIz1gmft9+oRm3G5UaUklULc7URiakkWqOSFO1BS1oSraVMIg1SeYc0JKSvI9dx0W1GkrGknfJ8LCkZYzJ2mLDDmFhD2tpwhHMZOEI8lpsyZMCYlkactR1aNBiToSjjkjT9kJ1DELZzSxLCJbPBK7LN5bWAJ+0ir5k1+uQEBr7DLPAkczxuF2lqyJzGhJjKBLGpIREBy6bBKe1yQ3rEGpKJJZOCh+w8BmFMxlBShuJd5A19STlv5xiR84idZ0hGX1KyshskGPo64kUuk5LxVd4AYJuMH26d4i+eaNMQ4R/qHzNHizlpoypcuhEThgXDcYi1sLGZ8Fceibj4yiOohQun3Pf46tWY7YkymAgNEc7Q5DWzRYuwBGIBRtz9PC8dB0Ao6Bi3ubckKTfLvSNQfmPLyenZvutKeMUpdjfh+p+VSaAE5FrgVdd8pyQvCaU+vBpPXWq3GpGQW5OJetwPAuWszsldH2uqMHI/FQiP490Zn3juUyv1opL3x/B/DyUklgbNUqLcYmmW9/Oi6RJgyoTasFPei6GEVZfaj2SBW/sb5XiUM+Xc689xkPjJj/z8iv+/wElvG4ShjhCEtFQxTDVnjEu+HVdw7OTWxcn/Nkq/pEILYolcJ0ciAtn1APHiGeASeV+o8FLCfj3zxQ1f9KiHlKOd9fCFkTo3xJsd+tf4jkbVPWH22mFnrAfg+ZRF5b9Sj2nAUy9ieSAyLcQR1MbCPACpf8b6+dxPIFK9x7sMjByPY90mDkLAvt2oVv0Y+0V99OpBXXx31NS+j+/7o8/+9Ioge1Uipt6/nvQfdjTqbuJ27+d/Nj2qVX+Nb3WrWoaM6EiLvjpDvQK3EWWaE0A1AxtJoyIK+urSWMecDpbZ0QFNSejrsNStgjlps60DmlIfv3IVuRYxbUnYYUQmLvHcYcQC0JbKnwAAIABJREFUbdeSF0uzNDv0Nlg9GZEQcVrnuGQ2Scojvma2ADipXa5Jj3lavN+eIUD4UnCDAEPUv8C8tnjVrCMj4c8nS1yZLLNuRlzcbnOhE3DeGE4vwm++toAA/cA5ZicakmFpaURkBBRiQpbVOaRvBENSCh6RNn1NiQjoS0pPxtUG0pMxr3OTU9JmSwZMyEmInDO6RmzKkIfsIp/Tr/OQOcXn9Dq9YMQPN+c4uZjyh+uWD8qjPFrM0Sfnc19PeOZ8zsVLJ3jiyat89asXWN02nF1y32sYKsunN3n6yjku9gtiEYoRjNR1VF6TVTJjCRAWtcUmfSZMOCNLRBIwYsJDuswN6RERsIYnpeeYqY061Yyc3Rnq+gZZryrWx0UMwkjHDpSUHY2A3eqfH0WYPsb0cWfFLAWXQ3E39nnO/dTFfzsAj+MuyNEPP1LjuVpV9xJBxP3ZkniPn5NP6udoMRGnPtUkZpshWnYMvJ9Gplm1dvt7LquNR9Y7DPvFT37k51caRIx0sidxd51Pt2oAdIjp64hV6VFQOOl3Uhaly6AsiLUlQbEM1Y3bqgh9O6zc0IGqk+NJ5tTWFC8ZXP89eclii2K0XmDZVdGrJ+Y+gfcgps4bqXsIQU0CuQQG9VEsH95v6HY5ix9tde/j19Jdl3hfoCnK7y7TfF/iul+f6wWf+ujrvY7X7Rf3W/znR5/96ZWjuka9qSDkKI9h3S5myfDe7iI57IXzoH03ps/5vh57iizmo57EH1Vkf5iuTD2p8hXpDNfSHjHe02oPxXUChmVrWLG0pUVfB66jYkecCpYocCZ2YdkdEYSYBts6IJSQMWPaJKzSI9Wcx1miL26DybUgk4I2MQWWMzrHa7JGgOGszjOvTdakz7JtsRNMyMhpacia2aFNwqa4zzBk4hSzzIgc1zr/qt3mNB0uyxpvmIhOY4kf4CS/k17hOy/Ay9eUTkMYTwIeC5rkqnyBMY/rIhNxi/+InC/bLeZJ6EvKpoz4Dj3Fmg65anaYsw2elnlCEb5p+9wM+oDrhizbFt2gyTfkJgPGJbBSlrXD861zfKk/5PGoyXYxIVLDo3aeDUn4ylpBd6vB952E7yPh/OkRL70+x+sDZ7RoFTbXFnnkwjaf/Q9zJDshD3dSVOHqlWUCA93AIAJJIDRzwyvaZ6wpb8h62bWSqtvVY0ChlpYkRBgiAro02WBnTxXNS2P6TbcOPPaLOhCZ6KSaXZ/VsXDEz5okZW30aj8wcjtux36PPeiYLsi8HcAHHAOQox6feO5TK0Dpc1Gq1JUjWN7XwpHSQzbsNk+Zh0glZ4lOed9a59tU8vp8t8MnxynO6M8rSRVYAnV7Y0MiJmU3INecjz/3syuRuPHNWBpMNN2T3PruxZ4xIZxkrieZJxJzRdereyURV4Aaa1p1NCJx5YtMCwY6JC5HWaUmvdsouyIhIVnt/KUcSfOeRwGus+t/Xo17Vqaqfvyp2HPOfq3LNCWSBlb1loKIX6v8Mf34Wv1pfg2dHiP1cYuIRV2xj9KnRXc7OPVxV89JrD+mtfP0hcV6oce9p70rP5fbxYP0QTrKa9RxJ+SAMYvb4RPHg3Yzpons08d+EDE9Dnbfj3+PycrdksUPE7/2W7/ywnTHYzo8mJr181nz7/XX+bGrQktiHOJkectqT6oZhfYJxNCzfWJp0NcRISFu0MgTkpUt3cFgmOBnW53ue0HBG7JOlyYRAVYabGqf77VPMJacN8oOxzwtWtpgVI44XTU7nLZdLpp11syIh3WJfqlo5ZP7seSct12sUXZkwro4HwqrykQykghGGZy3c7x4VXh0ASYZrO8Y3rOkLHYLXnxtmXPl7PRSGPCviissaQtROGu7XDJb3CxHxrrEfCVY5UfMw9zMMpZJWFP32oiAy2abpBy9KkquS0TAGdshK6BFyLeyEd9vzvL7XKNDSKYNfjP4On87/DZeWi/4C0/kpGnA5lhZ0wn/dn3CBZPwza0m3/9kzheyHrIOz2zN8d5lIc2Fq323+Y8LS6GG6zrhBAlPc56v6zUaZVUv16Jyql+QNhk56zIgx3JVN4gImZBWG5qRAFsqyMDuJl6Zh1GveO7tTOxK9jrlHq/iM2vsyvNJZnl+zLqGd5OJvWTP6lwecHFkOo6lv4/jQUTl2q2744lFKQFbqc8RMm86pWfTmAXa1T3aKKVlA1qs0WPMrqqUBwZj3fXdKCj2eAj5rotPsqtz4FYZWqe6l+7hlO2R/cXJggeYqhjSJC49kpyaYl52ji3KhJRcc9rSYqjjirPi19b6+9dVrgIMlJwPqwUijozvOzRGfQc2r87Mn99uFygnqHmG1MHWLHneOo9D1dakg7VaB6fXp3pMd2+9Mat3s4/EcScDhFxL0YDaMaZHtfx5FnjVrd1Cz/3ogOyXBx5UCfDtVKi5XbzrQcjtvsj73fGYrvJNE9cPM7p12Pd8EHGLbN09zoE/yKgDkVlxp/P21ZpZsoEhzschImRsR3RNx1XAdLJnkW9Li7xWzQkwpTlhwYZuM9a0WvDHGtCRFqE4E75Ci4q4fkEXuSY9evS5FPRIyfiWvUpDIs6xAEBTIxZp0yWuVLHWZMCjdpHrss0p7RIRsCF9xmS8HKyxoC6x3pIhfdy5d0jICogCeNy0WGgYNgbwymjCk62YGztC0gj4we4SX94ecz6KefpcwWsXT9AjxeKAzSnbISFgTQpGZMxrwlZeMCRnXSY8ZRe5IgPmNWaemCvGEdZOldXINRnw+eAS2fgCj0qbPzZvMK8tIgzfvhADMb3eaR47lbNzxdDbcRv354sNvhpco0mD67rA++0i2/2IZVX6ZPwel3n5xiI/dKrLVl7wzGJEbxzw+ijl8bDJ6ZZhfuc0N4wTGjinC+zIhJO2U93/r5t1FGWo42oD87PkXsnMiNPmH9gBUkrszppXrl9d/lheSlc1q6qvsyqP/t93IpVXBPWaC/rMa/5NBCCz3vuobbD1auLbtav/bgvPAakq7f5a9+NBZaLblIYbWZWQGKf0V2AZMuEpPYviPDW2ZIDjdhXkJU9joimhhG70SMHiiOIhblTT8UscAGqbVqVcBezx4TDl5R5KiGIrUnwkAUOdYMtKvO+eL9Dmum4RlIn2uDRtbRAx1oyGhAyt47oNdOiKGBKgNgWxVYd12uUdqNYtpagMWD1o8V0R75y+R0ijLLhMh3+uN0QMCECoPIVsOWLqvx8v+lGorRVi9g9FYUYuUh/Ncl/77rmC6zJ7Q0XF8Xam920vYMCMz/VWxlFbH+823vXE9Hup9s2S1j3se+93rIO8393E/bhw9xvjuBeH0DeDsH433RZPtrvd7Lsnp6eaImLo67Cao/Xzo2GpguWrb67K4urZgjC0I+akDbiqnKuq5bxqr9EkZtk4cBFgWJM+67pTzQFf1y0UyylZ4Ct6kdPa4orZYkGb1Xm+p3BSvJdNjyYNhpIRYVjSDm/IJqv0KLC0tVFV0xpEJBryJ1tjzi0VXOgEPHk+ZZArjzdjvjmcYAR+98qIx85OOBM2SAKh1w85G8RkYilEmZDTIODlYL0abxiS8QW5SY6yIUMiDPMa84rZ5BQJLSKGknHGtnm/XWRZ23zQXuCsNrmiI56ypzDAU3aJGwN3TX8bC/zBRZe4bw4Mn7uWk2hIg5Bl7fBXm6f48JmQyxuGJ5OEG2ZARsGHm3MszWcshAFWYT6BHhknEsPqUBHgzxeP8oie4Lv1JD/Iwzypc5zWFl81V9jUPhkF89LmjCzQFDdmF0tMS5JqQw3K60BV94zv1Suys8wC6+GBg+cpzfIFqYcfwahfy/X3uVPh4EFvdLcrAB2luN04w48++9Mrx6Dk6EZdJa7uXSEiJaXcda7HpLSJaWlMm7LboDmLNiHRkEgNC7g1OiMnJKiAi0vYlVhiIglZki5jUuojPKGEdKTlOhHle/tkWDBEElVgp0FELDFz0nLJOwGCVHK7OzpgCw+I8hKYuC5pTkEiEX0dleNUQQkCDH073FXlU1tJEIcSVkphRgInj44S4tarXHM3slaaHnqOhOeXeJL/tFhGfYRKZLdrK7KX8F3xNNRWHRnfrfId4EN/7zVFMr/G1k0bC7XUi0B+jfbfB+yqm/nXR4SHFhi40zneIh38AAowR3l9eteDkNvFQYnp9Zil6HKnOGob7p1iuvtR/9MDkfuhtnMUoi5HWq8c70fmBT+jn1cJYkFRzhOb6hghAQ0JK5ldI0HVJcnJHYG5XPjnpOX8P9QNbz1RLLuxIZ044FC6dn938RAW5eVgHYPwNXONS7LhnivOO2Rek+q8TtsObRrM0WKBNpsyYlGbpCUAadJgICl/GFwmy4TTizmrmxGqMMiUAOEroyExAS9ejGkFhsvjlH+9vsNLxTaPlkpep6378+FinrO2w6I26cmwVO5yAO4/mDUsSpsGL5tN14UpAdGAnI42uKAtrpRclgva4gP2JB/sJiSBsDpQVjXl69KjVxRcHVg+F1zl5cDxSq7KFheHGV+4kXN5lNHPHPjbwb3/H78a8viiUFgQgWVp8HuDNS7lY5rG8Hgj4T12ns+aq1wtxvyJuckfBZcZa0pDwko1Z16btIlJaDiSPyGFFtXm5qUwvTHatDzv9LUEVKo49WvNP79Sk2HXWGwP6ODW63RW7HfPPuhuyFHli9VjFgC5nSP6cbz18YnnPrXyiec+tVKXua6I0+Iq2w2iapy2JQltEhIanNQ2GziO2llZYigZmzKmQFnWNoKQSIOWJOU9aIglJiRkUToVMV3VEhK6+1d2uwAwWxEvFrdmNCRyEuAYMgp62icsz9eq0pCQmEZ5bJesJ9JgWeZoSozBjf2Cq/x7o0XYJYr734vv4Li1ahcUZbXug+/MeJUv2B2dyjQrPVbyPaNm027xHkh4QAgOBO1Z52pAw6+JFTBCbgtE9ow11fbheofHyRLv5ZZ4LycfObufw+/Nnjvi1dPC+zxANL3e364z/XZYLw8bb0mmeNR+kVr7b9bj9Zj29jgMQLmX590voDJ9/vczZjmY7vfzWfGxZz+54v+/ryd2yDgIgJqVzNVnSv2//WM7ts9Ah248yfaZk7arMqGMdUKu+a63iLrryieRV+0aA8YVl+TfyFfp65CCgk3tOyI7IZfLkaGv28vcVKfUNGDMtky4ZnaYLz1CFrVJoiGbZsy35CYtbVQqLq+YNZrEdGkSYmhrg4iQ//7mN/mnl7a4tAUTaznXFZ5ZaPDhuRYLEvLKeExoKAew4CnTZT4InIQvDSI1ZGJZpFGaKLaJCVk1fdZlwEBSXgpWGZDyuqwTq9twTomTy3y/zHGNEX1JuWkGbJHRI+XFnQm/l17jf7eXWJMR57VNxxiG1pFJr+smuRbM0+K3zTe4pmPGFFwqxsQa8LAu8X9NbnKtSHnysU3asXJqIePDZwJeNJf5bfMirxUjRoXymeBVLukqqzKmq65SajCMNSVX5+XyEm+wzZCWJMxJi6bEtKVZznEHtKVZjWD4OenbdTM8yJjuiNSBiJ95h915av+cvKbIM5P3UXvsQY9SHnSk9ajtD/tVEX/9t/7BC4IcA5AjGvWRJ1+ptygRTpo2K/l6BkNLYjokbDEgxBBi6NLkrJ1jKBmr4iTSvaLgPG26NEnKTsCCtDkhXbo0OSXzbOoOIoao9BFKSoAxIXX+I+X7x9JAMM5UsKyyn5ZFQkIa4jgYBsNIJw7glHK7iTimiid7Z+X6k2tBTl6BDd+pcF0I9zrf/YnE+aB4sY2s5Jo53oQrbGSalc928sT19aQip5eARmS30+C7GLAXXPjHfDdq1vcVSlh1Z6wWTOyEtCYbfJDwsr3+nLX0K/Ed5Ix8D/jw4QFIKCGJSRxYlWgP+KhzdO4lBLllTa4ki6cAx5tlofBWxDujXH2PcZgv+G7IQnUyu/+v3mU5CBh4IMpWhzzmLMfRWQkN3Hne/E7xVgKSu0nGRMweQ6U6URhcctiSZuUnsmo3GeqYdbtJQ6I9iaSv2oAjahdY1myPnvbZstuVOouXSvQmeF/hUrW5zEub7y4e4iTz1Tm2aHBDtumqcyO/Jj1iQuY1ocDSUefC29JGVYV7QzZZZ9upPsmIq/mEdZvxG1urFBYu9wuWooDzUcx8E67LiISAhShgaC0TKWgRMJCMCQVXZIhBeNQusGxbvKdY4pR2WWOHh+w8fRnTosG6jLgkG/Q040IY80es8lqwyTnbKSt2MJScS7JDRsGcxnTUbak3i4yhFg5ESZNADF2NAVg3Iy5Ln3kiTmiTMTljMsYUrK93+KPVlCTOeX1dmKOFwbBAxFbx/7P35jGy5dd93+f87lZrVy+v+22zkcOdoijZSiIpguU4QRIhRgDOkKAICaFgwYb0TyhQlglJgdsjBJYFxYLsPwwnBgwEcCAZlEk4UBDEkQVZQrSboriJy+wz77157/VW+626y8kfv9/v9u16Vd39Zt5GimcweN1VXcutuvf8fuec75JXhVpfUn4w2uLt5RZtsSaKXp0mIiTVjISQuYNJ7EiPgpKmJOQOclA/P46xyyenIf73laowS87TZX973k7bqrgfDZD6bWd5LT0KsWoBfxAS49+ONx8Gy10o1crNlmo9fWJszo0IaIg1c32yWCfA8FVzky5N9hhyw9h86SeZiZtyJNiNaIOYrrSYkdPTFokGTjyixHMIrUdUYm/XY96WUloSOoY1ZyLoi4KGI4HHEpFTEBFyoENXGNjNdYolsHvDW59PQkJ2ZJ2QgKYjsYeENB1xPRAL7crdtD6tqTha6FdYOauHElp+iJ9G1Io5OJYH987oS4sLCUicA71f1zwcyz+nF9ywpo7GTX0KvD+K/8x8rGoYK7p0T7LKaNDDpf00R5AKruanVqolqfsuCspzSS3fj1jcOz5qzZo3Gw+UmG4keOSIy2+G6L2qo7e4oK5SzroXi+yDVq6BO2FYyzggy2BK9b9ffPx54kH7isDq97eIpff/BhJU2z8/wfA68iIWdjMuJ05lI2ei06ogGZajamTsR/YiQlqmVXI3ErAhXfbE0vS80d1UU/uzwLvlMl/VVwlRbusRe2aTq+Uanzev8Zhu0dCQhsQMZcZAZkSEJEQ0CXlHuU2BMpGMDjETndNjnZsyIEXYLFtkUvBnwS0aGtKXlM+OlJ2yxSjL+N5onXkOl7TJe9sNXhjP6ZqAy2WTQ+a0NaJvZkyY86T2iBD2Zc6aKw6u6AaCcEE7bJZNXggsjGxfZgzyjL3AEuj/PLhBVxv8SXDIE+Ume2bMHkPaJGSm4DYhV8oO18yQa7pPTzqomyR0SDAqXNAmDWN4d9Diz8sbjJgRmYBre5d4exMOByVpqVzUNW5xxG8FL/GDxVOVIICq8h/nAyYmq2Q83+CQTDPXTyyJxU65Agx9pqQ6pyMho3JcnUt+ylHIcaFQJ6cvXi/LriEfy4zDFhdgf9vdXpOrVFuWKf0ty0vn+Ztvx7fjXobvZs81o+7ubRBmmpFTWHK3WbPqhcCXgzeYMmOHdUI1tCSmoKAvKQGGmzLAuBy1L2M7uSCofIyuywFrtHhDD91G104PCs1IiKsN91wz2+l3E+9YIro0GTBhTsaASTVdmOmcJjZHzsnoYOGyESFNYprEvKRv0JEmAQEjplyUDY7U5pmJTq1IiiOoW2incZt1JRfLvzhWphIiRwj3Iim4v7X3n5SqzTWvigkRqZS3SgfPEpRM5wTOs8Q3VQKXkytYmgSUrogpxN7jpxZLURVLcsyqWIR8+WKrdCtDQYFRC9vyUsUlpjr+DFwRa6rm3E98eHe3Jx1ulgeANXi8J4pZC/l32Z5xGTrnvHvKR9Ev5IFOQlrSdNrNj0an67SiYFXX7rSoL86nVatnjdlWTVLu5r3cz1hWaKyCdazqut5xsZ2zO/ugJiNnFUinHaMlAmZ33FfhXKtzw5xIwl6dw2C12+udo1gitujeiW92yT+WiO8stgnc6D4m4g/16wxkxkRTnucG29qgozEzcuuqi5VivOBuj1yS7Tsvkr6kdNVKAicE3JQBewzZ0CZKyaFMGcicocz4i/mEw1QZkGEEnmzG3C4zLoYxLwSHXMV2Cdc0IUJ4X7PFRDLeMCPeXvZICEjFHusGCd9TXKZNwoyCQpQRM450zIgZfZkSYNg3Ft7WIGLkuB1Pll1eMX1eln1i160sKDmQMe8qtsnFdihvljOO8oIPFJeYaMpm2SQOITRw+eKI23nG0+U6ESE39ZAC5T/Ri7y/vMxXeJ3fNd9gRs4hY14sr1vCKLYYzTXnoBzQL0fcKg64Wewx07nDdh/jjOveIf6cqjqMdwEJ9E7BdW+R0xSy6re9WYL6qvx2lkT5WbntUVkbvh3fvOF9QYAKSiMIpRZOtXCOqnUSb5OwLm22ZY0Uq9qUMieTgp4T9uhLSoSdCPS0xbo2ySloaWz/JWZDrXHsdd13AiTWvi+qkZjnWOiXzfF2ShuLNaTtMyGUgJY0mGvOTOcI4oQtTJVfbI6xxc2UOSNSQkI6NLldHtIkcUIZ+QklRru+qMsVDu5T8/1oONhrphmj0gqrzHXOuByTllMLh9KCXDPqKmN+guDXKQslLiuCe+KmTPZ47GMiQsvPqAu1uO+moKi4kD68jHjdtBBO5kip/be0SbyQ26LK0T6ouB+RO1c8H9M+r+XoeJiZP6cMViLZPof9Tv/2s//T0r3Jx5755Ln3LHfDp32UJ8d3Ew9sEvLjz/787uKo7FGO8+CXV3UFl01BTotVPI1FGd/zvu97XaCsUsNanIYs+/m8ilmrOrLLNkc//KGf2n0UYBDLjqvydFi4z28OPSE54lilo8TQcBrm6rxGvJFV/fHP63XAygp6zfNIIuYO8/pp+Vz1OnMyFOVLvEqhBY+ZC9yWlIlkPFb2eJFbXJKYdxSbjMj4UvAGW9rmiDFXdJ25gzgdOuL4y+YQgIva44YZcsSYJgk7tLlcdlgjYlooHSKaEQjwqgxp5QHvZouWCXi6XCNDSSmY5EpKzkhS/jOzw1ewXiV7MuUFc0RKTkcbpJLxuhyySYdEQoZMKSgZ65Tv5Am+JjfYokuLhFsyJDIBb0ifgY5Zlw5zMqY6s47zrmAakPBCcMAHi4u8ZgaEEvBycMQf7zf4zrWEySRmRskNGTt8s4WUBcBQZsw144Jp81SxzmEwoWvadGgykimFFtV3V7rzwS/Uc83o1CQ6c39e1xbXsxzIT0w/VvA4VsEklxXOy+6D1XlkWX46K+41tvlhxKPWPfx23Bn/wzN/b7cy3BNvfmeqQsAgdKVdGfpl5LyhB4SEXJUt1mmzz4DIeYSkknNZe9yQvoW5OoWqI5mypV0KKYmxnI+Z5I5NYDvqM51VhPCCwm1chbGmNLDKhy1pUKqyTrvqzCtKToEgTJlZOK6WXJKtiiyeOdivVd9SGhKzpwNarqiZ6Zzc8ToKhKmmeBEMb4wLtlnmO/uZepND+3hfENTXcX+9R4TERMyYo6pVviuwa5dXyPLqwwEBkYMy+SZboUVFcvchbk2Ma0R3D1Next+w3/MpypV+H1aHiVOcgMt5RTJfdCSSMNFpNUkTVyS1nAfWTOckWEGZsab0pMOYKQkxJSU/8sxP73oDxi2zwZHjeZ7XD22lcMhCcfUoSpi/2Xggk5CPP/up3cKNvR6FuFcL4VlKWHdLAj8N5nDeuF+L/GmuymbhQvcKPf7f88jrLW6uTnvdNxsnXNzfJFdlcerjj32Vjvni+/dEYrAQGi//5/XcpzpDKZloWiVgIwGZ28T6BdSPlDMHN8hcR71wi4/H+frO2w8WT3DdDCxZXea8m8uUKK+YPk0Cruo6OUoL17mSnH2ZVMZc3twwdOfXOm16tGhoyAvmkPUg5KvFkFdlyB8eTikU3qk92sbwwWab7SSgYwKGZGxKzGvZjG1t80S5QSTCe8ot3jBjpmI7bZlYR/iXZI8J1lX4HeU2c81JCCuX4EtscMCII8Z0afK6HBIS4OWQU7e4zsh51RyQUXDDjLihB0zIXTFQcIsjhmREAUzTkIsm5g/l+Qob/Q3p83WGPFWuEUnIVGfsyZQODbZY47L2CAmJHKHSnmNip1uE1c+jclKdB/48XPSeOS1WTTrqOOdlsex8P+3aOs/Ud9XfrMp1q3hwi1OVh6kC862qQPOXJTwnwXa4A+ehUdCSBttmgx7OFFUCuk44Ykd67DOo4D8TZmxqyxGZC1IyNt2ko5CSBhE9Nz1ukXDEmADDRGeO++FFIuw1mYiFUwUYNqRroaFOqtfKeecMmTJx8FywBVJHmszd5vg2AxR1UK6Q0E02YiJmmjFzvMCZc033fiH2dY838wUlkTNf9PBf8NwzuzoZdw1YcnhEROggxsfXxZxjh/Xc5dFSi2oK4o/XT+qBihBfTU04aZZYzwmBEw8ICKrnW1xjI0feX4zF67c+QYklqngnYKfkngeSSOwKk7h6Hs+f6Uiryg22YWg/+3xh4uTDfr5lVXyuivPCyBaP760IIj1qBPX7Ngn5ux/5R7sjneIdL/NKN+f+x3mqxLdaSZ7FDVn82f++6rH1+06Lh4GvPq2DWg9ZSBZ1idqIkEzy6rGrurWndTbulRni3T7Psr+vH2t94rOodlS/rZ6MRISyJnnok3PgOl+B2FEwUHE+PEbVa92LSDUJ8YWIUtKSZtXNyl036/PmNoKwx5AWm+zJiDk5LWK+FOzxzmITI0KI7fi1NGImNrnOyThk7Mon5Ygxj+smRzLlyKQ8XW7wfwWvEAUBQ6a8xB4703exFYb8SXHAX5lv8moxZYOYG2bE12Sf/7S4hGKNDAtVuoREGtAhZkbBllqipm+TBGoYyoyONFmnzZN63Bn0XcrbDOjR5LYOqgXDQw/mmjkXc6EtDWKX+l6RfVAY6ZSvBLd5st/k1tgu2gkxQx2Ta84Xgtf5b4p3c9HEfmnlT+VFNugy0impzK3qDg1SnTNx31NBaRf9WoWQAAAgAElEQVQqh532UxB/3txt+C7bWX4iy87Verwp8YUVzZGzctCy+88iV543N696L+dt6JyVh/3vj8r09dtxeoQSummz7V7PmGPFdZVtXWMoUxQ7TQgk4CpbGBW6NO0mX3IKSo5kym0d0KZBlyaHDgK6x5AGITfNgMd0Aysr3uCAEV0vpS5WkKItTUY6sQp5JEyxCk9tJ+0bEzHQCUaE2E0oBGuEWFI6pasC46YY/pyca87TXOLrXGOiU0vsFuuEXmjhJHXthGSkE7xzeyyB5ZzomDVpI86Q0TbFHDHbrSmxRKhqRcbOakZ9dsJkIcOFWO5I3XvDNsOO368nuvuiqP484lYVP3UAK7jiC4I5iuqdJqw2Bx7L9vpisx4Wcmaq86CgIJGEQgva0iQjr9AGLSdzP1XL/1GxeduoUAgMdUymOW1pMijHjvcXMS1TIvd5ezHohrFFzAk+sNsjLMLJT0yJz5hyL+ak0/agi49bvO1R4obc8yLk737kH+32yxFbZr3CtPsOgx/p3e+4m6nD/XqN0+5ftvCdB1u9qqi5X7EIi/Kbp8WL46xOrmpJIceJZKkfwQJpdtXz3Iu42+c5bQoEJ0nDZU2jfLEY87d786h6UWIT+7H7rN+02q5RQOZceQuHZbZj+QxPbvadHa1t7jwBsgBelFuMNWVNWnyRV9mgy3vKi0QEfFVuMpKMvqTYcbtdlnraYE7O6zKh4fpYBmHmOl+xhrwgt5kZe33PnFdGi4RrZcqOJvQ0IRJhT6bcYmolLrXFugn5AxlwtVzjQDP+LLjFxbLDzBFB+8Al7djXVKHlHN836RCpoaEhqWQMmbLDOgUl+3qLlsSVDOVAJ4ROCMArisXOc2OTDq/7yZBbQAIMvSig14Br/ZzM2OIhch25lIJUlYaxHdAJhj3tV5yMNWlxqMOavrxU36VvxtTPi9Ogim/mXPePCSUiX4AvvOWie0U+Wnbfab8v4phPK07O00haddt5IWCLC/uy+79VYA/fyvHxZz+1693Pfce/cFPIyMGQ9mXIvg4qU9g2CQ0NaWrEa+aQnrbo0eKAkYMFRQyZ0tUmN6VfbdjHMnfXf0aoholY2NScnAlzGkQW/qljWtIg14KxzJjqjEgC2tgN774OaYttWiC2S9+jxYSUwHENSlXaYrOv9zQKMLwme6D2Ws00J5CAJgkqylRTQgk51CFg15ZKqtcVG6lYjkdTGlYgxV0iqoqK4pErJVqJoBzDsmzR4j8PIwFCWRUgfr/nn68QO4WZ6qziwPn1zr8n37AB3AQ7JZGEQAyqTslKTk5KvMQ9eBjVZOm54RW+BKkc0vs6pC2tKm+PdUrbFSuJWFn6YTmykxC1O6+mNAglwKh9xyEBRqyEQCLWw8UgbJguY02Za1YhFequ7XAXUNbzwM1OyV3fDHFP4Vif+sgv74KtSi2WsTix4D4q8VYXlVUje3/7eYuKuz1Jlj3ng1gkFyccp8l9LsKVfKzq1C5T/XlYcVpnepUK2KrPwhdrJ6YfLHyONayrHznXx8uqXrVdq8W1us9BubyMI1BBsTrScr20k+fhpqzRJOGybPKOcocvmusVdngoMyIMftoRYRgzZyAzvJHVZe0xkBkNIobMaGFxw6/IXvWexsyYMOPpsMWeznmbabEWC09o14lPCqUov88tEg35hrnNNRmzWbY4kikttQtL5CY73lDxuhwxZEpLIw5lwpFMuSVDujRJmbPPwHYWmTDUCQfloPpM/eIXSUCuBSlzJsy5Lge2UYLtcEUa8MpsxvVRSZ+MdekQSUTHLVJ/FlznyxzypF5wn9txF64pCWO1BFYvx2kQ6lhmb2BZhy++lWJj8ZzzsQh7XcYnOe/zL8Zb5Xcsm1zU//U/P+iNvyz8V91eg6H88Id+avdHn/mZ3Y8/+6lHCs7w7TjpRQEW+gP2WuiaNj2ahATsyDqC0Cahq00ySnIpaZEwlpQ9hsSEbGizgqYOZUrupsaX2KCt9vbbMmBPhngIU4BhphY6mmlBQkyq8yrHeE7GkY5dkWObFVvSBWz+vKb7bEiHNg06NLkga7RJGKotQCx8K2ONlp1uYCcVqiVzMkdaLyoZWd+lr2/wY4noSLOCDBk5aRI4d/Au717ueRTWPdzCe1vSrIoTDzkNa9wbL66SSHysLCXm2JwXy7kI3PMBpA6SbKcipoKYJRKfeI/H/Mqymihb8nh9fdTqvDgqB6TltDIrzHTu1taygvVGEjJ3s/WutKrGqiXQHyM6PI/ETliSyjtkphkjnRBKyKicnGjMVkI0tQn2HXu5JfuLOsJiGQm/fpwnHrvkvmW5+qMf+sTuowDNuqdFyC99+meeEwxvl8vs64BArC635wZ8K8SqTf+qxXXx58W/vxeL7f2qeBfhUuedVKyCfyyDYS2+zv2K86hqnYrdPAcBuL659Od8vXCoJkkuedb11gs97jzVPST8Rta/t0BM1UUqtKymJZlmrhuX0VfrluuJkiKGmWZ8V3GVDxSX+YHiCf5YnucD5RVeMLaAeF0OuaC2Qzij4LYMmUnOE+U6EQFdbZBowJQZDWLeWW7SUmt2FRNxxJiUDEFIyXjbJkQYvl6OeHEyZ0zO4w5XvceQr3GDdW3ydHkBQZjInJHMOTBT2s6pPUf5srnBn/MKt7XP7fKQr3ODN/SAL/IqhzqiozFDUp7UC3SlSYCpjMH85xVLVCXuWEIOygEHOiDVjLGmxERcpMeWNvmcucW0LBmI3UwEEtCWBiEBt/SIL8hr3JYhG7RpO+lMg9DXESNnRjl22G7v37JoIFhocYfSS/2cuptYBW08DYK1rIC+F7FYPNQLjPp9q6YV9fsflS6ex/P7jUAo4UrH5J/7yD/e/eRHfvGhL+p/2eJvPftzu3C8wY4I6JkOl8Sq21np2hkjpjxRblqRCyLmknMgI/bEQk29S7pBOJQp+2oFOLw/0BPlJiXKvFbkt4idRG9c5dvU8fZiidiSNa7KFk/pNj3p0KVRTTzWsJDTOXlVrBiEAZaLt6cDWiTs6cCRvm3TyfNIRAxTTWmJ9Xma6JTbxaFVq3JTaT/htZ4bdr1pSZNhOSZxvkVNSehICyNBxYewssX2OL15YFMSyw/B0JTkDvd1OJ7og3VT91MYdZPgtmlVj4kcrNhzUESs83wdQVCoPS44LjQ9T8QXQRHHBPJ6WMJ5ViFwvHkhWDj0qJzYx4kQV4LLhkMdOmERdZAvQyR2uiU47ykMmdrPLnWGkoEzNuyaNpEj4PvPr3pPd7nXebMN2rMKkfrPD7sQuedwrF/+9N977qWP/d+7v5hdpyNNcsmZl/NHfqRdn2CcB0q1ClL1Zo7zLIz1efkibzVWnfDLioaVpKkV1ftpj12G53wYcdZ7OGvT5rsyPvl4bOyyv6v/69VTctd5qhcd9SIm0zkBAYmJjo0RKSmwSdV3u+oTmFE5xogdG/++eYGWNBg4Dfmp5FzXfZ6QbWbkvGD2SMmICempJeLdljEXtMOhTOhoQkbBFW1y3Yw4cDCtDg0mWA+QHV1jT4b8i73r/GBwmdfKERFtGhpw6DDIHRLmktEqbaL+j8FrXNF13lZs8EfBSzyuF7hYdrhhbJdxoinbZgMre+mvDdtdfJ6bFFqyJyOOdMyO9EjJUKN0aXJLj2iSYESYaErqOpUlvhNmoVZXyzUe0xYjmTPVkrkUXNF1buoBsYSMdWqNv8QwJiUmdHhtqbDOqcyJJWSiKS1pMneLsOHk1EPkTgf0ZefaqqLdP9fdyDner2njqkLibh63LN4MFGvl36449vN8JpnOSUxScbA2TPfEa//Eh3d3J+WUNdNlXdrkWvALH/mnu3//0//jI4G3vtv42DOf3F3ksN0L/4P7FR9/9lO7HpYTuGK/KQ1iIg4YVSRkaz7YJ3M+Szf0gPdxlYnMGDKlR4suTS5ohxvS51APaIjdmM5rfIYBEzbpMCalR4upg1/1mVi1KNc8akpCQkiXJkeMGcqUy9rjUCZ0aWBUSMU2jlKO1ajWnPHhTOdEYu9rSlLJrvcZ0aLBQCc0SZjKjJCAhiRMNKUgIyJEVSuIlYghInKmgLZwsV5TKU1pMNWU3DmIe+O+QgtHSD/mXXhYk1KyVxzSMA1CQtIypRDLy8NxFz3fI9OMRGIyN3GpJhRinGdJUE1fDFIdqxde8dMHPxXxoXrsqZVIfGISVt+L3cGfcJBp/3j/t9Y3xkLCZqX9PjxnMzABM51zSTYYMMGrmBkRhmpFCSwCoVmJCwhCRlHB4DwMO5N8Kb90kUu6eH/952odOSXfLuMmn9UYf1hxX4jpb/u1H3ruwke+sjvHjvVEDPhO4CNw8Mu+vFXFxb2KZYXEKlzg4snzoHDJ92qTssglWbxv8bXudwHy0Q99YvdeFXCL0sP+eKoRsRYgx9yOenJZLEgql1gCvGdIHb5TT8S+GBGRakRu3KTRRyF23J9REGHdcvuOv5FpztvNZW5wUC2UQ5mhZUkmBe/QHb7IazQlYY0WU8nYUEvIzNxrH5qpLTo05jXTp6UxM8noasKBjIgIuFJ22ZY23xNs8Gqe0pSQvmZcDRI+zSt8Z3GFA2PJlF8O3gC8MovyatCnVOWGHPFUuc6XuEbqFqehji0RksJOX1zHs0uTl/QNtziUzMmZMiNxHUqwEIaZE8bwijkWR227WVuyxtN0+IYMOZAxvxOMGDMj1TmJJNzSIyxJMrMyu9KkgZ3AJMSuQ2hNyCLswmgJowaUyjNm8TxaFV5RTuXshemsqceyeCtQsNNiMZ8t8jsWpx1vJeeeF1MNd4peLMtPpxV8bWkxZsJls02IYUbO7kd+dfdIxxXkeKopDYkcJONsNcBHLX7kmZ/erU9h6/GxZz65+2uf+ZVHqhD5sWd/dhc8pNUWIJlr5qxJi5ySuePddSRBKWnT4Joc2UaLhLzEHk1iQgwTN+G9Jkc0iBjrlJyYLUmq5s4X5DW2sU0WgBkZcywJvUHMDKyiFRld2qTMmTlf8g3aHDjJb0UYOIJ8ypwNugyZVtOLNgmIVboqpGTdPddAxwiW/N2QiEwtyR1gXE6JCJhh82niFJw8VAvsdGPNSRR7fp8vSApONrzqTTAvdTwuJ3RM2260pTzhD5JrTum8rkQE43kceOGWzOZvt14VWlp4am39Evcaa6bLoVoiuWoJzuR61Wbdrg+jGsx5dd4Be61Hxk5nCi1QsST+qc5oSEyKRRF0TIujog9YSNh13Wdb1tnTPgbrQh9ihQA818dO2OcVPA+O81sk0QmC/7JYPL7670u5tAvHdhon7kHyie8m7htG6h9++qefCwgq7epwAbP3MON+fgGnwbL87+dVN7gbiNeDxlGfBs9apXpVf5y4TsiDgGLdbdf0rPvrG5a6RK9PuHXyfj2h1EfVfkrik72fgNQ5IP4x/mevimWTs09MYpO+BBW5r564vOkhQMqcQkuelsv0pMNflK+yaXpc0I5bwEMu6zqRBoQYumrlJ29yZN+7Gt5WbgGwox1uSp8+Yy6VbTbpAHBgpvzkO9uEBr5q9hGFIRmZKo/pBjtYLHJXE6bMyChok/Ca7JFREIjFVf9+8CKH5RAvcxgSEouV5n2KHZSSiIADRnSkiWClE/d1YDuEOqu8UmbMadPggvTYMuuAxUoHjli4U3aYaMHnzevMyLmhBxyUfabY5yix37c3sVKU5/U6CWH1N36jMtaUTbNmccOuq7isk7VKnKEu6XhceJ6dN5cWIgsL78PIv/W8tCzvLUKvzspjy3gbJ+4/ZUJ7nlj8u0hiMs1oSZP3lZcQhLFOOdJxRX7ONSPVlBvFbaaaMizH/C8f+V93f/Ej/+ybAprlzf28yIX/GY7z18ee+eTu3Ziu3c/48Wd/fhdcx71Gmo7FQkPtxr5N7DgHJdbsNCZkUzsYhG16bOMNCu3mtqWxU7OzcMmQgEO3uS2xBPGZ43V5zkmDiA265I4XEROyxRpWXc82TTbp2PzEsfRri4SMnInOOGJMX0eWU0LOoY4qaC5QTUNmOq8Ki8wdd+pMDSMJq+aF52R4iFK9IeYN9ryUrxc3KR20yHb1A2csLdVxeifxiaY44VzAnh/ejd5L6hZ6DDe2+XyGl5PPasflzzEPkYolIpKIw/Joae5TBz/28DDvxTLVWTVJ8fvMQIITfC7/ufvGoYfuWRSGVSITpGpwWREYq0TYL+1E3quZ+Vw/0gmeVzPSKbf0yL5PyoqT4wvk+tq8bIJdX+8XBWvqn9WyWFZknBWC8K8/+0+e8/+f+4H3Ie7rqvRLn/6Z54ATMqR/mWMZoeisomVZnIeT8jDiPPCG+gV42gTktOLsvFKZP/yhn7qDeHW3hVodP18voHwyOa1TfQdGtVIWOZls/G11bK3/HOuFio+YyEKsOB6Ve/JfSxo0HI69bmYVuOnMVdni3cUW3sRrkw5f4XW21MIRcim5oC1iDeloxGO6gXeV/Qte41BSppIzZObU5oU/DV6noSEFJUOZ8bmXYnbawm0Z8h3BGhvE3Cxn7JQtFKWnDXbKNnPNmarFaj+l27Q0pkuzImxagQuL9+1Is/q5LxOuOoneAMMWawQEXMK+1xQ7Ot9irTLnGpOSOriYuE+6oOAym7Q14oA5m3S4oB0uSI9EEnrSqRbxpuN/5JozKC2c7UjHlg9SjrhZHpwoFj1UzheL9XNo2YY4EI8ov9NX5zSJ7GWTxepv6wvwKa/9VuNuctmyPPVWmienQdLOug/unJIsPtYTaLdkjS+Y17lW3uaw7HOkg4qICjjztaLa6N1mwC2O3vRxPYj4+LOf2v1bz/7cricH1zewPvzvj4JwCFhDQjgW5vC+PKolHWlSulza9rmEiCEpHaxYxETsdX5dDzjATm/bNJgw5xtcRynJsc/lncqnOnPy2yGJA49sa5eEiE1t09MGm3Rok9DH5oYhKUNSNrTFgAkZBRvaZMqMnra4xRERIYmDYXWkRaYFOTlr0uIi69V05/XydgWhbUnDmdVaMQwvguGJ5t6pPKOgIVZOfLFT7q/BQu1nCNZR3fuEiMt5nvTuCxGfl2Y6d3LypipAgtr1c2Kar1ZQxcv/Lm6wM47hSYnETlzl5HkomNr1ao9ly6zjeSXeANEXRH5d9ER6X4yoaw6q45kMS8sB6knbFnJYcrqH3jWlUf39XK3Gohch8M0SD+Wyx+3WWgInj2+Lq4LyhDqZPY47G7WLhX896lOqVTntNGjWoxz33THdV425ZsQSV1/Mt2qcxSdZtVC/mWp2Me72sfcaJ267+yeff3G8aLsZNvGehXuvx1uBbtTfh54B/VrFf1lMGgV3JoPFDc0yud4T78vBr+yE5LjgMMgJSJe4ZO9ler22ucWcxpWKll0wbGSauW7/ccF3k0P+6/J9/G7wIgGGhJgX9IYl3JV2U11IydxkXC57HMqMAxnTJqGlMWvSItGAXpmQmYKYkJQ5c3L6knKoQxoS8Xv5Ho2jkO/XJ2knhqwoyVBahFwJE4JceEEGXJZNDMJt+oxkTua6gBdlo5qSKMqatBnplC3pskaLC9rmhvQJMMQYBlgt/BscVFhs4wqPWI7dbWMJuSFHFFrymFzgkBHvKy5wKDMKlBt64BZC2xUEC+U6LIc0JbEjeyzm15M//aI3VQsB2zAbDHRipzAOd1wfwdfPpaD2HdeLybP4Ij5OwxGfOM8WCul7Hac1VhYhEatgsOd5/ur3+oanVlgtux7rxUYgQZWf6p9dfTLru5/1146JuFkeVJ48XokIqDrxoZtgGQnoSIuhTrgim2ce24OOH3/253e9O7Z3Fa/DQEUElBO3+c/kYRciH3/2U7ue+3bsk+SNQa18uVcsUpQxKSEhBugz5QJdXtFbrEmbttjC4Rr7pA5muS4dSrTyE8pcYydyG/SEkNT18vuSWsU9Am7JgHeWOzQlBIEh1lMkIuCaHBFiyMjZk5HlUEhGqhlrDlIViy1uIgkYa0ooBTc5Yk5WwUd9R92usaWDqVqzw6FOWJO29QghrKY4qc6Z1SYlYK+3sTNMPd7kW45d4Y7Xcx5EAhJC6yDuIIYn1f6KE9eRJ5BHEt/B4bBFjVXVshCwotob+vXZTnqOm2+CIXb8jfqq7dc0/x7WpM2+g0BZSJotJRNJas28k+EbfqH7ThuOkO8LkIKCNg2OZMDcr7ea0TAxpfqpRVlNlXx+S4hpmoRMi2MqQvW+nWKbntz7nLz/pDhN/Xs6lTdSy8F35MsV+8JHxfvovs/n/+W/+YfPWSWFqMKxP0rqJ/cy7maKcRovZVncj89rFVnzbuOsyYaHLfmuhDqcqu9UrJqQ1QuPN3P8v/7ZX33uX3/2nzzn4VKq1rjuvPAWr2B02uJrzni+VZhOralh1f8WXOcHb31kO+oV7KB2yVpiXVn9XFCQ6oxUZyROerBjWhj3XDERnwuuYbA+GX78/oRarHssEe8tLzEk5c/kFW6YAWNSnio3aRJyiQ1ekj1UqKAo31Fe5b3lJdY0YU3adLTBE9qlLynvjJtsteGHHku4KAkGaATCyzJkIDPeXVzgctnlqlqI12XtsSYtCgoOdEgghqfYsTwUsV3NfQYkGpBjnYxHTKtFMyaiQVSp2XgZ3ZMY6BltafCB4hLv1svsSMLrps9EMpqS0NcRXZpcpEeXBlOdYZCqq2hd0Y99QHLNnUOxkJPzavlGpU5WUFSY6aXnmHtP9cmH7yr6771+3fjz0f9eP3eqjt+K7n/9GvR/7yEL9ed4q3EWFnlZ1GEBZ0EDVk0Yl3HPFouTetGx7JqsL/r+/0wzbha3GZYjDorDit9z3CGdI2Lx4zvBJhuOoL4p3Tu8Wh52/NizP7u7uMZ4Uq3vMHvj0/p0xMN8Hiac2k9AAiwPLpSwUi4r1Hb952RssUaHhIHMrG8GShOL939Fb5G7wuFIRxi1uP1t1gjEsEnHwi/1zmL5SEfMyLmi61gp8pTEbVhbxOyZMX2ZVnmxS5OeN1wFxthNufUlStmSLilzGq54rV/L3tVcELZYoycdKy0shsNyyFwzBuWQo3JgN8vSpK+j6towrqtfYI1r7ZTV5otCi6qgSCQ+4eZef/1cc2Y6sxOMWrMMqKYf3svKnz9+bdo0a9Vz+UmLh8V5d3V1Iiz1sAW+VlBWg9CWVgXXsvnK7ptGOrGwZMrqfq/E5R3h60X08fsxxGJ5fG3Tqoofe9z2vy4NAgL6akVdxEGE/Wc1d5wiX7hsyhpr0qYrbXe+JXSkSce0aEuDJtYg0ft2+UnPYi5bxnUBu04sE7jxx3NH8+UuGrXnUQ2933HfJyEA/+Lf/M/P/cSHd3dH5YSmRMzJlpI1v9ViFedjGXGqfvt5n/t+xmmdr2WTgfp9PurJdZGo7YsRWN5pq3dO3+qxnvV+l6lVxC4RFq67s/h3votd75wufZ0aqc5/DlXnleBEcvf3ezWlRVK6X4A93jYhqZKTlee1iT2RmI6TQkx1TkNiSkquyCYzMi5rz3axRLgoGxiFLwY3LElehe8vnuK3zde5qYeEElACX5MbXGWLuWZ81dykS5PHdIOIgB1t8qdmyGO6wVRyvmL2+DtbV0kzYWc948uvh7xvU3jxyNCKYGvWpEDpEXFLxvwXXObL5YChzHhMN+hLSkNitlhjU5tMJaOnLfpiMbjXjHUyLpnScMZfezoglpC+78BJhy5NS1hXZUu6RITMJKPPmG8EBzQ15HfNdW7oAY9pj0gCnpBtruk+tzhiS9ZIdV5JHytaLeheUMB/d5HriAEUZFVhcSwJaU50448XoMB9h0HVAbNQh6BaiH0fz75eTuR4PovnTv2cqZ/L9VicVi7+fjexioi+Kvz9Z3Xf6oVIHU7pH/cjz/z07iKcarEQWZyC1G+rwx98Lqreo9ss2cKzUXV3TwvVklTnbIuF0EyZExMy4uzHPshQSnL1sqOB28RpBZXxBqpKSaE4HwcrarEMFvogQ0Qq521L8M0q3L73RwoIaGjEkcyJsOdnQcGOdrnJIX6qlVGwKV0O8WIXJQ1iRqRu8z6vjPe60qSJlcRNCDkQC4syWLPWl7kFSiULHhMSEXBB2zwvt7ioPW6JNUUt0Wqa0tMWh4zIyHmbXOINDivegHU+t9yU2/TJteCi9NgnYkpRKWmpKgNn0GffYwZioU99RmQ6p5CGmwpbXobf+PsOvsHDeo1r1jjOogQ0JCbTnIiAzB+zNKrrONecuboGjwSUVcFyvObb9SjB5zfh+NzzeareiKkXLTH2e77jb2pNbD+R9nzLsLZmNt31689zccVPYmIumk0mWANJ/35StY2oG3rgJIiDimDvw/69seeHuOmNhMzduut9WnJyBCHVeQXXKrUADx/EQrwWJ6/+WH0OOtGMcqUk4BS+7uTkvhkRo4c9EXkgRQjAP/+N56qD/IkP7+72y+GZ0Jhv9jgPRu88HJHFomXZffc6lhLOaxfyqqjjGpd1+Rc33KfBsZapOSw7Xr9REWTpBufXPvMrz33smU/ursJ/V6/nN4pYAvistgFZVWj5n1dtaBanQydUwdzCXt84+ueJCC1O1+HMjVjX9JY0q05/idpFVI6LlkAMLWna51e7VS20ZFPWeH+xzUvmiKtllz2Z0tFtXjb7vKfc4qY0+WN5ietmyPfpJbrSZOIUol5yXiLX2K8gJokG/PetHX57fMRvmq+wQYfHtMdMRvxQcJUkUr66X9JrBXzH4xmqwoUefPW64YppsF5aHsZjZZd9MiIMPRougZc8zgVChNeNJSlOnZxliXJB21yXIzbpYFSYyTEsoSExDWJySloak0nOkAlXS0tI3zcTNrTFB8pt/iC4VhVaX5LrfHfxGDEBEhhe01uVulYgAYNy7DqLDSc7mbhu3tw6qrsFy3+ndiNXVAVtoWWldrXYmY8ktgW5FxvgJDnYFztedcYXIBYTLQ6KZ4sij4/3cD7fdUM4fpwrgEMJbVEr51u07gUk524Xu2WTkf/jM9uyqewAACAASURBVP/4Od/B88VcoUWVS+q3LeN0AdX9/j4f9Tzli8rF4mZZZDrn1fINmk6taN0Z0D1KERNRiu1GV6pGHKsb+fPOSoabE5sh33B50PGTH/4H1QQkcpCrpjSqY/DrgVfRO5IxhzpiR3rkbirxiuwTaciGdGgQc8AITxKOCbnFEZfYACAhYuI5aRjmTrrWK+LdZkBAwJZ23WdVsiO2qTMn5/Fyg5fNPtfkiDYJ1+XAXb8wYc5l7RFJyKGMCbEqfaHaqaQ/32aaYTDOyTtlS9Z4VW87+e+T66onQHdME1G7P2hKAzR1pZntuAdqN/Des0MQWqZJ5jbxxzR2IUCq2yxkynpyRBITE1XiIUA1AfYNk4ycw3J4Yk2zRoUWHlZ5J9WuqcR5jngIl59qJ5JY/otbC1vSYKwTAgnsv66J53OvfS/HTTt/vH4i7fcQE51aARNJ7O2ijNXmT6/C6N9HKGH13B6mm+rcTTQMA8bs0OM6ByTAqJywZdbJgVxt8aNaVhwRu78wKEIsVFN0z2fxMvwlx9ejb9YuazotizezJ3yYhcgDK0Lq4QuSh22S8rBiESddv61++yqc3+L99zJOlQ5d8nr1jYnfdPsEYzsv2cox/t12YU8rQHysupgWJSa9JCUc+5T493PalK4OPTsNg17HfNY3RIsbObuwF3dsTG3XyG40Pfk1kYSe6XBYDq0aivvPLjxJZZgFcFMP6UnH2S8JT+gmvxM8z1hT9oMxOw4ylTj89J5MeEK2eVexya+bL7JOmy3WOJQhT5VbfN3cpEnM+4ttNoj5d8GL/Nb4kI5GPCnbvK/Y4rpMuM4Bv5c3uTjd5j9/ek67NafZSvmTL17ACBiBdmBYCw1H84x3RS0KBcmFvs7ZJKGnCa+aPn+l3OEPghuUKEeMiQkt0VRmpGQMmNCWBm0axIRckU1SMi7qGtfkiA4xO9qlIw0GzPjrXOaFIuEPg5e4SIctRxAd6ZTQddnWicnI6UmHVOesSZvcdR+nbkGcMQd3voQSVlr7vmis+B1yXFQkElYdTr+o+OLSLtJRVcDMyapOrIjdHJSaVxtFP0E5iRkOqg6bVf2yXc9ATLWo2o2bvT+WqCpeVhEi65OEZdO+03DI1TWxkOfudVTTwlrO8QWZL7yWHVMdjrU4qfTY7PpE6Sz+jSem/++f/aWHjrFeFaWb5Klqtfnx4SE3voi1MuBldY5bCVbDjz37s7u55vyrz/zyAzlOf954roJtRNhCpD4FiR0361BHpMwZOfiTl3uIJeSKrpNoAAaOGDt1rDmFF52gdL4dGRfosq5NXpTb9JnaAocxa7RImXNT+vRoOV5cRttNZF8zh4D1E1GUUpURU3rSYZ02t2WIJ5zbaafwmuw5LyP7Pm6WB7SkwSU2uCb7DHVa+Vg0JHFqbR2GjCu59r3iEBFhXdaIJOConFdk6LlmtKTBRFPUvW7sDQTRqqGSuq5+XOlfBW7DbL+DRBJGOiFxKl9GrCx5IMdwLVPlhGOvqqnO2JQeiL1OJsyr69Qg1TSlPiFJHLzOrp8WAbBpuihKR1qMCZgxJ9WUrrQZOf+OiNB5wwSVUtpi5Joz0SmPyTZTsf4rXWmyp30yzemYJgG2aLLCBgoUlYyvIPRo0tMm+3rEvgxZEytdnIhlJE7Vyi83SSiktJxBSSqjWq8Ypjqr0BKlK6B8nqk71fsGkj2nXY6rcQqr66USB7j7/eFHP/SJ3YehlPXQNXO/FbkhZ8VZJ8gyEvbiIn6/Pre7xf0uwo9OdA1rmOjFyYi/7W5J6as2M4sTpXosK3Z959T/XP/3xPOu+DyWOV4vw6XDyeOskvTCKLW+IfAdHsBhn2NCCWlIQqoZgQQ8LjusiXU4LyjZpkfsCoqCkg3pVgVJmwbfLzsUlFyQHlPmvK/cpKEWcvZU1OTHuo/zVLHOgaS0pcGQqe0A0uSitogIeHe54wo15R3lNhkFb5gxT5XrjMh50dwmIOCX/kbKvz3aZzyxm98vfe0ChzPl68OMZghbDeHl+Yx1Ih5fh7QsuRLFKHDIjGtmyBNlj6Ej/zY15AndZE5OlyYNDWmT8JRukzKvnI5bmjDUKYIQYrharjGRjA8WO7y33OTfygu8HNiiI5OSnbJtPQU0pyUJXwpu8HVzyCVd46L2mLjnSiSiI61qQW47JZrQFYITnVaLYyQxkUQ0TON4AdGighX4BTKUsMJm17uK9tw4XswL1znz43j7fRcVId4XqJa/ML8DZ+0f4zvedUhDrvkxpJDl3hmL52ido3L8GqsVr+5Xw8S/h8pLgGOele9a+oJtkRuyeEx1bPYinM2H32Ddcey1nx8FkudpkWlGWlphBSsdLdU5GEtU/V0kMZ4XAv4cPJnvvKzv/Y5//hvPPVdtyrSoYI8xEc0aNOigHFTFdkhgC25nMuglbSdkZFJyiyPeW15iRk5XG6TMmVqQOGua0CZhyJTrcsQ6bdax6klNJ3kxde4ffSZWqdBdFwGGJjEJoYWLqjW0AwvLnJExI2dMypyMvo5oS7N63yVaddlDAt7gkLGmzJgTETJ0qnxNabAubVrSrPKMkYDQSYb3S89Js+d57OBrfnrqVai8x03o8pFgKniaYL03PKQNbNfe57yWNCl9406tUIDnO2Q6P7GZbpkmGfkJuXj/nN59vajtFbyaVq65K4gMDWO/J9sUyp2ymC2GcvLqXB3phHVZo+uaR/6YFyHvqmqn3S731vlbc6f8ZXmAciLXW76JYZ8hhzJh0/QqxEGJ0pCINVpEThxgolPa0iDAShVbDsoMVQ+DNNX+wEPRoLYWLExBfG5bjGWwrLsNQR4KR+ShFiEPW5/4YcZZpPRlWOu3ohB17vd1Bslz8fZl99c7q3W40uL9y7gYy2LxgqoXFf4cqhco/v6PfugTux975pO7gvCxZz65+8Mf+qnq//rxnhZ1iEc9jjumbvOjx3Aqv3mrb47889yhvuOe228m/XPYjqRVRIltf4dMs8ow76YeMifjcdnGYL0uxk4Lv4tN+h7vGhPxf/IiT+tF/svibVzRdV6UIWOxG5L/t7jOjXHJ+4IuE8l4R7lTSVu2SChQtrXLu6TLO+Mml+OIS9qipRFXyg4jsb4efrP88ouX+KHOFmUJj7/jq7zt6pinNiCl4G/8ta9yaTPn8Sjh/b0YEXjXpmGrIXxH1GGbBpfLDhklz5sjupqwL2MLryKu8L0T5hzKpFowCkpuyBGBGF6W2/bzkpJEA/rM+Z3gJQC+rtcAOJIpU7HGhg2nqJMy5895hVflgIFM6RpbpBRaVjCCgY6rDl3b4a0bkhBLRFsaNCqZSfudtkzTbhAkrDYbDdOoFtrI9WoLLSrtfq+FLyIOlx9Uksy+GLECC1J1Di0pM7YbSLG6/4GYE2ZgoYM1+HMtcFCx2C22oYOV2ftq8pZukx65Yrh+7p64Vmp5aVENS9F7mu9/7TO/8lxVONQI+/WoL9bLeGB1/scy6CgcF20eNlmRRWs5YVV+fFQj19zllqCafvgNJFARv3PN2TDWW+dicIGu6Zwoch9UIfK//cYvPGfP79A5iYdk5KQ4UzjHX1CsaaTd/M2Yu2nOSCc8yTaZFLwi+zRJuGYGHDLkgrZ5h14kxNBnzEjmNIlZwzYdJswYMCEnZ0ObRGrz6Tv0Ih1XlORYcnSTmD4TvHRsQ2J6tDFYDkHqpk+pWn5ZwxHmhzqmRcxIJwydAab3ECm0oCWNapOdY39/vbxtFb7EehL1pFM1Gvy52HQwp7lrXIVuLh4T2dzjSPETTZnotPLN8Nf4MTzruOOujqsy0SkN06iKfrse2o10fSICMNNZ1TCrN1MAl+dcrpSmLV7ctCDAkJPTliYBhoNywEDHTHXG1E26BEO/HFUwMV/MePW6Yz8tQyKJvVbde+u6Qs44xTULAbOFzZGOiJ1kLxwX4j1pYxDaDjoMOGloZU1aCIZbHLFNj0wzNk2PQTkmEHvenBQTObmvKbQ8ITF8Yh/FcaNk1eT6rUBlH6aM70OBY9Xj1z/7q8/9ZYFlLSsmToMqrCpEzgODuKv3tYRQXb9tsTBZxFfXbwfuWNALLSq52rp0rf992c/1WKYo5guJZZ+hICcKFY+BX6VW4+FYy+K0zUXVzal9HqXc2SVdRpD1r+s/K09G9hCdUgsaxuqUT52b74SUSC0JPaegVGVfbAfwmrGeBGNmTHXApnRpSUKiAQcy5geKJzlkxv8XXLMkPMmZkbOhbb5XLxKJ8Ho+5yJtNoiZ6JyhzBiT8oYZ8316ids6Y29uP+sZpVPeFxoa8l69woFYqdwP/le/Q/DbP8jlKwf0967S7kz5a3/td5n/2g9xtL/Ny7dCrnRhNIMwEF7ul6xFgggECBGGSxJzsWzwsozYoUuiIW9IxpO6RUsjbsrAfYaGDglDUtZpk5FzQTu8Jge8aA64qYdcC47o64iZxHSk6XwBMvqyz1xzSkpmzDko+ySSMNAJU025YNYZk3JZNrlNn0jaWHGNhH45qlSwZjqvMM/GddAEqdyJm2LNE0u3gUs1dcWFYeoMt1JN8UZffiESrLTmVG2x6HHD4rDudeiRJxa3TNOqNokjdErgsMhKYmLrwuygEk1pkOqcpomZakpHmkzKKQEnBRJKLGTDXmfitl165qJXz033Y0qwuNmB07X2T9xf4zzUYW3+8zSug2rcMa96zhPwr0c8EomtwKy6wtVN3ErxEt8FojZXhQiXg23erZe5UDZ5OmjzH7jB8+Ymh2W/mjTd7/jJD/+DXXGFhv8XLL9lxpxQEkpK2s6pvCkNEolp1r7bhsTMsAV/k4QBE44Ys8M6L5g9WiT0tImIoc/EFhDEvK3c4oYZVoWIzzkNIkYyJyUjdM8ZEfCy3mRN2lYa3BkVFpSsS4cDHVj+h5vSRAQclfb5RITbDCpYkjfg8+fqreIAgHWzxpyM/fIIg9DXEZnjOsyw6k224z6v4JglWpk4TnRK27QY6QTh2HiwcHLiU1I6tKqi1JvxRRJTYs0aBzomwk59QoIT533L5Q842QBoSoO+jmpmr8ewIm86uGl69MtR9Xp+OjLWKS3TcKptdnO+JV2ul3uk5O4ztp+ZqKkmGBYCtsYAa/wYEXIl2OFAB4zLMSJWKTEnZx37neXYiXVfB6yZLlMs9KpEaUrEUMcgbXruO26R4GGCc81IJHR+Mgm36dOSJrfLQ1rSpKBgTkHkOCY+P8PxHsm4wj8QU3nr+dCFvUs9z9lreLXdwb3g8t3P+OZp37hY3HR/M8G56p3BZfChuurDqqnH3RzveavbpST0xYvgFNjVqvCLfH0acgL6sATStCzqn8vS93/KcXqi+aJiUL1AOA3rXS+2FgmqixMPf/viz4tdWP9/XQED/Odju9giUmGefWfMLhjHxkdtaRBiCZcGq5H+uG66JDljnyFdbTBgwo9+95yXgyMul2sMmGCRzjPeVq7zW+Y1fru4SVcC9syUz5lb1Xg5JuJ95QZ7OudK0GBTImYuIU7JeUMmfD64xhNlF0H4YHGZ4mCD/igiaU6ZTROef3GLvS98N0bg9z6/RWDgDw6mvP+pKdcGSlqWfC4d8v6rOW9vJiQEvHstYjuKyCh5Z7lBw014XpI9/tS8zHcVV0mwi+sBI+bY6dCIKbdlaPHNlKTM6dJkQ+z78+om13SfA0a0pEFCfCLpB1hllaFTnilRJpoyc4T0UTmpDCEjiYgkpElSKSk1JSHi2MTO315NRNz/9ahjpGc6p8QSiEdO0x8srKJ00Km6gZhXO/LuxFntXK/7PwzK0QmfJo8V9/4CvsixRHtTwcmqY6GsNp+Pwqa7/h6qLn6toPC/e3lZryrmY9Um2kM46gt/JavJsQyqLVAe3cV9MfyGvF40+Q72uBxXn4c/N/9m8R5SMq6ZIf+O12kQ8TfL97Fu1qrzzcc/+/C/3P3RZ35m98ee/dl72lBUlJyCNWlV56F3tW65SaTBkLguvydNRwSMdWqVnbRgRsaIlIycS7rORXp01IpYdDRm4BouDWI2tU1GwSvGbv7HeC5YTkbBY+WGNQN04JxruscRY0q8S3rJnNxNmmyOj7EwsozcNuXUGvVlrkHQV3ttFpSVX8ZU04p4biVuXd7VWTXZiJxc7qScUmhhN9a1aVVOToOYsU4RsVLjOXmVS+ZklZCJv2YmOq1eS8SQ6oyZzqpclLnipi5tG4hxm/Zj6OjxFNEw1wwvruEbfqGENIjtJl3LqmgMJWRUWq+lRGJSnVcwsYKCW3pkxQmcmltMVPGYADbpEGAY6JhEYmKxE46IwJ5HElvomqbW3JeIDk3WpVPtSTaky0inVbMnkZgL0sMgVVOwSYLnKSnKGi0LedYOmVuzk1pBBVTeLap6AnZbcdrEyh3XTY19PvK/13mj1XWyomG8Ku5Au9T2Vw8akvVQi5CPfugTu8sOeBG/92Y3oA8qzitNWf99mULWssecxnWoP4/W/lv1OmfF4ua8/m/9b+DOzfayQmZxQ1C/3XZ9715tZVmhthiLXBrvSxJKRCg2YfmRb/291+EoJ17zjMnPabFYiCw+h/8sKslM9Ym6rI6jLU2mOqMlTccRidiSNbbo8j3F43RocFsHjHTKRDI6NNhmjXXaJAT0aPObn7du5c+bWzSI2WGdgIDPB9fJyNmXCV9nyF8Pt3mi7PGqHFBgXX7/n+AF/ih4jYMiY6wFV6TBJjHbNGi6jcEtmXLEmD8KXuXf//vv4Qc+9pt0Hn+BW7e7/NXv+RoXvuPzvOfpA6a58sF3jPhvH4uJopy/+raMd/ZCvrfdpduZ8+RWQU9CboxL/kNxi21t8sfBdToa8V3FRb6veJIebQ5Nyqa2KpjUGq0q2Q+ZEomVuG2SMCbFat8XtKXBnGNvj6nOGOmEzOHHATehSMk0c7C0qduklxX0K3LGWzOdU2jBhlvA/GMUZewWMXselpW8su8IwjEWvxq5S1DJZ6pqtSj5RTqs6dPXF6E74DSEjrzrzzutxvxANdkwGLrG6tsXagmhFeTLcQZ80Vv3DFilW78sD90vrsS/+swvP+c3O4vqcj58obCM63E89RC8JLL/F04WeH7xr8NJ/Ge5ikfyqMVM5wzL0QkekbV1y6vJHNjv+arZ5jUZ09aEvqTcpE9GwVfNPh/gcdZMl/9Ov5u/8+G/v/sjz/z07kjshlQp+dgzn7wnm5hf+Mg/3X1cdggJGOiEsU5pEBN7b4ra+rKnfXt9ENKTNtYs0P6dxeebSip1LKnjv405YMSEjI5aGGVCSFPtNd9nyjXdr7rdF+haB3NzyGt6m01tc+Qc0ufYTfaYlBKli83Zlv1RVOIWBSUZBfvlUfXe6xM9cdO4WWknIS1j/SZa0rTysw5maaE9FkLUkLjKG967w3MI6ryZmIhBOaYpDQdri6pCyecW/xhfhMx0hid8A6yZNk1pVBt7HxbiV9sTuCvG8lxmzDVjprMqHxmkyoeegxK43JJrTte0q8myFaa1udcLJvSkfexCLo7o7+BYt+nTEbverZk2iSTWJJEZHSy0a6JTUk05KAc2J7sCsycdIom5Xu5Z5bBaQTfUaVWweL8Xg5BR0JEmN7BF6yuyTywRa9ImcJMvL3zTcHBZoILRniD0652S63WfkDok1/++CPHW2t7Bq3MtxlmTkY9+6BO7Dwqh9FCKkNMOcJks6+J9D3v6sQwmddbfrXrsWzmWRfnes17vjvuWwIVOk6KtY6lPU7xa/L0i1NbI4G+mg6gL/50npHaR5k7Gsa5wBccTmdzpdp94vJxUz1nkepz5+kuIrMe6I8ebQl+M+MRuEOdVIqRYsmJXmnwXT1Gq8p7yIi/pG3wj2GeK9QLpSJO/3X2SA0ZcLru0NGJPxnxv8Rhf4JCZU45KmZNowCb/P3VvHiRJdt/3fX4vsyrrrr6mp+fanb2Xu1iCIBYAAZDQEADNmzQJgKQiFJIohx2mjzDlsIMSw/RAEbZsh6gIKRRhycGwpLBkSwB4SSBFEiDBAwRAnMQujr1nj5md6em7667KzPfzH++9rOya3pnZ5YIC3sTETFdXZWVlvXzvd3yPFuIX0bq6Bf1P0h3O+MoYwBXZdxVUqnwseo5Nxuxpyqejq2Qoz5htalRINOIOXeERexpV+MpHv5/nPvsOvv2dn2Hr6mk+8eEfwNqIe1bhzm97jL1BxMa5y4zHEZGBVgKb2w3644iB5kQidDRhS0Zs2DaXzAF/GF1iX5yW/4gZz8sOa9Jxhl7a8IpebZrUqFFln77vSkSuTS6umnbgYVQTX2Wsey5H2zidrZFOPDTHsmsP2LJ7LlHRjIGOfKfEdT4qnrMxZEpMXJh3BQiVmyeu6hWqWsG4K/BCwmNhhOpdzTjFnRAgBm35MF/CvRYTl2QjowLrPdEZbdMkJqZmakWQETbhjIy+DtizvSKoC1jxChErpltU+Yq2v1eBOhpw3Px+/EZW1oJnwiIMq1w5DIlTWWkmjPLmHmBw4d6DUiLicetHAkYfTPzrX/8Hf29Rfe+bcYTPUJeaN2GbBz6ZZkWAGInhR+29bGiDL8uLvDE/yR26wrNynZflgKfkGo/qeQ7NtJgHj+XPHJmff5FE5Jc/8H9d/Gfv/xcXQ5LUkASD0DFNLMoJumRktKgjCHVJWJJ2YVYaBDlqUmVCSp2ElJyUnDVt0WfCpWiflJwYwxXZZUcGnNQuNXXQ1pzAQbOuIk/MHg4utKYtGpJwSbaYaEpbGu4e86pREYZrukfquyGbdofYdxSnvqugWN/NqdMxrWIe5limdkosTqJ2XZaK3gK4glQiie9gJK6b4rllfTss5nHku+Zl4YxQbAhrU3D6HlknQpKrPVKECyaEgTcUzPpqJWWtcH+F44XEI4gbAIWCVxCOCByQpveyakoNRemaFjGx507aomsHTlShulAkDLDWwM2LCTBWU5DXrSqnZIVVaRMTc1V33Xn7dSzTjL6Hz0U+2G9IrUAdzDQlx3WYcpxJ5b4O2NIDnxhPPFVdsCX55AjjVBShKEpZlNQXckJxI4zAATyC2OCoYEYQD4C5+thRjomLNQJ00R3jRjGNb7bxl3pmi52PmwXkr6YbctzxvpFjkZfxat473LzHvf5mx7rVY6+lI3QzzkcYIWg/TjnnyLmEhGZhSgWscfl5ZSL3sed1zGe5Gf9FSn/Kj5VHxS9uBTzjuOtZgkqVPxPcfptz8XWLxy4vPIttVSPB3TsqAp6K108/Lc5V/B2scycn+CO+7jco4Q35BvfqOjNSPncwLqA6I0lpa41dmbJlBoyYkYolJuaaHNLUKmNmnNElVrTOz2x02TUj/sC8SJc6MzKa1OjSYIjDDz8RXSdH+c58g89HL7OiLnC/bgasWbepRAYefvufce97/4DdFx6kP6ix1k3Z3Gqx1En57B+/nQfO9/l3H3uISsUynsF4Br+ytck/OniWp80+n8v2WFa3qb/NrJKL5Q5d4YvyoqsUSs4abXIc5CqVnCETdul7R+Shw3/rkDZ1pgQlKEsiFcY6YV261KRKRxpYVfp2SIbTgrc+IA/fj6v+W99erxGckWMvGxycgOueUzK3lNQCfnWkq6F5QRovk59DpS9gpcsjQGDcZpZ6lRtXYTVIgZsOju4h7SlDsEKAFqrGYeMuQ8RiD18KSVX4JOYYScj/2CMEvSGBCCMkJsd1Qsrwx5CYAIVpXwjOjcxfE0QiQrJYKVWBv1VGFaeGN/NKSXC0ixOS4Uwz/hmfYRfHE7ouIwYy87yCSaEu90n7tTkkxwd184qufc3JpzPsszTVkX9TzblbTrGMg53u4HwoUnJvxDqHxIRO5bYeUqNSJBJr2qLtZXSDimCTmudvVEhw3h19mTIjo0aFAx0SxDaqxNyla4x1yjU59BX1iIrnggF0pME6SyQ4JaVMM/a17++jiIGHdzplp4S2tLyka+S6C1Soe/J0w7gE67rd44R0aUjiIJJ+/qWaFe7pczGJeXGt5hO34JwuCH079Ltk0es8sjYkUiVTl9SMdeL2XBG3ZpEz1gljnbJnewVUOHQ8wjoRkvTQ1XDf5417fVjnIgxp6Ih49UjXXZif1ylZKTq3dUmoUingbkChBuYStCo139GOxa2X++p8YULXPPjLiC/srOCMbWtUaHqQXUXcdxHOpyZVVqVN4NiAI6QvSZMDHdLTEVVfvAlzOHyeql9Hm1Knckwn21kZZEe6IEXSIVJc4+MgsOX47JUQKcXvSt2Rb6ZxAzHd/je/fRHgvvuuk84qjEc1+v2EJ16ukMQwTJWZdRPtREMYpdCtQZrDp3oDPm6e5FE9z5fkRYZecWE/PyiOv0gkXvz3ZoHkrQg2r9fFvRWM6ZUI47dzXjfjtIT3PS5AvhUJ/dVA126VMC3q5pdHOfs+7vuYb9pHjQHLnZRXeu3iZ3m1Q8RA6WYLC00gYsLxHZjwecL5JCbxMno3TzxeKwmsKhVPQnTqNKHiUSgZyTxYNDhVj+vqHH//jTxBzQcTZ3WZB+0SB6R8h66wJyM+Hb1EX8dcNT1SMr6/vsZHJ5vUtMIlPSAxMQfqOgQxxnUOtMpD0uX/vH6NM9rhabPFd+Zn+Eq0SUOrGGAkhu/Nz/Mb5mt8NdouTAXP2DZPR7uctR3+j3/wcezLFV74owvED20DsLvTodWccsf9X+HjH3sHb33TNT7zxbNsPt2mGsGzmxX6qeVSNuYMbTZocU7qDDWnJhHPyD4isJ43edJc9+7Nrjo5wEEW9rWPilIl5podck7WGMqUAc7/Y5ue/+610MuvUmFX+0xxLf+4gNzYIqgIQbfzIUiYacpEpyyZNn0dFs9vSsOrkU1oSaPggOQ4WFTY6FW1MAsLAW9MXMj3hnk4smMSqRa68uoThsxvvDWqZJL5oMvJQDakXiQ9dR+wHNoBE50VFcwAKwn/d7CyiBydm55Jo5ife/aweC7++YudztsZ30jpWhc02+J+gqOVxAC5Sn1lrP5e3AAAIABJREFUtBzcwJyHFeBq4TuqSMzIOgUgF7TnxfUOxOJXdhP65hyBE5J66dMAyUHn63bohlSlwp6MecSeZldGPplTlqXlEnoPGwWI9KhXy2shwf7MT/z8xeVoiTXpOtiNWJa1ToOEhiTUNaYvY5alVRQeBMMdusLLcuBc0aVXKFQZjOMbaIUdGZD6xKZCxBjHm0qIaVEvvIX2ZMSIKSO/rgQ+ynld5xmuMpGUkyyzwyHLtDhBh0QjnmGTKTMOdcYefRQtugxo7vkLuUtiMSCOS1aV2Enveu+NyJgicA0FkFgqbKu7DxUt+A8hEM7UcT5mktIgpqd9DFEBX4olIlJDUGZ096/x64vr+mbet8Pt7RTrUS55IZhSpeJgTx7uNdUpdZ/UgSsGLHYjy/+W99IA7QoCGk2pMdIpNanQoE5F3Bq3qz0U57fhkr2chCpNMfR0VCi4IdA1raJ7Ai7h3rOHxBKzKq5TPhJnlmhRaqZOXRKGduSgddIm8+ub+nV9HlO58+4zZqxTZ4jrYau72qfmYXYDHdGSBgHmG1ANMzLqOI8VV8AKCAxT3Hs3IEhk7m4furdBmj0NJqp+fTMIeHGdW3FcX834yyrqx3/nA7988WF7ir/zqOHzX1/i3JrFCFSTCXkaE0UWEaWdOKOxYQpLVUNvpuQWIoHcwjSDHKeTnPovcJF0ww2M/1dHuj4CFeJo5+BWSlOvZrzSsW6Hy7H4u5slFjd73e0+/mrGbXWTjgm6w3U/LgG5VXcg3CBhIVo85u2O24HAOdLu0Y6Iqr3h+RYtFLECYTWVrHj+1E6Pvv4WicUrwdkWR6hkhBZvGX8edMhDIJmTUtUKI6+u1JUW23bfv1+TrrT4il7mIZb5QnSFqZfkfa99gE9ET/P+6E7+Tf4cH51sMiJlTwa0pMEJbbPNIanmbEmfCjFPmy2+xjUaVPnZ5TN8dE9411KLx/pKiyprts5+NOJT0cuO3KcJTa3yrNnmmWgPQXjPShs5sYwM9jj/7j9Eh8Lgi9/OQa/K23/k97j2qXfy7GHG/XtLPHR3n82tJlf2DUkESWSoTBucW4IX9uGhDcuXrsZcsRNOaZue5ozFVSc70uQuu8qOceZUDRL26RdJUSQRz3KNczgPkalkbNt9tzmIMwIc6riotgWuR6j8uSqbV5ZCvPKVmyMtn2yMPKExqN2ETcVtEG6jmNiJx2bP4UFBkTFUDBOcfn/YcMqbeKopqSdzRjKnQof535KGl+t0GOe+r9q2jHNZXpY2Da8YVNbgB4oKZqgmV4hIJPGGi0Jfh4VSTwhewvVQDyN4NXyun/mJn7/Y8NCLf/5rf/91S0j+2k/+jxfncM9jtPMpmUKyAKXCFBt6kC623ofC+DXEeFOxRBImpc9rJPIJ4pj/99f/4Tc9DCuMMKeAwh8k8wpDcwM0l3BOdMYXeJa/qm9BFDAwlYQRM7o0SDQqEtcj3fISXObVQECqPmDr6YjzrDMUB4kc+A5slxpt6mzTo1mYC2a8KDtUqXhVophN3BrpDAEzrovrDG9JjzYuUe/QoKU1rnlX8woRz8oWfR1hscXcB3dPb8oBsUY0qNJnTBMn9pGSs0uPmg/mYw+3CipViSS+UxkX8qwViej4znLoUAT+xRJNdugV93nw86hRJSMvChcpOUvSKTorAx+QG9y8Vl9ICec1I6VBHYQisO1Iky2755PSGSJOLS8cM0j7uu9RaJsmM82cWLGv2gcJ8fC8MAIUrBwvxP6+grnKV0vqpJrTpOaUt7zB48gT77s+4Qyd5a60GHkoWSzREfgTOC+WFs7Jfs8esmqW/OM5U8kY6pihHVKTGl3T4jQrPCEvcU5OsMeAJgkv223apkkdB3kbyRjV3O3Zfm3P1UkyOzlix8up4roWGRkdGtSkwr4OCv5OTZwEc+AihvmQew8YKBUCyH26KkXyUYa2lUd5z4iOibPK43Zi0sXHf/on/ruL4edvVDEpPqldOlpFZMZbH96jmsyIqynt+55k+vibWD9zBTjL6bO7XH5pjXS7wnd9xxa//ulVMhuRxLA7VvbTnE0z4F49yY4MXetUhKmmtE3LmSQJzIos7sYOQkE0k7n/wpEAb+EC3S4347WMxUTkVp2GxccX+RqvdnyjZdWOk+Ut/z9MaIeNn28oZSJUOREJN0wItHPxcrzMFaBuJiP3unwmf52PnLvmxFIpXNAjqZLrrAhZGlJnaIdUPWFM0WKOHndtjnv8lTwIyiPAQkI7V70UayDiufN1C5CrOrlKl/HHdt0LV0X5ofw+XpA+z5kdPmQe5y32Lipq+Iy5xB9GzxBh+Jf5U1R8BWVLepzVZaDLk1zFYr2M4xJ9XNU+ISYl57O7E35srcvF/cc4rycAeHO9zctTB3PKJaemMXdrm2v0qWtMlZh/dPgM/8n1fRjG6F6b6M0DXnr+HG//z36TF37rvZz/od/n5Jd/kuGoyiM/9Vuc/PJD3D/ssPLwYzz3J+/i9B2XefG583xsO2PnSpV3no6ZXkvoa0ZPM/7T5kl+ZdznHj3BNdN31TEidugXm9NZWSMT1ykZ4bTkXWAw45RZZVd7nJUT5OLWko40yLD0dOg30zFDj5/OyYvrHROTa84hLpGLPf46ksgp2BAVUpmBHBo6HoCvbipGIiY6KQKFAGVJvRN7qJyNdVqIE7j5pVTFJQyxOEMyg5BQ5VAHdMVxe0QMPTvkhFl2m50Imc2KJCRASzK/cbZMg6mmTHVWdG8cPMvBuqY6K9yRE6ninKpb7NlDHJ8lLe672ym0vN5QrsQr/Rh1KchxOOmyfHH5MaBQGnOyyJ6Iz1ydTHHO3DNNqfhqZ+gSWebV6G+VETo+FsVqVvBpYG6AGcQH8HPmX/EZd/1sVlzXCjFfJS/W1/L+EJJTEcPt8GT+xvt+4SLAyWiNqc6oS8JV9lmjTU+mtKhhUXZlRIyhhgvu6pKAh4tWqfCUXOMhPcOBDDnJEmNPlJ/hZL2flx3qvityh13maeOM6sZMaUqNPXvo4JWe09U2zUIcZKxTqhIX3IFQrOip67iE+zy4tnekwVUd4+Ri5+RvcPfghLQQwdjXPgZHnu8zYeBhPaHQcGB7DBkXEMvMX/dde1DsJQkuEahKjbokrgsj8+9CVZmRFkpaBuGa3SZXeySBaEndFWj8uubOI/b8t0lB1A5eIGWT1SMy/KU9LcyXkHiE6xTgZF3TYlf7VCRirFO60ipI/ks02OGQKbOig52RsyHLvhuc02Poiza573S6okpFHETWBfvwol5HENqmxZ1ykpycQ0asmSVqWqEijrPTNk36nrxf9dyasB+EpLDKXH49+EKd0i49GZGrZSQz2tRYkTY76rxNuqZF6ICHvUVwHe5wzSKNitgjFg9/9fdjKEQp7mdXOHCjDOvKS7HLK8VZx63VATGyuJb/ZXRDzP9wboMf2mhSa0yoNSas3Pk0jfYew+fuYzKuU23vcvb+r7B9fZnnr1d4+5s26R02scBbHz5gc2hpV4UT1ZgpOZvSc1+utthgmRPSJSYqSEjHjRsC+FsoL71ShSVcvG9E1+DVkKFfl27McbKwNznuq33P8vGPu54hoy5v6KGCWz5GSEDKhKnQIiwf9y+zRZh7vH1YhItgScwNcIzcV89TMqZ2WsAKXkkVbPGxm3FIyknK0evmlUg0LzDpASoTWq+OFOgCnQDHCRj2fyWf57I55D35XQA8Za7zbLRDJIZVOnRoMGDMu/I7eFauU8N1J7elT9NvVA6/6jbcFVp0tcEb89O8ayPhQ7u7VKmwYVuk5Py76ctMJCPRiK7W6WrCV8wuXXVOsA2t8J35aZgqspTx4pcfJX8y5g1/99OY9QprJ7dgGnGqq/QHMXarTe3si3z9iVM8/6ffw10Pf5n97XX+8GlXHXpJhoymhq9zwGUZ8KVok/9vdIUMy6p1lf8T2uYOu8y6drhDV1iRNn3GnNcTnNcTDJkw0ZQtu+e7Hk4Za8ik8PEQhJ7OeSABGhUMvQKUKSMj8dcMICjIdKRRBKzh8bFOKHeBjwbEYSOZz4cQBIZ7y8lvmmKOzFv37tUxkf8beyiZFP8GiBUcXQ/m5odurgZ4loMm2CPPPUGnqMyFc6pITEMSVqTjOj1+s46lUuKCyQ33SDA8DERJqzk/9/4PXvwv3v8/v2bS8t963y9e/Bvv+4WLf/sDf/+iU345Kll5RNnKX+tgNBb4OcabP4bXtUzDfVLVQoEuHCPATwBfba94HLurjL7ekrTfyKGqhUy0k2Aud9fK+8xcPCRIyAJ+vZgLaVhuNKN9tZCssPoFuejg3TDCEYEtypo2OWM7roNLXiThq3SKe61KhQkp53SNEVO62iDxSkZ7MuKMLjESJyF7zfSL8+1IgxnOVE78PRRJRKbufSzqzfnc3jAhZUsPSMmL9aBOUshyW1ynJfHmiQMdY1HauHV3qC6R2dc+Bz6Z6kiTfdv3yU6lUKgLCYJLNKo+8J8WXXODMLGug2twBPTF9SjI3Lrv2Mm41yRx4gRIEdwmkrCjh2S+01FAE4mctLBmBfckqPYJc05VWWky3HcFvNmvt6lmTHTGvu0X8VTowHVokFClp0Oa1AqSu/jr4GCQbkbsap9d+sxISf3zAh9vDbcPNKXOSKeua6XDgoy/IStscUCG5dBDarelR4MqD+UnnDGwvz+GPhFLyYr7I/jTzHyhRlFqVDiUCW2coEvTS/eO/boc/gaR7wjHIyp3v4P6YVijc7UF7CrcJwGuHT5vVOpSAzcUT8v345HHjomhyoiR1wtRdLvD3HPvNd5+4dMsnX6OtR/5LJVH91FrmE1rGJNz/dJD7F+7my88l/DmB3vsbC/Rbo946znl819fYmwtsYGvTYd8t27QIGHKjG3pc3++ylvyszTFkawq3p23PMof/AZysZgjFyNcnEVllvJ4NcnCceM4zsarTWxer0QIXhu+Fm6/Fb6Y5JWJhYvHioiOfdzh1u0ReFH5eEdgEK+jSsNxxwqBkZHoiOtz+EyL1zNUSMvEdbcAO0nfxfcpK1CEY5afcztdkZyc3CuFhI0mkPRCdSvzi36qTt4vwtDxeNOKVHhTvoHgyXcY3pGfo0aVN+UbzMjYYJnfjp5GcZAlRdnXQeGTsSZd9hnSpUGkhiZVakT82uYhQ5lyUjt8PbrOs3KdsWQ8kp/AAlPJ+Xq0zZiM07bFSFKWNKEvKZMv3cv0i3dz9/s/xs7jb4I0Rfsz2u95nOmz5zkcCdt9w97Tb+Brn/heDkfC0soB/+RDb2V3t8WZhlum//odbXILdWJ+rL3GebvMoYy4z57glCeFxxh2xJlu5ViWtVH4Y0QYL5E5KSpy23afDg3GuGDC+K7XRKeFOovi+AABwhTUfoIcZq7Or6AqMUMds2MPCyhVUFwJlbMQpFh/b+Tk3igwvyEBCYHdnORrj2CCY4k5tH1XHZQWqaZeQjjzkCI3V0IwEiqOYSzijV01Myh0SfEvwBYH/jNW6BgnO5xQZZUOJ7VDyyunVamwZDqF43qh+ubvucQkhaJO7J2uwSU+Ezvh597/wdcUvIfAOVRxc/IiaIags++ud+FpUsKiB0fmzHcCim6p70SNdExOXiSjIcBNCjLp3Gg0QCu+VcZ8rlraHupyHEcuBIaBQxCCsAANWSzkALwaeF55rJsVYokZ6Zhtu09TaqzR5UCdfO6UlKfkGo+ZywDEHqgCeNf0lHXb4rQuMZaMVXVFiplkjHHu5yva4Cmu+aNl7DHgtC7Rkjp72mNgR+x4A8CONBGcImGMI54bTEHIH+iIXN297Pw24mKOLOFcwTfZJ4ifGD+HtvSwmCsr2ppX2NUW8ygUP1wn1GA83AlginOKD/yMEOM4QndOyzSKgNl9h6ZIJIKgxkjHHq45ciIWAVpJzsROjiikgeOtdE1rnqz7ez+QyAFftPMpUWlfDCMUgMY+qQxFtSCi0aDqOj46JBJnNLinfaoS85JuF0nBnLPk/g0EcIuyapa8QaXjALk9r0qFmB4jYiIaUmdNukxIWaXNBJf0ZmQMdUJDE56MdgDn85T6rsDYm8iGAk9AKkw1JdOcqabs64AX9Hoh1TtgzL4OinVVxPGTXAHIc1ZKBS1bKpaG+CEodWZ+jZv6JLj8neelIufNZHhvVrRfjJdvFjsHYanXW7o3+l9/yX6QyRippzCz2H1D9d5tqpOU6WCF6SSh0Rqy0TH0+jVOntpjOGgwm8UkkdCMIh47nPJIs84wU95RW2aW1vgeTvGUHLCiNa6avsccJoxxjr1zFZKjalFBsWARBhU22eOC+9cjcVg81q3ex32xesP/X+sQDwcI1+Rm53ZsO23h58gncLc6t/D78P72yMT1PgX+ue47CRjPmPmZKkho47nHrFoPb5qfX/FYEZjf+vxuPm5MQCO/aRhMccYVnIzqcSodKu4oLgE4qiLRidpMPDQGQudHSpA0KX43/7058nihtMO8Ph4RYXwlSf21qZual3Q0zLyZVAgu65LQkjpTzw+ZMOMlc8BXZZOuNB38SHJOa5fPRM+zocvcYbv0zZQmCeu2jYpyHaelf6B9B8mRCgMmTmJWpqxqkxejQ0binIBjv6Hcn6/x59FVYgw9GbOmTc7ZDj2ZcZft8Lffs8cfPF/hx//6l4jP7LP/h4/SWd1EekPsQYJu1elv3sGw5yCaB/stnrwWce9GxvLKgPW4weZewu5IMWr4+kHOShxzNqpxaZjyHc0mTGt8IbrMi9LjLflpvmiucE6XsWK5Lj2suHviYbvOU2Ybg6ErTWbiyXykHOiQgR0Re1LoQCeICDOcWooKxfcS5koiVRpeNz8WR2p12PGcKjFd46BQGTlGjA+KlUSqjO2khOd1/Q/D/J5wgYoLbAXINSugqPh7PJYA5RBEKCqzLWk4WJm6EKEuiZvHElHxlVLByfSmZAUHKpxbGR5S7oYEKVOL5bycxIo7Zp8RB+JkitdMlw1cEGcFNswqVePgjE1xjssbZhX8+U/8Bpp7OEckTor6iY88deHTH/nihXf81KN/fLt3/I/99Lv/+Hc/8skLwYwSwh7ixhx25TDrAcZSMw7W45JSV/VLTOJ+Fltck9R/ryp4OdvEmbWRFvj3IAIw05R/+ev/+7cMJ+R3P/LJC0HmtCNN5wANxfUI6/ucQ+TWdVdIOtoxcwHOfD+Yj/nP7/+ZH7jp9/o33/d3L3akSc1LzS57yM5Ax5wQ1+U4p8scyLgI6No0EDHcqavMxMltW+PW8dO2Q4byouzQok5bE7akh4qwS48xU3b0gIbUeJFt9uyh/wRayL868npcJBljnWJEWDYdTyxOQNy60KLOUCesSocDHRYxzlAnVCSiITUft0DT80TGTNljgMV6T4oY/PoRukt1SahJlX3bp+ELJKHQAZD67sZUp1SlykxTZ4KI93whQsWnl1IONinugdRX8gMKIGLuo+LWLMdvSMndHipxYd4X5gOC7wy69wvQtNyfq/i11HVXUiw5Vi1VU8WIoW0aGJ/cHmifmiSc0zV26NGQOilZ0b0eeDn0AHMdMfUdmhmrZpkqETNyEGFP+9zHBqd0iRfYZkOWsaLOX0r7jJixLl2GTOnS4CRLVH3S2JMxqWa0peG9QzJASb2pY1BODHL6VanQkpozMJQqQ3WKciOmTJnRkSZTD9sqOt0YZsyoSYITQpm4e9DHDuE+Kv9sfVxSjsEsNyYei/Hccfdl8dzSn+O4zjeLMT/wMz9422v2rYbR4RRzLiW6P2fr0+/g5U++i+FnHmb/5XupNwfUGxOmowaqjld+sNchyyIio3xx0/LkYcpL0ufa2E3gE23lXbVlfldeYkpGjYgVbSE4RQNwAVlD6kXbv/zhQlsoZJChyrZIyDnuwhw5zkKGd7sjvOZWr70dlafbHWWJtdt6/m0kV2Uy6eJ7lcci72YRXlSucC2aEUGQDb1Rdrbcpg+SvMddq9eTGyIImabF3zk+2VfzFjoa4f1DVSJ0Q8LvDvLD4rOVr2PZHChUsY+DEIbX3PgdSNF+VZ+Qh25SAavRvAigQls9ImJPe7R9R6QhNSakTMl4WQ54wmxSIWZbejwT7dJVp/ySSs516RWbhIOmWIbqguTruk9ba+yYMSu2QULMgQzZoU+PEV+LNkmosCV96lRZt02ei/ZZ1oRf+pufJYoyfnx5hdFnvg2dgs0jqu+8AkbRwxaVd9e48tI6b/ueP+Puc30euO86b757xpv+2kf56lfP8uCjn+OOk2NOd4SR5nz3iYQfeO9jDFLls2aT3x3tEOGgBA/m6+zKlPv1JI/JS4wkJcNyQp0/yDNmH8WZaw0Yc1KWC2f58D2MfBdjpM6EsCutQg0ttMhjYprSoCk1+h6yNfISlR1pFqRvp0A1FzIIpM0AwQsKaOW5s1jtCvNIiiQmtOZdJbMMmQmeCUaEOq7L0DUtej6grEnVEyUrZBoUncyROVg4uPvgJ9w74bGpOi5NUPNRLC3qTDUtugqXuE6E4SRLJFT4nvwe2tJkTTqclTUGOmZJmox0QhmaEUYiFSpq+J/uPMVrGV3TWthkjx+ZT3wC2bhwgpc58TjgshtSp+NN0MI9GMj/4dqEwGGqsxtc77/ZRxlOe93uEcwZwzoU4J5hBKhtGItd3pt1tW9GYv0v33/x4t963y9eXDZtql5Cdk26rguMc0OfkDJkwtflZQShQ4MNlmlolYSYZ+U6B/jqOVX2GTKUGddMr+gogyN7V4hYkY6XwHWKTg9zjkCsTrxiWNMTs0deuKJctFLfqXaiFoaxTn2127KproMTkoz72GCiM/ezTmlTI8Oyp04I5LSsEBHRxnViDnXAsrRd8uDX+r4dsmI6R5zSnadIk+BbETiM1ifEoYsf1o2qVObV8hDX+EJa4EpNdXbkswbokGCoeAnwYBJYYS4RHtakrnEyw6FjUR7l/S8IQYQqvsEpSLmkMyhd5Yxk6nsYhnNyggzLinQAWJE2a9IhI6Pmky8R58cxZEKXBi/rTtHxeN7skGrKlh5yt64zJWVF2hiRYk3c1H0aVLgi+1yWHU7SJSNnoGNmzKWsY4mZ6LQwoAxrKLhCTlPqrOB4Hzt6SNObRAaRi9R3ELvGQZwdHM4UXenF++uVxq2QJIvIi1cTVxavKRcbXiEO/pmf+PljjcZfy4izx9Y4fPluVv/KZ+msXHdvrBH1Zp90lrD+PZ9i65PvRFVoNFIqlZw8N3Q6E05UO2zPMu7TLg+uug++PxQyGzTYDc+YQyoYmiSoWAY42d6YyBOHqq+IZUu80Zgt2diH390QzDLng3wj8WyLMrOv5fVw/IR7pSB58X1fzXsstsnLxPDF5CdsOosOnMe9LveO42V5ucXXln8u8KKvIxwrnPPidYk94XIxEVhMkMJrp3ZaENPdAhgz8c634bWLrwsjvE/5eOXfhwU4J6eCkPoNI5HkCOYTHNZ2yqzgTwV4jUFINScXS0KVsU55iDPcZ5f4ePQcp7TLngxIyVmhxXUOeNieIhhuRWpIxKlAzby/REsa7NlDzsgZtuWQQ8Y8mK/ygunxQL7K49F19nWAIEy9ctgj+SlaVHjS7FLTmF/80RcZXD2PMTnved/HMad7mDNNVr7r89iXYnYefxP15oDK5Hm+45c+RfZHIKJUkimH/QqjLz3EO97zp3zpT99BmgrbA/i2Zo3/Z/c6m7/zRq5lUxqmQl8m9GVCQsw5mvx78xQbukRHGnQ04Zrs8Rgv0iAhF0uTGhNmnNFVnuc6Q79hOKndig+u3aYeDAmrUvFQiAkVvz6NPRFScPPCeELmofaL76zg7/gKY9m4y80J12pb5EQt3n9W9MgcNQiJN0TEq7IYhIn3BZhoSVXFz7+YiFRz6uLgJBPmxNEqFfBk1UIS2s8v5SgkK6jpWBysYVenWJkU0BKDsC5d+oy5y66yLxNSLPfoCbakz4yMRzjHs2x5cYSjXUgRYd/2ebfez3ga3bKk8r984J9czLBs6h6Bq3boDSfd5zD+3poXqsJaFvw8XKDl5JSHjAja/IEDMNPUVbipMvHymjWpYrUkaRzUcfxneT2Vvv4yRphzzk08Kjo75eTjuG5xmCd2YX2rEGNFC85dGLdS0VmTLiOZEhHRY0SmOQ1xyUVPR3SlWezhHRxUpyFVjAp1YlJvRnrKtvmqedkXOx0uf8yUOgl9xkwl9X4SDkqz7hOdLT3ga1xGME6VyQ6pSsXLxFYLuFWQao6JObRufW1IjbY0ONRB0fVsSo2e75J0aHBZdhGVAh6aktPTIQ1JGOnUu7BHbOlBEaQ2vMyuVaeg5UxRI7btfrG+jJkys2mROAZuV4D0hvUnyOqOrEumCk8OMYhUEDVEIgWcEebJR0gwYonpSJOeDv3+57omVaXg5ARyfq45a2aJK/mmO4rM0RKAM1fUlDXTZd/2qXgIqetoGP+7Jfo64oBhAfsKbvQhYeh7ywdBOLD9IvZwHY4Bl9kuOnbXxXW+Kv75L8keb83v4EvRFSbqVBNT3DlckX0HS6bFS7pNTMSEWQHTVJl7nQVHescpcWtQTSoMdOQ5RM4/ynVqlb5PWg9sj65pFcawoSjiOFdHxX3gaAwXEv5Xm6SEe7Z8nFsdo4y2Kf+8+JzbESK53RH90s8mH8wur1N7W5+4s0nlnj1MOmb7uQfY+MDneP5X30176ZBsltDuDogiS5rG5HlEq2p4YCMjGyUsN5WlVsY0jejPlL08JxPLy3LA++K7OMzhLfYMmTdtERGGOvEX3xxZ/GKpeLMdV3k42nB3lwgWIFHfoOTjxgtdalt5aFE4l5tBqcqvv/F5emTylCt84bm3d2w3IjEe+nQU6rTY7lu8jgGOtXgDlJ8/D8RDwjd/B7sQgAtStPMpfY5QjfmLQNhu/Czz4SpH7lytzoEEAZZVKdSy5nCwinHkuZnH7Tek7jtxrjI39bjdG8AHxyaJWpDGjN/0HafAKYsYvCOzuGuFSD/VAAAgAElEQVTUMHUmOnWYbTJmmlHzCzc4YvBQx3yffYi7dY2nZJN9RliJqJNwXQ7p0OC0LvHdnGLsE4cT2mBTBjzKCXaYcsBorvAhFQyGttQ5qR0SYu6ny/PSo6YV+mbGRGZ0xeGc17XNW80qP/aGlLtma/zn79zjN//gXtYaFdbf9jl0ewnTmEA844XfeC+9l85Tb42o1sZUHx7DbIruZSzd9QI7zzxAbODPHjvFl796BxUxJBUly4SnBzMSjfmy7hOrYUObTLDk4lRHhpJTo8q29Pih/F6+ZK7SIKHPiDOyRoMaL9nrtKXBQJwiX0sarEqHzHedTpgl968sMcJ1g4KZVIZ1cpi4TlSo8FXFGY65+WELGc5g/BaqlYudWUsAfBQzpjx7js7ZAroXFZupQAFBUCgw1V1pFRCOkd/oUgLMwiDiVGhmmhYOyjExiXFV3zAPyn3fucmhkuKgYRPSAgrrtPpT7uKEMwQT4VAmDorh7/vT2iHxmPVUHEa6air0dVR83o5p05Q6y7TQQY1TP3X/TVv7n/rIFy8ExTPwkDnvLrxkOkx1Rs3U/P3mKrxVLyQgzGEHDuudEvvgMvewNoulITVaUmfsYRThytSl5qEtToygr0MU/ZZLQAA++uFPXAiwwLZxMJEypOPGueq+sZCkzNdVKUjY4PYCI4bICyD8xE9/3yt+n3/1J//7iyumg8V1KQY65jzrzCRjR3uAkIuDAHVpcIddYdv0XCeBPpm4c+hojavmkCYOynVal7gmDm460DEpOavSpqU1KhJzxW6RYdnRQ2Y49+3w2XOsK/6IW4ddwj8XMUn92h8S0xynmhagym2pO98SgZ6OmZFhMAwYc0ZWOWRIjvXQJtftaUjCkDFr0kXF8fUGOmLZdBjqGItlpJPifSm65Vp8J6lXkXIwTScvbYu1yAWJkZcJD+tbSKRnOvO/C8gTt0cF8rTjpDifEbceuj/4ZF5w3/lEpyAOZpSSeX6D2/9WzBIZOetmmaGHcoVOSsMrDy6ZtpM7JyP2AhZ9//2NmbIkLa7aHR9LwKq0OcTdg0Yilk3HeYN4afFYIhoemhrg2W1fbJoZN18Tzzvx9SFSMupSZcgUFWVNuhzijuUoBI4X0pIGrnfjEpGutGhLnYGOC27NkrRQYEOWOWQICDVxJp9O3dDJflssE6ZHEpBwvwFFrFGGw98qDiwSmRIUfv6zKR3jaFx4O1CsG97L//5XP/S7F24Fu7zVMNmfn6L9yFfQ/RG9T70J+0IdxgmVSkr2VZeRth94go07n0JVWD37DOff+aeoCvc/+AJRpDx855g4Vur1jLc9+jyNWPixzhrvkpOsaYuKEd5bW+Pz0TU2tMUQV9WsSbUgGIURefWOljQIJPRw88HRTO8bKmHLK3dVjstMb9b2eqWuwmJL+7hjHCcDezvvcdzPARpV/gzl6r7xi89xXYvyc19J2jeMoqpWgp4snstxXZFX0yUJ53izjlK4sctqP8edB8DEjpnYsVcnceohU526CpROj1aO1R45fjif8rB+AYQbq4sO1z9XnRnZcaGWFarUQ2985O6FOlWpsGVGbJkRp2WVJWly2rZ5p24QYdhjwDU55KPyHAbhUT3BC+aANglbOuWkbdHyi3FdEro4fKvDArvK0e+Y5+nLhB0zIlZT6NN/f34PD9lVVmqGg17CUjPnxUun+Z439Kk3Jpi1FLPaQ4eJv66wfuYKvf0uzR9+keyTOV/55e+FJMWcnPKFJ7pculons9CtCbMMntoSvtKf0jQRdSIesEv8t29NeSza4lAmXJF9alR5gqtclX1mpHzIPM6e9tnSAydNScoz9goViXlnfkdR1bMoq9okIy+IpxGG6+o8BWKv/tTXEYlUWJEOBvHwK5ek9O2QiYfg5GoZWd9d8RCq8Cd83yGACdCqV9MWzzy0oixzGUy4mt5ZeeQ9PYDCxT1wQYLWf6jyg9toHanWUvFmY6q2kAoNELSQOEUSeQfhhAZVmtSok3BO1orzHDMjxvDGfJ2uTWhohYbGdLXGlvQ5qZ2iCu0cgB2J/YftG/gJ+zA/e0eXqrl1NW1NnZtDR5okuGKBqqXrHYxbpjmHWokpvFMC/yWRapFIhesZOoIz0qJS3NMhU515XX9XBJjqjKFOmGlKX4csmzYd07yt7/GbbZRVkwa+Oxi+7zBfF9fIMMprWFUqXvhjDhMMQdhxpPUwfu79H7zogn/3Hg2tclpWSCX3ROGEiswVyPYZ8IS5RoThbfmdvhiUEUAud9gVwJGUr8khyzjZ1FgiVqVNQ93cHjLhlFn18yMqVLhCIh6Jox6PdExLGkU3NPhduOuTe2f1KhOdcWB7THzBKpD3RzopDAiXpOnhPgekmmM8ZKdKxRmJemWsvo4J4hqJVDnQfpHkZJ7LZbGeJB8X8MjiOy1EFRICpC58J+H3Ve/+7bhZtiClB4i79UlK+H9Ye8r3i2AIzulhL5x4xanjoPKC4UB7TtIaV/UPUudNqdGQmkMDeN5NrpYGCUs0aUuDZWlRpcLQE9rBeZusqPPImuiEuiTcxUk3J32BUVEyHNQ4nHuIMVNyHs5PFh4tQ50URoO7uO52h0YhWz9T5ykVHNYPtU8scQHvc0ie2HGGpEVbGm6vkTYv6RaK880LbuohwUvJmOns2JihHFMswrjh5nHgIux9UZUTjkeOLHKoX20x/y8KzYp+8d2rH4weraMvT6k2DyHOiR4cURtM2L30ENZGdE69THzvLskoY/fl+2huXGOpvU+1c0C3e0g2a7Gz2+D8Xdf51Ofu5Hvf8TwvXF7mE9MdVrWBzQxvWBe2ezHnpEFFE3Zl6Mk/uYcXzKtcXdNmSVrMyIqJPs/MXj8y+K3GceT2IpjHdwNuQSaHcjfDUO5slLPUMqn56Dg+CXolQmAgg4UuxfzxxfdyG/m8azLPnAOprPy8eSdkTrqek9iPdldUFjoE3IhVPK6L9HollTVTO0LkCx0HQYpKaFjYyt9f4B5l5DR9S9iRxhNmwQEXD/2TcqXixhu9uD5lcn7pmkcS+83cFFXw4BsCrvrdMU1f3VJOywovyDZW4APmXh6RVX5LnmEC9GXCKe1SJaaNwyY/J4c0qHLSNrli+qxpHfEb8F12lb7MaJKwx4CxOPzyI/kpnjbXqUnCphySEDMh5U5doUfKA62EF/eENz6wz6kzmxwedNnaaRFtrtN68BK630J7EVeeuo/Tb/8c7XjA+KuniVs9JnsbHDx/nuEzd/LVKwn3bWRcOTCcaMEX9mZctmN2ZMw7lpo8uGq4d0WII/jsVs5Z22ZsctZsk5lxOvAdGuzTJyiXKMpMnATkiumSCeTiZBinzBjJrEQC9XKE4kigxndlKziVntiTNSsS0zAO2zsjBXHzJ7Siy4nHnLZbbmP7ecW8E3izURYzCBtQLE5SM7i45z4sCvjzAAcTXHUaT66fkZGU1HLKx8995T8Y84XnTJh5fpTrvnWlQTA0dL93OP11OuRYOtSoErEjY3IJBoyuQn6HLjGU1GvmN0jFsmq61EzCu6OzrFcrNCrCuRXLmcc+f+G3/uHzFx557IkL0/d82w1Vtcc+9JULfZky8hDJIENaNwkr0mZK6iujruvoNP4dJPOMWWPIpOgqGt89HDH2AbgtFIAUpWXqOAKv9cRif22lUsyFgR3xmx/+/Qv//sOfuPDjP/2e142g+Y0eH//Ipy+ELkCmuYeOhOq6+1Pu9ob/x8R0TJvUd3JdZ26uigiOlBzm+vt++vuPXJMPfuAfX/ytj/zRhYycU2bVBXG0PMzF3YsDxqzSKb4HN4dWWaeDivCyOaBFfR4FiLArA87aZbalz9jLKQx1ynnW6cmYLnUuyx4NF6Jiccn4UEdFJd/g/HasKKumy64eFhyRdbNSJGs18ccQ6+8N14mMMIyYEhPRlSYd6iQeznmgA5ZMu+CTRUTex8N3MHAyq6m6BCeQtBMq3hHerSJ1qTHScZFATT2pvCVNr9I2Kzr+kcx9J9x64zoITmTBcV5MCFKZI0gKHpp/LHz3TpAjJsN997m6TnH4vsOaNl9nfOBvKoWqoMGQiyP810zVQYuxBDp9gLRlWA8hFZZo0pQaY2ZMdEpLGoyYoCLsa5+Zznij3M0V2eUBe5IrssdAx7SN62wGCd0T0mVKyoSZU0eUlJnkXoQgx3UqnAeSg2mF7qAUccDADqlJ4gqEvtBR8V3WCSlNatxn18lEfZfGdZXHOnHn7btjAztgSgmmyxxZoqWrN4fFHe2ORGJuuo+EuLD8vHK8No/jbh4/v1aY1WvtikS/+M6NDzLqEz1yAiYHSDtH9w1f/A/v4f7v/wQHl+4iQWC3RrR2SL3Wg0rG8196K0ur24wOT7Jy59PUtEW1NuHsqQFffuwOzp2c8lDS5t33ZDx7vcpwYni4nUBuqNmY6zJhw3YYGudI3PfZYsc48tFLdouBHdzk1I/CjELw9kpE9Vc7jhyngCuFm1eLL9b4OlABx/ABdfmvLgTrRQJS3MTzyeiq9lokBserXB0/kYquBuK5IPPnHT2H+eIx7zD58zxG2WkRggb4YDoq4FYhIGchmbjxWBSB+GKyc1yr8Ajk7pj/HweDC0pAYbi2cuyxqC5gscU53HierhMxpmkadE2bXbtP27Tm3KXyZ1m4PvNHvYNp8dmCe/w82XPEQMdH8duFhwFVigpSkJRVccf9r2oPsVQT/un0Cbo02JMhZ3UZg7AnQ1okxBg6mtCgwq4Zc0m2SaTKedvhmhlw3nZ5ymy5ahYzpppyNyeoEHEoTsc+BHZ1qjxjdmhS4/Fpnw1t8KXLVR5ch/39Jnfdc40nn97g8Ll7WD//IpUfXOXE2ucYf/VBwHCws0Gz1mP1R75GZTvh6afP8Kb7+ozGFbZ7hhMt5WRS4cLdlvsrLc6cHLF3WGWnF/HUZoXrdspXomvcZVfZMgMa6ro4l3D8taDqsi5LTDw84W7WeV62SaiwowdsmBX2tE+KSzCrUqFGhXO6ymV2UBxROiiPWbTAQwPF60JwFKAAGa5TUVZ9Mx5uEOAA843+aNHi+MLFvDgBUJGqr7S5RxKvsBLKDOAqneqnY4aTfG6bpoMbaYBXzJVvgtGeFXWkcdH5vegTK8UlOydl2fkEMJernZHR8cGdFejJmDY1ejJlKhn36RKXTZ/vqizznB2yog32ZcRJlvhh7ibWGk2tcKYZ8Se7Y5Y14U8uVchQHjqpTN49T0Ly//q3L/7R//31C5dNj8vs0JUmGywxNikVKvyofcgpoYl45Zy48MDpewPKAa6S6aAUfk4b17GrS42mD1pGTFmSNmOdeSU6h+muiXNO7kqTvo6IvbLXxBclfu8jf3rhh3/qwrdEIjL6t5sXiCq8197HY3KlWCNdoDhXnywHQgbhRLTi4DPigjMFr3JmimRboSgG/PqHPnbhP3z4Ty588lc/f+HXP/zxC00fOC+LS/b2fBegqhGpONM4xXXWnE9IXiSBJ22T66bHjJwT2sIKvCU/TV9cYWFXBoyYcl7XmIqD9BzIABCsuPXtit1hRk5fhwUPIJDAy/f2yFfXLcqycZVuB0OMfeW+4pPvBk1cgjrE8cZycreAIz4xmRXKWbm4AHfdLDFmylDHdEyLCVME8ZAmKSR2Q9fF+pKDiBMhCfdnWDeC31EoauWah5yjWKvCKmNRprhEHAmdesM8+XTcLCekETpktvgugqRw+BMX82VOnIf5Glf1XicW1z2JxBTKciMmrqvp17OIiH3tYbGMdcJEp9zJGnsyoE2dng4Z4fgtY2Yc2j4VcWarI2aIROxojxkz72bufJ8yLA2pkpKzZ3tssMyO9ImJvH+LZeqV76oS05Y6GY70r2ixL4+ZOp6muPuiKy0GOqYjTd/pVV5khwETp4ro4VbB66bn+UNlOHq5OByGqvUdKS3UsMoqmqGY6153Y5xU8D6YxxpAsV+U0URRKWa92bgZ9+O42OtXP/S7F37twx+78P6FQsTNRnTxgw980Jxskn95C+3V0JkQPbLEhn0Rc35Ep3ONqD6k8vYe0p6SXlpHsHS7h0yHK9Rbuzz3+Ju5utlmb6dLPbE8f63B1zaFu9YsX3iuRuwDr/2p5VI+ZpsJb5Y1vmy2aJLwoD3Jy3KAigtkz3OCy7p9WxcpXKriwvwFEo9XPsZxwT9F0OFu7DJ3o/TKhYC86D4sHLPIWsOyEYL90uuOnOMxbbUQVIfK641SsfMseTEYKj83JBWLHZ+jn29+jqHLEGRry8nEUUxj6XqUErQyz2R+LeaVljJn5ej1WiRvud+XE5BwUzrPDReEhPe2aln8zgKZNffJgIPljAvHaFf9csZBkfgWMzeSOV3HLCoCxEXMZ6iul3kDFuurc25+NE2dll8Y3QIYsTpd4rfTq0xJOWRYVHeWtQEIMYaXZI9T2mVMxnntsGumfKddZ0DGPdpljykDM6NBQosa13QXjKEvMwY4ucAlmkRELGudddqMZcbXeZl77QkylNO1GnGsTMc1hqMKj/7w7yO1jOyLgumMsXsd/vCP3kirBmvv+nN01xDFE1pRwvXNZc7escMdaymHvRrP70EnMfzJlZyNWpUr+4ZLoxn3dWN+Z3aZNnX6MqWiEVvS4xr7RZBfkZhV6fg6qDcMlJQWTjkMXBu94iGeY6bEYrA4F10jhrY0iImLzWNVOj45m9EyrrI51mkpmTCF1GuZBxTmdXmux14QoNxVLN+Ti4u4S0JcdyeYkg11TNVUmeqMjnGwsqLLIXMTrKnOCv5S1SvRDHTk+R1HFQaD7HDuO9GBZ6EooUY+ZOrx1cqElDpVDIaZ5DRxr/+O/DSJxqxpg7YmdKkyxvKsHRBhuFc7POmT2MjGvCXpYhC+7WzK5T3DpemUCQ5G8vb7Bnz9n1698Dv/+osXzn388gWr8NIo47LpFXLRuTjYRlMSHjdX3f2Lg8Q1qTHSKQc6J7/G4uR2hzqiIhXapkHPOrz32BORe9apLGXYApJT81KsmeaMdFxUo10FN2NGyr/4tf/t732rJCAAz3zoiQvfW1nnp797h3/+4s6R7l3grc2TkLlgQF3qBV9oygyrOXXjOsUZuTfDdAWpsCMkpkoiFWZkWFGmmnKOVUYy88agEalknNQ2OzIgqE8FPoXiupRXzYFT1xQXIG9Lj4SEmWQ0qDKUKad1mZm4oG9GTp1qQXAHPJncKQWGskDNw5dcsWfmoVk5MzLWzFKxp8Ve8vrb7VkyUQ50SC5O6Sojp+YLRlVx5qYqyoEOSMlomXoBC6r4hDgE5WGNn3qOmYOhR8zInMSrzmWJEXEcEh0XPDBBHFHdf6aQSAHF70NyEfiJIs4rxsF03F6TEQzztKj0BwGOIEvuDEHn0B4BAp8sdE7maAdXRGkYt291TcvBrfxsC5LDIya+eOA4XKlmqLgOggo+wI/YoYcz95tSMU623vmzVL3SmV+zxJmSTn1xIMPBjldoc10PWDItJ0NP23HLZP46J87g1r+xOlndCTPX0ZKIvh0UvjEhBnDvkXuGSF58D3v2gJFOeDv385R9CWSuPBbW/XIMFkYQeSg6TCE+otQduQEts1gcLiU54hXQSvOh/J5aHDm8/vjOSJhHx8fV80T2yGtE+NV/+zu33RWJfuF094N6deQ+dyMjOgPZZ2MOL99HdTZGhzVMZ4w5AfbpCtOdk1SW9ojWDrC9FltX7mYyqXD+7k1WVgZsXlvhYBhxpiOMpoa3vmGXxy8nvOk03H8q5bEdy1lp0I4iyCu8kRXqGnMpOmCsE5pSZ5UWL+lWsYHPP/LxdvOhDbzI4bjhuTdJUG713HKQWwT7Et7Vh5ilLPd40jolHO3RAD/yQWuAQ904HfSGYy0+5s4kIqQypqhfzsmuoXMRAvAwiY76gcwrYYtBE3BDZydM56IyVnr+4uc42ok5moBBGQ42D84WuzflYy2ey3HPcW6rWXFuQZbXyiIszv0bMOwznbFkOiAOgtE1bTJPGk/98fQVvq0Q0B2pbpSed1R9QglmhYAnwloaUi/w/fdzikgiHjfXaFErXjtkyrI2uWYOmUmGiqtWX5MeNSo0tOqNmwz3Vxoc2pwtGTulKdtl34zpM8ZIRJeGJyFaVrQBItxrVxAVmuqw9ajhTc0mn9icMu3VuOvUhLvuucrOpXtodXfZff7b0L0lGm9+gvPtPsPDVZYfvIQsK6Y7IdpLqEQV9ve6dJd7PP5sl+9723UGgzo1W+XSHpzpwMcn2/zx7Don1UE0rsk+uTgH31QyTnrt96mm9BjS98F24onbZ3SZl9ljRdr0dMSyaTuDKV+1TKRabNwDX+HOPHRgRua399y16MPGppkPTOduua/UBSt6ZHLjnD6uPV6eNy4Ig8RUGemItmkWz0o1KwihRgwVz+lwAVFSPD7Usfu9xCBSKEHBXG4yrKfzdcz935lBusRkVTps6aGDM0iDARM2tMu9doU1bXLF9JiIO7YVd0VqRNwtLVSFz0XXOKtOh39FE560fQ5sxviwxmolopdbIoS/spFQjeGZzQp9m3O6UuVL+zO2ZUJbE+61K1jxK5wYHsjX2DcTX603BDquyBzXHrpSQXJYoEgmg7Sx835wgVGEMxMLCcrUQ0GCMMFUZzR9cBVJxMc//KkLP/hT7/qmSkL+8ft/5eJvfOQTFx77yFcvPPuhJy/83q9++sJvf+SPL3zqI1+48LzZY8fmTK+c5AtskvqkdF5pdd/fXKZXfQBdIZEKO7pPmK8hSLXkbgUTF2CCgxB2TNPJPItznu5KixWaXGWfltS4165xIBOuyQEWJwE99sIDkS8MWCxr2mLTE85n4pK/kaSMxQkmNDRhTZvsygCDU/0SXDDb0TpbHNKSWkF0XjYdlqRFS2r0AzQJx/loe3iT8fepYyK5IlOFKrvigtEzskofJ0Hd05H3b6o5VTmp+VTIdY36OiIj80FtteCbhNIdgud9KDOdseL9SEISHDx+ejrwyfbR4D8mKhSWnNN3BDLvWLggdO4DY8T4TogtuqMhcsAft1wgDWtCRSp+bQhct1KMUMy+EkzZxyEKrttv3H01YUZNXDHDyBxF4ZK4KU3jBCK2/3/W3jRGs+y87/s959777lvtXd09PWvPRs6MSGpIWUs0omxZkQUL4ZIFgSEjQAwDEfIlnxxZGMrxB9uwJSBBbBkxJMGREom0JUeBoH2zSM1QHFLkcFbO9Mz0Vt2117sv997z5MM55763qruHQyO30Kjqd7nLWZ/l//z/2qcisRO/9hCpuc5ZiVz7JBJT8+ypD9t19mTISCeIiIdOuXF4Trt0aDKVBX110OMWNQeRY4b6sRLESwEWOK2VY9unKlXGOnEBR4kZ2zGIc04HdsRQx3SME/4c2TFGImqmytu6g4hhEaiUy3ZU6afcdhWp+jUrL9o32HRhdgY77c6MyNJWKjsaMXHhPpbvIVzz7N/3cjju+pqH8p++jzAODB80GxL99CfXP4exmLUF0bOPwmgA8zn1CzuYjQnREzl6INidGDFK0j0h29/AWGXvvcusbOxz9eoGmle58LGvkKRt2lVDu5lxPKiQpxW+66EJN/fqrHZT4nGTQWYZ2ZwvRdd4RfZ5UFdZ0RYHZsJFWadGzLFMuGg2ObL9922IO725uzfcMkYjH7yRS68FAztsbMFwDQwT4U7KBvMp56OU7g6eaXkgLqMuSaF4XK7FCM93Okux7PTAfx+YMSzqKTsdlt0W2YGl0SyF46ElI2n57GVWhbKhHp5Nz2QSllH90/d+NjtUbpuzmMWwKZxtv/J1yzC301mS0BbRqUxLyEZ0TJuZdYxsZeG4wtsPDiERbdMk05yOaZ5iU6lKlYGOqEq1cGxOkyacTrOKjw7erV1CxKNYVMRFmAxBETj3zB1VpuJoKGO/EQV8/jYrCMJ92uOyXWMiGRUSVmjwjN3g69EuPxKfY5IrEcLbDHmcHi+a62xpm4SIzLjx0tU657VLgypX5ZABEwZmwSXtkqH8cHWTJ1s1xgv4Kz3i7zylvPx2h6f+5hc5euch3n3jEba299m5dp7aqEH9o2/Re+wdyAQ9jpm9/Agn++fZePxlzKLJa69f4NLWgi98rcFo7GoEEhH+qD8gIXLiieaIfRmwRofbesT32YeoS41dGZD6edISp5a+IT0Xqdacx+0WkUm4pnv0jONvD3CrltSpENOUGn1fbJx5nHJdKoUxJCyVbRVFRT2ZwJ3RpPeLFnFmTViOg+VRHusxkRchc5CgkJFzLDlwXtYYs4SOKEqVCnMWhQEW1itFSUiKbEdoBytaRHlnuvDvuWu3TJ2mNFiQsq8npGRcMhvMyWjTYCYpY8nYNSPGMudhu0qDmAu+jipF+ZY5YV3rDGTBBdtmX8Y0tMIaNXokzNVymGd0JKZpIi5vZvzyGxnfs1FhVSq8NpxzW6Y8QJO/MNdIBY5kDAIP5D2+FF1h5EkD6lJh354QYQo4HSypS7XUJrl37gNEpCl1Z+CJE3bMxRlsGbmD0qBsyyqCMGHOVGc0pO6ixMR8p07IP/3sv3r+C5///ed+9/N//h05MP/yM7/4/J994S/fV9jxX37mF5/PUMYyp+bVo0eycEKLvsh6hSbrts7X5VZRcBv5YF/IQFSkwna0zokOcUx+FTakw7GOCpVpR3ddoVJAalw2qiWNwpEWgYu6zk3dJ8fylL3AwMxp4IX7xGkchei08bDBRGI2tctY5izERZ17NGlolYks6NIkIUaBoUw5kgk1KrS0Si7KgQ7oSZMDGXkWQPH97ihy9/SYvo7JNKMpDYwYXzs0py4VEhyt65yUW/aAHCX3DFAdaTL1hvS+nhATM1YHJ5upe4ZAehDU1mNfXO4MWefghUymg1BJoSCekftzTos9JqwtQf8kKdkSLjjiEAiZr9EJNRxhdQqBLrc/2+K7gckqQH5g6ZCaws3xwUExribIn0vhFAw1rG3BzjHinMEFC4xfZ8bBLkgAACAASURBVMBlFoNzGmHoWwfNm+iMWCLPULagKjEVYupU2NUTlz0SUwhJRhLxJBcZyZyROJ63QIvbkDqXZJMDHXBJ13hZrjInZV2cQzL2tXQrvq7nyPaLdkYcvboRw7pZYcfuFY6Vg9y5zEbqa0dVLVMNAtzOCR7pGIulJtXiMwGaTbGeByicOwwRKWkBh/XW5qn+OmUrFMdp+zdkP8oB6ZApXwbEwufL9lc5KHr6uHfwvmRHnkGjiAi/+eu//9zZ+rC7HV5xzWI+9CAsZkivh84TdFjDHlfJ384hyok/YtHMoHO3KRPntHt98iyhWrGsrPaZXH2IbJFwc7dBf1hhs5fSaqZcudammiiVSsrh3DVqVQx/r/I4n8gvESP0qHCfrDNhTlurbMvqHcxZ3+4I9RofrAHvfdztO7kuCS3DEUtyiiEk3IPIkpVmCcMJSqPOgQnCPeEzNakhGGa+QPYsM0JR7Oy/cxZyFN4r32NZoO8sE1f53s6+VxRGlQym8P+zbF3l7wV8ZzhCG7wfM1D5PSnSxsv2u1sblNm7zrZT+Xtni+FP8v4dbRDuu3zenJwTOyhgF0M7dilZXTCwIz+R7R3XvtfzhXYpt0F5fARWsjA2Ul8E6FhbXOT30J6wxwkfzc9j0YKt6O82L5FgmEqGAR6zKzxkex7Lb3k8X+e9xZw+KYc25dl4hQu1mEftFk/FHVbUwZYu6So5yi0zpKUV/lt9klVabFsHnxnKgu976oRO3XL/mqWlFV56s8351Zxr//EHabYmXH7sGqrC5U+8SK11iFxoI92I/MoWX/33P0Y6b1JtTPjy73yS+azG5tqMJMn5cLdCMxZeGcxZ5LBnxhzImLfMvmNHUcuenlCTCl+K3mFHTvhb+WXm6mBHdRxzVCjUrkqFb0X7RGrYFsegM9IpU+a0pE5MzL49JsPSlDqp5oWWyKLEGJP6uGMICjiGJxd0KOvDnO3T92N5u9tcOMvmlpN7mEbu60KSQuArVweTAgp2ntjPuTpVRz/tDZlMM1LNSCTyEV133cAIBW7DDUGUmXVYbatLXvygwH5TD5mRcoALCuViaWuVp/JzfC26iQIHusACj8SOJSZH6cuUa6ZPKpY9M+bpVo0rMuAmExIME82Z2JyjQUKTmC/snfDe2OWoJqT8B/MmYxw72iFDLMob0R4zXbAiLQeX0GlRu1E4HmoLcoqyWGOA3eSaF1kRcPogHdMsmOg2pFvALyY+K+bK8E1BiRwYy76TI2DxIzH8zGd//vkP+r2hzOlqjX/+2X/9/P/7qX931+9ZlL5MiDEcMuRYZhwxcnVFfp05kDFTtXyvXi6+Y73xGpiPKpIQuHzapsW6dNnTPhVJvAiboeaZ9cDBnbqmRde0mLFgS1ZYkzap5rzLLk1pkGpW1HZ1tcZVOeRyvsaWdhkxI8JQI+GCrNGjya70qRCzqW26ON2kQxlyTnsoypgZQ6YMmBTjei45t/SIi7LOhAXn1dU0talTc644fXXrd+ThN7GfGw5e5ebEVOf0aHJg+15UzkFnBzqmSY0GFXbtkRezdCyHTc/IVpVKIW4ZjGJF6ZqWM7RxVBp1qoXg5UJdkCvVtICEAi6y74MPYT0KjkgIjgT2LvCBUm8kB5bFZe2p17TCFn9H4gRSA5wxjIcwV5zz5tZAN5echVFkTUrrXDCAC7YwXRR6JeGa4TmnOivmXcs0GOm0qHmZ6IwFKR0avt7POgdTGoVzF8YowLGOWNUmTsDQFY+7WkrX9laUtoflHWifhIg9e4Rii2xLaKOJTjmxg4K2+0a+W7Rh2MfLa3R47ook1GS5NkR+7sw1ZPaiUh9FFHUyRMvzY/3IiO7aD+H/p+H9d+4zob9C/4fX7ta/cGcw7P3Ofa/P3c0mNAi/+hv/4mc/yDkMUU70dBtmE8gy8tf2IMox940xK3OiJ2pIJWf2e+fI9jbRSY14/QCdVai3DwGoVnKS6pz5tEFSnfPkY7v0OnM2t/pkmeHRB/s0apbFIuGpc/DdWzHtKOLaNOVB02BCxt+63020h+06Q5mzpydctbsfuHHKRdl3O5bo1zvhM+UowB2veQO9bNwu05C2UL4Oky/2k6Pc2WFglQ30XG3h7ValWrCANbyGiuP6P+0cnFWzLd9P+QjXLhyg0kCLJSme56xTFT539vVTbSXm1L/wvXCtIFwVjOryfZ51XO5GRxwG9enMwfJ3MNjDOcqUdKFfljCyu4vzfJDnDM8z1RAVNwXcwF333ucNf5edIKVc+2JKRaHL+y0vdJtmjQoJq6YLwJZZZapz/jh6q2AralLjeKb8d6vn+alLa2yZKimWNgmXbY89mdKXOQmGLakRIfSqQmTgce3xdjbhneiYc9rjKbtGgqP6nUjKy3rCtu1yYMZYlFWt8fZ7XYyBRSb8/UcbfPyJASejiO7qMQf7PfZvbzI86WLO9UnnbZjMyN+ss3f1MfZPYhort/j3f3IfaytzPv9ij9X1E3771YRGVXl3uuAt0+eLiyMesT2etOtct3ts0fV1Do46tUODW3rEF6Ob9Hyh64gpXWnR9w5ijYRjRiQYBkyKvqtTJceJTN1nNtmzRwWWt+lrb5w4lxs3IQgSaH7nOj+1wd/ruNe4Prtp3y0wEBEVNUlBeGyiU8RDrEQMx14sMaxjgeEL8FKbgYverXUD6wrsQzQ8OGoWJVWnKxI2zCqxg4D4KFpTaqcCHj+QP8KqNhkxYyhz3ogOUJSRpFw3A1ZNwh/aHY5lyjvmBIvyrux7A3nGaOFMkqfiDm/LgPdkyH21Cj939C6KciQTXuWYP43eoy8z+joi1ZyLusIz9j5mLDhUx4p22x5icNh4RZnqzKtE56641wuVlWlhQ8Q59kXsIdMUoo0jnZKRMWRaOL5t6kUhelWqdE2LptRZkPKPPvu/fmBH4hc+80vP5+Rs0eOirrGurQ/6VSIMb8otZxBi+eVP/5/P/8GnfvP5X/jMLz3/bz/9K8//j5/5x8+PZUHVw/NWabHwIqNuTlSYMEexvGwOeEp63giPC3HUiIiOaXNe1pgwL4zplJwZTln7wPYLjaFMM+q4aG+XJrlfw1wgxRUHX5A1HpJzAMzJeM8csSsDLuoKr0S3aVFhRZtcthusabO4ToMKVRJ2xI2hCXOa1IgRrqvTjdjXE6w6h2ROygkuAn3IkB17wFCmHOuIPU7Y1WOu2z2s5oVzum02UJSONIjEMPP0zCkZb9kb3ph2jHup5oW2Rdhb57pgqjOMOFHB1AcP2j5TVpcqY+ucpKnOS5k6y5EOvNEshZEeCEky8lM00xOdoVis5qSakmpazMlMs9I+752FkrhzMHSLPZGyjtdyHQu2Rfh/mC/l38LSYbnb+hcCc4lUXD1bySEa2UkRXAGokBTCfXXP/heyRSueevuWHnFD9wvh1bLzOLITvs57WCzvyX5xD6f36ByjQhVHwd6RJjf10DsZSlsaPKJb1LyjGZ7fBR5H3r7LC1usHFBcXq9eBHRy76CFwPTZgHERlCVAtMPcjk45cOH8Z2mz72ZPnd1Hyo6HiGDOBMNDvxd/f0Bn4/2OewW3P+gR/cz/cOFzDMfYtyfISoS5fAm9fYB5eA2Jc/RoQvTEOSYvrSLGEq8fMnjnMRbjLu0fvYK5Dds/8TWSI0vjgWtM97apt4bkqcM0JollZf2Aza0jDvZX2T7X5/rtBp/6b/6I9159BAV27IxLSY3x2MVfvml22M33HZTojuNehuPSySga59s0ximIVhlKU3JAyp9dfn6ZSouJfIrVpTqdCJYtDSZX71HzeL+UjIpUCXUAxg/wmlTINHMCNm7JWV5bAjO6u4Pir5BO85tqwH+G+y0coRIcKkT3QlpvCdso11W4IxRgn2awKgnplN+nxP6kS+rfu2Hmy/Uxob+WtHKnP3O61uM0W0R4v8wKUb5OOGdIEZdfC8/gsmf2zFhRKqaC4nC6sUREEjHRCUF7wWFOnfHiFtzTLCHlvgv9UW7jSFwRoPVYXpFlPznYX8S2rHi14Bo51glf6YSWjxpdsB1eZJfPPljjn759yE99PONXd4ZcpsNVGTORlPu1zQkL/vbDEetJhV/p36KT1vnoVkxPq7yTTThnW8yw9GVGLDGP2FVejW4zEidw+KnGRT75ILxwA559ZMzG+pgHPvISq594lQcuXefmG49z+6DOgw/dZmXzJtn+GrVH30FHytHLH+Vgv0e3mbNz7SLDSUwjMfzRoM/vXYNbMuHPpodcMwMu2A59M+N1s8cbZp+qJAyYOvVy5jzKNlNJ6UiDa7rHhvR8LUheCMoFOE4iCScyZqpzX1hcZazzQhBrLilNqTHSKSLC0BdSx55lJvXRoyBImHvDPIzE0zArivn4fuvTWfjhHWMFoYATssQEI0JdaqT+p+0VpZte5Mtt0EtIRc0b3c7BglDAb7G+bqZSsBuFNSyw8Ex8JDZoqkx0Tts02JQuY2a8J4cFHe+qNrkhhzxpt3lJ3uNYxjxs13nN7GOAfRkyYcZMFxwzpCoJ39BDKsTs2QUdrdAkYZFBojEvR7fZ1g5/zhv0pMWRjOnrkJapc0FXODATDhlgxDDSCQ0vLpiQnMpKTHXmp5IQoDixZ8zL/N9NqdG3IybqBC1Xpe373NKVJjUq5OJqFXb1mIzMRWvFBSCmOBanMTM+SHH6P/7s//Z8hdjDDBvMxEUs/+rz33ju2f/yI+/7/X/+2X/9PECLOgOmWHG1ADUiBuIoTacmZVWbzCTzxhDU1AmiznxkeKYLztGjpVVaWuFVuU1NqvyUeZYX9Dot47S5Ug+Jakq9cPIBetL2IneGnrQ4J6vMJWWmc9akSyoZa9Jh4cunq8Rs2y5XZJeOtJbaGEzpUKdJFYPQlwkpyjU5oOKFLle1yUCmrGmLFW1wQ44A2OGY2MOc+h5G15AaFWIOdOCcbh1TlxoH2i/qo0LNXWISMnX021Vxa3wR1cbV07WlgcHVOAVIpqK0pMGePaIiFY7toNASEcBgPItW1c8hZexpdTMPJQqF2UGroyIVxjotmKZCzUhNqgztyO3vSDE/Q0F7gF65/d8UQS23J5vC8A8h19hTk6ss2fRChqJskKq3R4oAIEvgaflfsC3OwrDC2peRuWAn4umO0+JckTioc50KI52y0JS6VMl9QMRBsSqMdYbBEOPaz63V4wI6mJOzZrrumcU4ghaf/Vho6rVAco5lUozHRGKsuH30RMeO3lhybtuDU3T6s1KGFAkF/sv9uQhoq/XEDJGn5J0Wmapgq50mp4m8jZh4x0V8kMSR3gQ4XrDxlCVbavjb2apL28+ddwmhCwGsAKNMi6BLYDc7bSOH4yxBytl97G5MWOFzZ+t3Ffi/fuPnPlAWBMBIuwONKuahKuTO84o+/iBY3yljIf+rPV74y0fIFnWiR8bkWUKzu0/6Qg+bV8hf6XH9rQ+DFVq9fabjNue/90v01vYxJmfY73G8v0maGtrdPkkEx1ee5K8WQ3KFj1TafOHwkI80msxlmeb6/+uQe/yE97SYUKcdl3L0vQzHKkdDLFpExd3nXFqtJrUidZirY10JnR8iquVsRTACKn5hg6XInqv1WEY3isLLklMkLBVew+vh++G18vlCRqH8rGc9WOdN35l6DZ59iO6H74bISsiyhHSv+6wWC2bxzKUIa4Ar3SuDUc582FJ/nf199vMhpXz2GeyZPj87Med2XtxfV9psmBX3fBLRlIbT79ClcFOZIetUqV4pw1E+ytC5sqBdON/IjnHLp+vThIgbelB8t6EVauqi1tNpzPMf6vKvXqzxj55Y49Pfc8IFbfBhXeG57QpVIr7yXszhaOl8/dbukF+Zv8e6bTCRlAhhJhkP5it8rOmoBz+cn+Ov548wz+CLb8f86OMpL7za4fCwza3XnsEe1pH6gq3zuzTrOZNRC5snVJ94B81cW7dXdtm+uMf+cYXffdfSqsCXrrsF90gm9GXmR47QlznHMvWGiGWkUyY648Ce0JI6M8m4rcfcUGesnDBmqjNWpMVYZx7j7agzu9QZ6KSYO9Yb7Q2qrEiLVHNmmhapekfVqg6u5SFXqi5F78aIKaJLZbGw8nGv1Pa9jjI0K0Qpg1EQMhXgxvBQR2SaeVEyt2Ef2yGCM8JmLJizYKQT1xZFVM2db6JTauJgDXNdUJVqYXCIGCfE5mtMAvtXDReVrZBwQw88/WXGqjapEnNdjqiQ8Lq5zTlZ4THd5o+id/nu/AI1D5GLCFxfxgvNOQa3Da2zZ8b8VbTDa+aIHhUezzf5sHQdQ45/9goJD+gGFSJuccRMnTEddHxiv6b1PVxyqs6RVtXCAQlHyCAKUtxfEV1FCuXjIx0yZEqLOn11elar0qEa1k9xML+WNFiTzgfq61VtssBp3EzEZd925AiL8vOf+T/eN5tiUYy6mpQxM2IM+2bEN6M95mRcM0d0aFAlZlNbdGkwI2VPBhwzIsfSoEJXmqRiWUjG6+aYZ+RBHmGb39brPGUeoi5VWp4Mo68jqsQFw9Sm9LA4WNGqdGhQYY8T6lTpmQ6KZaYpPW1yoP2idu1dc8AKLRYek3+gfZq4mq4qMTtyQkOrjGTGFg5q9WS+xYlMeMiuYYFXzA02cN9dlTZdmty2h85o84QLJ4zpSaugrQ0BzMDK5Axj139N4+qd+jpyUEccomGmcy966ljmnKifmyOZZt5grnHbHhbMdUHnBGBLVnzWwxnQFa/11BCn3RSgSKEWx0GIKgUUKwSxQk1guGYY5+H+xQdLUl0UUDpw+0h5HyoyF2dQBGezHQFmVd7HAkSofK6wjt4tkl6+7lK7yTKzM2rGZSdCbUhMzLGOijl8bIdF4MSwFFttSI2eNFmVjoe6OchlJBGZOsjWqrQRDMc6oidNLJaxh09HmIIZTQi1HDmHDIklok2NAx0Uz3E2G1T+u7wWl5/dIDSlwdCOCoh9gD+VYVflc7v+X9onGdkdGYvAkVgOWBWSuLrso7C/haOcjQq2Q4CulSF6ZdvzXpmLD2qDlxEu3+keCGD09i46mmFvTrHXBuQvvIXu3ABjwBjM5RVkJeeTP/rntH/kFaRdZ+Xyq0RtBwkQkyP1Oasbh9h+k+moRxRnHH39Y7z45cc4OGiTVFJWNvYQUSr1Ia8fZfzvf7rCD/c6XGwZHj+fM5eclyYjqhpxYgff+YZOYIO4E1r1ft85e5yFTNzt7zJur4y3Cym1SIyn92wUC4djW0oLB8AtYqfxnVWpEnDn5fRc+SgPuJAaDAtSglNrDa+dnQCA50m3xSAP5wnPV/7svSBddxAA+IW6UIz2Sul3g3up2rsqqRcaHJyGZZVxkAFKdjdoVni9fN6yoV9+tvJzlWF8d4PqWVwh725+wGa0yn1mk4ycD8sD/p5iz2x2GspXhu/drXalfK9l+Fi436ZpMPYQvYGOiTC0pM6m9Ohqg4ksODATfjh/gH/zzpj9wzp1In7x1YzPvQgJhpflCBEcTEuVNydz7rMd1pKIJ+IW9+c9LpsW92mL22bMtm0zk4zfmeyzog1umxEplq/M+9Qiw+Vnv8xnf+IvuHhpl0Z7jNmeMn/nAVSFx594j1ZnQOX+66Tfuo/+K0+j0wqDo3PMJnUqsZIgvD3M2K4mPCldKhozYc6lvEuG5ZK2qalzLjboUiFhQ3qsmHZRaJvhsPkPscXQR7N29ZiONGhInYEds9CMrtaLqCC4gsiZLtjTE8bMfNGoMyI60vT1EymH9sRRsYY6qPKGW9rQyxjfcjr8g9SFlMdFMTYlKoyBTJcire7eHcRCPMuVU3G3Pjs3K7DmAXrkWFEir5RcK2Blx3bodWmSohCzXHeXa15o7EQ+Y1T30ep16XKZczzCNgcyYuQ1Dh7SDT6R388hA67JERMWDCUlUUOPJqk6+kqDoUrMY3aTgcx5LGlQ08SvV84JeCs65PflOl1pOQOCChfMBlflgJfNTawGQ8C1W6jVCcWbAWFtNS829fCs1j9rTSo0PEGB0xZxUcmR1yGIMDwgW6zR4VAdNfBYZ5zoyDtUhjZ1ThiTkxdZgm931DShRsxIFlyVQ2rErNBmLAte5/r7fldxtNM1KnRo8J4cFs9zS/p0tUGiEVdk30HYmDBhTtVHTh2Tn7v3IVMq6kRIV7ROg4REI9ZtowhStaTBirTZ1z67ekzFi+clRDyiW8xYsK4tP78sGRkTFlQk5prs05EGA5+h3NIOh77APcfSkgaP2k0EYejHUOTX423bRhC+Ft1gR494Ua4wkQVNahwwpCk16lToaqNYY8N8yzT3GhwzlGUdF4QgpMtEdKVFXao0vHr5gpQKrtbFemelgtOZMbiC8VXpEEvMhnQK7aAwrgxLWutDdSrqiSSFIT3WGanf9wHPjEUxv1LvaMSeICIot4drAAXFbbAXUl8LBhTjPJJlAKO8ZsGyFiTs+WX4eMiahPVMvPOQ+jlTPkdhC7AMWJ7doyOiAuFQIcFIxMzOCohSrg6SFURhQ3vs5Uc0pV4w1SUSMWfBiY4Z6Ji+HVGVioPUevTFLT1iRw+pEjtBWl8TGO5r4qGVN6yDa410Sk0S1mhzjhVey97h0B4Xz5hpWgQBy20SanQTL8Qa7IfQlgOvZ5f7etJg45WPu8GhykHsQBxU1Bb7vSD3wamzzk84Ql+E8eWyfBVSXRTfC8/htNKWGeMwHsJrd4MH3w3+dfYow+vvVsv77Y7oH/6d+z4nlQTz4HnELGCxgDRHsgUYIf+a5dd+/m/z1HMvY1Yg+2ZC9OgYshSJLMkPzsm/2aa6tUv8+BG17h7Dqw8wGTXYWJ2RJJZ2d8Co38NmMV/75kViDA81E46nymuTOS/0J+yaESNZcNuMGDFziqFnGJHuDXUIjXpnRDu89n5V/+X3lpS1TrQncC6HtBi4TS9AjaqmQhFzkJKD4TGmiiu+KpwNEV/E5FJzdVMvCuozHE1gcBRiSTxnd+4jfssUX/gNLksY0nRLH3wJTwupvkBlGWjbcs2IJMb4H1Byr4ob7i+0wemB5mni7tIHAXJUZs24V7u//+G/JwGi4u7P6lIkcXlv92LKOntGL8imS071s8wQ4d4KRhIxBZVjS+o8oOtEEnNFd7Bi2TJrngt+ec7w/VPPcuaeTrNSLJ3AkKlY6KLABW/LGk65OuNxu8kV44T4trXNo0mTn/z4mK9eafB7XGdVG+yZMUcy5xNs8BujXTIrPJDUWFh3T+/mUx6q1uhR4STP2WfG6+Y2m9qmqQn/4HuU1683+MkLPT62Dc9uGqoSs/Pu/Vy4dIM4SWl/7yvI+R5yOKKyckTjUzm1p44RhjAw1J+6wugbT3Ht3W1u77eIIvjD0TEPmSb7acZXZA8DPGvPMxCXrv+KucpUFqzR5jbHTJmTktP1olBv6g1qUuUJLvAK16hSISb2UfaUbVkrClXX6ND2xtSentA0ToAuRBdb0sCIiy47FeWpp+91FM22rHnjx0Qw8ENMKcynAMlwML+oSEmX4VdnaRrL4yCM9qBEbYrtY/ldI1HBhuUynksjzI0XH0n11041K8E7ar6exBkJAULgtAgooCoZDrIQ1OJzsSxw4mmRRAxkyg6HLEqUo1USrpljQFjXNlYsXa0zNAu2bdsZjlJDRflYfh9fj25yya6wn6fEYnhXDnjUbvBatM8Y53jVSBiz4MN2m4HM6evY0XmKeNYgt2m6+paAp48J0VxkuSE6aIXbgOc6J5HE0bXqrJiTkUS0PatTW+osyJiyAAGrziBKyXiELQ5k5LUs3Frxc1/4B3fADv7ZZ3/h+e87w2L1yudfeS4X5UhGtKhzJKNirE2Y8+oXXn/uVz//28/9eAna9fc/8/zzv/v5Lz7XlBoDJhzrCCtK6gt3FS/wJwtOZMSTdpt35YCUjIEdM2LmYEdUCiPK/Y54xm5QI+YVc5uL2uWithhJTioO+lIlZuKLzCfM2ZZVzmmHm3KCIOxKH+Od1HU6xETeYTU8oOsMxAkIfk2vYEXZkh4pOT2a7MuIKQtEYErKHn026ZGJo4YeiaM1FYSpzIt+rlNlRspQpvTtkIoktKTOVBd+f3XU04UorWgBvdo0qySSFPM8GIpOT2I5nsLeOdYpubq6IlcvMmfCwmt5LIqCdESKzwVtkBxbUIAH/ZW6cTofcxbUfN2Cg3m6p3O076lXPg+BDse+GUKsy6zeEpZcUM+DZ+4MdLyegp8gYVAOpgSo1fLHkVuEtW4pWlne5+98bblHS1j7wt4uvj6zWEehgnP6Jz7AYnHQqcTDxerG1ezNvQE91Rkq0JQaAZq1hOlrAXGLJPJimG3PeOigbDExJzok05QJM+a6YKhjbtl9buvRHXCmssyBez1A9MvomYBCiZZBXgn7gCnaPvbZ3IpUyQPzFlq87vonCE1Gp+w0UBKpYP34KWfcDFEB+T8LIQ8QPLdWLiF6OY7WPcIxnIWsUqByzj2TWjE+zuxXd4fGLyFapwLK/jPfiVihkXOXIJwkipCVCtQiiAx6NCN6fME78xkS+5RLc46stIg+VEfHNU5+/RGkPQFjOfrDj5Nf26C9csDG9g6TSZWLD7/JaNBhOGhiImW0gL15xl/0xxylObdkzLFMScmcE6LHBWygaN4zEe67ZTDu9fq9Pnu3415wHXBGYa75qUyBi747A9kZEKXINrZgTZjp3BXxSc0VS4qLIAX8eUjjGYSZX+Bc6nCx9JolKrInIaph/CAGioxLueAJlp512asPboprH0umKeXU6/u1S4jWl9sBlhGR4JXfLZN11vnQ0s/djkiWEYUiFewnV1HsL6ehZd8uAh3OUYaG3dW7Rzyu1aW+V02XA9vnK/o269rifrPFmum5iJF+ZxDC0E5nIwshahEc2YBPX1EXeU2IOJYpF7RXqJvXY9c+/09+nY5WOTYzUsl50HbZswsesj22qFGLhcvthP/6mYxt6rwynRCJ0Cfl1eg2H7LbbGiN/2K7xZ9+L71qtgAAIABJREFUbZVzUmNrc8x4GvPwh16mEls+8Z//MRil+uw7YCD7C4u52Md0J+Tf3GH677oM/+Rpdt98iunXnuDG1fNsbA7YGwh/cjPlQ6xwM5/zhpygKLsy4KXoNgcy5rocsUGXHk0mzJnpghqVIk2/q8esmDZ1qfIK16lTJRIHcdiULpm6YuJtWSXC8Jpe5TbHXNN91k2XmS4KdhaAA3vCXn7EXOcM7JAQUXq/TNqiVKMWxmUZW+1oQ0+zqJTnxNksXljXwmsLPw/DdUOULPbnnXlqyoZncAqp/fD9io/qO+PGwULAwT+rnmWrEDdTF/ENkMIQkYyJC2fLqpLgCrj76uBOAD1p0pYGOZZd6aMoNRJGMuO784scypSaunWoRsKACU2qvBRd50Fd54o5QMXBoxIi3o6OGDLlfnVO5NBDjm6aAcc41qKFZkx84Tm4jO7CF+i6jI4L/IQAT9A7KfeXiIs8h0LSkF1seqMwImLMjBY1NtVBQB5ikwZVKiR8S27zoG4UY6NOlf/+0//wDiiVQfifP/svng+1HOEYe3KFB+wKW9op1r116Rbj7Kc+878U3xGEulS5ofsElrypzunS5IYecFMPaVChR5MKCe+ZIxpUeEy3aZumy4Z5uNGUBevapK01RrLg0VaFIU7U85YZ8g2zjxXl6fw8M1x9icVyW4/oiMu0rFhHab1Om0u6QY8mT+fbzFiwqg06WmVVWxyKq53ry4yWNOhKiyEzaiQcMqDPuGiT2PdBXybsyoAdX/sBsCbtAt7coeEdcJfRbJoGj3Kh2Acqfr2c+DqDsA/2TMfrLU2ddg6GhlR5iK2iNqhCTJt6UfS9Jm3HECYOotjF0bSP7NiRRLBEEQSDsypVQg1Z1dPsO/pWpzMzVscAVmFJXGNRquLG1kwXqIb6wiXBSqZLqE45A+GCDkuhTgfTubPwPFwrfL8M5ym/V4Ybnc0Al6Hn94rIB/hWMFrL60o4V0peZA0i72RkOAa/QK1usXRMszhvQkyTWtG/5WNinV7IrXyfmIgGVa7kN049U1h37wajCnbK3WBYZ+2hgCwJfR8kEcr7wFlYdsjKBmQKLAltwjVXTffUOcrZpmAXhprggI4J7XmKibR0jYUnGwnnqkmNAJEL2S9XgqCnnvNs9uZssfu3O8Ln/+/voB4EIPrpH0k+hwicnIAqVGLM5ibS7YFJSV9o85/90OvEf+0cYDGNDJpN8m+OkCSn+eMLJDuk/42n6Wy/x3D/PmrNPjfeeZj7HrrC7WsPs7J2CERYa3hnt8KHNoVHOwn/ZnKFubhN91gmrpAMV8y0lx9S9riCyXo2e/HtnIxydPvbN2IQ8QNK5w0R0cJjLRmPBb+6aLEQVT0uNZYIp8TscJEWS0dazFkWO1sscxYlL1qLjTUictE4LEaCyF7FuxChqN09W+qF2NR76YIs740y3Cc4MMZzebuCtdBGITNyVgU8ZIKKginv0Yf/h2i/whmj/N5Oxrfrn9MOyuli9RDNCQVlyynmXjfFfZ0+wusho7Ps6bJKe1io3Mafa8666Xm+d+VB3eAl+xY51uPvs8JpuFe2rhwBD/dcfp6wSIRoiCDcZzY50TFjcRm1B3XdfR9lXRvUSfjMUym/9dUONxlzn+3yKB1uMCZF6VDhf/qbt/nGO11GmXKysPzubacb/F31JvVYOEgzIhJ+IFnlJ3/4Ji++vsLMa4osxnUevn9A+9wOvaaSyJydKx+mqUNYgJ60iC6NMedrTL74CDtXH2Rt+yo3r97P8LjHzf0621sj9g+atE3Mt9IxD0R1XueEC7bDe+aQNnVGMisoUPtMOKcd1Bffhahg0PQIAluJRMzVGdS5aMG7f6RDLst5z5TjuOATYg70uFTk6KLejqd/qQUsLIsPQ7bPlMZnEAMLQQQ3asKm4mqyykrTQWTKbd5uHoYNJT8zR8L4jj2ExkU63TsZaRHJCgXWIdsSS1RkMpakFBAw0EH4MIiphXUlw8G+xBdeWixNH/hZaEokhi3pMfIFnhUvDjbUCRWpMNSJzxq4LFYQOHzHHCICfZnSoMqhOFhOhZgxM7o0eNiusWOGDMwcg2NSEoRMLBViFmT0aDJg4ktKl/jnsc5csSsZodB1Yqeehz/3yu/GjxGnsOzoezPw/SA+8NCUGqmPYjsWojnGz8WZpEQY1LfP1Gfmx7Lw53H3kkjMH3/hxefk1/ae+4MvvPhc9us3npursq5OYPKLn3/puT/9wpefG8icLjXutz1umiFHMmZVW1xnHxFXPA6QkvKbv/6Hz/3BF/7iOUfIMCNkxBaeCWkhGSYYeygdrXMiY3q0HEuUTPxK5LJCsV9b+jKlToVNbfJCfsBEUhL/XiAbmEoKAu+xT4i+f1Tv59BMmErGJh16WmNsFnxXvsU75sQXOhvfdzkH4hzHc9plKu73whfMT3XBhvQY6ISP2vvYNUNWaHFbj0GEJlXqOJjevofDjUqscLEYBjqmZerMJWPXHtEzbcY69Rlrt4erOEN626zR9MxIC3F75KY46t6RzJgHHS2/f8XEjHCQrpapU5UKO/aAqqdgDUDeHBc8dIEHZU7Kh8wD3LD7jL3aeU2qLnPis5QOquIK0GeeACJkTHLvGIesQCRRoU0V1puw14bMRdAEWx6n7aOlwenWhIBuKGdfy86UYIosUSBrCcXU5SwIcEeUPOxpwa4IQZKZJ3ZJ/Vpt/LMFiKCzU3K/9lLUfNWligp+fbcFS+WMxalnDkKRkUTseQIJWELj4zP3v2wpvN3m/lcpNFm8eCGOnMGhIWJv0yzX1IysoEYPR0CZ4O24SIzPKicFa6jLcrjAgAs6zwsbx62zARoV+tEdoc5WwDMhhn1JinUt2CGKy4aENTLYMoJQNRVSL8y4wAW54yITo8V+V6CAeD8Ux9k6SOXXfvPnvyMHBCD66R/rfs6srUKSgM0hzZBa1V2s2casHmLWxRWtJxWYzpBLD8HwFtLK0OMxmkO1dYy0przw+8/y0FPf4p03HqbTTpnPaqTzKotFwtaFHV58c4WDMaRpxGGqpFgaJLylO8x9hwx04jfI5QZ9qhG4E3Z19v27/X32KDs19zrfqWKbEszCsBTvCywbrvi1TkNqNKTKQCdFIVVg9MixRTHaxG+oLsXrcNMu1bZ0eAAqXmE0I/cDx+P5fCbFQbNCNka9QbwUFoqJsaLF5HIL0vLzwejNNfNCR/mSiaHU3rb02SwsHP79IHYGS+anwrgv/dyrrwp41Pv01ymIVyk1GNKhp10WZ7gE6BksBYBiokLdOfRhMDpdetQxjdWk6hSqEYZMmNgZG8YpNx8xRFGekgfZ0xOMLKMjZx245R0toVdlWJt7bgrDNkDnjhmyYjosSOnSICHiQ3aNDW0wkAX3a4vru1X+nFu0tcZMcnaZYsXydze2uDrOOLi+wdV8xv2VKpe6Ee2syoP1KomBTl3RRQzW0JCIl97u8MkPT3nhlvKXssdsFnPzdoO3Xn+Ar77d5dFz0Or0EStEK33MxT56q8H4yw+T1EZUK5ZK95iDmxd540aNRkW5cqNFHME0g0Fm2dEZEcLL0Q4d6hwzZsaCDboc+jZ9UNe4ISdFuwS6WEf7WSnYTtzcSJhrSlsaDHVKqhkH2icSU6T6R95YVMFj/F3mVb0DEjZbQWiZJrnmjvnGQx5WTY+FDxSELKewxHeHYEAsiZ93bt1yTkPix2d+amyWIXghwFEEIVgKVgbYhRM/i4uNszACcHz0xj+vituQQmF7MEZizypjcdHncD4I2hmWOS4Ts2K6pGQMmRQOWuKjuV3TwuBUiWcsWKHFlNTBbLTPBVbp0aBDnSOZMGPh9Et8kKBHw1Gwy8DDupxE15yMLe1wLBP6OmbMDBVl5tlzBh6K1JS6Lz6PfKZHCia7lqkXwYjUQxByzamZKo4ytQbisgtVSRjrjJnOmeuCdenyEJvs0ieRmIFOqErCfXaFW9JHPDRrytxpp3iox4wFbWnwgK5wJDO+afYQpBDZE0AFnsw3qGjEjhnSlykJMWNxee8VccXkRwzDQleM+RCcEJFThlGqOevSoYdTC1+QsSBjrDM2pMuQiYetaQHDWtcOO3LERV0hJaetVSaSsaaOFWtNGxyaCevaYiaO5KErTY7EQRxHMmMscyaSUtGYHTNiX4ZUSaiTUNGIW+aELe90XGXPsVPJgo/mF9k1QzakywwnCjkzlkeC2jUzB4/UOXPJOFEXMU8kpifOuXIMRI46OMOyZ4/YitbY9UXqznmOqBpHfduWBhvS5YQJ+3riDV7XhwcMAGFTuhzriNv2gJYvWG9LgwmOUnek4wI54FH6WLUYH0Cs+THl5k+G9c5cTSo+C7JUu17oAushjmUWS0Gw4s7pMh/LGsuga7ME7rqAQ1h3QnCjvM8k3nkJ4ZXgfIRdNhiaAfoVgpABWhocF+fMUdxnuIrBBWuWcOvlWlaIAMsyEKIsnf8Q3Q9CyiKO5jYlLWyhmpcsCDbQTOeMdOwL/kOgaHnNVBfMPBlHwczJMphsiudfHuV9eulohadZBlfDGlKRChkpFam6OXjGMLd+vRcPGzXiVl71AeQlssI7BWKomqqHCyYYMR7ObwqIn/UOT4DsBciVxYlMBhY8KQWgHHvY0vEUb1OGPnR1IbF3klw9a6Z58X317VEEqUVO9X35MN7uC/8Q+Y5gWMV5zPamY8JKF8iFB8Eq9uYu9t0b6HhI/qY6ByWp8ELrn/zsC5d+4Wc52Yc54GuJ7U4HaTlP9Yf+q98lPVlFRDk57DGfJ+ztt9m53eLFLz/GeiXm/k7ELyxe45w2eJMbLlqmTmhqaEfM7LR4yPLxQZ2L8P69nJWzr92zZqQE8bmDh9kXR1WlytzDrazmbIpj+OjbEefNOgmRZ79wuMChjhjrjIVmRWFplQriHRRXvG5K2QQp0rzlAqRQwAoOqlUuUIMl93RI14KLcJbbtMyEEcSLAs6xnHot/z+kagObz9l07tm/7yh0KjkudzvKtSzhd/FzpjblbDH3WT2WcgFeOc0ZdBCKa+qSnSqIwgGsmx4hLfq4uZ9L0TkaVLiUd3HMGw7bHdLPgT2rzAp26tlL916+drjXMvQsx3HFt6hyWc8xYcEt6fOeDFmQs61N5liObMaPxxd4RlY4pw0ihEu2y8c+eoVHq3U+/WMv0yVhpS70Ws5035vmRP722olhRs75luHhTsz61j5bUYUGCa9FB+zbObEI9ciwd2uTxuVvEa8eopMq5kITszGh9f3fZDHtkC4q9G8+BMBGWxnPhU9+/9ucTOEjDzt2uEvS4Jw2TrF+RUTsceJYm4h51dxipBPWaTPSKTGGkU4dQ5am3qhasq6FIutVaRftGhNz0Ww4o0bn3ticF5CiTLNi/Aee+ACPrEqFBAd7CinuhrioaEWcEkdVKsx1TtBayNUW4lxhDJahkSGNH8ZieSyILyotj+PAxAVLZ2eqM0Z2TO6LvTPNiqwI4KO17rphbWhKnZScoY4Kjv4g6jf1RbxTrzye4MZtEG6Mial4hqwwB3NyOjS4pE63Zl1bjHXKESM2pcctOeFt2WNGyhy3lsW4epMqCVdknxkZTWqFM7kgpU2NG3LsPx9R8Y7AgrQo8l3SoYf5JB6b77IwM1/wWqZabpg6Dpvt2I5iIsckpo6i2TljCUNmXJVDVqVNh4YvPLZYUVZpcY4ValIpGH0avs7C6YjUuCVjDmREW+u0tcqauqzSZbvCM/kmX41u8qXoClOv3TFlzhxH+3uoQ27oAQ0PzdmSleIZu9L0RltMVRKsqocmWcbMidRwv675vopZkHLIkDU63slKWdc2PZo8YVf5vvwhvmFu0tEqt8yQNa3T0oSu1ogxtLXK07rqBf5cVDZAusJx0fa4IYckGtHCOaMHMuI9c1is6yeejrdHkwYVrkV9Ziw4YsR57dHEFYK/YXYZMuMB2fTtHjPRGSvS5j5xOh7X7R59OyrUtU88o1EsMUe2X/xdlQodaRY1lpc5xz4DBOFxLnCZcxhMQWkdxlWY+0PrdEaOdECuua97cn1S7B/q1MCt5p71coaqI7dIyQrihxB7numi2KMKena/CoATC01LEBvj9+CymGiZJekUTCqQYHAanpN6AdIybPpukBrx9QhnYUhnYanl424slGdZkcqiiLEsWb0CpDzTjIyMuhcXjIioSpWpX5/LTGbgRBsL8pbwrGeIX87CssvrsMswnGYdLQcKy4X7SYmFzD2vWztmOkMwBYGJu+4yIxBsI9UQTF3C5Jd9GbSbnCM591C9xEO9FOv3MlvATsvnKrdn5O3KMjwfKK5V/rxBCpX5sAcAp+qJwjPAkhChqLHj9Pi5l87aB4VtnT2in/7U2ueYz5H7H4UoguN9zMULSLuOXt9Dmjmy2uGF1j8p0izX40/+2faXv/jc4r0LRCvHmPU5O7//g7QvXufg6x8jS2sYIuLY0ls9YXe3y/f8jf/IV7/+IL+cfov9GezICe+ZEy7JBgf0OdHhqeJi1/pS/F0UHX/ABz37uXs5MGcj9KcgX3d4+84jrEvNe/zLVF/iaz0avoiqIgkHts+K6RbKp9tmjSmp1yFI6RiH5XXvU/DEBLxn4OuPZMkTHtrD8UFnBVVsJBGpZi5SU/jOIaK6jICEoqoldKsUjSUqnj+cIaR/yxmTIqoi4ZvLoyjyCpGEkiO4LCj/T4fQnXUYQ1aqSK/6nwAfC8/rokdStEWReSllQcKiErRAFDepN6JVrtldDu0JD8o2qeRcZouW1JnIooh4Zd6xLFrlVMTmNJ+2v/ipsVVOgxpx9xtLwr4MeEDX+YS9wBzLdTPgphlxIjPeNofMsoi6xjQloqkVnu00eOKZV3nmMy/xm7/0Q3zfwxmtmuWNnZjHty3rdRcjWe1kHAwjNBd+4gevst51Bc8NbXDlGN4xByRUOMly1k2Fl3Ytf+3hQ0xzTvTYFLO5CQ1D/vUatY/foDKZMx+uUK9nbGwMefdmm/mwyxsnllf2IEPpmpg/NzsYhClp4XBBKPx2BkxXWg565aPCFUno65iuND0Lipt34f8T5h6O4QzVhlQ50ZGDS4g6pjqSIqoYSwziIkfBUAdoS6PA/1alQss48dCBjkjJaIvDh6c4mI8zzgL0cQlzUPxww+G6YwkwrUCb60ZgyLaEKKSD/J2uPQsRcLeBCA1TK4rT8eM+ZFZDJjMI9AW4VNAGCTVViBuHIepofBYvbJSRGFJNaZmGbzN333NS7mONm3LCmrQ5kjF1qRJjqOOE/S5oj6HMaWuVJlX6PpIeGITqPvPSoUFPG4xkyoQ5G3QZMvVib27uJsReq8dBXwKRgKIePqbFGJqE6Kkoqc/qRkSFOF2IJFss66ZLjQpbskIqblOd6py21Elw7FGXdJU9GTJxlAZYlArOOZqTUjcuo1Ih4ZYMaFHjSbvOoZnS9VoccyxjH8U/ljFzMtrU2dFD4tD34tom8ybKkfYLUoEpC2IimlJdRrbFwX/X6DCVBW+xgyCMcBTD27LCCWPWpcOIKW1pcItjFiLsmTGxN7ZScYKcKU7NflcGbGmbL5sbrGiDAxmRYf09GHIcI1eLGlNJmUlKnYqjPCUqIrMqjg77REc8oeepEHNb+kU+7AZHzEgZMOGc9DjQAVZchiLHZawC85ggTHVO1e+vTpCyUsBzYkyBeU99n0hYv0UKyNxtTshFOdIBF8w6c1x9WJ/JMrtE5muuar5OIWfFdBjpZBkIlEDK4iLJofA9F+dkI8LEuoBLTaqIODroua8RCeiDZZWGer2opaMf9kAIAaml/lhdqkWWL3wf7tQHcevCcg0Je3lhLPt9Bl9QHghplugAU+ytUlrXwmeKPc6Px9AuSYlARyWshELTryOFrgYuq1XzxegxkYcvubocg9C3A+ZepHCZPQ42yNJeKT/T8u9lOyaFI+CceQfTWiIkwudiH/nPvc0X+iolLbJJwdZzdshSPy3AoKSUAQlik6H9YokKuKeD4TkI7EznFLZJcHJDf/k6twBfAy2y2kWfFDvL0slVApIjpWIq3qZb3lNKXiABQtYkBEJzcp9Ny+84fxgDZVuxPOY+/Z+QCYl+5u898Tmp1mAxw775LczWBvb6DtJpwWKOrHd4ofPP7sB57Tz2N/7s9jPf/2f3DX/rORRa3V10nlAzcxoPXqVT75OnTaI4wy5q7F2/xGYv5xf2X+OaHDK0Y8Y6ITJO1CYID53FmBWD5KwBx9JYLRut5b/LhvPd4D7vC/3htAN0CiPu2QcCc4ZSSg36DXLMjIbUCrxdxzQKmkfF0pQ6Rgwz5h4eUHFiQgSspaFp6oC4onjv9ITJ4tppeR9zLyLloBPBk9XiCd3kC4aSEvDq5YEZJpZ7/iULV/nzoVfK7AlLJ2BJBxe+awsjeyleeLf+OvvavbJTd/abFphNlSDYk/hJu3Sa3DnLKUc5dY+mdA8Gw6XoHAMdsx6tEOHEucbM2ZIehzLiCbvOicy5ziE90yaWiBku0qqCp/90BuGSZME5JuUIylKwUYsUt4PmuMzYhCkVKoxkQS7CO+aQHg0yLE/bDXIMK1Q5ZM5HW02+kp5wPFd+4JEhUWPI2197nOEkZr6IaNegmiiVRPnK7ZxnLs1JiHn2kQnfeO0cO3tN/sMbMatSZW+Wc047zMW147s6pkrM06tNWk+/Q/byBubcAPvuCESxezWSTyyopAf89u88w2tXWzy2nbN7EoN1Y+RAF4w9tjYh4lCGPG63S5hVR6NYFaem25EmYw+9wY+5Kglzj583GHKxBGM8JSvqdkJW8ZxZ5UgHXgh04XQExG1MLqrkYAE5rm5h7uPzIapuPH45YHKnOqVmqgW+e2nkBliicyTqplZyHDybjQQa6GUtR5iXIYPhCB5M4UwEhz9oADmnxkX4w+boNkA3792G5uaEg2g5zYNlUEELGGeAbkYs60wapl5Auyq+wLYlPkLNjIWm3OakcHQOdUBTasxxNK05lusculoqPx8nMmdbu/RlyoKMdelwwgRFORH3e5UWN/WooNp08WF31yEyabGkmpJIUogv1nwdnjPOHf6+LU1fQFxiqhE3ShJJWDFtBjomkZghE5605+nRYCE5CzI+lp9HJGJGzn3ao0rCsUyoU/EOQY1jRpyTFVa1zVQWPGw3cJBKxQpUNOLQTJlKxq6HLcVEtKmzyzFVSRh57ahAJxsytBaXHQosSq6w3NHZTnEkAzUq7OoxRiIWOOhUyxOfRESMmDLSKTk5M0md6rukTDzV7l+Xi9zUCU/bdY5kTlVjvl+3ec0cMSelSbUIGEUYtrTDUBw19EQWtKiTkTNkVmT4ctRnLBtUSRARRjJnT/qMdMqH9AIzcXTIAcJ8zMgZnDoiI+eSbBIh9NVB+SY6oyMt9xw69xAiZdV0qHlRu5SMiU55xjzMIUOXMWVBS+osJON+XcMK7OkJFkf2cawDtswKI6Ye7pWhAhvSY6JzH6Bw0E0XLKzQMDWfJXTzpubZ6hrecR14Wt8Af0GWQqAuoLI0GsN8E/BZ4WXNRRCvC86/QXzQ0BZrHLgofdjfAtR7uZ9Fxb78/9H25kGSZPd93+f3Mquysu7qe7rnntnZe4EFsIsbGC4IkiIJUCJFwrJoknJYYTtCjnBYVkiy5BChcPgf2wwpKEfIliU7GA6JIABSBAmREEEAywMAgcUudhc7e80es3NPX9V1ZVVlZT7/8Y7Mnp3F4QBrApjZ7uquqsz3fu93fA/tcwd/ClHAmjiEunCN4HLxkemMqqr6/KHgC2DPMLMX3YTBJfNVKuQUnXXHa5npmRGmsHmNyyoG+dA3X82eKM5Fd1aWIeEulziMhCgKrDJMO6Dgzjm4JqU8oCIVM1GV0ELjC2NBx+9BFCJGDlqj/RmfkXsuR65zYlVjwcKvVcNJmxNIaNeD+dRdG4fc53HcFnfNi4KngGpXHcxWlHkd6zlTtbA9MIVLIFZFy/mq+bVV8VD6MiQQCkXGcj5YzvvK19dlx+4+uGntX/vER3/wIuQf/9KpX3U3WdbWIZ2hByP0YISs9O5YgJQfmy/8wXnQkFSQKCU8vU++F5ONOjS23kBP67z22jq1KOf1GxHpvMVILUBscNUzL9nmwDeHpiH+cXgi8laP71VklAuUOz3nTl14t/hdpxLsmE5VSUn95nXKNUvS8mPVtlWyqVE1rp8krKiuVc1KcfhLgLpE1kjMBCHTbS0cRwtIRO43l7JFiRLjTeJ4JY5YZrDtgSddHeJB2I3mpjrlSZPDSLpHQZg+7HJennq4z1EhpKixS8/zyXdx7W/n5dypELnTPfHPk8PQNUXgr5PDxhYBS/kutOtWuN/lJPEcZnXIhJPqCDEV9vTIy/+9oq9xRJZY1Q1O0+aEXmakFlzXeybwgCcUVqXitcWxB04kVWJlHM/rKvZJasETcbrdlrNiuxItiUkl473ZMSIdsqxjLqp9Ellwjg6/9DbNv71iupz/9GNX+Be/ew/vfdtl7jp5nS8/eZSqUnRizWSmuLYf8OARqEUZOlc8+1qdNIPlZs59K4r777/KfHuFlq7wvBWLaOgKfZnxvtWIZrRP5TENo4Txn9/D+NoxGo++Dvspai2lcvkkx1YW3P9jX2KxfZx6oIiDgNenc54NbnA07/Cy2jaBlyq3xGDh9/SQmlSZ6QUtFdOjyT5Da8RnFJ9yK2ThDmeTuCmrAW+4bG6KMCdloMfGR4eSipldz6GEXqY2tAeR63hqMetnbqVoh/nIFxALq7DkCLmRssaC4laxZqbn/oCoq/qhRkFVKvZwMIeikcfNfJzRlEflxbo9NFG07znHYM0FoSGxf7+Jnho5Rq+8p2wB7EjboW+QGKNU5ddsYtW0Jnli3rc1QQQDfWs5iJCdFk4NOIxIKrQwnDiXTNSocp19lmnxOrdoSMyGbtviA5pE5BhYXsLcJzOG+Bn6A9PFAjdx1LbocJK9iZ7abqvD5vt+AAAgAElEQVRJ3xxpuyaRVR0zHcaWNIgwGOwt3aVFnZvqgI5N0o/qHmNSLqt9ZmJaSq44mZMxxZC5N8TwKgaSsKpbtIm4ovrUiXhA99iXuVer6suEk7mR2d6VkZ/SVCVkpCdowTuFY2NEysKrBM303K9tgE1ZYh/DmXATMQPfjFiQM2RCpnN7Dik26NJnYqCk+RJjSXlAenyHPQ4kJZGUVDIOWHBdDli3Urxn8xXqRNySA5rEzMRAu9pYCV8xBXsDk4wbCGNET8dckz3WdZddGSIoutLglgxZ1U1TRBLQt5+hYTkeAG2pG/KxmM8WS81PIJ2YgjnPcsY6oS41OqphXOElZVU6JMxoSmy9ShYsxKzxY7LCnjb8k5ZtChq57tg7pe/qgZGIFUWFCrloK2/thGYqzPSMtmr6IheBYT6mo1pW1nru12pi+VoLW2Cae5v5gsHtZ1VCCLiGgek2c0im10B+xSphlX0+XLuv4Hxo+7vK+8jBmlzO585nnwOWSOvFmV5qPtqcIi991zdO7Z9AAg8NrGLizCRPQIw6nWClhUtFmCs6ylyLcrPOvb7jSrm4WGRytpiyTdFyYeQ4e6ahUSAVDhV91lvIvD/zfTf5qnoiufKkbscSqqiKb4AI+Kmmn0ZZzkVoifoBiq5q05A6u/kBR9UqM4sIWFCcCaFtZk2tJ0xghUqcUIH7BGZtmXZNXcW+6GhYGOpCL4hs06yjmrSk7sU9ylYOyvIWHVd4ZtewO3fcmeoaZm7y72Zl2p4H//+KkF888atoDfU25AtI5+jBiK+f+VefvFL90e/5C1e+dOF80B6QXt5CqQUiC/Qk4o0LD7H87uf4k9/+EPVazsrymKpSrMxbPL8Yc4IVrrNPRkbNkn1coi2I7xi/ufh4c5FS+Ii89eO7JbLuNW/vvjv4ULmj7hNjMbCpjmr5GxqgOCubXGefhkSGsIOiTkRKxrJussuQkTZmUhMLg6hLjRlzBnriu3iRVGmrBhOrvlSVCDd8LY8hPRTEVr8OcACFiVdBvjUJeygV24l1B7zpKht1hwBX6ZprpGzgcSPAwxriBcSrmIa4Dq+74k6qrhzWbr8Ht//7+//vw9MyNxEp7qvyzwtQrAQ9xnrir4kbbbqkz/8RYSc/YE8PyNEcY5kDSdiUZTI0W7rJhIwQxR/znDkY9YKGqjO1haQxnzKbuqXq/m8NLKm27wy5661scGmo2CsVxZZs25MWZ7NlWhbS0KICWvF/ffJPeTCusnruApefvputoMarr61xsh3w6T89hr55ip/9W3/IhSfvYjQTZgvhp3/sab74rXVOb8x59vWYR+8dcOFqhasjzV/9p39ApLY50UvRkyW+OZhwRDeJCEhkwTu7MfVKTvLMMtHWVbKrPZp3v0hw/zqy3CR7NmP1kQssbV0i322zvLLNd144xpGllPGgSj2Peaja5Fo+p4PBzzepsScTRISZxbTO7SGFmPXuuk4RVabMOS6rJFIk9ZHtGC3I6ErLHBo2TjRVfGjtuKmBOQxNR7+rmhzoIbHUjGKJ9QAIMB4BZbK4m3gYbLP5eUcmBWPS5ZTvDL4783wsJ2SROoiWjXOh7cS5zqUb7xfdvtzve5dcuNjkDt4FCxa2w+rUdcCah4r2WHkDyVr47qp7XgWj/hVZIYyUzMO0XIGF/QyxTe4dvj6zzw0JGDDmiO6SipkuadH0JeE4q0xkxrYMqFmbQg3c1PssSZuZpJYc75Ie53NgrnmVii/sXVHqpr2BGB+VhYWouqlTQ8XM7FRnZp3WjUpabvgCYgQPjuddVnSD62poZtkCp/JldtWEtbxJRIUGEWMx0KScnFv6gEgqLNOgp42R3nV1wJCMsZj1fU365GgmMkcL7OshVQv/mevUwE1sIuGguIjh/zi/FveZRnpiICuyMJMIyViSFsvSZg8DG0NgmRapZLSo09djlmhyg31DWJcZI6aQVVnSMcs6pi9TUnK21YipVdc6rVfo6Bovq20O9JgdBga6JykLydkVIxm+sMnICGOYOWJKKjlbusdY5nRpkDBnVw9oSZ2RzNjRAxLmdKTJMVYYyZSuNIxPjBjn+hkpE9tYSM3K9tASbQtvBLuGYVkZwntuYYZjPeMsG0awgil9PSSVjGOyypCEOak5owQzedMprrffUnXcuRpRRYlQlxqDfEwgyk8PDZRxZtdezStepfYcSPQMTW4L4Tk1FXlOWjnhNk3MEhnandUcNhUEyyexybtTWXIdf+XzggAHO3Rw6Nube4fKjHKi7yYIJU8jMzk2r5VbXoaPTSWosUYbArdeeKix2OaHg4K6mHg7bLtsUFw0dZX/nps+OcWrIjaa91Kx4juuSCm4oGXJYvHQdvx10jav0n6vFYk1nkfhGj9G9MB6xWFgn8pOjpwKoONlGKhd4R8nCDWpMdRjWlJnQYYD0Dvkhot7Lq9yfBYXE2uq5r8X2Zhm3qfyDVSnujZze8S+n1SbhoXZTZmf2Lmzyk3r3RTGxXtXuJVhuUVxUqw9AX7nU390/nd/60vn/+onvnft4B7BY3/r/5bL6vzjl7P3PH5Zf+Dxy+Fjj19Z+vj3/Qtuvv39j2+9/B/Ph1s76HHExcc/TG/1JjtXjnLlyXt57y/+HmF/le+8tMzBOOCz4xu8yk2GMuWErDNlwSAf0VUtEj2j6TavXXieR/CWMC3ln3snCM9bwXrulPwWG/S2zSpFcHAkMa01sYpJ9Mx2OUwRsIsxUerQoGpHgNv6ABFhT8zI2eEqq1Khbjtcrgp2+u4gVMTIQCoRe5jjx2wGLqWswkFBHNdWnaMqVZNsYLCQdamZgC2HB2oFIbooxDLbaUTEy5q6oOwmKcYlvFCd0KVrWeac+GBmuRcuOL1V4efu6e3FZtHvKF7nzdMs7f8upiJOTcQknA5X737CKztg1MGMukkB8dHAsWCNV/R1hnpiIHUScE4vk5LTF0MYn1upR/ceWqpBTaoeavdfqod4mh0GekIsEat0WKHNTC3YUivUVZ2GiqmrmOOyxkJyjssq63SIJeKebJUlIm7JlGtqhNIBH19vMXzpDBtnX+TyM+9kdz/m1XlClgsf++Al3nNXn6deXOb+t73C2Y0hL1w8QkXBPQ+8TLpznDQNWW1nVCo5j/3sl4h2TrF+/EWCcyGV9RvU+01uvrrOLZlSI+C99Q6PPHqBaPUm8QOvM//2CaJ7LpFfXUbC68hqj+zFBXoUIbWUg1fuZTrukk5jRpOQ997XR/c7Zv2nITnCq2qHHRmxrjvsMGRDuvSkSSoL7s2PULWyvRojTZjoORpNH7OXUhtQaxKZfSIV+vmQOSkNFXt+1UIvqEvsJ48T6zruBB4SHGwqKIK5nVYULseGHOwOHdcUmOmZl6B0B5mThnXwCSWFXCaAFjO+dnhgN5F0RXB5bbvD2HzfTPAMnMUcLGJhHkYf3sQAt8fd6D7EYOartgAqY42dSaNLhl3Ca/gCC58EOihAVaocWI6Mw3eD0Ja6x/HvyYjjepkUAwfq6xEzyfx7RoQdfcCYKQ2J2dEHuCjekSZzzGfpSpMpc698lGCutSsEl1SHqVVMmjJnQy2RMKNGlViZ2OqaRDm55/+YyYHxiOhZ9++7pY3KQ3bUmLuyZRJZsCNDmkQcz9vsy5Rz+SqpaAaSsCJtzuQr7KuEiaTsKHPNTuVd43klBySkHNEdEGHMDMRM/6YWuuaShZSUljLdfpecdVTTGK8BsYos589E4QmGE7EkLa7qXSoS0JWmT6AEIUR5vL4SRVcboYsWNTTCs8E1FmJeayAJEQ4DnzGSGdfVgBBT5McS0aCGYbWYmDphzhptqzRFqVhWxERcYYe5mF0WivH6uJzfQgTbbJuxz4gJU69WmFjOT5WKJ+0aRbyUmeUGJTohkogjaoWhnlCTyPOMZqSsSY8BY7TAgISjuscuQyKJ2Ncjg0KwxW5s0QehVZ7LbVKYWH4V4BPZBQvqqubPkomV4p1jkB2RVK18qjnPV1TPEq1NE6AqFesT5u5P4A0GlZ1KumaGb0YQlqDFhYS76Zyn9t+Zn5673rQrRuQOf7QUU4Zy89b9nGtUOAVQlyeYs1lKDcXDapImTmpvJ1BTNf/7XH6TkXnOo/scBW+0MIl2dglluHs5JpabygYGmXtYoMtunGKVOecDP9VwZ4mTBnYytQtdwN/c9EnZ5N41LJ2IQXkaJBa+6osxMQVTXRluW0UqVk1wiojQVU128j4pGQ0V2zOgTAjXhvwuheqoaYwqGmJMd93acZOlcvHhPKEcEqStmlba2FyYI2rZ8qqmLKmOgW1aldXM5noKA/Eqp2bliYh7PVeULDAcSmMQO/2BVLKCX/7lXz7//T75rR5XT/+Vx4+lv39e9ab0OtdIbhyj0Uq4eaNLPG3z3AsbrHQWrK/M+Pc7+4SiuCff8Im1UoEfRzsog08obZevnOCVk9RycfK9io3v51Fe3EWibCVe7UaIVVxAMuzNcPrR59RRJsyoYZQLVnWTvhjieaZz39GdWmUEI71rJhBzjGLN3CrLOJgBGByi6b6mVjpN2S6x2QAumJVxoq6qD1DMbZJV7ryDm2a48ZrF/9ng5KYaisOdWFdEafQhF2hXoTucJP4qln5GFxCot4TE2XsdlALPD3p/yzjyFdUzSkYUGNme6lrFC/E8HldY+e6qHccOdUJORiwRJ9UGE6as6Q7nwgaX9YRYYu5iw3fzQwn4Jf12TuYrPCobvCx9vqFv0CamLXW2877pzEpqYHrUWNdtbnLAOY7whuzgOBJd6jwmm1yQff7uI5o/vD5nOY/5hz8yYGOjz/5umyefOs0DD7zG//t8xR6k8MKrPcLJEkd6C7721bv5s++s0Y0Ul8YLlvIN6vGCe9/1DVr1jKtXV1jv7ZLP23Q/NoTFgvylkCgecuXFM9xdq3GqUWEw07zjV74KY0W+3SId9QjbfdT6AL1TR9oJ0pkgiwV6VCOSKU8/eQ/vfM8TvPbqUc6cvcpaN+dblyI+uFnhlUFGnSr35KtcV0NmYjg1Mxac0qts6iYX1E1yyTnOKgsvb2kSVbPshKme0bSdpf18iGCgST1pGkdpPcNhyl0BUZ5+ObUbN30oY4ERV+wb8qYWiKTi4T+CM8XTXl4y1WkBgyof8uIkuDNiqfl17jpr4JRzzPtzcCkp7RcXnxa2h+YbB/6zeP0tX6AY/kfupWXNVET7zpYT1XDCGE5fvszxcmpwjjOTk9GWJlWpsK8HLKsOYz3lLOvsyZhM5yzTYi4ZuwxYljZ1DEHygDFHZdnLJkdUvWyp4+6Y1zRFx0IvcHAFh2kOJaSG8f9wSfIRWSInNy7azEis5LvzlqlJxLp0DfmdmFP5CrsyYkO3qRHyvOwzkBkf1ps8q3ZY0w22dJsL6gYREYmk3rOnIgGbeZsDmXI675EDN+SAv8YZrpFwSe1RJzI8GdHsMiDRcwIxMrQVR4IlJ5KK5wApDO/JJT4i4HxU3BqpSMVC3nL2GLIibfuzRvmpRpWIkD1GgFiDPM0N6RMTsS1DMjHX5QZ9DqyUcoZmQU4TA+uaYrzBh3pCKCFbustNDliRNgFCl4Z/zgY9mtSssWPITFIMkXxm3KpJqUnV8CD1jAUZPWlaJUEh15rQFsFTPaWhYlK70hwcCoxpMAiIZqjHaJ2zqVYIMEW1u44JRmQgJeMmB7SkbvH3ysvvdqVlCjvRRBhotRM3yTHGhz3V9nCXHOMLNWNG1XI4XeIWoJjoBKfUV5eY3bzvG4RVW0wgwjSfHpJNLSbilqMlBX/BnWGKwpk7tMnloSmpKL+fXbOi/HDw7LIqVBm2BXiPNJ9XHcq5CjLyocaglAsevPeYgxrH1iulIiEtaTDKx/5n3e8qn/GuOFEEtmGKz0XKuUsBF3cTkhzn2+GbKYRFrmQfC9uUdRMUV3S4n9NYQQHkkGw69nWN470RpJjaSYODnyIF+sLB6d3PTplRpUJTxfzap//hJ3/mEx95/PO/9ZXzmY3LgTioV9UXJa4gA4gloi41C7NNiVTV+8+4dVRXMUk+JcNwCJ2s9URPyXVWEjcwSnhDPWGip95PyUgFFxwQ15wFd1aYgrMcU4t7aK5taJtwn/3UF85/v4XID6UIATi6+7nzb/zWR4iDlPq7X2RxdYWNzW3qrR1k0eHGTo0rO1XO5l02F12uKUPiGogZ4+7qAYmeMtNzI3daGvMJhYlgcWOKBVxORN9q8vH9fr/8KE9I3DiuJjUc9q+uYp9sxKrGiup6N9iBTmhLnX0Z06bBrh5QlQqj3Jh3uc5AQ9V8p6+MCQVNqkvqBEJhVqgzYlXz4zYnTzfTc9+Jcl2ABUXXwpGHygVHMRMpSHTuoQj8VKYoy5yzZ2G+5h5ONaJgg7j75EbO39tFHQqManGf3xq29VbTFMfJUYhXJzOfyaR0jsDrPm+hflGQ+B2USjAJzKP6LHUq/I3qGV7PEuY5zCXnorrFLRlgYDI57+AUVR1yf6PGzfmCJ+UKJ1nhnnyVp+UyR9QyCuFXwru5ms1IJGUuGau0WNENMsHLmnaIeUpu8c58nS9ezVnVMT9/MiadRTTbY/7Zk4pbswy1u8l2ojkVxpyoV7g6T3nPmSlZpnhtN6AZKpSCM0vC2uqQvf06x37lIupKytG/8gR7zz5IWFnQfHSE9FbInp/wjS/8CK1I86m9HVayOh99dJtakqCOHKCHMdEDl8mvd9DjCBYBamWKOnEckj0kTCFV1KVOEObc9f4neOXJt/PVCy3OLQut+oJv9Gd8rLPMN+Z9pmJ8Io7qLi0dExGwoxISW5jsy4jd3EwUU50a/4J84A9el0xre0+VKHbtBNIV45GKMNOKil+77v66qV8xS3OFqOFTNaxDeE0iqy4lJDrx/j6mADCvtRYsmUTT7h3X2si0SWViZdR3pnrmV605/Gr+UHFwDLfG3X52yYd7lKGg7nnYA9Gpm2hy6qrOvGRoWt4vrqPm4F2u+eCubc2+RsKUhTYJmcHlmy63c/1NmHGTvpG5lZhL7IAIQ51wYHvogQRmP9qOb5UKVYxS2VgnHFErJMxQKI5Ijz09sIlZYaoVSsCKdIwLuLQZYOCtTSu13Gfszboya2DoDvERU9s00mzqDm1tuFYK4ZhucUnts5w3iXTIphXiHUlGgwo1Qq6qPjFVmlSpUSEiZCoL9mTKu/MtXuCA94Q9Fgvz3B5N5ixsk2lREENFaFCjJy1OssJCjCFixco/H5EeewwJbHPKpWo1qVk4F7bhY9Sq9vSIlhhHlhxNjCGvl4vboU7YosdV9limxY4MWJaWKRYpOANjphzoMV1pkui5VS4KuEGfWCJvrDgjJaLCum5zQw5o6RpX2PWw0pt63xYGTtzBrNE5Rpq4QuB5G04yd10t4fgeYKBRGTnjPLEzO3MGuul2V7XJJOdAj5mSWp7QlAoh+4xYsGCip8Sl6UdDYl8IBSiqFgqeaAMHS63jepkjNrMkauOibtS5XPLqXLlNM9AQjJ3fh2sQQlGsuBjmVK6coahTkHNnkoNTuzPa8TpcF99FjqKQKAvx5If+LiMdHGfTxaTCr+r2xm45v9Ie5lw+k11xUvy3gyYZCJCD8gzzkeeVmXCr/DlfRki4SZBpghReaS4yuYLHqX4CNFXDxlI7OdAl8rqY9xtSKIBldhLjubNi46uFJSFFdHQGgoJpBnSlxYIFJ2SNG/muFS5JqUsNN60uVLGc3LiB5y6pNlUqfOQX3vc4wMc/8djjv/mb/+G8k3R2pP65Fd+oSwwCTal7iGLTnkOJneSrUoEV2cm7E31wzTYHK3OfL5YaEz0jVhENqRv4v6r6mOLPRis0oLUurdGF/13le+Cut2vkaTT//lNfPP/ZT33h/PdSzPqhFSHqXx+cP/rzXyF56Thqr4LOK4z6SxzsbnDzVpPrB4o0h1oopBl8W27RlwkDJqzQZi4LL4vnq/w7VOTuAhVdxNu66G9RYPygEr9v+nxSJAW55NStaZbDg5tFaPxB3KGzwwHHWAExnfmBHjOzEnQZOYgmtou3rszfBTZQ2wrVdJRcV14w5lwLXWhxRyoi0UmpaCgSl9Bv2GLkWUj33e6wWpCM3AK+naDuftYl+EXQw0+GyhjTgtdzeIr13e7V4eLmzTCst/rZ8tdWgiWv7e0gZSEBK0GPKQZCF1oZX+WvmRlEvl/dxxW9SyAh96rj3K2PMFUZQ5nyznyDm4uUt9da7C8yagRUifh7GyfZHK3zn/SOsTuBt9Xr7M/M/XldDfkRfZw/Cy5zr97kPJtckF2eyG/xE5zg5za6/ORmzANBj4eXqpzLlrg5y1nRTVZ1zEDmXFUjNnSTmIDjjZCHf+n3+Df/z6OsSsRU56xXKlybpXzi3QMqVOipKi9di+iPQpQIay3NT/2D36Y5aPO5r6/y8V//c175Z4/wzDNn6CQtZtMal95YoXNjQnX5Is9/9jGu7lR4cjclJuSBXoW77nmF6oOXUUsh0h5DDuQ5elQnPLWHTnImf3CE6HyMtAOYDmh+eJ+wcRXVS2lNoX9rjUDB9kHIhzZDru4H1LMqU63pq6ktCCATzYSUk3rJdJQl5TTrzMTgaHPJ6UnLd2nN9CKwSY4BEG2qFRxvwAXmqi+UC+GC8iozLscm6Z3brpfrwjuJWtd503Z/VaVK1X5NUIzyCS3VNJ3UoG3iROngdx3OQwegLzCKh4MElLlbHmdt/17YCYLbhy5OmWR34V/PcVIMNMdOOm3H1ZlnaXLmem6TI/EkYAGfNIl/55lPj0xxZgi3Gs2IhCmpIabaiVJXml4u1mDp5yTMTKeOmSHaWiiDK3wmViPLNVdc978mVQY2Se5QZyopy9K2ZU5GjPF2cLy9tmoytGpLOZpTrLNGiwOZsq4bLNB0dIRCuKYGNHWNG2rEkIwDmdMi4jW1y4pucne+zIGYBPQ1tcuybnBDjfiw3mS9UuHPuIosIq6pIbnANgdGDtd25+sSkYumRpUMU3is6RbXpc+mLPOA3mQkphjpSINUMqOklU8MlMtK9R7okYdvhBamNrfwuBlzDix/ZEcf0LBnU0fqXJE9QlEkktLEeJSkkrOuO7zBLQAaUmNBbgtZzaq0meLUF00SWyfiut5lkx59mbBuxQamVt1saguquZ1ntCQ2qlOYYj+UwE6EDHF5qMcsqTYjO5020GPlJ5+ZVdVzBZHx+ckPYeFNITy1HfDC+DOSqoVfZjRVnVxro56l6kyZkeiZV4FrSGxMBe25Z4xNMx8vnKypixEZufECsQm4awAaMQxnLmnORmcgag9CnBGeFs3C+oMVk4wi51H2emkfAYqkL7PiEwZ2HODMiL0YhY9zGe6890WO89nSh/Ot26HS4q5xCYZVnNUGmONEOtwEL8mN4fRUT5na4s4hWtzZ7qDa5dgFRcEBTnzDNE2dh5m5JoWBcqYzD3l10wEDVwq8B5hYqK6TO3acPDcVdvzBoomqfaJtYHKBz9uaqs4t3ffvY0X1rIyzuY5taYIIbdWgJpE/O8Z6wj//zD/+ZPn6/ewnfuzxz/3Wl887HoaD7tUsTNAgbyIfvx1PDPtu6yom1QuaqsFYT4zxo11LVakSS426iqgQ0pK6l7sOJWSsEw87XNiGt1N3C30BbugQDVX3ednCwkLLUx+X4S3sGWF+l7mmv/2p//hdC5EfWhFy+tXPnw839rn4lUdotRN0HnCw30Nr4fXrMe9/xw2eej2mqhSDRc6Lsse2PuBuNrkpA4Pjtp26cvfcJbCOfH7Yu+N7TzbeynX9B30UibH2CyuSKqN8Ysbgem6J6k060uA4y8RSY0U3SGRBlZBlaXMt3/FjP4cBNXAPMx5ziX2uLcTLQkXcJKYqFXKdW/dQUz07LKPDILrN6UnjYpQPqpa4WSQSJqzltjPgNiDgE3R35UJLuHLXwH09P6S0UUgVl4llZfzpnaYhd4Zl3cFX4/b78V0K0KqqWmKgXT+2yJroKSeCIwytUVVktdzd5w8l5DLb/novSZumjjiTL7EQzd3SRrTwwmLEnJwqRj///lbEr40vcG6+RldVeNvJOa/sKhpBwN/YWuJfjl4mJOBvNo7xyF1Tfndnj3+yfB/jmfC5wS6f3+/zx5ObPDWY8eXFVapUSCXjH7034+Ibbfoy4797IOZjH/sqN18/wUZnh1efP00gwmZU4YHTY+5fVjz3Sodv3sgJdcBqE44sLdCZYn055ekvPsjJE7fQox6dmzlLG1cIshaVSsbKyRfptVJaP/cGtJrMn+mSzmq848Sc/d0aKw1hqZtSlRG6r1DnYvTVjBtffx+d910gv94gOKrhWhX2dgnOLSF6TH4zIXt5jfCRJnJ9xInTlzn+0Is00hWmSZXdUcBWI2A0VdxQY2aS8YutLban5iC5KSPmZPRlwvvyY9xQRu1qpBMOtJH3bKo6jkjdkbonV9eJGGDUhyqENKXOUJtJZV1i06W1iZJRxwo5IkvMWJDoGQ2pW2iVScTdyL8sr1xxyltY5TMrszjVM7RoJvmU9WAF02k1MI66xKa7LYeDeCChP/BT27UNxClFFdAAp0pVVndzyUsxyTG8JiPP6ciDJZJmiZtVhkRUrYRxob6zsOothQ8KFHhrAwUJ6OsBdanhCPxzjERlz0K2nGP1UI893MoZB2bkLEvL8gJM4mjgO4YE6qTL56Q0VWyVn9rs2elyk9i7gkc2Qd5nZNy1VY378y1QBn5TkYB9GbOQnLP5MhNJeZv0UFrYZ8bdepl9W5ysEjOVjFsypk2NdV3nieAaR3WXG2rIhDlbueF7iA7oqBCyCitE7MiUvoyJibih9wgsRGRGateLRgRO6GVEhANJGJFwVfp0adrJkynChnaS6xIEl1SZGI310DDFw1Anvng4YMyqdJljOE8Lcjbo4rgXIYZHMmTCjp261KxpnNtPoSgGOkEDW7JshSHMBKQqITcx5PyhmNZOJBWGTKhLVEhiI5rFXpYAACAASURBVH7af0Qtkdh7vrCIALe+RnpiJiQSUBMD0UutD8dMzzkZGI+mdbVkSeymw2tc51sgRtBiSVrMKX6urkrSxXrif85NyqqWuzfSE8Y68ZLWThq3poxwjp+W2wlHSspcz61kdAUntWs4BcrDu8oJtlPeMkpOllyuC2dsjzCwf/A7PS/tPTuxLyk9YeOT26PlJDC3RZSUvlfEgOIrd2rsukTeEc/v9HDvMxcjGFCY+ZUFhLRvorqGSgGzKp5nuHi5RzKUJ8PuGmkc18O8ctU2mJxUuuPduAaOK44KWV4zFTciAuYemSIr91y5moqMESW5lzo3ggSKjjR9gdmyoieOcwZmup7oGYme0pIGLYk5xRr/02f/+0MFiHt87Bd+5PH/8OnHz9ckImFKQ8zUuqdaVKXCIB+zqnqE9lxzarJuDS8HXQZ6bM0fHZzWxH7T4Mlwyp0isMC4uNcsH23OgiNq2U8f3USnPBAw6zzEWQcUQgj5oVVRXn+uqQua3/nUF8//7Cd+7I6FyA+tCLnxtg8+XvmN/fNHTrxO4ydeZ/9bZ2h1Drh1c4ml9oJZEjEYVTnay/mL4ZhvyyU2ZZkTeY+X5SZDPSpBgYqF64qPMvegnJh+1256qZvgn39oY7z1o1DcKr2WrdodVtHxCACr8FVhaoN9mzqX2aGvJizpBhu6xUjm7DDwxKwl1aEmVVt8KJq2iHDYa+fWmUvuN6BbTM6Dwo3ARPDkKIODdKM1t0iMvGbgwVSlyZA90LLbAl35Kpnge1iK112T8lUva427ytrfzTu4rbpx8Z3v42FyupS6FeVOTfmhLL7Sjcs10FFt5tbpVEQxtJC5UEKOqTWGekKmM48FdV2SnJxdPaQjTQy2OiTUIfvMWSGiSYUX1T5HdYvJJOAXN9YIUJw7knJ9N6JbVWx2NNO54rHmCutJj0CEl28F3KtXGExhq6XopnWu6YT/onWSr6U7bOmuOWQI+KMrOT1d46Ru8di7LhOqOUHepNHb4cEPPctLT93Nd5IJZ5tVLl6PeOyDL7EWtHllW9GqCpOZ4u333+LIsSvc83N/wvzyBoODNqNBi3ZnQm/tCo1zF5HqgtqPDECE/OU+/+rfPUQnDHnqimKiNVstxXDQ4NjPPIdayVg8EXHrqXexdt/T5Nc7TK6fIHpoQLA5IL9SQ60lSG8JUXP0jqCWxuiBUc5Lbhyn2d3l4isb5LkwmGme0LsEKJpEvG8p5jvDGdsWstnTMRUqPKOuY/x0DVl6RXWZkdKVJm2JLRxHcVQvsc2AARPWpMvUyg3OmBNKQEvVPSF6qMf0lElaHKxigjFEc2RsNxFwRXvdmqmtqyUytJfkDDyHw8SNZdVjgYFdOd+KpaDD0DoAe5gATi3G/ClkaXP/tfIURaQgoy5YHJpsHoZn4KcuCkVN1Zhrk9wvdFbATWyh46ijZuxewLbc7zvUeHCxGaElDbQYuIhJBJyfQWj9Kia4JAQROwUwkw0lQsvKsnZoMNSJnyrNSX2h41xOQow/w4Ee05MWAYqIkF0r93oiX+Zo3uZBvc7Lsst9bHFat3lZ7dIgoqtj7s/XfbNkSzd4jTHHVEykQ8Ys6KsZm7rBJRkS65BMNLEO6RAxkpS+mrKk61SpsKbrHNdNDkhZVVW28zkBwjU1MgUEiY3pBnZrEv0qTamxRIsDmfDu/AgjyVin46dTLR0xEkMCH+oxjv/TVDH7emCU4krQrrFODuG0e3byVBWj6lUhpEODAQkRVbZ1n440GJKYRFkssRoDYZky8wVEVSrGk4MZQya0iJmRMtAT2mL20ljPSJjSkQZieS9OqWhBRsMaCk6Y0ZOmn36sqR6xRIz1lC21aguvgodSyGhniOU0bes+GlhTSwhYE0vjsj62c26T+IuFHxfFrXOwzsj9+R7YhNeZtJXJvg6CmaN9o9FAWozoQZGQKQttclMafYiA7c4rJxPrpilOOMLxQhz3sgy7dKl6YBPAcqLn/r8wOi17Yd0ZdeAg1bnzKrmtQVhGjbhr5IsBcTDut2ocHm4Wl/m7rmF5OGktozLMJyt4IYU8bJEnmJ8x8Tklsip9rqXiCj6x98PwUcz0uyaRf45Lqj3nTQLqKrYCPqbx4hqThotmfEAc+VrE8DQ0mp2871/fwHYzP/1wa+offebv3LEAcY/f+dQXz89JrSeOO2kMlHxV9djLD0hLvzO277OnWvQtD9KJeZj1YNXV9IJe0CZHsyxtAgn8ZDQm8hDFpsQeUmbk6UO75gLTYFORPXdyf+aYcysoSmW73l0D3LvbY3LBv/QiBOBc+n+er5zdZv8PH6YamdHr0nIfdEilmnNsbcp8HqImMR9vHCNPYr4dXKcpNdpWItMlxWXlBi8zZ//7dr8JONxNv2OX/LaJyPcqRpxM3eHfUci2nVPHyCSnKXVvYLSiOl7rfs+6yLalzo4MyUUItKKrWiypFj1poQUeyo+yrxJqUuUgH3KPHDO67XbqsGDh5ULdRjWKPpkfL4YSkFt5z4aqW/IetkApvDIUzhtDCvUFi7ssw0HKcrvlRQaHJxyUvua7KIemV29dgLi7duj63uGeuYDstK4L+bjvxgdRtFXLE7ySPPHTDvczrtI/0GNzSKo6HdXiUX2aa9LntNpkWToc6BHnMB24VDK6OuKaGnOcBgkZHR0xZkGuha+MDri3Vuf41ojXrtdo1TSDRPHqQc7DZ8ZcuB7y6OmUU+spL98y3exHzk3Y6Uf89LEae6OAe6XHz5wKWZ50Gc7hfukw15q/fX6XKB7yb3/3nTxw1x6Sh/zup9/P5STl5x/I6Q+qLLcz4kjz6httPvjwDr32go21MRdeWuHMPS+TXlkjWr/O8PpRbu1HnDxziXnS5Zk/fYT1Tp/Rt49RSXZQ6ykXvnwf739oj9mwwWiuGU6Fdz90k7h3hcWFZVRnQr3RZ3z9JNXuPnpepXJ0ByoB02e24OqC/I2E/FqFq888Qiu8irSnhA8uqH0wJEjeQPY2ubYTcWotJxo2IQ/40WaPXAv9REjRxFQYyIytvM22GpOS8dfz+7iuEvb0kLvYZF9GHNPL1KhwU0wh1ZY6iFAnomVNQw2evMISbSMFKwam5fbGTM98l8utzKmesay6zDCH1orqEErIirQJCRiSYBzaa37iEBLStGPymT0YDCY+9PtVY6CYSgqHX9etdIefICx06uOe+5pJSAsIlXNZL2BaxaQGCiWT3EJJHOHXESedaldgcdMFHLZwUC5iZxE/3cGb6JnpBFuxDucZ4jqdhQy2SQpcApdgp7nWsXxHDwzcweKeUzsBchw6hXGLT/SME7JGjOmAns0Myb1JRCCKpq4yJOUuvUpTV+lKhY28jUZIJWcgM9byBoKwJ1O2dIOhzpiQ8Xyww9vyFb6ttlEibKsJUzvJrmsjNuKKxJN5hwWahIwuVWa55u5qnSvZjDfUPgMmVqY7oGo7sVUJrcrU1EDJSKlQJRPNi3Ld+iss6EtCwoyZ5RmA6fCOdEJVqsx06osEN1UICGhKbNzHtZlCOIjPnIxdPUCL5nq+iwAtqbOjD2hJ7CfDValwoMc4joIjlJtiYkGFCvt6ZLHqMX099s2rmlT9eg8sLExjlH5GzmuD0PKDUrTASE8MUdZ2dUfaWFXO9JypnpmJIpqKVM0qtt32hqqzk++xppYYW6d4U5w5YzyTyqUWZjW1ggqZ5Zu6yUToIJwWwuKmIzmF7P1MGwlfX/SBl4E2hYeV07YTV1fAR9bjy619n3PYc93ti0wXZPECAi0+Hjh4UUE8f3OD1XgSFepZrhNdfrjU1sWSctPw8Flc7PEyPMtNRe5cgBTPP1zQvDkvcAIwwmHeizMedf4ot0O+zfUyMcTB88r8GVOIGREG505fIfSfLPcxtpgiuGvZUS1mFh6YWOU5B5cNbHx1E96QgIlOjAIWRnXUeLoZTobhkoZsqhX+18/8g09+9Bfe/5ZQJPd4/DPfOJ+jaUmDKTPaqmF9aUL6ekhbNf0k20z/TDNp5lSvwE/dmlKnoepM9NQ3xiqEZJIztNM+1xiIJWJGSiwREz2z0EtrBmyFEGZ2MtqwHBW3P6T0R6GsUIObyheCKm49/van/uiO05AfahGy9eofnidaEHV22Hn5HJVqyiKtEjcm5FnIN59bZpQEaIQkhfu6FfrjkPfJOhdkl1Ryg4MrTT3K2vbucTjpLbrptz9ur9IPP95itGgDhTO7Ca1igLKQjJCQ08EWdarGvErPGOuEWNU4yjJNiVmny65VLenrMT1p0tUxVQImktLSEdsy4IjucVHdJNFzxE5XhkwZkXiynnPgduo9TvlCBNvRXNjugnnPYz3xY9rCZRmfrLgg6jCaZbJroZvNoS5OUEpuytwYOOy86oqTsvmRu1e3F463c0MOd2q07bgUbq6ZNgdIpKJDQdsVFbf/t5bCZdTI4nWYaaOKcjrYQklo4VrmM6R6gRbNmnR5Vd9gVx9wnxzlHo7wOBd4QG+ZayqaFR1zkylbKuZFOaChKwQITwVXWZ526UiNb++l3LUkrC+lbHU0/UGN1wc5n93fZXO6xE994DLDnR7/85Wr/P3HdhFd5e0PP88LF4/w8INX6cYB6zT449E+//U7YHXrFV574V7Of+hJ6ndfpLK6y71vexV19RxPXaqQLhSjRPHCG3UevntAVJvxm1/r8pEf/SbfeuoU2eAIR+59gf5r91Crpdz7419B1aeE4ZTl7pggHvPtbz7Eyf9mjN7d5+33vUKY1Pjj73R5jgPe1a5zZH1E8yMD5t/pUX1sQfZKQHTqDRY31ol/6jrUY0jm7H/7LravHGflx19i92tv44lnV9l99TQb9QkMZ8hsG1kW9p8+xfZexHIrI0tD/mRxi5/YjLnWD+gvMlI0V4MhD+SrPBFc4eFsi0RlaK3YEeM1tCsjHsy3mMnC7hUzsRpheBgRIS0dMZYZR+ixgoH9zCwHrSMN2tTNGpEmG7rNQBJOscYOA58gVSzsI0ezIm1Ac1nveKEHB0NyylkzPWdJtVmTnhmhM6MtTUOgFOUP3NhxTiy225GQTdFQIp+6Boz9SkUqxhMH7WOCwim64Kcdjr9S8FDMXnHv27kEOzniN8fIO3c9Cyy3+b1GecwkCti9F0pIkk89xMthrG06WIJY5LQk9pCl0HZTzXPTgnSKEWtoWnM7Nx3ZUxO0CLkYbkeVgBeDHfZUwkwyRiyYSs6OTEgl53Te4/lgm4iQTd1gTs4bakCsK2xoU5zclDF1XWUtb/B2lnlO7TAQ4za+ro0viABrEnF3HDNc5NREcTGbcEuN2ZERYpNal0hWbEzr0GBDdwChRoVrqs++TJjbWH5U98iFEreiQqITZla8JEAZA009oWJjftMSuBdkKBE2LawQjDfJxHIlIqu+k2MI2hk5AyY0JGbCrOBS6Ixl1THvQU+tlG3K0DrdRzZpcWtjWVooDKQ4ZcGGLPmzYkZqi1Ez/XC4fIWipers5wMLi1zYdWRUIHtBx/slGXf0hLlOiS0EJsdIbbtz0RGnDTx65r2tnCO9O/ecz4E/K2whXvG/x0GDCv5HpCJmFk6JmJ3g9qQzF3ZdeydaM7YFlp82eonlzPM3zBSmgM+UkzjXTS5PQt2uDHFKftpzOzx/gcOkYfNZ3O8rVCqdyM/t0vnFXr8dcfLm8/p2yFVRiJSEhW6bspQLJPdeTbE693lXucFY8GEKFIR7H+65kVVeK+cDxrncokesMilgeSsWviZCTdVMwo4xqU2tMqnbIwbWlePI2TNSWtIgFEWF0K/lim1GNezU83/5zN//rtOP8uPHf+GDjz/x6WfOA34dj7WZLFYlpELFTz9N8WTeT1eazEi9cpqSgIbEBqpsm1nOy8Y0naCpjIG2W2ctqVMnomb5e6ksrFGiOatSndJQdV+wdVXLQidj0tL0pXxvXLvKTK6KpvVffhFy8QvnX/nCR1DjDv39DjvbHVbXd5lOGvz+X6zyo4/cZDquc2WguZrO+cz8DZ6VyzzNLXb1ge3g69sSWOU3u9tEHOIWvDU0606Fif/gEhTThdJGKpSZnDKXeQ2XjNdUzXcIT+cr9JU1PpKAW3qfttRpUGWPMQvJbLKRM5CpmXCwYIkG78iPcEy3yCWgS4MDSbw6R2bNzbwymB2zpxjpz5odDc70DEO+nPsNaTDjFRdx/XVT9uDyXBSHb/WBuCh0XLB22EznHZDbVwgIPEbddZjLd+JOcK1yt6WswHH7Aq6pmEwvqCkTvG8vFssFyFs9BHMNl4IuSZ6wFHTZzw9YUj1CCYmtkWQuJvk7FWwSqYi9/IBjssJlvQPAA/oof8BTLMh4Rbb5kD7NNWWSi4XkVHVIT0esSERbhZzKl3iDMZeGOR9YrdFqLLiyHdFuZtz/0PMcqzf48FqNJy4HXHitzb9JX+Lzf+85rn77QS5earOxmvCpF0POVDpkmeLt7/kGa/2zfOZFzZnaMqc/+iWYVUgunyRIhH/9G+f5sQ+9wDdfWuaBzZyttSmDYZWzp3a4cW2FyajKRj2mUwtoNGZESvj6N0+z0psxuHSS/hsnWP7Yy6j8gOn1Y5z9JzdBKaQVowf7ZDd79KTHhR3h5x7ZZ+1jX0N1O4THd9GDMVIdk19bIh13CeNr6L0Z6uwmcXyBTnOXa19+lK9/e524Cl/dnvHBd75OcLyPVEE2luksPcddx25w4Zkz7Ew017M5/9kHLjPcXeHyKGNT1RjpnH0xSlNzyXkoX2HMgpFKqVr4VF+mdHRMXyV0bHKYiPFn0MCejFnXbSoE3JABDR0xFKNS5zwDKhZfP5OFQaiLtpNOI7+4qZcYkOBcze/N1ukro8jkOmcLMkY2yTJJRm7gSCSIKC8lnPsDzRxuiZ76rqrYNf5mPlQxmSjvXWxyUDaUctBNFzNMg0HZyUJRlDiIpUs/vp+HgWi4gobS7sUnRc40da5TWqppeAJ6jEazorokzPy+b0rMmMT6QxgSujdZtEm1g88sSKlLTM9i/tuYBHUrN8Z4y7k5KF8L9lnRDepU2cpbVFDUCVnXdao6JJWcno6p6ZAzQYNvyTYn8g4BwoVgm5aOOKFbjGXBUM3ZpE6iNZfULsfyLi0qzMjZlykf6LS4OskY6YyLMuA7wXX2xcDtGkQkzGlLnUwyNugxZUFPN+jLBCXCPfkKF5WJNx3rszKSGSMSA08jsQ7ytpNu43rKggqBl6R1fhLOyG0sRj0n1WaNt8QU2jv5PrHUzBkiwpiEphieTd0KBrizIhezjpdVx7JTzP2oEVmTSvFd1D09ZKwT47as5+xbVIPzkakpY4DZULExurSFwUhP6KmW5XlNPEk7VjGhBAzyESJCSmox+E0WekFbNa1p5dwnnw1nGGjhJO68cvyV2KqngXglyCqVQ8qU2u4VJ6XtIFNOSjWz05TAqiLl2kGlTUHveJyphfPMtSswtO8sm+m+g3I7qNObCwbzb2c4WJ5DQnF2U8qP3AS1gFO7s7A8FQV8HCuavA5pcbgh6B4VKZTWDj/KcHVLLC8hVqDI41yT8jC3rIiFrvFgpnuZnWKUbQWsqStueltIGjs+hiuwcht/IlW1xYWBBRlD14W/T1VCRAyJXGN4VU7EwNkmOIllx82LpEJTYuZ6Qd2u/abE3kx0rhf8i8/8j993AeIe53/hPY///qe/ct4VmE6KPCQgspy3hJlfjy2p09dDa9JqJkROKtrxhky8Nv9rSd02pl3srdGkRkfHXJM9YowYiJHeTtGYs6itWsytATMI+9oYlxqIsUPXaH82lRvaxRll7vudHNV/qEXI0cFvn9974SzkirXNG3R7I9J5RFhJWY2rxPUZN261uDLOORFXeTrbZ1vvE0mVAz30JnllQrN7mMo9xHXEyhvirYqN71aE3F7IlEelYGX37He9aZKqEWF4Hw/r41xW+3wgO8EwMIotxu5+wUyM1n1TYvb1EOf06ZQdtAhTyUnRXFb7XGLbB/C2NBhZcqErQLzigR2HZhisnxvPRhYbXsaTurGlG+U7h2fAdhTNJvddOozj6O1JUGgVetz1znXux26hhD5xch0NoYwZv61zKmLVcgojSlcUufvlFCZcUHf3pW4dx8vCBXe63+6et1TTd7HGekJbtQgJWJUOPd3gQMb08yENVTeOw3rISbXOk/lFH/Be5DqCIZrN9RylYl6VW7zBHg/oDRTC2HaxhnpBQsZ/eiYiGUac2zRwt6V2yvZ+latX19ja7LO/3+Sn/6vf44Wv38cF3edvvn+bTnuH2qLLb/zJKn/3Jy/za1+v8bm9fR5Mz3Lu3le5p1Pn332rxp9/7SxPPnuCKFli696LXHzuDNvXV3n//UO+81qdG/sVunVNt72gt3TAi2+0WK6HvHo1pt3IuXl9hYceukSjfcCt66sAZK8uE2ULskWN6tlt9KUr6MGQ1z79Uf63x7vUpw1+5qEpWRbSPvI6emcMaYo6sYmEM0ZPnaFx9mVUNyW4dwvCCvrygPHL5/jOhS0GU+F3xjcJdMDDnTaNU5fJL7dRywkEGbpfYfu1U/xxf8h/frLN6vouly+v8uwkYV+nRASEKGIqnNYtIgmYkTOwfg0jmXIyX+KmGlLXFTq6RiIpE5nTosYbsstR3SORBankNHTEnoxQKOpifCbqVK2uUs49+ToVQl7lJgsypsypi4H5GFWjCm3qvKRuGAU8feALGJcIYgt1xBQmdSur6jpkUwuVGevEHyBGFrSQiCx3Dp0qketeFdKLhwma4Ey7CklQ9/CQCoxbOlLs03IBcjvfqrw3DdfscEx23W5Hanaxu2oTZ+xuNR1sU7BFmMPMXZuZ5dSsS48MzQxTiOznB16sxE1qUoxLeyQRJ/JlcuBAGV5DgDCWOYnMOZCEd+Qb7MuMTeq0JaQXVBjoBZvU6UmVBZqLDFnRdW6pCS1dZSoZKzrmqoytuaLmVTEwpjP5MmNZ0KbKiJRz0uaJ6YBd5jxQbXCQ5dxUI6YWhmR0+gPaNJiTMWNBk4iYClMxHcRn5SqP5ieZqswr7lQkpEXMhu5Qk4gRCYl1JM/J6ak2CqfUZhLjyEKvcpzLtuMOmVhqQE6p923Q9nUcT2Vur6tgoLq5aDN5wECpplY1TYA11StJNOd0pMFAj6nZrr+ZfOGLShO3c6PKpWdGgYoFbWlY5TMjNRxgJiUG829ivVEGmiNipG8jK5GrMNytjmqS2zPNdYjdHnLJq5tUasxncp4aUHAdU5vwOUEI58uVU8hfu7zAdeYDjLpSgJFVrUrVKFdaCHWOhWRZgroragw0+HAibvaYKv278OgpQ6PfPPUv0Abue2W380MKWOW9+6Z/vXUTotzgvCPi5LZG8e2/yU9CpPDkMJ+7KPyKd6P9hFRwUCjtiyZnHuhk140hrVlDYqFCgeXIKjv1aqq6EXGwE1pXvLhYGktEVSrepy3H5RJOKt3kbpFtKMf275kTXhDDgTIQwjn/8rOf/IELEPf48qf/4nyIM5fNmDB1F5EKgYdVGR+QuV8DDq7alLqPCwbGWED2nTBKRkZbGhhT0So1QvZlTNVKjg+Yet4XAs6U1l0vRJgz93etAGWZfxf+8CaHNEbe5vr/pRchV498/PEHK79+vnX0Mv3XzpLOIjorNxjsr3DzZovXr7b5/M6Az6inGWUVXuIqIz2xBjalpVvaLG5Ru03m/rwJ3vMDFyLF64UW2uC+HohJqU0Xz5CVV1XPKGSIcA+bXFV9mjrmfZUl3lVZ4SAVtnSXueR0dMz5/BQxNRYK1nSLU3qFq9InosKabjEmZUM3eH+wxnPsclzW0CLs5QdG2ceq7FQk9FrjCoNlDe3It2YPFCfdFlr8o5PrA7PRBbFkoiI5cQmTg1oZ4padPjioltVPL6QFnZpXYD1SbJCWkKPBmq/SHSSjkAzWJZf1otiLbXF1eLxbFCplIYKaRIzzsVUAy2kqQ4AsSHxG6SpUoZ0kGTWLST4hUgZSs5cfsKVW2ZEhHerkYhKhK/lNpnrKkAQn+3hvcNIqK2n+un4XL8s2B2LMfXLRbNBjlRpfCF7il1Y22Z5oHmjWeG1X8dGHByTTKv/tqxf56ZUuL10Pedf9u1y+skSznrL7wl18+Cf/hNrz76I1OkKrtUe1mvHI3Xv8758/TU+q3CVt3tgNeOhsn19/vMf/8Lf/jPXZMT76/guEKqLzyAt87ouneffJBeNxlXvO7LPRW/D6jRq3durs7LTpxtBpzXnkp77E3qWTnDx7idaHn+b6N95Od2mAzkOO/uoBSl6ietcejBLU5hr61ohXv/YA+bjG69M5961nnPw7T6BvpKj7l5AKyPpRsie2SXdWqH9cQwWu/x/3kT8H8xtrxO1bpONVFvOQZ5IxfUlY2t/k5tP38pWvnubUrEL8vhSmfRbXjpLutXnHfds8f+EEk5mCeciJSsQbWcI7qh22s5QhKUtS5apMGKiZdeVe4nW1y4gZR3WXsZjA3JcJE5kToPhAfpQX1A4beZtccjZ0mwYRezLmfdlJLql97s03/j/q3jzYjuu+7/z8Tt++ffft7e8BDwsBgiC4QdwXURBFmpJlWVZkQ44SzziVlF2VvbLMVFw15dHU1CTjpDJjeyrlqZlKZjKRE4mWZUWWJdnaIFukxH0FSJBYH/D27e5L3+4zf5xz+l6AoOJNHk+zHoH33sVduvv8zm/5LmTwqUufvgw5Hu9hTTWZk5rxH7D3523RHOuqSVv3mZMqO5iCpiZF8hLgDK7u4xBtZfTjnXxjTjJ06LFHTZGTwCZIVhJSTBLipHFHodBMJMe5XUlDwAZ41zlMiZ9s8qElDzrNehMPJEnQ4mumzO+GM944jurrvpzBl+vCxnZaaxLjvDIy5omTO9qqgkFVioaMLnlSYuQj3aZXljw9K4dpIFqG7A6jhGVGVTkaT3BFNXg02kOXmL4MSePxMW8fK3GfGOEOVeYMDQra54ruIsAuAxqEt79AZwAAIABJREFU9CRKpl57dZEmIQd1iXfULgEeOXzjHC51flLtJ45NnJqVgEPpLC/Fu/QlxkfxvN7kktrGYdkdlEIDHUxn1Sn11KVrExxhDzV2pcf+uEJd9dEC74v2cEZWWGEngWQl3XQbw7uWv+Tge85pXNlE38EknKJbREQan7aFdYkIDd2yEwafFB5tC6VycCmj1OZRVnlCIkJCFtQ02xbe5aBmLXqJ9HUan47uUlZ5HGne7dkhQ4qSZ1c3mZIKm7pOWeWp6zZt3aWp2wQScMhbYEc3bTd7wKRXTRpjHl6iehWjaelOIrDgkkuHpXdNOTMRCpPpi5ucDK1Bm3lvYXKOIozbt1ljplPvYM9uguSKk6GOrAP5CAZmkAuS7LVOyEKhEr6mK5Z08hpmv3YIg1FnWcYmn9eWCqNiY9TwuxYhMg5j19c8bnSMnnE0URifeL430iT5+Q2Lm7FXsNAvV1i56OGaoC4+uSami2em8WFyl7Q1ohyBMM25cfdYTETW5gZaGzgcOMsDax5om9waTVoZ/5pRDqNsgTqCpAtCTsxzpq2SFJh8qM/AupKbhsFQImpSepcE75/0ePzkQ6c+dPKhU7/71HdOhAyTfaIqea7EG9yuFxM39CkpE1rJeicB3aWXXKdFNU0ksREsEpMj5lWGkCFd+hxgmr6Yycg2TXzLV2vQpqV7NHSLtKQTVE5HGyFwN0Ez12d0zUbDAbEFpJnoOAlpQfFTn3r8R1uEACyc+/oJlCa/5wqXXj7GxPQG59/ex/x8nZX1HL/WfwYPRUuZsa8jdcOooh4nRl/PP3ALb7w4ub4QuSapvW7h3KiaTxae7Tq6x8UYsyRBcV+8n6JkWZcGR+Jp6qrPlM7zM7fCQx/4AS++to95yYH2KOmAbTEV7HRcYCgxee3TVzFHoilupswVaTGjc1zQbbalQ5cBZXI06JhukTIY6YoUGBInJKrImoWNJOxcoUGycGFUrLmuqOso5FXOEov02Ogztp0Cs5idLrlR4PETbXaHL0+Lz81qD9s0WVCmoz7AGgHZXoPrnJiSyBV2HhmVsRtDZDeF0fVN2+IrFjdadfeFIY516BHqATllxAAiYtPVZRS4SqoINpHb583R0l1qXtluzEM29A4Pcpjvx28aCEBs5C+LdqONLIH4Fua5xAbTqsZzvING8zf0/WSlQN6qmC2S5w5mmCsKZ1oDbqulWG7FFL2AyYkOLM9xeStFN9b8YMnn8eObfPOlKm+tK3Yv3MTNe7qI0pQn12huz/EPv5FngTz/6H/9Ku87+ja/8/Qi/ZU5nry1x8QDp3nu9+/gwIFVaj/9Fv/9P36cW7JG8vrev/UlvvLUPfQ6Aftm+rzvfe/w0puTPPnEC2SCGD/dZndtntfOzDEVZpj9Jxukl69QmlqFxkVoedAQvMOThN/q4S1GTLFDa20v9x3sMzW7QWqrgXc0RPYeQHwPwj7UtwmOrEM0IPxWmfy+SwS3LPH0f36Y188sUs7HnFnxeJ1t40UQlnit2Wc57nFnNUPl/lX0Vpf1N45wZStFoLOI0kyUQ9Z2U7SjmECn2ImG7EifPD7bGPWhIkY2+ZzaIiTijmieTdVhf1ymKQMWdZXDcY1YhLfVDhWdZVO1k27YujRZ1DVKOmBSF9hWXUo6Q1P16RPSVkNyVk3JeU4oFHXVo0XPdkAVdTpkJM2OblmHbrP6UsrnfdECA9FkJSCQNPO6wiZN5qlRly5buk5EZLtvTr9+pMoyDm9wa3U8vjnIyLUGriMumOmmjeAgriBxBcj1hNQfBsm6UVJi31wymXGTU21hnI5A7BoTroAqqwLO6NCZDKbwqFuZScBME2QUR1wR4ks6UcfqepqQiFXV5bZ4gqvSIkLTjjTHZYI5leH7epO7VY2e3TSzeOxVWV5Q6xyLa2xJj4AUHRly1tvgTbVJBhPzhhjZ0So5puIsu4QEKHYJqUiaIE7xSKnI64MmQ4k4Gk+zpHYx3hQBG/EOM1KhTZ9Yj6Bqbd0jEvPJ2mLgQx1lyO8+ivNqA43hQXRx5Gxz5gNJE5BGiZE7nlJV2rpLRgJckhnbGN2x0rqRxAT4o2aScI2PSmjjfNZOBk28N/dbzzqU9ywcwxFZzf1lioGKKrCPaZb1ViJx2tKdZNLV1T06uktR8kxJmUB8Y2SIeZwzXgODOnDSxIfVHjxl/ES6ukdWsqRE0bcFqpk+jIqfgmTp2CQstr9PJZ4QKUoqj1O5CglJWeKyKRzc6/ujBBjDO4nsVMTlChoD9UmrNH3dTxp8WrTl1Di4k92PbSHmpqSum+/e50ghimQduZmB26cTCPWYueC4AtU4VP1GMM5rCxIz0RmhTnTy2uO50/jv3jMuuHxpjJB+vZqpiV8eDu41DilzLt6akadZAuOzsrnjUz5HWvcllcRJo2ro/JCGZK0oSFFMIex4EYiQE6MQ6NAlSowRZceKa7j3pTAiQh3dpaQKowIWA0EsqCxVKbCjWwTi88+f+iefeezkg+9KsP+0x4dPvv/UT5w8ceqlp147UZIcGqjToa1CQonY1k22aTApZQZiCq0ZqdBLIKsFNrQxUHaKe1Oqyk5slPUWZZquDEiToichaUmxo1tMUmKNXTSaw2oPS/EaU141aU5gc1BzDfzEHmKk5zXihIyg+jpBzfzIJyEAC5e/cuLctx6jUtwhpbOE/Rwz8+sM+xmOHL3E2bcPsqO6tOIOO7qRdNxHuEiLnRwbHyYFR7LIXOXlZObevTmOFxrv+ffr/p05UQ4SlrKW9332qElelyss6BqX2KCuBjwRHWBWsrQaGZYv7uPYhEc69ri9FLDPz3C6b7qfO6rLw8zwjLrKoWiC894OW/TZG5c44ud5XW9TIktHBqzqbUO+tg7Fs1IjJraOxSbAeZKyEr59HB7PdT8SZQhGhmYuZXHFXH/MO8PAlUwwSifkOixZFlvYmI6Nwd56THoVQj2kjZFg9ETRpU8zbqExij9OStDpdxsd7hGUw3VGCypPX/fJqzyhDsmpLM24RaQjfOUnG5PWMR16ScA16jsBjjMSqCApbGJiiqpAlz513aSg8hhDqT6BpNnjzbAtHZq6w0McYVO1KEiO7bjOtFdjTk2wFdeZkDLbNO37NNjzB2UPX5Y3+IQ+wj4p4CtF3lO82QjZ4wcoLaz2I640NGdWfGqpFPfsi1jaFXyEEnn2TESc2dLcPhezvmOC69TMGjoKuD01xU/8nS8jWY+lL3+A2ycUDzz+PXzt88//z+P8zV/8Ol62yxu/+Rif+pU/4uUvH+To/jaDpXnUMMPVXcWhvW16nYKROe2VePqlWSYzOX7wRpVH7ltia32SSvlZvvXZH2f//qukjgt4ffRmhrc/+wCTD7xBdLpKe2Mv//HVgHKUJ/BSVPZeRs3HSKmMXr6CFEpAD6lVkEIRvbnLC195gkk15MrSDP2hMDsx4GurPbZUh/m4RD/WnFM7HNQlfnXtCj/34Q1kvky5uYJqzXHktrdJ4dPrpZnMwzd3W2TxaBBSI8Pt2Rx7/IDuECYIKEkKiVNMUGBLdZjQeebI0iDkEEWWpE1XhhQJ2JIWeR1QImAizhnDJu0R2AQgo1MJ9OuibHJXNM+qagLCTXGVog7QAjl8GtKjSoF1vUtRchzSMzSkx4SUyEnAtFQItHHSviI7LOoascBFNpiWClvSIotvTfuMDKvjaKTEJ7ReAuOxb9zjI/mZneSZgn20DSiMD0EsI2y56wZGYxPf94JXXH+4AuRGCneOt+aSDF98S+w18WbkSu1bKWOTZLStypMgVCTPit6mJkUaukObrimgbLKpRCWxIKuy5CVHWRU4Gs8yExdoS0iHCE+ED8gs70gDP/a4jPFdeoc651WdnkRoLaAFX6eYloCs9lmzUKz9uswhXSOjfTZUGxHYG5foEoFW3JHNE0XCsnR4VtbI6oA3Bi22VIciAVmdYlf1yGKw1kqMJO123CDEdOJ9y+kbWP8LjXEoj9G0rFO5iFh/jpE/SCBpiiqHh1HmMaaTniWiqyTRdgmcu8IOjtW3aUSojYpPQ7cs1NYY3vniG5I3hsvkS8om/kb8ZFHNEBLZ+ccwgXBpjKT8rrQwE3dzfzmisGk2DRMy64AhdSuR3ohbxiyOERehqPI0Y3PutZB4PZVVMTE+jG3nVxDTIcf4J7R0l9A6lbt9NOG5iPGIGlhSsSfGYNgVao4H42Mmjn09sEIKYg3trDSpbc5pdLJfmoTY8gu41stiHJI4riA0ErNw12q8+BDG5U9dgTRiXl1v5qsT+JnLkbzrGhnXT0I0Iz7I+JR1/HAFiJsOvHvtX+87dG1hNF6QaIuMcK7lMIK8uy66ixrm+sVJ4Za25nrumhiYkVk7eTGGk1krO6vE+LMUJG8+jZDkjK44DCTAKVgNrCGga9QM7cSqrwfJekuJ8cwZ2uavKw4zkmZCSvx3T/29P9P044cd7z9576lHTt576tGT950K/9PGiWXVoEwOTxT7mEREJTK7xoHJFBSm8eMl5q9gBCqOqkWUKHZpUaHA3qjMQzLNadkyMEWJGTCkGbeMCTZ9mnGbQ2qPmU7qOGkshWMN4PH/kmJWjICSuXZGYesvpAjZU//iiUw7IDhwhWx1k93L+5m49wXefOZOsmnhsX0R71yapCR5rloCsMOtiV08wDULzX04D6dRPVrITu7NKAqMJiI32mDf9b3tEORULknLEaOW4Az+JlSZGKhJgbMsMys1FnSFdemyT/L89b92in/xbB6/nWO5H3KuE/Lx+7a4xatS6RX5QLXISjvmM491qLZm+JvH+7x9qcx3vHfYimOm4jyRaIaieTg+wEGm+Sn/AKs6Yk9cZiYu0lNm5JiXLA3dZkqVuVcfZFf1jB+GXZg3eQvs0MQ5Lbug5TwPcpZkm7V/Cma8nFYmGUqLz7RX4yCz1KVNWRXJSMAeNUVR5alIIfl3e2SSFl024x16umfkOTEVslOhElFkVIBCJaNQs0gMTtZhfUM7RUn00kXIjUkNg6bkGQk9BwPTdiPKWSdP12VyGt0lVUgKMS1GUz4vWS5HK+RUjjlV4yIbFCTLHiYM9AqIRTOlKlxli4oqkpcMaUvmfUDN09Eel6RJJk4Tak1GKWaCFI3Q3IudSPPph3c4s5RjOeqzmE0zmxeqaRO8/vXSKqfVOt+tN7gvmMBPQWNrmv/hOfjGdpNDl+9m9q43KZZXqBS32bl8M6WpJVbOHuStl46Qbs5z6z/4DvUvHCavcnz9jYAH7lxid6vG3ukB03OrTBx7lVPfPYxPmnM7MZ+93OPmdJ6vvZXhd9fbHNk4Rr+fYrIywGvt0nzxGEF1izMvHGXvoXN4hztcffp2Zr0siwtNFm99hdSRbSTwkKlZMwkRQW/tMPxWCTW9xcrX7+fIx76BV2gylfaoFeBbrxUgFroS05YBz8lFK4nr8YlgD7c+9CqS9lClHeb/Sov03Zpc523Wzh3g8obPjGRQWnhaLXMrVR67vcVTS8Zv45yq8wt3QW+zhI6F270Kt2RyvBV22FQdBow8MpoyIIdPDKRsry2NR0mnDZzHn6Sk01ymzY6YLjWiyGOUqz5RneRst8e6arMrxichrT2a0qVAljld4hJbCTRyiwZdCSmTpUIOQdiWFgeYYl9cpaUMTGw53ky8EBxefGAVYlz3M2UhC+BgliqB9YzHNBcPkwmoqGvinZuQuu7heJz80x7jEp4uyTCJgIVgYia0WnTSZPCtKp1rVgTis61b5CXDtC4nghHOTd3glwsMdIiv0lRVkXmpUdQZ1lWTnsQMrJiAh6KpYyZ1lnNql0Vdoi59qjpLUQesKVP8tWTINFlyyqOlI455JS7oFnPkqHo+vvZ4R+3yQeYZariiGoDQCE2quKG6NKVHW4ZM6hwpFGk8hqIp6IBd6bBFk6GF9yjMZ3IQKdcQ6jFggJF+bWjH2hjS0T3EJkKOq+Dw5g4HPmCYdGU1hnvQjjtm4m09j7r0AG1NzMwdlJccbd3BcXIchKmne6RVGuxzOT+ZgDQhQyvbaSAZzuhT7B4aYlStevQt/tuzE5WULSQNNM/5hmQkzVZsuq1uCu98GMzE3xRWWetb0IrbdDH7nG9XcM/yhoZE9vnq13AjTRJk0ltDLo6TvQvB8k86yRow/h+aPn1CPbQeECOOVWg74c5Hx/iNmOvpHLidXOpo/q8TWJjrro8mmo68GycTytGaHDmmy9jvHKwpSarHFaf0aO1fy/MaK1yQse9HDQX3Xq9v0Lr86L1ixHvxQJIiQ0ZqmUlyKmI5dNrKHJtz62BoSpTl08Sj5qVdM0MiApXGwedcwVmQXMLPMbynjHsnKIyClTPOdPd1yt6jrlCeUJWkCHfvOSJOJs19QubUBCXJ0WXAokxTp83/+NQ/+pEVINcfRz917NTjJx869cjJe089dvLBUw+cvPvUwyfvOfXCU6+c8CVFRQq06TMllUTlMdRR8jn220ZCnyH79ATr0uCK2qGDkMWnTI4NMaquocTMSZVt3UQLbOpdW/yZSaEz5TUTOSOYkbLTKbCmt2BFNYYJNOsvpghZ+88n/MktdDdNeHWO7Y1pqFcpFfoM+gG/92yNb8lF8gQ0pJfcSEPtZMZskWHH/ONKMAar6l+zYJONDmBsIf2XChDsazjMqtPKVziTI4/Qat7XpMiCrnCFbWLRzFDmHiYppxSz2TwTjWnOdvpckiYtCVHr05zbFh69rclX3oGH5lJ8940yr7X6fHkp4kPlMoNulpIOOOdtM6nzrEidDGle8Za5pLsohKk4x47qsSZ1FnQVnxR/haO8JdvMxSXqyhQORclzDzdxRbbttqJtcA4IlFGCSkua/WqWyC7UWW8SX3yOyAJlKRCowGrYB4Y0KUIZUyDVddtCI7RVr0mxoevsxnU7SjXnfoTfBkcK71mtagSGjLDuGevePm6adK37pqamKkbRS4S+NpOYASFDHeLbaZG5wQ3pPqOy4AKZvZZGuczwX9bjLQIJ6GrTcSypPDUKiAgt6yBccz4Rogmt2dGkNrKuh3WNB/MlvjtcZZ+usOAH/Jv4RT5WmiejFJ/tX+SKtKhszjMRKPZmffKZmDuOv83G2gTHblvimfMFfnFyD3/1sM9d975Axk+DVlxZyfNLDw85cM+zqEKPeK0EfZ/8zef4wmcfZ2MQcdeeiLNXM5z5/cPsmeky6Kfxwgx792wxMbVDkBly9q1FVKfGTQsdFvaus78m/GDZY2nQZ6/K8ncfabG6WiKKhZse+x6pu1I0X55n4/xN3HLbGf7Tbz7K8MxhQMhnh4RhCsIixQ91kWwWwgG63UIvraLmp9CNHVQpojBzEd3IQKz42pfv58p6lrv2D/i3u0t0JKQpXTr08URxgnmeuGeb1GaaVHUZmSzCcAC9LtHpPNtr89xyaJOwk6fTF4rDHFO+z9ZOhkEo5PCo6Ayd3Ty+ElJacTHqcjnsM0lAH80DfoVvcplH9DyLFFiSFk+qBfoR1FWfjoQsqTofTy3y29FlmjrioC5xSdU5HE2S1T5n1ToLusw7vT77KHBZNUmTYjYusKCLbNjpS0dCSpLjYDzBkuxws55lVpfYVB2GEnNFttmjq7xfZvh9dZ6j0Qybqs2Q2JLUjfxnTGxNqEz31AlICDAyGDU/H2HgvQQO4jDVCm+06ds+o1tT0XtMjv84x7tj6AhG61xDHBjTFTsmtg+TbmtMTCA+aSvw4cjRgfhs00rctAd6RK5MScrGi8gQky10VBB2pM0hq1LYkZCOhOzVRSKErkRsSJuOhEzrPCWdIZSYjoSIFpZo05Yw4Yo0ZUg9HvKW2uYRvUAMvKq2WIzLnPbWqOocr3irXGSDBao8pGfxMBy9e4Iy3SH0JaYrQ0tCz9JjwISYZs5u3MSz0pcREdOqaiU3I+akxlCMO/q4geW4mIjrrjt+kSsinGmaJ55Re7Ik87yV0HS/M6Rxn6wy0wHnM6VsIu1ZBTdX1Lh7yCnhDImYUlUbG4cUrBSok2N20w+fFNNSSfgseclifAYyZG3cLqsiDW0MJl3xnFe5pNM91GbqMA6VclxBo9aYSu6RkCF5yTJkSEEZ9TnHe+pbjpI5VzHpRFLbrCuDKghtA0EliW/I0O7/YZKgu0f4TiDC8glGBoKmPeoS/KToscWIa2y6dePW50jVSjFeHLjHJCI5cj3pW1/3J8l6vBHMUtnm3fXr+fpmxTjE6oc9r4xNGK4vRPQYkiVFKnkV939s3oaQ3JvuPDtCv5P0BitsYeOkstDutO2we1YVKrLXwhUuborY1T1zne0937KeOB6e4aJpIxpuEBaGe5JXptAeMGTA6F53Dd9YNP/yqT++BO+P8nj/yXtPnTj5wKn3n7z31EtPvX6ix4BYYEe3SInHYWZpSo8uA/qE3BLPISKUyDCQiJrO01BGgbIlfa7GG3iiWNWW46YNFcDJv5dVAUFoxe0Rp8ayeoAERmuaz2EiiqT5C5Dovf+5X/hlBIgFNKSmdgn6abrNErW5ixQqW9x20y7He7ewvJNiQxl984yV/nITCWfQ5ZSYYKzLZhesL2mrKz2SmrvRcf1Cu17X2uHV3GjXEy+BAsQYgpoWzWU2yUuWw3qWT+RmuWkCShnh0nKBcj6m5vk8O9jmQ/4Mz/Ub7PMznLoCe/yAb9UbzHkZvirn+ai3lwudkNuyeVpDzXm1zRDNFTbJiZEl3KXN7dEs+1WeSZ1hIi7yaKbGQUr4otgTVQjwOKxrPJ5eIBvmecvbYEe3cHyMCa9CXrLmXGH0pRd0lT26ZiRLpUJfhhyOp5jXRd6StcT9tq7biXu4h6IgWQJ8o/SFl8CvHG7VKU+kbAfXkcPGPUwyrkvlRrMyMgFijGCek6wdoxuiaj1uMrTXIq0CtC1EhwzJilHGcBCToR5aN/vRNR1auWNfUskG65KmouQo6SxXZJNJKZGxmOkhsZGLxHRkJyihBa7Q4TcHr/IpfQcDYnbikL87txc/pcn48P5SmYcKpjO/2dG87+YW55YzbKxNMIyEXrvA15pblDpFvrLS565SlVy+DbHHh//6N+mvzPIbv32M+2++ytJL91E78Qrbz9yDH2V58LYtDv/9ZzmycIZbP/Qa+cf7vPN7Bzm4uAt4BNkW/W6Bwx//BnqjxsR9z/GtLz3Ia0sZbs9l2etnKKYVx5/4IwZb89z5ya+y+YP7yUYrvPn87ehYMewX2N7JklLCc0vC9zZCjtU8Dv7ct5HBACmVIV9GalPotRUgRvIh9a/eydabR/D6GU597QH2TptO//+2tEmegOPxNMvKdDM/GB9iKpUmNcyy79FnUZMKigWGpwTvjjJ6d53J6VXyBy6R6dTIENBt+9w8pbnzlk2uLBeYClLMZVI822myPgzZ0gN+9mDAWzsx99eyPL5P6HZ9Xg4bXJYmj2RqPB9tUY8jOjIkkph9cYmP5KZZ7Uc8GNRID32WpMNkbDrbDRnYUfeQLCkmJaDOkIydTGyrLkUyDNFUdIYVqVPTxpu6rE2Xf0XqaDR7dZXzssnrbDGvK6yrNjWdIxRzL+ckoEkn4XkJBnYU6ShJfDIqk4y0TTw0qliOw2Wi2YiU6gQnxuOcE5dwBf+f9Hgv6Na4Z4jr9LppzNBuYC72RnZi6oqUrFXHcphtF7/2qCk6DECMylJP9ymrAlUKlHSWEhnK2nBJpnWeTdXhw3qRKFZsSI9lZfDQKRRVbXxFQok5QokV6dAVoxBW0xne51eYJcvLssmu6tGXIW/LNjNxkfuDKi/H22xJmzJZtqVDTcwmvEvIm96m4YGEaVZUm7YMaEufIllKOsO2tPBJcSleTfws3J8DDCRrI95GRNHRfUDbzn1ycu0UOU6KCdefdgmbU4USRnwgxwsZPdYke26ikrVcDaeWZVAFFppoXagLKovzWRAZeQ50bRHVTeRxDYSmHXcoqwKb8S5bepeC5KiqooHDMmA93jIJoAjr8dY1ieu8N2Vgaxam5hS/xPInnDKSm6YoVJKUm707RdMKQBhshDKfT2WsspoD+4g9P9oW+UPr7dMHmzyFekjZmt2aoiSVkJgN1FASI1z3GVyh5JLhEbdCJ/e04y+QXJVx1IZKHjW+zhyM63p53PfMd8Z4ZOOyuzAqRsZzohtD1cfNBa951Xd976Yd1xciyfRDxyAjyWAn++9U3WLbsHCNQ2PoauCBPesBM9BmCuVMQl0zw1d+IuctNuE1MKlgJFWtu+RtXtVPGhvmvackxY5uJD5JGQnIS4Y+IXnJ0tUDfIxoRsfylkJMMfQrT/03fykKkOuPR0/ed+qxkw+eevHzL5/YoMFhZgGokmdHDAxyV7pUyHEwrrCt+lR0hpz2GUhES4x8ekd3CSz/zjU7al7Z5EjaqZXFSU6ekQA3bfTtZLNrC0Y3TfsPv/0vb3jO/tyKkCP/5jd+OXvwKmc+/1FmnrjEa//+SWYfepPz37mXQrlN8aHTRCtVwl6RTFrTXa/yc3ur7G3NshZH/E9f+Kef+ejJE6e+/1svn9iNGzgiVWJ2x2jaEeko4TDACOvH+MKz40QH8/lhXYPIJq9DbaT0tJhFuN+bJ7TBcE5qfELfzHfkHA/7c+QDzb9f3eZMv80LrQ5v97rcL1P8jr6IFng7bnKzlPhbv/D7/PpLAX+oL+FLivv9Kb4dr/BMvEoomnVp4olQlgJlMtSli4fiFbnMiyzzHJc5rzY5G7f5vr5KFPt8Q52lTIEvyIs8HV2h6Q1p0eUwc+xhgp6KqJJnXlfM5kafqhRYYisZRa7pXealxktykXdkg77uU5I8O7rFMfaSEo9N3cATjwPxJNvSTrp6DiKQFj8huYNO8LrmvJrrEFjJwiQw243DdS1GV0UzpWp06dGJjQpDLzZwgpT4ZFWWbmwKg5JXxMgySoL/daICaQlsYQMDPbQO8gYSUFDGiKyiCoAYOBkNBnpAmx4NzCaWlTRdBski2qZJgzbrehdbODwKAAAgAElEQVRPPA7pGTIoVqXL8602n+9c5PlOmw9WSmw1PPKB5sSDF/j57zf4YLEGQBgJt9+2xIVzU5yROrdS5o+WFC+dK/NjP/MdvvIbP8UdHznF3bMDrrxxBzu7WaYra2Sm15i75zRXXjrGRPVN2mduZePVW3nuPx4h7WkmJ5u88eYsUbdEu5Vl+p6zfPvzD/HdUzez0osopzwef+QCb16ocveRJhOPv0Nl5jy6HaD6Ab/02WO8vhuSaucpZ4Q48vit9V2u0OH+TIXn12Ie+fFzkM8iQQaUwM4GUi4itUnod8g+6pHbvUiKkJvuf51vf/sWvl1vMkRzWW0zq0usqAYp8TiqJ+jEMSUvxWzaJ1hYRtKCd88MaI3aX2H4vIeqtOivz1Iqdin7afYfWGPqxDPso8KhfbvcdvubtC8e5OZCmsduijn6wDPkNg6Rz0Zk0hETlSHl7SlmoiKb4ZBlm/h3JWR/XOKCqrMWRkyQ5nvRJh+qlbjQGfCmt05ZZ6mrHo8yz6IusEOIslyCJ7KTXBgaQ7qBRKxLg23pUCHPQ0xzkSaIcFatUyXPnrjCVbWbbO9dMcZtx+IpllWThiXwatsRdPCOjjbQGtdtNGaG13qBjCAm18Y1tx7HlWg0XKOc814FxXsd75XIjFbvSOXLtxKajpPgXIsdjD2wspaCgfJoNAWV5W69n4GKmZUql/UGB2WWnAQgZtogIlTIsyI7VMmTxqNAQFNCbo8nOEOdgcRM6ywZfKo6Q1kH3OmXeI0dBhKxJC3DUdEpbqNKkyHbccjrYibSJ4uzvNE3vIy9usgfxmtclC1ypBP/j2W9RUnM62sgT5q69ImIuSgbDIjwMBCH2BZRTd0mJ9lkQw+10eFv6g5pCzEy8rVGinyQQLdiO6XwkiLC8WQAJlSJ0PqoRETMqUlauptwNiqqiI/PUGKyBBRUzngc2O6/IHR1n5xk6ege06pKQ7fshMC8lpFUJvFMyEqGnFXZcZKdsY6Nq7M44zjTzNvSdXr0bQFtip2u7ifQ6rx1d56QEnU65FSGASFlVaIRt4z8qjXUNMpfPZR4VsxB2ym4SSAzloPo4dGy3BrnFRFawq5TKXPptyDGbwsDw3Zk9r41bnS8EKPU1U9QG8ryIcd3sERBknE4ojBuHuhWpGmc6eQ6jkPJxwuRBEyViEiM+FY3Pq6FW43zbdzz3wiq7iCUYHKicdXJJAaIM3L2zPsWSa6Nezb3ed1zGcniKClQAmVgOg5+6llYmys+nCea2KlRyior+db7zJ0bl3+4zxaIb6CrAoH47OpWIr3b1l1LPDcy6ca3Jkg8ZspivIyUbU72MKicASFVKZDFoDECy8fNkuaDJx94V0f/L9PxwMm7T333qedONKVPmSzr0iSNz2ZcN8Ie0uUVuUIoQ9akwRV2OKSnKeksl2XTKI3iWe6eD4hxhNcR9bhh1tYY6iUmtpPXIRnJ0IrbJJM+e2/dCIoFf45FyOxLz5xIxX0K6Ri1PaCzPUPt8DlqM1fJTq+hQzj3gweYue1VVt45xPHbr/CVFys8ebzBsV//r5MK6fGTD5368lPfPuFIc24S4iptZxSkxCOrMjgcpAsGnuWHjFfliveQnUyKHHPzOYK8SdR9m3SX6Og+gfhclQ6H42kWvSw7HeF0vIuPxyOpKd7RTbb0gIIOkkRjGAuNN46xNowokaUtfTphiligpvMs6iIHdJW6hPh41KXDjC6xJg3qupm85xhNVYoUyFLQacqSY1JnuaS2k0LrtniBlgzoyIA9usqWtEjjsyUtQw7CbGJVcgwkZiAhAWlW9TZllceXFA3b/Y8FtnSTCSnioQwRVxdoS89273w24m1CbXDCYINn4i8yhlfFYHv7upeQ8ha9eXbj+jX4UHd1erpPUeXp6T45lbNBE4YYmbmcyloogk1yXKdFxwQqg7bXzwXRvh655friGxdrKhZHPkwmJYZH45yfjQa/6yTmJJMkhz4pLqpdzisjt3xEV6jqAn977xTn11P8bnedw6kC25sV3udN0uwJzb7wyN0rpIMeTzz5Ig8Fs+xslNgJIwI8Hjiyxs13v8W5UyfwlMflpRp33fsync0FsvctEb0+w8SBS6hDPfzUNhdevpVOz2Nuukultkut2idID5nZcxUv1WahGJEZlqn5xuDo2Iee4Y5PvEa5soyuC2oqJF7LE3dzfPutCodUnl+LnufL25vs6cwgWvFzN2XYaHh8+vFLZB+OoT+AXgfxPEilQMeQzoCOEKWQzBaEIbqb5n95Ic+SqtMSQ4ddUQZ2UdF5pnSWvHh0Q3jgk9+l99JhUlNr6KUtZLpMfOESKttEPPBaacJBjjhK8cbZKYrtCWrz52jtTNPcneT4+05z5uw8b656zPhVjj7yPb74nf185NN/wNpbhzh+bJ2ryyVum/R4od1iQmc5TIl5P816bHTOq6Q5oAocXRhweSvFjC5wRTW5NZ7gWVlHaY+WhOyVPCGaZ6Mt+hLRlyF9jIeCCMzpMtk4zRtqjSIZPqD38qxcJkOahnTJY4zS5nWFoWguqV12MWpQzuzQuPr6tHQ7EVoYTxZGyclIptf93qnduOdxnLpxRcEbQTT+LMeNExptu8wj7kNkoTWOQzChqjgTwllVw5cUE5S4KiaedhlQlQIajLIUZpJaoUBGG5JqS3pM6DxZnaKpBsQadlTXfGYtLHtGVKAjQ97U5nnTOkWGFNtWTetZtcYVtcudTKG1okKad/o9dq1i1dtqixldZF0aOEM6hdCkixZoiZnerEuDApa/IEbp7OZ4hmVVp0GHGkW6Y6RqD9P59SyULiMZcx9heCJ9+qQtn8Ipo4Hh2gWSTjgNZjIxSBIx5y3QZ0BVlejaqUWIwcG3dYdm3KJDPykCB7bxJoKVqY0TyJiIslyJmJZuo0QS9TJXnNTjJpGOyKgAp4K4qxsUJc+MVGnRNdAqRkqMrrFYVRU6umup7pqaKlnDtZRR01IFW4ClElle4xFiioNQh5b8bRpaPd0nJSZxcudDWVIxaBvnXXKtcfK9TgLWrRnNaO8aJkI1DjKEJTX7dv8bQbXGCwm3wtwkwxHN3fpw0zAHK3RrO77mOcY9t94tpz1+XN8ccInf+Bp9L6i6ewZwJPTRux85n6sRJwXnxzEydh6pX6nkPLl35CZTTuFqSETWIh4GVn3NoCBMYetL2hon+0mxHigjkJCRgLzKJXmhj5mSGXWnZuJVdDcHucIWVSkQYWCrPW08cYYMKUve+ImALWA8Any2dZMZqVKjyDZN2vSsEWGXCSnSose/eOqf/qWcglx/PHHy4VNfe+q7JzoScls8R5Y0h5jhNX2RRZmmIDnW9S5N3aGocjysF3hRrVCQrIG6Ycy1iypHqEMrAdwnIwHNuE1KjDeUE8jISECHbjLhSlm4Z2RztU9+6skbFiHqRj/80xzFB16hcfkQfqaFHvgMhx6640EQsv7SPVz69gkO3f80z3/1MXzfjIzv/q2f+MzVf/ZX33VB/+8v/M+fKUoBuHbsOMKlmdHlUBtOQU6y5MRY0bsgJShykjXmLmMu224RjT+31kZS0SiQFBBRzEiVu9hPDoO3HWAMCB/L1tDAHw7MOPkNdZUvxhc5HFcIJSaFEJAiq326MiTrm07YitSZ0SVeUpe5IJusqSan1RYzynAwqjpDhjSb0uYmPUWKVCKpaMbZIefjZV5XV3mQaQoWFhLpiO24wavqCh3pU6fD61whJCKHGTM34zYbepeO7rEmDQCqFLmgV9HE1OMW9bhlSOoYw8WiZO2CNUTvZdlJEqmVaMPic3XSWRraZF9rCwOwPBSF0I7bye9iNJei5eT7gQ0MgFEq0UaZJCV+EuxLyhDiJ1QVIBnlu6AmKAIL1RLbURwXOggknZAt29YPpGO7X66gnZaKxa6naMRtnPPoQIdE2kgkO1fUwJrTrUidb3jn+YF3ibUdj6MLA9alxVpbszDb5tihOnsnIybz8PrpWV595SDPfenHmP6Jp3m70+fTxwf87Z98E6k1+Xf/x49RqTWII8UtR5fotWqUH3mR9S89iBS6pO5qUP/ancRbJY7/g68yN9nnwO0v0mqUaOwWWVsrc+XCfj77v3+EbruE1sLx4+eo5GN0O0C3NOrAAO/uIrojxI0CwcGL1Aj4HTlHXrLczBxzOY9HpgJu/9jX+eDdG5Te/xLh1/sQhuD76LV1uxhj0DH6nQ2G31+FToTu+WydvY17mEQhHI2mKJGjR0iA8QUKiQmU4mrU56XPfZRep8TFzz0OAUQvXQYlSDEmujhFcOgivU6G3XqGAwttJo++ArEwdfx55p/8Djr2+Pjf+yKf/OASez/6TXQ/zd/4yHnU/IBKpU23nePghNkKP53bQ400lZTH94fbPBpM2CIWiinFO8sBR3JGFa4pfbZsYXBR1XnQm+A0u2xIl/1xmY6EVHWWkIg79SJZAq7IDs96KzwQLbIpLZ6WFQ7qafbHFYbE3BpNUdAZNqQJQIY0oY7wLS65IFmKKg8wpvyGJW/aQkNUIiPpDtN39LjR4dbbeKxL4Bo3TET+eMfnvvirn/ncF3/1Mzd6nlQiczmCyDohCeP+m2Ir3iUv2WTyM6PLNOmauInBYQuKFb1NGsNTm9IlALKkKOmAHMYw7BVvGV97bKsuU3Ge2+IaKYx0cmyVYXwL4RlKzF5doEOfGCNCMakLFD1FQ/oU7LruiPUDwGdTdciQHks0sQT4Dk3dZYcmu3GDHWnTlZAsAXkynFGrTGnTONrCxNxObKbcA0yTSmtDyjYw0dgmbyaGDTAFSNGKgZhuvUri0cgtgcSlPGUlngNbKJTtHhpIOhELATNV6+s+kY4shNZI8Kbs53cy8O24jSamo7vWy6XAlFRoxE1i+3OAkiqStZOIPBluUgtUpcAubTpx1953pnvt4bwZHL/FkFybuoVCWI22SIsRa+1byFNf9xM5XvcZUhi599i2IAdWSa2n+5YjY4qLoTbO8u49hDokjemaR8T4Nn9w8vOxNsZ0Rp0pToRy3N7gph/9ZJpjDtMwUEkBHukoMTF1CnUObuW+d7mIWyvueo7c0kfqVlrH7wnB+i8dbp9zr5ey+7M5J+qar0hH17zOyDCY5N5w/879ftyTTBOPPp/9+XhDwvB8zXVIvF9EkVPZsXvYCNYMrUBAWvykABdRNOImTpI4EXmwnBHfwq5e5AIADcv/KEkew33yjCs6/USMYCveTaZ801LBQ3FOr7AoU8xJDYUwKSWu6i3+9VP/7P8XBYg7MqSN4agMOCcbNKXPnJpiWW9zRW8w0CEPyhFiNJ9Xr/NkfMAW5nEy6ezpAROqQkubhkLeFpAOegiWsyhi1pZVixtfPyNe4ruPP7dJSO333j5R/thb6Cs+3uIGk+87CwKr332EuZ/8LvHVKVTkUa12ufSPf/4zm/fc+0PHWT9+8gOn5j7fO/G6rCddhFiPpPFgpKQRMkxUl1yh4khgviWzOQzy9R1AhzlFTMeioLIUJcfReJa2DHggnuOTxT0shhN8cqbKh//F13n7Dw5xXy3LchPaKqKoA/6A08xJFUFxj0zQ15quDCkNM/xXxzzOrKU5pzY4qKfYljb74hpFAq7S5UBcYVf1SaG4L57lVbVKIEGi6uGI5s5kqY/HvekK7ThFrAyXpa27bOkGXd2jZ6FEA6XpYKYXJZW30owdtvQu63onwfu5o6OdkGNkpwFDWnYcF2Pcond0iwlVYSduJIS5eCxoGQKqOe9OsQoxBoRuYjGfmqasijR1BzDwqm7cpeqVEwJ8RERRFcjb8by2gX/em6JplWTMSNaQVJ2ClRlxm+5MihR5ySUdPgcda+suVVWiaPW0q1JkR7cSzXmDBRZykmNOTRBJxH6mbWHlU9cdspKmQ58d3eIO9vLV4SU+MVfi/mCC3bZQDBT5/IBmM2BhtsOxO0/Tb1X59Yu7PKqnGG5P8AcX4ZWzU+wJFzh2oE4m20VUTOX+F2AnD02P0pPnkVyb6HQFP9uEoYeaajJ9zzukHj/Ia//PAnf9yhnKK5vM/Niz3PXJSzRe3IuIMPuR77H4k8t4RwuoffPEFzaQyTxS9NDLMbqZ49GPvkr1lYcp9sr8q0+d5srFRSbLQ+Z/4hzZ/i7eIY067CP7Dhs4TRwipQpkC+jdLaScQZU1UisSXwQ1TDPYmaLdMhCV87JOCo+3oyUmVIVN1eVNdthSPe5OV1ncv0Rzt0r1+AW8O29GqhX0yjaS70IQU5xcpbe2QKMZMLtwlWGnhH/HJiu/+yjZfIvO5X1oLVx4+jiTe5ZIL15FNz0uvHSMH5zNcev+DqeX0lRzwmLR40vtdW6nyqtRg7T2WExlCLXmai/ky/FlqjrLrDbJ261UWZIml3TLiBfIgAf9GnNRge+py+QJOBzXuJMpzsgmfUJ6ElOzDtlLss0bcpXb9R5WlUlSyzpDKAZP7/hLbd2jrbtGBlIPEwnrQNJJ8mOw6Gb0PcQZk46Ub8ZR79d2S2+M6/6TTEI+98Vf/czP/OxHTrkv9/Of/tkPnxr/+q3Pfe2EHqtJxl2bPZvkGaWkHANC03HDwCA69MmR4e34ioU6pRIFmww+edL0ZUhZZ+jKkF1psxCXSYnHEV3hDbVOS4WsS5dt1WVGF01SoXMc9yosxz1aMuBVtcKMLpHVPj4eR3UVD6GlY573lllTLdKkKOrAKqhlyBNQIGBN6iahFYt5F0mEBTr06dKnQZuchTIoUVS02bC3adpmjYOY6CSRctKjkY4oq2ISv5wrupvzu+65Z7vTkTaqUw4JYKa2WRpxy7qdDwAzLdFiREUSzh4kyoWeeBRUlkbcTqYjYPbDjMok3eiYmF3dRAMTqpK8d0eU34nN9AkRLkRXqetWMhMwe7CReE/bBGacAzTUkVGlA3wMeVjEyKOH2smtaPrWjX5gSgwiHZFT2cRXAhvfleUZahmpQ0baCdxEyb4ai8kmnKCC8+ASwT5+5LMgOC7JaBrpoEjuM7oJvCcpjDy1l0y0HEfC4eTdarxe0tY9ZhxC+V5iEoarM+KguKmH+9no+1HuM7JDSN7B2Ne1ccL93U093H6eOJ9bqPzIVd4VG6YA8caaHua8mmQ1owwnybMTG2fIDFiDvEFCTjfqWSP56ZoqXwNXnJYKbXqUrMlpTYp0bBPJXe8ehlPW0T07HYyYlSpO8n9SSjaHMNe3b7lCbXoE+DTo8Gt/RiPC/y+Orz/1RyeKVsHxtF7iPr2Pt2SVu/Q+DjJFTuVYlwYRMc24zauySl23mJIyiDAvE9RpE4hPVUwDp607Sa404gRpy0yLExEI7byRbDP545/60I8OjnXv7/y3vxy2ywzPl8l8eAPd7dN94SjD1UkmPvYy/af3U/rxs8jukFf+2i/9sS/k6ufPnvgxfx8v6x0mVJmGxXk6LKXrZrjl4k6Mn3QohAlVsml4xI3GmY5v4sjcbW3cX8+xRlp83lE7ZHoFpvwUH/2HX+Stf/sEf3Q15ul2g4eyFb4+PMc6dRbVNB6KfbrE2zRYVS1u1lWellU+v7nOoXiCbdVliS1ETPfqPqZ4Wa2zoTq8L55mUmf5Pe9t9utJuhKSsYmuLylqYj7HbezlsmzzdHyVNercHS+yohq2EDOa2l1tVIhCTEKjgb4eGLdcKzXogqi2m1lGBUZxxIb3ZtxhQGhMAcXcWEOJKIpJIDq6S6SHzKamcNhu1+sxUo/mxhzJKpvzXvSK7MR1ulb1JKMytHWHmldJJlslVcATj0Zs8PWtuJ1wgFr0mPaqOPJ5n4GVNfWscQ7WEFElm4fbdI2gQSopRAzR0RRbHXoE4mOw64ZfMiGlRAJzKDF3RXO8JavslSnqtJmjZjxVBK7qTX6qMo8SzVYrxU7bY6YyJJ8PmZzZJDO1yme+OUFFB7xxvszP//3f5eFbVnno06+THzRZvngQT4SpDz8DXY/h+gTpQ1dBhqibFohOQ+r4FkQDvLsWkL37QXkU36nzwv91hMVjb6Lmeiz9h0fpdDIc+rnfp/P0UdI37RC92oThFt7Ni6iZfQy/uYJ3uEl4cYaT/+4gfi/La2qTe1OLtNpplrY8bn3gNbzFIRQLSLEMg56BY03vgVYDUj7sboOfhmab8DtFLrx8D6+enuebW20+MpvnS90l2rpHhz6OMNqky1BimnTxm1U+cPd5Lr2zn5m5C+jVTRjsoPZOE5+L0Oslwo1pBr08hUIfGWYoPHGWtd9+CKU0w0GWym2vsnrmKHN7VvBrW6jigI3vP8jBE99lb5Dn+69OMpEXlhualbbhCxQ9jykJyJDiUtzlom4zLVlW6bKlukzqHG9462zR4xcnFpgNi+SGAevSpRJlOaWWCRkyrUvUdJbnvTU+wU2cZZeO9HkgnudVtUpDt/l4fDs7ymyIZTKsSoMZXeIym0TE1KRoEyUjN+oSVDPJUzgjL5ciGdzySMbcHdcSQ9+drPxZ4FfjhccPO376Zz986qc/9eSpn/7Uk6e+8Lmvnxg3lHW6/jmVTaBNgfhkSLOmd8mJmWz0CSlLnoW4zI6VQy7rLB0JqWNIzbfGk2ay5m0wkJjLqsFCXOFYPMGW9BLn9LLOMCSmoH32SI5U7Cdd+KrO8rq3ys3UeEbWKOqAGV3gotrm/dEiNZ1lj86zrFpMxDkuqC1AbEw0e0RojbqGDC3vxVynDj069BiKtlPuHOvUE6iT6+AOdJg0gcQWpAM9IMKoT7nHGhK2SUizKktf9ymqAs7ZHLB+MxEdKzsLIzlZ9/eMZBLlGl982rpDpI1ZZk8PrPnYkIwyDt/TXs1MacTAWV2y37dQrZoUyagMgfhsxrsYcZmAUBsiu5uOx7YhtOBNsxxt0NVdMirDAZllJd6irIpkJUM9biWxN2OhOe24k3CMIh0lEqFiJ0kZlWGojVqW24ecQIqbTig7vXDNSCe76go3l8i7IsU8TsaKJ5cjuCusk8e5/cXBhkewM3td3cVlrOy4rtAwcCfHZzXfj8BcJD+78XGdtO4NJpzvxQNxr3TNvx+DX7l8yU1QXCGcGiPXu8/hijTzeuZ8JjLiYgqQ0HpyuNc0kLohGUkb2B2eKQi18QFzzZhYjxTR+rYxK0hinOmJZ9YjQ9JjsPJx+FlX9wFhQkq0dZcFNWk+D8bLJycZyjoLIqzpXapSoEtIS3eYkgo+Ho+evP8vNQ/kRscrT71+4qrepihZJqVEDFyVXbISsKLqNDDNLw9locXK+otY3g7GpNrHYzneTM6rW0M+Jud2/CuFsZJwkyxlp1wx+kdbhCzGnzuRSrXwZzeIzgcML80ShQFBaZPdF49SvO91IObZh379T1RJLn3+7ImneNsYr2FwqY6EHqOZ96aZVlW2dYOyKuIWat4S78SGcKdPrVCUvRJdbdzMEwMuMaCGvh2TG15CDhBujYwM49/55PPIZIt0o8BXzwV0ZcjX4gusx9sM9ZC2DJikTFp7rHttZnWRp71LlMkykCFZ0kzoPMuyixJFngxLtOhLSFEHTOosbSKO6imeV1esbnxIX5tuVkf3uI9DvCFXuU3Pc1pfps+AZVU3hlIYB/Ku7iaY4751hhWw8w3TAYltghNatQiH6xzYQO9KNYeBLogxFbyL/WxJi/VoO4E7GQOqcExa1IRpp4zlCgC38SawLawDrA1exrnXzLY6uktNlamqEi3dSXwUJjzTfWvpDj3dSwJ+1Raa48TXIRFFlSMi+n/Ze/M4y86zvvP7vOfua9Wtrau6eu+WWq3Vslu2ZGMkjMHGxhgvGIwzgPlgnCEwGYaQCSRRNBCTzBACJA6GIRnCMga8gWO84EW2LEu2bEvWvvW+1F519/2e95k/3vOeut3qloWtgD3krT+6uu527jn3Pu+z/BaGOGKcx9+WTJ5JKcYwizwZp/UebRwWS1kKMdyrQIYmXWYpsVsrrJsWq7ZKQbJkSbPEFmXJU9ic4YtbA166aChllKnpFotHv8SgOk1AyHcsJPjaCWd+dsuUEHYKvOc/vpSrF5TyZI1MvsX6A1dTOnKS1I09wscL2NUCZrZBsG+InrZIbgT1GrJzB3QaZG/NsPuVZzj/oSspf8cmnQdnmZiqkp5aIbV/HcmnoNsn2D8HoyEYw+hexa7meeLBG2CzzCeDU5TI8NqDIzRMMTsRMnfbOWRywsGjcgXotaFQRmvriKorSkSg3YbAUL3vSmYWztKtTzNsZvhUs0ZPQqYo0qCDh284UmmfkhT43vQC137vA9i1eSZet4VZnEQKObRaJTg8Bb01Ulc3CFbTdJplMtkuSVun9PJTZMIqveoMo2qFhR++j9TUEve97N13nN/5/Z+r3XzD587vfc3n6n+6dGspp6w1DM2RUkoGDK1SSRvqI8tUMmA+kWY4MrQZMa1ZRijfV5lgsyPs1ALHO0MWUineq0+SIcX1iQlSoxSnTY3Ddob7gyVeYfcQqpK3WQ7oJPcG51xQp8sJ2XKTRFEWbZGGGTCQkAYdXmoPUDO9KKirk6zFyUlvS1Zr7PvjGgdhjE/3nfFxHLn9BiEbz7aeaxEyvt70lu/93PujQsSbrvmkICNpchHJcSRhDDsdElKRIgUynJJ1Mjg514E4c9QKDpq0bjrssWWSJClrmi1ps99OEqIcoMQyXUZYipoiS4IvBktUGXHW1MmS5IxscUAnWTYtDukkS9JhS7rMao5V0+Jc5D+yLG0U5Zyp0abPhq1GyWxkAKiDiG8XOddHyVlRnLlpiy5d+mzRph0VB45EHsZFh7+GroHmrmXeZClKPpYMTUZd9USk8OfleX0HvqVtfDK67UfheZCRKS2JOJHbTghhu3Pt/Bq2E06lr04lqGyKMX8vKymSkop/X7VbkbollEyeqm3S1W4kOe1ifFISJCXJlq3H17+vfVZ0K2okucRyyDA2hvXvL8Q1yLpRvA8kIClJB+nBGRb6aWE3cr12xz6IiwjP49j2vXApdBB59LhJj9Vjg+EAACAASURBVLv3duLqP69egW67YPDKWgm25Un9v15q9mK4kJdDDsYKD0QuEIu4FOn8UhOQ8enGBfe9RDHyXAqUC5fGvJD4uMakga060ZdQnfeD92K5eD8fVyOT6IT6Qs3LP/v3MmCAV3nLmUxs2+Cl94H4uqYlRTdSzRpF0wzHBUmRIcW6rUW8IifiUDI5mtphypSdD0gk7jDCkpcM53WTGZmgRJa+uNeq0mQ/s1hxr9egi0V5xQ/d8m1XhGz96albF7XC18xZrrMLNMyADZrUI4xMQgKqtuE4L2Yijhsd7eE4eT32MsNInOhIXnI0I85iTjKxxLhBGOiArMngRTZ8Y9ii/D8feNdlc/9vmhPyklM/ffvKR19O99Q+wnPTSK5Puz5D4QdO0a/PxPf74qH/+288ynrJB77/jheHu7gt3M8MJW4yhyPeRxqDsGlrvChcJCdZBuoSdsHJ9b2Ca2JVq5QkHW/EOAWQ6WDqGaT0nMlSkBy7zCxFkycgIEuaM0Gdp80WiReuIAGYwH1QHzZnOReu4ke4I3UQgTQBi2GJ86ZBlhRnZZM2PVZNi5p0mZNJ6rbFCV2mLh3m1Zkh/nVwjBwBq9Ll7eZI/KUMsRRMjoop0xWn2nDMrDNlJihLkZLkyZCMqH2WkilSMkXKpuAq2GiD8NyIMIKv+b+lJInV0BUskTmgHQtBaUkxIXmmzAQZdVOEK4JdzAczkfNoEGM2kySYCMpkJUNKUhQk53CYJOKNKyOZmMMDzp29aArRZpuK3VFb2mXFbjBrKjjVlxmypEnhMOdJP2InZMvWHVRP3XF7AqPf9DyWOCVJCpIjhfvinLVrtOlRox1L+gVRp9mf+wM6Q5MeGVIk1bBqWlwfznON7GFCcyTVFZRv1Cs4VDG8ZmeWJ5YCrn/RIwz6Kexmid/965381geu4999apoqff71b36K//qRI9xzzzX8zP/8SYov+hq1jWmCRB8jyj3v+UEe+DevwOyoY6braKcPmSzmugkkqxCAPvUYZPKM/vIJ/utbrmfhhffxT37yJXzxoRm21qcAMPvmMQeuQcqCtlqQzmAff5zgwCqD9iSTEx0+bJ4mSYJJzZLNt3jqfJonzqaR6RlGd7eQIIEUK1CagkwemZhBB3202UTSaeyZIahSeeFX2VjeTTY7YkdBmFXng3NeNtkpU2QlE3cHAfoMuPXGdX7z374GVYOuVwm/sop9chmZmiZ8/DzBkVm6n91L/lUn2LHnSUo3P0DwArDrIcPNGYqTq/S72WeNIVOTXa7d3yEQuGJ+xCnb4b/1VjhcMXx10KAXKgHCT7xgxD+4VnnTxAzvra0yRYY5k2ZvkOXJbo+r7A7eXl7kznCNFiP+UeaIc23XLB8zx/mCrPDl4Cx7k1kypGjj5KCHjDivmxQ1TVX6LOkWRU0zIyVOBlVCQuq0aapTXvLeGQEm/jxfbvlx97PeJ+6q/t0tf90zJhN79gx05BRodEQOZ17XVMcdO88mM5QJseyykxdg78vqGgKnjPtud2RISbPcHZzkaVNlUweckXWmNce6adMnJE2CDkP6DKlJl0nyHDM1Fu0kf2WOk8CQJ8WadLnCzjKrRU7LJhvSYlPa1GhztZ13JGgdRVy2YdxNH0TTZQ9lrWqTLtscjI522SY5R2XM2LXNSjrmpgWRMpFT6XOO3m3txDh6j6/3UxLPZ/BrvJjJietAehlTD+9zbuPudfxj54MZcpKlGDVfUriYOS0lFAd5SUmSHUySlTRZSbOudQqSI1QHx9oIqwx1gOfpObNBG30GbPxaHe1SMHmKJu9w/NH7TUbk8E5UxDgVsMApXEk65nT0bC+ClrlENilJN1XSUYxTT0f7XsJ/l9D4+nl4sVUnPxyIiZ/L+52Mf24dF2fg9nr1Dug2vv1Scte+WPXP27f9C3gfECX0bPMrLrUuxwO5VAGiYz8Xr6/HARs/hnH+hoydG/AKfhF/ZOz9u+TV4ijeHpy1PQnxsaoUTfD8czq+ZjrmJrVsJ76eHsLlJ3S+IMlFME5FWdc6izLDgCGrdiuSlU3FUC//uWtplwlTIiBgxkxSkhxXhDMsyjTgBCFcEyggTYqcpljSTQCmKV5wDr7d1iQpbg0PEaIckzVSkohijqGuLeaMy5Gb2iYlSecEbyZYkAozUqYpTokuT4aabZCXXGx4vaAVUjiVskwkFNXRLmUpUJT8czq+xNe/y9dZ/ZAdb7ib8MkCZrJN+5EjJBLD+OanfuYdd8A7vuGnf+0H33THJ9/wodsvd/uH5H4mTZGW7SBRt6FuW8xrjn1mB2vUCKOqt609ylJwbrw46JHvXvjg1GPIfp1lQxxRrqwZ7tOnIRCGD+3k4QcPMqJBmlT8JXRV4NCppgAbpsPBsMKKadGSLlnSrOIgB50oORGcFJwnioeEJDEEKtw92uQaWeRBTgOQJkVNm5Qkx0hDarTpR8ongRgOmJ3cag9zl3mKtnb4Tq7hYTkbazcPGUVGPeJG8mi8UQwi1QPX1fPERBslCG6jPWfXAThu3Aa3pnUSEjBpilhVatqgZIoxnj2MoG9Fk6cb9rcDuziCn4gzripToG5bDKPg4jpRiVhQ4DrZw/32OAEBy3ad27iGO3kkJg2CI/Ei2wmZ15Rv2TYTZnvq5RxuRwxkyAR5mnSZM04+t6FtRjJib8TXQSCF26hOygavCa/gzuAUm6ZLT0Z8KTjDHp2iF+n5b2idbGDIppRa25AKoNOscMfXuvz+NQN+6ruXueNTZXbhvpQ6DOmp8p3fczcYpfO1I8xfez/1U1cy97IvsLLyGorFPmaHYKsDZHYGRiG6tIW5+gC6vgLDEVKaRrtneOuPfYrHPn0bS2aTj7b7ZE9Ns29SsE8vY1IZpJRFKtPo+ir2XAoz0+fsiX20OwlmtMiy1ClqkrXlHbzhn36AwYN7oDhJ4rVFMNHm1G1CmIF2w5HSkwkQg5k3DO8tEeyo0qznmNmxyfFzi5yXFqfEBfGzuhE7LfdszxlCkeCXv6gcVqVQrjP66hyJF6xirj4CqQzB0TL2ocdIX72GPW0IFteQ2Qz2ZJfm/TdQOPgkOjKwMc+9O//THex8ZmzYaiS490yGDR2wJwEb9QQ/fQM8enwHk8Uhh6tFDk7DEysDPv5gnrfctkSjmeb3XnGKP/yLozwUNqjYNNfms2y2ezxVGzGrOXabHF9rdzhjGpQ1Q0t6vETnyMsCv2cfZKQhN+geCprkLvM0gvCkLDsJUXUd/xWtMS8VVrRGQbJ4gl8Cb6zmPsv/5RIdpB974z+9PS1OLea9H/yNZ23uvOUH/5fLxs7nsjwB/RtZ7/3gb9zxI2/4+dvBxbe2bZM3eRSlrk0qUo5hpOBU6BIY2tqjHsnhJnEk89vC/TwYOFGEvXaKec1zxjSYtXnOGxdDC6Q4Z5pcYxcjk8I+M+Sp2BxPmVWSBNwc7uK+YIlTss6VzLPbVmhJn1VpcEBneNgsUSBNEQfPOCNblHCeLjmyNNXxJvzEYEGmWNJNN6mIkmlw8SMkpKfeRXs7XvkY6Um4RtKEOmScvOn3JFWNzb58wuanHQ3tsaabTJhSXMB5SKufDAQEFzRngkjuePy+IZYtW2e/WWBZt6iYsptAkGJaC5TJ0pQeB3WWhzkbS/yOY+9Xww1XHBHgyfWOzB/EMI3ZYIqVcIMkCaq2zoypMIgYe4olH/kxSNQw6tkeIkI/Iihvc1UkLi6A+PFOBMURkVVsnNj6a2LVKZH58xCqjXgAbh/3e8QI4om9iMaqUOMCN4KJ90iJmDbjTRZfPPi/XKrQiKcBz+L5cQFJ/FmaCRff9mxqoJd87rG/+6J/3HTRN2zdNSbOm9x3NHFBcTVkFBci7pyYuADtaZ+C5GKhBHdenciQ9wYZRRNAz30b6pCMcaI4HeugfBkxZEnT1yFrWouncWlJsmUbsX1AOPYePKcVYELyfFoeZSczbGideamQI00L5wdSl17MQ5m2eRYpX/bcfysvg9BgSI4ESQz/OH0t7+2f4Yyuc4gdLEs9UuIbUTFlB1PH8XZqOMPVLVrkSLNqtyiaPE3bxqLsNwt0cUIPoVo82+yA2UkCw2l18foPPvBrz7qHfNNFyBev/P07jn7kF26X9BC7WYj/fu/0b93BT36zz+7WKz/4g3e88jK3/fgb/9klN9k/NfczsiNnmCTFWJGko10CCZgOJp0qgg4JCOIE1c0VXCJwXTjP08EmFSnzpd+8mXxuxItu/jJLH30xd/WewqtgWDFAyDV2kceCVbZoUZAUy1KnapvUaZGWNCpKlnQ8aq9QIKMJmtJjB5N81ayxx5a5Uss8IlsUJEdAwKatAXCWNSpSohWpkjgH0QGb2uRkkGAn0/RkwH16jJRGPQlJ0rId0pKKVU5CtRRNPlYUSUkASjxl8ATBSZOjYd2X1rmhh/SiDdSqxh2mHWaaeqQtX9MGOclSkjwBhqLJU5I867bKy7mKz/MEKQxV24yDkEYQBS+fN7TOZfOL+kSkyuA2lo4MCTSgbIps2mq8wYHDK3slFnDdsbo2mZASfQaRIowjUZ7TdUomj1fGyEiaum3xCB3SOKhAweQok+e8XedD5lFUlaQEzGmJCjkmbYYNY8lrgqxJ86lwma8uZ3nbzhJ313q0PrfIP7miz1v/+CB/8sMnmeEoHwgeJU+Gd/7iy3n32z8LRrnzva/mFf/4g9TvupH//TNZXvOl17NndsjkdBW56hrM449AtQ6BQSo56LQwh65D189hH/syg415RoMs17ztI5h/dQtvmZpi144OwQuOQq4I1mKfPInkO9jTPYZn9pKePIFauOKqk+w/uZPXpRcZGqXf73Pf77+em372L2GYd3wPa9F2HYqTUN8Eax1HJJNDt9bp37VA5s0jVt79Yq58/cf5+O/8IIkArpVJzlFDyNIWt7F4eEZfnXDCw5zlEGXu+uIebr3FMlGsuddUi33oMbBg9hexJ5oE+9PI/E7sfZsMBylIhKTedgtzD9/LicvEhvJ/fvUdvPG/3X60mKM7gsXZPslkyPl2yOG9A5KS41xVeOPuHOc23ca7vJXgP3/gIEINRTllGsx2U7wwVebe4RZV0+WmTJGNjrAmTeYoYRA+YB5jp05hRNgpUzwpK7GpG8AEzoMnLSme0mUysXCDW4nIVyFJIhJs4JIFCDj1wMu85W+55QsRH3d8tzoVKfcVJEtTu64TToI1rVMRp4TVZxDBCBa5P1hx8ZshIiOsqFN8CdaZxkmJ96KJRMP0mdQsSTWcCerssAWmKdKkS8UkHSldJtiQNk3pcsDO0Jc8fRkxp9vJxrTmWZMmCQxr0gKFCVOkGan9gZMQBq+eFEQTixEDwSVJDCKeXi+aRLsGSs4XnlFjJhkpO3bUSfwWJMe6ViPOwwgTQYd8DG9bV3gZhIZtXlDAJKOJ74ZuRV1rNzFxnBOnAnTE7GGFKkWyHA/Pk5Y0zQh2smFrJKPXW5IaMxHJ/5xUmaVMjTahWjaoM05kB+ICRLH0ogaQX2uhK5iykmYjrDq+IdsNJT/dCAnpa/8Co7p+NIkQthXi/MR7GCks+smDiTgCPglNRjFnPFEOo6S7G/F7gPi1xidvPjkff6yqJRyjl443I8cT9osV6cb/72+/HFfD3/9yhcel4FjP9vfx1/e/X3yM8fFHEx+ndOegNlYuLJD9edr2I7HxZ82oxEalXhFvhCuo0xG3c4hTBvSFnS9SfJPUL+/T4gvOknG5XE8H9HXAXrODJj2a2nEIDJKUxO3tVgJ6DOL3taH1uGDa0Do7zQxZUizKDENGbkotWTKS5KSuRjLhym4t8ung+CXP6bf6OmMarEmTnXaCL/IUX+nPkiTBokzTxU2hW7jCcMu6yWZJcpTJ8XK7wH2yRjpCkmxJkwUqnJABu2QGg5DXFD0Z0MTFxEkpXvD6lzMoHF/f/CQE+PJrf/2Omz653XF79B0/+7e2ST5blfX2N/7S7btlhqKmWZUGXZwpkxFx+EASpE2aRuhM+Tq2SzpIcFa2cFyBBO+5OcfUwhJnn27zngeEn6sc5A/6J+hpnyPBfjaoM0+FJ+1Z7uZxcpolK2mGYqlogbOskZc8CQki4o8wKxMR+0F5nHPMMOGCqCinTZ095Dgtm3g5xxAbjdCFk5E8HbgvbsZkqGmDjrrOvkW5ml182T6FRdkRTJEzDq6WjyRAvbxxSpKkSMbumBIFjhAHbepEwSAkBIU1rTkil3bJSoZWtGnMyWSkD9/iVdzAvRynSIZ1daoLRTKsonyBJwG36U6bsuO9aD8OVl6SMR25tY+Aqq1zMFjEohxjFRTq2mQ2mGI9rMbdq47tOhhCJC+pUfCrRpAsf758d6ytPfbJDvpejjPCqVosO80Ma1qjJ0NeJAdpSp8TusKRcI4l02SIpWeGvETn+IQ5RVf79GTEcdngN5faWAPJkVA6k+MXd+/gP7x3js8Gx9kIq/ywfAcPyhb/7x+9gh95wz0MRoLsnuYLX9rHDu0QGLjxXV9FV1ZhY4rR10oE+7cw1x5g+PFVkq+bwR57BNl9APuVJZK5BqnKBiTgh4vz3Pq6TxMcrGHPTxPe3yD5/S/A7JlDm03MvizDB/Mc/9PXcNXrP07n4cPckM3TC5V8UjjyEx9FW2AWncsqQQLsAPpdJFtE82VobKKDHnRc0Em99BxaSzN1pI7Wshze16DRyKCaItNIUaPNkJCyFEiRoE6blEkwywQAr9gTMBxaioeeYHRfjsTwUUg7JTVz9AZkaifmYA1dPkX3D0KS+5rka5skX3cDEDU7nmXtzicYhPCGH/gCj375KIPBM0Peh8+4ZCn71V3c1+jwfaUpQgsPtDrUdMAjUudkuAnG6dJ/orvBsWCNEMuTssxVusC8ljlvauzVGdaleUHSkolws7NSpk6XRZnmlK5SkCz/8f3/Ij7+n3/zr93+b59nDfo/+9Bv3fHNTkO+2eWbPBnJRE7EvQj6k6WmbUJCipKnHXVBUyTIkmaKIn1GnDQO0ioYRjKiRI6D4SRN02cf06xKI1KvajBBnqQGdBgwqVkm1PXkjTp5zntYoawZNqVDWhNsypDzpkaHAUaKTNscj5plimQwwJyWGGJ5miUykqJEjjot8jhFwi2tR9w6h1HP4lQNRzoijEjUHTuKbneQkpIpumZYNKXweHenpmRoa4e2dpg1U9Rsg5AwUr1qkjd5mlFjyO8DvgCZMCU3wYi6wgvBXMSTcB4JBkPFON+rJ+wZdps5lnSTQqRWZnDE+0AC9skOtmjRxxWNGVJ0aLGprvhJS5JW2ImLBv95d7KrkQiJKZAlTV1bJKN9o2Gb1GjEnAnXUYesZJyK1xi0yT+nh0d5xaWe9mKYsYfC+fsNdYjVkE4E9wPoR3uCh3v58xZGCbaH+aQlTc86PtFz+Uz76UsssTsme+s4iEEseTsuiXvB/y8qQMYLA5+MX6xyNX5b/LiLio9nK0Yufh3/uz9+Lw3szw8Qw6t8sQEO+ufPnS8iPCJBVePvPZHSmIcd+s+5h0oFum3U6pAS7nEpkttTKJzZpr9+SRJcITtZp+HU8yQdnyPHXwpY1SpzMsmK3SQduaELQi7imgFs0GSOMud1gykpMYwmgLuYok6XfXaKu4PTvDjc/XU/E9+Kqy49CprhvKmxwDQ50kzbHBumw7TNYYGnjBNhypLhKhbo64i9doK7giWMipuOSpERISd0hbxkqWiOmnRZkfoFn+FvZP+Sz3zmM//qeXzP33Lr19/8u7ffFO7gcVPlSVlmhjId+izZDbLi5F8boQusmYgXsltmuCncQU4CkiJ85xVDduxcJ5nsU/zuh7nvt1/Hvz+3zGE7xfftSfLx0yPOSovP6EOxV0leMhzSHTzMWTd6lIAFmWJFq3EHB4gUSiwHg0WOhHOcCtzUI6kB69Jw0ARtxfjRcZyqD7KhWmaDChthlauCvaxRY8vW8V6tQIwvdpyZJE3bJhCH2x3qKMbsj3ee4EK8a0ISqOoFGE/Ps9iydSZMKd4UNHJSHUVKFy3tYCOS0t5gngQBK7rFgkxxLDwX60p7vHJIiIeclEyBum1wTXCAfeEk9wWnYs3qkPAC+JgPlglJUJI8DW1TlkI8TQEomwJN2479GDxELNSQguSioixNgIONZUhG0LwsgrAlbQzChOY4aCewwBNmk7p0OG1XOWL2cEyXmJeKc2KPVGh+XG/i/eYRspLmU2+s8+hXX8R1b/0IupHj//idl/NgsMavHpxneSPNK//NXyPTszAcoO0mtDqMvlog8V0GmZiCZg1tNgkfBrO3i1SKPPEbL+PwL9yDZLLY48tIIYG58WbCz34ec3gXWq9hn+zyl+/5Aa7a0+WKN32UP3rXG3jbz3wEu1Em+Z09ZHoWrW1idl+JdpuQyUG7DqMRqIVMHsS4vxmDLi9DIceZ//RClpaLNLsBgxFUu8r5wZCHzAbr0qQXyQ6A29BK5MiR5i2p3bz+NV8i+eLz6GoA6RASipnPofUuwa2vRDIFtNNg9NFPc/4Tt7Lrx+7BHL6BL9R/6jkFvPNv+9jtb/q1D7P+oRdx7Od+8hmPOfXWj90+CN0nvToa8XPv+AzrD9/IH91TZjGd4jODdc6aKgahoBkA3prbxWdbNQbilHrymiRHwKeD4/xf7/vFZ7zGL735392eI8UmTeap8Ivve+ff6jTjGy1Evhk4ll8/9sZ/entirN8VRFOfnKTpRfGmpR2mTZmhhszKBFWcw6+bfvTJkWZRJyP4gOtcz2qBLemwQZNpigwZOYUboC7dOFkqaIotaTOnJc5JFXCfwSJZcprktGywS6foyYgmzlhynTolclwT7qAhA5ZNI24I1bQde2SMJ3sOmuISJj/ljUU62E44fVfYyDZkZbyTPx7TfHI+vKiT7xNZD3sZ4mI4EHEgM4Rq44ZRNmrsNLTNtnKUMCEFNrTOtJRJRs+2pnWuZRd16bFGjQnyrKhT1Nm0NYqSZ8CQWgR7GUSJv39/gTg4mFO8cr44411zdy62k1yfwKZwTvGeZxgLyETJb06ydCNPlQvgt2MwKYg8SUjEe5i/3p6Q75eHEvnHCIbe2DT92ZaH4/n34d+fK8IuhDpdvC6lSPX1uB/jRO7x3/1tz0ZSv+DvF73W+JTm4gIpfq8R9MpxK4O4WRgQRKaIxHBBjT7vw0gxzkOzAnESFf56qzrOk4P/yPZkhG1VrUEE9fHHU5AsoVoGDB2sXvvskEk6Ef+qQScWs2hrj772mTMVlu0mWclQkCwVCpzTDUqSo6Zt9jPHATvJl4Iz5EhR1EwMxffn0SDcGC7w2g++6dtmAn3xuv3Nv3n7K8J93BWc4eZwkQeCNdIakCJBWgO+IqcICZmSEhPkyWgEn5MRRoW+jDipK1zFLjoy4Jff9zPP27l43swKv1XXL7zvp+8AF4RuCw9xIKyQIcWicaT5kuRIG+eQnpIk01LiYFhhiFJKBLz55asc+Z/+il4nS/G2h9j8q6OohXddX+KFpSyjkeExs8FdPBYn3QAt7XJOqkxInr1mnivYyYpWMRinUIJhRspuM0JY0xp3mafZG04wbfNsRt3Ua1iMx8XABePLFEkqpkxW0rTUOVWe101atkNecrGvipOwcxjaESMnrxtNO7raIxmN+buRP8ZQHQE+uCgY+XGrxxh7cl7NNiKzKrchDXTIIFL26uuAZuTB4TfRk+F5ztjViEfTizcSB0tIxZtVz7oAVbcNJs0EPQbca06QJDEWzNz588fkJE5t1FHsxpCxBTNNPoJAbIRVhoQcZiHe/BMEjhuD26R8wpEiwaY2yWuGjjgVlcPhLGXNkibBg8EaXwrOcsBOYlG+l+tYocoL2c+QkL06Q8d2GeqQk+IIq0WyJF/d5bq3foTw+Ay//Z7v5qPmcTKaIJMZcvjAFlt/diUyMQtBAq020daAxNE2MjGFnj4JmRzmqhdg18t07joMrS6HvutOZGYBpuYJbvsuKBfQ9XOEp8sQjgjvt5z+zG0I8PRZl6y99c13037iKpJHa+hGE9l5BVKJBCVstLln8pDOOqgUuGIknUE31pE9+5CpOdKZHh8+1+NLmz0CA+cHQz4ZnOCcOEWhZiSbeKXOx5+DDn3yKQgWHXbU7E/Q+doRzHQK3epAX5FMZFq6tUJwZAeL33cnsucAunLqOceAnX/86jvu3fU7zxo0t0YhoSqlIMAUezzy5AyPmi3uHGyQxPDCcCc/WzxEVxxh8s52jUnSPGFWuVbKbJouHcLLPv+73ve/3fHP3/ezd/z79/3SHX/bBcg3ssYNCb/Z5QuQYcQxc3EopKXdyN8pQS4iOgIs6xYJHJnUJySZyES1ps5Ar6AO7ioIu7XCtM1zwE7TkD516bKmdXoMKGuGVWlgUZalzgE7Q44UmYhM6Z2YzsomM7YQm5sdtXspkKFIkifNCgkMVW05k7wInpCWFJPGQcEsyh4zd4HhJLjkbtZU4qLETVxNrPDknsfdd9I4KINgmDAOknapAsQb/vnkbk8wz4QpUTaODJrGJYMhNiZ/Bxg2bY2KFCMyeIId4sxfX6KHCLGsa52AgP3McVI2OKWrDDVkSbdoaSeCT4Vs2lrMvQgjEz3XgXYwI988qtsm88FMXFj5Sfx4UeFjOBAJy7hz6feR2DQwmqSMd+I9odyTwLenMTbudHtzPC8ZOn4exycilzNTuxSXI+GJ7RF0aZv7wAUTj8s9/uICwe/Tfo2Ty73/x/h6LkITF0xFLprCXHA/f/3GCpBxk0FfgMSQ9UgkIOk92aJrGURqnE6+WSI1MGfYGIwVyx59MWGcVK43UTQ4GV3vDeILUz9V8gWIomQkFTc7ewzYrRUUZb/OxnDCXTKNV9ybNhNMSoEkAQ06jluL5Vp2MRLL/cF52tojH5nJKsqN4U5eFC5yfThPUbMkLlPUfbusDCnOSpsbwwWOmXo0BXGfkQfkDFcyH0+CCppiEDUI1mnQ9O6zDgAAIABJREFUkT79yNfprGxQpfm8HtvzZlb4rbz2vOXw53LvX7r1xqkkFc2wNhwxFMttdj9PmXWGGoK4wHeFLPBTe8sE/TS3vmCL48dnqD12iENvvxOza4Hc9euUVkLmXnUPle4E+eyIzy5Zlqk6ffQo6IVYBoyoaYs1u0VPRszLFGu6FSfnNW1GuuYBeckyYsSS1DnOMt0oGT7LBj3tj7nDakwmHOHkOlWINkIitY/ASRpGUnt9HdCLvBpcILYY3G1evtdJqdl4XBohQ/GoVN9tsdHw3UnjlulEWEyL0rRt9gbzVCO8sjdz8oomNvrZnVigZpsEErBla4wYkZMMfYZMmQma1pnDBbH+uOPyjMQ5rI+wTEoxUhSysQkS0fv3MoBeh1/EsG6rMWhAIn+QrnE64IVIUtMFVFDZJpamJElJspxjk4wkaUuf02aLLWmxRJUGHd5uruUD8hhnwhWOsUrdNjjNGjVtclbXsRqSNmn++fwVrDez/MnPPMSv/9Jt3PKSY5iZNv/sbujSx4jh6ZUshWaFRrXM7qs/D7U69qxg9uSdXO7sTui3kXyB1XflKH7H45jhgOBlh5BSFzot2FyFfAEzOcvwfUukfuJaJFvAPrXC6YevRK1hMBLCM/s5c2IPuw48hdnZwhy5yoXadgPSGQhDpDCJPv4AMrMDel1nUDjsO3neTgcxwujzy/z6nx+mzYi7g1Pc09/gtKmTJc2N4QIPyhmatk1G0hzT8xQlz5Y2SEuSNy2USXYnyKQ2kVSX1L4NJDCYhRm02Ub27IVRHzMxh8zuxhw6iEzOQTrL2c4NfyPJxK0X33jJ+0+88dDnzv35U7caYKhw9OanOPm1wzzUa0UbsdCTEXcOV3lhOE9eU2RI8AnzBD0G3M8KOyjztg/86B3f80Mv/ZaUcXz/n3381r/J/b8RWd7Lrb/880/falEHyyBkwhTpaI+cOCO8YdSVH0bQH4PBiqUkOfqMmMUpCFalzV6mOcMGm7TclFKTZEnyiJxj03Ro0aVMnkAMCzrBktQ4YufZkg5dhtSlS4cBbXoMCenLkCvsDtKkSBNQkAx16XKcNRp0UAkokcUKhOImHd1I0GSgAzKRop9rtnTchMdkSEmShrYpSt6JiphC3CjycddLuHqhkKGOnPKPJElKQD2CXXlzXoiSRvG+MIa5oMIA52K8V6dRcU7wc7jE62i4yFlT54idZ2Sc5HxeMqzToCJFlnWLg8yyKk1CrEs02IikfIUtW4ubND369K1LMr27uE/FjQTsDGYZRXyOoimQkhRVWycjGRaDOVbtJoqT6PWx1dsG+hTPe0d4D5Rs5FlicJLy3ujP+RQ44vB4weFke4kNi33Mtzj5+G0PFY0f69P8ESNiQ75o+WR6fPl9LS4evNRu9K/E/z77RAS8R8mF045L3u8S045L3Wf8Z3tp/O/lTA+dIeo2H8Z9PrenPD6ncdfaFc9Zk2HAAFWNvSCMeFWsbT8Jp3JlyEg6VrUaMIwaEEnKUmCEk+pOkogFcTJRcZ6TjEMumBRFyTJL2QnLyAR9hlSlQ4BhiSpZSTElJZa16o5FPJzM0NQOecmQJkWfIXu0woa02NIWRcmiorw03ENflGXTpKBpLEqWJBXS7H3L4W/J2P5c1pfed/+tK9IgR5qKZngiWOMHzF4e0yp9GTGJ58SVCDAs2hLLpkmXAelI8SyMMkJB+J4fetnzdi6eF07It8OqZIRzdaU9CpnWLCfZ4N7gLKG6YNe3A0Y65H49zo+ceoqyFHn9F6+hTZebCnmuuHMP6auPg4HC0Q2ad1/PH39uFkVZNmukScWkv35k/NSy7XhcWdcmvUgP2wc1D2fyF7htO3SjTqGfJnh8r6qN4UwQxh0kBytKRU6iJUK1VG3dJeE45Q4fMP342ktF+t9FTawW4jGX4zJ944/zKxBD1dYQTIxvnjAlVrXKdDAZkzc9z6QUOBx03/ZZsRsxqT4t6YjHYSNImCVncnQitbNtf19o2Q5TZoIQy5Y2SEaqW0PvPSI2jq/OTNEVIO1IzWGgxGPhEQ5bHoqNzRcF43DfkqMTSUR6PHRIyKY2mZRCBFsbxdfxy+EWJZMjHyxyPDwXXy8R40bS4jDhv7eyxr+4rsDaV4/y82//PP2n9pE+eIpZLbGpNRcMNM0tR8/SbefQdh9JJQj2CbreRCo59NwJen85Qea1q8y9s0X4QEBwsBkTumVhL6MPP0UQHEP2X40Uu6AW3VwiODgilxty6Krj/OHHDnFtbsDhV32S4EgSmdsPrTpki8jELKohki+jzS1njAiglvChJsG1efecKx1O/O5L2bHrNDcUchxvDenbBU6bLZIk2KLFx8xq3Anvao+AgJZ2GOmIBSY4v5Hi0JEavTN7yC0+jhRSYBUd9DFX7EHXTiM79qHhELEGDUcQjpD8BGw8f/HhJR/4/rjr/yVeS+EWeOdF9/nXb3737dtj+u1uo8XyU+//sW/56cbf5XIYcPfvMDKXC8RgVUFDAjEUJEtPhxhcAlTVFtkxRcE5LbEidVC4hkUCNbQYcELW2auzdBiQpECNNmVynJMqbXqcNw08AdpimKbIBq7jOcLyiDnHYV3gjNliWbdYYIodMkmfIR2GnNUNAjUUJUuDDsnIJDcdGSx6knNBck5VxjYpmwJzpkJVmxQkR4YUOcm4hpKkCNU1kAY6ZNpMUNVmpMjnmkue93FxAixi2BfsJMTS1T4pktS0RUlyrEqDl4a7+Wqw4iArmuWe4DQ3h3sAGDCiIyNWpcEi04RYpqTEZ/RRpphgWkqc1w0qUkSQ2MDQPXYYx38/jfeJrpGAKTNJ1TZjf5BmXEApPe1xMjzv7hslpklJMYygsP4T4p9zXELXmwi6wsxNyb2ql+cSbk8iPCF9mxvo9y3f0ffH5NfF05AACGWbxO08eyKuRAQ5c5Ajr541xuG46N/xdTEEK+Z4XKR+NV5AXM4P5NnW+H2NBM+AXl18bDEcKyrkAjER1M0bDgbx+fHnKsA1Of35nDRlqrYeCQlYcsZNwlz+4ojmBcnFPkijCJabjK5hk2iqhiVkEE0JHR82Q4oAJ9NbIEuBTER8D6jRpoRrIDbpkpcME+Q5zyYhIXMySQJDhwHr1sEwE+KU3660O3jMrJCJpoZZXGF8b3A2Jq8bhL6EXC0lViII2rfrSmvACWrspoIRoaRZPshJkhKwqJO0ZECONB0ZstvmeTBYZloLdCXFmtY5wBy7tMyGdPj597/jed3r/t4UISfbI7Z0wKt3ZmicT5EiQYMO01JiRatkJY1Hg4Zq2dQqdwdn2NAGR3kh2VtPYVcFEhb6AfnDj/PYJyb4Ak9Q0BwDHcZ+GTaCAIkIVkPn6B39WHWdBWdElY6JWw3bxBPA+/QdUSgiUgkmxsJ6zoNLpN20pK8DetqjRcBsUGHSlOkzII1xeutqY/iSx8TGpDG2AzFEGEgJQF1Q97J54/KIXpXK42BdkbLdYepqn7Jx0AZHiM/Rtk5y0xcYffrkTZ6kJKjZeux6PoigYHHgHyuGFEtNG6RIRgaGFokKpCQJrCojehFu1U2lXPHllnPVNdGmBWftGpOR2o3/ezd6rx7fvUmDrvQj1Y1crB0+KxOs2i2ykuFhs0QFV5xMmUk2bfWCTSwrGW7RQ7w0OcU/eORB5pjggy9v8u7/8nJ+/vZjPK5nmDGTdOnzq3/+CMu/vZdyZRNJJSBhHPzoiSeQhV2QKzIadBl8rET6Jyaw1TbGtJEgiey+ksGfPk7yDQeRfBmCJMm3fQf23s9gbryZ4Uc6XPHjn2D4xQX+4b/8C8585LvpPn2IfOVRZG8Ruh1UQxgOIJlCO82o2DgHoxCyGRK3LKAb6+hKDZlMsmPXab5y3zWkAphNJegP8nR0yIa0aasz4mtqK5ZOrpgSe3SaY7JKR4bcV+9w5Pwic4unkdkSNDvoyGKPdzATNVQhuMkglR2QchAySaa3oWJ/i+tiHOxr/9aP4NtzeUhVStJ0IghWQbIMdOSgFiZHX4c01GG6h4zIkaEiBZr0UJQ+IzbEJbZd7XHcONnwm8O9rAcNBuKSng3q7NJpNqTJgCGJyMm8Q5+ROuPSRSZZiho8BmGKIlvSoaZtsqQ5Es5xl3kag3BU93Fa1rie3Wy5ZyFJQFKy9BjEficLMu2w5zheRjXiQhQkR4iloZ1o4rytBOghQBu25qAqJkEtSuR99xmIEyKf/Pv3lZU0Q0JeogdYliYvCncQIFwTztJlxKppMaMlzkSeKkWynDDrcTMsR5o+IxbMNHNa4qFIEn5Lm8xImbq2mDRFqrbpEns85DUZJ+Hu+gbUbCNOZP0xwzMVo8KoGTZhSnHzCohl4r18rnfPBpzUu/bjvSsZKWLBmNcGXj5W4mITiAuVcCz5Hi9IfDEI2wpPTqXxmQXFuAHgpYqIS60Y5nTRlONSRPLLPsfY6/p1qcLkGTK9l4BePeMx0XURDEkkzhXGr6W7hiYmovuCRNXtvU1txypuBZOjG8ksF4zLixKRL4Wo0KVPRUpRE9F5UtRty8n2MsD5sRua2o5kpntczSIPcJIcadapc5A5dmuFTXHQzDpdJshjEI7rMoqyR+ZY0Sp5yRBg2G8WLvg+rZsWV4c7OB3UENxxdcYMURMYng42uSmcB4EHglVeddkr9O2x9sgsPYbkbIKRsZQ1wxBLN4qdGVIcCp3HWEmzTGqWVXFQ+5ymeNqsU9HC13mVv/n6e1OEvP4FHfa98x4IAm4JQ17xru/myiMn+P2P7eMPpUoyUrAZRRvjEGVJN+npgDPtEVobER6fJ/WjBbp/EPKmD5d5TL8WTwKAmF8AIf3xoDdGUPTB1hcsTp3DTSUyZGJDqqIUqNkGU2aCAUMsQj5SVVHVSEbYbHcwCBAR1sItB2WKjsUVChq7jvsAnhSHiXakbIm5Ff74/Mh1iHcCDuPxbBJBEUY6pBJM0rAtNPILSYozThzokKEOYyyuV1DpRtMJ/1oN24pxqG5zdhhyx8m4UPoQnOnTSEbOyBVDyRRIkqClHUIsk9H5Cm00PB4jLQ50QIoUlshACWUjkqULCaPpiY2FBEbRRlbXoQuYuA5mUhIs2Q0SkqBmG6QlxZKuxh2b8Y6YiCsEP8VDfH7okp7f2X+EjXvTvOpKy5t/9ShFs0WPAS8O9xE++mk+dd9RfvRdX4CpKej30Y0VAOyxY0g6QfEfXQmDLKOPnCS4qoe5+lpIptFei+RbrohldWViFnptzAteDCYg+SM7wc6T3rlJ+IiwcPARts4fIG9ATzyBVKaQIOmea2vFQbtyBaRYckVJr+MI6r0BMl2EZJLM9U/zshc8xWfe/Qaunx2yt5nCrEwwSYZBMGTJblCUAh3tss/sYEhITlPcxD6ulTIdsex+6WcJDlt0zXWbZDKHdg3MDhALtOuM7nwCs8dgbn7l30kB8j/WN78U3S4KtE+CwEE5VUlH3y+DICqR5HeKIhly6kjVPRnSY8AuMwu4zv6DwRK7tcJJ2aBCAUHIkGCCPOs06GqfVXFJ0xF2ck6qDNX5RnXpM4krdLwIhUW5PziHVcu8THNSNpllgmXqVGnhFJ1SkSeUUpY8m5EKoGscOQiJ9zjwsCrvayEisehFXwcMdBgnv92wj8fRDxldEPfKphRNUdKx5Omt4UF2mgxbOuRAWKKPJS2GH96f4k+OKz+QWuSu/iZrxhku3hbuo0PIF4JT9HAmjzlJUdM2RcmyyDQrWiMvGZZ1iwBD3TplK++C4Sbbw/h6+smFn4D45eN5Imr8IJCRVCyj7vcxD5fxyye3oYQYcTyQJA7y5vatVHy+tv3QtycsI40KCTGESjxJuXCitP17YgxuZaI9zRcl4+sZMrvPUjT4c8MlPDkuIIKPN/8uA916Ngnfr7eek7P62N4bxo/bvt1L33sVsZCQQIMLJiS+oekhhf473tVeLCIxwPEpZ2WCEZZ5qbCqVZq2zYgRRoS0pmhGEEb/fRlheVpWmMB50kxS5JxUyZJmSTe5gb2IGBIYlnSLQyxQF+cGvlOmYlGKdeosUGHSZjlvatRoYwPlvG6yV2aZswWSajgebJHAMGMLTGmGKgOsxQ/qvq1XRd3UqCcjpm2eddNixhaYtGk2TJedtsAZ02BDWhyw09Slx6wWWZMmVenyK+/7X/+7TPv/3hQh+3/5GBSvRDJ5tF3n6P95P+SK/JQco/lX13F3cJrT0YgxJ1nykmO3zFIhx4sXBHPNHpIzK2hzxMby1XRxZFofkAN5pvrHxU6qHqolInFnZpxMlxKJsKqGlnYomryDrrBdvLhCwFWuBZOjGW1wVsP4OVNs66unJBmbXcVKFRH8y8vohWrjKUnDtvB4XUHiQgm2nVH9Bili2AqrkYSeWx564zw53GRnXEbRYT4HIMEF8rkjHcaygG6D2naz9+fI66b7jmBKHIK8E+nNj3RETRtxJw1CkOAC9ZRB1EETDKpuGuNhZc7/ZLxDtu30m5M053WDRTPD8fA8O4IpqrZJyRTZCN10xIo+Qx3FYYUj5a9oDP3OU4/CKejrWXbKNHXb4mrZw5Jp8p5feT03LljMVUeg13YyuYAcPgydJuGD5wj2g0zMYuafxlxzo3udc08ju65Euw0wBsmU0ZWTyMIBtL4BgyqkMkgqSfjlR+g/epDk1DqVPU+hXUHSqW3i+WgUuaPPoqtn0I1NtDXAXHsEOk10GCJqGX0+5PyDryCVGpBKwPJGimoXnjZ1WgyY0xKb0nQOyZJjixYpkmyYNkVN8/pb+hQmtrCrE8jEClhBCorWu9jNnQS7WoRnC8jsFmZfBNcQA4mkM0z8H+t5X88XGX18uYQiiL0y2uqEM0Y4rH+IayqUpUDfAX9IkmBZt5iSUiSX6+JAn1EkS+F+2vQ4L8qCTjijUWBNnIx5mSxJCZggT5sej3GeeSqsSp1J8jS0Q1VaFMjSpU+JnBOhkAwZcfLSPuGta4sFmaJOm01tMi2lSPEtZIdMxl3XgQ7pa5+kJGlqiyIFcuImeB6OGEYwI9hO4vz/x0nOPpa4ye4gTignyfNa2c11eyyn1gL2ZwKGITzdHNLQEfVWhh89AL93YouODJm1eXKSZE16rEmLjvYIxPCgHieLU0dcki1ypJmUAi22lQcTJKJ2UOSOfYlE2U/rvadE0RQcXEed+AliI9O5UQxBjifeGKyGsSCIL0q8SIk/N33t413nA7lwb/VQLL/n+qIoGTnEX7zGDRXHm3h+Lx83JfTr6008nqF4Nfb7BepdfvJwUVFzufP6XEwIL3Xfy6pjjU1xfHPUw64unmQFBNvS92PnykjgyOHR+fIE9SB67kwEtfaFSs/2SJjAGQlqk7xkWdUqZSmwohvMmEkGOqLHgILk6NKPrnOKpCQdaVwyrliJ4kCPAfNSoYo7vi4j+gxYkwYNbXMlC/QlpCNDkiSYosSkzbo9GsvVdienzCbXsouhhhQ0yWPBOleFM5RJMsRSY0CJ1LP61H27rCQBPYZ0ZcQ1dobHgg2GjFg3LXo6ZENaLAd1Xm33sWTdtOO4aXF9OE9gDDlNfp1X+MbX/+/Vsfy6R375DgBtVpFUFtmxFz1zgvytj3FlNsOclshJhoCAKTPBLXqIISPeMT/Dby+tYM+co/3x/YgYVlbKnAqXo46L+6LFUwd1plv+C+vHxH7E6TcdcAoq45hUb9bk1Z26ES9hoMNodG3i21U1wuymYqhVQhIxnjYtKUpme3SWNzmykonI3uEFkr8GB1vyihQBASmSjhgYnZNSpNziCwL/ni6GTXnJ2/HOU9kUHV4aiYuAUN3rek5McBF+1Stm+FE+RCZPbKuwJKPOY4hlqI7TM9Ihfes6ihcbIenYj3//bduOTK3CZ+Jlo2RnoEOqtklHeyzrlpvgqDOADDBMBhOkTTqWJb5YF96f0wNmp8PbqtKPSPOndZWb5BDvfdtxjto5/uG//AsSCUXPn8QeOwX5UmQMuOb8ORIKwz5Yi3nxDW5yMehBrgC9CJZVrEAyjVR2oOvn3O2pDJIroWtn+NR/eCPJuRWCXXWStzQJbrkaOl03ARn2scsnXLE+6MLkjPPtSAcw7KO9Lroa0PvYHMFVTeZ2neDMuQpntuBP6st8dLDMmrRoS4/TsslshMF10qBOvz2lCY7JKr9yd5oHvnKYfruCXSliz1bo3nsFkg5I3nAO7QrB/jajuzNgleHnJ10hOvz2xuf+Xaz/HsXFc11t24kLkBEhWck4t23tMStlutp3kAjtY9V1S4Pou95nRFN6HGM1SoUdtElxhmO7dIpeRE6dUecrMmDEpjZZ0Zq7P504Jq5Ri8nlAFaVhnYct4L2/8fee0fZdtV3np/fPvfcnCpXvXo5K2eEEBIPhEVOkoww2O2Ae7ntMTNtPL3scbeXRu1uY7en3eOenjX2opfbY7BnBEi2MWDDIEABAQqA0tPTk14OlcPN8ew9f+yzzz1VqqcAltyW9HvrrVt169x07r2/vX+/3zcwEhrb7jYTUdMgS5JRKXFSz6GNYUQKnDVLdOmxpFc5bRbJSSaSmE2E6ocAHdOJ1AiBKIc7X4Q49MWtFc5bIhEqaCmEUVUmL1lGpcS72Eo1CDizlCCdgOvefBCA6bTPG4cy3DPX4b8daXJdahiAI2qROaniGaEiLYqSpaLrkTy7VUdsM6OXmNVWXXFMDZEIy714I2ij6NOnoCwcJh3KBK/doFoTWre+OY+qiATtyMNhs8zyNJNr/gYDNSxgMGFhLa8jLoncM901a5q7jK9N8XUrXhxsFHESeTw2ks3d8PaxwsCpUr1QuPVuze+sJZ5vVHCsV9ja6NJ9zlxR7KBybm0He84HipMeBZVHmyCCLYOdcLn31jYuu2QkHUFwd3ubo8e9mK2kQijhhCmxRU1En/sMA0U56xtm4Xm7ZIqm6TBGKVTIq3N5ME3JZCMpb58EO2SSKVNiXMrUpUvCKBax5OppXWRW1ZgLydnH1RLTpkyDLqdlhYPeQnTupv0UI5Jknyrw7rtuflVw/Zr0aEqPjEkwK03eJZspmDRZ41MwKUZNnj16jPtkhqNqlQ7WL+WMqnFMFqlL94Uf5EeM10wRAvBA/VeiD5RpN5CpafAUH/5f72KPHuKeG4fZ6k1y98/NcNubu9z1via7d8/xl39+BDU1Tv4TVqb00ydWbXdP7JdHRHGBt4Mv7LmWb1z2Jv5i23VcqfaHCbFnDXfCTk2kOBHCdNwX3iVHlxQzkiYraUZViYLKR9ORlmmTDj09HHHPqaxY/kkvGvW3Q8dmbQKquhZ29DMUVcF2M1Q6ej4iA+fbgCDE4PYjAlpV1yipIk7DHQaFSPyyawYwKtcBbJtu5NORUqk170k3LBw05jlj8LjRUzxh6/D6nulFBmDrw5LvTZRkI65LLNYXJS5pW1Ux+941dIN2KLHb1i1quk6AhXM0TJO5YIGVYJVuOMnp6LUbZPf4WZXhqJ6xOFc69MKOqTGas7KK6fr8+ie+jrpsD1d96kFkbBK1dyf4Kdv173SQTB61axSadUx1ERZnMP0OdFp2cjE8BdkCZnkWSWbA863PhyhYOIN+9jFIpinl++ilMuQEsxhq8e+7CMmVUBPb8S64FhP0LDek30c2bUOGS9BuWkWssYD0T5zlmf/7Rpq1Yfp94cFeJcLsH9VnWabOgl7hhJ7DGLsxaJoOedLMiZUCvUiVeGZR+PI39mNqWYxWpPceRS8F6GUfEgZJJ/D2LmGWuqR+ZQ8EPQs3C3q8Hv90woRT4DRJWqbNnF6mKFaydbuapCx5iqFbb4suVW39LPr0WTBVlAjzphJixT0ypEhLkkWpM0yeWbPMQU6zbKwcb07s9KRmmlTD//N6iRVdY8Gssmiqtr8rwqQMkcP6cVRDkuzTzNAzAW3TY8nU6IXT4aT4LJhK5GEyosqR+/uiqYQTYCt2kQrN2Zx6lssvCUlQ0bUIxhvfCJtYpzwjKXIqRy40E3S5SgGb0z7HG332bm3xjXvOZ9+2BmfaPaZGu5QlwS7JU0zBfj2EhMU/wAp1egQMqyJ5yUQ8vIykInlUgDm9TJ8+LdOJ/DrOBRdqawu7KakiKUmiUGz1piJ4U9f0MCaUsxVvTUHjig5gjYdH3/TXTBACsxZW5W4TLyhskyMRoQni9xl39h6YAepo7XWv7zk8ithm/jmO57G/uVjP24gXKOs9ORLiP6dZtb4Q2ggaFr3esJDdqOCI/7welrXR896oyHTNzsAMCOrtsLBzgjdu7+GJihAPKUnihwI7btqSCz1ejsgCtdCvrGQs5zUgiCBZXXqRGMK4GsbJxBYkywIVEig2yTBzqkFFmrwl2M0Zs2Snp8bjCHMsmxo+HqdkEYUwbco8qWbImiSjJk/BpKjTokmPZalzbbCdfcEoAQFlkny3v8Sw7/FMKK7waghHJt+rh1mVFt8PVvFQDJssbekzqfMMG7s3GzVZdqk8wzrLjFSigvTlitdUEQLwQOdf3m5aNaguIZM78K5+C2ZVSKHIX/wElweb+cyf38B/vc9qqI9//DC02jA2bTGe5TFuGhrlCtnFuBrBJ8Eb1XncoLfR7wvpTIcrf/qLVKRpIU1iv4i27z0gdPWNhRzENbndpStIbCHRi3C0PraI6MWKBaeJrsJNtlvwYEAIVeKRUzkCY4sLsAmmoRtRp8g9djxZA9FkJTKUCtVHXLhJRbx7MqSsG7aDVbV1yyrKmIEZIgyKDFcERL/HSJBRcl3fDQp/r2rruOw6Z/Fj3O03SrovFK5gcbd1ExwTckZWdC3StxdR5CUb8nLWeauEi0pN13EY5reY/fTCxbNlOnTo85kvvAm9kodWE8pjmOUFKAxhlmasWaHvY6orUK9j2i2Cbx/FBIGFbCUSSCqDaazayYdS6JkjmHYdyZVRW/ZjggB9sII+epxGy+PkMxcgaR8TgBSGEPGQ8gR69ij6zGFbgKSz0G1BbQWZ2oo+NoPx/3h6AAAgAElEQVQ+0UZGC+iZDnt+9muU9jzFvn1nuUgVGTV5npU5hlWJummFOO6BSeG4lDlhrNP4GEX+nlNcd36DHeN90MLKqT0sPXwVej5H79gWJC/0n8hgGopgJo/kykgiZQuR14uQlxznmoY4T5AfZVrysZt+/baP3fTrt33kQ//ynGaIf37Xf7gdYFSVqZsmZSmGXgAeK9Sp0GA55FW06ZHGp6hyYTPFbr4dYdZJfJ4xVhqtapqcNPNsllGmZTSaRlRNM/rc9Qki6VDrT5GOrtfGUKHJKTOPRtt8azpREZMJnZYrps6wFEkzgCUs62oEEamHhqQ2D1gJcNdZdpNWtykL0FHuH8BxB5NYyxnxyEqGy9nB+Wzm2mAX2xkPYTTCWMFw474+mUyXi/dWePDpLD91zSr1RoLpbIKnqJBP2/s+EGzj0mCKiupwTbAz9B6xLa+8ZNikRsOph4ryphNAcdOHjTbnLlKSYkVX6GP5LBVtTXYBkuGkPl4sODiWlSteC0teo1wl3mATLAO3bvdZiD9+9LfYfboOu8u78dvYxtRaZahzeXqs39jHi5UXIpS7tSreAHNNtfj0PX7pJmXrz4e7bbyYeb7iY/3v8cv1x8Qff9C4sxymQYPR+oA4iLb1B7HPM4FV67RFh7ZKVpIlgWe/37oSFRxJ8VnVNe7jKc7oBfrGKlm557SPTdFrc9yQLj3GKFEz1gOoLl1ypPmGdxiAebOKh7CDCcalRNb4HAj2UCBDnS45UjSly5I0WZYGOdKsSIM3Bdt4xDvDBGkuCzbRDAUduhomyZzzvf2nGHuCEWakwajJMqNqbNFFmtKjQ58SPt/xTlGXNo+pM/y/chAfxfXBNnaYURqvT0L+YeOB7idvf0D9tl1wleLgXe/mqjEfyQdslgw3XFohI4rluWmCR2rI5h3QqCDJDObgY3xleZUnOcXpYBaN4dbsZn7lg0+wddsCxaFlTDXNdj3MNbKfvGTt4hfCtOISvS5h+pKMYFpWEtCONPv0WdGrkWmg45S4xORuFzdhcq7hgMXyhklNYU0InSN4VjLR9ORc0TbtyPEcLM8jHULInNOsTyJKqu45rOoqZVV8zsKVUqnIk8OFSzTuZ3cZhzS55+gSk8GEj2tfly3MBl+Sc2mvv6CSSfjPJWIHmVjfbXKFlStM3DnwQkneje9bRUXLw+o4HipS+XpnsJt/9tF78G80mNk5y+kYnbC3m9yGFErI2CZkcisyNY2kM3hXbkI8D7Ni+Sjm7FHQVlZXkhlbWJQn7CQl6CO+j/fm3chwgWKuz64PfRXATlaMtnwQpVDj26HXQQrDFtaVzsHwBObwQdR5u5CiBqVYvv8N9B8ex3QT3P3dzZwJrGttmZwVXgg3a4NiTpMKr+vTJ2+SvEe2kC80KRW6BL0MtUqeejWPXirjb57BrIIaamCaPpVnz8e0G/Zzkc7D83QIX49zR7zQ+HENCX/u5v/lNrfRA3i+QuRP7/zd23//8//qdo31EkjGNvP1UDlvQa9gwkKkYdqkJUkq7Iomwu+iJb8GFCVHx/To0GVICpw2tus5IgWypNDhhNg1bDrhhHhcjVA3rRDlbr2Ixo2dCpQkHxYfKlKQ6hjLUSlJHpd9nN9RQhIs62q00QXr/G6n3lZ4IzBBJJUOdvPr+Hlxhan4Blgbu1HbwyRNeiRJsKiaXKkn+M2hfdx45TLLDWF+OcU9jxXJFWrc8p4f0m6nqLUUvoKs8fk/F87QJGA21H1s0mVFtdnHVCSPrBCqphnBabox3xY3PXFT6oGc69qNcdd0GfHK5CXDol5GY1jS1qS0Rz/07lAR/LgTTuiddK5br5wylTuXcSUmt8Y5W8L45jze2HLhpvXxQm99OOTB+gLFxUaNq3gRsJFq1UbHuvVkI7nc54VvyQBuFj//Gz23jTgp69+vl9KIS4Z7i27YRHJwNXD7kA7GGBq6QUM37JoccnFapk0lhN+WJR9OyfIk8OiaPhVTJy0phlSBVGh66OORw373zsoqIir83Ag50gQhkf9CNtvy2AgzLDPFMJtkmEkZQgOBaJL41KTDrLLPYchY6f9xk2ferLI7GOXSYJoeAd/xTnJ9sA2FsEnZ5sQFwRjP9psv+lz9U4mW2HX3sJpngSqzqoFnFNt0iSYBlwWbAKs4mCLBo+oUp6TOhM6xWZdetuf1miGmbxTfnvmIXYA/BfIvvnIbVsQD5blNr4BWoGJffm34jx9/mKfuu47//EyN37lWM/GeO5GsT6p3HCnlePDf3cjPTCry2T6nFib5ZPWbYRIRlAGNilQ4BAvLAkiI4Bkv6uA4laiMpEGgqS222I1Ce6YbatYPuj8GTVblqWtr4IMMSOopSVI3TVJi1WYiTfWw0+jCdcEMVgvfPb8+/TWdbSD6OSnJ6GdHLk+pVARP6pquc9iKju+aLkosUVPFOCDCwBcFBt0nERUpjgQEiFHUQ91+t9CIqLW3iyXeeGHyfBKHjrTnOCXu5/il6xI6In5N1yPSfXy0HscrJ8JirmHa9EyPtEqjjLUi+/nP7uPfP5Vhy3vuRtoNzNxZpNy2qlQzZ5Btu2F5zhYLiYQljw9PILUVzOmTyJ4LrIu4n4RsATW6xb6mdgPja2TnRfY2yuOyj3wJMuECWh7BdFqokWno9zCtGjK+DbM6bzkmw5Poo09iVrqY+hFkvEj/Ac3IW7/Hqb+9gT96SPHRHT2OH/OoSJsGHbKh38u8rkej/ILKkTU+ZcmzS49xPmXOG4dccZUjR0c59q3LuOKSs6TSTSTdIZgZwZz1OPz4Rey96HGePjTNaOsUKm+nbI3ffYY9lU/fNv57JSSR4v4TH3hVYHdfifiH4ofEC3/3nXih+LM7P3U7wP90y7+/LTCaHnZzWQyVqSq6ji9+ZEbqNld9Y0nsLdO2UqH0SYXeAXVaZMTKd7ZCH46e6dnmBD3aoWR4SvzIW6Nl2hRDB+85qVA1TbbKGKfNoi0U6FOQHH365EizZKpRQdPFypM7Hl7N1COifZokDdMK5cDDjraxbvEdrCFby7TPaWY35JURhFv0JRyWFa6XSf6Cg/zB9HmMjTT47sEkI/NFzjT7/MR1x5k6NY0oQ6+TY3rnUywtX8HhmQRpPG5Um/A84UxPsUSHIZNh1GRoE5AVnx/ISZJYVUhHDPclQUnSVHQ9gtdkJB09543CYKjrJtvVFL6X4ET/zBoHbrfOOCVGF67T7hplUQ5HPecYl65dweCmK3GOhxe7bRzq7O47Xow4Vaz439Z7dsR/jhpRGzTtNuKDxOV710OyNro+HuvFTdwjxoVaoiIjLGTihPPoea8rcl6KylaP/ppG4PoiLTAB7ipBRc3PHn3LG8Ewqxe5QO0gYRSHOE2aJElJ0NJthlSBtulSkjwJlG080CYRFr6jUrLTDynRpst2xlmhQVu67DYTTOsi+xnhoLfIWVbYxDCnZJFNDOMbDwQOcobzzTSn1SotOrTpMS5lnvBmALg+2MHj3jxtAp5Ui+zQZUr4dNHU6XP9Xa++NaUqXRJhk2RZmlyuJ8ng8aA3y85giBuC7dzvnSEZ7qk08KG7PvyynofXdBESj/Qfv/v27/JurroTTgHXYC9P8dPQCA9aAbZ/HLbDG0q/fNt/PrqNzFtPYJqEbtbDsLzKn5+s8b//zKOQ6uHdcz2bGxMsmypXmV1oZfg+x6LCwG1sm6YVeW84krjBkJIU3XCC0ZM+2lj8pesigU0Cjhfhxv/W3ClBW9cjlZBuLCE55RNPDH0jkW/JmlFrqGjjulNOb93xQtxI3UlKxqNrehRUng5rORKuKLG8mMFG3xhNn+dKCq7p6IRJ0UG3EmGi7kl/rSzuCyTbF+oIre8cxdVS0iqzZnrU0A2yKktB5SODrnjEJSSLkqNiahHxrq3b+OJzl3qcsuT4s4eGuO03d9gCJJe3MCxR4NuunmnUrVwuCQj6FoqlFDI8DEszyPhWzOo8krabK5SHhJt2+j1M394mODKB/64WatdF6ONPoXZN24mJ0UhhGD133MKdel308afQx2uYxZz9PI8aEpf16H1rivmFPL+wv8+dhwJSKGq0SJKgTiva7DVpoTFUdY3j3gJ5MsyrOoUgydUUuPvevZy/o8GWdI+lhSGq9XHOu0CzMDNJsVzlwp/7Es3vnEcx30ONbbXnoVllaXYznXaS8X4f4/m8eddXbBdea+4/9t5X3eLx32O4RkW80H6x8Udf+Ne3f/InP3WbCg36wE4RiipH23RDLwyPrKSpGpuALV7c5sVuyFdrSDuEP3k4YqyDnLomgYcVg7Au3B69kP+VI03NtKiYVVKStET2WO6tmQbDUqRiGha6hbFiHVhX767p4WPVBd0m3qnvqHAj6WTZPRmY6/kmsSZngZ0SO0GQN5qdPKNW+YnEBLW+JpCAZEKz7S33sPWiAp/+7HWMpTxajTyJREBp+9Oc/P4bqNW3U8h3eeP5TfLPFjlW73PNVrjz9FkuCyZJGY9vekfIkuQ9ehfPygJJEjRoRxPfBB5vC/bwjFrhKU7RMZ01z/lck/OmsSpFXXprChCnvrh+E+ver7iRoFvPHMfATY7deYtP+eOftYjTEU5IbGNu7edUrYN9RWqMJljTNBoUh+G69AJThI02/893/Pric6OJ/frj1herCrGqj5g1jbmNHv9cDbdzRXxyEw93nlyDMi5zPIDU2b+3dRtPqUhZ7SQLpBnApfOSpWU6oc9Oj5LkqYfCO4mQSwKQIcW8WWW7TFCVFj4eeZMma5I86p2lZKzioodHwaTYyyQnZZkpSmRNmobkmA7y1LwOQyiqYr3W0vhcGWxCEC4JxsmR4HI9znkln2O1gK7WfOCun3zVrSGeEcbJ0DVFNIZpXaCL5pBaYloXSeOxQpes8UmToCEdjqqFF77jH/d5/ezP/uyBl/1RXoVxZut779n81FcPqNEK6uILrbFcvYYMD/PmukJ30zzw9av58qkue80IW80o16SHuGWvoj4/yqQM0VaasuRDGILtxBiBrKQtr0NcEsH6T4jFZbq0CIInCTSatKToh+PLUW/Icg1CkqRbBLvYKjgpvh2xhtKIdiQ6MG2ySuEqhCx4UUFix6ODxGYIoVKx5+kiExK3OVcyju5hEBa6s5HUoYndz+DvOhzQ20VkHYfkHMl3Izzs4HGIdOiJKbZ4ovAkET1rQcirHAZDn4BJb5SmaVkFMpWma7poo9d0/QwW3qax3d/L1B7OmAUQu6D+4cRl/NRN30aVl5DxSUynDc0mvbtzJN44gj70NJL2LbQqlUbGtyCZPLRq0O/B6CbQfaRozYbwfOg00U89CL0WMjSBWT4L7SZmZRFv/ySSLaCm99iiRQR0gF48Ze8vmbYO6s0WZiVA/ADxDf0flFGjFR774lu59P1fpTmzmWdnkpyhyVlVYcXUyEqaummSlUxoXGUXLmtalaROm2XV5L7GCheHvKqZhQzlQo98rkuuUGdk6ji6n8GrQuaXyoyV70W2TVujwl6HZz+T5/xPfANKJVRhxJLVu23ET7K1ePDA1vLTB7aWnz5wsrL/ng0/gK/Hjx033XrjPXfd8bUD7ncDfPGOuw986NafeFHn/G8+940DbmLgYJkN04pgkADNcIOcCnOWCqfEfQJKKkfbdMgpK7GbEp8OPToxZaZeCH1KiR9NQsBKb9dNCyMG52dhsLCjTjjNbOk2fdE4uRABEKFjbDHUMz1LkldpumFuNGKnHkrsdtkSbr3QD8TmrA5OBdHmp7TKkJIkCfG42uxEEG5Ij/LN7hLHpMYQOeaWfThyAeNDPVLdIW54971kCivksm38rXMMXfksz953AQ8c98hLimzKkFUesxVFv5cgR4ITqsp5epxTapXDsswWM4wKJ7pd+oxLCV8SVFSXEmlSkmIvm2hIhy4DONRG4UkCI0IfTVM3B5tZcQ1ziSByQbhdtedUImibe8/czMLld+LHhR4U7i8Dsr7CZX93rCtY3OMOJiFuah5fW+z/NZ4ez1mHnhsvdrqw5r5EnuOPsvF9DfiMbh00GBAJJ/AmXGfsax807NaugS9lAuLOwdpzNLh03wMVFnBKVPReCAymNAh5ybKJMhVpRs9HiVA1dVKSJC1J2qYTwc179JmUYZp0SJIgRcL68YiwW4+xXQ9RlQ4pfDbpIvOqQclkuFRPMS9NpnSeRdVAC7SkRyZUd6rTCr/DhgYd3hBs4bveKRCPgkmSQMiIYqbdp2sMGfHYfOu+V926ceGtF91z+I6DBwIggeKwtxA2ChRFkyKBIo1HwiRYlTZN6bLZDHHVhy97Wc/F60XIjxFnL3zbPadH33/PlvTDB+h1oNVE0hmCgwIojp8YZ9TzafTg/XsUC1WPXZsb5NtFrh5J0a8UWVVd3m72clyt0sEqabnF1i3QgtCxKGKSkqRPL+SDWHlF12VzpLCCyoUmeqloQmG7jbZL16dPP8T9ZiQdabAH9HHJx0pJqlDL31uTZAZUP2s25VK4hUWt9RIZ8YYtvjPusBuqT9nX0o+SpFNPieuRx4sL9/u5EquJJcDnS74bFSLR70IkD+qWORUmZQ+PXgijM9gNhiceiRASVzV1FE4lZK0Rl+WYJMJuj09VmnRNHx1ugEZqm3jzB39ou1opD3NmEbVtC94e62AuW3dYqd1eF8kVrBGhn0LSWVtA9Dp2mtHvQr+HGpnGnH0GFubxzrsKfeSHzP/hMP/bH2znLTceQpV8ZGgcyeSt5G2vi4QyvvQ6mGOH7JSlVKbypZ30F0dI7ZnFrKQIjg9TKrX5nc9ezFWbA548m+RRb55LgynmVIO26ZJRqbDznIiMMZUo6qEDNmJ4n97LZMZjqSFcfv4SyWSfufkC/U6W0vAKh5/cy5b/MIQqjWM68wQPPIXauYmn/rnPxX+8jGzZbV9/u2kJ9EHfQid1YH/3EmwtHTqwtXTowLby4QObG3974JS+9lW3uPxjxs23vuOeu+742gEJ84UnCT5469uf9xx//OZ/fdvff/6+A0XJWVW52Hd2RBXJSZqy5BmnRDWcqvlYwY1hVaITznQToRCHlbzuRnKzVumvSyec1GZVmoZpU9dNcpIhLUlaphNOj4WyKlAxtahAMAx4ez1jc6ULgRg/wX7HgzCXOhWgDl2KkqdBK4S76vD1aZzilw43urbggoLkeJPZTcYkeNZbhG6KQAw7TJEPjpeYSiW57vofUl2aYO9PfQ013MK7IoOqLGOaKRa/cyVbt5/h6jccQnXLZNIB5XzA3u0VTpzJc5YWF8kQc7R5p7eZVN/Css6qVWq06BNQkAyLpsoKNQKxZpBPmVPsYooTZjbKhxtFRqUZVkXm9BJG4vw7Q1IlbYEXFpAu23thIWLXJonl5YFAitvg6uh31xhzsvcD2V13e3ese7aKwQbaPac4J2PtWvAjThBia4ps8M/e86CYiAqKdffhjlt/3foiRqMjFSkL+3McyRD2fQ5e5Au+jlgR5okXNdPclFGxdk32xAkarD0PSUnQpENL9bgh2MvTMocfNheyIfQyFarNKYRpGaElXdysZUYvsmoaDKsSQ9iGX1W6dCVgSeosqSYXBhNsMQW+450kjU+SBMvSJBmKLMyxyhYzzLxU+IDexxZdomRypPHIkCRrfA55i1zuDYf5RNEzhmvvev+rbgriYu+t59/z2OcePzBpsmw2RSZNll2SJ0+CJ9QSh9UiRTJUVJt9wRg/f+c/e9nPxetwrH+AeKDxq7ejgNHwip+zF8WfhyLQ+9jf3bbvwh9w8c8c5YE//hALdUPat1+3kknzlFpmWIqsmko0Gk5JkgBNUXIhibFPD0sOz6lcpJgFIQQr5EgIitlgKewWWohVfITusMy90EywHaqYOHysix5Wy10ZWfO3uNGTW6ifQ7gPR7dFVUAhNHSDeDh4UzeU+4vjcTca969JyrFROZy7W7XRz/HR+nMWh/B+nblg/PFcRB4nWFWdyMgs1CovqLxdGLUeGEHFJDcVwtvlYvoYvqUfj+43KxmepUrwzDiJ/fME32nxt5+5mZv+5jQmhFzRrCHD45iFs5AvWShWOoepVyxkq9dCJndCqxoZ+altF/D0b29n56mvQCrg8/dexs37e6AMsmUvprFqCehJqwJiWlXoda1XyNEGBE3U7hbfe/hKrrr8BPf+0U2MDnWYmFqkNHGCYyrHvY9uYndeqNQnuNc7RhofI5pGOAFJkaRLj5zKs6ytNG9OhX48iQQjBc3Vu+dYXiyTy7VIpwJWKmmKc1u54s9WwMtjuk2e+NSbOHgyxU2L/x8LSzewv91AQiNH0nnES1i54qBn379+CANMJKz7+9gWS7Rfec7H6/X4McN+302kUvR88Ws/+bu3AYxIkSVTjYQqGqbFiBRJk2TOrOCJYo4V+gSMqSGaxhavE6ZEXlKcNAuWvB5+vkoqj+MmuO+sy0dWBrwfcjg82qY3yDtR08XJtIaNFWOiY7ywcLCmgc4tXEc5L86nq2BV82o0IsM9Oy3WEf8iHpawnaQsOR7kGNtlnKuDzeTF492TGdIpDQSU8hrldZm44T5q915C8f1PY+aX8a4uQ6VGaXGGdqOMVEaZvvY+Tt77FjqdBMlUlzdu8rigVaacD3j4dIeJnNCuZuhpzaXBNN9QhxiTEjWsKdycXmaGZWsyJwkq0rQcjOcRMQHIkaao8mtMaJOSjGBV7v2ISPphf3q9+3b8MxTnigyKiYFkrEfM62rdpj7O91gPB4vzJaL14Bx8jZdajEQ/bwS72oAHFD2n53mcOO/DGR72TT86l5YjM4BGuffqpU1B1sLAiqpATTfspCyEwsXfm4T49E2PhAxEJty+ICEJOrpLYDQPeac532zmOAsIilVdpaTypPGp0mOLGWVRaghCiw5X6G3MsoTBilVMSYmCSTGvGvTCz8i4yTOj6qRMgouCKR70jrMiDYZMjo70adJmhCK+sY2Kb6nTbNdDjJo0HsIRtRjBOAMDPWMY8j3S+qWdr3+K8TN3fuz2r950121tArpo5nWbU6rKlM5z1ltlRVokjOKEt/qKPJ/XpDrWKx1KoNMs8cRn34sxcKbb4+gyrNChpFNcosds/S8DYzsns7uqq/hY00BHsnM4XS9E1jo+iBfbMDt1EIMmp7IxBSodEUpdkrbmWmsVSnzWmkG5ZO6FY9j17qpuQw6DjfuqrrKgl5/33LiN+kaTCx3H64Z/ez41r/XqIWsS9zmOdxOWcy4MoVwh2AXVQdamvfHoGKf0kyGFU5KJL4jGaC719pCSJB8oj1A2qUhMwGGjf/+GBr//l1fRfOg8JNfhg//pa5jZ45izJ6BZg2zBEsVzBQuTUhYLLOlcREY3s0dtUeIl7DQA2PNvj5K46Ua8/VN89O0n2Xvx91F7xqwRYa9rYVvKsxOYXhfJWQ6Jnisi44bgUcM9yw3+zd0FvjhXY+vOkwD8lzuu5q0yRSswJBNwSC3h47Fdj9AzQfi67FQoiU/bdK1RY7hRrJg6c70ej8/CoacneexohmePDzFUbnLF2+5l4p33sfhvW5ijjxHc93U6XcWN1x2jfXYLjY5i5T8l6H7mUfTJQ5jVOfv+1SuY2jKmusDi73RsIdKsIcOT1tRRvZ7uXq5wn2NjDL9w82+dUyXLKTKd1YtMyBCN0CtjkwzTI6BNl5ZpU8V6IuUkE6k05SXLktTwSVCSfDQ1dsIay7qClRLtPQfu4oxcA6NpmlZocGYnyEt6lb7pR2pMxphIvSkpftSoAaI86CIwOsqFzgFcRKKix2A7ySlJhYT2OGHbQj/zKsuEKWIwzIo1CntMlun2hUuvvx8Rw8U3fwVVrKPPFine9Cym0kYyPiQ8+gdTrMxtpvzTJ0lNnoFegrGps5z/kS+TSjd5Zl5IJgybp1e42AzzF9WzfJcFfBRPeLPslEkEYbMZsuczVPDKqgyTaoTTeoGN5MddCEJTN1miahk458ilzisr7ni+fmMbP7/xAiK+Brn1xuXQeIERl7pfX7y4OOcEIq5e9RILEBP7BxsXIO4x1hwXa4atj/VrlzPrddDt+HmQUILacUJ/lHBrW16y0Xs95g1MK+PhHtuiMHrRuXbvR003out26BEWpMZ1wS4+GOxnWJVIhFykPBnO1yOUyeGTYJsZ5TtyJPYYhoq0OKaW8I3Hdj3EZcEknrGvNU2CGVUjQwqDIYlHgzZDJkfa+CyoOlcFW9gfjFKRNgfVIgmEq4MtFEyGK4JJ2lpz2rS4YLrPttJrY42o0aMpfY6rVR72TnFKFllQDc7Tk+zTw5RMmjrtF76jf4B4HY71CkTxpj33jD3y0IGRiQX++pEhtqSTdLThwlyGEVLUgoBV6VGTdkjQtkowQQjL6hPQoh3KDYbyhVgoQc8tbFiJPHfpxv2uW5eWVNTJd3wPA7FkRozHIKEo3wBm4EbfDvYFcZbGYMxsH9d1E+3PCRmQT2GgTGK7T2sXoXg4CMP66+LJef2Ymo2uZwDPsrAgO7LWMfLhmrG3CNro6HnbKYZncc/2rOGFmxpBaJhWtLBkJU2PICQ+ChPeKLu9zRzWp3mXuZh37u8yrvP86u4J3pPezU2l7fzz/WkwwlsuOUv2iqdQm5METxpkQmHOVCCXwJw8iZSKmNlZW9WKsa7myzNIaRQWz0K2gJTGMKcPQyYP/S6SKSK5EhRH+MvfHuOKTz6D2rwHNb7NHlNfsZMQESSZRs+fgNoKwQ8E0zRIQvOdB3eRx2dUkkwkCvybR7oclxoztHjbaJ5Pr5xmyhQ4wjwLqoYBRqTAiqkyrUZZpoYFKIYGV2K9cOrK8ASLZOsl3npBg5GhNtt/7QkSN1yM2rKJ7E8U6P63eY594008eSLDSM6j30sxNVHnK9+e5oc/3MnF008QfL/Og5/axfCpEzzzma207lfMzY6w/JUkY29dQABJ55FMgZMre16HY/0Dxy0feec9N9164z133vHVA9YAFb72+bDMASIAACAASURBVG8fePeH37LmXP/qLb9zW9u0mVQjrBrrZDwsRVqmwyoNCmKVrsakTIChJDmW9Co5yaBEUZAMC2aVVep4oTdPN8yRdkMawjZR+GKdOBxkpWu69E1AIvRscv4eShQ900cz4H7o2KWT0VURpNRExzp+nM1xXjgRsbnPcUzcRKUXKk11TRdffFuyiYXRJEgwR4WrzS4a0mXE5BgyKca9JCee3c51v/5F6BsSB6bxdqeR0UnoN1GXvwX92GN4U4bi29r0v97H272Kd9EwqasV5DwS9Xlmnt6DEgh6SU6swhf5Ie80e/mud5ptepjHOYkWgxZY1Kv0QphpjyDi0ThZYlg7RRaEhPgYseT0cTVM1TSi/KwlzJeOy7NOTlcTWzsIlREZwKgUTppZR5Agx/sY8D9MdPyg8LACL64xZ6d1ska4ZH0BsH768XwFwouJcxU7LyVUTJrXcUk88ULOkb0/BwcGg8LDV64B+dwCKquyCCGEUFS0ZtqGHBRUjoyk8cMpYCaELo56QwyrEmmVohGaDdrPOKGypeOlDJ6XJ4q8yrIsTTIkOaqWaEjAOEWWpRE1LtKkWJEWLbqcZpGsZOiGSA2FYkKGOKHneKvZzTFlPUK6EjBkMiyKRVlcqCfoiqYhXZp0aEiHHCn26hGKJLnfO86yNCiQZit5fBTvGR6i0oas8iiJz7Zhjfo/XhuiJj/43GMHDJY7kydFgQzDJsOMqrEoLa6VcQ7KEm/78DUv+3r52ij7/juIg7/0P9yuvB6f/O2/5T0HniXrKS7ZV2G1r6nQI02CDCmuUnsjrwlH5gXbbXeyrr0QGmCdSQeqE06W0Omwu+ibPu2YnKZzjd1Ip9wp3vhh9yM+yo6gReGC4jpU8NwOYbyrtH7h6UeQhnP7lLjx+PpFYv0x8evj97Wm+xR1aQbHrVEbCTkq7jEdadQl6sh3xXRJih9NoCZkiJQkSYpPXll/jEW9zGY1xrg3ypQ3xjV6J2/XW7hIdvAL+5Mkkz1ymYC7n8hy4SVP8fSsYm6uyGMHJ1ma3YQJBFPr8M3/511IOoNsH0U2bUft2I6prBAcM5iTi+hDz2KqS1AawXRbmH7XTkDaDSgMWb5IrmzlfIM+1dtP8nOf+muo1O1EQHmI5yPF0eg8IAopDBM8doyThy9i9tErmX/kKgqSwEeYSvk8dCTN5WaMN5oJjskis1XhY9ktTJkM57GJJD6bZNhuMJWFeIxLOSrOMlivgJT4zLDMtC6TEuE7B/OcmSkguy+x5ov5YfBTqJEqn3/U56lWi7n5HP2ez0OPjZPyhLO9Lt++4z38zR030O4ovn3/xYxvWiRXrHFmIcl5v3YPBL3IXf3+I+9+TSww/1jhix/CRoNI4SYe/+UL1pupTssaFqLohVv/kuTDfGCo0iSNz7xZJUCzahp4KBphZy4vmUgau2d6NE07Ip4CgwIDicxW3YYniHENwHZ/8yp7ztzlOstd04vMB919DxS5BtNosPnWHq8HkxkGU24YQI/sefMYkSIzqsa79A6SKLZ7Ga66aIEd0036D04gQwpz9jT66HHUpt2YMzXET6G2TkJCofZeiZTrkWwqXgJ9ZBE13cBT0OwI23fO0jOGnGQ5JjVuYhcVaVsBE6M5a5Y4wPnRxt1D8WupSyIvB/d8Xbj8G4dZzeqlsNEweP/t+etGsvTxScdGPh7xdWW9oWH8tushWGsmVOFzit9m/WRi/fQi/rdzQXtfTLzY41/McW6tkpgHlTVsHHic6Og527U7iE3v14eb0BW9IhA7J+Fly3ToMphsZEjZ48lGRp0FlcfDoyQFYDDNin8+3PtXCRUj502FAM2KNJnSOfbpST6g95LAoy5dWtg9yxYZp4xVd0xii/VjZpadahOH1BJzUuGYLOLj8QN1MvqsFEiwVZc4PxjjYj0d3UcWu3aNmgJ79QRvV5tIiqKN5vtLXXbkfAJj2FnyePrsWsPjV3N87M6P3g7wNjVF3ljLho7YvN0XzSnd5sZg1yvyXF4vQl7BSOQrzN19HbVKmfOme2x5x92MpzzOqBoFkyIjKVakyYhXjhJvx3RIht2ohFjBSZ8EOZWNIFvrk5Abe/fo42BSa8z8wslJvPNjuSJ+dLzD67pJiYUZhItTzLQIBgksDik71+ge1nac4o7t5zrWPudBwXEu0vnzJfX435yBnnIGaKFPSdyMKe5w615bYKzOv4jQNC3mzQAzWQ/HzzmVI41PSiwZ8xf3ZsgoxfsT0zRaCUQZzrvgKL/4L76Kn65x6/seIdDCvp2rbHnf3ZhqEnzFDZ+8C33qLDSbMHcKMzcDnfA99AU64cK5umCLiaEJ6LYxJjQsTFmeh2SK4CXIv+8YatcO1CVXIcNTljvhOBUOjmW0dVjfNcp/fLSHiKE4skDN9HlcLXKy3WX3mOG0aXKvOsMmUybQhr9onuKo1PCNxw4zylmzzIqp0aVH3TQZMwXSJClKjqZp2QXU2O7ojKqQVorLdnb4xpku5sRTVunKaAh6fPPz72AikeS9230ePgt3PJzhaK3PM602EwmfB2f7HGp0eGiuz/FVw+LcCEPXP8x7PncM2b4PGZ5CimOYxiuDb30thxfKa3oo5vXSOY8LwuZHWpJ0TY9kCAdt02VcyhTJ4uNRkjxlVcAPXcY9PEalZEnmxvogOZgquI29CgUkrKhu07TWPLZzpPZjG6aN8sZ6mVJnXgqs2fwF4T+wzR0Hi4nDhByE1vlkuGM6poOPR5EsBTJ06fGILDIhKYZSikolz75f/ga9dgFJ+5z+zDWobZsJ7v0SpunRv/uLoLX1COp3UNM9TFVhKhUwGjWUoH9ojGuvOcT7f+9LnD45QTrczN7HIQ4GNTysj0fNNGjrNivSIifZiAPw6c5hKx0fmrE6/L/juSXCpsxmb5KsytjuuDHP2dyu+ZyE5z5+flzE17LB+6PWHDP4HA022hYKuNb1O74GrV+Pnk+4xKz7F4/1RPGXUqCsv68XA/WKr0vOu8PJ24N93f2wQI5k9AlIiL+Gq+HCFcVd03vOOQLbIKzomvX0wGfBrIacqIAG1pB2m0ww4pW5iC3RfbrLiJMSwho7psuNwW6GJM+qrrJKg2fUCguqTtMEXBts5UI9wrDJ0zIdsiYZfqN09B1rmTYXBuNUpMWUKbHVDDOt81wZbGNeauzXIzyhlkjhMUySFWmRMgneZKZ42JuhScBFeoyrlFWOzHnKNtZESFpLER5d6dLsv3juz6shSibJN/UsS6pJRZpcZIbJmyQZk+B9d91y+yvlk/J6EfIKxonHL2d5ucB3HxvljqMdlr79Rj7w7od5E+P4RnFpMM1FwSQpklGXzQ9ddw3W7Klp2lEicfwOh0V25oLxZOD4Hq4wgZDIF5PBBDcMj2Ni1ZrOhk0qLskPpiNxnXVXtHjrNvDrIw6nincQ4/FCCXr9dOOcvA+3WKxRvArvYwMpxnjEscgusdd0gwRW8akoOSvTiTUtcxOeVRp8MNjPJXoL//Vwi5oOmC5DNh1w4uQQjWqZMw9ew9LZ3SQvOcEVN38F5QXISAr6HuJ7SDmNZJNIeQgTBMjO/fQfTaDKPYKnhkCBWV2AXGlAXvcS0Glbl3NR0OvQu+NOgke+zpd/413oI8esgpYXW5zCzb49ESHPJJkib3xWlgtUl8boELAiTQrKo9FW9LFY/Lb0ySUFH8VDcoR5VadFn6ZpsUumGKFAXrLMSIUSWTKSipx/ffEYp8xWPcRDwTJffzrBteMpZGQTwbe+DmHRcMNv/Q2BMXz6xCrLusciHdJKMSst/tocp0KPOWnxLe8Uj/er7P3I3+FdezWLv28nQJIpWAK78055PV7WcBK35+IFuPwEkCeDEmtE6BodJoQ6rVDHQzGCnZismFp0OzfNaJh26EZuN6Due2kfx3Y146RxjaGLVczqhF4kNqe21uQ6t8GON1REBs7wLp/GfSmAaCLiDGXj3Xp3m77pRzySCMOP5uJgjDFT4KPlSXwRylnDpi1noBeQ+6ka+lSXia1P039gDhkbQ8YNMjmCPjKHfuyHdP/kQdT5F6C2pcEYen+zAKNDSKpLZs8z6GMBS6s+7zi/x/+YuIwuPb6tjvAEJ2mZNk3dJCEJjssCVV2PCquVUE413vl2m1sPLyxMEizoFWq6Tk3XI9jviwnX7IrzD+15DDfW5/gcufcl/t5Gt48Z7K13K38+t/MXamLFYb0b/c1BfeGlkdnX39eaxzM6moZsVNjFCzHHjXIohqzKkJS13f0A6yGWliRDXhmfBFMJy29Mik9aZchKJuJqlSTPiBRp02XKlBmnzAIV9hjrybGe0+Ni3LMb/ilvjB6aS4NNXCzbaZkOs7LKIjW+5j3LjDRYokNH+uySKc7IEl36IaNQwudQ4K/lB2RNimVpkMbniFrhAXUEg+F+7wQ+HqOS5EveYUZMlnmpcr86S8lYF/TjqsrneNpO4ZW93+m0z1LLkFKKkUSCav9H49L8U4333nXL7b2wWXJ+MMH96ixJk+ATX/jFVxQx8Don5BWIP735M7cduePQge/OBezLZji6AuNekmSQ5cvfm+K0bnPL5hyLVY+rS1myrSL72MRWGWcLI6RVhiWqVg7SWCWQXjgyNVicZ9d0bfIOv8Bu7GoVspL0wsXac3yQWFL2Q0k7gyP9DWQUCeX53Fbf4UfdBIHY9e5vjhdyrngpZD84d9fp+RaQaFlwRofo6Lq4xGC0sGwwuVkv8ZgQj1GvzBYZp0qTBu1QItlOnRLis1mNMWaK7JUi31fz/LsrsvTrWZIJKBe7YISdB+5l9qk9bNrxLNQTqGKLrGkjiSqq2MUsAWILg+CpJvoZBYsn8HYLkk0gyTp6KYWZa2GOL0BnHlaWkHQynGgYSCS57wNJfuPvi/zVF8f5rS9XUFPTdmIS9O0x7pRpDcojuP9LyPgUwT2P8cD9ezm4ZOivlPir4ChLpsawLnF+Kcl3mhVOsEhCPB7qL3K13sRRtUKbLrOskJYUi1Ro0iUtSTp06RKwqFfp0mVElS1PiSR79BDHvAqnabC5V+Lzf5rgbb9Qg4SPpLJQW+D3vpiiSJo0Hpdm8jzdbfK0t0iVJjXV453JSc6TMm8aS7N17zM88DtX0u+kmfhoFnSAKMtDed0z5OWNO+/42oGWaaHReKK4646vHbj51nesOed//bm7D6QlSY/+GoPTHn2KIcxqwVSsWaF4pPGpmTY+CWqmgcFCsHr0yagUYL0HeqHUdz9U9WualsWpC1HOy6ucpbEbK2+qw+mK8zuI8wDiPj8m/NldOo6Ck4eNy8dK2L6xE2kVya3bTOI2srYAyass02qUsskxafK8o1xmuQnvuGqZucUc+2/4Nt1Ht2Ce7aHyLUwti7ezgj7RRHyNmW1haj5mPk3iWh/9+EkwfWSoiOQqsNzA2xyghuCJz76T3btneeTgCL0APlTYwSPdVZZNja7pklV2clo3zUgJLK9yjEiBMSlRp820jNKQDkOqyE61CU95tEw34mqkJUlZFQnEkJE0aZWO5MpdeGGnXhuNCrl22gQR38SEpV08rCTtc9cME3uPwmsg+t39vH76oKOc/+PE801Rnu82L+Zx18OMN7q9iVanQSTFx0Dss20/f2mVxnGcwHJG8ypLhx5J8ambFkVVIC0p0pIkrzIEISR71dToEbCiq8yxQps+79D7WZAm46ZAwcszpobYJVMsUSOv8uxR09wY7OICNnMpE3TQnFF1tusSNdVnTi/TMC2apk1B8pRNmhlVJW2SJCTBqMkzw0r0XezSs7BL6dGhxywVhsiziTJVaZMlxao0aRvYroepqS6X6kn+4N2L3PmsRY88oc4C4AUpxlWK80YVz1YCaoHmQRbYIXku/cJrgw8Sj2s+fMU9D3z+kQNDJsOSavKJL3z8FT8Hr09CXubY9Ud/etuianCfepYfeKf59Kll/o6T9IxBCSzrHjsTGU4ve1y/yefvqitcUkzTJmCzyXK5GuaomQlH3CbqHLnw8GiEUKD141gHCdBY+Uywi73POswuz+0ADCQr9XM6Ug6C4In3glOPHyfiXaXnm3S8mJH4em5JPJlHST/WNVvjZhyeOw+PPBlSJkFOMoxKiZ6x6k9J8dmkRvFQtKTH980yb9Nb+P6hEt3w9DabPqOjNU7edz3FcgORACm06D6zjScevorqI5cSHB0hODuMFFOYBYO31eBtqaL2jWDqXfo/TGE6Cm8vSK6PaaQwSxpT6RE8fBpyJaQ0iqQy5DJ2WlEyaUx1ERmdttMOLwaBi0MWRsfQD36LlQev5Bm1wuPeAp9tnSRAk5M0D3mn+KvZKm+ScUakwD49ToDmcbWIT4KKtu61u5kgMAFJSbBsqgDUTJOMpEniM6+XyJOhQZt5aUWypQvdPpu8NLL9AmRsC5LOQ7tDjiQ/VKep0+db7WXK+GSMT8O0mWWFbSOaS7b1KGT7/MkfvpfHZ+HUXNpK84pn4Wbq5fucvh42HCcN7FTAlyQ/fdO/WqOUFRgrSJEIuQC+eEzLCH0CVkzd8kHEpyyFaMpgwuMAVGzTuRxKwa6fhgYmIBN6EXh45MVyPjqmizYBvvjRNKVjuqE63qAbr2NNiTWKSSE/bD33YCOuAhBx9xQS5WXXNS6qPBlJRRv4H3hz/FH1Gb7XWeXeH4zy1rd9j8NffTt3f+Myjj15Kd0jW/H2zIEnSClA7d8FHYW3SyG5DsEPGqipFN61b8GcXUZt34KMFeg9MIae77O4muTxJ6d565uOce2FNZabhhuDXTj+X1M3I4UwE05qrDmgokmHnGRYph5N4308K5EsSYZVMZIzboWQHfddXz/JcHxE9z5t1EU3mGhiulFzKA7Nes79r4PQrb/f+OWPw914KUXMeiL/C8XzrXcvFH3To2XakcR/XmVxEsaeeBQ965Zd1Y1IzdHxsbrWwhhBkSLJkl6lY7pUtM3hxhgmpczXvcNcbSaZVTVKJh0WO5rLZCeTUmZXMEzJS9Ckz/3qLE367NYl3lDMcnWwORRu6TOsiryVKZ705nB2BYtUOIQtGAIsqiIpPr741EydHn12MUlX+pyWFcZMgYJJsVOPUJUOGRKM6AyH1TK3f3ma7bqMbyxE9CK9iUky/LBX5dCibbR+T82SNAnu1fM/0vl+NcT//Plfuv2Wu269/Tc+/8v/KEXY60XIyxR/csuf3fbUh79yW6uRpUqTJb3KnF4micfjwVE+w5McPOvxzu0JZvtd5tsBRxaFoknydLVHAZ8ehnJSMSnDEd7WLWhORjdeQFj1kUF3MQ59iGOXIyKhOGugtaRJ+7fB9evDjbkdafNci8VG8VKS60ak9fjo+4Xuz0kZnguytb6AGUxNBscphJZpM6qGo8Q+L1U6psuqqVNWRTpY+IEj2k7rIhMmyxnTYv+WFhftbLB7+wqBFhYXC2ze+zizsyUee/gKgrlhnn7iAi66/n5WFkdIXFWjubAZU+mgZ8oExzxICbRbmFVB0j30chZ9qg8dhZTaEChIgNrqQ6OCqSwCcNn/dYabvG1sN3lkfJudeITcD5QaFCDKQrfUnivQpwI8r88nt43xAW8LC1SZMiW2mGFypDnfK/CkrrJoqhxUs9RMCw+Fj8eIKpOTNCti3dJHKKIQkvjkJMNICOcAqJgGddOiLl1OGSsX+jgrfOCaRczKLIu/OWd9TDZvZVVaGDQrqs0Wk2OGFloMZcnRMG2OLgifOHqY+VWftKe4cAL27aha+eJui/uPvff2+4+99rpc/xgR93IAq7bz8Zv/dVSIDKkCbdONDACbphNOQXK0TNty30hEzuZnzbKdmhjb9GjH1KWS2I1JPMfZy7XypV16ZCQVwWWcIIaDasVzpuMfrCdhw6AhE//dveb10NX4uYh7ifz/7L15lGX3Vd/72b9z53trruqqHtSDhpZkWYMtyQaPso2NsY2NMQQSIHkvvBAgJEAIvCxgPaFAyMrAlJeEtXjJIvg9A8bGxsSxTTzItoyFrHnoltTd6rm7uqZbded77jnnt98fv3NOnbpdpVZLcltOavfqdavucOrcM/x+v733d3BeJTl2ySRWlQpFRrXI66I5ZnWUIh5f9Jf4xY9fz73PFni6EVBfLfHDfznB4x/5XpY/+x3oqkfwuQWkEoHnYes1t+97rwYvh+yZpv8nwGCA2VnHnpvgTW//OtfuX0VEefbEKAVPeO1UgbKU0v0e6CA9lhVxnZELukY/VmAskuMqs4OBBjxtT6emjQ3bjrkKSYJVRRBa2r7oGA4TpjcjUAtyEcwoG0mi93zzzmZ8kOEE4HI78i8ltuKYXO42NiQ0YvDEo2iKlEyZUTOSFgXb6iSuLUpFylRimFXCiTIIK3aNvu1jxHVYcnh0tc98tMSqbVwkCiAi1GmzHK3yR/IwR+1ZWuKTU0Mklmlb4dpomnOmydd1kU/L4yzQ4EnvAj0ilrrKtCmkHMzFqE7LRvzC5DXss5M8bebxYq7UmKlRkDxBDJesitt/QTjJInn1qGuL87LGeVnjiFmkQp6veSdois8pWeawt8DbxkcYo8Dror0AXKDHI95ZlqIBZWM4Kyt05YXDB7fj5Y9tONY3Ke74W7d9ZfljR+96+nyeCVvjUT1BqCEnWCLUAJ8BPS3yfddFPH4uz3ntc9b2maLICj4Blmkp8MaDPq8O51js5+iZMHZKd9PsuizlemvaxPCAZAp20IF1+cr1RbnGMrWx+KJoOrxlHWmzSU7aNUjb3OvOsxt/Tgb+yyfiXaqzMdzRyLqqb3hfdhJCNiQYG6Faw1As97oXc2YEQ1XK8cJhijui3ayYLjnxGJMqOQw7ZJw+AW163Ky7uUZHmTYFRskjgyL1RoGHTxWZG1V2zHR45tC1zEx1GZ/o0KzPsufAKWy/zOTVR4lOVim/8TjaETpHDlK65Tz2fAGzpwzawy6U8Q60wDfYpRHMVA+ztwJrAXiK7D2A5Ao899PwH/7NOP/4tz7Dm35aMbP7HfzKxnAtic9jFLpH4zmZ3h0FSnuOsftDfda+MMtqu4gv7jqqUuDG/Ag1myciz4I0yUuOujin+BplqhRZjqGDNcr0CVBxsqnOA8KPhRVK7JBxVqXDT+ZuoRPCz99a4IuPjfOVj0zwW6fW+JG/l+cTH7qaru/RMD7HWeSoWaFlBizTZFXblCjwjDa50c7xF4MTPKpLPNrp82O3r1B81y5Q5Uz71dswrCsQf/7Rv7rLQUbcGDLtjTuSqynyvr/1tq8AfO5j993V0z77zRxdfEJCfJy52LgZwaUnDs7Vsh3mjFNaG5MKFSlRV3fNjZsReurjFASdFG9Jig6yGlfxnYkqqdxnAtUR1qXNXWLiuBmeGGyqqrQJj2DDuJcRk5V174b13xMAViJDS8pXKUuJNZyMbUHynGSJp2WRKWo8Zs5wTOeZN03OSJejps5jrQFd8fFWJ7h+LqQ2d56jX3obsjJJqXSW3NtniZ7t4+2y0O9gnz5H/mYfeyKiffjVPPHArYzVLGfPzPCNQ9Nct7vPubpH04dbdRfX6i4e5dSG8TvhqnS1x34zh4fQJ2TeLqeiG23tEmqACsyYSXrapyRFQiLa2gXWPSsSufOqqcYJ6EZFqpzk0nllXTp2Y6zPM7JhzkthctlzsmFOWF/8Xw6R/KXGi/1bw4W27LY2Fs3c/B2pWxE4RbaQ7DEYEDDQAdNmgpZ2ARiRKh16hITkJU9PnUJaV3sxVC6KpZWzW3L/nGmoC4tlSRsssMaC1vlQ7noeY4Un9ATnqOPrgD4D6trkDvZyPwvssFUekTNpEfPV7OHj/hkuSIOmdmhpN3ZRN+k6x7LOM+1qD5+AVemQjDWROCnfInlGKRFIxFU6wZwdYbFvmTMFOhoxQYEl6TOrIzzjrdC3wrJ0+JWP/8w9d/yt27bniG9RbHdCvolx45+9557XfPx99yxLb13hInHdVsuNdpJGo8prxlw16k2lca4uF2mIzyh5VjXg04dyPLDS5zadpkaZvTJDXnJc7e2+SFkqaaMnP6cVwoxe+laRfDYhhybb2arSlHqFbNH+3qoVfqnYqlo0zPFInhs2f9osNktIshNuAplIOi+JDLH7nsbBOFBujWa5Pl9htx1HEEa0zE4dY9bWmJFRrpY55rTCrkIeT4RDrHGkGWIVPIG8p5w8M0YYCRcWa4yMrVEq91Br6HXGsc0a3t4VoicnMVMetdufoPPVVyHFkM6fz4Jn8A62iI6OI2MRUvXRvode6CBzFQgsZmInMrkTa4UQxXzn2zH7bnJfPnZST037suervYY9e4Qzd49jbrwd2X2QekeoaI7v8ubwMFxnx5mqwmFtcqtOMkaZVW3R0T59BsxrnVO6yAQ1J+mo+RRGU6FAFGv0l6VEoBFndIk+AV8KF6mQ45EjNUYLwnFt87PjV6OrF3j/T/wlB71qrI7k9ruN82Vxyj452vSoS5dVbdHUDsd1ntLeU9BYdpCu7bgikVT+C5LHExP7crgx6pd/8Lfu/ic/8Bt3A9RMhT7r1UdPDFMyGsOXHI69goNAntXlmFcAa9qhJAU62qUTV3kNQqCh81KKTQUTeFTAumIQuO6M1Yhh+OlAg4s6ym6/Nhr0pRLlQ67XWTWm7GtZ9SfBpM7hAw2oSZlRqbKgjkslCE9xhhXrZIlb2uZ4dI5lbfAcF7gh2sGrJnI01qqEZ+e4/nv/CjGKOeBhj5wgd2uI1uvYx5/DHJgkelaJTs9QqtW57c4nWF6YIQgMnUD5xnNF3vn6BQqecEibjEuOt3i3bIA5hRpgYzXAQ/YEddrk8eL7NxY/iRfAvvWZj5boaZ8Vu0rC7PCtv2F7AB3buWhuMBnZ3kt11IcLXVmH783Unobj5ehqvJC4XN7JZnDhDVBhNhLiLy74EXtTuZ+LpsikN8GEGWPMjNJQJ/Sw28xQliKD+HwEOsC3PmGshmaG5tikU+bhUcoo0SUS1AmEvlWrIwAAIABJREFUD+AL9jyP6/EUkeG6LyVuMgdYok8glt2lPHfKwZinYnnzjiLv0X2EcbKRHDfNJLBFKaZKoKrKtBmjQJ4pGU3nF4DD6gjqY1oiENc594nYWfGYkgIr+FQ0Rx6PA9EEbRls4KVtx7cmtpOQKxCHzHy6WE4GEk8MXSK6fY/7Gm0mKbI6sKz4Eb5EVMTjGbPC//COAzBQy5hW2BuN8RoOkIsntYqUNwwcyeSQ/R3At/5Fg/TwZJBAERIdcmdCJOuyjPHfGYYlwKVhUS+Uu/F828jusw5NWtnBK4FbDH/XTZWxUk18L/1+WcWPAvm4uhqxIF2OBz0elVNUKHJeVnlW5lmVPm18JmwZH8vBnRHve12d/TrCtSM5JmuWG2ZgcrLL7HSPUjHi1Irh3KndTO47woWze6iNL3Dh1PVEZyeRiTb2LBAY8qU2wXN7KO44T/jQFIO/2Uvn3AEIQMoDzFU5ZKIAQYj3jvdALo8un+Paf3meR7wL2Ie/6mBXUei4IFkYVhj7tdTnkYrTjX/66BSfft8UBD53vKrOwVyV77pjhR+d3MEdE25CujM3TkND+gTslRlnGqdhjHE3rOJgGAvScFAsSnRxWvDJAlVRRsXJGa+JU4m/0A/5w/5xjnnLPFUPsYePkvvumyh6wrV2hn3MUKYYX5+WmlQcnEcjTsoSCam1SIHT978R7axtS/NewfjIJ37rHogleFXpaC+FVFiUHTJGV/ux2lUbIxIviNxCIFKLEWFOJgiIGDFVqlJKCyqFoaJLTSrAxsV+T/sxQdemMKmBBlRMmSy81MNLEw8v/hdqGCcL65wFs8nCbyvFpuxCOLuNdfiQ21by+wW7TAI9amibvvpE6hQPBxkX6pCQVdPn3nqbf/9sm15riujkDkZ3nsCeCYmOjrv7ulBAI0GKRbpHD0IuIujX6HfGabcLnFv1mKkIx4Iu9eUx7vfX2KkVdpVztGXAD/OdKcQHHKelqz0GsRfLsjbIi4PKTZqxDd99ECuiqVrWbHNDAnI58XxqWMMxfG62+vyVSjxe7N9MxVE2mR83VeMaSoIhMy/G19+MmWBGxhznKYYulsXxNy7YlQ0dr6z6W1alLLlHXGdiY1KVlaVOlDsf0qNYjdKkxFefjnY5bE/xeXmKCMtT/Q5X2VH2yawbxy301bJDR+lpn0ADDCbmizmuZZkioYYUyDNqRlIu2Yo2sSg5chzQaV4t+3iWee7jGSIsZc3xusoI1+zyGfU83j05RiRKSROvHrvBP207vjXx/CYN2/GyxLloI+lJxKlXXZev8PR52CklDrPGtYVpmgPL/miUP/OeIlJLz/b5iCy6KrDCEZNjVia42k6xU8b4G44wbSZZtQ2sxJKQoqmkbhY3u9UEmgx0SSUlIbEFOoi7KJn3Ps8koSgf/eTvbYm9/+EP/tzdW722VWTxr2mnYqgrshXpb1jSMEtAzy4oIgHwUiMtwcSmhE5+Mk+e/TLLSVlhngZ/O7rVJZCEHPXqFPG4zu5gxXTJW8OxCzWOXZjmjNTZPygyGgmvf8NjeOU2Uvb59Ee/i7ESrKwVqX/pTZxaMbTbt3Dzu7/I4NwuvH4Rb+8CZiokt+cCAJ1nb2Dgl5j8jgeJDhWJjk+Su61JdFQxswPs6SreW4rY44+j8+f4qX/0HfhmBZmegcB30ryBjw56rjsQGxZq4EN1HHvhOH/9E9fxXf/8E3z4lz+ItursvPk5fvxX5/jaT76Kd/ziJ/m1//PdHDNrvL+4iz1aZNaOclrqVCkyLSOc0xXmZJw2PhGWERyufF7rDHDV31AjmrbD1WYX53SZqjgllke8Pg/Hk9zPlG/klmtbeK97PfbwoywHeymqxzhlTrIYw2aUnvqUpEBfB3Ri3fuKlPEZ8A8fn+cL1XH33bfjisVHPvFb9/zI9//C3eNmgj4DAg2ZNGP01HdJKHlKFBwUSyNyeExIjQib+lUk3AKAUanQpENdmw7yJ2VXaSWkICXKlOhoD5MkKjEZOmtmaJDU9wJxakxBbGiYJAQRESUp4evgIljSpWRit1oQRjGPxcl3h4gmkNCQxagOQCMWbhj2XAKIBJq2BcCa16Ph9fnOaCcri0WOHbmK297yNcxuD7ukRI8LubfU0HpE8IUuNsrRmD/A+LVP8/gX7uL8Sp6z3ZBlHfBJeZTPPVtihxljd7SPp7o9dsoI93pHqdgyfe2nY2GyP2tRA088qqZKTUo0tB17UeTo2156LIa/w6Wq+cPHbXgb2W5H9j2Og8Km7/12iw0qjUOP2dfT9z8PGV/EMGHGiLAs6GqchAg99bnB28uR6MxF1f9EVj8Bca/fF5aBRilSwI/hj9muYdYawFc/lW1WXTe39NXHkxLH7DneIru5aSLPJ1cfwSA8uDzgzukCD9edOEQiIAGuyDAgoKs9JsyYU6DEY822mDLj5LF04+5nQOSsDUV4v70NT4Vzps3hbp/7jvQZJaK/VuRHd4/yxbMhXUJ+cHaUP75QfBnP5Ha8mNjuhFyBGFYAUXUQgZNBj1pBaGjIm/NTPNXrcusey201t3jbLVNMGQf9aWvX4SF1wKq2OW1WaUgfERNP3A564PQtLtZXH4YtDbd8s1WOYUK6zXwm+wjrE8lHP/l79zxfAgLwp5/83csiB2cnr02TJ4011DNdkc3Iilv9PqwQNqx601OfHTLOiFQoqkefgCVtctiscm2xxPfuqvAu9rDDVhnTIldHE9wi4wRWuTAI+CevKjNWgqMryp98+jbu+/yb+PAfvYOJqnKmZSmXIiIrvPeu48ztXGP+4ddRvOk5Vhf2Qa9A88uvwewZ0D16EC8/YGznCey5Car/h+Dtr/PAb78PuziOXcpDIUQDH5naxR///Hv5tTcFXG2nkJk98ReKJ5580SUgoXMS16UzEBscfvRck3t+8Xv4A/sEuniG9plr+L+/70a+fD7g9MfewZjkaUifU70BvrWEKCt2jR4Dzqgjw1/QNaYZIcSyqGss06Am5RRK5Qw0lXO6zIg4U7oKBWo4R/UcHgUPzs7XYGQSXR0w6nlcY2o0pE9FSvTUx4+Jy5Fahx/W9cp5WYqOi7J0dh16th1XLCbMOCFRqvXf0R6emJTr0cWdu2TsWdBVBgSMUCLQkFxciReEuracxLg6wEbDtpmWMSKN6MYwwJpUnEu7eJTjRCKBluRjx/QEcpV4e2QJ9MmiKnFaf6GL2q3GpWy4hfy6saEnhpKUkFhWONspGe7WJp8HWGCNonqUxFAoBNz2znvpLe9CuwFmpk24OE34wAq2UeUbn3s7UeQhRnn4r97OhXqe0bLyxn1ue2/lRtra5byu8EfyDcp4DIjo64BAAxIX8w3HIYbH+OqzZluxDHyQJiBe3DW/qFOd6VBvlYAMd6WzP2eTjq26HelMtsXz3w6x6dx6CeTAZtCzMTOCwTjvqli4JuF6HLVnU8ND2FiUzKq7Ze8P2NhxSpW2Mp3EnOQYaJCuG6KYo+P8ePxYGc4lOn8iT9DoESMVhJJ4RNZdIxVTdhLPWEJCWtqhELu0t7VLRYp0tJcK9HS0R4E8NSlzUpY4Jc4g9SveMbqEHJEFlqVPV0K+7p0iUOWR89Al5MduMnxpfsBps90p/1bH9gx9hWJ4kPHw+Kp3krxxN37BE37mzU0eOiPcsK/HT3u3EWKZpJYO8BZlRKoEhNwczVEhz0CDtP2Zncxg6xb1pi1e1smVCTckGVSynYThQf2F4HBfbGQnLxHj4GGxuk12vzeLzbDcyfMbpHgz20qOXbbS06TLgIAn9CRFcmnV9lm/y38/38O3ljfVHJTpOyuj3LpTWAsjxr0ca40C9661GM0b9k+sc8EXmsJr9lgOXHOek3UQsUSRYe7gk7QfvoWpA89w5MtvZeCXCB+fZuHcLtprk3gHlpCJNlRGkLkK1x08y6A1hdZHGJy6Cj11GPJFnvDb/Plfz/Aff+Fr6fdPFuPi5V0CohY9+VSckISEn/46XQl5xLtAhDteY3c9wrId8IB3nt9/yKMQH8MJL0cEvNpOU5MKVYpMSI2ceBQkxwVWAShLkRw5BgSMSpWBhtSkEgP+XDejqV0uqEtk3h5dQ40Sx5ohb/2NzwNw5r+9g/vsIl+Us5yWJQa4dn0ieRoQMiE18jFUJ4jFGHbpOEzMQKHMm/Z96rI7cNvx4mNA4DoKCLNmkhBXKJiQWgw/Wr/vAsK0u2FRZs1kXI/VNGEtSWHdfBDLsjZS4QhffRIRjYEGNLSVwp7ctSJ4YqiYMqoOq54sqpKuZwLHSvZnq8gWbuD5F4lJMcTtl0uK8uSI1KYQp+xiMMoUU5K/lW4rTuYWpMlZ7fHw4Ul65/axfGGORIAq/6rTYJRTT76W6ckeX3tgH/fefxWeUW65vk4QwZnlHM94K4xpkTvkWrq2R1/7/Fd5gBEt8AF7M3eagymBPt0XXcfq+9ana7usxbKtSSTJ1EXdaUlEiGVT2d3hbvWmx30IajRsIvk/W1xcFNzCk2TomHniUSCPr37aTYB1BbIEkg2bJ3xJ4p5VzISMJP8WCnBZ/mTymEheJ5+xGtG1PdraZRApd8nNADzDGgf3N9lvx9PuZSXmoSTF16IUyUuOpnZdZxGPZbvGmKnFEK6Ihm3Tj8echm3ziHcWxTKmBXIIB+0O1nSAJ8IPXF3gV55Z4vWTJd5p976AM7Id38z4n+8OfgVGgpVOQlF89SmR5y86i1xdKLF7wvLQE3MEqtTXihQ9Yb+dYIet8l57Mzu9GTw8Vm2DGRnjfu8UY1piwoykE3RSwRoeVJK/CetQps0m0mSiSLDQiRdI8vkN24irY3/6yd+9508+8dsvuMNxqW7JcGQhVFtNVJu9JzvYZiuaw61rIxulOEVM6gtSlAIT1Ag0YsKMcK2dYb+dZLcd4WveKY6ZVb5gzvBQu0NbBny5t8ofnq9zlBZftPP8+dkue6TMfD/k2IrysaVVzgU+9/ZXePWtT3Pv/fsYKQj5Uocjz01x6onb+foD13Lf597K6YUyK0sTYAXfzzEysYR/6Brs0hjUFyCKCPwiTz56Pbk3h5Tfu4DMHUA7axiEn/qNvyD3Pbe7L2UtWIsOemAz5PurrkemdoMxeG+7iQktukUhZf7Rj97EP/vld/L3Xhuwx45x1Kszb/t06PMH+igfk2f4c/MYNSnTwWc1Xg15eJQpxipYHnk8IrWMUiEvHh3tMWFGGMTqRONSxWK52k5RxGBU+Nnf/zzmVXdCFDC98wxdAgKiWH2rRKBBmsw447uIXTLFIHarVpSTskT/D1bRU0+jannj3k9uJyJXKP7g4//iHoAcHgER18ouyhRZ1TajVBiVSkx0LZAY0+XJsaptOtqnSIGS5Nkh4+lYVZEiY1IjJ7kNi/YCeZd44IxYnTWZl3Ygohiv3reOyJ68BqRjXOAsDFOybhLDsKINr12iyp6FZAFpUpV0qbOd6efblqqDx/Q1oIdPXfqMlhUv59Pv5ek8/Gq++P98Hw99+P08+on3MDbRZLleZnrUsuJbvnTS8oVHJ/gfy22+1nYY+rYE5PF4P7eTlwJd22XZdHnQO8dpqVOUAjd5B6iYCju8aUQMFVOhbCrrxbRMQrAZ3HczAv+w1G4Sw58f3s7w8UiOXeIkftExewFJ4is9NoMYb3WNiBhKppx2IHvqs2abrNnmhi5GNrbq9iVpSJJcJI9ZEZzkPspGkpRkX0u6HVFc2CuZEhER9wUrHDdLqCpTWmZ2zynGHJiKtnZp2nguEY+iFMjHyVVVnKLipIyk36sSw3ktTq2tbpvkJMeyNmhql4e8c3xoapLD5gLv2Vvk5jkYH++x047QHcBRWi/8pGzHNyW2k5ArHOuDi2G/nWRVepwbDIiscKwZMlfKcXxJ+Ky/wJc5zDfkOI945yiQZ8RUAahQpKkddmiJCWoMcK3QYX36DT4emeRjq0E6wYUmOObNtPJhHYZwJQb5JNnZbFLLvid5zP6chQhkJ8wtFb9i7CuQGhC26DMhNRyF2uO0WeVZbwWLckaWadHjObNKkRx9QvbrCHXp0hCHVW1pyEOyxHIUcLWOcEra+BJx5vg1NAdKEEF+vM41+xrMXXWGehcOLVu6AzhzocyFYzcxOdniiUdeRbc1SWtxLwQDdNXn6LEdvP4ffwqpjmBe/Xq0XYdgwK++/zjm+gOQL0CpCsagUQBeHm3X0Shuyfs9x5kwBvIFVsVPpYafMQusmj5/8WiFO3MT/J3SXt44XmWEMp4YFq3DtK/FC8de7GodElIkx+26jwpFqpQYkTI5TOr9UMARHZvaoc+AouTZrVWeMiv8831zyMgI0dfuBWMoXn0SQcjH1+KSrjFrJl0HkIiW7RASskYnhet0tO8WpdU2urKM5Iv/U1ZMX8lRjTX9AWpaoIfPlIxQpkCfgLx4KZTOILS0S1488pIK5zKI3catKoFGLNs1l5jE5zKH89wokE+7Ifm4Q+br4CKYiEFSLgiQwjqS1zaLFwrp2ex9WWjScPf1eROYzEIf3ALvZq5ijCo3M8HhFcsff+YmpmdX+eQXrmPntI9nlAdPC4ef3sm95wKeXRSORG0eN8uMF4UxCrRlwG47yud5kvv1WT4rTxDEHBinPuegkeNSo06bMVPDE8NObyYWBsinBZrhjvJwbDl3DL13uCK/WdIxfIw36xgNz20vqFv1bZakbEZQT4qEFSmlSlGKdUIBWPJxgTJ5f/bn5BzmJB/LWQ8n2huTkwRpkSTvCWrAVz+FaIG7XhM/nKTzmJdC6u3z/RPTdPEx4nFb2a1pPukdwmdAEN+3UXxdBRqwEq2xZpusRGvM2xXO2AUA+jrAiKRQXxMXInbKJOAUvM7qMv9q9Rmuszv49XOn+NK5AWtrZX7hNR7X7vI5ZOZf+onZjpcU2zPzFYrhDgTAlzjECb3Ao2aJc2vC3/+e49x+sM1EWViSFiKGvvqcsYusaouaVJjyxlmmxY/pa3jIu4BFKYuTu0srEEOTGKxL6g5rkGcH/mzVLtSQYS7L8GevVCTVz60msGFFmueLDZPa0M9JFacSuy0XyLNXJwmIuDGa5VFzmhEt02dAhHVKNkQsSJO2DPAQzkqbcS3zHt3HBdNmiT59Qk5Lm6l8jjI5btVJPvqU4XDQ5qFei0997G00m2W++OVX8ZbXrHDXdSGLPctoNeKhw5M8+OQMXd/QbVUZmTsFxRLhEzvZOdsBX6FcgcBHco58XrrhOQgGUKqBl3feAc98AzRyXZEocI/Gg0EPggFSKFNRNxEFGlEkh0/I58xxDgdtTnQCvrjWZEIdX+nVsj9u/Q9oa5cZM5EmGXXaPGHO0qLHCk0E4ZyuMCJVDM7Q0cRDT902idRSwuNtZo4j83n+9Y+8jsf/4H1gLVINuM5OsNuOOt6IhszblfS8JTCePTqFYX1S7mqfL376LqiUnQrYtmP6FYuf+YFfv7unfaKYo1aXDjVcxbKLn44rO2WSkhQYqCOcglvsLNlV8nHHYk7GY7aWWwTXtcWEjLgFMk7gI4FmDQhStZyiFDYsmrLdjwSGlUjmAilW/vm6H5vFBsjoJlXrLPT1xXAURAwVKXNKVviZ6kEKIpSM297/95Ud5ERYa+Z57kKeG6YNh5csNfG4L1rifTtG2G1H+FfdR/iIPMi9PMX/4HESqeKu9pzSEDmOscDT0UkCIta0TYUiXpwAJg7pNSkzYcaY9CaYMhMUMgvcLC8P1iV5s99jWP41/ewWfI/LKRxc6rgOn59hWO8rKbZKtGBdNjonecbNKKNmhJ3ejEsiVGnYZoqGyEku5mQ4eF1ynD3x0mvdJaDRBq5INknfuF+Oy5rMk14GRZCXAoI4ZTpNIOfu7kvgfaGGDAh4oN5PO6GdUPmdvzyIr0EKx0r+tq+DtMtRNRWKUnCCFDFPJE+OjvZZtQ2qUk6LjietE3MZlSrXyE58Qg6Zc/TwscBHj/ucPjfCU6eL3GR3viznbDtefGyrY12hGB7wQg2c0ZOxrJkOnxic4Z29ChcWK5xtW2Z0hOM6725cHRBKSE98ZswEVYo8IXUadJnRUTri06brqv7JQBvDqpJIBiYPjyw5M1G1yL4/qXRkCWrDE2ySUP3QB3/27suFWCXv/6EP/uymEBkTK9gICUdjY2XLZAbRTY91PLhupj2v8RHKVvKSQT3BspbiybUkBc6xRkjIY945uupzXlbjSlPIrEwwr3XGZZLTUseIsKYdrmWWEzbP2/NzfIazzGqN7x6Z4AvNNZrGZ9UW2Zcv8a472tz78BSvvb6J51lurvo8cmiK59oh3397iygSwrDGLbcd48EHr2PXnQ9AMUDbEVKBSqWPecMdRPc+zG/+y4P8sw8+TemuM/zwv7uDj33WR5tLyMikg2D1+tjH/xqZ2wNRgFTHXSckX0jVs/7tDzzLT37iAMvSoaSxGSEeXzZHyInHFKOpv8MplqjbBuB8Ibrap6M9ihSYlBH8GFuvKB36cW1b6GqPrvYYj52zEyjhcWlxQld4VX+WnoT8wXH49//1Qcw05BA8XU/iLcrvffxXNlxzP/+Dv3m3b53TbjfmCbzm5nmk5FyCib79VHO+HePHP/Qrd693Yi2hRqxIi1vtVRwzSzFnx+ABLXoc1DmWpc15rWPVeb8kbt15PNbopGOQh4evPg3alKVID5+iFOlpn7KUaNoWPj5ZPHuyaCLxLImH4WG3kGFC9eXGZp/bjGx82dtVS5ceVXbypWaDedPiddEsvloaBJwYhDw9X8AnotUfsEPLtAnJY/g3K8foewEfiG7iOdOgIX0adNmt4/Qk5FF7NIWJrdg1Qg04FZ0HIGe8eKwVetYtGhu2TdavJyBM4atJNdzJHMtFVfesmlXy3PD33PIYvIRzkk0+stuxGm31sW96bMatfL4ENnltxNQoiksOy1LkTDRP1VRZjlZTZatkDvfE8ZxUfUI2Qq+Sbz5cxLt4PxM1N5v6myWQxWQNUZRi6hUSsp7kRzE3b9yM4qtPQYp4eMzKJO/ek+NPTzeoSYWdNaHfyKVFqbwUMAhjpkbTdihLCcGpe3m4a61re5SlSEPbTj5YvNQjCNYRIG26XBAn536r7iUUyze88+Tx+Ie7LR9bjHhXbeJFn8fteHliOwn5FkR2wHGqLZYfKl3Fwnyf03WDJzbGM68rNkVEFClQtw2KZooOA2Z0lEAi6raxXrXQdXzmMBciJ/lU3WI4sq3YpB1qku6KsCGhGZYRfDGJyFbHZbM2evJcwt9IIA2bTXbJYzIYblVZc4mWc/INNcBIzg3wYpikxjlWUjJeoBFNupQo0NC2S94osEqLgjji9YRW6YrPuFQpaI7HvfMMglmuY5Kjps6zzRp10yPCctI0GQSWY/eXGSXgs49V+cDrVxExHGr5nJMuf/HwCDdM5OgNoFBuMjUeEC1Mkr/lLNETLbxbLHPXfxU9Pw1WOGrWaCzv5O///A5+ae8sUnbQrOi+L2BPlMi9/yDhZ57Gs2cw190M+SIa1ZHaGHr2KDK+gx/9xG52aJ6GdJlkmnP4jDFB3Ta40dtPh348wNu4G5T4LLjjVot5H8vaQBB2yiQ7dYwTsoQRYYoROtJ3+GAKVKTIgq1TMgUe5AShRqx6bXwNmJJR6BV54Pbfvefdn7j0teNrgCceTe1SlRK+Bhw5MsuOxmPfZqCLb9/4+x/65bsdbGfdDM2RxwNOGgffq2uTEak63hA+zRgCeJXMcEoXKEgpxpCvS4V68UICSBe8Hfqxl5FLYn310/Eh1DCFWoUaIiJpMpIUVhK8e5BJll+JkRRTVmgyZSpMaJlF9WmIzw2MUfUMX7DnGdMSfQk5KQEr0qVDn2vsDE3xudc7wTV2mkCiFNaYj+E1iQ9Ecr6ScX7NNjHiMUYtfi0gH8Pf3ALX0NUeqpa+OoWsiAxfcIhzsFWnevi1rfh7LzY2LV59i851Om+Kgcwcttn+ZJEKG3k17uezkYMkjUmNLr00KUihinEX5Pn4kMlz2ffkJJdyOWDdTyd5zJPDudQrZNYSzpw0BLwNIhIWi68DfB0wakZ4v72O1771a3zow69jpxR5y5uf5Df+O6BuTi6l/FYTdzu9dBvgrs+C5NPxAVyxwhOPIgX8WC3vBt3FIc6SI0dBhCOywJyOM61VlqTFbz464F++JeCX7lvgH76Ec7odLz224VhXOIarMmNmhKt0kpuv7vLZZzzeeHODehhS0Bw3m6vT9zp5RHczL+oadWmz09bca1jKUqSwiapJEkmrEtZVtIBUWStpryYqM0kCtG5KtPHfZsS5y43hxCU5JtkqVZbPkg0jG9vBw5GtCG1IVDLHIAsREBFKFPDwOK91ylKkRIGWdmlom1BDetpPK30jsaKPQegy4DgLsYRvgzXpsUvHaZkBHQKsKI8bJ2E7Y6tUNM+K9FiWHielxWFt8qkHJjh6YoJGrOgToeya9inlIejXuOktX+W5Q7dw5r+9A22ViZ52WNoz/+FmfvtfvY+f3jPDZ/5mlt95o8fOnWvY44fQKMCezZH7nv3oqaPkvvdmZNdVaL/jIFlq0cYyVEbQCyd5k87xU9eX+bnyTSxJi2XbYAHX7TilCyzYOgZhWpwaWD4mAQexvCfAQMOYWFxgjU76XF8H5NQd7z0yQ0f7lCmSlzw99ekzoCpleuqTw6OHz9s/Mv2Cr6X/9PG775kW56QbaERFiuy7ag36g3Xuy3Z8U8PDOIiF7aZjVYBzMy+SRxDGpIZiKcZQCliH1NWknBoXevEiua09xwkhTNWuYH1sGxBQlTIBETYW1MhJLl2UDRNlIZGJXodfXil+24sND0fMbYrPYXOBZ7xlFk2HZ2jwZNh0CYEE+ISsSJdcPK2fM2vstiN06HPMLNGiR5MuXQJasg6LEww1qVCQQgrPDQjp2x7Ltu6gqhox0EHsBWHSpMWLYU0b+BhDyUPCidnwXKYLvRVENvnsyxHJPuSG5siXO4YQV6dUAAAgAElEQVSFXpLHJDkTJDXZHI4sZBrc3DdmRtJzUpQiJSmwatdI4NfTjJCFTa/zdHRDEW4raFu2cJeTvLuHMtwSE3dUTOwRk8CUk+fBdcKTIkFWbsHDECaICvFo2hZfNGf4q0/dxa2VCj/x41/i7r/cT1sdimNUKgw0YESqrNoWHh4rdi31+lGUatwlTaCYfevGkBw5jDjjT6vKMVlkQmoAzDCKRVmTTgwRjWhInz/8yhQfqMxe1vndjpc/tpOQKxBZ2NEw3vMuewPvKc6yXC/x3Qctn3t0hEOmzkNynKfsyQ1JwEBd9bljO7S0R8O4hdWsmYwnfT9NHpJFdrLQzrqf5uMGWNJuzWqEJwNLIimZQpouMUn/0Ad/9u6t4FWXE8P46WzXZnhAHXZtH37PcMdnM0xywqkpiiP7t7VL23bSan8QSyD76qd66z31aWmPnvqs2hbz0VJs5CRYLMu0OCN1TsgyfQlZoMGcrdEnpCMBHRmwaDqcNWsEWN5WmWDRDtg92+O4WWNSixTE8Nz5Iqs9+K+fvQ7vmgaj4x0mZhbJ3dRlsDBH7/6D/OOHm3zWO4ZauHl3xGOH5vjfv+E4E3rmGN5tNWR0ivBBD/vgQ44rkss5TkgwgNZqTGCv8FM/9AAjI33+S+84K7QwCIsx/yLSiLzkCIjo4BKFBKfrrjvLgICC5NLre44JnuQ0BnHQNVnjIDvpM2CHjNOkG8uWrpMcr5IdXMMsu3WKKpdnJPUbH/un9wxwi6Ou+hw9MQHVMtprQr9zWdvajsuPZBGVVZwLidgh4wAMCGlplzkmqNPmgMyl72vSTcejCEtTu26bOGEKxx1x5zYhRysae8IMqEgplW1OOh1JgSUiSh3IkwVbUmAJNbhoTH6lRTIHnNVlduoYs9Ytrg6ZeZ7z6pQ1z1lxPJoBAT4Bk9RYpU1XAqqUqGuLHj7z0RKP2WPpHAEQ6ICu9ggI2eftYsyMxBXvmFETL56TYtiabdKxHaxG6Rg8LHyS/suS8zc5xpsmLENJzcsVijpD3m9iwjkMocr+rbKUqBhnwFcxFXKSo2qqjJoRKqZCUYru0RTZ4U0zYybpaZ85z0mht7SdmlyCS6YPRSc2oBiS+S1xsE/3a6gLkqiugSsmJT8HhOl9476Pjfkzlo7tUJB8DOs2FGMeiIcz9y1l7sFJM0o+5nJWjEscSqbkhFyaITkDn/rjd3KfdwwgNaCdNKNERMyaSYf6iLldCe8LEiilpWaq5CVPQMQOGaNtu6nLenLsDcK72cMbov18UK/lt94M41qlIV3+xpvn9hu3fUK+1bENx3oZ41KL8CyMCdxg8IM7Rzm6rNSqIVEk3H6V5d7TIW3buYgYBqSYzlXboGt2IAhzOs4FljPa3OukMl/9OLkgrvppzLnQdV1vXV80JCoooYZpJWUr9Gx2wE0mjB/+4M/dfbmmhMPbzEai7rVViz5bTdsa1xqlyl+wTrxLErOIiIENNhBYA0K6tkcwJD3oYGpKweTJ4zmsavx3V7XNmNSwKPt0isNyjpOmTh6Pr3nPsUenOGTOMccEN0bTPO7NM6IFvtJdw0P4T08pHyrv4S/98+yUOZ5q93nteIk7r+/R+uqtzF7/JJ35fYRP1CgeOEXv2LX84uxevjFv+dX5o/zmnms5uL9J5fEierqOuekADPro2iK5txaxR4Fi2S3IC2UolLBHnsXcNIZ99gS//Kfv4oJpc45lKjGkpmYqdLVHOTYB9HVAstAPNCSI/Q48DGNSY8Wusd/sdJA19ZiVCa6x0zwjCxTJ0RKfQQyBaWmXEangMyBHjn06zap0aYnlYDTDPe/uc+gyrx+nxOLRtj5B5M7z15s/+ZKhgttx6ejrYMMCxtcBRoVl0yAUGytTGSZthfOmjqL0GMTkaA+fkHw8NoFTXlOUkAhRp5DWZxCPSa6T0bQdAg3ISY6aVGhqxy2wdd1IzSCEGf+P5P7frIjxSgzHpeozJlX6hHSlRRefpnaYY5yjZpEIS1sGFMizTINFbbBfdnDELNLWHpMyQok8BS/PrI7xWHQs3XaaSIjhbLRAThzkpkDejZ8KRnJOylgjNHPctiJQb/XctzrZ07gqn8Chvll/I5t8JHxDQdJrNeEwJHNOT/upQ7iqUtcGO70ZImtZjFbijnN4EYl/wOAid/tL7l+iuhbPreuFzvXEJCGXp547GqRFSfezhx9z+gY4UnmgQXrdNLWbvi/UkEnjuucehk+Zo/ze7qv53SNNmnSYNZMctDu4j2fYZaZpapdAnAR3V3vsMFNO9ESICw4uqQmJuNrsos8gLaxaLA1tMyEjrGqbd9rruXWf5b+dXeZJQp746iSjWJ7lLLtkin/xUMCPv/hTvR0vQ2x3Ql6GuFQXYHihnnIcEPqBUDTC545CLqfkPMur7XRcce9vgCZlB82AkGc5y3Gd5wQLjMkINVNNpfESaEJyc0554+lnra6bDyVE9aS96yaldXxo8ncvNZlkOxg/9MGfvfuHP/hzL7orkmzLxFCqzbCsWbndrQbfbIUoGaSTTk8iOZgT5yybuM0nA7Gi9NQniLtPSaU+gXH01FUVQw2J1GFWs+S4I7LAXdF1DAiZpMabo2tYlQ632ato0WNV+giGtgR89+g4o7Hixxf6y7zaTnEi6vKot8iTawPC0KP2+sfRfgFVQUa7hGfnKM2e4w3/9FPM5D3+8PYdzEx1yRcC3qVXuXOxskj08BL22DGCz8GjH34/MjoFYYjWL6BnjkEzQudPo/Ucr6+MMC8NfB2walsUJE9P+7GLbcSKXWNSRpiViXRCTc5FqK7xnpMcc3aE10e7eK3dgaI8Z5ZdN0RHqWielnbp4ZMnxzjV9HwfZ5EKRb5HD3BzbpTabU/xBu9fv+jraPQ/f88990/9znYCcoUiGauy8JCAkJZtk4s7GjUpE4mlQJ5VOhTJUyKfJvvJZwCKkmeXTKXPJbjwRDHKwRbXYVdN7aSch4RLMqz0Y9PR7qWTxq9UZL/bKV2ki0+JAjWpsCYdSuQZo8yAgBWazDFBRMRJXaRKiZx4rGiTFVoMCDjJIjVTzXBu1sfZMO78FsinHeCc5CiZEge83RSliMSQt2xsdhyHPZguFVdKeTFZyJsh+NM3K5IiVcd20iJfApvyxKOvfVStM/LTwHk1mQoL0bIrkcVGkMncNezFklzLlwM1c4mRl27LopRiVUhw95tLetzvhZhPmsyPiYcHOChU8nuiXBVq6BTVTIWKlKlQ5DuiA7w7uoY+AzxjeUiOc5sc4M/fVuUdpWkKkmdBV5mKfUAmzShzZpq2dilJHhPDAJMkr0SBc7rMebvMaV1MO3YDDejrgBEp83nzLL995gITWqZInoJ67NAKb9bruaCrnNt2TP+Wh3zpS1/6tW/1Tnw7x4uFICUY0Q9wJx+aG+PhhYi37FcWVvN8qrHM5/WxDRW95DPZKosnHiPGEQd3mxnGqNBnwAl7AYOkWvlFKdCNyYNZd/VE2aIg+XSySUidCTEe1iuG2YlmeF+SGH7PpboiWx2/FzIxbEWAhM1lH5OKUbYTkvyeVQNLiPnDviuRrndUslC1xGG+IA73vstMU6JAhz4HdBqjQk8C1qTLuFYYEOEh7Laj7jxJRE3zHPaWuCOa42lTZ78do0tIEY9F06WieW42Y0yXDNWCE3uaqFlGawE3vP5+ZKzL4b96Bw+dKBCp8mMfeITCHadofeVWanc+Qf1rr2PqPd/g0H9+L9ff+jD5d7axz/bQ0PAzv/cGvjM/yYejZ1nRJlUps2RX0++Vx6MiZQKclGoYL/162idHjq72MAi7vFkGGmBEmGYMxTKtNZriM6pF2jKgonkmbZmuhJw2dRp06GifmpSdBwmGX9t9Nbfe/gT3//Vt/NlSnab41KXD//Wxf7KdULyC4+9+/y/dDVA1FSIsVSnRjCEco1KlrwNKUmBWx2hLn2VtMiZVIlxnA2BVnXlYAZcAT5gRuuqn6jk97W8Yd5JqrCcefdtPK7NeXIE14qVjXlJAeKlKWFcqsuN8UYrpom9MahiEZXV8rWkZA6CtPQqSox9z+kalygglugwokeeCrjIlo7EMb8slN1FzU3ncbCS+UYKJOSHeuvBHBt6WkL6NZF5/no7DC5lDvpkh8WJ6eD+3Iotv9vlhZEOyHWfs6LgZWfnb4WO9VRHtUt2NZDtpIol3kSzy8+57BjWQhWAn82Ae1/lKJHeTc52XQsobyeNRlpJ7H/m0Y55IZTsOTo6yFGlrj10yxa/vuZp21+OZFcueqsc77jrM3/1MzQm26AJVKZEjx5q2GGjANWY3C7oKOP7XjIzjYbigdQZxVyk5nhUp09UeFSkzLtVUSKVEgQoFZnWUU7LiDG8J6eAMdX/jY/90e175FsZ2J+QlxEvhQCR42RPeKifrMOZ5TIy7RGHZdDdgbbOPyWfBDfwt26YoBS7YFc7oMkvaZNRU8cRjwoxtqKAkuGhYX1hXTDmdpBNnVIcDdojKYVWs5HG4q5MdwJP3vVhY1jC2GDaaFia/Zx+TSHwBNjMqTCKphCZJRlJ5LUievBQ2JCDZ92chXQmMK9QgPUYDDfB1wF6dZI0Obe0xYgucNnWWpEVVS9Slw7RWGNESB0yVFdPjObPM4948gjDjFXiH2UkRjzyGCjmMClNa5Ou6yGIv4onVAZ9trvKn55vc+O7Pu530Ip49W+SaKTge9sjtO8/iZ95I7ban0Faeydsf5uH/+H76vkevuYPwvgK9Z69j5dE7uJExfj96nBVt0tU+KzHxMRslKaAo0zJKUfJEGjEiThihIHksih8vMj0Mu+0oPiF/Z3yO98pV/Js3wp5olDEtMZCIedNkUdfwNWAsJhD6hNS0xPm6R+l9y7R6whnTIMKySpt//oP/btv1/BUa/+BDv3q3iKT4710yRaARN5q9TMkoJfKMx4lIIG6pU5AcETH3Kr4Hk3s2hafEY1NixJZ0PjzxiLB4YlLeVqKApapxZy4iiE3bkoryy6G4dKUjMaJNE68YwjYqFTT2KgqIKEqBRuw2XYqTsTpt+gxY1iaJMaRBGJEqAw0oSGEDPwAuHlMjjdI5oiyldOGb/UzCt8smIHnW+WGbRXYB/3JxQC63+5JcE554652RTWG9ctHvwwlUmlggKXk/gTNt2MdNPrNh25dICrORCCtsFJy+dCSdmBR+Fd8jw/zQJMlIuaYZGf9E3lmQuHPi3lOIJUsSIr2iXCM7GdUynzjV59/Wj/GI1lnsue83po4rUpYiOXKp35QgLNMgF3dZDIZlbXDaLlBgXekzKRz6DNJ1wpp2KOHkfq+1O2jjsyDOQ2W3HaVPQIEcVUqXddy24+WP7STkCsTzVfXP6QrLQcjecfjKUyNMj4XM2tqmFZLhQTohBq7ZJgMCWtpxqiY6cJN9DC8aEBBloEiwbkjYsZ1U6cJ5cxg3cevW1autns9OJC9Fsnc4oUmrPpn28UVOupm2cvb/pdVXNuJgAx2kMI7keA2bQGZ/1nhBBM77xRPDgjRJDKCe8M7jxwN0S1zHoKJ55k2TR1jhrKwyrhW6uC7Bn/EsjSiigscOSkRYVkyXo2aVMS3xmK5yQlpcxwgr0kXbJTAKKhzvDjixAp81h1j8m+/k8w/M0nr0FrRZQa2w2srx2h/5SypzZ/j+j+zjBz6T5+fvU971qoC6baJoKm4QqSXUkFFxKlyrcWVqUqvpwN3TflopMwh123By0eRoSJ8aJb5a73A+HDB1zdNEojxjFnjGLNCkS199RqVKT30qFNmpY7zP28P3/don+atffCfLPaUYwwlrlF/xlev/VeNHvv8X7k6cy5OOxYKuMioV2vTx4m5a4i/RoMuSNlJ1LIPQsk44ILlnPXEY+FGpYDD0Yh+KJJLEP0k2km2HhGmBwfHjLg8v/0qIrcZYjcfsGmXq2kzHoJY6L5WGtlMTx8RoME+OtvYYENDWLgu6Sg/fmb5JnrzkqZoqJVlfjBmEiqmk41pCSE+gqdmFfgIVStUIh6rql/M9t+qMvJDjZWLicqKl+EIiW+hKPbLiOWazZEYy/4b3fzhx6cfXdpYjBRs5OBdtf6i7MexvtVkMYojiVka+W0VybpN7JZkvE8XD5F5r224K6U4kml2BUsjHRQSLxcRIABvDmosxNCy5xwNCfqx6FQ94p+ngs2jafEyP0VyZ4YQs42GoUKQbv7+vgw1qnxr/DcVBqBvaYkRqGAwVKeHrgEkZpSolJuLi2Jp2GGjIk+YMbe3Rok+FAofMfNp9bdF73uO0Hd/82E5CrkBsNYAoSlu77MjnuPnGBd595zLtrscNjLLP27WpI+3w78kCXNUtIDvazQwpmj4PbJgUsgvripQQkbRymLQ4HQb00hWq4fe80A7IpXg0sFHlakvPjyEZXlivqGa3Ybf4HtnOSJKcJb/nY77IcNUvSVLWK3mxD4tazmudvg7YLVPp31zTNh18Vmhx0ltjtx3lrKzSoU9XAsZxi/0Qy5NSZ5E+I55HgPJ+s5cgrgBPaZkzpkHJGEa1yIf/6B0ce/A7+J3f+27ee4Nl7wT8/q7b+d8eWOWvgzp//cDVyHiHC/e/gbd97718448+QPPs1QSEnNcVFqTJTzzzHKNSTSF7vl2X7+zHVdfvs7fyD/gO6tKhQz+FhyTVvqRdH2HZoaNEKDO2RoRyQbr8l//3rUxokTweB+0O9ukU+80cB+0s0+K6JrfaGb7v7Sf42790J3+ytsCf6hEu4JKvPkG60NyOV1aUxCXLecnj6yAdC5a1QUjEvNbpMqAbG1224+tMkBR+NWKqMXFdKMdu3QZDXVvpIkdw5HQHC8kR6IC8FGLDQt9Ji8fVXcGQ8ry2UGB6pcYGSKu475F4p0RYGtqhQH5D4rZs11C1qaFgW3u0tUsrVhkDqMSY/472adj2OnRISoSEFE2RnORTmO6wMmKkEb716dseoQZOgSmujifzUNIZTv4Px/BCPvv8iz0nWRhUqOGG7bzQzsj6PLpe5MruZ3abWSnjoimmcLlkX2C9wzKcACdk8M38rYZNdjfMd0PHK7uvm36fLebKbPKRmBenryUJO+v8yGTNUJB8ui7w8MhLDokTqoEGjEqFMkWcQWmI//+z9+ZRsmR3nd/ndyMi98za6+1b7+pFrW611pbUAu1iVTeLhRmBxfiYM/YZY2YAA2P3PGZgDBzGwMwwRjYMuyRQSxyzCySLlkB7r+q9X7/91au9KvfIiLjXf9y4UVH5st6r17t66lenTlVlZUZERkbce3+/33cxETUpMyY13ptcR4zm9zun6TOw1x9dDIbp/U/xE/XrCYkJ067KAEtoFySTfl/XbQppp7NAQFUqlCQgTo+vLlUGWOWzSVNln0xREJ+YhI6xhZAlvUaXQSYoM2Mam7hoO/HSxM6s/hzi45/6taPPlUznoXhmEPKn/zjL409P0xsIR+q+vQnlQknZUWFyC2hjDH6q4tQ1vbRKZQekggSbJmhHXG/qdlaxyRM4LzbIPdf3fSkom4NeuW5PnuORH7RHGS+6wT8fw9vId07c387AzIVTJHG/u5A0Dcnr7CepEMCsN0mAT1XKmU/GJDUmpU7TdBCEFj2+oc7SwpoXKoRFmsxLkyNmmoiEp7wVVpKItkQ8FDUBOKuadIgo4jOXhNRMgcU44v98tMM8fa6+8UFueNV5bnvnvfzcvit5RM0xOx7z1N+8i5OnJukv7mF6qsf//NkC1yQz6SKwzYpez0iIw+dzTGo0VJUF6fKuaxIW0gr2gIiSFLKuSd535inOMW0qNEyBn33/OWom4M63nudfvHueAJ/vnZriFjPNmClzTq0xbSwc6797yzonntlPS/qsSY8+EZFJmFN2ofoLf/IvdrC7L8NwhFRXDXWeRsZoFsxaBhly0D2n3++ia0L6ZsAgJUS7argzZitKQN+EGCyPzRhDW29I+g7SDqQ1TIsyqOTw/fzNGg4m63DvGpMVDDJZ5Fz1PEwTMk8sf0shRCama/oZ4RzsWO+haOcSFUdAduNePszQ16X+v52kYtRz3ee/qdNwkc/xYryTPERqO8fiYtiDyiVJRVXMhEqM0Wnim2SO4gClFJK4FVE8wB8JE3bhilwXO77LieFt5WX7nSCN63a4Oc29JkuuUBuwZBRReq9OqjEEYcU00/WFYlKN0ZAqMTFruskXvdMMiDkjyxSw979LMk4/dSPvvvMz1ChSxOdV7CXApywlAnwCfK41exhTNfpmQFVKFMQnNLY4VpMyV5ldVqo7VfBalBbrdBmYmBs5QJkNWJhOr62alFmR9qZrfydemthJQrYRP3jnT9z94bt+5oJF8wfv/PHssWe7II9MxNWlEnsqird+x2eoFA2egtuTg5nSxHYi77UAG8mEL/6mtuYffvJXjv7BJ3/5qBtUwPJCrOrJxuWQua5vkWy4AXb4/9uBYW2rA5J2MvJwrIvCqvIVrBF8kVHbyAiJKSY4T7KEzZ0jh4F2sr4ZyTUd2nwJqKoKVUpUpUTbdFOCqOasWaaAn3EfShSYZZzEaPYxlQ6YERWKrEiXVekxZkos0Gdd+hZqIbZydMpbZ3/S4MveGR715jktHUJJEOD+r9xGNAiI1iYJB4rdZpwfP/04p8+XufX1D1IcW+TQDfczq6sU8HizuTp7j3m+kIgikAKBWEztL828husY52+ftBPzpFhCfdv0MMZ6NbhrJxALvFmWHh+6usTv//WV/PM3Dvj4vbv4fz69n+uSae5bioiN4X3ePg7pSV6tp3ldcpDmepXJiTZH9BTjxpLUA/Fo0aPCRldwJ15+UcipJdVS/xgAn82L5ACP5bT74fxDNDp7ni8efmqApzIWgzV4a5suoQkJ0qqsuw9NWn217ugqu68dDyQfL+cOyFZh1fo2yPsF8RlTNcpSyt6fWyw68QwvLYzEJmZgrGWkQmjqVsYv6ZoeYSqrPDDRpm8Hy3EY+zxE1sXwudxqrsh3P7abpIxKTEbFpTooecSAn1bvt4r88WXnVVk1MMcZCXW4SaglL3frwnVst4KjDcxgo3szRFh3kZdNzh/f5YSDNbrktCCFbJxOTH4O01nn38rQ57ghOdij+91dhz1C2qbLtBrPPHxCM6BhmYwIisNqD6f1An0GlCiwYprWqJQ+5431PPkf/8vN1vCWAWdlLet4eKK4hj00TJECAVexx3YE0/nacUdOiZXBr0mZhISQmINmkqqUOCdrljBPgSk1zrJeo0SBPWaMMSrcxMHLOqc78fzHjk/INiI2EUUp8MN3/fTdkRmkN7BJJ0Ibz3Zy88Xnt6LH6UcR9T97B4f2tSiXQ05+aZJns0mN4Tc+cfdRgB+666eyxf4ffPKXNyUHzgskkIC+7m+4pKZqHgoB2ZCzddUfN3gmbPbvgGefgIyaSAwmOyar858jMeYSDbfgGOUlMqyeNUpNy08rf65Nno88hMtNKAGCGarQOWWs2MScNYvMqkkGJmJMFVEIBQlYMOuU0sRtnS4h1m/jtFnKPo85s8KAiAMyi28Up7x164LsdRGEedassodYM7Gi8VmQDhUCEgzFIOH06QnuvX+ag5OaK/Q48946J9c17b+9hYIHcx3NZ9XXMtKoI/Ra6UPXDdI0xMJjNIaPza9yR2kSbSAMQyakRl2qLOhlImKqUqFMqtYmVdqmy7p0+cOnQn7qHWvMnZnlO1/b4nMPjvFjP/Ql7v3TdzLfEgaJ4VoaHKr5hG3N+cUix5YrTJkep9Ra1nHxxefon/zYThfkZRQfvutn7naLFpdw1KRM03SITGILKOlt4nT9FZK5npekSI8BNakQpzw2d006HPmE1OkRUqXEKpZw7fhtriAQmxgPL8PG58NVyJ8L1OelCjf2bciyGhDr++R8ehzMVowiJGRM6vZvhBYb5pxujLKLUcEYg5GNDjlYwnlXdyHlRAwYbH790HjpijH22C6uNvZCnvvL2babt0dBrNzfwwv9UNtzPIwM2Oq1XuqfAVsTy0cVx1zkO1qXglyN2u5woU4PmfYaNEUpEqWfb77Db7chGRzP/W0X/SpLYIpSIEETmtBelwomVQODZkLV6dCnY/rMyjgJCWUpUaJAnwENqbBGJ4NP/qf74Xunp/jSUo0vemc4a5YZkxolscnOorSYlyYrpokSoaN71KSS8sx8jpgZzojlOTmEQhGfM7LKgIg6JcaULf4V8Xkr1/EkCyywxjhV9pnGts/vTrwwsZOEbDNiYupSYzmFBcDzY3bVM32OJ+cwaJbbN3FNaUCiFed0SM2r0Irb29pONtnmBqHfvecXt1y4OTlZ27K3JEUnvadMsmlxDxuqMh/95L8/ChtdoOdiTJg/9pG/5zCyjjA4nGxspSSS74ZshY91eGuFoIdwuXkCoZZRHSKFSas/joTpYCHufHp4lAhoYlu+ZYoMiBkQ0TH9bHKwA/oAQRhPF15Pqi4NKiRYh/LXJgd4xDvPpKkyp6ybeYENxaCrVBXPiwkjhSewe7bD8lzIsl7j72We9wx2UygLt18d8vtPVpmgypP6DDG2CuqECNz7i0n45+r13GOOcU41OTA5zi+cP8HRyu38Yu8+HJa8LjUaUmHerFBJpRBDBlQosioh3VaV9VbADT/050w/dRcf+9130ksMJ5IuYwTUlcfV+/usPF3k/vmE0GhKeFyfzDCXGtr9H3/yL3cSkJdRfOjOn7w7U9FLr+ECASEDotTgskAAZgPeA7ZC3DF9Culr11NzzyTtePYZMNBRKulboWP61KVMREJZivRMn4qUaZmNBbaTFR8eB/KQnm+2BCTrAucKLWCTDkFRpkiUws0cvl0htOlmSnWltDPSN2FWuQbbMema3lDRQVKJ9tGdd8cdSMQuYvMLV6uKFWyCI71cI+vmpDDffAx3eEZxMLaS081v13UVLuZflf9s84/BZvGTrWK4mOZeP3yM+esnX1CLTZwVFoP0fTo1LJeYus84z7HyUDARob0AACAASURBVNTEym+XUlU1ZOP9zzBGjwEdQqaknkH6ShKwYpqMp4aZV+ppvizPMK7qHGOF+5ZKPKOa/FDhKv62P87DcpoJaiyZJoloGlSIJGHZNKlJBSVCgyod06dEwFVmlpOynKrE2Z7fGJW0l+fRwCY+idE8oE4zSMUsBOEr6jgfvOQZ34kXMnaSkEuEW2z7+LSMTQjyE9vFBpvthDGaOIXzHNkV0WmX6XYDri8VuCfspNvffrKz3aTAJSChCfHx6et+yh8ZZBM7Q1rl+W27ZORy4tlIGm/V5RhORPJVz+HXbuWonq8UOUhW/kxnk5UbmMVDUKke+0a7fGAGFChg0NSlwcDElKWIQdOki49vsadSY920qaWV4Ulp0DTdbMHlODuOjIvADJY891XvlNX8l4hJU6FPxICEGVNlXrX5ul7h2tVpAF57bYuHnq7zBq/Kw6ZCSQd8LDlOpVVg5ck2K6bJGi37fo3ebIiZu5b/zJyiYoq8yeym01f89K7D/Nb5BWZlnAWzZvHQJCwRUZUyXpowTkiN3do6xy8t1dm7u82xj7+HwINmrOkbzZV+hcgYbtwFf/eEjyea73njGv/3F6v4CJ9VT1pTqx3E6MsqfvDOn7hbIVmltKQKhGZAlz6eKIpSpK27NhGRgADrJ9BjQxyjQJm26VGTcirOmxCnSTxYaKAjrHdNL+042/t7YKIs6XedkFFEXvjmhF7B5kWwXeRL1vEBUn6NzrobrgtreYF+1tkevneGK+LDqn9+SvbPk5fd69zzrSzqBm/ALnD1JmleuFC98OUUDuY7iofiYjsQ4Oy5WyQsW60NhuczT7xsrh21n4spPG4FS84fh00W3GcnWaJqXc43OiLuntqA5Yr1hTIxQXotufs41CFjqsaqbrFbxhkQ02OQCYg4KW2wRbJK6h+iEB5SZ8FYbsyMrtEh5qysUSvOclWvwdMUKRJkr8+Tx1umw9Wyn3W6BOKxRIeQiC6hJaxjuy5dQiZNjdOyxCQ1AnyStDjWwxaSB1gC/U68tLEzw18i3OK2m+LfCxJkg1ceE/qst4/JzKDOLQesrpdYWg84Mp3wLnPjJgWmS8XldCX+8yf+9dGPfOLnjrqJqJji/x253YWbuJ6Pjsd2ZXvzyliw9eBvhgbtkbCubUwirnLl/AeM0ZskG/OTsjOEcsT5bBJJ/7+mmxkZtG166cBYZFw16GDN/TSGMbGL9KqU6BFSkgId02PNtJmUOmUpUiIgMLYCVSJgQEzJBHSJ6EuMh/Ajh+v8zME9/Ng1Dc43hXZfaDZLlAP4iH6AxGi+yBOUTcApWeS0Xsi6PH3dt8T/9EtjMifdt+treX2ym1v1LAfLAcXAcMONJ3kdUxTxMz8QEUWBgAERV5ldANQpc6VpcIVpcM0NT/HE8QYzuxdYbMN00aNnNNVA+Cff9ggA335zj5ISHn58mteNlemR0JAKh2RXJhG8Ey99/NBdP3V3fiGrsQTx0ISsmxaRSWibLoEEtHWXmJi26dE0XUIToY2hJAWK+Gg0fRPR1l3aukNf9wlNmDktOw5DRJJV+i0h1ia+RSlm1yxcuOh9LqIZL6fIOqzpIjEiziTUrQrRIONzOVltsFj+OPV3cLyZhIROqqjlODV5ToBbkDpYC2yGpA57B+UjLwbi5sztktNf7MjzW4bVuixvQrLC1FaRyQIP8Q/dGAqj55t8QczxNDItyxHwq+EC2lb+WPnH3befduNdJ8vCIEv2c085IXZ9YX3BnEyzL34m22/QGac0zy3VGPomoiwlmqabdexjYkIGnNNLROk8WKZIlRIGw6JepZqqaL1K72bcFHnIW2TMVPhEc4EmETfpvbTpcyW7M2PBAI8paTCrJjPBFy/t5rwpOYDBME2dAbGFexGisHPRHclBAjx2ywQrpsWsjHMt+5kxDW7kwEWvlZ144WOnE7KNcO3xRBL0Rfwznu2Am5gEj4Qz7YRS4LHUgWOrwkPeOQ6zh+PJ2QzPCs/Ng2M4NuAMnh2EdD+DT/zhJ3/lRYPB5M/fcHIxKkZhct12LvnaiyiTjGqlu+TE8UaM2dCR9yXIJnh7HPZ1zuW5Z0IiiYlNQk3KGNmY0NdNl2mxhpIRMbMyTpseK6aFwZCgicQakY1TZZkmdSnTps81etYmsMowv1rgXMsAhpOtAWc6PpqEHqFduJHwmDlFYjSvl6t5nHPEomwVOuWEGGMykMXr1KuY0EW+41p45FSRgzMxnZ7HQw8d5kvM8/pkN3+kzhGkCflBmaFJl8Robk72csxbsQmW8igdPMl7fvYJpFbCu/cKmpFmQvmshAnnTh6hWo458s7Pcv+T30WzD6sDzePeMgbDbl1nTHbMpF5uocR6Cql0IRaaMCNGWxlYK9cLVuIZLEG9R0iDCi16VlnN6Kxi6pL7GE0X67Scr4AqlOWYpLd9fuGWj+Ex5JsxRiVQef6FFoXHxliTV+pz3SRPVFrpjjI4jtuGLULorIthE46EQAopz8RJk29OOPO/O4duL12sIpv5eZitFauG3+OL9Vldao4WURmEaJSgwVaJrStCOTXFrEvH6ITCz0EUnYT0BTDhLTr/o455K+6I63y43+11slFkdFArX2xRKes8pLLrA6MzgRsgU1ybkDotbOIxJba45uFlcrpgYX/Leo19aoYCtpN5mz7MF+UYK2mX80m1wE16L2t0qEuZ7/T2cdvVfX7k8ZNcYWZpy4CQjU6Fh7Ly8BQsEZ0i1yWzLEmfPUxyXTLFvLeeve8V6XBzspezdLlB7+EBdSY73q4MmKS2Caq4Ey9N7HRCthH5is5W0nmXO5BuqsGI4gpvH02dcKZpeM9bTnLDLHyHvpob9J5NA8HzHR/5xM8dBejrfqaKUZTiC5aA5BOo7UxEruNwQeUnV70atb3hyG9j1CCf7c+RXockGoexunkZRacsphAqUs6UfizJtkfX9DNoUVt3MxdoQVg3HUIzyBZkV5jZrBNSliJOwlch1CkTGI89Zow16RGhuedYzMMrEeNFoZ9o3ron4And4q/lJFfJXnbJxCa41VNyngERXWO9PlwCYqt61vH6/f5ePIREC0l6es6uwUJT6BFzUtrcyEEiE3FEdrNIkyolpk2V/+lW+MVXzfL+aw1vOZKqqTxVIXk85q4P/Q1e6iA8FnhMTK/y5LkCUosZJDBdM8wnA0JiJk2Nrgzwd4aol02468Sgs2vFcaosLEpnlfl+Spr28ShRoJTK+PZNRGzsItdxE+IhSIRTdbKeDzbd6JuQnukj4si/4QXFhG/mxONi4RSL8gvjDQ8Plf10yQbYBWaU46y585bnc7iOr/NDyrthD+8n/xpIF7WistcPu9GLKIa7DNn/XoIEZDv7uhgRPN+d3+RRlfZNwKpeZZ5WjIb/iqgsSRnVpb8UvHvU/7whn6wgV1t287kvPuW0UOCLT1EKWQdMm8T6bJgo5Vx4+PgZnNLBhF13qGW6KBRVKRESM8s4A6Ls20tlosdVgyZdBmlX80m1wO3mKgap61Xb9HhQnWWSGtOmyj3JKcKBxwxjAFSME3Lp0KbHOb1EmSI+ygpWmBJzqsUBU6VJl0e9Jaap80F9I+9PruOIniKUhG94czyu5rlB76FGCUFxR3IF3+cd5rZkF+f+yV9dNkx8J56/2JnhLxGz3nT2e166zsWzGUTdgOaJR13VrBoFCVdWCrz3dUvEUYGD+5t8Reb5ohxLDeFeuI/qt+/5haO/98lfOpr/+wXbGTYR+finfu3oxSBewyZTmwbpEV9wkUQmnVyGJxmVJoD5SdfBsvKfs5OrrKiKncZTbogvfjbgKoQgVQ3pmT4aTVmKBE5uEMW66VCWEj1jK0d+CrUbEKFESNA8LfOAlfFtUEEQ9ppxisZWhgMULQnpSsSS6nDIL5FgON9NmCp6PDxvuN87wyotnjbnOKXnbeUawzu4ER8/3b+d7AoEmbrRPzVv5sPmNr4SrTPtBTxyqsDhac1qy+etr24Safi3N0zwbY1JTsoyu9UUK7Qz2Me69Ol2C5TLA+47XqBSifiz330fqt6HyOf8g7cxXhT2Vjw6seaxR/exHCYsfOZ2Dk9r/m6xS4Tm6mSKSBJiDOEIv4KdePHjQ3f+5N12IauoSy1zKy4QUBKrQFOSAgWx11Ndqjivj5qU6RmrqlaTMmUpsm7a2YLM3cP5Mc4mIhuSsS4piU2cVavzVeBXSgJysS7I8GNOZtXFBkdjg2ycN2DNwy4NOlu8Wtjbxnl1ndk8RMgVZ1zkx9K8suBW85RzNH8pYXIX2/925ld3rvKdi3wS5uYOu6/NsCv3fbEkBUaT1i8VeWl12Hy9VKRsu91pN2LjOtCUpGTl1UVY0esYdOY/417rCqBO7tcTj67pp2O+ZlmvcdLMWwWtVBlsYGJrZotPz4SsGOtz1aHP19VJfKxUckTMNHVa9AiJadHj/3q6xa3Jbvt8GfCm5ADXm314WLnf/WaCPgOqlOhIn1ASzopVjxwzJVr0+Bt1giKKU2qVR9Qcr0n2ERKzoDqMmzIBHgvS47PxIkVR/HV3iZ//3v+0k4i8RLGThFwifmX2Vg74e4DnhwPitgO2gr5XTXOzXMGP117FW197nt/5xwaf+fIeHnxinH86uY8fNDdvqmy8kPFb9/z80WEp3xcrRpH7LvX7qBgpnZibBPLbyNSvhj5XV/UpSEA5dRn2xHIzPLEeGk4dKE/udAN8UYpEJmZN27bzwERWj58kg6c4jXNHto1MgoeibwaUCHKtZ58laXNOViwkSywRLyTi1mQ3a3HCO48IMyWPhTCh6itu0HtY1a0MHw5wjdrPQVOlQoHr1CGr3iUeUXq8BQkoiaKkFGIgMoZuookToVrUfPr+Oq8+3OeXvtHlc+stjphpFMIuM8b1Zh/79TjX6Al++/EB5+bqJAb+6htFTnZidKvEl//0Pez9lnv57p+5h+m65o1XRjy5KEwWPRbmJ2j1FPtViTo+86rNSbPAojRpSf9Sl85OvEjhcOWDVINGp5XUhqpSIGAvk5SlmEGC3DXuiKDLpknTdCw8wyQXFAVG3ds6fV5EvIk0u9XzXylxsYXocAIAGwtPl2i4DhJshm1t8oUwG55SW80x+bFzFFQ1P6YOj69bKRRu+t+LmJRkye4oNMOIa2kY4eCSPMcbvJhYynDiOOo6H5bHzf9/K7jw8HPccQ2Hm8Ni4lRwwNoMOCidE3tw25hW40gK3XVIgDhNSq16oiXzJ+k8VcTCKWtSocgGj6YkRUoSUJMyXUJqUqEuVRI0Ncp0TUggHlUpMSY1CzU2VQ5r69uxKj32+0VaEnIkmeDL3hlOynLKKbHn7Y3JESZNlRhNmz49YmqUuF5PcGOyhxZ9/sJ7ii4DDphJiil0a4V2JudrMBzQNb4mC3QkxEfxW3f93k4i8hLEThJyifjKeUuu3Epb/NmE21bP9DmtF1iRNl9a73Ly1DRFUdzfb9GLYLEtRCkEIuDiRkvfrDGqK3JB+z43GF/q/Oc/pzy3JC9huFXVTmEd5H3xCaSQghdsx8NhqMtSsslI+p1XkHGJSZx+OSKfJQAq+ibMDAt7pk9ETM/0GVd1NJoOfQJ8Fsw682YVhdAnwmBSuV5NnBIAJ0yFP1ePcqAccOx8wLHugFO6y0AbHlbnMgKvTXAGnDQLfFQ9wHV6V2aO6DC8t5hDjEmNiqfQxnCdqtPVCbNlxYkVOL2qEIGvHSvyRm+KVemzIh0+YK6iQsCSdPiB2SnGlE+A4siRRRJjuP0QBCL80X95NyeWFBQ0yZOTfPW85v97yuPpuEvZh/mVAnECbzqsqSiPCM2MjNEx/YyEuBMvTfz3d/2ruz9818/cbU0obTXULcaSlBeSpIuuedYBe++1TZdqmsC7yqfBWEdjs3YB58O9bpRct0kXTtnjrxAVrIuFe4+bpdJHh/Nwygzmcp3cvNO1g84B2ULTpGR25w2Vh5m647gYHyH/M3/c+d9HLvDzicyLNK9dzn7yEs+bHs+JlmzVyQDruyJivSyGO+6OR+M+t5H7zz3uoMHDXRWXOLru1/BnJ6hM/VBjMqW5gYno6z4BHgMTsVtNs27aGfzKQnQ33qebAx1P0WByRqP2sQERfQapCpWmZ0LiNGFpmy4rpsWSaWawTIB102aBNSJJWJIegVF8u7+PvXXbcU3E0MVK1x/SU5QocFiPUcTjgB7j9uQgNya7GUhMxQQ8rJZ5W3WcMgX2mwkmqXFO1viGt0BIzCEzxduTw1QpsaDafMNbopmiCm5NdtOWkEe+7y92EpEXOXaSkEuEEku2nPQmnrdtZiAiYw1/lsw6X/PO8pFTq8yZPk95K1y5O2J5kPCmvR4/ypuRFEv/fJLSX26Rf2/DSYR77FITSX5BstUkMtz1AEswL6ataTdIJqkyD1iiZ4KmbwZZguG8VYpSzCQqa1Khl5qtdXWPnulbfDYJBQlY0evExPRMaKVNTY+W7hCTsKKbrJkmbdNlQuosmXWqFGmaLufMMnPGdkO+R1/HDXqaD+gbuHJXzHTN8Jha5U21Ol9PVjlkprhFXc0+NctV3n4CKdA3IYdlFyfUKv+yegPfbm7m1XKEt3M915gx3pIcYTGO8EWo+4rr6gU8BdVAuHp3zERRUfaFBxK70IxI+IQ8ybw08VAc2Nvifa/ucmu5RqnSxRg4cuUcD5pVEgOdWPMb/+a7+dgfvR2D4bTp8b+8fZ0vtTqUAsO5tuYvn9E8bFbZq+vcnOxFiRA9D148O/Hs4kfu+tm7nX+Nq5o6SKqHoiLltItmF0MuiZ9RE0wqK7hQpkgndeYOzSCV6bSQylIKAxyOCyrQqbxoftGWHxNeqTGq2r7V8/I/8zKsGx4t3ugkRTaUsPL/G96fgyENJxnDxziqM3LR9zj0Wb+QcTmqaY6rCRdyLrbq+OR/dxDdfEfEdSEg5zeVJhTD+3Dbz3eg8jFq2654tvF+NUUpEEiAl74+NnEGmfTEoyQFVvR6Jn1dU5XsfRWlmHVNZmUcH8slKUjAvFnFF48gFaYQhKqUMv8xK4Eb0Ta9lNdoOSQ9E6bJT0xJClxn9uIbxZNqgav1JJ+NF2n2ITC2K/P65CDvSI5wTq3RoscTaoVdUmRZ9fAQ5lSLkvHZr62U/V93l3htso8FaaW+IQktetyS7KNPxDfUEk3T5fpkhhY93prsZ9yUWSXkFj27rWtjJ57f2ElCLhF3HIbPf3iRa9lLVVWzhe3zWb1p6ha3JpYM/BXvFE+bc/yHkyvc9cYVegNhzLMD1/Mhk/tyD9cV+finfu3oyLb5NiasYY7IqEkiTzx3A+3ARAwTPH3x04F1Q4HGbbujuxnWGezE5TC1Hl76P/u6yAzo6h4xsSX4Yg3CEpMwIMomLSc9uqhX0RjOm1Ui4sxT4Rlznk+ox+kS8+ZdBX7nZItjK4ZbzRSPtPscNDVqpkAkFvd7ziyjTYIvPmfMEkXjc7qd0CDgfWo/1zHOMWlyc6nGLr/AsaRLLzH0Y4gS8ASenPMpeDYhX5UebQmZNFX2mnFuTfby4eohWu0SZ+ZqXLEr5oEHjvCWa0Iee3QfHxif5mwUspLEHNcdagXhUCXg5kKN//i5cer4tPuK+WTAU2qNOdViUXVYVj36ZrCjXvISxYfu/Mm77fVoeU4eitjE1FWVqpRoSNUqXJHQN1H2OTkuQWgGxCZBiRCTUJEyJSlmyUeBAIWi4TUuGE/zf+cfH+bjvdJiWJo8H9sZ9/Kvy/uKwIZS0qhzaNBZJ2U4yXAL5DzvY9T+RvliXI664YsRlztnuw5G5ndhEvyUpJ2PUepWLvLn2yUOGSwud36GeR3Dv7vX53+6ecp1RPISza4vEpkoEygAO5+5+9NyFw0VKTOrJolMnJkVBhKg0ZTSpGLNdGzxjYCKWNlc10m3XR7bddnFGF3TZ1Lq1MWOFZYv6dE0HQJ8K9KSqretS59V6fCaZB+nVYsl6fCZ/hIVCjQlZJICJ6TF25KD7DXj3KxnEKCuC0won6aERKI5o5pcqSdYUh2eUaskaIr4fEtyBbtMgxNqjSQ9x5NSx0O4IznME2qNQQo721cYbda5Ey9s7CQhl4jX/psvErxrjN9814DvMrcAG4ODL34KwdmYMC93oNMYxlSDOdWhLzGLepWu7vEIZyjX2lQ/8v6jr/rj9x+9mPv5KzW2SroudY5HJYoXG9yd2deGnOHGYO5a3AEeBWznIzGagYksMT2t6rrwUrJubGK0SYjMwFayUCjZMFfLY7JDHWbGf6EJCbX1SzCpwk3X9OiYPgMTMS41BkQ87i1zvinMS5ue1pSVYlIFnJI2NVOgbPyUWWJhYkdkN2/TV7NfNygpxXnp0U4SzpkekWj6iSEy9riWkoibr+xyy3XrzHUT6kWYqGo+1T/HinS4gz3cqKf5325ocMQv0ygbTp4vcMstxygWEq69epHpXat0QsVtrznD996o+R/efZbv3V+lFMCZbkyjKOz3ShwqFolyzY63Jfs5Los8zOkXjQu1ExeGSvlCOk0aXEJSoYiHokyBddPGw1ZDyxTpmZAKRdqmm1ZmrRlmNYUw+njUpUqAn5Ggx6RGw2tQUmWc90JBChvqShfBvb9SY9TifTvziutK5OFXFwsHpxvmehizQVrPw3KGuSDDi+/tcHQuB1q73bioKuJF/rcV/A82BEqsP82GkpVTtso/bziyc5rjDNpjURcUs7J9jxBOGRV5NS5wkvKS3Z8mJZxnzxXJPD8sedxB6ISylAhNSMt07L3sEgUJiNMuS99E+Hi0U0WsWRnPYJXOKwpIFa8Mp80Se2WKdWzS0jcD642VygI7REGRAj0TcsbYzsRXvVOZita69LlOj9OSkBPSYkl1mfQsrO1JtcrjWKL755jjoB7nRjNBVyKeVCtMmDLr0mOMCgBnpc1pWeYEC3Ql5A1mN9+aHKaMx33ePN9WnsVDcZPf4BuDDs1kp/P+YsdOEnKJUEduQnYfprrrFJV0UfTRT/77ox/71K8e/cNP/srR/IBQVzXGvbHLktQN8PHxuMlMEqMpp2ZCMQmP/eg/+68u8RiOUfCzS0otDv3fDe757ke+22FfYwf/2MRZ4mDSRbkj2oZmQDElpec/Y49UNSXFx1qjKH8TX8R1Q9y+hiciYzShtgmI06vXGHomxBj706maJEZz2iwBUMLnuOlwKulzXHe5VU1wXrWJMRTwmZQGoQmZ1bVUWWtAYgx3To9znyxzhaqgjHBgDK6eFN4+XudtewMqlT7feGqMscDj0eaAibEB36J2Zzj+102WOL9YoeorioHm4OyAk88cYHbXKuVqm3u/upfFtuGJJ/Zy/GyFYqXJ0wuKdgiHqj7LfZvsHJzUGAPnpUeXiCdk3VbUZAJBskluJ17cKKT+N14q1+kS8yWzTmQS5s0qRQpoNJGxgguC0DI9K/eaQRcT+mZAXcoUHB7dDDJ1OKuaVaciJUqqzIyaTJP7IQgMmxevLyaM58WKYYWwZ7sNF8Ndj1Ewq7wC1vDjwzHMAcknItsVCRjmgrwQfJDLlW8fdS3lj9NJSDs+VER8ASQr/9PNLQ5CCKRciQ3fDpecbJV4DF8H7vmCyhTOwEKmEqOz+8OaEWoqqkyQdvDdsXii2KOmMGh8fPqp6aXrcjqeY2QiJqTOAZmhJAGzMkZZShyWXSyYNUJj3VAGJiYySSbDHRPTI2SVNmCFKQZElCRgRhrUpMJBM0Nk7DsoS3HTHBmnMM83m93cvtdPxSgUt+s9xMagjJ0PblbjFPDoyoCi8fiKzFMwthi8Kj2KbHQ0HlSn6Zg++2SKLgPuk0UeUks8qBaJSPiL3gJvZpZH4hYhCW+65zv/q19zvdixU2q8RPzD/A8cvX32D+7+6nf/4tF3fTe8iw9s+v+wmtQ/+56jd0tar9iOWVNCQkF8viDnWKeLMvbx3/jE3Ts3QxrD53E7xpAZFGsExMGSaRMkNY0KTZhBstzvsGHkWFFV2tpWiEKXoKAzuV5n1uYqi2GaOBh0NgkMzABfgszga5QZVvbe0kkplnhTl8bDtsa9tEr8O4OnaEqX455VxCoZn4ISynHA43KS680+lqSNLz63yiTHTZeqCfCV0AmFmikwVfL4b+vT/MniKm8tTjJRhlZP0W6VGSRQ8OADN0UsLpd53ZGY2/1DPHHa48oDbU6crfLG65uUK31m3/yPnLv3rTx9bJZiQRMoeHjQp7RWZjnU/M7/eyMlD3qhZl9dkfQNJ6RFf67Cq6sVNIbr9RQGw4qaZkKXWVLrz+Gq2YnnEg2psKD7+GniXpcqIbaqCfY6DdPEw2n+O2cIgyEyMfvVDAtmjQmpExJTokAX61gck2TP7dBnShqZgaGDbmk0bd1ls9LTKyvxyEf+3ndxOTyG7DVDpOaREKwhHsdwMnEpMvkoNazLiecz+RipiJhLRJ4NrHfUecub1Y5KxPJwN512C5xaolOYcofifFbcXLTVvl3nwyYvXsYrSVKOoTYJZSla/oOJMh+fgYmye8bLcRzn9HJmLOqODyz8ysdjVo0TpWnOGh3KFDlv1ihLkVNmkbIU6epeKvFbzOBgjvchGNq6y4ya2ICxoeliOWENU+SgmuE8qyl3yadAwICIRWmy14yzq+Tz8bk1jjDFLWqc0BgCJbxWT/L3zHEm6bOoulyhJ1lS3ey8XaHHedCbQyG8LjnAGWkTqYR16eKhGKfKnKwzIOYqM8suXeOYWqIoM9yg6izGm/2KduLFiZ0kZBvxDws/uO2EwCUP/80Hfuzu7SyWNYaFZIV9aopENL/+iX+1k3xcIraFjx6aeIyxTsPup/MogM1SiUUpZsRyK0fooXMLII3JSJ9u0ZBVtTAkaQcl76CepP+LTZQlIqOON89hce3zsirlKl9Wl91NIqvSJjGagvi0ZcCq9Hk6KtBTlmNy0DS4nkm63oATSZdbS3X+drDA3row3zZ8+/gET6zFJMbnzqkJdRzV4AAAIABJREFUEg2eMlSKhn7oc8WeAdPTTTxPMzW7gtHCZ7+0n6v3RDTGW9xQ7VGutfnIp/fxXWvvph/61Kox7Y7P+77rXm585DWIRDz2TIMTzYS33dDmgafqrPWgHiiSxHBFUOaRTp9rpcECITV8rkumaEnEW5OrOKual3Fl7MTzET9810/f7UzLLCzDEsKLUmRARJkiCkVRghRu4aVqbFBTlWw782aVxCQssc6Y1PCwij5N7MKhpTuZ6k6fAVUpgfOtMGmXWPk0dWsTCff5VCp8ucSW/hWXuVgffn5eyWmrrsV2yO/5bQz/vlU8mwQKtldkcs9z+9nO6zYd/xbPdc9xHMGtYhTxfljJzC3CHekbLGfKdgrjHLQquWA7w+Eec4UtNz8kJOSfXpCA0IRZ8aogQQb1BVJ+l1V9HKSyvV3TJzIJAR6TUgdsF8NPBSHGpUqXEI1VVxtTNQYmppDK/IbGik34eJSlyIpZp88g63bsNRMcZ55r2IOHsEwrvccTSlLIxF9KErAmXe7vtRHgKqmzksR4CA3f50wcMiFlSni82kzS8Dy+oEPmZJ03JQfRGIoEVEzA17zzHNDW9HCCKjVT4DHOEaSd3ZLxOaaWOKwn8XwIlPC2P/6unbXXSxA7WIcXKD7/+jdwi3/tJs6Ii/zfrgJfxGOvGX+xD/ObIp6NIthWcI1RfBDnFm79POyAWJYSvvgo8ejqHkq8rLrk1IFCMyBKOylOCSs2MYFsJrhpDIEUskRkqxAkIz7GKefEtczdJOYWgoFYjorD1p+TNY4zzxe9U1yrJylKkbOqTV1Zcl5LBjza7/IWNUPgGZqxZq5t2FPyqRVguSM8uhoziIVHzsPHnwn50jM+rWaVxYUx+t0ySexzxa6YanXAiRMzTOw6w8riFG/Z57P/yAkmxrvUqiGTYyG//dG3UCwNaLeLnG5pyp7iC4/UCBSMl6ET2QXKE1GXcfGZCjxuq1UJ0fy99wx7TJk6Adfoycv96HfiOcSPfs/ddzekSkJCQ6oZN8oRVQFCrELcum7TMyEd08uIyfbvPolJqEiJcdUgMQkrep2Tep6SFChRsMmLWFx5WYrUpEIjxXFXKdE2XRKsE4kz23ReBfDKTUCeD1PaUedmuOORr+Rvtc+txtDtcBdg+wnUMIfvcj7bUV3yrfZ7ySSLDSPCUNt54IKO9YgEblRHyI3hOh3zXefDmAs9b/LqWMMde/fTfTvPKheWk7Ux39hEoJQdd5TCrPz0a0ZNUE0hW3FqOXhY7aYiJXapjbF2QmrsYpxZxvFQTFKjKAEd08fHT+1pNU3doSplAAvRIqIsJUw6R8bErEgbXzxOyjJPqgX2GrvdIL2vG1KhKiVa9Jk0VR705lhUHR6SFa6sFNhd8LlhX0wFj7cVp1gh5Lzp81jcpmg8DuspnlIr7FclqsYS2l+dzNCWgS18GI8laXMFs8wwxveb6zimlrhST9OTmK/H6zwT7vhRvVSxk4S8QHH2pz949JHkeOpqazIVpUlvgnFvLHteQQKuVPt4E7NcpXeSkK1iVCKyHUyxw1nnddod2TBgo9pljMlUsGpSyZSynAFhaELi9LHQhPRNP3vcuTs7k6ow5X7AZjOxUdAwLyXiOjdaVx0rpCTDPG+lJEXKUuRK2YNgyfKC0E8nO0GISLjXO8GPcAsTusQDZoVTqsUeU6WI4sm4w4NLCVc0PBajiGO9kMWuYbwMM4HHl9e6HGgo9kqJkhIeeqbMnn2LfOGBaQBaXY9de+eJY8XJp66lUu0z0QgJe1W+8sg4y6tl/uIxHwU89tQUX3jap6MTPIGHohbtAZxoJtyXrDFrytxaqfIFOc/9gxa/3nuUv1SPMUaZWyeLvHt/wI2V8rO5XHbiWYZC0TQb3izFlJQepylBQyrWJdpYOEgm5jBk/OmLT9v0WNNNPPEsqV1KrJoWS2bdKmNJ1eLDjaZjeqzSpkSBc3qJAC8VZOii047kpQzcvlljEzk6D3nKfV1ObFXl38rD42KL8/z4+mxgV5v2f5Ev9/9nE5fLLXH73Aqyle+M53mEm/Z5ic5SvpvhpK2dN4uDUzmuhuusFyTI9q2y49roqjhSfGJ0RkR3XlQ1VU1l4lX2HiXdn0tSkhTWKFgZ+rKUGJiIdTpMSp0V06JGiR4hJQqcNctEJKyZDufNGjrX4Qe40uxKz6TmgExTlIDIJMzKWNqlsdyTebPKDGOUCLhKz3JGVlmjQyn187hST1NI/VTmpcktyV5mdJUb9STNgWGyIhyfDzhLly+GayyrHkuql/60XdW+xHyBeV5vZtmjG5yXLkXjpQItinFT4Q1mN3d6h3jYrHFjstuKZxjFB+/54NF3fPIDO12QlyheeSP6yygGZpANBgE+Da/BTwZv4Fv1q7LnhCbkmD5LQQnv/eRdOzfCReLZeqTkjb/cTwuT2iAKlpTtfDhJUtsuHmRQKGN01snITzLucTeBJiZhAz61MVnl5S6BC0jyvvhU0oqSwiqaOIPEiipTlyoAHdOjZKx5YpkidanYdrnpZxWuddPhb+WM3U9aPauLz+smSwQoXrvL44n1mJryqIhHV2tONzXTVUsOLAaGm2Y83n5zk3ZkePqpPVx/IMQvRFx1eJWHHzoMwKm5CidOTKGN8MWvHuJcL+bjJ7t0SWjphJtvnGOm5Fk990HEq4M6InBCd7mGOg94c3y52+Jt7OFz6klWTYt9MsVZs8xnVlpcfe05Xn/D2rP5yHfiWcSH7vzJu/smpCQFYhPTNJ0MUlHCui07UqnDrTuBhshEaXfQklHdPeSw5k4qNDHWQLNreizqVWpSpixFZmU8hXnZhdmAyC6iUqlrF3k4zSs1RDYU9/LdgctZpA8nIqMSnYs6cqfz1gvRcXqun92l+B6X83og7YRfSLqHCzsho8LPdSJGqbnl5x+XTLg5JMDyDu1dYvkc9hg3tpNJ8aYeINn8gM4Ska7pM6HqDEhJ9CjbkUgVFg0mMy1smx5t02VSGhg0DSq0TI8paXCWZUITscg609JglVbWeXFQspABFSlyWlaYVZMMTMxZs2whXkScNcvp+7XvxXY5LI/kSTXPOFUUQoMKS9LOJFqqFOkTUUDRlZh1IvpaM98xiEBHIirGp0vEW9QM69LnDr2XSVNKC3Car8gCV5kGZ1WTBdUhZgMS+CTr/Fl8htNqnVASrpcxJkzp0hfMTrygsZOEvIDxsU/96tH3qdsQUdzoXcFHr76V7/vBv+Nz6gkg5RAYW03/d/HnXtqD/SYJl4gM+wpcLEaZQOXDGXF1dS/TT3fqVPmqGOQ8SEZUEDdV9cyFzrquBe+kZwvppJKQEJs4w8baFr6FwZSkQE3KzEgj235bBjgddICm6eKLR4+QMazU6QJrPOrNE4lmr65xsOLz9ysdFPDkkqGiFDVfUVGKz8gZrt8FK124fbxKuaB5bEnzjafG+JZXtzh0cIUnzhT5q8/vJ459Hpg3aA1/ubxOtRJTKkUca8YUlcLaWwnawBe+theAhu8x4wdExvD2Wxc5qCqMBx4hFu97L3NMSZ0An+PmPLMyTl8S/urz+zk3V7/oZ7sTz1+MqwZjqoYgVKRMLYVJhMa6IRcICEm9AZBNajpAdv0CmYGZW4g4ud9ies27BGZRr9Iy3ey+WTMdilLEOElPFNokF3QFX0lwLBf5seMCifFtVvs3ycxeYozcTgfkUqpSo36/nGMc9dh2uttb7e9yu2UugRh2Nd9qe6OgV64wle+KDMseu9e47bux30GzEpKsK+/UFAMpZERzJR46m08EJ9bgTG/B3n+uS5mkRbZyKtdbk4r1vCKgQMBuNcWKabJHzXBGL5KgMxl6P0UJNOnST4+pKqWUy2IyL5GO6XPQWAiXu699rAx9QyrUpUJCwiyW7L7LNEjQrNIiJKZLyKJZY161mDJVEjRVilTwSMQmaS2dsBBFDBKYMCXeWKvxwcp+ImOY0hUCEY55q8zqKgo4I6s8qBZpmCLvMPtQ2OvlFj1ji3EUmTMrtCTkPpYJZUeS96WOnSTkBY7//L9+nruDd/LH33+Km/73r1L4geu471+f5pcr7+H75XYAGqqe3cQ7celQOfysG8xHTVxZFTGvf49HVVXx8ChKkelUEtRJ4LoJxT3fGXldkMjkJsJRk/0wttd2QTZa61mnhA0teS/Vpq+oMp4oylJkUhoIiha20wGW8LtHJglTHfY4bdPPMs6iaRKI/X3FtHhMzvEP3gnCGK4rVJiUAomBvtZ0Ys3rr4i5OZnla+c0V+2O+ePmPHt2t3nb1TEHd4UcP22TgNddv07BE46frnNFw+OzcwPaElGthvzHBzQtk7CWxBzxynSICcQmIrUinIsGttsS97nvkRnGfcUzgz497KL1Rj1NREJDKuySCb7TXMVhqkxWhJPLo+EQO/H8x69/4l8d7ThRBjRd06OpO8zIWIbvHpiIggTpIkdvStINOjUzK2OlRHUqyWs/55gY66wjjEmNAgEGQ0XswqBpugywvgQ1ZTt/ThI6j6F/pYXrODwf5PTtJg2X8/iW/LqhfW0nubjcRGqruBjvY7t+JXlxEldwCkbAYIFNvin5xGQ75ox5g9w8PNdBqfLzQba/dDEfmxhJE3EH2fLxqUuNspQopyIlk6rBwEQ00vtGEGITMyDK4JBFKVDCKkLuMWMMTESFAoH4xMSs0s44hq6frxD2mykGRCixalYTUmOGMSakxop0uIY92fmqSpmylLLPukGFkIj9ZoKW9DPDw0lq9BiwV6boM6ArEe/TVzJr6gSiWJUeD3hzNJTHAn2+1F/j+qBGuQCtgaHsCe8qzfAF5vmR+kFaEpJgGKNCiYD3+nv5GsuUCCgR8Iy0KOJT1wWuZx/v8nfxVm/mktfJTrzwsZOEvMDx5dd95OhUSfAOLcHYFNTGUNcfYrpmGEul8wTht+75+R0o1jYjSPGj5dS51cXIZGAoEfFE2YFcfEopydtKDm50LRxUKu+WmyfFuu1KmlwM7zPTdB+SbXRSu67lnmArvEUppP0DxbhqWFngdDBf1Ku0TZcCPuNYorAvHnNmhSWshG0xncwWWc8mthY9ilJgYGLapsc98SkeHLSYCDxCrWn4HmOBx3or4PXjZXaXfP7D6UXWpU8UeTxwvEChkDA1HlGutVlarnB4Nma9J3gKXlMvcZua5A/uL1DEY4UQjeE+vcpJtU7D9/h0d4l7VztMewHzOiTG8A/rHe4ftDilWtyRXEVXYu7zzlMioEtIgqbuKb71CsO73vsPfP9v/v1zvFp24nLCGM20WM5aRcoYDHNmBUE2kV2Hw3l7gFUD0mazmpXjW3VMD40hZEApvVa7aQU3zJKVBIVK+VIBIoLIK7sL8mLAy7ab5Iwqrmxn2xctBOW+LjcupyN0AVF9RFekkPKcRhlgusV+PikY5nhcbN/ZlxldvMr2k+t2w4UcEAcV9sSaG2bzE5IZ2JalSEkKjKkaLd1hTNXomP6mfdakjErFcxtSZYDtuM/JOrfJVRzRU0zLWMYp7Jo+nigik1CmiCeKkyxmxHaFEBLTpk+AR8UU6UvMETONh0cFm+w0TReF0KZHmx5L0qZFH0E4ZKZQCONU6RPRJ2JG1zhOm1Xp8ShrRCTUTYmKp7it1EjPBZxtGiq+cGzQ4+/6S0zrCr/ReoaqKfIetY8JU7Ydj4Lw3uIsd3izTOsyDVPgS/IM93tnuMVMc3Iw4Ia9Cd85NcZOvLSxI9H7IsTM777v6Jd5HwyAs4CC8m/CO4B3DPmO7MT2oiLlDCubkJtsUo7GVvrt1rHWQkEiE6Wa6BuOwPnEwz2W/Z4O1PkqWLb/3ONuAsmc0tnMSXHEeOdoG5oBnqhUrrBPTaxKkHOhjU3MeWP13QMJGKSKJ4nReKJoUGHZtBiXagbNAqhQoCQBHfr8cPUgv9p5jOV4kpuZ4kTU48pCmePLIGIH9mldQSnh2OkajSKcPl/m6sNNgmKPg4cW6bar7J41/PnX69z55mXmzk3we8+EKIRV1Udp4Y3+JCtxwmocE4mmYBRdrWkTMUaB1zbK3NOeZ0W6vMGb5s+YRyF0CBmYmJJopivCPx73uPGOPuIVeVP3p+/+YuXf7STpL0I4TlGQXUOWp6RTVbiiFOjobkayjYkRsbKdq7pFMVUDVIiFZhBjMNSknFqYQWI0kYlZNa10waepSpme6Wf3oIMoeiiKUqSvN9RrXiiuwksVIzuqF5HAvRxzwFH7uiR8la0TvlFqVBfb/rOV6b3YNi8WWz3PFZbyvECbNG8ky24mcAlBfn7JHtvCJ2TLY889d5hfEqTcKxcbSZG3IambbjtIhR1U2pH0ULR1lzFVy461rbvZMbtu5ZJeZ1I1WNJrmWzvq9RBFllnTtaZF+EaPcuadKhSYpBuuyGlTErbpWXWI2gjwekQcpPeTUsGHFNLOIjmTXovD6tz9FPopkLoEdKgwhoxi9Kiy4ACPlWsBH1fIgbE7NMN5lSLNTrUKPCleIU3ySQ3+Q1ODELOS5cD3Sp3zJT5wqJilZBpU6Ulfb6erPL+0m4+11/mk925lH+jmZQKBsMVzPKUOc/9soQWw2ube6kUXznjyDdr7HRCduKbLiqqTFGKhGaApBVTN/D6aZcDNvw2SlKylRfVoCFVxqVOX/ftIt9skNNHYXhHhYN1FaSQYX9dsuPhbeKDOLWrvFuxIycq8TL5XeuK3k9b48mmyvMBNZv5L+yWiUzSMCJmmjG6WAJ903S51RxihjGapkufiCI+FYr8Xvs0BXy+u7KLMc/jtkaFiZKwHif0Ekv8e9f4GNO6wl+urPGnnQXuW4r49P11zp8+wFfu38vnH2pQG1snEGHu3ATPzAe8ttDgp+7o4BnFJAWeiCy+fzbwOawbFFGMeR5TFOmT8IftMzwhc6zT5dOcwUexTJMgXbCumCZ/3lymrRPu+eg70Mfnaf3R7hfmQtqJC8JJijtTwpg4NeMMsgWon/oDdHWPfnoPtl1iglXoGRBRlwozaoJZNZli1+23SpMIJ+PZNwPaKRQrSrHxLgFxykKbCgOvoARkWJ7WxaUWttnvl3kuRnJDLrKNrY4vv61L8Tmey+c1ah/Df2+Cxua6AEVVzB6zXAY71uchR9k20nOaT0BcV9mJMOTHcGOsOEP294jjy3dF8uE6hpHZWKRb9bcNl/VAClYiPp0/YhNTlICe6Wev1xiapksh5xBekICiFGlIlWkZs74dusmMmmBMapYcrs9w8//P3psHSXLdd36f38vMyrqruquP6Z77HhyDGxiAJIAhwUs8JYqkKMmW5HAsbYXXto4Nb0i2BCFkBTesP7zhtR2rsKWNtR06SIikxBUP8RySIHiDg3sAzgBz9vRZVV1nVla+5z9eZnV1Tw8wAwIgAdYPMejuOjJfZWXme7/f73vo7ayaNg06PKYuDDw/DjHLPjNNQB8Xlww++RheWYoltLeaMr0YCryg2lxDiduibeTx8VhvSZDCizkvmi6hLZjRIELToccd0SwhEU9ynpvMBFXpALDDjNOWkLOqzplewNP9Jtfl0nSlzw0ln88tNjkynkZjuNlUKJg0KeNwomM7NAohYzw+kt3Go+o8VenQlpBDzPKh3AwZ4xFp8P7Pd42KWz/lGCUho3jNxU6ZJiO+9SAgMRS0HYM1l1hLrnNEUVQ5/NiLoEfIillFidVTz6nsmvrViyQg62QcN9FzTzTh7fPxRBMrqgMDD5DkX2ACRNa6NBlJ4+JaZ1nJUlA5xlWJZdMgiLdbpYlCWDVtipIjY6yLu8HqrZ9RVUIsMTDpxBTIUDYZdulx/qrzHKExKIGvra7yFXWOjCP8Xeccn6ov4aEYw2dJWpyTNiejNo2Gz/RYn0Zf027muXm7ZnbrMu//0FfZv0XzxFM2SXjYuQDAnOmyHEaUxCUnDo/1VzmuFokwhGLJlD1CzpolLlKlZbrUTGvQ8h83ab7pnOfHnQB8If+Gx7jjy//d/S/DqTOKF4kxKayT3lWs+X8kOHFfUghW+SohyEZYp/NEIcvBYdW0aJg2bdO13cf4P2do2kkWbxpDCg8vVggauC3HUsDOZWAtr/V4wQRgw2feDNrzQipYm+3rSrgdV/PeK33/S40X4pkMd5CSTvRwQSlxKU+ER14oNkrxWp+Pfrz9Nd5e8h24sazu1cRGHor9DMNzzlqSrxBccZlwxnBEDQpriVlg8jtA01jobTIHJl34Zqx4Na6KlMjSjvleRZXjuDo7OH7J9dilx1lZ4Yys0DEBh/UsHo6FXZFiJe5lLon9mZYUIRHH1AWaEjJpChyJZnlCzTFrxgbFhnGs2EU65oBda7ZSxM5PP3QuckBPsVUq/EiWKJk0O3SZ96a2Dj7jV53nmFNNlrqais7whdUaZVJ0evCu8TIC+DikcFimS0CfgD5vS0/yjeYq+8wUADVa+Li0QtBi+NTq4lV9f6N4ZcL5zd/8zaM/7UGMYhRXGn/zy39z/z49xjZT5oyqEZjekFnTWtUukfo0xDd1URQly0FmOWsWUSg6pktggsGUloTFn19efSXRdU+WSl7c5gZjq2MYkDUuSoJnvxSqkKizGESEcVWyU70YPHFZNS0CegSmx6yqEMSaXZFo2y0RezP1xLWGb2RibkWPJh0UiqpposWQw+eCqhPS5zQt6KT5rnOWHn3O6C4r0iRDiiXpYBAWpYkgvDVbIQwdJssBh/c26HZ8stkeD363xLkTe6kUNCcXFc+YVRyEMZOhgEuNkCUCAgxPOYt0pEdL9XnGnMfEVcZ2jBGelXGadHFQlCWHIy5543Nvscj+a0/gHLkG+nOcG3/fiCDyCsZHP/jH91s8eB8jyYLIsVAq00bFUMaES+XEQENjNJ64ZFV6QIQN6A35Fkh8nbpo1szakrALaRt5ydIlwI0LC444A4lRsGpbrwZ/4tWMy3I1LrkPXVpt30xFa/j54ceulBi+MV4OONRPGsPJ1sZ9rP1t77vJ/TZZBK89ZhOp4eeHj4mQQGjt356k0EQ4KAxDx1MkFtRdn1BsNub1Y13btz3/E3huAu0SSqpIjx5p8QlMSF6yhKY/6MQk4iU71DRGoGsC8ipLYj44ocYwYuJCT5+AkJS4cWe8xxs4iE+KeWooFDeZHfh4rEoHL+6o2GMEqxLQR5McuSQhqZkW26jQlC7X62nOqCo36knOSIMV6bJPT3BRrRIQscNUOCtLlMgxaQr0pE9NbNd0uxlnURpclFV26HFulwnEKEI0We1xjFOIyEC++Bxtpk2WQ26eFMK3elV2Olm+F9YpmRS/MJ3jVKtPXQUYDJ1QcU+xwKmgS1P1mDYFfiwLPKar/KsH/+sHbvmVG0fzyc9AjJKQUbymov+J00cdFHVCAmUr6xnlE8bYcy+GY6VUirT4dhIQ6JuIgJAVWhyQrczIGPNU40nCoEQNJiurwz6k1S9qQI5NsPDJxFxUeUIT4irXtvvj7ah4YVVQuXg7BoVDZOw4UyplJx+BnMrGhoWKNl28WMHEi+vGWgx9ND1CtsgYfTQp8ZiiRJ0Ws8ZOPC4Ovbj3ksFWqVxRBCZkmhI5UjQloEnAnGpyQzRDQ/XYbko0JGC3HueMquLh8NHSLh4NGvzLo0s0a0UKhR5/+YMUX1jo8OmFVQJj+I3bW/zNYy7fNYs8w0UmKLAkHR5Vc9wpW/iyOsVTMkeTDqu06IiVPzYYWsbije2i1E6gyaTZlpDb9Azf7lU5//BN7LkYkOp02bbwqaPnd757NHG8AvHRD/7x/TlJW8UqsTCphm4REg2gGsQLobXunk0KvDgpD0w44CO54mCAjPismhZFydEyHVysapZGDxZfSUXZE48wNgS1kC/7SrCLIE30ghX512pcfuG+vsMxDE3aFMK1IVm4WhL3xrEM7+elcjt+kqRkY4KwWcKVdEBkQ/IxSChEBoWg4Xu4Ntreq0nI4GulqOS9kHChdEzuVoMCSkgfiQtdmyVFm8LX1omYJJwTJ/4MEvtCaZLrymD9dxKeR0Fl8cSlIFkWTY0xKdChF1+bsF1N0KQ7GINC4eKwny20xYpkb6XMSbVASlLsZQoR4XlZxsTX9bV6KxelhsJevzdGWzmjVjBAKoZaiQhjZFmgTk/BDj3GknR5izvN41RxcWhKj62mTN6kyJFmqylRVR0mdZ67zSzPS4OW9OgSUiBNxeRoGFs07Inms+oEh9nOlMlzXmoss0pKUsyYPE+ZVSIjvGuiyBdqdWbJ0kMjPQ/POKwSMm1yZHHZknJ5w0Saa6XMM90OfTEIcO+Hj4zmkZ+ReH32uEfxug0BKp5DQ3rM6uKgA7BDTbPP2UZWMqTEaqFvkwlS4pGPHwtNn47pcppFnuY8u9Uss87UQNnHwYkhJVbGd9jfI6nGlFRxgJd34pa/iOCLT9kpDbYjWLOoKK78WkL82oIqMSIEi5stSJaOCeLqsn2PF2ORE3+GRI6xjJVhbNJFUHTFwgbqWGLiTlNh2TQGvg0hfVakzXmp0WbNzX1ZdejS40l1kSYBvTh5uSA1sr6haHz+/J+nqLcdPvtomqoEHJczPGcuckotkyvWeVotc06qBHErP9Fl/5w8R800CExA23Ro6CYt07WL2Bjz3zN2YmyYlpVy1V3apktAj884T3JSLbKgexTGLiBjTUxrZCz1SsS//OCf3p+TNHky9AgHBNcwJod3TTjwMNDYzluCUzfGJgXRIGWw53hyjQQmxMN2RrSJBmZqsAYDSSBdkJBgDaHpDxZyFi5pkNfpdGU2/LdZXOli/sXgWJtt93K8jyvlfLzU8VzN+zdCzgbJySY8vsQrwxiNLz6ueOs8OmA9/Coxl90oSLIRNuXE+0ocy/smvOLjMTCpjfkKye9ZlRl6zdr5nSQeyef1xKVCgZbpsmJWCYxdwF/LtlgBS1imERsRdlAi9OlTkAwnmANsQeAZtQBYzsWiNPBNbDoaX68XVI0svlVipM+jzgU01Zp0AAAgAElEQVTuiHbRjfkrfSLeHu0DYBdT3BpN05IeeeNxNgwYMxki0VwbTSIIS9KmYrLUpcs9ehZBWDEh23SJg9EE4+QZNzlCNG8ftypYt3kljkb7UUaomAzbzBjv1dfh4fCUWuGcqvJZ9QSfWapTU13mpEMJj1o/om5Ctpk8flzimK30eXIedm7pUjI+FZPlnXr3FX1no3h14vV5Vx/F6zZuevA9D9yxv8s2k6doUuw2E3xEDrBXTzBjSpaQJj4pcVkwdRwctpsJZmScGVVhu7L40L7pc14vciFawBirlpUQbjWGrrYEQF/5uOLiiUdeZUlLineYw4yrEqUYP+/iUpI8fixPODzZJUpZCXhLRMWVRT0gHTZMC1gze0skd5NFnVXMspNVjRartEnHrXMPhxWahPF+lswqZ2QFgFS8WNwuk5RMmq2mjIeDi6JksjQkYJ+eJIuPi+IZZ5EdukyRLH+1sGAnDOmyY7pDx2iyxqUs1lRqxTT49Ff2MmYyNE1nMP5TLOCgqGETix4hgQkGxFBtIltJjw3oevECNzHcAhgjzw4zyTYzxuFchounD6ImA9SWxit5av3cxv/+4B898L9+4g8fUAgHzQw3y55Yqcy6MfexfgVJUhIOca+AdYs3QegTDfw+tsgYGiu6kESCfYdYptqs8T9izbh147vEQO4nqK7/rMXGJOAnIXa/WOdjsyRjs0X+5d77anehXigh22gg6OGuS0YSqXULt11TPRyWwk14FsCApwdr53OyvcSkr2/6cRdcr1O62jS528DfSfY9zHfKqgz9GGpVEuvHFBExqcbWdXNcsV32i6ZmXxPzpJqmw7K06MTGoFbVKkuaFB0TkJU0LbrsZwtNY2XoO1i+Vs74hDHUTCHsNZMciXbTjYsEPcK4lyI84yxykBlujqz57Jy02a/HuEFPcFxZWfcsLl0iDukyt1PhpFNlUmcpmwxL0sI3Lt+UOXwcGoQUTWLWGJE1HrfIGMdWWqxKj1O9LuNY3tnhdI4IQ4OQvEkRSkSNFl0TcFAVuFtN8UN1lifUCtePu7x/jxuDsOH6fJr5qstz/Q6fflrYrXKUdZrPqlOXO+VG8VOIURIyitdcVP/0Aw948aTxJzdlKXgKzygcY0/nX4gOsaJXWdF1anqVJ81pTurzzOsV9uoJftu5mY8V3shWNcn7uJUDznZulN2kJcW0sg6wShzyKjvoYKTEY6dMUyRLiRRHzB4y4vMefZijXMtd0S5+ixu5i4OMqRKTzhgVVSYymo3qWBITDQeTkWQI6VOSfLzIs1Csum4OJrjVGL7UNB1WTYslU6dJJ3ayjtXA4gQsjBeBKTyypGjRZV5WWZb24BjOS520cfGMokWXLcZ2cc6oGkXjkzUei9JiSdrUGylWCTmvGlRNk5ZukZcMfxE9yjGeGpjYNU2bmlllXq+wpOu44hLoYFCVbJuOhc3Fx8E6+65JJIsIaUmxZFZZFqugcmh7l3bbBw0yklN8ReOBT/zOAxdllRVpkYsNxzqmS1O3CWOVLLALthRrKnRJQuLF0JKE0Ns1ARfMMompoSsuvvgxRMYMEk8RISdZ65cjDl5cDBheFA6LPrze4Fgb43JV/yR+0iRss87GZtCujXE5qNeVjOeVSBw3ksmH/Z2GY1gaPSGZK2SdQ/mAcxF3vV3xBka4rrh4uHiSwhd/sL0XcmYfNjUcJswn+3Bw6Oru4FppmQ5R3GWp6dXBdpL3+eKzXSZwcSionF2gs50FY6V3gxiWlSZFUbKMS5G+iQhNxD5dZj9buF5vY4exHYoMLiFR/K/PeanxuDM3mEtqpkUOO5fM6BIRmjnVZI+eZF6arEjA89IglIhtusRJVeWsWuWMtMi5il1Rmd3KSuNqDG3pMWGya98VQiia7brEjMlxx07Nfs9yW9464/OWXYqKSfNst0sKh/vGCsxJnfNmmTI5HBSPmzoX+z1KZHiXv4XFJvzr06e4e8rnjlKWyaLmR/WAW3I57ppMccuMMKeaNOlczWk2ilc4Rj4ho3hNxt2ffP8DABeBHfG/4fjyh07ev6irtHSLdrwI6knIQ/Is+/u38NRqh9/OH0AJ/GeTGcrlDv/4yG0s6IDjTpai8QkkIovHCea4lq2UIp8iKXwUUybDndFO9rpZpnM5TjcijuzU7KpO8FRjjm1mjHNSxaBJSxqDoR9j6e3iO5brNREp5cZQLIfA9AZVqMTwzRWXcSkypxcH1anABATik5UM05JmhSYaQytWRJmSMm0CPBymTZFzUqVFl57ps1Uq7I3GcRB+5FzgGr2FgIiGBNwYTXFCrYBAKBH7owrjY10y5xy60mclqiMozusFCmJVWtq6MyBNGmPoxrjkZPzJQmozz5XksZSyUKuyFFAI06ZoP0/H5fb/6h9QB3ZiVpZekXNpFGtRNBmq0iKNx6LpDbqEyZrUSpzGLtND1eAUHiEREpN1HWN/trVdZBmR+PwO1u3PSl1b1RwrQ+rTjyVUk8VeONQle70lIJuRrS/3+wtt42oW+ZtxPAbX4ybjeaGxXmm8Et9bIqMb6IBQrCqUXAbSNFBbI0LhDsQRkk51kogYNGH8WieG9SaS6tqsbe/FPqsaSpASpSsrO+0M9pF0WJJu4xanwpKuD+aGxOsjEVk5bRbYzyyLrOKIw2mW0UaTwx/AHFdNe4hjZenv33bOscUUKZoUz6h5AnqcUTXujfbgoXD0Dp5Ty4PjNG7ylCXHfWYrfWP4e3WCm6Kt1FVAW3oc0Vsouw6NSHOMFnOqQcH47NElPIT/1D/PXbKFOR1wgBLP0Yi76m126TJ7nSwrUZ+26bOVAn0M9abL3grsjMb524s13j8+hoNwdIvPrZ1ZvlZt8tv5g/xx+2EcyrjiUtY+n3NOkCPN/9s7RY0W/zp3mB8shDTpY+qGI+UsX601uC9VQBv4nZlZHrs4+xOeeaN4OWPUCRnF6zI8HBxxGHfGyEqGtEozrkq0TJu/VY/zHecMW8oRb7vvOIfffIwdd3+dt+yPqEiKo9F2JkyOcZ1lSue4w+xmvy6zhQw+inHHIyAiRLO9JDxVD5nXARMV22n4b1OHmdZ5GqbFTmeWnWqahMQIDFr6jiiUOCzrGi3dHlTAIqPRJkJEBpKnK2Z1IEGcSKB2TBeFsEKTHhZ774vHslm1jrNk6NGnKT0ypKhQZKtU6NDjhLM0IAk/oxa4hhIF4/MF5wRVaSMI72YH59UqO/Y9w0dutLKHZVXEoOmbPjVtsclrZo9rsJrIRPRi6eDhxQ1s7mpsK4RWCvamaCtlk+FGPcnNd32H/uNTmPmLkPJf8fPm5z2q0mLZrNKOFXqGF1OJMg8wqOAmfw/D6VJ4MRxrrcZljBnIaQtqSCXI1qeTqnTHdAdwExOfE8P7eb3FZryL4biSxfvlEobNeCbDicTGjshG8vvwezdu45WCZl0OmrZGQFcD+BWsnYcKGfiADMY5VPRIzjs1dB9OILEbPUCSSHhroenhiouFj/YGHiFr0K4Xh9TZ160R4R0clDg4sWSvgzOAtYJNQCTuOIamb+FmGM7IIjOmRN00aZoOe2SGBVPDYHkd/fizVWJ4lyDksV3NAh510+SQmWWFJh6K7ztzVEyaNCkqpkDF5Nity4REPK/bfEcWyOPTkhDXKG7Wk5yRFl/Rc3gi7I3G2RWVUQh1euTE4R5meN402eNlyCqH95YqzJosh/UkE/jMRb24OxJyx5THB6+POL4U8fH5Gl9ebPOu0hhfX27z9uksn7nYZLljeF7V+Fqztu77rUqXLVi4p4dD3TT5960fM0eHPC7fdc7zcK3NcecCO7d0eHI5Qv7dux+44RPvHnmD/AzFqBMyitdl/Oknfndwo9n6sb+5/+PfyzFHhxIpbsqm2T4ZkvZ7/PmnD/KGiRuYKIXMV1N89F1P06hW6PcdvnF8gjffuki+XKW2NMH5CyUu1qxqyNv2R5w6W2T/rjp7dwjbDz7G2ROH2butTbvtccjJ8tSpMaqmyUWzjBNPPAkJEhjggYdhWv14ggsxOMahg/Vn8MVnyqmwGFXjBbvFKrdMhz4ROcmwZOoYoympPEu6FkuiGnoSEhnr77Csa7jich3beVat0DAdpqTMCeqMmQxH9G4ed+bYrgt8wnkWTxz+4FN72KsyzLHCqra8DFvBUwQ6wBVvAKpKibcO8w+b6/pvjJR45CRNZDTXOgW2FxX1LuBGBNUpvHCFh4v/y2jyeBUiKz7TlOjGgg4L1OjF52pfLJ/HE6vEpowloYeEg+TUFRejDSEhvqQGVVpjDL5K0Y4XW4mjcc90MMbEviOGHn3EqEHCvdFp+vUQmyUBl3vdlcRmcrwvtr2NnZDhn1cTV8NZudpQsSFgAmlK7p090xsYWcpQcryu+zHUFTHouBuhBzDCQAeDZMIXnw7dwfsTDhtYT/WN52DS8bvkM8b7TMZg+Rx60FFMfDxELGyxosos69qgE+iKa5N/lOVriKDI4IgiQ44eIadkgWnG6BBwkSoKRUifacbYwTinZYkITQafm6KtPObMgUAPzS3sZqvOU8LnYecsAI87i+yISpxx6kQI59Qq+6MKdQmoSZsj0TbmpEWI5jxtgrhTuTXr8NVOjQltPT/qEnDSaComza2pEt8Oq+SNx3WpPHVCJvG5e4ci6Lm0A0FXwXMMf/l4xJsKLkuNFHOqRcrNsiBtnl/JUJOA74cRH0hv45vdGjtlmoPRBPOqyg1S5tMs0yXkfXofZ2SRGi2+55zBwyEk4pvO82gM//dTIe+sZC75vkbx04+RRO8oXvex97l/OprrTnHXVphbTnHXoTY7957mqWe2kDUetS7cduMcrdU8u97zDWSlSHn2ObaWPSbf+G1c6VK69Wmm/Sbbt3TZta1OrtDk7x/Ncd2UYWbHab557FYuLKV5cs7hiWXN7XvbfGKuTkBISJ+cZAbKPyRVq9hbZFgKGKx871oHQaONfV9ACIPHrVJJspBr6hYd06VPny69wcQUmB59ItqmgyMOLdMhI2mWaeCKyzbGuCBVqtJhv66woroE0ucZtUgvlp98Si5w1N3GE6ZGzayuETexGvxmkEjFFcUrWJTYCrsZLC5SkiIraXLiUzMa3Ulx78Eejk7jej1+8OY/GyUgr0I89vHHj2YlTYMOZZOlL5oULl0JcUSRlywt08EQ+xuIXVoasepViayoLykLeREHF4dEdtQRZ00EwiTSC4ZULOhgzx1Bm0SxyJ5Xr7cYThZebHF+JTCoFyWly5oMbbLN4a7Hi43hhar8LyW5eLGxDkvurknvOkP3yvXPJb9b41ode0et+awIieT62v+NQEp8OxKx99pkXJ54g8Rjs2TRDHZrLhlv8jqJkwjBJiPhkJGgJ258T+/Fpn99nDgBKUrOdkdiEZMJKRKiqVAgwtAnomtCipLFxSEjPloMWdIUSNOWHvv0JEYEH5euWN6Hh8uyarOkOpxXNW6JtjJt8iyrNm0JOagr1CTgkK7wqHORDCkyeCxLh7xJ0VYhF1UTH5fnZZl6T3G9rjAlaW5MFTkXdcnHXlMLUY9rVZEbi2keWumwLF2a0mehruh2HB5tt6kScE0+TbqXIu0JFZXi9mKOh5cDQjR3VNJkuxlOmxZv25rimlyaHzbanFLL3GJ2sN3N8JhZpkuPO2QLjzBPRnw6JkDFiWny+7Jqc3emwh/9P58++rYPv3Ekz/szFKMkZBSv+7h445uO5b721NFjJzyyjuK2w+cpvvMpdh14mkNbljm0s059eZJD7/wyzqzCdZaRbED9zE7ys+dwdnYIH9mJynTx3A7Hf3A9pVLAGw5W0dpFRPEXT8CsSrMSRnzKeZpvzglpUixSZ0qNUTfNWJI2qa5Z7fqE1CtDE2aSqmgT4Ygbt+1t0uKJt+a0HuvKr/kuyCApiYjox94pmihOSqLB6xxxyEmapgS8KdrNCbnIDjOOIJySRXL4tEyXomRYpc0jZpHf4nq+bk7FuOa1ZMjESdLwZ7hcJAsZRxS+SuOKi2Cx2jmxREY7gQjtpRz3fPTLOLrDhX3vHE0cr0J89RPfOdrGqpmNkUUAJYoULll8DNZZGWznoh8nqipOQCCW2ZUh404YVJ/DuCuS/EyS0YxKE2AlT5PEZK3SbC5ZRL+WYyP34nL8jOHX/+TE7ssTzX9aBPPLb8umCYnHx+D3uNgxMH8dem7tnZZrYc8dmxgnxRwHq5hlPTo0CodoUMqxile+SsfcvfXJ76XHKNmvHasjKk7M7T0xr/KxzHT8ajHkVIYIawSo4rEkxSKblNiOcJeAMSkMoFU28ejH5oiaHGk8cagbKyDRo49G25/xDlsSkjIW7pXGY0maGBEOR5OEGAKJWFRtIjEc0uP4xuNZZ5kb9RR9DNtMMZa0zbAo7Vh0QnGtHmdZulTIs6Ra1KWHNkI16pPGpSEhLekRYZiVDM90A2r0EBF2mTw1QnLi8s7diuerQthxmcgqOiE82enw5kMdPjffQYuh3XIpew7XpnM0OorvLgdcp8o8S4229HnWrLLEKg6Km5jmRzKPQqFFW3EXcXHEyhD0CHmi22W/nuSrn/j20Ts/fOtoPvkZiREcaxQ/F9H42C8+0PjAP9x/z4GQxYtTBJ+8nck3fwuU5uTTB2h3Xcaf3U9ux9M0n7qB4tuOc/rBcab2lnn8M3eiI+HwfV/DObDInbv+ke7Te+l18wTdFA/9aIJpE/DF7hJFk+K+aC8dIjI47GOcCXy+6Jyiblos6+oAgpWSFD2GcMvi4ElqQMSNSCRM1zTrO0Pwg2ESpn3NWiRQBGM0WhQzzjg908dRdlJumy4+HhMmx7J0+BV9A593TnJbtA2FUDMtlAjnjTWy6piAv5Nn7DYTUv2gA3IpvGSzSF6jxLEqSfESAXGI0LRMh6Lk2B2N8eu7clTGW0g+xffe8W9HXZCXOX7rl//g/oz47JUZHKO4KDW2mTFaYkUFuvQGEp6OUURiv2cnFvUsqwJ10yQjPn3TR8Ryl4alT21lOYatGIYS8GjgkdA2HZS4tmNn+nH1V8GQOlYEl+D1X8txCSdqw2L8qkjmG+4BG30zrmQsV3rtvtD4rpS7ckVdl/gzWNiSN7gHDvY1BF+F9RLOkYmIJBrAtFR8Dg7zkBJIrC8uXd2JH7MxLIIw2MeG4ylD99Zh5StffELTGyhfrb3eqr81dct2WCAWYUgNREj8WIkrMRhsEzBOnibW/dvBoUtIQM/yBOkPCk4+LiWyRER06bFbT9CWHovSYIUmOdIc0tP0MTzizA8I+PuiCeZUk2dVlf16jCAuUz2tlrldT3NcLXGDnmCrLrJdMvxQljlLotgoTOs8oWia0kMZnwXViscFdztTrPT7LMX3k5JJ0SXi9lyeM+0+xWKXX9rr0480/+n5gBZ9+mj++hGf/3JnlsfPO5zpdznfM4xHHvccCOBshlLGcMviLCU8vuCcxEHxvuga/s55BheX/0Jdx1/oHw3goplBMS3LPDXOyhIzMv6C598oXt0YdUJG8XMTO3/l0LHSQ48dDXsu+UKL3J1VFr9xGIwin+txcW6SifwFvFSbC1+/g4tLGb72vR0c3NYhkwnJOBGnvnGEcrZG6pqzuC0oTc9x3b2Psj3Yxj4pMd8QtrlpIm0nlBnl8z1Z4DSLNHTTYnUlbXWyYvd0Jc6gsxHF8KvEvT2ZcNd8Gcxl/q2PYbhCRtK0TJeAkLxkMFgvkiVTZ5kWSjk8LM+zbOr0HYcV0yBxAk7UvHoxzKtneus6IOv2+WKQkKEuiIgwocZQqIHLvXWH1/zRvllu+vA/UXCadJ+eZf7mN4yqVi9j/PoHfv9+g8GXFDOUiTDUpEWJLKticfE+Hl3p05KAAml6kkBTrBu6ndhzFvoXL8RU3PlwSQzQHJLUODRWc0gTxU7VEi87DApr+pnAYZKq7zBPQUQNXQOvr/jJugvr7wFr1fl42/FxuxIY1WZ/D3cAhhOWy712I2zrSrkuCaQp6SogEncq1uS7E+iqvYc4g0+ZQJ+GKfPJe1zc+OjowX/JUVtPUh/mxCSdmDUzzfX3tvXHO+kIJryTjErH4iGKnMqSFeullHR5IxPhS2og0269pRQpPBJ/kHHJU6c92G9genjxPXJer1CRYtz50DRMh5z4tOmxy1S4oGq0pDe4Bqs0aUiPVeng4jBp8iiEjoRcqycARYRhpykjwGlVoyuGhgRMmxx3lXJ8K6jSF83b01OciBo0pce/mNrCD1pNOhLioGhKQMGkyeNR1X1CjO2IkGWFwHqEhIabJly+9ZzDQt3hoWqHHC4NQt5cKHG+F1I0Pse6K5RJsS+TJucKT8wrMML/1T7JKbVCXfrMU8MRxb3uLN838wjCk6ZKSIQbK48VyRBISAqXFV2npPIYDG/58F2jOeVnJEbqWKP4uYqFP/7wA7U/+8ADyonAcylNzPGlx3LsfMeXqVQaEHjQc5l9w0P0I+E/f99xXFeTK7R4/Pg17D70BM7+BU48+AvMnzmIs3sR00kxMb3Addef5r5tLvdc16JOyL6Mz3FTJURziK0oWdOnTyBMEMOZkmRj6OeLVTQ3KuCsU7CJ35/IoqbjDktVN1iIrI9HS7dj8yrFsq7ioDhvlmjHxnIaw83sxhefhHScPL5xbFdCbHXEieEQ9rbTMh22SoUP68P8qr6RLTJGjjRhqAie3o2zY5H0jtNX/N2O4sXjVz/we/cnsqAODreZKbQYtmAlpSM0HrYzNWayA+WgrPHIGYtdL5NjD1N0CKhIcd32U7JmoqmQNTUs1MAR2hhLwrWypxGhCYeghBHroVhcdXX/tRQvB7xps27Iumr9ZRKQjdfqZtfuxuv6hVTuNttH8vMFOSWb+Ho4QwnIWiHGrHtN8lxyvmwkjztxMpy83ou7DclzGxOQS34fvi9v8lkt1MehF5+/VhEwpK07g+23dJs+0UCEBCw0sUeIwVBSeVT8n4mvS4Bxk4sX0dY7I/ECaZsu46qEF3NBIjRjkqdBlwjNirQZN7kBlCtP2t5TiVilTY1WDNc1VKXNKVWnZFKcV6scd+xC3sOlQcAN0SQ5cflSvc7RdAWAIIq9rYzH31+sM6GzlIyVoL9eTxBKxH2ZCrf7Jd4zY31B3nsQ3pAp88ZKhjohx5Y6HBxzeT6yKlYOQhGPHzW6HExnOL7aZcykuaWcZqZk2DYe0dAReycMdTr0CLnTTJOVNBrDP+qzVoCFkGVjFSZ7hDR0a2Diq5DBcQt5/QldvJZj1AkZxc9lLN9+27FzmXcc2/rEl44ee2KaN77rBEvHD7C6MMPy/CxOu4DSabqtEr2ehxIoFrsUdzyPPj/O1Nse4bv/dCuf+ef9PPj9aR48JXzjZJaD6Sx/ePo0j8t5fqRXmJMav509yEJP86y5EHc6otiw0MNX/gB/fDm1nJeiWJO8X2O7KqEJ0RhC+ujYpyQh/S6a+gCGENLHTTgfIizSwEHRJRhwUF5oYr7cWARBiVUIS4uPg0OIhfGsqJB/cSCHXipzu5rijbedofChOdTN1yKlFudSbx1VrV6G+Mgv/c79A5KvCLOqwjlp4RkLskpLirLJUpUWALOmTI4UeZOiKT18XMsVkh55fBDBjWEilnyeeCQocipDz4Sx23oYszxMvBCz/xI+ScJrInnGJPIGyTXxysjB/qzESyJ2xxyZzRO0zbuj619xKSRsY5JwNVyUq4KQXfLaNV6HGxO2FYlPkll7zWW6EcRnVtIhSbrICe1+TJVo6zYReiAIkpyPm91zX6gjtH7UZtDBNsPjjPkrw0lQSlzLrRIPT7xBEQAgL9l43IYxcmQkxXZdZpYyp2UZPejErEn3trBw2oTR0o+hky6KFWlRxLqV16WDRnNIT4ModulxjEDR+Ph4tAnJGo8l1cY3LsvSQRAO6HH2eVmO6xqrKuBsv8vNUmF3WXiy06FNyJucKWZdn5WoT1OFNAn55dIU+7Z2+fJ8wOmmJoPLgbJCGcX0eI+nlg1t+tw07nHtuMOzdU1ARAqH9+xxEKPYX3Y43dDcNgvfOqfZPQ7XTGm+fUaR0Rl+e3IH32jXuF3P8IxaomiypOPjaecYm3aUJMe8qTItZZoE9pzC4OPy5g/fOZpTfkZixAkZxc91/ODX/uSBu38Nvs374Y9h7I8+eX8YKsLAx3U11VqGg9eepLo4xdJigdOfewt3/dpn+A9/8gG+qucYI8MMGTytuG+8wFihx7sX9+CJsBiFPOLMc3hfg+L5Ev+4GBu3YcBojIitGEuagNhZfENV7qVWStctMjbwRWCtuphMYRDLWxq7QAVoaWt8lVSwh93Or3YsQqLHbyVeE+nKFV2noNJMTNYZPzXJb/67zyPjFWTnbeB4SHESzl395x/FpTG8YM0o6x1wMJrgSWeerPHJGJc+ZmBw2ZO188JB6Mf488S7o2DStCVkQooohC4hK6ZBn4hmfO4kBm3JmZZsL4nhjkdSxd8Yr+cE5GpjY9JxCSTyJXSNNuuKXAlPZDg2VZDa5P61cXzJ38mCfMCBk/XbXZccyZqRqxLHFnXi55NuiCNrCUAiK263e2n3Y+P2r+a+u5l8dMKDkkHabayMuenRMV0ExRZVYdW0yEmWtuniioNPijodKxmMw2PqIi3TpWO6ccfE3kMnKbJMgz72uPXpM0WZFl0umBWKkqNKC4/iYATPqxU0FmKlMTQFItFs1fbazRiXtlglx+uiaTI4ZFzhQL/Et5gjlIh9JYe/XlmgoFO0VY+Ho2XyfY9QNHt0kf2ZNF+rNThKgZqsUDY+t+ZytLsaR8Fq0+P9uxy6QZpWV5FyhbwYdmTSHG+3CXouP1gKEeAX9zp86SSs0OMHZzNcM2lTumecRXIZ65j+plmXz897rEiT/6lyiP+4tMBzsoSHwy3RVk46VVax96GdpsI5qTJOnpuj6Sv6bkfx6sQoCRnFKIai+qcfeACgOfRY53/7y/tXV7PMblvmUzsJrpEAACAASURBVN+cxv34e5jOCW9qTvPm6zr0epBOC5nsElM7T3DguiKpiQXqpw/wl1+eYeeRryLfvYv3LN3Gd+QUC9HSgFxeUWVWTWudk/RP2gXZLC43+SeOwRAThmOX3mH4QLKQtBOevqpJejgSk69kcSooAtNj0azy5Im9vPHaFmahjdp/GHF9UA7IqHX+ckayyO+aHiWyTGK7Uh0JSZsMMUiKkkmjEBZUizmps1dPUJUOVengGWeQkGSNhbgsSYscDgix2WGwbmF5uRg4VW+AI/48xNUs9JPrNTk+A0EH89Jga8OJwmaxMal4qQnJ8NiGx51ArgDMQB53PazscgWZyyVhCb8OYvgVLiLW8C8xTt1Y6Ek+35XGMMztcjGcSBmj6WE5E664OMb+TDgirRj6utNMMCd1DutZQjRfVic4Gh1gWTUG49UYeiagJi16hKTwKJBm2TSYIMdFqmTFx8elQYe2BHTjfbcIEIQlaTJtilSlwxZdYE4aeKLIGZ+6dBAURTzGlcfFbp/vywIlk+YNzgR/W5/nXmcKx4XP9i1BvS1W5e6CtGl2+tycLvB4rYePwy3ZHN9rNfmVaZ9Gx2Mq1yeMhP94cYXfmhmnHSj25T18D253shw71+O+XYqgp8hmO9xYyTFehNWWMD3RYazo8KmTEZ2uw5jxeWoB/vvMtTzUbPCjxYhF1eD+8WtpBcLDqy2WaFCSHC0CZigxTp7nzTwZ5fK2K/7GR/FKxwiONYpRvEisHLnlWPuTp476HswUBM/V3HTzj9mS9SkUWwQdn3S6T7ud5jvf34unCyye3kmtVuD9H/oq7h2K4IkJPn9Oc1jP8K+mr+HOaC/3yB7e4E1xG7OM60mWnC4dAspO0UryEg0m36td9CeTcfK+olOgTx+GZCVtWEiC9XNwY96KIozlUi1uX6PFDOAHSWzEeqshSdaNMbzoyaoM1oAxRUal2S6T3Fsqcfqiz75dZ3GO7EdSGTCGbz7/vpEy1ssUD/7t544mcJGM+KxIk1WlmdJ5mtLDRZHCYZwcNzDOCalxmgVqNJmWMlnjkTYeCARxZyNRz1IIGTxEFGXJ0aWPEeuknnTBkkikVSH2mbnMgu713gF5MfL2+thwLEQwQ49f6bG6mo7Hy8FXuXTvJr4HWeGOvglRsSJaUhB5ofG8kECBfV4GXRIthqzK0jStde/ZCDm73LZgjTCfEs/KmqvsWieXyMrsxkT6hFAPkFOWT5VRGVxxSGCHiSEnWBhW4hNSkxZgOSAn1SIrZpW2ExGJxsOjYVr44rFDJonQNE2XXKwwGEqfRVlFROgYK+9bNQ0adNgmFVaxvj4ZPDwcGhIAhlA01+sJzqsmeZNiWVrcEW3lCbXMPF3mpM0RprjOz1NICY+ENW5Nl/hib5GACB+XnkR8qDzFdbk0XpiinBYe7lV5a36chU7ErOvTCRxcBY2OPQ7byJHzDd9ZCLluCsK+4ju1NgCHKvDwWTi4pY+n4M9OX+TOXAljFJ89afjViSmmxgMOjTnMFA17tjeZv5jjpimHVKPI8+0+077LtJviVjXF96IF/pvUdex1c3wzukBKPFoScN+HR2InPysxIqaPYhRXEM7/8e4HtFY4SnPNvV9ncW4WP215FNoIWgs6Eupdw57rH8FxDIdueBRV6NL/ljB13zf46GyF3727iRL4yJ9+kt/42Kf55d/6PL/xsU/z5//jl/gP+68nKxnykuGXzM22khdPvJvBVIZj42SaLOwccRh3xtgtW7hPblonZzkcCQFZm4iu6cbuvuG67SlkU8hMMr68ZC+7sEogCW7sc5KVDDvUNP+Dd4ScSfH0vFDKGtzre6DjBYMa3Z5e7ki+u44JaOsODTp4KEoxtCpCMyd1miZiUVYH0qFZ4+Hj0pGQpvToi2UUecYuLHxcFELepEgZl10yRWSitWr3ZTodmyUgG7H6r+d4yXDLDcIVV7udpHAw3GHY7Jp9uWKdfEY89uT+Mkwy38wgcN12LkO03/ieZD8t3RokJcPxYoWdRERjMC6s4V9LW76UTUxSePG/5Dz3xScjaQC2OBUr9BCbc/qSIhMnDmCvQUHomwhtDD3T51m5SDpJFkyHKO6kFFUOQZinzoKpUZYcbQKqpkkKS2bP4DMmBVZpMyVldsk0Z8wiHWMV7kIipk2RCM0uPU6bgEedRQyGLSZLmhQXpM09zHBAj/EGmeIxU6UXGb7cqmIw/FNngZL28XF4S2oSzzh8sbqK58Aj/TrGwN1eBUeg5Dkshn2ebAY4CvoR9PrC+XZEO7D3oWbX/jxSzjLr+kRamPJdWu0Uz82n2KqLeK7hodPwrj0KbeD4c2kqlQbPzHl87XiRLWmH1Y5wRlrcUfGZb2tWuprJvOHfbD1IZOCpjpVi/t3Mdbwp2nXZ730Ur37IV77ylT/5aQ9iFKN4rUTxDz91/6F3/TMnv3gfyw988LJV+sr9D97/bx8PeE9pnLe+9Tu415xHinDhk/fw2NMTbJ/ucu1HP486cJDoe08iBQU9zcm/ewfzi1lymT4fOPl1IhMx5pSJ0KxGVvnjhaQyN2KoRRS++EyqsUHrvmZWCfQQ/GuD3r7GkJE0HdO9BMIFbPpYXqwU5Xy0OHh8OBFysDLEicLWW+RGHISbZIwPv/kC4/d9FxnPItv3IOUppGC13L/543eOOiEvQ/zqB37v/o3QJ1/5VFSZd0eHeFQtUTJpGtJFAz4OT3J+4OmhEN6k9xGJoS5dlqWNi2LcZDknVbqELOoqB9V2UsblvCyzolfpxQvN4YQWXhg6NEpA4ufjY/RSvT9e7LnL8The7Rh0ODZAqzZ73ZUociWvUeJcAusaft1lt7GJ2thw+OLb7SNDBrI9m2CIGtzjEr+cnKStczeJ5K+Fo7riksKjadrkJEOXHgdkK2fN0mDMrjhERqNEyJOhiU1M3LjrHBpLSC9IBkEokGFGF3hGLdCjb53YY95IaCJmZJwOAddHMzzlzLNDj3Ne1fDx0BiO6m3szHp8rr1IFo+0cZhXLQrG5x2FMT7enMPDoaBTKIQ7MyV2TUb87Zkm7xgrkXINnZ7wXCOirvuD8+2duxUPPS9cP+FQ7wgZD45Xe7xxxuXkkrBjDB5bjHjn9QFRX/HkmQwTeUO1JcyMR6RczadPhfzSXpfjZzxSjvCmG1b4m+/mmU15zJbg6WVN3lXcvKtHJ3AwRvC9iH8+CSGWEF+RFEumhxPfC7/tnOF//sTvjeaYn2KMkpBRjOIVjJMf+ez9H3rvD/AOneHcP7+FypYLpHc9D04EocuZh9/I1NZz+GMLOAeWiU6VefzY3Vx/7zf4g7+6g+POBX6/dJDfqX6Dtm4Ptruughl3IdqmM8A8Dz9XUgXuNPu4062wrSj8f9WLfC06vm6cm028iezk8Gusjr2VW00qfi4uE1LERfGsPscWNUGGFAbDIT1NSMSCavEetZ1aP6LsOhw93OBzP8px4xZhstLhwEe/ghy8HvF8SOeQXHmUgLwM8esf+P37hcQrwn7HSUKQVhl2qS28MdrBcWeB66MJnlTLrEqHOq3YddjC9Pr0CYyVFbULKgvVu0n28Zh5bqB95eIOFHzapksi7bwZgfdyMUpC4udfJmnizRKPjUWMKznmV8sNeSnxcu7jctu6LARr6HhfLgF0Y/NEB2cdxCsy0aBzYvexdj/NiL/mjm7Cga9I0hFOZK4dFDXTokNACg9tZTxIkyIr6fiaWzOubZuAfcxwmsWBeIjCcmBWTZuM+LRMh6z43Bnt5iHnJFOUASgYn3NS5a5oJ6dUjYZ0mDB50sbDx2EPeTKi6BrDcVmmojMohGXV4V5nkkfDBgU87hrPsKXS49RcCm0gl4JHqgFHJlPUO4Kr4B8bS0yZDO/f6fPQGZg3XaYlze1bFD+8qPFEuOdASKPpEYSKJxY1hyqKfDai31f8xdwCtzNJWglvOhjw8SeEkrhM+S61XkQ55bB9TDO/qkg5cO2eBucv5nlmydCONG/eH/FvfrzMPl0enBM/cC5yn97BvLG+SF2JCOgzYTK855OXLy6O4uWPESdkFKN4BWP8g/uPzd1w77Hzs+871rj78LHZx792tDs/S/rABZ797FvZed0juLkmF585TPGGMzz9D2/hmiPfwdm1wp3pIr+yV+g2C3yqdp6cytIfMtwCO6HmlIVBJc8NT7LWgVooS4F53ePIWIanGyGnScjxekOHY40rkkhdDj/miEtB5SioHGlJ0cNWu64xs5zgAl3T4y72sVePs6ha7NQl5lWLj2S3cfdNKxw98jQ3v/WHjN36LN/96iHe/ZbHmbzlR7j33IpkCuB4IMJDZ35xNBG8DPHg333h6LDMgYMCAUQoqBxlyTFpcjQlZFk6uCh+zBx5yYAY8mQIsElLy3QAQ2ACq5SGoUYLR2ylVVCUVZ626eKI5RUlVWHWnUfJaK5uEfx6i826mev/funHZDOY1UvtLrweY/NExGzyuxnwQhLZX4O9hJIei+XDWV6LRpORjHVJV5kB7GtajbNqmrFPkjVRBENWpcmTZsHU6NGnR4g2Ot6XTSjsvd26oYtAx/SYlBI96RNInykpUTVNtkqFqmnSJYw9RKxqmCsuPZW4q1sHn+26RIU886rFjM6jBarSZtxkaUnIBWnRNJoVrJpWDo9VCdhqcnS0IUBzSy7HFxs1dqosX6yt0gsV57t9dqd9ftzo81Svje453JjOYyJFXrmUPMW+XIqznZD9FWFPRTOZVSiBz5+OuGNXnycWhL3jMFHpcOzHLu+dybN3QvPEsubB5So1CXj7RIHzTc3bDnd4bM5hsQW37A750oUe9eUc8y3Dj6MWb9uR4nunHW7yizwRrVImhSAc9Sf5B/M8CkUGlyIpCqRwEP76wS8evffDR0ackVcpRknIKEbxKsbFG+8+tnjrkWO9f989uvedX4rnOqF6fjuZVcPsrz2K0EQKwHyG7G1Pc/8/bOGEORt7fQwnCnYCdMSlIDnCWDU+mTgHxF8MDemyh0nGwixPRHUa0qNtOkPE4KH3DGKNwJ4salxxyUmGomRxY8+Pt+mDzJBlQXVp0SVUUDZZxk2Wi6rF290Z3v7GM0y979u4N2jodHDuuonnH6xw/V2P4d1dQsamIZ1F/CyIcLZ+zWgS+AnDdkGGEtIYEicIDoqiyhFhaKmItoSUyLAqXTLi08HC9QpkaJouShSBsWIFRiAj6TWHdHHISJq0pAjoERFRUFnaujMQ5r1cRf/ndREMmxPTfxIy+HCXY7PtvNB+ht/zQnyJl5+s/tOJSz7/Jfe+4efWXqtwEGznQ+J7Z5KAOKJi/yVDWvlWtEEURqBFl4iIWTVB3TTxJUVftBW8FjWAcvliux7tePEfmhAjNuHZIZN2DLEXRp40DTpkSeOLh0LRlR6eWAPFWanEErUaBFwcVungIEQCLelZtUOBRWlwZ7SdrkREGHwcDlLkKVnhGjNOmz5Hc2M81l/l+lSexSjk7QcinHqBb6+2KeARYWgQcvb/Z+89o+U4zzvP3/NW6Op8+2bce5EjAeYcJShHK5AWKdnj3TOz1q69c9ZeW56xz86sMbBnbM/Yx+vxrM/K47V3xvaMTUVayYo2IYqiKGYSBECARMbNsXNXeN/9UNWNi8sLEBQpEhT7jw+N211dVV2h+33e5x/CJh4WM9Lg5kKOXAp63Jjsm08bhvtaPDYbsaPHxrYNs0sO3xpv4aI4NCNc3ety/1SLa0Yi/mx6hucqIZMLDu/ZqPh2eZGfy4/x/y6e5nK7yJlZF0eEx/Ui6yVHr3L5YbNMUVyu6/HYP2PYULD4x+ois6rOqMmx3nWZ9SM87WKjGJU08/gMSIoqIX0my/P3Htx94N79uy+7Z1f3d+jHjC4dq4suLhHs+s//aU/x5yZpfC6Lu/kkh//+3Wy56fsc/N7t/NzzT1LWlRfRoywsrlVbWaeLfEWe7Fj9tvn/7VAsQRhQJT6it1MxIe8ec3Aczc8df5hmYhMZJSGKF6KAuOKSVRlykuHPtm3hvoOKf/7eU9z7rU0sRCEPW5NcGw0xZDvctCng8k9+Nd6fkUHoHcJMnkSt2470DKKPPA69Q0i2B4yOOyG2w/deeH+3C/IK8bN3fmoPJNkvKwoRTexQNiQlmgS8L9rG16xDpEmxUw9zSE1RI6YpZEjRImS96aMmPsfMJE3jd5Kd29z2dmq0Jyk0Og4qTFLRz0fFerMWH22sRolaTqP8UTN5VtvOSy23shvzZsDLKaja37WGs+L55RRWhXQCWIGOjiouVGJHul5VpKrriCiy4lHVddLiMShFQjQNfDSGeb3EkOplSs+zXg0xaWJReDZJCM+QokaTIlmaBNRNk4CQrHixRkQUveSo0mKrHuSYmmXR1PDEIYvHiOmhqFNMqCoKyOCyVufI43BSamwgywNqnBujYXbkUnyuPs56XaRBSCCalLF4X3+eL80tMWTSjDkux4ImdUJyONzan8K1od4S7l+o8v7hLF+brPG23hyWMszXhP6cYbDXp+Ur9h2H29Yqnj4jHI8abHcznPRb9CqH7zJBVXw+zAbuN5MEaO5KjTHfimlYO0YCjkw4WAqu2FTjwLEsrg2PL8aFUAtNk4hNTpqRvPCflo6yd91avnosIoVi9zqLz5+sMyZpACIgMJrjUmWtyZJXNl+Vo3xcbWE+jLj58z/V/W16ldHthHTRxSWCLSN/udvMlhE/wNqlkJM5vMICRw9uQsq9TFsNtqm1TOhZIJ6h22lt4AOynl/72ON8+oAhJOzoNmwsHHE6KcS2WIhxeV9/gUpD8X/NnEKJRdlUz0myXg1tSkKf6knoXVmWpov8+k8/RWrjSerHtvFYtY4RuHttjvf/1AMMbH0BlfVRV14Ojgs6QhwX6R9BlA3FXsTLgOXw4MTH955c2rHv5MLW7szTq4C77nnPvrvuec++z9/7jd1tykisDYkpeu2U5ax4CDaSzOI2JKRCo9MBGZAiFeLzWqZB3TSwEjvV9qBZicISCzexH22YZuz4k8wKr3ZNvVkGuufDS3dBzn8ftr2mLnYQvbLIWU2Q/uLt/+TjYj6vSNzNUFhxVwMwAjE1VZ1DnYrQiY5EYvthIkBIJ5bkTRNnduRUGpfY8jcgpEydMnVssdBoeiSPi03Z1FCicCR2SRyUOJCwYuos6DJp5dE0PmVTo2EaBESkVYosHgumRoU6M1IhEk3NNChJHgeLssQd8CRjHI1h0GQ4LXVCNLZRGITr0gUerC8xpxqkjUNDQoomxW25PIeWQjbbGUq2xWBO8LRNGAlX5FMcK2sWG7DQMlxZTHFyEa4oppLjKUzU4tf7MsK+47Cr5LB2TY2vTrRwUPQphyv6bT5XH+cK3c+4qjJvQsrSpCUh+6NFpmiyWXI8N2d43w1zHD6dIQoc/FAYKEQ06w6jGYs5P7Z9ntUBD7UWmJMqv3B9na+94DApdbxaljx27PQngqcUj8gM10kfx6lxTCps1CVKyqFlNIfvPbD72L2Hdm+6p9upf7XQ9cDsootLBDOfvZaH/vgjWKNVwkcciqMvYJouV11zmMu9LPdes4k79EhneWM0z+tx/sw8yfjRXYQmTjS3JbZL9ZTHz5jruYHNfHroNiKj2a9Oc2YJpmqGf712DA8nHpyukry8fKav7QhTNjUiInJ49CuX8twoasRnuiyMSYbLTYkrrj6AdfUi9i0uavMaSHlx+vnQetSGyyEMiR7+VmfdD56+qzu79GPGuTkdhox4aDQeTpxLQJWiyQBQIH4clCI+QaerFhF3yhxxyEgaK6FjqYTeZYh551Fi56wvUGi8lEXqmwE/SiHWEU6/DLrUamL0Nxv96kdB+/vPTix24+7eWZc3nVBY2/a8FlYiQG/noMT5IjZWx8Qj1nwYmsanYVr0SI4+KSTrjoiMZtYsMWUWGFMD59xDASEN0+p0GCf1LHXTIDCxVXzLtCjrGoum1jEpUQgFMhRUNp6UwsbB6oQM1iXAweKoKhMQJc53LRwsHAvOqErnsxRMiptzOY7VQrbmHUaLkHaEJxZanPFjlVh/QbO1ZHGfOc5YTpFNGdYWhcs2L/H0gs/9czUmohYbehTfPWE4RY3TFc3nnkzRj8cNuSzPBw0+O7PAlbqfO4ZSaAz/y1hfp6vbImBealy90admIv7NQ8I3wkmWGsI3a/Mcm7WYCn2GeyLeMuJQEJvL0xlK2uOqaIRf/naamgRoMVSjiIaJj3JZR+QcxWbdQ11rFqRBDZ+NKsMzYZmDLHFGYovm+++8b8+Dd35pz4/x8nvToNsJ6aKLSwTzN127T96/bd/Y/Jd2By+M4lw1jrWphUzDWJ/wrUcGKNgWnu7lBLOJGFh4Ozv54bhwiHEc5ZBN8jocsblVxnhGZqlVUoyrKoEJOUKFm9x+1g83+YfZxjLe8FnxcHv2z0gcvNWnChRUliVdQRMHgU1IE//4GNfuOEFJlzg5keGuW+cpfeQAauMmJNcD2QKSKSDpHCiFpLLgN/i+/PreU5Vd+05VupzbHxd+9s5P7TlX4WM6w06TDKAKkmGD7sdOSHuTskQOj4LxsJPE6fW6j5JJc5J5hqREhdhSVCH4tLMeYm55PEDSWGJjziZAnLNfb/YuCPz4irDzdUgu1tL7laSJvxGhxFphvrEc52ridIeGFS/f1ooIcaGiiZCkD91+v0jcKWzoZtKRhlYiMB+SEi1CMrjMmzKWWPjGj0MHdYOiylEgw7yp4IpN2TQoSJayqXWKoRBNVmU6oaARmqZp0SIuTK6WjQyaHGWJHe1CIlwcUsZhrS6woJo4KN7jDnEqalKVgCGTxRfNejtN2YcsLkYgh8NJv8WNvR5PLbaIfAslwqmwxZW5NGs8Gz8U8mnNpqCXiXrEUE44vQSeOPQ5Nkt1YfeoQ8aLWJOxeNfWAOOnmKhrei2HUy2fW/s9/qE1wxaKfKs+x5DJ8XC5RoRBi+GfpjbzRDTPbek+7qtM8X5vmGzo8bRfYUpVuXNdmpu2NADF752YZE58Nls5KqFmh5NlIYrI4DCuyszj02/S5CyLB2WSMLC4c5PDvUtT3GiGuNzqwTeGw7LIiMmRwSaPw2fU0xyQab752e/t/tt7v7b7I/e8s/s79iOiW4R00cUlhtP9H9q35ukHdt//t28nPLyW0uAk2RueYx1ZRvtDtlpFWBxiwOrDUx6/saWfufkUj8sZwMS8fDEoFJ/83P+89467b9j3p5/70u5bog1cb0bYr6b5YHGQUtHn1ESWk2oRR2xIfsR6rZhytcvaQE1aZCXN5WaMNC6BMtjYLJoqWgzauHzlh6N84p4Hue6e51ETDu5b8kjvMJJKI/m+OHzQtkFrHjz5kb2nald0v7B/zPjEnb+656wg3KwQ0QqWWDRNi0WqXG5GmVNx7scsFd6pN/GMmiSQiFnKBKKJiAWxLUJ8Qhq0GJV+Fk2VjKQxYhIKVizKbQ+KzAo61pu1ALmYrsOPggs5jF2oGFm+Ly8lYv9Jxvm0N236acclMH6SdmHStitvUx0NkJLYllyJQhORkhQqccwKiO2tMypNSERGPMrUSInLgqlgJxa+IkJBcnHyu2jWmB5ccamYOoJQoU5gAlIqRY8qsFPWc4vegLZslLJihy3TSiaRFGPSz4Qqc000yryKdV4uNk3xiQScRC84HsbBiXZSRBVwORW0GFcVPByUkdi6107zVK3OsJXi65zmZ7a43Dc/z3uGM0SRopSP2D8hhBq29gnH5qEcRjy/pPF9xTflFCONEmtKEd87ZZifS3O6pnlQTbCgQ2wsrhlUWIsF3rVN89WFBRqEVFQLJO4kHYqqXB4Nsa+yxLRUOB42OKTmmFIVPpndRNYzhKHF7x9ZpMekGVdLZIMM7xxN8ehiEwvFM9Ykd6lNeJGLK3EIYgtDHoeZRYvDaoF/vjPNF6eqnJAqV+l+XlBLPKpO8LicpmV8qqbeuf+++Jlv7/7u5x7Z/Tf3fm33F+795u6P3vOu7m/cRaIrTO+ii0sY1q98ec+Oy45T+MBBTCtk5ss3U+w/w8yZjdRrHplsE7/lsHbrfh79+G9fkNa0/2Nf3TOcVRypBLx9k2HLjqN87/s7+HT5OBUa9JBliiUyuFgoytRpmoCrWMf/tqHEfUcD/k49w3v1Lr5vHWdWL7JdrWWaRT5hLueOzRFX/cY+ZGxTLDJ3Uoib5nsnPtylW72G+PhH//c9KwdX7b/bWQZtEW1JFbnObCSDw7NqAgvFkCkwLRUUwoSZxxMXF4dZvYglFgXJJHafLkumSkGyTOpZelWRBR3TNwLj084nWSlM/0koRF6uq9fF6g9WBva1t9MO3luJV8PmeLXuyJvFOvlC52VlbshyGKNxxU1ei0Xp7eBCg+mEdLZpiW2Req8qdrbbMC0cbJpJ18LDxScgMGGnoMlJhgwucya+H6umTl3XccWlpIq44tBPnhYBZeqUyLNAhYlohshE7LQ3Map7mFQV+hK65ZI0aRHgYJM3KaKElmmbuABpSsBWXaJKwLw0KUuLcVngSj1KyaS4Ppel3DJsH9KEkfC700f5rXXrefSkxY5+4cnpiCNU+JmRIg+cCWkQEaBxEspmn7hc1mvT8CGTMjw+G/B9a5xf7lvPD6cDNqZd/sY/wbDOUTIeARoPi/W2xxfNUUI0OTx+sbSWP1o4Rgqbeaq8N9rCgO3wXT3NqM4xKCm+pU5io3CNzb/aPMiXj2jeNuKyd+oIb4nW42FRIWBOWqwzWSqEKKAoDq4ofKMpm5AX1CIK4QYG+I/6u3jKo67jFHZbbH5N3c6fmEdomdhcQJuIv/zCf+j+7l0Eup2QLrq4hGHeu33fzHU37TN/Ut6drkTkr30WaVhU54boG5yhZ+QY+UIZVagyvu3dF5x9Gbx72779f3Vo9y98+ABj7/sBB79zM5dffpLeqY04zSxXSIl+XeC93hp6/DxabPrJ4xiLG0seRePhNHrY5X0nCAAAIABJREFUaGU4wBxDUiJvUpxhngXRvDBnMfzMNkY+psHLgdE8eLob/PRa4mfv/NSes7bLZ4XMlqjErrk98IrFs5ZYtFREKCZx/zEEoskZj2GTx1JxkrLTdtWSFB5uJ4/GkxTzeglJBK4BAe2hazsgsS3kPZ8g+o2Gl9r3Cw5sz0OLirHs+Cwb9LbTKZYXJcs7K6+k+FjVmesNfG5+FKz2eS2x0OcUfWfpV/HrCojt0Y2JQwVj++q4I9imPkUmxFEOKXFJSYq6adJMBqpN42MkLj7ibB2Lloktrl1xcHHISIoUNilxmDWLHbe5lEqhJE5Kd7DRaLbrIY7JDDPRQiefJxTDgBToMWkm1BKX6QFOqAXCJGuq7eIVomlKxJ3ZYWZaGh9NRXz2qzOsNSXOMM9lZoiahMz5ETsKLr3FkKWqzdONGtZSgYeY5n3rbTztstVL05MLWZu36BWXU82AKgFL4nNLMccDCzWyxuFg1SfC8PGhPo7MGj56Q5n/59Qioei4AyItbrb7OGIqHGCBQDQWiv8ps4HegmZH1MdOVeTJaJ5Z1WI/83yytJa3bA75i7kpfAIa+PzJtf18eb/LQbXAoUrAP+1Zy5PNKgesOfpNhhlVZ9hkcBNh/LwJCExMXz2g5kkbl7Umx9Myx7vMdg7KFL4JSInLPzE38EX1HPN6iZCInMrQND733fvt3V+891vdrshLoFuEdNHFGwDy98/vzuWbpLZOYW0x5Dc8j9s7g+qvwXyaR+/8vYsa7I/es31f6aGnd6d6pxm++RBzz2znymsOkK+N0ecp3n1Fme2bZ3j7Rx6k9fh1rCfHJ2+qoxRkU/DWbXXW9Wp+arCHgcUhUkGKstL8t/c2ef/VJ3Ftl9w7HHBSPHjmY90C5DXCJ+781T1tJyxgGc/dtJ/oLBs/a7CTwqJNBWmHEkZoJphnlBJp4+CLxsVmlvhH1hOXNCmqpkGLgLRK4eCQFjcW16I7w2adOAbBT87gdjWB92q5G+3HixGBL8+pOC9FaNn7flydigtRu36Szl/7caU1cefvju7jLC3LEtUJGXQkLsQdsTESW6EHBB0qVtQJlI2PY1qlk5yeROiOTUFl4ntPhIZpYovVMXzIS4aQiAY+dVpUacSid4nF7xlJ8w69gzlVx8EibVxaEhJIRFZlSCuPXbIBLdCSkGGdZ8wUecg6wUk9Sc00ccShKQE1aVEkjcZwU7bAl/yTFEnzhDrJdrMGEWHElMjj0JQIC2FQpfjrmTnyrTQ3ZXr4RjCBjeLtIzZfPhrhRjYTS4pDCxFRqGhqTY2Qt2V76ctptuYdhns0D5SrHLbmOVoNOWXq5Jf6eSxcoF9nKKsWgWgWo4jrrV6KUZqSSXNaLTHYKjJREf4+mOBE0IqdswjImzRrgjyfn6ryOzc6vG+gwPx4L/5Cnu/pGd7jDPOedTZ/OjnNojQxAi42AyaNjVBQNg8zQ59JkxObQ7LIoMkyRhYPizNSZSMFnpIJIE65P2GVWdDlWG8jDi18NFGn69wtRC6MbhHSRRdvALTecdm+6Wtv3ne6/0P71tz/D7tP/sMdlHa9gOpzMDM24zsu/ktu9vob9qn/vrQ7VVH03PAUz37nDnZdfYBNux8mv/U4cwe3U7z2BWafuIxcCq7+Dy/QM/YYwx+tk9/yHL3XnaDvHYtss05w/zMj5I3He285gnPLItnb6nzf/9W93bDB1w4/e+en9sTxZercbkNn8GR1rETbPHYAT1I4YlOSHC1CAkJCNAERtlgsSYNZqdIioEVI3TRwxKFFgJbYfrdHsviElE2NgAg/oZaoJEU9xllxL7zxi5GVXYiVOov248oB/YU/92pp3auH6L2S7scrwU9Kp+R8n+Pcv88mpluikuIiPtdW0kEUUUQmRImFJkoKjbiYt7A6QnZLbKJEB9I+d6kkv0mjaZk4ZjavMlRMvRMWqtGMST/zlInQZMWjX4qkxWONlPBwWJIGLjbpJFndFpvbo3UYsaiJT8GkUaIomTTfUYdQIvRLD3Va+ASMSh8uNhbCOl3kUKNBAQ/HKHKS5irTR5mQXRQpKYeG0dgohmyXb+jjnKbJdake3Gaa41Lm0UnBxeIFXeOmngxTDU1VR4y5LtcU0wwUIk4vWJSysRHKl6tT/EJ+Izf3eZwqw5Qf8p5sP8+GFSIMt+oRnrFmGI7yvGOj0Ki4HKHMp66z2DQQ8cSkIoXNTt3HuKoyYHKMqQxHTZX50yU+M1XhhFqi6Ge5Z32WZ+cjKlWbTSrHVW6R0LdJY3NClek1HidNne0UaRCxRMBGyfN36llEHEIDG02RCMMjHAOEm2Qbt+m17JcJlCh8E8TXBvH3b7t71i1Ezo9uEdJFF28wjDz+wO65qTX0eAtIf40f3vrHL7vjUL7tyn3FB5/dnXl7g6HNT2GVFsAIerpA8arnIBLWFhfYcvdjSNpF+npBWcjoBgibSDaPtcPirXc8xcGvX8YNu84gus4PNv1Rt/vxGqIdShgPbl5sihu79uizGQfLXlOJnWhRstRp0UuOimngitOx3d2sB+g3edK4nGaOtKTIikfF1PHEpWqauGIjKBxsVEIjikx4ThekvY8/CQPY8wUCrhzcLu+WLH/+ore1Skfk5eSDXPR2VuvMvIL9vtSwsoO0miPY+Whysuy+6difi0VgwvhaT6iGkhQmEREqEbK3La09SWEwOOLgiIUtFpHRpMShbpp44iYFhIXComJqBISJY2Gcon4laxkxRWoSEBCyQffyvJolhUPOuGSMg4tFRVosSgs/2Q8bRY9Js6SaBKK5XI8yKWWWTJVtagxBWBcVWVRNyuKjEMrSJIvLM+o0z8siw6aAGMUPZYo1JksOh+/paSoSh9xu0708oRfYbnp5W0+BrHaY1D6LdTghVT44kmUgZ8hnIsJIsVAXHp5vcfWwYXomwzWDFt847bPWSvOYTPNC0OCu3BAH/CpH1RJNfCZVgycXfe7c4PH3S7M8dSbFmckM7+zLc9uwxc2bm8yf6WUQj2O6zhbynDYNJlSF39o8TKXi8MhcwLpUiqVQM5hWfKE5zlqTY2cmTcu3GFQunrGpo5mSOj0kCeoUKJoUBVyeUTOMmRz7ZZLQhKRUikfkJFpi6panUgklFTIqHZMoxWCLzRc/8+2ui9Yq6OaEdNHFGwyPfvy39yplML4DLz9UuYPn/tdf2GtOzqN2rMe6cQtqVGFf38TaOYja3o97Txa1eSMmCIgePQ2Bj1gOavNVkC8hg+tQ17+FX/6r7wPw8K2f7hYgrwE+ede/3vPP7vo/9vyTO//FHolj1BKOOp3MgnZWQcTqaeUONukk+6VO7GR10syQlhQtkgA0WqwxWZ6UE5yS+bg4ESEkpCR56qZFWlIEJiIiTmoOTLisA7I6LjSLfykOeF+qeLpQYbLa/18pXu1CZLV9N8s0PKt1ed6oWP5Z259x5edq02ggLjzahaArLgEhgYkH6wohNCGSDKOW/+0bn4zKUFJFBlUvecnRMi3SpGiZIMndSdaLgydu5z5qmRYAJZWPNXlYLEmTJWkyrPNkcJmRGhni+9fBYiEpCHpMmroECEKrk94u3M4QRTJkjUOJLNfJZuq0mKHMYWuG42aaQCIGTYZ5qaExbDdr2KoHOaJmCNCcklneM+rxmJpmSsr8emEnRZPmW+EU89JgxHHxHEPKEkZNhpv6PNaZHPun4B9Ph4SRwnU0i37Ee8dcag2b56w5jkwqfDT7zATvs0a50vTyfCVkQeq8w8SF0jvMGLNS5Q+Pz7JR9zOravyPN1f5LwvjfOcoHHyhyAE1x8/cXGGblWPCNDmjyjTx2bDlBF8OT/OoNcGzrRoVHTLT0Fyl++lRNvXQoIAzuklKFBbC1arEkHJZozwiNNOqRgaL55ngi9YBAiKUWJzS05R1hZZp8X5zJYEJE9qd0EzyWyKj8U2AeSU/1j/B6BYhXXTxBsTMno/tffTuf7v3B1v/7JUN/B0wZ8YhW8QsBeA4mCCIk8yzRVAKohC1OYM+PQFOknzrpDCVBczMKR7K/O7eR3/6d7oFyGuA3/jYH+zRKwahtthEyUBp+Q9dW3QKvKhHYicpzBGaiqmxmLhaeTgEJiIjKRZ0hUetM2xgEIBB6aFhWvRRIINLnxSomQaeuJQkT8PEds7GnLUEXo7lA9vzUWEuVTem1UT155thf9W2uYouZLX9+XFgtfOzsih5PfCyu0kvcV5Wu+5Wurm1Awk1sXmDJx6OOAk9S3XuMRcHW+zO+1qmRd00UQij0seQ6mXRVIkdovLJwDTerm9CLGI6T58qJWGihgY+O/QQllFooC7xwFaLYbcejalhRjGq8yxIE9soPGPzXjVKyaTjz4OhN2XhGpsFabI+6mFUFyiZDNv0EGXqDEqR66IhpqXOiOnBMYqMcTimZtmgezHA5XqMr55pEEl8Xf7d4hwK4SOZ4TjJPKgwUxWeatTwRPHMfEBKFI4ISwSIGL57wrC1V/HoOPzp6TmujYb4RjjJUbXAmC4wWoSHZJLAaG6J1pJRig+bzXyXCQTFgjToMSlujUb47MNFisbjKTXL9VefZpvuJZOr4imhISF7d/aiMXxz3zZ+94oSY7pIAYdHrSkCEx/3so447rfI4wBwKAkktASeNoscNhVmVZ1rTD+ztGiaFnN6sUO1a+ckZSTNaVXGEFvkGxMr4lyJr4mcynQmiLo4F/ZLL9JFF138pELt2AyWjYiFidrsAs331f+5l2qyUBFuqf7GHoIK+siTyPA6pNAfLxf+i27x8Rrhl3763+5pz6CmJc7iaM++Qux3ZdCdQRHQGThFRBijO65LLdOKuxdYRCZKEptDxtUcQMdidMLMU5QcFVOnR3LY2EyaBQJCHGxCIiwUi6aKb2KRuqe8zmzuclxoIHupFh+rYTV3qldKNTufGH3lNl8vPcj5tvlGKh5/lNfbaN8/bQcrgz6nA+mK0xGuN3Wzc58VJItPQISbWPXGuSFDpkBFGhTJYRMrulxxGJMBmgSkcanSwGBoSNg5/y42GRNPBD0l83jGxkaIxJDDJZ8I1B+O5ohEkzMpPpQb4AeVGrYSQtEMmzRH1BI7dB8H1BwFMqRwOKjmcbAo6hQ2ioJxaJo+LBRrlUegNY9YpymaDFoMc9Lgo+4YZ+pxF/QFa55rojzTUicwmmu9PAM5w4G5WMxuKUPDaP7r/CTXmgHe6Q4y44dcrvsYdhy+wThfmAsIVMSjapo0Nk8Zn9/d1cfXDvj8+60j/NqRUxxXi5xCuMUM8+vXWfz+Yx7p3AS/eIfNb36nwLyMs84UOXA8wybdx1cb0xx+qkggTR5T01RoMK8DsmKzZAL6lcsRXSGNzbSqcU00SD3SzEqdXpPmpmiYv7MOs2SqaBPnZIUSdiZ8RISW8VlScUp9y/ikxKVhWjSJr5WGaSamBl2sRPeodNHFmxlOChyXB5c+uZerPnnexR7K/d5etsMt07+0hyjkwamf6RYfrwF++af/3Z6CZJgz8SxbaEJKKk9V14FY12HQqM5YKp6xNYlHTyxE1zjYBBJ2cgt0si6DQSfriICmbna6Jq7Es4P95Fkk7pSUJEeVBikclnQ1tug1ZSITYYtNzTTxTdApYi4Wr9fgerVtX8ghaqV+YGWmxo9aiFyoALnQPr3aaH+m5R2r8x2H5fvzau7f+XQbr+b6fxSkVCq5Z2JtSGhC+qRE2dTwcDv0mzXWQEK3iuijQERE1njMS5WcpGnhc4xZgDgrRCz6JE+FJh4uI6aHirSI0NwQjfGodYYrozUcsebImVg/kkqKhXml2WDyWAjTxuIaL893Wj6BaDLG5X8Y6qevp8nThyx26j6uKXoUM5oHp6toZdikezhkhWyOemlJxGPqBJ/KXM2ftA7wSXsnC76PZYSdg4rtOsfcXImMcfjFvjH2zh/ggVaGnHGIrIRaZgnHolkGo3Ws69X8x8lxdssa7t7icOi0w+PWSd5mRnlIpij5aarig8D+SNOQkLqUudvewM4xn+8ds/j4WyaoVULerddxfMLGweYPri3wfz+quPvWBf7moR5mZQqjFb/3QJrNkuWXdrn89rNlZhsZjqo53hVt5HvWGUI0d0RjADymptmp+1igxaik6MFFA5ujEr3KYSxrMV/p5+vWIR43mrKu4YqTTPJYna5GeyJHYxg388mEj+5YIMc5SToOqTQXpqm+WdEVpnfRxZsYp6Jb9p0Kbrxosdzp7Pv2nYpu6YrrXgM8dNeX9hxUs/SQpUxcdIgIWUnTMC1UIpq1ktlVjUmei6W0KklDFiSxE3UwxNz2iAgD2JKQtiReRiWJySFRZ8A3ID1oicW5NdMkLxlqpoUnqU5eSIuAIOmmhIQvGrxeqFuwcjB/KWgOVnY6lj+/2t+vlej+Ujg2bfw46Wgv9TlfrWuls55VXMjOIqEXJvQrT1L0qxIBEVoMRZWjbOL0cxeHOrE+Q4lQp0ndtHDFoc/kWJIa1+h1TMoSgtAnea6OxqipkIJJU5Y6wyZPhCGLS4DBl4i6BGRx2aKLbCTPAj4DeDSJCDCckRpG4ETUIIXNojRI43D7kMPB0ylOhy0y2KzNK56e0RR0mrekSzwdLrFG55lVdVLEHfGBMMegLvANTpE3KdaTI/AVn6tN8PODQzRrDl9qTHCTHmOrleVJmSUQjWccFgLNbjXKRi/F8SXNLqvI2oLi1JzFF/zT/ELvGA1fOG5qzKk6KSxahLQk4l1mjGdkhsulj3d8+H7+6EmPHxzP8I8TISelymRT42DRX+/lQX+WJ056rFEeTQ23jir2H8/zuJrmyJTLuKqgIod32GvoS1lsNUUIHbLYPK1mudEMMkmDRdVknWQpWjb7WaAsLTZIjnoQd63W6z4OyTQGTcu08JSHLRZpSSES56s4iZFHwzTJqHRyHQlIrAdpU/QsFB+65+3d384V6BYhXXTRRReXGB6880t7nlSzNCVkTPcwIxVcielPERoliijpedhYaNGJNWRcTMRZBg4iZ/M6LLE6BYruaEdiXYgrDq44+PgdwaxCkVYei9QYlB6iJCPEYKjTIiSMB12Jda8rDlHSU9Gc68X1ag2eX+tB+EuJy1/uYPjCg90LL/PjcMY67z6c53OvdM16Lc+HLPvX/vvVCGqEuLhvO8mtPP5tapUhvicQQ800EISMeBQlS4/k6JcCTfFpmCYjqo8MHkumjgBN8QkknjVvSJi40RXwcOIwQmODxIYRgybLc2qaNCk+6A0zEubR2uImuxdjhAnTZJPkGXZcxnUTC0UgmgGT5ho3T5+kqOq4m/XcPKx1U3ja5uohi95iQH/K4mglJGUsKjruQGw2BdLYXKVKPGMWMcAd1iDX5jPYRvFAOMdptUCzkuEDmxXfWSozKw3qBq6klxmaZHBZlAZnqDMRBhRwOagrFMIU3wgn+YWBEWarwpeCM9zBMNM0UcSi9FAijkqFd+v1HIqqfO3JNSDCr+zI8pX5RQrGY9Kq8qHUCG+9+QS35HuYmcpy1y0L6Nk+Dp/M8cGrGryllOO+uQU+aq9jKvKxjOL+aIoJ7dOHR15sjFE0iBIBv+IAC9Qjw2ZTYIvkOagrrHc8AgNfVoep0SCdOJzZyXesn9BRkZi6ahHbOLdoJZM8mrTEtFRHbP7i87+zt1uArI5uEdJFF110cQnhL+76qz0nE5EjIpyRBYakhyFTpIccZRoMSw9aDClcajSImeHE7ixJh8QRm8gkHY+EPuAmIWuwXGOgQSAjGULC2OGnbTNqIhxxKEkOjSGNS50WtihCNCEhUdKHEVG0jL+swDHnDOgupCt4vWf4X872V9ODXHyBcDaQcOXxWbnMuc+cW9C9lsfsQh2P1+O8Ld9mO7zv5RZo7WXbXUER6YiNwTBkDRAkJf/ytHQ7cT5KSWzF2quKiQ1ulilZokQOI7Hj3E49TEEyjJoSNfHZoYe5Vg8xoWoMmQIlk6YiLQrGw0IwAn67Q4nNu2SEo36TltFYCCd0g0UC3t1b4GCjwaIO+fnLXNxajo9sER6eDalGmh15l5NNn2ucHuZ1QErbpJRQ8GDfKU2zaXHM1JlNBubb7RzHdI2ahLjaoonmpnSR436LtLFwLOFoWGfQ5LjCKfCFuUXmpYYtFiXtcUxVqEvAVbqf9/aU6A9ynDJ1bszm+V40ze25Hh5tLfJ8LeDpaAENTNPiSt3HZpXD1Q4NCamJzzZ6eFxNMSc1bjbDPDDj829usJkfL+Ib6Dceqpnn7w8rmmiqswWGC4b7qjM8NW7xkRtP88UXHMZ1TGdbwCcSw02qn5aJr1wHxTY3zWIU0UuKy6WHZ2QOH8NlXhYrsqlEmn7H4uvRYULi70CfAEsUDdMiJS5N0yIg6OTGeJLCxycwQRJiCWnxcMTmA3fv7hYg50G3COmiiy66uETwV3f9tz0Oij6TZszkmJUmJN2MpgRUpUWZGrbYNBJhuk44yAaSnI42qz8uBtricZWEaCGCEsESO05pVmnS4qHRNE0LI7HdaFEVEISSKhChYx0IdRZMJRmgaUITxVxnWa4lOMvgX60b8qJZ6At0GF4LvBoD65dXiEC7ALmQFuScFPXXsUhb7Xy9noXjuduNr7k4TVwtK4Av/P72uTprYX32/KdUig1qGKPiIqftgARt+qKVdCIjbGwCiWhKQECAT9RZp5Y4GHTM5JlWNTTgGYcUDiksZlSNQZ3DQTEtNUomTQoLX+LCp6YNB61ZcsajRkhDQkbJkhObp8MykRiqcxn60ooHz5jkuwC2Fmwm6hpfwxbXY3OfUGsJj5QbHFNlFqIQBE5aS1ytSjwf1RkmTR6HBhE35XKcaoQ8rWZYS47vBrNMqyohmmOmyh3WICZy8HDYqQpssXIMRzkM8EBrgf3M8/NDw3x2YZYAzVwDrpU+drg5eqMMW1WB09S5I1/gS/4ZFqXJdWaAeXwsbTOuyni4jFNnJyXCapZv+lO8yxugHhnuuH6c7x7z2Oalue3KeQYHq3z1VMC8avDQ4RK/eUWO5nSRw7LIB9PDPBUuclu2SD2ATTmHKIyv3XkdMKFqeNphQtUI0VxmF6iEmiaav+Ug1YRmZ9r3a3IdREk6ekCIK27ne1ghpFS8vJ9YnxuBn7r7bd0i5DzoWvR20UUXXVwi2EGB3ekSNop8YqObwqGdRWCh2MYoPgEFyVCQDLbYSV664GAhidNOW4AOZ4MJATLikZccFopeVSAnadbJADeaTR1aVkpSKBRb1RhV06BJQM64WMRJym0b0yjhRAOEJk5dvxCWC5+Xuz2t1FWcL5vilQx8L/a9P2rmx4XW36b0XAjL8yngxWL112vgv9r5eT2xmrNaZCJWWle/1Pvbj55Kk1IpBq1+Lrc3k5cch/QJZqIF7jDb49ls7Dj/gbbrVYasZKiaOku6ikIokcM3QXLPuoRoZqXCE9Y4IRE2QlMi7MRqdo3Oczk9tIhYp4tkjI0GAiIcLALRjOkiAIHEeohey2F/Pbap9YzFgg6pB3B5j8u1+TR5bPbNNGKRdcblOb/Bt6ca8efE4lr6yOIwaDy2R324Skih2JJzmKDBbX1pXAuuHoivxYylqIvP+8w6+k2Wf7lxgOfCGptVjglV4RmzxL5omqdknifUDAbDx1Jr+fT0OABpbCZVbLOYsiFvW2woKm6RAb5VWSSFze0M84TEQv0zqoIQU037TJrDLPHIUgON4a3XTaMAN9VAATOtkD/8ocXvPOhyB8Nsj/rYKUX++OmQI7pKhOGpep2PumP8oFJDgOmGpq41T0ZL8XElYo4WReNxO8OcaYb02hb3qf3M6AUgnpDxTdDJjHGW2TCnxYsLD9yOGUfDNEmLR5/qwSd+3y/+9L/Zc7HX95sN3U5IF1100cUlAvnK0d2BhpI4uErhRSnGTB4bm4r4hEQ0xKdhfNIJ97iJTwuftHiERKQlhU+ISCyONcv+/Zcv/N7eD9y9e9/n7/3mbkdsBqWHy/Qa+nSanaoI4rGdNdRUyE5GqEgLXwJ26VHWmjxZPI4y3Znpa2tPFIKWs2qQGBe2dV3egXipgf1qRchqr61cz8V0XVZb30VrPC562VUKG5Fznr8YO9lLoQuxGjXs9cLyQhbiYk865gwv/T6IC+kITcv41Ghii816NcgCFU4zz42ylXmp4ZsAT6WSBPTY5jqbUBh9QoqSoUbsLmeLRZUGGs0uPUKWFGdkkXeqUZ5lnh6Tpt+kmZAGReOSwUID/XgUjceStGhJyN2lQZ5t1nl/oZcTTZ8zNAgxFHHZ6mSY1T4bsw7HKhFTrYisspg1PgUcokjIi82SCbms6JLFZiGM6FHx/bre9Tgd+KyxUhxqNXhnX55yMy4W/r+FcW5nmINhjWlV4zq3xKB4BC0bCW2+zySBROzUvdyaKaJ9m7dmSzwX1jgVNtllermtkOdpv4yN4gx1Fn3DcVPjhWaLjbZHEAm7rALfMRP4ElETv0NLKxqPK+hlZyrHY3qeD3tr+MujPseoEp1cx4N6inGpc6X08s9uaPLp8XlutftwlHBUV5lVdd5vjfE0C+x0czwQTbOOHCMZi+G0xX3BUfbLaRap8yF7A9ucHI+HSwTGcEo3eJbTyZViOnTTTlaMKIJlRFRHHBqmmVyHdDrSqUQjB7FhyFc/c//uD3ZpWS9CtwjpoosuurhEMPmZI7sjDQeDGidMnbeV8iy2DBlj02cyHFcLpHFIiZP8CBp6JIsrLkumiiM2Lg6tZAauPTts0Pz1F/6gY6v84XvesW/fZx/ZPW8qeOJRMCkOyyJ9OsMJaxGNYdDk2KxLNJWhN6GKCEJJ8ozLYkxnMTHVK0Kjje4M7fRFBO2tFBmvhvMVFivfs7KDsvz51Z5bbZ0vR+T8cgffq2k/VhYhF/u+9vZfD7zeRcdKrCxoY33Ii6+DlKRQYuGITc7K0UwGjTEMrjiUVJGQkIzyaOBT1fW4uFdpZswitlhJlzB2wEqJS0CALTaA9b7nAAAgAElEQVQGQ5k6BvCSyYEMKWwsbjLDLODj4pCKXOakSZ/JMCtNcsYhg811+Qxl3zAudYq4hBjuLA7w7YUydQkJmzaz0mRBGlxJLwohMDEF60wrYMEErLFd0pZio5ciioRnzCKbnQx9yuFYLWQkaxGEwh1bA5SfYvtowI5+GMgJm7IOfUWfP5+e4ZHWIu9SI3zHTFCRmJ55MmrwvKmw08nzQqvJLunhGrvEE3qBRd8wLjU+sNHmwfkmZWlRJ+KZoMKSNDvp8lXxMQJXSg+PRgtMqTo7nTzZwOOkKpM3KW42Q+yQHmxtM0OL+TBkC7EFcaxbaTEcZTlGld+/Ps38XJbPnwj4zetdjkykGMwo+qIMIybHP5pxLBTXZnPssAqkLcWpRshfB0dYNDUMhpCI/SzymJ7haj1MjZB/VIc6rn8Wsd7IxiJEd64Yvew7rE13dcVFiWBLXKykEjdCLYY//dzevd0CZHV06VhddNFFF5cI1pfguk0+I+Kx1mQ5thRxRW9Mskqh2KGHCNGkialRS6ZKSETNNFBJUdAwsWOOkwSoGTSyylf973/2X+4NiViUGoFopqXCTivP9dEa3hZtJGMcnrAmqdKkRUiVgF5SFHWKW83WeHAh7SRplQzG9IsSp1di5ez1aq+vxGp2vyszOl4q62O1ZX8UPcpyCtnKbZyz3DIK1mraj5fKBrnQMq9nOOClFEx4Pi3PynPoG5/QBASE+CbAEfec12O71fi67aOQFDRxIoRPyLDq66zTTdK1W6aFQnVMHwTBwaaZ0HI+IdtpEfKUzKGBnHE5JVU26CJZbDaZPJOqyiPWGR6oVLAQPtrbyxmpss5k+c5iGYVQMC4/sCbYaPLkTIpx08RTipOmzu2jMYVrVypLPdJs6jOcrIfcedsMdw/0UvKErYOGK3odfrBUp6kNnhew1DRMz7tkMj7jczbPzwjpdECLiNvNGh7Q00SiGdUFPpFax5I0GdRZ5puGIZUiayvectUSH8gOMKGqXC29/PlzTd5qDfIW1hCIxicklVDM2t8RGeOQtRRBomP7uj9JgMYxFrczRMG2yFrxcZ5QFY6rJaZMi3Ul2GoKXK+HSCnFbWaY/pFTfKs5w4Sq8LuPRmRtxVPlJgMZYSAVW4/fxABfWVwg7xm+0prg8+og02YxMTSPr51FXWZSz/IV6yD/oA7RSIJc43BJwROXMKGZtidcHCw8cc/2JZN73SaeBLKx8BO78jSpV3qZ/0SjW4R00UUXXVwiEIGlikOPYzFgO/S4FhMVQyr5kbtSerg+GmV91IOFok8KaAwDUqSoch2SgMGcw13+6y/8/t7VtvefP/dbe/WywbSrhJxY5MRivWQYNDlGTQ8AWRwmpE4Bh4/19fHH/bfyZ0N3XOCzvDQtBs7tQqwsJpYvs9ryK1+7mG2spDStfHwpitj5PtOLtAoXUWS0By/LH1fqR853TF4PXCr7sRLLj1ucZG5hi40tNk4iLDYYtIlo6gY9qsBae4SMygBgjKEgWbaqMRapAZCRNFdYm1hrehk0BdaqQZqmRcO0GJE+MpImK2lS4pJKhp4DUiArHn0UeC6ssc70cp3pZ9Ck8YyNAuqELOFz04DLdl3ihmiUOdXgpiGHr80tMaWqPKKmuD1XYJPKMKmqjOoCI67DrVY/61SalAj9pDg9r/BQuBb0OhZeSnPX9VX+4IEsZxahL695eFzj2ibJZocjJ/KU0sKmtRX27c+xdiDg2o0+f/i0z8/lxnhUZqgTUDQek6pKoOET7gamVJWCK6zNWawvwdJSmlwqLjB29inekivy/XCOI7pCkwAHiywukWiaclYr9kA4i4tF2jhUxaeF5n1qjDUZi/ujKUppwUlS5CvSoojDoVlNShSHZJG3XVnBFeG+r1/FtKryscwoJ9UCjoJdOQ9LQT4Fa3WBL8tRDqkpGr7wNCfYaPo7Ia0KRWBC8iqLoKibZnL9KGxinZ1P0NF4KIQgEaT7BJ3vFIXQMi1aJrYlsMXqfAv7JqCF/5reC280dBPTu+iiiy4uAfT8qy/syW+o88KxXhwVU3WmWyGOCEsmxEEY8CwO10NmVI1+k8NJ6AIvyAyBichLjrpp0Kd6mIpmscXmv37h369agLTx25/9lfO+fviu/74nY+KfiS12hm0DaYJIODxnuHlTiI6EP//8v3vR+3/2zk/tEeIuzMXgQp2RlUGGq3VFVnvP8nUvf7zQcisLotX2b3kX5tUIWVxegFxM4dLFuTAYSI6bJPqkCLDF7oTFhUl3Is56cFjUZUKVoUflGVS9zOgF/Dh+kCJphkyBaaniGRvbKEIVr9+TFC18bBQDUmCJRkLFcXASzcDbo/+fvTePjuw8z/x+73e32quwAw00eiXZXJoUqYVaSdHabEqyJFu25eNFPrYnsT0T25nxSc5MkrGZmXjsM7EjOxM73vexRMmSrMXOaLMoShYpihQp7mySvQMNoABUobZbd/ne/HELEBpCN5uSSNFW/frgACjcureAulX9Pfd9n/c5xMOmTk8SpmyJc/RZMG1mbYmK+pyTLm+vjPOJlTZ9Ut40UoV1+NhSm1SUn8jv5w96T/LB7iJvDWY43KthgUeiLqNkFZwlQm6plal3lU84T/Mv9AhPR32OeobfusfwQ/vy/OmpdXrnahwquYDy8imP+5ZT4hT6CagK9STmT45nQahNCXnRkTX+9N4YTw235ib5q/5JFvsxDsJb/FkcA1EKxXzKWtPnb9fW+YHSDB9dafL2qQpRNyEVSw6PFn1+vLSXj7TqRAMRYhCOMsKDrCMIVc1xhZ/ngajN//SSFqfumGSjD/ebZf7ojX0+8bnD3B6eIZVs8tR1doK/uA/6hLzuMnjgwQniFL5fDuEa+EynQVV9XlErcMo0WdBVpmWU/9x5iK7t8Yg5+7VzBgjEp6d99ptpzunalqiISSlIDqMycH5klRwhG4JQlAIuDiqZmPHwBiLEbomZlrYpSP75eAn8k2ZYCRkyZMiQFwDNls/dD44xPtLn8pmYK6YtBWNwJKtOpCiPdUPWTci8rRJKstUmUaNIXgJy4hEM/CFGnGx87jdBVX32mwLzpkDOER5dhmZXCK3lvhMe9gIXxv/qg79xUeFzsWrHbpWKnYv97Qv+S8nQuFj71KWa1y9WjXk27NamtTl555mOO2R3Np/fRGMijUgHo6M3/Qi++Pjib7VhueISa0KFrBLyIjmAg8MqG4TEFNRDsXQlpm46W4vTmpQI8FnWJi1CDug4VQpZkCceJc1xj3OWy+wonm5OrBNudibpkhBjKahLKae0JSaUhDvXO6zT5/qgzIvdKrd3z3JjOse69DACbRIMEGPZX/CYzXkc9Uv0YvCM8ENczsf6ixz0A44v+pwyTZ4857ImXfKOUO8pi03hnqWsAnC6nXLlfMhyvcC4m7WWfVexxo9U9vCf7oGfKM5zyI7wdDfiR4J9THouE77LRAk+3D1HNa8srrlEidCUkI+367zYr/De5TUSLO8oTfL24iQ5XD7ZamxVNV4lE/TJLqpYUV7hjPEaZ4I74lUud4p86St7KDhZVectzhyfv+swL7u6QSRZXeFqO87TZoMHzQp16VEud5k32TSv8bzw6737eVWuRhWP+xoh3+3uAWBd21iU7+el9LVPTEqoEXkJCDXLE2nSGcwXlCwPhM3Mo+w17g4EJmRT7DZbYAOyvJCuhjjinFdF8fG2gl+HXJihMX3IkCFDXgCs3f7Ua0eKyvzeVZ46VWViJOLgdETZ8bhiynLVBDyyCkedKiVcOqqsSYc1E9IjYlZHaEuEEaGnfRR9xirIM3H5D111x9oHjr1WEAIjNJOUkmtoJRarMPlHb73g/r/vh954x4dv//RrVWC3KVDb2592Vhq23779tu37uJhYuNiI392Oe6H7XKogeSbON5ifL16cQdta1lL09ROzhiLk0tnszxeRbGqbOFm7lATEJDjiMO7U6GuELx6TUqNNyARlOtKnpxGJpHQkZp02RbKJc31iSuQY0yKTVDjLGpNSY1XatOmRqOVynSaUhKN2ijOmRVkDFk2bKS0QW0hQGtInlBSvmyNS5VXFKo8lHaYp0Ektp20ICFV8xrRAkhh8DPNejtPaY7+f479GJzCJz2zgU8nBJ3p1rmOMe+wax+IePz05zWTVst7wmcv5fDA5zSuLVUqOQz+BayYND5xxuHK+x7k1j0NBnvGy4jnwhgPKatPnqpqHZ10qebi33eXyss9LbzgNZ2cRhMCDj603eENugofTJkeDMu0INiSiHiqnomwBH0qCg5DH44tylormOOQWOWNDOtayZmNOO01eUxwhTMAR4bPhGqMa8Kl+nUfOBuzXCtNa4kg+TyEJWJGQAJf1syNM5R1OdxP+KHmEn/ev5YFehxuqOU73I6rGYzwd5fXOXkbSCj0SnpblrGolbja8g8ycnmrKiKnQ1i4gFCSPSDbO3EpW/TCDUExHDFYtZVMkJKJkCgT4BOLR0i4pKY44eJKJkN/7wG3f1HvwP3eG7VhDhgwZ8gLgxhvO8MV751A1XHflCmtrZe49VmS2puRzMSureW4cNcxO9Pm7xwVXDEfTaU6bDV5iZ/iCc4pJrbAsG7u2SH2jXH37m7f2dWDwefYS7vcj3/dvfhnYao/ZzvbF/86Kx8UW3heroFysbWv7drs9ht32+UytVs9oYB9UPDarHJvtVrtVQrZvu5MXqg/jhYqqzSofeHQ1a5fKS46aVOhqDx+PCTNCS7ss02CaEc7IOoftBMfNKnu0xhlZZ6+O05U+r073AvBp5zj77R7Omg0AlnSdo+zllKwR06WkHqfMGidMkyvsCC8fD/jDtSbXlHLkPPjkeo9by2P8bWuFuZJD1Ao4003w1BBgODrqUS04QJ4vL0DOOkz5Lo5xEeBQUgJgxpYp4HKqk/BIp8G7xsYp5Sw3xOP8zXKT/Xs3OLNQZkVC1sIc73D2Uu8q4wVhv+eQpIoq3P9kkemKMjvR49cf3+B7ginWOkV8B1xHKfrCB9br3Foa53ONNqWH57gzWiWJlB+bGuOtozU2wixro+jDd5XyvG85ZMV0yatL3XQpa0BPEkZtjpoWGNEcIjCiASdMkxHNMZ9WeXQjYtR1udMu88O1aUTg842EW0ZLfHy9wdWmwkZkeVDWCDSrNDyqLdY6eT7lPMXr08OUAzjZb5JrOrxmPM//vv4wqZPyWQXrKF0NB1UKh772qZoS4cDz4Yq7laNkUdraHbRlJeQlRyQxBiFBs+3JWv0ArCp9IgLNQgs9celrxB//za8OxcclMGzHGjJkyJAXCC+77hzdTo6Jo/dRKvVRhcsO1imXuxRyKVceavD0QsBlJZ95LVLGQ1GaRBQIWJcOv/b+X3pB/Of3Vx/8jdvsoId6O7v5Onb+fPvnC223fV/P5ON4JrYLot0qIpeS4fF1t+3SYrUpRLaLj+3JKpsG6y2fyEWqMUO+ns1zIdKItnZJNSXWODMY00fEEOASaoSLwwRVWvSISTlh1uiTUJes/aotIVlwnvJ55zQxKcecVUa1gItLQsojcpaYlGuYoyUR++0o69LjpaM57q5H/Pfjs1yxt0chUMYJ+NRGg3+xd5TEZlf925qyX4ocHfXwHOXxJcPdZ5XXXpZw3ZjHdQdC8i7c3+vQ0oQ/C4/zlKnTIma+6PKvry7w9/UWTy4bwlh4x0SVrz5RZXnD8D8cyZN3hFdclRnK/6B1kmKg/P7qWSwwP5aysAHveazDq2SCWh5SC74LjU52/rUk5LPtBm/dU+D2pQZdiXmZGSVJs20/0a2TwyO18P7lBge1TKAOHg4vS6cBmLZFuiS80ZmhLj2+FDe4zClSwuc13hgjBCwRUnCyY/5R8zSX79/gbcEeuhG8xMsCG4+lHUrqU9WAQ5SpS4cPm4eoaxMDfLbR4nVmho+ah3nP+nEAOhqyoV3a2qMmZbKRugk1U6GtvS0fUKIJdW3iSDb1asSU8QeVjJ6GWRCrJjgYAgmyc4yv+YwCfKLBuOZYk+Fr9lkwFCFDhgwZ8gKgMnaWsUOPMr33JGa8Q6HU4U2vPs3GRhExinEUVeHgnj55D0Zdl0Xp8rZgljkpEBEzosVv969xHn/9wd+87eumRnH+aN3tn79RdvOIXEor026C52KVkUs5/q4/v1ilY3Cbh3ueR2TIs8eIgxEnE3eabgmSlm1vfSzpOntlnHGpcFZXOZ4ukJDQI6KAT4MOeQIsymvSOZ42mU/klvQgh9JR7jMnsVgMhlQt45Tpk3DOtLLHgPCJtQ1eMhowPRby+49GfHkp4VXTPjcVa3R6hsVuSs4IM67PTM5leizi6XXl5Vf08EVIrZD3LScWc/xh9zgvLxeJsbyVfYxqkdNmg42+8l8fgpvLFY7syYR+qyd8bqPF7FjCsTN5Sr5w9lyJ93cXeJe/j79YrvO93hyf1yX+34U6V0xbXuON8aYbNsj7yhe7G3T6EKfQi+GnR2f52asDPrjQ5vsnarw5mGYkMPx1fZWPNdYJJaGgHnc0W/RJMQIBLoE6nJQ2rhraEnO1W+bxuEtu0HwzVTCExBR9oUnEa0oVHup3uEHHudWZ4+xiiSv39tk/GbN/FB5JN2hLZpi/1ivzEfMED3OaW+zlW0nlc5Lnz/QROtpjUde2xjFnRvIcTW2jg4siLe0wITWqkg33KJnC1sWSSGM2bIdUU0ZNBV+8QY5MGZHM65GN7838H5tCxsGhJHnykqMkhW/D2f9Pk6EIGTJkyJAXAGZkA7wE1+9y9pOvZfTo/QAcvOp+Ou08J8/5hKHHZ4+5HJjt0rWWm3IjLIQJC9pjxlZxXoBX4N73od+67b0fes9t7/3Qe57RrL5dnOxW4dhuPN9tdO234grkt2ok7s62q92ExWYLyGaw5IUez5ALs3MYwebf1BN/q48/a9HyUM2ubLcJadChYbPWquV0DR+XLhHzOkqPPtene7jXLHONjnBNOsPDps5Rt0KAz0GZZlbGqEiBNiFr0iEm4ZRZp6wBGxJRLaT86uMNYrE8JOs8vqKMFpXAy2bGjeUMNx7u00uUdsdlLGfodj2u3wOfedLwfy2foR3B22U/f9Y9xStHC3zZrnGrO8uP1/ZwZMbyptkc66ES9g1PN7JF9FG/RLPt4ruwZyzhj5eXeYu/hwOTCeOaZ6aqLEuLlvTJBSmfjev8zj0uf75U5xWFCo4BIzBWUj5ab+L7Ma8pVTAC4yXls711Qom5pTDCW3PTvG1klLbEGGBZ+4jCiumiKFUNCElYSCI6m9PH1Cfw4FZ3D3e329w6VsUITEvAfNElMNnzt9b0OXjZaT5zLuSmYo0RDfi0eZQ/to9Q1yappnzWHMPBsELI+8zDNLUNZBku0WAwgaL0NT7PJG7RzMtDgiceAT7jpkakmSm9KHksStO2t4Z7tLVHogl97eOLh8GQ8LWqh8USaoQnDl3tPden/T8bhiJkyJAhQ14A3PM9v3kbgDO/xNT8McxEDxHL6sJhJvcsMjWS0u16vOZQSqvts6/kMDdqqbkOPoZXueNcbye/3b/GBXnXO37xl5+p8vFsRcVuFYxLHZm7M49j+36+EQ/IhY6x23E227K2Cw/F7r7tC1BYvtA4vw3va3/DzUDNilPBxyNn8iSacNau0LIdcibHuDOCiOBgGKVETfNck86wLiEvtpM8IKsckhI32mkWkj59ItpkY1gTLAftGBblxeksMSkr0uIHR8f58gIsS4sXuzW+vzzJQ+kGi01htWUoOgbXQKfncmAMTq0ZHmv3WVr3eHLZcEa7/FR5ng/0T/NhPcGP5ef5w8Zp3lYd5+hcQqMHJ1YclpsGEXhiRZgvO8QWxgtCswftPvz5qUxs/F20SNh3eO1IifFaxM/kL+cXJvbiGOVdY+Pc45zmFy+v8KVui5GixXXg9pU1fByOnazw8hct0w4lyzEapKDf3WlxZ7fB/7fWxMWQU5dVCQkla2lLyCaKTWqBRcmyV17qjHJ1UOTza12+GrW5Nihx32pEJ4Yz2uPvOnU+Hi/g+9ngi0cf3keXhLs7LVyESGNWbYO+9gHoag9PXFwM67a1VRUBBpkdLoJgBrOvNjNkVC0btkNP+1gsPe3T0i45CUixW+GvvniDykaeUamQk4BAAiqDSkdBcghCUXJYLEXJ42AueEFhyNczFCFDhgwZ8gLhnu/5zdvEt0iuz+o/3EirUWPy5s8TdkvEsVCr9Ti36lOrhvguOI7iSLbYOvK+W2+76YNve0H4QXayKUAuNm1qp0l9k53+ke3m9Z2TrS6V7ZkcOwMDN7/e+bH9Z5fC9tYqV7zz9muQrwsoTDXdtWIyrIRcnO3ngKolJd0azevg4OESaYxF8cXjsLOX/WaGvWaSAzKNh8u8mSYhZZWspSrA4azZYJ2IHgl3yRKPSYOnzQYVCrzZHqSsASMUeYU3So+Ix51V5nSEqha4fa3OXMHhoB3jznSFrzb71E2XuRHlwEyfkUBY6SrtrkMxn3D9oR59LLViyoFx5T7nDEbgjTJLky55H95kZvm75jpPnfPoJoo7yOyoBcKesvCJZoN+An+5kWVh1ArKD0zWyKnLuvRwHWW8mpAkhsvnQt67tM7vPNHh4/UNftwc4a8fT7i5WqZajqkWLD88NYqPoRXCf/yChwj81Uqdd5VnsCgBDi/2q7QlJibFxXCFVrFAVXPs1zJ9Sfmqs4RFqZsu/5jW+Uq/xWnT4ozZ4Ew/4kvOORb7MaGk1E2HWVvmD041+NRyl/sWYJwAD8MDzjIG4V/Ky7mFayiZAjdxJQB3mie3zofNSlik8daY5ojs+c+JtyUQgsG4Zh8PT5yt+zsYSlIYiI0ighBpQo8+BoPFEmuKM1g+b4oYg6GjPdZtC2fb/oZcnOF0rCFDhgx5AXH3y37vNl4G8//5L355z8GHkaIyMv8EN06e4JGvvISRcsLKL//AbRXAApcNPl7IbLZi/dA7fuGXd/7s2Uyy2vnzb7Q6sZ3dfBg7p1hd6n63T8Havs+U9Lz92cFV4s2vt9//YgGKQy7MViufWhAnG8mLgyOGCTPCmm1mV8AlC+c7ontYkg2utjO0JWK/rfIl5wyQZXJs0GXRtFmTNpfbSdaki4dDgYAnaHGLmeZP9WGORT1eyX7WJaQrMRvSoyldTnZqTGqeBbqMOx7jaYF7l1KuS31GS8pKT+n0DWeaLpNFj1FJWG46BB78h8kjbPTg45xiVms8tp4wk3N5EVWunA+59+mAQ9MRnmdZWMnhGOWU02QhbvFKnaGQra+5dzlhXAv80sEJnl4y/EO3SUv6jNsCb66OclezS1MiQqvUpcc/NmF+yqVUTFhrehw3DV5iirx9rEY7hNcFE3yh2cEzDlf4eRrJYEIUSg6HgjFM2wI5LfOwqdOWiEktUVIfD4cUSxkPbIFV06NLynxa5SQdRgk4pw6Lps2NTLKkfR7TFj1JuNEZ5eN2CRHDX8pDxKRcb+eZ0BxIVhHJpvBZnIE5PNaYQHxcyUSoIw6hxluvUQ+XrvaIRUChIAFt7SFiBoGULj3ts5kdYlUZlQoNbdGjn+WAqEUQOtqjYor0NcYXj6Lknv8XwD9RhiJkyJAhQ16ATF17L+Kn2GUfM90AAzNLqzz1Cz/5gqx2XArv+9BvbT32TUFyKUnkO7fZyaWKkd0qINu/d3BIv26gcHZ11e5INL+QmNkNVYsjzq5tGueJn11M/EMuzs6qmSBZVQlFsSQKDTayjBCp4uGwqlnFo0MfizJq8zzkrLDXjjCpeT7jPEWoWRVEUU6aNQ7YMUY0ICTFw3CnXSYnHo+bNa6yYzxpVnhluo8rGOFQLsfHogVUlB8pz7LaU9zUUNc+1ZLPF04INc9hz1hMsuIRxjBbcHEM/G7zBL92ZIpfe2yR75MD5FzBNTA7Yjm2EHPyXI6cA/eccDkwInxobY3vLo0SEnGD3cM/OKe5byNHSX3WpcfNMsOdx5WHqfPO0XH+dH2Bt0yW+djyBsumw6jN80i6gRFhSbp84IkiP/vqNp3uOAnKW3/4k4QLe/nkHUf4StgilJSYlJNRxAlpEUlCTl0ec1apptPkcLiiEBB2R/iSc4Yxm/krZrXAA2aFnLrkcXHU0CKmis8p02LM5ijgUdGAB1knFeUqanQ15U/0wS1vxqbv4z45SWB8Uptuva5zEhBrzIwZY9Gu4opLRYos6RpGhf7g+Q5waGqbwkAsxCQkmqWkOzgolnXbIpAsjDAQn4rkaWuPWBPGTQ0Hw5JdA6AkhS3BUpEiHsNKyKUybMcaMmTIkBcgZqSHjPZwjrjET80hhRTXi77dD+tbwvaKyIWSyHdOuXqmVq1LYTcBsr3darOVJ8s+zo7jDNKSDbLVZrFb1sdOdrZw7RQgqaZfN754yDfPdsG6mZRekDzjUmVNN1jRJjMySoolhzdIuTZ46lA3HZ4yDcYoA1lb1gE7DsBjZokvO2c5aZpYlP22wo3pXq6wo6QoN6RzADxtNliNUr432MNLdJIbrl4hVdgnBdrEfO6EUHIcEguPnnXpxfDHveOMl5SbX/0YbzP7+LePLRPgcngcDk6kvOLoGosNw2X5gF4M7VjxTTZF6x2jo3ykvQLAounQIaQubZ4y2fjce3WN+2SVU2adj641+LnpaT613OXnX6J8jzdDVxJ+8rICvjr85J4x3rzP59EnJvA9y7+anOXf/8Er+cIXj/DR7jJVfM5Jm0lbpE5IKDHztkJBPSZtkQhLjOWpbhZW+PJ0joiUAId1Isa1SCyWBMthreAg7PNyvELGWaePq4b9FCmoRywpeTH8vfN49nyKoWQyL4ZF6WiPNds8rwLZ1ZCqyfJUKqZIpPFgKtZmRdIO8j6yyVdlKZx3wSPSBMXS0czz4+HiD1opN98Dxk0Ni2Yme1OiakrEJBgMxYFHZMilMxQhQ4YMGfIC5O6X/d5tIkDgk0QF7rrmD297/Od+5p9sFe6p33kAACAASURBVGQn21uNdo7s3S2AcDvPVBm56HF3qYZAJja2f5aBmXW7v8CiWYvP4N/OfexsxdoucLb7PS4oXIYLmGfFbm1728na4ISmtmlom7wETEqNAJczss5L0zkWzAYnnAahxBTUG7QrFVGUpoT0JGZKKxxNZ5i1NRrSoU9KDocYS5eEkJS+pHRJeKUZp2NT7gjXmM15fPmhCVSh5jlMkCMn2WN8nx6jlypWlTfqPL9dP82n7jjC/XGT/2XvLG+WvVxzzSkC3/LYU6Ncc6BH3oOz3YSKLzgCS23lT9cX+O78BAAh8ZZX4SfcKzgj6yyaJmdknTfrQS6XzPOxLn2SxLDQz1LDGxvZQjtJBc9RKsWE20/0eNGLnuKwW2BuMuSIrTHj+rzdm6M7CO+btEX6Az/IOdOmTebBOJD3uaFYpIiLChRxaUvmy7jBqVHCo0nMGbPBepLwaV2gKzGXa5VPmhOEkvBSJliyEU3b3no+O4NgQV+8rQsCqpaaqRDakJz4bGiXujYZocSoqdLTLO/FFZdA/EHF06Aoa7ZJTnwMhkAyb0ioMR4uo6aKka+9R4WaCauImIbdyJLRt70PFCQgHBjj/+P7//U/m/fp5xrn3e9+92u/3Q9iyJAhQ4Z8PWemvveOM7k33rF0/Svv+HY/lm8Vm1WQ3RLLt1c7LjSlaqdoefYL900PxraKilpEBINznqRxBy1Ugfj0ifDFIxlc9VTs1mO0O7wgmfiQ8/YtIjhitrZl25GGbVffGBcLtBQER7K08b72iYhJsJSkQIM2EQkrpktdN7jBzjKvFY6bBgahKxFVKeJi8HHw1KFhepQJUGC/VjghbYp49CThhLPOqM1zg1/hK/EGa9Lnld4IVsEzwmwNZkcTTjaEquvylled4y9ONXmxTPM39gTXOiO8LFcj70E/dFhoChborY+Q85XxWh/HsdQbPp4Y+qnykeQMh6XCNX6FD/fPEkoMg0XzrXqAT9oFfrp4kEejDi0Jsepwy3gRx8ADzYhHzrnc7yyTV4+VthBKSinK8fl6nytqLlEr4MGnR7hhPuX2JxMWpcut+zzuqyfckC/zaNqigMu69Kng4wz+VnlceqlyOoqIsLgYHjF1ZrREBY8vscyEFnjMrHGVHUcQRjXPE87qIHnDwQqEVpmWHHdxgrzkSEixaimaPF0NkcFzLGLOm4SVw898HJLQ0i6uuHgDn0hCSk4CLJaIhJQ0m5omPi4Obe1hMHjiYjDk8VFRPHFR2EpRD8QnJqVLSEECXDF0NNwKxnzLD97yz+b9+rlmKEKGDBkyZMjzwnYBsv3zzhyQ7eyWYL7JsxUgm4v/89uxBNDBAi47oiC44g68HC52kKosgwWK3eEbMWK2msey42wzSQ++zwzTg2MNBci3hJ3nz/bbsylkhoQUUHyTtWWt6wYODn0iuoRYLH0Dqybk+nSKM6ZFSX2sKA063Kx7OSNt8uqxR4sowj3OGRrSY69WGcHnpGlwWEe5S1doSp9pLeJrZrl91TUbxJHL3Nw6brfMqU7K/3O6wX8XXEFk4XWVEf48PMGriiOs9+DFc0qv53J4XFjvCI2Oodn2WG1mbvMbr1nj/zy7xK2yl0/aBX70KsPHV9qw2VJEQqRCIpbH487WwjwV5fFuxNMN5Z17yry3e5L/eXI/M7aEpIYpk6NrleO0uL/Z57T2aNiYm/YpH1/pMKkFHmzEvH46z19vnCOPxzGnzk3MMO54tKyliMuo8VjXGA9DH4sAZ8wGM1rCweCpS11CrtIRVgiZNTlShQhYly4VDShpJiT2eTnu0xViErq2R85kI3QrJvNg5CRHpDGuOEQaD16HSkxCqH3GTJWmtvDExRFDXnKE2qdqylSlQIqSE5+e9vHFHcjU7OJCIB5t7eGJs5U5kpB5jUqSp09EgI8Rg49LiuV3/+ZXbhsKkGfHsB1ryJAhQ4Y8r+wMBLyQ+HimSselLOAv5N/YOQFrM7cj1ZSUlHjQfmE1JR3cL9GYvu2f5+ew6JbfY3uLyPZjD5PQv/VcSIBs3rY5rndz4ljf9mnbzpafINRsAtJVzDFva7QJecKsMWMrrEuXkIhRLfGArGaZETiEpIxowJV2mnEt0SUhxnJ1OkVIyl5bYb+t4mFYsCGt1PLZByrMza3RbJR4z/pxqo7DBl0+Fi5RdIVy3vJHrwlo97OQwHIpInDh5a/+Em97yz+ydzzlin1dejH4Ltz7yCjvuXaMT+sCoSTsv+7L3P6OJSoa8EsT+6lonliy3/mdpWlCScjh4uGwaDY4Yza4fWGDeTvKf1sMKfqwaPuM5wxNm/CW8jijmmNaC9RNl1/7ao94ILo9DF9eSrAoJfU4kk6yZPuEVpmTPDGWBRtmrWxEVPFIUQ6lo8RYHjGrBDicMmtYwMXgirBAl8NaIcDlrNkgFssJp4FnhL5GgzG6Hj3tU5T8lh8jIt5KJ9/0bgDkCZgxY8QkzJrJgRndJyJrl2rYDbr08cShrxF24BkZDM/GF5cNzbJNutqnIgUSUroaMitjdDQkEA9HDH2N6GjIb3/gfx22YH0DyGc+85lf+XY/iCFDhgwZ8p3DhYzpzzU7x+huH4v7df6CzTwPzNZi9kL7NAh2sPDd3pa127G3M6yCfGNcrA1r5zY7J64VTCHzimDwcHDF5UXs55icY1bHaEmPHP7WfcsaEKhLAY9rnQrnkmgwwjdmhIAAQ5uEVQkJ1OHmUpXf6T3ODeks7zrkc99Jj5InrEeWqyfB9y3jo12SxKFY6lEdrROMLPORD91EaiHnQpjAaDH7nW5+/T+CFR740sv4nVNr/Nj4BL1I+P3WCX77hhqfe6DKiahPDodj0sQg/MqrY/7HL6RM2RKLpkVJA5oSUsBjVbqMa5Exm898GFQ5o11+9GCev346ZEbyHGODH91T4zfOncGiTNoiy6aTGcdtjSIuXRJOmSYvsVMsE9KUPjeYUfrWclw7nHAaXJNO0CYmj4uH4GE4Jz08NQQ4pChVPL7iLHN9OskJabFfyzxq1rjeTvBp5wSLuoaPR0OzdPsJMwJk4r9GkQVdxaK4OCiKj4cRwWp2EWNUyixrg1hjPPHQgam8KDlStbS1SyB+likzuIiQqqUgOVIsCcnWOdXTPlcyx+MskBvkjAD85vv/7VCAfIMM27GGDBkyZMjzxrve8Yu7tmQ9V2z3XziDdo3NFqytVqxdH4diJAsnM5w/tev8bfW8fbLLgvi87Ybm82+ai1VBtm8jg+cbwIiTeXNgKyG9r5nPp06bLn1isUxrhRktsypdDMJZWWOvjpCiPEWLVQl51CzzEqZIsFxTDng6CjliyhzJ53m80+cmb5Lrx3x+c2GRdx/xObvqYYB9kwmfOy6cXg64/5ywulzizJkJLj94lrAxw0jJcsP1J3j8+AizYykbXYfGuVmefmqexXWHn7tlif/joT739pt4GDoLo/QV5nM+P/i6k3z2qQK/clOf3/pchXeOj3Jvr4WI0JGsAtCWiJrmcTGEkhDgcplX4IwNyXWLrKYJV5dy7HPz/MNaB4MwYYtUCOhINDDpF1GgLj32aoUGEUumw7gW2OMGPJl2mSJHXn1SlGPOKuNaYF36RFjGyWXZJJJwxJR5WBpM2SIOhhoBU07AaTosSJsnWUQAX1wgSzAPiUixuOKwrm1STamaIr745CWgR5+qFOkRUZQ83cH3IoaIGHeQTGEwuOJQlAJd7SEi9AfG/oSUvARZe5ZmZ1BLu5RNgb06yjlpkg6qJ+/5wL8bCpBvgqEIGTJkyJAhzwu7Jac/m0X5TkP3pXG+/8IRB7vNq3Gh/W1dPRcZSIsLt1IZcXatfjxT7sl3KpfynH9DQZQIgQSICJ54FE0Bd2Aytlg8XIw4BOJjseRNjmkZRQWmZYSQmCp5iurTMn0WdI1xqeLisGI6gylpmQegScx3j1e4vxHRI+VF1Ry/330SDx/fevg4zGuFybKSd4QoNvzZygpXe2UE4aZDlm7PpR5aWuf28lvnFmmv55lxK4Shi2qWD/J/Ly9wf7fNg8kGs8157uk1eamd4qRpUbN5XrXH5SPr69zzZIWG6fPQiTLff8Dn42dDUlFa0qcuLSYp4w2GT+fUpaAeL/VrfDlu0ifhLl3Cw6EVwYk4RIFjTh0jhooGLJgWV6YTnDJNSupzjVMFFZ4063g4XE6FpTRmUgJGXJcz2kOBcS2wYrpcSY0NYh516oxolh3yiKxTVh8RGUgLiBVQoW66XKkzrJkuqVpcXNraoyh52Do3ZDDxSnBxiIhpaZdAAjraZUZG6RBmHiCNEYQRKdEjYkRKdDRkkFWYtXyReUcEoSg5utrPvpfsOKla6qaNIya7PyG3/uDNQw/IN8HQEzJkyJAhQ543LiRALmVh/myTzHduK2LOy+a4FI/Glr/jYt6UTc/HNq/LNxOq+M+VS/n9jTh44uOKlzXDyfnBbxfbR87kKZg8M84EJVMkT4CD4aAzuyVA9ppJcvgcMrME+IxrCcUyonmutbOsSofHnGX2pTVSLAEuLelzs53jTc4ertOxzIyteRpd4ZVTPjkcPt/ocEO6h1trNZY05J5Om05qefiUj+cpD3a7XKdjALzz9U/x8Cmf93cXKDiGP1k9xxtkD//qTWdZXvPJ+8pyS/j91bN0JCTGEkrM++trWTuUMaRYxo3HnWcTWtKnKX08NSyYNu8/3mddQpalQ1si5nSEFn1y6m2N8Y3F8njUJSZl0bSYtCW6ErMsPc6ZNmfNBhblCTnHF5wTrNHmhGmw39ZoSUQ9jXlKNhjVAgX1eIgGZ6WNL0I9iRnVgCo+DkJFAxY1pC0xR9NJTpsmvjpEklCXLjOapysJ4wQsSocnnBXGbZFF06JlO4PzQgbpPZlAaGo7E5IS4OBsZXtklYwEV1w6ZLd1NNyqXNS1SVdDFmwdTxx62qcsBfpbMghKkmdFm1zNHKnaLU+YLy4t26GnfTbo8l8+8L8NqyDfJEMRMmTIkCFDnhfe+6H33PbeD71n1/+4L6XPH56duftCBvFny6UIkfO232X6lW77951IIAGOZB6Mi2EGbTclU8CRLIdh+30297FZ9dhOX/tAlvEyK+PUpMi0GaOieQLxMQjr2mZUypQ0G7sckeLiUpc2J80aipLHx8fhBt3PjC0zaYvUXIdmknJOQ8YDh30mj1XY6AlzTg4H4RGnTt5X3jRZZF1CZgsO56KY+bk1juQK/MirV0kVjj2+j+sP9vjtW1I+FS/zemeaomv4T/9tko+ubHBo3wZ3hGu0JORniof5qepe3p07QFNC1iXkq9rgu+wcoVqOSZN5WyFBSckC9c6aFm3Jgk3z6tElpi0hqWSiajPbY136/OjMCFXNsW56JFhGNUdbItoSMaIFyuSJiBmlRF9STpomfUm528myPWa0wJjmuEwrhJKwYVOekCYhKdGglbEtEUVcTppVnMFraIIcNc3Tl4Q2CUumxQlpccKsYRBOmFU6hLyCK4hJaNsuU2YUI5I9HlPBw2XVNrae/xJ5KqaI1SzPp6U9+hpTlBy+uJm3C2WfmRqM7c0uSMQklCSPPzivImIC8XhcFnHEUJECI5SINcWiXM3eoQ/kW8TF3w2GDBkyZMiQ55nz2pd2ERAXMphv/9l5+7uA+LiYIHg2YuGZvAnfqcJjs4rh4DDpjGbmfZTFZPmi9ytInhv1ICedNeZsjS/J05jB7KIf1mv5opyjgMed9qHz7ufh0rZdUkmZMlVSUq5OJ3nEWWJaxnAwbNBlVmsclzoFctRpcUDHcdVwwqziDXJBTpgGfUn4bp3nYNXhq+sRT0iTgniUAuglhkoO1rqQd4QrnDzVaIYn6oprhGXT4fK5Evc/Znn02Dg5F5LEZV8NHl/wCFyPTzw2w5j2uOFwxJMLAddSYrKs/Pv7lqibNv9+9gC/e2YJg/Aze0e5+cwMd7BIgvKkOsxrkb5JWKbHUWq0NGWRLk1JGdE8bYlYky5FzYzXLfpYUWY0E29NCbl9YYO+SZmxZdKt8MWIAgEL0mBKqyCwRAMfj/12jkecJTxcDtla5vswa8zbKoJwWjpYUXqa0JUEA1RtQJeEUS3yRec0VS3wqFnjtKwREfOIYzirqyybBh0NiTTOWsfEQYzwzvQ6Puk8lT1/tsOMGWNZG+QJCCQgGYiIzUqGLx5t7VKUHKpKR8Ot4MHLmQWFVVp44nBIpznLKtEgR2S/THFCl8gN2q9SLIlYprTAGerczJU8Iue+9S+W71CGlZAhQ4YMGfK8slkNudB41U22plddpIKxKVI208mfqdqxJWqeY2GwW5r3d2I7lkEomGyB2NWQNdu84N9eyJLpS5LHIByy46xJj5qUeIe9htfay/lb8wTv8PZSszkKkj+vXSvSiJQ0W9jakyzqGp8zx2jSoUyeSS0zzQgnZZUaRWoUeWm6NxNGprmVNn7A1vAw3JTOIUAngmOywRtzEzxl6tSKliumLL6rnOlHfCFe5StRiwnPpZ7GjATCq9I9fPVEjpr4fHRlg9OdhH93d0KlmJKo0ouVJe3zE0ey0bKphctnEq68bJU+Cb96cB7Xsfz42CQhMU+ec7l+zpKgvJYZ2hLxoFklHbRq3Ser1AnpSsxBW6UtWXtRVXPEWGZtBQehPBAgq9Jl3Bb4gZkKs7ZMST0SlLrpMqpFTukKOTzKBLTo4eMR4GZjdTHstVXyOOz1AuZtleOmwT5b5iqp0CZiybRxETwc6qbLiUEFpUQOgJb08XHpa/ZcJST0NBt/HWuUebdQevS511kih4eHwx4zTkiEqqVPREnydDQkISEi8320tENegq3vjQh9jUlIOS11zsoqnjgD0VTfqogUJc851lGUZHBbflBtOyl1btYjPGgW6A0qbkO+eYYiZMiQIUOGPK/snJC1nfN8Ijs8IJtfmx33M8gFx+Lu9IV8u7I6vlNasbb7ODxcXHFJNGGGUVJNSTTZ2m43zwdARMy69OiTcMDWuDbdw1ecZcYJeEN6iIIHJTyul4PMOdNfdx45OFvZLV3t0bZdzmidr3KSFTZY0xYRMQahP8jAKGsORXlZOkseh6IGNIm5X9fxHXhzcZz7em3+ZfkwDywqpWLCI6uWG6c8HjNLTJGjkWTX4u9sb9DH4gg0NaYvKQ1NmLNlvnrG4XNxnaP7Yk6aJrWRNn/8RMjfhctEsSHq+3Qlph8Znjrn8fcrbW6RGe6KGuyZyaZifZGsknRK1iioz6I02W8rLJoW69KjQTTwgLjk1KUrETl1MQgl9QnUoao5jroVvrCYUMUnxtKRPmUN6ErMIaYoko2pPaxTjFNmj9aISAmJOWs2eMCscCzukqI4CMfMOl+iTiwpK5I9lkXToi6ZIEmxhEQDn0tCUzu0bYdYI2JNCDVCUUZMjViTgSBJadAhj09ANmJ3Q7uMmRp20H62WQXx8Ug0pSolIk1wyKajubjkxKciBQ7oBDWylq1Nv4cZhIv2NSJVy+XMIgjTMkKkWSvaBFUcFeZ1lINMPievne9Ehu1YQ4YMGTLkOWe3yVgXYqcQ2dmetWku3xQpFv1arod4JBpfUlXkuWZ7K9Zz0ZZ1KaNqv1144pMzORwMk1Jj3Ba5TK/lH8wTWJQN26JiymzYFtGgUStn8uTEJ9GUn5+b4vBlC3z8zjG+HDc5mk5QNg5LNuGudptjzhpVzbFsV887P1QtPbaZlDUhMIWtx7WZ+7BKCxeXu53jvCG9ggNung/rCQq43GuWeYPsIe8I3Sjhro0OBVy+Z6pIapX3ymNc1biKgoHT6/C69BBTnkesSkENB9yAcgCVvPJwT1kwG5xkjYIGXGEKzCdlAj/lp8dnuOurwllzhp+ozFEth/zGV1Iw8P6TPY76JXqSsLdi+HSzzX+5q0ZseszYEnXT5aZ0Pz1SHqXPF53TlMgxn2Z28AktskcLHDMNrkonaEtMTfMAlHBB4XjS46xpA7DXlnEwdCRir62yLG3GbIGexFQ1N/jwqUuP70oP8nnnFG2BB5xFBMOMrXDGrA+EXYKLw15b5VFnBYAl2SDBEhFTIKBDSEIWDFo0RYqSo227TJgR6trkRXKAZWmxYOs4gwsJuWx+1WCSVTaRboMuI1JiQ7vEkr0v7NVRFqSxFbLY0g5VKdHSLuvSJSLBFw+DEGqEVc0S0zXFiFC1ORxjWKaRiSLN86gsUDAeBfX5qb/58aEf5FvEsBIyZMiQIUOeU3arfFzqlKvzjN3P0JqVLWUvPEHr+TaH72ZQ/07AwUGxOBgqUqRLxIpp86BzjjfYI7zJXsmcMwXAmFPDFx9BKEmBgOyK9em6w8LpSQ5PZH+3EfG4bMRhv5dnRnKU1GfM5gdXsh3cbYnZwGCBGZIOMh9mZJS9MskIJSYk8y/MaA0Hw4POMg8mGxxKR/mKWcbDcDztcVfc4G3TZboktIjJBZZfXzsGwMeW2rxfn8RzhMNBjkogqGb+kFoe6j0l8CwTJuDV6SzntMFL7RRfiFcpiUOj6WMEPtI7RyqWWtHiupZbqyNYlBXT5QvxKi3p87HGOrFY5kyes7JKy0RM2xJnpUOAQ56Aa9MZpmwJlWyA7awW6ZEyY0t4GFrSx8XwUwdKPG2arJoeISn7bGbSX5QOOfUyQa9wmR1FEPbYMtNaICThbuc0Y5rnc85JbkrniUk4ZMfpEFJRH4sSk3KZneC6dIaG9LkynaBDn5iUGa2iKGfsCn2NadpsAlekMSXyOOKwoV1STTkuK6zqBr54NG2bHn0WdY1lbdDXPooyZUaZlTGWNGuhKhIwKVXq0sYg9LRPSMS4VCkSMCYVQmJiUjraI4fHYabwxduqnFlVHjfnULJKyX4meVKWSTTllKxtGeuHfGsYipAhQ4YMGfKcsT0dfTuXWqWQbf/Ou32XVitgK7nccL6x/fnmGxUdl1opEgRn0Mr0QvOapKQ4OESa5TYYhHlbY0orTEnAlAQU8HmrvZob7L5se3G4yV7GNcxRJk9sIZ+PeXJFOESJNY24bzViqihUXIeculiUo85BrnUOM+GMnPcYcpKjaioEEhBpwtN2gUVdY86O0CFkgip1aQHgqcO6CYklE06TtsiK9LjOVFlpCS6G63IlJsc7/Nq+A/SJePcVPv9m5DCP90JO9yMmypZbrunyV+mTvPRFC3gC/dgQiHCg6PFTXA/AfXKSt7+ox/G6w18ur/KzeyZw1OC6llbb58qDG+hATjuY/5+9N4+S7CzPPH/vd++Nfc/IrTJrydqrpNIuJIEQsgEBarABY8DreOyxj8dMn7bd7TNn5vQMLY/tdvfxzKF7bHf3cZvp8dhtZBZhwAbMvkpCQktprX3NfYt9u/d+7/xxI7KyUlViE4LG8ctTJzMj7837RUTGqe+J932fh/dsL1OVDkfCUY5rnb06zpq0+s5Uab7unOOWcJqG9Jg3deLqMEaCDiE5PLqErEmHa+0Iq9Liv57uMGWzFG2Clvg0CEiqS1mTGGDSZqiaLh4GF8FBWKfLumlHGSM47LIlHnbmsf2VbtMCbQlo00MQYhqZ6YZiaYrPdXaKaS1yggWqtkGgAVVbwxX3UkUCn50yTloS7DfT5EixXUbp9lu0OhqJiTEpbKSmOxhaRHMkKUnQpEtK49RoEcOlIGlykqKmLSo0WdcGFW1Q1xYA69rgnKwQEKAoRoSdjDKjZa6102Qkybhm6OHjiOGgneBpM/vyvZD+ETAMKxwyZMiQId8X3vW2f/be73aDfHnSuRO13PRvi74ftGNd6fcrbEpF30hI/w7X8oMYJpcrfGz+2ebPl+6j+a5Fz0tNtLZoTWmTxGAY6wf+OWrY52ZIOYZdtsC45/G0rlMweTImTddYDtoSr02WmS5a9h06xVefLyHAkXycfSWh4wuPtZqMkuCsqZLA5c3ONEd1lbRJ9XMllKLJk5EUWUnRpE1C4ihwUVbZxRhnWWRCilHQni3TJSSGQ0kTzJg0C7SZ0w67nBQlifGQXyFczbPecDgYjtFuecw2FBfBFYk259NVXl8q8MjTZd7w6rPUqlmOr1s6IXxWLnKjU2I6HOEbC1A0HmkbY6YE59cNd2yD959sU+zk2RMUSQRxlqXNXMWhIl0a4tMWn/22yHNmibJmKJHgCGXefX3A7aMeJxdi/Oo1Hp9caeBgSOOyIh0sSpYYAbCNJOdMnT2ao03Ik84ch7RMQTzaanEQypqgSUAaj6YEGIRTZoVpLeKoYYQEF02dGVsipg7nzDqLUqOqDRA4J2s8w0WqpsOqtKhJl7p0mGKEqkQCIN7Pggk0wEgUJtkgqlwFYmnRIYGHFchIioKkaeNT0QaeuBTJAEqbHi3tMMM4F3UJ17jUtUVKEizoOllJkZAYCWIgwohkadBhTAo06UaVGHxGJU+RDOdZpiZdlqWGYKhKG4PhjnCGo84sN4c7uPZdR4YBhS8RQxEyZMiQIUNecgYVkBfbxF8tAX3rPIcRA7Jp+LwvPKK8iEsbd904X6Jh9b4Iucze9yqVlSuub+um//vM1a6zeR2Xt7TJC+7fDwOCoAIuDgjkJM2qNLjOjlNyXYoJIe0a4o4g3Th3eCO8JjnKDV6eE702t4+7pFMhc7Oj3HlkjZwkKed9tm2rYqyHNhOoFZakwzW2xK6sy/buCF/R09j+R97JUrV1spKiRZe49HNB1KchXfKSxiBUaSNiOKwlcng0CDicSpILEqTU49pRw1erTYrEeUzXKIdJCjGHpq8EqnxVFugBZY0TJ06367J/d4Unn95Gzxf+pPsUz8oq/zS9n73jIWcqSlE8TodtPmae5c35CXr1ODH1KITRzEatpxxlnRReX4D0OGBLTGqKFbpcNBXed0uOz8yGPOLM8eBiyMJimh6WxZU4bxhP80SzxW/c0eGV4x7fmI9C/n5+d5KH17uUNIEgrEmHQzYSIMepM0qSEYlzXhqMkiCLR6ufrjGhORLqMkWKk6bKITtCRXpYUXxRMpqgIz4919hhkAAAIABJREFUDajYGj16dPFxcOjg96siXRraIiFxPPGYlBLbTJkWUbVjRHIEEhJiSRKnrBmcfkZMnTYCJCVBiQwXdIWExFiya0yaMqHYqLKCS1aSzOsaI5LHwVDRJhWtMy7RYH1WkihKXLz+GqOKSnSNaCC9SYeGtrlRd7AqTXIkmdAczzoL/Pg77xiKkJeIYTvWkCFDhgx5SdksQL7dGYzNVrtbQwYHg+hbW7Ci7Uq/l3vLNbZ+f8VrfgfiYrMAuJqr19XS4L9TBvcxJrEXbSf7QbecXQ0Rs9FnPy5FXAz77CiHUwnSnjBd9snEIZ9U9ibjTGaFbFLJJJSdJsXxJcPSWoyxcpNPPzzKc3MO45Nr5EsruK7lNdfVKMdcfjo7zivGPM7XQ9KO2ciJEIR1W8cnpKJN9jPFdi0zIxNMmVFsf0B6VLO8NtxDWdM8bpa4ddSjKl3WuxYBbhuNkc/6ZPC4LpPggC2QcgzFJHxQT5J1Db9SnOZZs0AmJnS6hgvLHvc9rIyVemQSloqt89bwIJ4DyUTAVCzG9rTLgmnyP8iN/KsL57llR0gp57PSC7AKH+cc9yRGEYWUusRwaBFwQZq0JeAaO8l9j4S854jHtC1EA/rSxkFY0R7bxpvsJ8vHHi5irTAvdRZMky+ekb51r5IVh0lNA/AMFSrSxsfyhKxQky45cWkQsN/JcN6pUiROHIeYCNtthvOmzkWzzrq06dBjQSq0tLsxbG77rlQdenTpUbF1eupzq+xll4wzKSVa9DZcyq5hOwA5UtRsk5q2OMki69TxcMgRmQs0tM0KdSyWONEc0LJWCLF4OCzqOhZlp4zjYFjWCkXJMG6inJp1bZAist11cShJlpykiEuMpMQpSqZvJ90lLh4dAq4NJzlplnneLHJbuONlfjX9aDOshAwZMmTIkJeUn373m770ofs/dffW27cOpl85YHBzTohsaqeKiIae9bLvXyRf/VIr1hWqLlc664rtT1vO/VYtU9+LE9agnQkikbW1lcwR51JliL4drYY/FHMhguBJjLIpMm5KpCTBXjtKUj3S6vFc0KDVg6l4nLinVFuGpAfTY126PYdi3mdHQdkx6pNLB0xsn6OUSNBuxpkcbRNLtMnlW6wsF9k91WJitE0+6zORMTTbLifCDlmTIWeyrIZrOOLgE7BCjTlWWNM6d9g9tExAFx8jDtfJCPtiKZwgxlOtNtOkOaVNXrctwX9ZWuGGdJrHq10qPeULzhlu80aZb4bs0Dyf5AKldpafHhul3hGqHeHAdJcPVOa4zRmjmO9xT2yGI1MByZhSa3g0uoZ9kz0eXe/RscovjY3z3ILho2sVkuoxHnc56bfJBAkumiZzpsbvTE/xaK0NQEJdfvNGh7unHLL5Bj++0/KVc9Fm3CD881c3+bePCFaFi7Q5M59gRKN0dwfDOVNjRnMYEb5pFhnRFBdNnRvtGG1CrjcFcjbBl5yLTGiaWW1zUIucNFUmNUUXSw9LQl2OmQXO6zJ12lS1TlvbdLXLiCn25yyi7I2DTNGUHllJsiYtbg2nOGNWGdccGeIsUGGUHHE89tgRLph13myvJSYxGtKjqi0SffOCFh1i4kWzHrQiMwOJEWA3htuzJDmni9RpMyJ5ypoFEeJ4VGnhiYuDoUGUVbKiNUIsaUnQokOPgHEp4BNixOG8WSNL1Fo4Z2q85p23DSshLxFDETJkyJAhQ15ytgqRF26S9QVfR5UO2fQzZevMx9bN/YvVWS5vVbq6ANla4XjB5/6aNtvtXmm9A7HyUsxoGHE21u+IA30xZtVixGyIHYvd8pj94BCEmIkzagqsao0djLJqGqSJc50U2OWkqdmQo402lbrDci/kfMfn2jElmQj57AmHa6Z6lMfW2DZzjHajSHF0kQM3PUt7vYwxysnjO9h/+Biuq8zPjfDQiSQeDiCc6fr8j/ndPNVtsqIVfPUJNMBXP8ooISRv8iCCRVnTOkvSI+YnGDMxvmTOs2w65DXBQ406GY2RaqeYjsXJGwcbxAgCQ0NDxrwYR0yB+/UEb5vKYNTh/fULvCJVYKYzhhFhueKSiikikEoFtDsuPV8oZEK+ud6jYXwebzU5Es+QCGI8bdYoBykWtcvPH/D44mqTOC5BNcX/fO8FDsdL/ORtF2jWcvzh4z7/cF44fjZPSwL+97s6PHguzpkLOZ6XNW4wJeo2II4BhBIx1umxXbOMux4KFGyCsonRVEsOj51eAleEVRtwnZQoOZF4zLsOEjpckAZNCSgRxwJLpkmI0rQtfAIEIWsyZCTFQaawRihJhm2axxWPHVokFDhMkWV6LEqNpnQZlyJJPM7IEhdNBYNhwTSYY52YRNWUUCyCUJBMlDWinUiMI1S0QVxipIiTIs6srjIiOWISWfq64uBgOKMLjEgOl8jOO0eqLzQMOUkSYGlqlxHJ4BP9vXTx6RGSIsa0zXNB1rnnna8aipCXiB+uGu6QIUOGDPmR4f4H/t199z/w7+676qzDFYIIrzbf4PQ/BmwNLPT6sVeXzZIg33arkvSP3WgL45L7lNkiSq609pjENr62/faxred8J2y+H17/dwMbSdI/jChKV7vM21V66jMrUfr3otQ4PGoYTQkr0iGDy02jLm/YB2/Y4fHx5xx6PYd33FqjOFJFjOLsWyAMPLxUDdtIUZo6RaeVZtfMAieeO0A6v4znRYGAU+Nt8inLHlvg2YrP4XCUvc72F1gkx8Rjl82zwxa4K9xFUuK06PKUs0xbLW/Tvbxep7nFy/M6b4xxTZLyYK0bUguijXqLgKy4VIKQvWOWf71jN18/EePfLp9hjQb/6eIqaQ9U4T+3T7NSN/zB+Xkq1Rifm+sxmrM8ds7DAsdknhOywKOtBp5EaeZP2io/N1rm/c/38LHcrKO0NGR5fhu7b3yEv/z7g6QzLWZNjbyNsyIdlqTJB748ym/e0WVW2/wY24gZoSE+6/RYp0sHSxqXjDjUQ0vdRrUKB9imKSzQDi3n/C4tAmJGuBB0yLkOn7VzeBhulhK3uyVahMRxGNGolanQdyErmgKhhsxomUmb5QY7TYIY502FGA4uhimbAyKDgDJZksQJsRyXBUK1tLVLUTK06TIhRTr08MShQJo0cQxCghgOhhRxutpjWsqkiCyCo1aqNj4hNW2SIBa1aqlDUbKsao0LdgmL4qlDmx4x3H7YoUtZclHoYb+tzMMhRYw6bQzCTh35gby2flQZVkKGDBkyZMj3lXe8+41fese737hRGblUQWBTu5Rc1p7l9KsJ0edNGR99RywXF0tkxWtwNqoCg+PhUm3g8rauS9WMrWndTr/CYMT0HXt0IyjNkeh6m6sjBsETD5CNNQLETXwjGfy7JRJBg3WEJCWJJ1Fi9GAdgsETN7quXBJRP2inLCMGXwOyJkOUx5Gn3Yix1A0xCHlihIFhZiJqw7pmMuTEXJyxoo/nBagadLVApjiHxH2qC7tIZKokkk2wLtO3PcLc0zewspZk//Y2y6tJJsZapHppZgrCrmSMv24+t/EcDB4Pi1J3QtalzZJpoij/U/IQJ/0WnrrsiMWZC3p8kXnevTvBTTu6fOJ8wOFsnJtnunx0rcItbpG0Y6iEIR9szrMnKPHn/jHqtHhDuJ/X5vPUuvD6V5/kL0/3+CelEQ5LgUeXAzLi8NV6ndPaYN5UcSPPMFSEivosmga/PDrO0yuW8zT5ZzvLjCcdXjHTY2rmNL97/xEek2WeOpchT4IkLhMk+YO3XOSvjsdYny3S0JAULl/WBboSklKPPLGNv4jB0HnCOJSdqCISE8Oa9pijzbpEVsU5jXFRWmwzCS5qG6OGDB6nwhYWpUCMnMbJkGScHKd1YaNil5AE2zSLIKQ1RlGTJPEIsKTV4xFngZ06whlZokWHurbp0iMmHiXJUieyA+7gkydFUzuUJccSVdLEqWiTmHi0tUtKEuRJUaeDALO6QsakSBEnLynqtFnVOhlJkNUEiFCWHC5u34Wrh09AlhQVGijQosvt4U5qpkejn3FyxG7jjFnlX3zo14dBhS8hw0rIkCFDhgx5WfjAA++77wMPvO++v/7I/3UfsBE+eKX5kMEw+iD3Y8Dg5z6XNvmGqL1mULGIhMnlqeuDz5srGBu3X9ZqZaI5i00b+cHXW8+VfiK3Ky7aH4o2CD31v7cHahODClBSEnjikjAJYuLh4BDrW5za/rv8W6tDLxdbBV2gAUmJ3qWOOuwta/SoE/CmySQnpMpSL+DB51OUCh3aHZddYz6fOZohnoxmH+KpdSTVRTsxkpkaGrgYr0uyPMfJL9yNMSHpVMDaegI/FFZWk0yNdjly3VninpKS5GXrg+g5uxgucCac4/nwPIt2jT9qP0ldutycTjOagafNCteHYzx0yuP3nuzw84cMx2o+XizklCxy2m9zotemhs8bmeb6AxV+J38QgBsLcXJJy58Hz3H8+V3UtY1V4aO1FZ4163xVFqibHvckRnFwNqp3K9JgRVr8Wn4HuXTAIyyzS7MsrrtkMwF/+EwTx+3Qw/IfX+czRoIpTTEuCXpYfuvjk0xpmnnbpUSMJe3yaiZ4FeNMkeK8qaMoLQJKmiAmQsE1rAUBSUfwRFiVNjc4ea6XKIMj7RhucPI8HlSZsXmqElVZ4jjk8Aj7c0opdbGw8aZCT33GbZY506ApPsumSUsCAizXmwLHnVXuCqc5JvO0tYuvITmJhs536xirWqdAmgxRm52Hyx7GmWONODFWqZOTFIn+UHpNm9Rp4+EwZ1cINSRLkkVdx8FhlBy7ZAyfgJq0GdEsDk40x0KrH1oYY17XKJCmS4/dOsoJJ0q4TxMnJOQ5s8C//OA/HQqQl5hhJWTIkCFDhrzsfOgDn7wbXmjH+8LZBt10jG58D30Rg2JFsRpuVAR0cOTGOVzh8yUGm+jL5iyI5i8GNr+GqM3KFS8aGCeyoTU4hAQIUQXCXnbN7xG5NPju9RPBm7ZF0iT6SdO9qFqDszEIvCHmXkZBkjCJ6PHpp9XHJI4rLllJIQh16TCtBRZMg5Fuhkpo2eklaISWTiNBGBoabYcbd7dJJHr91TvUFqZJ5tewQYzYtWfRWpKT37yV4kiNidd9hQuPHaTTc6h3hErLMD3eIZ7o0W6m+FR1la76hISUnRFs38I1qnIJAQFTzhhvCvdzwqxyjY7wt60lHAx7Jct6GPDj+Tzdnstd11RZW0/xs9vznFz0yIlLznh8TE7zxTnL2bbPH103yh+dX+LmZJ6LTeEthzo8NBujVvfYJWluSuT4rD3HXlvmWFinoEnKNkWRFD+VmOaOTJ65unJ0NaSgce6ejPHESsCJNdgtaf7Dcx47NcvZcyUWbBcHiRLRnTiBFZI4zEuLZemwUzK01fIMVbZJkjIJFGHWNGgRcH0yQ2AhbiLZGijMOCkcA1nPsOIHXNQ2WY3hq2KAPHEumjr7JYeHIeM4oFGQYxqPrMmxSJWYeLxSd5BWjxXT4pAdoYdlzbT4pizQosey6XLQjjMnVUYlH/1d02GUHIiQ0yRr0qSHT5MOdemQIUlAiGDo4pMkRkI8rCgt7ZKWBAGWpImTIEaNFnlJc16XaEmXElkWtRJVTaRNHJeatihIGheHpMRY1HXGpUBDuoBQkahaVtc2/+eH/pehAPk+MBQhQ4YMGTLkZWejRasvRiCqSFx583z5pn5rK9cLh9w3i5WrC4LLrxdVQzzj9c+4FHg4aBdzxI2qLOKgEm34PRP1j5u+CNnI7vgOhcjVRIMK5E0WX32aGlUJQg3pard/jag9i0HrmMjG/d7q3vX9yD0ZtIVtdyY2xEdC4v1KjctuHWWXlsgTYzdZ5sIes6bOuKaYyTk4BsaLPq2uw6FD5/iHr+3CCROkUiE2cGnXS6wtj5L2lZNHb+DAGz7H+aOHiTcyuMbFdeC5BcNUHq6/6+tUF6ZIp7vk52b4ydQMXw7maWobT1zykuWXuIU3x/Zwnd2JqMe0SXGrW+YRv8rjcp7dWqajlmXpsNAJOdX0aSzl+OxqkxtKLmE7QTFmqPnKKamQJcFFUyG2OM4NXoFP1teI4/Kh2SZWlHFNEQKf1ovM2BFmJENJE+SIcc7UaEoPejEm4y7Ptjp8w5nniJR4pu6TNS6fM+c4Ly3e6Gzj7+UsmTCJ208uP2Vq7HXTjDoeD+oyr42Ncco2uC6WpRqGTEiStDGkHYejuk5O4xyQHI3AMpp0mO8GJIzhoXCVgo3ztK1TsDEm3ThTJkHOc3gurNEVSxyH/ZKjYy1Zx+Fxu06TgAumHokhTVMmh2M8HjMXWDddZmyJJWnxvLNIniQ9CWjS4c5wJ486F9lJmZ5Y5nSVsuRJESNONP80SoZlatHcjnZJSIwsSWI4bNcSoUSvrwCLESFLCitKgQyrWmNMCszrKlMywrKtUKGJxVKnTVw8arT6A/Ex2vTIkKArAQZDnTYTmseXkGktMq0lbnzX9cNh9O8Dw3asIUOGDBnyQ89lQ8b9Nq4XC+l7YYXlcmJ9y88XnIfZaK0atMsMCPsfit0Yku+pj4dLSNQ+FvnwvHimyOaqy7cKc4xay0w/ecESqE9Pe1c8PtTwsha3zdkrV1rPdytGtq49Jh5rWsOIkJcMXe1iEHbrKCdlEVF4xqxwVpscN2vcG5+gHHe4ULdU2uB5lkImxPF67J/0mdkzx4kT48zNF4gl2qxXksQOnGN8cgUz0iWV6lFbLzIyPo/vG+7c32XXdJ3T33wFsXgPI8rBcSWXVEZMgUB92tph3Vb5jDnL8W6bp6mwXdOcsg2yMeGuZIFtUqIuXf67Qw5JdXnKWcAgzPk98sRIpXsc3N6hlFEeYZmCpliRBj4hX2WRT/izXCcF7ogXWJIaJZvia855QpS3Ozu5zRkhKYa84/C0WeWALVKVNp9zTjGSC3nMWcCiPK1VqvRYtl3KmqFBhwfsOW4IJ7khk8Lruzv9ykSZFT+gElh22Bxdq+yyOf7BX+SoWeEUdXxVlnyfexKjrJk2HbVkHIf5VkjBcWmHlv2aJ+86XOfkCFRZDQKqQchyN+AAeW5yCuyLJXleazxpVjgVtjgihehvjkigGIS8xqnSomobtOhxwVSpS5eatpiXKl0CMiR50lkiTwpFWaFKWfIEWJaljgFWJMoGyUuaKMQwS1WbrFIjpXHaBPgEdPDxcCiS3WjPXKXGiGRZ1wYxvKi9UhxcXGJ49PBp0WWHjrKg65zVRRxMZN2LSwefAmnKNkVGE5yUJQoa/65eJ0O+Ne63PmTIkCFDhgz5/vCBB973om0O737bb37L5PUr8e2kiLv9FqdAfRxxok080axHJCo2u1wZPGRDbAzmL5KSoKtdPFx8AgyCj278vsHaB1WXq+ejXDpu832wYmj18xfsRo3mahWjK/9s89zNC2yG+farNle7Zra/Wdylo6SI8Zg0CQh5Xua40e5ghASnZI15U6crIVYj96iOtSSssH3XLEHgsXBhO4euf5owiHPkutN02mkKNz3OXmM58Yl7aDQ98sdH6bQ9uj0HkTEmt61Rr2XwYj7F8iqLc+OUyhVu/bGv8vhXXsn/IdfyWyst1sMKPXqRM5Kj3Gt3szfv8slai7gLH6ku8692b2N2JcbHnuuxQzyOYVgwTW6Ij7HaDUkkK/ybb4bkNKBrAjoSEGLp0GO3LfBz1wjxeJO/eSzBlClggHHNsTeeoOqHnLYt9jppWtYSimVfMsGDPeGn9SD/+cI6b5Id3Lxd+atzDQDeutvjT8447NEyBiGDy/lmgAGq+Dw6LzwuK7zeneCxoMkeTeEhtPBRFL8f3xigPNiuIiZq4RLxSDiGdmjxVUkZh1ZoybqGmBHSGAILjkA1CJkPeuxNxrlZC6jCo7rOCRtSIIavln1kWaJLTXrcEE7xjDMwioCsxsmYJGXNcE5WSBCjaJOcMsuEYtmuZWrSJkuSFeok8Khog3MiUZsUMda1gUXxNWReKiTwNh73FHGadAAICBglj0+AI2bjjYKS5IgRVTGbdBklx1mWGJEsTboUNM0FWWGcAin1OC9rnHMqFDTJKLkf0KTVPw6GImTIkCFDhvyjYyAYBEPcRO90CoZQLw2Yh5usdhWLiIto37lLdcMFyxWXUKPqSEi4sflxJKpORJWIQdCihb7NrkEI4XKXri3D8g5CoAEODoISbHLouhKbh7A3szXQcCBWvq00+4FgGTiF9e/HQKxNyQieOmQ0zmvjo9zSG6FlLcekSkJdnjUr3B5u46SpcqdO0gxstNl1DPe+ap4nn5xh23iTRiPGsaev4dxCjHYA2Ti8wtzCVx+Z5t43Psyzj9/Iw5+7k4MH56isFahW02zbXmd8qoYNXWLJOqMT0RqPfu0OGi2HlYbwy3ozX3cWOKpnaWmbRV3n4+YkP1XfR1wdnq702EOOxfUYf1Wf5T3bJvnr+Sq3h9soGY97f/x5/uDvtpNMN/C0zJPOPK8Ip/iic4YQS44UjzsL1J4p0ZGAHcS5kwnKCYcn2k0e6UaJ4yecVd46nuaPFmdJqcdIWun0fA6WDBdXEixol6dnkyRxuT6R4TOne1RNByvKa+w26oQ0NCQrDlX1WdEed8gYhqgicehv7r3vEHD3VZ7Hz7/9o+8tGpcFv0dcDEXX5clglRsoUrMhBDCZcmj5UAkDXBG2p12eaPQQoB6EBCgjmsAAIxLNjByTGiqKQ+Sg5RPiE6KiLEiFcc3TkYARciTV44RZIsTiYFiSWl9Q+JEAkTZjFDBE4YJzusbNupNZU2Ne12hrl7wk8XBp06VLgNevPyaJU9IUVelQ6D8m60SD7gN8AuZYIyepvkhTXIQJitRp05DOxu+7KOvst2OsmPa3fI0M+e4YipAhQ4YMGfJDy+ZKyaAq8t2yuUqgagkBESVOjKAvSqKsgfCyFiuINt4brVcbblvRJmawjx+ImoEQiVpBop8J5rLPhkvVEIPg4Fzm+DUg1BDb73/fKixerOXsxSoeW0XMtxIjg8ctLpFYsxpi+tklRoURmyKrMR50TrOrk2XCjTGW8Phat8n1Os0eMkxnHM4260wkHZ5stdjpJhhLGowTsn/vMivLeQ4ceY5WvcTMni7NWp6Hjo7y+NFpXnnzPJLoMjLSQEyGdH6V/Pg5es0ifjeJcQJq6yXGx88jYgmDODHPMlnukM+4nDmj/GR8mrP+Ms2gScXWwMBnZZauEzCvcXbYPKpxDtoRji0J86ZOaC2vKpf4+U8l+L0DLv/Pp3dRoMcvevv4lC5sPOfr2uAwUwBs1wwnTIUWPte1yxxzVqnSoqwZAP50YYEUHnlN8Cfr55nWIvevrnLeWePmcIrjQTNqyeo0OWuq7LMlYhhUYG8yzjfbdR42K2Q0xpu9KTqhsucD996356rP3iU8IuE3noxzf/siN/tj7CGHSlQt8YxwsRVStQGHM3GeaLSQVpwULs1A6ajFR3ncWeCucJq6hjTxKWqcFgFrpk1GvX41ZIFlrbJTxqhKizQJRjRFl4CAgO06wrLUsSgVbZKVND18xjXHBVZIS4IVrRETlzYBeU2QYYLnmI3+BjeqgpFRd4dO5IolNbKapCJRW1WLHoLBJ2BN6zgSvdGwR8s8ZxbIkySvCS7KPA4GDweDQ4uoFe64WaLUf+6GvPTI5z//+X/1g17EkCFDhgwZ8p1yJVHyrWZB4NKmenCs27e3HYiHwQZncFuo4UZwoWBwxcVqiE9wWRuW9Hv1B7dvPtchSkB3cGhrZ6Oda7MV8OZ2qc14EtuoPASb7H+vZDe89f5fJryu0sb1YgJkcxVkYAu8ed0OhrfpjcyaBodskbNS51anRKjRZM3ndI6iJtiuGVbpksHjrpEUF2uKKvzEq2fptJNcvFjg5ru/xslv3srMweeprU2wulTAOMqOvSeI7bzA6hM3o1YoX/c4J754N3vv/DLiWmw1jXbiSKxH2M4wf24P//4x5aenssytG5q+Ug1CbtsuvPPcZ/sOXh77zQ72hSP8WDbPzGSXz55weJYKuzRLRhx2Z13ONULqNqBFSJeQMRIs0uGoM09eU5xiAU8cbrQ7cDB0CclpjBQuFuWiqTMvVaa1SNbGeMZZwMHw6nAHh9IJ/t/OGbIapyVRC1VMXa61ZdbpMiNpdmVcHqm3yOCSNlGNrW5D4mJ4xYff8h07Np1499+/d90PGYu7dELl68EqTelyr7uNfEJo+VE4oytC3BGO9aJrF4xLxQakxKGqAXlx6aglIQZflRDlqFlhwmYoE+d5s06AJaUxQrE8K7OMUYiqDdrmCNvpEtAjZEXqpElgEIqa5CjnmZAiPpHd7qyukpUkWZJc0BW2S5kWXVLEadChpBkq0sQnpEQGo8KS1DbatgxCSdPMSYXtWuIECxxgknVp4xMwplkWpcak5jkpi2zTUj8OUfiND/3y0BXr+8hwMH3IkCFDhvw3ySB3ZFAtiSxyw41WpquxdSMeqI9PQNgXFoMBdJ9gY7MdarhR5fC1Fw2oqyUkCuBzxd04B6JN+kCADKofLu4LQgwHrVvmCtWIwfD3QOBsFSBbB/SvJMCuNoi+NUn+2xmQj/rso/OKJsuEidKjH3PmyPXfDZ/UNI7AdVPKctjj10emeHuxzDuuUX7rVuVXb+1x7YFlrtkWsn9USWZqXLxYYM/eRWw7yotYmd9BMl0nle6x+/ATuJkqdr7EF78xSb48j13PsOfIY0gs5KEH3kBzaTvO+BqVuT08/PUbmD78BL+4O8XFNcMDnTnGM8KujEu9ZZh0x3i93Mi/L9zNbxZneNd0Bqf/UCREOGmWGHM8HpQFQguPssItI3F8LFXp8guvXuWwl+H3t++mLm0KkmaKEU6bVZ4wFzlnVjntrHPCrPHLN1gO2xL/PH8ARw3LpskvmANMaYE5afFEs7Wx8QYoaJKfcKc4ZtaYJEnKMTxV77Iu0TxQ1QbM2S6J71KAAOz7wL33+aok3ahd8FoK3GBHWfNDnqx1KCSySuKdAAAgAElEQVThxm1wPuxggB6W7bEYq9anTkDKRNGgvmqUOi7C9qRHwXG5RccoEydvXOalyoTNcNosc05WieFR0hQhloCQuDpUpMW8VCiRIafxqALERUYkR5MuTTp06JGUOAGWWV0lJOSMRkKuTY8mHarSokZUbQGoSZsQS4M2u+wIddpckFV8ApalTkw8PBxWqG685rIkWZAaEKW5u5ihAHkZGIqQIUOGDBnyI8ELBrI3fWwcc4XqweavN2/qtzpwDdqzNocXDsTJIKBw4I41EAybQxS7/VyPARYlUP+ycMatONKfM9m07oEoiJLlL82TfDvD+Bup6puEy1WFCrLxeLniIpi+R5diVWlpB0ccfEIWpMGydMiIw5fCZUSUH9sWY7TYo5gN2LnvOMl0i8kDR0ll63iOcuhw1Fqzd988hbGzLF3Yzd4jj1OtRD38jzybp1mZJGjkMdtWedtPfo1PfuZGHv7CqzCjVerHDlFtGbrtJA9+5A0Udx3jrt/5KN4r64RW+GqnwjtS28ilLI/V25xaV37WHuGfpMs0e5CKK38xW2XPdJv/7dQ8T2mVP90/w99xnnvdKR5qNKhIm/+yPse6dDhr1vizrxSoByGhFTKawMWhZFNkSFAmi4fDdhtlX/z+0TY+lg+tr+BLSFrjPBJUOC9rPGcWyOAybjOcMauk1KNJj3O9HtfZMinj8Gk7iwHWpMUFaZIShxo9bv0uBciAV33kJ+4b/4s33Vfxo8H1lHFY1h4F47HahJNLhgOxJBM5yOIxVYAaPrdn02zLGMYcj1laXJePM2+7/H13gWdsjQdlgc85Z/hbOcW0Fpk3dZLE2WtHSRJjXqokiZGTNCVNIBja2qVBh1mp0KbLLhkDIEuCNAmWtEpLO7gYipJBUfKSoYPfzyhJ0KDNHh2nSYcGHTr4VLVBkSxnzSo9DYj1ww3XtcGkFjgjK7i4hFiWpE6dNk26jFFglDxV6XwvD/GQb5OhCBkyZMiQIT9SXGnmYcC3s1EfHLe1WrBZlGwWE4OqSNROdbmgULUbG3fFXiZkXmwtRpxLFrsYurZ7megYiBvbd+KCSwJj435vsujdyhWPu0I1xiB9kbTpXmjI7bobVxxieBQ1jS8hPUKmEx63SZla0yGdDMhkutz85s/guB1yI3M0l7aT3nGK4/MehYNP48XrnD0zwfriDBO7n2P+zGHyhSYilntf9xSB7+G4HfBdPvGJV/H61zzPq37nY9jVLIWffIZ0XHn06AS3v+kLOPvXsBc8wmcMu2YWKGoMR+D9c6u8dY/Drrzh67LAzGSP3eM+fiDc5hZ59ESCO8MpeoT83XMuv5DYxbFei1+6xuEe3c6YTVOXKJflUWeBigY8cL5Nq3/bObPKm812qrSYtHmuc3JkNcF5WSNAKWicUZtizbTw1LDPjrLXjlHrW8wGWDIaI0ucrDhsj8d4hGVa0qVFyD5bZKdmWNQOe+Slm0+49cNvue/2vqApisfOjMO5bpdzvR6ne10eWulSMh7fWPLZ56RpB3C02uXLusi8qfPB+iJfdc7iYGjhU5c2KeIEhKxLmwQeHXrMmhpTWqBNr+/XBY86s4SEFCTNDltiXPO06DLLKgt2lQ4+WZLkJEVJstS0hU9InBg1jYIMo9R5h+06QlGT1LTVtwx2KUmOOm3GNMuklOjQI9HPIKlKixQx9uoYTTooSgefHVqiTjty4rrCfNaQl55hWOGQIUOGDPlvnne8+41f+tD9n7r7ytkfW0TJIMxPzKbAw6jqoJuqHIMWqc2hh9Exm9kSlrjp9w2w/Q9E+uJGrjq3cfkao7WHGy1cg6qO9gMRL63A6Vc0BuO6l0TG1uBG3ZhNiRLfo2T4S4/FpTawwf2JmxiKYkTwcBERKqbDDXYHZcly2I6wTTMcd1bp9VzumoyRSliymR5LK2ns+gQ2SJG96RmOfv42pm58lnJY4uiDR+jUyxgD2w8+RbdWprj7GMlYG1RQdfjEF3Yzf2Ga8aRLNuYxesMTgI+4AbroMPPW5zj2+QMcfOPjdL65l8e/8Cq8TgHHC7h1Z48vnkjwmkKWP1tY5tXlJNd5eYwISxWX36s8S8Uqn7DHec/OCW7KJ/FCj7Gc5VzDcueBOp85Z6hIl5b49CRgbzjCrKlTkQ5JYmzTPCB8lYs0tUPcxDitdVZMg4KmianLv/mVb3D08T2Ma5rjZo22BBzQAt9wZklrnDMskZUUMXXoKbRDJa4u+7WIBWI4rNFjXbpMSZLJd+5/SYPztr1z/5e2v+vAl/yPn7x7thNggQSGOgG+KkXjcmjU8OHaMuekttGkWJVo4L6gSS6YKnttmYp0iOGS0RgVaZPVJB3psSZNAiyH7STr0qKNTxyXOm1q0uGIHWfO1MiQBBHq2qZGFDJZ0gw9CYnj0aZLVlKMkadKC4OhSIoELmlJICIsa41pRvq3e9SkTQefIhkmyTNjS8yaKjVpoxCJD+nQlYDdtsyIpuhIyKn7j919/buuG4YUfh8ZDqYPGTJkyJAfGQbD6lfLH/mZt//2ey/L5riCO9WLVSgccdg6TP7tDMMPrrWVrWJk49hNrVCDYwbX3lqlGcyVDOZTBmxe59bbBza7mysyV7sfaZPuD9yHJPquWClJ8MpwNyXiFE1ktHnNqCGTCgitkEqEdHuGiYkqKys50qke8XiPZLrF6dMT3Pzmz9C9MM0nP3eEe15znFY9R2nqFP/1w6/kF3/3I3zwd3+KN9x1mk99eTdvvfdRKkvTjP/iUYIHHcx4jU/88dsYyVpcV7nlLf+AGW2hCpJ3ECOEpzyqz13D6ZNT/Pm5Ku8cLaEKo8Uep+biXGyFfFCeZ8GuAvDHpbv4yNoqs6bGK8JJ3nYQ3v98j5r0yGkMixLvP87XxtJ8MpgnxDJpszzhzFLRBoZoZuYwU/T6f0tpjdOTgJi6xHF4ysxx2E4QoPgSPV9V6eCoweu7Mx2yRdbpMSkJumppEbImXX7mwz/zss4o/MVP/eV7S5rgpnSaRk/5ZDjLDptnTTrkNc55U2XGFpg1dVIaY9HUKWoyGsiXdbZpgY4ENIgE26DKkCeFT0CAZV0bFCTdr7EpK1plVAp08Glrl5yk8LhU/RvXHAtSpa5tkhLfEDLTOkJXAlLqMWmzkVObjtKVkHnWGKPATFjkG85ZSmSo0yFNnLBveW36/2bsCCvSJET57Q/92nAm5PvM0KJ3yJAhQ14Cwvf83XufWYKfuG2Vs//iF4f/ef2A+FbhhxvzHluqDyIC+u23aw028ZsnOa62id/I5NgkHrYet7UNStGNzJDB797IHNky12LE3RAcA4ERCatLeSibZ1Ng4PzVFyeDoMYrrN0Rh452GHfK1G0zWgtCoCGrps3zssSbdIaK9fkPyyvcd7jEeiVOreGRSgTUKmkO3/ow67N7WV0q4Psu46NNPvT+N/HO3/8ob7/pOLOfupuJ/U/x//31a/j5tz1IeKzESjfkyw/uxhFw0g3qtQwP//Y93PvOz/KJP34bla7iGINj4KEH3kBoBSNKLxBe8+5PghW+8c2dHNpT5Xd3dSlMfJ2Lx49w6nyWv2uucN6ssWordLXLAWcn+UzAr+Vz/MkZy9NmhQvHEhyQPI4KHQlp4ZPSFHeV0nR9GPFTrEqLWVOPMjFQypIjJGpdW5I6WZIsSZ0xzbJqWuy2JTLEWZc2FojjUJEuPgFZEmyzWQIsS3S4PpZlttdjnR7dfp7Fy81mr7ZPh3NY4IypYEUJ1OJLSF2iFqumdClpirp0yWqcsmYB6BJwOBzna+YUCYmxTQs4GBYlcohLS4KyZjgtS2RIUpJc3xUrgSORKOsSMKY51qSxYeublkTfTNdQIGoHXKLCYaY471RJk8DDIVQFgSYdnnBmN4bXo2pNgguyQpk8RU2SUo9FaRCIZXdYfNkf73+MDGdChgwZMuR7JPk7f/te11GuGYPy9AkO/cc/vWqexfQf/tV7k7/zt+/d93//2feUeTHku2NzVoaIwcPFFRfVF3HT2jRbEWq44RS1dR7jqudv2fwPjnfF2xAVl7VAXeHaqhZHnI3zB+/cikQZCEoUlrgRJCjyAmEymOrYGqp4tfUPbHkTksDB4Xb28057I/vYxmvtASBKBE8Zwzlp8Pb4NCcvpHBd5QuzPo6jLK0l+Mpn7uT0qTHEwOTOMxgT8qbXnKT26esgY/j6kyNo4JB2BffWBc49cQsJY9i9rUOtp3zuE3dzdjbNG+55lC9/7LXc+1Nf4Gf/+3+gE8A9v/pRXvlrD9ALIBG3/Nh7PkLvwhTajnHPrz/AMyfz/OWXxviN+w+wtJLmkyt1AP7X8j5+07mDUVOiRot/efEU7zu7ykMcpyMBp8wKgSrr0uWgZOlKwLWJSIBcbIZcY3KkNEZVWhy0EyQlzoimmdYieU0Qx2XSZjEIu2yhbxMb5/pwkhSRbe2dMo6nDglirEsLAfLEMAhP9Rqs0CVPjHd/5Gfu+7kP/+zL/sbGL334F+57y0fecR9AW6LKTiBRUnmIpYtPWwISRG5TVelQoUkLn0AsdekyoimecGbxxGFc8zzPLM9wgRvDqUhMkGBdWuzRceK4eDh9+TCwxBZ6+LTFxyVKYh+kn++zo1iUg+EYnjrs0XG6BFRpMa45GtKjJm1yRK5rbn/Lm9I4DoZaPxjRw6ElPi3xsXIpJHTI95/hTMiQIUOGfA8c/k9/8t7FxSLZtE8249OolCmMLjL66CN3L910x2X9xBO/d/97u12PfKFFo55h98nP3r100+3DnuOXkQ/f/+m7gb51rjCQCCHhxjzFC9FLx4vg9jMghE2haWqvcm7EIJtCUUx/ixPZ3V5KLn8xIQNg+qJg83UUNiodAnjiYUQ2AhU9cQFBsXjikZQEKpurQJfkzwttjRXXuMQlRg+fgzrBDidJ2iYoS4yuKmVN4qkhpzFOBi1um3T4+Nked44mOLPsEoTCjdcus33mAmN7j1Gd30k62+BvPjvDK171JH/9vjfyE/c8R/yWczz/5cOU60VOnCnx1t96gHPfPETCMSzVIekJx05s43Vv/xySa/Ppv7yXTgCyMMPY9cfZdeAkE6OLSKqLd7iCSJcv/flbOXJghQMTIY/OGtLdFEft+v/P3nuGyXGeZ7r3+1VVd3WanpyRM4icmEmISaQoeUWKIilbDusor30sh0ve9fHuwXKPj86u0wbJa/myJQfp2KLAoGBLFCmSYgJBEiABEBkg4uTU0zlU1fedH9UzGICURdKyJJJ9zw/0YLq6q2u6er633vd5Hn5rWZrPn59m3NS4RRZwb3sPB0sVyuIxajKUqFLFo6gMa3UbE6bGetVMU1SoBvCcP8mwqXBOTWHXYyfT9Wvvw5KlLD4RbMYlT4wIo6rI6qCDGDarYzFe0uM4xmIeSUpG02SinJNJOmkigYMB2oiwyo2T9QMW3rPyR/oZkbpz2VNX373lqSvv3vzUczv3bA/F3w5l8UgSZalOMyFlIlgkiJKuL/LbTQItBgebFDESRJhn2rjM9HDYGqXdJMnX7XSnpUSvaaYgVQQJNVSAj66n79Q7cASzxUheqkB4XvToFIMqxxnGWEBbmNkiPh4+ghAjSokqHaaJklRJmxi9pokJKWDXwwljRPjNB37pvmvu3vrUqnsua3wu/xBojGM1aNCgwdtk3h9+aUfNdVm4YJyz59pZv2UfOnBwukY49dT1dNy3c8f4jo/OXsHs6j+LsnysRJ7SeD+F6ZbX3afBvz4yu5gPiw8RG8tYBKb2fbed6UTM6C8U8j3tdS9lZjRqZtuwcJjpsrzeLWtuoCIw28GYIexuhFeoI+IwE5ZYrQvZHYnUuzZW3cHL0GqlGdETaAwRceo2waHU+NIiaCaY0DM+m2UJaSJMBB4dyiGqFBldYaWkeNaM8uF4D4t0hNdGYXPKJhn3+dvJCf74Cs2ffbeF6zo6WLemCTdWRGuLX/jU13nkcx8m5wckbj8LFZtbrjvJw48vocUVdv3Vh1m5coi9+/vRGKZKsG5hBUlW+OZff5DLluR46WgTl33sH0GDpDxU0yT7v/ghimWb6WI4qvX0Kx38xPv3M763nYMVhw3SzuqbH+O2odv4T6XnOSApfi2/Bk8ClgWttKgYSRPBNRYxLBLKos92OFWrUijadEdtqhL+Hq8N5vOcdZ60cUmZCPusQVpJkqVEmvjse6yKx0FrjA+xkCcqE/SQIisVviWnsSRcAF8RLCKFQ5cVYTyosSQRam+2P/ThH6vPht/d+YnX7c/Td35tx6jk6DJNlMTDQ9NpElQJqOITxwnzOYxFp4nzsjWIQohiEydKhiIKYUzC0TYXhxo+KWJ4+JSoEcEhIMBHE6u/70MXq4CcVDljTTLftDJAgPUGGR//4aN/vCOFy6SEnbBWEvgYFus2zqswL+TXH/iFH6tj/V6g0Qlp0KBBg7fJ8sGvbI+1j/F3/7ianqQwOdYFQZxkMkPH+sM0t4/S/OTJ7YtKf7M9+GJh++hQF5VimuxYDyIK37PpX36Q1FPHt09u3dK48vZD4K57b33qrnve/9TD9z+2HcA3/kUFyfdC1RfoM4XATMaAfpMaEhG5MIpFGG7o49e7KTP2vhcL0y84cdXHqsQQkeisEmXGzWt2vKqu7QjQuOKiUOHrqz9ORBx8NAmJodHYYtcF515dnBt2aFxxwyvP9Y6KLRYxFeNne9rZ1O9zaFyIKUUqiLIoZaMqUeYlLfrbPe6fmOKQV2B3rsh21c3GywZorXXyzaEKCyMpYjEf2/aJxLN0xS02rhjjb//X5Wy64Qj7/+kKbvuFb7Dr2RX8xH94mNPPbkQhrFpYYGA8yuolGZ56dCOrF+d58WgTd937JKqzDBaIA8fvv41i2eHMhIWv4dZb9rD29hf4/Beupcck0MBZKXD6pdXs9XKkJNQwPBsM0mqSdJg4VQKGVI4INnf1J3k+W6IQhAvfJUkHW0Gbn6BZu6SVTQafSVViQGXoMy1UJaDDhOF7ealyr72IKV9TEZ+sDrBRbFItjFBlgW5mUpVYpNsoSTjaVNCaBVaMg9Ui+Iq+e1b82H8uLLhn5VPvu/uKp174ysvbJ6WAIzZFqVHFZ63uYFSKtJsEBanRYlzWm06y4jEkWZLGRQQiOJSp4dbzPGZ6c37dcnfcZHHEZr5ppVTvgMwUKhFjk5EC//GB37jvg3dvf2rr3Rtfd8x279y7XVD0mWaKUmNSimQkHNualgr/187faBQgPwIa7lgNGjRo8DawfusbOzxP4TiadZteZXxwAb5vo5RhZCzJ1p/6OnoiyeMP3kxve5WhiSgLesoEgdDdN065GGc6kySdLuLGK+hA8VfPtHDDj9mVz3czP3Pn7+6YEWy/kTPWXD3GXBctCAuWGWepuSL3f85udyb53JIL4YYznZS5HY+Zx5hxw5rZztQLh5nMDltsasZDITgSISlxqiZM1/bxMcaQUGFKdUVXSKj4rLtVrO4MBDChpwFISIyCKV0ogIwmLjFqeERwuFmvYKEVY0mr4AWC6xgCDcWq0BQzNDd5HD4f4YnqBHe1tHP5ltPsfmkRa1ZM0rX0EPbScWqvLkBZNVRHFjW/THCkBT/bzHe/u5FYxHDdv/8a37zvwzQnND1dBRbe+jiqSzHw5WuIJUo89+I8rtg0TG46RRAoSiWH9T/1DShbPPaFf8ONH3mMZ752E2cy0JsSrth6ioeeWETcFr5dHWOZTqOBLDXaiTJMme+qY2w2i5hvUmgMNTQpbAIMTWLzgowRNxFi2FwZbca14flCnpV2korWCHDATNNl4pTw2W8NE8Vhkhz36Mt4WSZJmrArtVxSHDZZJqQEQE18FgTNHLCGcLBYotvZarWy6isfeMd+DvzhRz+3Y13QQRyb3dYw64NOvqZeZbF04xqbKDZjUmC+bmZY5clRf8+hMPXuBoQp5nnKWCjiRBljmjZSpIxLgKEsHm0mzice+Ldv6lj9Px/9sx09uokBlSFHCQuL/7bzU+/Y4/xuoFGENGjQoMFbpPfTX96RycRYc+1z7HpkO1fd/jjnXt6G4/goS6PEIMrQsW030/s3ogOLprYhzh5fRbVqs/oDj6JHWkFpzh7ayOmBJI9M5rgy3kTUhva/ua3xh/GHyL13/OYOW5zZ0MG5zBQHM4VEXGJ4xruoCHkjLk0lhwuOWo5EqOjy68afLh3tssS6yDp4JqPDNz6CCgX1dUE6hF0dW2wCgllNSLNqIm8KxMQlLu7sGBiExY42hgA9uwCMSoSsKdAkcbx6V8VCkRCXAE23aWa1buMT1+TJTidJJMucOduC4xiMhkIlfE0b1w+QnUoTT1Ro7hjk2MHLePyk8JNXZ0i1TOBE8wS+i9M1wq6v3cJVtzzFo1/fzqKeMmNTLuvXn+EvHu3jii6H637tIdDwzOfu5ImRCle2xti8foCWjS9jilGsxUXMpML4ihfu/yCTeYWvIWrDyaxPq2Mx6QW4SrFfTzMuRdbqDs5JHhebD7c189XJaTJSZoFOc1Jl+OlkP+cLAZYIgTG8bKYoSJW0cdkebccSqASGgq95njHW6jYcUTgijOsq41LhiBqh37SQNBHSJsoCFWM34zRrl2FVwJOAcXJsDeaz1zpPgigLdSs/8+DH3xXn/x9/9C92WEYxJQWi2BSoskx3ckZNcnkwj13WWTQmdLFCkyIGMDt25RNQohoGYpJgyEzRUg9rXB/0cNya5N/v/NV3xbF6r9IYx2rQoEGDt8Dluz6xY+jgChYtO8vQiRX0908SS2VIpaZJpqfITXVhDIyMpCmcX0jvFbuJOkVqhRZOneqmVLGpDs+nY/OriB0weXoRE1mH+U6Usg+vlWo8uvOF7Zvv3vBjP4bxbuHhrzy2/YIs+2Jxti02pt7YmAkvnE09/2dHsS6EIYbFh8HGgvqYSeiIdUEWbuq3lagw1LAebjhXLq7rIt2ZwMKYcuudEkVUIjhiE5Uotlj4xiepEkAoTo8SoUuaKVBGG4MtCpcICYlSxaNdmpg0WQxQNTUsLFbSiy+GtCSIE+HaYD5rVDPbFyh2HojwwESG6YE2+lPC0VHFsemApa2wauUIsWQO161hOx5uxwhfemI+916Rp3PlAfKj8yhOt5PLtJFKT+LlOmnf9ipLrzrMiV1ruOaTD/P0A1eRVBZ+IAzuXcXQKytZfdkg+ZE2NqzIUirGqYz00rThOAf++kNMHF9K5/pjnN6zCtuCQIedmqRtoURwlaKqDcfIsFa3E0XRQQwNDJbC32cEGxvFfJPidLVKjx0hGwT4GOZLnD6TpBMXS4SEI6QiwgvVHFFsmohgifAaBRRCQTz6TJq1tFAxhuvTKbxAWGol8QOFbSy2SjuvyjhZVWGJ7qAoHlkpc93dl78rzv2r7t7y1HM792zv12kqEpAyLs24VEVTkBpl8UJLYzQ2ijK12R6ijyaCjYNNuu5uNRPI+WGzhAMywe888CuNAuQdTqMIadCgQYO3wPz009vbkkcpDffhxqrs2dfP0mv3Y63MMfXKZQB0rzpAV+8QmdFekpZPcaqbWjVOsRBDKQgC4c++uYDHX5rPrnGPTS0RilUhHoH+pMUilcD7xsnt7neOb5+4/7Xt5/7hxPb0Eye2Wx9c9q5YnPy48dD9j26/kMNwSV5HPVXcEWe2A2KJXXfvmetSZWaF5DMp7JYobOyLCg1gdvuZERRVd8kKTFB3yZqx8Q3TzG2x0SYgIg5KLCyxcCUSajbqaQ6O2FRMDer7m5BYaG9qanSrVrppYVSm6aUVJUKfaSEQTZYSa808bBQr6KGTJkRZLDfdtNUdhDpMgrNqilWmnW19wqFhi5z2+d0rAp44D9ct9uhNa65ZP8YnD47zoZ4Elq357Lf72LYkz+lDq+iN23T3TFCZbuczT7Zy69WH2P3iMpauOcmu51YwuH8lI/tX4gfCuRdWMTANaTccbKt6QqEq5KebuO3XHmb04Eq6+4bJZ5vwh7tZeN2zDB9dxqHn1lCuCamYoVQVqr5gDAyVA2JKERhDn0nSpGyKJsARISU2PoYKmhg2LUTCpHA0RgttjsNhnWORE6M1arHbn6JfxWmKCtrAK940zcZlWSTG0aDABifNLhlhWkrETIT1bpL9epqg4tAft3m1XOaQmmRUFagYKItHApeEcchLlf+489ffVQvrq+7e8tQDDzy+PYJNTUJnqxbjUpOAmvh0mBQpXLQYksTIELpVjZgMttjEcJiiWLfujWChKBtDUTyuvHtz4/PwHU4jJ6RBgwYN3iRrP//fd4x+fhWDz12LV43SvPpVVizKsfsfPoT/cjtNrSN0LjrCsd1XYi0ZZ+n14d/IailO1C3R2lJm7YbDKIFN8QRbE0luSDSjjdCZ1ngBrF01xrx2H8eCY2NCYKDFFQanhfGf/VYjW+QHzMfu/O3ZY3rpaJVdd4cCCIwmqqKz2pC5wX9wcRYIzM0WuTgjxKDxjU9MokBo8TvTWZnJHJnZ3sGedbYSURcJ59MqOXvblQhhukHdlhebCjUsFGmVRGM4J+MsNB04RlGkwnGGGTHTbNTzAejQCZpMhBbjsibopiBV+iRGGocUDtcGCyngM1WwuHJ5hXvX+/zp8xF6xMWNVTk3GkNE89m13ex8vgURzTVdEZLzXmNgzGVoyiKWnOYrT/biYnH/Q1dx1bbTPPQPNxIYqPkSFhsVYaoQdhqmSoZaAJaC5rih4glULVrbp3n1wGIcx6f9uufJH7mMdT/xCO1pDy+AiZxCGwgM7CrkSViKl6pZXCUUdEDSUaSUTUwU/XGba7uiOCh8NFf3hvbLzUQYp8orfpasVMj5mj2lAlc7bez1p8mUDbtzRW6NdLMlluJgrchauwmAJhNlZdDJUtPEk5VJDIYsHhEbJqTMet2BQTOocqRMjADNhCrRYmJv7k37DuMPdv72fWVqZMhzWibISJkaAfN1C1Y9JT5tYoyQoY0wrLBFUnSZNHehbY0AACAASURBVAA+fphwbiLEjcOwyjdyPN4lNIqQBg0aNHiTOG6B9q0v0nfFLtpW70diNeat28vlH/42Tz16DVasQFBO0tUzwfTzm8HxcTrGqNUipK7aT/f80xhjsfbG75KpaiIWvFQo8u2RUKTaFDNkJlPkiharl0/QEhXyNcPxQo1KYJis/PBTk9/NfOzO395hjJ61yQUuhAfWF/0zPw8IqBkPYzQ1U6sLzK2LHi+qoq8rUGZCECN1zcmMjmPGInemcJj7/DPFkFe3IYVQT2KLTdVU8Y3PlM4RI0pMomEOgkRJSbIeaqholzRlqth1J/520pxghCkpcnWwmJX08gG9Gg9N2oQFURybsvh04rJJd1IwAS4WBTxiWCxyXHpaPWw74PGDcT51TYWP3zBMKp3lj7JHyUy0kkoX+OiVGUYGezifgdp4Fzfc8hw3XnOSP//aavoSFj93VY5iYAgCG0uBo2AwX3/NQdhfuuNnH+Gqy/KcKfpYCo5OaJqTAd7pPjrv2sPJceHZA61MPbuNWtXliS9+iMmczUApIB03PFfIc7xYpSIB5UCTVVVqGjocB0sgMAZHCceLVQanBRcLG8UjQxU8NDbCGTXNmITjVS8xTk5qLGiBqyItKBG2JBMkIzBY8dgST9IUEU56JRbqJqoSkFIWE1KiUydYbid4bDqLxjAsJSI4WCgC0aRMmGGRkfIP8N3944XB0E0LLSTq42oVClKjRsCY5BmTPC4RXOPQbsICJC9lEsZliemiy6QZkAxVCZggz4QUftQvqcEPgIYwvUGDBg3eJN1/cP8O163Scdk+8GymTq+kWo7iRHzaek8iyTJDr24m3TpFKZ+ic+sL6PE0EvGRWBWjhf2P3sCLZxWb5xlyRYvzWVjWbihWFel4QDLhM51zaG7yGBiLMpQ3tLpC19+9sVj9tXu/uaMcGK5fXea+Q9mG1/3b4Kfu/J0dc0Xml7piOdjU5mSIzLhWzdxnhpkuiaDw6+5XtjhEJULFVHAkgm/82SJlpgsSE5eSLhGRyOz/OdizuSLh49izxZI2Aa4KReYRHATBmbNPFVOjW1qYNHnmSTvKCDkps0x3UBIPxyg6TZwKAWkc4mJRMgFNYnPWlPDRLJAE1y+Evz9d5lVrGI+A9wWLWRFz2V3OsjWa5v3XnKeUT7DzpRRlo/m5K/P8zfMpKgR0qSidUYtkFEaLhmKgmZ+wSMUMr4z79LsOMQdUmP+IMRCxIVOCribD2Qx0JYWbfuYbfOnPPsjHf/kRHv7r21i3pEg06vPoKyk+sG0Ky9IoK2ByrJlsPsK5SQvXhpMFb/Y1jZsaS50YTwZjrNGtNNsWnjZ4xlA2mpSy8Ez4fRaP+Jwk+QphITYsZUp43JnuoOzBvmKJW3tdXhgOuKLX4oUhzZpWm3IN2lKa/zEySJdJMt8kuGGholi2SMZ9/uTsGA4Wt0e7qQWGb/nD2ChGJMd/3vnJd/W5+9/v+ssdngTkCC+6zNjv9ppmzssU80wrU1KiR6eYknIYdGgUo5Kj37RwSsZYZrr5lQd+7l19nN5LNDohDRo0aPAmeXR/ktaes5hsAm+6lXIxzryffIZqJcrImZWMHllPe895zp/po2P5QfR4Gl2NYS2fYuLgBnb+7fuZyDqc1WU+c36c6ZLQ3yRUPMGxDBsvf4VcwWEir5jOOfR3VvmJawdpTRpeu/ebF41irfrc/94x/4++uKMpKmjgz1/1mZLij+jIvLOZscGdwcG+qLi4KCCw3v1QyPd0xroUz3gICm0CfOPVQ9Z8DJqYuJRNZfa+Bk1apULtB9ZFXZpW1YSDRZvVjEJISoyFdKIkdLxKESMwmg5JY2HRIU0kTYQUUa4M5nFGZZink8w3KRbYLjmp4tTLpi4rQtq2eNUa5ueXxTlFgamcwyKV4Fa9mI+YFdzUF2FhR8CUKpOKwv1P9vLQS02klEVMFIFvM8aFq/npGDgWnPeqXNauyFQM+bLQ5zrcccczrFpUoOLD5ZvPk6tCPKppicM1dz1CwddUPVDtNUTgwLduYrqmefyQC0DCVjz9SjsvH+wklshTqtjYtmHd4jLnigE9UYeb5zscIAPAtB9wpXSwtcvmQJADYK9McERlyGqfp9UQAGdUFg0U8EmIRa/lEheLbU6aW9yO8PeQMNzeH6VQUZSNxrY1K9I2xSpEHfjT0QE+YPUToBmmzBfPFEjGfYrlsCvVpROUfcOUFxA3oc7h3V6AAPzWA7903+/u/MR9GlMPJPToMClOy8Rswvx83UyAoSRV8pRJGZeluhMHiwWmnQrej/plNPgB0hCmN2jQoMGbZMuZF7cnmqaJvG8cfTZO89pDUNEkIzmaVh1n8uQSRGyy0wnKmS5K0x0k01OQs3GsgJ0HU3xgbYEuneJkIeDVIMsN3VEeGawyUQLJ9NOSrhG1hUzBYsOGk9hOjSMnO7jthmOUd45uP/b509tvKO/cHlv6Gr/7YD+r3DgLWg0tQYxJz/Dszj3bt929qSHYfAs8PCtMD12rgnrAH1IvNERCl6t6h2RGQD4XW5xZ96uwaLnggBWgiUgEEUWkrt+YEaYrsfCNV9d+hCGDHgHahAL4AJ+ESqAx9EgrcXHJmfBKskdAXFxKVFmv55GXKu/TSzimxmkxCQpSJS8VFutW2iXCYRlnqWnDQ9OsHHokRpdr0+wo/j44STWw6NFN3J8dYqluYaJs+MBqn4XN4AQOfz8+yXU9Npc3x9k9HDBuajQpZ/ZInByO4tW/WeRGGStpjhdr/J+/9iQLrzxIc76PqWyU9ctyfPXJpaxZkmVqKkEuk+LWn/8G/ctOs2jNSf72L25lZYeQTmrMYA+HBqKsWVji1EgESwnjkzEKniYfaOalhanxFjZs28sL++azdzh0wko6QrWm2JBMsK9SwDewuS3CVyfCkaj1iTi9OsFRM808k6LNxKmiyUqVxZJisRvlhFfiiGRYbqVocYXnijkWRaM8MVWk23J5OlMkgqLXdZjfUybmQDKmSeVaGPc9DEKAIUWEQj7CQ8URfra5j6gXYXWvZle2xJgqYiFseYOAvXcr+79ycHuCKAlcotgYEfp1mpxUAAGBqgREsHGx8SU83xImwicefHOZIA3eGTTGsRo0aNDgTXL53l/aEbzWBcDwa5fRu+ElVFsRNd/B3xtl4tg6LNunqW2I6PZhpr62jv/5nQ5Wx+JM1zSuEk55ZVbH4vQ1a748kiVmbD7SH6fqKa66/nkAxK1RmexmZKCfyUyMjdv28MTjl5N0NRVP2Lr5NH//2AL2mQznVIa0ibFQNzOmSszTSV60BpmmyJ/u/L1/8R/se+745EUdGKnb1M79HsKZ7/sf/p/vyAXCT935Oztm8jlscfCNd1E+yD/X8RBRszkdwKzGY6a7MnfMK67ioa5kjg5kbhjhzPNCqAFpVWlqeBR0iahEaFEpBEXWFIgRpUNCEW+AZl3QSwybPdYAG4JehlWBKSlyZdDPWSlgIyyjib0yztV0kbAU457PBFUshI+vUrx40mXIq5Ghxi+uVXzlVYtfvXGcSilGIpXn9Ml+Hjrl8ZElNsWyzcFRuGltkS/us+mxolS0pjNqU/Q1NW3Iap8tbVFemKxw83yHTdfs4qWnrqazPSyiHn01Rm/cIls1jHs+KcuiqAOaLIsN/Zp9A/WASIGYLazor3H4fITJWkDCUtS04advP8R3nlzL5jUTfGV3MwYzOx41L2ZzpuSRx2NtPM75ss8hMqymmZF69ymOTbuKcNzkyUmVtI5iBLY4afbWskyoEjdZPRgDz+px5ukk/ZZLRAnHvBIfW27zj8fDzlisnhPSGbMYKwdsW6D5+mmPMSnzs33N/LfhAW6XeXxHD/GTiX725kscVpP89gO//I48b/4l/O+7vrBjSKaxsXCwaTExJqRA0rhMSJ4luoOq+IxKjhQxOnXiXZOf0uACjXGsBg0aNHiTFPauQ3VkyY4uIN06xeTRteiRNPp8DVOM0fWBZ2hZdghl1fAPuKRXHaJVIuwp53nejBOzhQQ2N14xwIZNx/lwWzNbYylOT1gsXTzJw9+4ikN7tzB0ZAOxpSeZmnZJxHy8aootGwbIFBWvTgb83qPNfF1O4RqbIlXeb/XxuDrGK5xhUqoMmykMht/66KfftpvWPXd8cselBchc5hYf32ub7/cYPy4I6g2DA98IU/+avW10GA5IOGo1U5DMFCCqbtsLzKakByYgMBfCDoP6mJYlFhFxsAgdsSrUCExATELB+5TOERDQKc1sMQvr41qCj+ZVa5hWIqwPeilI+DzzdQunVI6FJoWH5hF1CoUwqCtYAp0Rm6ubkpxW0xTLDgtbYUksyqe25xkYifMr26fITLTw188n+eyjPTx3RpGVGs+etjg8CiWt+c6rCebbLuNBjaVNNrsr03ja8P51JRa6UUQMHpqN2/Zw/OWtbL3+OXYfSbDrSILlrYrRsmbU81gYd5gIPK7qU+QDzfCUjTFw/ZoCN22ZZKIaFomZ2oXRuFUdQjHXxpqlOZJNOdocCxvhxFg4xDZSCaii6VBRnitlGTAlVtOMjdAtLiXxOa6m+Cc5xVE1yjbaOWAN4RHwSi3HEklyNV1UAk1FazwCRqTEmaBMKQhT1f/8RJGoKA6rKWrG0Bmz8DXsNVNMZB02JuP8dG8LLwwabmUeuSBgmW6lFsCkVFmkm39wb+R3EP/ugZ+/T2PwCTUiGSnX/y1iocioMlNSJG3iaEyjAHmX0ihCGjRo0OBNcuiX/4/7pLlC84ITpLbtQ5Th7L4tZF7cgtU/gR6OYV8fRdwa2VMr+a+fuREBWonQp5N0NRnu3Vpi155+XnppBVdfc4BaYEi70No1yEDFo3/+GACDu65h9ZqT7Dsd4fnnV7FrT5jgfM4UeU1NUKKGjaLXNPNEMEKrpNhiFpM0DjGJktNF0iTe1uv85wqHS8eQ5jKTh3HvHb/5jig+3oi53Yi5hcnc4mPm+xnmFiwz6eYzBYgtoQ7AFmfWoneu8H0GEUWTSmFhEVexuhg+LExi4iKi8I1PAhdBOKJGuEMtZF3Qw516BQYTCsJxqYjPmOSZb5LM00nGKCMI1wQLWKlbWR9LUNPwojeNr8HHcH7CJlcWrlgzzdhIKyIQdUvs3O/w01uqZPGoGc1CEgzoMhqwgJStGPVrCMLJnM/PLU6wtMXi2UNJxqo+xggOitcOreOp4w73P3ANa+Z5vG9DhmcmyixoUgxT5lipSrvl4EYDFiVs8lXwjaFUinD/7jSuUpwcinD7pgLvW+5x08YMz43UOHasm9aOSfbtW4QtQpdr4xlDyQSM6vAxp7XHlbE0rUQZMRUMkFBW3akqS44SW4N+juk8HgFVCchLjddMgdd0ke/IIEdNnqixOa0mOa2meZZROmyHK5wWXFEs0c20OTbnSj4xG25PdGAMVAMYnlZs7VW8b3WZftfh42vgu+UMAyrLoMr/oN/C7xg+vfN37gMom1D/MaN/UghVfBbo1tdZXzd4d9EYx2rQoEGDt0DtV7+5I+poohFNb98kxigs22d0uI2VVz/Lji9cwUeXOKxYeyS0JHUdIhYko3AyE7B9RY2pbJR1G06y/5WlZEuKTNnw+eAIv51YTVPMkIz7LFtxlj94pItfXKv4/UNTrNUdDEqBHpMghsUeNTq7T6OSxcVhSE+EugMcqqZKn+p4y4LXt1o8XDqe9f2YGdm6947ffN3zfPnh//Ejudr5U3f+zg64oPVQEo6IVE31DTs+M8xdIFliMTPS9b1GuGZctWZ+Ztcteme2s8QirZpQKCJiY2FRNGUKukiLSpM3Re4xWyjik8CmhqZXuTwox2iuF5zrgg5yeCRxCDBMSpkAw093tnPZ6kE+92Q7x1SGJbqZfdYoGkPSRNikO3nSOsv1wXwq6FlbXg2kcVACE6ZGr3IZ11XOqjxXSwftMcULhQLL7QRVrQmAy9oVay8b5Pm9/WQqJhzPqlsVb+i0eHS0xAd6Y+TLiuZEwJ4hGNVV/u1Gn0f2J2iJWJyv1Gi1bYqBJmt8XBS9EYdxL6ArEhZ2q/tr2LbhxGAUbaC/LeDlARjVNW7ujfIPw1k6TYyFkSgTns9ZU6QoHv8m1cFni8cJ0FTwcHHCbAoS+Ggy5OmkmU6doCA1zsokcSKkTZjcHcFiWkos0m1UCbjGbuOkV0ZjuHe5zfHBKIMln6IJWBaPknKhUIWIBQ8WRvlwvIuhUkBBBwxS4t6HPvaevsr/63f93zsCgvpQYvjeX266+cQDDf3Hu51GJ6RBgwYN3gLlmpAtWTiOJtk8RfuKA0TdEsvX7uPQM9cyKiWWrT7J//rqKhbEHQRIxwyBhsXNFofPxXCjASIaEejvrBEY+H97VnDrrbvQBtZufhljFHmp8U8Ho0SwiGPRaeK8ZA3ymHWK8zJBTsocMmdpJcmwmSKp4lRNNdxPU+WMHnlLr+3NFiBzF99vpQCZeY577vjkjks7C/DGhckPA+GCrkMQtAnHq77fVdi5Y1lzC5DZx71kxGvmPjM/84130XYtqhkbi6qpUjBlxvQkCkWb1UyAZpnq51nrLAesYbLU6FcucaX4rfhqFgctrAracVC0EGVZJMaaaILt0XbmmxRDWWHPvn5+7ooi3TrJPmuUy4IOxsmyUrcRV+G+Pm6doUIQpq5j0yJhAQLgIDwpg5xRORbqJnwTZthYCElH2Gum+MgVU9R84bsv9ANh0GZr1OKKecKYqTKaE27rifHVoSLZCrwwaJjQNaoEWJahYjSeNqQtm2k/1H4sjkTpchw2LKqxocNiabfPqWqFA+cdBsfDAmTTihzPnNNMao8Aw4vDmhYTZXPa5VitxFlT5LaWZtbRwtfzE2wMeuk0KaLYWCjaSTFChoCAXtPKFAU80UxLabYA6dUptBg6dQKXCBkp02lijHth9yuJjeNo/qk8RnvE5piaBqA5GZCrGZbNK3Nnoots1bDfZIiKYrFcCJ18r/LZB/7TfQZDxVSp4dFGU6MD8h6hUYQ0aNCgwVsg/fnb7mv6q9vuEzE40XCU4q++vYDPPbiZ/WcdrrLaefDbq4iJYs2iEkXfsGnjGVIxTb4CvoZ1V+4mEsvSlPRoa8vTnRLmzcsg8SrHJw3jg0vwahEWmhRPq0E+Gu/lQ2s8HlFHuCaYR4CmmxYcbBapbo7rAWomFDAbYyibyhsG4f2guLR4mFkwvJ2Fw9xiRERdlGD+w+JLD/3R7BXXmcIhKtHv2/2AcP8vKsqMrgcGWpduOqshscR6XafEGE3eFKhQo4ZHUmJ0qBa6pZnfsDfSIkluMfMpUuHmYBHdxCjogAG/GmZiiEevckmJzeJINBy1MobXKhUUMF0L+MvcOT79glCVgMuDHtokQhNxXrZGeJIh+nUz24J+ioQL+aSyiIjCr3cxzqo8t9LPMt3M5qYYjhKyeKRxOF2ucbXdzpN7OyjX4LVSjdN5n9N5H23g8HCoJ0lEYNewz4e6k1gCy9I225pj2Ci+vCdGp2Nz/bocN64p0ec63Lolw61XDzK/BfadjrBy6QTHhy26rAhXrSqSjgecK/lMZWJs6rBJic3t/VFKJmCxE+N0PmBrMsGNqWYKVThABoPhhDVFqEnQlKnRruOs1fNQCGcZp0qNKj69upkFug0thuPWOAUqnLGmcYwibiIMqyKWCNuawtyV54+73BrtZPOyEh9JdpH3DH8xMElPUjhyNkY8ahjxPFbTTLNtzRZ/73U+98B9933hwU/fV9EVfn/nr93XyAJ5b9Cw6G3QoEGDt0H1xlVPffu/FLe/sm8R2lBfTAgiUAo0Q6ZKvJLgYLnMl057tFWaSLtCMgpnXlvA3kP9HBsXvjxQZp7EaU0aImLz3IkkRwZdFjQ5HBgVPphu57PF43xrIoeIYkrVeE0PEoghYwqMm2k0BlciFE0JWxwC41OlhkJxxz03v2nrzwfuf2T7WzkGly7I/yVXL6Wul5jZj7vuvfWHaln6rZ3PbO+wWqhSwxUXV6JU5uR3XNjH1zuDzb0dFjEzhZUhIpHZnJHZ+0tdG4Jgi4OIoESh0WijUWJRMmUSEiOCQ5uXYliV6NZJNtLFP1onWScdfEedpcMk+JZ1jJW6EwxMUGMy8Pn9mzIcO5PmhmWawSmb76gBPrM1yf3DeQZlmpNqimEp06vTjEkeJYpluoUyAW24KCBrfComwMeQEIub21Pkq0JJa85Xw3GtKzsjGM8iGwQcNNOcDkpEvQgttk133GLTQo9nRj1KWnNLRwI/ENb1atyI5pUxTW/CwrYMrRJh0KsxFfisalX848EIH9o2xaN72hgZSdMUMyybV0EpKBVjpF1hPBNhzepRIpUmciWLiA2BZ9GSMJzNacpas7bNpq/D45lhj1NemQgWBVWjyURBoNs0MSlFpqTEGcZZRAdLTAcp4uTrOSpV8ekySeImSouJkTARWk2MgqqxWKfZZQ0SrcSxUXRGbaZrmmopwkTZ0J0UnEqUmieko0K2IlgIVWPYo6ewtM2Se1a9Z+x5vx9v5fOqwTufRgneoEGDBm+To0GedlfR4lg8bcL5+sCY2Tn6Qg0shD7dxLf9Yb6ZzXAyE/D5zAD7KwVO6ALDKsdtNx7GdT0++ViCgDC5eed+h2esc3wmfwIHmwJlqtQ4ZYYBKJoKFVOlaqoYNCUdhsTpuutSTNx/9dd/aadgtqPxNjsic//9YXdEYkQpmgpxiRETF5/ggoC83u14owLkn9OMwAWh+lwuDUIU1Gza+Qydqo0ATYDmn6wTbAy6+K51lsl6cvvjMoBHwMvWEBYWJ9QUGcLRqJs647jxHLv8ST5zPM+AKbMsaOOZfW1kKbFad5MmzhQFTqgxekyatHE5rwo4KAIMOTyiWExTIy0OeRPwwniNSqBJK5uPLFdc1qnYNVrju94EGWq0mRibpAWAQ36evYUSr5yJEGAYlTLPjoejgk+ehq+e8mi2LV6duhA+1yIOHprHDkfJGZ/n9nVw8+YpfA0rVw7x3NEYlYrDmhUTzO8qs37VJEeOdrN44RQLe0sk4z5TXoAouL7P4SMbqhyaDPi7ExWu7YqSkSoFqRE3DlmpUJAaBanRZ5qp4eGIxXmZ4ozKMC1l+nWa0zJBi4mxXw1yUo3RZKJYKLKqSspEGZYiK4N2qmiGpUyubsVdqEFTRHjf9v3cstJjafuF323KUXhGs9ykmZCLC90GDd5LNIqQBg0aNHibDKgcR4oVjIF5uomn9CiWCEvTFvvVOOPV0AL0GeskS3UzGSljgFOMcdSa4Gl1gmuDfo4fWsYf7gmdep61zrNHjbJfjeMT0GLis3avUzobiqeNoaIroTGsCRexAQEplbwo3fvvHvrDH8lIw1vVibzR9sboH6pGpFZPYrYIuxDWnD+PMwXIm9GIfC9B+szPZ27PfawZW964xBARXAlTtD0TsCroYnuwCBeLm/VCnrbOIAg5SrwvWIiDzac7V+Ni0y0uaRz+amKY3/v6PFqMiwY+dX0BgPdfc5ZfsFbhGosJ8twaLGOt7mW9bidpIlhG4WLRrhyWqSRpsbFRVEyATTh6ZUsYBrj7NYdHRkp02A79JsmYKlHEY0hX6YzadOGy2AlHtjYk4tycaqZZbL6TzRJgaJYIk75Pm21jjPByscgpU+C61gRndamus4Gh4TQ3XXUeYxRXryyTSIbFdkf3JI7jsWh+lhOnWrEszYEBG0eEhQvGiTiaXM7l6gXwwc5QE+NJwLVOG1UJiGITNw4jkmVUclhYJHBZE/SwMmhnUdDChBTZHMxjnxqgzzTTYZooi8/zcpJlOo3GEDEWMSy+pl5lTAq8oqc561W55aoBpmuaP/nqCva+5jJVEA5kq+Q9zbFyhaSy/oVnSYMG73waRUiDBg0avE0E4bCa5DvBCBtiCTwCdvuT2JYhZaIsb7YRhCWmi5NqmnHJ8/dymIIpcSg4TcGU+IZ1jCdPQ9pEiOKwKehFS9hNqVAjJ1XOmlEiOERwKJsqGoPGzOZOzOX/e+hP7vvSQ3903xce/PQ7eqb6h90RsbjgiJVWoVhYzelyzO18vNEY2szXpYJ7hcy6bkn9tsiFXBIHG1ei2GLTpzoAKJsKccJC5IA1xJPWKb5rnWVzp82H9DJW6i7WBD1ERdFuEnx5JEupng1SQ7NGt3HUGiOK4pga4eefLbPLOsXfPN7Lbm+a02oaC8WgFNEYTkmej7S1skW10q4cKkYzpmt4xrBUJTgleY6oDJ0S5XkzwbBf40RQpErAIT9Pk9isN21sjqTJUSPQhpWpCP1NQqYWELHAsSCqFFujaSapkrYsMtQ461V5ZbLGbT0x5pGgu63GKjvJr15T4HzZ4+iwRVPXGV58pZfW9mm6Vu/DGGF0uD00hug5z3BWePl4kmtXF7lueZVdr3STStZ46JTHi+eE1rTHnkqOXp3kcK1E3DiU8TnMIMN6khJVWknSbdKUxOdla5B91iBp49JClJrx8NBs011kpcI2s5gsHne0trI5kmae7XKDXhl2fFSBYSnyl092cMQvsMR1uev2fYyWNf/u/YOhbS8Bj8sgJyT7ug5YgwbvJewf9Q40aNCgwTuZ1bqN/WqckYrPR+P9HCmG4xUWivO58Kp4k4kSNw6nrDGmdZ6aCXOulQkomjKPqTPU8IkRZb81ShWPJmIIiiKV0EFJQscrW2w8U0Nj+IeH/vQdXWi8WT5+56d2zBWP/2tgoVAizJNOJkyOmvFmF4hvNHI1o+fw36AQlDkBhRYW/iUjWXO7JY44uBLBlQgTJoepi8BHTIZWSTFmprlNr0Eh/JeJo1xGD6NSoIcUk6bGPJNkTMpMUeCgZEDgpIxxU7AUD83WYD4HrRG2BQvZo8Yoi8/SoJVmiZEyEfokxoAp8+REiRYiAGSocU7l2GQ6KGpNL3E6JErcUqT9KBUCVjlJDnsFolj4xpC2LCa9gA1OmpaY8FK2TIs4FEzAYK7CfCvGVOAxObjW8QAAIABJREFUHQgr7SSjfo1FVpyBoML75zucGVfkqXB0IM4RP8+8I50sT8GmyyZ5/olr2Lx2jKhbojiwmN0H2uhvDXj2udWsXz3GgjZNoRIe77Epl3VL85w4m6JKnqMmz7USo4cYh1WGZTrNNFVKUqOTNEP4BEYzIXkitDCsciRxmaJARXwmTYX1Zj5FqbJHjbJat3NG5VhqxXl8soCHRmPIqioLdTPjqsSYKuIGNn3EaYrCr3+1nz5T4cihBUSUcHk8QSnvc8AaYlPQz1fu/PKOux+69z1xLjdoMJdGJ6RBgwYN3iYxbA6rSTbpTnwMAyUfB8XRKY2pazv6dIqjapRj1gRZXcA3PgYdpmKbgKIpMU0RCPUDg2aSKA79uolJPU3OlHDEIWsK9VRu/yKr13czph74Bz+cjog2hkmTn9VxzBQLM92NSzsgAcEbO2jVC5CkxLHFRomFusQtS0RhiUXV1MKwNl0kbwpoDAkVJy5RhvQENeNxTmXJSY2rggV0GpcpKXBYjTCsikSwGFQ5WkmSlQopHXZQHrde4ynrFAskThKXKBY5qXJl0EMExbgKsz0EiGMxoPIogXxdC7JQp7GAiAiLnRhT2sMSqEqAhVAMNDe1pvDRVNBUtaZkAiqB5oot5/nFK0q4SvGxjVUW2THWdofFzRgVrllVwkIYCircvdziufOazpTBQXGsXOE3tnmsWjbJ4t4qh463sWXbQdx4kcSSE5w73UdnU3jMaz48+mIH8/py+AFUqzZtzTUSyTK5utQiaRwePK6Zoka7jpFSNuMSnm/DZop+aadDmlhiOjjDGAt1C/06zTrdx4BkaMOlyURoNjGuo4e1SZeUjnDELzApFRSCJ2HeyLgqMSxZosZiSpUpEbBl3Qj/dXuV//jxPXxpMIcl0Nfu8b5UmsuD+YypIhV5vW6oQYP3Ao0ipEGDBg3eJst0Mz06RRaPdsemzbFwRWEBaRPlNV1kQOVI4DJqMlRNDQ8/tAY1Xn3uXRMnShWfLEV8fKp4HLXGCAjImwKe8ZjWOYBZzccbaQ/ejfjGD4+V0bOhgv8aKBEsUWR0lmmdo6iLrxutgpkOiI1VDzR8o8wUbQKM0dTwEISoRLGwLsoNmdGALLJ6sbBoVk3Y2CRVnIXSRcGUcSXKQtXNWZmg2UT5/9l78yjJ7uu+73N/7716tVfv+/T0bMAMgAFArARAkEMRokhBK0WQihMtWawoic9xnGMnPooT2OckTmLTPqHs49iJThzpWGG4iBRJUYRIgCBIggBBEBgAs2A2zPRML9Pd1dW1L2/53fzxqhuD4ZCAQNAihfrgzJmpLtTrqlfVM/e++/1+b00CTphNCmSY02Ea9Hiuv3DQotwTTxNIzK3xLPM6Qk2bXNIOd8VT3JTKsdeOcF0+xW25HGM2Rz2OyTsOi6bBnYxzhjqLpsGY8Zg2Pi9TZ017DKeFOddnJQw4kh5hwiQTk29X2rgYhiTxYjQIyTqGP/7aNBcWhzlqtzj5SpH7DvSYGGszhk+JFJ96yaHkuGRxyed7DLsuX1/vcO+4D8DRl0dw3ZgXL/jEFtaWdrN8cZqV795Nsdglm45pdg2eA7/6wBkeO1qi3oMzS2keOWX4xJMjHO20WDMt9rtZvulc4EVnjaPOCou2TVt6vDueY1gK7LYjhMScYJl36DzLps60Zjlpkh07l6RJiRRdiXhOKxxtttkneSIss5rjejdHTboctmPcYIfJk2ZEs/SIuCxt/vTbE/zPjxf46p+9m4/9ygUcI/gpyw17m+x3s9Sky6Z0flwf6wEDfqIZNCEDBgwY8CZ5xlnFQdjvZunEymYY8zI1hlIOeVw2TYeyNGnQoa6t5Mr5Vc2DI4YL8SoNbZHCI9SIy1qhThsg2f+h7Z39E2+X5uNKrmwGfvfDD7/ljcjHHvo3DzsYGraFL6k3fI6DflLV1c3KthxL+v/EGpIGx8O9InHLEhKzbDeIialpk4ykSeEB8H57I7MyxrpWiTTmy+Y4ABblcDzBomwSiaVGmwU7zC3xBC+YMjfKEBXToUfMXpnmW8456n3T/axm8Rw41wqY0CyCUI5CZmyeto2Z0zw36wgvaZUx3+FdqRGyuGx1lZG00CRiqqTsLTms2R4hysFUlnHfZdF22ONkGc8JEUoqZfnd/QWsQmwNuVyHX7ox5tcOwn0TPvftD1nIpPjy0RyNKObIRIa5qTYP7nE4VU+e70gW8mllYzOLEaU4XOWR5wu4jsVz4NC+GieP7yXrCvXQcrLZIyXCCar0iPlZd5JQldvjOW6Pp5jTYYZIcU88x7NmnQIZjptVHAy/aG/kgqkwbQu0iOgRsd+Os2YaPOdcxqLcJiOMSoq/MBdoS8SytPiaXWXWFjhmNplJebTpkVePfXaYCc0AsM/k6UXQa5d4JLjM4mWfF87m+ZbdYF88Qr4vgxsw4O3GoAkZMGDAgDeJQTjg5NiMImJVjlElQnmst0FBXPKaYk23cDEMm0K/GH2tLKdnE5+HRalra+frZVsjRZKaZK6SAV3rCv3bgVhjarbBb37ov31LG5EVKgQa4fT3c/ygCcirEcRJpK4rHo4411xgCH1TOhYHg4ub+ICu8ItYTeR1glCSPBERETElTdORiAU7TFpSFCXLXbqX58wltqSNRZnTZEP6PfE8B6VIWbo4GMo28Qu5CFn1mGGEdelQ9IVFaXK8FjDsuKQwrGmXSc9jS7q8ZDY5Y6rUNOR2Z5hWpHwlXCPEEis7Z6PaEr6+1WREUgzhYYBOpNyWyTORMQQRHB7x+MpiRMqN2TvTxXWTpm7PweNkswETIz2+ecZl/1TIdSOG+/cotbbwhZdSfONCci5fODHB6WrEH62V6fYcTlzMsLo0zQOHW7iuEkRCt+sxOtzh+pmAXQXDOakz4jn4OEyToRUrtTgxmj/vrLFgizzhXMBBuMmO4eGgKD4ePoaSZjksJZZNgz06xv3eKPviETx1qEkXR4Rz2qRGh1PmMqumTkF9Lpk6BuHRaJ15O8wHJ3OcMVvs8dJ8127y4Xs3eaRV5uTx3YzYDDdfX2GqZPmHt6fI4zJlc2/lx3nAgJ8aBsb0AQMGDHiTHIzH2CJi3PW4GHV5T2oUxwgnum2eNJfpSkgGnxxpVmwZy6sbs2NeLWytxvT6PpFIox0jc0eT311J/qreTln65Oc+/rY0sSYRuDEBiTTrjz/7z37k8/D3HvonD297MgxCpK/V51+deKV9KR1ALDBkiolUTi0iZmeKYhACDcmazI6HJysZ2ppIb1xxUVVEkuOPUWBDt7hLDnBKVtnFGGVpJLG+GF40SxTIsKVNlk0zmbA5ya6K27PTTNlhXEOysK+qjEuKy9rjrNmiJT2+0Nzg10fHWG8Il3oBPoa9qTRjWWGimiGlDjXTY6+X4XjY5K5sgXvsOMW04cneFsVmijvzORwDeVwOjzo8XY64cS7i6CWXyCaNSi9WzjYj7pvwefGCMF1SbrnlPF/6+l4cM87u0ZjLVYcP3XeZoYkl4tDnpaM3ECuUHIdyHNImJoo9PBEezI7z/FrE/qKL68bM33CU55+8h7mJDtW6z4nVpKE7FTaZp8CpoMMNpsifm0UUy0EmMAgLdpgaAYLgYTAot8WTPOfATfEYTzkrvCue5Qmzym8W5/h44zR/HDeZl2EK+PzG6Dj/38bmztTCw2HKFuhKRF5TTGiWOgHjpPnk+haeGE6GLT48PM5XnlEqssSptVH+7j0hkzc9z1efe4BiPoOHcsZs/qgf4wEDfioZTEIGDBgw4E1SwCNGqUQR1/kZxnLC54Mlvmhe4hfcWTao0dWAU/YSPU0WtTni7KQubRe2iUxrO3Y3QFXp2V7f+Gzp2SSW9+04/fhBvBXm/N/98MMPj1FIQgKId+RVV3Ktc77dlKhaOtojKxmyJguwMx2x6M4el+3HtLWDRXFIpmFGHHzxmZDEWwSwJnU62qMtPQpkcHFxMEwyhIfDDczSJiQgYt4O8S6Z5A87izzS2eDOw2Veqga8bDZZyLscTueoSZuc+hQ0xXPlkNkhRYAZL0U9spyqRTgY3l8a4r3OBJ6RZGJnoOAaHIGDUsSinGuGfL3W4KGbLKcrlnvHfZ5ZdNg9BGtBxL7JiFeCLg8eiliuQtaDXCZi4/IEh2ZD7ri+wbMrsNKOGb/vaYzfIezluLDhsNaJ2YqTBnDS+Ky3k/P+UrNLSoThvOULL/qsnz/E5xd7tDoeUSSshyE5V6hLQI+Yl80mTRtzJN6FYHjJSZZ7njNlsrgcsOM8Z9bpEPNdZ5W0upw0FToENAh5wMzw+VqZYc3y3ng3Z80Gjho+tnmOVdPgkqlzs51lSktcMltM2BxjmmFTurQl4oTZxFPDmM3yK3MZfE/ZNQT/cXEXjdjy8ad8/vn/9R7ee3ODTi+JKb6fKb74oc/8e13OOWDATwKDJmTAgAED3iRZcXAQnjVrhFb549oqBmFMSvyf8THa2qVhW0Qa7UxB4r5p+criNtZ4J/kKeE06004R+zb0gvwwUvLW6egb2tx5T661B+QHkUxFInKSuaKxtDtNRsHk2W9mGTaF1+yD2G46e9ojQ5KC9aJZYsqM0ibA7YsUWnTxcNhjR7nMFm0CFmWT/XaYKS1xxmzwXbvJ/XaG67TE//a0y/X5FCOa4dFmFVVwMZw2axiENjGPr3UpOS6bUUTFhjQ0IsSS9ZXzYZd8CrakS8aDbqwYgTXbY7fJ0CTi/kKB88tZprMOpXxELY6YnezQ0IjPXOiyx0tz9HzihUi50O05NBo+s7vKDI1WyTmGdx+I+MzHf4XyhYNkx5dwBGo2omAcfud96xw5EKEKoSoNQgquQRXeuwdOnxvlUCrL5y8EfHGlTZ2QJztVJjTDrElzt53iCeciniTv4eF4mvfGC9wYT9IjJq8eDoZvOxe5N57hBjvKfTqFj8duN8OXdYm8pthnh3nUOc+d8S5q0uXeeBceDmOa5RYzRF5T3BHP0pSAmgTUpEtWXXLq7yRmfXGpy2pNqLWFTpgUXD87WuA331Xl4nKB/+dinZ7EhKrMmfRb9XEeMOCnhkETMmDAgAFvgv/uoX/68KPmAo85F9iQBr8ff4+mBGxKi02t09OAtu30c4tev4HYbkTgtQ3HtfwJH/3Vv/1TddX0jRb1bxRFSZs0/+mv/fc/0nkoSI7TdomsZFhw5zDi4IjDmDOKL/4Pfex2s7ItlfNIphqJV8Rl3IyQlwwhEZfjzb73w+z4e2JiUuJRlCyeeNxgk0Srhra4TRcIiYmwpHCZ0izD5AkIMQhPOGdpScA+O8a05mhqjC+GpgR0IninO0pIzOUgJCTmRjsNQIuIDA4X4w4TXvK8S+ImEqyyZc71Odrocr83yslqxNmozdxIzDtLWeYLDmXp8sVmmT+plnm22eLsZY8aIR873maTLk0JWQoDnu3WORt0uVS3ZNMxT10w1KsFLrwyxTfjDTKZgNuua/Liy+NcPPYObt7X4qI0UeDvfy3DY6cdylHIinbxMDwfNPA8SzYb4nvKwqjiIORx2ZIe92aGOGE2+Q4btIkZ1wInqHJTPEEah3W6HHfWAKhJj5p0ibE84VxkTTqsaY8D8SjfjSt4auhJjAL3xLvoEXPIjnDabJHXJCXrJVvDwTBrEu/OAQrst0PEouTV46BTYK8WuDmToxFZTjUDxgvKwVHDY5tNfv9bOb67FlGVDmVpcV5brNvvn8INGPDXHee3fuu3jvxVP4kBAwYM+GniHzz0zx9WoEe00yIMS44Gyd6AjPi06BKR+D8iotedZCSF+mtlWj+Mz3zykSMP/foHn/jRX82rPPTrH3zioV//4BOf+eQjR97K48IPb0TeTJNixCEtKb7wqa8d+cWPvPdNnYf3f+S+J375o+97YvVTi0cccRgxRRp0dszmvWvIs66mYHLEWIwYFswkm1rr7wYxyURLBIQrttsn0xaLYjDMmfEkGljanNUV8ibLutQBYTvQ+bJp4WAokGWPHaUsiX+loD4OBh+HQ4UUu02OrSDmGVvhVhmmpTFVCcnisUfy5MXlMl06EtOOlRDLTYU05zoBBii5DmOex827A3rtFBob7rmxymfPKmd6XaomMb9ncAnEcj5qM0WGlkR0JaYjIbEmu0QumBp3pod4uQwvaZXdWmJ8tMPza8JX17tcvJzBi1xGcsKnTirvzJbIe4YwMIy4LlbhUC7NcpgY7duNFDlSjJQC6i2Xlbblwb0OB9NZPlVbxyDcruOkxDBDllekxu3OCCepcR0lMjaZUGyaDj/vzOFGacr99LqWRMneEtNgUgsI0t9/EiMIbYm5zx3lnDbJqkdDeghCjYgYBTXUJYljniXLknYIsYw5KS6EHQRhxHHJp5WlhnJ3Kct1Y8JW3SOtHl2J6Ynl8EcPv6U/zwMG/KQzmIQMGDBgwF+CT/zaJx6OsPi4JOGaLjGWgiZyihxpmnSIiJk0I9irvAtXp2MN+H7eSFMSakCgIS7Ojxzbe0KWOWEXWbRrOBgmZIj4qunV1TKt7dtZSTMlI+ySCQST+EMkw/XMMC8T1LRJt9/MpCRJO3NwSIlHSMSKVgiJCYnZa2boaI80KQ7YCXbpGJOU8HCp0WHM5jhuVknh0iZg2dTJ4rBFj3/XWqIaWNy+0f2obnFO6n0TtnBUylzWLqFYDkqBBS9DCY8Dc51E8mQcXgiajOXg4ppPZGE85fJHTxUYE58RkilPKDGeGiIsBuGM1OkS4SKkNZmuGISDdpRH2mXO2zbDmuaF9Zj//cWQZVPnv94zTFFc1sKQz52JuaOYpZCGXgQzro8jwlzWxTPs7EB5x5TwF2ttRsfqfGOjy5jjEUSGStPwgDvBATvMOZocZ4usMdxox/iaXaVJQPeKn8GC+nw72kSAeTuUeHfEklWPJHS4fyFAIBTLNBnq0mMx7JFVj45ElDTNh0rjyWJJ9RjrSwNvd0vUCPn5iTwPTGQRgZ+ZzBAQs9K0tHuGhZRPGMMfrlToEHN3psiR9AjzOkjIGvD2Y5CONWDAgAF/CTw1uP3rN00SWccoBWKUGMsqFfbqBCdYYjXewKJ4uAQEpE2GsF+QXhn5us1ftfH8xy3zutZrhu9/3Vfv3LjWYyKNaGmLFi3GnNEf6Xl1tEdafNraZdgUWNMtUuLR6TeMqpa0ydC1nZ1GZDtmdy0uU8bBEw/Tn3D0tMeLXGCvzJAmRUtbCIZAQ3xJ0dMA1TjZCyLJXpA1WwEDNzBHjQ6rpsa8HUZUGFafJiHfcc4DcE98gKaEiXQIy1Fnmd82h/CM8KVwhf9yeopPrNYwWNKaYUoz+OowTIq2xqxpj3F81ujyByeTtKsLcYdbUnke22ziYbg1l+WmPW1OHoM0Dj6GhvTw1GHNqTOtJWrSZdoW8NTwPecSH9FDvECSxmUQdmmeBiGT4rOobRbIMxr7/P75Kh6GgzLEf/WeMpncIpn5C3zuDz9AKS3MT/b46hnDfMbjl6ZzbDQM31oJ+L0HL9Ft57k1n+FUM2B+rsY3Lw5xzFS4xxlnRD1e0C2e0jJGhDt1kliVc1JnjxZ4yWwyYjPk8Fg1TRwMC3aEhvSomA4TWsDD4ZKpcSAewcPjhNkixnJRGpTUZ0qTBZWVtmIQbvVKXAg73JMe4uBMyFQ1h+dYxke7+F6act3QJubeAwHZbMA+hedPF0mrS0ziuRGB+z77S2/LxLsBb28Gk5ABAwYMeIP83kP/7OGnnfPM6TBCkiLk4dCiSxaPUQpMMUwaj5Lkk3Sk/n+OOCj2NWlJV/LXuQHZ9lr8MK41aQB2zOLXYvuc5STNf/PQ//Kmn/+QFFCUSTNCS7s73z8lHnnJUnSKAPjG3zHEqyYG9KIpUDQFMpJOtqj3d7544uJi6GiXrGRwxU1kWv3Fhb74uOKybjdJ9a8HpkgeO2+HuS2eYVZzdCWiJgFV06VEjjwZJiVNXj32SI66BHTosRFG/El0ERfDn660cFT4jZlhfE2kZfOSpUbIGVPBAi9rnWFSXDJ1qgRcNHUuBQFNCSlLhyCGY+ez3JrLMuI6nJHaziSkrHV6RPjq8KJZTozYRCzaNvu1yM86M2z295lEWGKU9w0ViVGGJMWETTaFPyvr/KMnMqwtz7Lx0m0cuXuJD/7nf0q96TGb9rjv1g22WoZyx9LDEoU+/+LxERYmA35uHzz94jgnzBZ3M8ZxW+ecbWKAvKaIxHJUygy5Dgco7kxsqqbLhmnjq8stOopHcn7S6nJW1pjXHA/KLi6aGj4OGXWTxkU9Djp5tiRgSZo8Gq0DMFMQFrwM+RR8/pWI5bqyb88mQeDwueUWKRcchPWKz9Mnivzb511ebLWJRfFxeKHdZrXzoye9DRjw08igCRkwYMCAN8i2ROe8lOkQ0CMi7OvG65JE8K5ohVWp9a+uJxIti+KLT6QRbv/K908KH/3Vv/3wW9GAvJ6E6o1G6l7ZfLyR426fz6ptvKHjX/MYmFebRk3ezzkZpyD5nVSr/WaGWC2BBgw7Q0w4Y8w7U+QkjYNhH1OMSJEhSaJ2Q43w1OF6swsRww1mNxnxdzapC0KsMdNmnGHNkZcsHXqcYoVn5QIvOeu8YuqclXXOmTLX2WHqtLkxnuI0NcrSoaoR45rmsN3F55wTXNQNJmyOPSbLECn+bLnD79zksiwtvmFW8ElM1x6G02adM6ZKLJbjzmXWpcFLpgwkn9eL3YBOqKRdSLvCfziZTJse0F0sMMGKVLkoFWZ0iLaE7NIxWhJyVuo8Gq9SVJ916TBGmkVtcaGevP9HpcyWdDloR5mxBcrSYnMrwxefGuOl47NoI83d73+CFzotFi+OMD8ZkHMNPzfvkZ9a5B/8R89ycjnF468Yjje75DXFS1ojrQ6hWDwcQpJ9OnczTsoIDsKQ+qRxuVXHmLV5pjXHt8wKh02JQ3YEC7zDzvO4s8hKFHCLHeeUqdCUgLv9IR4oDvEIS9zsFAkl5j4ZpykBj1eanA3b3HfXIhkc9o9tb4jv8dGFDHtm2wAEkXCu26VJyN98ZwdfHTalw7yTpmsHyXcD3p4MjOkDBgwY8Dr8qw//3w8/+umnjtRp4/WvVre1x6ZWd656z9oh0rg0pMeabhFqSEjUV+QnKUoxtp+A9aNNPT75uY//o7fClP5WTj9+qPFcDI6YHzrtuZbfYluKdeXtK+/3JJVIoEQpmByPfPqbRz7wkfv/0ufl8U8/fSTxHlh8SeGIYUNrABgxZCVNjgwN2uRMFoMhxrKlDSIsOclQlRZ1bREQUTTZJJJZoEKDYclzzq7g9g3vHe0gYog1IhZLLFCQDBGWomRRUaraoiZdGtoGEXxSZPE5bzbZlBZW4CBDHDcVOhIyQZH9OsFDswWeqrdZlw73ZoYYLVie3OzygDPNCa0T9g3XHg5NCdjeGO6RyIw6ElIizbRJcz7qsJBNsdKKebLRYF6LvChlehLRoM0uHSGvPlXpMqE5WhLiYECSq/8uhsumxUGGiFBOmiqzNo+Lg4ew0jfc+408IvDtRpOvPruLu8cy/NxdpxkqxORyXdbXi3xjvceZY/u4/X0v8pVv7uXnD0Vc3PR4cJfP+ZoyTpoMLr84k+V4M+Bd7hiVOOKYrVEjichO47EkTQqaYkRSrNDmFeqEKKn+RGSvHWbKpPiOrJFXn/f7k3y3V+PFoEFRfV6hCcC69pjXAh/a6/HMVpfLF8ZZ1S6/evc6nVaWf/d8ilPVmPuvr/P8xTS/8s41FnIZ3r0QsbmZ55UtcDD88kElR4rULx4YmNIHvO0YTEIGDBgw4HVYlwYjmiWF15eYRNRtg1gtLW3T0i4XzCZLssU+HacoWWLsTmSrQQg1iVd1cPpXw7+/6Iakwdj+da3n8pO6Lf2HyaYA5A3+c7PttbjW8be/x5BTwjd+P4XK2VnkGKvl7zz0j99wY/V3H/pfH/57D/2Thw1CgczOfg+rSlGyRBoRakTTtlnXKu+U6wBYkEm8/hJB1cSgPUSOQEO62kuM53hs2C1ypLEoP8ONhP3j5Uyu3/IoWUkTY2nRZYgcNW3R1ZCspFEsJcnz23IjXUmWCu614wjCRd3gEXOefXYIB0NJ0xyUAv92pUJPkqlDPbB4XkxJfZ6I12lIIhvcp8WdCU9Kk+Y4oy4ZEp/CYVPEAaZNmsMHyxRdwz6Tp0dMW0K2aLFXk2mIFSWLh6PCsE1kYqM2Q1Y9QmI8dYhVKWvAvC1wzmxRwiOFwwE7xP1mAoC1KGTVNJgmw3NH53nh6bvptPIM7TnFu+95hYr0qMURy998N/fPG1J+xAcOhvzJxWTS0CYmKw6qMKEZjoZ1mkR4OByUAhNkmNUst+gIStJEjGqGA3aEO8xI0lRKh+edywA7Ucrf6G0yTQYrSkcSCVpWPT40PMa6dPifFldpmICLtsNvHfS4vDwOJE2Yj+FffrPIgpfhXz8+xtJamkolixj4yEHDKD5/eDKm0nprI6wHDPhpYWBMHzBgwIDX4WA8wcvOOhalqW0CDYmJsSh5yeKIIU2KBh3aBDg4zJgxLtk1XHHJS5a2duj0/QZwbQ/I1UX8j7Ph+HF4QEQMeg3Z1bb5OnqdAZCicEWUsYh57e3++bEo+8wsS1omTYqS5NjUOhExKfX4+w997OG6tvhXn3n4mufv7zz0jx/uaI+S5OlqQFXriBhuYw/PcZ4MPoFGZCTNiBToErJpq7wgFwFY0jIhEb6kGJUiXULWtcasGSckpqJ1cpLGx9+R6z0t59glEzTp7PhOUuLRsC0ykiYkoqJ1rje7uKRlOtrDkSQE4aWoTsdEjGm238gaYmLqtFmWFitSxcVwykKWxHfyO7uHeWEJxiY3OfTyLCtRihOmjEFYlQ4TmqdMC4CsJs31Rakwp8Mci+vc4pZYinqcOjtKxhMuBCG3Zwp0uzFDkqEsLXbrKPO2wAvOOl0TUtA0HoaipijgcZ5g5/1alzY3MMQ+O8xCyudUkBT8vrrFTj12AAAgAElEQVR8gDkADtsx1ujy2c2QB8MRvr04wn8xl6a6OcoectwzJ9RqGYq5gHIlSy4TMUyKMj1qBHzoOuFLpywH0mmWuw6XaDGvOU5QZ5Ysm/QoaQohaTIOyxBntEmoSXPblpAb4nECUawoFdNm0uY5YbZoEzKqGWY1x1lTY7QYMV3JkoldxvFZyKT4yqmI2ycdSkMtGhiaEjKsPnNFYcHxue0d5/njr+6mbEM+cqPynjmHby/7vNzpsut1f7oGDPjrx6AJGTBgwIDX4ZizyowOsSbKum4SaIiDg2pI07axRnHEoaUdOvTwcKhrv7jUiC1bZdQMk5UMVVvfWUp4JUkB/5OtDb9SHrXN9u0rPRw/KM3qhx3z6sdcfT6u/J497RFIcrx5GWeFCiNSoEU38W0QMi4l/uFDH3+4Q8AhO0WM8rws0tYODoaAkFAjxs0wASGBhhTwCQgpmTxNbZOWFHXaRBojYvDEIdUv8gMNadk2+800k5rnMY7hiUNPQ9KkuMnOccwsMUqBVSr44rGmW4xKkbAfVLD9PFKSTFUcHJZ1E4BZGWVZNylKlotOjZKmOW82KWmGvXaUjEnRIaAlPbokuzTKpk2MpaRpLldcQo15+tldDKXgubiNQRi1GVZMoy8SlL5EMJEj3RnvYtU0GSVNrMqY8RCBZqAUxeWVTg8BSuozQZY10+KM2WLC5mhLsgfF02SSFWKpS49DdoQx1+NZ2+NZ1rlRx7gQ9OgRc3M8yeF0jjPdLh7CKVPFVUNTeny5LvyNmRJrFw4ys/c47926AzHQCxxu+OWv8n/8iw9y34LhP7l/kwuL43zhYpfHTjuA5V03V2i303zmWIYQy5RmiFGKeLSI6BHzjlSBxSCgRIq2tSDwiq7Scrp8h4iaNjmgM5wzZRbsCEaSEIomEa4anlyEG/M+zzdjHncWOdAdp0PEy2vw63aUDjX+1g1p6k2PLy22ed9Uhj/8yjwfuKlLaajBp58cZ8J3+IWburyyNIjnHfD2ZCDHGjBgwIDXYU6HOS9lGnRwcUn141gBIg2px3XW7SYtbdPQFjfbOXKS3imcJ50xAkLa2gHY0d9DX4r1U9CAwLUlV1e+ju3XcLV/I+4nSV0ptbqWx+M1zc0POB9J8Z4kkuUkzQoVsvi4OFhVSpLDw+VwPE2XkBQuL5hLnDHr5CTNvJkkI+mdX23tkpYUjjh8S07vTBo8XJq2jYNDQMiIFDlsd5EmlSwnxOCKy2mWeUrOkRKPmm1isYREXDJb3GEXGLVZXFzmdRxFcTCkJbUj2QMYp8QUwxxkhqa2uU0XKKpPSjxO2UtsUOcoF6jSYkLzNCXAU4eKNhJ5oMZ0iRjWDPvsMDHKn7bWKduQtZbimqQ5yKnPhmkzZrNcb4e5xY5xl06yJJssyiZtidhli7xsNlmNAy7YNt9bjzga1plMu4Qos2Spmi5LJknI2nlfMKz1m5sbUjnK0uVBZxZB6FnLu3SKO3SCl8wGRXGZkjSXTJ3HehvcXkrzstniocIks5pjwQ7hqLBaNfzTpx0e/tQt7FpY48afeZxeYDj6hZ/jN352Ec+znD03Qb3lkMFhd87lgT1Cyg/4H05UdhrbcSfF33r/Kg6Ch3C7X+SZsMY94yk8DKt0OGkuk5U0q1qhR0CMxUEoaobTZh1PDddJgbvzedK4HKPK+VbEBVPnvngXE5rmt6dHuN8b5fbbznG3P8TeQy9SyIY8dL3D0bWYhbzLyNgWS0uj3DkH+6ZCqnWfuYnej/RzOWDATyuDJmTAgAEDXodTsoqHQ5oUoYZEGn3fNEM1KXjykqUsbUJNEnpykqGrAQXJERPv7JKAVxsQ+MFeiB8Hb1aKdeXUYmep21UTjCujeAXBEQeDYPpfv7LZuNb042p2Jiz9+2y/kHcwhBrT0R6bWicgIiVev3g0POtcoqM9trRJ3bbYtFUEoax1AkJijSlKlpCIFB57ZIqS5Olol6V4jZCII9zAQTuFi0tNm3xXztGhxwGdwpdEwtTVANuPXn4fN+GTRPi26PGSWWHZVFEsJ7mETwqL0tEeWVJMSIlhKXBdPMpJe5El2UIQnpVXOCtrBBoybAo7Da9BOG5WaUsytZmQEmVp0tFuUuwTs2parJoaaXWZcVOUfOFYs8tNTpEpzTJuk+SsNA6rdBjzXHbrGHt0DF8d6hIwa4ts0qUhAet08XFIOUnBUCXE08TU7vRLiIb0UJQxzbFHchwLmtxiSvyRfZmiuKQdw6PmEjnjMKJZnpY1npE1QmLenx7nz+qbfLQ4xbdrLY5MZPAx/NpUibwPB0yBX5pLE/Z8qqdvZH6uys13P0N2dBlVWN4yrDWUfek051sRN932An/+9CRHmMYCDSKesxXCwGfIuBzOZrGq3OWV2GpLsvEc2KNjdDXY+Vyl8FiRKutSJyRmzTT5tLzMH3TOJl4XDC2N2GeHWJYWOXF5/rLys3evsb46xQ1zAf/yT27DS8Wk0yG7ci6tEE6fniG2wpnLhpHhDn7K0gsGpdiAtyeDT/6AAQMGvA77dJIYS1lruP09EPaqIjwpjhMZ1km9SFXrZCVNQEhBsvQ0QDCYflG+jddXxarav/JdIT+I7Qbi6iWCV/6u6E4DcmUzsX2utuVYV09DrubKpuxqA/+VZPAZk2J/uuDQ0i4eDl2CZE8LWTKSyKt8SYr/6+wkGfHJiE/J5Gn1i/eaNtkkaU4OOQsUTI6M+Dwj5zlt1pIQAiw5SVMiR4+YlnY5YOa4wezGxSEjab4pp+jQI9CQmjaZ1hJ12mzZBl3bpaEtFCUjPhtax6JUtM7TziK7zAQ9Aoomhy8eTe0QENLtp6zl+gb2qraYtgUcNWTVp0OPrGTYkDpHZZGLUmGfHcPF8AVdJIjh/kmfUJU2ES0JOWUqhFiG8Xk5aHPQjjJiM/QkpiwtmhIQi3KrGeaDE3keGMsRWzhralwwVU6aFdap8oqs4+Hg4RBh6RLyMnUumhoZR7g9nuOL5gwGmLB55vKGs2aDX/Bmd4r/r3creDicrUW0iSjmIvZ6GUaHekyNBkxnHC5XHf7iO+OsLo9Trb4qXdp7/Tk6kTKSFe462OCXb23z6KN3sHsI7tsXM248qtLjTmeEz39riqqN2OjGnAk6VCPLl9sbeAizkmFN6szICIpykBkAbraz7NExehpQpsEYBZokU4vDUuLDNyoLKZ8bZYgR1+GcbfLYM1N873SeesvjDHUcx9Jo+EwULTkPVrYMjlFuWQhI+QGfPRNx9vJPVmz3gAH/vhhE9A4YMGDA6/DVTz915EK8QqAhAYn23VztfZCkELdYjBgMhlZfftUjJCJmxoxR11a/kek3HZIE9ookxfanP/nlI5/55CNH3ooI3mvxZqYgV/o9ZGeq8eqEQiSZ6Fi1OOJiMKgkr0k1iSU2IqT7C/1AkvteM+VQQPvHSh53ZayviPRvs3OMOi2yksYTB0dM4rXAYVTzLFFGRclIml0yhiMOa1JniBwN7ZCTNId0BjEOoUTs0XGq0mZLm2TE77+XSowlIsYTl1t1NytSpUSGnkS06DKpRRrSxZcUB5lhgwZ5k2FWxqhJh7b2yJtMshYbpaFtBMHvLz1MiZc0qmS4bCtkxMeqMi/jNOiSFo9QY1xxKZBBRFiRKo64DGuWSOA6neAsl7FYekRMUiTGUlCfGcngGaHgGcq9pPTP4BGhHHM28HAZwefnZ7LYls9J2cLDYVwzPGUuU2k6DKkPwPG4RltCfFLkyVDXNmUaVKTJQTtJSwIQIasef85ZJrVIz1ie0VVusOM8FVbYa0d4Wtf5BWeO77HOTXacXSbDt2WNXVrgwJDDmS2l00rhG+HJSodKENOyluWKQ954RK1xFs/txlGfe+97jpGcx+LFYZbW04wWYqptQykfc8ehLWbDEe64YYtuI8cd8zGbdZf1OGTKTbGoLVSFE6bCrXaSk2adORnjZjvOhulwWerMaImUpEiLx5TNk8Jl1dR5UTbYVZ/hj6IzHNBhTsQNtkyXSc1y+54ABbxmjs8vd3nw5jq1ao7TFeWB2yuculDg+cvw1CWX2woZyl3L5EeuG0T0DnjbMZiEDBgwYMDrsEm9v+cjfs0U40ockkIYkqv/Pe0Rq2XGjDEpw+QlQ01bFCVHjEVIrvS78uok5CcVV9xXTen917gzAdLXnhNHDFHfuG/6cjPpS9B62sNqTMHkuDqy98oJyfbxLK+dDMUao2pp2zY36y7ykmWEPGlS7NUk6tXBsC518pIh0phAQ/Ka4rLdRLF0CUiJS0M7HDNLrOkW98f7WZItDMKkDNPVAEHYwyS36DwxMXtkirx6KJZzrNHUDrFadtkiOdK4GHKaIi3JPo+0Jle3s5Kmpd2d6UtesjuSrC4BIRFWlSa9ZDs7WRRljSpp8ZjVZElgSztUtEFLkyjfrHoYIKMeFkiRSNEslob0iFHWTJOlMODztTJGYKi/V6VNSFsiDsSjuBheNltk0zGrtseeeJhRm6EmAffE0/yNfT67R2P+vLeW+D6osq5VABaY4C7dw112D2umSdR/v26VYQ7ZGWJRFuIh3h/v4ynnIhVpc8opM29L/Km9yLvieValzddZ5T0yzS7P598sVmloRMaDrZYh3Y9NHjIu1w25pFylXPEZH23jeSFPPHovrUaOu9/7JHtnOiwsbDIxFHN5M0XQS1HIR3z2OyXCGDKZkPUgYkM6RFZ5jzNOTXrcZSe5aBrs1lHWqXLMlDloJ9mto4xqmjHNUlCfuvT6cchZxrVIK1T+4PYxIpSfHxki2V0jvHzJp9Z0qUQRQ6T4xDfGWa857B92+NdPZQH4uRuShKzzzYiMM4joHfD2ZNCEDBgwYMDrkBR48c4eCQcnKcyvkAtFfa+IIwarMUJSjF+y6yhKUztExDS1TVbSOGJ2tqjDFTKkH9DkvBVcPQV5I99re7P39p9tvxHYlmi54hJrjO3/P6pK0Snu7MHYxmqyjTzs71jRvil7p6lRu/PnWJP0qO2viZjXNDoihu/IWQzCutaoaIMzcpnrdRqAjvZoaodhyRMSsSxVspJcuS+R3XkfRykyLHkAAkLuiheo0SInGQzCpjQomxZZyXBB1zjqLBNoRFGy7JVp3mev58vmGBtao0WX02Y98Q0Rc0nKbGqditbxcGnYFk3t9A3pMRERNdsk0Ki/qDBCEFr0SImLh0uRLMuSJGR5uCwwQVo8GnQ5L2WmNMt/NrSL4f6kIi8ZtB+1u26aHIzHmHQ99mqRfFq5ruhSwmNWc+yjwLSkWZcmt+go1aZLTQJU4MHxAhl12aDHi5c8nl2BvKboEu00nw6GS5JE/talR0cipjTPOdngqG6RU48JzXDa2eCM1HhvvIdpW6BGm3NOBYOwLC1mNcvtdoJnbYXFsMuvlsaoEXKyHuA5EKN4GHxjSLmK6yhhLFSrGc4vjnLr4WVyhRYvPvVO9t/+XcobJfLZkNtuXiGKDJ2Ow3/wnnWu393isWNZRj2XroQsxz26VrnTjPKS2cRTQ0Cc+H+kw5imKapPHpcJzZDXFFM2z4TN4eMwpll+8V2rPHN8hL/5wfP8v9XLPFSYpGst7UipdWDIdSiJxx3ThvmJgO9sdvmNw0oho6xvpqlUfYquQyv6yb0AMWDAj5OBHGvAgAEDXocHPnLvE5/75FeP5EyWdH8B4ZWFN/QLehFc8XbkRTExriRL2wzCsBSYkCE8canZJgjJteurpEcAb7Uk6/VkWFd7PK51f0pSWLYLpv5z7cuMfPGxokQa4opLzmQJNMBekZgFSdPiSyox9surhmvtS7G2j+2Jh5XtgtfBYvv3K05f7jYmJWKxOOLgk2JTmuTxUYF03wfyDp3nLGukxeM6pjnNCiLCmBTxcGgTsGRqOBgumwZDZGnTI4NPQzs06DAlw7zL7uW8qTAqeVr9CcYxWdp5bV3t4YpHCpe6tpmVUbLiMy5DRGJJSZKq1tYee2QST1y6hH2Du8XFpSAZAEJiRimwoptkJE1Vm6TEpSMhkwxR0QaHmOE75gLVjkfFdBgmx2WqFCWHwbDPjjJNFl8MaWPIuYZ8WpnyPYaMx5fiJTpqmdQcR6bSjA/3eHqzl8TqegXumxXunBJObxi+zRrPyQX26gRGnP5k0GGcEpekQpE0w5qhKQETWiQSS0U6lKWNCmxKi6ZE1EwPF4dZWySjHoumgovHBVPjsI5y0lTI9bJ8z6wRI+RCn4JxeFnrDOGze1gp5EP8lDI+Uef4+RxhO0+lkmel4jJZcDj1ygh3fuBxTn7vJirVNO2e4dylAkHg4WBY7kYcdoscKHigwpmwTae/Qd4Rw4yWcHEIsBQ0xbTj85E72nxrRbECa6bJ/3hohKmoyOJygXYIz5we4la/yPRQzCP1LWZNloVR5ZZ9TTJxhtNlmBmyHN+KKcU5luuw3LCEoYNnBFeEkQ8PNqYPePsxmIQMGDBgwBvASLIFO9minSxhuzrNySD0tEeoyd4GRxxKkt+58l7WGps0duQsqoor7vcZun+c05BrcfVujysbEiPOzu1Ag/6mci+Jyr0iCWvbcO+LT0xMWzuMmZHvO0cFk6Nk8kg/Bnd7wiFidkz6jjiE/fhXB2cniWxbsmZRxs0wddp0NewvHGyQJcUeO0oKlw49DMIxs0JO0hTJcpY17tH9jFJgkwZNujvb1kfIExHRJuBWuwsHw5QMkxGfKi1ecFbZraMsaZmKrZElxZiUABiRAhlJ09AWZa0RE7NBnQpNQmI2bZVDdoYiWSakRIMOLbrco/t3pGp5fFr0SOPh4dIlYEjytLXLhAyhfQmXo4Y7dIGKtEnhUVKfJdlizhaZkCEAmtohrS5fdV4hZYRR3/BKLTmHYQzDWeWDMsfPpMe41S9wdC2577d3DdGQHtMjIel0TC7f489lkZYEjEuJoqbYoJZEHesIJU1zUzy9k+o1YjPUpMuSbAEwrBlymuIDdi9b0qZOmxZdLpot5rUAwDlTpi0hL5gyDoajWuH6eBRfHV6M63zLbnCQIt9lgz9ZanNsMU0UCetrJXIp2GwYGh3D4X1NTp+ewXOUzvJu7njoS7R7wjtuXuL6XR3OVZRyx3LXlMt3oi3O1iO+3ivjk3y+uxIxZXOcN5vM2yKHKHHJ1BlKGT7zbI7fu1O4NzWMj0u36zAxFLLVVb4WbOCJUMrAJ5YaFNXne1GVTiCcv1SgkI3JuYZuz+GOYpbdMy3mSnD9qGE8n/zMeQM51oC3KYMmZMCAAQPeAEXJ9SNdY3zx+1fjX1s8WDSREPU3WieyIkNGfGZllLxkWNBxRqSIRUmb9E607/Zj/yrY3v9xZfOzk3p11XPa3hYf9+VVV54DV1yCfoOWkTTrdnOnAdumpwFFsqTEwxOv7zfpy7CukLxB0tRF/z977x0k2Xme9/7e74TOcXLandm8wGKRQRCBWBAEAZAUI0hQJCUrJ8slWS6XXLbrwrzXKt97LVmsa5UtW1cqmb6yCCYwiwEEkYhAIi2AzXlmdnL3dO7TJ3zf/eP0DgGKFmnJomWjf1tbvdXVPdM93T173vO8z/OYYOt7WGJtrWZZxJdJcRiRAilJ0KTLd+UcSVwmKJMjxQh53qP3ERDRw+eIWmbFxHG9HkF/DJK+g0XRMl2OqhXSxIpXgTRZUngE+EQkcJlWI7whmqaNR1nyrJkaZckRmJBsf3Uq/hnFnSFTaoSnOUmNNi163BbNMkSeRzhKx3g42CyZKiEhNdq4xAlscT9JjjVT60cBG6rSoi4eASHbTJmKePHwh8WBaJy3RLsoSoaXrWXGTJ7nwhrz3YDtufjnmk9pIi1cNa0pJCHlwMNqkWQyYGSoze9fWaZY7GLbEdVqmpRx6OIzaeIB5zI9RYYkABvSpql89l4ypQOzuhCneEmbFRU/1g3T47JojDxpimQI0XzXWiaBwzvNDlp4DOs0C1KlKwFpbOqqx0+Ui0zpLCmlyJkE75rIUIzFIk4tO0QalEAhHbG6nmao5HHr7U/F79Mz2yjnQ06fmuD4Qoq0pcg6io+vbjBjMqyZOOVqWMXraQljcV7VCPq5XSeJB4pHvSrawKlzJdIOvNka44V5h+NLDsNp4cMjwyQs4Yubm/zkdI4P7UhSlQ4LdRgt+5xdtZkqGl5eFroBvHg6Q70r1LvCekuwFQxmkAGvVwbrWAMGDBjwQ/iNe3/nfg+fjCSp6sZWctL3VoQu0Y/tlfjg3RW3b0IXtEDLdFg1m9hi9deaDLZY9PqJW1v371/+91jJuu89v3H/px/46qEf9fY/TIV59cpU/wosUa/xcLji9PPAIkZUGY/e1tqWKy4eASLChBqiblpc6h1JShKErXQxgyGhkhgMjtgYICkJlFi8Ve9ntxliUTX761ABBYlXqXKSomCSRGLYGZV4UL3MTkYZo8gCG9xgdnCdnmRddbkz2sEVDNM0EStSwxUHTUTJZLFRLEuVrvHZyShr0mSCElpgwmRZUA3qpg1Aiy5jqkTdtMhKCo1mTIqERNRMm4xK0zMBt+ldPGqdAwwhmrQkCImwxSIysY+mjYcW/b2fshD3zeCzjwmyxuUUK4xR4LRa4+pomjwuKWzmpUVF2mRJsiYNrtMTnJYGJ3odJkwGS4RWD9Yaiit3N2i3E9y3y0Ypw8zuVzBRknYrTRhY/MnJHgsqVjXOssqyatCVAAtFhgS+RHQlYE2aaAEXiyQ2KRLs0UNUVJdro3GOqyrLqs6sLnOZLlM2GUxfAdLaYl212WaKpHA5ppZwJMGGtHnWq5M1CdpGs6E6OO00a13N+ZrwUtjgXM+j0QMrcGl4QsYVchlNdW2U4tQ8M3c8TymyePxYCVuEahCx38lijLBmPD44UeRIM8BC8ETjEjfXG8AnoiEe+02JvTmX29/8HN89OoESeOdt8ywtFSlnNflMSLNjszOR4tOVKtlujjNRmxfZoFgbYjQrfHpjk5epcOdwjiAU1ruafZMh31jvcPWQiyAk3zVYxxrw+mOghAwYMGDAD8ESRVnyLOsKORX3FCgEB/uv7PcQEQoqS1lytI1HApecyhAZzZgqU1BZgv6q0asVgFc3iH/wPb/51yoWBP5G9/3ec3jtfxOXVqbgtb0hDhb7rdn+SpqPQVNSBTZ0lVErTniyxGJElZD+2tq6qVGQ3Na6VWhCtqlxBLVlar+0+nYpfaxnepRVPm7bts5QJMOmaeGIRY+QkmQxaJakhofPvKpjMIzrLEtSY0jy9CTiO9YyLg4vqwqfUSeYV1UEoWnapEgQiWZRKgD8krpqq5xvXtZZNlUetc5RJovTVy2KkqNHSEoSzJgyRTIsmyoeASXJYqHQaB5WJ2mZLhf0Kl3j0TAdGrpN1/QQhK7xyEsGF4ch8vSIn79PQLI/1D4r57ld78E2igN6kqb4HFXxY91PgZui7ZRNmjkzDMA1qsw9mWEyLvghfLO9ST4BL5zMs9o0LCxnOL+YY+HUgfi1dCLKw3UuqiY10+5HCjvkiZO9FIJPyITOsT0qkjcp0sahJxHjkmTKZDhuVbghGueo2qAtHjdFMySNxTesszxjzbPHFIgwPKSO9V9f4SZ7CJe4BHIuKlGnw5pqsyZdduoiJdfiK+osAB0CZiXLNjdBpRfRDgyRFkQMQ6Nr6G4a2TWC30sxkrCwBMaSNjMlOG4apLG58urT/NQ1Pj9zY5seIXXx4q8tARuqzQ1mjOfVGoGGJx+/lp/+tS/zbFDn1Mlp/AiqLcXvnatQTBtWOhG3ucN8xV/mJka522zjoWiFhGO4O1tmUudIJSMsBVfPaF5YsLh3e4rluqD/btYDDRjwt85ACRkwYMCAH8IXP/nIoTVdBYkN04Hpd4WIRcRfXqHa6rrAxClSEq9bpSRBkTSBhFRNkx7xqlHU11UuqQXfz6cf+OqhTz/w1UP3fvDuH/ls6aUB5K/jL/lBjeYJlYjN+HLpuvgyrdKkVQpLLBb1GoEJcPu3vfT8i5LrKyOaospxtdlGR8WDxyyj9CSiZ3r9fhHhFvZwQSrERYTxmemo796wUPiELLNJQ7dYMVVm1RjjpkhVWpTIkjQOWRIsUiEtyXiFSyzq0iVLiu26wKKq8aZohiNqDa//OgRE3Gh2cpb1rU6ScYpYkUNFdRGEimmQFBeFomqaXGdmWZMm+5hg0hRoS8CGtNg0TUBo0+UyM0WeJEUybNDkANNcb7ZzQlZQohhXZZQouvSIiFe4XHG2VJakJLg92sVxWWWFGhrNCZaZZohxkwaEgkmSxuKcNNmhspR1kk18FlWTfVYeWwnHmz4mUlyVS/FwvUErMDxrqsyRxbFhpZLE+Clm95zi8ad3ccDOsysY5TlZoUCaZVNlLxN0JSQSQ1mnmLfqlE2ac2oDF5uEcViVLjnj0iFirymyIT1yJsHL1gpvjmaZMHkaBCypOtsZxohwUTU5Y1qMmCxt8UliUzBpQtGcVmtkSGFChYXDWalxUq1wmRnhm2aJgk7ysqkxK1keP5Zn75SPUgZV20C3c4wWYNuoz7nlBNtGArxaig9d1+XY8UnyOZ//9GwKG8VdmSHWerGp/kY9wU/dWmW0PkkQQdqFbzy6m4YOaTRsygnFWw4dpbi+g3ZPeLHXoigO94xlebbhMWa7DOkUCSwM8MZRh2/Na27c4VNtuIShQhmFF4AfQeF9AyVkwOuPwRAyYMCAAT+Et33gtkc/+8A3DikR/t/P/M5H33PfnY++5747H/3cJ795KD6L/ypTtygsrH6SVIQrLm3jkVZJBKFDjzFKtPBio7vE5/xDLkX1/uBBRETxmQe+duje++76oQcr36+A/FWDyPd7QV59+63L/rrVpbPgCosZe4KW6eAbHyUWrX4b+A57mgiNIlaBLsXlpiRBWRVomg7zskFe0rToYotNpd9EjwijqkjJpDlr4gN0jSGr0hRVLl5b65dC+sZHocipLIFE3GnmuCBNBMVFKuQkTSSGPXqEZanjikNIROnc/v0AACAASURBVBefs2qDCVPkGXWBYZNnnDwfTs5yszPON6NFkuLiYFM1TW7TcxyxNijrNDXpMEkZR+z4zL4k6UqAIw5VaXOSZVKSIEFs3HfEZkxKWCi8vnKQEJdRneFr6gghESlJEvSjm+OQAsWcjG+t8PmEOGLzHOfIqCSjErewj6giDhYV6cZDnERMkiZrHJ5Uy9TE51oZ5ohUWNU+z0TrDJk0DR3xkt+iLQHHrQopHJ4IVzje7fF0UGGoVWZ2NCTqpam2FCf8Lr4YMrhs0KRCC1tiR44limWp8fO5WU71euzWZR6zzrBHj/CKtcxeM8xXrBPs1aPMqwbX6HEOq3VAmCbDmnh0JUAQZnWJmnSpSIs5HStnZ9QG200p9tiIoYfhlFrnSj3GmupQ1HFfyrPWReZ0iboHPaP52MIG793ukr55CbOYIpluEwYpJkqaRCJkWxk2qmmKeR/LMpxatanhc9dOQ7qb45WwwTV2iY+f7/GcX+PKRA4RYXZYc+OkQfwEKReeeGWI/+id4NvhCp6KuFwVebze5g3ZLGu9iAhDGAljOfjz1U32OFmGM+AHiqv2V4gCB7SFAOl3D4aQAa8/7B9+kwEDBgwY8P999l9/9Puv+9PP/KvXXPfB9/zm/cZoEAvTLyQEKKgsnvFJS4I5M8JFqWHxvX4M+N46VmgCvh8R9d9kWv/Egx/76H3v+Y37BeETD37soz/53t+637wqThhem4h1yZh+6d8/CN8E/RZzF1ts1vXm1mPr6A4ZlSEwAS3T5Tqzg1GT4rssgoIiGS7o1fj5EW0V6ykUl0VjLKkKE1LmvF6mQ1wIV1IF3qUv5xlridv1DHUd8A3rJHXdomO6lFQxTrMyHmXJ4RtNxTT5VXUN/8Fs0qEXlwIS4mDhGEVSXPboIs+pCyxIhZbpchuTDLkWn/eWYwWDFjfoOZ6SM8zIMPPS5OpolBHLZVeU54K0eEk2OaoqfDA1w3wn5MtympLJsE6dFrFiMkmZBbNBTpKsSJ1ro2mOWKusmBrH1GLfVO8yJDmW9AaOOCTFpa5bHDPzFCSLT4CDTd20SIhLhiRNPLKSJkOSZamzW48yr6p08GnSIy0OCRNXRX5LLjKuc6ypFj0CHrHOkSVBiGZGl7hgVllE8Qa9k8soctTU+I6uMXZ4mo4Ph96wyPjxST6/XKQhPSZUGQd7y5OiRJg1QzxX90grh4p0cXF42rrArdEcR1WFLClOWxt4BBxXsYF+XtWomg7J/irb3UzTRXOcVeb0MPOqxjZd5GesvZRzwp81Otxpj/Fn0WkmTIEaPvujMV6wlojQvNPs4hRNIC41/LC1C7+3gtkM6HUKpAsrKGsYr+vg9WyKxS4jbkAYWkSR4q7d8OApl6PzKYwBB0U5JdzqDVFOC6fqIW/eHfHkGYeMleT6PR0urqZwlfBWfwdVfCYlyVheWNlMEGmYStlkE9DqQaRh2KS4Yiqi2bHxQ2FhoUSkheFCQM9X/5VP3YAB/2szUEIGDBgw4L8T937w7kc//cBXD8WrWLGX5NIakW8CspKmKwE+AW080pJgNxMsmQ3itmUHQ2xsf3VviCVqa1XrR13Jev8H73n00m3vve+uRy+Z03+Q6vGDUrFei+GSOhP1e7njxxWvo1miGLWGaJrY2+Bbhuc5Txeftuni9RWMX+F6njbzJPoH3A06rKkWPiEtutxtDtJThob02GFGuNzJsRlFFIzLJj5D5MhJho6EdPst5FNqGEF4glNoMTg6Q0myrEszPmBVXbr4aDGkjUsWF18Mt0dzrKouGwTskRzbJcui7lKXLgkcfntoD091azSlhy+GFeNxTtXxJaQpHr6ETPglNo2PiMWS1ChLlgwphk2OZakiIoybAnXpsqm6JHDpis+Q5GnRRaFo41FUOTx6GKCosv3gA01S3L5CYuGbgBDNuBRZN3UuMxNEAudkjY7pcY3Zxmm1yg49TEN8QiJ26hKIUJE2SVwEmNRFNqTFcbOIEA+FRime4CyhgjvUJFlHON8MyZkstm3YlUqQ6KZ4gTXqtCmRYbcZpaxTbKgOAYa68tiQFkMmCwJdiehIgEIxp8usSRNbbELRlEwKF5trzDArdNg0IYfVOv/bxBwnGiEpHI6oZTY1XJ7I4ncdvslFRk0On5B9psS8auJJxN16jq+reW5RY9y1L2S6ZPh3lYt85JoVvPlpslccQdIeiSjEcSzCwMV1I3zfxrI0ShnOLaUZsR32znis1Wx2SI6pkuFYLaLS0+zKOTyxFDGbcbh8e4/F1RRNT/gT/yRt0cyaHBv41DzooWkEmoJtcaTZw9JxittUwiHjGhY3FaP5CGUZHNvQ6tjYloF79gyUkAGvO+Thhx/+F/+jH8SAAQMG/K/Or9/7f9yflzRV00Chtnwll7o2AhO3UUdEGGMICF/TIg6xGT4yEZ948GN/SZX5UYlVEf0a9eMSlxQXW5wtRcYVF9/4r2k2tyRujLeJC/h6xifdT4T6gL6SP+FJdlnT5EyKk1ykY7pMqBG2mTIvcYG0JOmaHj9rruURdZE2Hk26HNTTnFdVIDbAV2iQJ00Kly4+DjZXR+OcUXEvxpSk+Ht3LvCLD9lcYJ1/nrmGT7WXGddZbi/m2D7u8dGTG3T68bFp46KAqrQZM3l+eWyMp5cjfvKNdT79dJF9JZuOD6daPh0iPCLOqxpX6BFOqk1CdNwTQUDRpNiQTvzaSex/KZgkSWPzDy/L8P5jLzIpQ1um+RKxL+baaIrHrLNERPgmjMsq0aQkQUO3mVGjNOjgm3Arsjhe7DGMSJGSyeBisSJ1RkyOBang9WOTAXbKBE269AgZJscGTdb1JnmVIYFLy3TpmC5TaoTdeoSG9OhIQMrYzOoiIZoIw7WJPO1QU0oo5sYCCvkeC8sZ/s/KKQ5E49Slx03WMM0o4tuywkWp9LtfLK6PZqhLDw2sqiYTOoeF4phaAWDSFLlMD3FG1SnpJD2JSBubeVXnFibYlrHIpQy/XzlP1rhcrofJicXjskxdOtwezfKy2uAqPYIh9lv9wqEKnVYGpSIeP1zmbM/jlw9VKW07SW1hF5lChfWL25mYO0p1eQeddpo/e94F4P0HNM2Wi6UMxy467BoPcW3Nes1loW6YyguTwz1sS/PdM0mumfM5u5RgOK/ZbClGiiGrmzbdAKbLmm0zmzzz8hAXOyFlJ/58+xqej2pcqQqIwFha8Wyjw7t3OIgYuv/6XX/tz/SAAf+zMlBCBgwYMODHwNs+cNujj3zqmUMRmp7pxZG2GFT/4N4Wi57pcSlrS/XTlHhVk7juDyU/ii/kv8a999316L0fvPvRzzzwtUNwKf3KvGYNy0jc+WH6je4i0m/TiFvhTf8ettiEROQkgyEenI7KKpNqGI+AqjQJCLmN/RzWZymoXOz/wAaEpznPzXoHyyo26Y+YPBkSjJkcPhGj5FmWGjlSlEyaDC5nVIVr9QRpLN44afPlw3nuKpWodG2+ES4ybnLUlMdJr8vzlYhAIgokSWITELFbl3mrO0EyTPBKq8eVuSSFtOHkms1CN+CxcJ1f2Jfga5UWG6qDi82kib0HPpq7UyNcmcgzbJIUwzQXpU0gETYWP+FMMSEp/sX6UcoSl/EVJduPaQaPgFAM6zQYljx12luJV57pMaVG6OCTJdmPG45joW2x6JoebXoUJcuCVNhuhulKSBtvy7ifkgSrW6tomp1mhCWJW9Q7xqNh2jhik1ZJ5swIBZPkvKqSwCESQ0t8quIxbbIEGta1TxQKrnZotR3anvCz+1zmV5Jck8oTGpgrWHyhd5Y0STSGNC7nVIUNaVGRFh4Bl+tRvmvNkyNWQBSCMooLqkpD+UQYTqg1rtUTZMTGGOF0I2RdutxjT/GmHZoHNtdoicevpPbQDAzr4rEkLRalxRWqyL7tLcLA4bHDRY76HX7+ep+R6VNUF3aTTHXodXPkSxV6nSLnzk4Qhhaun+C2PT7GCJZl2Gy4hJFgibBQsdk+5lNKCiMlny+fUDhBEiXC8qbNRDHuW0m5hpWaRcKG55sdppMO69UUtS5kbItSWoi0MDdsGI7ShAY2w4iSY5HDZqwQ0Wg7WG8feEIGvP4YRPQOGDBgwI+JVbNJy8Rnz1OSIKvSuDikJAH0y/rke8V9388l1eTD7/1H93/kvf/4bxS/+4kHP/bRTzz4sY8qZCs96tL3uFQOWFKFrRhi3e9Gsfp/IDbTawyOWHRMd8vnYDBMmxLaxGrLtMlSVHlO64toDDfrnfRMj5LK8RfqFWqmzdui/ayoJoe5QN64OCh26xK7zBgdejhYzEuVW6IZxmyHJ6wF/ny5zouyQSkXss3keB+72ZAOHQKulTLvKpW5zoziEZAzCWZ1kUNDGRZ7Pnll0ZKA400fx4mwgDoB/2BmhEbb5Tf35LlMDzGhs+zMOhwaTaGBZzpNHmrVSFrCeWmRNwmyxuW3pyd53K+wd9TwT7NXEvQX8S6aCh4+ddqEhJyX9X4BYdw87plY4dkv21gzNbqmh0fAumnQ0E3axiOJu/XaR2iaus2ixMNGXbdo9mN0q7oRlxyaALt/+7puUTNNOsYjIS5FyZAnzaJs8m3rDAejCRyj+IC7jWETD5NZbJ6VNb6ijpG2FGcaIc9vBCQcmJo7zWzK5ULX58ZdHqstQ2Q0HgEBIVmTpEyWTGzPJ0eSw9YqGkONNm08WuJxxqoSELHCJk2JU8FestZ5lg2W/YARx6YuXVb8gIdPK+6yJvnF1C6WOhE5ZfHu9Dhvsyf52fwMthJajSylkTUejBb4petCxradptcuM3zV8+Ruf4mhd75A5u6z9LwUu/fOc/AXvsTchI+IYXk9ycZmgpYnjOY15ULAPbefpFjoEmnhyIUkb99rsC0INWwrazo9haUMSzVFo2dwbcMOJ0W1LXR9Yc9ERMKGoXwcy3txU7HQDYmMIaMUZ9sBhaQwv+ZSLvh/k4/ygAH/0zJQQgYMGDDgx8Qzn3rhUF4yZCRF03QIiQgI8UyPjErh9TsxLvk0YhUiHkouDQYKa2uN6rMPfOPQe+9769/oDOqDDzx0SHiVUb2vdGQk1Q/V1djK3kq8svsKzqXbO2LTNT0KKosrDjY2BdIcNfNMqxEQ4SVZIikJ0pLkWjPLSWudMSnhE7GdUa7QEzxmneUyPU5RMpxVFfbqYcri8vevha+u9NipS/z+IcO/X2jwj2/tUl7bzomgTRqHZ+odbsoUeChYpWhStJSPo12+69X5ickMt5QzHKmF/F+/+AynXtnJrpLQ8hT3HQyYX0uQIcFwSnHTjOEPzzbYm8hwfNniPdc1eeaiwvcVd1xd4YX5JC6KBBbXjNhclklxTT7Fm8dS/PG5Ji+pRTbrKSo9TSTCsMlyj+zgGebZwTjrNMhKigJpHLFJERc3AvgSUZA0ShQREW08LBRplcQz8QG+QqjT7q/tGQIJt6KfL3mP4hSwEM/41JSHZ/ytEIBhVUD1/7h9NaqtQtalyWoY8R05Q1pSHFPr1KXLNENkogQ+hkesC1zsGj5xIkE11NwxmubBCz6/+hPH2LdxFc90a5TJEUhEmx6TpkiWBF0JAOGKaIJ11WLCFJgyBc5LhabpsptxrtKjDJscIyZN1ricUTWeZZUELiMmQ4OAM6aF5yt+6Z5zjGdd3vCRL3PZZIWJcsQN9z1CYXyRxZev4G1TLr2ezZEj2zFBluq5OYamzxIcHkfPJ2lUx0gke3ROT9FpZfADmzf86ueZ/87+fs8IYIQL88PkMiG5rM/CeoKLmxY7xwKyLrFSUzVkHWGhpblxZ8DnFjym3ARDGcOOqS6VmstIISKKFE9UumyEAdcPu7R7AgL7hyyu2Fdhz64VfC9F59DlAyVkwOuOwRAyYMCAAT8m/vyBLx+q91dipF/qlxAXSxS2WIyoIpumGTdoY72qk+O1sbmmv6YFhvf9DVazAD7zwNcOCYrIhP1r4gPZUatMSMSwKtIynf7A4fLP7DfxPOt9T0jcgWChqOkGPiEKRVEyiChm9RDr0iQrSSI0d0d7eNqa5w8v2057pciyatOTACPCZXqUE9Y6P53dxrwfMm7SrNBlzsnwXL3LlMky4WQZbQ7xwrm4G+OYqVHuDx1LQcAt9jDndYcr9BBHVIVINAfcPJ9f6lBRXR58YYSLXoTVTfBzv/clWkemuWr3JmcXCowWA75+SvGhPS7jo22GMsIfvmQYIsGQbTNd0lxYSdJDk8XmTCvi8XadG4ddjiw4ZLTLknicVqu0VGzKdrA4IZv0JGST9tYQd6mgck4PsSz1ODFMHBI4LOkNPOMzrso44tAzcYdJRlIkJG7XHlFFItFs6iZKFJ7pYYnaMrMHhAhCYEIykqJjulhiERDGyWamhiMObeMhImyaJk3xKEiGVbOJIzZzZpgRncYj4rC1SoM2dfHoSUBVOlRbDpvicbk1wT9bOMuIyfEmJjkqG+zQQ0ybLMfVOldEE7T7BvV9epi8SdCUgD16mKvMBCWT4qK0GSXFt60LzJgiXQkJRHN1NEYShYVil5Xhp+9YonDriySaNvXTe8i9fYGNZ3aS1h7txR0UyhWa9QJTc2eZGG+QSveYuvtRTD2Fc3Uda0eP7NA5bD9i6exO9r7z64zOnePLf/Bu9u/c5Mqbn4HOMJaCubkNikMV2s0cxbQwnDWMjjap1VM4jiYKLFIujKSFnTtX8FaH2TakCSLh5JLDhVZEzrIp5QMadZebJm3aPcXcaETgWxzYXedT38nhtMvkcz3atx0YDCEDXncMhpABAwYM+DHxF5967JAjNl18esbvN1G79EyPlunQI8Q3Po44fTNyHN1rMMTtI/GBZtwebhES/cjdIa/mF9/3z+9/5NPPHPrKJx871DU9QhNseUMAHOXSNXFqkwHaposlNtpEnJAWddMirzL8lLmGI7KGRw8liik1wk5GqUqbYZNjpylyXm1ioUji8B05z2841/DHKxVa/YElFMNpljkj61xpZvB6CsdYeETsVBl2j4fsCMt4vvBSRSNG+Pn3vMADr+Q5rBbZZsoohCtUgc+as+RMksvdLCd1k1ld5FTHR2O4XIqgFXXp8XPXhgRnhzh3foRmI8OuuSpDIzVG7AzZbI+HDudIis0VRZcjTZ+3zCr+6FjAneNpFluaA9kkT4YblEySK8sOX91oYRDWVIfdeoSTrDBMjiWpERASEJEQBy2aaCtfzHBRNslKit2Ms8AGa6YWD58YAiIciYc8JdK/R5ya1SPAo7elVuVUuj9seCDxKp8gqP57JCIiMPEA4vV9JiERvX5Km0IREtGkS05SJHGpSYcN1QYRFqhgi81BPUXJpNmnh/maOsqvF/bwxQWf/WaItw0VuNAOaUiAEsUJtc6Hrd1c0B06ErDN5Ji1UqSweEYtUiDNd61FrmKEvHFRCNtNiSSKUZPCoCiToEFAFptfft9zfPwr+3j2W/s4e3aMG+7+NtGJLCZIkdpxHuXZJHadJxkqlIoI/TTF256HtkX9lYPUX5oj0WhhTbTonZ9i/CcPc+RP72L83mV2uIvk956CnktxdJmkZXH45WkcSWCMMDK6iddNMXv1sxw/soPrrj9OUtIMlTyKeZ9k2uPY+TzrbeGmgxvgp9g9GjE+0mHXgRd55fgMe2c8lioOxYwmCBTbZqqsrxQ5dOsxNjeG8N68fzCEDHjdMRhCBgwYMODHxOcfeOiQiDAjI4RoHHHISYqGacemc4Swfxb70uBh0NjY/fUss9UvEisoFv/5s//3f1Oqzj95/+/e7+HjirN1UBsSfV8PiZBSsSFao4mMZqeaoichQ5LrFzEa/tHeEv95Yx4LxZgaomoa3Kl3cl7VqUkHlwQTJs9uPYQWwRab+ajLrC5wUbWoSguFIiUuV5oZJk2GUdthTzrBN6Nl7iyUSbmGp5bjXvqcsniMZT53PMtllEiZNCUSJLFwUYzqLBuqSy8UquKR6FdhFUmwaDocdHJYkc1nVtosnBtmuWX4Zr3JW/d0qFcLuE7EY0dyvP3GDbqdJKVCj+3JJH92vs1v39Jj/zsf4nefSvOeqSxHNjU/tztNGClergU8ay2SwOGsrPW9IBuERDhi0zId8pImIMIzPh5xhG5GUnTpUZMO9+j9rFixSmZhkRCHim4wpPIkcLD7a3iX4n0nZZgqDSxihaOHv5W0ZfVjBOi/voIwZBXpGZ+AkI7uEhDiE69qOf2AgUw/taxHSJsuPiGr1BiTIgbDvFSwxGZNdRinwNf8RTZVj3cUhpke7bFvRNNYz/ILByx6K2Vu3RmxVHEwCIetJUpRliuKCZ7w17hMj7DfDJFWijv3RixULPKWjS0KECYlyXTK4Zt6iZuTQ1y2b56DOxq43jB33Pk0kvRBIH3tWfRKFnt2DdNI4e5eQTwhMXMRNSpEiynSb1wgsRliz60BYBfqhMcKrC3OMLbtJZAItIW1u8XRL7yZl44Pk0sZ9h44RrdV5Oy5IbbPrhN1cgSdLI5lMXPjUxT3nCFDQK+T44a7n+TAdJVkus3oaAPHUswvFnjs+RnGssLxZZs37Gvx4tkkb7rhIk8/P8W7f+YveOiLN6FDC/X2QUTvgNcfA2P6gAEDBvyY+KPP/MuPBibknF6hSw8AD5+iymFLHHd7yfQtqK3Cw4hoyzieEHfr6/k/oNjwr+JX7r3/fp+Aa9kRnzUHtqsxLCxscbb+qv7A8wfbLqdnAjSGM/oi41LCwiKFS44UK5UkDnFcr4Vil0zyBesEl0VjuNhcUBUuqCovWstxDwo2IzrDsCQ4qIfp4ONgcbfewa/tyfCENc9S6HNgZ4vfGtnOzJhHKhVSsmx+5o4lAmP4w7cE/ccHaWPzkHWaHA4V45MWi1Gd5qJqckAP0yHguLVGGosiLkeCFgAFnaBLxL6CQ9EkiEIbyzL80YvCehAwesfjjI+2MEb4JxdP4KD47JNDrD1xEz/BLB8/22EPOV6+4OD1FGlsbohmaNJluxkmJykKKkteMlR1nWEp4PWLB0elyKgU+x0gXTzjkyfNK9YGWVJkSTEseQyGpLj9sF1NEoccSS6aDUqSpUZsRs9LhhEpEpiQhLiExGt1vb4X5JLqshZVscVmWBUYsorovuqVksTWe8FC4eKQ7Ht7Lg0wF/QqPgE2NmWdBmBDWnj4DJk0z9Q6/Mlxn+WNJPdd1yWd8fh7t1Y5s5QggeIdxRL/YnQPR1WFQ296mSQOO9wkJcsmaSlaHYdf/+0vMR91ed+NVb6gTvLTbz/Be3/jQf7L25u870MPIbkOTrrBVYcexz6wQvPMXtRYg+h0EWtnHYBnvvBWTCC0lmbBCMF3yogymIrGObiIZCPUuIuup1k5eYADb3mY3uEdXHzyFkwzBaFhZLTO7pkuI0NdnLkFZu58mOFyj1xplfzBl5jZvsr42x4DQG9mCP00jutjPAe/WyDoZbhwZpbhHcfYf9lFNLDU0ty8r8N3jufIusIXn5iiEwBa2DbWQw2OxAa8Thk0pg8YMGDAjxGFYIm1lSpV1y0ccQhNbDKO+hZjheCIizaxrdj0CwIv+QkSkuCPPvMv/5IK8pPv/a37L/WJXEpUEhRDqsiwFKiaJgWdIKFcevi06fUPRLtbA1BGpZmTcf7NhXV2McFZiQ9CG3QYIs++aJTr3QLHNzQJSeATEBBRo83PqwN8ktNMmSIdAkLRXBYNoxCaVo9ANG0TsonPNXqGbSbDVMrmsVPCG/UMEZrPHHb5B+98hRefu5zx0RZ5N8VXH9+OIxHf/PYcs7rNt6wFcibBmClQJ2B/Io2toNENmdV5KniUTZKDeg4ELtKmZJKkRPGWXJFNz/CNWp0CLu1mBjfh85G9Seb2nGb567fx1CtF7rntNP8pPcnUdd/mf//3b8KyQ5JKmIhS1EzI16LzvHF5mhIu77g85NvHoy2lymAYIU9D2lRMg1Ep0qCz9dqmSYBAmSIFk6IibTZMg6Q4ZIhTsxQKz/g0TJsJNUTVNNnNJBWa8YqbuCSwWTM1dqhJPHwCEzCuhmiYDnXT2nrPXTKvr0VxiaKLQ07l41ABydIxXdp4+CagIFmS4uCZgJ7pcb3s4iQruCiOqotUdYNxNUSaBMM6zbPWEh9QO/mL9RaZdZd3zGb4+IUW08bn3qs95j70RY7/yT38m+0pkld8h31fuIOP/MMv8E9/9w62mxSurXniT99Fih61zSwP3BKSuvwZXvrk2zmx5PL+v38WEhGm28FUc+iLGXK7jyMZjW5kkYWAytErecM7v86X/+17ue7AOtlWkk5livyeF5DZMsFDFrqbRlk+KMPE1d9Fb+Z4+FtXc/cvfQ4ZNYTPj1IeP8fw3iaNC7uJ5se4eHY/41MrBL0MdjOJ7QSYloXezNJc20Zh/xGsN2TRL7ZJjxzHtF327TyNaabIj8zzrts06VyVz3/1akopWG4ZrpoJObXs8NADd6MEJkc8Gn/rv3kGDPi7x2Ada8CAAQN+jLzzvjc/+oVPfutQEpeACFtsusbDiMHFxhI7Liok7uoYtYbw+2fRLxnaR6wySoS7PnDrX1rhiI3m8poOEkHomA4eAZYo9pox9ppRjNhs0CASTVpSqP5wNK7K/FJqFw+FC2wzJW4w01xhJjnIKJa2CcVQiUK0gQ8XttHz4kDWy/UYoYa7M6N8NbzAfj1KwSS5KZujFwg9Y9hustTwmVcN7k6OcirssDOd4I+CY1zJKNsTCTxtOHN2nBdrPo+uhCyFPodNlYNunpGcodqGfVJiFY+bGGPCdln1QxbDHidUlbaEXCUlNk2wlTS208rQNhobxXzP52zUxkJxRtV4ckWzxwyxY/cilh3y+Ue3cduVdb7+9ASnV5K8+NwuisrhjXc9SbIxzZcrLTaVRyTxaHhLMctTi9AgZFM6bNdDdCSIV9lEU5Qs66ZOpp+MpRASxD4IG0VL4ghiX0ICE7FhaojEg0PYf4/UTIukuIgIddOmS4+2YjyeSQAAIABJREFU6ZKWJONS4qxeoouPJVZ8icKIidUQE6FFbyklLg5Zlaah24REJMTFFpuEOABYovqRvk5/uOxiMGyYTWZlnKppMscYN+opnrbmAeE7ZgURxYhJ80Kth4tFi5DrxoXC0AIf+9Is2W6Bp7+1j3sP+hT3nOOOO04x0phh34f+gu1vOsGt155m7egeZj/wMLQU47efZM4yODPrmI6N2BpJ+bz4xbsYzjU5/8SNnDg+i7c2Ta7Q4OtfuY7bbz6N7YScfOEKKhsFnFqZlHuWl7/2ZkYmVlGlBqaTQpwQa8cGicoUWadJcHYUZ/sK4gSYWpbU7nOoQpv80CpRO0fmwFHEjXCLVY5+6a2sLkxjKYuXnzrAdOkVlp+6GauZxrY98FxMz8V9t8V3P345c29+hv07LzJRCrn25sOMDq+hW2Os1i3eeN0CYeDSvm2QjjXg9cdACRkwYMCA/wF0jUdKknSNhysOgYnTsjQGBxuNIStpfBOQkwx108JCkZD44HWEEr9270fvr+v4HOql9a1Lq1QiamvlxpgAV1xccYjQnFM1hk2a//LB8/yHT9zEGd3ipLXeL9PT/N7sTn7q3FP0TI9ARTh6hqxx8I0wShJt4IyqU9QuX6xXCDGkTbwmVifguVbEFTKJhdCVkGOtHgqhqjrsI0/BuPgScrjbpiMhs+M9fqe5nz9YXWIhrPAL9kG2F2FtXdEUn7p4/FRmBi+ESEOTkOPUOGjK1E1IVRsCNAoY01myOHzNWuC+5CznvXjt7XzUpYRLFZ9XrLWtyOOboknOSZMdO9fY3Bhiascx8q7geQ7tyGAJDCUVf9w9S/MP38J7r2/QFI8JnSMtDqvS4vlaBoBIDFdHkxy11on6ylVkNHXaW10wi2adgmT7np/4MTh9BapluhQki4ffb6ZXROittbu26aIxJMUhgUtggv4w4nFQZjnFCgEhZcmRIwXASbOIkTjMoGvin0XP+NgmXv+LPSD+Vpu6pz2SVikuyzSGjCSxUAQmYkSVWDJVspLmPGusWDXGTZE0DnXxKJgk37GWWDM1xqVE1ri4bop3/Nt9jKomkc7gacPsXQ8jwwazKUxsP4PkHEwQceQzb6Pj2QTfneL5p6/jhg99kfQbjxGdKaBGm0Rnxjhx+FoO3vwUi0evYnL7OeZuusijn76LiemIm65e4cuP7OLQNes02g779y1THD/H0mO30vEsTr58BbalqdQS3PQzn6fz7OVMX/8UJAKs1AbRuVGee+JGbvjIF0EMS996E8MT8ywvTDEaOpw/M0UvsNi9ZxHLisiMzbO0fCsbz1/H+Qsl6seGOLh3lKk3PU60VCb4usctP/s59HIx/sy3c2SGm3zjc3dwxzse4egDhzh3ZoKpyc2/nV8yAwb8HWeghAwYMGDAj5mvffKJQ1oMYb+c0O/7LkQUIsQG9H7k6qgq0qZH2M85ykiKtCQomhQbNOmZHtJfAUpLcktFAfpmc4MlFu/lBtZUmxvMHC42JZPkiZenmKfNaWuDq6JJblXjLNDldFWRUAkQRY4kAYacSTAiSRakwwVV5yozTJ0AB0VD9fiPHz7B0ZfnOKcajJgkBVzyOHhottspXqTKW6xxDus6NekxrNNUlMdt7jBfX+uSi5IshT579Sjv2KuZX3fYVVSM6wy5IM1QwmKhE5JWFk3fsNfKUtEhw8qhY+JG8g4R46SYlxY3Mc43oiVGTJrdmQTrQUgCi3HLZSjKUBGPPXqIJBb73SzXX/cyrqtZmZ9j/94VbNtQ2chy8+VNpsfaPHfRIW0c/t3KCttMiRSxIpTCYV11Oa7WmTB57pvO8kLTY551AiLKkqNqGpQkR9t48WBJQJkcLTy26yE2pUOdDi3TJTCxWjWqSthYePg4YlNUWdrGY4+aooW3laI1KgU88Zk2Zboqro/MSYouPk282MhuPHQ/ejkraSI0br9U0hUH3wSISByA0B988pIh6PtL6L8nLw1WVV2PU93wadKlKm0yJFmRBhrNHCNcq8c4rxpUFsv4Am9gmM/3lnAjl//nqTyfemiSJx7byz3XLmDqBlE+Q8kGhWzE4ecvx3EMy0f2MLHvJGcePsTwO9bxXh5hfP8R6CZ44cWdPPnKKCsndmNbsGvfcb76yH52jIU8cyJLwhL27DuLVaqxcmYnU1NVJmfPocMUuUzIi49cSejlWDs7x5kX9jG1bYEnv3qIN37gK4gYas9fTXn8HNbwJt7GGJ1WlrHJdcYn1nnhhR2UCj4XTuzGUoaRsQp+N4Nrw2YtRZEE33z4Kvbe8jKSjZCEz/qL1zJy89PoaobZiQ2s7RuMU2DuyhfIzixwcec9AyVkwOuOwRAyYMCAAT9mvvTJRw6lJUnPxClJQH9Vpu8pkPgMelHFB7ABIaEJSasUvvHZwRgawyo1tNFstyZo4RGaOPY2MiGWqLhUUBQGWFZNDpoZCibBCCk2pcebi3l8z2LUZLmgGhR1ikS/rdzFxsLmTrZRMyHvHy+y1NbstNNs6IAqPS6qJj2JmNY5vvnSBFV6vCM/hKMtlnWPC/L/s/emz5Ld533f5/mdtU/vt+++zr5jMNgBYhuCJCjulChSi+VUZCV2lLxJXHJSiWwz9KvYTiWOoli2U07kSLIUgpYocRNFAOSAxDJYZgaYwez73e/t23v36T7bLy9OYyr5AySygv68nKl7p6eq763z7ee7tBERrMTkyWyBc/0uA4l51qkwZ7hsxSEHXJcZ06GS0xQDj2NFB8sQokjx+s6Av/frP2T76n6ee+o6377u8F7YSv+PCVgoPn/C5/qGTUDCgnK5RJOydlnHJ68dFlWGRphwQ9q0JORLBxS1hs1DbpGMNrmgG0hs8viBBt958RhvrRg8tL/BX746x0uDHdROic2qx3YYU8SiqD3OGauEovElZEU1aIjP8XiGvZLj33WX2ZOUuS019jFNR/ooUWwm6f7GB6OCWXGxSVfBc6QBfy3pRaun+/QJ2Uka9zJEPd2nokrEJOTxiIjZyyRtGaCBvoRM6QKWWPQJqOk2ASFV3RxeYfQ9wavESAPrOqbPgLxkaesunrg4YqFh2Lal8XWfvGTxGQwFrh42tcVYYhESooEGHUIiEknwJeKsrDBNkXNqnS1pEmqTPbrEaWOF4/EUbQnIYPGNCyVefGcB9e5DNDbn2PPpU2S6Jba28pxe0XTeP8rCbJPv/ttj5M0M47+yybf+xdPkXOgNhMeOV0lCm8nZNd5+f57lJvzKpy8zOTbAK1RZvvAg9aYLiUngFyiUGrz81gyDUNCxQSEbUir1aaws0u7adFd2UZldob2+wIV3D3Dlvf0UcxFrGzneuFTi9asltnqa1+/YlMWhmI9488IEe+Y7nLmeQaHYf+A2S/NN1EBQu32IYrzSNmomxDhWQG93ePX//gxX7+Q59NFzAKzu+sxIhIz40DHqZBgxYsSInwLNpENOecTE5CRtHJpSY2k9r05vGY/q3UyoMgBKDHpJase5KVvEaOZlnIpRYjOpDa1W6SOuKdb/J5gO8PHkMJfVBleNHV43lvmN3TmuNCNcUeyxMjwlUwCsqha3VYOYhP1JGQV8sTDOH2/WuKO79BOdXlTMMR5NpphKsjgYrKo2R4w8a52E3RW4P5PlkC7x209E7MrYFDKaw1aW57NjdKOEPRMxG9LhjU6HyYJmcizgO/EqK+2EV+/AVDnkiqphHOyweypgZ2uSbdXDQnFXNe9ZmV4+n2NfzmLJcnmZNXLapicRnjZ5zCnhGoqGjihrhzmd5euXE07HNU5329yMfJ73xlmwbf6rby5iG/DcvoQrlxcZzwq7dJ5Yww/8Khuqw2ljg3mVGa6Nw12pMa5zFPG4YlR5US0zk+S5pRqUJEtfItrap5G0ccVORx11i6ZOH9h3aLGlm9ToMKULzFHBEoOMOGTFxRlWIae5DSfda0HRxmeMHG1J7VU2Jhu6zpa0aeMTkeCIRVnyKIRo+L7wdR9D0r2ZaDhOKShsMamoEj3tI8i9PZqEBFsstpM6WXGZHFb1KgRPUruXQjHQAzxx0m0ZssSkVrKbsoWHzYKuDAcbB2m2CUVdegyIaUqfnoT8fnuZZi99v77z7iyPPvUGodYc3NNg4ug5pgua+T3XePHvP8PJj9yi1hGKLrQaWYJIcevS/YxnhP1jiqvv7+ftc/NcPPswl24W8NyYhT03aDYzXDi/i3IGdk+FDELhjesu710rcOV2ntmpLrMLG+xcvJ+bt8a5tAVRDNfu5IkT4TNPrTLtmhRNgznLoZyPsKyYR440qNU8xjzh2P4GOxsLiEro1Wd453c+i27aqPk+DDT6+jaQ2goPLvgk9Sy64/61/Z4ZMeJnmdElZMSIESP+hvnMV06eOvWNN0/6enAvQK7RdHSP1FCjMcXEVRnqdO49iBZVnngYJD6o00+Tu/TJiMNxFllhh4SYtJtJg6QP6grhmmxST1ps6To13ea7zSqRmPzWMz3euZXhmX0RsW9TibKUkgwZTC6rGkECE+JSi2K6EvKuVLkvnmBv1ub5h+psrhW4RouHZZx6EnFkzOJ2HcazQjeAvVMRUd/h7HZE3lDsDBLeTuo8O2tye9viqXyeS/WYpG9TDDwemIWNjrCrkhDVcjw01mDqwA2qd5eY8suUkwxmbPH4uEMQKLQGxxA6kcZMLO6zc9xNfJ7Nlfhhf4dFI0NemXxyr/BeLSaDwVWjxpTO8exYlgutAVllEMZQFIvbO0JGGVysR7yvatTjmBNGiUXJcVd3WCRPnRBf0i0NEWE6yZOgeTSZ4YyxRiARbXzKOkcgcSoiSDMzjtgUVI6O9rHEZFbG0vC4QJMe4xQQEbaSGvZw/dwQg0CHRETsZZoCGVZkhwERznBDZkzyw92XdOhQoYYWLJP+MAuiUPfKDZQoDDEoqRwAPT0gIcHXA2JSASvIvR2RrvbJSiaVM6LJD21daabJZk7GadJFRLGd1JmVCnO6TEPStXZfQrZVB9C0JaQuPfYmFeZ0nv/5PzvNl47XyeoCp771EPmMZn15nv3jmtVNDzcq0WhmeP/KDM//rb/kxpnjdH2TfgR7l5psVbNcWLawDaHahbwjPP7EBQI/z7HjN6huVtjZHkcp2LV7k11LW9R2SniOxjMVB5Y6OIawupWhWS9w5U6eRAs6ET7+zFUmKwN+dCkLvSLdgdCLNEsloZSP+OZFxdZWll2TIbsXG9y4Pcb+w1e5cO4IO9UiJx57E2OpSbJtE9+cQIdgHu6z6/73qRy6jm5mqN88Qu2xB0eXkBEfOkYiZMSIESN+CnzyK0+fOvXCWyfHpUBVN+8NxnmSwRBjWLU6RlP3EARTDCpSABEUwr5knAdknFKS59fLiyjf4ZrUSdD3PkH/wPOfDCPqiY5Rw6xJpGPWqXHnziwb9Cn3C9zsBXzqaMi1LYP9GZdSlMXG4GjF4PE5yHQKrMYDQjQzkmFl02PMVSwoD89UvBM3ODXYJokNnlwQooGFjiyUgr0Vja0U8yX45G7NymYG3TdJtJAxFN0QDlQUczMdiobN2WWDvSWD77w9y16vwOInfsiZnxzm+EJI0nO50A7Yk7VwDKEdap46OCBsZsjaQhgoTG2wy8jgWcIbgyZOJwuJMGZaHFIl7i/b/Id6lacKBU716xSwWfRMkgQMESQRzrHDqmpwlx7vsU0gEVUGfFRNc54qk7rAA8k0DgYHKPFnxkUKeJgYuFjcZosJKZIRB1csugwwxSDS6TDgB9XGAqzpHWISqjQZEFJWhbQVTcyhBQqKKsc2LXboUJE8BoqC9iiTYU0aBEO7VIU8bXwsTPo6YEBAQbJklDNsSvMB8JRLLWnR1wEBAQybxJSkc4cGBpGOsMRCBOo6tdclaEqSxRCDRZnAEpM7egtPXExSy5lPwAYNiuKxTRMTg23dokWPvHj8n4+VeP74Jk/ub+McXuH6Xz3H5laOfDZh//5Ndh+4ytwz79C/u5urtwsc3FMn6wgrl/dRa1mICL0AOi0P29T4gWLNj3h8T8zsbIPX3trNzESfPz41Q61t0uqadH2TsO9xZ7kCCI4d49qadtem2rSoFCJ279pm0PPQGh4+WuXGjVmu3S6yUBIybsx71ZiyZTCIhMtbwq89U+XAYofxqU3yYxuMjwX80fcOkYQGJz/3Q979yZPMPnCF+MYkb/3kCaZKHeLlHOaJiPh8HhLFoFVm1/Xvnlw//uxIiIz4UDESISNGjBjxU+L5rzx56ptff+nkgABPXEoqjy0WLd29N17YGdajeuLQGzYZ2WIyq0vsczJ8+cEuhWyM33J5I94kIw4d3aOo8gQ6JKc88pK75+cH7i2xC8KkKvPzuSkWJ0Leagz48ufe4MWzc9SimGfmTB6cjzi/apIxDHZNB8xHRQ5lMpzp9DhQsLjUDPn1X36VV87NM4HLA2aZSWXz1nbEiRnIZSP8vsHhI3dZXy+z2lAEfRsReORAh2rdpR4kVFzFyacv0e/mybgRVuIwN9Xn7a2YT33yLXrX9nN4/yavvztNzgZbGxyYDXllM6CfJNytmtyMfSYNmzHL5AfhJkftPMt+xPPTLt9u1lgyPK5EXW7oNls9DQjngw6fzVc4GzRRgU3BMrjQ87mT9KiqHgfjCWrKZzYpEoumLX02dYAvIeM6x2vGLeZ0mVeNZSxMTsQzzOkc52QFA0VfAgaEw3B4KgYjorSCF4cMNl369/ZDDFT68E+MSSpME52ODoY6IpbUJqVFmNAFpnUOE4WDxY600/YsbGISWrqLHm7PhEQ4YtHQ6Uq9EoPusI3rgzID655gTUPoA4J7tq0PxG9OecMhRE1WHHZoU9dtNBASkREHnwEd3SMhuXeN204aeMrBFIOW7vEHKzX+7IbFG9fH+MIzdymVNwjr0wSBydKBi6zfPsS108ewrYSJ8oCrt4tkbE2vbzBWDNlomFRymtUW1H0ZXsQUOy2TlU2PvbMBL1906emEOc9k2Y/ohRoiE1Ole/KDwCBOBMPQFLyYWstiq5pl3+46q1seN1ezCMLhvS3GSj0u3s7iKQPTEP4guMkjVgUrcRn4LjMnzvFnL5ykUy9Bovjif/FNNt55lMU9tzCsHvVbh9j3yFusXztK4OfxZA0110ItDnCtGtbJiBX3+ZEIGfGhYiRCRowYMeKnyKe+8sypb339hyfTT5gTmrrDvJqgrXsEw04sT1x6ekBWMjhiM6vHWNR5HphU/OiqQ6fl0g41M1GZq2obTzIMCJhUYzyoF9FKMSYFTJXukNhiMdABnvL4Agd4f9BjT8bhqV0J33v5AIeKNp0BdHzFmU3N80cHnF+2+PhvfIuXXj7AsYUQL8hgGnDW7/D8I8u8++4eAq1pxjHLSZ8sJk5scfjAFlduFzhy9Cqd+iSeJSzMdogjkzgymJ/q8+ChGhubeXa2xrFNzSAw2bt/nY2NIosZm3//yjwbdyZZW5nENSHrapo+NLsmvSgNSl+XFhYG21HEvpzNSj9il+1iIyy3NU+Us1ztDoiB42aRs1T5bG6C5SDgR9EGjzNF1lCcCZqYKOYkww4DmjJgUxrM6iKa1NI0lxSY1QXqqs8URXakhykGh+NJbqkGa6pNPGw/+6BVKh4WEKhhFDMkoqN7ZMQhICIvGdr0MDAIidKQNwkZcQiJmVRlJqWcZkskFZG+hOxIl5r0qEuPEll8AgpkaNIlJEKQVJAORWhAQEnl6WofTcJgeG0B0l2T4ca6iYEzbND6wJKlRNHSHXKSxdf99D2qw/S6phNkKDg+sGhFxNhYtHUPgL4OyIlHb2gh/OHf2eDjUy7Nq/tYuXiQmcVVcrmAV35ylCurDouTIZ2eRRganNtMKJgmB/dX2fWR17j67n4sA9oDWA9CMsqgkhH6kSZMYHE8JGtYjFkm9b7GFsWjuyJMlQqWc9sxTx5psl3LsFY3sI20mW6zJejApd0XeqHGUkKv53Bt1eN2L6SfaFpRQqTBiix22gZ+z8LtTXF4/zYbmwWef/51dM/hpVOHOfrYee688RFeOTuOas3R7tjs+9RL3PrhRynmqrz3wvNMP/Y+UrSZv/HCyZXJz4+EyIgPDSMRMmLEiBE/Zb7wSx879dmvnDz1qa88c+r7L/zkZEO3McRgViq4YpPFTR9QRTFJkS9Zi/zqR1fJZhKOLXWp17Mc29Xn+UfvcObqNDf1BodkgYeSWU4bd7mbbPJPJ+/no5kpPpWd43HmORe3GOgAJRneVnd4oX2dt3YUn6mUOfnzL3Kw5KL6OWY9k9O3DcYzitrlg1gY3H/8No1amUEoTInLi28t8pWnt3jjdppQyGLiKYO8pcjZBiaKVm2S6Zk6cWSztpXh1o7wkUdvkst3yZU3uHJtnuWWJm+ltpn9j75Fb3uWG+vpZeOXfvPblKIxjp24wPXr85zt9tiMQn5uySKHRdBPH6QfK2TpBrBkunRCzVtRgzI2g1DwxOBw3mEqD42OybRt0QwSfi47wcEJzUuNNpuqQ136nFdbFMlQky5lcghCn5gxneGCsU4Wh1g0ljaIJOZ4MkGXiB3VY0x7eNj0JMQWi6y4JKIpSY4efayhZSkiJiDiOIvcocqY5HElvYwcYxFDTLaSOlnJUB/W/DboUMSjqXsUxOPxeJGGSrNFA6KhAOkhKExJhYSvB2TFpam7gMbXg3TMEI2JQUg4DKHbxMRorbGURaJjIqJhUF1jiUlCgqdccuLR0T458e5lTBI0WckQEg33ahIS0fcucOkuSZ+iyvPbufsZbC/gujGXr0yTzUSYJvzhKxMUTIOHDnQpj7WojLdQonjwQIN+LwNa8e+/c4D75mI2mwa3+wFFZTKRUVxsD5h2TW73AzZqFsSKq+0QhbAaD7hah2RgkjGFWj9hLm9wdd1kYSw1vLlWwnY3Fa2eqWiHmtkiXNvR7BnXXGqHxMAqPYo4VJRF3lScfHibKDS5cn2co4c2+dFPDrN6a4lnnrhOa2Oe2Sdex23Ok80GzC2uYQw07dokl88dZHqqQ35qheSWibHfYCU7quod8eFhJEJGjBgx4meIF1947WR++ElzRhxKZNmiSU68eyOG01GBipFj79M/IWxUOH01T9YwiQY5/un2u4ypAjeSVR5miS9kZzkXN/jqf3yGPb94lbGWwYl/fBXnzx9iLK7wlekiu3rTXJQ6B5NJPrFbM/lfWljr1/Crs2zXbbohNIOEjQ7sqYBlmOSzAQqDbl/hGYqdap49BYOdXrrXsaUHXA67nFzUXFvJcGfH4M5anoypcGxNxhB0kOXStSmu35hnLJswkYUgEhINXlJg/r6zLIxBv1Xk3BuHOH7iEjqxaO5MMmvZjCubIFQ0+9CKY7Jicv98xBtbIbciHw+TJ8oes3nhbNvnjGxzaxCw3RW2pMd2EPOl3Q5zkwO6PQvlO+yTAivaZ1xn2VQdfILUCiUJEzqLhUEomg1p4WDSUD4minXpIgguFqvSoCODe7kci9SCpCW92gSk44OpcAhZo4anUmuWBhYYpy9pe1YwHCS0xKRLumKuUPcqmDsqokXvnpXLxMQmDZWPkcPD4W6yQUhMqEOUpFmPDy4zJgZpR5Yi0EF6/RDSXBLR8O/Bk7QeOq0X9mjpLpBedEwx7wmPgBALC1dsenqQNr8J2GKTF4+u7vM/TTyMbaZ2qO9fzJBVBltNi1IWpJ/h4GIP1w155ew4OTO17oWBhZcJUUoz4xmsbNvMVSJqbYMvPFYnYxps1k26keYzR0OqdRvPEmZzBgZCJ9QYCNMZk8fu36BRLVBrm8RJGma3lKC1cHjJZ6NmU3SEkitUu0KUQMMXNuOADekyMRyC9HWCH2umLZdCscfFO1m2qzmKnqaYi6jtlJia2aZ67QBnrxaYrgRcvDzN7ZsLHH/0HTaW52m2XcKtecYeuoRkFXNXv3lydXZ0DRnx4WAkQkaMGDHiZ4i/+PrLJyNiJlSJhu5So01RsvQJ6OuAROCSVJlsTHN8oY5brvLyuVmKhonnav60tsa/WXyQt9shxxnnYtBlRTX5X940ePt79/ELT15n7dtHeOThizw4ZbP/idM8cPIC+649S69rkvQyxK95VMaqjO2+RSYqEfU8Ds1FXNrR7KtAq+WwsGuDjJMwlk+YKkeMFUOi0KDtG7zOJs+5Ezw+5rJdd1iaCqi3TT598jqTU3X63RyGErbqDp6T8MzHXmPx8fdwehVurWaZKEV0uy7rt5ZYWytTbStqfc2Dz72LMgOWr+6hHygq+YTXtgcoLXz+hE9RHFZrJjcCHwvFuLK41QvZ6cFtaWNhsqBzXFE7ZLFxMdFdF1NbfG37Kp0EXpUVFpISgSSM6QzjOouFiYdFIDE7qsd4kuUWW7TFZ0BEjgw2Bk3V5xmmuSUdAFzSdfst3cAVm0RretrHUy4TUqRLn+wHRQRE7GeaSDQ2BjYGVdKtDxODospii00Wh6puYYoiJMbDoU6HPGlbVYseJXJDK1jCHb11r67ZGNqmlKS5k4R0SR3AQKGGOyIiKh0j1AM8lUkvJmLeEy458WjoDq449yxcC2oCR2z6hIRE+HqQ1igLZMXlH+Ye5HvBTUDzcLxIGAlNX5jPKcIYcg6sV23CBFodi62ai6VgvBRy+mqGhfGQXs8mDE36gUnbVxSzCRfqMVdWM2w3TAwROknC7W2Ti3GHapBQ9zWbYciBrMP9s5pCRnPldoGSpxmEadOVawpxAkEsrO5Y2ErwQ2gNNJtBxHoyYExZNJKIPSpHVQ9YUBnKhkneMKi3TVY3PapBzMO7Bzzwhe9z/vRhtlsGST+PZWkWpn0Wnj1FuLFE3ot488weHnrgDhlX4/ccxkpbxFcL1C7dR/3xB0YiZMSHgpEIGTFixIifIb7z9VMnk2G7UEbsYWA4/aS5JFlmdQlXbPZLgVw8Tqc2xYxr89ZGzJ4ynGnG/KjVYDnZYkdFhKK5yzb7ZJYbbPLR6H66XZeZT7yFIx3e+cFJXRSVAAAgAElEQVRHmZld5ddf93nSnOYjx1r84EKGPWM2zfV5dqp5FueaZDIBxxcGnLrk8UJ7k9nWArv3LuN383R7DvuPvsfpdxfYM66hnSVvKsrZNPx94pF3OHzoLn5njH43x53VAr2+gaE0T37+Ja6cfoLxyha3Lu0nSRT5bEQuN2Df4cskQYHp8T6XNyz6tw/Q3Zzn/kffYm15nqX5Nu+tmTw2bfLOLYeJHFyuxbiYLNkOd6I+0XBc79cO2qhGnpu6ix4ugD/hlnkx2uLNXpNZXeSOqnEinuVdY2041WjgapO8dgDoS0RNumSwaEgPExOFsKBLPECFUAuvqmWqusUMJarSpkCGNv10cJIYU0x6uk8wvEz0CTDEYFyKrEuDDA4DCbnJJnnxaOseCZq+DjAxyIqLKSZt7bObKUKJ6Q4X1C0MMrjU6eAzwMWmJFlC0XjKGdrvFKaYBDoAIJGErHgkojHFQANTqkKXPjIUEclQagDDf7uHABHx0HalmZLyPREU6/TP59UkwbBy+v2ww7ZuMqYKvB5v8N8+FbK+XmRpekClGHF+TZEksDCmWW5A1hZcS1PIRVSymp5v8cpNxbhrEsdC1tWcXYWmDpkwbKpxSIKwP29hohjEEKPJYBCh+eR9PV695vDw4Trn72RIYsGzYTIPtiEogUTDkye2Wd/O8sjhFhfXLZa1z/vGNklkYqNo6ggXg5aOsHVaEf3Q7ohGx+Bq4HP/hNBZXWB120EDrim8dsMi6nlYzRm2qh77D9/GJsP0sXe5cu4wB45eIWhVuHn5EEls0n326EiEjPhQMBIhI0aMGPEzwm99+X/4aozGwqQkWVq6x7SUaQzXrA8kk8zoHOPaQyfCq9WQg1mH791KsEVYriv+xd9/le++vkRB5VjTNWp00Gg2dQ1HbD5TnOb4195h58+O8K3vnuDRB5axjT6/tl/Rq04zv7jNU5/7CZlCDUfFzD13mmB9BqXg3MUJ1vsRvzBb4HZNOH99nKefPMPK3Xl+//VJ9hdNbFNTNA32zAzIZiK+fzuhs7LEoDlFt50FhDubDvOTAUqEyVIby1SceeMYUaRwHU0YKXbtvcvd63vZ+/CbFKfXqd7cw7GDVdptl/r2DONjPuevFZhy0prfw/MBWS9isQQLeYVnKnzfoKFD6gRM6Szf87fZo/P0SYjQrMR9ZnWWy2qbI8kE87rAlvjsTyrs0QUOWTlqScQ0GW6qBstSY38yxU1VpUIen4AcLqFo7kiblgyo0WaJCSZ0liwu77OMjUVP9++1TxmimJQSanhxMMUgg50OG9JmR7fIqkyatUAxYIAtNj3dpyBZuvTp6T4Z5bCtmywygYigh3syAkzpEmU87lBFD4ct67qNQu5ZqBISKkYJX/cBTTC0X3V0j5CQvGQZ6ICsZPAZDJfSk3u1z5Bu0JRUnv+ucpDv+6soFBnlUJQcf/jEGN9cGfB/HD7AZ6ezfHOrzoCQF04cYH29zEbT4M3NmIrh0PCF7TBiua3pJQmLBcVYPmK7bqO14uvLXcaUzflWwHoHltsJXZ2gEGKdisqCYRIn0E80Y6ZJL9acKNssZE2abZtKRpifr7G9WcRLZ1t44MRdtreLKAVZB1rtDFttuLxu4xmKDCbHjTJlsRgzLcIkTbgsmA6bSUBGDHZaJlECb+tt9ukyOS+hko8YDCwybsLSWMLMpI9th1xf8XDw2P+Jl3jlhZ/j4cfOc/HdIyw9/2OC1TmiyKR3ciRCRnw4GImQESNGjPgZ4Z0X3jtpi8kvc4TLUqNNjyUm0AIxCU/rOeZth1Yc81DZ5e2gSbXm4InBkmfxy58+j/1Mwp/8eRlfgnsPrI2kScUoMSZ5Xq53+OUD1zn93cd48EiN27cnmXvgItaBLdbOHGJufgP38RrrLz5CbXuS0vg69Tu70cDGtsdmPyYaGHwnWuWRTJHq2jx3qgYPzwhvbEZkMXnovi3OXiliKYOtjrCrJFim5t27FuWMMF6MOPLcj1i+vBfPMSlP3sExMsSxxdqOxUxlQK7QY3L+FmGnyKBZYbwYUxzf4OLVWR79zEts3tjD8o7Js4+ucfl2Hh2nOQjbSuj2TM5vaVyleGZB4bcdTvk1xnUGn5iBxGS0yWfKZV4Najyl59jApyEDQklY1DnKhsl8QXHdH1CXgGtqa2i6UnRkQEl79CTAQLGh64yTZ13qdHWfSBISESISOvQxxSAvWfqko4H94UWipwd44pLBZj2pMqsqbOsmJZWjlXTJSoZDzNCQPhUp0Eja9+pvBcEWm6J4xKLZpolBKmYy2EzqLDuqR4U8dTqs6x3GVJG27qGHtw0Roa/T8PyAgFjHIEKsY0Rk2O2lCYmIdXTvihLpiILKEukIEcW/WjyBIfCZ8gQfc2d4hBkeMibwTJMnjBl2713j0rUJjgZzHNeztJsZXt8KaUQxx/IOmx2ItKZimxgIdxIf3bcY94TF+Tbnb2eYNmwsSXMbIan4WB7mcBwMYmBN95FYAcJURhFFwlhGkWj44U6Pnq+w/CKtvtAYpFe62UrI7bUsphIK2ZgHn3yD85cWsFQqbgBmcwrPFJJEeHpvTNRxiTXMOhaNKCajFEdmYxaiEvUeTBY0N1ZdLAN2LbQARbnSpNfNsHepTrOZJYdJKZ+Qn15GggKdW0t0Oh6OE7Hv9l+c3Lj/6ZEQGfH/e0YiZMSIESN+BviNL/32VxHFjC4ySKCjQjr4zFKmLyFH42mOeh6PHW7T2MmyeyLG6+SYdEzmc4pPf/lF3M+D3qjy1l8dZUt1yGDTl4iAiKKkIeVv/c5Fogtlrl7eRbOZYWm+xdrl/YyV11n6lRuc/cYjbL6xlwO/+ldk2gbmeJ18aZv8/DL7P/YuuwazvHnX4qhRoh/DF//JX9A8c4DlukE3TnjueJuvny7y3H0dZueqvH87x1IJ3l83sJVQ6xgUMrBzdxcicPlWnrztsbVVAC00uwbLOxYuHsQe168tsPjgWQoPXWXnvaMEA4epcp3G9hSOMrh8q8AjRxsU8yFnb7joyOLVrYBHpkxmyzFd3+Bip08em8cKWfZ6NlE/XQ0/PWhS1i4hmidzBZzQpktCCZtjEwa/V1+mKwHzOseaauNgszcZwxWbG7KJi01nuJqeCOzS4/gSUSJLG5++hBTFYyOpEQ8vB75ObU6+HuCKgyFCRIyvB/gE5JVHrNNrgys2VemQkNCmB0PrEwiO2DhiU9VNegwwRA1fT5+DySTn1DKLyRhKhCptbLHo4jMYhsvVsE5XkOEYomCKiTWscP5gfV0DCfEwI5LW+yoMAsJ71b3/6BMrfPf0PPumIuJEMVkOWdkx8PsmlULE9laRNzdiBlrzsYMRnZ7JvGcy7ZqECXRCzRP7AlZqBhfDLkesHOejFrdaGt3IszoIuW9auNiICEmI0YwZFrUkYEN1WJMOee2wpXocMfMsx30uhV3aOuaWH1DUDjeTHn2dsC9n0x2kYma6oDlz08NSQjeAjAXLtxfpB4Kh0mB6N9SUXOF6K2Ihr1itmTgmmCLkHdCxwhSYKcdkLNg718e2YyylmKz0yeW7LH78FMvvHMO0YvLFFtlsQPmz79G/Nkl7e4aJhWt4+QYTz5yhuP8Wb3/yn3/tp/aLaMSIv0FGImTEiBEjfsr8ky//zlfbuocrDp/Uu3i46PFe0OagngLSB8Uvlyc5sruD79t8/Jf+irA6w0P336XkWjz0pb/E/OQien2NP/pvvsDlpEVPQm7rjXQEUXJ8NN7Dv/rX7/Bf/+bTfOLJm9i9SdarLpNjgzRQvj5P0bnDwmcvMfuxFfRqjP1wB71uwsDm1W9/jEuvHmG8FLKxneFO5LOeDFhc2YcgnDi2zok9HZRoCnj8+WXFsWnN8d0dXruc52994R2SzgSTpZgoFm5tWGy3FQvjMZVKG9MQDn75e8yZJq2dMVwn4aULObKGor89T6WyxoU3j1EuBvzFTxa5u2Oy2YXdFc3hx05z7cI+On1FtafxRHGnk+Bh0h0IRbEwtWKpDFd2Emo6LaWdIEOXiJg0gPxIxaXdE3ZnbM40B8zqLIs6T5UBK6rBgi5zRW2xIQ1MTHbrcerSw8GiT8AyVVxxaOgOY5LHw2Fd13GVzbSM3Qtta0ivVNrHFou+DnDFpiw5qkkDWyw8cYcDgCE97dPXA5BUfPT1YDhAGKPh3g5JjwEDQrZUBw3cpUokejhwqOnpPjJsvoqG+x6CEOrw3veIdUyoI0LCoRBJUCJEOsYQk2+eOEJhY5GppMJV2eSflZ5gLG/w4i2Lg0WDRAtbdZvje7v8+d2AK42Ezz6yxelbHhOWxUbDJO8IDxzbYHM7B8BUDgwlWKJI+iaB1tyUFnvIcy30aROx0k7bxO5Khzw2vo7JYjKpPeZ0jg3x+XR2nLwjrAxCXAwmxcVAkVMGq0mfxzNF2gPhO4MNZrRHuy9MZNNQ+nQxYe/uHWwT4tBCiZAksG8qptlVTGQUu2Z87u6YnNjTp9Wx6AaQtYT9M1H6Uypw+OE3aW7PcOC+d3n77G5KuYTunQWUAbt//gdYRptMsYaUfPzrc5Sn7mLMVTF31Rm8u493fu6fjQTIiA8NIxEyYsSIET9F/tMv/cOv+gS44oDA0+YML/s1bG3yhcI4e80sTxby/FF9i7/98QtM7LuJrud57fR+VlcrmKIoWyHKv0myI5z41Rt0XzrBW8kmWtK17CwuGRw+9fwtjncm2Lqxl9fez5No4Ym//V36y4ucuVLg8DMXwEmQYh7d7iIVD7U7g663GGwscvjwCnFkslCJObeuyGNxaAKqDZu8pzGMhNfPzBBGin0lgxsrHrsWGtxaKTCZcxmfaFDbyRNGiqWZPsQms1MdtrYKjFU6xJvj1LYmsQwhlx3wwMEqqxtF8l5MpdCgU59kc8fBMxU3/AADwRWDyxd3caYakVGKjClM5YScoaj6moIjXO0OcEWx3oVWEjFnOjSTiMHw6mGiyGGSVQaHKga1HmzFIU0C2oQ01YBNaTKlC9SlR023MURRlQ6hTityHbGwxcTDoU/AOAVWpIolFhls6rpDW3fTh2LxAOgTkKAJdURASEO3yYiLRtPUHUwx8HW6AaIFIh2lFigEJWmrVV55aWxcp21YrjgEhNjDAcKsuJgYw3pfm6LK0tcB8kFV7/Dr0t0QhrkPnS63iwnDjY8EzbhR4pi/BDptHjuu56gPNH94O2CP5Ki1Tc7XIpLQ4AfrIVPiMmXabG6UmHRMVvsR908LV3c0c0VhtWoznk9o9hTrTUWUCN1Ic0W3eUDK1HSIMaweLopJi7RooSsRFgaPlzPc6ge0CNECN0KfRh9mlMO0ZbMRB+yx05+rZhxztGTR6MNU4hGS1vL2o1RoDEJFuexz5WYJ24TOIK3m3TPfZbPmcHCpi+9b5G0hlw0peOkA4kQpotq0yGZiglBh6yy2HTHoFbDEIpvrEwxsFvZfYP3thyk+fZ3o+iTmsZDgShl3/03efPr3vrYy+flTG/c/NbJgjfhQMRIhI0aMGPE3xK/9wj/46re//qOTf/H1l0/++BtvnfzTr//gpC02glCSLAaKXfEY+8wsv7DP5MerMR/ZE/HIV77Lx4t5Ms+soPZnMPaGNE8vknESVqoW9/2d0+Ak/On/+EUOnzjPfZ+7zv/1/QrTukxTehTwuClV/sNfztC4PU11J0MjivFjzaPPXaZ2aR/NlsPe49dQUybR25o7Lz9LcfIytLqc+ePPsmvvMr12gZkv/YTv/ckJ3k+aFHB47oF1Bt0sYWjS67qM5WNWdkwWJgPObMDDB2osjMd8440Sjx1bo9MscWHFZPd0n6nJNn7P5S+vCgfG4e7dCQ49epqrF/dy32d/wJsvPcy7OxH9ro3Rn2Dvoausrk5yvZ7gSPrQvBMkdCONiRBp2AojVvyIKBK24oDTQYOHMwW2wog13edoxsOz0t0IDbgo+iS8p7bxA4NXe03uc3OcmDC41U744myWP++ukMdlS9oExOn4IAlj5NAiDEjzN8FwoTwgxJeQRtLGwGBAyJjkCUmYVmPEki6Vp9eMEFccROSeNcoQgwEBDg6BTo1PAEMH1XDVPMYWi0CHDAgxxSQjDl3tY0q6/TEpRSZ1np4EeOKwo5sIKh1JHAqRD2p39VB4JEPR8UEzlhIDT2VwxeHFT5RZnN/hB5ezDBLNivYxtcKXhJ6OmTBsPnVfnx+vJ0yKQ6Q1Y5bJu36XL5zokE88gki43AnoNTze93tYoc3iRMSdBqwNQmINOazUHoYiRFMRm6aOhq8TAkkb0L4TrrKkC0RofIl4xh3jTFJnQmdwlaJomFhK2A5j5kwHhfBav8kuK8OtyOem7nDA8ZipBKzuWEQDlyMHNzGVwWbdItbQajuMFxLGxzugFWtVm37folQICCMDx47Zs7tKodjj3PUCB/ZWGfguph2hE8XsnktEgwJecQcduViNGL81SeYjmrOP/oOvrS5+diQ8RnxoGYmQESNGjPhr5u/+4j/+6qsvvHOyo3sYw0+wM+LQH1pePHFQKH4u3sclafC5JZvvXtfcX3LYt7tK9eoBJp48ze/9o8/xB98c43e/ZfP3Hm2xuVHmoQfuYtYixPLZP97kj/73j3OsssHqufu4pRo8F+/hd//zt3njzQNMJ3lOjnt86X99mVPfXATg4k+O8Nzf/Q77jl3j6//bFzho1zn7ypP8yys+yZkH+bffX+BYyWJrq0wUWvz+Hx+gGycoFC8bN3jOXeBH1yxW6wZHlnrM7bpDY3uc91YVH9mVkMQ233g7zxNzCs8VfN/hwrZweDbEsiLi2ODYXEih3KRWyyP9Ert2r3Hn7AmSREFg8sqght3OMVUwefmqzYkpg+91q4zj8q7sIImBKwY5w+DHbBChaeiQaXHZpTz2TWjebwf0ifESi7KruDsIhjvh4BMzp3McdbNEoeJA0aTTV1zp+5zudKhLj4AYF4u27qFEYWHS1L20dhcDJG2O+iBL4YlDWfJUdQMlippuYYlJOn8YDd8ZmgQQUmGRWrMGaamtThgQkACJju+FxDXcq9LNKJeu9gHNQA8IdIgWja99fD2gplt0JB07/Hf3L/H76ysEOrj3Om2xh99TD1/NB0IkwZRUCBgofst8gl8en+O1i2Wa1QqbfkKApojFDgFFbHaZGQaJZqmi2V8yWKsr6jqkFScMiHln1eSJpYRv3g6YNVxirdlMBriJyUpLKFkGYaL5yKKi2zW5lfTYkQENGRBozfvGNiXtksWkLSEOJr5EbEqPKe1x1dghCExuqxpNIia1R6A1NyKfCcPmWtylqG0WDJfjSyHNho3SBh/ZlXBx2eHwYh9DaV59v8DCRMjs+IC+77Aw1Wes7DP/6BuM7b/BuOmhtA0IuWzA+GQDw4r44Rtz5B3Yu3eNymfewY1bhO0KvWYFyw7oNcfx8i3cj65x9qNf+9qyOjkSHyM+9IxEyIgRI0b8NfKbv/jff1WjqSY1xlQpDfgSUJY8tliUJYeNxd5knHeNDZaSErrjkjcMtvsJd9fyPPMr30e8mOrZozT6sKG6/EfPX2G2UiX3+XXUWJ1T//KLRL0Kz558E/EGPHVimZtnjiLAdG8X00GRvY7L63Wfj37+Lte+t5+KbbAehLz2o4O8/Npe/pPPneet1x/k0c/9gPjSfZx88iZj/iR3qgZ5F7q+iRmbrEYDThm3iIkpVue4oJscdT2OH7vNG28c4sfbfY6VbZpdg0HfxhODdl8x6GWIE6HXM7i2bpMRl3K5y8Kzr+COVfE35nnpvTxb6xUOHdik284yPxlQ9gskCL97t06bCK/vMakz5E2DbOzgYDDn2LimMJPkWFAeF6jxxZkCm12od4VukrDfzPLgQsI7mzEGwpiycYaXEAVcj7oUsLnQ87nZH1BTfe5jjHXxadLFJ8AQxWDYKNWmR0GyOGIREN6zMqWtUwFZcYnRQ4FhExOTEy/NeDDAE49UCum0ilcPhl8PjthE98RHiiGKnPKISBCBaNhy9YGE4INLCgpbbGxlY2IQEfMLlWn+ZHsNSK8b6v/VgvVBFbBCocRAAAS0TrCVzX16lk5fODSlObcdY4jQJuLxcQcntLmd9NCJYsG1OLUWc7EWE2n4tQcD1rcy9HUaJl+wXaZNB50IBVtYtB36ESwnfe7EPi0iOi2LQGumDIdDboZMZNMaVgfnsLmlmjxAhRXpYmIMV+sHxGh6EhKL5mlmuKLbFLCpKItLSZuHnSKDRPNKXCXfKdCJExqEOP0ME3nN9GSX1U0vtX+JQbncZWlp+/9h781jLEvP877f+53tnrvXvnT1vk9vs5KcITnsITnkUBJFWSQUK5GiQArgALGBALKQwJahCAacGAHixEogOUQiwxEVkZRokRLFZbjNDGffe9+7urr2qrtvZ/2+/HFuF4eCKUVy2gGC+ys0qqpv3VtV9zbQ33Pe93keFIr5QxdZvfAw65ePgLG4ulii0XEIQ4ejT/0Q6Xvsno2Ym+6jBLw9a6iZEJ8mlY/eQtY9yg9ewJmo88qJ3x15PkaMGDISISNGjBhxn/iln/+N38papQ3a6GFJXcSUGsfBYnzYbH0inWUGnxlToiURy3rAnPhM5BRBCrvtMvnjK/yrP9/Flgz47RNVquUakg+RXJvkwhSff3mSc1vw4oU5PvLgCuLFfPnNaf77/+FZxieX+PJze6iHmv/mV1/kV//pKX7jmWW+c6lMl4QBKQmGxxYM371Q5sU39pO3LO4uj3GhnjCft7hc0ySx4nzQ45ZqcVRPsayaxNohb2y+rK9wsnWCajHhZsNwqGJxvaEp2opj+zt8fwke2xfxwxsOP/2+Gg+fXCXsFXHdhLQ+yfXXT3Nu0UcDeycMnXaBQj7h8h0f34UjCwOi7TInvCI5S/BtxZ0wwkExbtvUkoR+YhhzLN5IWnzYnuILnVVmTAFLhDMTNje6MZttxWWaFHFomYQp22FZD9DAbavJuupRMh4+NufUKv3h4dZGERITm2x9yRKLCSmxpmt44jIwWUKVIJzUCwxUnJUIik00nHjdS6EKiUlNiiMOMQkWihSNr3JEJsZXOVJSlKidOYXKpAHJ8EBuyOYolmTN54IMv3/m36ioIp44NHQLRxz+3nGP37/TAAEXm8gkOwLmXjJWURWG5YSGqpRQovh16wN83rzNnnQCIodGklCxLCZsh1e6XUJteHq6wL6SRT8SJmybtSTidNFnZjxkuqBotVzKYnNyb4Bnw7NbffbnPGT4vWdsl6U0YB6fBMMu36EWp6zHMTUi2hJRMO5OMeI6A550JrhhuiBQNC6RpJSNR4KmbwzHpYwtggb22T5Xoj5bOmJT+uyjhGcpdnsulwYD5lyHlc0cUSJU8ob3f/pZCoUm/sFF1i8fZeqJd0g2plhcGqdUiDn90CUOn77KrrlNnAfbSCul8MgN7KZDEvnkjm2SvDOBGEHtN6iwjapEvPro/zYSICNGvIeRCBkxYsSI+8SffPFbZ7Mr38HOwdITj5ZuU1IF9uoxtqTLqmrx2fIcL8U1EtEcM1We2Kcp5OChY3Vu3prine88wFtJk2Omyh9u18gvnuKBf1jnv/t7H+bShX28qFbpSEzRuLzx9n6i28f4+7/+Degqehcf4Ol/8ApPPHWFr/3uT/NLZyKmf1PzoV+s8+YXJugQ86q1RO/mPpZMnxYxgwQWo5ALapv1UPPxuRz9UPieXuMqKyxLE43hMbOLCM1d1eCj/ix/vhywYOX4P/qLfG52jCAWthvZof7WpsMHD0ds1Yp89c0x/mK7C9sTXL1T5rEHV5gsKD70+LtE/XF+eMPh6c88hzeYYe/uOi9fHOP4NEyUNIf3dFmYCvnDjQZHVYX1NMIA76oat3WfDdXluFSp6YSzEwXO9wJ251yaQXagLxiHFHh8zOfFfouTVplVAtoS4huHcePzmnWXDV0nVJlpu0eAwRAQ7XhCYhJSNHnJUR/6LWyxWKNJNLwtJIvETU2KwdBjgI01FBkWrjgwLBrsm8EwrSq7L5I1lJvhNMUSCwtraGbPwgw8cYdSwqDEIkUza03wx+/bw/+5ss3ABEQm4fOLmyQkmaQRRU55JCbFoEnR/DyP8ov5/SxEM6Bc/vn+/VQbuzg1I4x1pjmvakzpPDlRbOuEGc9ml+MxSA37qsJ31kKW4pBbSZ8Pl8qUfcNLtx3O1TLR9sReuHQ3RxJb3A4iXo3rbIeG0xWPemDYSiNSoCQOZVvRTzQhGhtFTxKOqBJlcZgTn21CVnWIAdoSDE30iqrJEUrKYVNBk8XozucttiPNsYLH7SigbDwaOmtZr0cps45LO4QbQcip6azwc7Cyn25tlmBtgfm9t7H8PrnqNnZ/ksnZNdyZdVpLR4iCErU3D1GaXUWNhahcF9ceoKp9rL0Rai4E1+bVE//yt5dnfna0fjVixF9C/X/9A4wYMWLE/5+p6QaJSejoLoLio/o4ADEpV9QGc6bCghnjz9s11qRF3jh0iFncdCgXImYeeIdmT1HJG+rSZ0l6HNXjOBak5+/whrWBEmFAyDNqF7+wz+cf/863+egv/xmmmcNEivWVOWpfewjJOzz+4CaT83eJ/+AWtGo8UHX4uV0FfvfYfmZdm75kaVBlsdmUPnXp8bpa5B9unGNlkNAnxMMlGdqj/8K6zLbqk8fjO5t9Dtp5Pm/ewaCxbcPDJzdZmI54JWpiCSxv5vCclA/uya5g9xLNsbmUSxd34zgJF996mOcu+XzyTI/mjeO8c73ID9+aZf9kyux0j5fuGq7fKfP8+RIP62kKlnDMz7o+AhKesqb5iF7gQtzlffY4xghj4rBvvs9DM9mqUYwhRrPRNczis5ZGVIzL6XSK3bpMiiEkK+Tb0g0CYgB8PHq6l9nCjSY2KSUpUNNNptQYE1LGQv2YOJmWKgCeeDjiIChCE2GLTWhCerr/ozWs4X/JajiZsMmKAfXQZ5KaLEY3S6A0LW8AACAASURBVMjKHqetOzhkPqOKFBlXFb71jEenled/nHwMyISMoHYEzAfkCP+s+PjObb7kOO4UGcQGR4Q9usK3bkJsNDkvZdOE/Kdjc3yfVb4ld3lHbQJw+kAmml5d1byv6uOQNYy/2ekDsLdocbKY45NHU/54MQDgq80aDoppXUAhfLvWYa4ofHauxIAEC9gMU5QI48ohIOUYZSzgpu6yULBoSUBDBkSSMGUKjJkcHUIuWRskaJ63lomNwVXCD7ttLpomqYZQUlIxnHaLvJbUeUfq3IgHWALvn8jR7FmM+ZBqoVLtMX/oAhfePU6yOo19Omb+xNvkH72I6eWoHLiCZSWUqi2irRlQIH6C/UQIKZg4E18vT/7Po+nHiBE/gdEkZMSIESPuE5/9jz753Fe++O2ztlgoUZyRA7zMNSyx+AV9BrB5TCYRbXHN2uLj6X4CNN9RVymEVb5eb3MmPcDVVYeSB42uzRTZgftOEHH9hZOctKr8G3OZHA67dBknddh+7TB/9OwBqs39TO2+y+Rjl/FyTUzL8Cd/+iBzZYer544yN/UW9SuH+Z21dc5vCX/BTQ7rSWrS57Z0OGLG+Fx1lrfDNlOmjGdcPqhmuCA1lAhzMk7ddMgNTfZ3VJOaSbDFpkUfpzlGbb2MMhYnSzl6odCLDA+fWscSxQ82YpS2WGwZ5nybxbU84+WUt7YTjk+B0TZx6PFmI+TMvGar5nN0OuXMgze5eGsCTxTP6S2CWHEq79OJhI7WvGjdJRZDmApu7LKiA1S3QBALL8d1fGw8FHf0gABNBYfbqs2W9FkwRV6y7pCiqekWu9U0DdMBoE3W5eKKw0Gm2aCFwVBUeVq6Q0CEJRaBCdFiSNFsmkbWgG6yRhKFwkjmATGYnUlHthJlYYlFTLzj2YBsZSnzbfyoYNAVN+vwQBERMaHG+F92n+a/eMDDcWNm9tzEMWX+r7XaTteHKw45lWOecc74RZ6PNnhMDrEgk9wwbdqJ4X2VPC/G2zygKtygy1fbm9n6Vt/hjmpTMbls8hI7tBs5lnXA2TmXP6k1OKgK9I3mM/sdWj2LWt/gWMJzawkPFvLMVTQn8nkmlctqlK24DSThoJfja9stEjHstfJoY1jXIePKYdJysZUw4VnEseJmFLDbFKlLyEFdZVm1KRuPuhrgGjt7jkRYkx6DFDZVjyesKYLUcJlGNtVKwUbxQXccyyimfcU7rZCysulGsGc6YnG5RNCaYawasLE6x7i9BqmidekEhQ9cxzQ8gsYkfrGBKI1VaqNmLUw/RHwbsyG8cuJfjQTIiBF/BSMRMmLEiBH3ka9/6bmzIBijiZVmQsoYIBGL42aMa7RpqICK8VlULS6pVUSEDdUjFs3LGynTpsBTj24QbEzwn//cm/iNvZSNx1hOcXA2IapXmDMlfuNz73Lh6gK2En7xM68w+1Nvo2Y00eu76W8s8Ot/eJif2WdzfalEN1DEd/dTyCfc3HSYwOOWavAud/i4OcR5tc41tcnNIOHjZjevqmVmTZk/kncoSz7rviDrsugyoEqRkJhn2MercpeDZpozdoUXkhr/Nlzkm/0V3tTbtLVQrs/hKsVzrQ7GCKf9Ip1A+NgH7xCHOaSfZ6Xm8sU7AbuUz80woNPIIVpR61j83mXDdhrzfXWHDWnxUWuBtTDhRWuJaVPExuayrBIr4ZbpcMCUeTWp04gMVVy2ZMD78mU24mzCURaHgdGMmRxXVA1bLGJS8pJjSzfomQExKY7YeLhs6RoDyT4HiEhomQ6JyQzofTMgMQm+5HDEoWHa5MXPph9kKVYCOOIAEOhgJ43qXqO6NRQnuaFPJCUdRvgy9JVEsPOZ8LUHT7L3wF0cN6LdHOOPv3OY15ctDjDDVdnAwuLnzEN8Qu3ji/oNfmlmNw/Fe9hDAZXYWVCvwIWwi4PFLbLnrWp8fGwGpNTVgAf1FIuqxRGqXNUdniyVudHQdEzK2VkPO3L483qbViCs6ZD1OGa/4/N20KWc5nix2eeNpEnZuLQlIsUQBRYaeMQrM5EXXKWYsByaSUpgDKHRtBPDGgN8LHqkjJscORQaYVm1ScXgDSdCEyZPV0VoMbQkYNUEFFOPO6qNb1wecytMKJfzUZfQaL6frPPLe8p4tlD2IY4VrmOwFBx+/CUc7RO1x3ALbXRQILi9i9yxO5hahdzJG9gLDawH9/Fy5Z//9vL4zz63XPmZ51YWPj1avxox4q9htI41YsSIEfeRxCTo4RXt0MQkaIric1fqXFVNKsajR8SR4R77ATONhcWAkE3TxMPmsQW4eHmW9x9v853vvJ8oEYyBmbGE04+/wqf32/y3/9V3ufjWw5zYG+DaBudsG73qgmvz7hsPUj77Nqekwht3bP5F9wr/urfEP72xxa9duUZIynetWzycLpAXn29Y13dWfPoS8XV1i5SUtkSUpMCMKTNjyjhkV567uk+PgIiY/503yeHyU+4cD+6L+bVdY6RoHjC7iEyWBPXPuu+y2XR2cp22wpRWnPKdF/cyCDLDc5AaPpCrcr0Tc95a5yvc4GI3oBcbLqgVdpHns+YwGsNYLjMsH9HTnLfWSUTjiEXF5EgkS2a6rbZJMVTEoWRcvhDe4aZqcMXa5vREJiYm8FjQZbZosaq38XEpqcLwtQvp6C6bujZcnRJSo9nWdWq6gSceKSk13djp4OiZARExFhaxiWnpNjY23jCs4J5Yuff4fd3HEw8La+f5b6ftnX9LWaAvwzSt7He+1/OxuVXA9dt4hSZfebXCpK/4A/UmNsJTnOAXzCNMistWGnHc2ofnaooeTBWFgthMGI9nKlXeb43zoIwzq4sc9D1CUq6rOiEpz7DAJVWnYFxcUZy0yrzQ6XBXBxy3i3xhvcHVaIBC2JQBDoop8Xhwd4IAK0FCgiYkYUV1KRsXxyjOqxqH7Dzjefheq43vQJAaXBFio1mkx5LpUcRGA+O4nPTz1Il4qlBlQZf5sJ4nlJQ5XWJJNZjUeao6N3zeDNuEPJBOYiPcikKeTVdJ0CxYOfbpKr1B9nwX/IR+qDAG1uoO4ia06hXSxOHWO4/iT66iVEp6exL/4csQK5JLs7wk/3g09Rgx4m/IaBIyYsSIEfeRf/vFZ88C5FSOh9jPIT3BhupwWM/QloBINId1FQuhaDyuqk08HFIMvrgsmCofmhdEhEHg8O31AbU+DBJDQWycZIxiIaZ+az9RZPEn11Nu9hLsN47j9icoTt9iQiUsvfgonXaOgiM0I+HXD07wxeYyj+g9vG4tclLP858slPlqZ5m85Hgs3c1N2cYSxbLeRGHxAbPAlClzUa2xzDZzMk5IAggxKQERvnjMmTG+p+/wAZmjWo7pbldYky51unQk5GfTY0y6FnfCiGvWFi9wi1+o7ubFTod6w2WLzNz+fFRjRXpUTR4DWMZCtEUqFq+puzg6hysOF+I2y9LlhtpglxlDISyxxQEzxS3ZpESBu1Ln/cwDkMdGjCKQhP16jM2+wUKIySYlm2pAiqFHgC8ennKJhk4PC2tnihGaiJzKZb+/iVBYZGloGldcHLExgCM2IVmsb0Kys2bligsCqck6QrTcaxkxJOjs+w0Tt+6lZJlhnK8edoncu/1JDnL42G2+9a0PMOkr4hSOJvNcliYvc40z7GJAyhvWOkfTSZ48EDNWjnCUoEOHVqz5erzK339E83trNc46U7wVdpkmx4b0aUnIXXo8wTS+9nhBrXDAVChic121GNM+43g8Me2xz/FZC1LyWEzZDi/XQnxsnph1SAcOWyaiYjwuW1vkcXCwKBqXfiTYRjHjW5Q94bVBBxDyWMyJz0PjHjcHIfucHK9GLU7YJRwR6knKk7Meb/W6TJs8Ph7b0mdT9XCxOamn2K1yvCvbPMgEK9LHxWZL9bC0zROlItoIg0johxaH9nSplEP2761h+gXWVydptwocOHqdrTuHmHr6FZrnT5I/sgIGXv/4/zQSICNG/C0YiZARI0aMuI/cEyFFyVPE565qIAhtCehLRFsCGiqkLTE5YzNpCjhik4jmoJ5i2vicntM02h6z0z1y/RLnwi53pcen9il+eLlA1M/R6rjUOjbvRG1OO2VOH+qQJBbNawf4zst7mSjCat3mpz9+icenfN66WiaOPeaMz3VVIxLDt7tblCQ/nCis4YmDwdAxfcZUmTfkDh0VU6WAEWHd1BmTIgXJDb0PZWwsbLGomw4P6V08txHxfesWC6bKlnT4WX2cF6y73AxDnrSneMWs4IrNz4xN0+xaKIQ+KV/lOo/pOe5Kl1XVYNaU0WKYEZ8ZfI6bKR4q5plLS3zTXOeEnsMRhy3psCUdWqbLlFSpkOdFrvB+c5BP7/b5QneZtiTUZUBPImZNkVetu3zCnUNr4XDRZTPSXGeVgQnQYmjo9tCNkU1APHHpmf4wsjcmISEnuaFQyT4OTchAD7JELRMMhYuNhUVgAhzlkJqU2EQAaMmmQoosclebdGgo/1GrOWTdHVnFoqFslTOTOxbfi1b4/CWLZ/zdxCm83u3xefMCn5UTnDTzzDgOHZ1yyFSxEc7Mp1y9XeGhR67S3JrklX6HislxquSzseXzutnmcxMTvNXvoQWqJsdeU+Yt2WJFdZnUBa5LkxuqiYViyvjcki7v9HuMJXlumx4lHLpaMyYO24TkIpfTC5oLrRgthqrxOWjKHLAKnJ62WO5qOiRsB5pOCLNWjp5JcVB8dJ/CEtjtenRCsFKLJT2ggssu12Usb3i5287SzfBwjEUOB984LEieczQ5aKqUlUXfZKlbiRh+aU8JS4RuKOQc8GxDr++yspmDxKO+PcbpT32XmZNXceZqVJ9YRso5Ch+MoZTjlT0j4/mIEX9bRiJkxIgRI+4j3//yq2dDsjSkvUxQwEWGV69jUsZNka4EzJoy+6TAnMqxYQLmdZkCDp87bOPnYlodj2/cEMYdm++laxzV44yRY7ZsOHZom2evuuwtK4qRz7txm6hWRqUeX7sB/9kzN/jCy+PsLVk8f2GcLy1F/JefWOKPb1iUjMuy6gyXqVIK5FiWBhu6TknyxKT0TYAtFuNSAqBFj4Zuk5DSI6QsefqEDExATEqbPiXJ8xfxNe6oJruZoICDJy6vqyVCYjZosUifeHjF/+Bgjs+bt7mt2jji0JWQO6pFXXr8vD5Kg4jXZZE35C6BKC6rbU6rCa5GA8Yp8ZhT5U/0OaZljFW9na1MKYtrZgULxTzj1Fs2a6rHE2aGW6rNjC5RxKFifEgVV6XFG0mdVdXEFZeEhK7pkZccMQkFlScyEe+tECwMCwff2zwemyjzewikaIqqSGKyPDEZNqvHJiancsQm86WIyI7AMOisUBCFGXaA3CssFBHAYInFP5AP8apZoqgKfIijPG72ZF0dvsJLHd4w65w08zxzVPPOpqYoFiXLIjXw2qpQCwxOZ5ooEV4OGhSMw4u1LPL2l2bH+Tfb2+SNg29sTlplYmNYVG1yxqaERyyaVAw9iVhXfWwUZeMxb3kc9fLkxaKoLFxROMbifXs142MDautF5pXPJB6zOZv5MnQGQj3SnFfb5IxDh4SOSYjR9Eh4vTXgRichn7jMloSDVUEFHsdmDBMlzXbb4no0wAB7pMAj5Tz52KNFghiYIoeDomLbTCiXW6ZDHofTFZcwUuQcyHuGek8xiIRYQ71jMT8RMXnmCtaRPFtff4Ti2QSiEH2lwSv7RgJkxIh/H0YiZMSIESPuI9/40vNnbbHZLVPclRqRaMomRyQpJZPFmm5JhyI5jlglbqR9Fq0GShSzJs+s67KylRWp3QxDPG1zjm2etGfYP52w3rAJ+j7nuyFBYKGAQzmf7SjlA8c77M55bGxM8EYr4GMHNfHA5dW4zrM3CjxiJtkgYF11mDEVanTYwzgNya7y75IJaqaNiOCJw5PpAd7gFgMTDleJhEmpsKK3EcnSnfbLLBumQUUKdIeeiLp0qas+saSU8QmIGZMi70sXeIWbPMg+XjZr+OJSNx0mKDFrytxkk4CQd2SNmuplC1EmpSYdEFhNEl6Qa6xIg+f1IgZD3bSxJNvvH5MSh5lDqeyq+AFK2NphQrncpYcSqMuAUFJOWBXOSY01qbOabmHEEJhwx1wemIDQRMNJiEdCghoWDwI7AgTYeS5y4mGLTUy88xixyQSLJRbRcArC8BF27k+2fncvNSsd9nlA1ppuMChRfMo5yHX6iAixaBwcPr3PoZzX1DqK43qeX/nYOrMLd/D7c3zmF76P09iDil0+eXLALt8hjC0+/FM/4GRwmOdrIZPGZ5xM0DbaNhrDFB7PqVWKxsMA66pN0eRoqsxEP2Z8BDirZrlKiyN2kQtBn0MFl61Qs5QOKIpNDofLKy6feqTFd5cNh/MeJ/cO+MZSwrtxh46JGTc+PUmYIsd+N8ekcgmyX52bVoMksTjge9gW5CzBsQwHD2zx0k2faZOnTsiHxoqsdjVlx6KXaArYbBHiYXHONNBa+Pn5EuXYB62IEqETCL1QaIYGW4RyDrSGJ/7uN0hvz2HNNTn3yX/y23etp55bzn/quZVdI+P5iBH/voxEyIgRI0bcR77+pefOesMUpDweJeNzi00OmCk8bEJJaEuAiKKtNTetGjEJ06ZICuz3ctiW4XZL0zcpkTbUJeKoVUYnFkvdlI0e7PWy4rgzc4Kt4APHeoxN1jh3dZJKIeVOU1jadlgLYyomx28+GfDunTyLqs2MKSECoST0JaZmWigU22TTjpL4CIo11UGJwhOHwISUVJ6myRKyDBCbmG3Toih5mqaDksw/UVJ52rpH1/TRAhOUadHjkqyTkLAtXZ7WRzhqxrkq23RkQCpCnzDzR5iEn9WnuChrOx4IDayYbWyxCEyEMzzsK9TO+7ppkVc+U6bEtnTZq6sYDBelwW3ZZCAxazSYocJrrHE5vcN+NUcgCbGJ8ZRLSMSCmmLbNIFMKrjDpvN7ssEYTUHliUl2Pk9NNvUY6AGOuAx0HxGFLfZw2mH4qzFDIcLONERE7Xx/SyzO0+Qz6XGuqRq/OXGcKe1zqy4cnImZrmj2jsPCoctY5Q7zR28gXsL0/ApHTtxGojx7Hn6ThYVVrF0NJo/c5Kd2D6jdOMhiMmDceJycVJzvBCypDkf1OCEpTRWwX4+zZDXxjU1fQtoSgAgbJmJC+4yLy7oOyaUOz7LCblPCkBnTP34kxnE10i4TpmBSmxeCBr6xGcPjkrVN2XhEaLbSGK2FgJQUw5TJU8GlE8B8SfAcQ5wKS2slZgrCoUlYoIABNgYpt5MBV1WDgnHRZIb+aXw+ucdlcdvi5J6QciHl/LowU1Bonf2MTxyKeezp5zhydAnT9XEe2eCV/aO28xEj/t9mlI41YsSIEfeR0IREJiEkoWY6XGOFiJiK8XBQ2UoLKXfNFm/LEi4OATEDSbhibdLow6vbEXWd8PRcjlvSwcXiVhRwvZWwqPu8axqk2lCyFIvbiuOHa/zgfIm33t7PzW7CRsPhdDHHlGMx77isqC5f/eEMLRPjGIuGBDjGGjZ5a0pSyDossEhNStcMqOkmfUL6JkCb7Ep+3wSMSSlr+DZZhKyFysQGBmM0RZWno3tYYu30XVzXyxTx6ZguGoMvHl9Xl/jX8hqQHRa3aPFxfZTAhHji8U3rCgAKRUkKWeqUWETDdaZHOICLQ+YqURxXe3bK+TSGddNgSXo0JGJS5/lkepTFdI3DzAKZQMwrn4v6NhExAxNSFJ8JVaXLAGMyIzhkU497yWAAIorABDu3Azs/mysuoQmxxMLBJhn+vO/9ur/MPYFijN5JVhP5UZHhhFXl45zmyfQQS9Ljk+lRvrzVYD1I+Dsf2uDwk8/TaLsEoYXkIn7wtY8ibgpOgpruIIWQ8p4bqLEeKEPzjYcwAxdr/yaf+cR5/u5Bl9e6PfbuadCRiAVdIiAlj022JCbs0mW2pct+PUHF5HGNzZq0aKiAbprSkICX9DaOUWwRUCPkLWuDJFUs3q0ggDFZMeC29PCwiNDsS6uMGY8KLquqQ5uYB3JZMMGm6lNRNpOujetqTp5aJIyFMAElEEaKkm9wbcPTx2J8LDwsbqsmGsOmZB6dZ5dixnxY3fboBzYPTCmCGHIOnJlwOPb091DjXeyHW1izDej9dYJxxIgRfxtGk5ARI0aMuI985UvPnnXFwRGbgOhHvgElDCRhQMSAbM3nFLu5LZtMUGZZavjkMJFLgqZBiBV4LJs+66pNU0V8sjrG3UFCTxLaqWagDZYRvn5X82sf2abTKjLtW7xWi7gThZwac3iz26dPwlWaNCWkqyJS0ezSJdZVh7rpUhKfpum8x+VgiE1MSRXpmQFFlUdjqEqROVNhWzpoNHnxEclWicqqgKDomQGuuCgUyXB6YIsFAsnwgK2wiEmoqhLJsJ/j8XQfL1t36JsAEXhaH+ca64TE9EwfW7I28cQkuOKwTA1bbJRkyVFruoYSxX6mWZEmH9aHeFYucJVVrskadSumY3q0ZMA8YwwkRgu0dJeH5SBrpo4rLht6e6d88J5vIzaZsdoXP5uIGE1RFYbrVQYRhSselmTt6a44xMOuD1ucnUnIvdSr93JPbOx8jqDEQpsUT3kUVYGQmCfNPu6oDr7J4oXn8PnYYcOtO1XmqxG9VpVm1+bqxf0sTEdY/QqmVWbl7dNUqtugFYhh49KDiAg5NaB94yj+2AY5x+HROcPtxXEOWEXuBjFVXAak5IxNDotL1iaH9BQF49CUbGKlBcaNz6J0mDA+nrHYVv3MEI5DSyIu1uB4IccgFgaJoR+BJC45LCo4rEmfI6rIY9MOiz3NMbvItajPgISmDEi14uxui9ubNjfvjBEmUPQyP8etbUGMECTCrS2bkmVT1blhYpbFYavIeWkQY3h4zMW1Db6Xcuz4XS7fHuPAXETBT5l9/CLYGik7rP/F41z4O/9kNAUZMeI+MBIhI0aMGHEf+bMvff+sM1zBmZAyIQmHmacmHSomTyQJDdPBwqIlwXC6UCRBUzE+H3Qm+B7LHDVj1HTMFD6RwJJs82bYpicJD5hxbBTXVYNjdolLpkWlPs8bWwmHJoRXWj3OuCXe7Ax40brDMT1FjOGyrOKIRZ8wu5ovMS16FCRH3wSk6KxlWzwSUiIToTHDsFqNLRbrNPHFI0WzT2bYMI3hilS4s56EgIdLTNamnhOX1GhCYmyxmZAyfUKOMEdOPNZNnW01wMOmYbpMqip5ctSkh4hQEB+AwIQAwwjdiNjEhCaLwjUCGk1ZlVjWmzSskKbuUFYlIEu8AlhQUwgKLYY8HhumwYqpIQjR8OfPigM1ejjpEFFUVImu6Q1f5UxI3EuxcsUhNOFOx3n8numH5kfTEhlOUth5FPNj7+/dnld+JmJMymFrNy16pGLzurnBo+zOvgYbO3Y5daTOy2/uYbGm+MLgLuNhkZs14RuLit+91SdYm2Tl2kFuXd1HvLmX5bUiv3/JMDfYi05d1pf2UK50GJtZpFwUVJrD6vucmYWo5+BicTSfIxcV6ErMPHm2ZMC2dBkQsUtXiETjYNGTGAtFXQLGjU9NAo5RYcZXVPKGJFXYShjE4GGxL++QT1z2lmzerEckGPZ5Oc6nbQo4aCASzVJTuJJ2eTfN/r1/4mPvsrUxjWcJ2gjbA0PJFdqxZsOEnJ3NEQSZuDtoFTju+2gt7JoZ0O66hIMCH/m572KHZfZ98CVkSiM2vLL39367+cSZkfdjxIj7xEiEjBgxYsR95JtffuGswZD5QoRZGWNV6thYNKRLQIyQFduVpYAnLgExZfIEEnHLZDWAN1SND6lZrtImlpQt6TBnqtxkAw+fd6xVXBze51d4fKzAmZPrmE6VeleRSxzOJW0m8bih6uwxVXoSE6qUDdPAEosx8kyaAktSY0yKaDGkaKzhmycuiWTCIy85NDr7mVV+Z3Vrk+ZOm4UMJxKeuNnEYihAAGISiuLTNwGO2HRMj6LKs0qdshSIJOGJdD93VIOElI/ro7ykbpEMD/kLMknNtNCSzRRCEw69Ei5mKI4EQaHYMi1Oy35umGVSUkITYcSgxMIaJn5tS5t5U+V1fRWFkAzbyeNh6lXmSGBnyqMQ3i9HuGu2doTJe2N073387/J+3JuA3JuC/GUhosT6MRGSUz593d/5ujbZStG0jIESTpopvqLOsar6fDNapLC6j0vBgK/JdbZNG2U8vsxbrKsuBfH4SG6K8TwsdzVFy+LooW3+cLXLoWScS6sur24mfOj4NspKiYIi1fEWC9Mh4xM9ju/tkrSqrPU1F6XBHVUnEhjTOZZUJtz6KiGPS944FHFYUW3yOMyYPKmBWeXxlcEq3bbHnrxNwTPsryimfQvfhYJlsdU3tHX2am8mMWecrI9kTnxmxOe8bKNQOFgctAvcXpzFUcJHfvnPaN46xCBQ7BrXrHehKDYXOgHzrstUQfAsoZw3XK6nHJzS+LmUOFHMPnIJq+tiz9eQPLxy6POj6ceIEfeZkQgZMWLEiPvIV7747bOeuLiSuQhSNE3TJSIhIBpOG2wKkt+5j4XioJ5kXdo7B9XjepbrtKiYHDMmz5Ya0B0a2mNJccgiYf/Rz58nbE7z/Lkqn37mNV48P48GPGPzttrEE4eGhCCQx6UvEWX8rKVdYnqE9AmZlbGd2F31nhUhDxdPXAwQETMjYzSGq1t6mNqUxciSrUaZNDOoD03W2qTkxKNPgCWKyMQUxEdjGJcSTXqc1Au8qm7TY4CNzQ3ZxhOb4+xigxabQ+GUmGR4OM+EQUw8TK7SRCZbERJRfNQc4rysoEQN7xcza03S0V0OyRyn9BwTJse7LMFQHGTxuJmvRUTIi7/ThO6Lzw2z8u9cp/rruCcmftL93vv3mYk9xWCy8kPlUpQ8ITGPmD3MUOaUX+C7yW32Mk0gCe/IOluqT89kyVXXWeWY2sMJM0dTAlTkcbhs8wf9u5zr9bm9VKFicuRShy+YKzRUxNHBDcBOzAAAIABJREFUfnzPorY5zuyJdykeu0Uu38T0i2xvjfHAQsi1LQsPF4VgxNCQARXy7NfjlI1HCYcN6TOvS+SNg4dFHpu+SXnEHmPadagHmk4o5O0sFvd2SxMmws2kz7yV41DeYytOKCmbMdfi3biTNZ8zxhG7yCw+4zkhTuHskxfoLe/FwuLMqTs8f24CTwklR9FNNI8twFsbmjiBMwf73N50ODgT0+872LYh2ZjFcRO8g+u8cmQkQEaM+A/BSISMGDFixH3k2S+/dFajUSJ4OKzrGvNqkh4BLg6u2PRMMDzE6+wQj2BE6BJgY1EyPj4OsaSEktKSiC3pDJs9Ulb0VtatwYBT9UcII4ubTc3d23v4VrLKFWlwTKqEGvabCtfVNl0JKZFDicWhdIIV1SQVTYcBnjjUTJs9MkWXgMjEZF0nFikphWFvRkJmuDcYBiakID6BCZlUVTqml60ikeIMS/cEARF6po8vuaxoT+wdz0XDdBCEpeGVbsgO5VNSoW46rNPkE/oEV2SNhGTYuZEd2h1x0MOpRbYeJTjiIAjvspT1hkB2PxQd0wWEVAlLqklDRWyY+lCACKlJduJwLVF8TE6xKi08cXnGnOaWbKNEdtrP/5/wk3wgP4l7kxRXXCDrHBGyda8bbLAhXf48vpx5UJSLxlAkR5Ecd80GVVWibwKmpMqKZMECN9QW7/YDfGMTSCbiuhKzbgIi0exPx7jSSviXyxvojUnCu4cohVVsDbevHUQpeO6mxcvWCnXJigyfsmbppsKHmeUufcbxSDCsqS4WQhWPHBbbhHSJ+ZUPdKnXC/wwaPKA72MpeK7V4Wg+hzHQSzXrJmBMXI6VXZJU8GxQsc0KfZ6eLPKDbouicejHhklfMV5URKFHodTjj767h4d3a/ZMJeQsxSce3aBWLyGJzUP7Qxb2LTHpFjn0keeZ3rvExPgWaI/Kf7yCmhrjrvXUaAVrxIj/AIzSsUaMGDHiPpKSYpMlINlYeOIRkaBQWbmfSckNrygPCAlNRE03adEHsjSkU3oKhdCWkDVp0Scih0PXDAhNjI09vPKt+P2VBqkWEgy3BiEb0qZOl5fZ4FXrNm9YazjY5HDpS0yLPkUcysZnyWwxMAGBidFoVkyNguSGB2GH2CREJqZpusQkOwIgMQk5cUnIWr7bpoeFQg/vF5gIGxsXJzOS4wwFVBZla6EwRpMTN/tDdui+19g+4Ed9Gt9UF7KZizE7z8+9n+HexxqDLfbOWlRW/Cc7JYNm53ivWU7XMWiupnd+7P733mdrZYauRHyUE5zlAboScUYOcELt/4mv+1/2d/xNuSfaXHFJh29qWHA5MAEDE1LTTSzJ0qrupGs4WEQkNOnh4tAzAdNqnGWzTR6PiJgSmZk+j0uPgJZkf0rGpWBcbBRNCZkyZRxR/K+NJc5dnMWZ2KJa7WEpw0+dDljQVRwsPqTn6aWaz1WnmPEtnnInsVAsqBy7dImGBNxSWbzxuupyWJX401fGudHO/Dbt0HC7lb1Ox3YH1OOUh6s5yjgUHGG5o7kdhrRDw7xv87g1gW3BB3NVdvs2Pa1xbXj9/BRTu27TaZV5+tSAxU2bSqXHuXXD3TszHD22RCs0lEoBXmWLQx9+nvW3HkON9Rg0Zyi//23Sd1qQJn+r12vEiBF/c0aTkBEjRoy4T/zqZ//Rb/niZWlQQIk8kWQdFkoy4/M9v0REjADFYd9ETrLAWUH4sD3NmHG5Ig16hDyo57il6lSkgCXWTmFeWfK42FzohByziyiEa9KgT4gSRTCUDmOmQA6bSVNg1pR5y1qmJp3hrZqSynOC3axQIx6mVcUmu9XGQonCxWFSVYZrVVkcbTo0ryuxiIgwZFfvHe6tY2XrWmZobre4N2mwdgRD5uTI/j4a+mUSNFUp0iOgrwcIDP0lMkyT+tFB/70f3RMjeniIzx5fdt7f+159EwyjfLOEL0ccElJ8yREPV7BmZJy+xKyoJkVyzOkiqRgWzfqPveZ/nefjvchf8WYPzfWpSVCicMQlJWtUF+4Z3M2O0T0dttfbYrGlmwhCSMS0VFk3dSalgiBMmzIFPKZ1gVgyudWR7DXcrUvcVA36KsZGWJc+j+oZXh90eSQ/iW0neF5KGLo8NpbjuXqfCZ3nwXGXpz76Kou3F1jsZ06ax6YdLvUCJo1PguG4U2Jbx6QG2ibhoqozbnxWdYA2QgmHT3/mBR7a2+Hww5fQy4eJEmHCF5aDhA/uEa7VDEcnBUsZZqoJYWRhG8WjDzS4uebjxONorTj82W9SbOxi/tRbnFroMzm9jeP1Obqnh+WkXDl3nGBrHs+Nsfsurt9GvBDJpbyy71+MVrFGjPgPxEiEjBgxYsR94Fc++1//lkLteCESUiakRNP0GFomsgMwIYlJyCsfT1wQgy+5YVdFxAxVfsAiLTRLbKNEsaxabOkGIlkE7oSUsUSxqRtMSAUjhg0dcVHVWRu2lx/QE7QlzDoeTIWOimhLgAHGKGAEWvTZpSYRhD/9nYv88TfneMjsQZRFVYpMS5UGPQ7ILAXJsW4a9E2ALx55lSMwIUYM+9QsDdMhL7lhNHHWsB6YcGcC4otHRLQjODIBkxATk1c5iuL/WBv5gBALi2SYWKXRIMNiwKFvInssdqYeFgolahgFbHZs4lnZodl5k+HXAzyhjrNLxtmkzeNylBXqOOJwR6/RlgHzMsGCLmEhdCRi0Wz82Ov+XvHxkwTIvds85SFkBYa2ykz7ltg/mtyYBEusbKozbE2/N2HKHtsgw98ZTGby1z0OWPNMSoWy5OkwoCB5CngMiNhnxvFxWLSarEmDVLLnYNqUCCRl0uQpG4+aGqAQdlPkmXmfvQdvkyt0GXvoHFWvTbEYs3FtNzZC1bYo2CWmxiI+9cvf5qFinmJec9QvUonzvBO3OOmWiWPFgJQnSkXuRCGP58YoGZfT45lnZPv2fn7wzgzd5QM8+shNJsuGqfGAh3bFeG7K0V0R+w6ssnvfXSammviWRzDIcWPFZ2FMs7DQ4MadCsXOON1unokjN9GdEkmYJ3fkJtLNUXrq4v/N3ptHSXaeZZ6/97tb7JGR+55Ze6lKpc1abEuyyhu2oTEWTGN8pg0H8BzO0Lihme6hD2emC093zwzQzeA+gLtpGAP2GLstDAbjBRvZkmXtUrlKVVJtWVW5b5Gx7/fe75s/bmSqZOOmB2Tose9PJ09mZWZFRGZF6HzPfd/neRjoCKOvfYbM7DLe9+exX5PmyZl//YG4BT0m5u8W++/7AcTExMR8JxKa6JC8mzAVEGKbqDcCE00tLAlp4xAQ+SQm1AgN0yYnKQZMgQU2uMAKB5gAwBMXD7tf8BfF6CoUTdMh0Te/u1j0CMkbDzRsqwQ9Ai6pLZJ4FEySVVVj05RJikcWj6Sx8SXkJmaYCXOcszZAhBwpfuvXHyM4m0ISPqaZ4D1/cJAPvUH4Hx4N967CD0uOUZNlXSVxcXjonVVu+RMYlQEW9SaC6of67qY+KcakwHWzzrwaR2O4ptdeXocyhgoNFKp/7f/lWNvIjN7fJDamfzC3+pOViL37ESEwwd590l9pgkio7BYqhn2t4IrDipRZ1pt8D7cyqpMsWKPMmyGeUwu8w9xCWXeoSuStsFDY4nxTAeGN5vNvJURSKrVXtBj1j0TN8M6e/yPc84LsPt4b3SfRalY0a3LFIdgTY5q8SbHABmlJYGMzTHZvve8FtcYAaQ6Fg9SsFmkS2EaxrurkTYKOBNSJxGIGl4fVMl/ZEH6nOszg4XNsP3UPzUaKwZEdTs4plos2h2fbUeTt/peQTIhl95h816Osf+idDOc0b2iNc3S6x/XLih+cTvHR1TK3SgFHwV+aNRK1KXraUNoxZG3FRk2o7Ayztp7lwMFNSsUBdChMza32f1+axPx1co0c988ssnz1ACPj2yiledNPPI6as8l/aQrrDg/XWmL7a/fjLU6SeGAJ0hnS71yh89k5Tr/3Fz/QT1iIiYn5e0AefvjhX/r7fhAxMTEx3ym85wd//pRC9tq6B1SOTL/8b0QG8AmxUeyYl5Ov2qaLQePiMKfGaNGjZlp0TRdHHCZlkEk9wBlZYlIGqdLCwWJZb0VX0bFISXRV3cGiYdq4YjNCnhIN0nj4hMzrIa6pYt/OHnlR5swQnrFYVTU+fep5yFj88D97Df/53z6HXrKwDoAu9lBjCYJnE6jhOm/9d0f4wk+s8GO/e5yKtPiTd69jTZYoPnM3mXwRHTr8wOcNH78/z7u+uk2GJAbDtXANW2zG1RCf+V74gc8qPnEyTblY4KfPrbKitwkIsIiu/huj8QlJikdTtxizhvcO7lXTwKBfITJCQkbVEDu6sidOdn0gFhY5laGqa6QkScO0yEiKhmlhYZEUjyk1gsEwaNKM6QxJbD4jZ7hV9jGsU2yrJi3xORwO8by1Ss1EB/vNcDt6DKK+qTX95QSvl0WJKy4JlaBrunR1d0+QhH1BEfYFhdXvf4dIlERTD7338+56W0bVEEVdelmc3cABawqfkCxJmnSYNgVq0mVIp1hSJW4LJ3nUusIRM8FFWed2PcuyquJg4RmLQZNiwHhMqgTv+4GzXDh7nN9faPOBt2/T6yZIZmqUNsfJFSq0mxl2ijnGJooMzFxh4fm7abUcKg2biZEOruuzsJRjpWYoeEK1a3gqKJM2DpOSpGc0AYZBZXP/oYBWxyIIFEOFDqubSTKpkFzGJ5NtkcnVyB47h6kncd4xgP+FMuXLxynsfwnE4LwtF/0CvASPh78Qr1jFxPw3SDwJiYmJiXkVMUajRWER+RYKkqFNF98ErJkinrh0TY+kJKjp+p5BWlCEEh2ca6ZFQBAlS+GwYoqsSQlPHFbNDhpDQLDXwdEkiqOdkiF8QjL9yNtVs0MPn1rfqHxdQZoEHXp0EcZMjnWpkiHBqM4geQcci4//1HlohUg+xHQV4cVxth8+wPBdT2OaHr+2/wDW1It85BeeovbcbUiiR7g0QmHqCgQW2CEfvuMEA+Pn+VeDr+fgfBUv0eG9j0dpWf/3rdMkj3yZP6zfhWV1mb/7Cf6v7v387GXDsMlw1lwnNCFzaoyiiUoFlVi83z2BZ0GtZ/hg8Bx109gTewbNuDXMP0/czr9qP0VF1/DEJiNpyrrKuDXEW8JDfEqd4R5zkEflPG8zJ3jIPEEo8A/MrXw8fIJpa5y3m/28JFU2pcGb9XFyxmVBlSlLixQeSyr6nYWi95LNhCjG18ena7ooscipLDVd3xNKu2tWu8+T0GhccekZH4XgSZKO6UTf218RC294Xtl9k3xENNVRCGVdjeKR+wJlV3wJihW9zWE1TYsuPiHrUiVEU1INCqRZUlWSeDgm+j22xGfLVEhLgrfrg5xWWyypCi+ieOFPZni97TCMoVYpUClncJxBRsa3CUOLDz2W4e2fevADtf4jvCv5c6dMOYt99ybi2ejlkHMfehevO9zh0lKSW2YCbrMyfP4ynBiyOPnglzn38APM7lvhwksztLrCm973abA002ePUtsZYeytjyJZxeO/9gPcmd3h+f++32b+jv7bN/JfH14WExPzd0zsCYmJiYl5FXnoE58/GfVkRDv6PiEN0wai1axu3zfh7y0o9QsBxY4K2MShYiKTeFI8HGwE1f+zGwkI08PqH0jv5whL7DCocjTpMGryzJlBLpgVEsolKQnaRM3iXXwyktzLiFqjzA9zjKYJ+e3fPw9KMOUWeD7WwTHCMzZ6OYc4AanJVailESckKI9g15JYdhuHHtXVA/jdDKlbrmB2MnRqowzOXaZXG2ZufhXbNlh2wDuGC/yD4QLJZJdUskr6pgUai3M0Nqao1ZL86E3CP3r7WT57eoacpPn1ozN8bHsZSyzSKsHjwTrvnR7CEYvPtZf3fo5xa4S6afIz6nUsdru8XmZ43FzjVusAb9cHuaIq/OrIHXy2vcVteppFKypFnDQ5zrOMp1zuYpbLqsSApJnSedZUnR1pMmIyvGBtMG6y1KTDa8IJTqtVarToGp+tsAhEAgNg2hqjrKuk+gEDGRX1ehgMCZXEFWfPyB/VOkZEscwvG80jURH5iXanK7tm9L03eTlBS/dP27uraTY2lthMqxFcbLoErOkiCfGw+hOTBh3mzCCCxZaqMyRZrss2CfGYNANkjMe2tLhVjzJjspy21rhk6jzgjTAz0kMEPvKCMO9mUSim3STh247s+SrWDr79kbWb3/TISuH7H5nhkZOm0eLw615icGqRm37wArkGNGt5bp4MKNc89t9+kdHxdRJjGwynfcJOjtE3XIGKIvFgnuwdG+hLIU/d8aEPyPcefmT9lgdiD0dMzP+PiSchMTExMa8yu9OQrunuHSYFwe/3YkDkbbDFxje9foyvjSM2qX48rdNvKtf9lKie8Wn1xcTrOYJPSMLY/IePvsS737uPYZ0mRJM3Hr/86av85LsOMxam8NG0JLoS3yHAMYphk+SL1kXu0wd5/59fQZ87D7qAKdaR0Rys1tCXN7GOW5hyDXEMpq3ovjSOQ5n8YInLLx5ktp1m4M7TVC7eQhBY5DaylNb3M3T0BQgVj33tGAdmGoxObNDtpEllGiSSFs9+fZp9tXs4+MAjpLIVNlemcRxNrZqm+JWTfPTkFtVSnvxgkaELAxRIY6F4u0yzvK1IOESRv2LoAd8fHuF3ZYdfDh/mvbweCxhRg+RNgnXa/Li5g4c320yQ4bpVIW0SJLFJYpFQSQ6qSa6aGm/Rx1iyqqxJi1v1CIuSpKhajJosDekxofNsSZsx8nQJuG7WEVHROpxKMix52kRTEI2hrVskLBdPPLr9AAK960kRC21ejoONVsZk7/my61kB+glgvGLV68a/N6oGWQ7X0RgcsVAmJKeyZCSJT8gL4VWmrFFm1Cg100LEw2AYIceCKkZBAXhsUCY0mjZdHCyuqyqCcFVVOaYHSeExaFIk+yeHL59Lsay2Obc4hjYJ5j/2jm+59vRE7lc+wK1w7D/+5qn8D13CbDRJTC9x6NAVOpcPMn/3KmjBOlgCBQn7OrUf/58+8CQPwjzRRMMD3vg3fFHGxMT8N0fsCYmJiYl5FfmRB3/ulNU/hCqERL+UDyLjM9BPgvJx+teBRKK42JykaZo2GUkRokmIQ88Ee/GxNzPDs+YKFx8+gOk0ETeJKW9gSkVkaBRSWfC7YDnQrkO2gNgeenUBlCC2i0zux5Q3+N9/4Ai/+KUN6LSg28ZUdjDrNSTvgqXQSz6mp5B0AG0LXU4jmQ5oRePSUWy3w/mzh7nje74Mvo0M1uldncOdWgM7xHQcnv/8mzl4aJXs8Ar14jRessnOxiTGCK7bw0tFnop2M00y3aTXSTIwsoozvknl0nEuvDTFVk3xPW98kSsXDvL5y/DaMYeOL/yLytME/ZUkjYkmTMbwTl5DFoeDboIdPyQqKDQ8J9vs1wN0CTGARVTSd12VORQO8XVrlRQuD4SzLEkThWCZSAwcVGleMnUyxuGyKqHFMKrTPCkLdEyXITVA1/T2yhK7poeFomFa0SRLBBubrulFAkOsPW+HMYYb44l3nx+7qBuEya4oiVrc1d5q1y43+kQsLAZUjrKuICimrbG9npAmURfMlAyxbLZJS5JhsmxTJYFLydQZlCwuNuumxP8odxEaQ1H7bEmbpLFJYTFjJ7gWtPGweOunHox9FzExMf+fiMsKY2JiYr4N7F617phOPxw2Ohz2jL9XtKcx3K2OkJIkGUnh9qck6X5Er4vTN653uNPs56P/eZXXyRGM34UwwNSKkC2A4yCD45iNJbAcTHkTRCG2F4mUrdLe1XQsB1oNfvHTF6FZhU4T/B7h2S6SddAbPnq1S/PsMfRGAXEMeitPc2U/puPw9c+8leLmCJbdZXiwRVgqsPDsPZimx9KVQ5i2i97JUb1yjLnZIrnpq1Q29rG9MYLRilbLo1RKURhbwbKiw7ZtB/Q6SV68OMLnv3g7xXO3EfQclkoKzwa/m6RSc3jzvOKFbU3a03RNLyo9ND7G6D0zewabLWljKeEiNeomZMN0uNOM0iFkGI8RPDSGYl9sdCXkoB5l0KS5JFVWVY2adMnhoBDOmgoJY2Eh1KSNxnBV7XCQCWyJxEXbdCKjeV+M+IR44iHyzelYuwlfodF7k7FdQkKM0TesYEUJXrvv4eXJyDc95/rPMVccPHFpmBaeeP2vGdIkWNNFXCIxvGS2sLFomBbLpohvomJNu+9Lypskh5nivK6xqbuMKAfHRNIni0MtDCngxgIkJibmb0QsQmJiYmJeZTRRAZ+giI6u0f9qox3/KPnoAXWC16qjfOQXnuofXDW+ibKcGqaNMbp/1VywxOJ3H+7BwAgf/tOdSDhYNqZahlYdcT1MaQO1/zhUtqHVgl4HE/rQ7SCZBMb3wbYhiFa6TLUM7RZmZZXw+Q0k06P5pYPQtTBNh8x9L+A3BjBdoVcZIj2xiCq0GSy0cZwQHdq4ns/W8n4O3PM4BBZDIyVMz0G8HitLYwzOXWLt/O28cH6CXs+i3cxx6A2PcOLkowB4qSrbGyOUS3l2ijkOzlfJpwzXF0eo1zLsHw05ONNk4fI0xYZgW5oh1+Ke+5/EEou6aez9zkNCRlSBceWhELa7AVMmTYUeNzkZOiakIl1ahKxIixDDsEmTNwkyxuEOKeCZSBAkjU3KONTwWVU1NlSDYTx2pIOHQ9LYGAwpHEITBQFEpYhROeLuOpUhMp+HRuMbf2/6AeyZ1XcFye4UQyF7ImN30nFj6tZftZIVmnDveQcwqobomA4pSXBITUe3gbBpysyoUTpEE5khySEIY1JAIUzKIDmTJCkeOUmxLlUU8DW5wp+qc/jGsKrqrKsGlggvUeW02n4VXjExMTHfjcQiJCYm5ruaux/96VPfjtu9cTXmPnUMgJ7xuVcd5151nI98MeBjf97kvb98Dyc5RkDI6/R+3sYtNE2bEM2WiZqvQxMSnn4aEQtEQaeF2VlHEkloNTC+T/DoGqZRhfwQMjCIXt0Ev4dptSCTRgZHkMI4ptOEkSlotTC9LsH5FKbtIgUhcegKxrdYe/xe9GqK5N2X0MU0ybsvsf7irZi2jdbC+NxFrl48DIAxCn9nBIBuJ8HmleOYrsvx+7+K6XgURrZIJ6OD8589NkFrcT9Pfu6NrC8eoLg+SzLZw7ZDNnc8MvkqRw9GRu9Gw8VoePpiilrTZjhjWNpyOTAW8PRjr408NdjYYjOjRjmq5viFzM1saz/qUVGKvLIZweNFv0GRLjnjUqW357+oSocuIXlcrukWoyaFLyFdCXGMYsEqcVQPcUs4Qg+NZywyxkUQErjsSAtLLCqmQc/45FWGnvGxxY6KJ2FPlOh+sSCA9Q2rVDfG7t7oA9kVF694Xn1DDPA3ChCAtTAqUGzoJqtmh5CQDB5jUuBSuEwxLOOKTZoEVdPgariKJy6DJsUi22RJkMQjgcOm1DjBDEeYJMDQkh4dAp6QDdZUhZ956CfjKUhMTMzfiFiExMTEfFdx8N//7qnXnnvfqZlf+eipe578qVOSDGj/1GdP/ciDP/eqiJHdq9iB8fc8Ah/+n59EUDygbuajH1/mo5/vYHbWQGs+9vEV/uMjIfeZw/zG6Sy/8ZjhbeYEgvBGcxMt08ETl1/6+e/B7B5KU1mkMAaDYzAyhaTSAJjlq5jSFqZZx2wo6LWh14NGE5QVrW+Vt6FeRgYKoDX2vTbdxRkkl0QN+2Bpxg+ep3HlCGgoXzzBxmffwPSDX6b4/J0srWU4/fSdrG57iBgy+SrLCwcx9RTJdJNmI8ny+dvobE5x8extPP7kYebntnHdkIQlfOWxw2QzAc2mx85Olu1iGhHD4QMlSttDfPm5EYJAOLPkkEyGHJ6IfuYXtzWvublIrWnxE+uPRSlPYtE1XX7UOcSvHB2n3DZU6TFj0pzWFS7rOvV+w3pXQoL+pKG7V+tnyOBSx+eQleZ7J5JM6SyDOkVLAjxj0yUkhc05VeSstUZNunjYOFhsmQouDqbv2WibLhaKjCSp62Z/EqaiMkJuFA5934aoPXGyOyG7EaufuHWj6LhxIqIxrzCt3yhsdmN6y7qCJx5V2lwNV/qTlsiDtGHK5CRNTmWpmgaPmRdJikcHnx4+oybLrBkkZ6JErcAY7g2n0GIYNxn+l0++PxYgMTExf2NiERITE/Ndw82/88FTygpBg9HC2qP3IwOGg/NVDIaZX/no31qI/BRv4LA1y93WMe5Vx3lAneAf/uodPKBu5ve+5mEqpb11KpRChiYB+O3nB6L1qaDLv38uyVu4md94JsE79M28TR/jX59JIo4HRoPfRfLD0R0ajdnewjQS0Z+7Xcxyhe6VeUy1jP9EDr3ahuoOhAF6ZZ3w6wsQBkg2jylV6bRy6Csdgq9PRCbzRI/cA6fRJZf88DIjB89T+8rtdNpJLAW1pkUhG7C9lSMMHJQyLF68hezwCsYIuYEazz17hGbLZq1uePrMOMNjO9xzUx2A8fESWyWXM4sOS9sOk/NXOfPSEMvraQ6MhSS8kJG08Ny1yLvw9e2Qu2YNC9eG2G4Ic2qMpmlxnBksLH6p+yhnriZJOsI9yRyXVZVREyVbeShSWJxQeeqqR0161KXDGWsNg6FBj+uqwvyAouMLY+KRxKYjPtM6x6pqcEVqaOCWcJK8SbAkJTapkhB3L+42KYm9IsUdXQGihDO//7kohjkSJLuTEEFFpYx884rVN5rOd9kVHbsC5cbP7f25H+m8uwo4roZY3ytUFFycvbCEpHjofunjqBqih7836fF3Y3+NkDcJLkmVr1mrvJ1p3vNH74kFSExMzN+KOKI3Jibmuwa/55FI1TFdRSrbJD+1wOaf3cfkiec4/Pw8Zy8MUXnP504dn+0xOlJl4doQE2NNAC5cyzL44W8dQbrLP7q7yTtrcxx57RNYt1SRlIdeb2G95ghmG2RgEOoVyA8BYEyIKCsym5ug0n1BAAAgAElEQVQQ3CSmWeE3z4xhmhV+9UwOdP+Q6iahegWGJzCbi7s/FHo5QI3WkewIptNGhjwSd14hPOvgvH0AggKmXodOG7VvDn31OqZYwlQ3Mb6Qm72CLqdRI1W6izN4+6+DqyifuR2/62E5AUYLm9tpbrttgWppkPOXB7n1pm16XRethcHhMp3aCGEoFLeGWKsKIxlFwRMyCUOtnGdy/iquO8PWZoG5qQaWytDuwYWzx7AtuP226zx/ep6UB40ujKSES+sWsylhpQi5pKEXGn5lfh//8rrLr74mxfueG8NGMV3QXNoWJjJCvu1RpkvOeAQYUtg8ajY4ogdpEImCY3qYDiFXVZkJneWFks+QbVM3AcMkKEubdVXnzTJJWQesYBOKwSNar3L68cmJfvlkQIgjNi5OZAjve1MEiUzzyqZnfCwUIRqRl30hu+lX8ErxsZeK1V/BulFs3GhU3yVa9wpfIWosUbRNt78SFv39vMrgm5BEf2WsoZskVIIx8lwxa1hiYTAg8JJZ4YiaZkbnWLCKvCGc4+ZPfV8sQGJiYv7WxJOQmJiY7xoKb3iWRmUQbMPQu89j31Gl2/G4/MTr+c2bZrnl6A6jWYMSg+872JYhCCx2yklsC373h/7gr52UtFoJ/uKizcc+dpIL/+ltbP3hbdC28D+9gdlYiQTFwAimuBGlUzWrmHoJ2rUozapZgRsmHqbbBh1G7ztN5OCtiO0RPrUESmF2iqgZG+uWMRieAD9Ar/jIaDpar2o0MdtlwjOCKdbRl65hthX+UwUk77D8uTfTK45i39ZFzUeliqaZIHwxh7JChmcv0ailKRXzDA506HUTuF5UVHdpYZgvPzfE6StpbMfnxXMHsG3NpaUkrgW9QLhpvsXhAzssrWbZ2ZjG922WNj0SyS7bdaHUgpWiQxDCn35lnuUKZDM+SiCTMDiW0AsjAdLsCqGBZ67b/NZdOf7xc1X+zcx+PnxfmkZHccVv8YVylVAMLQkYMymq9LgiNWZ0jnVpsqrq5IzLujRpSCRI5iXNaSniKGFRGgwrh7fZ42SMRzH0SYiiS8CyqrIlDW4LJ0ngYKOomgZ5yeBg4+HupVMl+n0vNpFh3cUhJQkMhsAEBCZ4hfAQXp6O3Ogf2U3P2vWC3Cg8RNQr3m70luzedmg0W3oHWyL/TGAC1sNtLImmJW3TxROXnKRYZQcXh6pp0CGalBxnhsus0ZaAA3qYa1IjJiYm5tUgFiExMTHfFdz50C+eQsPQ0RdY/PO3YIpNghfSNBoeyjJcWU5RraTJpjSOo9nZSZNOBWwWk/zFYoClDKMm9dfez5mFFPNpm3uPNUgkfZQKCVeGUNNVZHIOMnnM4pU9ozhuEskORtG6zUpkJi9tRGJFWdBpvHzw3P2+0EcyIfr8AmgDjo0pl9HnX8A0OsiAAcuChIVebWNq0FmZpfvkPvRKCrSwceU4wbkk+cES3sFrdL40BZbCGIuwVGD93O1kR5eobs6RTHYZnSxiWdHj6HVdlECxIdQDzUjG0GmlWS5aPHMpyYGpDrkEHN1XI51ps7o2wJmtkMWlAtVqEoBLC0N0AkM+AZfrPisNzcwAjKSFc4sJXnO0xoWiZjAFhyd8/FA4U+vgKsi7iu2tHL9zXwLPDVldHqbdgyTWnvfDRmgSsKkaKOCqquBgMamz1KTHtMkgBkZNhqoJyJsET3Ur3KkGua6j/pL77WFeUEWelE22VJNxnWHUZHjSWmRaF3D7AgMgRONI5MPwTYAgdPq9IA4WPaJemJCQpHh73oyQ8BUCZDcha/f97orXjalZN7L73Ng1v1tEHSS7nhBLFA52v6H9ZZGSJtGPCLbIqjQ2NnnSTMggNhYZkv1gacMxpllSFSrS5if/6EfjKUhMTMyrQryOFRMT8x3JbX/wf57a2Zjm/KUhHnjgNHgKEwoq32LmtmeoP3orAEGoGB0vcVu2xcpqgWdW4M1HoN6yMQY2a8LhZILniz1+aJ9L+E//7NSfL/YoS+9b7sXvn+gxMFRh4PB5TNdBTVTxz82h9m2A0QSnXZzBBuSHkcRwJD4gmn5YNjR3MJXNSIgk0tFV7noRMnkAxHJQtx3BXF1Ab/mo/Slk5gDidzFry5EoWa1hfMF/cRZ7eoPk8UtILmTpU29h5vu+xLWlAuMHExQ3R8gesWnWhkmoJTaWZ5g9ehbbDunVh2jWs5RKKQYG2uQHy1R2CtHPOFMnCCwev5ik4wvPnB2lF4ISYWE1QSeAy4s55idbPL0sJJXw2FrAbYMuxZZh0hZWez3mh2xq1YCdsMdr8w6LGy62gpeu5kjbhpSnKdUczpejw/i2H/L6caHSsDla2GJ1eZjl7cg7UqZHiMbDRqJtIlLGxRfNTXqQVWkySoJlqbKCsKpqjOksGbGYswd4Lqgyk1VMhFmeq7fookmJy6BJcN7apCodqtImics1VaRHNM2om9ae96NnfFxxSEuCdr8l3ZWo76VLj5Qk954nUXCB3hMkxoR7ExCD3luv+lb+EHjZD7L7vbsf74qj3cdjTEjQ71PRGK6Fa9hic0zmWDFFUuLRpEvBpMhKii1TYUTyLFBmnlF6+Lz/oX8cC5CYmJhXjViExMTEfMcwdOqhUwff9QX0VppG6zCdtocxUNmeIlsokmjuoLcGsA5so1f2s7U+im1pvESLpcURnluBQcei3nLYrgtJBwINndDQQeM6wjNXXXwMoybBtfd87tRGz+eH76nxkSczbNHhnaM5pqa3eOb5Gd6QrpB601WMVrTrQ6i/aBO2MziTG5DKYLptJJFGvCRm7SpkC5jKNjIyFSVZiYJGCQrjmMoWlDcgmQUnWvUh4WKKAvuBThOzuY6MTWCqZfR6GrW/hbNvFQDrrlnCZxeZPHiOcGGEu157ll59iJ1yksKLJ0ik6nQemcV1fUqrBxievsxzj9/DwUPrtNsuk/suUdma5uzlHLccqrG5nebMumE6I3QDaPYMSUdIu5BLaVZKCtc21BsuKaWYygmNsqLdg3uPNXnxaoafOFnk4acmOZaM1reCwOepepMOIe8cy3KxGlLbEDZ1izwOt2QTLDQCPrvUZUR5nKgOMT5e4enFIUqhT1sCvP50wukf5lM4XJZtDpHHl5DLUmFCZyM/h3HwsKgaHzdUlKXNdjNP0haOJlL42vCJcIPr7ABwTA/xrLXBoPEoSRMLRQcXVxy6WPhEgsNgKJs6oQn31qlMv6hS+q0glljo/krWbqHh7iQkIQk6ptOfiERfuzG6969iN1nrZfESoZC9LhKF7D3GwATkJcMGZYYlRwcfhbAsOzRMmxEZoEvAqORJGJszev1VfKXGxMTExOtYMTEx3yG8+8GfPQVgmjaiDNXSIGubaTzHIKJx02X0RgHdTtE9c5DM3BWuLKdZ3Ejw2JMHqTVtXCV0QoMOhZGsIZ/SVPwQW4GD0PMVLwQ12gTcP+axW4bdbCQJMdgoHtnqIMpw603bJI4uACCFLOnRZezDW7g3XyPcHESfvRhNPjrNqLtjaAJCH7O8DcV1aNSiKcjQFKa8gQxPoi8tYtYWMatX0Veu0PnCONa9KcxyBbRGBofATaIXyshgCzTUXziBmhFMvQrKYN+8gTVXxJ1a4/zZw+Sz0e7/1YsHOPPcLUzcfJpuJ4EaqRKEwgvnp6g1HHrtHOlchZsPNDBGeHZNM+RZ3HK4yrH5Fm98zQ5JB2zLUG8rQgPHjmxyftXCVlBqwUzSJuGAiCHlGZ4+PUkrMOQSsNIKeXpRMSQuNztZyk1hKmGzL2sRYijSpdg2LFBn0kpwx4QgyjA0eZWTN3VoEdIjZF3qUU8INj6aVn8NqkXIUV3AQpHDIYnNnXqULWlyxtoiZytSxqGnI/N7LzRc7XWilnsicXPa2mJYpylLmzk9SIYEDdOibbp7DffRqpXCYHDE3uvxkP66VEBAD39vzQqi5CxPvL01Kk/cvXWqG6ca8M2pWTdG9u6uZP1VaVs3RgAPqmiiVtQlUnjM6gI10+o/ToWLQ820cLAYMClK0uSAmvpbvkJjYmJiXon1Yz/2Yyf/vh9ETEzMdw73Dv/+qXvf9nsnf+3HL51cDu555Nt9f+9+8GdPvetRc/JTjQXeOzqP20lBx6Vw4gWS3QJjo3VSmTreLVcJVseolybY2Rxj4dxh1qsKz4LbjhX5/Ysh33tAMZHXjE/UOHM1yUjOsFqDQU+x5QdU6w5bpsthlWWxEeKKwteGYtmlp6MD4jvmHA4++DDNq7OkBrdQR3KYjTLWPh+94mIdTWJ2ukiyh9gdCHpRh8fyAlTKiBJIJaBahUYVLAERsCzEDqHZhFQKc72FVahCs4HpCmo0E7Wol4vQakPTBtHYbY013cE02ogxhCsZrHkDnYDO6ixL6ymGC106bY9ctktze4LCcJGgWqBWydPpKe6480U6zRyB73Lu4jDXNz3GUoqdjqZRTbJTdVDaodaymB/vMD1V49Jqks2tLL0QBpORqAtCYWq4h1JCs23jOaCM0OxFEyclwu3jijtuKtNsJLlQDZjNKY4VbI4VbC6WQzalTVa7vGZ/h2yuTredY3OjwHIFDloZzkspSp9C4WKxYTVwjEVbQlalzl1mhJyyuU6DMUkSALM6x6BysEKbJdMmbRy2Qp+nrTUmdZ5VyqTFY0rnWVU1csbjktqi3V//0mg8XBLi0jSRuV/trVX1/5PoY200IkJCPHyCyFSOINDv/RBU/3MhGksU9L9udt/6twG84r3s3VJ0z9FE5OX39G8pmrT08MTlJqa4qDbR6Gg1TARbbBLiksRhyuTZkSb/8pP/JF7FiomJeVWJRUhMTAwAH//BPzy19YmFk4ufuHBy/t1H/0bi4Uce/LlT7x9c5INP2Lw/V2Lu5ssnZ8xXTk6vfOzk7Mjlk8vha181UXL3l99/av2D+uQnq1f5VGOBMWuEH5kqIAJXLs0zYEOmsIHt+CTnFjE1Dytfp7I8x8LiAMWaxeSAxrbgqStJ3jDuYluG0bEaRiuyjs1XrkPGUtR8TU7ZbIc+ZemCETwsrusWo5bHVuBz74RNp21haYvV52/iqcspBjqT5NorqEyb8PwA9r02dDpIpoVYBgIfU61htrYx6wEynIRUAsnkMTtl1PgYpl7FrKwhwyPIwCgkPCTwUXOjSM6A34XAIF50e5LJInNzqP0jiONjDVUIzqewZgziCKZkEV4eQBI93G6CFxeGGCsEaK2wbU0626RaKrC2MkKpbpPPhGysjzI42KDdTGEri2zC4CjYbkLeE+6+dQPBIunAs9c8pgdDzq1b5ByFryHtCNoIw/mA86s2aVuxVlEc219nacvDtSDnCcYIng2ENmfXFXnbYmYoJJUMWC06vNhtYaMoSofSZhqrWaDVyHB9y6ERGFZ1hyOmgGNsmuJjBDakyj4zSEnadCRAtE3S2FxRFYZMiop0mZU0j5pNStIlYWw26XBXOkOpZ2iJzyg5QNhSdWb0AB0JKFInINyL3E1KAp8Ar+//cMTCAJ64GIn8KSKqX/oXovoTjmhqIXteEU88tAlxJPK5BOblyN3deYjVFymG3WQt2Zu02NhRvC6g0XvyJUrcir4np9KEaMbVEEVpsK3LpFUSEWibLq44uNiMmixX1Db/2yf/aSxAYmJiXnViT0hMzHcp/k9/9tSnN+rcn8nxcLOCheCjcVDct+8zp7D6u+y9Nl9bevCbDiH3zv7xKbOziiTSEATI4ARvVXew/989D8D+3xIW9FXourQ2biI1c413/3a0MvWJP/7g3+pQc9dn/tmppa/fxz9c/CIA8/Y0P2JuYmz6RXY2pjh89DpOokGrNkoqt8XaM/eQyjYxWnC9HrOTTf7ihSTzYyGOo7lQjDwW4+MVms0kWguWZXBEYUl0iHvUbHKHjBBgsFAoiUrctIn8B7W2Iu/C3HiXIBDeMOZTGC5jejalr95NtZRn320PI8MFaHaRbAK92cHUHdSkQUYEvdBChg0EITJcwJRLL//QpU3M4Bj0OqA1pl5FHAeSDqbeRV9tIiMSfS6RjiKAfR+9qQmKw9jWBgDWiRD9mKa3OEMis8P86ByLKznyWZ/raykq7Rz33rqD43i4NiQTAdW6y+b6MMubCfZPN6nUUiyX4faZEJGQy1dGub4jKBHG0lCuJDk8YDMx3COZ8HnyYoqTt1ZZWcsBkE4F5JsWaxsZRrKGeluYGelh24bnrro8UgzIAmNJl+tbNq+9pcozV126EvI6b4ClTo9QDCKgLMO1pk+RLqPikVYWtdDHQsgYh0GT5kW1wfFwnDVVZ8mqMhEmyZsEF1SJrPHYMl2UCCf0EC1CUsZiqx1yQvKs6S6XVYlBk6QrAUXV4opZIylRi3jDtEmKR8/4pCWBT4glUUpVmy6uJDAmEgG7CVVKrBu6QaK1q67p0jU9/L4R3cPCFvsVa1u73LietfvxN65p3ZiUtXtf0pc+22G534zeIzSanKTp9MsLLSy6pkebLnlJsalLxMTExHw7kIcffviX/r4fRExMzN8t8//2D04tXBui3FTctK/Bh88IPTT3ZrLkU4a3fKGf0kQkQsTx9grzTOhHH/tdTKNE8JmLoAXJdVCHc/zET57gL8PTe1dj/0ss/9Ewj6v/9b9KkNz1uZ8/pYYbmLbNE5/4Pv7F1nkGydChx/vcIwQaBpKQcAzllnB4qsuh4y/id9Nsr48zOLJDGNro0MJLtPjqEwcZSGsWisJr9nfJZLpYdsD1xQJTEw2uLOa4WA5p6pDrUmdLNTkRjlDHJ4FFHgcfw2VVYb/OM+N43D7rc+uP/xnh1QEk2QNl0Nt56sVp0vltus0Bsg+cASVIwiY4lwSlkYQPWoEWrBMg+Tym1cRUo6JEUQKDA8jAEDRrmE6b8EwLa15HsbrVHjiCmh3HdLvIxGz0b7W+hFkrEXx9gmZlgtxdpzFtm8b5m0nPLrB57na2NvMsbztstSNPQUtr7pyEVlfR6Aivv3OFjbURjIFuz6LWtAi1MDrYQ4dCuW5zphjyjmM+T19OcHhUs1q2mB/1ub7loARWWyEDtmLfsCHUgmtr2j3Fk1s93jRr88wKtHTI/pTL7EhANuXTCyzWiy7FpmE8B9W2cKHZZc51sZRg+k+vjZ5PVlmshJ1IDOJTlS4OFp6xyGBzWZUZMEk2VYMT4QjXVY1xnWZHOsyZDBZCmR4HrDSLYZtDTorpnPCVnSYNfHoSckFtMmeG2JQaPaKSwRZdABxs6qZFWpIMkmGLCh3TIykeWZKsm+ggH5qo0HBXkCTFi1KzTA+DISTEGIMtNq44ewKka7p7r4Nd0WGMjszt3yBC9goO++8dcQlM8Ip4XlccRlSBlXADjWHWmmA13GJIDQBQMTWOq328qBf5vT/6P+IpSExMzLeFeB0rJua7jKFTD5068OECyWdXufOTeYZ/0GP8KwHJTpr/7muG/T9K5EEQAaORvhjZO/WFAZgQs7UIKOb/SY1ffxo++FWXn7YLvHVWuHRtnqvmr0/T+RnLYu34m/6LK1p3fe7nT83y0ElTTIDSbDz1Ohr1BMeDcd53QnGHmeClSkBKKYwRii0INbzurivUyyMksxXajQEyuQqCkL/5LK2NKfbNbWOCBFNDAdlci3otSeA7XFh1SdkWj65HJXVV47OmGuRNkh3V5hDR1fwiXZoEdCTgsGT5oTcuM3PiLNZxh/JjR7ADw8UnXks20yWRqaCcLu5AGVNxUSNdzJoghQ7WPrj+Jw9QXZollyuhslXCy12ENsHzY6h8HdMwqIkCJNKQixrXxTQIL2QRr4XpCGIbyCWh3kB0AOk8YltIysE0d2gsz5GaXWHl4TfiJTt4w9tkD10lL8KfvpDjaN6hG8C1oE068Gj7wn13rXHp0gSHjiyzUxwg1NG0Q4Cdmo3Rio2aMJJQuLaQtBW1lsIPIQgs/BA8G/xQaIWaUksIA8W5UkizIwzYNmIUXT8qIcy7iv1TTV64lkYZxfWKYSSlyCYN7Z4wkbDRRiikoNY1jGeFdldYCjtcUiXGTJouIWXVoSodatKjKl0GTYrrqkSZJoMmg41iW7UIRLMjHQLgkMpQ8CxSxkaJsN7SbJg2BTzq0tvzVexIA7sfcxsQ4mCTJUGTLh26DEgGiPwtCiuaNqAJTIAnLoL0e0TAJ8AniCYR9LBQIIJG0zU9kGgyIaIICF4h60Xk5fUsLG70fEQTD4Xm5Q4Rq78eZtAUVJ4Nvd1PEbNRYiGAp1xEhLpuUjRVPOXx/T/8xm+7rysmJua7k3gdKybmu4yb/p/okDT2wZkonQm4+eMJbv7GyYXRUUTsblGeDjFhAJ0G+D1ot+h8stdvcg4xGA49VETY4fRbj/ILf3knX9DPfsvHccie44c/meI3Fj9yanm5gONo2h2Lmak6jYbHly7avP+ffwYdZuk8cQB3ag29NUBhZJ0wsAnCHAuLORZKhqQoBlPCcl1z36Eez1316HWT1GsZmo0UaxsZXLdHbmgbvZ0nN7SM38kyObtMtTTM9lYOYwTH0VT9kJ2aTUYgoYQh47KMQ0d8UsbhaN7h+UqIxnDdqpA3CTrGkB1d4gt//Ea+9+Af4ThdSutzBIFw5cJ+hoYaKEtj2wFDx85A19BbmMWZ3MA4LXo9hwPHTyPJLvgQLE5id7eRdJvSI3fTbqaYnnoaNTGP8bvQFxfWTXX0soeaC9CXEyjZRobS0T/f6lVkcITw0ibg4Hodio+9jlIpxdTNz6N3ciw9cS++b5MRi/nJNuFKkkLPpRMYBlPCF5+YZH5Yc+6FeWamamxsZVivKJx+rqIfwlY3YNa22SzbDGY010pQD0NGQptb5rs8d9VlbgCaXYtqx7DWDkgri5mcYqtpcCw4Omb4+rpEKVnrGZ5vNrmDNAOukE0atmsKA6w0QkYSFmlP83i5RGbH5VY3y2k2mNI57htz+dymj8Ywrwd4ydomZVzWVR2fkCwJNlWDaZ2jKCEdCXCNTYAmZSk2OwFTKZuVVkDdBKRxaPW7QHwJGdYpiuLsTR9cojheQTEkWXZMnUWzSVI8mqbDmBTYNk1cHELRBISE/deK1Q/ghUiMWHuFhYYQUH1/x25M742rVd/IjWlY0ddf2S3iE2CZ6B5DYEdX9iYkaZWiomuMW0PUTIuO7pBVGeo6aoKPiYmJ+XYRR/TGxMRE61X6hljPG7sI+l8zRu8JELOzhl5c49BDRXQ/gnQXg+G2L77Er73jr94lv80+wn3WCf7NxGE+9Lok3Y5LMhniuZpkIiQMo+SktBWtJ7XW5njsq7ciyR6q0CB572USqTYHDxSZHm/R0JpN3ePwXINqGGDbmkJCePLZORoNl2Syy/Bgl2Yzie22ILBoVcfpdTIYo+h2XIZH6gwP1zm/5DGRsrhaCUkpRcGLfg8touZrhVBsGVJiYaOwjeKQHuACFZ555F522obwyiAXXprn7IURHEezsOFi2yFbm3ksO6B65RjNJ45jZ6usP3c3esfj0Nsib4ukOwQXRrHyVWQgSllqVHOsrAwQnhHM+nXMS2fB95GpSSSfxrQ8wotJTNuj/cRhCEL8L/rgOPD/svcmMZal55ne8/1nPneMOSIzMiszq7KyBhaLIiVRlClZtlpqNGxYHgDJ7oUB7w3Dy140QNgL24B3NtwL2+3WxvCm5bFbFtSDBpKiyS6yKos15ZwZmTFH3PneM/+fF+dEZJFiA16YEkDeF7jIiHNvRNy85xzgf//vHRYzzG6Ham+LJw93idtj4rjg/NGbHO/dwRglyx3eWnVJEpc7N2asui738oTIVyIPng8MjlEGg5g/eWZ5OM9JSxhlytORcqwZAgxT5daNc26uwk7oIgL/+J4gAnkpPJtVPM0zRlryStdwbStlXlqKCg5Hhmtth/fPcwbz2pt0bb3kPLO4jjLOlEmmdFzDWlu5fyp81VnjusSsx8KmtrmlXc5mQojDLdtni5BXq1UCHK7aLn1tsaEdhrLgkTPkll25bFZ/R3rcLxa8u+kwzpQKZUzBSDJmUrClMbF6hOoy0jkVlnnT4xHhk5L/CDm4puu0JORcJ7i4uOLQlggXpz7PjVyqJRFtifFwaUsMQCRhPRGBehrS4MLT8fnvL2/XHzv++ecuYnkvJFr1PWqJJQRqImUQTqoBqa0jibdkBVdcTu3wJ97DSyyxxBL/f2Apx1piiZ8jfP21P/pGPdnQl5Kri69V68fF8bJZANkKHLeeeMwGda/F6RF3/5u/yf8yePIv/Vt/78GPLmDecG/yitnhv/1yn3//K8cYjTg/7/DsIObazoyqNKytT3Hdiod7bVZCwRle4ex0ld2dCecPXmOyf43u2nNO7r3O4VEPx4DJfa5FHvOFh2NdJlOPq+sFqMFzlbNBi24n4/6zDocvdtjeXLD/dJfDoxU67YK7n67TaUqsA+PgOcKrGxW9wPB/DodE6mK0lrK8RZ9fesUSqscohRzL216HfZsSJjHv7JY8/uwWjgN/cVRwMPBou4ZXrswRDMNBB8+zHB9s4lQxZyd9ZLpK+82nLJ7cxL9+RHGwg7t1hp720MLDWJ900WL0/BXi/RkyKsnuruG9laPPz9Cpx9nHv0C56PDB+7fxn6+gWUz4+gnS7pL9byG2Cjg73qC/MqcsAtI04OS0TRwVrKzM+OGTmHvnyvU+PB84ZFZZ91weTgtEhX5oOBy6aFXv3T8qErJKyazSEY9fvFGwP3ToBy4f7Bve3Kk4GAtjW2Kt8Dwp6DkORzZjQckVN+B84iIqgPBkXnCQlRQoMQ6DqmQ+9Ygcw/EM3r6iJJnhz7MhZ3PhtbaHar3YfrIoOJAZc0qCwudWFPBJNeEVJ2bLBGzT4poT0apCWuoxMjnvVBtMyOs4Whw2NSIUh43QcLywvLnq4hc+E1uxSsiBzCnE8r7sEYhPRcW2rBDhU1Ax0hkqemkGD8THvZRRVYSNDMuRWiLliUtJiYjgictcE0IToNQEpaS8TJ3+I2IAACAASURBVLa6iOC9kGnppaGdy4Ssi3jfywjf5jnTzFdeSrNepmQlmmIw3DG7jJjXCV5iMGI4s0NWTZ810+U3f/dXl3KsJZZY4qeCJQlZYomfE1wSEKl15xinJh2Xng8LptlRvVj4GNNMSATNF7D/EB2dI+0Of+fvRzzRo//Pf/9/fuNdfvfNgtk05nB/i6oyeJ7lytac737c53ziEYjL1s4ps3GPvSEE+FRWiMKKPPcYjCKeffg6/+gh3Oo4vDjziAPl+UR5NCuZlBXTQnkygutdwXEg8Cum84DVbkmeO1DGfPw0pioNq92Cp0cRhwOPLPF5MXRYZML2asnhwENyjxMydiWmqz5zKhZTn7PUcqwpAS5rxufLnZjVGP75QUGVuZxMhUAMA1sQ4LCYtBlP64XoP/8s4pfeGFAUHvtHHd74rT9DMwevP4LC4G4MMf0C00uwJ13S2Qo/vLfKbOFB1mV1/QT/q8+RdgRSImFCpDOGB9e4+zhivS2srJ/hvz5CjCDlEcXxJuPhCgcHqzw7jNjZTFBruPuoxc1rEz561kKAzZZBS4crkUtSwO1VQ5IZQg8ejCturzjExjDPYdLIlDY9j7ZnOJnDi4HDg2pOnEcMioorvsfH1ZQKaOOyanwcNXyczxjmlkWl9F2XQVnxahTwtEwomsFagGFuLb/1dobrKH9ymPPloEtphY3I8P5sgVrBE0NfQ9YIeb3rocAH5QjfepxqjquGt9YcBgnMKRlL1pAdj3kjsSsUShTJPT6yE/plwP1iwWtuTM+4PGKKh4OIw4wEH4+exlhRzpleRvG64tCSkIFOCcUnxmdBxoJ6wpCR4+ORad40ptdlhBVVU05oWWhKIB6Flg2x0EvycUFMatO5c+n5uJjBXMT1XpAQGsJxIdf6y94R4USHdE2bggKFpl1deEuu83f/4X+8NKUvscQSPzUsScgSS/wc4Ouv/uE3Lpcqtqq3Ti8mIhdkRO1L8iGClhniuHU6lhhIZuA4sJghxvDFx7scHG+yY9a5aXa4brZ41ezwtCEmv+J8gS1njS2zxrZZ53dfV4aDHufDkLwwzBOXzfU53/qwj1VYjeHDQ/jwcZ+nU0thle2WsL055+SsxYd7HkVueDSyuBgWaT21GCfCD4oxp5LQxuOLKz57SUmVueyPhZubBXf3PK6uVswWDjvbE9Y6lmtXxjx7vkqSC3d2M46GLp0Appli1OHusMCROir1EzPkqYxr/4e1jLTkhZmyqRF947HRhvfPSr68GvD+fHGpbHNFKK2yyGGRQ1EaRIV339rn//rmNd68nrD66mNO3/sqcfec4WdfpPXWHoQCvjD84G32n2+R5Q7zHEQdDp9fozcLcfM9TE/RiWKP+vzw7uuUVd3FYXCJ3OdIt2Dx7VcB2NvbwvMU34Uid+n3E84GIe3AsHfmsRk5XFnLOBl7uIZaApVArjDLYMV3uLWTkWQur60Iz6f1zvuK4+KZ2s9wWpTc9EOGheVq7PKDdEaIiwInWkuxtk3APRmxqhERDpmFlqmN0Ve9gO/ac3rqczMKsFbwrY+tDNOZy/1ywYrx0Ep4s+czz2DFc7geu7hqWImFo7ll08Y8lAmlKFdMhC+GsoJIXHo24tgsqFDeZZUNQjwMN/wA3xH+me4xq4SRyaAyHNqMsWT46tDTkBOZ0iFiIglDZvRoYUVxMWxqlzYBc8nZ0C7HMkJEyCnxcAnEJ6dEm9vMaWSPFybzSAISzSip79HLAsNLKZVeSqrcz5EQ+NF0LHg5EbmYglwc/fx8RAFPPBaaYMRQaYUjLhbLf/0Hf2dJQJZYYomfKpbG9CWW+HnAXzKYFzWx8IKalKj90YlIVTc5a5HV8qsQCNswPELiFtX3DvnP7yb8T3/326R3bxNc3ac8WcfpjfkPfv9tAP7H306Zjvtkqc90FvK9uz6Fpe7XaBRfnzzss9EG11GeDuGZTZrdXOji8dmp4eOTNqERUqts+UK/dMgrJasUk4MFxiZjzUY8NGNaI5cHZsRxEfC1oM/jg4jjIufhfsA7r03ZuPqIkxev4UcpUVCx0YWDs4DDRcVa4FAp/NPTBVdMyF0dMjYpXQ2wRnnPecEvVbtsSsDERtx1TlitdvmL05INx2eWwQsz5QElr9geJ2bOG3aF0yrnb78uPDuKeONayvffu0PLFc6HIdP//W/heZa13ZDVdz6gur+GrMwwWxnHh+tcuXrGO7/zL/i//4ff4WBkGOaWeXqLybdu8W/9G3/B2YvbLBYhW+sJrhMiooTxnKPv/QpXv3QXtYbByTaO05igSyErXM4nPa6uF+wfx7y6Cl985zn/659e5V99a86ffRJhgRXfcKWtvBhDN4RnRwH3JyV3ei7XvICDIudxkRCmMZELWXONjW3JD5Jz1om47cUMyxJfDPd1ysRWbNOmwLLhuXy3HPIr7gqHeYEnwra2uBPE9QJclW+e5IQYvrrl8eC4YmQLXuQlv1q0ObY5sYb809kIR4VvDnJ+3VtnWsFCchwNuNV1+N4o4bob0jLCgS15w67wXOZca7k8mtUTAN8RSlt7msaS8lhOcOQap2bGlITXdZPPzDGlVrxuN7jvnBLisakdhpIQah1ekFHUEzKNmEqLc53iYOr0KzUoio+HxVJogRGDi0um2eXc4wKuuFRUuLgUWnDhvnJwKJpJlL2cbJi6g+TH+kJ+0rG6Xb3udL/wjNgLgvNjHq8lllhiiZ8Wlsb0JZb4eYMX1ATE9WrCofYvv8ZWtf9jPgJAx2eQTLCfPYSqRK3w9//T70JhaoO3FdLZKpO91/h7v+7w3/2aixhLqzOlLB3ORx43dzJUYbNrWW1bPpxk/OKXXuA6SlYI94r5JQEpsIwpMAgf2CG+I0yriqdjy6M0peUJ/wdPafnCfp4Tq0eAw7FMOG0Wc29Kh++lY/otS89xWRRw72mH2WibIMw4P1lle3vE2kpGkkPk1Es13xFuezGrXm0PjvEopCKjpEPEmSR8R44YmZRYPX4gZ+zLnCstw2fzus/htl3huRnjYDgioUIpK0Nl4cVJRJLXMbOHI0NlhSRxMCtTdBqBW3H+wVfQkcOtN+5xdLAGrZJOVCdJ9X3Dn58vyCs4P3iNsnT44EGH7z+IuXci3PrC+wzP1nj+fIXqh3McL6MoHF599ZBOO2eSCItM+LOzhHsHHle3FjgG0kXEr7xaFwteaRt224b3kilXNlP6gXB3mDPP4U7PZbBQdrtCTsUrboRrYF7AF3sBmy2hIw6bNuaqRDwvMkpVxrZkQcGa6zbdFBHfLM+4obX/p2scQjHsSMhpXrKflvR9hwLLlJLAsxybGTNKMir+2XREgOF+seBIZhybGZmU7OUZC624bnukUvKd0YIAw2vr8LhI2JaAL/YCNjTkB7MFVwKPjnEom0HgG3aLDdvCx+MTs09fIxZk/NAcANCVmIfOGQ4OKxpzLguu2i6v2B5t9QnwCPGI1cPDpSXh5b9QL/ZzClwcAglwm7LCSEK6Uv/dWCJc3MvekKopOASatKyXnR8/bkS/wOfN6hdfOzhN27ppKIdFVTHiXP4eI87lhGaJJZZY4qeJpRxriSV+xvH1V//wG5cGdKhXWo7Ljxy7OF5kYBx0coamc6iqmhZkC5iPIZujoyliSsS3kDrYUZfzJ7dZffd9jj59E9e1GMfS2XmKWMPhi216nYI0cxnNDYELk8TQMS4fP+2w1oK0EPayvDHlWiqUHal3449JeFTNOZOUNQ15JBOcwmMoOXEe8URmHJopGxrz3Ix4YcZsa5dDEhamZKds88u3Uz44EvqeoRsZ1Brm85DZPCRNXXbWMj49ET4r5hSlcGgznlUJa4ScSYJFCXDwcEikIJWSfRmyRZexpDgYxqkwlIw36TMgb95Ti7kpCNWlm8e0AmWcCKsty2ghjDJlPBc8Yxjt3WBrd5/09CqOYwm/dMz045v0VyYEmyf0bciHD/t0A8GtHKal5V/7j/6YKCt59GgXC+xnBfHsGvOFh+8pcRkxOt3k6LjD2XmXvHBwjfDJoOKK53OYF3QIOZsKZRoSRxXvHysdxyGrYMsJOB15fDRP2Zc5r/oR54nyuEgwpcuZLZjaEl8d7uULXmQFtzseWjlcD3z+rDwFFVbE55nMuKEdBrZgKgWedbDACzOjSF3WXJdxVWGBpzKnVOXtns9BUlFgeTKxzKXkl7w+SaUkUuFTlxF+bI65YzfY1IiEiikFA5OyoKDA0sPndA6ROLy96vHt4eKylfygqsmxL4ZRUVGocmhmIMKCjEB8LEqET05FnxYFFdvaJaWiS8CRmXFmFvSImElOT0POTUKJ5bpdYWJSAFxxKKmIJayJBUIkAUZqadRVVplLTaLrXpAL+ZXWnSHiXlrSTUMkLr6r8fJ+vvCEfP54IEEj0aoDAaAuRqy0QqUmI3WxofLv/t5vLw3pSyyxxE8VSxKyxBI/47i++uA36kWMeen5uITWxnORevoxPQfHw95/H/FcJO6ih0+gLLDPniNxgEQB1Wce4lXoNGR2tMuTx5vEZZt2d0aRBYTxgvMXr3BysM08cTka+DwbCJEruEZYFJCVymokWCv0Ysv9aUEbjwplISVbEvKBDojwSKSkpwEzKnaIec8cE+DiqIODYSp5XSgnwpZ28XEYSMKbdpVvViekp11+45bFc4TX3/6UIMz58NNt3j9U1kKH6cLl03nGl+M2f1odcUVbnEjCTWmTae19KMXypl3luZkiCCEeTqOrT6RgTSNekZgjTTk1C85lxrbt0FGfmeTMM8OjRcG9cs7rUcj3h1lNxIoZW05Alhvaps33P7jGxkpGaM757//huwTpCjtrp8S/9pRHf/4mz+YlX9mF740SVp+9je/4PHjRxircXjE8PTNcWat4eOhzcrxCVXocDF3SXLhxdc587rEVG1ZiaIvLJ6OS2HF4OK748KxerGYluM0yNrewb1PumA7P84JDm/I3NmNGKZzbgl9otfgom5Nj6eCx7nl8Os9oiUNRGr7Wrn9uJgWvu2064nIniHm/GtGnXhS7GDwcVlyH93XIrrbo4OLj8EE1Imp8JZ46jKoKB+Ftv809O+WaicjVcGLmfN/s0aHFU2dAIZaFZER4tNVnRMENL8QgDHPLlIK3wpishFtRwKf5ggE5awQMJKuX7SK8WW0wMAktDVFRdm2PvkaN3CqrJxmizEhIpCDC54btc2gmbGmHAzNuSExAm4gpCwSpzedU3GKDShQRw0CmTfqVAbmI1bWIyOWExFI3q18QiZepWy9lVBcTkosOE0Fwm7LCl+Z1RdXiGx+3aXFv/ss44vBv/97fWJKQJZZY4qeKJQlZYomfcdQkhJ9AQJpjxly2ozOfgC0hmaGHB+Aa7NN97P6U6pMN7OMQZ2fK5Ltv4vkziuNtROCVdz4in/WZjvqsbh0wONnhTz7o84XbQ+4/65CWkFcQuMLBzOKIkNtawjPIalPvLINXowBjHVK1PGHGpsa4jYm2EMvU5FyjxaEsSKRkS1sIwmNzzo52SaTExTCRjBCXFzJly7b57ashnmtJMofjwy2e7a1TVsLtdeXhqYAKsbq8yArWNKZE8XG4FYZoaXhmJty0fSYUzCSnkIqehsyloBJLT0NcNQwk56kzIpUSF4c2Aa9I/X59dbhiQmZacTJXCixf3XbZm1pM5XC1B4tFwIMzWAs9dLrGg/2YldDwvfdvcK0I+Ohxn9Qq47nDyJbcHRW4ozUiDwoLwwQSq8znLgdpyUZksFY4ncNqCzpxxcnQ51tnKUczrcmGCE+KlFgc7suENUISKnqOS6nwfjVmjZDbHa822FPRtj6fZAtiHGJxiNTFUcPb7ZD704Idz+OoKFgzTcxuUeDhMLQFVgUPw1PmvON0GdiCO16Lj+2E0Ho4ajiVlD4+osJTZlzVFiNy3nI7fFsOmUrBG06XJ9WcsZaUYrGilGIZS8KWdqlQfOrF9UhS1jXitZbPooSyqtOwtjyP87Lk1Z7LflKygs/3zQkDmXNVexRim+lXbRT3cJlLwU3b45EZ0taa1GzbNirCCjHnMmNkanJyICNmJGRakJDTlohCqtqsriUtCXGl9mK1iZiT4olH0UxBCso6zlfrn9HGrF5Q4IrXGM4tFw6PC9JxIc/SxtTuSk0pSy2psJdRwUYcCi0IJeDC9O7i8A/+4L9YmtKXWGKJnzqWJGSJJX6G8fXX/ugbPyLD+nEYpzGtK5rNIM/QdAZFhp6PIJnDpOLsz36VePUU59qA6Tffpn3rAeJXiFVcL6GadzCO5fxknaODTcrSIXYMp+ctjmdwmlWAkFbKzFp6Xu2PeFguSFVJcrgWeTxJctZcl8JCgMtrXkxfPFbxeSJTXrEd5lSUKAspeGaGRPg4OAxNSkt9KiyJFCwkYywJK8Ts0uJo7JDkwtZKwXDmsruZM1247M0qWq5BRLhXzrjjx8Q4OGr4jj0jQ5lKzkhSOhpwYMbQLNbmkrGpHSosz82IrobkVNyyK6RS1rGrCk/NiNvaJxLDiab4GHwcTmYv+x6iZnLwcJGzd+6STzrcTed89YrwxycLHj9eo+M6OAhbLSGwHgtr+c03Mu4denR84XlSEjuG+3nChuuBFe6O68/0PAGn8ngwtEy05JdWQrq+oeUaPiqmeHrxGXp4GKD24RxJUidcFYau52Ar4eNySguXm35IyxN8Y+i7LodJSYrlzBYcS8pQC54VKREuT82Yd02fwBjOqoIxBceaYgWqSlgnwBWhZ1xmWjEkw1OHE0l4YIb8mrPJ/XLOwKRMJeVa1ee5zGvfjZnS04gt7VAB79g1RA2b2qKvIQXKNhHzAtZDw2poyHLhSttwmlp224aDhSXDcmTmjFgwkoR3qi3uOaf4TYZLW30UZSgpET7bGlOIZZUAwbCtMZ+YIyICAFJy8iZqVxB8cekQk1MhAld0lWec4onLgCk9WuQUTYmiAwIVtiYb8jIZC+rjF9eOxeKIc/n1xTTlIgGrokSAkupyylJfm/X05CLSt6LiH/zBf7kkIEssscRfCZYkZIklfoZxffXhb1xKsX4StHnOVjA5R7MFerQHs1k9pkgrdObw8Lvv0okrvN6A2YtXmB/vEF/ZJz/exu2NmRxdJ5l28YOCLPMBGM08ikooq1oQshYaPkhnTCm5EQbEnvCH1Qte1R7PZE5VGM7J2HR8QOiJy6NqwacyYl0jJtTpQE/MmA2NeWzOWNM2z82Qf12vMaakoMLHJWqkUp64hOrxTitmnCl7aUFHfKyFdmgZzhw8May2lLISKF0mVUXsGJ7ZhJyKRErmUvtVcqn7HBLJmUuGRZlJRoeQvNk172pIIiUxPnfoEuIwk5ICOCQhE8u5Seioz4CclIq2ePQ9w8lCcRCmWvIsyxlLzt55PRl4zW3xNM/IrFJVgtVaxmSyEM8IvVjpew7WCkmpzG3FYZmzoMJTh2lVkeRCZpVtz2eawUdJwmFeAHUYwEjq7u8+AanWi9lNam/OHgs+0wleI4GLcDirCqQyTMs6UvnP5Yhf8VdJKuVcUo7NnEJq0tjVgEQVq3BKBgJXNAaEM0l4xYnpuYZ5Zdn1QgorDMk5MFNa6mMqj4+dU75SbaMYSoUnZkCsPomUbNt2PYESwy1ps2VCrvsB08qykJI2HruBz6dJyjCziMDVtuHxon7uqKiDEB6Zc2aakJITSUgp9cK/rxErtj63AL66vCItzslwMSRSMpOCc1kw0hkdiXAw5FJiRBqztzBlgYdLhI8Rg5V6YuPiUGIRDDklrjjcYpux1P4VB4eOtEBoiEXtKXHEbSYeF/G89TTEYi+nJYLgiYelwhOvieJ18E1dvHghz/r9P/ivlgRkiSWW+CvDMgJjiSV+lnFRTmicf/nztqpf0+pD1WhVHAfph5SfbKK5w8bmBLUGTXw8P+P0ZIUX3/41/PUTJnuv0b/2kEdP6kQmq8InzwM+HZTsjS2nWcWtVXg0KxDgwEz589mYF/OK67ZPgdJWj02v9oO8KHLu6pD7dsaZJLhq2NM5BZb7zim37QoLKbip6zgYAlzGWvIlXa0N5OrQVp8VjYjVY0UDRgmcFxUt43BvWPLlt09xXUs7VIqqNoufJBWPmFKhqEIHr9buq0OoLg7CDdvDRXAxrGubABevcU94OJzKlIEssFATGLXMKXnV9imoGEuKonQ04JYXkUhZG6cdhw9mCadlwZnN6YrLhJyr2uJAFrTVY1TW0wGAZ2XKo2pBqpYPhzlPZnV6lOfArLAUzS55C5exZExtSWgMqlAB65Ewqko8DB6GhdTn5ortEKrLkJynMgXAk9r2vKkhbfXZNSHXTR2HPCLnMzvlITMObFZ/ElI3yf96sMaObdeFhhpxU1rMqCcgmVRsasSGU5u+36DHo2rOfl5gRPjT6phz0mZRLpc9Gbu2x5icr5o1koZ0rmhIT0MeNT6QW7ZL5BgeVnPuZwlnkrKQgq64DIoKQ024MrWczQQHYZhZ8maGcNtu4IsHwL6MLq8pr/GdeDjcsH0SKXh7xcOiJJScypwzWdAiwG2SrCosLUIiAjxcYnwqteTUxG/EnDU6zEhoEZKSYxBWpE2mBYXUbet906VtYjxxSDSlLXHt7RDBF+8yqlcbEnOBUstLuVeuBQ51QaJpph6mEXJdEJElllhiib9KLCchSyzxM4q6oBB+JAHroqAQfZmOddETYqs6HSutE7DM1V1ID7CHqwR+jutlSG44fn6DMCwJo4RsvE5/9xEHn36J95+EXN8oGE58AhdWQ4fVGAaJcrZQHjGjT8DH5ohXdZV3+wGLxDCjZF18ULjqhjyzC87MnF91NhjYAivK2GT0NKAQS6geU5PT0VryMpOMB+aM5zJnV3u01aOHz/vOPle1jyAMyooxBZvG5wc6IDjfQCuHT88s46rik3zOA5nwBfq8b854wpRzSRmZBI/a0DuXnFws+zLiivaZSAYIAW5dTmfbtQdAMjIpWdMWI8l44Azw8ehr0JjuCzY0piMenzGkEkWrukDuhcxIpWKfBSfOnHfNCh8zYE0jnsucPgGrxmfT8XmsM7YlYo85+7LgVbfN2RxOioJHMuGiOfvMpHzR7XJQZVQKM0pWTD0pEpU6nlZhJgUHZsrQJAS4vE2PCphSciQJd9w2hYXnLLAqnEtCS+sW+Fp6VnsR3o5jXOvQC4SPixlrGnEqCcckVGIpRTk0U9rq08LlnJxNExDhMiBnqDkFyqEzZV1jKlF2bJunZsRN7TGTgnuMEIRt7XJsZkwlY922UIF3vS7fLs8ppS61LMTyJVlhTxNu+iGjqmocFzAsK95uByxKxcOhY1yOqcs555LVxzRkWzvNtZbX/SbNBOdFUtLGZ5OQtgZ1n4xAIbaRP1lignqygWHCgjXpkFKQas6qdDAILcLGoP8yhheBGSkB/iU9uOjyCBrfiIeLiDQkAmhM6gLN2aj9IBe+EZH6YaSeZl1EBSssfSBLLLHEXzmWk5Allvh5wOf7QOznekHE1Mb0zz30xRnm5g0IWzi/eIvh89v4N/ZwggWT82u0u3PS1CNLQ06OVvn0//kaD5/2uLEKrmMJPFvLfgoYLYRMlYd2zljqmFJByKh4PK4YkmNRFlrxAx0Qu8JCCta1xbfsKYVY2upzw/aoRHEwWJQrtk2gDpsak1JwRfu01edcFsS47LoBHSJa6tHFQ4GFlPyJHrKjLd5fzPnj0znf5Ii7co5F2batutHbtnkhQ9ymWbqrdQN2qB4t9ekRM5AFHg4pOQcMCNTlE7OPg6FXO0rIqTg0UzIKhpLwQ+eEFQ0ZyaKOnK1qc/2JzHjPOeAzc86ZLMioLo3vmSo9DdkzYyLcugjRTnmvGpJIyUIr5lJwbKacz2Fc1AQmk5IAhwkFgTocFQUnkjAg50QS0kpJKstAc37xmrIpAQYhxuNG1W96KYQCi4ehwrKwlo/NGTPJ2TNT7tgVIlymkrOmEULtmXg4Lbm1Ci/mFbmUbBAQq8tCclIp2dGY61UPgIEW3DZt7tsZq24tR3rgDLijvUuJ0bZtcVUiUilZUCIKBRULKVhQcNP2mVF7WVIK/rDc59hM+cwc88gMmUiGUEvXPs7mKMqckhBDLA6LAq52DKuuw27ssKUx/4rd4ZquIwirGrGlEXtmRC4lLlJLzWRGgeWaCdmTOW5zXd+2K8QEDJjh4RBoPYnIKMmaacQqbVoSUlCSNPdAQwub68cQNl6nCntZWHhRdCgIDgZHHFKboqo1uWhkW5Y6bvfimCvuZc+Ig3N5H0kjWfz9pQ9kiSWW+GvAkoQsscTPE37cG3Ih12qOixfgfO1rdY9IWUt/Vl+5x/DuLyBhzvt3d3HckrNBSJL4HJ4FLFKXeSas9TMe7keIgVtXUnqxkpZKxzGcm4SvssEDM6RDxGfOGd/jjKjZ+T0nYyE57yVTEik5lhlps4AuxDIhZygJz+S8LqyT2rx7y62L5LoacNuuctV2aklXmVFQcmCmdPA4kQUp9W79qvh0qc3uX7IbrNv4Mhp4SkFAbfAN1aWjATOpyxALqdjWGACnWZj3qL8/kQkAV7RPRUWsHk/NOVPqBvgzmZE2i81d2+e5meBhiNVrJGkVJzJlKHOmkmJR1m3EfZ1eRvx6arhuuwCcypxIXT4zQ3a0xarGfDM/Z2BL+vi01WfPTKizxYR7ZkRfa6KRSskn+ZyhbbwNrqVQpac+t2yPbSJ+zVvjM51QorgIb9HnM50wk5QbtounhgNZIMC5WTCnYCo5JZZDTRjMBU+EXdthSsmimZTUkiGlh49Sy6J8I+wQMSorXAy3q1XGFGzZNq+7Lc4lZaA5vrpUKBEuPQ25ajuMJeWuc0xMgN/0uKRSG8FbhOzaLm/aFV7YmgBX1J6bnHoytuE5HGRFLdcrSzwHrrshgnAuU163m+xoi3azgN+ybRwMe2bEL9otung8sQs8NUwoiHB5ZEbcrFYa+VXAzCjBtAAAIABJREFUUOZc0T6Z5phG+mRR1rVDhF+/Tj0WZBRUzKjfa01Ha+O4YCgbElNRERNgLwhJM9nw8S57Phwxl9Ksovl79nNN6Pq576tlQ/oSSyzx14QlCVliiZ8HfJ58mJ9ARNTWLeqtPvgRTEcwm6DHh+BWPH68DU7F1Y2UJ4926Lbr8sHPBpaTkYNjwIhSWigKYW19wv0zxXOE1CorNsQRqRe5ts2u7fFlXUOod9tTKQnxyKmYk9PXCB+XPWeMKOw5Y1Ip6RFxzfb4zByzTsiTMmGNLvtmwq4Tsi0hh1J3VpRYBjLnfXPKiZlRSK2B/6EMOCLhlIz7MuamaeEgzKX2Kjw3E67rKl0NuGY7LMhZ1RhFOZQ5gtSkhJJYvcud7Tre1LAgr1O5tIWPWxuKNWJVW7VBWn16GrKgJMClpR7r2mZOSoDLUBYcyIg9M+ZEZkylzktaSMFHzikvnAmJ1CV8Q0nYM7VZfM8MOSMla7wNY0n5pvOEc0mYkrFvpnRwyagN1BVKgXI29ngkE8aS85E54xMz4HGecU1bPDFjzsg41BRHhR3t8ciMaKvPQBYcmjlrNr6Uy+2bKQEO92c5L6qUQzOv3x+GjIobtseCiriZFI0l57ws+a5zRKqWVXy+4Lc5l5QdbfGkTJhKygMzxFD7bCqUUGtydd3WE5VN264LA2WGry47totBKMRyRELQkN24SbnaJOKpGZNaJcPy2nZJKHUwwMJaNjyHPi3ekh4ehs3A5QvVJgBjSXmz2uBa4GOpJ2wXZK9q/CGfOMes0mbReHgOZIQvLmHTDVJhcZppRlt9PBxC6ilbHc1bETYEpUNISR2t61KTDYC2xM015+Lh4ImHfq5ZHWpZl98cF6mnJKWWlFoT0AtiuMQSSyzx14GlJ2SJJX4G8fWb/+gbqDbpV/LyAZ9rSr9IzbpYhDQdywcPKb95BsWI6tOI40++RJl7rPYm7D+/wnDqUVWGwLdc60FRGnwXPjpweZzmROoxGcd8f77AtQ4LW7EhAd/miKFkmMYIfSpZbUrHYyL1LvEaITMp2NUO55IwkYRSlKmkdDUkxr/sbdjSFhXKmsZ4OCys5dvOc05kysTkbGqHq7bLmVmQkPGKXSEXy5nM8XFJpMRBCNRl1w15rotLU/tb9DmVjA4eOZbX6fGKdngqk7qYrplWDKQ2KieNrdkKTEloERHjEePTJuC67XJs5qSU3NZeLQ1yTglx+dvbK/zFvP69Hi6rGjOTDEdqo3slSksDThppl9d0XxiEF3JOh4gNjTk3CalUBLhMJaNNwEJyOtSfW6AOJTCVjFwqplIQq4dmHo+ZcmSm+M0kwWAYSU4iBa80fogWHtsacSwLppKzbdsIQlcDxpI2zeQhqVRsSMhTmRLiUolypYnKnVLgUqd9+Y38aN/MoUnPisThaZUwNhkZlifOiKu2yw3tciYJTuPx6BLwnvOCxFRoQ/5SSjIpifC4ph0KsQS4nJsFV7XFnJJtE7LqurSNw6aN2Y4dRrnl/riibVxSq/wLTvly3GGe1jHNZ2SYyiHHNh0hfWJcHBUGWtDC41QSOvgcmCkTSSiouGb7jCWlRciUFB8XwbCjPba0gyOGEkuE9zLpCqFNSCI5XWIifCL1SaVghQ6F1FOOsc6xKKnW7epOQzwKLRAxqNZE1BWXggKhPuaIg8pL0uEbH4vld37vN5fFhEssscRfOZaTkCWW+BnEt578m//Zpc/jx6G29oVY+1KOZW3tB0nnUOQcvffLzL/zNpMnrxO15nie5eHHX8AIPB8raSGEQUmvl/DxecXhWHjnasVb7YCTpI6CbePScgyuCLkq69oiwMVKrX9fkHMiderVuVkQq8tYcgoqJuT1axsj98VC88u6wQ1afKXaoi8uBZYIh98M1/nluMOYOS6GG3aFSF1uSJsdW/d4jE2GUWFBxhNzxpbGdDWgb+qEowCXfTNllYDt0CVQh0OZc0u7zLVkrAVdDWo/AjkpxeVOske9c97TCKeRBRVYWo15/jvOs6bMcMa+LHhghkTq8RvOFl/7vX/Mv+NdIybgREd8VTd5zW4Qau0BOGFEIgUxAQ6GlByok5t61ERs30waMrDgSGaklAwlYUM7KMpAFqRSMjQpgtDRgAU5IQ6PqjkF1aUJ+3rVo6d+7RFRn5SKCQX3zIBx429pq08lStT4Hb5g18ik5IUZU1F7TS4+G6PCB84xZ5JQibIlIYE6l5MKgHVtATDRklV8xpJSYdm0LQ7MlAyL05DXWuo2agKbX5q1exqypd1GKqes25h1DYnUQ4CnZsya73BSFBSqbIUuz+clb/d8HpgRo6rkhU1YtRHHC8sKPiek7BBxoPXEKZMKF2FMjgIeBgfhqrYRIG3M4jfs2ucSq7S+NoiJ8VnVuI48lgHzxie1ohELKSikJiNb2mXHdkiac90hxMNhgx4rdC5v5UDqtvOetMm1wEidfnWRiHVRWngBg6B60YxelxcuU7GWWGKJvy4sScgSS/ws43N+jx855rj14wIXX+cJ9DdQFYanm8zGPT64e512Z4HrWqYLl6tdYZoKP3wc8+BZl6EWfCsf8PvPpuQVTGzFwdwyoSCpLOdaJwqtaO1V8NTgaP2eIlyGkrFjO8ylwKLNQs/wZdaI8LlTrePjcSQTzjTjKXMArrVcdiVi2wTc2irZSwoignrKQS03KdSyoSE37TrXbYcQlxVtYRCmUjCXgr/gCIA3pEOMx5SC+0nKjoRsasQZGQkVz8yUc0nIKMgpmmVhbV7vaYyHy7nMMQgnjDiTKWdmzolMMY3B2cHgqOAiFFLhCOg85Hf/w3/Cv2du8Fv2Dr95p5ZpXbO9Ot0IlwkLUvJLKU9KTruJfRWEmeScS03ABjJjXWNuV6tE6tHSgBkpE8kYyKJO+aJiRSNOJCHGrWVjzTnZJmJdAg7NlK763DMDnjojrtoOK/goloUUTCRjJnWPx6yRpvU1upTY5VTkVGRSklFwYubE6nKfMYa6NX0mOZs2RprUrgUlT2RKoA4VyrGZMZYF98wAD4dYPfbMmJ6GbGiXMQtyCl7IsC6opOCK7bAjdZN5Qd2kvqiDl/lBOqVEaTnCh8mc55rwaFLyrq7xUCZckZBfDnv8oBoRiiHAYc1zuSYRLfX5oq4S4zI0KU/tgh4ebVw64jKQlOu2R0cDbmuPc0kIqGV7KXljJnc4kznPzDlb2mVDOwxkwbHMCNRly7ZfnmOpRViFVIyaa/6UcROY4BCKj2muP20imS9ie0MJml4ScBsJmojUaViNSd3FbZKzlnKsJZZY4q8HSznWEkv8DOLrr/7hN15OOvjJbel87niTnCVRvcvaufJdgpMKWwWMR21eublP3Ep4srdCZSGv4DStuLVh6diAB3m9035SFJxKwiMm7GqbuVZ1+Z3Uu8ptPFwcEilp4XNi6rQiEWnSr4QDGZEay5ZtMZKcFQ2pBBaSMzQZRzJhJDm7VZc764attrB37vCd6pyF5ITUEhMjhk2N6IjLVYlpiUMHjyklu7aHj8OZWRCrj2vrxfyppPxGvMIf2Re1z0ASPnWOuapdEkp6hETNTn2Iz4KMXi3O4TlnJGQYESICFqQUzeK3xNYTEnFZ0YhcLC9kyHbZ59GHt/nKf/KCL776Ab/ciXnvo02uuyFvtAN+kI2ZaYKK/siO9VBn7LJKJfX5nZNyVfscy4QKy1QyVjTmqTNAEHa0y0nT+7Fre9wzx4wk4UxmvKFrTCmoxFJIxbEseCoT+hpxnTb7MsPDcFv7WJRjk2CpG+1PzAwfh1NZUIkSqEuAy2fOCRE+h2ZMJUpPI6Lm3J/KgndY4ZiUM7MgwuPIzNjSmEdmCAKR+rSaMIAOIXPJyaSqI4FFWNeYUF0qgUIqckoQaZrZB3RtzAszufTe+M01dy4Jt+nypEqZSS0NS7Bc9QKyCkrgTs9lujBYYESOWuFq6JOWMCTnVT9iVioehp64hMawpwm3pM2OE3CsObecFon+v+y92Y9kaXre93u/s8aJLbfKpaq6qrp7uqdn5zImOSIHIiSZlgzZlnzj5Y8wfOFLw4QAX/haV/aFYdgXpiARkgDLtEFbAjXiMuJwNs3ae+1LbpGxnf18ry++L07l9Iwhy7A47VY8hUJWZkZGnjgRmfU+530WN94X0nCgIyLfgJ5LRU7NX9a7tApXpsSK8pY9oENZe2niQkoSnGHdpXQNqcSVcXZiSYlYexN7QwteogeOhGzIhQKJRLS0PfEwPr65pUOBv/Ef/ZWtHGuLLbb4c8d2E7LFFp9U2M7JsX5WItZH/37kNjJ2ZCROSj73+QeIWFaLCbvjjs467mJEOLsKuXvsNh1GBRV64/UVNXNqAoRGncxkQtQnKrmaNGGsKQDn4mJ8rTce/5k5JcBwIQV/MTgkIGCqqU8UMvzP+jZ/dFpxuF/yz8oZP5QnJETEXiJ0bIfshSFr7VjZjpltWWnHVBNu4Uy9pbS8qhMaLLfTiEObcVlabtgRIcILs+JIJzwza2am4IWsCNSQErEgZ8yAhpa1lBgRMkmxKBkJjXYU6gbIha5ZScmcnJkpneSGKXtBxK2J0Pzvhu7pHoOdF/z1v/E1/oP/8GsEBkoaKhyx2lxNn+mKkQxYiksAW1GyouCWT27q6GjoeCc4I6fmTJaspOZT9gaZJj2ZSQj75KT3zTngrqRPNSX1srk/Cp4wl5yV1PypecEZFVNNWUrBXEofi7zGipMdzaXkVFYc6IgXsuBEp7S48sSGjgvJiTC8x4pSWl7vdplJQYiwoiH1xvkbOkA957IoDS0LcsCZqe+bK2rpONCMPR0yIKamYSV1L4PKNPZdJhHvmxkWZV8HPNCcR2bBuay5LQNumpRvtDNumZSjIOa9WcfdOOYwino506Kx7PpSwIumo6Ljy6Mha+14biuGftvwfuf8Jw+7gl+Ld0g1Iibiph1zYDNu2QkndsoNJlxpzQ+DF7RY9jTjpkkdIcEZ2ae+hDEmoqAiIaTD8kjPua27AL3BvPObkI00rabpfSKRhK4hnQBF6bRzvhFc5PU2nneLLbb4eSH8l99kiy22+P8drH3Zkv5RkuH9H8BPExRr0fUVWhZ07R6Tv/Qd3v6f/hrDYUEYduSlwSocTSx1Z2g6+PBZzB0d81CWHGhKRkCnSiyGK61dFK0JWGnNgSZOm28zrqRgrAmNOG1/QUUtrkn6TJZUtOwy5DfsLV7fh9FpzL4dEIqhFcur9oAFDYvVgEhrSq3JiEiJ2LUpMymZdwkzanaJeSEFXwwmPOlWvE3dd3GECA1C2Sm7xLzf5TSm83QJ3rC7fC24zxv2BiONeWDmDIiZasZ9Tumw3JJ9Km1IJO59HJt41ZmuGEjCgpwBCaeyYuK9Il84hv3dnN/9+19lGMH+2PLWZ57w6P4Rz5baD5XOQeGeq1hCVlqQStRf7baqnErh07gMJY0vwFMGJLyQBY0vrHthVuwxYqSxl0o558I9O+Udc0knlh0d8NQsen/CTAru2mk/NIf+WDaJYIEaKv887tkBE01YBM6DU0qNiu37KQ7skBbLLR1x3ywYacxzs+LSFBzYjGdmyZVUNF72NdSYjIQTnRIR8FiuGGrKM7NkrAm76giZM+0HpBpxISUGSDXkmAFL7+GZSdF3bHzJHpAF7nHctBk/ZMGbOuYHXHGvHpNKwB3JyLXjcGDorOFhbrmvK2rpGCWwWrUcScLAGJ517rX+pdGAD9fudbyvKaU2xBhmUvLV8IB/0SyptOVH5pIjnZBowD0dk9NxhwkXUjIi5g2fJNaYjrUkPJRLYkL2ZcwFaypaIomICKmoMRL5GGCh1oaRZKxpwZMOUWdg30TybjYiW2yxxRY/L2w3IVts8UmF7aBt3N/rZYX/NxG9m6+RdIjs7DP43DtIFhGGHbs3TsnzlLoVzgrL5cqQhvBP1lf8w/wFvzB21WoHJmI/CslpOYpDjk1Cg2VtO2ZSkojhQDNaLyep/KB+5QdogDHOV7DHiL/Q3eTzuxGPZ67E8INgxlxKPmWn/IrZYyUN7zwLOPb7kXNZkWnUD8SldmSENCiRGt5p16QakmnIRBNesxPOqdiViFnjvmaK24jMtGGqbrPRYXnfnDMkZC6536K4QbOjY0lB5MvkBMGqe9vRkUpERsKAhJSY3+xecSWAavjnTy03js+4X1b8aFnzL54r//U/nvB332/4ne49Cq0ICVhRAI6ojRnQ4SRIF+rJmox4z5xS05ISez+Eu0o+0pg1JUspWEpBphEDdQlaGQk/NBcc65RzKXgqlwQYrqTozd6+ZYKMgKGm3s/hrsq7iGHX2ZFp5COLDR8EM/atS/mKCLhr97AoV6wpvQyqpKPyBvqppow0JvFEZUcTEgJiH8V7xZpDm/HYzAH621V0fNs8dKWAPmWqlIYz4zYtnSjPKShpmIuTkZ1LzqfslCUNz7oaEbgVud6N73FFphFz3HM7CQN2TMizvCM0cEcy9r0b6NFc6VBuZSGFtdwMY/7iwYBBBJ+eRIQGvpyNOLYjDiThRId0CmMixpq4Ykg74Q2dckrJC0oemgX3dMQ9O2FOw4qGXBy5OtQxu5pxQ13jeq0NmTgym4nbJgYS9CRLkH4bstmYxEQE4uKAU2L+29/9W9styBZbbPFzw5aEbLHFJxXmZV8AbfOyKf26WX1DPjYfNwFkE8jGBH/hU2jecHLnIXU1YLFIqd0sQ6fw7qrmi8GESAOuKuWmZjy2JWkgHJmEJIRCLRaYmIBbdswHrLipGY2XDA28JOg5M5fGRMLUy7PudTvshSHfnlV8WFVcsvKJSp4sxIb3zBl/p/uQAGEgbshXTzju6Igd44jNDeO6FRosxww4xrVgW2AmJeMg4FTdTuBGGHFqci6k5BU7osFyQyeMGfSnak3JmpJ9GZNKzFpL3+/Q0m6uNHv5S+4lWQUVBRVvjiNmsuavBbf4j78y44P3bvHmMGE/DPlH8iF/YN7m98z3ea4zZ4D3919pzdyu6LBUWvcpWYnvqFir86DsqjvO0m8GHsg5kTe3P9NLLMpMcl5wxZLCeyo6fmSeM8CVM+bSMNakT7gKCCjoPFlI+w3NheQMCFlJ0ydXbRKyCml83K+4WGBv5o+8OXvlPw+OKN3REQupmWrKB2bmmsJ9WtRt3eXt4IIAQ6aOMJTUvJA5Q9JeujTUGFdj6aRhj43rl2mwZJr0vpcYw4qWC0qetzUfNAUDDSnFlVXOpeIHzKitcmsYcDML2B0qv3xsuBslrqyxrdgjZhBB6zdSnRVWFVwUbkd1PLUckHBfV4wJ2U2F22HCKzrki7rHjIpLah4Gcyrp2LEpc1y7/SOz4JlZEqn7Od7RAQc65Klc0dIykATBYERQFIMhJiIS7/sQ8e87Yhj5zUfg/9v/27/7X24JyBZbbPFzxZaEbLHFJxGbeN7rRMT6ZmS9RkauQ1/KtGQ4RYY7SBpy+uQ2X/v6XSaTkryBezuGsoWjKOKViSHC8MN6zURCvpAOuag7duOAb67XPNA1p5LzwtbckyHnsua5FGQas/TpSrk07DJiSUlDywtZUNKwS8LXuwv+1+AdPpQFY1JSDXmgp/xBcJ//sX6PU3Wyp38c3Cf0KUERAZ0oYxPy1FYcmZjv6ZxdYkppiXDH/IWxu4rciqW0lvtmziklqs4b8RYTLI64HOqQW3bCM1kzImVCxhgXx+sGvcBp8dUNwLFEtH7Lk0jEmV6xg4uh/dv5jzEI/+nf/BMOv/RNPvXmIyIDz9uaX+1uc1P2eItb/LK+SqstK3Wt66EEiBgudEEoIfW1q9s5tQ+DdYZ3QaioWVMS4I4tJWYoKeeyulaGF5D4IXdM2vtEYg1ZS02HI5FrSn4YnHElhT9/7j5HGrOQigEhu3bApRSOiPl0p6HGVH4jc6QT7tk9Kloq6XhiFv33vpScPwmeciUFiQacy8p5hlRosUQauMeiBitKcz0NykuKUo188pf7fkONObBDjjUjQFw0tDo52NeCxz7+uWVBw3PJ/cbMlVXOpaQVy4uu5rxQnuYdaWIZj1reOHQBCqdSshuE/B9Xc14dRry+J1ysYVZZik55WrbcvxAWNHwpnPKhDwa4bFtuxTELbXmFIZlP/fqSccWLnY9UPpMljbwsNXzfnHMqa27rrnvOteyf/8YTX+PT1AICKm2o1FVXirgztSGu24LCLbbY4uOALQnZYotPMn6W9Mp+ZPtx/a21SBBBOoJkgH1mKYuIX3xrznBU8KhoOF/DKIanTc39ueVScmZS8gOueLss2IsCfljmHEvC0tTc0TEzKiq1HOiQkpZSGlJ1sbBXrEmJiQmZU5CRkBKRYiikYUxKLjUBAQc2I5OkNx/vy8T3iuRUtFyyYi6u3btR5cQk7MQBQ404Dt3AvBMGpMawqqGkY9e6zUuH5YQBs64lUsMLrbiQkplUvG/OOZc1D/0V+okmFNQsyJnpsvdr1F7GU2lNo04y02iHIOSeFMx1zV27z5/+4a/xJ//wt5id7zNKlRtBzL/3miM1n7cH3PLdGaoWg/EdD5ZaG2IiOlwDfEntdyEuletK3bZkM5xvIlyHnjwlRIwZEHlJ05ksOZVFT6g2g2qLJadh4PtazpmTS8VjmbGkcOZnDFNNuZCcXJxxfk1F6DcoZ7IkI+HAZpzLiidm4VKigDkunCD3sjaDcODjkw901LeKN1jOzZqpZs7zo86RMvbllSGGmRS8MEuWOFlfoiFnsuTC5Hxg5ow0wagb5tdeIpZLQykNj82CUhrOpWDl44s3Q3pGwKpzA/75PODhi5hh1rJLzEgj7kxdjO83Vms+95nn3N23vDIxP/Ef62fijE6VEx3y++sL3pcFBjiVggrLU8k5sSPOu4YdYh6ZBS3qQw86KnGm8s1m6LHM2GNEIIaV5n1w8+ZnotCSwm/ONhurTR9Ipy6tbfM63WKLLbb4eWJrTN9ii08iWj9kmMD/vTYWfZSYXCcitkE3G5Gmwrwx5K3/5Pf44O//FgnwmWnEN+YFb+qAFS03JGJsUw41o6Tjmaw5aGIMsNaON3SKBe7KkJk23NKMCkutbsBc0aBGaaRziVJe8pMR8S1zypEdMZLYp2StuTQFQ1IaOucdYEighkdSorQkuCvvl6bg2A7Yj2LKTnkzGLLoOg51SBAIpe14XluupOINv/HYULMfmytu2RHfD85YU/Ib3V0ecEEljuykxOR+kL20c0YmQ7FYLKqWVl5GoQq2N6hf6YpGXw5/f+vFu1zpmi8/v0fInH93Z4fH54Z9m/G/BW+Ta0Xoe0AySVhriYgB7ehwg2kghoXmDMWZ0EsajE/I6tQykcwlInkSU9PS0DHTFRPJ+s6RAQkNLZ2/6r70HRe5VLRYYk9EWq4RWFyAwJ4OCXznh/PPdOS4GNqIgAONOTc5I025lBVGpC/hy6Uh0bD3MbxrznjV7mOhl2pdyooTnfJCFmQa+Y2X+iv7DSEBJTktls6ncEXi7jNU48mW25jUdAyJSTUi9xKyDpeG9tysKKnJiHtJ240oRAQOh8L5WrlfV0RBSiYtlVrenrVMiehQ/vm3T+gsPFi3BOIS4d6ua24HaR8csKspZ1Lwx80MRXkhBSONXJ8JsKBxyVhkfKW7ye8H71NSs8uYlJixJryQOff1lH0Zs6ak8q+pRGJqbYgkdERVIhptCQloaX1MryNYlVb/73+3bLHFFlv8f4TtJmSLLT6B0Otej+BnXGv4WTG94AhL54YazZfIeIpMI+595Y+4f3+fi0JJMFjfunzVdtzTMSsaMgJ+LdxnaV3i0hPJKenIJCAWw0wqzqmIfOxuh5IRcqBZn3I0ZsBCKlqURiyduMb0jV/kVJZEBLjKwI5LVl4P72QoipJ6g/qchmd1w2njHk+hlreCMbPWtSM0quxowkawNiBEcf6EZ7KmxA3Vz70RvfI9C6XfaFyxZmpGHLLTl8dtUOO2FRuPigsVdr9uAwl42zxnQU5Dyx/Lu/yffJ+vzwpuTDpe1wl39cANj9oylsxJvghJiQnEbSwsFquKEecHEYSlrgl6wuEe94Y4FP7xvOyWcANpTMSKgg7b3zYl9psKQ+NN8CkRI9J+S+CM95b7csYLWbCkQH0fSu4b7wEKcaPvjn9+G1oU5ZbuoCh7OnBX+bFkJJzJmlzqvgyxpmUuJWMGzKWk8J6PUtwxOeokTHFksPOPq8MyJiHRgJmUdOJ8KjUtOTUP5ZJbdsIK97kDO+RQxxhgV1OONOOdJmcSC0+Xynt1yRNZMyvgzjAkwXCmNW8OE+7ECd9fVOwOlcda0KjyWHJuSExqhMgIhzrgQGL2NSXE9OEEA7+VyWnZxW35Ug15ICsGJEzIXLy1Ji4FzCdgRbikq+RaKlYkAaW+lFwpysquSbyvaJOGlXhD+xZbbLHFzxNbErLFFp9A/NHDv/nSdLqRYNmPSLHAvb8xrat1vpEgcu9HMQQhOmuw8yHjUcNp3XI3Sfh+vWZAgBGY++EWIBL4tjmjQwnU6eFz7Xhgc5ZSUYgbQBNCbkiCAoc64MSOGOrLIf5K1kw15VzWTjZD69rWfY/EPmMOdMRN3WEm6/7rBOGRXJIScygJb5srfmRmhEb4sbkkEqFVRYCCjldMylMteWZL9qxrR39k5sxM4YfYAe+ZM0Y+ESklYqGOlDTascuYObnvThdEnAxpQNIb1TvtCMT0A2BMxELXvY6/VWfe/m5wytkiYEbNXzE3+S37FgATMoakJBK7q/nakGtBqa4YsVNL5FOQNgb4UhsiQtaUfsDvONUrZrokJWYgiUvuIuofS05N6wnHQEOGpOxrRkJIRszAb5kyn6yknpiNvLQrI+ZMF71Mq8WZyi9ZOQmXlMQasqdDIt/5AvgtScJQYxINqKRlX90GZ6KJ/74NjScbrqOkIKci04TYk881JSMG7DLmhc440Sklrtk9RMg0Yl8zWpSIgHu6zy8EU0aklDTc07FvPE8abs1SAAAgAElEQVS5IQknQcKBJHywbggEphJxW0c8LGvKFl6JY2ZS0Vl4Xjui/N3Tji/FI95nya7GKG7Dtuwsd6OEWAxvhkNmUjDWmE8Zt0XaGPhzWj5jj3nD7hFiuGN3uGf3MAiFtJzYMYEYEom5ZMVIMgYkvdl8k2QmXronSB/je/31l/LyZ22LLbbY4ueFLQnZYotPKkzwMqYX3L835vT+Nh/pD9mgqdznwpDu0ZT3v/tlxuOScyqe1w1zqbC4obfB8sysWNHyQV2xqylDQg4ZMCHimeScmjU5DZeSk0nAbc0YB4aF1AQY8j7BKPZmakOqrl3cmYVnfCjnDNSlK238AJnGJIQcsgPQk4RLVjzXkhXuCvGi7RzJaBsalAYl9wbm++aK+2ZOiOHt4BxFyTTqU5c2Rm31f1xjh2BEWFLQeonThlRsBkKXlNX2hvXAE5SSmkprOjpaOgIJCCXkXfuY/359n1dNxr//1Sf8578555454TPdDV7v9jhR9xg3W45NFHAszogM+E5yc80fUNHSUqozrrfaUlCR+RjfJSWZb5jfxLp2WCrpGGjEXMp+Y1J570FMxIkdU9HS0XmXScgVa4y4QTf05vXGexU2JM0AK6mpaHkmy2vpW+77nssKi/JM5rRYnsgV+zrkWCfkVJxy1R93SsxM1pQ0LNVFGM9Zo1hqGk7siNbHDKdEHFi3cZuLKz2casq3uisiDRhqQoRwISWfYep9Q8KV1lRqWXeWG1FAiHBflqxby7pTJhrztXLGkpYVLY+04Kq1vMaIm0HKbhRwMHCP+2gkPLUld3bok+E64HaY8Gvc4KZJuCkpXzJTFxOtKUMfe9xi2dUB57LmGFdUWGtLLBFXuiKQgLWWrGxOSEjrhVcdllhcMtwmxjeVaJuMtcUWW3wssCUhW2zxScYmHes6+fjoRiRKnGTLGAjdwELXIBK48sIi4Z2HA77z4x1umwGtKvd0zFEQMw4Mr0jGrqa8H1xS0vFpptRYMgkY+j6JIztkRwf8ih4xNAHvyoKLtiVQ6aUzjXSMiDnRKSc65UzWrKQm0ZDID1ZWlKkO2NOMXZvSSEdJwy07YcqAFQUtHWMGlD7K98QO+UBXvC4jAO+d6NgnIQ0MCSEhhoU4nXzgjc637IQ1NSc65WmfONUyEucXSIjdkEfUd3IA/fCnuHK4ROKeCGzK4kKv2++8aT3AJV890FMe25If/PAWy/mIv2zvEPsr5Q84YyIZN80BgnH35eVTNW474oRmSuKlYTERVpVU3BbJfX/Lmc5ZaE6lNee6ICL8CbP6OS7JaUlBSd2THItSULGUym0qyPqkrM33u26IF8TvkAznsuJS1qwpmZKRS0XpbdWX3hR+Q8dk6jY++5q5LhnpmEtJSYNgGPoNypKCAufNCXz6U+wfwUASPjRXjHDblUQDVtKwlIqIkJHGPia45kAzPssO78qCEGE3dMZ3EdiTmOcULG1H0SlDE/AmLsWqUfVkYcAb8YDPphljb7ufa8MwEF4/UF7kFgE++6krvh+cMs4sI01ICdiLAu7twi/ehHFouJWFjCJDTkeLMiQkwrCrA2ZS8EguvBjNMpIBtTo3Sak17bXX4KHZJ5SQyMdEl56Mj2VIuLWCbrHFFh8TbEnIFlt8UvGzYno/irb5qfdFDFos0XINnRts/p3f+jM+9/qSuW2ZBq6McBgI+6nhwuvRhxoTY2jVJfJEIkyDkNd1wrGkjDRibAJedDVfjfa5N4j5bDDhU3HK52SHXevkV2NNqGgJEPbsgIW4q9wJMRZlJX6gIuplQ8/Mkkx9bwKGhpaKFgNkhLwmI0q1zguCxQBDE3DVOiP8Znuw8RPsa0ZFRyvOlL2Jsu2w3ixu+r6OmoZWO2KJMH4QBnpTufFX+1ttiYmIfW9D7BvPW2377UmuJX8QvMf/cHbK//KdjHOt+KfBB/ypfADAlMyZ8TE+NUsotaFS/7xt0pBoqbTqyZEjRMabs5t+8+H+7dKSFuQscb4BRXkql+ww7B/PgpzS+0zOZdVvVzYE5BXd97IsJ5HKvL8B3MbsQh2xuR4Pu5Fs5VJxxdolN0nLHbtHgCEh4pIVK78dMwgX3hc0JXPbACKOcP6SI50yVuejWEjBkoqVT9/60JzzRK6Yaspzs2IhFZk6EnGmTrL1qo4JBN5pctatJRJhhHvdWiAzhnFgWHeWRpUKy4GJOG9aVo1l7g33MQGjWAhDJRChAx49mfDp7oB3nxvu6JiRBMQBvHupXK4ChrHwMG85q1pKOta0PJO1+1ki4Io1AQEzln1fjKIMJGEgifcJKZGEDH0xZuA9Iiey51+J8hNkZYstttji54ktCdlii08qNqWEH03DgpcyLRP0xnStipeG9qqA9RyaGrO/JNi5YrlMGEtAbFyS0Xu1MzhvCtC+rIc0WHI6bpsBr00CXhkbfvOeMAkCDkh5v8sJEPYHQiAQidApBP5+7lgnOWqw5NLwy7HT7E815YCxJwGuKXvt+yZG3kvSSMcOQ/ZljKIcaublVI58XNGQirvSnxLw0Oa8UBfnO5eSJ2bBWFMW5OzZAQ/MBWtKnsncJ0aVbrjHXXVOCGnpKNUN8m6j4q4ybyJTW217v0YkEQNJ3Md8l0MooX+8zjsSEbDQnB/LU/47vs7Xg4cMSWi0YSAJ37f3eU+f9oNkjeuC2ET4brYQhVaEEmKxFFSsbN6TiQ0JCAh+6qp4gCHw6VqC4YwFFyxc2paXOVVe6jOWAVe65oq1v18Yknqi03KBk1s1Ph0slZdehCWFJ0sdJTUXLDnyG4ZzluRScyorEg37YXvkyc3mOJcUZJJyxdpJ01AWUnAmy75YciUlN9QZ2jdm+AOb9a/bkUZ8M3hOQcubTFl74nopJaH3D9W4TUZpLS/aGlU4TEKeakFFR6VKrh0i8Ckz5DgLOAgidofKgzO36UhE+OYzy6smo1Nc/LRaHhUtnSqqsKyVRAQFpkSspOa+uWQlDeeyZkjKGNdgL2JY6rrf/gwkIZOUgaQkxFzq8uV2SCLeskdeqrgm3yZjbbHFFh8TbEnIFlt8knE99WrzoU36Vdf+JEFpqj4ZC4Dh1Mmxyogf/+FX+doHrp35R82KAKHG8merNXeClM/JhDuZM79+aTTg1XHA05Ulr2GZB1y0LXfjmJuScjOKebDs+F6e8912zu93z/ixXVJIS6LOR+BkNCGP6pojO6L2Q3BFw543LQN9j4LpZV0uutfJcoSZlEQYcu1YScNRHBIhRBjeDdyA99gs2LUDf+XdFda9G1yQkZD71oiGjjNdMPWFg412VLR95K7FesO67VOpNm87v7EYysAN92IIxFD4srkNNk3Xv6Zv8Gk94a45Yl/H7OuQPTPlzM5QrI9dddIqg0HEUGlFpy6IV9XJc1JxmyNV6wmJ+s2Muybe+T+bbQXAiKRPAau1ofADa07tDeghKc4vMyAhkoArddG2l7Lq76uho9CKkgZFmbNmxIAcJ+NakPvz5p7HXUZ9iWFHx3OZey+JYanutiUNU2/ST4kpaTjSKYlvg0+JWPCSbK0oONIJ980VMSExIffshPeDSwBO7Ji3whEDjfhsMAbcBuOi6bijI0TgsRbsEbOwHR3KnIZ51xEH8Evp2PlTtCWTgCxwPx839zo+f+TO5+09y/F+ydEgoFDLKDQcjYSbsduCBTjydp4rSSBMQsMkDBiIYaxOmrj0W6Ir1lywZEjKSAb+OXQ/37lPvkrFSQNHMmBISkTIieyR05CRMJEhExn+y39vbLHFFlv8OWBLQrbY4hOI33j9936b6zG9G7Jhgpd+EGN+OjUriFC16NNHsLiA0YTucpeqNhylIftxQIih9slSr4YDjoeGpe14mLuryKGB8UA5axvKVuksvJLEvHZDuTcK+fShcjMLODYJQz8wdqIspWIhNadmzaXknOiYrwUPGGpEJS2xvyo+0pgA4cK4K9FGhUYc+TjUMYc6pqNjRsVSCp7ImgpLogHL1vUxPKNgVwfcNzOsKEeaUdPwoZzT0fXxua4MsOlJT0bSxxN3dCS+OFFR1lr08qrNFWqLMtclA0m4sgtqdWb1lc1JJEbVEkhAS0dISKU1n2UHi3Lb7mJwV/0/Z08IJEAwvTneOQNsP3QDFFr2HSqFVoTXDOvua9zznFyTgm3IUuwJWOi3KkZcS3tM1Levz3TZS4OWFAxI2EQPbzYeM10RYBjJoCcxm4jhTa/HAVNqWtZasqRgqgOuWLOiZIcha0oSDXksMxJxYQPuYwFjTTixE24wIcQ1qt/WXRIiXtUb1DQ8Z8Yt3SdUwzOZ81SunDwJQ4hhTcmupnyvXXCiYyrV3itkgVNKGqtcSsmSltezmFOt2CPGCKxqpbPK7SBlR0JScbHVa9tR1YYHF4YnV0LVCO89GTDwT9GbhxYRGMXCRJwxvVHlUVNxv6hpFHYTYRq6523XpuTSXNte+ZhnAnZk1BNJwcU0Nz6yt6bhho6ZSMardp+YgF/qjhmS8N/8vf9ia0rfYostPhbYkpAttvg3Ada+3IZo57Yg+K3IJjXLExURA43XjYcJ8Vce8cWvfJ3Xj1rqzsXbvpVmfHrf8MrENY+/NY4ZGcMb8QBViEPLrTimU7h/pdzaUdpOsArTcc2LwrK2loyAL+shr+iQXR3w1Fz1jd0HmjLVjAtTOOkMMXs6oqFjJTWBGnKpiTBE6iJfT+yIIzvklt3hQXDFSFNe0zEzqZgS8aKrmVNzP7ji0Lr42UgN75hLxHtJNpKiJWW/WQFnOF9TEojbIpRa9+lOrbZMzJDI+0AAWj+Uh4SsbO4/1vZG/M259pbxfjPyD8yPeSAXPDYzTqy7Qn+og94QH4ojK512/dcE8tIcnknKUAb9+6GEBPLS/3HdRL6Rg22O2aJc6pKZLkmIXYeIFi9lP7h+iZqGlRZEBOzKiD0dMSVjxppKK2IfSDCQhNKXFq4pGeNKFXfVHd9YBsRELKX0HpWWBTmdWp7JFQGGISmhl9G9kAVLqVhKiUXZ0QGKcs9OKKj5VT0iI2GHIQeasRJ3LENSDu2QHQm52+1wolPmUjkpobr0qkrd6/GpFlTSUatyoAM+FWaUrZISkIjhVhpxkAnv1SU7sWGhLV86FoaxMNOGtoOrpuPtKkcV8gZUIUB4dhVwmcPeSHl1HBKJkIiT0a1oyUIhCmA3Fe+Vafhsd4BBmDLgmF32dcyJ7rDHiEPZ4RXdZyojInFdNQMSRgywwLFOmaqLi/7K3oD/6u/9Z1sCssUWW3xssI3J2GKLf9PQtu4nf7MoCTZbEgNdg0qAHOzDZB/79o/ACDKoOD6e80/ujzkxKZ+5VbMuQppOuDlVAqN0arAKgxi+/1xorCUU4Xho6Kwi/gryi4uE97sVCQEnJmE3DpjVHXfskKwLeWjmpEQ8khXHOuLcm3OdYXxALo0fp5VMYx/Zm7CSmmdmRU7DZ+0+j5ixw5RnOGN7SkAkwrsy4163wxOzJFCXPFV5Y/ZG6uP6Nuq+8G1CxsJ7D4xPs6ppKLXqB3jxfxSl0ppEnDHYtVi7NuuNOdii/ddacbdvab2MzHCpCwoqLDe4lDUvggWtdu4seHnVBr6rnU2LeOjTqmIiOp+WValrUt+Y4Dfnc5Mk5Qz2bZ+m1GpLK63PyhJyLckk4Vh2WVMyIMFScqFLDmWHK5961eGObeXPuUXp1GKlIvVpYkNSTmVJTMhUM+aSM6fwvSXOM5JJSq4lQxnTesmYbMz7VJzKlZOE+fOwwvlsZtq4/hAS3jYvHNFhwEBDTnRIjXJXhoxs1Hs9LkzBExuyomGXhFOz5rN2j3NqAoTKWr7TLfmMGXOSBby3arCF8lqc8qPCbb+ezw3vrR3Nu1wLd8cB5SIiDGCcwGRguddEXJSW2AjLQrgx6TjLDccjYXbV8qlowKuHLas8oG6Fz0RD1t2Au5OAi8VNHps5v9od86EsOdEh982CXR0QIMyloPSvzQtdOAmW1Pzb9g6dKLsa8/orK87+9f5m2WKLLbb4V8KWhGyxxScRG9P5ddjOGc83MiyDKybsP+9ZiW0hzRwhubCc/+FX+OD9Q0bDll89ihhlLc8vY5oOVjVMEuHucc3Tq4R3ipJfTVMetxW/vj9gXUFn4cEVvDKFTiGJlAMSDsKIylriAN62SxIC7piMlTYMNOTCFBzZAYea8khWVNJxIW7oy6Xirp1SSctUU2ZSUOOSrkTkJx52JR3HOiAzBgsspeKQIZeypqLlUMcMicmlJiNhTk7mpTulWlQatyGRlJmuKKh8G8dL0pFITK4lje9uaLT1MiW3DYmJKChdcaEf/J1USal0Ew3s0rfO7My3lDd8P3jGuc4dabElRn7yOd1E5zoz+caUXnrj+NAnZHVkkvbfB9xW5zrpmcqw3/Rs4pBXWpB4QrbZaKR+a9PSUVEzliEzln1amCMnSkpMTMiMFZEENNphxRGjkcZcydqliklHi2Xsm9hDb2RPCMlxMrKalh2GrKhosZzoFMQZyxdSMSDm3eCCEMO7ZtbHBDvplbCrAwpaHsuKI804JHHbM0xPZks6lqZmaF0nR0HHhZSU0nKgCbc1o0SpOnjEmoSAu0FMh5Jg+N664G6Usmpa3s3deT4IIsZZy2SovJjFlK1y1XbEIqxa4dZBxyQOuLlf8/bcpW/FoWV/xzIZl+xdZAwHLbO58FftHt9aDpgGIW/ZKSeDkKQwzGlYiWtysSh7jLCiZCSkGvK5/YAfXHQcRzFBsDWkb7HFFh8vbOVYW2zxCUSfcrWRWlkvwfpZjel9l4jtm9Ptex/QffNt9GrA73ztBqvS8O6ThChUZsuQy9wtUIpW+fo8JzCWo4mLLJ3lEGGYDDs6hTiEq9YN4weZ8Hxu+PxOzPFIOMkCqhZ2SXg/mFFa592IMOxaZ2IH/FXsltCblVssJ5IyUtfiPZeSqabsa8o9O+G55Ex0wE07Yl8TVrTMbctDmxNpwI+D0/4ULKSilJaUiJzKewbcwLbxgjyypzTa0dBSq9uAbLAhIq6d2jkqIgmpafoY3Y3nIyDopU0bbAjBhtZsPBoAz+wFpY9AFhE2RXybY9t87839XD+mQDb+AffWXrtdpVW/DVnout+OLOyatRZEEvXHUlH3/RunOu9lZjFRv93JiAkJmfkY3gbXwD4kpdSGWKK+wHAlNWstCQj6iN4NAXHbGcOCvJe9bUoPE0+OLmV9zatje59QTsWlrAl9ctZN3SHxvpyJJrRimUlFagz3zYI1LQfi5GUPzZx96yRiBzbz5CVh16ZE4ozkz7QgCWBASIPlvaKiwboeFzpqq3QoUxOS0zEIhDC0lFXAeGB5ty6YUfPKMOStfcPTi5jDiRIEyrlW7KfC04uYo6Mrus4wHLSs8oiDvQoR+LemAwaBMA4CBhHcSWMnEdOAHR0SYnir2+dIJxzZEb+ihzyZgwC/eKdhsjv/f/bLY4stttjizwlbErLFFp8w/Mar/+i34WUKVg/tkODa8tNeIyobhCHYju69IT/4nb/OP/27f5XjNOTJHJIQkrjjKhdCA/uTjlf34HaQ8uNHA44OSgKEN06ckfvFLOBgpOQ1fPUuZIly+7BmUSt1Cw8WHbtD5bLpeDMZ+B4OZUrMkJCpl+Y8lZwnsqb1Q3SLu9LbqBtYz03ex/Q+kzXnUvKhueivdJ9JyUoaTik5NwWpXwBvYnUbWgJ1hu/rg3xD18ecuoSslsoTAoBSayIJe+KxiUrdJGZt7qOjo/JFhZv26ora2cq18xKpwN/W9tuN0N83QG6LPtdK/LDuGtdb14quracUiojvPFHrk7iCPsLXot4jEqBq2bS2ryh8baCl0YZMkp7gbAjTTJeE3ntS0bqmdnEG+DOd+22MJRDjbovpN0YJIRUtz5lxxoJUYhpaFPWbJdNvgiICBiTsMmKJKynMqfuOkitdc6VrUiJKGhaa81hmzgyvKTkVa0qO7bA/YweekJ5LzrJzErULU/BYc+7ZCYJwJRXnUnCLDIvrlzkk5XYWcNZ0PDMrHqxbjiTlgJTHsmJMxI3IBxB0LV9IhwwDtwM7GgvvP024Wgfs75acScm+xNzc6yhq5/24dbwiiiwRhvFAmWaWOC0oiogsq/tsibyB3ZHlF+/VDELh+dpixLWtfzoccsuOGZDQoWS+oDHXjsgIO1HA7TunnD49+Ff7RbLFFlts8a8ZWznWFlt8zPDrd/7Bb1MVkAyQIEKbCkmHLm7XX92mqVy7+UfLBk3gNx/2ZVHhRoa1kV5di+WVKEGb6uXHJYCupT095BsfxJy3DTeiiB81K34rdI3j60aJA2GZG/JK+MKx8s+e1LyyjjgywnhcsGcGHO12HB6sSU6HJHHHugh4/c0PuJx/mmUBF7Zhtk75wg3Dd05b3tJ93jZX/ILukRjDg67gtGmIMKxNzciTkpkU3Ot2mJiAQzvgUvNeknUuK4508jIhSlrOJaeUhgMdcik5qbqNhKX1Hg2lE0vu5UZrShcBq65bwvo0KsPLMkDXp+GG+olk/ZZEN8lZ6giF9e3oBnHeED9otx+J5w0wtFp7KVnQby82siLXdGJ7T0mr7U9IszbkqaPDqjtK1BnfVS1WHCExfjPTadcb5GttnNlfcxRLJBG5Vj3hCkziH5tFRVloTiQBVl0B4qZXZETChVpaf+4b3wESitv+tLRYVTJJnESMhIXmCEIpDTvE5Kg3sA88ITOkngRd6pKRDOjEYlU50BELqQjEbU5uMGEhRU/kZt7sPtWUhdTsakJCwH1ZsWcHzExJiyUm4EAzajo+yw6BCLVaEgyZMRiBozjksBrSoCxp+HSS8a32BYfdgCwUdpuYUITP36n43Xc6VrR877IhxBUVhsEAKFlpR1mF1B3c3G9ompDVOiLDcvOw4MmLAc8eH1FUAVWdYVUYDgsOxynTcc18GXNrR1mVhiiAPYR5IXwlmhIVhgMSAis8M2tu2QwRMArTGw/5s+/c9k0sW2yxxRYfD2xJyBZbfEzw6yd/57cdQYjAuP4HwJGD63G7an/a7/FRGINc83tshtGfwHVplvp0rKYCa1ld3eB77YJ9Ep42NScy4MZuQ1UbJokrGuysIAJtJ9wbxMyWsJ/AdHfBG7sjjg/nDMdLyiLi4YsBaaQ8eXCHL37hMX/8jVe4EcQULQSFcCMOiRshsFMuqNmzMQK8K3Nu64hOlUCFEENnLG+YEaUqMyqmmlJIy4kdcx6s+q6PDsul5EQYShx5ASilJdEQK64pPMCV8hnEeTG0dZsClJKXCVgWtzkIcRuKECe5arTrJVKttr1J3HWCuK1JhyUWJ1/STTIWBsT2n+/oEHXbhLEZ9puZ69uZ63Am9Zfbob7vw99nTUOoYS8HCzAoxvtVXvpmAgyRBHRqeyN6qRWJOD9IpW6zFUqIVSXXgtBH/hp1O5YOywVOipVITKOtL2B0m5HNZmYqQwoqcq24IVMEYV9cAlh+TQL3QmcMJOGAqXetCCspyIhZqSspnJFjEDISAgzPmfXpXTu4LUhORaDCARlPPPlQgZkpGWlMI5ZGLWey5nP2gN0opOgsx0HMZdtykAT8YFnx+iDhtmSU6uJwf1CtORC3MXlatrw5TPjWes33HgypWLlYXR1QYnlrHPODi5Zddce2KITP3suZXSUUVcLXn3aMTcAP78e8v2q4tUwJBL74WsG33x8wzEbcn0FRpwQGdsctxzcqzi8HfOuZ8tok4DOvLXj/WwmNf63csWM+NYp4/WbFch0ho6LPn9hiiy22+Lhg+2tpiy0+JtD1Fbq8dOTj2hajJyMfxUf9HdBLq3oC4t/fEJBNNO9PSbEkcKlZAMaw98vf5I4MiTC8NUy4k4XcvvOCnUnFm7dLdobKnaOC1sJk2PC5OxXH+26IvDzfQUSxVti5+y7PL1JuHlTs79QMBjUiTkqynxiOxsploZSdG6aPwogjk7BSd1yv6oTMD6GdKIW03LMTGlUe6pqVNNzWEft24NrTSTkX1+AtCJnGjDR2X48rO7QoVrQ/ZZsr7hEBFTUWS6V1T0DADeqVuujgzQBvsYQ+Teo6Yol+os/B3dbdZrPNEAxWu55EuON9+Ty36p6LTjusPxcbgiDIT2xBNklZFkXEpUht/CexRC6x6tpxKkrke0Ji3Nu5XaH+PHRYMh+dGxGSSUKAi3+NxcX6psR+Q2R678vMLhExnmgpOwz91scdf+Ob1nOtmMiQK10zlgEZCWMdUNL4ikl/ftWRxAuWlNTsMCQlJpSAPUbk1MwpOGfZH8/Uu1P2NfM+kZq5qRhLSCktS1OzkIpnMifTkKVUDAjY0QEGYd1ZVrbjZGgoscxry31Z8rz0z4cnex3KrqYkuCLMRa00WL5RzWmwTDXhEWtuRBE/XtZ8oCsOJGEqIWe5Ml/GVI1hWRjGJmAjiBwaw7fLJaME3n88wAh8636EKpytlZsHFYt1QBBYPjgTAuBkr+FyNuDL+wlf2kn4peGQqUQYgVff+JA33nhOc3nAb3z1u2yxxRZbfJywJSFbbPExgQQRIgFarh0pMMa3ml/beqiFtkGbCu3c8PcTG46fsSFR7w3AWteIvhlgrydjgfODtG4TIjstO2HAU3JWtWIE7n9wzIdPhkx31qxKV8L2PO9YFyFvfP573H3tEQBR1PLmq3OG4zUPv/vL3NitmU4LwtBS1yHv/Pg2TeeM7WUj5J1lGAmHg4DSKqVa1rTMpCLBDYPPZOkNxjH7EvOBrpibionGlH6EW0vNVNPeg/CKnTBU9xg3BmhwkqWxJqwpqWgpcXKkK13TaMdYMn8V/6VXZuMtyHxXh8HQ0hERMvD+iVyLPva2oetTmlp10bf1pl3dR+0CiLi431bbfmMQSui9GS0tbX8MHZ33fmhPTIwEPQkJJUS9TybYSKC0JRHXRLKRYIUS0vqCu44NQXAbHsAb7AMK326ea+WTwmrWWvQFiMYb11Pia+dFWGlOp51vm+8YyaA/H8YJW7kAACAASURBVCU1Ia7wUlFCAs5Z8lyufPN6g/FpXM2mM8QfY0nDud+2LMgdmSJAseS4COBA/y/23uzHsiu98vt9+4x3joiMjByZE4ssVhUpldQttEoDXGoZfhAMC3I3ZMCwXTb0YMH/gV9cIPzs9/4DDBiGu9Voow0bNmC71KJa3aViqSYWi2SSOUUOMd+495757M8P+5wTN4LJkp6kLPdZAMG805nuzcReZ31rLUOAT9pcty3GBGp4X/YJ1Seh4JnMuapTBnjk1FQoN3TETFxvR4blMFUSKp7UGfd0SqaWDd8jx3ZjeDk1JZY5BR+US5ZSkkjJpDHD16JktWVOSYjHnZHP5dAns5anhz7zREgKCI1wd+Lxpes5l2N3bT6ZVxQ1DHyIPSH2hF96rWL/OGQcWz55NGEUCvc2PBaJz4+f+Hzj1z/kd775A7bHyttbPu+8cUp8ZZfl6ZiHH7/JJx+89dJ/d3r06NHj7wo9CenR4xXBe8d/9C5BCHWJBNHn/R5iHAGpq25sqoPxztSOC6SkG8UyxhEPbdWSZhrTGPdcVbnG9CpHV4aNSJibnKdlwf1lyZ89EOJAee8Hl3i8sPzwNCf2DLvHhr3H97j/0W1ublnm8wGeXxHFKZ8+npDlHj/+eIPD44gf3R+TFk35n3XmdCPCWzedYX1uK0pVrkhEQc3HMienJsZvlrRKhbroVDtg3piJj41rCq+wBOoREzDGZyVlt1gG13geE6K4u+yu6TzrUqdaRcTF0sbdSNR6wd9AYqJG7fDEsGoLA9uOjWYB7ItPJCFmrUhQGv9E95W23o9GwWjHtxxRqBrvinZ9G0BnSm8/W2vdkSQRodSyUyNyLdxIFQUiTuVoyVhF1Sku7eP2OFfq4oRXmpJoRqIpuRaN0dyQauaM6824lTt/03STuOM81RWVVgwblQIgbVK5TtR1vxzpgoqKhSb4eGQUlFozIsbgWsgLrViSc6oJS00ptOKFPcJrUrksSkbJFZ1SiDuHUmr2zIqhBmRSkYi7HjONAYjVZ18ydnTEETk1iidCYh2p+FmRkOO8HccUPJIVJ1XNtoRsGh8FIjxCPIb47JpTEilZSoGPsJCCbY0pVLnjDRjhE3ogAnu24M6VgtNcOc6UO1twaVKT5h4nheWNYEimFt/gend81zWySHzSQnjttWM+PK7YGCp3b6woKri1CelqSp5MmY1LvvHrHzKeLnn8V7/GdHPO3mHMdz8Nf+6/Pz169Ojxt42ehPTo8QrhvYNvvYsXdGZy8fxzRvIOLXGAblzrC8e21tQR8XynhFh7NpoFkKdQpOjRM/B88JVf/6V9LtshG8bdIZ54HqtceJ5VBM1i+WvXa4zAZ482SXOPN998SlkLUZwxP9ri5pWMk6XHPHPekdCDrVnJ1lh5517Cka0wwLODkDe2XcTpQAxe0yL9q2aLolk4b2nsRmk043WdspSSbR0wl4y5ZJxK3tyJjpjpgENyNjVmz6yaUauyGcmqeCYurrRNmKqx3QL6QOeNAd1S65n6UFCSaNpdslhCMi3INW/s4tKNEgV4pJp3I1AVVTfu1I5jgVNFXCu715jdvS4xqz2+dpG/7uNoH7eJWEAX/WsbpaSmJjYxmeaU2hjLG4JQadV4W5xSFEpArc634onXjW9pM34kTUqYbUhW2xIfSMBKMxa6arbjiG0ggSNq4nHCqiN8hZbkjbLirkvdkZZQ3AhZ0JC2QFpSV3fnaHEt7AOJu+9yypAZw448avN7SZvvNadix444MCsycaWGSykoqBmqz55ZkVPzmV0RiLBnVhxJRi3KGJ9JY4zf15yl1iTWsk3EGJ+BuAS2oFGHaizPJeVQUjYlIBDBE2EqPs9WluOiZlMCVqlPYV0zehxZ/vWu8tmez0lVs1+6MIijVElKeJFadjZq5onwlXunPNnd5K1Nn41xxWe7IzYnFRvjisP9Tfae7fD2b7+HGMt4Y58kCTk53ODJsXB36+X/PPTo0aPH3xV6EtKjxysGTU/RZeMN8YNz/R0Yz5EPazsyAU0cb0M25OKY1ZqvpFNEwJUR6vq2DSRL9OFH2IdbnJ6OuC4D9m1JbAyJtXxwUvIrVzx8A/fiiGFcM47gynbK195+wGh2yMY0x1qP93+6QZZ5HK4c+bh6OaWsYb4IOFwKSRIwMz5La/lkXrN7YggQnmrGytZsaMS8rjol4KrE3NMpudQ8knZBaUikZKgBAw24Zids2wETjViIm/+vsQw1IiJgpCEHzJv0LO9sAdwsNIuuvq41op+pD06pcOpHriWCUx1EnDKgKKEEpGvJV210bks86jUSICKfS7hyS3KndKjqOaViHe7YnRrSjnm18bzd70hb/0Lbml7gNWNh7XNVsz9wpMbDa5QVs5bMpY2p3ZnS2zG19lxUbZcgVmOZydgpSM3zp5o0xnavIzR1ozyVWnbXXjBsMGqKHXNCAvyGEJW0vwPbkY26iRZu+0cScSWXQ40I1FCLyxSr0YZolMwlw8cw0IAYn4UUJFJ25YUHtuCqHROqxxUdUKHcjSKu6ZBtCXkhruujQtkyAVPP40gyrtgR9+yMa3bCkSTUYnmiKXu24EmVM/Xc9xwa4e7Y518/rYk9oajhh7vCQis+SZ1KdEDOT8sl+2VFViuZtYgoN7dLwqjkeOHxtTcP2dpKCDzFM8qlSyvS1Od4HpMfXmH3wW0Onr7GztUjkiTkSV4Q/5Pfe/dv8M9Pjx49evytoSchPXq8Yvjz0z9+V5PFeaUCzsavvABaleTc62fFhMDnFJTPKSUtWalL8CO3/a0r1O9XaBby6NmQy4HHgWSERnhhcz6WOZ8cQForgYGNzSV3byTc+tLHeH5BsHHEYhny6f0dBiGcrgIuj5XLE8XWgmdgfyE8S2p+8sTn5tDdE9+JPH6WZlhgLjlXg4B3wjEvyLhrRhzIkkodGdnWmI/MHp4aElwK0+t2gwkRkXospeSajgjxeGCOnBdActyAjaXUmqWm1NSU6trEPQyxBE1nh1t4+/jdArw1ZceEWNVm5KnqRt3a6F44az4PmlGqdcN52ZjbW/9Gi5astGlS2pyrYFwcb7PoLhtFRcSlTbXKhhHnK1kfjwLcWJaeHRucmeQt2qg4Qq45ItKpQgtdoqjTjtQRllblaVUNaVK01tO7ci1d0haWlaZNxG7ISjMSzbq4YovtlJR5s6+2s2XYtLSvlzaOiLENwVlqiieGKUNCAhIKnJukJpeavBm/itSjEksqJRkFN+wG23bELbvBoUnY1pgKZceOqJtrssBFBb/GiLtRxEJKPspTBmKIxfA6YwZ47MqKxFoeVxlbGlM3HpExAbnUDDXgU++YOQUeQmaV0AgPq4yihoyaSeSStSpVDFBiSajZIWYpJd83+5SqhCKoCh8/Czg5HvOV1+d8dH+Lo6Mh13cynh+FZFmA8ZpRutNNdl8Meby7wenJhKI0/Paf/H5PQHr06PHKoSchPXq8gpDp9heTDADPRws3GtQaz7Wu3H8Xo3jX/CIdgggJIqeaGNP9J0GEVgYZ5jycWxaVJVDDSVVzWVzS1LKuCY1QK9jaoyw9jFdia5/Vs9vsXF6Q5IbAU1Th2nbG5c2Mz54NmA6UF2mNAKtaiQOY+h7jCMbiMRbXqP1X5Sm7RUGF5ZFNmOmA/SbC9ZCMYRPFukXEUCOmzRhQLjWfmUNm4rwAMx1SSu08BSTMJSVu2sAFQ9WM87TjWG2ZYNt90Rb8RYTO+CymGw2qqTuzd7tAT5tRozZlqlTnSam0wqpTLsyawmDXzO9Aoz8YzmiNPUcanFJhO2LSqh3t+9qODBE55y8J8Dqi0v2cmnQuD+MIjZ4pIudSu0TW1KGz59sY4o58NeNTy0YtGUjszPHNdSqbsTbproLgetQNI4lZaMqcVdPdEWBVqZr4Y1do6EbWQvGZMiQmpKYmxO8a7h/oHhkF+7Tjdrbz4AjCDR2x2aSlGYRLGpOLKyI8lIxjk/GBOWRJhWdgSyMOJGvM+sKtkc9pQ8ISap5KQkZNKZY9SblvjvHUnd2mHXCVATtewCObcGkgrjwzq7nuR4xj5Wrk/CUj8TmShhBiua5Dd71V2Yo8ylLYHiu7+xEHh0M+OLTUVliuAvZWyslpxNbmkrwwPHp4GYDJqOQ7Px5T/A//UU9AevTo8UqiJyE9eryKCKLzj9dazUUMEo/PyMM61vtFLkT4dkTErqkotu4KCjVPwVrMZsruj3+Vw6rip7pgiM9TTalQcql5beSzqGqySnn/pxv83x95/L//z98nS0d89slNHj7eoLbCMLJcvZwyGudkuc/ORsXBUvilK8LQM3gCH51UXJ8IT5cudahG2bIDPvYOed/sUYvykXfIPbvJsTilJJGKUH0OzIqJeNyxG1SqjDVkoD7bOuZDTjiWlDEhftO9kWlBgOv3yCmI8BnLgJSCAI+VZue8HW58ybhGdDnzY1S48bZKq67HIxCfUIJuJAk45+vwxe+SqVrzuPsuzwzprfH8541gORJkz0zpDQloyYg7Jkd21k3tRZM61aIdlYKG3DS/mVQzR77EjXu1xKnSqlNrKq060uU3VvXO49H0pFQ4klRqyUqTprVdOiLSkgBnlDeUWhNKwJgBx7qk7VWpOgJhGEhESs6YQff8GBetOyTkhBVxo1YNiQjxidTnUBcIhgOzokLZk5SJRlw2ATfNgB075Jodc0ljPDX4GD40h/gGCiyXNGLDd+NUp7l2HpA9SZ2KR9xcC+GO3WCmjnzt6JADckIj3DFDihpOxSlhtzbgwwNL6MFm6FGoZawBEYZxk2x2w07xRbixYfm3z2viUPnJcclPdn0M4HuW44VP7AmP9n1e7E3ZXwiHpz7D2PLZs4jv1kcv/S316NGjx6uAvqywR49XGZ3no0I836keDUGRcHiuB+Qs7eqvKTLUGlWDWNAqh2zlWtONQfceoYXh00dT5iQM1WdGwCkFH3BCLhXfXbp+jqA03LdHXGfKwyOfr6YRuwchWQWjEIynVJXh/sMZUaBcubziYD5BBCwwCQyr2hL6yvOqIKVmISU3GFJbZSkFMw2Za8RliVhS8kwSfIRSalZkPNaU6zLgPgv2ZMU3uEqp1hEQDZmLG9Npx5YWpKSa4+MjyNmiV1xHSNlE2pZaMpRBR0lsOx6l2vWFtNpAG8XrISim85UEElA241CeSEcC1sejarUYFCNe49BwkC+4P+SUi9YEfUZMz9QI023T4sa0bKPWeOJ1j9vxLprRsbY4sT02g3YKiFnbfltoWDVt86H4nUekaHwyqPPbLDU5dy7tNgotQZx5vR250tYU3yhIK01pY5BjQioqRsQUlGSUVFTkVFzTDfbktBkhWzGTMYecssmEQUNUB0S8UV9iz6z42BzzZbvFM1acas21MGBSBByQMSVgrAFlQ/aepTWnUjDWgE/L1PXV1MImEbVaHpslC5x68cgcM9SIbR1wVV2p4YyAXUk4rCrujgI+XOaMCTjVmqenhhNbsVjVDI2Hh/BGMGRVWyae4UG54o6MmIUuIexmHPD4CFItmYTCqoIs9/j+YcGdQchJYXnvIdybwOEKisrj/1gesdWkgfXo0aPHq4heCenR4xXDb93+F9+WaNCVFXZEY71osFU5vohwrPtDbO18H21PiBd041vr25NwgB4doUcTdk/gWDICDG9vBlwmZqYRNZaPzREPzDF/4X3GMSumGvL7VydsXXmOZ2B7rNy5nlIUhgfPYqpauHZlCcA4tiS5wQChB5lanp+6BXyBG/0KRBioz3U7dulEGnGiBQklx+K6OBJytnSEi3steGROuKQDFOW5uH1NGoPyIacMiNiWGSvNXLdFU9bnVAzv3KK/pQKxnJnV02ax2UbcrhcUSjOilWvRLejb1KpOiVjrCGnHpNpX2/6Plly0r7VoFQ04TzyAbl/gxqjWY3zb/bUJW+142UXy0vaWnBGQM5VjfZSsPa7Of9KoIq2y0RKdtvyxVX7CxmvjEsLcPjItiMWNRbVFg7kWHOgpqWbOM9JEHbsAgLprVE80c+Z1zcmltdY7RWVAyA4b7OiYpRQsSJk0akmb9nXNRFhRDsn5uEj5wBzyzCxYUlGKZaohgRp+pMcM1WcuOQkVRxTMKdmT1BFm8i4K2KJMiJwJXhK2CEmpidXjQItGQVEGeIRi+LTImBifm1HoRrUCn81IiI0Qeo6wbvge22P3PW6PldPCci+OeJbU+CIcLT2uBAGxD9PA/b0Jfcit8sPTnCNJWHIh5rtHjx49XiH0JKRHj1cIv3nrn39bs5XrCGlb0+H86NUFgrFuYHdpV/Xnycn6aJa1TgFpU7ba1+oKAh+zM+et6zWv65RnsuIogYSaGI+ZxmyrK/MD2NEpv3tpzGvXF7x44koIs1IYDt2CcRwrv/TV5wxHCY+ejilrYTKoKayym9QEIpyWlst+wGUiruFK74Z4XJaIBRUllueSkIpbUB1LxiUdkUnFUgqeSsIv11cYasADVt1pJk1HyICIAI9tHVNjiSVkQNSNELUOBR/f+RrW/DMrTRtzt6Wi7lQOwZUKerj3l824Urs4v5hstU4qWrTEx2sK916G9j1nLe1nPoyLasl6Kzu4Ua/10SxV99mW7LQN7O5345SPcyNbbfoVZ8TGFx/FEkjYbMORjbIhNq1X5XzilumiiT0MYxnSmvBbRSoQv+tpaY+hHfkyIkxlSKIZU4bdsY0kJqPgmm4wZsCGjAnwuWYnGBWOZEXUiP2HJqXEEuNTqTKxIQGGJSWXdMg9u0kplgNZsaKkFEsp7ryD5ug/8Y741ByTNaNmqZTMNCbS5uxUOJWCF2bJATm7smLXLFx3jRGueSGnlJRqKbGUqgwDGBNQWCX0QRXGIVwhRgTuHylJLhwshY/qFQ+ynGfWjXh9OC85rCr2UsvliXIp8vjguOKvyjmfySkW+Mw7funvqkePHj1eBfQkpEePVwjvPfqDd2lb0C94OkTM+QjWNRN6V0jIWgzvRSKy7h9pt20t2pQjarZCRmNks+bO3ecAVGI5KNyc/wfeAds6YFsH/FZ9iylDft+/ydffecr3PpySJG5M7Pp2zmoZce3KirfefMF44wjjWVRhZyvn0313XCPPcCMOGHjCJDCdtiAIN70YA1yWCL9ZjCZSspKCfTnlih3jYTgwCV/SKe8MhqRSUVIzwGesIQvJqVA2dYTrGRkykIhxQ3TANYXHzV11b+2fw7aFvF0Qe+I563dzjdsujfYzXhO1G0hwztTdmsbXCUO9RnLWn183fa9/vlUuzgzWZ4WHeoEoAOcUlVbZaMnLulJi10au4MzPAY7AtNv0GlN7S9Tcc6bzlaz3mlRaOXLWvD9r1KH2M23aWDuuZbGk5G40TgZ4YpoELXddBkT4uAZ5i7IiIxSfTIuO/Mw0ZqbDJivLYyUFj8wRI40J8NjRMdftmFKcErSvBXOTcyw5Sym5w4hTKZhowC07I5GKSJ369MjMyanYNwmBepRiseKSsHwME3Xek6FGnSoz1IC55NzQET6GhRS8KCoOakeK55RMCTDAfuquTWYtJ6kbU6wsLKi4X6TkqvzspGIvr1lJyTEFd/0BmVU+YcFPmfNRteL7+xV+81Oam5wQjwE+/+3/8t/0pvQePXq8suhJSI8erxriUaNQ1Gjb5UFLLtaIRVtWGETN6Ja98N/50R2shao6846UBVQ5Mpy6x0WKrpbUn8yYH88wApfsgL93zbBJyKYOiDDc8mLuxRF/FL3Bf/CN5zx6eJmouYsbBzCbpUxmCaqCGCVPRzx/eonbN5ZYK4jAVmzYjITLY6WwyqpUpsZj4nkkWvOiLqhRbsTO0F2KxVfDTGOuNEWFO3bUmIDhwzRjrAEPzBFVs2D1tBmTEjfqVFAzalQRD9OEutruLn5K3qVdKUqmJZFEeOJRakXZGNFb07Vp/AvrI0xtazmckYp1T8RFtQKgbPwVZwrA54nJ+j7az64rIhcJTEuigoY0WG17UepuW+uRwOvH1aZwXYz2NXLmH+nM91QY8TpTvosedmStVUDaMbZ2hG2pSUdUKlwpZ7vdpaaEBAT4hARMZNCNrAEUDVEKGuO8afxBU43IqQjUmbpnOqQWt81ADWFzLAklCRUZJZsaMZeMB6zYkxUDPDZxComHYUXBUzlhz6yYS8YYp/5klByYhJGGHInzvZRSU1ITqsdII4YEbEnAjh1RoXzEnJF4VFiWlJRYdjXlfp2wZQIOtOBJXmBVub8sO/Xv2sAjV0utyo4OuGuGjALhWZ2zoRELyRjg8WM55oerhH3NGaobI9yyZ2S7R48ePV5FeN/61re++Xd9ED169DjD4/lXvnN78+NvukdtAV27oF2DgvhuYYStXeGg1u655m68IzDajHdJQ0RKSJdOGbE1xEN0foj93ofYXaieXeW737+NZw3L2jKsQx6XOb8Rb/CrV4Ubm8o4hNd2co6OxjzeD7i2VTGb5kQBXL3xnOml55TZmL0Xm5R5TJr5zDYSnjwbgwoKlDVYK5QWlrUlMu75uVYk1NwOY57kJWMC5hSIwFgjphqRGdcF8TW7xQfmiALLvknw8BhrRCYVC8lZSuYWqk2HREVNRklCTqHlubvqcDaCZLGUlJ3CUTZ+hnapHjR366tmIVw0rxsx1I3vQ86+pkYNWf/23EhXu8w3GGw7RoXQZldJ93nF4BGI373miMTZe0zjD3H/b3tGpPt/e15tAph2W6ZZ0HtNKeHZD80dk9tDKEHjbXHej7ZF3TXBt6TA7bs1rnviYZselVACAvGb18AXj5gQFbeHjLM0soKqaUyH9dJEXzysujSvAZH7TJM8tpKcWiwbOqCSmiNZckM3WErOUEOWUjAgoEJJpWSgASNCPjVH3LObFNjmNUcyEacEltRMNaagwooyJKTGsmNHPDEnjBsvyICACJ/XdQIIu5JwanLu6oQBPinO6N6S4DYhbCY+h1qQULOvOSdSsE3EBhHLqvmGmzCHTd+lYT0rS7YlIqHGxyOj5mfePkupODIJHoZv/bP/vFdBevTo8UqjV0J69HgV4QdnqocffL5oEM73iBjvzHxOY2Jv1ZG6PBf5q4e7qNaurDAcOEXEGOpPL/H9P/k9Xjy+x3Gq/KBYcig5P0tyvhQMWJSWt3/5Y7Y2E16/d8APH8R88jzgzpWCK5dX3P61v+C1O08ZbLzAeC5BKvRrTk4jVqlPFOdUtRB4sCiUW5dqaoW8Vqa+obRKbIQdE5JSUVjlL80eE/G52RiNDyThyKQsyAnwWFIRq89CchIpuaJj3mGzUw2yNWNu3igfp7oiaVrNV+qyllJ1XRBR43VovQyZ5k2s7FkxYOtjaBURV95nO6WgbUFvx52AZkFuuu2sqx2tJ6RVSi6az9fhWsTbwr+zUSs4UzHAkYp1BcGNLq11hKypNOv7a30h7dhWa3x3RYln23TeEtdU3qId1RKk8Z/Ypp3dEbCWoEijjLSET5AmUNddP5dEZlnapPPkZE0iWa3O07PJpLteLzjhQJbdeFYuVXeOX7YbxATsmYQInzt2igFGhOyaBUGjrlVYKiyfmmMsyq45JaNkpDE1lk0dUKNcsWMidec8xOfN+jKKck0n7NhRo9IIAYZjSZnZiA0TcNsfsCurbnyvRrnlDZwqo5ZNQuaSMzfuV7qkYigep+oM8YnWZNTUqnyQZGxL2Kkec8lZSe7UPqnY1hF//E//q56A9OjR45VHT0J69HgVUZVgXR8F1r6ciBjvXNEgXgBe4NSPukTrEsocqsq9Lp5TS4yHhAOoXCwvzfuSvZs83Pf5k++N8Ixw2izsa5SnZcFPqyW29gmjktVyQG2Vq1Mlyz22r+2i8xHj6w+Yv7gNfs0nn15ikQS8duMYzyiffbbDILScZrAZC3FUc22jJlelVshU2Rq4UrgNQp5WBYryAzlCGz/AkICF5NyyM962W9w3xwR45NTd3f9cLYF6+BiGzQgNQIHzjExlRNQUD+aaU1EzkgGllsQSdkSkvd7tYnndy5Fr4Qr4Gp/FesdHa0yHJlL3JQSyfe7iGNW6Mbx9/axTwxnjv8hH0r53vUBwPT1rHZ3fQ87uyJ877jVCI8072gSvtjCxHWVb96C4z56Z0m3T/wE0LfOu6M8Xn0orUnIyzTHSkJRWNWnIVnuMius6CRvVY6oRqyYlq029ChoSU1J337urLqy6xK6EmgCPgToqeWASAjyemgVHJiWRkiNJKanZlwVjQi7rhIyKGJ/9xuw+1YgXkjCiHbmyhBi2NWbqexzj1JdbMuIjXZDbloi5iN89SXhSZwwb4jYxPqVYtu0A1+vixrXaJveEmrwhi3uS8iFzUlwwA8A1O2FbR2zogO1+DKtHjx6/IOhJSI8eryDaufpODZG/xmgOTQO6f850jjFnKkhdghhksunISrJw7xEDRcZg6xmxDwut+DTLWYobU0qoeN/sMZecTz++RZ6F7B8MeW0TaguHC4/o6i4HT94AKyTLMeBSsqbjEj8smU1Kvv/EHe84gpvbJcvEx/eVYXMeB5ozz5QXdUGMx6dmzi0744kcs9+0Vs9sxEhDrhBzTEGIT0nNjo44ZkWJ5QfmgD2z7K7LZZ1QUBHidz6CgURMGSJyVk/YFhoanDm67b9YbxBXXAt6O+ak2DNlQl0UbmvSbglBSxra5y3aLOLPG85bXFzUr6NoGti/qEek9Yesv6f1m6x7SlovSXteF30r6+SlJSrdPkQ6Vac9x7JRPNqyxFbRaU3mBuniewHihiS0/SCl1t04WutREYRYwma8y/WFhE1W1Z4sOp/JgIjLOmFLR9RYYgKOxUXzvu89YS4pAEsKHpo5Syk4lbxTTvZlQSnuOwjwWEpGhCM7rZIWYMipScT9vXhdp+Tixso8hLlkZNQM8FhUNW+YsfOcqIuVfmpzLmnMtsYkVFzRIQbYlghBeGYzhhpQNlfhmsQspeRjc8RSSh6YE+aSDpPtvAAAIABJREFUowpTDSmxLKQgk4px8/fhnp2yaWMWDTHp0aNHj1cdPQnp0eNVRmsurytnVFcXr6vaeDvANZ0DWiRn8bJB5Izr683rLTkJB40iYqDK0cUxeD7Bb845zeHvzWJeC93ib18WfOgdkEtFKTX/233lX/5gQFEZbt1YIAJf/+oB5AFpGnH69C5Pn004efo6b92bc/etn3B8OONoHpJZyyCumQ0tvm85WBhOlh6ewNAXlpR8XKQsqbgvpwDEeGzoiIG6O8aZVGzbIU8k4b45JlKPGiWRkt+q71CKU0Sc2Vu4qZvN4s75QTaZYBC2GBPgERIwkKh7f3vHvb2bvz4e1Y4ptcb1dkF9ES2JaA3eFxvQL3Z1nPu6Lygj7XPrUb0tkbqIi9ttR7/WR7Yu4mLK1sU/A+cJGNV5lYezlKz1dK32OrhI37bo0B1324TudYSpJUCWSCIiQlxPSE2lNYLgN10pkybZ7LRJ1mpjfWOCZljsbP8tGQrwyag4kRWl1BxJQiYVmVQMNWSBG81bSoGnhpkOyJvRtV05YawhqVQkUnDTbjCzEXtkbOqAAssjM8fHEOHxlITvmKcsbM2UoGlVF+ZSMMYnQEikwke41BAsD9g1y+7YxxqQq+VtmXHPbmJRZhpToy7UgZArOiTAcKUZAXsiCSk1mVSkcv731qNHjx6vKnoS0qPHK4j3Hv3Bu+89+oN3tSEfLQFZh3vNmdGpSjdu1TwWacazWuJR5hBE6OoEyhxNTl054eIYPX7RmNSV2iqPFjW3N6FCXTwpc67ZCYeS8BNzwM84ZTKs+OjBlMmwZvv6A5K9m1y/c5/lfEaSG148u0QU5/i3nwIQhZYbAx8jkOQGERdFusyEqxPhcVqyw4AlFV6zwB5ryAtJ2NSYRErerq+yo0NuMGRPlkT4xOp3d7EHeGzamArllt1kqhGX7IADSZpRnJwaF8m7IGWkMR6GqLkrH4m70+7jEpaCphOjVRfWuy/WC/zaxbeInIvnVey5qNt1vGzsqsVFgnJOhVhb9F/E+ujWOnFpPRxfRETWcTF5q93u+rmvn+d6l0h7nO1omofBb0jYegqZzxkxMQiluvv/tdYE4lFQErRFiWtjdhUVGQVxQxwjCZxHpDGK51IxImJFwVAD9vWUMREVllTOktBqamL1HTmVmis64836EvucNucLET4jYm7ohnusHttN1LNBODApgRoemjm5uO3l1NSixOrzF94zQgwLyfmq3eIuY44pOKIgVo+ZBNRAojUViq+GezqhFmWIz3NSpr7hS/6QSxpxRycIwmFV4YmwSciWxkwJ2DWnzCXjhST8F//sP3v3v/6n/2XvB+nRo8cvBHoS0qPHLxKsRfzIkYwm3UrCIVpXLqq3gWYrxAvQvYeQnjpCsjrpXm/fqyf7blvxCIzwceFGR/YXLvo0wmdAxFADCkpGGmEQ7j8P+DdHGR8/9/nsw6/x9PENgruPiYcpcajUVnjv+ztkH3+JwSDnZOlhmzWwZ5Sjecg4gp2ZW7Del1O2jM8mIRk12zqgpOamutGuVCoCDBk1C9xd7FB9suaub1s+NyXghp0Q4BGrz0pKTmTVJBK5caBtHbOjEwxu9Mc1qIdMZNiUEVpCguYOvE+Ad86w3n0VKKpnvejQxtjWZwqDnvdsfBHa8bv1salzr68t9te3v/55OD+O1e6zVSDWt9sWFa4TjHNxvMhZqeEFT8v6ObXlhOvjYV03ihjnA6HsvDI1Nbnm3XtFXNxvRd28vybVvPOFGAx50zYvCPs679SqGSOGElNSka+pQ0eypKBmQ0YsyFiQUlOzpWOmGlHiOj5KLBs25ppOiPC4rZeYNvG6kXpctRO2dIiPOOWkKaosxRKrT4THVR2zZYcMNSCXGk+FSzpk2444ouCWnXLJ9137ORGpVGRSs9SaUl0al1W4q1N2Qp+JBkR4bBBSWNgeCCN8BmL494JLzClJ1JV8tn6Rb+hVvq7bzHRN9ezRo0ePXwD0JKRHj1cY7z35R+86A/pZc7qqdSpIXbrCwiIBrdEyd76P9n3Z0qkoVQXZEl3NQS36+CPAERXSDMZTp4oc5lySkP/0K8LjtMJX48ZPGPLAOyHAI5GCF7Lk/0oPuG9O+FG24l99GHHj9mOqh9cBOF4aJuOMUQhlPiBNI+YprColCCwicLgUtqYVlzYz0sLdNd+3JVf9kGOTkUjVxZg+MwsKKpeYRc1zSZx5VyqOG6/IPRlzLXTEYYBPIq7L4ZE5YdWYkz0MQyJSKmICXsgprg+koFY3atT6QnKcKd4Tj5KzRfx6uR+cjUo5D4nt1JF1XHz/+sK/RVtEuf58SyzW31+/ZPstXjaO1W77ItaN5z8PF8sQu7Ew8c6M63KmvrRo+0LyJtWqI2yqTcqWM6e3SkmhJbXWVM0YVOu5aWN/Lc4I7+NxSkKqOStyXPitoRCn2LXhA8eyIiYkU6ecTHTQKWYxIceSspSMQmrmkrFrluzYEaXUBOooWiIFJTWbGjcpVCkRPjd0iI/hRHJyKsYacJVBV2AYqOG3/EtEGLYlorTKJ2XCjTCkpGasAR+bY04o2ZIAizIWj5Oy5opEjMXjjXDAsq7xDAyNR6FK4AkewkAMe5qzRchlL8QCX5r4/OM/+U96BaRHjx6/UOhJSI8erzqacawuKaslGlUFRer+Lx5kjRlbPPd8kSJb1yBqPCBh7BrSsxQt0sa4LpAmaLJAE48/+g8/oraGF5qx0cSSjjXkmp2woUMOZMmQgLlkHEpCieUrOxANjzl4eoeyCPAMrFYRt6+mhPGSovC5uWX5lVsl25fnJLngCVSV4fn+gNlQ+bo3Y0nJ0HOLuLzp3SixvF5vcSo5b5lJ0/+RNYlYFQaYaoQAHxcpBZZTKTiSxHWBUBITYlEi3AjOQlI+kwOWmpJq3hX6FVoR4pKc2sje1gheNAblNob2Iloj9/oYFHx+tGod50oo17/ubnFvPvda+1yrmnQKiHxeATmXrLVGXlq1pFNvOCtabMsazx/nGXFqR8xso660JnvLWQpYe84ldRdR3I2INeNpFqVQR/siCV2xYRNt3Hpz6oasBE1bPeB6Xhpis9KUYxZ4zfich0dGwYjIkUsKJjLglt3CAHMSTiVnW0eUVGzqkFPJeS6nbNoYi5LiFLehBhzKCoOwoiRWn00ddl6R9hrlUrOQglFzk2CLiC0iQk+47sUAPLc5UaMqvaZjrpuYe3bmvh8RAjF4IqRqGXqGUAxp7QbWPjmtKJrr+zyrGOKRqsVHKLA8q3M84Cj96wlljx49erxq6ElIjx6vOP7s4e+/ez4py3M+EN93Xg9j3OO66uJ6tcydAR2cYlI2IzBBiFy+5hKx1EIUoasFZCvM60MGX/+Qf3s/5IrE1GK7u8dfZsodO6PG8mtcZkuH/NHgHr82mHD7tROeP/wyQViSZyG3rzqjfJb7fPDDr5BkHk+PDTdeOyCIcmZDyyCEvBSyUvAMvDY13DVDfpKv+Bob3NEJJZYtCXnLm/CN+gaKW/TdsjM8dSpJgMdMQz5hSd0sIjMqEnJm6s5/gxEBHgEeE40Ya8xS06Zo0FBSEUt4zvDdRr9adQlP68bwNnK2i7VdH8d6idfjZUZ0VftShaJ9rcVL+2HWnvuibazv/+K4VXusTp04r4isb68ds3qZYvIyQ/v643b8ah3r/SO1ukStdjQrlICpDAkJuhGu9WOOCLuOjYFE1FhG4r7fOUkTXSwsNWVLR4T4JI1/pD2j23oJcClZQaOWuQ6QIZuEnaLiCBMMCBlryFOzwCBMmnGnlJqZhmxoxJfsjEos+zbnho4wwEAMu1nJ/XrFE03YlRULSo7Lmg0TEBshpSaRipWtuRH71KqEYnhtaghEXGw1rnOkUqVE2Qzc3/8dzyl+y4YQ/c4bNU/LPhGrR48ev3h4+W29Hj16vFJ479Ef/M1GLY7P/vibG//825S5UzwWxzDZRMRDgwj2d2HjMjIYg+ejp8eOkCxy/v1fmbNYxPybjwpmGvPr/iX2y5KvRSOOix0uxx5/MNzi9VunfPp4QpLEDAYF8WDF/otNxuMca4X9wyFXLq+4/3DKrcsVn322w8Y0xxgIfSXwlShQqlr4q6OCL41CTleu0dr1ORguBx4/K1JuejGf1CsWXs5UImqx3KhnABxKxjsy4yNdkosbnznALRhzKoZYrusGB7LkwCQuKUtrZ0EXw5F1Y1m5Fqi4vgkRg49PbOKOWAQS0PZN1M0dfk+dB6T1SawrDOtlgi3puEg+LiodcEYEfh5RabFeWtjucx3tNta3U7fBBWvb+NwxYLp44XVyth7ru54a1qZjrZO1VkGBM+KwHlkMEEnUqTTnyFCjhhiEXEv8RknwG2+PqiUQjxEjDphTUZFozkhiEkpcBWXFhg7ZNaeuM0ZDDMKhLNjSMUeyJBWPmQ7ZlRVbGjMnIRTXIVJhKahJpcRDiNWnFuVAMt5kwme6IkA4kBVqlLfsJttewP06YUciHsmcK3aMh/DULBALRoUrGuNjmMsKT4WvBiFBLrywOcvcZ+IbxiF8tnQlnkEtvKgLdkbCsvKZ1zXbJiQ2gifCjx8JGcnP/Z306NGjx6uInoT06PH/U3yOuJzCb0z/ybcJQrf4PX6BbFyGukIfn4AP5oZw/Z3v8Zf/5zf5FbvDxzLntKpZUbGqfN6RGU+zit/99ef85IMbXNkquPPlDzh+cYv50Ra+b0lWITdu77J3cJfr936CtW8zmqz48GdXsdZgRBkNLLV1C9sPXsCckvsreHMc8sEy57FZ8Ibd4JMiwwDv2yPuMeUDCj4wz5kxZEZI3SQL1QIHJiGRkk07oMKyLwsUJaVgREjUjFrNSRhKxFJTRJ2y0drLBekW6O3C28fHaooRl4olEtB2b1RU3UIdzrwWLzOYO0XkPPHwxOsW4RdJxxeRlfb5dj+qFrs2uvVFxOWib2Pdc7J+vu2xlyjmohdkLXlrXeloz+1lOPOTNERJaxDnK1G1eOKhKEkzAgdnxnnn0rGkWhJI4Frf1XlOWs+IVWUkIQUVPj4ZBR6Ga2wR4DHWkLmk7MoJUx2Qi4cVJSbgut3goTnEb1S1azrrCgC3dcQlYk50yFHjPfIRJhpSoNSiPJYlJ6x4TWdk1OzVcCwZS0rGGhHhMdYACyykIFafiWc4qYTfkB0OKPlsUXFj6FMmIUUNkwjiAK7FPrXCAHgjjLl9NeUoGbDIa67GHpEPgQf/6nj1N3D39OjRo8erh34cq0ePf4fw56d//O6fn/7xuzKaUv5Lj+p/f0D9b5+huVB/eAmZDcn3r/HpvkeC6wb5U/OUx2bJ+/aYn+gpsQhilFUubGysUOuxXAz58P4WqkKS+Qyme0RBzXz/Frd+889YzMdszQpWiU9eGqpKqCpBRImN8NVwiEFISxiLx8w6g66PcCcY8JqOSXBRuFs65oadstmYegMMH9uF8yWo4ZE5YkVG1ZjMZzrkqZyw1cz0t8lLrY8gJiQiJJY2oNd0vRTalAoGzb6MeFRUZ3G0TTvF+sjTy1SJddN5q0y8TAV52XPrn1n/3MVRqvVt/DxCIhcIS/v+7vjWRrAubr9VetYJ1nqc73oTfHstPLxuvMrtU7pttLG9FhcQUDRJWAbBF49IAnz85vq5Y86a0ABwvpNAnD/Ib76bYeMJ8dVwImet467I0GemQ0pqRhpzRydNCaLPM1lQYrliJ+SUROr+DuzYEbk4AzrAVYkx4lS2Gss13WCsIQk1H5ljAjz2GmWlxLItEb8sm7wjG3zNn/D6llCh/MbrJV8dR2wEHrOB8tUtn4Hvoqsng5rNIcxiiH0h8iDJ3D3DqfEoakiaS3rHG3Db61vSe/To8YsH71vf+tY3/64PokePHn+7eMxvf+fp1/7hd8r/cfHNIBnyyZ//ff7sL77E+PE2f/G913iQlHxgjnjDbvIWMyqFhSnY1JhAPT56OOW33j6lrj3S5QY//XSK7ylvfeUhYSCMLu0xmxRY6xGajMNnNzhdhGSFi+rdmhUsE58oUOrKw6rgiyGvlVyVyybiQ065RMSJrSix7MqKE0l4w24zxEWkLijZM658rh2V2mDIXFJGRIxlAAIJBb54rJpiunbZ7LejT83d/fXSwlJLfPEp1JUXKi7Fq9QKhGY5bs8Rj7MmdbrHSqtyCLhH3Z8V7YjC+utn73FoP382XiVrr59tr31tfRvdZy8cxxlBOZ9yJS/xsEj3Pl175oyotEdixF0Jdx1sk2zlDNhKMwrW7lW0cZ24bVVa4TWpW5GEGHEeEgWCpsHc68ayfNe03iglAT41ljEDwPkpMikZEFI3+7EoV3TC3DhVY0KEp4YRoftuxTCXpOkDMSylYNfMmemAlZRsaMyJyVA1eAiHJiNQjxkxqZSkUjfXDzY05m1/ymUT8tzmfH0jYmtg+GiVc2vkE5QB1zdrstxnawinqfDZoiY2hmEIh0vD85VlUShHZc2yUrZjg7XCsoRpKFQW0hK+/D//3rtX/vDN7/wN/tr36NGjxyuFfhyrR49/h7H33/3hu3vNn//0H/1P3/7Rd0N+ap5gjDOBLym56UX8ij/h/QxumQEHtiSxNUkSsXcYc/Vywpu3lzx9MWL0+sfMv/sPkLDCC3KmV3bR5YCi8BADr9895KP7l5htrHiyF3O0cv8E1QqewGFdk1HzRhyT5CWfsWApBdfsmEosMx1ybDImNiSl4sikeLg73qH6zGXFkIgNRpySMGHAIQvAjWFF+MxJiQlIyc+lWrnjqLtFsBWX4FS39/XVJUD54rt0KHH+BMvnG6pbsrCufvy8kaV1XBzDWveIfNH719URTzzqte1c/PzLxr7Wt33R6wFn6V/dcV0wnXcGfLV4claoKA1VE9w104aaRE1buNuf7VSmoBmz8sTQJlyFBA2x0y622RePnKLbz00u8VgO3FgXBQEew6bfJsORxgCfU8lJyBnhvCM/8w7YsWNycV0jV3TKgaxIpCTWgJiQKSGqMG8iefdNgmdHrigTi6dCLcINHTHEo8QyFI/tgbC7shxKRlkPiQN4Kgnf2/N4ZyvgRw8jtobwkwPLrZFHZi3zQoh9w35WsxV5HOY1G75HYZWiciEOX9uBeQKjEFa9H71Hjx6/wOjHsXr06AG4VuinknDDTngshwTq8YG3z9Mqp6iVd8Ix25HHP7wW8rvXXXnfZ4dwdBwjoty8tiB7dJtPHmyS7t2gzEbIoACj/OhhRJIZjo8mpIUQD1KOEnh/mbAolGVpqZrE4A0T8CAtuGpH3NQxAR6ZuKGfIQGeGsbNwrSlCIJ06V37nBKpR4VlTsIWrvAwamzNEb4zp6+NArX+jkiiZvHtRpRKLbF6Zqb+Iq/HxVbxi2iTpv46z8fLCMcXkY8veu1l5vS/bvRr3Wfy0v2sETWvGVVbVybac1Ys9YXSxXWzvi9+kyrmCMV6YlfQRO2GBKSad38uOEvR8sQQik+mBUubEEnIVIaciiOjKzImuKJLD8OeLJhLQltweMNOuK4bBOp8KCUVD8whC8mZS4KnhlxqIvVZScaWHbKkZKYhpViGhEw1okZ5gyk+wkmjrAzx2Al9huJhAd+DkW8YacCycCWdIw0wzc+jtMruqfLMZhykiifCvK75cFFQquIbeHvH8JXrNam1bE4qagtWhayCrHREpEePHj1+UdErIT169ADgP/6TP+yM7H/6j//7b48JmZPwiZlTlhOuezGjQPgHv/Me/+u/+G3u3Vny+nZEXhqevRjx+t1DADwDf/mXbzAdVWy+2GHn+lNmMSxSYZlFGIFnu5c5LS0BhsPS3e3dLXNyLLnWPDNLftluAxCoIVKvaTevuKETXkhCiMcNO2VCwHMJGBNw127zwizY1hEzjdk1px1JcYtmqLAkmnWJS64ITym1wjTqRamF8y2IK9PzEZDz3obzbeEXxpcuJFy1+25fe1lq1XqCVvv4i7Z5EeuvvYzAXPSGXFRYviiRa10NWScVX9S27uE1hMOd7bp/xF2DsyQtT0JHWnBdJL7xaROxqkb5qKkptMTDuP4OIlZkRE2YgPOAuHjdFgk5GQVtTaWPT0jALbtF1pQRzvA4kZSJDjiRFSkFEYHroFGPe3aDH3nPGeC7RDWNSSh4zc4Y4nEoOSdaYEWZaNSoXMqDIienZoDHPPOZhsKsCNjLK6aRz2UiJsZjkYEq7FclJZaDyvXRXA19douSNzd8fnhcUNmQt2//f+y9WYwl6Xmm93x/rGfPrTKz1q6lu9ndFDc1RVEUJXAEaWxytFgbDcEYEx7DgK8NCfaFDSLnxiPI8PhiYA9g2IMx4AttfeGbGV8ZGnisjSOKFNfuZnetnVW5n/3E9n+++CMiT2VXdVc3qRGrGU+ikZknI+KcE5Gd9X/xfe/75szV0OulrCcer+0JLR+mhTJKYfC234aGhoaGp4OmCGloaHgb/+yP/ru6IPnd3/xfvnyONv9G7/Ox6Qbf+cqnUIU3bq7z0Y+9wbe+cY1BL2WxiLAHmwBEgdKOc/b2O3jeeTYGBQdDpwe5en7BN2/FXF+BeBSRWmWUF/TEZ48JSSlAbxvDnk25qj1WjM99Jswk444ZsSdjNrSLQVjTkFWNsLgU9Yu2T1cDbhtnW3rIiJDKXleZa+IE7eKRqlvuBviICJlmdWp3iNMchBKU2g5XaFQjRx6nuRcVjxOKA6i8vUOyvOh/nKvVOxUmjzrO2X0ed8xHvcblx5ddvpZ1L4otOyAFZ0Xqb3sequKmGnez9UhW3QVRLbNY5HQ0CzfSlau7FoH4RBKwIGWhbkQrwHVEutIqnbIsPj5zEnItmElCXOaL+BgyKXhNTogI2LJd5pJRUNQBlR6GnsZMJeG+mbqumXoYEcaScNH2aeNx2Y8Jc48hKYOyALkgMbko91iwqTGC8FeLMRekxZoJOLIZx3OPDT9gZi2jVJkUBTnKgJBLUcjFFeV4KvzUWsDWxoxvn7g2x+5BxJqn5HnB5saMk2mXSQJX1yB/kvm+hoaGhh9SmnGshoaGd+SKXSHH8kBGnJDx6r2I7YElL4SDB5uIgcIaZrOQe2+tMEuEvaFhdX3MCy/d5vi4xd6JR1ZA6MPm9hEXVy3TBJ49p1xbFb7LCAFWNeQjsspH7QZDm7MqAQPj0/ENV2yfDdvhWOZsa5+uhhzLnAPcqI6PcCgzjmXObTNkJAkRPi0iAjxmpKRkdCR2o0Dq7qRX2oS89HkCV4B44tUBeeAW1B5erXmo7vpXo0Ze+QGP72JUi/qzhcU7ZYO8W17I457rcdud7ZSc7cgsFxWPcsdyY2VSJpU/PMa1rBXxlrohy58F485h9b1IbQbgYUg0IdG09J5SJ0Avi5GJzimwZORk5RYBHoc6ZkCnPn5LIjq461wVIvfkhIyCIyY8MJMyOb1gyoK2hmQUXNA2AR67Miw7Mqc2xjmWEI+7eYKWxcOAkACDL4KHcF5btMX91rTxyNQys87X62aalB0PSKwlVeUTKxE+wiCG7XNz9ueWOCo4PI4ZeB7rHWVvLKxGhqIQ7ux2OL+WMWhBv5Ox1j/tzDU0NDQ8bTRFSENDwzvyW3/8WzuFuEXmnJx5plgVts/Nub/X5dLFY97cjVAVBr2U4QKubqXcvLlOZ/MOo2nAhfWMl65N6bUsaRKSZEInglli2J8IL9DHiMum2Iw8LkchHfF4oAktz9D2XUYDQIDnguNQUgoycQvFvvj0NcIChzJ9aDGdlYF7la3qOXHPB5QahAWJJuVdc78eNVKUWCI88R7KDYFHaC94/JhUrQnh7SNVj+tKvBce1/l41MhXxdkOxuN0IWfth5eTz8Et+s8eq7LtXX68Kkrq11LpRDjVidTvBRceWZ13LTtYAH1pl6NalpiQKUmp+ciZa8JcE2IC1ko9UUHBjOShgvKICeu2RZuo/N2AbdvjnkzL0S9n1TwgpKcRl20fFVfovmqOyLAsys5YD5+JLZiVeS+huNGxHgFROYpXYNn2Q7aCgJZneMsmXOsEXFh3IZ1hOZOw1TakqWFjbc7VVVe0D2JYaSvWCu3YMp17bK+nJJnH/Pd+5clCTBsaGhp+CGmKkIaGhnflt/74t3Z+qrhCn4C9LEcVsszjYGQYjzo8f2XG+vqI8STEN9DtprRaBcd3nuNk6hai/cGUSxdGvP7GOotM+MiLe4S+Ms+VG32f2AhrhBhxot22ZzgnEapwklreEjdeddkOSpFwwDntYFEmZLSMoafuzvJ1u05GzoyEAssCZyPkY5hrQk9jwLlhpWSIGCKJ8MUnKMXTzoXJJy8LmNO7/c69yWrx0MJ6WYB9lmV3qbOL/Cpw8HG822jVe+FsEOOjWP5Z9XUVHggPZ6M8yiWrKrQql63qexHBq1zCtHQbw6tteTPNnR2yWqQ0CnDbFghCpjlVmGGuOR2Ja0tlQTjRCbEEhOLzjF1nJmldACzICPBpE9GnXeapu9e+RpeZZAjCfTNm23Zoqys+BhLgaRnQqDAiY03btPEZkbLHnNsy5bZM6zPxwCZMyDki5bbOCMTwkXabZwaGwAiREc4bp43aPwm5tF5QWHhrr8XmoMD3lMHKmPWVjL2RsNJxwZ63H0QMeim+p8x+91d27P/0i00B0tDQ8FTTFCENDQ1PxH2ZcdOMuKdzbh3DaBLQayl37rfY2Dokas949vk7XN7I+Tff7NCKM/7q6+dZ61oG/YT7uyvM5yG3juHDzw45Oe5xMBFurEMll/BEmOWni/St0OfVfMqDPCPAkIuzRJ2REqgb6bmobVYIGduCDW0xkZShLMgoanFydSd9QTXmA5m6+/WpZlQxheAyLkRMnWSealZ/XQml5TF/Oh+lCXmoE/GIIsWi7zhu9W48qpNyVuT+uNf5KLH7495b1cl51OjVsni9etyJtZcKFnUFSiBOg2HK4gMgLz+cY1WBovj4ZJrXgYam1IyMdEbPuNGrVDNSMgLx8PHw8eltQi9MAAAgAElEQVTRYiwLxsxZkNUdmROmZOS8VJwD4MQs2NAOF22fgoKxLMjI6eGcsVJyppozNq6ATUvL6oGGzCkoRJ1onYIjmdEWj7Z4jEgpUGI8Agw3dcKltQKrcJK5hPf1yON4oQznoBbW+jm7Q8GqkOaG117fZPcgZG9RMEsMw7nriry1F9Npv90SuqGhoeFppClCGhoanoh/9Mf/cOemHHJsFnw1HTFdGK5fGZHmMB31eOP1i7z63csYgW4gHBy12J0oF89P6PUnLBIP3y/Y7Ai797u8ca/FRlcpLJzM4eqKkKmz6h0Xlgd5xqvJnPMSMyOnpT5tDShEWdM2HsJAIwACDG8wYULGjIRdGdIl5lndIsDnIut1mrcp/+yluDDCSEIMwrr0AYglJNe8XqCHVVo6Tgfhi5udkTNC80dZ4dY6jKUF+6NE4I+z0X3oGO+gDXnUeNfyfu9mx1vlmFQ8KvW9emy5K1S9N68u4MxDn5epOida6jycLqeoCzzXGTFUgYnLVr7VnkW5fSVKjyQk14JUc/rSZqiTOrTyvA6Y6aK26000ZUFGl4A1uhzLHKNCWwOesWuMZcFFXeF7MmJNOwxlzjEpbQ2YiTNLKEQZS8YxCQMNedauYIEN7XCiKRMtWCfmxahNXI5kGYTCCnkBm7HHrTQlKZRb+ZzDxHL/xCP0LVc3LLsnhlkivLovfPskZyXweGukFAoPxspk8fgOVkNDQ8PTRlOENDQ0PDH/5A9/e+e/+KMv7RwZl/OxWARc2kzZ2+9yMPS5d+hz/vwJ17dyZonw0Us5W5dukec+W5tTjk9aiMDh2GO4gMlCCH1lmBWs9nJiMTzIU/ZtwoKCCEMg4kLhEBaSs6YRirKQAr9ctN6XOQcy5aB0xLI469RYAwI81mybDjFF6eZ0KGMMQlz2QCzKnIRYnCORILUzUzWaVS2cq8Wz6mmqeSU2P7uYrzhrZbvMuwnP343HFT7wcKHzuI5J9forKj3HQ9ueGb+qCpGqO3JqVXxagFQ/Xx5ZyzV39rycCtOLsoipOiTu+dzzxxLV+1VFSSQhY53VYnZfPPrSdo5nZQZMV0NXqIohJychJyqv7TEJ1+w6A405NDNyLH0N2dAuYxJysUTqs6FdegQEahCFG9onoSBSjw4BER49AvoaEqvHnIK7MmFGzmZPOSl7Oi+HfXZPDK1QWes4HZM7T8JJkfPvJjPuHYSMZoavT2dMU9cZHGvOpVVlXFhUYS/L8QzcfhA9wW9FQ0NDww8/TRHS0NDwnvkv/+g/21nkyngSsrY6obBCYWGRw8aFm1x5Zp/CCjeeu8tbt67heW4htXluwjPn5zyznfDMuuVgpkwWhusrHp5RWsb9SVqTkHMSYoFElahKotaAFMuemZJRcCwLAhE2tRzBESdAvqHnsCh3zTEhPvtmgiB0cXauUxb0pFNrDABmmmBVydTZ8hblHXSg7n4ADIwLP6wS0+HUGavuCJwpRpzO4d21He9UNFTHea+cFacvH2e5K3Jqmft28fpyAVIVF2cLl2r75ULFPcfDAY91yKQWD+1vEDzx6Eq7vi6JulGoqkBxWpGCRFPsUnfJx2OqC0LxKSjYNWP2ZVruY4hxxeiW9llITl9DEgoGGrNvprzmHTEjYygzWuozkYSrtk+AoYXPTHIGns9F7bBFzIaEzEor6QFhOUBWFr8E3BnCRDKuhhF7Wc7eoiAKlP2JlF5djitxyHNhi9dHOcMFbJuI2IeNlnDJj8gLuN73sKp89oqw9i8+v7P2Lz7faEEaGho+EDQ5IQ0NDe+L53//CzsZcBeIf/v/+nJWeBymBffeeJ6DozZXtubMJ12ms5As9egNZpwcdxkMpuw+6LO5MeO7ex2OZs4B6OaDkOMiZWB8jqyzHg0xjDWnKBfIgZr6c45igG9xQoeAGSnntE+Ez8BGJOJzIBN8DGPmZBS1W1KXltODLIUPurvoeT2uFRKQakZQFiA+Pgi1M1SACy+sLGtRLXULjkrrUXU6Coq3hQqe1W08aVL6k/CkgnZVW4vDH0dlR7zseLU8clWFMbrzwNK2lqI8rCvqHrY1tihWc5e7Ur6GygL57D2yjIK+dJjrwrlhlSGGBQFzSQklIMd1WTJyDmVEH9cBi9RjImmpUFG+6x1ywhSjPRaSM2HBT9srfMVLiNVd53UJ2deU636Lt+yEbxUjDMJtM+aK7QGw4Qcc5TmXgx5fzYdE+CwoeCOb08bnXppyNY4IDPz5bsal2P12fTed82LU5lxXeWvktFAXVywne4LvQa9liQNnI70xyFH1Kf7pLzXFR0NDwweKphPS0NDwfbP4H355Z+v/+PzO9b7HN17v87W7hiAsSNOQKCxod1IO9nvc3Yv45mtrBJ5ijCX2hIM0Z7VruTXNyVBiY7glE+7JlHvMGJeFQoTHS55b/K1pmxWN2NY298wJu2bCBV3hqh1wXnuMJWVFI/xSSF0lps/qTBHnklVZvVaaA4Op77BX3Yxq7MdiKbRgojNEpNQo2Icsa6tRpsdpNJaLj7PZHO9nJOtJ91kORHy3fc4GFj7q67OcJqWfbrucI3JWkF+Nsi3bHvv4qFqmuqjHtTxxnSUnWrfkFLiIwdORsZnOScmIceNymRb1qF1lwTuTjDkJu3LCAzPhPsdkFLxkN4jV56KuEOPxQrHJTDJWtYUvQkLBUe7E8ftmyqvePs/ZAXsyx6Ls5xkHJBxmzm1rJAkHMueSFxPjseEF7CYZ3QhyVe4tcq63XLDm+YF7/dPc0vcN59YXDIucwym0YzcKuD8R0tTQdD8aGho+iDRFSENDww+M1f/98zvHCyVR5fXbXTw/Ry3c2+1x/yjkq8cJkwR2jwJEwBMwIvS7GW1j2PLd2M2HdMCqxgxNQkpBjuXFoMPVFcOUrFxkemwbNx+fScGqtrhthnQ14Jr2OZQ5a9ohwCcqsygqxjp3nQmEFmXaOlktgq6E0b74FGXxkVPgiVdrGKpk9WoxXeki3k0Q/rjcjrM8SYHxXrsly2NYy12aR9kGP7Qf9qEOyOOsiKtk9OX9qn0eRdVZqTQgBQ+fs1Szen8pxeXB0tjb6WiXG7FbaIYRqa/GkBlj5nQ1ZMqCAS32GdYuXyvik0jBNTvgezJii5jr2ifD8l07rjVHA425YVf5bHGZn9gI6WvIFdNmxfO54bXpeR5XtUtKzvMMOCgypuScaxnu6IzXhwU3OiG3dUrowYthm1kipLnQ8w0X+vDq7TbX2yGDGPr9Bccz9/+H1UaM3tDQ8MGkKUIaGhp+oDz/+1/YefmPfnFn8L99fkdEMZ5y6cKIi+cSnmtHrLSVcQI37/ZICyU2LgMhV8UTYVgUriCRiF/rbNEh4OVWD0/g5oklE0tXA26aIZkqLxVbdDV0SdriBOxrxq9HuFxeiEtLrwqMApdF4ZLR3Z/BqgCpOiOVOL3KEim0KO/wm1qz4MTqHqa8Y+8Kl4BQQjzx3jYSVX1/tng42z2B738cq3q+6lhPkrp+9jmXc0LOhg+e1X7Uz3mm+2GWztVDPy/H1ypxf1r2OIJyrCpYyibxlgqfZX1OID6KS1AH14nqEONj6NFiTkJKRk8jPAwhzunsI/YCHSK+zjHPFmvsyZzndcBdmZUCd0NY6pDuMC1T2i0Tcv7sIOGG1+bIZvQDw7AoeKtY0DYeA4251vF5U0Z08EkL59x2185RhQEhaQFrbeHWuEAVOqEQB0row3pXaYXKnbd69GPwDAT/8xeaLkhDQ8MHkqYIaWho+Fvj9u/8w52jf/zrO+evfYvZwufCquXNY6UXwclMeD1dAM6i97k1w0lesNCC822PQIR7U9cFKaxbrM2s5ap23cJOjvkbTjAIAR6C8MliiwjDic3JxbIgZ0v7pGS0iCjUEuCzKl3WpEcgHjNd0BWnEamKAS0/Kgq1JJqSLI1wVcnqQL2Y9sUnlIBAgoecod7Jgvd0/x/cn+PHFTuP2q4KDKy+h7e7eS0HDy53QZbtegVTC8zr4y8VJFXRUelFnGbmdNuq65HjnMeWwyGBpf3dz2IJ8fFpSYyWzlgAC1IC9WipzyYrKMoDMykLiYI1umxpm6t2jTauMDqQGRfCgFUNuSnjcvhLGRCQiXu/Q0lIsYQYIk/oic9JWjAmY0VCOp7hRQZ8c5IwkYRNE3J/kTOTnBiPw8RyyY8IPejGykItV88vyAoYzgyXNhfMU3c+sgJevDHEa5ogDQ0NH2CaIqShoeFvne/89cus9BOsdfajKx1LWsCmCcmss+gdL4RYhALla5M5mSq37JwNIloBDFquU2IQ2ng8azd5y5wwkRSj4nJExCPBckyKQegQciQzYkLW6BJJyKq4sZmMAsGwIj2qPIpKPL48nlSUS+ai3L7qhlQIUlr3noYdFtj6jv3yAv+dwgPfr/PVk/KoTks9nvUY567qsbMBhWeDC6txtLP44j9U0Eg5KlUlp1dUoYZAnRtiyvPq0tJdpktWhkVWjwfi0SptfCuNT6YFM3FZMTMSurSYsCAsC44VbdPFdcpmZNw0JxjA4jouG9qiVRY0xzht0YdZ4bx2SMQVWX+aHQHwup0SYIjElVttz3Bbxly2A/e7KsJL0mfThNwu5txYE0aJEvoWH+HBYcQkU0YJ7B2599FtF+SFcH+vyydeGD7p5W1oaGh46miKkIaGhr91Jv/kP9o5+se/viMCvcBgLSxy5ULbw4hwbeDxzcmCzZbnBL0m5IVVp+N4rhsyz+DB2C30MizfMcd1KnqG5WeDDQDe0Ak3zZC7ZsRQFgxlUd91H+IyRFJyxjpjhQ4dItqE9eusckQKLci0ylZ3i26XqW4I8OiXid3V4rsa7yooyMlpSUSAV3ZFQvxybGuZxzlhfT+Fxbs9vkylC1kuAKr39KhAxYeOv+RwVXVDzupKPDxydT0FX/zyuA+PdFXi9aprpKpkmtWWvNUIWGUEoFhCCWob34iQRLPayndFesQSMiFhpDMWZAx1SoTPR+1FAjwu2B6hCBNJGEnCXFyex/00I8OyLiH3ZcIdMyoLIWGulhXx6WnAGzKmrQHrgcehmXNIwlf1mDtZwqv5FIOwQshmy527oc25PvDYlIi/2S+4m6Xsnng8O/D5//YT+qGwm2R870jptyz9bsa3T3K+s2u4eaf/rtexoaGh4WmlKUIaGhr+vWH+2T/YufJ/fn4nK4QLfeHqVsq4KLh6Yc6nNyJyC6Zco26tpmRYuhHMCstukjGxriwYaIyPsKIdFy7nCYkU7JoJM8mYScZAY06YMmbOVBdMmNfjQm2JyMiZ41K0rZ6OX1UL35ycUIIyv8LUuo+2tJxdb7mtE6sXpY7ktCNSjWZVXz80pvSIxf3ZpPPvl0c5Yp0d06o+F0uZHZW18DJVsbXc1aiKgmpUa9l21yAYOXXMqgTrjypWqkLFogQS4ItPIH7tTpaV1r4tiWoXLU8Ma9JjqBMSTevzvMpp9ktQXpeqU3Isc64UA3Isfy577MuYhWSkZGxplzE55ySkYzwu2h7b2uUcEQaYkDNXy4fCNuCc2nbTjEg93HAgzMrzMJKEy2FI5MNtO+NqHPH145TzsfuduRZFnOtZNgY5nxjEfOT6jFAMqVWGM0MUZby44hMY4c7x+7v2DQ0NDU8DTRHS0NDw7534n39hZ9B1YuKf2PI5GYZcvThFcFamRzbjYBiwRshfHabERtjThCNSUgqOZc4Ldp02AS31OUwKWurzIbuGp4a8nP+3KAkpRoSk1hy44MAxi1IMbeuOR7VArwL5KjF6Fc5XaRJO7KguUrIyWb3qkngYcs0JcT+rOwZyqr14v52P97L9k3ZDlrseZwMWlz8/KvX9IU1MeYa0PL9wKiJfDjOsOiJAvVdl8VuURU1VFDnNiqnPaXWshborN9fkofdzzJSUrHYu69Nmoa7QPJIpU8nYNWOOZMo1u1Gb/RZYZuS8qVN2i4TztOhqwJicGQU9fGbkfCUbsqIRQVnqBHh8pNVmU9sIrkuXYemHwneHTg+yVaanTzJl4HucZAWjueHOgc/FjdQFfgYeL6wbBm3LyUmL7bWUdgAvXChoaGho+KDSFCENDQ1/JxT/9Jd25r/3KzvnN2cMeil/8Z0uL15OCMTwyzcMqsKG5yTn80KZkzOSlG+bo3Lev7TIRbhrF6wRsiczUnGPH8mUa7qBh+E8a/SkTYCz3M3JGeA6GmHZ1ehJp17gtySux6wqMXWqWbnwdWNg4AoPT9wYUSQRbWnV788thXPa0qoX38GSa9YyP4gRrCfZ/nEOXFUxspyaXuekyMPaj+rz2dDC5bT05XGtqjipsj2WuyGZZog4F7JqfKs6XqIJueZ1N8QXvzYFMBiGOll6PYa+tCmwdIgpsLSIyMtOyDW7wUBbHJoZR+JGpqpxvqQMOJxIykScg9o9ZrxujrlphhzLgp5xxgdvmCNumhMEIS3f3zfnMzIsR7IgwuNFu8a/mh4QlHoQEfjsSoeDPEMEnul5dGPFCLx16EYBBzFMFkJhhbsHAfcOQm5cSFhdmb+na93Q0NDwNNEUIQ0NDX+nHO78xk63N+dWkrC6NuGlVZ/v3Q/ZHStz6xZ6R0VOIVqK0gO2tM9QEi7YLi31mZRz/QbhvO1x3a5x3g4Y2IhVeuWiNGQdF3bYpUVbIxTLhDl9aZNqRpUNkpPXYz2CCyOsFtVVAdKSuNYiGPGwWDwx9d3/vNwuwK/1IlXHoHJygnd3zvpB8qiMkncqgJaLkorlrgdQj2Qtu2TJUufibUGFSwVKUTphOcG61OeuHh8rC8GqCDw9t3ntpmUQetJmRlJfw+q1W5Srus5EkrqYaePGsw5kTEZBh4gFOX2NaOFzLCknZsFl26etAR0NeMsmHElCSwNiAoakjHBF6YiMhIKJpByIKxpi9UnVcqHt8eqhcvFcyvV2yPme8GCqrK8ktAK4NSoQUV66MeJwoXxtr2Cta9ley5jOfEaj+D1c3YaGhoanC//dN2loaGj42+Xuf/Of7HwOuAMkX/pXX/6r6ZSf32wzPzScFCkxhk1tcSAL2hrwSW+TPy8OmZKxSYu/lj3WNGLPTGlrwHU7wEPKTJGQVW3xmtkHIMInwONYpiw0w2IREdIymT3AZ6pzp/Uox4IKCgKcxiPX3IUYlqNalYtTphmxuDvbLYnJyPHxychPR5DE5WVUI15Vx2E5Sf39umQ9yX7vJGR/lA6kLizKrkihBaGEDxUd9TFKXYcpC4xlIb47VlVYVPoUAS01JAoZTpCeaOlsJm0WmhBJSIDPRGeoutGpWCJ88Uk1w+JyXBaaIghJed4jfGakzpSgTDhPyenRwqJsaA8r6rI9yoDCVAtSKbgvI2Lx6WuEAIsyMHND23gYxpKyqW02tEWEx6I8H7lYJprzjHSYq+XurGBsc24/cIVPO7JkCq1WSjuK+PCm4dVdjxeNUqjyyQuG44mhnQvxP//CzuIJrn1DQ0PD00rTCWloaPih4ty//PxOhmVtkFGoEiAckDAj50XpuVGaouBYFiwkJ6XgBbvOa+aYNdvmM2zWOQ9ddYLle2ZETFiP67SJSMtxqUgCci3oSFyH2RmEgXQJalcnixGPVDNCcT5Znnj0pVPfrc8omKlbNmZlJ6UKPazsad3YkSGWsM7nONsVeT95IdV+jxvzejcNyqO2Wy4y6tGrcjTr7HjWMsvFR675Q9tX2pCHti+PWSXPV52VvFRs5JrXBaInznEsL3UjoQSsml6ZF+IW+tWY1op2GNBiKAsCfHwMBZYhzrLZirJh23hquCsTvmkOODAzBGFb+3yIARekxSdi1z0L1HBNe6yqe54Yj0H5+2JRPmHPcSQzPIQDddqlE5uxoODOpEAExnPDUZERRhmqEIWWRaFMZj6fvpGxtpLQaylngucbGhoaPpA0nZCGhoYfOn71lS/uzICbv/YHX+4S0MbnSBL2bUYiBVPNWSVmVWO+ZQ65ale4bPsEGEZa4CP4GIaSMJYFN+wGf23uskKHnkburr4UGAxJ2Q2xqnSlxUTdSE2qmcuWkABrlBynW/Dxy8wQqYuNqkOSalZ/35aITDPaEjnnrfJuf3VsOA34O8v77Yg8ap+zx3qn4y67aFWdEFWLlcr1q6hfb1Ceh+p9uMekdLkKsXraLanE6stakkoHsmzbW+WreKXAH8qxqrKYyUqNCLhuU0FBpgWeuLR7gyEsR7amsmBBRg+fMXMWpOX1j0mkoKfORnkiKSlKj4i7ckxfIz6m6yRqOecHfGcx44K0aBtDPzDsJ4bX5Ig7CCsa4SHMJGc76GAKIcBwVHbsrkibuzpnI/IwAtcvTdmbtrl/f8Brh4onho2WMJoL166OAej2ZmRpwP33fPUbGhoani6aTkhDQ8MPLb/6yhd3Miwplkg9LgcRP+Wtk5WC8TY+HUIEd2f62aDFHgvmFAwICfBoa4So0xfEGnBXjnkgI6TsSChaB9klpMQSIGJIyQjFjVNFEhISlOF7lq606/18PFpExBLVC+eBdMm0oGc6zDShwOKJR9u0XMfiER2EiveqEXk3a9/HdVeWOyfLz7msqai2qzoU1c+qbocLbjy14gVXptilUbW6mCm3qQIMT/Uf7udVAn3lnJVrTiTRQ2L4AkskEWtmQEdiOhKTaMKiDDCs8DgV0+dYYgKm6joiAR4ZBYG6z1PcGFfPhmQUhOpxrBlHpOznGW18LsY+bxYzAgP3NSEmYCEZHXxGknKVDn+RDWlrUKauKG18NiOfD/kdRpnFE5hOQzqBMFt4PL8hfGhTubKVkuTw5s019vf6PHjQZ/+w/cTXv6GhoeFppSlCGhoafqj5wiu/vvNLr/zGjoewl2XcyudYcHkg5MzIGBDyPXPCuLCcp8W2xByU7kcBhgMzY0CbE5kywAXdtQkxCC2JXIEigbN+rXUglpAARcnLJHUAi33IGnZBSkJaLrOVSELmmpCRE+CTaUauOYWeWs9WFrLeUnHwfh2zvh8tyJPuv+xqVRUd1ddAbc1bFR5V0VGJ1B+Vjl4sd2ewZGV3yKJ17keL6DRxvrxWthzXWmhKqjkihkQTZpqwLSulJW/KjJSRzjhhikXZklUu2QHgdEFTSXhgJoxkTldDVCiLC9d1OS8xbfFY8Xy+Np9yz4z5m9mMbYnYth3aGrBp3IjfUDPOa4uP23MMJaGvEc/4MYMYCoXUKlbhYBhQxoVgRInCgsnUp7CwPzbc2Q8JA8tw0gwpNDQ0fPBpipCGhoangl955Td3Dkm4J1MCDFu2w6EseKZcWP5a+zwnNiMQ4etyxERSZuLukD+QEVNcUbJpu8xLwXJMWDpfnS6oA/GIywLlRCdYlEKLuuNRjWNFhC6Tos4J8YgkJKdgVo50DXVSZ5AE4pNoiqp11sDlQjuQsB6ZepR71ffDO3VBlsXwFY/SeJx97FE2vMufa+H50j8vUvdEzEPFx/I4VyQhoQR4cprPMtM5qWbOBhl1NsmasdCMmc6Z6Ax/SYPyjF2jLx1iXCHYlzZx2eXyMSQUbNoORt17mpPSJWYoC1bVdUIua5cNibivC3Z1wevFlKGkvGTXuGXGvKULVgnZ1Dbf0CEFyrqE7LHgchxyQ/tsEBEa4WCmZKrctU4r9GDsipELW1PuHBvmC48otEQ+tEPwDLRaGd3/9fM7T3SBGxoaGp5imiKkoaHhqeFXX/nizn/+x//pTk98+gSc0xZtPF4zJ3xnkjAiYzXwWJCxamN6GrFh22xqj0u6ygodNrRFh4iYkBCfvrTZlAEFBVuyil8+VhETEklIJAHnpF9rDrT8kPLDw9SFyorpk2mGwRCXgulUnftTRkEgrpCptA/vlqZ+FnmHDspZHlWInM0HeZSo/exY1tteQznydNYpq0qHP1u8PNT1KTsnvvi1JqTAUmjh9iy3TcmIJMTHwxefSMKHMlx60sHHIyg1OG+aQ2a6YIGzTq7iKKvnLMRyYGZYUW7YVbrEGBW2bZcJOVvax0MYasaumdAj4JK0WJATlML2+2bKfeZ4CH0NSSRnO/bZM1NO0oKbMuFzVzz+NDtikltWfMNnVzpsrxRcGMCPPTvi9dtdWj7cPjL4nuUTLx3y3DMj7o0tf/btzjtez4aGhoYPCk0R0tDQ8NTx6T/+pZ0tLyzdieAn2eCrZp8WHoXClu2yZ6Ycy5xXvX0SKZiTs8+Ie8YJgDPc2M2CjAkJfdrMy9GqKYta7yEIAT5WlROmpJqXoXpu7MgTj1YpPvfFIyVjXupAqnGsWKLyLr+7a7/QtC5AtNRHvBcx+nsVmj+uI3J23+ViYjmE8JGv4REakerx5WNVblfFkuak6pi0xOVgRBKWgnan2VC1BKVAXVECXGETlJbH1dhbX9pMdY6iDO2EAx25kMhy7C3E5XoM1PWwtm2H+zJiQUobj48U57CiXDNtAlzifSweU3Jesmu0xeMNnZBIzk9d8PhxPcdzdoWJZK4bJzEfset8az7nZT3HiXVdsdfuG1Y1Zs+6YujZy1NarYJOyxLHCcMFBB7MCsulK3s8eNBnffOAn7ias91/7OVsaGho+EDhfelLX/rc3/WLaGhoaHivbH7x+T+5+/uvfi7HjbigwlgyDouMAI+RJGRYcrGsa5uppHSIuC9D5iRYICltei22HNYqmGvCuvSZMIfSZncgbYwY5pq6DkZZTLjMEA9fPIwIVp1+ZKGJy40gZ92suONrii8eOYVbaJf6iWBJoB1JRFYGIP5g0Md8TfmcprbGNXgIy8XDqWvV8p6y1CFxHSAPhVIYLmV/qBrNqj6W97PlvuVjZVChG1fLak3IhgxQXK6KEUNbYmZ1Iod77oF0GOqUUMK6a7IiXbZkhSFTPPGI8GkTkkrBQgou2xUuap8RGTfNCVaUro05koQXWWFPE2aSM5WcQpUeIb+5PUAEnt8suNLxuWq6aG74yKbhIxcK7h+5cvVyK+SFuMV6F9KFx89dFUJj+MtbHpdWlH4v5falxjYAACAASURBVDtvrNCPFVUhNMKl81OKLKTXnxAFyuy//41mFKuhoeFHgqYT0tDQ8NTy06/88s5/8Mqv7SRYugT0NWQmGSkFC3EjNAN1d9vXbZuuhrVGY4MeA1oM6NR36UMCQtzIlI+H4TTEMCYol8/qckIwtKWFh0emxUNah7a0iEuNA0BXWgRluF71WCTutUSEtVBdkEeORi3zfnJEHoeeEYdXlrzL2KVipH4N5QjZssVwJTivOh3VflXXo9KQBBLWx0s1w2pBppnrfuARlF2VKYuyoEuZa8JcEzw8+tLGovSlw4gZG2aFQDza0nIjWrQ4YOwsfcldqrkkPFessydjrkuXoSRclQ4b2uFKMcAgXNUuPeMxEXe9L2q7dGDzKKzQaeUUVrhwfswkgZevFMwSIYpyPn1Z+NRFwTewvZazvpLhiTCd+6z2cgIjTGY+t97qsLWacWl7xvZaygvPzHnzzU2CwF3/dm/8A7u2DQ0NDT/sNEVIQ0PDU88vvPKrOwARhnVtOVE5U7Il69gb2udAJvx88SybrHDAmCFzFmRclg22WSUlwxPDROdLblCGKQtGzAAwGAbSocBiROiUI0ULUlIyUs0wCCEBAT4pWa1LqO7w++LXI10FFlVnUZtpVo8oPU738W4jW++nSFkOFHycfuSszqMqLOTMPyNVUbK87bK+xCD44hOU6fLV82YU9X8Gg9VKdeN+7go5r36+qS7KgjGnUFvreHIsCSktiVijW7+Gt8yYi7rCPZ3jYUjK9PVtWgzEZ8MPmFvLdbpc1DYZlp9dbzPwPL61b/ne/ZBXdwPy3D3/N+/6HEyVf/dah70TjzcPPCap8q9v5bx2L8QASSbsnQRMC8uDkaHfKdjYGDOehFy4dMiND3+VH//Mn7H7oMNi1iZN4vd87RoaGhqeVpoipKGh4QPBL77yGzsW3FiMtHlG1zHAQGM2bZtvmSM8PG6aEWPmFFgu6CrXdIOuurvzHqbsfwi2vKt/UdYp1Nny2lLnMSdxWRTqFu8r0iEkcHkgpWB7ru5O/sTOGOsUg6ErLY51XKelhxKQaIIvfl18eJgy7LD1tkKkGgN7J55UV/KoYmN53+WU9GUr3rOZH8Dbvq8dskQecssKcMWXqtadoqgsRloS1YnyAf5D+S1XZJM5CYVaEk0ZSJcV6bAgZaEpA+ngYdiUAYqlgxOvHzIuC5u8zIwJ+HDQ5Tl6TMm5oX1WPJ+e5zEuCuZq+cnzHp/ZCvn0RsTxDDwRBoFHbuGlyynfebPPuZ6y0RHW2sLlVUsnqgo4eKEV88YsZW4tD8bKV44SXt42dCN45sohSeJseFcvv8ZisoFZHfPRj72B8Syqj8+PaWhoaPig0RQhDQ0NHxi+8Mqv73zhlV/faYkhIScmYEUj5uS0NWBd29ySw3rk50gm7MuY+zJiyIyivLPviWFd+i5PRCO60mJFevSk4+68YwnEK5O6LT7e6UhVubj3S0vejIJUM5Iy1yLALdQNhpCAlsR0pV1rGiKJ3IK9FGYv4+ERlov298PZjso7dU2WuyMuIf5hF6+zXZHKftcryzigtD+2GPHqkMLYlJ0epB5NM+KRl2nonngkpYVyNQp3yIireg6/NAHwMFzS1fo8BuVztonq7tXzdosfs5cosLSJCNTjsnbI1NnmnjMRHWP4+EXle/kMCwQivL4v/OWDnHOrGWsd5c18zmZP8Q3sHweowovP79MKlbVuwbWrR7z43AE3tnI+ejnjYzdmPNcNaRmDEaeRCUPLej/HeAXPfPirXHvuTe6/+mO0zt8m3T1P7zNfYzGL6Q4O3ve1bWhoaHjaaIqQhoaGDxwf/6Nf3LluV4jUczayouyZKR0NWcHlSACc1wE9jckpuKJrdIidbgGfKQvOibMqsigJTpvQk7YbAyodmCY6Z8K8XrCbsotS6T+60qYrbTzx3J17UubqciOqPBFb3vWvjuFhSDWja9x+VXJ4IIHrLizpRt7L+NWyNe9Zqsce5YZVjV4VFA+NVlU6GMXWFrzu9UldQPniuwR1dZqRtAwlTDWj0KIet3Jp6OFDz+86Hj1CAoYyJybkQ3Ybg9QjbhvSxyu1JJF6RKWu59jMOTQzfqq4Wr4v1yW7ky9YUPDCmqEAvnvfMJaMgeexEfjczzKutgO+dSfk9gl8etDm2qUJnRCuXZowz2AybvPs9UPyQtjf69NbPSTPDZ6nfOtmm24EHzonfPRqQls81lfneEbpDg5YnGwRxBOs9bj9V58iWN+nuLlCpz8m6B/zyT/4b7/8xBe0oaGh4SmmKUIaGho+kPRKabkrIAoWpIwl4UoxYEFKgFcv+hNy9mRCQk6LiIWmFGrrcaCJzokICcSrOwQznZflTMCsDMbzy9QPcIvqXHMC8ZiWoXsxYT1ulWlGgS3HtvLaKavKwAAotKBjXG6EL36dKr48KnV2/OpJipLlLki1f/X5cbkgtWZk6Z+NSv/h4aFaFhNq6wBHLfUu1TkTDFYLfPFd10ncuYiX9CEWZaEp69Ljqq6jqBOca4sN7XEic57RdYYyp01EgWVBSlj2Q/KyWMoomLJgz0xLu2SPa52AERnHpHzzsCAQYTfN+Gy8QmIto9zyHz4HL78w4iQtOClyfA8m04jLWwuyzOeFywnGWHqDE86tL2i1coZHG/S7GW/uRlxcz9kYZJxbXZDnwt9/3tLpzrl0ZZ/hwTazcZ/R4QWyzOPSjW/zF//65/j2//tZHtzbxKxOMKuNOL2hoeFHA//v+gU0NDQ0/G3wM6/8ys7PLH3/X//m7305k4KeRJzXAccy54GMAOgQMWaOYpniuhRVF+RIJnRpMSkFzXOSemxoUQrZM82ZaUJUjhclmhFLyFwXTHWBonSljSD1Ar1r2kzsrH59c13QlhZzErrSdmnhZLU/rocHQt1JeJz2470mrS8XMo9KbodTzUe9T1mIOHtep8QBF0hYvb96hEs8bDlqZdHT0asymT6SkEN74nJCKNPStSAkYFeGtfB8XVtkWO4bp/MICThvB+yaIZfsCgXKsZlTULCpPdrqipLb5oSEjEANfzmd8NPtAX86G2GB59aEu8OAg6Sg5RlWImEyFbodYSX0IIUblyf85Xe7vHAh463DAN+DNI/4VHdGHGesbT5geLTOd+5EXN4o6HczpjOfvDAsEo+VOKXVmfDGa5e5/twdjJezv7tNkvi89o2PYUSZznzmqaH/lU9xcNCHX3hPl7ChoaHhqaTphDQ0NPxI8Lt/+Ds7KTk+hgUZ67aNV6ZgVzqNTVYACMQjJOCICSOdlRoF57rkYZjqHEGYa0KmTvMRiRsDmuoCU45T9aTrktTLhPRq/KoSmLckpmc6buzI9OvuSVWQhAQU6nQqlabCdUzC9zSG9STbVq/pnbYtyhG05aKkGrtaDiQEV6iEEjy8rZh6rEyxZOTMNaEtrfq5WxITScgBQ0bMMAjPslVnv6xqiyEzF1AoC54tNrihfVY1psCyqm0+bNf4tL/GgpyBxnyyuMh5WrTxEYEBIRdjn36nYLMjfPicIbVKYUEV8txjN8nIVBmOIm5s5fR6Cc9sJxzN4FMfu4+qQUR5cO8CYZQS+fChF27z7Mt/Tq+b0mplXL16SJJ6FIXH2tqMb3/zmfJ9ghFlbW2M8ZRWq+C560d0emOu3bjNtf/xXzYjWQ0NDR94miKkoaHhR4bf/cPf2fFUiAlY1Zj+kp1vTs4hYy6wxjar7HHirHbFp1BLLCHPcxEt07pTzQjEIxDPCcoJGem01HZYutImKbMtWkQEuBGkAouPz1yTOl19rgnguiGVbe9cFwQSsGYGBHgUeqrHqPDEe9cC4+zPKz3J49LSz1ryVnkgpswwgVONCLjioxKee3K6V/UzVXXFh3gU+rCupDqHofjEEtKRmI7ErJsVzrNGi4gVOmzZLrsyZiYp523X6Xc0ZK28fgpsEZfhiYYTMr6RTTivHX5WtvhQ2ObFfsjLgxhPIKHgxnbG3QOfNIdL5ydMbcGgBd/egzjO+AcfztiOfQ6GATee3aXbm5FlhmsbBa3OhPu7K3h+znwesPb8N7h2PsHzM+ysxYUrd7h0/TU2f+bf4hmlt32Lyz////Dih28RtkZ0elMuPnOPw8M+WWa4eGmPbn9EGE+Iu4e8+V99qQksbGho+MDTJKY3NDT8SPHh//jH/uTlL378T179/W997pY5JscS4hMTMichJkQQpmU6t2BAnHB8lTYjmeOJYaYJPemQ62kGyFinWApiifDE6VGiMvHbiCGWiEzzOnXdokQELEjJyQnFBRdWyeVtiVmQwpJTFSIPdVeq1PFHJaK7zWXpsyIiS+NX8ojxq3Kb8ojuP62zzy22LoWq771aX6L1q2ibVq0ZcWNYxtkei9CVDik5Wr1HTbkk58q4Q5cN4iGsapeJLAjFFXaX7YAC5TkdkCvsmxktAvZlwZ6ZoyifYZv7OufjYZ9pYRlqzlGR8cKKT5oLD2aWv/eMx92DgN1ZwfUNuHU/5hNXCqZzj14ojCcRs3nAT3x8l/Gww4O9AVEAeeHRigvm0w5p6tMfzPE98LKYCx/6JjZpMR2eo7NxD2/thOzWRfqDCX40Q1DiH7uN+AtarRFSGDqdlD/46w4fOW/xg5w8i/ibf/TbTQHS0NDwI0HTCWloaPiR5E+9W8xJGdCuheAhAZG6jkUlpAa32F6nx4FMXKdD1WVclGNcofjk5ARy2u1YqAvMs6qkpSaiKliysnioOgKqzjUqLjUlVd5GSkaLqC4SQglq4Xq1wD/N5Hh80vp70YlUx1i2Bz5rzVtrQsrxKlXnlFUVPOC6HIJBRMr3ntfFyqIca6sKEFuKy921KHhWN5mSsKFt0jL1PMKnEOVVb58/N/dp4fOMHdDXkFRyuhpy1a7w7MDjx4MBmx3hF64EtErT4KOpEAXKgyKl28noxspnn81pRQVrXes6Fq2C65emPHf9iE9+6hsc7q/SjguMKG/e7TKd+ZyMQuaLgPMXjjn3ia8Qt+Yc7m1AKyHavsfRwSpYw9F3Poq/uUfYGpJNVtDUp3hjhfnrzyKDKf7GAe3BfXZlxv/9l+vs3z/H+GTlia9TQ0NDw9NO0wlpaGj4keTnv/iZP/mFL/70n/zFH3z1cxNJSMidd5UYEjKiUt+hcmqbGxKQl+F3FktKQSwhgkvwFoSMHCNCSyIEKce3fGaalAvqnFhCBtJlRkJOQc+0iSVkZKeYUpvhXKEKeqbDrNSg2LL48PHINAcUXzwoB6YMBpXTrgflkU6LE62/Py0YtP7aFStVJ+R0PMuUXR0pxeVVjrnB1N+5Pgnl45XDV2lbLB6B+Pi415qR4eMh4pLlU02Zk3JOBoyYMZeMLi0OZEJOwUIyNrXLUBYMaJFIzifNOmsmxCr8XH8FLw346c2IvzksmBSWy33D1+8rJ5qxaULyQlhtwYVWwPVr+4yHXQprSDPDyz/5NR68tYUxysb6mJPjLuubexzvb+D7lsIaXnj+ARcu7pEv2lx7/g08zzLdvcB42OPC1Tfx2jMWb13m6GCN9e1dRodbyKJDkbVYzHokw3Uif4Yfz8Ao6YNt/v/23jzIsvM87/u931nuPXfp2317m15m3zAACRIAQXEBCViQSgllRbZFseSlrLiUKKnYlhOnksiRbKgTWVYkK3YcW6JNW1V2SRWXAJAll7VSJgVaHIiLiG2wDDAzmH3tve9+zvne/PGde6YHJh2aEQFS+H5TXdN97z33nu6+NTgP3vd5nvjYZR4M9rA4qQwGEWsrH/VTEI/H87bBT0I8Hs/bmr/5xI+u7G46Bxev22dELBHHdYEZWnQZss9Ouk4PiajgPAyT1BFMGTMLbnKSak6meVnIlxa9GEHxz+6WdghxBvi+DuntMqMHGCZN05m9yUmLzhHX6h6UAkEKYTSeSIw/IsI72tXf2Ib+tRAxhMX5jo/bPV3ZHd8bEJCT33GbRW+vZhUlhWNRNZ6mNKRWPrZCTFOS8vMbbCEIm9qlpjEjMu61SwxIGUhGhrIqXSzKep6ymWdYoFWzDKzl0gYcaYbsSyKubwvfdc+AD7RrnJgx7G0Jy3t6LM/16XdrXF43tJoj5mZ6XD53lP4wIE0NJ5+bZWsnJmxuMT23SaWaMjvdY+bDTzMaVjl41ytUly8iYokrA5YPn6Zy7HUkzhgNE9rtHbrriyy87ySNEy8y6NdpzV9g6n1fwiyvYtMKUhuiGtB9/m4ApudW6fzsn/ECxOPxvK3wIsTj8bzt+fnH/+eVn33c7eLnxaV1UDRxX5R1ahpzUGe4YNYZkTJJnT0yxY72uKmbRfGemxFEBESEDNVNOba0w5btYFEScW3hFmWkKRbrVrk0I9WsjP/VIgUrIGDHdoFivUlM6c0Id3WKZJr9Bw3nb2xb/2qrWmORoWpLAfPGY8bRumPG5zWO4P1ajFfKxj/HtBAsfR3QNq2icDBjW3vUJeEQc2RkHNM9RBLQk1GZBnZQZ4g0YL9t0dCYD9slVhkyGYTcN1kB4J3tiO3MsjVQ9rSUqQRW1xPmJlMmGxkT9RxjlLiSkWWGR957jas3qwSBxVphaWGHQ4dvsj6wDEYCkYsanpm/CcCt3/8A7Q98kbC5BaGlfvg1akvnCeodzN4AafeZvP8Z9nzf51i/NQ3DCF1vMn3fl9EsRCbALGSYpIeZGlI9cJ4gHDLRvv71vk09Ho/nTxR+Hcvj8XgKHv3YB576/ce/8EiGZUTKsra5JdtEEtGgAggbdOnQJ5WcjvaIxU0ihqSMNHUCQgyRRNSkikHoMUSAlIxJ0wSEpkno67C4NNfSvD0uO4yLnhEXc+umL+P+kRFZWfrnjhOMuO4SZ+q+vW5lxBSTk93G9dtrWeOvx+dtuV1eOJ5q2MLvMfaD3E64UgxBsYh12z7vDOiu82N8Hra4ryoVBjpk0kwwInXnijAS155uBQak9HE/m46MEDHM2TodGbFPJ1iOY+5pVGlVhCyH3ArbA2hEhrkmRKGyf6kDGLY6EXv3bvDCuQaNWNjpxrTbXbY2JljYs8303A3mHvgKlVxonniZe+YHLC/fImhtUw16xFPrTEzdIqlvcemL72Py+GtInCGVDHKDme4AGRKDriXYmzXa730Jc7yKme8h0YhgeQcMSBwipofUFallGDtErPLcX/5bfgri8XjedvhJiMfj8ezi7zz+Yys/8/j/uPL3H//xlSuySa62TMqKi1WojJxlnWLBTNPTPj0dMNQRADVJ3EU4wo52CcQZsMfrXuNo3jLGVnMapla+flakRm1ph5ycobqJQKoZDakx0rR8rrG53bktgrKXw6IuGrdohf//MqyP7x9PHsDF/46nKYEE3GFIHz/vrqhecNOP8dfjWN+gEC23p0CWhqmRkZXPryhH7BwW5bpu0qYBwCJtDtg2OZYNM6CqIVdMh5dGXUY5XNjOudGzDFK4OcxZbjuz/r7lTU6fb7K4tEoUKqfPTNPPlMnJPvUkoz1/hQPHXmH+0EtcOrcfHUTEMzc5/9TDRJPrxJNrvPa5D2PmNzF7tjDTO5x+7gE2NxN0EJFfnkFHAdeffQ/51Tbps8vojoEwJzi8hV1L0NUdNM2RWog0Ysz+ZQDMUoAszWDuOYxp9TF7Nr7xN6vH4/F8G+Mb0z0ej+dr4EoKtUy8uiHbJLj1n1vssK0954MA9phpVnWLjIyQkFBcP0hfhwx0iEVpSYOUDFVLh1558d8v4n5HpMREDBkxsAOmTIue9mlIla726WmfUNyq19hrMja/Z0X0b1isd+U4sRDhpiUqQq45oURkRes6fHXfh6rFyriB4060nGm4zwVDIOaOosJxk/p4egJuncz5Wgx9HRITlffFEnJR1gkIqBdrXLO0CNQwkIw+I+62s1w3XWoacV/S4EovZ6QWayEZCes2JYoC4lxI05AL2zkHb0xhRNnuG773octcvjjHoaMXiWZvgrFIMmJx3zXs2iTXzt9FHKdcffE+RCwb2xXIDGd+47ux1nDX+0+ycf44DGN3bGvEzOJ5pDJiuDFHmLq1qrVPfwft7/gyGIFBjh0CWU6wGCP79sFwgO5sY6+cBRPwxYd+yU9BPB7P2xL5zGc+81Nv9Ul4PB7Ptzr/90f/+WOnucactOgxKszkrgE9kohUUxqmxlBTIkK3HoUhVZem1dUebdOiqwNa0ihKCgelr6NlGqSFcb2jPRKploJmvCLVsV0iiYqW96AUA2khKioSF9MIpWf75blHEjHQQblK9bVM6mMxsntCMvaEjCchu0XInUWG44QtLfpRYgJxJYuJVNnRDkERl3unP8ZSkwojzQh3rZMtahsrSg9X5LjPTvGhaJrNzPKibnJYJzjWjNgYKFNVIVeYrCk3d4R7D/SJ4pzXLze4797LDPoJo2HEvvu+BNZw5dT9LD3wh4hR8ltTmEYPHcRcePldHP6h3yF/eRqzd92tUAXK6IX9XHj1BEe+5/fonz5KcuJVNDOYiRQdBGg3pvvaMRr3ngIrvP5738mB7/wsZtb9XnQrwCwUQi80IIK9NuQL933CCxCPx/O2xa9jeTwez9fBX3/iv1r5x0/87ZXxZGQ8lQCYFudv6OqgiKF1E4oNu0NHewx0SEUqVIhLE/hQh+UFflXcxCSSgD5DomLaMU7TslhSTV3aVLniZKlLUl7MRxKRkpcm96qplslUrgDRlN0iu3ljv8huQ7uqvcN8/tUECHBHpwq4/pChjhgWTfBjseYmNO7772m/nISMU7UUVwo5SZ2+pFQ1ZEprLNlJ9mqDVlVYqgVkKM0gIDBwcBpSC+995yoP3H+WJITZPWvMzN9CRLl2ZZbpPZe5er3B6ZMf5PwzD5KmAWami92qc+Pi0WLlapu5hevkZ1qEDw0wR1qkL+1D5qrE77/E0oELmL1Cdd8FzKEKZjJFFuvIlGX9hXfTePB5d/vBgH13PwtWkCQEhfz8LDQSdCfFXh+h3SFm+s7gAI/H43m74UWIx+Px/Ccy0pRK0QMSSsiWdgmL7da+Duhoj0xzKhIXBYbOyL2u2wAMdESOJSUvErVC18GhbuIw9omM18DCInLXas6I1PlAJKSrfddgLlIY3J2hPpaojPp1kw8tJyDOcB58zXJDt0Zlyo/drefjqQc4z0coYekDSQtPzO4kLVUlI3NTEVxc71jEZeo6U24ngbmVLmdWD5jQCimWKhFWlNkg5sxORhLBcW0xFRummzk7fSEQ+NwzM3zxy4c5utznmeed/+IDHzjF+lZMGHeJY0ujMaQ50WGyvQlWiN67zsJdz2OWAjBK89Hn0W4C1QS2OsTvv4QOUsiV2sOnoVZFJgbQqCMLTfdNRobp930RqSmkGTSbmFaX4P4JdJhjjuwlOHALe3EbagESKFh4et/H/RTE4/G8rfEixOPxeP4T+KnH/8bK//XET6y4acaIkaalMLjtcYiwaGlWDwhIpEJYrFCVqVoYAgxDRmVrOzgjeL/wkbio36zwc4TFRMOSFyKnOr7AL167Is6z0tM+itI0DURcd0gsMVIUD+5uax8LE7gtIsbPuTu+V7F3vFam2W1fCEG5rpUXyVq7G9TzotjRPbfBSECmrrhxpBm9YmoyTZM522Bbhsxpne+QGff9WMuWpjyzMWQyDJifUKwK+/cMWWjn1CM4cXQNMfD+971CY/8ZXn/1KAtzPWwec2szZGMjYfb9f8jqzWmkrdi1nPCBbcgtZv8QMsUsrrkTbjWQqQZSjSBV6FnsuS5mbwz9AQyGUE2QaoRMWagE2JspUk0I7hoh9SYyVUO7O8hETnDXArqVI7MVCO6MQvZ4PJ63I16EeDwezzfAP3riJ1fG0bkWJZKIkJCoiNTNcSLBojSkRrdI0MpxF/wd2yMjK83oqm7FK9fbBYARgZuAFOG34/SrtpkgkpCKRORqy4jcsecCKI+xKDVJqJvaHaJg98RiLEx2e0XGRYTj+6Twcuw+bryOtrtfpEy80tuPG69jjQrvSkpOIhUCCRjoiIycaZlgWWaY1SZXzCYNjWloxGf1KnfZSS5ql6WgQoThoeNDOgNhu2c4c6XC6pZbzcqzkKs3q5x67ih2Y4JazYnA7bVFAB4/m/L67z7KkYc+R/5yi/z1GbLn6tjLoNcN9lbE4LUj7ufzYgd7poO9MkRT0L4Bo+j6gPxUMfU5u4q9NkS3QVp1zHSAjgboIEO3NyDLoT/AvOM4+SvXwArUEqRV/wbfdR6Px/MnBy9CPB6P5xvkl5/8mZVfeuKnVsCZw23xByiidS0hAR3toWrLlame7ZNpSqbOqzEiJZSQmiSlGXwsRnJ1q1yRhKXY6OqAVDM6tkdHe1QkLqKB3WrXG6cVo+K1AKJdoYjOUG7Kiccb17N2i4tx9O5YeIwlDnDblL4rEUtEnA9GKqVfJJSwWBITpmWCCnE5IaoQMqEVrsomd+VzdGTEnFQ4btuEIlwzOySBUDOGPBdyC4MUeikszYx4x9Etnn1lkunJlJ1+QJ5VOXNhgnqjz9TRF7nr4A7H4xp5buhdOEywbxMzt4n2qsgei73ZQoxS/4EbZJ8fgFHIBXNiBglwXwO6GSITI2RmGjMVYg61MEda2DMdqMYwHCJNN42S9jS6OYBqHbPcxByeAjE8Pf0P/CqWx+N52+PLCj0ej+f/J9/3sT/11K//2r97JJSAnvbLrg+4ffEeSURf+2TkWM0JJCCS+LZYICeWiJ4OqEtCJkXhX+HJyMgJJSjN6dVikuBWuVwpYCShS+Qio21ad7S/W5SQgJQMgyEu0qvG6VwiYyEiZVHheDriShFt2eYeiiso3J2INUbE3V6TBKVohi9Wx8bnmEiVPiMEaEjCkJSBpK7IkBREWLYtvhLc4JC2iMSwRcpeU+MrdoPNtRo2F0Y5LE/CxbWQa6tV3nvvKltbCVkuHPrQl+hePkg2Csm2p0nqAx5837MkFUvzkRfQFMwMmMUuet2goxCpj5D5GvYcBEdSDkciEAAAFkVJREFU5GDbiYqJilu/soI52oDREN3oQGZhbYCYHNlThziGPEd3+pjZWfTadWR2Ehn1kTDk6cbPrlyufPdTb+Jb0+PxeL5l8ZMQj8fj+WPgXzz5d1d+8YnHVlTd6lJNEgIJaJhamWB1O87WUC3a1MdrT4oyIiXCCQtwfo2qVFxRYrHmNRYC3SJdyhYTE9n1pyUNtrVLSxrl+lS9mLIkUqEibgIREZSm+YiQcNw9UkxExgJp/JhxJ8j4NneOWoogJ2Dc+he49Ss3xYlKf0xFXA/KiJQZmSArEresKlv0OGpnsSgXgy2mtcZNBmxpyrzWGFnloDZZqobsm4R9U9AZCGmu3HdshwsX21xbDzl6aJ3VP3qQu+/7MoNhiBElCDPyfoPRMGHr392H3qxDIZ5kPie4p4NMW/TiKmZuB93MkDBGanWk3oSqIbh7D0QxMuNM6dKqQiLoIEOaLXR9C8IAqVfQXheZnUaaLZiY/ua++Twej+fbEN8T4vF4PN8EfuQHfuKxcQQvUE4cRrs6PWKJ6Noe4CYlDamVk4Yd23UTAx1QkQoBhp72sSgTps627QIufjeRKjERsYR0tI8pYn0NYzFhylUxg6GrPSpSuWNNa3d87/i8y9JBbveFjNOwxtMPEWFUdKOIuPb28RQolJCRplQlZqCj0uMyLi48KHuINOCGbJGT0yAhIS7XznoMmdEGLa1ywazT1jrH7RQfvUfp90MmJgb0+zF/dC4mCYUogNmmJanmxJHlxIf+Petn7mFjrcXRP/vb6HqCJCOknaHbAWZvSH7WEn5wGd3Zwr6+jjnYRlc3kYkEmWyDtWAM9qVLmPuOwnAA6QiiGIIQwhA9fx6MoDsjiI0ztLem3GOyDGpNTvZ/zK9geTwezy78JMTj8Xi+CfyLJ//uClAmSwVFGtTur8GJj8qutay+DogImTQTdLVPSl4mXVWkQl0Stm23jO8NcIWADUnoFMb2ng4QhFTT0qtiMOSaM9Chm9JgSqO7cjsBa5xyNU7D2l1QGIjBqhMr41WsTLOinDEgkWpZNgiQa16kX7kJj4hhUposmhliIq7oGlVC5rXForapUaHHsDyXBlXeYae5aDb5z/Ugd9s2TQl5+tUqL1wO+b0X6vzuq4bNzJ3T5lDZt7zNdjdkz+IahDmTsxfZd+Q17IV2KUCkVcEsGHRrSPhAG/IMej2kCjK76CYkvaETG3kG7Xnk0AwkTXTQR9MUjHH3W4vMtJHlvZhDS06A1Jvo+ip65ZJ73KD75r75PB6P59sAL0I8Ho/nm8QnnvzplU88+dMr4FaTRFy/R16UCmaaUZekTM3Ki4v9Le2wo13G6VuZZgwYldOI8YX+WLg0TZ0N3bkjuWr8XEC5xpUWq18jTV3HCQHVYlVsjGLLFTH3Wrb8OygihlXVpXKJFL0fOamOXNdJkag17kgBF1k8LinsaL9sQReEm7KDILTVeUiGZKTkbNHjcD7FF4PrHLJtvsQtTptNBmrZyXIiI3zZrmMVAhE++O5V9k/C7NIFcgvPvrBI78JhtteWiPdfIjiyhkxkYEG3h6RfmMLsnwdj0EEfe2mEbhvo7yCzU8jstJt0AOxsILU6euE16HSQKHITjrEQCUIIQmR22X1eqbpkrJGbMp3Uv+WnIB6Px/MGvAjxeDyebzKfePKnV375yZ9ZydXS1wG55ghStKz3y2lITk69SLnq66BckWqYOqqWjLzIlrrdSQLQ04Hr7FDLpJmgJtVy5QlgRloIQiIValIllohKcZ9BmDZT1CQpk6/GCVtj3DrY7UnOWJTkRYfIOMJ3qKOit8Qw0pRUMwY6YqAjYomoSZUJqVElvr2OxrBc3wKY15ZrkSdmR5x3ZEP6NLXC9yXzPGducUtHjHLloXCav/TQBpEIRpT9Sx22Vhc4emCbD334GZLJG+RZiJkdggHNQYeCTNUgC8hfuomuryNRhDmYELxjFqp1Jy5qDVC3iqU7W9izF5GZOQC014MwBGvRQd+JDkA3rkMYQLWO7D8CgUEvXvwmvrM8Ho/n2xcvQjwej+dN4lc++fPl/xF3nSBu+pHhVprc53khUkxRAmjIyKlJQk8H7KhbxapJUja2p5oxaSYA6OuQblGGKIUpfFW3SkP5WCgMd3V2pGQ0TK30h4xFxljuABwP9lMzCTk5gbhywrw4r7KMUUL6xSpYIAE5tiwoBDeR6TNkQVss6CQjTZnHCaRpTXh3Ps+O9BkwIiWjJymH7QwBhu9vT9GowPdXltgfVhGBo3PK1PxFPvrIFS5dmaI1tcX89/8BS+/4CvG957jw8ruoNbfRFLBgpkKkJRCFRB9aQ6YErKKra1BNnLiIq8j0HnRzDd3YcF9PTiNzLfJnzyJTU5CmaK+LvXwNqdXdNKTXAcAsHSm9Ik8v/eLK03t/yU9BPB6P56vgRYjH4/G8ifw/n/w/V4CyGDDTjBhXeFiXKj0dlKtLEa453RRTE6AsLTQi1CVB1SVnbWmHHFd4KBicVT0kJKAuVSJCGlKjITUSKkSFgdwgxEQ0SKibehkbPBZB42lKFzfBGeM6P1w543jVyyA0pQFQek5a0ihjigEWaHNDtgHYI1ME6v4zNC8VEgLelS/QJOE/s4fYkD73BhMctZOc3sjZ7MMgU+qh4c9/9A9YWtjh1FfeRaXaZabdozV7EZlpIe0d6IYc+i9+l9rhM+hORP/LxyAwmBN3Q28AUy2kEiLH7kb2H4Th0MXwGoNeOIPMLrppRq2JXr4AYjBTIbq1hRx7B/R6mMMH0Y11aM+7yUgUo7cuQ3fLCROPx+PxfE28CPF4PJ63iPG61Yi0nIwolkQqpS8jlohc83JKMfaI2DKdytCQWrniBU4AtKSOESEjp6N9ujpg024zITU2dIe6VMu43rTwYQDUpMqkmSCUsGyDnzJNBoyIJCKSmEyzMpZXURqmXpQQuuLF77YnyLEkVGjToC419socuVpCNQzJWJeOE1eSEWGYDAP2RBFbMmRKEwwwpQmRCANyLtoea8Oc+w+OuDZMGe1MU036zEz3yLOYA49+FrOwzs7je7n5h+8nv9omf30GafcJjkVU7zmD7ozQS+fczy0IoVaFnQ0IImR+0YmRrTXk4DH08nn0/GtgDLK8H2nPIstLUK2iLz+PbvTQ1ZvoTg999RRkGbp+y4mPah1sjsfj8Xi+Nl6EeDwez5vMv/7UP1z515/6hyvjBCrXKu4ExLhTJJFqGekrYspJyG4/ybgLZDxp0MJMXpek6BBxJvZa0Ulyrxzghm4w0pRbdoMt22FOJmlKjW3tlvHBHe05YYNLt4px05ChjlzRYpGg1ZRGYZPXwghvmZImG+KM53WpEmvIorTpM6QldW7IFjViYkIsyoftEhVC4kA4lXb4UDzNYduiagwztsaLaYcZiZmhwv5mwPpWzA8+uMOwX2dq/gKrazVqs5cJ3jOPmR6S7H2d2Xd+BYkzJE4x+ybQjR7SVmS2CWmGLC5hL19H9uyDah29esH9YioJ9sx5t06V55BZ9OVT7r6xuFALjRqShGAEs7gHmZuHpIbsPYKq5WT/x1ZOZv+TX8PyeDye/whehHg8Hs9bxK9+8hfK1awc68SFpgx1RF8HBJhiwiBMivN8VCRGUTLNSqGwbrdLo3oiFVIybuomQ5z/wyAMGHFR1qlLlQlTB9xUZYc+EQF7ZZaG1BjokIrE1KVKXWocY4k9OklfhzSkBlCUC+Zs2x1SzZw4QWlJg0M6y8PRNO/mAO/Ol7gqGzS1yl8ydzGvE+zVNlXiMrnrNbbZkgGLLWVThqRWWWfI63mPE2EDg7CqI1pByPLciAN7t4nijInlc3zqN76Do8cvEB65hX3lCoQQnhgQ7O8QHFsn+kgLmV1wjecioBZpT0GtSXD/e5HWDHS2kPlF9wsJQsy970QvvIbsPYAcOQ61CvbV19DNNTfdKBKzZO9epJqga6vutjiB4YCna3/Piw+Px+P5OvAixOPxeN5CfvWTv7DyiSd/eqUhNUakZYRvRSpFx4e7uB+RMiF1BjosDemRBAQEzoQut/0b4/6QmIhUc0JCWtKgo72ywyOU0JnUiUtzeiQBs2bKHVeY5delwy3Zdr0jZOV5344btkyaZpmKFWnAqVGXo3aKKSp8b36Mj1aXWUszIgKmtUZEQETIlCYco8XH4n388q0bnKDFF7INDEKVgOmqYV6qNAmZTwzL+24wv/ccvW6VK6fu5+BchrUB+aUW6allZLaJXcvR9RAE9No17Iunkfk9yMwk0pyA+b2QZ867AcjCAff35BwSJ+jaNUiqTnSM+lCrIY1q0ffRQxYPFF0iFs1z5NBxaE45IaIWj8fj8Xx9eBHi8Xg83yKMo2pV3aTDSEBFYtbtFoKwrV1y8rJ/o6sDtrRDU+plYeFB2VMKjY726OvAlRiSk0iFrg7o2F4Z4dvRPgBtreMihIekZAQETEuTIRm37EaRqDWiUUxRwPV8jF9rWWaICTltrtOTjIUgJkJomZBP91f5reAMm9LnRXONhsZEBNQ15hU2uTrIqKkTUBMa05eMB1sJrUSpihCJ4cXOkJtXZ7hx6RD1Ro9q0ufAgVvE1Q6jW/NImDH87TY3P/sQMpujA8jPCObofmceNwZml5AgQq9dQmaX0Z115wdpzaBXzmBPPwuAWTiEWTqMPfs6MjWPtFrQ6bko3nSEbt6C5hRSTdzzdreQIIJq7c15o3g8Hs+fAIIf/uEffuStPgmPx+N5u/ORjz381L/9td9/RCnWnbBFN4glwJX/DUkJCZg2Lba1QyQhAYYhI2pSJSVnKCkL0maNbWKJqJsqFYnZp9NYUaakSYcBijJh6hxiDiMB12WLRGJysbSkzgGdZlP6hf8kY0d7CLcnKHWTkJIxb9qMNGOLLnfpAgkxH45mmaoaLo9GqApPBWcZaMpDdj9DUW5Jh0ftfjJVjpsmxyZDDgQ1DrcNR+oxvU7IZBTw8lbG3dMBDxwc8VurHfbpBFMTI/Ycf4GkvsUffekE1aBCliYYAno7U8z/N2exr/eRFgT3LqDdDrJnL1JroNcvFklXApUIggg9/7ITD3GC1OpuRWt7za1uLS6j1y/A2iYyN4N2dpBKBZloQxCiq9eR+gTSbPP5jR9ZuTR8z1Nv9fvI4/F4vl3wkxCPx+P5FuHjT6ys/LMn/rcVg5QJVIotuj+cdyOQgE3dYdpM0pAaqabkmhNJQEZGR/tc1w0yzahJhQYJx3Ses9wgIqClLry3Jgmp5rwut0jJmGWCPkOsKj2GXJFNIgJiXBTvB+UuMs3oao9f+eTPr3z8iZWVhtSYos6H7BGWZIYqId9lFvl8usanu+vUC/P5PJMsSpu9YZWepPQZURHnYTmlW7y6mdFLYX6mzx/cGHGsGbE4nZGpMsqEf/Wi5TsrM9xzbJU0Ddi4eIxXvvIe7jqySqU6YOb481Tb1wHIn1sFQKZq2LOXIE3d6tSFM8jUNLL/ILJ8AMIK5BkyM3+75Rxg0IUodrcNurDVQa1CtYZMTbs1raJzRepNF8ubDt/094rH4/F8u+NFiMfj8XyL8ctP/swKuKlDphlaFBomUiVXF927brfY0g4WpSYJXR2QaobBuNhcqWFRtrXHWbnFjExgUW7JDoqSSIVEKizSZk6b7jYqBGJYpE2NCgDb9HiX7iv7PP7lk/9Habw2CNfZ4ETYYFprPGsu86SeZULdsRelyymzxj35LH8hPsD1bMT+fJIH8338tlzkv3638nC1zUfuHZBaZXW9yj0Nd+wTF9ya2JUdy4ONOpGBwaBCtZpy7tw8n3pV2dmpuTLCfoXuzb0MB1WC9x/A7A8hCJCJSmk6l/1HkIkZqCTu5NXejtFNR+7DBFz435fdbUGIPXMG2TOPOeL6Qggr0OshtQm4ccn1g3g8Ho/nG8KLEI/H4/kW5Fc/+QsrsuufaFUtY3zB+UfGhYdG5D84fvfjxuWHXw2L0i8M510duIpCvfOx49f9V5/8uTuSn/7REz+5YlWJjdDSqvOrECLApFaY04QDtkWIEAVggQ8mk6RiedTuJY5Tbg1zvnS6wdE5N/U4caBHrvCn9zSoiGE1T5lvWQ7OpwyHES+dmeQ93/tpfvShbeYWbnLz6gLRI11ajz7D0o8+B8OBEyCLB5DFZQgiiKsQhs4DAsjUHmR2GZnfj8zsdZOS0y+BGPb9jefdMcZgjh13JYQmgGG/+IFZEOOieLf/25Xxx9fxK/V4PB7PLuQzn/nMT73VJ+HxeDyet4af+8GPPxZpwEgyrugasYT8/cd//Bu+qP65H/z4Y0fzNhGGCgHLYYUTv/aRFYDV//K3HosCaCaWX7m2wV891uBL52Lef2zA2csJR/b2uHoz4eVVy1w1YLapJNWcpJJz/noFI/A9P/Q7MIwBOP/Mg1grHPuR30P7GdoTpKJoCsG7D0MQIvVJVHOXbDUxfftETeBidQcddNTHPv8MZnnRGc2TJuQpeuF116xen3DrWUHEyeF/7wWHx+Px/DHgRYjH4/F43hTyv/obj9WTnF8/l7E/qpKr8mcfvkK/WyPPQl4+2+LR7/k8F15+F4d/6He4+m8e5rmXZzi83KM12WHuAyex1ycJ3rnO2m++l8lDr/CHv/mdPPSTvwmATM9gX7+CObrfTT+qdaRah6gCxqCrV53xXIxbxxr2nf8jqqBXzzoBYi26voq0i9Utm+OLBz0ej+ePHy9CPB6Px/Om8rk/9+uPfe9xpdXqEldGhHGKiEXVcO3yHMcf+CLBwVusn3wv/+CzU7ynUef7/vynwQrh/QO0N4JU0fUQmc2RmSYyu+Dicy9dQu6+1wmK4QBptNDejnvhdAhRxfWBaI5MzAKgNy8gSRP70jPI7KwzqVsLlYSTo7/pBYjH4/F8E/AixOPxeDxvOhP/66ce+8KZCg+/o8PE5A5JfZvk6Gv0Xj3O2dMHOPHu57lx8QiL936Z7uVD1JfPQZgTnhiQn43QboXww8alX4UBRBFYRdpzZXoVxiBx4hKsNm86AdJso91NAGdUT4fOlN5oQTrCPnOSp5d+0QsPj8fj+Sbje0I8Ho/H86YzfPTEU7M/ePSp+lMvPlKpjlA1mF6FZ758gnsfeIFhd4rp46c4+W+/i0bNsnZxP414CN2U8O4UMz8ClPyUxbRSdLuLRAHSakNcgf4OVGogwM4GMrsXiROk3nJekNUrbiUrz5DWLKDorYs83fh7XoB4PB7Pm4BPx/J4PB7PW8bNv/OxlTM/9iMrp/87lzBlVRCTM+jVCA5u88CDL2NEmd1zg2yUcOozD5M+PYldzTj3Tz+I2T8k+1IdqVUgzZyBHNCtDehugbVIfdKJjdrE7VjeAr16vvz8ZP6/eAHi8Xg8bxJehHg8Ho/nW4Jn//KPr0S/+JGVr/zFv32HGEjTCFVDsu88C4vrmLlNtBuy/0/9PubAXoZr8+hqD73ZcwdsrZE/v4NevAh5ho76YPOyVFBvXeKk/ET5Gp+//AMrn7/yMS9APB6P500kfKtPwOPxeDyeN3Luf/grK+f4K3CX+7r1T//JY+bogNb1a1w9+UEW3vVHSG1I+jsbJPdfwN5IkMbt5vLgoCD79iOTc+j26ld9jZPdv7bCxJvx3Xg8Ho/njfy/AB3OcGJXEscAAAAASUVORK5CYII=",[[10.38333333333,34.14166666667],[3.50833333333,42.9833333333367]],0.8,null,"pred_lr","pred_lr"]},{"method":"removeLayersControl","args":[]},{"method":"addLayersControl","args":[["CartoDB.Positron","CartoDB.DarkMatter","OpenStreetMap","Esri.WorldImagery","OpenTopoMap"],"pred_lr",{"collapsed":true,"autoZIndex":true,"position":"topleft"}]},{"method":"addControl","args":["","topright","imageValues-pred_lr","info legend "]},{"method":"addImageQuery","args":["pred_lr",[34.14166666667,3.50833333333,42.9833333333367,10.38333333333],"mousemove",7,"Layer"]},{"method":"addLegend","args":[{"colors":["#040404 , #4C1258 20.0734959237676%, #A01975 40.1469942909764%, #E55457 60.2204926581852%, #F7AB2E 80.293991025394%, #FFFE9E "],"labels":["0.2","0.4","0.6","0.8"],"na_color":"#BEBEBE","na_label":"NA","opacity":1,"position":"topright","type":"numeric","title":"pred_lr","extra":{"p_1":0.200734959237676,"p_n":0.80293991025394},"layerId":null,"className":"info legend","group":"pred_lr"}]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[34.14166666667,3.50833333333,42.9833333333367,10.38333333333,true,"pred_lr","Zoom to pred_lr","<strong> pred_lr <\/strong>","bottomright"]}],"limits":{"lat":[3.50833333333,10.38333333333],"lng":[34.14166666667,42.9833333333367]}},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n if( strip !==null) strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null},{"code":"function(el, x, data) {\n return (function(el,x,data){\n var map = this;\n\n map.on('keypress', function(e) {\n console.log(e.originalEvent.code);\n var key = e.originalEvent.code;\n if (key === 'KeyE') {\n var bb = this.getBounds();\n var txt = JSON.stringify(bb);\n console.log(txt);\n\n setClipboardText('\\'' + txt + '\\'');\n }\n })\n }).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> ] --- # Spatial Prediction .pull-left2[ - [Random Forest Model]() .tiny2[ ```r pred_rf <- 1 - predict(model = fit_rf, object = bioclim, type = "prob") mapview(pred_rf) ``` ] ] .pull-right2[ <div id="htmlwidget-1fb1846e5a1724559068" style="width:504px;height:504px;" class="leaflet html-widget"></div> <script type="application/json" data-for="htmlwidget-1fb1846e5a1724559068">{"x":{"options":{"minZoom":1,"maxZoom":52,"crs":{"crsClass":"L.CRS.EPSG3857","code":null,"proj4def":null,"projectedBounds":null,"options":{}},"preferCanvas":false,"bounceAtZoomLimits":false,"maxBounds":[[[-90,-370]],[[90,370]]]},"calls":[{"method":"addProviderTiles","args":["CartoDB.Positron","CartoDB.Positron","CartoDB.Positron",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["CartoDB.DarkMatter","CartoDB.DarkMatter","CartoDB.DarkMatter",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenStreetMap","OpenStreetMap","OpenStreetMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["Esri.WorldImagery","Esri.WorldImagery","Esri.WorldImagery",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addProviderTiles","args":["OpenTopoMap","OpenTopoMap","OpenTopoMap",{"errorTileUrl":"","noWrap":false,"detectRetina":false,"pane":"tilePane"}]},{"method":"addRasterImage","args":["data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAyEAAAJvCAYAAABsy95bAAAgAElEQVR4nOy9ebCt6VXe91vvN+3pjPfce3tQS6glIUgqiLgwSaqCy2WnylUUmHAHByopcEJJGEKZkoophnjTxgxRZGRsl9xCkVBaQytSd1NCCo4LW0kUEwPF4FAYEE1LrZ5u3+lMe/zGlT/eYe9z+kpo7OH2+3SdPufus8+39/6+d5+znnc963nkE5/4xE8TERERERHxReLtl941BmilI1FDJQ1zSgCG9Hi0e4p7zDlaOgAEwSB0KM/qPh0KwC8/9A/ue4FeQkRERETECwTzQj+BiIiIiIiXHjwB+Vz4tF655e1P6Q2e1f1ASBIMb7r09z/vsSIiIiIibj9I7IRERLy08PELD42/7ZFLcec44nnBOy+9dzynppWOnqbMpKJDKTRlKksOmbHJgAEFn+qexCCIGFQ7UklptKFDSSTBIAB0aPg+EDolQLiPv1/skkRERETcnogkJCLiRYpPXvjoeErDBimH1GQIc1qeNEe8oTvLX33kv4zFWcTzgrdevn/8aPdUIBUAiSTU2thuhiQIgqLhc05GTUNCQkNDqy0AqaS0dCQYWjoMgsHQ0SFIuA0gJaWhYd4teOCRt8b1HhEREXEbIZKQiIgXIT554aPjDqW0e8bUKH+YXOUN7XlalAxDTccOOX/lke+473cufmw80YY5LQK0KFtkkahEfNn4Z5fePd6XGZ/trgaSkUjCpgw46qZ0KJmkdCgpCQZDRU1ORkuLurkPgFZbEkkAOx+y/hmgW+uIeCy1onMEJhKRiIiIiNsHcSYkIuJFigShRTmm5g+SZ1hQ8hlziEGo3W5xTcf/ceHh8RVdkmPoUP7v5NN8wvwZ+1T8+oWHo9Y+4stCTceT3bUwv/EKc5ZXyx1sMiCVlEzSQCQ6OnJJ6VPQ0NCXgoIcsF0NT0DgJPlYJyqn0ZOcnukF6VZERERExO2B+Fs9IuJFCAEUu0swImOkPTpRXtFtkiGUrmhb0PKYOeDebocnZMY9OmRIwV/uXkmOYUmL75IMJcUA3/zwt4fd5N+6+LHxEzrjbz3yXXGHOeI5uP/Sr4wNkElGqRW5ZE4y1ZKRcqfs8ozeJCdjR0aOqCRUUrPBHlOWqChXtaInGbUaK9WSlLmWQbqVkNDSYjCBkKxLu+bd4oU9ERERERERX3FEEhIR8SKEYodya5QM4XXdbiAmf24OeUoOAChIrRWq2efV3TYlHVMW/H7yFP9J+0oShKm2PGoOATjfDfmtix8bX9EFT5gj7tUdfi95ku7Cg+PveuS7IxGJOIEEQ0ZCQ0smKWdkkwOdsiF9+hgMwitkj4yUkhqAlpYEQ6IGIwII98g5Wlqexa7bV7DHXEpu6DEtbXi8jg7j/mtpVyRFDKrKf3PhR8fvf+R/jus0IiIi4jZAlGNFRLxIkSAkCD0SEoTHzRGPmn2ekgNaOlIMJXZIuJSGz5hDJ+HqaGj4v8yjlLTMaPhjnmavGwBwXUuW0vIpnmFGzUTnL+TLjHgR440Pfe99foh8T7bokXFOtumRoSH7w9CsDZN7HMrMDZ3bzonPBLlb9sL63ZPN0FkRN6DukZMBdiYEQERIJP7JioiIiLhdEDshERHPEz5+4aHxY+aAH37ojZ93J/dXL3x4PCBlTsMWOUtafjd5mjkVDQ2CQekCAXmWAxIS9mSDIyq+pX0t/8r8CYqSugH2u+VMKBL/TfIZZrqko2PfLPir3esZkPKDl+4bv467MMDrul1aFAGiHfDLE++49J5xguFYSlSVq90+X2PuDORjqD0e5xrbMkQQbugxmSRsMqBDrVWv65ZAS0LCHbKD0tFCuM9AClo6Sq3p6DgvO8xYUjkXLli5ZEVERERE3D6I20oREc8D/vWFXx0/Zg44kDk/f/kdtxwWf9vld45//cLD45KWx8whv508Qet2j313w8PbmLauuAN4Wm/yyeTP6VCG0ich4d8kjyHAk3qDipbPmEOOdRb097tdnw7lcXNEQ8uneIoO6661oGFKzSMXPhyH219meMel94wBMk24qgcIwt3mbEg771COZI6i9NzgeUPDQkuu6SE39IhrekhFzRLbyfDzIhJMeYWMlLNssckARSnImVMx0yX1KdLRl17ojkREREREvPQROyEREV9l/MaFXx1PqWlRKhqyU2+7n7r8i+NNBryy3eJxc8RTcuDu3XFAiR1FF3bY4CbHeH8hr6VfyWKEWlueMEdut7llro0lJtrxjJnwaa6GDIdc0tAd+WOeCs/HQCg0W1GelCM+eeGj47/yyHfEjsjLBD3NKNxKq2mCfKpDyUgD+a1pKKlpXcbHOs44qdURM3YY0VCTOgKidIg7XkvLtg5B7Br2pAfscHpfCkqtEIR3PfwP4xqMiIiIuE0QSUhExFcR/+Plt49NIryhvYt9sVRkQcmbL//cOHFF2F3sss+USbKgpbPp0hjAhAHz13R7XDETEhKUjpQUpaNx+QmF5GxLj4zElYT6nPyFJ+UGndpiMZeUpdZcMRMe5VkaWlptea25h3PdiAUNBQlPyBGP8iyZSXj2wofGvyF/xF/jP2SgKd/xyOVYEN6myFyQ4L83V2i1c7fZLJAn9XqY3Wi04Sor0pBgU9E3ZcA2Q1o6ChdamLk1C3aOZB2VNCypXSqOuvsIldYUYjstpZsNiYiIiIi4PRBJSETEl4GfuPy2sZWkZGzpgLc89Kb7wJIPgB45C0qeMZPQ3VhSkbvuRkPDVY7C8SpqeuTMKYNt6W8nT2AQdhmFIs5mhGjQ1e8yCsfoUBZaAqv8hT3Z4ljnNLTsyAY5KUtqnpFDKrXBco0Ij+tVCklAcNIbKLWklpanzDEoXJEJmRjefuldY4DXd7vUdCxo2CTnWx+5GMnJSxxzqTEIfXL2tUPE8Lhetd0PtS5YPqRQEDpHngVhU0b0yGjpniOpOk0+PGon5WqcTNCjkDyQj/c8/HNxXUVERETcRogkJCLiS8SPXP6FsdWoC7s6ZCJLfvTyW8c9MpbUZCQsXOb5ocxJnO2oLdYUA07GYolC5vTyJQ0JCQstGUqPkooEwzPsc55trnIITorV0ATXoSu6TyKG1nU7fM7CkF6QV/k5EiVhKD2OdRZeT4fSak0nsC9TdnVkjyGGR3k23O9RnqEvBQkJd+kOj5mDUHT+oTzDtz5fFyDiK44HLr5/XNOhojwmN9hlZNeNdlS0zgfLrqNEElL3J2RDBi4rXUhJ2NAeR7II3ZPkczyeJ9V2VsT+rD9+4/qC7374ZyP5iIiIiLgNEUlIRMSXgB+5/AtjwbCgJCfjukxcwV+wcIO4lctNEAxzquAWBK3rZDTkTqqSk4X7JyS2IyJZ2B0eSh9Dxz5T1JVufrw3xXBF9xH33+px7X8TXWBE3OMLcy1ZUpNLah9La5CMBENFy5/xNEN6PCM1U52j2tGKpU+JJKjrtAjCo9gQuZ7kvELP8A3tXc/7tYj4ymEudu3UtIwoGGjG15tX0tLxZ91ToftxXnacZNCic05snkBMpSJxhMJD6cIcie/y+XyRa3rkiHJH6nJJrLtb/BMVERERcbsiumNFRHwJSN2IbU7GWTbpU7CnI+7QTSpqWlonubJvMetutdr19cTDF2K+o5GQ0HduQ6nbRU7dPrKXt3TqNfOGzg2ge9lLqx05WdDWK0pNQ0pKIXlw0upcQdiTjFwydzxLVBptONY5Rzo9QTySNdNV+3paR4jsbEonyg7F83cRIr7iyNWuuQZlQ3vMpGKfKY/rVTJJySTlDtkF7IxITUtBRkaKYIL7Veq6fjUNI81ROobaC+vdd0BqGvZ1YmeSXHfPr93KWfZGRERERNyeiCQkIuKLxA9f+tlxj4yeswvdZ8oxc/ZlxsKRAR/S5j8SDA0NBkHpAvFIndOQdx0aUjgnojTMc9iisKHU+kQgnC/kWvdZUTKxshY/9L4aGDY2/VoSJ3mxAXQpqdux7qi1XrliaYtq53IcemzIkJEM6ElO5giYuoHlVlt2ZIRR4W88ciFKZ17CWIqVEWYYHpfrPK5XuaFHVFpjMGzJiIyUzHU5CtJArtfROJJrECaypKHjUGY0jkjbTkjHDNvpA4Icq6W1c0qSUWrF91z4sWgRHREREXEbIva6IyK+CPzI5V8YK8qURehUJNgCak5F7mRNKWmQQ/lkc98V8TKVDj0hwQIrgwFcf8HQl4KZLkjISN0us58HWXfSsqnWimBoqd3OdBuetzorVEFIxZaHnuR8rhC4RBIKcnesFaFCoNKaRFZK/w36vK7b/Yqe64jnHwZhSY0gzHQZbs8kpaMLxPvzzXl4otw6et0CBzoN39+REdA4qeA83O7nQRSlkJyOjkzSOBMSERERcZsikpCIiC8Qf/fSPxzvyIhEanJS7tZtnpIDdnXEgczISDjGFlW+wG/pyJytrmBYqiUqZo0irFuS1rSh0Gvd7IhxXY1cVqnRrXaBwKjT4felYKGlk2jZfAV/P096rITmJOkYSI+lVoxMH4CpLtiQoTt2R+66MtsMucoRBmEkA450SoLQlx6JGr49Jqu/5OFJ8J/yDAmGPdkiwbCgcuvnJPVYd7vy2SFWhmW7ezf1mNTJD71Zwk2dhJ9RJ0HsXIfEE3ddM1KIiIiIiLg9EUlIRMQXgB++9LPjQnJqWjISdnTIDZkGKcoGtoCfU3KEdZxqnQQLN6PRuMC1xs11JGKCgxWsOhIz1+G4FWGwx7WPmZNRuuLQuEwFg5BJZsmH2p8VZ+F76J7XGTbpa8aBzJixJCdjSRW6MRsypCClpKGHfc2v1F1uyJS7dIeJLLimR2F+pKHhzQ+/MRKQlzjee/F94yvmiKkuKLUkkYQBhevsuVwQOlIIskJYERHb9bDObC0d+zqx80Lue0k4RrMmE1yRGk9gPCqtGcngeXjlEREREREvBOJMSETEX4AfufwL40RMkJcAHMuCORUpxn1dMqekpeMc21Ta2A6EKktq1HUyErHD3Zkb9vbFmJ8LWXcTqtzPJWtFnv+6Y9XhAMLucUVNrS19KehLQUODosxYkpGwQY9NLcgwnNMNF36oFGvD6X4AvXAyrDt1iyUNx8yDrXC2JsUSMbzt8jujbv8lgAcvPjh+4OL7n3OtPnDxg+MrLstmoUtSSdmWDTufsebyZrNjVmtU1gbQfZekoeVY52tyv/aEk9bpZHUrPLTfX39PxC5IRERExO0N+cQnPvHTL/STiIh4MeIdl94zfkL2aWiotDmRwdGXggy7UzynZIsBE2dXm2CYsKTUikLykM0BPoujI5HVfIjHWayz1oRlICU+rRpOyrYUDe5U64WbIAyk4FW6x02ZBXlYTkZBygZ9llQ0dNypW3xKroQsB2/h2zrtf0bCOd3gMbkO2MTqr+EcAFfkkInOaLUlkYRt2eCcboawxogXHx64+P7xn5qrAGwztOtWB9iwzZRPcYWSKqyvnIw92XQD6NmJLgWs0tFLampajnQWhsxrNzO0Tjh8qKH/2soI121+fRCnYaklPSkotbJEWpUHHnlrXFsRERERtxGiHCsi4hT+3uV/NE4wJGICARGEVp1MysmoDEKhKXOxgYQ5GYfMKEidu499e50mFD6zo3PKd8GgdMxYnngeip74ej0DJHNvXX+719CPpE9KyrGU9MlZsJKAZSQkaijFzZWsFYSJO4Y6xyPvrrWgodKaDemzJX1QSJy8a8IMEUOrLSMpeGW39dW9MBFfFgRhQclSK+5ki4kseFpusnRrtdTKdTk05NhkpCyd5K9DGWmPqSzD+m1pOWBKqx2LtUH2DmVbhmFGqXOr3ZOS02vbr8VV129lvuDxPRd+bByJSERERMTtg0hCIiIcfujSz4w7OoZi5zsWlACkktBo62RUJtjqQsaxLChcXkLt0g4m6sZy1RZdvuux6mTYf/tCzrgSraQJ2nt7PyERoVJv7at8Pqh2LKnIJA22wBkJjTs+QBYcuowbmrfD7j6x2j/Gkpo9NmjFyssK96uilMZZuCb46EUkYag9fjd5kktf/mWI+CrBINzNGR7nGgBHameEKmoa9Tk1dl0YDOdkG1gRg4yUA5mFteAJdK1WwpU417VGVzbVYOVYqh0ihsR12rw7m19v/r3gSUrhuiAAZ8w2BhuyGRERERFx+yCSkIiXPX7w0n1jsDIQ4zTuPpbPF0IAuUswt05VGaWbtzjLJlc5YkjBgIKlVGQk1NqSif1sZCVLabQN8idf+AG00rGy0l0lq/tj9CSj0ZOBbutIJbVZH6IUat/andiMkZzMSrGkYUeHdldcGieRUQRlQMadusWBLNhnilFhQMa+TGnJHLFJKN2w/HoX5hk5YKYL3nb5neMf+cj3x93qFxEevPjg+KbMKSSlUEsUclJexx18Rq7zWmzK/T5TjnUWcmT8WvRD6M/qMSUV98i5NcOELswdnZcdrushqaRsypClVvY4joB4eHIiztggldQ5wLWWxmvLWbODinK123fm1Ia3PfQTcV1FRERE3EaIJCTiZY0fuPTTY1ilha9r2L1blR8mBwIhACsV8V0Qg1DRBBkTnLTpRW0RJk7mkpIE/fx6roIhdXr9CnWBb4JBxHdNVsXcurzFJqvb7y20pJaWnku/7pzcxbtj9SQLhd0OQ4aaU4eB+5J9puwyopGOm8zJXLBci3VHamhp1lyQWm05YkrurIV//cLD42995GIsGJ9HfOjCg+OrZsoNmfIzH3nziXO/cFK8koZSWlDC3M95ttnSHn1N6UvKIdY+16+zxnXLrui+NUrQjlqsTKul5Zoe0bou4Q09otaGRBKOdIpx3Y4OZUTBXBe0QuiC2E6f76hZ0r0jG2zKAIPwP3zkB+MaioiIiLiNEUlIxMsavohP3YC4Ebvbn2AoyFHRMNNRaR1mQRIMG/RtIjSLkCDtLW19x6RWv5N8kpj4r0/LtbK1ToOH/XlhqTUJhoqanuSUXUkqNsPDJrJbUpNJwpQlGUP22OAqRycCCQ3CgIJMrdtR6lyPplLxLAdhcH6kOfsyCxKdDg074H73G2yH6LzsYBDubXcwwMcuPDSOuSHPD/7ZpXePZ0lF6eaFHrnw4fE1M1u5WIklHZ+VG9Rq3a5KR5y3tU9LRyktG1qwZ7a42u1jxHCgUwQhlywQEH/MBSXHOqd0EqnXm3uoaek7ieKT3TUySWm1JZcsrBtP9L0UyxJru552ZIOzbJKpsWQpIiIiIuK2RiQhES9bvPnyz40NhkxW8xA5VnJVYglKT3Iqp5dfzXAIIwoWlIG8+OIsFPUitGqJRe1mLrz7j7+/P/6665VNUbeZHSP6FGTsM7UERzJaOnLNWGqFuAH5VltEDJsyAIGUlC0G1LLqtCRu+D0hIXcZIEhKT1MyTXjWTDhgEobnr3OMygYDLZjK0pE1DRIdn0HiHZRG9KhpuWkWHKthKhVvu/zO8Z/rM6FD8mrO8+aHYp7IVwr3X/qVse9udCgDClo6njRHlNKypOKQGXeww3WO+Ro9S03Hn/AkfVL+hGfoSc5dus2j8iyv506OXYL518h5ntDrVFRUWnOn7NKTnD/vnmZfJ9Q07MkWd8quJbWaoaRhBuleYyVe3sr503olPG/vnNWhvErOU1GHrJpMDT8c10hERETEywKRhES8LPGmS39/nJPR0dGq3Z01ImQkLMDJmGy+QSIGUXUDuxKGyGvaIEnycx01jXUT0lXa8+lcBLCSlEwSZ9+76pJYcmJIMGxqn2Oxtr/+WPb5WLtc1Y7OdVH6FO6x7L/nlPTISdXQiQYHLpv/kZFpQoahrxmlNChdcAEDGLrjLaQOj623eB0VNdf0kIGcJ8GwpObIWPLzNDcxrkOjKE/IdX7p0rvGPTK+/6G/HQvNLwP3X/qVMUAtHdc55k7dZoOCkoaKlkITWkk5zzabWmBki2fkkC0G7LFF1iW8Ss7yqD5DLQ21m1NKJeFeuYuR5twtZwA4Ys4GfXpuzmihS/rSY09HdEBBwhOyz8J1RVrtGEqPmobX651MpOT1vILatBwzJ5UkSAf7mpFJwo995O/E9RARERHxMkMkIREvK/zU5V8cH+qMc7LNRBdkklCqlbGkmtCI7QT4roXvjlRSk5GFDsBqjqMLcyAphjnViWIeWOuWrCRYltAkIWHao6XjVXqGmVR0oiyo2KDHkpqKes1Zy5zIbcglQx1J8h9zSs4wDBKujJRdRhTq4wiFTpSpVMydla+HQcg0YUDGkSNC9jWo7aYILLUiJaHnslCOmIP0eUpv2GBGN+jvz2WtLS3K3MmGIr40fOjCg2OMJaRTlmzQY4MiDJ0Xbg1nmtBT24Xa1ZQjM6ekpkeGIDwpN0BhrkssIVe+Vu8g15RaWvZ0CMAOfTqUq3LMlozYZWSJshoS7Jo41Am1c3EDgnzrj+Vp7mGPTS1o6BiRUzqbA/t+qSMBiYiIiHiZIpKQiNseP3jpvnFDS0rCOdnmjGzYAlmETQZc45Ce5LRqU8j9zr0PElxqRSYJPXIQwqB34mYp1mc4Utc56dR2NwrJg8NWT3IWWrqh3o5O1UnBDHtssKCicKGCRyzpacpdbDORJTkpA3KW1DR0DKTgSE8W8+fYJtOEI5mH5zSXmiLY8Hr7VZsjcsycTQZUrLodHsfMmUnJHbodbvPWvAmGGkG1w0iGojypN2jpmMrCDq5r6whKZi1cKXmFnLXyL5LPObz+novvG98wM57SG7ya8yj6sg9AfPuld43nUvGTH/nv77v/0q+Me2LNEIAgwSo04YqZBCOCV3bbPGH22WVEKx2JGkb0nDGC/dlX6lkAPsVT3C1nSNXQ05Q/N9dDZk2fIqzxO3WbLQYcyIwZS5ZSsaNDPsv1QEB2zRYdykRn1CjGGSp838Pfcx/Auy8+MK6lJcXwdx76b1/W1zUiIiLi5Y5IQiJuW4wv/+OxL7isdMl2CGYufM33A+xchR26bbTFz4nU2lJrS1+sNClfSxb3Uiw/X+GLeC/FWg2i205HJnbGYyBWMrNu/ZuRMNCMqSzpUK7JFLCdk1pahtpjLhUjtSRo6fy4/OvqSw913YaJLNA1kjRlyYheKCprGo7dsP1SK9pbDAAbl02yIz0qaRhqj6XU9DRjKS4DBaUQW6D6OZKGhlZXx2toAxlLMAw0t+dPhWOq5zzuRy98ZLw0lhCNXDAiwM9ffsd4T4e88aHvfdkVrR+98JGxGuWcjvjFS788bqRjTk0tLbmT/Pkz7q81wDPm2Dq2uWBKK8mz6z8j4VAWXJUjvrY7z67ZdA5pLYkYthlyTQ9JMJTUYT1V1GSSMNMlLR1zlsxkybxbuE6c0iPjGDtXYhAWWvKsHITX48lIRERERESE+YvvEhHx0sP48j8eHzMPicuppKEIq6hZaEmlDTf0GHAFsyu0bHp0HWREpVaUWjFh6QbPkyBxqlzHYaGlk1qZNdeoNGSK9MipaBzp6SgkD7vEPTJKaV2HobG7zFTMpaKiZi5VkF4lJAxdoVlIQSJJeJwJCyrsEH3mujhg50POshV2wO3x6zCD4js7p5G42RTvvuXR0rHUyqbJO5mYT3Jfl5dZG+IVQTO4ThHCkIy3Xr5/7I/50QsfGXcoSxon87GP20jnCu2Od15675iXER658OFxTUeCUKhzb3MSOiAQwp5mYW37a+YH1eHkTJJRe79SVt27mS55XK7zabnGY3KdnqZhxsh3Bf375qib0riuR6stk87aPm/K0BJHoCBnQ6yUq9aGM2x+lc9URERERMRLEfKJT3zip1/oJxER8ZXE37v8j8YLSvoUVNQsdSU32pIhC6x9ri2kbWHu5zYaWnrkrjB3XRSUvhRsYQurhdvFr7F5Cf6+iZMr2cH1ipwsFISemCQYBuTcZGKH10XYYkhBFroVnhgMyElIKJx8y7tsAZRONjan5A52sAKsNnR4vKWuf9136hZTqRhoxhU5oqKm1S6EKPqOxfq/c8nCIP4uo2BH7CU9DQ1TXdCXgoWW+GyJnuTMdUmKTW7flQ0MwjndINeUuVQYhEfl2XDN3tDdw1xqPi3XuFvPBAKSqqGRLgzST6Sk0JQt7d22u+oPXHz/+EAW7GifhTTMqEgQli5cMlVL5OZiLZtTNZzVIX9mrrHFAOA5bm7rt/mvl9jr8IReXyOKhkIyWrXrTVFG0qdHTknDVOf0pSDDkvojnbIjG+SkISfHk9sFJdNuzkD6LCj5S7yaP+Zp/slDP3VbXreIiIiIiC8OUY71JeDsfR8ZXx9fjn9IX6TYwO7ILigZ0iOXjGOdWycsOuc+tcrsMAjbMuJAJ2tFWhekWgPpMaQXhsRrt7vvCYH9OgvFV0EaBtAzEiYsrIRLW3pi5zpatcf3Rduc0slfVp0D8ETH2p6WNBTYbojP9vCEICOlYZVJ4gvQbo38bGkvDNAv1ggIWPLRqZ4gI5WuuhudDMlcF2ZDCw5lzq7uMhFLh0rph25KS0dfCkqtSUg50CnnZNvt1sOn5Rpn2GShJXfJGTqUI7EEbJMBrRusLjShdkSkdiF7/tw00vHgxQfHjbumBSlPm2Ne6mntb7v8zrERwcuufChkLR3b2mdGRS0tiSNiCYYZFYqypGbk7r9OQIy6NS26us25pJXUnJNtalpqGiptmOnyhFSxR07PEWojwtDNidS0bMmIHpmVcjkSagfVp6tOoiPtV+SITpV3Xnrv+FAW/PhHfuAlfa0iIiIiIr48RDnWl4Bf+8OCD1z84Pg3L/zay0oe8lLA2y+9a3ydI1dk5WsFmS2IclJnvtsGmUpfCpbOHcrftu7qtM2QykmPDpkFmQtYkuCLNS+tSjDu/yd3om3voqWkIZXkxOxG7SIRey5To3A7y/4+I81DOrsdEE9Dl6SiZkF5QnZjX4OEzz21haInR550nMbpzoj/+UNmoctS0zGl5IockWlis0J0xJ5u8HXdeXJSCvJwvAVlIEctyl3sBrlY60hE7YiHP7dTluE2j3VZWKGWmPlzdSTLIFl6qeJDFx4c73T9IHdVdoEAACAASURBVJU6ltKeH+kwKsypTxCJnmYUmrClPRqUV+mZEx0Qj040SLhglWljAzkzhvTYZECjLS0tqiuHuC0ZhS6hIPSci5aX9hVuH8ubNBzpjEOdrrpy4j3ShKt6YDuT1DzLAb946ZfHb778c/F3aERERMTLFFGO9UXgfRc/MDYIiQqb5Gy4P8Df8sh3xB29FwF+6dK7xmAdoZ7VA7ZlGIql9VmFxpVICy3dcHlGLhkzXazJphJat9O7QZ9j5s4iV9x8h7XLbWgoyF3J1Tm3qYaUJMxntI70LLViIAUJiSUkrhsxpEfj7HV9ce5/rkdGRcMGfcTp8nOsTGrpQg0LTSmlCYnZVr/fhGPmpOzpCEW5ITb40IfSrXdD1omHJyv+a7BOX/74PTIOdBpuG1KwpGaDPimGCQv2dYLBsCMjm9DuBvBrWj4rN4I1LMBAehSkDOmRaUIrHZkaDmQeimo/dN2hLhxP2VLr+LQv9vXU0r0kuyFvv/SuscGaERjs67DBlOmJ0Mmdrh9kas+aic3ZcJI/g7BYm/XwOE1KfHYM2PdF46R+x8yZ6gLVDhHDnmyRO1EdcKJL5yWGqbOZ9o9zRfdDF2VDBgjCXJdB2tWhDKXHTJcMpUepdVhfc7VW0AkJ73n4515y1zAiIiIi4otDlGN9gXjfxQ+EHbtWlFo79qnIYjPpRYFfvPTLY4AeKbuacSi2YzFhwZAeExaBkAiGTXp2wNpJRQpSJqeIykB6jChQOnLSMCNiuxdJ2JW3hZmhBTcr0QX3rIra3XOt2Mfa/6oomwyCna7fYbbdkM6NGEsoEq0O35Bpwlyq8HoaMaHTkJNRc7IQbWjZl5mT3zQnhtbh5DyI/9p3d2St6PSOXrbAtMV/5YrIxs0rHNDwWj3PgIxOlBt6ZK2N3Xmbi+0mTXURiJRBgivZscy5hz2MCh0EIufP88BJgSr32tftkmtXDL/t8jvHLwUi8uDFB8eJChOpaE1H7V7nSPPwWlo3D1NLi1Ehw9ro+nPSSBe+vlUXZL0bdnomxEMdpajUkufGLVVvgKBOCugH3+3PPvcxEixxqahJXXo62E5jrfY6qnYssKGGPtzQr7HCudC12vJ9F39y/O6Hf/ZFfw0jIiIiIr50xAr6FvjkhY8+RyJwuoicS0NJyzQGr70o8JaH3nTfWx56030G4VAWbLuQvh4Zc0qX/2HlUH6WopCcHdlgQ/oc6iwUQ36oOyFh6eQj6+GE62TEOzcpypCCxHVH7KM1wb5WMGSkVGrTyTOsjCk7RVAyR296TkpW0jBwg/IehSvuei435Jg5jXvOS6rg2OULz6UdQ3dhccXJtXyKgKx3RtbPRZjFcK/9kJnbC1cSl9qekdIjJ9OEoRYMyIO0x54D+3pu6rHdbT/xnupClkktLZ1o2Dkv12YewDo8rf9sKc6tyeVh7HaDL2bpvCD4wMUPjo+lZC41AzI2tAjn08MTLEVthgcpDcoVM+GGzCnc4P861mc+Tn8v3MedW/8+aOjIyTgjG6G7Yc0NWirqIAH0kqt0bfi8cbMgPoDTzw2tE93MdUv8elwntuvdNn98/5ojIiIiIm5vxE7I58BvXfzY+D99+NvvA0dK5CQRadxu+OfC+y5+YJyp4RuzLX6jfYYO5VGe5T/iHt7xbdd48tOv4d1/1PKdj/ytuNv3FURDR09TPis3OMMmc9fp6LtQNy8d8cRCXGHkE54zsfIgWwy1WN+sFVcX7IB5Jy5JnZqek2MtqYNG3hZpNsncIxHDUitQSMSH/rWOKFX0yMic/GVBxcC5e1kyVAW51dS5SxWaYqTPhMUJYgS2I9O5Z+GlM3b9Jgy0CIU7EF4/6HPmRE7vnBtXgPrkeIBWO0bSZ6ZLkJzGSYkaOkZmYDMnxNAjt/dxWB+gD4RJ67C778mNfV1WzpaIAYFdHZEgTKVyRMliS3vMpORtl985TtTQJ31BQ/EevPjg+JpM6ZEFElVKy470mVOzlJraSc/mlGzQp6Rlp+uzkPrEbNGMiqlxr1dW3SGPz0VIPt/3Pfz62ZMtADdonoT3TE7m5qicA5obRrc/27kOyUrGlzgyAfYd5N8r/v3gDRnWO24nSWkkIRERERG3O2In5BQ+eeGj4w5lXyseuPj+8YcuPDg+cOF2HmFXXJRWbv3HckcLXmc2MMAb9+4C4DXcgaJMDs6yuTmjRW/ZdYn40vFDD33ffcCq6HEWuhlJKMwzkhNSoyUVO7KByCrjQxCWWnGoMw50yowltbYM6TGUHj43AdYcm2goadwe9EoGY5xsqdF1CVEbnltBRo+MFJs9MndyFfs8fNGXU7j5CS8hq6Wlpyk9Mja0H35mffDYd2A26LOnQ2YsacTOmpzVTXYZscXAul5Jn74UX9BOtH+MXFJLrly3oqJmRsVEyhNF5lyXHOucuS6srEzMGrFZSYhKLbmmh8ywgXgjN6jeYmU8R+567MuUfZmxFJtkAbZDYmVqtvg9nW/yfOFDFx4M72n/uiqXuNEBmSaBkNRipXtHsgiubtaRKrXzHpqQqZXgDcnZ0T4tbSAAnwunCYgPiVyH77x4m2nvcDXAdrFatxKA0EG0P9cFO2r7vY5qTa7liYu/f3aKyK+vrVWo5+pzT/IT5D0iIiIi4vZEJCG3gNWqW01/K8pUajK1RdO6A09Dhyj86oUPnyAS//LCI+MlLd90F3zza5f8B6+/ypvO3M0P7N3N3947z/v+nzOYpOXr0xFTGn79wsPjf3sxOm19pTARK0A6cC4961Ie707l4UlCjywMSfviaUMGDKVHITavoy9FcLHy90tIKN3cgqy9nfwciP/PiJwIfktIgrTJW/76Y2ckFM7ydx3ejrdycq8pSxdymDCTZcgoMQgb2HmWgZN1DTUP5+CQme2kkJCote7dUzuAv8WATRmcIN3eYcy/ZkHoiQ3IG9Fni2GwHG61I3fFqbcOHkhvrdtCsBKGVSbKOua6YE7JjKUtkJ11sD1HVubmiSFAhiH1hPIWg9nPFz5w8YPjD1z84FjFdkD87QNyjmRBJ0orluDVa0TPX+9E7frJHWG2eSrOaEAaUrUFvnUis5k1tzp/pwnIrXAiwBAJa9E+Nzt/dJqMrq9vL1f04YiAc5BrnN3v6n2yHmZ5unt8K0LirasjIiIiIm5vRHesNfzbi782rlVJEEo69l2oHdik5wEJc1oqWlpnlTnUjDulR4rwLa9W3vtpW+C9zoz4a19fcebsAf3hMZODM2yeuc7xzbP89h+e5Rted0xVZXz836f89Xvho59uSBH++iPfeR/AH13+38f/xRumqMLf/cMbHDDhbR/5iSjd+gLxlss/PzYIO2yEXXXwxZMtcNazPGpajnXGhgzY744QMfQpuEt2OWBGQ0OPPHQpfAGma0Xgam6kPVGwBTmNc//pSR4IT+F2of1xBuRBgz/UHq3Y4fmGjgFF6IJ4966l0+yXWvE1nAsJ41OW7OiAfZmF5+0LQ79jPSBnS/sh9M6/jmc5YKkVKcmJYlVRcrHD8d4Na4sBBmHhBsxrWnubCjdksuYMVuMT4f0OfOs6Q7Iuu3K3ZWKTUM7IJsc6x4iE/IqV5CfhXs6TuOe4LzPmVAzIGWK7Vb6wB3jzQ2/8qrx/Hrj4/rEngP56N3QUmoTfE/sy50DmbGif1snNfA7KeodiSM51mXBXt81EVtK1Dhi4NeMH/O3tz+3Qfi4S4h+nkVWnCOxmil9XHplzdwOC5bRgXC/HYv3fp8mQt5ZOXddxTsVCSzp3m18H9jhyy87b/Q/dF3/fRURERNzGiNtNpyDAxCUqbJHTJ2WHgg0yBm6/NSch8X/Q6Zhry0aa0O/bHe1SWu7dsjt+SVpjkhoxSlZMGGxM+Etff0jRqyiKmissKCvDiJQMwx9c+vj49y59fPyaXeGDv19w/x8Ix8zpUN5y+edjt+QLxFIrNhmELoOXjvivU1dYAYGANG7uQJzb1FwXJ4qlJRWtrmaB/NB2s+ZuBasdY6Wj0pqlVsFZCuwMhT/mOgEBa4Na0lhyITYRXTBhGNhLsmoaDplxoBMmOmNJxYHMaOk4Ys6CKgwE20A72x2qtA5diQlLDmQeEuDDAL+2IRvCvo5VsehJheIT3S152dMROzrgvG4y0jzMLPgO0VB6IWk7jDivkQ/V7sQQe60NpVahe7PQklZXu+tAsEz2MqdDnbHQkgOdUlFjVKjFPtdWuuCg9pXEey6+b+xfp//s81hq6YJN8rb22dPRCTe9DksM9mVKJ3bzo9CEbR04mVRCi1JKS0FCpgkTKZlKFbpvt5LOnZZh+Y9GOhrxK251Tf3PmDUy4C2tV/08Szhs0GdyoqPhb/PzTP69tT7r0yMjk4SUhE0ZnHjs07hVdyciIiIi4vZDJCFr+M8e/pv32V1d63o1koQBCQWGAsORG2buUFRWO9zH1Bw3LdrB15oR37m7y9ntmuFwQdGf0bUZw81DutZalZ69+7P0hzPStOHALPkXT5VsJAl/4zXwV17X8M33KL1c+XfmGv9fcoWahrkug9NSxF+MWq1r1A09PlHQrKc69928QUVNT3L6FJRUjKQfCiEv3epTnBiWrnTlmJWc2hEGQsFm9fJ1sM0tJCNx8xBemuU7FLmTwPjHW5d9NXQsqGyIHy0zSibdjEpram2C9ekhM47VZ20rQ2dFbJ+TOlmYL2DbUMgmapiy4BDrErbuXLS+U91qR6td6C7tM2WfKRUtN2XGREoyTZiydDKjlfeFf3w/Kn8rnN7B990j3yHxRMV3dI5cPkiQEUnipGonwxttB+zzS5S+WDxw8f3jz5qbHMkyzBt5Ynos5SocUgmFeY3tfKRqaMUW+j50sUdGi1KQMpeKEmuA4IMJ01OSvfWulqLO1th+Xu+urM9bnD7vp8nIaemd/6ysSxBNcLJa74asZkVSMlJyF67pZ0T6FIykT07KpgzZkY3nPIdKa3Zkg5GsZpwiIiIiIm5PRHesUzjyDja03FA75Jq4oqxkFRrWoSQqqABqOyj/9E8X/M3tPl/76iOStKHolSznGww29pkenmUx26BrEwZnnqY9PEt/OCfRHk/LjLLteOYxw67ktCjP6oynkpss1Q78pqQsKPnhSz87/qWHfjLKFP4C3GvuwmB3mQ91yl2yG4LVvCNVQcrSdUWAUPAN6dGTnEpWQWotHbuMmDvZkYovpVfEcH14N3E7wR3WWcjISufuMxTs7FEddozXd/lrF6i4ns7ud/i9FOuku1HHVBeBMPQpOJIlM5a2OHbdFz+f0mqHitAn5wYTrsoRtbbBotfviq8Xp+ufe+TWlcsd95ocU7rU+UJSzuiQnJTPyk2809iKoLn/bkGqT0t6aq3pTiWn29mThr4MuKHHqFi5j7rZkYSEuS65Kq7AFVvgz0X4B5f/yXikPd7y0Ju+5PfQey6+b+xtkg90ylLqMCOjKHfqVphN6avtAAD0NF1bH8IBSwqSIF+raFiKXVGVNGSaUGhCTsq+mQcp4Xq+TbbWs/Khhj5V3Z/D9c/r53D9+loyvCLShueSksz9ucjUruTWredaWjJNgpsXENbu0rm8lW4Dx5O1HtlzSG7o2ukynLOIiIiIiNsXsRNyCkduBzLFMJWahTQcS8VSWlSgkpbaF6Di5RcdlSq/KX/Grx7e4J//bkKW1VRljnaCSWoGG4cs530W8x50BlXBJC3fvXeG7zqzxw1Z8EfJDX5brvK7co0nzBHbDBlIwVwXLHRJqWUgIi/0eXqxY0NtZsddus1Q+hyxcCkhtmhuaJmwIMGwQT/sYhfkoUPhszpyUhcgiJNyJYzoB+LguyYGm76+LscqxMtQ0hDal7LKQ/DHhdXuPoDfW1/vDCwoWWjJVOeUjpz6n7P2tlb21WpLn4JDZiwoaWjcZ1ukJhiMWJFNprZb4YPjPE5bpnpy4ovFnhuc94Vs6YpiP5jui1EfeOeJUKcaOi2n4c+hrhXB9hyYcN06lMbZA2eSkEvKgU457I7taxODzRxpWWjJUisql6XipV2C8KOX3/olvYfeffGB8USWHIhdO7mkYTZnRunMEDoGmrGUxknqvClCF85rgmFEzhU5YoIdWi+lpXREYkHlbKBT+ppSaMIxtuuzpT12dBDW3dKRvwmLMPuzpDpBak9f09MExH9edThOfm/9GDah3ZEdscSoEyXFkKj7wFBo4jzfLNnN1si3nzk5TW5TSSm1ZNrNv5TLExERERHxEkIkIaeQ6K1Dvqz2vztRQPnPfVISV6T9jnyaf2n+mKrMSZKOrrPFWL5xk9HWEcYomA7thN7gmNe86ph775nwX+2exah3abJ/rM/pBnfpLufMGZa6pNGGRpu4S/gFoGYVsmcLdls4+0JrfSfWzzXkpBSkpCTh+wmGwslLfKhbQRYK+pyMVr0sJnlOdyR1KefGkRmwc0R+9qN2Ew3ra2o9PwNsh8UTktVOc0ujzYlCU+kYSJ/CuXiVWlGqLYDXuw5+xxlswnyf4jmynM8Hv/58pog6guG/nkgZXoeymiHxBAZOFrefywnJXxefAuKJ4npI5FIrls5COyNlkwGpI4qZJNwje2wxtF0ehMxJnQzC2y+9a/zWy/d/TjKybrX73ovvG99/6VfGUyl5Rg65IVPmUnMHO2wzDK5dAFfl2HW5KhY0oUNx1Uz5jLnBZ8wNnjJHFGpDKbcYhOI904Trchy6c2BzRYZacFY3GGlO4RLTz+iQLe2xpX1GmrOrI3Z1yBHzMONxK5x2xlq/3cvG1gmJ/Z45cf9aTkr5jIrr9LhrpyasVy9FtNc0ce8fuwZ3XECit1NWlFTS5xCoiIiIiIjbD1GOtYYPX/jQ2Od+tFgtt5foeB22HzQF+wf5vPb5pt2CPFV+6OY3c7/+Pj74S6SjaxPq5YhcWkbbz5IkLc18k9H2Psv5Jlu7+yymI+69Z8LwIOf7X3GGdz51k9r9UR6Ss8uIJmnY745oafn67i7+3aWPj7/xoW+Lsqxb4J9e+l/GlTTcYIKKLVR3GAG4GYnWTUWsdpILR/zAyk78QHvfyY4St9tdrQ0EB1me2JLPkp4VbGFs10pOGoITm7WirOZkgbc+i+F3pBMSlJpd2SSThM92V7HzJk14PE98/LEn2lBrQy6Z1eNLbzWb4poQd8gO12SComzKgIkuCHMbelLq1eqKLAjCEQt6Yjs7Szd0739mT4dBdjN1DnPBqti9MoMNHay1CdfBnzP/ORgCaPWcgjqVlB1GPKnXSCTBp2bcqVsMpLC78Jow0IyepNzgmCUVZ3WTShq2GQay99bL949TZ1XsX9++mdMzKT906WfGHR3fxL3ckCkTP+BPTSUNX9ed50gSarGdtR0ZUdJwRY7Y1WE4ZyUN1zlihw2u6gEzWbIrAwpS9roBB7Lkpkw4qxtkJME97Rk5tM9pLcn+DEOuyFEgGud1k0Itgb5hZiH00hsJrJM8/7vs9OyHHzVPMW5GZZVpA1Bg6Gnqck3WHbJOdjJWjwMNPq/G/t60w+orSWtKGgbs+xQkYmjVzlDFwfSIiIiI2x+xE7KGVmy3w394QpKoLaBGasUFZ9Tq9Hua8Lp+wWtfOeHOs0u++V4r+OlJTlnmqBpEOmbH2zTVgK7NqcoeSbok7x+iahhsXqMYzBkMlvx3587x6WtJ2AVM3c5oS8sZNrjTnKXVjqfNIf9Knn7BztOLHTU2NT3BcKzzE7MggBOGmDA0a2dE8tDt8knmfmjYkw7wQ+L2Otc0zr40PTG867Fu2Vu4oXMJpePJwnB9QP20fj9zVrmeyHTanijKO23tbIYqlVoHrIUu7d68Niy0DB08n1Vin5eVLeVk9CmC5ApWQ+RGZO11rLJAKq1Dxw6sDOqMbJBKwp3dkKWbaVnf0c4kYUdG9CQjdxa84IbGJSGR5DkSM/tcVl+vd4g21HZ9XiF7JK47M9CMkeaMtLAuXe78AVRqbbCDa5NoGBLvRJlKSS0tMylp6XhS9vGWsla+ZBPvBaFWK3maS81Ac+7ULc6wyQZ9NuhzzJyBs9t9Sg44kCV9Cs7ogPOyQ6OtO/4qPPNYrQRpV4ccMmOfKR3KhAVHzDjCBjVekSOOdeY2KXpkJByYhRuGb8Ic0bqszf/79Nr059I7xoEfon/u/pQ/962sfj/Z2906DPdbW0O66qgUmoRr6buGADuyQU9yNujbNakNmzJ8zuNHRERERNxeiJ0Qh/dd/ECQXoQBXvcxJKNRO6C+pz1e3+9xXPVIRLhjS2mahLo2jIYll7tv5NeTP2W5zOgPDGna0TQpXZuhmrCYDdjaA+0SujZhMTlL22T8n793lrJTfrO5bncMnWWnd6BJMeyS8azc5LHuab5RXvtCnq4XNc7qkH2Zs8OQzBW2M5bsMKKkDkXaerqzlf4ktC53Y91RKsPKZmbYnIo+Tu7kdPfrkpXTO/qtIzjrj7deSFdqnbk6LIFQlEJyhNV8ROd+9ogZ025+Yi5C6RARjKsvQxdurcDvtCEVS5QqrUkkZyj98LoSLNkN8kIpWJxIbbfuC4bVULsgLNZydKw7VcurdI+rMmcuJT1y1l2Vthmyq0NKhjwpN8Lt3ga2c9fhL5LiqCNq53SIynl2uz6ladjVUZDhNbIiLn1N8bW4z9eYU7q5jB5TWVI4qV6HclNmVDRhTqZD7fA1NrDSB++12jGVytrnOicwo3Yw/27O0NKxQZ/H9Rq1tPQd0T2nI1Q6rumRzXUxwkhztmRIjbVYnumSHRmxpGYTO4C/J5sgUJDRk4xzusEVOWLuXaikCx2Uk9du9TvNd6U8obwVbPdNMGSUzgjBBmXa7BM/twJ+SN0TO2c7jCcsBMLSUy+FTNbeG/Y6Dp3znDcw8I5yERERERG3N17WJMQTj5OF5OqPtg8cq+nokdAjoS+GIoG7RobtYcfR3DCcZQx6DWna8W2vE+598g38b5+aM2KT7/7Lc7KsZjkfYZKWs3d/lrbpUS+H9IfHJFlJ2yb8v9VB2H1PEdCUM9rnUEp21NpVlrTcYc6gKEfM+enLvzT+Dn0N/3GUZZ3AjhZcNVMSDOd1k8flOjNdsidbtM6lZ0ntNOi2y+XdhjrUZXNYt6KJs4g9I55gdGGmZEBOi7Wr7bvwPltEr5CQBAmY74Ysqah1dS8vDLOdiOdapM4pUTQQEKWj9kWbIx9KRyop6iRRvqnhC/qFLsNOuM94GGjBQmy+9cgN5IPdFV9g5VW+G9Kq7XaMWIXtWfeqirvlDDW2O3NOR1yTKbs6ZCJlOC8ZCa/p9ljQcGAWNGtBhb5wbunoRBG1BW174kyePKc5GTtaBJnkq7s9vGPVHBhpzlQqUjUniu0pSwau6J0wo3IuVktqdmVEq53LurDuaCkJS614Sg5Cl+OalvSlQOmc5E+5Q7fpaRpIzowltbSO3NrzdcSMc2zQoWzrIBCj1nXuRtLjBpNV54aGHhlndEgmKZuuA1s6QuSlTKU0YZ5ivdtxK5KhrKRxvtvkOyCn7y9IyKXJ1HW8MBhWAYueJK8TkBPXSg1IR+aeX0biBulXSevreSIjGbh11fC9F398/L8+/D/F320RERERtyletnIsT0DWQ+M8Mrez2dKFwLEtMiov0ekgTWDYb3h60pGmStsZuk4o8pb//Bv22ZOcf518hrpOaJqEps6pln26NiNJl1Rlj2LrOm1tCws/xDnQlD3tk2HduRRloBl3dDbobIM+2wxZOhPXP9ADfufix8ZP/tf/IjpmAR+78FA4D1OWIZG8dRImf53XnYJ8p6JyEiK/qz/TJQub1sBNnQRZVEvnwgBTRxy9tKt9jgRmk0GYJRK88UBGLta2N5eMyg2P5y7uzctzcvfcUww31aePdyekXKfRoWF+wCB02obOgXdT8sXfQuyMyLr0xnc4vBwLLBnpS0FORs+lkQ/crj7YEMeKmiUVj5kbbGmPQlMGmvF13Xleq+c4r5vUtFw3U25inaz8MHIihkwSNyewGor2ZMlnUXgpj9/RLzAM1BbJfSeJK2nd49vjbGhOGohAR4+cmoYRPYb0ONQZpXMPAyvlG2iBYKi0ISdjw83L7DN1EyFNmNuY6iI4hXnXM0W5qRPmlEzc7MwWfUvq3Prx8ruShiPnemUNCyoqF5A50yVbOnBOUjU1dh5j4rzO5mK7equO2cmPdXte/+/1LkS5ll/j18vp2RvfpUqxrljVGjHsRMPQvf35k4PuqVupmRq3qbNuv7wyWvAkyLuu2dknuya/9+KPx99rEREREbcpXpYk5IGL7x/rqT/K61gNY658/Ws6pjTMtWWQQz9XqsZw18iewtkipW3N/8/emwfZmp/1fZ/nXc57lj59uvsuM5pNAwgDxhViVzBlpyr/OFVJquy4uHMvtisugWXjAsoJQRiBZODomkUGhLCMI8AyGLBjBLMQQhwncaLEleQPCuMKEBuBJaTRaJa7dffZz7v9nvzxW973nO6+M0ISmZl7nqmp2336rO/7nt/vWb4LUWRQjXjRddABJFLmsz5RXJOvBuTLQ8o8o15ZsrSpbbL1Fx8e8dTDI/rEVCgvy4y7smQia+ZSULhueoQwcipAn4zu8q/lLv9mteTXn/rVB3rDfu7aL41zahS4agahi9ulQyYdckcsz10SuUeXoUtfvTqWd3qe6wrv8VFTs3LwnTasxTuM+6Ihxcq1NspTtrjxhof+dbpOf8hj5dtcjLYZXEZqO+rULHW1AcNq3z+YxWmjzOWVpXwkktjCB6GgJNeCKUsLh3FTAx8xcTBlNKp0pROKOXVJsE9Yu9Lhns4A6zx/R6dkJORSMdIufU24avo8YUbcE+tAP6BLRxJb1IgtanzBkRCTSGI5IlvLk1cZ85H7z6pNcZk47oG/X0bj1+INDQ1KqjFDenTEyhPHRNzFfo6l2OLBK6D16ASDyakuySTFczkAO1kRW7imTp72UPYChM9fG21Z3JiIKUsmuuBU55QOtmRUWWlO5kj/VDXDzwAAIABJREFUfrrTtW4z3JKJ4yw114FprWMbXKGtBkv7eihdoXOeKEA7/G0VSkFN1fJs8XyPeuu1a3SDvH5W5jfa+Nk7rvv7bl+7u9jFLnaxizdnPJBFSLtbuH27/1cU9jQlViEjZklNl5g1NbcWymwl3DpJ6HWUz9zu0EkMeZ7Q7dqu9pNRn6/VtzGbZTz7ayNE1Cpl5RnrpS0+qqLHcj6iPzwhVuFPfPW/4au+6pOAxVKnRMQIcyl4OZqxdC7MFp9d8Sfrx/hP6y/hE3LnD/HovT7jmWu/OG4SJsNIMzK18Kc+HR6Vy5zoHO+n0ECSUkoqJiwAO40Y0CXXHOO6teI69N7V3EuYeqK7T0a9/lOb02D9KZoE3yfsR+wFSIqX8fUFSkpMH1s05VTc0cnGZ90mwLev25gYVbWu6NrwQ/yEIZGEQu3UYqbLMPnxELRtUrx1OLcKYoXaFN5zJey00AoxWFBXzcNyCBCUpuZSspSKtdQ8aoZ8eX2VP2oe5mEO+ff0cR7Xyzymh2fO53nfT3+MbfGT8mI0x7ug+0KkozEzV1D1HQ8hc1CirtipSI1hLmuW5KFQBJjo3N1ehMLBfs7EFQG22PReMh5GNHeGkFXrPAzpMWPlvFtKjplTUXMqq6BsVasJxYSfjvhCpaamIym3Zcan5ZiFc6Dfp8/M0f7vF75I9M/b/t+/nrhJxPZx9vezohj+8VCJCWR040Q7Ukfq345cKiZii1UPUUvYdHI/L/zERtVseO7sYhe72MUu3nzxQHJCfEfbJxHg5VRtouaVsSrnfZC6BLHAcJmM3ynnnE57vG2YUNbCqG8wCmUZsV6nJEnNn/6imqOjJYt5xn8ffYL+b38ZK9PjII35T77mFoP9CaoRt14+YrXo87UP7xPFFb/+G1/EkpV7XdsNjtUmTrFE5FTc45RSa+7Jmq/o9KGCh6LsgeaGXH/uL9wEOw0BuB01ZmcdUkbapZaajJQ7TOhpymfkHkeyZxNeVe4yJZGYPhkiVkY2lgijapNCN1kpncmah+x5jkiJlf7t0XNwOfs3L+tbUTHTBanYx60050COmGGlcddakMomcXdIl1OdnjsF8dHuIvtQtX4LlVau0IlJ3NShFsPKFR1TXTKQLhkp+9JnorYYq9WEz15JtSHZeyADUhJOWTgITYeZrrgi+ww141PRCY+a/VBIHGjGVHL6pAxJqVXB7LOnKVe1zxEdPhOfUEtNqXVIQr0ilY/A25KIffqMTOMm7x3KjUSUUiMKmSPb+2lIrSbAh06YA42ZohDxqFxi5h3mqTmUPTJSDrVHIhFLcvrSZaW5O4cRPefJYv/WQKNWDqJm33dETU1CzCucIAgvyimilqhdU3Mi9jtfUZOSsNaCrnS4rad4FTE/nVppbmdw0sjojpzhpi+cGn6HhuKyKTacwzyGS+w7pTc/qWtfV40ss6FxSq/cvYwoOI+Q9jQkDgUQrKRy35dNzlGJFxho4IUxEZVrsgyjASvNN667XexiF7vYxec3/sb17xtXVM4hrYHGAvz9Z77nC55TPnBFSFsFCxpSZDuBS123r8TQJyFzE4k1NQdxwqquOdaClxYRX3kVBr2aT7yS8mWP5RgTUVXQ61Vk3TWzWZecil/hUxDBX+t8MaoRcVqgJiKJDdNplz/5p/4f/reP/nF+fbJiShFgHVe1xycj6xWQaUwthkIrVrrmcR1wt6j4kWff9cAWH9tx7bmvuwnw/hs/NQYYmA4rBz3Z0y6VGA4ZYkSZmDkrydmTPvvS565OmJoFK8k4FOvXsMBK3RZahkS0pGboHNOzlhmhL0p6jrBeUDHXNXvSo6RgoWsEYaFr1pTkmnMqCwdpqYnETleKliIR2KQ7Umv61+a1+PCJXBuSI26CETnp25GTPO1ioVWvuAS5oCSnZF97HNGnKx2mLC3kTD0ssUNXUk50zoqCAwZU2OOoGCauiPKToZKKuRT01XIlCmoua8+qZlEy0oyuxqyouEqXQRRxqANqMVZNqfXxGu+UzY54SsIlMl6SZeBieHibV2yqUaZSkBChalhL4VTShtzVCTExR+wxkC4xEY+afV6Mpiydu7wvQMBOVRC7XhQStdYNe43YayFFaSSAK60DDMvHWgv2ZcCxzkjETq0iEiYsuar7PCJHvKKnpCQUruj0x+BUF2FzKCg3EvS+dDaI3rTKiTbvbUVOTsVcl4xkDy8K7f1s/PTEXjutyYlYU8JcGuNMH17qGNreIPYbsaagT0aJCWdwe9rmw0+pwBaHVrY54u3X3jX++ed++DWvcd994wNjgO9/+p3hMe+98cFxQcUPPv3t4bbvuvH+ceJmnd/39Lft1tBd7GIXD0x88/X3jqHhx3rkh4cvR/zhTKEfuCIEmk3QH/C2VKtxGP1UI2pRVO12PpCYA7EUyj+S9fhMXvBSveaxZZ8kVkTAqHD3OOPKpTWDvRWqEf/sYxHEVi2nS8rdtbFE9aKLiCGKlaODGYvpZX59smJJhQpc0h4CHNLhUyExsJv7kQxZScYjUQe5P7rhgY6h2s70oXaZyJqVlJyy4Ak9YiUVo2iPu/UJGinIgD3p2y40hXPejjjWWUju5w7f76cHHspirworc3ssc+4yI3OGh5ZQ3HE+HjbBavtnnJopl6IDBnSdGWIZyOlTXbJkTa21m7wkG1wAccWLTXptIeK75V4K1ROoMxLWlI5o7xNPm75HCKXUjOqMOjLckymKhknIoSs6ALpOtnVFweN6ZKFoAhPBTVysIhlYFacrpg9An4SuUwfzKagvFBbG8JDsMRcrBJCQuC57I+/rCem+YRC7rn3pADxd5yLeISaXiqVUZBqzpylraQzy1loylaWD1qUcaI9EJUj77mtGn5SZ5Aw1Y6AdJrKmQ0Kl9ph06QQiuyDWlV5gSI+7zBg4CeiKmqH07aRBG48az0vpYAubwnm5HMucKzrkJQypxJRq3NmxJZgXPShd1+pABoHf5K+PdoF6Hk9kqssNCNiMNQMyvDdO5bYgew01m5CiINYbxKt+1ZgA0bLnq5nARGqvKVsUet6IvW37GvbhrzH/njPXm0sk4R1PvWdcacVrKUb8mj6+8XfHdqrVGDa+98YHx3799wXI5yvec+NHx+0iZxe72MUuXo/xzhvvG8fE5JqTiN2XE+INHzGzBfv+QoV89KMffe8fyiu9DmJbEQuaDcvf1tGYHgklhpVLZBIiRqQcRSmdSCiNcpRZCMyoC3EEZQ1XDyoWq5huZnj46pz/7jcG/B/Ri9zmlEvsExOxpx3+I3kIA+ynEf/+F624+shd/s9fe5xRX0lj5X9+eU2XmBThRZZMopy161wvpeRRs8+Xss+fevY/321494n/9ql/OgYnuyvWw/u2zLiqQ16SU/bp8zHzaUuEdtCalIRjM+FSdECCNTvsScZMl0REdCWlVIvXz1o1vHepvsuMic55SA6ZqqWuQ5Oc5VoEmFRXMkot6UuPK7LPnJxjndIhJZWYYzMJUCuwkrWVViG59EVJTEPEjiTGaE0qKT3X4Y9EyNx0xk9NKir26HFPpzwiR3RIuWwGPB/dY8oywLEArnIAwIQlD+mIiSwpHMFcMeRYj449OiH5TDWmEsNbzZAKpUfMvZb3SEpkfSeISYnIqfm9+B4nLJjrynJaHAQtkLmlkYn9Sh7nS80hL4qFjx1oxkLKAMFaSsWhZtyNVtyWOb9vXmIgPfakx7Fada6v1i8hRihdl3/tuAspMXOKwGtZS2lLN7HyyhNHzPeqZ1NdcCRDNz+xi3iHlKku6LoJRcf5kADhfn0yFqwxqiEx35cBU12EDSElYSBdd/7tNKxwx8Tf3iWloHL+NVVQmwIrFmBhnTEzVpzoDEUZyV44rn06AYbqvTwKylC8ep6GIKEIgaZo8JyoyHE+BAkyzmAnuGux77mgPDPVahdF7fATl7Xz45mYOa9WiLznxo+Oz3OIh2ath2YPaEuy+2hPUF4t3nPjR8f+dfzz76Yqu9jFLl4v8bdv/L2xQbmrUy7JEG/QvNI85A/bcvjt5ozB8JPP3PyCrWkP1CTkPBjAnjoFI9e185jmkqaDuJKKkaZUqqQqPG9WZOWAQSrMc1uE9FKII2XQq0liw2zWpUa5x8zCKgTXKV9wpRfx46vf4ZHigK/uDKmrhP2e8sVPTPl3n9rnUFKWWqOIdS/W2CZCros4kTV3TfcP89C9ISPA6sSwrxk5FZexogA9Mm5zanHoWoEkdlrhEtG5rrgkQzpipxJD6XNsJlRULqGvmWnpOtY1uTTmgaqGucP4ly1ITSKJTdC0Dokd4PgZESvNHfm7QtQaC2YSYdz9fVHQjEwd9EUiavXqSlUDofHAGlV8RziVGGl9D7wfTgebbJ/qgkhkYxEqqRhqj1I61GITwxF9DMqMwsmsCjkV+87Icagd5pSsqEmJqFrvtxYl0Yi145rkCPukDLRDKRW5FNRaWxgV56ssLaWkwPC47jGnZO1gQqWbYvY1sYWN3OLUTF2SXTOizx09YRTt8YQZ8qLMbXLfSpptV0hCUWKbFBVloPEbUmfyWDquR+JgWT0yjnWKiE3mS3fuLoltQtxjFgqVHhkzXVl1NLXckVOdMZQBC6eu15OMPh28TPE9ZniOx1SX9vqjptCSUmoGZMydxLKXkx7Ro6Zmrqtw29BxYfxn9p8tcVtCifc42XSxx028cndGvTy1L8graaBatiC0k2RfsMjWOa1aU8G2epg/34IwoGuv/yhireW5/iHfdH08/slnbt78wae//eZ33/jAOEAZ8XIRgqe9tz83rnBo8wM/1/ieGz82rjHspiK72MUu/qDxwesfHn/rM9943zXkm6+/d+y5ql79sUMaTIVj58Pl10DfzPZrnucQGoRSy5Cj+LXY//s3rn/f+AvFD3mgJiFg5Xmh4YKM1OKVc6mDG3BPEwpqVJpEdhS0WoQTCkakVr1KhEe7CQ+P7KbWzWqODtb809+005RfjX4bgMfkCn3t8J/Fj9JP7BbZTWBVwVd/+ZSyTPj4C32yVOkk8D+8MmdEhzU1L0QzXpLT0MXz3c0pS07MjKVLWH7huQ/sNr1z4peufWRsUD4TT615Gjbh/ph5PkwaOmK/qFflAIMyZ8U+fTqknDCjQ8odc4LBOpr3JQuYfZ/wrMgtXMbdPpAeC11tdF3bBGF/PlNJGMqAYzOhdJ4hnhydSUbu1Kh8x6LpusYhoWue036eTDpUrsvhP1+tNT2x8rg1hj16rMjZczLFD+k+n5A75BTBqNCockX2N6YunqRvFbGst4qHYCVEXDZ9+iRMKeg5TlWFMncLYEZMTs1CSrqu0/6w9vn9aMKxLDlmzkotbCwkqOrMGN2ieFlGfE39OCO3AN8hR4ClVAEC9LH4Ni/qPXItwrF+W/Qov2c+w1uiS3xN/QRTKZhJvgEFgqbQ859piU3yrXSvYQ/rJTJjxVxX9CQLifixzhplLbGu9PtYE74ZVmq5pOYhRsxYBRUtgL5kXGKfF9Qq3g2lH+Bda0pOdX7uNZ44+N2e9FjqOhSmFVaxbEHOQtcY7G0PyWGAtUWugCyp6bqJzZqSDgm9lnpYM4GLw7n3Uzg/DfERq0UUl2KLjPbxrUPBE4Xi56Lw58Er01XUzNR6r/zQ099xE+Dt1941vhof0aVjzSUpg3dLW/GtXfifpwjWjvY05DyOCTRTkPb79IWTv253hcgudrGLzzY+cP0fjL3AB+ByVIsOWLBmris6kth/3R4IFgIcYX23PGdwu6AAuy+ttQzNuW3olV2fmwaU54x8ISYiD9QkpB2eRLz0qjpYFZ1KDQW2C1updUrPiJlRMiQlBbdpG3IMmUZAwqeO4ckjUBU+8psJ/2P8OyGxUJRTFvTp8B//8RN+498e8eRDBbUR/tWnYxaLjN9+PuOTec6X9jJqhbXUpFpxIjl72mEoVv3Gu2cblLVaM0OfIL792rvGJdVrKkZ+4MZ/M/aSo9/59De/qTfKWtQVFmv2ArSlGT9eiQ6Z6oIOKWvHMjh0ExN7XxNI1wOx3hKl1qx0HaBV/n4xER1JybVg7aRefff8zPty/6ZYboi6wsOHNxv0rAD/N59A+eestXZqXrKRtPnH+O62SENcjonYo0vh4DsltXPtzlipu8bUSvT6Rc4n1qesiSnokpK5pPWqGRAhrKRybBoYkLKiCqIOvviI3IRvqB3mUrKnTScnJqJPBxVDTUyulq1hpHH1jrEKZpekw4mW9IkZuvfoC52UiFf0lELt+TyK9piYeTiuJTULSmIVYonCVMMnx3Y9SELBkBIhKvScAtbQubXPZOWmYwMSl4gPxIoWDNxUaI9u2AC8GpeHIHmJYBUN64VBuSIj7uiEha7D9GGuK6xfub2iBmInBCvn4L7SnLn3J9LG1d4XID58QdaeOqzIKdVCzuz7sowhD9Hyx8VPLiwn5KwniT++zTXeFCAarknzmiYP/nU9F8a/7r70Kan5rhvvH0cIh9HIymrjleAala3YFQXt4mNbiGSbi9IuNr7nxo+N/eN90fGDT3/7zXYBsv352+HJ758NxGsXu9jFgxnvv/FT42YPsuvKHZlyxF7wOVtT4AVuaq3B+Vb5qKiotQ4NIl9k2DzATbXVruuFy3f8nlBpRSzxxvro/aRqrXnHU+8J6+HPPPuDn5c17YEtQtowhK4mGGk6We0OaI0GhLFB6UvCTKugnAWwqJTKKN3MEAn8L/HHKR2ZWFFKrTjVKW+Tq5Rlwp/4ilP+798+4MV1yZ/54pp//ckOv1suiBF+c7Xg09GUQ3oOZlJxyfR41Owzl4JTWYSk2HpZNPwAn6T+9evfO/4Hz/ztMxfI+258KOCXFUOBISbmfTc+NH7309/ypt0kDcqxLMlIw2QrpktfeuE+hzKkQ8KUJTExI+1xIkvu6SmpxMHVXJAwBagxoFWQYo3x6lgJiOV/+FDVYEDnbxeXXPrks6Egb753r76lbCpjGayUbSzxmSTQaO0IZ819E6zakKIcOo6SJTVH4ArvoXa54xzNh9KzUBsHq6nF8j9WmnMoe4y0Fzrg+75Q0Yi7sqLjvDlKR1z2E0YVyB1EaZ8OMwq8OtNl06cvKbDH70W37eM3PoHiifQeRgmwdtChPjEPa58ImDtRAM8ruMKIhfMGEbGfuxYl05jUvT87om5I7LFGpEAhFUO104g+aXjOjCR4pqydalhK4v6PAxfCj74TjfBre0pmpW3d62bOF6bUmrsy4Um9EuCC9hpQKgf9gog96VFQ4iWBLYjPdsF6kqFu0nVXp8x0SUUjMd2TjK6DFPpYOl8cD+sbuXOPm7q2C1+DULpr3pbSFrDm17satYpZyEYBYu/dTD5KGgjs/SYTvliyxz12xy8O18a+9IMhaFMgmXCe/H9sre8bCl+tv3leyTY8zMd2AbIdytmC7LtvfGC8K0R2sYtdXBQfuv4z45TI+kiJcFmHAJzojEJssVA4uHUsEWjb884E76gggCOmsRXQikRsvrrRJHJ7cSIxBptLtHlyNY2gTjsE4a8+9bfGEcKHn/3+z2lde6CdoLzzde3MtvxUxKoFmeAvkGPcFCRirjUpQkpERsSjnQ7Tyg7fqyoiTQ0DMrvRS8cmJGI7qrdkyic+vc9q1eF31yv+r/hFfu/llN8qZywoWVI5KUt3ETg42KnkvBhNrUQvjdJLT7p2E5fYEpLRAL955433bWyUf/PG3xkDwTivpGZNSUm1oVbzZoxjWeIxkd57BSxE7iE5pONUmyKES+wzxBK65zSu95EIqaTMdRkmDY2zt03kPYTKu1F3pBmTinv8eR3TSitKLdgmh20nSW2SOnBmcWjzRMCmhv4xlVa2i64W99/XTugc165rXGGYS0Esttu8T58uKSey3Eis+tJlpH3rjE5NTs0dWfOKLJlKwT1ZcSeyhHFvFNjThNwRmksxwSF8pBlLqehLzGUyHtI+T+qQx/WII90LY+J2MihYA7yXdX1mAbsSpVyJO+Ezt2MgXe7pjAih4xoIudjvboQlXQelKeeJ0fBrmtH2Nh8nIWGpjVpWXzuOwG0NASOVYPIXJITVO6d7R/SUTFI6krLWglsy5ZLsk4qF3FXuO1pRcyAD1G1InqS+VKvcVjmuhk+qu5IGo8pw3aAUlGGKNWNNROQ2JOMkpa35ZO4kAmqst4xdD8swNfLXvD8e/jb/WPt6xr33hv/x2cT2xMQXLv5//5z+fr6z5wsRz+Q5L86bjPhj9Grv46LnO+8xuwJkF7vYxUXxwesfHufUrKViqgtO1HL/SrF78x1zwkrzMNnwzSIPu4YmH/CIh1rr8LfYFRlAKFZs8WHz0+3YLkD8urbdlDEof/Wpv3XfpsyrxQM7CfGQC21tXJ6o4w94hS1EtmNAQiYR3SgijYRSlbyuWBcpcRwHI7A+GacsQgL8kt4jjt7GL/6/Eb8Wv8BdnfLP8x49RwaaSE6mMSPthvfT1YRPxMeUVBzRJyayyYAqfemy0BUpUejIKrb6VZR33njfOCVhonM6pJzKIny2dejl29+/48YPj3/k6Ten34j9jE2SVFAzlRUpCQkRh9rnt/RTlNEeKQkHWGhRoVX4ktdOLvUwGoYvdkIcOBwRYpWcZDMp8lCsxPlF+PuLiCOc102SptXGJMQ/Vlx3uyddCkomZoaqQSQKvJD2JExaus0+GfMu3DERlxmylrLl4eA/jyUNX2I/TDj8sUtdAYMkrLAKSSfYrkxXE45lyUzWHGifS85bY48UK8wKhUuMDUpPE6dWpq4wjFhqzSwkvMpbzZB7knPbmda1C+VUYq6YPTchaHg2cyoiTUDhqMVlAMgpGdDlRGeWH4MhdeT4lIhMY+bOad0W+0rpyOqCBDxuRsKKisxNvQ4YYMRCnmJiUo2DNK2/BmDTS6Od4PqiuM3RAbirEx6Ty1YowHXy/SQLrGFhR6yTu/eVWWlOQdlMNTDB/NLDuADylkzx2vnXxFg/mYH0Ap/CQtRKFuR0XUfNSj9bWWDPJcGtpX6KZD/jZrHhC5DtRP68onx7MnLe5ML/7iccXm63xuDNZX1h7a9l3Xqe9vNeBCdrP+5+3JXzPgM0BNBd7GIXu9iOD17/8BhgLgU9TZmwpC8ZU11ySyZ4KBQ0BYDBsKYOKIhaa4xogGT7fbb9N8BN0r0YzSYPxBY2DvrtnseH9ym73zrWLkR++tkfODeP/JbrN8dtOPFPPPPem/AAFyG+Q1v4jdMd89DhVHGQi8jxP+z9LkuGoryl20CxhnHM2himKyGOLHbbj7aA0DV/TK7wwin8r/HHAwfhZZlwlaHDytsRWuaStFQjUiIWrHmrXgrSmAtdO5frhIHjisx1ufH+jSqxS0aDN4EjKlVSUboLzog6QzXDd914//hlc4dt5Zk3ekycUtWakiEZc1mzICdz8JYTWbIvA4b08DKoL8vEKjdpEWAuBSU9slDwDaSHugXDc0E8REoksnwErRGRcJ7AdV+1KTLakDofZiPxqRlGAw4YsKRgyYrSTe0SSSxvxBHRPKTLP28sMRkNoTty18SEZYA1zaUpwo90wG2ZOV+RwknxZlSouzZjN7q1HXFrzKg8pHsh2buqPTye1fNBVj6J15g+CUaVFY2S1yusKNz1PUd4UveoVNkjo5IKdTK2EVbt46ordGr3Gn6hnjs/Ds8FaEMp2+Rpg/JyNAvme5kmjLTrElmlcpOa0sHULL/ArhtLYeOc9TWlJx08fyHRKBRHPnwB4qP990QjCinZo8tUl3RIKSi5q1M6YiFfNvm3U9BjndEVK6t7xB5LcjJSTpzKFkCF4zJphXFFqCAbXBNvsuinbHvSo++knGMiFk6K2K43h0H611+n7cLQ/7wNr/IFsOdoXLSRbRcG7dsb/kijeOX/tfwe+/fzeB/bz7ctydvetKOt59+Wcn8tPJZteeBd7GIXuwDL93is3udfRr+LorxVHgLBoVFqSqkCR0PVcMqMBMs5VVUiaXkqqUMxuP3OFxA1hCIiJqIWwn2AoHxZakksMaXjgNTq/UFaE3NXeETnwLHuFxcVJD7b8eFpAw9cEdIeJ3n1gVrUSeE6SI1oKADahz/DGoglIiwqyxUZJMI6N5SqTHLl+UXtcN4xU10wkj2m2uCh/3n9IghclhFrpy1UY7glEx7S0camWYohouYtOmJPO9x2k4yBdBkxcNyQ6gzEwSfL/jUTd7GlErPQNSuzdjAiS7aNXPJXY+hLj2+9/gPjkoqFLt8UBclltSTz5+Uecykcn6ZgII2pnJexe0QPOJZFkD/1E5CupOQm51SnIbnvOTs1ryLhibA5FR1ijqJ95roiwwoL3NbTjfd1Hll9++++oKnVyqMmRIgji6srOvD+Ga1FqiddCi1RNXSjLqXWIUGasWKlOQOxekKR4xYAIck9pM9abKe+oKIWw5KcLoPgb9Enw6BMZcWXm0uMtMOJ5OR4/wjr/+ELELCJ2dJ5gBRS01fL0/EFiO8cL9y0wSfIhhJBXPc/Dd8Smxg2YeFByl0paEe7C19jmOqSCQsqt/jGxHyVPMGepkxkvdHB7qi3o/QJK0RuunCkliTd19RCu9zEY1uZycd50xD/vmOiMLmINWZNwRNc4SUsVM+uJQsqairHOVtLEYqCLimJRE4u2AKpcjV4Mn/tIFbhvSDWGBLDQLp03XcArDrcWgvrV6IViUShuPXvty3d66cSXtrX/83zPvxEzU99Ys4igc8rUM5L/LcnGv5+BnXTvGYz9Z3EdlF83rFvv5afrJ33Hl8ttt/TLnaxiwc7vvvGB8YHDDAos6jZl57XW/Qko9Q6SP17boeHVHleo5V8b2BWvqETS4w6krlIFH42jvvpuaS+0AgNHPHwcROm4O1iBbYboX+weMdT7xknklA7b7V2RAjfdH08ft0WIf/i2i+Pc2r+7HPXP69JsCcmL6TcMPUSbQ52XxtIllfzAciwRPVSlbIsSUXY15hhEjFL1B6DAAAgAElEQVQr7AnMXbLou4+HDCyhVwumLBnSY6U5f9b8Ee6SkxHzb+O7dEkpHXkXGlWZWgz3ZGFJ6SxYaU5fulTU3NUJMzO3SggS05UuHkYx1yUpCZlzd/ZqCZVWtrJGqbBKCkMZ2GRbTEhoMkmJWgpRb+T4lmfecfMj135hXEYHzGRNRhKMCVfkdEhYOifxWgxdOhzIwMqUOrzkgjWpU73yClkzXYQOc88VNGCJvx1SRvTpiJeyrVma1YUdWrAdkTYcMEzliIMMc40Jyhb293ojOYQGphITWSyoajC2E1ck+QVq7ZL7kXa5KwuWUoLaYqDr/CksjyEilcTClzQmlZi+ppQYRs7HIUI40CxMOCKEuZQh6fNcDv/ZIqyalpfE9u8dYEnNgV0eQ3JeqnUlP9Q+aSvZq1CS1u9ragut0gaONscWgyOnkrVkFRbeRBIOZS8Q/XxY5TkvU2hd1WMsfKxwnf3gj4FtXMRYiNf25OO88NMW/9aXTqHKOqrbv81Yh/PejoKShIR7zBg4cdqEyGlaiXtPCV6JyrurH+s0lM1ePaV2SfeCNV5RzSuklW6js+errXSlwayxcJAs3Ka2Dbvyj/H8Dftcm6Tv8+BXF0W7MGn/3Ex1Gv6Npz0qm8XMtgREe+JStd7/eQXPq723pnixz3fz6f/6Dd/I2cUudvHZxV+//r1jIOy9FcbCYFv790LXdOmQaxm4Gj6syLlx3M7I5YRNQ8+rWJmWf1gk6Qbnwxcn/v5Ns9QiNdqNlvB8aChILspVXi3aXMo2d7Ut9uHhs69LYvovXfvI2Cf/v3Lt6bF3Ov98hpeb3PYHsBt5FDgj2xtcjqF0G2muBlXYz4R+FLM2ylfsd/hKHgesxv+frh9hQEYsEXv0iIA/Y76MLx92GNEhRviy+hI9OmRqFV/uiSU/35IpJTWfNC/xcfMid8wJE50x1QV3dcLEzDBYLKAQkUnHchi0YqkrZmbOUlf2YhPrguwlS3NnjLfSnKXamYyV+FwSiVhSsqR80/Xx5/3Y//8Ra6kYaZcD7ZOScMQel7RPDyu12qXjplfLYPgzIKOPVRLq0uFIhhxGQw5lj3369CTjUIYcyB4HDDhijyP2nEpSzCkLFrrm2Ey4a043Eqc2DAsaGEv79/aXt6DkxEw4MadUWlJoYQnwqhi1RDXV5vlWalWgvHGRx/QDFFqGhWAiK0bapa8pz8tdjpkHBa2uOqNGtR4YXU2DXG1fM66YAQ/pHk+a0cZnS4mIEcud0Niqz20lgX5cHCGBe5ES4+CrpET0xBZXnTBvsp/HcqbYeC5viqeu271NRraSyjmX2G+69G4UvSd9njSXGJoOuSsuSqzruyeUGxojvr526GCVrybiRZ01eGJ4L5qLwoiFe3lWlid5n7LYKEDAKqP4TeVYp6HI8KontdqpjuVA2OOVu8lWl5Q9VzD3yBjR40isn8u2d8Zcl8x1xUJXAW7oO2f+tYSIgooFefAVyfz7cMWHn0i0//fnyL9ee2O7iDC+cby27uOL7PO4ItvfI+stsvld24ZetR/ThnK17/9aNmJfZLWvPV+AvOfGj46/68b73xRr6S52sYv7x7de/4FxpVYqd2oW3NMppzpjqktqrcP/0EBz/X7v156+a2aKRNYKwEnv+rXF7p+Rm2h71UDbFLtItKYNk92W4o3dyurvf15scx3vFyJn98EzyADk9ckJ6WON/lJ3gDON+eVrvzT+2ue+7nPuKPmkx354VyGqn3TYjt+SKrx26urRnPOhMzNTM5/bTec3dcKVaY8/xiV+P75Njw4TSv5Y/TBLKdnTDn1inkx7rCv4zegOE1nyH9ZPAJacVFDSJ+OyGXCLE1QUNcqaRud/otMgt9aOtZPsLVvSsBZi1SWTdMMrwHc2hYilWXEpPuDEzFCMTULEVunbCkNv1PBfmpF2Q4E50A4VhmOxacOagqH06GFvj2mkSb1r9cM6AuBUlhyytwFzKZzSmCWgWx8RT0T3sV14tH/3vdltUlilFtTSXljaJLSmoIkt10TsbZVWJFGPI/asGlrruohEWNKY9E1kzYCu64YL92TBVR2SadMdSRAnjWqYyorLYoUSJlIw1NROCd339uLP2MBd+mp1xbwqHdgmAGqbA7l6iUGbVKdij3UVEujN4+qLkS628IFmgnAoe2z7sCjW3O8JPULcepBhpXVn0qhd1WKIVZxUsfUTQjuB2pdhmwexSiiEzpN2bUe7CPCeRRESuB6FVmegljERA+m2xvLWEddPLdqfrX3d+mLPYD1YajGc6Mwdn5qH5Sick8JxO/wa0JeMuPX+1hSstaAvGV3Oqr21pXO3//Zqifz9um4XFRwmHOuzr3Xe6/src3uy1C6OtjfY9vt6LZ3BNo/oPTd+dHy/6ecudrGLN0+846n3jGMiMsmotGIQ9am0IteCTKyISCYdayhLRK0lHsVgzWKdubBr7rSbNdtwqcrJ7vp1zXM+I5WwE3gPsXbuULvlbXuKcl5D6H5r+PY6eabR46DAsDmFacdZU4LXUWShshP2nDzuv7j2y59TJ+mZa784bh+oWIVYZSNpAssVyYiYO21mg+3MligZDd75mIKXdc2/ktu8qCtmsuaerJhT0aXDW3TEp6IJAF7HZUnNv6xv87HVmoWseUWP+d/j3+cuM17Q29zRCUty7kVLKq14wdw6t7u4XRwohlxz1sbCdiKErAURMnr2MbaqtgnsWgsqrbgaXWJpVizUklbPK3beiPH2Z//yzZSYhIgD0+Wy6RMhHGqPh9R6ZvTJ6LTgUIphpF0uaZ8jHXBZB1w2/Q2PipqaGWvmrDg2E47NNLjYeywnNEnRtsM5nI9t9wmlXzy2Oxva4hnVWm/83o61Wh8OD8HqSErqtMAVa3T3kpyyp52N0aw14stJiTmRFWWLWFy7Iv7laMaxLMOA+K6s6GINCUv3OZdivW58EpZpTErsVKHsdy910rgeKtkjcXwSwyXTdymc7cgfat/5u292d9rHUFEOHckc7Hcvc6pnUxoBh8SpSx2onVCWrtjwUxq/LjTdo0YxC2BPM6esZXkq7eO3rWrSXnwjbfC9bbL8IUOuYI33MumEgkNb43uDsk+fDimxRFQOZrmdVLePh1eyAivO0P7dv1d/zv059jwSfy3WGJYU1GowmCBsYZW5Gv5Mc27P+t20C6/27+dNAbff2/bt/rhud+bMOc+5XQD483suZ4f7T2nu1yE8b4rSjr/z9N/cwbJ2sYs3aXzDU+8e20ZOSe6myT48DH7lpNSbFbCZIG+LWvg11hPQ/drTnlZ4fl2F3QcAEhJSSQOx/Dyfj0A6bzVIPAxru9g5L8J7kejciYeHgcUSOxXPswa5Pl53Geb/dO3ZUGj4acRZkdzPPp659otjsO7ZsQqd1rP6hMmgrKVG1E5jbHfcXggjUqbOxaPrtrBTybmiPUoME8n5YnPII1GXYy15RA943AyZUvBr8fO2Eo46wahuGdkCZ6VrXtIiJKce533KIhjktRNSr7h1UYW6fUEUWrpq2YTnOy8WZoliPU4eia+Gzur7n/6uN83G+Zee/Us3f+naR8b+uopcwtnThGVUstwiM4+0x6Fa92ufnJ5Ea+Y05oL+uOdaUlAGZaM20nF72tEuRCKEbU4HcN+FwHcUwjjXdTqCo7pT5DIoS11zInOb2jq8qCCBeC+inOqCmqPgizKkx5EOAn9jTUGPlNQlbbHGZGJJzEfa51AzXpElt6M5j9WWR7QSW/SErj0mTBpyrUOTIUwbVegSs8Q6rAPskXBZe7yMLeRH9HnCjNgjCZPJswkmbjrRHP896TNhRV+zUEz5xb1ym4GVvq7oiZXg9ZAqr3Zl4Va1m/ysXdHkDRnrwAUByKnYU6sadl6i68OaoTZQtL52KMWKJPjbEULh5DHBKT0yrJzjitxJTcfhWmKDDN4coylLq7QlXlLXHsM7OsHrwnvJX7uRxRROXcuIUmkdrp+CkilLhIjMmTN67tN2+KLHFzn+Pm0o7Hkk8PMI5OcXIo2PS/t2j6N+LZOI9nO3C4r7nb+Lwps7bvNQdrGLXbx5o72HJ5KQa8FSVwEmnWtBIgkLY9fziqrl7yYb6473c1M1tpksjUcVF+QGEWKn49ikvzGUbXIEj6AosZMKI1FQ1oKmWeTXvftN9P20I/x8zhobhHUwATIGbDz/624SckfWpEQ2qZEVJ+Qb///sU//4c5qG+GQy/I6FYfgEaS4FKpbcuqebbP72xOSEgkvaZS4ll7WPEWVIwh2TcxSlvM0cAPDJ2JrMjGTATBdBgu2ezOjS4Wp0icNoyFE0sqmExFaWk/xMd9BjAwfR4Mzn8vf13UuDhmq80obM1P6ieC4JNATnCQu+SC9ziWEwYHszxdc99xdvBhgeVs1KgIe1IeH7icWR9sO0LEKYSsGJrFiIhbV5jLzt/ib0pEsiSeBpXKR+deaLesH9Lor2wuCVndrKGDV1o5yFLZDA4kVz7/Eg9uvvuycvRxMMSp+MA7U+KSPtYv0ruoFn0deUBEve7WsaoIx3owVdTZlQBEjTdsRqSdDtZC8CZ/zpJ58RJZZ3VWMnj15h6a31AYeOR2VVjhq31e1jOnULcEdShmKd3b1eevvYVVTccxLOfU3oaxImLRWbJDpfdAROidQh+V5TEqnQ15Q97WDwE5jN41CJaQjp4OB7FvpXSMWawhbGZFjIaBIWd1to2mLOTu46QcjA/3uWE2F5PDNWTHRO4Y5LuzPVlQ6XZJ8e2ca15Te0BFuMVJZmGDaPtZYtRS3/eWwh0p4EAXTp0HHFirb+g8ZDpH0O/Ya7MUGimShtQBY3rqfz1ce2sc51+Ja/eqGz/Xr3C/+ed8XHLnbxYIZqYyzcl164ve3f5X/260TlOHaqVla90irwdyvHURUsT3R7bfVT8Io6rNGla695I0IPy/I/n5db+ud5NShx+Jxs8kzat1sBnbrZt1wDyze+PvTM+OZPPPPemx96Zvz6k+g9liUJEaeyQhASse7GbWjDT13/2fH2JvNNz/yV+3bs/UH3CaiVajVc1T5LsRfAQPuATUa8P8jUdccrV/FdIuOEgtvRksumx4qKwnVAPy1zJrLmRAdMI1tE3MNq/ffJKLQMMmW39RSkgQZc5SCYnHXp2GREYtCaTHqkkoRJRruz5yvN7Quh2YybCYi6CyORNmzEwXjc7aXWvCSnPKT77MvZYufNEH/uues3f+Xa02PPDUmI6KuFZsUu6Z1JTqpRuF5OJedWZHH0bez8gjWl60z0JGPuuhzikvw2BO7VipII2/XIHakczsKwtiNgLompxRZGm10Nw1LXvMQ9BzFKOJBBSEL9e5o7k7x9+qyldDChmlQjSioGzvwvQsip6WnKVTMIi9BlMyBzZO2oVej77rd/bDNSxhHY7ezCQx5TrAKdL/hvi4W1HbHHw/Rax8oWIgZcUaRhCgLWCT1CyLDO8EOn4JVKHFSwYixh/QW5SymHPFkfUGK9UBBYuXO8vSgP1MIcC7Fu5f49jbQbJqx3ZOEKqpqO49HYzaEmI3F+I5sqUr6Y6GrKWizMacE6PH8sMQO8l4lVsutIEhoJvgD1z+UTar+5ZO4cDuiypghraq2GjiTsi4UoLhy8y8IBnOS3ZAgJtVrn9LayoJ0iFaG75QsTD9OqMcxYYbBmj/66jGlwwpUrruw1s7mWtSV9G/ncs4VIG+7Vvtb8+2jfdj9YVTvaXcFtyNd59/fY6nYB9dlOUnaxi1288eLnnv2hm1//1HeOAeZBLMTxKcV6sRmUSKzBcORg0XbfahSw2usjNGgWIKhdgpvSE7fUsOw6X7g9N5bYSrCLolq5SUl9ptFyPwJ5e+3aNl7d3hcNesao8B1PvWcsLo+ttSaShA89M964j3z0ox9972s+yl/g8O6RFpsdkSCB6OrJlRYKYLu5543KLypGfv6pfzL2m/dIM+ZSUlBzWXu8Ei0oqbmkfaaSs68ZI+1QYnghmnLZWHrnvWjFVdPnrqxCN9M7JIMtoPa0w4tySkbClKUbjdnpxETnwVMi103ozxPRQ5zonIFYw7KamkJLBtJzF5TFf0914Zy225Vwi2zrIFvCZpLppTdVDYmkZ7gD/nGZZBxF+zypV9zjlf/ymb/2poFkXRS/cu3psb/WcmomUc5l00OxBetdWTKXNZ6IvnBJ+0LXzJ1ZYSqJ5WA4OBTYBM9HgNxtFZTtRWEY7TE3yw0I3v0KEkVJpROeX4iIva8I1jukPfDsSMpX8Dj3ZBZGvv5a2qdPjw63OOURjliSM9QexzLnsu4RYSV1E424rH0eVVuk5m7hLLGmnioNXrX9He1rGpLjHgl9JyHrO8dtCJV/X78Wv0yqMX/UXGJAQomSIhQYx3Jx0DQa0rFgDRL/WfTbeDJ3h5QuKWtKXjH3wiLdpUNJRUzMk3KVt5hhmO54wnyPhCUlRprPBNDThInYImGoXfbc51Mh3P9EVuw72eJSbJdqpF1yKtZSBTnY9jVgPYFmpMS8rMe2a+/G8ldlFBSpSirnWA6Z9VMPn8Vfa1aGuqBHh7kTt+jS4TN6Jxwr74MzoMuQniWfO+nmkpo75oTDaIhguTt3zAmZZBjsiH0kg1CUp8R0SVlSkBKTU9El5RU9JiHhslh1Mv/82yP/bY6Hbwpsw7X8fuCj2bybZtX2z9v7xXmNG58IXBTnFS/t4mTbEBOajfz7n37nm34d3cUudgFf/9R3jq3BYBwKiu1GiM9nPWTUr3U+2ve1giVRKDQEYakrMsmotQ4TD1+wRG5dj1zeV2kVLAYaGNRmIdKGgLfXye0m4nkwrYtc0l9LvK7gWCb868f02oJnuKQMJZfmYLXhEgA//dTPb8C1Pnz958Yfuv4z44msOZUVSymZiT0RKREnsg4XwYmDZUwl50Rylo5Qa8VbHeykVYCA7d7OJKenCU8Y20mt3IZsVIOGv2J4SA4xGLouafTYvY7YTbpwKkAdsZOfWGIuyz4lFXNd2RGbS3Bjh+07g5F2UxE/KDtv09wuQPzjarUX68McEiEU1Bub6ps5/vxzN25ef+4v3AQrinDV9MORm8ia3F0LStO9bst7KkqhpRttug6oNkR0H0IUlC+2Ey4h2lhEgFediERy9vy0uT/eZ8P+HAWPmO3ELyZmpP2QqGUak5GylIIh1gG9xJCold+1uHccmMhhXqmp5WI50+3bS7ekKl7dw/4fI0wpecVxVB4z+1yJMjz8yj+LX8Tb58Pf7m/zx6KgZO2NnxA6kpJg3d+trGrNXWYbxYAvGsDCqPxjE7XfvfZn75EwiWxhOpeCoXaINSLVmCUltXt8RxO6mlCKfS7LBdmEfU4lP5tUO3Kff73SKcYP6YXzV7M5ZvdFSeImFV06ZCQOeumPnX3eQm1Bs3TFdYeEjIQ+Vva7cBO9buvaLZ3Uc+Q+hxV+sIXHipwpS5aOiGk7gg3s6qKR/3anzU49Nrep7elRGxLQvq//jOdtqBdNju/HG3ktcR40AXYFyC528SBFTBz2Zr+vK5uogJqai5oiPm/wq1uhJblaZUK/riUk7nk1/O6bM2ttTbrxcry2UXpGocrlfttiKtviHRetjZ9LAQKvM2K6utM0ZRm6jh0SRvRBaUilKmdMsxKNAub7J6//o7GvKn0SLS6pBrgrS1J3gH3Xy5MJLQjFsHTKWDER9yKnOKVCKY3rZKKWpPoi90jFdglfkLt0XCdwKBY+0qVRU2rDFOLWbVZm076XrnTcaygz1ix1jdE6kJgidy3YbvPZgkLVgDSVtz1OroKW8zd0IQrSbTNWdJ1CUYXhx65/ePxtz3zjA7GJ/vnnbtz81WvPjOdSsqIKUJ92+C9jSuxgKi5ppWKtOX3pBfLZeeGLzW1CesdNqISz8LqLIqZN9jp7biu15LfgyYEhdx3+7bHssczJSBjQpXQJ88pN9+z9JTQBTlhxpBkW/9kkgalGQUHqvM5zhKD20qbE0G1BcmosQS8WAbXF4GNmxGW61KrhnRp31Oz6EDOjZCkVfSfLOyTmU9GpnQ04iGEqcVgPOpLSlU6A0SUuebbn0JBIRF8TTllTiQlCBDGWQF+J5cP4It2vHSeyIhJhTkEqkTPGjFnTliB2MsLaQEy97DE0+N4BXVbkG12zVOIgL22f63z38ZpmUtomf7ebEpmkQeI3dxtZThEgUd5fIyOlLxkLXQdPnUwySqqwlpSt4meBXcfyViFshQ9MgJf5SY2/Xr2L+XZ3bntD3DZBbE+lLorIHe+zv29Ct9rPuf3485/z4olI+/ebT3/bA7Fu7mIXu4D/4tq3j/sOvdJktBad4J3J/dLh0Sm+gbS9l/s2kY+KKtwO2OmH87oCC8dKJNlYJ42bouPk7YFz3dHta8pGIeJddLcnIa+VM/Ja43UzCXn/jZ8al1JzT2bcMsesyCmpeMHcBuyBKF0nzZNkrdFXq7rUzYPWDi8J6g+i37h9Zy9zxmxwsTmVESt1Wrc6wp+RE2IiXpYJn5ZjOqR0gkq8hY6dsiAl5q5TofHyt96srA3NWmnOxMzDpj3TRUhUPdm5xHsInL2QPMlo431f8Hl8CJbwHhOTSMKpLpjImpVUQefpx6//w89JEOCNFH/uues3J7KmFmOdtzmr3tMltQZ+pBzKHpdkyFD6ZNIJk652REiYcFRqF5OOpBuLjDWbzCi1CNyhi+I8HKcPn+SFRLTFS/FTgpSYffobn0exhnldUuZiJaRHjie1dM7npdhr1hcmm+7UbBCxz+su+0mJLzj8c6StBLRW5ZAOb5EeT+iAvtipSyJNQpm673BGxD1Z82I05YVoysvRnFuy4jNyQkpCSkIkzVTEGx4CrNQmxF3p0HGLdyk1J7Iip6bjipoZeYAbvRxN+H25g/qCiYhYo0DG/lR0j1Isp8i4zzYkC74lloO02V2qsPBT31yp3X/LLXEKIEyE/Qwup3Gg3y5C287f23yIQ/boi+WXGCy/pNTaOcf46U4nPG/a0t/qilXvSrACBwtdsyRnSc5Crby3JyFW1KzcdEWxClsDsnDez5sq+vfuwxMdt+FV23Cn9rXmN9N2Eef/vn1M2+vj9jr5as2AzclTAxkWdk7pu9jFgxTf8NS7x6l0Qk7XnmYk2AnE9l7si4q20XB7HWz/7PmeJvzXCJFYkZWmqQZ239/2KTuv+DgPZdHms20jbtrv7XOdgtj3+zoKi+UuGEV7XOWAGsM9Tjf+7mUvfYfay62WznDOoPQ0xYieSZB8ouUxxrVLLGKaKYpP0C6K9t9qlCFW/eCEGV6uckSfUxZu87RJ25qSfRlwbCYkkjgJtxwCXMYgLQJ+pRVd6bOmsJWy1OFCvagT146LNlV/wXmpNh/+50QScgqe11vsS59HuURGwj25uLP/Zoxveuav3PyFp35hPJWcWsoAI/HFob+2thONd95433ip64C/bBYOTyIr6EsPkYguHeZOfnVbPtl2RTYl9oANmT1vNNTuKl8UbWDYCbYzfYV95qwQIq7qkLkUrClCorugZECXuRQsWJMxpOM6+J2WiSF4VavoDKnYd4S2sfIF1pipxLik3F6BEUIk7rvuFL7a8DFfrOQYUrce7GuHuyy4JVNbCIgJo+6KmoQ4TD2m2lzHqSSM3Pc3pyIjYSY5E5YMJWPiCrF7zOiR0afDS3ps7++KsRUV62jJodru10QXlvwuTfL8RD3iOLIFwVJKRmpFvieypq9pSJgrMexrxmfkhJIKj+vtOaPRQit6rohLiYOSlr8G/LGFV1dz6pNZeJoaMjrEErFW+3y+MZNqzLHM7SbqYGt+o7OvZcnrBkPkHuM/s38vG9erO4++oGtI5k0BclER0DYZ9JPk+xG+z+OZNO9v83Ht6VL7/vdbY/17beO8xd3/+3bFxy528TnFO2+8b3y7PuafPPcjb5jvUqkFXemeSfxVrbeWYmjzRFTVQbc3/aj8z37Na/PU7L64WUh4RI+qQaVp7AFBbt1oZaFYErkm6BYxveXEHigOYo2D/ZrbTIk/P9BVH6+LIuTHr//DcYqFcdh5hZXDVLFduDtMAwcid5vzw3rgDpq97VhsYnWgAybRjMs6CIlXG0stWLWtdkxkzWVtlKDaMK52nMiKS6Yf4Cz3oiWX1ZIya6np0rFkdI38eaJLh5muWEnOJYYsZI0HgtWOmBvTXHRtbHNBSaWVuwDOuiHfb9O+KNrumT5xbQqiRs43QtgTa+JWYbitp3zPjR8b/7vqeT7yy3/3DbMwfC6xoiLVmMz1gGup8QvG9z79X517DD7w9Ltvfv1T3znuiZW3bYsI+EVqFO2FbnHTcWgmYokkrFru9u1oJ3Seo+IfD805batkefypv22ic2Ji7smMK4y4yyz0ZLxMa+S+J7VYNSS/qGXYZNRDqvxEw7+fYouXAAQjQoNi1DYOEhopXs+7St3jKlUysbAto+cnlI0zu3JERlWP+HQMd5gGbkTseBTtZDWVeEPNzB+TyPXa7zClpOKOLHhZTp3oQIxhzVoKSi15JLocnuNUFkxZspA1BwzoS9eeQ41YSk5XuyylDFDSmoqllGSasJQcI/a4eUJ5Ix/drD8NlM4KchSU9LXDUlr8ldb6cBGHwvJY7HWSY9eVrnQCgdxOd5rHTmXl+Gyxa4ZEYTLSIyOnINecVNKAVYamUI6J6EU9areR1dTkrcKpgZJtbsL3m1q07wfNFMRPlv0kYvv+TZHTQK7Om4hsv277u3be62/f//t28KtdvIHj/Td+ahxrFMR2Uo3puXzp1RRIP5v4hqfePfaJONjvUOamCIqhL71W2+wPP95+7V1jr0jVTFcNP/fsD505Bm+/9q6xV8P0BYgXhPGfoXLqVP7nOMjlGmKxxUjpfETM1vrdfn11a1gqSShGFGcyiAG1+UPs/Ue0Mab192lg0udDX8NtbZd1zv78M8/+4OflenhdqGP9/es/PS6ouS1TKipOzIyHogZOqVUAACAASURBVCMAVuTMzTIYvfgT8nh0NTw+JuK2Wgz4vvS5ZY75D+RtFxYTPmaSU1Jzm1Me5RJDtdr8S7fh+sdGCLFGvBxNeYvZp5CKriZ8LLrFW3QEWHWaFRW3ZBI2PN/189yN1JFCawwrza0eNNXGKC5smOI9CWyXu53MepWrbUjOebfdLwKERrKQ9A6jPSqt6EjKF8nDGKzm9Et6j7UWrM2KL03e+sBstj/71D8el06I4FiWfOfT3/yaPvc3PPXuMRDOm1+M3hJb1bEX69u0VYwarL69Bv35uOg8QzMdaSdIbblW4EzXxftOJJKwJ33eplf5uNxmQMa+9qxZnibckxlDbHffm+UN6ZGqbRJ8hTkKohFgv4NzB0qqXJHiO8XeCd2/J7+p5dRBmvWy88U4r3+fSEMwrlQpMK654JNZ232/I2t+N7oVEn7viaGu+BlIlwhhpo7jJZvwzbZkrb8tpwju5LUa1hR8JY/TJeVYljyvt0LH/8uix8kdEXygHZZS0lcL28upmMiajCRA2V6U03Ds/MRmRREKxooqQDcTt3bsy4AO1um9DbkraLpb/jifZwDYTK6sWEZKQknFjDUrzTmUvbDueQWrjIQJK0ZOInntjutcVxTuPr6gEBxhXawq2R49LN/EFj3HOnVmjPYc9V0h2F6nz5tEnFeMbHOO/LW0fRW1Cwk/zbSTySg8xl+r7c14+zXvp5wVITvy+S7esPGT1//ReCLrMOk2osGwtd+acpbU/JZ+CsNZOdbXGr4AgUY9MkJIJaXU0nJf3b6ZSMLPPvu+1/w6f/nad4wjhJ9/7of/wN/Ft1971xiszL5XtYImF2u/n/Z9PcfOr2d+rfCCNQYllqg1AWlCiMg1d8ehE9aeZj31MiI+Z+uEhrGGtcu+v46koUCJJd40KfRSva2J8kUNlu1J8+er6NiO10URAvBDN35i/HHzIpeiA16qLQ8kkYSr0RG3zbEdIW2dCL/ZdCSl0opMMqZmRjfq8tV8CbdkSkrCULNwsjxf45ZMmbDi1ExDQvYQI0baDRtt33sjqDgeioVMeCO2j0W3eEKPOJEVCRF3mIQkDwgJT1c61NSsXZVcUYWq1F88VkKtE758cYDfbI78YZNg2o7zktPXAjNo368n3fCluRQfkNHhWKcszYqDaJ+YiH3pM6RHSR02fUVfc4L+RowPX/+58Tc+8/Wf1WIImwlQJh0Oon3muuTk/2PvzWI1y677vt86wzfd+VZ1VTfZbBOiGAl2HPshsIHkMQ958wO76yFwYAmK7QSOLdsULZq05Ku2aYkiLco2DVvQYCSCDQdokq/Ju/NkA0HsIIM1i1N3VVfXHb/xnLNXHtZe++zz3a+apCh2l6RahcK99xvOuM/ea/iv/7+72BkwTmUyqIJsByF5dkKkSIQFwCCo8Uy6T+j+e6poyIgKY19zp/JlPeFa1mkhWka9kHdipeSeHlJTcKoz7qnBmPLm4E0cl4us8d1/Kpr6IursGfbm9CPqNCodlpVbKb3DuVQTMtyG5ATg35cPk7ZGEz/l1/8FjnjCjfVTxQVjF93gSjfUMVDaBfk5kX0OmPI1fcyNLvAKwg8UH7Lzi4u3N+jfDVZlfVIsOAlTFmL0tb8pb/OKnlJSsBATO/ymPGEVe1eMAcsqrJWUdBrYRFjnCNMesmto19vHmic+HFbl7+V9FkAUPSxZ0zCP/ScbGg6ZJeriOWsOmLDChAld7PIK48Ff6jrOzUWEvXkmsGQmk7gdq1DsMeaxXtHSsSdT5rrkVA56Ac2nVD22F8ntoGI7ePBtbS+oORFJfr3y7eSf30ULvIspxhNO/vmfeuOv/6GdB5/bHy7zXs+akjflkpf0KMFEa0paQhJkLRAeyTVXLGi1Y6mrlOiZyoRf+NLr33Lc/9CrnzzzCoE74+7om45ZSJoannz9dgKKv/TqT5z5fA9QUd0KklzDo9OwE+b15z/2Y2e5fwl9Ei+HO3tFxAOQ7TmqkioyCjapspMHHu7TeUCy/bv3jnqz+kab5A9WEbLvSZJhhaIbHKdX0P34chSM7bO8JVy4Pa/l2/9DH4T87Qf/8OydcEFDN2jeGcsoLWwikm5mjq3zCz+Smo02zGTCPTnma+ERh8UeJ+yni3lX97mUJb8RvpkgSAXCYXHAnkz4AX2pzxaqV0J661CuxL73UC65r0e8JRfpYRzFJdAzborhpjtCxOeVGJba4A+ubr3RDePCnAqDYN3mm3d7WhACQ6c1Dzo8UHNGhqeZZ8lbbblbnLJmw2W4Zixj9mXGR3mRVRRp29CYZrMWCaL2qTf+yvMFmD4IcdsvZuzLFKHgm12fPYceIreLuezWvd8KQvK/Xf+lQAbjJ20vBisjqVNW954cs8ckTWBOGVtRJA2Ma5bMGHGgEybUvBBmHDNKVRC3DQavWkpWeqbPdI11yNjWq6QLB9Qs6GLDeZ+FLiX2hmT+aaPKgiGu1bZT8H8Wj3giN4OMt0/mM0Zc6DzNEw7NKkQIqqkykotBwe1KSSXm5N7oMj3HlVS8JKfsMUmkFWvpmLPiWPc40QnXsuau7vGW3ABwKQvu6QE1JdesWUrD45jIaGlptGNDkyoKAJc6j/ew4pgeQrqNJ/ZMlx+z3++ertlYvRas49zUMqLiUueMZcQBk3hP21QNWdGwpuWYPa4jffKGhoWu8KbLfAwfyowp45RBBVIl+JAZ53qTNEa2Kw/bPRf5OeyyPkHT9x+FGEjsalDf9V3f9q4M4fZ3txfrvE9s+72APg9Mntst+6uv/f0z1/15PxJ4rssGhuR4JNfc1f1ExuIQSiBVTL+h73Ak+7S0iUDHn5WR1CyCzQuzYkpFxVW4TkHEf/uxv3U2lhFthJ76vFxJharBkX2NanRDLaPk3zW6idpEmqoM/nz7fJ7PeZLWH0OadHSD7Tnka6MNjTbMiik3YZGCARjS32/DMte6HlRJtn0tPydH4yx0GV9TnL1wLOMBwsUDjRzG5dfJA7IkWpjNNf0xbQbHnCdRnhaMuOWveWIxF8j+XgUg8AwFIZ988Pmzx+HiFo4e+psDffnOBWC8gRNIA1qQpNXhdGmBwIiaUzngd8JbKQBxG0nNkRxwX06MOUjHLLAmUkGSgjtY8GHHMFTF7ehotGMv4sK9qXyuS+Oy0o6pTGhpk6idH3+nXVrAyx1Zwe8kEMltLBbYTGXMRbj6lp/3cuBa1+zLzDK9GjgsDzmRA+7rEU/khmP20gNWqLCUvhnr77zxPz5fcLFJd1bYRDeNQZwSeLMzobjkOD0lGNnO1Lq9m4q6iDEZ5RAw6CsjIpLGxJHs8wKHzLRmLhuuWaYM+0Ec/4m2VycoygFjjsKYMSX7VDjG3kQeAwvapFYOPa32VCuMitWdQ0k1QwVOGXHOhjElE27DzAAKgXWsgnhPSIsypqCJr/3b8uu0dKl3bEOTggqAPZkYxCkGHtu2/UwLQiHCHiYi2ld3QqIAvgpzaqmoqJhEVrMXOYmMURv2GbOnNicc6pgbWdOIMYz5cS6k4THXLHXNR3mRG9lwzjWX4YaZTPkw92ik48pUR7jWBYeyxz7jlIRxBz9foJxFcEI9cOCNhtmYq4x6NzL1RV6rmjJqhdSJMW0R+zkOmSWhy47AE73aOZaPZB8lsMckfTdftFe6oZQi0RHn+jl5hSNnznq3OS//vN8/zwY6cMFFbv19/17+mW1Ck10BzGB9ip+vM6fELT/m54HIc3P7xIPPnoEJxM4Y8x/D1/ilL3/mPRsfn3nwxbOjWM0+lwU1FW/pOSeyzz4TLphzofPkJ90vTnmsl4wZ9Q41miDlnQY8wFC1/o5SSnvGKVK/41hGg2c6T956osLhyMZs2DM9OdRpex7wIMS/O+hrQJNQX56QWeuGOrL8Be0s2R0TeNvwq12+WKObFIQ4fD7v+8jPv0BY6xqn6t1V/YChn+tBRw7Lt+2OabVNiWIPdiqpWOgyrQXuH5cUAyFCp/PNKyL+uvsev/ilv/eezlPvS2P6j7z66TO/kbWMGEnNn+QVnnCJItbhr0PnSyhShSThCXG5+j4azLP8EgebVxtGRc253tBsqZWr2uLTSMtDPTexQDniIRfAHQThkXxrB36jBp/wh8mjWLeWlrWus0Xv9oLqZbgiuzUSr8e3Y9sZ9amMd2bFfV+ekZcsCveH6UYX9pk4iF/kmI30GWgv0zqzWK5P8NzMKkyXYq1rEFhEHZFF7EuAd6fbfTfbzmhI5CLfFYDkVcNOu6gWXlNryXWs7B0yi9xYgTkbLmXBhpYJI+ayMiy/BmZUfL245iPhmBpJHF4WDFifSKlCJ7e1QiR9tsT5OPw1U1D3BSDvj7JmdSIssomOosNrrDel450oPOrnP3K4krQpebHHhHlUDi+2cLnBFweGcM+Sgpf1hF+TNW1iJrH9f0jv8qQwBfS5LrnRJQUFN7Likjl3OGDBBhFjzruKdLU5y5qxePUip2MqgsJVbC5saLmWJSVlFI5UELiKTFwNHUfMsKbF4QJs80mf3cqZqVZsEvGutZuXLNj0MFQkQb0csifZaxvavu9GTRArxDGZL9wrhmwxBRLZvXoq3UY7RlJnQd5uwo1d8NJ3s+0KkVfm8mqJn1cOO/Esq31mNxX6tm3rlmxXdv7eg39y5uPzj0pP3XPrm72nVLwpl6wiY9E4rvGFClOZ8FMP/vHZexGofubBF8/AGP4KlQSRVJRHesFK9lIVxBMXj/WSTjsmhQUhbdSn8DmrFK+0GjNghyFA8mqsa1gAhOhAC33CpIlsku5AQx8wFDgKpqec9zUtd/4FGcCMfD3JETWu1ZEHRtbbZiiRqUwi0UYR18yetRQY+I+OHLHqaZeOKaB0Eda+0YYcJpUqr9nabZCt3t+tok/g4tSOABpRJzi/M2WVUsbkSb/294kp09LyGcwFi7dNNbynQXBu73kQ8t+9+nfO4Dat6CO5HtyUPAts+G1zcq2527OpJi0IfmM7RAv6CFaTo19g9L+qvt8+6g5SpAduP/ZvPOSCpa55Uy5uleJys4XasIilFIyoWOsmNWxOZMRYahtE8XzXUQfCF/+EadZ+uXPWnICmACR/ODv67HYZade2F24XZROE6zB/aiZx0HMQr51f2zwLuJE2OU59lvU2a9dzM/uXX/n863/p1Z84Oyj2Uoa80YZaavDm6K1sa07FG3YEJ9t9P4qmbImNp/658iyOiDlYZcSpgjFlPOYy3dMRFftMuKv7zLTmN4vHNHRMGSU4lo3pCY9kyXGYJGc9t5qCOiqKOzTMlNUDBf1x9s6fmVP1+hkL0KBMY6BQS8FGzcUuIAtGjK63IXAta051nydywwf0lAPGXLPmoVzSxcy9N9k7/CrPimtcPvJsWiE9l/t9jllLr8uxomFJwwf0mDUdT8Sa0BeYWm0Vz8ghYbVaw+E60olfs8blqO7qHhtpuGBuC7J0jKgMRknH18IjPlTcY6o1rQRe1hN+SxpuWDKi5pqlibrGfL4FL31Fyp3f3GaY6KBrjYyThKswjs3jeQDh21qyYY+xsQLGJv4yhwHIEEoq3A4cSop0/Z0owOfBJrsHeRAb2M3ikm97V40wrSNbx5C/59Y3tr+7tlIepGxvc/tzu/bz6Qc/d+bffx6Q/MG2n33wz8+OdJKQAUlUOfoGftv9mTRIUZGqjvtMuC8nFAhffO2Xz/7al/7i92w8/Mirnz47Lg6YMeaSJbWULMPKHO84R17oNU3mqJYUSWPpOlKcu5OdBw2eFJrKmIVa0iRnr3M4pbrfoKQKhUNaOw0paQYRTkXkOM0c/D7oGEInPRBSlFIqlmGdKgz55wopqWKAUMYqBhATbb1v6PV6l4bIUTduft7ec6nxb69k7Job8nXa56YQr0GeQPQ+Eu+H3tDE8+j3XWAN5z5v+Tb7YMcCTD9O9x3y83y/AhDb/3tsv/Llf/B6q7Z4FYgJ8Gngd7s3ozT9ehCM+MBRHQ48d7jsJPpSvWfju+xfKQWFlMzDPDaS7hJnKZLIS24pqk2PT/8/N1+UBGsovQkLFrrkJixMsZkqYRmNjnWdAinVMMDyQV/R0bgg20NsD6LrjIxlRCUVp8VR5nD2t7SiYp8pS10PMu8Ox9llnfbNXfkgXeqaG1aMtWSKK2g76403sFXPIQeZfeLBZ8+Oiv3BhHBcHFqzsUySIGFPE+uNayH9VO2dx22olo+b7UpXrtLq42omU07lkAPZS/ta6yaKXgaWsQZyKStKCk7VsmEnOqNAONApK5p0jC8ypcoCAbcJpS2w0genYy1pJNDiSuqkfhJ/ihzKtYqOs+KsVlYF2cRr0sTJNGDsWrbQG6nhD4ZTPhyOeEmP+MFwhz+lp3wgHPCiHqdj8Sw+WCCSgg/tNUWg7w+w11seyhUfCIe8Ek74SLjLK+GUD+ox39DHfF3OuZA5Hwl3ua+HnLLPS3rEfY4osH4K1yaa6Ygy0hUvpWEjRtlbIBzrXqLMLRCOmHFXjtLxNHQspaGKje93OOAOB5yyz5I1SzZZcibik2OeaUTPLBjSNbRAyioadl18jurouGaVHG2vPHkVY84aY+uaUVMxZsSYUbp+Pm87JCDPVIJVR8bWUcYkCnzmDss2ZMp/l+y9dB93zMl9RWxYQdkFqcoDCj9Wz+DmELZ8H0UWHOUB0/Zx+Wd8W/5afhw/+eDnB/1jz+0Pni1w0plhsF9k4/hclsnR39CwjGxIgjDTERO1SuAXXvvF3/fx8KOvfebME8AbbbnWJdc651yvU+AwwRiXcnIdsISXzxc3YZ6SDL04Z591twCj760SokivGGnFSOrYN1smOFSrbSIWmso4OeKtGrQrzWfa9wsHNIk3p7/9tZgY8UDK/JM+Wd1E2QNvAN+oVab8HLroc5UUg4SK+1clJZNikny0la4Gc0gX53qn3Q1YojBfq5OKelyjvYJRUqRjCWjq4/V7sgqruL5HMoz4XdNwatPan6MhfL7frsyWUvIrX/4Hr7+fAYgdy/tgfvM6rCfCnaE8ONguo29/180fgu0+ivx3VU2DYXt7uz6/He16yXE7ePHMBlgEvdGWpa4Tm0HOj++wBc9Eu0p53vglMtQ08Acrp3PzoGhEzVQmHMoeR+xxUPRNqu5k+v+rcJ22VSBJy2CXiRS3ro8fK8AywrEW0Qnx4yyQ570gmX38wc+cNdqx0TaNC79eR7LPSXHAgeynz/vdgjjpS/97cuDkdkl3O3AZsmOUCXbyYsy0fVDuUMfGuZKCy3DDlc6Zx4m0JbCiZcaIPcaspE1H5hWwyoOYFEj02ekCCw7yY6yyIKtflG3BXsZapWXA7X9H33i+JrDCKh2eARqUxgkpf38gprPx4XBETcFCO+bS8NFwkgK+3CkopTAIVqyK5HrXvbto729o4zEQtwNHOuFY9o09jg0BKLVgX0cx4DCGGf9v7HyujNFfC6+KjClTr1WhQqHCPT1IZBdXER7ZSMeVWBJjEre4z5QNLWNqeqFDSY6vL6x+DRasuWY5SOAYG1eIjfIWgDhNr99HX2wDyjF7TBkzlhEnss+hzKgoGTNiynhAu+vXNXfYHY4i7Hbin1ZdyK/bruAiP9ddwcz233l1JrddNL35e9vbywOO7c+5E5c3ye9iXntuf3Dscw9+4exzD37hDPr77GNve/yBBR4+J7ujOUlqR8Ox4JCp3w/7mQf/7Az6CuVSVylQcOG6S71hLJZcHGWwKUFSr5Y7vBttLCEQERi5jpngVZMIJc2E+zwh1mf8NQUVIpL6Tzq6QSLav7Mr8atqT1RCAbhYn/QokfyY8zkwFwjOn3Nvdvfzy63Tzqo32jv70CeL/Zr69TMWVOdyzCDSriOit+cljcGU+7ZKYF9mA7RLKSX7Mhusq3llxRm5UoN7vB45G9Z73fvxNHtfekIcOtWX0PoBvH2z/PV8AG6/7yCK3fsZZj+f9hlr2LXFfuOBRXwY19okPnsYlrpMv6QcVGD6ARFYRufOheoMOhVSRnEVG9Jd+6OOE0GXro9NWK209nBryawY08UGeBMva7knx2ihXIcbDgpzbo/lwPCn0t/mVtuEhXxaMObX253eVm0blzrnknmk3qzZ0FJRDh7A52ZZJyBlX1osg1NTsYwQnImY8ziRCStdpbGfjy03m2xz/KhB9/q/Q2o+K2UoQnkosxQIB5SpVtyRQ/uiwNt6wURGsSoQONAJa2kZa8V9DnlTLtNCOmPMTPsGZw9IepiL/xSLMsSqIM6W5Y3pbh0aAwgGT/Qy7mFDF5doszUh/a1x376dMQUrNbpfP75vyIKvFk9oNSSRP7+2I/HMV2+e3d5OVtRSZI3NBY9lTgDu6JT7ekgpBae6b4ENlkEr1JzskoIgmva9kjaKshaMteQm0vVawNUx0ZqJ1qzEs3gFJ7LPOaZZMtOaG7E+tzrqj4wjvrymikFkc6vnwWEgfp6bGKhUcf6wwGSTAkZBmDGiJaQqSQ6N8u13dNZbREWHNZofMouZOZ/LbwfP1pNyu1ckVzO/nQwZQhfzJvRtx364Xgzvaa4L0u+3n8N2zYv5MfbByQ72um/xXb8a/b6f2x80+/nXfumsk36uHmll4n5qEJ8+Ix4TJ2pj5q7u81V5wiRWAlTsGQjkY5vUY/l7sc89+IWzP9O9yP9VPOZ35XFKWAKpql5TJCQG2JjdaMMlxtp3o8uBDoWfC9r7Pjmltlc8c7PvWbCxCisKKVMiqJQyIV76JJo9j8tIFe7bdfMM/xDCZE9t38NXpLXQ5nILoPKKZCm2CnnfmsptwouAgrpP2geT1oNpc+Ey6ydJlR8pU6+LBy954ObVB08AJois9v5vzgAYUE6LI56ES14oDrmWOY4i6uioi5JFyAUIh8m/XNIiR1QA3xad8ntl75tien7jvUejx5APH8KSPvDIM729o77bvDG8V/DtG8VzaMtYxsxkgquUr3TDJAYDrhnQNyn1/SNWDitY6hrHgDsdpUeyikX4QQ0bOsIwkF6C82PoI1wrvbloT4U1ra11HZubS/ZlyjI2uDbxaEZU3JcTpBDuy0kqKy7ZsBchEx2Bd/R8AM3Kr/dAZyI2mDpr11rXvB0zFXeKEwoxilJBeFFOvsO7/4fXfvS1z5y5w1lRMpaaoIGR1Nzowqpi2nBcHKKoZThoBxmX/J7cCj62bJAJdpVUSBPhCxHOc86cD+oxDYH7eshcNpzqjCsW7GGBx4iaE51SUrDGCBbu6B43shnAqwJwScOMnmkotxkVV2wQJWKfhVp7lfTc0awo0l/CdiBg5A25KGL+3ZqCXgHb2LncSTSmJyuL/4Y8oqWlyLRGJoxYsB44sl69tOMa0gkfMKUVY4/6LR4xlTGHOuZAJ8wYDRxqVxyu1AKNMSUbaVhJS6cmAlZCDPi6RAXeSaDUgk6smXAhDTdsONE9VKxptI7PqFc81rQ0tMwYs6LXN+nJMYZjavtnGjv0WiI30REo4z+DTlXRoaniPR5xE+cghxp1hFQBcV0Rl+q0+7t7rvZ7sB045Patvuu2vXZsV4D8vPPP5c3qZcxL++c7bj9zDtXwytw2+UJ+XDkWe9da9W4Bz3N7/+yLr/3ymZEwhBg0Ov2zoRUKFYIolRbMqLlOn1M8pdJm494ryYnliIoP6Wl0RBns4xNv/PffsYP49x78k7P/svsQF+Wcr8oNV7JmwohvhscIkgIAg0vFZ2CLfckd9i6SRHQS6LThRhfJOXYHGjDhOxmSOSiBCmNCdMalTmz/Q2hxzw7l5n9XkRo2n39zy5PO5mArCDHRNISSl2LhkQcC/p1NPJc2tFlTvTnsnqSdFBPQbvCewbwsoKikSmynYPD3BoOVrXSVfM2SklYbQvSj6gg9dXSOZuuPX0vTTxGmUbz4EvO1HD2z1g2PuicGC9NmwLjlumAO24e+4vV7FZj8Xtr7EoT866984XXTUXBHvcfA5824PrDz6sIuSJRHxf4dIJUDC6zxvKSMTsZttfF9mQEkMUHfTpMG8vCYVrHRJ6BMYmNVpx1FDCCCLqmlTjzLS10NqhGFlInOcQgjgHVYI0XBNt/zJnNyWu3Yl9ng+21c7k/kIGU2wTDtjrvebGUe/fq55axjY4ymd1soT6RgKmMehic02nC/vPt8Ic1sHCscc11Zj0F0MK+DZZkmcdLyxj2EpAvxra7jLkcsBSlSpmC+FMuojGVkVLvScJeDlAUDu+/e/2GwnojPxXojOgmDbLxjmd3ZeiQL/hM9YkVOCRuvAcOKYa3FIJBwZxf63hAL7r2PoEjjuYif8Wx1R4+9bempelP5O3NKZ2oUs3PWiY7XnmVN9NktXWqQHjRRxsncYEMjXgqHXMiSc5kzouZlPaFEaCSybqmftTF/ef/GNEKyRtQxILFF/IksTBBV63QNAkonLQslQZUUZULFITMecWFQOR0TRGmy++E/txM5Hoyk80ruTl8BGGXLQMiqRdcs0+8uXOZJFoOUeIXa9idI+pwdSz828mPcZbvey6FUSg8D2/W+37+nVRL9p6Tr1OPW8/3nYyC/bv33h7CxXQHI087t9Tf+xutgTel5Rean3/ixZ84x+KNk//S1XzmDntLZx8au+9oQkpp4HaudHpQ0EgxGmY1LIFVH/8qXfuTWff7Ca794Jgh//Ut/6bsaAws2LOl4O5xzITegcE+OY1JKEhwJIoMUkhrmHSDqlpixEsNUZ8++KkhJF2FlLiBt3/Gkb/9sFWrzy1jGKSjwhm23XEzX/LuOiYxT5QD8ueuTwN5P0brautzu5U3PfNRxGiVimL6q69tPxEKUiPSJNdcC8QRJSc+yZb7fmBCDLvc/3cn/Cx/78bM2Bml+dKoBpEwsYAbHakD6PpF0D+K5vRUec1wc8ii8w7EccqFX8fwtkOuya5mrsbseybEc0GnggjX/85d/9pmcZ963arBDT/LyWo5/337PL3a+wOSDdIjp66N7E2CrBpm9vrGqjMFKvyjXeVOSqQAAIABJREFUjp0jJKe9U2v68fLZSGrGYkw386hunR+XByBVZLcpKZnJdHCsecktLzMCg6x4QKNWh6brs4xR9kabBDVbsGYVm4uXrFnH1ycyYs3GGsvD4l3viWcIVQOreF7bmQjVwEW4Yh7mHBeHvMBhbFRd7drkHzn7/Bs//ro7vBWl3ZNYogVjx/JGM1dG98Y2t3fTAdllZZbNAhuv+zLjw3Kfidbs64haC85lQbsVtB+zR03JXd1nrBUraXhYXHMdx9ANK1ZsGFNHNiVz+r0Evs2Q5ZnhacqYV0xiwD2XpndQZegwdmj6b+cyNK88euUj36/3k7Txv8O0vllcJ4V0MKYrn9wbtWxS3qvg7wm9hohnsC9lxZtywZUumDHiUPueB69e+LPijeYdgUbMYZ+qaapM1CBUlmV1Z94c2bGW6RwvZZWOxe6fVUAfyzVtDHy8V6RAWNOwYJOqGP39GGb//Xy2//a5MP/urkqJEVEULGJQ6oGNBwpCTwPsQcvTrK+E396nO/fbkKtvtb13E2LdPhe47Wj6Prypdvu7uQZLwm/v2IYHHO7A5AxYP/3Gj71eIHzmjY+//jwAeXYsH3uCVWEdVe/zWg6VmlDHz1lg4rAr718Dmx8dWrrLPv6lv/z63/wuApD/5WP/+uyTDz5/9k644KEsqKXmJsyZ64JzvWEqY9ZqbH3+bExlQhkToSUFUzENKAtUApMIMcqTCF2sYnjA4FAtr2Q4RMj7EIL2aBGvbpcRkpVXImtKRPoqClhC1asBefJ24Cv6zJb1y3Y4s+cw8SHRfyz6b6XXiywA8CqQ9xgbo2k/TxQxOTYpJn0Ao3a3OwL/9Es/me7jr37lc697gOUQMU/o/quv/Nzr//Irn3+9o4tN/0VCofh19T6SjTZGQETFPTlO9wgMbtXF4DBBr/x8Ip2vz1HPagAC7yMcy7Ky1WBA5nApjzx9wbSm7SEka1sLIdAOyk/b+gg5I4IP+hE1BU5pao6Ioqzig+uQq1Us0e1nD0UVxWg8Yk7HIn3jGRCDljEbGjaRIaLTbsCxbcdM/NnEbLZNEq514owS+8WMTWwI82uVHir6RrKUDdWWa70ZCCI+DfKT07flTepeBSkQrsMNldR2/myY8XS2rT9M9ldf+/tnezLhWhfp4S4oUhA7ombFJjULj6VOuhIjajY0VCJodu/dBvDEHfofsDs4yQUuG9pIcNCXcR8Xc1ZsuCYGrrE/wxzgiru6T0CZ6YgnxYKHXNERWImpp49jif6cOQdMWciGWidp7I0xGFTuzCnKETU3CAtaxhjzXKO3e15cHd2Dmzo6720KdgwapkCe7+pQNvFzRcxXeYB/ScNaOq5kOdiXLTpEyu+CkPmOXsr25z//3pUuWMqaI/bopOu1OgjUapCwXVWsPnNvYoErGhqxBWtfR1zLmonWtFjDujsqjfRZwxJjGmukSwKDc1Yc6Ywg3kHXw1I9u55f5+1jaxlCg3yu9d/zbW03a7vjkvdeuINWICzpA7vc8mqFWx4I5PvNcdh5ILINvdq+zpbFHtr2ePNEUhlnfc8U+/GHrX3kv+vWdoYJs/7cPOB4N6bA58HH+2uuFl6zq5ehz+YnqCaRYDwOgZpi8JzMNGba6XCaEe8vfRqU8Luxv/ra3z/7s/p9PC4WXOuCtW74nfIdZoxZyIpWW5ay5p4cWz9p0vNRgnhl2czXIg8qcr2vhr5J3J+PERVI3ofVJ5RFhoiKKnaZ5cGEb8sqDwWiJqcADHpInC0q37cfp6dRnELX6H07yh3JiyTEh0HEHOqVP8MGaa8H0CviMRf01N8WMhTUUqeKSE9XMjTzSa2iNC7GTw0EvAqTenB0mPz9evcWM5lyxSLBbbsoUGjHWaZrkOiUpUy6Xs+6vW9BiGEFuxRQ9Fm4vi8hNx+EDt+SNBx67Qzv3TC4h5UK7eHoBypKUlU/D5eMCy+plSkYcM5lH8yrmDHt8yJRJ0A3ceB2KRvt2wrxnwcvGxoqSha6ZCaTFOU6ptEDLT9XD5pmYhS77igcFnusdMNVuOZucdpPENJPfHm1xcV3bPEdqsS7jYtxwksaRKi8Bevwe1bKyBqOZcQ74YIbWfL98oGoUfBs29mDf3TW0H3HDsAnH3z+TLLJClzB1RiPPLB082DAbU+mSfhxHdZUYsxmrbY0O/RdoA9EtoUM8wBlO2D0oLuk4O1wziOe4DSDnuVZSGXQIOlplmtK5rLmIZfMGHHAFMto2yQ7Z8U4lpxzlio3EykM0Rm3/yVwSM2SLj01+xEMtqRjSWBMrxuSO6CLDE64jjkhTxW0KHdlzG/pDd8srrmrM1RH7EUcckPgm8U1c9mwZBPdW039IO4gB+2DjlY7qpQRc8eyd8gDJqZ3l306CRypZcLW0jFWu7a1lqkK4w3nTTxHC/JsfmuimvxMaxZi9LiNdLFB1StovbZIScFN1B1p6LjLgY0xMedmxpg5q8RIZlC2pxNx5NVj/9t+H/bXlRSR1csb+Ls0v2h0RDaZ85LGZ+aYb8NanlbFyJ2L7cpr7tznzlxe4cnNj297W/2aMqRg9uPKoV65c5l/posrgDsqeWDkjspzivJn37742i+f+djxcdDT5/fUOFU2vvw+z+iJOQqVVJU0n2MI8XOSB7B57Auv/eLZx7/0l7+r8eE0u86M9P/o7zKVCSLCb3ff5AfLV1jImgUrFmFJWx7yoeIeXwuPTOdJJDnPXhHYXoO8J0JEEkUs3GYp7Zu+JWXlXSfD/Z2OwErXg2fFFc59TSopLKiJwnzmiPe/b88JQNJda3STVMSNFre4lUjwY/dz9XvURaX0eVgYRF5qGl1RAJXUdDER7TOFM301sUbmMNpfeUqT97/8yuff9V7nFWmXi93ujfafC13yjW6TFOd7+LElp/dlltgeTResGsxxz7K9L3As6wcx56nVJjEV+EVPeDn6sqaX0/LKCQxxdNBT2Dq8wF7TFInnEbk3S3kAstJNCjhyuk7DOlcDx3ITsYZgDrlXH6BnRcoHvD9Io9hD4iqgPcd239NSUsYHskhsER58BVWuwzxen/6fa5QETNegoeNSbwgoa11zlNHBbj/UM5mma6GEgX7FtvmgX+uGTgOrYCAwFzx7Vu3swT862/X7t7JPPPjsmU86JQWL2Jy/zgJTmzTsbrZqDd5NFHJTDVzpnGVkwAKrdIFNjPMoIvmdQrC2zbfZ6CYtpk4N7UJTngWupGSPMQc6ZqYjGjpzqKmSgJ33EuxHgb0Z41jh6bNCl9k9rykSrGiXiGGOOp5msB/FAyEbuyZa6Owm9n6JMKFkEr93qS3XxYaFrDmXJb9VnvNQFpyz5pEY6cIispBtO6nbFZtCdjiykb99296W61Q5MlX4YY9CJ9ZT00Ot+kbDpbTcyJqJVrEfzGBcS2mptEgZWe8FGWuv69GJ3bd7eojrhfj9LCnYo6fcXu+osOU/2x0Lk7/n82WBDNhv8oqr/+1z8XafxnZAuasqsmv/28HR06yiSNdou0qTBys+e+cwj13b92Aiz7j65/JrNcyI364QBfR5APIHxPIxuX3vPfg+1HF6zZ9NX2u9GthKH7Q0MUSFvodt+Fw8nVzh27FPPvj82Q+/+qkzP6bv5z7fxz7/uXw/M5kwljGlFIkswgksHoUn7KvND7l+UB2bqq0CnGlhZDS6ufRByrpHaxPtfA+5gr6qUiBJ18P7Uda6ppQemu4Joryqmvdv+HaAlFDOX3PrmUlDCi7KTCTQfUc/B0/M5agRjUnjdB4ZJL6N/3K68120u9+p5dd3e1vuwwKpnaCjo9HGfAwnGlCjOd4M1uIqrf9NPPJn2d6XSohj5HIbZNwZqqm7gzaAcSQIUwuUqRnHIV3GLtA3mbnlmTV/GEopWOvQid4usflNb2kj+0Nxa6KxY4/RvRjXtKujewAziUKBpqRpwUa+jYAyjg3ti7BEMdpeBwx4UCLi2QYboAtdRzamEQtdM5aapa6YyYTT4oi5rnYGcX4NcziZ9yo4s9jt+xTd3JipX7DhQm84e/CPzhwL/TT74Vc/dfY/ffln3vPFOr/v28f4tx/8w7PPvvGJW8f0o6995kxwNVSfAI0drZY6CSw5pK8QYRlWHBX7TBilMbWOolSJilqKAUd5XgWzfWQQxKzqkWuHDD6b/S1E7vasSc0DxzK+N5ERNyyZyoiFNByoKYuPIrvHnk74mjzmT4cPcSkrJtRcYr0QCEzoUC14JEtqLdinsqy/FAQdqlZvZ78FYZ5VOnKzxnSNlRXjDFGs0VtRVgTOZc2vF2+na3QpS1Y0LIp1ykausEXPe0GqLJuvKLWUNNr1zer0cMxC+uP0rPqeTGKgbW7pWEu8d2UVFee9v6OTQKHGfLUQyyi6M7uWjk6tauKQq1pN3PEywuWKSMGLjNICNWDMYs2Khj0meCYzd4jz7P2ugMPHWz5++qxwFxfmqt93Cq9d776vmLiD5gHrbccrx2sPez12HYvbsLrQVydGWSLI33fqYdtXn43t6x5Gr5wv6rn5+XeERJWcV26cQMT3me+b+Jc7T8/t2bYvvvbLZy2aqpP981MOaK3HWnGgI5axt8vHQA5PrNSCkDwwB5A4hw39GY3j7Duzf/zaL50log3pq3wg/OnwApUIf4a71KHkm8WYWiq+qU9weHktIyYy5k25IKhyIHtc6FUK0McyotZVqka0RGE8qfpnKqqJVxidLdr3lnilVuOzZ3NBb87wBCTGK1+bnAV0G/ra0TGTaarA2Dpo2xjLKKFCwKutxD6UDhVJ6BJng0rPrBQcyiyxGG5orH9GFwkVUFAwiT00eTVlHBvl/e+pjO04Mnax34vlqBqfYwtu+7q11AQ1v7CJVSojQIrzbmwLSPedyshlCANdkGfV3jc4Vr4g5BFgrhQO/UDLzasFAESWgF3brqQiqE8kVSqj2fb77zhk6tuxddTM8J6NeaS7dd7+DU2C6bgyZ6stc11wWByk7fjEFjyi1pBgM/7AWvQ+jhOcndM8zAGYMeUmLNgvZhQUXAVrQoO+bHgg++kh3yXy6AvzMmb3t5mw3CoPwGLVKmxBhZasU9D2Ew++cLbHhE+98VdeB2OC6Qgcs5cetj//sR87m4g5Ue+FWudPPvj5M38Qt6FYf/vBPzwDq3gIBZ9/48dfB/hbDz6XIFjutLq12jKVCdd6k6pCsYZETZVonccxCLVrVRsTBnbtPdDbDsbhW4tp5kwh9l7/rHgwrqoJmuX781LuD4b7vCNL7uiUc1lyKSuecNM7XGI9LCMtGYk7fgY9vGJBIx13mHEcJjyUBapT9qgICmMpaFSTBKGdDzhFZUdgj4pNDL5bQmSTCul3483vx2hDYEbJEuW3i3di2F2wYc3LesJbchWpqvtMlV+PkjJj/ipYY8wktdy+1tuOZBGb/e7rEUuxe3cpC45iZrHAlJK9WbWMY2RfRky04kgnPJFFKtsDvFVcc6STdIxjDLpVxeB2IevoXNsiPFa7NoIwl17fx+abUXTC8964vpbsreHvNqZcST2vekwYpeAmYA23hfRBtG9rRJ8d3ZWQyfczLLr3UC277g5j8TnRHEVJI93gX1OsQX8ha/aZJLHFEIMFb5r3oCAPKLbJSvrjYvB7D+Xqm8rzffi2tyFkf/d5FeQPjOXJOA9k3YzJruRJsUxBxzgmDeyeR2IL8cBi2L/kc1zSgMChWt9eoPrF1375DOC35BEvygkjtbTChJpSSl4WY6P89XDFv5PfRFH+RvVn+bet8F/v3eGzy3PW2iJi1MFFDC5qMYZMFA6KPToNvB3OEwRqRB2TVnlfTJcSlA453tAkoT4wcpwaJUhfRQWDLI0ZZ4GTJXsdhrWNAPHvdhpYRFY+iWsC2PNbSUWR+Wq+nbQNi8/MQY/rjzW+930Sh2J9myhMpGala46LI5a6Zi9Whhw+5uvlvsx4opdpTnHhwDUdvx8J1WGFxytRvfq5YtDhqYxjw3+XhB29ob2kZBUDtElMWrvv3BH4oVc/efasNqe/p0HIn//Yj52BT9whLTp5jiBfEAOaaDIlVhZc4MabnpzmFKCTPivmk4FXMEZSJXjU02w7S3e7ubLH4XUaBroDRnVpn99owyRSueUUdFfhOm3L+Z5T2VNIkfAi9DoeJUXqJ8ivkWt9XIcbjorDVBnaRHFCz74IwlyXA+Eh30Z+Xg5AS+eeOceVGMvXdbDKiAcg/vMmLMxZFNOWqGj4xIPPnh3Tq7gvWLOJToIHYxt9byBcjufevp+ffvBzCZZVbT0KXlY2/PsoBlrm/Pm9G1FbwCHm5F6FG0ZScxWu2Sv2qLQ0NiwpElwqbzL3Po48I5JbXuXIsyUQS7npHtly2KopZq/ZpAWyjv1P65gpGUnFJStOokaFf3tCzYQRDS0rGu7rEZ0YzMAntAvmg0V3IS1TKh7Jko/oIU6xO5ICNCD0tL0BmERn1J3MNR0q0KnSiDV5z2JAQnamAKdlzU1n92RPjc1FJfCRcMxMaq5kzdfkycAx7ujhFd6E2Dvo5uTm1MjbjdCCMGHEsU65q7PILNZn9RsJzKh5k0sAFrLhgEmiJPbMahWdkJnWPJFNYr4aU3GD6YkAtBJSP1EXlw9/3cZkYE8nrKRhFWkyOzrWsbLksBC/Bi5EmMbSrvGFZ3jLtA93sv1qjKKDvytT1xIyqt7b237aa33vGjjnv73Ww26d2pS4jyUbFtIHHJ6tzIODgmJAubl9DD7P+Xfz8ZyvHT4O+vHbO53bRAR/940ffSYX9+dm9sXXfvnM75fDPrd7goD0eoFEQdAQK5VTVrQspeGOzuI6sttPGDa05/OKJRJ+/rVfOvP5paLP1DfpKTB7kRNuWDGOVQnBGJ7+HN/Hf9AL/nf5tZQo/Gfd/4GI8GfWp5xnPkYhQkvHOhgM6mF4Mkj+jqSmzeaNXDPEk1j9delwWgqISBS1xGQjLcRgxXyPWPnVdfLZJjKmjb5aQVQrl15/zXtbnT2roKfl9bU5h0h5lcUCpypVTox2vU1zYy0THL3wOFzwQnHCdZgnch+HVbWRsRD6vpVD2YtgJiOS2ZMZgcBaN5RS8qtf/tx39dz7tcyTOz435Yl2Zx1bs4lIl2V6z/1L78kBS6o7vN4SkyX/4ks//czOUe9pEPID5R/jgjkPu8eAYwCH0Kt8oXJGA3d4vVwYUFBFIsuDNzpVsRkn/w6p2bxXwHSMXA7DyLGJu6ylTfCaG10yYRQHce+Ur7GMuLNfFVIylTEL+qDCqzaeWdhuIs21UEIMQPxa+fVx832B0e6B4R+nUZm9U2uMPw+X/fXcqnaoeAZwqKjp++q0Y6VL6qIcCudlv/uif6E3rHSNFqbIvYiUwSUFj/WSpa6ZypiPyEtcsuCr+hY/+eDnz0qK7yme+jNvfPz1n3jwhTNF+fSDnzvzY9puSAX4+IOfORtRM6JmyZpRDDhs3NgYc7VZD/Ta2P8BxP4Lyxw5zXFNNQiQAz0OdxtTuu28eZCSB+tG+2zaMXeK43Qe3q/iwa8XeJvYExKiY3YuC0ZUXMiSiVaMteQeB1zJmhnGqvXBcJAChZW0jKhY0TCiYqYjHsucIyasaDjSCZc0lEjCo3olo1/07Uy9if2EEW/KklLj9VBhTJn0RAqIHGOwomMeOt4s5tzVfY50wrks2VOjkpyoTWMvcsy1LHGxPDCRwQMdoyhr6bhmOXhvxYYNPabWFvtA7U3/BBay4UgnLMQw0Ps6Zi1tZO1SJox4xAV7TJhgpf1GAgvsuo/jtasp2dcJN7Lqq7VR2NB7qpyNzMfAlaxTENXRcRnvXcAIA/KMF0Q4p/ckEegXNHPwt8eZ02HmooZNXMT9uW6o0jEEvGJii+OG3Ymd7YTH0yoxRmLQC1T6sbp5f5V/3iEHFqxFUgAqlpHCwAOwEB2PbSbFKgJYt6FqQ6hqT+yRQ7vyjKUzjD2t+vPcnh1bShspsjXBFfN754KoYM9bi1UyrlmyFnNOL8UE+zzozhOlPn5z2wVD9VEdYgDytM8GYI0hIb6qb5u2WXSOfy3c8G/k/8NISKzSfioHHOmMf9L9u+S0m7O6SpoUDtuupOI6zKmj35A3ZtuxhLSG1RSoVNFnKQf+hpHxFDivnifFXIAvJTUSrMroZ105vdU2JYVaQvJX9mXGRbhKx2L3x7bhsEqvmICthZLNH+7QO1vWOK6VYEngNRZcnodrTotDNChzXaWe2bkuGUnNiRzE8VLxdjhHVTks9ugI3Oji91VxPA8MHTodMLrdVluKODxabRkVU2pGg/Hn987vzUab1HSfN7E/q/aeBiElJd+n93gTw3R7AJKLESYBl8x0QJuZqXPqkCGglBLUKNkMrz9K20gUdViDlFAk+NLTgo/89ZuwSBS1q7BihcFbfKGvFZr4gLijP439AjkFXpOhJjeuGZFh/fxaGOPR1sQWoVAOLXPq35WuLQMRIV1LXTFlQoM1j/n1GskoLehgWfmctnebFtYn6SpmJ3ZeIw2sYzPcRi3YWemUkdjDW0vNgczMeZMZezJhX0esZJPoUOv3YBh6IOKTmE/qHrS6eVBhgcKwITfEsQM9farzdOeTQiUVi7CgkjrL6hqFtO8jr/iBN6n1dIf+mgfYTlftvU5TmVBR8SfCB7mRNWMqHsk1X9fHKfjVGL6IFJzIASvdcK1zXuaECzEF1gDc1VmqeADMdMQRIw7FlM9DdLQDyokaC9qGhrkULGKP0qWsONIJ+7Fx29s0Z5Ss4sJgNLxl0hmZaA7JtPcNlqXMqDgqKlYhcE3LW7riWtb8iXCXBS0f1ZO0qP92eU5A+VPdff5t+XXaGGSOqDjRKa+EQxoCC2l5U4RGrukIzHTEPiPelmtjAlOlFNMK6u95y2/KQ76PezQEviaP+TAv8Fhu8CbxuxzwIicc6DgtrA0dE61jj4hwEIM7FWWJKZCXaoHwmp6tZl9HXMoyVUOamInzHg2fJ2ucFMEVl/uqxlgt6PVeFW+k3hYu9HP0QMIcj36/m5gdnGFVnBvZ4A76SCsWolSUKZDZniPyoPtpllcl7BiKW8+GCU5aQD/JCECs6teTPDgvfm7b8Ku8Gva0RAT0Ge2cCesn3vhryfEwcoshrOy5PXv2sw/+eYLV+j1fYfh+Z9ODno76IVdspInrtiU+9xnxNR5TIBwwpSEkHSQY+gkeoLaD16zi585wHcefVx29whriti5lwdfCI8DG9YvFHd4JF5wWR/xv4T/EEazclQP2ZMJYK/5c9UF+tZvjpDf5M1kQaHRjMO8QYVW6SM+FiKT+Bl/LrBe1R5q4PsXaKw6ZvkUSx9M+SKikSkmCDkN3hBjsWe9dmf7uhaW112ajQ9RFCWNvXy74nK23Ss9r1kaoWCUlG4ro/xQcyh4LWXIZbixhF264iNfHYVhrXfOkO+f7q1fi+DCouVU+nJK+QqUnAvluzf0Fr2aIFvF6dnFe6iHW0PejefDlPcceZJpP45oolqjMSZOeRXtP2bEe6QX/N18zZxrvzeidrndjGxBkwKLlFtA0SJ01IFeR3GXepJtvOzfd8W+tG1ZhRROj+bxU2atxOhd3SE7dQpfpQcyPOT9PFzSEjJ5XeqcDIv1wfBD2CnMExzGI2miTmCfAAogVGyYyYl9mabLx7IdDqmoqKmfFehddCtec+HbtnXDOIgrwFXEi2GjDXTm066UV16xQAo/1kg0NP/HgC2ffesvfnX3mjY+/DkPohfcJ+fmlEmiswjXasdGWJetblaRRFmQUUiYBS78XDhvMeddt/09/7KpYorag2kaD76eIqugbGua65BW5B/Qicgs2GK3zhBM5iO+ZRscH9JhSCgoKrsXuzVjLxLxUU3Iv7HGkE6ZUHEYYlzm1di5TRoyo2NMxe0xYsmGPCTcRYhRQbuLyW1MwEtMQsWZQz0ibdSh3GTOj4jAu+CMKZmJBylFRUQJP2LDGqIFfDoes4u9gGcyrWG2oKfmd4pIZ43Stp4xYRgf7G8U1Xy0uuIrsWYJwLUvu6l56Vkvpm7BzC6p8VZ6Y7ogqv85bXOmCho4P6AmnOuNYp9zIhgUN83gfNmLViLW0rKRlGbOqY6pbAcE4Qh3WsWI21RrPxtt167VorAG0TVWXHt7Rw7qAjF433HLOdznd1mdiY8yc+op9JpTZcfh430ibEhr+PH275hWHXRWSPADx+S9/fqx6485bSFWskAUTfQP5MPjIm8xz0Ub/+2nWEXZWav27H3/wM2efePDZ7/n89dx+b9bDC/ukk9/vCqtKrGkpVFIV9Rv6Dje6pNMQyTlGrLThQuasxEgqnPnOx2SDwZE3dMxllaodPqY9+ZWP4RzhsJKGR3KVAhCAk+KAPxFeQsR60zzTfygz9pjwJ7t7nOqUf7+5YY8JDiPt9Paa4yiLta4Nnu1CevG58X6DyoMPel/BWa7A/IFcMb3VNq6j9kzVMkrw7zL2JSaCFDGZglbt80mENgZB7l95oONUvFbZCCkYqKWO5D63GbNmxZSZjKP/M+VAzF9yWPn94pRZMWOp5oN4X+c74Rzoexed6t4rqkaOsuFK3130+du1NKepE/3YOPFKVIL6a0j+pjfc2z21YK+isn4RjO2xjhA+D0ZaWv6H186e2fnpPa2EfJQXWUnLw2rGtc4RhItwlSAnbrkT7Ass9MxT/n4VS4UeVftDXsfG7tzy7eQVEn8v/z2fGCCDQolBElJJUruE+3ZH1GFgQTtudJ725QO5d341xrlF6gHxa9A3JPUNYLXUbMLG+KuxAMKDr7GMkqChX5dWW27UFFS98b0VY3Pyysx+sYdilL9u7dZ18/19p7bSTV8G1TmttpzrDYUIv1Y8jFTIBaMsy/le2q7sZ4Vl9lNuRQ2u4lmljr5Po1CbvCbx4Z9EVVObzCMkD+tb8gnXcapFPta5DQ8AkliTj02nKKykSlWJSRTIctz8mg0vySkNJqhXSsGVLkw1ly4xaBg5OYcRAAAgAElEQVS7UpmOwu5vwbVsWMiGmY5YqzcECgdMWEStipnWPCxuaOh4UQ9ZxKziERM2dDySJfd0ylGkoq5jRqaIi32uQjzDWKZqiqQZ0qhyKjWL0HFaVRwEm6LmNBxjDXeTWF1pCTwqFtwL1nuUsm5isMwDHXMv7HEqI5pwwO8UHa2EdD8B9tWc+k08D88cFgiHzCgpuZA5LSYpeE+OeaQXTGXMfT3iUMfJ6b9maUQZKDM9GSQfQgw+Si2YUDGhYiUtDifyKsZNfBYWso7B6LBXaxX7fUqMKSsfzz5uV2xiVaNM+bGO0AcpDGFaFhD12gc9E5w5NK4AL/Swrhzasj0/5E7X9uu7+kfyuTl3Kpwu2uFzvi8PmnuWHu8T7H/6Nkw6zgK0ERVE59Hn7T7J0/eVbJsnL3J7/Y2/8fpPPfjHZ/bdip994289s5jrP4r2uQe/cNaTEvTPUf5MOvsZ0CcN2RgLIHBPjljR8DZXEaJYcMXCegGlpY6V3JKCuWwSxG9Mxdt6xRHT1PfRVy17jSVPuXYSaFAeccFlMFr9Wcy2HzIzBsJiyq/pNywjLvBRfZEnsuDflL/NXFfcKQ64YoHTzDZ0NFEGAGLfgQ6Fnu157Cl4vVpCdnzWiO6JgZgYk4JOG1ptmEXotYvnNXHd72Gb1hjvmfxW29TsPpNpUnPPKXhDZBdttY3MoFHMjxbVfk5JMNUtBs8JI4SCA9lL8xbAnkxSVeDF4o415qMsdc2TcJmS1yVFhO1uEs1+KYV95vfgCz3NcgioBWS95pffr0GyXnqNo043pg1Cz47Y0RG2YGq2pj3b1dr3tBJyL+zxcnfIf9V9hP+MP8ZdOQJu9yP4IisxE1xKOeCyzj9TR8yf34gmCsDlXNPw7hzdzmbhlGZAqtaEFPjYAuyVFtWQKh++DeeoTvvMNAi8cRiGMIU2ljk92+f7ayOLhAcm3ojueMpKKpa6YqkrFrq03hNdsqFhqWsWuuRGF1yGq3RMU5mwLzMOij32ixkm7yYcFHvMimmCp5WUgypIirwzitj8v1+v3JyW9kYX6RzeCResdMPXu4dc65xKKq60D4D+wsd+/OyHX/3U2V/42I9/z6L2vPdnE5t7fbJ1JySojcgmumv5RGYOTTkYX75gBEIKOr26NUrF9+Gj5hn3fNL3EvcojemyL0VTpX2PZcQP8BIA54WXuUte5IRJFBs81DGv6Cknss8resrv6MMUDExjteBIJxQquAjbO8WCmpKlNJyzYSIlnZgA36GOOdUZiiYI1v2wRxD77jfkgneKBdey5qEsmGvLQjtqKRiLLS8jirQolwilCDNKKoRTRhzGPHyLcknDogup8jGNy+U4biNde625rzPu6TSpFruWxIfCIaeMWWnHY1myH3VRJtTRIbWKzCGzSLMcaV/j8/LHu/v88e6F5Kh8JNzl+8Jp2geQtAIAXtIjXtYTPqDHyfFfioUv/vmF2KLr1as5K8Oe01NA5k600S3m7FdlGksOOfNx3S86Gp33DSOqpKsxoaaKgYkHIF5Nse9Yz4nPWZ4NXdNEeFZjQUpy3KtUVfbt+XHvCkJ8P08zTwDln1lHSFiu56HxWPtr0jMS+esOKfNG9IqScXRAxnEsreIT7sdVpCvYX0cYEljk9lNv/PXXO8LzAOQZNB8HtZbMYqLBA9Qcvpg/W4vIUnegUz4kd5kxZkPDSjdMGDGSmr0Icw4oN7LiXBYspOEtzrlmyRULLllwLHvIIOnS9zz57wtpaKTjMdfMWXEV5mku/6Dc5UBmtr1izct6Jx3nWEb8R3mTr+oj7nBAS8vvhoe83Z1zExYsY6UDtuYLKZJWhv/cfka9Cu8MnZ3GCqnU0bEtUiAhUrAK1t82kpqZTGKiLWQ9Jg7prhjLONH/+nvJCVeNiI4NrrFVi3UF+v6AwXd9nrTeFEvYjmXMpV6z1HXa1iZyCB6xl+bLAuFOccxLxZ1E05vEfwk8CVe8Ey5oI7GMCzxutEnMlt+teVN87vs5gkIoUqWnwOUniuyT5m9OZZwSnvk19Vk5T/A8q9WQ9zQImVIl6sVXwiGP9XJntiy3nh62ZytStWBhrWtcSEazwalxsOyCIfhnoA8+gFuQpEDfc9LE3g0POto4sEUs45BUPxlWUhyjvNZNasRyy53SPgLvA61cDTt/zQOPpa52angYXrxMPSL5d90JdZiGZYSL5Cy7Q5w72B0hBYCp72aH7aKazc/VoS6Or/QAq8A0TjaxtOv2w69+6uyHX/3U2Y+8+unvyYPTxewKWBDs1QWIkzWGj/er5pU1H1+2wLRxcgqx/G1n5TCqSgx24+fuwVh+70dSDwIRZ1HxitukmCQ9G79+vbClZ6u6tOie6NSanem4lBX7TGIm23jSD2VmziTCRlpKTMV7LVFDQ8vkdC+0Y6YVo6gGPlXTA7mr+7wcjriRJjFBuZP9UK5YS8c7rHmbFZ1646aZNYEKYxyPb2xaldhUWYgFeieM2Khd5wkl+3EPTgY9ic70PZ0a9IuSRgyyNaLmSO2aXbLhnI01kkvLiU440mma3GsKPhSOOGSWXOftXiCHOVhAEZjKmCtd8FV52xTP6dXEwYXCbNFw0bN+NrJzWsdsvLHJ2fVeYU5QgzFe+SKVN1Lnc4hXFVyZ2IObXMk8oIlQoIzXzPfncMTcdgUJTuSxomEpFpBsH4cri+SVm+2Kcj/GdfDTLU8UCcKSTRqLLhCXFusYRAAJ0pEnCvz+eqDWEbiJjfx+3b0y45/fhs3k5/C0QGSb8vu5PRvmY24WkwtDhEPfWzSOQb41n68oKXhHrim14C3O6eLz7iQNeSDRYXDAt7lCEPYZc8iMJeskINpTPQ+rfB3KO1zxDte8Hc4515v03geLF6gouNI5H9YXOAkTvi7vcCqH3CmPuQlznoQrWm15xEW2NoWkT+YsU+b8943M/vpYxhHZ0aV5xhMKgkGCvE/V6F9XKUAayygFMSL9czfXBQ1t7Ec038rFn/2Y/Fjdoffj9mPwY5wV095foIz7KdI+PTBxh9vfm8gYVeU8XES4Vd/j4uycfryuC/RicYcPFHfT2LmIyI11WCdky1hGOFPWruTK78UOiwMmMXDbDggdgp1boyY87MQ5Xvkd9pF6tdqvad9L8qzaewrHOqamiU6TAnfkkAsiE0JsunZLDdoMBfPyi+lOt/8Ow0ZE47AudzrI6mxZDp+ixyJ74zoY7Can2d3uN/Ho1H/3B6OvdPT0mdvm0J68wd6PKeT7yXoRcvG63AbaCFJCjOD9szm1ntEcF1lTab+tMrIq9YFHD31zx/s7odb1e7jRhqlMWOoqqsb357zQVd8Yl2Wm7PcSV4oN2vGrX/nuaPH68WPK8lX8BxZcBvV2bmWgvyB9H1Fi4tAeE+wQuFJKnKXNJxZj6mkTrtWOw7a11jUVFSo2bnt60MCpHA7GpFfbPsApDSZeBx2NdIgKE61wpe6FNMzUSrJz2XAoM+6q9YmspaelvZENKwxaEFCuZE2B9W5c03DCKDaUFxxQc84qVRxqLbiLlbxvZEOtJUcYCUEXSRXWEbYmQCViuGsRq5JowUQKFGi0h//4xFnHhrwZJQs6XirGPArGbnJAHeX9NIUPJzo25i2d8mLYY0LJDYF9au6GGQtp+JDu800KLmWJUDCn4UhHfDic8B+KRdx/hElKw6GO+Wh3h+tyyW/II17hlI/oC3xVnphIYtQPGWmVGHhqShbS4Jx70wi3qmMPDkAjHXNWaYw4e5bDtkzXpG+a9THjQcOYmiUtFUZUcc0yOeMFxgDowQmYI+GsV4oTBdwWGXQIV0Nk2orNoe60ew+VJ1gskOmTOvmi50FykY7JWd6G2gr+TOWQrDomARxy5lnsNW0KTKAPtPJFfB3Pcxfk0oOXPpDZMHXSAJRc0yUnrXgaeclzezYt16fIacb9+QQPwrs0dqZac8SEh3LFhViFfsp48AwZ3LEhsGGhK16SUy6xHoFFrDweMmOmNY5jKIGNtORqZNcs2WjLXK/j879JjFaK8hjL5l8XK5rSIEf/RfhjvFnM+X9LSbAt70/wOcuqAcaK1EYK2qlMWKgxPfna3cV+kLzp2yFRniBMkHCpQF1kuk4JPPMrYhVUKm6CaYikQMN7HaSg0CgsGlEknUAl01RtyZvdPcu/ynynEVUUbCzT9n3N9XtZS81NBi33uSan6vZ7ea1LChH2maa5xf2x83CZKiAzmYLA/eKU3+neTHPMD736yTMwLTbbbsc//9JPfUe+yaHsgcCb3duJ1MiPO8lAIMkvqqhoYrUIzK8bUSW5CPfxTMS67ylxKFtA+ZFXP332L778bNH1vqeVkP/0eMRRYboCv1Wc8xvtV1OzuR3MMBoMaHLG8+Cjkjo51o577Ol2h0JU3wrDpxoim0TknZc+m+nl/XzRczpWb5hyx7CkiJoQfcbb//aG8DzQ8d6LNPC0G1Rj8syef+5pFLlgKueWOVjfCs7669tTEueBR4j/TE17EjMKE4vSxR56Y8dQ1mF9a9/vZktdDbIfwADD6XjTHBPq5+ufS9lQqfnkg89/V5URfzDd4fW2cSDhRXtyUx18vnfy+jHWN6D3sI46q0J5wDWSmmm8tp7l8Iy7UxD6hJ7fd8+oBTRtY51lr51NZJ8RAZhE6EFJwYGOOVSD072ipxzphAPGjCOlrWfbG9okuGeN6tYLEjBY1JiSfa05loojHTNTW4heYMJYS2o15/iuzvhohCstaGKzvDmtHUqj5uQttOOSxoIMoBRhJM4iBkFhJJYtbFEOiopTGbFSEy08oKYWYY/K4FmuqIvBtT4crJujxnoBDqTiRaac6IQFHQ+LG2YRmvW4MPjROitne2bqq8V5HH9FzJi2PJYbChU+Gl5gxogjnXCsU5zlxvo+hFp7J6h38AtjzML6NryK4Y6QL4THuseUMZsYZDgtqCcpbFFxmGCR2LVuVZEpUq+DQ0g8wPax5sFFbrlTv5Y2Uhk3yQGbMEraJiOtUkZ4zG2ohW8vn4dy287Q+Weawf3oK0EWnPS6L3m1xaEWs0ju7Nty+JZXvp2C1ZvwvaHY/68igKOhTZSe/pw9t2ffXIvDEysJZhvH0jpWGifUaVyWmLjfkU7Yx8REjyN8xyGNJQVzXTHXFTeRcMYgjTU1FS/oIXd1n5MIWzX4rDFwBZS3OOchFzzhhid6nYSOK6k4lUNekCNKCn6z+wZvdm+jmC7QkU74KC/y28UFvyYPeVnvRKbJKatgFQpDftgzM8Ig47PCqsHec7pWI6spI7yoZ1zy4MXUwle6wqHCYxkzlnGERpWpQT33zQLKIiwTpGke5qzDmkkxScfU0KWgqBSDe6/CKjXH+3GsI4mNm/sFEuc6EwoeJ7Kc/cLg5QAvFXcSDMyrN7NI+eFzkycbLvWGN9tHzFnhVa2j4jAGOX24eFoccVocxTmjS9fKYWvQtwb8zQc/vdM3+W8+9vGdr08iyYhvI58n/X46JKukZF9mzIqpfSYywK4i5GwdP19SDNhk+8qIa3g9e0xZ71kl5Oov/q/pRvx68Ta/3X0TGGb0Ox1WAlzrwM0HmEONvD/CJoOssSlbAB3qsg212l4Qc5Ebb/7ym9Zk1Y1cnd33v46BUEBBe45s326f7bMOA3de8yb7/Dy3Kz/5gr5tOWOTZxH8+7fPsUisT3lg8v+z92axtm3pXd/vG3PO1e7mdLerW3Vdbsq4oQgJURLEC684D5FcdXgJRBEoIBEFC2LT2MTb1yljY5sg28FgokiJkkgJ10R5wbxEKLKUSIkSSIgLgqn21u1Ou8/ee3VzzTnHl4dvfGOOtc++17ioqnuRPI6uzrlrr7XXbMYc42v+jR9VI02uoLrsb6c9u7ilTu3U6/fs/YZVOw6x7C5vW15Hf5iyegYVLdYdcNjWTJrs9CoIf+H+z5/9zBs//HVl83YdD/lHY6A2ti/9mpTdEPNVSZ2KdN29I1IlUYJW29wqzV4Q1FQunpBUtK57GFglJwVxaoR2xzELo3vrXKY85oJX5S5BjVdwrLOsTrVPXY2KwE6s/fxyPOJSWkSEWgNLmbBNVXwnRR/rlB0dO+nTomdJQEfkBabUIjzVjhlVfn1CyO9bMOVIG16QKW20bsx7Yc1RSnjelg2VjpU2l7hsxKAKO428y5bXWBLEbPqiwiKBh3ybOqfjpTClEaEblIknIFXgaugyaf1IGt7THXeZMiQHd//ZQhtejaYD/yCsUeDb9IgvMHY1J9QcJWf0NlVSvQ0Oo9ywV+YrgmHKVWjDwIY9BsEYq7JX0nKqM9bpefBncMvohG7+LWtc8aui4oodx1gCa0mAPcEzJul4KyIOnfDnbSxC+NoxLYIuT7P9GSgThpHwnaSBVbIZVpUS940MLHWWVLKGg0ShfLbK5+76/9vcvy4KMhaArtjm83BImEOrxiBvhGhtCjib3xs7j4h3OcbjCBme4e+J+QgOnZx7ev7aGz/6kaoe/s54fvz8/V85CwVx+ZjpAXcISPyo/qA46R26isBbwSDidUr0YSyCOczRO4A+ly5Ym8CK7DjWGW6M69TgnVigWFPzLF5yFBaWBKBMZcJSZuzpMkSslpqjpOh0V+dZGOL/0i9xR075vH41cxQCkoneuUiRjAQ9mPfuqfM8LMEI7HT0K/LgFUYzaU/O9trlWMilelVTYUNHCLPtpTX7tBZ5uG4Vfb9mgREdsc9we+c55DVRbb8aUpx3lGDEV7rmNBzZ86rKbTliSsMX5G1qqkMECeOaakWiEbZ5Kkd0sued/gGfqr+NgHAsCzQoz+JlLo67Z4mfj3eIrFA7Tde8okmx6Z/47I+fBYS5THkyPGOve0QCf+QHf+RsEeb8rV/9ybyOHGMJxa1wwtN4AcVcvR67+b19QW6xwnm2VY497T0RFeOVtElkQFG2tLkz8lHs6H5LOiGf/8N/96ztoO1N/ebTwyuZwJtbdmUFTUfPgtzeT/4YrqQlqTJocnI3O657QK5qlSz/z7sengGDSwCPPJCSaF7eaCi6LDqM7U2G/CCVY69dbhl6kuCEyetEbx+mFjG69tqxxRvfX37OM+ZW9+kaJh3voipYQiBKyFHEVIMmNEQim7jJSUOdeAs5cLihE1ImXn5cTuAqM/CD65jajCFBmMrKDKTgnZAfKF/U6q9z2v75+z935gtJeb28QuoEXiOxjgpqeY7IGKTGlIjstcuVnSFdcw9oOicHerdE3Q9H8jnmxC7dpZqKucyYinFUXC3pBTnlVbnLMeYPcsWWB3KZ6tN2PCc6pSeyErtvj2VFoyE7ozumf4M5/1aYp0TAkpMFEyZqcgUOwWplYE3PhXZc0fFKMMnWV3XBmjFhuRddNtpVUYQX48IMCTEOxFo6rhL5cyUdrUYexj2dKm/KmlYGnuqeiVivqRJhIRU7tRnbo7wSpnSqtNE8RaYhcKk9bYy8EKbsUsJQI8zT7LfnXJlTsaBmqjVH1DlYcH+SUxb5XlQEPhaP82y9p0dMqHNFvCFwrDOeyjo7oAOW2Kl1TpxAX3ZqH4uRT490woY9qwQ3chiYVeK7HOQ4HGmDqe94sO+Y67I44V0C/7c/91798+Gkaz+uQ5f5mN3IvdtyKduD37Ojs+NMUDRPmNyfpfyecbY/3/Goi6dhpGMemhXC2A3xrpEfm3cmM2S0SIQ6hvw8m5yqVWH3CfZmz7xTQhP84VrA+lHcsH9nvP+IKJ2YAp4XlYCiWzE+G/7vmsBcR3z9aeo8TGmYM2GaCkrTJGYxk+agg+1QL+sabzmXDZeyZZMc13fSs6HlGWu2CaWw1fGZn6V1fq+9JTOJ4P2y3OK2HPGl8IR/xJtcSctcZrymd4wgLqN8NZCPSYlcJYK7DxeaCUhGTJS2AE2CWO3pTPo2PYlGwm7pEg+xUxeJqDJ/0zmNvfZsdGtCQakg5/shmGzuVCYHhUwpCOW2n1oEVUvNIvlgeXzn6+5UJjyOF6x1x0wmVjhjz+1wTI91fxppmMqEWRKM6Rk5kzs6trScx4tcoL7QNR0DW22ZZWGZSZ5DHUM2jfbikXVazX5gpRs6HcxnKr0+aDQeSxGfXV/X7sWFzTlZZt7OuAqGfO0c2TKTCR0Dx2GZ7RkcrePn5wVz5/XsUuxarmV//DM/9pEiqH/TOiHbP/lrZ90wbjwPt5FdVP4/nrEKe9bJWMcDbBhxwTlITslI5m0kyMheu/QZm/DXYQ/lA+g3xxOA/Po1XkN3DTLUa88eIxl7tltmwUB2xLaHcKx8+xj17QtHTOeWpEvjycVz3Q89NNHz9zqZ17soZXLi1fUMb1JrZ5K7DkJMsrNGTjcfDzBYz+VwyZ3qtlVLJBgxKxjRbRPtISyPdZSclYRn3XJ9+EPli58lX5bwOV+mTgmXE+W8w9RIw2k44iquuYprWtnzQrjNjptdmn87o6x4+jXriUwKjXR/oMtzGXQwiV5GCNbYjQu5MhXEOmtBjaA+FN+3TkZRZddKUda64dvDxwgIV4kvsEnk3CMWCMIz1vm7eyJbbXlHzpnRcMSEKTVXtJn3cSUtG+m4owsuZMcG44qcyzab612wYSUtrQypUr9PQb8FpFsqHoYNjVbsdc4pDS80Dbsh8iR2bBn4vXKbZRNoB+W7qyOiKjtV/jHPeKx7gsBFOpaHrPi+4QW+LFd8LVzwrwwvcZJ0xBbUDGrQrYkI72mLQHqKhJdkyoV2NAS+fTrlWTfwUjUmjLdkwnEV6KLy8TBjEyOdwozEPSFyW2eWeEjkRKdps1E+NdzlH1TbXBTZJKflmsC9uGBd7Vmxy0naggkLHdWlwJ7djXTciwuehm3eIEbjLiWKbW5LZmi6xi5TvaVN3cD+oFLvv+eUOW2iSFIQzmEk2frxlFLQ9n2BAavAjp3Qw67IOE9jmofbXE30n03Td3rXZEqdVaw+aFzvhngnwz83IRwkYM6N6Yqq7ChzOsoLL4qCQflcjhyuw/XqWVLkMylih9OOZqL+PJZ70Q999qfOfuFXf+x3uiEf4eHVbh/2XEcCJj0dZZTJ9QCzoWKpDU/ChkoDL8alrXuhZ/BnQgYarbLa5QVp/hB4wiUzLDjca0cvNodOWLASe56f6GXuJgxEjpiwl84SEFUaCRzJnBkNJ7LgXX3Kbw5fA+Cl6h4X8ZIvSMf3ymv8U95lk/xL/FnqtaNX25t77TO8dxHmJsubYhjfg4GcMGx0awVAkhcIXQ60O3rMvHS8vlVaVTKMmkPu10aT0aruWbFhKlM63SfV0u5gvzUEwSRz1Zz/O5GG83hBRcVRWLDTPXfklLsc80zWPIrnvCYv8JQVF3HFS+EOc8yc9yW5jYiwps3rxT5FYoJwFddZVMhkhRseDo8zvN87OmYgqJmcb3GWeZMYOd+7IKPB8di7DjkWysVHqWmo+fP3f+7smBlLnXGbGc9Yc5nUuTa6zQmiD+fhCoG3h4fMZcrvlm9jJXu+xsMMg3MObyBgM7eMWwzRMZcpO93TSM2f+OyPn5VdmQ9zfMOTEIddNUU8rgqbGHmgLf8z/yiRot4HXsQhcdFHJWMSUFGhIgfVeXec9HFdEvW3GjElHGWb030e7LjGIDJQZ3dLb0367yghYQ7TGc/tZi5FmUhkmE9KwEqip1+b6wmI/z26rFc5AfHFwrxEOqbSjFAoJMMcwLP/w+Rnl/Ck14/PrwWQuxTZ6O9aRdG/y3Gm3jI9DIQ0d8WWYcEOqzQ9HJ4emCVe/QsYBZX3J5PuGDlAPZGVjmZ2XmUtux8RJSQzSq9XgCt8kK7VSEjv0qK7146FzA74MN41cfiWYBCRY+a5iutt1w0G8/Lv9esWRFhgFaEraVlqEptVS0oupeVIJ6zYU2lgJ3siDVM1nPO7ckFF4JFcsWTGY1lbYCwDom6EOCTMf8Wl9pxKw7N+4E5doTTstOLOtOLuUvniObyyCDzYRJ7GlrnUfCWYvnonkQs2zGh4M1ywSp4kD2TD93GLXpUH7DJs6xVmXMqeudasxFRWiNa+HVDWfWQTI8sQ2GpkJoHjKlCJQLBOSjeYCzpq9++46ID4fOjEnHanVFmsAbDzlT17DF6WeRQyBsJHOhk5OlIo74nmIPlCthzpjK1YJ7ZNXI8ZDQud0skocGD/OYxoyMG/V+99rrqb+Tj/NHcJSk8EI3fvU0JrFdjRG8RmdYlFLp+TOn2Pfae54IDp8O8Kmm1EUyA2wlxKIrA/39d///U9oEkpyQ5zlPfviUW1eZaegSl1TlhKGNYkdU+7NGd9eAc0kzVz5Vjzs1f6VX2U1WR+Zzw/fvb+3zwrE5C5NnT5flsCcn0eOjfuIrRcsmGaEutnYUdDxblccU+PWOiRPS8Kc5lwmYjo1yWqEVLXvmLDPotNeNJvgjGW4C9lxnvDE+6E07Tu23uf6GVGIIiMcKCOga/w0HzVJBysFz48eAay5G1DxVY3yevCuqNb3dFQZ98w/3/rGztv7FCQxrsVkaTYl6Dare6ZyzQlKwlOrW1+/ltt7bsYzU01oQc8OfL4pNOOKAkFEiMkyHpM6qO7ZI7bBStZrXSLQ9W7tGY41wcsfpgxYa0mYeLKki8nJaz34uOcgA06QOK/esfoSBa5AyISshRwlb3fHPlin3Oup/Pv3EHe4yX7nfM0d+y1K91yoVf5HjaFh50UBRVflxppOJcNd3Vp8YQMnIYjlsyymMc/jV9LsYukxHxI59Tmdc0VEj8K4xuahGz/5K+dudFVNwhtajb0qqx0oEK4lcxtNrrN1fX3S0iudwh8WKW/PsD/WXBYfngMMO073p9M7d2CMjj3v/1z/rCX3Y6R5DlW2SgqhKUq1XVZy+vyuWVFnNQFEQnwPspg1z9nC+zYLvTr4cmVkc5sQWgSRrJLgYx3gRwi55wH5zH48V6HjZUjovnzZRXA/w4yIerATCasdIN3a2BUoTFYlgU/C5mz1R2V2CIzC7NEiBu9XH67wwIXM0n061DzQiEAACAASURBVOXVHO8qePJhG0dIFY7xPPo0I0LxPpsDVV40vKnqi4A5oI78koDhSR2+UmEusjOxBXOR9NFnTLIzr0MHHVpgFb2Yieu+8F1JmzfFk2gu3hY4mxrVcYIbvB0ujRfCPgd2F2yYUnOqC96TFXOpWeqEnYxeDQsqZiKsY+RpP1ABL08ajqcwnyizIEwbqIPwik4515bbOudSWjbpTGoCl9JaYC3WCfiCXnEV7LWgwoyaxxo4D1tgzlfCUzs/PeY4qWCthikNgV00l/W5QBeVEIRdVJaV3bfTquJqGOgUFiFwEY3902hgTk2LrU0r6XKiXxGMcC/RzAgVXhtO+WI18EAuaah5KR6lQL8yyd1kXjaIuSwvdUIrPVMaNtImIQBLKGZMrMuQ5rcrqpV8DCCBsJQJI/fB2+3+3AViItA2ObF2bfyOIW/K3t2gWEfKIOb6v8v1ypKD5L2Q1z0HUY3uvG4O6L/DRQlu+o7rY0/HIqkRVYzGoaUqnfNaJjQJNpa6kWkNKY0NXXbU8eONVBnDHvKz6mvQQFkAy8lk0ZH+oc/+1FkQYa07PipVxN8ZNsoYwtawGStp82rs99rnn8/tpTY8kjWT9JmnwfYuk9c2wYmWnk4M/tdiPLqQJGN3us8JTpWq0KLG73PPrjoVLffacZJ4jZ0OzGXKSjcsZMZFMvLdJWXOgZ6g0LLPhbvL1AV3E78uJytF/KCa+ZUzmbFOalG+b09lkhEcZSzTFcUG523A6FOlXuBI6pp1quwDWa1pIXP2dHTpVtjvH7KkricqMXEpzXMtpuBeaRL/dhO3LMIi2ywYhzJyxY6XMPL+e5wfcEwfxQteCLdz0L3Xzp53lFtixPVKQk5SLMZRJqHJ8PysTprKXJ40+vBEw1zdQ1b28mXS14mGmjWjP5qm94hYIuU+Xq0MfIqX+Qes87m02maoFYw8GjtPZactX4hvUVevccKCZZhhUFXb269kx0ym7LTNMZTfQzNWHg0z/4PP/KUzgP/i73zuQ13LvqGckPmv/MDrVVCqoFzt4HynPNmZlfxRmhCf5hN8Dx/LEwbev+pUVt3tfUVSURDEffKUm3T58+sJiGox4WSsZJeqCB7UjdCvUnFryFmufy8kYyARMyBMXJSIE7iG3MFwvod9r94Y4OduyDXuR/m+6yQsryxE9Dn1rStdpRaib9b7fAxXcZX/c55LJdZydOyo8xrCtXvl18b9Wjq1RVp1TAKDjOZ+rvogSVLOFS/AjBGdkBcQ5mIeGVMZ1W5+O/LA5fjR+3/1zLGVPspF2FVwJjRMpWGWMZrh4D5PkhM4jO1p34CMPG7X43Y45iQs02JdZdK5X7+X5Ta35ZhX5A49PQuZc09OGSUglTU7pqk67AulXeshLbZ23RzaM9MmK/sEFc7DlqnWXKYA+Hbyx1jJnvNkkOWLtisL3dIlT2XFhWwy7t45D1O1OfDluOEiBXvHdfIU2cOzdeBkKqjC3ZkwrwLfLyd8kiULbfJGd8E2f/dUR4+Ld+QZ57LlPGz5SlKmek8ueCtccMWONS1vyTmPZM0/kK/wOGz5x9Vj3pY177DhSezoVBlUDXYYrQvSxshEzBwR4ISG29IwpeKUhoZAR+Sr4WmRbE3ZyUhE9TXqlXjMt8U7GZbg8yIANZKUxEw166lYsm1GhOFAqWWNVVuvku6Uv+4bu89VD+73xb0uA3OHlRylSpi9x5JGlxf1SqwHY9c7JjcNL2T4WCePDfMwGY1U/ThC+plfL+8wlL/j+jrvZqk+IiMHpiQUl4RK//m+SGjL4ec0QrCsg+X+CGAwLMdwl3/K7/PhWPR8Tvr+RZA/9pkf/aYarf7OuHn8lft/46xPyf1cG451mpJ/8yXz58Lno6sFdTJwEVom1Lyox6yS6/monmYp9oVsucDku80MdoZ5b1fckiUDA1e6oWXPsSyYyYRLXRPVhEQaqfIeGpNwzZ6Ol8IdXgymJnhPTqmpbZ4mx3Croluy4zGEJ/++9zvXw4VuXA5/0CEnID5MfWpvLuTJfNlJ7aVQBozoBh+5OKIWYbW6z34hBje2n/eJN1JLwz55fNSpy2Kmh5O877qcsHdPtChs3A23OA5LXgx3OA0mg+vQYUV5Fq9yLLDSbU6sIsqOPSvd8HB4ineAXeGv9kQR4SQcm6fJNTEhh0kvCg8PH26ouIrrA2UvGNekNhU683WXQ2XDhY5rUUc8iHnKYyi50dkiIv2eL8f3uK2LrMa2SH5UExqOZZHjrabwIOuLuWE/s/f8yP2f/VDXrG84HGv+Kz/wevenfu2sDnA0EfYDvNfteciOX5P/h0arXAmH5+FXJRkbSvKw3XBvC77f5unDNvFEYE/Zto8yaXHYknEVLDmppUbjIRHcstmRA+ET1SdYrqrr4WcGhlFmuIBZ+ciQq0Ka186/4IaUpPVrnSGREV+ZIVNc/4wmPKv9aWSKxpi4DWTImFfb5zJqo4NVECrGBwcMpmUtylEByjsqfj29EtngSmCSeSCOKbcuiX22046TcJwDoEhkoztrOxKTY+vXB5OwQN6CphLvXf7tChdeLS1lOWNKFqOEXGmApL6WHOyb4nFyidU1O07DEWvd8aKc8kCf5kR5mfwJTuWIc72iY+CuHOMQMOe+eAVvx55BI8eppXt9dOIQvkgrptq0ZJIkdCPnsmXBhN+UB7gk8Fp3SK682HC1mE3qDGykY6YNF6HlKDZGLtcZjQiLWrizgFUL2w7e3vb8qy9VTBt4soO9Kl/VNe9Ul2zY43rnezoj3Usijsaau3qcFdWm1HwpPKMn8oBn1j1iwi2WnMuaGQ2tGmfj3WDt7DDcZknNJtrvOB967tQ1T/qeRswUEeCSjntMeVGmnKf2+5Xsaag51tFRd6IVexlSYOvmgLZ+TLXiQna8qMsEBbKgxV5vM39tR88iqfQMDMnDImR4lXehthkqVWUPjnmC2ZUJwZ6euxzjMr9TLPFs6bL7uRO0h9QF2acOzCRBmDxQ9+Cs7B6U872EM1m3cFw3/e8dFmiUniNl56McpaqQ//91GJcHiQsmbGifq1x7l8ShNxNq+gTHciI9xXf4cU2p2TDgnfp8nqq5Eps7m6r5fdN0fCElhPu0Tvn445/5sTNTo2lxvtu/94N/7uxf1M/owxo/9NmfOoORPwjwUebCfO7+L535XnMvnqCoGZMyME0hpBZzyLuBJc/h+rjeJVswZUOb5+4z1syxqrJxQXpqquTynfxpZIFi3fVKgr2eZLt7emYywTlRO/YZknskc/bikvjWc/d9tIx7vKAjEhJ5u2KjmwyvAgvynTQ+YKTtTdzS6T7v9Qa3ivnM3YfDnx/fs+1aPc83K6HXA+ZyLvSJ19mxT99l6VyCtErDNnWCAK5SsuSwMEc9TJKYgO3DQ4opkhJe3DANU9rYsgqGnBjFBmo+Fu7xdnxERLmKazay45Yc0zJ6j8yk4TxeJX7IuNYtZM4Kc54/CUecx4s8L/w6eHfCh8Hc2nxOZog6DjuOFVUINNzjSlpmNLysR+xCxyOe5s4QjDHbuGaSX3dY6blseEVPcSiuC9BMciHrEFrvRfEqx9Yh0xw+zPFNIaY3v/wDrx/9iV87u9gKT/YDK3oW1Hxf+Da+oO8QNAXNHOKCy+TDg1tf2KPeLPHo7ynx/fn3qYLYotOnQNH9PcoL77wJTcG8G/H4ZM2KWSnQv54E5U4GY8Z7PXmAMZu9aeHzc3cIVsn3KA0KbzJunMqUTcGV8Pp9ZDTRc4m2iFoLGXOuDpghj0hgF3e53VgRUtJkn/INGGCe/EMiSpOq/349RIQ2thl/CtauvR1O6ehZ6TpfJ1UlJMypH6vf20ky9nO1iX1aWBcyu/H6fdD4ifu/cObQkIgRXr0a7KQ6r3KWD+2UScaENlIjiWDucLE9HYIwLXTLF8xz9fmEBQjMmXAkc+p0rT3A8fb0nAnIcSYvzzAZSQ+2WvYZTqIiuXJeJuQBIxHf1iUr2bFMhoIbOnbS8Wo8YSMdX5bHOG51yIt28rlhoMY4Jr7ZqpiK1i6pPr0ra17VJe/IhtPYMOyUW/OaN9cW6Fci7Ht4eCXsY+QiLe63dM6Emo3suWKL82EcRvPF6ilrdlm2cKYNV9LmRLDG9Po3GGRrRczSlV4F/WL1lJeHORMJ7NXkZDdDRLAuxaBKh8n+kl7riFzKni+GR3w6fox7OmNDz5RAj7JXo6Hf1il3ZYIoPA5G6p8lZbGVGFHejQuX2hgZUozL4p4TEU3B9Z5TFrkbNBRrl71nTLYXifQacd4IPGXFq3qLjbRMtWYn1sEtZaE9SPfOB4CDp3xsDuzTbPia59K3/hq4aph1Zbz716QCkb/XeEsOOzs0ch0DO82vmf9JWns5JJ37c+7z3SGQnrztUqJ00zHbs9Pn6+oJykCCvsmQi1Nu9uXj+mut7lnKnG2ChwSp+GOf+dEz4+zYkc1lxk73TGX6L6WnyJ/+7OfOXAbcIalB7J7/8P2fOfv5N/7CRyoR+dz9XzqDce/3xL0VhxWF3KWDEV7o3Tx/zeGRPq53ABVTslvQsKOnE+s6C8KlWlXeYX5dgs629Kx1a0Eeoyy+otRS5UCzkyHP+R37VESomCUEgicTYHPSkgkoPSqck9rSHlTc/WfGdbRnxFEIVixL3ZaCS+ukdu+0ZPl8Ri5n6RjeJo+KkPbAvXZMZUKTnoE6JSLl8QxYh8H9w/oEcLZ4pM7HcTfc4z09Z0rNpUZUhJ3uCLK05zrBofw+vhxup/1r5Lu9HO6y1h211Fla3Y99T8fjaHLM3gkZVVft/jyLl3x/9e1csqIk1Ns5j+aQ5dzx79lKi+gYh/mYy5QuxYHHccJeBmba5PszT4peIcVtPX22pihHqy3/bHiTV8KnEcwjqUviTaPSYDI/Fh2L1JmP6xzHD9835JumjrX8Wz/w+rM/8vfOjqvANFVuX4qvspCG/5XP3xhIjg/qCF3xwHSSCNUOA7D3H6qf+L/LDd117R2iVXIzrLsyEpUdC+hVhhI+BRwkBNFhVTlztQfFjiERX691PZ47XwnPQapKc8brn7+pC6Ia2egmt2NhDEBElDlTukRM9RYqWCJxGa+YBZv0hqFscEKZJ2F23S2QcaxiRLM6RPngigh1Irw1iQPi5PjzeMHtcJrvUYVhMUv300Uy8+sZbEEuiL5HssBx21/v8CDKyast/Vhp0JEj44uh/8w3inmqNG9puS3H7LVLCcmMOxzRaMVdOWbNjlu6JAC1BDN0k8iKXep0WODW0XOsc/bS52rbnp6nrAp50X26X9Nc7fXKR2kO58fbyTByfsQ5JxZQ/998hUWCEuzTxpfnWvo9K9nzarzFu+GCPQZ/qQi8qMds6KglsNGBF3XOY1ru6ZSLLbytW37f7JjTGbxzqcwqmAThk9WUT+gUVRhUeTh0/Hr1VXqGA6K0q0I94oIJDY/kIp+XdUEaTnWeg9kjZmykxeEJJzrlXLapI2Fdjwqr6M4I7DRyFCquoiVaOx24UzXEQXkQVibhqSGvGeYQX7Gi55Fs+IQuiQqnTDjXlgiowIk2bOjoiFQaWCUJ4ns6zwFPFGXH6M8zo6FNBPWAOaTbvKsOuhZ7ettc0grW0ODyB49lBcBTWeVgq1zbmkS43KQ57TA/77zA6MTuZog+Bxwa5oR2n0P2HSOJHkbCvL/mwdSQqrueqJfJiBcDIiZBrZhpoxP87fdc5bWvLPj495UVTU9Alsyyyph3e4Z0xUZD0BFOW0mwY0wdP4e5mfBJR1RNn4+sdWtQ00Q+zWR2qTiRJWvdcZLkWwcd+Pc/8xfPlMh//Xf+ykcqeL9p/Mj9nz1z0u04/8c9uKPnz97/6TNFPxJ+KT95/xfP/N4HIg01d+MiJyAtA/O0Bl4fY/QwjhL6d1MXz/fTGTUzNf+qPTZnegzW575SO3XPH1unj2TOpa5pqJnKJO93jVSsdMskBcjvDA/5ePUS53rFqRzRsmeruwNYuQ9HdTgPsaenUpvZJXzcz60845DioUGH5BsyciR9z/bCpScdC5nT6j7Pj6lMc3fFE5Ct7liIcWhWuuVWOLHdLHWaIRVL44ZP1R/ngssceL8Y7vIwPuEomQ522vFQL3hV7vFVfQDAKUd8p9zlHZ6yioakceGfe+E0rwFlh3SPkeFPZMmVbnKnuCJwrlfG5UtkdU+iYDR+jijHOuOF6jbvDY/ptWMW5mjcEyUwScUKL3ZXjFLPVUrcXF2rTG793/uEXPiaPGYms2QSmdbvwg3+eoxYpwItwLtywSfibSZU7KSjTWv5iSx5Gi84Dksm2tC7Ammxpn5UOpzfVLPCV//bP/T6O3/075093Y1Y3Htxwa1wwvnwLAfr5cjEQLXNY1CD/SxkjoomOVTL+D0/HAOxESrlicigLqtbGlU9P/wB8++G9yfG589c+1nZNcl8FwnoDRhiTzIOj+GGxMy7IT6BkyfF9S5LRWUk12uyxp2MWE13UPWFa2DgKq44Dkf0sWUeljhBrCRRRwnEuGcWZvn8SunjEeJk1ce6wCFmopxGHsYnOP60rNj4Q5uvYRFkd/TclSRmQMtWW/7Gr/7EP9fD87n7v3Tmi5Om4MUr/oERhlUT2KK5iusjV3cTdvJK1zkJbuRWrqwdM2eqFrjNNXFACOwxYzxBqFWIMqVR60htJW0IKTnxpMODUtefz92u9JpLmo7qSIatrzHOwVgBlkyURmAj+1x9ejHcyde7kQkXuuZEFkSUU53xMKwyJKskdD6USz6ut1mkze/dsGYSA1/ZwtvhisW25nuZ8+JSiGrP+6QCEegjvLexJvbLesIWc2nf07FM3ic+j/06+D2YMeEomfX5Qvo98S4B4Uvhgj093xNv86Y0HEvNSgemBIIIU5F0RwO9KstQsYumpLWOA12a57d1wXnYsckGX5FTnXIebEPaMTCVwE4HFlozoeJcdpwysfdJyxQzOo2iXLLnVV2ykY4tPZOk/bRKXJAy4C8rti65O9eGh3LJhFF0olTEKvkWk+LzFRVrdkmwoDqAKUU0B/o1gWPmXLHN3+vQL09AfNy0RldFYuMbfp2esV1KXra0+b0O7RqT4j6rYLUMCYv/vOKXf6Zcuz1hV6SAYI1QLlPJqlJya0GW5u5Jx5KGbfLS8SqxK6Z5l9a7JZu4zfNuFmbUVMxkQq9D7vh45xZgn+Cm3Q1r/kdt/Oj9v3rmXdQpk1zwCOLr8WEV96MwfvL+L57BIRLils4z/NHWe8lFptGRehQvKHlEZZewhAmWEPGyW+L8uCdcFXffu+wjTMcLqk/1MuH05zzUZ0xS8W3CkiZ1RXqxYPm9+MQgSaHmQq8yX2NUlTJJWee0TsM0wX47WvbsY/HMyohw8EA4ZpSHZO6nFxszoiF1P9zDq9JRla8cE2lok++JPeOj2TMkvyFtM1zMuyQbNszVMBgdA0ey4GW5zROeZYPkSiqWMmPHnntyyoP4lCvWnMqcTeoONTIhpiLWebzihXD7uXsHcEtM2exYFnSp6GjeSA23wjKpke1zwF+52mgqSA8o36Mf4119CCTOSxIc8i5Rqcpqcant+fMkT1zGO+8Oj/hEuIeb2FYq3GLJJDS8F59k7i1YrDdC+UeDZ09AKgmc64qjMOOleJTEGGy9mzNJhenINnWDXKWskupA3fDDHt8yx/RtNGOxgPB9fJzPV8qz4eK597nU7E0jkrwkXLv52iLpmH0AxywedkUOF6/rCckHJRzXE5IPfG8R4JaJRvn6bzWuJyQjXnnI7bkyGfEqqDJ2Mfxa+DXYJLndINWB/nWvfVpUDKY1v8YRKM+5hP/4gusdjcAo04sUjsw6dqu8rWuBwdja7emZMsndDjdua1KV2PwApixTu/K3M4TAJAUi5mQ9QrMqygUk5i7CPpFvS2f7qkhdegy/O2fKXV3Y8alhWKtkVNWlinat5qY9V+9cCAu10NFcyy0odbf6KQ0bWgZM/arUtveEbc5obrRMzt5XSQq2k54hdRP9PO/pgoqKWmpWak7UrvQzl2kOAt+Up9m0qQyWW4xfcKJTNgzMMBnKz1cbPj28SEfkbVlztK2pQ0Pbw7Q213MUnrXKOkYu6Ph0vMsDdjyprrLrt6J5o/cEyGRh7f6s2HHESUroAnMqvn0+4e3diu/QUxoCv7+5Yx2XPi3WKH3yKwFz64mqXGHQshV9ltu9owumWrER81HZSs+l7Gl0DCp6Vd6VNb3EnFw+kR13dcZtnbKSjmOd0DKw1CZ9jymABYygDrBOwXmTkrmX9ISnYjDFgDDVmgvZZP+LWcHHUkbYAkWC6OvFwB6Xsyx/5hLUMCq7XbDJCZDL3Nq9PlTys+re6Kw8FL/H56UgTGlyJ8Ir6f55X7+6fLfHhMFHVMNpX68YNsW1suMbTRLL4UmPJY1dPs8pVS48OEem0yGr5/ixlmtuuZbW6b6YaMXEgkipLXDXPT39qL6V1i1XQ/qojT9z/y+f1bmUMXao9nS02qUzHQNwC8WeD+4+jOEJSDkiSqNVLrY5N2tUghvf51CdMimHMQEZ94RRjQ3I62BIz4wnmf7zmUzY6T595/iM2p5s44ma1G5IwalJxnacyDLfg1aNd/IknuNQWxhyEFtJRZ8Sh4qKXdyxl46oxvdoCzVLg2hvWSR4a0m8PgykO0QMUtjpwOBFuBRnhPycxpxM9NonnsuEXvt85drYMQ3GxDmRBYMMdNrl2AAMZr+SHXfCKY/iU76teo239QlKTDYCe+6GW3T0nOsVo+CLJu+skBS49jjoyAtTPZFLXXNXTnDzQTcBBLiIq1xQdDJ4OVy8IgsMxY7f5G3+Df2uHFN5pzMrXxERRhh5rz2IrX+rAiJfzrdHcslLesJUK1ay544ueZFjQhBWus1rDGAS5dfi4Yzk0cheOp6x5lRmTLXitXiLB8G65C/IrSQoYtdoxz6dh3Ecf+T+z5793Bt/7kPvhnzTk5CP/Td/6PXzP/x3z+qMgw+8Eo/5ON/P/8D/lt93ADviUDXLoE7KLm6ppclBYUnGzp91/kb6/6pYN683YsvOxXVlKlediAUv44MWYkWf63g8l0gUXZEPgqOV31HCs256rUyG+uKzfm4qtniU3SGvYvjYxM1hkhXIiYgvIFOZGM4T40WU1ZLDc7Suh8slG0m+zpwerzgEOPj8oJEoY1DlFaBaRkdz11z/5x1/6Y3/6HUYyYseXBmZdcidBYBaKnp11SnrQlQpqQquRiZ2hwAexqcZu9toZRrxMtCrYYgdx95LNI5HOuyN7IvgTHH33i5tNjUG34qSOi7UXLBJ5GYDY53onInDUsQImBdiKkuPxUjaz1jnzXVHx5FM6GNPrz2ttCxlwTpuEqGvpmXPZVwn7HviBuGk3oaddJzonCey5Tw5aLv/xW9Uj5hrzQs654Wm5q2NhbsvS82dBbx9qVQCx1XF3bBEFV6QI877e/QSuUrqXa/GEwLCb4aH7LHg4J4e26alS2ZqcIhbibOy7pU/0NxlWgn/dLvja/vIkVR0qsZPQbiKA8/oEOA2E3YMbBjYs+dx2NBQcVvnnMuWe7qgxwzPTnTCRnoaDWyl511Z8x16wiu6ZKOWvFgHrefNcEmjFa303NaTnGx+LVyi6AGp1YdLIc/S83Sssxz8XMgud+2ud0y8G+I8HiCrqfWp4+JBf03IHQ4Lxus072Ked1ManmKblq8/MyY5oLdndfx7hLKOiYgT1rNCDjGT3T0pmBYbdYdjuS0x+Ctv/MjBRvjn7//cmf8+wSSpJ4mAf72bnYszjNBJ8PXV3mfPu0us2hpgML1Jdocvyct7X5OIzJhQF5jsIa0N9j2j4EnLnk1MGH8dDu71R2W4xLCP8h5utcWVgAYdjYEnKZiG95fS/1aMmxIQsLXJxSwCYtyvdK9dihWsiDIO65bEtP56x9fnd/nsRUBQHsuKBdMMrdoXiXCrowGopr3rVJY81os8/wcsyfhYuMtA5MvDO0yk4TKu+P7q23kHyTCo0gS4fE3LZ079++1Pl6ryAVNvm4Qm31eHYE1klvd6uwoxdypy1zFBjOx5Iv+sLAxUiY8REGYJQg0W37SxZRmWBgOXgQfxkUGYUswgBL4aH/B75JNcyBUv6QlrsUDZPVEexifU1DRSs44bZsG+Y1Q5G3ICZGuO7VVXumbQgVaMTzmXKRe6Yi6GELDjtcLOXjuuWNNpn5Iyyb/PucM+GoIVQ9N7ynvh19HhXAEj9m9pc6xznSf8dnzInXBEm+DS9+KCZ2HHHY4O4KRzmZofChtcUdR+j6FLgsI6bmhCzdtS8R16l0YDL8Uj1rKnZ8aeJe/IUzba4lBen6MB+UgkIt+STsj3/+1/+/XfuP93z0Tgu+slg0IbIyfhhMvBJl7mRxQqUVAQtvPmN97QcnxQZ8IfoBJu9X7Jy02fy9XwawnIP291yDdKd0H/oATkt/P7S/K+G+7YZ0a3eSeJu9yu63A7ptK/hyIZ28UtGkay3x4zNZokaTl/aMvvdmyoq3rE9HoJzWqKqkSbWq9AxmfvkvFRRchVDOc9eAD29YyxohRycDRuMiOh1jdl74bY5jTkTUmQLONcYbjL74ov8ky2vKKmvLFQU48SJCUS5t2xpedUZ3ljs2tnJF0/Bk9AOjEo2lNW3GHCMkGRpokE+0AueIETWnou2aByxAUWUK91R8ueO3LCE31GQ81Wd7SyP2jv9inpmdA8ZwC5kHkOmq0jZe7dM45Thd5I+Ec6oxer/N7VBZ9qFkTgtWXNEGFSwXtXyiRAp7CshHkjXLTKZoj8gfoe73R7FlKx04GPNRNOpwJr+Ep4miFpx2pk/wvZ8XbY8weHT/CdRw3PWmVQeNqaa7o/0edDx2Waz0fUNAjLtNSt6BFgL5aA3o1zjmn4crhgSQMKV2FPiDZPXxM7p8faMpdAp4EpE/qUVL8tK7unySvgi+GcTmJKRCM76bLDeEvHlj3z1P1ZsqEx1AAAIABJREFUJtI5HKo/HekElZja5wZvelGPeShXOal4RU+5kG2+Z14V9Ep/zyjFW+fKcJVfW2IywZ4sO5RKkNyqPySTc/Bdpj04eo9ENHNIDuFRh3yQGRP+0zf+zAduemVS8sP3f+ZskyBVP/7Gn34d4Cfu/8LZmKCNSZqfWznqtG548WG8Jnb+3hXw4RyWOVOe6CWLoiv8fomF70/ulyApAf6gPenDGlEVlXF9dviZMKoDJrtTgMQfG43mPmrjXjQeAUKGVkZGMzgYIVfK2Pk+7OiPHWafU5bImEiIqQpOM3TRYWtAdj0vXzuSOU/0kgpTAh3EAnBVKwi9MzzEhF5s9f+N4YvACLOx0/E9ffQicwVPKNQ3dUgwbM0cBJHAOq75ePUybw8P7XdpdyBe4/u1SeNG+hTY2nUaBS5g7Dg6Qd2J8X1yiDep3WlOcDrteKwX3JUTHiYEgaeAJ+GIqRiP7hPyEu/wjI22TKRhE43MvpQFre65Jcd00ie4sUGIptLQaUMtNRsiQe2+XemaOiXMF7riSOZcxTW11FzGdT7nVfKma3WfoWdKzKqmHqO0jEVaIDuaN8kuwJ91vy7e9TQZ5j7HRAOMnmuZW2xcuI4Ft3XGNsUNX5EnPI5P+UT1Cg0N24SGeD7esr3buyUXemUeKHKLCvO3qhIX9Qjo5JSv8ACHzc9lylZH5cEPOxH5lsGxfvcb/3Y+yX/yh3/tLIrwaV7j/wz/LBPGbwq4x+SjMH7xnxXV+/fjb1yHa/lkOIBWvU+wn9+T3D1vktf1z3/QKE0Hy/eWicdNCUh5bNe5JY7DdKUGILVr+3ycrmA1SxK3gw4H8pKexIzJy3hubWwPXGQcimXnM8om29/W/vWg1Z3bK6pcUXAt8IXMs5mhw0/Gcx27TR09c6a5+unt7eGGe+zcj4aav/jGn3ruYfrxN/706z99/5fPHMZhxGDDCzumfZ0W6JlMcuIR03VRjQwycld6euay4B7HLLRGmXEhO+7EOZ14IjO2/p0k3jGwk56JpiRHRvUfrzwfySxVi20jdG7MhjZ1N6zS8p6cW5KkkffknIrASre4n8ijeG4LGMnsMEGw+oTdNx30OSvdMJcZsbj2ZVU4psStoeKJGFn4im2qEAcc+vM4rPlfhoE7cc7v4YRFbfLcy0Y4msK+h7qCZylufhQ7PjWZ8V31lCFCFysicNEq/1Z9h092x/wf1Xt0MlCru1lrhoMNsWFZC4/bgVYVVLnbVGwHZU7FnsgpDRt6ltTUImx0JGS7ktURNUfU3IsLWoxLsNCGBTWfDHPeGnacSsOVdLzIlK/JigHlNJlNDYzwjQ0dnViQ29LzSC65xZJTtecvyIhH92rt9efeg6JTHflbJ1gg7I7ARy7trHMukuxymQC4G25EDQ6I8UKsM9LkDdLHjIYZE1ZYB6YUPHDhjps6EFx7rYRn+TGVXQqAH3vjP/xtbXY3qTL9xBs/9PpP3//lMxc2sKRqQps8BPw1Sc8QjEWHEWI05Pt2PbnIHJDCGdmcld8/EG+ok9lal9fGj+qwxNKf8UO1RZeCJXcSRgjuh9Hdeb8OiAfJK2k50ulBoXGeknefe7Y3hFxMKmFX2wSZ8iq7dzS20tNoxS4VeuBQst3hbK9whzd5lEUYljIzj5D0Zy5T6ziHOQMDXxveNXy+js+YF+NKeVxXscoJUgpg+9glaf1DlAZp73fBGD/e0Q9tlPJXjczCkk47XJBHhGxmbO8XOt0fFBGdtG6KS5FeSRxdZS6zA0XRddzw3eFVvoiJ4Owxf4qdtpzHC9rQ8fv02/l1Pk8bW2ZhTiUVizCnpiJKzbvxETU1t8Qcwd/SR/m+35ZjzodnHIejtEeaH0ubkpKVbpOb+wxX07K1sTYuTox5hvj99K5T5gml+GmqFUfVgnbY4+BTH24b0GmXYO6maOW8nfKZKYszXxje4uVwAopxCYG3BiPhb7XlVG7becj2oAOCRBppaJI3i6u9ruKGh2HFixxxrBMWSXHLeVATmjQ3N6mrVCfYtcHT/tRnXz/75V89+1ASkW9ZElKO7/3bP/A6wP/7mf/u7LvDa2xp+UL/5s2BfnrQdnGs+uWgNeEWS9Wqm0ZJFj9ISjyo5hAGdT3RuRtu8yxe5jZ9LU2aAM93bfLninO4CVJ1/ftvev3g/29QxnIPkpCqE8rotK5qmt3+eX8AutTV8PZovOEYvPPTxpZJMjYqcd2qo1He3rHXYkFJXRyLD38YPUjydmbAcNPOEyml6BRlrbtMVj1OkrgeGPhwpRQ/v5++/8tnNyUi11/73P1fOtsk/Ly3PgciO90/5w8SUVCjrlZi3YN/TT/JFTtUMBKsCo/Dhpk2RFHqpAl/JS3HauTBp2JSxxNqVongOBBzAAlktaAde9a6y/4RF3p10CJ2zHm5sAH52o4JqoVdVeGU652qjW5xGM9euzwHtrrLWFoj8Rl8raXnlDnuyF0nMzyHCl1Ky6raM/TKq53R1ycifLKq2fUwdNYdaSN813TG6QyGaP9te2E3KJ0q+6i8Omn4/ftX+PXqLW4zT1we+EQ84V5ouNwrs0pYVoFd31MhPNz3RRBsNc9jGpwb0hK5kJZXdckr8YiN9HznZE4f4YtDxz2ds8nzOvKVIaltaeA75YiNDsyoUbEOyU4jX5M1qAs4KHUM5rAO3MKEHqZa00rPLZ3jZoMeBDkv4vozb4GIHlR0PTHp0Vz1ndJwRxc8ljUdPbPkrzGh5rYueCyrAxEJsKTXoIiVqfakDkFINc5ZgrvZcxCem2NjUjEGdFz7mfuRNBh3qISWfSOGc6Z8XLFNx38oPDF+Z2SdYHF+LTxEFUJKMpK5rA50bM0rAw4q3c91hVKAUCeVrI+CetT7jUZGkrEHvFXay5rUrfPul0M/l0xZMmWHyak6mf1zb/zZb8l5Xk9+IfEqkgT5gLJNvDqH7HjC7X974jFW9EdhhWey5pYuKSF5EeUJV9ySJU8wYvmWNnNmtPizp+NI5hwz56v6gJ12xFSIK5MMg04n8ZaULOxTEjAUhVSHThlk0PivXmwCmMjkua6UxwKaEAUVFbfCMU/is7zXDxpHCJdUeS/wpKJNHAuHQHrXAzhQsLz+vX6OrbYWc4Q5ne6ZBSu+HIcjHg9PWIRFWgPG0UvMMGUlchyO2MQtO3Y5QZhIw2XqJjlXcU/Ho3ie7n+k0z3LcIeGikus62GcR0siPJ6ZhwVHzHkvPkmeHAOTsMzoDfdSuYqrnIxfxRU1gVe5y7v6MHePPGHxPdhsJKy0ZJ5sRSHvGjfXr/O5bHmJJXMqNvSJFG+CHkc6IYgkhcSkMpjI6b30Geplc8LOZUvLQ4FKTngh7RUr6TjVGVNepMV4cC09g0REba2rJHBXTvjR+3/17C+/8R9/y9evb6hj+tczPhlv873x5Rw4Z1JckhTLfhkeLEsY34M8lzDk96T/fAErk4zrRB//XdchWiLWZvx+Pk4jjQWgBaQIxm7C9d91/fXfCob1XOfj2h9/r4+DzkRR0XPCmcOqGqxC4pO2oWYmExYyT9jHD+4e2eJiWf14D6xqkFuE4ucw8k588YOxauWKGO7C7gQ3w6YeVlk9OI4op2KOqacsOeZmYroT+UrNdx8/df+vP1dNc9M4x7RvknLU9W4AmFxeJeZ4eypHfB8fN7dtndGnIBVgljbGSkN63XD3OxlVQ2YYyc5lV0vJ05hedwKxojyLl6x0MypjpD9u+uj3y5O78TqPPy/nq8v7uZOqf/dMppyE5YFfQskJMGO8Oge0fp03tEwTJtcDw0qFSzomIry6qBkUlhO4PTey+nEj3JrDVQuPNspqrzTBXl9UgVkI7KLy8qTm3xw+xqf1Di/FIxTli9VTbjUVF4NJYw46HmclhuefScWx1PkOCnBBx5TAK7qkJTKjotHAe/uOL/dbW/TT+ys1+Na5tFzJnoeyoxLhXdnyHXLEa7pkl56/V3XByzrnNV0y1YoZNSc6TYGSbfC9xDw3PFCaYf/vna6esWrv7xGEntHhGVICIuP6dcWWjoFTnbFIHROHSPXFHC5lfP17PNhcpU3fh0lFdwcBoMOYvHtTdhjKOVYqD7l6lv/7JpWdr3eUhHYvQtxUrS8N1sAD0DEo8GBnRnMQEDZiTs+lEg3ATWaH/uejnID4qDF+3l67rIJVUaU1OeSCxKAxd4oNimkyET7cp+ObPX7ijR96vQzoAsJCp1RI/m8sbhW8zvSawRIjTp/2URYC3C8oomylT94+PU+4ZK995iS5r5NiSooR5QLrPj/mIj2nFsN4vFB+3yLMmcs0owYqGQN9n1+5c4GpWjn0yZUtg1R5/4RCPTPN0177zPdcJsNEN7jz909lmp3OvaDoxSp3Xc+w3cy7GJ+hIf3OhjF+iCi1jJ4TDrN/Re7kn5fP/wtySq3GITE/ETMEbeTQt6JjsAJevDDIpfa00dYn71QAXOqax3rBMiEtNrodXcZTQrjVlkf6DIdjxhRfxqQUZn5qNU5M93vyD6u3ecRlvh5wmJhZb6SceyNsTQiFkMi43wqBN/UhV7LPe1FNzTIsuJPUQButeKarjHAph6pmqN4u7ljKnCvdsmPP07AZi5JqyfaCSY5HApLlx12m/Bgze/wL93/+W/Jcl+NDTUL+6N/5d18/Spvz/Jrq0XUlqutdipvgU6FIJPzB9MC87BBcD+rzNnItoQmIacBjyheNTKw6UXRB/L1mzmfVi8zVkLHyko/hfZKLm47p4HrcsIgeQsrsfEuJ07ogFzdJ+cEfml77zPm4aZTHbtfz8H0jwVyyf8ugHkqPak5lhd6rB1vdHSy2UmyEPrJiiR4qmcBhi95x4mXn5Wfv/80zsK6Ib5Y/df+vn33u/i+d/eT9Xzz7yfu/eFZqhpcBjV9bV4mxFrJViRxLabh5IycPRHYyLhABM6Fay54ZNYvEo7GKcGvQq+RL4MHiVlta3dOmLsylrtlpl7XcHRPq8Ldx2z18fA+S4+J9Vt21RXNCk6+zw1MGtap4W1TKwBTLXJnLNemHdFdHBSt77xVbBiIzremSGtStpqKP5qT+ZKs8XivnO+WqU65aaAdlEyPv7Dsu98qzfaQKMEmnNSh8YjLh5WnNH5jfQhBmWvNo3/Oubnln3zEJcFpZ1+O0qvjkZGqmhGpNc0e+HuEqazH5CFQc0/CMjhXmoN4Vc2xNzzIpYK1kzy5GvoujfN1W9MZlYeCCjg0Dpzrlrk55Rec0Grilc16Jx3lO1RqsU1ZsyB605wSIQCsDG+kYZIRrdWKBlJNwoxic7yU9yfM3YqIIAEudsZMuz+8Kcx6eM2Ge2UWCt+ZLEzKfPw4rc/J2+fP362pM0uZevt+/5xtZZfuZN344B6fX15rytbL66MmdB6slWd4JoRNpMjciy2bLuDYPGg8SKsXw+FFvvh4fpeEwn0pCTkD69GeWeCCdWtAXkinqjIYFE2pCvh5eJPqJ+7/wLUtEPPkYE5Bw8F/Qwz2ivEd1sVKW8N9OIneKLsiV7HjGmnOu6HVgp5ayP9bLTHAGcgJSp65zl/aIioqFTHF4TkUwk1sOi0ye+LopoAe7vm9eRxIokRflVvqMSe+ehOM8p3P8IWMHY0LDRbzMx+rXZiKNdcBSEdCDfhOBqalTguMu7E5gBg5QKh5HlMMRDVOZJqlZeKZrKqnyPtElH5OP620uxSR8F2HOQubJh2dMyjxOGb1VGna6y9KztdRFUG/H1WMeQCY/LDnO8Pc5Dzam+eGoh4XMM9/jKCzHYgQ1x5g/lSRFMF/byv23lNKNmhAqWCLj17YclRjvdab2+j1mZvRIzcf1NhNq3pGngKmaltD28nt97vi92NLygIt8zxqCGdpi+8lDvUhzys7B4yBX4jzledWwb/b4UOBY5fh3/sf7r//KZ/+rs81geN4yCC8r0jf9u0wsYDQXPPDqeB/OSNlxeO41CZmD8Cg+papCbhvu4rZQl0gkbYyAVEl6aF1rm5hfh1Gxq+y4lJyW8iG/nozcdF2yu7ovtkUQ7wtZp/tMthqwh3OrO5xY5nC2clwn7u8T2U0wTGX5MPQJpjAk2d0hwcD8IVRiPu+2UGBxqJgvTi6B66ozYAHZPlVCvArn5/Gf3P9rZ2PFeMSiDilA/un7v/y+G6QtIhYoOzF3W1zr6/KOM5lwKkfc4Shtx2ZmlxW/MCWRnXQc6SRvfi1jS96rbhcJX1wTMjE2K+xox7PhEpFxgXPDKPSwnVt2wbzJ7UlKuTnkxUgaolo1yN14VTVJLRrcqkmV32lamHwOucmdB7Qe9La5SupBp6nIHOuMJ7JjtXfZZmFOxV2ZsNPIhoE7fcM8BJ5Fg4HVAd7c75mEqfFJEu9vOyjtoFQi/EFeAWClAw/Cmivds+hvMRVTOzpqhHd3PVd03JIJVwlitke5KxPmIdBrzVtxxyphwS2RClzS8VA2/F65wz/WCy6k5eW4zInVjoFAxUoHjqTiXdYMqglza+NFZmwYqBB+F6dcas8eU//qU0JRVuJvGgEhqK1LTZrXzl8pVYD8Hi/VAqN5Cv6XOqGSwJVs+VR8gbfDJR0Dt3XOTE+YJ17JU9myTb/3ni65kB21hgwTdL6FH/MRMxbacJlc7MsOiaIZg186tS+YZajhNwO+M9Z3D8ch72PkxwAZ3ggmf1nyN0oex6ARktT2kB6nidT0RRfdq+lN0Z3/l2XIwRoirNiy0z0zmSAqeQ0IGJ681kAU5RlrDIb0rSGq/2ef/VtnUZSlzg4SiXJ9ds4dHCbHHSOEeFIU54Z0zjO1tfwqyZy6MIoXXhxyPJcF2ySH21DzopwmeJoFsidicLULXeWKvlevPVB2yI+PvJbfoDA5JLQAjGT1Lw5vGSydHTvdcRpOKCG3pcjLJ8JLPIhP0+86/N1e7PPg27saqsZZmyR4XqstU5kaRFph0D0SJHEKD8UcfFhHYc93VK/yZnxAxLy1pjLFTRP3us88jv99+HxWOXXI1JDg2d7pHxiMfB4MQTCTGYqa90UqLAWEXdrXIiUhXEHS3pvinFrqJKs9FkE1XW/FSNuvcpffYI2qcre6xb8+vMzfr76EG8BWVAeJo8c4JZnfExWHa6mOvKqJWCFwQsMrOkeBKwzqN5UJU61ZFUIHU5nk6weHBWAfxlNdEhCWjBypKRVbSXYDGjiRhRkQi3VefH5cseWUBcd6sz3DN3N86HAsIFfvrgfa3l34oI6Bvx84SA7K99xEWv8gHocF4SPUyxUV4rUkx4+n7FIMDHRqTqG5fVngQ8tOTXn81///+rFe74KAy+WVLd8xIYFxQW61zUpDPnHnMj2Alh1cB+8YFYmIt4AH7Np4ouXVEmBs8WKmR9cr9X7Mjot1Toob9JTHMOiQH7aLuOJCV89dAz+/nIQxOqb6+ft7SkfoniHDROAQ2tFfqwg78exxfMYX9V0eyEV+UMtgMDJ2b/zvvfRspMuBu0Ebury5CcJabfPf6Z46tYFHwv4IZysXOIO/3Qxtyepx6ey9A1aaSJbB1jRp3O+0xTHOG93m3y9FBdQrhj72ifTuxHUwI7kL2fHV8JSnYcuXqnM20tOjvKM7vixX+Xc8Hjq2ntAM9szvo0Gsng12JyvB+BwxsooDD2LLr1dv8VJc8qoueXlWsYmRZRV40tqSeioNp5UR3QPCLZlwp6mYVsKdaZIhxTgir8iMF5lZhyR1tL6LYxbacCFtDnjeljVfYsVD2fJPuGBCxXthzZXsuZCWx2KQpimBDQMPtWVNz9vhKj93ZfLhc2yb5sI0cTNiCpgawtglKaq8Xjmbqxlj7lLHJGJGkwEjrN/WBT3m4j5jwkwbplRcpnNqpedIzQum0YpX4wnulO3qcK6aNWPCsU7zeSyZ5blQGgleX6dKw8Nvxhg9hQ7hrOP6n7qAuMTs9T1gdJ72EZBUSKno9HAt6LUsKsTcJ6wIB8nJR31cDyBN4tX2h53u8zVcMGHGhB17Wum5YMOcad73yj34J+//4lnZef5GDUGo1CBT3vUoYaLlKI0rSw6jP0tedCqPu2XgKSvcN2ml2+SXMd7PbeYAmEFlS8+T1B250jVrdrzAycGxeEHUu8d2Lr/1flgmWMblGyFbzsGIKGvdsAzm/dTIJDt3T2XCHT1iqy1NcmgvP29FGYPblX4XYN4ZnriXBPe8byaFL4cT+3BYlqNOSh7OVncHHQ2RwO/nd/EP9UsAuWviHYoKO9+pTHKhMiRopM9LD+SdV1p2QhYJUeMJmq9Zg9oebDHL2EFzjulGt2zi1u61Bl6s7lBLzRFz1ozcWZtbRgj36+HfV0LKvat10ziSBTU1kwIy/Ei2zMS6jkGF9+RZWn/GmKKcQ9OEygGLu6ZMWDJlnp5Z34uO0nXrJHIhGwQrKpSiHA5L37LnSlr+88/+l99SSNaHmoT8/R/8n87+9g/+92fXgxsfY5A5/jy3HQueSH5PgmCVyUfpQFm+5v+WG34mCSfn2fNKN0k14f2rP2Vlyc/nOqSsPMbytRKudXA+xcN1UyJ2/TqVQbnDswYdzG0+HVUtdaqEt6bYcY00f5Nviv08YcqTu2j290h/nBDtvJBW24OF3CFXjZjhl1eIvOpUJgwOG6sSkdK1yt3tuazulJ8xR9TxZzUhIZlDeihTq5lRknSfEoMqXf8Ga2VPaJgyyRtZJRUvye0DzLtDWvZiRK+pGv9D0bwQb2hZYwRzq5RZIPNEDW/cacdVXLPRLZdJZtArW6ZaIs+1clU1V2M8OSwDKr8HhivuEnEtZoPKsjq805Gsa2aJdW7r///UvWmsbVt21/cbc621+9Pe/nX1mir7uSuDjUwnEEqUSEQoYLseIhJJSIMU5Usg4YNFjC5EwSISsXGiAAFhEkhEEhsHRUrqQyRihSREBMpgG5erylX1uvveu/eee/rdrL32miMfxhxzrb3vfY2bAtd8ejr3nLPP3quZa87R/BufS3NWXLFk09vAHYqldPAUP99TuUYIPJEFp1xzLWtOZMki4a0/kAU/zzkn1JQpIThpGyqEy7blcm338EmzSdUku8eVBN6TBUc6ZkbFiILVRtkvC1Zt5NbIzmsvFHk+vToYUgDrCB+sG5pW+dbBmMPUQxuIMA2BV8KEIx2zVwQjx9OpJ3lA09AmKJclAEO1vt5EK15hRkC4TnO0IfIozPNaEFGCSk62fd6OUzXKN8U+jMn/PcIML4MKz8c9ggq34pRbcWobaaro+oZ8kDxHGiJjSl6MB7lSXKqRxZ+P+4yomOmAx+E6dwQPdMSeDpkmFsBNnbGvw61nzlv53nn051hRVtoRuP133ygS865yll/XPhysn5T3g4J+4gFdQGud7ORVJC7bWeTzraRI33f/fbMM70L3ITbglffOK8INYiPKFUtO9JIP9CwHniM60YoffuNH7//ZN/6r+/3O7A+/8aO/LkHMj3/ur94fUqSj6ThHXhQB96mJufjkc9LuP3gZ039u59vBaB/KBS1G6m20TRDUNgk2DAgE7shRni9TGfFQzzI0K2Ld5a/rB/n7JgXbJWZe6YU5H7twK2AL/lTJYCth2egmCwf4/Ws1chXnzMKEgDAJY4Yy4JVwl8sk3e2FIeeIVJQsdLklaGLXo2UUxtlcz4VkHF7tkK9Sygzf6nMUPPgOCDOZsKcjS4IIHIUDwJ6/w7DPQdhjphZ/SNq/fP3w6+AxiO+v7msW0hrp5+VQM9sL7ZlsUwHZYaZNrxjso0rGzB5f+bUHONdLzmTODfb5TnmZioJjBlt+Oc7pcV5sJ3PtfNzu2laUW/t3H3J+m8OtGHeK8foEYcwgm5+6WaMfo69LM5lwIHvZeNHjjrGWjFMhq8H4bkfRQLjmDVUmjpNdX1cgXbNJgPHIX/zcT9z/y5/76/9MkpF/LnCsP/T9f+z+XpjxB/neFDzGTuWKDm6kqTrTDwQdfpI9LjRu+R/0h6s19JW1PMCzIKokhMJ8MTDolr/LNQv2wozL9hLVuFVhz+/PNnwsS2NmR3dNROD2Q/+mf6798cxOTz9hQrbet+19zZ0SKfKDuNY1ozDOixjSOajvnhPP8FGBtMBno8iuupMTkqTOEbXNcLZ+MODH4g+pJQZukNX2fE9cQs5M86oEWRqmAH2QAjehU9rqk+L7poad220X1PmxlRS5KrBmQ5MSiKikhEZzgLjRhpf1Rm7rg5HbWzoIV5vMFksNWap1Tp2TNJdcXOmaZdJnd5jZoEdUBiO+RW2ppMoE/X5wWuzcn0ICm9RubnQDuMKKVXpcQ76bP3ZF+sMX7HHPgCqmBRYMojNhSMQM8oYJbW0bQZcclhQsqSkIphojT2glcohp+l/IipqGu7rPhRZMqLipI4YpETxtbbE/Y800dtvlnUHJqDni/+IhT1jxBDhdV9yVIcMQaBVG0pHUXysnVIWgDXxxc8074YKD9jnmseW5QcVdKk6aDWMJzGPk9bCHiCUxkhKGSktqcWihKcBZ8iHc1DFHUoHYc+MQLz/im3HCo9CZRm4SV6bfkaT3TPs99jnQYEnKig0TrCP31eKM4zjJScdBHNKKIgozKpMcFeEgufHejBMuZc3dOOVS1kzSxj3RimMdcSFrvjUecZE6g42Y9n6ZkptrWbOUDWMtuZI6HddmK6ib68qgHFQEkVSFterlhsgPv/Gj979RiYhfM9/Mt7uT2xyR/uhz0owbEbd+3mrkUKbMxaCrS+ycomqCrZSsUyD2LBnh34jD5pxViG0t2lFZSnNxIGVKQQOP9JzDJAyypuGr8QEtLS+FuzlMXibmlafCAH/qjR+7/3F+MB82/uLnfuJ+n+fkz48Xk5xI7Z2PfvfDvqbgEoPrWvEgUmIGq4LJis9ZWQU48fz6e/MLchOAx1zyrj5mLMNUoCo50QuucT5kZCDDDG8aULHASNE19TMTDusSdAmyDw/+h76nJUEYL975sdoaYfduEZc0SUHpXrhFRPlS+1Y2m91cstUIAAAgAElEQVRok/fVrmNpa/owDLkVjjiJF3xWXuErvMc8LnKF3eBTLYdhn8to6oxBrGjUcRWN6F5T07Dhd8l38H5K7K5lze+L38nf0L/H7eIm/0r8Ni5p+D/DV2hjizNO3HOjv7/l/TuZElqXrs5KUHnfTDwRIHF0kgJZ8hYqxaRo+6N/n/vEe7Dn/rGecyz7TNjnd7cv5bnnSUD2FnFYmhh/MSA54RNCdmDf6Cb/eyLjfK7nzG2PEeFJWHBH9y0WSR35rhMSGYm5TQlCzTqffz8uWNFwqNM8x8CETILa7491wiJxQwoNPJAnjFME1hcYcQTBP6vxz7SM86/9wH94/w99/x+7PwkTPisv877MWciG07CgyBNItjkT6b+QDPf2wjRV9rc3le7B2f75aifQ9rbnQCruFDfZF7tpu/As1cg8zp8ZjPvY7VL0ITQ+clekd1y73z/rffvwgmclJP61zwMppKBM8CEgYTGNfDYK4xyQO9HLsvltNQp4ugvS1XJdJrB65iTdaJNJ6l6N8OOwgDjdo6Ts4BUWh2T5MXmwXknFTMZMZMR+okf6de5fk34XZfeadRU6yW3HCpcmNVUIT5I8seqTTqGrmp6FJWvM62MpRkx3OdYqVcUDwiYlI47PHJDIgElKcUNr2Gs6aUS/7v0FAQzr6S3ffqDlx5q7Ruk/h8h5stnomrU2NIk46clLQcjdH78+ucpFn2jf0eCB3I0aUrJinTthBstaZ46Af4arLI0ZcM6cBTVXLBlS8YFccp2MHGdSsBcKJiFwb1Blx/NSbJFeamTRRg4HVsmZS5MDzlKEaSGcryPTMlCIUAi0qnyw2vBIa36xeMiEgfE4VPlgveG8aTnRNe/GFV8Ij9gvA3Vrbuuf0X02RIbp3q/Y0GD8jlfDhBklB1SM0jW80AZz1g2saDkLq+zIvnUdVXJXw2FWT+O2E9k2Xe9RqgX7657TiRki9jaiRiLGvSmpNFDTciPa3J5oSZuSJ4Ch2jpwkiqmAwJ7VLyoM+7GKa/pPjfjhBfVgs9JIuj7Zu1VtP6cHKQkyRORtW5y1fqjODC/1rEbyPU3Zcdvw3aAE3vPtXeIu2KJFR32ZYJT9xs2DJKqnM9t7/jEZxSnfqOPfhC7/XO7T3NdZc7Dvkw50ytWrDmLV4xSB8CD+/571myYs8pr359648d+zZXUZykI9rtw0ElO94d3T9wn4aEYIbfGEA1nek3A4FdjGeZzrygt4VATEvEAeam1/Z84RXUye6ukMohWWsfdm0PzHvL0/PBuiydsuXjWg0evtfP48HO91sUW8sDO3eR+Ad5rHzHTEU5i9iQByK9xiFNH0FbuhGOWYh156AfZIUMzvSvvxHp/jYnhbCvknXENwM/q1wjAUXHI9+inaFCupWGQUBCTYHGcq3GJ2H7YJKWqiOb4zQn+jW5MpTLxSgZUTMPUJGp1lZNBf6q9Y9MvUu9yKsYyMt5Tj78D8DUemQQ7HTTOodJOwvd75Hs7dOtKnovp2PvKnUMZcMCEDcpIAkc6JmLCIs6NdXi0D48lna9l18UFkeyZvpBOGcsV4fa0I5/PGCQfrKQGSMzdEed8jr/Ba/bueKoTsvr3/rf7AFdLV0SA/bEiH5MYPboSosK67YLBJgVP8zZypRtmMqFmzW+WVznQEe8EY/GvMA6F80AA2CFMByQpGBS8354/Rai2isSAdWqzPYtwDR1s6qVwl5s64wv6y1tGPp4EOcbxk7re+vvuVtw+7u+3uBg8Lde7+9pd3LVrf/sw9RMLRpdxwTSYV0FH+k4PjDqM4ukOzLMc5fvtYTcXkvSQu9QfpNasqkn4yYBa66yL7g9/5yisnYEk1kkaYhLCDZssOerDFw2HjrgrxLNeY+f2NNHcuyIByYo4VjXr2qYNy/xz1ZYottD+sr7Ht8jzuSrtxwFGIl46SVBNljWizBixoU2dijYfi6mpBDRJKje0qDbZud0rEn69NG8aId/DXSKcV6f8flUpIXVPgGmY5DlUJ/lHH2MZsVSrDB7IbGujc/LuhCF9aWG/zl2HybCo/aChZpPx2GBQjmWCqD2nh4y04raOGQUn4iuqwkjM+fVBW3MzDBj05LZvxDHnUtOKUqipcEVVCjFuSQSaFq7alrdlzleLEwoKXo77PGJNi/KlcMpvibd5IktOZcmEisuNOQcHEfZCwRU17sexlIaJGjfiUlsqAhc0FDrgQhsmmMxugyUDd+OU98K1JRrJlLL/735CaX9jHJuMYabb3GtaZhRGeteAS++eJ87FUAv26XwTZlTJRNM4J0MKxpbSsdbInIYZFWeyYhIrTqhpUY4YcFeGdmcV/mHxAVPtNrwDHfNE5gypGFJt8ar8ufLnpp/g/pmf/GPf0E7B7prbBaruu9Bt4LvYak8yNK0knnj0ZZGz905vTfSg9ZsJjuXDxCueTkLM7K3jUqxomDFkSc21GlpgQ8tYDDayTj3a3U5U1+375ONHP/dX7leErX3KKrLbCaSPfgLin+fJpM87X0f9+1psDXvEeX7GAiF3QkYyYIpBGa9kyRpTcnRo1ZqGeVzmubVW863aeCFGjbjt3Y/+Feh36/2rd0TAeG9FT5AkF6boYNBeRPK9Wz3mSHvDJIyTN8eUqzhnEsamrJSgVJ44lMnoDuBRe8pni9f4ueTa7ntNq7bH3A43eL993DNUDL39tCCKMpUR6zA1KXsNnOkVrsYH8Pvjd7Gg5UvFEwoNPGpPEczVfSJjSkpWrAw+TucX1i905i5OKlo+ied5f1rEZQctV4PRuuLfGvNi8Q6TDyei+3yxvbJNscqAeequ+BhahGD31++TxBTnGFqhK2K2GW3j99rnBVqzF6Z5HfpKOOe3cpO9OMgwULCuhrmeu+mgQSZNqbN7TmyttbnfLxhWCHU6t30qrhNvs6FlqCVnYcmIKiMWRr3ExsVlhk+nB9+QUZ7+W5+/XxZQNzAbfXh13hUIU4z5sUlJfxQiVBr4vfpZ/lf5J+yptYC+2L6ZFzAPjnyyGTa3I9IKIQVJXTur3foM09Jetz3ISZ9EvkO2njGikcjzcpuH8RRFWaag1GFhuwaBHze89alsdzI+6XhWQrL78z40a4szIp2k7WbnM/sLdF91omU7SXKFLvub7c9TInWsc8JWa804ObEDPRWNPlks4mpheZMSr9IH1tJVLXysaUANe2vSots40A0xVdtsEziSWV6slEiDcT/6WMv+RhR2Pm+UXuubkndMIM07sXboUIZGXgsmWHqkY8Aq2huJ1D3vhijdfPbrM2LACRe5E2eOtG2Po2Nzz1UwXOXKkzXfvHwT8uPrq2H54ucLqUuz+jCcbpETFrseVtsZJdyt411X2jBIrvcemK1YZ/haRcEVXZdxt3LiUDS/f51fRMcraYnc0jFvhgtu6pBHcW0kUqmIwL6UpjClkcOipEiT8hZDJlpwzYZ9Kt6t1zw3qChEuGpbQuqeXNHwdjhnRcMr8YBrTIT3XGouZMHPhyfcjTO+lQMLvkKCNarSkMzdkgxzRcHzOmVGSU1MdSS46HUV17TZg+TtcGmBbEo6igTR61fdHXzqwzt9u+TZIZ3B1M044ZEseV6nXLCmEbv7tcaUrFgy4pXgInFfFOVEUpKpA57IypTcRLmpA4YE6jRX16kA4yR4nycgGRJjSekgk89N4rfMz9yQAU5OdSlXRX9dE5I/8cafS+/bdQV9tvqe0X/W+7w9B+1Al2Q7VGVIyTXLnHwYDDVuSfXa50X+ebkM/2pGB/frKsG5apq4amVaH3xt2WDysA0bVKzAcMwsdRQ63pSvd91a/HRB7pMMOyaPA7YLT/01JubXkz/P1bAC5CLVSjYEFUZUvK2PuSn7DKiSMqFQUrBizW05ZIrxoR7KJQtWzNU8eEgyuoJwL9zgMnEnLnVunEI3HaTrvBks2uGpJaae5OT4TuXRTXp9j+yLkbhKlr+/r9mTMMwF0z6caBGX1GHDkeyxlJpVXGU+haMiJox7e2JgEoa5Q+p7i8O/DsIsd3X82Nc0GSbmCdKIAb+T1zmROQ/lirqts/rThJI35Yovh0d8vX2Q9zvfG03hs02JUUx7W48Hka5LNlbWNglmWEKgGEk9E/t7/C0h4C7jdt3tGR/KIH1WS5W4UOavsk4JhnAoJt18woJPpY6wF8IVpdENiuAy9qT7nUUAcMXOkOeA0vm4TRgw1QFDLWhQvrPY5x/GU/ZT0WcopcU8YgnyIEGfHZIF3jEvcjIxY8yBjhlQsqDNz0YEZlplKJajN3xfn7NijzFDteMKiHXaY8Ff+8G/cX+qFSvZ8Ef+9r/+DVnryqIXO1yvns4sgkDTCss1VAWMB8q8FsYDxf92vZH82iLh46Bb5CYhMAmBYy25jt/GE1lypCNuFUecxStiwvw5NyKItURrXTOUQXapftSe0EiTzG2arSRjP+zlFtYWdyK9xpMWr7x/wBm3ObQbmdQZQnqo+zyUPlcFnuZq9L+HbcLW7ngWrKr/u49KNnbHboKyyynx0WL4Sccq2gbTXYfYqzDnKs2OmliQIitbOM+m7xa6VpMuFDHuxDAMczcEnjaHNJPCgSlwaEtwoyWx42olZvOgoQw5zJC5wCoJ3y51yZABp3rFREZMMHhT1bt3fr79zUsQLpizYcPM6F+0REaE3Nm4lmWew6qRoYypk+vzW/Ehh2GfuzrjUmoQkqtpF7B5dcXnh8EFNrlylu+rpFY0HTHOWs5GkOxXXvuVtSzfSNeatwRUaaXNbXM/lj4mtaHluXAzdTTaTDKdJ8O6WuuckNRacxwO2GPMJQug4JgZ58yJae45Rn5J3ePXNJnj4o7Zh0wTwX3FQSokvC8XXBdGivsCj5kx4AWdMSyEk7WlYy3KXjAFrFWM7MWCWSg4kJI6RtaqnLGmagLHZcFcW6LCA5mzChv2dMiJXBFF+Wo4Y0+HKAb5W6Vg7Imu+cfF+/xefQVFudSWXwgnmFmbwaFAOaNmQmGQPCyMHRF4M1xyO074IMwps3FlSYScxPQlRBsiVQrQXS7UA8SgkisA/YQFoNLATR3jLupVSoINCtNyxJATWXGczBL9Pc7oVL5GWnCWuB2VBmZUllAJDDWgGBfnLKw4kwX7OmZEmZPXW7rHicwtgNEBjbRME4fIj9XlnLNqHoEl66fWyl/rmDJKvCyrMvdhg/bc2/Dj6JPTu3APCtjSy19Q0+eDZehjUgYai3UBfiUFpt8Iw6VEt6vsCWKXOC5+fRZpv1ixZl8mmfdhxZSrXNjxAqIP+3u7bp+EE/Kn3/jx+4pyzHQLumjv1XXYXGjBj7js3WcvuhnkxR3fW2ppeUdPGMmAM70CyBLD4+RYvi8Thgy4oXu8J2e00vJEL1mkopCvu0utuRWOONc5DsHblykb2hwwr3TNRMbUqbPiw+GtLSR4WNhy2oaez0Tag1varG41lAGNNkxkxFm8oNaau8VNTuPFVvcb4Eu8z6f1Dg94ZHtLKmC12nEyR2FErTUTGXFXjniLEwKSTQ1HYcptOeSr7btZ+MQKj13BARJBWuGRnvMt3OZe3ONhMLfxViO/R76diPJ5/QJ7OmOUipa+N1b5ubS9q8Yk/P2auJ8JkBOMqC1tShRajWykM1WErhjmJny1Gn8iiGQ5YE37nGoH6bO4J3meicnSjxmwn5Qwj+OEu+FGfv939JHdr55S5bZATNcxKqVklERu/HeO8qil5Qs84bPxmJVsuKlDhhpY6Q0qDfxSeMge41wkOZUONh7E4N4tkXFSxgI4iiNqWiaUNMQkL29r40BLHoYrRloZHExaNikRURlSaGCPIagVUwt1Fa1v3AjjgaIKRfjwFy3XlmAMK6VpBVVY1EIboY1wuuheG7EuiYiZjlXSrz4ZvnjGgAi8rLd4NdzjXnEr/94XNg+SDIu54nE8ZRTGrFNiMkjGLqMwpqDgMl5x0V6wyynpu2YCDBIWf8aYFWvWbLgpBwykytg92OaG9MeuKtfWa38Fm9Luhvxhf7vbHdlKsHrfP+vnlkB1aiHQ24CkqxJOZMxQhoxluOUyLxJya7Q/3AOkv4iuEyZWJGQ8a79Cv7vY5o6XdEpGfdUwMCzlOJk/mfPtJi/2EWWZnJ0NZtFtYP0Ezs/FlbAc7rbSJmvdOyTwCoMcjBlSiSuVdOZ+fqxXOueJLHlTHrOg4evyOL9XzJ+gLNlwxTJXhJWeoAJdpdBlA62lvw2vc46ND6989StRfbWyvqmkuyB7NQxs44x0vCCA9+MTLuM8V3ocyjWWEQZVCzmxskVqzIomG7TZRtcFJT4mKS1xDLQHi03+l2Zy6FBL9nTAWIyX0aJcYDyL99s1D2PNIM2hOkn2HlYFL44rbooZFL7TGKxowYa3wiktkXflDLBK6lEccyoLHokFJK/HG/xC8QGnsmJPR7Sq/IKc8cVwyoiKFRsKD/B1zKsyw9X+D/Ckzojcj8KCUi00KzAoYCvduXrlE+ChXFjXRFrel4sc8EaUKF1bfcO2olNAeDtc5u/vyZh7jDnA5HcbIje1ExXI84CCksBMK2o6GF0jkcvUyYjqc0Z4IYx5Ls4YUrFM3JujOOIojjiIQ/bV/l/LBsc6ewexryimdGZgirKvY370c3/l/kd5+Hzc+JNv/Of3f+iNP3//T7/x4/f9mXbhBvvcNm/Y/v02FGZ7bfCqpK8BEUtQ/XyaZ/iCONG1oODf/9yf+TXzHv55jb5hJUDZW+tdZKVJPA+vUruaoHO//G+9Uh521oCPGzNGSeWIvM73oX1gMUUfouWj3/Eve3M+AkvZ8LY+5lCmZK5Sut8rXVNR8DK3AbiDqcctteaBPjFDPbZldQ12ZUe30pqKkjUNezJhodYRHsuQa108xWl1B3p35l7FZRfI9zrV/YA2B7MSOA4HVJLMB5NQy5P2nHvhFvfCLRyEE1H2ZUItnbx9p2KZuItpLxMCl/GaX27fzQWjfvD/oh6n56AHQZTOUwe8mBi4itf8I3mLSgMvt4fZB+MlnaY1vFO8cjXFiLKXuBzdfbPnayyjLVXTJnUoYpJI3ugmJ3ZTMSNBj1O8M7Hrl2Uebt3z7/ukEnGvMb8XA6rcLdxIZJBiPp/fnQBCdw+dM+PDExATtbHr5F0bRXnIRQIG23G8yZxKjRM5SZzYFuWYGTfUhEimOmIvFfAAxgw50Elec0dqXA6Hnq17V8D3qoWsuR1nHOgoqyi6WlbzjMJKiaDyK4ttf6UjRLXkYjL86A8ZVpZ41NsxEpvWqmhgCUnfODYkgmjhbWyB14sZr8dDWrGb/058xHvtQ7xq7GoLYxmyH/YYSMVvkk+zF2Z8a3gRgHmcc7M44rniNi+EW0xCd2P6kwUSKUgM4jEMQ0SEw7CP0mldO4bRN6ndrkduNadMfBcqtZtQSO+//vdbCYWEp173rPFR3ZP+sfbHKIwZJRdSbw968O8k5qid67s/oH2CbH8R2F4ok7KGDKjEBXAlu4n2x26wvPuQeuLhn1Ukt/lSzKugosxOvgBLDzl8gU0SxBd6jZs27V4fv+4NG1Y0nDGnpWUm49w561dHrzCs5ICKsQyZ9PTSoetC/Lx+nWtd8kXe4SrOeSRXTNQWGJeMdHO5a12adCAhb1C7XJw+sdKvUz8B2UVY5+pueuBcLaMvB2gGWY57tSDhthxQpsTEvQ2qJIcMULPmSuccyx4zGdtrtWTKkCElp3JNjTvYpsRPrCpVs87KHSvWGbJRUqZOio2IsmCd5+6CmqMUPP+snHDS2nK4ITKhoMakcR/rmvfaFa0aeXy+ibQRrlIF9z1ZWBVfaj4Vj/mqPE7V24pHckUtG450zG3dQ4k8kRVDTCXq9XiDdxP8wiupFYGZDhlS8LxOONV1hmE9YEFD5J3kA+L3qZGWRiLL5A/jG2vNhicyZ01rnSVZcso1IypqaTO5tv8MjrXKhEFLiAKfiUdWXcPI8KNgHBFLRJ5mKHRVYpin+zbWkiIpprSe9KRH5yq2rNUMGO/EPV6MBxmWVRFoRZmqiV0f6IgxA2vlUzJhyCQJMUxwNaEqd0GuZcWFLKhpftVu212yYfeoVZc3LnIw1q3l27Ko9lXysdk5FV1QoZ4kb5KYxHZX22VZ3eiskk6F75ttlGle9deVTc8XxaFZJcXWdfCEWlNF24sTu/j9TzoqDcnfBha9IhE8rZoTcDgtmasV6SC2/vrHcklLZCLWKXsSzxmJSa4vE0dxxZoncsVdPWCoBQ/0CZHIUute4mzzIcOHNLJIvhdm2Bk4iecMU1dlQJXVmzZsWOmKOiUsPheHyZ/L95FIx+nwNdzOVWh0jarysD0xVUkSHyQVw2osmeqv+S/HG9TOPyB5TEnIyogbNtTJ9wRsH3o93sE76SMZ8UK4xZx1Fpfpd9Y9uPfnzKvyJ9F8oUz2vOJWOGIkgTfDOaMw2ireeuDv3SbfzysMqrWXkA9BisQf7ToM7uEGthcfMLbi4E4nadi7xv3uixUowtZ7u/KYj1LMG6klUmnBSls2KV71xMLhbH4P/bOfNUops9fchk1WxqwwBcJSAzd1xJ4O2Kh1/qsEkbsRJ9yMk2zWOUrSvZW4EA4c9RKRfTW1RB9Cx9NqkspjLRsWsmYlTRYU8U73pdQpAXLDTiumTrXiL3/ur9//iR/8m/f/4ud+4te18FI6Af3jxvIZvlN1I5QFTJLA0vUaBkUHzQLT6Y5tggBFS0RmoeC79Zg3dc53h1f4J3ydK71GJKR25pp74R5HOuGQMUGFfT7N/95+AUWzisGGDb5NO4YS7EE/DHtcxTkHYcalLgh0hNsZY2ZYa/CKJefxcgsylKFhvWSkj97+MMjX7t/1/3Z39OWIP2zs8j4+7j0z98NNtZI+uVMFnSju6U+RFMfAguC+G7yPPjl5l2zncruTFND3cdW7FR0fzkfIv5dOItMf5pKQiNeBJXU2zLrUBVMZMZYRK9a5axMwid0h20pf/SRwhHEkvIJyrldmIhmUOxxg0KSKczU1jKmMqJAUGDaMZdhTxNEshTuTCTfCHnd0Py92pzJPG9WGM71ioUsmYkpFq7hiKIO8mRe41rkRDV0Na3cjd7yupMrK7rSpdZ0VOlwecK1N7niUUjKTCb9XX2EeW34xnFKz4Z/qO9yUA07U1GP2ZMowSatOGHCsU0oCDUOOdMzX5STzO+xz7F5H2rygLbUmiJ2Lw9haYv69c0pWNAwomVPzs8WDbNb3S0FY0HBHZ9Rqi7WTCocERiFwPCx4XLecNi0fyIKxlnyGfa7Y8BKmm/5VTnheDwH4sjzkoWhus7dEvhwe8VvbF3khjFipcqFrjnTMmSytSqTCEUNUlSVtSgYiXwzmRpxndqoULaRbJC+lZl+HXMiSBWtGaW5eyhL3r5knmNSKhpvscSbXHOiEic9j6Z6ZEQkzTUdYV5R5bLkdBjSqXKgpeA0JOThbJGGAksBKWgpNIgqyoVK7roa1FmoiczYcqHWX7ol1hwDeZcnzMuZtXfAZ2ePdaEpxh4woCZwEM8La7znuXkptib90QWxfbvWnf+B/ug/wAz/9Bz8x1vjP/eSf+DM/9Mafv7/COjQuBezJtlc7LbmQVA5QJplv03VVp2kPaGiN8CmSK/6eaPT3AJdH9Ttfa8Nf+qk//U3DCelXhyuKrPTka5FDsvoBma3D3egnKf1r04cNftJuyH/yxn9x/4BxFq7odwz7n9NKBN3dlyTP8Q5eJmlOTik15CJQrWu+PT6PolxLzRflXb4zvkAAHoU5VyypdZ3N+7ZEVkhBspRoWvccgjyUgRXjKCHJ9AaElTaE1Env1vqC47DPWjeMw5DzeJk5kf3iXOYxpNFPFgokxzlK5El7zqlc8N3yGl/k7UTCFr7UvoXDz1taREO+921ST9ReojLRDt772fAahQa+wnsGuU7nNJKRXSPaBJkGxSXy7fd/L/4Ct8IxfyB+lmMZcKkbjhjzXLjB19oHuRfu0OB1cj3P0sQpFrnSuQkDqSEoKqmoxApbnoh5YjjVEVfxeqtQOpUJC13ldbKi2FIKu45zJEmKO9fTYc1BCvYSTNsk9G1d/C3c5PNc2eezSYmPAh3sajde83npHE2HVw9SIXghDQc6YqQlZ9R8JsxYxMg4BEq1a1vTsknHMdEBK7F1y58NfwZu6iztQQ134pRL1kypUAwCeBZWTHXAWEs+CKZetmLD3bjHUhoeyiXOm7mQTkXRpImFhWw40FEW3/n1HFv0d38W9BMWMgYlnMxhNiBBs5QibB9g3VqFQoEySK5ijkLgdd3jRjtkUTT8o/hFSHyAl4q7vBZvUqhk9+C/H5J6Q5rw3kJsicwTUUzENvc9mTJhyGGYUlAwlRFzVoxT233EIJGxzC/DHtDtDsf2dQnQMxnMwe0Od+KjuBvPGp+0A7L7Ps/6u/7v+wRlgFJC9ggZyyhDevqmdcu4YLeiZe+7Ix8qIZPEIia9utTaDAhTMOCB8LZMYcdncHyne4N0lS9TKCtlYA+4dNe7xa5/jfEqRjowN1axOlhMnwtPw7L8HCsK5igLXWWpv02CTJn/hWbVKd+gIeGR+7AM9Uq5Hfs9PSCosBKrVu/pmFYij7m0eSrdRi0iWbHDk2Y3aFqzyZBA9+zwSlTGgfrmop1ajx93f7TabvmCWNLechU32Y/jS7yHW2AdyIxrFlzpHGTKlCFHOqGiYKYDrsU6G4PED3BC8oSBQarSeY0T1nXIAIcZzRKnpKuudBClqyQGMWPEK+0RF6HmdvLBmFAwkYILjdZKZsO3lTMu25bHNdlY8NPtXrpGaqogGmlToAtwIvMc7AM8r4e8Lae5orXUyPu64iWZstZIzYahFjwvYwThXNcJa66sesFpJ5PZspDmqWfwQpb5OXEuzEWC5zlvwiQRB1yKmUFuv4dV4lrxoMy6HhEjv1cE9oIdw7u65NUw4TRaS90rxgcMOJKKh1ojap2PWlvOZMnzuscsdTI9aJJLqE8AACAASURBVJxSsqRlmjYiVzh8TsY0qtxjTEzfv6uLXDw4jLYxam/52NchC2kod4oMAIc6oUD4rbMZv5pRUWT55+05vo0N9/VsQGkQQsxBe4QbgHVymG2CFfbFH7wrAl2Rx3De35xdEL8eDh+FpyVFfezuM07M92vuSV//Z151/Sg+yJ9/47++X7PhWE25cUPXkW4xr5qil3T0/+1BXpHXz+6YrBt6ykW85lY4ynNERPh/5Su8IneJKAfM+Pv6JX4Xr7NI9mzQdaOdG+DD5NtTkEonXNJoQyUVC10ylGH22BARjsKeFaSiBXcVBRfxmkpKlgrXusieEWCmsk0K+LN6lXaiPGBrPkLirQZGYWTFMg38br6Nn+EXjScmqWCogaEMcvLQV96KeMdgyBVr9mXGWbzgdpzy9+WXuYxXW3PDu18FBa34/OikhV2i+FLnVJhM+ltc83f156Dt+CNepHRoXJN4H4IVT1WEtSbSdJpvC10mQn2kosxmxyMZ5GA93zdi9hPxAlyt6+QqnuBl0imr7UnJtXZQzhvhMEOT3HAYYB5bYqGEVASayTjt1wWn8RLvxPXRM/0nyjuvURURBVr2dMggdTy8ezEKIcfhHsNchTU3dcojuWakJUtZsx3bwG2dcJKKnzVtWtVhQsm1NJQamGrF42Cxsu+N11Jng2XS3K60YC4rGrHC20QHBCyZAbMj+Kuf+2/vA/zRn/o3f81FmGdqcEXtOhkfNVaNvU4VNhFGpdDsrGWqEEKqcqhlzy6dWggchqRUISVDMdWse3pAocJdxuyFklaV366v8b9wkrF/rnRQpS7Ia8XzPNATSgqek2MqNbjAUhrOJTJlxJFOGFJyKTXXYhVa90tQOlPBzFNw6FVfQaof3O5wGD4sifmwsaXe9RF/+3EJiA+Hi7nCkYkExKyIFXrSfKVUjMKIVewqBpAqXNqR07Nkchp9jW0L4OpesuHVyB08JkWu7msKxH3zz+3dRBrzIHxNYy3RviqNBDa0LHTFREwPvdV2i1y/qxjlP+9aqYZfr6RkEZcMQsWlLBgxYMSAQ2yDtqpA59DuVSZFk5yfzZ25LhlSsMEgN07YPmeez0NR9mXKSmuOwxGP41k6rtRW15qSzviPneRJ0wYoItkHxFviLQ5N0zwHlEjtfiFJrtmvxyNW3NAh74YzJkl4sM+bOZK9nIyOqLgZx1xIzavtEWey4mZyBn9fLlinQGIha/pmhaNE1FxqnbozkT3GGVtufAuDKbjC0hlzBqHktk75dDnhg82aazaM1ByTG8wX44PNmseseI0ZDxtDpu+F0rhnqTK1VwbeXNdm/qSmUDVI8JMJQ96XC4aU3NF9XFL3kAHv6ZI9Kr6DQzaiRIWr1MmZs6FOa85KGiZasZAmJ1Y+9/rDVa5GVJYAssLBcfOU5AaEZeqUHOsMV7LKHBvpS4520sd2TezJa9U6aVexpUJY5+OJOKhymnRVFhn6YLK+BV3VbohV/QKS+T4DETZpg1WBB7rkTjKWPGbIA5lTaff8VRoy8f5K1qxlkyp6m50A0quHHz/+8uf++v2B2hNyIStK6eR1KzqYRVftjbm4BIbxXtHkz7U1yd7jUhdWeJDAdVzkCudURsTEp9vtRvtn/PhP/cffNF2Q/sjFjXQN3PNn9/f94WvCbsejf238OfiRn/yPPvK6tMTsi+Dfd2uQ+dnUYnLUSuJJaQdb9kTQBRoM6ldzxhWXumAoQy51wfNyg7f1yiBQVPxyfMBUxoxlyI1wyJMkO3yh1zS6SUlLp/bYT8597ox7UvOFFDngNS6IZFPDi3i9NWe8S94mXoMH4Nlt3H2ytBMbUXRrvffqP5JMEeOSOXMu5Jrvk8/waXmelW74LK/wC/om0zDhU3KbX4xvoirPXKd+s7wKagmVx1NX0YQn3AtDpMykdB+Zr8GG0ON5DqQiKrwbl/wMP58/q+1B/frXw4cLZAPZU0vVYgUP4P26r2lotOU7eDE/zw6pqqTiMl37gKBONkeTS3rHnRSBCUNO8a5+1xECC7qvZc2RujeOxU9DTMymD4PzToh38iYy3vL4KHeKrZ7QTjGD2bZ3BRatrdtLNlQENmqJ+i5kHmAtG0ZaMlKD3QWEuTQcJ3jzkMBES6J08deQMgugrGlZbHVETUxlokOuk5qiXRNlpiVjjG/VJITCr8fYSkLadH5Bnk5E/Ge7rx8WBsMqAwwKU8aaN8q4NJyxiJHeC4S6NcUSqzRYgjIKwu9rX+NvFUu+N36KsZZ8SiYZ6/A42vblbtRH4YBGG97ZvMcwDHkx3OFWOOKKJceyl+TK1hQywiAxLTd1lhDccCbWejvUCS2RU5mzJzMKCSx0yVW83iJLwceQw5OsbR/6s/s3/b/tL9of5iHSJ4MLbPFQPirJ6b+fKyKtenrXIpI9PoYUPd+O8NQx9Qnk/eEbu0MThjJgpStWCSu7bQ5mgbOpc3WLmKqykU1OOr3L4Q91jbXGa4ShVPnBKymM4qwt1yxzsmQVmAVLqRkzzAE0kGplId+3m+wxEpO0I3Q4b8eGVyko32PMGQbN8op+IYFau4p3rWuOwz4xasZbnsmcNZtcpfXzbmTDc+FmTh7619akMA2G9VJxl4fxlEYg9jw/HL/qHjD9QKAPGwgYTte4P7aJT4IptlzGa36ueI8X9IhjnfFElIfxlBfD7VzdGVAyY8Rr7THDlLLdSxXLQsdcU/KyzgjBrudDMaK0BXprHC/tFWXBOhC+OPeryQ7N2cMWzBs65oYOWaeFZpbUPS5o+HQxZRUjJ7qmJPCervju0Yy3V2saVc5jw3GomCaFjb2k0x6l5W7cA+CdcIZzwSoKHsol38ohJ7rmC8V7RJTX29uZZ+B49yeseLe4zPOwpmElndrTFnQkvWaJJZa7pm79Dpj9jc3fBWuiWLfuSCdpXpiPgFecKwreDBe8Gg/YYIlHTP4Cx9mQSnNiYcG/JfFDDYwlUKiwYMNzccaA8NRqounvV2qzcyTWjo+YDPAxQ1ZEvsIl5YfotPvmVBB4Lu4zlzULqROswKBhF7Jkwk0Gn0CK3sNlf15u6R4ROJc5d3Sfh3LJHINUFRIYySBf930mnDNnxIBJ8ryw+xY506sc5AFZVGMi46xQB2S1N6u22x37ZkxA+mt5TMFIt2/tcmhS8KGdF0G/A1L1QodnJSIfNv7TN/7L+xVl4ktZwBNRxlpSS8tMDb7k3Y+NRMpUSHBkRP8cAsJCGs644lqX1FozDSOWWnOKwXRuyL7tC6n49bY+ZsOGJ3qeFc+29nQCExkRMQjpUldWjOspQoLtHYu4zGvtIHFD6sSBclUrl58FWMVVJ4+ecP0+z/qQcA/Ei2QsantPzUhGlJQGQUpxwsvhLmcsOdYJlVoX4rvlFUZU/D/xi7lrXtB1cTo/qcS31Jr9sMc/lffpLU8YTHnEKsniZzgWZMl0v2aHYcofjt8DwH8v/8DQBhQZPt+fW57gSS9JqMQExV3ivhWH1w9SnGXzrtXIWCoeyiUnekGQwvghMuO99lFOjo/DvnmS9Iqe9n7DPNfNZd2KoRu1OTENowwNHmrBLRnkWe3XYM6KTYrL+rwy85SxazxmmAnpXkytpGDGmA0tAy05lzqbx/rMfksXtoYKac5bAcjOq5+4KdesGDHjYZhzN862iq8rWvZS8Wms9rXSgqlalPNI5vYzOtlff0/Ekq6JbotGlATGWjJDuJaG/+YH/+b9X6t0b15JPOmI+nSy8XFjb9glMADTSpg3yqgQhoWpaImkvMISMBRrI6lCKcL3xk9xoANuyICqt7k9lhUn0pFaFeUqXjOQAbfDjRxcLnTFTMbUuA+GQWb8wk8ELmTFUEvqlAG6s7VP0FmYcK2LrcXgo7oQioLGp36+O/oJxCeBYKlGRilw7FQl9KnXPuvY+p4cz9oQBjLAVGviVst5GJJkXKzz+3mA35/Yfa5H3xXdfw/kSg9A+wz+ArAFp/Nz9Ip9SJ/tFfUNhtO2pd78RQIC4h0B8wWYMmJExYgB66TK4WZaAZM/dalUTwTmumQkAy4SXKglsp84BU6Kv2TBhjapnNjfu6Z4QcGpGCFvRMVd3edNeYJxN9bm7Kotp/GSQbjBS3rMQ07pm1pZih4YpE3JwQnGO/Hk7emqQ74GW/M15opOoDM8tHtf8TCeMg+rLKcbMZK4K1mBBY9vFue83B5yEhbcizNmlBzLgK/QUIgw1QE3dMQ6bGjEyNaPaanZrpg5n2etVkX0Bbrofdb3tc9zzpp9BtwNQ/bKwMN15EAqTnTNtxRTpmXgrXrJTRkw05L9UPBk3TINBY9jzZ0wzOf6ztr6T4+DCU7cZEqhgSEVt+KMTYjc1hkP5JxHrKilU3K7CDW/JI94vb2dTJ42vFtYorVIMqVG4rNFumGTTQbdT8TJ+H49R6kS75/h9903Hp/znqxsUOZJscsJg/SqxQMKWpSjULHSyAAzdxyGwEW7oaHzWGjR3NXYqLIvJTMteciKCxoOejyqSoRafV2RvDUpsOrNMZ+JL+mMB7KgSTLEE63o5IetK9KKcqAjPpDO6+e72ruMKPiew9FHqjL6KFOn5YksKem4AM/pIUMtuMmMgVTMZZV9fw50QiuRoRaspOFcr7Nc8nWSa/UObH99K6XMXViDmw5767YS6Lo+32zD5+Yu9GoXtvorHbvJ+IeN/+yNv3Tfu70r2RjU0JN26brOYAIPZeqEbCTm5MTvhcleR0ZqohcLrWm15VY4si6EBK50ngLhwNvxA4IUvBBu0fcqKpL8vNJXOhLcKdxdshvdUEmZpOqLVNSZMwqjJL5g+0Ota8YyzAnCWMysdhGXWYHTkwqXTN6TGQtdoskELyZEQPYQgZws94VUfA/7cvtOuo+R7wuv5+djQZPFFlQdTdD5TrW0vCVP+N74IveKWygx8aTaHJQL5mLe59w6t8JRE6Rz/f3xu5iGQBWEZrNO1zdkWJwT79t0hG0vBvGilCV7VjrM5+nWAJQEsc743XCDh3pmMYOaKWHukIrwYri9JYYCHRS81poXw528VgxlSK113pP8efDPL0VYxMhQS5Zi8LEBFSqa4FWBQp27N0if1fc9MSjjEFNYmzHiKHZiSm66i9hauxSDA7t4SJPgY/75fmxW8FsDM65lzU0mW52LiClMeom5pmVPjZKwyc9Z5FinPJKrp+KFAx3hyI4BBSomCDHTihEFtbYg8Dd+8L+7D/Bv/O0//KtKRgJ0SUc/+diFY31UYtLGp382KgQR+10TlSbxRapCqIKkpMSrMDauaXhHF3mTa1R5P1yxTmx+gLP2HEUZBaueLqk516vUBrVF7Tk95EBHPJRLTsVgMdeyNu1+UWZqgn0zHXCUOiKX8eopl8xnJR39Bds7Fr7g7JK6+3+3Oz7stSIBkcAqLp+Cge3+e7cjMw3TnOXvcgTs9YG1a30nQzKH+PQJ5B/WcfGJ7/yLrjq4jZ/t6/J/2MbUkcpl61p0gZap03ilYSbWvh+JJRlDBswSprZPkHSYnhttBUz+tSVSyyYb7BXp0dyk1yvKijUrXXPCRcaErhJ/YyKmPeTQrFEwONiahqU0PJYrTmXBWSIeF4TMkwkIk56x481wmDXZc3UpBaVvxvdTxyBscTqckO4O9S6XmEUIMGyuqm5xdnyTFQIHYUatay7jPFfXfe7aeRkk4eX20GA1ErmUmhNZ8pnxiFojL+mUANzWca6s3dQZj7nEFXP8nlgXpNuM6tQpGTNkRMWUEd/d3uPVaowKPJEV/1jP+Nq65kzWXOqGEQUP2zXzTeSODIkKgzRf1hpZxJY7Yci0CEwLw9Nes+HtJMP7QjxInBBTe2olci9a9XxIxVlY8fXwBIAhFSdi3a8vF4+5pOGrhZHQZzrAOSRgjraXLPDOiicZzv0ZUeEGa338vT8X0nuv0Ltmxi8xAua1mPpN36gzIMzT+zWqjGRbEeuocGCfJREvFCPGwSBSV+nvgljae8SAYe/ZaxKnxqq45u0UdrodDTGdqfFj7uqYJh3XzUSYtBTGOiKjVOWrKLmnB9zRfX7bbMZ3TcfU6bJc/Dufv3/5737+/vUf/fx9njFGlFZAosiVXDDY5LUYlG1fh7wWbzFiwDhZey2oeSRXrGnYT1KtS623kg4fAaHRDa1ap3WunRGnF3xGCfIxlAF//I0fuf8ffO7PPvN4fyOPbs/txA18rHbkwe11QpDtgtez1vSPSkDuv/EX7t9/4y/cB3pFiLAFNfRuM8AiBVpeJJxo1w33BCaiPOYyFxtXWnMQjF/U0FKnpARInA0LDi91kT1CDNbo/ky2G81kwkCMEyB00r6KSe2WKRHxhMCSkSsTyZEy+5pdxwVzXXAaL5jrIsHNB7R08rkTGXMU9syVHcmFCxHJflJKzK/3Nd1QDp0SpRUbLHFZiBnSjXrFBSHQsMGVJfud2Au9ZkjBZ+Pz/I721a1nY1fqOnNAKHMnTFKSNgljppgfx/mmzfArtwgoc1LXKXHuvrdiJoD+jPbtFdwuQInMZMLvbF/KXSjvtDS0TMOEW+GIFiPuu7u5esKExT7vxZME0Sy4I0eME3LGr7NBPbtuX0NkysC8kVJs4N3+ElPYMq5KQbllzizZs+RlvcGYIUdxnLnO/nk2x6xQ5MUbvw+KIXqWCf7rCbLvrReypGbDozCnlhbB1t9HYc64t2JWWJcmopyHFQdqfCLrim6L+gRMYKcRS9gNttWwTLDkBRs7B+3MpP+HH/hbv6r1sPwkXQ9PSD7Ja72jMt9EpmVgvjFrn2khrFrrjqhCFbweKEZcisK3hr0s59uo8mW94mfjl9P7ttk40MfjeMZaG2bBCLSOO35LnuTNf0DJWowKO8EqdYO0qXlg8SRh9C/ai61zeVbn4sOCc++KQEdk/7DXP+szPBDP7p+9z35W92T32HyyKjEb8Pni5mOTyNgFRfaN2Oi2lK1tOtstb3//frIREBa63HpwPanwRMTbzrYANTnpGVAlKFW5pVoh6WHt9LeNJH6m1wzSQjOmMwrL3ZREcIdOutOTjYDQJm31BTWLxEW41Hm+HrZIt1ky8JA9rlixz4QJA86Z503Q3M1tk7sRDjlkSktLzYYaU514TW/xrpzlyngrBfO4ZJiCGL9mAypmYZI9Ojz56BTNPFCIWxLL7rhur/UEJlARqKSg1nU2hPT7XFLm5OMoGDxpSMlNOeC1eIuTMGdFwx3dZ0LJbZ3xSK75PfF5foFzfmb1hH95doMHi5ZpEXh1XPJT1x8AcCrzvGENqLKiiiW9nZxvhPy7iPLb2xfZoLzV1Pxi8QEOmnuX84R5nvCy7vHywLockcBA7YkaFYHDoqAKBv+sgvBwbVjV98PcnOyx9nGb4FcAX5FT7rLPHd1nqAWPwjzfj1txRonwtfCEF+IRXy1OiZi884Us8nFXSCLk11sdDkVZpo3Bv65ocickQxByINYJQ3Q+Fm063pID7cy9wCrZivJEljyvM1ZqFcVCLFB0Evlh4aGebWpBhJtFxTIWKJ5sBBa0PFcMuGzbvLYbLhyutWWPgrU6z0tYYZLJXxZbJx/InM/oAQB3EoSskQhKVt0KCDOt+Pb2FsPUwdmkzrhgRaoP219O/sjn7y8bkxR+FK54Pd5hIQ0zHVCnjmaLGYstpOEsdcwjynuY8ICmpNfXaHda9gB0SLkFn3EjukY3JoQhnSKeb+euRAfwx9/4kfs/9pN/8psCmuVJtCmDVXlv6u8Ru7jzIB0/CLrOwO7oc0V++I0fve/Xq5+cdD/rSOB+TQVhqKYYZM9Yp9wU0cRLUq5klThkVxRa8J6cc6WLfA4n8YIDmdGwYZUMV+8Vt8iGrkQuoq3TLooykkEWKvEx10WC/QzZC1MW0faTfvHNYTb7qbhzEa3z6vyKQ9nnTC+2nndTgzLp3WkY8UH7JM8lVc17pnttlZRb19yhuE5e92KiryU/1/4yd4ubvM5zlBhHZblV5e/dW4Tfpp/hQbji/9MvIxJo2v5877ofdn8NtRBCSgaYZNL3vxo/yywUXLQb/ir/d28/bvOzNEhKYe6b4epdtn62bFRyQuH8DiP/Fzh/dyBDfkd8Ld2/kKBhSXUr7WeK8kQvDQ6X1n5PHltaSxwS97RNhaIX5TZf0/dQzJ1ciTwXD219UOUdh1mnWLKPRBn1EsJ+LFWm94kod9I6+R3tXeYJyVGp8eYKhHFKeD5QI4oDySTX1v1LMQPVhTSsWG/xOJ7oFZUUNGKS+C/qARWB23FKBPZSaWlFyxNZsa8DjuOYB+GKRlomKZFo6IoJnpS8Lxe8qEdE0STm1CXDAsyoUIXrVDj4qR/4H+8fMeBf/Onv/8RrYvlJEoxn/c6rXbvfqxrMaqPW/fBNcN4qg2CJiKp1SrxtOC6EF9tp+izNMl2+UHmbqCNMt1zF6/z9gcy6RSG1WmOamC2RB3KebmBNzYZzmTPHpFIXusoYR9hekL0j0HdN3/p9j8CdE4ZeQP0szkf//fqf5yRwr677a3c5Jh8G5xrKkKWuMiFKCIxlBNIlEkpkFa21+ixlL3/vQSJeKzFjYPubTERzMNyvrHkg5Uu5Yz1FespOiTTqn9kSIWHw19pk8l0kMkhSvhbgmm/HmV4zlmHmWvg9aLXllEurskvFXE2xyVvADW1OXqy6XCbVoV51W9cpyajZl0lKDDqCr51DJyP9qD1lE1q+hXtcJRKXd1j8mjuxNWCb+dtyap4KwWQAp4yopUlclyZtOlWufrm6yVPzKHEBVATRflVTcsvfkxqH9ASEvTCl0ZZpj1z/m+SYv8MTBOE9OedGMkS6rTMaVT7FjD0p+T/m57wue1RiQhQTrbiWNQc6oZYNB0x4zEU+bw+vfc74zz0Y+QfFg3w+DW32mhhryZ4OeUn3GBKoCqFpTWhhEAJHQ+HhMjIphatGMzb5lzinCd1zVVHyIFwy1REP5ZoDHfGZeIs3wxlK5DW9yZ04462wxs3XIrYAm+zh1Ah8wJvhSZJ5tgDWieVjBix7tlD958SHz7sO0iNbr7XnK+Cyss5L6ptrBYRSA7VsuKHjRJKHAYE6tkxCV/FyZawALDWyicpBUTIMgVWMDEQ4omIkIScgfc6fk3M3aILGWoLzQK55nUMGFLhPw3ssGFKkhGTf5g0jHrFiTInDuSB1X6SyLniCHqxTN2Q2UupGOPu3P39/sYZh2ak0Xsua16LxqYpUuas05Iq43+81G8YMOeOiE7Og6zZF4FCmJkOp1/R7SE4UbdJzUzmnCM1rTBeMW/D8Yz/1Q98UyYePrjD37AQEPrxo1u+I2Xq8Tebp5vGHBxIOKzRItHVdR2oFwYoiV11j2r0FMwqtCDyUC2YMeVsf85Lc4nEqxBik1O7XShs2umHBklmYmKmgDHi/fQzANEzznjeTCZUULNSCO3cGb9gkud6YC0NXcU4lFRs2eT+pkkR7IYFVUod0hSiAWZhyoddZscn/TlGGyfgwy9rrDq9PXGChZSQjM7CVrgjmcNvr2JkqxhT0BoRH7RNeCrc4UpPVdaWoXdl8IXBLx/wdfomNbqhSktDdYysk2TlZt0LEVsmhDHlN7vHF+DYR5bYMLb6LycdDvXvfUEhBixkae1zivJA+edtFVPx8+sqZA6rcRXmeCf9z8YsMYrXl6eLr84Y2K5V5wmadq24fvSfHuYh5oAMeyiWbBCMfUmUvrJoNBfCyzvh5qRlpSS0NgjLDFLJcwAZS/IqbEnZxz3tyyrfH5w16hXVtYvp3V5zqut1eNItophh4x7m/J+QCOP0YkS3H9M60WpklOWYvUwVM1TOkfadmO96bMOT9YHv6PT3A+YYL2WTCfqQTk9ikWPjv/sDfuf8v/PQf+ETrY/ksJaw+Mf3DkpPdBGTdGkm9372ft2Z6FVUZFYFVD7dVBtsCfHHbKwrqGKnEHpHT2NBI5FY4Zq5L5sm/ocBkdX1jGCYzOZ8I3qoaUHKh1xzIDKdfLnAZuQXn8TJX/PsLcsY79sjgfeL51jVwVaYkNws8Fdi7YpW/f78t2Feg6mMTu2u8nYB81FjqKrtxeoDqVWifVBs1wvBa10/BwQRhGIbUsc68ED/GSgZbrtyBpyFc7v2xZbAnQsnTnAUfjteVJPs7Su1qv04rXSezQAtc5zpnJpOt6ox1MgSkUy+5SpKx40RELQj5QRaEK10ykVFKsAJnepUJak5sNN8RA7bMUlPT3axXako7pZRc6BWt3Mmbj399Tg95S57QsOE6LpID+TBrfD8nN3g+HlKosJIjvi4nXNJ1Dvz9vbtFb1ECW7BLKdlEl2H2oKndOhahqygVYnLVkqqdMx1wT/e50A2hEI6ZWZVFGvZ0wEI2nLHmlXLMZdvyL+0dogpfvFqztyl4Xqe8SaSWlmNmPOFqi+vj9zIkhRNTE1slArYpRI0ZcM2S5/SYKMq9uMeLOmU/FEzKwEtH8PgKnjuA65XNt+UGbo8DZYDHdSSq8ty45EWd8TW5TC1mw7TOZc0LcZ+VbHgQLnmtPebVeMz7coWkiv2n25t8uXicAiC4F/cYacnb4TFIl4S+EA94GK6p6TZKV7b6/5l7txjLsvO+7/etfTn3c+re1T09PTMcckhJpCgpSuxABszIgAUJcQKTYgQDQQzYCBAgQR4MOAFkGRMjNoI8yAGC6DEP0UscWUoC20ASA4GsBLYjWLJFkRIpcq49l+6uruu5n7P3Xl8evrXW2VUzQw7zlN2oqanLObX32muv9V3+l4hTX3PH8T7cPbtvuwTE5riBEuL3osRxvG8R/13gcIGc61HedTfkOF7xExZh0x6RcfcQgXGWcV03zJqGDZ4Dl8cTS50Tm28fDSC9AqIJ9vV5JlTqeZGBcUFoeECfSj3vypxleC4/kKUpsmBGV50Q4OcIA5fd6nzEPWZb3wmG1e5z4+1ZGAbDxLlsGWrJldyGzs5YsaAlwsFOujNCNQThQqcptIpJ3t3ESqeDaQAAIABJREFUsf38xHU9Yr09moog/+kv/u3X/9vf/JX/XyUiv/z1X30d7NzLEJRuQsALJF+emjrJFN9O1H44Tkh7X7ybiMeiSHx+FqyZ0E+moW/LOa9wdOuZMD5VUK0MPjaxGHkZOs3XLNKatpWKF+WEKwxOnYuR3q+CxKxV1G3/Gkg3iRF4lINQNY9eFTGhiZDX9jkVoYuRSZa69YWYGedGtyx1lWRj535JRwrWuiEqICrKXJepmJWRcdFcpz2/Q4c6+GA4oku4CwG8pyslS7+i40pmPsRC4qhb8Ua8f1mA+nhg7EZEqFaMLWKCte8mFN7uT9u765OO+POMjJfkHqXm9KTDIzmh8crb9Yr/3X0bUUtJDRa+U7+Mr13pOvE74lEGlICKJL8QI+1bx8qSRMelTtP7tGFencB1jUUhg755uuG95sErLsYsUWI+w3Gg5jJ/Lp00hh0KChwL2VopJTwWHXIOdMClLFocl53JbBQ/2HW2d5Drb7n3+Qn/YvJS2kjDIHT6huTGPZGaUrOUrMT7muGYySY9b7vOVgt6jWMjDW+7a171+2xo6FnJmSm7WDA+bznCULs4FZaypRIr1EYubJyrkR/nRUHtvGNvrgoDc0DJlZouowO68tH96JOO/OMSje/HEfm4pGRdQze3+xT3tba+lANWTczC7VjUphbgsIB5XDiut0ouQi8TJj7np+SAM5nzPX3fHM+lZBEewDI4dh+4SaoUb6nYk1Ea6GjqZg7WNU+bC06zQ250hkfZhhZrO1GIHY+P61akpKUtzdtKQNpHG5L1cZ2Uu52IGHh0pMMyZPHxb3xsAtRauGGn+BArKbGiULdwvom7Eh6KeA7xb0WH7ZiQxPfY6Ia+9FjrTtnD3jv6Whh/IW4+hZRUuqUvA2Z+keT2XDAWi4uyVeuhUJL5XxbGM0Ijlrq5tXAnzonY/e4EB9GcnGudcR1Ip0PpsWJj/h4BUhEDO5P4C/jvMFM/417gUuY891fJ90RaD2EH45UsWBupXBwE6dy11KklHCEINyFIysjoSZe+dHlZD8P3LAnwEjgAVNzTCa/pPb7jDJbUlYL3/Fl6j3hPXWseboNOvbLjgThMUjXep0JyBtLjUrfsRSfaMGc+lGs+r6e8L7a5v+DH6WfLUGV+Ne9z09gYfWO6ZixW3R7njtKX3NRW1Xouc46xSngMOp4zpZBdy/2YIZU0yVm9Q8mpjjkXx4/6Q14te5w1Na8MCpzAwcBkcgclLDYm//3GYktPHKM8IxPoO0fp4MNVzVhyMjWuV4TtHPl+8m955K2K/8fuGT/W3GMptWnxi/LQ79t4hcB0HVrbn/PHPHZXvOqPeM/d8KKf8Afu/fQMxCQqPlMxWG0rYkWSeExUYvCb9P9pkr8KkAje8V41eBCHU0ku9cYfsfpZhmPmm+QZErvPlbfPk8ygcE2jXIek9Tgvws8ds6ah7xxz75NXRgfHnJqZVjx1c77oD+lKRkeEt3TOa4z4I655Q6a8pIbF/0AWyVQynn+EYMVOxPNmy37T3e0n4ZpXYZmK8KzpRtl6eLteMXfbMK99qESaqMhTmbKvfSrxzAP8csE6BQfdUPH3KDNd4kPQEse/oia6uteh1BHX06rlEu6Q1GUGWqaFP/zxH3z1P3s9Vr3/h9/6rz91AvPLX//V1x3C3/77f+0TX/N3vv5rr7cD+nbQBR/t0sX5aD5et/ei9Jowl+KR4MN3rr+dvMQqa+x8xM/t6m6FmQpeBS+dAjFFLDEhgj4FNZ5zmfNUrywJx9NoY0IhYb079zfMZWVmk2HPH7o+HUpudE5Dw56M2UrFjZ+jeA7chJWuudF5KvoMpc+NztM1xD0nFgBjIazRwAmUkrVud7yF0CnouRFbqVgEKHS7gBil02PAP3YjlrpiKP2gFmawpB4dKmzdnzZTg0kFcvgsQHcrbP8qyBM0d8dV8fxz/8f8nPwkf8Z/lj1KvpWd8yZP7DwCF+fn/BdYsvOTivewbUwcob5358afbh4gCD8jpwycY4bn7/G7OL/r6tgccyEd2o3BTkrb/m5braovvTRmS12halycUTYIiaBBSvuUXHAbpZCR0deCm1AgsfdYp/Nu81Uv/DWXCA/cEcfa5aTpoZma8mEoVj9213y+OeKcintZSdcXbKVO8u5gcKXbc39nEB07qzEazsh4x13yheaEE7p0cIyy3Zr9ZrMEMeGRM1lgjNIAAVbjCiWFrwAvj885mHP6Suqk1OkQDqVkpg0TSmahyxE9m+75IWdugYqyESvkl+R0QlGtq7cV8K5kRY+cJXCgfS7EktpCHSWO+9Jjq545NR0c/+Sr/+vrX/kU3RAHn+wJEr9/V6q3fbQTj3h8gnLj7vfD/w8yoXTCxnsab92SSm069p1NrHt+xJflFV7NXmAZyIJGEttSacWNzrn0NwFrvqtsHanhNOe6SotiLjlzXWHKD9WtbsDHEb/b+D7gVgcDLKifuHHrut2tz+3X2NhlH/l+VGcppGTihkGar5WAfAKB/e7hkBTo36qEt86paZGY754z2GJgZktm1lVImR5agyiUt157NwGJR1T3WPhd9SES8mL3InUztEkLRhuLvMNm2/Jcq1WqVqGac+PnifsiOOa6IsIrPMpcV3SwzkoeAo2CPEnsevytsXnGDSN6fN69aAkqFWd6nRavqJxl42pyiV5NEWVPewzoEmFXTahNTOgxCHj0lW54KlMqmrThRMhBJKktZcuEHseMuVLTOo8dkbhRZFjXKH49cUMij8chDFyfoeunzbEv3bAZe6791CAC4ZoE4Q13RoXnJX9IpsJICwInzoj5oTs5VdMsb4COOC6qhvfrDWduEbwgbCwn2k3BTbwXx0x4qPs8x6pYPTr06PCav4cg/NnmEQ+zLg44LXNOJ557E0+eKautIGJwnauN5z1Z8Idc8UG14apq+IPmhuva87Yu+CbX9DVnKtskLRiJ5QMt6GnOQir62knV+ajstK8dnrs5a6l54mZpHpaahSDVc+IHbAJhtV097oZ5VrBzNY8B193q4t3gdVdddEHauWYlNX0KCt2tF3d19QXhAzfnHReNsozYuA2LcTt4rHWnkBXb/1u/+7lHeb9ZM8l2rvR9Z+aM38qecikLBpJRqadSRcVgXrG7cfdorwUxAdnQMMc6sataWVQhuPMkuXZVuNg0zCrlre2Gb9dzPnBzcjWSfoRfrah5X67oUdKgPOUqjU3clDvsXNSnIQG5O/4Olzro/s59ilDR+Dzf4kmEwOCH7RrcPf7y1/7zH4rI6VF+5et/9/Vf+frf/cTXDejSo8MQ63jm7CB9QBKfSB1VJHX2fpjjBxHV23M1PgsApeapSAA72E+U5O1rQRfrZqyl5qle4UMRsQprdySVa3jOKt11ufuuh8OUsSYyTBXz2KkYyZBVCEyjUSuQuhQZLhV22rCXeGSYetVK10S8f0wCMjFT21xy1n6dpHdjcTBK5UYUwTJICkcifTzafIxcih2hOozVVre3SNNbrSjIkolf5GP+b/ovWUrNhWw48H0mMgzX4/gL/ktcyJp/4r6b/qZd38dXsNu8FkFY0nA/LxllGY3CN+XSAl/JkiJXWwWw/RH/L/7NtlzwNqA11mq8mYggiOaudYgV2l1PsMLANTvBFYdw5CYpGE9+JTQtPxAjqNcoHRw/3ZxyoIP0ng0NZ26REvVj7Zm4EUquLnEkogJWW2653ZWKX/ex7sxGGmbUNpICmQhrVc5lRUczVmKstYVsuJAVV7Lm0i1vrUHt/SdvxXx9jdIi0MVgcCPJuMG4J3OxBGvmtgxDxyPysPqhC19zey2MR09zVlJzIQtmsmHNDo4fn8hSHB0cN1rxaRIQaBHTv19H5JO+p1gF627S8YMc1yO4xIkwq21yTGvPOHesMEm0adjw/infYd7MwzlmCTIUHcAXfpEy61FQUDIMqQU8QzEoTY8OlW6pdMvEjVnJ+iNBfqzCR5J57GC0YVp3k4eOFORSUGv1EQ5I1uqS3O18pC4E5gDvwoMX24iVblMtNV7zJx13ZSYjJyGXnF4gYuZiymBrv6J0XVahiuLR9FDWat4dbfiUk4ws3NCYbMRWqteGmtsJCISqUJDVjFWRmMDE4NnUoZpWhX/3UG3xuKCI0ZYDbIK6xwyrBk11kTacGChGDLcXa906hI1u2VJxLHvkZKwxrXOP2thozkEgM0YIxqVO6UqZFrEaM9xrsKprSUHuLNFq8IaXFJfkWruUZOq4CNJ3cVMz86MehWasgpt0TJJrPEc65Pf1TUsEMWT/RjdGRFR/y/el1pobP2coJi2t4lOiPnB9Zn7Bhb9O1ZkKQBtzNw8+MhaIN1zIkkd+j6XWVOIp1HFMlw1KR8y93KpmJsE6p+axm7KkoiTjvh+xDgvwtSySMhShinMucw4YMmPF5/09tjR8Vsdc6ZbTsqCfC43CC3uexgt5ppyeLFi+N6LMlcbDHzaWxN3Imu9JQ18LCjK+qzMQGGoR8K07d9fY3Zm3umGv+QOmbBlqwVwq5rLFs+GlZo/H2Q2vNge8lV1ZciHKqR/yrrtkHngginmbrKkS4fxuhTlyXGLCebdCXd1Z6GOlta1qc0/7PA7rmENoZMexaQd577gpn/d7LL0F/eAoJXpr7I4MYZQZeXTmG/YyC2SGLmPpA+zItQpBZLzqj3jsrlHgfVmSqZCp8K7MeUEHfChL3pV56iAZNt2lln1cGwrdBcSVKo2C1LD1yqiw+X+5afiOn4G3BPjKrbmUJXv0TNFQHUup+NBdU9HwjGuQXZAYJadL8lsbto2fVdItMIhdKXtWo1hC+yiluFW0iepKOwiXvf9/8ov/5ev/3W/+zU+12f7Vr/2N12OHsq029GmOu4TxX/76r74ev9cNBZYo+Sq4hC1vB385LsGxYhC7DhCg3Th9TJfjTjfk7u+09554xPnskJT0VDRcyyJx8gzyUaRkNgYyGzyz0CFxrcAzXn/kWGy0Jnc5M79IUORIeu5LjxudW5dQzS/E/KWCUEY4h7EbMNcVS11RSE5fzGMkFsxuP2+2V0aX74YmyfFuqdhqxZLVLTGQ9t5ogXlOJlZ867sBHlPYsrGwgHWpK3rSTclV3pJc3+qWXIr0ORY/4nqb4VDxRAeS/1P/gK+4HwfgzzQv8113ybnMmFPxLfchc7/8yL1rn6/dg11BKcOEGf6B/AH/sf/T5EEQ43f8t1L3PRPHxld2nXeS1fh13M8iUTyiMHLJWeqKRn26pyIZW2r2GKSx/YvNj/Lfy/+D14ZCMvbdhJkumUmXqEw5ps8l0zBbd3YDscsZ9+Xfzd7j32peocDxkp9wne2kfdfUPJUlDwLodqwdVtSsMXhvKrapgkRRoN2aHz/36DDC4tOBxljFfPX6ufC4rhhRUuPpac6NRM60rcUNxqVaym4PMg5Mk9LGpVTc94YEivM8U2HiAjdE7Rmr8dz3g2A8sCtudTTDi0nqT7TLlsakebFOSVdzeponOFPbs6dBudAt+5TE4umnPbJfHP/Mf2EX9clHXH8ihrf9u0VmRoVwOyFZ+eC+GIjmsQPSBBfMRpVl03LDhmRStvTKud/ypkz5E32/lShoCuqtGlzRcV0yyThye2FxjJPLFGpmuqSUkn2GOJdx42d0nIk9RsfSlBzILmi+y8MwXW/CzzV9nLgDJm7IVBfh4jW9x8SNQ9Boi1fHdYxkRuBQhLHcc2OO3ISn/nlw2N5VWCAuvreJWu1j4Pr4ENArEYdu/8zVdR3O3SoZmWShAqg0WjNywwD52F27dWdytrpJC/BuE9SgMLNbYu5uTBLugZPASyALrW/b0AxI0lL5COdrih8NKrYgWaIYuws2Jj4lgsKBjFiySee20R2pvyJ0PUTx6llhBOQmdDBUlL70eNU9CEuUcVhO2ecZVzhxbKWmj8k/C8KQrlWbwhiWUvCm/5C5bHnIAc/khkIKRtqxuSgZM1ZpPB1CLcqe9gIxzaVGfFwqG2fBQUxcbKwgk5ydhEJ0YTV1jkJyBq6Hw/Czi8BD6UvP5p6U9KRD33XMuC0YRXbIg+O5Y6SdUCXZ8LKO+KY750h7zLVhQc2ULcfSYU7Nu84Sglo8B9rjy26Pb3OFijKkw6VY9+Uee3TI2GfAM5nyk81Dfna4xy+8kvOZPeELk4LjkTLsKNvKsT9q8F4YDmr6/YrtxojM710JH/gNWxrO3JRzmbMRz5Hvo7Fji3WVonxiLeZdUZKxkh1XphYFieR1x6WseFHHbGiYuW2QYiyYyoapbHgqNyG43wX/FsxtU5U3b1WXrQjQYKCQPCz1O5x8n5Kahjp06WIXrcY8agToUXAt0QB0t6DHtny7G6MoF7KhpzkfyCJ4KljHSiBsYjDM7OsivH6rtkYvvWff5TSq9DNHJ0C09rKcR1mPvXrAdaDuv0CfU9fhGWtmUvFQh6yk4Vh7jCm5kU1afzXck6lsKMmta4Yi3tmcaiyh3XqD577trRr9+9n77DMgQ3gsl4zpMdEOC6l47K64ZE5JnmRU4xoUP8ckJA9V8LYbeGTgRCPN2O0wedSdoIPD4cQC4J1iT3tj3akJ/sK/92d/5yOL8p3jr3ztl1+Pa178DPCb/9P/8ZWv/tKf/76v/5tf/29ej/c5nkMc2/i9GC7XRAlPwtexL7sLiqIPyiop7Hgcu+50nMPtQ9I+Hdb1TwgycrJbnRUr+kS1vKgaZLAagIGWRD7SIsyTJ3LNpc641Cki4MMesNBlihcO3QREGIqRgzfBbM9gtMLUz7ifHWN9hiaNT7yPgnWNK6rAJ7B70pcul/6anuumqjYQIFN1Upz07GDMO06FjfnA9RL5PO5fhDGLoi+IsNENk6BUWAfxilpruq7LVrd49RSuTMlyO/4oXZl8R6pAxw7yJnTF4owydEbe1A/5gCs+xylj7fCjesRb7poPuWShu4Dbt+ZXe4WJpY+CnEKKtO/8jHtE7oR/yDtc6k2K07bBQ8u3rt0KxbZHFzGpEqGUMpj+7dTKDNER5/Sub/Ln/ef4jnvOtat5pBMeZzOWrCml5NrP6EsPEcfL/pCts+B6qssUDz1wxyyCoqdB30tAeUGOAMceJV0ytpi0O2K8qoNQNLyh4rlbUounETP79AJL2YRYx6VnqR+kwuPao8BIuxwF/smEDhlC3zmGhfCsqpiLSf+upA7rT0xidp0oL1ZM2kFclR4lJXlS0XrNH1CSsaTmoevSccJYS4aSI+pYS0Mlnq1Yt7GLCUVUEpEZObV4epjwTC27ZyciN25kTUme9tsak27fYjzwn/ufv/qpoaafOl3xgSjo9TakKh4R06tKav3fberEikr8eS42yS1YC5M1fN7iOdIeEze6ZdjXro5HYng3yNC1uwAVTeiSGBToKVfJxOjGz7DWUZmI7HdhTxGWFI+PI57nUvBW/R5vNx/wUvYgBTmqnokbETsoVtVueNGdpK+Ps30cYg8OAUIQiFHxuNs5iYSt9pFJlmRmXchAI4QHSBK4sfIQq/8FOX3pkUmWcJNed34VQOLaROJ/u1pz92hX3NIYIsmM6DPuQVo0jTjvzOhJCnNzDfcyVUACFjR2jNpjkeT/tGGqSzoBhx9xwfEcomdGxP3GjdghiU90JJO0MUcyJMAX3cs0akHikp07doPnREfp/2P15tpP6QfJ5ydcGn5SQ7tTV5zKHscBRzrRbuCReDbUYZv0ad4+0L20AMOuCgu7xSheX+xaRTf3gpyOFGlDjgTcIpDSV4FjM2VJF+O9FOTsqwV6l27FC35Eg/Kn9JSjrGAiOetQFXmiaw6kTNj8rpo2/Tf8NQfaC+coqdI6VoOj9bXgkR7QJeNHXl5S10KeK8t1xvU8Y752PDrZUhSeTunZm6x4fj7g7fOMs6mjCdNqKhuWbDlgaDwdMXhOhJC1519fiyTlmmNcmRpF1Dgb77kbZrLlnh8yp0IFHjZj3nFXfMc9Y4WtKV0Khux05Id0mLNbb2IFPv1dyiBVbPCqu1Kli2B6GEn8a6okhJDuLdY1iwt8es5EcWrGmykxD8H0O27KSmpuqLjSipX3LL2lrV4DX0iVqY8YZbjxdcIlu9A9WXrPKMtYeEsURpJxIh1ekQFOYK0Gg3lFR+QI97XP+zLnA5nbvAhBnzmjr1P3qsE6bDXGAahQLn3Fs2bLua8YYK6+L+heguuAmXRV4pnLlgudMaTXcj7/6KFogm1OdZGeo+SMfud1GtbELVUKlCLsR5CgnuhJXgvhaW1DYn7QEedHG4Yaj3//q3/9h4Jltc8/JqhxDWkfFXUgodv9GtAlyp4vWN9KTu7CBj9JDOXjun4A0aMr7kHxZ3c9cuLfb/MjwQQH5mx4Ty5svwmqjFF+3YU15cBN+LK8ylxXbNV4HnFs57pkrktmQZXqub9KJnRxH+pLj7Eb0OATNPVJ85wbP2Wpa878JQNn0rPx/DzKpb9JsCEnWYK/xn+NNnhtkov63C9NnTIc0c+iomHfjehJh7EbMdVFWvtNfWrnXJ9JxtqvEncxQpxsbA36Y/4ku469R5N0fuzmxLjiH7s/5ombcUPFu3K+2/db8+D2HLg9J2IxMN7nq6am9so5t3kRqesb9vSOdG5J76pquh+xa+Exvo2NwW1OSZyP/zh7g7Vfpxl46acIjnWIU250RkXDQx3y55pX2Nc+R26PgwCZt7HYdTQnMuTQ7THSTlpvluH1J3QpNafQjGvZUAbSuEM4kzlP5MYKYYGX6lrJVpz7Bt21q+4GojvAvnY5kyXnbDjqOS42xmsp1KXf6WAeHPsa48O4r0Z3dSsgDlpcwvY92OJ5JANW6jnqOoZZRqVW6Bpqwb6WnGo/gNx2cfOOq7uLA/sByhz/hqKM1ASDajxLqdhIQyPGU4wCJZ/2+FRJiFfYNNDLP54/che2dV3vWlS3dcb5SALigsZ96YR+5hhk1iV5y13xveyS6+YmBe7xwbfANWMofcPky5BBMINrPyQd12Gta5415zxrztPGBDt37kKKW1yNeNxNAOLR1hI/yQ7Sg/Zu8yH3sqP0u3O/5FgmHLp9CnLuZUfJ+Mh8OipGQV98LH2WWDW6K90Ez4pt2HQeraQmnktcgDJcIk7W1Le0qwuC+2oYu/hgb6mYuDEaxywQvMEWoJ5YxygLrWto63rfPtoL2N2fRzObiRvyorvHYbbHQPrkZCw1QJsCxMsCpdgKNyztSjfp3t3ltVShghT/TiKpte6TuaRuAy7XXr/RLSvdMNUlRbjGioaFrFEs0PsJeYWFrtIGawuFyZMO6X1k0T53C/YZ4VU545oLtyRTuy/X7NTdKprkANweo7hIvysXAc+8U8iKi3hbGAAIEpD2PgtdsqVKi3KEuZVS0KFkqkuG0uNcp4zps8aUhvbCQlfh2dcec6kocFTqWXnPRn3yfajxvB8gX0VwNF5S8W33lEuJWvpWURnQpWZHxP7X9Igr2fD73+vT71fUtXAw2bI/qrl3sE2uvuPRhqrKePTSc7o5XG+UWeNZhXt9X/e470c8aiaIQl/zdC/imM4C/KoXfpZgQUTDrYxR4K9sQkXoicx4O7vCowzo0g0QvyVbboIxoUOSyl6EXa0D6Ap2gVdF9I6p04Iex6VHSaxix3sVN4F2tTxX+yudlphDesZk97vt508x9ay33DX/Qp7zTDdceqNHbkJC0hMX0oTbR+xO953jWWNBmFel5wyQYcovwge64lWGfMCSP3HXvC1TXtExR9rjMKi/malhUDUSC7Ri12hJzTol3qSxyREmFEy0G+SIGz6jx2zCvVnINiRKt6FB7a8Lcma6YiQ7R+KdH4tPxP428TgGkvH/47bcCWtrLITEIDRxEz4GxvVJRzzPdhEnKg7+oPdwrXvcXuviut8eW5uTNVOWLNjwzF+iaFjDGu4mZITxX2t1az1qJwixm23fd0T4893fdUjqerQTkfjZvAiatI5EztOVLFlgSeqln5qCHrGQafN86Vc4yThkZFXaoIo5coOU0B24MRHmFoP0eMTgs5DM1Dqlk8xuY3GsIKMblM8MEl2lAlZUHyxbe3L7mYydCEHYC3vqukW8joUKxSfxk5nOzWxWm+CjUaRCXIRTx/0+8jxKKZLwS6MRdiUpbmgf1o1obp33n/CELRYLDKUfrts+IncTInTMpZ9HjxOvO8+ljji+6adMg1hQnJfxiDFUnL8Rkm3JWJ18uuKzEWVtSwrjleiuaGgcHOVBdsKRDqhQjt0+sfQZZWg3urW9C+VEhxRkxg+VDs/9VeLFgBk1FmR0NOPQ91hQ2xokBYUIHTJOtMe+dnAivMYo3cOb4B02Z4ViBaVYVOoH1bmY3CvmRB+LzXFuH4cEwmF75kaa1HGPz3ShLhWntlIHyeAs/Dz/2LXjG9kzVlLzvi55VxdcrpWl97xQFpxgsWWNJQyv+Akv+FG6d3Ed7GvBQmIXR9O5G9rAxKAzTD1sKRWRPGDv/f1V1u4eCY71SYcTWFQwKD6+A+LV5Hl9MBFTYB2ykh20KbSPA9wpdjsE6GWOYeGoPHQyYevNQC/3BQupeI/z1NLcc2NyKTh0e3ixd/ZYa6wnHWJrOg8L9FD6DN2AnuuyocIH+EOjddoCa5rwsGjQHTe4g2vBryJ3JHYuGhoQa/mGC0WBB+6Ia53j8UyyMZ/lHpnkHLox/XDz11LzyN1jT4bU0nDs9nige6xkS43n1O0zkB5zVqE96SmkoAzytR3pJOdVWyRsQehKl1nQvl/5FVG4rZQSJ9b+7bgOudhCu2sfe9rN14aGgeuF8MAcxWOS8v0ge+2fZRhJjqjmIMqBmEFcJhlTXVJKThmUm0qxZW7PjZjrMrWjDbJirUkgJIzRMGxDLjvFiErrABHbbdZ1eKRLKexcdAf8cmJ65Td+yhnXnMg+G6yTsJWKpWyt1coGFVNBiptmhhgESHYkdC8ekYz7OuEquG4vWPMCe7ysh3gRJnQZaYeoU1yCAAAgAElEQVRMjNTWCyoUIjDWLhnCkA5j7XGiI2auYqkrJCzQHmWrbW6QcJztp+csD4vgMkgQd12HkfS50QUbKg5kxH3doyMl3dDC3dc+CBz5PjeyZSXWESox3sclW0ocXTKmVLyRXbIN7dy4/ClQSc1KtsxlHRyHG47UuhVf9IfkZPyp45J/86TghcOaq5sORe4RAVVhMNhy+sI548mCTqfi+mpIXRV8cFEyrSwReiwzTnRIN7SeuTUSwky2GLjD+BMb8VTSpGpPh4xaPF5iV8xcZS9lGch2Vhlcs8XjU4diHAB5DcYLsjZ7xZh+4kLERdqeIYPP3YUJxT6hyZAGGJfsgscE88BRi+dFP+G+Dphoh6nsuFePdMS1bBLs7G6g1w6mL2XFywxRjEy+Uc8STz+YR5biLPlQT88ZzOLDZkOJYy/LyUQYFY5lY3+9QUNCCofSYUrFqfZZ0gSOkAkY5KHwoMB9HdMNW2a8Fxs8l7ImJr8VyofO6vPDQE7OQ1fyXXeJij3Pa7ZJwz8evjX+ijKQLlUItFahixnHthAzJwRo6/tXWpMHnybFCKXb5M+zC4gzyRJUpKTg4yR6/8rXfvn1f/eX/twtiNU/+o3f/kqsMrrWuhpnzT/8jd/+yj/6jd/+yr/zSz+bXvd3vv5rr/9ff/93v9K+vvjcxuIQIcGI0KvY+XymVyx0iQIj6aegpqKhxtOnZBXW2AiFTZCf1pwCblWmI5TJEX1tTBY1Bh63glBIezFh3vcoQ/W4wIsGJaAZ5/7ako0gNe9jEC/RQNfW6yvmzFkbBw5l7pe8mJ1yo7MAPd7iwl7t1SdTuvvuiKkumPslS10mKFSHgo502JMRGyomMgzyurYGZCHwr9QKdmvMid2FjoXDhY6Z7U9btmTBMLYrJVu2jGVI1+26oyMZsKWiDr4mxovIQuJhwqxxHM2Bu+Awm9CTLiUFe27IMsjQn7oji7lYp7UjD148xkmxsLlSK4Z0peSJm/ILzRd4x10xlqHJ1wanb4MAGQSxkBwfuLE2Fi7N/UwyPnArvqnvpkKgC/DPXSitdFw3QNOMEB7FOzpS0nPd5A220a3xT0NSnQcYeYx7wCBx/3bzGseh4/62u2apawvKg29MTcM0a/gRDig148JtOJARRzLmUqccyZgFGzJxHMkEQZjLhhu3oRJlXzuU4jjMc06ly2NdspKaV/I+gvA+S/qUXMqCVTAt7VKmAmiHIsBwbQ5HmG6HghMdhoDdTHTXeF4sLC581mxZS0OHjA4ZJRn9UHwq1IL9DrnBV6UJMMdtQjPEBCLuAx6lqwaXKtSERrq548N6Q5eMKqzjWzwFwlO3SAmIYFDmuA7U4hMcva8lfQoasURFgUESAxcaMeGSL/3Sl34gRHW3pny/H4olGbXGKfXRJMSJyfPGjsb3O3xQaYm/1c9sc9s0yqrxppoSpHwfuC4v+UnCTGZkTGTIkUw4ZsxY+hy7fcZuxKGMLdsPGdqxjtOwKFa17kgncAGM0NXuMqROCzsVBY/eggFFIvNcgzxgi3AOcOIOafAcuj1yKdiXEXPZcqJDokrJvvY4EnPOnGiXPQb06PCWnDEN6hz3dMx7/hlbrRKJG0LQTc46VIkKKZOxXy55UvyIMLM6VLY2wTW20m0idU39jJlfsNFtqlbGoyQYGwUoU1tu7tNU/e7+Tk86nLqjAGOwytGp7NMlBsKhOyS7turSr9ImGys9sZKz9uv0N7ZBbQdMirbd/o6/05HyI9WseJ0xiWzUs2TDWC3BjAH0t5t3UtCwoeJMpsxljYdU9YqVjUY9r+hRgNDkCf5yKUZc76m14iuxtOXGbbgQM3orQ6uzQ86Kmo3UfMc949pPGQTIXBRMiPWInU64/e1KoyyfYxDwy7MARwQ4kBEdci5lwUR3QVojnonvcCVrcsTI3moJyCqkdVdsaVD26bCvvYD9bDiXOY/lkmtZcE/H7KsBam5Y8oLuGcTLW6B6JCUPT5d0Ss98kVMWRkBvGkFEObn/lE53Qae74PmzfVabjLc/GPBsXVOp8hZzjry5chdkCX4FJIfZDEnSwn212l43QHnWYsFxrF4qylQ2rNmGD3tetiF9r2joUyYyYYZjTJ8566TMMmPFnE3yfolrUFshxWAkWVjEXapg95OUr82jDjvpWDBs/+PMuCgNykuBcOhR3nUzct1BwDKNa5dPleYdt0G40opSHL0YnGHS2AutEeCZbtjPcs6bisKZCMGDomRcmkHfxbahFKEQIxCfs6EjjqU2vBjUZN5zJgIROzQn2mNJZXhjduo3a6mTQksMVdrXcSNrnrg5Wxq2Yv2lHh1q/EegPe1N02GdtyE9KhpTywneUpEfZxCr3Xu0uyDxaJvI3a3Gt9e2MhQQ/trX/6uPhVL91a/9jdf/w6/9SvpZe52N1dT4/u0gv/0au0aXApxu6KDFuWVzPs6rLCV6NZ4TsWLDgRvf+t34Ws9dsrlL4wTWMWgbPpYhfTSJVZe6hNGXIb5vnOttmFjsfIBBDyObcInxrRahsxohWO0OQ6M+IR4OZESGoxdgyT3psufGyZz3nuwH7ubu/u25EQ/dMe/5Mza6oQ0xetI8D89Cw+PmKWvdcqNzzr3BizpSMpR+6sJM/SwpXFn3wl7bvieCY+pnQYlzSV96nDXnXDTXXPsp0YtkIsPQ2SjpOltj2klJTIBNwMW8kDKCAhc1L7p7nLhDhnSC+E4W7lth6pTkxpmV2/42MXkDOGGPu/DAkiKpXrYPx065KnbYh3x0vxVMoj/6n8XvASn+iITz6GAe53n06Yq/k0tBEeDbqhqSnd262qXgRXfCQ3ec3uMFd2wJrsJIch76CUfeDEpFLEk4kgl96YbkwODWc9bM2PCWm7LWhmXjyR080gFfYEITYtehlvyJPEFRCsnYY0BEepiE8E5xL65JJTlrDFpbqiXFR9rjUZDMXzfKjWyTkaBdS7SW8FTiQxfRp/2jog68rm0ajzYaoMFz6VZ0yfAKc78rEixpKHFsaDiXFe84Ez+JHftcb8OHwdbKXuBXgkG0YOfqDmbsKAp/6bf+0qfmgwDI//jwr39igdvJjgfSy28nIO1KHxgnxKtyXftbMKz4WcNripCsNKrslxnbxpaFdai0eex9plrz9+T3mfl5chJ/KbufBipuIPd0wjO54YAhfbVq5bnMGdLligXbUN026M3C1KGkxAW8Zdf10sRvw122H6N2BQbzMYm9He4R4JXsBfqUbDAi84wVBww58n3O3CJtQpF74FHu64TnMkubtJlHed7yT1j65S1IEVi3I3p15FjQHduUgqlARZLdRjcJTwsGYxq5oRHfVRMOM5KY+663S2RahoOZuFvJz6c5Ynt4rWvGbsSrcj9glq17M2PFiB5j7fAGz9Jmd+PnaVEayZCZzpP8XRmUQDQIG8QNICZjUbq27ZsRTaYityUGEkWA8y2jTwzWZbPxyDlgyHOdcq3TlAbdcwe2qTEgV8eZTLnWGQ6XsLWn7pBX9IgbWXPGNQ3Whj3VCXm4501rIYkb842sednvk6mwlYYa5dvuCSvdJEfYPLxqpWvWofpjm+yYqZ8xdJYARCWZ+JosOD1Hj5BuCCcaPA+Cid/Il9y4DX0t2NcOFT75O/RDBeSZrBAlkcAvxOSXH8slL+gefS1ZScVYO9zImj3t0dOcDhkPXZdh7vjXX11zedOhKDy+EYrCM+jX9PsbXNZwcPIhN+enLBY93nxvyKaCi7UVJt72i1vzLy6SbRWg+HkZFLLi4YE97bCSmo5mvOemKMpKTBO+bZC2DGTz5OWCcRsOdMhUVvQoueeHfNedccAwGai116R4HqamttOmn7GiT5mMsuqQ8MSfR9hVP1S3HBKgBw2P/Jgc4T1nGPjP+AlvBSdbhyRn95gExO9XNHzJH5o7evQOwiQpC9wtA8OnfsNLeS9Vyx71CmoPmYNn65o8KLZ4VbaqfE9uOFWrEFb4xLt5212nIlA0nAMLSDfSJHwx7AKUKL+7DcWKeKzFVo4LZreSjjanIAZ/0YMg/nyqVt2riUptO2d060TY89hokxyz65AURnhdhkuQUQlVz3hkYnPmub9KcqLt5ztCSO9i69v3B7gFqZ24ISfsATs1qDiH2lCOu0ec/7H7EzsO27S3WHeyzT8zEZC29PgukGzDqEfSY4MVpKJLNJDmcuzHtQOXON5rKkpyttQM6N6SVp3Q5zk3XAWOpt2ruiW9LiY6IxOugv+T4eINzpOTcRO+35EOqj5V1GNQPXQDpsEf5G4iKTjuZ8cJsjPVBV0p6UuXJ81z7mfHnDWXlFJw6g553JiHU0RMRNn5CJVq39eNbtLXVv23ve/Q7eFEuPQ3iRdhQj0+eXqVkSPRSnDi34vrwsPMDHLzMC9mrMlwnPnLJOgSzRMBpn7B2FnAXFLw082LlDj+gfvDVjdik/aVeA8rrW75jkVIWEnBfyQ/xa/pv9hZJwSLgK6UzPw83Zdc8uTYHpPqKnhcxfGK8VEsvBrixCe10FgE/sv6b6T7947M2EjDY7nkQ3+OQ3jZ3cchfKk5YUJBLsJGPe/IjDfdOf0gmT9jRZeCDgVv61MmMqRLQZeSn2le4Ims+PFszNYrpbO5/Y36hg/djA/lkgjZtfub3bpPbT+cuL9UNLyge5Tk/Gx+wnXVUIhw0svY1PB/by/ohv1yLlWCGM+lYkPDNsj2RtjTXNYmqav27G1lZ1rYnuNfbE6o8TyUHve6OR+sK25CMtmgrMP7veOuqcXWgaGWrKnTXtJer/ta4sW6LDFOiN0alR8+AYEfAMcSgcLZJvT9Dq+m935VR1LfbShC/H8BBnlGIUaQ7LjQvlNYe2/KKcBCG26o+GP5kFJKHmQnDF2fYx0nJYKoBjCkQ0cKpqyopWEWMP2LQFw885dstWLFJiQbYfElqi61COcBXx9Vsz7uQ9mpONgYBQlXsQfmNX9CX0uuZUlJQU9L+lrSJadPyYQeI7qMA17+qRgGdkNFLhlnek1UzokYzzzpltuGYYaEjmXgAAhmQDR0/fTgroKsK0TCm5AFCFaDR2WXGBrESZPkX7xvMSlrL6iwUx+LRxvOFtutphzVweN5ptc891esxVxr56w40TErqS15lE4gm1ZJJtESUnus9t0EJ46VrmijlR3WwYphiy10dj1R2cZMGSUECI11iQIeVTBRA1M92SBYImTCswYD60vPFtGAsz6SMSUZz7hGcBzJmJkucTimOucdPePLPCLDpOwWrLmRJSouVREsUN8Rm++peX3UYrjQEsdU1vjgYWG4Xk/fdRlJn2XYyLMgZ5gFhbHYKm9v1HG0XuWED+WKh7qPojzQcfAFKVlJzb52OaHHuazpkdNLnRbrMKxpOKbHTCqTOBZbwMZ0Gap5rA7pkKvjZR1zgHlxPMw6bFXZKzMKyZiMKlCT4BWBwWDDwy//HllTcHNxwu9965jz65JtDbOt/U7llQMpOXEdRhScuA5HrsOhK7kIm9ZKdsTukoxOUO5oxGBXT9ychVScuTkr2bKVqBukaTOPYxWVTWoaSnIe6B572uNA+5zLnBtZpyrwnDXDoIJlnT2TtZ3JxoKUVJM1+d1FUD2LEJtovhX9ZFwIMiK49b4fceFWrKVhTzvpw6NcBwddJSQfosnwLbbkBXgmS+7R54aaN9w1xyFxiElwjvm/KJhXiAozGpa1ctnUdCRj5ZV73ZzGw9w3LGi4T5/vuRtGQeGoQnnHXYerNY+VWjwZkjobO8iAKXYtZctMbEzWsQsl1jGMmyJYJzImF8Y1CsUKetR4jhmzpuJFPaAWHzg5O/NV2MG2YmEhigaYsIfBte7C28xLxCrtheQpuE73T3KWuiFCVNr1w53aVJs/Ee9wq8shuw5FV8oU3Eg4B5ubMGcd8PkRFNXeW20fiuMUobT9oMITk+QY7MXrjBDp+O9ARiZ+EK43lyyN45CdiMpuLF2CFtq6Y2MZeVFgQVovOPNYB8UMS1dU5s8hu4R/ID2KoOZnHAk7D0WZ6YKahpWuQjfF05c+W4zvZ5BYu46O67DVKhTPfNrP4tyMo9d3Pea6YqUb9tyQaz9joVaojMU8EeHa75IgJ+ZN4jF4dN9ZYrbvRqwDoX3fTYKfif3rSY89N+LMX4Q1PTx7klMHmHAmjtJFqLWkOw2S9rN45+esWeia5/6KXrAr2GfARAZ0pGQkfV7gkC/5+7ysh7zMMZ/XE17VQ35Uj3gqS07p89M8pJGCofS50BnWCbO1xxIADckDaU9utGGjW17W+7wll6jsuKIiIZkVR1TsjHO5/d/IBVFC9yY8ex0pKZ1BlosA3zqUMadywOc45QF9mrB+Tij5Q/eUDiX7MmJPRlyz4Ik/5y13QSWO+zq0pENqpmKJWk9LPuePGNLhSpapkLzHgL4WXMqGc7fkm5zzJXdA5ZVh7lg0FqOupaaTGBn2zzgheejAOyqJIg0ZHQoGwYNjoCU/3h8wrZQH/YxlpdzUnqesGNNhJpU9N6EwO6AIohwWn9QtBautNHGRaI2ujf8DP+bEm8Kg3Tfhi4cZ10voSW6+TOGeDsiZi3Uhu+Gq+mrJXyyMxf1kK5ZQqUAfI6xX4hPX8os/BAwrHj8wCWla0ebpr//83xr+xc/9zvx/eeMrsFPK2tRWHVt7Td2PdhISx0kxvkejZkyYi3BRNWzVVGuW2vA+S95xUz50M576c9uUqJnrCnVWKVmGheFByCyfy4wDHXIU4CBLMfv4G51z6Pa49rNWfU1DpcEHMlbYFsIG0miTgv7dFLstNyfILTleI32uGbgBhzokR6hEOfGDsMH7sLDvyKeKcikrnvoLymDIs9A1Uz+jokoBvAQuA9gCkGN+Hwa1yAKXYsxaN9TUCZIU+0XtNNASFHvgu66bKmA+jYzViyKG0yqcFW15PYAykAJhx7/Y+XnsqmpRvcMhdF2XlV/x1J/zJXkZgDOZ0pdOMthxIgykS09KPss9vBP23Ij77POQg6Swc5IdWkAgGStdEbk7iP19UwzJcUGVrCMdolyvqXBZSzw61ioGG9tSpYCklIKJG2CY6h4D6TKWPjUNPcyVdSx9MlyAGASFfnE8kRsecWQwoOCjsqXmVT2iwOTt3pcLLmXOtSwDjrSkE0jvK6nYo8+VWCX3IHRpDIa3857w2qRAKDrNxgDqRCYsWBMN2R6wz7nMOGHMge+BmK54h4xN4E0MyFlSU5IxDwmMVY0qjjHOylTMF6QWq8rkOIaUFpiHhXctnj45HRyZCIM8o/Kw1xNGwwqvwiYQz0+/9K/wNyO6X3yTzspx9uSUIoMih00d5kTuWDe2EdQKg/AZ4FqtQmQyvHXihIAp7oy0DNUlE35YhbUh3hOHhEDW8OoH2qcM68Kr/pAJPT5w1wy1G6pfHaay5jV/YmZN4kKHo8MNy+ANojTiU5APBEnZnYJRWldSEuRSMhQXfUG4dEvy0OY3CWU73nWzdJ3xaK9XtkbtqukDLXkvmDDuBbPGbahk5RIr8hbw1GqmUz1n77j1Rk4vRHhSbfm2u+JlTEGtr0WokVs1La75OQ6Vdogcgtcgjwy2oZrhlX3LY4lUBOM0GOl/ySYlbsCt5FqBQ0Z0NeceBj1qxIpQkfsR17gSW9djZ2GjW5AYjob3Dj4T7XF0YolINxRHDHfvwjNpa2qcS/FfBJ/Gq98Fwju5+fjehPsexVY6Cfp0u+tREnl8QbIbl2BqRj43DkDDrtC2CgDDTdj/2r4AJmW8A4Y5EVaBlJ0FGdZ47mP6t+Zt5KG0E5LodxSDxBrP0FaBBEmpQlh7qbbPVVozcYPg9m1r1YYqSe0udUWUnq20sgQ77L0e2ASZ4bg3n7hDRmJJ9jZwu+4mlgduP3X2Z37OYbbHWrcsdMXA9ULhbUsZ/Dhi51zEEvw6nOtuHxkylD7n/opBkAwW2XF0bD5UKSkzVSHrzqmE0qLafh05LHHXzsI+ttVdx6kOCYvHyOxLXbPUNU/9ReC5lLzmj+lR8EfZE74l73Pm5kzdlm/I+/wzfYN35YJ3shuUgi/4A95xV+SBCwMmVRyTKAkxzN2k4S8Un+VcfVD720H5VKyjYa+r6bkd/DeORxGgaBZjNjuZZLGk1uSAc1wobG6pqUV56EdIWF8yhHPZUEmdnqen/oKKhj03QgU+zz4NcCpd3pclNTU/4e9xTJfvZle4cG0xTjO4qF1Pl5L7foQHjruOdSW8K1OTeG8925ET0tU8qVcVIRGJhyXsBjsfVV0aVT53BM8XwlvNkq00vOaGbNUgYhtpGISUfiOWlKyoUbmtSFdJVDqMQhBmCLqnXbrkdMl4Ne9z7WvmK8eX7wmLtaOfGUxrgxWJLsQI9jkucV9FdvsT7Aoe0bfEGJ+O/VBAq0V/KC5IPL5vj8Pv9gtOf/3nU5vl9Nd//m95teQjJiBREcte16780HqgTBd5lDtmdcMmENiL0DJ7V+b8c/cWv6dv8IfNG6lNPtclS7/kg+aMp82FwXRoWMiWt+WcAx3igJVUYWGquVQzkslwDN2AkRsmVa2+65kek+QMXX9nskMWEpBdcB4/2maDOyxvqFqG7890wbvuAo8ZnHVbqj0RmgDwVKY8lksWrDl0exTBQbwtsRjVL+pgKtcJ7uXRmyK6vnuUaz+1xEFr1sHXownt2PbRvq6I343XUgYoQfx5Ji4t7FGeNx533V0/ztXdY5wa2+w3LP0q4VxNntbUZSoaZmJBxkPdZ08HPNA9nosFTEO63MiSN+U5n+U+P+IeMaJLLhnjADGK8oV3D8OwunS+WUhK4j2d+hldZ0nbloqJjJjIMG2qDZ6BdBOhU1EjcgOf1/uM6adxiVXB+HffkDOi7gcY9GYVQpPHchnmmwtwCE0LYKxEvC3ntmSHhaAvXUrJ6QUujcnuFqHLkydRgr50wuaodFvJ5JqKRj1vyFk656lsWEpFXwuWUvGUlW0gYcOJejn7mO9Et8UpKALMJn4dzUEznMG2sE5J6YRCzJDJnhklzzzq4fpyzPM/+glUM2gc9bZH3QhlrhQZnI6VRwee46Fy0LGF8aBjo9zL7P1eywYpgC/1NnwxKpDlOI50kFrZUQ3GFLDME+RERwy05EqW3MiKiXb5bvacCs/L/sB01rXgidyQ43jTnXOiA6aYzv4LfswB5tAMcE/HKMpn/DEe5ZHfp6RgRI8BnVvSir0WRMvm80fn8ly2fCe7SAH/S37Ey37Mi36UglXbCHzqiji1Z7yrBW+3oFtvuRvmAVJT4ZlpzVLtr/pgVGgdYeVat1SqjAvHLKzxE+1y2sl5R+Z0yXjX3fCuuyFW6CLcNH4sQ6cjFk+AlLDGlWMltZ17K5GO59s2/2oHlA7hmDGl5ngM1rhgyyVzZrpgT0ZpfYtY/HhEMm1cm+JHDH5rrVl4w89vtaIvvVRUUTSMU+TvNEnZKBZlomt0/IjnG/8/OmzH78duyDoQ6ePeFT8ijyKu2zGBtQA9wrZ2v7MhynTsEo/2vOpRhgDKEp5b/BDZqfpU1CkBia+LNeCc3bp/t8BmMMNd2JkFDlNNw5XOw15lc/7STxMxO94rKzwuOXATExTRJhCXs8DHcclcty1be+2nPPXn3IS/Ye+XJYlcgFULWpxJxqWfWtcndFKnzZSedNLv+JAcZDiTlydLMr2xS3LmLwDoRO6ibtNakIWEtdItp9lhKuC1fVlMNCb6Z2TpXKPBchtyaqIJVYAkW7d8E4yEr3TOO/4Jf+De51+5x5zrDYpJVp/pDZf+JqE3GvVcyYoXy5KJ2vzOcBy4cZgvHy+1GsdVFRP+CGaNQHCsj0GrD9dQp/Fs8PSkS06elOokdvBDApKT0Rfjd8WuYE7Gkg3/0p2l81jS8FP+hB9r7lFSGO8ijNuZv+CJXnKpWwSYa2NiAHi+kZ1RiKRiXjxioaQ9pz/UFftFxnyrvK9LuhR8xh8kflaEZw61vPVaMPO/nuZ01BS4uhSc+gG5iBV4BBaND0mLuZvfd11OpEOmQoG0VARtbS+CYqIXZRskg7uakwWlykKzRG63GMtzXlccuRKvyqBnfJcPqg0XumVDw5I6+GnFOFhSPNOWjd/FsrvnfS7GF+2TM9GS/y/HJ3ZCoimhcjsBicfoq5/7nf1f/NzvXP/WG19RSF2QeHwccrXBsqjKK9dag9p24rEHdaAFL+oBfyJPDZsrEjCCGhZpTRvcQldcM2epa8654Ywbxm7IY54z1wUeU1hYhSqayfGZ7G0nQC1MutUM9OK1dqQMvIloEOjStexMhyTk4TB2IyrMpbHWhp/klZCpGxQkwh7KAAFoRFOF2+EYEKsEtkmudQMYTGjiLCDeagWiqU2bWqatKl4VoAwD1yOqnWxbWucQqoFBYaoK7xU3zfjP3r9J+M4sJEhduW3e9IOO+L6xEmVdHYN91UIyxlmy5kN/zrUsyKXgqVzxAZc04nmoZrpTUtCIZyorblhYZU8rnvtLPBEHHiESdtSYZnsuhvPuSCclWooP1+VTpcsC9Q1rNvSlm8iP/RAsZjg21AzppECoDPWwK+YWUMgOqz7XFWd6xZd5mXf0GQ2ez3LCPe3RiJk0TbTPRmqrmotyLUa6P/A9XtZ9nrgZnUAA7WJKFGu2HMrIyLesGckQhwvqcDYGx8H7pEPBOsAzLpjjEPZlwLEOEeB77oylVIzo8o675Ef0iDUNB3SYUxHBJdHXYUnDHqX1dsRIaTG4OdYhU9kkvOhCamZS0fUFtULl4TPHDY13OKfUjcM3jvW6w8XZPQZVyWoxZjnr8+jRNYcHS17+zAec3j9Hmj7bdcmoC+tKKHOh8ZA7k/YeasGR63ClFXeP2O156uYMtYMi/Jg/ZiU1n/eHnOqQQq1S26Pgnh9yqiM6ZJzqKBiKWpXHKksFQ+2GTds00gsyLmXJI79PLyg7veMuyMl40U9Yupqeluxrl4l2OZNZ6KBtGdBhjakFxSphhNuAcTOD8gwAACAASURBVHiclb8AOHNLzt2Kk5AMP3YzCrVwr7lTeVMJBZPw2nY3YSYVEy0ZSU4pGUOXkYmw9D6thR7Tq38sC1yd8ftyTqYZM6l4Je8zrT0zKirxjLUT1FuC+pXUKSkH2ATMcpRsjhrzGkMVgVzNP7sRTWtYQc4yeLIM6bGhShVZR8aCzf/L2pv9WJLl932f34n1brln1l5dXd3Ts3O4SUORItSSZQmm4IFNC+KDQVDQq/8IgbBf/ST/B4YfSGtsGXzQgwxTQ8CyLIpDc4ac4bCnt6quriX3zLvEjeX8/PA7J25kdQ9JAROFQmXlcjNuxInf+S3fBUNzb2IrWBw/0+g2bzj4GL9qbawnr11QQkz6RMimLJacWoIrpJL0jtqCuVbrIN+Y6yL4MdknoxRunDAYpDFh2D21acumyIhwrpjErmnJ2ZgnRuWjGLNjEVJRs8WY4dRieAzjv/Sx3gqTOpQwaSgcJ5R9IigYqbYIinvmdm+rKEKwejhvuB91mFAJEvTk0n6ycSUrXvkzzvVqQ01X35vudZhyWR2KsJEUrKhogry6wWrtnUeJ5Djtj9fP0ARLEklptCYLJq4NjUFV1d792I0MTqUdiaRM3Jhrv2DsysBBtZ9Za83UTam0CrzSyhLoUDDtuhkFNkFvAs7e7mEe1J3WPd/DJvcpZ90FMzfhMNnt77GThJ+RxzyUQ9QlXPl5mOZnuKD4FPONDe/M8oU2tCVsn90cFQ1Xfs46kN1bNg3KWCiuMZGeL+sBt3TCG36fNzjklVtwqhe9Z1c0WLYJhuUPucv5W3KfD3XOC86tURkUtmzK3PRrP3JZyoBAif5VMxnfKLq3ZMwoICH84FmJk5EEoZKGXZ0wFZviWRd/zanMOdc5c131BU9DS5MkPHdL7umUD8Kk9h/4N3ifOcfB5yP+DpNOd2HlJqSa8MvpAa0qrcKB5Ih3jEmppAuNpGjwaJK3M7UinbBvRDXMkWa2Z2rKtmR8/Y4wKjo+vRDeGZW8VZRcNconfsUJa7bIWdBy5ipTnML22xpPJ55CkzBJs9gaC8tREPo5UjO9zRAeFyV3po5nVcvDLcdylZLjOFcTa8kQxmQUmvbkcy+WS8UJZ05KI1FKBApsn/Mo64B8yHE8/o0v//TgWH9ZATI8Lr7943cjF+SvUgdWYD8zfHEpVvnlgexYiLBS8wJ4T17S0PYwoBh8osTqxrU8OsL6INfquA54/k5bEklZeXMLN9nQlpW30aX5YjhGruw3Dev+NowkSNRK3OyEOCKNycHMTei048DtMHUTnDhKKchcxm6QkEs1cDHYEJFb8Syl5oHucSkrzGG26cfVa2q23YwKO08EfBhFJwGjG7VehvdJibyWjUGQKUNtOqy5FKy0ChjbDWSqh0pgyPAN+XsjTTwct/5lx3CDHHJpInQtlZS5nxPFIeOUR4G11CzUIEQK7DFlogVeTDu8FTMnvCt7dOKZunEg5sOtZL93AI5HlPiNJPbY6Yklpw/BMUp31lpvJBkxyc6CjNiddQiXsqITk7i0RN9cU5twj2KiZ+Q04ViuWFOTScqxm/NDecmVrDjWS1Q2xneL4ChxV7eppONPkmd82d/mnt9m4dp+nH1Hd/iBPiELEL5hZ3hHpn2idKyXjKRgJDlLKu7LAVsy5qVecMiMFMcn7pyveTPZXEpLqgklKVPSvge2R84lTQ+vysWU3pZB0zxhY5D5xJ3xQHcRQodMHZfSUKptGLtZymxiiWCeeeomFI/ecXT3OaPpBQcHc2Z7r8iylvPjWyRpRzlak0nGqkrJE6FIYVGbqkjmhFdtw8QlnAd1nSgCED9uMaO7+zrjUA1TnGnSO8ci4NSKyg7tJys9QV+UiWZECExJyhY5IwwSuKMjWlEKTXnmLljImod+n2upGGGcsDS83kQz5q5hqPRShjUWE6phqtnhecPvcOUC7CTc61xTXrolj/02M3KO3ZJrqfsiIBakw+fSft5ixRf9buiwh7inNvkwsqKnouu7aVtkvKRiKQ1HOuZKah64MT/2c34u2+bEW5JSh67smIxK2kESsvHCiOcV4Q+x6JLB12K3fyU1Vyz7hPiaVf8a9q9tlDNGXMiSUk1N55qKi0BYBvpnODqLGxn9pkpWlCQtAkespulVfCTsCzFWgkFOYiLYhIaZEiXikwD/pJeXjwliJM6/bnYX7/9ISlbB53tEQYRLxTj1uv9GhHdZYaf9+4tTkOEaiPc/FjSbCdrGxyB2hyPPIycS+X1/LePv3pRY8XyiSamGpo29WkPHpc77IsWHqcbIlbTBIDdevy405CK3I4qKWDxOe2hyfO/RmdsKiXnPg4wJ+g2PE5GQBAf4iLa9Z4WiPfzKzt/u05ZMe/O/RBJqNf20XbfFhc4Zu8KI2LokFbvvce9oAlIhTrxiEVjpmmtd8nZyny2ZcFv2uAyw3AkFzqVM3IhD2eHcX4fntCMW3nFWGZ+V4b+KSeXGgkO5WYgO185RsofD0XjjgR25ggtt+C4fEvmWDivCI5+y1oZUUnbdFj/DISda96Z9js0EL3roxLUdCxSwXGxfZkT+aoSjK9oX3pEHlZKYXQEmCnHPb3OLkcF8nd2jSxoupCKThPMwVYucsV/xb/GWmiLpmdS8qw/YTVOe+hVXAd7tQgyKxtU7WtKIJftZm5OrY5w4Rqkj14RXuqYOPydiipKNdL157lzqvkEXeW2xOPjV8Q7buWN31rGqEm5NhUUtnFaeK98xDXDeKLgyl6bfJxJ1dMHpPLbBNTzU8Z6uw3mgxu9oUdYdvKo6dpKUw7GwVSrP58qpRpVCg1COSAK82vJDL7E9b+s2crdinM5IONARk57WID/dIuRW4H/8VS9w8e0fv7sIylbyE76nC1/tVFl7ZS9LULWxzlqVYsC1yHB8Tz6lwXD9Q0waIv1oM6bgsdPe0IQgZ4u80nUIcIQuQM06YByTkOTaqL0OZOVIYnSstQLZdJwkVLox5MZu2J7boQzjeyfCkWxz129ZIqCGTXZqCzVKhXpRpljXeiVNv5GN2JCxGlpqrZm6Sd81iuTxSAwz1aMNIdwuj41QC1cQO14jKRFcryCFbLTeAUopLPDKxp1Wb7zmzc3qrzri9YkBcPgVq9ateo96+3FSBISOknVsalrWruNErimlQDCFl4VYwn7IFgsqSldQhk1lzWbA6mQzokds87eJmF1TDRjc2JU0uGASzmPTnVkFX4KxmOLZKKhLRSJxguMjjrnWRZ/oWDEnYR3WAecfJCp1zVptbLxibQRcDVA8Ud7xh3w3eYoAj/0+HyTnXLJkxohV0KAfKtzckX1yMZf0Ds+VLlkFLPSaliqYYFZSM6ZgToU4u/av5IoM63x8KlacvO0mtAoFjhUdJQmrcFUTgneIrLmSda8CclfNY2RLRz0kygX8tEf715EmYZo5VMX+YgWIE6hXM3w74uTlIaOyxfuUT54egM9YLsbcuf8pTTXla1//AOnGXM6zXhZ8J01p1Mjrd9KCwyTnzLeMQiKRIOzpiIhVX0iLYW1bMnV9IZXheOUWbGvBIzfmB3LGthY3C2tk8PQQuvmmGpLhmGrJVAtykuD9smEBfOquONQJ21oydw1v+X12dcy1rPtNc5iadpge/IWrbiQUToVzV/FFv0uHsqBlIY0pq4kPXWulpqPB9/h9DZtbjedC1jyTOWdSsaslHVDRkWAKVzHZvQ5PZ40nV7ueV67mk66ikoZHbkrVKZV0zDRn6Rq+yDYXNHQDrfnNH0Is2ZAdz2TBPpNeJdCLspB14DgoI4qgwmR/RmGunIZCfyFrthhRkvaqdbHZsCnIfOgoSz9ZiIeIdULj52MSlMrGM6QbPNMGcTRsfKNNSLw2LuD9ZDok23H/MlhP0jem4v2Gm0VFhDVlAX4Ui4bh2ohJiv2MhER1U9jG4i8W5PGI1z1212MR4cNa3kzH6R3WI5chKgsNG1+bImoDHxMk+HIpa1pTpdTa3lUoymLjaSIjvGifNAvmxeVCItvQ9O/dpiXZZ/YhM/WzgiUJpnXG+7Q7GqFew4Jk5Zfh3jsarY2zqOteNXI78Cyv/TVJmGLEfdfgTRVGbC8599dEr484dZjJxJ5picbIPuwL9B5kJ/6cC13wqX/Fmb/kBWc811PedHe4qzvMKClcwTnXAQKlNzgBN69/zJOMfxK/Gn3AhveLsHY1rP0f6if8SF5yoHvskPM0uabSuvdD6fr1ngboYcq2TPi6HPAdeUInUYa5639mWKDGaUiExe0GI16DktnUxKOMKfu1H5+FOSvW2rKgssmumADGjhaABJEjeOYueaFnQaJ4w7v7Oe5zJ82pvHJbJ9zKMi7bjmMq5tL0XK5RgFU5zEqhCBK0iZpox16SsTcWqhYufcs25pexouuFHCJKpyTFqU2iE4zLt8L2nENGvLGvfHSScLaApnNULVy1HafUXAQO15XU/XRjGB/i+otNm4i2GXJ2dnXUN7Pi3xrP330kqBc+Pkv4cbMiRYJsRYwNcWprvmBRJbIOnFGbrKS0Ax7Khrtiv++D3/nhux/+zp+/+59SjCT/3b/6H2T6X3/hO6///eu+wO4//sJ3nv3uX7z7k77eoT2BHWCWJFx1njZgj+MEhHABtpKUv5ALFrrEh6DjxPUfbzbrJBCo2/DouTACrXs969jdijje2HWzCx4S+NBNiQFKMOiDVzMqiqPhNQ1TGaNo73Reac2ZXho+j9bcVqVjR0eMA+kPYO5qFtLwxJ0xpURRTmTOAjNXW1CzYs2FzvuuTAwQMd2MhdCws0KYMsTFF4+1r6zw8ibnGvXCfShi4qbahKLMrv1mA7Xrs+kmvT4FiRvu5x09NnhA5M8CmT7KTKaScOT2GEvBREoqaua6DFOokqWuaLVhritMJaygDLCAsRQsqLjSJQgcsM0F8x7TOxbbTCK8Ya3rvrBS1YBdtXPPJTfyrtokbSQlNTV5L2dpymOF5BzrBRMpSUnY1QnjQHh2ON7neb8Rx1G5GTDaWDaXjDJIDZZS9OvZhTVrJktWaL4vxyShWPvQnXCpiyCR2XCil1RhhhQ7rrmkQc2rs40jpAlRdCFyQu7JPs/1jJqGiZQ8lVNLGiTlQznmtu7yWLdYqKcQw4iPSHnGklkIMlOxpO9amp67kqiwDBOmVrRX24lr6KFOGJNSSsJZ1/LkSvnk0rGTpuSZUhbWEarWKaPC473j448Pub7cwqvQ1CmqwquXe0ynNXnRslqMqKqUSQ6zAjofnFwTIXH293aaMyJhSsqltjxKxrxUcwMR4BvZNkeMuNSWLyVTDpO4URmpdSoJhc94MxlzonUIziZr6BAqsYQ9J2FEyiSMp2NEj0wABzxx59zyMx7rTp8o7OmIRjyfuKv+fg6nF45N0hCfvpho3NMtLqXiTCpeuSVbWvDKLVCUh902x27JKsDsoqeLD5vUOqyHWPg34jmVFa9kyTM3p9CMRYBO/dCdcuyWzNQmbv8hecLa2bTECdz3M36s1zbKx6QlG/E8leseGraZ2G5M9Ri8lw5lj3FIYkIMEktel6zZYcI1q56sHWNd/PhC59Q0nHPNCdc2dQwAo+HEOKaf1nRIegVEsCR2iOeO07P4p8E4IdFEz5oISZ942pQ66Zs4sUkVlYF8UF40LsGmjI0QrhsEdtng/EWEE71kTUshNokbwrJazAwt7mnxWY8TkWHxEw9hk7hGxkZ8vWFMn1H2hVsWYHbG86p75asIl4tKYzERjQqJZ3rNhV73e3arDXtupyfS11r3jcNWu97raR5UH6M3UizjXd94C3tNiOsmw3tNLLRTyXrzyZgH9Ep4YUoQ4bNp2Auj0V481yO3i4hjxZqpm4QpSN0XoKUUqCgFBZNQwMQiMKIQUkn7BleEUtm6T/pmnO0BKZnL+33qhEve1+d8yhlf14ekLmfBGiempJaEnCbuta9P0zZFova/e1g2OnHBM82S8E6N+Py+vOLXsjf5BrfY9rv8OZ9yJzlkrksKyVkHePd/wTf4teweaw9/ygkLrVj6lUH5gpJZlB+3dWbv9zDZMdngoD5YkLFgHSDHXbAv2BTt53pNRR2k6g0lMZMxBzrllVTclxEKXGjDibP54V3ZZ0dmgRzfgsvZ7UZE2GCtsNKOHyQn4dnYGPE5HNtaMibjy2xxH3Nmf5CM+KPugj1f8rMPG1bXBVuSsp/kPHBjFkG9aR38qjQUi4UmfEG3eTMdod7xN0bblIlwsN1yNk/4Tn3KHUb8oF7yocyDH8hmorGWrjf/y8NsLjZYDRIZYJxYwTNVa5DW0jHRnCkZu5JxlOVc+pZmkbM3ge2R8s4s42zuOEgzrryt+v71kTDHNTjuLgWTwAQbBz6lxRxDMa3EJpzj0BRq8Lz9G1/5a9cQf4X47l/v+Jvf/i8/A9mKJltg/JJ4FImQB2LOyDkaVdJ+Y7HjNrt9RdwOxub1APPdatP7WgiOIuA/7U1Z4Gm0php8j+L7rgmYPrvib5gCbpzIN0TDuVrXJOIMY3dsy01xCAu/YOEXfMXfDmoK9r4jcc/pZqryVM54LraxPO2ec8mqJ5WZH4cVN622/fuN5x7f29DEL55XPCJJfji2jt8TMdExITdye3sjaRx2IqIJlmlfbzbq2NGKxPNhR254DvHf+PqKjV/HMmKhq37z3JcZTXjP6zAijBJ+a12bKZxUnMvCpgGY3OsEgy6Ukgduj43tZ27C2I16U6WcrIeY9eswEPmjVnsk9Xfqe5Kd+anYhKqUgjQkN01w4o5J5lTGbDszpRzLqO8G+pAkrLUOujA+mBZtvAwWWgXsrV3fkRR9kXjANlMxEYWX/ozRQMY1Dx2lE70KXSvfK3/FZGKTlHR8qC+I5o7HekWnpnjzUs/ZYkwnZkroUdbqCZoRqMAVTQgw8IrVZvPToIQink6UE7e4AQWJCeNzWVFpxwtWvGDFmdacLcTUOWrHaNSRJcr5xZjra/MREVGKvDOz1NYholxeFfzbf/+Ae288wYnFlTxVpoVJiZcpTDLzNHpa1ZROOPMtsyAguEvBl9yMHJvEfkefs4MptCRiRMDHI9tgTn1Dhql7vXKLEBvsoqykZawpy2C81z/nGD8lTk+ib8YvdgZ5q0In/pCCKSm5Jjz2e/39SnA88vsAvOn3e0z9m36fh36H2MV/39kGetcbpOHD5Lx/DRV43O32nUYlSt4afyXCA/zA7TFueIry3F3zgTvjB8kxKzFlqj9JnvOD5GXf6Wuk456f8dwtyEi464xD9BYW+0Zq+OJhARJ/dhOP7BhhzugGN4QU6b2WxuScMQ+qUJufjQnxiV72ilAxkR5OOWKhIyEGxG7i64TlBHdj4ht/Rzx3wVSaVmochagaEye3/fdJMIwc7FMR0hrj3/DfNEhrr0MCGxPvSmtWQX512H2HGJOjO3Pav88m7FvxiLF4qNBmP/9ZaE5MaOKEdyioEeN+gmPJxk8rvn5Dy9B5HmBNE2BZ3Pj+THIu/BUHbrtPmlttSQIRe6mrHsaj+J6IvrmWG4gvWHNHRLjWef97LAm2gjt6TEWCdWzeFbIRhYjnvvZrdt02Yzdi6sZc6ZIHcmAFVSDGx2sy5Ee6wPkYCqPE9bWBvW3ei0NoByZ9MZY2WtvkLFzXSD5fS8tDv8vX5JHlJdwUm/m8HODzvrZ5Fn1/XeK12ZDgGz5oVpRO2Mf8PW7pdt+Ui74eX0+3yBLhB90VDSZDHqd7EQ4cf5/d44aRFDYxCr/7kiVrml6gIyFhETzPJmEiAvTE7yhw8IF/zkfunGM3D+es3HcjFDPo7MKfC3/Njtvisd8Jqn/hfqvnfbmiUJPTNQnrpI+tx3LNTDMq9dSqfDEfM0sd30x3WXfK955m/MLDjknqSMSUXu/KKMB1N8+WYtzIR6OcTIQ30hGJQOqg/h+/9dsP/xejOfzfzRmXUpOGvQPgWuqN3Ho41sE/LMORqilYCcJULU5Gv5Dhmor71XFj+/efNXPWzeYcj7KUp20Vmj/QfM4qsqmuNXNKzMX9QEvK4B2iaI9+6AY//Xu//i//+WcW5E84fipFyPAYFh/x6C+OwFXradQMYJJBUihAKsJF1/J9/wFAb7gjoVsQj5jcxuR45iZsuyl7snXjof68Y6jsZF4Rrv/YzmOTUEReQwyosaBZacWbyV12ZMqD5A6Pk/t8OXlEQ9dv/kBIfiLpJwlE4ZprVjz1ryik6EmS135h8rwhGPUKGuEaxGBsiytqaWl/PVS9ubh+TjACg6St/bp/r7Ggsp+/GaSGXcG4gUSDpqEaSTw82psJ3ei6iQtVc9IresVOIdCrY12xZNttEZWs4ut7rAhQTHJzFXgT46AQFU2AJpTsy1Y/8hWEpV+heLLQwYnvU1X7QiNOmqIS2NBQKW5oWehO7UgUpDNo3UhT0vA+dmXKhDIoa3luO0sg92UrTCusM2eJqSmJ1Gp/17qmoWWpFVVYC1E15poVV7o0c0c3IZIxS8m4xQ4Har8vqqYlOB7KIXlPJr3pWAtwKNtYR1A40SvuywEFWQ9LuuXyfjuNwSfFUWNmdHeDUk4WAmTMZT3K426XXJN+/Ru8w44XrDhixB1GHEhO3YUiIvOURUueh005UVSFNFWyzJM4m6SmqXL79iVffbQiH11yuRIuV8IX3n5lZqqZxZBO4apW3hjlvGpaHpcFb48KRonwdlFy6hv2yJlkwj9M7vL2qCB3QuGER2XBi+DQnuHIsY3m7zgzBstwJhagCctQiJhEbYxHVsh0KAdqngopwhwjAO6Scyg5S2IBK3zgzvhyZ6//0O/x1J0D8J475qG3rvGajj9zz3nkd7nn7Tlp8byfnPX39Y6f0dDyxF3053Muq77oSNQFmVYNBoAGDR0a2IEppkXH6Sh2MWfVNzGq4HPyY3fOnJorWXPlO+4y5kPmTNVgCpFm/3qSFIuCqCsfN1PDgtsx1Zwla+4ELHfsIEdIIMCZmsFdbBjYeutuNF+SkLSZcWnbx9AYD3r4kGygUVGKdUMwToN7sxVCDmEm0xsQrKi8FJs6tm9sYqTqxjMgJmF14AvU2vRd6TipbbU11bwQp878JVWIlSnuRnISORc+XKV4jV9/7j/viMVHjEvx+/NQ/NrnjdsR+TjDRlUs1IaHD7H6uZ6xK9PeLC8eThLOg+dGIo4dt8XSryiloJCiv34GqcpuNI6GR/SvUt1wO5I+3rn+e4bJf/x8E+Cw8fO55OSSc+ovqHzVc0+3teSd5EGPJoj3MvpMAZz5S+a6RNioZyqeC2+GqFfBtG947VLJ+u+N8sM3k/ZNkfx9nrCUhkOdcNcdcN8d9lNAsGbj66nj8LkbQnKGezxwo/kIBtF6z52z6JTbac639Oe452d90dbhmboJa++5qj0vnanQvb5nN68ZPccJSTQeHD7HcfK01prKV6HBYJPAmYyDZ9jNnORH3ccsMd+MQoQL35CHXOuWbnGqJl9eknEuFbV6DrOMsUs4o+ZSKhKER36HFh/KIVvvYy14353znBXPWNIpXLeeyiujxN7ln3+aIYL5XKWOa235WjblnjchoeiK/rVki6pVJplREb76xoovP1r27+Offvs3fzvB8U5o3lTSsZRmcx2lw6lxQaL61Uo2BX+hST85HbNxWy/VOIsNnhda3VgL33lVsW6EthMe7MKDtPzMGonTFYeJ0nhigbJZZ1sEHx1pqaSlwTPH5NZjbP+9X/+X//zf/Pr//lcWI8lv/dZvvftXfdNf53jyuz96d/goRAjWBhlo/zpi90BZeU8Syd+BvFep5z/yERGOlAfsKERvDLv7ecA0Pkhuk0vGlS5YEk31NgPQeOmGoWyjfW2j1R4nG7CbeeBUmA63KWltjINs5LzjZqRhE4rB+plc0okp8qQ4FtJwyIgmLBLB8QP/MabKZJrnaxrqoP5hh23SiSSsfRW6boZHjqE0vi8fxv8R3tP/tEQwhwUAEXtvpRvdIEp2GqY6EjHjdl2G93E42o3n1WKkfyOah2sgcaRPT0BPMRnDoUpXEyY8BiVIWdP0k4aZmwTtcEerkaDoWPpVGNEbDCBubDGJqAJe3Lo1lpAb/Cw6sIaBtUjAwm6gYjaajrhhg2UlYW2UYSqxI9O+G7gXoFhxA05x7OuEPR2zLzO+oEfs6ph77LKnYw7ZYuRKbrHDVMZ0YnK7RSDkz2SCCwWehOcg8gyiq/NYCq502Y9AD2WHJWs+4RRBeCiHrKTmn/ivMfMlL9ycKgxUN9ALg2+Z58Lm87fZ5q5u8YZucU3DlRp/w4LzJihOSKlDcFoRVdpsVPxAJ4FP4KjFb+QKBa6kodCELXJOpGKG+UosO8+HF3B1lfHJacYbt2p850gSJUs9ed5R1wnOGdyq6xyLRcHe3pzRZM7BNOXWXsPZ6YzJyPPVr3zCgztzDrc911cjFrVyf5owzuFX/+ZTPn62ze4Ibpcpt8uUq7WyVdg05qzx7BSO91drjvKMWWJqUa98zSQ0P/5IjtkJ6lYOoQ4GTcOEL46zJ6RsScoRJV6FLXJyHLsuBRHGknAWEuE9NUjBvk54P0AE4mZwLHMU5VoqBDjSGU/dJW/4XTPZQrjvtzmXFadBFaqhY0rBlpZMg+SwYLwJU3DZOIX7EAsjjHBNNFfbNBMuAtdpRc1ESk71mjkrLllyztwkNNWxoO1VVQgxdUJOJRv/jHjI4A+GDyLVBMQS21NZcFu3qQNMZkLBCpuQXgeVqhjrm0CUbbWlxnDs7fD5xvXnFDdRc9Xu2yv9PhAjXvR+iupZ0SwUCBOLmg0PwpL3SBz2oYCD4STYiqHhZAigo8Wr7zl8rbaUQX3JIMLRH2LDoxsFadCofBb3qR5a3O8Qm0l8jJVRTCNOPzLM1HPYrIgJblTzityOOO+OMT7Gw3UfpWytXKiZ/c11YVA5bZi5KVsBwuzDXqrh3qypmblp4HBGIWIdmNxtFL9MLEX7hlinHVtuRqWVL1sWZAAAIABJREFU3c84ce+LAQ0bv/TNrwfuFle66GFwgjBxY35O3uJClqYmFaZT6hIe6B7v+097/xpzRt9wO2DDfOjXvnpSZ2pam/ux4a+CkkhqPk8D7mk3aNrY5NCgf5/oKe/zgr/NOzaZcaaSaA00K46Hap4x54kwwNdzsLgf2Huw/W4kBf9Av8pMC15qxYO05NOu4g/ce+E+G7fwv9Jv4NTxTCuupeYSk7Ve0wT+UZhECv06UaDWmkJytmTCQiumUnKkM66ksvUgULo8cLsswW3xxiUSaGggrHAVZUdm/Izb57qznPCBzjiTNR/KCZmkHMo2GQkLafjIXfADPect3eGNvOCwm/BFt8WfyAnXUoUGi/mCeFG2KPmq7HAoOUUijBJH6xVEKFPhuvF8852KZ2cptYejzHLReec5kwovykO/hVe4Vdr+8YtvVXzvoxH5//StG6ihn/mNr3/n3/zuf3jXIaRqst0ZCWPNmASlxRFpz/WLeceNRoNYM6eRjlLNQHBKxpQ0uHvZcKDEvKGeLz0PJilpovzRVRWeb4MW+7BC4pp0bCYckTMiGExrSsZUc3YoKEmYhWlzzYZekeJ4+js/evfRb3zpJ8KzfuqTkBsvLjc/dmIyvZmYGksZDLDiXw98171iQ7KykWs8hqNZCyJhtBfGsQmmHd4F2MMQTjTs3seu0fD1YgdNxFH5VUhkC9bh4YkTCI8pmFzrioaOj7tPmVKyxJRpzphz4Mf9+cxpmAV1loj3i927Tj3bMv1Mt8ejfWIca9So2d6P9NVGvHFUbNc4GQSkzx7xZzuNii6W5Mfr8fqE4zM/F4Lz8HOvT5CiB0M8hhOpOH2J73euy/71rDAxbGHlq/7149fN8d22yJgkrWn7JKmmYSoj1rqZJEQIwFDzPSpyRKhbhDGBdc6i2aOqwZ5GUvRd4gLz0zBsdBu2Ac8aIwB/LKdE2dENDCThwI+ZBEL7Hd3hSLfC+tj06OKGGMUTIi68lKw3zxOETBL21eACdk09zznjm92bKMqazmQ7Zcwd2eO2GIznjuwRibm3ZYdETIu/kjYUHC075BxQcCvNGEvCK1nRouwQHbEt4Gc4Hop5pCQqvGBFE9zeHRLMDx27mnOkJTMyzln337vEpg0AF22HCDRNmPR54XqekSQx8bC1E6cilxcTfvSnX+XkZIu7X/1jDo8uefDGcy7PdllXJVnW8JW3rvjGo5pRrnzti6fUq5K9MVxVFoOmpfLlu5Z4nq477o0TEoF7uU1IVp3SqXV0ppkjdVZgvJeckeFY01FoLNw3QTqqha3xPRTrIEzyBOHUN1z6lo/9ctCBtn87UR4GaNY9v3NjOhHX+4qWR36XD4MXUfz4nt+60e164s7p8DwJU5UIIb3nd/pnIEpVx98TJxyxpx5/945M2JEJEyl5pRdEF/hoBPhKrnjp5jxz15yHDX1bc+ZiLIGp5uzqiG0tb2ygsROahi5filBoypKGmY76c6ioSdRxqFsYSbrlLHQ6Y+xZ+GU/3YhJXaV1HxcSnJkQxj8Dfw7fJ5aRbxdgWAFGFWNpHmS845TRq0nbDq/VsFsbj42J681uddxLYowfy6j/3R2+J4Vu4lJxozh0ulGTiscQErSB9bjeu2WDzrcucRaaDcOjxQdOR0csCezzGzGMeF5rWta01LRc6bI3IwSTrs8l4zDZZakrLnUeOuTmWbHWmigve61WbA8FVrwaZ7HfXwZfs0lgkFvGvKDi5KgbcBHjPtpozVrXrHXNU/9qUKBaAXrpr/mP+mO+yH323DZTZ/v3XE2SQ8T1kTpe7+hTomqF8+t7eMw9wPbluM5u5jDB3FE3Pjjxe+L7jFDzkRScyIKxZtzVnX76EPOY4V4e31tcT8P1MDwsX/Chuev4Pfku/1q+RyOeReu5dGtWWnHuLykl5x/pN7iflDyRBUtpuRSDT81DgyzK3sdrH/8d/u6Y+9S0XMmakoxoz7nFGGED++vXP0lARHjGMmImUw51Ru2Nm5aJ4/eTj1lLx0rXzEI+lpPxze4uGSnbOqIDXtRhgtW13PdbzBj1+2uc9h34Eb8vz/g/5Rnv1XECaMeyVRNX8fDNd1ZMMiFPYNXdnPReSs1hltF4uLvrOb8qiBCs149/+u3f/G2bfPgeYdCJsg4wWpuuJExCYbKtZvDsMfhqFqYkpW5Mtstw1c+peyUvMMGZDBODaTrby028xMpSW+Pm8aU3IspQ2dBea0zCDlkP1Y6z1A28eDN9+7e//q9+4kTkp1aE/K1vf+szF9jrzUIkF0cZCg6AROwEGlWuu44/5Jgf6pP+go2kNE+PEIiGxoJdgP886V5YEhf4CfHrG1LYZxPyuPm0wcxuJOUGt6ueMhjuxAR4HZQ9IqxoHYhtn3QvERyXLPm4+5QX/hSHcOKWtFjXcRmM+aZk3NEJWSAsJ+J4lNzhgRxw5AwHHjfQ4UM7PP8IebJrl+Bf53289v/4M/barg9uw/HbsGAZjvOHfz8PfvWZ34MfOLtrXzQNx7TxiKPTta6ZuQk2YYlTns0GDQGTPMAGx05tTC62ZNwnBjNGFIHLUwcyvmMjVxyvUR0wy622tqbCxw6h0baHYSx0GUhxJgQ9x4KuFw3+JYH4GBLzd/xR/wDaNdkQRAtNOPITDnQcuhsFb+oBM0Z9NzWTyL3ZiCcsdU1CwmO5jZNNErQvs3B/bAN+wASvhICS8s3uIbf9jB0dk4gpfowDv+UNv8ebekghGU/1hCfugjVWOMRn80/klBEpO1qwxlRMol/3AQVr9dxm1P/N1JFoxIl3zAIkp8FzIgZj3CJjLR3nUvNMlnQY/2TRetrO9SPu7z8X2jYxrobzqApN42g7R9M4Eqes64TVq3tkxZrLs12OT8d0bcpqVXB1XXJ4dMk7b53hu4TOO77yzgmPb7U8uFXxztvHjMuOVWvmh/f2W/amylXreb7quDtxVN7zKC+YN55MYKoFb3d7vUfKB8k5a+msW89NWEOBKarMtSMTYUvSQA63QmVGxi0p+qDtEN5zx/acIrwfPs4whZJ50Cf7gXvWJ40xIX3k9z7zLAJ9ARITSY/yxJ31iSSwUaMaJAwR+tTQsaSmouG52s/FOFzpmkrNZWJNTSRCe1GW0vRCBdeyZi51/9pjzUw3n00RAAR3eSvgxmSMyTiXVX89RqRcihXdR7LTP1cJn799tcEDYq1rKl+Z67ZuNOViUyp6U8QYmEnaczxiAfJ5RqxmtJYRJYaH7wWCcMdP+NkIZnKYClQZzFBrmp4gHyG9S13178chJgcerrOXjbpY/J2vcwc35+t6mEtOFgwKkzCNdv2airFrGXQG4zVWIudk4ysVv7/WhpWu2ZJx39jJJSPD4DeN2kS5045p4Oo5hB23Fe6vu8HLi+axdg/kM8l9gu17qtp/bxLgPq/v9/FaR0i34Po9Mv7OCGVutOZM5rzFLR7IEZGXt6ZlLKP+dSM0adjEXGvdv16EHPc8kHBOsZk6hIf13exBoTosopIgkbtWI2f/kb7PXGq+4vcRHFuBh9j/3tde9/V18Hn/VzVExbXOSTHo4b+T94zzoBGeKPwCjxFgK3P8ofuIfy8f8Eovepn4TNJeAS02gYfnYqbFdQ9HjMXJjo57i4IdNRGKYQMmXreUhMmgsXYmCzxwkKU0MRfUY+a6xGPGx293e5xQ8cvdXX5Jb5EA+1nKbmZF4X2x5kgUSgCDOJ1JZd5EakIeJ3XLdddx3Xa0Xlm0nj/+YITvhJ99a8miUWapFYCCsO0LfjbZZqewAqXIO+Rf/KPPLUDiUWAQ77V0lJr0xcg4qKpGX6JVmCxvkbGvZYgBRlyPMKptDPIbRVQSDBKcIPzKUcEvHGQ957Mk8sFsr14PpsLDzK9j48HW4PvPNeGv7Xl2FceDJnRs0B0km2HC68dPDY4F8Mnv/uhdoH+DsKmeDBqz+Te+oSp8nIhwizGJjPiUCzJJ2XZTrnTRd8CnbkwdHMTrMJ6PFXgqJtnY0DJxlnhVWtF7jbAZjEYOA2zgCKWUG9xnDF+BjxHhW5lYvfhmcpe/273FL/CQqdvCCxzrJfeTI7axpLqm49qtyTXpF86IhDc45E2OeCiHHOiUkab8mX58I9gO8b0xMVBiUDKndEWNWC92xm7Y9QjGjhGKZZjmrIcXRA12e9A1GLJBFFiEm3jROFKN1w5u4o3j+DeaJnUYia0bkN5LKUBMOtHIm0YgL0KHMT7AIkb8MwOrjSfMnttmIiMWuiRCqzp8ryseR79RV3/XbaFYEFvT9D4ucUQOBgfoUDtPhG03Y+rGXOuCbTdlKqbao7IB9EXjra1e8hVO3JKFNEwGjqFDsjHhp+PHIzIOdBpkXAtq8Txkj5W0eDFImEkHOvZkxrFespCqd5C9khVnQUltLAWPOeJIxxTimJDxBd1njWchDb/AAWM/IcFxX3f5kj8Ko9mMlbT8on9IRmL646EjdpBmnHQtZQiKU1JOZU1OQil2zV5SsUXGCWv2JOcidLgOKbmWhpV0nEhFK0ZgjwVTgmMrqCnF1GCtyifnjourjB8cw+PthFGhZIlnPKkRFZo2YTJq+vvQdY755RanpzPqOqXzwnyRM5/neBWur0fM5yVVlaE+Yb3KGY1sPV5cjHl5VlCkkCfQdsL3j+1pmKUJnTcTxMRZgBeBvMv6CPLYTfiy2+bC27pRCDZXEhLpjQ+PadkbV2XHZdRqK+OZLBmTci0NEzKeu2suZdUnfjERnQ+w+AU5Z7KgCsXDgU75yJ2xq2O2tORaTOa0vRE1IoRG+3ONcTlu9FWQoRZMDehSF2RB/enEX9rziqkYjV15ozud4LhgwVwqLmXFsVwhYuaPvXuvJpy6JQUpDTYlslI7FiOmf+9FeSYXHOkUsI05whSXoZg5DVyQKGLxebLhMcZZot0xcaMgp2vyqRYPk34CEk1gE3G9mlEkTA/fpwtVchbUlhwbX4QYg9LgHv26sW2MoRECmzuDGRvfqumT2AidjSaxghnsTmRM9FbJMYdkLxsPkQhbinHUJha2D7jP2QMjvDmus1WAmK163Svtpx8dG7NUawC1zLXqlQ0XuuJKF8xkQiE5q8BzazG4cBV5mGKGvi3dxr9KYOLG/foaCqXEaUCckkSocF8oq0nZz9yUQnLmusCFZCzuV7b/D7kQG8PGqGoWVTBrMWfvipp35C7P9Yx7ss+ZmBmlotxODrjWRVxpxGloEkQGNFzvKLSyIbXbFGXkSlZaMXFjU5AcFHWxi+5D6pZIwu3kgCUrO08cpSvY0hHvyatgnliE6xyVkhwM7vNwv44AG1vrQeEoIAy8ev5bvsm77iF/L3tAnggHvuQKx0oaviVv8bgs+W51zY/luI8vC1ZsuUl/T4eTongORTjHVNL+2WjpuGTJfd0LHM+Ml3LZ5yUlGUtsUhIlcPdlxq6O0QAxe4cdSiegwlKF1GXmP0PBtZii3s+nO3g1P7pxknDSNiw75Q+SJ7wn5zTiuQ6ebIVkLIPXUiMdJRlnUvElt80ocWROehGTVIT5IuNgp+GT04wndc2JVPxSuk/jhe0k5efeqsj+xbd+u/uHX/yJUKR4/OHv/n/vNrKRwheERizGzIJ+15qOGXm/5mo8K2kpSHrVwx01b6EVHd9Pjjl3VZCKt73ph4s1Hy4bfryouZsWPN5xfLRoeWdUctyacM2IhP3QwJ1jim8RxROnJhkuTDpM2r3BpOLjPlMPGjQZjtIlfPw7P3r33m989lr81OFYwwLk9SOV2Om2x65SJRehcHYa20nCF4IKTKyAy9B58NgYdyYTHrgjRrLRlI4PbypJL4caD0uITQVrOO604JP0G82QsA43x5mxHlzrmgTHN7q7/Wb6fZ7yI/+UPbfNl4M61sNumz/273HKNa14Uw9CuQyJWopjrBm7oSv4yN3hgbvVb0Txd0aC9FCJyhSmin6T+owS1aA42Hxuo9DR9ydkc43jv9GnJJOs77hH0nZUtcrDpGEIdSsCd6KQoieh9WN8bcP1j5viUD3EFMHiNn0ZSHyCsIowLBwTGZscsr9kz23jRKhjVzb0mNdac6yXZCTsi3m1XOuiJ99PgwGlKWvZe4xj/3guF3rVu/OeheRrxZpX3VkgIK56GNSpWzKXdb+BAGH6NSDkhYA6/J5KoiiAdYtalMd+j3NZMQ2qII2azGUZzNJKydhnxoKKfR1zyQIXBBNqbXsC+StdU9HxoMz5ejnhV9IDrrTll0c7jDVDAsQoTma+EgjRRzpmSwt2yJmS8qRe8yXZ4oGMaOg4loojLXuzwmPWZDheUnFbSlPTwsbHEKQC1TEiJVPHESP2KZhLwwEFDUb8fsGKl5g6R4eyWwpL7ZivQb2R1JsmQZzBsYqyoeukJ7HXYSpSt47JKCTGDlSFzgsiRm5XD14F7x1t63h2knP3sKIJg8PEwd1RVNaz/48yoe4UB5w3HY9GOffTgq+VE+5PEo7bhiiInGCdqW3sfq3xvDmxjWPhbRqy7VIq7xlLwixMay+lJlGhDM/FwzDViOvjWld4VXwQU4hmnCmu71Q/8nt84E75yJ1x3+9w3+/0cXHIQYjNBYMSNv3ajtAHH2AyjXaMpWSu0RjQBxM3I0bH5yJ27ePzVat5Mb2hB3Qo12GNJDjO3IqJFpzJkjjyN3WnQeNEhVcypyTnVVDdqmnDFMQcqw90yr7M7D5/zpQB6GOXhMIgQi8bNd+pIfQqJomxZGu0tUIlEJ1j3LXntOtj61Cx0b62+X9UbIxfi0fXtywInLcNtDbyE8duRK2NwWyJ0KekFyqpwmTKITfUzaJQSdwv4u+Oe95wOpCGuB0nZAbFNEXGa11RBzjrOnwucgHimlxQ9ROwBWuWuur33yit69Fg/JhTBe+NUgoT4QiGok6i94rB6SI/AjYqWDBs4tg1i8T1e8lR+Lo1H01GtrghYDMsQGzN2WSi1YY2FCDD6dHCL/hh9xEfdc+5kjU/I48AuCcHQX0z55PuxWtFjaEubDqzmUJFcQMgNN7s/lz7OQ7h2s8pnYFmhvDlDS8ziEvoNbcjIV0STrnu9cuGx1jMIHnYjPy8yUhcE3Gi5UOBVIgRmf81T/hOfcJ57TnvWra04I7sUTph3SnLQIxeacVKK3ZlRq2GFIiF//D8BUetDVM3JgsiD0XIN/ZkxlrsZ7e0YJsxE0rG5EThhyH8b0nNLT/l7W6Pjo5zbzO87Szhro553O0yocBEBca8o9tctB1JgP/Pu45SHMtwLxo65lREZbrn/pQTvaRDg+oefMXv80lX0Skctw3Xne9VrgICizwRjkKn/1XT8IXSJnTn//2v/6XTj+Hxz779m79tHD07VsHDCmKS74NIRBdwJFYYjEiDIJKpa12HZk2LJ9coTW25yRpPTnRIT8lTqFthRkrnTUhpTMqjvGCSOK7D9CSuaYUg2JIwp+1Nbe3rUJIQp/1ZHzOMsH7WNSw/B6kTf/andvxlBYjDSKAxkHugDIsDoHCO3/cv+D/cn5FLxmO5w0rXfQenlNycRXXOGAtofdc+PFA5GW/LXZrwYEVVraiCFTceoO9QRAhRpx1jNyYND4iNRrt+tAgm6Xvb7SPAFHOj/M/8F7nvDjmQLf7YPWGqmZkUikEpiiBfdupWnLoVZ4FkCpaENHju6k6PpYVoSLN5iIEbeM+oWhOD3XA8Ho+IEQV6/GgsIBI2I/A8FGxR9SoGqFRSxm7U//742jkZpZSMpGDsRmw5k9eNBYzDTA9bbfqCZK3r/tzrsAEcJiYjeuovSHDUAXIQ+SpjKQMvw4L0REZsuyln/pIrv+hhAluMyUh6w74I02rx7LktOswIcqkVOVkvMzgMzFM34Si5CWvpBhuKBdCMceCLXKhJ0TbiOXeWrI01oxPPfABBiR27jWSvQVJqOv7cvewDCkChKQ/9DjPiZiLsMOG+7rLLjEtW7DGlJeCeB+dfBm2bJ+6KR3mBKqhCmQh304LOw1tuwhsy4W8flBxQckDBV9NZP4b1KNvh2hwmObPEUal1SgtNWNJRim0HhxTsBunEp7rkGUsKEh7oBLAAeIcRLZ4j7JrtuIyZZr3cb4Nnn5KChJcBevPpwvNcVryoOlSFk7MS5zzT2ZI87VguC5Ikkj+NsJ4kSuLC1MkpeRqKcg/TScO6dogDJ0rbOp68LNidepJEyVN4uVC+f9yRJxaTLtvgaeDhRdPwomlYq+dZ1fD/dmd8t7rm+dJzN8uZkPFQJrzpxiQIx6y5oOaMNWeVp3TWLVr5oJaXJEydrdtdzbmlIzpRnsq8h1UZ92NtYgu9eIL9O2XEOvCmMhLec8e87054yx/wOEj7xk1sCFcawq2M++H75yR28xUNiP6ubwCc++sbUJn4/A/VeYxAuiFuP5dLXskVEgrsvtseOEPlQLp3WCQ9cxf23NGxZM2JLIj6/VPNOeWKtXRUNP05vH5koWiPz21sZkW/KItLbf+39454LaGLBUhMaB0Gzeol0wcFUBu4JhEy9foRY2osdqILtUl529REsUm8IIyD3Kji+0YbwLVfBAlTj1PZqNOxgeMoQ1+Um6DakrxPdhtaE06hYsE6eDRtICnDw4cYXNEEBb+659tYk8fU1SJnptKaLZkE5T+LgytdM/cLDEI0Y8dtBUBYcmMCFJtjQ1RAhAPFe5qI4yjZ43l33BciNU2A3q1u5AXGubGiY1iUx9e1tXdTNjl+/FRfkYUk+EinjGXE19yj/jzidY3fLyKUbtT/3k1jb1MUjAdfz7B9MSo0xs/HyVx83cqbVPMWY96RexTk3HVWhhiSoGAn7HPDY1iMDY+YBw2vhwt707flu3ypO+DvlQfs5o69NGVXS465ZJQKH9QV/4+83xfao+DJFbmiESI/vI5AgI3Znwg9jvlLoTaRWUrDWDNGaiujomFEboa+Qaa6o+N7yaeUJLzTHfbcBK/aQ4Bu6RYP/S4/749QoHSOQoRx4tjJEl7qmo/cVYggQknen899Z8qS52IiR514vpccc0HNH/pTlnRU2lF7Zd4oTWcNr599c8UkdbzJlMM0o+qUL/3Or/21C5B4/LNv/+ZvgwkbOYwnuBLjni6l6UViGuKszI59HTENvmXbWrCmQwVuq133HyYnxrkLseg/v1Pyd24V/b75N44yfryu+KU7iXFGEmHR+f77I7dDMA5Kh4Z/N2id6CviUZa9TEfg5cbG509oHP1U4VhPf+dHn/taQ7hNNAV02IY5Tixk/KE/49/xo6CWMaYkZywFt2WXLZlwIFts64i5rLliGcjHcTAlLHXJlpuyZM0C86AwLkeENm1MfWwqcDN5T4KqUlTASkkDj8Ie2lRSvuge8PPdXX6QHHNXp2Q4JqQc6IwdHaFi2u17WlC4EW/6PZ4lV4yCasArmXOgkxCEXHC7NDjPE/8ywH7YnBMbk8Z4/kP4gcGrNjc2JqamImKBMWJzh/4Rw9fZjLl9CBKh2BHbPp0YMS7iblXolUgUeonhQgoySYOhZEsWHNpzyWmCwssqKNskkrDSirXWHCZ7rLUml9TMkMTObamr/vy9mNOwI2GhK47cHiqhQxwUdeZacUussKloegJcKRbIRlLgxDgfinWOaloQ2AlBciQlJ/6CmZv0kIt4vQwj7HuIxCULtmQc7lWEKXRsackoEN0AFtL0hUaEyqQ4TmXBnk5sTKrCFHNVXUnLREqmjHjk93oX7JyUseZ0KCMpgou0JYunzHnPnXIuCx51B0wT41eoQpGG6aOHO6OUqhGuGs8vHuTsjaFaGf50QkoSppJJeEb/QucIwiRwGATjORiUZ6MrPiLlKEwsX7CiFs+51NzVMSnCO5OCw5Hjcm3PvIEiNvC5Bs+UjKV2XEjNPgW5ZmQJlIVS1xll2aDqyLKO8XjNclnYfRx1dK2jaR1l0VGWLdU6pcw7xuOaxAlZ1lI3KSLK3qzh+CJnZ9by4izjojYFmZ3CpjyZCM/XLYU4pknCwntKcZxrwzdH20w1o/KeRmEmds3W6slEWND106AxGZPE4YG1KkvtWKnp6pRBAWVJx7mscVjH94k7syIkxK2oeqcoWzLu4+ibesCpLELC4jnSGS3K++6YC4n+9jGObLhJ8fsbjbR0BcEmb5IGSNYGXgmEZ3XVG78NE6yxlH3yao0im0A81D0mmofnxuJEJS1bWrJyba/C8tQZr+ZSVn1MyjGuxb5OaPFcy5oXcsmVn/OpP+mTK89NP47YXNhAkTYwHjAzvGGC9HlHF7ZODe+xn0qowZJyyUy1L+wdEQ4WX/f1qWeMpxGm08OM2MBhhmyMNKgUDpUSO1pAmLoJKQlbMu6hUQhEg7z+/faF0AYgmwcVQpvSRRUjUzyLBWeMdXX4PoVenrwNHjIVdRAQ6frCc8fNqLUJqkwmq1rT9HyWvWSbWhv2km06PNc+CIkMzCI3RcdG4QtMAKYKDTSvGyJ/z5URKxQvuyuTgJcN7Cx2bu05GniNEEu1wCsKUKxO217tKgll9rFc80xPuSU7lFIw0ZxP9Dgk0K5fM1Fyd/jaeRSz0ZbUBbiXNsHLTBiFgiQ+5xpyDpOktdfvJ0OieIH7usstnbFU5UC2OQv+RakkzANsPf687TVdeCdDiFZYWRLWjMRzNelhTTK6OiPzCftFwv/lP+Xvd49J1PFC1zyRcxJJyMIkNE6+orLZJieLkygjk0dTzOjvVYrFh3f8PitaVGwNXoiZVK+DxpIPDZPYMpjKiF0d80gmpGI2D06EsUtovZBrwl1MNGUkjv08pVWl8sq88+y5jE9ZsRLjhRaaWrLPmnO9NnUsGbOjEyKE+tIZ3mJbS3ISdpOUcSZc1Z5pkvLJSc5VrVz7jj/VC/7+//qP/5MLkHj88e9+713p75a9Hy8DhIsY5yKa7pqJoMGfPKaAZW7nNmluRaml40jH5BgE+nvzig8XLW+M8n4tfLRsydqcL+w6lrVw2rRBSt5QCkV4zcgNScIiVF8uAAAgAElEQVTHN+H6trdbUbuB8hc4tlxKJsKdf/LOZ+BYP9Ui5OFvfPE7P6kQiVK8hXOMnQXfUeI4bVv+Z/kTfhxcp3fcjC2sCIn+BSKmlewQtpmQSsI9d8gZc0zbx8Zrhmtv8Oo3jpnEke6QOGj/j2ZRaUiYzWzHOmctHW8l90xe1e3yBkfc120q6XjL75DhqPBc0rAdenZ3dco0UIuPdMyEDHAchEpVxTqYkQgUZdEm5NxlH+9ko/QVAmcim5Fky2ZaYiPtELADtErE4QJ/xrTrbeRdBJnjIZEybp5OTCM+blYiA5k9bnqSpPGcSJjKqC8MUkmpwrRDUSZu3EM2BOld762oMSWPmoYdt0UpOSpKRsYoKAmNJDfscJhEjWREGQLlWEZ4OhwWCNfa0ErHlow513kwEfT9fZ7rirGUJDhWARoQC50tN2EsJUtdcyBbpgojceO2rmpGykjMtyBe/5SEh3LEMZcsqCjJSXGUpHTiyUlYSsNKjKB7KWvWYsnXn7uXfCJn7DGjlo4VDdNg2JThKAN5d0sLg3dJx1wqEhwLqUglYYUV4oh1yX0oTP4b/2U8StUpO7njqvGUiU0g/zf/Eet1yv284O1dxx8cr/hg0fKrdzJ+7mHDqCv7Mf8sc5w1HR/JNRkJBxTczjMqr3TAfpKxnW4M/Sak5GLwLEG4xYg9rEhwCNtpwvZIebLseC4L9imoQupmCh9GjvtUlozV+ibPqpZdl7G/3fLx8xGjXHlxUtI2CdvbFUXuGY8a0tTjHOS5ZzyuadsE3zlW65Sd7Yp6nVKOGrz//6l7s1/Lsvu+7/NbezzzHevW0FVdXT2RTTYnKRos2WFgI44dJ5FJJf3oPCT5F4IAQQgaCIIAeUiQpwgwYAdIYhoSkThObMCK7I4lSJZoiZQoit1kd1UPNdcdz7yH9cvDb619zm01NZkSlN0gb9Wpe+/ZZ+01/Ibv4BiNF+R5w7DnA9k95cc+McUveniFd+YVR2WKU8Oef9CsebFXcN605CRUHu76BR/InEes2KPg37zjeXJuYqiv9EueVA1H9DrccOnsd41dQnSlXYQu2wcyC2Gv8NhNuaIjzsWcf+vQ8ejWuwQiNQnPZEZUmvIoj+SCp3LRBYd19zW6WDeX1I2MTxYVeKTz34kBvKpnqSvysH4TSRnJICQcUYbWRBMiCTmG34VkXMiKU1lwpKNwWAorqVmFYPbMrTqX3ZqGaDgYDQUbWqaBT5SR8ECPt1T5lKHr2z1vcVOiW3nsfmSShc/vw+dNu730o1dXIOsCYLqgehNUZqED0HaiFTFY163/ADYy6NLtuS5wTTTIpnqMa2ZSubFoYb8tduOr6GchCUdulz0xLR81GHyXbMa7gA0WP/5eBeaY/8KKOnTZas51YSa8gYOiW/NEUQox/5uFLlmrdUG2ZYhjsarSOuzJPsC0aspQkIpGrykpT/0JsdgXxxCkO5YjBEvVs5dMEFxw4ZZOkGXHjY1zEzoxrbYB8rXqnm1MDoSNlPBGGjc+1814dRLVgW8Zn2ZNQ6UNDTW35AoCnMuSY70IQT7dORNXVgzzHcklKXuPdeAyZ8+150rWuiaXnLWu6TlTg4s8R7BuVU9KQ2KIkbpPZMGHcsIjN+Wvy21+hKvMcTxjGtzXQyLfde1D4ht4lbL1n+v8uiLU0FARKyruyjFOC/a15J7O+Uyyw912yQ4Z+zrmrjzDIFwZFQ1Lv+rWJhiqwhAkJvoT/bYarNgxVdtx9mXEDR0xlZr77oyl1KxCUlPTUpIH80oz57XEBZ66OXt+aAUOtUL247rmIMkQFa5mGa0a/L9MHGdNy06WsPCet5liEEorBraiTFmaWmTgI10NxcxGfJeoveB3bX/DMUlSTmvPIDV47ydvLxgkKQNSfup/+/KfOAEB+Pwbn33zc2989s1HX/v+F8dkXIj1GNYYYT3yMPKQBPRIeSJL8w8JyUo/7CclCVe0xzUdsMTzbfeMqwGt8NN7A8pcab2gKtxbGkLhcCj89pnxrQeSsA47YE5CnyTAuVynuGUGs67bvWJCEv++S84oJCB7RcLkyy//6XNC/qArEaFSz8xHHoO5McOmfTgPrb45plDQorggDXDLT0gxk7VFOIQ2sADP3M8NjkXLwi87dYl4beMjt1vXsGmFyta/3/MPKckYa4+rgSx5z50yDve67ROQ4DjHiD3nbGRpd7TgQipyHKWm9DVlHFB9BUmoBLe8657ywD+7NF4iQhoe8ccpTsT7jl+3ccA2vkpfeh0EILZdtyua8ejtMKXhAIswhtgFMuWYOuCma+a64rg948JPqba0+idu2I2Hw0icscUc3YljoFDrpmMRsccjMZ3wHTe+BDuzbseyM4aa6ryrGG7zV6I5ot3Dxpk8ISEnSiVvlERiJXDFpqpqSZxtmoYpb6wCIyOGYZwytd/Rw9yxGzGJ3hVNxwuJFamHcsYDOWO1ZTR0wowHnHTY7gbfqRX1NaUg5a57xls85KlecMwFZzrnEacsAhuh1pYr7HAg45BACwdZyl6WcFZ51qosG+W315bY/m7ymN+cz8kSC34KHGdzxzsf9mhayBK4vQdHYyUNB+K5rBg5w8dmElXllMor16XkOVfigJW2XJOSI8oQ0NjnGruEPDHsaV8SbumQQhwTMmrMXd0hnLAm6uQrylAS1g3MFymq8OBpiQhUjaNaZ6RpQ3+wIi8qVIXFIiPNatK0pd+v2d9dIeJxiSLiybKG6Xmfpkk5PS/JsobrV5Y8fjogT5VXbi14vpfTKvRSoUyEPgn3lhU3yozHrJj7TRPcAQOX8PaDnIPSsZMnZA4+mQ25mmXsZwlr70mdCXHMvGfkEsrAf4vBuRc7SA79sFt/LZ7oQQTWcYiFiDVNCCg3qlNxrsevH93n4hUDnChRuwgOxXFfaAOuXcQZFDNWhiUJ6+7y3pPgGEjvkmGg8RaqjvMUscpDNW21BmWslnSbUtbmvmua7nPFe4/Vz6huN3FD+uQMAjRSgmJchILGwDhWy2NAvPmdtu9t/y9+pvh3ICj92PpNxTwf4vdFCFZ8Xttfu/cJgXO8p/g9cS9WfFCWWnVqQBGSG40MIw/Ca6xoWzFu+3s7iVYLfbuEPkLvIrRvysqejdbMgjN5/Czx2j4PrVNt0KbkUifBd2qCGUkghC+6xCeqki380qrXqhz701Ao2ShfbUsLdzCsEI6c+xktnr4zOdaxMyXAWUj84ljGhDp2HbafxfZn2YYZN9oE7kjWGVPGn9leK9uqjt/hPh7Y00HHhYLNWtsem/i+O25ERBjEBDlK+qr6TmWrL72PldRPSLqzKd7HQpesdM25n/GmPuRJW/PTcsQN2e86iDVt4DWaxH+c3y4WHUS2uomXE/IeBV/0r/A5fZ4vuD1atW5TJsJNV3JOzYfuAoBSMqrOmNMKjXEOVlsdyjjPYpK/CqgIj2fKcqO4qENmLFlTcRGe84Wab0tUjQQrAJp0d+g4J45Woe9sjtYoicBelpA7U6fazRKKxDrWsRDTdv8zEvyAkpEbMJRe90z7uoF3PnIzdsh5sSw5aVquFAmjXJhWyrOTEvkf/92vHv69j5fg/ZNcf/XrX/rqX/763/yqQSGbDnq2mfehoEXDVe1zqD0KNa6XcUWs1L6goUbJiZ07SyjePWv59pPNvMtwnDS2F9zpFaQiXMlS+iTdM4pqW9MtuFXJhuMIXBpbh1CKlQXu7DhGxcd/1h9qJwR+MCQr1h4KZ49YRJj6lrfcKfuMeKAn1qJFmMiAD/wTKlquMCZBQktpwDNZ8nl/g6mrGTmTdIvmQYIDJ1z4eRgQM/mLVZFNF0C7OzKznw1cK9b949/PmXFV9hiFanWpGdPQBDdTN+2IQwcUTEMgWYQgYo1nQMaKlkHomNBtYsI8VAT3tM+7PLFsVoQb7go/qi/wAocsE1Mvie1e6wJkCIRuxKaCFxU7rEpjSUDcfGLbN6piuLg5S9JBG+L3bipsdB2XRJKgpuGY+UU3pn3X6w77qS5Y6Zr90H6Ph0Ma8NqJbNRLzPAxZUjJMkCymlBRK8k41QskfF9Da+35IJi7L2NTvukgIXYvsbI61yUVDUPpd4FtNEVcsiYlCdKAFnjMdImIY1eGrKlDR8yZgnlQ+4lwrBZPJU1wN69NmjSoy5zLnEM1I8tEHHNqzmSOIDzA/BtcNGMU4VinJC5lLhUTLTmRJSPNOXaLYEZ4OUHuS8FKq0Cst0DwRX9Aj5zWC3sup5cKvVT4zfqCX+EhM6kZkFOJZ097XHEl/3fzIXNpuC1DWm/eGd88q0jblHEP7s88x1S8pBMO04wyNf+MvTTtVk8qwlI9NdpJACoEAp1thleylCwRerny3MhBnfCJA8ftiePtaU2szE2lYlct2D6ViqFmFDiaOqGXK+tauHuunCwg9wUXFyXj8RoQer01zkFvsKKuMoqiIcta6jqlqlLKXgU4XOLpD1YUWZjXTpmM1xzsL1guCqbzlPcWNbPWU4jB0gDu1SsUgvSskQF/vNjhg3rNfpbywbLmpd0AeVBh2iijzDHJXWdu5VH2spSndQj+UJZiXgznsmIhNUPNGVLyLGzzHu3MvSIsYRD8ImK/L4ZBkfgaj4c4riV51wUx2InvumcNLRVmmNdVcj+C2xUx357c5YxcPxCRg5CIGzKNanUhyFCsO9cjZ6Ql33dPGdKjJGUhBvdaiAXZPTJWYmt3vZXIxCsqQe3qkKUzye1dGXKiUxJxLELBKnYdPBqgo0kHHzVuwrojnZeu6IL8WBVPg0BJrU1QyDK1xTjOhP20UesSmMpP9CLaVmOyKxEXqs90r3dQ2a0gIlYNi1DhjbBOh3WnNXRlE0kYy4CxlgZBUrfhC4U9PCapGwiY54QpsWs106XBVAMMattZPgbzLjxH82PZQPaiEliEfcXCVhT2iNC1Umxsx26IE8fMzxm6Pg0G9RnKoOsabMbEdpNt7mHuTExgID2bb6HTv9J1V+yKP19r0ylrsTXGmy5IhFxvuKNN+P5YkIoJ0DYkK57/DseL7jo79FjSsO/GTFnS4OlJESDcNgd8MAx0khg0XJeAGUua/G1OVI/sfGKIXjDZJcGWbv0hFK4gennEDtTC1Zy5mmFbclPHLB2cY1CmWETsSdF18OKz1TAS8e8RBhml8D+j1/jJ/phlqzxoV/yI26eXOp7UDf80eZv39DG55KaCxppVWIOtXt53IqSskBwn1smo2IjEVNR8guu0CkvXcBFkcbch8puurXmPNQEu71FqgVsMWavxE9LAL84QJnnCsoXCCZW3Yvf9dc2HYslNC11B0IwQLQYryLqupBkI2zwaBvPEXS3ZcQmDxJE4IXXw2Tsr1v/df/BDSz4+er3+xutvfuGNz7752Tc+8+a3v/btL24XncFk7wWT6H0sS1RgjacMXdCYLGQ4HsuCHS1R4FgrfuqooPW2Zl4YJTxZKFd6jt87r1ng+eRuStIkrFp7HgOxeDeaEQ5IKUKBYhW6aZs9yH7v2Nn99RPH06Vy8B/+/k7In1kSEq9KPaMkoVble2Jkw78kV1mS0HNlqJjk7MiQnuTkZNz2O9xzJ9zSHY5lySM3I8WxpNpI+IpN/FlQ64gLYnsj2nQ56IL5WEnqu34npWetxgKP55XkJo0ouyHTLEnJcVyTkgalT8I8LK6xZKzx7IVJGx9WysaFXICzQFxe0VKQ8N3kCfs64DkOuMMVvqC3eE4nOISVtNzQCbc5pE6EBStGbsCh2wlq7htyUC45BGfX+NlbNqTBJhwYMRGJ8sMRihG7IVGppNWWofQpJbdOCA1zv+wOmojDNXIX3eItJGeuS0sSdc5IhnZYigVFMUhY6CpUNQyH3JeCipoTf0EpBU4ShtI3xbNAhO5JwSBUhtuQaOWkFKESNNwidkdVkhRr/Xs8Vahibld1s1ABLUhZUneu6SkJ+4yCrr7NiyyoUQylNHx1ULKJMIYWT5+CqVtTkPI2D8Nh7skkIRVH03m32HM7YcpVJnxT3icRR4+cCSV7MiSTjIGUzLHNPpeM6+zyih7yur/Kvg7wAp9ghxfyglbh9r7y4VR5X+dWjSBloFYReyYL3qrMLO9z/pDTpuVqL+HJDO77FXUDI5fyS/XT4Iqac6M0DPraGyE8dULllaV6jrViQcNugKQ9YRXwo7YRNR4Oe47HM3g0VyaFY3fg+Wf3LficSW0qWmGEMxwraZnT8L4uGbcFmThmFXyrvuAnD0vmK3tuJ6c9jk97TKc91lXK06cjdiZrvHcMx1PAkTjIs4Y0azg+HjLZneFEybKW3mBJfzjj5NkuaerZHba8uK8UbYH3tlt4haGY3OyAtCsyPGor7rpzPmjWXKXHcu147daaUanc2vcMMjjaa7h2sGaSZKQ+5azy9J3jTC0IGpKxkpYUx5CCKvBJKrF1aol3QSEZL3HEOiirwabLVwVZgXQL0hMDWUtKNpVuwr63ogra+HSdDysMbarCgjDzcyKuu1HDbcc9A+g4dyPXD1yCzdWK56lMcTgeyhk3/S5TqVhLQ6bmkLyWkNBvkaJr2lDVta8rKk5kTp+CUnKe6Bk1DRdBSc/WryUbMcEANQduttWvbN9vQqBpqn0m2+vRIJ7hu2SlDhj3NHSAgW6NR0WwGLBYIGmY/m1uxmV4QtxxLLCNFd24d29gxXSwOsFEM/bchF2GjClZSBV8hjLqkJBED4FCDVZ8IQse6WmX3C10FSzLYmfGd3uSFa028CqDw22ggNv3b1DWyJtwLMP+XWsdAs2ctVZm5utn9F2fcz/dCJSw3koJ7Kupf/mQP2iQ+F11zzByFUwHaQMPToNPi4bzykQJtivFkXkWhV60O/e372DDgdLQIbCEaBvGdyR7wVneIDA3ZI8X5AjvhBkr498RCppi9/OcO+SZnnfvTVirxHOaTVdtxXrrjqx7F3sqDW3HN1Gx8yWTjEpNKe2+m/Hj7oiXGbMi5ZlYAWPkBtzWA3YYcCxT+lJ20KYIkSqDmpiIBFjykjZJKauSQhy3yoLTuuV/8r/FNd3lBd3nu/KQHTeipQ1cjYZW/dYTDZLFuFAotS5JIXknHHMgo9ChcR3seKIF57JiHdZaTPDMgsERpaJ75Ozp0IrC6riSBNivBLUqEVat0k9sfM8bT+EExZKUlRjUUyBwci3hz8kYaEEjBiMrNKVF+ffS5yibnGNZ87l0AmKd8n4OeQr63/9+f7w/revTb7z+5qffeP3NT73x6Tdfe+PTb37yjU+9eeeNT7759te+80U7Nz19NZf0FiuAr/FkGITrUPtBkKJhRMbbs5o7wxRVwatw1EtwgR/yUlkyyOF8Bae+YY0pO+6mpqTlEA7TjDzAjhMcB0nGmTb0MRgXGG9SgYfrhoX3PP8xzul/JkmIVZ1ihd4qqDVwJxkwaksGiUPalEqsulCSk2OZ831OQBLGlDyVJbf9hIXU5CQ85NRwtV3VqiESAQ1be1miLxIE46ERD+00kKlFNnClSBIfuoEFhVpyNznlQPsdSSeSbKMB2QMW7JJzQR2wdHataElDUDan6ZQcChLOZc2ZmIRlL2x0M6kCnyRUIjF1hEMd8TLXuIlpazfOIBUeq65fcXtM/Xxrg7WuRhIIjzV1VylcYXKJn3F3uK/POmL+kdvlTGdBmaXt8MstPiichAqrlCGRi7U4O2Ijt2boeqzVVK6cRJJcEo4Ta1H3pdcFUj0pzFU3PLuh9DuTwDJoVkdyfXT+3ibbxcpddBp2oRoYD/wqQOUOZUIrnon0O238jVJK1N2PWvJKEUB3CQlz1ixZd52iGNw0ukGPO3FMZcmcFc9kSqy0bisc3eEKT5nixIh6ThyPOAtEX5PnbcSgShMtWUjNjCW7MiTBsUefM1nxEhN2JOOKFLynC14bloAw6SnOO07Wyoksue5HnLkVU6mIZPCClEPtcbuX82vzKWfeoI2nVDxYN6GT4bjvptzSIY2HWevJnXDRWou+nziuJDmfGhd8Z7VgSMqMJhDzoUK53cvJE/i/Vk94jxl/YWfIdOmoKuNGNESlFjNOipWbtbQm86sp6h3njWemDYdS0C+UqhFWdejaKfyLBw3NOrXuQ9lw7719RoMG55T7D3fYmSzZPThnvSpZrwuyvEFEmbz4XXKfc3Y6Zryz4O4HY+6de/ZKm8+1t5WUiTDTlhEpFcqEjBs65L6bcah9DouE81nKZOAp85YbN49ZLUpQGA1rTs9z1MMgc3gvrEIieiQ9zqk5lgVDteBkoj32dMA1drgIymFjekzocR7+HvfV2K+M87al7SrhscLZYopqmaQQPG8UpYlFly1FopQN16SDSklC7jay59tytQALXXd8MTBCc/z3GGinkjIgZy4VldRdNyVWhLf5aubIHvaJ8LoBvMLc2PIHyUIVPu1EMdpuHcdrOzmKld+tHfISwX0brmrdvqTrLina8T1gE2i5EGQj0uGyY9Ab98v4bhvC9LZ08oaQvbk/gy/loQAUyyYIYS/3NOKppKVQOzcWsmYha6YsO+lyh3Tu9qoelQ2T5fLoXIbrwjbUzHW8j7Va4SUmG2XYs2LgvNBVSNya4HdiSIPIO4zv13FAiFwQ7SRenRgOf8eNWeiK2MmGTdfLuvL2+iQZUwcURRznmAhG6GFMMqIAy/anT0InIyYk8ZxJSHg1uUlCwkzW/Kb/Hi/Jte73lWT0XclUzccjQo8ScVxxuxSuCOfxZj5aoS5Bt8YidrvMJNdgWHE8t+dPNHqMkvmpJPy19hVKceTO0W9zvsV9DM/fY0DBjpYcuwWf0Zu8zzNGMtia+fafCcNYgvAX/AvcTEt28oT/pbrLL3OXFs9dOeZ3+KAbOw1r17zY3KUxBUuk4nOyLsZGzOCCBS9zFYCbOiJBeOCmPJOLbv6lWyIDVpgrugLgPgNyTblGj2GSsPSeceqYt54rZcpF4+kl9tx3C9cJrsxaz1waClIu3AbqluoGVp+TGupBPD0ybsuQnTShbHLy0P3rZ0KrcPB3f3jwq3+d66U3Xnvz9hufePP1N15/83e+9jtfjDD/Bk8/7ByrsBOvaOkFBE+BQxqDF+Yp3DvzvDMzlM9zpUn4Xp0oD6eb+VI4YT9LeW0/4WJlry28Z5La+4wlZaam3iWIeXwFL8AW5fafdhLyK1/6h1/5uNdji15k03wdOkel2rVtbuaFOUr7AW+7YyoaHuqJuWuLoyTjOT9GEC5kzQt+By8JOzIkk5RdGVKL79Q0Nq3HzT3E/98oRdnmHyto1om5jM/cC9yEG37CDR2R4+gF7K2DAM2yQY+E8xKTIO1juskNUeYwmGIhXSLznjvnTBYcy5yDwDvxKCXR2MtUDnqadWR2xLwYruiIm3LAa9zgmuzzXf2wS0icCJ93L/NQj5m4Ic/JATc4YEeGTFnyujzPyA3JSWmcZyJD7nCVldSMpc+MZdeajnCOPTe2yrzr0dBSSt4d+kAIgJKuUlh0XhxGhAXohSSoq2LhKchCK3arghLmSk8KVmpyeUCHd66oGVJ2GPmYcMTUxIiZSZeUxFawcU/s9XWAheQhWx/TY0VNSsKQgjG9gM23JGdAAUj354ht3W6fRypsIo4JA2asuoMQ4BV/FS9qTuniui7QkexQkhkeVqAfEvFaWmppO3ng2B2bUHKVPneGKbulHUBehd0+LCvhdy7WfDN5bPANEppgfBSd3l/2OzxhxVnjed9d8FSWtJj/yVQqxhRk6lhLy2v5iMbDIHEUiWOYOioPo8wxyIXfu6iCYocl4zVWhdmRjN3ccbxUbrsBg7bgIE/53nnDVFtWtIzJWdN2+0B8xmVIyJ/Kmh3Nec/bffWbgukaciesW1g38HCmPNaK47bmelbyex+UvH/heflahXOeew8GXD1Y0DYpTZPhveAST70uaGcTHnx4lbOLnP29Bb/xbg8n0Eus3d5LLRGpVVEN0uIkVCg385zzxjOM1DwVJiWAGR2WZU3TmCqXb1I+++oZrjUpxUVlbfNnrGnEs5KGSloqaegH7pJgjsJjSj6Q02BmCM/rPudiuP40kLqjRGuj7Vb11S471E2EI8o7R4hGTJxjcNwl+gGuswpB5UrXZJLRo0BFqUNVdWPUp1u+StLt+fE+XvVH3T5ZS2NzOwQmcU/e7NCbbkycFXFfmer8Unenq2yLXkqOPi4JifCr+D1JgKE62fBvXBD32FTMN13Oj7qgl1J0+9h24SeOg9/6t2hYGPeGy2Ct+OdNRd6Joy/90PW1PbKSxsZMWhYSBZU9uZigxoqaM52zYBVUrJrw3DdQu8hP2Ixu7KzF57hRSYzjlEvWFY4sIbQ9tAo9ihg0zvzCyM0hiG5pmbhRUDr6/RKyca5FQ8K+M8XCUqy6nUvWQZp8d3eePHi/xLuvtO7GOSbVEZbkQlc7Qqw3cJ+NgMA2L8qeIQzdkE+4W3yoz7jrH/BMzwBl5Twjemy4iikDV/LYn5KSdDLRU5YcyQ7HehHgfVlYY9trxTo/TfjMJpudBAi1zZhNAkyHYogQbI/yHXnEuzLlC+6QwyLlV9tHZGIdxMec8aP6HC/pAf8yuRfEazaJ2Stc45gZr8gNTpghAkPp87wMKBLhnzRBjleisqMVOPpuU1iMkGv5yH+pmG9YEUQiIpIChH0Zs68DENjTkm8mj3kkp3jdrM00QCKjEWOF2SwA7DGgJOWa9LridumEQbrhaWXOXn9WeUaZ48G64RHLYPCpTKXq3qvbM8TUNh0GoZtoydAX5M5xWBg3UYB1C8//r398Cd4/i+tTb3z6zZffeO3Nh197+4tVmNEOOmWrSGxXYCIZr+w50iBes9sT7uwIWqWsW5iUcPfYjHuX3neJS63m0fX8LgxzYV0LNydwslRDdyiswnpf0XYFzX/r6z/zsWP2Q0tCflAC4mSjcx8f9gpPpcpajcA6TBJe+dpf/+q1/+iVNx/9g+998YEsqGkD9rLlWC9YSs3v8gs4rLkAACAASURBVAGrxA7NQ+3zobvgPX3MjCUnesEn3E0e6fHWRE4D5MWq61nA3sajIQsYzUqNFP1Rkt7ryYskOO74fUYYRn1Ow04Iks9DV2MS/i1BOGbNhdRU4hmThZZYy//ON3iLh3yGmyzCz1V4+pqFCueKR3LR6ex/Q9/hbR7yezzgQ3fGUzfnkHGnJiLA/yO/y3055bGbcS4rPqfPUyXWRbglVyhJua/HLHXFYz3miZ7xolylEs+3/T3mLHlfn9CTkjOdciZz5qyY6sKqewFgcFV22Zcx5tW8gWLEwCBCDLY7CQnGuYgynlGNp1HfYbBNss8OybEMSMSxI0MznSTtWqNj6XWVl4qGIWWXZMZqX4oLgKBNByR6FGySEvs8MWnxwET69CmoaSnIafDsYclZ9HhpxXdqRFd0RC12EA3UICJXmTCTioj0dTgmMmBNwxJTDTMuiFCQc6B9XtUdntcJC1Gus8uP+iPed1PWUrPHEMFa1Q7HNT/kwlmS1iPnpXaPUlOeyZojepSpQaR2+xaUpw5+sXpCiRHcMxJW0jDQgn+7OOIL/SHTCt53Mx7IlImW7FByLAbTGmtBromZC2qPn7ye8oVPHvPOgz7jPIRPISD33joRt/oZb9cLBqSsaNmTnEmSUKaWLPRS4dYw4clcycTIhEdpzrGvg+OrZxkOKSAk9MJCGi60YS41R9rnQ12Y1nu94sN6xQ45321nXeVlunDca1Y81CX/8oHwjfspj1ctP/HigunFkAePhvTKFtTxW29NGOYpZxcF42HNaDLnhaMaWfdJE1jUVjjJE2tXF05o1GqYfUn4Tjvlp/s7eG+KYjYujpOpiRasVxkXs4w8U4aDhum0pFXHeNAitckD28aeYAaONu+tE5p3wWMrnh36jOmxQ5+78sx4SxgHKw8wkrjneTxeN53gGN5G+GMRigexArsxhLM1VW/tg5HEGt+jpmEdVOZqrfFBPjqKT0RFnia8Q3TOfubmLKVmQo82BDVxfXo2YhoGv4lrOunufRY4XhHSEvfoTDJyl3chagxYY1c7FmS2PVdc2H88FlVsE4OjGlOE9hjmP0CSOinZzdhYp9V1ClMR/BVVrdJg0BY7U7EDX0hO6cogfFFd+p32LOy+11QUUoTPbv4ekZNgXVNTvbpgyWN/YoId2nbnRB38miIMrRCDuhWymV8xfAe6ZxkhxLG7VocAtKYJPDo7O4vQqbnwUw7cLllQllzrmj23w7lOQ/BoycbmmW4lkCEZaGjYdxPO/ZSJG3Hiz4NssXYcSIejDgpcNXWAv9l1WRwgPjP7GZFNF6lDQcjlztTmJw26/FCfmWJS97pjx424q4/IXc4eA3pqMN6RG/K8HPKQ48C58lyVPTKXc+5nVLrGi63FnpRUuu6C9ah+lQdIWU3TJRy5y8kk6zgXRTD+zUh5lWscMmZEyfu65LT2/Ez2AjeaA5zkXGeXb7oHfDG9xtVml5XAT/pbXNcd/lryPGmbsXTK5/0VTtyaF7jCX5QjJrntB7/SPqCmJpesu780mFBu7yUJDi+Xk8zMZR0fc67LLsFraJnIgJt+wsuMWWJnfCUb3qslkvbcYqITpf8TEvYZMNCMG2lJ5qzSnojNsuOqZZwliMBp7dnPEz5Y1hxTGXQ+QFojJ60IxZAynJNWAMu4GQreO5JTeeOdJGLx7LgQhn/z93Mb/jxdt9/4xJvvfu27X0yxwuOhy5hISqKOfZfhVKhRni2VJ0tPTxLKzARkJiWMChj2PY+nBrfqu4QreUrhHE/qmmnbslw79vswLuDuqUkUO8wfatdlnGodzjY7515947WPHbMfWhLyURiWk46bB1xuUTs2rukK/NjPb3B1z73x6pv9f/Doi9f8mCduxUwXJDgWGLkwSgze0QP2tc+LeoXKKQvW5pQujufdESupu8x94PpbniEQXUkj5ncz8a1i+HxylT035rX2Crd0xAM3YxIUXfYoELHK6BkVo6DZbskN9EhYYHJpexSdisPTZMFcl3yWm3wrechYe8ykYoeClbQ8kgse+WN+Sl9mJhWnOu3GDYyvcSILHsuUQx3iRHiLB3ZA+wWfk9uoWJVgjwEFKd8O7VMv1h0ZuD59seD5jHl4Tq6DbJkDt5Gy46HlMdL1jgw4DfLBMXHwYTOM1aWoYBEJ5ikJQynpS8Fc111l1JSxTEJ0pWtGbsBaaw7FeC49ck51xkh6pLjgEuwCcSyCrTZV1tipsAWw6YgQnkuszMbuyDO9YBg6CzEhOdQRc1mzr6PusKikNb8HDA62r30uZM0etrmWpOxrPxixNUxkwBwjMatsyGNFkEt2CGcypxa4w4REhCMd8C33iO+4J1YFJDfoghinaBK4AhMtOZMVx0wpJWdfexxRkorgvbA/UFa1EcBXtfAbzQkl9hx+yl3hSPvccn32SuF0pbzTLsgDNt+L8iNul8SnHTq/T8ZSGl6WEX/lJ97nl7/xHL3UfrYO500icFy3ZM6xaIwjciMt6JHQTxx7pWNUwvFK+bBec61MKVPhZO2Za0uOYy/JmHlTOzHpv21HWAvwYjcwxzTQU0wWuEF54tddsAQwD7VDCZvvTdfnRpYzTAoWy4xVldC2juPTkiKD+SKjV3jSRDk5GXIxLWlbhxNL7IpMKVJhXhnmOGKLExEuqLmWlAwzYV7buC1qGOTCB6dC7izhyFLPxSxn0Gt4dp4x7LU8vkh4v1kRoTVzV5OHmbsKkAEBHrgLhhgZ/QM54UTCut3qCOaSdoZeMcCKVf+FmsN17E6CBTmx07HBwod/08scknjFbsdKV/Rdn4UuiF4Gng3h+uOUdwRhT0Z4zFDWZLQ3vkdx3W6TLg1GZB2TqPBk3KuNLO8gKHgt/JLIhYlnTqv+Ugdy+4r7gf8BDr5A1wWOQfp25Zzw5zp4ZsTOxbYUbAxwNxCzWMkNhN0teNt2EhK/v+c2SlIWeFoS03aFHgDtxk2BikjctddKycncRgUslYQ1lUGfkI7rt9JqyyNj44tVU1vwK9YFqmlwIgxl0MmdL9QKhQPXYxmgwR5lz01MAhjpzlu6kYTObZyU3OVsoGg2PheBz2bwK+MrNaFIEYP3SF6OiUSEB9vvt/ewzxVVJd1WorgZ749CieIz38zPDYdwzpJWW+7IVZNNJe1IwA6hcaas9hPyKgnCO/ogmBJu5mKEIA5cn2XoNBrPxc6INHAprGPiWAeURseLCMpaZ7LgEWcspOZE5pSSc+T7XCsyXk1HSJ2xEs/39YKfyPdom4BGIOXCt3yy12O3HnCrLBjVQ16RETuFKQK+16z4njzpRGlcELLJMKXMGBsAHVdsu4hrnNPgM9NFezbnZrriR/Q6AA9kwQfulKdqqlvxeQ7F9J1qaYjdH8GS5B36jDQ3R3CFndSFdWSWA4WzJGbeeBatD90SgxqrwIVYApiRMNA8QBSjiWTCFW8+IwcUXMuz4J1l8d0oF47+5z8fMKw/7Hr5jdfefPtrv/fFkZhELsDLk5QHyw0iJ8oJfOJAaP0m3hz1PG0rPJ3Di/vCTg+mK+HGjnK8MOPdV8LPAMwqYeU9/cTRTxIqVSpV5jTdef7KD0hCPt5W8094ucuFom4hAV2gv2n/Cn/x6x+vKvD5n/8bX/2VL/3Dr2ywzdodGC2ekfSo8UQTuE+1R3yWa6FKf9Vk5OTASJAk3POPiHJ5dl+XHW/t9YQX3Q3e84/xKAc6ZBTeoaINnQuDkPRDovG+OwcPBxQdXtk0nBPWtJxQIRj3QxBGbsA35D7nzFmLTYTQ6OdT/hqf4lpAPm8O421oxZnOGATy+JqGlLRruf+yfod9t8Oz9hQXTIsG0mOGUgSH+RbPPX3MQHqsfcWum7AMihRWyazoSy/wHjbvu9QV04BFd0gwiiRs6Ha/MRnJJbXOgrRdhfWCBVn0OxHXBUQpCSNnhn+JOJ5xzkorJjJkV4ZdpVTxLLWmESOQneqMXbFuBTSkoRqYYu3bmrYL3GKnJOpnGX7Z+Daxu5Fire9J4Pug9sxsnFuKALeZBQWnWjw72mMhZvz1UM7JMUztHY54KGfsYAkJsoGWmPSq45wFF9pwIyl43FpyvIMpvUV1JMVzg31mUjGnYoQR9UahYzFLKv4dniMR4fl9z8nMUbWwXghPVy13/C6nzuQ/T9uGgzTjaCCMesqqEV5JBqxV+QYrrvoB91hyLhUH2uNDN+UvlwdkyYj9ofLWd29xfb/h/nHK4UB4tjBoUuKMK1E6w8i+mve4V625mubsFo5VA/NK+UW9j0+Ul9e3eLpuccBhmvE7zQW7vujmWY15rEQ5zbhPtGi31u25OabU7JJzGvDuGRFOYut1GDhVz3xF6Qvef1xwurRD6s6hsKysBT3pe44vUoalo8g9RR6rs0LVJLRemK7s8KlaQStPG5SzbtLnrGpJRRikjnnjzRhrLlzpOe6dKnmACKzWDsgZlp40VVMkI+Wc2vYXzbr5JpiAgBflmh93UMSbukeL530xU8OZLhgGw8w1S9ZKJyHtUSr9KCn997uD2+ub4C2ucRHXycxuX4UU/NzP/+2vAvytL//nXzHi+pbLs3p8CLiiAlMhBTOW7DMmDc8204Rl5wAcFQk3vIkmBPQr6kv79fYVuSBA4BPEBEF/YAICIV1R/YH/Hn/H9ucyPtum2ruBnjkWuuzIvo02nbR5l1Rp232GTPJOBhgIlfbN2RQhrUDnTJ2SUgY4z7a7NoDDM9/itOSBBFxHBTAiXGmjxGTv47ugceQsqchD8Skj6XxZ4mdsMeWwnCx0t0yFUNWECaLqUxzfM70gmgx2iVsYd4NIJfSlx1RnrH1l8FUZdXN6rUZ77qZfkNatQwe+J2XnyL6BUYXEUQSnZqjaySmHjl8hOV7txNuGDl+GY7nuswAcJruMMQO7sRYsXMVAC6ayYioGPDnQPhUtOzKgkppSU+65YxrfBPjbRvTB1CgrVO1sWuiSvvQuSSHHe4mwyQh9LKSgoe1EERTtJHHflxNeaXc5WTXcKQteKApecjeY10ri4NW8TyrCeW2f7dnauKpVq7zUKzhZmxjCOTX/1H2XlIR56IBEOFaU2c0lYyR9proI6+VyN9Hmve23UXLc41lpxWd5nkIcjSrPZMETPScKJRSS4VWZsdxK4FsI6pkAD+WcQsxk9woF81YZZ46zdcthnnaJR4sVvKfaGERefNepBJiyZhhOnEyFXS0RLB4YkTKQxMjuWAFKhP/fJCDxKklYqycXR6WeRZVwmKUcN41xHCWlEOHXH7UcZCnPTcw7ZLF2DPstn7kBT89SdgZWakoT5QtXHe+fOFq/2UOvDQWdphwNhAczz2O/psZThvMhSgx/3PVDS0I+moB89NpgdfUHJh/b1099/d//avmz/+grZfISbzdz7rspv+a/i0d5ome86Zbsy6j7nQ7h9fZq9/MT7XEqC4oQjHS11ajXHTahuOn3XY/vtx/yefcyL7W7JJhPwIqWz/kr9MTRw7FUzyzotv+4v8pjVjxhxT/iN/kx9yoHvs+YnAvW9DGlgh4Jf6V9hWey5Bf9b/NKcrML/D3KFe11pjAzqfnAPzGXVzUMdy4ZJ/6choaFrvi2Ez7lr3Hk9oKWtkF+TvyF4ZlDO36upmRVyEYbXFHO/EXXjm+1pWGjlrWmYig9zv0MxDw4epJwoQuuyg5nzBGVzlm30roLhICglGVVhrNYzSJqXRvOs93asCKuvVXfSQoCLKhQzMAsDUSxqPeeiONUZ+zLiMj3cGwwqzmxE2Nb95qmq/BFla14r6VuAoIMxzpU4uOc6gVib+yYlJqyEgsOdn3JSqx7saf9gJE20veUZRdMe5QJJRPt8Sl/wDkVe5LxdjvnO8ljMhKmLBmEBOSAEWtqruqQTB3OCX3NuK17eKAW6/r8Ks+4UQ9ZPioQWl7aTVjV8NTXrKXlZwZXEIEyg6v7a5brhPGw5tF5j1qVQkyy9b6b8gU95LvumNYP8CivXK94dJLz4gvHvHN3n4fnQi81uFeR2Gp2AodlQtUCXrloDGDxYbPmQSPczApWrQWT7+pDvr084K9eK1nVwrD0/O6H5vQaDZDWtKHebWM/IusU5uJaiUH0hJzj4BjvEJZhDhiLx37vdelxphVvNXPePrfE8poreHKecHfWkIlwVCf0Qht6OLD3KnLP3Yc5ZQaJg0nPHOcTsQBltvJkmPP5v9ITXtMd1rUFfEaAbJn0hH/j0yc8ezri0UlOkSnThSNL4PypVTJXtCwxLogPa7CShl3t8Uzm5AEOeC4rRlqEz76pWA+kZNuboC8lM90Q17evtivibK5Eku71NARkmaQUYY3MsADj7/zCf/2x+/Xf+4X/9tLr//GX/4sOkqvqO8ilyQcXnDBjIRkHOsRDgBLFqlz0T2lD92MTYGaYRK0gPNLjS/ew8MuOcBwTiw3xl26P377+sAQELJAaBZf6vhQ8ao9/n3wx0J0jMRCManwd3wbBBfWulrZTIIznYdzP4nONweo0qH8JjjO9YMeNASP9e40SzZ55CAL70usCvVjwKAOfLt7HUteXkrPIEVlrRSk5tTZE9alIFJfQ0YkwoKWuSWnZkTFrKkSkU8mKzyvu0waH2+D0WyK52pQRpzpDsCpzDK7zYDIZr+hlFZOiTPKQWLgu2YpXt0foxodEwl6OWh9hA49rqbTu7vOjyfb2vHnUPoPkgHf8/fjQAbjlrlKS8a5/yOflDgc64LPtEY9lxDO34ETtPC6C6V6rnuvJFT5sH1NKyTL4sAAsdMlIjBPaaBP4MBX94KwOBuOy52dn6NTPu/n0Ekeg8I+Tt3lRD/ml+pQLXfACR2Q46rXnXBb8rfQV5t64aBmOCypu1gMe1yuOKGFt4/ifyOf5O/rNkOytOyPQpa4YhC7FOkjHVFqbuuEW9CWRhFJyVgHqvi9jnuk5gvAF3WeqLd9IHrFgjVclGg14jYmgMKbPOXOib5fg+PH2FksahmQ8n5ZMW88oNWPenTRhFRKSJ+uWhbacYR5tT2TFE5nTimfBmqFap/FUllwNxof23M3sb8el9BMTH1KF3MGLf//PJw/kD7pShBrP1DdMXMqwgCtjWD1NGIRix7S1ef5BVdGc5rywZ0Wy2cL+/XCnwbdC7ZX3TxwHA7g6NnGYNFHKTHnnHF69ojQt3Eoc19s+D2aed9pFx5H+wff4Q7h+7cv/58fyQS4Tw+3Pf5QEJF6VelYNfDofcqPu8eu81S3QfRnx6fYq30keByiODVismJ7Kwg4wTbjprnDPP6RVT61rzKHXXdp4Zn7Bp5IX+I5/j1ViVaTX2kMKEnYl41grhpgQZux6OIFfkreY6oyhDDgIbbw1LeNgUvjP3Xt83t/qDgIR4Xv+QzKXcuSHtChPQ1C7owXfd0/xXjeHqGyIn/GAeuxPeMwJpeYcyARkwIf+aVd16lRHkA7PGQ27InHSY7KU0UwwJSGqSM10SVTUicpVFZYErNQkQddaMZS+YZsl58Sf0w/6373gqWLkwJTIFfFYJW0ZgkfBzBhNY9426pL80nPx4eBLxAwK12SMpc9C1pcw0SNKVmwki4GuOxKvaPKWhWp7XzN2AywOrDpoR7nr4F0Suice5cD3OXdrRgGaV4thHW/5HVq8kfq14IGc4sLijyaTMQGZuJS+JjzQFdek5J4WrKVhRI+ahj2G7Gmf+3JGgyngHPgeM6nDRq3s+0E31x3wxFfcSkvqFn79YkGfhFcY8XDhuT5wrGp4dpZz89qcs/OSMoVmpezmCcU6pU/G/+vuk6rjXMz1/NYLD3n5tTnzi13WtbVjjw0JROKCOkp4TKmDaaPsFwlPltbZWWFu7e/VFakzAuc77pT/4bHnS/lN8tTxWjLi2+0FGTmm2GWblQsFADP0u2wIt52I7FPwlJVJLYZ7qUNQ3uB5qEvWmATuhJwDl9FLHA8XJhN+0LO2/dO5ciuHNPF4LzSN48Xra86nGe8eh2qch6OB4AQmSUIbuCGf8DskIpz5mhtZgSr0nONwt+L+gx3uPU2sla/CvYuWXuLYKeBpFRNWw8hHHsRErdOV4ExJCoPixbWgKM/pLu/JMXNdMZDSxivAC4fS6xIRj3YVys3+47rXYqIQncqj87ntbQJ/eKx+6fq7v/DfdHv7f/az/9VXLLhrOzPRKLxwgx0uZN11KTcVaOlUEWMg7cL6cwgnesGGZBzmnlw+wmzv2xSYtveSP0ryEa+B9FGUuS6Y6yIE5htieuxefLRL45CuKBTN+bYJ3nHuxs+w1qoL1MESyZhYbF9LXQX5WoNczfyc0pl8QyJJZ84Y781ry2yrExbFUZIA9QS6IFwQFn4ZXLSjQaWEAo8lT616Rm7AxA07M0HDzI9RlAudXxqDeEXJYjtj0gCdgx03ZqaLrhtjML8eBTnH/gzFlN1MSjmnL72ue+PC3Iidg/UWnK0j/7NReYtfYyepJyVLbbs5sj0H7dm0lz4DhEQkXBF2fKIXLPyS55IjZlSMwjo90B7f5REApeT0pOBpexr8Zeru85ZhfsTOTKtRbSq5BG2K14qKCUPjRohjKH0u/JyJGyIa4e6ed+QpJQa/+54aZHtfxnyhfY4kh3+SvMOSNUN69Mh5FubbHfoclgntaiNzj1r3Jc7pqKLVpyQnY84qdM6qS8n/wi/pJyVZ4Ltc6IJrsseUFajt0z1NWciaHRlwolOiQWvci5ZUofPWUmlDTxJ+NXmP53WfK77kbrPkdtJj7c3f4nhl+8Vp5blSJKyWnkMKHgd5+4KEE1YUbMHdtjpUHmVfcnriOhPE1EGjsPJ/zM3wz8nVE+NW+kCqr1twYuNVpGZQ7NWZIqSk7JVC02JiKvFMrW2M8kS4qD176gi0oSDvCy8eaOc5kiX250aNyL4IXLYfdP1Q4Vh/0OXVuht/nJ8pxdFLHbPGstwfaV/mW3qXRhs+0Cd8KXmRV+UOv6Dv0A/O1TWeV/weC8b8TvKI236HdxPltrvGBQt2GfJ2+0G3WIBucb3lP+Ald50VFZ9tr9OLG5ma/fzGJdLc0Z+w5NifAqBhE47SohUt33dPKcj5V/IeiTius4d4OyAec8Z1xsyCSsPb7glfaG/wyfaIddLwoH3SdSsc0qlD7LlxdwgsdcWDUGH76LXQJaqGO40VjOiqumnlWyU8wbGioQiVC9jwZuIhsp1YDMTUsSrM/GrJmokbstA1KMw5JwlJTbwEYSAlFzpnJH1qTF98QEErbUcAraipg5GfqYM0Rs7UnIkMbcMLPzuXFYdMGIfDJCdlQdV1PWJ1xbGRao4E9SM/ohVPje+SxkVIRmJyY2RM6+r0g8/GyFuVOFave4HYlmvCbd3hmSzph0pygmNXrUt0LkvT51Y7Hr+VPOzGZgM9svvc9SVrN+w25D4pfbVk7qmsOJYVGY5dLdiTgutlyk4PmtbqqROXcq1vtZ1ermQJDHots1lO4pS7s4Z3dcY7K/BOOWbBZ9pDvp0847AdMCDjvXdzrl0/Rb2wrGDVWDu69VbtAOgHvXRVeLTyeHU8n5W8Vc95JR2wapVrScHvqnmk3OUprXp+rp7ylx7d5qoreC0Z8367DKpavlvDC9rg1pqERHCzXmO3Knaa1rT0SPnZr79xaX/5B1/6+1+xTpZiyHDIBCNGCzxcGIH7xiBhuoTkrGDQa1itE46nGb3cVkKrsFcKyxoerVpqVUoRysTxpF3xTJYgcF0PWXhPAnz7/QInkCcGSVs0cFAkzGrl3txwzrsh4b6vi654spJNkDVQk7R9yDnP+V18gHC+J8eBxJ51Ccd28gGbzkfc37qvagFWJL7GAHm7YARQ/wF8iT/K9XM//7e/+p9++b/8SiEF+zJCEOasTaBCI9zOuiTxPmIncxtGaevCca4zxtLnVKfdnhSD40vV/T/gsNuWgv2DEpKj5IBKa2oaBtLvPEm2OyFxf/xoIhJf2w4kI2letpK8CB2N11gG1DTMQ2C+Dd/xKLXW1AF+FrsulUbS8Lr7fsV/bAC7SX7aSx2jjxvDaIkrKIduFzAZZpPlLRhKn8jpsYLDR13uN906oPNkWQdo0bXkkMf+xMYkVM8rrS8lIB9NIs2PpOo6G4vQ5d/uNlvKu5kD0cAzXjb/7X4FRxo6QR/tgnw0Kfno63UgSEfTyw/bxwBck9e7zm5JRis9+hTcbe8jOA7dLk/8cdjjNmdwEoQ64vwpJJwfklgwH7ohOVko/vU40Qt2ZMQwqDl9j0fmoYV5aEU4U4YlPodqqJF/vjrmXOZMZECL5xGnHMiET7aH7PUSvMKVIqFqlXHbB4Gnem5zPyRuVSjLxWRtsZUE2nNPaKUN9gFW4Iwy0V/wN1mpp0fCqSw40Wn3PT0pWAbYeE3DmD6rkLTF+btkzVvykMo1vO4PeNhWHGoGJFwbOM5X8HBdQZUxShK+016QhqLiuay6AhXBr6gmevMYvH+hLTtJ2sGvrgyEJ3Ol8X8I1OfP6TUMnnxe1RIrD6taGJVq3ipeOFvCOHOowqxSBoXQeiFLlDIYBeepMm88icCg8FSNJR++taKkV/uZxNnvvT/1nRhIhuNvfP1nf2Ds/6+dhPzqlz9eFSvpNj1bXX/cBASMG/IvvvR/fCUVoVTHj+kRd3SXd90pa2n5Z/qQn9arfNZf463kmG8lD9nVPp/yB3wiHeKaa9R47rS7vJvAgQ6p8Xwyuc3vtfcubXZ33DVyMl5q91hLa5wNNUOxx6zYI+8+U4XnN5IP+X77Qbc4Kq34dvqQkowX/D7vuzPutg+C6Z/nMNllFSBGAMf+jN9yMAqysCUZv5nc51PtVV7312mShhN/EQ7kmr70WGsVjPxyyzB11VWmooqOkbos8J2xICFhwTI8C98F59vJiKfFa3vpgAJbhBFSMQpt36uyy7FOGYf38FLSqmcRjKUGYhWSPnm3gWQkndleIRnzgHXuScEjPengC9HLQ1Fa3zJ0Vum56a508Iw1DX1yTJjwZgAAIABJREFU+mKwr0JTTmTWfa4iHPQFKaugngKEqkfKgfY4lSULsY4IYhWStViLv68ZC6m7ROTA9wwmFCBQMajtadpV3b04xuTc///Ye/cg27K7vu/zW/t1nv3uvn2fc+fOaPQcxAgQEpLBYMfEsTAgCWLHZUyFlKucopxUJVVJClddpgJx/nD5j/wRkyI4/iM2jtGocKgYBxzxtIUEEgwajaQZjeY+5j769vO8z9mP9csfa619dvfckYQYhCRYU1O3+3T3Ofvsvc9a6/f7vsyYRA3ndZWOJpzXLndkzHntEiN0JaYCJlpy1W54zmTMS+aAxJ+DGTn3zbjejHdIWGoihG1tcSy51wQJ33FZOBzCyQz+w+wEgDV12Ry9lnJxd8Lte11ElOEkIS8hEcuejDlne/RtSmVc6nmkhleioXNsW+1S5DGLRcpGzzJZGO6OlE4s9DPXQckSZX8kzp1LHFz7YuG6avfKnArlUpyBLjUKbcnY0j6r3gnm+WrIit+Mh4LQG94ypmSDrC5Apr5QdcWfE0Svk3GfV3eOwwh+6SsSkxjhZu7Qhctxi35sOCoqChtc/JT5IqKVVfRLJ7rbbAuRgb2x0k2EK92Iz48LZqokKlwxHa7gPguFKie2oCMRrcpN6tPSFWmXV4XJAlpWsBi6nkLQjoRr8Rq/OTvx7mDUn8+eL0LOjrDRC9TJ16JgNZsNgXoT5itFa4QC8IWe67KHf5tFyh9nHOqIdd9AsBie5w6q6gXyy416KDwumu164+Deg9KVFrnvggfdmnMAc00dIxGIPTWnN7UW9fs843D1sILkgXWUr67XJqyYHjFR3XB62Ab17HAb7LjWJDh+ukufDx3jgoqu6TC0Iw51GSYYUrxj4nrjDp5ipNQuRZWnBXelw9hOTjeXWGpLHL3JBcS2pYWI0yE4iqkrSAMykZKwIGdN+pSUtMnY1xMqrVg3feYaAupcAbXwjkZung2b/1DC+ILLG5lE3qXrwJ6wIl0GOiLXgm2zwZgpY502aMOWBEe9CvdI4vOimkVQoGO5ezyutTwiguiySHROZg71iSRC1Z5C1E7dEzheRVNDGp4nIeRmVcw0oCguUO/j8nneLJdY1RbvrC4TIbxojrDRBSzKVd1EzfL+HFi3bpVa0jXduqhaaF43BFdMn6Yl9ZycmIhtWavPb4VlXXqMmdU06Zi4UZQJd+WET5vbAOzIGhGGt1TnuEiHjThmaixW4QvzBQlCSww/kjzBc/mEW6bHH+jLtKXFf5N8K/+w+CgjnbgwUY+AOEF97LWnDm1yJglOR9KRjNv2AZWxvCt+IzcXjg3RkzbfXl3hJXPMF3TP388ZLRJm5HWDdU7uqd8OEbspB7ydbVqIc2WaOWF0ocrvRvfoaOLCUCNbOwk6hkOgTBpKcY2ecP8EHWhNowRmnu335Xzmv1ZHNxIqFVqxsNFTOq2Kycw5iKWxZVOEkQ8AzitXSARmwWRusApprDx5QSkKj+rGzgAHXCQAeCc6EdLE8ui6e2x+mPKgoS972HhdkJAltPXqBevdz/zxEiX/woe//+nXKnSApeq/2uBlc8JAZpyQc953o4M/slHhvO0zk5I9CZs8xfkmGD5v7/J2uUafhPO0GGtFgqNe/SK/x3/Be6jQ2sWn8ItcmPBFDDeqezwWXSRSYU7OTrTBvnXUnMPqhN1o/VQn7qA6JooiP1m4qvzl6JhLdpV9e0xKgqWqKUuBb2xRKi1QLMd2xK7ZJKfg0J4AEQMdsSYr3iLPpb+HSbe5ODRHEFUGTUmgCKxIh4EdM5e81nhkktIlY8CEjLSueHNfTDlkpSCkHhdU3g3LTdpjnZIQMdYpj5hz7OugXg6cZ7txEDPCJeN4oW2yWj8SBKtAjZik/ppEfuILhd0ygdn69GXLlvZYsy0GxkHMFqWjKXMpmPrsEIPQ0piR5LR8wRF0CwD7Zspl22dBxao6vvR52603ylvaxgAXtceIgvPS8hQepSsxPU05Ly1+1dyo4f0xc3q0+CvReT5ZDJhKSaKGQ5mzq5260Ar/doh55TAijWCzq6zPMo5ZMNCCta6hrIT5PKHbrjgexVTeTvfqSsTvDVNGJucdbPIxZswo2dIOT8ka73ysJM+9ANY63ufjV0ZUN/ocTZ3Af7KA8ULopg7iLVVJjJAglDhx+FvaHf5wNqGMbI24Vf7z0zcRb92FwZ02fRM7ATlRvREPbjMDchKcDeeCihUSgp2owRlB/O1n/tZrzjF5o9v8hXLmaV7KK+WCK7S4ZafsTSK2JOVwbrjQMzx7X0lE2Wq7zpEIbHWEooI4cjzbEmVYVaxEEZ8px6yTckcmvFlWiUUYlq7DtpG4QveVoXJtA2YDdw0Kq2y1DHuzimfzibcPXZp4lP7r/+4X/t6r3tt/+0P/8/VzusoJI/7RL/z3X3R+/bEP/MR1x5O39eYr0ISC0HRJSXD0m6Yg/bX0IF/O+Nlnfurpv//Bn7oeqFXgwlVTievCqU5pV7etvGx2uG0fYFF2zSY0EI/Cm3A4h6dXC2HD3NosQM7+GwqOMAd/MUTEYIgkYmwnrJh+nbERxmuhIeAD8DSkby8NPjrSRn0nHzhVhIW/AydGLxo6kwiXu1Bv0tUniWtZFxap32i5Jkxaz2MZaX3clTp8IfYIbih4Rx7Jz3EbvbCZ3dNjVC0906HQqqbldKVdO1hZ3yRpos3hXk59mGS9Wcdlsxyp22BmknFinTNSeG8LXXjK1NwXDkEvUGEk5sgOWDMr9WtYlq5slZ8/gu4w/G3QgQoGo24NC+e+SfM8u9lsXtewqQ80x6bgvFLLtlkjUqcjEP97b7KbGCNsaIeRLOr74cAOKNWxDzrSpidtVFocVie0TIu5ndM1nZqiPNM5fdP1ZiwOEXBUvKX5S582E+a+8bJMvd+QPiOdcU7WOdYxD/SE77PfxOeiQ17mmEfLdS7S4TcXJ7wr2WBUOTrTrFQumTbrNuVFc5+SipuLBbFxKN7Y07h60mGk47owVdXaKjwmYkU6VFgumC2c/bEyouRAXZP12egeOc4QIWSFTJgjnKZeBtv9Li0mzPnN6BbfVV1hI45JjTC1sMecN1VbWJQbZsBUFvUezT2nOyeZ3xNMyME3kQywZhJ/PZ0DYhbDKF/uM7/eRmzcGnmu56jHs1zotNyatvBumr2WpZ0KcazcPohqGpZVR8tKDETGPThZGNZ6FSIKM9doq6xDQ8LfzAupvwe+KAoCr0MR8u5n/vrTD9OEvPOZ73vdRDxfrJB57of+n4cWKL9i73CRFRK86FVKV4AYR2W6Fl3khr2P9XCs1YrclMypKNRtODNx8Nwj0XkSKyxUWZWECGGss1OTEjieqyDsGQffH9gBlXfAMBLxvL1xqvNmJOKBPWRV+swpWJceC5zj0q7Z4r49qAsIcAtU4jtgIdiowsGemaTsmi3mmjPWKSN1nbEV6Xr3KDcpzXVO4i0fBVOLx8J7aELkBuHADlAsB3bg6FB+MXugJ2xInwlzXwQ4akjYJIDjugZnH4ut6QTbZp0OKfs64IGe1JaD4Pi2QRwpmBpBmbHgIpvc59g/4jjmc4pa47F8jpAD4rq9c3JaXhxYUPFARtyPhuzqCiWWtiZUYuvuSbD7nPsiwPoCu8TWDjmbts0dM2bXdmsa0ZZkHKnQ88GSFshwUUGlKutRzKBy9JPL0saIcMmuc2AmdDThPfYiMyq2esLbJ6u8UE44lDmb6uwZF/61V9Shcg+Y86hNabeVwUyYUnKRDneYsto3DEYJkbHcOcyIBHZWK56778KHVm3GAzPhpp2SScwKCbdlwYvVhO/pQlV5N5+kot2uGA5bTHPniBWySJII8hIubJbcHAv7i5JNSdnThcvHqZS3t7u8sDgi8Y44u7qGRflVvct7Vzaxd5yupUKpUFJPSYtY2mY2xesJhhJlTMlUSnoNY4Gz44c//Dee/pfv//nrjo5o2SBlREHLb/Tulk6cOaXkC5rzJCskEVzuuZ+3Erg3UrJIOJ6pC2YqHb++7fMnIv9Z2WfuEs/FISKbScSscn+Txo4W121XbFcR+UDoJsI4X26iphTsqLN+fkWcPfePPvO3HzrvfanC42HDonRJmTFfFtlebBxyEepUZ0+9eD1QkP/lQ//g6f/qgz99Hai1YMG1K3w2IYjkLbftg/pv73tEYtusO9MHLRFZip7DHFYXAo2NwtnCoO5qNwqW1xqhEFh4CpLFmXl0TZeJnbyKlnX29ZpIDCxF2QYhkZixnS6LIgI6vZzDWqZVN4tcKJ/r5HekzdCO6Ji222p6p6m5OttdK84coSvtegMPXrB9JvslzIc9abNvj6nUIOIymSps7WzUIq0DZ935MxivFQzFa4LhnNkA4J49PNXk2pCVU2YC62bVvUdJObAnFN4SvonSZ349CAVMKPsqKgp1rlzBUWqoo7pACUF8oQBpis6bCFksWY2WGD19j7vzvryvmtcvnEtHN3JftyWp9w83q3vcl0PeKW9g7AuvNjFP2i0+a465IYec1zX+UF+uqWWOzjSjT9cxAhpF7sRO2YrWqXwR2NS7BNTfWc4P3B8IzHRBV1r15t2Iy9sJxUpFxYp0uG2GDJiiKK+YiHWb8aj2uVvkrJqY0u/9t7MIs4DzssF9PebXo5sYdU2//1jfxm9Fn8dzJWq9y4y5c+30a/k5XWFV3T29ri2ObcWJLLvjU3Jm6lw640byvHN2yp3uTZe2zAUVW6zSVheMW1ilGxvm6hpc25KRiPAKp21/Q4FuEGfvK5aui8MmI6oLkG5sHB0tc2h4Fi2taL/exk5X2FgpORrGbK6UDCcR80Vorimz3CCi7I+Fy+uWnZUl3brWhFSOarVYOE3kcBIh4hwywRUgkXn46385dcDrgoS865nvO4VWvJ4FyJcab/uFv/b0217jZx96//91fUcy1uOYTnGeQxY8Vq1Todw2w1M+8RbleXuDRVTUnenvtpdJMXyffZwKpSMuAf0XzQscVU73cCXa5W3VeX5RP8ahPWbIyG20ZZ1102e/8hC+L3Yin8sBYZI0THSGoqxIh4SIT5t77NkjBHOqU2YQ72Bi64Uy8ohCQelyUqC2ZzQIsYn8RO66CImkJEQkXgDZ7PyECTAsjrkuedlKxYgJ6DJozFEVnEg6JaYtGQkRA88dNrJEybZllanktDwF5EQnKMo5WWfo6TRugnFhWBv0mDr1R+10taBknT6HDElJag65+y1bL4qBSuW+du5VAdbteOveCENfMxe/KI6KFOyS17XtYjLFuWFlRATj5EqUFXWhl7u2y5akfJ4RPU24pZNaJH1NnAPQ3Nvg9b3QrWUMhSqxuI7QO2SdfrRJbl2I5+Uk5fPDkiN1yEBH3b1YorVuYnnvKKWFwUzYXbWsT1IuZSlPJCkPjnxhvD6m96DNSrfi9kHEpCpRNby3u4rqKr8+O6ZDwoakXJIt3vvmBWk6I88Tjk/aRMZRlEaziHe8cciD/S69bklZCnlpuHsYU5ZSe7VboGPbnJQVN/IFb2q3+KHsMv8ir/g77avMCnh5saAjEfvHEQcM2aDHKgmpCEdaMK1xAPx7dwL1pT5kyeP+vi/RZQFnIHBfZ1SiRCqsk536eYnl29JVeqnLBclL18m5ObCsZ4b1jpukj+Yuh2IzjinUfb7mleU8LmX9Mwx4VDqkZ6wCZwXe9jCmstBuzLpPfeh9Tz/lv/6l93/o+pfqGv1Rx88989NP/9gHfuJ6MKMIG7zSb+iWLk0hbPG1aSpf6YjEMNRpTSsJaKb7OvKhh26j3URswzU+sm6TFcw1xnZKWzLf4FluMgNdx+lLlp3vpl4ioCUPG2tmhYnOasSob7pE6nj5iReVr0drjOykLj5gSddpNkJa0vJBgVIjIoplZMcEC9hgiRt0DSFtO5ivhPffN46/P9YpLeMQgogIbWSRGHE0tZDTkUlWH0/Qnkx06lyotGTiHxvomEwyViQgrVGdOVM2CtEQOgmuENmWNVSUPXvEhlmtO/SZpOzImqPeYBnrjB2zwaE9oaLgxOdBhA940LskkjhLeZ8LFihFjuKT1/SzZRZVQiLO1TI4fAVBfnC/CoVmKEDcPZTW60RXOgz88YTmVxjN6xsejYioNKdq3D5B7/JO8wY+y10GOmJByYPINSK/pdrFIBzKlKu6yR05YUtWObADCl/gtry5xKO6xUhc2GTL58QoSl/adbEeqM4WrcM7IyJmOqfUsr6Gid+wx8SMmdUoyaY4l7V7MmDokYy32fN8MrrPgoJDHbEpfUblnEd0k7/Mea6tRvzVwRW68VViERZWyYwwr5RftTl/l2/lf9Xfqa9VW1o1Q8Kq5bPcAXG6p21WeIRLfI49Vr2N/YKcTelzwqQuINekyxyXgTT3+4nCFzYFJVfsGn8h2+DF+ZwTW3I+jriYpkzKmM/oiEOmPJARFZYOqWNj+P2C08oYEjXs2i47kiG41PWWEbqJCyR0WgfoLQ01v+7GWrcizw1xpOSFIY1dMywvDWlsORgkGIGLa8t1D5YFiBGXA7MopBae5yVeL+kQlWY/Z+V//6NbGMtHPvKRn/zjvMmv9fHRD/zf19eimIOyYJ8FqyQMfaX9YnTEnJwXqtv1778pusJ3VJcJUfdrccSwqmqqWSKOg3jAgjVSehIx1orfiG7wQnWrnuxSSdg269yr9oGlSC50JEIXq+6kqXIl2kUQDnVYO6Q0XT4Sgie/PdV5CwtRha1dcoZ2jGJZN2u0JCHXkqFOvJWeYW7nr+ZHNygLp4oTD2c203zDJt91zzoIpvZ4D929tFY6iHfRiGvB2bE6RGrNB/wJQp92nbcRJo2EyAvgS1KJ2WaVI8aE4MEFLtgtFCJ93ITuUlDTeiFe4DpTTpTe444ZsqM9V3yIZeFpVOE5V3SpReh73mgl7ryE7rtB6HuNSIJhhvNdvytTnjJr/Kbu8V45xyftEQCP6QrnkgQLzCpLOzL8anWX7zHnHUVHlZYPRjoqHSK3Ecd8uhyxQwsL9TEVWM5JxkYScW1bmefC84eWx1cjttcKBuOYohK++a33+fizu3z7U/f4rd87T17B78xPSIl4T6/Pb4wHvCXqExvhP/qWI0SUskjYP+jS7+WMvJh9kRvGc8PmSsndwwQR2OhVHI0j8tKF9BVWuVvkjHCLYYeYx7MWkYFRYeknhrdeWTAcJ6jC83swshXHWrDqC6wBxal7L8Y4qpTmZP7+DxStH/zwD39ZE94//8C/uF5hSTXib3z4b77qb/7l+3/+epuYDVI6JmItNkwqZT01zCvl8W3l7rEPxKpgXimDyl2fcKR3ZMpA5nw72zz1ofe96jUOfvSXrxtx1LW8gsv//KvrN//3P/hT189STUK3EpZZISFPKIx/+sz/9Mc+zh//4P94HZa2qU0B6lmKU03pIPDdl1arTguXMtYpTack4BSNNMyH4T2+lkakiYYYcTbAc0/1ElnS1BSXIp9KUlOHmgVHmKPPjsTbjYemEFDbCVdqaVoLG59jEcJc3XsOWhB7Krcj0F0qKlalX9NUw/t1WpIFscTMdEFbMm/DmzHVOT3pkIgrNkJmUnBDFN+Z75kOlToKqzMwcTSuTFI2ZYU9e8Su2eSO3eeq2eWuHnJBNskpT6Eh4bwHOlTiG2V1YLDXIjbzU9pkTHWGiKmpXBHmlAtWJimZZKxJl3v2sC746uws4hr1Dw1AcAVxON896XDgrZcjonrdal7bps4mfN90SQO31j9mLtL3FNzH7HpN2cyI6BAx8nuO345uck5X+DS3GdspmS+cHzHnOG9XKaTid/XztWvljjjkuKDiim5418SSHm2GTDFIXUyEc5VKQkpS7zUEqSlMHTIOdEBGyoKcLVllQ3s8kCE7usJCSnrqCrV36DabifsUtGL3/ieFUqieoieJwI18wa9ELzLRWV0oRuIyRbrSZqIzDIa/ZN/IW5Me94uCEuVD5lk2pM+b7S6fMDcBZ50fmqbOGavgomxyyMiFs3p3q/O6yvdGF5lVlmurEc8dFwwoWFDxUnTMfY7rPVLmyGt147JFSqYxq9riO5MtKnV0rMQIHQ+uh8Dt0kIn/co22F8rw/64YwwtiqZ+w61HkXGOVuHx6cIVLeCs69PY5cyksWU0i4iM0v7fXj+74q+aO9af1nj3M3/96c/88L+5DtDH0WQ2yThgwbVqndtmWG+yAb61ukCGqWkWo8rS8h/muVoSXFLyBWkz0pKpVhyx4IXqFqoWFak7XQFWhKXzUf216NJSF+PceuxhvQF4mL990fi+6cUe+K1zzck8+hIcN0Y6JpFVF/SjeEFiWmtAXqsr2PTZb/4bfpaHxZOIgTqB3bpxYVNB9JpIzLSR5ZDjHFAmuGCsmS7q0MUgZA9aj3CuQgHSlowIwwGjugDq08Z6vCS45Yy8ZWDqtTP1OSIhUSdA3/PC703b5tDMsCir2iKnovT0sRKlFMuu7XIs8xpKDlQhl2ae0JKIWzphixYFlldkSimWj9lDEPio7qOipOpSdQdlReJTpveLgjVZ+sCD8yM3IlxsxaSRc6t4a9zn5XJGx5d0cypWSYhFiIyQpSX9rmVnnCEC42nM1UuO0rOYt1iUMJt0SIzL+Qjnd1HBE6bHVtvwhosLBidd5ouIfi9HRBmNU6xCmRtmuaGVOOu9NHbdEBMpG72KygovHQh/WIycyJ+KVVIuJxmRcfzof2fv8q2zc9z4nLARwVbbMKgKOsbQ0qimYzV1ZRXqSQruPQeTiw7xKbesLzX+1jP/2dPgio3X+p0Kx1MurDLNhb6J2Ogq47nw4gPnchUJZDHs5xUXWm7q3Ju7/vB5j57x8I8TW//sT3cBm/hAPYPUX4fNahiJJI4ao4vXpfgII2R6hzmk1GUgqz5kUwfLDXgTZai0YuatfQNaS2jS+M0sig+mkxp5eS2RenO4Oc0Hngo1Dz8Wh9qknjLUlpYP/1vmFsFSU7ds1Cy1d8HlcHk+nDNTOJ6OaTPX/BQCFIqxRJbXKGyo2iat9SVFo5Ac6bjOmgi2qh1pOZdCcWhXR1quyMCldc90zorpYlVZlx7HOvZ6jAUpCaqWNbPGvh4TYViVHg/0xN0rRFwx5wDYFUfHij0VdtdskhFzqCOGdlQ3xKaesju3cxCn8wkaJXAFSHhMWFrqjhu2xaERF3Q1TfpXYAmUvhESCpBwrnPv8CQsqYfNta2pD2l+H67xwp5OP3foiOWGvc9Tcg1BeMWMaGnMima8YkZctSs1FegJu8NUCvrSZSbzOqvnhAnnWeXb2OYFc59SK+bkTFnwuJ7zgbhxTVuLMS5XKlCx6vfvBPNOI9Sqz+mhDmttiLsH5nSkxUhnLMQ18g5kxAY9SpSrdoW1JKK08Gv2Pt9tXQbbdkc4njuN3JuvDRmPM371RcOnzD4TnTHXBepVzZFGS4QVdQiL5AzKik+YB7zFbvpzaPiYfKG2GT5hzKasOKTf2xFfqVZ5EJ3wFr3Iy3LAnJzztk8pykpsOJgoqQhzrdgmo6hWOYkm9Z4pFPihIEs0ok3MjnY4LlzI4VrbCa5nuZBEbnNeWUc7/iO4e39NjryUutCorCCi9W7S6TqWC1eWKNOFYwUYj3SUFZSeov1a1KuvdHzDFyEAb/5X/8nTH//AL13filwH926R153Xq3aVUXSJz1U3EQy/F93lu6orBMmZ9f9PbeWF7I77H65DJoZPmXtQuQktLEa5VjUVK/BUg8gu16LRpfAezA0BX1jAgh7ETYpuNLUbpSpJ4M1KRIsWD6pDT2tQ/zu2Pg7F1ouTYGo6QK1TkSUK0uRTB2HkWevHdbPGiR3Wqe6CYV369Qc/p2Dh03qb2pPQCe1Jm0Xgg4plrgUr0mGi81OWkW3JmGtep6TPyPlme4kXzT592mQaMZAZFQkjnFYn9aiLYnzqspBJXAvVC0rGkpP5QLggYA6T00JcinWFsqkup2Pi9ToFMBBnR3zCgsv0+Kw5wiA8kBEPdEBiHHWog3P6eEy3SMW4Vwl8azF8e7zuikVb0TKG3EIkjjJyvFCeL8aEAKUpTnMxVUc26UaGKxuWveOEWQFPXp3xyoM2u9sznn1xhVnhNs+jQilLQxzB/Tw4lhn+w+yE72iv8fiFBaurE2azjPE0Yf+wTahN43iZIv7IlSOm0xZ50WW7VdY2fi/dT/hUPmKVlKtpxrP5iDe0XB4JwD+aPcucgpvRASkJ53SVbxpv8US7xYFPUO+aiBO73KhlmNrG8hWdsU7KgIIOEX0S5mcK9C9nPAwFCY9/+P3/6vojcYuVxPDy7LQjlQXW23Dk90GXOjEHc+fEcltnlFh2aPGfv4aG42thBDF0mFuauRKh61h9CReTr3SEkNLA+waHeNSZG7JEdJWg93BzrbPpdnagDl11+QhTT2Htmg4TewY19sF00EiJ982Fs3a09flR73iEszKfS14fVziWZtPnrLmHsKRTnX080D/Dew1he5GkdbHR3OyGYibXgtI7CYamVDjO2G9IUxJakjCwrnhIxVl5t8noygYjZkx1fkp3d1m22WfAI3KunquNCPs6qOk6Bc5ByBUxhvPGbRhTYjqyBUBf24xkeR/dtHv0pFM3mKa4QMLNyLkxTdTpB4P98bZscOLFyYGSFhCQBBcq2KXtrrsklIgP311a2AI1ghLOcUDhHYFseR9ERCQBXVLLUEf0Tc+9rp3V800YzWLE4vU5clr3E5CMheb8Pl8gGCrc9dqmK7LLVVZIEDKJyLRHi4iX5YBNs8ZYZzXCM5PCWalKxogZK9IhJuaWHPGYbvOC7NEiYUzJTbt3qil5dkx91ldP2mTEZJJQYulLm5SYrrQY6IRYIiY6Y0HEujhmwHnbY42EzAi/k5/wRl3nt9nnphwyHE/58egd7E+Un/+DCd8b9/nBp0as/eEl/nF1i0xSrsgOJ0zqNPUn7QVeMA/4ruoqT/YzPjGaskOXX4u+QJcEwfO1AAAgAElEQVQ231Fd4VPRPnsMqKjo0a4/E6Fo2MbpXNZtix2u8HvRK5ynTS8xtGM4mFtmarksbX5X9hnJgj5tRt4ZVBBWtUOMMBW3Bq7ZVt0zWvjmUxrDOIdusqQavd6b7j+N0W07RyznarV8T8F4JfwLoNYVIu69L/M/gjj99R5/JooQgH7kkQiFBKEgVMcVnQZH/EX7Ci9wm0fNBd5nHyNXS64usyRIYDvGMLeuOCgaJXLoisDS/9tZDHroGRdfH4mbciNZWikG73e3ELnJr/ScbTcRLh1mIHQIqxqGDsLGJhc5WBSG46mg7qqFibvpO6/qKVeqpwR+zYIkLPQVFSd2iLIUnLteta0dsTIvagwwf0tSFj5d1YhL8G1J6pAcSlqeBx2oAsYLIlWWiJJDWWL+wLxSn6+EDi5oragT0ecULHzYUeiELbuvtoaMS1zWx0DmrGqLmZRMqcj8z4eyYF1bRCrEGFpETL2O5Jg5pViOdUHIFdnRPifinWa05Al2uSMn9bGLh+kjxXVubBAnw9Ra+pGp0YCT0ulhUuLalcsqvDHpcliURAJbG3NmeZvRQmi1Cx67UrCYJy4FvIJpuSxgd9ZKnjtR3mj6LPx9e3W7IstKZrOMldUJDw58orNAUQrWGkoDWxsz7t9fY2VlzrntCaOR+8xM5xGrbXh80eVeteBWvuBx02V/UfIbxV3eWe1SGJ9N4el5Bm+/21VenJUYHBWtIxGlL54TnE1mTrDsFdZIKXDZLimv78rw/ga163Lj8S5wzn999Dd/+frZaTj05P/ih3/ga7YACSMUIOHflt+YOj79w13zXo8RS0BASjJxLnchNDHyVJ1YYgoNzmhLa+AmNSYgIhHLZk8oQL7UCM2VWqB7Rrwc+wZFTIyI8Y56Vd14CfNk83igSdc5PcL82pzXw9+mklBoSYQrSsL7DrlG7viWNLT670iIjPutkIy+0EUdZGhwWVQW5YgxGTGjxvkH2KBHRcUmKzVSHzZ7mz540IXILgvDMFosdSpnqX137D49cW5OcztnjqP7llrWmp6mNieTjKFOnH2wd/gKekfnkrdYGriw1L6E89q8V2NirKg3R5nWtOXQjHPnfqnZScQXKGq9LFkYMPLX4HQ+ydlCJIxQ3Ex1dgq9EkydG1JRcau6zzNyxNvlUd5mN8nEkIjQImFX13iZPabMKNTNc4HmlPpjjIGrusl7zTZjm3MoI4Z2UqOBZ0coUkOex4mWYKBLy9/fESMcsr8lK66QEfe+CyrebrcBuMeMe4sZ983YW9oXrNHliBG/XR5yYCbcYp+35Zt88nMrfPc3DamefTd7Vc6FOONn9Vm60ibXgt+Rl/ivzbdRCRzOleeiA4w66vNfqR6nwHKXozoHZSbe+QuHUg11ykejV3hf9WY+bwZcsD0etVsojgK8kQqtXEhEuKlTRsbtcfYZ0CIlJaav7foattRRqI/MjFwrdsjIrZLFQlk5y/lAV0KgkzlE/Ot5fOK2YER52zlIY4dsWF2iPQHxaKUOBXEBuy6MMBQiIhBHSvpPXt/k+D8zRQi4FFjVYP8JAxwX/Tl7o17UKnWT5EvVHX4lchPUe6qL9eQwrzfm8LvRHacnqZZFB7gk6bCANWlYYaMeuLD1FOLv7xDOF74LEC4EmNnZz9ZhWH5zjbpFNfjHR+JRC1mmWp8dAY1purJULHnSAbmB0wFUZ51Ggqd9KJYCnxOW9LG233gEfjK4MLTUUxdCqFHLUx7CRsC9blU7tQyZ0iEj9nSr8DoDcdzYGFPHZuUUzDX3CekxJa7z2iH1QvdQwBiG4lLQQ3L6VApQaBFjwZ9xralb4XpUPjE9Fi/691SEC2ywx4C2RJRquaTriLosDfe3QipOTDq3wV3HFaiqUODC8ABvB21ZJ2VMiRFHC9pNYyIjFEV0CkqdTDI+czujUqddWM+EVgLjSca9o5i/9qY5z91M2V1RPvGgRK0wGqdsbU5ZzDNW+wXDse8sGsWq0E5LJhNHzRpP3PUbThK6bXev7o3cOdkyKXt2gQJrScQPcpnnK8ffb0nCFi4w6/vTS6guE9gvpikHRUkqho6nZg0pat/2AkuGMw5YJu189fHxR39+Sam64P/9lq/6UXzlo7lxa+odAmcbTm+6Xq9RqgsjDfSK5gjzWkBB3GPLzBBHuFoWMVa0dt9rmdapHImzwzVnQjPHzSdrssahPak3jvVmU6taw+Hc1JYOP0DtkhVcq866X50t4nKPAMOy+LMA6rav7n0vf7/pRBZJROHzQdzrOFRgqjMitXWGREJU079iIq7Idv0crthwn5RzrHHEmBU6VH6LLSxtmOsA2Lph9fDxsAKkpy2G4ihWAx1jtaJnuihKQsyRLguQwgvMlxRf4zFq4wspN7cEnVCBc8LqSAvr3b9KLYkkcs005ZSOZEF+6t5tFjHBjt+t05FrQgl+hdF6PTslTn8ITbA5mvdQeE/NwqDWI2oOAg+Yc5kOhSrbfi4UhO3IBUJaVT5vJ2yaPgmGl2SPdbqcs13iGDo2oa3r7HFUuzQ232ewpwa3/wiF7IPKoUerpkcI/gwUvYKSqc7pS5fvqh4lw/Axc5+UiCOZ0Nc2T9pNPhbdZ+E/uy8blx6fU7AdJ0wK5df+sM+7H8/5+EsZqYF5mWPqz4ejm7/jSsHv30rYss7p64pd5YScj0Y3HJpEfsrFKziVhabti+aEseS8Kd3hTjniYpJgRJjmLtF8N014Pj8h1ditm9KlxLKqS8pzuGYdPDLkHQ23Ws3AUacDSaMlYpB+ne+Uwyc2jpUqF58B5wqPoH0xAvP8tL1uZJxgPUv+5Nbbr/NT++WPqbVMbeUXDOibmI5GdCPDVd3lZXv3VX/zgr0FwHu4SCRuyXlgS6ZVxUejV/hsdav+3UBXato0GsRpLnS5sIefN3M5wsShLIV1QWguuKCxSJvJu80uVVQvjrkWpzjeTd3Iw73z47pjFI49LNxhqC4nWFs/n6kn3s1ozWtQlCM7ZNX0/AIZ13kAq95GsvI8V2f1GwTWTnDuCBS2Xq6bFoNhEVe01piEc9ynVaMfirKlGYKwJwMSievix3CaBgfwkjlgSx2HWkW5J27BvG8PWTU9LrLJlnZYSEWkS5Eo4EwIva3vQBxlok9GqjFHclRf77tywlvteSpRPsOAS9qjLxEztUQqtDyqBtA27thaxgUCrpUR+4WzQzwmp0NMhNBPhV4Gl3dnDEYphxO3gD33+RUWpZtwWrET2fVaymNXRtzb6/GWayOyVs63ZCWTacr5QUqWFrSykrI0GKMMxwmjmaHftmxvzjk+yaisYCJlOInpdyqswmq/YDJ108eFVbhxLGxmhvk0YWwruuqoZ89H+6TE7LDG+4zDGM71lbsDuDlzgYYnRUUqbskyCD0v9neCzpJVT7+a4pChYLH75+PLH81N8ml0QU9p117vAgRCWJ7/WstTm/Ww8YuauR9+hjtrERzmKXDZBEOdOJes0AzBu2L5TVlM7PQGsiwspjo7RY8N6dexpzEFuk8qySmEOiAw4fPfpMpCsBhuuiu99qLt3Ldc57zQ4pQta4lFfAie6wQ3BNAkNZ2rJSkpyanCozlc1oylR+Y2W7QIwusmyiRIbcqhjfOdNrYGZ9EAg9DTVn2tgLoYNAhDO14asPh1xiA1FbikrAuOlrT8eXevtdAFSMbMOt1gJc4uOGgaW+LzT8RphEpKLssOBuGOHpI3jC1aplU7QzqUxa201s7qc3Ex2mGsM9bMCid2eKoIaY4QYPhaP28WH83zFUT3d+WE2Bh2bIuORDxlt+lHEW2bMCXnu+Q8v8ZdDMIb7Dp3zJguLUY4pCUxbj9wV064KNvs6fEpSqWInHL7Ontsc52zqPL6+0wyuj7R/YJscJ9j/rX5FG/gAvfkhIXmnGeDa3aNEwqfrRKyWCxDH2z5T/U5osow0wX7n3sba5HyvnfvMf/338peWfBh8ymuyg4A7XbJfgFX6HMuTrhdzukS85eqx/j/opdIGyyLXB2ZLpOUFMMhQw5kwN+Vb6ZSeIvdIDLCVhf2x/BCOaEqldK4XdJEcjqa0cJlcg1labhhEHZsl5yKdVLHSLEQJTB3/UcqC72ucjSRWgf59TwExzYoCqkteEWWFKzmCHbEweQxFCCtn3l9EZAwvs5P7Zc/Jtad+RA6FzZ+kQjvKi+RRDEP9IQDb42rWJ40j/P2aoe2GEZV5VKVEVYkZsR8WUicKTDOjlCMNN1bypp6sKQ8NUO9miMkHTetecU/58IuiGuI+tW2kfhiKCAZTYFmOIaAoDgkyC6LJx4m6jydyTHwC05tJayuW1VSMlSXxK5ia3u8lIQTHdUuXhB87LVeuDPvaBIKkMrbdqYSu+PzFK+OZMwp6JBRUHFPj1wqMBkZMVNyIgw9aXuL0MpRwEjpSIsZC/bFLdYJEcc6dvaX4iBgK4phi4yYDgkdTTiWsr6HInWp5laUAVOOmbBBD4sy9zzfrrT4rNljS3u0SNhjRo8eY63oScTCunSGlShcJ6XwMOlKKpwrM1xCe0RbDCdacrKImOSw3k+4sR8xKS29xJBEzg42NjDKlXe9cYqqkGY5m+tz8kVMUcTc22/R71S84/Ep02nCdB6TlxEb62Mq63zFtzen7B92MJF6RISaI2or8ba9vmtlYVSVDGawncSc+FkujYQnix3KyPJUdY5KlM2WoahcGNR2nBCJuN8POhljyNWyTead6Mo6HyVB6JLSkojq610p+FUeTmO2LNZDlzJsmFJxVIjXU5AeRoWtw91g6eLT7KiHhkw4WuBUIRLmxjD3hM85QEfaLDSvw/0CogEuIBCWrlEG8YYargBZN6ssNK+Ll1CMBKvc8Hrh3M10USM3VtzGNIjmg7UvOHplU3wf/o0wp4oV9/jpeRuoUfCQCwXLMD+RiMuy7WitKpSypJo20Yql29OyWIob1x1O61WCqPxsAdUsQNa1Q9UoZBTllt3zCIdznlozK4x0fOraBiewZaigo/62cY5j4e9DcRJ+L4xcy1pDiDq03IhzzNrjmKvs1OcNHApvdEnfinTpfhUacRHOgAChDs98rRGcvZbXd4miNVkBZ62awaFsE38fPmDOY9Jj4Z/nm2WDT+kxt+yMd8suxxQUuBC9uacTH8gM1d4p1HBFumzKCnftAVaUy2aHOQVjJg9tOIb34I7VXxevmXH5YQ65O5IxC81J/fW4byY+W2voNUIZJzqmJSmlOueqtmQoygLLIyvu/E8ry4vG0ZDvc8yd/BwiytV2yhsv5fy/Lzpd4x5zbpkBq7QZ+wZj4ZFTxdFWnClNxBpdbpYz3ph12E2SugAprHJe2jzPCSNZkKgh9Y6WO7bLQBYNa3vY0A5bZLTEMFfLxFaMC8NG1wm088oZkAxnwmr79XWC+tMaa2nEehvA1va7+H2Gqgvhfdj4auhivgEkN1/e+M4Pf/+pG6nAhb8dlyWrkpARsyZdBMOboiv8CO/hqWqHDZPwsk5YjaIaIVBg3x77jv2SahC4q5GnEtX2kmfKTdfJWULpDm0wfumzfjJbLgRznXtR51KgCW7BSv3ieLZDEyae0GlsToxnvfNdKJebACJxi1/dTRS3cCeS0kz/De/jLBVirFPnXe4tM2OJ6ZsuG8YJHqc6c8JPXfqeByQjdGRD99Et2Lb+OlAh3HtwgrWwWO/jUIxKnTf9iBkjnfjFxm0OVJ2t50RnLqALy5GOmOrcJ81WfoMWunvuvJVYDmXGgUyYSE4hlU9Wd+nZBRU9WvRosScDFCe+TyVhoi47vqcZsTqR7UItPa9/mKslEaGwSuSL3MwIs9w5WQlwqZUwpWSulsezFsOy4qS03HgQMypclgV4xyovMkuMcPNul+1dZxG9de4QEylZVvDyEexsj7GVsLo6Y3d3wNbWEGu9l36krgCR8HxCWRmyxBUjwWWjm1mSSFmUTt8S4TZfm0lEGgknecX3nm9xyJBfjj5LaoThQrkzVK5uwGFZcliUZCJLqpr6ggdlbi0RS2TObUSVE83/SO5Yfz6c1W5AG42fa2zjHOZ62hr59RzB3hWoN3EP6yg3N0tAY56T+nMZHgsWvwGtsDgzhzBfdb3rXCgoQgJ5JhnrZpV1s0omWS2APnW8WB8SKHXxER5veZTkLDoQyenmjGncs2cLqfB1GFKvHA3tnUSn0JWF5uTqEnKuyW79e4W4c7v0C1tSrMJxBKP0h2kbzKlXdzz9+AzS2Hze5rGHufiy2SHC0JE2W9F6XdCpenMV/wyR/68OmPQNr0iMz/0IOgBTz+vNVPMt+kQYdmTVz8tufp6Tc4MHXJTNutCNMLXuKLyOW9eWTIJYYl6x+6eaRmftlpt06pDD4c7vskgKBciVaPdV66Ng2IrWSSSixDIwi/oqF+rmuHeYDR6N23SM4ZxJuWhaPGrXuKibnNdVvs1sYv0eYU7BhEV9jtZMn2vmPBkx+/a4fs2z633zmMIxH1YnDOyYO3afhS7ItWDshdxtMhKNGMicPV+AuAZZxYp03ZX0aNdYZ7Ql43yU8mCijIddKuDTvEJJyY/wJBfTlDiuuDHL+eSNhO0k4ZxkbJFx1a7h8DH3XypJQz24XPsLSjrE3F7kHJUVk4UTlE8qy12d0dGYvjoXzY6m7FhHx5pJQSEVI5nxQEaMZMGAgrk659OuiVjNpNZ9RH7dS2O+IQoQgMTAvbFSNEJuQgFiFYxorRMJIzQeRU4//nqPPzNICLhC5Nff/4vXwW2WEoSFWtZMzDuqc3QkYo+LnLMtWsalLGciPGZ6jGzFVCs6RD4RelFPWGGi/Tv6bvexUfg/5KP1BjqMs8VIcDtxXM6l0Cy4n4CbfIPADpboRYmpi4tmavDDRigemqhLWBRC3FeAysMcKo0CKfBLM8nqBf8UPN+4QUM3sqBkM1pjaCeUWjHTORWWdbPK1NOjAhVkrjmZJHRo1aiHg9CrmhesfgIUxInWEY4Y0yH1qe1RncgMMLNzREydHF+7mij1oj2wy26d6FL8GCgZJRWf414dwjjUKS1fNK3heM835ZCUhFU6dSHZNBtY+LT2sHA/Tp8hJZkaplQMyXkj/XoT3k2cT3nlOamdyLDSgu4sdu5QRUTLGNqRkEYu3TVMFnkFq20YzuHSmmW1nzM4Wmd965DFrM327j6TYZ9uIqgKa+tjoqSkyBNGwy6LPCJLLUUl9DsFw0nKaj9nOE7odQrG0wRVYVE4epYxDqqOxN02u62YeaV8Ih/ycnTMgYzYetCvO8CPbSn7Q8NW33L3xC9qWOa2YsMk3u2tsT0TaGlEiLab40TpFmX6GrSIPx+vPcL9HTJBmsF4f1Ki9DDOduhfCzU+W4iEpPHme2gG+eUUtL3tbG3Xi0s4b+YwCYYV02doR7WRh2IZ6cRju4633vZzXCg+QiESHIyCODockyEEyLrC4ZR966lT6n4/9rq3gAbFPg8ivOdAKyu0pCtthkx8J1jrTbQVJVMnxg5IxmkLYCVTAyH88MzPvtR4WLGyqh2CW2S4VwqpSDSiT5tXdJ8Ky8w3swKy4Qq3Vm2ZG2FoeWG+0whVdKTtVzHzqmtrcNbuqpa7csQmK7Q14bxssM+AEkfxDY3AlITSuzUmXrAe7PITjSmkPIVadKXDGl2mMmfss0zCz/BU6aY2MlD3WpIytbO6mA7GBoGGfSk6R0LEoY64pg6leTfnGNmSbuICaytVNuKYhbX1/WFEuFcteFPc463SI7cuILVSyDRmlzXebre5lKR8uhhzwwz4lL5cf65CDgpQZ6S4a2lOfRZqOmJjPTcScWxH7JpNFGVPBsQYTjztETy1Uh3CX2s2cfbWP6t/wG61jvn0VTZi+GDxJAJ8/3sP+ORzO/zC77f4regVrpUbXNIOu2nCSV6y7RHNprYmlogO7r4xCCkpW9rjfOoypt7xaM6zNyKGZeWROTgw0/pz8Ij2WDMJL9kxq9piIjluFY+4JwOuskJLDBZ4vB8RR7DwrNTSujC+bySw/eL/+Vef/ux/+m+uh3TzvFwiHKLUug9VmBdfXRH+n6kiBOAvfvgHnv7I+3/xusEJIMELslHW4ogbRUmlylAtG5FLRj6xS870AsuuZFwzl7hl75NozCWzzXurR+hLRCLOhQrllA1vyARp+suXXyS9Fxxc6rzawwJU1dAvLBeL8H2YPJsox9nipBnc1VxkpnaZWh583Numgzag3aUTjJz69+zomS4DO6SoCjqmXXvaD3TsN/JpLUp3E6ArOCoq72bVoF5harescHxhMxXed9fb9y1YPmcN2weamS/smrz4pm1m1Qi0giWFoKKqE+1dkNcCK8qcnDVxScaFR8NKgqhueQ0yUi7omntcLM/rCX3SZeaHlPXmO7fKeuzE6WkMszlc9vD2HEubyOlFEsPRomJWCd1IyNVRn9Y6TkSmCpO54YknDvjc5y4wHLXo9+YkixRrI6alUhQRWUupipj5LPMBgkKWVE68Zg29TkFZGmwlRJG6IMPSFSDHo9iJ5I3SyaAcK5XCcVHxLekK5DCLcu5z7IpxgbWVnFYWceNBQi+DC2XK7WLBpkmYqxIBpadlRXhurjrB/sif3QpllaSmaP35+PLHz3zo6af/3gd/0jdh7KnHv9Ln/Ac/9I+vg8s4+rlnfvqhz/Nzz/z00z/6gf/h+mnNhJ+bGiv9w9LMgylG+MwH7RssCwKLE2sLjqYy0HHdDGjmjAztCONNO2pEQgx9PzeBQ4RSSWr0pUlzaqIJ4edF47kSGgWF14uEznxAgJrFXrP4CIhtM5MkEsPjcoG7esSWrNIh4wvqtIuFLOerZgHS9kGqpbj3vKCq5/+zwvIvpv95WAESRlNfVEjFlAUXzBZ3rENdAyoy0DGF5mSS1Wueor6hlWKxxER1wdciYajT+rUMUjfjSinJteRABnSlRZuMTVY4kAFqLTkVt3G2uMGCPfey92B3v2DZZAuZMlPmfEHvsWFWmDLDNpp7DzsfweJ3Vzb4Anca78uSEXM1Os/jdpvIGo5kyhXd4FFdwYBzXRTDoKwofAHyseqQuZQkNqKQii3b4VHTZW4td+yci6aFVUhj4V3RpjsnMbyYz7hlBtySfexDdsthr3EawTpdiIQRUCLBoqqc6AiDIZOEjA4LXZBI5AxifGEZWAtB77XQgkwS3lNdxEbwvd++z2za4uOfWeHf//4On5hM+O3oZf+KG8QiTEpnOKLAo3aLW+aICYt6XZ/pomZmgNNX3sg36ElEu10AKXO1dE3E82af4Hp5zroCxABz8cG5mnAs0/r+XZOUVATFUZhT6018FPoth/JPFt9YRKEnttULzZeZX+DZDgpl5ZywmtQr8yeMgsCfITpWc3zPh3/g6aalpgE6EjGvLI9Il0SEGVVtv7thlnD9qom5rwtu2fv1Rv4vV9fYlBQjwpF1m6Vds1VDz4qt+c7h+4BonO0Ihu/DwtK0UQyb4jAC9BpGoGo1ealLMeKSC1x3epqv2+jgVR5xaHJogZpe1aSEnX4OpyWZ64JVs3Iqw8Txr/s+Xd1t5qe6oFLrJjiWepew+AdEZKoLjCy7cxZlRTq1E1dO4ZLTz1DTLMrULrm+oZA4C5kHvnoijpoRNDJLe01bH5MTuJfMteC+njDTBX1ciKFLel/S01o+pOymODeRSN057hCzJ1PueSvfRIRCXcersA7RgKUwzAg8nrW4lrXo+Blipx3RjoRe6pCIwiqDme+2Giew+/jvX2IwNcxmEVvnHtDuTml3p7QiodX2AWhGMUbZWJsj4iaospSagqUKG+tzBqOUlV7Bat/TFowrSsZz97uxCLNSWU8iVrIG9aPOnRFG44Tb+66TdTR1MHrAPRLvFhb+plClVCXDJdFnGEpfgABEfHW7Nd8o45986Cfree9nPvT0038cDch/+cGnr5dYRt6m9Mc+8BPXX+t3/9kz//DpZpHymkiIp/Gc3SQF17+QxeRyNhxttSUpK9Il8/kUyyJlyYsPFFiryw1m0CkMdEwqCR1pOTMLLWrkQ9FTSd0uvdxRo0IBEuhGc/94cHBKZIlmh2MP800T8QnvtUlzionYpM8resCG9OngaCaPm4s1ktscmcZkuuwphvkzxmVuhCLnK6HcLTOqvJGBhLXL/denzZ4e05E2maREEnGiQ9qSeb2OszAPuS/gs2N80XZFN7iiG2xoj0BljSXyyIi71sGwpVJ3v2W+iWN1WehZlCe4iOAoaEub4+XrNu+pcP6b62Wz+Gieq0Ala5kWW2YVRfmm6DHea97CXzBv5Tt5K4/aLd5oz3lSr/Kk3eZRXampo8+bY/rGuVpuJzG/Yfd41tzmgYxcRgjKY6ZLxxh6seHRuF2v0sPSMveU3Urho9Et7soJVt393xyhkD/7GQqPnf3shQI7JMkDbMoKYzvlrj1w923DaWzh1+6BHWMwNX0vwvBvo8/zr6vbPPv8Dp3ulO98ap9JoTwXPaj3DS2NGGjB0Lrv22K4pn0yb5ecklBqRSIRK55hUGHZoEeLpYX9zTwnQjiyBdfshnfBdPrObrT8TI3FaUM7ZDxRbfMGu02u7upa3HqZV9S2tOPFVx8N+GqNRWHod0q67VczCZpZIWcLEfMneDrkIx/5yE/+yT3918/4nQ/80vWWCHN1uQrHWrBjlh/uI4+GrJmESh389+/Mbd5bXaypNMsgMOfkEyEMJOff8gf189QuWuI+bLCkB5lGdz+MIJaMJWbqXT3Ois4etrCchdTPft9EVFQf3lVOJXUwd2MyjyWpF9BMMifk5jQf2sHVDmKd6JTY867D4hAoVgUlBlMjPrDsrLgF09ZULEFO/V1fnN3kUKdcki1OmNRWoOGcTu2s5oiH9x5MAcJku9R/LIsYEdfdykgd0K9LN5dUYspaKOo2QSt0mOBshgOdrCXLaxv8zh/XHYw2ubrC47pCIsJKFFFYZTNzx7Xecemt47kwzt0EMSld/1oLF4kAACAASURBVNoA223DMHf3ajd1YjrBTRZ5Be9805j7DzpMFq7rkSWW7a0p56/c4JWXr9FuL+j0J+SzFmUZMZtlHA9aZEmFGMcRrSohSysms+T/Z+9NYyzNzvu+33Pe7a61dVX13tPD2UkOh6RoibRkhjQlRaasjRQlBAgs20AMG0gQILFjxwjSGSHxBgP5YDsGlER2giyQJcqRLSmSEEkkRIkiKYsaitvsM7137bfu+m7nyYdzznvfqpmRbYQzcsg+/aG7quve+251znme/8b+cdzoTQAubuVcv5exN1PSyHW1Uj9TdSKhm8A/mD7LMe7+bzLkx+KrDFKYV/DKvOByljKunGXC2NbN709HDLlHhwAW1jKjpsSyISmFWmdXDBxTskrKv//zH/um4O7+/2n8tU/8vWurdBkxb7Ig3ggNOT1+4uN/vSlY9FQnV0QaEbn72jTC7+VrbMP5Dz/T5v0PTI9DO24Qz/Czq2bAVBdNc+R0x7tdWJzetJ/uIrdtjsPc8nooQ6AhhaLEaTuWOSCB3lX6OchieUDOMmVBl4ybusuDcq75nPZoFx5hVHJyPm+H2rmEddOUTmGE413VLmNZ0PMug+AQ3EjNCfTDHfvyHMfMuaeHjWNVoFjNdNGgIeGaJQG9wq0vmSSNMcmK9Jr37NNhl1FDwdqRY3qkLChZZflzr+oOITelIynbrHFHD5r/dw5ghaME+wIwIBiZpPSk27goBsZCOxPm9dbX0zqiEB4Z3vO9POjE8QjPyh2sKo9xno+m55lWlt+0d/kKNxhIjyu6wZ/kHK/aGSmGAstjcZ+XqjmvmhEfMef5jN11WSF1zWoc8Uv1LQzwiuw2epZggd+2wp9r7ovipb7yj0LAgIau2L6/IdV+Tfoc64y5OrpzB+dWNtcF62aIRVmjz5ScC7rGj2SXiA18enbEM+YWAD1SOqS8s97ii9HdJmOrpOKYWUNlXpcBlX921+gD8GfNZWpV3v9QyUu3M16eeJt4zflytMNAUy7aFTpEDIk5wAVT77DgqvQdDQ63tnTEZbVdXTWMF9BNXBHSSUBE6f/UN4ceJAz7H//StRA4mJeGLLHkpaNzh6wQEVjpV0zn3kDCvnmuWGF8y9Gx3mi8/5M/8G91ob/+47987UPVJUSEtTjiqHLJzwtVPhvd5HvtVcZasanOIjIE+A2kx9yzWNvw+Gm3mPZoL2ztEuWNNCDt17zR99rdH9tazBu6Uuguad1YHaraZkEJosGudMg1J7hvhc8p/SLfHm07UCNCrJEXF5rm+2HBbvPWS29ZHP4NMNJJw98+kmUBkpL4LmDxmo1A+DsUdapLK+DY2wSDowW0jyksoIlEFD7tGGgSWUMgYuY5yPs6pvSdnNKLcmNRF4hIh0QNZ7TLXTNlpCWJCmWlrJiIeaX0E2Gag1UhjmBRuTs9sZaOSHPX1zLhKFdq64OHfCenm8BkklJb4am377C/t0IUKWcvukWgrgzd/owkyemv7LF/9zJ5HjdISEhI73YrxpOUvVHcFB9Xzua8cDvj1m5GHMFWXxgtHL+08p2U/bxm20Q8arf5PfMKimWXEYGPGht4sJtyWCydhAA6IpQKuadmBSTSCAyIsBpRquXYc8VTImopGGjMr3/sn18L1r3f+fM/+E21ePy7Ovq4eWCkE9bNkAP7WnvQNxr/yyf/7tOhEHE5Q6ZxDwwjWJTji/7TDlSB8uS6uLZJvM4kpdSannRYaL7slHu0N8GhKiHLKRQwEDZojh5aUZ3QlQS6WNjEnnDragqS11JdAw0p2BO3CxDFCafxQvSpznjcPMCreo8H5Kz/OWn9vXzfdgEiCD3fyHFIkjPMmEnJwDsFuQZGythbiq+08hOCXHxfpsQYJrJgqF0UZcy82QS2hfbtMaTLLd1troNzWErINT+xmY+9o5VBmIv7v+Ac1SHlWGfEvgM+YkatllgMr7LLVa+tSCQm1ZhCKuYUxEQ+B0TYZq25LmEDm7AMwnROjpZA82uou+E++QK4vR4tr9FSIxK0kg2qIJWnTTsk/vM83xR/l2WbTZ/bcmtRMowi162XDmOdcl2cJjE2Tmz+AdmktK5IvCmH/Jq1S6MBcRkPV+0qj8cD/qAe8Iy5xSo9brFPVzKuylle1R0WLX1OW+D/rxuqSi11w0wQpLkeRzrFIHTFof8LCtRnlE19Q/JYZgiGd9pNYgPTUjmvXb7qi40ayz5jvhoJGTGP1WcQ4DPRK01R075/EYYZOeva48vlmEvS41eeNZxLXTM4ESdKfLze5J6ZMpeKSIXVKKZjDUaEHQv9yLBfVSQibCcxpbo19N5EWe84G97CExm+2QoQgCTxrqiVIYkV8UGFkfPGd/9uoR71G28vv6HjW5KO9Y0Yj//MR5+OEDIRxlVNcAbqiOED9SUEF4b4TLSDYBiaPk+YK/SkQ1c6ZJI1i5M23fLX3o6w6IV/B3rU6U7W6fFGnNbwf6dfL2IaSpawtBMWnA1woCy1PeZNCI3yo82ljSVmoUXT6ZtrfiLdeKEFFbWPHAzyeuOTMITYIyGNVR8tQaQX04WNRq2WCmcXOCd3Gg61JC0BaRjBz77NCw8jXH/DclOizXIVFlJXuJwTFzA1pEtNTYeUvnbokXFO1p1WxG+eQpcNYILLxnjVjDAqHEnOzPdea6AbC6U/rNrC2qDm8hrMa1eAuKR5d+cOF+567OfuBf106Y519yClqITJuMfVR15CREk6E4rFADHQHRyTL3rcefUhDvaH3opP6HcrIqOkaU0c16wMvWtaELSVhu0Vy7Rwqauzwn1mbBylLNjzjnJlpdn8OHpNXmtTsACc7RpPL3C6j4UGlI5GFzJTl0sy11AserQFl1zf05gZNT2JiETIsfzKxz557Vc+9slrn/34v3hDetD98f99/Dc/+58+DTQW2Kkk/KUf/a//ra95g+z6uS7Mi2G0tRjwOgJwX2QkEpOICyatqFrzROVRypqRnTDVucur0KWrYdCE1VgqKhJJfEfY+PTmk26GgcZjW3NTcOtqU33aG/DTSEYwBHHhhBWqtmnwXJJN7uohN3SXnl8rQtFjEDKNaRcDr5kj/Qaup0nzmhjXwBhqh3VdIglhVCgxhh0dsaMjxjLneW4jCEeeOlrK0kGwrZmZsiBQXQOyFa5tGwUJx1LjKMnhvAEW3pZ2oQV7esyQLudknRu6hxHhZe4xljkdTVhI2SDjqSyvRbguFTUdnyJvRJpnJPbUtOU5V8x9WO5T8iDikfm2+1XYjLdZCm3qM3DChKYrTqMYqH8jnTLQjA/LeddkE7hsh5xjnYF0meiMm3LIXTlmQcnL9ZyX7ax5Vsbiju8L9QGFWua18o5kQGzgUOZ8f/0IwYr+Id3iT9YXmusfnrPQeAvrfnCSe71hJOKc2TxRcBqkcbHckCGRL9zD9wJlMewt2r+v3/HonOsyae59SLS/yyFvrzeJEFZIGHjKIcBQuq7RiWVD+3y4foDv0LM8ZPo8NIj5gXeWfPtjMwZRRKHqA20jzto+BmFbMqwqXWOoVdmkQ2Fdo68rhnFlUY8ArKQu0DckiJffpJ4n9X+/bLRbC3nhHC+7nfo12SBuH/DWHNd9OtY3YHzh4//yGgQnF7dx6kvEqzrjkvR4mQkDTZhIyXVzxCW7yk0zoqTitoeNcy2aRSbQtZadMjdpBLF0CBYM+Ryvh6S0J4PX+36bzhD+DpaQ7e8HHcjpiSWWmABpBy/+1GtAAoe70ooV49yR5rpAMCREdKVzglq1dEZx4YZBnB6oWu64HToCjs4Qi9sahOvWlazhageoPKAq7WIjXF+gyWxZUtQCRJ95/nHtCxlXIIbckkpr1mXACYEmNX06LCiYk7NQ5+MU+LIVNU/xAFu257rHkhMhfMXc4R32PJva5aLpsJZ4x7XYIQuD1E2MtYXDuRdtG3hwu2I0ibk3VgrrUINLA8O0cJzhb3tkysrqFLXC/v4Km1sjjg6HPH+jy3vevk/WyVm/8hyvfunb6A/mzKYddvf7iCjntidknQV7u2sYUV663aW2Tq+SRrDacy4aB1PxmR8wKS2RCHeqAgPcZc6L0YGn3LnF5931eZ4z7nn/97hA1PqdEZwTWChiHLVMKLyNYq7KgRb0fNdzRtWEFs6oWCVlQU2HiIKaHnFjwx0h9Ig4oOCHfv4T33Qdrj/u8Vc/8XeuHeusQSp+6ud+8o+8xj/x8b9+rdZlEjJw4vewTYVqWhR+rgvWre7nlnkVQcwNTm9QUTfHE+aoQDEJ2Ru135ypWi9idvNtVzqNriRQNxVL7Tu+YU6J/fs0dNFTwYNBTB/G6fk9iK4VbVyhrsgWN3WPB+VcM0e2h0WbTv+m9gjqkkCRSjRiKl7v5Tf77VaLOfFe7utg0CIIr7CDokRimOqCmIhtWXNsfHXnEz6rXSi+YG8hYuhLh/3a5UP0TLehxIbrY/25hhHWiVhiUhL60mGhBduyxj7HTX5Hl8w7oblMinOs09GYhVTklA59pmJNBlzSdcaSc1sPcEYrLiw3rCczO28KiLARD0Xmo3KRr9nrDSXPPXPunrb1fmFkktGTDj3JeFjP8iWusyYDHrJblOLCVR+zG5yNUv4Zz/Ij+ii37YJXzYgX5R7gGnI1lie5wp5MsChTFgzpsqVDXpV9ci3YlBUWlJzVFb7XXCT2FquRQGHhD+oR356sYoHPl0d8Rp5rml/heQzreFi7Tlvsg2t2rsqQrmSs0ONVvde8tkPKFdliwoIEl63WPNe+SOtIytvUBUieswP+8++a879+Zo2vmyOGNuWFaI8Z7px/TJ9wz60Pg75eL/h09BJDOg1qskKPt9XrnKfLhIouzgDoB9+VMzrOePZ2wkFZsxYbbhcle+Rcki5rSURinP39MHW/h/PSracHeU1mDJ1IiI1zlQxj5X/6M9/Ua4T+J790rayEbqdmvoh8WrpiPVV8dVhSVcJ8ERH9o+9/06/FfTrWN2D8iTegcv0J//crH/s/r62SsEXGDUZcN0dNVyV4YScSY7Qm16IRfzeiRWlZ67ZEm2HD/3pUrjb96PWGiBMsllIRaF7OpvLkz7e1E0HUHmgM7rjTBseIiOhJzEIXqCo96TYFSNhUNCnwiLdgdMdRUFKoW2zaovyAlBS45Nuw4AcKQyhS2om94WvjNRttLncQqC47mab18wnBKtGlvkcNKuIm2mV+izt+99lzcnItSfznhUm59p8SLCG3bM9trv1nvGB2MQiP6CprcURhldwq/VgwAqsdV3wcuxqPrYEwK2CQwb3DmO21mpvH7vg7RrhyNufZmxlGYL5I2Dw759WXLjAc5kzGPZ673uWhiwvqKma4fh28KH0+66AqXLxwhK0jFgvnlHU4SlmULk02hBltDCy3Dl0wolVXlOzO3UYnM849ZkrFwDvUfKC+zJ4sOKddfi+6S0lFh5Rvv6R85rqS+o52oBmsxRGjqiYWITVCWTsXuUKdk0qJkhMQJkuF5QxZI/4M92dC5btjhhEF69Lnjiwtme+Pb9w41pnrYP4biBf/3Mf+i2sibs5SVRDb0FjazljteavGQpNOvszZcPORe03oxDoNmNtaBw2G+MaCQRpBeRhhDl2Kw5d6j5K6EbMnkqC4Bkcw7ggFSNhIO8Tv1FzzmnNx29uMpOnOBxQmCNoDDavttmUQUl98nNEuudTsy5xV7TTPfKyGqeQNx36ZEH1yLMQZ5vY0bfQfAFNxmgmLE9mHubjdfDpdgITN7CPmEs/aG0x01hQf4ToHd8KAiIT7G2h0LuTQFaUFTgtwxNSlZnudjJv/l3TYkcwQ6dPXlLkUztRCXWbFyHfRV6THhHmj4XMOTw5xM+ptaxuLfOeWNZEF75AH+JK+3KxpYYSmWZuWtSJ9LMrb7UV2zYT36FVQd53erZsoyq9GL7PwFNI9W/CSOcTi1phgqnCn3uVr5haTekrPdJ1eRmruyTFTnZMQM9COC8aVOV+pxjyVDptj60TCe8QVIM8WM74U3SK3S0OF5TlEDI1DCw68jXXQfAKcj7YY0uGm7vEX9d38JnfYlNVlMSgZY58lMmXhMl68oHzXOnOCWi035ZCKik16iFg+eCmiuLnCx59Q/sHXN/iKucNZXWVma9ajGBEXHJ3Vhu+tH+a3ouseUxRiNWyQcUjBWEqu0idXyxeeHfD4hZL3Pjznd57tEBmhZyIuaZcaOCrrxshlUS1NT7qRsJZGjUVtGjlU/40C+77ZhvyD73+av/LL18rSEEdKVQtFLaSxM8dRG1yxhLfiktynY71FIxHhts65bFcpqRgxZ8ScRKLGrrbGNhQoWG6swzgtXA+LZ0A2Xo/v+UfRskSkcU45Pdqiz/Z7tSkGoXMYFvjA/00kde5SPgOk3e2MMOQUnmLg/nQkZSA9T4NYOmSFAmT52d4XXyIP1bYFlzU90/XXsW42NIEKdFr82obPA9oT3mfmJ9xA+Sgoyb2T12lxX41tdCgWpaRmlT6RL/LALV5XZIsKywoJqT+nfT3mYd3mgIL9qqLrkYDDwnKcawMPRwamlbI/dZ2cvSmMc7h36Dt0fhP3yt2MNHKaC5dsHvHQo69y9uJtosjywLmCTqdk68J1d+JRjbWC8cVIpzunqgz9wZy9/R79bk1VO3rYvHSIhypsDpbXcla6rJKOEe4UJTXKlsmIfeH2XZsdnjSrfPcl1/1MSThiyvXdmLbDlVWlsM7m14grPKa1Zeyf/5KWkxaGBEOIDstbm60cJ1ofSeG6rkRs0mHcChq7P76xo62tAv5IlyyXOXTSLXD5Pu53tVbbbIzC+4Z5sDHTaKEhwAk6a2hMBLOLBK//0vqEQDz2bni1p+6EESgrDY3Jp6hnkjboaOLdugIaEktMz3QJLkru2Jbugs35eNvw09a94Xiu607TqW4/r4mGhopSoWQaMSA9gUY444/ltiF7nR6jQbjLYaMT2dERc3GoT08zLI7a2vGi8YqKntf+hNeHa9T+3BrLeXMGgJmde2pdmBMdch8CIOderB7eJ5OMVTNgTfpYVY6ZOZczlmiUQ3RcWO+a9BnSbRo+XVLWGTiBuz+ude0yYd6gTR1JyVuuZ+1zaRee9/SQCOH98qhHv909NhI1gm2AVTNEMA3S+9t8nRfsLb4qt9iTKe9ni1qVZ2SfQ51QUTHVBXdkyj055ro4O+MrukVKTCYpEzv1z4grIN1sqvQk86iY4an6PFNy9syM36h2+Fo5ddayQGaET1W7/Fb0MqlfPcMI4blD47Q9xan5MJO0oR8uKFmTIZkxfI9c4Lvqq1yUMwykS5+MnMqH+1o6pE0GzEB6DnX00YMdUg7MnF/+3bNcvbrPn3//lDSruOrT399uz3A2SXjvlbpBdIYSk7B08nT3RfiKcQ6TG5pRqXIxTZlXyt4o4fPPdyhVKWrXrJqq0+f2IufumLSaG/NaWVTus0RcAZJEbn37VhqR8cVHJVh1DcVg7w9OmD/svRYle1OO5Sd+4ic+9JZ80rfweOePP/npm//s2Q9ZlBejQxa+q58Sk5Gg4H3NvROViC8qnId2sC4VTwcyskw8DqFMNe51brNO83fo/IAXZIbuAtGSFyoOjh6ageNPa+04n7J0MnGLckQIO3Pf945GHj1xYk/r9RhO6B0TOQG5d8KSZlF33NmKmjkLd/7qtBMlles4iWkg9FLd9wJnO6g2Qv87XBc3pPketKyFPdVsieK442gLUy2WjmQYMd7DPqGiPpGcHL5ecq0jcn+8gULSJXPWmpKyLkOu1KuskbHA0YM26PCEnqOrMUNiehIxt8qZJKIXG44qV4gcF3BlDa6Pa0q/Sd8tK8AhFL14mVQuCGdXLeOFcDyJ2dld4d7ddfZ2NqiqiDSxxLFlfes21iZUkzUWsyHzeUq3l1PXMecfeI4ksbzw0jYH44gLZ0r2xhEdTw87njt0IjauKJmWymFVM6pdN/dckjK1Nd91LuNJ3eL2xG+T6ojDHA7MnJySH946w52xEvmCw/G2XQZK4gXqqTENjz0R8YWIK0Jm1MQYMr9ghfDCym98UgwzqahRBr6jvKld3vbjT3z6G/m7fX/AD/74n/70D/zYhz/9L37m1z8U5qRf+tlPf+gHfuzDJ671n//4f3lNgMhb40Kro+7nqIKyQTvBhYwqeDteSFuuWaGpEeYUi+9ys8ykCPoAI4bUp51PdeGQZz9Hus93lr9O1F75AiNqksvBNxw8lSi4IbUpX0t0piWAx5ENjZ/LYoncGXgHpiU9xl2Dq3KWgBSHa5NpzIZ2yUjIWOofeiSUWCqxxGpOuFa1r9HpguGG7rItawiwSp8b7LHPmK50GDEhIWnORVE2ZLhcUxzgQIzxqDbNPUiJQQw902Fq5434P5WUmgpQKk+/DdqXTNxVmpMzI2euOU7EXzoKskeOwmcbhFosfTpu3RTLQDPm4uIKRRwdb0sHiEQc6oRV6dMhofA27iVVs2Yt6XMKvsm3w4g9GfMuucp5WedIZt6+OaEvXac9IkbEsNCFE4CL8KQ8yG0OqKTmPXKOnzFf4UW9y4rpA0JfOjxuN3nFHLLOwCEdHDJm7lZYHz7pjk0bF8iruskZhgjChnb5U+YCF+lxnh4X48wFGuJWvQdMn1u24FAmrMmAic59keiejY/YJ/i++ArPsEup7l6E362h9PnT9hG+05zjMd3k2y5bRlPDcW05rwM+GJ9jperzgtkHlCt6hgOZsEKPhIgcF/IZCnaLsq1D8hpu3VzhF29UHO30udBJGBRDzkjKn3nvmJWVOZ+7ntAzEXfrgq+afUYybwqxXCqmUvButkn8nmhWK4UqO3NLpfDBRyoOxzGRGrbSmI4R9sqa7U6MEXG0ZoFhKtTqrlVeK73EraNJ5Jp93+xUrDCqX3zhQwBxpMQRrPZrep3aCdabmk2ovvexN329vI+EvEXjA590jj3vrc/yffZtPFVfwKLMKBjrrOn4h24+LNOFQxc/hBbCUqx5QrDJSUtJg5D6zTK4LmKJS0g2Enn6lPu8kO/R7nS1oej2e4YRUJi6VeiEYw0dp4qKmS5c6rjXlvQ8LD/xVn+1epcaTvLCLUquuaNzSeTpUdJ8RtgchNwTx9VeOs+EkfgOWQgqCyK9cD6nu6y1FxQuvBd6rk4wGYqPM7LSdCVTYhaUpBIz1hlr9OmR0fUuH++tL3C1XqOQmgW1j2JyQrogyEswHGhJoZbEK8M2Er8wxIayXn5tgb5xT8uk8oJO6zQatcJ4bqh94uuwa12GR6R0sroJHDTrx0xHZ7jx0oPkeUwc1wxXR9jaIKm7b+OFsLVSkxeOelVZh4JMK4fQVBb2FpZaXeG0GSd0JKITuc7K7lg4WLiiKRH373UcYrZKj6/egye3lo4v7tlxnalp7RZV6+0UwYUYtkfCsnAs/LUMPzEgZiE1mUbcNRMmvjP+kZ//kW+JBeaPa7TnnqCpaI9/+sm//TQsQ+jalFFnbX1SERqE6m0BeLAzD+LoYKhR4zRsUbM5lqZBY1pzQUXtEuLVOjtej47ERI3IFtxGdiBd52rYdOK1+VNQNvNEcOlyKEncICwhF6SdUxGKmoAaG78JPGs2TqDZywDEkIGxXKwHmlB6iu5MCgq/sa/8MYbXtu+HhUZP05Mur/hOfOLnTIvljg9FtNjGfvySbC6vnViCHLv9/u3P2fAuUI3WQhxa3hZBSyha/Ly8oGioW0HbE/JenHi9aAo+py1xAYkL72o10JSMhB4pfTqc0T4LcdfkrKyz6i3UQwHSFs+fpupGGFbNgLdzia4mXJcD3sUVgtX6ivSaxlRMxJqsYCRiID1e5h5dMp7QC3y1PuZYZ5Tq2ACKpUfGK8ZZWR8x5QHdZFvW2JAVdyz+mDKvbyy1YkePeFX2uWxX+JCc45xkRAKf4R67Nicxwqf0Lp+rDqnVuT19NLrEOdYbdKIjWWOZ//noOoVV/hxPsmFWGXhL5L50meqcz0U3+JTe5dPc5gvXhccv1DzW7XA2SvnOx2f8+DstQzqkJDxi10+gSH1xCeddWa5/L5s9+sT8vDzPF6LrFKp84MkD/oN3CD/0bVOqyvD//N4ZziUJN8qc34muc0dGrQafa7g+aDdZaM1WErMexZxLYzZiT39WpdsteM/DU77z8TmT0lLYkwjIwdyF8C2qpevTIBWmXiPSSb65EtL/dSP9x0v3L1UwkaIqzBYBoZXXUPPfrHFfE/IWjtRrCj4rd9kzYz5YX+WXzdcwmMbXG8BQos6uaOnM4TmzqjWqJ2lZNSc99E84uWCJNMJy0qLvKXkbtVi+Zq+33t+LQQNVwovj3aSYLzM3lOa9LI4THfzVRaQpliKW4vpcHT2m69NzM0kptKQvXUoqZrqEzkOnb2EXxBKTeSveIFg1XgviMkTc8urOVZtjbVPAGqcSf16VVo2IHmgyTEI2QXC5CRuHsBFOiOn7jXSgWlmUqadv9b3fvOvSuU1SiaVLTFdjEpxOJPJ/L6jZkJRSbdPfLK2SRcK0cijBqKwpR4aVTDgo3eY89vqJrjEYcbBzaUNH2Vn0bqyUXHlgF1sbVA1Hh0MuPnADE1Xc+/J7GKwcs74x4mBvleH6hLqO6A2n5KMtsuE+40KpDyKyeBliZBUSWaaoriTO3WpSKgdVRSROBPip6AZvT97GrZnTdiQi9BPhTj1tuppHVc14nriQRn/guSqVhdQIi1rpRYa69u5Z4pLhq9bGJ0aar93vl/PYL1ESNXSIuOw944Nm5P5480b43QkaqDcaFsWeyi4IIzQUTjc7wlzjNrzabM7bOSGqFpXlhj/QhBJiJjrDino3ItelD9a/gXbZLnQajUlTBCwFve5cnamCbeaGpX36CdvfE/Oya3AU2Ndcn3095qo5f+I4EiI27dLFKhQeI8mxKBMpmvOE4NC1RGKWOj7TJH3X/vw6dJhIwT09JAQE5lqyr8ekODfDlITbesAjXMCeygYJQYhBO/Oy3m3u/CpwbgAAIABJREFUx6oZUGjJwPQaK9v2vQyjLeo3CFM7axDnWGJyzYmISHxXveNNQjqSMNIpK9Ijp+SemdDTBCtKTvkaStqygAxl2rJp5tY999ytmSGX9Aw32edL+ooT2vsgWkczjriiG7wgOzzOBb7CTd6ll3lGrp943h6za3wqepXKVnRMh5yCiA4jZmx6q+Oamue5S0rM2E6J/JpmFJeVYpw+8jvtQ4BzA/x1vY0VWK879HxQ72eKA26bI24LUMH36gaRwEP1Om9nkz+Mdpp7I+Ko0IkIr1ZzhlGHMTMeNhfZ4Yg+XUpq7sqIC7rGuV7EpUs7nD/vC43Iki8yvtc+yFgrLicZPZsypyAYSKzQY+FpdGv0OWTMx99h6X31Yf65+Tpf5Yg/W2R89rkO770asb014t1XO/zuS7EvSMvG+rt5Tvzdq1F2S1dEjmqnm+ybiKmtefaVIW+7OCdJKrJImNfKlX7sjFP82jgpoBvj1xbPGBHXUJvkrgH2rTREaFLTJ9OYfrei13E5YfO/+9bZ3N9HQt7CcVMm/KvoHlfsKsc641PRy/ywfScdSZjojKD5CN2JSEwjmguiSos2xUEoNAImEokhWEQGZEBVG95qW2j3ZX2FZ+0NT59yv6TtpOG2jmLJw7bkni4V0A+DnBC2ndZMnA79CgtO+KwFxYkCoOkYnlr8U0nIPK812GY6jnHoNgZ/+nyJlGBOaGqsui5oJMbTBJKGXtHu8AV6AbR441pTUHJPD5vNUKCNbcsaa+IW3txbCnd0ibQkocDAJX7n1PRx3NeF1m7TjCER06AbpSqbHUNmTFMADOOoWc6DPiON4NyKO+7j0jLJXWfn0qV9xqMBo6MhcVIgoixmA2wd0+nNuXPzPIO1A2prODgYMty+TjHvcLi7zfTgAp3IFTqz0n22qpuw57VDQualkhgY+ayPSISeMeyXNX+h8zZujS3O7ABy637mqg6JMDxUb7CVxA0svmieGdheUSqrnOtFLKzj+Jaq5K3N6pyaVRK2TEBWElZJeKLb9ffF2TUCbJCSYei+JRK7+8P9fr5xMneDVLa6lGGcFoe3u9UBFQ5fh25/QEICorBESdyfgrKhUQXt1nIeNQ1Ns42WhLlroTnBTSvyjZOG4slJm9rQyXdBhM5VKSAlIbk7/HwbfbUo27LGRTlDEK1bX2QZlaYBUqNEapqwU4Mwo2iQj7EsThzbcrt98j5EGB7jPAstuKMHjTnIxM6agitoLWosD8g2tbxWBxdQ6xDItyK9Zo7cs6OWRfvyPof1KqAdjabGf5ZFmWtOEMOrWiotGxv4gIkFPcOM3NNzLTMpWdUOj9fbjGTByGeh3OWQXUYUWlF6JMVdh8glhLM0JejT4TluMbITSq+LAUeHe4RzPKLn2JMpD+s2sRqHqhHxJJd5B5d4n17lfXqVCOG27jfOa6syaNblcP0MzpjlwB43YZwxMYkkHNhjzso6V9lmx0z5lHmWXzZf4+vc4mXu8YLZJUb4bHSDL5lbrOHE8S+YXQ5zZS0Tvms45Imsx/dzha5k1Fo7FIeUO1XBhajDWV3hR+y7eKo+yzoDPmwf5X31Zb6rfoAPsM1Tjxzz+We2ieOKqkz41c9t8cyzq/zgU3NWJWYtE/p06ODuZ0pM4luPF3WNnqa8v36Qm/c6PLoS8xfMO3jO7PDTf2C4Veb8b8/njEYDsrQmEugR8afqB/lAfbm5VrEvbm6ZI+7K3D9HrgBZi2K3ZiQx4xy+9FKX3/jDPt/x6JzUCDtzy7H3u1/vCF3fcp/XyrxyVryxz60KQYXfSiP5H06iIdN5jImU2d/9obeUMXAfCXkLxn/2ib99bWynjlNKl4Nownk2iDH8gvkKhZasev5mEFMOZdAs2CHtNYgkK61IJHW/8GIotcDZTxpi/3XcurVucx/5Rc4JBHum2zjDtLvJbTqWiCz1FDhRtutMLREFWC4uVmvPqS6XVsJyMkU4iMkLnEtNQAtgSUGAkM7u8IGZnZNIQumP+zQdIHy2SwOOKXGoSRVoCf4cgktL6KyG4Qof2xQbkadtuXMumu5crgWPmcuct0NeMvuAW7ATIu7pmE1Z9c40Fat0nEsKlik1GREJhtxvkKdUXqAu3vXJci6Lnd1iraynhjSCfiLcW1QclY62lHgUJHQ2b05roplgVUmMkFeQGLhze4O8jPjV6yWrco4PPqicTXMmR+uMxz2MsezcugTAxsaYarbC6uZdDnYuUOQdkogmhT1M5P3YUKqjfh1VljUM/dhwMy9Q4HY957G4z82ZQ0WGHqHpRYaDvOYZs8sRU87jfOyPc6cFuZAkHPk0+K/sOEqa4BCQrhgOtCJBGFHS8Yv5wESsJRFbpsdX5zMqlBfmrpiz1CQIHSJCjGZbBH9/vDkj0I/C+I8+/l9d+x8/+d++ZkELCc7w+kLnNrp58nvub1XFiiMnShNo6cXmuHyhULaEjXWDXGqg9y3tzgfSO2FGEey/wzA4MXtXMmbqNvsRzhghzBng80swjbA49ZbnijMOmVOSkriNvzcgSSWmS4biRL6NlkMd/78Ur8dTWRYDKszkZBhr5TfoQdTbvqbtnwvz7bascUN36ErGgR05tJmySTHvkHrb4tdqSwJVxmOYzMjpktExbp68Y/e95sbQk4xde+iurZ9ng5NYWLcCcuaQfm1s1lNJPW3Y6XXC2rh8jmqOmbGQkk2GzKTkIJoyYt68R+mvf0HZoGahSG4oxP55mzBv0sfbom4rysuy65tnFSOZ8RBbbMsaX+BlX6hGWHFFzZe5BQqrMqCWmhpXYDygZ3hV9n1OxpLGFBCzhV0g4vQyGzrgRe4SjAIslkuyyW094Ein/K68SKwRPelwhwPOs8FHuezm5twhx1aVza7hLy3ezXM65RnuUFJTUPM978r5iO3R7U1YzDP+UnoGE+XY2jCfG4yZ8398MeWz0Qv8X7+9wstyiyEHvHd8jqOvDrjYFb79yT2+rVrllRtXuDMSnp0vGpT/E08o/UGOSAUckqQVRZ5y6wtv49ejl0iJebzeZjAsGI96TGvL0ERciBOeL+b+ma5bz7HlZbPPSp3ycNSjEwl7RYUR4bCoeKCbclxa1jPDH7zY408/NcJa4ZnnHNXtKFc6kdBP4ULfMM4hipxOxLvis/lPvzX0IO2R/uOPPl38lV9ujETe6gIE7iMhb/r4a5/4e9eC08dMF0x1zoEeMydnwoK+dFiVATlF42DRpjScRBFMQ7UKi4LTWqQnk4a1bjpy4fs1deNaFbpzQUOh2OWm4FTKebt75cTqUbMItYuP8HXbKz8gNmER7EhGl6yBrYFGoOjO76SvfuhqdkynceAoWx1EZ9lrTyAxwTIzIiImpmM6wLJgCt2v9ns0Ke7+16HNAU8laV570Wx5l5LIb3CWdIRNWW3caATDvsw4awfuHAk+85aapcXs1CNU22nMVpwwKS2HheWoskwrJa9cEXDWi+vaHNfI/zPxVrYdb0WYxZAlEEVKXhgeSDI6xnD5gXvceOUCi0VGUcREkSWKLA8++iL7+yuUiz5V0WVj+zZFnlLWcOxRjtI6ati0siQiHOaWi72IUmG3qBiYiL6JOC8d7lUlC3U/F0mg1Sn92PCI3WCVHp8zd7ldFvQTYSU21ApbWUStrrysvb4lEeHSwG0HzqWux7bw3eJwLSJxfPcalx1iUW8PrM33af19f7y5I2wwA8J4OrzQOUMtl51A4Qyj3ZmGJRrSRkdCcySMdoiqew+XKRT5LV5AGMKmZjVk/HhXr7E6V6KAbtRaN5z/smWoUXh0wx3Pcv5LApqq7jWBzhrm4GBHrqrNz4KbUy/JJoptNtgWZagZfdKmAIGlHgOcDuT08KHHdDV5zf+F9w0jIWJVO+46qdKXnjvWBrFZ0tlij768HrIVHMmO/PUThB09aq5xoOC2qbFAs+aIvze1b16dMWsUWjT3K2kh1aqWvnScg1aDjsck4rKi5h7zGjEn14LE05uG0iUSZxQSUKPw+tOj9pqc0ie6R2Ia7VKtlpnmVNTkFHyZm+zpsb9/DnEqtPJhmAuCGLxWy2U9wyV1AbZndOjE1hQNGyCsxWFsyir3ZMREZyccIK/rLhW1d2ssmnsSYThiys16wYvVjF+t7vCFfERh3frxoSfHvDPtk5HwvvoSl6IOg5Up/+q5AZ//w02GqxMODgfY2vCLv7fKr3+lw69/ucdvRzeYUTCSBQkRmcacjzJ+q9znMLd8+vc3yToLHn14lw++Z5ePXEj56NsMH38czmwfcXdnwK/+/ho/94UVjg6HRHHFDz3q0JNtHfI2GRAZyy99LSb3iPftwsUTF1R0vPVvQuSzw5Q1EmIDe0XFWhJxJo2IRTgqLP046K7g97++iio8cskhYmuZcFxa9ufKwVybjJUXJt9itlivM9J//NGno3/0/U+bf/jmZ4K83riPhLxJ4y9+/G9e2zBecMaSVlBqSUnNHd1ny6w3G/rAw21vyENgX8SSZhUKh5raeXL7TXjHdJZCSeP0JZU6kVhFcGuJGu9701oY26JBg2B8h7A9glD9tPA9BI41nGkJaevxCd7zhlmh0IqOpBzp+ARFIZO04WCftt60aBPOmHr3rfA6Vwg45CO8JhQWte811V4LEhY929pEWK0oZCninOoM1eDsEUL0hJ50GUqXDik1NceSE6wM2y42KQlTFqz6DubYFJyz/QbpGXo3mynBycmF6U0q7/pkhLy2VCgbJmJRKZFxtnlZYzmMt2R0nxmbwG1WFlapPb+1rl36+buvKjsHKWJ8F9MoVSXEsUXEMjna4Oz5faoqwRjLYuae2UsbltFdR7fa7kYcF8paajgqLMPEcJwrN8rcFQhRym7lULVVE7PiF4NYhNrTR6aeZjanIKHLAQVFnbGWOSF7EN0/sWk4mglZDFEujBZKRyJ6sWAK915Xoi5ne4ZJsYTQz0uHPXVBiWNKxP+9TsoHf/6t7+58K44OKQtxm+8QflpoyV/8+N+89tOf/FtPQ1vbpo1+TEReVx/S3iw2AmJvk1rLklrjMoMCLbPlisey8+3ex1FWF1o0Qtym8PCbU7xerNKKQhwykLM02WhnggRh/KIV1uooOknT2AhzUdjgzlo/e142mrnfnaOyqp3m/JcCcygIQY2t834dBLv92qBhOb3pDmvOY1zg69xqSGA1wkznTZ7Hedngeb3NIx65BLcRLnzmxVlZZ1V7DMVpCRJ/pVNJONQxAEd27O+RUkvt15fU02bdNYolaahJrzeCZk8QumQgLpumxnKGIWNZMGLKKn36ZPQkZUbBOVZxlvAVtbj74hDuEjwdKzxPPen6QN1WE0wdgnIkc4JmJfYEocBSGDPnWKegNGYENZZ1GeJMADImFIyZN7SlcM/TVvEa7nXYbE90jlWn8Cn9ZwVDhUgiVs0qZ1llw/Y4pz22JCU2ws/I10mIuBEdcM+u892LC3zxuRWeuFjygzceIE7he963x29+/jxfns94e6fH//0751hYpWPW+V3uMaOkozGZJJQ4g48DqZnIgnFdMzU5X68n7OmM6e9sA3Cub/gT79rBRDUiyi/+9nm+VB5jUS7T58bdLu/aGNHtlpzRHseS86JOqK3hnesJF7dyfuN515ibUdElY8rC6ystc4+2PScjni+Fh3WFTYkpaqcN7EZCbR3NKjToPvOVIX/yiSl5DQ+fKzm8GVNYR3M+LhxzIPXMgvvjj2/cL0LepPHTn/xbT//VT/ydawCZh7YH4hJuuwgjHXOsU4bSa+D9hRd4OdelpOHrdqVDVzqMdcJEZ4BbSObkdCRtHLUCJSHYlha4gidQpAJdAGg29l2TNpNg4EuH94dlGFWYMMMCDDTFUYRprG+XlLDSIxLh/2qSVnItOIveBQUpSVOIhMk9vG8oLALSsrCLBmkJcrWmW+gDCsuQdtymD/hQpnbBcxr1CZ1Kd50chSIYBhzrjAc4w4yCFc24J476sa49DmRKjaWniaNhAdvWdd4m4vjn57TLUGKu64wtOsypGUnOee1S+m5vrMJUazaihGllm/wPcEVIJEJl3dk6JAxAG1ePM2nEahfy0rlb7I0S3vHoPhcu5ty87gLQxuMOq6sLqspQVQlxXDOd9FhZO6auEu7ePsMTH/4UN29+N5vdiKKG48I5kIDQMc7ScFxbhhIz05qptxvpisGJb30BZxyKkhgnAsxx1IgrdpVHoz5pBDtzy8IqEfDurYg7I2G3cAVwzxiOK2WqFbXGPJx0SSPHB741rVlLI45Ly4Nxl4Oq4p1dFwZ5a1FxTxcMiE9QDe+PN38UWjb0mRDCt3yKQzHhNnGRvlan80Zakub1fjPraDzhfU2zuQsaDfdZoZGxTA0PepGpzgmC7EqrRtsQhNKhoMm97kLVegF70EvUjham2jRiwrlb7+SXa0Hhz39oBo1LUiYZF+RMc65hkxkopKcLCuvRlODQFLJCAG9hvZzn5lLSOIchr3mvgKCHDvPjXKRGeU5u+XyPhFxzMsm4owdERLzCDpfYbK5XSsxMF9ywO8zMGmdZZcKCW3aXs2aDO3afSCJWTL8pwiZ2xqoZYFFmnlqM0FzHFLfhjcQJ0Z3bUrc5t4F0qag4ZMyALpuyQtZ0x/s8Vm8ylZJMI16JjpqiZcycmpqxnTbIemiawdKp66TexTQ5JSkJ9+SIXEtiYu/26N5zTVa4YXcIWs5A5V2XITU1A1xWyQynaamo/HlWHNjj5llQtFnXIom4Y3fJJGtCGrvixPELu2gspB9ii5yKL8iL9E2HNS94P1BXyKU4Z6//XY+xC+X7Xn6YT7xnzOFRl9/64ha/kN9iL5rwSrGCQbgrx0zsHASm6hgaZ1hhXfveydEwZsG/jJ6jQ8Jx5M7pF3jF3Z9Jyu3PXiA1wm/Vu9yS50kix7qY2Yozx2d44be3+OH3HzDUjGPJmVEChmkBv/qcYULJQ2mXLxZlY8c/99cuxjUS98yUnJKxLnih6FBKzVQKLhRDviNboxs7OvKiUqa18uKNAe9+aMaXX3Z7r7M9b+YSu0ZXGsXfMra8/66O+0XImzj+/s/+jacB/vKPXrsWRI2GJdex8l0tRTnQcTMRtjtpIRMDnE5kzoKK6oSYPHSKwoYdgj3va6F5g7AhK+zqIbEXxF0229zUPSKMc6mSGFVnY1iJT4ht0R/CpBtcS8II2grnJOJEoEF0l5rYJeD6nmPI2XBdzCUFICZuNgJhKJZKl98LSecB1QibAJeYTpM4n5E0BUlwRAlIUNgYBSRmamckkjSuW2FBca91rjpWlR4pcyou2XV3vUVJNGImJT11KbexGiZSMNCUPZnyPt3mmIpDLcmI6EvMri6wwC45l+higVyVjShxG35Pv8r9RgevrUmMc5bqmGUmSt3yFhx2LZurNVadq9XNm2sMByVGXCBhWRqOx53m68UiYz5PufLU85BUfO3r380rv/udbG3O2B8PmZfQj4VRoRQ1pJFQWViJDUelu/53dEGPmLHWJCrOZliEeaWUqt5PTFkh4YP1A1QoK6lhUSnrmSGvHLx+Z+R4vgIMooitnjAvYTKveXVesJUkFLXzMS9UWdQO/TmTGh7MEg5z9+xd7sbszYKw8X6b660aLtnZ6dRin10UNBz/4cf+2rUwv51GWV9vhOLFYvAgnjPC8Hqt9m1VP49YWbpoBbvcgIxGQSvi3ZrC3DiQHke67MB3JCMmopKo0Zdoy2zDUayWXHVHQW0F4EnWoN2AR3hgYmekkhC3XBCdc1d6okh7bQGydOwK+gGLEMO/UYH9etSjcI1yak8VdevQwPSpqFlYh9ZkkvlzXRaLmwzdptojXvv2iF0OWRVHPd3Ro6b4a7Qmfo06tOPmnrSL0zYdL/FZLjERFTUr0udYp8w157JscpdDaiwZWaPNU5QvRrcaC/fvqC+xI3OeN7vN9xqkCxqnxNgfi6JNURp0g24N8B147TDTBYqlKx0G0mXPjniCC/wrXiQE1oZ7tNCCoXSZkjcMgpqaqS4a3Uig4IV1NZGkeb5UlZnOGZgeGW79zLXgSnSOCpeTlWvFgUybVPRIIsY6Y1NWedJe4FEZslcXXIwyRKCXGY6P4Ys3Iha25o6MWKPPjowZ0CEjYcKcAV1qsazhUtUPZYqibOiACOGeHLOg9Ib1lhVvf3wsM67X6+zZOS+bPUKI5Ib2uKpDjquaR1Zjamt4u6ywXmfsyRwR5amHJ3z9mYTPRTf5vToijgwFFX0yIiKmLJq0+gVzUmIssJCSnNr7hAndxOkYF5XTGq6nwq1j5aHLltrCY9vK0dQ9a93EuUO12G73xx/TuK8JeQuG9QhDyEMwHhAP4seph+gb7rHvwIRJKoTmhbTfrnR84J/zX5/rotm4B6jdvd/y38E5BuBIJ42l5VVznkMmzc8EClVj8avabBra7wE0+oy2TebrDUU51pnTr/gNfqklc124xdC7n6ShcHkduoGIUGrZcKoDumE8uhOcryBoS1zHMJxH7JPhHcS93FS0i52QtByOpUNK4c9xjT5jyRn7zgw4nnakhkxjep6LXVJz1fN+Z5Q8ZbeIEFYlYUVitvwm5AwdVjRlKiEXV5ulPugdRGBsgwbGX0uFVKRBT2p1G/1+ZFj3ETN3DxLuHiS8clyzsb6gLA1FaShLQ15GDPo5dS1kWcVslnL56k1uffm9/P6vfZg0ttzd6VGW3uffOAeujY6hm7iv3bVzm6AEJ6zP/PVej2IWVlnUltxaMmNYWIsR4WqacSXNqFFemucMUmmSa40XUgJsZTELa5mVzk4xE8OUirtl0UDtPWOY1o5yN60slXUITDd2QYoAx5TM+NdveO+Pb8z4nz/53z0dnKpCu0H15EZZXmfJCQ0P05q/An2yLYgO4/SmOiAvIa8j/Mxprn0YCS5sriNp040OTZtci2V2UQtlrbFNAVJp1Yjrg7i6bSVb+uIrJKifthzelFW/kV6GNr7RaM/lIbyt8EVD3Pq/5lqytC9u/1/7GgbnqKxF9QmvTUk4E61hUcY6Yabzpvg5ZEqEoUfGCr2mmLBaM9Zpk50SYZzIGuNoVzhXxRqXzRK+F9aakFXVlYyQ2ZJJQsc3qlak77QfVPTpkBFzya5yy7jicUHJlAVjFhwxJUFY+GZSmxIcMmcCHXkgPQbSe80zEppoA9MjlYRjnToLegwZKSM74QlzhetyQND8hOewsZenaAqgNkI3spMWzW6J8JfesS0wDRp2g///TFJ29IizuspFPcPz3GVfx01BPNIJM3WKubsyIRHhyX6Xj7xrwnbX8P63H/Ps7ZS7VcHn2GFIlzscAHCbA3Y4ckU1hsu6SYeUQ6YECnhga2zpSnNcKTFzcvp0EITfiV7lObNDCGIeMeNl2eP3orv8mnmFLx7lFHnCR96e80DcYagpqsIL1wcYYF17dDVhwqJ53se+6FCcJXTqf3djhEydw+S6dnl3ssK8hHHpUM+wTr3jgqWqIt73+DHbmzNU4d7UNbIi41we748/3nEfCXkLRq45D5mLzWYfQjffemg9cIxdIWIwjDynNuR3LKyD8gst6Um3ydmY2mlTiAAnHETOmg321IUjhUW5Bu8s4/6/4Vyrt1QVoVbbFADBGSqERYVJKQRftREDcDS0N7oOf+MTf/8auGLFaWAWy66c5o3AtL3xCHA9QMd0moInTODtrI/g9mVbm4dw7g0FC2n42QhN9yuRhMS7cVVaMTDOJnnVQ+pd9U5d4rpAFZaFOGF5JZYVzSg959kCV+1q41Qz0ZqhRFRAqZaZp370iDmvXRBnydvxe4lKlRq3qV4zy1/RkIy7GkeIQBY55ywBRlXNahkzHxn25o7KtWMLdva6JImnq9VQVcLuXo8ksUxnKUaUvXvbXH7iGQ4O3s8r+zHDjvLizS4PnMvZOUhZlMLBzOlTilqbQMW+cWLyqXXHkGIY25qhccenGjQrwry21CocVDUzKq5EHY4LZZgKnRiGtaETCw92YqcDMc5R63wv4qh0oWqXU6fVmddKJD5TRVwC7nHhxPALb7/4WNrjD4sJHV5L+bk/3rzxUz/3k0//5R+9ds3NNS07Xl3SKi0n3emCgcUJsTlLymUYqgqyNNsIG/jaIxyRd1Jqb/Lq1s+o2kY/5gwMesyZk5HS8xk/4bgS37wIJhWhKAoNmfa/g4lIQEQSSVFVJjprGkXnzVZTHAQqVHP+mOb9R7LwovHXL75yqYl0KRx31+Ek/cqeQpLDRnjkbXxHzHiVHQAuyiYDTXmEC3xNr6PiipigmUm9ffyL3CEh5gXGnJMNAJciTswRxw3akbdQIRuaVCy1Oa44relKp0XBdQhDRc0Fs8muF7e77ndNn4zMFz0dUgaactOMuKvOdcuIkGtJQsy6DNgjZ107TDTnQKZNsRTu2YrpeQMBQ0ZMKjGV1sx10awj4TwE8a6GrvgcMWFVBtzWA2Y6b9aO8Ky1n9sl3a7i2E6bZ2tiZ/55cW6PgtAxncYda9usM/eNORdCWSNiOCvrHDBhT0fMdM55s9XQwzLJeMJc4X31OX7syZoomvKVF4f8ky/GDCj4J1865H11xu9Gt0mIuKMHTHRGLZaRjj0lOuMW+6zKgIqKbdaosRwwYYM+NZbbcohBmvyUEBgZYxo3SrD0fFhhQUVByRW7DkBRJAyGMw6qmFVS6sryxEMjbnxxyFbdZ52MjG3+0OxxKLNGRwM0YvUFBRkxuVS8057hkV7GrHLXuhcZsthlf0QGXt2NSA+6KF3e9fCYK2dz6jsZRqCoYLVzsqFxf7z14z4S8haNY2asMzjpDIM0KEhK4jmwJ5ODA9WqJ52mixi6cm2kIJOMlKTxzQcXgAVuEbogZ7hgNrlonLXgBTnDGR0y0JRcSx4wZ5vXOYGdnJhcl44kiYfpT6bn/tTP/eTTP/VzP/lHciv/zs/+1afn5E0ycJufHI4/a1G8AjQdslLA5wKwXIQjMd5x73EPAAAgAElEQVRRrG4m+UAD+X/Ze7MY27Lzvu/3rT2d+dR0b92h7+2BzeY8mJFEUbIBOlA0QE4iqykTSR5o2IAfgkBBkAchUuJGK5Ye8hAjRgwYNuKHBHmIRAmBYimCo1iibEGyKcmUOHRTze6+zTvXfOY9ri8Pa619ThWbUpOUuo2gvkLz8t46tWufffZe6xv+Q4jNzXztZryGVkTeHThcU4syt0usKgM63NYrpN70SlEKavZsj3c121zXIWPttN3Cp+yYDoYBMRmGGuWOzJhqTUecF8iAmC2J2ZGUAtsa9p3YqlWIshsd5PB3AXJVzuqGRqFoPC5chFiEyEAvdZOFA1s6A0QBtdDr1OSFobFCllriWOlkNc+851UWi4yHr3yIVx8l5DW8fqp89P1HPDzKmK6EZencZecbXaYkcp/CK3aO9ef4kJVzU1enprW01hcf7jwXjaUjwquR68Alxk0tVhXsdIVB5gqXSl1R1osMhytLqeoKN3FQsKW1zBrLwEPSChv4M8r1gXvPRaMt8f8y3t6ofDc3JkZEWohLiOB/FOLitORibBYsjdpWjW8zHETVrr2OxKzhlGpbQi/gp8huXYuJW+K580BqWghpWPuc54c9B7uCdaK5do12X5twM8E1XM50xqGefUNxcXEy1EKG2r69vun3N9feN1MV3JwqhehrSoPlhm6564Ly2CfyEWufFUHaBtFKC3LNKbVi7qfZj/WUR3pCQuxlfL1K4UbjKkx/2km8hySF9Tdc28Ab3JEhT8pVEiK2ZNj6T2zRZ6RuKpB5paoDmTFjxbYMMOI8pgoticSwoOAL0X16RK2DOji1rp50GJkBmSSe2+IadjsMiCVquRfB5b3yTa2udOiZLrHE9KTDROdUPrk2fu8IEWDFYfLh9uq12mK4xqkkdEyHSh1PJMLQNz3Hk/SFmaLM7YLc5hRacN8e8sget/d47EvPrnTokvGYM97V6bDKE2azjPuLhi9Fh/xbc8CCnAXOgf0Rp0x05jiWODn/rnQocYVcg1P0StQV9VcYcSRz7skpimVfnRR9Q0NFw4xVW4AEX48ZeQtHsyh3zYQjcv7Zl4OJMvQkommEKK45aip26fBc1uVWmvJeu8uO9umS0iElxtAjpaJpPzc3GVR+dznltGr4k2JF3iinhTuXaXn+2Tk66dHtVjTqPEIi44qVy3hn47IIeZtiiz5nLNhj3MKPgnoVBLUnD2tC6EsP8ROHhTeS6nrYwHno0Vr+MCzoYcMNCXWjDY/0lB4ZXY+grGla3flKK+7pEddlh0ychG5K4kmCbtPYxHqDGz07vfM1mf2tRIQj2q80b2EQ4fwtSqVVO30xSDuZAa8iopvGiWXrzt7om5tzbUbYKMNrQvex8p4Ba8nN2HdxKh7oCXc44K6cYBDG2qHwE4+l1IjCU3bEVdvjlh3QI+JVmfKaTHlFJpxRMtKUKRVftTOC8+vC69d3Za0ktWWc+8fcNqzUJfwz2ziuSBbRiYzniVhq6wjp4ziiHxmu9xyJPImUYWL46KDLc90OSWJZFoZHxxlVQ0tiXyxjrt18xP07T5KmDaPtU6alUlk3qv78F/d47ukJCiwqZVr6EbbPaUKxcY1uQP4zIiH1BVGjylYc0Y0CJr4hEeGuzamxvNosOS4bHixs6/Z+sFBOc/f/P7jv4VWRsKTh2bhHGuGgXf7zXFnLIDbEQe/fv7ftzE2jRl6N7Hd+/FfOycRexl9sOAM5/+ULkjebbLyV+GZ8hk0Jc3Dr4Ka4hqqb3AaIVpiyBsWu4Mi9nhA7L4dNaE6jTTudCWtF+F2AJw9n7fu6aMDoEtgMEcMV2WLLKyaFEAwP9eTcMbe1+w3vOcRK1sXNm0lOb/7cN5ukDNWtue/hRnvt7sghAM9xk+fkJistGJp+awK5uU6vtKDQkkprTq3ryLcTICJSScg1b4sOi9KTLj3TbWFXTukwaafqHUmZ4CYWGQnb9BnRa5PPMBk6ZEpJxZwVlT96hGGmC2pqZnaBYskpmePEBgaaEhHRkZSYqOV65FoypMOzepWgnhZLxFgGfg9cSztnkraJb5iQFB6GF65zgMHVNHQkxaqSqxNdmesSu4EY6IqDL0UYBtJjpotzhYz178vi5OljiRmKI/VvTo7u2UOCQuRN2eUHm+f46NMrvv445e5h6u9b5WXuIwhfiQ65Jm7KErP2whmZfuujsi/b7DPmRBacyJxYDQectZ/vVbaYyopHnLb3lEHIfEkUTDKDUlpQkVQsr0enVFhElKe6KaVa/tWXhzS1k6gfS8ysttwtS+6YKRWWjsb+WO5rTM+JPvhn4VVzxkQKcrV8sNelGzs1yUWlDBPnC9Ko86TqddzPPPdETmTcJORyDvLOR/SZz3zmk+/0Sfz/Pf76p/+Dz/32L/6bTy4p6NFhKD0WHr+5dtIQjyN1vQSDQcV1DfvS49ieek+N86Z+iOskRuIQwqGTEyKk20+Za62W+qnO2RHnXj2TnBkrrFpmuvTkT+P8MHwyvi0jMknpSdZOaoIL+z/9pZ9/8cc+/QOfe6vX4q/9jU9+7v/+hd/+5OZYPiQIwUW3lesVjxUXgxGn/e+w0BFBGWrtBr/+CgooFyPIHlv/ewRvTuY3nNgr+qSSknjDsqfZpy8d+pq1C9ZYu5yaFSupmErJsayYSsGpFDyWJbnUzE3JTApWUtHTBAXGpDySnK7fvCqfvJd+C+qJgzN1xDmlDyNDoUqEMIgNh2VDJEKpSmYMV3uGvHEThqJ2/In9IUxWQlErt3eVvDRMVkKWKEUlVI1T2SprYToZueveGLAJrz/K+Eo1Z2WVHjHVKiPyPiRGhG4slA18sVhwWFcc+iTO8UEcHKsBtpKIzBhqhU4kxAKNCltpxP264IO6S4NyPUkZJIYsEnopTAtX2Lx7T3j5ACIjjDIhtq7QyBtHSK9RUnG/s1S3kWynzmF9VcFZ6UQITrWiS0SN8vX/46uffOrT733L9+llfPvxY5/+gc/9n7/wG5/clLJ1K5H7T3H8ss0MQH0Sf7GzvJnYh95/aE6AtEdl4zescfju74mshSZqdXTuAIWtvOwuuA5+5k1RKxoEJx4SCpAwQYE1RyOTpF2XwhRBcO8jlYRds+XIzZ7wbnBeGmNvRjuULnvaJ0KYS84YB895IGcMWJPYwXHQlDXRPNqYgKyT4fW1Cp3yieSU0rCSilIaKmmoxbLPiKmsmNg512S7XUd3GDKQLtsyZFdGnPiuOTjjXKuO75Jr6SBwEtE1HQ+ro1V5EjH0TRcVZyzoGj0Rsbi5cUpCV1KuMHZcD409tLVDKe76V9Qce/XBFQVLLaioOdMpc3XFSK4FWzKib3rc1B1G9KikYd8OODALFhTsMXJSwtKwJX12ZMhAMxIi5lKypKD2PKBwr7hC1Sk21tSUWjmEgG++xRJjxJy/X9nwtEGZ6JwYQ2IS51guCbE4KFgsETWWyhdMTjzYTV9yCrfXScTQ9D20T+majruGxB5O7IrHv8pNbmcZZ/OE355N+X/rh9zUIS/LMVOWZJJyrFMW5BRaOmK+51dck222ZMBV2SKnZMrK76EQS+TWWb/PhsJ2iwEnzCg992ZIl5iIJ+0OI7oE3VtB6OG4oH+5ucmSmo89UfPFu25Ctd+NuHl1yeFRj7tVwa1Oykv1nFxqeprSJabGkhD5/wwpkeNj4uF0OF7oa9WKiVeVRKG0TlSlsW4vmS9j9ndz6jribBbz3qcWHJ6lDP76uy/3hXcwLjkhb1MsKdlm0E47Khp60qHQgq64BzIhKIe4UXBIxnMpiSVm6jXXQ5em7QQK3plXWkhV7EflEYZG4KGeUFFzVbZag6WCdZLg+CDBEMy2mGVVS+llH2Nir1WubVfx24mFdydWtJ2IhL9v/gm0Hcpmg/wei0H9hpCRUWjROiIH2Id7T8ET5M0HfsHTZEVNz3SdW7DU9KXDQnOfgLivpRT01BVhM8k5lSUTFlTa8Az7VBt+I5k6wudTdpuKxm1CGDpEjDX1hG4TdM4Yk/BElnJY1pQ41+OZrdmKU/aSmKOqJq+Vndgl2l1xZO/DpSOoRyIMEsO8suSVMxUcJQYR5WThIExnQS3KQFG5IiQyEWls6XZr8jxhkAjXGue3UTTKPHeJfjdxk49rWw2vHUZsk/KIFV1iHsrCG1m5fx+amEYhFleARC4rIfFmhx0idpMYU8Np1TC0EeNMmOXu9ZV1ql77PcO9RcPVKOJxXVFheV+3S1645KtjxPFGKgfTKhulG7kuWK7KwrrN/JGs2NPOJSjrbY4AsTIEkYj184F/pkMEv5DA+Xiz41x8ljdfG/yL1twDB0t1ctHr6akzsXOkVsV1qruSrh28vYhBjZMa3+R/RGLOwazCtDaYxl3kuISJypnn9kWYlusQIDshoSv87x3Sbde+6zremNgqK6lx4hXrtaxG6W5wS4JxaoxhKmsBDevLsXCeh0ywKCsKInWlzExyhtrB6RsKRt3xFOU95pZLQEV5xd4jMYlvk0l7TcL7d27zxk85tD1nI266HtzFc63Y9qaRPU1YSkUhzgvlVFaMvIzrlvZ4QxwEaUVBqRWVltyI9nnYHLY8wJUUdEg5lCljesQqpBg6GnNFRiTqElhkwI722mt7JItz519pRUnVQrwySRmaPhM7d4WXzSnFS+vq5j3t1qWwZ/ek28rpx5Iwkn57Dy/UqT2F/SXcS+GzzrVsVSLXIgl16wv2IW4z1g5P6ICShi1JGSWGTgyPlo3zqGLFr5lXuapDjpi2AgQrCt94c/ycjJQDzhjTp/DCuEArnR9gVp3gX+IVtKYs6ZIRhGQSjVqu6URy3tPs8nJ01N5zTzVbGIEdUiKz4EGTcyPq8OzNAjHKonb38u+tJqjgJlgqVL7YiM/d95brdsiprLw/jWGHlNek4Cn6RF7kpFLFKswq55ZuFb78qiPWv/vWipPTTktgv4x3Li6LkLcpOhuE8eCBUdN4LKvrDPUko9QaQ0TuVZiCb0WxYWTY9TyRYFRo0ZZIXWntzAap23Er4LGf7u+v6yM2jf0CXCHEZkFiccZJgawWiHKhu/jthisS1PNObAsvCIv4ZtIRCpAAWQuO8KnXtP9WI0yKLkqFztXJaG7RZ0cG7bWpsa4AkfXkZaRdMkl4g8fu3NRt0rEaruqAno0paChEKaShpzEFayhRjVJgKWgYtRhX1+U/8aZ7FnhUOq0dEYed7Bght1CoZSQRlVU6RkiMI4Efzl3ivzdQDiaGsoHGKhIJi9qNqCMDvUxZlcLZImVvFNPr+A6xukV+0jREuZcFLrx09Dyil8BWHTNvEgoaXjaPec7uEyHUKAsPu2pUSY3QTYWigdSbLt7wRpqR3/QLa7kyNNw/Eyqr1Ba+fgqRKN3IkNfBDVqYVY58PrE1fZ/YGHHQq0bd+wwRrvVV7TKQqFUTu4y/+Pjbz/+ME6DAUGtFKhHWFyKtWtM3WTsCvPIcD+RNmglu7fJQTQl+D8ZPGBsir7a1yQsJZHOnbOUgOuCmMl2fIJ/ptC1ANhPr8Fs3z2ulFxP9bwyL8xiZsqRRZzBbUTPSjCOp2NMhcymJ1TBhyYB1QnoRUrVZgARZ0gAvjX3BsEl4DwXN1/WAxDHEWhJ3UJ+aWpeE39MjetLhab3CHTnglu75a+4STIBccq6ZXSa64AnZ42vGwXCdr4iTOY/8xMhBeh08KSF202Q6xH5NtaJknni+lLVYgEEZqYMEHzNlITlDOuQScWan7Zr9uDlq7w2DM5B03EE345rICoMjQ29rd83J8EXOe5pd7htXIJ4yo/EwrtarStaE+tbXBNg249bwsmO6LD1fpisdyg0o8VzdtXAcRsetScXxLQbSc9M1FWa6IJWEgXQptfZKl5bUSxv3pEuxUZTsyIiPNFcYmojtLKKxynPXG6DhwXHMP5d7qChDuvQ1ZSElHRImOncNSW3o+3swFMOVNhwxJZWYDqm/j1wOMcM1DDsOkE2fjpfP7ZzjYRZS09GEqTh/lTvmjE80N/iqOWNOyYFZsGc7XI0SGmvYlZSjpqLbLbFNxEF1HtIdqZDhpjDb2iWn5gkdcCArllKx9KqSEUpHYx6T09WYJQ2VKlsS06hSNLDfF2aFk++N1MF2B8MlSVJzPBu+6XN7GW9fXBYhb1OEjmBQkRhIj20Z8MAcs9KcnnTZdB6PMJQoibi+UkeGnOikxamaC8cOm91mMbEjQ050RlCMsVgO9IygFBO6h+LdhkNnBzinu69qOdOpx/QqHUmZeVWu//JTP/fC//TZn/lTCekX43/5pZ97EVyy4jggTo8+cEvaDcUT2DchGbUn+WWSfUMi41R4aH1O4HyXVDBkkpJr3iYtm8cN2N+EqFWgacRNhWqxHDNjl6HvTFZ0NWFHRq4b4zfqueQ8QrhKnz3t0NOYmVT0iLgjMzJirvqO4/uSPrl1iXtk4HqWMKojHtUlBZavlAsihJyGZdm0iftYEiocQX0YOanak2XNThxzJy/pScTRPGZSOl5JeJcGR9br5q4QeWNq+dA1SBNLWRsWlZuqzKm4RodZ03BPVxyYBTu2y/tnQ15vHAnxwCx5RR7RYFlIwS0dcEpBQ4JtlP04pZ845SoBholwWjiSeVkHDwXh2Fa8fpwxyuBGHxaFcLRUzmrLkwPHc1nfk+5PC3zgF370RYCn/ff+5NO/9kLj1bgifxsvadgj5lSrS5Wstyn+5vP/zQsheVMPewyF+2YEkvK3Ew1N++wGp/VINmEwFuOf+8Y3KyyKVQcxDWvkOjlMWOiqlejebE44Zb2ixf+DW2cKLc8VCQZpvR7Cv4VGTnDRDt5JV2WLJRU9MioajplxjS16ZBTULKVqndNDp9kVFF65C0OFpUvMVIp22lBQryfYfrKTEPOEXGkTasGg4tb+U+bsmDE1NRM7p9Kal8TBrdawVuGr3OMJ2WPCyk0iqLmjj+mSscIZG/alQzfKOGpOXeNCYiocP8eKk+ads2o/r33ZbidAzk9pzlUdciwLat/JT4iZ64oFTrwlQJA2m1Kx3wd64qZIQzoscCpgd8wERblmnZHfsazoacJ9M+UPo/ssKSm1IpGIheao2lYYJUwe9s0OZ7pgx4w40SkrCtdw8hMLgL70uCpb3NfDttgJ+02XmJ5k7T50aieMZeA5ne68u5Ix0TmlVnQkpcuavyQ4MZhKa/6mfhdbUcQwEw7zhrxRSqv85huWN2TOPTPhEKeG2SHlu+11/q/oK60yVzDCzCkJcMJg45mScIUxE5YOJk1GRNq+blt7WGAmq5YfkxCzb4eUUvNQJhg/AbQ4P61TSt5vt/mqTHgfY1ZYVtZ/diIstSbrlPzxy7vc7sBhUTPSBBTenXZ5UFaOvK7KfV0xIKZSN30ppGFPndFzn4Qezn8LXCP22XHsBFUyWi6j13EhjeD0ZMBiGfPep2deJ+4y3qm4LELepmidyn3v6oq4sWBwGQ+dB/da8WPtBoNhSwZMNZhddVlat5iHRTl09Kz3/hD/MB7phMA3CQRMoFXQilgrx4TiZxOStXYJdo6lFTUdSVlp0U5eCq34Lz7137/wP3/2v/uWChGAAKBwSYXb2IOBVMg4Q8czXKvIn4dRB7cKkorunPVcARJ+fv37bOvkXmtN37jNqdGGgfSoqNmVIZEaGu8BUtGQU7L0XbKFJMxYMaJH15tK3tdj9mWbnJKSmp5kPJQZua25qX3G6lWwxFJ4xZCChru1+3S+O3ZqNV75lhtxxmFdcUDOCkufhC9GTn99qBlXbI+njXMHV3WeGfOm4XFdESPc1xXXGbbd/wqHXFk2zrejaoTTlXK9Z5it4PaoYLZMKKylVJfcDKOISdOwkIo/4g0+Ic/yRTsl8ZvMTIqWm3Rfzngvu+x6DPvYxAwSPz2JIEpg6YWF+r5oEmAYGQ6t5W5V8L60w6vHkNuGfmTIxDmj3/jffuTFG2/hXsqtK8wPq4oSyxVJuasrjim4Ll2W38HU7jLeWnzm+Z96IUwyYolbY0BnSLomkG8q14UObyPNOQjXZlz8GQgGpmtJ1BBBlSsoL6G0GP+gnuUgQXHrcK5qz0GwWpEMzkON2t+9sb4EQZDAzdgsShwHxLSJ3q5f88fa45GccU23yKXmCiMMcMaCiQh7DHksU67osD0urA0LQxTUdDVmJXXLJyxxfhPBI8OiHDPlCuONa2RI/HHP7JRYYgbSY6ErlpoTYbhvjl1ijpKRcsyMheaMpMdNdqlouKeH7UT6xE5p/FfioTuweZ2Mm8arA2ktyB0sSSKQYTsRyal8o66m8s9sRc1S83MiARC1/IxSLCt/3pXU1P51RoTrOuaRWfCMHbHQmC1S7jN1Tupq/SRV/d5iWsf3TLLW8ySRiLmusH4SkRGzIGeuqxbBMGVJ4ASpL5I7krbQ3gbLD9oP8EdRwsebW3whesgBpUc4OONCJOx3hlwLxmZAnw7HOnXNSM+xOy4sn+cY2ygfx02sXjfHLHCQNHBwvs9Fd1gF3p4XF1hpTkri70l3X2bt9CPyCArHJspw7u5b2qdDQknNULtuIo9zuK+koaMx+4w4kBlj7XFTt5hLwV1TE9kxBjjwU8MfejKiqWMqVf7jJzMWc+Frk5phHLHyxW+CobQwjmIWtuF6lnCSRxz5vXOgKXsa8VAWDDVlKiWVxogKt9IUrZR5AaMOTHNY1UoaOfPcTiQQw2yRsLudI3I5IX+n4xIR9zbFZnfQqV5FnDD3REnrja8aul7+NrzW4klZEtHxZk6BlxHG7c78r243xCCf6xbNISPpn+vOhQIjTE6CnGX43rnz9q8L6iIGoSNp2wXqSOo2km8j/ukv/fyLodMSVLGc8ZhpIVPBcflid7Ly5VzABAcZ3yDpG6Ynmx3AoNgSjtVeB/91S66QkXAsM1fkiOW+HnOssxYmdqwzSq0ZaZe7ctTqpd/VAybe2bfB+o5mieAmC5X/PGJ1XJCBJvQ1oadxi121Cl9oJjTqZH2X4t7boay83GzNTArOpOArOuUNu+J39Yjf0ke8aiZ8xRxzX5YspaabOJL30loiXAFyo+tG1KeFUimMe0o3VWbLhGGv4nufslyJEz79ZI9alT80B/TVdW+/aB7wu/IqlVjumSkPOGHAWjrzNTPBAiucm7uDRzkZxHnhuCixEUaJYSt2Y/ZFY7kVdzil5PGqIbeWWp0Lej82HC3f+gbx4V/80ReX1vIY50S/lfhOOA7edhlvTwQjOMCr9UXnTEzD8/hnTUEuwpsuFiKbalvh9wVR22D8VmhBvTGFCccMUNIgoVrR0Khdc0BEqH1jI/Kk14uFSCwxiSRtAZJ5MYvgfxF4ft7zvf25ioZHckZYdSIMR8za74d1+4r+6TCRBLMBS7EtJ+6RnnGgE042PKl2GZFT8lBPOGSyhtD4hkw7NZG4dT6f6tJBk7wR4cTOiYko1U1B7uoBKQl7ZszMui5+YQt60vVKVfP2GudatkaGYa2de4fypRYce9WrU9z6OWfd4CipqLRqP/sAzUvEaSZ1pUNfejTY9jxXmrPSnClLTmWFUeG+LLhGlz7O+G6Lfrt/1l4drSsdZhvO6WESsdCckop9ryx1qBNHilfXlDu1MyZ23pL0gXaPPLUzPsRtRtKjxnJbd3gq6vKj+jRj6XPLXAXgGfb5K/oe9mTMvmxzy1zlrzTP8tfsu3kvN/kb9i/RMYY08ga1NHxYd7jWi3jJOOXLmS5JcbK79+WMQ520hsIlFTml92NxsjBBFMeIcFN2mZMTE3uyd8JAU2cg6GFzQCtH77gYEUHtMaemS0olTeuPVWEZEDPWrH32Pv91wZiG778N+1cW/PZLHc60ZlqvJ/0Nbt9qVFmp5dU8Z+gL2x4xJQ1TSm5qnwFu+jjG7VMGuJY6IZjTFZR+/GHEGRc+sa3c2qvZv7JguUyYzTt/6nN2GX/xcTkJeRviZ3/iH7zQIxQXrpN8iFvIglmRVbchzXV5DnLU81haQThiisXSkYyp97gAWHkoEQQoU9CtNxzrlKCo0nDe3bzRBnzXaHMScvE1ke8EjaTXdnvCRARcJ+UnP/X3XojE8Pd/8ae/5YnIWnZ3bRxYU/vR/TfCrFxB4eEeLYF+LQMKTgaxoibXvE14XPG2FgYIMTaDlqBXeQ+CKUueYA+LbWURExzh3CC8IYdtwVFS0SGlom67jzGGuZTckzlWlIFN2LVOfrNDxO2oQ60hKXJ8CavKgSz4t9YRURONODAL3mN3yLXiRBYcMePdusOx5LwUPeKGbhFjWoLltnapaJiXTr73rG7ox4ZhKjxcNsxsw343wgjMc7i9XzCZJSwkYZlHpAZ+8w3LKzLhq/KQV8wjMlIWuuIT+qxPkPrsaNer7tSc6YLrvtPqlI9g3HFFyN64Zg/oZg2Pjt2kpF8LnTLmUV6zm0R06ohnxmuDw8Y6/G5qvzUBxe/5pf/wxeb5X3nhSEselRW9Vk5aKLmchPxFx9rhfN08iMS08JUwYXgzaNZFrxD16Xt4xi9K24r3h9gsTkIDIkCvWrKxOI5ISPoDj0L8fDoUH5H3IMrtyk+K1wWVRemKS6b2zQ5TXTKS3jn4VXhv4d+mumTLN4DClBRgRI8ZKx7ICdsMGdOjQdnXseNKaHTuuNVG8eUUFWMWUra/c/Pa7oorXmaseKSnXJUtHuqJM+SjodaGr3PAbblKTMRUF4xl4K6NOEfwFQVOSr3vYWQRlTZM7awVETASsdRV6/sBYLxBnap67yZLblfEG9y9wLkYm2Hbka89rzHYQjoFLmWmC4IAyya8NkRHMq8U5SZOYaKeEJNIxFSXHMgZGMh0Pa36vuYWvx89ajkasUSMGTLRGara8jC3ZciUZXudD/WslbYdy4hjppzZGUPpc2od9OsvyTN8L/u8rFO+Zg75vuY2103Gh5pdlxyr80eyqvyn9j1MmgbD0wwjt/Z+l90OwlIkERxWDR/Xa9zux7yxrM3ie8sAACAASURBVFgsG/5YTni33eb/ie6ws+y7e07dDz3yks8riva+dsgCt2+VUpH6CX6YrPXpkGrMTBz/o8B5lwRxg5FmTMTB4xIiPmh3edmcsm07HJmlUxzTLgsKjmXBXFJ2tNc+C7dNj4U3sW2AX/+DLX7oY1P+8KUtTpuSHlErPT8gpmMMZ7ZiScMN0+GeXXn4YUROQ+rvgcCrHWvqDSUtg1Q4XFlGqSExTilxnDgu4/Y4J4osDx/3+eod95zcuvqtc0ov4883LouQP8f4uZ/4hy9sYnkLcTrlCC3EZ3MT7pB6FYrgTxG5jhxuNBxG77EaENiWAcc6xWAYmQET68wII9aeIKGICIm3ot9AIN/sTG6GGwWvN1NV60yVfMGx1ILMbziVNm0xEoz1rOq3xREJXcOV5i2uOkSAXWwS1DeNsYJHROzhbKFICRtO3/TPwTHCddnczBq1FLJ2VA9kynt65DfnyEn+URHhpRsp288PaAsQgIKKDGegWEjNbTuioGkThR4RqRHii8mYQoxwz0zY0R6NOAzvkeQcyIzrOmYPpxefS8UVdQohimVH+5zKivtmyp7tM+5AUXtfBAsPl07ed2gi3lhWPN1PSGNY5jGr0tDvNtQNLBrlDZm37u8BP/y9+iwV9tw5n8qSPhkLHBQiQbgRd9kfCIOOZZ4bullD0whWhW5qMQZOT5wbes8YGuuu+cFCubXlJieTpXCaKx97svK0yLcehVoSnOhBuMt/6Jd//FsujC/jWw/3rGqbqAJtwtpoCT6pb6eUqq2D+cXYLDzWSfbaWdzxQL4RpnUxXLHh/hfcM1zbmsxPlZ1vgVODuhib61BXMmKJW0hVKvG5RkYr49sWvoYt6eP8IKQlhjdYTpnxjF5tGxpzKTmVBTs68Jw/w5GHY53Jkp6upXo76sn12JbzEd5nQdVyQmpt2JYBB16Ra0SPoXTb77+hj8m1ZCwDJxGrFRkZSOk8VsQlstsypKFhT8YU4swcU4mcgpSHtRovnBIkfp3MsiuPUkn99ZUNblDkJukCS7uiknptFIhhbpeU/r5ISVqIbhBhMYg39nUfWo+MnmSc6YIt6ZMSk5Fwqo5wfigTdmXEA5ky1IxYB7yn2WVsOijKV+Whk8JlwJIV22bIVJdMdcFI+v5etTTgxEf8fpsQs2OG3LOHGG9C+BL3OTAzntd3c6RLrkeZa8pEMYlZ30/Tyt23+6n7/M6qhkpdcWLEqR4+0Yv435vX6ZDyicV1ljT8RvQ1tujzWnRIRc2ADu+3u7xkXPFxca8PQg096RAUzQJkr0NKScXSS0jvq7u378lpq6wJtHtpRc2RzHnFGD5kd4lEOGLJ++0OL5lTCnGTroE3xWyAN2TOM3ZIRwy1l1dvVDBRw9cXNYnPRQrVdi93cK+IKRUPbE6PmKEnmkNFgVNZbLDsScZE3b85DhgMEs/nVBjGhl4CN3YrDo+7PDxzjuqrCjox3D3IvHzBZbxTcQnH+nOIn/uJf/jC3//UP3khISKX8+P/pV9MF5J7mcCIVNfye+FBL6ko1Vn9uK5bxy1yDCjELTY9Mg+vcnyMWGL6po/x+ve11ufJ5RsFSBhvv5mKy2YHL7iZbx4jwtCTTluAgNtk1+oyTvM8vI+f/NTfe+G/+omff+GtXDuL+kXRep3+tN2AWlle1iTUUICE8w3dnlxzGnUJhWJZaeHkFrXy6mPB5Mx6mUdnFFlpTSoxU7tgbpfeKdi0ncGEuL2msU8kIp9sWH99g0Rw4n9PUIAZaoeBph5fG/HIzMmIWNKQGGmN/yKBwjpJwafsFgbhlh3yfrvdTl8+0lwnVrfNvxqdOuMoHfC9zQ3ET10SvzA/NFOWpXOF7afClZ5Qq1J4uFNHDMe5ZdRv2B7n3LiSM1vGfO5wxRvNilxqahw+t0PKR3mKBqWShh3tcF377bQI1p4JDcqyscSR0sksaaysiogoUgb9gjhS+j0HFevHBouDiUUIpVWGvYZep+Hx0o3iJ/OE6j//tbd0H71Z/PAvP//iD//y85cFyNsUoeAIwhJGolYGuzX6u2Do92ax6a6+uV6tGysbk4GN9U7aJGY9jUglaZsSLsmNSCRtYavhWEYc7CoY712c1rgpw4iCmiOdvOm5btGnoqHAyemG5K107hPsMaSmpkvm14S4bTbNfbltUR7KhAbLY5m277+VQQXuiks456xas8MwfV1fq4hTnROJ4Zpscarz1vTtQCc8IXsMpEcqSZvQL3XF3C4ZmwHbMqSLI0w7KJZpr2csMTtm7EpKf81TSbxs7XpisWu2zn3eqSTeQyQkiQ0d43yiIp8gG//6vvSIiBxxXmush0wZhL7nI6Ye7ndDt+iQkEpMl4w9ddK/wRRQUQ70jEMmvC5HvCZTBsS8z26TaMR79QYGYUv67JottnCNq5UWTHVxjhvp7j8l9YpfDmng7qEPy9MU6iZeRoS/ynX3XmIHyPut4ohVrUwry8TWfEWnHJY1Z1XDr8kbfN4e87Kd8evcBeDEG7cecMaEii9Gh0x0zmM9pUtKqTVLKfiaOUNRRtJvIWEBFmbRFkpd47hDzgy544vPmkZdYVxIQ/C5cfuy21tnUrCkbJ/hioaX5Yw/YcInuMoRhVOFtH0+0txgrJl3t3fP0SkluVp+8P0Vz/QTghJm6aeT338b/qOPeBUucd5Y19KYJ8RR9J9MMm51Y251E1Kc1HKJJfVtv2CWm9Pwx4slkcBOz0GAz2rLvWXD49OEs4UhNrC/XTPIIK+hOC+QeRnvQFxOQr7D+B9+4h+9AK6DnWnadscSIhJ1i2vhO0AdYip15UCEIZOMpa7oyFpv21zYSMKx7uoRW9Kni1OS2pJBO4J2Y0lXhNQ4vGxIviv//VDshImAc1Rfb9qw2XHcMORS56y+UtctCRtrGoidLfTLJcCpx2Y2at/SVGRTIx2cTnuQJURAvPxiUFW5GIEXsgmdcK9zCYbTC18voEFFJSjjJBJzamcYhKHpExF52WTji5Ki7d62JFocBnuhq5YrYkUR1VZnvasJp7IkIXLTMB/Pxj2n1mHANngyosP5JiK8J+nxRHOLNHbKUh+xO1SqfMmc0NOEgpqOxn6YrjySFd/f3OSIgokU3DWn/nyhE7lu0L15wziOiPzvuj4UyhqWuZIXPT7/uCan4A0z41BmDkqCk5hcqiueO8QsqXgq6nKvyamwfE/zFIdmwTEzXpcjtugwZMCwZykrw839FbYRBsMVnZ4jAd65N+T9z8xQFX7ryz0e1SVjSXjvFcgLQ1kLNwbCzqDBRJY/umeoP/XPXnh2O2LwT37kzywoPvnLP3ZZdLxDYSRak4dVaajbBO0igfybxWZSD+fXJi50ecN6EAQrgHN8sIg1TwwJ0wqXqqvngIQ1L5xrMCvdfD+xOKLuksI5eUvCmB5znN/DQFPmUnLGAuet0bT7QOjuhwbILkOu6JCJ5OzaHnfNKRlJqzh0wpzrusWJOEnVgXYIwiLueripBkDueSsVNTGGA18cXZUxW/RB4EAnbNHDinKoU3a9a/uBTrC+IVNStcR8S8OJnbaTByMRA+lxwrydEIBryFwx2+0kuPRNHXA+UD3vOr5rtjjzMK5EYu97YRmZHjFRWyQoSiwRfTpg3FQ9kZgOCWMz4NTO6EvP75cpA7osyNlhQO6T5civtQZhpBk3ZZeEiJ4mfE0e+0ac5SXziJeA99hrvEtHoPAMY75ijmhQZ+Rodtx1sift3umaTk7B6g37mD2zxUd5ioSIe+aUf19vMKbDyzyisJZeZFCFX6jeoKKhNBUfaLY41pJ/E92nwfIqRyTEHDHh2EwdF4OMf8VDsPCAYxpt+FXzJfdMacOKnJVkXJcdRupg3nc4AF1LQgfxGeNFZ0Jjzz1jbj8PXKaKmiFd5uRE9MiI6ZFxwpyUmIbGE9PddGQpbpJSYXhoC1IMt3XInnGqjakIr9slz5mhvx/cXvUvXkr5gQ8s+EjSIKI8/zFXePzqF/oMHwxIve9Po8r90v2OMQnd2PlHRQI9iVhqTYTwbNfdb1c05mFR0dWICksSOTf0VeWmScsKZh51lUZw5yAmiRzcrbpE6b7jcTkJ+Q7if/zUP35h8+F2m43bOEPiHyYQQ0/OyojJiHhW988l/MA5s6yQ3B8zI1LDticMJn6ZSInbiUhwdw1ESVhPQUKHqV2UcBAd62FGQZ53E/oQoiWsb3QFIxw5MWxejSd219SkkvjOo3v9W4FlhaR908BrpQ6TnLKGZtVavymZtS08LkAjIgkb/1pRJSQZsZfprPz7cG7Npn0fgjCxc2+MGDv3YxxOOOCfS6q2WEklaRf5QHaspKFHxlIqFlJ5BZuaP6onGKBqnGJHad0IOfJ+F51YeKIf0YsM/UgYxRHDKOKWHTLSlHfbbd5nd3jGjsmpua19HrFiuIHhBfiDxYIvzHKqBq50Ih5VFY/KmqOqppdZGguzlVA1cELJa2bKY5ly7DXzNydnVtRvOg1PbRmGJPRImJmSiazYlSF9MrrE5GppGuH6/pxeL2dv/5TR9hnzqbt/R/2G4XhOkrjPJcWwE0cMejXGwNFc2N+uGA9L/uCuUITOqlEWf+fbn4pcxl9s/Gc//l+/EJL6EJsTi83i4E+LIJ3tVpXNqa49d+yGpl0jv9nxG7W+GGra9SSsTyKmJdE7sYrqHJSlYzootiWeBwJ3UBFK1Mmt72mfiaw4w8mgtmIhvkkQGkQjeiTqxEgUZVu7TKUgI6GnzpvokZx5N4b1BCUk6QHzD05F60DPPME47BmWHRk6IjcNRzrFINyWKxzJnCOdUlEzZUkqMWPpU9NQ+CKiKx0G0mPHjDy0MmJg+u1eNtE5gmFbhvSl08J8Nj+PsBb3vameQVrVrJqauV22cFaL9W7c2rqoh2vrnNQztmXgJzwxI9OnI6lvEjnBjj4OxltIzRUdcoUR29ol0YiCmp4mDDUj05gb7AQwIBGGZ/QKBviKOeGhrMgwfFyvsaNdvr+5wXP2KnsMPe8kbptrI3FTku/jPQSjxWva40fs07xsZ7yfLd5nr3Oz5/aqubVsa7e9fxMRrhgHfZqy5FhnHDOl0Mo7tjs+RiEND+SMSgN00O3Zfel6yWcH9b4vZ3yVBy3xPPVGnR0vr7t+Fhoy/28tHFxSgqBw4aFdGRF7OuAENzkL6/629tr9OPNy9N/NFRJMS0SPROgZw5mteVfUx6qSW+ftZBC+57ayyhN+448G/Mq/3sZaIY5rMhFO6oal95gCWFLTl4ibnYTawnHZ0KhLWLtE7Eh6DklwLU243Ul5Ks2YlUrVwChzCotXBpDXSmwc37CbuOKjsWvZ3st45+KyCPkOwi1oET1NPcpSyfwmspnYh4V8c0TvTJ0Cdja8zpyT6W3UtnyLcIyg5tIlZYs+Yxm0qlXgOyFom2gHXfzw8+tjr/G1F2MTmtWVrCWgF1qx0JXbcGSNsQ7E+pq6lcv9B5/9b99SR3rtOrupfGPa7s2ma3GQMw5/D+834KJDERfhuDXntP0JalsuMQgJR/icRtJri49wjZZ25SEM6wIuRFAjU3WTmEKLdkO3KDNWHHDmsdCW+2aKFeXM5E5OVpyTeSRCbcNnDMtaqayXtvWQrUjgmaTLdekwFoePveJN/xqUpdRcjzLOPDl9X0e8Yk74kjni86uZcztP3Bj8epZwPDP8y9mMfzmb0Vh3fk7y2DK3y1Znf6k5N2SXgrpV99oe1tzIEuZSYlTYU1dcnDLnD6P73JUFV/eWxHHN1t4Ro90HrBYD4rhmscjYGucY09AbLBgmhg+MUz5yu2KZx9w7MZSNMh4V/N7XOmuJYVWqRlD9s5PYy3j74zPP/9S54vDNiOQhNrkimxFeH5oF4ecuPu+CawwkxDSes/ZmU5bG87+MOPhVVzqtX0ckEbnN23VG/PMd/gO8lK+fNEvUckHCuS6lYl9HKMrM+1hsrvkQzFXd8a/rmD4pz+k1D6tSBqQcMuGxTMk816Okcg7QOviGtTmoBz2h2+fM5hQHxTrSKYJwoGfOI4o5mUYc6bRt9uQ+2Z3oop1ErI9v2onwlhlR4xo2c13SE2dON9UFc10x02W7LxiEsQza5lGCI7KvtMBqQ2GL9vNuaBhJn4y05fU4AdiYMf2WLF16Xt2YHnsMyUgZ0aMnXTqS4gjoEUPNGGrGSDOu2j4DTdnXHgPN6JC0/k2JmlbNr6TiRJbMpeShTKix3JEZlVpu65BxFPER2cayVpf8CLfZMSP+sn2WD8qT3NQ+3988xe/LHU6lIDOGB2ZGrcr7GPPHiyV/Ui/IRBjatL1OAH/IMY9lSqPWi6dU5xAQgbfR4DgWqcTsycjzY1yRNtcVd+WYubppQpikOV7k2vuj3fs3FCxdMeEMFxtt2uIip8TijB4tDnJWe3jhkcwJIhGC8F16lY4RbsQpO5KwZxImTU0kQoXSBG4LFuu9onZ3lvzOqxGFKgtr+c0v9anrmJlt2hzoqHFKkimOxJ4YOCrrNg/aiiM6RCx8XtBYp3yVxe61446wapSj3OJpNzyeKWNfkIjAaa4OiXBZgPw7EZdwrO8gZrJiypJb7NIhJVZDJWsORlBU6ns4TihEEiKMCs+yzwmOXO6cyF3nqyddju0Zfem1CXYwtgrHcJudsseQbenzhrfcUTZkazc6cyFc4UG7KDXanNvsNn1CRAxWte0ChSQfXEESzi1sRo1aumRvaQICzqzQIFRakbQKK0E+slx3N/XNu6shNs9rs2jZTGbCnxGmLZRgvfGG4mMofZbeuEyx6wlH60Hi1bW8DKhB6JkuC7sgp6QnnbbruCdjVlLR85thJQ1zKfmcPuSH9QarRjlpKvbjFFXI1bmf19ZNSkaZ0KgjdQMME3e9v7LMue8lOF+VKR1ivmBPeUK2WVLS1ZhSao5lwR2jfKQZOC+OfodJoVwZQDFvmEjOb54ID8yMwOvoSqeFeQDsap+P61W+56ZQN8J0IRyVNblUpBKTaMRSSo8rntOwR9MYsm7B/Gybq+99meRsn8W8y2BQMN45BWD05Mt89GiLG7fvcXKwT/5owBM7ljSxfO3rA26NlaKKuTe33BhETFauMBu8lRvrMt7WaLxwBHje1gYvIHTIL6pYbU4uLzYL3swTJHwv8MCC8WjNeVC3m8w6ZcEQscTej0FbSJCRqJ32uXuf9v93TIdCCwLRXNVyjHPrzqWkKxkjerwqh+0x3uw8wzSnS9au2aey4ioDYjWcmhXP6TVOZc0HERxZ/kTm7aThsUwQnOfFQ5lwU7d4Nzd4hQcYDIcehhWgogbhSJ2S4msctN3uyPMCAudtswCxfn0vtXLwKztxXXcEFWVmnXxuLDE96dCTjCM7IVI3LS+14prZbd/3XJdYbSi1JJUUI5Fr0Hn59QHdtjEWml7BEO/ingWw79X3huIKiSVuz7krJ3zQ3uCRzHksE3YYkGnEQB3v5kkd8JqZtkXJmB4LybEoD+SMI52gxkkiRxie0gFfb3KeiDrEalpO5oHMGdDlvdGQd+sAMTCzDf+JfpjME8k/odcc34+YM0oOzYIjVpxGKyoaFuT8c7nHJ+1NEnXiA8FHJniFlFpzKNNz9+5cV8xZMZAutTZOEl+dOExQD6vb58xN7ULRGcQQDNI2yWLPwQn7tqJM1e1fIoYZOQ0NV73wSb7RXBtrylg7vMKUrHbF9EIqnrNjOmI4qisKGk6s5zEhvGuQcHO3pqoi5rYhEUMkodEJP/qBil/9spPV3TIxC2u9zLu2SmHHWrKnMd1YuBUn4Iuu2NAWG0Yc4Xw3M+S1K0Sudg39VFDWkvGdSGgsb3KXXcY7EZeTkO8g/u4v/uSLDZa7csyRLJhLQUFNSX1hVG3bDSJ03wpx7rZh81U/doa1GkXQ8C6lbsfxDZZ6Ayqz6dZ9cWMJESYfm4v7m01A/rSouYjVNh6K5MiBQQZ4xVuTvPvbz//MCwCZH3dvwrGsn+S4czfnYRd+0/pmqjibncjNSVQscXvMILkbOpVBVQxgrssWZhZUeDY9BMLPwzrpWNoVmWQMpOdH9j32ZMyQLl3P48ilcsIEaviYvcq0tqRG6PhicN54eFIkFE1QG1u/r3vLmlmlpBH0cATV7za7AOTUrKTm/Xabj+s1PiBbHMuCGMNUVnRiB/XqJnB7C37leMJr5oQVNa+ZM87E6ekf6aQtusJkqkPCIIrY213xwQ+9wasnyh/IEX3NSDTidXN0LhFcSc3XH/bYvvUKuzdeg1XKaP8Ot9//BW59+A+YnW0xOdlGVxn9wQoRi7XCeFg60nqnJo0VI5DGrjPmrjXs/69/NifkMt7+aDw1FGjFITa5YiEuTj82fz58wfnJSYjNyefmmtBoUMVxz+nQ9OlIZ0MFz7RmrK0/SKvm9+au7Q7GGp1bIxMvRNGVjKXmPNbTc/f95nq05m+4P8OamBCxrY4EvpKKRA0raiZe4EGx7DDgWGbUOP7IY5lwhRF7DMml9pNw67yMMN5DY+0ZteafNOcaR246H8qRDY8pbPtzfc+XW+iqLUAScQnulhk5iKs2nNoJD5tDFNt6ghRacGBPeKN5wIE9wWpDRU0qKR3TcTwO4yA9lbqEfMaKBQVnLFhSUol1JrE4wzorSkpEzyecA1I6GmNUmJGzpOQKI+6ZCUspKSjJNKJDwkoqIjU8kKV/f0Kshm3t0NOshRoFX5TKK1fekTn3zZyTpuIH5CbP2R0SIm7ZMR9urrfd/swYKn8NT6xbo+7rigkVPWN4Uvp81F7hRJbc0cec6YIG5wR/oAUHZkHquUYBOl15/45cS+et4g0UG23ouDlIy6G0vskZIFXxxvQxcBYzL5Ps7kV3HEGY2QW5lu2eW9MwlgENlgW5Ow9tuCPH9DRhX0deTtlJx2fecPJdZoAKfEx2UJzUfI4lsH1inErhnUXN730dfv1LGSWWSp3wyOO6JM0qoshNS/omwogwjCKvhOWiVGVXUo7KmiABH6BYeaPkjdJYx3BNIzftGGXCdmpauffSpy9hT1XcdGZVX5Yi73RcFiFvIT7z/E+98Lee/+lvwKP/red/+oVCnarVKTMy7zQaIvUbF9Ca+1SetFjRcCqr8wUHGyQ9TwgDX2zI+SJgM9kuxHW3woIajhdiM4H+x5/92Rf/0WdffHET/3xRsjdwSIKeeEn1DQVOSeUXy/MSm//4sz/7ZyaKf+dTf7e9lhanwlR4NavQFXOa7+tEw2Gy11yPgBsP55tuKHdtqmddfF+bsQmfcBKMlr7vtF3kmGwu8Js/FxKuju+8jqTvyKt+MlZITUOQz3TvZyt2uuiz2iUK/UTo+s5PWHtTv8oerdwmkogwrRsWFVxNYz7EFmeNI+hF/r4x4h7onjHs6aAthO+uaj406LAsYZ4Lr5ojFMtCciay9HCr2okQ+MlYJgl74rqPH3+24Pa7v8zsbMxv6SP+Pd1rpXoLap7VfTJJ2ZYBiUb8zvGK/OQa8QfuQVZhrkyI3nWIljHXnnyFpjE8+JMPs3P1AY/v36TXd4lCt1Px8CjjXx+W/P5hxbDrrs+8dB2vy/h3My4+V5vFxkWiOWwUEhdgVK5RsoZjXYxwrACDzCRtC6CGxk1G1JnuqTq1PREnAJJ4bkdJ5ZS7voUmjOOp5UQS0SGl0aYltZ/ZKWc2TLMdFj+sD41nhgTn78cypfLFQUbMQDNybzb4WNw0I9pY14Lh4CHu54Kp4V05agnikZ/ohPU5cB42Cw1tVzA9n/TqmsfmuvAVPa/K6LglNQvNybXg0LoJ5o3oaquQFeBy6qGvDQ0dcVMkIxE96ZJJ2nI8gDbxXmpOqbWDBPnP7zGTtjlUo4y100oKV9JwJAsmkrOSih0G7dTjGKdQ5eDKyoKyff8ljZvWYLiufc85cdcsxrDP2E1B1FDSkFMzp+ShrEhE6BHzrmabD8kWT5oe4yjm5WbGL+rX+BfRazzQFccU/IK8xLFZ8YxX7vqSnPJlc8IHmitYP1Ef0OVpu8dXIif9vs3QTZOJ28ldKC5cs7Fp99yckqkuHA8R06oxhgheVQlx61nlCg33KcVEZF5YRoLqo2++xf8fe28aY2l23vf9nvOud6tbS3d19TorZyPFkShK1BIrsWwrjhFbCil9CBQohgEZdpAAEZxIcUBpNLCkGEKQGLYhGRCsDwqSfBCp2F9E2ILj2FAg2JTMnSI5JGd6pnt6q+qqW3d9t/Pkwznnve+tGWpIhcNRlHoKheqqrrrLu5zzLP/FH4+h9FhRbgjAWKCgaXObioaplBTS8O84YqgJC+t+f2rd9b7whe+Shkycp9dSLUu1pGFPZw0F/53P9CiwrKxlbhumTcNB7poDNxcVF5PYSfuy2ZhrFE5ry7xZg7mLxsGy4shNPcChCtriw38N3+fRN9aMPY9vfpzDsb6O2JYtTvSU//xDP/OCYHjUHDj5QwyZJOwwZFedJnyNEpZngIVUZLomeHUjdBRaB29ZFx8ONlA4rKesi45u8dH9/kC2Wfku9j/+6C++CPA3f/TnXwi/E372ZhGgV2GU7ha+dTctECy705ygJd+d4Pyjj7z4dRUgjTZtR6fSqi3Quu8nwDJU1wVF4HqkktB4KeGw4QeFnJConE1+as/hqKE1HTME28O1ik1YhIPk5MKunSoCjyYodQVH6JEMWg38ua646PHjlTSu+JCIpR9pRxg+prd4V+PUV/rE1EtlaCK2E/daDE65Y1Kok9at4dC6ZKVaKbdZEGM4kYJSasaaU2E51ZrHkpyTuuGWcVrv2wz4fbnP7y5rvre5zEsyITVJC6NaUXCiU7ZlxNgMmanDA19mm4SIb9ddLl2+i8Q1ZZnQsCARYUHFK3JEQ8NlOyQR17EcasLTeY/e9ZfRwz7Ru0r3ZtIh9vWCKG64+thXaKqMwzvXAUiygsvX73Lr5hX2tmqeWmZMCnUKOX7TuvK/nE9B/iTFT3zwp1+AtT9OKB5CfITvuwAAIABJREFUcyDCQSkjP6W1enaSupbMBXe/ijcLfLNCpNaKzKwlYC3qEzL33PtmjyUFcUhaPbTVIC00SCTYE67XGZf0N05copOUByhrd3pt1HLIpPViKr2x6q6s3c1DEVCzNi6NREisExTJSTjx8CvEmQq6Isqtea/oPQIBOrzOhqYlvjtVv/UaF3xI3DE17bENE5CQjIcCpZ18yGYBmErCRGds0cdIRKlrQZA9s+1fo2WmS/bMNhERc12yJQOMcefyjn1A4CNWWlL4YzzTRbsf7phtpjprIXXhOggu88dieCA1A3LmrFr39EuMucMJQ3rs6KA1eqzENasmLGmw3JdThmT0NeNUCgxuyuHgTwt6xOxoj8wn8ZG6Y7bjncDD8fmKOSSvo5Z4HYkwaSoOKXgpOuJIp5RU/Cvzkju+Knw3F1CcstdDWWBRJtGKi2wzY8mpLvhDcweD0COlT8Y2Tlb3iFNK3TxHoUEX9j+nflkD+QY6QPFGmrq+XgNUOkCwAHJJ1hMND8XKSbkkO/46VK7rBQzwmhwBbvo0xSE83HM5vgbqcpv7suACKTNt6OP4Gu55LRmGiVYMiNvv59TknlnSoPzOH+zyF75tznKZ8NtfcsVCLA4udTGNuVtWnNQNVt304/aq4mqecFw2FKrsJRG1hVltmdWuaEpKx1HdTp0pbxqJPy/OG6SysKqU8nwI8icizouQryOMd+wWnBKSQdjxJk+NWo5lhhFhrD1i1opXJTVDTVlSY1h36MHdLIpyw1zioU79tmtbjOdCl0SY1gG3fS1sKmqFEAxTL40Y4lc/8vNfM3HrTj/ObkihG9X+f2eqEiYdP/mhD7+gavlHH/2Fbzg5NKyliEPyH0Lb743zqvDGZ22hxtq8MPIzgLUSTXi962LkzTqehZZeGWftbeKSofXv9iRnamdkknnvEAc9aDxZv8AlQLXWTJhhEIbSp6FhxgpFua0TnuQyxzJvC7qUmEotX4yOUJQnml0AcjVMPUQrM8Jp4c7xSpWsUe6Ik/+M1FBIw4yKShwUb0GFUWFIzMfrE67rgL/UvIvfjl6ipOYrco8xA343us0TzS6f5hZb0neFjE5ptOEhp+zIaMNs7a+Y6zz/aMViNmR8teHGU5/hr/zh9zGv14IJT+kBC6m5pH3GJNxjhRGgjqBXwqBH87kCyReYaw3Fp5507/fgFsOih/ok4OUvX2M8dsnZ1dKgDyMKP2R79H87L0D+JMVf/dDffqE7bay9HDhsckK+VnTXsPA4LmGv3/S+jYiIzRu3KvX3fl96XqZ2vY65hkPTekxAcFTfLIgqdQRg23nNgTe3bsasC5GuTHBJhaptE7jwt9syoGYtEgJwKgW5v7fGmnMkC4449WtYIJivJxrr97hei7s/DwVI4HqEXnAAtXV/1v3broBJVyzFFRR9BNPyQua6ZGQGzHTBUPoISt4RAgiFUoNlpSUH5kL7PA+skwoP0FWAUkMjJmqPi5NBrim0pKYm9xCiUxbtY/XJWFDQqJve7uBc6I+ZskWfGkdcjsRQ4PySarEMyZmwosEylj6ZRiypeUp3eIVpu++MNcfJ5q/3uucap155zeTcsiu+1MwopOEPzV0sykh6HGm1keQvPJfhjl1RRTWBxL2rQ4wIU1m6Yyp9TllQ0pWItl72Pric27ZwDOcveNvEErHQFX3J141K1tzIkEOEx3AQPucJsidbJETclpIdnJraBR3QJ/XXmxN6iYl5VPc4lDkFlVeCc5CtE3GCLfvWnf/7FGQYptQM/TWetQVmuF7dqxkQs6DhuV6Pl5clDyoHxxqPl/zwuzPUwse+4IoPYZ0rGd/wHBjDrHJ+V3tJ5AoWcS2LUeTUSRNxRyzx5HPFTUOMCPPKTUNiERDnz3Ue72ycFyFvEX/zR3/+hXCTA63/BjRk4vCtbvHaVIQBqFESoE/STinWECJogMwrXtTePyR0PtbuujMOOp6eNW8OVQhj9v/xN/+7P1bC1oUlhQKlwW64jHeLml/7YxQfP/mhD78QJIELLdoR9DfKTzmLGQ+bonucdQGyLkaajY5r5BHfQMuBCIpmCUJPMlbeWyR0axualvTa+OKo200ttGTGgr70eKhTIjFOWcRzf3JG/vlqEtaJ2mvmFAO8z+5z0tQ8YjImVUPfL6iNKp9sJtzQIQ8pmVI5iAFOKnFLM0oaKrH8TvQyJTVfwDCmT+5VTx5qxUPca0pMxMirxHRhZY3HaYeI1fC+x0quP/o6/a37iLHUVY9nr1b8/k0HK7yq2+zbAduklDS8dyelsSlpDNGTE6hALlxHxl+heWmf5Psn5O//Ms2XdykfXMIYS9MYbBOxu7tgMJryB5+6Rt3Ad7/7hK/cHPPd++t77zze+QhKWF0ieSymLUS6nh1fTzECLnlKPNwjItoQmgj/n5JsdM9dglvQEyfT2uWstQatZ6CijiS/2XBJJPEFyCb8ssF5YYREPUQkUQtZDFyTJetE0MnReniKzhl6I8B+Zw+ZyIq+phyKss2AYw+zEt+J7/I6wnEK69WpLtqfd+WAQ6wnOG/8WbdQ6hYg4gu2hRbkkpJIgFY5SdcApdoUEHCPOfeGu+F3aq/2lEmKwUHheuJMDxvbMNNFO5UOxrQtiV+1ha4p6jkoysJzLSPZJNQ3WA71lFhcxzu8rjkrnNpT1fJ2DALSI1HDbZlzXYft8Ygw5GqoxLKQCgNckx4W+IKdspKajJhjWbaTsxrb8oQaGq7JRQbGkBnDHeb8kH2UqdZ81hwylRUpCSNgwYq5OuK3c0Z3XwO3qe6c27P8TvHXXDC2dHuUyxMKrelL3v4siNyE6wkcr2nOiku6xQUZYxD6usWCikYshW8CLBCe0IseNr6WgF5IxVhz7skp1+w2hTREKmx7qeRV55oL07k+zpw35DVTai6bjFeXFTdyV/gk6ZTVMmMwWFIUKbWfnsZn0oLtKOawrrgQJ5xozTYR09oSIMgijq6eRk4Jq7LQBOVJEQqrrBrLIDYb7vXn8c7GeRHyFhE2l5CICoaxuptuSM6pLOnhFCMKv/24qYf7WhM0UtYEnPBv1yXpc5djlrpqYUkqoTsSRrszdhl2JixV+7oCBAK+fllcWE80/saPvtAmFYGg3ZLlO9jQP2qq8vXGr330F1786z/6cy+4DeiNBUj3/cWdSzMUQUE2M4SRiMqPlUMRGAqREAGuIeIcdGfWbeIJSTuSBkBqYmJWWpJLSmFLN5vSDg4d6zXu18TWhgaU1k8gyCsaNQylxx19SCSGPUYkRNyRCVd0G4CV1OTqNvbbnpw6qt2k5Mt2ToQw9h2q+75AaEQZaer/1h2XGEOsbgS/pORUF5yyYFsGOHsuSw2MGfBQZtS+u3qsThVrbMZMPcY8dE9/LLvOpct36I2OkLxAdmbEwHve/3H+7c3vcRAwby713DDj38xm/NB/+Vs0L1/g6JVnkL0BcukK+vLLmAsp9rUS8gwZK+aRhySrlNViC9tE1GVCnFQs532ef+4+d17f4dVbW1zZX7JYJJ3S6DzeyfiJD/70C+H6d8Nhi1F336aen9F6fOi68D8bQfEu3EeZZBskcaObhqkBihIi/F8uuTNb08oXKF7+3Buy1ti2EDJn/I7AFSAGJ0l7Yk87655/bdIpqMLapFVLag6hainFJdNh3Zrpsk3MR/ScDC8JtZ92h5dyyqJdtwKvYeALqxBhbSsoKbTgotlpITtLr/p1durR5YN0v4ZjepaYrmqpBRbq7rZgTLvQgr5k7T4z1xU9ydrJ/kDWMKbA9dmSPic6w2KZ64KFl3UPULtYYqcwJoN2zTQId+wRE+/s7qYXw/a1B2PcJ/USC6na1x/8RawqS1b0cFPq8NqCjL4gfn91hcx9WZIS0dPYyY2LcF0HVKrkGPLI8PnmlJejYypqcr/H56SkKNftmJGmLKTmQHvkRExpuGmXPM02E1wBEnaFxOcPfcnbaUfpeUVBerxqCybTnk/LugnoCrPoDf/flwxL7CWSnQJamOaFhmbmoVgDHHR3y8PVCmm8GLDDEwS53mNZtk2B0MTrq8s5nmsucSIFuUb0iBhKxLFWRIgjlwP3rePlDIhJMGuJd0k4tjUxwp2iwir8k3/r9sMfereQZev7PBVDboSTumEcRUyahlwM9+qSa3FG0Tgfku04aj1EGoWyY/5hgZOqoWcMvUjoZ5HzB7FKZITB2UrnPL7lcV6EvEWUVN6hte/IeurcaQscKfhYZ2SynlSEAmQlFbkm/rvNCGl06E5F4qQgUVqpWvf/lonOMDLy3Qx3I7/4m//12wZPCV0TRf9Y046vJ8IkJPwbXAfNFVZrtbAAdwrdsq5resDEttjyjloOrCceoSNYa83MdxHD+Hto+sysS7773mCrsG7OkEnKwvMjFMvADCi0aHHjJmwukrYbdoxTkgnzkrCh93EF6ityRELEocy5oIOOdr5lapyyyy1dsEPKLhn3ZEmjRWsO5XDp7v32NeE1c0qfhDklhzLlKbvPoZnQk4ypzrH0ifzxGUufBsuSwmn0+4mOquWUudswpEfui9vvevaUrd27mKQgeuwQ+/oWGGUxOSA3QmQNuXX09O989oS//O0fB6PEzx9y8H0fRw4eQU8fooWfRD15AnaEvWmhJ8TP3cX87hVqFbL+gvnpiNuvj3nmuVfZ20vpL1O2tub0egm3346L8Dy+4TBeihV8cSAReDUjVW2TzPWEcN2lD/8nGPDQmZDsB9hGkNCOJGoldUPT4Q0iEx7+5eBU3hdB3LSk0ab9Wfhd9xxvFOAISnghMsnaTn1wWu8WHGHtCMlzmCYa9bwOEe7bh+wZl1gpDlq0z3bbNXf8wLq9D99sEhygWRGGLfocM92AjAVp20T6PNTpG/7+bMH1ZrBX2Jx8d2FywVFbURatH4jxDY16o0nUHgMcJ67EMJI+EcYnyMqJnYK44+t4CZZTnVNrza4ZExFz2ezxanOXUtx1MrEzRjJov6Y45/MwyXUKhmsFMlXLgmX7fsI5UiwrKnqkRBhm4sjwI83YIWWoMUsa+hLxijpVwZetmxwPNWUi7qwdypw97bOg4jUz4T3NPgfaY8sXCQ+0IsPwB+Y+77UXONAhr5kJO9pjJiV9Mg71dL0nEYRnunyezWs0JiaICrzZeRWElboUPxTMGWkLMY5wCpsxMamf7G1rj4U4GFmAaoX7JYD5VqxIcKaPmefRVH7vPJGCbc2YSMFltmlQ+kT0fQEysTW7knDk4Xex3xsHJuLErgVaKlWWNKyskGJIkoaqisjF8GceU/71y8ppYxlHEbERxv6eH+LUsyIRInFFyihyyloWpfTu6ovGMooNozhi3li2IqeWJX5Kksi5WeGfhDjXnHmLiIiwqtyQiwR99aqzeFyXCw6O8zWBUm8sQwzSOuO+IkfsMqRsfSc25WgDQTPRt+dUdcnk3QLk6yGZ/3EiTGAs2kKhgI1NLbyOrku6RVtzK/DdSnXdtdCx6XZLw+bTVclaY2VdETS1M4K6ThAIqL0TfMVaplMwbQESnisiYteM225nkCzudhxblRy1To8eZUXFkoIH4hKH4BtT+eQkwTCh4oSSvsYU0iDAA7NwBEmESiwTKdjRnJXnhsx1xWfNbRq1LLXAYDjUCYd6uj5+WMY4HK/gDMZETKv0M9clxzrjJ9NnGIzmRAMPExkLqGD2JzRNxAeeWvFfPdNDBb5vOCJJKnSZYR7dxlw7QJ56DyxmyM4l99yvpxAJ9isLmrs76GnK8vefoioz1ApNnXDw1GfJUsvx4R7be4dcPHjA4eEWVfXm3fTz+NZHl5flXMbr9rMLgTybVJ81HOxGmKxYfx+06j+StipMZzkkIaHuQoNiids1tGE9wQwu6t3pZff7gfQ3iowQ3Z+F9Wij6EDWnkobvxt1iNxrhb++OkLwjvbZ0T4nzDegSAstWuO5ME3aVEZaUHgfh+5rEYRdGXmFowbjP9zvmPYxutOP7nFsE3YPkx1I3k5i2mkDoQniIUi66UXl4EAlSy2cD4k4YrxbUxwUaSjOaHDnDIl/14ydk7sumPqmT611u+9NdU5f1iaDjVpOdN4el5mftJS+GK1ovAzzWsZeMOzrVgtB6xaoD1hxQsljps8XOeXYrDiRgtvmlFfNCfdl2jaz5qz4Hi7ymN2mouaS5IwkZqWWcRwxkZJTKmKcWMAr5phMo1ZGP9OIR9knQOBM5yOcr3BNx2xOPKTz0eU+BegVbMLyUm8AkMgaepwRE6shIyIhoqfJxv7YJ2OkPUqq1o1+RA/BkGnMwsvMDzVhLhWB17pUS4Vy0y542S4osI5zgeNjLHF72MI2bJuE/SRhSUOFI6tfid0e+rFPDkiShh943PL7N9fvcWobjuuaSdO0163Fydpn3qW9F4fr3CkpJkYYxKadkAA8KBpqVcpGyYxQNUpi4LUf/9gblE/P41sX55OQt4ilrhhIj0wj9mWb2/qgvQnAFQfh390pSHDA7cKzwr8jDFMpuMcJSy04YUpN7TdE6xefsAG45Wghm/jmb2aEgiOoab1dBUiIrlLXpvTxJvEyRDcJCcpVZ40YnVJZ1nZqu2TPUMyFxfZskuRUtxpKrYi9KtbczhEMe2a7xdWqhKmNw0sHg8MgFWq9zC9Aj4yFuq5cgAdMdYHFMsfJIt7lBIA9GXFNd3jFPGShIyqcJKIgHNgB982Cl+QuC7PHXFbsqiNmfok7zkhKaf0PwnG5bvaZs2JiZ2ybLQzCqc45Zc4lP7mbU7AjI0f+R7mkY344vsb3fdtDdq5+GRksoYqhtMjWAtlp2H7fJ9jZbvi//8F/wt/7mX/Gv/i1H0GMYkYL7FcXRN++g6Q9dLyHHt1BRhmUEfZ10OMhxfE++vCAqsxo6pjRziGnRxeRrGQwKDm48TLT432yfMnlqw+YTzeFGc7jnYmf+OBPv+BgjDEi8UbB3w0RQdSQvEl3vxuBOxIRvC6cjGkkpmPKKl6a28Fzaq3bwgRcohbWgKUWLYwnwC/BrQNhsgGb4iCJJCx0Ra31hrlhCItuGJu2cLFOAeKeQ9rHLbTAiVT0WFJ4lSzDLTmmoeFYhMd1HycRG7fqYrmkLHTFwDuTN2oZSO4l0h3pvtCyFUYRhEM94ZLssM1gY2Iz0yUGx63ZEbeezDzcMviFdJWguhEmIO74xuTiDPQS4lZBzIhTHyqp2vWmJ5l/L+vCLJCmC0qcULkzxQsSwQHC6qYty7a51208Rb4B1Jcejdp2TUWh8PCjqtPF7zazgkKUIOS+eOoqny2kwrmzNyzVEa5zEuZStI8XYFgzKfmu5iqVKHtkPNdc4h4rjmRFT2L+hb3PgWyxlLpV1Nr1U5MDHXIocyxOdvg9cp2Chpe5t3ENnS0S3bXWEZBhzYnZbFYKCY60nnhHdff+I2osfVIu6RaZRl7Ras5cVtRYMhInEexVs/okZLrl7yV3T8ykpBbLVbvFXTPjkDnb2mNsMyzQF+dgfkV6lGqZUVOr8nTW5/XCiVKvsDzTyzkuLbcrtyMaIgYS8XpdkGJYWMvvfHLEDzy7ZCtNKQpFFHpmnWOpujwqEsiMoVDnzl6VSi8y5JEQ4SBZaSSsGmVaN1xIYyrrIWpGqFVZNsqNXShrOP5rH3th59fPBVDeiTgvQt4iIomY6YJTUxBj6EvPjynX05DwNSwLma4XV7for2MmJTvao8ZSd/TmxSemc3VSgz1xLrulVlTacCSn7LPN6m1EyH8zeB9/3Gh84gFh84k464a8KSHsoguLAAfJCJu4SxbiFm5xtiMboFmhu+I2UrfxxCZqtdR7kjHX4GrsIAAL7xugGopGDwnBPf9MFyhKpHiYxvp3atYa/SKGI51yKgu3SYrr1BXUbDOgLwkvyyFzXfGqPGClJa/zEIC5LtiAuYAfdTtSZuioTnW+Af14qFP6kmMQBgR9euVnblzm4oVTLl69iaQ1kleQV2AhegJ05b1L/vn38ty7X8NcNnzbs/fZ/57fI/rOi8AWGANNDfMJsrWLzuaYd61oPj0Go5ioolwNqauUvD8nv3GTk8N9jr70Hg6u3uHB7UfY3jvEWtcJG22ffBOurvP4ZkTLi9Ogb/dGiNPX8ghxMKY1NCtMJFe6amEkTr2qC8c0LbQqJMBhAlL56UsmKYWWvohZ3wcpCRVOetdqg/Gk+aHpU3kvEYOhLzkTXTtUu8cr2jWoKxUc/t19z4GM737u3ufSwzCXumLofSNC8l6p5aty308Q1scnEIoD5yKERdkm56FOSPy0x/liuET+iFOnkudXzy36jKXPQ2ZY1VZy2x33TVJ5OC/hedZftT1fC3VGeImHtCUSUXnjW6uQiKH0ohZpp6hoz6EYCm+o6M6J2+9CATLRGWMZEhnDqS6otKRRB7ettSY1PXp+jVpSUPjjFJgkofAwCBUwkF57/MP/xWcS9i3N2kZihSX35nrb2uPEe3e5x3OKi2PN+QD7HFKSiHCilpejYyIMJ8ydrDHCRFbubxUiEYJ/y1hTjmVJimEiKyKcUlaME6QJBPw3m8gBLaQKoFLrz49pG5Zdda0uD0sQdhly1W5h0VYaOkbY1j4rqVlQtAVIeA9bmpH4iUkhDUNN2dKMiXh4sSbs2z4LqXmopSsU/PW3pGFMQmYMXyqWTKl8cRPz0tIVdzXquVHCo4OYl2aWBQ1DYk7qhiiy3F3VLNT5jEwaxwmZWcvQGApr6UeGxB+TTISldXdlYx0ca9Y0XIxiprUj/j+sGnaTyEOxlH4s5JHw8pFycSCksVL9F7/9QvIrf+m8EPkWx3kR8hbhNN/hiFMusc3j7Pvlb3PKAespSDdyTdqf5Z7Y5QiMhlgiMrJ20VxRrjt5/qOdAGA5Ykr6p+yU/fpHf6m96X/yQx9+ARw/pOsH0F2cuyQ9WHcl1RdzAW4RzA3PRsCsR0SMzJC5daP9nuQsdMmucR4fLgGyG+NueJMNvN2cE6d6ouVartQXAd2NPqiNlawVdmqp247r0ndSBaGgpDQVD/UUQTi1841uWUzs5IYRENuaS/Y8ITOwjkqt2DajtkAKo/aSmhUVK2/udXAw4eDdn8BORsj2DMkbSCzUoJMSXQr/9Fd+hOceXfDYM5/BvqYc/I0voLcbGLjjRrmCKIZihezfgHKFVCU0d2k+s0ucLmjqjEyU4TOfhzoiSd17P7y3z97+IWIscVRzerxDkrx9E8DzeOv48Q/+rTdAFdbqTG8UlVjLw679fs6G+vTpzcIp0K1VsIK3R6NNa0jqBCpCUrbmhITHBrzZqmFlVySStPekRdtkPqwlIzNsuWHzjgRvgG923eAd5HJzXTnbCAFabliYOAQ+m+kUWeE9zFsYliMQh6ZJTsKhTlrH9aD6FRpXkUSsvKLVLkNOWTBl/Vjd5km3ex7kfddCKbpBhg/r1kpL4rZYrEjEtsegUW0LkkzSdl09e1wi1nCj2jNiutMScJObofQYSo8FS++/ZBgYZ75nRPza2rSKWo7EH0R13R6QqHvObbPV+rS457WsqOn7wvRUCrY0A7Sdnr2qCybGFRFTcWtwToqi3DanPG4dSf4P9CEzU3DNjvmMed2Z9nZkemNiSqn4c3IF1CljftWckHklrZSI1+VkwwA47DOJRC3vJ0ybog7kz7Z7ois+Yg/pNiKIOmldJzwfGlIRe7ZPQYMBtrVHjJs0TmRFpMZN4Cja67dHyqkU7WMEo8fb5pSx5l4AJaJB6WnM3Mv3lliOtOCZaIQBalUalEOzYNcGA2Cl8ADoUBgGE9pwLfz5J5SmMVTeI6pWZRzFFKr0jYeVGYMqXNkSbk2VRWNZqaWulVyM3/ngQVEz1YbriRN3mVQNmTFspcJxERoKMO4rqpCmlq2f/a0Xjv/OB88LkW9hnHNC3iLChjq1c26rM/DpTjdCIbKSipVUbdIc9CbCz2BdoBzKjIqaK+xu4I2D9GNK0t6UfckdoVodxvjv/OZP/am9QQIRXlVJurALjwuPPQykC6faNWMuyLjtkBqJCGTyLoa9OwUJf19owZYZMTIDUonZNiN3nP1niEIrAnb3zRR/gnu8K0bca07FdZcySd176WDlBWnNEHuSb2CYIUxTLEtd8UAnVFq30tCR7/6VWlFoSSC6hoIkIqLSmhOdMtMFuWTsmi0aLFOduw2fnKu6TUnFn20e50PNu/nVR97DwXOfxFw5xeydIL3aFSDLCIYGLEhP+csf/L/I84r0Bw+RSw1f/Lvvh6VCkiG7l5HLTyCjPchyyHqQ5iAG2R4SPX7EarZH/4kvM3r+09ijLaLnhTRbMRwfcv3ZTzF8/Eu8/uo1AEzUUFXnEr3vVPz4B//WCyHxDYVAKKrfQBRve9OOi9V4bH7tfThKLdu/7yarmWSuY92BOnXhOOF+DbKzgTvlEvvIO5BvTjpjYhpt2gIkuHYnRG1SHZJZi20bAOGe7b6fcH914aCNOkGHcM9GEm2oCjY4VagLnpw+s4t2ihDiLEm8yw0Ap0515NXrggFgt5hS39BIPaTsFb1H4GcEN/AIw5YM2JYhZ/kFQXkqnM+lFu1ny2ORjEzS9YQ6JMEe9hnkdJda+OJiDY8DWgnaMEVx58aB8EKTbSxDdmTIA3vMiXXvt9baKZ7RMNEZp3ZOoQVXzAVice7iwdQ38FoGkjM0fSJ/Hrqy+k7tac5dOWUpdasMleuaj3RklkxkxetywhQnxbujPa7ZMY822xRqmVLxmjnmlhzzaXObpRaUWm9+UvGkXuJ39b475+q4F4U3s40xpMSkJG3hEYlZC62IE2Cp/IQkTPTDOY8wrYyykyYOinCmbQ64xpL7fM0c87o54VhWHjquLbS7FPd6e95pPfPHbM+66V0QRAHYtX12bM5Yc/qaUIg79g/NkhpLSeP8OlS505R8wU65bWaMNGOPDMeldA707vw4yNqXp+649H1z9fO3UvK8Yhw52eVcDFcGTiUrEeHZfbgyMBwMDK8/p7/aAAAgAElEQVRPlWljnRWCGJZqmdiaPHJHbBRHjE3MnbLitLb+elTurhqujgw7meGZC0LdCGlqW5Wy8/jWxnkR8hZhsVw3+/Qk51TnvCZHfFHuAK6oaMQt7RZtJx09TTp/v9nR747g+5pgRBhIjyCRB12FFvcRFvY/rgfI/5fi1z76Cy/+xm/98ov/+KO/+GIoOFopXDYThUAbb7AcmD3AY839gh6fwa6HZGVkhmybLfrSYyA5e7KFouSk7aIeupS5pAyk126kThLRdQ3jrzGVCslJKq6YzCTB4EzAgvrZQpec2ikrStf581jeWmuHm/aqNSu/wYcNSIKSmn8/oSiqOth1h8desSMjBtJruSvgNuTnm31u2C1+sHmKf/9Sxge/Y8n+pQmre1fRwz72aBvJLJIp9BrM1UtupehHkFVc/wv/J8QRcmHMU3/1n2He9zTSHyGDbXT2EJ0eITeec8dxsA1xjB5O0cIwePoLmBsrZKfBXJigh3NElPTSXTAKRrnx5Evk127y6ms7nJ72vvGL6Dz+X8d/9sH/9gXwCbYvfEOi/bVgI2enHiLGS4q6TnU3UQ/FhoNYRRswrginTpeJo9d2f67tStqVpDV0YVghcpPTk7xN7gPvQzvcrRBD02fbjNgyw42fb3oQ+Z/JWpK26UxOusWKYFhqwbGdMjbDtjjo/q76AiDwNNzfucQ0xngH+Lid3FZatYWcIES+4VJ6uFNYI8LrmOmSET0MwrYMMKwT1ZkuHQyqfQXSrnlTnfuE2a0zzcaxXifEa98RN90IU5ezsSbHu73M8R97bl0l4oGetK+9/RuRVvSg8tLsQZUsvKYARe2RtYVR4lWg1uRvryZI5vgP6o7ZRFZ8IrrFiSyZSclNOWLKkiUFJzpjx/bYsTmZl1C/z4q7ZsbzzWXvhVL6Vtfa30MQLjIm07jltxybZXu8LEpBjbP/C+qNjhukvpEUJu3tNEOi9vjHvmABWgjaetq0Pi5d/iPAwFsKBEn1GttK5gZFujC129Ye1nMfC1lLGveIW77LjuY0ON+QXdvjoVlyKzqlxnJiayaUbJNy3Q7JNKJGifz7MtDK9qYYbvTd9b2gJhXhQVlT14bvftxJbvcjw6MHBZEvIOaFITLw+dOSSt3jLbRhpQ2X45RMDA8qtxc+qCoqVSosC9tQef7I1X5EL1XGPShrIY7c+x32K1arcyGUb3WcFyFvEd/Bo/y55jFqjy+utKH2G88dfchreojBFR6vygPHGyFpb/zuptldHPqatYvLyrtvG8R3zqOW+PgPP/KzL/76R3/pxW/EA+RPS/zGb/1y+567CU5IuK0f61q0JbOmkqDqjK+6XJCgVLNlhmQ4+d3CGxICnqDo/FeMx/MG86dCSxq1Lbmy1oazmO5uiKw3TEGY2BkLXZJKwszOWXnC5pYZkZMSiPE1btPte0JqeD+5pG1yE4qqgBvvvoaQnAQs9F171BYuE521CUyN8mcv9vjhGznPP/8Kw605491D8r27qBWiG/fAgOxkYBTZv4FWIJEhenpG9N5LmHd/ANm/hnnXE8jF69DbQlcz9NWvQD6Exp0jPXodnU2xDxKi9z+CLjJk1EPGOdKr0bmQ9qZQJDTzEfZ4yBc/8yx/+C//A1Th5PR8EvJORuMbAKaT0HXhhd3pRrcweDOVrA2BCRoKLdopSEXtoFcibVLdYDcU9LpJakTkf8+SezhQMBIF1wAI8MP163UfYTKpaql07c1Qas3cE7gTf891FfUamva+C9OYrtRtV4EqwjDRKTtm1N6XVpWZdTyuWSdhD5OQ8L5S1pPT2nfQu8eyJ7mbAvkk3XHUlhzqhJKaMa6TvSX9VlkrfA8wkj4NDVNdtIn7UPpsyYCxn5qEAiQYEbYqgLJ+HanEG98nxFS6dp93hZJp18sgIxsSaHBd+77kbgrtz11PMrbFNYl6kmH8BDghIpOEHhm5pBvHr6RqC5ARvRbukxCREbOgYKw5GWvT2rmueFke8BJ3qWhYauEmN8QMNGGfHIvypBmwkpqXuEuPiO9tHm+fOyhVZZLQk4xd7fOqecjL8oBEI06YY0Xpa0pPYy7ZYVtYJ/4uCf4wIT8Iqlbuda6L0+714X62VoYMPKt16SXkJIy1z74O2NMecylZSNXuTW6fcHtVt9gHZ6jZYFlJTUrESBMydV4hd82cvrrJ4oCEmZRE6l7LjJr7Zs6xlNyWeVu4NP4eGIiDchlgheVz85WX5XH7Ut8Y7j8YkGc1Q+Me8zM3M959peHa0MGw7s2VAsvDpmKqziohl4hZ4wQGYpF2V9yO3VFZYR3ES4R+Ckmk9FJLZJSyFiLjPYvSN9/Tz+Pti/Mi5C3iL2b7XE3dghcWzr//kQ+/+FMf+ckX//5HPvxit0u01IKv6B2+LPdw9OA3Hl6LMtKcCKHCOpKfh1qBW3T2vLv2P/zIz/7/rvA4G4Ez0t2EA2bYdevcJnOiwXVYWtnPblJkEE9KdYtwgCkNyFjhipGclB4pPT8+dvCuzcmIg1JtTkC6RPnuaw3Y3b53TQ6JTMCjL3TJXJctEXalJSLCXBftRhPG9mHTDbj4cBwCZKRbjISucoThVOdrOAXKJdnhqsl5fQJ1LYgoUVyT793FXD3GjJcOigXI7i7Rtz2Cfe0lou9/Hwx7SGxgtIPkA+TSI8jlJ8EYJIrR+zcxT70XVjNHTq9K2L6I/fwEqpjm46+iy4zmc4pOC2RXkcSrwH1gRtSbUS+2ePKZr7K7N+XJJ+6zf+HNu6vn8fbHWY+OUIBsdPzFbMihhq+bjt2GswVJiEDkDs2Drs9P+Nvw7+4Epnt/BzjVWVJu+Dvnm12304YA9ekWEYU6I8CYmKR1Cs/bx4xwQhUVa9ndwIPoTkNCuCaGm+Kd2LUcd99PvYf+/yzqOR6bBdd9PXEFoP9c6ooAI7M4snjkDRgDhy4Q2yMMF3TUec68k5qu5eFHvvCofQKeEJHiCPunXgY3HN9QGGakHkqaunPjhTlKrT3p3BWC1p9LByWL2kIrvMcuH6bRtfu4iFund2RIKjFjM2THjLhiLnCic3pk7VRhXcg4LmVF7V2+m5YsHuB7OzqgwfKoHVPIujANhWP3+lSUT0a32Y7cOv9/yJe5Y9w5vC9L/l10i+tygee46qBUnfvkJbnbSggnGLZxSoaVOC7KQmqesvv+mlrDe8OkrHtvJX4yEn5vDXdcT0paL5DORCQUFCU1BzpsH7PGen8zQyN2Qz0svO9wbAMXJFM3idmXjKfMkBEJY5txTfpkGnEiBdftGAPclzlfMg+ZSsGxLFlIyUpqDlmRe6he5nkd91n5yVDDRUl5b2/gFKus5TN3hF6vJDbC975rxW4fvnAn4tKO25eujALs0K0AYcrSAD1xV3mllqGJOKpqMjGMJGYvccfl9qkzW80zb38QKY11x+DkF8/5IN/q+NPFcn4b4vr/+h+9+PJ/+rEXfuUjL7zpxXm2UPhrH/rvX6hoKKRpDX6msla0cpu4U6govKqJJcHqeiEUTOuncR7roiN0cCqtEJ+ML3FqKSERiCRq/x3gWIETsrBLts1Wu+j2yHio05ZYuaJsseND6bUbZphMNN6rpfTyol2XYlgv5o1PEgDPy0jbhCmWGFUnXxleVwMb595qQ884+F/SUZxZatFKdILXgm95Im4Dq3yBc9HscNceIRjGMmSiMy6Yba7qNoUqw1S4uLcg7U0ZbB1THO+TAebqMcSKee4RSFJIe0jqpnpycAN2l5hHv81BrtIeJAn2K5+gefUWkifI5SexX/i4+93lFPuJl9AyJXpvjH2pJHn/MfXn+lAsYRjRvDYkTufYmzUyWpDun2APx+wPHnLzi++l1ys4j299KJZGmw0pWtgsBEJ0Sd9rqGRErZX/+zXB+6w5Xvj6Rslf08Iww33cN70Nnhe4gruRhp7k7VQjkXg9ofgjXm9Q3ALYMoOWu9GwnjgG2NhSV76X26V0r7kj3fff5X4JxsGx1LaT1vC6hG5xZ9iSjIyYutO8UJSZn1i4yYgXBdAzcLROjXdLD11XniHHOPO98Hwj6VPR0JeMlZaMOrCouVdedAWYg3HNvWhKUAncPEPrhsxKqzapLb0Ht6Jkul77gsBH4KCE9TXxxdSu2aJRV8DMPTk8HMeJzlnhptLh/QvSQmpDM9AgTqmLmAs6ohbLnu07WJTCZ6MHNFge6CnbMmDqJ1KnOmfoJ0VjetywO1jR9vFOWdBg+bS5xQ4jp1Aojh+zJ27aNcdJBueSMtUlr5kJY815XU5cgUhBLgnP2EttEbG+ntz7Tju+OGFKFX4v8W7mQYAkkPwd4T32ao524xo8lDkX7aBVvy87zYHwGkZe+aorGNCIdTLI0hCp4ZYueToasKsJuyROnao2PM0Wt3TJmJzb5rS9H8L5KKhpJGWhNT0iVtbyXD/nuEjZyQxHhZtoPCyEOTU7JmcQG/71p8Y8vguTacr2oKGXGqaLiE8fu/M/IKYMTQhcQfXsdswfntQYYGQi8sgR3C+mMdPaUjTKpb5hVUPdQLmIiAzsjEtmi4TZ4nzq/k7EeRHydcRj//vXrx8dOvf/zY/93ReM7NDXhL5mLKRg5CFaQe8704ixDDhmRuUX5PPi482jm/wkPhEPm/4kTEF8VzZ0tlTX+GyAgZfLbNrFqyaTlNwXHgHe9UBPWejSER19gh9SfWBjErLwxoCwhnQFRZgApwrY25QEhE43cP13Z/HiQQFmqSv60mOqc3JJfedz/btPmKusKLmmO1yxQ/5V9GXuNw/Xj6sNE2ZseQfjUym4nsfcW1ryWwMuXNqmWGwxPLiJuXICqYVaHKE8SpzkbpLBau6mG4D96idhfoqdemlTq5gb19DjhzB96CYgxkCcoasIc62GbAudWuTSFuboLvT9e3/qFD2eoaVB0gZdpkhaQa9g58Ixq8U5J+RbHUERK+oQqc8WH6GYMJ2k3LBZlIe/75quhgIk8By6ohHhnm5N/sQSeXKsyBrKFaSCgZZg3p1GdPlcKy0dnFHVqc11uFNBzjsiYmYX7T1otVlLBau2BclZ76FudHlqwYC0PQ4YLA0XZMycVQtPSsSw8qZ/TpLdJdYZMUvW8NB2iiHOMyM0H4J88VkTvkRSN8nwiWft4TZrjwnXNUdoPYnqANHxsFOrBcYbF56VDQ6x0rXrdpCRXWnZCqsEBagQXRhWQUngiAS+R1DemukS9Qm548KUTsHPm+JFErV8IkE8l8K2109CxJ4OGGtOrcotc+yk172f05SVI4ATeDcO4lRqxRXZZaw5qUZ8mhMu0OOAHQ6Z0uAgSlOWrdJZJIZTFuSk7Tx6iz6xRExZcqAjz7FQRjhz2ZfMg3ai4Tr6awhbuH4jP9EIzaUY0+5R4RgGPk/ui5MbdptbZsJAU+ZStsT7Q7NgR3vtYyuKUWm5H4oy1LQlrC+pW+jVQtw5PJGC1+uEFQ1PJ30ahRTDKIqIaid7fN9fY917waIkauj56VulyklpedjURGXCpKl5JMsQgZNVxK1mxXXJUYVVZTi4WHB0nDPs1zw4SXhIyfeNhojAg4VyfSz8/lHBOIo42C15fZowa5wsr8FJJU8q9/1OZqgsDFJoLEQGlqUwX3ohjL/3H5/nXu9AnMOx3qYoteaL9jU+z21SIgbqfBkyvPMoDhsZOizn8bXj1z/6Sy9WfsPpdh4NTme+mxCFiCVu5T2BjQkIuAVyiz59UjK/OSrablYHZo/MQ7Ng3cXrSUZExMIroxgcZjoUJl1CJMDCLlF1vi/B2XdLBi0GGxw8JPy7K0PaE0eshbWsZq01iSQkktCTnBt2m6fsPlftkGsm5880T/B4dJUTnbYJwA25SI+MET12tMdnFgu+5/Gaw7ly/84+ZZFTnuxhHs+RkSH6jhvoyZGbhESxKyiixBUXANMTdDKBZYlsbSMXLzmI1uXr6Ml95OJleHAb1KLLGDnYQ5IE88SS+t8cITsGyWKYNegMjj/+nZgLFYeffB/2aBt7OqS8d4CIZTCafpOvpvP4oyIUIK2XzptAjUIXPhQga/+MzUKlq24V7tzUX7eZZF7xKSWW2EELfXIZvD3AFR+q2nJAzkaFm4J0SfDg7tMujNGIkJEyNkMycV/d74X7qzNZ6I4V/Pehy9w9NmcLkS4nJLiIAxzbKUaEI+9HEiRdre9bJx2IZYAPBY7DIkjs+gbLOmFvWmWwkMiGDnSlbg071XmbwJYE7siqXUNDgWNZGwvmkrIro7ZpYdG2+RHI9JveKWtJWVhD4krvDxEkbMN7dfy9uiVjA0x0xlyXVNSugFHLXJdERE65SSJyAkfHtu7oQXQgOM+nxIzp0ycjJabC8ro5aTkPgXNRUzv5W39tGowzWhTDSDPGmvOl6AEvmQf8XvRq+z4DeTuYAg7I20JihVNH7KoqGoRjs2z3l8BbbLBeIctdUymJ4zDJm3fiQ/GREHNBh9yw2/TJ2rtqrDlXrTMjfKzZwSA81uxw2Y7aayJSac9e6guCoaaMNGtfKzgfsz4JuVdoTNSQ+3M1oaSgYdFYROBa7IqHIXG7s6Ya0+8I88QYxv4ZL0QJj/YT5wPjC5IG5W5Zkcfwrn7GJZNxq3bT70FmWRUxW8OK8VbBhXHNf7g/4MpezeXdmvdcbfj8w7p9LDHw3mtN6x9yY8cluGFV6iXuM0uUyIBVSGNluYqoqnNlrHcqzichb1P8/Y98+MX/4cd+5YWhOpTtTTnyfIAaFC7LritE/Jj+LOzhPDbjN37rl1/86z/6cy908ecNFmtLt4lrA7L2Jwij5VjiNhkI6dBl7xiedYysKmoG5Jwwb7tFfVIWlEQYZlp6V9+aYBRmxbKwS2ayaLtwuaQsdUVPcpa6ahOssIlVWlJKSqMueaq1picZpzp32HBt2DVjSioyUgpKKq3pSd5u2gkxV2SXdzcXeb434LuePcVay3D0gLJIWS6v8lOfN9RiuafH1Fi+v7lBguF6kvGD7z9m+8IhtnmMiweHjHZeJ378DjLadytzsUIuXnGTjwYol64IATcV2R0gWY4cPOaKFEDvvozOT5GLV9EHt9H7D4muPI5sF3B4DI89hh7fRfIG6WfoZIkcDOB4wfbzn8Dey/jk5w4Yv3wRMbC345Kv/ByO9S2NUFR0Jxdni4vuhCNEV6kNQlITlLFcZ/dsdGFYAXIVig6X8K3/ptbaS/dG7RSl8Ul4l6sSXlvo0qv/3RCN95II3gxdz5Hw++G9hUJozbEK8rhvnIS8GeclQKkG0mNmFy1HpEdGJE49y6n5RT5JdUniDGeYF4mhT4/CFy2xZES4xsdKi9aE0cHcXHKfeOhYocLYDFEcD6DQEiuOU9Ij45QFc11hsezKiJH02uOgKA/sMSKGCzIGHCwrFCOBqO/4EOE8qJOVZa1SFpQLwcmhXzQ7nKhT6nOFhaNmT3TWHveHeuohpu7qWfmyLPPrauQT9aH0NvgQl3WMIDyQKds64J6cYkS8R0jVFhDgJhUInDDnWa5xUw7b921xCIWclJFGPGLH/F70Cg1NSxZXLEN6nLJo94rwuEdMmbKkpGKbASc4fkiYXg/JWFAyoseKkh4ZpedpuALB8Y5C8dAnZcVaiGUiK/a0z2PNDvfMjAjDgR1SSEMl7l64bIfMOgT0PglT42DLVtZcnbYgUreGX9CYQCAfasJKnChCQcPYGzw+E42IxDmWH9cNO3HM9SQDMr5ij3G8oYQLts/Is1oez3I+XcwoGktS5g56LjBpnNRLrcrxSjlpai4kMfdLeOqCkKYNn3wl4fkbnvNllJ1R1fI3PvGag0T/e1djPncXPnUz4bueWPE9j8bcvJ9QVG4YqMClniFL3PO296w4i4X0V88NCt/JOM9838Z4wKTtJs11yVILFrp0yWnowoi03fZzKNYfHd3jE5yKQ3cO8PwPt5GIJ6h1ZSQTiRhIziETSiomfhNJNGKsPXoac013GNFrMbYlFXfsA+Ze1WZuF8zsgkgiFna5MR43CDO7JpX3pdd2Ypce0pBLzswuKLRk6buSJRW5pAxNn77pEYmbrpQ4QqjBO0T7pOhx9rlgB7xvMOAHvvM+p9Oc7b0TkqxgtD3h0ac/y89dfYyHfhpiEJ7r9RGEZ6807F66S5wuuXzlmCQpMOMpJJbmC3egqt3kA8D6rl6cuWIjSWExRdKeK0CaCp0+RE/uw2AMyxXMJnA6RYyg915FEpArl9HDe0TfcQNEoWlgqej9OSwVc6HiD/7JX+QrJw3//FbFqjAkSUOWlVTlOU73WxVBlvfriT+KbB6JQUTeMI0MERLCICIRhCQCX0o8zOVsrAm766KiJ3nLvzpLLl5PJdxHl5zeLawyT0QPxO0gWwpuCmIkaj8d9OeN773LhwkRCqtYIg7MHi0cSmSjMApKVDlurdiiT4wzAjSyhp0G1Tvrk8sgYxuKpAbXFGkLMpxq0lKL1mQxvM8t+mz76etUl0x1yakuONUFFmVkBu0xCHCxhGhjrTvrKB88L0oc8T9MRwKvp8EykB5bMmBA3q6TI/86gM5zBoNDN7WoqZ3qoBdyCVyJET0u6RYJEYcyQxAmsmhlcLsxps+QzE9KEi4yZqgZ32kf4b32Gk/rZXqaIMCCgoWUvGJO/OvqKk8Zll7RMsIZDDZYTnEqZKForGi86Mma/B4KJ0ekb7zQQeD4rb1ywrEIMLkI00KyEjVUYrlmtziwQ+e5oYaZlKyoOZaClbgEf6gpRXCpx2C8F8aIrJ3MuOd0CXnk79aZVPQ18W7pKUupXTO1WXKzXtEoHKQJx3XNUV0jAkaFBGeGeGSWrGg4pmTRWN6TDnhPv0fjr9VKlZGJnORvFHPS1AxMxP2ypsTSyxv+5U3Lg6pGVZhOXY40GhUM+jV51rCylg9cMfRz18hoFIoy4mQWk8YObvXIVsTlvuN+GHE/i/zScjg7n378SYjzIuRtjP/pN//2i5/Qr/JVuU+hJWMZsm1GZOLGoH1N2GHovULyt37A8+DXPvoLL1qUlV15+INtE4wQbizetAUJwFwXTOyshUJMcRr6EEh/Ts98KgVLCk5ZsMBBFAbSJzeenO031qWuaHAOviEijCexFiysg14t7JJ5pwtaaNniq0sq+qbHSJxscPistGGlpfdQWN+iIRkJeu9PXyuI4prrj9wh70+pq5Te6IiqGPH44/f4ny99B09xlV0d8E+L23zKPOB4GqPWcPfVx1ArbP3ISyR/vo951w7m6gC5chW54iUo5xNXeCSpm4okGeR9tFy6n4lxPiBVAafOyFMf3HNfraIrr5E/OYFVgd6/5zknFtmOsQ8yiEF2+8wWEQ9txeNpxvUrU648/jnqOiaKzqGK73RsJPcdKFb42i0yzhYPXehS+Nzwg/CFSFdyNUTXh6c74Qw4+TAxyWQNK2p0ndSdjXUyaN70++77arCYTue7K7EeGkvhMQIUtMt/CKpX7u/1DX8TpiBnnxPwyWm9wRUI5OugpOeI4vm60BO3FopIS1o+1TlTnTOUYEfnpJAnLJh7daLwvNr5mOvKKQVK1hYla76ObQuiUIA4GfOqnT7lkhITsdTCnzP3MdUFc10y1xWFX68raj8PcXyCMJkaSK91ii8oW0f7cHyW6ojrU5bkJCypW9J+RtIm9CU1PVI/WWjaUvPAjti3Q3Y15ylGXNCel5013DB9d/4RUiKe1P124tINVyR4npyqQzn4fwcPkNTzNUoqlhQs2vdUUfl9az3hdtdb7uFQXbUrtz+515T7x55JxVJqFniZWo2pxTr/Ml+8BE7HisodI7F+uuGOu1HpPL/xhYghRphI0V4jinIqBXM/VTJAZd0UwyqowgXtU/vfr2i4bxYcmxVHdc3rZcVpqVzsGRIjPNFPSYwwMhGTpuFQSyKBCRXXTc5nb0csaYg60Mher6auI6rKUFWGD1wVellDY4XCe4AkiaWfW/a36/+HvTeLlS077/t+39pTzXWmO/S9PZJsWqRkNiXRSmQbhmRJiWInkUw6UoBYgyEIeclbkBgJBDQIBAGSGAgC+CFA4AiWDSSWTIJ2EiqAYmpKIImiSFGk2GQ3u9nTne8Z6lTVrj2t9eVhrbWrzmVTUWyRbIrnazTOuefUqdrD2nt/w3+gyBRVmAQ6Uyw+WusH/k8cPSqGcRnfjLgsQr7OccXsc64l+2bKnBHv0mvckANekrv8Ia9yoOP/7ze5jAvR0XGY7PFU8hhjM6bTrsfiJhiumH2Ur5YJPTR7XGefmxxyjT0KTTh0I6y4vmt2wopzLSm15lRXlFpRatUXBJFcmZCQSd4/lGOSoX0i4U22ouxko23/4Gxo2Tdz9mTWJ1oNLSv1/KCGLf8lES/FGc29JjLiLqfcMQs++XLOJ37vBs4mJGnNwbs/jyk2DB5/nYPv/Czf/xMf53/52Rd5zh3xitxnrgN+62TN4uSIW3cmfPaFK3SfzLFfuEv3GyBPPYuen6JtDdUaObwB5dIXIGJ84VEMoVqjd78C6rw873jP80WqGgYDtGqRyQBJEmSaoYs1ZCmoYvYDfKtzuDsHyDTDfbnh3lK4kuTc3IO79yec3nuG1aqgLL+aEHsZX5/4Jx/97z4MF83/dosN+NpkdcG7Pu+aDsa/202kkrd45OyqAQ2M54vsum/vcjRioq4BomMQam17b40sGIT+SdEE0V6AQWgIRF7Grvz3owXWW223F8DY3j92C6BYuswZs2Dt4UQkrHTDMsCSRDwfAbad8ATDVdkjwU9u92XKJPhaj2RIguGGOQrbn/dTlTj5iCZ/sQlTasVCV5zpsn9tbMDMZMxQCmYyYigFk6DItaFGUSYy5IE77SFC8RzuHhev/uePeaWNlxoP5zQKdfjj5V3ba61ZhamLDcaRYxkw6ZW6UsZhWrHWasfLxUOUcsmYy5gn9ICrOuOBLDmWJWsqTnXFfc4417KXHC7IGFOEMswwUV8aPKlTKrF8mRUnUvGMTng7AdMAACAASURBVEkwnLiWd7urTDTnRNakuvXjiGtDwn0cPB/Gczp8MX0oUyyWDTW7JpVOlXMtWbFhxYYoeAIeGlyQMQqTmgE5o7DNCR4eFvf/RKrw+X6SUknHUppeBQ08/KuStp+0+OvP7zt4X7N4ze2u2SwU+R1KKQ2tODbS+YapDniHTHhXNvLFiniok0W517Y8rhOecvP+GMX9vk/FgpbPdgu+tGp40LW8Wrbc7hoGxnuI5Bhap/zojQFnriM3cGRyDPCHb6Q4hapKaJoEa4UscwyHljy3GIFChKOB4Y17A/LUcbZK6Cw8LJXEKJOBcrIWTtZCYuBor6FpLtPft0NcnoWvc/zXv/KffljV9bfu2PVpNfhFhFPw3//Kf3kJxfpTxneYpzhiHuRm6/5G3oSkP34fH8a7MdOCqRYkGAYBDmBxvCmnvC4P2GhNKkkwCHSstMSp9e8VJSYD92TXfNLrsHvuxtR44vnADDzZUSbMzLjvVI5ldKFjHN8jk7T/jNjpjNORRAwHMuW67LPPlJEWvNCteFBb3njtGqqGza2nwBkoWkgtMuwwjy34uR+6hyDcNmfck5Jbt/e4cX3Fe965AGcwVzOyH8l8wVFu4OSen3qEAkPyIRQDX4hEcno+RO+8iq49XEGrDcxnyHSOeepJZLbnIVzzKXI483/bORgPQRW9L6TvvYN7Qzn5g+/ls9WKE9vyrqdPefzGOZv1iOGw5YXXLtWxvtHxaAIer6k/KSnvixV5lBdyMTHfJY/H6H0+xBsIql70S9iN3YlC9MmAi8TyPykiD8Tvl+unIAZDtiPZG/f70X2OvJMIJwP6BB240PyI0xGD9BONlZbUWgfY2TahTQKMJfIoEgz7MrmQ+O/LhJmMyMl4qAtysh7G5gs3009KLLafxnbhe/Dd9lKrfhIS37/FMgjvV0hOStp38YsAxYrnJBLB432325k+eV6Dv68OyC9ICXt/k5SpGTOTcc+zcHiPpMg1SSWhouVc1/358MfKn4e5THhM59yWM05l3TeOYiMqEr2zAKkFPx0RhKkWXHcTBppSBl+RuF/LAFta0DDWjLkOuKYz3jCn5ME+ckR+AabmdDuhG1IwIKMKxycWKmuqfttmMsKpBlPEJBj+Ff3xy0iYakGqhkyTHoZ16EY8aw85cmP2g8qmEz8Vr8Wvt5U0FyCL8dzG6V0s9sda4ERDcXUxCRxq2k9CrrgxU/UE9SdlxDuTkVfDCpyQxkEmQt03BpR3J+P+eICHaLXi/PsxoA2vNQITSZjlfhpiEB64li8/8BYGy85xz9XkIrznmqNukp4LIgZUhTRxdNaQ55YrQ++k3nSQZY489aTzJ/chTfznjXIoUjgrfRGzP2+YjS+nId/suCSmfwPif/rIf3VZYPwZRk3LTd3rlXFK3VBITqstrbac6jLccA3Xk0MeugXXzSFHwQRyqCmFJqykYS0NI83oZMQpywuqMIh3xF2whDBi7+iYyxRBmMmIha49qZyu75MpSiFFmGSY0AEdcWj2esnQXcx6hyUSPONDM5Xoqpv25mKHOua97pCMreHT33huzWzvnGYzZTA5oTq/wuj6CZJZD/LFsP99n2L8G+8hIeEr5iEv3jmi6ya8+923ac8OMA9PSJ6cofduwzhM5uqNn4iM574omcz9JMRZPxlZHEOaIpKgapGrj3sZ3zTdcknUge2QwRA9P0GXLZQtr3/kh3nh5Tk/8nf/Be7uPscPDgBvPvXFlw9433vvcvJwzmKZYx3c/qlfff7GP/7Ty2Rfxr9+xAS1faQQuKiM5NP4VL76MbLLefDrXPqiodkh3iZshX2jyp1v2pj+Gq6CwAMEiV/1sJpIXo88j5ikXpyAys42+IgqRMLW0NA3hraSsrsQJL9tF1WgsmiIqq4/DrvFVUzaY3G0JWj7ZHt3uuMeaWbkpD0ZOf58wZo526l5FK3wxOaWoRSs2DBh5EnoBKK6wpoSIwkbrWiCcWqLZY8xt9TDKPdlwjklM0Z9UZVgKPEmjid4AY1d+V3wkLlcUkSFOvAfdqV598yMpa7DDMIXSXsy9saDslVmysWny2MZ9IXO1kF9EL4WjIN/yZty2sO1WvXFTB28SmbBMd6hzLRgpDnTMAEoSLgmA17inDqc+6GmIFDieQ8J0vMpWhwb2fI61tR4mfSOSM5XPN+ixfZfJwxZU/dNpUS8seO5lhjZTh/ipKbDkWmCEw+dysP1k6p/7UIqntYpU/WF4b4WnKDUYsP53CIBdo0Ph5rRiqXQBBemi6U03sE9PGsiNCsWY4UGoQRxVOrPwj2t+QtB4h7gtPWAvCgeMRBDo47UCM+2+7xiFuxGjeNYam6qn6IstOV94xH3NpbcCMZBieXL7QbvLJRyzRQcu5YiTwLkS7BWmM8rzpcDui7whrqEYQ51C/OR0nWCCfLD0Yjw4Uq4uW9xKpyXhtU6o2kNw+LSIf2bHZdFyGV8y8VVnZKo4aFZc4U9VknBJMALNoHHYZI9ajoe132eles02nmsrXqJZI+7dkS64SlLTtw5GP/wWrr1hSQiTi56THFU2ArJSIzoDQKw1DUm/Lulo3QtE+M7mcfujKnxSUXEEscH8LbP6m/wM0YMNWWsBa/Jimd1FrpchixrePHFG6wrww/++CsMD069O/ntORgFJ5gbx3zkh3P++PNP8U9vnfMJfcD+6ipfeOEmf/nH/jhseIaMJ+higZYlMp/3ylfYDn14F8kyGE2g63yBIQb3yheQx59BT+9x9ktPMP+hzyA3j7zBoQOZ+uJF1/fAgB6n/MqnRjw9gs//bz/Kg9OMTz9sgYYaxwsnHe9cjXjsibusX3wC8AOUy/jGRIRUxQR8NxGPylmxAPjaClHbCUUiiYdJBZEHRRkHflR0AY9cgIxka2a4Iw28W+RYjQVMTJR9YyDbkXz1W/jWBUlM+GNSGbdzE0z5tr5AF79KIJM/qmLotzUqZqU9D2BXMONNfdhz1KKXUJwyOLQnpMdodvhtMTGcMGTBmkq9O/iQgihPO5ERyzA1eMJc5ZZ7QB34GJFXkpDweHLNFy5BvnhN1Tu3A4wZsGRz4biMGXBo9vrz+8Cd9oIbfmrsC7gmFClOtwWKl5VNOJAZFsepLknFqz+Ng4t7Q8vCLXpFwFJrHF6CuJCCGtf7wBzKtCd/t2rJJaVTy0AyhoFofUVm/STve+x1lrQI8LpZ4ICbbsqbWoJAoUmfxBu8ktZKGhbiuOrGCHCgA/6SfZKXkmMWlH3zKJ7fCGeqAlH9XEtaOoow1RaEMb6IasVSaoVV7ZtMFkdNG8jyCYWm3uAPx4CMpdQUmvSNp3Uof1rxnA+jQi0dSJhCqS/3WvEeMp045joIhaU/VwNNw+d6zS4Tz1UPOxOeNCMMIAKFmVI7x6KzdCiHaco0SSidowkTnnPtyDActx0DDNfdmDtmRSeOzHnI1b4WjMJ+n6rji+uaG1mOKtxIC253NTWOGodVZS9NWDvD775qeMdewnTo+JevOn702QF1Y5hNWqoqIcscR3NYlSmJUZLEO6LfPzdMCs//sM5PSIyAEQ/Rcg6qv//vXza3vslxCce6jG+58Pr4yoF6Tfgj5lzTGSMKntIjBmQc6ITHdI+5DvoEfkVDKS2pGlock4CzBa/lb7GcuiVLt+47cb1fQbhUInkwjtJPWHlVGEzfyZsz5Mydh0TDmxSWWlFIwVqrXkN/6dYkJAFLnPUY4d0k6TGd9593U8cMSXl6mGNR/spsgnMJSaJ873e/hi5DB/B0AnnHZ37n38AcnqMnU0aHt/ie7/ssf/8nvsBzesDpRvmDexY9nbL6vec8z6OpPcMQcK/fQ8/PoPQ+LNgObWp0cYqul+hqha6XyGiEPriN/b0Vpw8PkZtHsDhHF2ceouUc9lOvsPyt59CTlLM/fD932PD5csM/f8lxshbuBWiIRbmnNcvVgMXJAcvSH+vzxvHiT378T63cdBn/ehGJzhC4HjtTjV1fkN2InfPdycNQvAJSRnqh+xsTt9jh3ipRmZ3pwm5hn1zgmvRTkdCRjn4gqSS9X8+Wi5Je2KZYnOwm2uCnI8pWVW8XwtLLzgaT1Fg8qLoeLrTrexGJ3rvQUINcKKZ2i5lJmE7svnd8n0iQTjBBAtcXaqVuInXdQ0ZDwn+qqwuQtSNzwL6ZMzMT7rpjZvhJb0dHHTr/ETbm5VwLxgyw6hiFxD4aHbZYMsko1asCxqlxL4dO2svYxmN4ouf9tszEE76XWvbFWBMUDheBI1Nrg1WvptXRsWdmAIzFS+JGAvyejHvyfE7GuZbkknKgY/Z1yIGOeMmc8oZZUIpPtpUtfGkYknUN3fySNrwuFtywlIYXEz8pOtIxT+vhhTUepZ7HDEhJPXws+LtEVassFKYNXb9W49/HaaALU5eCFKPeS2wUnk2Z+uP7bnfAKhQgtdieg5QgO/uxFVxIMP15K8PzJjbVEt3+Huh9PZpQkNm4dooEC6yspVVlqZaNOu62LYlA6SwDMexJ6jkdYV8yMTxphv17f2AwY0rGk8mAUv0EZURCFfZhnAlL6wuc+NkrtSQC17OMY9fyxdOOX7/V8ldvpDgrZImiDpJE6ayhqhNm447ppMNaobPCwVg5r2BVe3J63STUjelhXePh5RTk7RCXk5DL+JaKf/C3/+HzDliJJ3kf6Gjn4e+xyVMd9tX1UuoeU50StdeznoBXSktJjVH/2LQ4JjIJhMkd52QcU5kxlIIjpv2E5Bp7tEE+Mkr/NrQUUrDSkqmMUZRRSLhUt4nFTXPFj+zVMpFhj7XeEgkTTsTrzKcY3pQVH5xf4ZnHNrzXGdJ0g4hy9WjNeP6AcnEdFpAPl4g4VIVP/R8/wnPf+0dkT94iKY4hc/zHP/45TNJw65X3kH7fPUZf2KBvrtDWQqdQJMj+CDYN7OXQVF5W1ym6XELrePDx7+fqf/RZ1ClydIXbv/cXeeP2lKf+6AStU0gqxGxwJ457L/w1qk2Bcwn/w/91BUdLSceJVNQbSxGgAAKkCHVjeHg8YjbuKOuM1hlOm8sHxtc7fuZDf887pRNVoS7yMnanCoaLTucXuBrh+91pXiwUyjBx2L7PVo70a0X0D+m0w0jW87w8h2ArBJGT0dExkJxOd7fV9K/b+oBsC5A6EKnjNkRuym5BADyyT1E16OL7Rpfr3df0fxsI7FGMIr5mF8a1+/3uMV2wxiDMZcw5hlI3Pd+iw3lOiHgH8BvmCIvjZfsmC10xkREWy0gGfVERf17TsdYNhzL7qvOwxp+rJnTzK1qmMqKiYUTOkg05WS9Dm2Aog0LWbuFXBnnylLSfvJzoOSMZMJSCoRScuXNWbPpj1WnXKwrGYx55FWMZUAbi/JCCDTWZJOzhZYVvugkKHMuGO6biHqt+nx7KmsfclFps/74G4aFZswmGg4/pnIXUzLRgQ0OBV5G6Y5bh+GzPuRE/UUpIgiFtRR7kiON68Otzey3tSjTHn23omARTP5Qgdev4Lj3itpRkGEq6fn1YtH/exeNtcf29NK7+uQ56JTKDeOhxKG4zEoqwHUNNmZAyCmvzdVcyaifMUl+2n3eOKbuiFGGNRlihejgVeB7IIEnAwQ8MDnhYWwoMtSpz498/T4SqczROSayX7FWUDKFGuZpkvNrUzCRlJAm1Olocn7rj+GvP+OnGYnlxgmidsKkTRoOun36oei7IpoU94MF5kPP+B3/zcgLyNonLIuQyvqXCdzMJeFRP3jNAK449HTHSPIydTej/KK1YMjUQYFIL2UIvplqwkopnzeOU1AzJ6XDclEOOWfbqLhOGvbrKbrJV0zHSglo6jpizZMOEwmPGRTlh1T/Y62BL1dJxxez3DwIk72FYEeceC5X4Wa1YNrQcziz/6AWLwfFT7zF8+oU9nnv3OeoSzwlZHVCXM7Jiwx/dEn7yR17hk7/zfv7qO18HJ2AN+TtfRyYdT4w2uJeHmKMl7jbInkIDMhSYjNHTB4jtYLVG75Z0X77Cr/3v/xZfWnS8Zy/lR3/2C6z+2U3Ojq/wi58c8d0z4bf/0Y+RJspTT57y0isH/MH9jgrLAypucJPbcs4r5kFPwm1Dd9GhvCmnPKkHrMorHMx8h3nTelLhYZHw4k9+/Pl3/9NLY6mvR/ydD/5nz3tTTbftBot4RN8jxYhBgmeNDdOLlDRAqKI+XORDeWy8l9MuA0TIyJbQ+9XvuU3MYtLvPSb872JBUkgRhCC2ztHdTrKVylZdC/Vb5DkuXZ+41dpQSE4eJH6954aHg0VuCmwNDw1CGmBlVu0WqhngZ/7fjt3iIW4P0MO79JHfLdVDoCaB72AQBmScsd5CwsK2rNgwCNtca0OHDf93HMo+jbac6oqrMuddyROesI3j3C0RDMYYMkmC6pLflquy18vbxvtq5GuAVyOMhWZGghVLRUunlpKKPZnSiBdZGcmAmoaUlJWWfcF1rmuywB+xOKYyZq0bX9Row9SMMfiEfiB5r5oVpwdRMXAkXjkqOkStdMOzXAfgO9whd2TN6+Yci4chHemYJTXn4gucI51wz6z6NVBKS+vGPGsP+WxymxbHqWxopA1QP8sLycMLRcWaqp88NNoxk1HPcZnKmBF5X5jUYV6VkfbQpXjeo+RuQdbzMwzieSwkDEiocexpQYmHO0UgGPhnQpxEGfVGhD23Qz1xPSo3ZhqeQWJJdCuoEK9Hi7JHzjTx18meplhVxomhtUrpHCNjvDM50DhlZBLWzvbX37bIAqvK+/WIu1XHJElYuI6F88+CEQm18xzA0nn/rh6GFfbwRbtmTsZtrShCuTiTlJvDlLqxfPq2J8V/4ClLWSWoQteF67pN2Jt0iCh5mpEmyh5Q5Jb9MTTdpT/I2yku4ViX8S0VUy2IfAiLo5GOlTQ9RCsWIBbHUiq6gJ1txXlsrZp+yhBNnG7qHqkahkH5ZKJ+FD5lwBFzDpkxYUCx86BIMAzUy/WWUtPSsaH2BUiAgMRkYkDGnBEb3equn+h5/yDbNV/bNQR79LN+0FznC3cMp1Lx3HDMF14fMsjAWoOq4dc/8QHuvPkYn/r007z5laf5i48pv/bb7+IvPHsP7Qyf+cjfAAfaGdxZDlmH1hm6LnB397Avz3EPRtiXg0FgC+7VO7g3NnRfvsL5nWf4rbMVt3WDdUBnOX1wlU//8SHHVNQdfPGh8vJD4bMvHPLx+yvuU/Fls+B3zCvcpuRFc4/7esa5rkOS4bvD3tegpMXy6Qcdv/dqinWCCFyZKK1V8uTy4fH1iJ/+4H/+/HYiEQVY31oJK8NzoHalehO2CXssHIbinZGjOlR0544TwQvwrLcwJoTtJGZXjSjdgVuNxMNgIuY+Df8l/X9b6Ezcv2yn7/aokt7X2oaL3Wbb/7x3le+7/lsOyaMywUlQgfICFlsSe4Rd7k5ZJCSoE4Y9DCv+3GrgSIQi3pOivYnifT3jSbnaeyGZMH3KJGFuPFfizJ2z1oozd05H50nt5FRslZVicyT+n5KyZEOKn7TEZkkhXrkvxXDfnnBfz4jmgvFeabE0tIxl0PMg4r1xHFza92VCox2Vtl41K+xjQ0urnj8SfWRa9VCkMsBac0mpxfJud8AgHMGKjmNZcx74FFfUT6M7LCeyRhBOZE1GQkXD6+aEY7NhQE5Dx5oKPxW3XNEpayrKYH64q3wVIWCC9/naaE2jbf9csdgAL3M9TyZeL4L05oP9MQnFwUoa5pJxkGRYPOncu1L5BlWH4mS7LuN5m2hOohK4HeHZEf6dk6BC/32uCSPNmIa/2Q/Nu0yEVpVMhEQE67wK1jyJ2wknAYKYhSlIE6YUcXpzrA0ntsOqcqItS2u3cDgcC1oa/N/cGKRUztGEq0YI4gEMqHEspA7PSMO5drxU1vyLNytedmtaVerGMCgsmwCzMqKMhx2DokMM5KlDFcajjrpJMAaK7E+W8L6Mb2xcTkIu41sqfvYjP/Xh//Fv/+LzEeVbhy5d7KAspd4pVJIwezBYwAaTv4H6B1clXZBqbBmRUQc8rkNBIWHQT106cUHSMGFK4btlWAaaYUOCEVVO4jSjpWPKkCUbMhJGMmClm35f/MNDA5l1O8LfLUzizxayYZQYihQWtuKzmzXXpeC1dcmXzkbwR99JJpZPnHZ8/0R49faIByvljabmu1cjzB9+L+//d38N7QySWzQoi0jRYq6VuNTi7u0jgxY6Q/fbG8yBwd0aYfY3aFXwmc8+Ts05BuE3F0v+7U8pZTngTum38VPlCocysilvBA32301eY60VKy1ZmJpFwKx32rGi5EW28I1MElDYSxNuTAx3Twydi862inWXD4+vR/zSR//bDwP87If+i+djMgz0fCjYFicOJQ9StlFiNpGkn2LE13Sh+5tgqILqlFXLio0nVcufXIDEiMpa0ZjQJ78Flfpu+1hSmpD0xeIp3hMgAWyvTLS7LxGeFWW9Y0kV92lXlMK/V0z40x7HH6Oj64ufLRTLbz3QT1mcenjn7ueBV8LbhY95WIp/NEcCdCKRE2L6YgQuFlWQcEdPuBZEMxIM4yAb60QZJUX/flasT6LJaKXjKnuc4RP0OLkahh50dAAvqXsSvaDUeKW/FsuTyXXWWvHAnZKQMJVhD08CWGvFWAahOKQvLjUcZz+57tioV5+ay+TCOojH1juNdyH5Nxwx5Tl7FYBPm/ucyLpX97I4FlJxpOMdKd6Wh7Kiw3JPzvvm1T05JyNlSM4wQM1arCd9hzVQ0oRSQHHqiz/wQgIWT5QfUXhNLLX99GbXiNPIRcGHNKzZVE2QKhCG6onfp9ZPP2ostVg6ddig+mRU2OlVMdhZBwZvXBhnWVaURm3glwgaXjMlY000jVRaVRqnXq0KT0o/a8N8T/2VmIgw0ZQ3bcVB2C9fjAhOYUnbFyNdPyGBqSQ06ouQeuf6+dKmYiopGYZNOLr1jrDBQFMqLOfSsK9eqGAlLaJwy1XUtwoaVf7qje31XVYJwwISowyHlpNFRlV77mSaOG4fZxxxGW+XSH7mZ37mB77ZG3EZl/H/Jz7wE9/9m5/85c/8gL/1bTuqGv53ojTiu14L8Y66ZYALDDTrb8L+Zp2RivG4arZKV3kwJXQoVvzNcEBKh7KnA6JKylNuTkbC027Ong4YkPFAloy16PkkBRmJGu6z4EBmGBEmMuqldwHivlh0B0e87bq2WF52a/64O6eUmodmwxuy5p5ZsaDlS+aU+4Hg/f7piI+dnnDXNpRYXrk74FOvDvnO0Zzh0f1wkIwnsmcdrDN0NYQuAWsobz1NYpXy9afplnvIecEf/O4HuL0wvNFVZBgqsXz+U+/m1Qcpr7qSSvyD8jPJLW6Zc75iTkkl5SW9TRuEXh/KkkobHFFCctvxzCUjEcM+Y8SmuMYwzQ11cLddWZ+MXvkPnv3Nb9Q6+3aLH//JH/7Nj/3yv/wBf6SFLnTbjWxnc0oUU45TuiQkk5GQ7ddyNJVbsqHRBsTDs6LnTYRjddigHBXf/5EJAp7AbCTBhAKhCy2HVJIe/lOQkwdyuME7knsn9dC3FoKruJeujZOPXLIgkR14LGJwGvu2uvNzf616s72WR8n5JrwukwwXCpH497FQ6x3SxfSTpAjDBJ/MTsQrVK10w0a28LWN1lRaUwT/Ii8N6/9+TyYepiQ5NS0NHa10FMGvw8PQLHmYymakPbcD4K4esycTxgxYU2MwgVxt8T116SV2DRJM+Pz7NbSkpFR4aNtUxr0bOHjVsYkZUUhGpS2peLNGz4vwUKKFrgNnpaCmZRA8PiJhPoog14GTMmfE++xNntVDwHDfrGnDRKiVoCpFFQjjhlIaXzgHaJeynZYDDMM8pw3F61BzzqQM9ykN5137AlAwqPiVmoovjtZaYXFsaJjKMHB4vKhDEsUORHpTzcj7G5BTaEZBSo6fTgzxRO9ZmoQpgS/SzoxfD504wtDErzlNeqJ9FBqI664Vx0hTmuCUnmHYJ+dQCg6SjLlkjPFFwEGakohQOxfWFmRiuGe9rl2DUqllSdeTyAfhuqjVb2d8hrXhaVxhqbBs8AaI2/Xo+q9rup6krnhodSxkUgwTUhxwJjUtjikZDY4BKQ2OUxoeMwOGhXK6Tmg7w3RsWW9S0lQZDryfSJFZVmVGWQuTv3X5HHm7xOUk5DK+JeM/+Wc/9ydyA/7nD/3j568wZR3IhnmQ5p1oTitbcvhcCwpNiLooG+kopaUKXBCDUGjCng45kZK5DphpzkoaaizXZcChFuRiaNVDvhY6Ya4D7soKRVngJSGf4iolNedaclX2AqFT+87q7gQkwkDiA0VRVmy5LANyHrJkQsFDWfH99nE+k9xnojkfP16wNk2fdD2k5Gk3p6lz6AwkFsksZlqidcatz30vjz37OWTQgBPatiBtRohxrM722cs35JljkhuWTUOrKdd1xG1KltJwUye8IQv2dUhNw0o9Rr2W7oKqUbVDWo2ykHF/AG7qIVfdmHdlQ65NhKYD65SNVYaJXEr1fgOjhwft+FmICOiWH+IulB5byFOcBEaFORGvEJdJcmEi0dL1ExR46wIEvEdHElyprVo69WvbK0N5laioMOVQ1uox+xtqnGqf8AoJIpZOLU400ICl3+4eIrXjVeIe2abmLQqQLZHf9YVGLEAiyCQn8YUUckHdy4TjkOI5LMe67L0n7M61Y9Vy1RwwIuehnl/4jIX6CcahTDmQ6YXjONEBK6k4YNJPaXyH3vTci+GON8eUISPNuSXH/fThCjMecN4fq1hANtLiVKmlwalS4VWrnpJrLNn0U41zt+6P7Z6MWaEUYaLiz1/HVMb9ayptQdhur/pp9g058OpQdBwFCN4TOuHT5i4T9d5LHY6hZoH3YliGQu5xNycTX/Tck/Nwr92ed8VP1eeMaANp3RFlnrdQw10IXmxSJWHKHadS8XfRU8WpV4yKJo5xXXjlOL/KB6Qkup2h7WUJqQh0XgErlum7K88rmh+IeQAAIABJREFUaXloVYphI12/rXF7R5pRhmlOimFEytSkZCIMk9D66mAgfqKxts5PRFSpNRYjXs0qE2EV1K0yDE34rOglUmH7Y+KbBUqB6be7whPUo4rWlKwvPgoMqzCVmWmO4KW2hySs6ChIKDGoQBmLefzkZULKZxYV3yMDNi0kxpPU758lPPNYx/FZTpF5oZbZOPiOcBlvl7gsQi7jz2UYIFXDHkOseN5B1IXPNKGWjonmPMZwq/ABvKorppozkqwn8w1JwzjYq7VESMORjvz4XIQqJFdHkpO5Q1a0vELNTIccMeWeLMg1pRZvLBa7kLELG+FjMTpiN8gSya5J+LfB69IfMOnVbj6T3MehnMqGRAxD9WTbUhrmOuCGKXC24g//zx/iuR/+DRhXkDraO0fsX7mPDBoefPF9vP7mAc88c5/V4oh7dw546h1vUJczmtbwcKPsuwGfSt6g1CPmrqCk4XeS19lQczt0d8sAOTvRrWHVLs/lrfDyqSTsuwFXGTBIhXUDURBr6SwrJwx2ztNlfP2i045UUlJJabT1nVy5CAFqgzeHDaTonAy7c14V7RNyh1Lj6Eg8/Cd4fVRaX0j04/QgfoXosJ4E1lDaTxu6MM1wQWxil1cR3yMmy9EgNHaJo0qXVUvkNvSu7WxNE6MnCURVu23h7Ld3K4ka41GYUWwixALcf8ZFCNoFOWLxZOdI7I/HXMQEI72Emfj7zikrorpWhPnEcxK3cyVVOP4eMuTvHa2HfwauQk7G/XCtXpU5pTTsMUYwzLTgWNZc0zlOlIcseZqrnIh3KfdJ9nZf6h3p3zgNG8uAhVtRSMGxnjOXSb/PuaTkeDneuK8TGZKRcKznzGRMJikbrbmms56AbVH+7+S1PqE/DwawACeyCvfCkiTwPhosp1JypJP+PtuFTnsVyuUWywjLAi95vLtNcVsrbfvz22EZ4qGBseDupx6YnTXs5+5xfe5ybowKEy1I1HNEAN5TjHilrnjnYMDUpDgHSscouJejpl/jmwAp9sVXykqakPwnuHC+fdOspQup921X8Y50SJEIx7VlnBhOOgvWsZ8nVFbZWIfFQ6xmkgZlSV8sZEE+2KE81LpfvbvXxiAc44qt7G/ks9h++7R/XfTsirDqDMOQhA5lH28aeaQDTtnyKh0wCvzLEstvna2osfylbk6eZr061qIUDibCMFHK/+bHLoVN3mZxWYRcxp/L+NmP/FR/s/mHH/ql52N3MSYpR857arwqK4bqB+QJwlMy9g9VhQUt+0E554gcp975dSiGx3RA1Y+gYWK8d+zaOaaSMlDDc/YxXkpOMAiHTDkRz4e4wszDVAIxdJdwOpFRDycBnxQcs2TGqH+I7euoJ+MDfSECfpRtVVmamlFw111Lw+d0wfknJ9RYPvOLP8TA+MTlcAzP3FzzeFozmi6ZjKacncxIU8vDs5zxPY+3VoXjtqMVr1jzktzlaXOFN+QYh7JwXgazkIgZ3yaSMXZ/Fh8kikPV8RRHTDQnE6G2StV5BZZhIiHJUd7/kX/v8gHyDYhdP4uYCEcfDSMJiqOgQHF9QmLDFCCTrE+xLa5fD512NOoJ1bBV3BL0q9ZJEiYGVm0/KSmk8FOQgDT3PwuY9FCceNx+faEoqAPheuXKiz4dOx1bnzwG6U4Et8NTiQ2HrfrVW4fv2m//bleiN7IRInQt/j76lkSIWIQgJRgSMewxxolyyz3gitlnQEYkvyeYXu52EtD/Xo2p7bc3Cj6Ab3aUgVRusT15enssvMrWGev+mFbasJQCRamkZaM1iRjuyYLHdZ9KWhr1pPN14H5stCYXvyfRfHWjNUMZUOE9QCZS8KY+7E0dYyHhcFyVPfaDq/bTetgrSEV+x5msecYd8f8kr/upFyWC4TU5JiNhzKDfL2/66CFYt8wZirIQz3c5Y90fixkjyuDkccZ6a7YYpjtWLTMzZqWb0HQqvFEjCZU2vumDP06lVgEuuC2y4nmPBW1KQkHGTIv+Hj4Ma3hIQpHCu82AxMDa2UD6lnC9CFaiFK43XKzET4q8KaXBhfMelbI6HDd1xENqrpiMjSbkRjhvXL+ip4khE0EVMoEkNSTWF/Ib9VCqTpVUomeNIxPDUP00sQrPoK2IhJfsjZK7gpcdrrCMwzrfk5RUhIXrYAd+5cIE5bsmA353tSLB8wVbVUr8hGsVns13tOK+WfOYm3AqFRkJv18vuPdwxZGOeeWlMe8Yw+nasDe75BS+HeOyCLmMb4uYu4KlaXjCTbgvG8ZkPAiyjcOgj+5QhmKo8MTRA3ISIBXBISQCIwwOvOQgDXNy5iZl5SxVeFhEhas3zTlxgF/RMAkQghM8TCu6H0elm047FqzYl2mPx+7C2D4mVhkJr8sJI3LmOmIhZUimCqZaMNGcO2YJ0MPPjqVkzw34kjnjqhvxYtcyIWOAYZTlPDwZslx9B6NBR9sa7jwssE5IjPLy6zOeuL7hbG1wdAw06NCHDmEbZFnjQzd6LsDFzjZc7Ph6KIhFAsQmI/HnQL151b2N79B9oSk5IOevfPTS2fYbEbtSmxDkZ0NSHpN4y9YfYzd2YUa7fhyRy2DDNCAm9oavLlTjNsSCxarr3zdK8MbPnsuEc10zCCaC0TsC6N3ZIcj6hvfMwnvlZFhxvarXhUmdXty3WIA8emyie/xuYeSPj39tLCj8zy7CzhQPSWrDPkVFPa88ZujUciYexjQMSf5UhkGjyfX3mJhk+knqdh/WeKWmPEyNAGr1RnpJgCUdyrQvxM50zUgKVrohDXyQXNK+QNpjzEAyKlpWuuFu4OeMZdB/jefWnyt/X2vCNGwoBV7S3G/De3i8n5zlJLQ4hqQ9+d6hvG5OWLJhzIApQ/Z1yJNuTobh86wZUoSuuT8OV3RGKxHek/bFzYCcEt/UaemY6ZBaOlb4oiIWXnWAgbWh2B3JAOssYxn6JpH6htGKspcwLrVizoRKG+pQsLRhYhsnILFB5FByUooAw/JwqZycpJ8ACL4QyBMoW0LfX31Cr4ZGLFOXY0Wp6KikI1P//rUoE80wIv3UwyCU0pEGUvqbruKqFHQOJpkh6ZTShgln4rlT1kFpHaPEkBmDbb0CVmTEgOeKrHUrdx1hVvEqaHEMSHpp4ajwFcn4Bybz6lvqeSO+sPYcEYAVHftj5QfzCSelb4K9VteMSDmRmqlmrNTL/bZY9sk5DZ/gRL05sTT8ASV3VlO+K5vwtMK9n/7V56/90r9z+Sx5G8VlEXIZf+7j5z7y0x/+5Q/+r88/6aYYhKnmjEmZqzfoEmBkQidUhAwPy0jZCpB4nZ3Q4XGWYfBAyEIn55aU/UTlc/IQhEA89921rYzigE46zrX0n7dzYz80e6R475GB+A7zfT0jJWEsRZ9wdHSc01GKV2t5So94TR7Sypg35KRPmjo6FpJzLbiuz13RJx0LGjIKbpeWQZYgG0O+ThkNHJ0VikypW2EycHzxtSFvlpZ3jnNeLD1sY0PNG3KCqrLWrckYXJRUjUfwIo/AR3TMRuA1OeY6E1oc19uUoyKhscqR5Kx2eAOX8fWN3fOzqzgVQ/Bk9NjB7kIKDFtsfVzPqaRUQZY6Ckhspw9b0ncsFvoiR62HWrHlmkS41G6c6zp8nofURPx9LECikV5clwah1Y5M0t4Y1EhGESBdlTbU2nCxCIsO624nxboYu6aNu5LAkQ/TadfzRmJBF4nK6HYqk4WpU1QOU7yq0YFMaeh6daouTDIMnt+xDkVIGfhvFtfD6KJk7lSG3vcFbzzoidGezNzhOJQpIwpOxLuP++KxZSD+uNylISXhqsxppOVcS+Yy5qpOeVNO+6Q6eoTEwic2U9ZaMZEhLZYVFUtRlrrxghRhHcV1dYUZt/SYJkj2xonOPTnnNMl6PkXkXVgcUwakCMtwvmNx0uDCBMiT7RXHuXhfliEFp7r05oda9zC9VjuG4l3j4zEuyNkzU5bOS4tX2mDCeT9xC6ZmDLpzTwsRC9EIMUp31lAUJsk0dv/9GpkU3h/ppI2Tbr8/fgqtdNJhxZHo9n0jnCmu3RRDott/b+iwohSacF9rxgwhNNYKE9akeA5eZqRvttVOmSaG0tKX9Q7PA9mdOA7YiqwUGGocOdGPxIV7gD8eNZ53cs/V/OWDEe9k1L/PF087ltqxouP373W0OP7NqzlvnMG1NOeLnZ+4l4HM/tB4nmZClBg3vG7OONART7gpd4LZ76/b+1y5f41BBid/91efP/jFy0Lk7RKXRchlfFvET3z0P/zwJz74secBjiiw4aaowNRsuRhntmNqkr7gqNRPRaLBmhFhYjyxrVXloWu9LKEazqTmxJTcdDNEYZ+C+7LhSZ3zcnKCxbIRn0zNZYLFUmlDq20wZPMp2TqYl2WkFOIx9Me67LfRj6Zt7xT8ppx6NZ0At/Dk14THOOABnsS6Fi+daEkoNGFOzkAShonhC2ctjw8yKuvVXpwqlVPGqXBeGRaNIwE+t96Q7UBVNlr327obB2aOxfYQrUchWLlkTGTIs3qdieacSImoxzcfakFihMYqy86RIvzIR//W5QPjzzh+5kN/73mrHsNeSNF31XcLjpjMgU+YWmxINbzKju9q+m75FlITNeouFhPVDpY7lpRF6LSnpJS66TkknXY9ryEmdblkqDo/advhBkX4YrVTqEYI0kYrEkn8NECjjC99RztGq1tJUNhOWiTsU0T5Pzr92f1ZTKDzXSibcuF9DeKnLFFpSyOHwzGQrC9EwpuTkjLcUdHyswUPJ9r0PXLFqZchjgVITsZMRj1/LAnvecisV4Dy58RPpqqQ5i6p+v1pQyNkrRUzGTMkZ0NDgncmR2CqQ+7LkgEZLZZTXTKREQmGtbbsyYRCcg6YYBBWVGyoueuOuWGOgumjh2E5XCiJhPtyhsXxHdzkwI14zZzgUM5Yk5L2U45YiEX1r3uyJc8DgZuS9PfSGPF+tAnTkRN3zkAKrDpa7UgkCf4pBamkZHjjxUjUz0mCGEJOoobEbDkau5ygCJuL537kvddxqJeRD8RyK0oa1opBqFrv8v2YJtxb+/McSedF2IYWxQWzwjI8UxJNqSTysLxpYEFKoQmltL00PeqhrrZ1JLL1Bll1yjAxJPhCpLLKMPX34qvDhDsbS6OOUm2vdDUTP71ORJiEIqqOU3K2X6M0cIQ9r9Qyl4wvnnbU6ijEcEcraix/82jGJx5aFrTcMAVFpjw+9+f09x82jDRjFMq5MnidvBqgzk/omJd5yNNuxn3ZkGG4bZZcdxM+fVZxIy0YZ8IBl/F2icsi5DK+beKvf/THP/wbH/zY82m46VZ0XDEFiic/z0zC2Hh317EkGPFk6Eq9Uo1XdKIvRjIR5qRMSVlqx1xzJjZjTs4wQAwyNZyYTU9ELKmDi3Td8z5ETI8zNqFDuaFmuOMfsMFjkFOCxCWGDtt3iA0ezmXFJyFdgDhdkRlrqXjMTVBhxy3XcaINi1aYS8bLm5oSyyBs94SUgREwQqfKSXC7rYNyTJTnfCuYylLXHMiMxc6x38XIg0+qDp3HtD+m0x4+kGBorH/dsWvJv0b3+TL+1ePnP/QLzyf4RIugqRsLkJg0xYQ9fg/0yVVM4COPQnWr7hY9IOK6yMKUIXppdHjFtFTSXsAgJvP9RCEUwrE1YLGIxsmC9wwREdJwTb2VOSD466rVjt3C6lEzwTg1fCvCOWwnIG8FP9vddvCF0m5Eg8K4b1FrKCG58Lpdda42XO8DyfukNrqZR9hXhPdETkMmhpVuPOkfLypQeSFigCBV68/3gNzDpAInxJ8rD3vbUO5MF5Q9xqj43y9YU2vLmopMvIHhPiMOdBLEL0bsyZhCPbToSKacsKKm4S6nVNqwJ5PeP6TFF6iJJFyVPRTlTNeew6N+Ld50M15NzihDsZSQUGqFStEXGzGpj+dwo3VfXFyT/X7SEwsuv//BUFA1wMQSaq2pwzQtChJspO55SLtCC200thQPudpoTaddX8wr2sPPds0IoyHhhJxUzYVCGGCEb24lxntczIfA2nMpJpqxlKa/zgqNPlJKK35FV9KFyYovvCz+fr8JhYkVJVFfBNxyFUMSrqc5lfMTxDddxbvMiKiI5fDPuf2BcKf00wvwz8A0bPtKLYMwE/JeNp43MtIkyPR68niEZVmUr5gFN90U1YQDk7FS5Z76tff+bMYfH1t+8HDMuobre5ZFaahb2Bt7+foOx4qWE7NhrDmHFCyoKUh5SRbs64j7suFBMKYEeFJG3NeaYSqcNY4n3/JqvoxvRlwWIZfxbRU/8NEf/zDAJz74secd8JJb8t5kRody7mw/3vaOzn5Ufd61jE188EfDKV+MJCKIeglD8J2hTUjKXjHnPfEQfJKxDg/I3QlCglfEqbUOxF4vwdtox0SGW4O/QB6F4NWgW0ItbI3W4vvcllM6Op7QI15LzninPcAg3DZLMhJSNRzpkFSFN2TFkQ57Z16HctJaJknCsWvpws9el5Oex9KoV/qqtekLEcX5h7gI+2bKiTvvty8maV5pyHFsNlx1I54xI8ap4fWmZiYpi85yrB5UU3+N5O8y/tXi5z/0C8+LGFBHRsrAbOEjDu3XkOwkXfF3uwUIXORO+KQnFCLiVX9EDCZcC1YdhGTIy+sGXwCN6RL91x4GJhH25ad7Ha4vQJId+Meu2lSEWcWwBAjUjsoUj2x3fI9HieexuPhaBUiMWAyIyoXrcbewiZ+hKK12wbfCK1NFgr3nrHgFsRHeBLCmI/pTjMhDF9yrMnm+Q8aKTS8Rm2CotaGiuVBQgk8SK1oyEupwRcdjvaFlIsO+g5+GyZdV15vHAQzEw5/WuuFL3OmT7WGAR5XSMtcBRgXDlIQZFuVNOeaBO+WGOWKpG29cKAkDyRiSs6dDcsm4p6d0WL6Dx/lccocWb/qXkLAOLulbkY6LMLiGmpbOu7P3crkmlFmuXx81HQtdMZEhpVakpEGWuaOQnHUQMRDVfl3vrst4jHoVMxISSbwnioyIPJoWyPCNlmEoPKZBRCDKM8O2MQRwYLaFrBF4Jh1SOcerrmSuBZtQWDrZTlvawKmI0YhX0upQFqb26yQoaCEGG0wDR6S0AX61do4NHXe6hqMkY2E7nhjkFAlsOihd5ABelAqeSNKro0VJ6TjZcD3fKvw+HMcDN+SWWXLTTcmdIQ3TkRTDg9Y3uz55bJmblMnAsKphnMMLD5Tg1chNRlTh/lTiZfVRuGMWPGcf44Fs6MTxtN3jlllSq2OfnPWlzvvbLi6LkMv4toy/HoqRX/rQP3k+/myXtxBVSXIXTcj8g+LM+g7i3KScaMdSW9bS0ojHyF7VIV8xZ32HtRbLc/YqL5oTNtKx7DX9Z0SiYpyKpJL20ASrllTSHvcOAWeudmt0Jl/tYxATyTrAvBzKK+JVhu4mp0BwJyflCQ5YkbKi5fPmFh+wTzEL+v0Njkodd7uaDC+tWAWX5Zqm7wbuYvV98pcwNWMc2vNewCd7nmRb8ETwq41Cog9cy93Gcc0USPg3wONmwNJd8kH+rOLnP/QL3hFdL3b+d6FYuxCsVFIvV8uWt7ELOwHf7d/11HAohDXq5WB9SZsHuJF/D9cLG8QE3OJ6h/ZEDNGZ3P9ux4tEEuYyYUPNOCgubXSr+gT0ExdP8r7o5t4Fz4mvLji2ULRtobX1iXirQiQqXvm/2UqyAn2DoNLGT0HUouJfH4npuwR+g/T8iDEDopSul5BtGVP0KnhrKpqQMHvBi2G/DxkJa/Fk6d3ze6pLJgy9GIYkF6Y9rW6T9AjzFAQVfz5n4vWMDnTCiooRBQsp2dcxA/Xn6WGQxfVKWn5GOsYbL57pkvfpU7xuTrinp3wnT3CfJQY/wWmxnErBiJz3cpNT2XCP836bUjyHp6PjSOb9vcUfs5QmEKRL3fCEudpPeSB40eB6xauVbkglodGWZRA9cATFsTB9Ag8JjAX6Lj9pO9nyn5dK2q+vLMALu1CQKsq5lD0sS0SotaMIEMaZDtiIVxx8BxOuD1IOx4p10NnIpXNkIjybjCmd477WWJShQh3W+1jHrAMkq5SWTJN++jHUlA5DKw6jnn/SYMlJWNCwcA2pi6Ry4T4bjsg4SH1qWFuorZ9KWiAX76/SqrKXpNyzjff40G3zqiBhpU2AjvlGkgmFRpTnbbF0OO4Fg915eO6cBE6PQXgmKzgr4cpUebgSrg0NtpwiQKPKTR1zW0oGJPxwcp21ddRqeSoZ8rgO+LRT7ps1T7gZZ7RkCIkVDtKLk8jL+ObGZRFyGd/W8dMf+TsX+Aa/8cGPPR/H0R3KVAwL7VhYf1OfGi8VeMd599ZaLHtaUGMZBefZDuW73CGfMw8AP05/p9vnS+akh12d6ZpcUkbkjMjJJOGBOyUJePGto/IjOHS52N2M8SgkarcbvNGKTNLeNCzyT96QE4Z6DfDJx5htF261gx1vw+f5zmxzAVO/y/eIn7l0a0rxRNSzwGVxKI16Sc+N1BRhnzOTMHZenSwR4Y6tmUvGjUHKeeM4SC5vUX9WEScTvuvbetx/+N2WVK1fYy35cxuhTBcUsIK533Ya5iEtu+Wjw3MX4hQjQpWAfpJmwpqPEJZdFa74Na6xqIykYVISoUfAhe+tbuVSd49D5Gu8lezuo6/PSGhwPf/F/53t/71bwMUmgchFbxAjX534DCTvoUdjCiY64ERWVDSMwv4BocvfsabqidhxP8cMKHa6/ks2/fZFxS2LYyJDEjw/Jv6957F0RAWpKUOOWVLEiYvGos4XIDkJFS37OmKfMaeyJiPhSCeMKDjQEV+Rhzyhe7whJ34fyTiQGadsuKl7vJMrvCmnvWHrfT2jpeN97gmcKF+W+72vi0NZ6KqXvz2QWT9NAj/VmDNmod6DJMKy/HSgvVDIxWlcKgkn7vyCJ1MZ5IUnsiVID4wvBJvQyNm9JmIhCVyYFlq1bLBMzRirri9GLB1L8cVbJPAX+EIhvvdDbRg1hmFqyMMtr+lgLzc4BatQOGGmI47bjhMaZmQ95ClTb+TnxVEMtVoa6ajFUtEywE9GWvHnvsb2E5j4vPt/2XuzWEuz677vt/Y3numOVV1DD2qyTdKUKEqKI8kZHNMODASWDMlN2pngyICBAEYegvglAYyk3E5i5MkvyZODBEYEWzQttS0YCQwYnuPEmRxb0ERaFMlmd1d3dQ13ONM37ZWHtfc+371VzaYsqtWKzmo07q1zz/BN55z13+s/xHpj2PA91QyAba9kAvPMvvcuh4FMhJmzicqUjAYfwgttfxoGxvYOQ3rP7F7jSCco8I4z3WDh3ZVj/OliRu7golUOe2FSwKxSyjyjH2BaKZfbnJfaiv+7O+ez04qyyfih9gaN99yuc/4AN9n2ynu+53aZ8422ZVCb/Ozro1P7b/h97WtUka41rr/96l+7V4tLXulv6YYDLA8h04LnXc3ae5zAWgd+yN+iw/NJf8KldOQIN6Rk7Q9Qp7wtjzlmwSMu6PEsmPA2jymkSKBjzMkf1/UGMd4Wa0yRSSuJkjOXCVu6pONoMCegbzoTnKoqS+moNUvPdyHWHIpilr6a42W3Ij4GHteFzYMOXOrqqe2OK+INLWjJr8gDFpRMmbLytpp2lNuI/7Dc6UP29euvkiIkUe8mA7Cz1DX3oO7KeWwDZacgS837FSASVvxTtkZwzWm0tVVmjVMMAymqFgIXww9Vnr7Ko/hccEl7tKNhORra5I4ElsXQa5/u240aw1jjfYqNYy55AiPjGkb7HB87bv5h14he1zpF8XmcnownDuP3btQIZGI/FxoJTZJWzk1D1oZ3mi1OlOEr2wetRk6WLGjrAMZ2Ookh7Xd03Wqw47TVNtEjPZ5SypRqf4ND7utjCsmYUHGqCxzwnlyS4Xgkq0ANNWrYfTk31yvZUpLzq/Ke5SKxTEBpQ8NtXeBFeUGPqaXkRKc8hzkW9uJ5LGtOmPNNHpLhuNSlEcxE6NSz1A2HMmPFlolUXGrPI72gxZLbI53rhhzyRC85kQOiaP3M27aju+u+koqBYaS/set66iYJUM5lykX4HBtPvp41SYwTw7VuiZbrKpqS0ZdsWVBRqOnuonC7CMSyjfdsese0is9tGUmT0gBJ0wurzj4fp77GA7UT1oPnbVWcCitgLT2T0SQxbq8X0wIdak2kHXuURgZuaM0jDIx1eL7SbPhUPUkN+6yAx42ndi5NRRpVXLj8OzwlLoB1wZEF692dBjFqQt5w5+FIT5iqTUDW0jNVW4zaMlBmwrJTZoUJ9YvM3LvWrdkXX24NmExL4XddHtIOdvsLs4zMgVfoPeROqPOCSQ55KziBfC8z/EjVHoTsa18fUP/m63/4tX/46s/eU8y3PY6OUyqsetYMVGq2htvw8T8lZ64m4VsGseNdv+COLBCFj8sJoiYYfEEP+EZ2xtf0HW7JMT0D7/rHRom5ttp6vTx6BXwYfcxCsmIg2Pf5F1mJrcz+PG+yDvaY7+oTCnK+lxdDYrA51CzFmtGvuAec6YpaSj7rn+d36yf4JXefS12x1Z1Q0iEcuilP/GX6Mh43fpHGoOo5dHM+obfJ1VFqxmlYjz8P1K5Zbk4qTuCVL/7BvTPWd6iaYIQAJDHu2m+YOqPztHTk5DTaUErNUte7pjlaywYgIiPw8KwJXCVlmnCMb0+8et01x0CgxexoYJHjPtZUxcdPKS1EEW/heOEaj9O28XU33r6FzHAiPPGXidoY6/r0Y/xeG+tdnkXjGu/XeHujY9ZusrMzXygpRiBmJ/SNVrLx/vbYLDldXX1fZeRBE1AFO+KaMoGQipyOgRXbZPubj57bhO7Ci3IzTRhe1BusxVz3bnOMRzmTVZoyxH2+Mv3Bp+eMGo4L1pzr0nRH4vmYv8EDt6Sjp6KgoeNX5T022oRtuME7+sS0ZDK/cmyNLjYwlzkdA50OSWju8UkQDgbu7vtHDHjOuOSWHLMcWYhXUtL4FZW4VO3XAAAgAElEQVSbstEtlZRskj3vJoVixor24wAzmeJwrHWTPvMmUrPRLRFkDwG85JInZ7dMHCU5pzpjRctCKrZ0TLWkFzMveSQNjXoebzImm4xXpiVlRvocLHP7P3dC5oTM2e0i0HSO+WbGoMrXB5hpwYqOOgCRgspcsYCZlpzLlkOt6fHcpOYBWx7K1rI36BI4agebGvRqEHiaOSa5sOoVVcJUJFhYq7LBgghXYSISTU4upGWieaJr3fQzVMy9bK4F2zARupQ2aImE+03H7bJg1ZnuUhVWrTAtoQ+Aw14Xbs6EIoNVA4uJ0dkGLxRqVseLykDMy5PSnm8PQj5StT8d+9rXt1G/5/Ufey0JZrn6xvF6tRErsOCl7aghMc/6geeYIAq3mfCSzniRGR9nzssy5/uHW9yQQ3Icp7rgljtJSdOflOfptH8qSC1+YY5fP6X1UvAyz3FHTphoziv+iFJ3qcydGhUjl4z7YqtTa+lZSkfHwK+497hkY1ML4CvuAW+4x3yvv8uhzK2ZkoK5TLntTvkh/7HUqEVwMt6uQ1kg4vgB/xJzLVloyUSt+VyxC75a9fZF0u01hL9h5VFyKZ5q1je6DXxtcy/qtccHAP1+E7fx3yI4yUKyOtgKspOMXOzaq6SkkMImG2Lp4DvRr0/XUBfbljAFsefSELZnQuWoFwBSUz3WtniUG+6Q2+6UUgrO/TIBHB39BzstSJwimAHDbjIp16Y2192NItCOxyJOM+Jj43sh5pR43YU27sDILll7nAUypg9NwyJIBD21FimMbxJsYGvK0AiWCXjMmaB4Vrq16QeFpbJj2Sql5jRieR0TKupA7bFsj016zWdNYsdgs6KkxzOXKROpWLHll9zbnLHiCUuWbAMJzEDYRCq+qQ85CPa+A55zXY3oZEOaMKyDOH2npTFAl5OnaVUpBXXQGp3pKpl5TCT6OAVdXFzg0ZFJgnoqKVGUAzdL1D5B6LRjE6YcM5lQBZeuidTpmp6GFPtaSkrJKSXngCnTIEgvgykAQC92rTQy0DFwLg0PZMMFHV9eN9xfe95ZD2w72LQGOmaV0ZLeWSp5prx1oUwr5bgW7s4dv6OYclsqZhTM1SYwteYUalS6dVhc8hg164GYJkPUpp6D2PWYI5SZ2UhX2e463w7KdvBsvc3sV36gUfs9vzL3M/rVgGWTZEjSrXTiE1iP4Mi0KxpAb8ONImfVewZVcgfzWikclLmSZ9FBzF6nzBURAxdBG0/mlHntmRS7v09yuLsQjqf76fpHqfaTkH3t69usz73+46/9g1d/9t743+9337/36l+/91X3hE/449CMCKfUnNEyiFn+1mIf2gNKJcIhBT88vJRWot7I3iOTjNtyyoDn2C1MyKk7vUb88i/CSvJYJAz2pXfDT3mOmgbPXZ2ivMRX3Ls89maiG9OZX+KYDT1fcw95xd/giRo/XdUsLtvwJfKP5QwNn/YTKv5lfZljX3FDKn63fJJ/pL/MVGrWarSJkoKGln9FX+EX3TvMfcEhBevwRWSrzLbKe0NK6kwoMxun7+s7U3/i83/6XnTTibQ4gC5MKypK0/uEFiXy5OP1FXn6DoFoPRuoWG3IuYkJ3VEjEm1pJdCLzOGK5OajuhPBW59jgunovvasqsOqd2zOSwoGHSikoMf0GL1aOyM4DmWeRO47N6iny7YjuPe4A6Lr0UN/loDCGPDHqUQ8RuPpxLg6hpRTAlGUP6RV8rHIH2ChRrNppU8AJlKA7H3UU1NQBiesIujQTnRGJ553eILH7HXBqHdTSqqQwhF1Hg7HTQ5YY7bgU615Vy5Mn0bOjDrZcUc62u44cWW/7XVMrB31Fxs144ANu0nVSrdkOM4xYFCJ0b9e0hPuyzkXumYRAg0FoQvWuPE4x6lFToYZJ189ky0dW79NgvJGm5SLEutSl4FuZYGDU6nTJNCJ2T0PIWBz6decuiMe+TOUXZr3ocyJ7yUXUr/j8SnDPlXYe8Ws2XO2tHRixgIx4bwLizuHWof3hl2Dj2XLOY71YKTDxxe2n89tCm5Ng8DdD3ztiQnE3zizkMsXDxzHlV2DlZ/QeM/XdMVWeioyVgksOi7FQOsNP6WRgUHMpjdT4TPFgkKsoT+eCBeNhRi+23XMXUaju2tgwxBoVp6bruKx31nGFwgdO5AabYUrza58d73rlglkzyk40IJmMOrzYWH0qk0rbHuYhqkGgIgBkqazpPdJqVSFkjklmuBVhVIEzmVdKG1vQbztn/xf7lWF53KTMf/v98GFv5klf+fv/J0/85u9Efva1/9f62+++jP3sjAZ6fBUuLRqFN1CIm/2MnztHoYvtDfF6DCVZvxq9oQ39L3UnPVhijGVCS/KDRo6HupFsv4tpaCi5NPcJcPx/XrCKqxoZwj/c/bPec9bw5JJxrEsmFFxwZq1NhzJnN/hn+OX3H0e+bO0cviD+krShUzVVtJf0hlvyIqPMee+bsIkxQDP190Fh1pzIQ13/ZwnsmWhZVDUkEb0d2TC1FlbcXvm6D3c/p/2Xw6/3vqJz/+n92IuR5xOwSiwTbccukUCti1daph3Kd+7bI7r6eCZGK1LiGF5QwKosGvYoyD96sTOX3k+4Ar4GE9AYh3JAaWYhW0Uaq/UUjHA9CRjatU4vM6e36fbx/c7dPMkyI7he/H+PT1PvBksxNBGOz47m9RISxprn2LehEeZyYQtLX1wcqqkpBJbwa8puKUHrKVjrkGQLVvOWHGTAxTlnHU6nrHRj/8+DoDjPc7pdOCunLAOzX+c6MRtXLIhZsTc5pgm2N9uaKgpmGnNSrZG61LHI7m8sr/Xa7zf47T4eHsEI/F4RzC70m06L6XkbNXcr6ZiDfk6UAfHQONZU5gxGOlH01SAlV+nnJpYpRQpn2YqExptqaVipetr6/j2GbtwMxrdyaxP3AHLEKwZ9+lZuTm57CYztcRs+t3xyXAsmJiVdXjl8ZrLodYUOAo1Z6tCd2nrBlj8lWuhIuOQglt5ya2ZaUkGDw+3nsvBc64db8oSQWilZ0PPnJJtmCJOtWQawOJEc04o+fikInfQDvCg6RnCvl5qT4FwSY8AFTEPx8xcutFZ8ihLaTnWQLmVHSV0riUZwkPZpL8XOO66moMg3KjCglQmu+nHxRYOgqNGHgDG0XxAwvdS5hRVYfD2f1UMnC8L5tOe5Tonc5FiarStaT2wbRx1FRzi/vwf2n/vfIi1n4Tsa1+/gfVvvf75Kx9o//DVn703biDiV2aHZ0FhAj5xtOpZaJFCDxXljpywpeWh7mIAo73kC/6Yqav4sn4TkR0HXRCmGla9USpMDPtdesoLcswv8hZmC1rxhGVarTzTJf9Mtkyo0gok2lOT86Kf2Zq17KhoZv3oWUqXviw7NHGP51pSICy0ZBo+dlb01Nio/nZlt01yox3sAch3pgSXxM+OncXuuFkbgkWrI2NQW6kvA52loyfTzDQYagnLNq0YENyVFX5/bboxLlVlEAMoTqL5QZYa20ipGt//WRWnH5FSstQNmTh6HSgpkiXv2FJ47MoTG8frGpA8tIljq954HRdUPMFASHQYi88dm/vx85nRrq1sH7oFHuXSr9KEp5IqZXpkOG7pgdEidQcuFM+COmkfFky4ZLObfAZB+hkrlgTQgLlfPcQ0L0c641zWHOmMS9kEoBCaO8pAv+rCa00QhLW0TLViSsHX5VFq/GMo5HUg4sKc6yoI2x2HDTZFiwF+GqYkRQikBMJnTrBe1oaeAQtj7RPAMHBqNszjsMd8BGxdAoVhUiIZheTEYNdeexptmcqElV+zZoPgWOn6Sl7N7vnkCgBRPJd+lSZ5SQMSKGDpGgt0QScCGumpXTgmOQPQ48locAGcm1NdmXQ955huQ8Xer534tGAz4OnE0wSRuWKOVGt6fA+Pzx1HecaLB467uWPdOg7bjLb3PJINteZspGdJy4ScpbQUDPSaUaizdHYtGLx9HmcCQ2Pbcaa74Mi4iAY74XnHQI6jCRMSjzLVIm27R6k1T9OPWjOOtUaApXQ8r7N0hR2UQjOQPqlWwWwuCs+dmJVxN8DNIrh+tY48V5rGMQzCpLJzenTQ0ffCyWHLpsloW0c/SAIkZbmDgMWf+hv39kDkw6s9CNnXvj7E+j2v/1gKS3SQpiBgU4FLegt7wrzb77iaX9YL3tJH6cu51V0gW6cdg3immtNpxURqVrph7ub0as41L+oxb+uWY0oEeE8b7uqcB7LmX+JlvuzewSGcskBEuPSrYDHaJJ99gJlMGNR86x+p2R0+JxVvsEbUQqNiM5bjOJPGpjIa49aECnMVupvVfGXoiHQ0sJH/ppc9Des7VH/i83/6HnClcfNh8hYbxqgr8DowyFVdBwS3Jzxjk7Lrjle7pm13pwhaYsVpSqRDZam5HAUHjiYf4wnI+PadHW9BTckFa7wOiaIDT4vMx/Wsv0Utxxg85QEg9Nduj6vcMTvCh4Z0bNAQKWpxH6dUPOaCSPGyTAprVHNc0oE4fDovAIc6IQvUmYEhAZE8PG5gYBa0BnF1fakWXAgwIaelTInaAEfMQuLGwEqMcic4nrCipmBKRYbwQC7DUSGdJ9vH3TEbT5jiNlzNPnFE2tok6CfAaFWtmq5jLhNzNdOOTfi8iZkvMe/Fzkeezv/1cxenIS4ckzhNi7qQCCyj4UHcF6dAsKydu1mahhzIlCf+jAh6ZHQt7DJjnOl8wrQrucSFfe/pcSrkYp93MdhQRcnJMc1gd2WydsZqd1wFLtlyrLsMmMeyMYAQtv9Aq0SrsmPjktHJaV/x9uOQZI7wfUcVsppy1lt4YaRQ9Vh+SFwsAgtPLHFkDladAZGJODZqC2WXAVBVODZErzwDIjku2PZeDek1vWFHHTSJxzqhDvSsi7APL+qcd2XDMQveaTuO65KuVSIMqTLLLalzc8k6nNi0p8xhs8mYTAaKTNlsMorCk2dBoF4oTevYNhlNq/SDUFcDbesS+Lju2lv8qb9xD/ZTkQ+j9iBkX/v6Tajf/ww9yd979a/f82GcPYQvo0vfM6fg2M2pKfmavpOag1N3xCf1FsdDzQ0q8DDPJsxlwl09slAq3SVLbxh4IEbXuqETXtA5K3o+rubd/0gvOJAZIo4hNJrb4F4zlTo1WhZ26FjS8HX6lGL7RMbu8Mb9nYQv3KgBUZTPzqa0A3zGLWi9MsuiIBjwyst/ee+I9eut//AL/8W9UorQAFmuSxF0GzEYsMf0GhpW6EOk2mg+t6vUpKu5V8X1bsGBRuqSuQK5UZ8fheiNtmECQhI7x+eNdZ16deX1VZm5KQ6hpOBC10HQbg1oE1ZoC3n6K+1Zk5nx9gGJLjOwmyBen3CMpwDjxOsIHOL7MmOn9VjplnNdWkJ6cH+LCfDxvbSmCy5JGTmSTCJqMUpUbEQNkGxo6ckpEUJwXGjUjzD72oaWnJy35IyoSwBogzOVCY+tYV7ohAOt2LiGDS1bOirJ2Sar5qualzgNuQ5A4jEZ/zRaWp7creIEqwhTggjexoGnY5MA285dgGsEmdcnHmNKlj3Orou4rXHyFyeB/TMsnKMGSvEsdZ2og6o2uTKql0vnz57bzBcEozr60fMqwlaUTF2aell+jk1FMnFhKuIZUiNvf+/FXqsK1/JCay5ly1JaNsGAwDQmPQU5G0zDUpJzmdVUmrEMTog3/ZRP5TP6AS77IfzNU4XXXtMF22BHL56SOEVXLlrP8wvH+XYXVNji7bNcB9YMCAY+eiyBfU3PGtKUZC0dUy2SIL6VgXPZcqQTHMIGo3m9pAtOXMEtKXk89NzMC95d2+JHqbAobCpSZqbzyDObYpS50ar6QVitc7JMcZnSD+FcOQMlbe8YPKgKVeHpOgMgVWm/v19FMAKwmLc8/rOf3383fYdrD0L2ta+PSF0mNxyPABMsCGpDzw8OL3DfrZjzEr8g30QQbrDg1E84osRjoj6HcEsPyHHc9nbLlDyNxyvN6GQ3aZmRc6EZS9nQaccFq5EmwD6c5zLlM7zAW3LGx3VOFrbtoWjiKm+kT6tfcYUr0lnuugonwqnbfdyctQOVcxyV9uXw8l/e06++k/UXfvrPvgbwJ7/wZ+5Ft5+cnFzMgrcT6NREyXHK0Y8MDwgrwLBb9Y7WsXYfF+4/btKjsLxPQERErlCtotgargKQD6q5m9EF4btpLGpzhpKStW4TH/+D6lm6ArCV63xkbxunRQUZ9/2jsL1KJnnKIdlpIK7SvDrtEy2s0dbup08L7eM0KUOotWRLDwg1BZdsKdSa/QZL1a4155YecF/OOVHTgcTmzoBIzl054W19DEIShW/puK1HXMqGSzYsmCTB9hD0Bpe6YSETGnoug25krNOJJgMxGDFSkt7vuMZ/58EmeKwHiuLocU3EJjoGknf3vw4y4sRjDEoEuULdShMx9YjsJmTx8VGz02qHU7tG43TPwOTuXI3NFaLmKQL3+PcIdsaaIDea2MTrKYLxeL8IHo3maJPDjh40XFtiYGAlDVvpaemSC9qgnnNZUWFU2Uwcm2AysB1pt+ZS8qjvueNKFnmG6yRNQcA0GKd+wpk03NQJPZ619DxRuEHBuWEziiDAj9d5h2dKxjJQajs8W2wqFPPlPTYFf8tdcMvPkxXvDZ3xQJYcaMkxFWt6DiXHYxqQflBEzImrEKH1yqBC7mwmsm7MorjMbRICkIXAEu8Frw5VmE0GMqf0GGBZNw4RqCsoMsv16nuhCKAkc8rg338hBOD4P3/9HsCT//LV/ffVd6j2Fr372tdHpCz8y1OTMaA0eNbYJGRNz3N+yif9CT6s5n1N3+Ftd8manhU9/zS7byuoIVhwQZE0F1PMp/0giN4f0dCjxDXtpV8z4NkGZxhBOHRzTtwhp7KgUMcr/gbf1DWPteMbsiQLnu4ZQqZGtcqRKzajL7iaOnNs/a4R6D20gZojwMd+ag9AfqMqrgJHkbLHaCMRVOSSp5VYINFhYkM2/gmka88/o6l2kqXbx3SdqO+IK/Lfii71rJq7WXBTqijCtscmaqNNciIadKALguHr9rmxvtVUJGZxWFNvtqnv+sfptTLclVX7OP2Ik4H4mnH/CjGL2SxQckopUrMd7+exsLi4ct2jHOqEGyxSY1uRU2ueXuuYGZdi79Na82B3PWGhFoRXS5kmV/H/HOFEzY53riWHaoF873HBO27JRCoazJErrvDHczYWbDf0bIOH2nga8qx/x2MUaVEbbTiQ6VNgZTJyPBvTuSLQiNOQ8TkdC9bj9ON6GeC+Op3tGcI+2XPN3DQ5lcXtjv+nx6gFQEbQFa2rjVIWpzNy5RoAUmhlKTZ1iBPJ8fkf1GYgcRKTjd4jPT2dDHR4Gjq2dAzqk91vTs5Gm3R9CsIZKxp6ngRz9YaBb8qKt5d2nzvYBCI6VXmUC2k40JILadnKwHMhzRyMpqQKg2pyxYqBmnHa59i5otl7ApbSshajDS+05l235DzQ/x7IkoXWrKWnw3OgJZkIi8yx7D0TcawGj1cDIdEh2ImJ5DMH3WDTj8ELq0Y4XwttLzxZ2aKWV1hvbQJSFJ7MKV5tX7aNo+scTetQFfo+UkDf/7Nh/Df1BkaO/vTr9973Afv6tms/CdnXvj4i9Ydf/6OvgTlqLShY0XNCyRNa5kG03uH5/f7TbKXnH+mXUVG2DKyl40infExPmWtBxc65Z03Pk/BFMydnoiZGRC1YscDxvLvJW/49PMqBTDmWOS/5EwThpk64ISWPtOX/yt7kk/45PsUBPy9nTMgp1JnFY2hwPMoLwfayEOGgFKowBdkO9mXwQz+z59r+Rtef/MKfuRfFwIN6aikDHcSRS84lS1SVSipiHkevHkZThesA5MDN08p0p92VdPNI68rIKEIGSSYZa93gxBLXczI63RKdsT5oGiJh1TumeV9PrI5OT53u9Ej2uPdf0by+Tw7h3C85dHM8ypm/SJTEYdRkRnCefheHqP10CD7oRDLiY4OTUbg9D3ClCMc3NtxxOhlzI1o8D7lkxil1oDBNteShW3Hqp1f2M+odDkf6gNt6xNd5QCYuTaHW0nHOmlt6yIPgePWCHrOVjkqNHuUCzSjS0p5V1ojvJmTjug5Y420Ztko/Fm87JE1fxo+JepBxPkd8npyrk47dcz379khBFHYaKCBodrbM3dSOjd88E5zGyQdElzTPoJ4Td8gj/yRt77MeN64ozI8gxtzAWiopkqnCQmYJOJYUZgQCXLLhQKYU5LQscWLHLROXQjtbOk6wgMc1LQUZT4JW8Ew2nOiEtffcKXMu+12GTtSARNqUbXvGExruMEFsGMOiglVvi1VrP5CLHc9tAppP73epu+My14JJaDVFoWJOpRkFRu8CWGSOd7uOm0VBrY7lMDDPMtbeNIgXreewtLlrnhkYEVGaTphV9p3S9pKyQvLMQg03y4zFRNi2wqQcu2ftxOiqgojZ+maq5Hl4z4dx7mab4wS6LkMViiCCn85a5v/NX7oH8OZ/9u/vv8/+BWtv0buvfX0E62+9+tfunWMZDnUQ+kVb2y0DnRhNax6crwZRDrVky0CJowoj8j8Y3Ll++tW/cq8JAtUjrViF5mQlHQstOaTkXVnzz917/BvDy+l1TrBVqjd1zUZ6jrWiIuOhbPkYc36BJ5wEe8WX3JRv+DUCfCKbkTsb4d+oHc1gq1N72tWHU//JH/lz9+IExMLurAmIzU0fqEyKcu6XwI52A5YfEl17Yh27wytNdMycsKR1o71EMBAnAJELHxvF61OQZzkurdVci8oAZKJtK3AFuAx4Lvzqqeccr8q/X3P5frfH6cO4cb0euumQp8T7uybaXdEcdAxMpU78fwNmltdxLPPRfkmaTtnCQcOpzhJQ2aVrZwxizepDWXGsk5GdsPJYlslRK66Ug4m0O3puccRDLslwKVE9mhPE1xqDiWelxFdBAzFuOmND+6xj6xBWumUWdGUADS0z7HNjxTb9/lgvycl2jnzYdTl2dYu0tzghedZkTRC2uhNtRxpfnFasdcORHPDEn1+57o/cAQMDl3711HNG4JyJC+C9TMd+fB5tm32a/GWS0WlPJSVTqYj6D6+KE0lamXitbWgSRWtClShODS2DeiZS0dFzwJSX/Ak5wrls2UjPA87odOBUFun9e6xTXvZH/ODMgMo/WJ2xFdNiRODrVELIrksBsj9QLpgUQp3D/ZVn65W1Ny1Ih08LW2+5Sz7uD+nYOWXF6+G+u+SOX/CuW3LHL3jHLXneL9iGkMKOgTs6o8ZRiKMQYZY5KmfWvG9venMZA45yx6QQBg/ziqDx2LllqUIVwgmdwPna8kPK3Cx8y9zTDaNp2rV1ChFNgASgKj1N6yyxvhxo24zFoqHvs3T/WNPplq7L6Tr7nN1rR7792oOQfe3rI1xffPWn7pWBHnOLmkt6lnRMgs7Dha+9AsePvv6Fb/nB9xc//5P3CrLgfmIrUTG3I9YQ6CEDnlf0gFKEb+g6pdw+pxMeyIaP6YI3Zc1cjdzwRBqmGsW9yncXc4Ywvv+eL/3I/gP5Q6wIQGLjZo5mcfU9WOKqiWNb7UPTv6M4FWQ02oYAwJ7GN5xkx+Rkgc6SB7rIkG4Dkjh8rM/IydJ2wC6lPPL6r9e4iYuN3UJm1EHboHha7a9MPz4IdHy79azniK5KkZLj0bR/44Y7gr0IqGLCtjky7fQV27CwIAinckAUaseGPwrHS436GzNsONCKXszGN57bRnbOddYUmg1wBBYFGSsizce2q6Y0a1cmLGl218i3mEiN81zGye1doDZdr2eBkLHIP57jIlwb/ehcL8O1WEuJICx1kx5/3f0sCtPtNex6WgeLcYBonZvhEi1rIlWghc3CPvQJcCzcLGWIjLNyxhqmU3fMmb9IzxUngXGCM869SedUyrSdR7JIoK/VLtkWxyrIEuDwauYEcZp1wJQtHU/0kufkiBVbvs+/yO8tb+BV+X/bS/5Z9jYXuua2HLOmoSDjRGfMteT75ZjKOd7sG2ocb7FmLT0rsYyYTB2DmJ26KPxwdUTuYFbCVy56ChFWfuBmXvDVfk2LaQr/9+yb/GvDSyzpkj3vhbTMteBNd0GBo5GBQh1zragDiAW4GfJQBOFOXjIvhFW/Ox6X/RBMDOCozKhzAxxZABd9GNlFy14JeSJODKR0g/1+uvDMZz1d55IG5IMqgpLMafp9Nmvx3qGeFIg4nbVstwVZ5unajLg+sdeNfHu114Tsa18f4fp3Xv93X3v19T/6Wk3GY1oydvzb4qnB/7euP/4zf+w1G/ebMXDklA8ol9JyLk1qCAoyflUu+ZquaGQXVvaOrG0lEftSuZCWQwrmWvDArWkY+Jib4rCVqT0A+XDrP/7Cf30vOj151TQBiRSMijz8ZrahZbAtBRLv3aMpdVpwVM5yLSwRO2MiFaXktmovzpKvyZnLlLlMmVCRYdQriNoHf2XV+jrHP5alNezC7yLvPgqhu5HmxJq7Ha3qX6TiO2jX6F51wxqDCiBNd66/ZhNW3TOiCa+jkCI1ylvaNKEAOBDTZ5g70k7YDDZVWMmW6EwF4GUHtFrp2UhHx5DC4cAmCnGbO3oadlqGKKBesaPCxX1Y6iZNHa6eC8+YLhfPg0evAC54OuflWcfXEt4zmgDEIgCKGrLx8dkGQGvUrIHrEw/TI+xASfx9IhWF5FRScijzlA3iw7022qB4VmoUrAg6wCZwg/oQRGjHbAxAjt1RAiCZmMPbOMBzJlNzNMPojlG7tNUQHKlD2tcqvHfGwCwez2gPHcMeo9X1mZqNsmoIn1TLwMkc3JwLPzw5YIqZgKxpWAYD3fPgrHWzzjiphRfyio9PS747O0gg3qi0BYU6ltKS43jcDVcAQVxUut+33HE10QHrjh6ypucXsneTpfVUc5uC6IKZmoZlrhVriedCmakBxLdkxceqirX3bAdlM3har7ReqZxjkQUGQHDQcAKT0iYek9J+jwMyZYUAACAASURBVFONdrBk9DJXugFmFRxOlaLwSfsRxehguSLvV6rhvae735smfwrAbLfFM/Ukd//cF++d3vvpvW7kAyr7iZ/4ic/9Zm/Evva1r29dv/JXfulzUbR+KiXRg78Nt/3IB0xBYn3fv/3Zv//Lf+UXPpfjmJGzwb4YJ+TMKBBMxC4IKqACdRjPq8BCy7CSBU34En4oW2pyKs2pyTl1BZkIn94DkA+t/vwX/sK9f/ylf/K5tbQBaIAXn9q32MIN4ukD1aOjZyYTs1FlIBejwdRh5TaXDCe2knsgMxBNtrgWxmYtZkYGYs1gIRlbWqJYOFqzRorTTsK9E/GOhcVxghAb+NvuZLT9ioqy1DVOdinlsZKYFknPGZvd6xVpYnrtsbFi81wEN6wMl7QfAH0IRFTUjoOYMD4e13jcTQ/iEREOZU4lOVOZUJKHY+fS65sIvaAN/kJOHEXYh1Jz012Jcl/OwzQK3pbHdDKYg5K2FBJXmc2lS0JQ5SmLK85Kg3i22gZaXmUhkIH+ttQNpRTh/OyO0DqEDCo7PYv93NHJdk3703/b/e5oQuhkPANrGmpK2piaHnKKplLT0Qejb7n2ulcnK+PfbbozUElpTmUMzGVGKQWboElqg9B7fL7jNt7MjlnrlhN3uJuO0JOLmTnEkNj4X9T9iFgwYqcdpSvTti7cLLwn7RXiZKcMCwYxh0TDNbHRNu2VIMxlwoaWlp7aVfQMPC+n/IC/SSGOiwZaDwfDhHfcmjkTKnL+vewTfF92wj/3l1y08L/17/F/cJ//dbhPNUz43OSEnx/OKXD07OhqCEw156TImVfwaGswLxqmnGvHESVfcU+47Wf8fPYuU6040ppfzN7lli6YUPBAlpzqhEtpmVBQkKXp2XtuxQ2d8kQaqt4yrErnaFUpnYXhxvNxUjmOaiHPbLpRFzb1aHtJDllAELEL3WBakcXUUxQ+gJTwni40ABKhaR1l8a0XMLIR5vA+CGUQnDMXL5A0KXFOyTKP9850JgKTv/tLn9v8vu/++9/yRX4b156Ota99/Rarv/nqz9wDOJWSHuWBNvzY63/k19Tw/41Xf/peh9mD7qgDV1eml3QMojyvUwaUEsdX5BxB+DSHvKUbE9TimWvBpbR8mkN+cC86/1Drv/oj/+29uEK9oUliWWFnjxs59AMDrfYUktHpwFFYkd/SXqENQVwlN9e0EzlIDVVs/CPAaLHguQmV6TR0lV4zbkf8ORYRRwrNOBU75pocujmDeqrg9ARwoWtUfdJZ9IHS8n71L0LRiuBkrGnptU+i+pjL0AcXruuzyLFWxCxi7d+HMqciT1z5qKvYrfHvXLbiNs+omQTN1wJzr3pbznhBj/lVeRC2N6aV27mw4xhbXMvS0AAao77HhS3PMHHzI718Jl0vulpd6jppGZK4m+zKv8d0u2iZG7dnfC5i9QEEzLDE7/jvXk2bciBTO99osra9Kngfg88dnBy7clWUV4wFBOFcl4lqtXBGybr0K07cYaIXPvbnSfdh4DK4o8nVFfC4fyKORhuq4PK19hskaD0icM3JqaVgqx2l5JSjOfbV42rXX5z8rXSbJmWRutXR86Le4FQn/PjxKW9fKr/YLflUPuO4cvzT1Zo7ruLWJONJ46md2XD/j/4XqcgpyDjHaG6f97+TfyIPqTXnXLa84o8D0Bh4SWZ85iTHK3z5SU8bJi9rejYMbGXgy+4Bv2t4nqV0fNk9wCFMqTj2EwocD2VNgeOOX7CRnonmvOOWHOskXfu3dcrLxYTL3kTvk8zReqV2QpmZNmRWGvgAm3Yc1EaXGrzQ9jZ1t3NhoKHtDagsJuH9VnryzNMPLgEG2NG4nlXRsldEr9xHRMmyq58pZTmkaYhzymZTMJnYZ5MPtr+PXvv2Fgp/u9WejrWvff0Wqx5lw8BKBxr1v2YAAvCHXv/CawWOY4zS0uFp8TyQDTmm8WhlYKI5ldhXeC7CJ/SQT+gBGcJ3yZRXmCeKQYffA5APuf67L/wP9450mmg9MVna3G92Aum4Mhyb3QxHKTmXukkagugGFCcPBRmHMudEDtLq5S6YzxNtRDdqq/Bjh6Mx8BhPPaLzUaeW222hckPg00sS8Apm3boMKdYr3TKhYioTCjKq4LT0LE3C9cY47u+4ER7TjMa3J9pQoNNoWN2eyyRMdnyiw2TB8evQzTlyBxy5A1Q9hzLnUOy2+PuYanMdzMV/jx2LxuetF28OVpjA+w15TE3JSrch/dxslXMsMT0CkHiePJoASB8cv/owrcnIAoUo0shkt99IcnYy0HI1+yL+jCBqDKjykXjdozTh9aPDmKJJrG/UvR01KT5XrEZbcskSUIrX8dVgwzBRCoAhAuh4/gHOdclUak7cYQI2rXYcuQNyycjJuNDVFXAQAci4KqkQzJWqpTPKoBp4HgJdUDC61qHM03XVXgtJjMc60muBQLHbBRgWknEqCz7NXaaUfFxvcpNDfrfe4kcPTjnfwIOuYyktW68MHr5/NmWeO84b5eeGC36uXfJO1/Hj/lP868N3MdWKj+kN/qPyM5Qi1EF/dBj0GecSJ2bKu5cajjlc0rEOFL+ldPyqe4yifMNZuOYn/E1e8TdwocF/JBsKHIdq1K2ZFjQy8LI/5FgrbuiEAaMVb4d43gxQHJeOMhM6rxTBkjeK0AG2nVGk6kI5mOxoWRGMOLH7PF462t4mHpae7hMAifd7VkVAkbkdABEx56wxABmC0N2cs8J144Wq6tPvsfbUrGfX3qJ3X/v6LVY/+voXXnv91S/d8/zaMxfG9Yde/8JrX3z1p+7dZYoAhThuakUhjkMKvJrFrgBvyYpPcMDMWQxXtPDMgO+RA742rPkTP/Mf7AHIh1A/9fmfuncu29TYRSrHTltwNatjLAoeO/J0mOizIONCGw6CiDcKmiNxIoqdM1zIsLGGYR4yB3LJmVOxprWJmZhwfRsctoAEiAYdkECl8iEcUcSl5vGuuwHAe3qWLHJbtbyB2lnjPZUJK92+7xTkaYvYMSgJzjdSmTifqIfZNQt5sHX1GKC41DW9DmxCloMLU5LYYMqokTxxhyyoWYcsjTpQjiIdDaL4eGf9GpvyqAMYO021YsdtLR1vyGNQuNAVU6mYhXwL+xzwNJjGByw9/YJ1OvfxtXOJCQ8GylZs00RnFfavljJld9RSUlMm0BTdyca6kuvHfDwV2a3y73RH8RjEx3mUFQYyV2xTlsoYZAwhc8Mem6fjZ8+xA11x8jS2Vx7wiVLlcGxpmUjNUm2VvgyAMxOH9wNH7sDArzg6bamlTqYLYPqfQX0yLCC8Tj4CljEMsdGWiVTBGttCNsdamh6frIe3tPSB0gew1Q2f4QU+6Y/57EHFe+tbDCg3J+Y2uGph2SkPabillr+y6pWLfmCRZbzXdzTS04unU8/LzLik444ueEGnDB6+4Tf8gekNvrR9kxs6ZUVvInIypuLInHC5jbkldozfkzXfcI/5vcNLnNHx/2RvcSZrFlrzvD8AIWSPVFxIw7lsue8u+eRwyrFWNAwcUqbzeyA5deZw3mhYmUCdw6aHgzK8ZwINq+mhCIcvc8qqEcpcyTNl0wqT0sBH5riyPLHcOqrS8+BxwcHMQgzHFXNEMmdp65nbTTu8Cu59MkTicKzrDOTMpu0z77ev9689CNnXvn4L1qshU+TXW1VYt85CIm4ljk6V2llTlYvQqDVRX+WS75VDvO4+kAsnfM+XfuS1z34nNmZfH1hfevWL9xCYakEnJkzOiJQUjxs1d/FnpLiYmHgnVZ5SsmKLRzmWOS09RxhFJQKPaUgRz3FJ5C4IuToey5KcjDk7qs44XyJqQQaGoJewyUFNGbjxu1VtRo5Ta2yFeqcvMNehC79iLlOcCBOtyCVLIW/PqnE2QwRoKTBR/VNTlBN3yGN/zqkc8FgvmUp9ZZV93OQeySKs5mcBnJVhyhOzHvIrCwR2fK6el/F2RiAwjP49BDh5nfoUaVKEv+XhXJ5x1VY27nOnMTAvuukFIXUIvVvRsNImbW8EQ+OQxijs/nZqnAheUCVQ1eOTLXB0BPN4VrpOgKqUgqVaNgaMDQECONYtLlyHPbvUdg35NhHMKprE/+sgQjetxTZdk3OZ2r4Gq6MLv2LuZoGCOJpoBLB7XT8SgzkdgpOMOrhlxduiLbCokktmWTcjEB+PSwSKBTkqmoJFGzoKzfiBo4rMWUO+7Ul25+1gAK4NU5NchblmfJUlflAG2RkeVJR8lWXSetxnw7vNljtM+NubR/xI/jxvdi0P2VJpxlJMk9J55bAWZCMcUPBOELt3DLyFTZq+e7jFz2Vvs5UOHDyRFYdMuSDagSsNtpBwX1a8oHPelGW6Xjbq2XaeucvYDsosd2zD0GhQo1VtOsiDFe9Zo7xwYOGEdu53iwsSNDciJlAfvGlL5rUd64PZ0wk40f0qhiKOAQjwvgBk/Lc892kqEule/gPS1/dltQch+9rXb+O6LXVYwYuNkX1w/q6f/tErIOeXP/+T9xoGzoeehcuoAjjZu199ePWzr/7Ve7F57UOjOiGjCZLW6FrUabS1tAYkNusRiNRhCtIx8BxHtHSsg5i81oL35IKaghqhoWeOrd4+ZsktPWApLReyZoIJmo0U5EdNf05HTy0Fq/C6hRTJznfcUMPOZnUqVZrkZKFlj82g4snEpgoXfs0s0KO+VV1v/IdABjp0cy78ipvuON33kT/jXJecuiMudUMtZQJRLZ1RsCRjEaxdD5myYsuCSTr2krbZJ3rVuHGP9xuL5eMUJGZBxGa5p2eNULKz+L2lB7wrWCMu0Tp2CI32lkx2TlgepVHLhzExuR8dk+im1YyA3lV61Xjbrk45HCvdME3Tip325Hq5cP2MV/4jMIpTvDzQmgoy2kDFcwgbmgRA4vZ5zMHqxB3ShSna8IzjG7d9o9ukHwKjdaUpXgCzQFpUmckk7NuES12ykDlnenFlorg7hgY8ZDS1cjgO3IxteJ0I9wf8LoAwHDc73z0TqvBe7BnUM5OaZZjaeDzP6ZR5rTy4EDoPW6+UmQQAAlnYh5oMH7ZtqjlPZJuoXnmgWAmScqXOXcNcSwqEF3TOr3QbLoJrWReu4Xd9y6fcxOxu1SaKnezS2bfSc6x1om+ZNsncsjIcK1qOdJKS0iscUwruy4oB5XfqEVtsIeVWXjCEyXvvoQw4uwmYIRMDI+0Ax5UEcbqdj21ngvW6ULZdoH162LQWXhgnJBU8NQER0WT5u5t8PHUpv2/l2e766/ucvs8oiv5bPGJf12sPQva1r9/GlSM4EX7wGui4Xn/8Z/7YawB/79W/fk+Bz/zVPfj4MOpvvfrX7nWh0QOSD38RmguP2e6a0NmcqaL1p+DwOtAGgBHdiKxhtt/e4+LK6uwjuWRDw0q3KaF83ERGu+aSPNjOdmnVW1EqLBQv5iYUYqbSMZwtf2azatSgLGzHOJsh0saAtFrea59WtKdSh5X06VNTkagD8QyUYtamp+6IHEflykSDEYS77gbmDFdxX4xGdq7L1JzHxy2YpOc/YjbSQYxXyaP+hfT85Wi/4wRlTFuC3Yq4hqY+wxkAVNMXxAbvvhCOib3CggkXsrZmWqwVfTICIGM6nr3OLpl8G1b5b8hB+vuW7goAiM1+pHAZWMwSQLyuqYlgIN4nJsdLAFpxAhV/HsvC6H5BxJ2J2c+uAyBo6RAVDmTGIlDg4nHVMGHz4bHDaHt9ugZsAjKXKcsgdo9J5ed6GU8KR3Jg4E83ZGQ02iTKlQ+PiSGCg3oEz0x2CfZRgD4L+TuV2HRsQhV0TDYFivbG8XqPgvyOHlXlOTlkoTUFGS+42vIuPKx6z3rwDAqN90wzC/cbROnUcxyCZW9JzUZ7VOCOTnhlUvGPNmfMgpB+KV26pr6uKyoyNvRsJbjAIRSa8clqwmEND1a76zNTISPnM8NtYvjt+PMJ4F25QBB+5/Acv5I9ZKE1t/SAC0y7svAlK+mYOscUR6NhUULUpvGZpOnHNHgbDGp0rEzgYGJgI+CvBEaiU9asGoFmgSKzicj1GryBlDi5GCLgyT4YhUTwUZS7yUqW7xLY9/Xt1x6E7Gtfv43r1yok/9zrP74HHx9SfenVL947pGQgZnxY07wLqRSy0ap6FihT0RFrLExXlDKED8ZVWFsBHmLPHMDEEFZv+7SCO9eSHs8tDvAYhe++nHMYwtPGtqkbbZLVaxsmAs8S98aybRmYyIRJ4IkXoRUyS9Bdc5+Tc6mrBDgUC6ebyoStNqkBjqvPMVSw0J1TVWx8dza+tvMRXDxmmV7Ho0lUrigHTMnV0cmu8YiUmkhxqskS0IvOVHBdp3K1YsPviOAuNqkOp5KO9x09xATXG/IAEBWf8kFmYkGEm2Cle/01xrSvOMGopWCO2b/G+xlNz8wGoj4j/h6B3kQyxpqQlW5YBArVeGoQjRIiuIrakvF5GCC5NsWwwgYLy2xoU6L4Y73gRA5Y6oZG2yQun8mUSMfJJEvicIAqUNcUE6rHf1/6FSLOph5+ZdcSlqJ+phd2bq8JyVV3xgu55GE/di1UKUWiF0b6m1G9DNgV7BzKcnKacMybAAQlvBcEE3N/ikM+eeoYPBzW8ORCudCeerDMqFtDxUNtKXBMyclE6L3RM48obYUf4cZc+fh2zhu6pseTqfBcuN6fSJNAREHGFtOQfJdf8JVmw0tDzYuHwoNHcJ8NR5R0KD+Xvctnhud4w52nHJBD7PzPsMWC99yKmpIbOuPr7gnOCXf8jDU9NTlr7ylDEKGZAIyuV7GJRO/t/zLbBRNGGhZYVkg/CF5DiKEjgRMnkGeaMkHsecPkL4QQxt8ZOWY9bdh9tfLMJ/Axfj4Xns97IctMwD4Me++nD6o9CNnXvva1r49YfenVL96LjkGxoWswS+VKs0SLGCeLJ1Fs4J5fBlpHXJFWfLLpbbS70pQC6W+7JtqAzVtyxvN6lCYmb8oTWjrOWXNHj3giK9ZhhRh2mgLgmQDketK63dbTh+lKScHCzTjzl7Yqi2MSpiBj21WAqdRstaGSkoI8uRtFbQwQMhquOh1FF6wJ1pR6lMcsGfAJgAApWfyEOQ5hRkmnA5fS4AMYhKtC6yy0+UVoxh0WxHc9OHI8QWgCcHQIdcgKiccnrux/mfvmkiR1Ogbj51nTcMQsgdCo5RjUJ8vbTOy58jBHA1hi14mBCk3UvPjaYA12tMt11/4GRmUag5ZIg4stWEwTj7OnSFeKzW+cdpWS0+lAxQ489OGa7LXnkZ7RhWtZEA7cgjZM42IpylymRscK2iJ7HRdAy8QyWMSmURtpmMuUC395BSw6yfBBKB4ByPi9ZnkmV9878RrLJVyBAg0tcyZpahjB9VbbYNVr4vheB0rJOdYJP5ydcjb09IOwas31qRCjSl2E8/uGrunEQMUTGlBQn7NRzxktgygX2lI8nPOpw5yj1YLfcVP5S/fPDGyE89MBx1qhWI7Qoa9YM7CUlmmf8bIrGFA68azVprC3/YKfzx5wqlPOZMN36SmKZdjE98ANnfFIHuOZo3ju+BlbBpbScaAltbProR9pDL0qhXOsg757WsKmNXqVV5jXO7eqKkxAily5WLs0Edk9l1nzJuvc0YAinmcXwE4UpX+rKcb16YfI+99/GFwCJbC35/1WtQch+9rXvvb1Eaq/+PmfvHdIlRoeIzRJar0TaFCHC01ldNkZIAWgTaSip0+i6QgsHJIa4kiTqgN1pNdhl1ytoKIpAb1j4LGs2NDgVWmk5bEsyXAciGkknNgqdnThihUBT0efVnwjXWgeVmVXIbAuTlQO3Tyt4Ns0wtsKv1ruRlxVlrA/G90m0XbMIBmCPoZA8xnw1IGWUlFQaIYXAyAbbWzbUW7JMRWW53Acwjtj+nScPI2F7ZVeNQSw32FBwYo2TYdi8+3YWdXalMtW+8cNdYbjXTnnlh6yocGJ8Cu8yyf0Nr8sb1NRmpYiGA4MdJyx4lQWPNJLfKQ4iSV0x3Nk52PnbhWnNbufOwF8BDlRpzKefsT9jVazVwXzUfmi6dpzgSCVGkDMkjhOTky/kKcJVjy/qCWhn4fHdToEh7T1Fere+PVbOkRcmKLYxKwKAGmlGw7cjHO/NICiAxeBlhVBy5E74FLt7+4ZC+OlFHTaMZOJWfSKT9cFMJo0EjQ5Rpt8qOdUlCnPZ8B0IAWZWTFjeR1/179DRc53c5NNp9S5cFg6qmFC72HtPYMqb+omCP4z1vSgO+tfp6bb+BpLvjc/ZJILX31oTmu/L3uOn+su+Wyx4B/3j8lxnIWpSCM7Z7H3aCjznCU9mQo1GQ9lw1vujFt6wERzlq7lbc440hmf9M/xlrsgV8elRBMAz8f8KQPKMSV3dMLUOdbe81yZsww0s8rZ59d2MN1LJtB0Zr0LNgnJnIGPrhe8t+DBrjOHLJGrkxGA9TajzENgYab4QexnL+R5mFz0IRjxA2hU0Qkr6j6u3/+6EH0vTP/2aj8r2te+9rWvj0j9xc//5D0LWssCDJFAWdpxr3s8U82v6Axe0pP0+3ilWnBsg2NPSUFFTk5OKQWFZJSSBxvWglMO0v1yMmoxPcZtv0gTl0M1wBBtfiMdKQKXQT1dsEcdRk5FQAo1tOcuycRxwHQ3NWFIguVJEKlnmCOSiV7tuZe6phDLConORRvdMndTcnJL2Vajic3dNK2Eww44zNXE9n2YKK10SxmyR+7KabhvzlQNqEy12DW7mrPQikKNLlVrTsxCWWjNQmvmWnGgFWs6ZpQcas1CKw61Zq4lC62YanFtEmVNc0nBgpqGngklT2RFRcmMmixQs7zumv947MCoMA8DnWinDRIes2TJJp2L64Lu0LYCBE1PBChXhenjDA0Tv1+1JI20q3i+Ii3tunA9BhQWIxARAU3M34ip9JnYNZyTpwmJXSM1vfZEy+BooTyXaRKuF2KQ79gt0vbHHJqpmMW0yE7HMtb3LGTOgZtf2W4fJjKqniIASydy5RhFYJ+JYyET6uAw14QAR3P6ypNmaQiTwToQEW2RwKY0qxZuH9hE4Bc2awonzAqhdrYoEV/3OWpuS805LZcYkDqk5JMc2rWcwa0D5f/szrjpp0xy4QfKBbcXwo8dndLh+VeLY57TKada8erJKd8jR3iUi43jXGyy8iRY7n73cItcHY9kk647Rfmqe8iKLXf9gieypiCjkYGp5lQ4LuhYZHYt3K0KzrqBXpViZBKgCpvOrlSvBkQMjChVobiQRF4USuaUsvSI2FSjH4S68ilPZEzH8kMMHlTy3H6qf1qs7r8NTUfXhammlys/n1VF8bQj1752tQch+9rXvvb1Eaoo9hxrBWw12v49DZOOuNopYaU63r9LK88SSBeauPmC8Ire5NP+DgCf9S/wGX+X7x/u8kPDHaZSU0rO/8feu8Zat531fb9njHlbt319b+fic+xjjm2MjbG5GqXgNFLbUEWqiCEhNLjqlyhV1KgSQamgOhyRSHyoKlFVSj71GyUBm/ZDVKlUqSChAWzAGBvj2/HlHJ/zXvd1XedljKcfxhhzzf2+7zEmUHNi1rO1tPfa6zbXnHPt/fzH87/MZMwxe9zSA87MmgsJLldjLbYrvISAvCWbEE6nW9tdj6eLP7V0bKKRaGrC9hj3dsBDYa/i2ZA48oYLXVDFxjO9v4mMYuMcJgtTGTMzEwpyKslZ6YaZmfRAJu2rESUzQlPYxtXeXLdNcApnNAgzRow1UJByknDekMfQwJyQolLFYzHVsI1TzZlq3h+rKQWZmjC1GoDDdJlqwSSCEo8yZ02KcSzJWBKscw+ZcKRTntSDfv80tD2NCQIAHDb1QTwtPQCYMgp5FRrOh9RsJ23J8Hp63mHI4yZOCZZRjwOBPvWw7fCQypWaaYP02o4unhnhXPH9vuifR0MTn57H6/D8CNqmla7ZaMPYjNiXWT/l2PgNHY49E+x511rT0OJ0O3HJo1FCFz3dSikpJGdsRszMhEwyLv2cha44kllwwZIttU5EKKUki1OoBAid+h70hfMpCNE3NMwJIZIjKcnJmEoA8yMpI30yIydjX0dMtCBXQ4bw6qojt/BgpexHkNy6kCZ+nFveZqe8w864y4Z37IfbOzxvlSkXNDxZZfynewd4hVfO4AeqQ95XzVh2yigXLtZBAD4hY1oIf/V4wn98c0TTwVFu+WsHe1gDb2Han98FGbfNHC/KmJxzAgXyTJaM42fsS/asn4IAvTvXmdTs5eGMOm0dSqCZlSaYESQwMimEeaOsWijzAEYC0Eg0KsU5oWkNXWcYFeF11o3gnJAlp6uItfvAwdjxJmbmw9OMBFL+pFIfJiLDyrLXBxu3/sm/fOFPfNK/pGU/9KEPfeAveiN2tatd7WpX8Mlf/tQHAq1KqRg6KgValmMbrNdKWGl28Z/mmax6yk9qHJNrlZcQaffzv/KPXvzOH/2O3/jor3z8A9eZMdacW5FwdSsrqNyYzgjP+WPe4g/4ojnpAc5NPwGBhWkoYiJ2ajsbWoyYK4nfOvjKyTmWPcZSUpJHKtLWESoE/oXUatf/VjGYEPYWBfMi6XG+t/1N3P5eoB+vO/V98zihYsYIq6YX4lsMTjz3OOdQpr2gdqaj6D5m++csY+NaYBlrgZPQMl/z475Rtxi8hOMx0oxJfJ8hj9z0wKUb2JwOJyEFljqGE1ZRHP4mPWRKSUHGA1lwKktusEcrQdfjIw0oZcHU0sYJSCiR5D4WQvpM3H8qW+H8MMRx6NA1dO8aUaASKFu55PH+YYISAEkAkEn1ETJJTH8sswiOhJTfkpyzPMmCGGBNHZ9Zr4CbBCCCcBtyycjEstYNDV2gR8UpWhtpY8nFq4vbUtNGm14XaF+RsldrHUCzduSSsWcm5JLT0HKpK26ZYzIyGhqMWMZSPeLy5tGoETFB8yTBetrhGFNyoUsyySjI+onHRMJxLQiTtUmk1001TGr2tWQsGU1jVmlfAgAAIABJREFUGGfCExPLnZXjmf3weXReUIXMCJVaLhtFVPi+2ZTnjuBYR322xksXnpOuw6hwp21Zek/nYFYY9kbKVHOqHD5+VpP7jM8tGg5yy+8tlnxx3XBX1pzKiqmW3JclJ7JgLhsMhlwyrBpKyVkQQOBNnYEIYw3OYCNyFhKWRw59RfKQyySdY+F9eIXKCpUVZqWQR0F6VQSAkVuJwYLBHavIgwA8BAwGsJHbMCkRwJgwDREBE4FJYGdKOMOjkD2BkXSbXMUm4e+FDZMTNIAZYzzeB+2HCJiYL/LwJdX03/zRBxY/8K7fePSZ/3LXbhKyq13taldvkFKUGsdB1EZc0kYm/VYf0kaQYXXbwBqEp/Wwn4J0BJ1IWkX2qvyPv/KPe3Hk3/vwf/XiWHNU4IyGFY7PdUtyDO93T/CczviiOedd7hZ/xT0FBNBjMbzFX6PDc8S0F9g+LEAfAhCPciwziuiAlCqtlG6iDiNMVCyTSESzWKyYXmidKF5WTAQdvneBGtJ9ho5cSU8yi+nu6XnSBCSIzSvyCKoOdUwrbgAMsl5DMtOKfQ1ApdKMa35MK9vE6xzDkVYcRLegJgrXU3aDR5lozqGvOPIjjnxY9R5r3k8DrumUgpw6zkMupaYWx1fkJNi7ajAEqGmYxDDFRP0pYuI9cMUkYPg9VbBr9v2ELOy3bSBlOnIhi6agiRStRFcKEacJgGz1JInSJP0+sb1RwXafBnDWaNfT7YBea2REegA0BGnb68nbTNiXGV4dK1335gVOfUwrb2MuTsFcF2z8pt8Pq+jClba5MhWZZDTa9holjcAECHbNGoBGoCFu9TbpHKxp+glPOvczsj4QdKNN7/yW9cA17McyTtQKDeBoqgVHcQL4xEyYVYFS9fTEYo3GwMKwrzfO06G85JdhseDQ8X/f3vDvlpf81mLO/7uY844jw5dlTmGEGs+CjnPf8YVlw2uXsOyUL80dt2XNF9Y1IzF8YrPggax5IGtqHFWcZE0owqSQkgtZkalhIRuednv95/22XPCcC2qqrLeY9rxN91l5z35uOcgtRgKdzUqcnJkAQCCCjzxMQowEcbrqFkxYG0IGu04oIuUqjzqPzgmLjUEVNrXFa5heDCcpVy42XOShjjiJ0TPriZmW4fHGY4xiTJwe2qt//4Arzlh3fuZvvXjnZ/7WTpz+mNqBkF3tale7egNV8O0PPlUGYRGb8HCbIY+XMjZ4mRqsmr7BUXykvPi+KRiKZlP9+Ef+zouwnago8CZTsW8tM2t51u/T4bnLhg/4pxhpRqmBgvROf4sLVmy0iU2EiSLh8EzXZJ/rckBGxg056BvYdEk0jRX1FX0CwDqCktTQeU2OU6/Pu073TWAkNTcQ+OpN5MnnkVKlKKVajvyYCRUeZaoFtTjyKDJ3kRq2iY/NEBbSMNWgranFUaplXwsOtWSkGVkEiRA1OXG3b22VQ9ZKLY5WfN9OVxGk2L55z/rJTKUBGCQwtqG9Im5OTXCjIahymDKepgkJjGXRDCC5VQ3PC2UbDploXIqyYN1Tm9L+TW5rwYfM9sf14XDGtB822gSQMTj2yUnLStBLlBRUUvS6Dx9Bak8viy5ZYyn77U7TD1XtNSEQhOGVFExljCBMZMzEjHvB+vCcUzwrv+7tey911dOrEgAKUw7b78tS8gCIMWy07YMik4Yl6KoyumiZnN6PxTDTEYJhphUaaXepDDDSjGuUVGJ4+34WKULQdLDpgkXtwQiOxsJBYciNcGDD+XNBw//+8pozaSixnEnNe8sZRQZ/Y/+YpfM8X1W0eC5oWdCx7Dyfauc88A0tjmeLgi/pkrMYMjjTaJ2thlfNJStaMjU84w+YMWIhG8aUfMmecaATplQ87Q+5Z5aMtaAi41ArKs1oUcbGUHtl3vng+BXpbIWBygjrLhzbURHoUXX883c4UarS07bRY000Agyh66QHFF4FY2A28iTjrU1tHwEYr1d9CvoAgMAWgPiBHW/6/jgr3scBk109Wjt3rF3tale7eoNUELA6vCgbHLmaIAiNORjHfVpIWHkfaYYXZSVB9P2sHvNFucfQNQjgf/7wzzx2Fe7HPvJjL/7S3/ylnq+cm0DxMMDTpkKocEApwrN+TEdITr6rNX9b3oYC/yt/+AjIsRhOdR4DD8Oqb9AZGNbUjCj7xPFLVixYU1KwYN2vogP9Kn+ymgX6Rg+4shoPW/pZuC1QtJKmJFNDLY59rVjRYDGspCHTAOjW0oX9GRv2BEzyuD2zOLWwCEaVGWGCsWFrpbuO0O8WI15jxTEVRoNTVqWGlkB9GYIDQbgrK6ZasJCGazphLnU/dVlK0983F9ubq6ZpUCpFOZIZFywHU4stpWk7LbI9yBuG+w0pbekxQNSPaLTp3epGhvczCAtdh2Z9ABy6OEFJ7mtJs2IiISdpT4KwvA0i7+TWNgABgdIXjk2anEykYqV1oOip9ja6qVIWiOp2ewSD03CMkhPacP+lbc2xbEzT067KSP1raJHolgbQDLJEGu368zJpX9qYWXIoQRg/1YqcYF+rKFZD2GGhWb/vKyzPVOGcdR5WtbDZGoVxugouUc5DZqAw4RPzFpmwUU+Dp1bHGMubdMof1HNeu1Mxp+UDN0aA8umNocXT4tnE/XZblow156PtBWvZfo7m0jDTggtTc00nrKRlTk2pllwDBRHgUEdx4gEbaZlpyVIarAoX4rmp43gMYN65aDkcPuO1DxPO3Aj7VZiAiChFFkDBYiMUGXTR1aptha4zPSWri9khw0wQgPXGogqjygUq1aNZqQBXdCCZ9XQPgYqhlsSI0nWWLHNY63sAYoz2AvW8aGmbvP95V69fOxCyq13taldvgPrlH/4XL4zIcHL1H2lawTcEKkXw9vc9xz6POoe0go+krI6WQrIrNKzH1Y995Mde9/Y//tH/84WN93SqjKPlzFe7mjeZir08XP+ffvG/f+Tx/+0H/8kLgjBlxJINe2ztYWc93WULFtLvXFxTTzQmI3FFWrar+w+v3qdyA/pZujUlhmcEJ6xSbaSBhfDHdQxOm0tNoRlr6bb7kUB/sxjmUnOoFXW0w62wZD29KB2bILS9QYVFuDVIVw86BhOD9IQU9dcSshokXj/WMWs6nHjuy/zKJGBIqUoAw/fbGUDHJauespaqo+OQKctI8trmxqQJg14BIP0+lK2IPmR7BLJfymoZ5sFI1J10GmIux1Ky0pqJjFjquteKbGhj6GETs10sjbYYCROdghwnnqVuegAiSGz+BU/QnUwkOIVVUpBhWbOh1qYHOEWk0EEwaki0qkyy3obXYRlJRS4ZKx+E7lMZ99OgIs4Wgw1ymFiFPBB/5VgkC96UcZIyWVycSE5lRI6lIGMtbXBVI7hb5XHalBNAQXBU64iMJJyHxsXEcB/OFQ/UEZSICL/XXPCdxT5HuWXjDb/p7/MtPrhizWlZSsuKjhbHJ+8XvO+W4QlT8qrfkCF8jgs6UZx4cjVcyBqjwpicz5i7PK2HfMbcY6oVK2nwwNN+j1fNJReyZkxwiks23nmcyi6lYaYlz+iElpCbk4uh1atLFlmcWk5yITOBcpVZDXa8hcdtgt5jWQt7I+0BRwgDlP46sLXddVenpqoSpyJxUiLpXBceFqJ3zvQOWUPwkX6X3K6s9XES4h+ZhLguoygbmrpgV1+7dnSsXe1qV7t6A5RHWUtHobZfkV/JNsSulqvuK4mWlRrFTrZuSI5tDsSfdZtG1mwFpAo3TUFlDcvu9V1k0uTFYphQ0eKYUDGmZEXDkuC2tYm2vclFqdam5+Q/rhJ9aVjp/V65nwiF5Aim12wkh6qwLUFzM4oORpVmlIT9vp00BRLYSDNu+glLWkpsBDCWFqUl0LSuS8FTMuJpGWERKmMi0BAqCS3rRtPEJO1byON7uaEjbuoYq8IDE+DCgU64pfvc1GCdPAyATFSgh52lnHr2ZdJT8QAOo7PRWuseujysswCuALhCchrdruB6lCkjRlGvk5yqLCEAcK01Yylp6TCDKUwiaSXHtAAeTAwg9KFpjTSnldYsdM1SN/S5NlF3sXVQiyJ73X4urBhEDKUUjM2IyoQwx+CKFQwNCskZSYmPU5BMthOvkZQc2wOumX0mUfPhI4CdSNXvq/AOwn5PUw8rpqfNbalu4Th1MRMni+LzpHs6k1U4v9SSa5gUjjTrJ2Qe5aQeup6B84OVehFyASvh9+/O9vjt9izqU4T36DFP5SUzyTigYBI1Rxvp+BxzHswNF77jq2bOGsdL5gFjzZj5gjOpmfkiTFdpmVCxoOGaTrmpU05lyRN+yl2z6EHHhibSRhsqcp7wMw59xURDsGUphkOTMTWWmQ2mCYmG5VSxIhRGuGyUdQeZDZqOYdDgqFCmleK80LkkIg92vkPAMNR4yABc1I3ps0OGoGNIvUqXdB22LlrpvkYU7w2uM/3kIwjSIwVzkJquKrspyNdROxCyq13taldvgGokrDU7CWAkUWQSOLEqcQISKuWJ7JFjNAhAnWxXzoE/cQryJ9W3/fJ//mL6J5F61CEgedMv/vXXff5f+PBPv7hgzUkMgluyYUOD9sqR0Jz2TTIhcXtJ3T9HomNJajajBiGVYHpdQG/lKhIzJbK+QU+ZIDmW635CGYGekPIW6KlB5SDDYapFvz8PKBiTMSbjyOTclJKbUnIsRWwMwyUlQY+NZRSTsyuxVGIpxVCKYRQvlVhyDPuSkyFcmKCRyckYkbGUJlLtPMlLKzW/KTcFYI9A6zmWGQvWOPVMGXHIlBOdc6JzMrGP5LikffbwFCQBkI02vag40M3qqEXZAo2kpdhoy0QqOjo6dX3YYZpYQAAzC133122ENMH+1/d5M2nqkSh4JuotxlL29raXuqTRjo0Gkb4Vy8ZvqKMGI9HIhpS0TLI+9T2X7Mp+GFGSbJ/TOdDQsiIYLxSSUUreT+USGJxr2N9lnJIsdcOamoyMfZkypmBfgzHCmKKnv71HDskxvNMfUmJ53kx5ghFvkjHP7lmOJkputwAkTQw+Xs/ZeKXzMO88H3OnPM8eyy78bTjILLnASh0bHF+0ZxxGK+gfqA45nCjffa3gg9NbvCOb8m3uVgyKNBxoyUo6Zj6A9EwNe1qyoQsaKp1watZ0KPdkzg2d9Z8bA9TR9tgTNCRH0aRh4R1z73AapoKFETIRbNyXnVcmuZCbIEjPbJh4tK3hy2dbe14ReueqpA2BAECqcgvcjASR+WTkqMqQI9K2BufksTkgnTN0ziAmAJDH2fSKCZfhVGRIwXpcVoiI8spP/Zc7QfrXqB0da1e72tWu3iCVKC5De9RElWnFU6hie3qOUGLYAGNy6ph8YDH8/K/85J/bP763/csf+vd6rn/4wX/6QktHKTknesl12eeunvVNZGpkl1GwnMBJyE3IQx6GbPcBbJO+E7hQuiuvaUT61foUhpdjozWv7x2tghNR1IGQsaJlIW1vjzqcLsA2myVsQ6DFDBPhr2zD8HvUOBig1kADS5MRDzQaxLlOAyXnrlz2z3NbLhCEZRTvp9T44CA1BAwdl7IKK+g651CmqARqliDsSeDiX+qqP5ecBvrQKNrHpolDKq9bm95am5i2XrNHeK4VDbUG0FiSsaaOmRk+CrJddG/ylJJT96GSdU+XutQlkwFtLNjgtldS4xOJPwRTOhoNtKKOLoZuJrNdwanjwOyx0FVvw+twjKSKNDgAuaITCW27IVkUZwSb6TYK9wM1y/Vi87RP2ijWT1Mcp8qKhhEljbTUsUltaJkS9tGhjrmQNQcabJ0tcKBlaMQValWeLHOsCcnfKXDPRp1W7ZXSCN9iJ2Gy4T0f5QHfK9d6qqTTsC0bVVY4LqThmh9zSctKWn5rfQFr+JGnZhiBT1zW7EXq2oKOV80l7/bXuMcGRbmUNRbhSEfcNnNOJeiN9nXEoY5JGUUdyp6WeFEuTM2+LzmkZEpGrZ6psQHsq5KJ4DSkvk9MtJOWYM9b2uCGBWGRI8+Vp/e2x2cIDnrWoITAwYeBgxEFG6YgZeGoG4tqMgff1hCUtK25oisZTkKSO5ZXIc8d3gt1nTMaNY8Akbxod+Dj66zdJGRXu9rVrt5A5QnOTakSBSlRsgzCHjkOpcZTYGJAXvhz/ucJQP4s9Qsf/ukXFe0tcOese6qLxbLH+AotaChm3lxpRL92DWlEyUkrPVdBxoJNPyFK+1YQVIJGQRDWEYCkfdgO7HWH2+fwHKdV/MeAkK+1xUqYiDTq2aiPQCQ0tUbCz9/lnuRJf8BN3WPGiFu6N9BGhPcVaGYBTMxkxKFM+4nBgUx6YXqtLY12lGQsWA+2MTpyUXChS/Zl0r/vh8Fvet8r6j7fIlDswtQgx/bZMBUFYykpJOshZYal1nZAzgqBfQsN21Nr06eudwNAmcTjXVxlT+BlFClfW52M7cXhYxmRiWUmE0opY16MYeXXWLH9Ba4mxk+k4kAmjxzvrf1wyGFJlMHkUrYFIL4Ha20EP5ACRm0fLplE+XNJoBuetSNyEd5RjHmyyjiewMEInBdOl8LZOgDO2gfg+JvNKZ/0FyFLwxi+h2tkIqxcgFNWYO4dX/BLNpHStqclK2l5s98jJ5gznC2CqPv7r5eMyZiRc8csoo321gTZ4XkgC6ZacCJLPMHoIWi8DeeyijkscCk1RsNjGnG9ZmpqLIWR/nNamAC6NzjaSMeyAsvOs2iUVy4948qT59oHD9rX6VTTR/BhQXoqI8po5DACZeGCFe9DYKXIXE+/8iqop7/A9nuaghhR2tZS13mYikTgkWUdxuiOgvWnrB0I2dWudrWrN0glfUQtrm8Wt+Lg0Bxf0rBK1qQPrepl+sb6k/7PPvyzL4YmOwjMM7FYLOMYxrfRQPZZRFvU7YTD983esDFOtyUaTaqUL5FLSPtOdKqGjjFlFMxaSmzvKgYh+6PF9YnnWeS5H+qIfV9SRRG6QzmUvKdeeaDVoSweHtcGpYmJAIUEmlYhhipOAMZxKiKROz8mYxmbuZmG7b7O3hX9R6MhAfxIZizZcMmKA5nwZrnBic77aYXBcCwz7uslrW61F44g9LeYXkQ9oeoTvx99D0KnjtTWpt9BoNhtLWi112qE6z5qP8JzpqTypPmAYNObnKQytlMNEdPrTtJxThOdFGqZzo2OLp5HnlN/ybleUmvdi8rHZoRHabWj0RZVTybh+NfacNefctefhnMEy6leRsBh+nML6CcwSY+T6FgB+IyCu1PvoCUUsqUDPpAFAEc6YV+r/nP67J7huT1LboSjMayasKq/qAOg8MC/3ZxhCML0d7DHPVny/zT3aVX5uJ7RqjLJDBvn+WrbcKKBwleLo8ZxJiEf5auyYC4NG+n49Ys5/8e9Cz52v0WAJ8ugG3lNznlJLrlBhaK809/i3e4JPmPvcahjrumUTA1zCT5wYy05YNIfq1Q3NRDPWjwCbLwyMtsQzYk1XLdFpGTBYWkYW8MzR/Dk1LBcG6xRsswzKh5Pj4LtNKRtzWNpVqmazpBl+lgNSNKSFJmjyFxPuwKwmb+SDWIzj1dhNGquULsCLctis+5xL7+rr1E7OtaudrWrXb2BKjV4w0yFFE5YiyLiOdeGKXm0j/WMsVituC3Lv8Atf3z93K/8d/1k5id/5OdfUEL+hotfK90G3aXVYwhEnOHKdJpwCAavrrdzfVylSQYEYJZhuKZjWjwWYUrGCTWbaG2cyqMcR4E4bI/FNSmwclXqb4BOlWG8suP1V/bSbCuAjqvVqKeQ0MpNtOQ1c86+jijIuM1FfO5EEWrJxfbajw7Hpaw40knfqD/QSzyec10ykjJQn6g4Yc5xtItNKfV7Mu7382EMoDxjccUFqsOxz6QHhACHMuVMF/2xCdsYwyvxjCgpJGehK6YSpl6tdgwD/ea6IjleJSDio/4kG1Dg0vYGjUrXb8dKQxBgonAl4blBWOoKEeG6TLnnTvvjW0iOVRAxvVOWx/FAL9iTce+sFYDyBo10uS5OYGYyukJtC/vH93kzeaSj2UgFPNU5yJRLqXm/e4Ivy4I9LXjzfjgjqlzJbVjxd16Yb4TGwWc3a751NOId3R6/2Z3wPjnkY3Kf9+stxsaQi/A95ogu0v02qszEcq4hFadQyx4Fs0gjq0mOei3nEkwKLrXlPzqc0HTgWs/b/A1uaTBXmGnB5+0J73I3+ulHTjBgnmrFLFpIozCPmSJtNM8IeTnDz274mBTxs5KbEEy4cp693DAt4fpMabogQh8VntXGUpWOzoV0eBuTySGAj6E+5PUmIV6FujFUpXsEyAxzQ4Y/D0MJ29ZeeVyeu2jNq4xGYYJnTJiMVKOtls2aXUbI11tvrGWzXe1qV7v6S1o/8ZHAIRaNonPdZmw4PE6CE1P6GcI/+3EUNzuUJ3TyF/oevlb95I/8/AsbbVlpzaUu+7yH1MgNU7hTw7d1RZK+uewGWSDKVZCS7rtNpA5pzp147suSijAN8RBXcvMrj4Ow/z3KaLBGl6hXW1OAUNljgJAngBPPo9ORNBVJj9ioxxH0PQAvy5Kp5uQxXT5Tw03dw8YvgEry2KxvX/cpjllIcCc60wX7MiFZ2YYGfZucfqYL5sEImA7HhS6DjbKMOdF53+APLXgzbGjI2QZOGoQDmXAgV8+5RKEapqQ7HEvd9AGFAFMZD57vKhBJE8AEOAKQWl2hiQHRjSvoQTaR1hWOQZyaqfLAXwQxulisGDrtqDU4OpVSBKtgyXHqaNUxlVHUhGwBcaImpYlQsooeScmMEQ0tc11TDATt6WeD4UznnLNkScd7zQFPmxF5RKWtE0SUk0XQSiwbxQq8Zzri363P+ZzOeZ8c8vt6xnv1WhSeB3epLuo/li58VjqUfXJu6pg9Cjo8FzTclw0r6fjB/Rl/fXbE33v2gP/6yWN+9Pmccan82uKMZ/w+nzf3WdDxgJrP2xPW1Nw2C57ye/0534qjE89KGi5kw4VscCgzwlQrTRXzCNLaOKWEMM1pvbJxIayw9cqyUy7WAVgk2pVXoW6FxSru/3jIE/Aw5gr2p23N605LyteZpAwpV8NKACQvohXvYBriXKCx2ayjKFusVWzWUZZN78bluoy2zR994l09tnYgZFe72tWu3iCVgAg8qg1JgvWcq4lbqSn7G7/6wRd/6Ff/5htCD/Jw/cMP/tMXNtr2eoG02pwoOpUUfTtqI/c+NbBDrn6rQfCc3vOwIU0p1en3GSmUL1C7jnTEgjbKvJVDrWKSc86RjployS0fnLoSADmSgiMprupOBheIrkWD95ruO3zMMME9bF8AIvlgCfaTcsqFbDg1ayrNuC1hArKSsFrfBt+p6G4VGqSpjDiQKa9xyj09j9kUAVh4PAtd02jHscyQ2NR3OGaM+n20LxNmjMiwjKTsgcfD1CynnjlrLln14CHt6z0ZM4uGAxOpmEjV6zcUZaMNtdY9UBCkp3Fl8Xgn3zdPcAeDAEzT43WgEQGugNFcskc+F2G/217bEbI6cnIJl2ABbMklYyIjJnH762g1GwwBHFNG/bFLxgmJhgVbV7WRlJzqnA0NMwnJ4RDO32PZ41Qv+aw55VPukpkNlrGtg7qFdSO9hWxuhY+t52w6eKtMg1VuZvguc8QXmEealuJVcUCrnrlz3Img/oSaEssdWfHdxT4/ON1nonmg9h10fdbGphHunBT8+p2aN+uUT9o7vMvdwkUg85w/Yo8xhWa8ZB5gCcYJ13wIWlxJixMf9FaimDg9rMUFnZqxTCSjGXwOsgie2vi73AS7Yafw6rlwbx60MF85Mf1kKBzHuFBhA0VreLFW8Y/HH72t7rC8Cp0zXC4LOmeuAJEscxjjybIgPM+yZL3ryXNHWTa95kMkABDXZYjRIHqPvzP2qp36rl6/dnSsXe1qV7t6A9Xf/tUQHvgvfviXXvA8+t/VIF8zYPCNWL/w4Z9+EeAffPDnXijIe0CV+POwBVk+BuoNwcfD4YXpMYmSlZLZiavvSggoTGF/HuWBWfGcO+y1FQbogOs64kw2jMiYRDntlIwV7jFt7dVK3krE736wTXDVNnh4HwaPua0b7pkVc+rgLkQIunN4XjL3GRMA2lADk6ZjC13zjFzH4bkjZxG8hRyOA5liMSGbItKmUvL4ic65JiF/JKzcd1GUbHvwdyQzHujWrSvpMxLlaziNMghnGqiAJUUPHKyEHA6nnpaWDtfrOVy0/h1WRUFDSxsnHibqVk70steXDAXzqh6Rq2upw/Mm0bO8RKqfWLx2/TkgCJUUfTBhIgkmY4SCMLnpXcVwVOQUknGil5FXl9Gp6w0Dkj3xRAvmEiZO8xjYuJGOC1Nz3jlukeE8dDGQcL8iCtGFt2cTnFf+kDPW0vbah/eaAwxhMteizKzhNb/hgJyNOD7LBW9mxgNqxppx2Xlu7Rl+4smCuhlxvrC8fA5OhWf2hfM1PJANN7RiQ8tn7D3e4W6wwvGMTvgs91hKxtvcdV41c2aMuG0uyQiToFKzPr+oHeQYGYQL33FgMjSe+xuvvY31sKwR1p2yXwaDhlkVGvouitJTbalY25yQ4fevt4wo69ry4MLydPl4sNB10ZWtbGlbi/eGzdqQF+Fcy3NHlgcwknQgIbfk6zPT2NW2diBkV7va1a7egOVku9rcESxcFf0PDoAM63/58P/Qb/vf/+DPvgChwWvVUUhGox0eHycj22kIpFDCQJMpJGetNYVkEO85zIMILk6WRP6p6djQ0OAosLR0UaQudCgzLahx5BiuUdKivMlUVyBg99Bkww9oJv3vH7rfw5VAilMlJZV/0ZyzpyX7VDyQZd8Au/7LB1cxCS5NECZCijKVEQs27OuYFscZAWwcy17fZHfqGEvwtjrRy0jVkoHtr16ZOkEQgV+wHGz39j4WwwXLXgCfal8mnOmCha77qYaNoZCZWFD610sOWQV5b0BQkIf3o1xJSE/H0rMFn56HAFncL5UJ4YKNtldO12uHAAAgAElEQVQ0GxA0IEP7ZUuYxJUxu2NMEfUlXQANIsyouKdhIjWWiiUb7ul5eJzkNBomS8GKd8RB3AdFr2gRJjJioSuekmPmrNmn6icBnQevAaxebMJ7++3mnO/J9/nt9px3c0iO9Na7RoSlU0YxUv20c0zIWNKxkIbn/T6vyoqVtDzn9/DA4ayj6wzzVbCo3XjHxBp+97TmM+aUipyP2ttAcIv7rH3AO911ziId8qafcmrWKMqcdaAJklzkBuYQg8/fp81ZMFfwBW8zU2ofrKhPXcuRDVPQlAq/7oLWwwrsjcL0I4GPzgm5VUxPhxr8PbBfPwBJ7lepTi9L9see9dpSzAbTtc5ijMcYPwgkNFdud0761PQEPESU+eWE6SwEUe40IV9/7UDIrna1q129QSuLYuz8m4w5+w8++HMvBGpZ4P8fy4xzlj1dK7WLqRkHEssfCFSc1NBWUmwnI2wT1etBGnsZk9E30rGhY6x5bKBCsvmYjBtUMX8DcmCSBSXFJnI9KkxwxBrQSSBSs2KDaNiGOT7chiT6jAEcyqfkDKuGQw0p4hvp+uDF8PjU1HUs4+p9eO8Og+Ga7LGvY05kzlzWoDCTEedxIjFn3dvXZgRr3euy3zfnW/pS2GcL1lRRjJ3ct06Zh+8xcFJRznQRskgeM6U7lCknekkpORtt+tc/lCljE+hKHS5MDshJmSFBEB7C/vZlwoUuEYSZjDjTRa8ZSZV+7iStQpuYjh4qk4yND/SkylSoelR9bydsxfYTo46OKaM4GwuTmolUbGg4J2So+ChMTxOTmoZMg4i+kIIRJad6yVgqnpHrnEcAd1P3aHBYDjljxVzXXMiIl2XJt7oDrAHn4KPtBd+V76MK3273+HgzZ2Ea9qxFFbI4QrMiZBLOoaX33GPDEzIChWM/4rasaXGMNWdBR+EN/9uXNhxRcmh8ryX5o3bBMSXv9sf8hn2Zb3HX+JS9zbvcE9w2wUXrUmq+zd3i8/aEqRZcyppn9IivyEk/ORPZfu42dHhRrKbPZYYX5dxXeKDUEM6ZPiutV3Ij1D59UiwiXJl+JDF6EqInetSwyuLR33kVjOgV8NG5cM5k1vPU9XqrM3noT2tZhnNq1RVXfl+NWrI09TCPnvvjcR3PxT/dZOYve31z/Wfb1a52tatvkvrxj/ydF9P/M4Pwdz/y4y8ONSP/IVdayU72vXM2oZHRhiQITuGFibKT9CCwDdNLTWFa3U5i9uEEpY30GofnZXOGJ+gswm2eFs8o6kcswsxani5DA1Jl0gfBTXLhsDBU1vS0EsNVDcjDehEGt8PWIevTnEdL2GAoAPCynPKE378CvJz6fvrRRS3LsezhcBzrhBzDuS55oJdYDKc671PTG+04lCnHEUQoIcBwP4bltThmOmKmQQsxZRR+r64HKvsy4TQGIIbjtj0eSR9ySVj9NQgXuuRIZrSRkpSE62e6IMNyJLNeh5JcugwSk2S2BgEJgCzZxCmM4UAmJOcsoN9HqZIRQasdTh25hBX3LgYctlylCyXtSRFf28X9m86ZPIIiS0io93HyFMCmifbCaeJjep3TXS7o8JzIihzLu/113q+3uKV7HMo0TKfw/MHF1k3p3WaPj7bniMDvuTO+NZvyXVzrpwUAlQnbsHGeeecxAocUfJpzbsuKIwoupeZYRxxqSYvnATUnZs1XZcH73+x5Zt/w9oMMFz8rv2lfpcPzVJxqNZFSdSZr7suce2bJ2901AG7pHgsajnRKhqWhu6IP6sQHKqF4qri+7VFuy7o/x4fA1cTQwpQEv+6CNmbdDLRTX4PdVAwAyWODCh8qYxQTwUOeD3Ul7splswnHsRpdzfvIixYxirG+BxqJfqUqV3QgX/7Jn/im+Dv9jajdJGRXu9rVrt6glfQh30z133zwxRcgrGYHWlTeN5TjmBvRRl0IhCTtYSLHUHOxiand2UPraWkakug+SdCf9CIeYSENFsNYM16VFde0Yt9keFXGOXRRFFtYyIzhy+uGm3mOU6WMDWHr9RENyPB6aoVaDXz4u74OGS8ShNIOzx0zp6FlQ8vnzb0rYCqJsY9kxrkuB2nzlg7lQpZYDdqXY5niUc51GfI/WPXTjuuyj8XwJr1GhnCkExocZbx9X8d8ibs9aBlqcZIGZHhMznTRZ4woyhnheofjUlfB3WygHzmWGRppckcyw2J4oJfsyZiFhtX7M130Avq9aOmbUtfTY1K5gT6kd+ESSxNzSpJ+w6N02pFLTq0NldmubrcaVu7XWvMWuQUDEDKmCPkxBCCUqqHlmBkrCe5aF1H7kkuGFcNGG0JIY4YR4QZT3n8cBOqTixv8hg8AeSMdG3UUFv718ozvyw943gdQtpKW33fnAHyvHvavnRlofJjQqcIfdZc8J1OceE6jBuRZP+OOrCK9r6DF09CRYTg4WHN8vORzX9oH4HfsbTYE0P9v7FeBEDg4lzWHOuY97kk+YV/jSWbclUu+3z3Lio4zs0FUuC9BLzRnzR7hfJtEMf6wFtJwI9r5Jhe49HlIIMsCjVNOV3A42iKPzgnWhInIw9U5cwWIPFwPU7Bezz2rKDqcMz1AsTaJ0cP1stwK0R8uicJ3FylbO13In752IGRXu9rVrnb1DakEQDqCg06GDeFvkfJixJKyQlIwYUpYt7INrku6CiU0kTMZ9av3w8lI0pBUEYwYLPfMght+2ucYnEnNfbPgjmZsXGj6XlsI35KPmORbIPKyLHk2O+Qr65p3jitO6kAnUR+4IpaQE5KAiNPkHrWlHBxJwR1WFFgmPmchDcs4pwnhikEPkpP1TlLXZb+nJGWYmLzt+4lPACAzkvnvSEoudEmH455ecCjTIBxnwTXCRCOs8G81HktZss+k19Mk+l9KdAe4IQc4QvZIS9e/RiUFG2041TkGw1RGrDF9QyoIpxpoXUAfSHhN9nD4fjqQtCFwVZuSrp/pAo0ANa2+p/2ajn0Rpx8OT+evBselgEKD6S16VcOevC8XHBCshhNV7ZLVlXNJCVS885gcDjBiazlcEux+k730pa645kf8q5NzSix/7WiGORdOZMU5S3708AbOw3dmB/zb9oTvz44xwLv9MVaEMoZiQkgTL41hlhvubjo+wyXvMQd8wS3xErJlAO7IikOteq3Gq2bBExHc/OLvF/xnb864ddzwrosZr7hLvts/gQFOCedageGBXfBWf4iN7/s1M+cZPeJ37CsA7DPmNGqP0jkyZ92fO1dT54N4PdUqar8sREqjXJn2FFZoBwOuzCqdC9a9RhTvBGPDd6z2VKo+NPAhCtbr1ZUUdJUecABXfh5ed114H6rSi9IBnDc9QHn5H/3db7pFo/+/y37oQx/6wF/0RuxqV7va1a6++etf/fKvf0AjvQboefkGExvIGEaIZyg49/h+uiASdARGQgM+ltTspmY0NKHJhWk4kzBRaTKXmoU0LKThUta8oveZs+HM1HzRnPCyOce7gnmrXLSei0b5lL3PZQu/Y1/my13NV5hTuAJVqMSQiXDpHac0rNSxwbHBc4cNSxz3ooXqRhwTzfmiPWMpLZesgTBpCLoRj0oQSCeh9FiqkHWBcF1nIMJaGk4iVepUF1yTvTgBCML+FTV7MsFiONQpe4wjTEnC/W3A4CucUEjQ5xRY9nSExCyMoeA/2M+OGUvJOq6ih9uln1gsdMNMxgjCha6YSMk5Cxo6JlJxXy9Z07CkZiQFJUXv4rWhZcGaDS01LWsavHgWuulb2+H0YyiYNxj2ZUqI6gvvUUUppQjnhHa0GnQ3Hk8Wz7eZmTChCta7hByQJTVelUOZsmJLm5LkziZE2ho9VSvQA0NCzZiSWxzgRNlIx1Ja7q6CGcGInNuc85Xa8T3TGZ0TDn0VAC0h+6cQYWQNnSqdQmkNmQlWtrkxdE54oA0LaXGiqCjfXxxx1jn2KVjjqLBUmkUnq0ARvH9huVlZnjx0nJ6V0QHO8Hlzxg0d8wl7l5lW3DZzXjML3uVucV3HTLXASsZduWRDy3N6nVNZovGTlY5Hi6Ohi4GN0fhACE5e5HQR1OYidJrOHIn7MdCzageFkT5HJbMBhCiCzeLfAJNuU9rWYKOOJP8ak5GHazxpaBtL52wvNP9a5ZzBe8EYUDWYXnwe/vK88lM7APLvUztNyK52tatd7eobUv/swz/74j//8IsvJuej1BAOq4vqh0T9OWLaa0cS1amN9xk6IA1D9iA0RhWBfpOa1ZqOmpYsUsFW1FywRhBWuuaOntLQUtPwZXPGPVlzRsMDah4w58vmjIWuGZOzkpY/sHdxKOe+4yW34owGh3JJwzkN92RNRkhsL7GcScNtM2chDS2O+1yieBpt++32KK0GYLZd8ZeecqYoBzoZNN/CoUyj+1cQDB/rjLfKE1TkHOqEEsuYnEup+9dI+0hR3iTXWLLp6VslGSPNmGnV79ccG1t12+eQeHyf3XEse1H8vkeywE1BhkEH4XiglxzKlH2Z9BMaFxUK12W/p1l5QrbLscz6iVB4vmkPYIc5JSlpPdwvTNhEtjoNgIkZU0qBYHrvKodn4Vdc6ooz5pzpgkY7JlRYMVfCCtN3jccnbYNTHwILNTiwLXTNqc65lHXvdGaiDuTNOuOvmOv8uH47Uy35vx4sGOdQme04oLRCZoKAvDDbSYEqfLQ9Z5IJL5lzKizvswdUmvGc3+Ppw9DovypLanG8LHM6PHfMsldI3dYNWab88asZJ1KzT84n7D2u6ZgnTIXDcS5L3uaO+V73JJ+1D8gxfMreo9KM9/k3kWH4ipz0587QNjlZS3vZgn6HZyENL8klBQFYnfsu0LEIExERaLwGQG8F1aAPsQZc/POQ6FR+4JDlnfSTj69nApKqKDuMcYwnDaNRw3pd9JdhJfvd5Mo11ICkElFe+alvDq3eX0TtJiG72tWudrWrb1j9/Q/+7AupcVGUJ+WYS1YYkZ7yIiJs09O3YmiAXHK6mP8QwImhIu+b0rS6qmzF3GkCIPHLx3srypQRFyxAgp1tTs5S16gIN3TGV+0lOZbX5Jy5htyHtXRcsmTBhoqK37WvUlHyir2gIONle85aOl4158yoeNlesJCGcxOE1mvpYmp5aOTDe+fK5GESpx81DWPZ5lXsMeJEFniUQjIudcVYSh7oJStqSskZUbCWhpmOsEgkQIUph40r4xmWudQxh0RRCVMWg6WIdLhTWfavqxDJSWGfh+3L2RB0EHsy5kwX/YQjPc6jfXhhJWHqcaFLalpGUvYTjlOdc132GUnJSELa94q6n9wIghXTA9CCvAdpycxgTd1T1QRhozWZhDDEDTWdOkQMuYRsj3Q+OPE9ULJYDmRKhmETXy3db48xIympY+yjIabKh8VwOg0p9HsRfL3GGTfZx6rhJmOeKUqemAVA+a3lhKkvUA20q6Gzs9etyxok0wW4LhV/3CwJCeUFd3zNPgXvnY1YNUKhltf8hid0zIicc6kDSKXEh03k8+eebz/KOVsKczq+yxzzfD5mZA2+LbihUwosHZ59rfhDezdMrqTlrf6Al8wJHZ73+TdxRy77z/HQDGIUrZkTqA1TIssKx31Zc0zFWj2VBEhpJQKv6ERXd+F3VR7S0ZNjlpirblZZpGRl9vGp6K9XWe7YbEq6zlIUHU2dkcY6KSk9bLjg3PYFsziJsVkXXbt2E5A/a+00Ibva1a52tatvSCUAkkoQ7ugZGZZa2z4pOzQ2HolNXsoI2SPkV1QSQu0SZ7+mI8f2zVCakiTwsqHrxe8FOTNGnHAZphqyTbZGLDUNViwdHZ8xd3lGj9hIh1PPOlJzzqNlLcAfmzu8SY/4rLlDS8c9c05K41Y8nzZ36Og4lj02NDh89OraOgYV5L3WwcVGeK01ezKOEwLPTd3jRJZ8Qe4ypuwnJKMo5j+UKe2gYU6aidQIXsgGQRhrmA6cy5qZlghCScYt3UMQRpFWk2E41DGnsuRAJ5zKouf8G4Q1NRUF12Wf+3rBPT0nuVgBnAz2EdBrJZItrxXDHT3llhwBYapwqnNaOvZkQodjIqM+WR3opyLh/p6ZTFjqun/+tW5I+fWKD/SpRAtSJZcAXFQDUMkkQzXcnl4vHziztbhI1RomuPve2eucJXkMeRSEC5YYDGtqJlQ8JcchTBMoMXyyWXBDZ4zz0GgDOB+O029357zPHpCZINR20ZY2idSnmvO+0ZRPblpyLPOooamwOIVpCZ9btBxT8Z69irc/u+QzXz7m1aXjj7jkWEvWeF4xc97+LStOl9d4adXwA2+rcd5w/cYZq9+6wUnX8dZxwayCp67XvHr/Of7orKVWH9+l8G3+Fq+Yi/78TZOQ7bFREFhHi2ZDCDMsoi7nVV3xFGPOfcdYLGvnGYkhN8Ky8/E5DKtWeObI07mkzboKNDpnwAUQkpLQHwYkNvOUZcdqWeBVqKqWLPNIdMjrOtMDj7YJOSCJ3mWsw6pc+V2qnQD9z6d2dKxd7WpXu9rVN6SGAGSYuD2kZaWsjCRMh+CGVJD3drANbXROUprY0m8T17fi9FS2J8UEa9oVNa26aOd6NfguvTbAmpovywkP4kTA8Gjj4XB8Vc7CdqinVRdXxDtadf32nbOMdsHNAIBs318S44fXD1qXDEuL46bucSHrfgKQQvfSe1OUC11yQ/c40iklln0dBfAU+fozLUlJ8ilIL1n1pklDssfN40o4wIFOoqPW9Mr+3EQKmY0aHoPhUKZ9w+5wrHXDStc4HBkZE6nIyCijmL2NTertSINr6agkhAaOpexpamHqk/fnRhKEL3XNTMY9RUvi9oRAvfAVNAg5uYRckkpKRlKRSYZTx8xMmMqYI5mx1poWxyKCzaHuxMa91EVQbOJkJLmvLVgzlVGfEJ/AoKJc0xGVGP6T41l0ewog4/fdOa3CJ5oF75S9PhPDxY/Jx7ozahz3ZEmN49c29xlpOHr7FKykZYPjaKK8462n/PB7Gha0jArlxhP3+N7vuEOjyqGW1HiOKXneH5AXLW+91fKWUcHZZcmzb/0K49klS+f5obcpTx56nn/2kl/7nOFXzx7wQ+/s+MGncn7o7cpfdc9xzyxZUfNt/gkmlLzT37rymZizxqj0kz0PEWIrI81oxPEqK74ocz7NOV7hc37B/a7l1Hf8vp7ReGXjlMVm26YOqVgQJ0YDGtbjKFnBmtdRlB1V1SKidDGI0BjFWiXPHXnuqEYtGkGHc4LrMmzWUZRX7XqTEH1Hw/qz124Ssqtd7WpXu/qG1D//8IsvwtWJyMOaDtiueN+SQwzCKQuaGD44TCrv1PWC7dAsppwHT8qNcHg2NP2KraIs6KhpGEnFStd9eN0VIXbkx5wzp5MuNvAZa92A2LCCLpaFrvvGM6W+C3IF3ISpjKOl63UQCXRspwvb7JRZtKhNk5wTWfa0oCxup40C6DlrjnQaHIpEWRAsVnMsVg0XsuFYx5zJehBQCHtaci5hipCrJVNDJwkIhpXhhTTsaxVBi+/DM12cFaRpR0bGscyoKNjQcKYLOu2YJEthMTynN7juJjhRPi4vB0AV3awslhtywG09xWIok7BZQru/psZFJ66FrtHBvkW2YMGrozB5b2WcpihD3ZGiWDHc4CA6YNmod8m5FvUsQBDSD9yv5qyZUsbzac01ZsxZB+ApYWIy13UPqtfUIa9FZryFYIu7bqDMtzqHd5t9Pt5d8APjA1oP/7q+D8D32WN+x50C8F45pFXliywYkyNApZZ78XgKwrIWirKhHNVc44jjvaB5ODvd5yCzzFvDd07HjAo43usYTy/IsiOef6Ll6WfuM7r1Cl/4ne/nv/jOJWVV4+8e8LHP7PGKXoTj9dKI391cMovxokmsv5aOFQ0vmQdXPucOz1kE7qkOdcKajjE5a+niQgE8MCsqlzE3DaesuS0XODyvaZgAfXDxVp7AMK38I8GCEKYhef76dKyiCMB1KD5/eIqRtB90Gd5Lf3vXCd7njzhmeWf46j/+8R0A+XOoHQjZ1a52tatdfUPrYUpWqjQREYRKQozcfQLtI1n5MnisFUPH0Iq16AXcKaAu0bRCwOG20fexkRZJtqJXV1EbWgpyCnIWug6J3GLJJa30hyqj+L2l6zetiLkUhWQki+E+gDDRgwhheWmq00R9SAIYqcFMWSHD/TZjxJw1NS1LNtQS6Gh3OAsNIAtuyAEAhzqKRCiPGwCzFS0HMagQoI6NYaamB3nHMdgwgaF9HXEpdX89bS/Afb3guuz3wMSK5Xlu4VEqDVOIC1PzpJ/SxsT0lLx+KNN+emXj+/5e9wyfsQ/Y14p7Mu9F3oVsndFO/QWXMV3dYhAROu2Y65KRBBn9SMpgdiBbI4NkApDsgVPGRQbUdCzZUJBTIaxoesetOoK4tdZciGXBmn2ZsKGlisnvyW45Taju6yW/YRtu6h7j1ZN8wl/wPDOuFxmFEb5d9vAKrVe+Q45YesfvuTOUcP3LfvX/sfemsZKl533f73nfs9Redde+vffMcHoWLrNwGIo0SY2ofVdiUkZgJ7ADw1AkW7GNJAhiwaOJDfhLYiCQYRtCPiQxDDuiJCgRmMgyQXG0kBS34ZCctXu6e3q7fbvvvXVrrzrL++TDe8659w5HipYZyULqPyh0dd2quqfOOdXz/s/zX6hjOUUDFC7JgBPa4Jw2ucWUHZlSj2qIUfZ3e/zNj/apNaYYm7F58i720jl+9EHDqdO3Aag1xswmHdbXh4hRVu5/mW8991E6nTm9tT3+1b8/TyzCa/mIGMsI+Nxil7nJ6GP4j1ub/PuJPxYdjVFR5qSc1zWuiydOZVLW0clhXyZYDKNioifiJ2+nXY+PNnr83/O7/nk6Zk5CnZjvcQ/xe26XDw7XOCeGlSirYnqrfwNMedHAkwdrjn+PpSDWbyYeWWaqBnaXW2yQFTcfx3t0WuJTsQqJlsnfsjF9iT8ZlnKsJZZYYokl/kxRJmQdnTwAxXVWv8AsE4jAE5BywVoukK0YVorkLDhsUS/lTlGh1bcYWhxekS+TtsAvoo/G1YI3X5dTiBmLqr+iXnhHHEq78D0ALEiYq5+0JKQkpMd8DOXkoxSEAUzVL+TL9z5qvLdY+jqqFuUBljVtV5HDXvozZ6jTSj7Vpla9R64+dayhnhwNZM5A5sV7G/oyw2KoETAQH4sbqKkICIApFmyhWoxKtX+8JCwvpFum2p7S1F3uU/DTkYZGnHVdtlyTKQmnXIu+zKlJRCQBfR1zVw+qCU05wcgL6dCj+QaxBpWM7IT6ScUp7SEIq6bLopiaCUJc+GPKwssZC/Z1yFCnlUxOEFrUmZP4SQZFuhUpi4LQlsfSez4sgiHTnBkLhuolgUOdkqurygz7OqZBzArN6vNE+G0f6pR9mfCrvM5XzRsstFwY+zjaWeZ9IU1rWAsCFpLzflklFuGCaTCmTJMSuhqzLjEDUlqERGr5v+4OuXz5BCfPbJMsIi6/dpab184zOujxyOmMs+e3afXu0V7Zob55i53tNabTGoODFjqPuO9dN3HOoCp84sN7jFzOWBL6MudJWeX7Guv0XI1YLdb49LQaITfMgIcKKda2DKpz+FBWqNWtlAVOSar9/ER+ilRyfmHxIrFazrg2j3KaXHMeZAsB7poxn9M7qEKSmGMEJDxy34h+W6Fgo+nPDecsxuQYc/hvytHpRklQpHgPG2TEtYQwSnFFaWn5elXzlsWFS/zJsCQhSyyxxBJL/JmilGP1CoNvSUYWhWyqXLjf0X71mgGTY5GrAKNiAVhKp8pW76C4el8SlNJQXuIoSXhzF8aadI55LspFVKn9V3XUCOmZTvUeR/0oR70V5WuPvo8taE6qeSXZygtfxoZ0WZM269KtJFE5rvB4NFjTtp906LggIwsiQqYkOFXqxFU6k58Z+VnFitYRDC2NaGmEUd8Y79/bT3ZKAuJEq4jV8s+S4CzI2NUhZYxuiZLMAZyWdbakx2lZwyJEGCzCSdcu5lxaye0iCWhJg35RflcWTIZYdlnwnqjJw9rjvflJBGFTW3xSH+JRt8Zp7VEjpCYxC00IsIXh3C92M82O/Vl6VDLN2ddR5SuZsWDEjBGzY4S4vD9hjuKncIkeTt2sGCIJfGCBLo699qR6+VXjiJyrr2P6jMk056qMsMZ7QlTha+mAz6d9VD0xMSq8rCN+W3fYdgvOSIMrxT6qYRlpRqtwvPSIGElCf2xYzOs0OyPe8+QLNBoLemt73PfAbdrrN6vtSA9WubsfMRrFBEHO/OAEN984xWgU099do9Ea8pFzwrulx7rW2XELHj034zQNzmmLf3dw4H02BOwz5qrZpUlMcmQiWSNktSjGrB1JMZsW3+/yO35PZrzELYbqCyADhK7W+GF9jAuuS4DwofwUA5lze+SjfEuUBCSK3lpmFcVZRRaOko/q+Fl3jEzkWUCW+n9fSjIiotU0xFjn+0Fs/sdK4lriD8dSjrXEEkssscQ7jp/6xDPPAJyQleqxQynNoVyphKLMWFRX5Y0elsKVUqoASyAWUxQYgr8iHhFUxWlD9QlGVgwTnVeL5ZIo5Di60ip+p19Yr0u3umpbPj7WGS1pUCsNzoRYaR/xEKRMdF4RH1PITfIjC/Sq/Z38cBKifvrSlFpFPAAOdEwsIRbDJblDVMQQJ5oSFrGzp4uJwE3pV/sMoEHEWJLK7xBiWdE6I1mwqg32zJSu1o5NYACG4hfT7WLxbI4s6koZUylhKh8ThDOyzi3dY0f7vI/zgJ+6bGmDDGVf5pzRFldlSI2A73QP8g27zaP5Cb5or2Ix7OnIx/gSkpHz1x8OMTLn/IOXuXHlAX79Ww+wGQV8+JEJv/HNenWsF5p4o3txrJrSYKrl5McSiDeo1yVmRHG+ifeZWHwKWp2YOUnldTgqEUw0q4hkuSccjog6TbwcqTxui8K3FBFwwa0RYbll9sjUCw036GFE+Kbc5kfjNR5Pu1iBgVnwIdkA4Ctun76ZMfE0wDEAACAASURBVNWEp3STPRJqIqw5Xy4YYrghY7a0wS0zZs3VcaLkDlrdXUb9DYZ7p3jtapfO3RYPvGubN15+jAuPfQV1hitff4o7Q8FpSH8UkqZnyXLDa9shq4OQ6TQijhwfOi/0ug5jlGs327xvRfhGP2HbjHnQbfANc6uacCRkPOS2uGp2SciYkTAnpUF05Dvkv7ddvDzwI/k5ftNeZuJmfJx3s6l13teq8fzYnztfsdvMSfjO/AJP5Sf5DXOLi3rq2+RYJZxKRSps4EgWAWl6uMSNouPm8iBQRByqRYywSuUNOToZKdvRjShOPHG59l//50s/yNuE5SRkiSWWWGKJdxQ//YlnnymnBHf1gBzHunSrRXNL6nSPLG6rhfwR2Q9QiKOyioCUBAbKKYh/bkxYkJC0ktY4PYzvPVrWZxAaRIXEJDpGGEIsMQFjndGTdjU1KMlQeGSRXyMixx3rlQAq43mZsuRQQrEVCepKqzKiT3RelA4exhWX8Clb3py9RpuH1E8HAgxdGhgR5iSsHSkLTMkqeZrF0FZPLjoaM5B5RaAycWTiKgJiVI4RkDIpa19HHOjkmNbf7/u8OF6GsSTEGviWbQxzchaSE2NY0Ro3zYAbZsCEBZfsHk+4cwTFZy2N34Jw8akvceV2nb07p+l0Jvy1j/T5rsf7pIllM7ZccD3WtFn5b8rjllXbIsXkw58XE51VHhGfMGaqhCvDYddFeb88zoH48ysUW91M4enp65iIkC4NujQIMNSIOOla7JoJD+KnamvSqQoSG8Q0iNidKqEIX0oH/lwqLvM/JivEaokIuMKYW2bMa85PQaZkJDhaGnLNDGi7iLNS53vCTZ54ZJ+ot8eNG+vs3Vvh1gBOnRyQLmLqjTkYZdHfJE0tTpU7Q7hxAAeDiDt7EZ+d73J7qHz6FcPeQchgHNDpjag35oRWeWPo+LLZYS4pL5s71AsvVLOIi75i7pFU7iNPTrpap0FUPfZX9VHOuRUezU9wTUYM1ZvX92VKjOH6JONls8+CnEUhVQwQYgx7MsK9RSxumh6RUpaTu6yIaXb+FoYZWnhGVOXQC+Lssfd6s8zKiFY3YOkFeQewnIQsscQSSyzxjuG//MTPP3P0ynmJA51UV9LHOquMvKXUo5Q/AZgjP3szMjJifKlfKZvqM6mIR70o+pvpoiICQLUwL++X5uYZCTNdkJEfW4yWU4MAw54OAU+WNqVXTQn8th5uo5c7+W3z7wNlClbZC1KSlKyQ/KzR4YAJtWLaURrvy0lPhwab2jpGarpaI5c2d3VALo5Jsbju65gLbHJPRqTkDJmyTpc9hmzQrbpDOhoXaVshRoVUcsLiCnEpycrwBvKyP6PE0WNWfqaFZIRqSAvC97rc4zFZ5Sx1TuSnGZDijL8K+rrZ9WlY4hewq9qiqzUkusUshVdeX+XUxhznhHv9iDQX/tLju1z7Qoe+TGnRoCERE+YMdFyZ/gOJiYvSvBmLIu7YG/iPRu+W/wUEx6ZxZRoagMWRq49N9uTO+0LKiUqA5aR2mZJyQ3b5seA8N3XItPA1dWlQk4gZC27pHpvS5YtpnwDDe02HLxTyNCveIyLOkyGrwpOscVnG3GHKljYYkjKWlC3XooblZQZ8lDU6KwfoPGJra8Cv/n6Xh1aEmzd7vP9jn+fg9gP81q9/F3GoZDkkDubOcbZlWV2Z88J2nQe0w0GW07UBl/aVC13vEXn10gav7SoTl3NRV/m63SbAMiPhve4U18x+8b2wldfje/J3cSaImasyzx2/aa/wE/ouXtQhz5vr5OqqaWZT6my4JjUx5MBPhGf4THqXj+XnecHc45p4f9T9boN7I+Fkz9eYVsfpCDFxKsekUlF8KBE7mo715tdV39fscElsbCGHdKYyu8sRQrLE24NlY/oSSyyxxBLvCP72J/7RM8CxRTqU6VT+f+zmiDHcy1xC1qRNQ2Jy8f0fDYkrU7nFsCk9FuJnIoLgxFUpVRZbLfoVkOIKc85hk/bRBbwAoQQVCXIodYkYMSuK6AIyfO/HnAWzQtdeXt2dsmDKgly8Ibz8bOVURIvnlIb4chrjr7ALofgr5AtS1vA+k3O6Spc6PRqc0h67MmaVNoJwwa0QYslFWZAVcafCmrYQ43tF5ppU0qCRzJio325faJgx0wUn6RJiGcqMNjWUonNECmIhUvWMjGRBjYCF+M9n8L0YIQEnpMcBk2rft6VOixjwXRY5SigBfRIu2AZPnjQ8vGLZmvd4d9Dh+XyvIHj+2D7mTvC3Hzdko1X6/SZf2p/TI+bSjuXlUcJGFNCMhb1+zNfxiUoxAXlFGB1G/GQqkoBQAhaaYMQQEjAjoSm1amLiSYpvQC/DElrUi6v6DinOnTKZrU29ksnlb5rIGeBHeICWNVyULk1jucyIBSmr2kRFaEjEhra5bQbMJOM72z16aR0jwu9n+5wxDba0yUka3GDCtPANNQlJUe4anzL1Pa0VwtzS1pgHV4Vs0WQ6XGdvr8lwHJA7oRYK6XiD0bDBb99wLBaWL47GXNIR522DYaJMxzEHqWNfU54395ipMneOL80HTG5ucG8KiSq/aa5x0wwxCBfcKifUn6sBlr/VfIA7C8d5t8IFXWVBzmfkKg1XZ18WfJiTBCJ0NcJqzA3ZpyzpTEjZpEeucEDCNFeaGvJFe5Pv1rPUNOQVu0tPazxar2MEolCxR3Q8zgnO+UJBRVCEuHY0NY+qbNCTjz+oaFCqmzoDcvgab3z3zzr48GPP/QFvsMQfE0sSssQSSyyxxNuOn/vkP30mLcRPJcrF+1GJFRyPeu1JiylJpfFfkw6zwnhtxRRX2v2UY0M65KLVQhD8JKJHiykLetJiQUqqeSW3ORRKeZKi+KlEUBCRvo5oSo0JPsVJRKoELcFUn0eBGlFVoGcwxXb49u2G+IW9X9x6ZFVXiKk+cRnrmmhGW+psaIuaBoQYmhoxMxmrtLgrQ87pKgoVOZBiwbQQ33CeSM6clKRoiF+XLtOjjeOkzEmwUki4MNSL2YDfPkfZA1La9UeyoKVR5RfRYh+PmeFwZEVHRiwhCVnRiu19F00NWRT0YCBzzmqLR04n3DuIOL+ZsNbOGe612NIOKzTZ0BYNLCuuydcuN7k+zdllwWxmuZF7YdnjG5Z6PUOykL2pT5J6PN9iYBIO8P0kdYmPEEKtiKgvZ8yYkyLij1+JmLAinCGWOhGLIhbYoRgxzDQhkpAVbZEUhGysPmHsFD1WtcHFuMF6U+jUhK9MJnxLbpGQ8YN6Pw/TY5eUoczpaI035B4fr5/kC5Mh58Iar+cTbuuMbWbcY+7PT1G2aHDNDBlLwoNuhYEktLMa57uWM21ha23Og+95gRe+cR8v7wgvZxNiF7A7Vd7YDfme7/oyNy9fwAB1DdigxlU3ZdVEvLyYsmFDflOucp9bRYAX7Db7MiHIa7wqB2xonRXX9Olf2uBVs8OmtvnxlVVOZC1ems15l2mxpwlNQp63txkx46R2ycQx1pwdnbMrnoyfokdL6vxH7ixP6CnOmSa/Z27yQ7WT9DM/TTunPX7f3uFjtTUepMum1DjZAWO8n8Me+yfEfw+OtpqXkqsShz/7ozWdOye43OJyC2qw9ohfa0lC3jYsPSFLLLHEEku8rfi5T/7TZ8r75k1JVXAo4Tn6WK8whw91So5PjnKqVUu6ESEtIlbLON4DJiSaVh6N0sx+V32CT/Xa4kp1dCSpp3wcoFnE7yZkZJqxqz5u1Onx7SydHKW+XxBqRL6bu5BPhWKpE2OK+xFhRboUZVO8ObksMFzTJkOdYMQvdlsakZBT0wAV6LqYUC1NatQ0IJW8mghNJSUpihQjDRgwZUHCunQIsFWKVrkY9yb8Jl2aFfEQpJJXvTnN60D8sSijecvnpeSVh6dFnV0d0KFRmPAtEYda+1QcqTjaWsOIcPNunZ2R8sL1kK+8EXDG1vhAs8XT9RX+q8csF8I6vVbG1XTOgSYIcKAJc3JylKu7luvbDXqtnB9dWWHCnDqWmnq5WllOWcYGWzHUJGJF2jSkRlz4fnybfcaIWWVKn7KgQcwUL917wp2roqINQkf8QlxRDtTH9zalVvzOlBNaZ5w6vrA/I83hkbCJw/Hj7t2shZbzHcuTusGqNniNbTbp8fn9KRdNi4Xz+30uGankPFYEOJR9MQZh3TWYkvGDjQ1iYxCBjZWElbURYnKyXBCBd9kmY5fzu+ywXjN86ysfYJQ5Jrnyio6oG8NUMl7PJ5yzdT6tN3gqP8NtMyLC8kh+AodyzR7Q1RpflXt8oN3gu5srhGq46Da5avY48F2X7MuCV3SEQeiL//6lmvMNe5tXzQ5XbL86p56yK6TieK/b4PP2DT5n3+AzcgOH8iuLm1wyB9yTOQY4l3fJHLQj4f4V/9n+KHKoWu24Cf3NUiygit59K5Q/C4KMIMiOpWu9lS9liT85lpOQJZZYYokl3lZ8/Cc/9NxnP/WFp2MJqUlUyW1KS3h5Nb0rLeaFVKlc4HakSZ2ICQsMQlvqlZTKii+kM+JV4WWxYVkgeLSzoictprqgLQ1mJBik8pkIVBMZwZOPBUnVop4VV+8NBiuGXlFIBxSyG9/fUW4PQE1iVspFqhzpyxBDjo+e7UqLsc5JiuZ0i6HPhJbUmWtCVxrMyBibBV2tMTQLmhoyNklhgPfTjqQw4S/E77OaBswkBfGfZaILOtIAkYrcCNCVJlHRxRIVROooyuXdG7IHcuiV2WPEVBe0pMaAKTVCBkWyWS6+/T2SgA3psulabGgDgES8lCtUS4ylpSGjObznlDKYGuZOWTjHKHXcTTNu3I25lc+50TdFeSNEmILG+W0dZ447s5ydgeHCClwea+FpsUxMSkeaxcSMajIzdH5b+zqkYWq0pEYmeRVEkBU9Lpk6cnF0aDBlzrYM+JngcV7XCQk5Y51Sk4i5pNQl5kAn1CWiLlHRFu94JG7yS/oqH29uME3gUXeSjSAgDoRJoryUj3i/WeH3ucWMhO8NzvIVt89NN2MuWTE9c9xhTo2Ada1hgA/Vejy5GvNQO2I4F+a5slIXQgurawNm41W+dKnJ1XzGS9LnFA32JeHBoMnVfcEI3M399+Dz5g7nXYc1YnLgDlNOaJPbZkRXa7xkd0jJiQiYkbGpLYI0JM2h7zJ6RHx//QSvzObczhfsmilfl+vMTc5tGXCRLe4yZMqcRzjFx9hiRWI+strkf51fpS8TDiQhF0edmHXX5BG3xkN0WdEaA0m4bPs0iHhXXKcW+qZ5EX+LQncsrreEtUocZ8ejfN+iKf1o+eBbQdX/PM+9ZFL1UJoFMPhLy0nI24XlJGSJJZZYYom3Hf/4U3//2UMZ06EHxB5ZUJYRveATsspityFTHN5jMWZGqjkt6jj1ZuGjfSGljKssDDSFFKicgpQdIRl55Rspt+NYUpbmlRE5KiRKoVhydYyZVb+rkl8VscArtDghK4UPAgQvdSrfVzCsSweDYaTTIpHLFD6TwjNSRAfvMSQtiMXA+O0eio87jQkq0jGXjLF4uVppILfFtAP8NKAkdUERTwww0lm1X8aScIk73JMRinJZdnhD9tiVcZV4VB6nLk16RUFjrp74rRSTq9JH4VCsGjoa0SWsjPyCMJeUFY0JEHLg5p5lkOYYYKaORJUhKfvOk8RRkceV40OTmxJgis+Y4hiTcUNnWKPUNWBEykdqvaqlOyhM8hbDvJBQzViQ45vaMxxNauzpEIup0rIUJdOc09ojKfpFfifd4+ncxw6XhLosKPwp8wR9HRMTsKUttrSBFXjEbfHruwO+mB7QtobQCNNMSdUf17oVTss6ivJ8NmAuGXM59DAIwpO6RopjREqKY2+RE1rl1XtKr+5DDrZHytmzu4hRrl3b4HTbsEpEX2ZcZsRHzAZJDr/j7vKNfMiaCYuLAMo1M6BpDPua8KTbJMFxPu+Ri099e8RtccZ1WdcGVoU33JTPuNu8andpEHBnnvORjZiBLOi5mvcfMeeMrvB+Xedv8gRP6f1ccB22GpbTDcs8hREz79fBctr1WHdNGoT0JKJpDRtByHuly3fraR63XYxAM/Z+jMAqYaiYP2QYUSZcvdX0o8RRAlJOPUpfydG/H31cy9LCpTH9bcWShCyxxBJLLPGO4NlP/d1nn/3U3322XLiXV9bhsCSw/PtY/UI/J6+K5BRloX4CEROwIR2yIoGplDQJUpXfgScjAQGZ5oRiq4K5UhamRxbqJXxsblgUBnaKSYEnOkZ80tThcx1r0qZLkwYRZ9wKimNdW9zn1gDYZ1z9Hosppjqm+nxVx8kR2VJemNl3GbHLiIb6PpK5pIwlYVd8NO68WFyHaqhr4DsiCsIQE9IrCEObmu+xKH8mET1pkqpPydrRPj3xsqyb0vcL8GJCYxAOmFSG8/JzpOT0pFl5Qrz/psmchH0dcYnbDGTBbZnS0ZhbZkhKzlSKY2gMncDw6LkZ3dAydQ6L0CchxLDAkaPERzwqC3IsMCVnTw57WAT43ZuOFa0xlISnHh7SIKJNnXXatKRxTO4XYIklJsAw1TkZjlhCYgK6NFAcsUS8l7M0NOQT7r0IhrPa4j3dqNo3c9IjxFX5GfN+AL5q3uC37VU+O9tnJAt2zJivmje4nM74l9m3inMQPsg6v5Pusa37JKTMiljbKo0M5YO6yTc5YC4pM8m4I1MmzvHiXfjAhYzNXkoowtOPDWj19rn6+knGU8vzBwsaxpdSpjiuZ3NGmeNxXeeujAmKqdiUBSddixxP9EZk3DETXrF3uWWG1fftthkSqeWq7bOQvCK+KY6JyxGBi3RZo8achPe4U1gMgQhf1z6p5FyIYj43PuB/mV1GFdrUmRZTzova5SHtck6btIzBihAYaAaGE3XLSmyIAyoZVhC4b4vRPYr4SBpWln378vatHoM/eDKiRx4So7h8uWR+u7GUYy2xxBJLLPGO4od/8unnfvgnn37u13/pt54uF+NlUaAv4fP/GQxdaRULxpBYIppSY8yMKQmZeK9IRo6IVPKhNWkzZl4Z3CMJsIVHIy96O9akw4gpAQFH07rCwicCXmr1oJ7AGa18ASvSIiLkLKvVIswnLMVccKt0NKZDnbZGpOJY0To9bXBXhpXUpyRNil8Ml1KwKhpYDIHYYtt8ueGKNlFRhrKgrTGCMJEEJ55U1Ao51VwyMnG+JV4UEE5om4iABhFb2mFPJsx0UbVaV94PEUY6RcWTvbItvtwXYeF+SfGN7kOmTHXBmDkr0mJOQiCWsU7JxREQUDPeDzORhAYRBzKjrTEJjqlzDPKc+bDOd71/j6/cCMkqI7yfdLSKpKspGQGGTYnpa0qDgIfDFrfdnAYBCY5TNmZb5zwVrtAKAgb9Bj/U3GC8MFySHea6oC41EjICAhJNyVEycgLxE5CBTmkUnqAFCUNZcEP2eB8n+IH6SX7FXebhfI1H3CZf4hZBldJm+Sp3+PGVTX5tdpWEjCf1PI+ZHokTppKyw4Dvs+f4juAEryZTrumU60wIMNySAxTlnox4rzvJTDJycXwHJ3iBPlNJ+BAneChqclLq9F3KSxxwxnUIjPDYgyN6a/tcuXSBb94I2J44vib3OKVNtmVGjYAdM6HuIp43d/mwblEzhhc54JzrEWJ4Wfq01U8Haxqwb+YFOVe2ZeAlk9TZkwljSTjtOuzKmB0zpaYRr09SPrpZYzwzdLTDSBY87FZ4/2bAp2e32ZMJr+iAbTNkTsbryYKxzGlTp60xq9RYsQF1a4itl4wZoBH6+4GBXtNhDIShn4aEb1FUWKJWJGIFgX+NtcfN6eYtOMShN0QK/4erbtY6nDPF4woC1/+b/2xZVPg2YknrllhiiSWW+DPBL/7y//DsP//lZ579F7/8888KgqqruiXATxmGOqmM4QMdc1AUmjkcU11UEwOnWkmELJbNovwwloicnERTEk2rKNa+jqtiQ2/0NVUzemlsB4ixnHG9qpV8yJQFKU2NqskCQEhQeS1qao8ZuvfMtPg9piA8lnW6hFI0vHMYSVwa7QG61LEFSYmxWD28qi0IodpKRhZiGcmClIyUjJHMidVW5CkofrcT5aKeYF06hGIradaKtHhAN7BiqmnTUY9IWLhfyj6Vu8UxOS1rrEmblJyONGhT50FzhvNygk3pAn6qs+rqVeHhVFL6MueaGTAmJVcYjxr0TFDsc1Mdg0mVNiasi+/5eChusGEjalY4KTWmZNw1U1Zr/nWvphOu3ws4b+t0G8ojQYuMrPIKZepzvyLx062FJkx1wVAnPoK4kNvJEd/Qb5gr/Iv5K/xlfZBcITRybP/MSWkSszcR/pY8SYAlVMNabPim3eZp2cIgbDSEry6G9GVBW0NmkvJu6VUyxTkJu2aGE+XDbHqJmqTUCFkNLdbAlXSGAy9bajqCwNHv1/nn/+40AJEVWoHhrOtwRxecdm0AGhpy2XgyEYiwm6Vsugav2LtcNwN2ZMgVc0CG45oZFOEDfv9fdJuccT1eNNuccT1SsmpKsiDldbvPVDL+9b1dTjcsm1rjolvhkhnwtbs5ExbUCCk7cBpE7MuYE9ph3TW5qF1WbYAVITKCAZqhUC8IiBVo17WQYPkJSElA8rcwlbfbhylwIkqef/tz3jxFcU4qn4n/u/UTj6LEUIwSBBliFFXhxn/715YE5G3GchKyxBJLLLHEnzl+7f/4zNPgezyOJlB1pEks/upsLCETZihKr1j4mmKqMScpkqFgXvgmyoZri6EhcdXPUergoVxo+wnJlHlVKGixnJAVOuoL6WaSMWWBAnWJWNUGQ5nTkgYT5iTiPSRrWkfxKVClXGpHBkVELIUcylS+knICJAg9adGQmDkpRnzS1KKQ/fS0UcXAZjimkhARkEnuywlxTMXXAYYEnpiJj5otQwBKCVpCxgoNrFgSMhpFgWOAbwDPxXlDd0WO/ITmng6ZFfvZTw8sExaeDIonhZEEbLkOLfUTkCYRdQ1Yo0YqyorW2ZEhRgxOlBoBYR6w3495aEMYTYVMYdVEZHpIhFYkomksL2gfzQx9TdkIQlYjS54ZRA3n4oiui7msI26nCR1CArUkDr6iOyhKUpCRkIC6RCRkNE2dqc45ZdYwYioZ17xopS+jfBekfG/tFJl6s/KTbPF17haJaIYDnfAtNyDMa3yMs/yWvcrJdIUJju9ot/lcss030hFr2mRkFuTAQjL21Zcalolt/4l5gBNa52va55ZMidTyhKzxUjamn+W0CDggIcZyKow5uTHj7n6N1SBAncHlwjiFL3OPDOV1u8esSFHblymPug2+Kvc4kIRrZp+AgPvcCuvaZN/MWNU6DQ25ZybVd2QqWVFmCWNZcMb16MuU826VntbpqJe2XTd92osWHz0Zcm8iTMk5IGFfpoRYYkICDAsyaoS0iLlfO6zbkMB4+VV5NTwOPCmwAoGFWghh6KcZRycgb/aE1OKsiuT1fx6P6z0KTzikun+0tFBVQA15LgSlp0T9Ni0JyDuDJQlZYokllljizxw/9lc+/tyP/ZWPP/f5T33t6UjCopPBkJASS8hAx9QlrozlSVG+pyiLIqmq9JSsSos5KTXxC+GyrM/TjUPZ1krRG1KaqctW7DK9a8qcC7qGEd8AvkmHPRkz1QVbdOlpnVVtsEmHVJR7MmCDDhZDXKRXhVha1LgrIx/dS0BapXL53xuIRcQnbDWp0ZAaDWK2XIceDZx4P0RP62RFYaNKERNLjWmxLzrUKPtJArFkOFoaMZI5iWRQeAByUX91XQOa1HDi6GqDmaQ0NOJAJr5RXWc0i34Th9KUGnWJqRFVC3mDoS4R63QwYmgVGVNntUWglk1t0Cmayl+ze/RlBigNjagTkkjORdPh4rrwyMPbmFmHsy3LyZYv5Hs98VKiBMdUc85KgwEpt80ESQM+/vCCbFIjSYUvpwMerjX4/nMBnaTJtWTOi9mIWa6s0WFNOuwxLtKvDKaQvQH0xE8L5iQ0iOkzYagTQgmqqYwgZGmNB2t1fi/pc8bUucSQB90mBzJjyoI5KbfNgI9Hp3jKbvJFt0dKzs7csUmHntZpFsRwIoknegJOhCfdWW5KH1yNi1GTF12fHKVByEQL8iEJ++JJ7zo1zjYCQiPkudCqOX7xzl3eSBacNDVu6pSBmdPQiEwcc8kIsIwlxRTHdCAz3xdjHC1ibskBY5MyMHNAOedW6KmXS3W1xkj8928iKedcj1fNXba0w00zYE88adk2Uy6Ncp6stzghMVluiIj46RMn+X8m24RYtrRLjZCO1jgpdSLjpzO24AD1UKqJhClISBj4TpCSgBjrJxLGKvV6hjXeJxJGeeHpKEsLj8uwjuJYw/pbTFTK6Yi1DnXipyBOGH7kfctErHcASznWEkssscQSf24oE7RKlClaR7s8VqRNphlhkV511OjuJTyLSmqVFFOFtWKRKUiRTiXVNCIj5/16X9X3YTCsSAuH8oY9QBQ6GtPQEKeH3Rk1DWhpSEND7nerlbndIqxWje1CiOWibtGhwQpNyp6HiJA12rzfnec97gwX3SYnC+nMWdclxhKrZdO1SMVPfXyaltAqtPteiuX3mY/69f8jDwvp1lRSBFPJugCM+kb2kcwZyLQq6WtoxEK8X6JNvTKfG8THzxJRL676J3iJUIOIHk0C9SlgsVpaGhWelZQQw5SMPotqmlAjwuElXudch15kWOkkDPodksybjpv1nFvTnAYBg6Lno1xK3pUZF1yHOTkHw5jQwmYYEKvl9ixjPA1Q9YlRHY1Yk4gnbY9ADR1pEBTip0Qzvy2q1eRjgy4j5jSJuU+2sHhvjiDk6tiVCb87GfB9zVVE4Cf0Ab63ucqQaTW1memCX0xf4guLg6JF3RTnqTfOgy9rrGlASk7bRZx1K0RYchxfkMtQ7J8Qy6xIP1uTiE2tkeI4oQ3e3apx5cCxNwwQgYcfvY7FsG1GvJyNGRWFkotiCtLVGXs18QAAIABJREFUGg/mq8VxzhnJgify04RYzuQdrph93puf5JzrMSPhvFslJuA14w3q102fc26Fc26FAyZV0MBVs+dLQAupY0LKdbPP9ZmX2t1lxobW2B+boufE748FOffTIi3kh0ejdN883ahHXooFUFSo4HJPQFwuqApioFAXoioEgZeq6f9Hl8fRFKw3w09HIE1CxJTk5w8mNUv86RD8eW/AEkssscQS///FP/rU3/tDZQ7//Sf/p2f6OkLE0JUmuzpAEFpSp68jwDLROXXxfo1cPTXpyxgrxi8kdUhOTqaecBgMFuE9nMFieF3uUSOiLjGr2vA+EVVuWq+nb9PEqp92gPeNjElZp1tt5z5JtegP1dDTGrZwwq5rk20z4Izr+cW4GixhJeEqfSkWKkJ20rW5ZYZ0tcbiiFa/9Gg4fJRq+TvhMPGroVHlB8nE0dCQrtYZyZyO1pnKoiqOtlg26DAqZG9jZqzQoiMNdjhglRYpOWt0/GfXoJLOxeq7SkJnKt/K83aHc3mXy3a38ALEGBXWtE5LfXXjIlfO3XeLNInQmy32x0JglY3Ysj8TYqxfwEtIjpJKjlPljK1hjU9lqofCx6Iun57c443tmLEkLEzOWddmqBlttTTVp4UdLa1MSFmRFvuMOa9rdLXG05zhEkO+v7nGL01v0daYp8wq/5KvsStjbto+PxR3GSZw1U35ldmtysfjU7MMY53xuO2yUuvxq9Nt9s2MUP3nWEhOR2MylDVtMJWMBiEfbrV5bmr4oN7PNxZjnpR1QhF+jx1iLBtxQL6Ak+q9T60Y0tzw1JNXSRY1Lr92loHcpqkRr9rd6lwqYZCqKrMswrxjfIHl63afc64HwBWzXz1fFN7lNogLz9NNM8ChPOK2sBhO0+G66fvwAfxk55zzxYqfMzf5WHqGVBxrRLw2SplbL5PcFceqNrmlMy7a1rHveD18K/+G7/2wRsmdJx/giUjtSAqWf64ea0h/qwStox0hqscJ0OFzDrtIymQsdYL8IWb4Jf50WJKQJZZYYokl/oOFrczdhj0deomNDqvFVk7OSVkt4mRt5f3I9XBRYhB60uFAfcxtXWKcQqQBIYZ360kU5SzdaoEfYLxRt4i0jbVMtfILvVANF9wKUvgYZmTUCYqGC1CBlkY4lIYGtHO/AAeYS06stmoHPOHax8hIKdvaY8Rp7TAwcwI1NIn86zgkKwBJkQA2LyRXpabf+1IMA8kJMLS1xliSaqIEhxOnOWlF2vKCHOXq2JMRXZqsa5NYLRl6TEIRkrGQvEi4Uh7MV73cJrdcsnvEaqkRckIbNLDkKIlTXnn5PFHguP/siFevtXlhW1i4rDimyrZMSLSBRQgwdCVkO1/wpTcimoHfBmuEC9rmlkzY0gavyj4xhgzl4rqQ3mtznUEhKQvZ0yEozCQhJuCm9Lkp8Bp3PYGbwBVzl58Nn+C5ZJefse9n7vxs7v/c7/P+oMddM6WvY9alQ1/HPg6aDLD8a32FE1OfMBZrQFN9J4k3ZythcW69bLa5qCeYZz7Z7UnT44v5HrvMuODabNEiw3FjnrARhpgsQoAv7S3omoBb108SBI5mPeUHOMNvs0NOzj/41M/+oYT+3/zlf/PM62aXGQn3uzWumwPAT+FeNNu8bO7QpEwK85LHh9wmrxaTEcUV3o6IKTPe7U7ykrnDthkBfkJ5gwnfWVtlmilfyvb5B933cGek7GUZ35I+r9i7jF1CSyPeE7ZBYJYqzeiQFZSdIHDchF6SDxs47wsyEAQ5QeDIMlOkYuW+XPAtUE4/3kxASsJRko8wzAijjDQJltG87zCWJGSJJZZYYon/YHF0UvJTn3jmmaOL73KxPmRaLIEPiUcglga+1drhqj8Nlrkm3DYHCIbTrlO9JixkOxG2IhMtqRMXwi0FQnwDeogB9X9PC8nUYSqVJ0dARS4aBAwkoVUsTBsERFgmklInQNSb20vCdcuMyItWEU+A8BMJrZPhyEWxxdXduckw6qcURoQpKW2NGYtP1JpLSob3CHhiBLk41l2DXTPFqPfVTFmQiiddUpC1sCB2CzJaRDSK6UcpxWlrzL74FvWaBuyaGedocJY6tzXivPPTonUTYYCH1wxvHMAr28IPfvguL7+yxTQBp54+PtqocX2W+v2B467MycSx73w88p5LeXIrRES5sx/wxmJMjOWGGfPXu2f4Qn9KiGGyEEIRzuVd9u24MuSnmjPROaFYWtSZMKdFzCnXqyKY/7fsNfoy5pru87DbpEnId7dW+MJ4xAo1/oZ5gv9dX/Cxy0ekP+/LT9E38yLSWGgSck9m1Rn7qtnBoZzSFVJyPrvYZVPafNnts5CMVVcnKvxFm/iAhBvpgpZYAoS6GEYu4+XrNb7jfXvEtQX110/y0//2v/gjmaZFvdeoRkiklrSIQS6J9yNui5fNHTIc73WnuGTu8ZK5Q0LGA26db5nb1ImYMKdGyCVzjwYRi8KjNCflst3lZjLkQt7jP91a5Yt3/D5tGMtJ12THDrls7lEnZpwlvJse7cAySQ6JiP4Bg4dSeuXct5OCchJSJVu9aRoiRXz1W76v+AJDYw4LDrPMVn6QJd45yGc/+9mf//PeiCWWWGKJJZb44+KnP/HsM4pSL9KeABLNqqbz6rE3GcMFoSNN2tTZKjwZtiAL5XMitVWLdenJCDFVgV6MrbT/RyVRJUpikpDTwsuKBrIgxFJTe2zysSAvOj7glhmS47iHj8R9wp1jR8Y4UbpaY83VESj6Lg7JTyntmkpKqIappMzJ6GqNYWEuTiQj0qCSlfkixYSFZOTk1YTkDn0CAprErGiDHKVJREdjRKlImpf6CGNJqaufAl23Ax7N1zlhYm66GRbhtKnxA49NeO5bbd53JiPLDZOZ4dTmnDDMuHy9zTSB18YJ9WKl+VXZ5WG3wj4JE0kxCGsac8bW6ESGRgjvfmDIC5c6XBonPLEW0Ygdi1T47XtzQgz7LLjPNHmO22zLAYpjrimhWO8LEV+cd59u8CPhaZ5fjPi8vQJARMicpCqtDDA85E7Q1JADWfA1uUZOTk0iZrqgXcRFr9KiqzXOug4ZjrGkTCWloSFfkWuEYvlo/gB7Mucps8qX3R5OlLaLmEjKo3iJ1FhzzoYxl9MpQSGrsgiPtGIunEhotxfc/Yc/+cdObPr5T/7Pz5ROqFLi93C+yct2hyY+de2M6xKrJRflFbMDeKlfSk5YhClMmFdeG4snNBMWNIgqv9KGtomLc+rvnFvn829Ay1pWa8KvTXe4JyMcyoNugyeCLplT6lZoRUKnrjTrx6N56/XsLT9TGB7tB8nfkojkxTSjlFy9efohAmGUVs8vycfN/+6vLlOx3kEsScgSSyyxxBJ/4fH3P/lPnpnpoorEzYqoVThM0VqRdrGYbWPAL5jUECBEaplKxprW2JVZJZ0JsczxGn6gIhDjYmHcLSRXeUFEFuSFkdtfVQ0w1AqPgzeaKwk5ifjtK6Vd4Ps0vmFukuNYo80p1yORjJSchkY0NaRJWHRse5TEyCCkxfRmLAmpHJKwxZEuloZGxTLx8ArvXZkQYhiIT1YayazoKgk543yXilWhR0SGMiblFA3/HDFMNee2eN/F9zZXuT7JuK8VsN5xOAdR5LizH3D/qQWfflV4tBPx5Lt32d7u8eIty0GWY0XYdb45XVFeMntcdKvclDHrWqcvczoa8yMnWmxtzLDW8fVLLayBk72c0cxy35kxt3eafHUn50X6fHJlg18YXOEH9Dwv6gG3zJAB0+KYpnwov8AHG21GqWIFEqd8Ne9zxeyxo31a0uCinmDN1blk9zjjujxuu+xkKWfCiP8x/+Kx/RgRYsWwhU+Y+i67yZ00ZdUGPO8OqGtQHZscR1MjJpKQknPB9bhhhjzkVtgKIubOe4Qm7jCoIUc5G0U8fl/C6J/8xJ94cfyPP/kLz5SFlQGGkICUDIsPGbAY+jLjwXyNl+wOD7lNXjTbFfkQhBY1xsypEXG/W+WSuYdBCnIaMGNBhwZrrgFAWyPmkvnplhlT05DvM6fYzzJekwEDmfMj9gyfye9wznX4QKtJFMBaJ6dRywis485ejUXq9/eFU7Pq88znllrNf9++drnuj4WFDzw6qIiIqhzzgxz1hZSEpCQzAFGccOXv/Y0lAXmHsRS6LbHEEkss8RceU51XiVBlQWGOI1O/sCivaJ/SHg1CppJyXfaPvUcujhylqzFTSVlIzjXTZyY+CShWP/0YS0pDfTqTAaZFt4dDqReG3nLacJQw2OKKdoSlW/RqTIt+k1S8sXuFNjER592qN2SjFbnwbSZ+gtIlpEdU/a4WAY0i7ctisGqKNC+fhBViaWiERahrQEcP/SUrRe/ImjbIxdHVOivaoFUY3MupzRjfy6ICYzLm5KSq3JEZKY5dmXJnmnN/O6A/V5qNjCt7wu9eV14ZJXz6VWFGzuVRxme+ssZLty07acqOLth3KUExhXJAjZB9WXBSm8zI2NIGNSzDmTCehFy92eLhs37Cs742IwqUW3eabG1M+dBpIRPH1/YXbLk2WzVLX2accz1WaPKAW+ecrvJY1OJb0xmbDeFasuCL+R5PmhXWtcVTej8LTfgmNziQBScL385X8z4JOXOnqJbHJUQQbNHC3i26Zq6kfhK0m6d0NGIoSSVl8zdHWHhm7pgJNQ3pSEDqlET9edMwhl7gvTSPtGJqgfypCAjAz33q7zz7Dz/1s8+W52yfCRbLOdfjPtfjnOtwv1vlqu0zJ628I+/NN/lgfo4aEWPmrGuL9+Un2JYRp12PBRkJGYqjTkyghhreG7NJjVoRarCuTXZkQOKUj5wO+PH2Bie0xW/ld5mT8R2dJv92erMiB4vEMpqGfObulH82vMI/G17h+dfrvPxGnWu363zjhv9uq4Pnkl3+VXaJu/P8LT/7WxGQ8n6WHToUkkX0p9nFS/wRsfSELLHEEkss8RcePtLVoeqwcrzBPCZAUU5op5qOrGidVRqsaMxIEg7MnFXnr6LumRldrTEjYyJz8qIgUKAyGg9kQUdjpoWU6oAFsdqKcOTF4j0oJhU56uU7BTnxC1BDs2jfQB3X7ZA+I97nzjAXv6BbSE6tMNB3CyKVF5OdVRNQV8OeJtV+qGERhbGktDRkQc4ajWpfVLGyBXEyCAvJaamXtLWImFFeEdZivyqLwk+yICdUw67MaGnEuGjSTsmpa0ggwu2J47GTyucuGyBnVizW5+TEGO7pArKYRB0zcnqEzHEV2TogOYyzJSDF8USrQa5wYiWjUfM3GzjatYjBMObeSHjyoSHbO63/l703D5L0vO/7Pr/neY++e3pmdmZnZ08sFyQIEBBJieKhq6RIkSxZVSZARgwtW7FlqVKVOGWXU5FjW0OkHFeq4iqXykkUl5XEsa2SIh5xxAOWZbssShYlkgJJECQB7ALYa3Z37unp6z2fX/543u5ZkJBEQiRIEP2tmpqze/p4u9/n+/y+B7cPDd/JMs/IgBDDJ8ZH/KfhOknp+AzX+Ql7lkHpuJIlXDVHfHGwx7I0WNEGn3N9jHgS8wv1N/MPk8+SkPvnX2FscgqtsVwzMAZTTcxWZIE9HXCJk4BPQNuWCaLCvqQzsphRzNroOxqxbUYz4jiSlIXAMimVs7WQ7cRPiFThgU5MLYTFTs7xs/1nwy9WRvb/7l3/88aIhL4kPG/2aOixvLFFPAtLuGwOGEnCojYZiCEXx2fsrap00R/Xbeo4lAfLVW6ZISWOM7Spi+E8TWrGkDrvcxqr4/2bYzbNgLfoKl+UA77LrfJPRs/xo5xjlMEy3py+d2T5lH228ibF/Fr+PItZg4yCH7anmEwC6vWCH20uc320wAOrx3vsU9/Oi6Vf3U1K/jgvyhzfOMzLCueYY4455njF4yfe/QO/84n3P/4DSbVEm64nFCUWv3g/ge8LuS19LrhF2hrRqZwNTQ3pEpFVBu6s6unwbdo14mrKEFU9DlIZj/dkMp0P+DK+agoz/f9TiUpZTTsswoEkiPjrUo7N69eNb9ewEmArP0kpyrI2WdY6jWoSUquKEXuh/z5Vn75Un2n0Dd2qm2MiJcFd8cJR5WWZyrdq1f0JMJQytfc7BGYljCe1waGkiHhJ1gmJURXvNRHfR9HVGj2tsSARD54Umo2S9bZwaaXk4DDkSEsmFLQJaUvIVR3SIWSXlCNyfzsoOZQMBI4kpU7IriScoclh7rhnwTBJLVEAm7sxW/sRK72Cg0HA+lKBtT4B6ezJhCe2vBflivR5R7jEh9xVLtHltFvklkv4vNnlC+YOmRTsyhGplDzAEtdlQIDlhGvQk4ikDFGBTEo+Z26wpgssECGlYUEXqEuN+90aT8oNCkrGkjGUjDtyxJK2EISz0qBHRI+oepximhpx2wypV03lZ12XI0lZc016seUod1zsGfoJ9MuSSQ7LDaHdLMl/+HVf1+K8H373O37nB9/9tt/5w994/Af6MiEQSyrHpZ4gLGi9khB6gt8mJqvIqzenlzzoTpJKyaGMmYijriEjydiXlFKFnvHThcUoYJU6pxqWjou5QJteaFmjQd0Y3hovshgbTi44Wo0Ca5WiMDw3Lvmp6AL3SY93NHpckBZv63R4zVpOHPno5pVezj1LjsXeBGM8uXDOVOZzqjJDxRj/vaoQxTlG9CtKDju/9/kfmJcUfmMx94TMMcccc8zxbYP/8pH3bSjK//6BR19UsvL33vWPNgAeKk/RJiRASHG0CBhScFtG5FKyV3kclrSBIKy7FjmOGMvtykMx9Yz4AsHKrK1mNgGZ6u4PTELP1UirqNyiSrwCn4hV14AjSblpDjlkRI82F8tFds2YjJJLbpEalpZYOtaSO/Wt7lUjOsCo9MRhoq6KgxUSHGOOzfXTSUOBo0tEQslQclKOpSuZFLPbDd5Dck5bFCgD8tlExiJcliMWNCbC8NqowbBwnO9YOnW/eHx+y0fLdurKk7sliTr65CxJRMda9ouCLZLZ/2oRMqZghRq3GKMCbQ0xCKsmpmEMizXhvvNj7uzUGaWCNbCykBOGDlXodBIAfv3TdXIcl80hC67Gj/UW+PjBiG0Zk0vJtgw4YEipjkgC1nWJZW3Mwgjebpa5USSkuNlkaN9MsBi+x5ygExo+MemzbyY8LbcZOn+8BBIQEbIiC6y7Dj2tsW3GrLsWp0zM43owK6Jsu4hdM6auIYvV371NTrBSN9wel9y3bEhz4cmDnFCEt3zwz7+sHoX3veuXNoAqQcvS0zpnXYdnzQE+Errg9eUyT5m9ilD7ickd6dOkRkJWFVUq685PIRMp+OFohWboCUJ2l2oqrIYXZ1f8RoIYsEaxxlE6w2gSEIcl1ipOBSOKMYq1ijqo1fOqOd1DRL+iId0YnX02piTPffFjEHyl6X1uSv/GYy7HmmOOOeb4OuCJd310A3zU6Hd84CfmJ69vEn75A+/7Ex/7GhExwcws3sK3fXvj+HHy01TXbzG0XUSOY18SelrjvLaZVJr+vIrbPZDUexrEm9Op+kMA9mXMCRqMyWc/K0VnpGVaJGexdGigOEaSs6QNUgpG5CwSUReDFcEYITCQljqTkUQiJKo0jaVUpa/H5YZ1bEU/zIxwjCkoqWJ+haogsCTSgFIcNZ220zuuyZA6AV2NaFcToSEFDQ3YNEdccAt8Ou9zniahtewMDAsdZaXjuLZn2Jn4hWEshlANE3VMCsfbVyJCG9BuFXzhesQfZIf85FKX392dEGBoqJdiTU3/Y+dwifD0tQarvZIkt6jCbj/EVovPSWqpxyUP9SI+cnjAqmtyb9BkmMJrwwbXyj5bcjRLbSooWaVHT2u80SzwxXLAgUn4w3KfDhExlh6+CLPjImpY1tqGvIS+SXlabjNR702xYhGEJWkzYMK2sfRK37txwwxou4CRSX0HvQqHxhOmQhz7JJxwDYyF7YljIbSAMkxhIbCMym9ua/eYlJiAy2afEp0FNUwJuaJEBCTknNYeglDiizYHMmHLDOkz5l63gioMMwgMWOPb0gMDoeflBIHOpFHWON8HYhyd5jE5UXcc2TvF3QTkxTD9/fSzqsFaRURxzmKDYpaKJX/Kdc3x9cGchMwxxxxz/Bnx2Uc+sgFgRAhE+OwjH9n444jIkxVZaQaGC7/2Y3Oy8jLjoXKVoCICUUVEzkQRk1IZO0euSl0DDkzizdqVT0TxBm6LzJKgTmpjRkQWNGYk+WxXf1ylZ1kMK67FSHz8Z0tDHMq28QWIKsqujOlpnRXXwiLk4nzbufoFeIcIK0JsjiOBSxUCIxROSVWpGSFQwYowdo4uAVYE53yqUg1LQskKNSIx3NQxDQKOyGhocFccMaBenuYlUQG9yvuxKxOO1JKLYyCpN1eLY1sm9DQmEKERO7LC8PHLAQPn05DaEjCoSNGajXnc7bOuLX5z+4iLpgVbFij5T1oLZAW8rdcgL2Gcw41JzltOBnxpxz+u9y4LgXXEYUloDcNEOLVU8NQty2JDWKmV1Go57XrID+U9Pjec8KlyHybwjnCJhka0pMZZt0BhHAcM2eOI0pQ8wU1O2C539IA3c447Zshr3AIhhkv1mHropzpJDjeHjudll1RzDIZAgqqHxhATsqwtll2DA0k469rEGFbDkLW8zZ6Z8HoWuOHGbJsR+zJmVdvcMgPedbpOlhmyQrlzJHzHhQmH/+M7vynvE+97/3/zKPg0LUVxoohO44INsXrS1dSYvDLcN4h4SBd5miNiteybCS3tsi9jFmnxJrNIocpCLEwKn2IFnoB0Go5GrXhhgtVdRGNKPqbp29PPtXrOl+PuKch08nH374CvmJJIaZEqVe7FWtfn+Ppjno41xxxzzPFnwJPv+uhGUMWrOFUK9VKZ6WTkbvzBwx/emDjHxDmcKs+/57Gv+Js5vrGY7qovSERLLMs2ZFg4xs7R15wDSehXRmKAA0kqYy6zfo4FjVnSGiEymywMJSOrUo+mHg+pfh9iZl0Re5LMTOs1DRhX5GQs+ax3w6j/XYmjrX4KYwErUKpiRSjVt4aLQCxCWS3cAuO/j0QwQE9CupXsLMZQE0OpWnlGfJqXik/P8p4VQ4jhqOo0EfVm9141MQowDCQl1oAVbbHimvQlYV8S9jRjkglXjgq+71JB01iOyLmpE6b7+LfLlIaG3JAhS8ScblhyVW65hM1RSTNWAqvcGTnuJAXnGyH7w2p6pEocOYLA8fxWhFO4MswRUUqFrID9fkQcZ6jCIPMelzVt4oBP5ocoyl9pnOcT8iwNYtZlibeX98ykWQ6lKTWumn3OuA47kpDjqIew1HaE1S59LzK8qTyNwXCPnMThqEnEmizS0JCahgwlp6sxmzLiTBRhBZaJySlpGstYcmJ8gtlYMk66FqpCYJXVpZR713Js8M2dgIBP0/r77/+bj/6t9//8V5ChAKGhAQ0N/TFUJabVqzCFhobklEzISMhoB4ZmKCQFtGM/BTECjdinqYWhv78vZhLXL3sowqicSbBejGR8OdxdxYN3ExBVb/5XFW7+wnsfvfkL7330+n/70/MNopcB80nIHHPMMcdLxJRoGCAQoRn4fZ1B4c+WT77roxsPvP/HZyezVrWTXahWEZ/Ksz/1sY2Lv/7n5ie8lwlTcrEaBVgDTqF0ymHhGGsxIwU+RUtIpSDWBs3qdFmitMQy1JKa2FlSlINKgqKUFDhRjAq2KhC0FREBT1h6rsaRZEQa0MV7QryXw3DHDDhTNY3vmjEnXYuwIhpTAqJ3FUCLgMXLvyal77yY+kasCAIYMYwdbOqEFWrEVfldQklDg1lpIkxlZJ4QqcBYS54ye5x1XUIMa641a42v4ZOU8qq88PK+421n/SIyV+WsafCsGwKwKSOWtU6M5aK0WG/6bfD7FgJeqwGfOBxzawt+9FTN3ycRzp4ouLEb0IsM1oCI/74e+sXrhWbIqJJ7XVjLWDnRZzSq41Q43YXmuMETozF1AlIKxpKRO1iVHiWOlJxtM+J7y9ewY8YcMuGCLrMlRxxIQiol+xg+faS854zw3GadU0sZbi/CTIQ3cAajwilZpEZEz9WZSD7b4Q0xlOL4ZN7nbfECD3Ri4sEJrrkJTnyKVlnJuAC2D73xvhaXBIGy9+gj31LvDX/jA3/tK27Pr7/z1zYUpVEZ0Y3AORp0A8sfFgcsuwY18URYBErnJ4uqXo4Vh0qjVmIrv8YUgf3jCVgYlYThi8fwAmz+7fe84Hae/Pv/z8afJte69d//1LfUY/1qwJyEzDHHHHO8RITVBCRxjlCEcelL4zrh8ZD5C+/+6EY7NBxmjsgIk9JRq8gIAtYIz7/nsY25NOvlwV/4kG+Zvv7exzYEONFSdoZC4vxzl1LQ0hhBWNE6u+K7HgbkNAhYNF4LvyiGgSvYNMMqmtcvcBa1xoGktF3IwGRINdXIxeGAXtUP0pfUFyZqyIFJCNUSINw0vindL0q1Mr8bSvwkAIXIiJemVF9PvSHiL0KpzKRb5V1FCA1jGWpOrJYTElWJWj7iN8fRqYoXpyjFe0ZiDBddD4AWAQ5POhJKAoRFrVX9J4ZPsQc3lohNxEPLcOMQzmqTyxyxonV2xPsgnneO/WHEmo14fHTAw0uLnLU1clWe3hbaodAz8NRtZbXlpxxRwAsIyPrqBNmuz+Q72/sR6rokqaXbztnvh9QCWJSIsfqW+4Gk/NvxHgdmQIkjIuRAJrQkoq4BUMU0M2BRmsQaVCTN77BfPD3Gld4QX8Oy4hrcNkOaVTqYA/qSsO7a7JoJXQ2puw43ZMg9KyWfvyXc0wy5NvQBCHUNaFfHxIFJuDVqcaZtaLUy8swy/ga9Dr6e+KkP+QX/v3j4Vze66j0wJXBYlFyiQ2wERzWhs1A4L8Nq1fxxGwWOsioS/HK8mPcDeFEC8ieRiGM/yHGj+t0yra2/95/N33+/CZinY80xxxxzvAR85pGPbNhK8tKJDEfZtNnYLwwbQSVMo7IdAAAgAElEQVQhccdlWJPCn/CsQGSF0inWCLnzP69Z4cyvzsnIy4Xr731sox3DIIW9tGTgSvrkbJoBa66FQdiWMb1qYbVAxKIJyFQZasllc/iCxmyHElapV5kUhJVmPqkmISdcgwZeolSv+i8E2DIjVlyTI0lJpWpaV8NQUmqEvIVlOoEhc55UBEZmkwaDP+ZK1Rm5HZYlRo5v18iVhCIcacGWjFnWOraSZ03hiY7SJyfC0JGAsfpeE1Nd1dP0SaTgrZxg4EraxnKyYTnRdmSFsLKYce1OzLVByQMrhuv7sJOX3NQx7ap8MK+a7KeytZZGdAlZMiGLsSUvlUsnHb1uwvObTb544L0kmTpe34k4u5qxuROx3C24uh1wetGRZMIkE04t5ZROiKOS1bV9bt1cYjgO2BkYnhqm9MnZlUk1qZBZZ8xNOWBCyuvdOj2N2TZjAjU08P6dk1qnQOlIwPecFepxSZYbbu2FZCV8aZRwTYa+uLKadk2jmTsa887FJfoTeODChMN+xDg1/H87fXJxjCXnhPOTti0Zc0HbPLAQ0fs/X7nvA594+Dc3lsOAojpeIyPkqrRDQz3wJDkw3l8DlaQw8n4Qc9c0xBjF/DHSqukkRES/Yurxx+HUP/j1jS8nIXf+7px8fDMxn4TMMcccc3yN+OwjH9lwUOXDwCg/PpkarXafq8Vp6rx0phlWvhHUV07jpyBGoBH4ndXSweZffGxj/V++chcgryScrQhf9HMf2xAspDB0ftFrq8Ssi9rxzeAoMYal2C/KL49LllydoWSeBIifpExjSgsc7UqqlJBTckxSG7NkJjcjMRklHY3vSq/yC+WwKnVr4bsOcqe4SqUylV0psBRb9tNyFrZbVtvKvmDRT1B2JUHwxDmr/rePHTbYauoyLUQ80sJ7VMRL1qwIm+aIZW1yx6V873KdcSostR2dVk6WG4pCOL+WEIcxX9pRRq5kTzPahFX0sU/dsio0iCnRqlclJFHlwkrB5l7A+qlD/s2nF3ntiqNrLa9Zho/dSbAmYns/4jVnRnziqQa9GlzeFkIjLNThYBDgFM6vJ7iyikAu/eM7pKBW9axMY5UNwrKrMzIZsQSc1IaP8KWkjp+AdAmJxXC2HlIPIY4yGo0MmUT0mo4kF+6jxuZ4xKo2eFYOiTXAibLsGpyTBmvLKffUM8bjiDuHlivDnLEUtDQkVEO7esxTKb3f5BVMQADe9sGffBT8FFjEk44pAan2W+g2lKI6WKNASTODiJ1dR7Ne4JygMn2vdC+aivXVEhCYy62+FTE3ps8xxxxzfA34o0c+sqF4aQH4RWWuSt1WJ8vq56NcGeSOmhXqgTCupiANe7yYPMqPNc956U/QSaHceO/csP5yYpIJm0nBrsu5XrV/b5ph5e/QWSdIjiOyvtsgR9k1Y2oaVNMInTVLx1iWtMFAUvZlTEujGdnoSUhfMjqENAhoELDufPBtVrWR28pLsqxNsiq1KnNK4ZS6NbQD/7FfFCxEFitCUiiZKonzgqpQhLRy8oYIE0ocSkcjIixRFdc7JT0DCtKKKE0N+PukjLVkQO7LBTUmVMshGU/tO54YeGnVaByg6iViH38m5NkDfzsEoUPIASnbZkSsPo3srbUFfmS5xXfWOqjAtiS0reHZrYDSwRNPLfGO101Y6qV892tS1k8O+cn1BteOSpo1x+ZWA1uRo24srHUVI75N/EQvZzDw5G91tU+vnXOQKN/dbmIQYgJC9Y3lDmVfEi66HveXq+zKZBYacCQpfUlJcRxpQRxAYOG52xG7+w1ElChy1CPlqVE6m5TUNCSVgp6rEWP5zlOGhd6Qm7dbjJOAla7DVJHIpSg9YrZlwhjfz/Ilc/DyHfjfYNz/G94PVw+EegBJJakDOBwJgfWekLz0nR+TxJLnhiwzs2lIfxCiCqbqAoFjg/qcVLzyMZ+EzDHHHHN8DYjvkrmA38mZEpCa8cZg8Is4W2mgS+enHeNCSZ2X0Cw3fN7S4QTqkZ+iFM7H/DqFrb/02Eap/rLekMxcqvUNwrhK+LzbD7HuWjQISCi5J6yTlCEPnhTA8cnbvgRw0dXpG98RYRCWnJc5OZQtMwLgrOtiES5om4axJM6xQp0hBVklx2oREmFYMRGJOnY1Y0uGlKq8zvV4Vo5YLurcE9Y5qraPU3W0jSUp/fHUL/ysJaEE9YlTGY6RenIRYmhpOEvrSqueE4dyUC28M0pGVbP6RAqGkjHUjH7l47joenQJyXA8uGK4uh/zuTuOh04KAcr1rYhDl3KpEXOYwR+W+9TFd4mYagIhOD6e7vHWcpHfLXZoa8RQcp4pRtwvLSIrnF1NuX67wdm1MUkaEIYlQeB47ZLlc7cdr13yJv0rw5yVKKBUIS3gzKqbFdft7nTJc0PpZOaX8X0sIYU48rsKGvckYUXrfhKkMUfIrPn8grbp2YB/f9TngjRZDC3qvCehY5UwLPjurMXbzRL1yPF9wQK/dH0Hi7BpBpw7O2F7q8v5M0cYU7K72+aGjhkbn5a2L2k1TfPH3s9/4Ge+rV7j9//Gjz9666cf20gK7+PJqk7AUmGcvvC9dGpUBzg4ijCitFvFn9r/MccrF/NJyBxzzDHHV4kvvftjG4E5PnFOFzeT0qddTQnI9He1ipyEFvJSaQZeCtONhVZN6dQdi40q3ci8sLxL8aQEvImzFsCdvzSfkHy9cfSzj23kpY9y3ZQRZ12Hs67LlowZkNMlZFB4v8jmgaFRK4lFaBPSNyk95/0iK64BwKYZANDVGquuSY+IAGGXFIsnCY2qn6SGZXkm2SpZa1giMUwoaGrMstZZMAH3ywJnbQ1VT24FP4kBv3AbOkcJjKoujgE5fXLSqkixQUBKSSJeZjSsLj1tdrcIEylIpOBAEvbMZNZzMp0MNIgYkvOm5ZAfWIsIrLKbF9St4XBkqNcK1ha9hOuT4wGj0jGQhDGe4ZU4xuSMJedAJvx+scdEcnLxhvgLpsGJpnDveooqnDk5YTCMOLE8ot0eYwTWlhPecs5x4xAeOF1wph4SW+HaOKcRwnAcsrh8SOkMSWrZ7YccDALO9/z06s2dOhMKTmjNR8pWnp1MfJ9JSyOG4gvx6lVT+xYJu2XOW2pdLrQDTnWhKAVrHXluEFHOr49YXx1z4cIOrab3/+yaCTUN+K1PLQFw7WaHw8Mmz9wOeVu9y0nXZFlruCpVqqPR7Fj6dsOpf/FjjyqVXPUu3pGXxx/9sSeLU/mcEaU/NhjxHhFXCsYev7/mmWWOVz7mJGSOOeaY46uEVD0NIt6M3rA+xjQwPgr1xQIlrcBhqvTq4pN66t7/YY2/nlbN+cWlq3YC5djIbuW4RdhHlr5sd/VVgcnPf2yjHimXswmFKqX4iUGDgFVt0Cbk7euWM82AQPzz9txWyHJsORdFNDRk1/j8ojtmRIRh3bXpS0pDAwIMQwoWJeK8abDvillpoEFYlIgcJaGkh++RiEVYp8F9dFmVGkaEUMQTDVUiMXQDy6IJ2HM5t4uMUnXmARlTUlRxu1MJmcGnWQGMyGe9J0U1IVGgU0nGFisTvu+6CGgQ0tUaNQ1oEWINZIWQF8JSEDApHXkJv3c5Io68dGsgGZ+QO6RSIghrrs2yazIUPzU647oMJSMmIKHgjVGb158wFCU0mynNZkoQlBSFcPVGl+euLhFHJWJgNAm4kk+4uu19FFcmKUMteWaYEYclt24uMU4s13YCtga+cbw/Fr40SnjyKOWCNClQYrWEGDLxMqhdM2HXjClREslxolgMR5IRYriRZPSajovnjlhaSDFGCawjy0LSNKBeT0mTGs3mhEAND2iPEuWZwk/E2o2CJ6/HlA6+/zvvsCwRdSzLWvMETYoXhBx8O6KfKoepMsh8jHRaQD9RsvKF5CQrhKwQ4tAf007Ff3wZEZnjlY95OtYcc8wxx1eJyz/1sY12JL4ErToXOjw5aAReRlUq1INjiU879gTDVAbNpVYVFSnHJ9hJVpmTK8mC06pnoWI1RuDkP39xKZb9Gx/eyHPD5+4oibqZKXSOrx5X//PHNrbTgkR9+3lHAgqUIy14Q7NOt66E1hPH0gmf3S55ij7LWp+VH4L3UkxN58+aPsvOd2IYoCshuSoTSkqUG2bAumuxJWOaGvKA7dCNvBFcBPYTN+vKmMqtgoroWvyk7Gk3YKWKlE0q0lGiTCgIKs/DVOZz9/cjyZHKRD/1u/hW9YBlrZFUt3GBiMftFjklIZbvLk9yIgzZywsiETJVtkgQIKXkh9oL/KvRNg4lq6Y5pTgGpDSJyCioEdLSiFWtY4A7THhD0GG1YTjRKWm3cg76EQAnlhImk4D9o5Dzpwc0mglXnl1mlBo+t59RE8ORFpwJ/TTp/vWSxd6Yej3l2eeXmGSGx/cyBOiTM6ZgWJVCTic9U1mWQYjUP3eJ5NQ0pE5ATe2MHHzfQos33LtPngcEgSPLAiaTkMA6itLgVFjoTvzOvRN2dht85hY8/L3bXLmyyuHIcLlfsBBYLvgBCc/vwe8Ve9QJeM8Hv3qT9SsRX3j3RzdCEQ6Lkud0yAO2w/PlhAL/unudbc/6YNIC1hd8ceVCJ8M5IbAOY5X9/+Hhb+vH6dWEuSdkjjnmmONrwCT3I+Rpp1YnFEa5cpfHnKyERgiTwp9M62Floq0dJ8I4FWqhj6CcmjVLJ2QFVSkbhOaYiOz85cc2Tvzfx0Tk6Gcf21CFxY7/fttlnAu/PeUc32iowvecV3YOIz55kFACNTE8uFRjksFSp+ALNwPO9ITf2hpzKCmLWqsSn0rWXAvwC9mWWBJ1rLlmlUAFJ01MropPVDMYlBXXQIG2Rrwx6nC6C8/uu9nfNYwPMFiuC/tVGV+uSsMYclVCEZqE7JKwjJf1GIQxBU2mbd0RfckI1cxa3B1KT2uMxPeF+KZ2Sy45uZZ8wt7gHeVZvmh2eb1dp12com4MqSrrLUtSwH6eMVE/QdgxI6Tyezw+mOCMkpDPmtczCk5ok5FknHcLXDN9zmiLjJLX1xvcZ2MeuDhkNPYegOE4ZGV5wu1tb/6W6rVQFJZr15fYHxq6DeWNSxG7IxgkJTfylHviGk7hqec7vOHefcJQuXUIY4pqOuTjcDMpyMCnjklWpYVVfT+SV/KoaDYZWqHGei2kV4c3PnAHVUOeBzRaQyY7i3S7E8bjmKN+iDXKaNzmaGw4vZLSahbkanGlJQodNwZ+02K5IXSaGWLgmTs590n3VbF5MDWq/8HDH94IMAxcyYEkHFSBAIkriCaW74q6GBG2joQTbdg7jFnsZhSlwbxYmcgcr1jM5VhzzDHHHF8l/LRDfc69HHeB1KyP2p1KCkr1X9cDiAMYZcx2uGedIZmXHDh9oQTDViV04NN4pr2HTj0R+dK7P7ax85cf2yhK2B/DU7ctH741JsNxJR/zr9/5wblv5CXgS5sB144ciybk7WeEB1YMd448iRRRPpcNeWxrxISCgaTkOCZV/8e0fXxZIjqBZVKla9WwFCgnG5Z2YGlby7INqWE4kHRWRLibF/ybnRGFVn4PPZ5gTKqJWt0YtjXlZpkwcCX7ZcGo8luAJ0BDcmIsR5LR0pAhOaK8QH6Vi5ulcE2JS1cjbkufJa1T4vic3eZNboXPFH0utAMCI6zEls8NJ9QDeFOjSS6OGItDCTAoynUzIKmihQF2ZMRQMk5pg4d0yUuQNKzKDpUbk5y3v/k2T19tEVhHvZ7T6yYcDWLWVsZY6xgMPUN/frPJQidloak4hcAqkfUk4564xs004+PXPMnf2u5wev2AZgRngho1LLm4mQF8xTWZSE5EQEMjulojrCYe09JJh9ImJMdx6WSBNbC/2yUIM9rdIWURUKvlGFuizm8gpLkhL4SDiZeNnTpzhyc4oCwNN3YDBqXva7k5dL6MMDfEWA70+Hl8NeCLHHLHDNnUiZcMiuNNboVdGRNh+ffFDp9NB2wmBXeOZCZVFYGjf/AXvu3J2qsJcznWHHPMMcdXieff89hGyytFZibLOFTS3C+6pgvGOPAnzUnhG7mnmCa/lO44Lz/NhaW2H4/sD70BRBUasb/evDw2qAfGF+vFgZfrbJUZSybkpktmsqBaZXqOsfzwh/7sJ+x//MivbEwXCqEabkufvg65KGvEanma22TVzvcvf+B9r8gFwv5/8djGQtPx725lLJqQ9Ybl0nrK3mHE49slmTq+ZA5mC9TiLvdPiOWSW6AhlpoII+cYUsyejxDDj54LeH474MYkZ1ylUk1hgEklCbo3aLIQGfJKvvfcJGXRhizXDAep40oxZpmYHDe7nimRyXEE1fI/q7weKb5sMMYyJCfAMCInq7wa07SsAMNZabCpk6o1vuSNUYenszFvaja5MspYCUNiI9xKc07FIb+V36GozNwHVXqWorOCP/ASrRbeOxOrvx1nTI1MlbGWnAhCLi0JnWbBPZdu8NzlM1y89xq7WyuMxzG1Wk7/KObGTngsU6wet/vOTRBRfusLESfigMOsKnkU4aHTjuXFMc4ZJpOQ52/HPD6YsCkjxpL5CYdr8ri9yQVdxqjQ1ogQw5YZUeDoao0cxwMskKuXqzWNJVOHRWhaw/e+LqHVHmNEuXNngTQzDCaWUQqfGo74rlaT+86PGQwjFroJv/w5/7pcJOJ7ThuCQPnXVwt+6OvwOn2l4R8/8isbIZaUgp8MT1Oo8h/zPQaVb6itMYkUrLkWD9VarC8o4f/25151j9O3O+aTkDnmmGOOrxLtmMpU7j+c+qSceuRN5ifaSiv2C0hrvFdklApJLrO/h+MErCmJKUphMLGzCYg1flIy9ZK0Yn9d/UQZFL5nIDLCETlX3Gi2MLNf1t792+/8f1/yVOSXHvmnG7/0yD/dANgRn/hkEPb1CEW5obs8zW0mmqDqOyH+2sN/d+NnHv7bs//5c4/84sZfffjvfMtPZo5SaDULWmIJRRhlyuXNmJ2BsBBYxhSzdKfb0ifHcU32qvQoR4ky0oLF2NIwhha+O8QAMYanbvsm8N3KLr5phrOF+ilb48Goxf1hi17s9fBhVVy5HkUkzpGV0AkN9wZNDsjYJpldfirBUph1fvhCPi8zSvBmcYOwJxPqBNhqcjGUnFKUy3aPtrVctE0ejFtcNC1KhdOmRlb6acO1POF3sl22SfhMOiCloKERJUqLiBhLm5i06rvoaZ1lbeBQEnxq1gKhL1YMLd+/HvLjb9mj0yzIS+HKM2dYWhrxxScv0OqMiKKCzTtN2q2MtIT15ZwHLg4BWO06nrnuvTCB+Pb4HZfTDgxWhCwzWKsY4whDx6TAJ5KJ97Y4lCPJKHE8I1tcNtv0TYoCkfom+1FlSH+SQ67KkKsy4At6yG1N+DwHPFEesbNXY3e3Q6M98GV7VeDEJ4cjEkr6qXJnp06zniOivM62KHBcbIUsdDMC6/jxe16dKU//9Qd+9tECx88urrPWVUaFY11bLFbHTFdjvt+scl/YpHRzAvLtijkJmWOOOeb4KrH8z7wnY2ocN+IXi1nhicj0c+n8VKQeHl92epkvRy1U8lJIctgdH//BlMhIla1fqo8CfqYcMsjU+wMI6BLNmrencpKg+rrkxfK6/nT8L4/8HxtxZRl0KB0aPKd3eJrb3syrBWOd4HCEEiBiSDWb/b+fe+QXN37m4b+9UWr5J/2bbxmowuZOxGs7Iec6hofOZyQFnOw6/qg4JMRwXfZ5ji12tM9VtlmgyS055KzrUOJoSsBiA+7pWhYCX4Yn+NLBUeE4KhwrEnMgGSuuwZaMuWEGHJYFnVg41fFdF7fHJeNCfRO6QtMattOCwvlkNoVqYV9iEWpYGgRkdxnTpw3cANtmhEWIMYSVVGtaqkh1XT7hzRPbcel9LNO0rceTAWeCGpfNAbk49iVhu+pA6UtCKgUDUgzCgJQ11/Z9IAqH4pOimhrSJWQ1DFkILUeF4+a+Ic9DgsARWuXU2iE2KChK4eiwzf5BjYsX9gE/AbyzHzIcRrzuzIStvuHiesLhUUzTGq6kCTmOxCn1QAhDJU19geJkEtCK/ATlpGtWrw9DnYDX6AoAE/XSuD0zqYijJyoDSRlLxq6MGEvOthnxh/Yaz5gtrApX94Q0M2zdWmWhm9Fq5AQWvqvV5P6gzZP5kFt94ePPxGzeaWFFuDdoMsqUJLHU6wVR9Mp4jXwj8F994K8+WguVj20PueJG9Mm4RJu21vjBXrvyxX17J4a92jE3ps8xxxxzfI2w5oVfq3oiouonGJPCJyU5hW7jOEkLOZ6GiBxfbnco/Ltkl59oL9OqVVKWXKhHjklmcAoLTcdRKkxcwSfyfVa1QXaXHCfDUQD1alE6pHhJu0xTmURexchOF6mvYQ1BeJqbKIqVgFR9p4KiBBKQVdp2VU+Kls0CAP/kkX+2UdeAXRnzNz/wczNzKsBiYFltCbcGyn2/8c3Z7WyEnjQOs8oAXsYc5Y4bB4Yz2iTHUVDQkQaHOqQpNfY4ItGMTTMkVkuklht9yxvPFlgTkh34BXfulJ3cm6PbErCoMWMKcim54LqkOJ4dFnzXScPZRdjfgoO8nLWdH5LzUL1JtwY3DwoE6BCRV5IrgKumz7JrEGIoUa7KgFxKFKWlEWMpqLzt1DXgs3aT7yjX+azd9LIsajyXJexKwlvCBa4VCaVTFgjZNiPaRQgGckpKcbP/3dWYrsZsGe+VsQh946c9EZamxvQ05rrpc8o1uJVn5CjnwpjttOT5612a9ZIwdOztdegPQs6fO6BWnxBFBckk5qmrTZoRnFjI+dJmxANnU7ISLt/0IQwiyrIJ2XU59y4LgwksdFJGo4g084/PE/2UsPLtTBPCHEqK71UpcVyRbRQHAhbLVCqUaY4RoUODNe2yrCd5wtxk14z5D3nGf7xu+KH6Ej/49mtcfuY0WQHtmvL7wyEFjsOspGYNX9pRnCrPuTFviFp86mpA3Yac6lLlm706Yf/XH380ffjXNp4z+6y5Dlvq37++me8Hc7x8mE9C5phjjjm+RszkVsZPMkT857vRiiEKPJk4mgjDRGYEZDpFuVue9cjiMostR+mY/a1TwRqlFirWKM/nyWxhmeGY3OU9AKp41pK0irzMeZHRy5+AKQEBXhDvesiIsNo9zrWY7aJH4nfcCy0otfSLOPyizqHsu76/vyhjyelqjY+984Mb/+Gd/2pjoAWJluwXJVf6Jf2y5Ml3ffSbIt3KKt/NxWXlWp4yzGAhMtQCuCJHPG0OKHEc6ABFOdIxiWbUJCKnJJ0u+ENhNLGsLGa0AsPZRcf5RVgMAtZsRNt6E3uA4YLrsmoiFk3IQAuGieGpHT/56ASWe1sRZ2oRF2x9dpz0eWGBYFiZ4i+5HtHU+yEZBniN6876PXJKYiyhmpmf5Yt2C0GICbi/PMGajUko+HzmJU+3zJCnzCEO5VlzOHtOp9OdpsbUNOC66VeTOJ/8VNOAhoazqcJtM2RZm0RiyFFiDKVCzQg7A2EwthhRgsBRi8sqEUvJsgDnhIWmcjCBo1FA7pTffMb7rM6vZnTqytUsJTKeXMSR49LZEZNJQLudEoaOhW7myRE5fclmkrWckmuyR0lJXWKKSnKXa0lOwagKKo4kxGI4VO8VaWjIOV1m0dW5YwYIwtVxjjGOTjMjtPBvd0e8NmjiKhfRk/mAL5ZHKN5PVDhYrhv285Kn917atPLbCe/54HseLSm5Yrb5jL1FLm42iZvj2xvzScgcc8wxx9eAxf/LS7KOfva4vTwKdNb1EYfKych7QaLAm8tbNZ0lvIBP0gGdpWn1Gkpg/c+tgW7DMUyO94h2BkLu7F2L+Xi2Gx1Uu99TwlBWUp27ScTXgmlvAjAzo0+vaygZPWnTd36hOp2EBBJQUFCXGpnmFUnxl9l1h9RMSIkjEItTXxR3JOksRems6/BgvUlsYfMvPrax/i9fvBPlG4Uzv/pjj15/72Mb7VZBWwKsQFYqT09S7qFNqIYrdotV6TEhY6QJptrDS6QgUC+98sRSuLwZ8YbTOZPU8twe7BQ5kRhGWnDCRuSlsmhCGtbwfJ6wZmM+c5BysR7P/ELDzPt/broESggTMzONT+VCz8sRtiIlF7TDbRkTVkTkBkNKlFD87UwpecbuANCkxqI2uCWH5JRcNgf0XcaqNggQGgRccn6KddkcvuCYAOhpnQIlqQr2fN+IpaYhiRS0NAItsBjaGntipPCGVo3PDsesdyJqIZxdP+L5G20mqWV5+YilEzmDfovtrS7NZsbl601Or2TsDCLuHAmZc3z/WsgzOxAFAdcOYVcSThPzPcs1VpYHZJlf1jxzrYUqJNsh+wyJq26QsPLDTMn7lEza6Z6seOowje4tKTmtS9QqL01a+UpGknPe9TiQCQcaE4QpSRoQBso7Oi22xo43R13+KOujAg/aDplTviPs8GQ+5EfiNg81LTuDudwI4Bff/9cfBfiFd/3DjZ/+4HvnE5BXCeaTkDnmmGOOl4DOr/zYo1nBbMoRh0ocel9IkntvCHjZ1eHYFxImuSACw/Q4ond/dFxUODWjZ4Vf0GYF7AyFdg1Kp+Qoi84X5EX4RemEYpaGVdz1+euxj2jVkIsjp+RpbvG0u8GBGyBiZtKrQILK/2CYaEIs0YxcTD0iW3oAwA23zTOyxVNmiye5znXZ54ts8nm7zWHm+NRwxJPjCQd/5bGXfSJyoqU8fTPiZBxQKuznJd+7EnsZjwgRIQk5mRclcVZOcFqXOGTETTng4/Yavz3Z5RN3cg4zxxM3Am7uG95yyZvIFwPLycC3ol81Rxh82MCPrMec7xou1mNqAZxZLlhfKnjNiiM2/hQ9kJw+GdfNEQ0Cfsc+x65MuG72SaTgGbPFVRn4mF7JuC77NDSkqzEOJVLLk/Y2Z12PEse663Bb+tSIeKg8RYtoJusThDtMyHFclwHjavqy6lpccj3qGnJe2ySS05eEv37qJCGWhIJEcqwaRH1qWEbJktZ4wKi4e7UAACAASURBVC2yrxmfHY5pELDUKajXSrqLBziF4djy259epn/gS2/WTvnjZX05Y78fkpWeFAI8u+v9K79/u+BOnrFY3cfNPljr2NmrUTrh4ukxJxaKGdGf3NUg39UaTpRVujQk9ub+yhdyRpdZos0FPYFDsdhZD0pHY0bVNGUi+ez4HpLzhc9fol4vWFsd0ms6IuPlmQ8FXQC+UAyIjFAq3GubiCivvbTDAxcmL+NR/q2P/+n9f2tOQF5FmJOQOeaYY46XCCNQqzzA47SSKAVKFOhMspUVXnblyQX0x0JSeAlQPzkuMvQ9A35aouonKvVIObdU+khfIK8M6OAnHmMpOEmdXBwjclSYRa7WsFVG05/h/uGJyJouzIznJY5M8xdMWqb+EcGTk7t3zhUl0YxbbpcSR1+H7GqfTHMKCnrSoqTkZpmg1X0cZ7Dwdz70shKRa4dwkDlqAUwKpWENaycS3rpm2ddsFs9rMdzLOm8t13iTW+F7ynM4lAkZ57XtZXBOuZVnlApPPFfH4Z+/zPmOme/QRQCOCsd23xAFyv3nEhaaym4/ILTK7UOLFbhsDnjabNMiZFsGbMuEjJwtM6TEcVv6WAy3zCFPmS225IhL7gS5OLbNiCtmhyftbQC66vvbuxrzxvKU90VISdf5ydq2TBhRkErJASllFXKQU7JvJvQkpEHILca878EW722d5tzZfQocJ7RJTUNiLK3KGL+qDRJKasZwLqizKBECnL9wh1NrfW5cW+OhB27jFK5nKcY4Dg7rqBN29+vUaiW3+/54X20Jp9uGB88U7OUly0HIgIJlE7FV+NDjW7fbnDo5ZOXEgGZrQhw51pcKViQmrY5Rf3+mfhtvsHdoNQ2x3JJ9ulpHEO7VkyiOLenzrOxwvZKnndQWOW4mZxuYjN+8nvDPv+D48GcaTDLDuZ5/ziMDF2jxhqBDKH4TIrZ+4jka1kmSV2c61hxzwJyEzDHHHHO8ZLTrikhVbqbHRCQr/ORD1XsNJrknJYXzKT0HWcmnhyN+NzmgrIrOVD1pycuKZFQ/DwJlmHpiciAJptLkA1gVVsIQq146IgoT8WZbA/z5Dz3ykncVd2TAjgwoxfEUNwklmJnUw2r2cTemv5vuDt/d0A3M5C2FFrNI374bcsvtclN3ec4cctUc8qw54Nlxxiev1PjwOz/wshGRsmohF/yEohcbnrpW59ahoVd5X1J8stSbnU9VisWwLRO+v7zACTo8JwNuiJeqnQwjslI5d6LkJ+9l5pnolwWLoSWqtuiNQD1ybO/HNOsF6ycyJqnli8OUK/mES67Hm8t1tmXM28ozPGE2qRMDsKodT4rcCufcIqva4ZxbZCw5VgWL4cHyFPe7taoxvODe8gQAT9ld3lyepKaWDiGlOHoaI8BJbXCSOuMqSctWnSNW/PNZihKGJTeGJWKUS65HRyMyKWaL/Vgt+5IwlIxcfZrb911QXteOONxfoNU54vTZLTZvLAPwExcNv/eFNklq2bzVY3lxwhPP1XkmndAMvTH99lC5vR9yoRlwq0gRYL1h6ZqAzdx7QCaTiKIIyLOQPDcMx5aO9fKpqRHdybTHpEAwWCxlFQTQw6d7NdRLtB5w61xyKwQYGhrS0JCckhVtEqulpRGx2pkE8gkOeN3FfcJAWW0Kh4Xjqg5JnOOPCu+RSksv07yz3fxGHtJzzPEtj7knZI455pjjJSIrvPk8CpTDkeDwkqqp6Xz60Yy8DGMvdVwpR1wzfcY2o6Tk6rDN2WaAAt26N7lP07c2D4XSWa5lKVv/P3tvHmxZdlZ3/r59hjvPb8yXc2ZVZs2TiiqNLoENGNwYVLawTTPYjO0O6HA7TAd2mGpFR7e7oxvcdti0jXFAN7gZhAQIW0LCRhIIDTVQ85hz5suXb77zdIb99R/7vJsp+w+BVEVLobteZeTLzHvPOfucfevttb9vrSVjRhLR0DwjSQjUEMyUCTfxlfZTHwQTWpS29tmlS6opSbYY1awC4mXL6lTTWT/9QbXg4Di3IhAnaj/4twOHLUFJNWUj22WOiLmNJpsxNAkJ//5Hnoh+5jve8haNamAohbA1VGo5IUrh3uMTXricpyCuzlMiz5SEvBgCEQ4VPY5piyujmAVb5KRWyYuh5Bt8EaZW8T3L+Y0cozSlY2MSlEIQshAKvQk8cMc+n3mhyVpdOb8ZsFRROiPhSBjSid19f0Z22JUBC2mBB+0RwBHQVJRlW3bp5eqxokW2ZUxFc64ioS5Zc0rCbXaJqjo7510ZZ88BivhcF1cRWDcuD6avAQOJCPE5aWtclT6BWC7YAUaER8M6P/NUhwJj/slnQn78PicoH40a/LNzPWIsh6VIK/C4HkUs53x8A5By35kunp+wu7VIuTLitjvPs7CzzM52lXfeOaC1tEt3v0mp0ueexDA8n6c9UV675LRWF3spT7FDUQLe12xhFRZTj7sqhmajT7k6oN8tc+FqlVf2E6o+nE9GjExERXP0ZUrFhlz1HCHI4ZNgCcQjJSUi5nW5gUUpSZ6zusxAIppaxpvpSjxy6oIoH/BrvBoPmEg6Cz38l096/N2He+j1KqYHDwY1PpPs8YjX5J61lEZ9zF67wMaez7ObHg+91ZN7jjm+SjEnIXPMMcccXyZUBSM204DAOILAYyZCP7DvLYTQHsP5dMiG6dOXMRPcYuZlOtxXajGJnZDdqmCyndrnJ0PyeERYPBVWtYKHUNGAvsT8tQ/9zQ8A3P4mjeeAgOzQo6fDmfBcsq+D7w+EvYKQZLvJAd5M7OthSLEz0uJhiDWhaWrs2c7sfLEmLJoGe+rcl1zvvc+L3jZthjxuz1AqxlSe+M0n9j7w5Vd1/rTYHinVUOhFSs4IVzadDWykSiqWCq5NJxChEXocWYy5tBXwKl1Oa5XL0ucuqZNYOLvmCNdOJ+DGKKVvU65kVZJ4aLk7LXGsZQnCBE9gvS3kfDdnFiuWvR1YzPn0YkvZhhS0wWe8yzyYHsbP7GYHxCDMCOmuTPBUmEjCAiX66nHRdDht61jghjiXs7HE3Jcucc0MKGuIAfpEWLL2CHGVrLKG5DCEeLPjWpSXpkMK+IxJiCTl/30+z4+8q8dvPZenSZ5Vcfetl1iO5XKcXExJrXDk2BZilN2tFq3FfXY2W5Rr+1zfqPPihvDIyZCwU8NaoVTbY9ivkDMF+ollOQjoJZYzDQ/aiww15RN7ff7GiTzlvEeUCMXSiCuXl1hd7bBYjxjtKV7qhPsVzRGLJcSngE9OPRDwMzvqWFM8MaxqjVVqvMJ1mpQJ1aNMOHN3O6j4FdVnCTfOAh6xWgYS8ZrZAuCO5+7g3iMJBc/nmaRLKpbIKs+tGxo7FR65Z4/Aq/L6ReVzj3/kibd/6K0n2nPM8dWGeTvWHHPMMcdXAEceYBLfrGD4XuZUlblvDqau7SYiZSQxEQkhzjp13XTwPaWUszMCIgLGwDXTY1+mdGTKrhlx3fSwKC0J35KxGIR9BrS1T6LJzJ70oMJRM2WapoYghBJkwW9OmB6TzojKwesPXIYOWrT6OpxVTQ7erygiNyspu7bDmIgFKuxpRC4X89rlEq0n3trWrEHs8lx6kZKqcuZQTLVg6cWWfCYQj4g5blsAnFhI2e8F7ExSTmplRjAAyoGwue9Tq0R4Bo5XPU4UQt7ltziuFY57Bao5WF0eEE1DVmrKWl1Zrlq6I+HVbUdeqzkoeE5sflwrnLXLgMugGZNQzSodqdysOpVwgvQbjJmQctrW6RBRI8iyMtzz3JEJgRoG4shHWXMYIE+AIBTVOZpdlj4xLs0anGPanoz5ntsDCviE6tFmyniUZ0DCj94Pp6s+Zxs+Dx+G9zywR6M+5e57LlOstClVd1EVyvUdjp95g8vnT3DP/a8QZT7Ew2GBMBdx5dztdLoFrCpHKoZDVeF0zSNKICdC0/P5GyfyHDu+xZG1Hv2xsL7uns2gX8TzlLe1cvRtwp4ZY3FGCRNi2jIhwGOsUwaMiUkoSZ4Q12oVk5KTkOvszdoPD9qwFjXPES2xQI4TpQBPHDGfSEKoHke1yb3pIe49klAsxngCD/k13mEWeFbbDFNLL1I2N2uEYcr7H5xwV+OWVNM55vg6wpyEzDHHHHN8mXAidPf9rbkfSeoEqOPICVBHidKLLOtejxFTQnwX5qcVBMOv3GhnbVjKH92IMy2IE6FfNm32ZURXJqzaCgq8SpeyvvkLl+vSoU6Jsxx2InR1fe53y3GapkZIQFcHBOKTqstQaJgKmu0OJyQsmBqLpsGyac6qJgcVk0hvOgrFmswsfFNN2c0qJE1TY1s79Blz3nT46eeGvNif8u9fCln+n379LSMigzRFBJZLwgNHUoZjn3O7wu9ymd/gdXL4CMJZrfPIMUujPiVK4Dlt4yGc1ApnqXHO9unFllTh2maB9siR0Yfv6HGkDrcVchQ8YaGWUihO+NxLdfYHwqe3ppw6sccohkSVN9IhGwOLKlyRPXaYOHOCLCywgI8AC5onJmUgMXn16MiUHB6XvDYveZt8xrvCRBJqnhNAH9cya7ZKS/Mub0SFrkzoyRQLM2I4kpiRuPYxlxjfp6whLVugqjkWFnqMM43FCgV++9kiBuGF8xXWB5a9IVRLEevrdWr1ARtXV/GCKX6ly+HjV1i/eBtqDbff8xzrl05ye9Pw0TeEz76e5/pGnZN3vIBnlIdvH3NkaUouUD7S2SdOnePcobLhjRsBo0EZ46WsNJ0T1mjisblT4PJmDt9z1z4iYihTYlKO2tosZyUUnwI5PDxWtcZxbVHTPAtaIofPGi3aZkJBfdKs+nfRdNlgRIxyz8khxxpQxucgX2fBFgEXouh5bh70U8swtbw7aPEKHYap5bl1w5MXcvT6eU6sDd+qaT3HHF/VmJOQOeaYY44vE3KLNrsQ6swpy/d05nLVj5TIKq+lA3ZxPfcWnbUfDZkwkpjuyCACl6XPH21GfOqKnS3Yp+Lcpi57HXrZInNP3nxrz2WtOTIhlqapUjNlQgmwohzShsuEUEtR8tRNlbpUaNv+rLrh4eFni3XBJaYf6D/kP/txoyiLpkHT1GYLX4B928VimZKwI30GEvGUd4MnZYsvvFpl8mMffdOJyOce/8gTZc9jklraY+X6bsBmV5ikliET9rXPe9MTrKgTn0+mHmGY4Bl42GvS8HwCEUqe4QglUoW1VkKQGR+pQrE04o7bdlkoK+VQGE0M00mOh8/2iFJ4zexxfaOBb2DDTsnjsZVGPJV0OKYtnvOuOyte02aSCcDbMmVAPBOO75ox+zJiQsqxtD6bYymKbxzZsCgdmbItI3oy5X5pZiTSLQjGJITqz3Q//oEJAkJHpoQYQgy/+jmXI7JGkabnM9SEsnh807vO853v2MY3UCxFDCaGaBryh2/k+MIfP4QmHp29RdZOXGB/+xDiJ6weuc4D962zFASMrOXQahcJI5ZW9jl+5hXq9SGeUe6WGleGCYcr7rNy55GI/f0i3U6JZn1Cb+jPrLBVIfCcIcSyLVPTPAnKVdOlJ1PWxdkA93REyk3NU01ztGXMitZYs1WqmmMqKSEeiTh736HEGMD3U1r1iG86CWelgo/L1PnhMwUWFvtYKxypQSUrkapmJFKV2Cr9JKU/8Gl382/2lJ5jjq8JzEnIHHPMMceXiSi5GUAYpzIToo8jlxMSp3AhGvP5dI9tMySPa3FJMicdgAWtkFefFzsR/bETvnaIeNnsZwJYn5rmmRCRU4+ROAtc7y3437ePEKhhj36Wj2CoSRlPDalYFrXKHXIUDy/TgNwkSgfYtvvsWLfAc+na7jpNRkxutfb1MOzbLk1TpWXq7NoOTVOjIiUUZVs7jJgyYMIFtpikypPbMWv/5FffVCJS93xEoBZ4nF6yRCn0YsuH5BwAFSmSx+M+bXGfqXNoZcD5K1XOnOhxpGpo5JxXmCqOpFiLETh5rM1iRVlupDz9wgrXNxpMYlcdC31lfaNCLh8xTpTjts40MoSe+8EcYJhi6cqEdWnPWrGO2BpBZr/ckTETSdmXEYrO5sWBbuF2u+TajiRmL05YtRUqxmNNS7RlQlVzlDxDLOlspx9gIi4DJq8+CUpfpoyyv4uxjEhoa8whisQoCwXDohdy/4LP//57h/iDp1Z4z0PbtJZvcHR1RBT5HC4bfE/51MffzavnWqRxjsbiFluXz+L5U3rtGqkqh4seufyU7Ut3kC8M2d04zmiUpzsylDzD6apP6CuvdxP6A5/RxEdVsFYYTp3NdegrV4cpT21aztNnxwxpyxgfmVV7mlomwCfFEmTUeULMphkS4wIJk4wMDySiKxOO2AoTEu6hQcX4eJ6Sz8fEqXCk7LEmBQzC77/hMegX2W/nudaFWJWJWizwIC0COXBGE8rF5M2cynPM8TWFuTB9jjnmmOPLhKrMckDA7bweuFvFqRB4MMq0EreKtA92/UcSkcOny4RXJWWwX8EY4aLXxqgQ4ATHAIe1wa4MWbI5fuBD3/uWilhXtE4sKYG6rfyD7zelQ19HTOwUX3w0szVNsf+ZG5bOLHpXTYt1u4NiZ/oRRWcp6w1Twc9+FDVM5Yusf6tSmulnbmOFT6Vb9Lwp+afXOPtDH3ui+gtfebL6H7/vI0/cXgko5pQTh/v8m+eEZ72NrF2ujEG4Lh3+0FsHHJlKnztCIJYoqTKM4KEzfXIXKyxWU0o7TUIPtjvgmQr1csJ2J6BSUPK5lIs9oRV6HC0lDIY+T764wDvvHPDG88IbO8JGPOW86dCREQOmMzeyV83mzJ2pph4bpk+egD0ZzYheTn1GErFthl9EEvdlyPNG+G8PLbHZ8agnBW7XwxQDSBXq4wJplkGTZL9blG6mnchrwEgiDLBtRtRsFhAoI5Y0z41RSiDC53YirpsB3WmEeWaJSm6JVjVlNDG869GL7n5/4ST3nt3jqafOoAp3nd1iZ+Moe/tFAhH2J8ozzx/i8NKUMBcxHuWJIg9VWCw5AlfMWb75TEIUedRqriI4nQRUC05X9fRWQldjplgOUaQj49n92JcBTS1TwOeYtpiQgMIV2eWYLswE64oSqKFvppQ1JJaUCSlLWsQzwsOHYTIJSBLDQnNE6Od5qafc5Vd47J4+3W6BwcQwTi1nW4a9Hfh8sscjXovQQDUnXB4k/MLFIaeo8OhXOpHnmONrEPNKyBxzzDHHV4hSzs6SmaNESK37w/ZIs/RyZUhEnnCmkXDpzcksMG5EzDXTZ09GBGqYSuLyHjBsSY8RUbYIjd6yccRYppLiI+zRZ0u6xFkr2JZ02bddJjp1Vqa3hBKmpDRNdXYczRayVYqztp6WqWf1Hzf+hjjy0bUDFKWrgxkZSbPj3ipqb8uIV7hOUQNagcfFTspXil96/JefKBrDqbUJSSq8frnCMVMgIeXt6RG2pMd16aCZu9KEiD5j/oNcop0mbI+Ue447u9UohZ2eRzmEUysxuyOlWnUtQiutiLvvvkqnFxCp5UjDYkRZbI1ZrlmeP19m3fRJVXnFODH0iIgiIQWc+PyAaCzYIgmWquYY4QIk8xrMWuCSjLQMiGb3z6Lk1OfKnsepQ1NUoZKD1WaKKjzCAu81K1iUqroMkoNqi6IMZTq7Zwaha6b0xc3eHjFJlgNyVVy74ZSE18cTXu8mNGoTzp7e5Xc/eYK97QXuu2MX33fnvdhWLlxcYjQMCQLLQlHIe8IL7QgRRVU4dOwSIlAv2VnVcXlpSLEYUa+PqdZ7TCcB13fyWBU+vjnimo444ufZNiMumh6HbXU2Vx9I12ZtZymWCjkq5FiiPpv/BiGvPkOJaWiBUB0Ri0hJcO1USSIUCu6zqCo0myPeuRLy9tsmJIlHZ+DmsicuM6hkPNc2Zy3PRn12x0rZ8zhJGQN8/vHf/XMN55xjjq8GzEnIHHPMMcdXgEJoiVPB93QWNAhOjNzKi2ufkhQrSp8xzoPHtYFEJLRsMUschyEREyKmkjLBLXC2pU+OgJHEDJm8pWNJxC34XSq0Iw179N0v26GeEQ3BoLe0Yfn49K0T1y6aBgBbdp8eIw5rg0XTIE84E6unWPZtl4SE02aNlJRq1oIFkCckT0BH3W7/hNi1F7EKuF3kK/bN0cSERmgt9BhM4f/sv07JN/xoeAcXTZfH9QwABUJ2pE9MSojPe9NjGIRnky75QszVHZ9TKxHdieJ7sLTc5R13DCmVx/zB1pjt/ZDXXj1CoxZxTzNgp29443qOly+VaNan/PPJywjCVuLGeUl2M/LmEWTVowM8721wxezjZbbIBiEVJ1RvaZ4cHjl8igSMiGfv8xA+O+7w6rUc73lgj2/73o/xtr/6cTqR5Z5l4V13jPjFH3iFVS3N5oB71jITXU8kmZHCgUSZc5RhXcdctxMmkmDUaVCuyYCrOuS3Xww4d3GBcap86sUK1cY+v/XZFqsLEyKrxIn7zIzHHg/ds82ZQzF5MSwsdmisnWdvc43DxzaoVyOKOSVJYe3kq8Sxx2TshOkAt5/o8vHNEWsmT5mA2LpKRsWG5PA4Y5vUtcBQYoqE9GWMIDNy0rJFigRuTGroyxRBOKzF2b01CAU8Hlr2yOdSxuOQSmXCaBQyHocsL4wplafc2HbidFXBqvJ6zz2HiJS1ksddfoVmXthNYrzM8jm4VWA2xxxfJ/C+//u//7H/vy9ijjnmmONrDYMf/ths51IPggntgSuWS0evFpRkFHJV+iRi8fAYEzFiygldwBOPiSRMiIjEtR2l4sTEEQm5zDK0pDl6MuaQ1slrwO//5mcfe9f7H/70mz2mP/7g048B+BjOs0lfh6RY+jp0NqQ6zaoZdiY0P1B4OKesKh3bY8m0GOqYgY6xRrjNLtPUEj1xf7dg6pSkgI+Hoqxqnb7cdA3zMOyp21V3Vr6QYOnLBE88Xo+G/OXyAp/45Wcfu+O77/qy78POr1987A3tsX2twULB8N88AIdbKZt7OR6ulPnF6TnWtE6egCIhY4mdQxZNutkC/2SuyEI1QQwMxz73n+mAwisXarx0pciSH/K2u/YIfHjhUpGtoaUSCs2SUi0qlXLEh7f3uN+ukqDsyZgiOaYSk5By3DaZiiXOdu4PqklDiSkQ4GOYEGfXZriLBntEWFFiLCtapaYFIklpaYFDYch+u0hNa5QOX+P+05scvvsN6s0dJB/x8rO38dPfcYm/cmbAv3+9zM9/1zrt10/TzoIm8/jks+c0kYQRCWNxeSGJqDtvRrojsaDCxUGCj6HgGS5cXkBEmIwDjtScjurY8V2SOKTe7PC5lxp8y4N9Ou0yybgOKkSTPMYonV6OcSQsVHwEw3gcUihEdHoFwsByJCxQDw3pxOfuFWEyCDmbK3E9ndDOdC2uvVGoaJ4bpksZ11rm8lZcwOPQOBe3EiE1Qr71cMhoEJCocmehyErD3Yul5Q42Nex3ipSKsSNGk4BJ5DGJhP0RTFIlEGE573E2V2JzbHkjHfJgK+CBYzG3LaXcvhoRJjmC/+q2N/0zPcccX82YV0LmmGOOOf6MuJWAJKlrv0ozAgIuRV1wupCuut1tl1EQEZNSIc+uDBkxpc8YRYlIGMvNnes8IYF6VLVAAZ9VrZFXn10zJCbhf/7r//JNb9/4yQ/+2Ad+8oM/9oEN6TDSsbtmnTp7XWKirCLhZTvjAKtmEQ/DgtRm49zXHpo5gO1ql0m2+FvRGuAcsDran+lDiura1MZMSbL2oYaUWZQay1oll1mgGoSrukNPpnzDfdu0yPE77/vgl30f/tKHv+sD3/eh//oDAN2J8vkXFiiWRsQW7rx9l29KT+BhuMsucI9d4L70EH8pPUHRGHrEKLDb9dnYC9ncC9keuypBsTJkuZFwOZqS852LkrWGe4+PWS4aji5FNGsRYWD5lec9qhT5rHeFXRlzVfa5IS49Pp+NuqZ5vCzjA1yVopbtzM9yV/A5qRVavtu19zV7bUYKBhKxKDk6keVcN+GZF5cAkMYAMcprTz6C+JYf+9ufJBpX8MMh//YHXmbcb/Gk3WcoEQ3NU9aQvkyJM0JkRWmoc3fKZRqiA/OBnHrsmjHbMiJWpZOkrE8jrsVTUgv7Q+HRd7yAWicsjyYFbl9N8fyEhcUONvXY2nbVt6deq5IL1AV/7rt5dHUrh/EsqkKpPObo4X1OH2/zrQ91GEfCe0/AsZZlXyaMJSHBEqpHKtYRJZQt6Tlxunrk8emaKUu2yGFbpaohPWI+d125b0X4xuUCp5YTcqFlcWFIe7+GtR6vbxqmUx9rhSCwjKbCOHY5L+AE6NfHCb1IWSoYHi1VeGNHSFPh+laBze0Si423rs1yjjm+WjEnIXPMMcccfwYMf+S/tIidxDddslRvhhQCJAeOQ0RUKLCmdRIsq7aKh0dHh9S0mDlnpbMe/wCPquY4bKuMSZiQsGUGLNoyTS3R0MJbNsaDzI4DkbiqotkA0+zrVtergzarfdtlyTRpSpVQAspSRNVyQ7oMJZqRjaap4WdhjYtaYSwxeQIK5OjqgGWtzrQj+zKcVQA6OsTiqgJXrra4sxrSJOT33/dbXxEhe5kOT0VdXhyOubHRZKWq5ItjzkmXa7LPi2aHtTDk7qDCYuATGuEuv8KD+YpbTI+Ujb6ynk7Y2yuxfnWZC1s+MZYPjzdQK1zbLDCa+HzDfdt4nuJ7lkI+4Q+8CzPx/Ql1wvwDF7WmlmYZLAfajgMyEuDNrJ7zuODHC9LjubhHgGFZi5y2DQaZliivPrs65Zm0zfOyD8CrH/tmXv29v8RP/1/v5p+e7/L3fu7t/MN/+wh+OKGzfRydhLz04jHKGmCz6stQYooaEOI5G2ZbwEOy5PWbLUV5DZhmC30ryjUZ0teEnBjK4hF48Oi9O+xvrVFp7FCtTlArlAoJk3GOJPGpNtoU8glilJWqst0zfLi9S5oKGzcq7I2VJA5YXBjymT9ZotstkytMsdajVU3oDX0GE8M9NGZtVxNJZsL6w9rAxCehiQAAIABJREFUomxLn0hcdcNXw7YZMZKEcuaaBU7rtbYypNUcEQTuWHFs6PXy9BLLbjtHFBv2Ozk2+sru2HJjaJlkeplW4NHIC72p0ouUh09GtLshl9vu2O3eWxNAOsccX82Yu2PNMcccc/wZkA+c9W6UODtQzzATpYNrywr9m2TkuhnMMggCvblwvGF6LGgJXwwDJtniM+R02mTDuGC4nkzxEBa0mImVY1K56V70ZuMHHv+pJ8C1QEWzCk6KZCWeW7M+DEIz03/AzdwPD8OWOovekuRZkxYTIpq2wEQSfPHp65B6lquwIR0CPI5aR2Rik/KyXmOJOklGdva1j6I0pMKiVuky4p+u3+ARXeFtiwG/urv3FY17JG6s6wz41xcNoXrclfh8z+IC/fEinoGVZsxg5LPRFcohpFZYrFo6Q8NaHf7dzi4DM+VnL3v86PIyvzO9zlFqPCYr9PtjDi1O6PZDiuUeYaeMiFItj3m/3sHEWq7KkEvS5w67wkhiNsQRQZdo7hbMG1mFBJwj2UEl6YrscVSb9GTKWBK+rVFjHMH6MGVoy3TNlACPy6ZLVXPk1OO3ejt4PWEqKWUJqdkcXTOlJ8o//91T/OBf2Ge8v0pqhTWK9DWaubul2RwO1KMtUwr4eHrTAS7AY5KRlYkkBJm1dFsNU00p4rM9Uv74uUV6keKbFscasNCYctvdz7O/eYI48vGDKblcwuZmjZ2+sFRV3hcu0O1bru0bzqyk5AtjomnIo/fuYVOPNPbZ2imRJIaXdywDm3IoDLgzavGK2cs0Wpac+lhRjmqTCa7N7hlzhbvsGr4aZw6BpYzPQ8sel/dhbVkoVyb4vk+10WFv7wi+b2mFHr5nmUw9UivEqngCE6u8Qod3eS2aRSH0lW4/pegZ/uCcR6wpeeMSWoKf+7a31PFujjm+GjGvhMwxxxxz/CnR/cGPPRElghFnx2sV4vSmRW9qXXZI4CmVguX1bsKEGEWpqUuatpC5ZFn6MmWaaQuSTPC7Z1x71sH7wC2SuzJhwRYYENGWMVO+cneoW/EDj//UEwdi86lOUWxW8fBmmSEeJhMje1m+gjez4j3IpohIWBDXLnNQLZmScFxKjCSmKVWaUp3tmnsYYlJ6MmXfjAjwWJI6i1phQsSyVnmA4yyJCz48xwYLWqYrY56WbXYHwn3S4Pfe96GvqBrSzoTKU0lJRXnquVVygeIZWKy7XfI7zm7yFx/dIB9AIYT9gSEXKJf24XatuURzUgYTMyMLp5rC0uo2R46v02pMuHjuKGHoAvq2t2pEanlNOvyJucblTHAOsKzVmW0uOPvdA1e1o9okp24P8eA8Nc3P5svtJ7rcd6bLD73vaeqE+OrIQSFz0XL3XbLFuMcEp9nw1OCr4XXToVju0tlb4J3f+QlOlwMOa3l2/nx2bh+hwM3rCNTMbJ1dXgmz8wXq5k5XIorisRcnTBLl9kXleBM8ozxzMceffP5tnL+wwMrx1/DDMWlq6I88miXlua0UI47gn1hK2Ot57Gw3SBKfMBdxY6tCr1eikHOfjbJvGJFyclF57FDA+wqHZhUkICNShjwBZQ05rcssaIEgm+c9IkrG48TRNu+6q8/vvZSjVO0wHufo7LUwXna/V2Ne2YLnN5VKMcETCEQY2pSW5olViRJ4ec8RkECEiVo8ESbW8tzOm/tZnmOOrxXMKyFzzDHHHH9KHGg+DtqtPAPTxFVCosRlLlh1VRLPQE6cU5BkIWkH9roxCWXyzr2HAsMsybqowaxtpKQ51k0bQVjSEn2ZOttbzVEmZMSb00P+A4//1BNWU3zxv4jW+Lgcj4MWoIMdK0FomfpscZxg8W/RKgx1QjVrw0Kgz5h77CEu6oBEbi6qD94Lzg62oXkGRDR1gUtmF4DjukhfnE4kwGNbOyxJnXO6SawJFQrceXRKdClkPfri0MQ/CwxCEZcFoShbMmB7VMf3DFf7KRsDw6mmsPHsKqWcUi2lhL7l2MkNfvtTR9lJYgYkriKA4Ze7G1jgHZUyoW956cVjdEeG++/cZfVwh/buIgC33XGBYycL/Mc/PsaJcZX/6F3gZXMDgDwBa+qIV0EDppkz1SlddGF64jHJ0tKPaYu2jNmSHifswqwSYMxtfMtanp+/4UT+iVhQCDMS2bB5ds2Ioga0xTmvxeJsaH/+o6f4W+9sM7h0O+9950XSz5zkP43GxFgmJiFQR7SSbCHvY5zmAg9RVzOLxYnoRxKTU59BlotzWQduvkRF4t2Av/LOG4yHRaDKxR2P4wuW8y8+QJhz5O/I6pB+P8f+TsLFjuEbjqVc3Ao4uRyzsNjlxkaDOPYoFRKWD21hTEp7d5FaJcc7cinPnTO85+EbrHTLfOE5RzKum4GzyRZLzeZIRSlrSFsm1DRHV6Yc1wpDm3L+coOFesS33j3l//69UwB0bMz3PTyi2y0wnnhYoGsTnrkaEBo3/rwYTpkiqcKzgxEPlIvsTizD1LLoBwzSFM0ySeaY4+sRc3esOeaYY44vgeGPfPSJ6CPnH4tTACdAj1NhGEEpB9MUcr4jJ7msFWswFV6bjmcLsZO2Tl9cS0tPnOi7RYmIlJFMeVt6mBtmgOIWxSE+PZmwqGVSlIq6nvFILD/0oe/7wKPvf+grdtK5tfqByC2hik5Yb7IvgENmwSWHS46xTunqgIo4K1JBZtfti9uxHxNRFCdYXrM1vqla55VowECms9feKqyuZ1aoAR45AsCRmnaWBg5wjAVEDI9zO/fpKnd6Ne47s8+vX3IOTU//xnOPPfjd9/2Z78vTv/HcYyYbw3XpUKPAZR3y8mTEZfqk1nBP043LM3B+R3hmL+F3Lgnn6HFDhnhq6MqES7JLR4Z0ZUR+UuUXx+cIe3VenA55Zj3k0WMxqPDUs0eZDmrUG33yUuDyniFPnu/wj/CUbs3GvicD+jKhRI4mJQQhVJ8ky2+JMxJY0IB77DKHpcSFjmUwMhRNjgcfeZYPv1aipGFGFoWJxDS1wECi7Jm7Xz6GioYsa4kf/9Z15xgVTvHDMc1iwHS7wVU7ws80JpHY2UI+ltQ5dUlCRUOsQCRpll2SkohFMlIeZ5bCChTw+LXLCelOgwfv3OfIYsTq2i5vXGzyynpIwfjkQkuaCqU0zyQBTTzOHB2zsZvj8KE2gket0adaH5DEIdG0QL4wJp9PaK3coOwXWDx0FU1z3FXLc2k74NsXq7wynFBW54A1koSW5klEWSTPihRZDQNub3gcXxuQpgYR4eRiyqmlhDsXIY5danohl7pASuMhIiSqjFL3ySp4hlThMkO6kbLoB64SYl0lpOJ7iMLK+2+fO2PN8XWHeTvWHHPMMceXgKrgGZcBMY1hMBE+1R7wnya77AyVcs5VRXzjiMjeWKnklYdKJSo2pKghws0WldXMJWpd2jQ0T4k8L3hb+Gr4e7/5wx/48d/8oQ/8nSwVXW6pHFiUv/MmpqUfEJADzYdmAvCD9iqLEojLNDlwrRKEshRYNk2AL2rFspkeAKAp1Vnbi0GoFpR+Fnp30BJz8L62DmbWxCmWUB0ZqWuBXFawX9Yq+zKkoQW+69F97mx5fNujO/zmZ1szu1gP4dfe96t/6rase3/xZ55o/OMPP/ETx1p8d32J72ksc1SbM13DkAgfww0z4MUbQj5UHnzoHBOrNIzPUJzb2UXZpm+izElr1el+1PJJ7wKK8jHvHCOJ+bR3no/84WHK9TZnTvS4se/I1urhbR5c9OnKhH+RPufCIG/J+DimLYoZCU256ezkCJvHmCTTDCVcoM956ZGoEvoWkZR34XQm5ewYB8ShqjkOAg4TlIiUqaTckCHGSyiUOnR2F+i3F2mtXOevvPMGXlbVS0XJZc86lpRYLKPsGYBrvzpwSAvwZvMC4G7qHJUiCxLSsQkNzfN6NOKjTzb51PM1JqMC9XLKUkm4tm94+lye2+48zziGZl5YqKXstXMs1WOSOKRUHrG92eQPv3AEz09QK+ztNKk0dhn16zQX9xj2FkgSn+ZCm7/9F/Y4emjAmpZQgbZEjCTmqunhqdAlZksnPB8N+IPdEf/xxRIXrucpFqfEsSGOPeLYEAQJ3X5IuTLmSB1iVQJxFdFA3Cd3mFoupSNuo8LZvKuKFHyh5BuMCJFVgltFZXPM8XWEeTvWHHPMMceXgO8pSeq0IJWCMpgIm2ZIRMIwtQymHuWca8uywHJZUAVP4DZT4aodUSVgHUVRtmVAgMey1tg2Q3qMsspH8kXn/ekP/sRbJlb9O4//wycAPDxS0kyETiY8dhTBwyNWt8B1C3N/RixubSE5ICCn7RL7ZsQ+g1nblUH4xloVEZ2916L0dERZnMNXXUpcpwPiXl/WPBWcv+khW8dHeNncoELeuWS1a0xj4aOfX4Rb2sEMwpYZ8Bvv+7UnJpJwYL/7n+N33vfBJ+7Ll4lqd/CJrRHXzT6HbJl31UtMSZw1rhry4tO0BdpmwpN2n9NxC+MlnGoIRoSzLHOlA59OfS7IDgkJqVjKWYtdQjIjkc+ba5zVQ/w/9lX+alTCGKWUUzwvZdArceb0Htv7fU7qEtvSn7lhGQRPDVEm8BZcOOGujKlpnqKGeGLYkzFdmSAIBQ1mT8cUx7z9uOVzl5k9txTLQCIgpKUuWdyoEOK5oD4z5ec/eoqVMODd9+3T7VRQNVSbWxT1KG0zoaB+lljirjGXXVtEykBiinpzrhyEPBp192KkKUXx6GtCiKGCzy6OoD58aoLnWe556Dk+/B/eBsDxlrJ57QjvfmCHa9cbRJGZVSNFLN1OhdZCD9bzjIdFomnAoaPrqDWMh0WMl1Ku9tjdrdKo+ywfPY9Nj1KiTpuInkw5ZitclyFTSZmSZm5uAQtaIFXl0/0edyeGWm1Mu1Nkpx2wDEymhu2dCqpwqGzYHTlR+jC1CI6MWBRPhChVEqu8OB5zR67IlXTEgoQzB6455vh6w5yEzDHHHHN8Cag6IgKw3ReeHQ9Qo+zJkCd1l2+zy3hGyQdOD5Ja2B/D1iShGXjEcYGcuNajqSTk8JmSMCKeaUBi0tmu/58HXPOVIc3Of/N3Vx0Js6DEQ6bFjm3T1v5MHA5fXAG5FYMs1f3g3xMs53sJ0gOM04EYhJZUiDNLYovLSalTml1bw+bZM2MeYYFndJ/TukSgrt3nlasFXh1O6BLzSLFCPC5QswHXZEBTi7OKzS89/stPfGN+gYIPtaLy4a0u+2YMBjajIWZHsMYtpGMsW0PluK2xLxNGErMvQ9a9Nse0hQE+uNPm0sfu4Hv+6jPsXD/BF15sMU5TjtkKr3s3CAl4d3qMP/Gu8xPB/fyz+Fnu0EO8xDpGhHOy6UhfErC7V+See67y+adO8sA9N7h+bYH/5ViN0cQDVvnoxphPexcBGGbp3e4/IVCPFa3wmtnEE6crKmhAjFIiJMBQ9g2l0oSkX+Pud3yO4PJZAKbiiFFMSlvGgCOCDc0zkIi+cVqjc/T4Ww+nJIlHr5+jPwj55NNLjGTXuV0RZ/nigqfiggAzUXpZXYXngNAeEDFBaGmeAMNULU0TIIACdXwupWNO7RZ55kKeb3qb8NiDO7z8xgLXOwbfK3JoNaXd99nsKysV4aEzG9y4vsjy6j5p7HP60JSnX2pxYjWi1K/ihy5AsJSL6LUb+J6lWB4wHdbxg4g+CTEpZQ25LkMumJ2ZC1mMC1w8pYvcUI8aOTY2y5w6sUe5NMX3LJ98I8BqyjuOCcWcpTv2yHvCbpzQ1pijXp6CJ5zQIuftkBOUuGAH3BtUeXk6xKIsBD7D5MvXM80xx9cy5pqQOeaYY44vAf3oucfiVGiPhO1pyo46K9S+TBjIlNfTAeGwxM5I8dW41qOpUA9dy8VywaVFv54M8DHE4hYdY4kwCAVyjImoaZG3vwlajz8NPvwbn3iMW6oZ3sz7SrCZr5HBUJUiRckzyTQefvbKMnnyBES46o0gLGqZHAEirhVHMt0H6tMgx0gtHRllZzIsa5WBTKlRwqIcsXWKGjA0EakogXp0NSER5e1eizfoUtM8/913Pssbrx2jhM/nk332mTLK9Acpljo5+hKzrEWeTzu8Gg85E5T5nndt8VC+xUs7NxfNBwvpiSTYVPgfvv0ah6eH+JsPdfnspRI70mdVqwwk4h+crfCed76EiFJpbFMLS7z7kVd54bVDLNoaJ7VJnYAXzSYv2g5HtMU/urPKNxdXOTte48l0hxCfX7s65ve7+/za5Yi/vlZnPCxgjPL3L13k6eGQY3GD9WTKNdPBIPRlyo706MuUBiViSQnw2Jehm0dEHNY6oXh46ojuJTsk3qnxtofOo5Mc33oq4nfeKBLiEUnKoi3hYZiQoOKsgCuaYyARD+gCP/nN2wCUqj0G3Spi4PjqmE9uT2dkWh0vmj3nWJyd880/21nbHbi2v4iUEI+q+BgR/vL9Q25biTmzNuX+ZTh6dJdTR4ZcvrJIsZBw6vR1VhqWwTDPK5eLCLBcdTN3c7uKZ2BhsYPnp9SbXUa9GlFkyOeUNPHp9wsMBgU63Ty+pxQKCXGUYzIu0N8tQ+qI00gSNqTNLn0CcZsBE53Skyk1CmybIUvjGqePDImjgHw+YSnvca1jSKY+b7SVUAylEIwa0lSo+h5jqywWPEziEaMcDwrUc8JKEHIoCAEhZwyNvzZPS5/j6w9zTcgcc8wxx58Coa+IwNPssm2GWSXBtSmNmPKs2eF1eqTZut4TqBdd01KUQmBg0Rbxs6C5mJQJMUXNkVefPAG70v9zGcsPPv6PZroJwbBmljKz3SwXJPteb9FtHGg84qxZq64FBkxuLjBx1r0lDcmpR5Q5axXJ8b5Wg6dl++biNBv7lvRY0RrFbOccXAXlQE+wYwa86m3RkTGVwLApXd6ZazDqrmBwSdQWWFNn/zuRhMtmny0ZcUTL9G5xEPt4u0uuMOToqfP86Jn87JwHwvhzZpt3LxT49GfOstX2CYIp/9t7Er4jPcP3rjQxCD/72pB//Fun+bWPPMRvfuQd1Beci1eiyq6MOezl+Zbb4NH0BLfZRa7IHj/86kX2ewEP3j7gPelJJhw4pDknrp+4eI7nLoX8H+fbjJmyS58PTdd5xdv5omfm31Ily2tAVyac0IXZGA6c17xb9Dw/9P7PE/ca2DQkKPbwM2KgKFNJWdMSeXw8NYTqM5IYi3JNR/zrjx/m5z65wB9/7naSxKAWfvaFKGvAcsefZgTU4JLZAYw6onFAtA8qIQf3uawhMcqGTujahE88X+byeoVieUBraZckDplOcqwd6tBY3KbfrVNr7rLV9rk8igk8p7tyVr1CpRjjeSlqhc3rK/RGhs2u4dpGhRtbVXqDgOcuB+x2PZqtPnHkE0c+T71S55HbxxQl0zBpnlQtoQSzCs6CVGlSZl3aDJjwTNLlZz9T4AuvVtjdK7J6aJ+/eEfkWjGBTpSyNbJMU6UV+OwlCceqhhvjlJwx7GpEIydMEviT4ZAohXaUMklvbgbMMcfXE+YkZI455pjjS0AEemNhHDsbXR+Xh3AQ3nbg9ORjGMSW7b7w2njCzkCw6hZMkwR64nZWK5ojwCPEny3eIpIvEu++lVCUMHOgWjQuTbpmytl4vFmL1a1tNLeOMyJhQzrZcW5qA0ThiJYIsqM4shHx8d0BP9haZce4NPQcPn5WTdmSHjvSZ0HLJCgvmxuZ1iFg0ZaZEDOSKZ8fd7k3PYQRuHjuCGNrSVXd7ntGXCzKSdsiltSleBNwSqu0bIFTpkzxHa/yR5+5h3pjQFVz2X1PiSXluLb47O6Ub3zv83zDfZsUj1ziM08f4r//vs9y+tQuTVvgDbPFjhnx2/YqfzjdY3tjFVMYcW8jpEjAL/EC/+p8nwVyvGa2OKXOivfMyQ6fernIfYUSP7N6Nwk3tT8+hn+VvsR16czuvQE6DGevOXi9Rbkm+8SSzvI5DtLSb0iXS7LLJbM3e34vfOEbZsewsatyTDOdykQSchjWMley711ukVefHD5tM+Gc9LguA952/zrTyDCNPaIsPPJgzs6OjSMfHoZELFacyD1QMxPUH1zTnhkzvEV0b3CkYjIuMJ0UCHJTqvU2rZV19reXGfSLbK4fJrHw2HFhsR5zpQOT2JlFtHs50tTjxZfXGI58Th4eYgSWWhN2uz7rbUNghMNLEZ5nMZ4lV5hy/219fuUl2NAxFQJSlJy4a+3piEhjlrTCsq3gZ2GaXZnQl4jXpyOOn9xEjGKMpZy3PHQY1koed68otdDQSVJWQvcZq/qGamDoScSVYYIqvKNWIlHlkh2xkUy/nI/xHHN8zWPejjXHHHPM8SWw8+vnH/voYJfrdsowszVNxZKIkmSVgRouEfwaI4h93jBtzqUD7gwqRKkjMmES0LAFBsSkopC1LEXZojkk4MKvv/bYxV9//bE7vvuut6Q943/86//siZqUqUqJkU6ISRjqmIZUqEiRihSJslEtmgaCUM0SMA4SzP3M3cq5fd3UheyYIQXy3EODl2WXAiETYrbNgM+O25ywTb4pXGKS+OzLKHPcsixp5ZZ76lyWFKVCjp5Ms8TwGiNJeNLucKc06U6UV+ihuFaaBMtxrbqFMJZ108fHo45bWN5dzfEL/+EIL0wG/NG64Vvqdb7thOH7v+Mpzv/JWf7XH3qS97zzAu//d0f4zNWAf/NcgcWoxiPvfZnXvnAv337XkL/77a/x/FO309Qiz3jXeLtZ4w+fOsa/mLzKDenyDwr38tvpBa6ZHke1yZSEroz5xI2IH72jiE18quWEwW6di2Z31j520LDkvrOMJJ7VG8gI4EGbnAIdGVPORPoV8qxo7f9j782DJbvu+77POXfv9fXy1nlv9gUDYACC2AgQAAcAKYmkLO6gJSsyJctWxFiJKlFs0bIDwWSiVBIlpShSRVJsa7GVACApkdpIiiQwIPYdxABDzL69/XW/3vuu5+SPe7sB2KkiKdF0JPR3aqq6evr1O/fevj2/3/l9F1piMJ5elcmh0TzZHvChq7cI/RJRkOcDV7ZZaO2jHhQ5rwb8nbkCT/X79ETI37tpmy+fN9iSfQraIRIJd1rT7Fvq02oVAHi2OxhPwka2zLFI3a/eSMkykGgBeW1hCEkoXj9KQUr96okQoSUuJh0fnr5ksreUhikKAZ1mHcNMCEOLUrnPzFTMjqU1bAvM2CNRgtlqyEbLYtAp0h1KDCFIEgOVSHzfxDSg5EHOhlPrBlXXII5NHCfkqeM1tqKYITE+6f3nCYfLNHmvOkyJHKFImNV5yngIGAc2NoTPQ+dsrnZL2HZCr2+z3TOYLiWc3JBshQkSKFqSC72YomkwV9LEA4uKZVByBC90fHbnLF6LBgxJuPrjRyZ0rAnecphMQiaYYIIJvg1sA8raYSCicaGX7vTHY+1Dghrv/p+W7YyukqR2nTLlzs9YJiYCJXTmwmRn1I+EASFJJpId/nsuWd9LOFhY2SRil5xlShSZldUxXQagJkpAukuf0qdiHG2yU6e2vAKBiz22iR2dgxEu6SFvTxbZo2qYSEJiQmKaYsDpwOfvLpS4Qs0SZ2J4A8mW6LEuOtR1gUVVpqpzXJZt9qk6O1WVgYg5IdeoKI9/s73M+6/vcqOZNklbYsCiLnBRdFmTfa4VlfHU6UW5SQ6TP+5s8a5y2uysyx5/0m7w2ydC/vAP7uYXfu6rbL7yNqLL82zTx9MmEsHTYgNhJ8SxZPrI8+BELBouy7JLQsKTKwnvv/0yP5TsJyImVvC1DwZjytK2GJAmbw/Y9/anueGGU+w7eJZPHIF0niYo471ZN5FRgUaPgXHDZ2SzklzWWI2u2ejKRSREJKyLDo42KWoHrQ1Ukk5NpBFjGppnozY5bfH8muJTt0R8pDjDuTPz+CJmSnvjSceRXREnT84RRYKvX4zHJgKj9UnE69bBGc1LZveCQqe2xVq8yRp4NFEDGIqYpg45qXrMmjZSav7k6Sk212pYdkwSm9Smm1hOQHGqhT/I4eaGzNQH5F1Fp2ex1tO0+gYlT9HzJUEo8WzFeif9HcstwYUWWFKwvOnieQH9Xo5IwSff1eLOYhk3m0BWlMso1b2gbZZUiQSNoQVT2mGb4E0Bm9ttm2K5x0x9yP6lAUEk2T+t2Zk3yRmSdT9mV96kFSWcaabPTzkCx4JF06Hha8rj/JYJJnjrYfLJn2CCCSb4NlAaeiLC02lmRk5bb9IxFEidhUYZG2nyuSIUMZtBgtKpkNYyBFPSwtEGFe1haImlJTYWVQrEKE4ZjXGR973GJz7yqXsv6HU8bZHXLnnt0lCtcTHrZhStlu7jCHu86w2pq5J6A40rzNK6I5KxWB1gWXa40ioQiIS28LPiOdXBbIgujxkXybnp5GIqC99TaGo6Tw6HDdHlnGyMf2+CYlP2xo8DEZPXDpaVUnr26iIV7fKMsUwOi5uoc1n57EuqXBJNXCz+0jiLBB5v94lQbNHlZbnCN40VtqKY5SdvZfrgcZ576loSFBflNjtVhRmVB0NjW4qzj9/Gi1/4IS4mPg0xwMbivVfEfPThLn9unMLFJkxAGqk2Y0v08DP/r8/UruHM8zcRDHO4tTX+5+NDbEwiYlr0MwtkhX5Dcf/GIMdR0e5ipY5KOtUpjM7RQKTXYnT9BoREIqErAqQV4OTaAPj9Kd52/Svj6dVpepw8U+em61ZYWtrCQOJpk3mV59fePWTPvos4tmLP7ianZJt81kyMkL7P6xjll4zWbWRuZlHWoI/0QI42x+sG8DDxleKh4zlMIXj1okfgO9hOei5zxSaO1ydX7ODlW9RnN9kx32V+tseeKhQ9RXsgObi7wyBI7XsNCattQcWDoiXYMaXZs9gjikwcN+S2a5p89cl5WoFmQExCmux+hCUq2s70T2k+ThmbiDRg0Rs1j9rm0KE1pEzI5VNb4WEoME3FdqAJlcYUgov9GM+Q7K+lbXrD18QJzOYFidbZdf3+0DAnmOD/b5jQsSZCVwvmAAAgAElEQVSYYIIJvg1yHzxw7Oz93zp6xCwRJyk1ZiAi2mKYTgW0RU/4IAQOJnEmyu0Jnx6KfWYeU0KUwFYcI5FsicHYLhWR7mL38FnQU5S1w0sPvHz0yPeQovETH/4n9wohiYjYopMmegubOVHFxRoLiHO4VEQBUxi8kfkvkdR0jkbmyDSrS3SEj0ZTwCMkPa6QmCAx2EuBF+UaIQkp8SwtfK9J5rmmYvFcK2BddNFoSrhsii4BMdO6wHk2WKJKUTs0xIBFXaYtfGYzulVb+FwtppkvKzo9k6ZKKV9tMeS42OK83KZGgZrOM5WlYF+UTdZkjytUnSnybIsh+9U0z8oVvraacOzFRT43uMi1agcmBnltcdpo8uBjC/xJZ42f/9hLbJ7ey290XmNRV8jj8GgjoEaBbTEgIuaCCvitEylFbZeq4WHRFgN+6T2rTNUa2O6AX73/7VznlvjEvgKrWwXWRHc85TAzWlPqPuYQZOd0RH2yMFjQUwDM6OLrYnAENV0gj0OYOWeV8LhVzHBoISGJXMLAZdAtYFmK2xfhTy+mDfONNZvLKyWW14rsEnleTtr8ynu3ePmlvWxsVPjt5QYPrvSyxijVfHREgIeFqy3IJoNJthYTA1eblLSTTUcYi9Q9bVLAQaEzLQu0ZEBL+KwyxFewqUO2o4TdnsPjJ4qculRiyizguiGWPSQOPV45voduz6HXt1mc71EshHS6Dv2+zeU23Hj1FivreSp5Tc7R5BwYhoK1hsPCzJBex6M+t0VBury6IRiiMBAUSU0VLNJQUQPBneUSU9IijAUFTPrEXGeVObpbMBx4GBJWVqd49qzDpq+42IZp10BpQV8pDpRMVocJdU+y3NWcjYeUhc3KMOFQTXKqn9I7D338ygkda4K3HCaTkAkmmGCC7wA7ZQ5bCpzMYjevLco6hyQNeTORBMQEY5VIurd9QJXxY51mjUgoSYMoS5FOd41fp3LZmLTEkLYIEP8RDHM0akzxGVGuRtQZN6P46Mz1qIg3FpjncUlIWJYdPGwS1FiYDmlxuV9NExLjE2Fpyd37Ne9T+8a7+CPRvYPBK5dNVmWXK5KZN4nx53UZgeBqluiKgIGImNdFYhQRCS0x5FtyDYGgXh3wzcsmp+MBB2QRC5OqzrNb1ZjXZV6T6wxERE+EVLRHDps+PndOe2yJPou6goXBQTXD/qSKhcFhNce89rggGpw2trg2maWqcwB8+tfv5FfPNfiv3WuZUXn6ImRddOgRZp8BizW20WhMJGfkJnntcEjP8yNfzPFnX7kOc2aDJdvmmt0+v3qyw27yOG8IgIQ356vYmGMRvybVH62LDgMREYlk/DOjfI6ctlhUFcIs/+JxvUHge0gjxs2l06Rep0QcWvywO8tn7hpw5dWn+f3GBkrDzukYV1s8++wBOkNJdzj6TEdsiwFKaNrCf9NnauSMZerXqWO+iNmWr7/O0yYFnVIPDZ026gkKG4OCtnGypkWhmcVlwXBx3QSt4cqFhNkdK0iZMOhW2FqfYX42PZYgErxypsTZS0UMqTGkpuYJGlslSp6mNRD4kWBpocvijM/uuQDLisgXAob9Ao+cdHjfNX72+TfYFqlAPMqolDlMHmv3uTiM+Mi1IZZIJ1Q37AsQQhNFkiCwiWNBO0nnlzvzJtuBYr4gODxlkigoGgY9X+AZgg/vsZkuaPKG5Pz261OuCSZ4K2LShEwwwQQTfAfYVTCwpKCTFdmRUOMv0C3RywxpFVsiLZCijJplINIE5UiTaAi1JoeJT1rwJaR5GC36YwFyQw7YkoPv6fplZkU6ovyMxOUBEQOCrBFKnxs1VBbGuGAEqGiPA0ktcwIzmNNlFHp8zAAOJieNTer1DhXTpEoqah4Jl4fERBrerZeo4HAkmQPSycq2GLIlenSzYtDTZpounzlf2W9oWCwrJlSaO6sF5lyDvapKRXusyS4F7XBQzZDPCt95nWcqCzF8aHPIRjaBOSk3uNWu8LRxkYp2OCHX+KpxFhuTK5IZIlRq1aqLPGFcYlW0ea7f5x8ekfz69SXmdRkLyXQ2oblO7RxTkSISTshVzohNBoT8lv8av/M776YZJRy/4FJTHs+KDSLSyUWSNVqjcz0yAXhdlJ7aIxtIBgRYeuSklRaxvojoixAJeDg42kQg+I1jZe7/yn5+70v70tcNHTY2y1yzL51oaZ06vU3lFSoR/P25KpahmcorpvLJm/QKW9kUTKNwdOrslmom0twQeL2ollpkU0JzrI/yRUxbBmlIp1BpNguaoYjS8E4RsY5PXyWYZsL7b1vGshRJbKGUgWWHmKaiNrPBVVefR2vIO5pECaIkbTj8TE4VxoKimzrTnTxfwjQVU1N9EiVx3LRBuvvqAa4XcL1XJELhZOd0SEwBiy4RAQmWELQ7DlOmwV4zDXeME8lKw+Kl0wVObwru2p9+H5zrx8x4kn4Ip9sJX+lt81Lc5uv9FrNFwXbXROuUinUq+t7e4xNM8DcNEzrWBBNMMMG3wfZP/cW9AjjRD9gQQ7RIi62BiFAiFXunepDUdncoIgIRUdcFrrenxnqQSGk8Q2JoyTpDTCR9ETIQqZi5ogscUdNM6xw17fLFBx89+tcNL/yHH/nn9/7Fg48cjYjGgYQAbd2npXs0dRdXOKzqBjnhIHndilcg2KtqNMUQnRWLNZ3HwxkHNe5X07SFT1sMcTI3LIHgsL/Iy92Aae1xWXbGOoSbmSVQmrptcs+tDR65aHJeNumJgCuSGeZ1CYQgRpHDxhcxQxGP11TC5WY9xy1HVqnZOTa2LV7qDzlpNFkWLao6x7YcsleltKUpbB4yzmIIgyvVHJtywGE1zS++Q3GbM8d7funP+IkrtnHOX8mT/Q4+qV6lhMeLxjJ9EXCtmsPApCsCujLi85sNnl4x+cz1Hv96bY1FXaGic5yRm3hY41R4gENqjilybIkeayLgZ650+NMVn10yh1AGWqTuY2U8GqKLh0NIlLmRqbET1aiJ26VqlLQ7dqEq6PQclbSDg8lQxDiYtOSQHDauNrl11mJPXbG09yJR6LGwtEwu77OxOkO52mS+twSAH0kcSyMlJEqw0jK4HAXj5mOkYxEIStpDC00sFJFIxnqPkUlDJBIQ4IuYWCiSjJKV0rPSWaGrTfLaoqQdBIKejAhEQqA139iIeOpcjlebijNna1wxn9Btl6jUG8SxzaBXoFKKKOZjBgMbTaoDkUJwuWHytkNtmi0XKaCUV9TrXUwr4fyFGqVigGEkbG4WsUzNpU2HS4mPgaAjQkwkOUzs7C7wUSw6DtcdauNoB9dJcJ2E8xsWd1y3ydJ0TK9vs7uuqJgmBVfTHAi0hmmRTnZK2Mzm08DTkqfJmZILfsQdlTwyMpm55+CEjjXBWw6TScgEE0wwwbdBomAYwYJls0PnsTLqSVG7Y7cgA+NNwuJR0X0pCOknmkRpLJkWJlUr3dF3tUVFpzurs7rEumjzorFO7w05Cl/68Ofu5a+IX/7Yr927Q04zI6YwsinCG12sZmQawrehmpRFmhMSZUV4RExCwlnZ5HAynVGlJA05xMneK8nsZK9J5lFo5lVpfPwH9jb5R7d1OJLL8fZkcTwh0MD+kkk/VmysV7jKLDLKLdmWPj9YL3CbnCbMnLMWVYm6yhGhqOjUScoRgnazilaway5kS/iEROxRdS6KJgrNhhhw3FhjmIUCTqsCr8kNLogG62LApUsVrjp6jM0/uoFzX7uLpR1dWvRJUDiYNMUQgHckO7m1nOe8bFDROYraYUYXiUj4xef67NPTXJRNTshVQmIW1NTYutjCwMzO925do0Wf7ZbDLC5fE5d5xriETRoSOBARdyUHiUiwsd40BRllygC0hT/WguS0xSijJU2AT2lbjjapZpOfIhZ+JDh05RlUYlKZXgfAtH0W95zn2acPs7SjRWeQukoBDAJJlAiWhzFB1mAIBEPCMVVsRMUDxlOaN7p8jZqSES1x1JyMmrPUFS5dd0cE9EVEQdsUlU1TDsfnJEJxii4nTk5TLHcIA5c4tFCJxMsPkFKztNClXo6wTY1takpuui4hNNsDgVbQ7eRptwocOLCGSiS9bp56bYBlJVy1M8R4w7ocDHwS2oQMiZHAM+sxnY5HpRRgGBrHibhmd0AY2qxtFEiUII4Fu5c6lAoRJQcWi5KiKal7Yky/2vATZus+39juc0spzxPbA8SEkTXBWxSTScgEE0wwwbdB8MXTR7/Wa9FJUnqKT4LOdnn7IsDCxCfEw0GjU8GuSKlJRe2QaCgZBoM4bVMGiaKjE/oiRAOhUAxEqi3Ia5dEaCraZVp77DI9zvw/3zr6wv0vH/1uxKv/8mP/+70xCg+bS3oz8//hTQx0XwdMy0qWEzKiTaVFpZMVwhExPRmxU5Voy4AZlWfF6OKTZll0hc+G7FHWOW5gmjOildrxXpphTha49ZZXeOa1Ood1jfOyjatcdjoO7zzSZnk9T9kRzAwrnBTbxELjDvLcslvxULtDR/gUtMOmTB2kApGwLQaYyubSSp4/bGzy1VabO+QsMrE5azTI47KgSpyXzTRUUXYA6ImAui7QET5d4VPr1dlTyLG2UiOfDzHMhPalOc7KJjlspkhF+Jdki5+/QfGlS2nK+JpoU6dIQ/QpkYYqSiR1ChRx0UJT04UxRa8ngrEYu4jL/907y9+fWWBfMsWJpEdD9JBI9iVVtrO1xZnj2BvT6Gd0MVOQpBa+q6JNVecBcDEze12XBZ1nCpuqdmmJgKp2OTAlmd+xiZCKF5+7gqU9FwgGJb5y7CCn2gnvfPsFlnZsM7ewSRwU6PYtHEtTNAx+/IYuf3wpTUofmRekxgwhucxiuqY8IqHGGS0jndFo7cC4aclrm0Sk71XCGZsW+CImEDHmqFkWYmxX7YuYtR5cWXZ56IUqr17M88XLEe3lOt9cMXnbnh65fAjaYLo2pNl26Pcd6lMhfpDmhczOdmi1cpTKA/q9HKWpHsOBy2Yjh1aCVttii4B8lnqiGYV0CnoipWcdmtbkC6ljVxiYhKFBu+swVQoIQoMHzvu8sGLwXCPiTOBzQ92i60tiBWg4GQ24vurg+yYX+jF7CxaNoaatYvZ+/PBkEjLBWw6TJmSCCSaY4NvA+8CBY8cfOH7UQvL+z3/0vqs/fuTYNR8/cuy5B148OhAhrrYp4hIJhQAKGZEjLeIVIRpPW5jZlmdfK6QWVLSLoy2acojKdolDEaMFtEVATXsMlcZHYSM5ef+rR/d/B43If/+x37jXxiQgwsWiJPK0dBdFGn4nENjCwhQmZZGnLApIJArFut6mILxMq2Ci0YTE1HSejgh4nzdLJwSEwBTp7vyQiFDEVFSed1tzrKiQb8lNzrQkvbO7uXVJUDIs3lOpkItc2oFGRC5KQxALvuFvs0OVmdIuVWFz/YEur1z2MIQkry0O6ily2uaCbLM/qeGLmGXRpyMC7lJL3LRT84lPf5W7tvdjrizwqLiMT0QOexzkp4GhSPfrb0yW+LHrA3rdHFf8nS+TNGsIoK5KnOlCWwzHbk8P3FHiRx9t87u3lPni5ZAZXSRB8ccPNnny/nn++Z09/vXFDluiRyBicthooamSRwpJWXtjOhlAjQK7KfJ4v8OmTG18AyLWZJeWGGBhkMMhfEOqekymnxAwpT1sUpepSCgioVJHLG3QkxHbIkBqSUVYGNqggMnFruK5kzW+cNJjoyt5/MQsD56Fi7HPNiFfPOXy8KkC4cWduIaBKTWFfIxWkm43R6vlgZA0xQAzs2VOp2AhHhZ7dYlV0cfKKGSa17Uho4Zk5CI3EFGaYYJDREKcaZBsDErawcWkKwMcbRAJxc/sKbPWtLAxeHZd00giHAz6JGgl8KRkR0ny0DdLHFxMp1e1qYC1hsvANynnEjY6Bs3tHDO1gPMXK9iWJgwcNhseYSQxTc23Wgn97JzHaBaFxxYBCZoZXHbaNs+sK1pbOQq2QZxIHjgX8Ei3wxMbEc90hpj69SlKEZuXOyHX123OdxTnYx8Pk2vn01yTH7h6gCElu8sCK3IofvjApAmZ4C0H8z/1AiaYYIIJ/ibgRz/3o/f9+8/9zGc/8abnfuujv3tvUwxSQbRI6UcWBlUcIq2JtMYSWROQUXUEAkebRCIeC48NPaLhSPrErMgee1WZiO/MMuuXHvwv7vvlj/3avXlc/umDP3vfH334gXuHRsBltUldlulrn5oosaFbb/o5A8msqGS/O/3vYbSjfcZoEhDxwqDPO3NlEl3mYb/BKZkWzhEJW2LI41HAnXKBdhLTJ+YrySpz3QV2LwyIIgOj6bBUFoQxGFIzjOBGo0ovSThNj6plEEUmN9pFLEPwrus3mPuxb7D5wHX8Xw8v8Kyxjk/IPlVnWuXokXDg0DLqguIPHi/zd29s8xfP5ZjGYFN0x7v3KWUoYp+e5knjAv9YLLLV9NhvaKp3Ps3mX97Kfatn8UXEHl3nsthmn6pTve0YX5qdBXOTP/3pBL2+zQf+2XX0ftflV/7Xr6IuGliPHOSwmuO4XOGCaJDDpq4LDAgYiIAiHoFIcLTBqujwcM/jgtHCwqQAY0e1BEXM6/SnkWuahRwHO/ZEQEE7ADjaGBf1OSwGOkIJzYbss619DCGItCJBsy19LC1ZR+Nmblqn5SYCwW5VZSAiVvyYu/Zu8PIr8yhtIWV6jX7+tgH/4jETU0j8LB8GGK/JE6+L8YHx+mU2V1NZEz7KmtFoBhnlMBYKqdPPXiQUPaKx6L6uPVYaFhuiy81mhXaccEkPuWFB0l+2ubZikyhY3HOeHyzOIIWm3/eYmV/j7KUixZyiXAxJlEMQCc5czlEtJFzeSLUiQmg8WzNdG9C8kN4D11hFnolbvHMXrJ43GYiYTe0zDBNCFITgbjgMYo0kROo0fLSmvHG2yMd3lDi1Ibhpv89zZ8AzJCjYZdtc3hK4Fhz7ZomZvMA0NGXvP4IV3gQT/A3ARBMywQQTTPA9ws989hP3GZmdqkJTVDYxKs1Cz4LJIj3KUxBEKMys2ZAICri06bMu2rTEgJq0uCQ77FAFzme0ou8Uv/zgf3Xfpx785H0AH/r8PfeNisK+9ol1uuNbEcWMpvU61eaNlqEjjr+BwZAQA4NnjWW+PNzi9DBgXucz0laqWzgnG2yKPi+rNlXT5IDtcZOeYb2nOX4uR69vUi9o5usBhw80KOQSPj9c4UTc45zuc2d+inpeEMcGP/qP/4QbDnWZuf0xzv7O7dTe+SQ/dfs2P1tZ4qpkloq2KWIxa1r8H1/ewa/8Lz/EgISlD3yNv5dLhdZWlrsxogLt0jUiEmZ1GTc35IrDlzjxR+8FBblCmw+SWgr/g7k6n/tBwW/e+zAnHnwf5hVrmPs3SZ7x6R27moCI9/65QhiS4akDPHC3zXvyVe7Rh7Ew8Ikoa5eyznGFmsXSEqlFusOuK7RlgIFkSIhPRJjND7wshd7PCnSZrf2NwY0SwVBE2fPg6NQI4ZTcxEAgdepIlb5esyEHdESYOVglDIlpCp8LsgGkVKgzcovLYpsXxRb3HfO48cZT5HMxQSTo9A1efGWGd7v18TkdfTZqushOVeJF0Ry7aI3+ffSZeKMGadSk5N8Q9Ono1x3PStpO6YyY2NogyF7/370z5h2Hhgy1YhqHHfNdfBJebUXcfstpmutz9HseQmqE0HRaFbRO197Ydti3d4tECQwJ7YGBITUL0z6GZKzHKGPzj65wacQxZW2nUzoS9lPAxiBBs0fmqBoWR/YOiFUaYOpiMqNyhFnzCFAqRNy032d1y+WWK3v0E8WCdCi7AsuArT5oDb3gu7qlJ5jgbx3E17/+9V/+T72ICSaYYIK/7XjiI1+8F9L0dZkVPoFW+CSclW0SFG0xJCB1NwK4I9nJhcz+drcusEmAjeSHP//R/2Aq853iMx/79XtHdBobEwuTLsOx7a5E4GaZGjYWSfbakdBaocnhcCCpcr1X5PQw4Iq8wzP9HnUcAhSnZIumGHC3WqKtY3qZRPldxRL7dvhc3nBJFKz3NQfqmt27ttnaLFKtDhgMHEwzwcv5qESitaTZzFGtppOU3Xc9xFd/90cYhvCBX/gcYsblM5+8m522w4/9+NfZOnmE2Xd/AySIHSW+/N++h/+h/QoFHETW7HUZ8tVPnqfxzeuYft8TiJzF5ueuZ2rhDMILMPa3iZ5bRJgxmAlypoVcyHI5IoXuCeScxS/93F18+t4vcfKz72N6bovTpxY4s25gG/DbwWtM6yLrokOY2R1P6+I4Od5EMsgyRkYBhaNpyAijcw6MrXwLuFR1bmyhvC46VHSOnggJiDCRVHUeR5sEIsbRJhdlMzM+6IxF8zmcTM/yeo6Low1y2uJmr8T1h7rUZzc5/vJuhqHANsF1En7zUgNPm9xkVpHAD968yc8/HmYi8wCfiFFq+qwu4WCO1xoSp8JvbVLV7jiXYyTe/+SBAmdWHG48ssWrJ+uEcXq/3HL9ZZQyWF+tMjWVOnWNks//4pkqd1/To9dzKBQCBgObWr1DFJqcPl8hiAS7F4aUyz2Ovzad/j5DEyeCvUtdtBZsbOXI52JMQ/H4KZs7DvtEkUGx6PP55/JI4LpZg+mKT7trEyeC+dk+l1cLOJYiTgT9QDIIYUct5sSKwQ37As6uuKz1FVfOwmpLctWuIY6bNpiPvlLkbOhzW91luQ0H73/fX/menmCCv6mYaEImmGCCCb4PWPr4oWPLD5w8qgEfRZApNCSCPqmNaSDSct/GIiRmRhcZiJT+MiPczKEo4fDHr/or88fvuOfmY3fe845jX3/wiaMjGtVoUjAKLQyJyOEQvUGXMArkE6T0oU3Z51w8ZF7nSRJBoskoRJqa9liVPZSWlLHIY/KysUngW5zfMpgyTDYHmpU44EJPkQ8LFHIJnhdiWWkRfvr8FGtbOZ49b3Ptvg6vnq6QdzX+6iLXvvsYFVEmv+MyG39+E++57WVm3AJnjh/k3OUS6y9dQXRxJ82nllCxwXrHBgQzOs//eIPLf/6OVYx6h/yRC4i5KeTBa/CsJ5C1Pqe/che1w6dJ1ipIz0fkfIy6j7z6IMI2IArBUIiZKuZj+9l1wwmCSzuYvvpFioZF1C/z2e0GG6JLWwzZqatIIXGwcLI2LxWfq7GLmpWFXUJKS0onU6lWwsMZU7RGVCZfxFhINjPxeyzUWEMSkdATAaFQ5LC4KJtIBB0xJIcD2dTLJ6SAS4xiSnsYpEJwB4OisLhq3zaNzSr1Wp+NRg6lBEEouXve5pYZiyv3dtg9GxKGFl9bC9gU3bH2o6YLWMLEwqAvwky+nk7ZwszUAVJjhze6gG01HHqRpr9d4JpDTXbt3GYqr1GJQRzZuG5Modxh2M8Thhb9nsvBHT5aCy6ve3S6DpYBpgGXlkuEcXpOdy22aLcLdPsmQoBSAkNqKuUA37dwbMWldYe8qyjbknIp4PxKDlNIDkwrDs4mtHsmL102uOmqJoY06PZsolgyOz1gedOh5cMw1izVYyxh8PsXe1xfcTGRvLat2F+DZy+aXNiwOb1uIxAcLls80wwpGAb1j000IRO89TBpQiaYYIIJvk+4cP9rRwdZQTn6G2fNiAaaop9qRLAAzVXU2CQNVqvjUpAGfZ2QfO780fvvf+ToTfe8/a9cuNx1zy3H7rjn5mNPPPj8UQHjnfiRM5CrLWKR4GC/oUlReDiZJkAQioRFXSLSafK7m2kDHCE5wBSXsgDGd824JD2PTTEkQvNy1OEVthGZHerTgw77rQJPn3F5ZcWm2/J47898gXJcZsa1eOJEkcM7ff7ttxQfvOdhjD0BZx85QriyiJSa8g+f5f/8rcN86IOP8adPLnKpq7n5im3mr3qRqD3DcLPIto7JaYsPf/RRzCta4AuEqRFlFzGzEzqrCEdSf08TuWsJY2aV8NU6xkwT4+pper+f4+QfXcXs+9s8/St3sfz1JQa+wbRyMUyF7ud5/KkD3HTzCeZ7SzzUa7BH1zkntpjTJQIRj4vxPA5lPPoiGOsrllQ1u/6MKW4SQVXnGYqQBDWeREXEY32FO3Ko0vnx+xVwx2LvRICHPX5dDx+dNQs94VPERQmwMWjLgJbwWVE+/+pyky9sbvPsis1PXN9nujpko5FHK0GiJKFvsdnw6PZs/mywgsyap6ouMBQpHU2Taj5GYYzAWJ8TZA1IT4Roka6nIXwCrTkbDrl1h+Z3HytzzY6YODaIY4MkkSyvVMnnIhwnplQesLZeIp+LGA5t+oFkca7PylphTL/SQCmfoLWgXvUZDmwaPYFjwaUNl9lqyMqmy3w9Qgg4vWYyX41ptG0WZgf0+jZhZBBEklpOIDE4t+Kx0kqvke9bPN+IeNsc3HD1BmHo8Oqyyft2G5imJkkkYSRIEplqdJQmZwjmioKNvuZg2aToCJwfmTQhE7z1MGlCJphgggm+T3j1/uNHEzRhpgUBMJGsyQG+iLFJrX19QlxsejoZJ5YXdRrIdtBziTSc1T0efvDpo++854a/VvHyzAMvHPVFNBZC+2PqUAEPm0DEyKxBSZGSmhxtYgjJsuxxVm7TEiFT2mOHbdNLFGvap4JDGYvX+iF5UvG0jcE8OZoioKhtuiJKk9F70FEJO12bkgvHn7qCS6tFWj2Lm67sUCz3uPvIBo3Th/B6AU+9sJuvnIffW+5w7fkr+aHbj7N66mr+YjVkWfZ59LzNay8cZDFv8P7/7THEQ4f5L/9sk837d2A2LOjb6L6J3GsiSlPEjy1jvONK1EtnUSd6yF0FrGsUeq2PKCnsAy2e/8JhqmuKXYdfYXa2wdmzC2ysT7HvwFke+cYRnt726SzPc3BXl5OrOQYi4meL+/hydImQGAcba0zAEuSw6YghFgYd4eNik6AwMbO8EJOeCLCxUJm2KKXLWWjSxqGs3cxNbUgBNxWya2PsduZm0yuBoCuG2fOaWV2iiEuMZkW0KOJySmzQF4TRtvoAACAASURBVAEbosuirlCjwB5VYnm5zG9ebvGK3+fF/pBnOn0+dGXA+lYez1X8ZwcsHlmBEi4C8LDwtE2YuYJZGBQya15gPBExkExpl1mdT4X12YQnj8WXViIUgrMrLg+vxby8Ljk8ZWEamoureTYaLtutHDM1n3OXi4SxYGkmYKvpZdQowXw9ZKtt4g9t5mY7GGZCMR9TLWgubVnkbGh2LGwTkkSy0TIpe5q1ps01B1tcWilxfssgjiRztYiBb7DatCi4mqv29ljZcjCkYMY1ON2EHB4b2zYLUwqE4OSqRc4WaJ0mue+dTmj0JYaAmaKmPRTk7fR7YNKETPBWxESYPsEEE0zwfYKVOR3JTJQekGpCFDorMCHJ6FENuuSwqGiHrgiwheQ0XVb9mNPRAOBN+oG/LrrZxGWkEWjIAQuqwLwqZpSZ9M+sKnKH2sGiKuFqE1NLctqhLYaEJGxHCXO2SQETG4MIjYtBgKJNyICYbULqKsdF2WZL9FmTvYyaBqaE5gC2fc3qIGEQZfQZM6bXrjJ/+6N0mzt4vNflZbnJXSzwzEmPr/zlDZy9WCJGUdBpqGTVNLnqlif5yE/u4UvtFp++e5rfPlbBffcy9sc8omYdlCJ64AXUVhl94jjJiXl0z6XzhzvQ3Q5yr0n0cJ6Tv3EHt936LS6em+e5R27lyUevp1qKuf09j/L1h64jjOE12eT5fp9/93w6PVoTbY61u0DaBBRIjQpGWRqONrMU8tQOuaxdwiwkclFXEJkrVj+bXqS0LWP88xEJHRFkrmoGQZZmPkoxh1R4HmSPR5SvEW3qkmiyKTokKIKMzhUSsUvXMJFUlEtDBDyuN2iJPoNMEG9pg3/7aIXPbbYIQ8m/eDairnN8+ppS6mqlcnzqbTYuFp8oL2YBh5qCTqcxZe0ypb30+LVBiCKPSSJS7dEg044UM2n7LpHjllIe140YBgalfEIppwgiwdC3iBIYhnBx3aHVlyQKbBPOrDi4FviR4My5GqfP1hBZI7RYS/BsTcHVaA1BJCi4mr1LXVwLjp+aYhgKrtntU3A1jZaVJbKnOpWnThTJ2enjw/va5AzJ+Q0TKWB2ps/JVZO5kqY5gDBJ09wvNAx2V8AyBJ2hpOSk4aWGnLhjTfDWxGQSMsEEE0zwfcK5+791VAMWqVMSQI+IhhgQiDjl8mfF4D41zS1WlZf0NmXtUtQW75vP8WLX57xsYZO6In3twcePHr3nHd/VLuoffOTf3fvEg88dfen+l49uy/7YdjUgooiHQFDXBZZll72qTFsE6Q660BS0g6tN8pj4JIRZojZCsKiLmEJQcwzakSInDAwhuLZi0wvgrOziapPzssVeXeKsbGFkTdmq6FPTHhtRTN2y2AhjapbBvllFY9ul4GnOnK/y4mOHufKq0/zJt8o05ACpLJ7TW2z5Gid0uKAGCGBTDtlOYr7yzVl6IqQnQvLaZkpY3PqTfaIvNzG8AcahPGJeYuwDkhhjf8Sr97+LV07O0HhqD8uP7mdmeptLFxao1jqsb1SIYknPl3iORsZF1rdyXHtFk6eXDTxMTsgma7KTanhEgiNSel1X+MQZ9ShGIQTkSDNmtkWfngjGtrdapInoLjZCjNLmdeaKlTqZzekysUgteAvaHl8LW5tEIhWyD0Wm6xEgkYSkVtBd4Y9F6gB9EYybkylyJChOyDUcLC6IBjGKg2oaAUzrHC/LLeZ1gdv3RexOKlxXcvnyacmq6BMLxaPrCZ++WfJrZzrITPfxL+8IyW/NcSoaZBojg1+6LeLypRIV0yRI0kY9QeNhEqNpEFASFkcWFJVKn7wXU8hHVCoDGts5glBSziu0kigN9XKCZ6XHYUiBEBrXgmZGwSrkEorFAe2OxyCQJEqwc36A75s4tubimkeiwI+hVlQ0OxZRIqiXYxYXW7Q7HlOFhJlywsJsn3bXplIKGQ4chhEs1mJWNz121GIWZvtEgQ06dcUKEvAsKHvgWKAR5ByNZWjk+w9OJiETvOUwaUImmGCCCb5PeP7+bx4dkuZnBFmCOTA2xu2JIOPUh1R1HjMxM5ejtKAMfZNQwWXZpaAdBPBPP/vJ78pV5/MffuDeKNspN5DEQhOTEJHgYBGjMDGo6zxT2sXDpCl8ctjMqgIeJhZpknRfxHRkQBGHBM1l2WVG5dmOEvbnHPZVBRXLIIyhFSoMbbAlfLoyoCl8EqFRQuNi4WFhawMNtOKEs6KDSExebod4sc1m06XoapY70Fpb4NXhkGmVR6EpaSdNM1cBJpIV2aWmc4QioSEHzOgCczpPHYcGIU88MMd8uMRXHz7EVx+Y49Yfb/Hsp25kxwdSG+SZo8u0Ht+BSgSbbYPHj89SsiX7bniBX39olmvrJic34e7bX8O2Iy5cqvKvzg0YipimGGbNZOqCZAqDFn1sLGISAiJ8ETEU4Tjsz0Qyp4u0xJASOXI4mFriixhfhDhYlLSLFAZT2qOAQ0f49EVID5+QGCFkpiORY7G3Jk0et7SBL9L1pPoQi7LOkcfBEzYeNk6mRBm5tHWET6AjIpGMU8wbWSBhX0QoFIFQDDdK7KppNtoGu4oGP31Lmx/YpbljzmJ1dYoXWyEuJmVt88eXYm4u5znXTycdHRnQuFhjb9GkF2o+ck3EO3bGNNeK/OQtXQ7mPRbI846DqTPa5dUClzIBehjYGAL27GzR3PbYu7ODawr8wCBKJIvzPfbuW8MSFr2Bxe6FIaVChJcLGA5cdixtQuyiEkk+F9Pt25iGZhBIpvKKHdMhnb7J/t0dcramVPax7YhyISGfi6hNtyhNtZiuDYkjG8cUXHvVKrlcRKmQkMtFhKFJrZJOUirFmCg0iRPBob1ttrZdHOv1KcikCZngrYgJHWuCCSaY4PuEj37+4/eNdp61gJAEK3NLEgiKeKyLNgLBabmBKwzePZ3D0QY3lj0GSbrrXVe5sVj8u8HvfPT37m2JgCntUNYusVDUVQ4LgxI5bFJ6kE9EhKKsbbaEj4mkR8hAxBQwWTBcgHH+RZeAAjY7k/L4mAaxZrMrGKa1L0t5k6qwqWmX65JZ9qgpbG0ypb3xezkYzAmHH9jhcFCX0Wiuz+eZLsBsWfGVFZ8P3Xn+Tce0KfskInV88rSJo1P9gUSwKrsk6DQbRLg8LzeIUHSI+Oxxg0GiKEiZTkCk5g9+/Eb+zT+4g+hLcODQRa6/7Uke6XQxpUAI0H2PWwtFVhom1y4qXnj2ClqNOgd29ilrhxXRyrJf5JgyNSBgp66mYnJiqrrADj01LuwFaZPQyexqbW0yo/JY2ZTCwGBA+m+efnO+8EhgDtBhwIAASxuEIiYSyVjno9GZtigZByAmQuGLCENLfEIsnSa1F3DI46DQ2MIa55ZEJOM8k4iETdFlWbR4hFW+tSqZKafUwOdfXOKFby5y5lxKffrFax0+Wq/wgYU8n7oy1W64GPzi7UPmVB6AvKM5NC145rUCL5+c4oTusrVVwvNCpisR3a5Ds2ONrYIP7d1mq22yd3cD00zYudhh0Lc5s+LQGUjCGNY28px4dQmtBQuzfQrFAaVyn8C3maqmIZ0DP8s8URIpNPXqgP1LA6rlANNU7NvVBsD1InKFPmFgI4SmXG3i5npII0YaCXEsWdq9jGGkUyfHDZAywc3seHO5kEIhoOAqlmYCul0Xx8qyX5TAc5Pv+l6eYIK/DZgkpk8wwQQTfB+hBYg39A8JeizUhbQo1ZlwuK0jhqGJi8GxdpfdMsdey2EjGiJR/Nxnf/o/mIL81Ef+2b3TssKWajEvaxhINnWbG/ReXNIivYxFoNPpSiBSPYFPiEBkORMFIpHQ0xEz2mMgIjxMusLH0gUqtmRzCNO4rJLmmATE7KRIWVi0dUQuluRMg7Knea2h2F8xOOIaGA0XUwgCbXIZc1yIOkiqwqZqGXSHgoMFm5Kn6QxT3n13KHnPvMvjT+9ha6BZkz12qBJVneNtRplX4i49EabOS9lkZ14VyWmLddnjJRooNK8a6/yA2s26DmjGqRPYk79wPe/4n54h/Cc3UqsMeeEbtxLHkhtvX+WHZwsUcjGPnBfcFOQouJrPN7aZ6jpsiiH/TTWPEJq7a3nOb+dxMGkzGBsKjAIfZ3SRDdHFyKhYdV1gU3RTx6usiQKo6xzrsjd2wwLoMhzTpkYWvpDa8abCczGmWa2LdqY5SqiLIpY2iEXaiNhY+Fk2SYzCIxWPJ7w+GUsdy9IGaEhISITI9ittTNZFZ5xZkoYrwrFkk5tKRf54OeY9tSKJgmEo6A4tlrcsTAO2eoJmL0fe0fzgXI5XX8vxU0cGeF6Hl09W2VMfUilJLq47XGOU8LwBL5wscWgxwHZj/MBgz1KfKDJYXilzeG+HMLQ5db5EMadIEsF8NWGrbbBnx5CLqzmE0HR7NrXqgE4rz+a2y2x9SHOrQrW+zd69azQ2q7huxIH9bdqtIvXZBloJhNRYVoA/zBMMHQwzIl/qIoSi3axSKKdTsyQ2mVu6RBy6BEMPIRSON6TTnmF2YZ0Tr+zCdRKKxYCpUkguF2I7If3hFKYWTJUCtlvO9/hbZoIJ/mZgQseaYIIJJvg+4sjHjxx76oEXjgoEiNTCNBaKDdEdZ3Ds0BU0mor2WA1i3lnJYUYWMbASh7REQEk7/595IV944GtHPeEy1AFFkUMiGBAQS01TpFMDQxtUcShiMyBmKOI0AC/TDxRxGZI6ZkVCs0cXibRmQReoS5vn4jb7zTyeMPCVZkkXGJKw9P+y92axlqXned7z/f+a93T2GWuurm4OTVIcxFAUTdlU2xaEBLAgOKB4lSAIEOQmgIBcJBdBAiKAc5tcJAEMBIaBIIYBCoZhZJAcyYJaNk2JEpvNqdljdXdNp06dYc9rr/H/cvGvvZvMTW4kkkjvt9A41XXO2XvttVed+t71vYP0GFmLVeGRFqxreD9vKFQJWkvVCCeZYRgZjlLDR22f1AXUDt4wEybUNK3hVuIH19MFvL2uOF87nq5b9sOAOFTOc2Xp/Cje4Ph01qOtDVeUzKXkQpaMNaOQhgBLTMBCSnKpOdAecxpOSPmhueR1c8FgPuad/+MOez3lk1/+FvX8iKIMeOetu0xy4d88qzljzf7iDnUrPM4d37GPuZIV67Mx06uEVQXa+hK+Qhy3dI89TWlEuZQlSym5oSMCTJd8ZUkIyTRk2W1BKmkppCXq0q0QHzEbYLePMcBvofbIuJIlChzogFD8JmNfMyIJKagpqIklJFTro4AFNulmLY4lBetuvzHHhx3M/dVCSsiA1D9GJ9PbnO+YgEMd0opjrN5D9AeXc373o0P+yeMFr5Rz3lgXfDTMcAofvb3mB08NB4nhxRdmvPkoJYtgsQo5v0rpJ0peBDy+9MlT14aQpQ3GBVzMQi9jcoazy4RVHrI3rHnvSY/ICk+uAiIrvHEOgVqqFlZ5SFFDFsNoUPP+kx4XsxAQijLg9DJicjUgNAHWKs4Z8lXK0bVz1Bm/3agiVA2utYRxTb4Y0DYBrgmoqoDX3jgmi4S6iqirhPWqRxjVqLMk2ZLheIqxDQcHK8b7c+oqYbmMGY5yXGsYDgu0tdx/nAJC+Fu7dKwdPnzYbUJ22GGHHX4OqLpBtKIlpybs0qdSIj7u9llQE2OIMHx7UvBUcj4po+33x1j+69/5778+1RVj6VPRcOVmWAxXboainLpLTsw+tdZcMOea7FGL64ryYEVLpiHHrkeLkkgE+LvtpTRcyIo7bo89G/CkhX2JyF2LEeGVdsqXwzF3XQ+LMNWKgpYRlkFgudumXLmGfeMjYg97Qt361KJ5Af0A9nvKfk9YPbPbVK5TWfHtib/3/tL1iNdOfZ3iWho+JxFO4bvOy2m084P84coX8j2nA95GGUjD2+acu3qA60r/NqlMFsMb5owLzbmUBbd1n0eu4G7coygN7333V7icpNS1+KSjkSMNE/7lpORfPi54IUp5w15yS8f0NeKJ5LgPOh35wjCjnis/tM9YUXQeG//8T2XOgfZw+GEePtiE1Z18q+quhUoaQrW04lhRYDppFfhtWU1LQMCJjjB4GZfBJ19ZNYTiiygvWbAnvkE9VP++hmykVwk5m5Zzy6rLagPIqSg76rFkTUxErl42qOJ4KBdEhKxNRUbMY73kf3lrAMC5LDikz5c+f8qPfnyNVR7wm5/yLeSvvr5HP4bDvYofP4zpx3C1EvZ7Smj9BqWohemqx4v3FgzzCBF4fB6TRsrlUhj1Dape5vTcsSFLGpwmnM7gL8oZMZbfPOpx+8aC7701ZJAo4K+/VeklXXkpvP+kx3jQMByUiCjf+/4djvdL4riTVcUVTROgCr1+znLR4/5D/xrTyPHW+0NUoXXw/I01dW3JsoLlbB8RR9pbYoOKxeSA8dEzwnBMENXkyx5FHnpTe9+xWBvSv/KfMDvs8IuP3SZkhx122OFnjG//3isv2S4FKMIyNyVDTViJH/siQo5ISLA44NiGnOqad2TGdc0YEtOgPDQzCmpS8f0Sa/V31H/SLbLWkkACvqDPYxCed2PGRLRA2j1+iAWBRL0R3mIYakymERZhn5hALWMbdEWGljUtD52/210r7BGx7PwgrSonqSUVS+2UQIR+KNQOzlZKEgjjnqOsDVUjhBjOmoZKWg679m6LYT8IeT6L+Ox+yGGbMSmUVQkPnY+sTTTgjmSgQi4NF1IQd50UY822RXkOv625b6YMNGZAgqKMSHkqM75ir/Hvf/VlTt+7y2QecbRfUDeWLGk5mwZ85uNX/IvTNUON+RMe84n2kIVUJBqQElDjeGgWBGo5MQl/qI+wGHKqbSzusQ7IiDqfh2zfn4iARnx7+kRyYiKMeMLRimPCCvCkxcf8+i4O798JAR+r7Doy03Y+kZQIFe/fKKg5ZkBFS0LgNy5UVDSknf9jjx4WQ02DYKioOzmZvx4rfDxvQUUmCStdU1CCwIoCK4ZCHOeyxGL5tDviu+9n/PonVvwXbzzl7uqE++eWb68XLAvhX18WXDUtn94PMCKc7NfcuzPl4ipj1HMEBp5cxEyWAXVtSSPFGFgUws2jih8+sXznieHuyBAGyncfWX5YL0jw78nzw4Czq4TjvYbrx2vOJxF1C0cDR9l4QgJQ1obpIqIoApxC01gCC3HcUJYh/cGa19/Z5+y8z2TuZVOt863sq0K4dVxRlJbJImQyj3n3SY+TcU1ZRARhS1WmDMaXNHVM2ltQFSlVFTGdJfSyhnfPQvZ7bmdM3+FDiR0J2WGHHXb4GeNb3/jOS7W0Pl5VvLzlQry3wqGspCLTmH63rM6sYeEct7TP374Z8WDhyAh43VxQ0ZBIyExXGPFDJHzQTO3jYFueyYLn9WgbfRrjk6gSvCEcoE/Ihcm7PgpDov7jpZbsE1OjZMYy15brkhBrwB/zmEphSs2JJIxDy6OmZCheJjOIDMPIbxWmpfJOveYoCHHOm70Dq1gRbpmUX0p63AgjnlY1l5T8oFzyyTQjDpW2FR7nLS/rGf2u/buSljExj2TFoaaU0vLQzEg0pBUv17qpfS5NwVRKxpp2WwnhK/aEP+cJL7hDTrXgN+5V/F/fOWZRwH5iePPU8spFw3erBc+eDLlGxvuy5J7b41V7yk03pJCGU7NkKgVf5hojQv6wfcI9N+ahmQBeSrVJnFpJhRGD6WKJwZOJITG51GRdRtVaSjKNKaUm7sICSnxp5IiUEEvSdYw4cUTdddICtbSU0rCQNTkVEUHn/XAY8c8ZdhK1AQlVZ2CPNaAVpUdCTtE1rFtfcqg5Df7rQgIWukIFnDpAtoRmQY4Vv9V6bOY8MnP+4GmJw3FWOv6UR1yanAdmTp+EqSm50fb55nzJ968ano96NK3BCPzlWcvL1SXnheN2FPMnF2tWK8tzY+H7DwP+1D2jQinmMaaOeGdd8sDM6WnIE7Pitvb5g/mEOM9YLmOiAOoWXr7K+Ug/ZJgqrROaFozxGxKnQlEJ6yLAYDi9SHh81sMa/73P31pxPon44bRiZAOOhi1Vbbl2WJCvA6pGOBg4ellNmpbkqx6D0RxjG2zQUBUZ2WAOWJLYG9ePRi1J7Cj+zid2JGSHDx12JGSHHXbY4WeMb3/jlZeArdkYYETKgAQn3hcyMWuu6QADnLmK79tTnpqcXw72WZTCWluemCUL1tsyO9OV23ni0Wx7IAThnrnOI5kwNQU/Nk851hEzqegREiKMJUQRLqVAxQ+qRoQeIS3KgpopFZfqPSmHJOzZgMJ5c/e5rGhUuGETWgeztuV6amkdXJb+Lr4qXI8istBva6JASULFGk+aXpkV3OsH/GC9wiBkhNyNY86XwnPHNet1yHfdJReyJCIgwDChwmIopOU9c8Vzbp9KWpZUhFg+G4xYtcqLDLnE+0LWUqOt5UB7lNLS4Hjl9RNuBBHvNwWvTRt+4wXl1cuWlIAW5TUzISFgLQ0rqVmYigrHHTeilJbHrHgkq20qV0XDgQ7Iu46VTXzuipK5rOl13o5KGgIMfWLWUtOIY0rOAT3WXayuL7NsULT7PqWSlqyLNq6l9WZ8aQnVG92LLiK4oOJIh3gLknQ+GUMuJRYv9/Lnr+q2NyVLLViQIyKstCAkIJaQmqbrsVFEhFgimk6y5ftvAqwIU13hxAcEDEnpa8zUFDS0PNUJBzJkKRWK8t12wsQUHGsPVjEHPeWdC+Hf6JmPjhbHD8sll7LGuICbUcSreU6ifgv0mBX3goxX6xlVF2X9MYZ87EB4vIAH7ZofVUtcEfJ2VfAr/R6HQ8f7l5Yk8CWCeQUv3l0zX4WUjSccTWMZpC37o4ZFblGFqgoZD1p+PG3pS8DR0G/z1oXfnoQBqArvPk1wdcJwUJIve6gLSHpzitWQKC4IwwrVgDBqmM96pGnN8iu/tCMhO3zosCMhO+ywww4/Y/zq1z7/8re/8cpLipfSzKXcdjRsSuR+vb1DTyyRGP6tfYKi3HIjXlvnfGnYZ1HB2zKllpZj2WMkPWasOhGNT7wCX1AHMNEFK10z1RWKcl/OSSUh1pAIyxpHhKGvETktY405NQvoSuX2iVlIje0StJa0GDUsqDHINhbXtcJhEPLElUQuYF47rmeWxsG7ZUUmlh+t1wwIKRphUQiLUnAObqUBeQVVLfTw0q+oDRhGwq1ra5bLiFHRx7qQUlpaUZKOJAjCoettCVTQpU41rXTpY8rzpsdfyikihqVUjDThDXvOgIQ3zDmfkgNGJuSB5rx+Cd+zp2TE5DTc1j4/thfcdkPmUnGoGedmwVpabjs/VJ+4PokGjDWlEcilJCTEdT4LRQk22wVZc8TAv0Z8RO82KW0bsWsAxXTvZUlNXxI6h7nfZmgECBOzJlaflOVEGWhCLd7hkXZen42kS2VDGuyWnGyI0pqKWEISiWlpCcST0ZUWGAyZJChQa4MItNpixdCTlLITcWkX4wsQiuUhlyxYM2OJIIgYFhSI+OfNNCSXhr8x6lM1wr/KJ5TSkEvNNdfn3Ky2PTJfum64mgX8yFwy0oS5KXmrzllJRY2jT8S7Muf+0nFDUl4zV5xojx+ZS6ZScFo1DIoev1+f8jE7QNWnr91/FpCGQhJ6YjIeNFzMAxa5JbTKIFVEIAwcD6fCjdTy3fOW95ctd4eG6ydLmiZgnlvGPYe1ShI37B1MiOIKVUsUFyynB8TZiuV0j7S3Ik1r+sM551/44o6E7PChw46E7LDDDjv8HPCrX/v8y69843svVTgSAnKpyaVkrD2uZEUuji+mI87rhrs6ZCJVN1zGvFMWfH6Q8aCq6ZHyZXeTlbScMSOUgFBCrPgUJoGuHcJtCYkVQ6U1p0xY2Za5NFzTjD0T8Hwac6wp6oREI87Mil+PDhkHAT0X8YAlvc6LkNPynPT8nXUaSmnJpeU7cs5/dP2A04USitffB0Y4CAMEYd8GtMCkdjyoS+oW+oElr/0d9lQsX7wNso54Wtc8qEqqWY+HuW9Rf+S8UVuBUnz3fNVtNCrxd89NN5hbDK/ZM1oRLnSzlTAMup6U59yYN805I82QNuDK1ewR8baZ0IjyVOaEEvCmuQCEPjHXuzb5Qmp+2V1j2R1DRsgTs+hKBv0m6pYbMZG1L/ej2SZMCcJKKo51QCUtj2SCiJBLyZCUC+ZbP8eaDUExLPFliL1OSjWVkrjzvdhu07FJXsspqWk6uZaX5zUdSYgIKLutSNltzTbXS4gl6QoMF7qmoPK+EK2IJKSkJhBLTITr8qZLal+O2F3ftTaoKEsKaprOXaIIkErMAQMMhn3Nur4TeJS3vFxesJKKhJCEkLXUpIT8dnbColKGxPxxdbEt8WzEn9eexlgxvMgeD2SBILwrfjtyZdb08b4pJ8rrbkaI5Q59/mQ9IaxDvqUX7FUZ945a/unZhHen8JsvVjybRIwyZZ4b/rfLc/54NkMxvFrPOJeCf+9wwN6w4f6jjCxx1I0wWRn6sdLr1QShb3O///YtstSxmPdp65jx8Rnr5ZBsMEOdZfznr750+cV/Z0dEdvhQYUdCdthhhx1+TviVr/3yy9/5xqsv9br281ocKSFDUhxKW4U8o+AzaY8/c2eU0lJKy0hjrtmEb7pTbuiQMTFXUvIUX67WdkOuJyQBicTbu/GbwjrFp2B9hGtkRMyoeEpBUxsWruWFNGbfhDxra5I2Yi80ZIFhUrutCRpgQcPdIOWZ89ucS5NzzfW5XBiOwoBhbEgCYVI69mIhMBBZIbKCcwIqBAindUXeKj1j2EuEs7nhhSPH6/OGWzYhssIX7jZcLAJed177/8ys6GvEpckppaHuRt1UQ94zlww05sqsec7t88hMsVjmsu56Ofwg/tDOuOaGTMyaiIBPBQMeujWKMJc1e5pxLout6EgQZlJSdzKmU7Pklhtuz0dfIyIC5l3IwELKbSP9ZkDf+HWOdIjpGs0tlitZUnaRuW0ncfJkS3Fdk73B4HAM8NGuMZZWdFuQqOJJju3I1tjfcQAAIABJREFU1obA9IixnVxvSclIM1S0236U+GLDtiMS0n3dmpKagaSU1Bgx289vShI3x+P9IuFPkQ3EH3uj/uxtvj+RmFO9ZE5OnxQB7roRj82CvkZMzZqq6y0xCKkG/LBZcCkFPygXnaysxoliMdvzfccN+bG5JCJgaryscEnBjJwrWdKKMiQl1oBCalaN3xq9Jlf0NeI+C+5PPAn7rWt9vv8gYtXAcV+5dT3nX5zPGJJQSssTmWKw/M29AdOF9+RMlobQCtbAxz9yyenZiMAIZZEQBIoRGB9eEcUVRT7wpUEIdZVgjGNHQnb4sGFHQnbYYYcdfo744tc+//K3f++7L7mu1XrjE4kI+LgMeFNm/KidsZaaT7cnDDTmOekxa1rmNERYrknKOQVzU3blch/IsPqSEYghlRiF7V1nxd+RNmK3sqY9jWlRVjRcNg0/0CmfYI9rUUjllMAIi0YJEQaERBgKHM/HCctGWYuPHb4yOaKGRet4oR/y1qImFMNZ2aDOcNT3UayhERIRUmuoHOQ4yhY+fUN5MhOMGvZtyJOq5mZmCa0nLcXap3Pta8qFyQkwzGSNESEjohUlI2ItDbF643emMdNuM7KnKRNZMdCEuRTkUnPiBuxrwiuc09eIpanY14yJWbOi3CZQHeuQm9rnQDOuTNFtXxxDjTk3OYPO/9CiHdlpWXciuc0AT/dxIQV7ZFTScKAZKsKamhrfbxJhiTpZmqKcsEdKREIE4jcv4GN+l1JhMFTiB/6+RrTiWFNSar31plgMKwpCCbZkwW9C/HNvkHfllUPJyIhR0c6bUhMQdIb7lpqGnqQUWtLSggitejITiPWyLaRL7moxYrBiiCXkphzSJ6LBsZaWhZQsxZvcexoTd9K1gUZcSu6lZNBJvuhcMspUCs5ljpWAmICCmrc5ZUlBjjfGX5MxmXoyXoiXilXiushq7yUREa4kpxbHX64WTFzLZ/sp68rQVAH/fPU+h/iI3qnkpETIosdJajkc1dw4qlisAl58Ycbj0z0u55ajcclkmvHoWczxfomxymo+JMlyXBuQDeaUeR8btOz/+SsvXf3qjojs8OHBjoTssMMOO/yc8YWvfe7lL37t8y9/8Wuff/n3f+9fvxRgOXE9BoQ8bwb8xv6Qf1tcsUfGXekhwLlWTKXgBfU9HqWDffpcmhUVDX3JSCVmTI8BKXva44ABRzJibAYUXRlhIy0XOucZc87NirYrtFtJg0GYUfOkLZm3juthxDtNzkK8D2QkIUsa3m3XJFhCDD1CfmiectwNa7024npqSawwCCxHfb8BuSyU2sG0afnIvqEojfeaaEOVh3ziuuPZwnDYV0Y2IA4htMrNk4JHFyFPumExwJIRdg3kS1rxEq1GHPsuw4hsiV1PY0Yas5SKQ+0DcN0NeFHH3DM9TvE9GDNTsJSKZ+IN5gN8bHBGQqoh79spK6k51B4rqRhpyl7nHamlpZCGWtxWorTp3lDo8q88QXQoU8nJiD1hwnJInyEpKTEpEecyIyKg0BrbJWt5KVLDWDMqaTGdtOtQfd9LLT4quZCGhJBSagqt/Gas88u87862ZYjrriukovmJlnd/bs/cFTNdUWjVNZk4HC2hhJRaIl2csBHjtzDqr72KGhHjiQjNdnuyIcAg9CQhwDCXgrkUJIQ80SvWlIzp+/cRx7lZEWK324/NuXwoF/RJecA5DscFC54x55Jlt0PxnhMrhoKGheSM8a32Gxlai2OoCWGXFhdhecOcck1HNCivlzlvl2sOmx7fbs8IJOCRXKEov+Zu87G+j+1dri3aWurGsFzGNI1QNYKopap94tdqFaNNyIMnfZLQIALzyZj5PCPrleTLPou/tTOo7/DhwY6E7LDDDjv8AuGbv/edl2ppEPHJT7dMyj9ZP2SsGceaMZCAe/2AujaghgTLfhhwZCNumIRX9YKKmp4k9Iipael3cq+UkJ5GjDQGY0kl4q4eYsWSU/pNgiTsqe/RSDUgxPBxM+Bt5ixquGcyehpyPYg4jAPOGn/3fEzEc1HCm27Bpax4KnNv6m6VN+ucd5qcXzuK+dGFIzaGUew3IbPacZ4rM9cytJaVczzfD3g8Fe7sK1XjB7jWwV6/xVplNg/R1sfMtqI0OBIN6BNzzfWJCVhJRdqZ7unM2oIf/HtEPDUL+hqzT0KB41xLEix9IqZSdOlRhpCgkxsZxpriRLnn9ggJmEnBSkpKaTufSUQjDiewlrobbgPqbpg2mI6CGBTtIgRa1lLTIyYmIOvihSMspTTsaY+prDyhEWVf+zTiGGjMqku08jItIRBDRfvBZqi7pp66K/om3ZriHcpK15RaIZ1EyqGUWtMTH2OswEoLL9mSjdgMAglAxH8vgoj4zUwnG8skZaXr7mstldY/tYEDv4FLJCIiZCEF5zonFe9BmZMzkh6RBMzIu0hj2ZpNrPpXVYvfhpwx7SRh2j2PlxwGnZn/WEY44LrusUdGRECivpul7trf+10fTiu+WHJf+4QdjXnbnHOsA+43Oc9kyVSXmM5QPzU1eWn5+CCiqGVbhLiuhEnuvVD9GGa5Yb4W0hDOJiGtE9QF5KuYtx6lHAwbev01dRWx961XX5r/2md2RGSHDwV2jek77LDDDr9AUBxh1xvyZXebP2+vaI3X+P+lfcw1HfKDZcCn2eOw6+7ohcLrq5K+WBITMpADnumMtZSstWTCkgMZYNWgoqSa8Jwb0+Doa8gN+rxvMh5yRUnDqVlwqx2yR0SCJTGGr5hD/qB5wtr5Aa1sIioX8MmoB4ABLuuWGseIjCPX5xP4u8m/cdDjOxc1QaDczCx1C0UDq9oPpoU67qURp0XDQCzjvqOfCPcvhb0Y0ghEII5anBM+es3xFw8KhkQcasJTybkyayINuCUpr1ORaUj4ExuHU5mBwIkOeSbLLs0LahyHxHzLnjIjB2Bf+xTU9Ejoa9RtBiBUy3vmktL4vvOVFDiUsaYIwsQUjF3CWnxXy7rbJtnurvxGbld08cEtfotxxJCJrBD1d/8Bxpryljwlp0RV6UmCxVJLS0657XYBmEnh29XxsrC5lKREhGp5m1MAEqLORwIL1oxMn7nmaEc+QgIyiZnogkr9puvIjGlxXLgpqSSUWlJrQyi+ayaVlLUWWOODDjJJWOq688cItTbdteHPQSYpy+45axoumZMQ4XBMdcWkS8+KCbhiyYiMM6b+fSDpZGpepFXj44c3vThhd243PpJI/PbIoZzoiFYcqGFNg4hw7Ho0OIruPQowrKhx+G1I1aV79Uj4przlNyr6wfV0zICbbsinwj5GoBfDn18W9FYBnz6wTNaQBMJrZ3CQwLWRQwSev1Hw9DLhyZXlY7cKXrzrSfzVxR7D4YrFIvsr/omyww6/uDD/31+yww477LDDzwqfb290Wn3De7Lg1MwZacLYJRxojxfdmEwDKlUGxnIShPxgtaYvlpk27GuPEx2yLwPKbjBscTzT2fbOedkNbgGGEMNcKqrOst7XyKcySUONMqHinSYnC4RbbsgP7CnfsY9YUJMYw0HqNxqFUx7rmlparrsBB5pwGAacU5KXwsf6EY8uLad5y1vrknWtnFY1N9KAKTVJAAttyKzhdGpIk5bP3K6JQxBRliW8e+qlL6+dGp6TPn9zP+M9M2eoETe75/ymPOVEM55zoy2BALjlxtxxY85kzk03RBCemRXvmSmvmQkFFQAjMq5kSUpMrAFLqbpzJeTd7/saEyDeD0JDScORJtx0feZSEavdRi6XnaU9xkuhNlKs+idilB9z6bdGUnkvCfDM+KZ0g+GmHGCx7NOnoKJPQi41I/XysFj9BkcQSmmoumSutdQcyx63zTFTXVJQdUlXQSf3imnVEUtIrmtq9aEFPfFk5UJnTHQBQKklVvzxVt1w77ptzrLbfBTqz8/mz8F7X7bx07qixbHWgkZbWvWENZGIqvOjROIlWfv0u61VsvXj1LTk3eZpRk6u5fY5/PcG3JZD9mXATT2gJwkjTamlJVYv2xtq3KWDGb6UjvjtvQM+FQyIMFtfVE9DRhoTYniqE2IJPZES/96FBExZ8VI6ZpwITn23yNtmxqcPLE/mPnxhkIAVf3Sz3NA64d3TlL1+wyefy7FWOT1PKCtLFDU8fDxmtYr+in6S7LDDLz52cqwddthhh18Q/Ohr/+fXc+f4d3tHnFUtSym5kCVGDIjw9+LriBr2JUKA/Sjwnoos5qJu6Ivly709LktH0KVi9UkJJSAQy0D98DpUX5SXaMCFrGlRMkJv/hXDNTfEocxMSaYhA0ImdcuhRIiGHGiPhVS8zZxfigfcX9X873KfPTLOzYpTmfE5jninzXls5tzSAVkgvLYsuRGHNC30Q8sosMQWXuhFfHO2okaJ1fJ2teZWFKNqGGQtvcQxzJRpbnh8FXDcE95Zl7y9LrmtPa4oiTpN/4GmPJWcjIATSZhQdSTAd3HcdWN6hJyaBSU1h9rnfXNFn4SBxrzgxtzSETd0QKYRKSHH2uORmTMTP2zPpeC6DinEE4mZ5IhYFlJtPQ+eIEQUUpNT+t6SLiK32UbW+kE9Iuz6SEouZE5fUmpa9rUPAhUNMcFW9jTDN5gj0hmxG9/5gSeWfY19FK+w9XgkEnWCJcgpiAlpxdGXlBZHIp6w1VpT8dMSqk2ssBGfrrWRkzU0BBLQl3SbxhZJ8FMbkEACIom62Gi//QEv62ppcaLkWgDgcFTUWCxTVgRiu3yuhokuWeJb4BesWbJm0+C+cao0tExZsaZiTI+buocRTy4S9USipyExATOpuN/kvFdUfG6QchJG3LYpzyUxP6oXNOJoUO6yz4kOucaIT7oTQol40R1zqH3+or2gKAOGGvHqck2DcpYr99IIBeaF8pEjJQmhqL0XapA6elmLc4bJLOJyafymDyEMlPE4Z/G3Pr2TY+3wocCOhOywww47/IIg+7/feelmGlA28JlexvfLJTd05EvYNGLYxhwllrxRWmDWtKTGsGqUWAznWlHVwpyGuZQkBNTiuGLBie5xpD2fFqUxj82csSYMiRgRUYojl5qBxhxpRiPew7AyNZ+wA4Y24GFbcN9OKGhYmpKVlByXI5ba8oKOed1e0uD81kEyLrViJTWfjYdYgUWtPGkqeiZgPzYkAcQBBFa5HUeMJeLNOudTUY9eDEVtSCJHUVrO55aTUcvxXstsZXm+H/IH1Skflz1u2oS5a5lIwQumj1PBIiRiOCLhgSyw3ZYg6KRRC+MN5TNTcMONGGnSDe3KUmoOSWhQQixPzZKFFNuODUE41H5ndl53d/u9hKeWlrgTgs2k8J0hVFsCUNPS4ONnG20p8VuFUmqOdcRC1hx2vo/NY6XdluOSBVfqyVNBzVAy1lITdRuWSIOt76IRR4+IlZS0OC51TkGFE0fW2eMDrPfTEJESMWOFiKcdkfgErpZ2m25lxG80Qgm6jyER4fa1bXwugQQ4WmKJCbsznut6+1iRhNuv/6BvfVPmGHS0ST0J0YpQwi1pczifNaauIzUfbFy20dRdB8qYHnMpKaThSlZdlLHFIjwxc8aa8qIMsRgGsb8WGwdN5X1E3neivKBDjiRhIAGfjUYEzvLZfsbHzZBnTc3YBHxiL+RTvYRbSUQcQF6DNcLRqGWytBzvNaxLb0YHIUsbruYhJ3stWdISRS1ZVmGMY+9b33tp9muf3RGRHf5/j50nZIcddtjhFwD/01f/0dcPXcZvH42IrCACqYbc0QEOONSUjx0Yni2gBUp1DIwlsYZV61i4hh4BF1pto2kzQmpakq4x2yCcaMaZ5Nxzez5GFeGJ5KQEvOAOuZScZ2bFocs4JmGmNTVKFhqOJeaxJjySCYfaZy0Vh7Hl9WLBG+YZfRIshkJqfqRzjPgm9bdXVXc33t+t3wsMdeslLGmkWAOXKzgeKPvriJfrS76mh6jCMrfUrW+yXhaGZ+eWa0Pl9UvHR90+1kDeOgyQaciFVtwwifd7qPIGcxZSspCSv9HeoMRR4OU5JS0FFQPdIyUgVW/dvi4pVuARy63s6UKW2xbwASk/NE8YklFQYbuErrxrjy+kZt+lIH4zERFsk6cK7QiJQCwRtTYE3XkppeGEva23QToJUktLRUOlXrK0Sa4C7+9Yi/WbnM4cHqploPFPG7qxhGKZuSVOlLH4dLCkE4lt5GgBAU785mSpayIJabXdXj++lyP0nSLqKClJJdmSAINse1HCjiBsSEUsESEBq+5xN90km9ws7SKADUJE6IsWJaDSevt504VPGwn9xkY9GQK/UdrgiCGPZQpARkxfk674MCDvpF8G4XWdQwm/lY1pHUQWYjFkGmBUmEvFvV5IP4aqASMQ2cBfuyF83mb0Y7jK4XjgP3+VgxXfvB6GjtvHJW0r3LuZI6IkSU3bGl64U1PXltHegqbx41gQNNz/z//j//av6MfKDjv8QmO3Cdlhhx12+Dnjv/ud//nrSZeM9LEsYboW3lhUvLQ35Pmh5XODjI/0Q3584cidYoCh9b6Qvcjwbl3g8OTksSxZSUWAocYxIGbYpTq1XUnfsWZ83z7liZmTEDPWmFxqPiUjXpABQ5fwkTAjFkNPAi5dzbf0GQXKQkrWUjORnOf0gFfbCRkhN9yQuZQ4Ua65Pj+2Z6RErKVmKhVjTVjQMJaIaduyah1ndcNAAnqx19ArMDAB32umPC8DVpXX1BvjtyVFLTzKG767ynnCmkdmzkJb7tkegRoOTcRcGy614k2ZMqOmFJ8W1SdmX2MmVJT40sdj1yMlJsLyzKx4QYd8LEnpB4bCKSttuaN9ZuI3Ops79yU1EQFNV++nwB0d89TMfXKUNFzXPoW0LDuJFtCZs+Nt43iApejkRhUNBb7PI5WYSC0LKTDAkhKHbjcQvqvDgvjHTIhYUrCUkqWUOPEDthN8bK/AnvS40iUiQtFF6+4zoKJhzopcfRrY5rhyvETKJ2f5DQMiJOJ9OUaEGm/qDsUP0H4r4YmKCl2biNnKtrQ7V5t0qY0cDT4oQNzE6rY46o64VV0NZattt6lh+zWB+KJHf24TDmVITxIaGp7qhDUVSwr6knLdDXhi5t6zIsqahpHG7BNzKwmZFhAYGASWVa18bphww6QExpOLRQlOoWiUUSpbImKNkoZCUQu3TwqWecD1vZYnM6GpApyzXMwDjvZLxgdT0t4KaxURGB9MEANB2BCEDW//7n+yIyA7fGiw24TssMMOO/wc8Q9+53/8ekK07S7oJ47QGu4eBBSV3xKczoTb+46la4nF0AJ9A8b5G90JljNZ01d/dzmXmhDrS+hUMND1IHjZyw/sM8DfRa9xrLtErJM241RWWDGs6oQDiZiqL7IzCJkG/G1znUdtwR/Zt6hpuekGPLAzQjVc1wG51rxpz7muIy66aFmDcGVz7rVjanXMpOQFHXJsQ9KwG+xqn4KlwBf0mAd5wwNd8XeiEX90teDvjgfEoTLRmhDDTArmsuaGG/BusyanoaRlZgpCLDMpiLq72b5lPuGx5Pim8oQZJY/snExDbmufay7FCDwrG47jgEduzTEJ92XOTHzpXYujoSEiJMSS4E38ay15ZGYkROSUhAQsxfduGATT/VNbUNN0FXuVNj4Wufu1Kf67wTE1LZXUDDXloVzg1G8mci18ApXLt30ZEeH2HHvPiC8fbMWxrz5pKVRDLjW35ZAFay6Y+TQsDcnFE5xYIla6ptbmg14T9R83Ab0bUz14c3ZDs92C6JZiwFoLYom36V8bD8mmQNPHBH9AQBTFdmlhm8fZbIO0+9hutyobt4r/c4vtHsuw0oIVRRe+UG2lXddln5qWZ2ZFqJZCfG9JRkiPkAb1mzmnBFZII/i4DSkbf21mkd+C7GeQhMrTuaDqCQvA1UoYZ7Cs4McPErII+r2Gk36MCOSlEIfKgyc94rhitH+FDRqG+09ZzQ958z/7T3fEY4cPJXYkZIcddtjhZ4T/5nf+h6/3SVhT8ZX2Lq/YMwAOXYbF8KvpkEG/YH0VowpJpNw/98lATyaGsQ24aH1f9J0o4LRquR1aqnXLQL0U5VhTAiespaHq7vi3OEadKT2XmiUFNS0RIYXUZOr/KVCUUlrGLuTH5oqvcJ2ZVliEsaZk3T8Zz2TNEH/Mmzjfd+2EZyw51v42OenY9bnoUp5udGb3ASGvmwucU77APt+a5sQYvjBOCC0MEuW2CzgeKkdXQ/7X5UMOJeOfTS/46viQL41T/tn0gpqWQ+2zR8T3OlK17kRPG6nSA3PFUFPuuT2WUlPjOFAvGatp+aX2EIvhoSwJugSmocaElR/eG5Tr2iOXmnVHQgRhpQWRBF32VYMR4SkTDhnR4kgxXMhqm+q0MWP7ZKhy+2ebwbztJHOFVkxYMabHgtJvPtQbrr2CS2i6ZLOWlgbHgjVTXXBLDinxUbuK85Ijges6oK8xGRFXkrPokqy8xGtzDP5xw+799VRp0/HO9tpwWhN3WwyAuNuKVNSstaQvWZciZbcyq42P5ifxk9KtTbLVZtuy+VrT/f6D7YgQdMQGfCkigIj8lMnf8NOhnwGWCQuGZFx3Qx6ZCXEnU1zTkGG5Hfq/b7EVzlbKraGwrLykalq1GLHbbcjKCbH1vTXzUrk2hF7kE7IiC6NUqVshCHwk73M3ctK0oihCwrAl7eUsZnsMRlO++x/+VzvyscOHGjsSssMOO+zw14x/+NV//PVLybf/vxm07rQjJqbAINzWHkkAf/lOzBdeKHFO6PVKDvctp88yrlbwbXdF2JW3/UqUMDSWvIZrJmbhWvrG8ope8SV7wB+5J/SImMqaO26Ph2YGwFBjvtje5pv2PUZkTGTNdfpcdwP+xL7LiIzatDyUC/4pl/Rssh1GWxx79Mgk3DaOn8uCUzsjwPBxd7jtqDiXBXv0vEekE9QsTMkjmVNSk2nI6+2Sv3+9z7oyRIFSNcI/v7riIwzoxyE3Roq7Ut4UTzL+0bSgFUdtWladXOipnW2P7yeHWu1M5ffcHimWnIaUgLYzT3/E7dEjYEWDwROOa5rxVHJ+qDVGhGW3YfDGap9wFYh3a1zojBMZU6vfYMQSMmVFS0tMsPVibIZmYNsXwvZ4zZY0teqH8LmuUFFWWoBAQ8tAMua6otHGS486D8lcV9tB/b47Je68Py0OVUcsMfuSYVSwaniPs064ZOiT8j7n222CoIRittKmQqvta4gIt9sQ79kw23Ned+RFu81MRU1EyFj6zMm72N+WRhtvVBfrXxvQaosVnxr2QR0i2y1Iow1W7JagbbAxxddaU3Yem82WZnO+N+doc5wJEQvxqWAOJdOIj7oxd6KY+1XBKE6JLby1LrnuEvYzTzT6seUiV/YzwRplVQqhhcu1EhqhrCGLlbwUpqUySOAzL54zn/e4d9MTcBGHqjDan5Av+zz8L/+DHfnYYQd2PSE77LDDDn+t+Idf/cdfd91AvClpKztJzjVSbrsBa2l4IEv+YpHTj4SitKRpzelZn/ef9DgclzjdiFD8lkG184AU3i59RsG3eMbnZZ/CKX9XbmxL7J7Kcns8S6l4JjkG4RlTUg35M/s+r5szGrxEaAOLJaeioN5uD3LK7TEEajjRITd1jyEZ94y/E76kIMP3MRy6bJuAFGtAqIaMmAd2xgMz45WnDhHv96hbP2D+mZwxKyCwXkrU4EiJuZIlVyxZUdDgtkO9H6T9P2cjMnwTueNEh11srmDV/7dHtP3azf15B4w1ZiIle12PxKEm1HhPx5qGRCLspr0bOklWsDX9l1qz1JxCK3IqNs3km86MzTXgn89tyVJKTCxhV8TnCULaHeNV19GxGcAzSelLSk+SzvfgB25B6EvWkQl/Pvom444ccSErammZyppEIvakR09SH7UrsjWbWyxRN6AnRF1zuj/ejVSsoiYVb3hvu1cleG/IxrzuUNadLKpHQssHfShDybavZbMx2ZyHWpufivbdpGy12m7N/A1eRhV0f5dEDJGE3uQuBiueoMXixY1N9wsgp+z8Kc6TYmnJCHirWtMnIAq8HPD5OMEpnIwrrIHpGoaRUDWwKoXG+etynAiR9eZz4+0ynPQ8QQmimiwrCMOWJKkZ7M1Ikpq6jHcEZIcdfgI7ErLDDjvs8NeE3/3qP/h6ScMTmXo5STeMjenxUJZcC/3Q9xE34lhTLmTNm6uSd89CXrvf53JhWFfwkU/9cPuYghCr5f25IxThVhLSqjKTEoPwbbyHYNI2ZBqSEfJL7nAru7nVDrkwKw51QECAIFzTve2AvKJgJv5O9ZCMQVcW1+8+HusAQTjQDNsNsAkhoVr+lTzmLXMOwJHrM5O8G/AzHpoJp2bGRHJqfKHhAC/n+bOLkvvTltbBQjzJeVo0XC0Fh/ezfLI94rqOuKP7CELCBzGvm82SINumbYBHMmEuPgT3efFN749kyRvmihrfgbHfkYgrKWhxlNLy0Mz4gT1nKRUzKXgsl4TdudrAit/wzHW1PY7N5y2mazD3nogVRUdAP9iKGMyWfAhCKvG2DG9OjpUPfBRDyTiWvU4OZlhrycwtSYg4lBFAJ59yHMjQJ0tpyzXX55I5b8sZtfhhvqDCICx0vSVKG2q7pvRbnO41+F+e4PQkIZUYp4oVs43mrbXebmfaLnoYYK45PRKuyz4HMiDs0qwEQdX5VDC8WX3z92JjcN9gQzQ25/YDGVhIItGWrPykbGzjN9kQFoOhUh/NPJWcCYtt10jRSc5uJr67Zrr2ZEIVHp5H3DouOB4ow1QJLEQB9GP/eSPQi6CfOE4OC9JIuXut4PpxTlOFZIMVQdCQ9nLWyz5RVO1Sr3bY4f+FXTrWDjvssMNfE37/G3/6Ui51J0ZSDrRHSkhGRCOOoA15Ijm3JeNOHENjWdIwkvD/Ye9NYzVN0/uu33U/+7udpc6ppau6u7p7lnaPNxIvMw7xxEGIgAJxIlv5ACKCKBBsxREgBYMdtUYEIZAlBEIIPkSY+mbEAAAgAElEQVSJIr5YNsYy8IFIFjY4GIPxMjOepXt6767qqjp1znnPuz3rffHhXs6pHlsee7otYj//Uums7/Y8b3Vf/+f6Lzy1r7y1VIwIf++LA2/0ToqUIGQknGhLqglf67dc0JNhOBU37L4nW2osL+o+J1Jj1KUVfcwe8H8lb3Fd5+QkPKuHvGPOOJVVHE4FoaFHcFeP95kxo6TQhHPZcVMXrmBOOloZyEmdERrLY1kzYLmle7xpHlOQ0orlQnZ8zB6zlY6pFixly4mseSwbviQnnJuO7ykOmBZwu5/zT/Q90JTvXFT8H/VjbuiclbT8xeIWz5sZm95woBWPZUNFQes9DG4oVTY00UvwL8mz/LY85jZTZpJxjZI1PYMoh5LzChdRJDXVnFYGJprzmjzgEResxbd7Y0nFibMaWm7IQfSguAZu12UxkYI9pqjAXEt6cSP+4Id8BTK/ATBiog9io9tIUkIiVE5GrQ2FFFRe4tT4qsMgvdqTCVbcfZZScKqu7PCm7POGPGKjNYKwYkclpevZ0D5udZTLBvQeixWNEcCV5JRy2SmiQE0bNymEjYhotItbL7FS4Ewv2NIwlZJBXLO6iyR2hvHEy78GBlJJYlyxk2ldFdcRCXMwqw+ePvXqtorpFflVOPcZKQuZ0tKzpeWcNaEw8kBmHKnb3D3sOx62A9fSjEUB7QB1D6tNSpZCNwgvfeycWWW5eX3D9WtbTs4q7lxvuHa4o2kynrp5QVm1JImSly153lDN1hhjefXH/sbnzr9v7P0YMeKDGDchI0aMGPERQZDY7TCnwiDM1V39n2jGe7JhKy2/Lo/4zWbFko6HsuPttuHLD2EzWC5aG/s+guyoE+sjRp3x3JXkOfM5ON/Hiyw4p2MlDa8nZ/GKsSC8K2dOd6/iWyISCnL/c4NiCcGnA5ZEXdzvHT1g7cvvZuquRHd+CLei3NAFR8xd/K0u6BioNMWivGIesqOlIqUkY04Vty8rdvxSfUpilCxxEqEDLRms2yIc2QkdA4+bAavKu2bJK+ZhPM5BltV5IhKMz7d1n/tDw0v2GpkIj7Th1MuBwsCfkbCnBQst6LGkGELlX+6N4Z2XY838cw5X5EsyrvtNhDNam5hWNVGXoHWgFVOcr6bw6VVhWxCawhva2HXhvCyudbymZeYlbjUdHT1bramkYF+mTKTknp7SaBvPb0rKMQv2teIabityLAtauicS0hSlomCnjrBNxb13BrU+cSoU+5k41A/ev7JVR3ZbOsRvb0KSViopuT9KwZcRPCap35ikpFT+8YAoWQvHMciwBh/LO+gQjecWJffN7MFI7563+70glwuvc627uCkJsrCplEw0501zzhvmnJUPLfhqvWPtrTAzd6rYtUKeKmfnEyYTd766LuXpGzWTSUPbpsznNeWkRkSZ7S0ZupTdZkbbVHzp3/7RcfsxYsTvgZGEjBgxYsRHhP/yZ3/ic5lcDvOHOqEW55C4rVPeMct4VXdCQoGhIuVVc84X7AW/Jg9Y2YEX7QGpNwNvxZMazTj18apbCVGwDp33VbxqTrG43o6OgdfMGRkJHQNvyymPzIYNDTt1EqhrLGIZ3T5TXMpSQ4qwrxWHOiHzA+pKnMb+QKsoERvQaIK24uRHS3H7gvA304RjnXOokzgotvRspOVLjwcqP1e+ak5pnZyfN5NzOrHMUsMvdSfcsnPADbchkjX1BG3wREIQ3pTH/EryFv84+Rr3bUOP5b5sKDVlpjn31XlLtj5JrBNLqQlfMQ+8Udwd0YyUFhe5C6Es0LBmR+IH+1xSZlL5jUfLibgMsq0f/oPBvRTfby7ihmZxUiyLay4PBKqQ3PlFPHHpA8mQhH2mzlxNTiFO0peTccScZ+SYPe9nmWlOLpnbnqjzdDTakUVj+5aEhIVMONNVlDft1En7dr7pXVHW7Niqa40PfwPZULWRjLj3nztOYXtxwZY7cuy/l7geFEljMlj4PhA3GyImkparZCaXlCklvQ5PGNqjUd7fPie74su5LE20WI5ZcMvOeN4e8HF7wGcnLrygwFClcNG6+9p0zvOxaYTTZUbbZiRpT5Iot+48IC/cv0URJcsbysmOanZOXtUsDh+M0bsjRvw+GNOxRowYMeIjxDF79J6ArKXlQhqu2ym/mLxOSUYjbpjaMrATN7x1DDz0ZvKvseJjMufjdp+vmaWLkFWDAoUmZJ6cPDRbajrmWnBithzYip24r3uUM9myoYmbGYDHsnGyH0nZZ8opa17QYwTha/KQfaY8ZkUtHTdkj4lmNAykCB2WRhoObMltnbOWjo24LcOOng5XErhkF43MOSlvmJMosun9cCgID2XFa7Yk28xB4FTW/P3Nm1iUczZYlPfalt5Y3pDTKKsZsHx6uMOvJG/FCN3uSrysoiyY8IXkIS8Nx67ZXDoWWrDxV8APtGBN5yJsJWFHw3XZd14OSSnJudAtnQxMKXxHhmVGxXt6+Xq2Xvq0o2GQCTtpya6YxRNMbE4HZzQf9NLT4EoIHYUKx6YgZUNNqz2NtEwpOWNF6yVVORk1Lce4COSn7R4X0vCaPHCERi2ZJMzMhI3WZKSkpMzIWMuOYxasvWSqVlcKmJByruvoRwFnoEcuTfKO7OH9LOWlfAy38ei0i1uckox39RGl5Gy05tQuI7FQ//oHBh9JfEku7JX3h8FgfFGhIBzKnCUbDCU1LarWERb/c9dkb2IUcPDXZOLa4d9IztjTko/bA355e07iz2GzslQkTNKcMoWmh5fubul7wxvvznj6ZsL7jyqeE2W6WJEYS17VDEM6ko4RI/6AGDchI0aMGPERwslaXLLUiWzY0rCShjkVLT1rXJLQmdSspY0yoauyksfa8mVzysfsHje0IsPwxeQhjQw8kh0nsovpSxZI1FCSkpEwtzmPjPNqzCnJJInpUae6wqq7anzOJsqjwlX7Bmcezkh8zO7A03bBoU441AqjwpmpGVA6LLmmbHHN4it2cThfMKGiYE5Fj42DeCANhR+830jO+B29iP6ElScwnScbv5i8wRtyEj0oz+o1Bgb+SfJ2PNYhDUmx0euQYjj1fgCAAaVloNAr8iRx8qqbdsYRe5ywZK07turSwgKRCIlU9/XUbXZ8IR7gj1FPgmGtu3iVvyRnjwkTCg6Z0XJJBMPn4VgNnkDdkAP2ZfqE6drFAQ9MKUnEYFXj1uGZYY9vHY6ZaBqN3E4a1kYpX05GJYXfvBUcs0dGyk6bSIwMhrlUUXKWk0ZSN6WMzzN4M0LiW4jtDe/bVFL2qKjI44Znpw2DOvneoC6216IkkpB7OVriI5AvY5cNGU5+tcfUSwczCjIOmHEkCyqKSGrCJin82wnvi6t4abjBRDPnY6ImU8MeOQcUPJOW3Moznj4aSASeu9HS94Z3Hzp5IMDN4x1Z1jFdnND37nFHAjJixB8c4yZkxIgRIz5CJJg47Iars/dlSU0bG65ftDcAsKospfYSEpiR08hArU5akyC8Jhd04obbjoGJpixNQ+YfoyKlJuFMdijKVhIyTUBgRU1CwkxSGh8jO6gzTB8wY0XNlo47dsE0KaKMzPiB1gJLaZhqhpGUQ62wuCFvXwseGhcHa71fYErJhJyJ5jEONvR3pNF34IbjFMOOllfNo/j9yyvi4s3aIfHIDalvyWNS0iiTan2/hzvuCYO4I3nKmoGBhoFEhYlkPq3MkbUzaVhKTYrhkdmy0IKHXvpU+AF6LhUNPTMKamlBnZwJnB8BeZI4zmUaE7wqzWLD+ZJtlFg10rq0KQyNNq5A0g/j4MjLKevYzJ4grHQLMvGeC7eRKCXn1Oy4aSt+Ifky+0xx3SY1BkOj7lw7A7z3NcjALbsgw/CePGaPGSdc+BJGJ2EyGE70wj0Xyejhic6O/koaVjCzB4Q0sEAIbssRJyyjT8qdw8tjNvjbh8cFS+pDiw9kFiOfJxTctDMeGtfJEmKSKwrOdRXfr4MOWC8Ru9r0fp19HpoNz9g9Cv98L2h5Pi8pEqFKYVA4WSbkKbx/mmMV+gGeu9W46N28Jy93NNs9ykk9EpARI/6QGDchI0aMGPERYqIZBSkX3tAdhvAJBc/pES/YI05kw4lsnBcB65X2RO/C22aJQVjSMdUsbglyTVCBmeZUpOxpwZ7mPG/3MAhraXnLPAacATsMxaUX5RhvSE5JOWONQbgv5/xO8j637T4r3IblOXvEnpYc6YReLJ1Y1tJS+qb1rXTxanOQwtzkgEOdMsUZkG/YOZma+PMdLRNynrNHnONibms6ar8Z2OK2RY3verg6rArCxA/y4LYJYTAO7osBZ2JvtWenDddYRGKTqYnblkS9B4OUHT0bWkd+PGlqtGXnOyYGBpbs4msCqKRgz8zodfBCJ9e3sdOGDa6I8kJ2sV9lj0kMKQAnc3LpTimVFCzEeWUsypItGQk91m0nxMU8t3R0OrDRmsEbwr/Ee/wP5oscs+f7Qgwzv9FIxd1m4mVTBhexvJWWp3XKXa7zQM8YGHiKQ8505V6HT7EyCK32dL6zA/CyLRO9F4LEIsNgGL+vpzzUc+7pY1o6XtSnuCPHZJKSSho/TqWkJPeODRf7GwiIouxouW6n7GvFbbug1JTb1pnuM03408PTvGhvRNI6l6k3wAcviSO8pWTctgtu2CkTUu7LhgzDNUq+0m7ZK2HXu9bzuoevLQfqDj729IaXnl9zfH3J8e23OLjxtvd+rPjaj/31kYCMGPGHxEhCRowYMeIjhBuietQPbUCUpyRq2EpHj3KormTPZS85HX3LwJ7m5CQcaUWH5dTsGLA8pft0YhF1mwh3lT5lKS0d6v0bneuc8JuEIE850zUlGRutWesuDpO5JyY1na/LE+Y4b8m57LhnLqjU/U6hbsDb14KJ+pI7T5qetddI1PgtSBavODfipDgp7mp7iNJ10q/OC4cun+fW91YEg3PuC+jC74UhNSWNGxCXQpVSUWBEYvcGwCE5nVh6LOemppaejXSsxRUypgiJJzhTSrY+OarVnpqOjJSBIfZ4ONnVNhKwXNJohp5KGdOYZpQUZKxxW67HXNDSxY1IeJ+UktPpQEEeyRg4r0kwn4fo2h1N7C7pdKDGkYMTVq7QUXcAXJc9vt3e4Y4c09NH0vB53uZV3udL5pScxLWeo7ytj8hIeY4btL48MBjRg/QL8CTBbd8KyaJvIwQtwKWsD6Chp9KUu/bAETeZUZBzIHOuMedAZqRXxBktHY0nhDtt+E15m3tyzkPZRKngTHP2tWKPnFs6pSRnX+bcYI8bcuBDAEw0pHc68BXzgIaBU2mYa86xyfhYUfJsUpEY+OJuy8VOuGiVaWI4nClZNlBNavJyhx1Shq5AreG3/vUfHwnIiBHfBEY51ogRI0Z8hBCEB3KOop6EGJpwtV9aKs0wQvR0zDTnseyYkJGT0GG5phOW0nImO9bS8piV32YYGunpxHJkJ5xJiyhcSEsjl3G1r/GAgpwpBQmGM1Yxqcmq81G02vPtepvPm/eeiHI9Z8NKHPE5Ys6Z7ChxevrrOqUR563YSEepKYU3bRtc23lJSaaGE7NlohkrSZlQuE0Ol/6FmZaspGZgQP3guqEmJ3timAU33G79EO7auq3bXIiTHDX0THCvTXHleuds+PXkAXeHfXospab0aCRoAQUpX5X7dDowl4kjPN57EbYOwWBtsVRS0GpPKZnfwFhKcZuQW3IIuO1NaI0PkqKSjBU1RoRGey+b6iIZ2eiOfZkBjtzsPGFr1G0bKgrWuqWSkkYbMskwIrygx3zKHvK/JK8yx20OPiX7WAsv6TFvmyXgCCFAYVN++OCYnz5PeU0esdWaF7jBgEv8umw4N1Eq5YzzQ9w0hOMMlz6Mq+//nJSt1rxiHvKcPeK77F3eNGeU4goPH+kFoRslJaGXQGAcgZxIyU6buCH89mzOo67nodZUJLS4dLlUEg6YkqnhB+xd3pENXzT3Yknip+xtbtkpNQO9WvbIuWcb3mmUDOHeY+cXurcdeGE/oe3hzq0Lui6hmli++iN/cyQdI0Z8iBhJyIgRI0Z8hKg0o/QehC0tufc+5KRsAglBOJEtz9prXEjDhIxUDS0DS3HGaIvGbo7HsmJCwXtyzpHO2NLwtnFD+R2757YUfmYPw3tPzz1dx++l/qq+YjmzKwrJ+S3zLorlk/YmgvCcHvHbvE3pfQonrNiTikxdf8SFtGQYerHef9JjcBK068xIVFj535lp7r0ghnfljH2mHGhF4r0Z9+U8Sp6uPu8Uw8pf1Z+LiwOuKFizo9Y2bkAWTBA/HIMjMImYSBxaOmaasxZXyngmNSvZMdOSBKETi1G3xVjgpEoTvw9KxHChW18yaLjGnE48+UGiBGynjesWwVLT8ohlNKWH7VbjB+IVNZW/f8SZzgcGZxxX548I0jzxnpGeAVUbI3wTL7NKJeXbeQaryhd5l++UI/5G8hK7QelFeWaeMN1ecwO8zLwXAywwr4TEKD+0uM6yvs7/2j7gK3IPgG+zT/Ml815M4mr86w3H2L2v3OehqTz0ibhz+KTY4kK3vGPOuKV73LUHvGnO2FBTSs6ZrlyTuieEzs/hPl/rjmsyx0VD13y5c2lp35rP2PSW1+2WLfDZ4WPxvXNCwyvmISvdYjA8I8e8Yh5Q2tvsac73L+b0Fg6nyr2l8IXa3edL+ZQbM+H2cUNZuosF+4cXvPa3/82RgIwY8SFjlGONGDFixEcIK64pPaRPva+nbLSmY+BQJ/H3whXkPS0ZsHQy0IsrBeywXkak0YdxxoaSnHtyTu0b2Qef+FSLk744/4RhImUcbMNwv9INW3WDumu4vkw2umfOAbfJCN8LJvN93xVyYEtmmlH55xOu82ck8Sp76v8X0/nXEX73jh64eGF1P++xMdUqPF6QMlm/yQjIyZ642l7hyh8b35oe5FA5lwZvq8qMii1OenUiWwywwRG8jISJZqyl5l05iwllhzpl4k3hQYrU07Oh5o5eYyolx+xxLAv2mUa53U4b9mRGSR63Nku25L50Mng+Wnofaes6RgSh1jY+Vuf3J+5xQyP8ZanfoK6rY8DyJd7jRNa8xG32koS9UigS4R1bM1i4VgqHlXBjLlyfCp0/hInAuhYGCzcXyl87us5nhueZUtJKz6eH59zx9dHO/RWCEVKxMlK/lzPej3K5xVBvcg8pYGe6Zs+6mOlvGY6d90XXsewQQHybe3jtFkvtvSw5Ge+bDRNSUhH2soQcwyeziadrwiezSdye7cuMhUx4nzOe0n1PtOFXLza0Ayy3QubfXrekZJoJn7h7wXxeM5tvKcqWNGsZMWLEh49xEzJixIgRHyEGLCeyjgNo0ORH7byEQjzDIDYOwKG9+0xcmtFS6piwFGJID7TiTFYkGGo6SjI6sf5rFxU7IY9bgQQX67rzw3doyQ4f3RDcsRVXUreShiNZ+PvOY0lgQcpKXOJS5uU5A8SuBXCkwIpyoCWNDGylI8FE38iF7xTpxEZpmcW1eNc+VynBdXYMakl9BOvgCcuUkl6s676QhIYWQxGvrAWPBIAR4YYuOJUNe1rxFo9YiNucWFFQZ64PKVQzSna08aq8M3U72dWBzDjTNRey5Xm97nw6so4StU57CnKWuuZA5hhvijZINODnpNS0tP7+EwyN79cI58HF2DoyutGdf26XnozQDm6xVBRMpeS6znnO7vHIdgybFBH4avKYVzen/Fm9RWEM+7mhSKC3ioqw66EZlBszoe2FVQ0/MN/jB9jjf1qf8Of3FvzaOseqM+d39K4H5EpSWdhGXVViBelZeC2pl8651DJ3zDMMPT37MqcVJ0Wzqr7I0cb3eS4p15i7BnnNObClI+bWpch9IpsgArdNyTQxrAbLnIwX7Q0mml4R+AHGvU/vJCXXpsrpRrixUD6dTOn8ZiRJLHnRMts7ZXl6NMqwRoz4iDCSkBEjRoz4CHFflkwpGdiSYEh9+duCCT02xucmGIwKK2nIr3gyOhm8ZyJnKTUdPQdMecSSt+QxF7rlwEtV3tdTvlVvUvh42rv2kFfMQzZa+7jdIRq6Byy1tp4cueE2+BlyUnbiSMwde8DgpUqux8TtPEIjOkChKcc6pdKUE7NFEFI17h5FqaVnKTWH1iU/raUlRehxDesqLqkqI+GGLnhflv51ldQ+XjbxKVFO/pT5zY8r4ktJyZFYmBc+NrQk/qp6aHp/IBegl36TiWYspeZAK5ZSs2bH2idgDWJZ08RjJwjnuiGVhGP22ErHA84Z1NKKv1IvOYNab4hWclIKMk5Zs2DCil0cynt6enWm71SSKB/LJWOjOzJS1rplwNLo11+NFzF+Q2I41gVbOn7HnMSY5Dfs+zwnN3ld7/GWPGKnDbO24lP2diQHa9vwOg/5F09fZF9yltrxUlWRJ/CvzI+oe/jMcJc3zTm9WB6zcvRMk9hpMqOI4QJuk3UZ22uufA6O0H3FPOA5e0SuCZ8dPsaR5DzUhsZvex6aDV+TB+wzjcTtIedcY8GRnVCR0mF5p2voUT5RVBzP4GSTsOuVe7bmaVPSWUvNwMSTuw7l+7nF3f0EVchTZdcrZWHZNAlpAvPJQDXdUk0v+MJf/3dH8jFixEeIkYSMGDFixEeIjoEtjYsexW0DjnVBQUKhqd8muAHZisZtQpAVdX4w24m78gxuSxL6HlyZX8829j9YV0CoCa241KQgiQm/H65gu+fnCIjFkohLzJpKScdAQUorPT2Wic/OWnmPysDgW8PLaKBvpSHXlFo6XMuIS/8qNKH0JvSddGS47pJ75pwGlzxVkHKhW/6CfYGfTx4zkZILXA9HLlksTAyvtySP24cg9XGdIV0kKILQ68BMKk5kw5n3xGSSRKnPUlxZpBHXUm5VmUuFRXmPx3Q6MJWSQ/Y4FzcQZ/7cdXIZWWvUUErmCwWvSrea2GOyYvdE/0mtgbikkRxalI3uotzJvYY+/iy8N8BtQwpxCVtf5h1Kcl7kKU5Zc6JLKgre0PeZmym9Dgw68JQcspbGH0cbjfn3ZMubcsGX5R6vNjf54clTGIF2gO+bzflzyZwvLzu+Iue8ah5ifWwxwJmunpBgWU9Uw/MM3wOXDNfSc1MnNAwcSU6KcENK7uuOWgZu2zlfSx7E1xvO7TPWJcJ1tFz3HTW3kwKryulWOKjgOIFk6TYjtFCS0KPskbGi5227Qy4qAK73CYsC1MJzt3ZYFdLUjgRkxIg/IoyekBEjRoz4CBGGfSc1coNz5VvInYadOGCGwT94K67iQCufsOVjX/1GRXCldHLlcTqxqMBOXLcDOC1/2K6E1KXwdZABdd5j4PwKrp0781G8tXRs/bDfeLcCwJqalTRs5bIQLzS319LH8riJupyr0B5fS8cz9oCSnMJH9rqCOxtlZAsmMWZ1o96HQcmCSRxQO4ZY+ujSldy1tY3uoqRnjwkdPZnfNqT+GAPMteBY5257g4uY3VBT01KrK5fceSnSITP3V2dYcT0eVwfvWrvYlRHOxwc/dgy0vvfEYukZ2GrjOzguI4qDuTvc9upAbhAySSkkp9OORptIzl6V93mo5y61y5+vld1E4tN5V8nn9U1+R9/iNR7QaMOp2fIbvOEkZVje3Q483imZgYtGKTLlhVnGdZ3wvcMzbmuDjWWJBkMlRXyeiQ+bDs83DX8k4XuHZzFAh+WhNqTi/h0Unoz/WvI2N9iPMsBj9vjM8CwTTak05aZW1Ayc0dCo8rhzx2p/OrCqhaf34WgiPJ9VfDyvWJDxvTcyF5BAynfcEh/f7PIbto1hNt9x7eiCsuxGAjJixB8Rxk3IiBEjRnyEaPzV7EvjeXVJOsQN8sHPEYhI7v/THAb4QEhyb+zeSM1at5Re+gPEq+YPZePkWFhel4dRxuNSlZyE51ydRyWYjBMxTHwB3z7TmBjlnuPgmsU1ZWs6DnXCO3JKScZUSw605F1vZO/EExcSrATPgrIVlwjV+eOR+ecnCDftnBNj2NKQSsI/Tl7zZMIdo5BuBPA6D/ik3uK+LKM3BJ5sxB6wzpQujmQFudkDPeOaLNjgNkaCsNYdS6no6CnIOMcN64H+VVL48+Z6UUyQkOHIxJyKc9bx3A4MLMQZ1Hc0TCndhgvXdbGQCU5Q1/vIWbe56bUn9R0jjhBeBgi4cyuRpOWSMehAqy4VS8S4c6sDlZR06lrMnSQsYSYTlqzjJuJ1e49MMmZmwoXdsNEde2bGV+077JmZS7CSE/6+3iPrU/5y+ylOaFjUC1ID3z2b8uvrTewQMQgFOalYvnd4lt9K3uNUV0x9rG54fx0w4VCnWHXbCfd+sTQMbG3BKS3fWU357Z3y2eE5Wiw91wA8cbXckYoz7XgqLbjXN+zouWdrMoSDIeF0nZAnzmifJnBjKmQJvFCkLLfCS5OSeQn9oNyZCYmBxMCdm1vOz6fc/dhbXLvziHt/4H/lI0aM+MNg3ISMGDFixEeI0vdcCMKBTlhJjQtbtXFABdj6Cj5FYxIQQI8rHgS3jdjI5RDdeT8BXKZGDWKp6XlXznyXhXsMEXMldcg+kXIUiMxTHDLxaVOZl1g9lJWXN2lMs0pJcGp8Zww+0hmFLzpsGdjQ0tLH1xO8Ix1uQHbeFstGWt43K9bUJBgWXvSVk3GhWzZa+8Ql9zeThDfkETVt3J6EYxc2IHpFnhVuN9GCSgoudItVJfUpTpk3u4fNQ3e1mV0tmX8u4MJiXTywe5wlWzbUFOJ2WiEhqqWLr7PnsnxyKmV8nEHduW+8eV7ERMnc1Y2Ha3W/jCx257yPCVnqI3PDe6mmjTG/QXI3+DCE8DiVlMxl4soG5XLblklGTsZCJmy1juWFP2e+yE565qXSDm5z8OnFlJlckulbusdnhrv0WP7scJe5TPjTw9P+Xa6u3Z2aldQsZcsFLe+YNUupOZEt73nZ3aZXbpuSmoHMP/8Uw5Hk5H5cabE8sw/fOiuZkZEh3DAFrVWmhSICafFSue8AACAASURBVOLIxWCh7tym4+nrDVUGTQernWAVnr5R88IzFyz2NqSJpW0qNqdP/UH+eY8YMeKbwEhCRowYMeIjwn/6w//Ny8EErihL2VHTOXM0brCsvU/EePN5+P6KJqYz7aSn1JSJZl7ABaEFOiBsFl7T+9yTU59EZOKgGkr/3G01tmCHZCz1Q+yamq001J5kzLWKmwyAVA0Hvt39XXlMipD7OOAQLRxSvOBSalbTsZFLc7Xr9HAJXy29z8e69KqkkjyRBjWXKr6GOZX30jiJTxHlbZdeD+NJ2oScM9n4LULqChrDZoIJNW0kgL0OsZEcXIpV4sf/IBXrGOjEcqwLbugi9nsA8bYf7DppfNRu4wsPQ6fGB3s0wuM+sQXRLhLIkDiVxPeAMqgzzOeSXRIdte78i3HhA+KIk/P69HT0tNo5IiEJvS9mDGKwuUyZSulpj/CaOWFVu+eUJbBq4DPDs/G9c8PO+FgypRNLi+UH7SeZkcWtkHtthses2NBwbmoeyAVnsuOu3WOPnB09ZSLsZwmfzKY8lbhtXElCLoY/d21KJsKfmkxRFc4a5dN7Ez57XDFNDHfmhl0r7E8tn3h2zbVFjyocTC3f8vHHVFXHpFAmhVLlSp5CVQUSaMlyd86+/Dd/ZJRijRjxR4RRjjVixIgRHwH+8x/+b1/u/LXgJF7F7Wk8OchISNQwkZzaX7m3+M2JCjvpaK9sRlJv8gY30LnkpcT7N7o4lLd0XKi7sqxqSXx6FLhtSU0br4wbhAvdIAgTyvjcSj/uW5SClK121OIkYxfSYHADeknGQ7PxzegZG1oaOuZa0aMEi3Iw3s+1cB4CzdhKy0ILOiw3dZ9BLAMDaxpa7wVo6aPZe0tDqz1HsvBJTJeEJhyLUHYYNjShKR2g1pbrss+5bmLs65YWxdLp4O3jA5VvMz9jTYJhzY6cjHfkhD2mVORk6s7oTpy0LCwvQgHkhIJTXZFKyo6d95V0X/cemUrFRnekPg0tkJNwfbCnj3IrgFTS+HnYdIiYKB8LbeotnUv0CkWN6kieVSUjjWQrI2UhE+8D6Z8gP+e6ZioVE3L+ZX3B3X/vtgtFCt+VzEi2z/LLyetUpDw1NdxND+PtVTOSs+f5jeR9TlnzmeEu/3fyNofMWIt7D1aakmKoffjC/aajU+VWnnF3z/CsThFxj3nzWgMU3DpqmEwabm4K3nlYIAJvdDU3pcIIbBvBGMvji5RJoTxz5wIR5cGjCbdvbJlMa+yQMAyGJLFcv/MG6XzJ6z/4n33u0Tf+z3vEiBEfAkYSMmLEiBEfARZasEfJfVlGEhF8H+AGZ3z0bTD5Oq+Bxq1I6AiZacFaGr9PueyWCLhMzbq8Am8w9AIFefQsAE+U+IUr3SkJF7rhuuxzoJNYNgiXnpa1tFQ+ESpVw4wyyrOWssMq9GKZUHhzunnCUN3iyhcnmtGKOx47CduWgjO/jcj87iH0fCQksRk8k0uvhJN+uQ1DMKaXZPTeMh/8KIEEzmTCBdsYgzuoRcVtDQ5lzpKNH9LdSF+JI0xTXFJYSsqMkkJdGaOokGlC4wlkLrl/bon/fdcO32PodGAuFefq07UkIdWE9goxCTHAiX/9oXCxpfPt4RD6WEQMxhPMqwhE5oNm+Kvkwl7ZpATZ2gehWArJWOsWFct/L5/nb/f/DJMM1q3CAFUmPJNUvGhv8lI+pemdBKrMlG4Q0kS5nRS8Z+d8Uo94Sir+2eEuHZY3zDnP2wPeN2seypbr6ooGT7SlYSDvDIs2pcygH6DIFKvCM7e2iCi7XU5R9Nw6NKy2KX/hTs7+3o6Hj0uee+YMgBefX5JmHdP5iuXpIS9+y3vU24r53jkm6bFDyu/8W3/rc+/9rkdgxIgRfxQYSciIESNGfIj4737oH74MlzKk1sua4NITkZFwLjsOdfJEAaGTZVka6b0h3UlwLqTxXg83mLeeGIQBP3RiBCP7oAPGlxMGA/fV4RR4QtYTTO17OnHJWqjfZDizucEP3FfISeFTszISSnK20vqr2h0ITDT4EogbnEydIT3ThOQDg/EF2yvJSibuNWptKX0M7YwqDtQhrjcM7CHmN/RvAPG2A5aKzLsunCwrDOyJGCpylmyiR6Qk9/6PnBU7SrJopoeEU9Zk4prB3dYojTKnkF428THHJTml74HJJKHRjkElngPjpXi9JyBOpnXZFh+OYYB4QiJiftdzCsT7TMSAfv3/5mO5YDj+qhjhCfLm3lMdS7tmbqb8Rr3iO/IZr7Y7epQ/M5lyyxj+khyjCrNS2bXOEL6uYVLASzfgvXtTjkzOyvY8k1RkIgy9kmN42i5wSbqWHJdcBTCocn/r3mufvGYoMmW9SXl0llHlStMJzzy14dq1DfN5ysHRGSKWNL3G/tEJXVswv/62e3F9MkqsRoz4/ylGEjJixIgRHyLekBPu6jUSTUAu5Ug5KTfUSYmMuqbuAcuOnoqUhoFT2TLTgkwS1rS+UTpE1w4c6oxBLK/oe6QkPg5WWMiCVrs42GeSxVbrBEPuS//CcAo+ypce44fjgtx7I9wjduL6O3JS5lpRSxfL6QaxcfuQeKIU7hOcXOxCdiy0Yis9hR+EuyskprtyG4NwzIKGnnM2lN5kj8BOG/fcxZG0qTrfwz1OnRlckrjtCJsUcFKmqVQolgGNaVU1bdw6lL5N/i196KOAE+a4tKxAQFzR5OA9HR0iEh9H/XNv6ZhSsvXdGY70mcsWdR+9PKi9jEP2RCnzBntXOmgjqWi9V2jARrmWO95PEgggnuvwedyMeb9IQCCeVj1ZuUJGgu8luIxyb0yvfMrVL5lX+D+HlO+Qpyk0YemWdOx65ek9dz//77Lmn/8Hf/lzx/4+9n/i517+wSphsdiw3RZcrCFJLJ/oZpysDLse8gRe2TQ0WD57O+ML72d86jrsWkc+nr97ymS2Znl6gCpUVUM1W3N2cg1V4dHLP/yEjGqUVI0Y8U8PRhIyYsSIER8y1tJy6Adv4zcc13UeCYggZL6BPPVXnlP/e1ZcClUqElOQMgxbBloRzthQkkfpVdighG1IkFfNpOJCt+SSRekSEDcBrfakJBS+h+O27vNYNoSEq/C7A5Zz2UQZV0HGmprGS8yuXv13z8eguBLDXmw023d+SxMienNNSRGWsiMhI1NDJ8I+U79VGJhTsmbr/Btau+QmaZlpzlwmLolKB6Zy6XG5Kj1KMLS4QbxXJwcryWno4+vp1EmnMu/nOGHFHT2gkYEFk3i/IWVswDW7N/S0dG5Q94lirfYgTlrVakchaWxvr/25AWJQwQfbxMNmo/ceiavbqvD7IXI5lywSlHDb1HtvPpg4o1ymsCW4jgznF5FISFrtycUlfC11zb5MKSWnUxc7nJOy1A2/Yd5iRsWfMXuIwK82S24O+9xfwZHJnnjc8//kr3wOYAfc+I9/+uXJpCHwnuNrKY8el4jArCh4c2k5OtzxbVTceeqCt99dcP1ox2xxQVbsuH57Q72dU05WfP7f+PfHzcaIEX8MMJKQESNGjPiQsWTLnpQYFUrJyHAdEIGAuE2D+g1IjwRpEMZtSYCpFmylhSsJWABbrUnE0OvwhL8DLofYQ5kDbgifkMchOPghws/mUrHP1G1kvAQsEJ+GnoKUCQVranb+Kn/oqwg9HuFKf+gzCab5xsuTUu8N6SXc0iUoqShoimBI1VBLz1wLttLFxyrJYynjnszIydjTkpaBnTbMpMLIJbEJcreCNErTcjJ22hCM3P0HjmcmCTlZ9GPkpGylixuaho7CH+cdDftM4zEoyQn7IYtEsjQXd0xXuos+loQEK+52TezYMDGsGS49HYFwhO3HJeG89PZcJSCdJ1Uh5vdq8GXYnAQiFb9/5b0QyFHiNyo35ZAtLY12UWr2SXuTt4x7f/yp4TYXrbIeBr5g7vH97T6vtzX/ws/9ld+THDz4u3/1cwA3/95Pv2yMMpnU3M4v08/MWwsm05pPHDlPx91nB975O//a5x7/Xnc4YsSIf+oxkpARI0aM+BChKDNf/CcIt+3+E5G1imJFWdGQIHTenD7RHCuKKwl0Y2SQFoVo24aOmpYpJddkwVI3TKQkwXAgM070glxS9nRCLV2UMM00Z5A5A5attMwpacVtGzoGHnJOguGYPZbem5GRuFhdalJSDvzwvfSdDj3OhH7hvx6umMBd4aLzabTSc6wLVuw40Ckr2WFRjrSglZ6JXsq6zmXLVEsa6ZlSUGnKvkzZ0ZCTxjStpWxZMKGiYGCIEbitl48ZJPaz1HTMpXLxw7pDEOZSkZKwwzWi7zNlQx17PTp6Wi9lCxuQDTVzKrY0zHH3d6Fb5lKxoWZQG3sxQkmfXiFpIX1q8ESj1x4R99GiZJL6xxq4dHtckVKppfU/C4lnFtdlchVhGxIigAOpdD4bL7lSnogiDj8PRAwc2bXYKHc7MRu+Y3iKt8wSg/AL+haf0mNu6R7/sHmdn/i5H/2GthPv/+Rf/V1/bwa8843cwYgRI/7YYOwJGTFixIgPEWGjsaKJXxt1f4EYfSuebFiUqRbx9okX6bhywISNtP4+E070gtBbAXAo80hUguxrnylL2ZKRMKPiQCfU0nOgk7gpONAJN3RBRc5EMw6Y8awegX8+rW+MCFKgILsK5XETH58bPBAQkqkuPQ1hEzHxpvXMbwJKMiftkS07WmpvWjcqbGlZypZ9phjEJ4u1kSwJElvZEwwLLTjSGQmGFONJiY3bifAaglytkoJScmpaNv78hOd5TeekGBZaUcSMLmFGGcnNHbsXk84y78npfBlgiLwNZZCZJCQ+HMCRujS2mSckiBha3wGSSBJTq8Ljmit/4ntDEjLJ4veu/uyDvpzwnnDfu9z+hK3KVSRiMOIla55K5eKKGrfacJMDMk34anLCVlpHtGhZ0/GGPOInfuYbIyAjRowYcRXjJmTEiBEjPmTsaUmil3Kay6hU14AeekGA6JkICOKcsCUJ33vEklxSKi7bsAsyTlnzlB6wFLeRyEhp6FhTU5DSyOATnlJyUmo6Ok8WKnLW0lKQ0cjAjpbek40gR8o9afCtFJFsgEvpmvgkKRdLa2j97YNpe+sTveZU8ap8kHKFwdsA1t+3u1/no5jEcdxtBVoGLtjGBvaHsvKvOWFDHU3eJRkdwxU/RuefY++lcY4AJBL6OCxrWXOoM1e26DdBHQM37IxTSTjWKa+Yh0zI6bGcsY4+EnAJU1e7Sy5jcp1RfVBL6rcKT5RMqt98hNSrDxAHg/HGc4uqxYrELcjVTUe4jctXe7II0WJjl0yQcF19v4XjC65rpJCcGRWt9LFzZiM1FuWhLtkzJSd6wX0z4ZDZB9/+I0aMGPENYdyEjBgx4k80vvsX/s7LH+b9KYpRd8U+SGpC07VBGDyxKHziUTAp57jywrWXXlkCIVH2tORZPaLWjjlllEZtaShIacXJh3LJKNUN3TUde1pxwsoP2S2ZTzy6kB0GmKjzipyypqZlQkFoHQ+PnfvBPsQD914CFBK/AvkIV+DD7cLfnJQJzvzuOuCVHsvWlwO6Y6AsxZGLDQ0rdlTqCMljLthQO0IjzRNDdEvPGtdC7/wZzscSjrd4adjVcwNemnTFExFgxSVgdVdKJd82ZxzrNN7WtbQbppQUkrO0azbep+POm9t6DHrpgRGEwveIJN4HMqh77Ve7PvQDf9z9OUKRSoivvWxot1f+XMWl4f1JGWBI0Wpo4/MLBMn455iRMqMgdLXkktJjOWXNXCtySdmI+/47csJP/szfGrcgI0aM+ENh3ISMGDHiTxS+6+f+w5cBxPjBzcD3/Mq/87KeT/l//uJPfdMD1VO6TycDJ7LmGGcQ39ByqBUW5VS2TDVn5csHJ1qRIGRqWJC7KFxPVDJNmPvkpQluiH2kFyxkQum/buhYsYsbgEdyQUhVCl0fofwwwcluOj/+B/kPwDkbppRMKTjRxktyMjY0hODWkMqV+ivw4bZhs9NjvZ9CowwpmNZLPzgHkhKSsjrWWFH2qDwtcb//rjyOG5WENBYT5r4HxCCxB6Tyz8slhPXx+Trzdupp3sCgrmul9MN2T8Ogyk6cNGtNzcyXEwZJl4sFzvjN5D3XHk/DxLeqW9Q3lntCqY5SdtLHbpNOB3Y0LGRC54lHQMJlG7qqZYBINpxnxHgJXEJCBuI2JSEh62oSWIhlhifJhyMZg2tv1zr+vDIFgyqVFPFc1bpjIgVrGpa6ZioltXY84Iw7csRGanodeCDnKMp/8TP/0UhARowY8YfGuAkZMWLEnxgEAvJ1aNxV9+/55R/9prcijQwsxZmcH8mKU9lyKmsGPxqvqdlKF8vtzmTHTHMKEnY+RjdRQ6kpVpSFFkw047adMZPK/RzDYy5o6KjICdfcQxRrKDIM2xeLciZbOixzLXwrutP+BxM9wIqdK/aTglwyLtvZnRF68F6MILHqvCk8tKqHksAw+AMM/k9NF29Xu3wsMpLovljjiM+CSTRHh5q+koyV7mjpomEciISq9p0a4QgM2Jgc1ntiIxhS79Oo1XlUEjHkklLT8kl7g0/a66zY8YglA5bae2P2yNhQR2lX53vZY4SyLyEERwx6Buev0ME/fxd129Cy0zqay0VMJDIBV+VVAKFDRP2G5XfzdKSSRtHak7e18efRoO7Tuga99O/U2rHThrlM/Lk2cUtnvT9EUd7Xc7cZ0WEkICNGjPimMW5CRowY8ScXRsFYtPX/KWxTvud//5GX9WICWY8YxT5esLu4zna94I1/76/9voPXnpY+haphriUW6KTnxGzY0bKh5pbu8YZcsNEdpeQ8I/tMNOXc1NT0DGJ5yV7jy3LKMzqnZqBh4AV7zOflHd9V4WRBiR8/TeyIuIxkPZFV3Dy09DyWFYc6oxG3VXgkKwQhlPHttHFt5xRsaRjUkglkXk61ombiY2kDQYmma0Lj92XsakLiiYkjCyV5bDpf645rMqehp9Y2pkwF43pLx4Scx/45hc3CgPWv1sR+jmDODz+3qs7boUGWJSDOl5KR0MnAhIINNTMqegY20vKC3Scno2TgkBmnrJlQMJGEQ2a0dGQkkUyBT6MKj3OFTNS0qFomnjjuvBE+HLNwngYufSEWxaiNnhtV603rTpRlvX9j8OdXxG1SWu3I5ckYXot9om8koNOezG9bQlnhwGWpYZD5VVJwkwPe5AFbbdh5CRbAf/WzPzkSkBEjRnzTGDchI0aM+BOFIMNS643AVYukA7K/AaPoLgdRxChq/WBmE9QKd3/qH/2+m5JME+7aPb5tuMWelhQk3Lb7LNlS03GDfc5kxzN6yFQqntUjHsiaB2ZDqSm9WHqUNT0dlvuyYS0dtQzcN0vmvoTwKd0nI40yqooibgiC78JtBFIU6xOhau7LOaesOWHJloYzXUdfiiC02nOqK2pt49XwC92ypY1m7fAYAUNMoxri11c/hq1MIBDhCrtFudCNI200pL43paUnJeVcNy66VzdxowKgWEoyWjr2mZKREEoErbrHafzzD83wABu/2xiwrNgxo4qysDfkhAkJ/5x9hk/ZW3zHcJ2SjBeGQy58r0fvj2foVslIXKGfN7ynuM6RxJvJRS7lb5X3WQS5VfALfbBg8YMIEq8BV6o46EAiCYUUUcpluIz6Dff1wY1JgokkqdUuFj0CTKVkKk4uGNK7FkywKFMpKSRjzY5We2rtGDFixIgPAyMJGTFixJ8IfPfP/wdPEghPMEgHSL0EpuiQ1CKlH7T6hK6e07UF3yiua8Wh5DHZyqJx87D1mvwN7uMLeuyuXvsekaXUziQuHQfkvMlDvmIeMFG3aXhpuMEn7HWMuMG1VCd7avwV+kMfVxviaTMSClJSv/RutGOjOza6o9GOWlsslkY7ppQ+EFa4mq4UZEY7bVh7SdROG9cOzqUvIWxGwlB91QQePg8xv50OMdlJcQ3uoWU8JGQF/0oihkwSKopItEpn4+eYPToGztlgUVa6i2lPgJeleUeI2pjwdTUwoKZjShFJ1cQYPs6cfUn5zPA0982KX01cg0XwpQTPzYku2WodyyjdqzCRkBiERjsSEhovQQuv7SqublCuIra0X/GShO2HxT5xu+Af+b3Qa4/qk0lYrffy1NpFL0543+RkvmwyjRsng/Bf/+zfHbcgI0aM+FAwyrFGjBjxxxJfRzo8wgYE46/kWwNZD12KVC1KB7sc3RVghe3qAFVBjDIMht/8of/55b/0vWfYwfDuj/+rXzeQzUh5gzWPZUvLwFpqZlrGYXvney9i14dOeFPOXBKTPOJ5vc6XeY+3ZA/AJRmhfMU84NvsU3zJvB89F4HoLLSikZ6Fv9pekMbOjiU7buk+9+X8dz1OIdL1VFf+c40RsBZ8glLYaBB9DuBM11e9I8HDERKhgjwovFb1W4lSXP/EWndkpLTaYxgoJOdM1yRiaLV3RXlqWcgsbmQyuWxl32caB+lw/0+cay4TrUrJI9kIm5OcnAu29AxkJKzomJMwTxISAXr4Pm7yP/JKHPBDT0hJRkHOVncYI1hVv4m47FpJvViupYu3d5I113be0j1BJK6SCBGD0UsT/2XMs9AFn8iVdK+rUqzfi4yEnytKp30MNriaprXThqksYmt95k33JTk/9bM/PhKQESNGfGgYSciIESP+2OB7fvHHXtY6g+7Kf9qMXm49PgCpGjDWGdOLDu0Nuikc4VjNUZsgRsGCiOsx//Pf4iN0bcLdn/pHL4tRzh7PqZuUbhAaLI9ly4DlvpxTkPJQLgiFgzvviai9MfyWnfHp4Sl+KXmTlJT7sqQk5zU5iV6H15JTBiy/bd4DoCDlRNbxivqcgh0dZ7KLrd4JNiZTWVG+3d7m1+R1PhjnOlyR7QzeAL0nM07tkkQu5T/GbzTCBkEQcnFbiyfJRyjSE38V3xX3+SPuJT/u6z1xTeUah2OfauVv47Y6PTta9plSS3dFitVT07pWdu2xEmKBrbNWi7joXFybfPDGpH5XE+RjLkXM7Tf+t+R1vm+4yx4ZT+c5RoSfl1cjoQrno/EbHedpab2pe/r/sfemMZal6Z3X73nfs98l1oxcasnael+8dLe728u4QXwZDUZGiBEMWGaZgY8GW0ICDapBRpZAg0aMxJfRDCAhDcLtGQQjG+wZj7Ghbbm73e3eq7vWzMolMjLWu571ffjwnnPiRlUZBuxqS9XnnwpF5I2IG/eeczPy+Z/nv7DQFeXG8fXkrWmfuWDb+6l7d4e/TRDq9iWq6ojFbx5qqbFAV1pYt+Q1lKAnER28BO8yGKA7/j2JEQMtgdyUZnUktNaGUAJSienifLvjfsqCcpBhDRgw4M8YgxxrwIAB7wm88Lf/3ouEtZdXGb18s5dDdh/L2yGqwTpkVHiyUYSIUWRUYLcuCHZPyCan7fcqTR0QBA2qQp6HON0YAhVOZwEBvlTvkVxtN+/8CWE7dj7glDf0EWeS811zxkeaG9Rt+V+I35j49KqUQ876FKktMqZkV1KtCmr2dQT4ZK1uYB6R0LRxs++0JVCUWusrb402zHTZD7BdgtMmugrBUqsrMix/v+7K19u3SI0UJW8jZtcU1NqQqx/kuw1CSdV7O3ZlwpO6Q9EmYHXflxLR4Fhr4Ru/+02B6X9GJ9sq2nZ3RVlQ9D0gVbsB6Qz+DuVCCg5ZM68d/7N8j5muWLQRyNL6R7pj3m2NGm1Ya9E3pqu6Pi5X1fUSOaD3pzQb5KSTcb214Twg6FO+SqorPpfNzUf3c5qWZHRk5Mpx39iabMYAd+fEEyJ/fLvHOyGhwJ/jUOzb7nPAgAED/jQYNiEDBgz4M8Wn/rdffPH8lQ+xdf0OX/7L//m7Lt944W//vRf3PvuHnMqnkK0SCRwaNVAbtLZ9/G4vwwIkqrwMyzq/JXHiTehxe7XXqJdoOUO4e4ycX6OpA4xtGI0q1uuYKKpp6gDVhroxPD4PWRRQ4LiQlTdJo8x0xTWZtsZuaXsuyrb5IeBCcu7JGXesMCJBWgkMQNRe044ImZAy0ZgQy4XkFDT90AteJtR93y233cqCoDQVpyy4yYRUYgr13oSiHfq7srzNIbXeuMqu6loi0kmupB2EHYmE5FqRSEgD/ZbD8PaYWd8VErEkb1Om/H8/sURUeum18M879H4QDD/S3GJCyG/as97K3UX+di3g47aNfbOJvitdNEjvxRHxnoc1JYKQtzKpLirYodzUEe9LEkIDkyJtgwL8sF5Ss9aCVSsrC2WrDev15Kkf4sUSYL3Mq90WwWX8boDtSUrX5N5th7rlRidF6+OX+6QsvWJI30RHGN96+1VSaHsTe5eM1m21uuCBkpqoPT8pEQvW/M3PD1KsAQMG/NliICEDBgwA4Ef+h1950dgS10R89ef+/3UAPPe3/rsX9baXkrgq5qN/979+MV+N2Xnmu6wOn+Jb/96fXbvyZ17+ay/qieFUPkn50jMEoR9AZbeGuUPP06uyLCdQWzCKTFaeoDhBgsuBWXqfiL9da/+xaww2qKlrS1WFrNcRZWkJQ4eIMluE/MHZCgecScE1nRBgeCgXRBK28bWeXMxYEeGH9kwS3uQE225HOgMwwDYjMg190pPUfcHhrmbsOO8BsRgCNRybJdfcuI/QzfRSjuPjgI9JNOBJ3eGBnPebCGhL8Tb8Bv3hQqGNjgXagdUhGwPtSosr7wEyiftB2GyQhrSNxBWEVOJ+wDbtmN39jE4O1HWdvGSPedJt8aTucL/1tXQG6gkpuVStJMsTiqAd3sO2ET2n9MlUIpzqnJKKLRn3P7OhYUrGmoJ9JpxRkDcxVoQfb27x6/ZlkraLJSLAScyJO8eK5ZGe9c8zIcKK6bcIAb5PYyIZM136x9xK2zqCAvRkpGkjjqO2BDEmohHXksbL+F54u5G9i/h9qwTryjkVC9r0jyHaCAzo3vvzY4gImLfhCduMBgIyYMCAdwUDCRkw4AcUn/j7f+OKcdsRoWr7z0lUHa96MAAAIABJREFUgjO4KuYr/8bbE3E++Q//4xfP3nwfu8++5DcJteVEP8DpnfcjRrk4foIgLFEV6ott6jrkh/77//JFVcvX/+1f+lMNNZ/+4r//omKY/fEPEYQVNl1Qn19DQtClQNNuNlrDOcZBHvkNR9D00buooLXxt1cB2kq3JGqQUQHL+NLADogoy0VMWVqOzkMmqcOIUjfCmoZTybkQP7xVrSypM0ELwjnL9lj72yekzFm3siBD0PoTwMt9cikZaULc/qoeaUSohrjt5VhRMSWmwLesX3fj1jTtz+OxrLkrXk7WiDKXnBFx31nhh/nLtKe+0A5zdaBVB63kaTN9ahOdRKsjJF2vh7blgktyGnVMJPWSJvHDb9MO3BU1pdYcyFZ/nAzCrmZ8UKZ8har9er8BWmtBLZceiO5xRYTU4u8rEEuiXrZVasOWjBG9JFVxmyj2PrfLoVnyadnnS3JCbISdDBqNCCtLQsScNXHbXwK0xYUlMRGlVFgxrLXon4sVP+R3X99J2/wGpuy9H90W5HKL0sboUpERg8DazS7JYPv5bkO1ucHq0HeOvMN56uRiKpcpYYlEvYwuRFhSUG+ctwEDBgx4NzCQkAEDfgDxVgIC0G1BjG2vkm/Ilz75q3/98uuDBjGOeunTm+qLbVQNrglpGsNyMWJre46q4dHRAWVtefnl6+zv5iTJNuOtGXsv/tqLRRHwkX/lN/jDT/2dfyZC8mP/+BdeJK78JuOmI//C80TJgnw1ZX16kyRb+C9sDIQOiRp0GUBYo8ukLyaUoPWIOIEy8PG83SCnAs6gtfqo3lGBu0hRbTOPVJgtI+6d+CF/e6ws14bjhXDfzDmVJV3DdY1jTNx7CAzCLmMslhNm7DHFoWyRAZebhhDLms734DcjI/X+j1RDcvH9IfvOf9+ZyfuhM2p3Kqv2Z8YEfbngXXPOoZ7zPm5wj+N+SO3Ix9XeD9cPxb6Ez76NfGx6TC49CpeSrZXmvek5k+Tyvlr5k+IICFGUSup2T2Q40AkzWbVyNX+cP7xn+NKJ41nd56FcIIgfjtWxJSMWrEklpqYmJmCh6z4auKJmRyYsdA1AJCFJu20IsXza3eAgCvgwGaNIcCtlHMHutOLNi4CndZc5Bbl4Gds2I1ZyOZg3OEbt8wsIsGIYMWZNwUgS35beUr3ueFgMiOmlUKH4fpNEov5YNjSsKIgIidvn1h1r7bZILRExvWROr5yPtyJom8+7c6S9fM73w8QSERH0KWQNjl/5/J/ugsGAAQMG/EkYSMiAAT9geCcC0kFMg6pFpMFVMSYsEGn8wG6umpOPHz6NiHL/9Rd4+GiEMXDjYEEc19R1QBDUfOtOyndXBRUFn1hkfHtW8rmnx3z4o69y59XbuNPs//XxfurXf+lFGedAACrIpIQKkp98DT0W1l/6BONPfL31duB9Hi1/krhCa4skJbpKkKj27ehl4G9bJt7IHtZQWySqPSlpfSLdc05HC+raa/u/cg8eu5wEQxpGPF4qh1XJuV0x1oSZrFuzsd9q2HbD4TX4/v01tlhRtDKoztdxeXxHrSHYm6alvWofsKUxizZda9IO8HfxzezXXNZvQCocJ2bNWCNeaPY5Niu+zh0Avql3+k6J7kp6Rzia3ifgoN3eAL3h2cilB+Wtg243HPv79n9PJWStvnndiiGSgAVrGnVE4gkSwIyGSh2RWL7DAxIithkx1oi/enOf+Vr42ewGaQhfOJtyKCuO5JxIon6oXmvBc1zvN07dEN35ZnZlwpqSmpqcy6Sr37Zv8BerZ9mPAp7ar/m51TUmWc3j85DP68v8rDzPvSbnG9ZL6jpi0D1fK4ZrbLGmJBLf9r6lGUvJ+4b47TYFzGKo9GpIgH+tBG2j+2XYwJwcX1upLcGyvbzsSnyvuneM+e0IT/czerLRRvJ2nTCd7K1DTkUolkqbfps1YMCAAe8GBhIyYMAAAFRtG0l7tWlZohJXpBjTxtkCbpUCsFykWOv4/MMZN0j5V/cNQVATJzmrxRjbzkZzKs7yhJGxNI0fkiaTnDt/+ONc/+L/9GJdW7JRThSX7H/u92EZILs1OrM4M0LiuvVwGFhblAaxSvHSc/4xxgqhQgESKhhQ4/xvuLL9NRe0ZGpjw4NR7xMJ35Im1Ep2tDbgDHUdkq9SmjrgWzpjj5hbUUxRw6pxTIwPX62k8Vfl21K9kKCVswSU1Ji2Ods3b/vxuBtqO02/L/rb7NXwnpDMRcykpJKGVAPG7a/vZ9w2XUP6kRQ8p1MmBOCgkIaoLSzsNh4ipvcQ9FfUkb4JvYuR7X4+0JvX4fIK+mZ3he2+vpX4dFuTEQlrCla6JiQkkaiPs1UcZyy4wQ65VtTUlFp5AqQwF8ORnHPn5AkapxgRxjH89H7K509WTGXEQtdEEuLUP57P6g1+29xB1V/h92b1kkprrstOTzyANpHKsqbgJWZ8uNribB5wY6/g+DyicbCvY/6RvMETMmVXs15qF0lApQ2B+CjgGkdGTEqEIGxpwpYmVNKwoymvm+OebHQdMd1WCGCXceuMuSy49JuaoC947M5f5y2BjUSyTUlWe666RvrutXX5vH0HTJd2ZTG9dK2mplHHddnhWC6ukJMBAwYM+LPGQEIGDPgBREc01HXJSPbK7RLUUIPWARKVl9uQQGmWEy6ObyKinF0kFKUhwkuh5vOYNLWEYcVslnIwdby08EPVYVnxoWnEhz/yOsU649qtu5we3eLsdIwxymqZkK9jrllYffd9JM+9hsQ1ZmsFaQNlDUbR0iKhg0pJfuJV6t9JcY8yzJMrTz4aEAcSNmiF334UIRJVaB71RArwxIY2kKjb9GxE+uo87ZOzmjrg8GjMsTxkqhHrRpmvHK+7FQB7JqOi4UC2mbNmREJOSUJ4RcrUae/jtpF6SzMWkhMQEqqXU3m5zuX5KqiYS85YY1INyDRkTsWeRDwnGV/ihCNZMGfNU82Yp8KYsDY81DVzKbnpRogRppKx1NwPmxvSHTY8IJvDcUdCaq37WNdNOc/bXldvMUMfqTeS+1SnSwN6Jy1aa8GFrEglZq7+qn/QkoeSioiQv1t9hxwvEfw3zz6KEeFpnfAqx0wl88dYIhoaLlzFbbfL0+xQq38uCylw7el9IOd0nR87mlFIgyFhLgU3U0scKk6FUeo3gv+8vcEX61MSDTiVdd9Eb7GegCBMWzldqJZSakK1hK0ULGgb7TuPzYQUFV8ICPTFi0C7LZO+uXxETEjAjFVPBjaP+WYx4WZkr2x83JGXzXPakcyAoE/2sq2Jv2nJSUPDaGMrM2DAgAHvBgYSMmDADxB6w3kLcYrWAcaWPSEB+kFdTHO5OQga3Drj5PApANarhHsnltPc8bFozP2y5Hdeh79wOwTG/Pb3LJ95SrGtAbhCKRtYL8acnEz5wKde48b2CfrSxzk/H/HwKOPOGTz7cMT/8lsf5kdfeJb3/8xvIuMarQSzW6IrC6FrZVeAg3T/Aea6JwKEQAOqQKBQK6Rln4wlSdknZOGMj+oFb14X/7U+FctcmteN4pzljTe3+I3DJYVteNPMsI03dR+ZFYH6Y3QkczJiYgJCtaRk1CjnssTik5PyNlY2JiQipJCauPVDJAQkGmKAY1kRq+1jU+eSs5CSPc34uGzzR3rKDYnJrOHC5RzpORU1v2Vf4Zd3P8Irj0oyAjINeNmcoSjP6QEv8YCCy9cA8I5m87eSkUYbEHsl5an/fm16idf/E/I2HrgbwiutOWNOl4rVd22IpwpTMuasCQi40AV/hz8iI+Zv3f4IX7mX8cFmj//Tvu476NVxyyb8xZsxD88tgYFnbxbUtbBcB/zeg4oH9pyMiISIT+sN7rkVBuEaMS/cKomChrK2rHPLdNRwUAZ8rtwD4DcLH2lc0fTN611gwL6OiNVidJOEdbK1hi3NWElJRc01ptyXEyJCpmStWKyVvXWkAMuSNROEhJCcqieIpiWoro1RDiTo+z46XBKUy61fZ3j3RFCoqTeSuAIqaXCt8b/sH8dQJTZgwIB3D/bnf/7nP/fn/SAGDBjw7qPzgoiolxtp63tQg1h3RaYktoHGItb5bYhpIHCc3n+e1TLFNQZVYRzBdmy4WAmNg1tJSGSFf/y64yE5xxeWUAwxhpyG928FjBJlPFmTxjkmLpjcvM8kdlBnLJcRx689RxpCUxtuHjzC3MihViQGsYpYkLR9oAoSF7C2gKLnIXq4jdlbt4lX/jlJYz3JcAYtw8tjgFweC6NI2CCRg0YQo+g6gjogv9jjf/9WymMpejIxVj/AnUlOLd5X4QSmmhBgKeWyjG4uPpq2avcPsbcwA7Bsi/eMCqU0TNttByLEGrKjCUupWEtJiGVLU562GYELyFV50635OLu8ai5YU1BS89z6FhGGQ3L2ifmCeZ2Kmkdc9CbpTS+AlwF1hIN2hN6Q8EjgbxPBdR0Vchmua9vUp0YbzMbHIvI2YtJ9DvGSpAZHINYfFwn7UsEdGbMk76/ohxJQUBJIwE/Ym1wsAx6YBU/rDtd1ihjLv/VCxiq3/KfnX+UPqiM+zS3GWYMIaBHxleaYmIgtTfhoOOW6iXkijHEKz+47kqTi9QcZVSO8eiw8yhucg9rBPbdmWxMq8UcoJiBsRV0BhhERsVpUYCElIw0RoBQHAkspWND5hSwHugUCmYatYEqoxbV+HP+ccypqXFtsqP0WptsqdU32ItIfT3PFH3LZii7ivUZd63pXTtgJrja7UgIMMSH/2ef/g8GUPmDAgHcNAwkZMOAHAFfM6M77HAS93H44g5gGEfWExDqoA+9OEB9ri0C1nDK/8Cb0KKopywB/WdYyDQ1P7DV87VA50ZIIy5YEXI8D1o2ybQKe3IKiDFivIpLIYNQSTGa4VYa1hheef8jjo10mWYNT4cmf/GMkBiqQSLzhPBT03CJJKyd6Y4fVG89hl8LJSx9jeXyDND6DBiStEcH7QhrriVVYt9Krt1y1N4oZFd4HAj5la52gq5jTR09QzTMOi5qchgAfR5thWXM5bMdtVGyAoRaHCgQIaylbH4i3mft0KFoyk6Ci7GjGSEOilpxMNCIlYCU1uVSI+KF0KQVxk/CBOOPZScDdvOLf/ckZ/+DeirwtQXwoJR83u5y5ihEBx6ZkTUmpFZu5Vpdk4/JYvNM2oxt6hTZqFunJRvfH3+Hl37sy+e5zmyTFiMGpa70REIglEh+769QxNWOu6RQReqIj0Bumfz8/5Zd/eEp6tse//CHH1nKLoEhI64SvHFe8bi4otOL+2vC5GxFODYkVvrOoGLeenE9lU/bHMEngxljI0powbPiVNx/ylfKC13TOBTXiLC/r3JvSpWJPM+ZS8ITbItGQkfp+kLB1xpTSYDBEWEpx3ickJSkRdetRGZMQtlbxscaspOpPSGcmjwj7cIIGbc9dFwhgerKx6S8RudzEdCK7Ln7ZoYQSYDBtk7z3mCQ9BZJ+sxNgefHzvzAQkAEDBryrGHatAwb8AKDzfHR4q/kc8ETDKDJaI8YhUelvi+reL5GOZ0y3l4gor9/dwhhllFY8c2vNMzcKrFHOXM2UkJsm5lYa8L4bNRNreN2tOF8azhcWVeHocJc3Xn6O8tENgsjLqc4eHxBHDXt7S/Z21nzjV/9F3L0EPRqh5wIV0ChnX/oEemGhALM/o6lD5sdPEkQVp6cT7n7501x862NekhW20rKggbjsSRjGef9H92YUAkUihwQOd7xFdb7LenaAiLI3cXxmPAbgVNYcmSXPRylP64SKpvUxXMbnZhpi1VCI19dvNp+HBL3PISXAqHCgKZmGdEuakoZnTcangx32dcTHmgOO9JwHespX7QO+VFxgBD4Ujvjatw94Une47NGwPKpLTiUnM4YPN9eYShsFvCHL6a6GX3oHXPtMtH/r0H3cxfl25ujNpvNNAtOZ7OGSiFixl1fhxfZvQC/RiiXGIKzEE6qSigBDRd0XAp66C+7cn/Ajz69pasMvr77My+ac/2LxTUoc+0wYScLXuUuSVEwnOUGg/BQ3+JfSG/wUN4gCH8O7v13y5M05aerPh5fMlTyQU87MmrtmTklDScOFeGHUE25KgCFTTyrH6qN1VfxGYVtjgj5xzGHUm/X3dcIOEzKNcdKa/hHWUlFJQ6SeBHRt8raVpFlML6fqztfmliMgaIMObP+5oP3KANufn81Es32d9IEFXVxxFyedbZRmDhgwYMC7hYGEDBjwA4BNA3qXgtXdJqa59H4Y50v7ypBmPb56J044fXQDgL2DY7bGFWXp76MoLY9OI75x3zIWy64NmLuGm9uOa/tLEuMjaxsHTx7kOIXlOuDeUcK3v/4hXBPxve/d4Ow84/F5yMnJCBHl9CLke//kn+Ol3/kcx7//GXRlcfczonhN+d3b6CLg9I8+wWI2Zb0c0dQBbzxM+NJLU5o6RGdtxOhmvLBtWgLi2u4Q/yZJBbWgpUFrw/LoKYrVFFVDGFY8c/uUUQw3NGNfM0IsWSDcDv3Atmp1+wA7xOw6rxnrkqQ29fUOZUdHANQ4EgIatC8s3NKIAMMrbsnjqmYlFRm+aTtoJTMfD6fsb/mh8TdOz0laedhIEioavmYeE2GpVNkh8hGx7cDfEZAupvdyW/En/5ewWVSofwJBubIV2fhc96ffkmwMxbXWOBylVu3Vek88Zni/xlJzTnXeX9Ev2o3Af3t8yHIVsMoDQrF8k3ssNedQ1uy6rL3Cb/jmy1tY6zjYX/KTL9S8/4mCzzzjuL5TUlSWOPbH0FrFOcOPNteZaMo1tpiz5lRW3DWnzKSgoibG9s9RBbLWfN7gqGj6lLEOHbGocdTiCXEtXTiBJw1bmpBpSEpATNj3qGREfYHkiIS4jcyNxJOHRCJSiUkk7MsRu+Prz9FlCIPdkGl56aAPAUjx3SDduQ1a4jdgwIAB7zYGY/qAAe9xfOLv/40X1Xjy0RnQxTT+fVBfSYtq1mNsuqBc7jA/22eyc4wNCpo6xtgKMUoUlxTrlP1rc8oipK4tR0cpVQM/9nzOg8cJu9OKb90PuX5tiXPCs/vK/QchTiHL/BXnh8cxf3C65jOa8lv/5EcAsMby1bOCn0pCoijk9hNz6triKsvDB3vsO3jjCz/JaLymrveIz64D8NIru4QWRmnD0UqZhMLrrx9Q5J/l5ie+6Dc6tZfLaHB1SJSWoEhaovMEXcdgG+oqbJ/vmjhbUeYp+1sVP7TO+PbKeG+7wigUgsZLWypxWDU8FcZQQa41D2XeD6nnLHtjtEWogZVUxGrbr1EO8FHG39OS65pySsmKkokJSCXuB/RpJHzjvuWsrjk2K17iAaqOSPwQ+0DO+ajbp8Dxe/ZubwoHTxZCCa5EvV5eJe8iXi9jeDd7LTr0ZvUWTswVCrN51f2taVvd34PWZG0wRBIS4yN8O2nRuk2+WrhV31kxkYwLFsyl4Fv3AxJ72bNRacNcCo5lwYwViYT8zcW3+cXvfpiPv/+C6TTHOWG1ilAVTmeGURoQBA3OCUURELakAIW7xr9WQywrKXsjdxdp7DcVEYlaChpWbZyuQymkO7YQYjilICRrIwiEHZf2m6hMIwzeK1TjqPBRyl5e530i+0wwKjyUc0bELKXoI4ctITP1BY+bcLjer2P7LYm/IBCqYSwJDY4xngQVNO3jvXo/AwYMGPBuYPCEDBjwHsetb/wfnzO2be1OCySsvTRJxbefWy8/ojE8fvMFJtcOObn3LM4Z1osJ5XrCcj7FNTGrRUYY1izmI4o8Yu/ghPnFmOmoZm+7IgwbblxbEATw3K2cIHAYo9y8dcKo3GJvWqNqePq5u7zy+j53i5IpEX+4mnNWKuU65A23YlQlGLVczGMCA2eziHFWM3aGb7/0BFmsHD0eExhBBB4dj3g4g3sXwht1zsOqpFnFjG3ITlIS3jrxqVjgzfgtJPCmYayCKBLXNEc7SFIiVUQ6PiGMl+SLHcQoe9fOePnODtejgLFYFCGwcFjVzKXsdwAftlMqB9sa84aZYRDOWeBwrLUklYi55L2QaS0lBzpmRECB4+koJm4CHPDRUcrDquIb5pgFa0qtaXDcrg4IRTh0JR+RLebAmaywYvls8xRfkbv8tDzNHV3xNbl7ZXPRm8dxrbdg0ycib3t/6cu4uum46h/RK39oTdJv3YwIQiIRNQ2pxIQSEEuIFUtBddXcLo5cy1aeVpNI1KdFVdJwTwvu1GsuZE1EyJqCueSeEGgOCImE/Eb5Oh+4eJ7nbp8hAkGgWOvYnpaEYUMYOqxtuH845g9WcxqUc1ljxJBogBHDDTcm1ajfhCQaoAKfyabsBgGpBpQKUWv2XkqFAHMpsRgmJJ5oiiPVgAkxUesNirDEamnEJ1UZkd5bkhAyY8mYlLAla5lGrFoS0m1KArEY6WJ6Pcl0KJF4yVjQ/iyD7zWZkGDwP8dLsmBHvVcFgU/+5R/+3f/vv20GDBgw4J8dwyZkwID3OHyyVYMYT0S0TcGSpPTSpMb2g7k6wbWlfMY2GOuoqpDlIqYoIvI8ZLK1YDxZ8uprB2TZhLK2uEbIsksJh7WOOCkwtkGdEMU5H/rwmzR1SJItWM62ublb8+wq5Sv5nKVUFDSElbCWmvOqYXkipFZ4PI/YSqGqDG++9ixZ4lAVqsrw4HDM1qRib1pz98Jw3HT5UzCNhFFWk3zkFU+4bANYn3rlpN+AENaXsbyA2ZkBEB886EsLw4uSskioypi8VkIj7KRCHCr3L2h9HMpNneBQ3igLXjOz9iq/736ICP1AjS8WXFMAcWsOjogwrGm4Z2Y826QU6nhFZkzXO7yPKeKEe3JMJIGXeCnMGn+1feYaflxu8MD4mN5H4j02/5QHHNkZTr30ptKqb0kvcZdm8Q3C0UmtLrcjHuYtBMRHuG5sTNr77T4H9NfTN7cqAIX6mOJSa2pqUol7mVXnG4nk8r+nLhK4pGKhaxoaFrrCiuFQaoINj03X9t6lj60o2JYxX1jM+aH5iCguCcIK5yJM+xowtqEqQ/6vR2XbHQI3dcIhC3Y0ZU3dD/yx+o3alJBIDdZAbOCkUJbURK3HojOyG4SFlL3cKWxjl/t/KyptR4hiVbAIFYYYSykViumlfH23iyi5liQScqBTHsgpCRGG0G9oxDAiQVtyttkZYjF9BG8Ho9JvYg5lwX/4a39tMKUPGDDgXcfgCRkw4D2MT/7af/Iinfyo2wC4Npq39X9c2QwY5fH95ynLAHWCqmCDmvkq5OTMexzevLtPVYXUjaBqODqNMFYJgoY8D1mvY6LYD4Tq/PfHo3OieI1r/K+cqgyYTgretyfsE3NDM2ocr4onAF/kmC+7U75TrvjSes7hXHnjUYSIssoNxycp1igXK8OXXov4/Ttwr8lZ0VDQMJOSP1ou+c6dFC0s1D6Cl7CGoN38dF6QwEHQxvGuI78l6g+I35CoCmFUIOJ4egfSELYyx/5WxbJx3CLjebfD84x5PxO+bA85lRXHsqRo9fUTkl6Xv2Dt+zIoKKi56SbsS8xNSSmk4U69JhHDnib8sZ5xoRUfky0EodaGShsWrRQqxjAxlld1wTYjMmK+YR4AcIfHzHRJSsxEMgIJGPf5xpfYNJe7jfddseImNk3om5/bbO3uZD9X5FqbX9v+sS01WWreJzl1jd55S0oUn+qUtsHGAKl4H05JRd62rK+1IJaw/xmpxFyTKR9xT/Cp5in+ynMJzgllEeEaQxSX2KAmCNvUKSf87AeVUmpyqTmSBXua9t6ZUA1Z67tJsUwlYDfwj8cIrJzrJXVVu51QlKL1icQakGhASLtd2Th+nUQqwNCI9waB95GEakmICNVSSUOinnCFYluyc/nvt6LxmxJiFCVtDebd+TCI3yjhj1PVelScKGONEWUgIAMGDPi+YdiEDBjwgwCjLfHQyyG7fovu23kDtrGOsogIoxpjG5Zzb6AOrOOV+zG39mrOz8Y0DqrKejIwDxllJYcnEdtjy2SyYr1KUIUorqge3iaMC4x15KsxYpTZPCYOHc9nEUd5Ay7jm+aYAx0xl4ILUWIXsKbmd5s1yTLg2WLMfC2MUuXu44BlpfwjvUsghkgsT7gJhTQcy5JUA37sY8f+uQW+SZpaIK58DG8VeFLSXQ2f5IAnTlKE6DrynSJRheu2R0Z54ZkzlsuU8WTFcpHyE8/D4UlEaEP+m9M3+8Gy63eYserbp0MC6jZ/yqmiIuyQ8kG2eawFzwUpc13zRXufHc34Ib3G75pXuMeYJ902I5NwrBcA3JU5Pyw7zLQmV8ceCdPGe2TeNHO+1raG7MqUbUYsyRnJLjEhF23DfYeOWLzV+9EZ1f8kb8hm30i/TdnYiIiYnohYuUxp6u6rpMJiKbRk10ypqbFYHrszAglYaUGhpfe5iCUmIJYx57rkQLZ5qKek+DStWmumsk1CRFe6l2nMszrlLz0vvPIw5GPttq5pDGHofSCqEIY+nldVmGjMt8xDZrrikzwDKFsuphFl8+k/OwkoG4jaf0aRCCh9z0ZBQ6iWmSmubIEyjfooXKBP0RIuzf/dMd9Sn2i2q2MMkLQm95EmzMW/Xh0wJiWnIm4joC+LJj3R67Y4DY4Qy3WdklMRalt82JKZf+0f/usDARkwYMD3DcMmZMCA9zpa4iFRfWXr8VZIlpNkK9bLjKpqk7NEyUYrVIXGCfsTx2JlscZxsFtw/3BMXglGlItZ7KN3zwOOHm8xmyeoCq6xrNcxp8c7NHWAawyrZcKjs4DXjy331zVf1TMswoGOvFFWvQn5QnIMkONlLfcfR9xbNvzBXfi99TmPqopMvd0XYCEVx7Jk0foz4iRH4qYtKvQE43L70fj3Tnwq1samSPMQRGmWEzT3j8UYR12GLOZ+kxAnBaPxGmuUg+2Ki7XwhJv2EbaKI2+3IEtyFqypqD0JUUdFTaUNAYZEhOeClNwpuzpioikVjkgMe0z9tkRHvOAOeFL2CbB8xxyP50eiAAAgAElEQVRy4ioqHBYhRMhamY9B2JMpWzKmomFHU/Z1zPubaz6VSaI+6nUztar73nfqCume1zv5Qja/fnMjsvnxZuxvJ/lqcNRtLKx/HF3fhlxJaMokodKGfR2TbUjYppIxEk/wrPhm+Y4EJERkhLwhcwD+x+Iu63XAo+OUpjE0jbQkxEvzRBwiyl9IdqhoWkJw2bEhCoU0ZAR8MMlQhWnSHhsH+5E3lQOt2Kn1Zqgl1lZCh5dbWRVEIVTTezTiNvUs05BILVn7bwB8jO/mcTbALmNCLKXUpET9ZinEEhG0UdDejr6jGRkxuzruiU13/hP1fpbLMzVgwIAB3x8MJGTAgPcoPvmrf/3FfvPxDuRD3dv/+cfZBVVlcc4wu/CRt6qGuha2JiWBVcra31fd+MjdJFQCqyzXAU6hbuC1hxH3jiJWq4iq8r0gq1XIepWwmI8oywBrIA3goeY8MOe8LjMKfLHbkczJNOSenPlG8lbW8nvnC0IRfsfc49vmkD+0h2xpwpqac1lx15yTS81UY0IM0737vjVdxZvPnXjZlVGwDncyxs0T3OkYd7SNFiEUIe5ighYRYhry8+tY29A0/ir0/vUz9g9OcI2hyCPqWihKQ17DJ4ItYNMTYfrB0alisSQSXZHhCMLcNRzWJReuZl9HVNJwLHNKdXyouUZJzUvmhBjLR5sbhARc6IIv2Dc5lZxKlQuqXsZz4DKecXvsMmaLlC0XM9KYY7Mi0ZBMYqxYUknYNVOmZnTldfBOZvJuCH5rwtWmzCqUkKA1RHdvIqbvJjHI22RbjTY96Qix/WbDIEwkY9tMaNSRSEQhDQUVqcQc6hljUnYZk2tJQNDufipf8qeWNTUf0C1my5Bnmm0OTyO++1g5O/eEuUNTB37zZxRr4AU9wIphq22vPyDFIMRqOZOCslHWNexMatJIyWLltGo48K0vGIRCGm9CJyAj7Mlb95rovq7zl8Qt2emOi0XI8LG9V+Od6QlDTEimIYn6rxm1ZvOYkLFGRIRsacZEYxINGBExwpObrh29ab1B/84/+LlhCzJgwIDvK4Z0rAED3qN44uXf/JyIep2HSm+y7qHi29A7xBVnd14giisEQ1kEgCEMa3Z3VozGK46PJwBYA0nUcDoPCXyYDjevr7iY+wEnr8AKBMYwympAOD2PUWeYL2PiqOFkFnCeKy/rnBrlxCyoRfstxkpKznXJTFYccsZUMl43p3yPc06Zs9KCTBIK/BDrRLlgRUjAM26HbSI+tBeT3XgIlUXPRuh8hC5SJC3QRcr6wW1Wj29BkbA4uUnkKihDLo5uY9RQlylNHdI0IXUZYoMaG9SEUcl6NeLR0YRVHnDvzFA0yutVzn0zb4mHHzDXlL3jIpGILUaUUvXm6wxPEO6YGQ9kwb3WXD7XNe/Xa4wJeNPMuMcpr8kRK+Pb60sqTvSCOxzzUW4RYjihZI+YCyoSAgIs192YQhpWUvFYFhzJOY06Uol4nuvc1j1CIs5lCe3VetNKpzz5MG8jHpvvO2+DIEzNmJEkLHXdE6wuItbnZYGI4NT1/hgjBhFhLCk7jLhghe87SdlhzDWdcMoCBLZ1RCPKdZ3iRP3rgxWRhK28q2ZJTknNSkqcKBNNmS8tBY4Hec3YBIwCy3TsSYeIYq1igwZjHAfbNZ/dyfjN4zk/Ez/J7SjmVhLwapWT4gnWzDmqWoiakNs3csZZg8sjrscBq1I4E3/OK2mYaEymIblU7Osl2Zu2TeUWYdJu8hRPMGocIkLa+kgMQiWul1gJtCSm22YBIiQaUovrNy8JARkRhdRYDHFLVpwogfrW9RLHL/zaXx0IyIABA77vGDwhAwa817G5Bem8IYB0/SAbt+3e/p7/XNDw6pc/TVGETHcuWM4mREAcOR7PAkapoXGGJFTOlsLtgxrnhChQGidME7jIYTquqEqLGNiaVHzjTsy1MYyzinkBJ3XNXAq2NGEma/K2E6OTkaTim6dzSl7jCBT2ZNJLcfxw1rAgJyMmbUv5ZlLy0+Opf/rHW1Bbzh4+55+uadjZmyFBQ3rjTeTREyS33qS+8wKr2QE2qBhvP8Y1IeoMzhlcYzG21eobh6ofoB+dWaLAE67SKV+xh1ekSWXrcSi0xLa9D7d0m5SQU1lgMTzttv3QrRFrqfvm6j2Z8Kq5wKrpU40CLI/0jEQiRpJQqpepvSlLxhqyRUhOQ4ghxmDUS33WUlPQcEOnpBJRSc3TbodEA1ZSMcaX3uUIIX6AXdL0BvJ36vx4J9TUZER9WhZ0fgMfAwy8LUEraJswwG+OampC8Zb1E2ZUkjEm5VwXPBLvh1FxFNQ8IXucsmjjeGklST5Fq1HHSGIKGu7pigkhH81SRODadsk69z4mEcXYhrq2RHGJqrBcxmRE7Ga+XPONhwljDfvkqgkBRkAEorimLALGibIshEiEok3TCtU/jwjru0e4TMMKMUR4100gQqPe8K5chgKoeCVhTIBVn7blvR2XhvTuOGYaEWI40LE36lMDStV6kKDrLLmM9e1uGzBgwIA/DwwkZMCA9zI2m8LBG7Eb83ZTulGoAmS0pnp0g3D3mDQtaRrD4mLK6WlGEse8fBiwm0EUeP28NYoRYTTyw9vNgxWHjzPKGiIr3H8cc2u/oMwN949DvlUs+XQ0YrkOEIFjLXlSp3zbPga89+NMvYZ/S8Zc6KI32pZUaCsfmeuaG7IDQEXNTFfsMmYlBU/qDjc047OfeIANKs7vvUBdhgRRhYjz8axnYyQtIKxJ3/cyqBCEBXUVE6UzTLqCMiBv9gGwQeUb2PXqyJbFXopT1obICHETULWSoU7rnxBSUOJUMQLPuW1mlIxMREnNTAq2dEKljrFEZBqyouJ9bod/al/2Q6c6DmSLh3qKoqy14JbsccGCQAJOWdHggwAKadhq/QR1Gws71ZhcKo5lgcNvB+BqoWDdGshD8UOq7evthFwLwA/5sUQIQk1DQsRS1/4YiaXShhPmvTm9IyKbQ293m6ojaOOGLYZtRjzGp6M16jcBY1LK1jOSSMSp+s6VWmpKrXmSHU5EySkJ2kecSkypFWHbDl+JY08TtiTg+RsVQeAIrPNywsYgolRliCq4xhInOdPpmmfdHk8eeHKTRsoHwoxl47jvlG0TUKmSRtrLup64vubbb2Ss1XfPxGoppEGU1rPjH0+MbXcgQiaWsTGcNTUhxpcLdpslbat8WpN7QBetu1n2KNQoAULS/ne+STg2je7SblOKrtFdIGiN6QMGDBjw54GBhAwY8B7Ep379l14kaKeL2rbRtA04A+0VfZxBy8AbtY3rL4nef/15ipc+QFUZrh3MeOPOLnUjzJYBqrAoYLS2hKHDqXBjt6Yo/K+SsjLsTEsu5iHWwLoUjs8jGgefXz7kXwhv0ChcLAJ2UtjNI47ImWrMkZSc6KyX/pzr/IocqMExkoSEiEQqLIYlOU/qDheyZo4fhj/NAR85MKSju8Rbjzl/8DxOpScgRw8P2HeGdHxOdPvNvicl2TtkfXzLExB8j0pQrqnK2JvSN+Rs3eB5fbekrg2NGh5UJU+zxX0zo5CqNRsHrOiaypWI2O8XBN7ntrknC142j9kjZYuIzAW9REdRXtDrvCKPKLXiUM+otSaQgFKrvkHcYjmVJQ/lgs82t9nVmCU1ZWuu7uRS192YfRldMYbnUlOJ41RWVFpzINv95iKSkISQBscDLfrbU4lJiMgpGZGQU9JoQyYJF27xNpO6baNkO3Sfr9rHYdoNz6GeYUR4Rq/xGkesNGdHRow04lgWbe+FstTcExDZZ0V1hUiFYn0jvYSMSQjVcqAp123EUxODiI+OjpMK8pC6Nq0k0ZPq7rxGcckv/nACeHJ9bUe4tgOPTiPCs5RrqWFWanv+LbYt5fSPQfik2eWsqTnXinMpuKEhhTZ9JG7WZmJdjwLGEcwXwl4Q8KAq+00WQKWuN7uLwhZJ3zlSt68pA9Qotk2+qtoAga59PcT257toCZ1/XVgKuYzrHTBgwIDvN4ZN7IAB70F86S/9V1c13o31BAT8JqT72PmtiDqD5hFilNFozWsPEg5PI5aLlEVuSJPLvgfTzpiNE6LAMckqRKCuDct1QF0bosgRBUpoYVUIRmDfZYRGWFcwy+FsDXMqcmkYadxLaTpkkrTkw/9sg/CU7rOjKTd0m7F66dWeS3lBD/ih5iYfdNf5mR9d8MIzZyR7h5idBUm2oMgjxtt+23L4OKMqYsLd48vjABBXJNuPLmVqQBAtMcZhTIMNakT0qo8GKEpD2cAFFRmBb7zeSDK6UkyH4bGseZ4xBzbk6+Ye13XK1+wDjsSTqBPJeShLDmXFM26LD+ktn3KlFakk/nq/WN7QR/1W4Z57TEFJQcOcikK8Sdr/TE9CGtGeDHRX0huUM1lzxpKRJGR4grGlWU9ANhFL1De2l1pzoUsifNt5d46s2P5x9a+Z1pDem/RRrFgCAiaSMRWf2BQQcCYrrsmUA9kiVH+/aSvx2mGMaU3sU405YX7l+RRtZ8iElJtuwpNuSoxh6Rp+53zO2Tzk8VmEc4YwaggCX3zZkQ/npJXeNYRh3X8ujv3wvr9V8fyufz1//KmKphHquo3YFWUcQyqGp7cMOzZgm5CRhsRiGBMSq22pQytFaw/TyFi2YiHEsE3EVvsWtp6PzsgeYsk0JMS2aVoeipJT9ylycWtmD7EEell2GLa7re48xBr0m6YBAwYM+H5j2IQMGPCDACdQtOZX9w5yrNqidYA6vwV46lrJ6Szk6DilqODu44BbO468AWuEi5XBmIAsqVnlAReLgMDC0UxY5CFGYFn6DoWihjSCvzDe4ixXTuuaqBQOtWBEwIqKN80ZKTErKWjUexEyYlS0j7MVhIXkiAoBwoEbYcx1Cmm45jJmUnJbx6SjE8oi6ssJs9uvUb/5BMHBEckRfOQj9wjD4urzNwrGXSkqVGdQZwnCAlVDEFQUtf+VWazTnoycL706P8bwHXOKgSvDftb6VBocY2Iacdx1K57VEY04LiRni4yvm/t8qnmar5q7NOqIJGSneZZ9l3JhJ4TGskXGA04ptWqlO17CA7DWgpX44dyhzKloRFlRoyiLtr07aU3LHVINSCQEQiINCFHGRJyzZKYrIvHdE2G7sVhrTiiWQgtEDBEhEd4YnkpC1Zbh5VJiMZdyILma/tSZqq/rlg/lFWHc9qlk6tOkTmXJRFMKKsaacJcjAgJCsZywoqTigG1qqXHqJXsVDYEaTmTNDgnfkAURASkBXzyquC8r/ko8YXcnpywtcVxjjEMEbLslTNIlrrHUdYCqIiLt55Wb+zn3jjzBa5yQRRXW+k3Kkwdryjrl1n7Booip15bKReyHAVFtmDvLgpoVDduENArbI2WcCI2DgzLgflni8GRm1cr/3MYWJcan0Gm7yXJcRibXqG+gb0lIFw5gFKyYvrXdtc6TSrTdqAwYMGDA9x9DOtaAAe9BfOp//Y9e7Nq+cQat201IHaBl5D9+6xtAFTB+7nV0ts/5ecYiN3x5tua0atgPQ+4sawzColKK0lDXlry0vHrmf45T+OYi55mJZVEIRTvTR1Zwzuf6PKormnZIVrzm/a45o6T+v9l7kxhJsjS/7/e992zxLbaMzMqq6qqu7p5epkc9+3ATLwIhARwIECCAAwEEBrrppIMAQeKFGA1000GAjroJOggEBAiCJEgHnQgRGoKkOE3N9L5U155LZMbimy3vvU+H98zCI7Oqu0VwWIUa+xcCUeHuYW5m7h75/vb9Fwpc7nqIbGh4U+6njojss7hhxwU3nMgSL5FSHfOcHvSh2fCHby2Z1Z7l0Q3l+ZPUA2IjZ1/6GeIi9nhNtbyiWN6AKFL1Y2qYqKQCQ0i3xZwephaNFt+XiYwUKRlLBB49WbKcBy63hv9D32EjDTvpxjQoIHsYIpHIW9znW3rGE2m4pOcDueJadxzJnALD2vTs6UESqbjPEQ7DQ11wpgvu6ZwfyCOK3CrucPR4jBh69fQG7usSBHaSzu+QnrSWlrW06eo4BgUa8Sy15EznrHRGjcOKwallRc2V7NjoHvLkopKSRjsUxib4np5KSgqxHMsileZJQaMtc5NIyQDNqVgGk19rTyGOJRVLqmzWdrQSKLFjqZ/NRYUf6DPmpmLJjA/0IiVyyTwtqkXxBFo6dtLSSeCpbFhLQyc+lzT2CNCuK86KguWix1q9lVKVPWXdIkYpqwbUIiZ5eYxRnItYq5QOnFVEoCoDImAMWBdZVDCfd+z3JaUYVsaxLIRXl4Yax94nsnAkjtPK8NU3txyvevb7gpMZfLCJnNmCB5XDh0T+1pKaR4ym9xIonQT6TEEcMvaZBIljT0lHOo9eUvO6l4GyJL/SIGX7q3/w2//wL+Yv0YQJEyZ8MiY51oQJn0dEScQi2FsCEg3qf8Hw0wXUG7q25M3X17x2L2nUN3g+XCc5z5UPfOg7rrvIT68CfYBvnAuLEja98oFs+cGzyKPGs/WRqND20HhQhTeKioVY9uJZS88zs+eY+ViuFvPCyiDsaLmnc76mD0cZiUH4sX7In/M+j82avXg+Mps0Hbm/pu8KZqtnSJkZ0ECwBi9MlltJ3d+VY5X+ZSO/C0i+Al1We4wNdG2NzdvYd+n3raQ29IaePWnKMvRARHQs4HssNzzRlmtp+K55REVJKY6GjjUNz1hzzIz7HGMwXEtzkCJl2ItnJhWlOEqK3DiephER5UKvWUuXivCyDMwTcRhOdYbFcCHbdBoQrqUhDKlVOVJ26AKJKAtqZlIBjElcAK12aJ5wOEkt8EPx4gO57UoJWYJ12KA+BA20JH/Go2y2D0RaSa9ZrW4sR4yAVcM2x95G1TRZ0sDD3Jp+ozs69XckfVttskSto6JIKVES6Yk81Y63Lyy7XYH3NsmwotC1Jc1+xubmiL6vmC9vqOo2+YlyGAPAYt4xm3W4/J5Ksq00KZnNOoxRzo/7JM9ysKyV+ycdlYUjYzmTEiuSE7YUayPzOnC06LnnHF85FeYF3HOOWiyicK71+J4akreG120oaBy8JJIN6yG/pprJCTAWOg6v5X88xfNOmDDhU8JEQiZM+Bxi8HhoU47k45MIiKpF84JmkGmFYOg6x2zmeVVqVhQ87dMEI6L0RC5Cz00M7HuwRrlp4Id+y4Xs+L95wve5xkpa6Ow91A6WuQT6J7rhQnb8yDxll6+oG1K06bAgBXisl7wSFzyIC1ay4ERW3JcTVrLggRyzJvVR/DnvJYJ0tcDaiD1/dud4kJT+hU9XtmW1T63oh+heOD+ZkNhqh+RY3qreJj9JlYjGg7OOGIQ+wnlcoET6IWUKR5Gv5M9Jk4JvhlcwCMdaj83fJQVrGq51Q6Md1+zZ0bKUGZeyo8CMZuNAZEH63ZlUnMqKV+Q0kRJJsqgPzBUfmQ2ijJGuqTBeKdTQ5+vkm2zSHjwjXuLtwl80+wx63DCNwHAZkwdjIEaBOAp/Nrq7c18pxSgTOiwrPIz4Ha7K78UT8pX+ntv9gLTInuWpSXrvea7jhpnUCAZPoKPHE/Dq8bmJ/vA51ux5KmuaXOi4oad2yc+z3ztUhRAMMQq7bU3blGyuj/JrvstFhmGcmAyIQ9x1JifGBObzhqLscU45XXkWFTy812GtsqyVV+eWe4XlyJnUpeMiznlOT/ZUZeAb96F0qYW9tMJrdSpL/FKRpGqVWgpNU5WhHyegYxKayXLFoefFj3G9jGR4ICB2WgJMmDDhU8TkCZkw4XMIbapf7nF66w0RE1KCVu84Pb/k6tkJAF+YO/5s26P56irACkdA2eDxseR7T2ATPReyp8ax1JKNdHRR8VE5qw17n4y4QZUPzZooyhO9Zi63+1rloFWvnlfMOTbLhjoCr+kpnXhKdRxpxbFW/NReciFbHsopQZVd47j/oEO3M6S+YbzOEvJxDglhgCxT/OpAVHRfpUb14X4T0xTpY9A2NcE7YhDaPj1+lWNxh9QqGDoZ0oK4pKDGciF7vmcecY8VSpLObHSPwVDn+FvNV7a/FO+l3UfHq9xnusQhXMueV+MxgvC+EVYyY0HNRzznQ56B+QJBIoXOCaIj+eklspOeJ7IeDd8wyLZuF6gtnkZTEpOIYUbFRncHka9pf1IHSjI8D1fZF9TsaEficIiBmKW+i0BJwXM2POAIQWikp1JHFMVmT8SOHosdJ2WKspQZV2zTzxrxwmiQD4Rxga2kroyGjpXMUE1eip9seoSChyfQPptxvOp4flnTecO9kxZrlWq7xDrPbLFB4wpjI22TXl+RJLsaCIgIFKXHFR2xqVmtGpwrWS07iiKRgC8UkY+e1ijC8UxxVqnqlv2upix7rq5rFrPArrEcz8FvhNLCF8uSSErV2mTSPpzXQk2OAh5kWQfpZOMUTWjoqXM48B6PQ6YpyIQJEz5VTCRkwoS/pDgkIAAaLdKlSrmTb/w57sdf5+rZPU7nSreNNNncXKiBnNzTErjpIj8MW44paI2nUsc+a9jfDXteNzWXbeRebXjWRFThrXjCT+3lOP0YFr5r9kQiK7Pgr4c3+Km55kOzps8L3wvWIDDnAR/IDWv2vMuG34pfpJfAg/MtxobbgsaXihpNlqrZ0Yw+Pi5KijJuy0RABqmWC4iE3H1h2O1qmqZgNuuICpfbvGiXwCynS2k2jVs1WElEZKN7Zli+bx6z1QYE7nPMY65Go/C1bvh1vshKEzFzGBpJC3vyNONUawzCic5QkrH4S/EeW0ktEjYbwL+r7/AbfIkiy7qWWlJgeRAXvGeu2dJyqvMxetdoKipss3znqaxR1dEAX4ilwBG0uxPDaxCCJhnXlW45kUUqXHzhcQBGCk5kQU+gwHKpG45knntJUo/FUECZzml6b1xnE7oTl0hPNry3muKBh0mLIJQHkbNRNZVEakcp6fZnsudUaxoC72yFo5nlnSv4cl/z/YskIftKlzpXvlqkVvXFSqjmu3wMSt8XiES8d1gbKYoeW3jq2Za2meP7gnq2p57tUTVoFMQoVd3hXICPFrz52macrNR1mqwdH7V4b9k1lvVe2Hll54fPayL/N3Q5ISxQYcfej0Q47k6ZDInALyhp6LmWBqNpevKf/I//0URAJkyY8KlimsVOmPA5h5jw0m13JiCSFtkjvEWbguWv/ID5ckuIQoHwgVnT5u6JC2loCCwpeBJ6LqXhuyZJoHzWnl9Lg0PGGNFnTfYQGOFGWnoCVswd4/IQSnrGkvfMhnfkGY/kmp/wiDfiEX89vMWbesZ3eJ/35ZIHumIhNT8xT/FEvvuTI1zRpZQrb1Fv7hKRQ+Ro1URGNE1Jskldo0FyWtZhZC8k+QxAWfVsdi6ZvwVOtWJLQ0fPXCtqdXeSskpxXNJR56DU+xxRqOU+x9SURJSZVLwtF1zIjqeyTW3Zmno29nJwnnITdxKvKdfScC3NWEZYU+LEcS0Na2m5lH2S7uR9GWKEV1qPvei9pASmHf3YvzF0irTasdWGXv1YYCgIThxzmY371ebG+7fjo4P3lxm/35dj3oxnnLFkxYxTWXKmS05YjO+TQ4lQILCjpaNno3u8ppQ0EUNNMb53ht8JWcp1GC1ssRhJE56Gji53o7REGo1c74UuKt+/UHYx8ji2/PPnLd+96sdtNLsZvi8xJjJbbjA2YGySUdXzPUf3njJfXo9llrPFDmMjxsaRgIz7YyNvfWGNMUpZJUlcPOig2WwLdq3h3W2gi4qT2/uuDqYgw+fsMOlsIOs9EZe9IyXDdCjJ8aIk2duECRMmfNqYJiETJvwlwEBENNqXCMiLjyFKMrQ3UNU7ZmW6Er7UEouhwvJEtoRchPYFnVNi2aA5hym1MhdkAy7pavlz72kIHInjq3pMFR3nsuBaGjYkadQxcwqx7Oh4LDf8TniDUyrekw1LHN+czSn3hg/sVU5TsvxOeIMLs+NaGv7bzdv85nbFkVG0c0h50IEQ5a7x/M5kJJOPprx9jNGXCEzfpQnFyekGVeFyK8T88IbAfY7Z0nCmM9bSknKMIg09ko3l3wyv8I/Mmiu2vMEZ1+yopCSqUlFyrRt+Rs+RzDmXOQo8lS3nOr+VGB3sVi+BQIoxPtY5p8y5zp0jDR0XElhq8hOEHOh7rDWteK5kz7HWI+HYSMMFaxSlJEX3Rhxe03mcSZ1vS/KukiKVCKI4sWx0z1q3afqjgZnUdPSIJKP5iS7GxbNR4YQFnfg7xMPmhfIQ03zBmjNS4lchjhvd0mmPinIkC0KeFKX+kuQPSYWPSTbVaY8R4Yj5GFG8o+NSWloNfHejWe6WiMomx0HPsbnAMHmk9tt5/swo1kRs4SmrBuv65MFSgysbvC/y4yLBFxRVS4xZ8heFouyJwWKdxxx0zohJBvWT44Zvvz/nOnouYuQsFCiJRNRYtvRcm3b0Tt31eNx+H0hnRTb5i1Kr4wV13IQJEyZ8aphIyIQJn0PcIR3RIi4tIjV7I0TCyxOSwwV6FDBC19a89cVL/kZ/xv/7zPKncsFcHTGbXp/JLhnC86JXc5pRkMircYUAAbgKyXj8RBqOWPLGrOAr9oSo8MHO87+YH6ddQPhmfIhV4U1Z8ObCESJ8y9b8r7snfEvm/LWjBd/Zpubvb8YzvjQredQsWMfAn9gP+N//yRl/+NoKe3b58VOQgYwMx/tJkxJIaWFNSQxpYbnbLCnLjv2+IkaDEQiaRDC/Xqx4pa95W9ac64xLaUZzd8xnpiPgxHAmKzyBe3HGh/aKmgKV5A1RlIXUvKlneJSN7Pk+7/PX+Or4B7vFZzt4mgKc6ZxjrVOHBMqppunEP5OfMZea1/WEjoBkqZOivBGPec9cc6JDqaFhR5s9Ax2BgMPh8VixzKTiPsccZ0IzSKIiipN7eCINHTEbpANwKkt6Aje6o5aSa3Ys9YRCb1OeGno8HiNCladHhdoxzemBHgEwp2IvPVGULXtKHK/k+96Wp1kuZtlqQ0U5GnPx9UcAACAASURBVLPHt3d+PoOwyHK3gPKcFk9kls9ujeVMCgIQ49CorsQo7LdzYiYSVdFRVHv6dkYIjqJoERcoq/2YqOZz0WXfV4l4FB56R7OvWFYdGgVjAxoFjUJdt3z40QlB4dwW/NP4jA+QUXblRGglsMsG+0NInjq6g2MusGzocNmwfngeJkyYMOHTxkRCJkz4S4DYV3dkVx8n0UoPTAt09ema6skXf8TjH3yLr3/5kg+uT4lBed/cAORlahylPsOCGFLZ3JXs8boiRuVd2fLr7ojXzTFn8xRN6qyiKixKx29fvs5aOj4ya1Za8AVT84WlJUQwNi3yf1PPGJQpvxbuU2D4+rJkVsLxzNF5x59s4U/jFX+wOWNxdvky0TichhyQD42SErQyRgmWt6lXJd/mXOCjD0/Zt5bepymIFcCkJKP7FGi/4oaeY62THEr33JMVgchGWvb0nOsyTS8k8q3wKq0E/i/5ATOp8Oq51DX3OeKUmnflOWiaeJAX51YNRTaSF3p3MdrkK/kAM6lo6fCSaFCZO1VClnfNNS3WmyEal5I5FR/QJt8KiVieyIoKx5km/0at2XchYPIU5SN9nk4rimpkKXPmVBxrzft5EgG30xhyolOBTfG5BMgysUMpmCWVK7bi2dNS4ahkNfpZhv3e6H6c0AwSpYGIHPpEWglYepaxyBOTtMifY/EoFQYnMi7xh9Z0EaVtC8qyp+8KfF/i+5J6tqVr6yy7iuN7JfQlrmiJwVHVW5rdEjC5jX34HCoiekeutW8sX7snfO8icqo1rQRWWnAhe/o8VTMwUltIUyVDIlVDRPFwLgpuCyNTUEKk5xM+/xMmTJjwrxETCZkw4XOIf/Yf/Jd//Lv/4O//0a3EKn0TEz55AvDCz+qTeXuxWrO+OuZ8LiRfuOAwbKTNC9pIQClzelGLZ6YFx1pRGsO1evZ4aiMsS6EulG0rbDuhMBAV/upixaZT/mEIdEQWLk0ZjIU+gDXw+tyxKOFiq/zBVwusUZ5eKlWhrBY9IQhum66wd23NIhjUm1RY+AlInpG83FRBXLg1pJuI+gJVS9ssKIoWV3Q8uSrYdfmU5VPp81NURjixjjbE7G5JaVlbWiLKh3LFinqcjlQ4ShxbaXlFTseekVpKUHhsNlzrBovhSvac6wKjtz0eA/lrxFNpWswPBMQh3OeIa3aspWWlVbpKnuVOWzo20jDDUajlwmxZs2dGhcNxmlPLegJv6T0KtbTZl9JJIjpOTZaDJa/QkSwTMZAkrbsfl/QS+KLeo1I7+lpMfksOdCSlYdkxLjgV7aX28148LZ4Oz4ySJXUyVx9M3e7rEXtpQeMoQ0rJajpKyw5TutbSUonLiWYGi3Ak6Z/DQpKocGENfWcJ0eBc5OJ5RR+EV+9DWfajPMs6T9+lxnlX9IgowRfJnD7foGowtkeMooFsUG/HKQgwTk7EKK+/uiFGYV7P+ep2RdND6+EHO8f7uf29oR8J1qEnRJBxQjWkgiUTu+ZktMBieB9MmDBhwqeMiYRMmPA5hUY7Tjw+dvLxYjHfi0lS3iKup5rf8MG7r3L/xEOqiRgnHpJJCDBq1HsiX9Ul35zN2HulV8tjs+a4PmdZK5tGuG6UXmHu0jRhUcDDlfC7V/c4n1mC3i7wjaRkIFWYlQpbOD3ZoVle4r1werah7wqOteZIS3zn0a6ArkBON3ePbZiGRIG2vE3IyqRLiOO5ERMxtqNvk5wmhuG4k7Tex7SfZhi0aI5NRbiQlKY0TAAAZnmCsKBmOxjJJS0WvxFf4U/kJ8ykYkHNWva8rxfjVfx39Akn8kWOtGIjHQ0+GY0hkZq8neE5ChzHWlPj2ElORmKGVcOOHi+RORWXsseIcMGaFTXnuuCcRZ5CGHakBvhh+zanoiWClU32uiJK8nAstaRQS5QUK7ylzSbyOE45IE12igN/0lmc00gqX7yWNIUpKfhIn7PXhqXMWUlNi6cSh6iM5YrDoroUR1SlEEuvgZlUaDaqz7QYTdqHC/d0rtLr6kQSAc63r3cFhU2+kA+uDMsyGcePTIoFFlFuLo+xNqIqtPsZxt4lvckbUmGtJ3iXpm4wEpBkcg/EYFEVZvMGjcJrZc/9Pv0T/ec/PuLr8wp28CO5Gj0gQ3TzENFruI1aDtx27wAstGCHcE9r/r3/6e9MyVgTJkz41DGRkAkTPqf4RMnVL/O7ZZ+WaS7Q7o7Y7h2XazcufjZ0nGsy+j6R7bhoiygPdcmvlDVnC+X5VjAeznTOrLwlIBEoBNqgSUaigg/w1lGSYF03io/CrLglI42Hi43w1r1I3yfT8Pn9a7y3FEVP8I6FljwyW/b7+u4BfYwsS+8QkxeCAqOky/Uud3zUe4IvWN8s8QenNWZy9OKs5TrLsYJElrk/ZE2LICwp2dCNiVgdPWe6pMFzKksaelo8JyyYUdHjqaWkwHFNSsE61RlG01XvYZowkMEomqYO+DEp6lRnfE8+pJMeK5Yrtuy15Qtyzi43vJ+w4J7OUZKBuRE/hg8YJMcNGxyGjgDIGLUbiJzrkg/livmB9CnJqEIiZ8KY6OUyeXWYMf2rytsxKklyhqWho9VkwrZi2NDi8rvNiBAILLROBYcaU7u7QKcp2neeo2kX1FhuC/wKLAst8jQJzuRgnxXOlzmoQAWRSNtZfuVhn2SELo7lhtYmiV5VJ9I0yKqCdxRVm+RZIRERM/i01OT3bESM4lyPdR2+r+i7Cut82n40VHUaufUR3t33FNx2saTz7vHcpl1ZzHj/UFQYSRcLduJ5Ted3fDITJkyY8GliIiETJvxlwycYscfJSRTIEbVEw+LkI954fcnND89HnTnANzjiqXY8lk0y8QKvxgV/ZXaEFdh3MCvggTr+pr/Pvss+hSJftbZpwbfp0mJ+28Gygh+vPV9Z5pZuA6VRnIVNK6imUjgRZTZrs4zFYmykqlu+JfcojLDd9i/7QIYErKGAcLzfJA+MiXfPjbdoNMRQEnxBs6tZrytKB11IjdbD/gD0MScpqbKioFaLqtJm8/bA1M7jDAzstKOVQIHjqdygrMYW9WHRX0jaxhlLBMMNOzyRIypMXrQPE4vbhWlEcGNq1GBCXlDjibT4FN0qC3a0PNAVAHMtiaK0pAjboYF72HaQJLO7kfZOIlMgUqnDS2TFbJx0DCV6RoUm98bUWtBIz1JTSaKXSK2OYvC2SJqQRFJZ46VusGKxWGpSz0mSGwXAssgm+QtZE1VHr4MgzKQiojTasZAq+yVu/RG9xCxhi+w1sjIWK8ncXRXptXQ2Ym1KfHNOqes+J2Ylj4hzkbLqMnFIL7B1KUbXuZ4Y3OgTGXwjxgbswfvM+4JytkZMJEaD70tElBgsP/jpCW88TKEP7+j2IBVLsXkKkmRpMvpEHDJG9Q5yrbfiCQVmfC0nTJgw4bOAiYRMmPB5xc9Jf9IcGfritOROklZXpEnI9oyowqYZpFdJruJEWKnjRvY81CP2eOY4zuZwvYddnxbqAAtnKB3cO/J4LxirxJD2q3tu2fdKaYUQ4V3d8kY4prSJnFjAGuWDJvC1I8fRsme+SJIV7x3WRNqmoqw6/t3fvQbAuZD8HV0xRvVq59K56N2dc6MvTkGGc9EVxL6i3Z4QvOPqaoGIUjolNkKfj81I2s+gsA+RXpUHtqBVpdPItaYl4YN4a6Q+jzPeNR1ODZeyBeB9ecZOG36HL3MhW65lx0b3oxxrT8srmlrFh44VnxebgzfHqcHnRnS4lc019LyuJxRq8RJHr8DgmzjsnRgkPH1OjHJqaPNUJIqOkh9BmGlBL4EaR9CIyxG8XiJW7Tj9Gfo8FDdGPbf0VBS0BGY4rk2biUsiSUPnisNxJHNe0xN6Is9lw7kuuZY9jaSJ035I9dKWE1nQsB0nJqUUFLhRcjV4URrxOAxWhSc02DhjYQyqwvUu+ULOjhRjlKKIGJPkV2XZj5G7s8Uuk5LBcN7iyj19u6CotqhaYsgN8d7hvWOxWtNR03cFRXnbRaLRUFb78eeuc/z3lx/xd+OrvLPruUfNdfYWCamsMPmObv0xyQeS7g8SMZrimH//wZJ/+iRFF5cyTUImTJjw2cBEQiZM+Dzj5xCQn/s7+ffERH78/S/TtJYfbjoq4+gJWYMOx9bxVjzj6xzzkTbpinm8lVA5k6YhRuDhvY7S3ZIeHwzvPakIeQEPacLwW+URIuAsrGaRm50hKrzNhq9xwnzeYWwgRIcRxRYeCZauLZktdgTvaPa3BXq6rRG3hTZLbg68MMNURFx4iZj4ZonvZnRtzXYzI+aUpKEhfYjnHbANkaCKzYu8QlJs1lzTn9kdnv3HpBJtSVG+UZVzOabRnnf1CYJQUWLF8IE+4xu8ng3X8pKkZpgADClYw/2D92Ewa7e5k2NYoL+4jWEKE/NVdp+Jx7ANg4wxwMPzFWrZSX9bhIewoUMl7cdWGgTDXEvWsucr8Zyd9FiSDKuXwHGsedc858t6bzSlX+kWQTiRBStmBCJr2XOpG2op+UifU1JQcZ+oOk4+bL7iP0wBiizrSnbs29QrmycXbiQnmid6mkVaYK3S9wZrw9hu7n16PePwGZE4tqIPKKptvi8lfqkarPNY53FFkr/FkHtIouC7WZJs2Z6iNFjXUpbH/A0esJpF/vH1Y5ZSjlOcZOy/nXiMr3F+rU5j6n7Z0PFNTvi1X33ErHzAO8+maN4JEyZ8djCRkAkTPof4nf/hv/gjxd0pJvw4DL0hdxAhbpfYxQaA7d6x74S35YrX44o9np30LAqDj/B1jjl1jtfdilUFmzaZtqPCcQ3LWildpHSB03trQjA8eXyMscqsVHwQQhRql3wfR6VQWDhbBs5OG252C9pe+BWWiKTiuEHWIkbHjgURxfdl0uo7fzvhiCZNQe4cY56AvEhIvM33WfabE3yf/BxNU2AkyV6GhnQrdzneOnpWxlFlSU/Mk5ABNfYlEnIpW4r8Z9iK4Yt6j7flAquWQixLcoeHWJ6z5UwXd9rA4S7R6A7ieQ8xEIthcjGQiHFBjsGg9MQ8MUhG7r0MQbA56lYdzcFttxKwQMgSLodhJx1OUzysYDjWmpkWySNCYKklO0qanJY1GNgBPIpgKKVgr+0Y1/tctiPZek+f5D1WnsgNMyoUxePZEllIfYdQLahfPCV04nGaFu5zdTgROo0cGUfjk9TuZlOwXHi8NxSFJ4Qh1wuMUULvMFXapxgtfV9hi+TjsK7N5KNF1TJb3ozPXVR7YjQUZUuzWxJ8kUoQM0FRtdTzPf/+v9kk0vMoTc+G4wmkeOOPzIYjrbiWhmOteS47jrXmdV1g1XBNx++84nBFx7/xGz/ix//n1/nKP/j9yZQ+YcKEzwQmEjJhwucQwadF1ydJjQaMfRgHjzU2t4xnH8Vf+Zv/mMfvfp1/p7nHo12gMEJthNOZsO/B9o6jSihtWpzvfJLsnM2F82NPYZWyDJydX3Py4H0A3v/gtyiJfPXLz/nOD+8Rs7eisLDplPOFcHqSFmTWpJjer5866kLpe4uqGc2+PpQ5RjWVvlkb09XppkrSMhNfkmAN38WF8di1qfDdfJTQ9G2FsZFmV2NE0TzhmJfJ72JNmtwMZ/AVlwhLyAlZRgSfR0Ifp8OPQIeno+chp8nITuRGd9yTFTvSYnZPy4yKJ3rFGYvRiwGMC+0hNtkTx1LCoTci9USkhfyQcDVMDAaPz9DxYkhxue0QwZuNzuPzSRyPZTB476RjpgVRlB09XZZ69URWpJjfY02t6gtKGulRVWoKKk2TtZ30bHSXDOYSCQTmlASZsaflhAVbGt7S+wSJdPQEjVhJ79mWLk2NMDgc9YE53qEUmoRpg2dmmJak6VCKAna5e6X2B+3tl5ZXY8HZcTemsUmeNlgb6boyTeJEMSYgEunb2TjtsC4Z1n03J/h0LopqS/AV89VzjE2vccgt64ef19liTbNbUpgk2frd2SqnsaXn7yJU3vKAmp9ww6nWXOmeWh0bPGdU/Fa54stfvEzvR9tPBGTChAmfKUwkZMKECXcgJmBmu3F6IHXHo0fHvPWg5+Idy0lpmJc5MrcAkURA9lnePlwrtgZKF6nrQFn2nL7yzvgcrz1M3o2Te0/5lTdLvvOTFUaSWf1G4Wge88JOOV56mkvHrFSMKJfXJWdnAsYgRvHe3k5D8mTEFIrv5hTl1UvHp4Pp/jCi2FuCr+ma5cF5UIJ3bLczfDA4G+kx1IWy65J/ReTWE1JbIaqy/yVDyY60opcVXe4M8SQvxwkLvqCnvCvP2bCn0Y5aSh7IyZiIBbcEZPCFDMlIwyRkIEc9MUfTJnnOWMI3TkMSeRnkVl7Sdg8nJh7Nk5IwLuLTtsMdUjT4SgY511wLZhRZStWOJOBaGnw2hlsM7sCUfqw1G2mwGAoce21B4B5HBCIP9YRnsqYVj88JYIO3xeE4Zk6txUjCbD6uQ+JVYFnFMvtf0jFt6XnAjHaIWRah8crTteBsQVn6u++jTEqa3SzJrJxHjBIjBF+iaqiXe2Iob4n9Cwi+ppzd8LMffJMvf/PbNJtzjPUY2+PKHcEXVPMb/pZ8i7ceePreEGJ67xkB+3xJYYSiO+YnkiaXj80Gq4YqnvLWvYLF0Zpqtv3l3pQTJkyY8K8REwmZMGECcDsVMUV7R2f00z/9Pb7/oeWoEmZWEYHOp46MyzayLAz7XjF5UnBcC5suPcZaZT5vqed7xHmk7sBEzl97D+safDfntbd+ynd+8huECK/d85wfCw/Ot5jcJn1+tiOGBbNZYFA3RRWcBJzrceM0464MaVz4RYNGvU3+4paIDFee1Tv6ZkHfVbnXwSFG2W3TRMkYpcv+kU0jdzpMPsnmO0xEPglnWhNVuRahoWMtLU+55sv6gMdyw5I6kQoRXtEjStwd4jFMMW4nHyGThbT4Tk3hNsfpptvyK53jBW4nNIrmJm6h4dYsfUgwIoOs67Zk0OWf+9y4vtSSnoBKknb1BFYkudBz2VJkSrCn5UZTj8oDOaajQhCeyY6ZJiO5w7Cm4VSWYwLX0AAuCMfMaPFc65alzEaZWjK8O2L2SFhNk6IhoneUpuGSKV4dy/xP4UwMhSQyaUmSu9O50nTCvimY1f2YhtY0uY+l8Oz3c0SUew9Sa7yq0HcVrluM77d0uyUGcuqawxYtvpuz3pSEfjaWFsZQEPoK63pCX/F3/q33iMFSlB0hWHabZZYgnvLoGkoRimj4ht7jFVPRxMibs4Jf/eZ7VLNt9qZMmDBhwmcLEwmZMOFzil8kxfokxD7JknxXIxL5R9+bc1Zno64Rtr3iRBCBJ6HHmZKT6laqdLryHDWW42VPWQbq+Z7V6RNk3ox9HMXRJRil6Drwljfu92z3lldfTZML5zzGRmIwWO84O7U4FyiKFIVqsiQmBMt8eZO7Fwp8VyAWjAlJ6hLNHaP9ywcr4C0xlLRNKhVUNXRtST1v6Lq00ByfLwpl9q5ETYtUIJcr3iUdXpVCDI1+8gLwPM4wIjwzyYxtc5zuUFL4QI4pSdMZHacStylWqTwwE4FMNoYJiCGZrkOWaEXR3DR+S0AisMeP0xH/gldkmB64/P/DlCXkxTJ6e99AWAosVtNkopc4luVFlKd6g6LstUEkJTs9Y80HPEMQPtALChwnLDjWFSJH9AS2NDSSShHThCRF9p7rkmuyCTw//56OuVQ4Ndhc5mjhzv4ByZuSHTBnUo5TkyJ7egBeWQkPzlpUwWeZ1mBI3+2KMeWtaR0iylFb4YpuNKq3u6Mk0zKR/XaFdZ7lyeNRmqUxSf++9o2f4bvk/4nBYext5K+qoZptx9tNcJzc29M2C77x9Zan/+xVmhwL/QVX8btf9JzfXzNfbu4QkEPp5YQJEyZ8FjCRkAkTPmf4jf/uv/ojDHeu9H8SRmkSt6QlUnB18YC3f3aWUqqy96F26cp/bQVrUvzuF8uSZSmcLZNUqiwjpYvMjgNV5Tk5u+Hk/rvJm+HtWP43+E3EBTQavvZrP+L544e4MUGox/cFcEs4nIsUpR9lV5C09PX8BlWLdR3BH48TESm7sQUduNMFMjwv3uK7BaGvxu4H1bT9ZlffmbxYo7TK+JghGcuaJE0bMJIAEfQXTEMgTUSesaOk4Bj4UK4QTdORp3rDWzy4I8EaFtuHnQ9xJCaHEw+4lD21FneeL5GZ24X24T7rwbQkbU8Obkv3DwRkKBVUlZGE2BwZG4iUOAo17CRS4XhdT0DgsSaPQmoczw3nObkqEDEi7Oi4loZ7Ok/TESo6+mRkV7hkw5oGwwnzbEIf0rAW2YcywBwcc6kOl/dzjuNMylE+6JBxmmdEmFnhdJUI1GGqbQiGmD8rNk/IRBQxsNvWWFshoiyO1uncRgvR8rOf3ef1165YnYaRUIQ8Xevammq2u309gjtI3TKopO8hJplXOVvjfIlG4Wuvt9y/KfnBU8PCGTpv2G5mHJ1cjgQkSbw6JkyYMOGzhImETJjwGcPv/c//+R+Fbbr6bYoW9Q7fpav0rtzRt6lc7uMkFqoWxb5c4f1zEEPx0vauLpd8+7FSG6GPkRsPR8GwKCR1eiCcLZRdK6xmSlVGqiLgnFKWnsVyT1l1nDx8O0mwyCTAc6c4EFI7exmvmC2O6NuKarbHuh5jAq3Ok7a+7lMresiLPxQxmmNNPTEAJk1AIpbgHWG/zP0NPSYv8O4kYkWh258Q+pK+r4jBYp2n2c0oip6L50cpljdC5w2li3SdyWQlTUEGMgZp4dpHHRe1u3hIB34xktTI0bEdJyIAj+SKBRXHOhsX+BZz5/dcNqcfltSl6YinkLStoVUb7pIOi4zyrWFacuf9kScmkKYtVk2SX0mWc8nwGGV+QHhSW3siJZeyz8dnUU1SMiuWoUPEYbFiUE2Tl7227GkpxI6N7Q7DSisuZIvDYTG8w1NOWbKlHc/InOpOEECm2DnXDGpNEcOnJAJiRRBVCkkkpDSpFHNVJuI5FFIam8hnaksX1juXiGmf5HnzSmm7OTEI673l61+GsurGycgPP7KcHM047eaUs/U4+bCuJfij9BpZP34Ox8+lSWRkuFhgM5mo5tcYu+Crv/ZdtjfnfL0riMEyX6356Y/eIEaL72YU9RZju5c6gSZMmDDh08ZEQiZM+Izg9/63//SPkn8hLxptd2exnhbb5V/Y8w9xvkayAVaVD33PK7ZkZdOV/22vqdfDK6crz7yylEWkKCKzuqcoAsujDUXZMls9SxKsoYtjmMwcEKTDac3RvQ/4s//nt3ntVTg+f4SYiPcFqkJVN2M/w4ChZ2E4N8FbjA3EaJOfY32GtQFXNpSz3e25NJrieKOh3S+SlKu7XTzvdhVlaWi7tN9R4fmNY1lH9p1JXpdUAQLcdoVEPVisq34iAamwtB/TFzIsnO9zzKVs2WtLR0+nPQ0dViynOhsX00BOthIMFp+TpQYSoShzKowmszmSiMhhh8hAVg5jfeWAlHDgGRke4xDCwaQEoMUfTE0S2WjxtBJwCO/qE2yO8j3c7+Es9fix2yOiFGIJGnlfL/gKr2Qi07LUYz6Uq7H7w0iKJR7O30DSXowpHnwsnXjQAhXYqGepDiEVbw5n1QB1kYjFkNomouz3jtIFnl9XqTzzSgDhpo9EVU5KS2FSp82TNhB+fMKXXt9R18lj89oxzBcN++0Jy5PHuDKRYlu0WOcx1mNdi+9md6J9jU2/H0ORJyjJJzJOMNVQ1VusrTAmUNRbfvU3/nyUc7ly+4u7gSZMmDDhU8C/nGh8woQJ/8rRPz8nrFd3iMdAOgaT9dC+/ElG019kQFW1d74+/jGG19+44EHl+DPzlNcWhkUpHFcpynXuhNoJTWuZVYGqDCzmHUURaNuC1elj+q5KCx/RNHnIMiwp+5djgcfCwER+fP45hiKXxIVRfvUiyqoZ/9/YHmNSbG/6fUsI9mVJ2uAD6ZNsJ3g3Nl4PZGTwgkRNErSqUEJMC9MQ0wTESJJhmYP17vBMTfz4UVR1Z4Zx+Ht3F82nLDiSeZI/ZWqzpTmI1E2L7UOfxtDbESV3eByUDN4eujL8N+BFuhQkjo978Xafn+NFuDzxGBK2Bi9JkEib35MdPV49EcWJu7NflpSSNZAPSL0pw/YKLHMqWkIiVpnADO3xhxjTwXK7+3CMmtO9dtLT4tnhx2Pps2wuqrLNo619r2z3DtVkNH/7wvLoWc2TG8M7zyybPrLpI02MBOCyC2x75Yf7hqDKn1/2XN1U/OhnR2w2FV/90iVV3RKDpd2epNfdenw359t/9nqSW/mKrp2NEz7rWqxr8lfuEIlmJCYicZRUVvObsaPEdzNCX+HK7JcxYSIiEyZM+MxhIiETJnyG4Ls5UncY2xH62Z37Yvglygfz/Yck4+eRDlX7EnERiaxOH/Oo9URR/vSm4TubtNhflEIfUiv69c5wcV0wm/VYGwnB0DQWW+w5efAzbLU72OjdhauYeEtGcjmgesdXfuUxR6dXxFDkxVaKPbUukTBrI875cT+t6/L2kscjGdIHI65ibRh9INqVYJSwXxL6Gd3+iJjLGvvO0bUl2+3tObcmlSxWZcCaW0Jihw7EfJV8lGUpdDFNQz5OiNUSmWM/Vil32L0xYEmNy8PqHs+elp307HKvxU76kaQMcqiBzPjc52H0dtowTEii3BKU4b7hezwgPS9OR4ZtxbytIl9pH2KDy5xedbidYd/O5ZhTWXFslpyZY2zufR/PdTbQDyWFg8m/lIJrSd0XRlNscEtPORQ8Yg/EYrx0LIc/QzLeb6RjIx0GYYun09SPHkmSOiFN/LZB+ehauF4XXN0U3PSRHz5T9j08bwOtKusYaDSyjp5djDzznh+ba55rRyHCexeWn10lGVc1aynKFpGYQxTSxCP0JW0vdO2M/eYoEWnbHxjTh89vmo4cfnbExPEChUZz4AHpx+JDjP6FTlAnTJgw4V8WEwmZMOEzgm//4d/74/oL74K9Jsjm1gAAIABJREFU7bDQaMfyvIEwHJKGZDi97SB4kVD8ItJyiGHbYgK22jE3aWH4nmzY4GkDVA4+7Hq+1+7451cNP72KGBNZHm3Y7wuqMoBRZN4iiz1oTqYKFgqfyIgL6WuYbrhA8DWqluPz90ZiYWyPSCoeHMjCEGEKYJ3HFm0279osXfGYvEArqz3lbE15fIF2ZXqOpqJvF3T7FV07I0ZL6B27Xc12MxunMABlGfAhybI2jXC1E/qQihN9TF86SHZIBOTnYVgMz/nFr4knsmY/Gs49gaAxRfmyZy2JFL44lTAqd70fB4TjxccPt3uJY7/HMMUYt5cnES5H49qD7xYzyrusmpwzdfscESjVMVPHmS55RY95yClf0nNOZEEkYrFjY/xhulghljKH8Q73e0mRvw09x5r9FLmc8OOODxinMAPSNCT1vW+kpx9/znK6vA/rEIiqXHWRxzeGR9dmvO+6DzSqNDHyTDt2eJ7QsKbnx6zZ0vFIdhQiPG0CtRE2W8d+O6fZJflf8ElaFUOBsZ6/9rspgjd4h+8LdutTuv2K4KtMyO1IyofJ3uFnPX1uh+lJg5iYihKNgomjp2zChAkTPkuYSMiECZ8h/JN/+7/5Y4KBbEY9XGADHzvRMLa7vRr6/4N0vAhVmz0aaSrxt3/7BkH4Fie8IjXrPvJoo3REdnie0fKFZTJqr04f473h/sNnqaHchkRA4NaIfiipGhbGY0RukboRfJ008kWbPTBuXJylfcxXhw8Ssg7lVkW9yc3Viiv3uHqTDejHxFDQt4l89H1F8I6+K9jvb9OURHT0mewbx+Xa8ezG4UMiG7suTTyEJMsKmr6UW6tLzFfWBzQEmoMSweZj/CDja4Cyz0Tjua5H4uLy5KDLEiJPxKoZ06iGZKqhQ2O4bViUDxMKp2YkKYfekQFG5SUiMp6bg+0OcqgoisskZZCDHT5fIixpv+qcljXs60wqCrE4Sab0of18wOF06Hk2tj+XLT0he03cS/sPsKO9c9uLRAwYRXHzPJMZpiAKBMjTrPRa3nTKulf6TFI6VYIqu/y6PpId16blhp6nclsK2Gik0eShev+5Zb2uublesLnJkcsqhL7E9xU3l8f89KcP8d4Ro6VvK/quxHc1IcsGB4P6UGYoeWISg0NMTGRFDTGUibw7D1GI7SxPViY51oQJEz5bmIzpEyZ81mAjUnp0Yz62afnwCmgyqv6ri94UiYgJ7Nf3ef58zjdDwfvs2YnHBGGP5/eX55S2JkR485WWEAzGdjx8eMXJl35wV3o1EASj0LtEUABUkjncW0I/G2NIx8JE24+LqgFGUhu6LSPGRuzBVWHVJEUxWYJSVg1ufjM+vy3acTG3384pq+42jveAMTgX8N7SNJarjSUq+HBrPpfsA+nyYQzJWKM5nURIEinJZCaTjmtpeajz8ecXMXgcrknpT2ey4pne5Kv92SeT06xcXkQPhCHysofDaEq7GmRaY0/IwfdDsjEkZTEEFOTbe2KuGBxSsMKt70MT6flkG/7teajU0UvgiaxZs6eixL9gKv84POUmReZSjoWKT7k5iC0Oee9AiezomB/E9L647UOfzY5AQeo0CQrd4DPRiCF1jPR+KHtMBZRDCeUlLb1Enss+GerNLdlZ0+Kj4iRL9PqDaOdg8oRP8b7g+vkp3/vpEe/cBB4+cBhR9vtqlBWGkMzpSZqYppUxlHniEeib1fg5SNPDkC5WeEcMJcFXGNvzz//u3//jT3yRJkyYMOFTwDQJmTDhM4Z/+rf/6z8mykhAjO1GU+ohDq9s/jJ+kRcxyK8Ofy9NVTo2N0e8/bjkiILv2Me8bZ7xM3PJO+YZ397saHxqNzdGsTYiZcf9X/t2IiAqWYY1EJA8GYmCdgV4m77f2feX/xRpNIQ8BUnkSMfph7EB6z7+6m6SonTI0KTeJT188GV6rnjbB1LVLWWVI4RzT8j1uqRp7Wg+7/MUZEBQ2PrbG/qoYzQvQKsfb96+luYlAnIpLZfS3jFZt9pTYClTcC0FbkyMmlOOC+rh+QaTOjA2pg8yqUItTg/6RA6Ix6H/Q9FRklXkycWwyH/R4B4PnrfIXo4XEUkTksP96yUQUHak891ylzwP5vT0XOaFHhTlht24H8MkZvS7fAKB+ST02avSk832mszpvSo7Dazx9Ko0GtlrpNFIP3wR2R28jlFSd8pabj+jgwxMELwqryzyJMqF9HkxSp/fl9fXNY+36Tg+/GjFfl/RdQVdW95ORdoF7e44bdP5ceKB0VGSaWw/knCRgO/mIzn/9h/+vYmATJgw4TOHiYRMmPAZhHYFYkKWI5UjETkkI7d9AuUvRUBe1JC/+P/DFVaMcnFxxJ9tGr5nLvFEGnpWWvGb4TUupWPba54SpDQr7UoIB39OBm9FcTDJqVKiz1iQ+EL88EvnIJtxjX3BeKyCRsmFbi9PFQ63pd7R7k7Zrc9omxldW1NW3ZiC5bsUATxgvy+5XDt8SF0Rw6QjJSRlCVYEH2/TlLqoNAcspdOXF+UAx1qP/3+dyceF2XJhDiQ8dBQ4rnTDFds7i+tBhiXZJzFIsYAxmcpxW0J4p1jwxcjaTCwOs7oG0/kwWRlgDwji8PhhEZ/M53pnGtPnScnhhMRiaPF4icwpCZqOYzCuDwlYbpRv3faDWAxVJmQvGvFTOtjdf8Y0n6OPIyaHsqyeSI3N52wgWrDLyVlNjhe4NbhDQ6QhsCG9lzfyyVNIg6TJCfClV/cAacq2L+j29egNOTpqeDAXCiN873EqQmwaS9/fChX6rsT7JFn0TZJzBV+Bidhin75c8gn5bp4mJiZy8eEb/Iv/8D+bCMiECRM+k5jkWBMmfEYxyIdicGmRYfb559Wdxw2xvT8PPy+695DABF9hqx0/+qDkX9h3WWpJRUE1moQND7Rm7oSbreVH7wmdGv5W/5ucnG5om5KjkzWnb/4wkYyYJyImCZWk7PMO6UhUho4P1aTO12gYvNvW+twcXUIcGqQFU7x8PGJiWpS5luAr1Ft8N6drZ/iuwNg4ekuiCn5f0bblWIDovaHtDNYoTS90XvCBcTk+SK76qDiTignToj1h/0Isb/vChOBUayLKtbQ8lz0babB54Q2JBBRYainptMdrwImlU08pjpqSQKSmHKcS/uA5Dqcaw2I7EInyss/jUKJ0ONk4/PnjGi8PZVe3XSK3jx72wXC3q6PIL+hMk5RsK6kD5ePgMoUZfm8gG0OC1u2+HO5fDi7I53JHy5J6nDC9WMA4oCFQZdmVIU0uhslUVKXC0hM5puQZDb3EcR9aCXemHy+iJVJh6DTy7uNkpD8/7lktO3a7moVR9ruaGA2v3euBgnfXgfc+WrDeC68Fg3OR2SJNgKz9/9h71xhJsvy67/e/Nx6ZWVXd1T090z07+yT3weWKpHZJrUxyaVGWDIOiIcOEJAuSYcrQJwEGBNuAAAOWRwPLEiCDMvzFFAwYFgxZEGiJ+iJbtgRZlCAZIHdFUjRlLne5O9ydndl59FRXdWVlZjzu/fvDfcTNrOrZ0XNnRnEahcrJjIyMiKrpvifO/5wzMuyOGMeaxeoxVbNBKhfGGgFZdNBXjP0yJ2NdXBw98fhmzJgx49uNmYTMmPEORjJnJ5RRm/806scUh3vzNsnw3izP2Z7f57z3+c77sU6f+cv2mzytJ+w2t2k2hl80r/OaecxTL36SH1rV7HY1x65oJS8Xf16gHidDeZc6UPq9cbKUGBR6Pxq8N2F8CtlLxwrt1ZaqmaKApXLYo0vMrkf7JiYLSRxLGUnib90MPHrz9t516HrD6AxGoB+FvhBnfKGKDKpYEYZoOh5UsQQjcsIOtzd6Vd6VfyQ7RvGcc8WChlOO2BAWjcn3YLGMuDiSZbBYjlhkxeDQQD4Wo0nlyFVazKcCv8MIXYsJ41qi1BoW40kZmMhJjK898J54UWz8+dq4lSOkWIkKJhMWT6sVOxlY0bAARE95SR6ikQyVDfEwkZ0xxvyW53oYyZvOxyZvB44rOla0+ZzLJvj0eBn/+dvi6KO6ssByJhsGCb/3aUTO+yNeMZdYhEFuVroOcUHPMywYVPn1RyOVCKOr2XWW+09vWF+u6LqKl15b8PTpwJ1jz9cv4dceegZVzjvL/Wd8JOGGoW/zWGIiGmmsUb1FfOgCEeN55cWPs163bHbzP/EzZsx452L+G2rGjHcYvv8v/6nnUwOeGAeuyp0hT1I9Dr0dh7hW2PekbZqRx+d3GHy4K58WqmmxeKILLmTHL9gNt3VBS8WKhk8+N/KBT/0y0vaTCb1MyPImjGaNFi1SslKB2pOOXUyIAFbr8G7/HLw3wZhfd1P6llGkGmC09Fd3GGMKlojH+9TcHgzui8XAdtvE66cMg0EMe50gMCVgiYDziioQDeqDKr0G4zZMqklXLJzLhX/YJvw5Ycm5XlGLZUOfl/hl+V5SQWosK232CMKhqRymcaPDZCzPRISMSh7TSkpJdeDJuSkha7zB5zKIo1Kzp5AMOJCQxFW2ru9k5LZfsJaeZdQ70ihXcxDvm44tnJvHRwqUsrnS4/S9vAbpPWXLe5kI5iU8P8ZxLIdyLh2DOFZaZ4qzlj57azpx14z83wo7CTJaugKjKo+2ylVvcP4II4pX4WwD/VhHpc0xRpL7zFFhpo/Hf3lxi5PbawZaWqC7fIr26Cxs0zdsL58On72t+bWXWi56z29520c8Y8aMGf9qMZOQGTPeQfiBv/Inn8cExaNMdDrE4aL98L9LdePttKhDSNbRXcvqeM13nD7FLz5a8NBcUWtoyxYVWiqcKksqHsmWz7r38Z2c8qnf+qtBbXEWkiLgbIgaHi14wSy78LgLRYR5NKuf/hoq0668m8zr6Q5weBxKCJ2z+yrR0GIXfX7shgYXTe/qBVHdU1KWRxt2uzoqKkJVhXjeo6UClkdXsteGrpGMVGZyIfTqgzcmLk+flPL0KPd6TEThhCWdjJzrVV5A96yxYjJxuCUrFtTUVNSkVnItVIbrOOz5OFw47yVmEbK8jEhUL66fQxik06wgjOJz1O8gDpV9T4iL1KJmgRPPMippC63yAr/0nRwma5Vm9HztC0KSiAhM41cQUrKOWAB1fO2tzeobCUlbO0YeS4eT4H1yxe9I+FkpmyeMjr0VBhyXDNnoX4vQ+5Cw9ZU3Q8JaY+DKOa7GoKwJIYWrBu7eGhmGioUavvSl+3zw/Y+5uFhycnuNiKe7OmV7dZxDHWzV8eVf/yCvn1fUFu6s4Ovbt/5/f8aMGTO+nZiN6TNmvIPwhT/4X78A7JGPMJp03QB7E7koywwPiw2fhLSNavBQHN1+gx/49EtxUdZzKVvW7KgwrKXnI/6U7/JP8R3+Lj/41IJ/69mG6s4ZGEX7Kvg94vhXSsPK41kmej5Gm03rSQ2xVVd0lYRUH/XBqAuENnQTUrFEfC4lpHJ754E3uKHNi7NEPEoCkghNVbnMmSrr6XrLZmdxPsbsaviycT2bCgnLYsLDKF6P0hYRuhBIyJlsOTPT6FjHSBV9DqkrY8QxqKORipW03NdbLIjpXnE0ae8Of/wu7Mfx5q/Ycn6oxpSPUwrVZEyP1+MghSt9Ts8QigPF4XAMjHSM9AwMkY7VVLHdfKTRCokq2iCOCsOOadatiqrI21EZDk3oCQtqFjSsaFnRcsJy7/XUFZK9MhLO+FI6tjIRjyeNWl1KN41zoU/0mAB7253TM8QraoHOezbOs3OeK++4cp4LP7L1no0P42yG8LvX94ZXXj3GO8OvvKZ8/Ru3uNraSJoNm/UtvLO8/sqzfOnXPsL26pRHlxVffez4u29e8c1L5Yd/9vfOpvQZM2a8YzErITNmvMOg3mYV5EmpUeX3w9feDvG4+XMNpg53fG89/XVW+t0g8H3uWb5oH7Jh4GPuLp9eHuMVPnfS8rGPnLFY7fb7QIyGlax1QeVIhKRoI9exQrxBdy3eNfu+joPjN8bjfShutFU4PjE+L0elcmhfBWNugdS2HrZXjJ3m66frJdgq7H+zqzi/MjkBC2Lnh8LOhTGsTjWv0svlarhfPi2iF9hsTH8ku6wklAvtx2y40qCQ3GLFkoZHcsXIyBGLQCwOVIEcwcs+ebgJgQyYa/fwn7TYLz0eCRWSx7ASoQlm+CH7OML+3F5SVzq2GksvI7Xa7M/ocGxlCESFkTZGEKfRv0OksavpWvtrZOSYRTbpl9ftraJ7PcqOkeZtNNinMa60z8Pyw0z+DoogAVbxn9lUflhHP1GF0KunRvIA3pkfMMAtKtY7y1cvHM89u+JWZXjjUri9VIxxeGcR8Wy2Ky4vW77yas3F+v082sCDhYXdgl/t13zsW57ZjBkzZnz7MJOQGTPeYfhHf+hPvfADf+VPPp/GkVIhYTJwvxXeDgFJ5OUwprdabPCu5vLsAWcP7zCwpaXmh06OObmseciOf+eZI1SV28cj73//m9x99qvI8TbuJDaFD9UUv5va0tOXUaRyyCq8xxxvkLHDb1eM/Wo6nqSGFGQjHa0ba5p2i1gfthttVkMwHrzB1l0wuPuyS8WwWO0YxxoRjxvr2I4uqIfHm0BAYhcdXqEbg0dkPXo8sPMejMGr5m0gLEJXcTFdtqN7lDPZ5g6OpGJs6bnUDQaDw1FTcVsXiASlYaUtXpRBPLUa2rhQfiiX3NNjRhS5gYAcKh6eKVY3LZSHqKdUNyz4S5UFUidHuEYpDauKBMMCNRVb+hita/N1sGqo4/EMBD+FQWi0ygpbOu+SaNniOMpz8k9QQBI2dDTUpDLHt4ONBHrW65P/n8kk76DNPfl2Wq3wErYx2QMDbYwXrjB0eDr1HMfWe1WlU08b93nOQKOGCuF12bLUihWWXzsfWYjwG1+7hRF4uPWcroIK8vJL9zg56bi8bHnxtYbHg+dLb/Q0GI6N5cHC8v1/6Q/MKsiMGTPe0ZhJyIwZ72Cku/ZlKtZN+KdVP9QbMHFky3i8q7FHa372Z36U2sBXNj2/aV/mnh7z85ehx+L7m1ucrBwiynd+7BscP/jatPi3U++H9lVQRrxMr0MsMCQQhXYMZKUekbZnvLiX/SBip/EwY0d8XC5nFUT8lPYF+O0Ke3K51ztSXru8nbPR+zFkhaSqPLtdIHspDctFM7HzcDV6RELy1VrD4t37wySnfSQFxKM8ki4vUiEs6jd0XNFlAlLG7S41DCetqEGnCN7yjv6ZXIVxI51GphLKO/YQjOMGCV0fMnVy7L3nYBRqakZnz3tio08lRQUPjHCgIgQDfSADOxlZaMVORlYaekISsdrSURYapvfe5Ad5EpIiEtKwdgUJua6AHCZjJZQjWiXKzy8VlrKhvjT6IyaTn5aKRkOYgEW4oA/HpQtaDFcxOe2ONhiBLSPn4qjVxBSzEHjg8axMzW88cgyqXKnj6U3L+vExL77W8OqLlu84tXxlPeAi+TIIr/ueT/+ln5gJyIwZM97xmEnIjBnvQIz9fr6/G9snEo3S0/F2yUi5iM9m8F3D3+hfCXe6jbKkoaXiS+YMi3A5DHxSjqgqpYo9BLpZIKtd3JFcIwIZ3oQv64JnpEzr6hps1SHGMcQituwJ0TB2km6Ee2cx1mWVRL1h7FdYLovPEsZ+dU01Ci3VLSaSnKbd0nWxNyQet48EBMII1qjK1nte0k02Mn9YTwildmE/y2IsqRzBAjgzm7ygTQRkzXY6JoQTWaF4djLmWNj02mFZ4V095ku8zIkss4cj4SafQtmunn4zUmLVdFzXjejm4HGK7nV4jIayxK30uUAwjWQda5P3u6PHiwZXhChLrRhw7OgZ1FHLk8egDpPFvhWSb6Y857dCIiOJtCXDvR6QoGslj/H51GWSflZpjC0RkLRtKnWEENl7i5o1I1vG0EFSxDoPMo2jbXHBs+JMjIMOCWwKvPTKCV7ha37L47OGR9F3cocGh+N1mX6/ZsyYMeOdjJmEzJjxDsTbidSF6wrITaNWh88/iaiM/RE/yH3+lnydH3bP8SvmIQOOp/WIS+n4ujnn7PEpT5/2nL3+gObiLk+9/8tTO3p5zCkyt0QiPun5eAda+wq7XIdRrYORLMHjqbMBI6Rmsb+NuL3P0zGQMWNHNHpJUrqWGyu8Cy3stuqi1yQsIJsqqCFeg7qxHj2jKmt1vGgfsaVnRct9F45xh2MRCYhB2OLYxVGnREJgGi1SlC0hYKDXEYOwlGXsArEMuNwsXhft4ImM5O4MHelkZEm9R0LCZT9Mvgpw7KdRpcVuisF9Owv+rFKIxiStsP2CpliSSx7pWtDk/pMNHQgstWITr0EypJfHUWNvVH/S+ZQpWel72m4bP6tnyB0hqQPlJrXj8LqV51kWI5qCbCQIQv2EWOleghnf4TEyjcd16ngM2ai+ZmAnLuQ4HJCZTQwteJ0dy/g5FsNVr7AWnIcWw6+bc3YMjChbDb+XZ7JhxowZM94NmEnIjBnvUnyrFvS36g4pVZOJoHh+1yd6hi9+gI/eqvi1taVnpI+L41Nd8n+8ecEHz4745Nkpn/nebyKLPhCA0U7EyZvrBKTEgVoiVVA28ELVbBj7VU7GSsb8pNzYasC7aMyPasjeflPSVt3lMbO67XJTelJBIKhLbTugvsarcHrk6AbD4OBiOzVc7GICFMAl21xsl5BSsdLzj2KLdiINFsMFYWHokh9DLA01K5ps8B4Y2UUbeaWGQcKoVnhHXKyL52m5w0AgIeWY194lPhhnSnf5E1FIi3NXHGeKD67UUP5muRv2D8TYYMsi3vkfiuQug1CrYSUtGzr6eG6tVKx1i5V9EpSubyIxJQlIJncbDdw3lRVeMZG+dM0AKrGsKJLmmMbVSsKWXgs6mN/b19QBozdeB5g8IIlAGlKvisvnoAKdulyd06lLPHwvEKCOBLHFsovv3eEYcbzU9xwNll6Vr5lLekae9Sc8NBvOowLydtWjGTNmzPh2YyYhM2a8A/GP/8ifeAHg+/7in3s+PZeIw9sduXpSelYeZSpeT4rB6AyfuF3xhfMdgwnL307Cwqpj5FfsK9xyH8JrxZ3nfgPdNaHvwyj0tlA7/GRKh+vekNFCG3Ob6hEZKhSDWW6gXxUkalJXrHW5QT4kZYXFvo2jYfnzjGLaLboN+6jrDvWp/FEziRn6lqoaEVNjvLJcOtrGMzjhcldxUhkuR89CLTUVK7VsZdhb4O+4/rO4OEjDCgpIx6Aujy49JSes4tLV4Rnx1FR0EkeaZFrMO6ZULIC7HE9mb53Uh5tQLrIT+RjFX0vqSot8RRllUhfSuFKtwQ+SqJkhmLIXhbEcJhO8ITSLL6IR/RFrALbaMeIQFXq53r3RE0be6ujsKNWb0ux/U+RweR02dDgNSkRLXSzM5QbisX+93g4qNXlMbUmV078QskKSvDUWkz+nnJh7kjenxuBQVlgWGGoxeRxrzcilhnEug3CkLc/Jkq2OLLXi9/3sfzB7QWbMmPGuwdwTMmPGuwTfinwcpl19y5LCYnwqKQq//o2WdTctxo615VK6MFoSF00Dyq9fjPyt//1H+OqvfhrdTfP4UrlMRC5f/dD0YaX6IRr6Q4yHrs7HImbfcJ6fj+eSVBFjx9ATYsdsqMdo6B2pHLIMpMS7inpxia17qnqgqqdFr6rknpGmGTBWsUapqnj3XWBRCbdry4lUfMidYhE+4G9fu45l8V5SQUaZ1JKOAcGEfg1iBwgtK61ZxA6NChOMzGoY8XRFj0ZYxAYk30er1d4CvRw5usmUfUhS3A1qQjmmtWdUL1bOIQVqIiRpuwFPFclDGl9Kn7vQitscIQhLaamweUxJUZ7m9rRIL65lSpcK3orxiUSrHJcqiYiVkGJ2wYZ1oZTcRGAShiIooI4KVfm7X6mh0mBCTz+X9DMEcidK+pxMAPX6V/q9qSJJsfF8k8ncirCQUHLYYlhhuUNDjXCLmjt+wYf1BIvwjC65z+LG6zNjxowZ71TYn/zJn/zRb/dBzJgx42a89lt/+O89+OV/+KNgYpxsQFAxzA1fCfv/HQhJKu/T/F29ARVEFDGe03bJ3SPl8w9HtjIiEpZUPZ7HsqWm5hvmgt+QRzzcCWdvrPjeD3VU7QZGiyx6xITm8e7yDnXVI4TuEKnH4ngAb9GuQQTEetTZcCxKiNe1Lvy38TlKNxMVleD5UEt1tA5qSuUCGXEGxgrXHVGdPMbaHh0bjA0LTFuNgNC0W4wNJYXGKH0f2tNHZ+gHg9OQlOU1dj2o5R7L3IA94HkkO1bUnEnHVsY8EhNGnYIOsqOno8fhWUnLHY5ZapPv+NdYKgxtdklIJibJKu3i9z13goQ76ypMikh8cW95XXASi9kbQ0oL7EQoEoWYSg+nJX5a5jcxxSqNDYXsMqWJqkh6r437r7EsqLFScUTLUhoMhlOOWVBzR1dsZaBnLM5OGOOx+TziZVCUCpuvxP7xTaQonQcCHh9b3KEhkIREwpLnRotrlvw3Jj+vQemQ/c9bRoUlOVuk2IcykaOyPLLG4lBaLBaDxXC72E/4PQiRzI0IlUg8bsPCGBoRVIX3VS2tVtytKmoRlmL57X9tLiacMWPGuwvzONaMGe8ClKrGt/J73IRy2719xYWkeoOK4X0f/xVef/GT/LJ9k/fpKRv6nIy0Y+BD+hQdI6/J4zB2pCdhLCp1dRTt6CcPvoa7OgEbFRLIsbzqDewqaCIxqcfwXi+Y5QYbvR2K2UvCMnbA2JGhO8K7KpxL4fPAGxgqxs2tGEOsyKLH7oJCkd5vuiOMHVidnLG5vIsYRS9CIlltlVsrz/bC4Lxyqzb4XnkutnCPGhaoG8ZsQL+QHa5QP1xc+EIYDeo0lPMtaWmoGcSBWmqIUa6GWm1sIZ8W3wlp9Aemu/1JoRjF7xGNMqb3MEHLxVGrdMUS0TlM4ZqISFIl0nbpfRNJSqWEoRPEk7pBllrnz/MIdzS1mLespWeldf68QzO6yxQu+UJsvi4204T9kbJk3y+O54EZAAAgAElEQVSRlB3BsGPgmEXwvnCzEX9SP0q/iMnXNWFBTa1mb/ws+Eem0bEQjbyvithI4lJHSxi5sjTFfpqoEtXxuwVqExUqVY6N5VYjLH2F88pwQ1TzjBkzZrwbMI9jzZjxDkfyh/zzQr2Z4njVXiMxqpZhfcrrr93OXoWWijfkMff9Mb/TfYQL2fGD3Odz7kP84Vvv48c/NdCcvhl2MFq0r9G+Cp/Th5JFkkIRj4Ghik3qobiQygX1IpUZGo+tt9h6O7XDF6NjZfSuseO+9wRgGdrmx6GNnSWCrXZTD4lxVE3Yd9Vcxd6QMIrVNA5jlW7YX9QtrGEpUxldiTPZ4qSIWsXlRXTHyKgOg2EhIfIY0qjRlLCk6NTpwWTaTl/hPXEc6gk/X59GpaK5HK4byss0LEijXtNC+XBEKZu4C6IxeTYmWEyM7o2ECssoPnd/JMWnjalRrQZScSU9D2XNGMfPDntC0vGecpQN6omepGMO5zkt4m1szLDxz2G7elI5kvKT3leOaR1el8Nr2Wq6FuTRtGTGT8pWrSYrHCsqLGGs6ig+rhFWMaZ4ZSyVCAtjqKICAmBE8uha+m+bSIqBxgqtERZ2JiEzZsx492FWQmbMeBfgn4WIfO///FPP3/R8MqcD1zwYAPfuXfLHH32MX3rU8UvmDX6H+wi/45kwb/6Z/oN8z8fPUDU8+4nPo7sWHS06VrhxQdVcAQY8uGGJbTeFYTzO/ff19GFewCraHZQxVg68oVmeo95GwrBB1eKGFhFP1WwQEz0orkK9IM0IXnBjwzjUuO0xxvZINWLciJjkLwE3tGCUqtnR7ZZUUa1pKkc/NlQGjhqJrehhFGc7hsXyjkAYnHjWBCP6ipAEtYs0pIljNsEL0eQo3nR/Py+AVRgkDQSZfId/WoinpvK0SE6XLnZdFIvlKdOLnIAV3hOISScjREWhXFwfLupTb4bDgxgatfmYmmJRX0eyWsYCV1EhGMRF4uD24mwbqjx6ZTH0RTLWISGykUwstOZSwohWak/PsbZJzUMhtpKPRZFiUlMEwyaWJCZYDMcHXopAfEw28SvKgiZfo/BzTOe9rwq5YtRtFcfTHKGUUeP4VSAj6UiFWsgDZnsDlZFsnDaTZjN6OGlh2SgaxyhVhbM5lXfGjBnvQswkZMaM9yh+5T/+z68Rl+/5n/67569F2x7gqQcv87s++k+w/9eP8MVHFZ9YLDlajjy8qPjdv/vnkUWHtD16uUKMQ/sG7xpEPN41GNNNOzOhSV0x0BsYLa5bBXKS1AtX3FX3EprcK4eOhMc4xBukGdBdE/wiDEg1TqlbSSkxGtQPQhKWqgnkaHGB9dGvEdvnjR0Yd8eBlBhHXTuGwWIrz73bA6Mz7DrDthe0C2b2Ctkbb+pi9GwXW7HXbOPi2rKiQTA0EshHhcljR+lO+pjHo3wcgXJ5sVsmYh26fcrIgbJwz0DuPTl8/TAR661gEYgxwQnJ3wGTib1UE8pm+PzjzN6KQKSc+BABLJMB/ogFO3pqSrP9fkxuGnWzWJrowtjFUOA6Lu6n6+bzct4VVyqM0PU3nu8xCxyhLLDVKiSCYXJ3Szm2ttQKjeQCQlRxwhDJqcHk16fzIOsyRzKZ89PPti4Uj/To3sJwaxH8Wy7+/3Ky9JiD3pOTJW/zJztjxowZ7xzMJGTGjH+N8P/+0f80R//Wtx7BaOmv7lzbzm2PeXgVFlV/v3uTL3ytZhDPy3/9t/G5j3d87Af/H9TboEREiHG4oQ0xu4D1W7QPC36pwt1uHau997wVcn8IgIkjXXGsy9ZdSMJKPhRAqmnhaqs+p2lN+0qkh7wP9Raxfq8/xBjlaDXinLDZtfhoTu+8ZqMwamPzdRfGjFjlBe4RC1a0LLSml5E2Lq7T3f2gJoT74A7PkFWPsE0iH048Rqe7/odxtBCIhVWDZ2rbTspGJh1CJjpJASn3lZSXkgwl9SUpGKErZhoVC++fXCowLcADISqJyr6PJREbg0RC4iMRmdLLDslSKj1c0uRF/y7G+Q5FF0c6nwSHp6HmJqTrsGbLMQtGQqRwqkFM5najQYGpmKhgJy57RLoYl1uOuCWfSwoxSEjZWabwe6SjTY/T688sDaerSMo0BBCcLB1eBV8QzUNCMmPGjBnvFswkZMaMfw2Rxrt+29/8z56XzS2+8sWP86WXWz76vh7vwgLnb/ev80g2fMY/wwZHrcKd1tD1lq/94mf5wEf/v6wqiHGRlPgwonVghHfdCltvwzhUYgGJVNiJSMDNbfESFRUqh612BamxSNvDUKGjCUTEeJrlRVBnjMO7EAcsyy6oLh2ZQI39kqrZUjcdthoZx6lBfbur6EdhdGGSrBawlWE9Ro+GhgXwfb2dDeUtFccswrhO7MkYZDJrL6IZu4p37oc8/BMweSImI3QyiZfbXVNDEvEofAtG5VoyFuwXLZZjX1M0Lgzipu4LpoX9gMuqwLSPaMCXgVZtJgNpZCkZ7qui8yUNGC2oQKGl4pFcZWJhCuJWGsiXNPmYE7Fx0UYu0YuSCJrF0MY293R+KbEr+VZ6RhywZkfy4wwSfre6PVI15ZIN8Rp6iSlgKnSRWNVq9o47DYiNpLhmT4uhV08jJl/FQZXWhMhfEbjTTgQEoB+Ftla8CtZO6XZN5dn+t//enIo1Y8aMdyVmEjJjxr/G+PyP/fm8gPm1n/jrz3dfX3A+OL7hd7xoH3LCkj/4bzxmt205Ptnw1Pu/zOXD9/PGq0+javGuRqIpOxULunGR284DCTjwe6Q29aJTZA8HjeoYRWPqllQOFl1O0gKQxqFbwnONQyqPWW3Rq6kJXtdLzP1zdFdHj0nwraRjrtsr6voWfTfdNb/cWC53cN57btWGRSX0ceVfS4gSPqJlRc1WhRNZsqHb8z+saLjUjkdyxR09ytGsyZ+RFIe0UE6L1bKcr6RkJRGxGohM7u6ICoQUC+cyKcuqiZG17roBPflIDgjG9PkptcrvjWKl5wbx7OjxUoOGjo6sUkggL4ME0tBQ7akvi/jP0CIShg19JBPh9bsc520TkdrQ5X2E5CwHxehUijpOP4mk0pRnFM4qjM5t42eeyyZ7eVLYQK0mvz8RqkYregnDdTUWFAbxmXiBoRNHm5UkT4NhzQDUiAaK0hYjWIMqtUiM4Q3PqQZS0lRKU2l+/mgZiFJVebbMmDFjxrsTczrWjBkzAHhQNQxeWVrDL9lXuMsxO3oWyy3v+9CL3PvoP0FWO1YnZ3zwo18OCgNhIV82m0/Fg0EdAaKB/C3GRmIy1lu9lnws0gzZCyKHTeypqX3ZYU8uowm9z8dBPC5jw+iPrTrGfplHxFQlKyGXW+H1neO1cQhjMjcEEC1o2DDgJN7xx7GVIZijs49B8p3+gX2PQj7FvCgPODQpJzjx8b5/eJy9HjeM5JQ+kGCanmJw0+hVObqUFu4QVIxRfH5cUsWkzKTkqk5cLlcccKylZ8fIWvr83IBnwLOWfu+8y2I/gIYaLdQai2Gh4chW2mYF5Kb3pvNKf0qUZv8wPlcdRP4GwrSm2zu+8LOZfkY2vpaSzRSlw+0VG3rC+Fq6No4pglgJCosCg3q6yCoqEZwqXpPKEQjI4ITRCU0VPCEigYAsFgOLxbDXHTRjxowZ7zbMSsiMGTMA+NTP/HhWRXa//6eef1ZvcyEb/pefu88Pf+BZnn7qig9+5hdC2pRxjP3RXocHhLEpVRtWbpY8DrU3gpXgZeoXOYzZzduYJ6olUjloB3TT7BvUjUcawHTIpoVmRJYderHKyVxJwVENI2Te1Xhnc+KQMZ6nTjxv7gwXPnhCNPpCLvzkNTEIWxnY0uFFudIdK2noZWSR4mipqPWIF+Uhg3i8Diwl/NWbfBZTDO+UblWOEXmKtnJR7IHKUaLczkelpCwgDJ87fWb5WZWavUhYSI3hxTYYtrnB3O8RKxc1iWBS9wwyKT/p9eShSOby1FtyrA1r6TlmGRf3I8cs9saf0ijVSdxmzY4dQ9SXJOaShY6WROMW1CxpGHFUMZ0sKTllKlgy2W/osJGoBE+Nh3j8aeyq9IT0MqIaji2NaKV43hHPRkZWWqGEXhBPGMuqEZw6DBYrQh3jeJ0q3SgMLigiHvAqtI2nsp6Tk9BNk5KxZsyYMePdipmEzJgx4xr+zP82JWv92d//Pzz/1ZdOufX1hhc+tkKqET+0AJmAZA9IXNfaustKSWg/H7gGb1CviNeYhmUnRcTLXuJVSs3KhMQbqHqkGdHLJbRDjudNAq96QVZTUpffLHHjIpAPb0LyVlpUbm7R9w1NM4aELOs5vdXzvt0CLhtU4XJQts6zjkNTQoy+JaQzXbHBiLCgoYv3vsvY3BMWnLHmrhwzqNvrokhjVv5gUXyjaqDluFYwpieVYy9uV2WvvwSmDox4YQGyIpBQqwmjRZk46B5ZCucctJqhIDLl8aZUqiGOOSE1tVpW2jLgaWN/SFJ1gGjiTmRi5IRljvotx7fSuddquS0rlKvordGpb0QVIx6nnoXUtNSkfhAXVZxwjQO0OPZAagKdWhBGCZ14iEpXhcljZgmdjOHYdUo8S9dSZfp5eqYmeQc0WDr1LAjkz8cizIvB8bQEwmIErFEWrUM9eJ/GteRGdW7GjBkz3i2Yx7FmzJjxlvise5bP1qf8Q/ub/Nzf+aFpFCp7QQpTeS4XDH6RXI7o7f5OvYSvvgrlhrsGqVwgEn2VU7VSxG94XKRDNcP0XEzJkmUY/5Fljxx3mJMdcrQL+xiqfMzeVahaxn6JsWN4PNRU9YCtRuo6pjeJcv9Ozwduh/GYPhSGcLtIWxpwXLDJyVhl+V3Z6B2iXi3nuuYy6ggOH/WEshn95lWl4cAbIhPRKN9/GJFrI0HaG1nS/XQsW+w5qCfTvqaUrXAOY1ysp0K+t/oHJCktY3ShOPE0xaeV403p81Za58X/sTb5+dQpUiZgjZEohUjefQN7UnMqCaWIG7qskhziME0rfS/TusrrlI4ljWClIkYbe0VKTxCALa73Lo6mjeiUgsYURtCrMmo4oodXQX1LKpx6GJ1hGCzjGL6G4eD/qxkzZsx4F2EmITNmzHhL/K6f/fdf+JHfcomi/J031wyP7wSfhYYGdlt1e9uLuNxqrmqp28toBA/EQ8cowBoN5CSWHeIF7Su8awKZKcewbmhFpx73XlcvyFEHtUdajxyHNnU5vUJO15jbl8GnEheJbgxkwlYdi9Wauhmw1tO0PU0zYCvPajXw9J2B2sBJFZa5K2NpMDyULYpnQU1LxZKgDnWMtFrREBbAPSOXsuOcKxbScBXLDeG6+lF+h0ldSHfwp9MtYnjfwg9iDsZ10lhT+lMiLZ6TelLuIxUswrQIL1FG/6a28oSkFKX9OKYxMcf+sSTvRzrflEpVqaEuChOnPhHNSWMQPCWC0FDnLxdHxlKDfcfIEMfCArHy0bfi8rZp+4FgLl9pTaUmjtZZag2kI5EPW0QpG0KC1pYRlXDNNzKyliEecyAebbxGhv1WdEcgJFcphU2CN6SLhKMcwXrzhd83J2PNmDHjXQv7kz/5kz/67T6IGTNmvLOx/jd/y99b/5WXf1QQhlc+wMc+8XVcH5um85ooLMSa5WOMcWFURCVuIBg75GjcVG6IGlATIndV4ohWIClmtQsKhoSuC6nH8Ngq0o7T/sfQ9YFRpA1N6/Ej0YfH4Azm3hZUceenGONAQ6t61YT5elt3GBHaxSYs+wWGvkEEKut58yI8FiT0hSB0qlyYjp6Re3qMEcOChhqLF2UZF8QDnjfkgkFH2jgatKTZ0zwSHUgjV2VUrz/YJmFPDZHp/eXzJUHRg+9p+0oNIrH1O/tIJgN6Gikz8SdZxYW3SPJExCjbqEgko/dY9HeYuO+kyqRz72TM23umdvheRoyE0r/UMp5/neJ2BsnHsIvOEYOhispEaVBP5xAa110cK/PZAh+OVbNZv4rvVxQV4YgGJ55L6egY2ZiBrQyhMV6CypPOrZMxNK1LiB6m+NkANJjcOt/Ec6tScaGE6+xUWVjDrVYwJqZiqdDUoXXl4r/5iRe2v/O7/x4zZsyY8S7G7AmZMWPG28KHzQorwt9cv8mPnt+nXlyFkStns7cCQiO5vX2BrJeoHk07MJoKMHLJhWp8b/Z6lMUWbkrCEp3GwEpVZM/obsAJ1B48+JfuMl6cUn/oG6GtXQVVE/wpUalJio01jnb1CFWLsQOubRnHiu1mgTHKs3cdr5xZWiv4QWnEcFdbXtKKToZ4B7/O/omdjNmw/aZc0mtQbbba8YD70yEfXONyTColWPnCgHyjulEQjUMCkrZNPoWpNPH66NYgUyrVYaFhOtYwPuTDYlmFWuwTtg0pVB1jXPgHFaApKv9S0lUgPvtXIo2NhdSxkM5VRw9KSdQ8joaak3xeSh99IYfK0mE/Sno+aTylUd3F/e4YwnMSyNSOno30LOJIXmpyP9EWid0qqWU99cDksa3otUk/kwHPKr628Y6VKdw6Ipy2gpGghMRJQJwXun4ewZoxY8Z7A/M41owZM94WPv1X/90XThvDY+kwdsRWKaXH7flCxn6Fbha4YYmxA7beBhVkrJBFB8ZjbA+Vw9T7o1wYDfvzNhQL1iNElYMbSgy1j9tkUzrhuzPI0Y76wWvI7T68f7TUtx7tESY3tqHPRBxuDMpO8/RrtKtHtIsdi2WHiHLrqOfZU8f773hOKkstwlKCJ2JDz8tynvd5Kbu8UBaES91SSbg+Vm72UYSxrLTQL8axCiKRCMYe6WAiHemrxOG4VklE0jGmKOH9hK6YlHWDR8XhwyhTLGgUhFYrairKYGEbM6tSw3ka0XIF0UlEq4wLzl4SiV6SYnsbj6mNgcLp/G/rkgUNLXVMw2q5xSqThbEgOYfljOVol1eloWZQRx+JU8fIFTuu2OEIzektNTUVW3ocjp2MnMuWS9nRiWOTYprRadxLfCg2xLGNBPWSkSEewcY7xugJscBuDORjcIGMNJVnGOZ/smfMmPHewfw32owZM942Pvi//tgL/8lf/aMvNMuLrGIkAlIu7sfdMd5VgWwYjRG4TUjT6ptJwTA+lBmOdorXNQ5bb9E+CrVdDc7sqyTp/aMNLem2uJOemsbvbpF7mxDnWyu0A7QD5tY6b+pdhW03ANSnZ9jTc2TVR/+IUtdDiO01cOe04+7tjpNaOLJChfCcv4VBcpLTVgbOWMcywjCmdEeOOWGZR5W6mPbkisVvuluu0bAM+3fuwynvkxOYFIOQXnX99cP3lwvvssE8EYBKzTUik14fJCy2u/h9jL6HspujiiNJyQcSFus2R94agqKR4AoVIn2Oiz6NFKWbyFJoMA+li7UapCAxiaA0WrHSlpU2rLTOHpHys8pzKtUPid4Mh4vfUzhyICLp/U2M7g2G9CqWJXoGRjYy7PlNnPicIJYKD3cSzOk9nh0um9X7qDAFzwg8HjyjgzGlT2voDLnazkrIjBkz3huYSciMGTP+qfGP/tCfesHFmF6YCIitOkQcIj6XC+KDH8TYPsTwQhi/io9tvQ1KRuwLkUWPrLrQA6Ihslf7OntGMowGtaRyYUTL6CQpAIwyPc5zLh5Z9jSnb3J0+k0Wx2dTx0jbx8+yyCqkXakajo62GOOx1oe+hgoWldAaw3Oy5Hv8+3hGj2Mh38hOp5I+gGMWrKJp/ZSjeEkmg7gX5Uyu8mlNikDR91GOWR2OPoneqFgc4poiUhjU7R6V2O8g8RIISDJxJ+ViiIlXyXCeDNu12mxcT2NJKb42OBqun0NFSLkKSVeGUqlpsVSZSGj+EadY3Rqbidu0P9mLD4Z9k//htU7XoYolhunYpUjkKtO5XDSvp0hhgDqqM+lrIwNr+r2Y5EF8HE9TOnEIoTNkE39XvAZy6lQZVBncNIo1jELz07/nheanf89sRp8xY8Z7ArMnZMaMGf9M+Md/5E/kxdD3/cU/9zwEQuHdydSgHiN0zRjvNlcOYtmfLMJincpNRCCVF4qGYXiI6kh8PFSoFxg1EJl6ROpYdmh8IB4ApPentrfoJWlcUFSWHbLsMFfLcCxND12DjjZ0ltBz69kXGR7fQYxndXLCbrPCPbrF/TsjV1tL7wxd7/kwR7zuKzamZ0vP03KbRi11vAatWgaEp+SEjpGmiPgVBKvCmaxZS8d9vR07Ka77ONL2ScE4HMs69JeU/SFVkd6UTOYOBYnP67Q4d4U3pNx/en86nl6S92JScWz0m1gVnIz5fTVTfG2Pi9vpXvdIicOEsFQOWDahH0bh5pE2CWNVaVzs0PuSzh7IDeVGhIoqkowqH0MTHyeje4nUhRLO3VNHhWTqBHH0MoQkAGnShnueFhe/2oJIhR6RcC3f2HoeHBm8wnImHzNmzHiPYSYhM2bM+OdGIiSJjGguGiyWxqmQkEhGIPg5VALpgGnMKo4EaV8Hc3ppQHc2lCZ4CWWFqXOEUFAIkXCUyVAKVAqDhxQRHAkHXuCoQ6+WuTRRaDD3Lqjj/qrVY9rVcYgl1hNUhQdqOH9T2HnPMRUtFQMj79c7QEqFkrxIP9YFr8hZvtN+T0/ywn0vpjaZyveM5NeLC/cuSbHQTot1A7npu0QViYgneBRS50YVSUsiBYfqShh9Cn0nh6QByE3uVaF0pOby3Kouk9dkOCAfNVMZYCJbXpSBoLZ0MrLSOqsgNioOqW8jHCMMaDCAI5kmhHPZb6b3QC1TYWNSOeo4XpV+hgA9Y25ZL69LSURGJtKoUdEBuJIdbVSHwrEGo3tqVE/fV1i26jkylsGHbdbeY43h6H+cCciMGTPee5jHsWbMmPEvDKU6Alzv98jlhSYQEmf3yMJe9wfE4sI6+0O0r/b3RyQczoBo8IdA2CZ5G9LfcjKZ26fxLY+Olsdf+a5ASEwYI9PRopdLzl76GGcvfYzd+X3scs3p0y/zlW8seemhZdEoVqZ41fv+iO/1788jSIedFiG2t6VniIbt8BXuhDfcYrVHRMKhX+8Okbd4nIJp03tSyWG6BFWxfSI+o4Rls0R/RXpvecf+ySWK07iYj4v/dIc/vT9oAi6PMJXelUmZCaSjKhb6e6pHJnSSzyEb6LM3JHx+8JVMje+Hx5sIQiAy4U9Fla9HMtuXsBj6WC3pUXaxSaT0mZSEJOx/8ruspSf5alI3Svq9qOLvAMAWx0VUCgdVahFeXV8nfDNmzJjxXsCshMyYMeNfKEoi8gM/+188nx5LoYTQV9M4lrPI0Q7d1aEXpFRPmji6dbJBjKKbBVO+r8RxLyWXW6dCREAI5ERiA7oC0g4odVBRFuFNctSx2p6BdeAsD7/63Tz14S9y9huf4vzRLd54c8HPffOUP/yZBzz97Cs0VWhQryrPSVXhh3BXfqGWZ3TBTh2P6DEIr5o1HS4slFU4kSXHLKjV8lDW3NdbuT09X6esYpAX9zeRgLSYH0RzjG1pgblegphaykPUbdp3+uwh+noOF+DAHpm6/rzbe83J5KFY0OydW1IWRiafS/KTDOLyuWcSpuG/L2RLKlo81iaTrUvpSPHIiUwlDOKiKT6ceYoEBrKXQ4prk84n/bfD7bWmJ9IRUq8mo/qShh3D3s+optpTYXoGHseXb7NAVKgAF0fHPMqakWMq+uwlCv+vfPJnZhVkxowZ703MJGTGjBn/0vCFn/izLwD8wM/8l89nAlK50JLuDVINQZ2ox7CE8yPa1ciyD0RhGYlKUki8BJO6N5jbV1CP6G5SU9QHYiJpVsmDdgY58mGECxCG0K6eCIwo9YPX0G0Lbc/p0y9z9rWPs748QlV47bHh/7RfZv2LH+EnPvRRvve7X0XVMPQVj6/u0G4tF52y8x4BTsRy4isudUT8MS+acwL1MXSMHGvDgN8b++l1oJXpr+M0LkQyU2upHkxkI9SuhG3L+/5pTMmJp03pZXmsS/fM0uk4tFikH8b1phGj9Lh8vmxLL9WA1JJuo8rgI1kySCY8PhKQ4BEJ248SFvpOfI7kXdOFEkgUI4F0JPJRIqk5U0+Hz76TcDWVBU0mQ2kUKo2ZhW08Lh5biulN52up83MOxyISkDF2k6SUMxgjEZnOc0dPGxvcjQit1lgVhvgz6nC0mPyzdej1hsoZM2bMeA9hJiEzZsz4l49C3ZDKRRIQkqp0NEEBiUQjRO4GVQLj98a15GQTSgf7evKT7CGOZOUyQwLRyC9HkhLJB0bRdTCk0/bIYqC6c8b5lz+BtR5jPIsqkIRfsq/w6dc/zvd+5pvYo0vG9W2urpY8GA1vnjd89cygCpWBe4uKq8ECLd8c12xk4DEdVaFC3NYlKSJWUVq12d+QTMqHcbnm4DvcrFwkJAUjLcr39hW9JwutuJKYBpZGxyK5SJ4FLXwiaZGePA9TvG74Ge/5RqJY1clIq9M/N1bj8UhhxI7PpT+m8JekKN4ay4aOk5g2lshESsoqr07yvQyZYilNNJ+nRLDyOkkkWmnb1JieyNuKNntb0rjVEMfM0nMdI21O2BqxWMrIgBFPh8t+oeRd6Qvytosja7UKVr516tmMGTNmvFsxk5AZM2b8S8cXft+fyYpIIiRysg0KSCXorkbSTW2bonkP8p6GCjneBUN5O+y9JLVDh9AZor1FFi4kZVWaTe1iQWsPg8kEBGcg+kzOf+NTuLFiHCtefWPJVSe876me2yvPpx9/kO/SU1oLY7/E2J7muVf4oDe4seXZbsmtFx8AodW6sj4FT7H96n3+vr7KhWy4q8dhYa1ajAnBsSx5nUtWtHjCyBEQF/tSkIMJpdcjpCxNo1xJBYEQC4tO701G7zT+tIspVodkwseFtmPfdD2NLO0TlsMWckcwhhuRPEqFklvDS3N9KGUMhGTMPpHpfO7pCZ2MLKLyYRC62FGSE7iKbpZwTcIxt9E1sqPPi39XHHNqLwnX2HLYKJKufegKcXu+klL5SZ/ZMUbVJpCfUNbo4uuOjfQstWIbPSYLKgbxLLVizcI7kTAAACAASURBVIhFuGJkteeemTFjxoz3HmYSMmPGjH9l+MIf+NMTGfGSFRBp3JSS1dWhWDCpAPF5Hc1kKE/pWqJTOWHtQoN6TMZSH5aLEoKRQkyvjclFVkNilo/7U+H87BTnDN4Jn/yuV/kb//ABTVVTW/jh+imeORKWjeJdxdivsO1r1LceUXthuex4+mqFiLLdLPBeWCwGbt89Y7v9EMMr9/m7jNQYXpPHWAz39DicHsopR7zKIxyee3qyN0IE5CQomFSS0MxuskfCE+J1q+ivSLG76TWY4ncnsqBP7NNIj8vnE9Jw1mHvRkkKICy6PVMCFWL2FBJJhCAnegm12pzglQjDgoqFptysqaBwiB6NZFav1ZLM9gknugifIcLAfvDBipYrdtkQfxhFLMV1GmNClsXG/bi8bVKFkmIDyXAeFJHDRK0L2XHKcm/UzqM4UYxamj2ta8aMGTPem5j/ppsxY8a/cnzhD/zpF7SvplEs44MioZJjdss0K4iJVuk1b/ZGsfJ7TBzHGm74q60CGkJ7ekNQXgYTErdGy9BbHjz3KrbyrB8fcasRnBeuOkEVHu/AeXh8fod6cYletZjbV5jbV0gzcveZV1geXSGiWOsR8Rw99yJ37+z47qcMv9094CU5Y0FDS01PUCJqDBs6nHq2dCyShwLyAjvZqD1THO05V5xzxTflYo9GjMVi2sv+KFYiJmVz+j658HtqRtqmJC2w33WRcKiGlCrBli4XHebkMFFSg3pIv9o/35LgdDFZa8vIhoFL6VhLz1YGNtLncy6je8t9NFharWip9whBGlWDKVK5JBSHSPHFafwrnUvIvJpStRSli0rHYYpWOJ8hv3dkUqwkqkFt9JYcljDOmDFjxnsJsxIyY8aMbws+/+M/9QLAZ//2H3+evoLKh7GsUhVJnhCjQR0ZYkSviQTFxoVuMqRX/uYPG5nml8JqFx2ASkMyF/D+j7zE4vQ1PnzyBlKNeP99vPr6itceGz4/nnPmNvxI94DPfvYNzP3z8JmVQ2rHm1/4bdy+9xK3n3uZkwerMN71ynci7cBzH/4aVfV+bh2tuPzN7+A3zUU0PbvcOn7BFQ5Pr+EuuUdZaUNKtCrTrhJOOeKSHY/1ChXlgZ7uRfum9KudjBgN5YKpfyMpIiUZgf1oWdgftUo4JBuH6V375YopDSsQrPBcIAhLGtqYc5WUDSD3cUBond/JyCXb/N8L6mxSB7iiA4GWio6RCkOjFU48jU7Fg2mcq5MhkwWDRHN58MQk2jQRkXidNCgpUhyjECKFk48k7W/FIkb4Dgw4mlhi6BjzY4DH0nFHl5GoBNVlRRV+J/DUUjHoE36fZ8yYMeM9gFkJmTFjxrcVv/Bv//cv6BhN6KOdiAWA9TntShoXiEiJcmTrEF5COpaCBmMDe9M4XqD2yHJETrYsH7yENCPmzmPkaMsHPvpFPvj+xywqOJMNj2TDp54xLJ7+ZjDFGw3qTe3ZbRZ41yB3N9gPPcQ8c86dD/86NCPN7YesjnbcvbPh+49XfI9/imf8UbxjD2eseYowgrWQhnOu8qIYpqSrEgY40gUNFTUVPQMvy5vhUsTxpvReH8eTxngPvxxVSttcu3QoqUujNM+Xi/fy/TftIy3k0x7GaOQecPSMrNntERYbF/VdbNJweHYyxuwpz7leMTCyY6CLpu8dQ1Yi0jULPgxDrSYrClMBYxjBKo97ofXeeSVFJBxToBtWTB7FKtWSchthan+/zQqLzWqQYzLIp5/Rjp7LqJcM+PwVrlO43tVsTJ8xY8Z7GLMSMmPGjG87Pv9jfz53IXz2H/yx5yEQDy2a0CUqHUpURCCoIcbnLpDw3DSapaNBRg9V8ICU629pNaohROcBYV+LAe0t1YM3uA+89PJnWFzW/Jj/CM89WCOna8QovreBGAEvvXzKg4//amAH2/C83Hscxs2AZrGl7xrunXjWfU07GL4uF9RqONc193iWE1lyrleBAIjEkR8TF8ilNyOe3sE17HXkFTnjfXo3Gr0lN6FD6M04VFVSC3oyWadErHSHH548grVfoGiyalAiEYOegYY6+zzCGFQ4lw7HkmDUBuhlRNWylSEv7lc0OPFstcOKjSlVfVCMaAp1xrPSRSQYk1riUZbR1N6qpZMqv6dsqy/Pt1RzDD5H+JbnmEbdyqSwnpEVbb5e5bZ9VEPStp2EvpEz2cQf7jFLKlD4wb/2e+d+kBkzZrynMSshM2bMeEfhFz730y9kA7qXYFRP8DKNXJXpWdW0fSIuuSvEmanE0ElQReJbpSa8r/HIyRY52QZzuzfoaLD33uQ7Pvwma3p++MPKvftvQFeHJK7GhX2eL/nuT30NOdoGL0okP+Gzw8J16FpMVHisES51xKrhUna0UmMRbrHiSBb08Z75I7liIwM9jrIbJC2aPXA7vkcQGqnodWQjHTt6HskVj9nwmlzwujyORmmXk66gVDzC3pM/oVxoC/st7Ico06KSupDSqILisWXQKco2YcBxIRvO5YqHsuZKdvQy0jFwERflyUGSygATaXLxueS3WNOxoWdNl8lF6cXwBO9J+FzPSps9k/oJy3yudaFtJK9HOpZ0Dil1qxxNK69NpZMqUhIch6cvPndg5Ew2bOlZab237YwZM2a81zErITNmzHjH4ed/6C8Ev8g/+GPPa19hltN4UkjAKoiIhPzZvPCHQEpGmYhIiuv1IaqX1O5nAhHRTpDjHpwJze2JuFSOp7/rV/iv3vwczzx4yNGHv5wjf7EOTIgXPvnYF6Fy6M5mkqSR+EgzMAw13a7h8dbgvLIhtKivtAWJi2S11BLSnbba0TOwlY6ltCxowsI5xtsm1GpYSM0uqgIAl+xYUHOhV1RiGdWF71nZ8JEyyB4xuKkRvYQ5ICJp8V2OJqXHiYAkP0V6LSyyhY4ep55eBpx6bMwzPiGoQUey2CsUTGSilSYrNhDM7120pSeEgsKQCKZovmZlozyQ29wtglXhNisuZJOv0nWfi8/n11BlD0oiG+lYFwRSWZIXmEohHZ4VTd7XJnplxjhGls5hxowZM97rmJWQGTNmvGPxC5/76ReSupFJRi6XmP762iMgEEhJNrBPz+39jZduSKfnKg0ekcUQmtorF0bA2oFP/u7/m7sf/9Xc7p4LFKVsZw9JX9pFEuOmbY9vPaaqPNbAohJqhFNdspWBBQ1nsmYjfTZlj4z0DPQ60hN8ETsZsHG0ysfPD/G1DStaeh2pxLLTnqvotxjV0UiVtZObUrH2O0L2I2rtwT8R6Y6/Qaii8lESgJh7lUnDRFg8YzwPjzKoY8Sx0z6oGtqz1S50f8RekaQYlFG5NZY6Fg4eqjjheni2klrOyeb8UTyduDCSJqFDpdaQmOWBTkJBYFJE0nFXTMb2Kn5unXwiTE30tjCo94w8kg0epaGO36f3NlSZEJqovNRYLmXLmxIM+FvZjxKeMWPGjPciZhIyY8aMdzQ+/+M/9cIvfO6n83z8tXEsjX6RVEoYx7ImciDTd6NhJEuJPSEUJCUmbK0GaIfwOWksDJBVh1SxyT2pEUM1jY2lrxQ37E0oTQSO7n0DYxynx47GQoPlWGue87cA2DFwzhVdXHi/T55CURqp2GnPinaPEBidCgwrNcGgHlWPsKgPC/G04LcY1nTFiNCUjpW+UgfHTabzctxIrj27/8/IvjIiWeVw6hnV0XHYzh7+LKSho6eKY05V4cFoqXIqVjDj2+ATKdrK0yBYzxALGNM412G/yWS2H8Sxk2AD78Rlkleiin/KMz5ikV+vsVSYTFJClHD4mVUY2mLgIJney2b6OpYpGiQkpcnuWqTvjBkzZrwXMZOQGTNmvCuQiYgXpHaTGd0W40/Fd+pEVlLBoU5EQ0BSTO8YSgzx5LEtaV0oUTzpkGWPnHThc5pxUkKi3yOMbXl0NOGzR4uOFt00+DeO0W0L3tC0Pc4JxwvlE6uW76qO+aCe8Kw/4Vk95S7HdPQsaTnnihRdC2FUq9KYViXKRrq9a9NS8xQnHMsSj9/ryDAiscm755LtjQvcw1Grm15LKKnLIR3pGPeSvcL2ShVN4h6l0wGH40jCQr6VYFhPi/2kcqQxrGMW3NYVRyw4YckJy9wsD8QRpxDdm57f0tNLUJOGpCpFHab0iozxvclD0kvo9hjxeeQqfSUSWGG5YkfHkFUMYZ+oJe/NcOCDKc3qadtGK5Zak9K1Okb+o7/2H86m9BkzZrznMZOQGTNmvGvw8z/0F15IfhH1sjcOldKzMpJaYTWMRg0mE42shNwETyAqR0MgMpWC9UjrgkpCVGPqSEhgUlviY0mN7kOFVI7d46dZHq1pG8etI8fTJ8qDY2Ephud0xWtywSqmNxkR1rpFUZz6vTv5KfXKo2xlYCsDa9lhMay0ZUHNqRzH04h323XyZRgkR9wC1EVS02HqVRrJ2jepX/8noyQhqYwwqSrp88ccVevy+YS7/yaOKVXRcB62S23jRyyyB6ZSg9HQc5L2fUQw5a9os6E8JHw5NnRv6XPZyhDfE6J0y+6RNHK1oGFBE5WXNidbAYy42J7ussLUxK73iir6Tcy+ghWvSyJaQUUJhZUrbRjxrP//9s4tVrLsvOu/b619qapzTp/p0z09N3vixIAxGCngxBGEXESUhxhjJ2OTFx5CwkMMkRKhSDwQRKcllCduQRglPEQIIUC+DCIJSAjFKFK4xCEPCCkEgRPZztgzPT19O+fUqdp7r/XxsC57VXXPZGLs8Xh6/ayjqrNr36ramrP+9X3/7y/DA/daqVQqb0WqCKlUKl93JCGyIzr227Ag/BdOJVQ4ygJAap0i5YhE4/o+nphVwlxNYa625OySOLUrXVtj+juNQydLv7rD8uRLXHv8lL5zHK4mDnrlDx22rMRyiRUWw2UO2cZWKkF4Qi7TiOVlub/zLXqa1ORwnBOmYY3isjG6k7CMthKqJ2XQXhICKTywFB37RvPXQrIv49WnZ20ZosfDRzv8nCiueDoJi/peujyTKlUiVvT0aoOvo/jZypQrDBbDij7msc/3UoqnLRPbmDWSRM6WkQu2+TwS26iA3O5VTr2y8e56WvoiLHGIU7tSxonD56lZ4d66B6pM6Xw9DcsYppiS4h9WiapUKpW3KlWEVCqVr0tSe5amsbxFPgiQhYcOZm7RcvE/eckLkr0c8Xl6PRnLIVRQJpmPg0JsRH9IGg88NOhkY7K7Qc/i6NdFWPAfn9zmyadf4drjp7ztqTOePJk4sIb3uidZasuGkW+SJ3mbXM1Viie5zBHLYFxXk1uxUltRS5h8lczOLprAYbf9J9y+MmhYkHtCYN7+eFnl4e1aa4Yi9dxkczqEHJA01jZdB2DUYD4X5twTRVnFViync/6GjYt4h+OQnolgJi/f65ot29haFe41CJaeJo/xdcXnkIRF+ExCUOI2jvYN13N52lYb7yNUVZqdqkU6R6psdLQcsdypkqQqTKjMdDsTsJKjpPw8WzU7IZTps/upT/xYbcWqVCqPBHVEb6VS+bolVUS+7b989LqmDJEiVEMns2tQTyZymCdnNcU2D/m7GdHwNHtLCEGE0f8hjUe38T+hjQtmdBVwEoSIUabhAOs3SBdabPqDu4jxdP2G03vHnDwmfLMs2Hy+58I77uslvih3eKc+jhXLHU55Rq+EMD+ZaBBGHOdssFgWMQAwLLznaVIHLNjKxMSEFYNXxcb0bSsmtk3JzuI7iJKwuE+TnBJlGN+GcceYbeI9LQhjbzcMLGI70n3mdrLkm1hKTwj4s4BjZMKpz3knS/osTMbYJpUM56WfxWKYooAoPSdpnyRqUutT+idsYvsXzIKrbMMC6LXBSHhfS23zxC4gCz/NlY1wrgXdzjXL0b3p8ytT1BNtIcJatVQqlcqjQq2EVCqVr3uSGCkFSCJXShKumJaVHtNxaVsawVuea5pHBWdhk/aLZnS8gShAmCzqDd516BB+ABaHt7DNgDWe5WrDE9fuc6WzPGtWPOOPOKDn/8pLXNIwdtcQ2440CI7H9CCnj6dFbph+dbFjum6x2QsSfm9oi0lP5fEGmXM9kkmbMB44CZNFHDebKi4bBi4Ycs7Fmi0NhgVd/OiUhXSsZMGhLMM0KiYEoY8L9pSL0koQEaklC1LbmGdgzFOt0nnD656m8IF0NFnwJD/HvF/YJ7S92Z0QQpg9KOn8abTvPH1L6LXJIsGjdBpG7aYWsHIYQKOGNIWsrICkylESHg2Sz9mrpa8ipFKpPEJUEVKpVN4SlNOzdLTovj8EZlHhivasKXlGZG7lUolVjWhoh2BWb3yuhOh5X2SWFK1cgPRDHiHsXYObFuFnDMe0izOWh2ccHJ3SdhPveXbLuy5b/uzBMX/MPcG7/dOstOOaHAOhIjAWAuOEw1yBALiIYYUhU8TFmoHSSZAdnbR5klMycCfWDGzjcclUnsRG2bZVTno6j3nlqcUptCU1pJTx1AoVKi0NHqWhYSFdbnVyeJb0UZAEMbKKwmGM95PuI7VQ+eL+0v9gt90qkSZmpTaxJprIy3MnLIaFNjs+EoNwpEs6gjiwUU702gSTPLDUNoRMxp90Li/KMg0aQHYmbC3ootgL4mYST6dzHkmlUqk8KtR2rEql8pbh19/38yFp/Vd/7Hoex2t0xzcijd+tfrRxMZrM56VvpPyaZgwp6joZpHOos+E8qRpifaiExJHA0jjEeLxrCy+JZ9ocIuIxxjGOPbaZuHRpw73Tlr4V/sT2EqMqn3MX/DYv8hgrPisv5XamUR3fpNc4lhW3OeMiToFKE6YaHsylSAv39BjermQh4/A71ZFyIpcUYkej+XuK+4fWrzCZKomfUFXxpIC/cB+WA2lIAYgDI8t4XIvlnC3LonJBFBxSVD/2KyFp39SONuFZ0DLEaksi3Wf6fNJCP7WOQUieD/e1iRUjGysowfeSKjFtbCGbP9890aCGJm4b8VmYGQ2frRMfqiSi+f9aIw4nnpW2VYRUKpVHiloJqVQqbzk+810fu4HRULF4IE19bs/Syeya09Ou6bl/yHEqoQqSWrdUYDnM4sXGBbs3NKv72Ca0K6lavGvxrsVNPePYh2qNeFaH53RduNjTR8K1peFtZsE3+7fxBbmdqwWjOtYaUrVX2nLEIrcwjTw8ZXtnXO6ewEjtROk1mIXGazExT6hKAiQdn865P3UrCaJE8mqk0bYw+y0Sqb1s32xeekM0Hn8Y51Kl2kdqK0uJ8g0GW0y96tVyrEuOdZnvsaPNn+2hdllwCcIo8yCAdI/lcwjtVek9TTGVvdXgaWljtcUirLSNvhJlxLOWkXVMev/nH/4X11/zw69UKpW3CLUSUqlU3pKIUXSnqhEX2VOsVkyxXWs0s/k8BRaGPeM2CfukqooLz3XdIf0YjnEmZIb4MFVLL7o5of0hKdzqw8Su9Jqq8NQT93np5iWsMThvOLCGa+OSKxzloL2BEY/yf3iRd/EUCzrO2EYjt3uokAjb5kpCi405IbvJ5hMjyhwUOKpjKT1AnkaVRviO6gBHL02+t6m4/ojL5uzkIUlVkNSGtfN5FIbysupS3l8SGwNTNqunqVMjU/aAJNP5BQMtll6b7PE41C78c4nP7yeda8Q94MlIFRGHx8ZqSRJSSexMhXBKpnSApc6CS5gHADTF575F82jlVNWpVCqVR4UqQiqVyluSbFaPvO/X/ur8DXOsakiXQgVNmJIloZUqmc91sHP7VswIkeWEbizSxcqDUfS8y+elnZDJhgX9xSoIDrXx0SBxAey9RURRFbyz3L+/YpoE54Wugc4KR1PDu9wVfsfc5Vw2GAnf1l/oBpEiUT3WQ0YcDSYv/B/W3pMM0465SmKiWfy+ntNLi4uiYEsYr5tCEwWhFXLauRDCD9NC3Odayyw+EvMUKJsX3UmUJNLI3WSdT21i6bHBMBA8G6vorVhgs/F8yxS8FxoyUlKoY7r2WkYOtcOoyQIjLfwbTL5uuhcthJvgOfHLnc/1trkgjR4O2+cpXaWgKKslrVom2f1serWhfUsNHdWcXqlUHg1qO1alUnkkyMb1ktiKldLXd8f1Cmzb3f0nQQcTtpemdB9Ei3RTmI4VcWP4xj+1ZEHwhYjxLJbnGOtya9j5RcN2NKy3wsUAnYWVMZzQcUl7nvLH9HR0tPkbfIAjlrkCYQsBstbNjghJ3+h7lFEdE1OePDX7RjxOfZ6q5dRnAVK2N6VAP4jZIzH8L+V8pO0eZUlPR7szOtc+5E9PaTyHuRLS0eQfCAv6niZ7UDYMbGOlxhGS0s9lw5aJRk0ULmPOMknjh8sxxKldLAmyJM7S7xOea/6ABsOShjYKlqt+xYmuGHFc9asdv0qZv3LFL7nilzzuV1jmiVjLKHtaLF0UUxbh3z73idqSValU3vLYH/qhH/rur/VNVCqVyhvBC89+4Fef+dy/+24giAgVRECsQqc5FV0ktm3Z+I21C8tJTaJEiInoHjYdTBaxPozptR6mkBmiYw8qmGbETx0i4ZrqLeOwwDuLGOVLL5zgvNBYmJwgIpxtQ5p7L4Yj33NMxySCkxD29wK3uSSrWAEYd1qxACYcViwjExsdswgJ20N2iMjsndjomNuiQuVhrlK4Yh5VL11+nkRPCiEkHq+ErJIuNi2FBXl4TIJm151CseyXKF46FgRfxlC8v3Q/EATYPFo3/LN0tCy1AyEmjoRckI6GRTSKWwyX/YJeG9YycuKXLLRhI1N8F8R7Dfd11a+iUBE80MX30mJoMXTa0BP8H0fa09NwqB2H2kXvTpgJ1sVPYUHDgbasZUKABstT/jC3mH3w+b9YAwsrlcpbntqOValUHik+853/ZCfgMLM1oTZsPUgYxwtRjDiLph5/0WA+901OSc+tWV7A2xheGFqvxIYJWcaOiPG4sWfYLvHFOafoXZE4aWszglelM3Dh4KRpuOfCt+2tWF4wdznTNb8XF/R/mCdZy8hdzgGiT8PlZX5qZ0o49ZgYXhiySCQG/5mctg5EIeKyJyO1YY1xWxIIENqsJqZcRWlkNoiXfpAy1RzYkyJh0X9QeEbGcFZaLEO8tzKzpMwEgdDalDweoZIxl/wNwolfZgEBcOJDPCKEigWEKk5TGOdbDC5uG+PUrLBfYEkwnB/Exi2nmqs6HmVEaRHG2D42xvu+7BfcMmtGHHdky5KG5z71g1WAVCqVR4IqQiqVyiNLTlgXRbehKoFEQ/lFgyyiZ8QLD3SvGg0G9L1taUpWGNGbjgfbbZiGFQBtu2XwC8DgppgRIeA1VEKU4AkZnNJbYfJwaAxPuAXb2O5kEM71IiSry5rLusLLivusGTU0Nw2MON31HySxoSpYMXlbohzx66NnIgmQFHToY6tRKW40+yuUI1k+4EcJOSA9W8Zs1C6rG4rGNql5+zq2eCX/Rlfc2yoLFYvRWVQkL8ZSQ3jhia64Jedc1QOsCt3eYN0kQNLzCZ/3sLH6oUAbRcm+yyZ8DikZnVgtCcLFABd4WgxTPDbt61FWRVvbIFMWWZVKpfIoUEVIpVJ5JEnG9W/7zI9eT5OvgGxSl87N/g8I4YOTBWx4bjxiDHRTECSbwj/iDbruUV/4Q6YFYjzWbDF2ZBx7/DQvaX28/KpXzrbCQQfTRnBeGVWxAmscK224rCtekSUXbLkmj/Gi3uYOZ4wa4gWfkMusGTjVUBkxmB2buA/jwvDRGzJP0vLF/j5XKXYFyJwzUoYDpolXKppN5Yk+BhamNqxSoKRzzjGGAnjWbEmZIyl0cFVUSJb68D9fV3S187tV4SoHtGqyUIFZDCRxUb63h5H2Ty1ZIbxwPkd4L/N+6UpJjJh4XOl7MQjX/IoRzy2z5qOf/OFaBalUKo8M1ZheqVQeaX79fT9/g20QCznUMCWob9vCqF7859KbIEj60IqkQ5M9JiGw0KPeIsbhph5jB5ruPI7mTa1XHhHFxFwREy9jRFm2cD6ElqxRFa+zebzBcKwLDmTBiVzih+0fZdSJC7ZsdMtjcsQresoRC5y6wq8RaPcqHTCP9DXFn4TymDmLZHaHTNGELvGY5P0oDd9JeKQfV7RxpQV/T0sbJU7yp5Sp7i2WBV1472pYasNSmx0PSRJRV/WAXi2tmvwI5EdXvKf5nnYDEUuBkJD4epff6249JLV8JZER7juY4Jf5s9ilrIpYpAqQSqXyyFFFSKVSeeT59T/zczfySF8XMzxSkKHROdAwVUvKAESjoSrSuGxk1/UC77rsBfGuw43LfMiwOczPX37pMl7nSoiPZvmumNRqZPYZABxrzx095Vvc2/lv412sWJyGBf5tvY9BuKX3OTIHURqEyVfJbN5g8xQr4AEBksQChCpIKQiA16wYpNeSAbzM1thvv+pjOGBZhdgysmXKguaIubVrLdssOkqRcFmXnOiKVk0+T5my/rB7Nnvn2H9eipFkVB9ypejheR6meG2uLj342aQWr7KqVKlUKo8aVYRUKpVKJI3xFaPgogpo3CxEplDloPFhezSWJxO7NA76IVdPUiaIasgIGbcHDJtDnGtQNdx66TKTM0UVBLajYCQ833iliy92RqKvwrLA8q36Tu6YDXfkgm+UJ3ncXOaSOcAgbBjYMDDoxFJ6jmSVJ1VZDJ00DxjCXR6Sq3sCxOYKBZCnYrnoDFnQcolVTivvc4TgvIjfn9wFwdOx395lEJbxTk/0gMu6oottVws6jnSxc89X/YqrfkWv9oGgwXTOlP+RRuvOCe4SqxWSR+OmRxs/gbR/2i/9vsXh2M0C6bC0GFaxahRaszxjrDd59qeBzeLjQ3UaVqVSeQSpnpBKpVIp+Mx3fSwvCN/3Kz9+HW/mlixTLCMnm6djAUg3hQrK6exJEOMwMuCmBdv1JXzRiuWmliFmioiENqDtYPAKowuVESuC0/Bt0cYrj5sep8qpTlzWntts+KK5y1N6zIqWMxn4Ajc5kUvc1vsMjDzJYyzo+B1eZCWzp6KjJUyRfWmpEgAAFsJJREFUCmboNDQ3GdHDuNgmb7fZmC0saClTzluandYsLSoGqY2rrGCEiVHTTmtYqrYYFbyEay5oOWPAamgqSynlQB6bC3Fi8kP+LW00jaf2qVerOMjea2VbVvrdFsKqbOsaY5vWGA3oSYCka7bxtX3PSbqv/dauSqVSeVSoIqRSqVReDaMP/j40wYzubQ4alG4KYqRx6NDhXZia5V2LiGW4OMLHxPTEzRdPAEjDq2y8Vpxwy/1BOWqFu4PnsDVcTNEbIsIUPSLP6hEnLlQHbpo1GyacOg6l515c1L/CKVc55mk54UW9C8BS+vwtfkuDEQn+kyhIltLnaVX5rRdtRuG5xAlPfVykW1Ks4X4bVFkxmOst4Zh0bqv73hUbzz8HHR5qx1pGFtrklqZXW8KXoiEJgFKs7Fdhdv6Z42v7/pAwurc0mM9CJ1VALBLrQrNXJflF0rnG+Bm0GN7//IdrFaRSqTySyKc//emf/lrfRKVSqbzZed9//InrpET0xs0G9LMFshjRTYtuO/zFKk/FCqb0ke36EhAqHp//3WdorGeYLEYU58JCdbO1TE442xg2I9zZKo/1wmYKrVnno7LxYSk7qnKmLvfTnjHlb9vHOGL298x97skFFstdzjnhkJGJVzjNYqaJFYxEKQKS2DiMgYDJo5EW5yk9vczUGJnywr4hBBmmR5grJmEB3sT2p2j2LqZXKcqB9rH9rGXCM+DosHkqVllBKMfkvlZlYd9sHu5TCoEA+xOsSvZFyzyKN30eQYgsxLBRzzb+WziUMV4lPXqUH3i+ZoJUKpVHl1oJqVQqldfBZ773Z29AbNGKbVQ5GT0Z1SeLeouxA9OwYntxgDFh0WmbkRc+93a2g8VZQUwwoasKxirGKsPWMEZNcNQKo5unZnmUS43h/uQR4LJpOPVzZkj69r1FOGXiqobovDMZeEKPuZCR25yF/WXO9kitVg0NE9POaN15MR6qFCMui4YUEKh4JtgRM0AWHmNhck+VFVs+j+KjrDq0aqOnImRDOvFM4jnxixwYuE9a7D9MQJShiWkIcBvfUxhWXBrJd4Md9z0ru59LaKtysSqSXtsf05sqJmMhSqoZvVKpPOpUY3qlUqn8QTEaskG8QBuqI3q+xG1XuKnn9M7TaDRKe2+YxpYvfeEZztfFeFwnqJ9T0scx+EFcbs+CyZMnZx21htPJc9gYemNoRbhkLBc4VnFB38Zv9VuEQ22zgRzgFvcYdMKpx6nHF9O2BMOWIVY1mjyVCua09VQZaYo/G+XCvgwZhLmqko7raWk0mNebKAAMgpPUxBU8JotY6fDx2HMZuaYrHvfBazPhX3X5vu/tgN22q0QThYDE9ilbvKcwCWtut0riwxfComEOJNz3h0D495J8vlnElDkl1YxeqVQedWolpFKpVP4AfOZ7/lGoiPzqj11n6sIULevQqcFNPZv1Ed5Zxu0S7yxfeuFxvArTFKoeXmGYDOcXs99itfC46C8xEgSI8yG83Qq0FjYTNCL0Vlg1wpiKAUPHXT/RYtgWM5g8yhW/5JgeJ8pattzkHidyxD3Od96TixWOZZxYtWaIBvQuLr5DdodgGBizXyMcZ/dM6bP4AHIEocMxyoOVhPT7kQbTfAr1S9WRK9rntqaHVSVKSi9HOao3Ta1aieW2DlEYzfsloZZcHMGUP7dmNXvXSx99ygBJE7AmHNu4PZnOxyxmwnF9TH+vVCqVR51aCalUKpUvg89818dupBG9etGzvn8NN3Z4ZzHWYYyj6zdsB8t2MFmAfOFWw0t3Gn7+1gu8ciYMk7DeGKbYzSQy/6RWLCGIkcu9oTFBpHQWWgNXesuJDXkbLaG1KY/YjQLgUFuOdMFVucQZFzj19LGdyuv8Tf4Q09OPWGAQOhr6+F2VYHC4Qpg8PGujNKGn6VdJpIxMjDEDJIf6qdlpZxKEPlZL0vn2Mz8ehsDO2N30WfTx9xHlpm45kS5/VumekyRI1Zn98cVBYIRtU/HehVAVWWKzUEnTsGCujNjiGj62v1UqlcqjTq2EVCqVypfJb3zg794A+JaP/63r49Dj3by4HOkR8aGdyglq4PdesfzG+Rk9hs+al/jY9j5/ZXo3xwuha2CKHhBVOBuCyIDQltXZebJTWuanSsnlznD7IiyWt9kEHhbDq5iPsaDFqgGBtQyxraplK0NORF/EukUylDt8bH8K2SOHLLLXw8RlP5CrISWpAgLgMXls70p7tjIFQaNBPB3R5+pHg2HNiCA8qfO443ISVilIyhancgpV+l2iF6OPQmGjob1s2pl+xQPnmP0cs6hKxyTzeyPCoLvHmih4Eg8zyn/v8z9QW7EqlcojTxUhlUql8v/Jf//Bv3PjnT/7C9dVDS/fPMY5wVrFGA3VDKtcbCz/ePNbfIO5ksfUftD/EVxsvZocdA0MU2i9ak0QGapwc+N4+6HNfhEI4iMtzI2FA4m+EBVamZfVn9fQenWoHS5OmVpJz4YgPl7QCzYMXJPH6GizmNgwsqRnYNwZxetwRRuTo40eEghelJEJyROjQgUkTM9KY3KFhbYoDZe0ZxLPhpGFtPRqOdGetYxc02VewB/GZrB1zjR5cDRvesepErLvSbHxrrbxs095HunoNDb4Yecknqm8RrtX9VnRcIHL10nenDHerUWq+KhUKpWCKkIqlUrlK8Bnf+JHbgB0P/mL18/WDYeriSlWRtQHoXFVj/iffAGP50/yjp1FbwopHIuCQvKHXO0fTAOHOaTv1oWnlZCbsZKQvqHAVLRaXdMlp4x0ajmXkTBPS/icvIQg3NEzrsnxA36NjpaBMbdGbdFcORmZ09NtNJsnAZLoabIHIrV2KcpxTD9v1Yacknjdewy8TQ9xKEssY6zEpLanJEzWReVlEWVQ2OZzI1ca62viNecKx4PViQZhyvtpfr/puFz9oAgsVB9FTqjOpPvb4LJ53cZ7n3jtdrJKpVJ51KgipFKpVL6CDH/vgzc6wP31X7oO4JwwOeHWmfB++zb+gxOu6SHXdMGlxuIJgkHjZCyn0QsSW60AFs0cYrhfBRjdLDY26lmIwUFceMMlWh6TjrU6jml5GccojgPtGHA5uPAJHoutVp4Rl03qW0aOWDLG+k24B4+lpY/J6RBG8aaxu702rGVLmLo10WDotWGS0JiV0tOdeFba4IsQxyv0LKLxvRGhxbJWF6o7OsubFZZ1zhwpfSizJyaY2+OoX4IZ/dUIAiVMzUrZIUn6pTYrCC1ZwbQuuXpiCmlT5oDALFr+3PPfX6sglUqlUlDdcZVKpfJVwP2Dv3AD4JVTw4v3JAQOOs/79AnewQEH0tAZQYtxrsmI3tnwU+Lj+lkIQuXWxvPyhefeEJa7jrDgFeC2H7nlR041yIpznWKVQlCBlYal+io2FRmEMzbRWN0QMkGCgbyPieUjwcdxj3OmIksjtWUpnlYtiseLsqDL43hnE7rliJ7LukBRerWMODYycU2XPKOrUNWQ4LdI7/tAwvdlCzH08afFcCIdV6Wnl2BuvyodB9Ls/GFLI3RTRQPmP3wTylSMB54FxCwiJAoSlwVYOGbAMeKZUC5wrJnytRbYOBXr9ZnqK5VK5VGkipBKpVL5KuH/4QduXP1n33fDKdzZek79hFf4tPkCTpW7o+Oum7gIVgeGohUriw4Jz+9sNf/cH8ICd1LdabkyIrzix9wGlEbbjsUCeqE2J59PeP64f4YF3U4b0jKkemAxrLSlVcuCjjuc4TWknW+jP6PMGnESUtTP2ADQaDC5B0dHaG2yatjIhBfNYujt/pAh3uvtOB540iCabrLBqebsjfTTimCBXoSVMazEhuBA1WhwF7rozUjjclskP9/PNCnN6HMq/BxEmNrOSh9Iej2JDx+3bfE7f1y/u1ZBKpVK5QGqCKlUKpWvMu/4l9934z2f+PM3/re5y78y/4OresAaxxBFxFkM/fAaWq+8wq0Lzcb02xtl8hpatjQ8jlF8GBFsXJBb4DHTciItDmXBXE5J4X+XaFlpQ6smBw9+g17h7XrC3ZgfkqocvVrWMvJFuc1dzjkg+Dhe4X5RV5jH96Zv/YcoUARhpV3eL6WUH2jHiGPCB5ET72OLZ4HFqXKLLU6Vq/SYOK54QrMTZFTlto5s1LP2nns6McUAxmCAT9csx+POP2mE7xwmOO+TKP9APiylPeV/pBG8pccn5bbURJBKpVJ5ONUTUqlUKm8QP/rJv3zjRz78N6+3WH7Nfo67es6H/Xu45xzdJiybz4Z5ROzL6zlwLz3u5HLklqWiGgLc1SACRvzOwji1ZLUYlljuMDCKi5WOhmc54UyGaBBfg8BSW0Yc79anmcRzS87zGN8yLyS1HxkNXg+j4ToDjpW2rLRlIxMXMnLse3oN5vZjOi6iIDln4oCGVoSrGsVHrFys1bGJRnMIrWc9hlFDe1QfJUOZYL4vAMrxu+m1NlZLkuOlHKm7f3w4dteo7lBscc5UeWqwdRpWpVKpvAa1ElKpVCpvIL/wqZ+58b/Ml1gzcMGWX7G/yxbP4DV/S68a2rBgrnzYIm1832PgCJUBJfhBysVzSD63eZGcMjM8sFBLq2FRrygr7TjWBfdYs9YN52z4HC9zSVYcaMsQHRRlS1K4fqhP3OYMIJjJZYvDcyEjKRBwwnPVr1jR0EcxYQgJ4yG7w7NmYq1BSnglNnLBBpcrF01xBz62Q82Pc7WjpNzmeHAcr42VkRQ4mNq2bDTRl0GQqd2r9JukKVkQBNL2gTuoVCqVSkkVIZVKpfIGM+jIhW55pzzFTb0LwE03MvgkOIiiJPotZK6GGAS353P2GiZJ3fbjAwKhTARPU562UTS0GFa0XNUgAxY09NrwNr2MIJxr8Ha8w1/JrVshmNDndqyBEZsFheBFeZd/IouCS9qz0pZBJhyeJQ2vyIYtjkMaVmLp4uI/VXnWOC5wLEy45iaOwi0nX2nx3l5NeMyfwcOfvxbpftJP8pGY6C1J9+GjUZ3i+oLwsJDCSqVSqczUdqxKpVJ5g/nZT/7UDYC/9pEb18tv9k+d46RpZr8HAjK3YeW2LFWMCLfdmEMKIbUDwbE03NMpL+qTuTpRbrc6n3fEZcP4e/kGfsu8SBPN6QssX5J7HLNiwmOxjEx5ClaLxahwQEerhhU9HRZFuS9bAJ7yR3iUA22Z8BzQ4BVOmXaqO+W9WhEWGJwGn0eDPLDvq/k4ZsO4PHRffY3zlCRfyTwRbPezNPm+wjm/p7ZhVSqVyu+LfPrTn/7pr/VNVCqVyqPKRz9y/fpf8n+KVgxHNvoaYjWkbMtK2yGIkDFWPwBu+ZGrpsUD9/y0M442kTwTayZGlIu48E8eC48ySjhqjD6OrTjamOFx05xzLkNORE+kCkkQKm2cpmVwolzVBRscRzT0YviSbugwHNMCcJshjgq23I+BiAbhSek5V8dCgohZq6PD7Eyh+v1IoiKN1J1bpuYKRrNXrZCixQp2/SGlcT2d26HZsG4Iqenf+fyHqgCpVCqV10Ftx6pUKpWvIT/3yRs3vuP5D904tkEMjF6zALnjJm5PU94+qu6Y0Etu+5FTn8bmwr5vZB7ZG8bzOlE2MuWRtxahV8tKGxbahNG6GvZdy8ixLrjNGRbLUlt6GlI2yJYRi+HIh6lXBmGlISl9wnPKxE3dBqGDZ4PnLmN4X3jW8Zh0n1Mcc3tXB0Z9cMrX62G/WlG2T7lo2N9PMdeiYlT+cSzFShJ4YRSvy+fcb4OrVCqVymtTRUilUqm8SUgL8VPnueNCxcKhvDyFBXsrkkfUtkWJ5FgaLkmTW7P2PRAOzylj/AZ/XtQvdLcjdyshgG8jU97WY7lpzrktaw7oucs5ZxK8Ih0tXpQ1A2sZOaBlFJ9N3CEvY84pScbvUDWw2WuxYjeZ0etuq5Tbs5G3r7Lg3x+3C+R8kHbvz11ZFSm37RvM2ygvkvB4mABJFZxqRq9UKpXXTxUhlUql8ibg3R9/f27jcar8trnLb5lXWOM4Y+K+j2ZyEUwUI8mMbUVoRDhXx1EUIm0M12sxHMTgQYvkSVRH2nIYrd4bCZOnFnFS1kpbDmhpYgvUM/4SX5S7cYztPC74LuestOWEw7ztSX8Qrz37WspxtmH878j9OEC4FEV9rNIYgQMaFti8T6rWLMXkfZMYKfM6Xs2Enkz5wE5VpMwQSZWQNBELggHesVsx2cbKTZqk1WL43ud/4EYdyVupVCqvnypCKpVK5U3Cuz/+/hvf/MkP3AC45lc4PP/JfpYRP09g0vnRF+1Zd/zEgVhO1eXFeFrcGyR7QuZFvKXHspaJQ205Y8zf7kNYaA/x95U2fKt7lg0jLZa//Ykfv/E3PvHRG0/rYzzjj3jWHfMud4U+LvPLsMDjKHQgTZIKy/6m+PNj9gobZ3FEbwoBtAiHxqLAIt5/qgQlcZHG6uZzFucLo4nnaWMmi5fXbqFKAui19msLMVSpVCqV108VIZVKpfIm49uf/+ANE03SKzp+297iNHooUkBhU7Rj3fVjyM+QID56CZUIF70VI54DYzkQG6sonh4TvvXXmI4eRUlaeIcKSkuPLcbhKj/ziZ/M3/Zf1iUG4YAWj3KfMdcMymNGPMe0XKajyXkbQaBciiZ1CInoy6I1yxTVCAjCY6Oek6bhSmt3fCSlWPD52g+fhlVWc0rvjEV4zDTFSGPZ2X+/pauO4a1UKpUvnzodq1KpVN6k/P2P/NPr9+WClob3uGt8o10BoQqiwBSrIPdjQvqJabnnJ3oxrDV5FoKH4sjM/o+X/ZYVDSOeV9hylQW32MQwQBhwLGlyKscQl+XPPf+DD7Qb/dJzn7yeFukXTFyiA+Z2sBPp+LyuucYiV3PuMmARnpYFjvBt2KlOrMRyocE0nhb8FmFlDPei6b6sSiQRMBLSzptCjKRt+xUR4rbkU5HYorZv5E/XKlu2UltWasd6//Mfru1XlUql8mVSRUilUqk8wnz8uX99/SoLbnKRqx4PExuvl3//3KeuQxAhHuWq6XhvbDH7rx/+xesNwoUG8fOsWeE0Jp1HDZDEA8Bl03DPT/n3/YyQXgxeYRunXZXXTenw6biREHjYxOlbSywX0fJuMdmPkoTLNvpp0vFTrCxV30elUql8ZagipFKpVCpvCL/x4V+6roSRwPd14sS0bFWxwBCrOq0IpzpP5yrzQWC3mgGwEstaXRYdqaqRPCAOxUbxMVc0QlDiBo8rKiJtbFFzRfXDIvy557+/Co9KpVL5ClNFSKVSqVTeUH7zI798PT0f1OcqSMLIPMHq0BhuuTFXJgYcC2weATzgdjI6UkXERsP4Jr5u4rjdNEb3srRsNdRVLqIBPwmYsp2rVj4qlUrlq0MVIZVKpVJ5w/nNj/zy9UFn2/i+ELHReO9UY94ILMQyqMcW2SMuhg6GdqpQDenjpKxkzE9p6Sss50wssKzEstWQY+LQLERaTK18VCqVyhtAnY5VqVQqlTec937yAzf+9Kc++NDFfieCZTaQh5DFUDWBeTxvi+FAGjSKjUOZJ2Y1MgcUpkqJi2LEoWzV55YtgRyYWAVIpVKpvDHUSkilUqlU3hT85+d+8ToEEZI8IjaOHU5hgiVeg0ixRfq5j6OKE/d1imnmjiU2m9LT1K8JzQLkO57/UBUglUql8gbR/P67VCqVSqXy1efbn58rI0mQQBAiTj2dzMX7QT1GYsq6lvulMMdQMWli+nmLyROuhCA4fuW5f3MdqvioVCqVrwVVhFQqlUrlTUcpSCCM903tWF5hYYIgSWJkUqUslPjYelWa1vf5nmo6r1Qqla8Z/w+vIIH0aFwlxQAAAABJRU5ErkJggg==",[[10.38333333333,34.14166666667],[3.50833333333,42.9833333333367]],0.8,null,"pred_rf","pred_rf"]},{"method":"removeLayersControl","args":[]},{"method":"addLayersControl","args":[["CartoDB.Positron","CartoDB.DarkMatter","OpenStreetMap","Esri.WorldImagery","OpenTopoMap"],"pred_rf",{"collapsed":true,"autoZIndex":true,"position":"topleft"}]},{"method":"addControl","args":["","topright","imageValues-pred_rf","info legend "]},{"method":"addImageQuery","args":["pred_rf",[34.14166666667,3.50833333333,42.9833333333367,10.38333333333],"mousemove",7,"Layer"]},{"method":"addLegend","args":[{"colors":["#040404 , #040404 0%, #26123D 10.6758423008113%, #51125B 21.3516846016227%, #7F106F 32.027526902434%, #AA1E75 42.7033692032453%, #D03C6B 53.3792115040566%, #EF6243 64.055053804868%, #F69322 74.7308961056793%, #F8C047 85.4067384064906%, #FBEC85 96.0825807073019%, #FFFE9E "],"labels":["0.0","0.1","0.2","0.3","0.4","0.5","0.6","0.7","0.8","0.9"],"na_color":"#BEBEBE","na_label":"NA","opacity":1,"position":"topright","type":"numeric","title":"pred_rf","extra":{"p_1":0,"p_n":0.960825807073019},"layerId":null,"className":"info legend","group":"pred_rf"}]},{"method":"addScaleBar","args":[{"maxWidth":100,"metric":true,"imperial":true,"updateWhenIdle":true,"position":"bottomleft"}]},{"method":"addHomeButton","args":[34.14166666667,3.50833333333,42.9833333333367,10.38333333333,true,"pred_rf","Zoom to pred_rf","<strong> pred_rf <\/strong>","bottomright"]}],"limits":{"lat":[3.50833333333,10.38333333333],"lng":[34.14166666667,42.9833333333367]}},"evals":[],"jsHooks":{"render":[{"code":"function(el, x, data) {\n return (\n function(el, x, data) {\n // get the leaflet map\n var map = this; //HTMLWidgets.find('#' + el.id);\n // we need a new div element because we have to handle\n // the mouseover output separately\n // debugger;\n function addElement () {\n // generate new div Element\n var newDiv = $(document.createElement('div'));\n // append at end of leaflet htmlwidget container\n $(el).append(newDiv);\n //provide ID and style\n newDiv.addClass('lnlt');\n newDiv.css({\n 'position': 'relative',\n 'bottomleft': '0px',\n 'background-color': 'rgba(255, 255, 255, 0.7)',\n 'box-shadow': '0 0 2px #bbb',\n 'background-clip': 'padding-box',\n 'margin': '0',\n 'padding-left': '5px',\n 'color': '#333',\n 'font': '9px/1.5 \"Helvetica Neue\", Arial, Helvetica, sans-serif',\n 'z-index': '700',\n });\n return newDiv;\n }\n\n\n // check for already existing lnlt class to not duplicate\n var lnlt = $(el).find('.lnlt');\n\n if(!lnlt.length) {\n lnlt = addElement();\n\n // grab the special div we generated in the beginning\n // and put the mousmove output there\n\n map.on('mousemove', function (e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() +\n ' | x: ' + L.CRS.EPSG3857.project(e.latlng).x.toFixed(0) +\n ' | y: ' + L.CRS.EPSG3857.project(e.latlng).y.toFixed(0) +\n ' | epsg: 3857 ' +\n ' | proj4: +proj=merc +a=6378137 +b=6378137 +lat_ts=0.0 +lon_0=0.0 +x_0=0.0 +y_0=0 +k=1.0 +units=m +nadgrids=@null +no_defs ');\n } else {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n }\n });\n\n // remove the lnlt div when mouse leaves map\n map.on('mouseout', function (e) {\n var strip = document.querySelector('.lnlt');\n if( strip !==null) strip.remove();\n });\n\n };\n\n //$(el).keypress(67, function(e) {\n map.on('preclick', function(e) {\n if (e.originalEvent.ctrlKey) {\n if (document.querySelector('.lnlt') === null) lnlt = addElement();\n lnlt.text(\n ' lon: ' + (e.latlng.lng).toFixed(5) +\n ' | lat: ' + (e.latlng.lat).toFixed(5) +\n ' | zoom: ' + map.getZoom() + ' ');\n var txt = document.querySelector('.lnlt').textContent;\n console.log(txt);\n //txt.innerText.focus();\n //txt.select();\n setClipboardText('\"' + txt + '\"');\n }\n });\n\n }\n ).call(this.getMap(), el, x, data);\n}","data":null},{"code":"function(el, x, data) {\n return (function(el,x,data){\n var map = this;\n\n map.on('keypress', function(e) {\n console.log(e.originalEvent.code);\n var key = e.originalEvent.code;\n if (key === 'KeyE') {\n var bb = this.getBounds();\n var txt = JSON.stringify(bb);\n console.log(txt);\n\n setClipboardText('\\'' + txt + '\\'');\n }\n })\n }).call(this.getMap(), el, x, data);\n}","data":null}]}}</script> ] --- class: inverse, center, middle ##### Gabriel Carrasco Escobar, MS, PhD(c) ###### [gabriel.carrasco@upch.pe]() ###### [@Gabc91]()